-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Drosophila Gene Interacts Genetically with and Shows Female-Specific Effects of Divergence
Animals need to make gametes–sperm or eggs–in order to reproduce. Gametes are produced from a specialized tissue called the germline that is found within the testes or ovaries. These organs contain a small population of stem cells that are able to both self-renew and differentiate to generate gametes and are thus essential for maintaining gamete production throughout the reproductive lifespan of most animals. Surprisingly, some of the genes that control this process evolve rapidly between Drosophila species. We find for a key germline stem cell regulatory gene, bag of marbles (bam), that its rapid evolution affects only female but not male functions. We further report that the endosymbiont bacterium Wolbachia that infects insects and other species interacts with bam and may be contributing to the wider pattern of rapid evolution of germline stem cell regulatory genes.
Published in the journal: The Drosophila Gene Interacts Genetically with and Shows Female-Specific Effects of Divergence. PLoS Genet 11(8): e32767. doi:10.1371/journal.pgen.1005453
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005453Summary
Animals need to make gametes–sperm or eggs–in order to reproduce. Gametes are produced from a specialized tissue called the germline that is found within the testes or ovaries. These organs contain a small population of stem cells that are able to both self-renew and differentiate to generate gametes and are thus essential for maintaining gamete production throughout the reproductive lifespan of most animals. Surprisingly, some of the genes that control this process evolve rapidly between Drosophila species. We find for a key germline stem cell regulatory gene, bag of marbles (bam), that its rapid evolution affects only female but not male functions. We further report that the endosymbiont bacterium Wolbachia that infects insects and other species interacts with bam and may be contributing to the wider pattern of rapid evolution of germline stem cell regulatory genes.
Introduction
Population genetic and comparative analyses in diverse taxa have shown that many genes involved in reproduction are evolving under adaptive evolution [1–3]. Various selective pressures have been hypothesized to drive the adaptive evolution of those reproductive genes including sexual conflict, sexual selection, pathogen resistance, and avoidance of interspecific fertilization [2,4,5]. While population genetic and comparative approaches have been valuable in identifying adaptively evolving genes [4,6–11], a combination of population genetic and functional approaches is needed to identify the adaptive phenotypes and to determine the contribution of these selective pressures.
The gene bag of marbles (bam) is an intriguing example of a rapidly evolving reproduction gene, having experienced recurrent, adaptive evolution in D. melanogaster and D. simulans [12,13]. Unlike many other reproductive genes that have experienced positive selection, however, bam functions early in gametogenesis, making it unlikely that many of the selective pressures mentioned above could act on it. Surprisingly, genes involved in germ cell development and cystoblast division are over-represented genome-wide among those adaptively evolving in both D. melanogaster and D. simulans [7,14].
bam regulates germline stem cell (GSC) differentiation and germline cyst development in both males and females. GSCs are present in a niche environment that is required to maintain their stem cell state [15,16]. When a stem cell asymmetrically divides, the daughter cell, a cystoblast, moves away from the niche, which relieves repressive mechanisms and allows it to differentiate [15–17]. The cystoblast then undergoes four synchronous mitotic divisions to generate an interconnected, 16-cell cyst. In females, one of these cells will become the oocyte and enter meiosis while the remaining 15 nurse cells will become polyploid and provide nutrients to the oocyte. In males, all 16 cells will enter meiosis and give rise to mature sperm [18].
In females, bam is the key factor for inducing GSCs to differentiate and is thus transcriptionally repressed in the GSC and derepressed in the cystoblast [19–21]. Bam expression is transient, as its protein is present only in late cystoblasts, and 2-, 4-, and 8-cell cysts (Fig 1A) [22]. In males, bam is not required for GSC differentiation, as bam mutant GSCs differentiate but continue undergoing mitotic divisions and never enter meiosis [23–25]. As in females, Bam protein is expressed transiently in males, as it is present only in 4-, 8-, and 16-cell cysts (Fig 1B) [25].
Fig. 1. bam transgenic constructs. 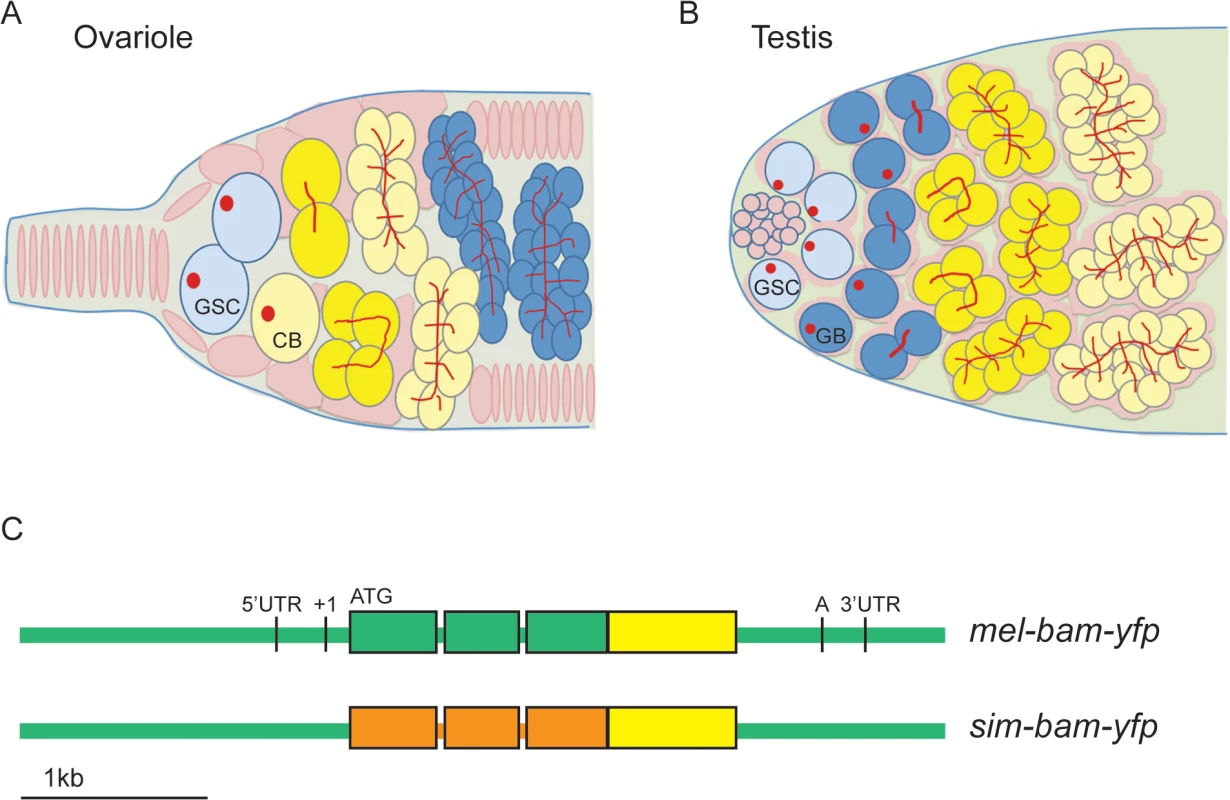
(A) Diagrams of ovariole tip and (B) testis tip of wildtype flies. GSCs differentiate into cystoblasts (CB, ovariole) or gonialblasts (GB, testis), which undergo four synchronous, mitotic divisions. In females, Bam expression (yellow) is restricted to the CB, 2-,4-, and 8-cell cysts. In males, Bam expression occurs in 4-,8-, and 16-cell cysts. Somatic cells/somatic stem cells are shown in pink, germ cells in blue and yellow (when expressing Bam), GSCs in light blue, and spectrosomes (in GSCs) and fusomes (in cysts) in red. (C) bam transgenic constructs. All constructs are drawn to scale and contain the entire bam open reading frame (thick bars), 2 small introns, and non-coding regions (thin bars). Green color corresponds to D. melanogaster sequences, orange to D. simulans sequences, and yellow to the YFP coding sequence. ATG denotes the start codon, and 5’ and 3’ UTR sequence boundaries are from D. melanogaster genome release v. 5.30 (Flybase) [106]. The transcription start site is denoted as +1 [21] and the poly(A) addition sequence is denoted as A [23]. Bam also functions downstream of GSC differentiation in both males and females. Bam also localizes to the fusome, an ER-like organelle that interconnects the cells of a cyst, mediates the synchrony of the mitotic divisions, and likely determines the future oocyte [22,26]. This localization requires the gene benign gonial cell neoplasm (bgcn) [27], and bam mutants show a reduction in fusome vesicles [22]. Bam also has a role in counting cyst divisions in females [22,28,29]. This function is more clearly established in males, where the accumulation of Bam to a critical threshold is required for cysts to cease mitotic divisions and initiate spermatocyte differentiation [25,30].
The molecular function of bam is not fully understood, but Bam physically interacts with and requires the function of bgcn [27,31–33] and Sex lethal (Sxl) [34–36] in GSC differentiation in females. Sxl has been shown to bind nanos mRNA, downregulating it and allowing for GSC differentiation [34–36]. Additionally, Bgcn is related to the DExH-box family of ATP-dependent RNA helicases, leading to the hypothesis that Bgcn functions together with Bam to repress translation [31]. This has been shown directly in males for the target gene mei-P26 [30].
Because bam is essential for fertility yet is involved in the early stages of reproduction, theories of sexual conflict and sexual selection that apply to many other rapidly evolving reproductive genes do not readily explain the adaptive evolution of bam. We therefore explore here interactions between bam and the bacterial endosymbiont, Wolbachia pipientis. Wolbachia is maternally inherited and manipulates host reproduction in a variety of organisms [37–40]. One report found that Wolbachia infection partially rescues the oogenesis defects of Sxl mutants in Drosophila melanogaster [41,42]. This result is an important motivation for examining possible interactions between Wolbachia and bam because a subsequent study showed that bam requires Sxl to function in GSC differentiation [34].
Wolbachia localization and activity are highly dynamic among Drosophila species and are controlled by both host and bacteria [43–48]. For example, in D. melanogaster, Wolbachia is present throughout the germline of females but preferentially accumulates at the somatic stem cell niche, a microenvironment required to maintain somatic stem cells that, when differentiated, produce follicle cells [49]. In contrast, Wolbachia preferentially localizes to the germline stem cell niche in D. mauritiana [46,49]. Transinfection and introgression studies have shown this trait to be primarily controlled by Wolbachia strain, rather than host background [48]. Wolbachia can rapidly spread through a population using a reproductive manipulation known as cytoplasmic incompatibility (CI), where Wolbachia causes the death of offspring from matings of Wolbachia-infected fathers with uninfected mothers [39]. When CI-inducing Wolbachia from D. simulans are transferred to D. melanogaster, their ability to induce CI decreases dramatically [43]. Conversely, when strains that do not induce strong CI in D. melanogaster were transinfected into D. simulans, they induced high levels of CI [50]. Additionally, some strains of Wolbachia do not cause CI, suggesting that both Wolbachia and its host control the occurrence/penetrance of CI [50].
These studies suggest that Wolbachia may be inducing species-specific adaptations, yet no studies to our knowledge have identified host genes that are candidates for mediating an adaptive response to Wolbachia. The critical function of bam in GSC differentiation and the striking consequences of bam divergence in females that we document in this study motivated us to explore interactions between Wolbachia and bam.
Results
Transgenic constructs to test for interspecific rescue of D. melanogaster bam mutants
To identify the functional consequences of bam’s divergence, we developed a transgenic system to assay the ability of a bam ortholog from D. melanogaster or D. simulans to rescue the female and male sterility of a D. melanogaster bam mutant. We generated strains of D. melanogaster containing transgenic copies of either D. melanogaster bam (mel-bam-yfp) or D. simulans bam (sim-bam-yfp) (Fig 1C). Each bam ortholog was C-terminally tagged with Yellow fluorescent protein (YFP) and driven by the native D. melanogaster regulatory region which has been previously defined [21,23]. This approach was designed in an effort to attribute any phenotypic differences to coding sequence divergence. Each transgene was integrated separately in the same position of the D. melanogaster genome at two different attP sites on chromosome 2 (attP16a or attP40), and then crossed into a D. melanogaster bam transheterozygous, null mutant background. PCR using primers designed to the Wolbachia wsp gene confirmed that Wolbachia was not present in the transgenic or bam mutant stocks (see Materials and Methods). The nomenclature used throughout this study is described in Table 1.
