-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase
Influenza A virus presents a continued threat to global health with considerable economic and social impact. Vaccinations against influenza are not always effective, and many influenza strains have developed resistance to current antiviral drugs. Thus, it is imperative to find new strategies for the prevention and treatment of influenza. Influenza RNA-dependent RNA polymerase is a multifunctional protein essential for both transcription and replication of the viral genome. However, we have little understanding of the mechanisms regulating viral RNA polymerase activity or the innate cellular defenses against this critical viral enzyme. We describe how the E3 ubiquitin ligase, TRIM32, inhibits the activity of the influenza RNA polymerase and defends respiratory epithelial cells against infection with influenza A viruses. TRIM32 directly senses the PB1 subunit of the influenza virus RNA polymerase complex and targets it for ubiquitination and proteasomal degradation, thereby reducing viral polymerase activity.
Published in the journal: TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase. PLoS Pathog 11(6): e32767. doi:10.1371/journal.ppat.1004960
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004960Summary
Influenza A virus presents a continued threat to global health with considerable economic and social impact. Vaccinations against influenza are not always effective, and many influenza strains have developed resistance to current antiviral drugs. Thus, it is imperative to find new strategies for the prevention and treatment of influenza. Influenza RNA-dependent RNA polymerase is a multifunctional protein essential for both transcription and replication of the viral genome. However, we have little understanding of the mechanisms regulating viral RNA polymerase activity or the innate cellular defenses against this critical viral enzyme. We describe how the E3 ubiquitin ligase, TRIM32, inhibits the activity of the influenza RNA polymerase and defends respiratory epithelial cells against infection with influenza A viruses. TRIM32 directly senses the PB1 subunit of the influenza virus RNA polymerase complex and targets it for ubiquitination and proteasomal degradation, thereby reducing viral polymerase activity.
Introduction
Influenza A virus A (IAV) is a human respiratory pathogen that causes seasonal epidemics and occasional global pandemics with devastating levels of morbidity and mortality. IAV is a member of the Orthomyxoviridae family and possesses eight segments of negative-sense single-stranded RNA genome. Replication and transcription of these IAV segments is catalyzed by a heterotrimeric RNA-dependent RNA polymerase complex, which consists of an acidic subunit (PA) and two basic subunits, PB1 and PB2 [1,2].
PB1 is the structural backbone for formation of the IAV polymerase complex [1]. PB1 contains a 14 residue binding site for PA at the N-terminus and a C-terminal domain for PB2 association [3–6]. Since the activity of RNA-dependent polymerases is distinct from enzymes found in host cells, these viral proteins are promising drug targets for interfering with viral replication [7,8]. Little is understood about the natural defenses employed by host cells to defend against the IAV polymerase. In this report, we analyze PB1 protein complexes and find a host interactor, tripartite motif-containing protein 32 (TRIM32), which directly targets PB1 proteins to restrict influenza virus replication.
TRIM32 was initially identified as a protein that binds HIV-1 tat (a key transactivator of viral transcription) [9,10]. TRIM32 contains an N-terminal signature tripartite motif (TRIM) consisting of RING, B-box and coiled-coil domains followed by a spacer segment and a series of NHL repeats. The presence of the RING domain is a sign that TRIM family proteins may function as ubiquitin E3 ligases, catalyzing transfer of ubiquitin from an E2 enzyme to form a covalent bond with a substrate lysine. Genetic mutation in the TRIM32 NHL domains causes recessive hereditary muscle disorders, often with a neurogenic component, including limb girdle muscular dystrophy 2H and sarcotubular myopathy [11–14]. These conditions are phenocopied in knockout mice that lack trim32 [15,16] and knockin animals that carry a disease associated TRIM32 mutation [17]. In addition, mutations in the TRIM32 B-box domain are responsible for Bardet Biedl syndrome, which has a pleiotropic phenotype often accompanied with retinal degeneration [18,19].
TRIM32 is a ubiquitously expressed E3 ligase, which targets several proteins for ubiquitination, including actin [20], PIASγ [21], Abl-interactor 2 [22], c-Myc [23], PKCζ [24], dysbindin [25], X-linked inhibitor of apoptosis (XIAP) [26], desmin filaments [27], p73 transcription factor [28], STING [29] and Gli-related Krüppel-like zinc finger protein (Glis2) [30]. Based on this broad substrate specificity, it is not surprising that TRIM32 has versatile activities and is linked to diverse biological processes, including innate immunity [31,32], development and differentiation [15,16,23,33,34], regulation of microRNA [23,35], and tumorigenesis [36,37]. However, the role of TRIM32 in intrinsic immunity and viral restriction remains enigmatic. This report characterizes a role for TRIM32 in intrinsic cellular defense against influenza viruses by targeting the influenza polymerase for ubiquitination and degradation.
Results
TRIM32 interacts and translocates with PB1 following IAV infection
Mass spectrometry was used to examine the physical interactions between IAV PB1 and endogenous cellular proteins. PB1 protein complexes were immunoaffinity purified from HEK293 cells stably expressing FLAG tagged PB1 derived from the influenza A/Puerto Rico/8/1934 (PR8). Two independent purifications were analyzed by LC/MS-MS analysis. Controls include our laboratory database of 200 FLAG-tagged non-viral proteins isolated by identical procedures from stably transfected HEK293 cell lines [32,38,39]. A well-established computational algorithm, known as SAINT, was applied to the dataset [40,41]. Twenty-six proteins had SAINT scores above 0.89 and were designated high confidence interacting proteins (HCIP), including 18 proteins (S1 Table) reported to associate with PB1 or IAV polymerase complexes [42–44].
In a preliminary screen, 6 available GFP - or FLAG-tagged HCIP (TRIM32, STUB1, IQSEC, GALK, PDCD6 and HAUS6/DGT6) were expressed in HEK293 cells and examined for their effects on viral replication using the PR8-Gaussia luciferase reporter virus [45]. The most potent inhibition of IAV replication was noted following ectopic expression of TRIM32 (S1A Fig). Interactions between TRIM32 and IAV proteins had not been previously reported. Thus, we first validated the protein interaction. Following PR8 IAV infection of either primary human tracheal epithelial cells or HEK293 cells, endogenous TRIM32 binds to viral PB1 (Figs 1A and S1B). To demonstrate this interaction is direct, TRIM32-HIS and PB1-GST were purified from bacteria. In vitro GST pull down assays confirmed the direct association of PB1 with TRIM32 (Fig 1B). We next examined if TRIM32 can associate with PB1 proteins derived from different IAV strains. PB1 proteins from 6 IAV strains [PR8, A/Puerto Rico/8/1934 (H1N1); WSN, A/WSN/1933 (H1N1); NY, A/New York/1682/2009 (H1N1); Aichi, A/Aichi/2/1968 (H3N2); A/Vietnam/1194/2004 (H5N1) and A/Anhui/1/2013 (H7N9)] were co-transfected with TRIM32-GFP into HEK293 cells. Co-precipitation was noted between TRIM32 and PB1 from all 6 IAV strains (Fig 1C). Furthermore, the IAV ribonucleoprotein components PA, PB2 and NP failed to interact with TRIM32, demonstrating the specificity of PB1-TRIM32 interaction (Figs 1D and S1C). Taken together, TRIM32 physically interacts with PB1 polymerases derived from multiple IAV strains.
Fig. 1. TRIM32 interacts and translocates with influenza A virus PB1 protein. 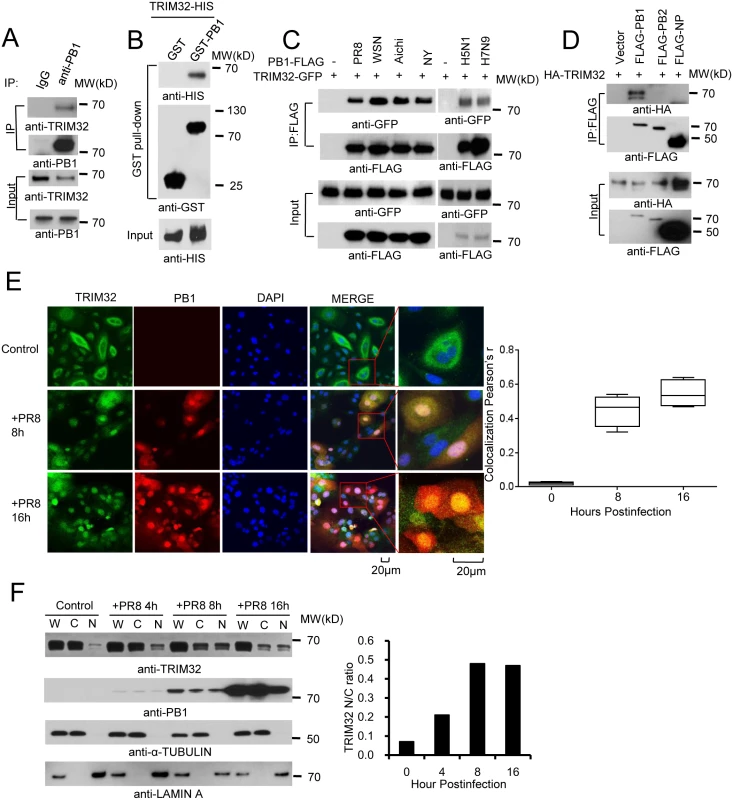
(A) Primary human tracheal epithelial cells were infected with 0.1 MOI PR8 IAV for 16 hr. Whole cell lysates (WCL) were subjected to immunoprecipitation (IP) and immunoblotting with indicated antibodies to detect endogenous interactions. Molecular weights (MW) are indicated. (B) GST pull down of bacterially expressed GST-PB1 and HIS-TRIM32. (C) FLAG-tagged PB1 from 6 different influenza A strains [PR8, A/Puerto Rico/8/1934 (H1N1); WSN, A/WSN/1933 (H1N1); Aichi, A/Aichi/2/1968 H3N2; NY, A/New York/1682/2009 (H1N1); A/Vietnam/1194/2004 (H5N1); A/Anhui/1/2013 (H7N9)] were co-expressed with TRIM32-GFP in HEK293 cells. After 48 hr, cell lysates were immunoprecipitated with anti-FLAG and probed as indicated. As input controls, WCL were immunoblotted. (D) TRIM32 fused with HA epitope was co-transfected into HEK293 cells with PR8 derived FLAG-tagged PB1, PB2 or NP. After 48 hr, WCL were immunoprecipitated with anti-FLAG antibody and blotted with indicated reagents. TRIM32 can appear as a doublet on Western blots. (E) Primary human tracheal epithelial cells were infected with 0.01 MOI PR8 strain IAV for 8 or 16 hr and stained with anti-TRIM32 (green), anti-PB1 (red) and DAPI nuclear stain (blue). Right panel shows quantitated TRIM32-PB1 colocalization data. (F) A549 cells were infected with 0.01 MOI IAV PR8 strain for the indicated times, whole cell lysates (W) or cytosolic (C) and nuclear (N) fractions were extracted and blotted as indicated. Right panel depicts the densitometric ratio of nuclear to cytoplasmic PB1. As influenza viruses are intrinsically sensitive to the antiviral action of interferons (IFN), we speculated that IFN might regulate TRIM32 expression. To address this issue, we infected A549 lung epithelial cells with PR8 IAV or treated the cells with IFN. TRIM32 mRNA and protein expression were then examined. However, there is no evidence that IAV infection or IFN stimulation modulate TRIM32 RNA or protein levels (S1D and S1E Fig). We conclude TRIM32 is constitutively expressed in A549 human lung epithelial cells, a model host cell line for IAV infection.
