-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Unexpected Role of the Steroid-Deficiency Protein Ecdysoneless in Pre-mRNA Splicing
Steroid hormones perform pivotal roles in animal development, sexual maturation, reproduction, and physiology. Also insects possess a hormonal steroid, commonly known as ecdysone, that was originally found to promote ecdyses in growing larvae and their metamorphosis to adults. Since the discovery of ecdysone-inducible puffs on the polytene chromosomes in the 1960's, genetics of the Drosophila fruit flies has substantially advanced our understanding of steroid hormone impact on gene regulation during development. In the present study, we have solved an old puzzle of the “ecdysoneless” mutant that has traditionally provided investigators with a steroid-deficient animal model. Unexpectedly, we find that the Ecdysoneless protein (Ecd) does not primarily regulate ecdysone biosynthesis but that a critical steroidogenic enzyme requires Ecd for splicing of its precursor mRNA (pre-mRNA). Ecd physically contacts the immensely complex pre-mRNA splicing machinery. Outside the ecdysone-producing gland, Ecd is essential for survival of dividing cells within developing tissues. Despite vast evolutionary distance, a human homolog of Ecd can functionally substitute for its counterpart in the fly. A conserved role of Ecd in pre-mRNA splicing might underlie a recently described involvement of mammalian Ecd in cell cycle progression and its contribution to malignancy of certain tumor types.
Published in the journal: Unexpected Role of the Steroid-Deficiency Protein Ecdysoneless in Pre-mRNA Splicing. PLoS Genet 10(4): e32767. doi:10.1371/journal.pgen.1004287
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004287Summary
Steroid hormones perform pivotal roles in animal development, sexual maturation, reproduction, and physiology. Also insects possess a hormonal steroid, commonly known as ecdysone, that was originally found to promote ecdyses in growing larvae and their metamorphosis to adults. Since the discovery of ecdysone-inducible puffs on the polytene chromosomes in the 1960's, genetics of the Drosophila fruit flies has substantially advanced our understanding of steroid hormone impact on gene regulation during development. In the present study, we have solved an old puzzle of the “ecdysoneless” mutant that has traditionally provided investigators with a steroid-deficient animal model. Unexpectedly, we find that the Ecdysoneless protein (Ecd) does not primarily regulate ecdysone biosynthesis but that a critical steroidogenic enzyme requires Ecd for splicing of its precursor mRNA (pre-mRNA). Ecd physically contacts the immensely complex pre-mRNA splicing machinery. Outside the ecdysone-producing gland, Ecd is essential for survival of dividing cells within developing tissues. Despite vast evolutionary distance, a human homolog of Ecd can functionally substitute for its counterpart in the fly. A conserved role of Ecd in pre-mRNA splicing might underlie a recently described involvement of mammalian Ecd in cell cycle progression and its contribution to malignancy of certain tumor types.
Introduction
The insect steroid hormones, ecdysteroids, regulate growth, stimulate molting, and orchestrate tissues to undergo complex morphogenetic changes during metamorphosis [1]–[3]. Temporal control of ecdysteroid synthesis by the larval prothoracic gland (PG) is therefore critical (for recent reviews see [4], [5]). The biosynthetic pathway commences by converting cholesterol to 7-dehydrocholesterol by a Rieske oxygenase Neverland (Nvd) [6]. A short-chain dehydrogenase/reductase encoded by shroud (sro) [7] and cytochrome P450 (CYP) enzymes encoded by spook (spo)/spookier (spok), phantom (phm), disembodied (dib), and shadow (sad) then catalyze the subsequent steps to produce ecdysone (E) [8], [9]. Once released from the PG, E becomes hydroxylated in peripheral tissues by another CYP, Shade (Shd), to yield the main active hormone, 20-hydroxyecdysone (20E) [10]. Reflecting the necessity of 20E for early cuticle formation, Drosophila melanogaster loss-of-function mutants that are available for sro [7], spo, phm, dib, sad, and shd [8] die as embryos.
Understandably, Drosophila mutants that display reduced steroid titers during larval development provide invaluable experimental tools. Among these, the ecdysoneless1 (ecd1) mutants are homozygous viable at 22°C, but exposure to 29°C reduces their ecdysteroid titer and causes a developmental arrest [11], [12]. The ecd1 allele has been widely used since its discovery in 1977 [11] to test effects of ecdysteroid signaling on a number of processes from morphogenesis to reproduction to behavior. Yet why these mutants lack the hormone has not been determined. Our original identification of the ecd gene [13] has revealed homology from fission yeast to humans but none that would illuminate the mode of Ecd action.
Mammalian Ecd (also known as SGT1 and hEcd in humans) has been shown to stimulate cell proliferation by interacting with the Retinoblastoma (Rb) proteins [14]. Conditional deletion of the Ecd gene from mouse embryonic fibroblasts stalls these cells at the G1-S phase transition, suggesting that Ecd normally lifts the inhibitory effect of Rb on E2F-dependent cell cycle progression [14]. High hEcd expression has been correlated with malignancy of human breast [15] and pancreatic [16] tumors. In a mouse model, Ecd has been suggested to promote tumorigenesis via enhancing glucose import and glycolysis in the pancreatic tumor cells [16]. These observations illustrate the emerging importance of Ecd, but an underlying mechanism for Ecd action is still lacking.
A systematic mapping of Drosophila protein-protein interactions [17] has uncovered contacts between Ecd and proteins responsible for pre-mRNA splicing. Among these are members of the U5 small nuclear ribonucleoprotein particle (snRNP) complex, including orthologs of the RNA helicase Brr2, the GTPase Snu114, the Aar2 protein, and the highly conserved spliceosome core component, Prp8 [18]. Another global proteomic study [19] has detected a corresponding interaction between human Prp8 and hEcd. The spliceosome is a most elaborate machinery and pre-mRNA splicing is extensively coupled with transcription as well as with posttranscriptional mRNA surveillance and degradation of incorrectly spliced transcripts [20]. This network involves hundreds of proteins whose individual functions are often inferred from work on yeast, or still remain unknown [21]–[23]. Genetic studies in Drosophila have shown essential roles of several spliceosome components such as Prp8, Prp38, Prp31, or BCAS2 for tissue growth, cell proliferation or cell viability [24]–[27]. Proteomic dissection of spliceosomal complexes has revealed a large overlap in protein composition between Drosophila and humans while suggesting novel and/or fly-specific components [28].
With the hypothesis that Ecd might be a new player in pre-mRNA splicing, we have verified that Ecd interacts with a complex containing Drosophila Prp8, Aar2, Brr2, and Snu114 orthologs. Consistently with a vital role in pre-mRNA splicing, we show that Ecd is required by dividing imaginal disc cells for survival, even when their apoptosis is blocked or cell growth enhanced. In the larval PG, loss of Ecd compromises pre-mRNA splicing and abolishes protein expression of the essential steroidogenic enzyme Spok (CYP307A2), thus accounting for the systemic steroid deficiency in ecd mutants. Remarkably, human Ecd can functionally substitute for its fly ortholog.
Results
Ecd binds proteins within the spliceosomal U5 snRNP complex
The Drosophila Ecd protein has been recently associated with the U5 snRNP complex [17]. In order to verify the proteome-wide data, we expressed Myc epitope-tagged Ecd in Drosophila S2 cells and using mass spectrometry we examined material that co-precipitated with Myc::Ecd (Figure 1A). This analysis identified three proteins of the U5 snRNP complex that were previously shown to associate with Ecd [17], namely orthologs of the budding yeast (Saccharomyces cerevisiae) Prp8p, Snu114p, and Brr2p that are encoded by D. melanogaster genes CG8877/prp8, CG4849/eftud2, and CG5931/l(3)72Ab, respectively. Our mass spectrometry did not detect another Ecd interactor, identified as a product of the CG12320 gene [17] that is homologous to S. cerevisiae Aar2p. For clarity, we will refer to the Drosophila proteins as Prp8, Snu114, Brr2, and Aar2.
Fig. 1. Interactions of Ecd with proteins of the U5 snRNP complex. 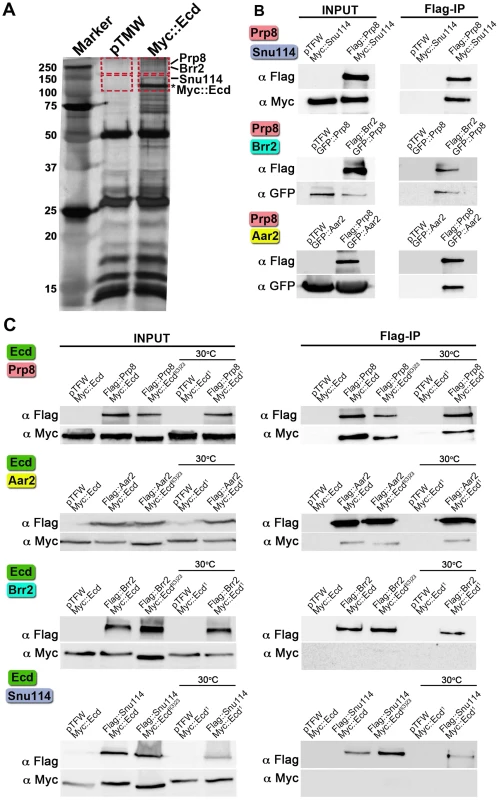
(A) Extracts from Drosophila S2 cells expressing the Myc::Ecd fusion protein or the Myc tag alone (pTMW) were affinity purified using the Myc epitope and resolved on silver-stained SDS-PAGE. Mass spectrometry on the gel sections (indicated by rectangles) identified Prp8, Brr2, and Snu114 proteins associated with Myc::Ecd (asterisk). (B) Drosophila Prp8 interacts with orthologs of known spliceosomal proteins: Snu114, Brr2, and Aar2. (C) Flag-tagged Prp8 and Aar2 but not Brr2 and Snu114 proteins bound Ecd or its mutant variants as assessed by Flag immunoprecipitation (Flag-IP). Cells expressing the temperature-sensitive Ecd1 mutant were upshifted to 30°C 45 min prior to lysis. In all assays (B, C), the pairs of proteins were expressed in S2 cells and their epitope tags were used for Flag-IP and immunoblot detection as indicated. The empty pTFW vector expressing the Flag peptide alone was included in control assays. To test the individual protein-protein interactions, we cloned the above Drosophila genes and performed co-immunoprecipitation with pairs of their epitope-tagged products expressed in S2 cells. First, we verified interactions between the fly counterparts of the known spliceosomal components. As expected, binding occurred between Prp8 and Snu114, Prp8 and Brr2, and Prp8 and Aar2 (Figure 1B). Of these four proteins, Prp8 and Aar2 formed complexes with Ecd, whereas binding between Ecd and either Brr2 or Snu114 was not detected (Figure 1C). It was of interest to examine interactions of mutated Ecd versions that occur in Drosophila. We chose the temperature-sensitive ecd1 allele that carries a single Pro-656 to serine substitution and the non-conditional lethal, ecdl(3)23, where the Ecd polypeptide is prematurely terminated after Ser-649, thus lacking the C-terminal 35 amino acids [13]. Both the Ecd1 (tested at a non-permissive temperature of 30°C) and Ecdl(3)23 mutant proteins retained the capacity to bind Prp8 and Aar2 (Figure 1C). This indicates that the steroid-deficiency and lethality phenotypes in Drosophila result from a failure of the mutant Ecd protein to perform functions other than binding Prp8 and Aar2. The above results suggest that Ecd interacts with at least two well-established members of the U5 snRNP complex, of which Prp8 is evolutionarily the best-conserved core protein of the spliceosome [18].