Tab. 1. Nomenclature. 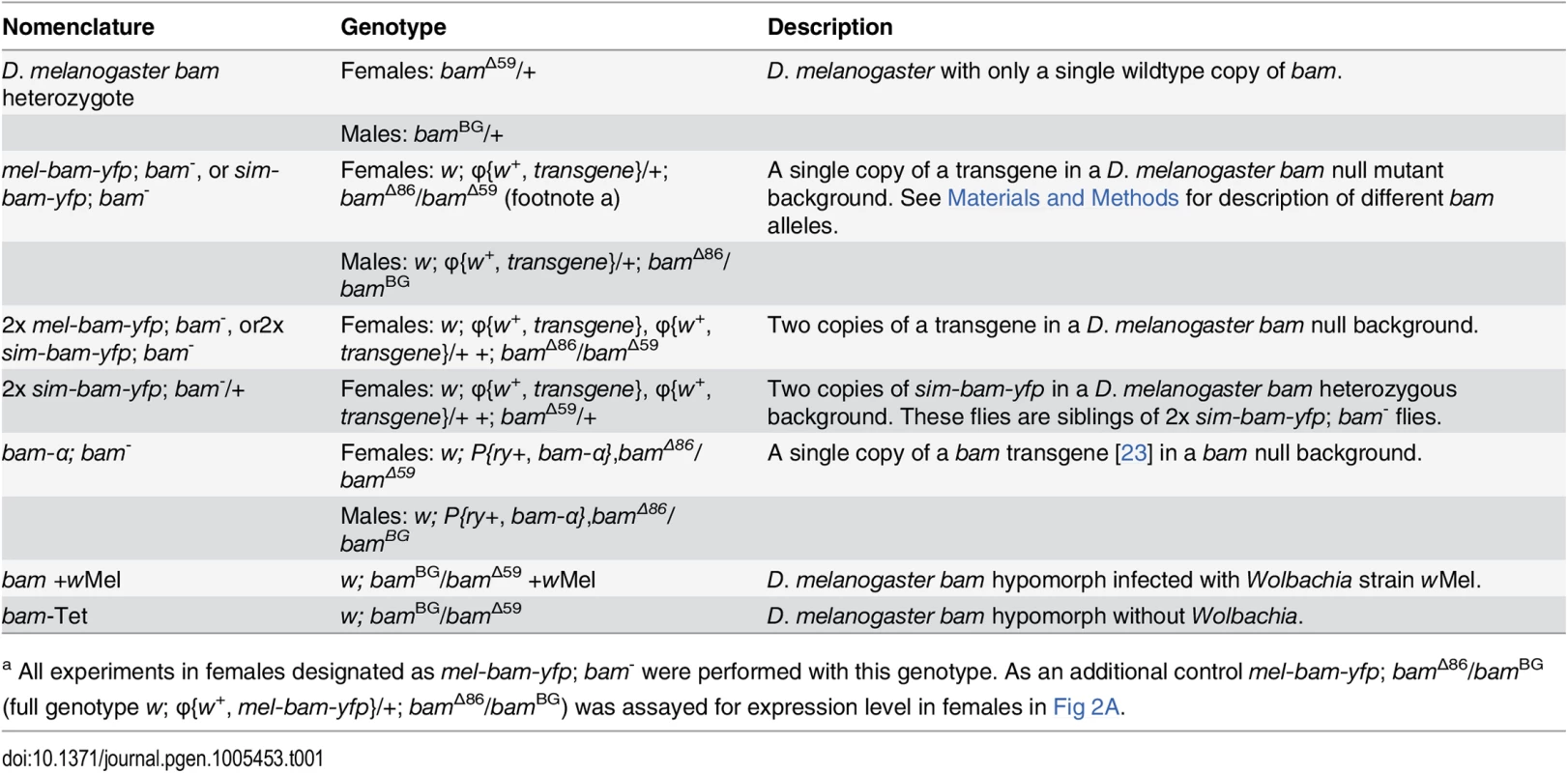
a All experiments in females designated as mel-bam-yfp; bam- were performed with this genotype. As an additional control mel-bam-yfp; bamΔ86/bamBG (full genotype w; φ{w+, mel-bam-yfp}/+; bamΔ86/bamBG) was assayed for expression level in females in Fig 2A. qRT-PCR analyses from ovarian cDNA provided two unexpected results. First, bam expression levels in mel-bam-yfp; bam−(mel-bam-yfp/+; bamΔ86/bamΔ59, see Table 1) ovaries are 13–15-fold less than in controls with a single D. melanogaster bam allele (bam heterozygote of bamΔ59/+) generated from the same cross (Fig 2A). To determine if the unexpectedly low bam expression in mel-bam-yfp; bam−is due to a mutation caused during transformation or to a background effect, additional qRT-PCR was performed in which we found that the results are consistent in different bam mutant backgrounds (Fig 2A) and across different transgene insertion sites (Fig 2B). We also determined that bam expression in the stock from which the bam allele in mel-bam-yfp was cloned is similar to the D. melanogaster heterozygote (+/bamΔ59), demonstrating that the particular allele we chose is not defective in expression (Fig 2C). Additionally, we found that bam expression in the heterozygous genotype used as a reference is not an outlier as it is similar across several genetic backgrounds (Fig 2C). Finally, we compared bam expression in mel-bam-yfp; bam−to that of another bam transgene, bam-α, previously reported to fully rescue both female and male sterility of D. melanogaster bam mutants [23]. We found that the bam-α transgene is similarly under-expressed relative to the D. melanogaster bam heterozygote (Fig 2A). We attempted to perform similar qRT-PCR analyses of bam expression in males, but could not generate reliable results due to its low level of expression. Overall, these results demonstrate that mel-bam transgenes do not express at a wildtype level in females. This is likely caused by the lack of some regulatory sequences, although we cannot eliminate the possibility that bam transgenes are particularly sensitive to position effects. We therefore designed the genetic assays below to assess whether mel-bam-yfp is fully functional.
Fig. 2. Analysis of bam RNA and protein expression in transgenic lines and control strains. 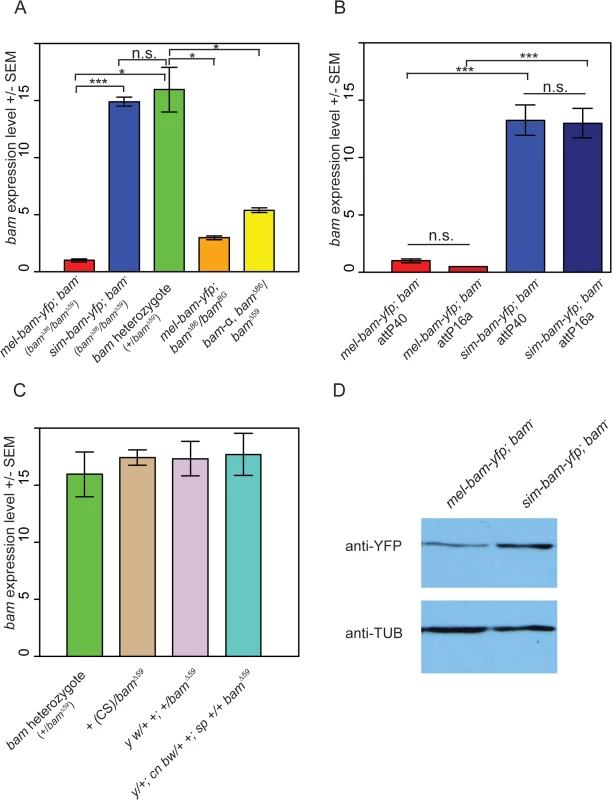
(A) Underexpression of bam RNA in mel-bam-yfp; bam−ovaries is not due to genetic background or the YFP tag. Ovarian bam RNA levels from mel-bam-yfp (red) and sim-bam-yfp (blue) in the bam mutant background (bamΔ86/bamΔ59), and the D. melanogaster bam heterozygote (+/bam Δ59, green). bam levels of mel-bam-yfp in a different bam mutant background (bamΔ86/bamBG) (orange) and of a different bam transgene (yellow, bam-α; bam–) are also reduced relative to the bam heterozygote. ΦC31-integrated transgenes in (A) are docked in attP40. (B) Transgene expression is stable across different insertion sites. We compared bam RNA levels from mel-bam-yfp; bam−and sim-bam-yfp; bam−ovaries in two different insertion sites, attP40 and attP16a. The bam−genotype is bamΔ86/bamΔ59 as explained in Table 1. (C) bam expression levels show little variation across strains. bam RNA levels were compared between the D. melanogaster bam heterozygote shown in (A) to that of various wildtype or marker lines (Canton S [CS], y w, and y; cn bw; sp) that were made heterozygous over a D. melanogaster bam mutant (bamΔ59). The bam sequence in mel-bam-yfp was cloned from y; cn bw; sp. For A-C, N = 3 biological replicates for each sample. Significance was determined by t-test, * P<0.05, **P<0.01, ***P<0.001. No significant expression differences were found in (C). (D) Western blot comparing sim-Bam-YFP and mel-Bam-YFP levels. 20μg of total protein was loaded into each lane. Western blot probed with anti-YFP or anti-α-Tubulin antibodies. The second unexpected result is that bam expression in sim-bam-yfp; bam−(sim-bam-yfp/+; bamΔ86/bamΔ59, see Table 1) ovaries is similar to the D. melanogaster bam heterozygote and ~13–15-fold higher than mel-bam-yfp; bam−(Fig 2A and 2B), despite the fact that both transgenes contain the same D. melanogaster bam regulatory region. We examined protein levels by Western blots and found that sim-Bam-YFP accumulates ~2–3-fold higher than mel-Bam-YFP which is considerably less than the difference in RNA levels (Fig 2D). We conclude that bam coding sequence (CDS) divergence affects both RNA and protein levels. We were unable to assess how protein levels from each transgene compare to wildtype levels as anti-Bam antibodies did not work well on Western blots under our experimental conditions (monoclonal mouse Anti-BamC, rabbit Anti-Bam) [22,51,52]. The difference in expression levels between the transgenes does complicate the ability to attribute phenotypic differences between the orthologs to coding sequence divergence. We therefore expanded our analyses to include the D. melanogaster bam heterozygote as a control, since its expression level is not significantly different from bam levels in sim-bam-yfp; bam–, resulting in a three-way comparison: mel-bam-yfp; bam−vs. bam heterozygote, mel-bam-yfp; bam−vs. sim-bam-yfp; bam–, and sim-bam-yfp; bam−vs. bam heterozygote (See S1 Fig for crossing diagrams).
sim-bam-yfp rescues the male sterility but not female sterility of D. melanogaster bam mutants
To assay transgene function, we crossed each into a D. melanogaster bam transheterozygous, null mutant background. Sibling flies that were heterozygous for bam but did not carry a transgene were used as a control for comparison in fertility experiments (S1 Fig). We found that mel-bam-yfp fully rescues D. melanogaster bam female sterility to the level of the D. melanogaster bam heterozygous control (Fig 3A), suggesting that this transgene is fully functional in females despite having a reduced expression level relative to wildtype bam alleles. However, sim-bam-yfp; bam−females were significantly less fertile than mel-bam-yfp; bam−at every time point in the experiment for both insertion sites tested (Fig 3B), demonstrating the sim-bam-yfp cannot fully rescue D. melanogaster bam female sterility.
Fig. 3. sim-bam-yfp does not fully rescue D. melanogaster bam mutant female sterility. 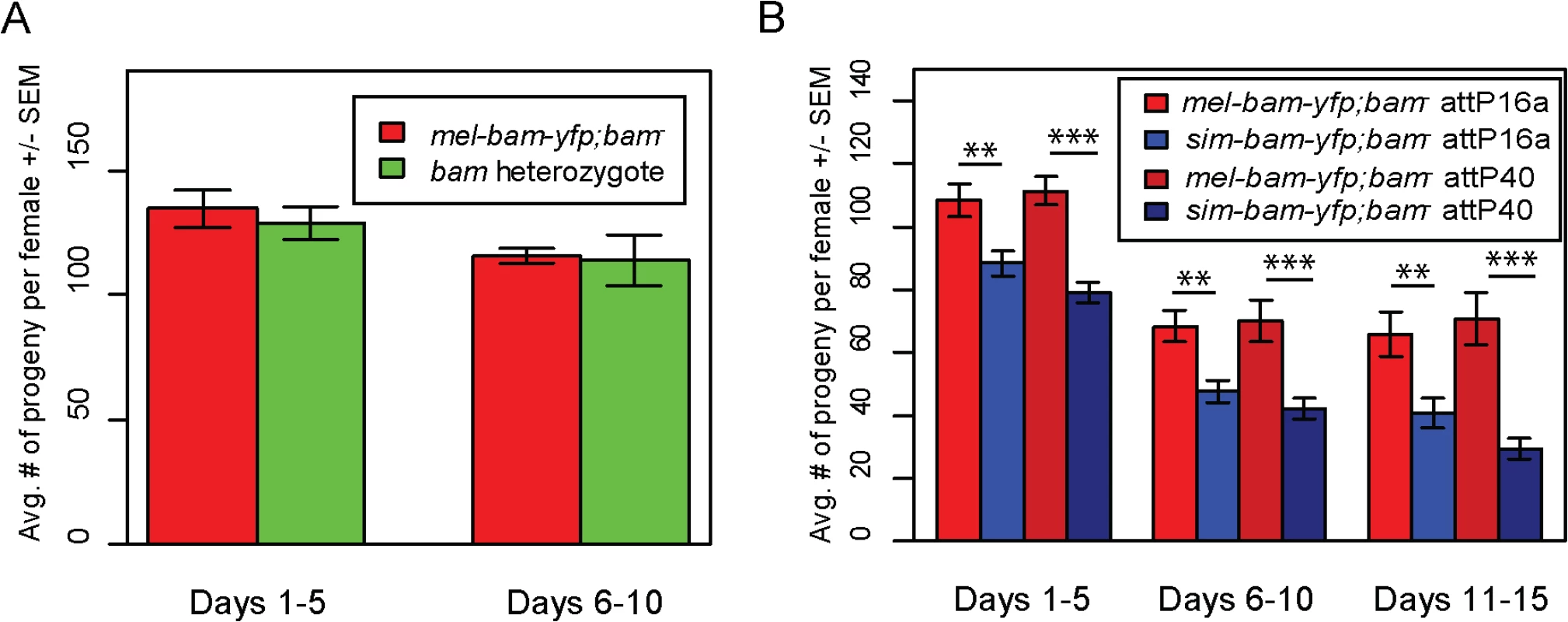
One transgenic female (or heterozygous female) and two tester males were allowed to mate and the trio was transferred to a new vial every five days. Fertility is shown as the average number of progeny per female +/- SEM for each vial. (A) mel-bam-yfp rescues D. melanogaster bam female sterility. N ranged between 22 and 24 females at start of experiment; due to female mortality N ranged between 17 and 18 at end of experiment. (B) sim-bam-yfp cannot fully rescue D. melanogaster bam female sterility. mel-bam-yfp; bam−is shown in red and compared to sim-bam-yfp; bam−in blue. N ranged between 38 and 40 females at start of experiment; due to female mortality N ranged between 26 and 33 at end of experiment. (t-test, * P<0.05, **P<0.01, ***P<0.001). In contrast to female fertility assays, sim-bam-yfp; bam−males were as fertile as their mel-bam-yfp; bam−or D. melanogaster bam heterozygous counterparts (Fig 4A). To test for more subtle differences in male fertility, we used a sperm exhaustion mating assay by providing the males with two new, virgin females every day over a five-day period. Surprisingly, mel-bam-yfp does not fully rescue male sterility, suggesting that this transgene is not fully wildtype in function (Fig 4B). Under sperm exhaustion conditions mel-bam-yfp; bam−males become sterile quickly which we also found when using the bam-α transgene previously reported to fully rescue bam male sterility (S2A Fig) [23], suggesting that D. melanogaster bam transgenes are unable to fully rescue male sterility. We therefore compared sim-bam-yfp; bam−to bam heterozygotes under sperm exhaustion conditions and found that sim-bam-yfp fully rescues male sterility. While we were unable to accurately quantify bam RNA expression in males due to its low expression, we found that Bam-YFP protein expressed from both transgenes localizes in testes (S2B and S2C Fig) in a manner similar to published reports [25]. These data demonstrate that sim-bam divergence strongly affects females yet causes no observable defects in males.
