-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Genetic Determinants of Long-Term Changes in Blood Lipid Concentrations: 10-Year Follow-Up of the GLACIER Study
Although large cross-sectional studies have proven highly successful in identifying gene variants related to lipid levels and other cardiometabolic traits, very few examples of well-designed longitudinal studies exist where associations between genotypes and long-term changes in lipids have been assessed. Here we undertook analyses in the GLACIER Study to determine whether the 157 previously identified lipid-associated genes variants associate with changes in blood lipid levels over 10-yr follow-up. We identified a variant in APOE that is robustly associated with total cholesterol change and two variants in TRIB1 and APOA1 respectively that are robustly associated with triglyceride change. We replicated these findings in a second Swedish cohort (the MDC Study). The identified genes had previously been associated with cardiovascular traits such as myocardial infarction or coronary heart disease; hence, these novel lipid associations provide additional insight into the pathogenesis of atherosclerotic heart and large vessel disease. By incorporating all 157 established variants into gene scores, we also observed strong associations with 10-yr lipid changes, illustrating the polygenic nature of blood lipid deterioration.
Published in the journal: Genetic Determinants of Long-Term Changes in Blood Lipid Concentrations: 10-Year Follow-Up of the GLACIER Study. PLoS Genet 10(6): e32767. doi:10.1371/journal.pgen.1004388
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004388Summary
Although large cross-sectional studies have proven highly successful in identifying gene variants related to lipid levels and other cardiometabolic traits, very few examples of well-designed longitudinal studies exist where associations between genotypes and long-term changes in lipids have been assessed. Here we undertook analyses in the GLACIER Study to determine whether the 157 previously identified lipid-associated genes variants associate with changes in blood lipid levels over 10-yr follow-up. We identified a variant in APOE that is robustly associated with total cholesterol change and two variants in TRIB1 and APOA1 respectively that are robustly associated with triglyceride change. We replicated these findings in a second Swedish cohort (the MDC Study). The identified genes had previously been associated with cardiovascular traits such as myocardial infarction or coronary heart disease; hence, these novel lipid associations provide additional insight into the pathogenesis of atherosclerotic heart and large vessel disease. By incorporating all 157 established variants into gene scores, we also observed strong associations with 10-yr lipid changes, illustrating the polygenic nature of blood lipid deterioration.
Introduction
The implementation of genome-wide association studies (GWAS) into large, well-characterized cohort collections has spurred the discovery of hundreds of genetic variants for complex cardiometabolic disorders [1]. Of those variants, many have been for blood lipids, with a total of 164 common single nucleotide polymorphisms (SNPs) identified to date at a genome-wide significance level (P≤5×10−8) [2], [3]. These findings come from large-scale, cross-sectional meta-analyses with sufficient power to detect variants with very small effect-sizes for the corresponding traits (OR≈1.01). Although demonstrating cross-sectional genetic associations is important (e.g., for elucidating biological pathways), from a clinical perspective, the discovery of genetic variants that predict a worsening of lipid levels over time might be more relevant [4]; to our knowledge, no large prospective cohort study focused on the full spectrum of established lipid loci has yet been performed.
The purpose of this study was to examine the predictive ability of 157 established lipid loci (as defined by 164 SNPs), singly and together (genetic risk score (GRS)), on changes in lipid concentrations over a decade of follow-up. Replication analyses in another Swedish cohort were also performed.
Results
GLACIER Study participant characteristics are shown in Table 1 (baseline only) and Table 2 (longitudinal subset). The cross-sectional MetaboChip genotype data from the GLACIER Study were combined with many other cohorts in one of the prior lipid meta-analyses [3]. Malmö Diet and Cancer (MDC) Study participant characteristics are shown in Table 3.
