-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Structural Insights into SraP-Mediated Adhesion to Host Cells
Staphylococcus aureus is an important pathogen that causes a range of human diseases, such as infective endocarditis, osteomyelitis, septic arthritis and sepsis. The increasing resistance of S. aureus to most of the current antibiotics emphasizes the need to develop new approaches to control staphylococcal infections. As a surface-exposed serine-rich repeat glycoprotein (SRRP), S. aureus SraP is involved in the pathogenesis of infective endocarditis via its ligand-binding region (BR) adhering to human platelets. However, little is known about how SraP interacts with its host receptor(s). Through structural and functional analyses of the BR domain, we have discovered a specific binding of SraP to N-acetylneuraminic acid (Neu5Ac), in agreement with a recent report of the trisaccharide ligand of SraP. Further mutagenesis analysis showed that SraP binding to Neu5Ac and the trisaccharide promotes S. aureus adhesion to and invasion into host epithelial cells. These findings increase our knowledge of surface protein mediated interaction of S. aureus with host epithelial cells.
Published in the journal: Structural Insights into SraP-Mediated Adhesion to Host Cells. PLoS Pathog 10(6): e32767. doi:10.1371/journal.ppat.1004169
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004169Summary
Staphylococcus aureus is an important pathogen that causes a range of human diseases, such as infective endocarditis, osteomyelitis, septic arthritis and sepsis. The increasing resistance of S. aureus to most of the current antibiotics emphasizes the need to develop new approaches to control staphylococcal infections. As a surface-exposed serine-rich repeat glycoprotein (SRRP), S. aureus SraP is involved in the pathogenesis of infective endocarditis via its ligand-binding region (BR) adhering to human platelets. However, little is known about how SraP interacts with its host receptor(s). Through structural and functional analyses of the BR domain, we have discovered a specific binding of SraP to N-acetylneuraminic acid (Neu5Ac), in agreement with a recent report of the trisaccharide ligand of SraP. Further mutagenesis analysis showed that SraP binding to Neu5Ac and the trisaccharide promotes S. aureus adhesion to and invasion into host epithelial cells. These findings increase our knowledge of surface protein mediated interaction of S. aureus with host epithelial cells.
Introduction
The serine-rich repeat glycoproteins (SRRPs) are a family of adhesins encoded by Gram-positive bacteria that mediate attachment to a variety of host cells or bacteria themselves [1]. SRRPs typically consist of a signal peptide at the N-terminus, a short SRR (SRR1, ∼50–170 residues), a ligand-binding region (BR, ∼250–500 residues) followed by a much longer SRR (SRR2, ∼400–4000 residues), and a C-terminal LPXTG motif anchoring to the cell wall [1]. The BRs of SRRPs from different pathogenic bacteria have varying primary sequences and bind to diverse targets from carbohydrates to proteins [1].
In addition to having highly variable sequences, the BRs from different bacteria are composed of distinct modules. The diversity of BR modules and combinations contributes to the multiple functions of SRRPs. The only four known BR structures to date have identified five distinct modules [2]–[4]. The BR of Streptococcus parasanguinis Fap1 contains two modules: an N-terminal helical module and a C-terminal CnaA module [2], whereas Streptococcus gordonii GspB has a BR of three modules: CnaA, Siglec and a unique module of unknown function [3]. In addition, the recently reported BR structures of the two SRRP paralogs (Srr1 and Srr2) from Streptococcus agalactiae defined two immunoglobulin-fold modules, which specifically bind to the host fibrinogen [4]. However, the module composition and corresponding molecular functions of most BRs remain unknown, which largely impedes the understanding of the pathogenesis mechanism of SRRPs.
S. aureus is a human pathogen that causes a wide range of debilitating and life-threatening infections [5]. S. aureus encodes a 2,271-residue SRRP termed serine-rich adhesin for binding to platelets (SraP), that is involved in the pathogenesis of infective endocarditis [6]. Moreover, the BR (residues Phe245–Asn751) of SraP, termed SraPBR, mediates intraspecies interaction and promotes bacterial aggregation [7]. We determine the 2.05 Å crystal structure of SraPBR, revealing a rod-like tandem organization of four discrete modules: a legume lectin-like module, a module with a β-grasp fold, and two tandem cadherin-like modules that create the rigid stem of SraPBR. Further structural and biochemical analyses reveal that the legume lectin-like module specifically binds to N-acetylneuraminic acid (Neu5Ac), which may mediate adhesion to host sialylated receptors. These findings increase our knowledge of the diverse BR modules of SRRPs, and provide structural insights into a novel surface protein that mediates interaction of S. aureus with host epithelial cells.
Results
Overall structure of SraPBR
Each asymmetric unit of the final model at 2.05 Å resolution contains a single SraPBR molecule of residues Thr251–Asn751. The N-terminal residues Phe245–Thr250 are not visible due to their poor electron density. SraPBR folds into a slightly bent, rod-like structure of 160 Å in length that has four discrete modules: a head-like N-terminal module followed a stem of three all-β modules (Fig. 1A). All modules have a dominant β-strand secondary structure. During the model building and refinement process, three peaks of electron density at the 24 σ level were observed in the |Fo|−|Fc| Fourier difference map, indicating the presence of three metal ions. Atomic absorption spectroscopy assigned these metal ions to Ca2+; we thus termed them Ca-1, Ca-2 and Ca-3 accordingly. The structure also contains a sucrose and a 2-(N-morpholino)-ethanesulfonic acid (MES) molecule, which were introduced from the cryoprotectant and crystallization buffer, respectively.
Fig. 1. Overall structure of the full-length SraPBR. 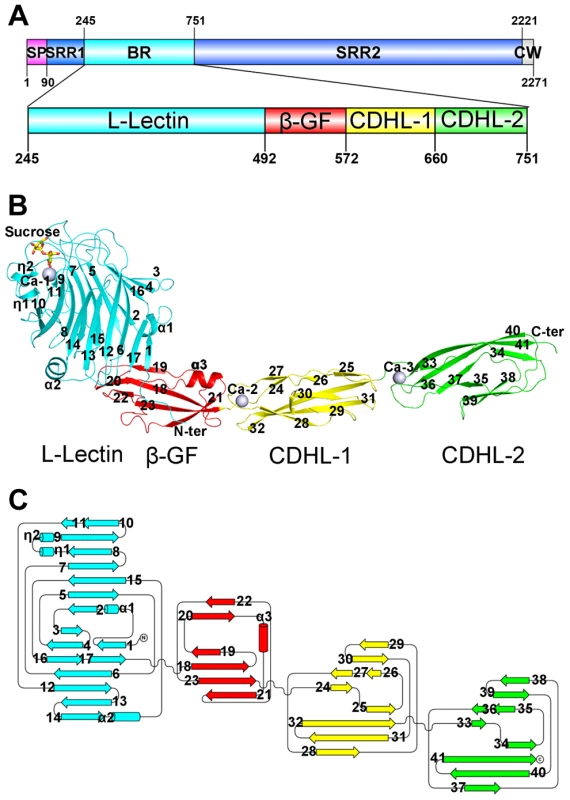
A) A diagram to show the organization of SraP and BR. B) Cartoon representation of the overall structure of SraPBR. C) A topology diagram of SraPBR. The L-lectin, β-GF, CDHL-1 and CDHL-2 modules are shown in cyan, red, yellow and green, respectively. The sucrose molecule is shown as yellow stick and the bound calcium ions are shown as gray spheres. The secondary structural elements are labeled sequentially. The N-terminal module adopts a jelly-roll fold with a β-sandwich (β1–β17) core architecture of two antiparallel β-sheets packed against each other (Fig. 1B). Beyond the core structure, two α-helices (α1 and α2) pack on either side of the lateral of the β-sandwich and partially seal the hydrophobic lateral openings between the two β-sheets. A molecule of sucrose binds to the protruding loops at the distal end of the N-terminal module (Fig. 1B). A structural similarity search using the DALI server [8] revealed that this module is most similar to legume lectins, despite sharing a sequence identity of ≤20%. The top hits include lectins from legume plants such as Pisum sativum (PDB 2BQP) [9] and Robinia pseudoacacia (PDB: 1FNY) [10], with a Z-score of 22–23 and root mean square deviation (RMSD) of 2.3–2.5 Å over ∼200 Cα atoms. Thus, we termed the N-terminal module L-lectin.
The second module possesses a ubiquitin-like β-grasp fold (β-GF) in the immunoglobulin (Ig)-binding protein superfamily [11]. The module contains a twisted four-stranded mixed β-sheet (β19-β18-β23-β21) packed against a two-stranded antiparallel β-sheet (β20–β22) and an α-helix α3 (Fig. 1B).
The C-terminal tandem repeat of two modules share a sequence identity of 56% and a similar overall structure with an RMSD of 0.81 Å over 81 Cα atoms. The two modules are linked in a head-to-tail fashion, indicating duplication of the coding region during evolution. Each module consists of a β-sandwich of three β-sheets (Fig. 1B & 1C). Structural analysis revealed that the two modules resemble eukaryotic cadherins of known-structure (PDB 3UBF, Z-score 8.6, RMSD 2.3 Å over 85 Cα atoms and PDB 4APX, Z-score 9.5, RMSD 2.3 Å over 87 Cα atoms) [12], [13]. Thus the tandem cadherin-like (CDHL) modules were termed CDHL-1 and CDHL-2, respectively.
Specific binding of the L-lectin module to N-acetylneuraminic acid
The L-lectin module is structurally similar to legume lectins (Fig. 2A), a large family of carbohydrate-binding proteins with diverse activities [14]. Legume lectins commonly coordinate a Ca2+ in addition to a transition metal ion, usually Mn2+ [15]. In contrast, the L-lectin module has only a Ca2+-binding site. At the apex of the L-lectin module, the Ca2+ (named Ca-1) was embedded in a 7-coordinate geometry (Fig. 2B). The seven coordinates are from the side-chain oxygen atoms of Asp365 (bidentate coordination from Oδ1 and Oδ2), Asp382 (Oδ2), Asn369 (Oδ1), the main-chain oxygen atom of Tyr367 and two water molecules (Wat1 and Wat2). The two water molecules were further stabilized by the main-chain oxygen atoms of Asp330, Ala349, Oδ1 of Asp382 and another water molecule (Wat3) (Fig. 2B).
