-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Retinoic Acid Signaling Regulates Differential Expression of the Tandemly-Duplicated Long Wavelength-Sensitive Cone Opsin Genes in Zebrafish
Tandemly-replicated opsin genes are found in genomes of humans and zebrafish. In humans, the tandemly-replicated long wavelength-sensitive/medium wavelength-sensitive (LWS/MWS) array underlies trichromatic color vision; defects in these genes result in color blindness and X-linked retinal degenerations. The current model for regulation of tandemly replicated opsin genes states that stochastic interactions between upstream enhancer regions and gene promoters result in a preferential association with the LWS or MWS promoter. Here we provide evidence, from the LWS1/LWS2 array in zebrafish, that a trans-regulatory mechanism may instead control expression. This array is orthologous to the human LWS/MWS array but arose through an independent gene duplication event. We identified genes that were differentially expressed in zebrafish embryo eyes in response to treatment with the developmental signaling molecule retinoic acid (RA) during photoreceptor differentiation. LWS1 was significantly upregulated by this treatment, and we demonstrate that individual cone photoreceptors were induced by RA to switch expression from LWS2 to LWS1. Experimental reduction of RA signaling inhibited expression of LWS1, and endogenous RA signaling domains spatially coincided with a zone of LWS1 expression in individual cones during zebrafish retinal growth. Our findings suggest that RA signaling within the retina regulates differential expression of the LWS genes, and therefore that tandemly-replicated opsin genes may be amenable to therapeutic manipulation.
Published in the journal: Retinoic Acid Signaling Regulates Differential Expression of the Tandemly-Duplicated Long Wavelength-Sensitive Cone Opsin Genes in Zebrafish. PLoS Genet 11(8): e32767. doi:10.1371/journal.pgen.1005483
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005483Summary
Tandemly-replicated opsin genes are found in genomes of humans and zebrafish. In humans, the tandemly-replicated long wavelength-sensitive/medium wavelength-sensitive (LWS/MWS) array underlies trichromatic color vision; defects in these genes result in color blindness and X-linked retinal degenerations. The current model for regulation of tandemly replicated opsin genes states that stochastic interactions between upstream enhancer regions and gene promoters result in a preferential association with the LWS or MWS promoter. Here we provide evidence, from the LWS1/LWS2 array in zebrafish, that a trans-regulatory mechanism may instead control expression. This array is orthologous to the human LWS/MWS array but arose through an independent gene duplication event. We identified genes that were differentially expressed in zebrafish embryo eyes in response to treatment with the developmental signaling molecule retinoic acid (RA) during photoreceptor differentiation. LWS1 was significantly upregulated by this treatment, and we demonstrate that individual cone photoreceptors were induced by RA to switch expression from LWS2 to LWS1. Experimental reduction of RA signaling inhibited expression of LWS1, and endogenous RA signaling domains spatially coincided with a zone of LWS1 expression in individual cones during zebrafish retinal growth. Our findings suggest that RA signaling within the retina regulates differential expression of the LWS genes, and therefore that tandemly-replicated opsin genes may be amenable to therapeutic manipulation.
Introduction
Color vision in vertebrates requires the differentiation of multiple types of cone photoreceptors in the retina, each of which has a different spectral sensitivity. Spectral sensitivities are conferred by the visual pigment present within cone photoreceptors; these pigments consist of a specific opsin protein, and a light-sensitive chromophore (11-cis retinal or 11-cis 3,4-dehydroretinal) [1]. There are five major types of photoreceptor opsins in vertebrates: RH1 (rod opsin), SWS1 (short wavelength-sensitive; blue or UV), SWS2 (short wavelength-sensitive; blue), RH2 (middle wavelength-sensitive; green), and M/LWS (middle to long wavelength-sensitive, green or red) [2]. The human retina contains rods, which express rod opsin, as well as cones, which express one of three types of cone opsin, SWS1 (blue), MWS (green), and LWS (red). In the zebrafish, by comparison, the retina contains rods as well as four major classes of cones, SWS1 (UV), SWS2 (blue), RH2 (green), and LWS (red). Individual RH2 cones may express one of four RH2 opsins, while individual LWS cones may express one of two LWS opsins [3]. In humans, the LWS and MWS opsin genes are arrayed in a tail to head manner on the X chromosome, the consequence of tandem gene duplication and evolutionary neofunctionalization [4]. Similarly, the four RH2 genes and two LWS genes in zebrafish reside in independent tandem arrays [3]. The zebrafish LWS1 gene is orthologous to the human LWS gene, but the LWS/MWS gene duplication in humans is independent from the LWS1/LWS2 duplication in zebrafish [3].
In both mammals and teleost fish, cone photoreceptors are patterned across the retinal hemisphere with both short-range (mosaic) spacing attributes, and long-range patterns as a function of retinal eccentricity or dorsal-ventral position. For example, mouse cones show a regular local spacing [5], and pronounced dorsal-ventral gradients of MWS and SWS1 opsin expression [6,7]. Human SWS1 cones also display regular local spacing [8], and the LWS and MWS cones show a central-to-peripheral gradient in density, as well as in LWS:MWS ratio, with an LWS:MWS ratio lower in central retina than in peripheral retina [9]. In zebrafish retina, on a local scale cones display a geometrically precise “row” mosaic [10,11], but cones expressing opsin genes from tandemly-replicated arrays also show larger-scale patterns. Within the central retina, RH2 cones express RH2-1 or RH2-2, and LWS cones express LWS2; within ventral and peripheral retina the RH2 cones express RH2-3 or RH2-4, and the LWS cones express LWS1 [12].
Regulatory mechanisms for achieving photoreceptor diversity and fates are the focus of intense investigation due to their potential applications in treating retinal diseases. A transcriptional regulation model is emerging, in which the photoreceptor transcription factors Crx, NeuroD, and Rx/Rax promote photoreceptor-specific gene expression [13], and the transcription factors Nr2e3 and Nrl promote rod-specific gene expression and suppress S cone development [14,15]. The nuclear hormone receptors RXRγ and TRβ2, together with thyroid hormone (T3) promote the differentiation of MWS/LWS cones and suppress SWS1 cone opsin expression [6,16,17]. In the zebrafish, the transcription factor tbx2b supports the SWS1 (UV-sensitive) cone fate while suppressing the rod fate [18]. Also in the zebrafish, RA and the receptor RARαb favor rod fates over cone fates during progenitor proliferation [19]. However, the choice of opsin from a tandem array, such as the human LWS/MWS array, is thought to be a stochastic event. In the current model, during cone differentiation, an upstream locus control region (LCR) becomes preferentially associated with the LWS or MWS promoter. This association then becomes permanent, resulting in each cone expressing only one of the opsin genes of the array [20]. However, this stochastic model does not explain the spatial gradient of the LWS:MWS ratio [9], and does not explain a predicted genetic factor that influences this ratio [21].
Key signaling molecules known to be present in specific spatial gradients during vertebrate retinal development include the nuclear hormone receptor ligands retinoic acid (RA) and thyroid hormone (tri-iodothyronine, T3) [16,22]. RA is the acid derivative of Vitamin A (retinol) and in vertebrate animal models is synthesized locally within the developing eye, with high concentrations ventrally, medium concentrations dorsally, and low concentrations centrally [22–25]. RA signaling via specific receptors (RARs and/or RXRs) plays many critical roles in eye development, from early eye organogenesis up to photoreceptor differentiation and survival. For example, ventral RA is necessary for optic fissure closure [26] and for the development of ventral retina in general [25]. Numerous reports have indicated that RA promotes rod determination [19], differentiation [27–31], and survival [32,33], and these findings have been translated into promising approaches for promoting rod development in retina cultures derived from human ES and iPS cells [34–36].
In one of our previous studies, we observed that treatment with all-trans RA (at-RA) over the time of photoreceptor differentiation in zebrafish promoted the differentiation of rods and red-sensitive (LWS) cones, while reducing the differentiation of blue - (SWS2) and UV - (SWS1) sensitive cones [29]. In the present study we performed microarray analysis of genes expressed in the eyes of zebrafish embryos subjected to a similar at-RA exposure regime. Our original goal was to identify genes that may mediate the effects of RA on photoreceptor differentiation, and the microarray experiment indeed revealed numerous candidates, including components of Wnt and Bmp signaling pathways, the RA receptor RXRγa, and several additional retinal transcription factors. Of particular interest however, the microarray, along with qCPR, in situ hybridization studies, and the use of a transgenic reporter line, revealed that exogenous RA strongly upregulated specifically the first member of the LWS array, LWS1, while downregulating LWS2, within individual LWS cones. Experimental reduction of RA signaling correspondingly prevented upregulation of LWS1, and native RA signaling domains coincided with an endogenous LWS1 expression zone within growing juvenile retinas. These results are the first to provide evidence for a trans-regulatory mechanism for the control of differential expression of tandemly replicated opsin genes.
Results
Microarray identification of RA-responsive genes in the zebrafish eye
An objective of this analysis was to identify eye-specific genes that are regulated by exogenous RA in zebrafish. The zebrafish embryonic retina exists as a proliferative neuroepithelium at 24 hpf, and after 48 hpf photoreceptor differentiation occurs concomitantly with lamination of the retina. By 72 hpf the zebrafish retina has formed the neurons of the ganglion cell layer (GCL), inner nuclear layer (INL) and outer nuclear layer (ONL) [37,38]. Microarray analysis of embryonic eyes of groups treated with 0.3 μM RA or DMSO (controls) over the period of embryonic photoreceptor differentiation (from 48 hpf–75 hpf) identified a list of 174 Affymetrix probe sequences (S1 Table) that were significantly differentially expressed at the 10% FDR (Fig 1A). Of this list 63% were known while 37% represented unconfirmed, potentially novel, gene sequences. Analysis of the list of genes was performed through the use of the gene ontology clustering software GOEAST [39]. A summary of this analysis, listing specific biological and molecular process categories of interest, finds that most differentially expressed genes were upregulated by RA treatment (Fig 1B). The broader categories overrepresented in the differentially expressed list included those related to development and morphogenesis, stimulus response, and metabolic processes. The more specific categories included vitamin A metabolism, transcription factors, components of cellular signaling pathways, and surprisingly only one gene related to photoreceptor activity (Fig 1B).
Fig. 1. Microarray analysis of eye-specific gene expression in zebrafish embryo eyes after 48–75 hpf exposure to 0.3 μM all-trans RA. 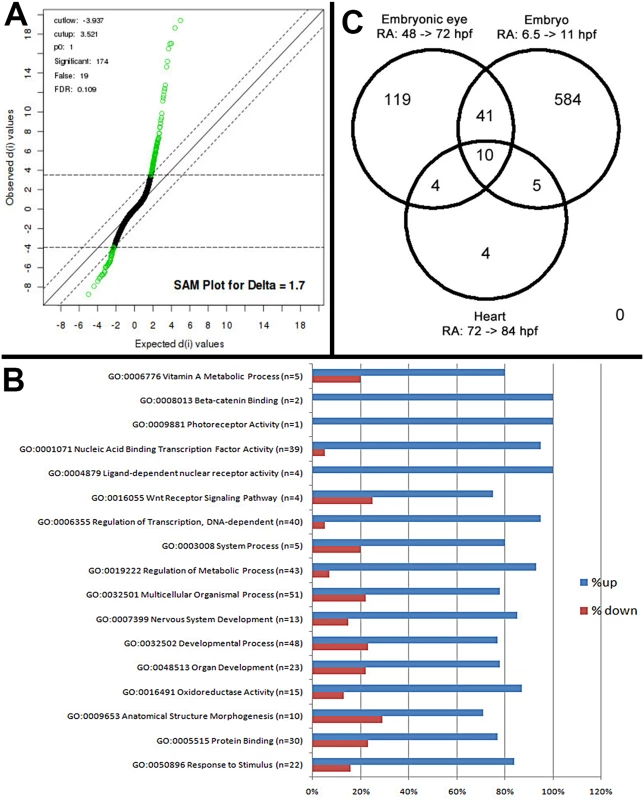
A. Statistical Analysis of Microarray (SAM) plot, at a 0.109 false discovery rate (FDR). B. Gene ontology (GO) analysis; selected GO categories shown. C. Venn diagram comparing differentially expressed genes in eyes of RA-treated embryos with those of RA-treated whole embryos during somitogenesis and with those of hearts of RA-treated larvae. Genes associated with vitamin A metabolism (retinoid metabolism) and retinoid signaling (Table 1)
Tab. 1. Differential expression of genes involved in RA synthesis and signaling in eyes of embryonic zebrafish treated with RA. 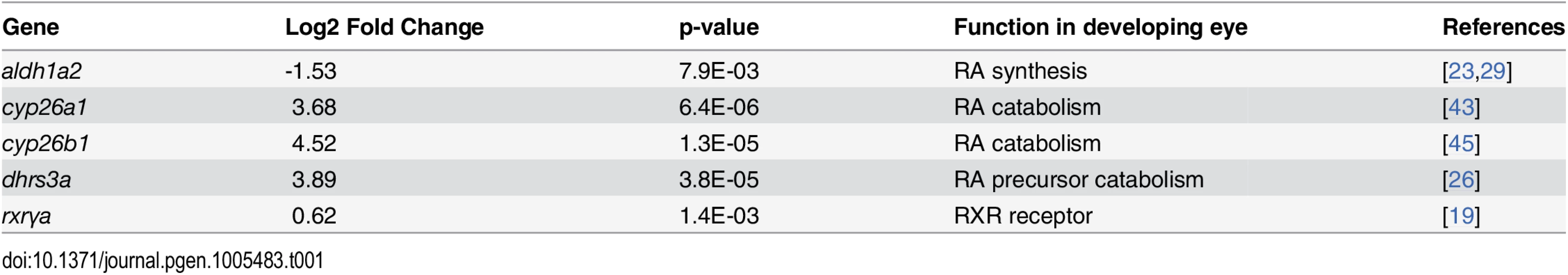
Exogenous RA upregulated cyp26b1 and cyp26a1, which encode enzymes that catabolize all-trans RA [40], and dhrs3a, which encodes a dehydrogenase that reduces the amount of retinaldehyde available for conversion to all-trans RA [41]. Significantly downregulated was aldh1a2, encoding a retinaldehyde dehydrogenase that converts retinaldehyde into RA [42]. At the 20% FDR, the RA receptor gene rxrγa was significantly upregulated. These results are consistent with exogenous RA regulating expression of RA metabolic enzymes in an attempt to achieve RA homeostasis [43,44].
Genes encoding transcription factors (Table 2)
Tab. 2. Differential expression of genes encoding transcription factors in eyes of embryonic zebrafish treated with RA. 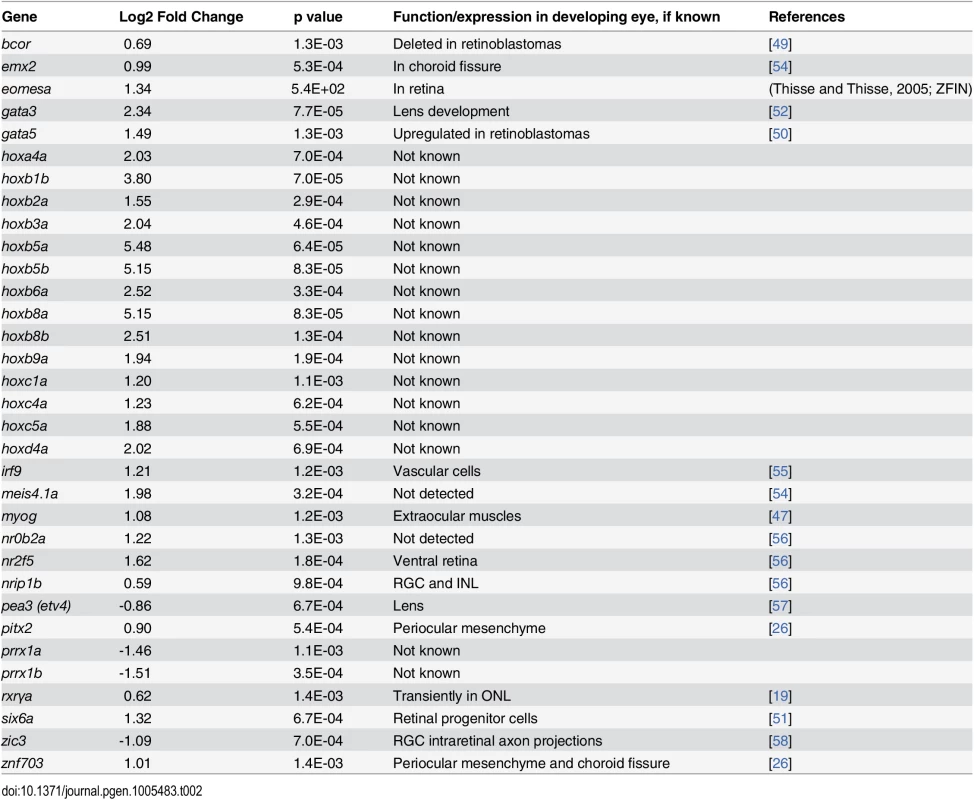
Eight of these genes are expressed in zebrafish embryonic neural retina (emx2, eomesa, nr2f5, nrip1b, pea3, rxrγa, zic3, znf703). Two additional genes are associated with periocular mesenchyme and its derivatives: pitx2 and myog, which are both known targets of RA signaling in development of extraocular muscles [46–48]. Two genes (bcor, gata5) are associated with retinoblastoma [49,50], the homologue of six6 is expressed in retinal progenitor cells [51], and that of gata3 is important for lens development [52]. 19 transcription factor genes (irf9, meis4.1a, nr0b2a, prrx1a, prrx1b, and 14 hox genes) have not, to our knowledge, been shown to be expressed in ocular tissues. Surprisingly, a large number (14) of hox genes were significantly upregulated in the eyes of RA-treated embryos (Table 3). This finding suggests that either these genes are not fully silenced in eye tissues at 48–75 hpf, and/or that an activity downstream of RA signaling acts to modulate chromatin structure of the hox gene arrays so they can be activated [53].
