-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Gene Regulatory Program for Meiotic Prophase in the Fetal Ovary
The formation of haploid gametes from diploid germ cells requires a specialized reductive cell division known as meiosis. This reductive division is enabled by chromosomal events that occur during meiotic prophase, including synapsis and crossing-over of homologous chromosomes. These chromosomal events involve meiosis-specific genes that must be expressed before they act during meiosis. Using gene expression profiling, we identified a set of mammalian meiosis-specific genes. To understand how expression of these genes is controlled, we examined their expression in the absence of known regulators of the chromosomal events: 1) retinoic acid (RA), which induces meiosis, 2) Dazl, which is required for germ cell competence to respond to RA, and 3) Stra8, which is induced by RA and is required for the chromosomal events of meiotic prophase. We uncover two key features of gene regulation. First, while the genes require RA and Dazl to be expressed, they vary in their dependence on Stra8, thus creating a regulatory hierarchy. Genes induced independently of Stra8, and thus early in this hierarchy, may encode proteins that are stockpiled in anticipation of the chromosomal events. Second, Stra8 induces its own down-regulation, and may thus prevent repeated induction of meiosis in a single germ cell.
Published in the journal: A Gene Regulatory Program for Meiotic Prophase in the Fetal Ovary. PLoS Genet 11(9): e32767. doi:10.1371/journal.pgen.1005531
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005531Summary
The formation of haploid gametes from diploid germ cells requires a specialized reductive cell division known as meiosis. This reductive division is enabled by chromosomal events that occur during meiotic prophase, including synapsis and crossing-over of homologous chromosomes. These chromosomal events involve meiosis-specific genes that must be expressed before they act during meiosis. Using gene expression profiling, we identified a set of mammalian meiosis-specific genes. To understand how expression of these genes is controlled, we examined their expression in the absence of known regulators of the chromosomal events: 1) retinoic acid (RA), which induces meiosis, 2) Dazl, which is required for germ cell competence to respond to RA, and 3) Stra8, which is induced by RA and is required for the chromosomal events of meiotic prophase. We uncover two key features of gene regulation. First, while the genes require RA and Dazl to be expressed, they vary in their dependence on Stra8, thus creating a regulatory hierarchy. Genes induced independently of Stra8, and thus early in this hierarchy, may encode proteins that are stockpiled in anticipation of the chromosomal events. Second, Stra8 induces its own down-regulation, and may thus prevent repeated induction of meiosis in a single germ cell.
Introduction
In sexually reproducing organisms, germ cells undergo meiosis, a specialized cell division program that produces haploid gametes. The reductive segregation of chromosomes depends upon a complex series of chromosomal events that occur during meiotic prophase. This chromosomal program must be supported by expression of a large suite of genes. A genome-wide description of this gene expression program, and how it is regulated, has not been available for mammals or other animals with specialized sex cells, or germ cells. Indeed, the best existing model for such a gene regulatory program is that of budding yeast [1–4].
The chromosomal program of meiotic prophase, including events such as laying down of meiotic cohesins, synapsis between homologs, and homologous recombination, has been the subject of intense study [5–7]. Investigations of these processes in mammals have relied principally upon identifying mouse orthologs of proteins that have demonstrated meiotic functions in lower eukaryotes, and that are well conserved among sexually reproducing species [8,9]. However, not all proteins involved in the meiotic chromosomal processes are well conserved among eukaryotes, and identifying these exceptions has proven challenging [10]. Identification of a gene set specific to mammalian meiotic prophase would provide an orthogonal means of discovering poorly conserved or even novel proteins involved in the chromosomal program of mammalian meiotic prophase.
Studies in a mammalian system are also required if we are to understand how the gene expression program of mammalian meiotic prophase is regulated; the regulation of meiotic initiation is poorly conserved. For instance, between mouse and budding yeast, the regulatory logic of meiotic initiation appears similar, but the molecular identities of the regulators are not conserved [11]. In both mouse ovarian and testicular germ cells, meiosis is initiated by retinoic acid (RA) [12–14], a signaling molecule restricted to chordates [15]. RA induces Stra8, a vertebrate-specific gene that encodes a putative helix-loop-helix-containing transcription factor [13,14,16–20]. Stra8 is required for all chromosomal events of meiotic prophase assayed, including cohesion, synapsis, and recombination, as well as the preceding meiotic DNA replication [12,20]. In mouse fetal ovarian germ cells, induction of Stra8 by RA requires the germ-cell-expressed competence factor Dazl [21]. Dazl, which encodes an RNA binding protein expressed in postmigratory XX and XY germ cells, is required for germ cells to gain competence to respond to developmental cues, including RA [22]. Thus far, the roles of RA, Stra8, and Dazl have largely been assayed with respect to the chromosomal program of meiotic prophase; their potential roles in regulating the gene expression program have not been examined systematically.
We sought to elucidate the gene regulatory program of meiotic prophase. We used the mouse fetal ovary as a model for two reasons. First, germ cells in the fetal ovary initiate and progress through meiotic prophase with greater synchrony than in the postnatal or adult testis. All germ cells in the fetal ovary initiate meiosis around embryonic day 13.5, progress through meiotic prophase during subsequent fetal development, and arrest at diplotene of meiotic prophase before birth. Initiation and progression of meiotic prophase occurs in an anterior-to-posterior wave: expression of Stra8 and meiotic prophase genes begins in the anterior portion of the fetal ovary before extending towards the posterior [23,24]. We therefore took advantage of the relative synchrony of cell state over time and space to finely dissect initiation and progression of meiotic prophase. Second, the roles of Dazl, RA, and Stra8 in meiotic initiation are well defined in the fetal ovary. To determine how Dazl, RA, and Stra8 regulate the gene expression program, we profiled expression in wild-type and mutant animals. We used whole-gonad, genome-wide transcriptome profiling, to obtain a global description of gene expression, and followed up with targeted single-cell, single-transcript measurements to precisely quantify elements of regulatory control at the level of individual germ cells.
We identified a set of 104 genes specific to meiotic prophase, as assayed in fetal ovarian germ cells. We characterized how Dazl, RA, and Stra8 regulate this gene expression program, thus complementing our previous understanding of how they regulate the chromosomal program. From these data, we discerned two elements of gene regulatory logic centered on Stra8, a key inducer of the chromosomal program. Initial induction of genes requires Stra8-independent and Stra8-dependent pathways. After gene induction, Stra8 is required for subsequent down-regulation of its own expression. We propose that these elements of gene regulatory logic account for how germ cells prepare for and ensure a single induction of the chromosomal program of meiotic prophase.
Results
Identification of 104 genes specific to meiotic prophase
To identify and catalog the gene expression program of meiotic prophase as it occurs in the fetal ovary, we performed genome-wide transcriptome profiling by RNA-seq on whole fetal ovaries at embryonic days 12.5, 14.5, and 16.5 (E12.5, E14.5, and E16.5). At these time points, ovarian germ cells are in pre-meiotic, leptotene (early meiotic prophase), and pachytene (mid-late meiotic prophase) stages, respectively [25,26]. At each time point, we assayed expression in gonads of wild-type and germ-cell-depleted (KitW/KitWv) mice [27], in order to identify germ-cell-dependent genes. During this embryonic period, testicular germ cells do not initiate meiosis, but instead enter and remain in mitotic G0/G1 arrest [28]. We therefore also profiled expression from wild-type fetal testes, at the same time points, to identify ovary-enriched genes. We defined “meiotic prophase genes” as those meeting the criteria summarized in Fig 1A.
Fig. 1. RNA-seq analysis of wild-type and germ-cell-depleted fetal gonads identifies a meiotic prophase-specific gene set. 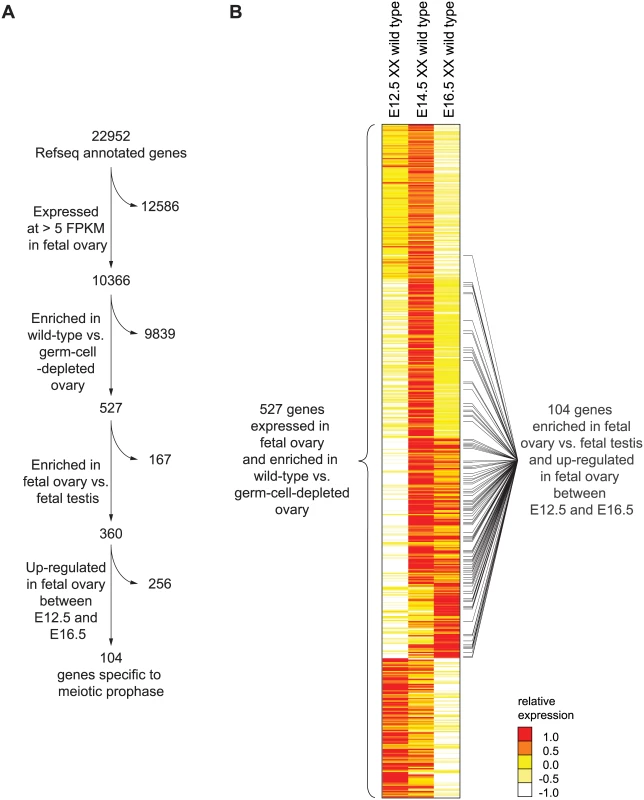
(A) Four filters were used to identify a set of genes expressed specifically in meiotic prophase. First, genes were required to be expressed in wild-type fetal ovary (E12.5, E14.5, or E16.5) at >5 FPKM; a total of 10,366 genes met this criterion. Of these 10,366 genes, 526 were also expressed more highly in wild-type than in germ-cell-depleted fetal ovary (KitW/KitWv) (fold-change > 2, FDR adjusted p value, q <0.01 at either E12.5, E14.5, or E16.5). As explained in the text, Rec8 was also included, for a total of 527 genes. Of these 527 genes, 360 were expressed more highly in fetal ovary than in fetal testis (fold-change > 2, q < 0.01). Of these 360 genes, 104 were up-regulated between E12.5 and E14.5 or E16.5 in the ovary (fold-change > 2, q < 0.01). (B) Relative expression of 527 ovarian germ-cell-enriched genes in E12.5, E14.5, and E16.5 wild-type (Kit+/Kit+) ovary. Gene expression is represented as log transformed and mean centered FPKM. Genes (rows) are organized by k-means clustering (k = 5). Black bars to right of gene-expression heat map represent the 104 genes that were both up-regulated between E12.5 and E14.5 or E16.5 in the ovary, and expressed more highly in fetal ovary than in fetal testis. Source data for the 527 genes, as well as all other Refseq genes, are provided in S1 Table. Since genes involved in meiotic prophase should be expressed leading up to or during prophase, we required that genes be expressed in wild-type ovaries at one or more of the three time points (E12.5, E14.5, or E16.5) at greater than 5 Fragments Per Kilobase of transcript per Million mapped reads (FPKM). We found 10,366 genes to be expressed at this level in the fetal ovary (Fig 1A). Additionally, since only germ cells might be expected to express genes required for meiotic prophase, we also required that gene expression be at least 2-fold higher in wild-type than in germ-cell-depleted ovaries. A total of 526 genes met both criteria (Fig 1A, S1 Table). There was one conspicuous absence: Rec8, a meiotic cohesin, was not germ-cell-enriched. We verified by single-molecule fluorescent in situ hybridization that Rec8 was indeed expressed in ovarian somatic cells as well as germ cells (S1 Fig), and added it to the 526 genes.