Tab. 1. Baseline characteristics of the GLACIER Study participants (N = 5,862). 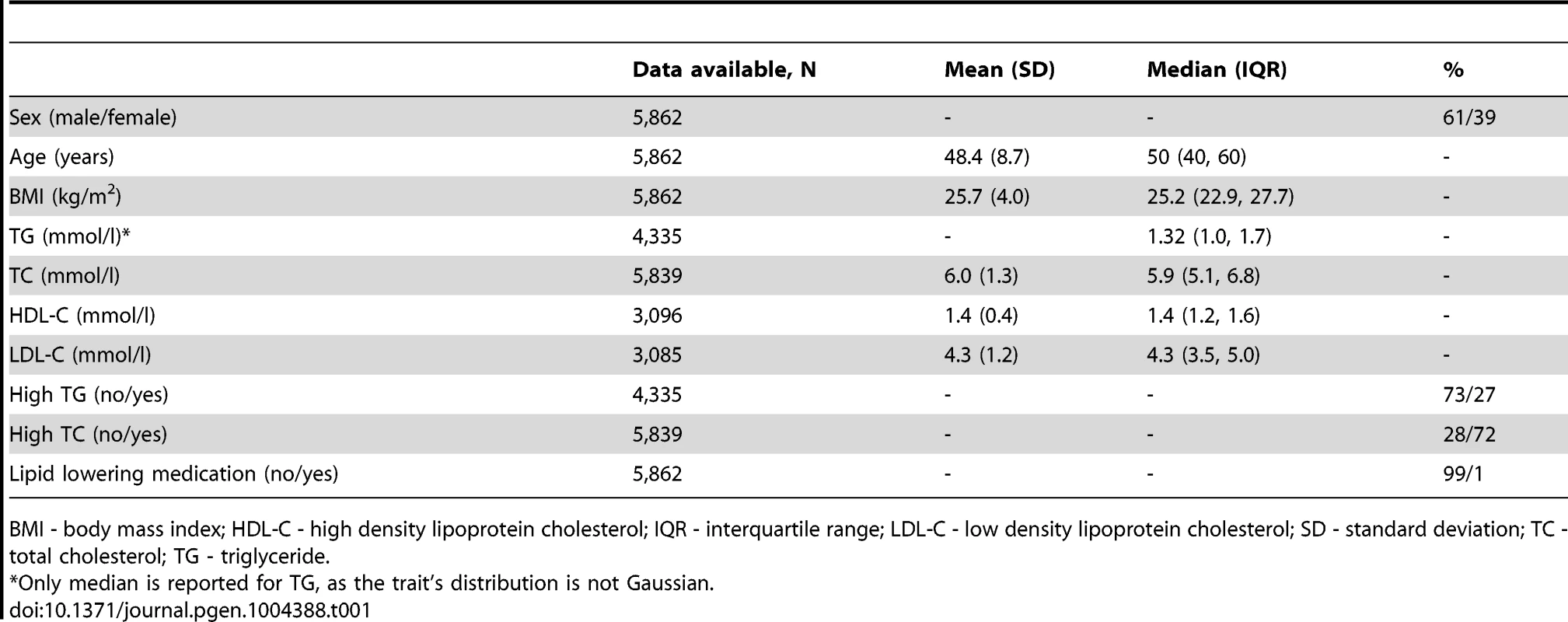
BMI - body mass index; HDL-C - high density lipoprotein cholesterol; IQR - interquartile range; LDL-C - low density lipoprotein cholesterol; SD - standard deviation; TC - total cholesterol; TG - triglyceride. Tab. 2. Longitudinal characteristics of the GLACIER Study participants (N = 3,495 for TC; N = 2,211 for TG). 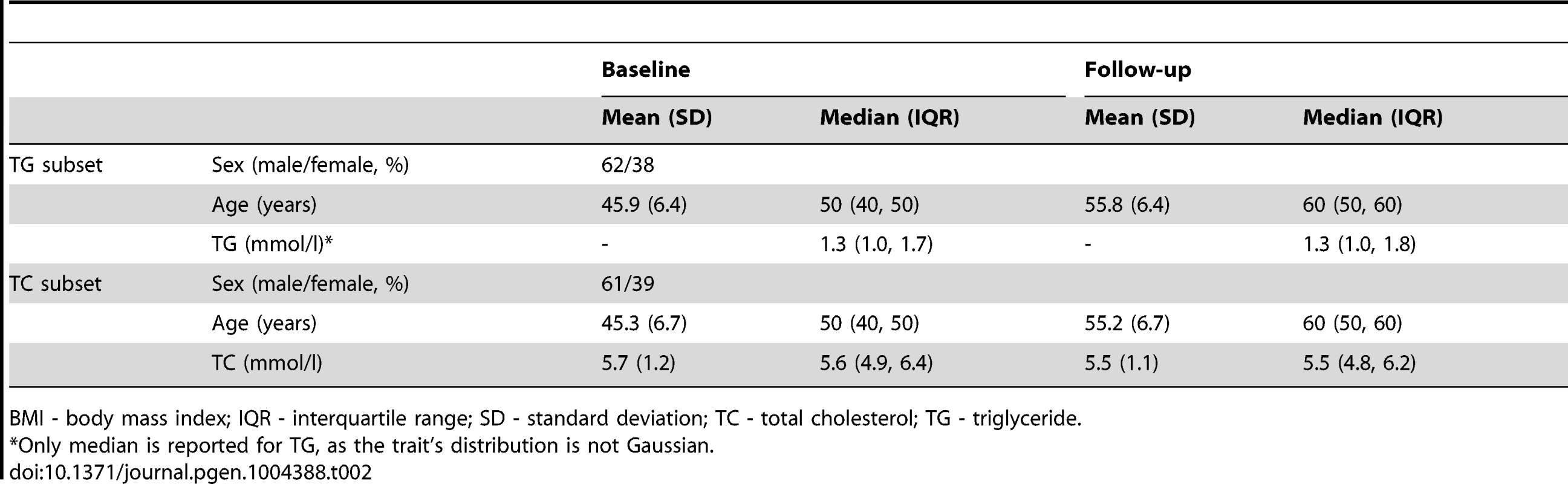
BMI - body mass index; IQR - interquartile range; SD - standard deviation; TC - total cholesterol; TG - triglyceride. Tab. 3. Longitudinal characteristics of the MDC Study participants (N = 2,943). 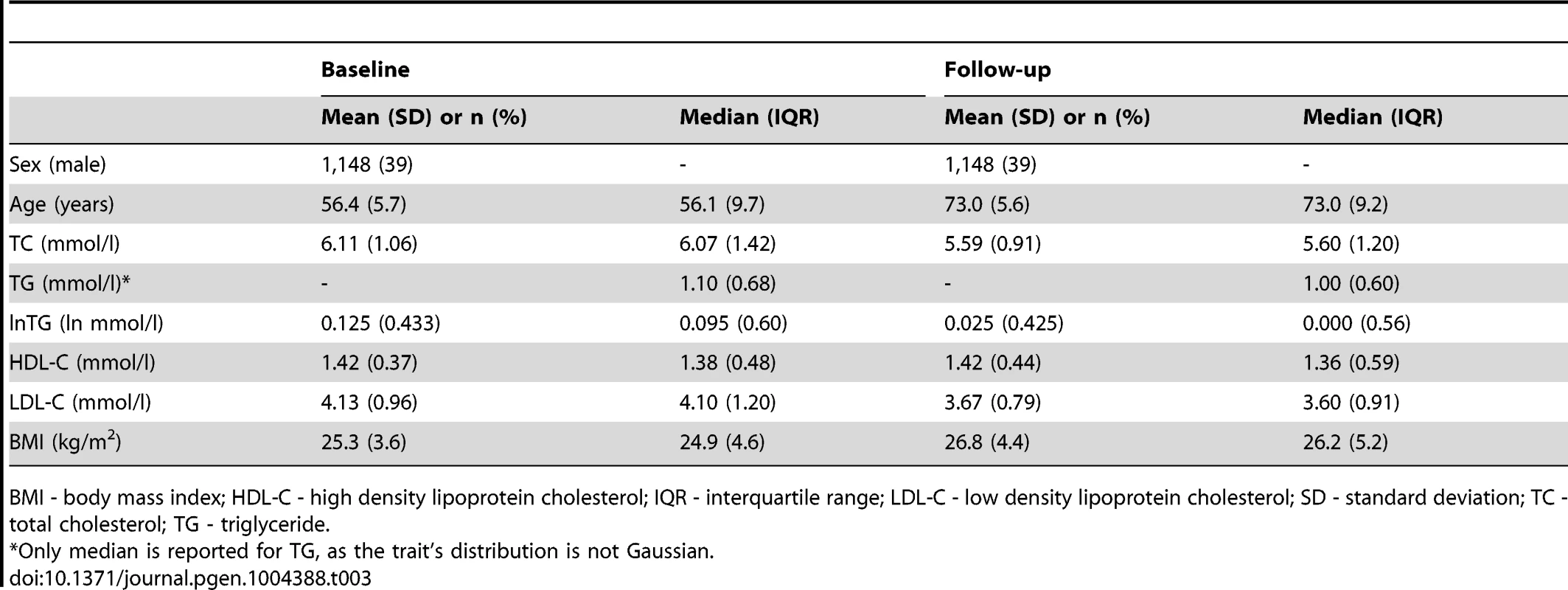
BMI - body mass index; HDL-C - high density lipoprotein cholesterol; IQR - interquartile range; LDL-C - low density lipoprotein cholesterol; SD - standard deviation; TC - total cholesterol; TG - triglyceride. The results for cross-sectional analyses in the GLACIER Study are presented in Text S1. The established SNPs explained 8.8%, 4.9%, 9.1% and 4.8% variance for TC, TG, LDL-C and HDL-C, respectively. The weighted GRS (wGRS) (allele counts multiplied by previously published effect sizes for each SNP) explained 7.0%, 3.9%, 6.9% and 2.6% of the variance in TC, TG, LDL-C and HDL-C, respectively.
Longitudinal analyses
A statistically significant overall decrease in plasma TC concentrations between the baseline and follow-up visits (mean change = −0.18±1.12 mmol/l; P<0.0001), but no change in the TG levels (mean change = 0.02±1 mmol/l; P = 0.32), was observed.