Fig. 2. Binding of the L-lectin module to Neu5Ac. 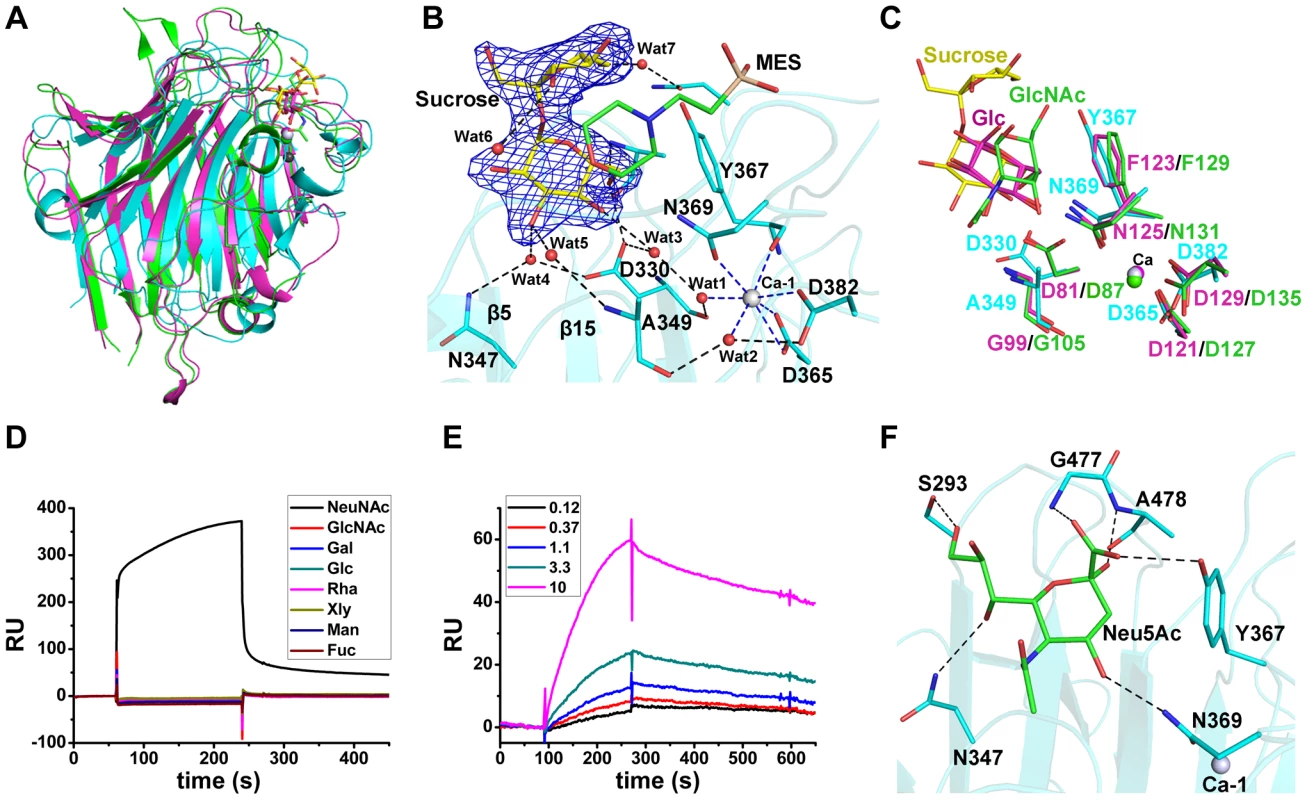
A) Superposition of the L-lectin module against P. sativum pea lectin (PDB 2BQP) and R. pseudoacacia bark lectin (PDB 1FNZ). The L-lectin module of SraPBR is colored in cyan, with the carbons of sucrose colored in yellow. The pea and bark lectins are colored in magenta and green, respectively. B) The sucrose-binding site and the Ca-1 coordination. The |Fo|-|Fc| omit map of sucrose is shown as blue mesh and countered at 3.0 σ. The sucrose and MES molecules and their interacting residues are shown as sticks and colored in yellow, green and cyan, respectively. The water molecules are shown as red spheres and Ca-1 is shown as grey sphere. The polar interactions with sucrose are shown as black dotted lines, whereas the coordinate bonds with Ca-1 are indicated by blue dotted lines. C) Comparison of the ligand binding sites of L-lectin module (cyan), pea lectin (magenta) and bark lectin (green). Residues binding to the ligand are shown in sticks. D) SPR sensor diagram for the eight tested monosaccharides (D-glucose, D-galactose, D-mannose, L-fucose, N-acetyl-D-glucosamine, N-acetylneuraminic acid, D-xylose, and L-Rhamnose) against the immobilized L-lectin module. E) The steady-state equilibrium curve of Neu5Ac at a concentration varying from 0.12 to 10 mM. The binding affinity of the L-lectin module towards Neu5Ac was calculated by using the 1∶1 Langmuir model, the fit of model to the data was determined by an χ2 value, and the χ2 is 8.98. F) A docking model of Neu5Ac (green sticks) binding to the L-lectin module (cyan). Polar interactions are shown in black dashed lines. A molecule of sucrose is fixed by a cluster of loops protruding from the apex of the L-lectin module. The sucrose molecule has a “bent-back” conformation with the glucose and fructose moieties perpendicular to each other (Fig. 2B). The glucose moiety is inserted in the pocket, and adopts a conformation nearly parallel to the two β-sheet layers. The sugar ring makes hydrophobic interactions with Tyr367 and the morpholine ring of MES, whereas the hydroxyl groups are stabilized by hydrogen bonds with Ala478-Nα, Asp330-Oδ1, and three water molecules (Wat3–5), which are further fixed by residues Asp330, Asn347 and Ala349 (Fig. 2B). In contrast, the solvent-exposed fructose moiety is bent through interactions with MES and two water molecules (Wat6 and Wat7) (Fig. 2B). The sucrose binding residues, especially the stacking residue Tyr367, are structurally conserved in the legume lectins (Fig. 2C).
To identify the favored saccharide of SraP, we first detected the binding affinity of the L-lectin module towards eight common monosaccharides using the surface plasmon resonance assays. Among these monosaccharides, only Neu5Ac bound to the L-lectin module (Fig. 2D). In consequence, we determined that the L-lectin module has an equilibrium dissociation constant (Kd) of 0.54 mM towards Neu5Ac (Fig. 2E), comparable to previously reported values of legume lectins [16].
Afterwards, we attempted to obtain the Neu5Ac-complexed structure without success. Therefore, we docked Neu5Ac to the structure of the L-lectin module with the sucrose-binding pocket as the search grid. Neu5Ac was docked at a position overlapping the glucose moiety of sucrose, with a shift of ∼1.5 Å towards Ca-1. In the model, Neu5Ac is stacked against Tyr367 via hydrophobic interactions, and makes direct polar interactions with the side chains of Ser293, Asn347, Tyr367 and Asn369 and the Nα atoms of Gly477 and Ala478 (Fig. 2F).
Structural similarity of the β-GF module to Ig-binding proteins
The β-GF module adopts a ubiquitin-like β-grasp fold in the Ig-binding superfamily [11]. DALI search [8] suggested the module resembles the B1 domain of mucus-binding protein type 2 repeat Mub-R5 from Lactobacillus reuteri [17] (PDB code 3I57, Z-score 9.1, RMSD 1.6 Å, over 68 Cα atoms), and protein L (PpL) from Peptostreptococcus magnus [18] (PDB code 1HEZ, Z-score 4.5, RMSD 2.5, over 55 Cα atoms). The B1 domain belongs to a family of Ig-binding proteins [19] that have a core structure of an α-helix packed against a four-stranded β-sheet [17], [20]. The major differences are from the helix α3 and the two lateral β-strands (β19 and β21). In addition, the β-GF module of SraPBR contains two extra β-strands, β20 and β22, which are substituted by loops or α-helix extensions in the B1 domain (Fig. 3). Mub-R5 interacts in vitro with a large repertoire of mammalian Ig proteins including secretory IgA, whereas PpL binds to the VL domain of Ig κ chain [19], [21]. Complex structures indicated that formation of a β-zipper is necessary for the binding of PpL to the VL domain of Ig κ chain [18] and IgG or IgM [22]. However, the corresponding β-strands β19 and β21 are much shorter in the β-GF module, which might not be capable of forming a β-zipper.
Fig. 3. Superpositions of the β-GF module against A) the Ig-binding proteins of B1 domain of mucus-binding protein type 2 repeat Mub-R5 from L. reuteri (PDB 3I57), and B) that of Protein L (PpL) from P. magnus (PDB 1HEZ). 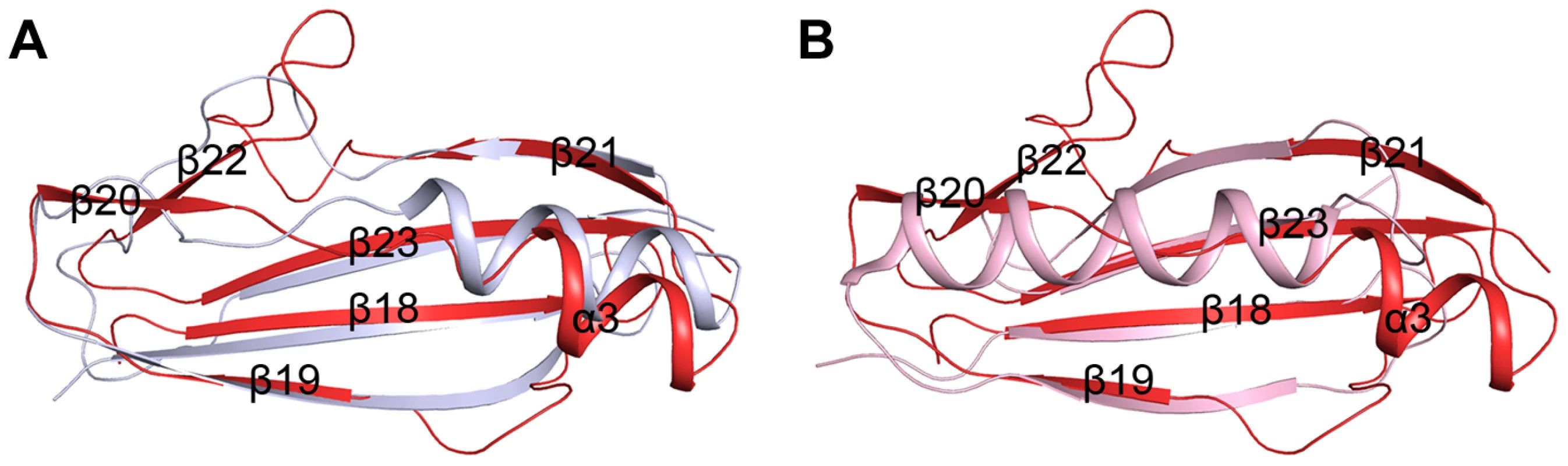
The β-GF module, the B1 domain and PpL are colored in red, grey and light pink, respectively. The tandem cadherin-like modules create a relatively rigid SraPBR stem
The tandem CDHL modules resemble eukaryotic cadherins in a superfamily of calcium-dependent adhesions (Fig. 4A & 4B). Unlike the eukaryotic cadherins, each of which coordinates three Ca2+ ions (PDB: 1L3W) [23], CDHL-1 and CDHL-2 binds to Ca-2 and Ca-3 with a 7-coordinate geometry, respectively. Ca-2 is fixed by Asp573 and Asp601 (bidentate coordination from Oδ1 and Oδ2), Asn602 (Oδ1), Asp645 (Oδ2), and the main-chain oxygen atom of Lys575 (Fig. 4C), whereas Ca-3 is coordinated to Asp661 and Asp690 (bidentate coordination from Oδ1 and Oδ2), Asn691 (Oδ1), Asp734 (Oδ2), and the main-chain oxygen atom of Thr663 (Fig. 4D). The coordinate bonds between Ca2+ and corresponding residues have a length from 2.2 to 2.6 Å.