Using k-means clustering, we identified several distinct gene expression profiles (Fig 1B). Of the 527 genes, about 46%, including Stra8, were up-regulated between E12.5 and E14.5, then down-regulated by E16.5, suggestive of functions restricted to early meiotic prophase. About 22% of the 527 genes were up-regulated between E12.5 and E14.5 and remained elevated at E16.5; these genes include Sycp3, which encodes a synaptonemal complex protein. About 11% of genes were not up-regulated until E16.5, suggestive of functions later in meiotic prophase. The remaining 21% of the 527 genes were highly expressed at E12.5 and progressively down-regulated by E16.5. These include many pluripotency markers, including Pou5f1 (Oct4), Nanog, and Sox2, and reflect the down-regulation of a pluripotency program as germ cells enter meiosis [29–31].
To winnow this list of 527 ovarian germ cell genes down to those functioning in meiotic prophase, we required two additional criteria (Fig 1A). Since testicular germ cells do not embark on meiosis until well after birth, we required that gene expression be at least 2-fold higher in fetal ovary than in fetal testis; 360 genes met this additional criterion. Since genes with meiotic functions should be up-regulated as germ cells enter and progress through meiosis, we also required that genes be at least 2-fold up-regulated between E12.5 and E14.5 or E16.5; 104 genes satisfied this as well as the earlier criteria. We refer to this final set of 104 genes as the gene expression program of meiotic prophase.
Of these 104 genes, 54 have previously been implicated in meiotic prophase by independent, lower-throughput methods. For 33 of these 104 genes, loss-of-function mutants have been examined for fertility defects; defects in meiotic prophase or fertility were reported for 32 of the 33 genes tested in this manner (S1 Text). For 21 of the remaining 71 genes, detailed descriptions of RNA or protein expression patterns are publicly available, and all are consistent with functions in meiotic prophase. Thus, among 104 genes implicated in meiotic prophase through our systematic whole-genome RNA-seq analysis, 53 (of 54 genes tested) are substantiated by prior studies.
These findings suggest that many of the remaining 50 (of 104) genes are novel and uncharacterized genes involved in meiotic prophase, representing a great opportunity for future study. Review of the published literature indicates that our RNA-seq analysis captured most meiotic prophase genes that are expressed specifically in meiotic germ cells. Of 21 genes for which mutant germ cells have been reported to arrest at leptotene, zygotene, or pachytene stages of meiotic prophase (as cataloged by Handel and Schimenti, 2010), 14 are represented in our list of 104 genes. The seven genes with meiotic prophase arrest phenotypes that we failed to identify by RNA-seq analysis are either ubiquitously expressed (such as Cyclin-dependent kinase 2, Cdk2) or are expressed in both ovarian and testicular germ cells (such as Piwi-like RNA-mediated gene silencing 2, Piwil2). The design of our study would preclude our identifying genes that are expressed in both ovarian germ cells and somatic cells, or that are expressed at substantial levels in fetal testes.
Meiotic prophase gene expression is wholly dependent on Dazl, but ranges from completely Stra8-dependent to Stra8-independent
We next sought to determine how the meiotic prophase genes are activated. Stra8 was previously shown to be required for meiotic initiation, as primarily assayed by the meiotic chromosomal program. Dazl and RA are germ-cell-intrinsic and -extrinsic factors, respectively, required for induction of Stra8 and initiation of the chromosomal program. The roles of these factors in regulating the program of gene expression are largely unknown. We first determined whether Dazl and Stra8 regulate meiotic prophase genes by examining gene expression by RNA-seq in whole E14.5 Dazl-deficient (Dazl-/-) and Stra8-deficient (Stra8 -/-) ovaries as compared to corresponding homozygous wild-type controls (Fig 2, S2 Table).
Fig. 2. RNA-seq analysis of Dazl and Stra8-deficient fetal gonads reveals Stra8-independent regulation of meiotic prophase genes. 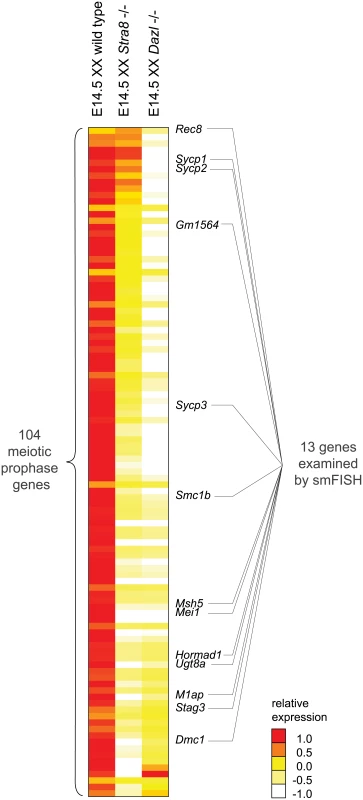
Relative expression of 104 meiotic prophase-specific genes in E14.5 wild-type, Stra8-deficient, and Dazl-deficient ovary. Gene expression was measured by RNA-seq and represented as log transformed and mean centered FPKM. Genes (rows) are arranged from least to most down-regulated in the Stra8-deficient ovary relative to the Dazl-deficient ovary, with the exception of the bottom four genes which were not significantly down-regulated in the Dazl-deficient ovary (q > 0.05), and were expressed at < 5 FPKM in wild-type, Dazl-deficient, and Stra8-deficient ovaries. Thirteen genes, listed to the right of the gene expression heat map, were selected for subsequent smFISH analysis. Source data for the 104 meiotic prophase genes, as well as all Refseq annotated genes, is provided in S2 Table. Dazl is required for germ cells to acquire competence to respond to RA. Dazl-expressing germ cells respond to RA by expressing Stra8 and initiating meiosis. Dazl is also more broadly required for the processes of gametogenesis, which encompass meiosis, the sex-specific cellular differentiation events of oogenesis and spermatogenesis, and the down-regulation of pluripotency markers [21,22]. Given Dazl’s broad role in competence for gametogenesis, we predicted that Dazl would be required for induction of the meiotic prophase gene expression program. Indeed, we found that expression of practically all meiotic prophase genes (100 of 104) was significantly diminished if not eliminated in Dazl-deficient ovaries (Fig 2, S2 Table). The remaining four genes were expressed at < 5 FPKM in both wild-type and Dazl-deficient ovaries.
Stra8 is required for the chromosomal program of meiotic prophase, including loading of meiotic cohesins, such as REC8, and assembly of the synaptonemal complex proteins, including SYCP3. However, although the REC8 and SYCP3 proteins do not localize to chromosomal axes in Stra8-deficient germ cells, the proteins are nevertheless produced [20]. In fact, Rec8 expression can be induced in testicular germ cells by RA in the absence of Stra8 function [32]. These results suggest that while Stra8 might regulate the entirety of the chromosomal program, it might have a more limited role in governing the gene expression program. We aimed to clarify the extent to which Stra8 regulates the meiotic gene expression program.
We found that expression of the 100 Dazl-dependent genes ranged across a wide spectrum of Stra8-dependency. For slightly over half of the 100 genes, including Dmc1, which is required to repair meiotic double-strand breaks, expression appeared to be fully dependent on Stra8. Expression of these genes was reduced in Stra8-deficient ovaries to levels as low as in the Dazl-deficient ovary (Fig 2, S2 Table). Expression of the remaining genes appeared to be partially dependent on, or in a few cases, largely independent of Stra8. Some genes, such as Sycp3, were expressed at lower levels in Stra8-deficient ovaries than in wild-type ovaries, but still at higher levels than in Dazl-deficient ovaries. At the Stra8-independent extreme of the spectrum is Rec8, whose levels were not only undiminished in Stra8-deficient ovaries, but in fact were modestly increased.
Thus, RNA-seq analyses of whole Dazl-deficient and Stra8-deficient ovaries suggest a model of gene induction whereby Dazl is required for induction of the meiotic prophase gene expression program via at least two pathways: a Stra8-independent pathway, and a Stra8-dependent pathway.
Stra8-independent and Stra8-dependent pathways act additively in individual cells
Whole-gonad RNA-seq analysis provides genome-wide breadth in characterizing the gene expression program of meiotic prophase. However, because this method averages across a population that includes a diversity of both germ cells and somatic cells, our observations may not accurately reflect events in individual germ cells. Specifically, we wondered whether our observation that some genes appeared partially Stra8-independent by RNA-seq actually reflected a partial reduction in gene expression in all Stra8-deficient cells. If so, this would indicate that Stra8-dependent and Stra8-independent pathways act additively in individual germ cells. Alternatively, our RNA-seq observation could be explained by a subset of Stra8-deficient germ cells retaining wild-type levels of gene expression, with other Stra8-deficient germ cells having greatly reduced levels of gene expression.