In individual SNP analysis, Benjamini-Hochberg false discovery rate (FDR) corrected statistically significant associations were observed for the rs6589564 and ΔTG (β = 0.31 mmol/l per allele per decade follow-up, 95% CI: 0.21, 0.41, SE = 0.05, PFDR = 6.6×10−7), rs2954029 and ΔTG (β = 0.09 mmol/l per allele per decade follow-up, 95% CI: 0.03, 0.15, SE = 0.03, PFDR = 0.009) and rs4420638 and ΔTC (β = 0.12 mmol/l per allele per decade follow-up, 95% CI: 0.06, 0.18, SE = 0.03, PFDR = 0.002). One additional SNP (rs2131925) showed nominally significant evidence of association with ΔTC (β = 0.07 mmol/l per allele per decade follow-up, 95% CI: 0.03, 0.11, SE = 0.02, P = 0.002, PFDR = 0.083). Seven and five additional SNPs were also nominally statistically associated with ΔTC and ΔTG, respectively (P<0.05), but did not survive multiple-test corrections. Nominally significant SNP associations are shown in Table 4 and all longitudinal SNP associations are reported in Table S1.
Tab. 4. Nominally significant SNPs from the longitudinal models in the GLACIER Study (N = 3,495 for ΔTC; N = 2,211 for ΔTG). 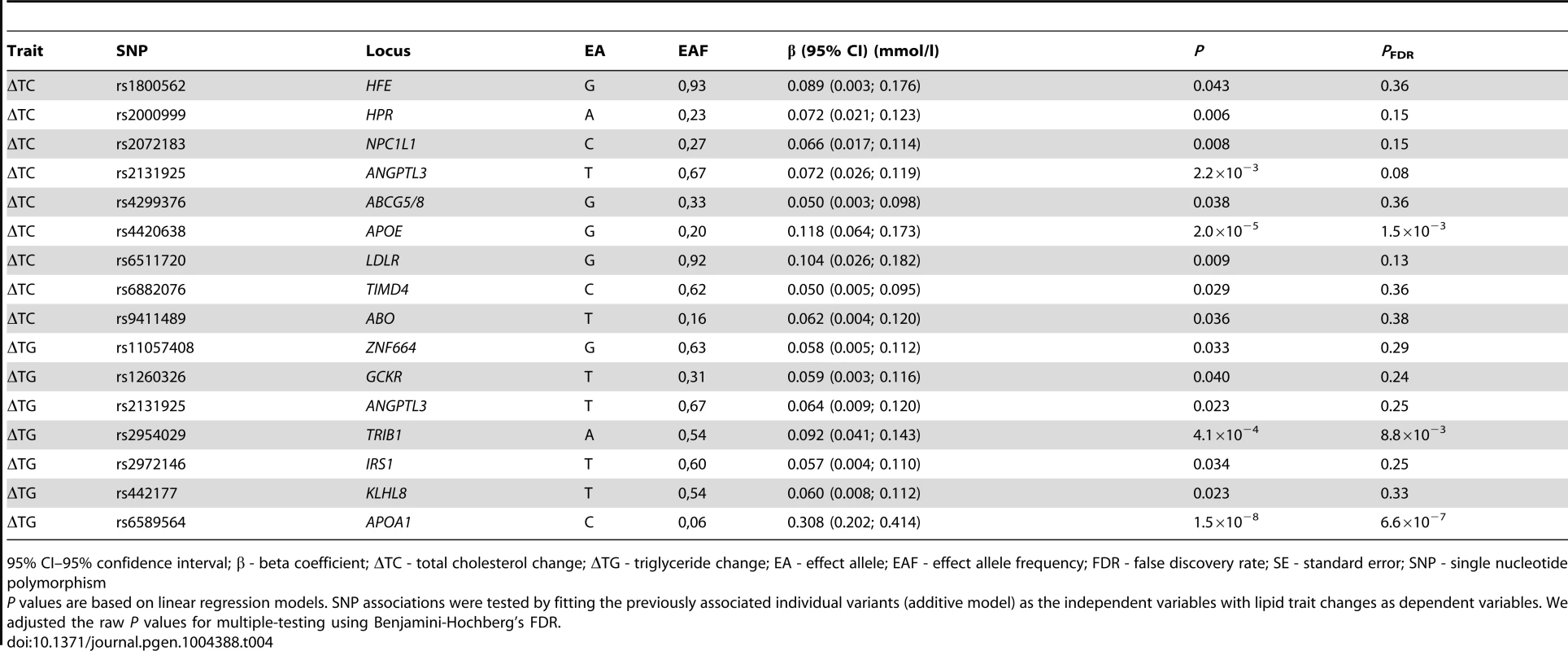
95% CI–95% confidence interval; β - beta coefficient; ΔTC - total cholesterol change; ΔTG - triglyceride change; EA - effect allele; EAF - effect allele frequency; FDR - false discovery rate; SE - standard error; SNP - single nucleotide polymorphism The GRSs were strongly associated with their corresponding trait (β = 0.02 mmol/l per allele per decade follow-up, 95% CI: 0.01, 0.03, SE = 0.003, P = 2.0×10−11 for ΔTC; β = 0.02 mmol/l per allele per decade follow-up, 95% CI: 0.01, 0.03, SE = 0.005, P = 0.0005 for ΔTG). Using the wGRS increased the strength and magnitude of the associations for both traits (β = 0.02 mmol/l per allele per decade follow-up, 95% CI: 0.01, 0.03, SE = 0.003, P = 9.8×10−18 for ΔTC; β = 0.03 mmol/l per allele per decade follow-up, 95% CI: 0.02, 0.04, SE = 0.005, P = 6.5×10−11 for ΔTG) (Figure 1A–B). The difference between the highest and lowest quartiles of the wGRS was 0.037 mmol/l for ΔTC, and 0.032 mmol/l for ΔTG.