Tab. 3. Differential expression of genes encoding components of cell signaling pathways in eyes of embryonic zebrafish treated with RA. 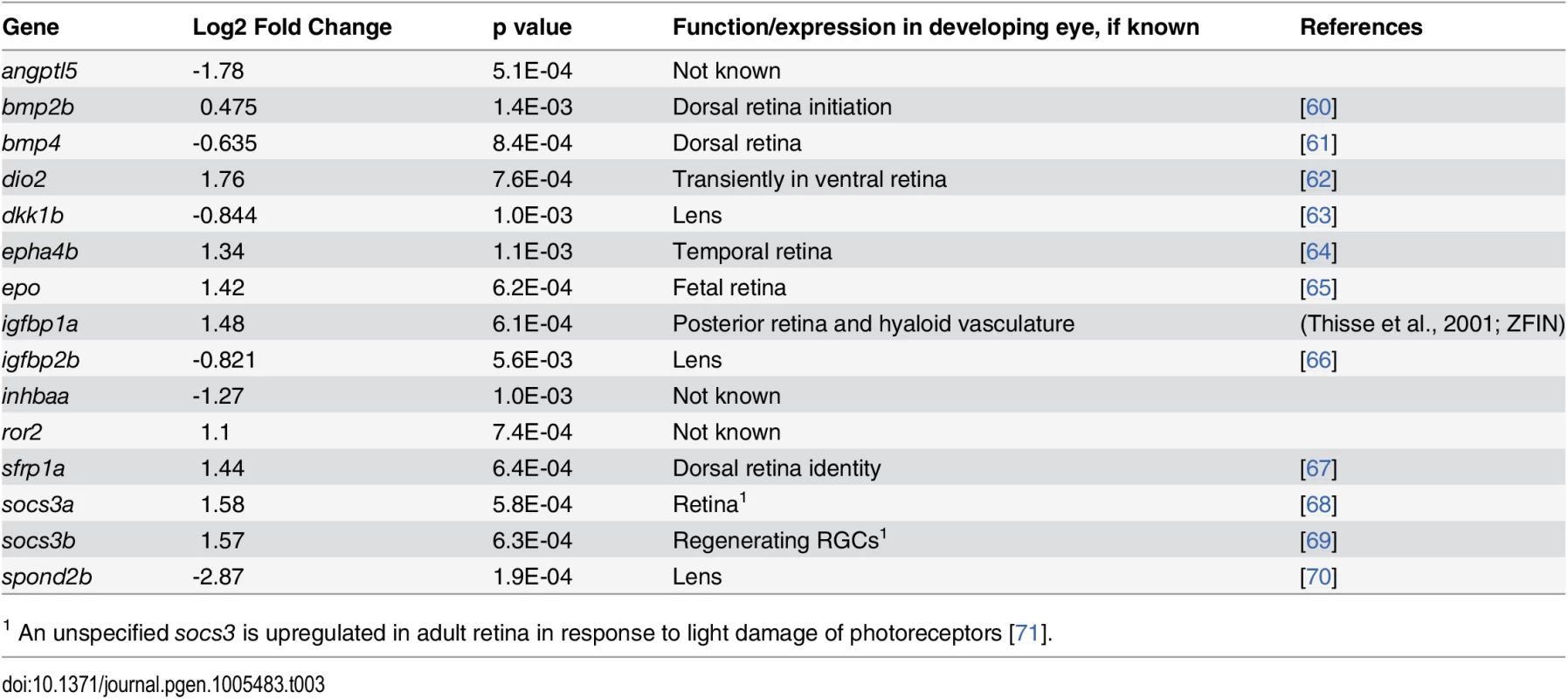
1 An unspecified socs3 is upregulated in adult retina in response to light damage of photoreceptors [71]. Genes encoding components of non-RA-related cellular signaling pathways (Table 3)
Seven of the genes differentially expressed in the eye in response to RA encode the extracellular signaling factors bmp2b, bmp4, epha5b, igfbp1a, igfbp2b, epo, angpt15, two encode intracellular modulators of signaling (socs3a and socs3b), and one encodes an enzymatic activator of thyroid hormone (T3; dio2). Two genes involved in controlling hematopoiesis, epo and angpt15, were upregulated by RA. An interesting subset of differentially expressed genes encodes modifiers of Wnt signaling (dkk1b, sfrp1a, spond2b). In addition, ror2, which encodes a transmembrane co-receptor involved in the non-canonical (Ca2+) Wnt pathway [59], was also upregulated. At the 20% FDR, wnt11 was significantly upregulated.
The GO analysis of the microarray data indicated upregulation of only one gene involved in photoreceptor function, opn1lw1, the first member of the long wavelength-sensitive opsin gene array [3] (Fig 1B). However, further database searches revealed three additional genes associated with photoreceptor differentiation, as significantly upregulated (Table 4). RXRγa is expressed transiently within the ONL during photoreceptor differentiation in zebrafish [19], and its mouse homologue is suppresses S opsin in developing cones [6]. Aanat2 encodes the rate-limiting enzyme involved in melatonin synthesis in photoreceptors [72], and atp1a1.3 encodes the α subunit of Na+/K+ ATPase, which is enriched in photoreceptors [73,74]. Unexpectedly, rod opsin was not detected as upregulated by the microarray, even at the 20% FDR, although there is evidence supporting roles for RA in stimulating rod opsin expression and rod cell differentiation in the zebrafish, using alternative methods [19,29,30,33].
Tab. 4. Differential expression of eye and photoreceptor genes in eyes of embryonic zebrafish treated with RA. 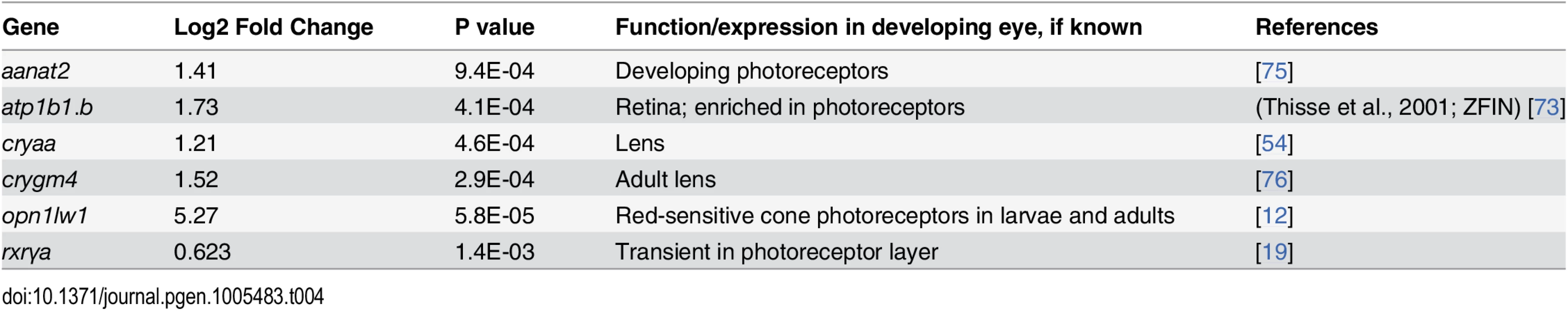
RA activates a restricted set of common genes in the developing zebrafish embryo
We next tested the hypothesis that RA signaling targets are tissue - and developmental stage-specific, by comparing our list of differentially expressed genes with that of others obtained following treatment of whole zebrafish embryos with RA [77,78]. A comparison with genes upregulated by RA during early somitogenesis (6.5 hpf–11 hpf) [78] identified 50 common probes (S2 Table and Fig 1C), including cyp26a1, cyp26b1, dhrs3a, rxrga, and aldh1a2 [78]. The transcription factor-encoding genes nr2f5, meis4.1a, nr0b2a, zic3, nrip1b, znf703, RXRγa, and 10 common hox genes were all upregulated in response to RA, in both lists.
The results of the above comparison were then compared to a microarray dataset of genes differentially expressed in zebrafish larval hearts in response to RA [77]. Among all three data sets, 11 probes (eight genes) were commonly upregulated (Fig 1C and S2 Table). Of these, three are genes involved in RA metabolism (cyp26a1, cyp26b1, and dhrs3a), and two are orphan nuclear receptors (nr2f5 and nr0b2a) that may be essential base regulators of retinoid metabolism [77]. The relatively small number of genes commonly to all three datasets indicates that the current dataset predominantly reflects eye-specific transcriptional responses to RA exposure.
Validation of microarray results
The following genes were selected for qPCR validation: cyp26b1, hoxb6b, socs3a, dkk1b, sfrp1a, wnt11, opt1lw1, opn1lw2, and rho (rod opsin) (Primer pairs are listed in Table 5). Each sample was tested with one or two separate qPCR experiments, each with three biological replicates. In brief, dkk1 was significantly downregulated in the RA-treated samples, while cyp26b1, hoxb6b, socs3a, sfrp1a, wnt11, and opn1lw1 were significantly upregulated (Fig 2A–2G), consistent with the microarray. The results of qPCR for rod opsin expression, however, were ambiguous. In one qPCR experiment this gene was found to be significantly upregulated, but not in a replicate experiment (Fig 2I). It is possible that at the selected sampling time (75 hpf), changes in rod opsin expression take place rapidly [79], and minor differences in relative embryo age in each clutch may contribute to this ambiguity. An alternative explanation is that rods make up a very small percentage of the total number of cells in the eye, and this tissue heterogeneity contributes to the inconsistency of qPCR results. However, the qPCR analyses in general confirmed differential expression where detected by microarray analysis, as well as the direction of expression changes between control and experimental groups.
Tab. 5. Primers used for q-RT-PCR. 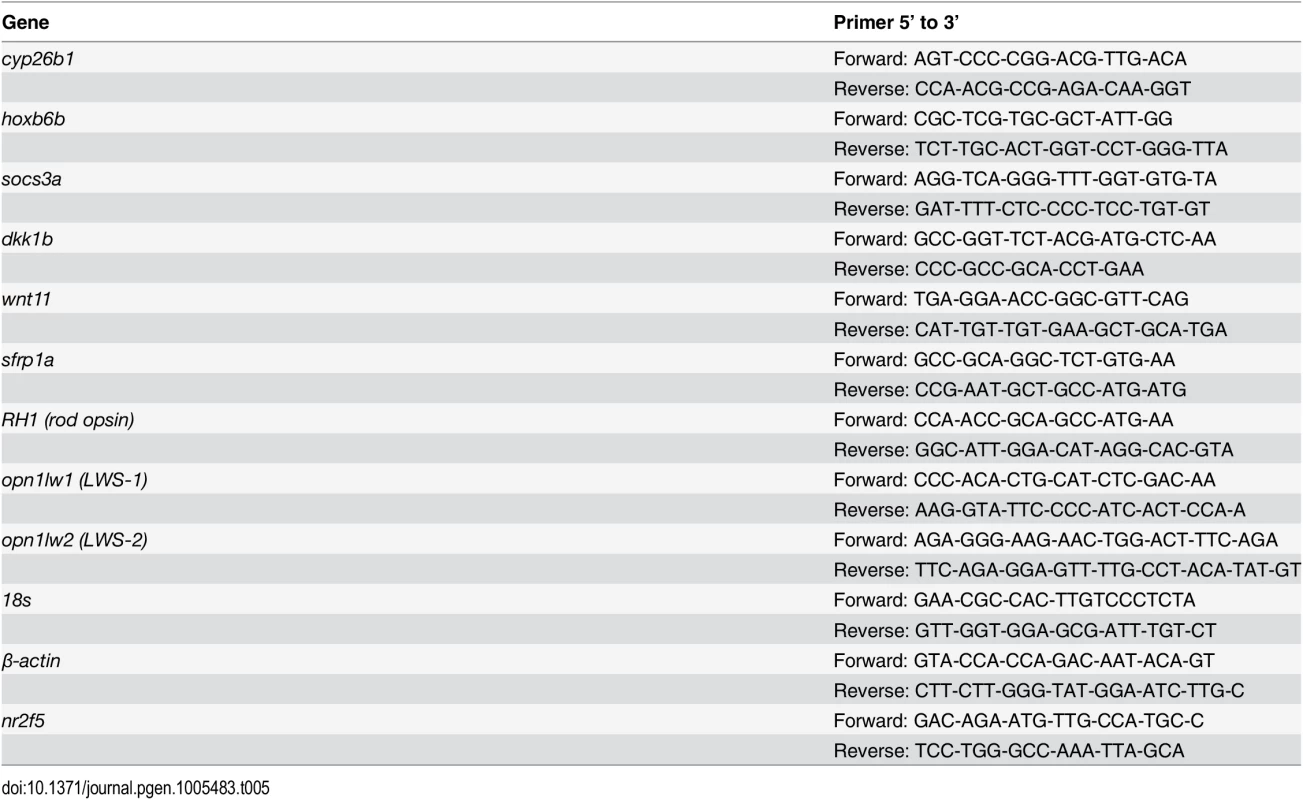
Fig. 2. Quantitative (real-time) PCR validation of selected genes analyzed by the microarray. 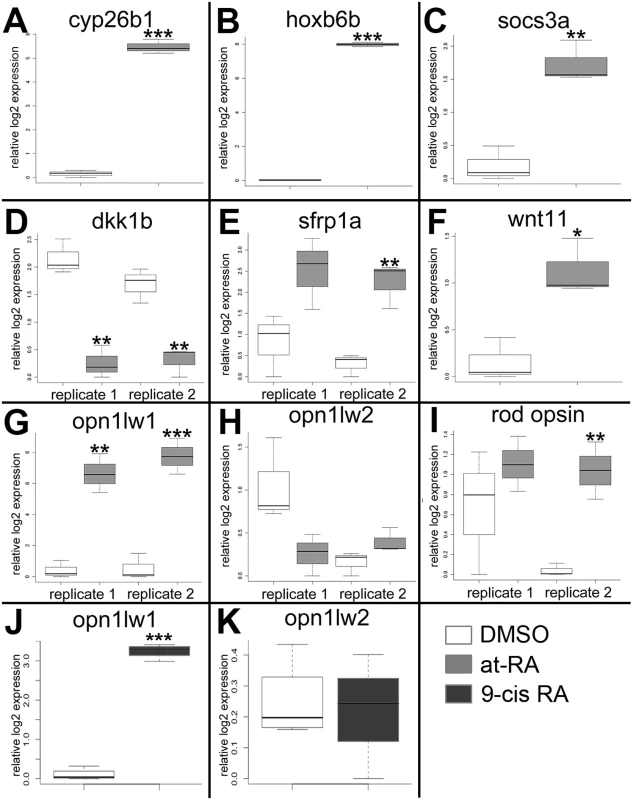
A. cyp26b1. B. hoxb6a. C. socs3a. D. dkk1b. E. sfrp1a. F. wnt 11. G. and J. opn1lw1. H. and K. opn1lw2. I. rod opsin (RH1). White boxes, DMSO; gray boxes, at-RA (both in embryonic eye tissues); dark gray boxes, 9-cis RA (in whole embryos). In the boxplots, the boxes demarcate the 25th and 75th percentiles, dark horizontal lines designate the medians, and whiskers represent the upper and lower limits. Genes in A-C, E-G, were all identified as upregulated by the microarray; dkk1b was identified as downregulated; opn1w2 and rod opsin were not detected as differentially expressed. ***, p<0.001; **, p<0.01; *, p<0.05 (2-tailed Student’s t-test). The very large fold-change in expression of opn1lw1, as detected by both microarray (S1 Table) and qPCR (Fig 2G) was rather striking. Opn1lw1 (a.k.a. LWS1) encodes the first member of the tandemly-duplicated long wavelength-sensitive (LWS; red-sensitive) opsin array [3]. Our previous study detected increased differentiation of red-sensitive cones in response to RA treatment, with the interpretation that the increase was related to expression of the second member of the LWS array, opn1lw2 (a.k.a. LWS2) [29], because this is the only LWS gene expressed in embryos [12], and because our in situ probes were directly complementary to this gene [3,29,80]. Therefore, we next determined whether the RA treatment affected expression of opn1lw2/LWS2, using qPCR. Surprisingly, levels of mRNA expression of this gene within the eye were not significantly affected by RA treatment from 48–75 hpf (Fig 2H). In separate experiments we tested the more RXR-selective retinoid, 9-cis RA, and 0.3 μM 9-cis RA treatment from 48–75 hpf significantly upregulated LWS1 (p<0.001) in whole embryos, but did not change expression levels of LWS2 (Fig 2J and 2K).