Distinguishing between these two scenarios required measurement of gene expression with single-cell resolution. We used single molecule fluorescence in situ hybridization (smFISH) to quantify gene expression in single cells in situ. smFISH involves multiple short fluorescently-labeled oligonucleotide probes that collectively bind along the same target transcript to detect and localize each target mRNA molecule as a punctate signal [33] (Fig 3A). These punctate signals can be quantified to determine the number of transcripts per cell volume (transcript density) (Fig 3B). We selected 13 genes, spanning a spectrum of Stra8-dependencies as measured by RNAseq in E14.5 ovaries (Fig 2), for examination by smFISH in germ cells of E14.5 wild-type, Stra8-deficient, and Dazl-deficient ovaries. Selected genes include the meiotic-specific cohesins Rec8, Smc1b, and Stag3; the synaptonemal complex proteins Sycp1, Sycp2, and Sycp3; Dmc1 and Msh5, which are involved in double-strand break repair; and Hormad1, which promotes homolog alignment and synaptonemal complex formation. We also included Mei1 and M1ap, which exhibit defects in meiotic prophase when mutated, and Gm1564 and Ugt8a, which are presently uncharacterized.
Fig. 3. Single molecule FISH analysis corroborates three classes of gene regulation at the level of individual germ cells. 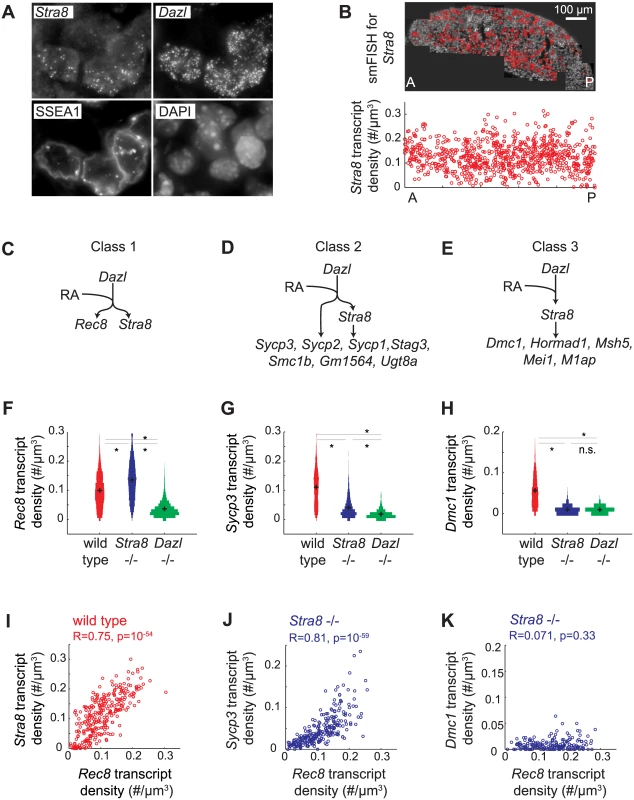
(A) Detection of single transcripts of Stra8 and Dazl by smFISH in E14.5 fetal ovary. Single transcripts are visible as punctate signals. Germ cells are co-stained for SSEA1, and DAPI. (B) Top: Individual images are stitched together for an entire E14.5 ovary. Red signal in the stitched image represents Stra8 transcript. Bottom: Scatterplot of Stra8 transcript density in single germ cells, along the anterior-posterior axis of the ovary. Transcript density is the number of transcripts per cell normalized by cell size (#/um-3). (C), (D), (E) Models for three classes of regulation, and classification of 13 representative meiosis genes into the three classes by smFISH data. (C) Genes that are fully Stra8-independent, (D) genes that require both a Stra8-independent and a Stra8-dependent pathway to be fully expressed, and (E) genes that are fully Stra8-dependent. (F), (G), (H) Distributions of transcript densities in E14.5 wild-type (red), Stra8-deficient (blue) and Dazl-deficient (green) ovarian germ cells, for (F) Rec8, a representative of Class 1, (G) Sycp3, a representative of Class 2, and (H) Dmc1, a representative of Class 3. Asterisks represent significant differences between the means of the distributions (p< 0.05, t-test on average transcript densities of biological replicates). (I) Representative scatterplot of transcript densities of Stra8 against Rec8 in E14.5 wild-type ovarian germ cells. (J), (K) Representative scatterplots of transcript densities of Sycp3 against Rec8 (J), and Dmc1 against Rec8 (K) in E14.5 Stra8-deficient ovarian germ cells. (I), (J), (K) Correlation coefficients for biological replicates are provided in S3 Table. Our smFISH studies of individual germ cells confirmed that all 13 genes were Dazl-dependent, and that they ranged across a spectrum of Stra8-dependence. For conceptual simplification and ease of discussion, we will describe this spectrum as comprising three classes: Class 1 genes—fully Stra8-independent, Class 2 genes—partially Stra8-independent, and Class 3 genes—fully Stra8-dependent (Fig 3C–3E). Rec8 fell into Class 1, fully independent of Stra8 expression (Fig 3C and 3F). Expression of Rec8 in Stra8-deficient germ cells, as a population, was in fact slightly higher than in wild type, an observation we later explored. Dmc1, Msh5, Hormad1, Mei1, and M1ap fell into Class 3, fully dependent on Stra8 (Fig 3E and 3H, S2A Fig). Their expression in Stra8-deficient germ cells was reduced (compared to wild-type germ cells) to the same degree as in Dazl-deficient germ cells. Sycp3, Sycp2, Sycp1, Stag3, Smc1b, Gm1564, and Ugt8a fell into Class 2, partially independent of Stra8 expression (Fig 3D and 3G, S2A Fig). Their expression in Stra8-deficient germ cells was, as a population, significantly lower than in wild type, but significantly higher than in Dazl-deficient germ cells. We always observed a unimodal distribution of gene expression, which is consistent with gene expression being reduced in each germ cell. The direction and relative magnitude of gene expression changes as measured by smFISH and RNA-seq are consistent for all 13 genes (S2B Fig).
Single-cell correlation of Stra8-independent gene expression with expression of RA-induced genes
What is the role of RA in regulating the meiotic prophase genes? It was previously shown that RA induces Stra8 expression in fetal ovarian germ cells [13,14]. Therefore, Stra8-dependent induction of Class 2 and 3 genes would depend, indirectly, on RA. Does RA also regulate the Stra8-independent induction of Class 1 and 2 genes? We previously showed that RA induces Rec8 in the absence of Stra8 [32], and we now demonstrate, quantitatively, the full independence of Rec8 expression from Stra8. By extension, we hypothesized that RA is responsible for Stra8-independent induction of not just Rec8, a Class 1 gene, but also of the Class 2 genes.
An ideal test of this hypothesis would be to eliminate RA in vivo in the fetal ovary. This was not technically feasible, so we instead sought evidence of RA regulation by analyzing gene expression in hundreds of individual germ cells, and using endogenous variation in expression of an RA-induced gene in these hundreds of germ cells as a read out of cell response to RA. If variation in expression of an RA-induced gene reflects the individual cell’s response to RA, then expression of two RA-induced genes across hundreds of individual germ cells should be positively correlated. To test this, we examined variation in expression of the two known independently RA-induced genes, Stra8 and Rec8. We found that Rec8 transcript density is indeed positively correlated with Stra8 transcript density in germ cells of E14.5 fetal ovaries (Fig 3I).
We then employed variation in the level of Rec8 expression as a quantifiable read out of RA response, so as to determine if Stra8-independent expression of genes is due to RA. If so, then expression of the gene, in the absence of Stra8, should be correlated with that of Rec8. We quantified expression of each Class 2 gene alongside Rec8, in hundreds of individual Stra8-deficient germ cells at E14.5. Expression of Sycp3, Sycp2, Sycp1, Stag3, Gm1564, and Ugt8a is positively correlated with Rec8 expression (Fig 3J, S3 Fig, S3 Table). As expected, for Class 3 genes, which are fully Stra8-dependent, residual expression in the absence of Stra8 did not correlate with Rec8 expression (Fig 3K, S3 Fig, S3 Table). These results are consistent with the Stra8-independent pathway being regulated by RA, either directly or indirectly.
Stra8-independent pathway enables maximal and early gene expression
Our model of gene regulation inferred from E14.5 fetal ovaries led us to predict two consequences for gene expression over time. First, we reasoned that, for Class 2 genes, both Stra8-independent and Stra8-dependent pathways might be required to attain maximal levels of gene expression. If so, expression of Class 2 genes in Stra8-deficient germ cells would not reach peak wild-type levels, even after an extended period of time. Second, we considered the possibility that RA-dependent, Stra8-independent induction of Class 1 and 2 genes might function to induce genes that are required early in meiotic prophase, in anticipation of Stra8. If so, Class 1 and 2 genes might be induced in parallel with Stra8, and before Class 3 genes.
To determine the temporal dynamics of gene expression with fine resolution, we took advantage of previous observations that fetal ovarian germ cells initiate and progress through meiotic prophase in an anterior-to-posterior wave [23,24]. Stra8, Dmc1, and Sycp3 expression have been observed to be induced first in germ cells in the anterior portion of the fetal ovary, and only later in the posterior. Therefore, measuring gene expression as a function of anterior-posterior position in addition to time should provide finer resolution of events than would time alone. To formally test the hypothesis that the anterior-posterior axis is a proxy for time, we compared expression changes of 527 germ-cell-enriched genes over time (between E12.5 and E13.5, anterior third of ovaries only), and over space (between posterior and anterior thirds of E13.5 ovaries). We found that gene expression changes over both time and space were indeed highly correlated (S4 Fig, S4 Table), validating our spatiotemporal approach.
We assayed gene expression over a spatiotemporal axis, using Stra8 expression in wild-type germ cells as a reference, as follows. We measured the transcript density of Stra8 in individual germ cells at E11.5, E12.5, E13.5, E14.0, E14.5, E15.0, E15.5, and E16.5. For each time point, we calculated the average transcript density along the longitudinal axis, from the posterior pole (germ cells at least advanced state) to the anterior pole (germ cells at most advanced state) (Fig 4A). We then joined these average expression traces from consecutive time points to create a continuous trace of average transcript density along a spatiotemporal axis (Fig 4B). Using this approach, we quantified the Stra8 pulse of expression in the wild-type ovary, which was previously observed semi-quantitatively, by whole-mount in situ hybridization [23].