Fig. 1. TC and TG level changes (95% CI) over 10-yr follow-up by wGRS quartiles. 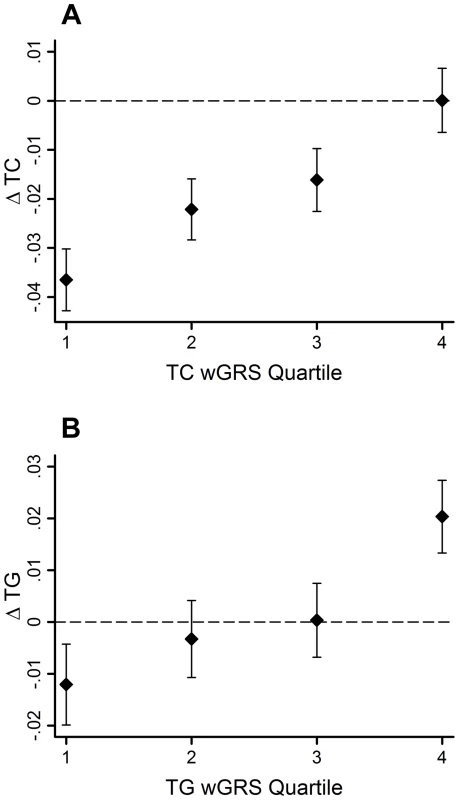
The TC wGRS was robustly associated with TC changes over follow-up (β = 0.02 mmol/l per allele per follow-up, 95% CI = 0.01, 0.03, SE = 0.003, P = 9.8*10−18) (A). The TG wGRS was robustly associated with TG changes over follow-up (β = 0.03 mmol/l per allele per follow-up, 95% CI = 0.02, 0.04, SE = 0.005, P = 6.5*10−11) (B). The variance in lipid changes explained by the wGRS, the baseline lipid measure, sex, age and age2 were 33% and 25% for TC and TG, respectively. However, the wGRSs alone explained a small fraction of these proportions (<0.05% for both traits).
To compare the predictive accuracy of traditional risk factors, genetic factors and combined models in relation to hyperlipidemia at follow-up, receiver operating characteristics area under the curve (ROC AUC) analyses were performed. The specificities of the predictive models were above 95%, while the sensitivities of the models were below 20%. The ROC AUC curves are shown in Figure 2A–B, and the pairwise differences and classification statistics in the models for high TC and high TG are shown in Table 5. The lowest ROC AUC values were obtained for the basic models including only age, age2, sex and BMI (62% and 65% for high TC and high TG, respectively) and the highest for the combined genetic-lifestyle models (66% and 67% for high TC and high TG, respectively). The difference between these two models was statistically significant for high TC (P = 0.011) and approached nominal statistical significance for high TG (P = 0.052).