To examine TRIM32-PB1 co-localization, we infected primary human tracheal and A549 lung epithelial cells with PR8 IAV. In the absence of viral infection, endogenous TRIM32 is diffusely expressed in cytosolic foci and lesser amounts are detected in the nucleus. Following IAV infection TRIM32 accumulates in the nucleus (Figs 1E and S2A). Similarly, nuclear accumulation of endogenous TRIM32 is noted in A549 cells stably transfected with PB1 (S2B Fig). Influenza A driven TRIM32 nuclear translocation was biochemically confirmed by isolation of nuclear and cytosolic fractions (Fig 1F), indicating that IAV infection triggers translocation of TRIM32 to the nucleus within 4 hr. The evolutionarily conserved CRM1 (exportin 1) receptor is responsible for nuclear export of most proteins [46]. To examine the role of CRM1 in TRIM32 nuclear distribution, A549 cells were treated with CRM1 inhibitor leptomycin B. Addition of leptomycin B causes accumulation of nuclear TRIM32 in the absence of viral infection (S2C Fig), thereby implying that TRIM32 physiologically shuttles between the cytosol and nucleus. The combined data suggest that during IAV replication PB1 retains TRIM32 in the nuclear compartment.
Domains required for TRIM32-PB1 interaction
To determine which segment of TRIM32 mediates its interaction with PB1, we generated a series of TRIM32 deletion mutants (Fig 2A). Initial experiments suggested TRIM32 constructs carrying residues 140 to 265 were involved in PB1 binding (Fig 2B). The 140 to 265 segment contains the TRIM32 CC domain plus part of the linker region. Deletion of this segment results in the loss of PB1 binding (Fig 2C), while this segment alone is sufficient for PB1 association (Fig 2D). Thus, TRIM32 residues 140 to 265 are necessary and sufficient for PB1 binding.
Fig. 2. Domain requirements for TRIM32-PB1 interaction. 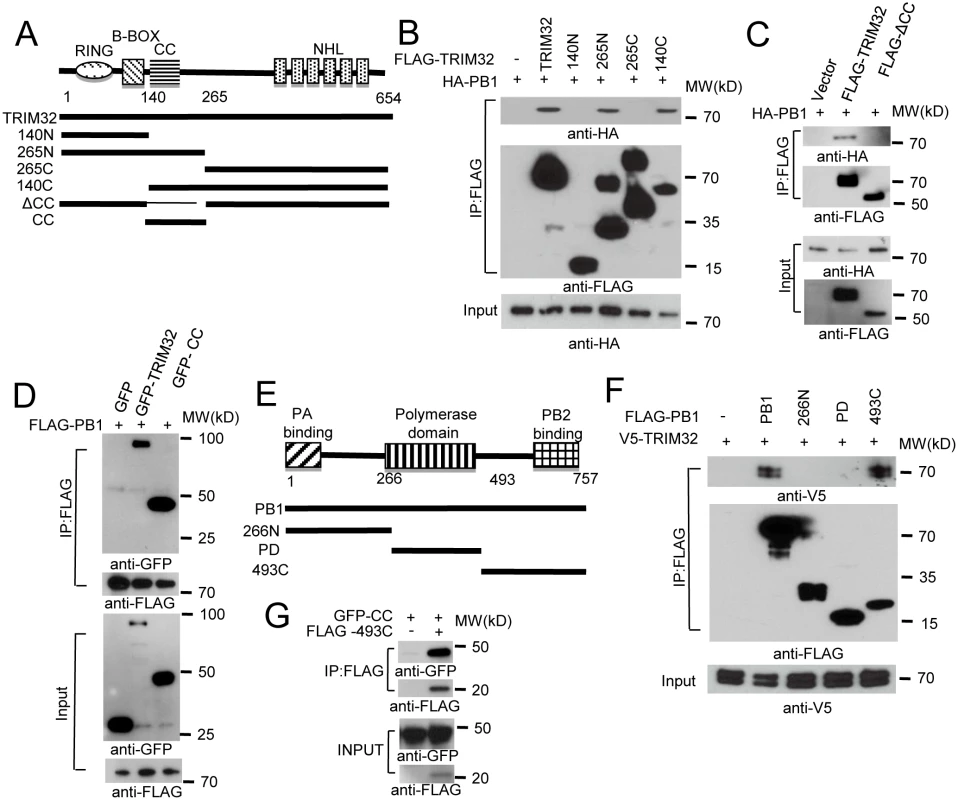
(A) Schematic representation of TRIM32 protein domains and the individual TRIM32 deletion mutants investigated in this study. (B) Full length and various TRIM32 deletion mutants were fused with FLAG epitope and co-transfected with HA-PB1 into HEK293 cells. WCL were immunoprecipitated with anti-FLAG antibody and blotted with indicated reagents. (C) Full length and TRIM32 with a deleted CC segment were fused with FLAG epitope and co-transfected with HA-PB1 into HEK293 cells. WCL were immunoprecipitated with anti-FLAG antibody and blotted with indicated reagents. (D) Full length and TRIM32 CC-containing segment (residues 140–265) were tagged with GFP and co-transfected with FLAG-PB1 into HEK293 cells. WCL were immunoprecipitated with anti-FLAG antibody and blotted with the indicated reagents. (E) Schematic representation of IAV PB1 protein domains and PB1 deletion mutants investigated in this study. (F) Full length and various PB1 mutants (PR8) were fused with FLAG epitope and co-transfected with V5-TRIM32 into HEK293 cells. WCL were immunoprecipitated with anti-FLAG antibody and blotted with indicated reagents. (G) TRIM32 CC fragment (residues 140–265) was cotransfected with C-terminal PB1 fragment (residues 493–757) into HEK293 cells. Immunoprecipitation and immunoblotting were performed with indicated reagents. Assembly of the heterotrimeric IAV polymerase requires interactions between the PB1 N-terminal domain with the PA chain while the PB1 C-terminus associates with the polymerase PB2 protein [6,47,48]. PB1 truncation mutants were prepared in order to define the PB1 segment which interacts with TRIM32 (Fig 2E). The PB1 C-terminal region consisting of residues 493 to 757 was sufficient for interaction with TRIM32 (Fig 2F) and the TRIM32 CC-linker fragment (Fig 2G). These findings led us to question whether TRIM32 could compete with PB2 for PB1 binding. The failure of TRIM32 to block PB1-PB2 association suggests TRIM32 is not a competitive inhibitor of IAV polymerase assembly (S2D Fig). Fine mapping of IAV polymerase interaction sites suggest a stretch of 15 amino acids in the PB1 C-terminus are most important for PB2 binding, leaving an expansive “thumb” surface available for interactions with other proteins [1,2]. The combined data identify the essential peptide fragments for TRIM32-PB1 interaction.
TRIM32 defends against influenza virus infection
Four assay systems (reporter assay, Western blot, immunofluorescence and plaque assay) were used to evaluate the biological effect of TRIM32 overexpression on IAV replication. First, FLAG-tagged TRIM32 and TRIM65 (a control TRIM family member with established E3 ligase activity [39,49]) were transfected into A549 cells followed by infection with PR8-Gaussia luciferase reporter virus. TRIM32 selectively restricted IAV replication (Fig 3A) without significantly impacting cell viability (S3A Fig). We then determined the effect of TRIM32 on viral infection by examining IAV NP protein expression using Western blot and immunofluorescence. A549 lung epithelial cells stably transfected with TRIM32 were infected with WSN and PR8 IAV. TRIM32 overexpression resulted in viral restriction as noted by decreased levels of NP protein detected by Western blot (Figs 3B and S3B). Similarly, HEK293 cells transfected with FLAG-TRIM32 or GFP-TRIM32 displayed decreased levels of viral protein production (S3C and S3D Fig). Microscopic inspection confirmed that TRIM32 overexpression inhibited PR8 and WSN viral NP protein expression (Figs 3C and S3E). Plaque assays were used to determine the effects of TRIM32 on production of infectious IAV particles. Stable or transient overexpression of TRIM32 consistently reduced viral titers (Figs 3D and S3F). To compare the relative susceptibility of various IAV strains to TRIM32-mediated restriction, A549 stable cell lines transfected with control vector or TRIM32-FLAG were infected with 0.001 MOI of 4 different IAV strains. After 18 hr, supernatants were transferred to fresh A549 target cells and the relative fraction of infected cells was determined. TRIM32 transfection results in 70–80% reduction of infectious IAV particles (S3G Fig). Thus, the combined findings indicate TRIM32 is an IAV restriction factor.