Fig. 4. Spatiotemporal analysis demonstrates role of Stra8-independent pathway in inducing maximal and early gene expression, and identifies Stra8-dependent down-regulation of Stra8 and Rec8. 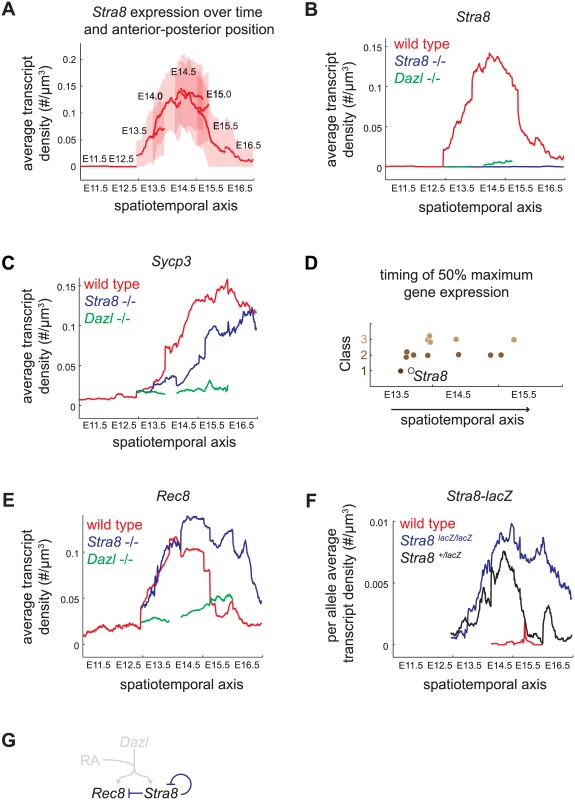
(A) Construction of spatiotemporal plot of Stra8 average transcript densities along the anterior-posterior axis of ovary at E11.5, E12.5, E13.5, E14.0, E14.5, E15.0, E15.5, and E16.5. To construct a smooth average transcript density trace, time points are overlapped based on Stra8 transcript density levels. Bold line indicates mean of distribution, light band indicates 1 standard deviation from the mean. (B) Spatiotemporal plot of Stra8 expression in wild-type (red), Stra8-deficient (blue) and Dazl-deficient (green) germ cells. (C) Spatiotemporal plot of Sycp3 expression in wild-type (red), Stra8-deficient (blue) and Dazl-deficient (green) germ cells. (D) Timing of 50% maximal induction of genes for Class 1, 2, and 3 genes (filled brown circles). Time is represented by a spatiotemporal axis (x-axis), same as in (A). Time of 50% Stra8 induction is represented by open black circle. (E) Spatiotemporal plot of Rec8 expression in wild-type (red), Stra8-deficient (blue) and Dazl-deficient (green) germ cells. Rec8 average transcript densities are significantly higher in Stra8-deficient compared to wild-type germ cells at E14.5, E15.5, and E16.5 (p< 0.05, t-test on average transcript densities of biological replicates). (F) Spatiotemporal plot of lacZ expression from the endogenous Stra8 locus in Stra8-deficient (blue) and Stra8 heterozygote (wild type/lacZ) (black) germ cells, normalized per allele. Normalized lacZ transcript densities differ significantly at E15.5, and E16.5 (p< 0.05, t-test on average transcript densities of biological replicates). (G) Model representing Stra8-dependent down-regulation of Stra8 and Rec8. We applied this spatiotemporal analysis to characterize expression dynamics of the subset of 13 meiotic prophase genes in wild-type, Stra8-deficient, and Dazl-deficient germ cells. First, we asked if Class 2 genes indeed required both Stra8-independent and Stra8-dependent pathways to attain maximal levels of gene expression. We found that, in Stra8-deficient germ cells, Class 2 genes failed to reach expression levels seen in wild type even when given an additional one to two days after expression peaks in wild type. For example, expression of Sycp3 in Stra8-deficient germ cells had begun to decline by E16.5, without having reached the peak levels of expression achieved (at E15.5) in wild-type germ cells (Fig 4B, S5 Fig). Thus, the Stra8-independent pathway is crucial to ensure full expression of Class 2 genes.
Second, we asked if Stra8-independent induction of Class 1 and 2 genes might enable early gene expression. We found that induction of four Class 1 and 2 genes–Rec8, Stag3, Smc1b, and Gm1564 –indeed occurred early, and contemporaneous with Stra8. Half-maximal expression of these genes preceded or coincided with half-maximal expression of Stra8 (Fig 4D, S5 Fig). In contrast, all five of the Class 3 genes tested reached half-maximal expression after Stra8 had done so. Thus, the Stra8-independent pathway is able to induce expression of some Class 1 and 2 genes in parallel with Stra8.
A Stra8-dependent process is required for subsequent down-regulation of Stra8 and Rec8
Spatiotemporal analysis of Rec8 expression in Stra8-deficient germ cells unexpectedly revealed that in the absence of Stra8, germ cells expressed Rec8 at modestly higher levels (Figs 3F and 4E), and Rec8 expression persisted for at least a day longer than in wild type. Therefore, a Stra8-dependent process is required for the subsequent down-regulation of Rec8. By our measurements, Rec8 and Stra8 are induced and subsequently down-regulated with nearly identical dynamics. Therefore, we wondered if Stra8 down-regulation also requires Stra8 function. To measure Stra8 promoter activity in the Stra8-deficient germ cells, we measured expression of a lacZ reporter knocked into the endogenous Stra8 locus [20]. We compared this to lacZ expression in Stra8 heterozygotes, where one functional copy of Stra8 is present. As with expression of Rec8, expression of lacZ in the homozygous Stra8 knockout persisted for at least a day longer than in the heterozygous Stra8 mouse (Fig 4F, S6 Fig). Thus, we have identified down-regulation of Stra8 and Rec8 as a novel Stra8-dependent event (Fig 4G).
After down-regulating Stra8, germ cells are refractory to further Stra8 expression
The observation that Stra8 expression is rapidly down-regulated after its initial induction led us to wonder if, in addition to down-regulating Stra8, germ cells become refractory to subsequent induction of Stra8 by RA. If so, wild-type germ cells that have expressed Stra8 once should not be able to express Stra8 again, even if they were provided with a second (exogenous) dose of RA. To test this prediction, we administered exogenous RA to pregnant mice at E15.5, by which time most germ cells have down-regulated Stra8. We then measured expression of Stra8 a day later, at E16.5 (Fig 5). Stra8 expression was not increased in ovarian germ cells of fetuses that received RA, compared to fetuses that did not receive RA. As a control, we tested if RA was able to induce Stra8 in E15.5 testicular germ cells. Since testicular germ cells do not ordinarily express Stra8 until after birth, we expected that they would be able to induce Stra8 expression if exposed to RA before birth. We found that at E16.5, Stra8 expression was induced about 20-fold in testicular germ cells of fetuses that received RA, compared to fetuses that did not receive RA. Thus, fetal ovarian germ cells that have down-regulated Stra8 expression are refractory to re-expressing Stra8 when exposed to RA 24 hours after initial down-regulation of Stra8.
Fig. 5. RA is unable to induce Stra8 in ovarian germ cells that have induced and down-regulated Stra8. 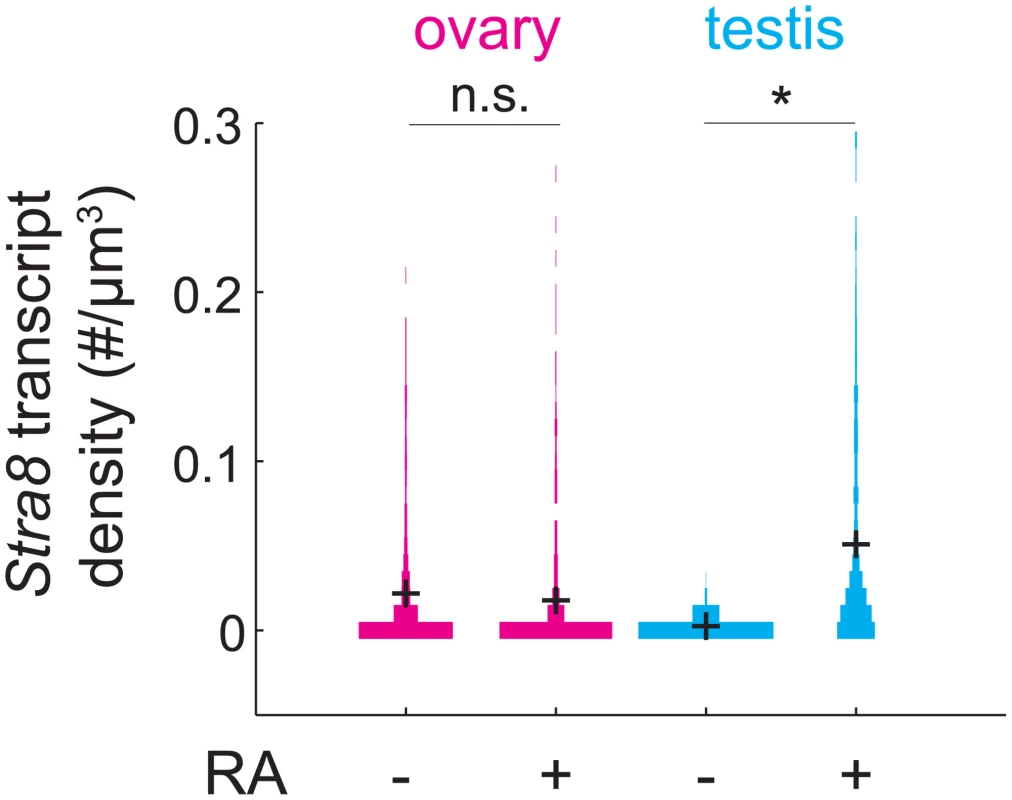
Distributions of Stra8 transcript densities in E16.5 ovary (pink) and testis (blue), with and without exogenous RA administered at E15.5. Asterisks represent significant differences between the means of the distributions (p< 0.05, t-test on average transcript densities of biological replicates). Sycp3 induction precedes negative regulation of Rec8 along the anterior-posterior axis of the E14.5 ovary
Earlier, we measured expression of Rec8 and Sycp3 in hundreds of individual E14.5 germ cells that lacked Stra8, and found that Rec8 and Sycp3 levels were positively correlated, implying their co-regulation by a Stra8-independent pathway (Fig 3J). We were initially surprised to find, upon performing the same analysis in E14.5 wild-type germ cells (Fig 6A), that Rec8 and Sycp3 levels were not correlated in any simple fashion when Stra8 was present. We reasoned that these differences between wild-type and Stra8-deficient germ cells should be due to the two Stra8-dependent processes described earlier—partial induction of Sycp3, and down-regulation of Rec8. Our earlier results suggested that induction of Sycp3 occurs first, followed by down-regulation of Rec8.