Fig. 4. sim-bam-yfp rescues D. melanogaster bam mutant male sterility. 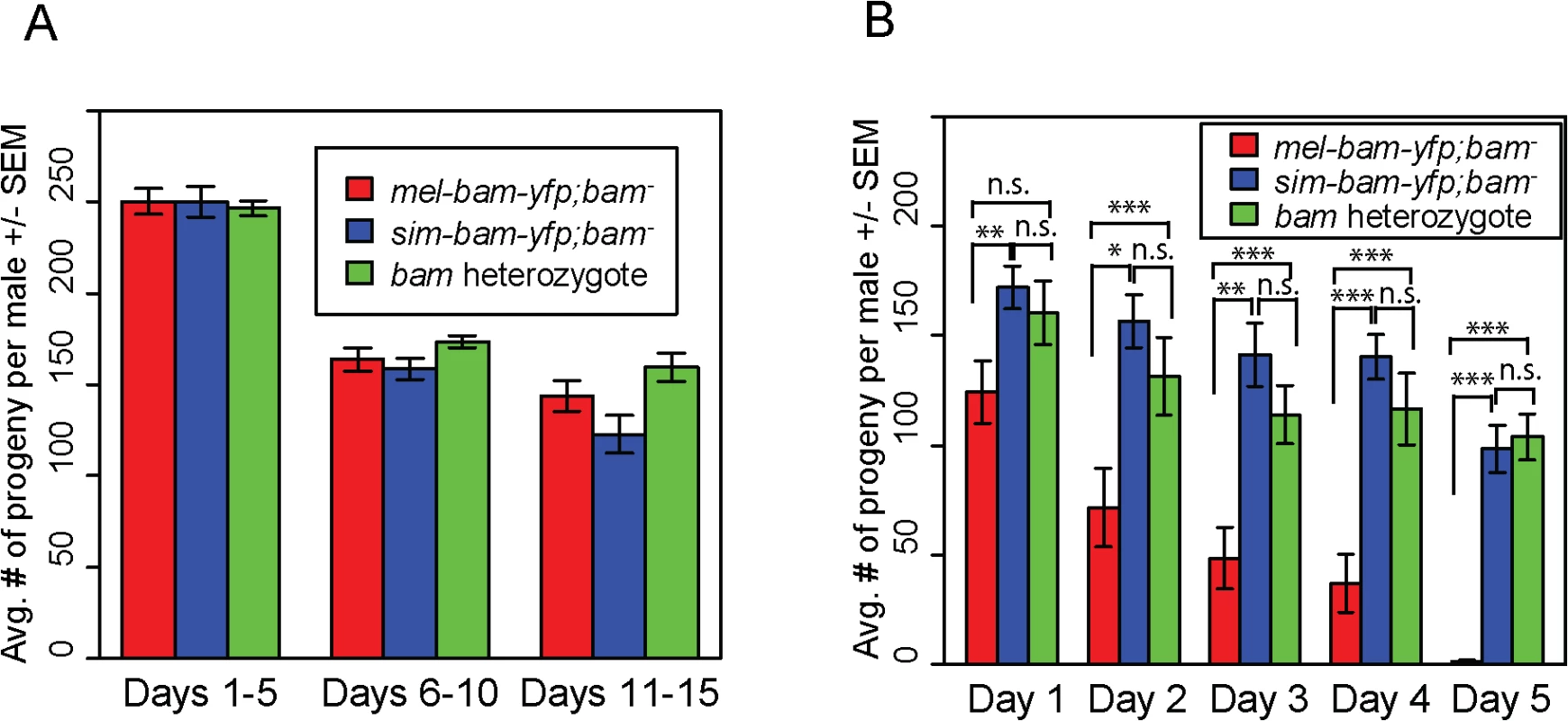
(A) mel-bam-yfp and sim-bam-yfp both rescue male sterility under standard fertility conditions. One male and two tester females were allowed to mate and the trio was transferred to a new vial every five days. No comparisons are significantly different. N ranged between 42 and 46 males at start of experiment; due to mortality N ranged between 37 and 43 at end of experiment. (B) sim-bam-yfp but not mel-bam-yfp rescues male sterility under sperm exhaustion conditions. One male was allowed to mate with a new pair of virgin tester females everyday for five days. Male fertility is the average number of progeny per male +/- SEM for each vial. N ranged between 28 and 33 males at start of experiment; due to mortality N ranged between 22 and 28 at end of experiment. Transgenes are inserted in attP40. (t-test, *P<0.05, **P<0.01, ***P<0.001). Ovaries from sim-bam-yfp; bam−females show multiple defects including GSC loss but not a "bag of marbles" phenotype
To determine the cause of the reduced fertility of sim-bam-yfp; bam−females, we stained mel-bam-yfp; bam−and sim-bam-yfp; bam−ovaries with antibodies to the germline marker Vasa, the fusome marker Hts-1B1, and the YFP tag in Bam-YFP. The ovaries of flies with mel-bam-yfp; bam−show wildtype morphology (Fig 5A and 5B). GSCs were identified by their spherical fusome (i.e. the spectrosome) and their location within the germarium. mel-bam-yfp; bam−ovaries had 2–3 GSCs per germarium, which is comparable to wildtype levels, and Bam was properly localized [22,53]. Furthermore, the vast majority of egg chambers underwent the proper number of cyst divisions giving rise to 16-cell cysts (S1 Table). In contrast, ovaries from sim-bam-yfp; bam−flies showed multiple ovarian defects that increased as the flies aged (Fig 5C and 5D). First, they exhibit stem cell loss, with an average of only 1.5 GSCs per ovariole when young (days 1–5; Fig 5F). Second, as the flies age (days 6–15) they appeared to have a reduction in the number of ovarioles containing mature egg chambers as a consequence of GSC loss, though we did not quantify this effect. Third, many of the egg chambers (24/100) that are present have an improper number of cyst divisions and show mitotic synchrony defects (S1 Table). Mitotic synchrony defects are typically seen with fusome mutants (e.g. hts [54] and α-spectrin [55]) suggesting that sim-bam-yfp; bam−flies may have fusome defects. However, sim-bam-yfp; bam−ovaries have both reduced and increased numbers of cyst divisions while fusome mutants have only reduced numbers, suggesting instead that sim-bam-yfp cannot properly regulate the number of cyst divisions, independently of potential fusome defects. Despite these multiple ovarian defects, it is important to note that sim-Bam-YFP shows a proper localization pattern (Fig 5C and 5D). It is absent in GSCs and present in mitotically active cysts, suggesting that the defects are not due to gross misregulation of Bam. Furthermore, sim-bam-yfp; bam−flies never show the D. melanogaster bam null mutant phenotype of tumorous ovaries [23] (e.g see Fig 5E), suggesting that sim-bam-yfp is capable of rescuing the GSC differentiation defect in D. melanogaster bam mutant females.
Fig. 5. sim-bam-yfp; bam−ovaries have multiple defects. 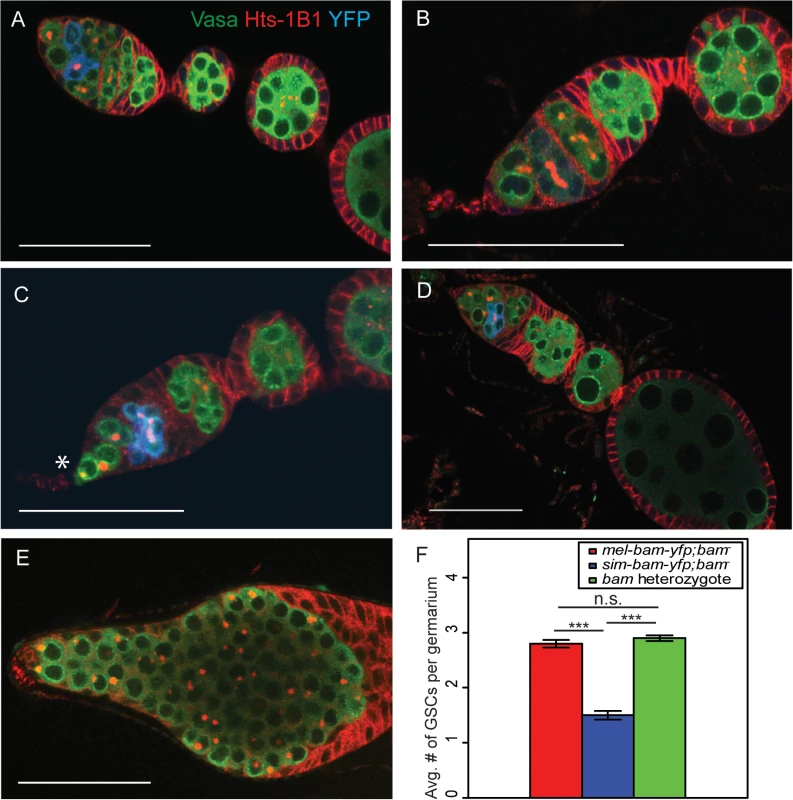
(A-B) mel-bam-yfp; bam−ovaries show wildtype morphology including proper Bam-YFP expression, correct number of GSCs identified by spectrosomes, and proper numbers of cells/cyst. (C-D) sim-bam-yfp; bam−ovaries show reduced number of GSCs (*) and contain egg chambers with improper number of cells/cyst. (E) D. melanogaster bam null mutant shows “bag of marbles” phenotype. (A-E) Ovaries are from flies aged 3–5 days post-eclosion and stained with antibodies to Vasa (green), Hts-1B1 (red), and YFP (blue). Scale bar, 50μm. (F) Average GSC number across different genotypes. N = 50 ovarioles. (t-test, ***P<0.001). sim-bam-yfp; bam−ovarian defects are dose dependent and are partially suppressed by D. melanogaster bam+
The above experiments suggest that sim-bam-yfp does not function properly in a D. melanogaster background and may be acting in a gain-of-function manner as observed by the loss of GSCs. We further explored this by asking if adding additional copies of the mel-bam-yfp or sim-bam-yfp transgenes either improve or worsen the fertility phenotypes. We found no significant differences in fertility when comparing mel-bam-yfp; bam−(one transgene copy) to 2x mel-bam-yfp; bam−(two transgene copies, see Table 1) (Fig 6A). However, 2x sim-bam-yfp; bam−(two transgene copies) flies showed a significant decrease in fertility when compared to sim-bam-yfp; bam−(one transgene copy) and were nearly sterile by day 15 (Fig 6B).
Fig. 6. sim-bam-yfp; bam−female fertility decreases with additional copies of sim-bam-yfp. 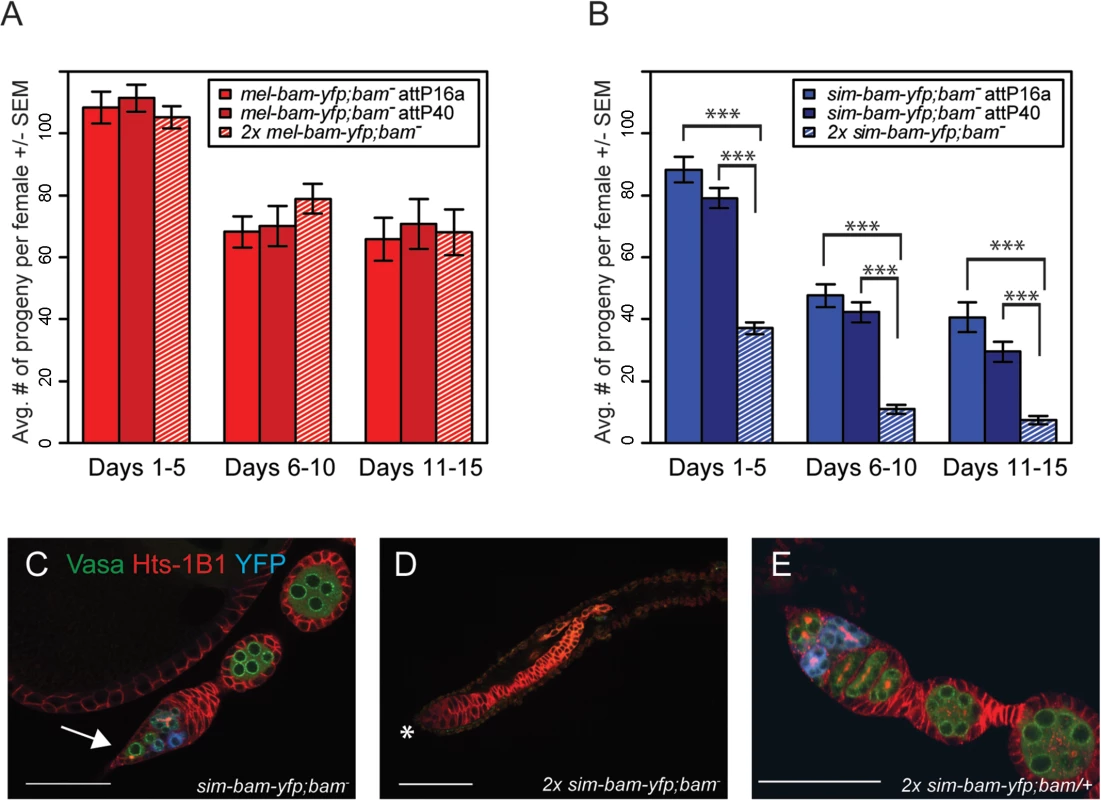
(A, B) Fertility comparison of bam−flies with one versus two copies of mel-bam-yfp (A) or sim-bam-yfp (B). For A and B, one female and two tester males were allowed to mate and the trio was transferred to a new vial every five days. Fertility is shown as the average number of progeny per female +/- SEM for each vial. (A) N ranged between 38 and 40 females at start of experiment; due to female mortality N ranged between 26 and 32 at end of experiment. (B) N ranged between 36 and 40 females at start of experiment; due to female mortality N ranged between 32 and 33 at end of experiment. All mel-bam-yfp; bam−comparisons are not significant while all sim-bam-yfp; bam−comparisons between one and two copies are highly significant (t-test, ***P<0.001). (C) An ovariole from sim-bam-yfp; bam−that has only a single GSC (arrow). (D) An ovariole from 2x sim-bam-yfp; bam−showing a complete loss of GSCs and germline as indicated by lack of Vasa staining. Asterisk indicates anterior end of germarium where GSCs normally reside. (E) A 2x sim-bam-yfp; bam/+ ovariole shows a more wildtype ovary morphology compared to its 2x sim-bam-yfp; bam−sibling. For C-E, ovaries are from flies aged for 3–5 days post-eclosion and are stained for Vasa (green), Hts-1B1 (red), and YFP (blue). Scale bar, 50μm. Ovarioles from 2x sim-bam-yfp; bam−flies showed accelerated rates of stem cell loss, even in young (1–5 day old) flies (Fig 6D and S2 Table), as compared to sim-bam-yfp; bam−(Fig 6C). They typically lacked GSCs and in some cases no longer contained any germline cells, as seen by lack of Vasa staining (Fig 6D). This phenotype contrasts with sim-bam-yfp; bam−flies, where GSCs were almost always present in every ovariole though often reduced in number (see Fig 5C and 5D).
We performed qRT-PCR comparing the ovarian RNA expression levels of bam from flies with one or two copies of the transgene. As expected, doubling the dose of the transgenes results in an approximate doubling of expression for both mel-bam-yfp and sim-bam-yfp (S3A Fig). Notably, however, bam RNA levels of 2x sim-bam-yfp; bam−are not greater than in D. melanogaster wildtype flies (S3A Fig). Additionally, sim-Bam in 2x sim-bam-yfp; bam−ovaries does not show aberrant localization when present (S3B and S3C Fig). Thus, we conclude that the 2x sim-bam-yfp; bam−defects are specifically due to increased dosage of the functionally diverged D. simulans bam, rather than to a general effect of increasing bam dosage or gross mislocalization.
We further explored the nature of sim-bam-yfp-mediated defects by asking how they are modulated in the presence of a wildtype D. melanogaster bam allele. We envisioned 3 possible outcomes. The first is that if the effects are purely due to increased dosage then they should become worse with the addition of wildtype D. melanogaster bam. The second is that if the defects are purely neomorphic as a consequence of D. simulans bam divergence, then they should be unchanged. In other words sim-bam-yfp will be dominant over D. melanogaster bam. And the third is that if the defects are due to a failure of sim-bam function due to divergence, then they should be rescued by D. melanogaster bam and thus be recessive.
