-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Molecular and Functional Analyses of a Maize Autoactive NB-LRR Protein Identify Precise Structural Requirements for Activity
The plant hypersensitive defense response (HR) is a rapid, localized cell death, usually occurring upon the recognition of specific pathogen-encoded molecules and consequent activation of nucleotide binding-leucine rich repeat (NLR) proteins. Rp1-D21, a naturally-occurring mutant caused by the recombination of two NLR genes, confers a ‘lesion mimic’, HR-like phenotype in the absence of pathogen infection and provides a powerful tool to investigate the molecular mechanisms of NLR regulation. Here we report the results of a genetic screen in maize that identified novel mutations abrogating Rp1-D21-induced HR. To characterize the function of Rp1-D21, we transiently expressed Rp1-D21 and various derivatives in Nicotiana benthamiana to observe the resulting levels of HR. We furthermore examined the protein-protein interactions between and within different Rp1-D21 derivatives. We report novel insights into the precise structural requirements for NLR function and determine the function of a previously undefined motif. These insights enable a better understanding of how NLRs regulate the switch between the resting and the active states.
Published in the journal: Molecular and Functional Analyses of a Maize Autoactive NB-LRR Protein Identify Precise Structural Requirements for Activity. PLoS Pathog 11(2): e32767. doi:10.1371/journal.ppat.1004674
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004674Summary
The plant hypersensitive defense response (HR) is a rapid, localized cell death, usually occurring upon the recognition of specific pathogen-encoded molecules and consequent activation of nucleotide binding-leucine rich repeat (NLR) proteins. Rp1-D21, a naturally-occurring mutant caused by the recombination of two NLR genes, confers a ‘lesion mimic’, HR-like phenotype in the absence of pathogen infection and provides a powerful tool to investigate the molecular mechanisms of NLR regulation. Here we report the results of a genetic screen in maize that identified novel mutations abrogating Rp1-D21-induced HR. To characterize the function of Rp1-D21, we transiently expressed Rp1-D21 and various derivatives in Nicotiana benthamiana to observe the resulting levels of HR. We furthermore examined the protein-protein interactions between and within different Rp1-D21 derivatives. We report novel insights into the precise structural requirements for NLR function and determine the function of a previously undefined motif. These insights enable a better understanding of how NLRs regulate the switch between the resting and the active states.
Introduction
One of the main plant disease resistance mechanisms is mediated by dominant resistance (R) genes with a major effect [1]. Many R genes have been cloned from diverse plant species, most of which encode nucleotide binding leucine-rich-repeat (NB-LRR or NLR) proteins [2,3]. Based on the secondary structure of their N-termini, NLRs can largely be subdivided into two classes: one containing a Toll-interleukin 1 receptor (TIR) domain (TIR-NB-LRR, TNL hereafter), and the other a putative coiled-coil (CC) domain (CC-NB-LRR, CNL hereafter). Both TNL and CNL proteins have been identified in dicots, while only CNL proteins have been found in monocots [4].
Each NLR is capable of directly or indirectly recognizing the presence of at least one effector protein, usually produced by a subset of isolates of a single pathogen species [5]. Recognition generally leads to activation of the NLR protein and initiation of signal transduction, resulting in the hypersensitive response (HR), which is often accompanied by the induction of a rapid localized cell death at the point of pathogen penetration [6]. HR can contribute to the halting of pathogen growth. When the corresponding effector is not present, wild-type NLRs are held in an inactive state, largely through self-inhibitory intra-molecular interactions, which may also be facilitated or reinforced by additional host proteins [5,7,8].
In general, plant NLR proteins contain three separable structural domains, an N-terminal CC or TIR domain, a C-terminal LRR domain and a central NB-ARC (ARC: APAF1, R gene products and CED-4) domain. The ARC subdomain in plant NLRs is further divided into two separated structural units: ARC1 and ARC2 [9,10]. In the NB-ARC domain, several conserved motifs are recognized; in linear order: P-loop/Kinase-1a/Walker A (henceforce called the P-loop), RNBS (Resistance Nucleotide Binding Site)-A, Kinase-2/Walker B, RNBS-B, RNBS-C, GLPL, RNBS-D, MHD [11]. Among the different motifs, the P-loop and MHD motifs are known to be very important for NLR function.
In their inactive form, NLRs likely have ADP bound to the NB-ARC domain. Activation of NLRs involves the exchange of ADP for ATP through a structural change that is thought to result in an open structure of the proteins [12,13]. The P-loop motif within the NB-ARC domain is thought to be involved in binding ATP/ADP [13]. P-loop mutations in the NLR resistance genes I-2 from tomato, M from flax and RPM1 from Arabidopsis impair ATP binding and ability to confer HR-based resistance [14,15,16]. Mutations in the conserved MHD-motif lead to autoactivation of NLRs, which is thought to be due to weakened ADP-binding and resulting structural change into an open conformation, favoring nucleotide exchange [15,17].
Autoactivate NLRs, i.e. NLRs that can be activated without the need for a recognition event, can be generated in vivo or experimentally via recombination between different NLRs, or through specific point mutations. One example of an in vivo recombination event leading to an autoactive NLR is from the maize Rp1 locus. Rp1 is a complex locus that carries multiple NLR paralogs. The number of these paralogs varies widely between haplotypes, some carrying more than 50 [18]. The Rp1-D haplotype consists of 9 NLR paralogs; Rp1-dp1 to Rp1-dp8 which have no known function, and Rp1-D itself, which confers resistance against the biotrophic fungal pathogen Puccinia sorghi, the causal agent of maize common rust [19,20]. These Rp1 paralogs are more than 90% identical in nucleotide sequence, allowing them to undergo unequal crossing over and occasional intragenic recombination. An intragenic recombination between paralogs Rp1-D and Rp1-dp2 produced the chimeric gene Rp1-D21, which was identified on the basis of its ‘lesion mimic’ phenotype in the absence of pathogen infection [20,21].
We previously demonstrated that many of the hallmarks of pathogen-induced immune response, such as H2O2 accumulation, increased expression of the defense-related genes PR1, PRms and WIP1, are associated with Rp1-D21-mediated lesion phenotype in maize. We also demonstrated that the Rp1-D21 lesion phenotype is genetic background-, temperature - and light-dependent [22,23]. Recently, we deployed Rp1-D21 as a tool in a novel enhancer/suppressor screen to identify natural modifiers of the HR [23,24,25,26].
Different NLR R-genes appear to have a variety of different structural requirements for proper activation and functioning [27,28,29,30,31,32]. The autoactive nature of Rp1-D21 and the fact that its ‘parental’ proteins, Rp1-D and Rp1-dp2, are not autoactive makes it a very useful tool to explore the molecular requirements for NLR regulation, specifically the switch between inactive and active states.
Here we report the characterization of the activity of Rp1 proteins in maize and in Nicotiana benthamiana, a model system widely used for characterization of R-gene mediated HR for both dicot and monocot NLRs [27,29,33,34]. Using a combination of genetic, molecular biological, biochemical and computational techniques we derive a model for Rp1 activity which provides a better understanding of how NLRs regulate the switch between the resting and the activation states.
Results
Characterization of Rp1-D21 sequence
Rp1-D21 is derived from the recombination of two NLR paralogs at the Rp1 locus, Rp1-D, and Rp1-dp2 that are 90% identical at the amino acid level [19,20,21,35]. Sequencing of Rp1-D21 showed that it encodes a protein of 1290 amino acids (AAs) and is a typical CNL with an N-terminal coiled-coil (CC) domain (AAs 1–189), an NB-ARC domain (AAs 190–527) and an LRR domain at the C-terminus (AAs 528–1290). The N-terminus of Rp1-D21 derives from Rp1-dp2 (up to AA 770–778), with the remainder deriving from Rp1-D (the precise breakpoint is impossible to define since nucleotides 2310–2333, corresponding to AAs 771–777 are identical in the two progenitor genes, Fig. 1). Thus, Rp1-D21 is comprised of the CC, NB-ARC and the N-terminus of the LRR domain from Rp1-dp2 and the C-terminus of the LRR domain from Rp1-D. A conserved EDVID motif (EDLLD in Rp1-D21, Fig. 1) can be identified in the CC domain of Rp1-D21. All of the important motifs present in the NB-ARC domain of typical NLR proteins can be found in Rp1 proteins (Fig. 1; [11]).
Fig. 1. Sequence alignment of Rp1-D, Rp1-dp2 and Rp1-D21. 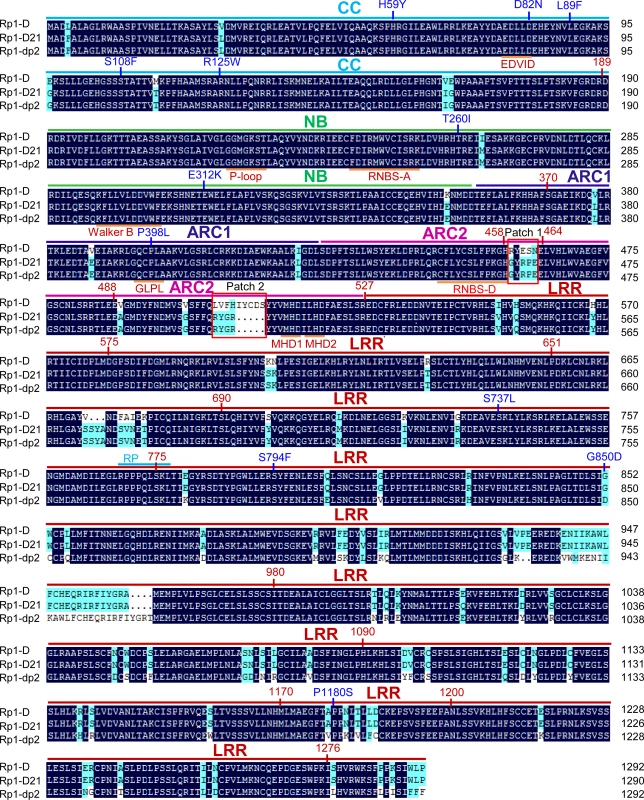
The CC (coiled-coil), NB-ARC (nucleotide binding), ARC1 (APAF1, R gene products and CED-4), ARC2 and LRR (leucine-rich-repeat) domains were indicated by bars with cyan, green, dark blue, pink and red colors, respectively. The conserved motifs (EDVID, P-loop, GLPL, RNBS-D, MHD1 and MHD2) were indicated by orange bars and labeled below the sequences. The recombination point of Rp1-D21 was labeled by cyan bar. Patches 1 and 2 are the major difference regions between Rp1-D and Rp1-dp2 in the ARC2 domain. The positions of the mutations in the intragenic Rp1-D21 suppressor from maize were labeled by blue color. The landmark positions of recombination points in the constructs listed in Fig. 6 were labeled by numbers with red color. The black and light blue shaded regions represent 100% and above 50% similarity of the amino acids, respectively. A targeted EMS-mutagenesis screen identifies intragenic suppressor mutations in Rp1-D21
We performed a targeted ethyl methanesulfonate (EMS) mutagenesis screen in a maize line harboring Rp1-D21 to identify mutations that lost the autoactive HR phenotype conferred by Rp1-D21. Putative suppressor mutants were easily identified due to their robust growth compared to the stunted, lesion-mimic siblings heterozygous for Rp1-D21 (Fig. 2A). From about 23,000 EMS-mutated M1 plants, 12 missense and 2 nonsense intragenic mutants were identified (Fig. 2B; Table 1). Among the 12 missense mutants, five had mutations in the CC domain, three in the NB-ARC domain and four in the LRR domain (Fig. 2B). Notably, one mutant (nucleotide G244A, thus D82N in amino acid) was in the conserved EDVID motif of the CC domain, and one (C1193T, P398L) in the conserved GLPL motif of the NB-ARC domain (Table 1; Figs. 1 and 2B). The mutants T260I and E312K were adjacent to the conserved RNBS-A and Walker B motifs, respectively (Table 1; Figs. 1 and 2B).