Subcellular localization of the spliceosomal proteins and Ecd
In S. cerevisiae, a part of the U5 snRNP complex that contains Prp8p, Snu114p, and Aar2p assembles in the cytoplasm before being imported to the nucleus, where Brr2p replaces Aar2p [29], [30]. Localization of Drosophila Prp8, Snu114, Aar2 or Brr2 has not been previously reported. Using the epitope-tagged proteins in transfected S2 cells, we detected Prp8, Snu114, and Aar2 predominantly in the cytoplasm, whereas Brr2 occurred in the nucleus (Figure 2A). Expression of Flag::Prp8 in transgenic Drosophila larvae revealed that Prp8 also resided in the cytoplasm of the PG (Figure S1A).
Fig. 2. Subcellular localization of Ecd and the U5 snRNP proteins. 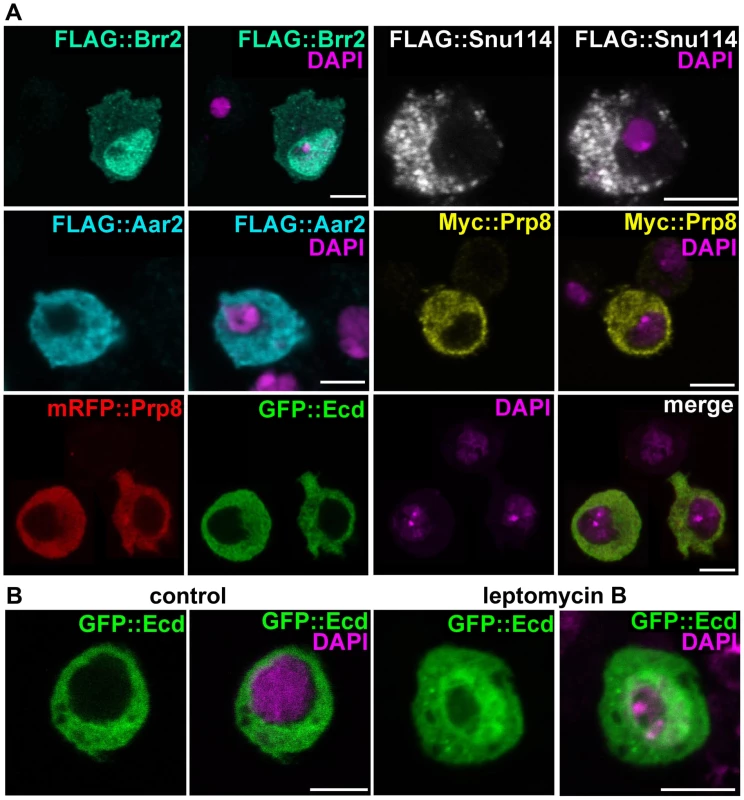
(A) Brr2 was enriched in cell nuclei while Ecd, Prp8, Snu114, and Aar2 were detected in the cytoplasm. The Flag and Myc epitope-tagged proteins were expressed in S2 cells; mRFP::Prp8 and GFP::Ecd (bottom row) were co-transfected. (B) Inhibition of nuclear export with leptomycin B resulted in nuclear retention of the GFP::Ecd protein. Nuclei are stained with DAPI. All images are single confocal sections. Scale bars, 5 µm. The Ecd protein was previously detected in the cytoplasm of Drosophila PG cells [13]. We confirmed this result with a new antibody raised against the N-terminal part of Ecd for the endogenous protein (Figure S1B, S1C) and for Ecd overexpressed in the PG using the phm-Gal4 driver (Figure S1D). This antibody also detected a clear cytoplasmic signal in the wing imaginal discs (Figure S2). The cytoplasmic localization of hEcd in human cells was shown to depend on leptomycin B-sensitive nuclear export [31]. In transfected S2 cells, GFP-tagged Ecd co-localized in the cytoplasm with Prp8 (Figure 2A). Only when the cells were treated with leptomycin B, some of the GFP::Ecd fusion protein was retained in the nucleus (Figure 2B). Therefore, together with the spliceosomal proteins with which Ecd interacts, Ecd primarily resides in the cytoplasm. Like its human counterpart, Ecd is subject to active nuclear export.
Cell-autonomous requirement for Ecd and Prp8 in imaginal discs
If Ecd plays a role in pre-mRNA splicing as suggested by its protein interactions, its loss would be expected to disturb vital cellular functions as has been found for several Drosophila spliceosomal proteins including Prp8 [24]–[27]. To track the fate of Ecd-deficient imaginal cells in vivo, we employed the MARCM technique [32] using the non-conditional ecdl(3)23 allele that behaves as a genetic null [13]. In contrast to control, no mitotic clones homozygous for this mutation were found in eye/antennal or wing imaginal discs of third-instar larvae (Figure 3A, 3B, 3E, 3F). Rare and extremely small ecdl(3)23 clones could be detected when cell death was prevented in these clones with the anti-apoptotic protein p35, or when their cellular growth was enhanced with activated RasV12 (Figure 3C, 3D). To achieve a milder loss-of-function effect, we generated transgenic flies for RNAi-mediated silencing of ecd. Observation of wing discs two days after heat-dependent clonal induction of ecd dsRNA revealed frequent ecdRNAi clones. These gradually disappeared over the next 24 hours (Figure 3G, 3H), indicating that the cells autonomously required Ecd to survive.
Fig. 3. Ecd is required for survival of imaginal disc cells. 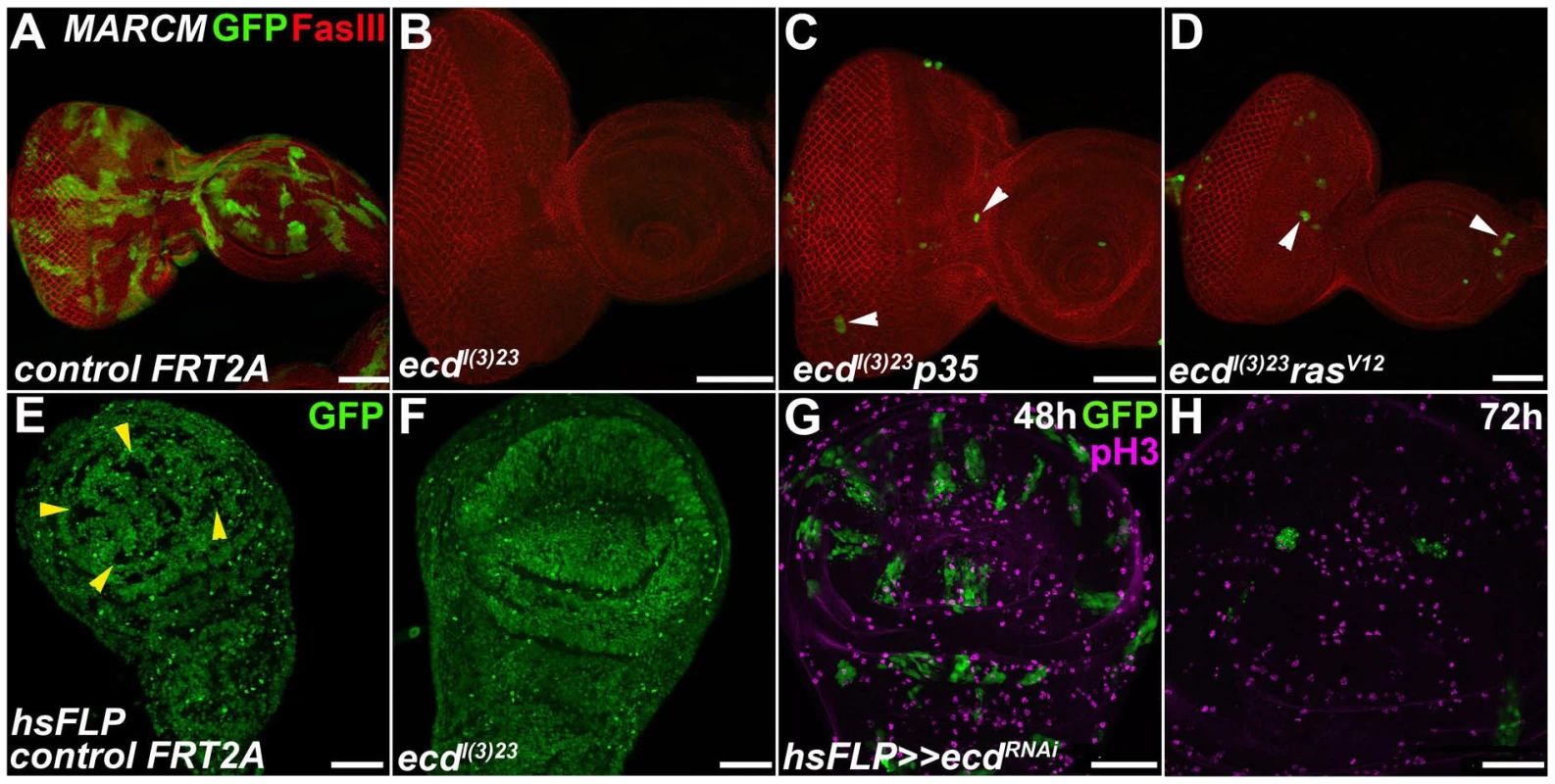
(A–F) In contrast to abundant, sizable control clones (green in A and GFP-negative in E, yellow arrowheads), ecdl(3)23 homozygous mutant clones could be recovered neither in eye/antennal (B) nor wing (F) imaginal discs. Simultaneous inhibition of apoptosis with a pan-caspase inhibitor p35 (ecdl(3)23 p35) (C) or cell growth enhancement with activated RasV12 (ecdl(3)23 rasV12) (D) yielded rare and extremely small clones in eye/antennal discs (white arrowheads). Fasciclin III (FasIII) staining of lateral cell membranes reveals the overall tissue morphology. (G–H) ecdRNAi clones (green) were eliminated from the wing imaginal epithelium within 72 h after induction using the heat-shock flip-out technique. Only rare ecdRNAi cells labeled phospho-histone 3 (pH3) positive relative to many mitotically active surrounding control cells. Scale bars, 50 µm. To see the effect of Ecd knockdown on morphogenesis, we induced ecd RNAi in a restricted region along the anterior-posterior boundary of the wing imaginal disc using the dpp-Gal4 driver. Figure S2 shows depletion of the Ecd protein from the dpp-expressing cells. The loss of Ecd disrupted the regular pattern of the dpp expression domain, and the affected cells underwent apoptosis as assessed by active Caspase 3 staining (Figure 4A, 4B). Consequently, the adult wings of dpp>ecdRNAi flies displayed reduced size of specific intervein regions and loss of the anterior crossvein (Figure 4E, 4F). The same anomalies were induced by RNAi against the components of the U5 snRNP complex, Prp8 (Figure 4C, 4G) and Brr2 (Figure S3A, S3B). Overexpression of Prp8 in the Ecd-deficient cells did not suppress the ecd RNAi phenotype (Figure S3C, S3D), suggesting that Ecd plays a unique role that cannot be substituted by surplus of its partner protein. Strikingly, apoptosis of imaginal disc cells expressing ecd dsRNA as well as the adult wing defect could be averted by supplementing the human ortholog, hEcd (Figure 4D, 4H).