Spatial distribution of opn1lw1/LWS1 and opn1lw2/LWS2 after RA exposures
When analyzed at 3 dpf (75 hpf) as whole mounts, control embryos showed widespread expression of LWS2, and no embryos showed expression of LWS1 (Fig 3A and 3B), consistent with previous findings that the onset of LWS1 expression is delayed until ~one week post-fertilization [12]. In contrast, embryos treated with at-RA or 9-cis RA still showed widespread expression of LWS2, but this expression occasionally appeared weaker in ventral, nasal, and/or central retina (Fig 3D). In addition, the RA-treated embryos contained LWS1 expressing cones scattered primarily in ventral retina (Fig 3C). We scored these expression patterns, using modifications of previous criteria [79] (see Methods). This analysis revealed that eyes of RA-treated embryos reached higher stages of LWS1 expression, but RA (either at-RA or 9-cis RA) had minimal impact on stage (distribution) of LWS2 expression (Fig 4A and Table 6). Parallel experiments utilizing cryosections verified these findings, and demonstrated that the “RA-induced” LWS1 expression was in the ONL (Fig 3E–3H). We quantified our findings by counting numbers of LWS1+ cones in each section. The statistical analysis revealed significantly greater numbers of LWS1+ cones in 9-cis RA treated retinas (Fig 4B). These results are consistent with those of the microarray and qPCR: an abrupt upregulation of LWS1 in a small number of cones in RA-treated embryos as compared with essentially no expression in control embryos would be reflected by the statistically significant and high fold-change increase in levels of LWS1 expression (S1 Table and Fig 2G), while a slight reduction in LWS2 expression in a small number of cones may not be detected by quantitative measurements of LWS2 mRNA expression in whole eyes (Fig 2H).
Fig. 3. Changes in spatiotemporal patterns of expression of LWS1 and LWS2 in response to 9-cis RA treatment. 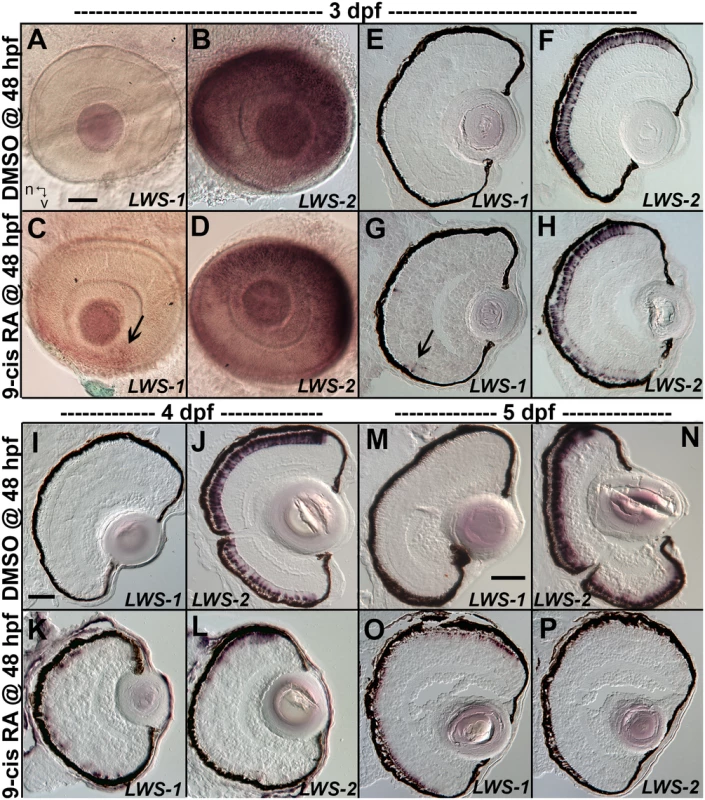
A-H. Whole-mounted (A-D) and sectioned (E-H) embryo eyes obtained from embryos treated with DMSO (A,B,E,F) or 0.3 μM 9-cis RA (C,D,G,H) from 48 hpf to 75 hpf, and then hybridized with LWS1 (A,E,C,G) or LWS2 (B,G,D,H) cRNA. Arrows in C and G indicate LWS1-expressing cones in ventral retina; n, nasal; v, ventral. I-L. Sectioned embryo eyes obtained from embryos treated with DMSO (I,J) or 0.3 μM 9-cis RA (K,L) from 48 hpf to 4 dpf, and then hybridized with LWS1 (I,K) or LWS2 (J,L) cRNA. M-P. Sectioned embryo eyes obtained from embryos treated with DMSO (M,N) or 0.3 μM 9-cis RA (O,P) from 48 hpf to 4 dpf, and then hybridized with LWS1 (M,O) or LWS2 (N,P) cRNA. Scale bar in A (applies to A-H) = 50 um. Scale bar in I (applies to I-L) = 50 μm. Scale bar in M (applies to M-P) = 100 μm. Fig. 4. Quantitative analysis of expression patterns of LWS1 and LWS2 in response to all-trans RA (at-RA) or 9-cis RA treatment. 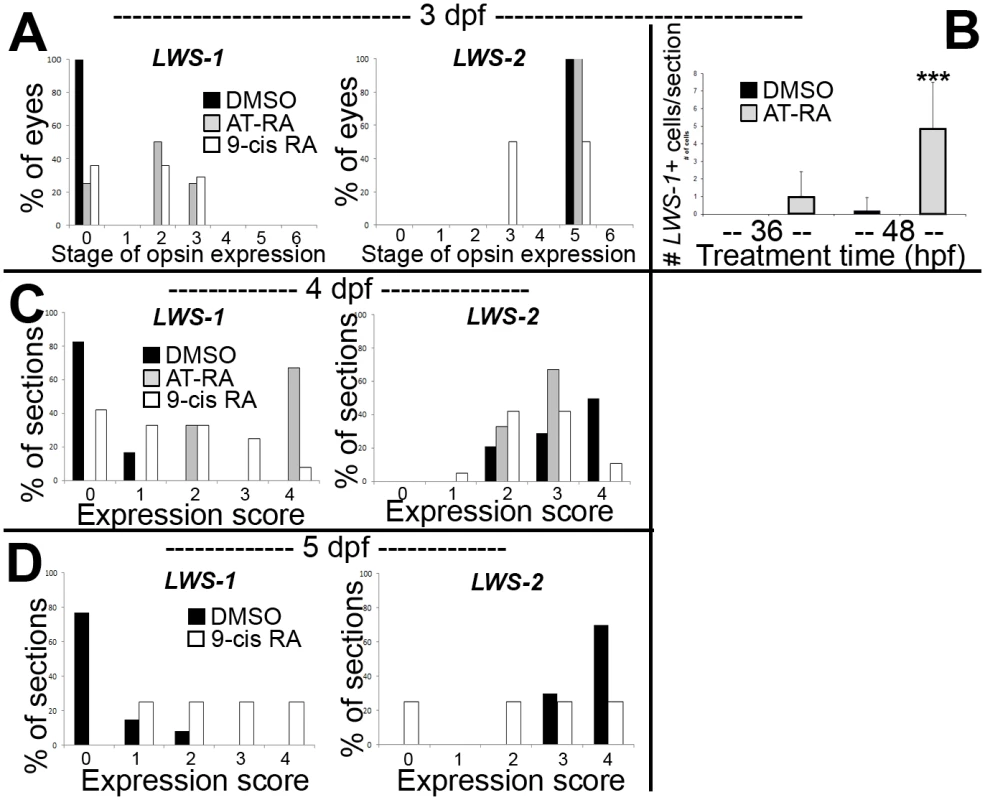
A. Scoring of LWS1 and LWS2 expression in whole mounted eyes of embryos treated from 48 hpf to 75 hpf; stages of opsin expression are described in Materials and Methods. B. Numbers of LWS1- and LWS2-expressing cells in the outer nuclear layers of cryosections derived from embryos treated with at-RA from 36 hpf to 75 hpf as compared to treatments from 48 hpf to 75 hpf; ***, p<0.001). C. Scoring of LWS1 and LWS2 expression domains in the outer nuclear layers of cryosections derived from embryos treated with at-RA or 9-cis RA from 48 hpf to 4 dpf. D. Scoring of LWS1 and LWS2 expression domains in the outer nuclear layers of cryosections derived from embryos treated with 9-cis RA from 48 hpf to 5 dpf (C). Expression scores are described in Materials and Methods; statistical analyses (Fisher’s Exact Test) in Table 6. Tab. 6. Statistical evaluation of LWS opsin mRNA expression scoring. 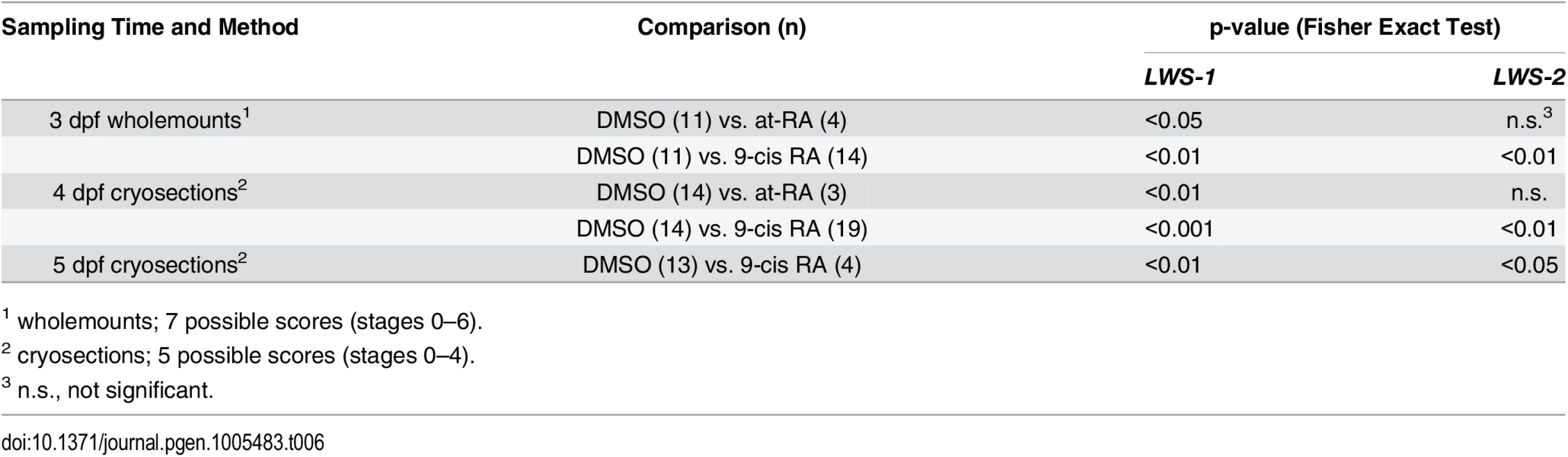
1 wholemounts; 7 possible scores (stages 0–6). We previously demonstrated that when RA exposure takes place beginning at 36 hpf, well prior to photoreceptor terminal mitoses, a rod fate is favored over cone fates [19]. To determine whether this earlier increase in RA signaling also favors LWS1 expression over LWS2, we treated embryos with at-RA from 36–75 hpf, and performed in situs for LWS1 and LWS2. Interestingly, this RA treatment was by comparison ineffective at inducing LWS1 expression (Fig 4B). These results support the hypothesis that photoreceptor-generating lineages experience states of shifting plasticity, such that increased RA signaling during the progenitor period influences rod vs. cone fate decisions, while increased signaling during differentiation influences the choice of cone opsin [19]. Alternatively, because fewer LWS cones are generated in embryos treated with RA at 36 hpf [19], effects of RA on LWS1 may be more difficult to detect after this earlier treatment.
Sustained RA exposure can cause an LWS2-to-LWS1 “opsin switch” in embryonic LWS cones
We carried out additional experiments in which at-RA or 9-cis RA exposure was continued from 48 hpf to 4 dpf (100 hpf). Control embryos again showed widespread expression of LWS2, and essentially no expression of LWS1 (Fig 3I and 3J). However, some embryos treated with 9-cis RA and examined at 4 dpf displayed a drastic increase in LWS1 expressing cones, and this appeared to be at the expense of LWS2 expressing cones (Fig 3K and 3L), suggesting that individual cones had switched expression from the second member of the LWS array (LWS2), to the first member (LWS1). The use of whole mount in situ analysis to better quantify numbers of each cone subtype (as in [19,29]) was not possible due to failure of probe penetration in these larger, 4 dpf embryos. Therefore we assigned expression scores to hybridized cryosections. These scores were specific to LWS1 or LWS2, given that in any embryo, LWS1, when expressed, was predominantly (though not exclusively) ventral (Fig 3G), and LWS2, when expressed, was predominantly dorsal (Fig 3L) (see Materials and Methods for further details). The at-RA or 9-cis RA exposures increased the LWS1 expression score in nearly all sections examined, and this increase was statistically significant (Fig 4C and Table 6). Treatment with RA correspondingly reduced the LWS2 expression scores observed 4 dpf, but only for 9-cis RA treatment (Fig 4C and Table 6). Because the effect of 9-cis RA appeared more robust than that of at-RA (Table 6), suggesting the involvement of an RXR, we treated embryos with the selective RXR agonist bexarotene [81] (0.3 μM or 0.06 μM) from 48 hpf to 4 dpf. This agonist significantly upregulated LWS1 and downregulated LWS2 (qPCR; p<0.001 vs. DMSO; S1 Fig), consistent with roles for RXRs in regulating the LWS array.
We next performed experiments in which the 9-cis RA treatments were continued up to 5 dpf (124 hpf; at-RA treatment for this duration resulted in a high mortality rate). In some of the 9-cis RA-treated embryos, LWS1 expression appeared to have nearly entirely replaced LWS2 expression (Figs 3M–3P and 4E, Table 6). Collectively the results of the LWS gene-specific in situ hybridization studies indicate that exogenous RA is capable of promoting (or de-repressing) expression of LWS1, while repressing expression of LWS2.
A GFP reporter for LWS1 is upregulated by RA
We considered that a transgenic tool may offer additional insights into the regulation of the LWS opsin gene array by retinoids, and utilized the line, Tg(LWS1/GFP-LWS2/RFP-PAC(H))#430 (abbreviated hereafter as LWS:PAC(H)). The genome of this line harbors a ~100kb PAC clone that includes the entire LWS array, but with GFP-polyA inserted into the location of the first exon of LWS1, and RFP-polyA replacing the first exon of LWS2 [82]. Expression of GFP and RFP replicate the spatiotemporal expression patterns of LWS1 and LWS2, respectively [82].
LWS:PAC(H) embryos were treated with DMSO or 9-cis RA at 48 hpf and analyzed as whole mounts by confocal microscopy for RFP and GFP fluorescence at 96 hpf (4 dpf). DMSO-treated embryos displayed RFP fluorescence only (reporting LWS2; Fig 5A and 5C), while 9-cis RA treated embryos contained some GFP+ (reporting LWS1) cones, as well as some doubly-labeled cones (Fig 5B–5D and 5H), often located in ventral (Fig 5B) and peripheral retina (Fig 5B and 5F). In the transgenic LWS:PAC(H) line, most retinas did not display widespread LWS2:RFP cones at 4 dpf regardless of treatment (Fig 5E), in contrast to what was observed for native LWS2 mRNA (Figs 3 and 4), suggesting that reporter expression may be delayed in onset compared with expression from the endogenous LWS array. Even without widespread LWS2:RFP expression, retinas from 9-cis RA treated embryos contained LWS1:GFP+ cones at 4 dpf (Fig 5B–5D and 5F). In addition, the numbers of RFP-expressing cones were not significantly reduced by 9-cis RA treatment as compared with the numbers in DMSO-treated control embryos (Fig 5G), suggesting that the response of the PAC clone LWS transgene array to retinoids may also be slower than that of the native LWS array. It is possible that an important regulatory element may be present within the first exons of LWS1 and/or LWS2, and/or that the presence of the GFP or RFP DNA sequences is disruptive. An alternative explanation for the persistence of RFP—when native LWS2 is downregulated—in RA-treated embryos, is that the RFP protein may be more stable than the LWS2 mRNA. The fact that nearly half of the GFP+ cones in RA-treated embryos co-express RFP is consistent with this hypothesis (Fig 5H). A time-dependent increase in doubly-labeled LWS1:GFP and LWS2:RFP cones provides further evidence for a switch from LWS2 to LWS1 expression in individual cones upon RA treatment (Fig 5H). Finally, treatment with 9-cis RA beginning at 72 hpf, when embryonic cones are postmitotic [38], resulted in higher numbers of GFP+ cones per LWS:PAC(H) eye (Fig 5D), and a more robust increase in expression of native LWS1 mRNA (Fig 6A and 6C), indicating that postmitotic LWS cones may be more competent to express LWS1 in response to RA treatment. Interestingly, this later (72 hpf) treatment did not result in significant changes to LWS2 expression (Fig 6B and 6D), similar to the effect of one day of RA exposure, but from 48–72 hpf (Fig 2H and 2K). Either a longer treatment (2 days of exposure) is required for effects of RA on LWS2, and/or the plasticity of LWS cones to downregulate LWS2 in response to RA is developmental stage-dependent.