We assayed our transgenes with the addition of an endogenous copy of D. melanogaster bam. We found that sim-bam-yfp; bam–-dependent defects are mostly alleviated by the addition of even a single endogenous copy of D. melanogaster bam (Fig 6E and S2 Table). This result suggests that D. melanogaster bam is dominant over sim-bam-yfp, but it is unlikely that sim-bam-yfp is simply a loss-of-function allele as the sim-bam-yfp; bam−phenotypes do not match bam loss-of-function alleles in D. melanogaster. We therefore suggest that sim-bam-yfp has both loss and gain of function attributes. Several hybrid incompatibility alleles, alleles that when expressed in a hybrid background result in sterility or lethality, show similar properties [56,57].
sim-bam-yfp defects in females are not due to failure of interactions with bgcn
In D. melanogaster, Bam and Bgcn physically interact [30,32,33], and like bam, bgcn is also evolving under rapid, adaptive evolution in both D. melanogaster and D. simulans [12]. One might expect that if substitutions occurred that reduce their interaction, compensatory mutations would be selected for to re-establish a strong interaction. Therefore, independent and compensatory substitutions occurring at Bam and Bgcn within each species might render the protein partners incapable of, or less efficient at, interacting when brought together with the heterospecific protein. To determine if sim-Bam and mel-Bgcn interact with one another, we performed immunoprecipitation assays from Drosophila S2 cells. Cells were transiently transfected with either mel-Bam::HA or sim-Bam::HA, and with mel-Bgcn::MYC transgenes. We found that in reciprocal immunoprecipitation experiments both the conspecific and heterospecific Bam coimmunoprecipatated with mel-Bgcn::MYC, indicating that sim-Bam can interact with mel-Bgcn (Fig 7A–7D).
Fig. 7. sim-Bam maintains interactions with mel-Bgcn in immunoprecipitates from S2 cells. 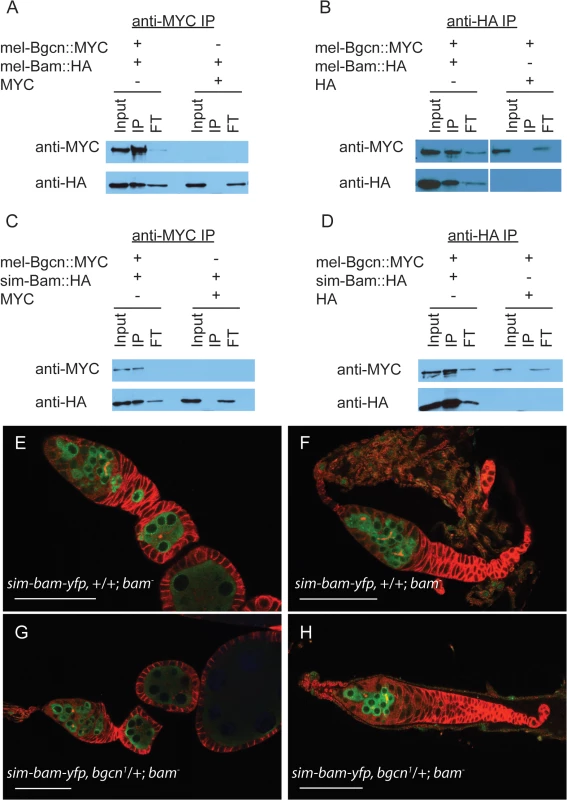
(A-B) Control experiments with mel-Bam and mel-Bgcn. (A) Cells were transfected with mel-Bam::HA and either mel-Bgcn::MYC or MYC. Anti-MYC immunoprecipitates were analyzed by Western blot. (B) Cells were transfected with mel-Bgcn::MYC and either mel-Bam::HA or HA. Anti-HA immunoprecipitates were analyzed by Western blot. (C-D) IP experiment with sim-Bam and mel-Bgcn. (C) Cells were transfected with sim-Bam::HA and either mel-Bgcn::MYC or MYC. Anti-MYC immunoprecipitates were analyzed by Western blot. (D) Cells were transfected with mel-Bgcn::MYC and either sim-Bam::HA or HA. Anti-HA immunoprecipitates were analyzed by Western blot. Gels are loaded with 25% of total input (Input), 100% of immunoprecipitate (IP), and 10% of protein that did not immunoprecipitate (flow through, FT). (E-F) Ovaries of sim-bam-yfp;bam−flies show a varying range of ovarian defects with mild (E) and moderate (F) examples shown for comparison. (G-H) Removal of a copy of bgcn (bgcn1) does not enhance the range of phenotypes seen in sim-bam-yfp;bam−ovaries. No tumorous ovaries were seen (N > 50 ovarioles). (E-H) Ovaries are stained with antibodies to Vasa (green) and Hts-1B1 (red). Scale bar, 50μm. These assays involve gene over-expression and cannot discriminate whether the protein interactions are reduced in efficacy. Ohlstein et al. [31] showed that bgcn acts as a dominant enhancer of partial female sterility caused by D. melanogaster bam hypomorphic mutants. Reducing bgcn dosage exacerbated the bam phenotype, causing sterility and giving rise to completely tumorous ovaries. We reduced the copy number of bgcn by half (bgcn1/+) in sim-bam-yfp; bam−flies and found no exacerbation of the sim-bam-yfp phenotype (Fig 7E–7H). Additionally, adding a copy of sim-bam-yfp rescued the bgcn-induced sterility of the bam hypomorph (S4 Fig). Together the co-immunoprecipitation and genetic-interaction experiments strongly suggest that sim-bam-yfp; bam−ovarian defects are not due to an inability of sim-Bam to interact with mel-Bgcn.
Wolbachia infection partially rescues the sterility of bam hypomorphic mutants
Our transgenic rescue experiments suggest that sim-bam-yfp has diverged specifically in regards to its role in the female germline. The bacterial endosymbiont Wolbachia pipentis is maternally inherited and manipulates its host to ensure transmission [39] and could thus provide selective pressures on genes in the female germline such as bam. To explore possible interactions between bam and Wolbachia, we crossed a naturally occurring strain of D. melanogaster Wolbachia, wMel, into a heteroallelic combination of bam alleles (used above in bam genetic-interaction assays) that results in a hypomorphic phenotype [31,32]. bamBW/bamΔ59 flies lacking wMel Wolbachia are weakly fertile, giving rise to a mix of tumorous and wildtype egg chambers [31]. Thus, the number of nurse-cell positive egg chambers (i.e. non-tumorous egg chambers) can be counted to look for enhancers or suppressors of bam activity [31–33,58]. We compared bamBW/bamΔ59 flies infected with wMel, denoted as "bam +wMel", to hypomorphic flies cured of Wolbachia using tetracycline, denoted as "bam-Tet". We found that the ovarioles of bam +wMel flies contain significantly more nurse-cell-positive egg chambers than the bam-Tet flies (S3 Table).
We then assayed the fertility of the bam +wMel and bam-Tet females. We found that the presence of Wolbachia increases the fertility of bam +wMel females to high levels (Fig 8A; compare to Fig 3, days 1–5). The fertility increase was only observed in bam hypomorphs and not in combinations of bam null alleles that result in complete female sterility (bamΔ86/bamΔ59+wMel, N = 20). The fertility increase is not due to effects on bam mRNA levels, as expression is not significantly different between bam +wMel and bam-Tet females (Fig 8B). Fertility assays were also performed in males. However, bam hypomorphic males were completely sterile, and the presence of Wolbachia had no rescuing effect (bam +wMel, N = 20; bam-Tet, N = 20).
Fig. 8. Wolbachia increases the fertility of D. melanogaster bam hypomorphs without altering bam RNA levels. 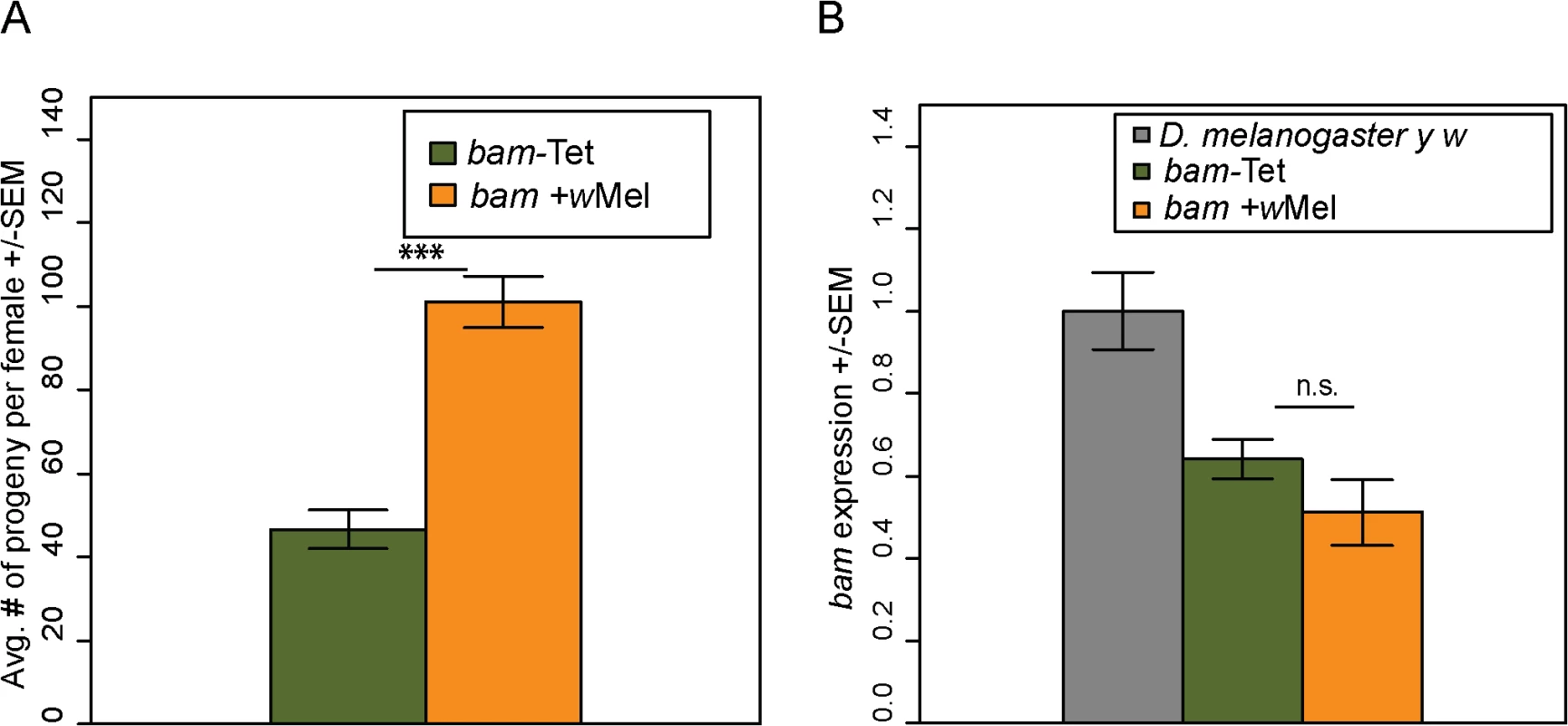
(A) One female and two tester males were allowed to mate and the trio was removed from the vial after 8 days. Fertility is shown as the average number of progeny per female +/- SEM for each vial. N = 20. Wolbachia-infected (wMel) bam hypomorphs are significantly more fertile than uninfected bam hypomorphs, bam-Tet (t-test, ***P<0.001). (B) qRT-PCR of ovarian mRNA from D. melanogaster bam hypomorphs with and without Wolbachia. The D. melanogaster marker strain y w (grey, two wildtype copies of bam) is shown for reference. There is no statistical difference in bam expression of the bam hypomorph with and without Wolbachia (P = 0.253; t-test). To ensure that the fertility rescue of the bam hypomorph was not due to a difference in the gut microbiota caused by tetracycline treatment, we repeated the experiment by controlling for the gut microbial composition (see Materials and Methods). The female fertility assay was repeated and produced very similar results showing that bam +wMel females are significantly more fertile than bam-Tet females (S5 Fig). This experiment demonstrates that fertility rescue of bam hypomorphs is specifically due to Wolbachia infection.
Wolbachia interacts differentially with mel-bam-yfp and sim-bam-yfp
Female sim-bam-yfp; bam−flies have reduced fertility (Fig 3B). We therefore compared the fertility of mel-bam-yfp; bam−and sim-bam-yfp; bam−females with and without Wolbachia (wMel) and found that mel-bam-yfp; bam−fertility was neither enhanced nor diminished in the presence of Wolbachia. In contrast, we found a significant increase in the fertility of young sim-bam-yfp; bam−females (days 1–5) infected with Wolbachia, a result which was consistent across multiple insertion sites (Fig 9A).
Fig. 9. Wolbachia interacts with sim-bam-yfp; bam−in females. 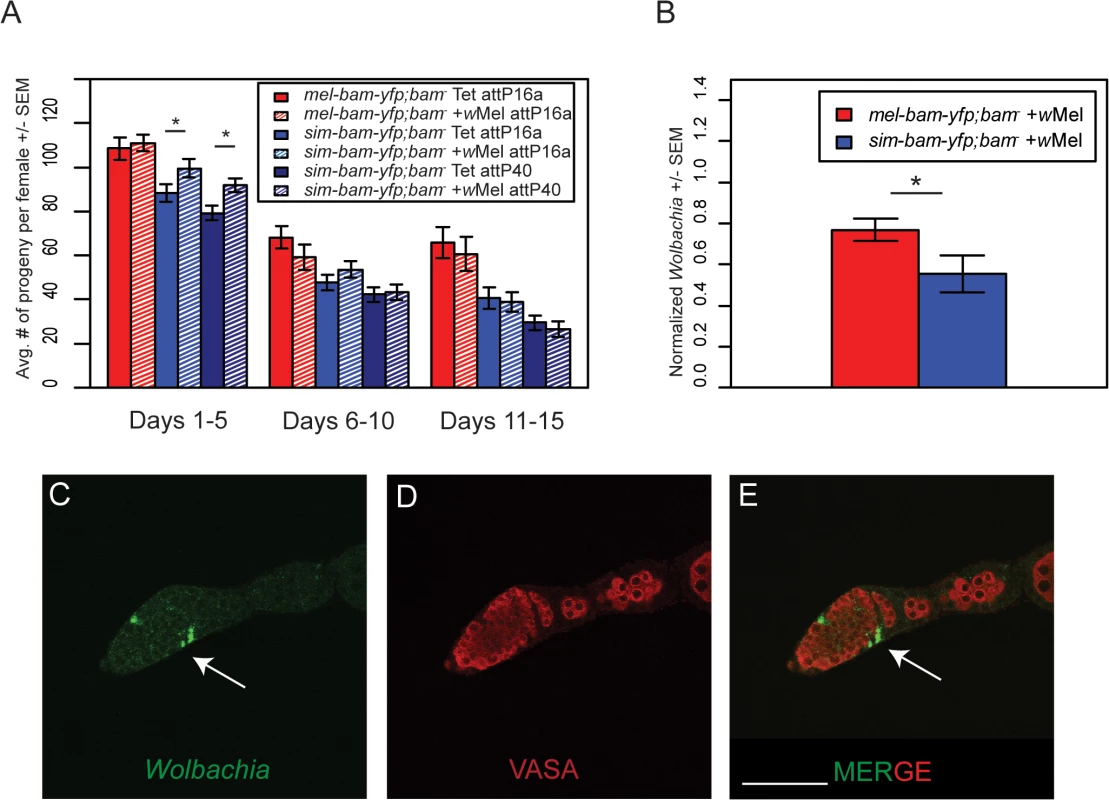
(A) Female fertility assay. One female and two tester males were allowed to mate and the trio was transferred to a new vial every five days. Fertility is shown as the average number of progeny per female +/- SEM for each vial. (t-test, *P<0.05). All comparisons between mel-bam-yfp; bam−+wMel and mel-bam-yfp; bam−Tet are not significant. All day 6–10 and 11–15 comparisons between sim-bam-yfp; bam−+wMel and sim-bam-yfp; bam−Tet are not significant. N ranged between 38 and 40 females at start of experiment; due to female mortality N ranged between 26 and 33 at end of experiment. (B) q-PCR for wMel titer was performed from ovarian DNA from the indicated genotypes using Wolbachia-specific primers. (t-test, *P<0.05). N = 3. (C-E) Wolbachia localizes to the SSCN in sim-bam-yfp; bam−flies. Ovaries from sim-bam-yfp; bam−flies were stained with antibodies to Vasa (red) and Hsp-60 (green), which recognizes Wolbachia. Wolbachia preferentially accumulate at the somatic stem cell niche (arrow) of the germarium. Scale bar, 50μm. If Wolbachia has co-evolved with bam, one possibility is that Wolbachia levels will be influenced by the species-specific ortholog of bam that is present in females. To test this, we used qPCR to measure wMel Wolbachia titer in ovaries and found that Wolbachia levels are reduced in sim-bam-yfp; bam−compared to mel-bam-yfp; bam−ovaries (Fig 9B).