Fig. 2. EMS mutagenesis screening for mutants that lost the hypersensitive response (HR) induced by Rp1-D21. 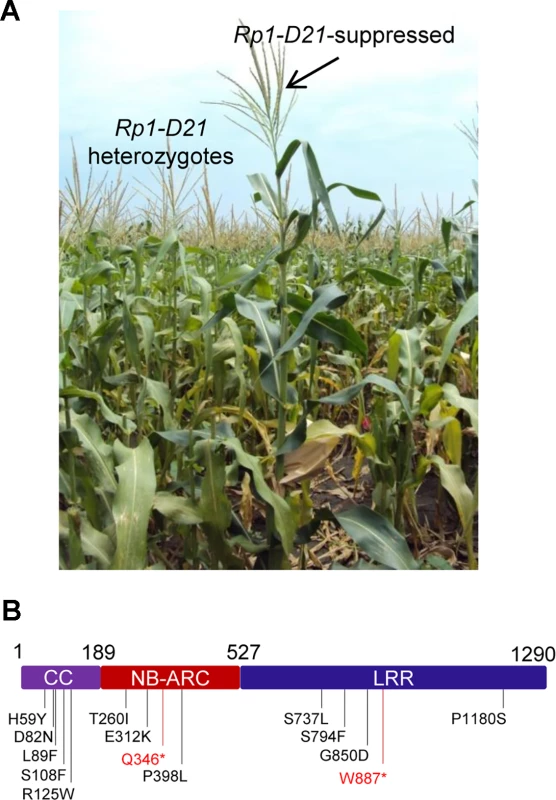
(A) The growth phenotype of Rp1-D21 heterozygotes in the field compared to the Rp1-D21 mutant in which the HR phenotype is suppressed (arrow). (B) Twelve missense (in black) and two nonsense (in red) point mutations that abolished the HR phenotype of Rp1-D21 were identified in different domains (CC, NB-ARC and LRR) of Rp1-D21. Tab. 1. Mutations that abrogated Rp1-D21-induced HR identified in an EMS mutagenesis screen in maize. 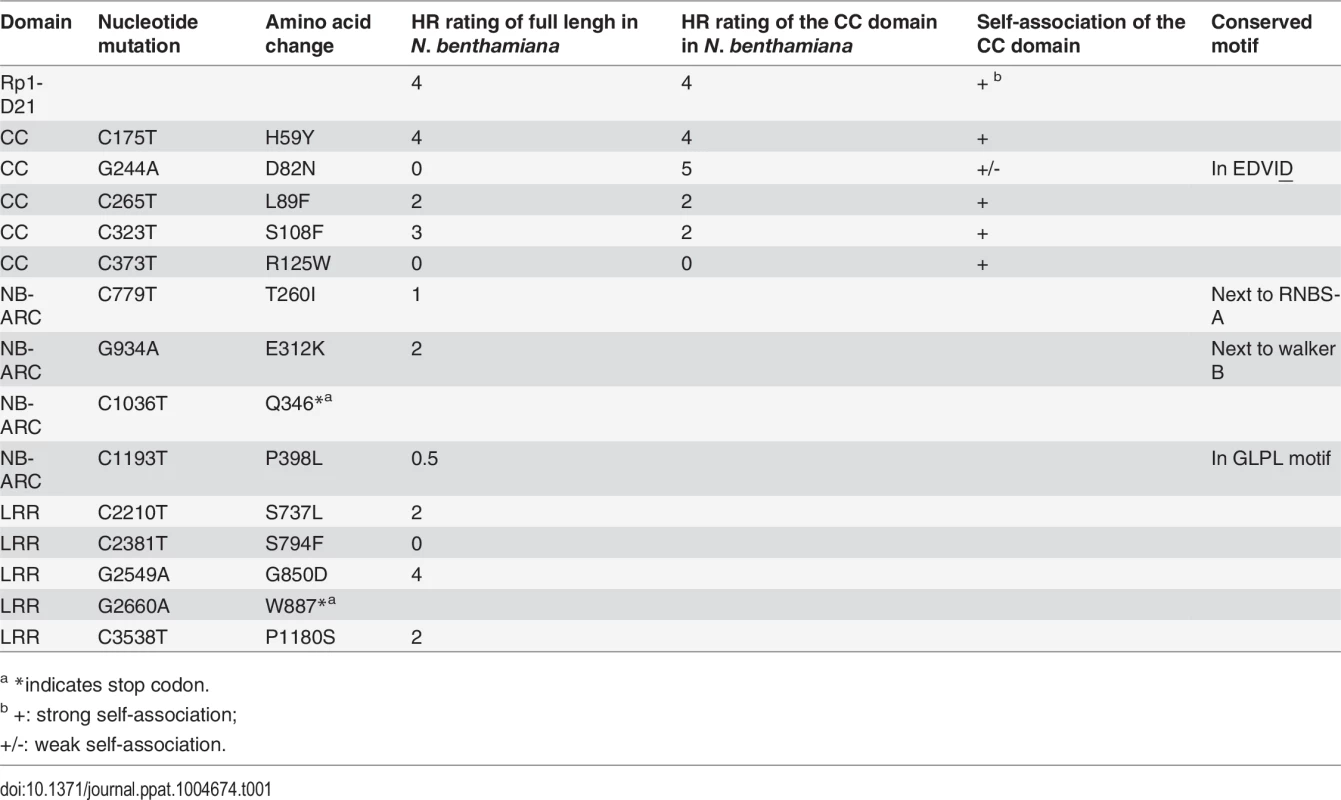
a *indicates stop codon. The HR phenotype conferred by Rp1-D21 in maize can be recapitulated by transient expression in N. benthamiana
We used Agrobacterium-mediated transient expression in N. benthamiana to investigate the structure/function of Rp1-D21. Rp1-D, Rp1-dp2 and Rp1-D21 were either fused to the N-terminus of enhanced green fluorescent protein (EGFP), a 3×HA (influenza hemagglutinin) tag or not tagged for subsequent functional analysis. When the three fusion proteins were transiently expressed using the cauliflower mosaic virus 35S promoter, a HR phenotype was observed 3 days post-infiltration (dpi) only with Rp1-D21, but not with Rp1-D, Rp1-dp2 or the empty vector (EV) control. The same phenotypes were obtained regardless of the tags used (EGFP, 3×HA or no tag) and regardless of the differing levels of protein accumulation observed for the three proteins (Fig. 3; S1A Fig.).
Fig. 3. Rp1-D21 triggers a hypersensitive response phenotype when transiently expressed in N. benthamiana. 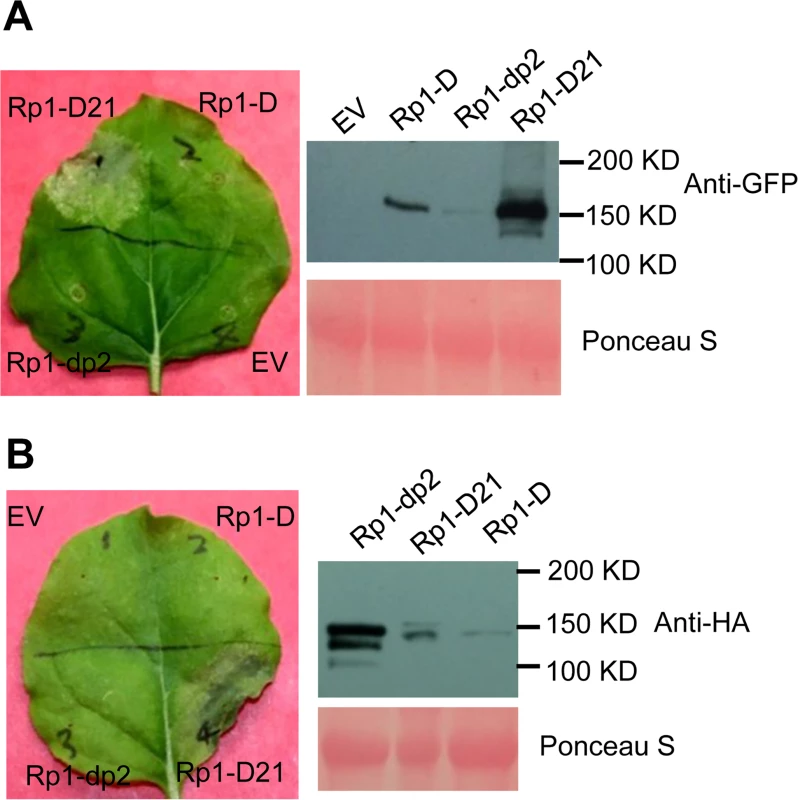
(A) Rp1-D21, Rp1-D and Rp1-dp2 proteins fused with a C-terminal EGFP tag were agro-infiltrated into N. benthamiana, with an empty vector (EV) as a negative control. A representative leaf was photographed at 3 days post infiltration (dpi, left). Total protein was extracted from agro-infiltrated leaves at 30 hours post infiltration (hpi), and anti-GFP antibody was used to detect the expression of the fused proteins (right). The sizes of the proteins were labeled on the right. Equal loading of protein samples was shown by Ponceau-S staining of the Rubisco subunit (below, right). (B) Rp1-D21, Rp1-D and Rp1-dp2 proteins fused with a C-terminal 3×HA tag were agro-infiltrated into N. benthamiana, with empty vector (EV) as a negative control. A representative leaf was photographed at 3 dpi (left). Total protein was extracted from agro-infiltrated leaves at 30 hpi, and anti-HA antibody was used to detect the expression of the fused proteins. Equal loading of protein samples was shown by Ponceau-S staining of Rubisco subunit (below, right). These experiments were repeated three times with the same results. To further confirm that the phenotype conferred by Rp1-D21 in our N. benthamiana system conformed to the phenotypes observed in maize, we constructed Rp1-D21 expression vectors carrying the missense Rp1-D21 suppressor mutations identified in maize. When transiently expressed in N. benthamiana, Rp1-D21 produced a strong HR phenotype, rating 4 based on a 0–5 scale rating with 0 being no cell death or chlorosis at all and 5 being confluent cell death [36]. The HR rating is based on the average results from at least 10 individual leaves. All the mutants we identified as non-functional in maize, except for H59Y and G850D, abolished or greatly reduced the level of Rp1-D21-induced HR (Table 1; S2A Fig.). All constructs conferred high levels of protein expression (S2B Fig.). The concordance between the maize and N. benthamiana systems is very good and we expect that our conclusions based on the transient expression N. benthamiana system are generally relevant to the endogenous maize system. Thus, we used the transient expression of Rp1-D21 and its derivatives in N. benthamiana to further analyze the molecular mechanism of activation/auto-inhibition of Rp1 proteins.
Rp1 protein activity depends on P-loop and MHD motifs
The MHD motif is a highly conserved region of the NB-ARC domain involved in nucleotide binding [15]. Mutations in the MHD domain confer autoactivity to a number of NLRs. Two MHD motifs, here termed MHD1 and MHD2 (actually LHD in Rp1-D and Rp1-dp2; Fig. 4A), separated by a single amino acid are apparent in Rp1-D, Rp1-dp2 and Rp1-D21, and in a number of other CNLs [37]. To investigate whether mutations in either of the MHD motifs could cause autoactivity in the non-autoactive parental proteins, Rp1-D and Rp1-dp2, we generated the following mutations: Rp1-D(H517A), Rp1-D(D518V), Rp1-dp2(D513V) and Rp1-dp2(H512A/D513V) in the MHD1 motif, and Rp1-D(H521A), Rp1-D(D522V), Rp1-dp2(H516A), Rp1-dp2(D517V) in the MHD2 motif. Transient expression of the Rp1-dp2(D517V) MHD2 mutant conferred an obvious HR, with symptoms appearing at 3.5 days, about 12 h later than that observed with Rp1-D21 (Fig. 4B). HR was not induced by transient expression of any of the other MHD mutants, including the Rp1-D(D522V) MHD2 mutation (S3 Fig.). However, Rp1-D(D522V) expression was not detectable by western blot analysis using anti-HA antibody. Protein expressed from all the other constructs could be detected (Fig. 4B; S3 Fig.). We also generated the MHD2 mutations V1(D517V) and V16(D522V) in V1 and V16, two recombinant constructs that were not autoactive (see below) and found both of them induced HR (Fig. 4C).
Fig. 4. Functional characterization of the MHD2 motif and the P-loop motif. 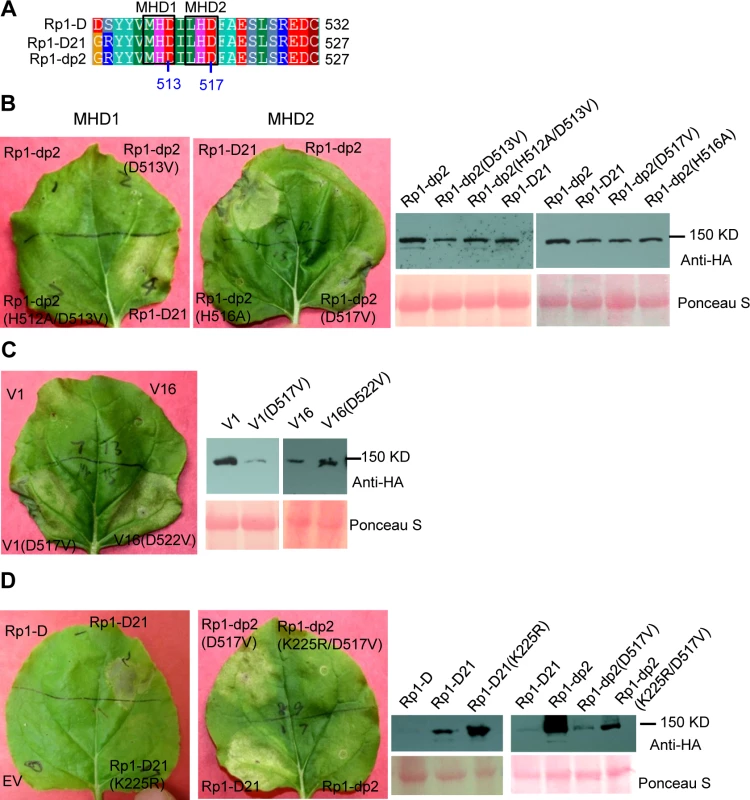
(A) Multiple sequence alignment of the MHD motifs from Rp1-D, Rp1-D21 and Rp1-dp2. Positions of the MHD1 and MHD2 motifs are indicated. The numbers indicated the positions of the aspartate (D) corresponding to Rp1-dp2. (B) The MHD1 and MHD2 point mutations in Rp1-dp2 background were generated and transiently expressed in N. benthamiana. (C) Investigating the MHD2 point mutations in V1 and V16, two recombinant constructs that were not autoactive as shown in Fig. 6. (D) Point mutations in P-loop motif of Rp1-D21 and Rp1-dp2(D517V) caused the loss of the ability to induce HR. EV (empty vector) as a negative control. The picture of hypersensitive response (HR) phenotype was taken at 3 days post-infiltration and total protein was detected by anti-HA antibody from agro-infiltrated leaves at 30 hours post infiltration. Equal loading of protein samples was shown by Ponceau-S staining of Rubisco subunit. The experiments were repeated three times with the same results. The P-loop is a highly conserved motif in NLRs and mutations in this domain often result in loss of function and loss of autoactivity [14,15,27,34,38,39]. P-loop mutations (K225R) introduced into Rp1-D21 and the autoactive Rp1-dp2(D517V) mutant abrogated their ability to confer HR (Fig. 4D). This indicated that the Rp1-D21 autoactivity and presumably also the activity of the Rp1 proteins are P-loop dependent.
The CC domain is sufficient for inducing HR
To examine which region of Rp1-D21 was required for triggering HR, different domains or domain combinations of Rp1-D21, including CC, CC-NB-ARC, NB-ARC, NB-ARC-LRR and LRR domains (hereafter CCD21, CC-NB-ARCD21, NB-ARCD21, NB-ARC-LRRD21 and LRRD21; Fig. 5A), were fused to the N-terminus of EGFP. CCD21 and NB-ARCD21 were derived from (and are therefore identical to) the corresponding domains of Rp1-dp2 while LRRD21 is recombinant between the LRRs of Rp1-D and Rp1-dp2. The transient expression of CCD21, but of no other Rp1-D21 domains or domain combinations, conferred HR (Fig. 5B). CCD21, NB-ARCD21 and CC-NB-ARCD21 produced higher protein accumulation than NB-ARC-LRRD21 and LRRD21 (Fig. 5B), excluding the possibility that lack of HR phenotype seen with most of the domain constructs was solely due to low protein accumulation. We also tested the HR phenotype conferred by different Rp1-D domains and found that the CC domain, but no others, induced HR when fused with EGFP (Fig. 5A). Surprisingly, no untagged or HA-tagged domains from either Rp1-D or Rp1-D21 induced HR (Fig. 5; S1B Fig.).