Fig. 5. A GFP reporter for LWS1 indicates a switch from LWS2 to LWS1 in response to RA treatment. 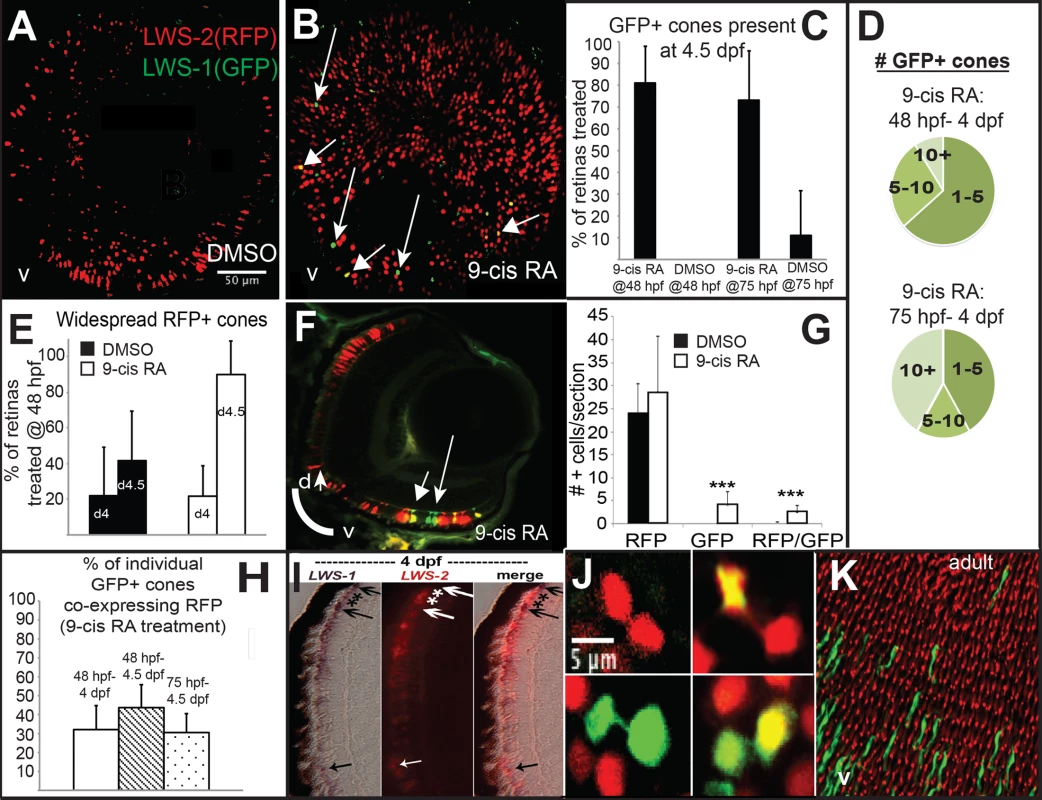
A-C. Whole mount confocal images of retinas from LWS:PAC(H) embryos treated with DMSO (A) or 0.3 μM 9-cis RA (B) from 48 to 96 hpf. LWS2:RFP+ cones (red) are found in control retinas (A), while cones expressing LWS1:GFP alone (green, arrows) or co-expressing LWS2:RFP and LWS1:GFP (yellow, arrowheads) are found in retinas treated with RA (B); LWS1:GFP expression in transgenic retinas treated with RA tend to appear ventrally and peripherally (B). C. Percentage of retinas examined that contain LWS1:GFP+ cones for DMSO or 9-cis RA treatment 48 hpf to 4.5 dpf and from 75 hpf to 4.5 dpf. Error bars represent 95% binomial confidence interval. D. The pie charts show the frequency of 9-cis RA treated transgenic retinas expressing the indicated number of LWS1:GFP+ cones for treatment 48 hpf to 4.5 dpf (top) or 75 hpf to 4.5 dpf (bottom). E. Graphs show the percentage of LWS:PAC(H) transgenic retinas that contain widespread (covering more than half of the retina) LWS2:RFP+ cones at the indicated time points following treatment beginning at 48 hpf (n = 15, RA 4 dpf; 7, RA 4.5 dpf; 9, DMSO 4 dpf; 4 DMSO 4.5 dpf). Error bars represent 95% binomial confidence interval. F. Indirect immunofluorescence image of a transverse section of an LWS:PAC(H) embryo treated with 9-cis RA. The arrowhead indicates a cone co-expressing LWS1:GFP and LWS2:RFP. The arrow indicates a cone expressing LWS1:GFP. G. Graph indicating the numbers of LWS2:RFP (RFP), LWS1:GFP (GFP), or dual label (RFP/GFP) cones per section in retinas treated with DMSO or 9-cis RA (DMSO vs. 9-cis RA ***, p<0.001; 2-tailed Student’s t-test). H. Individual LWS1:GFP+ cones from retinas treated with 9-cis RA for the indicated time frames were examined for co-expression of LWS2:RFP. I. Dual in situ hybridization for LWS1 and LWS2 after treatment with 9-cis RA from 48 hpf to 4 dpf. Purple color reaction indicates LWS1 expression; pink fluorescent color indicates LWS2 expression; arrows at top show cones expressing LWS1 only; asterisks (*) show cones expressing LWS2 only; arrow at bottom shows cone that is dually labeled. J. Confocal images of pairs of cones at the end of mitosis from whole mount transgenic retinas treated with 9-cis RA from 48 hpf to 4.5 dpf. Apparent daughter cells of cone progenitors were observed expressing the same LWS opsin as well as pairs where one daughter cell also co-expresses a different LWS opsin. K. Confocal image of adult LWS:PAC(H) whole mounted retina showing isolated GFP+ (reporting LWS1) cones. Fig. 6. Quantitative PCR for gene expression after retinoid treatment and reduction of RA signaling. 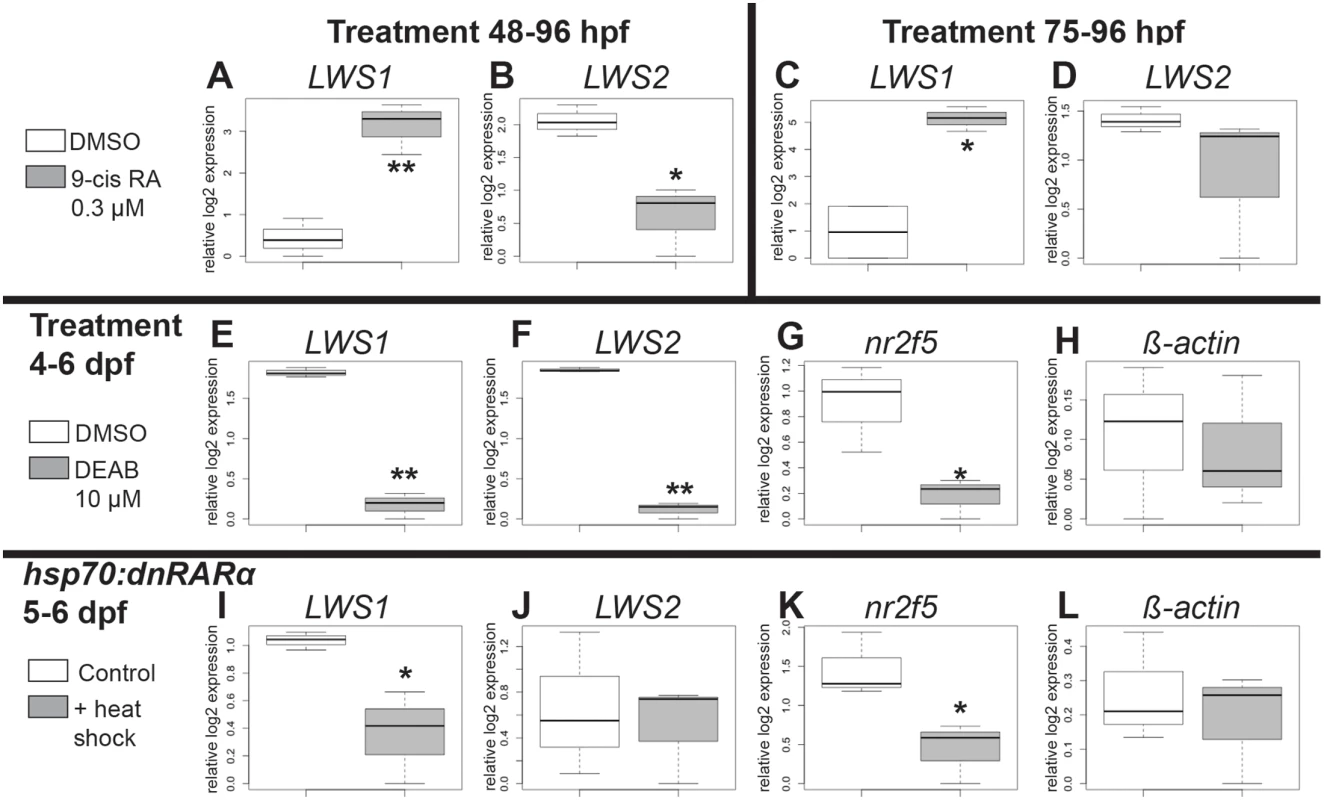
Box plots indicate relative log2 expression of the indicated genes. SciH embryos treated with DMSO (white boxes) or 9-cis RA (gray boxes), for 48–96 hpf (A,B) or 75–96 hpf (C,D) were examined for expression of LWS1 (A,C) and LWS2 (B,D) following treatment. E-H. SciH embryos were treated with DMSO (white boxes) or DEAB (gray boxes) from 4–6 dpf. Gene expression levels for LWS1 (E), LWS2 (F), nr2f5 (G), and b-actin (H) are shown following treatment. I-L. Transgenic pd5 (hsp70:dnRARα) embryos were examined for expression levels of LWS1 (I), LWS2 (J), nr2f5 (K), and b-actin (L) at 7 dpf after control, no heat shock (white boxes) treatment, or after heat shock on day 5 and day 6 (gray boxes). In the boxplots, the boxes demarcate the 25th and 75th percentiles, dark horizontal lines designate the medians, and whiskers represent the upper and lower limits. *p<0.05, **p<0.01 (2-tailed Student’s t-test). In order to verify that individual red-sensitive cones switched opsin expression, we performed dual in situ hybridizations using 9-cis RA-treated SciH embryos as cryosections at 100 hpf. This approach demonstrated the existence of some cones that were co-labeled with the LWS1 probe and the LWS2 probe (Fig 5I), indicating that an individual cone may contain both mRNAs, perhaps during the “switch” triggered by 9-cis RA exposure. During such a switch, the formerly transcribed mRNA may still be present while the newly transcribed mRNA is accumulating [83]. It is also possible that cones expressing both LWS genes transcribed LWS1 from one chromosome, and LWS2 from the other. In contrast to the situation in humans where the LWS/MWS array is X-linked, the LWS array of zebrafish is on chromosome 11, an autosome [3].
It was recently demonstrated that embryonic zebrafish LWS cones are generated by committed, TRβ2+ progenitors that undergo a terminal mitotic division to generate a pair of LWS cones that maintain a narrow cytoplasmic attachment as they begin to differentiate [17]. To further address whether the decision to express LWS1 or LWS2 takes place in the progenitor cells or in the postmitotic, differentiating cones, we visually inspected confocal images of eyes derived from RA-treated LWS:PAC(H) embryos for the presence of pairs of cones showing this cytoplasmic attachment (Fig 5J). We observed pairs of daughter cells in each of the following combinations: 1) both expressing LWS2:RFP; 2) both expressing LWS1:GFP; 3) pairs in which one daughter cell expresses both LWS2:RFP and LWS1:GFP while the other cell expresses only LWS2:RFP or only LWS1:GFP (Fig 5J). These results suggest that the decision of LWS1 vs. LWS2 can likely be altered post-mitotically in differentiating cones by RA treatment, although this finding does not rule out effects on cone precursors as well. In support of post-mitotic regulation of LWS expression, adult LWS:PAC(H) (untreated) whole retinas showed many examples of GFP+ cones that were surrounded by RFP+ cones (Fig 5K), consistent with the daughter cells of an LWS cone progenitor adopting asymmetric fates, or with switches in LWS gene expression taking place in fully differentiated cones later during animal growth.
RA-induced LWS1-expressing cones do not disrupt the LWS cone mosaic
The presence of cones co-expressing LWS1 and LWS2 (Fig 5I), or GFP and RFP in the LWS:PAC(H) line (Fig 5B and 5F) following RA treatment suggests that RA promotes an opsin switch in LWS cones rather than LWS1 expression in other cone subtypes. We therefore hypothesized that RA treatment would not disrupt the patterns of LWS cones. We suspected this was the case based upon our prior study showing that the two-dimensional spatial arrangement of LWS cones was not different in embryos treated with RA at 48 hpf, as compared with controls [29]. In order to perform spatial pattern analysis with a sufficient number of GFP+ cones, we treated LWS:PAC(H) embryos with DMSO or 9-cis RA from 48 hpf to 4.5 dpf and imaged whole eyes by confocal microscopy. At 4.5 dpf, many retinas from embryos treated with 9-cis RA contained regions with 10 or more GFP+ cones (Fig 7B–7D), while retinas from DMSO treated embryos only contained RFP+ cones (Fig 7A). The developing RPE and iridophores at this later sampling time made it impossible to analyze whole eyes, so instead we obtained high resolution images at 60X magnification using regions clear of RPE and iridophores for spatial analysis (Fig 7A–7D). For this analysis, the “GFP+” category included cones that express GFP only or those that express both GFP and RFP.
Fig. 7. Two-dimensional pattern analysis of LWS cones in LWS:PAC(H) embryos exposed to RA: Retinoic acid induced LWS1-expressing cones do not disrupt the LWS2 cone mosaic. 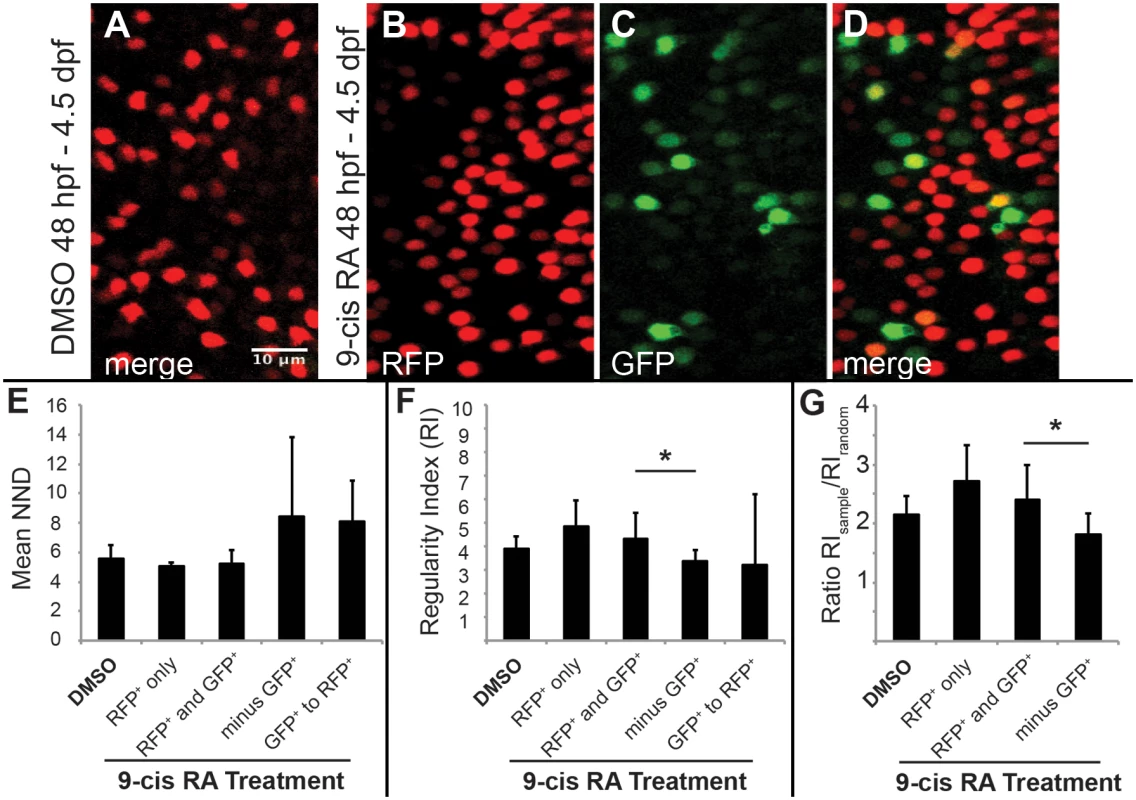
Regions from confocal images at 60X magnification obtained from whole mounted LWS:PAC(H) eyes treated with DMSO or 0.3 μM 9-cis RA 48 hpf to 4.5 dpf were used for pattern analysis. A-D. Representative regions used to determine the mean nearest neighbor distance (NND) and Regularity Index (RI) of LWS cones in control and 9-cis RA-treated eyes. Regions from DMSO treated retinas expressed only LWS2:RFP cones (red) (A), while regions from 9-cis RA treated retinas (B-D) contained RFP+ (red), GFP+ (green), and RFP+/GFP+ (yellow) cones. B-D. Images showing RFP signal only (B), GFP signal only (C), and merge of both signals (D) in regions of 9-cis RA treated retina. Graphs of NND (E, left three bars) and RI (F, left three bars) corresponding to regions from DMSO treated (DMSO) and 9-cis RA treated retinas. 9-cis RA treated retinas were further divided into groups: regions of retina where only RFP+ cones are present (RFP+ only), and regions containing both RFP+ and GFP+ cones (RFP+ and GFP+) when all labeled cells (regardless of RFP or GFP expression) are treated as the same cell type. Graphs also indicate NND (E, right two bars) and RI (F, right two bars) for 9-cis RA treated retinas when GFP+ cones are subtracted from the RFP+ and GFP+ mosaic (minus GFP+) and when the pattern of GFP+ cones to RFP+ cones (GFP+ to RFP+) is analyzed. G. Graph showing the ratio of RI from each indicated region (RIsample) with that of the average RI from 1000 generated random samples of the same number of cells (RIrandom). The asterisk (*) in F and G indicates p<0.05 for a two-tailed Student’s t-test between the indicated groups. Error bars represent standard deviation (n = 7 regions from 4 eyes, DMSO; 6 regions of RFP+ only and 10 regions RFP+ and GFP+ from 6 eyes, RA). Average nearest neighbor distances (NNDs) were similar for patterns found in retinas from DMSO treated and 9-cis RA treated embryos even when GFP+ cones were included in the analysis and treated as the same cell type as RFP+ cones (Fig 7E), suggesting that the GFP+ cones did not disrupt the LWS cone pattern and therefore constitute the same cone population. To further test this hypothesis we calculated regularity indices (RIs); the RI is the mean NND divided by the standard deviation, adjusted for object density. To ensure that RIs of selected regions could indicate patterns of regularity, each region selected for analysis was compared against 1000 random simulations of the same number of objects (Fig 7G). The RIs were similar for patterns found in retinas from DMSO treated and 9-cis RA treated embryos even when GFP+ cells were included in the spatial analysis and treated as the same cell type as RFP+ cones (Fig 7F and 7G). Further, when the GFP+ cones were removed from the combined GFP+ and RFP+ mosaic of 9-cis RA treated retinas, the pattern was disrupted as indicated by an increase in average NND and its standard deviation (Fig 7E) along with a significant reduction in the RI (Fig 7F and 7G). These findings indicate that LWS1 expressing cones observed in RA-treated retinas are in the same cone population as LWS2 cones, and that non-LWS photoreceptor types are not recruited by RA treatment to express LWS1.
