-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Crossing the Interspecies Barrier: Opening the Door to Zoonotic Pathogens
article has not abstract
Published in the journal: Crossing the Interspecies Barrier: Opening the Door to Zoonotic Pathogens. PLoS Pathog 10(6): e32767. doi:10.1371/journal.ppat.1004129
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004129Summary
article has not abstract
Zoonotic Pathogens
The number of pathogens known to infect humans is ever increasing. Whether such increase reflects improved surveillance and detection or actual emergence of novel pathogens is unclear. Nonetheless, infectious diseases are the second leading cause of human mortality and disability-adjusted life years lost worldwide [1], [2]. On average, three to four new pathogen species are detected in the human population every year [3]. Most of these emerging pathogens originate from nonhuman animal species.
Zoonotic pathogens represent approximately 60% of all known pathogens able to infect humans [4]. Their occurrence in humans relies on the human-animal interface, defined as the continuum of contacts between humans and animals, their environments, or their products. The human-animal interface has existed since the first footsteps of the human species and its hominin ancestors 6–7 million years ago, promoting the prehistoric emergence of now well-established human pathogens [5]. These presumably include pathogens with roles in the origin of chronic diseases, such as human T-lymphotropic viruses and Helicobacter pylori, as well as pathogens causing major crowd diseases, such as the smallpox and measles viruses and Bordetella pertussis [5], [6]. Since prehistory, the human-animal interface has continued to evolve and expand, ever allowing new pathogens to access the human host and cross species barriers [5].
Species Barriers
The suitability of any species to act as a host to a particular pathogen varies due to both host species – and pathogen-dependent factors, which define the species barriers. The species barriers separating nonhuman animal species from humans and thus of concern for zoonotic pathogens are the focus of this paper. However, the proposed conceptual framework is applicable to any host-pathogen system.
The species barriers separating nonhuman animal species from humans represent a major hurdle for effective exposure to, infection by, and subsequent spread of zoonotic pathogens among humans [7]. Accordingly, these species barriers can be divided into three largely complementary sets. First, the interspecies barrier determines the nature and level of human exposure to zoonotic pathogens. Second, the intrahuman barrier determines the ability of zoonotic pathogens to productively infect a human host and effectively cope with the immune response. Third, the interhuman barrier determines the ability of zoonotic pathogens to efficiently transmit among humans, causing outbreaks, epidemics, or pandemics. Zoonotic pathogens may cross, more or less efficiently, one or more of these sets of barriers. Only pathogens that cross all barriers have the potential to sustainably establish in the human population.
Identifying the factors allowing pathogens to cross each of these three sets of barriers is essential to mitigate burdens of known and future emerging zoonotic pathogens. The interspecies barrier, by its nature, involves ecological processes driving animal and human population dynamics and interspecies contact. Prior attempts to define these factors or drivers started as early as 1992 [8]. Recent contributions in this field underlined the importance of landscape change and ecological alteration (e.g., [9]–[11]). Here, we build on these earlier studies to focus on identifying the factors affecting the interspecies barrier from a more holistic perspective, with the aim of developing a simple framework that classifies factors into a limited number of mutually exclusive categories acting at distinct spatial and temporal scales.
Conceptual Framework for Pathogen Emergence at the Interspecies Barrier
The emergence of zoonotic pathogens in humans is dependent on interactions between humans and infected animal reservoir and/or vector hosts or their environment (Figure 1, center). The extent of such interactions is influenced by the prevalence of zoonotic pathogens in the animal reservoir or vector populations, which is in turn influenced by these populations' health and immune status. In addition, the population dynamics of humans, animal reservoirs, and vectors drive ecological processes that govern pathogen abundance and spread, both within and among species [12]. Increased exposure of humans to animal pathogens can result from changes in the dynamics of any of these populations (Figure 1, inner circle). These changes can be divided into three categories: first, increased interspecies contact between humans and the animal reservoir and/or vector; second, population growth or aggregation of humans, animal reservoir, and/or vector; and third, their geographic range expansion, at least where this expansion involves overlapping ranges. Changes in one aspect of human, animal reservoir, or vector population dynamics may affect another; for example, population growth may accompany range expansion.