One possible explanation for this reduced titer is that Wolbachia does not localize properly in sim-bam-yfp; bam–. While Wolbachia is present in low levels throughout the germarium, it preferentially accumulates in the somatic stem cell niche (SSCN) in D. melanogaster [46,48,49]. As germline cysts pass the SSCN, high Wolbachia titer and prolonged exposure via somatic cells that encapsulate the cyst may allow it to efficiently infect the cyst and ensure vertical transmission [48]. We examined Wolbachia accumulation using an antibody to Hsp60 which cross-reacts with Wolbachia [44,59,60]. We found that as in mel-bam-yfp; bam–, Wolbachia accumulates normally within the SSCN in sim-bam-yfp; bam−flies (Fig 9C).
Discussion
Using transgenic rescue to identify divergent functions of adaptively evolving genes: Utility and caveats
A detailed comparison of bam function in D. melanogaster versus D. simulans is not possible due to the lack of available bam mutations in D. simulans. More importantly, such an approach might be insensitive to functionally important amino-acid changes if compensatory mutations have occurred in other genes in either lineage. We therefore designed a transgenic construct of D. simulans bam and transformed it into D. melanogaster, along with a parallel D. melanogaster control construct transformed into an identical place in the genome using the phiC31 transformation system [61].
We designed our constructs to have more non-coding DNA than a previously used bam transgene [21,23], yet found that both sets of D. melanogaster bam constructs have lower mRNA expression in females than a wildtype bam allele. Despite this expression difference, we found that our mel-bam-yfp construct fully rescues a bam null mutation in females. One possible explanation is that female flies are indifferent to large differences in bam levels. Alternatively, Bam protein levels may be controlled by a feedback loop that can compensate for differences in mRNA levels. This hypothesis is supported by the fact that differences in protein level between mel-bam-yfp; bam−and sim-bam-yfp; bam−genotypes are considerably smaller than the corresponding mRNA level differences (compare Fig 2A with 2D). We were not able to reliably quantify mRNA levels in males, but the inability of mel-bam-yfp to fully rescue male sterility suggests that it also under-expresses in males. If so, it would also suggest that males are more sensitive than females to lower levels of bam or that a feedback loop involving bam in females is not present in males.
Our goal in this study was to compare the effects of bam coding sequence divergence, and therefore we made the sim-bam-yfp construct using the untranslated regions (UTRs) and non-coding DNA from D. melanogaster, expecting that it would express similarly to mel-bam-yfp. Surprisingly, we found that sim-bam-yfp significantly overexpresses relative to mel-bam-yfp. One possible explanation is that sim-bam contains diverged regulatory sequences within its coding sequence or introns that affect transcription initiation. A second possibility is that these regions affect mRNA stability. Finally, it is possible that our sim-bam-yfp construct contains an intragenic incompatibility affecting mRNA stability between the D. melanogaster and D. simulans portions of its transcript. If true, then a D. simulans bam genomic transgene might have been more effective than a chimeric gene composed of sequences from both species. That alternative, however, is not a panacea because even genes that have similar expression levels between D. melanogaster and D. simulans can mis-express when placed in a foreign species due to "cis x trans" regulatory divergence [57,62,63].
We have performed several controls to minimize the complications arising from the differential mRNA expression levels of the mel-bam-yfp and sim-bam-yfp transgenes. First, we used the endogenous D. melanogaster bam locus as an additional control because its expression is not significantly different from sim-bam-yfp expression in ovaries (Fig 2A). Second, we have shown that the YFP protein localization patterns in both ovaries and testes are similar for both transgenes and resemble wildtype Bam (Figs 5A–5D, S2B and S2C).
We also note that female fertility levels do not appear to be highly sensitive to bam expression level. mel-bam-yfp; bam−and the bam heterozygote are not significantly different in their levels of female fertility even though they express at different levels. Furthermore, sim-bam-yfp fertility rescue is significantly lower than both genotypes despite having a similar expression level to the bam heterozygote. These findings provide confidence in our conclusion that sim-bam-yfp has functionally diverged in its female germline function.
bam divergence strongly affects female but not male functions
Reproductive genes are strongly affected by sexual selection, adaptive divergence, and intra - and inter-sexual conflict. Many lines of evidence suggest that these forces affect males more strongly than females. For example, hybrid male sterility evolves much more rapidly than hybrid female sterility, demonstrating that functionally relevant divergence between species is more likely to occur in males [64–66]. Gene expression of male-biased genes diverges more between species than does the expression of female-biased genes [67,68]. Finally, genes encoding male reproductive proteins are among the most rapidly evolving classes of genes [2,4,5,69,70].
GSC regulatory genes also are over-represented among adaptively evolving gene classes [7,14], which is surprising considering that there is no obvious role for sexual selection or sexual conflict to operate at such early stages of germline development. Selection to increase gamete production could occur in either sex, but would perhaps be stronger in males where energetic investment in gametes is less than for females. We were thus surprised to see how clearly sim-bam-yfp divergence affects female but not male fertility, even when males were assayed under stringent sperm exhaustion conditions.
Only in females does bam function in GSC differentiation [24,71]. Forced expression of a bam transgene in GSCs results in their differentiation only in females and not males [71]. Only after males are exposed to a longer duration and occurrence of heat shock are GSCs lost in males [72–74]. Instead, bam’s primary role in males is regulating cyst divisions and entry into meiosis [24,25,30]. Elegant studies have shown that increased or decreased levels of bam result in cysts with either less or more cells per cyst, respectively, which give rise to elongating spermatids and presumably mature sperm [25]. Therefore, males may be less affected by sim-bam-yfp divergence because either they are less sensitive to bam expression differences or females have additional sex-specific functions of bam.
In our fertility assays, we found that the bam trans-heterozygous mutants used in the female fertility assay resulted in reduced rescue in male fertility assays, presumably due to the accumulation of background mutations that affect male fertility (see Materials and Methods). Therefore, transgenic experiments in males were performed using a different combination of bam alleles. We consider it unlikely that the different allelic combinations underlie the sex-specific differences we see in the ability of mel-bam-yfp or sim-bam-yfp transgenes to rescue. mel-bam-yfp expression level in females is not significantly different in these two bam mutant combinations, arguing that the different genetic backgrounds do not cause a general difference in bam expression (Fig 2A).
The molecular nature of sim-bam-yfp; bam−defects
sim-bam-yfp; bam−ovaries display a range of defects but never the "bag-of-marbles" phenotype seen in D. melanogaster bam loss-of-function mutations. The increased severity of phenotypes with increased sim-bam-yfp dosage also argues against a loss-of-function effect. Furthermore, the presence of D. melanogaster bam does not fully rescue sim-bam-yfp; bam−defects, suggesting that it may have both loss and gain of function properties.
Since Bam and its interacting partner Bgcn are both adaptively evolving, we hypothesized that these ovarian defects might be due to an inability of sim-Bam to interact with mel-Bgcn. We provide three lines of evidence against this. First, bgcn is required for bam’s role in GSC differentiation. If this interaction were eliminated or reduced, we would expect to see tumorous ovaries but never do in sim-bam-yfp; bam−flies. Second, sim-Bam::HA and mel-Bgcn::MYC reciprocally co-immunoprecipitate with one another in S2 cells. Third, removing one copy of bgcn does not exacerbate sim-bam-yfp; bam−ovarian defects nor does it cause tumorous ovaries. This combination of biochemical and genetic data strongly suggests that sim-bam-yfp; bam−defects are due to incompatibilities with D. melanogaster genes other than bgcn.
GSC loss is one of the most striking phenotypes we discovered in sim-bam-yfp; bam−flies, a phenotype that was enhanced with additional copies of sim-bam-yfp transgenes (Fig 6). While bam is transcriptionally repressed in the GSC in wildtype D. melanogaster, there is a small amount of Bam protein present in the GSC which must be kept inactive (i.e. not cytoplasmic) [22,32,75]. One hypothesis to explain Bam silencing is that all Bam protein present in GSCs is localized at the spectrosome (i.e. round fusome), rendering it inactive in promoting differentiation [22,76]. This hypothesis is supported by data in which a subset of antibodies show Bam localized to the fusome. Bam itself is required for function of the fusome, and in bam mutants, the spectrosome shows a reduced amount of vesicular material [22]. A second hypothesis suggests that there is a small amount of cytoplasmic Bam present in the GSC, but that other proteins antagonize its activity [32,75]. Only after Bam accumulates to high levels can it titrate away antagonizing proteins and bind to other partners to promote differentiation.
Based on our data, we suggest that sim-bam-yfp; bam−GSC loss results from sim-Bam-YFP either (1) failing to localize to the spectrosome, thus leaving it active in the GSC cytoplasm, and/or (2) preventing other proteins from localizing to the spectrosome. Fusome-protein components change during fusome growth and assemble in a hierarchical manner [27,76,77]. Based on our dominance study, we also hypothesize that the fusome cannot properly form in sim-bam-yfp; bam−ovaries but can when D. melanogaster bam is added, thus allowing proper fusome localization of sim-Bam-YFP and/or other proteins. We favor this hypothesis since sim-bam-yfp flies also show mitotic synchrony defects, a hallmark of improper fusome function. Moreover, proper endocytic recycling of the fusome is required for GSC maintenance, as rab11 mutants show GSC loss and have defects similar to bam mutants [77]. We have been unable to fully test this model though as Bam-F antibodies which show fusome localization [22] are no longer available and anti-Gfp antibodies used with our bam-yfp transgenes do not show fusome localization (see Fig 5), a result seen previously with different epitope-tagged transgenes [21].
Wolbachia increases the fertility of bam mutant genotypes
Although bam is essential for fertility of both sexes, we only detected fertility defects in female sim-bam-yfp; bam−flies. We cannot of course exclude the possibility that an unexamined aspect of male reproduction is impaired; nevertheless, it seems highly implausible that bam divergence is being driven by a selective force operating in males if the functional consequences of that divergence are so clearly deleterious in females. We therefore sought to identify selection pressures that could potentially drive female-specific functional divergence of bam. We examined the bacterial endosymbiont, Wolbachia pipientis, due to its maternal transmission and its ability to manipulate the reproduction of the hosts that it infects [39,40]. We found that Wolbachia infection increases the fertility of two different bam mutant genotypes: D. melanogaster bam hypomorphs, and sim-bam-yfp; bam−females. It might be unexpected for a D. melanogaster strain of Wolbachia to partially rescue the female fertility defects of sim-bam-yfp. However, sim-bam-yfp at least partially maintains many of the same functions of wildtype D. melanogaster bam: promoting GSC differentiation, regulating cyst divisions, and interacting with bgcn. Therefore, an interaction with Wolbachia could potentially be maintained as well. We did though find that a D. melanogaster-specific strain of Wolbachia cannot accumulate to high levels when only sim-bam-yfp is present, suggesting an incompatibility between D. melanogaster Wolbachia and D. simulans bam. The lower Wolbachia titer might also explain why the level of rescue seen in sim-bam-yfp; bam−(Fig 9A) was not to the level seen in the D. melanogaster bam hypomorph (Fig 8A).
Wolbachia, bam and Sex lethal
The gene Sex lethal (Sxl) is required for bam’s function in GSC differentiation [35]. Intriguingly, Wolbachia partially rescues the female sterility of Sxl mutants in D. melanogaster. This interaction is allele-specific, suggesting that suppression is unlikely due to a general increase in germline Sxl expression [41]. Additionally, microarray studies showed no significant increase in Sxl expression when infected with Wolbachia [42]. Sxl is expressed in both GSCs and cystoblasts, while bam expression is repressed in GSCs and is active in cystoblasts and mitotically-active cysts, though each requires the other to promote differentiation [34,35]. Therefore, it has been proposed that Sxl partners with newly-expressed Bam in cystoblasts to promote differentiation by antagonizing nanos and likely other genes required to maintain GSCs [34–36]. Since bam itself provides cell-type specificity [78], we suggest that the increased fertility of Wolbachia-infected Sxl mutants is a result of increased bam activity driving the differentiation of GSCs, rather than a direct effect on Sxl activity.
Is Wolbachia driving bam divergence?
bam has experienced recurrent, adaptive evolution in both D. melanogaster and D. simulans [12,13]. There is evidence that current Wolbachia infections in D. simulans have been present for at least 8.8x105 generations [79] and possibly predating the speciation of D. simulans and D. sechellia, which would be > 2.4x106 generations (assuming 10 generations/year) [80]. For D. melanogaster, however, the association appears more recent, 2.2x104-8.0x104 generations [81,82]. Therefore it is difficult based on current evidence to propose that Wolbachia has been the sole driver of bam divergence for D. melanogaster. It is possible, however, that the species has experienced recurrent infections resulting in the replacement of old infections not currently sampled today. Wolbachia can provide fitness advantages to its hosts; for example viral pathogen protection in Drosophila [83–86]. Therefore fitness benefits combined with cytoplasmic incompatibility can result in rapid displacement of less beneficial Wolbachia strains, an observation that has been reported for both D. melanogaster and D. simulans [81,87–89].