We also performed cross-correlative NND analysis of GFP+ cones in relation to RFP+ cones in 9-cis RA treated retinas (Fig 7E and 7F; “GFP+ to RFP+”). The cross-correlative NNDs and RIs were not significantly different from the auto-correlative metrics, again suggesting that the GFP+ cones are in the same cone population as the RFP+ cones.
Reduced RA signaling prevents upregulation of LWS1 during larval development
To investigate endogenous roles for RA signaling in the regulation of the tandemly-duplicated LWS opsin genes, we treated zebrafish with the RA synthesis inhibitor, DEAB [84] (10 μM), from 4–6 dpf, the time of initial expression of LWS1 [12]. DEAB-treated larvae showed significantly reduced expression of both LWS1 and LWS2 mRNA (Fig 6E and 6F), suggesting that endogenous RA signaling is involved in regulating expression from the LWS array. To confirm that DEAB treatment reduced RA signaling, we verified that this treatment also resulted in significantly reduced expression of the RA signaling target nr2f5 mRNA ([77]; present study) as compared to levels in controls (Fig 6G). An additional control gene, β-actin, was not affected by the DEAB treatment (Fig 6H).
As a complementary loss-of-function strategy, we used the transgenic line pd5, which harbors a heat shock-driven dominant-negative (human) RARα transgene [85]. Heat-shocked pd5 larvae also showed significantly reduced expression of nr2f5 as compared to non heat-shocked pd5 controls, providing evidence that a known target of RA signaling was affected by the dominant-negative effect of an overexpressed truncated RAR (Fig 6K). Expression of β-actin was not affected by heat-shock (Fig 6L). The heat-shocked larvae showed significantly reduced expression of LWS1 as compared to controls (Fig 6I), suggesting that an endogenous retinoid receptor [or other nuclear hormone receptor(s)] is/are required for activation of LWS1 at the onset of its expression. Interestingly, expression of LWS2 mRNA was not affected in heat-shocked pd5 larvae (Fig 6J). Together these results demonstrate an endogenous role for RA signaling in upregulating LWS1 at the onset of its expression, but suggest additional factors may participate in the ongoing regulation of LWS2 expression.
Expanded RA signaling domain in RA-treated embryos matches the expanded LWS1 expression domain
The transgenic zebrafish line, RARE:YFP, permits the visualization of active RA signaling within individual tissues and cells, and has previously been used to demonstrate native RA signaling domains predominantly in ventral, and less evidently in dorsal retina during zebrafish development [19,29,33,84]. It is interesting that these native RA signaling domains anticipate the native expression domains of LWS1 [12], consistent with potential endogenous regulatory mechanisms. We treated RARE:YFP embryos with at-RA, 9-cis RA, or DMSO at 48 hpf, and fixed embryos for cryosectioning at 100 hpf. In control embryos, LWS1 expression had not yet commenced, and LWS2 expression was found throughout the ONL (Fig 8A and 8B), similar to the situation for wild-type embryos (compare to Fig 3I and 3J). The RA signaling domain, as revealed by YFP fluorescence, was confined to ventral retina (Fig 8C). As shown previously [19,29], this signaling domain included some cells in the ONL. In RA-treated embryos, LWS1 expression was extensive, although not found throughout the entire ONL, while LWS2 expression was more restricted (Fig 8D and 8E), similar to the response of wild-type zebrafish (compare to Fig 3K and 3L). The YFP+ RA signaling domain was greatly expanded, particularly in the ONL (Fig 8F), and appeared to roughly match the expanded LWS1 expression domain (Fig 8D). Therefore, cells located in the ONL both increase RA signaling and express LWS1 following RA treatment.
Fig. 8. Spatial relationship of RA signaling activity compared with expression of LWS1 and LWS2. 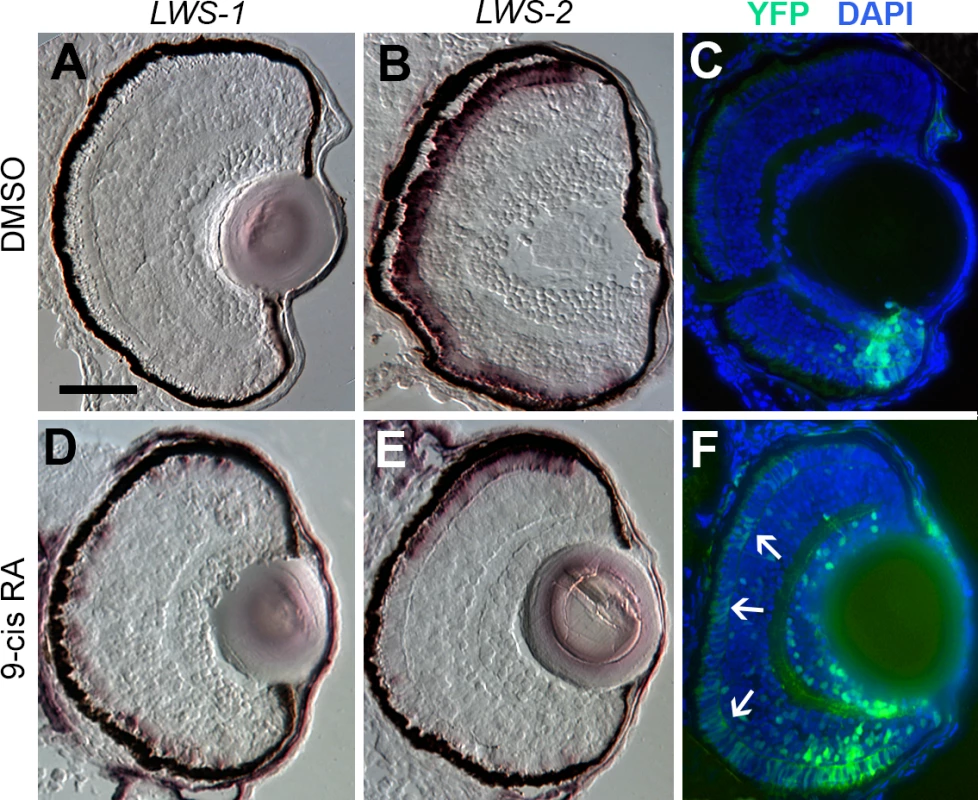
A-C. Sections obtained from a RARE:YFP transgenic embryo treated with DMSO 48 hpf-100 hpf, then labeled for LWS1 mRNA expression (A), LWS2 mRNA expression (B), or YFP immunofluorescence (C; counterstained with DAPI). D-F. Sections obtained from a RARE:YFP transgenic embryo treated with 0.3 μM 9-cis RA 48 hpf– 100 hpf, then labeled for LWS1 mRNA expression (D), LWS2 mRNA expression (E), or YFP immunofluorescence (F). Expanded RA signaling domain (arrows) is similar to the LWS1 expression domain. Scale bar in A (applies to all) = 50 μm. Endogenous RA signaling domain of juvenile zebrafish retina coincides with zone of native LWS1 expression
During larval and juvenile growth of the zebrafish retina, the relative size and shape of the domain of LWS1 expression in red-sensitive cones enlarges to include the ventral ¼ of the retina at the dorsal-ventral midline [12]. The expression of the LWS genes during retinal growth must therefore be dynamic, as the circumferential germinal zone (CGZ) adds new neurons (including cones) at the periphery [10]. We verified that the LWS1 expression domain could be visualized in juvenile (one month old) LWS:PAC(H) transgenics, and discovered an “LWS transition zone” in ventral retina in which many LWS cones co-expressed LWS1 and LWS2 (Fig 9A–9C). This finding is consistent with opsin switching in postmitotic cones as a mechanism for altering the relative sizes of the domains of expression of each LWS gene during retinal growth. In some cryosections, single RFP+ (reporting LWS2) cones were observed at the farthest ventral periphery, adjacent to GFP+ (reporting LWS1) cones (Fig 9B). Dual in situs for the endogenous LWS1 and LWS2 mRNAs confirmed that the most recently-generated LWS cone—located closest to the CGZ—could be an LWS2+ cone (Fig 9C). Therefore, LWS2 expression may constitute the initial or default opsin expression status of newly-generated LWS cones, which in ventral retina then switch to express LWS1 as the retina grows and new cones are added.
Fig. 9. An “LWS Transition Zone” exists in ventral retina of juvenile fish. 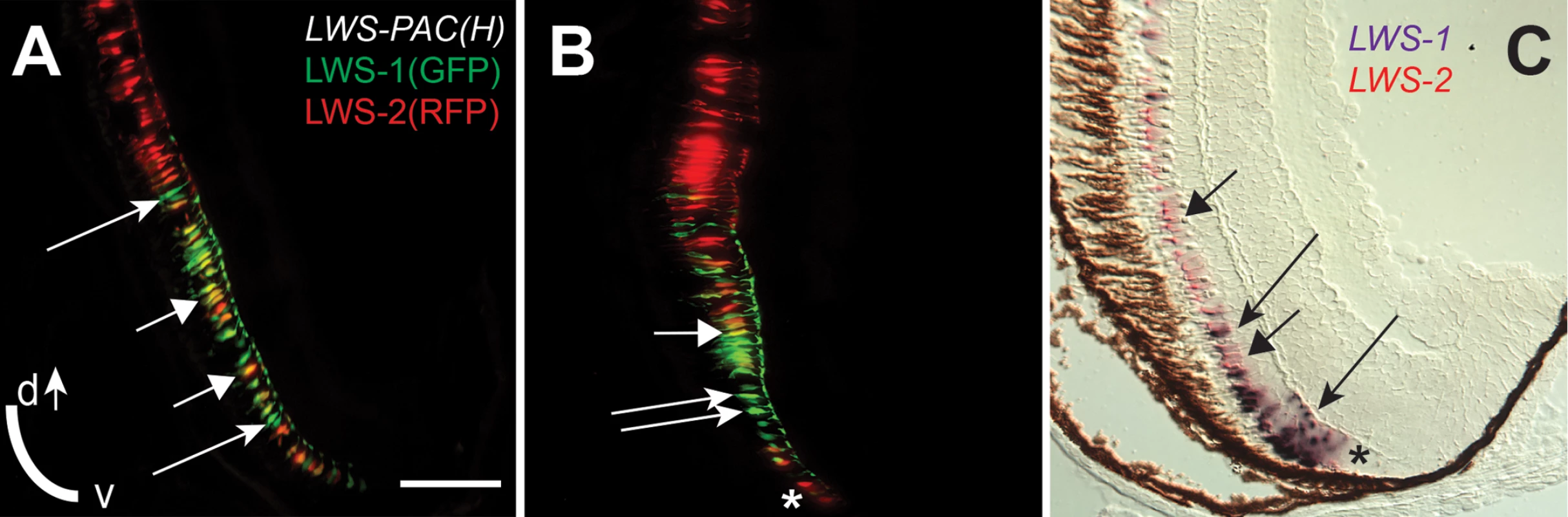
Sections of retina from one-month old juvenile retinas are shown. A and B. Two different sections from separate LWS:PAC(H) transgenic fish showing LWS1:GFP+ cones (green fluorescence; long arrows), LWS2:RFP+ cones (red fluorescence), and cones co-expressing LWS1:GFP and LWS2:RFP (yellow, short arrows). B. The asterisk (*) indicates a cone expressing only LWS2:RFP at the ventral periphery. C. Dual in situ hybridization using cDNA probes for LWS1 (purple) and LWS2 (red/pink). Cones expressing LWS1 only are indicated by long arrows; cones co-labeled with both probes are indicted by short arrows. The asterisk (*) indicates an LWS2 singly labelled cone at the ventral periphery. Scale bar in A (applies to all) = 25 μm; v, ventral; d, dorsal. To evaluate endogenous roles for RA signaling in dynamic regulation of the LWS opsin genes during retinal growth, we used the RARE:YFP transgenic line to localize RA signaling domains in one month old zebrafish. In three fish sampled as cryosections, a YFP+ retinal region was detected in a radial strip of ventral retina, located near and sometimes including the CGZ (Fig 10A). The location of this RA signaling domain was confirmed by in situ hybridization for the mRNA encoding the YFP reporter (Fig 10B). The spatial relationship of the RA signaling domain, as compared to that of the LWS1 expression, was determined in one month old RARE:YFP; LWS:PAC(H) zebrafish. In these fish, YFP fluorescence coincided with the most peripheral region of LWS1 expression (reported by GFP) (Fig 10C and 10D), consistent with endogenous roles for retinal RA signaling regulating differential expression of the LWS opsin genes during retinal growth.
Fig. 10. RA signaling continues in juvenile retinas and the signaling domain includes red opsin+, LWS1+ cones. 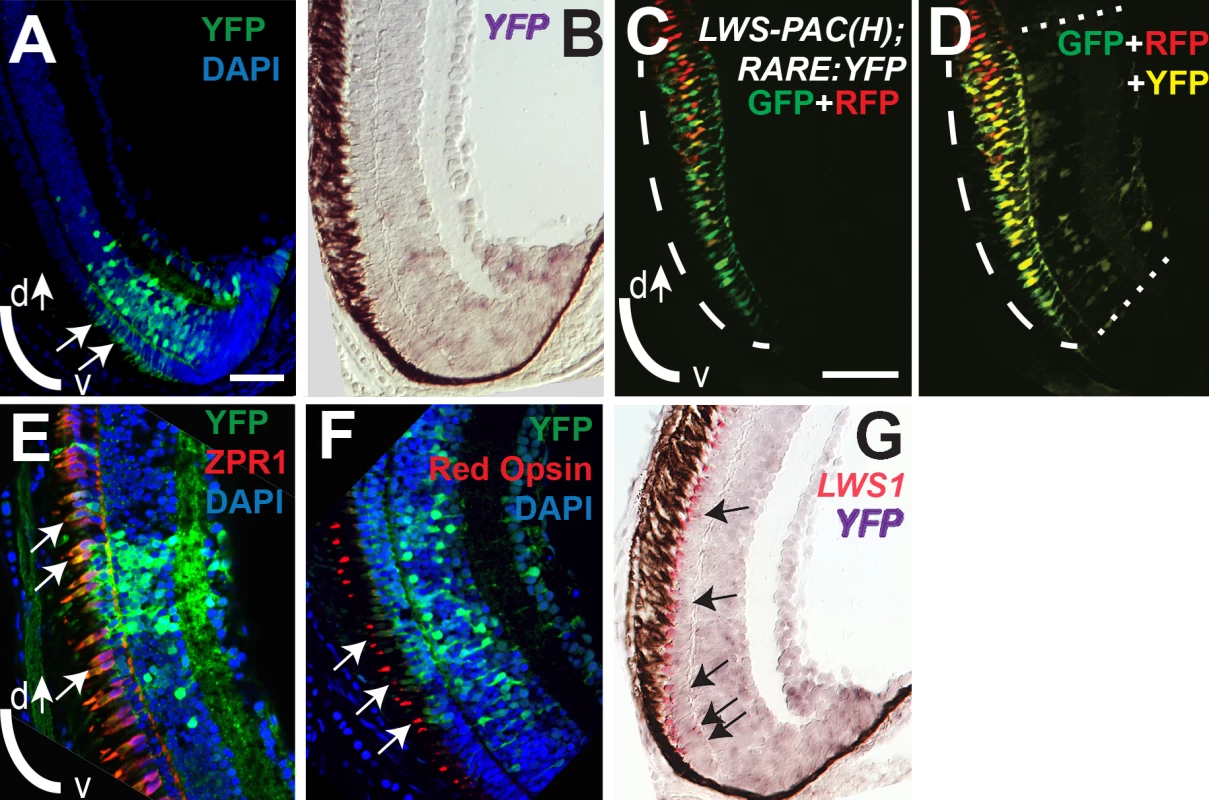
Sections of retina from one-month old fish are shown, with ventral pole at bottom of images. A. Immunocytochemistry to reveal YFP signal and DAPI in a section from RARE:YFP retina. Arrows indicate YFP signal in the outer nuclear layer, corresponding to the location of cone photoreceptors. B. In situ hybridization for topaz-YFP mRNA in a section from a RARE:YFP retina. C and D. LWS:PAC(H) were crossed with RARE:YFP and doubly transgenic juvenile retina sections were examined for RFP, GFP, and YFP fluorescence. Although YFP and GFP cannot be fully resolved, the YFP+ signaling domain coincides with the GFP (LWS1) expression domain. The GFP expression domain (reporting LWS1) is designated by dashed arc (C and D), while the YFP domain (reporting RA signaling) is between the dashed straight lines (D). E. Sections from RARE:YFP juvenile retinas were stained with antibodies for YFP, ZPR1 (labels red- and green-sensitive cones), and DAPI; merged image is shown. Colabel of YFP and ZPR1 is yellow. Arrows indicate ZPR1+ cones that are YFP+. F. Sections from RARE:YFP juvenile retinas were stained with antibodies for YFP, red opsin (pan-LWS), and DAPI; merged image is shown. Arrows indicate red opsin+ cone outer segments that are continuous with YFP+ cell bodies. G. Dual in situ hybridization for topaz-YFP (purple) and LWS1 (red/pink) mRNA in a section from one-month old RARE:YFP retina. Arrows indicate examples of some of the colabeled cones. Scale bar in A = 25 μm; scale bar in C (applies to B-G) = 25 μm; v, ventral; d, dorsal. We wished to determine whether the cones engaged in RA signaling (YFP+) corresponded with those expressing LWS1 (GFP), but the YFP and GFP fluorescent signals were challenging to unambiguously resolve (Fig 10C), even on a spectral scanning confocal microscope. As alternative strategies, we first confirmed that some of the YFP+ cells of the ONL were LWS cones by colabeling experiments in one month old RARE:YFP zebrafish, detecting YFP with indirect immunofluorescence, along with indirect immunofluorescence labeling for the double cone marker ZPR1 [86] (Fig 10E) or a pan-LWS antibody [80,82] (Fig 10F, “Red Opsin”). Next we performed dual in situs for the YFP mRNA together with LWS1 mRNA, and could detect colabeled cells in the ONL (Fig 10G). Collectively the evaluation of juvenile zebrafish retinas supports endogenous functions for RA signaling in regulating differential expression of the tandemly-duplicated LWS opsin genes.