Fig. 4. Depletion of Ecd or Prp8 causes apoptotic phenotypes in developing wings. 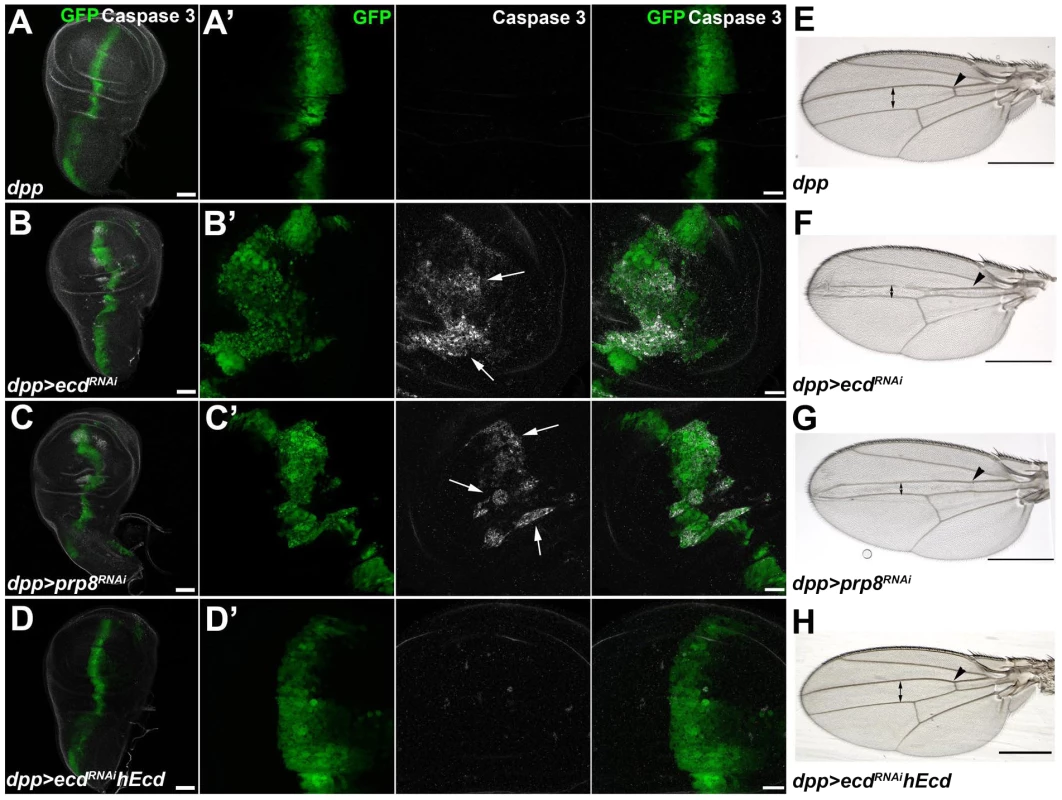
(A–D) RNAi knockdown of ecd (B) and prp8 (C) induced with the dpp-Gal4 driver disrupted the regular pattern of the dpp expression domain (visualized by co-expression of GFP) in third-instar wing imaginal discs and caused appearance of apoptotic, Caspase 3 positive cells (B′, C′, white arrows). Expression of human Ecd (hEcd) averted the apoptotic phenotype caused by ecd RNAi (D, D′). (E–H) The intervein region corresponding to the dpp expression domain was reduced (double arrows) and the anterior crossvein (arrowhead) was lost in the wings of dpp>ecdRNAi (F) and dpp>prp8RNAi (G) adult flies. The ecd RNAi phenotype was rescued by expression of hEcd (H). Images in (A–D) are maximum-intensity projections of multiple confocal sections; panels A′ through D′ are higher-magnification, single sections from the corresponding images in (A–D). Scale bars are 50 µm (A–D), 20 µm (A′–D′), and 1 mm (E–H). Loss of Ecd disrupts splicing of spok pre-mRNA and eliminates its protein product
To address the putative role of Ecd in ecdysone biosynthesis, we removed the Ecd protein specifically from the PG by triggering ecd RNAi with the phm-Gal4 driver [33] (Figure S1C). Affected larvae reached the third instar but were unable to pupate. Unlike in the imaginal discs, depletion of Ecd was not cell-lethal in the polyploid PG, although it reduced the size of PG cells and nuclei (Figure 5A, 5B). Using available antibodies, we could therefore examine expression of steroidogenic enzymes, namely CYP307A2/Spok and CYP306A1/Phm, in late-third instar phm>ecdRNAi larvae. The strong Spok staining in control PGs was completely lost in the Ecd-deficient gland, whereas the Phm signal appeared only partly reduced (Figure 5A, 5B). While both enzymes are indispensable for E biosynthesis, Spok acts upstream of Phm in the larval PG to mediate the essential conversion of 7-dehydrocholesterol to ketodiol [33], [34]. Thus, PGs deprived of Spok cannot synthesize ecdysone even in the presence of Phm. As was the case in the wing imaginal discs, expressing the human Ecd ortholog in the PG compensated for the depletion of the endogenous Ecd protein. hEcd improved morphology of the gland (Figure 5C) and restored expression of Spok (Figure 5C′).
Fig. 5. Ecd and Prp8 are required for expression of Spok in the PG. 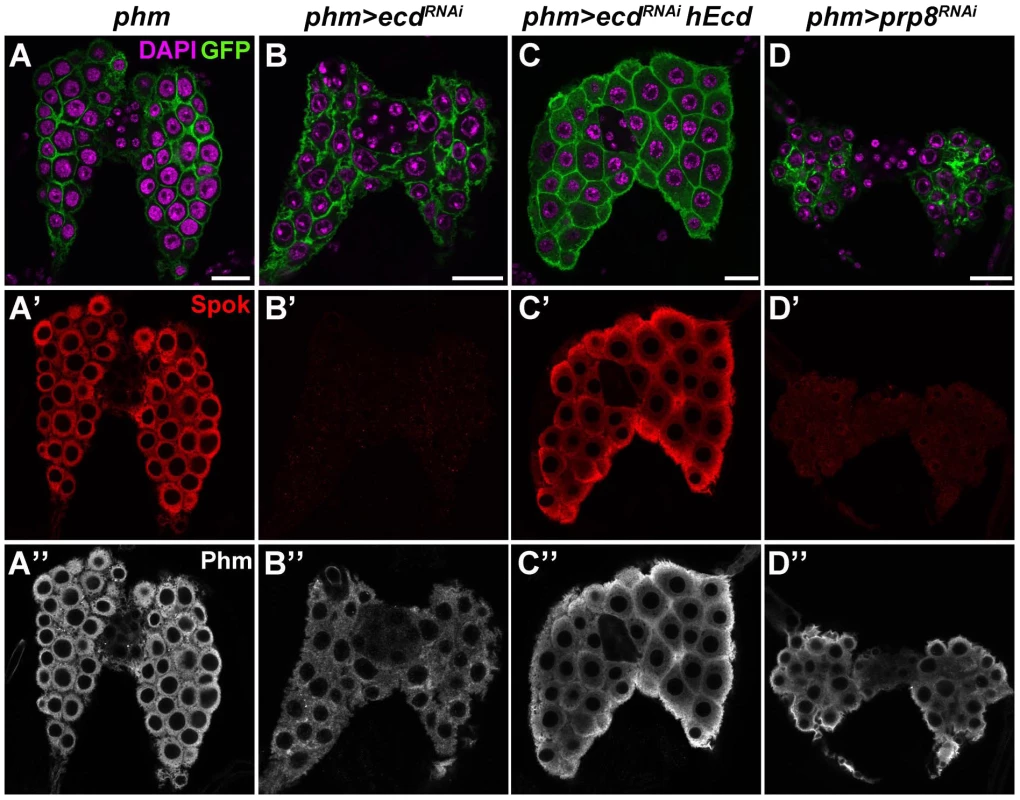
(A–D) Relative to control PG dissected 6 days AEL (A′, A″), expression of the Spok protein (B′) was undetected while the Phm signal (B″) was weakened in the PG of phm>ecdRNAi (B) and phm>prp8RNAi (D′, D″) larvae. Note the moderate reduction in size of PG cells and nuclei in phm>ecdRNAi (B) compared to a more severe PG deterioration in phm>prp8RNAi larvae (D). Expression of hEcd restored Spok and Phm expression (C′, C″) and improved the morphology of the Ecd-deficient PG (C). Cell membranes are decorated with CD8::GFP; DAPI stains the nuclei. Panels show single confocal sections. Scale bars, 20 µm. See Figure S1 for RNAi-mediated depletion of Ecd. Similarly to ecd RNAi, knockdown of the spliceosomal proteins Prp8 (Figure 5D) and Brr2 (Figure S4A, S4B) also reduced size of the PG and eliminated the Spok protein while only partially reducing Phm levels. These results suggested that expression of Spok, but not of Phm, was highly sensitive to perturbation of pre-mRNA splicing. Interestingly, the phm gene contains two small introns, whereas spok lies within the heterochromatin and its single intron separates the two coding exons by nearly 30 kilobases (Flybase, http://flybase.org/) (Figure 6A).