Fig. 6. Sycp3 induction precedes negative regulation of Rec8 along the anterior-posterior axis of the E14.5 ovary. 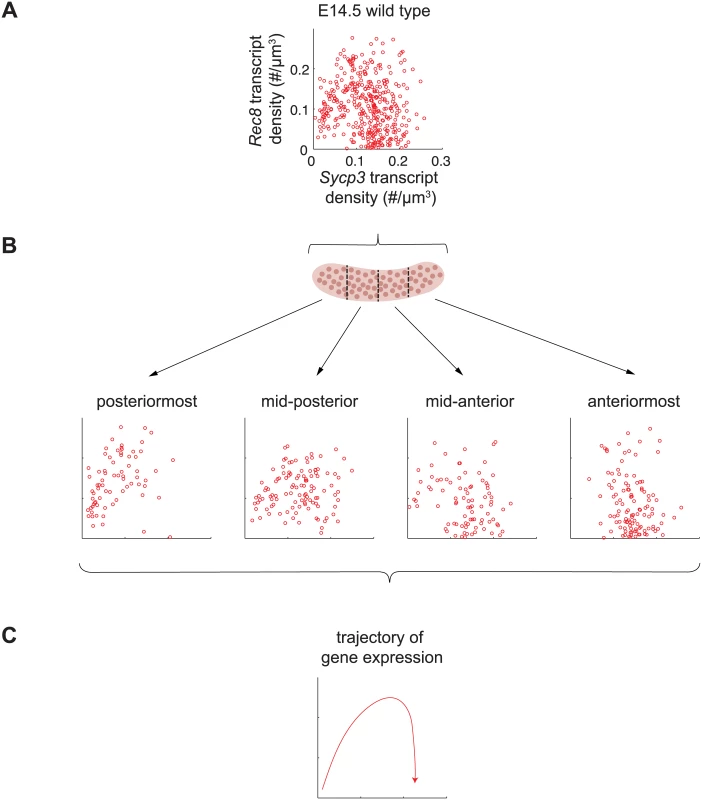
(A) Representative scatterplot of transcript densities of Sycp3 against Rec8 in E14.5 wild-type ovarian germ cells. (B) Scatterplots of transcript densities in germ cells from (A), divided by location in the posteriormost, mid-posterior, mid-anterior, and anteriormost quarters of the ovary. (C) The trajectory of gene expression for an individual germ cell as inferred from gene expression at posterior, middle, and anterior positions. To corroborate these understandings, we set out to finely dissect the time course of Rec8 and Sycp3 expression by separately analyzing germ cells in posterior-to-anterior quarters of the E14.5 wild-type ovary (Fig 6B). In the posteriormost quarter of the ovary, where germ cells are least differentiated, Rec8 and Sycp3 levels were positively correlated (Fig 6B), suggesting that the dominant process at this very early stage is induction of both Rec8 and Sycp3. In subsequent stages, Sycp3 continues to be induced, but Rec8 is down-regulated. For example, in the middle quarters of the ovary, where germ cells are more differentiated, many germ cells had increased Sycp3 levels, but Rec8 expression was decreased, particularly in those cells with high expression of Sycp3. In the anteriormost quarter of the ovary, where germ cell differentiation is most advanced, all germ cells had high Sycp3 levels, but most had very low levels of Rec8 expression (Fig 6B). In summary, fetal ovarian germ cells progressed from a state of low Sycp3/low Rec8 expression, to a state of high Sycp3/high Rec8 expression, and finally to a state of high Sycp3/low Rec8 expression (Fig 6C), reflecting initial induction of both Sycp3 and Rec8, and subsequent down-regulation of Rec8. As a consequence of down-regulation of Rec8 (but not Sycp3) in the most advanced germ cells, expression of Rec8 and expression of Sycp3 were no longer correlated. Thus, the conclusions arising from our earlier analyses were corroborated by positionally informed, single-cell correlation analysis in the E14.5 wild-type ovary.
Discussion
The regulated induction of meiotic prophase genes is a prerequisite for the chromosomal program of meiotic prophase. We report here a mammalian gene regulatory program for meiotic prophase as it occurs in fetal ovarian germ cells. We identified 104 genes that fulfill stringent criteria for specificity to meiotic prophase. A quarter of these genes have been shown previously to be required for successful meiotic prophase. The remaining three quarters remain uncharacterized and represent promising candidates that may play similarly critical roles during meiotic prophase. Meiotic prophase genes are induced initially by RA, in the presence of Dazl, via Stra8-independent and Stra8-dependent pathways (Fig 7A). Subsequently, down-regulation of Stra8 and Rec8 occurs via a Stra8-dependent process (Fig 7B). We propose that these two elements of gene regulation enable germ cells to prepare for the chromosomal program of meiotic prophase, and to ensure that the chromosomal program is induced just once.
Fig. 7. Summary model for induction of meiotic prophase gene expression. 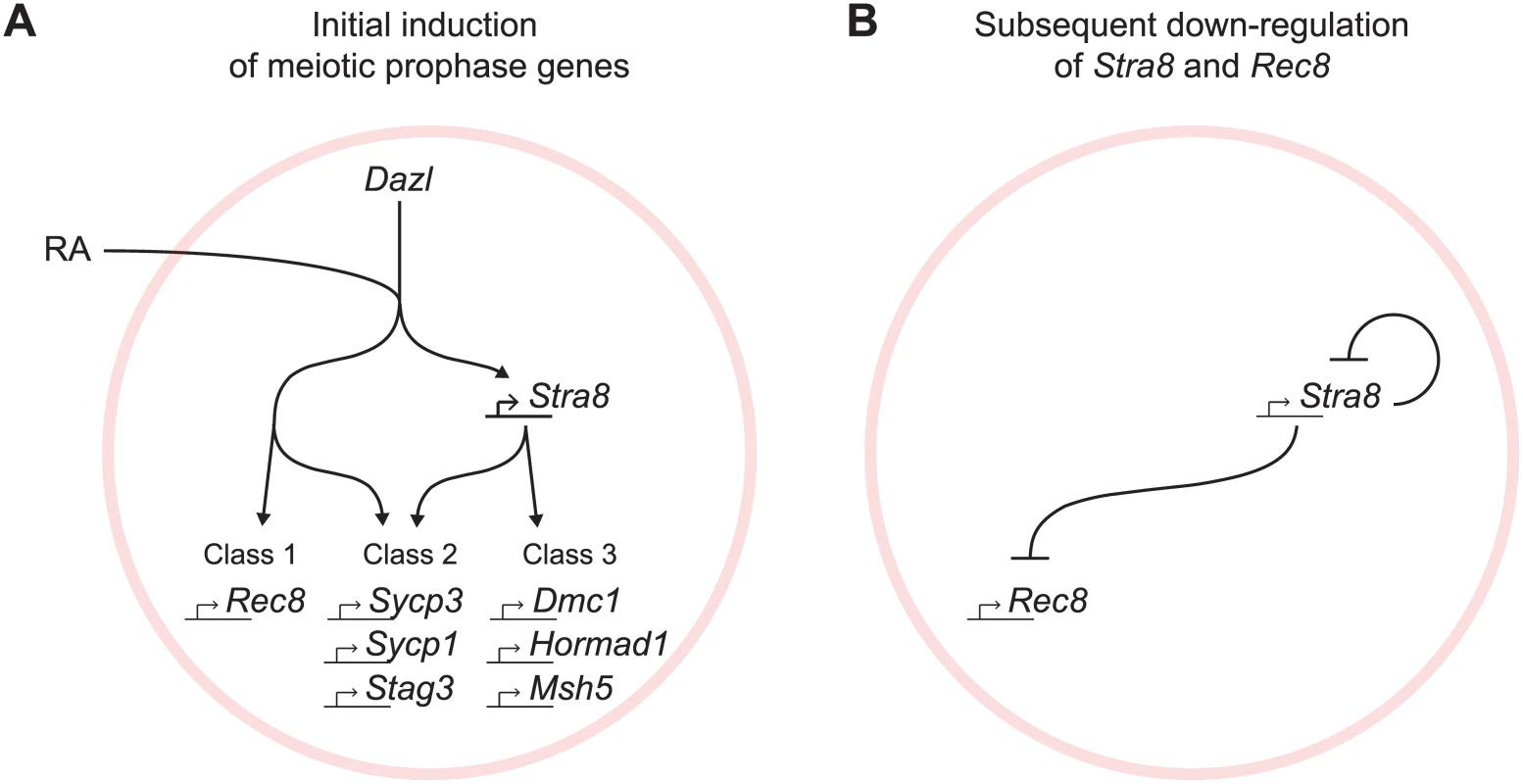
(A) Model for initial induction of meiotic prophase genes by Dazl, RA, and Stra8. In the presence of Dazl, RA induces meiotic prophase genes via Stra8-independent and Stra8-dependent pathways. Genes range across a spectrum of Stra8-dependency; for simplicity, we divide the genes into three classes: Class 1 –fully Stra8-independent, Class 2 –partially Stra8-independent and partially Stra8-dependent, and Class 3 –fully Stra8-dependent. Representatives of each gene class are shown. (B) A Stra8-dependent process is required for down-regulation of Stra8 and Rec8 expression. Gene induction via Stra8-independent and Stra8-dependent pathways represents a multi-output feedforward loop that enables a temporal order of gene activation
At the onset of meiotic prophase, meiotic prophase genes are induced by Dazl, RA, and Stra8, organized in two branching pathways (Fig 7A). Dazl is required for the induction of nearly all genes expressed specifically during meiotic prophase. While the results highlight Dazl’s crucial role in the gene expression program for meiotic prophase, the mechanism by which it enables gene expression remains unclear. Downstream of Dazl, gene induction occurs via a Stra8-independent pathway as well as a Stra8-dependent pathway; these pathways function both separately and additively. Expression of some genes requires only the Stra8-independent pathway (Class 1), while other genes require both Stra8-independent and Stra8-dependent pathways (Class 2), and yet other genes are fully Stra8-dependent (Class 3). Both the Stra8-dependent and Stra8-independent pathways are regulated by RA.