Discussion
Using microarray analysis, this study identified several eye-specific regulatory target genes for RA signaling during the period of embryonic photoreceptor differentiation in the zebrafish. Among these targets was opn1lw1/LWS1, the first member of the tandemly-duplicated genes encoding the long wavelength-sensitive opsin proteins. The results of subsequent qPCR, in situ hybridization, dual in situ hybridization, and the use of selected transgenic lines that report LWS1 vs. LWS2 expression and RA signaling, collectively indicated that increased RA signaling was associated with increased expression of LWS1 while decreasing expression of LWS2 in individual LWS cones. Loss-of-function approaches confirmed that endogenous RA signaling was required for promoting the onset of LWS1 expression. The coincidence of an endogenous RA signaling domain with the domain of LWS1 expression is consistent with a role for RA signaling in regulating ontogenetic changes in LWS opsin expression during normal animal growth. To our knowledge this is the first report of a developmental cell signaling system regulating differential expression of tandemly-duplicated opsin genes.
Ocular targets of RA signaling in the zebrafish embryo
Our microarray analysis identified multiple targets of exogenous RA that encode enzymes and transcription factors involved in regulating RA concentrations and RA signaling, including aldh1a2, cyp26a and b, dhrs3a, and rxrγa (Table 2). In most cases the transcriptional response suggested a metabolic attempt to restore RA homeostasis, and several of these genes were also differentially expressed in response to RA in embryos undergoing somitogenesis [78] and in larval zebrafish heart [77]. These data demonstrate the existence of homeostatic mechanisms to maintain RA levels and/or gradients, and that these mechanisms are conserved within the eye and elsewhere.
The strategy of using whole dissected eyes as starting material for the microarray resulted in the detection of several RA signaling targets that are ocular but likely extraretinal. These targets include genes involved in migration and differentiation of the neural crest-derived periocular mesenchyme and its derivatives [46,47], already known to be regulated by RA. Genes expressed in vascular and hematopoietic lineages (epo, angptl5) are known to be regulated by RA in other organs [87,88], and lens crystallins (though not the specific crystalline genes detected in the present microarray) have been demonstrated as targets of RA signaling [89].
Components of several cell signaling systems were differentially expressed in eyes of RA-treated embryos, most notably two bmps (bmp2b, bmp4), extracellular modifiers of Wnt signaling, and dio2, encoding a deiodinase enzyme that locally converts thyroxine (T4) to tri-iodothyronine (T3; thyroid hormone). All four of these signaling pathways—RA, Bmps, Wnts, and T3 –play roles in dorsal-ventral patterning of the optic vesicle and/or retina [16,24,67,90]. Changes in expression of bmps, and of modifiers of Wnt and T3 signaling, in response to exogenous RA suggest mechanisms for coordination of these signaling pathways in the eye during photoreceptor differentiation, as well as the potential for interactions among these signaling pathways.
A final interesting set of RA signaling target genes within the zebrafish eye were those encoding retinal transcription factors. For example, genes encoding three nuclear hormone receptors (nr0b2a, nr2f5, nrip1b) [56] were upregulated, along with the eye field transcription factor six6a [51,91]. Rxrγa was upregulated by the RA treatment (and is expressed transiently in the ONL of zebrafish embryos [19]). Interestingly, RXRγ in mouse retina is required to prevent expression of S - (blue) opsin in mouse cones [6]. It is possible that the observed decrease in differentiation of SWS1 (UV) and SWS2 (blue) cones in zebrafish in response to RA treatment [29] is mediated by RXRγa. Alternatively, or in addition, RXRγa may be involved in mediating the effects of retinoid signaling on expression of LWS1 and LWS2.
RA signaling regulates differential expression of the tandemly duplicated LWS genes
Excluding genes encoding RA metabolizing enzymes and hox genes, the “top hit” of the microarray analysis was opn1lw1, or LWS1 (S1 Table). LWS1 is the first member of the tandemly-duplicated LWS cone opsin array and is normally not expressed until larval stages. In the adult zebrafish eye LWS1 is restricted to ventral and peripheral retina [3,12]. In the present study, however, treatment with 9-cis RA over a 2–3 day period of embryonic photoreceptor differentiation resulted in significant changeover from LWS2 to LWS1 expression, suggesting that RA signaling may act as a molecular regulator of the LWS array. The greatly enlarged LWS1 expression domain in some embryos—occasionally spanning the entire retina—exceeded the size of the LWS1 expression domain ever observed in native retina [12], indicating that the effect of RA was not simply an acceleration of the normal developmental program. Dual in situ hybridization experiments and confocal analysis of LWS reporter expression in the LWS:PAC(H) transgenic line indicated that this switch takes place within individual, postmitotic cones of a single cone population.
Reduction of RA signaling, achieved using two complementary loss-of-function strategies, consistently reduced LWS1 expression at the time of its normal onset, supporting endogenous functions for RA signaling in promoting LWS1 expression. RA signaling domains in untreated embryos, RA-treated embryos, and in the growing juvenile retina, which predict or coincide with LWS1 expression, provided further evidence for such endogenous functions. However, the RA signaling loss-of-function experiments reported here do not demonstrate opposite effects on LWS2 expression as compared to RA signaling gain-of-function. Reduced RA synthesis (DEAB treatment) downregulated LWS2 as well as LWS1, and overexpression of a dominant-negative RARα caused no significant changes in LWS2 expression. It is possible that LWS2 expression is regulated by additional factors or by a different complement of nuclear hormone receptors than is LWS1, or that our assays were not timed to capture effects of reduced RA signaling on LWS2. The requirement for sustained RA exposure time in embryos (2 days but not 1 day) to decrease LWS2 supports this interpretation. It is also possible that unliganded RA receptors act to repress the entire array, while liganded RA receptors preferentially activate LWS1 over LWS2. Identification of the receptor(s) mediating these effects will be required to pursue this latter hypothesis.
In the zebrafish retina, increased RA signaling therefore has roles in 1) promoting the rod fate at the expense of cone fates [19], 2) promoting differentiation of rods while slowing the differentiation of blue and UV cones [29,30], and 3) favoring the expression of LWS1 over that of LWS2 in individual red-sensitive cones, although not promoting LWS opsin expression in other photoreceptor types (present study). It is possible that during evolution of the vertebrate eye, co-option of endogenous RA signaling emerged as a versatile mechanism for regulating multiple sequential patterning events over developmental time. In support of this idea, the spatial pattern of the endogenous RA signaling domain in embryos matches that of the initial pattern of rods [19,79], and anticipates the pattern of LWS1 expression [12]. The tandemly quadruplicated RH2 (green-sensitive) opsin genes of the zebrafish also show concentric expression domains with a dorsal-ventral gradient [12], but it is not known whether this array responds to RA signaling.
The present study informs our understanding of the transcriptional network underlying photoreceptor fate determination [13], by suggesting a novel mechanism for choice of tandemly replicated opsin (in humans, LWS vs. MWS). The current model for LWS vs. MWS opsin gene regulation in humans assumes that during cone differentiation, a stochastic event favors an association of the upstream locus control region (LCR) with the LWS or most proximal MWS opsin gene, and that this association becomes permanent [20,92,93]. In humans, the LWS:MWS ratio >1.0 suggests a simple proximity argument: the LCR is closer to the LWS gene and therefore more likely to interact with it and promote transcription. However, there is evidence that this stochastic model may not be entirely accurate. For example, the LWS:MWS ratio varies as a function of retinal eccentricity, with highest LWS:MWS ratios in the periphery [9]. The LWS:MWS ratio also varies as a function of ethnic descent [94], a variability not explained by any differences in LWS vs. MWS gene order, distance from LCR, or promoter sequence [94,95]. These data suggest that a nonrandom trans regulatory mechanism may instead regulate LWS vs. MWS opsin expression. Here we have identified a potential mechanism in the zebrafish, in which RA signaling regulates expression of the orthologous locus containing LWS1 and LWS2. The LWS array of zebrafish contains numerous consensus retinoic acid response elements (RAREs; S2 Fig), suggesting the potential for direct regulation of each array by RA in conjunction with an appropriate receptor homo - or heterodimer. However, direct interactions of any RAR or RXR with an opsin gene regulatory region have not been demonstrated, and so this idea remains highly speculative.
The observation that pharmacological agents such as retinoids can control photoreceptor fates [19], differentiation [27–30], and now choice of tandemly replicated opsin (the present study) suggests the potential for pharmacological manipulation of photoreceptor phenotypes in conjunction with anticipated regenerative therapies for human retinal disease [96]. Because nuclear hormone receptors in general respond to pharmacological treatments, they are considered attractive candidates for manipulating photoreceptor phenotype in the treatment of retinal disease [97]. These findings have already seen applications in the generation and differentiation of photoreceptors from human embryonic stem (ES) cells and induced pluripotent stem (iPS) cells [34]. Regulation of human LWS/MWS expression in concert with regenerative therapies will be important for the re-establishment of high acuity trichromatic color vision. In addition there is potential for correction of color vision anomalies or X-linked cone dystrophy in individuals with defects in one or two genes of the LWS/MWS array (that could be pharmacologically suppressed), but also with one or two normal genes (that could be pharmacologically activated) [98,99], as an alternative to gene therapy [100].
Materials and Methods
Animals
Zebrafish were maintained in monitored aquatic housing units on recirculating system water at 28.5°C. Embryos were collected according to Westerfield [101], with light onset considered to be zero hours postfertilization (hpf) and embryonic age timed accordingly thereafter. Embryos used for whole mount analyses were kept transparent by incubating them in system water containing 0.003% phenothiourea (PTU) to inhibit melanin synthesis [101]. All experiments using animals were approved by the University of Idaho’s Animal Care and Use Committee.
Wild-type embryos were of an in-house outbred strain originally obtained from Scientific Hatcheries (now Aquatica Tropicals, Plant City, FL) and are referred to as SciH. The transgenic zebrafish line RGnY was generously provided by Elwood Linney. The transgene consists of three copies of retinoic acid response elements (RAREs) derived from the mouse RARβ gene, a zebrafish basal promoter, a Topaz YFP sequence, an SV40 polyadenylation signal, and a small t intron [84]. The endogenous expression patterns of YFP in these fish are consistent with known areas undergoing RA signaling [29,84] and YFP reporter expression increases in response to exogenous RA [19,84]. We refer to this line as RARE:YFP. In the transgenic line Tg(LWS1/GFP-LWS2/RFP-PAC(H))#430, the “transgene” consists of a PAC clone in which the first exons of LWS1 and LWS2 were replaced with GFP and RFP, respectively, each followed by a polyadenylation sequence [82]. The spatiotemporal expression patterns of GFP and RFP replicate endogenous patterns of LWS1 and LWS2 [82]. We refer to this line as LWS:PAC(H). The transgenic zebrafish line Tg(hsp70:dnRARα, cryaa:GFP)pd5 [85] was obtained from the Zebrafish International Resource Center (ZIRC), and we refer to it as pd5. The transgene consists of a heat shock (hsp70) promoter driving expression of a dominant-negative (truncated) human RARα, and a separate cassette containing cryaa promoter driving GFP constitutively in the lens [85,102]. When pd5 transgenics are subjected to heat shock, expression of dnRARα causes global deficiency in RA signaling [85].
Retinoid and DEAB treatments and heat shock
Stock solutions of all-trans retinoic acid (at-RA), 9-cis retinoic acid (9-cis RA), bexarotene, and diethylaminobenzaldehyde (DEAB; Sigma, St. Louis, MO) were prepared in dimethylsulfoxide (DMSO; Sigma) and stored under nitrogen in the dark at -20°C. Prior to treatment, embryos were manually dechorionated, and then stock solution was added to the water to result in the final concentrations indicated in Results (DMSO was at a final concentration of 0.1%). The 0.3 μM concentration of RA used in the microarray experiment influences photoreceptor fate and differentiation in zebrafish embryos [19,29,30,33]. For treatments lasting longer than one day, solutions were refreshed every 24 hrs.
Heat shocks (to induce expression of dnRARα in the pd5 transgenics and result in RA signaling loss-of-function) were performed at 5 and 6 dpf by transferring embryos to 37°C for 1 hr.
Microarray and analysis
Three sets of parents from the SciH line were used to obtain three clutches of embryos. Each clutch was separated into control and experimental groups, which were treated from 48 to 75 hpf with DMSO (control) or 0.3 μM at-RA (experimental). This generated three biological replicates of control and experimental treatments for six total treatment groups. At 75 hpf, embryos were placed in scintillation vials and snap-frozen by immersing the vial into liquid nitrogen. The vial was filled with methanol pre-chilled on dry ice and embryos were stored at -80°C. After seven days, eyes were dissected from embryos using fine forceps [103,104]. Each treatment group consisted of 64 eyes. Eyes were homogenized using a roto-homogenizer and total RNA was extracted using the RNeasy Kit (Qiagen, Valencia, CA) and quantified (Nano-Drop 1000; Thermo Scientific, Wilmington, DE).
A total of 80 ng of RNA from each treatment group was analyzed to verify quality (Agilent Bioanalyzer, Agilent Technologies, Inc., Wilmington, DE), and amplified using the NuGen kit (San Carlos, CA). Gene expression in the six eye-specific samples was determined using Affymetrix GeneChip Zebrafish Genome Arrays (Affymetrix, Santa Clara, CA). The amplification and microarray procedures were performed at the Genomics Core of the Center for Reproductive Biology (Washington State University, Pullman, WA). The raw data were adjusted for background, normalized, and visualized as RMA (Robust Multi-array Average) using the Bioconductor package in R [105]. To identify differentially expressed genes we used significance analysis of microarray (SAM; [106]). The results were analyzed at False Discovery Rates (FDR) of 10% and 20%. Probe data were annotated using the Zebrafish Genome Assembly version Zv8 (Sanger Institute). The microarray data are available in the NCBI Gene Expression Omnibus (GEO; accession #GSE63873). A gene ontology (GO) analysis was performed using GOEAST (Gene Ontology Enrichment Analysis Software Toolkit), an online tool [39], with a focus on biological processes enriched in the set of differentially expressed genes.
The data were compared to those obtained using Affymetrix platform-based microarray analysis of RA treatments in zebrafish embryos, over different developmental times or in different tissues [77,78]. Raw data were accessed from GEO, were adjusted for background, normalized, and visualized as RMA expression values, and differentially expressed genes were identified using SAM [106]. Genes that were differentially expressed at 10% FDR in two or more datasets ([77,78] and current dataset) were considered potential common regulatory targets of RA signaling.
Quantitative reverse transcriptase polymerase chain reaction analysis
Total RNA from each treatment group was used to synthesize cDNA template using the High Capacity cDNA Reverse Transcription kit with random primers (Applied Biosystems, Inc. [ABI], Foster City, CA). Gene-specific primer pairs are listed in Table 1. Amplification was performed on a model 7900HT Fast Real-Time PCR System using SYBR-Green PCRMaster Mix (ABI). Relative quantitation of gene expression between control and experimental treatments was determined using the 18s ribosomal RNA as the endogenous reference. Graphing and statistics were performed using the R statistical environment [107].
Histological processing, immunocytochemistry, and in situ hybridization
Fixation and preparation of embryos for tissue sectioning, immunocytochemistry, and in situ hybridization were performed as previously described [19,108,109]. For immunocytochemistry, tissue sections were blocked in 20% goat serum for 30 min, incubated with primary antibody overnight at 4°C, washed in PBS containing 0.0% Triton X-100 (Sigma), then incubated with a fluorescent secondary antibody, and then mounted in VectaShield, with or without DAPI (Vector Labs). Antibodies used were rabbit polyclonal anti-GFP (1 : 1,000; Torrey Pines Biolabs), chicken anti-GFP (1 : 1,000; Abcam), mouse monoclonal ZPR-1 which labels double cones (1 : 200; ZIRC), rabbit polyclonal anti-zebrafish LWS opsin (1 : 500; gift of David Hyde [80]), FITC - or Cy3-or Alexa-Fluor 647-conjugated secondary antibodies (1 : 200; Jackson ImmunoResearch). For in situ experiments, cRNA probes were generated by in vitro reverse transcription of cDNAs. For single in situ experiments, digoxigenin - (dig-) UTPs were incorporated into probes for detection with anti-dig antibodies conjugated to alkaline phosphatase and visualized with NBT-BCIP substrate. Dual in situ hybridizations were also performed as described [19,110,111]. One probe was generated with dig-UTPs, and the other with fluorescein (FL)-UTPs. Dig probes were detected by anti-dig antibodies conjugated to alkaline phosphatase, followed by the NBT-BCIP phosphatase substrate, resulting in a purple precipitate. Sections were then post-fixed with 4% paraformaldehyde, and the FL probe was detected using anti-FL antibodies conjugated to alkaline phosphatase, followed by the Fast Red phosphatase substrate, resulting in a red fluorescent precipitate [110,112,113].