Fig. 2. ROC AUC for high TC (A) and high TG (B) at follow-up. 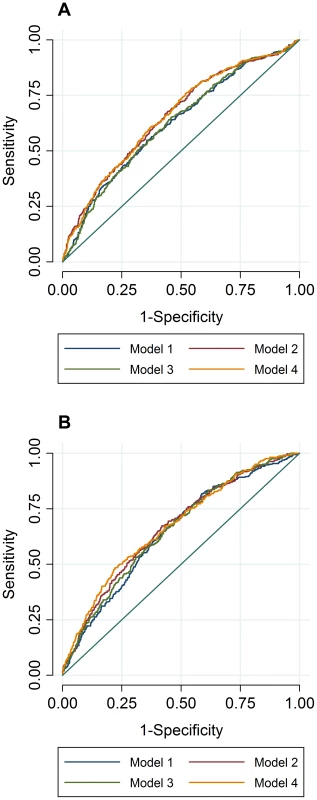
In ROC analyses we excluded individuals with hyperlipidemia at baseline and compared the predictive accuracy of four models (age, sex and BMI (Model 1), Model 1 + trait specific wGRS (Model 2), Model 1 + traditional risk factors for hyperlipidemia (Model 3) and M1 + trait specific GRS + traditional risk factors for hyperlipidemia (Model 4)) in relation to hyperlipidemia at follow-up. Tab. 5. Pairwise differences between ROC AUC curves and classification statistics in relation to hyperlipidemia in GLACIER (N = 1,257 for TC; N = 1,660 for TG). 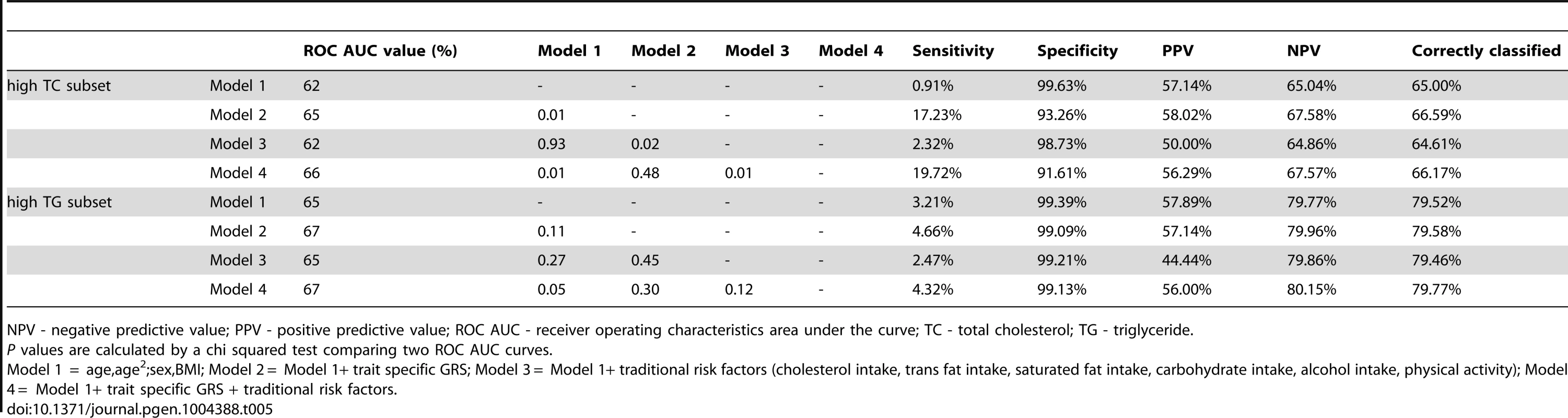
NPV - negative predictive value; PPV - positive predictive value; ROC AUC - receiver operating characteristics area under the curve; TC - total cholesterol; TG - triglyceride. Replication and meta-analysis
As described above, 15 variants (16 associations, as rs2131925 associated with both ΔTC and ΔTG) were nominally associated with change in TG or TC over 10-years follow-up in the GLACIER Study. Results of replication analyses in MDC are presented in Table 6. Associations for five SNPs (rs2131925, rs2954029, rs4420638, rs442177, rs6511720) for ΔTC and six SNPs (rs11057408, rs2072183, rs2131925, rs2954029, rs442177, rs6589564) for ΔTG were nominally statistically (P<0.05) significant and directionally consistent with GLACIER results in MDC. Furthermore, four SNPs (rs2954029, rs4420638, rs442177, rs6511720) also associated with ΔLDL-C. None of the SNPs associated with ΔHDL-C in MDC. All three previously associated variants (rs2954029 and rs6589564 in relation to ΔTG and rs4420638 for ΔTC) in GLACIER replicated in MDC.