We therefore propose two models for how an interaction with Wolbachia may have driven the adaptive evolution of bam, while acknowledging that other factors may also have contributed. The first model assumes a mutualistic interaction between bam and Wolbachia and is inspired by research on the parasitic wasp, Asobara tabida, where Wolbachia is required for oogenesis to occur properly [90,91]. Pannebakker et al. [92] proposed that the initial introduction of Wolbachia infection suppressed normal host apoptosis that occurs during oocyte production, causing the wasp to adapt by upregulating apoptosis. This response, while beneficial in the presence of Wolbachia, results in hyperactive apoptosis and oogenesis inhibition in its absence [92]. Thus in this host, Wolbachia has transitioned from facultative parasite to obligate mutualist. While the precise mechanism underlying the Wolbachia effect is unknown, Wolbachia infection in insects alters the expression levels of numerous RNAs and proteins [93–96]. Thus in D. melanogaster and D. simulans, initial introduction of Wolbachia may have changed bam expression. Because these expression changes could affect fertility, strong directional selection would then act on bam to restore its proper expression in the presence of the bacteria. The result would be a mutualistic interaction between Wolbachia and Drosophila where Wolbachia provides a constant benefit to host GSC differentiation.
Our second model assumes an antagonistic interaction between bam and Wolbachia. In the ovary, GSCs continually divide, and a host must receive cues such as nutritional status and age to balance GSC division rates and GSC differentiation throughout its lifetime [97–101]. As a reproductive parasite, Wolbachia is reliant upon host oogenesis for transmission and wants to ensure that oogenesis is continually occurring. One way in which Wolbachia may increase oogenesis is to override host cues and cause GSCs to continually divide and differentiate by increasing bam activity. Wolbachia may act either directly on bam, or indirectly on antagonists of bam activity or on downstream differentiation factors. However, having too much bam activity would be deleterious to the host, as forced expression of bam in GSCs results in premature GSC loss [71]. Therefore, the host would respond by limiting overactive bam activity caused by Wolbachia infection. This conflict between host and endosymbiont over bam activity could lead to an evolutionary arms race.
The first model predicts that bam RNA and/or protein levels would be different in the presence of Wolbachia. The second model makes at least two predictions. The first is that both host and endosymbiont proteins involved in this interaction would adaptively evolve. A second prediction of the antagonistic interaction model is that each Wolbachia strain will have coevolved with its species-specific bam ortholog and that the transmission success of Wolbachia will be reduced in the presence of a heterospecific bam ortholog.
We examined the predictions of each model. For model 1, we found no evidence of altered bam expression at the RNA level, but we were unable to examine protein levels. In examining the predictions of model 2, it has already been shown that bam is adaptively evolving in both D. melanogaster and D. simulans [12,13]. While we do not know which Wolbachia genes are responsible for this interaction, the Wolbachia genomes of D. melanogaster strains (wMel) and D. simulans strains (wRi) differ dramatically. Ankyrin-repeat-domain-containing genes have extensively diversified between the two strains [102,103], which is intriguing because ankyrin repeats are known to mediate protein-protein interactions [104]. Thus this divergence may allow the different Wolbachia strains to target different host molecules [103]. In examining the second prediction of model 2, we found that the titer of D. melanogaster-specific Wolbachia is reduced in sim-bam-yfp; bam−ovaries. It is important to note that sim-bam-yfp; bam−ovaries show a range of defects, and thus could have an altered Wolbachia titer due to cellular differences from the control strain rather than a specific interaction with Wolbachia. We specifically used young flies to minimize such effects, but are unlikely to have fully eliminated them.
Further support of model 2 comes from our experiments testing Wolbachia-bam interactions. First, we find evidence of Wolbachia increasing bam activity (either directly or indirectly) in the bam hypomorph experiment where, when bam is not fully active, Wolbachia infection results in increased bam activity and thus increased fertility. We would expect the host to try to limit Wolbachia manipulation of bam and find evidence of this in our transgenic experiments where, in mel-bam-yfp; bam−flies (with wildtype fertility), Wolbachia infection is incapable of further increasing bam activity (i.e. no increase in fertility). Our data suggest that D. melanogaster has responded to Wolbachia manipulation by utilizing or perhaps developing a feedback system to regulate bam activity. The feedback structure limits the ability of Wolbachia to overactivate bam activity, thus limiting deleterious effects on the host while still allowing increased bam activity when beneficial to the host. It should be noted in regard to GSC differentiation that this interaction does not suggest that mutualism has been established because the wildtype host shows no decrease in fitness without Wolbachia. It is only under specific bam mutant conditions that we see a fitness benefit to the host. Such conditions are unlikely to be common in nature, thus limiting any fertility benefit of Wolbachia infection.
Overall, our data are more consistent with the predictions of model 2. We note, however, that the predictions of each are not mutually exclusive. While altered bam RNA/protein levels are a prediction of model 1, this prediction is not incompatible with model 2. Similarly, the predictions of model 2, adaptive evolution of the genes involved and incompatibilities between Wolbachia and host proteins are also consistent with model 1.
Our discovery of interactions between Wolbachia and bam from D. melanogaster and D. simulans suggests that bam and Wolbachia have been interacting (either mutually or antagonistically) for an extensive period. We speculate that this history of association of Wolbachia with D. melanogaster and D. simulans has had major consequences on the evolution of bam in these species. Furthermore, infection with germline parasites may explain the more widespread pattern of adaptive evolution of early acting germline development genes [7,12,14,105].
Materials and Methods
Drosophila stocks and Wolbachia infection
All stocks were cultured at room temperature on standard yeast-glucose media. The bamΔ86, bamBW, bamBG, and bgcn1 stocks are described in FlyBase [106]. The bamΔ59 allele was generated through a P-element excision of bam1 (D. McKearin, pers. comm.). We sequenced this allele and discovered that the excision deletes all but the 31 amino acids from the C-terminal end of the protein. All five stocks were kindly provided by Dr. Dennis McKearin (HHMI). All stocks (including CS, y w, and transgenic stocks described below) were confirmed to be free of Wolbachia infection by PCR using primers wsp81F/wsp691R [107]. The wMel-infected strain of D. melanogaster, w; Sp/CyO; Sb/TM6B +wMel, was kindly provided by Dr. Bill Sullivan.
The wMel strain of Wolbachia was introgressed by crossing wMel-infected females into bamΔ59/TM3, generating bamΔ59/TM3 +wMel. The bamΔ59/TM3 +wMel stock was then cured of Wolbachia by feeding the flies on media supplemented with 0.03% tetracycline for three generations, generating bamΔ59/TM3 Tet. Females of the bamΔ59/TM3 +wMel stock were then backcrossed to males of the bamΔ59/TM3 Tet strain for at least six generations to generate genetically similar backgrounds including the mitochondria.
DNA constructs
mel-bam-yfp transgene
We amplified a 4.1 kb fragment from genomic DNA of the sequenced D. melanogaster strain, y; cn bw; sp, using primers 904 and 905 (S4 Table). This fragment contains approximately 1.7 kb upstream of the bam start codon and approximately 1 kb downstream of the stop codon. The PCR product was cloned into the pCR-Blunt II-TOPO (Invitrogen) vector to generate the plasmid p{mel-bam} and verified by sequencing. A three-piece fusion PCR strategy was used to incorporate a Yellow fluorescent protein (YFP) tag into the bam coding region at the C-terminus. Two products were amplified using p{mel-bam} as the template with the primer pairs 906/907 and 908/909. These products correspond to parts of the D. melanogaster bam sequence directly upstream and downstream of the native stop codon. The third product containing the YFP tag was amplified using p{w+mC UAS-Lhr::Venus = UAS-Lhr::YFP} as the template [108] with primer pair 910/911. All three products were gel-purified and used as templates for fusion PCR for 6 cycles, and then primer pair 906/909 was added to amplify the final product. The final product was cloned into pCR-Blunt II-TOPO, verified by sequencing, and. the insert subcloned into p{mel-bam} using NdeI and StuI restriction enzymes to generate p{mel-bam-yfp}. The full-length insert was then subcloned into the transformation vector pCasper4\attB [57] using NotI and KpnI restriction enzymes and verified by sequencing.
sim-bam-yfp transgene
The bam genomic region was amplified from D. simulans w501 genomic DNA using the primer pair 904/891, cloned into the pCR-Blunt II-TOPO vector and sequenced completely. A three-piece fusion PCR strategy was used to incorporate both the D. melanogaster regulatory region and YFP tag simultaneously. Two products for fusion were amplified using p{mel-bam-yfp} as template with primer pairs 926/927 and 930/931, corresponding to the D. melanogaster 5’ region and the 3’ regulatory region including the YFP tag, respectively. The third product for fusion was amplified from p{sim-bam} using primer pair 928/929. The gel-purified products were used as templates for fusion PCR as described above using primers 926 and 931, and the fusion product was cloned into pCR-Blunt II-TOPO and sequenced. The insert was subcloned into p{mel-bam-yfp} using MfeI and StuI, generating p{sim-bam-yfp}. The full-length insert was then cloned into the NotI and KpnI sites of pCasper4\attB, and verified by sequencing.
Transgenic fly lines
ΦC31-mediated transformation was used to generate transformants in D. melanogaster [61] and was performed by Genetic Services, Inc. Correct integration was assayed using a PCR-based assay developed by Venken et al. [109]. For the attP40 site at cytological position 25C6, the primer pair 949/1125 was used to check docking-site specificity. We discovered that the attP16 stock contains at least two attP docking sites at unknown locations. Southern blots using a probe designed to the white locus present on p{Casper4}\attB were used to determine that p{mel-bam-yfp} and p{sim-bam-yfp} both integrated into the same attP site (S6 Fig). We refer to this attP site in the attP16 stock as attP16a. All transformants were then outcrossed for at least six generations to a y w strain that had been inbred for 10 generations, to make the genetic backgrounds similar.
Fertility assays
All crosses were performed at room temperature (22–23°C). Prior to crossing all flies were aged for 2–3 days post-eclosion on media supplemented with yeast. In female fertility experiments, single transgenic females were crossed to two wildtype Canton S (CS) males. The trio of flies were transferred to a new vial every five days for a total of 15 days and then discarded. Progeny from each vial were counted for 8 days after the first flies eclosed. In male fertility experiments, single males were mated to two wildtype CS females as described above. In sperm exhaustion assays, single males were mated to two wildtype CS females. The males were aspirated without anesthetizing into new vials containing two fresh CS females every day for 5 days. The females remaining in the vial were transferred to a new vial every five days for 10 days, and fertility was assessed by scoring the number of progeny that eclosed over 8 days.
For female fertility assays, the transgenes were crossed into the bam mutant background bamΔ86/bamΔ59. For male fertility we found that use of bamΔ86/bamΔ59 resulted in reduced fertility of mel-bam-yfp flies relative to the D. melanogaster bam heterozygous control, suggesting that background mutations in these mutants reduce male fertility. It is also likely that combinations of bamΔ86 or bamΔ59 with bam1, the chromosome from which they were derived, will share these background effects. Therefore, all male fertility experiments were done with the transheterozygous combination bamΔ86/bamBG, which are independently-derived mutations of bam. In this background we found no reduction of fertility of mel-bam-yfp; bam−relative to the D. melanogaster bam heterozygote under normal fertility assays.
Gut-microbiome-controlled female fertility assay
To control for effects of tetracycline treatment on the gut microbiome in the bamΔ59/TM3 Tet line, axenic versions of the bamΔ59/TM3 +wMel and the bamΔ59/TM3 Tet lines were generated and the gut microbiota from conventional (i.e., non-axenic) bamΔ59/TM3 +wMel males were introduced to both lines. To generate axenic lines, embryos (less than 18 hour old) from the bamΔ59/TM3 +wMel and the bamΔ59/TM3 Tet lines were collected and dechorionated with 0.6% sodium hypochlorite. Sterile embryos were then seeded onto standard sterile yeast-glucose media. Embryos were allowed to develop into adults, and to ensure the lines were microbe-free, 5 adults from each line were homogenized and all were plated onto MRS agar [110]. Axenic flies of each line were allowed to mate for one generation. To introduce a homogenous population of gut microbiota to the two lines and to control for genetic background, axenic virgin females were backcrossed for three generations to conventional males of the bamΔ59/TM3 +wMel line collected from a single bottle. BC3 virgin females were then crossed to conventional bamBW males to generate the bamBW/bamΔ59 hypomorphic genotype.
Fecundity of these bam hypomorphs with and without Wolbachia was then assayed as follows. Prior to crossing all flies were aged for 3 days post-eclosion. Single bamBW/bamΔ59 +wMel or bamBW/bamΔ59 Tet females were crossed with two wildtype Canton S males. The trio of flies was removed from the vial after 6 days and adult progeny were counted every other day for a total of 8 days. To ensure that Wolbachia infection status was accurately maintained, each mated female was homogenized at the end of the experiment and Wolbachia status was assayed by PCR with primers designed to wsp (wsp440F/wsp691R) and dprA genes (dprA483F/dprA663R). Female fertility was only analyzed for females whose Wolbachia status was consistent with the status of the original stock as determined by typing with PCR.
Quantitative RT-PCR
Flies were aged 2 days on media supplemented with yeast. Ovaries were dissected in 1XPBS and total RNA was isolated from 10 ovaries using Trizol reagent (Invitrogen) following the manufacturer’s protocol. Samples were treated with 20 units DNaseI at 37°C for 2 hours (Roche) and purified using RNeasy columns (Qiagen) following the manufacture’s protocol. cDNA was generated from 2μg of total RNA using the Superscript III First Strand Synthesis kit (Invitrogen) and oligo-dT primers following the manufacturer’s protocol. Quantitative RT-PCR was performed on a Biorad MyiQ cycler using iQ SYBR Green Supermix (Biorad). For bam, primer pair 1160/1170 amplified bam from both species with high efficiencies. For rpl32, primer pair 844/845 from Maheshwari and Barbash [57] was used. The standard curve method was used to estimate bam and rpl32 levels. Three technical replicates were performed from at least three biological replicates for each sample.
Quantitative PCR
To assay levels of Wolbachia, qPCR was performed on genomic DNA as in [44,111]. Females who eclosed on days 1–2 were aged on media supplemented with yeast for 2 days post-eclosion. DNA was isolated from 10 ovaries using phenol-chloroform extraction followed by 2 rounds of ethanol precipitation and rehydration in water.
For Wolbachia, primer pair wsp440F/wsp691R was used [111]. For rpl32, primer pair 844/845 was used. The standard curve method was used to estimate levels of each product. Three technical replicates were performed from at least three biological replicates for each sample.
Co-immunoprecipitation experiments
D. simulans bam was amplified from w501 ovarian cDNA using primers 662/661, cloned into pENTR/D-TOPO vector (Invitrogen), verified by sequencing, and recombined into destination vectors using LR-Clonase II (Invitrogen) following manufacturer’s directions. D. simulans bam was recombined into pAFHW containing both Flag and HA epitope tags (http://emb.carnegiescience.edu/labs/murphy/Gateway%20vectors.html). D. melanogaster bam in pAFHW and D. melanogaster bgcn in pAFMW were kindly provided by D. McKearin [33].
Combinations of pAFMW-Bam and pAFHW-Bgcn or empty vectors were co-transfected into Drosophila S2 cells, cells incubated for 3 days, and then lysed in lysis buffer (50mM Tris-HCl pH7.8, 150mM NaCl, 0.1%NP-40). Anti-HA (Roche, 3F10) or anti-Myc (Roche, 9E10) antibodies were conjugated to 50 μl of Protein G Dynabeads (Invitrogen) in 200ul of PBST (0.01% Tween 20) at 4°C overnight with rotation. Antibody-conjugated beads were then added to cell lysate (80μg total protein) in 200μl in lysis buffer containing 1X protease inhibitor (Roche) and 1mM PMSF and incubated at 4°C overnight. Washes were performed following manufacturer’s directions and Dynabeads were boiled in 1X SDS sample buffer to elute protein.
