-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
SHOEBOX Modulates Root Meristem Size in Rice through Dose-Dependent Effects of Gibberellins on Cell Elongation and Proliferation
Little is known concerning the identity of regulatory components and signaling pathways that control the growth of meristem cells in plants. Here, we report that rice plants deficient in the AP2/ERF family gene SHOEBOX (SHB) exhibited a severe reduction in the root meristem size. These plants had shorter cells in the root meristems following germination and fewer cells from approximately 5 days after sowing, suggesting that SHB regulates root meristem cell size and number in a developmental stage-specific manner and that cell size participates in the control of root meristem size in rice. SHB is positively regulated by GA signaling and encodes a direct transcriptional activator of the GA biosynthesis gene KS1, indicating that GA modulates its own biosynthesis and consequently the elongation of meristem cells in rice roots via positive feedback regulation on the transcription of SHB and KS1. Consistently, application of exogenous GA restored the size of root meristem cells to normal in shb and paclobutrazol-treated wild-type plants.
Published in the journal: SHOEBOX Modulates Root Meristem Size in Rice through Dose-Dependent Effects of Gibberellins on Cell Elongation and Proliferation. PLoS Genet 11(8): e32767. doi:10.1371/journal.pgen.1005464
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005464Summary
Little is known concerning the identity of regulatory components and signaling pathways that control the growth of meristem cells in plants. Here, we report that rice plants deficient in the AP2/ERF family gene SHOEBOX (SHB) exhibited a severe reduction in the root meristem size. These plants had shorter cells in the root meristems following germination and fewer cells from approximately 5 days after sowing, suggesting that SHB regulates root meristem cell size and number in a developmental stage-specific manner and that cell size participates in the control of root meristem size in rice. SHB is positively regulated by GA signaling and encodes a direct transcriptional activator of the GA biosynthesis gene KS1, indicating that GA modulates its own biosynthesis and consequently the elongation of meristem cells in rice roots via positive feedback regulation on the transcription of SHB and KS1. Consistently, application of exogenous GA restored the size of root meristem cells to normal in shb and paclobutrazol-treated wild-type plants.
Introduction
The size of a plant, or part thereof, is determined by combined activity of cell proliferation and growth during development [1]. Cell proliferation in plants occurs mostly in specialized tissues known as meristems, where new cells are produced to ensure that plants continue to grow in height and width throughout their life. Prior to mitosis, cells in the meristem must double in size by undergoing a slow but steady expansion in the direction perpendicular to the previous division plane, which enables them to divide and keeps the size of their daughter cells constant [2,3]. A more pronounced growth (denoted as post-mitotic cell expansion), however, is commonly seen in differentiating cells that are displaced from the meristem. The extent of post-mitotic cell expansion is generally well correlated with the magnitude of organ growth [4].
Cell proliferation and growth in plants are influenced by genetic, hormonal, and environmental inputs. While little is known about the molecular mechanisms that regulate the size of meristem cells, numerous molecular players, including members of the AP2/ERF family of transcription factors, have been demonstrated to control either cell proliferation or post-mitotic cell expansion. For instance, the Arabidopsis AP2 transcription factor AINTEGUMENTA (ANT) promotes cell proliferation by maintaining the meristematic competence of cells [5]. ANT activity is activated by ARGOS (for auxin-regulated gene involved in organ size), a novel transcription factor acting downstream of auxin signaling [6]. In rice, several AP2/ERF genes including OsEATB (for ERF protein associated with tillering and branching [7], SUBMERGENCE 1A (SUB1A) [8], SNORKEL1 (SK1) and SK2 [9], were reported to have roles in regulating internode elongation, which is primarily post-mitotic expansion of differentiating cells displaced from the intercalary meristem near the node. SK1 and SK2 were suggested to trigger internode elongation via GA in response to rising water level [9]. By contrast, OsEATB was found to restrict GA responsiveness during the internode elongation process by down-regulating the expression of the GA biosynthetic gene OsCPS2 [7]; whereas SUB1A limits GA responsiveness during prolonged submergence by augmenting accumulation of the DELLA family of GA signaling repressors SLENDER RICE 1 (SLR1) and SLR1 Like 1 (SLRL1), thus restricting underwater internode elongation and enhancing submergence survival [10].
GA plays an important role in the regulation of cell proliferation and growth during plant development [11–13]. It has been recently established that GA modulates both the rate of cell proliferation and the extent of post-mitotic cell expansion [3,14–16]. Inhibition of GA biosynthesis, either genetically in the GA biosynthesis mutant ga1-3, or by means of chemical treatment using paclobutrazol (PAC), an inhibitor of GA biosynthesis [17,18], reduces substantially the rate of cell proliferation in the Arabidopsis root meristem [3,14,15]. GA was proposed to promote root growth in Arabidopsis by increasing elongation (expansion along the root axis) of both dividing and post-mitotic endodermal cells, thereby indirectly controlling division and elongation of other types of root cells and the overall root meristem size [3]. However, how this process is regulated at the molecular level remains unclear.
Here we report the discovery of a novel GA-dependent size-control mechanism in the rice root meristem. We show that root meristem size in rice can be regulated by the extent of cell elongation in the root meristem. SHOEBOX (SHB), an AP2/ERF transcription factor, plays a key role in this mechanism. SHB directly binds to and activates transcription of the GA biosynthesis gene KS1 in the root meristem, leading to the local production of GA that promotes elongation of meristem cells following germination, thus ensuring meristem growth and phenotypic plasticity during early stage of meristem development. At a later stage, SHB-dependent and KS1-mediated GA biosynthesis also participates in the modulation of cell proliferation in the root meristem, indicating a developmental stage-specific function of SHB.
Results
The shb Mutation Reduces the Length of Meristem Cells and Consequently the Size of the Root Meristem in Rice
In a rice enhancer trap screen we isolated a recessive mutant with a short primary root phenotype (Fig 1A), which we have named shoebox (shb; based on the shape of the cortical cells in the root meristem). Analysis of median longitudinal sections of root apices of 4-day-old wild-type (WT) and shb seedlings showed that the root meristem size of shb was shorter than that of the WT (Fig 1B and 1D and 1H). Quantification of cortical cell number and size in the root meristem of WT and shb mutant plants suggested that this was not due to a reduction in the number of meristematic cortical cells (Fig 1H), but was rather caused by a decrease in the length (but not width) of meristematic cortical cells (Fig 1C and 1E and 1I). Consistently, EdU staining indicated that the shb mutation did not noticeably alter cell proliferation in the root meristem (Fig 1F and 1G). Moreover, the average lengths of cortical cells in the root elongation and maturation zone did not differ between shb and the WT (Fig 1J and 1K), suggesting that shb has a root meristem-specific cell elongation defect. Notably, root growth rate and cell production rate in shb were not significantly altered in 3 - and 4-day-old shb mutants but started to decline at around 5 days after sowing (Fig 1L and 1M).