Fig. 3. TRIM32 attenuates influenza A virus infection. 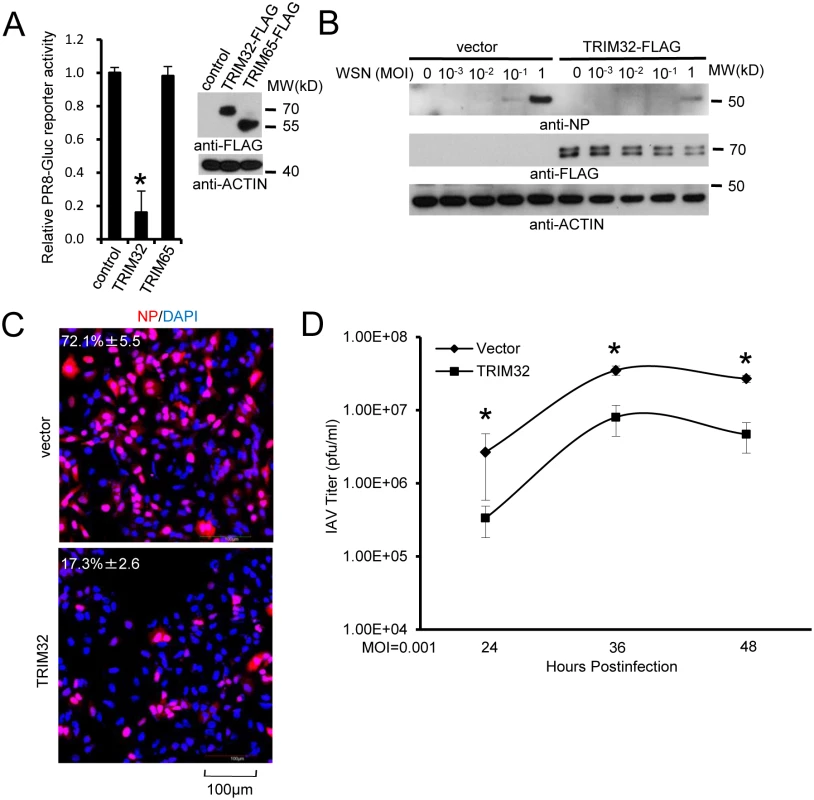
(A) A549 cells were transfected with control vector, TRIM32-FLAG or TRIM65-FLAG. After 36 hr cells were infected with 0.01 MOI PR8-Gluc for 16 hr and then Gaussia luciferase activity was examined. An asterisk indicates P<0.01. (B) A549 stable cell lines carrying vector or TRIM32-FLAG were infected with indicated MOI of WSN strain IAV for 16 hr. WCL were blotted with indicated reagents. (C) A549 cells stably transfected with control vector or TRIM32-FLAG were infected with 0.01 MOI PR8 for 8 hr and stained with anti-NP (red) and DAPI (blue). The percentage of NP stained cells is indicated. (D) A549 stable cell lines transfected with vector or TRIM32-FLAG were infected with 0.001 MOI of WSN IAV for the indicated times. Supernatant was titered on MDCK cells and plaques were enumerated. Asterisk indicates P<0.05. TRIM32 deficiency increases susceptibility to influenza infection
To complement the above overexpression data, we depleted TRIM32 with siRNA. Decreased TRIM32 expression correlated with increased IAV reporter activity in primary respiratory epithelial cells (Fig 4A) and also in HEK293 cells (S4A Fig). In addition, silencing TRIM32 enhanced IAV propagation in A549 cells, as detected by plaque assay and increased NP levels (Figs 4B, S4B and S4C). Knockdown of TRIM32 also enhanced IAV propagation in primary tracheal epithelial cells, as detected by plaque assay (Fig 4C). To exclude off-target effects of siRNA, cells were transfected with wild type TRIM32 or a siRNA resistant TRIM32 rescue construct before infection with PR8 reporter virus. Cells transfected with wild type or rescue TRIM32 constructs displayed comparable levels of TRIM32 expression and both decreased viral replication as demonstrated by reduced reporter activity (Fig 4D). Importantly, when combined with TRIM32 siRNA the rescue construct restored antiviral activity, validating siRNA specificity (Fig 4D).
Fig. 4. RNAi depletion of TRIM32 promotes influenza virus infection. 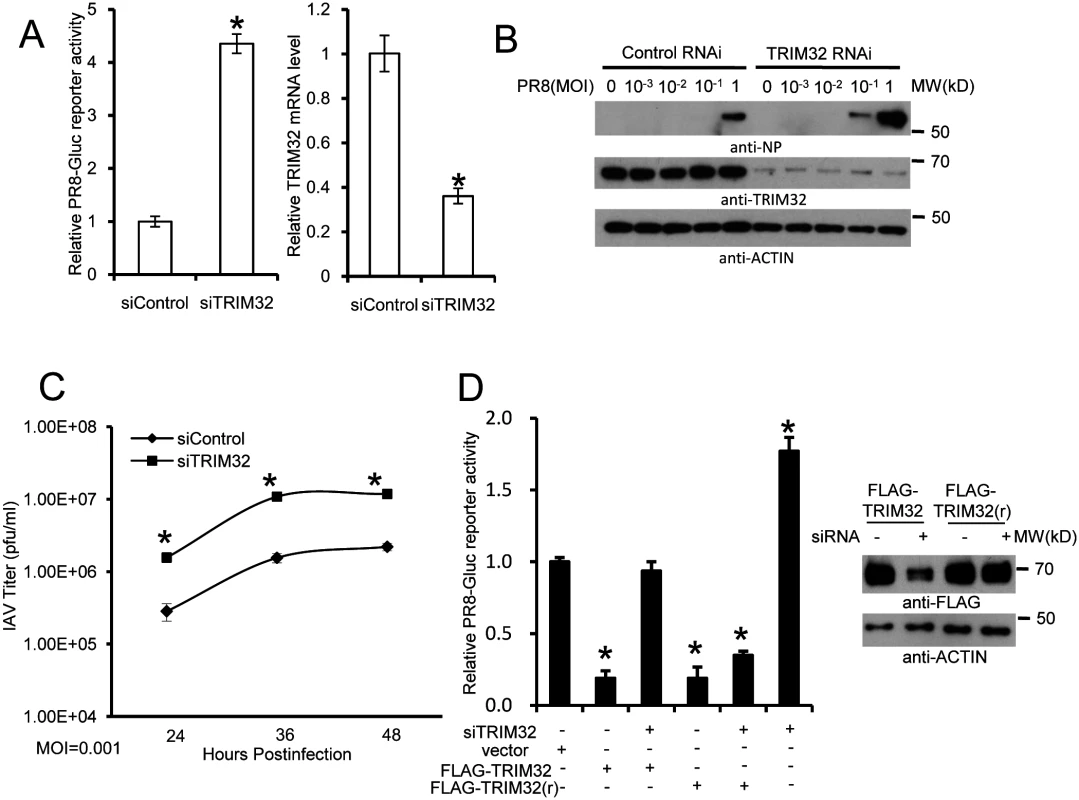
(A) Primary human tracheal epithelial cells were transfected with scrambled control or TRIM32 siRNA duplex #1. After 24 hr cells were infected with 0.01 MOI PR8-Gluc for 16 hr. The relative luciferase activity was examined. An asterisk indicates P<0.01. Right panel displays knockdown efficiency by qPCR. (B) A549 cells were transfected with control siRNA or TRIM32 siRNA#1. After 24 hr cells were infected with indicated MOI of PR8 IAV. WCL were blotted with the indicated antibodies. (C) Tracheal epithelial cells transfected with control or TRIM32 siRNA were infected with 0.001 MOI of WSN IAV for the indicated times. Culture supernatants containing IAV were titered on MDCK cells and plaques were enumerated. An asterisk indicates P<0.05. (D) HEK293 cells were transfected with TRIM32 siRNA and wild type TRIM32 or a TRIM32 rescue construct. After 24 hr cells were infected with 0.01 MOI PR8-Gluc for 16 hr. The relative luciferase signal is shown. An asterisk indicates P<0.05. Western blot shows knockdown efficiency. We next examined IAV infection in TRIM32 deficient mouse embryonic fibroblasts (MEF). A 7 fold increase in PR8 luciferase reporter activity was noted in trim32-/- MEF compared to wild type controls and antiviral activity was partially rescued by transfection with human TRIM32 (Fig 5A). In line with these results approximately 10-fold enhancement of viral NP protein expression was observed after PR8 infection of trim32-/- MEF (Fig 5B) and significantly more trim32-/- cells were stained with anti-NP antibody (Fig 5C). We also evaluated the impact of TRIM32 on viral propagation using plaque assays. As predicted, trim32 deficient cells produce more infectious viral particles than control MEF (Fig 5D). Finally, TRIM32 deficient and control MEF were infected with 0.1 MOI of 4 different IAV strains. Supernatants were transferred to A549 target cells and the relative fraction of infected cells was determined. Trim32-/- cells consistently produced 3 to 6 fold more infectious virus than control cells (S4D Fig). To establish virus restriction specificity, MEF were infected with Sendai virus. TRIM32 deficiency did not alter the level of infection with wild type Sendai or a Sendai-luciferase reporter virus (Fig 5E). The combined data using trim32 deficient cells indicate TRIM32 is an antiviral cellular factor that acts to curb infections with influenza viruses.
Fig. 5. TRIM32 deficiency increases susceptibility to influenza A virus infection. 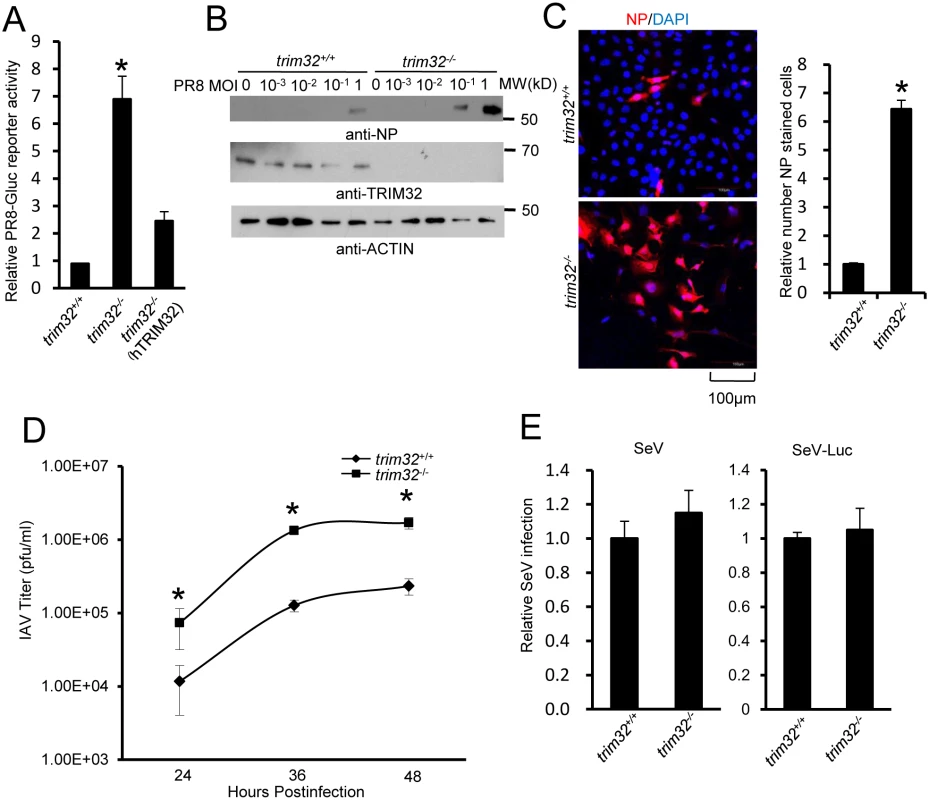
(A) Trim32+/+, trim32-/- and trim32-/- MEF transfected (24 hr) with human TRIM32 were infected with 0.1 MOI PR8-Gluc. The relative luciferase signal is shown. An asterisk indicates significant difference (P<0.05) for trim32-/- vs. either trim32+/+ or TRIM32 reconstituted trim32-/- MEF. (B) Trim32+/+ and trim32-/- MEF were infected with indicated MOI of PR8 IAV. After 16 hr, WCL were blotted with indicated reagents. (C) Trim32+/+ and trim32-/- MEF cells were infected with PR8 IAV for 8 hr, then cells were stained with anti-NP (red) and DAPI (blue). The right panel shows the relative ratio of NP stained cells. An asterisk indicates P<0.01. (D) Trim32+/+ and trim32-/- MEF were infected with 0.01 MOI of WSN IAV for the indicated times. Supernatants were titered on MDCK cells and pfu were enumerated. An asterisk indicates P<0.05. (E) Trim32+/+ and trim32-/- MEF were infected with wild type Sendai virus (SeV) or luciferase reporter Sendai virus (SeV-Luc) for 16 hr. Sendai infection was detected by staining with anti-Sendai antibody or luciferase assay. Relative numbers of Sendai virus infected cells or luciferase activities are presented. E3 ligase activity of TRIM32 is required for IAV restriction
TRIM32 was reported to regulate NF-κB and IFN driven cytokine production, which may mediate the antiviral activity of TRIM32 [29,32,50–52]. Thus, we investigated the capacity of trim32+/+ and trim32-/- MEF to produce IFNβ after infection with PR8 IAV (ΔNS1) virus [53] or treatment with the synthetic analog of dsRNA, poly(I:C). Trim32+/+ and trim32-/- MEF show comparable levels of IFNβ mRNA after IAV infection or poly(I:C) stimulation (S5A and S5B Fig). Furthermore, similar levels of IAV reporter activity were noted in MEF derived from wild-type and NF-κB deficient p65-/- mice (S5C Fig). The results suggest that TRIM32 does not alter influenza-activated IFN production and indicate other mechanisms are needed to account for TRIM32-mediated IAV restriction.