Fig. 6. Loss of Ecd interferes with splicing of spok pre-mRNA. 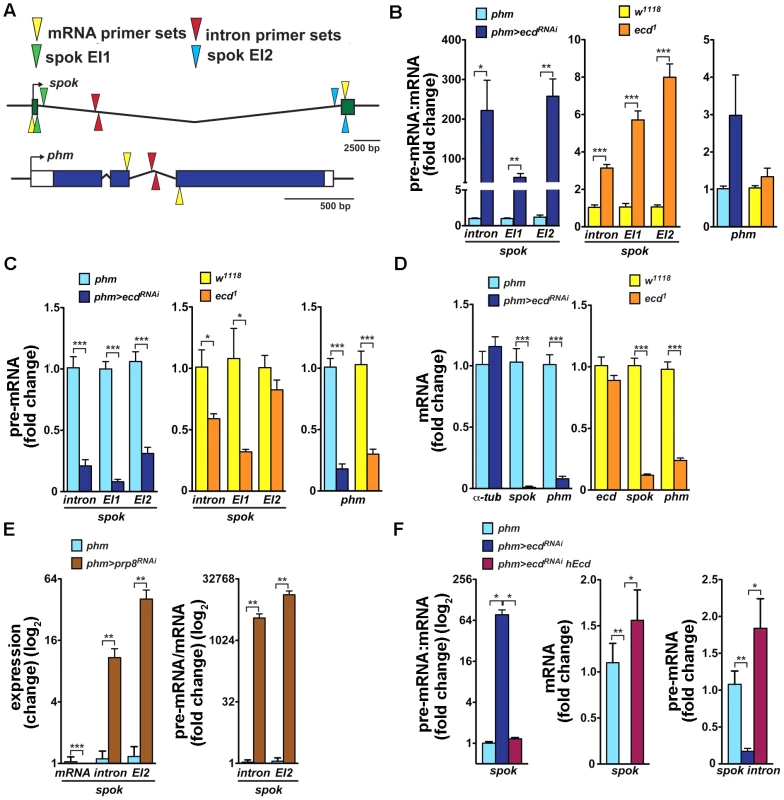
(A) Schematic of Drosophila spok and phm gene loci. Open and colored boxes represent untranslated and translated exons, respectively; black arrows point in the direction of transcription. Colored arrowheads mark the positions of primer pairs used to discriminate between pre-mRNA and mRNA species by qRT-PCR: yellow, in exons separated by an intron; red, within an intron; green and blue, spanning exon-intron boundaries of the first (EI1) and second (EI2) spok exon, respectively. (B) Pre-mRNA:mRNA ratios, determined by qRT-PCR with primers depicted in (A), were significantly elevated for spok but not for phm in third-instar phm>ecdRNAi larvae and in ecd1 mutants at 29°C. (C, D) spok and phm pre-mRNA (C) and mRNA (D) levels decreased in phm>ecdRNAi and ecd1 larvae. Levels of α-tubulin 84B mRNA did not change upon ecd RNAi and expression of ecd itself was unaffected by the ecd1 mutation (D). (E) RNAi knockdown of prp8 in the PG diminished spok mRNA, whereas unspliced transcripts accumulated, causing the pre-mRNA:mRNA ratio to rise dramatically. (F) Expression of hEcd in the Ecd-deficient PG restored normal spok transcription and pre-mRNA splicing as judged from restored levels of pre-mRNA, mRNA, and their ratio. In all experiments, qRT-PCR was performed with total RNA from whole larvae 6 days AEL, and levels of rp49 transcripts were used for normalization. The pre-mRNA:mRNA ratios in (B, E, F) were calculated from the normalized qRT-PCR data by dividing values obtained with intron primer sets (A, red triangles) or primers spanning exon-intron boundaries (green or blue triangles) with values obtained using mRNA-specific primers (yellow triangles). Data are mean ± S.E.M; n≥4; *p<0.05, **p<0.01, and ***p<0.001. To examine whether the absence of Ecd affects spok pre-mRNA splicing, we determined the pre-mRNA:mRNA ratio, a standard measure of intron retention or splicing defect (e.g., [35]–[37]), in phm>ecdRNAi larvae and in larvae homozygous for the temperature-sensitive ecd1 allele. mRNA levels were measured using quantitative reverse-transcription (qRT) PCR with primers positioned in two separate exons, and pre-mRNA was amplified with primer pairs spanning exon-intron boundaries or primers within the intron (Figure 6A). All RNA samples were free of residual genomic DNA and qRT-PCR data were normalized to rp49 mRNA levels that did not change appreciably between control and ecd-deficient larvae (Figure S5). Because spok and phm are expressed specifically in the PG during larval development, we were able to use RNA from entire larvae.
As assessed with all three pre-mRNA-specific primer sets, spok pre-mRNA:mRNA ratio strongly increased in both phm>ecdRNAi larvae and in ecd1 mutants upshifted to 29°C (Figure 6B). The relative enrichment of unspliced to spliced spok transcript was markedly stronger upon PG-specific knockdown of ecd than in the hypomorph mutants. Unexpectedly, loss of ecd also reduced levels of spok pre-mRNA, again more substantially in phm>ecdRNAi larvae (Figure 6C), suggesting that in addition to splicing, either spok transcription or stability of its pre-mRNA might have been affected. Consistently with the absence of the Spok protein (Figure 5B′), ecd RNAi diminished spok mRNA (Figure 6D). Levels of phm pre-mRNA and mRNA were also lowered in PGs of ecd-deficient animals relative to controls (Figure 6C, 6D). However, in contrast to spok there was no significant increase of the pre-mRNA:mRNA ratio and therefore no appreciable intron retention in the case of phm (Figure 6B). These data suggested that Ecd was required for splicing of the large intron from the spok pre-mRNA. For comparison, we examined the effect of Prp8 knockdown. As expected, spok mRNA disappeared in phm>prp8RNAi larvae due to compromised splicing (Figure 6E). However, because the absolute amount of unspliced spok transcript accumulated upon prp8 RNAi, the pre-mRNA:mRNA ratio rose dramatically more than in the case of ecd RNAi (Figure 6E).
Finally, we tested whether human Ecd restored Spok expression at the level of pre-mRNA splicing. In a separate set of experiments, we confirmed that the loss of spok mRNA was accompanied by intron retention (increased pre-mRNA:mRNA ratio, Figure 6F). Importantly, expression of hEcd in the PG of phm>ecdRNAi larvae restored levels of spok mRNA, pre-mRNA, and of their normal proportion (Figure 6F), suggesting that the human protein could substitute for Ecd in splicing of spok pre-mRNA.
Systemic effect of PG-specific loss of Ecd
Because the PG produces a circulating hormone, knockdown of Ecd would be expected to affect the entire organism even when restricted to the gland. In agreement with the absence of the Spok/CYP307A2 protein (Figure 5B′), third-instar phm>ecdRNAi larvae on day 6 after egg laying (AEL) showed reduced ecdysteroid titer (Figure S6A). Compared to controls, they were retarded in growth (Figure 7A–C). Without interruption, they continued to feed for another 10 or more days, producing large “permanent” larvae (Figure 7D). This phenotype corresponded with effects of lacking ecdysteroid surge and inadequate expression of steroidogenic genes including spok and phm [38], [39]. Although feeding phm>ecdRNAi larvae on day 5 AEL with 20E was not sufficient for pupation, it induced wandering behavior (Figure S6B). Importantly, substituting hEcd for the depleted fly protein in the PG increased ecdysteroid titer (Figure S6A) and restored normal growth and development of phm>ecdRNAi animals to adults (Figure 7A, 7E, 7F).