These genetic insights lead us to two speculative hypotheses regarding molecular mechanisms of meiotic prophase gene regulation by STRA8 and RA receptors (RARs): (1) The Stra8-dependent pathway is mediated directly by the STRA8 protein, a putative basic helix-loop-helix transcription factor, and (2) the Stra8-independent pathway is mediated directly by RARs. Transcriptome data from both whole gonads (this study) and sorted germ cells [34] support this possibility: fetal ovarian germ cells initiating meiosis express all three RARs (RAR alpha, beta, and gamma) and their heterodimeric partners, the retinoid X receptors (RXR alpha, beta, and gamma). Potential redundancies among the RARs and RXRs complicate genetic interrogation of the roles of the RARs. The possible roles of STRA8 and RARs in directly regulating gene expression can be tested by chromatin-immunoprecipitation-sequencing (ChIP-seq) of RARs and STRA8 in germ cells that are initiating meiosis. We predict that Class 1 genes will be bound by RARs but not STRA8, Class 3 genes will be bound by STRA8 but not RARs, and Class 2 genes will be bound by both RARs and STRA8. A ChIP-Seq study in embryonic stem cells identified RAR binding of both the Stra8 and Rec8 promoter regions [35]. Of course, it is also possible that the Stra8-independent and Stra8-dependent pathways are mediated indirectly, by germ-cell-expressed factors that have not yet been implicated in meiotic initiation.
The branching regulatory model described here is reminiscent of a motif termed a feed forward loop (FFL), which has been shown to generate a temporal order of gene activation [36]. An FFL comprises an upstream regulator, in this case RA, which regulates a downstream regulator, in this case Stra8. Both the upstream regulator, RA, and the downstream regulator, Stra8, regulate multiple downstream targets, in this case the meiotic prophase genes. Genes respond to input from either the upstream or downstream regulator, or both. Modulating the activation strengths of upstream versus downstream regulators can generate a temporal order of gene activation: genes with greater input from the upstream regulator are activated earlier, and genes with greater input from the downstream regulator are activated later. Consistent with such an outcome, we observe that a subset of Class 1 and 2 genes, which are fully or partially induced by the Stra8-independent pathway, are expressed earlier than Class 3 genes and with timing of induction close to that of Stra8 induction.
We propose that Class 1 and 2 genes may be induced earlier so as to prepare cells for the meiotic chromosomal events triggered by Stra8. Indeed, we observe that Class 1 and 2 genes include almost all known meiotic cohesins and synaptonemal complex proteins, which structurally associate with meiotic chromosomes and may therefore be required early, and in sufficient quantities to satisfy the stoichiometric requirements of the chromosomal program. Early expression of cohesin and synaptonemal complex proteins, prior to initiation of the chromosomal program, may be a common feature of both sexes and across species. In mouse testicular germ cells, the synaptonemal complex protein genes Sycp1, Sycp2, and Sycp3 are expressed as early as in mitotic spermatogonia [37]. In the C. elegans gonad and the D. melanogaster ovary, Rec8 and synaptonemal complex proteins respectively are also expressed during the amplifying mitotic divisions preceding meiosis [38–40].
Induction of Class 2 genes by a combination of Stra8-independent and Stra8-dependent pathways may also contribute to fine-tuning expression levels of meiotic prophase genes. Precise regulation of gene dosage has been shown to be important for meiotic chromosomal processes. In the mouse, heterozygous loss of function for either one of the cohesins Smc1b and Rec8 perturbs formation of the synaptonemal complex and affects synapsis and recombination between homologs [41]. Therefore, although the Stra8-independent pathway is sufficient for partial gene expression, the two pathways in combination may serve to optimize levels of gene expression and chromosomal function of the Class 2 genes.
Down-regulation of Stra8 via a Stra8-dependent process may ensure that the chromosomal program is initiated only once in each cell
Subsequent to the initial induction of Stra8 and Rec8, their expression declines rapidly. We discovered that this down-regulation depends on Stra8. It remains to be determined whether this occurs directly, via Stra8 activity as a putative transcriptional regulator, or indirectly, as a consequence of progression of cell state. In either case, we propose that Stra8-dependent down-regulation of Stra8 and Rec8 may serve to limit gene expression to their appropriate window of function. In particular, Stra8-dependent down-regulation of itself represents a negative feedback loop that prevents prolonged induction of the chromosomal program of meiotic prophase. In addition, we found that ovarian germ cells that have down-regulated Stra8 are refractory to re-expressing Stra8 even in the presence of exogenous RA, which may prevent re-initiation of the chromosomal program. In yeast, an analogous negative feedback loop is postulated to restrict supernumerary rounds of DNA replication and nuclear division. IME1, a transcription factor that initiates the yeast meiotic transcriptional program, induces IME2, which restricts expression of IME1 and destabilizes IME1 protein [3,42]. Absence of IME2 results in prolonged IME1 expression and additional rounds of DNA synthesis and nuclear division [43].
Implications for the gene regulatory program of meiotic prophase in the male
Based on similarities between ovarian and testicular germ cells, it is likely that the gene regulatory program as inferred from fetal ovarian germ cells is shared, at least in part, between the sexes. In both sexes, RA induction of Stra8 has been shown to be required for initiation of the chromosomal program of meiotic prophase [12,14,20]. RA and Stra8 could therefore also regulate gene expression in the male. In testicular germ cells entering meiosis, Stra8 is also rapidly induced, at pre-leptotene, and then rapidly down-regulated, by leptotene [17], suggesting the possibility that there is also negative feedback on Stra8 expression. However, several aspects of regulation in the male remain unclear. For instance, in the male, RA-STRA8 signaling regulates not only meiotic initiation but also spermatogonial differentiation [44]. In fetal ovarian germ cells, competence to respond to RA and initiate meiosis requires Dazl [21]; in testicular germ cells, Dazl’s role in meiotic competence remains unknown. Thus, studies of adult testicular germ cells deficient for Dazl or Stra8 will be required to determine if Dazl and Stra8 govern the gene regulatory program of meiotic prophase in the adult testis in a manner similar to that reported here for the fetal ovary.
Implications for in vitro germ cell derivation
Our findings have practical implications for in vitro derivation of germ cells and gametes. First, our results provide a blueprint to guide efforts in recapitulating the gene regulatory program of meiotic prophase in vitro. Second, our findings substantiate previous criticisms against taking expression of meiotic genes as sufficient evidence of meiosis [45,46]. By explicitly interrogating the regulation of the gene expression program and the chromosomal program by Dazl and Stra8, we showed that the two programs are regulated distinctly. Specifically, the chromosomal program of meiotic prophase requires Stra8 function, but a subset of the gene expression program is induced independently of Stra8. Our findings thus highlight gene expression as a preparatory phase for the chromosomal program, and underscore the insufficiency of meiotic gene expression as an assay for meiotic progression. Rather, both the gene regulatory program and the chromosomal program are essential for successful meiosis.
Materials and Methods
Ethics statement
All experiments involving mice were performed in accordance with the guidelines of the Massachusetts Institute of Technology (MIT) Division of Comparative Medicine, which is overseen by MIT's Institutional Animal Care and Use Committee (IACUC). The animal care program at MIT/Whitehead Institute is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC), meeting or exceeding the standards of AAALAC as detailed in the Guide for the Care and Use of Laboratory Animals. The MIT IACUC approved this research (no. 0714-074-17).
Mice
Germ-cell-depleted (KitW/KitWv) and homozygous wild-type control (Kit+/Kit+) were generated by crossing C57BL/6J-KitWv/Kit+ (The Jackson Laboratory) males to WBB6F1/J-KitW/Kit+ (The Jackson Laboratory) females [47]. KitW and KitWv alleles were genotyped as previously described [48,49]. Stra8-deficient (Stra8-/-), Dazl-deficient (Dazl-/-) and homozygous wild-type control embryos were generated by heterozygote matings of Dazltm1Hjc [50] and Stra8tm1Dcp [20] mice respectively. Dazltm1Hjc, Stra8tm1Dcp, and wild-type mice used are of C57BL/6 background. Dazltm1Hjc and Stra8tm1Dcp alleles were genotyped as previously described [20,50].
RA treatment
500 mg/kg of body weight all-trans RA (Sigma-Aldrich, St Louis, MO), dissolved at 30 mg/ml of corn oil, was administered to pregnant mice via gavage.
Embryonic gonad collection and sexing
Timed matings were set up by housing female mice with male mice overnight. Noon of the day when a vaginal plug was evident was considered E0.5. For RNA-seq analysis, embryonic gonads were dissected away from mesonephroi. For RNA-seq from germ-cell-depleted (KitW/KitWv) and homozygous wild-type control (Kit+/Kit+) gonads, Stra8-deficient (Stra8-/-), Dazl-deficient (Dazl-/-) and homozygous wild-type control gonads, whole gonads were processed for sequencing. For RNA-seq from E12.5 and E13.5 anterior and posterior portions of the ovary, ovaries were dissected into thirds, and the anterior and posterior thirds were processed for sequencing. For smFISH, embryonic gonads were dissected with mesonephroi intact to provide anterior-posterior orientation. For embryos E13.5 and older, the sex of tissues was determined by scoring the presence or absence of testicular cords. For E11.5 and E12.5 embryos, sex was determined by PCR as previously described [23].
RNA-seq sample preparation
For all RNA-seq experiments, total RNA (~1 ug) was extracted from embryonic gonads using Trizol (Invitrogen) according to the manufacturer’s protocol, and hemoglobin transcripts were selectively removed from total RNA using GLOBINclear (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. For KitW/KitWv and Kit+/Kit+ embryonic gonads and E12.5 and E13.5 embryonic ovary thirds, libraries were prepared using the Illumina mRNA-Seq Sample Preparation Kit (Illumina, San Diego, CA) according to the manufacturer’s protocol. Libraries were sequenced on the Illumina Genome Analyzer II platform to obtain 36-base-pair single reads. For E14.5 Dazl-deficient, Stra8-deficient, and wild-type control ovaries, libraries were prepared using the Illumina TruSeq RNA Sample Preparation Kit. Libraries were multiplexed and sequenced on the Illumina HiSeq 2000 platform to obtain 40-base-pair single reads. RNA-seq data for KitW/KitWv and Kit+/Kit+, E12.5 and E13.5 embryonic ovary thirds, and E14.5 Dazl-deficient, Stra8-deficient, and wild-type control ovaries have been deposited in NCBI GEO under accession number GSE70361 and NCBI SRA under accession numbers SRP058992, SRP059594, SRP059601, and SRP059599.