Tab. 6. Replication of lipid associations in MDC (N = 2,943). 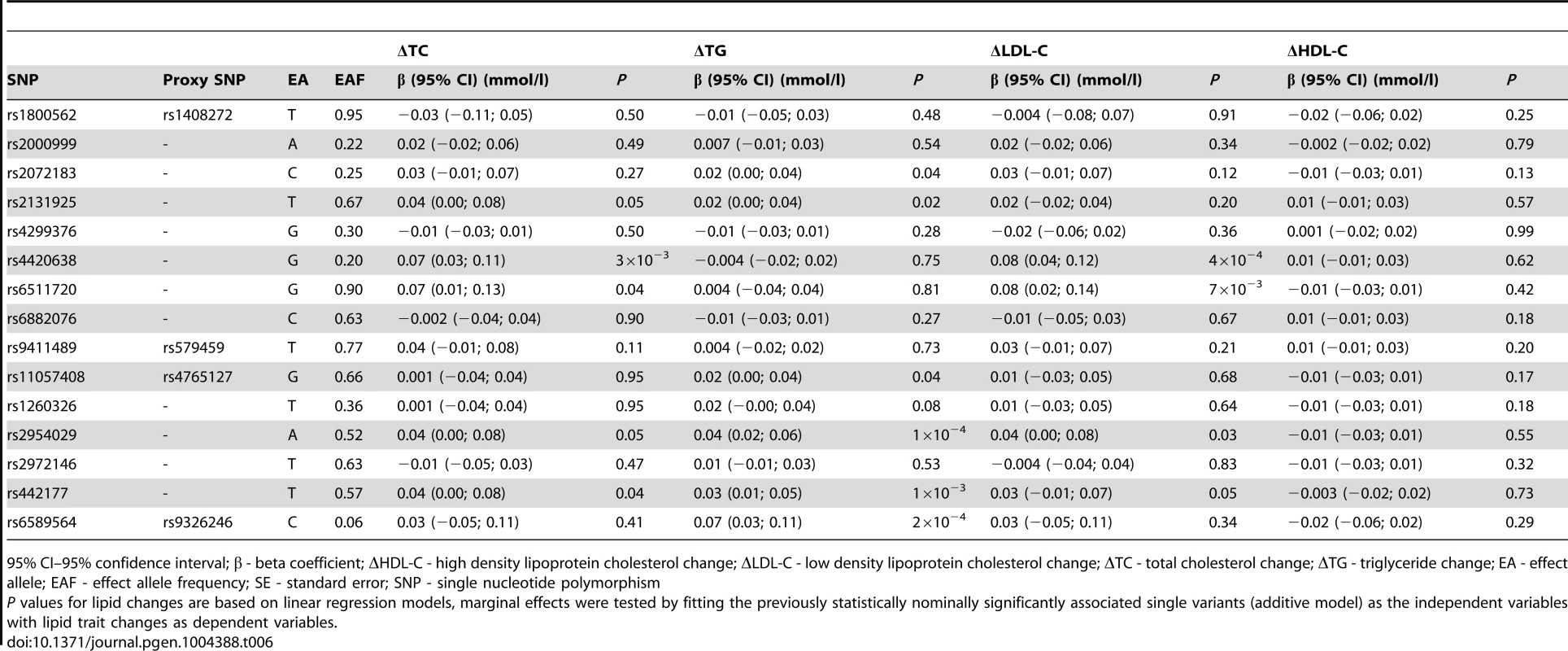
95% CI–95% confidence interval; β - beta coefficient; ΔHDL-C - high density lipoprotein cholesterol change; ΔLDL-C - low density lipoprotein cholesterol change; ΔTC - total cholesterol change; ΔTG - triglyceride change; EA - effect allele; EAF - effect allele frequency; SE - standard error; SNP - single nucleotide polymorphism Meta-analysis results for the 15 longitudinally associated variants are shown in Table S2. Three ΔTC associated variants and six ΔTG associated variants had statistically significant pooled effects (P<0.05).
Discussion
This study extends work reported in two recent large-scale cross-sectional GWAS meta-analyses for lipid loci [2], [3] by examining these variants in the setting of a prospective cohort study (10-yrs follow-up). The trait-specific GRSs were strongly associated with their corresponding lipid traits in both cross-sectional and longitudinal models. Three previously associated variants yielded statistically significant main effects in the longitudinal analyses, namely the APOA1 rs6589564 and ΔTG (PFDR = 7.3×10−7), the TRIB1 rs2954029 and ΔTG (PFDR = 0.013) and the APOE rs4420638 and ΔTC (PFDR = 0.002). We used rs6589564 as the best available proxy in our panel for the APOA1 rs964184 variant (distance = 24.8 kb; r2 = 0.688; D' = 1) [5]. Tentative evidence for association was observed for angiopoietin-like 3 (ANGPTL3) rs2131925 and ΔTC (P = 0.002, PFDR = 0.083), which given the high prior for association, likely reflects an additional locus that influences changes in lipid levels. These statistically significant associations in GLACIER were successfully replicated in MDC (P = 4.0×10−5 for APOA1 rs6589564 and ΔTG; P = 5.0×10−5 for TRIB1 rs2954029 and ΔTG; P = 0.01 for APOE rs4420638 and ΔTC; P = 0.03 for ANGPTL3 rs2131925 and ΔTC) and 9 of the 16 nominally significant associations in GLACIER remained significant after meta-analyzing the two cohorts. In ROC analyses, the combined genetic-lifestyle model had higher predictive ability than other models for both traits, but after Bonferroni correction of ROC AUC comparative P values, this difference was not statistically significant.