Fig. 7. PG-specific loss of Ecd results in systemic effects. 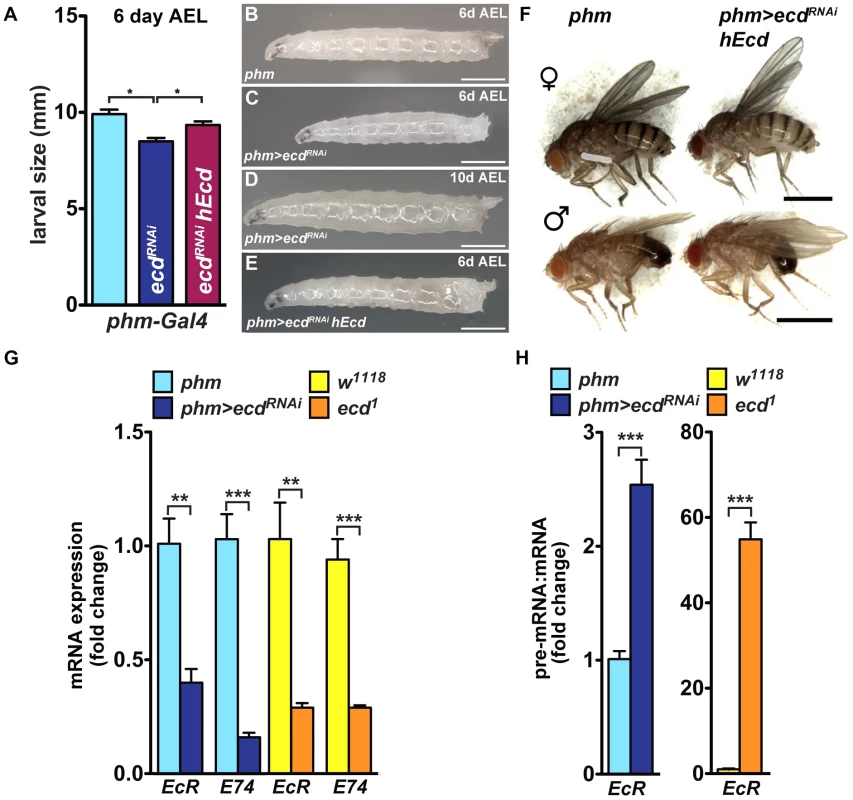
(A–E) Growth of phm>ecdRNAi larvae was retarded. Although reaching the third instar, phm>ecdRNAi larvae were smaller on day 6 AEL (A, C) compared to controls (B) or larvae supplemented with hEcd (phm>ecdRNAi hEcd) (E). Deficiency of Ecd in the PG prevented metamorphosis, generating permanent, eventually lethal larvae (D). (F) Expression of hEcd in the Ecd-deficient PG restored development of normally sized adults. (G) Expression of EcR and E74 (as assessed by qRT-PCR with primer sets detecting all alternatively spliced mRNA isoforms of each gene, see Table S1) was lowered to a similar degree in phm>ecdRNAi and in ecd1 larvae at 29°C. (H) The EcR pre-mRNA:mRNA ratio increased much more dramatically in ecd1 mutants at 29°C than in phm>ecdRNAi larvae. Data are mean ± S.E.M; n≥4; **p<0.01, and ***p<0.001. The PG-specific knockdown of Ecd was accompanied by lowered mRNA levels of at least two 20E-response genes, the Ecdysone receptor (EcR) and E74, as assessed in whole phm>ecdRNAi larvae (Figure 7G). A similar reduction in EcR and E74 mRNAs occurred in ecd1 mutant larvae at 29°C (Figure 7G). However, ecd1 mutants could have lost these transcripts for two different, mutually non-exclusive reasons: (1) due to the lack of circulating ecdysteroids, and/or (2) because Ecd may be required for splicing of the EcR and E74 pre-mRNAs, both of which contain multiple large introns. Indeed, we found a major rise in the pre-mRNA:mRNA ratio for EcR (but not for E74; data not shown) in ecd1 mutants, and a much less pronounced increase of this ratio in phm>ecdRNAi larvae (Figure 7H). These data suggest that EcR pre-mRNA splicing was sensitive to loss of ecd function in tissues throughout the body. In support of this notion, we observed that dietary 20E did not induce EcR mRNA expression in ecd1 mutants at 29°C nearly as efficiently as under the permissive temperature or in phm>ecdRNAi larvae (Figure S6C), where ecd function in the peripheral tissues was unaffected.
Discussion
Ecd, originally discovered through the “ecdysoneless” Drosophila ecd1 mutant [11], has become known as a product of the CG5714 gene [13]. The steroid deficiency results from an autonomous failure of the larval PG [12], [40] and of the adult ovary [11], [40], [41]. However, some effects of the ecd1 allele are autonomous to the imaginal discs and other tissues, and therefore not attributable to the lack of the hormone [41], [42]. This notion has been confirmed upon molecular definition of ecd-null mutations whose effects could not be remedied with exogenous 20E [13]. As no causal relationship between Ecd and E biosynthesis could be established, Ecd has been omitted from the current list of ecdysteroidogenic factors [4], [5], [8], [9]. Nonetheless, the ecd1 background is still in use to demonstrate specific effects of 20E (e.g., [43]).
Our present work shows that Ecd indeed does not primarily regulate steroidogenesis. Although abolished expression of the essential steroidogenic enzyme Spok in Ecd-deficient larvae inevitably causes ecdysone deficiency, Ecd should not be regarded as a regulator of the E biosynthetic pathway. Rather, the absence of Spok in the PG reflects a more general role of Ecd within the complex network of pre-mRNA splicing. This function is required in the PG as well as in some other Drosophila organs, and is likely to be common to flies and mammals.
A role for Ecd in pre-mRNA splicing
A proteome-wide study [17] has uncovered interactions of Drosophila Ecd with multiple U5 snRNP-associated spliceosomal proteins. We likewise found a complex of Ecd with Prp8, Snu114, and Brr2 using mass spectrometry, and interactions of Ecd with Prp8 and Aar2 by immunoprecipitation. We detected Brr2 in cell nuclei, whereas Ecd co-localized with Prp8, Snu114, and Aar2 in the cytoplasm of Drosophila cells. In contrast, S. cerevisiae Prp8p resides in the yeast nuclei owing to its nuclear localization signal [29]. Although Prp8 is remarkably well conserved [18], this particular sequence shows poor homology with the fly protein. Nuclear import of Drosophila Prp8 might therefore rely on another mechanism. Nonetheless, our results conform to a current model from S. cerevisiae, where Prp8p, Snu114p, and Aar2p preassemble in the cytoplasm, then upon nuclear import Aar2p is replaced by Brr2p as the U5 snRNP complex matures [29], [30]. Nucleo-cytoplasmic shuttling of Ecd, which also occurs in human cells [31], corresponds with such translocation.
Phenotypes caused by disrupted pre-mRNA splicing match those inflicted by loss of Ecd. Cell death of Drosophila imaginal disc clones lacking spliceosomal proteins Prp38, MFAP1, and BCAS2 has been reported [24], [25]. Our data show that knockdown of the Prp8 and Brr2 proteins caused apoptosis of imaginal disc cells and recapitulated a specific ecd RNAi wing phenotype. Removal of Prp8 or Brr2 from the PG eliminated the Spok protein, confirming that its expression was sensitive to deficiency in pre-mRNA splicing factors. Similar to Prp8 knockdown, spok pre-mRNA:mRNA ratio strongly increased in ecd1 or phm>ecdRNAi larvae, thus evidencing a role of Ecd in spok pre-mRNA splicing. However, removal of Ecd also affected transcription as judged from reduced levels of spok or phm pre-mRNAs. It is important to note here that pre-mRNA splicing is not an isolated event but that it is intimately coupled with transcriptional elongation [44], [45] as well as with 3′-end processing, quality control, and nuclear export or degradation of the RNA product (reviewed in [20]). Indeed, there are indications that hEcd may be involved in transcriptional regulation in mammalian cells [31].
An intriguing question is why would dysfunction of Ecd impact splicing of spok but not phm pre-mRNA in the steroid-producing gland? Surprisingly, even the relatively few and simple intron-containing genes in S. cerevisiae have been shown to respond differently to mutations in individual spliceosome core components [36]. In multicellular organisms, spliceosomal proteins such as Prp8 are not ubiquitously expressed and not equally required to splice every pre-mRNA [46]. Instead, many perform exquisitely tissue-specific, gene-specific, and even exon-specific functions. An RNAi screen in Drosophila cells has uncovered the necessity of several core spliceosomal proteins including Brr2 for inclusion of particular exons in different, alternatively spliced transcripts [47].
A striking example of how remarkably specific physiological functions may be affected by faulty pre-mRNA splicing is the human disease retinitis pigmentosa (RP), where degeneration of photoreceptor cells leads to blindness. Autosomal-dominant forms of RP are linked to mutations in Prp8, Prp31, Prp3, and PAP1 homologs that contribute to the U5•U4/U6 tri-snRNP complex [46], [48]. One plausible explanation for the sensitivity of the photoreceptor neurons to defective splicing is insufficient production of mRNAs that are highly expressed in these cells [48]. It has been argued that ubiquitously expressed pre-mRNAs carry universal splicing signals in order to be correctly spliced in any cell type, whereas transcripts with restricted expression patterns might rely on less robust, tissue-specific splicing signals that are more prone to fail in the absence of individual splicing factors [46]. Although the mechanism of Ecd action remains to be determined, we presume that spok pre-mRNA might be particularly sensitive to the absence of Ecd, Prp8 or Brr2 for similar reasons.
Ecdysone-dependent versus organ-autonomous consequences of Ecd depletion
In addition to abolishing the expression of Spok, loss of ecd function partially reduced mRNA and pre-mRNA levels of phm (Figure 6C, 6D) and of two other genes (nvd and dib; data not shown) that are likewise required for E biosynthesis. However, the lower expression of phm, nvd and dib was not accompanied by increased pre-mRNA:mRNA ratios, indicating that pre-mRNA splicing of these genes did not depend on Ecd. These results might suggest that Ecd exerts some effect on transcription or transcript stability. Alternatively, Ecd might be required for pre-mRNA splicing of a factor acting upstream of the steroidogenic gene expression. One such candidate is EcR, which has recently been shown to mediate a positive-feedback of 20E on the expression of phm, dib, sro and sad, but not of spok [49]. Elevated pre-mRNA:mRNA ratio indicated that splicing of EcR pre-mRNA was compromised in ecd1 mutants (Figure 7H), and the EcR protein was markedly reduced in the PGs of phm>ecdRNAi larvae (Figure S6D). Therefore, whereas the absence of the Spok protein primarily resulted from disrupted splicing, the partial reduction of nvd, phm and dib expression might be attributable to the lack of EcR in the PG of phm>ecdRNAi larvae.
A failure of the PG to produce E inevitably provokes systemic developmental defects in peripheral organs. However, an issue arises with the ecd1 mutants as to whether any observed tissue-specific phenotype may be ascribed to the lacking hormone. Because Ecd is required cell-autonomously, effects of ecdysteroid deficiency and those caused by loss of any cell-autonomous Ecd function cannot be discriminated in ecd1 mutant background. For example, low expression of 20E-response genes such as EcR in ecd1 larvae likely results from a combined impact of disrupted E synthesis in the PG and compromised EcR pre-mRNA splicing in the peripheral tissues. The problem of 20E-dependent and 20E-independent effects of the ecd1 mutation may be reflected by a recent transcriptome analysis, revealing that of about a thousand genes affected in ecd1 background, only a minority were regulated by the 20E receptor, EcR [43].