RING domains are critical for E3 ligase function in TRIM family molecules. Thus, we evaluated the role of the TRIM32 RING domain in antiviral activity. The TRIM32(C39S) mutation disrupts the rigid cross-braced architecture of the RING domain and destroys E3 ligase activity [29]. Antiviral activity was measured after transfection of HEK293 cells with TRIM32 or the TRIM32(C39S) mutant. Wild type TRIM32 attenuates infection with the IAV reporter virus. In contrast, ectopic expression of the TRIM32(C39S) mutant protein failed to defend against IAV infection (Fig 6A). Reconstitution of trim32-/- MEF with human TRIM32 restores host restriction of PR8 reporter virus, while reconstitution with the TRIM32(C39S) mutant retains high viral propagation (Fig 6B). Similarly, the TRIM32(C39S) mutant failed to restore antiviral activity in trim32-/- MEF as assayed by Western blotting and immunofluorescence for viral NP proteins (Fig 6C and 6D). Thus, we conclude that E3 ligase activity is critical for the antiviral activity of TRIM32.
Fig. 6. E3 ligase activity is indispensable for TRIM32-dependent against influenza A virus. 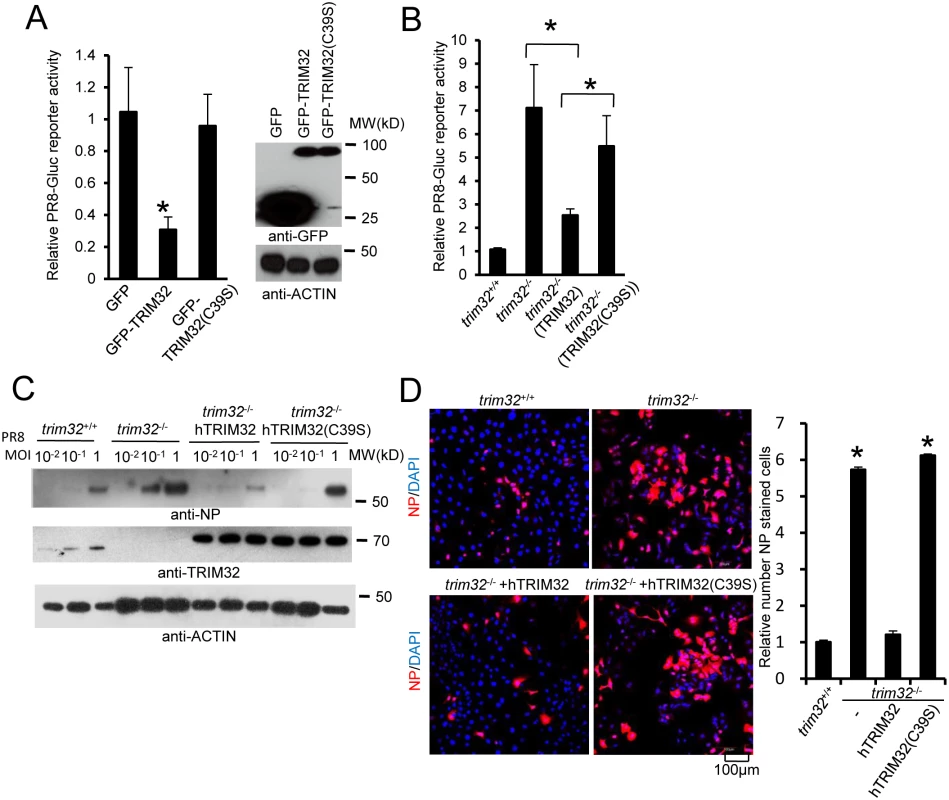
(A) HEK293 cells were transfected with GFP, GFP-TRIM32 or GFP-TRIM32(C39S). After 24 hr, cells were infected with 0.01 MOI PR8-Gluc for reporter assay. The relative luciferase signal is shown. Asterisk indicates P<0.05 (GFP-TRIM32 vs. either GFP or GFP-TRIM32(C39S) transfected cells). (B) Trim32+/+, trim32-/- or trim32-/- MEF reconstituted with TRIM32 and TRIM32(C39S) were infected with 0.1 MOI PR8-Gluc. The relative luciferase signal is shown. An asterisk indicates P<0.05. (C) Trim32+/+, trim32-/- or trim32-/- MEF reconstituted with TRIM32 and TRIM32 (C39S) were infected with designated MOI of IAV PR8 for 16 hr. WCL were blotted with indicated reagents. (D) Trim32+/+, trim32-/- or trim32-/- reconstituted with TRIM32 and TRIM32(C39S) MEF were infected with 0.1 MOI PR8 for 16 hr, then cells were stained with anti-NP (red) and DAPI. The right panel shows the relative ratio of NP stained cells. An asterisk indicates P<0.01. TRIM32 ubiquitination of PB1 leads to its protein degradation
To determine whether TRIM32 can directly couple ubiquitin onto PB1, bacterially derived TRIM32 and PB1 were combined in an in vitro ubiquitination assay. TRIM32 is able to heavily conjugate ubiquitin onto PB1 in vitro (Fig 7A). PB1 ubiquitination is dependent on the presence of ubiquitin plus an E1, E2, and an ATP regenerating system. The TRIM32(C39S) RING mutant (RM) fails to conjugate ubiquitin onto PB1 (Fig 7A). To examine the role of TRIM32 on PB1 ubiquitination within cells, trim32-/- MEF stably transfected with either TRIM32 or TRIM32(C39S) were infected with PR8 IAV. After treatment with the proteasomal inhibitor, MG132, PB1 was immunoprecipitated. Ubiquitinated PB1 showing a characteristic high molecular weight ladder was noted from trim32+/+ MEF or trim32-/- MEF reconstituted with TRIM32 (Fig 7B). Ubiquitinated PB1 was not detected in trim32-/- MEF or trim32-/- cells reconstituted with the C39S mutant. The requirement for MG132 treatment to visualize PB1 ubiquitination suggests that PB1 undergoes degradation in the presence of TRIM32. Proteins modified with K48-linked polyubiquitin are classic targets for proteasomal degradation. To test this possibility, HEK293 cells were cotransfected with PB1 plus wild type ubiquitin or a ubiquitin construct in which all lysine residues except K48 were mutated to arginine (K48) and a ubiquitin construct in which only the K48 residue was mutated to arginine (K48R). After MG132 treatment wild type and K48-only ubiquitin were conjugated onto PB1, while ubiquitin molecules lacking the K48 residue (K48R) were not coupled onto PB1 (Fig 7C). Co-transfection with TRIM32 increased the levels of PB1 ubiquitination (Fig 7C). To examine the role of TRIM32 in PB1 protein turnover, GFP-TRIM32 and GFP-TRIM32(C39S) mutant constructs were transfected into A549 cells stably expressing FLAG-PB1. Ectopic expression of TRIM32 decreased PB1 expression, while TRIM32(C39S) showed little or no effect on PB1 expression (Fig 7D). To further examine the role of TRIM32 in PB1 degradation, wild type MEF, trim32-/- MEF and reconstituted trim32-/- MEF were treated with a protein synthesis inhibitor (cycloheximide). After 2 or 6 hr, cell lysates were examined by quantitative Western blotting. The data indicate the presence of TRIM32 has a destabilizing effect on PB1 expression (Fig 7E). Finally, we examined whether TRIM32-dependent PB1 ubiquitination had an effect on PB1 polymerase activity. TRIM32 transfection inhibits IAV polymerase activity (Fig 7F). As predicted, silencing TRIM32 with siRNA enhances polymerase activity, while addition of a RNAi resistant TRIM32 rescue construct reduced polymerase activity (Fig 7F). The combined data indicate that TRIM32 mediates K48-linked ubiquitination of PB1 resulting in augmented PB1 degradation and reduced viral polymerase activity.