Fig. 1. Framework for the classification of drivers of human exposure to animal pathogens (interspecies barrier). 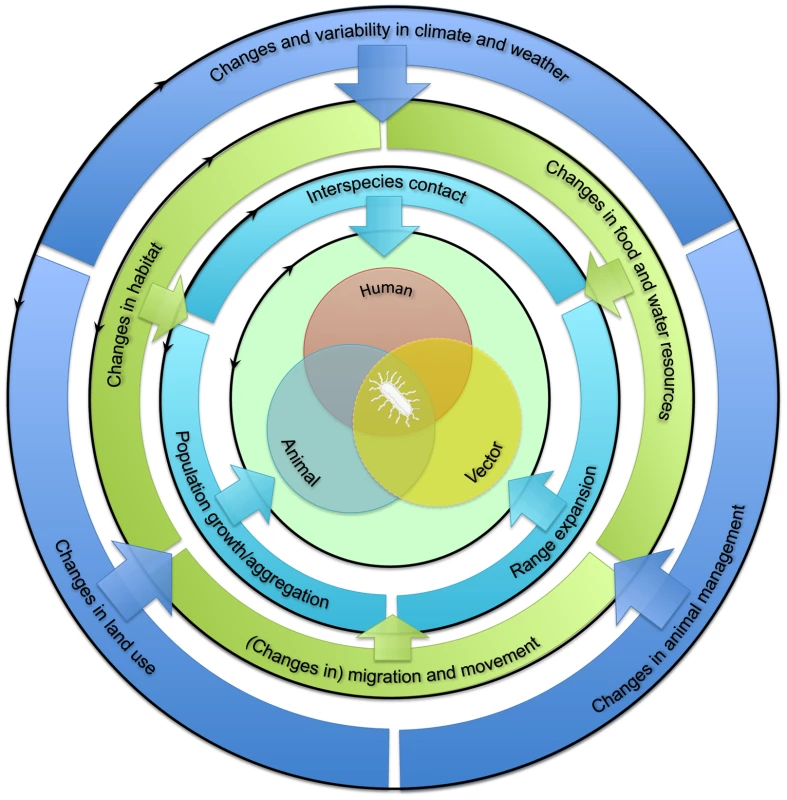
See text for more details. Factors influencing these changes in human, animal reservoir, and vector population dynamics may themselves be divided into two sets of drivers, acting at distinct scales. First, “proximate drivers” occurring at the local landscape scale are direct determinants of changes in human, animal reservoir, and vector population dynamics (Figure 1, middle circle; Table 1). These drivers may include habitat suitability, food and water resource availability, and short - or long-distance movements. The extent to which changes in these proximate drivers affect human, animal reservoir, and vector population dynamics depends greatly on the ecology of the species under consideration. For example, changes in habitat may favor generalist species but may drive specialist species to local extinction.
Tab. 1. Drivers for overcoming the interspecies barrier. 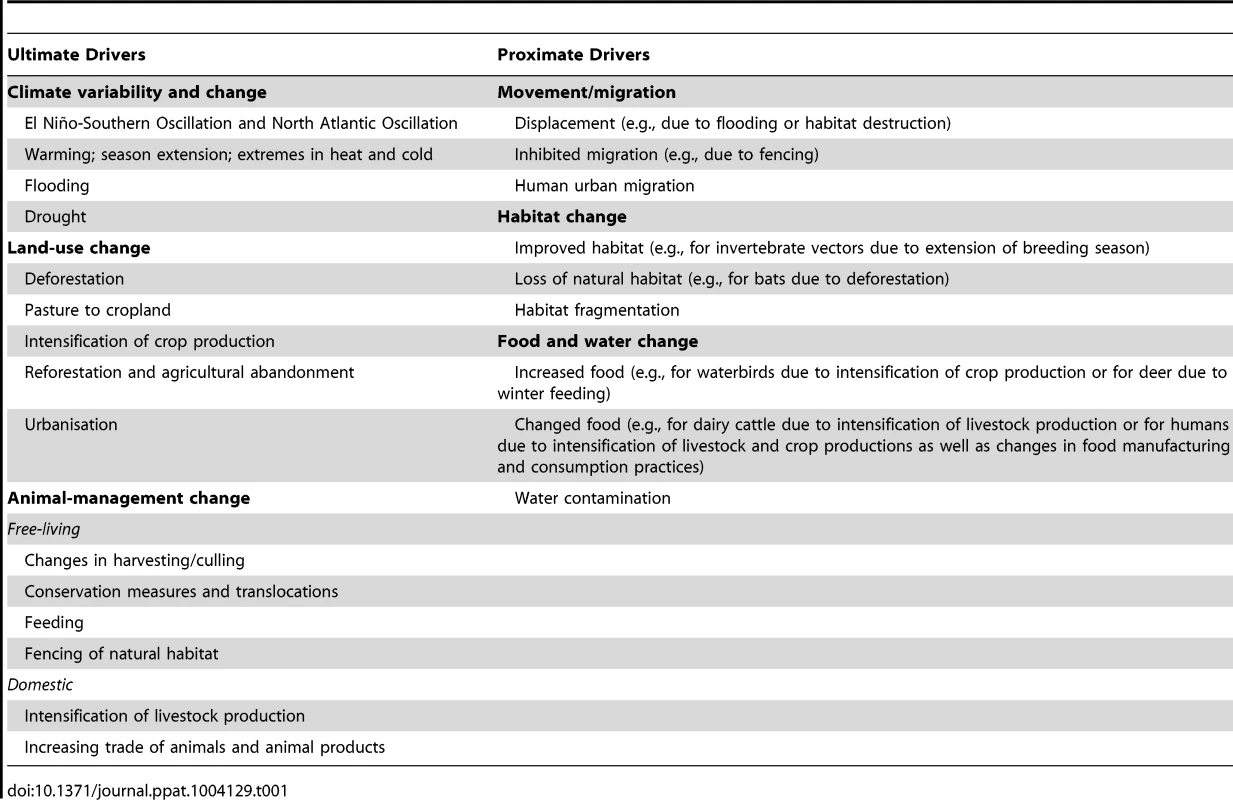
Second, “ultimate drivers” occurring at broader (regional or global) geographic scales temporally precede and govern changes in proximate drivers (Figure 1, outer circle; Table 1). These drivers include climate, land use, and animal management. Changes in these ultimate drivers may be either anthropogenic (human caused) or “natural.” They can promote changes in one or more proximate drivers. For example, changes in land use may affect both habitat suitability and availability of food and water. Together, the above framework allows the proposed underlying factors affecting the interspecies barrier to be categorized systematically (Figure 2).
Fig. 2. Examples of sets of drivers and ecological processes implicated in the emergence of zoonotic pathogens in humans. 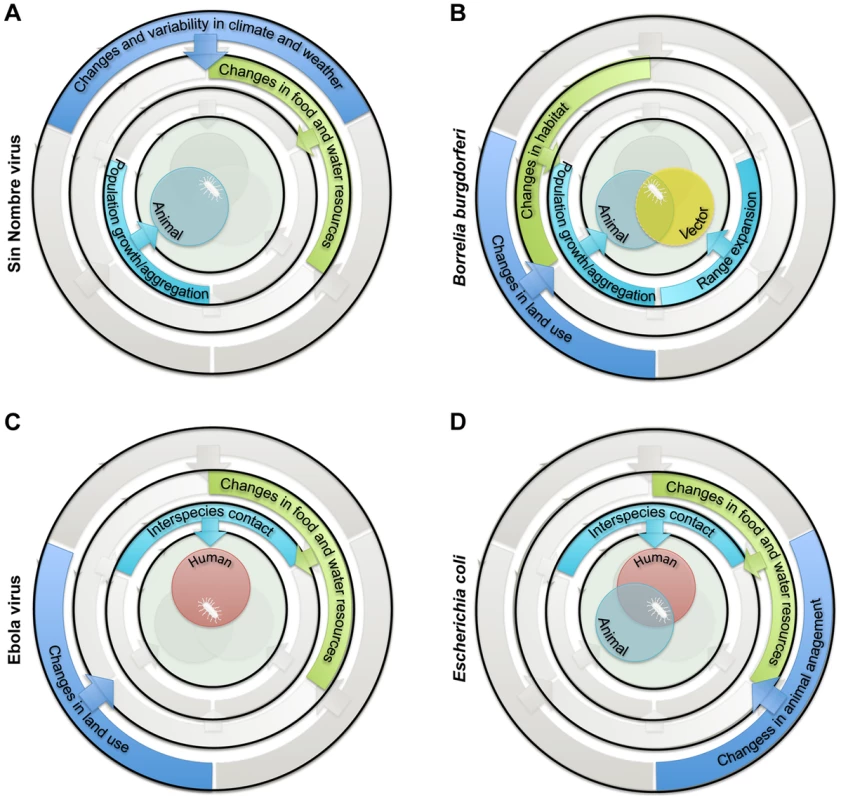
(A) Emergence of Sin Nombre virus in the Four Corners area of the United States in 1993 was attributed to population growth of the rodent reservoir (Peromyscus maniculatus) following increases in food resources (mast) associated with El Niño-Southern Oscillation events [16]. (B) Emergence of Borrelia burgdorferi in the eastern US in 1974 was attributed to population growth and range expansion of white-tailed-deer (Odocoileus virginianus) and black-legged-tick (Ixodes scapularis) vector populations following increase in