Fig. 1. The shb mutation reduces the length of meristematic cortical cells and consequently the size of the root meristem in rice. 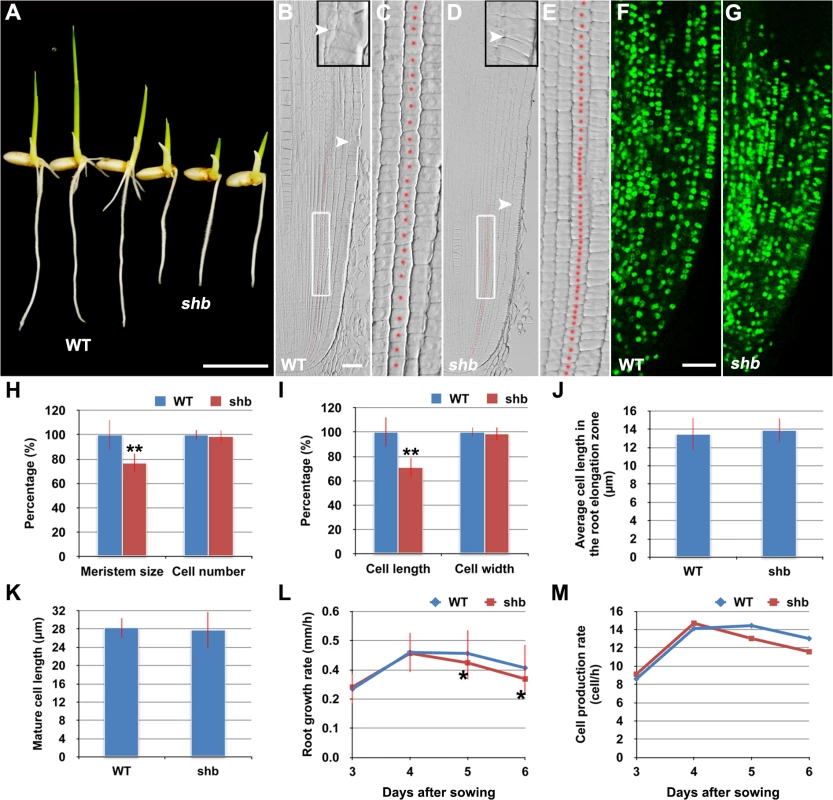
(A) Phenotype of 4-day-old WT and shb seedlings. Scale bar = 1 cm. (B-E) Median longitudinal sections through root tips of 4-day-old WT (B, C) and shb (D, E) seedlings. Arrowheads indicate the proximal end of the root meristem. Insets are an enlargement of the regions at the proximal end of the root meristem. Red dotted line marks the 4th cortical layer selected for the quantification analysis. (C, E) Boxed regions in (B, D). Scale bar = 50 μm. (F, G) Median longitudinal view of EdU staining in the root meristem of WT (F) and shb (G) seedlings. Scale bar = 50 μm. (H, I) Root meristem size (H), meristematic cortical cell number (H), average length (I) and width (I) of cortical cells in the root meristem of 4-day-old WT and shb seedlings. Data are expressed as percentage of the WT control, arbitrarily set to 100. Error bars represent SD (n = 15). **, P < 0.01, t-test. (J, K) Average length of cortical cells in the root elongation zone (J) and maturation zone (K). (L, M) Root growth rate and cell production rate in the root meristem of WT and shb seedlings. Measurement and calculation (cell production rate = root growth rate/ mature cell length) were performed at indicated days. Error bars represent SD (n = 15). *, P < 0.05, t-test. shb is a Novel GA-Deficient Mutant with a Mild Seed Germination Defect and Its Phenotypes Could Be Restored to WT by Exogenous Application of GA3
The aerial part of shb mutant plants has typical characteristics of rice GA-deficient or insensitive mutants [7,19,20], such as dwarfism and short internode length (S1 Fig). We thus hypothesized that the root phenotype of shb mutant plants might be caused by a defect in GA biosynthesis and/or signaling and examined whether it could be restored to WT by growing the mutants on medium supplemented with bioactive GA (GA3). 10 μM GA3 had no apparent effect on the WT control but could fully rescue the short-root phenotype of shb mutants (Fig 2A and 2B and 2G). Average length of cortical cells in the root meristem of shb was restored to that of the WT (Fig 2D and 2F and 2H), producing a root meristem with a similar size to that of the WT (Fig 2C, 2E and 2I). These results suggest that shb could properly respond to GA and GA deficiency is the primary cause of mutant phenotypes. In agreement with this suggestion, we found that the levels of GAs were reduced in shb roots as compared to the WT roots (Fig 2J). Particularly two bioactive GAs, GA3 and GA4, were significantly lower in shb compared to the WT controls (Fig 2J).
Fig. 2. shb is a novel GA-deficient mutant whose root phenotypes could be restored to WT by exogenous application of GA3. 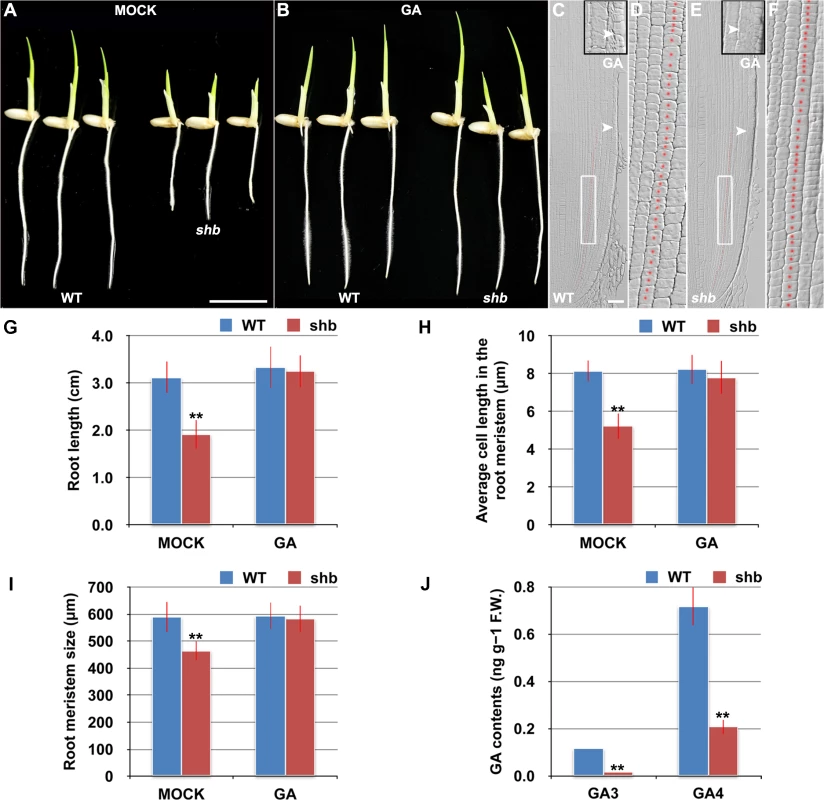
(A) Phenotype of 4-day-old mock-treated WT and shb seedlings. Scale bar = 1 cm. (B) Phenotype of 4-day-old WT and shb seedlings grown on medium supplemented with 10 μM GA3. (C-F) Median longitudinal sections through root tips of 4-day-old GA-treated WT (C, D) and shb (E, F) seedlings. Arrowheads indicate the proximal end of the root meristem. Insets are an enlargement of the regions at the proximal end of the root meristem. Red dotted line marks the 4th cortical layer selected for the quantification analysis. (D, F) Boxed regions in (C, E). Scale bar = 50 μm. (G) Root length of 4-day-old mock- and GA-treated WT and shb seedlings. Error bars represent SD (n = 15). **, P < 0.01, t-test. (H) Average cell length in the root meristem of 4-day-old mock- and GA-treated WT and shb seedlings. Error bars represent SD (n = 15). **, P < 0.01, t-test. (I) Root meristem size of 4-day-old mock- and GA-treated WT and shb seedlings. Error bars represent SD (n = 15). **, P < 0.01, t-test. (J) Endogenous levels of GA3 and GA4 in WT and shb roots. Error bars represent SD from three independent experiments. **, P < 0.01, t-test. Because GA-deficient mutants often show delayed seed germination [21,22]. We next compared WT and shb seed germination and found that shb germinated approximately 12 h later than the WT (S2A–S2C Fig). As a result, 4-day-old shb had a markedly shorter root length compared to the WT controls when both seeds were sowed on medium at the same time (Figs 1A and 2A and 2G).
shb Root Meristem Contains Shorter Cells following Germination and Fewer Cells from Approximately 5 Days after Synchronized Germination
To exclude the possibility that the short cortical cell phenotype observed in the shb root meristem was caused by delayed seed germination, we next synchronized WT and shb seed germination by sowing shb seeds on medium 12 h earlier before the WT control and performed a time-course analysis of various root phenotypes. We found that the sizes of root meristem (S2D Fig) and meristematic cortical cells (S2E Fig) were significantly and constantly shorter in shb than in the WT during the period of analysis. By contrast, meristematic cortical cell number (S2F Fig), root length (S2G Fig), root growth rate (S2H Fig) and cell production rate (S2I Fig) in the WT and shb were essentially identical until around the fifth day after synchronized germination, from which they started to diverge with significantly lower values in shb compared with the WT. Together, these results confirm our earlier observation that shb root meristem contained shorter cortical cells in the root meristem and suggest that fewer cells were produced from around the fifth day after synchronized germination. These phenotypes were not accompanied by changes in the lengths of elongation and maturation zone cells (S2J and S2K Fig), further demonstrating a specific role of SHB in the root meristem.
SHB is Expressed in the Root Meristem and Encodes an AP2/ERF Transcription Factor
The shb mutant carries a homozygous T-DNA insertion in the 5th intron of the LOC_Os05g32270 gene (Fig 3A), which dramatically reduces the expression level of LOC_Os05g32270 (Fig 3B). A genomic fragment containing the LOC_Os05g32270 gene and its promoter and 3’ UTR regions could fully complement the mutant phenotypes of shb (Fig 3C–3J). We thus concluded that LOC_Os05g32270 is the SHB gene.