Fig. 7. TRIM32 limits viral infection by targeting PB1 for ubiquitination. 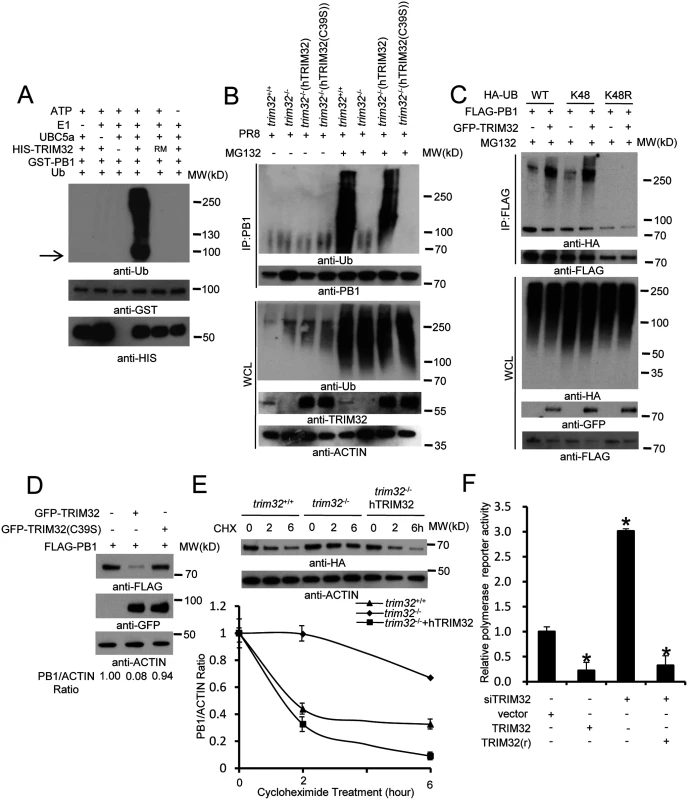
(A) In vitro ubiquitination of PB1 by TRIM32 plus E1, E2 (UBCH5A), ATP and ubiquitin (Ub). GST-tagged PB1, HIS-tagged TRIM32 and the catalytically dead TRIM32(C39S) RING mutant (RM) were purified from bacteria. The arrow indicates approximate PB1 position. (B) Trim32+/+, trim32-/- or TRIM32 and TRIM32(C39S) reconstituted trim32-/- MEF were infected with 0.1 MOI PR8 IAV for 16 hr, then treated with 10 μg/ml MG132. After 6 hr cell lysates were immunoprecipitated with anti-PB1 and probed as indicated. Whole cell lysates are presented as input controls. (C) HEK293 stably transfected cell lines carrying FLAG-PB1 were transiently transfected with either HA-tagged wild type, K48 only or K48R ubiquitin along with GFP-TRIM32. Cells were treated for 6 hr with 10 μg/ml MG132, then immunoprecipitated with anti-FLAG and probed with the indicated antibodies. (D) GFP-TRIM32 or TRIM32(C39S) were transfected into HEK293 cells stably transfected with FLAG-PB1. Immunoblotting was performed with indicated reagents. Quantitative Western blotting was used to determine PB1 levels, which were used to calculate the ratio of PB1 to actin. (E) Trim32+/+, trim32-/- and trim32-/- MEF reconstituted with TRIM32 were transfected with PB1-HA for 48 hr, then treated with 20 μg/ml cycloheximide for 0, 2 or 6 hr. WCL were blotted with indicated reagents. Quantitative Western blotting was used to derive data presented in lower panel. (F) HEK293 cells were transiently transfected with a plasmid cocktail containing PR8 PB1, PB2, PA, NP expression plasmids plus a polymerase I plasmid expressing an influenza virus-like RNA coding for the reporter protein firefly luciferase, Renilla luciferase control and TRIM32 siRNA along with TRIM32 wild type or rescue mutant for 48 hr. The relative luciferase signal is shown. An asterisk indicates P<0.05. Discussion
Humans evolved a broad spectrum of defense strategies to limit IAV infection. Among them, the best characterized antiviral defense systems are the broadly acting TLR and RLR pattern-recognition receptors which detect microbial nucleic acids and other cross-species biomarkers. These innate antiviral defenses indirectly inhibit infection by triggering signaling cascades that lead to the production of interferons and other antiviral effector molecules. In contrast, as demonstrated in this study, TRIM32 operates in a more restricted fashion to directly sense an IAV “danger” signal and to restrict infection in a species specific manner. Thus, TRIM32 provides a form of defense often categorized as intrinsic immunity.
Examination of IAV polymerase interacting proteins by AP-MS identified TRIM32. Associations of TRIM32 with the PB1 subunit of the polymerase complex were noted using 6 distinct PB1 proteins derived from IAV strains of H1N1, H3N2, H5N1 and H7N9 origin, suggesting that PB1 has not yet adapted to avoid TRIM32 targeting. Preliminary data suggest TRIM32 conjugates polyubiquitin at multiple PB1 sites, thereby limiting the opportunity for PB1 mutants to avoid targeting by TRIM32.
The functional activity of TRIM32 was demonstrated by a combination of overexpression, mutagenesis and loss-of-function (RNAi and genetic deletion) analyses. Comparison between wild type TRIM32 and the C39S ligase defective mutant demonstrated the requirement for TRIM32 E3 ligase activity in antiviral activity. The efficacy of TRIM32 in restricting IAV infection was established with four IAV strains and multiple cell types, including a human lung epithelial cell line and primary human tracheal epithelial cells. The latter represent a model for host cells which are naturally targeted during IAV infection.
TRIM32 is expressed in all tissues and displays broad substrate specificity. TRIM32 is known to bind at least 10 different cellular proteins of highly diverse function and localization, most of these interaction partners are also known substrates of TRIM32-mediated ubiquitination [22,23,25–29]. TRIM32 is a remarkably versatile E3 ligase; it participates in monoubiquitination [20,25] and formation of K48-linked or K-63-linked polyubiquitin chains which are covalently conjugated to target proteins [29,30]. In addition, TRIM32 can form unanchored polyubiquitin chains [54]. Here we report that in response to influenza infection, TRIM32 targets PB1 for K48-linked ubiquitination. The K48 polyubiquitin tagged PB1 molecules are shuttled to the proteasome system which orchestrates turnover of target proteins.
Depending on context, some cell types predominantly express TRIM32 proteins in the nuclear compartment, while in other cell types it primarily resides in a cytoplasmic niche [23,24,27,34,55,56]. However, TRIM32 nuclear import and export sequences remain undefined. Leptomycin B treatment causes TRIM32 nuclear accumulation suggesting that TRIM32 shuttles between the cytosolic and nuclear compartments. TRIM32 trafficking was previously studied in neural stem cells, where TRIM32 autoubiquitination helps maintain cytoplasmic localization [24]. In another study, ubiquitin conjugating E2 enzymes controlled TRIM32 localization in nuclear or cytoplasmic compartments [57]. The current study suggests an additional mechanism capable of rapidly regulating TRIM32 localization. We propose that the increasing nuclear concentration of PB1 during IAV replication combined with the high affinity of TRIM32 for this substrate affords a mechanism to trap and accumulate TRIM32 in the nucleus. It would be interesting to learn if this trapping hypothesis also applies to other settings where TRIM32 accumulates in the nucleus.
Many distinct types of proteins can regulate antiviral responses and they can act at different stages of viral replication [31,52]. Among the most studied innate or intrinsic antiviral factors are members of the TRIM family. In fact TRIM32 was among the first TRIMs shown to specifically interact with a viral protein, tat [9]. Although TRIM32 binds to the activation domain of lentiviral tat proteins, the role of TRIM32 in viral restriction was not studied. In a separate report, TRIM32 was shown to target STING (stimulator of interferon genes) for ubiquitination [29]. TRIM32 also stimulates NF-κB activity [21]. Both of these pathways can lead to potentiation of type I IFN and other antiviral genes associated with innate immunity. However, genetic deficiency of TRIM32 does not significantly alter the levels of IFN production activated by flu infection. Many TRIM members require interferon to achieve expression and function [50]. In contrast, TRIM32 is constitutively expressed in respiratory epithelial cells where it is positioned to provide early and direct defense against IAV infection.
In summary, we demonstrate TRIM32 inhibits IAV polymerase activity, targets PB1 for proteasomal degradation and provides intrinsic cellular restriction against IAV infection. Exploitation of this natural defense pathway may offer potential strategies for controlling IAV infections. Currently approved treatments against influenza are losing effectiveness as new viral strains are often refractory to conventional treatments. Understanding the molecular mechanisms controlling intrinsic antiviral defenses may illuminate novel strategies for preventing and treating viral diseases.
Materials and Methods
Cell culture, stable cell lines and viruses
Primary human bronchial tracheal epithelial cells and supporting medium were purchased from Lifeline Cell Technology (Frederick, MD). HEK293 and A549 cells (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in 10% CO2 at 37°C. P65+/+ and p65-/- MEF were generously provided by Denis Guttridge (Ohio State University, Columbus, OH). Primary MEF derived from trim32+/+ and trim32-/- mice [16] were transfected with SV40 large T antigen for immortalization. MEF were cultured in DMEM supplemented with 10% FBS. For generation of stable cell lines, human TRIM32 or mutant expression constructs were transfected into HEK293 cells, A549 cells and trim32-/- MEFs cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as detailed elsewhere [32]. Two days after transfection, cells were treated with 200 μg/ml hygromycin for 14 days. Single colonies were picked and expanded in 6-well plates. Protein expression levels in each colony were determined by immunoblotting. Influenza A viruses used in this study: A/Puerto Rico/8/34 (H1N1) (Charles River Labs, Wilmington, MA), A/PR8ΔNS1 (generously provided by Adolfo Garcia-Sastre, Mt. Sinai School of Medicine, NY) [53], A/WSN/33 (H1N1) (kindly provided by Peter Palese, Mt. Sinai School of Medicine, NY), A/New York/18/2009 (H1N1) (BEI Resources, Manassas, VA), and A/Aichi/68 (H3N2) (Charles River Labs). Influenza A PR8-GLuc virus containing a Gaussia luciferase (GLuc) gene inserted downstream of PB2 was a generous gift from Peter Palese [45]. Sendai virus was purchased from Charles River Labs. The Sendai-luciferase reporter virus was kindly provided by Charles Russell (St. Jude Hospital, Memphis, TN) [58].
Purification of protein complexes and mass spectrometry
Mass spectrometry experiments were performed as previously described [38,39,59]. For protein purification HEK293 stable cell lines expressing FLAG-PB1 were collected from five 15 cm dishes in 10 ml TAP buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 100 mM NaCl, 0.5% Nonidet P40, 10% glycerol, phosphatase inhibitors and protease inhibitors) [32]. Cell lysates were precleared with 50 μl protein A/G resin before addition of 20 μl anti-FLAG resin (Sigma) and 16 hr incubation at 4°C on a rotator. The resin was 3X washed and transferred to a spin column (Sigma) with 40 μl 3X FLAG peptide (Sigma) for 1 hr at 4°C on a rotator. Purified complexes were loaded onto a 4–15% NuPAGE gel (Invitrogen). Gels were stained using the SilverQuest staining kit (Invitrogen) and lanes were excised for mass spectrometry analysis by the Taplin Biological Mass Spectrometry Facility (Harvard Medical School, Boston, MA).