Fig. 4. Structural comparison and the calcium coordination patterns of the CDHL modules. 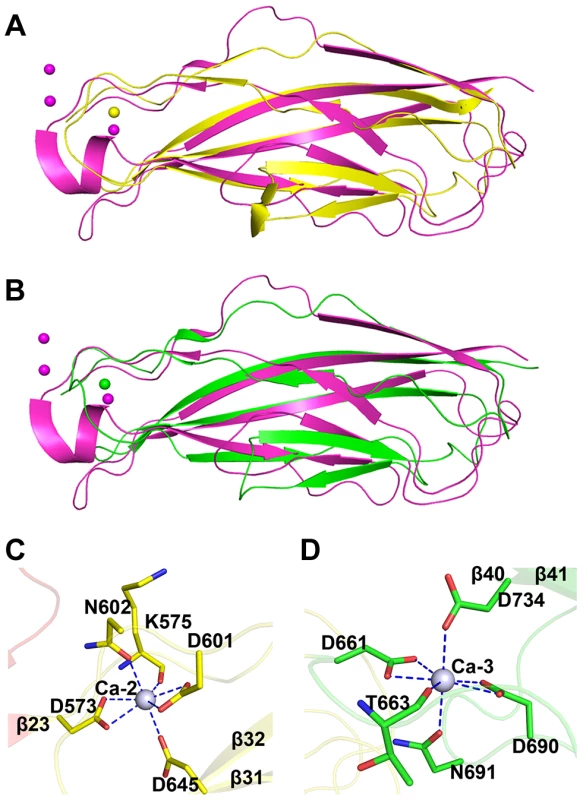
Structural comparison of A) CDHL-1 and B) CDHL-2 to the classic cadherin (PDB 3MVS). The CDHL-1 module, CDHL-2 module and the classic cadherin and corresponding binding calcium ions are shown in yellow, green and red, respectively. The coordination patterns of C) Ca-2 and D) Ca-3. The coordinate residues are presented in green and yellow sticks, respectively. The coordinate bonds are indicated as blue dotted lines. The small-angle scattering of X-rays (SAXS) is usually applied to address the flexibility and conformational states of biological macromolecules in solution [24]. To explore the role of Ca2+, we used SAXS to compare the overall structure of SraPBR in the presence or absence of Ca2+. A difference in wide-angle scattering curves of SAXS indicated that SraPBR adopts different conformations with or without Ca2+. Compared to the Ca2+-free form, the envelope of Ca2+-bound SraPBR correlates much better with the SraPBR crystal structure (Fig. 5A). The larger discrepancies of the Ca2+-free SraPBR are mainly resulted from the tandem CDHL modules, indicating that binding of Ca2+ makes the tandem CDHL modules more rigid, and thereby facilitates the extended conformation of SraPBR in solution.
Fig. 5. The relative rigid conformation of SraPBR upon binding to calcium. 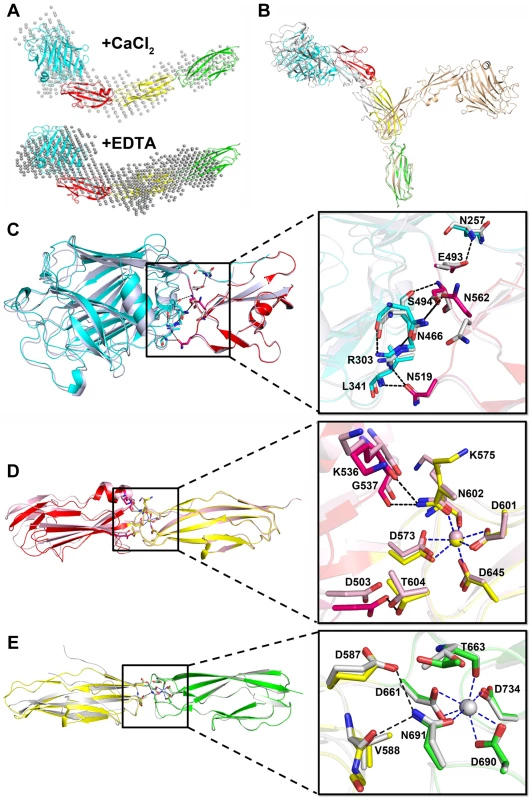
A) The SAXS data and envelops for SraPBR in the presence or absence of Ca2+. B) The final states of the 20-ns molecular dynamics simulation of SraPBR in the presence (gray) or absence (lightpink) of Ca2+, compared to the crystal structure of SraPBR with the CDHL-2 modules superimposed. Superposition of C) L-lectin&β-GF, D) β-GF&CDHL-1 and E) CDHL-1&2 to the corresponding modules in the full-length SraPBR structure, respectively. Residues at the interface are shown as sticks, polar interactions are indicated as black dotted lines and the coordinate bonds with Ca-2 and Ca-3 are indicated as blue dotted lines. We also performed molecular dynamics simulations of SraPBR in the presence or absence of Ca2+. Ca2+-bound SraPBR remains extended and shows slight conformational changes over the simulation time, whereas removal of Ca2+ caused the curling of SraPBR (Fig. 5B). Geometric analysis of the Ca2+-coordinating residues revealed larger fluctuations at the two Ca2+-binding junctions of Ca2+-free SraPBR, suggesting the importance of Ca2+ for the structural integrity of SraPBR. Moreover, the increased RMSD values indicated the two junctions in Ca2+-free SraPBR undergo dramatic conformational changes. Given the CDHL-2 modules superimposed, the other three modules adopt a more curved conformation and are projected to the opposite side upon the loss of Ca2+ (Fig. 5B).
The rod-like, four-module structure of SraPBR has three junctions with a buried interface of 700, 400 and 220 Å2, respectively (Fig. 1B). The interface between L-lectin and β-GF is maintained by several hydrogen bonds including Asn257-Glu493, Arg303-Asn519, Arg303-Ser494, Leu341-Asn519, and Asn466-Asn562 (Fig. 5C). The second interface is formed by residues from β-GF and CDHL-1 via hydrogen bonds of Lys536-Asn602, Gly537-Asn602 and Asp503-Thr604 (Fig. 5D). At the third junction, Glu587 and Val588 of CDHL-1 form two hydrogen bonds with Asn691 of CDHL-2 (Fig. 5E). The second and third interfaces are relatively small; however, they are more stable due to the contribution from coordinate bonds of Ca-2 to Asn602 and Ca-3 to Asn691.
To further investigate the plasticity of these junctions, we determined three crystal structures for each of two consecutive modules: L-lectin&β-GF (Phe245–Lys575), β-GF&CDHL-1 (Ser494–Thr663), and CDHL-1&2 (Ala576–Asn751). We superimposed each of these three structures over the corresponding two modules of the full-length SraPBR structure, always with the C-terminal modules aligned. The results revealed slight twisting and/or translation with intermodule angle changes of 9.4°, 11.7° and 14.6° for the three junctions, respectively (Fig. 5C∼E). In detail, residues Val496-Gln498 in L-lectin&β-GF, Phe571&Thr572 in β-GF&CDHL-1, and Asp661-Thr663 in CDHL-1&2 undergo slight conformational changes. Except for a short disordered segment at the N-terminus of CDHL-1&2 (Fig. 5E) due to the deletion of two Ca-2 coordinate residues Asp573 and Lys575, we did not find secondary-structure change in any module pairs. Moreover, merging these three structures via sequential alignment of the same module resulted in a total twist of 5.4° along the axis of SraPBR structure (Fig. S1), suggesting that the slight interdomain twists of the three junctions are randomly occurred and could be canceled out. Together, the results indicated that in the presence of calcium, SraPBR adopts a relatively rigid rod-like structure under all conditions tested.
SraP promotes S. aureus adhesion and invasion to A549 cells through sialylated receptors
Bacterial attachment and colonization at the surface of host cells have been thought to be mediated by specific binding of BRs to glycoconjugates [1]. It has recently been reported that SraP binds to the salivary agglutinin gp340 via the Neu5Ac moiety of the trisaccharide Neu5Acα(2–3)Galβ(1–4)GlcNAc [25]. In fact, gp340 and homologs are also expressed in lung epithelial cells [26]. To determine if the L-lectin module mediates SraPBR adhesion, we incubated a monolayer of human lung epithelial A549 cells with green fluorescent protein (GFP)-fused SraPBR and individual modules. The results indicated that full-length SraPBR and the L-lectin module, but not the CDHL-1&2 modules or the β-GF module, specifically adhered to the A549 cells (Fig. 6). Tyr367 is a key residue to make hydrophobic interactions with sucrose and the docked Neu5Ac, we thus constructed the Y367G mutant proteins to test the adhesion to A549 cells. As shown in the CD spectra, the mutation of Y367G did not introduce significant structural changes to SraPBR or the L-lectin module (Fig. S2). In contrast, a Y367G mutation in both the full-length SraPBR and the L-lectin module almost completely abolished the adhesion capacity. Moreover, the addition of 5 mM Neu5Ac completely inhibited the adhesion of SraPBR to the A549 cells (Fig. 6). Quantification of the fluorescent signal of three representative frames for each image further confirmed that only the full-length SraPBR and the L-lectin module are capable of specific binding to A549 cells (Fig. S3). These results demonstrate that the adhesion of SraPBR to A549 cells is mediated by the specific recognition of the L-lectin module towards Neu5Ac.