Fig. 3. The root phenotypes of shb could be fully complemented by the LOC_os05g32270 gene. 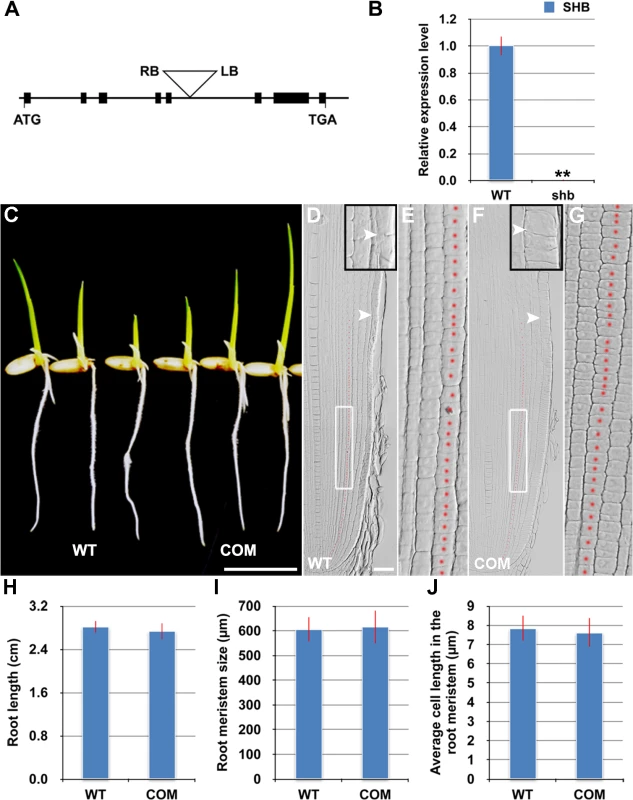
(A) Schematic diagram of exon/intron structure of the SHB (LOC_Os05g32270) gene and T-DNA insertion site. Black boxes are exons. Introns are the open areas between the boxes. LB and RB represent the left and right borders of T-DNA. (B) qPCR analysis of transcript levels of SHB in roots of WT and shb mutant. Transcript levels from the WT were set to 1. Error bars represent SD from three independent experiments. **, P < 0.01, t-test. (C) Phenotype of 4-day-old seedlings of WT and a representative complementation line (COM). Scale bar = 1 cm. (D to G) Median longitudinal sections through root tips of 4-day-old WT (D, E) and COM (F, G) seedlings. Arrowheads indicate the proximal end of the root meristem. Insets are an enlargement of the regions at the proximal end of the root meristem. Red dotted line marks the 4th cortical layer selected for the quantification analysis. (E, G) Boxed regions in (D, F). Scale bar = 50 μm. (H) Root length of 4-day-old WT and COM seedlings. (I) Root meristem size of 4-day-old WT and COM seedlings. (J) Average cell length in the root meristem of 4-day-old WT and COM seedlings. Error bars in (H) to (J) represent SD (n = 15). **, P < 0.01, t-test. The SHB gene has been previously termed OsAP2-EREBP-049, which encodes a putative transcription factor containing one AP2/EREBP DNA binding domain [23]. Based on the sequence similarity of the AP2/EREBP DNA binding domain, SHB was classified into the Group-1a of the AP2/ERF family, although all the other genes in this group have double AP2-EREBP DNA binding domains. A BLASTP search revealed that putative orthologs of SHB are present in both monocots and dicot plants (S3A Fig). The subsequent phylogenetic analysis suggested that SHB is more closely related to its putative orthologs in monocots than to its dicot counterparts (S3B Fig). Mutations in DWARF & IRREGULAR LEAF (DIL1), a homologous gene of SHB in maize, were reported to affect internode length, leaf shape and possibly root length [24], implying that SHB and DIL1 have evolutionarily conserved functions.
RNA in situ hybridization showed that SHB is expressed in the root meristem (Fig 4A), thus supporting a functional role for SHB in vivo. A fusion protein of SHB and GFP {SHB-GFP; under the control of the Cowpea Mosaic Virus (CPMV) promoter [25]} co-localized with a nuclear marker SRT1-RFP [26] in tobacco epidermal cells (Fig 4B), indicating that SHB functions as a nuclear-localized transcription factor.
Fig. 4. SHB is expressed in the root tip and encodes a nuclear-localized transcription factor that directly binds to and activates transcription of KS1. 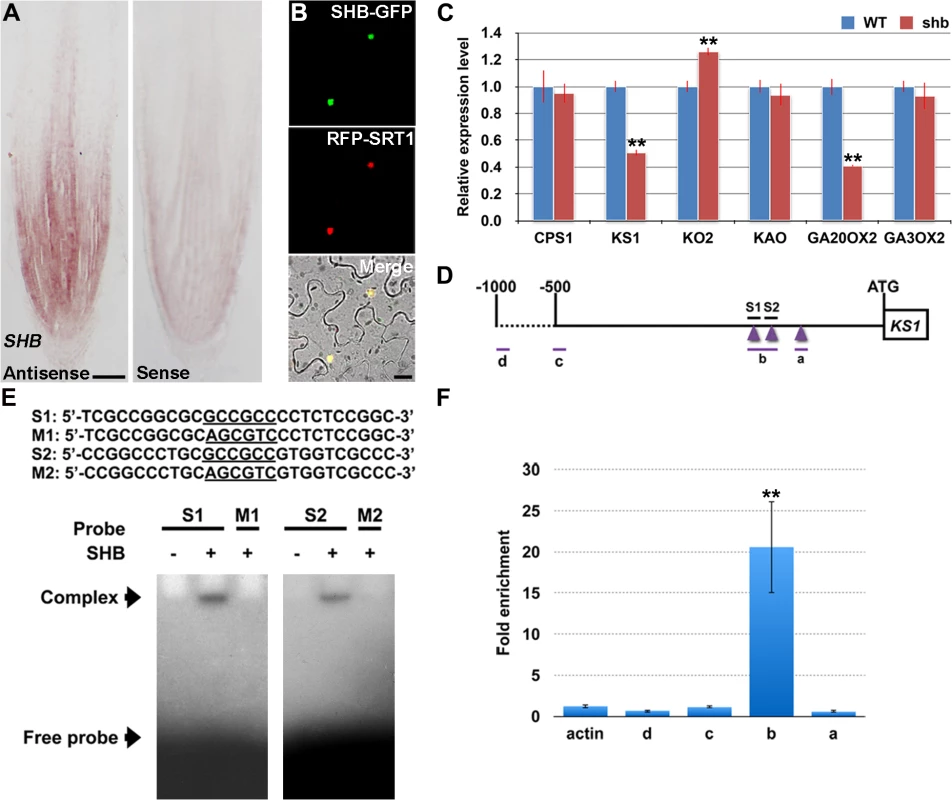
(A) RNA in situ hybridizations with an anti-sense probe (left panel) and a sense probe (right panel) showing SHB mRNA accumulation in the root tip. Scale bar = 100 μm. (B) Co-localization (lower panel; yellow) of SHB-GFP (upper panel; green) with a nuclear marker SRT1-RFP (middle panel; red) in tobacco epidermal cells. Scale bar = 20 μm. (C) qPCR analysis of transcript levels of GA biosynthetic genes CPS1, KS1, KO2, KAO, GA20OX2/SD1 and GA3OX2 in rice roots. Transcript levels from the WT were set to 1. Error bars represent SD from three independent experiments. **, P < 0.01, t-test. (D) Schematic diagram of SHB promoter region. Regions selected for EMSA and ChIP-qPCR experiments are shown by short lines and marked with letters. Purple arrowheads point to the position of GCC boxes. GCCGCC motif and the corresponding mutated DNA sequences are underlined. (E) Sequences of oligonucleotides used for EMSA, which indicates the binding of SHB to S1 and S2, but not to M1 and M2. (F) ChIP-qPCR analysis showing high level of association of SHB with fragment b. Error bars represent SD from three independent experiments. **, P < 0.01, t-test. SHB Is a Direct Transcriptional Activator of the GA Biosynthesis Gene KS1
To determine the cause of GA deficiency in shb, we next examined whether the shb mutation decreases transcription of rice GA biosynthetic genes, including CPS1, KS1, KO2, KAO, GA20OX2/SD1 and GA3OX2, by quantitative real-time PCR (qPCR). Among these genes, only KS1 and GA20OX2/SD1 were found to be significantly down-regulated by the shb mutation (Fig 4C), suggesting that SHB modulates the levels of bioactive GAs in rice roots through transcriptional activation of GA biosynthetic genes KS1 and GA20OX2/SD1. Notably, the expression of KO2 was weakly up-regulated in shb, perhaps to compensate for reduction of KS1, which is involved in an earlier step of the GA biosynthesis pathway.
AP2/EREBP proteins are able to bind the GCC-box, which is a short cis-acting element containing a core GCCGCC sequence motif [27]. Analysis of the KS1 promoter identified three GCCGCC motifs located at 205, 184, and 131 nucleotides upstream to the translation start site (ATG; Fig 4D), whereas no GCC-box was found in the promoter region of GA20OX2/SD1. Electrophoretic mobility shift assay (EMSA) indicated that SHB could bind to GCCGCC motifs located at 205 and 184 nucleotides upstream to the ATG (S1 and S2; Fig 4D and 4E). No binding was detected with the third GCCGCC motif and SHB was not no longer able to bind to S1 and S2 when the two GCCGCC motifs were mutated (M1 and M2; Fig 4E).