Based on the elimination of ecdl(3)23 mutant clones from the imaginal discs, we conclude that cells lacking Ecd cannot be sustained within proliferating tissue context. This was the case even when the mutant clones were protected from apoptosis with p35, or enhanced for growth through RasV12. Small ecdl(3)23 or ecdRNAi clones were replaced by surrounding cells without phenotypic consequences for the adult. In contrast, ecd RNAi delivered to larger areas was lethal or caused visible defects, such as the aberrant wings in dpp>ecdRNAi flies. Rarely emerging MS1096>ecdRNAi flies had vestigial wings (Figure S7A) similar to those induced by depletion of the BCAS2 spliceosomal protein under the same Gal4 driver [25]. Interestingly, imaginal disc cells were not affected by triggering ecd RNAi after their proliferative phase was completed, as in the eyes of GMR>ecdRNAi flies (Figure S7B). GMR-Gal4 is a strong but late-acting driver expressed predominantly in the post-mitotic cells posterior to the eye morphogenetic furrow [50]. Intriguingly, deletion of Ecd in mouse embryonic fibroblasts has been shown to cause a proliferative block and to reduce expression of several cell-cycle regulators such as CyclinE1 (CycE1) downstream of the transcription factor E2F [14]. However, we were unable to advance proliferation of ecdRNAi clones in imaginal discs by expressing Drosophila CycE [51] or combinations of either E2F with its partner DP [52] or CycD with Cdk4 [53] (data not shown). Therefore, the lack of Ecd in proliferating tissues cannot be compensated by gain of individual factors that stimulate cellular growth or cell cycle progression. These results suggest that proliferating cells are particularly sensitive to loss of Ecd. Such sensitivity corresponds to the fact that splicing factors prevailed among genes positively screened as being required for cell division in a human cell line [54].
Evolving Ecd function?
Unlike some of the core spliceosomal factors, no Ecd-like protein has been found in the budding yeast, S. cerevisiae, in which only 5% of the genes contain introns. This contrasts with 43% of intron-containing genes in the fission yeast, Schizosaccharomyces pombe [55], where the spliceosomal protein composition is more akin to humans than to the budding yeast [56]. Interestingly, S. pombe has an Ecd ortholog, Sgt1p, which resides in the cell nuclei and is essential for the yeast growth [57]. Conditional Sgt1 mutation alters expression of genes involved in virtually all cellular processes including metabolism, and impairs growth on glucose media. Sgt1p is thought to regulate transcription [57], although its mode of action remains unknown. Expression of Sgt1p failed to substitute for Drosophila Ecd (data not shown), likely reflecting a remote homology between the two proteins (21% overall amino acid identity). In contrast, human Ecd that is moderately homologous to fly Ecd (31%) rectified pre-mRNA splicing and expression of spok in the PG, permitting development of phm>ecdRNAi adults.
In mammals, Ecd has been implicated as a positive regulator of cell-cycle promoting genes, of cell cycle progression itself, and of cancer development [14]–[16]. Since hEcd has been independently detected in a complex containing human Prp8 [19], it will be of interest to know if any of those effects involve splicing of particular pre-mRNAs. Considering the evidence from S. pombe, Drosophila and mammals, we suspect that the role of Ecd in pre-mRNA splicing may correlate with the evolutionarily growing importance of splicing in complex multicellular organisms.
Materials and Methods
Fly stocks
The following Drosophila strains were used: w1118, ecd1 [11], ecdl(3)23 [13]; a gift of Dr I. Zhimulev), FRT2A (BL1997), ecdl(3)23 FRT2A/TM6B, y w hsFLP; ecdl(3)23 FRT2A/TM6B, UAS-rasV12/CyO; ecdl(3)23 FRT2A/TM6B, UAS-p35/CyO; ecdl(3)23 FRT2A/TM6B, UAS-prp8RNAi (VDRC, 18565), UAS-brr2RNAi (VDRC, 110666), phm-Gal4, UAS-CD8::GFP [33], and dpp-Gal4, UAS-GFP [58]. New transgenic lines carrying UAS-ecd, UAS-ecdRNAi, UAS-Flag::hEcd, UAS-Flag::prp8, and UAS-Flag::Sgt1 constructs were established with standard P-element germline transformation. If not specified otherwise, crosses were carried out at 25°C. Crossing all Gal4 driver lines to w1118 background provided controls for each experiment. Mutant clones within eye/antennal imaginal discs were generated using the MARCM (mosaic analysis with a repressible cell marker) method [32] with eyFLP, act>y+>Gal4, UAS-GFP/CyO; FRT2A tubGal80/TM6B flies as described [59]. To induce ecdRNAi “flip-out” clones, hsFLP; act>y+>Gal4, UAS-GFP/CyO [60] females were crossed to UAS-ecdRNAi males at 20°C. Recombination was induced by exposing progeny to heat shock at 37°C for 30 min, followed by incubation for 48 and 72 h at 25°C prior to dissection. Temperature-sensitive ecd1 and control w1118 larvae were grown at 22°C until day 4 AEL, then placed to 37°C for 45 min and to 29°C until dissection on day 6 AEL.
Plasmids and Ecd antibody production
Coding Drosophila melanogaster DNA sequences of prp8 (CG8877), snu114 (CG4849), aar2 (CG12320), brr2 (CG5931), ecd (CG5714), ecdl(3)23 (amino acids 1–650 of Ecd), and human Ecd (hsgt1, NP_009196.1) were amplified from respective cDNAs using the Phusion polymerase (New England Biolabs). Schizosaccharomyces pombe Sgt1 (SPAC1002.10c) was amplified from genomic DNA. See Table S1 for all PCR primers. The fragments were cloned into pTFW, pTMW or pTGW vectors enabling expression of proteins with N-terminal Flag, Myc, or GFP tags, respectively (T. Murphy, Carnegie Institution of Washington), using the Gateway cloning system (Invitrogen). The ecd1 P656S mutant was recreated using site-directed mutagenesis by CCT to TCT codon transition. To generate UAS-ecdRNAi, a 497-bp cDNA fragment from the ecd gene was amplified and cloned as inverted repeat into the pWIZ vector [61]. A polyclonal rat anti-EcdNterm antibody was raised (Eurogentec) against bacterially expressed Drosophila Ecd polypeptide (amino acids 204–458). The corresponding ecd cDNA fragment was cloned into the pET28b plasmid (Novagen), and the antigen was purified using a hexahistidine tag.
Drosophila S2 cell culture and immunoprecipitation
Drosophila S2 cells were cultured at 25°C in Shields and Sang M3 insect medium (Sigma Aldrich) containing 8% fetal bovine serum and antibiotics (Pen/Strep, Gibco). Cells were transfected in serum free medium using X-TremeGENE (Roche Applied Science) according to manufacturer's instructions. Expression of UAS-driven genes was induced by co-transfection with a pWA-GAL4 plasmid expressing Gal4 under an actin5C promoter (a gift from Y. Hiromi). Nuclear export was inhibited by incubating cells with 5 ng/ml leptomycin B (Biomol) for 4 h. Cells expressing the temperature-sensitive Ecd1 protein variant were upshifted to non-permissive temperature of 30°C for 45 min prior to processing.
For immunoprecipitation, transfected S2 cells were lysed in 50 mM Tris-HCl (pH 7.8), 150 mM NaCl, 1 mM EDTA (pH 8.0), 1% Triton X-100, 0.01% Igepal, and protease inhibitors (Roche Applied Science). The lysate (300–500 µg of total protein) was incubated overnight with 15 µl of anti-Flag (Invitrogen) or anti-Myc (Medical and Biological Laboratories) magnetic beads at 4°C. After five washes in lysis buffer, proteins were recovered in two consecutive elution steps, each with 50 µl of 0.1 M glycine-HCl (pH 3.0) for 5 min, and neutralized with 10 µl of 0.5 M Tris-HCl (pH 7.8) and 1.5 M NaCl. Upon SDS-PAGE, proteins were detected by immunoblotting with mouse anti-Flag M2 (1∶1000, Sigma Aldrich), rabbit anti-c-Myc (1∶1000, sc-789, Santa Cruz) or rabbit anti-GFP (1∶2000, Acris) antibodies, followed by incubation with corresponding HRP-conjugated secondary antibodies. Chemiluminescent signal was captured using ImageQuant LAS4000 reader (GE Healthcare).
Mass spectrometry and protein identification
Protein extracts from S2 cells containing Myc::Ecd or the empty pTMW vector (for control) were resolved on SDS-PAGE. Upon silver staining (SilverQuestTM, Invitrogen), bands of interest were cut out. In-gel tryptic digestion with 12.5 ng/µl porcine trypsin (Promega) in 10 mM NH4HCO3 and further extractions were performed as described [62]. Collected extracts were concentrated by vacuum centrifugation and desalted using STAGE Tip C18 spin columns (Proxeon, Thermo Scientific) [63]. Eluted peptides were vacuum-concentrated and resuspended to a final volume of 20 µl in 0.5% acetic acid, of which 10 µl were used for analysis. Reversed-phase liquid chromatography (LC) coupled to nano-flow electrospray tandem mass spectrometry (MS) were carried out using an EASY nLC II nano-LC (Proxeon, Thermo Scientific) with a C18 column (internal diameter 75 µm) coupled to a LTQ Orbitrap mass spectrometer (Thermo Scientific). Peptide separation was performed at a flow rate of 250 nl/min over 60 min (10–40% acetonitrile; buffer A: 0.1% formic acid in water; buffer B: 0.1% formic acid in acetonitrile). Survey full scan MS spectra (m/z 350 to 2000) of intact peptides were acquired in the Orbitrap at a resolution of 30,000 using m/z 445.12003 as a lock mass. The mass spectrometer acquired spectra in data dependent mode and automatically switched between MS and MS/MS acquisition. Signals with unknown charge state and +1 were excluded from fragmentation. The ten most intense peaks (threshold 500) were isolated and automatically fragmented in the linear ion trap using collision induced dissociation (CID).
The search algorithm Mascot [64], implemented in the Proteinscape software (Bruker), was used for peptide and protein identification. MS/MS data were searched using the canonical and isoform sequence database of the Drosophila melanogaster complete proteome, provided by the UniProt Consortium. Oxidation of methionine residues was used as a variable modification and carbamidomethylation of cysteine residues as a fixed modification. For Orbitrap data, 10 ppm mass tolerance was allowed for intact peptide masses and 0.8 Da for CID fragment ions detected in the linear ion trap. Peptides were filtered for Mascot score ≤20. Protein identifications were based on at least two peptides.