suitable habitat due to reforestation and management encouraging high deer densities [17]. (C) Emergence of Ebola virus in the Democratic Republic of Congo (formerly Zaire) in 1976 was attributed to increases in interspecies contact between humans and primates following increases in bushmeat hunting and encroachment into undisturbed habitats [18]. (D) Emergence of enteric pathogenic Escherichia coli at the human-animal interface was attributed to increased direct and indirect human-animal contact following changes in the food chain and in water quality, often due to the intensification of livestock production [19]. Gaps in Current Knowledge of the Ecology of Zoonotic Pathogens
The proposed framework helps identify essential gaps in our understanding of the chain of emergence of zoonotic pathogens in humans and, in particular, of ecological processes underlying crossing of the interspecies barrier. Major gaps include characterization of the relationships between environmental conditions, especially climate and weather, and host and/or vector population dynamics, as well as exploration of pathogen survival and propagation in the environment. Recent studies have aimed at addressing such issues using novel approaches [13] and are essential in order to detect and predict associations between drivers such as climate change or weather variability and pathogen emergence [14], [15]. The current focus in ecology addresses primarily single host-pathogen systems and needs to be expanded to a multihost, multipathogen perspective. Interactions between host, vector, and pathogenic and nonpathogenic infectious agents likely play important roles in the dynamics of zoonotic pathogens at the human-animal interface [14]. Lastly, systematic assessment of actual human exposure to zoonotic pathogens, e.g., by serology, is lacking, calling for a more holistic approach to understanding the complete chain of emergence. Most evidence for the role of anthropogenic changes, e.g., encroachment into natural habitats, on zoonotic pathogen emergence is anecdotal or indirect and generally biased towards developed countries.
Future Perspectives