Interaction scoring for FLAG AP-MS
Two independent FLAG-PB1 purifications were analyzed by AP-MS. The resulting data are presented in S1 Table and were compared with our database of 200 controls from stable 293 cell lines expressing the FLAG tag fused to non-viral proteins handled in identical fashion. The SAINT algorithm (http://sourceforge.net/projects/saint-apms) was used to evaluate the MS data [41]. The default SAINT options were low Mode = 1, min Fold = 0, norm = 0. SAINT scores computed for each biological replicate were averaged (AvgP) and reported as the final SAINT score. Fold change was calculated for each prey protein as the ratio of spectral counts from replicate bait purifications over the spectral counts across all negative controls. A background factor of 0.1 was added to the average spectral counts of negative controls to prevent division by zero. Proteins included in the final interactome list had an AvgP ≥0.89. Selection of the threshold for SAINT scores was based on receiver operating curve analysis performed using publicly available protein interaction data and the FLAG AP-MS data set as a list of true positive interactions. A SAINT score of AvgP ≥0.89 was considered a true positive BioID protein with an estimated FDR of ≤2%.
IAV propagation and titration
All influenza viruses were propagated in MDCK cells and specific pathogen-free (SPF) embryonated chicken eggs. Monolayers of MDCK cells were washed with phosphate-buffered saline (PBS) and incubated with the respective virus at a multiplicity of infection (MOI) of 0.001 at 37°C. After 1 hour, the inoculum was aspirated, cells were washed twice and incubated at 37°C with DMEM without serum supplemented with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (1 μg/ml; Worthington Biomedical Corporation, Lakewood, NJ). 48 hr postinfection (p.i.) virus was recovered from supernatants. For SPF eggs, 0.2 ml stock influenza virus at 1x103 TCID50 was injected into 11-day-old SPF fertile chicken eggs. The eggs were incubated in a stationary incubator at 35°C. After 72 hr incubation, eggs were cooled at -20°C for 30 min, then clear allantoic fluid was collected. For viral titration, plaque assays were performed as described [60]. Briefly, 1.2 × 106 MDCK cells/ml were plated in 6-well plates. MDCK cells were washed twice with DMEM without serum, then serial dilutions of virus were adsorbed onto cells for 1 hr. Cells were then covered with DMEM 2×Avicel RC591 NF mix (FMC Biopolymer, Philadelphia, PA) supplemented with TPCK-treated trypsin (1 μg/ml). Crystal violet staining was performed 72 hr p.i., and visible plaques were counted.
Antibodies, chemicals and plasmids
Monoclonal anti-FLAG (M2) and anti-HA antibodies were obtained from Sigma (St. Louis, MO). The polyclonal rabbit anti-TRIM32 was prepared as described elsewhere [23]. Mouse anti-PB1 and NP antibodies were obtained from BEI Resources. Anti-GFP antibody was purchased from Santa Cruz Biotechnology. The anti-lamin A, anti-α-tubulin and anti-ubiquitin antibodies were purchased from Cell Signaling Technology (Danvers, MA). Anti-β-actin was purchased from Abcam (Cambridge, MA). The anti-GST and anti-HIS antibodies were obtained from Bethyl Laboratories, Inc (Montgomery, TX). The 3X FLAG peptide, HA peptide, MG132 and cychlohexmide were purchased from Sigma. Poly(I:C) was purchased from Invivogen (San Diego, CA). Anti-Sendai virus was obtained from Charles River Labs. Anti-V5 antibody was bought from Thermo Scientific.
cDNA encoding full-length human TRIM32 or TRIM32 mutants were subcloned in frame into mammalian expression vector pCMV-3TAG-8 with a C-terminal 3XFLAG, V5 or HA and the pEGFP-N1 vector containing a C-terminal GFP. cDNA encoding full-length PB1 from PR8, WSN, Aichi, NY, H5N1 and H7N9 IAV (S2 Table) were subcloned into mammalian expression vector pCMV-3TAG-8 with a C-terminal 3XFLAG or HA tag. The cDNA encoding PR8 derived PA, PB2 or NP (S2 Table) were subcloned into mammalian expression vector pCMV-3TAG-8 with a C-terminal 3XFLAG or HA. The sources of all constructs are provided in S2 Table.
Nuclear and cytoplasmic protein extraction
2x106 A549 cells were prepared for nuclear and cytoplasmic protein extraction according to the manufacturer’s protocol (Pierce, Rockford, IL).
Immunoprecipitation and immunoblotting
Immunoprecipitation and immunoblotting were performed as previously described [32]. For immunoprecipitation with anti-FLAG or anti-HA antibodies, the cell lysates were incubated with EZview red anti-FLAG M2 or anti-HA affinity resin (Sigma) for 4 or 16 hr at 4°C. After washing with lysis buffer, proteins were eluted by incubation with 1 mg/ml 3X FLAG or HA peptide for 1 hr at 4°C. For immunoprecipitation with anti-TRIM32 or anti-PB1, cell lysates were incubated with antibody and Protein A/G plus agarose (Thermo Fisher Scientific, Cambridge, MA) at 4°C for 16 hr. After washing with the lysis buffer, SDS-PAGE loading buffer was added and heated (95°C for 5 min). For immunoblotting, protein samples or 2% whole cell lysate were run on SDS-PAGE and transferred to PVDF membranes (Bio-Rad, Hercules, CA). The membranes were blocked in 5% non-fat milk in 1×Tris-buffered saline and then incubated with diluted primary antibodies at 4°C for 16 hr. Anti-rabbit or anti-mouse IgG antibodies conjugated to horseradish peroxidase (Pierce) were used as secondary antibodies. An enhanced chemiluminescence system (Pierce) was used for detection. Quantitation of immunoblots was performed using GelQuantNet software.
Immunostaining
Cells were fixed with 4% formaldehyde in PBS for 10 min, permeabilized with methanol or 0.5% Triton X-100 for 15 min, blocked with 2% bovine serum albumin in PBS for 30 min, and then incubated with primary antibodies at 4°C for 16 hr. After three PBS washes, the cells were incubated with Alexa 488-labeled and/or Alexa 595-labeled secondary antibodies (Invitrogen) for 1 hr at room temperature. Cells were counterstained with DAPI (4',6-diamidino-2-phenylindole; Sigma). Automated imaging software (Fiji ImageJ) with colocalization 2 application package was used to quantitate colocalization.
RNAi and rescue construct
FlexiTube siRNA oligos against TRIM32 were purchased from Qiagen (Valencia, CA). TRIM32 RNAi target sequences were as follows: #1 GACCGTGGTAACTATCGTATA, #2 CACACGATGGTGTTAGCTGAA and #3 CAGCACTCCAGGAATGTTCAA.
For siRNA gene knockdown experiments, cells were cultured in 24-well plates and transfected with 30 pmol siRNA and 3 μl lipofectamine 2000 according to the manufacturer’s instructions (Life Technologies, Grand Island, NY). After 48 hr, siRNA transfected cells were analyzed. A siRNA resistant TRIM32 was constructed in pCMV-3tag8-HA vector in which the No. 1 siRNA target sequence was mutated. Mutagenesis primers for the siRNA resistant rescue construct were as follows: 5’-tgaagtactagtcgctgatagaggaaagtacaggatccaagtctttacccgc-3’ and 5’-gcgggtaaagacttggatcctgtactttcctctatcagcgactagtacttca-3’
Preparation of recombinant bacterial proteins
TRIM32-HIS was expressed from pET28b vector (Clontech Laboratories, Mountain View, CA) and PR8-PB1-GST was expressed from pGEX-5X-3 vector (GE Healthcare Life Sciences, Pittsburgh, PA) in Escherichia coli BL21(DE3) pLysS (Life Technologies) induced with 0.2 mM isopropyl-1-thio-β-D-galactopyranoside and 200 μM ZnSO4 for 16 hr at 18°C as detailed elsewhere [38]. GST-tagged proteins were purified with glutathione Sepharose 4B beads according to the manufacturer’s protocol (GE Healthcare Life Sciences). HIS-tag proteins were purified with Ni-NTA agarose resins according to the manufacturer’s protocol (Qiagen).
Real-time quantitative PCR analysis for gene expression
Total RNA was extracted with RNeasy mini kit (Invitrogen) and reverse-transcribed (2 μg) with QuantiTect Reverse Transcription Kit (Qiagen). Human TRIM32, human IFNβ, human GAPDH, mouse IFNβ and mouse β-glucuronidase mRNA levels were quantitated by RT-PCR with SYBR dyes on a LightCycler 480 (Roche Life Sciences) as described elsewhere [38,61]. Primers for hTRIM32 were: forward CCGGGAAGTGCTAGAATGCC and reverse CAGCGGACACCATTGATGCT.
In vitro ubiquitination assays
In vitro ubiquitination assays were performed according to the manufacturer’s manual (Boston Biochem, Cambridge, MA). Ubiquitin (5 μg), E1 (200 ng), UBCH5a (300 ng) (Boston Biochem), TRIM32-HIS (0.8 μg) and PR8-PB1-GST (2 μg) were incubated with 2 mM ATP (Sigma) at 37°C 2 hr in ubiquitin assay buffer (20 mM Tris-HCl pH7.5, 5 mM MgCl2, 2 mM DTT). 1X stop solution (Boston Biochem) was added to end the reaction, after GST pull down the sample was washed with 1 M Urea for 60 min to exclude potential binding of unanchored polyubiquitin, then the sample was placed in SDS-loading buffer and boiled at 95°C for 5 min. Samples were subsequently analyzed by SDS-PAGE followed by Western blotting.
Reporter assay
Cells were aliquoted in 24 well plates for 24 hr prior to IAV infection. Human primary and cell lines were infected with 0.01 MOI PR8-GLuc and MEF were infected with 0.1MOI. After 1 hr the viral inoculum was aspirated, cells were washed twice and incubated at 37°C with 0.2% BSA-DMEM without serum. Cells were lysed 12–16 hr p.i. and the luciferase assay was performed using BioLux Gaussia Luciferase Assay Kit (NEB, Ipswich, MA).
Reconstitution of influenza virus polymerase activity
HEK293 cells were transfected in triplicate with vectors expressing PR8 PB1, PB2, NP, PA and the indicated TRIM32 or siTRIM32 oligo in addition to the polymerase I (PolI)-driven plasmid transcribing an influenza A virus-like RNA coding for the reporter protein firefly luciferase to monitor viral polymerase activity [62]. Cells were lysed 48 hr after transfection. Luciferase activity was measured with a luciferase assay system (Promega, Madison, WI). A plasmid constitutively expressing Renilla luciferase was transfected as a control.