Western blotting
25–35 ovaries from females aged 2–3 days post-eclosion on media supplemented with yeast were homogenized in lysis buffer (50 mM Tris-HCl pH 7.5, 10 mM EDTA, 1.25% TritonX-100, 1X protease inhibitor, Roche) and centrifuged at 14000 rpm at 4°C for 5 minutes. Total protein in the supernatant was estimated using the Bradford assay (Biorad) and samples were boiled in an equal volume of 4X SDS sample buffer for 5 minutes. 10–20 μg were loaded on 10% SDS-PAGE gels. Primary antibodies were anti-GFP Jl-8 (Clontech, 1 : 2000) and mouse anti-tubulin T5168 (Sigma; 1 : 120,000). Secondary antibodies were HRP conjugated goat anti-mouse (Jackson; 1 : 1,000 for anti-GFP and 1 : 60,000 for anti-tubulin) and were detected with ECL Western blotting substrate (Pierce).
Immunostaining
Immunostaining was performed as in Aruna et al. [112]. Primary antibodies were: anti-GFP (Invitrogen A6544, 1 : 200), anti-vasa (DSHB, 1 : 25), anti-1B1 (DSHB, 1 : 4), monoclonal anti-Bam (1 : 100). Anti-Bam antibody was provided by D. McKearin. Secondary antibodies including goat anti-rat, anti-rabbit, or anti-mouse were conjugated with Alexa fluor dyes (Molecular Probes, 1 : 200–1 : 500). Samples were mounted in Vectashield containing DAPI (Vector Laboratories) and analyzed using the Leica SP2 confocal microscope at the Cornell University Core Life Sciences Microscopy and Imaging Facility. Images were resized in Photoshop (Adobe, version 11.0).
Supporting Information
Zdroje
1. Panhuis TM, Clark NL, Swanson WJ (2006) Rapid evolution of reproductive proteins in abalone and Drosophila. Philos Trans R Soc Lond B Biol Sci 361 : 261–268. 16612885
2. Swanson WJ, Vacquier VD (2002) The rapid evolution of reproductive proteins. Nat Rev Genet 3 : 137–144. 11836507
3. Turner LM, Hoekstra HE (2008) Causes and consequences of the evolution of reproductive proteins. Int J Dev Biol 52 : 769–780. doi: 10.1387/ijdb.082577lt 18649289
4. Haerty W, Jagadeeshan S, Kulathinal RJ, Wong A, Ravi Ram K, et al (2007) Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics 177 : 1321–1335. 18039869
5. Clark NL, Aagaard JE, Swanson WJ (2006) Evolution of reproductive proteins from animals and plants. Reproduction 131 : 11–22. 16388004
6. Swanson WJ, Wong A, Wolfner MF, Aquadro CF (2004) Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics 168 : 1457–1465. 15579698
7. Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh Y-P, et al (2007) Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol 5: e310. 17988176
8. Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, et al (2007) Evolution of genes and genomes on the Drosophila phylogeny. Nature 450 : 203–218. 17994087
9. Kosiol C, Vinar T, da Fonseca RR, Hubisz MJ, Bustamante CD, et al (2008) Patterns of positive selection in six Mammalian genomes. PLoS Genet 4: e1000144. doi: 10.1371/journal.pgen.1000144 18670650
10. Larracuente AM, Sackton TB, Greenberg AJ, Wong A, Singh ND, et al (2008) Evolution of protein-coding genes in Drosophila. Trends Genet 24 : 114–123. doi: 10.1016/j.tig.2007.12.001 18249460
11. Dean MD, Clark NL, Findlay GD, Karn RC, Yi X, et al (2009) Proteomics and comparative genomic investigations reveal heterogeneity in evolutionary rate of male reproductive proteins in mice (Mus domesticus). Mol Biol Evol 26 : 1733–1743. doi: 10.1093/molbev/msp094 19420050
12. Bauer DuMont VL, Flores HA, Wright MH, Aquadro CF (2007) Recurrent positive selection at bgcn, a key determinant of germ line differentiation, does not appear to be driven by simple coevolution with its partner protein bam. Mol Biol Evol 24 : 182–191. 17056645
13. Civetta A, Rajakumar SA, Brouwers B, Bacik JP (2006) Rapid evolution and gene-specific patterns of selection for three genes of spermatogenesis in Drosophila. Mol Biol Evol 23 : 655–662. 16357040
14. Langley CH, Stevens K, Cardeno C, Lee YCG, Schrider DR, et al (2012) Genomic variation in natural populations of Drosophila melanogaster. Genetics 192 : 533–598. doi: 10.1534/genetics.112.142018 22673804
15. Wong MD, Jin Z, Xie T (2005) Molecular mechanisms of germline stem cell regulation. Annu Rev Genet 39 : 173–195. 16285857
16. Xie T, Song X, Jin Z, Pan L, Weng C, et al (2008) Interactions between stem cells and their niche in the Drosophila ovary. Cold Spring Harb Symp Quant Biol 73 : 39–47. doi: 10.1101/sqb.2008.73.014 19022749
17. Chen D, McKearin D (2005) Gene circuitry controlling a stem cell niche. Curr Biol 15 : 179–184. 15668176
18. Fuller MT, Spradling AC (2007) Male and female Drosophila germline stem cells: two versions of immortality. Science 316 : 402–404. 17446390
19. Song X, Wong MD, Kawase E, Xi R, Ding BC, et al (2004) Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development 131 : 1353–1364. 14973291
20. Chen D, McKearin D (2003) Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol 13 : 1786–1791. 14561403
21. Chen D, McKearin DM (2003) A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130 : 1159–1170. 12571107
22. McKearin D, Ohlstein B (1995) A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development 121 : 2937–2947. 7555720
23. McKearin DM, Spradling AC (1990) bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev 4 : 2242–2251. 2279698
24. Gönczy P, Matunis E, DiNardo S (1997) bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development 124 : 4361–4371. 9334284
25. Insco ML, Leon A, Tam CH, McKearin DM, Fuller MT (2009) Accumulation of a differentiation regulator specifies transit amplifying division number in an adult stem cell lineage. Proc Natl Acad Sci U S A 106 : 22311–22316. doi: 10.1073/pnas.0912454106 20018708
26. de Cuevas M, Spradling AC (1998) Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development 125 : 2781–2789. 9655801
27. Lavoie CA, Ohlstein B, McKearin DM (1999) Localization and function of Bam protein require the benign gonial cell neoplasm gene product. Dev Biol 212 : 405–413. 10433830
28. Hawkins NC, Thorpe J, Schüpbach T (1996) Encore, a gene required for the regulation of germ line mitosis and oocyte differentiation during Drosophila oogenesis. Development 122 : 281–290. 8565840
29. Lilly MA, de Cuevas M, Spradling AC (2000) Cyclin A associates with the fusome during germline cyst formation in the Drosophila ovary. Dev Biol 218 : 53–63. 10644410
30. Insco ML, Bailey AS, Kim J, Olivares GH, Wapinski OL, et al (2012) A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell 11 : 689–700. doi: 10.1016/j.stem.2012.08.012 23122292
31. Ohlstein B, Lavoie CA, Vef O, Gateff E, McKearin DM (2000) The Drosophila cystoblast differentiation factor, benign gonial cell neoplasm, is related to DExH-box proteins and interacts genetically with bag-of-marbles. Genetics 155 : 1809–1819. 10924476
32. Shen R, Weng C, Yu J, Xie T (2009) eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc Natl Acad Sci U S A 106 : 11623–11628. doi: 10.1073/pnas.0903325106 19556547
33. Li Y, Minor NT, Park JK, McKearin DM, Maines JZ (2009) Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci U S A 106 : 9304–9309. doi: 10.1073/pnas.0901452106 19470484
34. Chau J, Kulnane LS, Salz HK (2009) Sex-lethal facilitates the transition from germline stem cell to committed daughter cell in the Drosophila ovary. Genetics 182 : 121–132. doi: 10.1534/genetics.109.100693 19237687
35. Chau J, Kulnane LS, Salz HK (2012) Sex-lethal enables germline stem cell differentiation by down-regulating Nanos protein levels during Drosophila oogenesis. Proc Natl Acad Sci U S A 109 : 9465–9470. doi: 10.1073/pnas.1120473109 22645327
36. Li Y, Zhang Q, Carreira-Rosario A, Maines JZ, McKearin DM, Buszczak M (2013) Mei-p26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS One 8: e58301. doi: 10.1371/journal.pone.0058301 23526974
37. Veneti Z, Bentley JK, Koana T, Braig HR, Hurst GDD (2005) A functional dosage compensation complex required for male killing in Drosophila. Science 307 : 1461–1463. 15746426
38. Hornett EA, Charlat S, Duplouy AMR, Davies N, Roderick GK, et al (2006) Evolution of male-killer suppression in a natural population. PLoS Biol 4: e283. 16933972
39. Werren JH, Baldo L, Clark ME (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6 : 741–751. doi: 10.1038/nrmicro1969 18794912
40. Serbus LR, Casper-Lindley C, Landmann F, Sullivan W (2008) The genetics and cell biology of Wolbachia-host interactions. Annu Rev Genet 42 : 683–707. doi: 10.1146/annurev.genet.41.110306.130354 18713031
41. Starr DJ, Cline TW (2002) A host parasite interaction rescues Drosophila oogenesis defects. Nature 418 : 76–79. 12097909
42. Sun S, Cline TW (2009) Effects of Wolbachia infection and ovarian tumor mutations on Sex-lethal germline functioning in Drosophila. Genetics 181 : 1291–1301. doi: 10.1534/genetics.108.099374 19171941
43. Boyle L, O'Neill SL, Robertson HM, Karr TL (1993) Interspecific and intraspecific horizontal transfer of Wolbachia in Drosophila. Science 260 : 1796–1799. 8511587
44. McGraw EA, Merritt DJ, Droller JN, O'Neill SL (2002) Wolbachia density and virulence attenuation after transfer into a novel host. Proc Natl Acad Sci U S A 99 : 2918–2923. 11880639
45. Serbus LR, Ferreccio A, Zhukova M, McMorris CL, Kiseleva E, Sullivan W (2011) A feedback loop between Wolbachia and the Drosophila gurken mRNP complex influences Wolbachia titer. J Cell Sci 124 : 4299–4308. doi: 10.1242/jcs.092510 22193955
46. Fast EM, Toomey ME, Panaram K, Desjardins D, Kolaczyk ED, Frydman HM (2011) Wolbachia enhance Drosophila stem cell proliferation and target the germline stem cell niche. Science 334 : 990–992. doi: 10.1126/science.1209609 22021671
47. Albertson R, Tan V, Leads RR, Reyes M, Sullivan W, Casper-Lindley C (2013) Mapping Wolbachia distributions in the adult Drosophila brain. Cell Microbiol 15 : 1527–1544. doi: 10.1111/cmi.12136 23490256
48. Toomey ME, Panaram K, Fast EM, Beatty C, Frydman HM (2013) Evolutionarily conserved Wolbachia-encoded factors control pattern of stem-cell niche tropism in Drosophila ovaries and favor infection. Proc Natl Acad Sci U S A 110 : 10788–10793. doi: 10.1073/pnas.1301524110 23744038
49. Frydman HM, Li JM, Robson DN, Wieschaus E (2006) Somatic stem cell niche tropism in Wolbachia. Nature 441 : 509–512. 16724067
50. Poinsot D, Bourtzis K, Markakis G, Savakis C, Merçot H (1998) Wolbachia transfer from Drosophila melanogaster into D. simulans: Host effect and cytoplasmic incompatibility relationships. Genetics 150 : 227–237. 9725842
51. Chen D, Wang Q, Huang H, Xia L, Jiang X, et al (2009) Effete-mediated degradation of Cyclin A is essential for the maintenance of germline stem cells in Drosophila. Development 136 : 4133–4142. doi: 10.1242/dev.039032 19906849
52. Xia L, Jia S, Huang S, Wang H, Zhu Y, et al (2010) The Fused/Smurf complex controls the fate of Drosophila germline stem cells by generating a gradient BMP response. Cell 143 : 978–990. doi: 10.1016/j.cell.2010.11.022 21145463
53. Kirilly D, Xie T (2007) The Drosophila ovary: an active stem cell community. Cell Res 17 : 15–25. 17199109
54. Yue L, Spradling AC (1992) hu-li tai shao, a gene required for ring canal formation during Drosophila oogenesis, encodes a homolog of adducin. Genes Dev 6 : 2443–2454. 1340461
55. de Cuevas M, Lee JK, Spradling AC (1996) alpha-spectrin is required for germline cell division and differentiation in the Drosophila ovary. Development 122 : 3959–3968. 9012516
56. Maheshwari S, Barbash DA (2011) The genetics of hybrid incompatibilities. Annu Rev Genet 45 : 331–355. doi: 10.1146/annurev-genet-110410-132514 21910629
57. Maheshwari S, Barbash DA (2012) Cis-by-Trans regulatory divergence causes the asymmetric lethal effects of an ancestral hybrid incompatibility gene. PLoS Genet 8: e1002597. doi: 10.1371/journal.pgen.1002597 22457639
58. Pan L, Wang S, Lu T, Weng C, Song X, et al (2014) Protein competition switches the function of COP9 from self-renewal to differentiation. Nature 514 : 233–236. doi: 10.1038/nature13562 25119050
59. Ferree PM, Frydman HM, Li JM, Cao J, Wieschaus E, Sullivan W (2005) Wolbachia utilizes host microtubules and Dynein for anterior localization in the Drosophila oocyte. PLoS Pathog 1: e14. 16228015
60. Serbus LR, Sullivan W (2007) A cellular basis for Wolbachia recruitment to the host germline. PLoS Pathog 3: e190. 18085821
61. Groth AC, Fish M, Nusse R, Calos MP (2004) Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166 : 1775–1782. 15126397
62. Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL (2005) Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171 : 1813–1822. 16143608
63. Takahasi KR, Matsuo T, Takano-Shimizu-Kouno T (2011) Two types of cis-trans compensation in the evolution of transcriptional regulation. Proc Natl Acad Sci U S A 108 : 15276–15281. doi: 10.1073/pnas.1105814108 21876147
64. Masly JP, Presgraves DC (2007) High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol 5: e243. 17850182
65. Tao Y, Chen S, Hartl DL, Laurie CC (2003) Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics 164 : 1383–1397. 12930747
66. Hollocher H, Wu CI (1996) The genetics of reproductive isolation in the Drosophila simulans clade: X vs. autosomal effects and male vs. female effects. Genetics 143 : 1243–1255. 8807297
67. Llopart A (2012) The rapid evolution of X-linked male-biased gene expression and the large-X effect in Drosophila yakuba, D. santomea, and their hybrids. Mol Biol Evol 29 : 3873–3886. doi: 10.1093/molbev/mss190 22844069
68. Meiklejohn CD, Parsch J, Ranz JM, Hartl DL (2003) Rapid evolution of male-biased gene expression in Drosophila. Proc Natl Acad Sci U S A 100 : 9894–9899. 12907700
69. Wong A, Turchin MC, Wolfner MF, Aquadro CF (2008) Evidence for positive selection on Drosophila melanogaster seminal fluid protease homologs. Mol Biol Evol 25 : 497–506. 18056920
70. Begun DJ, Whitley P, Todd BL, Waldrip-Dail HM, Clark AG (2000) Molecular population genetics of male accessory gland proteins in Drosophila. Genetics 156 : 1879–1888. 11102381
71. Ohlstein B, McKearin D (1997) Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development 124 : 3651–3662. 9342057
72. Schulz C, Kiger AA, Tazuke SI, Yamashita YM, Pantalena-Filho LC, et al (2004) A misexpression screen reveals effects of bag-of-marbles and TGF beta class signaling on the Drosophila male germ-line stem cell lineage. Genetics 167 : 707–723. 15238523
73. Kawase E, Wong MD, Ding BC, Xie T (2004) Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development 131 : 1365–1375. 14973292
74. Sheng XR, Brawley CM, Matunis EL (2009) Dedifferentiating spermatogonia outcompete somatic stem cells for niche occupancy in the Drosophila testis. Cell Stem Cell 5 : 191–203. doi: 10.1016/j.stem.2009.05.024 19664993
75. Jin Z, Kirilly D, Weng C, Kawase E, Song X, et al (2008) Differentiation-defective stem cells outcompete normal stem cells for niche occupancy in the Drosophila ovary. Cell Stem Cell 2 : 39–49. doi: 10.1016/j.stem.2007.10.021 18371420
76. León A, McKearin D (1999) Identification of TER94, an AAA ATPase protein, as a Bam-dependent component of the Drosophila fusome. Mol Biol Cell 10 : 3825–3834. 10564274
77. Lighthouse DV, Buszczak M, Spradling AC (2008) New components of the Drosophila fusome suggest it plays novel roles in signaling and transport. Dev Biol 317 : 59–71. doi: 10.1016/j.ydbio.2008.02.009 18355804