Cell and tissue immunostaining
Drosophila S2 cells grown on cover slips, dissected ring glands and imaginal discs were processed as described [65] and stained overnight at 4°C (tissues) or 2 h at room temperature (S2 cells) with the following antibodies: rat anti-EcdNterm (1∶500, this study), guinea pig anti-Spok (1∶1000) and rabbit anti-Phm (1∶300) [33], rabbit anti-cleaved Caspase-3 (1∶500, ASP175, Cell Signaling, #9661), rabbit anti-pH3 (1∶100, Cell Signaling, #9701), mouse anti-Flag M2 (1∶500, Sigma Aldrich), rabbit anti-c-Myc (1∶500, sc-789, Santa Cruz), and mouse anti-Lamin (1∶500, ADL67.10), mouse anti-EcR (1∶200, DDA2.7), and mouse anti-Fasciclin III (1∶300); the latter three from the Developmental Studies Hybridoma Bank (DSHB, Iowa). After washing, samples were incubated with corresponding secondary antibodies coupled to Cy3 or Cy5 (Jackson ImmunoResearch), counterstained with DAPI (0.5 µg/ml, Invitrogen) to visualize nuclei, and mounted in Dabco:Mowiol (Sigma-Aldrich).
Image acquisition and processing
Confocal single sections and stacks were acquired at room temperature with Olympus FV1000 confocal microscope. Maximum projections were generated using Fluoview 2.1c Software (Olympus) and ImageJ [66]. Final image processing including panel assembly, brightness and contrast adjustment were done in Photoshop CS5.1 (Adobe Systems, Inc.). To allow comparison among genotypes, images were taken and processed with the same settings. Adult wings were mounted in Hoyer's medium. Images of larvae, flies, adult wings, and Z-stacks of adult eyes were taken using Leica M165 FC fluorescent stereomicroscope equipped with a DFC490 CCD camera. Images were processed using a Multifocus module of the LAS 3.7.0 software (Leica).
qRT-PCR
Total RNA was isolated from eight third-instar larvae with Isol-RNA Lysis Reagent (5 Prime). cDNA was synthesized from 2 µg of RNA treated with DNase I (Ambion) using random primers and Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA). PCR was performed in triplicates with the SYBR green mix (Bio-Rad, Hercules, CA) using the CFX96 (Bio-Rad, Hercules, CA) or the 7900HT (Applied Biosystems) real-time PCR systems. qRT-PCR primers (Table S1) were designed to anneal at 62°C. All primers were initially tested by qPCR on serially diluted DNA templates and those deviating from typical standard curves were redesigned. Data were normalized to rp49 transcript levels and fold changes were calculated using the ΔΔCT method [67] or the Relative standard curve method [68]. At least four biological replicates were analyzed per each experiment.
Hormone feeding and ecdysteroid measurement
For hormone feeding, 40 larvae aged 5 days AEL were transferred per vial on food containing 100 µg/ml of 20E. Control food contained solvent only (6% w/w ethanol). Total RNA was isolated after 4 h (for ecd1) or one day of feeding (6 days AEL for phm-Gal4 experiments); wandering behavior was photographed on day 7 AEL. To measure ecdysteroid titer, larvae 6 days AEL (15 animals per assay) were homogenized in 500 µl of methanol and centrifuged at 20,000 g. The pellets were re-extracted with 300 µl methanol and supernatants pooled and vacuum dried. The dried extracts were processed using the EIA immunoassay system (Cayman Chemical) as described [69]. Ecdysteroid concentration was calculated from an eight-point standard curve of serially diluted 20E.
Statistical analyses
An unpaired two-tailed Student's t-test with unequal variation and one-way analysis of variance (ANOVA) with a post hoc Newman-Keuls Multiple Comparison Test were used to determine statistical significance for changes in gene expression in all qRT-PCR experiments, larval size, and ecdysteroid measurements.
Supporting Information
Zdroje
1. ThummelCS (2001) Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell 1 : 453–465.
2. MirthCK, RiddifordLM (2007) Size assessment and growth control: how adult size is determined in insects. Bioessays 29 : 344–355.
3. YamanakaN, RewitzKF, O'ConnorMB (2013) Ecdysone control of developmental transitions: lessons from Drosophila research. Annu Rev Entomol 58 : 497–516.
4. OuQ, King-JonesK (2013) What goes up must come down: transcription factors have their say in making ecdysone pulses. Curr Top Dev Biol 103 : 35–71.
5. RewitzKF, YamanakaN, O'ConnorMB (2013) Developmental checkpoints and feedback circuits time insect maturation. Curr Top Dev Biol 103 : 1–33.
6. Yoshiyama-YanagawaT, EnyaS, Shimada-NiwaY, YaguchiS, HaramotoY, et al. (2011) The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol-metabolizing enzyme. J Biol Chem 286 : 25756–25762.
7. NiwaR, NamikiT, ItoK, Shimada-NiwaY, KiuchiM, et al. (2010) Non-molting glossy/shroud encodes a short-chain dehydrogenase/reductase that functions in the “Black Box” of the ecdysteroid biosynthesis pathway. Development 137 : 1991–1999.
8. HuangX, WarrenJT, GilbertLI (2008) New players in the regulation of ecdysone biosynthesis. J Genet Genomics 35 : 1–10.
9. IgaM, KataokaH (2012) Recent studies on insect hormone metabolic pathways mediated by cytochrome P450 enzymes. Biol Pharm Bull 35 : 838–843.
10. PetrykA, WarrenJT, MarquésG, JarchoMP, GilbertLI, et al. (2003) Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc Natl Acad Sci USA 100 : 13773–13778.
11. GarenA, KauvarL, LepesantJA (1977) Roles of ecdysone in Drosophila development. Proc Natl Acad Sci USA 74 : 5099–5103.
12. HenrichVC, TuckerRL, MaroniG, GilbertLI (1987) The ecdysoneless (ecd1ts) mutation disrupts ecdysteroid synthesis autonomously in the ring gland of Drosophila melanogaster. Dev Biol 120 : 50–55.
13. GaziovaI, BonnettePC, HenrichVC, JindraM (2004) Cell-autonomous roles of the ecdysoneless gene in Drosophila development and oogenesis. Development 131 : 2715–2725.
14. KimJH, GurumurthyCB, NaramuraM, ZhangY, DudleyAT, et al. (2009) Role of mammalian Ecdysoneless in cell cycle regulation. J Biol Chem 284 : 26402–26410.
15. ZhaoX, MirzaS, AlshareedaA, ZhangY, GurumurthyCB, et al. (2012) Overexpression of a novel cell cycle regulator ecdysoneless in breast cancer: a marker of poor prognosis in HER2/neu-overexpressing breast cancer patients. Breast Cancer Res Treat 134 : 171–180.
16. DeyP, RachaganiS, ChakrabortyS, SinghPK, ZhaoX, et al. (2012) Overexpression of ecdysoneless in pancreatic cancer and its role in oncogenesis by regulating glycolysis. Clin Cancer Res 18 : 6188–6198.
17. GuruharshaKG, RualJ-F, ZhaiB, MintserisJ, VaidyaP, et al. (2011) A protein complex network of Drosophila melanogaster. Cell 147 : 690–703.
18. GraingerRJ, BeggsJD (2005) Prp8 protein: at the heart of the spliceosome. RNA 11 : 533–557.
19. HavugimanaPC, HartGT, NepuszT, YangH, TurinskyAL, et al. (2012) A census of human soluble protein complexes. Cell 150 : 1068–1081.
20. ManiatisT, ReedR (2002) An extensive network of coupling among gene expression machines. Nature 416 : 499–506.
21. JuricaMS, MooreMJ (2003) Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 12 : 5–14.
22. WahlMC, WillCL, LührmannR (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136 : 701–718.
23. Will CL, Lührmann R (2011) Spliceosome structure and function. Cold Spring Harb Perspect Biol 3 ((7)) . pii: a003707.
24. AndersenDS, TaponN (2008) Drosophila MFAP1 is required for pre-mRNA processing and G2/M progression. J Biol Chem 283 : 31256–31267.
25. ChenP-H, LeeC-I, WengY-T, TarnW-Y, TsaoY-P, et al. (2013) BCAS2 is essential for Drosophila viability and functions in pre-mRNA splicing. RNA 19 : 208–218.
26. CoelhoCMA, KolevskiB, WalkerCD, LavagiI, ShawT, et al. (2005) A genetic screen for dominant modifiers of a small-wing phenotype in Drosophila melanogaster identifies proteins involved in splicing and translation. Genetics 171 : 597–614.
27. RayP, LuoX, RaoEJ, BashaA, WoodruffEA, et al. (2010) The splicing factor Prp31 is essential for photoreceptor development in Drosophila. Protein Cell 1 : 267–274.
28. HeroldN, WillCL, WolfE, KastnerB, UrlaubH, et al. (2009) Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol 29 : 281–301.
29. BoonK-L, GraingerRJ, EhsaniP, BarrassJD, AuchynnikavaT, et al. (2007) prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat Struct Mol Biol 14 : 1077–1083.
30. WeberG, CristãoVF, de L AlvesF, SantosKF, HoltonN, et al. (2011) Mechanism for Aar2p function as a U5 snRNP assembly factor. Genes Dev 25 : 1601–1612.
31. KimJH, GurumurthyCB, BandH, BandV (2010) Biochemical characterization of human Ecdysoneless reveals a role in transcriptional regulation. Biol Chem 391 : 9–19.
32. LeeT, LuoL (2001) Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24 : 251–254.
33. OnoH, RewitzKF, ShinodaT, ItoyamaK, PetrykA, et al. (2006) Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev Biol 298 : 555–570.
34. WarrenJT, O'ConnorMB, GilbertLI (2009) Studies on the Black Box: incorporation of 3-oxo-7-dehydrocholesterol into ecdysteroids by Drosophila melanogaster and Manduca sexta. Insect Biochem Mol Biol 39 : 677–687.
35. ListermanI, SapraAK, NeugebauerKM (2006) Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13 : 815–822.
36. PleissJA, WhitworthGB, BergkesselM, GuthrieC (2007) Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol 5: e90.
37. StanekD, Pridalová-HnilicováJ, NovotnýI, HuranováM, BlazíkováM, et al. (2008) Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol Biol Cell 19 : 2534–2543.
38. McBrayerZ, OnoH, ShimellM, ParvyJ-P, BecksteadRB, et al. (2007) Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev Cell 13 : 857–871.
39. GibbensYY, WarrenJT, GilbertLI, O'ConnorMB (2011) Neuroendocrine regulation of Drosophila metamorphosis requires TGFβ/Activin signaling. Development 138 : 2693–2703.