To confirm the EMSA result in vivo, we performed ChIP-qPCR experiments with transgenic rice plants expressing a functional SHB-GFP fusion protein(S4 Fig). qPCR showed that fragment b, which contains GCCGCC motifs located at 205 and 184 nucleotides upstream to the ATG of KS1, was greatly enriched by ChIP with an anti-GFP antibody (Fig 4F). On the contrary, DNA fragments covering other regions of the KS1 promoter, as well as the negative controls, were less amplified (Fig 4F). These data indicate that SHB can directly bind to two closely located GCCGCC motifs in the promoter region of KS1 in vivo.
KS1-Mediated Local GA Biosynthesis Is Required for Dose-Dependent Effects of GA on Cell Elongation and Proliferation in the Root Meristem
RNA in situ hybridization revealed that KS1 was expressed in an overlapping domain with SHB (Fig 5A) and that KS1 had reduced expression level and domain in the shb mutant (S5 Fig), in agreement with our finding that KS1 is a direct downstream target of SHB. A null ks1 mutant with severe GA deficiency [19] was found to have shorter root length and smaller root meristem than the WT control (Fig 5B, 5C, 5E and 5I). 10 μM GA3 could fully complement the meristem size phenotype of ks1 (Fig 5I), suggesting that KS1-dependent GA biosynthesis is essential for root meristem size control. Quantification of cortical cell number and size in the ks1 root meristem revealed that there was a significant decrease in cell proliferation compared to the WT (Fig 5J), which was accompanied by an increase in the average length of cortical cells (Fig 5K), indicating cell cycle arrest [28]. EdU staining further confirmed the cell proliferation defect (Fig 5G and 5H) and 10 μM GA3 could largely restore the number of cortical cells in the ks1 root meristem (Fig 5J), suggesting that severe GA deficiency in ks1 is the underlying cause. Consistently, levels of most GAs were significantly reduced or undetectable in ks1 seedlings compared to the WT and shb seedlings (S1 Table). Thus, we conclude that: 1) KS1-mediated GA biosynthesis is required for dose-dependent effects of GA on cell elongation and proliferation in the root meristem; and 2) The different root meristem phenotypes observed in shb and ks1 mutants result from moderate versus severe GA reduction. The latter conclusion was also suggested by much greater up-regulation of KO2 in ks1 (S6 Fig) than in shb (Fig 4C) and in agreement with this conclusion, a higher concentration of GA3 was needed to restore in 24 hours the root meristem size in ks1 (100 μM; S7A Fig) than in shb (50 μM; S7B Fig).
Fig. 5. KS1 is expressed in the root tip and has a role in GA-mediated elongation and proliferation of root meristem cells. 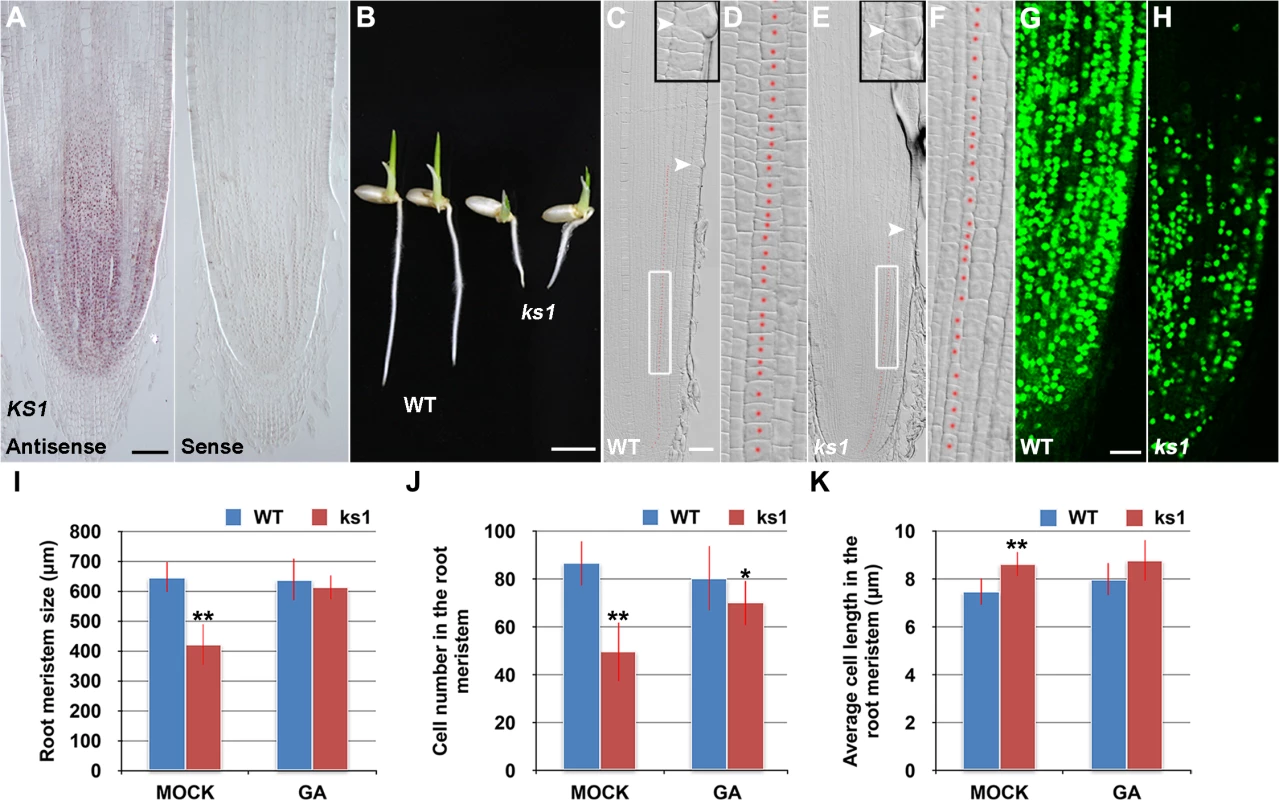
(A) RNA in situ hybridizations with an anti-sense probe (left panel) and a sense probe (right panel) showing KS1 mRNA accumulation in the root tip. Scale bar = 100 μm. (B) Phenotype of 4-day-old WT and ks1 seedlings. Scale bar = 5 mm. (C-F) Median longitudinal sections through root tips of 4-day-old WT (C, D) and ks1 (E, F) seedlings. Arrowheads indicate the proximal end of the root meristem. Insets are an enlargement of the regions at the proximal end of the root meristem. Red dotted line marks the 4th cortical layer selected for the quantification analysis. (D, F) Boxed regions in (C, E). Scale bar = 50 μm. (G-H) Median longitudinal view of EdU-labelled cells in the root meristem of WT (G) and ks1 (H) seedlings. Scale bar = 50 μm. (I) Root meristem size of 4-day-old WT and ks1 seedlings treated with mock or 10 μM GA3. (J) Cell number in the root meristem of 4-day-old WT and ks1 seedlings treated with mock or 10 μM GA3. (K) Average cell length in the root meristem of 4-day-old WT and ks1 seedlings treated with mock or 10 μM GA3. Error bars in (I) to (K) represent SD (n = 15). **, P < 0.01, t-test; *, P < 0.05, t-test. Our results suggest that cell proliferation in the rice root meristem is regulated by a dose-dependent effect of GA. Consistently, severe inhibition of GA biosynthesis by 10 or 50 μM PAC significantly impaired cell proliferation in the root meristem of shb mutants and WT plants (Fig 6A–6F), resulting in a smaller root meristem (Fig 6A, 6B and 6G) with longer cells (Fig 6A and 6B and 6H). By contrast, 0.1 μM PAC had no obvious effects on shb mutants and WT plants (Fig 6A–6H), suggesting that PAC has a dose-dependent effect on cell proliferation. PAC-induced phenotypes could be reversed by co-treatment with GA (S8 Fig), confirming that they were caused by inhibition of GA biosynthesis by PAC. Notably, 1 μM PAC markedly reduced elongation of cortical cells in the WT root meristem (Fig 6H) without significantly affecting cell proliferation (Fig 6A–6F), whereas the root phenotypes of shb were less affected at the same concentration (Fig 6H). These observations together suggest that GA has a dose-dependent effect on cell elongation in the root meristem, which is regulated by SHB.