78. Salz HK (2013) Sex, stem cells and tumors in the Drosophila ovary. Fly (Austin) 7 : 3–7.
79. Choi JY, Aquadro CF (2014) The coevolutionary period of Wolbachia pipientis infecting Drosophila ananassae and its impact on the evolution of the host germline stem cell regulating genes. Mol Biol Evol 31 : 2457–2471. doi: 10.1093/molbev/msu204 24974378
80. Ellegaard KM, Klasson L, Näslund K, Bourtzis K, Andersson SGE (2013) Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet 9: e1003381. doi: 10.1371/journal.pgen.1003381 23593012
81. Richardson MF, Weinert LA, Welch JJ, Linheiro RS, Magwire MM, et al (2012) Population genomics of the Wolbachia endosymbiont in Drosophila melanogaster. PLoS Genet 8: e1003129. doi: 10.1371/journal.pgen.1003129 23284297
82. Early AM, Clark AG (2013) Monophyly of Wolbachia pipientis genomes within Drosophila melanogaster: geographic structuring, titre variation and host effects across five populations. Mol Ecol 22 : 5765–5778. doi: 10.1111/mec.12530 24118111
83. Teixeira L, Ferreira A, Ashburner M (2008) The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol 6: e2.
84. Hedges LM, Brownlie JC, O'Neill SL, Johnson KN (2008) Wolbachia and virus protection in insects. Science 322 : 702. doi: 10.1126/science.1162418 18974344
85. Osborne SE, Iturbe-Ormaetxe I, Brownlie JC, O'Neill SL, Johnson KN (2012) Antiviral protection and the importance of Wolbachia density and tissue tropism in Drosophila simulans. Appl Environ Microbiol 78 : 6922–6929. doi: 10.1128/AEM.01727-12 22843518
86. Martinez J, Longdon B, Bauer S, Chan Y-S, Miller WJ, et al (2014) Symbionts commonly provide broad spectrum resistance to viruses in insects: a comparative analysis of Wolbachia strains. PLoS Pathog 10: e1004369. doi: 10.1371/journal.ppat.1004369 25233341
87. Riegler M, Sidhu M, Miller WJ, O'Neill SL (2005) Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr Biol 15 : 1428–1433. 16085497
88. Kriesner P, Hoffmann AA, Lee SF, Turelli M, Weeks AR (2013) Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog 9: e1003607. doi: 10.1371/journal.ppat.1003607 24068927
89. Chrostek E, Marialva MSP, Esteves SS, Weinert LA, Martinez J, et al (2013) Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet 9: e1003896. doi: 10.1371/journal.pgen.1003896 24348259
90. Dedeine F, Vavre F, Fleury F, Loppin B, Hochberg ME, Bouletreau M (2001) Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc Natl Acad Sci U S A 98 : 6247–6252. 11353833
91. Dedeine F, Boulétreau M, Vavre F (2005) Wolbachia requirement for oogenesis: occurrence within the genus Asobara (Hymenoptera, Braconidae) and evidence for intraspecific variation in A. tabida. Heredity 95 : 394–400. 16118660
92. Pannebakker BA, Loppin B, Elemans CPH, Humblot L, Vavre F (2007) Parasitic inhibition of cell death facilitates symbiosis. Proc Natl Acad Sci U S A 104 : 213–215. 17190825
93. Brennan LJ, Keddie BA, Braig HR, Harris HL (2008) The endosymbiont Wolbachia pipientis induces the expression of host antioxidant proteins in an Aedes albopictus cell line. PLoS One 3: e2083. doi: 10.1371/journal.pone.0002083 18461124
94. Xi Z, Gavotte L, Xie Y, Dobson SL (2008) Genome-wide analysis of the interaction between the endosymbiotic bacterium Wolbachia and its Drosophila host. BMC Genomics 9 : 1. doi: 10.1186/1471-2164-9-1 18171476
95. Kremer N, Voronin D, Charif D, Mavingui P, Mollereau B, Vavre F (2009) Wolbachia interferes with ferritin expression and iron metabolism in insects. PLoS Pathog 5: e1000630. doi: 10.1371/journal.ppat.1000630 19851452
96. Kremer N, Charif D, Henri H, Gavory F, Wincker P, et al (2012) Influence of Wolbachia on host gene expression in an obligatory symbiosis. BMC Microbiol 12 Suppl 1: S7. doi: 10.1186/1471-2180-12-S1-S7 22376153
97. Pan L, Chen S, Weng C, Call G, Zhu D, et al (2007) Stem cell aging is controlled both intrinsically and extrinsically in the Drosophila ovary. Cell Stem Cell 1 : 458–469. doi: 10.1016/j.stem.2007.09.010 18371381
98. LaFever L, Drummond-Barbosa D (2005) Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science 309 : 1071–1073. 16099985
99. Hsu H-J, Drummond-Barbosa D (2009) Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc Natl Acad Sci U S A 106 : 1117–1121. doi: 10.1073/pnas.0809144106 19136634
100. Drummond-Barbosa D (2008) Stem cells, their niches and the systemic environment: an aging network. Genetics 180 : 1787–1797. doi: 10.1534/genetics.108.098244 19087970
101. Boyle M, Wong C, Rocha M, Jones DL (2007) Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell 1 : 470–478. doi: 10.1016/j.stem.2007.08.002 18371382
102. Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, et al (2004) Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol 2: E69. 15024419
103. Klasson L, Westberg J, Sapountzis P, Näslund K, Lutnaes Y, et al (2009) The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc Natl Acad Sci U S A 106 : 5725–5730. doi: 10.1073/pnas.0810753106 19307581
104. Caturegli P, Asanovich KM, Walls JJ, Bakken JS, Madigan JE, et al (2000) ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect Immun 68 : 5277–5283. 10948155
105. Flores HA, Bauer DuMont VL, Fatoo A, Hubbard D, Hijji M, et al (2015) Adaptive Evolution of Genes Involved in the Regulation of Germline Stem Cells in Drosophila melanogaster and Drosophila simulans. G3 (Bethesda).
106. St Pierre SE, Ponting L, Stefancsik R, McQuilton P, FlyBase Consortium (2014) FlyBase 102—advanced approaches to interrogating FlyBase. Nucleic Acids Res 42: D780–D788. doi: 10.1093/nar/gkt1092 24234449
107. Zhou W, Rousset F, O'Neil S (1998) Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc Biol Sci 265 : 509–515. 9569669
108. Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA (2006) Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314 : 1292–1295. 17124320
109. Venken KJT, He Y, Hoskins RA, Bellen HJ (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314 : 1747–1751. 17138868
110. Newell PD, Douglas AE (2014) Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl Environ Microbiol 80 : 788–796. doi: 10.1128/AEM.02742-13 24242251
111. McGraw EA, Merritt DJ, Droller JN, O'Neill SL (2001) Wolbachia-mediated sperm modification is dependent on the host genotype in Drosophila. Proc Biol Sci 268 : 2565–2570. 11749711
112. Aruna S, Flores HA, Barbash DA (2009) Reduced fertility of Drosophila melanogaster hybrid male rescue (Hmr) mutant females is partially complemented by Hmr orthologs from sibling species. Genetics 181 : 1437–1450. doi: 10.1534/genetics.108.100057 19153254
Štítky
Genetika Reprodukčná medicína
Článek Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer PopulationČlánek Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2Článek Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA ReplicationČlánek SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from DegradationČlánek Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors inČlánek The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 Regulation
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 8- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Putting the Brakes on Huntington Disease in a Mouse Experimental Model
- Identification of Driving Fusion Genes and Genomic Landscape of Medullary Thyroid Cancer
- Evidence for Retromutagenesis as a Mechanism for Adaptive Mutation in
- TSPO, a Mitochondrial Outer Membrane Protein, Controls Ethanol-Related Behaviors in
- Evidence for Lysosome Depletion and Impaired Autophagic Clearance in Hereditary Spastic Paraplegia Type SPG11
- Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer Population
- Trans-Reactivation: A New Epigenetic Phenomenon Underlying Transcriptional Reactivation of Silenced Genes
- Early Developmental and Evolutionary Origins of Gene Body DNA Methylation Patterns in Mammalian Placentas
- Strong Selective Sweeps on the X Chromosome in the Human-Chimpanzee Ancestor Explain Its Low Divergence
- Dominance of Deleterious Alleles Controls the Response to a Population Bottleneck
- Transient 1a Induction Defines the Wound Epidermis during Zebrafish Fin Regeneration
- Systems Genetics Reveals the Functional Context of PCOS Loci and Identifies Genetic and Molecular Mechanisms of Disease Heterogeneity
- A Genome Scale Screen for Mutants with Delayed Exit from Mitosis: Ire1-Independent Induction of Autophagy Integrates ER Homeostasis into Mitotic Lifespan
- Non-synonymous FGD3 Variant as Positional Candidate for Disproportional Tall Stature Accounting for a Carcass Weight QTL () and Skeletal Dysplasia in Japanese Black Cattle
- The Relationship between Gene Network Structure and Expression Variation among Individuals and Species
- Calmodulin Methyltransferase Is Required for Growth, Muscle Strength, Somatosensory Development and Brain Function
- The Wnt Frizzled Receptor MOM-5 Regulates the UNC-5 Netrin Receptor through Small GTPase-Dependent Signaling to Determine the Polarity of Migrating Cells
- Nbs1 ChIP-Seq Identifies Off-Target DNA Double-Strand Breaks Induced by AID in Activated Splenic B Cells
- CCNYL1, but Not CCNY, Cooperates with CDK16 to Regulate Spermatogenesis in Mouse
- Evidence for a Common Origin of Blacksmiths and Cultivators in the Ethiopian Ari within the Last 4500 Years: Lessons for Clustering-Based Inference
- Of Fighting Flies, Mice, and Men: Are Some of the Molecular and Neuronal Mechanisms of Aggression Universal in the Animal Kingdom?
- Hypoxia and Temperature Regulated Morphogenesis in
- The Homeodomain Iroquois Proteins Control Cell Cycle Progression and Regulate the Size of Developmental Fields
- Evolution and Design Governing Signal Precision and Amplification in a Bacterial Chemosensory Pathway
- Rac1 Regulates Endometrial Secretory Function to Control Placental Development
- Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2
- Functions as a Positive Regulator of Growth and Metabolism in
- The Nucleosome Acidic Patch Regulates the H2B K123 Monoubiquitylation Cascade and Transcription Elongation in
- Rhoptry Proteins ROP5 and ROP18 Are Major Murine Virulence Factors in Genetically Divergent South American Strains of
- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- Regulates Refractive Error and Myopia Development in Mice and Humans
- mTORC1 Prevents Preosteoblast Differentiation through the Notch Signaling Pathway
- Regulation of Gene Expression Patterns in Mosquito Reproduction
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
- The Spalt Transcription Factors Generate the Transcriptional Landscape of the Wing Pouch Central Region
- Binding of Multiple Rap1 Proteins Stimulates Chromosome Breakage Induction during DNA Replication
- Functional Divergence in the Role of N-Linked Glycosylation in Smoothened Signaling
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- Coordinated Evolution of Influenza A Surface Proteins
- The Evolutionary Potential of Phenotypic Mutations
- Genome-Wide Association and Trans-ethnic Meta-Analysis for Advanced Diabetic Kidney Disease: Family Investigation of Nephropathy and Diabetes (FIND)
- New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation
- SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from Degradation
- Satellite DNA Modulates Gene Expression in the Beetle after Heat Stress
- SHOEBOX Modulates Root Meristem Size in Rice through Dose-Dependent Effects of Gibberellins on Cell Elongation and Proliferation
- Reduced Crossover Interference and Increased ZMM-Independent Recombination in the Absence of Tel1/ATM
- Suppression of Somatic Expansion Delays the Onset of Pathophysiology in a Mouse Model of Huntington’s Disease
- Protein Composition of Infectious Spores Reveals Novel Sexual Development and Germination Factors in
- The Evolutionarily Conserved LIM Homeodomain Protein LIM-4/LHX6 Specifies the Terminal Identity of a Cholinergic and Peptidergic . Sensory/Inter/Motor Neuron-Type
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of .
- Retinoic Acid Signaling Regulates Differential Expression of the Tandemly-Duplicated Long Wavelength-Sensitive Cone Opsin Genes in Zebrafish
- The Formin Diaphanous Regulates Myoblast Fusion through Actin Polymerization and Arp2/3 Regulation
- Genome-Wide Analysis of PAPS1-Dependent Polyadenylation Identifies Novel Roles for Functionally Specialized Poly(A) Polymerases in
- Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration
- Regulation of Mutagenic DNA Polymerase V Activation in Space and Time
- Variability of Gene Expression Identifies Transcriptional Regulators of Early Human Embryonic Development
- The Drosophila Gene Interacts Genetically with and Shows Female-Specific Effects of Divergence
- Functional Activation of the Flagellar Type III Secretion Export Apparatus
- Retrohoming of a Mobile Group II Intron in Human Cells Suggests How Eukaryotes Limit Group II Intron Proliferation
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome
- YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers
- SmD1 Modulates the miRNA Pathway Independently of Its Pre-mRNA Splicing Function
- Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