Fig. 6. Contribution of the L-lectin module to bacterial adhesion to human lung epithelial cells. 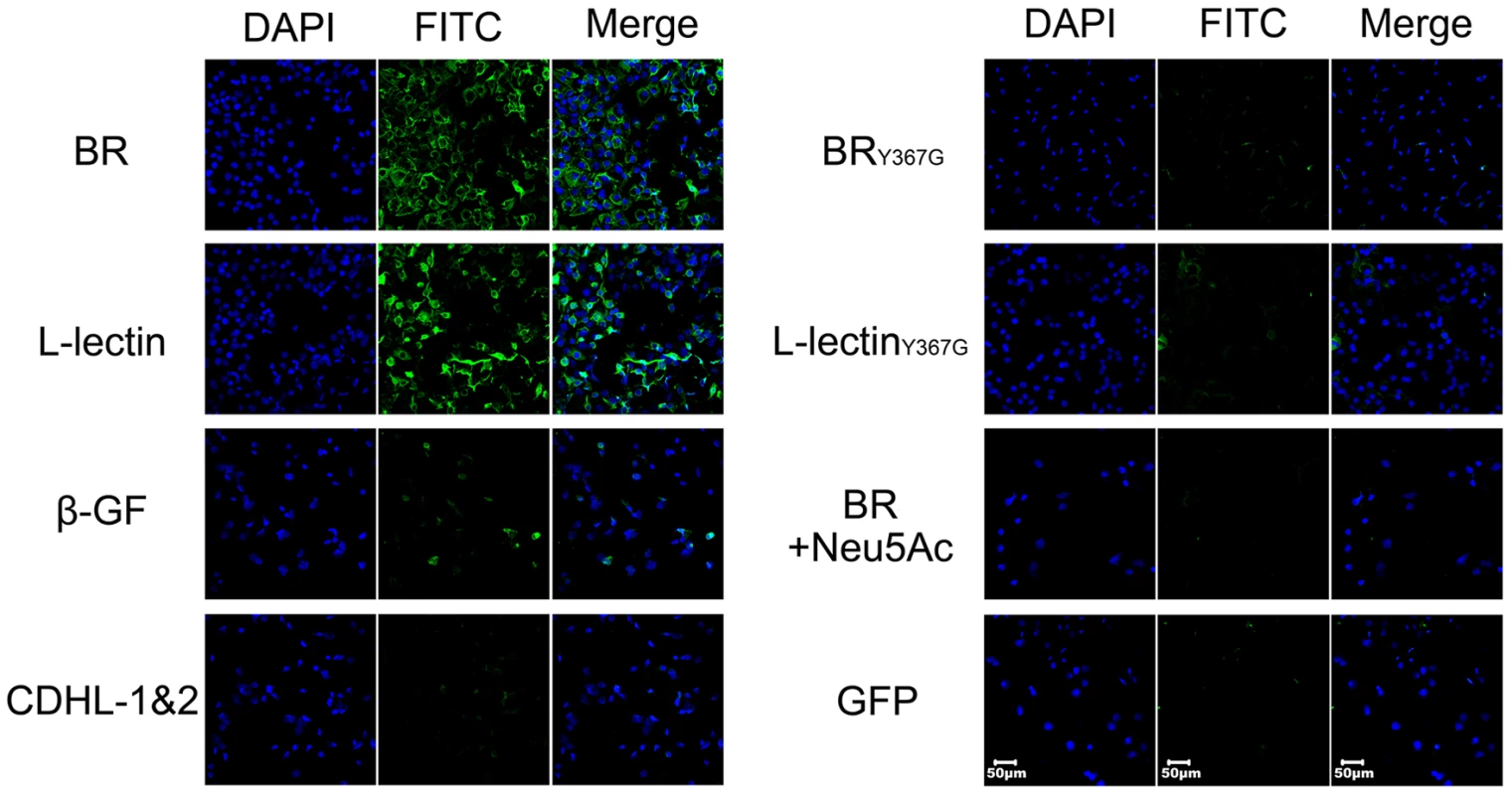
A549 cells were incubated with GFP-fused SraPBR (termed BR for short) and truncations (L-lectin, β-GF and CDHL-1&2) or mutants (BRY367G and L-lectinY367G), respectively. GFP is used as a negative control. The nuclei were stained with DAPI (blue) and the adhered proteins were detected with anti-GFP mouse IgG, followed by FITC conjugated goat anti-mouse IgG (green). Furthermore we performed comparative assays of bacterial adhesion and invasion to A549 cells using S. aureus strain NCTC 8325 and an isogenic ΔsraP mutant. Deletion of sraP resulted in an approximately 40% decrease in adhesion, as compared to the wild-type (Fig. 7A). As a result the level of invasion was decreased by ∼50% (Fig. 7B). These results indicate that SraP contributes to S. aureus adhesion to and invasion into host cells.
Fig. 7. Importance of SraP in bacterial adhesion to and invasion into epithelial cells. 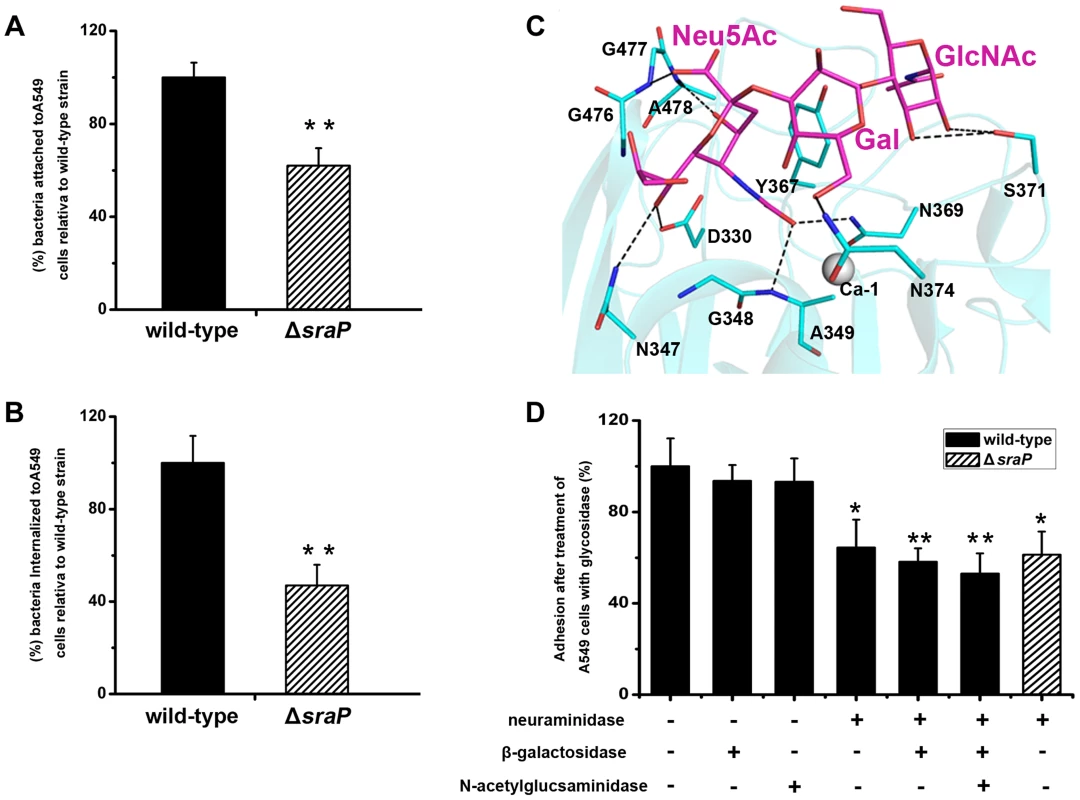
Contribution of SraP to the A) adhesion and B) invasion of S. aureus to A549 cells. Experiments were performed in triplicate. Results of representative experiments are presented as mean ± standard deviation. C) A docking model of trisaccharide binding to the L-lectin module. The trisaccharide and its interacting residues are shown as sticks and colored in magenta and cyan, respectively. D) Adhesion of the wild-type or ΔsraP mutant S. aureus to A549 cells pre-treated with neuraminidase, β-galactosidase and/or N-acetylglucosaminidase. Experiments were performed in triplicate. The standard error of the mean (SEM) derived from triplicate treatments are indicated as bar graph. All data were normalized against bacterial adhesion to mock-treated cells. Statistical analyses were performed using a one-way ANOVA. Together with the recently reported SraP ligand, the trisaccharide Neu5Acα(2–3)Galβ(1–4)GlcNAc [25], our results strongly suggested that the specific binding of SraPBR to the ligand promotes the S. aureus adhesion to host cells. We initially tried to determine the complex structure of the trisaccharide with SraPBR or the L-lectin module without success. As an alternative, we docked the trisaccharide to the structure of the L-lectin module (Fig. 7C). In the docking model, the Neu5Ac moiety of the trisaccharide adopts an almost same position to that of Neu5Ac (Fig. 2F). In addition, the moieties of Gal and GlcNAc are stabilized via hydrogen bonds by residues Asn374 and Ser371, respectively (Fig. 7C).
To determine whether the interactions between the L-lectin module and the trisaccharide contribute to the SraPBR-mediated adhesion, we used neuraminidase, β-galactosidase or N-acetylglucosaminidase to differentially remove Neu5Ac, Gal, and GlcNAc from the surface of A549 cells (Fig. 7D). Treating A549 cells with neuraminidase alone resulted in an approximately 36% decrease in the adhesion of S. aureus NCTC 8325. In contrast, the digestion with either β-galactosidase or N-acetylglucosaminidase did not significantly affect the adhesion. Further addition of β-galactosidase and N-acetylglucosaminidase to the neuraminidase-treated A549 cells did not significantly lower the adhesion level (Fig. 7D, the 6th and 7th columns). These results indicated that Neu5Ac is the moiety at the non-reducing end of the trisaccharide, and a major receptor to the SraP-mediated bacterial adhesion of A549 cells. Consistently, the wild-type S. aureus and ΔsraP mutant showed comparable levels of adhesion to the neuraminidase-treated epithelial cells (Fig 7D, the 4th and 8th columns). The adhesion level of the wild-type S. aureus to the neuraminidase-treated A549 cells (Fig. 7D, the 4th column) is similar to that of the ΔsraP mutant to the untreated A549 cells (Fig. 7A). We thus concluded that SraP is the major S. aureus adhesin that recognizes the sialylated host receptors.