Fig. 6. PAC has a dose-dependent effect on cell elongation and proliferation in the root meristem. 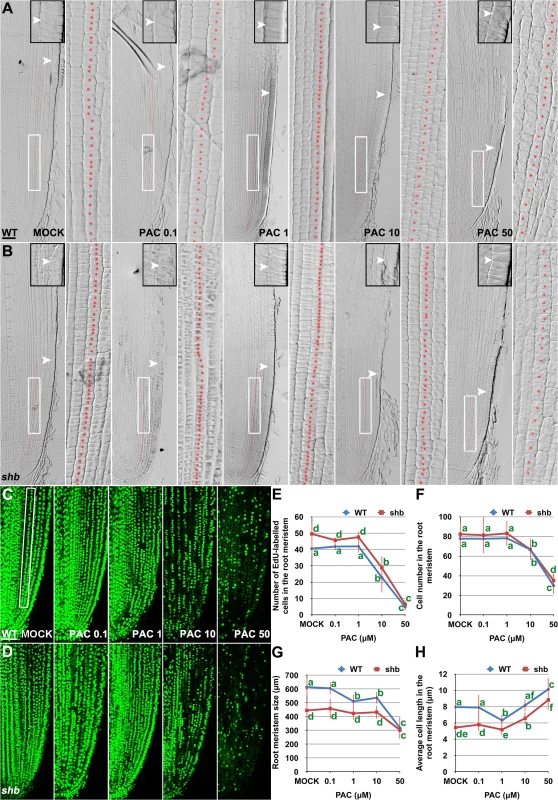
(A, B) Median longitudinal sections through root tips of 3-day-old WT (A) and shb (B) seedlings treated with mock or indicated concentration of PAC for 24 hours. Arrowheads indicate the proximal end of the root meristem. Insets are an enlargement of the regions at the proximal end of the root meristem. Red dotted line marks the 4th cortical layer selected for the quantification analysis. Scale bar = 50 μm. Boxed regions are magnified to show the size of root meristem cells in the 4th cortical layer. (C, D) EdU-labelled cells in the root meristem of 4-day-old WT (C) and shb (D) seedlings treated with mock or indicated concentrations of PAC for 24 hours. Scale bar = 50 μm. (E) Number of EdU-labelled cells in the root meristem of 4-day-old WT and shb seedlings treated with mock or different concentrations of PAC for 24 hours. Quantification was performed in the 4th cortical layer, in a selected portion (Boxed region in C) with a length of 360 μm. (F) Total cell number in the root meristem of 3-day-old WT and shb seedlings treated with mock or different concentrations of PAC for 24 hours. (G) Root meristem size of 3-day-old WT and shb seedlings treated with mock or different concentrations of PAC for 24 hours. (H) Average cell length in the root meristem of 3-day-old WT and shb seedlings treated with mock or different concentrations of PAC for 24 hours. Error bars in (E to H) represent SD (n = 10 for E and n = 15 for F to H). Bars with different letters are significantly different at P < 0.05, t-test. SHB Functions as a Positive Regulator of GA Signaling
RNA in situ hybridization and qPCR showed that SHB transcription was induced by GA3 and repressed by PAC (Fig 7A and 7B), suggesting that SHB functions as a positive regulator of GA signaling. Consistently, the level of KS1 transcripts was positively correlated with the level of GA (Fig 7B). Moreover, SHB and KS1 expression was found to be down-regulated in the GA receptor mutant gid1 [29] (Fig 7C), but up-regulated in slr1 (Fig 7D), a constitutive GA response mutant defective in the SLR1 gene, the only DELLA gene in rice [30]. Together, these results suggest that GA regulates its own biosynthesis via positive feedback regulation on the expression of SHB and KS1.
Fig. 7. SHB functions a positive regulator of GA signaling and meristem growth. 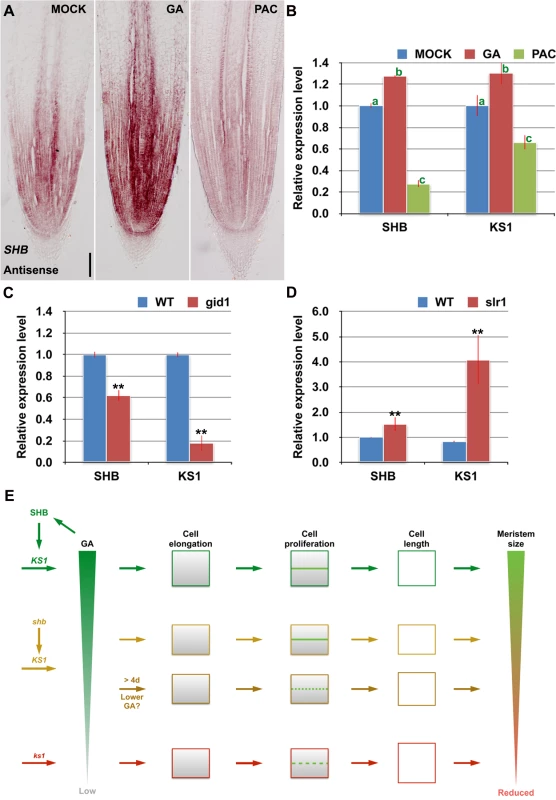
(A) RNA in situ hybridizations with an anti-sense probe showing SHB transcription was induced by GA and repressed by PAC in the root tip. Scale bar = 100 μm. (B) qPCR analysis of transcript levels of SHB and KS1 in WT rice roots treated with mock, 10 μM GA3 or 10 μM PAC. Transcript levels from the mock were set to 1. Error bars represent SD from three independent experiments. Bars with different letters are significantly different at P < 0.01, t-test. (C, D) qPCR analysis of transcript levels of SHB and KS1 in WT, gid1 and slr1 mutant roots. Transcript levels from the mock were set to 1. Error bars represent SD from three independent experiments. **, P < 0.01, t-test. (E) A model for meristem size control in the rice root. Root meristem size in rice is regulated through both SHB-dependent and independent GA biosynthesis pathways. SHB, whose transcription is induced by GA, promotes GA biosynthesis in the root meristem by directly binding to and activating KS1. SHB-dependent and KS1-mediated GA availability is well correlated with the ability of meristematic cortical cells to elongate beyond the minimal cell length requirement for normal cell proliferation, allowing optimal rice root meristem growth. Loss of SHB function reduces ks1 transcription by half and consequently KS1-dependent endogenous GA levels, resulting in a shorter root meristem with apparent defect in cell elongation but not in cell proliferation at the early stage of meristem development. From around the 5th day after sowing, the root meristem size of shb further decreases with fewer but longer meristematic cortex cells, indicative of cell cycle arrest. Loss of KS1 function, which strongly reduces endogenous levels of GAs, results in a further reduced root meristem size due to a more severe reduction of cortical cell number and a further elongation of cortical cells. Thus, GA controls the root meristem size in rice through its dose-dependent effects on meristematic cell elongation and proliferation. Discussion
In this study, we have identified SHB, an AP2/ERF transcription factor, as a novel regulator of root meristem size in rice. We provide conclusive evidence that, during early stage of meristem development, root meristem size in rice can be regulated through SHB-dependent cell elongation, without the necessity to alter the rate of cell proliferation (Figs 1 and S2). This indicates that meristems, like organs, are able to adjust their size independent of cell number. A similar example can be found during Drosophila melanogaster imaginal disc development, during which the S6 kinase (dS6K) regulates cell size in a cell-autonomous manner without impinging on cell number [31]. From around 5 days after sowing, at which cell number in the root meristem, root growth rate and cell production rate start to decline (Figs 1I, 1J, S2F, S2H and S2I), however, SHB appears to regulate both cell elongation and cell proliferation in the root meristem.
SHB is required for longitudinal cell elongation but not radial cell expansion in the cortical layers of the rice root meristem (Fig 1I). Moreover, loss of SHB function did not affect the elongation of post-mitotic cells in the rice root (Figs 1J and S2J), indicating that SHB has a root meristem-specific function and that elongation of meristem cells differs in mechanism from rapid elongation of post-mitotic cells. This idea is in agreement with a previous report on the Arabidopsis STUNTED PLANT 1 (STP1) gene, which is required for elongation of post-mitotic cells but not elongation of meristem cells [2]. STP1 was found to mediate the effect of cytokinin on the elongation of post-mitotic cells in the Arabidopsis root [2,32]. The identity of this gene remains unknown, but cytokinin has recently been shown to determine root meristem size by controlling cell differentiation [33]. On the contrary, our data suggest that GA controls root meristem size in rice through its dose-dependent effects on cell elongation and proliferation in the root meristem, which are mediated by SHB in a developmental stage-specific manner.