Probes for in situs were generated from the following cDNAs: LWS1-specific (3’UTR; [12]); LWS2-specific (3’UTR; [12]); pan-LWS/zfRed (corresponds directly to LWS-2 but hybridizes to both LWS transcripts; [12,29]), and Topaz-YFP (cloned from the RARE:YFP transgenics [84]).
Photography and quantification of embryonic in situ hybridization patterns
Images were captured using a Leica DMR compound microscope with a SPOT camera system (Diagnostic Instruments). Fluorescently-labeled tissues were viewed using epifluorescence optics, and in situs were viewed using epifluorescence, and/or Nomarski (differential interference contrast) optics.
In some experiments, individual, labeled cone photoreceptors were counted within a subset of non-overlapping cryosections. In other experiments, individual, labeled cones were not consistently discernable, and so sections were scored based upon criteria characteristic of LWS1 or LWS2 patterns. For LWS1: 0 = no expression; 1 = one to five labeled cells in ventral patch; 2 = one-10 in ventral patch, plus one-five scattered beyond ventral retina; 3 = one-10 in ventral patch, plus >5 scattered beyond ventral retina; 4 = widespread label throughout ONL. For LWS2: 0 = no expression; 1 = one to five labeled cells in dorsal patch; 2 = less than ½ of dorsal retina strongly labeled, weak label elsewhere; 3 = ½ to ¾ of retina strongly labeled, weaker label elsewhere; 4 = strong label throughout ONL.
Labeling of cones in whole mounts by in situ hybridization was quantified using the cone recruitment stages defined in [79], and modified as follows. Stage 0 = no labeling; stage 1 = fewer than 10 cells express the cone opsin; stage 2; 10–20 cones express the cone opsin; stage 3 = more than ¼ but less than 1/3 of the retina expresses the cone opsin; stage 4 = more than 1/3 but less than ½ of the retina expresses the cone opsin; stage 5 = more than ½ but less than the entire retina expresses the cone opsin; stage 6 = entire retina expresses the cone opsin.
Confocal photography and quantification: LWS:PAC(H) embryos
LWS:PAC(H) embryos [82] were maintained in system water with PTU (see above). At 48 hpf, embryos were treated with 9-cis RA, at-RA, or DMSO through 96 hpf (4 dpf) or 108 hpf (4.5 dpf), and then fixed in 4% paraformaldehyde sucrose solution for 1 hour or overnight, washed once in sucrose solution for 30 minutes followed by three washes in PBS. Following fixation and washing, embryos were incubated in PBS at 4°C in the dark for no longer than 24 hours. Immediately prior to imaging, whole eyes were removed from fixed embryos, the sclera teased away by microdissection, and eyes were then coverslipped in glycerol. Imaging was performed at 20X or 60X magnification using an Olympus Fluoview 1000 Laser Scanning confocal microscope running Fluoview ASW software. A z-series covering the entire retinal hemisphere was obtained with 2 micron step sizes. FIJI (ImageJ) was used to flatten z stacks via max projection and adjust brightness/contrast. Images from samples where GFP signal was not resolvable in all planes (due to the developing RPE and/or iridophores in residual sclera) were excluded from analysis.
For pattern analysis, Z stack images of whole mounted eyes acquired from DMSO or 9-cis RA treated LWS:PAC(H) embryos at 60X magnification were flattened via max projection using FIJI. Rectangular regions with dimension of 40–90 x 40–100 microns were selected from images obtained from samples corresponding to DMSO or 9-cis RA treatment. Cones were identified with an average cell diameter set at 4 μm. Regions were analyzed using WinDRP software (downloaded from the Massaman Lab) to calculate the Nearest Neighbor Distance (NND) and Regularity Index (RI) of the cone mosaic [114,115]. For each region analyzed, patterns were compared against 1000 random simulations of the same number of cells.
Confocal photography: Juvenile zebrafish sections
One month old juvenile zebrafish were anaesthetized and decapitated. Heads were fixed in 4% paraformaldehyde/sucrose solution as described above for 30 minutes. After 30 minutes, heads were removed and corneas punctured with a dissecting pin, then returned to the fixation solution for another 30 minutes. Heads were then processed for sectioning as previously described [19,108,109] and sectioned at 5 μm. For transgenic LWS:PAC(H) and/or RARE:YFP fish, sections were mounted in Vectashield, covered with a coverslip and sealed. Direct imaging of expressed fluorescently tagged proteins was performed at 40X magnification using a Leica DMR compound epifluorescence microscope and SPOT camera, and a NikonAndor spinning disk confocal microscope (water immersion) equipped with a Xyla sCMOS camera. For immunocytochemistry of sections from juvenile fish, sections were stained with antibodies as described above. Imaging was performed at 40X magnification (water immersion) using the Nikon Andor Spinning Disk confocal microscope and Xyla sCMOS camera. Image analysis was performed using FIJI.
Supporting Information
Zdroje
1. Nathans J (1990) Determinants of visual pigment absorbance: identification of the retinylidene Schiff's base counterion in bovine rhodopsin. Biochemistry 29 : 9746–9752. 1980212
2. Yokoyama S (2000) Molecular evolution of vertebrate visual pigments. Prog Retin Eye Res 19 : 385–419. 10785616
3. Chinen A, Hamaoka T, Yamada Y, Kawamura S (2003) Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics 163 : 663–675. 12618404
4. Vollrath D, Nathans J, Davis RW (1988) Tandem array of human visual pigment genes at Xq28. Science 240 : 1669–1672. 2837827
5. Fei Y (2003) Development of the cone photoreceptor mosaic in the mouse retina revealed by fluorescent cones in transgenic mice. Mol Vis 9 : 31–42. 12592228
6. Roberts MR, Hendrickson A, McGuire CR, Reh TA (2005) Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci 46 : 2897–2904. 16043864
7. Applebury ML, Antoch MP, Baxter LC, Chun LL, Falk JD, et al. (2000) The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron 27 : 513–523. 11055434
8. Ahnelt PK (1998) The photoreceptor mosaic. Eye (Lond) 12 (Pt 3b): 531–540.
9. Kuchenbecker JA, Sahay M, Tait DM, Neitz M, Neitz J (2008) Topography of the long - to middle-wavelength sensitive cone ratio in the human retina assessed with a wide-field color multifocal electroretinogram. Vis Neurosci 25 : 301–306. doi: 10.1017/S0952523808080474 18598401
10. Stenkamp DL (2007) Neurogenesis in the fish retina. Int Rev Cytol 259 : 173–224. 17425942
11. Stenkamp DL, Cameron DA (2002) Cellular pattern formation in the retina: retinal regeneration as a model system. Mol Vis 8 : 280–293. 12181523
12. Takechi M, Kawamura S (2005) Temporal and spatial changes in the expression pattern of multiple red and green subtype opsin genes during zebrafish development. J Exp Biol 208 : 1337–1345. 15781894
13. Swaroop A, Kim D, Forrest D (2010) Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci 11 : 563–576. doi: 10.1038/nrn2880 20648062
14. Webber AL, Hodor P, Thut CJ, Vogt TF, Zhang T, et al. (2008) Dual role of Nr2e3 in photoreceptor development and maintenance. Exp Eye Res 87 : 35–48. doi: 10.1016/j.exer.2008.04.006 18547563
15. Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, et al. (2001) Nrl is required for rod photoreceptor development. Nat Genet 29 : 447–452. 11694879
16. Roberts MR, Srinivas M, Forrest D, Morreale de Escobar G, Reh TA (2006) Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc Natl Acad Sci U S A 103 : 6218–6223. 16606843
17. Suzuki SC, Bleckert A, Williams PR, Takechi M, Kawamura S, et al. (2013) Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc Natl Acad Sci U S A 110 : 15109–15114. doi: 10.1073/pnas.1303551110 23980162
18. Alvarez-Delfin K, Morris AC, Snelson CD, Gamse JT, Gupta T, et al. (2009) Tbx2b is required for ultraviolet photoreceptor cell specification during zebrafish retinal development. Proc Natl Acad Sci U S A 106 : 2023–2028. doi: 10.1073/pnas.0809439106 19179291
19. Stevens CB, Cameron DA, Stenkamp DL (2011) Plasticity of photoreceptor-generating retinal progenitors revealed by prolonged retinoic acid exposure. BMC Dev Biol 11 : 51. doi: 10.1186/1471-213X-11-51 21878117
20. Smallwood PM, Wang Y, Nathans J (2002) Role of a locus control region in the mutually exclusive expression of human red and green cone pigment genes. Proc Natl Acad Sci U S A 99 : 1008–1011. 11773636
21. Deeb SS (2006) Genetics of variation in human color vision and the retinal cone mosaic. Curr Opin Genet Dev 16 : 301–307. 16647849
22. Mey J, McCaffery P, Klemeit M (2001) Sources and sink of retinoic acid in the embryonic chick retina: distribution of aldehyde dehydrogenase activities, CRABP-I, and sites of retinoic acid inactivation. Brain Res Dev Brain Res 127 : 135–148. 11335000
23. Pittlik S, Domingues S, Meyer A, Begemann G (2008) Expression of zebrafish aldh1a3 (raldh3) and absence of aldh1a1 in teleosts. Gene Expr Patterns 8 : 141–147. doi: 10.1016/j.gep.2007.11.003 18178530
24. McCaffery P, Wagner E, O'Neil J, Petkovich M, Drager UC (1999) Dorsal and ventral retinal territories defined by retinoic acid synthesis, break-down and nuclear receptor expression. Mech Dev 82 : 119–130. 10354476
25. Marsh-Armstrong N, McCaffery P, Gilbert W, Dowling JE, Drager UC (1994) Retinoic acid is necessary for development of the ventral retina in zebrafish. Proc Natl Acad Sci U S A 91 : 7286–7290. 8041782
26. Lupo G, Gestri G, O'Brien M, Denton RM, Chandraratna RA, et al. (2011) Retinoic acid receptor signaling regulates choroid fissure closure through independent mechanisms in the ventral optic cup and periocular mesenchyme. Proc Natl Acad Sci U S A 108 : 8698–8703. doi: 10.1073/pnas.1103802108 21555593
27. Kelley MW, Turner JK, Reh TA (1995) Regulation of proliferation and photoreceptor differentiation in fetal human retinal cell cultures. Invest Ophthalmol Vis Sci 36 : 1280–1289. 7775105
28. Kelley MW, Turner JK, Reh TA (1994) Retinoic acid promotes differentiation of photoreceptors in vitro. Development 120 : 2091–2102. 7925013
29. Prabhudesai SN, Cameron DA, Stenkamp DL (2005) Targeted effects of retinoic acid signaling upon photoreceptor development in zebrafish. Dev Biol 287 : 157–167. 16197938
30. Hyatt GA, Schmitt EA, Fadool JM, Dowling JE (1996) Retinoic acid alters photoreceptor development in vivo. Proc Natl Acad Sci U S A 93 : 13298–13303. 8917585
31. Wallace VA, Jensen AM (1999) IBMX, taurine and 9-cis retinoic acid all act to accelerate rhodopsin expression in postmitotic cells. Exp Eye Res 69 : 617–627. 10620391
32. Stenkamp DL, Gregory JK, Adler R (1993) Retinoid effects in purified cultures of chick embryo retina neurons and photoreceptors. Invest Ophthalmol Vis Sci 34 : 2425–2436. 8325750
33. Kashyap B, Frey R.A., Stenkamp D.L. (2011) Ethanol-Induced Microphthalmia Is Not Mediated by Changes in Retinoic Acid or Sonic Hedgehog Signaling During Retinal Neurogenesis. Alcoholism: Clinical and Experimental Research In press.
34. Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, et al. (2014) Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun 5 : 4047. doi: 10.1038/ncomms5047 24915161
35. Osakada F, Jin ZB, Hirami Y, Ikeda H, Danjyo T, et al. (2009) In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci 122 : 3169–3179. doi: 10.1242/jcs.050393 19671662
36. Mellough CB, Sernagor E, Moreno-Gimeno I, Steel DH, Lako M (2012) Efficient stage-specific differentiation of human pluripotent stem cells toward retinal photoreceptor cells. Stem Cells 30 : 673–686. doi: 10.1002/stem.1037 22267304
37. Schmitt EA, Dowling JE (1999) Early retinal development in the zebrafish, Danio rerio: light and electron microscopic analyses. J Comp Neurol 404 : 515–536. 9987995
38. Hu M, Easter SS (1999) Retinal neurogenesis: the formation of the initial central patch of postmitotic cells. Dev Biol 207 : 309–321. 10068465
39. Zheng Q, Wang XJ (2008) GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res 36: W358–363. doi: 10.1093/nar/gkn276 18487275
40. Pennimpede T, Cameron DA, MacLean GA, Li H, Abu-Abed S, et al. (2010) The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis. Birth Defects Res A Clin Mol Teratol 88 : 883–894. doi: 10.1002/bdra.20709 20842651
41. Kam RK, Shi W, Chan SO, Chen Y, Xu G, et al. (2013) Dhrs3 protein attenuates retinoic acid signaling and is required for early embryonic patterning. J Biol Chem 288 : 31477–31487. doi: 10.1074/jbc.M113.514984 24045938
42. Ang HL, Duester G (1999) Retinoic acid biosynthetic enzyme ALDH1 localizes in a subset of retinoid-dependent tissues during xenopus development. Dev Dyn 215 : 264–272. 10398536
43. Hu P, Tian M, Bao J, Xing G, Gu X, et al. (2008) Retinoid regulation of the zebrafish cyp26a1 promoter. Dev Dyn 237 : 3798–3808. doi: 10.1002/dvdy.21801 19035346
44. Dobbs-McAuliffe B, Zhao Q, Linney E (2004) Feedback mechanisms regulate retinoic acid production and degradation in the zebrafish embryo. Mech Dev 121 : 339–350. 15110044
45. Zhao Q, Dobbs-McAuliffe B, Linney E (2005) Expression of cyp26b1 during zebrafish early development. Gene Expr Patterns 5 : 363–369. 15661642
46. Bohnsack BL, Kasprick DS, Kish PE, Goldman D, Kahana A (2012) A zebrafish model of axenfeld-rieger syndrome reveals that pitx2 regulation by retinoic acid is essential for ocular and craniofacial development. Invest Ophthalmol Vis Sci 53 : 7–22. doi: 10.1167/iovs.11-8494 22125274
47. Bohnsack BL, Kahana A (2013) Thyroid hormone and retinoic acid interact to regulate zebrafish craniofacial neural crest development. Dev Biol 373 : 300–309. doi: 10.1016/j.ydbio.2012.11.005 23165295
48. Diehl AG, Zareparsi S, Qian M, Khanna R, Angeles R, et al. (2006) Extraocular muscle morphogenesis and gene expression are regulated by Pitx2 gene dose. Invest Ophthalmol Vis Sci 47 : 1785–1793. 16638982
49. McEvoy J, Nagahawatte P, Finkelstein D, Richards-Yutz J, Valentine M, et al. (2014) RB1 gene inactivation by chromothripsis in human retinoblastoma. Oncotarget 5 : 438–450. 24509483
50. Livide G, Epistolato MC, Amenduni M, Disciglio V, Marozza A, et al. (2012) Epigenetic and copy number variation analysis in retinoblastoma by MS-MLPA. Pathol Oncol Res 18 : 703–712. doi: 10.1007/s12253-012-9498-8 22278416
51. Dixit R, Tachibana N, Touahri Y, Zinyk D, Logan C, et al. (2014) Gene expression is dynamically regulated in retinal progenitor cells prior to and during overt cellular differentiation. Gene Expr Patterns 14 : 42–54. doi: 10.1016/j.gep.2013.10.003 24148613
52. Maeda A, Moriguchi T, Hamada M, Kusakabe M, Fujioka Y, et al. (2009) Transcription factor GATA-3 is essential for lens development. Dev Dyn 238 : 2280–2291. doi: 10.1002/dvdy.22035 19623612
53. Kobrossy L, Rastegar M, Featherstone M (2006) Interplay between chromatin and trans-acting factors regulating the Hoxd4 promoter during neural differentiation. J Biol Chem 281 : 25926–25939. 16757478
54. Thisse B, Heyer V, Lux A, Alunni V, Degrave A, et al. (2004) Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol 77 : 505–519. 15602929
55. Gomez G, Lee JH, Veldman MB, Lu J, Xiao X, et al. (2012) Identification of vascular and hematopoietic genes downstream of etsrp by deep sequencing in zebrafish. PLoS One 7: e31658. doi: 10.1371/journal.pone.0031658 22438865
56. Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, et al. (2007) Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet 3: e188. 17997606
57. McMahon C, Gestri G, Wilson SW, Link BA (2009) Lmx1b is essential for survival of periocular mesenchymal cells and influences Fgf-mediated retinal patterning in zebrafish. Dev Biol 332 : 287–298. doi: 10.1016/j.ydbio.2009.05.577 19500562
58. Zhang J, Jin Z, Bao ZZ (2004) Disruption of gradient expression of Zic3 resulted in abnormal intra-retinal axon projection. Development 131 : 1553–1562. 14985256
59. Petrova IM, Malessy MJ, Verhaagen J, Fradkin LG, Noordermeer JN (2014) Wnt signaling through the Ror receptor in the nervous system. Mol Neurobiol 49 : 303–315. doi: 10.1007/s12035-013-8520-9 23990374
60. Kruse-Bend R, Rosenthal J, Quist TS, Veien ES, Fuhrmann S, et al. (2012) Extraocular ectoderm triggers dorsal retinal fate during optic vesicle evagination in zebrafish. Dev Biol 371 : 57–65. doi: 10.1016/j.ydbio.2012.08.004 22921921