40. WarrenJT, BachmannJS, DaiJD, GilbertLI (1996) Differential incorporation of cholesterol and cholesterol derivatives into ecdysteroids by the larval ring glands and adult ovaries of Drosophila melanogaster: a putative explanation for the l(3)ecd1 mutation. Insect Biochem Mol Biol 26 : 931–943.
41. RedfernCPF, BownesM (1983) Pleiotropic effects of the “ecdysoneless-1” mutation of Drosophila melanogaster. Mol Gen Genet 189 : 432–440.
42. SliterTJ (1989) Imaginal disc-autonomous expression of a defect in sensory bristle patterning caused by the lethal(3)ecdysoneless1 (1(3)ecd1) mutation of Drosophila melanogaster. Development 106 : 347–354.
43. DavisMB, LiT (2013) Genomic analysis of the ecdysone steroid signal at metamorphosis onset using ecdysoneless and EcRnull Drosophila melanogaster mutants. Genes Genomics 35 : 21–46.
44. FongYW, ZhouQ (2001) Stimulatory effect of splicing factors on transcriptional elongation. Nature 414 : 929–933.
45. BiebersteinNI, Carrillo OesterreichF, StraubeK, NeugebauerKM (2012) First exon length controls active chromatin signatures and transcription. Cell Rep 2 : 62–68.
46. FaustinoNA, CooperTA (2003) Pre-mRNA splicing and human disease. Genes Dev 17 : 419–437.
47. ParkJW, PariskyK, CelottoAM, ReenanRA, GraveleyBR (2004) Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA 101 : 15974–15979.
48. MordesD, LuoX, KarA, KuoD, XuL, et al. (2006) Pre-mRNA splicing and retinitis pigmentosa. Mol Vis 12 : 1259–1271.
49. MoellerME, DanielsenET, HerderR, O'ConnorMB, RewitzKF (2013) Dynamic feedback circuits function as a switch for shaping a maturation – inducing steroid pulse in Drosophila. Development 140 : 4730–4739.
50. HayBA, MaileR, RubinGM (1997) P element insertion-dependent gene activation in the Drosophila eye. Proc Natl Acad Sci USA 94 : 5195–5200.
51. DuW, XieJE, DysonN (1996) Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J 15 : 3684–3692.
52. DuronioRJ, O'FarrellPH (1995) Developmental control of the G1 to S transition in Drosophila: cyclin E is a limiting downstream target of E2F. Genes Dev 9 : 1456–1468.
53. DatarSA, JacobsHW, la Cruz deAF, LehnerCF, EdgarBA (2000) The Drosophila cyclin D-Cdk4 complex promotes cellular growth. EMBO J 19 : 4543–4554.
54. KittlerR, PutzG, PelletierL, PoserI, HeningerA-K, et al. (2004) An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432 : 1036–1040.
55. WoodV, GwilliamR, RajandreamM-A, LyneM, LyneR, et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415 : 871–880.
56. KäuferNF, PotashkinJ (2000) Analysis of the splicing machinery in fission yeast: a comparison with budding yeast and mammals. Nucleic Acids Res 28 : 3003–3010.
57. KainouT, ShinzatoT, SasakiK, MitsuiY, Giga-HamaY, et al. (2006) Spsgt1, a new essential gene of Schizosaccharomyces pombe, is involved in carbohydrate metabolism. Yeast 23 : 35–53.
58. TakaesuNT, JohnsonAN, NewfeldSJ (2002) Posterior spiracle specific GAL4 lines: new reagents for developmental biology and respiratory physiology. Genesis 34 : 16–18.
59. UhlirovaM, JasperH, BohmannD (2005) Non-cell-autonomous induction of tissue overgrowth by JNK/Ras cooperation in a Drosophila tumor model. Proc Natl Acad Sci USA 102 : 13123–13128.
60. StruhlG, BaslerK (1993) Organizing activity of wingless protein in Drosophila. Cell 72 : 527–540.
61. Sik LeeY, CarthewRW (2003) Making a better RNAi vector for Drosophila: use of intron spacers. Methods 30 : 322–329.
62. SteinfeldtT, Könen-WaismanS, TongL, PawlowskiN, LamkemeyerT, et al. (2010) Phosphorylation of mouse immunity-related GTPase (IRG) resistance proteins is an evasion strategy for virulent Toxoplasma gondii. PLoS Biol 8: e1000576.
63. RappsilberJ, MannM, IshihamaY (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2 : 1896–1906.
64. PerkinsDN, PappinDJ, CreasyDM, CottrellJS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20 : 3551–3567.
65. KülshammerE, UhlirovaM (2013) The actin cross-linker Filamin/Cheerio mediates tumor malignancy downstream of JNK signaling. J Cell Sci 126 : 927–938.
66. AbràmoffMD, MagalhãesPJ, RamSJ (2004) Image processing with ImageJ. Biophotonics International 11 : 36–42.
67. LivakKJ, SchmittgenTD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) Method. Methods 25 : 402–408.
68. LarionovA, KrauseA, MillerW (2005) A standard curve based method for relative real time PCR data processing. BMC Bioinformatics 6 : 62.
69. OuQ, MagicoA, King-JonesK (2011) Nuclear receptor DHR4 controls the timing of steroid hormone pulses during Drosophila development. PLoS Biol 9: e1001160.
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 4- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- The Challenges of Mitochondrial Replacement
- Concocting Cholinergy
- Genome-Wide Diet-Gene Interaction Analyses for Risk of Colorectal Cancer
- Statistical Power to Detect Genetic (Co)Variance of Complex Traits Using SNP Data in Unrelated Samples
- Mouse Pulmonary Adenoma Susceptibility 1 Locus Is an Expression QTL Modulating -4A
- Transcription-Associated R-Loop Formation across the Human CGG-Repeat Region
- Epigenetic Regulation by Heritable RNA
- Protein Quantitative Trait Loci Identify Novel Candidates Modulating Cellular Response to Chemotherapy
- Genome-Wide Profiling of Yeast DNA:RNA Hybrid Prone Sites with DRIP-Chip
- The Mechanism of Gene Targeting in Human Somatic Cells
- A LINE-1 Insertion in DLX6 Is Responsible for Cleft Palate and Mandibular Abnormalities in a Canine Model of Pierre Robin Sequence
- Interaction between Two Timing MicroRNAs Controls Trichome Distribution in
- DNA Glycosylases Involved in Base Excision Repair May Be Associated with Cancer Risk in and Mutation Carriers
- The Myc-Mondo/Mad Complexes Integrate Diverse Longevity Signals
- Evolutionarily Diverged Regulation of X-chromosomal Genes as a Primal Event in Mouse Reproductive Isolation
- Mutations in Conserved Residues of the microRNA Argonaute ALG-1 Identify Separable Functions in ALG-1 miRISC Loading and Target Repression
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Isl1 Directly Controls a Cholinergic Neuronal Identity in the Developing Forebrain and Spinal Cord by Forming Cell Type-Specific Complexes
- A Synthetic Community Approach Reveals Plant Genotypes Affecting the Phyllosphere Microbiota
- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- Determining the Control Circuitry of Redox Metabolism at the Genome-Scale
- DNA Repair Pathway Selection Caused by Defects in , , and Telomere Addition Generates Specific Chromosomal Rearrangement Signatures
- Methylome Diversification through Changes in DNA Methyltransferase Sequence Specificity
- Folliculin Regulates Ampk-Dependent Autophagy and Metabolic Stress Survival
- Fine Mapping of Dominant -Linked Incompatibility Alleles in Hybrids
- Unexpected Role of the Steroid-Deficiency Protein Ecdysoneless in Pre-mRNA Splicing
- Three Groups of Transposable Elements with Contrasting Copy Number Dynamics and Host Responses in the Maize ( ssp. ) Genome
- Sox5 Functions as a Fate Switch in Medaka Pigment Cell Development
- Synergistic Interactions between the Molecular and Neuronal Circadian Networks Drive Robust Behavioral Circadian Rhythms in
- Chromatin Landscapes of Retroviral and Transposon Integration Profiles
- Widespread Use of Non-productive Alternative Splice Sites in
- Ras GTPase-Like Protein MglA, a Controller of Bacterial Social-Motility in Myxobacteria, Has Evolved to Control Bacterial Predation by
- Cell Type-Specific Functions of Genes Revealed by Novel Adipocyte and Hepatocyte Circadian Clock Models
- Phenotype Ontologies and Cross-Species Analysis for Translational Research
- Embryogenesis Scales Uniformly across Temperature in Developmentally Diverse Species
- In Pursuit of the Gene: An Interview with James Schwartz
- Molecular Mechanisms of Hypoxic Responses via Unique Roles of Ras1, Cdc24 and Ptp3 in a Human Fungal Pathogen
- Analysis of the Genome and Transcriptome of var. Reveals Complex RNA Expression and Microevolution Leading to Virulence Attenuation
- Genotypic and Functional Impact of HIV-1 Adaptation to Its Host Population during the North American Epidemic
- RNA Editome in Rhesus Macaque Shaped by Purifying Selection
- Proper Actin Ring Formation and Septum Constriction Requires Coordinated Regulation of SIN and MOR Pathways through the Germinal Centre Kinase MST-1
- Interplay of the Serine/Threonine-Kinase StkP and the Paralogs DivIVA and GpsB in Pneumococcal Cell Elongation and Division
- A Quality Control Mechanism Coordinates Meiotic Prophase Events to Promote Crossover Assurance
- CNNM2 Mutations Cause Impaired Brain Development and Seizures in Patients with Hypomagnesemia
- The RNA-Binding Protein QKI Suppresses Cancer-Associated Aberrant Splicing
- Uncoupling Transcription from Covalent Histone Modification
- Rad51–Rad52 Mediated Maintenance of Centromeric Chromatin in
- FRA2A Is a CGG Repeat Expansion Associated with Silencing of
- A General Approach for Haplotype Phasing across the Full Spectrum of Relatedness
- A Novel Highly Divergent Protein Family Identified from a Viviparous Insect by RNA-seq Analysis: A Potential Target for Tsetse Fly-Specific Abortifacients
- A Central Role for in Regulation of Islet Function in Man
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Sequence-Specific Transcription Factor c-Jun Targets Cockayne Syndrome Protein B to Regulate Transcription and Chromatin Structure
- The Mechanism of Gene Targeting in Human Somatic Cells
- Genetic Predisposition to In Situ and Invasive Lobular Carcinoma of the Breast
- Widespread Use of Non-productive Alternative Splice Sites in
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