Recent studies in the Arabidopsis root have shown that GA controls root meristem size by modulating cell proliferation [3,14]. GA deficiency, either in GA biosynthesis mutants or induced by PAC treatment, significantly impairs cell proliferation in the Arabidopsis root, leading to a decrease in cell production rate and meristem size. Expression of a non-GA-degradable DELLA mutant in endodermal cells was sufficient to inhibit cell proliferation and block root meristem enlargement, indicating that GA controls root meristem size in a DELLA-dependent manner. A reduction in cell elongation in the root meristem was hypothesized to cause reduced number of cell division events and thus block the increase in meristem size [3]. However, several evidences from our study suggest that in rice GA could regulate cell elongation in the root meristem independently of its effect on cell proliferation to influence meristem growth: 1) shb had reduced levels of GA3 and GA4 in the root (Fig 2J). Exogenous application of GA3 to the shb root could restore the length of meristematic cortical cells and the size of root meristem to WT (Fig 2H and 2I). 2) Treating the WT root with 1 μM PAC could phenocopy the effect of the shb mutation, resulting in a shorter root meristem with reduced cortical cell length but unaltered cell number (Fig 6). 3) Higher concentrations of PAC (10–50 μM) significantly impaired cell proliferation in both WT and shb root meristems (Fig 6). It is also interesting to note that while Arabidopsis root meristem size nearly doubles during the first 4 days after sowing [34], rice root meristem size shows only a 10–20% increase over the same period (S2D Fig). The increase in Arabidopsis root meristem size was attributed to a proportional increase in meristematic cortical cell number [34], whereas the size of rice root meristem (S2D Fig) appears to be influenced by both the number (S2F Fig) and the SHB-regulated length (S2E Fig) of meristematic cortical cells.
How does SHB regulate GA levels and consequently cell elongation and proliferation in the root meristem? Our in vitro and in vivo data suggested that SHB directly binds to and activates transcription of the GA biosynthesis gene KS1, which has overlapping expression domain with SHB in the root meristem (Figs 4 and 5). In addition, our qPCR analysis indicated that SHB could also act through the GA biosynthetic gene GA20OX2/SD1 to modulate GA production in the root meristem (Fig 4C), but the underlying molecular mechanism remains to be elucidated. Intriguingly, our qPCR analysis showed that both SHB and KS1 expression were induced by GA but repressed by SLR1, the only DELLA protein in rice (Fig 7D). These findings suggest that GA regulates its own biosynthesis and consequently cell elongation and proliferation in the rice root meristem via positive feedback regulation on the expression of genes involved in the early steps of GA biosynthesis.
Taken together, our data suggest a model (Fig 7E) in which the root meristem size is modulated through dose-dependent effects of GA on cell elongation and proliferation in the root meristem, which are mediated by a developmental and possibly cumulative process that involves the root meristem - and developmental stage-specific function of the AP2/ERF transcription factor SHB. SHB, whose transcription is induced by GA, promotes GA biosynthesis in the root meristem by directly binding to and activating the GA biosynthesis gene KS1. SHB-dependent and KS1-mediated GA availability is well correlated with the ability of meristematic cortical cells to elongate beyond the minimal cell length requirement for normal cell proliferation, allowing optimal rice root meristem growth. Loss of SHB function at the early stage of meristem development reduces the root meristem size by reducing elongation of meristematic cortical cells, without causing apparent defect in cell proliferation. This phenotype can be mimicked in WT roots by a moderate reduction of endogenous GAs in the presence of 1 μM PAC. Loss of SHB function at a later stage of meristem development (from around the fifth day after sowing) impairs both cell elongation and proliferation and consequently, further reducing the root meristem size. The reduction of cortical cell number in the shb root meristem could be correlated to the increase of cortical cell length, indicating cell cycle arrest. Loss of KS1 function and exposure of WT roots to high concentrations of PAC (10 or 50 μM), which strongly reduce endogenous levels of GAs, result in a more severe reduction of cortical cell number and a further elongation of cortical cells, suggesting that the degree of cell cycle arrest is related to the endogenous levels of GAs. The lower the GA levels, the more severe the cell elongation and proliferation defects and consequently, the smaller the root meristem size.
Because of the importance of plants on the global level to food security and environmental sustainability, exploring molecular mechanisms underlying the control of plant organ size and growth has become a high priority in plant research worldwide [35]. Given that most agriculturally important crop species are monocots, our finding that SHB and its putative orthologs in monocot crop species are closely clustered in phylogenetic tree is of great importance. Future studies on their functions may lead to the identification of evolutionarily conserved mechanisms in cell size control and further our understanding on how meristem growth is modulated without markedly compromising cell proliferation to influence organ and body size. Consequently, rational design of crop plant architecture may be enabled by modulating the activities of SHB and its orthologs, which may improve the ability of crop plants to cope with adverse weather conditions such as rain, wind and hail [36], ultimately leading to a second GA-dependent ‘Green Revolution’ in crop productivity.
Materials and Methods
Plant Materials and Growth Conditions
The mutant line 03Z11ER89 (shb) was isolated from a rice enhancer trap collection [37]. The ks1, gid1 and slr1 mutants were reported previously [19,29,30]. ks1 mutants set no seeds and therefore the mutation were maintained in a heterozygous state. For field studies, rice plants were cultivated under natural long-day conditions during rice cultivation seasons at the experimental field of Huazhong Agricultural University. For the analysis of seedling root phenotypes, seeds were sterilized and sowed simultaneously (except for S2D–S2K Fig experiments which were conducted by sowing shb seeds 12 h earlier before WT controls) on petri plates containing half-strength Murashige and Skoog (MS) medium (Duchefa, The Netherlands), supplemented with 1% sucrose, 0.05% MES and 0.8% agar and adjusted to pH 5.8. The plates were then placed vertically and incubated at 28°C in either continuous darkness or a light/dark (14/10 h) regime, depending on the experimental design.
Plasmid Construction and Rice Transformation
For the complementation of shb phenotypes, the genomic sequence of SHB gene, together with 2.51 kb promoter and 386 bp 3’-UTR regions, was inserted into the pCAMBIA2301 vector (http://www.cambia.org) to generate pCAMBIA2301-SHB. To construct the complementing SHB-GFP fusion, sunlight GFP was fused in-frame to the C terminus of SHB coding sequence, and subcloned into pCAMBIA2301 under the control of the 2.51 kb SHB promoter. The empty vector pCAMBIA2301 was used as a negative control. Transgenic rice plants carrying each of these constructs were produced by using Agrobacterium-mediated transformation of callus of shb mutant. To construct CPMV::SHB-GFP, the coding sequence of SHB was fused in-frame to the N terminus of GFP and subcloned into the pEAQ-HT-DEST1 vector by using the GATEWAY recombination system, under the control of the Cowpea Mosaic Virus (CPMV) promoter [25]. Primers used for the construction of these vectors are listed in S2 Table.
Sequence and Phylogenetic Analyses
Protein sequences of putative orthologs of SHB from the other plant species were obtained by using blast search against the NCBI database (http://www.ncbi.nlm.nih.gov). Multiple protein sequence alignment was performed using the ClustalX Version 2.0 [38]. The alignment was then manually refined. A phylogenetic tree was constructed using the MEGA 4.0 program [39] with the following parameters: Poisson correction, pairwise deletion, and bootstrap (1000 replicates; random seed).
Quantitative Analysis of Root Phenotypes
Root length was measured with Image J software (http://rsb.info.nih.gov/ij). Root meristem size was determined by measuring the length from the quiescent center to the first elongated epidermal cell. Cell number in the root meristem, average cell length and width in the root meristem were quantified with all cells in the fourth cortical layer of the root meristem. Cell production rate in the rice root was calculated as described previously [3,40]. Briefly, time-course analyses of root growth and mature cell length were performed and root growth rate was determined using a five-point equation. Cell production rate was then calculated using the following equation: cell production rate = root growth rate/ mature cell length. Cells in the fourth cortical layer of the root elongation zone and maturation zone were used to obtain the quantification data of average cell length in the root elongation zone and mature cell length, respectively. For each quantification, at least 15 rice plants were analyzed.
5-ethynyl-2′-deoxyuridine (EdU) Staining
EdU staining was performed using an EdU kit (C10310, Apollo 488) from Ribobio, China, according to the manufacturer’s protocol. Briefly, roots of 4-day-old rice seedlings were immersed in 50 μM EdU solution for either 2 h (Figs 1F, 1G and 5G and 5H) or 20 h (Fig 6C and 6D), and then fixed for 30 min in 4% paraformaldehyde, followed by 30 min of incubation with Apollo. The samples were next hand-sectioned longitudinally and EdU images of the sections were then captured with a Leica TCS SP2 confocal laser-scanning microscope equipped with a 20× water immersion objective and analyzed with Leica LAS AF software. Quantification of numbers of EdU-stained cells was performed in the fourth cortical cell layer of the rice root meristem, in a selected portion with a length of 360 μm.