Fig. 5. Investigating the functional domains of Rp1-D21 and Rp1-D to trigger a hypersensitive response (HR). 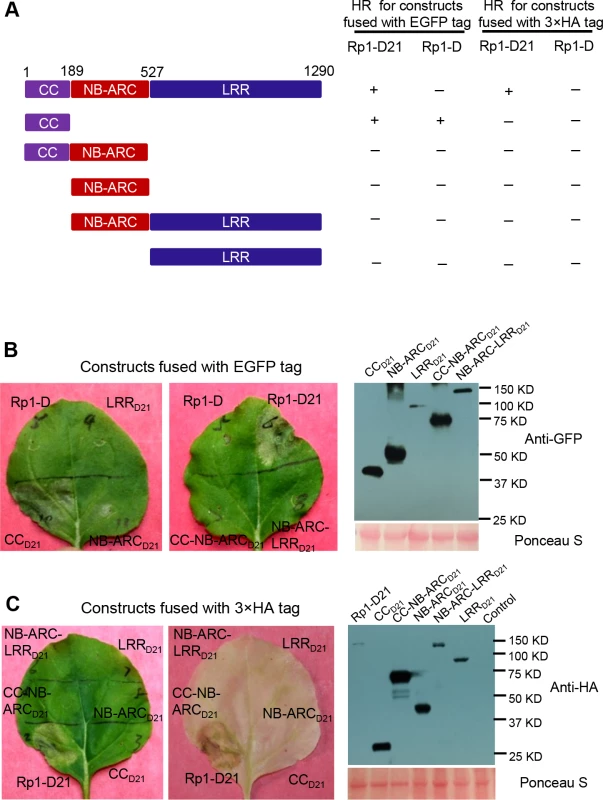
(A) Schematic diagram of the Rp1-D21 and Rp1-D domain structure and the derived fragments used for agro-infiltration of N. benthamiana. The different domains are indicated using different colors: CC (purple); NB-ARC (red); LRR (dark blue). The positions of the amino acids that form the domain boundaries are indicated on the top and the abilities to induce (+) or not induce (-) HR are listed on the right of each construct. (B) The HR phenotype of the constructs fused with a C-terminal EGFP tag and transiently expressed in N. benthamiana. Representative leaves were photographed at 3 days post infiltration (dpi, left). Total protein was extracted from agro-infiltrated leaves at 30 hours post infiltration (hpi), and anti-GFP antibody was used to detect the expression of fused proteins. Equal loading of protein samples was shown by Ponceau-S staining of Rubisco subunit (right, below). The results shown were from different domains of Rp1-D21, and similar results were observed from different domains of Rp1-D. (C) The phenotype of the constructs fused with a C-terminal 3×HA tag and transiently expressed in N. benthamiana. A representative leaf was photographed at 3 dpi (left), and the same leaf was cleared by ethanol (middle). Total protein was extracted from agro-infiltrated leaves at 30 hpi, and anti-HA antibody was used to detect the expression of the fused proteins. Equal loading of protein samples was shown by Ponceau-S staining of Rubisco subunit (right). The results shown were from different domains of Rp1-D21, and similar results were observed from different domains of Rp1-D. The experiments were repeated three times with the same results. CCD21, but not CC-NB-ARCD21, induced HR, which suggested that NB-ARCD21 can inhibit CCD21-induced HR in cis (when the two domains were fused in the same molecule). Consistent with this result, no HR was observed when EGFP-tagged CCD21 and NB-ARCD21 were transiently co-expressed in trans (when the two domains were co-expressed as separate molecules) in N. benthamiana (Table 2). LRRD21 did not restore HR when co-expressed with CCD21 and NB-ARCD21 separately, or with CC-NB-ARCD21 in trans. However, co-expressing CCD21 with NB-ARC-LRRD21 retained HR in trans, suggesting that LRRD21 might interact with NB-ARCD21 to regulate CCD21–induced HR in cis but not in trans (Table 2). However, since LRRD21 or NB-ARC-LRRD21 are expressed at substantially lower levels than CCD21 or CC-NB-ARCD21 (Figs. 5B and 5C), we cannot exclude the possibility that the concentration of LRRD21 or NB-ARC-LRRD21 is simply too low to affect the NB-ARC suppression of CCD21–induced HR or CCD21–induced HR itself when co-expressed in trans. In contrast to the in cis result of CC-NB-ARCD, we found that NB-ARCD did not suppress CCD-induced HR in trans (Table 2), suggesting NB-ARCD might interact with CCD in cis but not in trans.
Tab. 2. Summary of the HR phenotype conferred by transient expression in Nicotiana benthamiana of specific domains or domain combinations from Rp1-D and Rp1-D21 in cis and in trans (i.e the two or three listed domains either fused in the same molecule in the order presented here, or expressed as separate molecules, respectively). 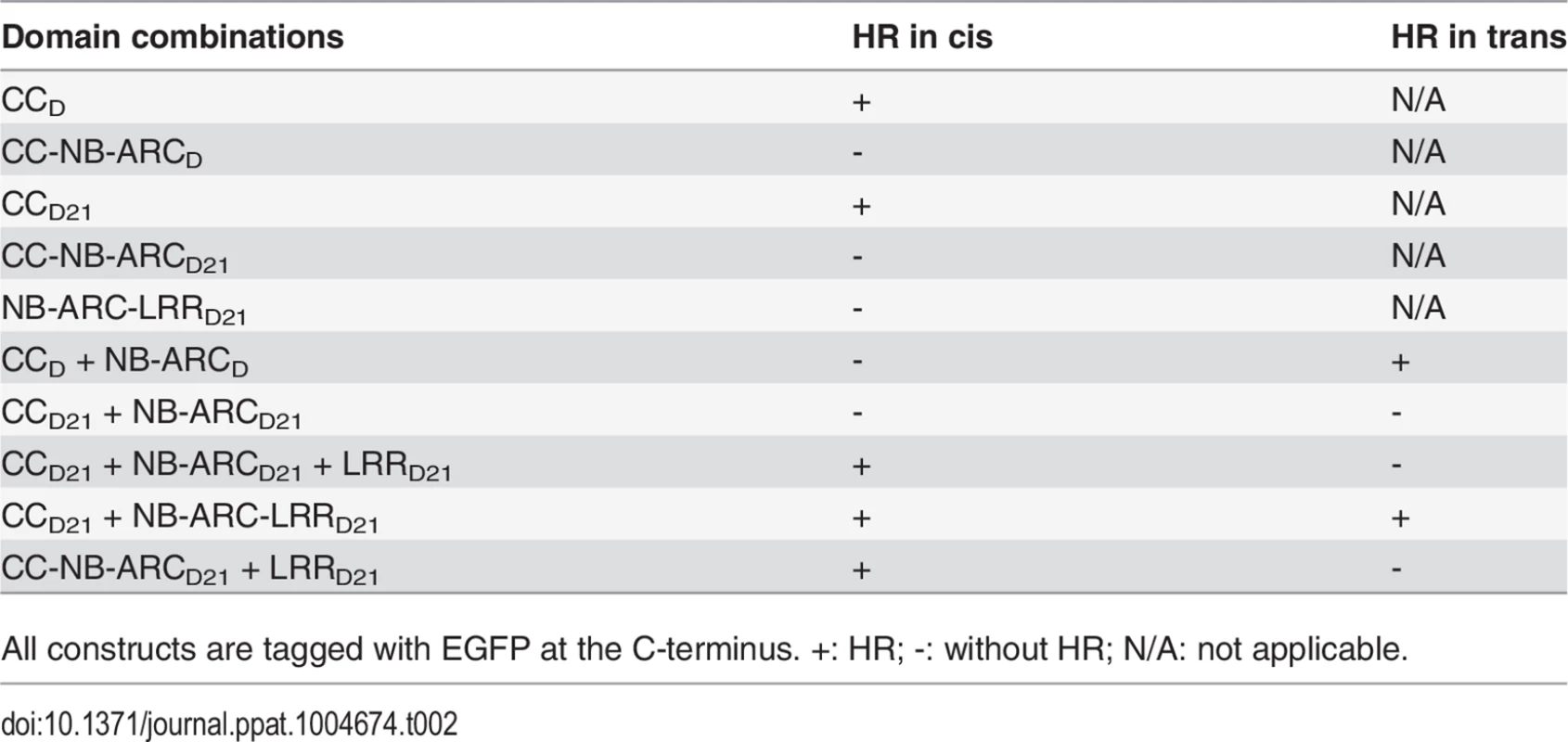
All constructs are tagged with EGFP at the C-terminus. +: HR; -: without HR; N/A: not applicable. To further investigate the inhibition region of NB-ARC in CC-induced HR, we generated a series of deletion constructs from CC-NB-ARC (S4 Fig.). We found that extension including AAs 190–260 from the NB domain was sufficient to suppress CC-induced HR. Consistent with the data, the nonsense EMS mutant (Q346*) suppressed the Rp1-D21 lesion-mimic and stunted growth phenotype in maize (Fig. 2B; Table 1).
The C-terminus of the LRR domain has been demonstrated to be important for NLR activity [37]. We therefore investigated whether C-terminal deletions of the LRR domain from Rp1-D21 affected the HR phenotype. None of the six C-terminal deletion constructs (ranging from 34 to 639 AAs) induced HR after transient expression, even D21-LRR27 that lacked only the last 34 AAs of the acidic tail in C-terminus (S5 Fig.). This is despite the fact that all the constructs conferred high levels of protein expression (S5B Fig.).
Domain swaps between Rp1-D and Rp1-dp2 delineate precise requirements for self-inhibition
To delineate the functional regions that cause Rp1-D21 to be autoactive and keep its progenitor proteins auto-inhibited, we generated a series of chimeric constructs recombinant at different positions between Rp1-D and Rp1-dp2 (Fig. 6). These proteins, both with and without a 3×HA C-terminal tag, were tested in the N. benthamiana transient expression system. Previously, the chimeric construct Rp1-dp2-D2, with the N-terminus deriving from Rp1-dp2 and the C-terminus from Rp1-D, with a recombination point at 980 AA (Fig. 6A), had been shown to cause HR in transgenic maize [21]. The identical chimeric construct, hereafter named Hd2, induced a strong HR when transiently expressed in N. benthamiana (Fig. 6A), consistent with the transgenic maize result.
Fig. 6. Schematic diagram of domain swap constructs between Rp1-D and Rp1-dp2 used for transient expression in N. benthamiana. 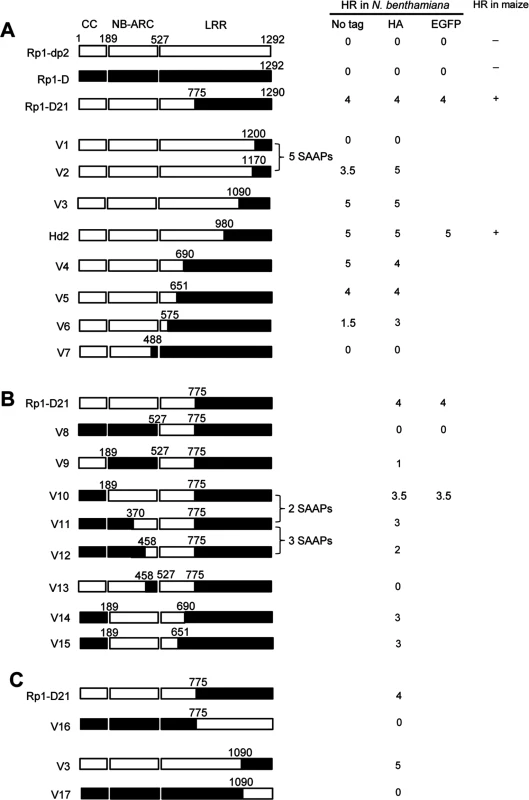
For ease of interpretation, the gene structure has been divided into CC (coiled coil), NB-ARC (nucleotide binding) and LRR (leucine rich repeat). The amino acid position of the recombination site of each construct was indicated above the construct. For each construct, either with no tag, 3×HA tag or EGFP tag, the strength of the hypersensitive response (HR) resulting from transient expression in N. benthamiana is shown. HR was scored on a 0 (no HR) to 5 (strong HR) scale according to Slootweg et al. (2013). If there was no score recorded, the particular experiment was not performed. Where known, the abilities of constructs to induce (+) or not induce (-) HR in maize were indicated. In some cases the number of single amino acid polymorphisms (SAAP) between constructs is indicated. All experiments were performed three times with similar results. (A) Constructs with the N-terminus from Rp1-D and C-terminus from Rp1-dp2. (B) Constructs designed to delimit the domains of Rp1-D or Rp1-dp2 important for self-inhibition. (C) The reciprocal constructs: Rp1-D21 with V16, and V3 with V17. The results of the transient expression of a series of chimeric proteins are summarized in Fig. 6. The HR phenotype conferred by the constructs fused with 3×HA was largely similar to that conferred by the constructs without any tag (Fig. 6A). All the tagged proteins accumulated to high and broadly comparable levels (Figs. 7 and 8; S6 Fig.).