The identification of a limited number of mutually exclusive drivers of zoonotic pathogen emergence and of current knowledge gaps is essential to improve risk assessment and prevention measures. The links between pathogen emergence in humans and their underlying factors are typically speculative and associative and usually only account for a short section of the chain of emergence. Overall, knowledge of causal relationships between changes in population dynamics or interspecies contact, on the one hand, and pathogen emergence in humans, on the other, is fragmentary and incomplete at best. Existing studies in this area generally are limited in scope and typically lack quantitative assessment of human exposure to zoonotic pathogens at the human-animal interface.
The above proposed framework helps in understanding the common mechanisms behind disease emergence by linking pathogen emergence in humans to distinct and well-defined proximate and ultimate drivers. Hence, it may be used to further identify and quantify associations, causal relationships, and risks between ecological changes and pathogen emergence. In the full sense of the One Health concept, it can serve to help optimize efforts to manage disease emergence and spread in the interests of humans, food safety, and biodiversity.
Zdroje
1. FauciAS (2001) Infectious diseases: considerations for the 21st century. Clin Infect Dis 2001 32 : 675–685.
2. MurrayCJ, VosT, LozanoR, NaghaviM, FlaxmanAD, et al. (2012) Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380 : 2197–2223.
3. WoolhouseM, ScottF, HudsonZ, HoweyR, Chase-ToppingM (2012) Human viruses: discovery and emergence. Philos Trans R Soc Lond B Biol Sci 367 : 2864–2871.
4. TaylorLH, LathamSM, WoolhouseME (2001) Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 356 : 983–989.
5. ReperantLA, CornagliaG, OsterhausADME (2013) The importance of understanding the human-animal interface. From early hominins to global citizens. Curr Top Microbiol Immunol 365 : 49–81.
6. WolfeND, DunavanCP, DiamondJ (2007) Origins of major human infectious diseases. Nature 447 : 279–283.
7. KuikenT, HolmesEC, McCauleyJ, RimmelzwaanGF, WilliamsCS, et al. (2006) Host species barriers to influenza virus infections. Science 312 : 394–397.
8. IOM (1992) Emerging infections: microbial threats to health in the United States. In: Lederberg J, Shope RE, Oaks SC, editors. Washington (D.C.): Institute of Medicine, National Academy of Sciences. 312 p.
9. PatzJA, DaszakP, TaborGM, AguirreAA, PearlM, et al. (2004) Unhealthy landscapes: Policy recommendations on land use change and infectious disease emergence. Environ Health Perspect 112 : 1092–1098.