Quantification of Endogenous GAs
Quantification of GAs in the WT, shb and ks1 was performed as described previously [41], using [2H2] GA1 (1.00 ng/g), [2H2] GA3 (1.00 ng/g), [2H2] GA4 (1.00 ng/g) [2H2] GA12 (2.00 ng/g), [2H2] GA24 (2.00 ng/g), [2H2] GA19 (5ng/g), [2H2]GA20 (2ng/g), [2H2]GA34 (2ng/g), [2H2]GA44 (2ng/g) and [2H2] GA53 (2.00 ng/g) as internal standards. Roots of 7-day-old seedlings were used for comparison between the WT and shb. To compare the levels of endogenous GAs in ks1, shb and the WT, whole seedlings were used due to the severity of ks1 root phenotype.
Chemical Treatment
Rice seeds were sown on medium supplemented with 10 μM GA3 and cultured for 4 days before analyzing the effect of GA on the root phenotypes. To determine the minimum concentration of GA required to rescue shb and ks1 root phenotypes, 3-day-old seedlings were cultured on medium supplemented with 10, 25, 50, 75 or 100 μM GA3 for 24 hours before analysis. To examine the dose-effects of PAC on rice roots, 3-day-old seedlings were cultured on medium supplemented with 0.1, 1, 10 or 50 μM PAC for 24 hours before analysis. For mock treatments, medium with ethanol at the final concentration as for chemical treatments was used.
Quantitative Real-Time PCR (qPCR)
Total RNA was extracted from WT and mutant roots using TRIzol (Invitrogen) reagent, according to the manufacturer’s instructions. qPCR analysis was performed in a 96-well plate with an ABI StepOnePlus Real-Time PCR System (Applied Biosystems). The following thermal profile was used for all reactions: 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 1 min. The melting curve was determined under the following conditions: 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. The rice ubiquitin1 gene (Os03g13170) was used as the internal control. All primers used are listed in S2 Table.
RNA In Situ Hybridization
RNA in situ Hybridization was performed as described previously [42]. Briefly, roots of 4-day-old rice seedlings were fixed in FAA (50% ethanol, 5% acetic acid and 3.7% formaldehyde) at 4°C for 24 h, dehydrated in an ethanol series, cleared through a xylene series and then embedded in paraffin. 8 to 12 μm sections were mounted on RNase-free glass slides and in situ hybridization was then performed using digoxigenin-labeled RNA probes transcribed with either T7 or SP6 transcriptase from pGEM-T plasmids containing part of the SHB or KS1 coding sequence, which were PCR amplified with gene-specific oligonucleotide pairs SHBinsitu-F/R and KS1insitu-F/R (S2 Table).
Subcellular Localization Analysis
Subcellular localization analysis of SHB-GFP was performed as previously described [43]. Briefly, lower leaves of N. tabacum. plants were infiltrated with Agrobacterium strains carrying CPMV-SHB-GFP using a syringe. For co-localization analysis with the nuclear marker RFP-SRT1 [26] the bacteria were mixed in appropriate volumes of infiltration buffer prior to injection into the leaves. Expression of fluorescent proteins was captured 2–3 d after infiltration with a Leica TCS SP2 confocal laser-scanning microscope equipped with a 40× water immersion objective and processed with Leica LAS AF software.
Electrophoretic Mobility Shift Assay (EMSA)
To detect the binding of SHB protein to the KS1 promoter, EMSA was performed as described previously [42] with recombinant SHB protein produced in E. coli DE3 cells (Novagen). In brief, the recombinant SHB protein was incubated with an [α 32P]-radiolabeled, double-stranded DNA oligonucleotide that covers the region containing the putative SHB binding sequence (GCCGCC) in the KS1 promoter. For control EMSA, nucleotide substitutions were introduced into the putative SHB binding site to produce the control probe. DNA binding reactions were carried out at room temperature for 20 min and the separation of protein-DNA complexes from the free DNA probes was done by non-denaturing polyacrylamide gel electrophoresis followed by auto-radiographic detection.
Chromatin Immunoprecipitation (ChIP)-qPCR
Chromatin extraction and immunoprecipitation were performed as previously described [26]. Briefly, roots of the SHB-GFP plants were first vacuum-infiltrated and fixed in formaldehyde. The chromatin was then isolated from the nuclei of root cells and pre-cleared with sheared salmon sperm DNA/protein A agarose (Invitrogen), and immunoprecipitated with or without an anti-GFP antibody (Abcam; ab290). The protein/DNA complexes were eluted and crosslinks were reversed to free DNA. The immunoprecipitated DNA was then purified and qPCR was performed with primers against KS1 promoter region. qPCR was also performed with input DNA purified from the pre-cleared chromatin, and the rice Actin gene (Os11g06390) was used as the reference gene for normalization of qPCR data.
Western Blot Analysis
Rice root nuclear proteins were extracted from roots of 7-day-old WT and SHB-GFP transgenic plants. Western blot analysis was performed as described previously [26]. After washing in acetone and dried, the proteins were resuspended in Laemmli sample buffer, then separated on a 12% SDS-PAGE and transferred to an Immobilon-P PVDF transfer membrane (Millipore). The membrane was blocked with 2% bovine serum albumin in phosphate-buffered saline (pH 7.5), and incubated overnight with primary antibodies, such as anti-GFP (Abcam; ab290), in a 1 : 5,000 dilution at room temperature. After three washes (30 min each), the secondary antibody (goat anti-rabbit IgG [SouthernBiotech]) at 1 : 10,000 dilution was used. Visualization was performed using the Super Signal West Pico kit (Pierce) according to the manufacturer’s instructions.
Supporting Information
Zdroje
1. Johnson K, Lenhard M (2011) Genetic control of plant organ growth. New Phytol 191 : 319–333. doi: 10.1111/j.1469-8137.2011.03737.x 21517873
2. Baskin TI, Cork A, Williamson RE, Gorst JR (1995) STUNTED PLANT 1, A Gene Required for Expansion in Rapidly Elongating but Not in Dividing Cells and Mediating Root Growth Responses to Applied Cytokinin. Plant Physiol 107 : 233–243. 12228357
3. Ubeda-Tomas S, Federici F, Casimiro I, Beemster GT, Bhalerao R, et al. (2009) Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol 19 : 1194–1199. doi: 10.1016/j.cub.2009.06.023 19576770
4. Powell AE, Lenhard M (2012) Control of organ size in plants. Curr Biol 22: R360–367. doi: 10.1016/j.cub.2012.02.010 22575478
5. Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci U S A 97 : 942–947. 10639184
6. Hu Y, Xie Q, Chua NH (2003) The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15 : 1951–1961. 12953103
7. Qi W, Sun F, Wang Q, Chen M, Huang Y, et al. (2011) Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol 157 : 216–228. doi: 10.1104/pp.111.179945 21753115
8. Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, et al. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442 : 705–708. 16900200
9. Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, et al. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460 : 1026–1030. doi: 10.1038/nature08258 19693083
10. Fukao T, Bailey-Serres J (2008) Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc Natl Acad Sci U S A 105 : 16814–16819. doi: 10.1073/pnas.0807821105 18936491
11. Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59 : 225–251. doi: 10.1146/annurev.arplant.59.032607.092804 18173378
12. Sun TP (2008) Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 6: e0103. doi: 10.1199/tab.0103 22303234
13. Daviere JM, Achard P (2013) Gibberellin signaling in plants. Development 140 : 1147–1151. doi: 10.1242/dev.087650 23444347
14. Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, et al. (2009) Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol 19 : 1188–1193. doi: 10.1016/j.cub.2009.05.059 19576768
15. Lee LY, Hou X, Fang L, Fan S, Kumar PP, et al. (2012) STUNTED mediates the control of cell proliferation by GA in Arabidopsis. Development 139 : 1568–1576. doi: 10.1242/dev.079426 22492352
16. Nelissen H, Rymen B, Jikumaru Y, Demuynck K, Van Lijsebettens M, et al. (2012) A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr Biol 22 : 1183–1187. doi: 10.1016/j.cub.2012.04.065 22683264
17. Hedden P, Graebe J (1985) Inhibition of gibberellin biosynthesis by paclobutrazol in cell-free homogenates ofCucurbita maxima endosperm andMalus pumila embryos. Journal of Plant Growth Regulation 4 : 111–122.