Discussion
The specific recognition of the L-lectin module to sialylated receptors may be a universal mechanism for staphylococcal adhesion to host cells
Structural analyses combined with epithelial adhesion experiments demonstrate that SraP plays an important role in mediating bacterial adhesion to host cells by recognizing the L-lectin module. Legume lectins are a large family of proteins primarily found in the seeds of legume plants that have a similar fold but distinct carbohydrate-binding specificities [14]. They typically adopt a quaternary structure of dimers or tetramers that enhances sugar binding specificity or affinity [27]. The architecture of the legume lectin fold has also been found in the animal calcium-dependent lectin ERGIC-53/MR60, a mannose-binding protein involved in the export of soluble glycoproteins from the endoplasmic reticulum [28]. We report here the first legume lectin fold-containing protein in bacteria. This prokaryotic monomeric L-lectin module mediates adhesion to host cells by recognizing Neu5Ac, usually the non-reducing terminal residue of glycoconjugates of extracellular receptors. Bioinformatics analysis suggested that the L-lectin module might also exist in other proteins in Staphylococci and Streptococci (Fig. S4A). Moreover, the residues involved in Neu5Ac binding are relatively conserved among these putative L-lectins (Fig. S4B). Therefore, we hypothesize that this adhesion mechanism mediated by the L-lectin module operates in other staphylococcal and streptococcal species.
The L-lectin module is projected outwards mainly by two cadherin-like modules
To scan the host receptors, the L-lectin module should be projected outwards from the bacterial surface. In addition to a long region of SRR2 that crosses the bacterial cell wall, SraP has a relatively rigid stem of three modules: a β-GF and two CDHL modules. The classic cadherins in vertebrates bridge the intermembrane space between neighboring cells by forming trans-adhesive homodimers through membrane-distal extracellular domains [29]. The extracellular domain of most cadherins often contains three conserved Ca2+-coordination sites at the interdomain junction [30]. In contrast, each CDHL module in SraPBR binds to only a single Ca2+ that does not superimpose on any of the three Ca2+ ions in eukaryotic cadherins. Nevertheless, multiple-sequence alignments suggested strong conservation among Gram-positive bacteria of SraPBR Ca2+-binding residues (Fig. 8A).
Fig. 8. Homodimerization of the tandem CDHL modules. 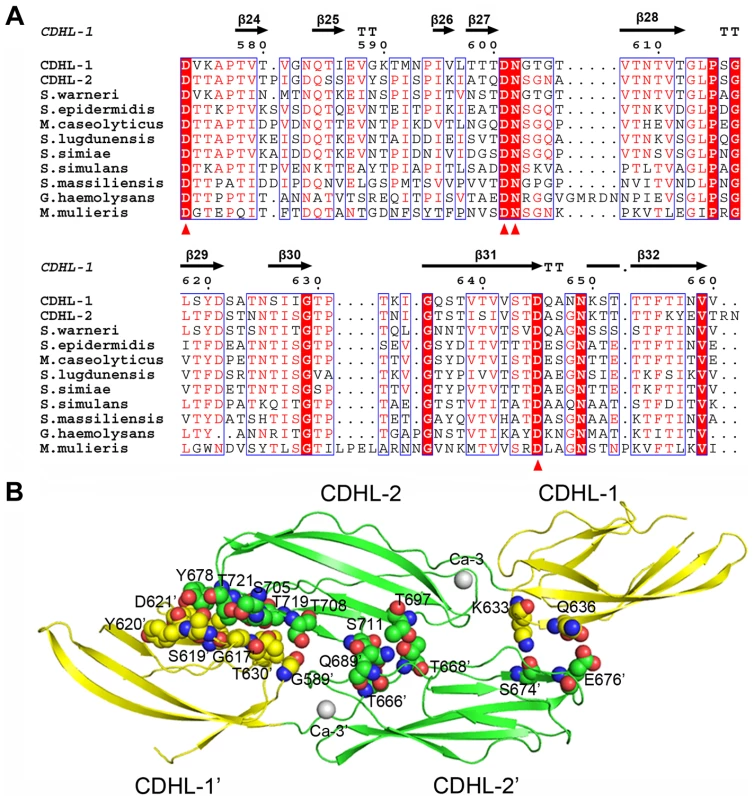
A) Multiple-sequence alignment of CDHL modules and their homologs. The Ca2+-binding residues are conserved in Gram-positive bacteria. B) The dimeric structure of CDHL-1&2. The dimer interface is shown as a CPK model. Unlike the monomeric form of SraPBR, the recombinant CDHL1&2 exists as a dimer in solution as confirmed by size-exclusion chromatography and chemical cross-linking assays (Fig. S5). The structure revealed that the homodimer of CDHL1&2 buries an interface area of ∼600 Å2/per subunit, as calculated by PISA (http://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver) server [31]. The interface is mainly stabilized by polar interactions, in contrast to the hydrophobic dimeric interfaces of eukaryotic cadherins [29]. CDHL-2 from one subunit packs against the junction between CDHL-1 and CDHL-2 from the symmetric subunit, and vice versa (Fig. 8B). This dimerization pattern may explain the result that SraP mediates intraspecies interaction and promotes aggregation of S. aureus ISP479C [7]. However, we did not observe significant decrease of aggregation or biofilm formation upon the deletion of sraP in S. aureus NCTC 8325. The different results might be due to the variation of S. aureus strains. Sequence homology search indicated that some surface proteins in Gram-positive bacteria contain two or more CDHL modules, suggesting that those proteins with multiple CDHL modules have a high potential to mediate intraspecies aggregation.
SraPBR possesses a unique functional modularization pattern
SRRPs have been identified in a variety of Gram-positive bacteria and function as virulence factors in a wide spectrum of infections [1]. The diversity of these infections (e.g. endocarditis, meningitis and pneumonia) correlates with the variability of BRs. Furthermore, the few BR ligands that have been identified to date range from carbohydrates (such as sialyl-T antigen) [32] to proteins (such as keratins) [33], [34]. In addition to the five distinct modules defined in the four previously reported BR structures [2]–[4], our SraPBR structure identified three types of unique modules. Notably, although the β-GF module resembles the Ig-binding proteins, such as B1 domain of Mub-R5 and PpL [19], [21], the binding of the β-GF module towards human IgG, IgA or IgM could not be detected, in agreement with our structural analysis. We thus propose that the β-GF module, in addition to the two CDHL modules, functions as a relatively rigid stem to project the L-lectin module outwards.
The two structure-known SRRPs, Fap1 and GspB, undergo significant inter-module angle changes, which are regulated by pH and ligand binding, respectively [2], [3]. In contrast, SraPBR appears to adopt a relatively rigid, rod-like conformation when colonizing a host, for the concentration of free Ca2+ in the extracellular space or blood is stringently maintained at 1.1–1.3 mM [35]. The rigid architecture of SraPBR enables the globular L-lectin module to extend outwards from the bacterial surface for scanning host receptors. This strategy to expose the functional modules has been observed for other bacterial adhesins such as the fibrillar antigen I/II from S. mutans [36], and the rod-like surface protein SasG from S. aureus [37].
Materials and Methods
Cloning, expression, and purification of SraPBR
The genomic DNA from Staphylococcus aureus NCTC 8325 was prepared for gene cloning. The DNA sequences (GeneBank, the accession number of YP_501439.1) encoding SraPBR (Phe245–Asn751) and other SraP truncates were cloned into pET28a with an N-terminal His6-tag or pET28a with a C-terminal GFP-tag, respectively. The constructs were overexpressed in E. coli strain BL21 (Novagen) using LB culture medium (10 g NaCl, 10 g Bacto-Tryptone, and 5 g yeast extract per liter). The cells were grown at 37°C to an OD600nm of 0.6. Expression of the recombinant protein was induced with 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 16°C for another 20 hr before harvesting. Bacteria were collected by centrifugation at 8,000×g for 10 min and resuspended in 30 ml lysis buffer (20 mM Tris-Cl, pH 8.8, 100 mM NaCl). After sonication for 2.5 min followed by centrifugation at 12,000×g for 25 min, the supernatant containing the His-tagged protein was collected and loaded onto a Ni-NTA column (GE Healthcare) equilibrated with the binding buffer (20 mM Tris-Cl, pH 8.0, 100 mM NaCl). The target protein was eluted with 300 mM imidazole, and loaded onto a Superdex 200 column or Superdex 75 column (GE Healthcare; 20 mM Tris-Cl, pH 8.0, 100 mM NaCl). The purity of protein was assessed by electrophoresis and the protein sample was stored at −80°C.
The selenium-Met (SeMet) labeled L-lectin&β-GF protein was expressed in E. coli strain B834 (DE3) (Novagen). Transformed cells were grown at 37°C in SeMet medium (M9 medium with 25 µg/ml SeMet and the other essential amino acids at 50 µg/ml) containing 30 µg/ml kanamycin until the OD600nm reached 0.6, and were then induced with 0.2 mM IPTG at 16°C for 20 hr. SeMet substituted protein was purified with the same procedure as the native protein.
The oligomerization state of CDHL-1&2 was analyzed by Superdex 75 column. The protein markers are bovine serum albumin, ovalbumin, chymotrypsinogen A, myoblobulin and ribinuclease A, which have a molecular weight of 67, 44, 25, 17 and 13.7 kDa, respectively (GE Healthcare).
Crystallization, data collection and processing
All crystals were grown using the hanging drop vapor diffusion method, with a drop of 1 µl protein solution mixed with 1 µl of reservoir solution equilibrated against 500 µl of the reservoir solution. The proteins for crystallization were concentrated by ultrafiltration (Millipore Amicon) to 30, 38, 20 and 20 mg/ml for the full-length SraPBR, L-lectin&β-GF, β-GF&CDHL-1 and CDHL-1&2, respectively. The SeMet substituted L-lectin&β-GF protein for crystallization was concentrated to 38 mg/ml. Crystals of SraPBR were grown at 28°C, whereas others were grown at 16°C. Crystals were obtained from 0.8 M (NH4)2SO4 and 0.1 M MES, pH 6.0 for SraPBR; 2.5 M sodium formate, 0.1 M sodium acetate pH 4.6 for the native L-lectin&β-GF; 10% PEG 6000, 0.1 M MES, pH 6.0, and 1.0 M lithium chloride for the SeMet substituted L-lectin&β-GF; 2.0 M (NH4)3PO4, 0.1 M Tris-Cl, pH 8.5 for β-GF&CDHL-1 and 1.8 M (NH4)2SO4, 0.1 M HEPES, pH 7.5 for CDHL-1&2. The crystals of SraPBR, β-GF&CDHL-1 and CDHL1&2 were transferred to the cryoprotectant with the reservoir solution supplemented with 50% sucrose. The cryoprotectant for the crystals of L-lectin&β-GF consists of the reservoir solution supplemented with 30% glycerol.
All crystals in the cryoprotectant were flash-cooled with liquid nitrogen prior to X-ray diffraction. Data for a single crystal were collected at 100 K in a liquid nitrogen stream using beamline 17U with a Q315r CCD (ADSC, MARresearch, Germany) at the Shanghai Synchrotron Radiation Facility (SSRF). All diffraction data were integrated and scaled with the program HKL2000 [38].