RNA-seq on gonads from KitW/KitWv and Kit+/Kit+ embryos was performed on two biological replicates for each condition. RNA-seq on anterior and posterior thirds of ovaries from E12.5 and E13.5 wild-type embryos was performed on two and three replicate pools respectively, where each pool consisted of ovary thirds from eight embryos. RNA-seq on E14.5 Dazl-deficient and Stra8-deficient ovaries was performed on three biological replicates each, with paired homozygous wild-type controls.
RNA-seq data analysis
Reads were aligned to the mouse genome (mm9) using TopHat [51], allowing only unique alignments (option—g1). We counted reads mapping to the Refseq annotated gene set using htseq-count [52]. Fold-changes and FDR-corrected p values, q, for differentially expressed genes were calculated using edgeR, using tagwise-dispersions and normalizing for library complexity [53]. FPKMs were calculated using Cufflinks [54]. K-means clustering was performed using Cluster 3.0 on log transformed and mean centered FPKMs, using the Pearson correlation as the similarity metric [55], and visualized using Treeview [56]. Non-coding genes were excluded from analyses.
Single-molecule Fluorescent In Situ Hybridization (smFISH)
Probe design, synthesis, and coupling were as previously described [33,57]. Probes sequences are provided in S2 Text. Gonads were fixed in 4% paraformaldehyde (PFA)/PBS for 2 hours at 4°C, incubated overnight in 30% sucrose/4% PFA/PBS at 4°C, then embedded in O.C.T. compound (Sakura Finetek, Torrance, CA). Frozen blocks were sectioned at 8 μm thickness, fixed in 4% formaldehyde at room temperature for 15 minutes, rinsed in PBS, and dehydrated overnight in 70% ethanol at 4°C. The hybridization procedure was performed as previously described [33,57]. FITC-coupled anti-SSEA-1 antibody (BD 560127) (BD Biosciences, Franklin Lakes, NJ) was added to the hybridization step at 1 : 30 to identify germ cells. In all experiments, germ cells were identified by either smFISH for Dazl or Oct4, in combination with SSEA1 immunostaining, and/or DAPI nuclear staining. Counting of individual mRNA particles, image stitching, and data analysis was performed using custom Matlab software as previously described [33,57].
To depict distributions of transcript densities for each group, we pooled biological replicates in one violin plot. Comparison of groups was performed by comparing means of at least two biological replicates from at least two litters using the two-sample t-test. To depict correlations between pairs of genes in individual cells, we show one representative biological replicate, but calculate the Spearman correlation coefficient for each biological replicate. To depict average transcript densities over space and time, we pooled biological replicates. At each time point, we determined average transcript densities from the posterior to anterior of the ovary for 100 windows of size 0.2 of the total length of the ovary. The average transcript density traces of consecutive time points were joined together from posterior to anterior. Using average transcript density traces of Stra8 as a guide, we overlapped some time points by shifting along the x-axis in order to maximize overlap between the average expression traces for Stra8. We determined shifts using Stra8 expression, and applied the same shifts to spatiotemporal plots for all other genes.
Supporting Information
Zdroje
1. Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, et al. The transcriptional program of sporulation in budding yeast. Science. 1998;282 : 699–705. doi: 10.1126/science.282.5389.699 9784122
2. Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, et al. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26 : 415–423. doi: 10.1038/82539 11101837
3. Smith HE, Mitchell AP. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9 : 2142–2152. 2664470
4. Kassir Y, Adir N, Boger-Nadjar E, Raviv NG, Rubin-Bejerano I, Sagee S, et al. Transcriptional regulation of meiosis in budding yeast. Int Rev Cytol. 2003;224 : 111–71. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2882377&tool=pmcentrez&rendertype=abstract 12722950
5. Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet. Nature Publishing Group; 2010;11 : 124–36. doi: 10.1038/nrg2723
6. Cohen PE, Pollack SE, Pollard JW. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr Rev. 2006;27 : 398–426. doi: 10.1210/er.2005-0017 16543383
7. Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5 : 983–97. doi: 10.1038/nrm1526 15573136
8. Schurko AM, Logsdon JM. Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. BioEssays. 2008;30 : 579–589. doi: 10.1002/bies.20764 18478537
9. Villeneuve AM, Hillers KJ. Whence meiosis? Cell. 2001;106 : 647–650. doi: 10.1016/S0092-8674(01)00500-1 11572770
10. Kumar R, Bourbon HM, De Massy B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 2010;24 : 1266–1280. doi: 10.1101/gad.571710 20551173
11. Van Werven FJ, Amon A. Regulation of entry into gametogenesis. Philos Trans R Soc Lond B Biol Sci. 2011;366 : 3521–31. doi: 10.1098/rstb.2011.0081 22084379
12. Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AMM, et al. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci U S A. 2008;105 : 14976–14980. doi: 10.1073/pnas.0807297105 18799751
13. Bowles J, Knight D, Smith C, Wilhelm D, Richman J, Mamiya S, et al. Retinoid signaling determines germ cell fate in mice. Science. 2006;312 : 596–600. doi: 10.1126/science.1125691 16574820
14. Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci. 2006;103 : 2474–2479. doi: 10.1073/pnas.0510813103 16461896
15. Fujiwara S, Kawamura K. Acquisition of retinoic acid signaling pathway and innovation of the chordate body plan. Zoolog Sci. Zoological Society of Japan; 2003;20 : 809–818. doi: 10.2108/zsj.20.809
16. Oulad-Abdelghani M, Bouillet P, Décimo D, Gansmuller A, Heyberger S, Dollé P, et al. Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J Cell Biol. 1996;135 : 469–477. doi: 10.1083/jcb.135.2.469 8896602
17. Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, et al. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod. 2008;79 : 35–42. doi: 10.1095/biolreprod.107.066795 18322276
18. Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, et al. Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol Reprod. 2008;78 : 537–45. doi: 10.1095/biolreprod.107.064337 18032419
19. Ghyselinck NB, Vernet N, Dennefeld C, Giese N, Nau H, Chambon P, et al. Retinoids and spermatogenesis: Lessons from mutant mice lacking the plasma retinol binding protein. Dev Dyn. 2006;235 : 1608–1622. doi: 10.1002/dvdy.20795 16586441
20. Baltus AE, Menke DB, Hu Y-C, Goodheart ML, Carpenter AE, de Rooij DG, et al. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet. 2006;38 : 1430–4. doi: 10.1038/ng1919 17115059
21. Lin Y, Gill ME, Koubova J, Page DC. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Science. 2008;322 : 1685–1687. doi: 10.1126/science.1166340 19074348
22. Gill ME, Hu Y-C, Lin Y, Page DC. Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc Natl Acad Sci. 2011;108 : 7443–8. doi: 10.1073/pnas.1104501108 21504946
23. Menke DB, Koubova J, Page DC. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev Biol. 2003;262 : 303–312. doi: 10.1016/S0012-1606(03)00391-9 14550793
24. Bullejos M, Koopman P. Germ cells enter meiosis in a rostro-caudal wave during development of the mouse ovary. Mol Reprod Dev. 2004;68 : 422–8. doi: 10.1002/mrd.20105 15236325
25. Speed RM. Meiosis in the foetal mouse ovary. I. An analysis at the light microscope level using surface-spreading. Chromosoma. 1982;85 : 427–37. Available: http://www.ncbi.nlm.nih.gov/pubmed/6180868 6180868
26. Borum K. Oogenesis in the mouse: a study of the meiotic prophase. Exp Cell Res. 1961;507 : 495–507. Available: http://www.sciencedirect.com/science/article/pii/0014482761904499
27. Handel MA, Eppig JJ. Sertoli cell differentiation in the testes of mice genetically deficient in germ cells. Biol Reprod. 1979;20 : 1031–1038. Available: http://www.biolreprod.org/content/20/5/1031.short 476239
28. McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262 : 1–15. doi: 10.1016/S0012-1606(03)00214-8 14512014
29. Pesce M, Wang X, Wolgemuth DJ, Schöler H. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech Dev. 1998;71 : 89–98. Available: http://www.ncbi.nlm.nih.gov/pubmed/9507072 9507072
30. Yamaguchi S, Kimura H, Tada M, Nakatsuji N, Tada T. Nanog expression in mouse germ cell development. Gene Expr Patterns. 2005;5 : 639–646. doi: 10.1016/j.modgep.2005.03.001 15939376
31. Western P, Maldonado-Saldivia J, van den Bergen J, Hajkova P, Saitou M, Barton S, et al. Analysis of Esg1 expression in pluripotent cells and the germline reveals similarities with Oct4 and Sox2 and differences between human pluripotent cell lines. Stem Cells. 2005;23 : 1436–42. doi: 10.1634/stemcells.2005-0146 16166252
32. Koubova J, Hu Y-C, Bhattacharyya T, Soh YQS, Gill ME, Goodheart ML, et al. Retinoic acid activates two pathways required for meiosis in mice. PLoS Genet. 2014;10:e1004541. doi: 10.1371/journal.pgen.1004541 25102060
33. Raj A, van den Bogaard P, Rifkin S a, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5 : 877–879. doi: 10.1038/nmeth.1253 18806792
34. Lesch BJ, Dokshin GA, Young RA, McCarrey JR, Page DC. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc Natl Acad Sci U S A. 2013;110 : 16061–6. doi: 10.1073/pnas.1315204110 24043772
35. Mahony S, Mazzoni EO, McCuine S, Young RA, Wichterle H, Gifford DK. Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol. 2011;12:R2. doi: 10.1186/gb-2011-12-1-r2 21232103
36. Kalir S, Alon U. Using a quantitative blueprint to reprogram the dynamics of the flagella gene network. Cell. 2004;117 : 713–720. doi: 10.1016/j.cell.2004.05.010 15186773
37. Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27 : 422–6. doi: 10.1038/86927 11279525