Statistical analysis
Unless indicated otherwise all experiments were repeated on at least three separate occasions. Data from representative experiments are illustrated. Methods for AP-MS data analysis are detailed elsewhere [41]. Other statistical analyses were done with the two-tailed Student’s t test. Data are presented as mean ± standard deviation. A P value of <0.05 was considered significant.
Accession numbers
Identification of the genes and proteins used throughout this study include: TRIM32 (BC003154), TRIM65 (BC021259.2), STUB1 (BC0075456), IQSEC (BC010267), GALK1 (BC 001166), PDCD6 (BC050597) and DGT6/HAUS6 (NM 001270890.1). The GenBank references for viral genes include: PR8 PB1 (EF467819), WSN PB1 (J02178.1), NY PB1 (CY039907.1), Aichi PB1 (CY121123), H5N1 PB1 (AY651664.1), H7N9 PB1 (CY147058.1), H7N9 PB1 (GISAID # EPI439508) PR8 PB2 (CY148250.1), PR8 PA (CY148248.1) and PR8 NP (CY148246.1).
Supporting Information
Zdroje
1. Pflug A, Guilligay D, Reich S, Cusack S (2014) Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516 : 355–360. doi: 10.1038/nature14008 25409142
2. Reich S, Guilligay D, Pflug A, Malet H, Berger I, et al. (2014) Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516 : 361–366. doi: 10.1038/nature14009 25409151
3. Ghanem A, Mayer D, Chase G, Tegge W, Frank R, et al. (2007) Peptide-mediated interference with influenza A virus polymerase. J Virol 81 : 7801–7804. 17494067
4. He X, Zhou J, Bartlam M, Zhang R, Ma J, et al. (2008) Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454 : 1123–1126. doi: 10.1038/nature07120 18615018
5. Obayashi E, Yoshida H, Kawai F, Shibayama N, Kawaguchi A, et al. (2008) The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454 : 1127–1131. doi: 10.1038/nature07225 18660801
6. Perez DR, Donis RO (1995) A 48-amino-acid region of influenza A virus PB1 protein is sufficient for complex formation with PA. J Virol 69 : 6932–6939. 7474111
7. Cheung PP, Watson SJ, Choy KT, Fun Sia S, Wong DD, et al. (2014) Generation and characterization of influenza A viruses with altered polymerase fidelity. Nat Commun 5 : 4794. doi: 10.1038/ncomms5794 25183443
8. Furuta Y, Takahashi K, Kuno-Maekawa M, Sangawa H, Uehara S, et al. (2005) Mechanism of action of T-705 against influenza virus. Antimicrob Agents Chemother 49 : 981–986. 15728892
9. Fridell RA, Harding LS, Bogerd HP, Cullen BR (1995) Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology 209 : 347–357. 7778269
10. Tissot C, Mechti N (1995) Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J Biol Chem 270 : 14891–14898. 7797467
11. Neri M, Selvatici R, Scotton C, Trabanelli C, Armaroli A, et al. (2013) A patient with limb girdle muscular dystrophy carries a TRIM32 deletion, detected by a novel CGH array, in compound heterozygosis with a nonsense mutation. Neuromuscul Disord 23 : 478–482. doi: 10.1016/j.nmd.2013.02.003 23541687
12. Guglieri M, Straub V, Bushby K, Lochmuller H (2008) Limb-girdle muscular dystrophies. Curr Opin Neurol 21 : 576–584. doi: 10.1097/WCO.0b013e32830efdc2 18769252
13. Saccone V, Palmieri M, Passamano L, Piluso G, Meroni G, et al. (2008) Mutations that impair interaction properties of TRIM32 associated with limb-girdle muscular dystrophy 2H. Hum Mutat 29 : 240–247. 17994549
14. Schoser BG, Frosk P, Engel AG, Klutzny U, Lochmuller H, et al. (2005) Commonality of TRIM32 mutation in causing sarcotubular myopathy and LGMD2H. Ann Neurol 57 : 591–595. 15786463
15. Kudryashova E, Wu J, Havton LA, Spencer MJ (2009) Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum Mol Genet 18 : 1353–1367. doi: 10.1093/hmg/ddp036 19155210
16. Nicklas S, Otto A, Wu X, Miller P, Stelzer S, et al. (2012) TRIM32 regulates skeletal muscle stem cell differentiation and is necessary for normal adult muscle regeneration. PLoS One 7: e30445. doi: 10.1371/journal.pone.0030445 22299041
17. Kudryashova E, Struyk A, Mokhonova E, Cannon SC, Spencer MJ (2011) The common missense mutation D489N in TRIM32 causing limb girdle muscular dystrophy 2H leads to loss of the mutated protein in knock-in mice resulting in a Trim32-null phenotype. Hum Mol Genet 20 : 3925–3932. doi: 10.1093/hmg/ddr311 21775502
18. Blacque OE, Leroux MR (2006) Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell Mol Life Sci 63 : 2145–2161. 16909204
19. Chiang AP, Beck JS, Yen HJ, Tayeh MK, Scheetz TE, et al. (2006) Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc Natl Acad Sci U S A 103 : 6287–6292. 16606853
20. Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ (2005) Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol 354 : 413–424. 16243356
21. Albor A, El-Hizawi S, Horn EJ, Laederich M, Frosk P, et al. (2006) The interaction of Piasy with Trim32, an E3-ubiquitin ligase mutated in limb-girdle muscular dystrophy type 2H, promotes Piasy degradation and regulates UVB-induced keratinocyte apoptosis through NFkappaB. J Biol Chem 281 : 25850–25866. 16816390
22. Kano S, Miyajima N, Fukuda S, Hatakeyama S (2008) Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res 68 : 5572–5580. doi: 10.1158/0008-5472.CAN-07-6231 18632609
23. Schwamborn JC, Berezikov E, Knoblich JA (2009) The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136 : 913–925. doi: 10.1016/j.cell.2008.12.024 19269368
24. Hillje AL, Worlitzer MM, Palm T, Schwamborn JC (2011) Neural stem cells maintain their stemness through protein kinase C zeta-mediated inhibition of TRIM32. Stem Cells 29 : 1437–1447. doi: 10.1002/stem.687 21732497
25. Locke M, Tinsley CL, Benson MA, Blake DJ (2009) TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum Mol Genet 18 : 2344–2358. doi: 10.1093/hmg/ddp167 19349376
26. Ryu YS, Lee Y, Lee KW, Hwang CY, Maeng JS, et al. (2011) TRIM32 protein sensitizes cells to tumor necrosis factor (TNFalpha)-induced apoptosis via its RING domain-dependent E3 ligase activity against X-linked inhibitor of apoptosis (XIAP). J Biol Chem 286 : 25729–25738. doi: 10.1074/jbc.M111.241893 21628460
27. Cohen S, Zhai B, Gygi SP, Goldberg AL (2012) Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J Cell Biol 198 : 575–589. doi: 10.1083/jcb.201110067 22908310
28. Gonzalez-Cano L, Hillje AL, Fuertes-Alvarez S, Marques MM, Blanch A, et al. (2013) Regulatory feedback loop between TP73 and TRIM32. Cell Death Dis 4: e704. doi: 10.1038/cddis.2013.224 23828567
29. Zhang J, Hu MM, Wang YY, Shu HB (2012) TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem 287 : 28646–28655. doi: 10.1074/jbc.M112.362608 22745133
30. Ramachandran H, Schafer T, Kim Y, Herfurth K, Hoff S, et al. (2014) Interaction with the Bardet-Biedl gene product TRIM32/BBS11 modifies the half-life and localization of Glis2/NPHP7. J Biol Chem 289 : 8390–8401. doi: 10.1074/jbc.M113.534024 24500717
31. Rajsbaum R, Garcia-Sastre A, Versteeg GA (2014) TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol 426 : 1265–1284. doi: 10.1016/j.jmb.2013.12.005 24333484
32. Li S, Wang L, Berman M, Kong YY, Dorf ME (2011) Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 35 : 426–440. doi: 10.1016/j.immuni.2011.06.014 21903422
33. Hillje AL, Pavlou MA, Beckmann E, Worlitzer MM, Bahnassawy L, et al. (2013) TRIM32-dependent transcription in adult neural progenitor cells regulates neuronal differentiation. Cell Death Dis 4: e976. doi: 10.1038/cddis.2013.487 24357807
34. Sato T, Okumura F, Kano S, Kondo T, Ariga T, et al. (2011) TRIM32 promotes neural differentiation through retinoic acid receptor-mediated transcription. J Cell Sci 124 : 3492–3502. doi: 10.1242/jcs.088799 21984809
35. Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V (2009) nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell 136 : 926–938. doi: 10.1016/j.cell.2009.01.053 19269369
36. Hatakeyama S (2011) TRIM proteins and cancer. Nat Rev Cancer 11 : 792–804. doi: 10.1038/nrc3139 21979307
37. Horn EJ, Albor A, Liu Y, El-Hizawi S, Vanderbeek GE, et al. (2004) RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis 25 : 157–167. 14578165
38. Fu B, Li S, Wang L, Berman MA, Dorf ME (2014) The ubiquitin conjugating enzyme UBE2L3 regulates TNFalpha-induced linear ubiquitination. Cell Res 24 : 376–379. doi: 10.1038/cr.2013.133 24060851
39. Li S, Wang L, Fu B, Berman MA, Diallo A, et al. (2014) TRIM65 regulates microRNA activity by ubiquitination of TNRC6. Proc Natl Acad Sci U S A.
40. Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, et al. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328 : 1043–1046. doi: 10.1126/science.1176495 20489023
41. Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, et al. (2011) SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods 8 : 70–73. doi: 10.1038/nmeth.1541 21131968
42. Bradel-Tretheway BG, Mattiacio JL, Krasnoselsky A, Stevenson C, Purdy D, et al. (2011) Comprehensive proteomic analysis of influenza virus polymerase complex reveals a novel association with mitochondrial proteins and RNA polymerase accessory factors. J Virol 85 : 8569–8581. doi: 10.1128/JVI.00496-11 21715506
43. York A, Hutchinson EC, Fodor E (2014) Interactome analysis of the influenza a virus transcription/replication machinery identifies protein phosphatase 6 as a cellular factor required for efficient virus replication. J Virol 88 : 13284–13299. doi: 10.1128/JVI.01813-14 25187537
44. Watanabe T, Kawakami E, Shoemaker JE, Lopes TJ, Matsuoka Y, et al. (2014) Influenza Virus-Host Interactome Screen as a Platform for Antiviral Drug Development. Cell Host Microbe.