10. WoolhouseM, GauntE (2007) Ecological origins of novel human pathogens. Crit Rev Microbiol 33 : 231–242.
11. MyersSS, GaffikinL, GoldenCD, OstfeldRS, RedfordKH, et al. (2013) Human health impacts of ecosystem alteration. Proc Natl Acad Sci U S A 110 : 18753–18760.
12. ReperantLA (2010) Applying the theory of island biogeography to emerging pathogens: toward predicting the sources of future emerging zoonotic and vector-borne diseases. Vector Borne Zoonotic Dis 10 : 105–110.
13. MolnarPK, KutzSJ, HoarBM, DobsonAP (2013) Metabolic approaches to understanding climate change impacts on seasonal host-macroparasite dynamics. Ecol Lett 16 : 9–21.
14. AltizerS, OstfeldRS, JohnsonPT, KutzS, HarvellCD (2013) Climate change and infectious diseases: from evidence to a predictive framework. Science 341 : 514–519.
15. SemenzaJC, MenneB (2009) Climate change and infectious diseases in Europe. Lancet Infect Dis 9 : 365–375.
16. EngelthalerDM, MosleyDG, CheekJE, LevyCE, KomatsuKK, et al. (1999) Climatic and environmental patterns associated with hantavirus pulmonary syndrome, Four Corners region, United States. Emerg Infect Dis 5 : 87–94.
17. BarbourAG, FishD (1993) The biological and social phenomenon of Lyme disease. Science 260 : 1610–1616.
18. WolfeND, DaszakP, KilpatrickAM, BurkeDS (2005) Bushmeat hunting, deforestation, and prediction of zoonoses emergence. Emerg Infect Dis 11 : 1822–1827.
19. BeutinL, HammerlJA, ReetzJ, StrauchE (2013) Shiga toxin-producing Escherichia coli strains from cattle as a source of the Stx2a bacteriophages present in enteroaggregative Escherichia coli O104:H4 strains. Int J Med Microbiol 303 : 595–602.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Recruitment of RED-SMU1 Complex by Influenza A Virus RNA Polymerase to Control Viral mRNA SplicingČlánek Systematic Phenotyping of a Large-Scale Deletion Collection Reveals Novel Antifungal Tolerance GenesČlánek The Contribution of Social Behaviour to the Transmission of Influenza A in a Human Population
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 6- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Fungal Nail Infections (Onychomycosis): A Never-Ending Story?
- BdlA, DipA and Induced Dispersion Contribute to Acute Virulence and Chronic Persistence of
- Morphotype Transition and Sexual Reproduction Are Genetically Associated in a Ubiquitous Environmental Pathogen
- A Nucleic-Acid Hydrolyzing Single Chain Antibody Confers Resistance to DNA Virus Infection in HeLa Cells and C57BL/6 Mice
- HopW1 from Disrupts the Actin Cytoskeleton to Promote Virulence in Arabidopsis
- Ly6C Monocytes Become Alternatively Activated Macrophages in Schistosome Granulomas with Help from CD4+ Cells
- Recruitment of RED-SMU1 Complex by Influenza A Virus RNA Polymerase to Control Viral mRNA Splicing
- Contribution of Specific Residues of the β-Solenoid Fold to HET-s Prion Function, Amyloid Structure and Stability
- Antibody Responses to : Role in Pathogenesis and Diagnosis of Encephalitis?