Fig. 7. Investigating the functional regions of amino acids (AAs) from 1170–1200 and the C-terminal end of Rp1-D21. 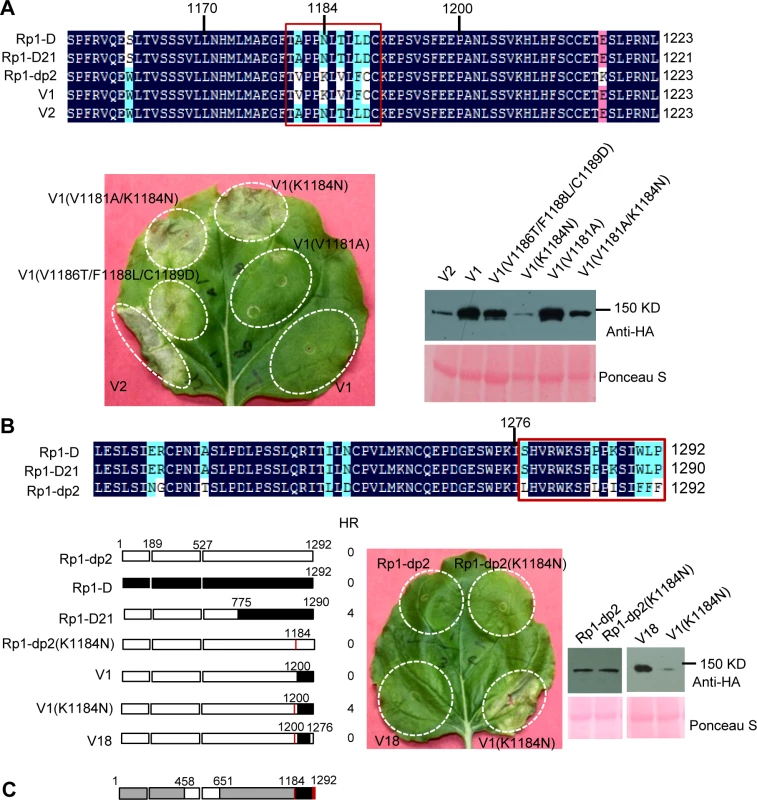
(A) Investigating the point mutations in the region of AAs 1170–1200 which are important for inducing hypersensitive response (HR). Top: Multiple sequence alignment of Rp1-D, Rp1-D21, Rp1-dp2, V1 and V2. The black, red and light blue shaded regions represent 100%, 75% and above 50% similarity of the amino acids, respectively. The polymorphic amino acids are boxed, and the position of 1184 (corresponding to 1182 of Rp1-D21) is labeled above the sequences. Bottom left: The HR phenotype of V1, V2 and V1-derived point mutants as indicated. A representative leaf was photographed at 3 days post infiltration (dpi). Bottom right: Total protein was extracted from agro-infiltrated leaves at 30 hours post infiltration (hpi), and anti-HA antibody was used to detect the expression of the fused proteins. Equal loading of protein samples was shown by Ponceau-S staining of Rubisco subunit. (B) Investigating the phenotype conferred by various recombinants and point mutants at the C-terminal end of Rp1-D21. Top: Multiple sequence alignment of the C-terminus of Rp1-D, Rp1-D21 and Rp1-dp2. The polymorphic amino acids are boxed. Bottom left: Schematic diagram of the constructs used. The K1184N mutation is indicated by a red vertical bar in Rp1-dp2(K1184N), V1(K1184N) and V18. Bottom middle: The HR phenotype of the constructs transiently expressed in N. benthamiana. A representative leaf was photographed at 3 dpi. Bottom right: Total protein was extracted from agro-infiltrated leaves at 30 hpi, and anti-HA antibody was used to detect the expression of the fused proteins. Equal loading of protein samples was shown by Ponceau-S staining of Rubisco subunit. The experiments were repeated three times with the same results. (C) The final model presenting the functional regions important for autoactivity. In this model, white represents regions derived from Rp1-dp2 and black the region from Rp1-D that need to be present in the same construct to result in the autoactive HR in N. benthamiana. Regions in grey can be derived from either paralog. The combination of AA 458–651 from Rp1-dp2 and AA 1184–1292 (especially N1184 and the last 16 AAs in red vertical bars) from Rp1-D are important for HR induction. Fig. 8. Investigating the function of Patch 1 and 2 from the ARC2 domain for Rp1-D21 autoactivity. 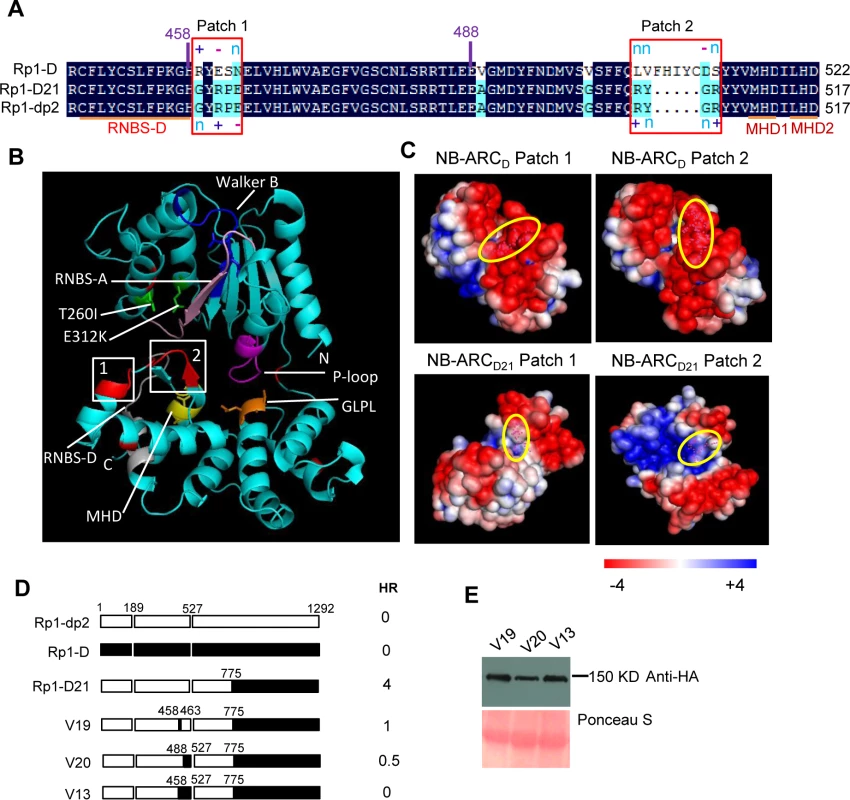
(A) Sequence alignment of Rp1-D, Rp1-D21 and Rp1-dp2 in Patch 1 and 2 regions from the ARC domain. Positive (+), negative (-) and neutral (n) charges were labeled. RNBS-D, MHD1 and MHD2 motifs are indicated by orange bars and labeled below the sequences. (B) Homology modeling of NB-ARCD21 based on the crystal structure of APAF1 (PDB: 1z6t). The P-loop, MHD, GLPL, RNBS-A, RNBS-D and Walker B motifs are color-coded in magenta, yellow, orange, pink, green and blue, respectively. Two boxed regions (Patch 1 and 2) are the major polymorphic regions between Rp1-D and Rp1-dp2 and are labeled in red. The positions of the suppressor mutations T260I and E312K identified by EMS mutagenesis are indicated in green. The N- and C-termini are indicated as N and C, respectively. (C) The surface electropotential of NB-ARCD21 and NB-ARCD. The positive and negative charges are indicated in blue and red, respectively. The positions of Patch 1 and Patch 2 are indicated by pink dots in the yellow oval boxes. (D) Schematic diagram and the hypersensitive response phenotype of the constructs indicated. (E) Protein expression was detected by anti-HA antibody at 3 days post-infiltration. Ponceau-S staining of Rubisco subunit was used as loading control. The experiment was repeated three times with the same results. Constructs V1 through V7 were generated with recombination points between Rp1-dp2 and Rp1-D ranging from AAs 1200 to 488 (Fig. 6A). V1 with a recombination point at AA 1200 did not confer HR while V2 with a recombination point just 30 additional AAs N-terminal to AA 1170 produced a strong HR, indicating that AAs 1170–1200 from Rp1-dp2 are important for auto-inhibition of V1. V4 and V5 (recombination point at 690 and 651, respectively) also produced strong HR phenotype, while V6 and V7 (recombination point 575, 488 respectively) conferred very weak or no HR (rating 1.5 and 0, respectively, Fig. 6A). These results suggest that the AAs 488–651 from Rp1-dp2 are important for the autoactivity of Rp1-D21. Together these data suggest that proteins with a combination of AAs 488–651 from Rp1-dp2 and AAs 1170–1292 from Rp1-D replacing the corresponding sequences of either parental protein are unable to maintain a self-inhibited state.
To test this hypothesis, we investigated whether swapping the N-terminal region (CC, NB-ARC or AAs 1–488) in Rp1-D21 affected its autoactivity. We generated 8 more chimeric constructs in which we exchanged the Rp1-dp2-derived N-terminal parts of Rp1-D21 with the corresponding parts from Rp1-D (Fig. 6B). Construct V8, in which the CC-NB-ARC (AAs 1–527) of Rp1-dp2 was replaced with the corresponding Rp1-D sequence, did not confer an HR. Similar results were obtained by exchanging the NB-ARC domain in V9. However, V10, exchanging the CC domain of Rp1-D into Rp1-D21 conferred a strong HR (Fig. 6B). These results suggest that the ‘parental origin’ of the CC domain (AAs 1–189) in Rp1-D21 plays only a minor or no role in its autoactive phenotype. V11 and V12 were generated by replacing AAs 190–370 or 190–458 (NB-ARC1 plus part of ARC2, respectively) of V10 with the corresponding Rp1-D sequence. They differed from V10 by only 2 and 5 single amino acid polymorphisms (SAAPs), respectively. V11 conferred a slightly weaker HR than V10 and the HR conferred by V12 was weaker still (Fig. 6B). Construct V13, of which the CC-NB-ARC domain was ‘reciprocal’ of V12, did not confer HR, confirming that AAs 458–527 (ARC2 region) from Rp1-dp2 were important for the autoactivity of Rp1-D21 (comparing V13, V12 and V8). Constructs V14 and V15 were generated by exchanging AAs regions 690–775 and 651–775 respectively of V10 with the corresponding Rp1-D sequence. Both constructs showed strong HR (Fig. 6B), similar with the corresponding AAs exchanges in Rp1-D21 (constructs V4 and V5 in Fig. 6A). V16 and V17 were ‘reciprocals’ of Rp1-D21 and V3, respectively and did not confer an HR phenotype (Fig. 6C).
In summary, this set of experiments indicated that the combination of AAs 458–651 from Rp1-dp2 and AAs 1170–1200 from Rp1-D is central to the deficiency of self-inhibition of Rp1-D21.
Defining specific regions (amino acids) important for Rp1-D21 autoactivity
There are only 17 single amino acid polymorphisms (SAAPs) between AAs 1170–1292 of Rp1-D and Rp1-dp2, and only 5 SAAPs in the AAs 1170–1200 region that differentiate constructs V1 and V2 (Figs. 6A and 7A). However, V2 was autoactive, while V1 was not (Fig. 6). To analyze which SAAP is important for the self-inhibition of Rp1 proteins, we generated several mutants based on these 5 SAAPs in the autoinhibited construct V1, including V1(V1181A), V1(K1184N), V1(V1181A/K1184N) and V1(V1186T/F1188L/C1189D). When transiently expressed in N. benthamiana, V1(V1181A) was not autoactive, while V1(K1184N) and V1(V1181A/K1184N) produced a strong HR (rating 4), and V1(V1186T/F1188L/C1189D) a weak HR (rating 2; Fig. 7A). The results indicated that residue K1184 from Rp1-dp2 plays an important role in self-inhibition, and the residues V1186/F1188/C1189 play a minor role. Interestingly, one of the Rp1-D21 suppressor EMS mutants had a mutation at residue 1180 (P to S), only 2 AAs away from N1182 in Rp1-D21 (equivalent to N1184 in Rp1-D), emphasizing the importance of this region for regulating the activity of the Rp1 proteins.
To investigate whether mutating K1184 was sufficient to convert Rp1-dp2 into an autoactive NLR, we generated Rp1-dp2(K1184N). However, this single point mutation was not able to ‘activate’ Rp1-dp2 (Fig. 7B). This result suggests that additional SAAPs in the region of AAs 1200–1292 are also important for self-inhibition of Rp1-dp2. There are 12 additional SAAPs in the last 92 AAs between Rp1-D and Rp1-dp2 and of these six are found in the last 16 AAs (Fig. 7B). Therefore, we constructed the chimeric protein V18 by replacing the C-terminal 16 AAs in V1(K1184N), the autoactive V1 protein, with the equivalent AAs from Rp1-dp2. Interestingly, this chimeric protein V18 lost the ability to induce HR (Fig. 7B), demonstrating that the very C-terminal 16 AAs are very important for the self-inhibition of the Rp1 proteins.
In conclusion, these experiments further defined the combination of AAs required for the autoactivity of Rp1-D21: a combination of AAs 458–651 from Rp1-dp2 and AAs 1184–1292 (especially N1184 and the last 16 AAs) from Rp1-D are central to the autoactivity (see model in Fig. 7C).
Homology modeling of the NB-ARC from Rp1-D and Rp1-D21 reveals novel structural elements required for HR
As shown in Fig. 6B (construct V13), AAs 458–527 from Rp1-dp2 were very important for the autoactivity of Rp1-D21. There are two major regions of polymorphism, which we termed Patch 1 and 2, between Rp1-D and Rp1-D21 within the ARC2 sub-domain, from AAs 458–527 (Figs. 1 and 8). Homology modeling based on the crystal structure of the NB-ARC domain of the ADP-bound human apoptosis regulator APAF1 (Protein data bank entry 1z6t; [40]) predicted that Patch 1 and 2 were surface-exposed (Figs. 8A and 8B). Surface electropotential predictions indicated that the surfaces of both patches in Rp1-D were mostly negatively charged, while for Rp1-D21 Patch 1 was slightly positively charged and Patch 2 was mostly positively charged (Fig. 8C). To investigate whether Patch 1 and 2 played roles in the autoactivity of Rp1-D21, we constructed another two chimeric proteins, V19 and V20, by replacing AAs 458–463 (Patch 1) and 488–527 (Patch 2) in Rp1-D21 with the corresponding AAs from Rp1-D, respectively (Fig. 8D). Both V19 and V20 conferred a greatly reduced HR compared to Rp1-D21, while V13 (containing Patch 1+2 from Rp1-D) did not cause HR (Fig. 8D), suggesting that these patches have an additive effect on the autoactivity of Rp1-D21.
Intra - and inter-molecular interactions of Rp1 proteins
Self-association has been observed in both CNL (eg. barley MLA, Arabidopsis RPS5) and TNL (eg. tobacco N, flax L6) proteins and is important for the activity of NLRs [38,41,42,43]. To test whether Rp1-D21, Rp1-D or Rp1-dp2 could self-associate, we transiently co-expressed EGFP - and 3×HA-tagged proteins in N. benthamiana and performed co-immunoprecipitation (Co-IP) analyses. We observed that full-length Rp1-D21, Rp1-D and Rp1-dp2 all self-associated (Fig. 9A). We further showed that all three of the domains (CCD21, NB-ARCD21 and LRRD21) of Rp1-D21, were able to self-associate (Fig. 9B).