Structure determination and refinement
The crystal structure of L-lectin&β-GF was determined using single-wavelength anomalous dispersion (SAD) phasing [39] method from a single crystal of SeMet-substituted protein to a maximum resolution of 2.10 Å. The AutoSol program implemented in PHENIX [40] was used to locate the selenium atoms and calculate the phase, which was further improved with the program Buccaneer [41]. Automatic model building was carried out using Autobuild in PHENIX. The initial model was refined in REFMAC5 [42] and Phenix.refine and rebuilt interactively using the program COOT [43]. The model was used as the search model against 2.05 Å SraPBR data by molecular replacement using Molrep program as part of CCP4i [44] program suite. Electron density maps showed clear features of secondary structural elements for automatically building the C-terminal tandem cadherin-like modules using IPCAS [45]. The structures of β-GF&CDHL-1 and CDHL-1&2 were determined by molecular replacement using the corresponding modules in the full-length SraPBR structure as the search model. The initial models were refined by simulated annealing using Phenix.refine to reduce the phase bias. Then the models were refined interactively using COOT and REFMAC5 until the R-factor and R-free values converged. All final models were evaluated with the programs MOLPROBITY [46] and PROCHECK [47]. Crystallographic parameters were listed in Table 1. The |Fo|-|Fc| omit map of the sucrose molecule contoured at 3 σ was calculated by FFT implemented in CCP4i. All structure figures were prepared with PyMOL (http://www.pymol.org/).
Tab. 1. Crystal parameters, data collection and structure refinement. 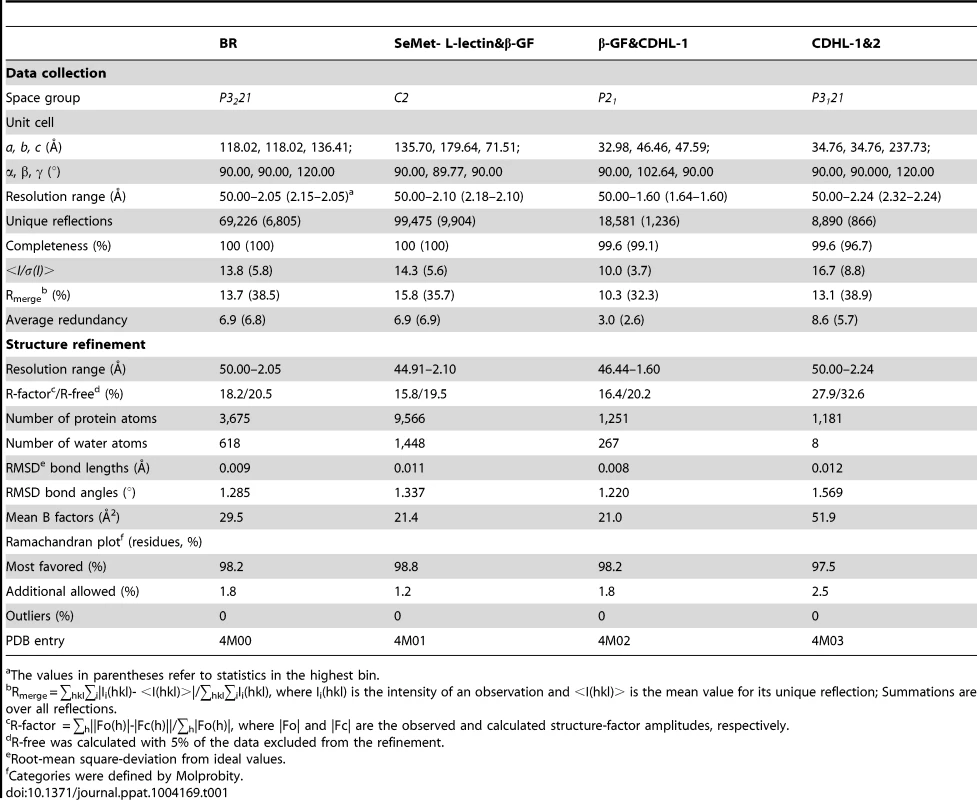
The values in parentheses refer to statistics in the highest bin. Determination of metals binding to the protein
The purified SraPBR, CDHL-1&2, BRY367G and L-lectinY367G in 20 mM Tris-Cl, pH 8.0, 100 mM NaCl were concentrated to 30, 38, 30 and 21 mg/ml, respectively, and applied to the analyses. Briefly, 500 µl of protein sample was subjected to digestion by the aqueous method using the HNO3 and HClO4 (4∶1, v/v) method. Afterwards, the digested samples were diluted with deionized water and analyzed by atomic absorption spectroscopy (Atomscan Advantage, Thermo Ash Jarell Corporation, USA).
Surface plasmon resonance (SPR) assays
The binding affinities of the L-lectin module towards varying monosaccharides were determined by SPR. SPR experiments were performed at 25°C using a Biacore 3000 instrument using HBS (10 mM HEPES, pH 7.5, 150 mM NaCl) containing 0.005% (v/v) Tween 20 and a flow rate of 5 µl/min. The L-lectin module was covalently immobilized on the carboxymethyldextran surface of the CM5 chip. The chip was activated with EDC (N-ethyl-N-[3-dimethylaminopropyl] carbodi-imide)/NHS (N-hydroxysuccinimide) solution, and the L-lectin module in 10 mM acetate buffer (pH 5.5) was injected into the flow channel. At the end, the sensor surface was blocked with 1 M ethanolamine. The blank channel was treated in the same way without protein injected. Each monosaccharide in the running buffer was incubated for 1 min in the flow-cells using the kinject mode. Both injection and dissociation steps last for 5 min. The sensor surface was regenerated with 50 mM NaOH. All analyses were performed with the BIAeval software. The equilibrium responses were plotted versus monosaccharide concentrations and fitted to a 1∶1 Langmuir binding model using the Origin 8.0 software (OriginLab Corp.).
Computational docking
The docking of Neu5Ac or the trisaccharide Neu5Acα(2–3)Galβ(1–4)GlcNAc to the L-lectin module of SraPBR was performed with AutoDock Vina software (version 1.0) [48], which uses a unique algorithm that implements a machine learning approach to its scoring function. This docking allowed a population of possible conformations and orientations for the ligand at the binding site to be obtained. Using AutoDock Tools (ADT) 1.5.4 [49], polar hydrogen atoms were added to the L-lectin structure, and its non-polar hydrogen atoms were merged. The protein and ligands were converted from a PDB format to a PDBQT format. All single-bonds within Neu5Ac were set to allow rotation. A grid box covering the entire sucrose-binding site was used to place Neu5Ac freely. The results were sorted by binding affinity and visually analyzed using PyMOL.
Chemical cross-linking
Chemical cross-linking of purified CDHL-1&2 was performed using formaldehyde and bis(sulfosuccinimidyl) suberate (BS3), which is a homobifunctional sulfo-N-hydroxysuccinimide ester analog with a spacer arm length of 1.14 nm (Pierce). Briefly, for formaldehyde cross-linking assays of CDHL-1&2, 20 µl of the recombinant protein (2 mg/ml) was mixed with 20 µl PBS containing 2% formaldehyde, and the samples were incubated at 25°C for 30 min and 1 hr, respectively. For BS3 cross-linking assay of CDHL-1&2, 100 µl recombinant protein (2 mg/ml) was incubated with 5 mM BS3 at 25°C for 30 min and 1 hr, respectively. The reaction was quenched by the addition of 20 mM Tris-HCl, pH 8.0. Then the samples were separated by 10% SDS-PAGE and were stained by Coomassie Brilliant Blue G250. Molecular mass markers for SDS-PAGE were purchased from Thermo Scientific (Wilmington, DE): beta-galactosidase, bovine serum albumin, ovalbumin, lactate dehydrogenase, REase Bsp98I, beta-lactoglobulin, lysozyme, which have a molecular weight of 116, 66.2, 45, 35, 25, 18.4 and 14.4 kDa, respectively.
Bacterial strains, media and growth conditions
Staphylococcus aureus NCTC 8325 and its derivative strains were grown in LB medium, and when necessary, erythomycin (2.5 mg/ml) and chlorampenicol (15 mg/ml) were added. To generate the insertion-deletion mutagenesis, approximately 500 bp of upstream and downstream fragments of the BR region of sraP gene was amplified by polymerase chain reaction (PCR), digested with PstI/SalI and BamHI/XbaI, respectively, and then ligated to either end of a double-digested (BamHI/SalI) erythromycin-resistance gene (ErmR), which was amplified from plasmid pEC1. The three fragments were ligated with the erythromycin-resistance gene in the middle, and then cloned in the temperature-sensitive shuttle vector pBT2. The resulting plasmid was transformed by electroporation into S. aureus strain RN4220 for propagation, and then transformed into S. aureus NCTC 8325 for allelic exchange. Mutants were screened and further checked by PCR and sequencing.
Cell adhesion and invasion assays
Adhesion and invasion experiments were performed as described previously [50]. A549 human respiratory epithelial cells were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS), 5 mM glutamine, penicillin (5 µg/ml) and streptomycin (100 µg/ml). Approximately 5×105 cells were seeded into 24-well tissue culture plates, and allowed to grow in 5% CO2 at 37°C. Before use, the monolayers were washed twice with PBS. For adhesion assays, S. aureus grown to OD600nm of 0.5 in LB medium were resuspended in cell culture medium without serum. Bacteria were diluted to a concentration of 2×107 CFU/ml and were used to infect the confluent cell monolayers at 37°C for 1 hr. After incubation, the infected monolayers were washed five times with PBS to remove non-adhered bacteria, and treated with 200 µl trypsin (2.5 mg/ml) at 37°C for 3 min to release the adhered bacteria. A549 cells were lysed with 0.05% Triton X-100. The number of adhered bacteria was determined by plating serial dilutions of the recovered bacterial suspensions onto LB agar. For the deglycosylation of A549 cells, The cells were incubated with DMEM media containing 0.008 units ml−1 purified Clostridium perfringens neuraminidase (Sigma), 40 nM purified β-galactosidase (Spr0565) from S. pneumoniae or 0.0025 units ml−1 of S. pneumoniae β-N-acetylglucosaminidase (Sigma) at 37°C for 4 hr in 5% CO2 [34].