38. Christophorou N, Rubin T, Huynh J-R. Synaptonemal complex components promote centromere pairing in pre-meiotic germ cells. PLoS Genet. 2013;9:e1004012. doi: 10.1371/journal.pgen.1004012 24367278
39. Joyce EF, Apostolopoulos N, Beliveau BJ, Wu C. Germline progenitors escape the widespread phenomenon of homolog pairing during Drosophila development. PLoS Genet. 2013;9:e1004013. doi: 10.1371/journal.pgen.1004013 24385920
40. Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 2001;15 : 1349–60. doi: 10.1101/gad.192701 11390355
41. Murdoch B, Owen N, Stevense M, Smith H, Nagaoka S, Hassold T, et al. Altered cohesin gene dosage affects mammalian meiotic chromosome structure and behavior. PLoS Genet. 2013;9:e1003241. doi: 10.1371/journal.pgen.1003241 23408896
42. Guttmann-Raviv N, Martin S, Kassir Y. Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1. Mol Cell Biol. 2002;22 : 2047–2056. doi: 10.1128/MCB.22.7.2047-2056.2002 11884593
43. Foiani M, Nadjar-Boger E, Capone R, Sagee S, Hashimshoni T, Kassir Y. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol Gen Genet. 1996;253 : 278–288. doi: 10.1007/s004380050323 9003314
44. Endo T, Romer K a., Anderson EL, Baltus AE, de Rooij DG, Page DC. Periodic retinoic acid—STRA8 signaling intersects with periodic germ-cell competencies to regulate spermatogenesis. Proc Natl Acad Sci. 2015;112:E2347–E2356. doi: 10.1073/pnas.1505683112 25902548
45. Oatley J, Hunt PA. Of mice and (wo)men: purified oogonial stem cells from mouse and human ovaries. Biol Reprod. 2012;86 : 196–196. doi: 10.1095/biolreprod.112.100297 22402962
46. Handel MA, Eppig JJ, Schimenti JC. Applying “gold standards” to in-vitro-derived germ cells. Cell. Elsevier; 2014;157 : 1257–1261. doi: 10.1016/j.cell.2014.05.019
47. Mintz B, Russell ES. Gene-induced embryological modifications of primordial germ cells in the mouse. J Exp Zool. 1957;134 : 207–37. Available: http://onlinelibrary.wiley.com/doi/10.1002/jez.1401340202/abstract 13428952
48. Waskow C, Paul S, Haller C, Gassmann M, Rodewald HR. Viable c-KitW/W mutants reveal pivotal role for c-Kit in the maintenance of lymphopoiesis. Immunity. 2002;17 : 277–288. doi: 10.1016/S1074-7613(02)00386-2 12354381
49. Tanosaki R, Migliaccio AR. Engraftment of normal stem cells in W/Wv mice assessed by a novel quantitative PCR analysis. Br J Haematol. 1997;98 : 1017–1025. doi: 10.1046/j.1365-2141.1997.2983115.x 9326206
50. Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, Saunders P, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389 : 73–77. doi: 10.1038/37987 9288969
51. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25 : 1105–11. doi: 10.1093/bioinformatics/btp120 19289445
52. Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31 : 166–169. doi: 10.1093/bioinformatics/btu638 25260700
53. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26 : 139–40. doi: 10.1093/bioinformatics/btp616 19910308
54. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. Nature Publishing Group; 2010;28 : 511–5. doi: 10.1038/nbt.1621
55. De Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20 : 1453–4. doi: 10.1093/bioinformatics/bth078 14871861
56. Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20 : 3246–8. doi: 10.1093/bioinformatics/bth349 15180930
57. Peterson KA, Nishi Y, Ma W, Vedenko A, Shokri L, Zhang X, et al. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 2012;26 : 2802–2816. doi: 10.1101/gad.207142.112 23249739
Štítky
Genetika Reprodukčná medicína
Článek The Chromatin Protein DUET/MMD1 Controls Expression of the Meiotic Gene during Male Meiosis inČlánek Tissue-Specific Gain of RTK Signalling Uncovers Selective Cell Vulnerability during Embryogenesis
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Retraction: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Signaling from Within: Endocytic Trafficking of the Robo Receptor Is Required for Midline Axon Repulsion
- A Splice Region Variant in Lowers Non-high Density Lipoprotein Cholesterol and Protects against Coronary Artery Disease
- The Chromatin Protein DUET/MMD1 Controls Expression of the Meiotic Gene during Male Meiosis in
- A NIMA-Related Kinase Suppresses the Flagellar Instability Associated with the Loss of Multiple Axonemal Structures
- Slit-Dependent Endocytic Trafficking of the Robo Receptor Is Required for Son of Sevenless Recruitment and Midline Axon Repulsion
- Expression of Concern: Protein Under-Wrapping Causes Dosage Sensitivity and Decreases Gene Duplicability
- Mutagenesis by AID: Being in the Right Place at the Right Time
- Identification of as a Genetic Modifier That Regulates the Global Orientation of Mammalian Hair Follicles
- Bridges Meristem and Organ Primordia Boundaries through , , and during Flower Development in
- Evaluating the Performance of Fine-Mapping Strategies at Common Variant GWAS Loci
- KLK5 Inactivation Reverses Cutaneous Hallmarks of Netherton Syndrome
- Differential Expression of Ecdysone Receptor Leads to Variation in Phenotypic Plasticity across Serial Homologs
- Receptor Polymorphism and Genomic Structure Interact to Shape Bitter Taste Perception
- Cognitive Function Related to the Gene Acquired from an LTR Retrotransposon in Eutherians
- Critical Function of γH2A in S-Phase
- Arabidopsis AtPLC2 Is a Primary Phosphoinositide-Specific Phospholipase C in Phosphoinositide Metabolism and the Endoplasmic Reticulum Stress Response
- XBP1-Independent UPR Pathways Suppress C/EBP-β Mediated Chondrocyte Differentiation in ER-Stress Related Skeletal Disease
- Integration of Genome-Wide SNP Data and Gene-Expression Profiles Reveals Six Novel Loci and Regulatory Mechanisms for Amino Acids and Acylcarnitines in Whole Blood
- A Genome-Wide Association Study of a Biomarker of Nicotine Metabolism
- Cell Cycle Regulates Nuclear Stability of AID and Determines the Cellular Response to AID
- A Genome-Wide Association Analysis Reveals Epistatic Cancellation of Additive Genetic Variance for Root Length in
- Tissue-Specific Gain of RTK Signalling Uncovers Selective Cell Vulnerability during Embryogenesis
- RAB-10-Dependent Membrane Transport Is Required for Dendrite Arborization
- Basolateral Endocytic Recycling Requires RAB-10 and AMPH-1 Mediated Recruitment of RAB-5 GAP TBC-2 to Endosomes
- Dynamic Contacts of U2, RES, Cwc25, Prp8 and Prp45 Proteins with the Pre-mRNA Branch-Site and 3' Splice Site during Catalytic Activation and Step 1 Catalysis in Yeast Spliceosomes
- ARID1A Is Essential for Endometrial Function during Early Pregnancy
- Predicting Carriers of Ongoing Selective Sweeps without Knowledge of the Favored Allele
- An Interaction between RRP6 and SU(VAR)3-9 Targets RRP6 to Heterochromatin and Contributes to Heterochromatin Maintenance in
- Photoreceptor Specificity in the Light-Induced and COP1-Mediated Rapid Degradation of the Repressor of Photomorphogenesis SPA2 in Arabidopsis
- Autophosphorylation of the Bacterial Tyrosine-Kinase CpsD Connects Capsule Synthesis with the Cell Cycle in
- Multimer Formation Explains Allelic Suppression of PRDM9 Recombination Hotspots
- Rescheduling Behavioral Subunits of a Fixed Action Pattern by Genetic Manipulation of Peptidergic Signaling
- A Gene Regulatory Program for Meiotic Prophase in the Fetal Ovary
- Cell-Autonomous Gβ Signaling Defines Neuron-Specific Steady State Serotonin Synthesis in
- Discovering Genetic Interactions in Large-Scale Association Studies by Stage-wise Likelihood Ratio Tests
- The RCC1 Family Protein TCF1 Regulates Freezing Tolerance and Cold Acclimation through Modulating Lignin Biosynthesis
- The AMPK, Snf1, Negatively Regulates the Hog1 MAPK Pathway in ER Stress Response
- The Parkinson’s Disease-Associated Protein Kinase LRRK2 Modulates Notch Signaling through the Endosomal Pathway
- Multicopy Single-Stranded DNA Directs Intestinal Colonization of Enteric Pathogens
- Recurrent Domestication by Lepidoptera of Genes from Their Parasites Mediated by Bracoviruses
- Three Different Pathways Prevent Chromosome Segregation in the Presence of DNA Damage or Replication Stress in Budding Yeast
- Identification of Four Mouse Diabetes Candidate Genes Altering β-Cell Proliferation
- The Intolerance of Regulatory Sequence to Genetic Variation Predicts Gene Dosage Sensitivity
- Synergistic and Dose-Controlled Regulation of Cellulase Gene Expression in
- Genome Sequence and Transcriptome Analyses of : Metabolic Tools for Enhanced Algal Fitness in the Prominent Order Prymnesiales (Haptophyceae)
- Ty3 Retrotransposon Hijacks Mating Yeast RNA Processing Bodies to Infect New Genomes
- FUS Interacts with HSP60 to Promote Mitochondrial Damage
- Point Mutations in Centromeric Histone Induce Post-zygotic Incompatibility and Uniparental Inheritance
- Genome-Wide Association Study with Targeted and Non-targeted NMR Metabolomics Identifies 15 Novel Loci of Urinary Human Metabolic Individuality
- Outer Hair Cell Lateral Wall Structure Constrains the Mobility of Plasma Membrane Proteins
- A Large-Scale Functional Analysis of Putative Target Genes of Mating-Type Loci Provides Insight into the Regulation of Sexual Development of the Cereal Pathogen
- A Genetic Selection for Mutants Reveals an Interaction between DNA Polymerase IV and the Replicative Polymerase That Is Required for Translesion Synthesis
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Arabidopsis AtPLC2 Is a Primary Phosphoinositide-Specific Phospholipase C in Phosphoinositide Metabolism and the Endoplasmic Reticulum Stress Response
- Bridges Meristem and Organ Primordia Boundaries through , , and during Flower Development in
- KLK5 Inactivation Reverses Cutaneous Hallmarks of Netherton Syndrome
- XBP1-Independent UPR Pathways Suppress C/EBP-β Mediated Chondrocyte Differentiation in ER-Stress Related Skeletal Disease
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