Fig. 9. Investigation of self-association of full length Rp1-D21, Rp1-D, Rp1-dp2, the separate domains of Rp1-D21, and the interaction of full length Rp1-D21 with Rp1-dp2. 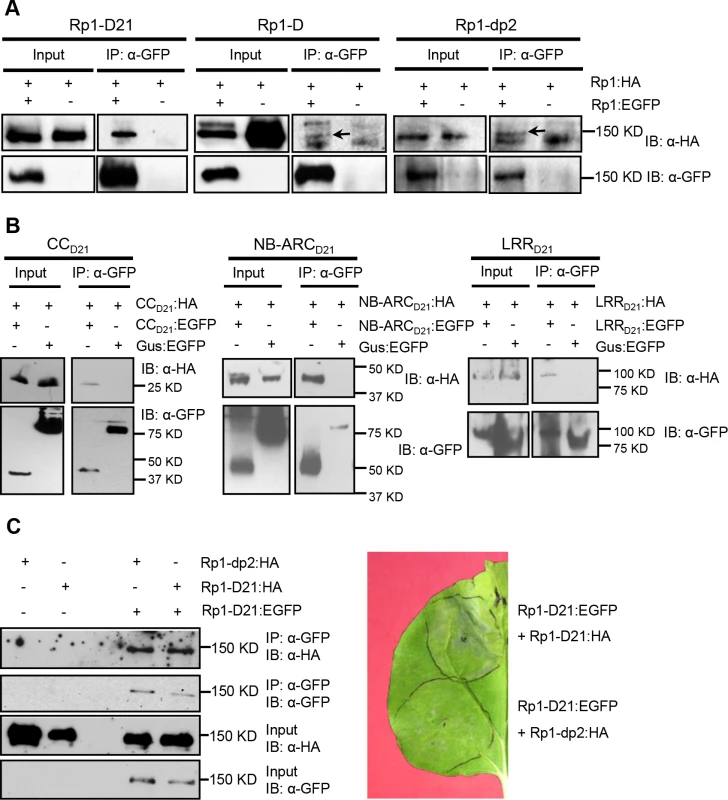
(A) Self-association of 3×HA- and EGFP-tagged Rp1-D21 (left panel), Rp1-D (middle panel) and Rp1-dp2 (right panel). Arrows indicate the target bands. (B) Self-association of CCD21, NBD21 and LRRD21. (C) Left: Co-IP after co-expression of Rp1-D21 and Rp1-dp2. Right: HR induced by co-expression of Rp1-D21:EGFP with Rp1-D21:HA compared to HR induced by co-expression of Rp1-D21:EGFP with Rp1-dp2:HA. EGFP- and 3×HA-tagged constructs were transiently co-expressed in N. benthamiana and samples were collected at 30 hours post infiltration for Co-IP assay. Protein extract was immunoprecipitated by anti-GFP microbeads and detected by anti-GFP and anti-HA antibodies. The experiments were repeated three times with the same results. To further investigate whether Rp1-D or Rp1-dp2 could form heteromers with Rp1-D21, we transiently co-expressed Rp1-D21:EGFP and 3×HA-tagged Rp1-D or Rp1-dp2 proteins in N. benthamiana and performed Co-IP analyses. We found that Rp1-dp2 interacted with Rp1-D21 (Fig. 9C). We did not detect interaction between Rp1-D and Rp1-D21, which might have been due to the low expression of Rp1-D (Fig. 3B). Consistent with the interaction data, we observed that Rp1-dp2 could partially suppress Rp1-D21-induced HR (Fig. 9C).
ARC2D21 is important for the interaction of NB-ARCD21 with CCD21
To investigate whether different intra-molecular interactions are correlated with the different activities of Rp1-D21 and Rp1-D, we co-expressed pair-wise combinations of different domains in N. benthamiana and analyzed by Co-IP. We observed that NB-ARCD21 interacted with CCD21, but not with LRRD21 or LRRdp2 (Fig. 10; S7A–S7B Fig.). Additionally, we did not observe interactions between NB-ARCD and CCD or between NB-ARCD and LRRD under our conditions (Fig. 10; S7A–S7B Fig.).
Fig. 10. Schematic diagram of the intra-molecular interactions and HR phenotype of the constructs indicated, summarizing the data shown in S7 Fig. in which the various domains were co-expressed and co-immunoprecipitated in trans. 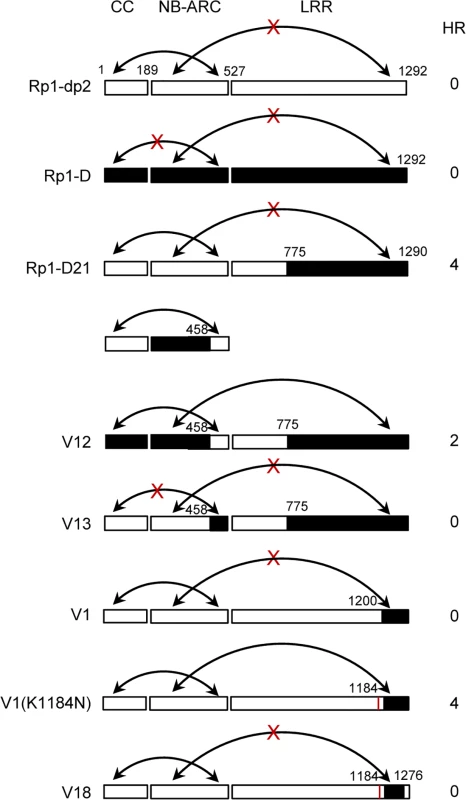
The gene structure has been divided into CC, NB-ARC and LRR domains. The amino acid position of the recombination site of each construct was indicated above the construct. For each construct, the strength of the hypersensitive response (HR) resulting from transient expression of each full length molecule in N. benthamiana was scored on a 0 (no HR) to 5 (strong HR) scale. The ability to form or not to form intra-molecular interactions among different domains was indicated by arrows or arrows with crosses, respectively. Since the CC and the NB-ARC domains in Rp1-D21 and Rp1-D have different interactions, we investigated the regions required for this interaction. As noted above, the ARC2 domain (including Patch 1 and 2) is the major difference in the NB-ARC domain between Rp1-D21 and Rp1-D. In order to test whether this region affects the interaction between the CC and NB-ARC domains, we performed a Co-IP assay with transiently co-expressed CCD21/CCD and NB-ARCV12/NB-ARCV13, and observed that NB-ARCV12 interacted with both CCD and CCD21, while NB-ARCV13 did not interact with CCD21 (Fig. 10; S7A Fig.). The results suggested that ARC2D21 (AAs 458–527 from Rp1-dp2) is important for the interaction between CC and NB-ARC domains. In other words, ARC2D can suppress the interaction.
We also observed that LRRD21 interacted with NB-ARCV12, but not NB-ARCV13 (Fig. 10; S7B Fig.), suggesting that NB-ARC1D is required for the interaction between NB-ARC and LRRD21, which is consistent with previous report that ARC1 is required for binding to LRR of potato CNL protein, Rx [44].
N1184 and the last 16 AAs from Rp1-D are required for the interaction between NB-ARCD21 and LRRV1(K1184N)
To determine whether, as suggested by the recombination studies, N1184 and the C-terminal 16 AAs are involved in the intra-molecular interaction, we performed Co-IP assays with NB-ARCD21 and LRRV1, LRRV1(K1184N) or LRRV18 and observed that NB-ARCD21 interacted with LRRV1(K1184N), but not with LRRV1 and LRRV18 (Fig. 10; S7C Fig.), indicating that N1184 and the last 16 AAs from Rp1-D are required for the interaction between NB-ARCD21 and LRRV1(K1184N).
The effects of CC domain EMS mutants on CCD21-induced HR
We investigated the effect of the five CC domain missense EMS mutations on CCD21-induced HR. Consistent with the results from full length proteins, CCR125W abolished HR induction and CCL89F and CCS108F reduced HR, while CCH59Y had no obvious effect compared to CCD21 (Table 1). Surprisingly, CCD82N induced stronger HR than CCD21, while CCD82N in the full length Rp1-D21 did not induce HR (Table 1).
Co-IP experiments were performed to explore whether the CC domain mutations affect the self-association of CCD21. CCD82N but not the other four point mutants reduced the strength of CCD21 self-association (S8A Fig.). To further test whether the mutations affect the interaction between CCD21 and NB-ARCD21, we chose CCD82N and CCR125W, two mutants that completely abolished HR in full length, to test the inter-domain interaction. We found that both CCD82N and CCR125W greatly reduced the interaction between CCD21 and NB-ARCD21 (S8B Fig.).
Discussion
A genetic screen identifies intragenic Rp1-D21 suppressor mutants
While 10 of the 12 Rp1-D21 loss of function EMS mutants isolated from maize also reduced or abolished the HR phenotype when transiently expressed in N. benthamiana, two, H59Y and G850D, did not. It is possible that these two mutations reduced the activity of Rp1-D21 to a level below the threshold for signaling in maize, but not in N. benthamiana due to the higher expression.
One mutation (D82N) was in the conserved EDVID motif. Previous reports have indicated an important role for the EDVID motif in NLR function. In Rx, the EDVID motif mediates the intra-molecular interaction between the CC and the NB-ARC-LRR domains [30,45] and the residues flanking the EDVID motif affect inter-molecular interactions with RanGAP2, which is required for Rx function [30,46,47]. In this study, we found that D82N abolished HR induced by full length Rp1-D21, but not by CCD21 (Tables 1 and 2), similar to what was observed for a mutation in the EDVID motif of MLA10 [27]. We further showed that CCD82N and CCR125W reduced the interaction with NB-ARCD21 (S8B Fig.). The “mousetrap model” for NLR activation [48,49] suggests that, like a mousetrap, the NLR protein must be ‘set’ in a primed state, ready to be activated. The apparent paradox that the D82N mutation causes loss of autoactivity in full length Rp1-D21 but not in the CC domain alone may be explained by the reduced association of CCD21 and NB-ARCD21 and the consequent loss of ability to set up this initial primed state in the full-length protein. It is also likely that the mutations in the CC domain might disrupt the inter-molecular interactions of Rp1-D21 with other co-factors.
Three of the missense mutations were within or next to the ADP binding pocket of the NB-ARC domain when the NB-ARC was modeled onto the APAF-1 structure; T260I was close to the RNBS-A motif, E312K was next to Walker B motif and P398L was in the GLPL motif (Table 1; Figs. 1 and 8B), suggesting that these mutations might change the ADP binding state. Consistent with our results, important roles for the RNBS-A, Walker B and GLPL motifs on modulating HR have been reported in other NLRs [14,42,44]. Mutations in the Walker B and GLPL motifs can also affect the intra-molecular interaction between CC and NB-ARC-LRR domains as evidenced in Rx [30]. Thus, we infer that the three mutations in the NB-ARC domain might also affect the interaction between CCD21 and NB-ARCD21.
Finally, four of the suppressor mutations (S737L, S794F, G850D and P1180S) were in the LRR domain (Fig. 2B). Loss-of-function mutations in the LRR domain have been reported in several NLRs [16,50,51,52,53,54]. According to our model (see below), the mutations likely abolish the ability of the LRRD21 domain to destabilize the interaction between NB-ARCD21 and CCD21.
Rp1 activity is P-loop - and MHD-dependent
Mutations from H (histidine) to A (alanine) or D (aspartate) to V (valine) in the highly conserved MHD domain result in constitutive activity of multiple NLRs from multiple species [17,27,37,41,55,56]. In the Flax NLR, M, ADP is bound to wild-type NB-ARC domain, while ATP is bound to the autoactive MHD mutant M(D555V) [15], providing direct evidence that the inactive “off” NLR binds ADP while the active “on” NLR binds ATP. It appears therefore that the MHD motif is important for maintaining the NLR protein in its appropriate state of activity.
Rp1-dp2 and Rp1-D as well as several other CNLs, but not TNLs, have two MHD motifs [17,37]. We have termed these motifs MHD1 and MHD2 (Fig. 4A). MHD1 is more conserved and is the functional MHD motif for many NLRs as defined in previous studies [15,17,27,38,56]. The aspartate (D) in MHD2 is quite widely conserved throughout most CNLs [37]. Of the CNLs that contained two MHD motifs [37], the effects of mutations in the MHD1 domain only have been reported for tomato NLR Mi-1.2(H840A) and Mi-1.2(D841V). Both mutations activate these proteins [34]. The possible function of the MHD2 domain has recently been examined in rice CNL-RGA5, but no functional effect was observed [57]. Transient expression of Rp1-dp2(D517V) and of V1(D517V) and V16(D522V), in which the MHD2 was mutated, conferred autoactive HR, while no MHD1 mutation had this effect (Figs. 4B and 4C; S3 Fig.). It is possible that the MHD2 rather than the MHD1 domain is functional in Rp1-dp2, or that the MHD1 and MHD2 domains coordinate the activity. This is the first report showing the functional significance of the MHD2 domain of a CNL for its activity.
The P-loop motif in the NB-ARC domain regulates nucleotide binding. P-loop mutations abolish the ability to confer disease resistance or HR induction in multiple NLR proteins [14,15,27,34,38,39,56]. As expected, the HR induced by Rp1-D21 and the MHD2 mutant Rp1-dp2(D517V) was abrogated by the introduction of a P-loop mutation (Fig. 4D), indicating that the activity of the Rp1 proteins is P-loop dependent.
Two minimum functional regions in the NB-ARC and the LRR domain are important for regulating Rp1 protein activity
There are several examples in the literature of recombinations between the TIR-NB-ARC or CC-NB-ARC and LRR domains of different NLRs resulting in autoactive proteins. The recombination of CC-NB-ARC from Gpa2 with LRR from Rx1 produces a gene conferring autoactive HR; The combination of Gpa2-ARC2 and the first two repeats of Rx-LRR region is essential for autoactivity [36,44]. Domain swaps between RPS5 and RPS2 [29] and between Mi-1.1 and Mi-1.2 [58] also gave rise to genes conferring an autoactive phenotype. In contrast to these artificially-constructed autoactive NLRs characterized in transient assays, Rp1-D21 is an autoactive protein that occurred via recombination and was identified in its endogenous genetic background. To identify the precise structural requirement for its activity, we performed a systematic structural analysis of Rp1-D21 using a set of artificial recombinants between the two ‘parental’ alleles (Fig. 6) and showed that the combination of AAs 458–651 (the ARC2 and N-terminus of the LRR region) from Rp1-dp2 and the C-terminal LRR region (especially N1184 and the C-terminal 16 AAs) from Rp1-D was critical for the autoactivity of Rp1-D21. This combination either destabilized the Rp1-D21 intra-molecular interactions that cause the inactive resting state, or stabilized interactions resulting in the active state (see below). In other words, these two regions appear to be important for maintaining the parental proteins in an inactive ‘resting’ state. Thus, in light of the “mousetrap” model [48], this region of the Rp1 family NB-ARC and LRR domains is involved in a precarious autoinhibiting conformation that is easily broken by alterations including exchange of a very small C-terminal region of the LRR domain, or presumably, in a wild-type context by the action of the cognate effector protein. It seems evident that the NB-ARC and LRR domains within each NLR must co-evolve to maintain the NLRs in a suitably auto-inhibited resting state in the absence of pathogen infection while maintaining the ability to respond to the cognate effector via intra-molecular interactions [36,38,55], but that precise mechanisms of auto-inhibition and activation may vary between NLRs.