For invasion assays, the bacteria in each well, after incubated in 0.5 ml DMEM for 1 hr, were incubated in 1 ml fresh DMEM containing 14 µg/ml of gentamicin (Sigma) for another 1 hr. Cell monolayers were washed three times with sterile PBS and lysed with 0.05% Triton X-100. The internalized bacteria were counted by plating serial dilutions of the recovered bacterial suspensions onto LB agar. Experiments were performed in triplicate. Data corresponding to adhesion and invasion were compared using the Mann-Whitney tests. Statistical differences were determined with the t-test.
Immunofluorescence assays
Immunofluorescence assays were performed as described previously [51]. Briefly, GFP and GFP-fused proteins at 10 µM were suspended in the media in the absence of serum and antibiotics, and incubated on ice for 2 hr with the monolayer of A549 cells grown on the 12-mm glass coverslips. For inhibition assays, Neu5Ac at 5 and 10 mM was pre-incubated with GFP-fused SraPBR protein, respectively. After incubation, cells were sequentially washed three times with cold PBS, fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 2 min, and incubated with GFP-tag mouse antibody and a fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG antibody. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) reagent. Slides were examined with a Zeiss LSM710 confocal scanning fluorescence microscope (Carl Zeiss, Jena, Germany) with a Plan-Apocromat 20×/0.8NA objective. Confocal parameters set for immunofluorescence detection were taken as standard settings. The excitation wavelength is 405 nm and the emission wavelength is 410–492 nm. The confocal images were collected and processed with the software ZEN 2009. Experiments were performed in triplicate, with three or more replicate wells tested for each experimental condition.
Molecular dynamics simulations
Molecular dynamics simulations were performed for SraPBR in the Ca2+-bound and the Ca2+-free states, respectively. For each simulation, the system was placed in a TIP3P [52] water box with a distance of at least 12 Å to the box boundaries. Ions were added to neutralize the system and to result in a concentration of 0.15 M NaCl. The solvated protein was subjected to energy minimization employed the steepest descent algorithm and conjugate gradient, respectively. Simulations were performed with a parallel implementation of the GROMACS (version 4.5.5) package [53] using the AMBER03 force field [54]. MD productions were run for 20 ns using a time step of 2 fs and the NPT ensemble [55]. Covalent bonds were constrained using the LINCS algorithm [56], while the cutoff distances for the Coulomb and van der Waals interactions were set to 0.9 and 1.4 nm, respectively. The long-range electrostatic interactions were treated by the PME algorithm [57] with a tolerance of 10−5 and an interpolation order of 4. Structure visualization was performed with VMD [58].
Small-angle X-ray scattering (SAXS) experiments
SAXS was used to investigate the overall conformations of SraPBR in the presence or absence of calcium. The full-length SraPBR at 1.0 and 5.0 mg/ml was analyzed, either in 10 mM calcium chloride or in 50 mM EDTA. SAXS data were collected on the 12ID beamline of Advanced Photon Sources (APS) at the Argonne National Laboratory using the Pilatus 2M detector (DECTRIS, Switzerland). The scattering patterns were measured with a 1–2 second exposure time for each collected frame, and twenty frames were taken for each sample to optimize the signal-to-noise ratio. To reduce the radiation damage, a flow cell made of a cylindrical quartz capillary with a diameter of 1.5 mm and a wall of 10 µm was used during the data collection process. No concentration effect was observed. All SAXS curves were measured at the room temperature over the range of momentum transfer 0.006<s<0.82 Å−1 (where s = 4π sin(θ)/λ, 2θ is the scattering angle, and the X-ray wavelength λ is 1.033 Å). The data were processed using the PRIMUS [59] program package and standard procedures. The forward scattering (I(0)) and the radius of gyration (Rg) were evaluated using the Guinier approximation assuming that at very small angles (s<1.3/Rg) the intensity is represented as I(s) = I(0)exp(−(sRg)2/3).
The program GNOM [60] was used to calculate Dmax and the interatomic distance distribution function p(r). The particle shape of each measured sample was reconstructed ab initio using the programs DAMMIN [61] and GASBOR [62]. The scattering patterns of the atomic crystal structures for SraP were calculated using the program CRYSOL. For the ab initio analyses and modeling, multiple runs were performed to verify the stability of the solution.
Supporting Information
Zdroje
1. LizcanoA, SanchezCJ, OrihuelaCJ (2012) A role for glycosylated serine-rich repeat proteins in Gram-positive bacterial pathogenesis. Mol Oral Microbiol 27 : 257–269.
2. RamboarinaS, GarnettJA, ZhouM, LiY, PengZ, et al. (2010) Structural insights into serine-rich fimbriae from Gram-positive bacteria. J Biol. Chem 285 : 32446–32457.
3. PyburnTM, BensingBA, XiongYQ, MelanconBJ, TomasiakTM, et al. (2011) A structural model for binding of the serine-rich repeat adhesin GspB to host carbohydrate receptors. PLoS Pathog 7: e1002112.
4. SeoHS, MisanovG, SeepersaudR, DoranKS, DubrovskaI, et al. (2013) Characterization of fibrinogen binding by glycoproteins Srr1 and Srr2 of Streptococcus agalactiae. J Biol Chem 288 : 35982–96.
5. LowyFD (1998) Staphylococcus aureus infections. N Engl J Med 339 : 520–532.
6. SibooIR, ChambersHF, SullamPM (2005) Role of SraP, a serine-rich surface protein of Staphylococcus aureus, in binding to human platelets. Infect Immun 73 : 2273–2280.
7. SanchezCJ, ShivshankarP, StolK, TrakhtenbroitS, SullamPM, et al. (2010) The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog 6: e1001044.
8. HolmL, RosenstromP (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38: W545–549.
9. PletnevVZ, RuzheinikovSN, TsygannikIN, MikhailovaIY, DuaxW, et al. (1997) The structure of pea lectin-D-glucopyranose complex at a 1.9 Å resolution. RUSS. J.Bioorganic Chem 23 : 469–478.
10. RabijnsA, VerbovenC, RougeP, BarreA, Van DammeEJ, et al. (2001) Structure of a legume lectin from the bark of Robinia pseudoacacia and its complex with N-acetylgalactosamine. Proteins 44 : 470–478.
11. MurzinAG, BrennerSE, HubbardT, ChothiaC (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247 : 536–540.
12. JinX, WalkerMA, FelsovalyiK, VendomeJ, BahnaF, et al. (2012) Crystal structures of Drosophila N-cadherin ectodomain regions reveal a widely used class of Ca2+-free interdomain linkers. Proc Natl Acad Sci U S A 109: E127–134.
13. SotomayorM, WeihofenWA, GaudetR, CoreyDP (2012) Structure of a force-conveying cadherin bond essential for inner-ear mechanotransduction. Nature 492 : 128–132.
14. SharonN, LisH (1990) Legume lectins—a large family of homologous proteins. FASEB J 4 : 3198–3208.
15. LorisR, HamelryckT, BouckaertJ, WynsL (1998) Legume lectin structure. Biochim Biophys Acta 1383 : 9–36.
16. DuvergerE, FrisonN, RocheAC, MonsignyM (2003) Carbohydrate-lectin interactions assessed by surface plasmon resonance. Biochimie 85 : 167–179.
17. MacKenzieDA, TailfordLE, HemmingsAM, JugeN (2009) Crystal structure of a mucus-binding protein repeat reveals an unexpected functional immunoglobulin binding activity. J Biol Chem 284 : 32444–32453.
18. GrailleM, SturaEA, HousdenNG, BeckinghamJA, BottomleySP, et al. (2001) Complex between Peptostreptococcus magnus protein L and a human antibody reveals structural convergence in the interaction modes of Fab binding proteins. Structure 9 : 679–687.
19. BjorckL (1988) Protein L. A novel bacterial cell wall protein with affinity for Ig L chains. J Immunol 140 : 1194–1197.
20. WikstromM, DrakenbergT, ForsenS, SjobringU, BjorckL (1994) Three-dimensional solution structure of an immunoglobulin light chain-binding domain of protein L. Comparison with the IgG-binding domains of protein G. Biochemistry 33 : 14011–14017.
21. BeckinghamJA, BottomleySP, HintonR, SuttonBJ, GoreMG (1999) Interactions between a single immunoglobulin-binding domain of protein L from Peptostreptococcus magnus and a human κ light chain. Biochem J 340 (Pt 1): 193–199.
22. GrailleM, HarrisonS, CrumpMP, FindlowSC, HousdenNG, et al. (2002) Evidence for plasticity and structural mimicry at the immunoglobulin light chain-protein L interface. J Biol Chem 277 : 47500–47506.
23. BermanH, HenrickK, NakamuraH, MarkleyJL (2007) The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res 35: D301–303.
24. HuraGL, MenonAL, HammelM, RamboRP, PooleFL2nd, et al. (2009) Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat Methods 6 : 606–612.
25. KukitaK, Kawada-MatsuoM, OhoT, NagatomoM, OogaiY, et al. (2013) Staphylococcus aureus SasA is responsible for binding to the salivary agglutinin gp340, derived from human saliva. Infect Immun 81 : 1870–1879.
26. HolmskovU, LawsonP, TeisnerB, TornoeI, WillisAC, et al. (1997) Isolation and characterization of a new member of the scavenger receptor superfamily, glycoprotein-340 (gp-340), as a lung surfactant protein-D binding molecule. J Biol Chem 272 : 13743–13749.
27. SrinivasVR, ReddyGB, AhmadN, SwaminathanCP, MitraN, et al. (2001) Legume lectin family, the ‘natural mutants of the quaternary state’, provide insights into the relationship between protein stability and oligomerization. Biochim Biophys Acta 1527 : 102–111.
28. VellosoLM, SvenssonK, SchneiderG, PetterssonRF, LindqvistY (2002) Crystal structure of the carbohydrate recognition domain of p58/ERGIC-53, a protein involved in glycoprotein export from the endoplasmic reticulum. J Biol Chem 277 : 15979–15984.
29. BraschJ, HarrisonOJ, HonigB, ShapiroL (2012) Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol 22 : 299–310.
30. BoggonTJ, MurrayJ, Chappuis-FlamentS, WongE, GumbinerBM, et al. (2002) C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science 296 : 1308–1313.
31. KrissinelE, HenrickK (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372 : 774–797.
32. BensingBA, LopezJA, SullamPM (2004) The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ibα. Infect Immun 72 : 6528–6537.
33. SamenU, EikmannsBJ, ReinscheidDJ, BorgesF (2007) The surface protein Srr-1 of Streptococcus agalactiae binds human Keratin 4 and promotes adherence to epithelial HEp-2 Cells. Infect Immun 75 : 5405–5414.
34. ShivshankarP, SanchezC, RoseLF, OrihuelaCJ (2009) The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol Microbiol 73 : 663–679.