45. Heaton NS, Leyva-Grado VH, Tan GS, Eggink D, Hai R, et al. (2013) In vivo bioluminescent imaging of influenza a virus infection and characterization of novel cross-protective monoclonal antibodies. J Virol 87 : 8272–8281. doi: 10.1128/JVI.00969-13 23698304
46. Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, et al. (1998) Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res 242 : 540–547. 9683540
47. Gonzalez S, Zurcher T, Ortin J (1996) Identification of two separate domains in the influenza virus PB1 protein involved in the interaction with the PB2 and PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res 24 : 4456–4463. 8948635
48. Ohtsu Y, Honda Y, Sakata Y, Kato H, Toyoda T (2002) Fine mapping of the subunit binding sites of influenza virus RNA polymerase. Microbiol Immunol 46 : 167–175. 12008925
49. Li S, Wang L, Fu B, Dorf ME (2014) Trim65: a cofactor for regulation of the microRNA pathway. RNA Biol 11 : 1113–1121. doi: 10.4161/rna.36179 25483047
50. Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, et al. (2013) The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 38 : 384–398. doi: 10.1016/j.immuni.2012.11.013 23438823
51. Uchil PD, Hinz A, Siegel S, Coenen-Stass A, Pertel T, et al. (2013) TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J Virol 87 : 257–272. doi: 10.1128/JVI.01804-12 23077300
52. Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W (2008) TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog 4: e16. doi: 10.1371/journal.ppat.0040016 18248090
53. Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, et al. (1998) Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252 : 324–330. 9878611
54. Streich FC Jr., Ronchi VP, Connick JP, Haas AL (2013) Tripartite motif ligases catalyze polyubiquitin chain formation through a cooperative allosteric mechanism. J Biol Chem 288 : 8209–8221. doi: 10.1074/jbc.M113.451567 23408431
55. Ichimura T, Taoka M, Shoji I, Kato H, Sato T, et al. (2013) 14-3-3 proteins sequester a pool of soluble TRIM32 ubiquitin ligase to repress autoubiquitylation and cytoplasmic body formation. J Cell Sci 126 : 2014–2026. doi: 10.1242/jcs.122069 23444366
56. Zhang Y, Liu J, Yao S, Li F, Xin L, et al. (2012) Nuclear factor kappa B signaling initiates early differentiation of neural stem cells. Stem Cells 30 : 510–524. doi: 10.1002/stem.1006 22134901
57. Napolitano LM, Jaffray EG, Hay RT, Meroni G (2011) Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem J 434 : 309–319. doi: 10.1042/BJ20101487 21143188
58. Burke CW, Mason JN, Surman SL, Jones BG, Dalloneau E, et al. (2011) Illumination of parainfluenza virus infection and transmission in living animals reveals a tissue-specific dichotomy. PLoS Pathog 7: e1002134. doi: 10.1371/journal.ppat.1002134 21750677
59. Li S, Wang L, Dorf ME (2009) PKC phosphorylation of TRAF2 mediates IKKalpha/beta recruitment and K63-linked polyubiquitination. Mol Cell 33 : 30–42. doi: 10.1016/j.molcel.2008.11.023 19150425
60. Matrosovich M, Matrosovich T, Garten W, Klenk HD (2006) New low-viscosity overlay medium for viral plaque assays. Virol J 3 : 63. 16945126
61. Wang L, Li S, Dorf ME (2012) NEMO binds ubiquitinated TANK-binding kinase 1 (TBK1) to regulate innate immune responses to RNA viruses. PLoS One 7: e43756. doi: 10.1371/journal.pone.0043756 23028469
62. Reuther P, Manz B, Brunotte L, Schwemmle M, Wunderlich K (2011) Targeting of the influenza A virus polymerase PB1-PB2 interface indicates strain-specific assembly differences. J Virol 85 : 13298–13309. doi: 10.1128/JVI.00868-11 21957294
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage TraffickingČlánek An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived NeuronsČlánek Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPsČlánek Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African TrypanosomiasisČlánek Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin ReleaseČlánek HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T CellsČlánek Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 6- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Introducing “Research Matters”
- Exploring Host–Pathogen Interactions through Biological Control
- Analysis of Bottlenecks in Experimental Models of Infection
- Expected and Unexpected Features of the Newly Discovered Bat Influenza A-like Viruses
- Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage Trafficking
- Recombinant Murine Gamma Herpesvirus 68 Carrying KSHV G Protein-Coupled Receptor Induces Angiogenic Lesions in Mice
- TRIM30α Is a Negative-Feedback Regulator of the Intracellular DNA and DNA Virus-Triggered Response by Targeting STING
- Targeting Human Transmission Biology for Malaria Elimination
- Two Cdc2 Kinase Genes with Distinct Functions in Vegetative and Infectious Hyphae in
- An Model of Latency and Reactivation of Varicella Zoster Virus in Human Stem Cell-Derived Neurons
- Protective mAbs and Cross-Reactive mAbs Raised by Immunization with Engineered Marburg Virus GPs
- Virulence Factors of Induce Both the Unfolded Protein and Integrated Stress Responses in Airway Epithelial Cells
- Peptide-MHC-I from Endogenous Antigen Outnumber Those from Exogenous Antigen, Irrespective of APC Phenotype or Activation
- Specific Cell Targeting Therapy Bypasses Drug Resistance Mechanisms in African Trypanosomiasis
- An Ultrasensitive Mechanism Regulates Influenza Virus-Induced Inflammation
- The Role of Human Transportation Networks in Mediating the Genetic Structure of Seasonal Influenza in the United States
- Host Delivery of Favorite Meals for Intracellular Pathogens
- Complement-Opsonized HIV-1 Overcomes Restriction in Dendritic Cells
- Inter-Seasonal Influenza is Characterized by Extended Virus Transmission and Persistence
- A Critical Role for CLSP2 in the Modulation of Antifungal Immune Response in Mosquitoes
- Twilight, a Novel Circadian-Regulated Gene, Integrates Phototropism with Nutrient and Redox Homeostasis during Fungal Development
- Surface-Associated Lipoproteins Link Virulence to Colitogenic Activity in IL-10-Deficient Mice Independent of Their Expression Levels
- Latent Membrane Protein LMP2A Impairs Recognition of EBV-Infected Cells by CD8+ T Cells
- Bank Vole Prion Protein As an Apparently Universal Substrate for RT-QuIC-Based Detection and Discrimination of Prion Strains
- Neuronal Subtype and Satellite Cell Tropism Are Determinants of Varicella-Zoster Virus Virulence in Human Dorsal Root Ganglia Xenografts
- Molecular Basis for the Selective Inhibition of Respiratory Syncytial Virus RNA Polymerase by 2'-Fluoro-4'-Chloromethyl-Cytidine Triphosphate
- Structure of the Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome
- Activated Brain Endothelial Cells Cross-Present Malaria Antigen
- Fungal Morphology, Iron Homeostasis, and Lipid Metabolism Regulated by a GATA Transcription Factor in
- Peptidoglycan Branched Stem Peptides Contribute to Virulence by Inhibiting Pneumolysin Release
- A Macrophage Subversion Factor Is Shared by Intracellular and Extracellular Pathogens
- A Novel AT-Rich DNA Recognition Mechanism for Bacterial Xenogeneic Silencer MvaT
- Reovirus FAST Proteins Drive Pore Formation and Syncytiogenesis Using a Novel Helix-Loop-Helix Fusion-Inducing Lipid Packing Sensor
- The Role of ExoS in Dissemination of during Pneumonia
- IRF-5-Mediated Inflammation Limits CD8 T Cell Expansion by Inducing HIF-1α and Impairing Dendritic Cell Functions during Infection
- Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection
- Crystal Structure of USP7 Ubiquitin-like Domains with an ICP0 Peptide Reveals a Novel Mechanism Used by Viral and Cellular Proteins to Target USP7
- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- HPV16 Down-Regulates the Insulin-Like Growth Factor Binding Protein 2 to Promote Epithelial Invasion in Organotypic Cultures
- The νSaα Specific Lipoprotein Like Cluster () of . USA300 Contributes to Immune Stimulation and Invasion in Human Cells
- RSV-Induced H3K4 Demethylase KDM5B Leads to Regulation of Dendritic Cell-Derived Innate Cytokines and Exacerbates Pathogenesis
- Leukocidin A/B (LukAB) Kills Human Monocytes via Host NLRP3 and ASC when Extracellular, but Not Intracellular
- Border Patrol Gone Awry: Lung NKT Cell Activation by Exacerbates Tularemia-Like Disease
- The Curious Road from Basic Pathogen Research to Clinical Translation
- From Cell and Organismal Biology to Drugs
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
- A 21st Century Perspective of Poliovirus Replication
- Is Development of a Vaccine against Feasible?
- Waterborne Viruses: A Barrier to Safe Drinking Water
- Battling Phages: How Bacteria Defend against Viral Attack
- Archaea in and on the Human Body: Health Implications and Future Directions
- Degradation of Human PDZ-Proteins by Human Alphapapillomaviruses Represents an Evolutionary Adaptation to a Novel Cellular Niche
- Natural Variants of the KPC-2 Carbapenemase have Evolved Increased Catalytic Efficiency for Ceftazidime Hydrolysis at the Cost of Enzyme Stability
- Potent Cell-Intrinsic Immune Responses in Dendritic Cells Facilitate HIV-1-Specific T Cell Immunity in HIV-1 Elite Controllers
- The Mammalian Cell Cycle Regulates Parvovirus Nuclear Capsid Assembly
- Host Reticulocytes Provide Metabolic Reservoirs That Can Be Exploited by Malaria Parasites
- The Proteome of the Isolated Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components
- NK-, NKT- and CD8-Derived IFNγ Drives Myeloid Cell Activation and Erythrophagocytosis, Resulting in Trypanosomosis-Associated Acute Anemia
- Successes and Challenges on the Road to Cure Hepatitis C
- BRCA1 Regulates IFI16 Mediated Nuclear Innate Sensing of Herpes Viral DNA and Subsequent Induction of the Innate Inflammasome and Interferon-β Responses
- A Structural and Functional Comparison Between Infectious and Non-Infectious Autocatalytic Recombinant PrP Conformers
- Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria
- Human Immunodeficiency Virus Type 1 Nef Inhibits Autophagy through Transcription Factor EB Sequestration
- Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes
- EBV BART MicroRNAs Target Multiple Pro-apoptotic Cellular Genes to Promote Epithelial Cell Survival
- Single-Cell and Single-Cycle Analysis of HIV-1 Replication
- TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase
- The Herpes Simplex Virus Protein pUL31 Escorts Nucleocapsids to Sites of Nuclear Egress, a Process Coordinated by Its N-Terminal Domain
- Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections – A Prospective Cohort Study
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells
- Battling Phages: How Bacteria Defend against Viral Attack
- A 21st Century Perspective of Poliovirus Replication
- Adenovirus Tales: From the Cell Surface to the Nuclear Pore Complex
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