The role of Patch 1 and 2 in the ARC2 domain
The ARC2 domain is important for function in several NLRs [36]. We identified two major polymorphic regions localized in ARC2 that differentiated the NB-ARCD21 and NB-ARCD domains, Patch 1 and Patch 2, and showed that they are important for the autoactivity of Rp1-D21 (Figs. 1 and 8). Replacing either Patch 1 or Patch 2 of Rp1-D21 with the region from Rp1-D almost completely suppressed the autoactive phenotype (constructs V13, V19 and V20; Fig. 8D). In agreement with our structural modeling, these two patches were surface-exposed and carried largely opposite charges in NB-ARCD21 compared to NB-ARCD (Fig. 8B). The prevailing model for the activation of NLRs is that the off state binds ADP while the on state binds ATP [36,59]. Patch 2 is located three AAs N-terminal to the MHD1 motif and is localized in the exposed surface next to the nucleotide binding pocket [40], suggesting it might affect the state of nucleotide binding. Patch 1 is adjacent to a conserved RNBS-D motif in the ARC2 domain (Figs. 1 and 3A). Mutations in or next to the RNBS-D motif of PM3, RPM1 and Rx affected their function [16,36,45,55]. Thus, the sequence differences of Patch 1 between NB-ARCD21 and NB-ARCD might also affect the protein activity through disturbing the function of the RNBS-D motif.
Interestingly, we found that ARC2D21 is critical for the interaction between NB-ARCD21 and CCD21 or CCD (Fig. 10, compare V13 with V1 and Rp1-D with V12, and see S7 Fig.). This is, to our knowledge, the first demonstration of the role of ARC2 in determining NB-ARC and CC interaction in plant NLRs. Our modeling data also suggests why we detected interaction between NB-ARCD21 and CCD21 but not between NB-ARCD and CCD (Fig. 10; S7 Fig.). In the CC domain, the side chain of the EDVID motif of CNLs is largely negatively charged, and this motif is thought to mediate intra-molecular interactions of the CC domain with the NB-ARC-LRR domain of Rx [30,60]. In Rp1 proteins, the negatively charged EDVID and the positively-charged ARC2D21 are apparently very important for the interaction between CCD21 and NB-ARCD21 while the ARC2D is more negatively charged (Fig. 8C) and thus may not interact as strongly with the CC. Interestingly, we observed that AAs 190–260 from the NB domain were sufficient to suppress CCD21-induced HR (S4 Fig.), suggesting that ARC2D21 is not the only region which can regulate Rp1-D21 autoactivity.
The role of different Rp1-D21 domains in autoactivity
CCD21 and CCD domains alone were sufficient to induce HR when fused with EGFP, but not with 3×HA or on their own (Fig. 5; S1 Fig.). This phenomenon of “tag-dependent activity” has been observed in other NLR studies [30,61]. The functional domains required for HR induction vary in different NLR proteins. The CC domain alone is sufficient for the HR phenotype triggered by MLA10, and also by NRG1 and ADR1, which contain atypical CC domains [28,41], while the NB-ARC domain of Rx can trigger HR [30], and for RPS5 the CC-NB-ARC is sufficient [29].
The fact that transient expression of CC-NB-ARCD21 or CC-NB-ARCD did not induce HR suggested that their respective NB-ARC domains repressed the signaling by the CC domain. Since full length Rp1-D21 induced HR and full length Rp1-D did not (Fig. 3), we inferred that the LRRD21 or LRRD domains are structurally incompatible with, respectively, repression of CCD21 autoactivity or with CCD autoactivity (Fig. 5). While interaction between the LRR and NB-ARC domains was detected in some combinations (e.g., see proteins V12, V1(K1184N) in Fig. 10), we did not detect any interaction between the LRR and NB-ARC domains of Rp1-D, Rp1-dp2 or Rp1-D21 in trans (Fig. 10; S7 Fig.). It is possible though that these domains interact in the full length context, or that their interactions are weak or transient and cannot be detected in our Co-IP conditions. Consistent with this, LRRD21 was unable to alter the suppression of CCD21 autoactivity by NB-ARCD21 in trans (Table 2). However, we cannot exclude the possibility that the lack of suppression in trans was due to the relatively low expression of LRRD21 (Table 2; Fig. 5B). A similar finding of cis but not trans interaction/autoactivity was reported for potato Rx and autoactive MHD mutants of tomato Mi-1.2 [34,44,62]. It is notable also that the CC-NB-ARC fusion from MLA10 and Rx can still trigger HR [27,30], but not in Rp1-D and Rp1-D21.
LRR domains have multiple reported roles in NLR activation. They are essential for the activation of the NLRs Rx and Mi-1.2 [34,62]. In the case of RPS5, the first four LRRs are the minimum region sufficient to inhibit the autoactive phenotype conferred by the CC-NB-ARC domain [29]. Conversely, the activation of RPS5 in response to disease requires the entire LRR domain [29]. Our analyses of Rp1-D21 C-terminal deletion constructs and LRR domain swap data (Fig. 7B; S5 Fig.) further confirmed the importance of the C-terminal LRR domain in regulating the activity of the Rp1 proteins. Similarly, the importance of C-terminal LRR domain has also been observed in flax TNL, L proteins [37].
The interaction patterns observed between NB-ARC and LRR from V1, V1(K1184N) and V18 suggested that K1184N and the last 16 AAs from Rp1-D are required for the inter-domain interaction (Fig. 10; S7 Fig.). No interaction was detected between the NB-ARC and LRR domains from V1 or between those from V18 when they were expressed in trans, but a trans-interaction was detected between these domains using the V1(K1184N) construct. Assuming that these interactions are maintained in the full length proteins, this result implies that the ability of V1(K1184N) to induce HR is due to the interaction of the LRR with the NB-ARC, which apparently is then locked into an activated state allowing CC-dependent HR. This also requires the C-terminal 16 amino acids of the Rp1-D21 LRR domain (Fig. 10; S7 Fig.). A further inference is that the C-terminal 16 AAs and N1182 in Rp1-D21 (corresponding to N1184 in Rp1-D) are crucial for the inhibition of NB-ARC activity that is required for CC-dependent HR.
A model for the autoactivity of Rp1-D21
Alternate “on” or “off” states mediated through intra-molecular interactions are thought to be one of the major mechanisms regulating NLR activity [51,62]. For example, intra-molecular interactions were detected between CC and NB-ARC-LRR, and CC-NB-ARC and LRR domains of Rx in the absence but not in the presence of its cognate effector, the PVX coat protein [62]. Similarly the differentially-activated states of Rp1-D, Rp1-dp2, Rp1-D21 and our additional recombinant proteins are likely due to the specific intra-molecular interactions found within each protein. While Rp1-D21 and V3 confer HR, their reciprocal constructs, V16 and V17, do not (Fig. 6C), indicating that the autoactivity of the recombinant protein is triggered by specific combination of sequences from Rp1-dp2 and Rp1-D. Our model (Fig. 11) explaining the intra-molecular interactions underlying the activity of the Rp1 proteins is as follows:
Fig. 11. Model of the intra-molecular interactions controlling auto-inhibition or auto-activation in Rp1 proteins. 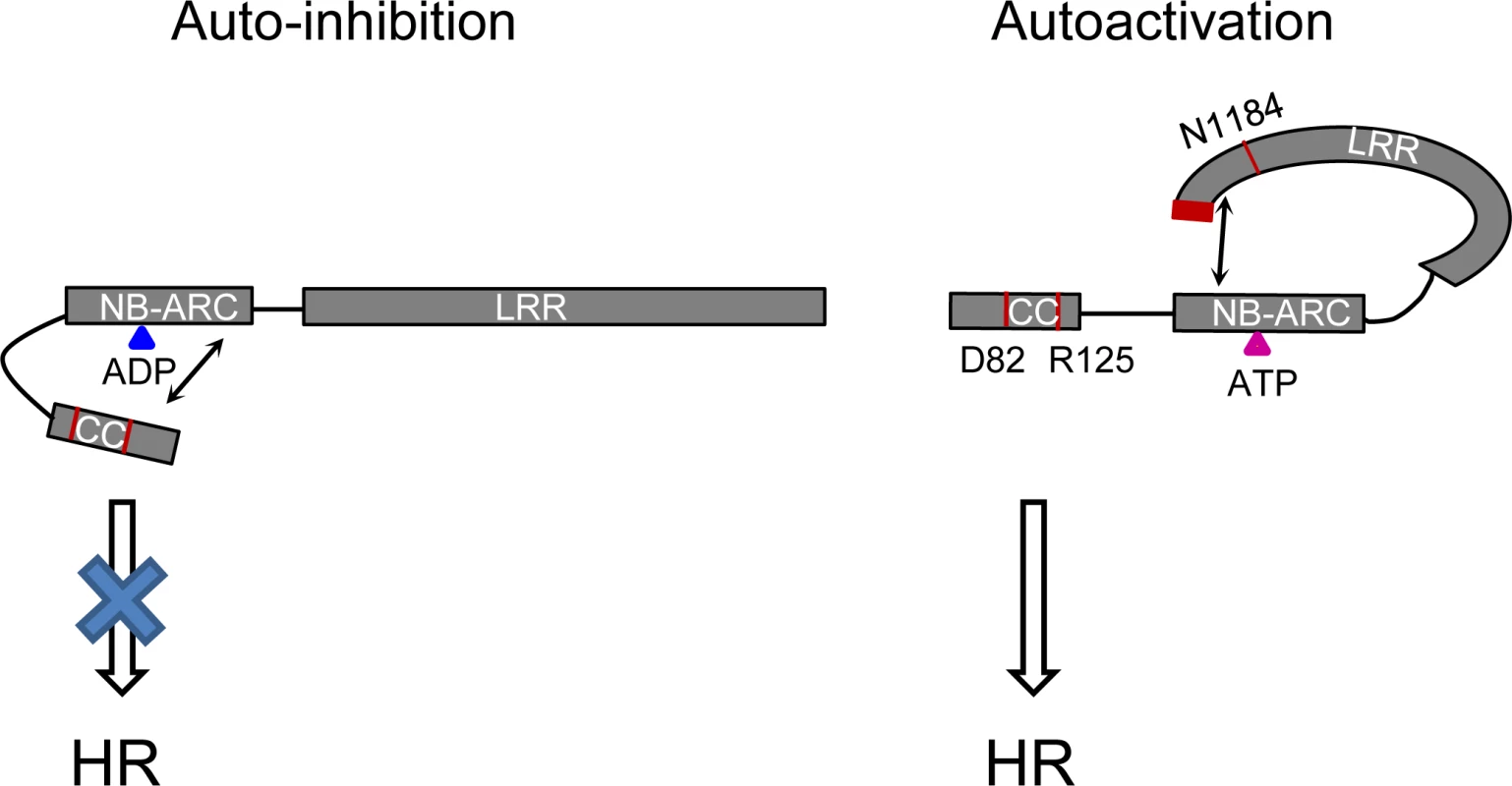
In the auto-inhibited state (left), the NB-ARC domain inhibits CC-induced HR via intra-molecular interaction involving the ARC2 domain. In the auto-activated state (right), the LRR suppresses an inhibitory effect of the NB-ARC on the CC through direct interaction with the NB-ARC which destabilizes the CC/NB-ARC interaction. Blue and pink triangles indicate ADP and ATP, respectively. Arrows indicate intra-molecular interactions. The positions of amino acids (D82, R125, N1184 and the C-terminal 16 amino acids) which are important for activity are labeled in red. The rationale behind the models is explained in the main text. Self-association and heteromer formation between Rp1 proteins are important for their activity.
Self-association of NLRs has been shown to be important for activity in other cases [38,41,42,43]. While we have not been able to show that this is the case for Rp1 proteins, we have shown that Rp1-D21, Rp1-D, Rp1-dp2 and each of the three major domains, CCD21, NB-ARCD21, LRRD21 can self-associate (Fig. 9). Based on these related studies, we propose that this self-association is important for function.
Heteromers formed by different NLRs have been shown to be important for NLR activity [57,63]. We found that Rp1-dp2 and Rp1-D21 physically interacted and that their co-expression suppressed Rp1-D21-induced HR, presumably due to their physical interaction (Fig. 9C). This is also consistent with our previous study in which we identified a quantitative trait locus mapping directly to Rp1 which regulated the severity of Rp1-D21-induced HR in maize [26].
The CC domain is the signaling competent region of the Rp1 proteins.
This is demonstrated by the ability of CC expressed alone to induce HR (Fig. 5).
The NB-ARC domain inhibits CC-induced HR in Rp1-D21 via intra-molecular interactions involving the ARC2 domain.
In Rp1-D, NB-ARCD can inhibit CCD-induced HR in cis, but not in trans (Fig. 5A; Table 2). Though we did not detect direct physical interaction between NB-ARCD and CCD, the fact that NB-ARCD inhibits HR caused by CCD in cis suggests that there is inhibitory interaction between the CC and NB-ARC domains in both Rp1-D and Rp1-D21. Data from the recombination (Fig. 6) and Co-IP experiments (Fig. 10; S7 Fig.) indicate that the ARC2 domain is important for controlling both activity and intra-molecular interactions of Rp1 proteins. We propose that the intra-molecular CC/NB-ARC interactions in which ARC2 domain plays a critical role are important for setting up a ‘primed’ state as proposed under the “mousetrap model” for NLR activation [48,49].
In Rp1-D21 and the other autoactive recombinant proteins, the LRR destabilizes the CC/NB-ARC interaction, leading to auto-activation.
The best evidence for this comes from the comparison of the interaction patterns of V1, V1(K1184N) and V18 in Fig. 10. These proteins differ by only 1 and 6 AAs, respectively. In V1(K1184N) the LRR and NB-ARC domains interact in trans. This is correlated with the ability of the corresponding full length protein to confer HR in the transient expression system, since neither V1 nor V18 confer HR and their respective LRR domains do not interact with the shared NB-ARC. Consistent with these data, truncation of the C-terminal 34 AAs of Rp1-D21 eliminates autoactivity (see also #6, below). Thus, 7 AA changes from Rp1-D in Rp1-D21 in a protein otherwise derived from Rp1-dp2 are sufficient to de-stabilize the LRR’s ability to inhibit ectopic NB-ARC activity and consequent CC-dependent HR.
Although we did not observe Co-IP between LRRD21 and NB-ARCD21, we propose, for reasons detailed above, that there is an interaction between these two domains in cis but not in trans. This would be consistent with our data that NB-ARC-LRRD21 is unable to inhibit CCD21 signaling when present in trans (Table 2).
Destabilization of the CC/NB-ARC inhibitory interaction allows for the exchange of bound ADP for ATP in a P-loop, RNBS-A, Walker B, GLPL and MHD2/ARC2-dependent manner.