61. French CR, Erickson T, French DV, Pilgrim DB, Waskiewicz AJ (2009) Gdf6a is required for the initiation of dorsal-ventral retinal patterning and lens development. Dev Biol 333 : 37–47. doi: 10.1016/j.ydbio.2009.06.018 19545559
62. Trimarchi JM, Harpavat S, Billings NA, Cepko CL (2008) Thyroid hormone components are expressed in three sequential waves during development of the chick retina. BMC Dev Biol 8 : 101. doi: 10.1186/1471-213X-8-101 18854032
63. Untergasser G, Martowicz A, Hermann M, Tochterle S, Meyer D (2011) Distinct expression patterns of dickkopf genes during late embryonic development of Danio rerio. Gene Expr Patterns 11 : 491–500. doi: 10.1016/j.gep.2011.08.005 21889616
64. Marcelli F, Boisset G, Schorderet DF (2014) A Dimerized HMX1 Inhibits EPHA6/epha4b in Mouse and Zebrafish Retinas. PLoS One 9: e100096. doi: 10.1371/journal.pone.0100096 24945320
65. Juul SE, Yachnis AT, Christensen RD (1998) Tissue distribution of erythropoietin and erythropoietin receptor in the developing human fetus. Early Hum Dev 52 : 235–249. 9808074
66. Zhou J, Li W, Kamei H, Duan C (2008) Duplication of the IGFBP-2 gene in teleost fish: protein structure and functionality conservation and gene expression divergence. PLoS One 3: e3926. doi: 10.1371/journal.pone.0003926 19081843
67. Holly VL, Widen SA, Famulski JK, Waskiewicz AJ (2014) Sfrp1a and Sfrp5 function as positive regulators of Wnt and BMP signaling during early retinal development. Dev Biol 388 : 192–204. doi: 10.1016/j.ydbio.2014.01.012 24457098
68. Yin J, Shine L, Raycroft F, Deeti S, Reynolds A, et al. (2012) Inhibition of the Pim1 oncogene results in diminished visual function. PLoS One 7: e52177. doi: 10.1371/journal.pone.0052177 23300608
69. Veldman MB, Bemben MA, Thompson RC, Goldman D (2007) Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol 312 : 596–612. 17949705
70. Higashijima S, Nose A, Eguchi G, Hotta Y, Okamoto H (1997) Mindin/F-spondin family: novel ECM proteins expressed in the zebrafish embryonic axis. Dev Biol 192 : 211–227. 9441663
71. Kassen SC, Ramanan V, Montgomery JE, C TB, Liu CG, et al. (2007) Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev Neurobiol 67 : 1009–1031. 17565703
72. Haque R, Alonso-Gomez AL, Chaurasia SS, Iuvone PM (2003) Diurnal regulation of arylalkylamine N-acetyltransferase activity in chicken retinal cells in vitro: analysis of culture conditions. Mol Vis 9 : 52–59. 12629487
73. Wetzel RK, Arystarkhova E, Sweadner KJ (1999) Cellular and subcellular specification of Na,K-ATPase alpha and beta isoforms in the postnatal development of mouse retina. J Neurosci 19 : 9878–9889. 10559397
74. Madreperla SA, Edidin M, Adler R (1989) Na+,K+-adenosine triphosphatase polarity in retinal photoreceptors: a role for cytoskeletal attachments. J Cell Biol 109 : 1483–1493. 2551908
75. Hensley MR, Emran F, Bonilla S, Zhang L, Zhong W, et al. (2011) Cellular expression of Smarca4 (Brg1)-regulated genes in zebrafish retinas. BMC Dev Biol 11 : 45. doi: 10.1186/1471-213X-11-45 21756345
76. Vihtelic TS, Fadool JM, Gao J, Thornton KA, Hyde DR, et al. (2005) Expressed sequence tag analysis of zebrafish eye tissues for NEIBank. Mol Vis 11 : 1083–1100. 16379021
77. Chen J (2013) Impaired cardiovascular function caused by different stressors elicits a common pathological and transcriptional response in zebrafish embryos. Zebrafish 10 : 389–400. doi: 10.1089/zeb.2013.0875 23837677
78. Feng L, Hernandez RE, Waxman JS, Yelon D, Moens CB (2010) Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev Biol 338 : 1–14. doi: 10.1016/j.ydbio.2009.10.029 19874812
79. Raymond PA, Barthel LK, Curran GA (1995) Developmental patterning of rod and cone photoreceptors in embryonic zebrafish. J Comp Neurol 359 : 537–550. 7499546
80. Vihtelic TS, Doro CJ, Hyde DR (1999) Cloning and characterization of six zebrafish photoreceptor opsin cDNAs and immunolocalization of their corresponding proteins. Vis Neurosci 16 : 571–585. 10349976
81. Tooker P, Yen WC, Ng SC, Negro-Vilar A, Hermann TW (2007) Bexarotene (LGD1069, Targretin), a selective retinoid X receptor agonist, prevents and reverses gemcitabine resistance in NSCLC cells by modulating gene amplification. Cancer Res 67 : 4425–4433. 17483357
82. Tsujimura T, Hosoya T, Kawamura S (2010) A single enhancer regulating the differential expression of duplicated red-sensitive opsin genes in zebrafish. PLoS Genet 6: e1001245. doi: 10.1371/journal.pgen.1001245 21187910
83. Cheng CL, Novales Flamarique I (2004) Opsin expression: new mechanism for modulating colour vision. Nature 428 : 279.
84. Perz-Edwards A, Hardison NL, Linney E (2001) Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev Biol 229 : 89–101. 11133156
85. Waxman JS, Keegan BR, Roberts RW, Poss KD, Yelon D (2008) Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev Cell 15 : 923–934. doi: 10.1016/j.devcel.2008.09.009 19081079
86. Larison KD, Bremiller R (1990) Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development 109 : 567–576. 2401210
87. Dewamitta SR, Joseph C, Purton LE, Walkley CR (2014) Erythroid-extrinsic regulation of normal erythropoiesis by retinoic acid receptors. Br J Haematol 164 : 280–285. doi: 10.1111/bjh.12578 24383846
88. Su D, Gudas LJ (2008) Retinoic acid receptor gamma activates receptor tyrosine kinase Tie1 gene transcription through transcription factor GATA4 in F9 stem cells. Exp Hematol 36 : 624–641. doi: 10.1016/j.exphem.2007.12.016 18439490
89. Kralova J, Czerny T, Spanielova H, Ratajova V, Kozmik Z (2002) Complex regulatory element within the gammaE - and gammaF-crystallin enhancers mediates Pax6 regulation and is required for induction by retinoic acid. Gene 286 : 271–282. 11943482
90. Veien ES, Rosenthal JS, Kruse-Bend RC, Chien CB, Dorsky RI (2008) Canonical Wnt signaling is required for the maintenance of dorsal retinal identity. Development 135 : 4101–4111. doi: 10.1242/dev.027367 19004855
91. Bernier G, Panitz F, Zhou X, Hollemann T, Gruss P, et al. (2000) Expanded retina territory by midbrain transformation upon overexpression of Six6 (Optx2) in Xenopus embryos. Mech Dev 93 : 59–69. 10781940
92. Wang Y, Macke JP, Merbs SL, Zack DJ, Klaunberg B, et al. (1992) A locus control region adjacent to the human red and green visual pigment genes. Neuron 9 : 429–440. 1524826
93. Hayashi T, Motulsky AG, Deeb SS (1999) Position of a 'green-red' hybrid gene in the visual pigment array determines colour-vision phenotype. Nat Genet 22 : 90–93. 10319869
94. McMahon C, Carroll J, Awua S, Neitz J, Neitz M (2008) The L:M cone ratio in males of African descent with normal color vision. J Vis 8 : 5 1–9.
95. McMahon C, Neitz J, Neitz M (2004) Evaluating the human X-chromosome pigment gene promoter sequences as predictors of L:M cone ratio variation. J Vis 4 : 203–208. 15086310
96. Karl MO, Reh TA (2011) Regenerative medicine for retinal diseases: activating endogenous repair mechanisms. Trends Mol Med 16 : 193–202.
97. Forrest D, Swaroop A (2012) Minireview: the role of nuclear receptors in photoreceptor differentiation and disease. Mol Endocrinol 26 : 905–915. doi: 10.1210/me.2012-1010 22556342
98. Neitz J, Neitz M (2011) The genetics of normal and defective color vision. Vision Res 51 : 633–651. doi: 10.1016/j.visres.2010.12.002 21167193
99. Carroll J, Dubra A, Gardner JC, Mizrahi-Meissonnier L, Cooper RF, et al. (2012) The effect of cone opsin mutations on retinal structure and the integrity of the photoreceptor mosaic. Invest Ophthalmol Vis Sci 53 : 8006–8015. doi: 10.1167/iovs.12-11087 23139274
100. Mancuso K, Hauswirth WW, Li Q, Connor TB, Kuchenbecker JA, et al. (2009) Gene therapy for red-green colour blindness in adult primates. Nature 461 : 784–787. doi: 10.1038/nature08401 19759534
101. Westerfield M (2007) The Zebrafish Book; A guide for the laboratory use of zebrafish (Danio rerio). Eugene, OR: University of Oregon Press.
102. Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, et al. (2011) Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell 20 : 397–404. doi: 10.1016/j.devcel.2011.01.010 21397850
103. Stenkamp DL, Frey RA, Prabhudesai SN, Raymond PA (2000) Function for Hedgehog genes in zebrafish retinal development. Dev Biol 220 : 238–252. 10753513
104. Kashyap B, Pegorsch L, Frey RA, Sun C, Shelden EA, et al. (2013) Eye-specific gene expression following embryonic ethanol exposure in zebrafish: Roles for heat shock factor 1. Reprod Toxicol 43C: 111–124.
105. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, et al. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. 12582260
106. Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98 : 5116–5121. 11309499
107. Team RC (2014) R: A language and environment for statistical computing.
108. Barthel LK, Raymond PA (1990) Improved method for obtaining 3-microns cryosections for immunocytochemistry. J Histochem Cytochem 38 : 1383–1388. 2201738
109. Stenkamp DL, Frey RA, Mallory DE, Shupe EE (2002) Embryonic retinal gene expression in sonic-you mutant zebrafish. Dev Dyn 225 : 344–350. 12412019
110. Nelson SM, Frey RA, Wardwell SL, Stenkamp DL (2008) The developmental sequence of gene expression within the rod photoreceptor lineage in embryonic zebrafish. Dev Dyn 237 : 2903–2917. doi: 10.1002/dvdy.21721 18816851
111. Nelson SM, Park L, Stenkamp DL (2009) Retinal homeobox 1 is required for retinal neurogenesis and photoreceptor differentiation in embryonic zebrafish. Dev Biol 328 : 24–39. doi: 10.1016/j.ydbio.2008.12.040 19210961
112. Stenkamp DL, Powers MK, Carney LH, Cameron DA (2001) Evidence for two distinct mechanisms of neurogenesis and cellular pattern formation in regenerated goldfish retinas. J Comp Neurol 431 : 363–381. 11223808
113. Stenkamp DL, Cunningham LL, Raymond PA, Gonzalez-Fernandez F (1998) Novel expression pattern of interphotoreceptor retinoid-binding protein (IRBP) in the adult and developing zebrafish retina and RPE. Mol Vis 4 : 26. 9841935
114. Rodieck RW (1991) The density recovery profile: a method for the analysis of points in the plane applicable to retinal studies. Vis Neurosci 6 : 95–111. 2049333
115. Cook JE (1996) Spatial properties of retinal mosaics: an empirical evaluation of some existing measures. Vis Neurosci 13 : 15–30. 8730986
Štítky
Genetika Reprodukčná medicína
Článek Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer PopulationČlánek Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2Článek Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA ReplicationČlánek SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from DegradationČlánek Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors inČlánek The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 RegulationČlánek Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Putting the Brakes on Huntington Disease in a Mouse Experimental Model
- Identification of Driving Fusion Genes and Genomic Landscape of Medullary Thyroid Cancer
- Evidence for Retromutagenesis as a Mechanism for Adaptive Mutation in
- TSPO, a Mitochondrial Outer Membrane Protein, Controls Ethanol-Related Behaviors in
- Evidence for Lysosome Depletion and Impaired Autophagic Clearance in Hereditary Spastic Paraplegia Type SPG11
- Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer Population
- Trans-Reactivation: A New Epigenetic Phenomenon Underlying Transcriptional Reactivation of Silenced Genes
- Early Developmental and Evolutionary Origins of Gene Body DNA Methylation Patterns in Mammalian Placentas
- Strong Selective Sweeps on the X Chromosome in the Human-Chimpanzee Ancestor Explain Its Low Divergence
- Dominance of Deleterious Alleles Controls the Response to a Population Bottleneck
- Transient 1a Induction Defines the Wound Epidermis during Zebrafish Fin Regeneration
- Systems Genetics Reveals the Functional Context of PCOS Loci and Identifies Genetic and Molecular Mechanisms of Disease Heterogeneity
- A Genome Scale Screen for Mutants with Delayed Exit from Mitosis: Ire1-Independent Induction of Autophagy Integrates ER Homeostasis into Mitotic Lifespan
- Non-synonymous FGD3 Variant as Positional Candidate for Disproportional Tall Stature Accounting for a Carcass Weight QTL () and Skeletal Dysplasia in Japanese Black Cattle
- The Relationship between Gene Network Structure and Expression Variation among Individuals and Species
- Calmodulin Methyltransferase Is Required for Growth, Muscle Strength, Somatosensory Development and Brain Function
- The Wnt Frizzled Receptor MOM-5 Regulates the UNC-5 Netrin Receptor through Small GTPase-Dependent Signaling to Determine the Polarity of Migrating Cells
- Nbs1 ChIP-Seq Identifies Off-Target DNA Double-Strand Breaks Induced by AID in Activated Splenic B Cells
- CCNYL1, but Not CCNY, Cooperates with CDK16 to Regulate Spermatogenesis in Mouse
- Evidence for a Common Origin of Blacksmiths and Cultivators in the Ethiopian Ari within the Last 4500 Years: Lessons for Clustering-Based Inference
- Of Fighting Flies, Mice, and Men: Are Some of the Molecular and Neuronal Mechanisms of Aggression Universal in the Animal Kingdom?
- Hypoxia and Temperature Regulated Morphogenesis in
- The Homeodomain Iroquois Proteins Control Cell Cycle Progression and Regulate the Size of Developmental Fields
- Evolution and Design Governing Signal Precision and Amplification in a Bacterial Chemosensory Pathway
- Rac1 Regulates Endometrial Secretory Function to Control Placental Development
- Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2
- Functions as a Positive Regulator of Growth and Metabolism in
- The Nucleosome Acidic Patch Regulates the H2B K123 Monoubiquitylation Cascade and Transcription Elongation in
- Rhoptry Proteins ROP5 and ROP18 Are Major Murine Virulence Factors in Genetically Divergent South American Strains of
- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- Regulates Refractive Error and Myopia Development in Mice and Humans
- mTORC1 Prevents Preosteoblast Differentiation through the Notch Signaling Pathway
- Regulation of Gene Expression Patterns in Mosquito Reproduction
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
- The Spalt Transcription Factors Generate the Transcriptional Landscape of the Wing Pouch Central Region
- Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA Replication
- Functional Divergence in the Role of N-Linked Glycosylation in Smoothened Signaling
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- Coordinated Evolution of Influenza A Surface Proteins
- The Evolutionary Potential of Phenotypic Mutations
- Genome-Wide Association and Trans-ethnic Meta-Analysis for Advanced Diabetic Kidney Disease: Family Investigation of Nephropathy and Diabetes (FIND)
- New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation
- SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from Degradation
- Satellite DNA Modulates Gene Expression in the Beetle after Heat Stress
- SHOEBOX Modulates Root Meristem Size in Rice through Dose-Dependent Effects of Gibberellins on Cell Elongation and Proliferation
- Reduced Crossover Interference and Increased ZMM-Independent Recombination in the Absence of Tel1/ATM
- Suppression of Somatic Expansion Delays the Onset of Pathophysiology in a Mouse Model of Huntington’s Disease
- Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors in
- The Evolutionarily Conserved LIM Homeodomain Protein LIM-4/LHX6 Specifies the Terminal Identity of a Cholinergic and Peptidergic . Sensory/Inter/Motor Neuron-Type
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of .
- Retinoic Acid Signaling Regulates Differential Expression of the Tandemly-Duplicated Long Wavelength-Sensitive Cone Opsin Genes in Zebrafish
- The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 Regulation
- Genome-Wide Analysis of PAPS1-Dependent Polyadenylation Identifies Novel Roles for Functionally Specialized Poly(A) Polymerases in
- Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
- Regulation of Mutagenic DNA Polymerase V Activation in Space and Time
- Variability of Gene Expression Identifies Transcriptional Regulators of Early Human Embryonic Development
- The Drosophila Gene Interacts Genetically with and Shows Female-Specific Effects of Divergence
- Functional Activation of the Flagellar Type III Secretion Export Apparatus
- Retrohoming of a Mobile Group II Intron in Human Cells Suggests How Eukaryotes Limit Group II Intron Proliferation
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