18. Serrani JC, Sanjuan R, Ruiz-Rivero O, Fos M, Garcia-Martinez JL (2007) Gibberellin regulation of fruit set and growth in tomato. Plant Physiol 145 : 246–257. 17660355
19. Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134 : 1642–1653. 15075394
20. Li J, Jiang J, Qian Q, Xu Y, Zhang C, et al. (2011) Mutation of rice BC12/GDD1, which encodes a kinesin-like protein that binds to a GA biosynthesis gene promoter, leads to dwarfism with impaired cell elongation. Plant Cell 23 : 628–640. doi: 10.1105/tpc.110.081901 21325138
21. Holdsworth MJ, Bentsink L, Soppe WJ (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179 : 33–54. doi: 10.1111/j.1469-8137.2008.02437.x 18422904
22. Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, et al. (2012) Seed germination and vigor. Annu Rev Plant Biol 63 : 507–533. doi: 10.1146/annurev-arplant-042811-105550 22136565
23. Rashid M, Guangyuan H, Guangxiao Y, Hussain J, Xu Y (2012) AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots. Evol Bioinform Online 8 : 321–355. doi: 10.4137/EBO.S9369 22807623
24. Jiang F, Guo M, Yang F, Duncan K, Jackson D, et al. (2012) Mutations in an AP2 transcription factor-like gene affect internode length and leaf shape in maize. PLoS One 7: e37040. doi: 10.1371/journal.pone.0037040 22649507
25. Sainsbury F, Thuenemann EC, Lomonossoff GP (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J 7 : 682–693. doi: 10.1111/j.1467-7652.2009.00434.x 19627561
26. Huang L, Sun Q, Qin F, Li C, Zhao Y, et al. (2007) Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol 144 : 1508–1519. 17468215
27. Hao D, Ohme-Takagi M, Sarai A (1998) Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J Biol Chem 273 : 26857–26861. 9756931
28. De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, et al. (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 : 1653–1668. 11449057
29. Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 : 693–698. 16193045
30. Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, et al. (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. The Plant Cell Online 13 : 999–1010.
31. Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, et al. (1999) Drosophila S6 kinase: a regulator of cell size. Science 285 : 2126–2129. 10497130
32. Beemster GT, Baskin TI (2000) Stunted plant 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiol 124 : 1718–1727. 11115888
33. Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, et al. (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17 : 678–682. 17363254
34. Ubeda-Tomas S, Swarup R, Coates J, Swarup K, Laplaze L, et al. (2008) Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol 10 : 625–628. doi: 10.1038/ncb1726 18425113
35. Gonzalez N, Vanhaeren H, Inze D (2012) Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci 17 : 332–340. doi: 10.1016/j.tplants.2012.02.003 22401845
36. Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, et al. (1999) 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400 : 256–261. 10421366
37. Wu C, Li X, Yuan W, Chen G, Kilian A, et al. (2003) Development of enhancer trap lines for functional analysis of the rice genome. The Plant Journal 35 : 418–427. 12887592
38. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23 : 2947–2948. 17846036
39. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 : 1596–1599. 17488738
40. Erickson RO (1976) Modeling of Plant Growth. Annual Review of Plant Physiology 27 : 407–434.
41. Chen ML, Fu XM, Liu JQ, Ye TT, Hou SY, et al. (2012) Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J Chromatogr B Analyt Technol Biomed Life Sci 905 : 67–74. doi: 10.1016/j.jchromb.2012.08.005 22917596
42. Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, et al. (2007) The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol 144 : 121–133. 17369428
43. Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1 : 2019–2025. 17487191
Štítky
Genetika Reprodukčná medicína
Článek Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer PopulationČlánek Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2Článek Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA ReplicationČlánek SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from DegradationČlánek Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors inČlánek The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 RegulationČlánek Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Putting the Brakes on Huntington Disease in a Mouse Experimental Model
- Identification of Driving Fusion Genes and Genomic Landscape of Medullary Thyroid Cancer
- Evidence for Retromutagenesis as a Mechanism for Adaptive Mutation in
- TSPO, a Mitochondrial Outer Membrane Protein, Controls Ethanol-Related Behaviors in
- Evidence for Lysosome Depletion and Impaired Autophagic Clearance in Hereditary Spastic Paraplegia Type SPG11
- Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer Population
- Trans-Reactivation: A New Epigenetic Phenomenon Underlying Transcriptional Reactivation of Silenced Genes
- Early Developmental and Evolutionary Origins of Gene Body DNA Methylation Patterns in Mammalian Placentas
- Strong Selective Sweeps on the X Chromosome in the Human-Chimpanzee Ancestor Explain Its Low Divergence
- Dominance of Deleterious Alleles Controls the Response to a Population Bottleneck
- Transient 1a Induction Defines the Wound Epidermis during Zebrafish Fin Regeneration
- Systems Genetics Reveals the Functional Context of PCOS Loci and Identifies Genetic and Molecular Mechanisms of Disease Heterogeneity
- A Genome Scale Screen for Mutants with Delayed Exit from Mitosis: Ire1-Independent Induction of Autophagy Integrates ER Homeostasis into Mitotic Lifespan
- Non-synonymous FGD3 Variant as Positional Candidate for Disproportional Tall Stature Accounting for a Carcass Weight QTL () and Skeletal Dysplasia in Japanese Black Cattle
- The Relationship between Gene Network Structure and Expression Variation among Individuals and Species
- Calmodulin Methyltransferase Is Required for Growth, Muscle Strength, Somatosensory Development and Brain Function
- The Wnt Frizzled Receptor MOM-5 Regulates the UNC-5 Netrin Receptor through Small GTPase-Dependent Signaling to Determine the Polarity of Migrating Cells
- Nbs1 ChIP-Seq Identifies Off-Target DNA Double-Strand Breaks Induced by AID in Activated Splenic B Cells
- CCNYL1, but Not CCNY, Cooperates with CDK16 to Regulate Spermatogenesis in Mouse
- Evidence for a Common Origin of Blacksmiths and Cultivators in the Ethiopian Ari within the Last 4500 Years: Lessons for Clustering-Based Inference
- Of Fighting Flies, Mice, and Men: Are Some of the Molecular and Neuronal Mechanisms of Aggression Universal in the Animal Kingdom?
- Hypoxia and Temperature Regulated Morphogenesis in
- The Homeodomain Iroquois Proteins Control Cell Cycle Progression and Regulate the Size of Developmental Fields
- Evolution and Design Governing Signal Precision and Amplification in a Bacterial Chemosensory Pathway
- Rac1 Regulates Endometrial Secretory Function to Control Placental Development
- Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2
- Functions as a Positive Regulator of Growth and Metabolism in
- The Nucleosome Acidic Patch Regulates the H2B K123 Monoubiquitylation Cascade and Transcription Elongation in
- Rhoptry Proteins ROP5 and ROP18 Are Major Murine Virulence Factors in Genetically Divergent South American Strains of
- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- Regulates Refractive Error and Myopia Development in Mice and Humans
- mTORC1 Prevents Preosteoblast Differentiation through the Notch Signaling Pathway
- Regulation of Gene Expression Patterns in Mosquito Reproduction
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
- The Spalt Transcription Factors Generate the Transcriptional Landscape of the Wing Pouch Central Region
- Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA Replication
- Functional Divergence in the Role of N-Linked Glycosylation in Smoothened Signaling
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- Coordinated Evolution of Influenza A Surface Proteins
- The Evolutionary Potential of Phenotypic Mutations
- Genome-Wide Association and Trans-ethnic Meta-Analysis for Advanced Diabetic Kidney Disease: Family Investigation of Nephropathy and Diabetes (FIND)
- New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation
- SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from Degradation
- Satellite DNA Modulates Gene Expression in the Beetle after Heat Stress
- SHOEBOX Modulates Root Meristem Size in Rice through Dose-Dependent Effects of Gibberellins on Cell Elongation and Proliferation
- Reduced Crossover Interference and Increased ZMM-Independent Recombination in the Absence of Tel1/ATM
- Suppression of Somatic Expansion Delays the Onset of Pathophysiology in a Mouse Model of Huntington’s Disease
- Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors in
- The Evolutionarily Conserved LIM Homeodomain Protein LIM-4/LHX6 Specifies the Terminal Identity of a Cholinergic and Peptidergic . Sensory/Inter/Motor Neuron-Type
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of .
- Retinoic Acid Signaling Regulates Differential Expression of the Tandemly-Duplicated Long Wavelength-Sensitive Cone Opsin Genes in Zebrafish
- The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 Regulation
- Genome-Wide Analysis of PAPS1-Dependent Polyadenylation Identifies Novel Roles for Functionally Specialized Poly(A) Polymerases in
- Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
- Regulation of Mutagenic DNA Polymerase V Activation in Space and Time
- Variability of Gene Expression Identifies Transcriptional Regulators of Early Human Embryonic Development
- The Drosophila Gene Interacts Genetically with and Shows Female-Specific Effects of Divergence
- Functional Activation of the Flagellar Type III Secretion Export Apparatus
- Retrohoming of a Mobile Group II Intron in Human Cells Suggests How Eukaryotes Limit Group II Intron Proliferation
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