Mutants in or next to P-loop, RNBS-A, Walker B, GLPL and ARC2 lose the ability to confer HR (Table 1; Fig. 4; S2 Fig.).
The autoactivated state of Rp1-D21 and various other derived autoactive recombinants in which the CC/NB-ARC interaction is destabilized, depends upon the combined presence of AAs 458–651 (Patches 1 and 2 in ARC2 and the N-terminal LRR region) from Rp1-dp2 and of the C-terminal LRR region (especially N1184 and the last 16 AAs) from Rp1-D.
The data for this is discussed above and detailed in Figs. 6 and 7.
While we have not identified a cognate effector of Rp1-D and do not know how it is detected, we hypothesize that its presence is necessary for activating Rp1-D via direct or indirect interaction with the LRR domain. This model is based both upon the data presented here and a review of the related literature. While plant NLR resistance genes share similar structures and appear to perform very similar functions, it is clear that the molecular mechanisms underlying their function and their appropriate activation, while sharing certain similarities, vary substantially. This variation likely reflects the intimate and unique co-evolutionary processes each NLR has undergone both with respect to the interactions among their different domains and with their cognate pathogen effector proteins, and/or the host targets or decoys of those effectors. The molecular mechanisms underlying the auto-inhibition of the maize NLR common rust resistance protein Rp1-D and its autoactive derivative Rp1-D21 show both broad similarities to and distinct differences from what is known of other NLRs. Furthermore, we have identified several unique, or previously un-observed features, including: the AAs required for HR induction identified through EMS mutagenesis screening; the functional characterization of MHD2 motif; the involvement of the ARC2 domain in the interaction of the NB-ARC domain with the CC domain; the observation that N1184 and the C-terminal 16 AAs are involved both in the LRR/NB-ARC physical interaction and in regulating activity.
Recently, several NLR genes that have been transferred between plant species were demonstrated to still confer expected disease resistance specificities without obvious fitness effects, eg. Arabidopsis RPS4 (Resistance to Pseudomonas syringae 4), RRS1 (Resistance to Ralstonia solanacearum 1) and barley MLA1 [64,65]. Genetic manipulation of Rx indicates stepwise artificial evolution can be used to reduce the costs associated with disease resistance of NLRs [66]. Rp1-D21 is an autoactive mutant conferring nonspecific response to multiple maize rust species, including P. sorghi and P. triticina [19,21]. Presumably the resistance conferred by Rp1-D21 might extend beyond rusts to other biotrophic and hemi-biotrophic pathogens, however, the severe growth penalties associated with the expression of this gene make its application in agricultural production impractical. In conjunction with suitable promoters, it may be possible to engineer maize or even other plants with some of the chimeric constructs characterized here that confer a weaker HR phenotype to achieve an elevated disease resistance with fewer fitness consequences.
Materials and Methods
Plant materials and growth conditions
Wild type N. benthamiana plants were grown at 23°C with a cycle of 16 hrs light/8 hrs dark. Maize line Rp1-D21-H95 [25] was used to isolate the genomic DNA sequence of Rp1-D21. To generate material for EMS mutageneis, Rp1-D21 was first introgressed into A632 by 7 backcrosses (BC7). A few of these BC7 Rp1-D21 heterozygous plants were self-pollinated to generate Rp1-D21 homozygotes in the A632 background which were identified on the basis of their enhanced Rp1-D21 severity compared to the heterozygotes.
EMS mutagenesis screening
Pollen from the homozygous Rp1-D21 plants in an A632 background was collected and treated with EMS for 45 min before using it to fertilize the ears of an inbred line H95. The resulting M1 population was all heterozygous for Rp1-D21 and showed an HR phenotype with a relatively stunted growth stature. In this background, Rp1-D21-suppressed plants due to EMS mutagenesis were easily distinguished from the rest of the M1 siblings because of their non-lesioned and highly robust growth phenotype (Fig. 2A). About 23,000 M1 progenies were screened in field to identify intragenic suppressor mutants that had lost the Rp1-D21 phenotype. In total, 32 mutants which lacked the HR phenotype of Rp1-D21 were identified and 14 of them were characterized in detail. Gene specific primers for Rp1-D21 (S1 Table) were used to sequence the Rp1-D21 gene in these mutants.
Plasmid construction
All primers used in this study are listed in S1 Table. In Rp1-D and all its paralogs, no intron is found in the open reading frame (ORF) region [20,35], thus we amplified the ORFs of Rp1-D, Rp1-dp2 and Rp1-D21 from the plasmids gifted by Dr. Scot Hulbert (Washington State University), and cloned them into pENTR directional TOPO cloning vector (D-TOPO, Invitrogen). After sequencing, they were transferred into gateway vectors by LR reactions: pGWB2 (no tag), pGWB14 (with a 3×HA epitope tag in the C-terminus) or pSITEII-N1-EGFP (with EGFP epitopic tag in the C-terminus) [67,68]. Rp1-D21 was also isolated from maize line Rp1-D21-H95 using the primers listed in S1 Table.
The different domains (CC, CC-NB-ARC, NB-ARC, NB-ARC-LRR, and LRR) of Rp1-D21 or Rp1-D were amplified using primer pairs listed in S1 Table. The resulting PCR products were cloned into D-TOPO and sequenced, then transferred into pGWB2, pGWB14 or pSITEII-N1-EGFP by gateway LR reactions.
Site-directed mutagenesis
Overlapping extension PCR primers (S1 Table) were designed for generating the site-directed mutations: Rp1-D21(K225R), Rp1-D(H517A), Rp1-D(D518V), Rp1-D(H521A), Rp1-D(D522V), Rp1-dp2(D513V), Rp1-dp2(H512A/D513V), Rp1-dp2(H516A), Rp1-dp2(D517V), Rp1-dp2(K225R/D517V) and 12 missense EMS mutations listed in Table 2. The site-directed mutations were cloned into D-TOPO and verified by sequencing and sub-cloned into the gateway vector pGWB14 by LR reaction.
Sequence alignment and homology structure modeling
NLR sequences were aligned by ClustalW (www.ebi.ac.uk), and edited by BioEdit software. Homology modeling of the NB-ARC domain was performed with Phyre 2 [69] based on the crystal structure of human APAF1 (PDB: 1z6t). The three-dimensional structure and the surface electropotential were mapped using PyMOL (http://www.pymol.org/).
Agrobacterium tumefaciens-mediated transient expression
Agrobacterium tumefaciens strain GV3101 (pMP90) transformed with binary vector constructs was grown at 28°C overnight in 5ml L-broth medium supplemented with appropriate antibiotics. The bacteria were collected at 4,000g by centrifugation and resuspended in 2 ml resuspension buffer (10 mM MES pH5.6, 10 mM MgCl2 and 200 μM acetosyringone). The final concentration of the bacteria was diluted to the OD600 of 0.5 using the same resuspension buffer. To prevent the onset of post-transcriptional gene silencing and improve the efficiency of transient expression, a strain containing p19 protein was included at OD600 of 0.2 to all the bacteria strains [70]. The solution was left at room temperature for 3 hrs on bench before infiltration into the abaxial side of N. benthamiana leaves. After infiltration, plants were put at room temperature with 16h-light/8h-dark. At least 15 individual leaves were infiltrated by different constructs, and each experiment was repeated at least three times.
Protein analysis
For protein expression analysis, three leaf discs (1.2 cm diameter) from different single plants were collected at 30 hours post infiltration (hpi). The samples were ground with prechilled plastic pestles in liquid nitrogen, and total protein was extracted in 160 μl extraction buffer [20 mM Tris·HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA (pH 8.0), 1% Triton X-100, 0.1% SDS, 10 mM DTT, 40 μM MG132, and 1× plant protein protease inhibitor mixture (Sigma-Aldrich)]. Samples were centrifuged at 14,000 g for 15 min at 4°C, and 12 μl supernatants were mixed with 2× Laemmli buffer and loaded for SDS-PAGE. For Co-IP assay, EGFP - and 3×HA-tagged constructs were transiently co-expressed in N. benthamiana. Agrobacterium carrying each construct were diluted to a final concentration of OD600 = 0.4 plus p19 with OD600 = 0.2. Samples were collected at 30 hpi, and proteins were extracted by grinding 0.8 g of leaf tissues in 2.4 ml extraction buffer [50 mM HEPES (pH 7.5), 50 mM NaCl, 10 mM EDTA (pH 8.0), 0.5% Triton 100, 4 mM DTT and 1× plant protein protease inhibitor mixture (Sigma-Aldrich)]. Extracts were centrifuged at 14,000 rpm for 20 min at 4°C, and 2 ml supernatant was mixed with 30 μl anti-GFP microbeads (Miltenyi Biotec) and rotated for 3 hrs at 4°C. The samples were passed through pre-equilibrated MACS separation columns, and washed four times (1 ml, 1 ml, 500 μl and 500 μl) by washing buffer [50 mM HEPES (pH 7.5), 150 mM NaCl, 10 mM EDTA (pH 8.0), 0.2% Triton 100, 4 mM DTT and 1× plant protein protease inhibitor mixture (Sigma-Aldrich)]. The proteins were eluted by 100 μl pre-heated elution buffer and separated by SDS-PAGE. Proteins were transferred to nitrocellulose membrane (Fisher), and analyzed by western blot. HA detection was performed using a 1 : 350 dilution of anti-HA-HRP (horseradish peroxidase) (Cat# 12013819001, Roche). GFP detection was performed using a 1 : 8,000 dilution of primary mouse monoclonal anti-GFP (Cat# ab1218, Abcam), followed by hybridization with a 1 : 15,000 dilution of anti-mouse-HRP second antibody (Cat# A4426, Sigma). The HRP signal was detected by ECL substrate kit (Supersignal west femto chemiluminescent substrate, Thermo Scientific).
Supporting Information
Zdroje
1. Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JD (1995) Molecular genetics of plant disease resistance. Science 268 : 661–667. 7732374
2. Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411 : 826–833. 11459065
3. Ellis J, Dodds P, Pryor T (2000) Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol 3 : 278–284. 10873844
4. Pan Q, Wendel J, Fluhr R (2000) Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol 50 : 203–213. 10754062
5. Bent AF, Mackey D (2007) Elicitors, Effectors, and R Genes: The New Paradigm and a Lifetime Supply of Questions. Annual Review of Phytopathology 45 : 399–436. 17506648
6. Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E (2008) The hypersensitive response; the centenary is upon us but how much do we know? Journal of experimental Botany 59 : 501–520. 18079135
7. Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, et al. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315 : 1098–1103. 17185563
8. Mackey D, Holt BF 3rd, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108 : 743–754. 11955429
9. McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7 : 212. 16677430
10. Albrecht M, Takken FL (2006) Update on the domain architectures of NLRs and R proteins. Biochem Biophys Res Commun 339 : 459–462. 16271351
11. Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, et al. (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20 : 317–332. 10571892
12. Maekawa T, Kufer TA, Schulze-Lefert P (2011) NLR functions in plant and animal immune systems: so far and yet so close. Nature immunology 12 : 817–826. doi: 10.1038/ni.2083 21852785
13. Takken FL, Albrecht M, Tameling WI (2006) Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol 9 : 383–390. 16713729
14. Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, et al. (2002) The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14 : 2929–2939. 12417711
15. Williams SJ, Sornaraj P, deCourcy-Ireland E, Menz RI, Kobe B, et al. (2011) An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol Plant Microbe Interact 24 : 897–906. doi: 10.1094/MPMI-03-11-0052 21539434
16. Tornero P, Chao RA, Luthin WN, Goff SA, Dangl JL (2002) Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell 14 : 435–450. 11884685
17. van Ooijen G, Mayr G, Kasiem MM, Albrecht M, Cornelissen BJ, et al. (2008) Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot 59 : 1383–1397. doi: 10.1093/jxb/ern045 18390848
18. Smith SM, Pryor AJ, Hulbert SH (2004) Allelic and Haplotypic Diversity at the Rp1 Rust Resistance Locus of Maize. Genetics 167 : 1939–1947. 15342531
19. Hu G, Richter TE, Hulbert SH, Pryor T (1996) Disease Lesion Mimicry Caused by Mutations in the Rust Resistance Gene rp1. Plant Cell 8 : 1367–1376. 12239417
20. Sun Q, Collins NC, Ayliffe M, Smith SM, Drake J, et al. (2001) Recombination between paralogues at the Rp1 rust resistance locus in maize. Genetics 158 : 423–438. 11333250
21. Smith SM, Steinau M, Trick HN, Hulbert SH (2010) Recombinant Rp1 genes confer necrotic or nonspecific resistance phenotypes. Mol Genet Genomics 283 : 591–602. doi: 10.1007/s00438-010-0536-5 20443026
22. Negeri A, Wang GF, Benavente L, Kibiti CM, Chaikam V, et al. (2013) Characterization of temperature and light effects on the defense response phenotypes associated with the maize Rp1-D21 autoactive resistance gene. BMC Plant Biol 13 : 106. doi: 10.1186/1471-2229-13-106 23890100
23. Chintamanani S, Hulbert SH, Johal GS, Balint-Kurti PJ (2010) Identification of a maize locus that modulates the hypersensitive defense response, using mutant-assisted gene identification and characterization. Genetics 184 : 813–825. doi: 10.1534/genetics.109.111880 20176981
24. Olukolu BA, Negeri A, Dhawan R, Venkata BP, Sharma P, et al. (2013) A connected set of genes associated with programmed cell death implicated in controlling the hypersensitive response in maize. Genetics 193 : 609–620. doi: 10.1534/genetics.112.147595 23222653
25. Chaikam V, Negeri A, Dhawan R, Puchaka B, Ji J, et al. (2011) Use of Mutant-Assisted Gene Identification and Characterization (MAGIC) to identify novel genetic loci that modify the maize hypersensitive response. Theor Appl Genet 123 : 985–997. doi: 10.1007/s00122-011-1641-5 21792633
26. Olukolu BA, Wang GF, Vontimitta V, Venkata BP, Marla S, et al. (2014) A genome-wide association study of the maize hypersensitive defense response identifies genes that cluster in related pathways. PLoS Genet 10: e1004562. doi: 10.1371/journal.pgen.1004562 25166276
27. Bai S, Liu J, Chang C, Zhang L, Maekawa T, et al. (2012) Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog 8: e1002752. doi: 10.1371/journal.ppat.1002752 22685408
28. Collier SM, Hamel LP, Moffett P (2011) Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact 24 : 918–931. doi: 10.1094/MPMI-03-11-0050 21501087
29. Qi D, DeYoung BJ, Innes RW (2012) Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol 158 : 1819–1832. doi: 10.1104/pp.112.194035 22331412
30. Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, et al. (2008) The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20 : 739–751. doi: 10.1105/tpc.107.056036 18344282
31. Inoue H, Hayashi N, Matsushita A, Xinqiong L, Nakayama A, et al. (2013) Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc Natl Acad Sci U S A.