35. MaurerP, HohenesterE, EngelJ (1996) Extracellular calcium-binding proteins. Curr Opin Cell Biol 8 : 609–617.
36. LarsonMR, RajashankarKR, PatelMH, RobinetteRA, CrowleyPJ, et al. (2010) Elongated fibrillar structure of a Streptococcal adhesin assembled by the high-affinity association of α - and PPII-helices. Proc Natl Acad Sci U S A 107 : 5983–5988.
37. GruszkaDT, WojdylaJA, BinghamRJ, TurkenburgJP, ManfieldIW, et al. (2012) Staphylococcal biofilm-forming protein has a contiguous rod-like structure. Proc Natl Acad Sci U S A 109: E1011–E1018.
38. OtwinowskiZ, MinorW (1997) Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol, Pt A 276 : 307–326.
39. BrodersenDE, de La FortelleE, VonrheinC, BricogneG, NyborgJ, et al. (2000) Applications of single-wavelength anomalous dispersion at high and atomic resolution. Acta Crystallogr D Biol Crystallogr 56 : 431–441.
40. AdamsPD, AfoninePV, BunkocziG, ChenVB, DavisIW, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66 : 213–221.
41. CowtanK (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 62 : 1002–1011.
42. MurshudovGN, VaginAA, DodsonEJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53 : 240–255.
43. EmsleyP, CowtanK (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 : 2126–2132.
44. Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50 : 760–763.
45. HaoQ, GuYX, ZhengCD, FanHF (2000) OASIS: a computer program for breaking phase ambiguity in one-wavelength anomalous scattering or single isomorphous substitution (replacement) data. J Appl Crystallogr 33 : 980–981.
46. DavisI, Leaver-FayA, ChenV, BlockJ, KapralG, et al. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35: W375–383.
47. LaskowskiR, MacarthurM, MossD, ThorntonJ (1993) Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr 26 : 283–291.
48. TrottO, OlsonAJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31 : 455–461.
49. SannerMF (1999) Python: a programming language for software integration and development. J Mol Graph Model 17 : 57–61.
50. ValleJ, LatasaC, GilC, Toledo-AranaA, SolanoC, et al. (2012) Bap, a biofilm matrix protein of Staphylococcus aureus prevents cellular internalization through binding to GP96 host receptor. PLoS Pathog 8: e1002843.
51. NelsonAL, RiesJ, BagnoliF, DahlbergS, FalkerS, et al. (2007) RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol Microbiol 66 : 329–340.
52. JorgensenWL, ChandrasekharJ, MaduraJD, ImpeyRW, KleinML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79 : 926–935.
53. HessB, KutznerC, van der SpoelD, LindahlE (2008) GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4 : 435–447.
54. DuanY, WuC, ChowdhuryS, LeeMC, XiongGM, et al. (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24 : 1999–2012.
55. BerendsenHJC, PostmaJPM, VangunsterenWF, DinolaA, HaakJR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81 : 3684–3690.
56. HessB (2008) P-LINCS: A parallel linear constraint solver for molecular simulation. J Chem Theory Comput 4 : 116–122.
57. EssmannU, PereraL, BerkowitzML, DardenT, LeeH, et al. (1995) A smooth particle mesh Ewald method. J Chem Phys 103 : 8577–8593.
58. HumphreyW, DalkeA, SchultenK (1996) VMD: Visual molecular dynamics. J Mol Graph 14 : 33–38.
59. KonarevPV, VolkovVV, SokolovaAV, KochMHJ, SvergunDI (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr 36 : 1277–1282.
60. SvergunDI (1992) Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J Appl Crystallogr 25 : 495–503.
61. SvergunDI (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing (vol 76, pg 2879, 1999). Biophys J 77 : 2896–2896.
62. SvergunDI, PetoukhovMV, KochMHJ (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80 : 2946–2953.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Recruitment of RED-SMU1 Complex by Influenza A Virus RNA Polymerase to Control Viral mRNA SplicingČlánek Systematic Phenotyping of a Large-Scale Deletion Collection Reveals Novel Antifungal Tolerance GenesČlánek The Contribution of Social Behaviour to the Transmission of Influenza A in a Human Population
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 6- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Fungal Nail Infections (Onychomycosis): A Never-Ending Story?
- BdlA, DipA and Induced Dispersion Contribute to Acute Virulence and Chronic Persistence of
- Morphotype Transition and Sexual Reproduction Are Genetically Associated in a Ubiquitous Environmental Pathogen
- A Nucleic-Acid Hydrolyzing Single Chain Antibody Confers Resistance to DNA Virus Infection in HeLa Cells and C57BL/6 Mice
- HopW1 from Disrupts the Actin Cytoskeleton to Promote Virulence in Arabidopsis
- Ly6C Monocytes Become Alternatively Activated Macrophages in Schistosome Granulomas with Help from CD4+ Cells
- Recruitment of RED-SMU1 Complex by Influenza A Virus RNA Polymerase to Control Viral mRNA Splicing
- Contribution of Specific Residues of the β-Solenoid Fold to HET-s Prion Function, Amyloid Structure and Stability
- Antibody Responses to : Role in Pathogenesis and Diagnosis of Encephalitis?
- Discovery of a Novel Compound with Anti-Venezuelan Equine Encephalitis Virus Activity That Targets the Nonstructural Protein 2
- Activation of Focal Adhesion Kinase by Suppresses Autophagy via an Akt/mTOR Signaling Pathway and Promotes Bacterial Survival in Macrophages
- Crossing the Interspecies Barrier: Opening the Door to Zoonotic Pathogens
- Catching Fire: , Macrophages, and Pyroptosis
- IscR Is Essential for Type III Secretion and Virulence
- Selective Chemical Inhibition of Quorum Sensing in Promotes Host Defense with Minimal Impact on Resistance
- The Glycosylated Rv1860 Protein of Inhibits Dendritic Cell Mediated TH1 and TH17 Polarization of T Cells and Abrogates Protective Immunity Conferred by BCG
- A Genome-Wide Tethering Screen Reveals Novel Potential Post-Transcriptional Regulators in
- Structural Insights into SraP-Mediated Adhesion to Host Cells
- Human IGF1 Regulates Midgut Oxidative Stress and Epithelial Homeostasis to Balance Lifespan and resistance in
- Cycling Empirical Antibiotic Therapy in Hospitals: Meta-Analysis and Models
- Rab11 Regulates Trafficking of -sialidase to the Plasma Membrane through the Contractile Vacuole Complex of
- Mitogen and Stress Activated Kinases Act Co-operatively with CREB during the Induction of Human Cytomegalovirus Immediate-Early Gene Expression from Latency
- Profilin Promotes Recruitment of Ly6C CCR2 Inflammatory Monocytes That Can Confer Resistance to Bacterial Infection
- A Central Role for Carbon-Overflow Pathways in the Modulation of Bacterial Cell Death
- An Invertebrate Warburg Effect: A Shrimp Virus Achieves Successful Replication by Altering the Host Metabolome via the PI3K-Akt-mTOR Pathway
- The Highly Conserved Bacterial RNase YbeY Is Essential in , Playing a Critical Role in Virulence, Stress Regulation, and RNA Processing
- A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees () Prevails after -Mediated, or , Transmission
- Systematic Phenotyping of a Large-Scale Deletion Collection Reveals Novel Antifungal Tolerance Genes
- Ubiquitin-Mediated Response to Microsporidia and Virus Infection in
- Preclinical Detection of Variant CJD and BSE Prions in Blood
- Toll-Like Receptor 8 Agonist and Bacteria Trigger Potent Activation of Innate Immune Cells in Human Liver
- Progressive Proximal-to-Distal Reduction in Expression of the Tight Junction Complex in Colonic Epithelium of Virally-Suppressed HIV+ Individuals
- The Triggering Receptor Expressed on Myeloid Cells 2 Inhibits Complement Component 1q Effector Mechanisms and Exerts Detrimental Effects during Pneumococcal Pneumonia
- Differential Activation of Acid Sphingomyelinase and Ceramide Release Determines Invasiveness of into Brain Endothelial Cells
- Forward Genetic Screening Identifies a Small Molecule That Blocks Growth by Inhibiting Both Host- and Parasite-Encoded Kinases
- Defining Immune Engagement Thresholds for Control of Virus-Driven Lymphoproliferation
- Growth Factor and Th2 Cytokine Signaling Pathways Converge at STAT6 to Promote Arginase Expression in Progressive Experimental Visceral Leishmaniasis
- Multimeric Assembly of Host-Pathogen Adhesion Complexes Involved in Apicomplexan Invasion
- Biogenesis of Influenza A Virus Hemagglutinin Cross-Protective Stem Epitopes
- Adequate Th2-Type Response Associates with Restricted Bacterial Growth in Latent Mycobacterial Infection of Zebrafish
- Protective Efficacy of Passive Immunization with Monoclonal Antibodies in Animal Models of H5N1 Highly Pathogenic Avian Influenza Virus Infection
- Fructose-Asparagine Is a Primary Nutrient during Growth of in the Inflamed Intestine
- The Calcium-Dependent Protein Kinase 3 of Influences Basal Calcium Levels and Functions beyond Egress as Revealed by Quantitative Phosphoproteome Analysis
- A Translocated Effector Required for Dissemination from Derma to Blood Safeguards Migratory Host Cells from Damage by Co-translocated Effectors
- Functional Characterization of a Novel Family of Acetylcholine-Gated Chloride Channels in
- Both α2,3- and α2,6-Linked Sialic Acids on O-Linked Glycoproteins Act as Functional Receptors for Porcine Sapovirus
- The Contribution of Social Behaviour to the Transmission of Influenza A in a Human Population
- MicroRNA-146a Provides Feedback Regulation of Lyme Arthritis but Not Carditis during Infection with
- Recombination in Enteroviruses Is a Biphasic Replicative Process Involving the Generation of Greater-than Genome Length ‘Imprecise’ Intermediates
- Cytoplasmic Viral RNA-Dependent RNA Polymerase Disrupts the Intracellular Splicing Machinery by Entering the Nucleus and Interfering with Prp8
- and Are Associated with Murine Susceptibility to Infection and Human Sepsis
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Fungal Nail Infections (Onychomycosis): A Never-Ending Story?
- Profilin Promotes Recruitment of Ly6C CCR2 Inflammatory Monocytes That Can Confer Resistance to Bacterial Infection
- Contribution of Specific Residues of the β-Solenoid Fold to HET-s Prion Function, Amyloid Structure and Stability
- The Highly Conserved Bacterial RNase YbeY Is Essential in , Playing a Critical Role in Virulence, Stress Regulation, and RNA Processing
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