- Discovery of a Novel Compound with Anti-Venezuelan Equine Encephalitis Virus Activity That Targets the Nonstructural Protein 2
- Activation of Focal Adhesion Kinase by Suppresses Autophagy via an Akt/mTOR Signaling Pathway and Promotes Bacterial Survival in Macrophages
- Crossing the Interspecies Barrier: Opening the Door to Zoonotic Pathogens
- Catching Fire: , Macrophages, and Pyroptosis
- IscR Is Essential for Type III Secretion and Virulence
- Selective Chemical Inhibition of Quorum Sensing in Promotes Host Defense with Minimal Impact on Resistance
- The Glycosylated Rv1860 Protein of Inhibits Dendritic Cell Mediated TH1 and TH17 Polarization of T Cells and Abrogates Protective Immunity Conferred by BCG
- A Genome-Wide Tethering Screen Reveals Novel Potential Post-Transcriptional Regulators in
- Structural Insights into SraP-Mediated Adhesion to Host Cells
- Human IGF1 Regulates Midgut Oxidative Stress and Epithelial Homeostasis to Balance Lifespan and resistance in
- Cycling Empirical Antibiotic Therapy in Hospitals: Meta-Analysis and Models
- Rab11 Regulates Trafficking of -sialidase to the Plasma Membrane through the Contractile Vacuole Complex of
- Mitogen and Stress Activated Kinases Act Co-operatively with CREB during the Induction of Human Cytomegalovirus Immediate-Early Gene Expression from Latency
- Profilin Promotes Recruitment of Ly6C CCR2 Inflammatory Monocytes That Can Confer Resistance to Bacterial Infection
- A Central Role for Carbon-Overflow Pathways in the Modulation of Bacterial Cell Death
- An Invertebrate Warburg Effect: A Shrimp Virus Achieves Successful Replication by Altering the Host Metabolome via the PI3K-Akt-mTOR Pathway
- The Highly Conserved Bacterial RNase YbeY Is Essential in , Playing a Critical Role in Virulence, Stress Regulation, and RNA Processing
- A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees () Prevails after -Mediated, or , Transmission
- Systematic Phenotyping of a Large-Scale Deletion Collection Reveals Novel Antifungal Tolerance Genes
- Ubiquitin-Mediated Response to Microsporidia and Virus Infection in
- Preclinical Detection of Variant CJD and BSE Prions in Blood
- Toll-Like Receptor 8 Agonist and Bacteria Trigger Potent Activation of Innate Immune Cells in Human Liver
- Progressive Proximal-to-Distal Reduction in Expression of the Tight Junction Complex in Colonic Epithelium of Virally-Suppressed HIV+ Individuals
- The Triggering Receptor Expressed on Myeloid Cells 2 Inhibits Complement Component 1q Effector Mechanisms and Exerts Detrimental Effects during Pneumococcal Pneumonia
- Differential Activation of Acid Sphingomyelinase and Ceramide Release Determines Invasiveness of into Brain Endothelial Cells
- Forward Genetic Screening Identifies a Small Molecule That Blocks Growth by Inhibiting Both Host- and Parasite-Encoded Kinases
- Defining Immune Engagement Thresholds for Control of Virus-Driven Lymphoproliferation
- Growth Factor and Th2 Cytokine Signaling Pathways Converge at STAT6 to Promote Arginase Expression in Progressive Experimental Visceral Leishmaniasis
- Multimeric Assembly of Host-Pathogen Adhesion Complexes Involved in Apicomplexan Invasion
- Biogenesis of Influenza A Virus Hemagglutinin Cross-Protective Stem Epitopes
- Adequate Th2-Type Response Associates with Restricted Bacterial Growth in Latent Mycobacterial Infection of Zebrafish
- Protective Efficacy of Passive Immunization with Monoclonal Antibodies in Animal Models of H5N1 Highly Pathogenic Avian Influenza Virus Infection
- Fructose-Asparagine Is a Primary Nutrient during Growth of in the Inflamed Intestine
- The Calcium-Dependent Protein Kinase 3 of Influences Basal Calcium Levels and Functions beyond Egress as Revealed by Quantitative Phosphoproteome Analysis
- A Translocated Effector Required for Dissemination from Derma to Blood Safeguards Migratory Host Cells from Damage by Co-translocated Effectors
- Functional Characterization of a Novel Family of Acetylcholine-Gated Chloride Channels in
- Both α2,3- and α2,6-Linked Sialic Acids on O-Linked Glycoproteins Act as Functional Receptors for Porcine Sapovirus
- The Contribution of Social Behaviour to the Transmission of Influenza A in a Human Population
- MicroRNA-146a Provides Feedback Regulation of Lyme Arthritis but Not Carditis during Infection with
- Recombination in Enteroviruses Is a Biphasic Replicative Process Involving the Generation of Greater-than Genome Length ‘Imprecise’ Intermediates
- Cytoplasmic Viral RNA-Dependent RNA Polymerase Disrupts the Intracellular Splicing Machinery by Entering the Nucleus and Interfering with Prp8
- and Are Associated with Murine Susceptibility to Infection and Human Sepsis
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Fungal Nail Infections (Onychomycosis): A Never-Ending Story?
- Profilin Promotes Recruitment of Ly6C CCR2 Inflammatory Monocytes That Can Confer Resistance to Bacterial Infection
- Contribution of Specific Residues of the β-Solenoid Fold to HET-s Prion Function, Amyloid Structure and Stability
- The Highly Conserved Bacterial RNase YbeY Is Essential in , Playing a Critical Role in Virulence, Stress Regulation, and RNA Processing
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