32. Hoser R, Zurczak M, Lichocka M, Zuzga S, Dadlez M, et al. (2013) Nucleocytoplasmic partitioning of tobacco N receptor is modulated by SGT1. New Phytol 200 : 158–171. doi: 10.1111/nph.12347 23731343
33. Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, et al. (2010) Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22 : 4195–4215. doi: 10.1105/tpc.110.077537 21177483
34. van Ooijen G, Mayr G, Albrecht M, Cornelissen BJ, Takken FL (2008) Transcomplementation, but not physical association of the CC-NB-ARC and LRR domains of tomato R protein Mi-1.2 is altered by mutations in the ARC2 subdomain. Mol Plant 1 : 401–410. doi: 10.1093/mp/ssn009 19825549
35. Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, et al. (1999) Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11 : 1365–1376. 10402435
36. Slootweg EJ, Spiridon LN, Roosien J, Butterbach P, Pomp R, et al. (2013) Structural determinants at the interface of the ARC2 and leucine-rich repeat domains control the activation of the plant immune receptors Rx1 and Gpa2. Plant Physiol 162 : 1510–1528. doi: 10.1104/pp.113.218842 23660837
37. Howles P, Lawrence G, Finnegan J, McFadden H, Ayliffe M, et al. (2005) Autoactive alleles of the flax L6 rust resistance gene induce non-race-specific rust resistance associated with the hypersensitive response. Mol Plant Microbe Interact 18 : 570–582. 15986927
38. Ade J, DeYoung BJ, Golstein C, Innes RW (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A 104 : 2531–2536. 17277084
39. Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, et al. (2006) Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci U S A 103 : 8888–8893. 16731621
40. Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y (2005) Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434 : 926–933. 15829969
41. Maekawa T, Cheng W, Spiridon LN, Toller A, Lukasik E, et al. (2011) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9 : 187–199. doi: 10.1016/j.chom.2011.02.008 21402358
42. Mestre P, Baulcombe DC (2006) Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 18 : 491–501. 16387833
43. Bernoux M, Ve T, Williams S, Warren C, Hatters D, et al. (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9 : 200–211. doi: 10.1016/j.chom.2011.02.009 21402359
44. Rairdan GJ, Moffett P (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18 : 2082–2093. 16844906
45. Stirnweis D, Milani SD, Jordan T, Keller B, Brunner S (2014) Substitutions of Two Amino Acids in the Nucleotide-Binding Site Domain of a Resistance Protein Enhance the Hypersensitive Response and Enlarge the PM3F Resistance Spectrum in Wheat. Mol Plant Microbe Interact 27 : 265–276. doi: 10.1094/MPMI-10-13-0297-FI 24329172
46. Tameling WI, Baulcombe DC (2007) Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 19 : 1682–1694. 17526750
47. Sacco MA, Mansoor S, Moffett P (2007) A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J 52 : 82–93. 17655649
48. Moffett P (2009) Mechanisms of recognition in dominant R gene mediated resistance. Adv Virus Res 75 : 1–33. doi: 10.1016/S0065-3527(09)07501-0 20109662
49. Collier SM, Moffett P (2009) NB-LRRs work a "bait and switch" on pathogens. Trends Plant Sci 14 : 521–529. doi: 10.1016/j.tplants.2009.08.001 19720556
50. Farnham G, Baulcombe DC (2006) Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc Natl Acad Sci U S A 103 : 18828–18833. 17021014
51. Hwang CF, Williamson VM (2003) Leucine-rich repeat-mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J 34 : 585–593. 12787241
52. Ellis JG, Lawrence GJ, Luck JE, Dodds PN (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11 : 495–506. 10072407
53. Axtell MJ, McNellis TW, Mudgett MB, Hsu CS, Staskawicz BJ (2001) Mutational analysis of the Arabidopsis RPS2 disease resistance gene and the corresponding pseudomonas syringae avrRpt2 avirulence gene. Mol Plant Microbe Interact 14 : 181–188. 11204781
54. Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19 : 4004–4014. 10921881
55. Bendahmane A, Farnham G, Moffett P, Baulcombe DC (2002) Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32 : 195–204. 12383085
56. Gao Z, Chung EH, Eitas TK, Dangl JL (2011) Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci U S A 108 : 7619–7624. doi: 10.1073/pnas.1104410108 21490299
57. Cesari S, Kanzaki H, Fujiwara T, Bernoux M, Chalvon V, et al. (2014) The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J 33 : 18.
58. Hwang CF, Bhakta AV, Truesdell GM, Pudlo WM, Williamson VM (2000) Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 12 : 1319–1329. 10948252
59. Takken FL, Tameling WI (2009) To nibble at plant resistance proteins. Science 324 : 744–746. doi: 10.1126/science.1171666 19423813
60. Hao W, Collier SM, Moffett P, Chai J (2013) Structural basis for the interaction between the potato virus X resistance protein (Rx) and its cofactor Ran GTPase-activating protein 2 (RanGAP2). J Biol Chem 288 : 35868–35876. doi: 10.1074/jbc.M113.517417 24194517
61. Krasileva KV, Dahlbeck D, Staskawicz BJ (2010) Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22 : 2444–2458. doi: 10.1105/tpc.110.075358 20601497
62. Moffett P, Farnham G, Peart J, Baulcombe DC (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J 21 : 4511–4519. 12198153
63. Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, et al. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344 : 299–303. doi: 10.1126/science.1247357 24744375
64. Narusaka M, Kubo Y, Hatakeyama K, Imamura J, Ezura H, et al. (2013) Interfamily transfer of dual NB-LRR genes confers resistance to multiple pathogens. PLoS One 8: e55954. doi: 10.1371/journal.pone.0055954 23437080
65. Maekawa T, Kracher B, Vernaldi S, Ver Loren van Themaat E, Schulze-Lefert P (2012) Conservation of NLR-triggered immunity across plant lineages. Proc Natl Acad Sci U S A 109 : 20119–20123. doi: 10.1073/pnas.1218059109 23175786
66. Harris CJ, Slootweg EJ, Goverse A, Baulcombe DC (2013) Stepwise artificial evolution of a plant disease resistance gene. Proc Natl Acad Sci U S A 110 : 21189–21194. doi: 10.1073/pnas.1311134110 24324167
67. Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, et al. (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59 : 150–162. doi: 10.1111/j.1365-313X.2009.03850.x 19309457
68. Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, et al. (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104 : 34–41. 17697981
69. Kelley LA, Sternberg MJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4 : 363–371. doi: 10.1038/nprot.2009.2 19247286
70. Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33 : 949–956. 12609035
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek 2014 Reviewer Thank YouČlánek Characterization of Metabolically Quiescent Parasites in Murine Lesions Using Heavy Water LabelingČlánek High Heritability Is Compatible with the Broad Distribution of Set Point Viral Load in HIV Carriers
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 2- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- 2014 Reviewer Thank You
- A Case for Two-Component Signaling Systems As Antifungal Drug Targets
- Prions—Not Your Immunologist’s Pathogen
- Telomeric ORFS in : Does Mediator Tail Wag the Yeast?
- Livestock-Associated : The United States Experience
- The Neurotrophic Receptor Ntrk2 Directs Lymphoid Tissue Neovascularization during Infection
- The Intracellular Bacterium Uses Parasitoid Wasps as Phoretic Vectors for Efficient Horizontal Transmission
- CD200 Receptor Restriction of Myeloid Cell Responses Antagonizes Antiviral Immunity and Facilitates Cytomegalovirus Persistence within Mucosal Tissue
- Phage-mediated Dispersal of Biofilm and Distribution of Bacterial Virulence Genes Is Induced by Quorum Sensing
- CXCL9 Contributes to Antimicrobial Protection of the Gut during Infection Independent of Chemokine-Receptor Signaling
- Mitigation of Prion Infectivity and Conversion Capacity by a Simulated Natural Process—Repeated Cycles of Drying and Wetting
- Approaches Reveal a Key Role for DCs in CD4+ T Cell Activation and Parasite Clearance during the Acute Phase of Experimental Blood-Stage Malaria
- Revealing the Sequence and Resulting Cellular Morphology of Receptor-Ligand Interactions during Invasion of Erythrocytes
- Crystal Structures of the Carboxyl cGMP Binding Domain of the cGMP-dependent Protein Kinase Reveal a Novel Capping Triad Crucial for Merozoite Egress
- Non-redundant and Redundant Roles of Cytomegalovirus gH/gL Complexes in Host Organ Entry and Intra-tissue Spread
- Characterization of Metabolically Quiescent Parasites in Murine Lesions Using Heavy Water Labeling
- A Working Model of How Noroviruses Infect the Intestine
- CD44 Plays a Functional Role in -induced Epithelial Cell Proliferation
- Novel Inhibitors of Cholesterol Degradation in Reveal How the Bacterium’s Metabolism Is Constrained by the Intracellular Environment
- G-Quadruplexes in Pathogens: A Common Route to Virulence Control?
- A Rho GDP Dissociation Inhibitor Produced by Apoptotic T-Cells Inhibits Growth of
- Manipulating Adenovirus Hexon Hypervariable Loops Dictates Immune Neutralisation and Coagulation Factor X-dependent Cell Interaction and
- The RhoGAP SPIN6 Associates with SPL11 and OsRac1 and Negatively Regulates Programmed Cell Death and Innate Immunity in Rice
- Lymph-Node Resident CD8α Dendritic Cells Capture Antigens from Migratory Malaria Sporozoites and Induce CD8 T Cell Responses
- Coordinated Function of Cellular DEAD-Box Helicases in Suppression of Viral RNA Recombination and Maintenance of Viral Genome Integrity
- IL-33-Mediated Protection against Experimental Cerebral Malaria Is Linked to Induction of Type 2 Innate Lymphoid Cells, M2 Macrophages and Regulatory T Cells
- Evasion of Autophagy and Intracellular Killing by Human Myeloid Dendritic Cells Involves DC-SIGN-TLR2 Crosstalk
- CD8 T Cell Response Maturation Defined by Anentropic Specificity and Repertoire Depth Correlates with SIVΔnef-induced Protection
- Diverse Heterologous Primary Infections Radically Alter Immunodominance Hierarchies and Clinical Outcomes Following H7N9 Influenza Challenge in Mice
- Human Adenovirus 52 Uses Sialic Acid-containing Glycoproteins and the Coxsackie and Adenovirus Receptor for Binding to Target Cells
- Super-Resolution Imaging of ESCRT-Proteins at HIV-1 Assembly Sites
- Disruption of an Membrane Protein Causes a Magnesium-dependent Cell Division Defect and Failure to Persist in Mice
- Recognition of Hyphae by Human Plasmacytoid Dendritic Cells Is Mediated by Dectin-2 and Results in Formation of Extracellular Traps
- Essential Domains of Invasins Utilized to Infect Mammalian Host Cells
- High Heritability Is Compatible with the Broad Distribution of Set Point Viral Load in HIV Carriers
- Yeast Prions: Proteins Templating Conformation and an Anti-prion System
- A Novel Mechanism of Bacterial Toxin Transfer within Host Blood Cell-Derived Microvesicles
- A Wild Strain Has Enhanced Epithelial Immunity to a Natural Microsporidian Parasite
- Control of Murine Cytomegalovirus Infection by γδ T Cells
- Dimorphism in Fungal Pathogens of Mammals, Plants, and Insects
- Recognition and Activation Domains Contribute to Allele-Specific Responses of an Arabidopsis NLR Receptor to an Oomycete Effector Protein
- Direct Binding of Retromer to Human Papillomavirus Type 16 Minor Capsid Protein L2 Mediates Endosome Exit during Viral Infection
- Characterization of the Mycobacterial Acyl-CoA Carboxylase Holo Complexes Reveals Their Functional Expansion into Amino Acid Catabolism
- Prion Infections and Anti-PrP Antibodies Trigger Converging Neurotoxic Pathways
- Evolution of Genome Size and Complexity in the
- Antibiotic Modulation of Capsular Exopolysaccharide and Virulence in
- IFNγ Signaling Endows DCs with the Capacity to Control Type I Inflammation during Parasitic Infection through Promoting T-bet+ Regulatory T Cells
- Identification of Effective Subdominant Anti-HIV-1 CD8+ T Cells Within Entire Post-infection and Post-vaccination Immune Responses
- Viral and Cellular Proteins Containing FGDF Motifs Bind G3BP to Block Stress Granule Formation
- ATPaseTb2, a Unique Membrane-bound FoF1-ATPase Component, Is Essential in Bloodstream and Dyskinetoplastic Trypanosomes
- Cytoplasmic Actin Is an Extracellular Insect Immune Factor which Is Secreted upon Immune Challenge and Mediates Phagocytosis and Direct Killing of Bacteria, and Is a Antagonist
- A Specific A/T Polymorphism in Western Tyrosine Phosphorylation B-Motifs Regulates CagA Epithelial Cell Interactions
- Within-host Competition Does Not Select for Virulence in Malaria Parasites; Studies with
- A Membrane-bound eIF2 Alpha Kinase Located in Endosomes Is Regulated by Heme and Controls Differentiation and ROS Levels in
- Cytosolic Access of : Critical Impact of Phagosomal Acidification Control and Demonstration of Occurrence
- Role of Pentraxin 3 in Shaping Arthritogenic Alphaviral Disease: From Enhanced Viral Replication to Immunomodulation
- Rational Development of an Attenuated Recombinant Cyprinid Herpesvirus 3 Vaccine Using Prokaryotic Mutagenesis and In Vivo Bioluminescent Imaging
- HITS-CLIP Analysis Uncovers a Link between the Kaposi’s Sarcoma-Associated Herpesvirus ORF57 Protein and Host Pre-mRNA Metabolism
- Molecular and Functional Analyses of a Maize Autoactive NB-LRR Protein Identify Precise Structural Requirements for Activity
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Control of Murine Cytomegalovirus Infection by γδ T Cells
- ATPaseTb2, a Unique Membrane-bound FoF1-ATPase Component, Is Essential in Bloodstream and Dyskinetoplastic Trypanosomes
- Rational Development of an Attenuated Recombinant Cyprinid Herpesvirus 3 Vaccine Using Prokaryotic Mutagenesis and In Vivo Bioluminescent Imaging
- Telomeric ORFS in : Does Mediator Tail Wag the Yeast?
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