Two large, recent cross-sectional meta-analyses identified a total of 164 new variants associated with blood lipid levels [2], [3]. Whilst these studies highlight numerous, previously unknown biologic pathways underlying dyslipidemia, they have focused exclusively on cross-sectional data, which may not be informative of the genetic mechanisms underlying the deterioration of blood lipid profiles. Prospective data is clinically more relevant, as knowledge of loci that predict change in lipids over time may provide information for clinical translation and risk prediction [4]; however, the extent to which clinical translation could be realized depends on achieving a high level of predictive accuracy using genetic risk algorithms, which at present is not the case for common cardiometabolic diseases [6]. A small number of prospective genetic association studies for lipid loci have been reported [7]–[10], but these studies have focused on only a handful of the 157 established lipid-loci. In the present study, we show that the ability of these established lipid loci to predict incident dyslipidemia is low in these Swedish populations; adding the wGRS to the risk prediction model incorporating the conventional risk factors for hyperlipidemia (comparing Model 3 and Model 4 (shown in Table 5)) increased the AUC values by 4% and 2% for high TC and high TG, respectively. This is comparable to the 3% AUC difference for incident hypercholesterolemia reported by Lu et al., although they used an unweighted GRS of only 12 established TC variants [10].
Teslovich et al. reported ∼12% variance explained by the 95 loci discovered in their meta-analysis for TC, TG, LDL-C and HDL-C [2]. The 62 lipid loci recently discovered by Willer et al. explain an additional ∼2% of the variance per lipid trait [3]. In Lu et al. 's report, 12 candidate SNPs explained 6.9% of the variance in TC, while Sabatti et al. attributed 4.8%, 6%, and 6% of the total variance in TG, LDL-C and HDL-C to 4-11 GWAS identified SNPs [10], [11]. In cross-sectional analyses in the GLACIER Study the variances explained by the established SNPs for TC, TG, LDL-C and HDL-C were 8.8%, 4.9%, 9.1% and 4.8%, respectively.
Aulchenko et al. used GRSs comprising 7-11 lipid loci; the variances explained by these SNPs were 3.9%, 3.0%, 3.4% and 4.8% for TC, TG, LDL-C and HDL-C, respectively [12]. Lutsey et al. evaluated the explained variance by trait specific GRSs, in which they incorporated the 95 loci identified by Teslovich et al.; the explained variance for TC, TG, LDL-C and HDL-C were 6.8%, 6.0%, 6.0% and 1.6%, respectively [7]. In the GLACIER Study, the corresponding wGRSs accounted for 7.0%, 3.9%, 6.9% and 2.6% of the trait variances, respectively.
The TRIB1 locus, which harbors one of the variants (rs2954029) strongly associated with change in TG in our study, encodes a protein with a regulatory effect on mitogen-activated protein kinases (MAPKs) [13]. Studies in mice suggest that TRIB1 plays a role in the transcription of lipogenic genes in hepatocytes and thereby affects overall apolipoprotein B (ApoB) particle accumulation, alters particle composition and regulates very large density lipoprotein (VLDL), LDL and TG levels [14]. In humans, TRIB1 variation has been associated with blood lipid levels [2], [3], [15], [16] and increased risk of coronary artery disease [15], [17], ischemic heart disease [18] and myocardial infarction [18]. An in vitro study suggested that the protein product of TRIB1 is in control of vascular smooth muscle cell proliferation and consequently may drive the development of atherosclerosis [19].
We detected a statistically significant association between rs4420638 and TC change. This variant maps to the APOE-APOC1-APOC2 cluster on chromosome 19. APOE translates to ApoE, which is the main apolipoprotein of the chylomicron, and thus crucial for breaking down TG-rich lipoproteins and essential in maintaining normal plasma cholesterol and TG levels. APOE variants have been associated with blood lipid levels [2], [3], [20], familial dyslipoproteinemia [21], polygenic dyslipidemia [22], elevated plasma C-reactive protein levels [23], [24], coronary heart disease [20], [23], and myocardial infarction [15].
We used rs6589564 as a proxy for the APOA1 rs964184 variant (chromosome 11). Both variants localize to the APOA1/C3/A4/A5/BUD13 cluster. APOA1 encodes the major apolipoprotein of plasma HDL particles and plays a central role in lipid metabolism. The rs964184 variant in APOA1 has been associated with blood lipid levels [2], [3], [22], polygenic dyslipidemia [22], metabolic syndrome [25], coronary heart disease [26], and myocardial infarction [27].
An important strength of this study is the inclusion of replication data. The findings of this study would be enhanced by further investigation of lipoprotein subclasses and the analysis of the effects of lipid lowering interventions in randomized controlled trials.
In conclusion, the trait-specific GRSs were robustly associated with baseline and longitudinal changes in blood lipid concentrations. We detected three novel longitudinal associations in relation to TC and TG changes over a 10-yr follow-up period. As these loci have been previously associated with cardiovascular traits, such as coronary heart disease and myocardial infarction, their associations with lipid changes provide further insight into how these variants contribute to cardiovascular risk.
Materials and Methods
Ethics statement
Ethical approval for the GLACIER Study was obtained from the Regional Ethical Review Board in Umeå, Sweden. The Ethics Committee at Lund University approved the MDC study.
Study participants
The GLACIER Study (N∼19,000) is a prospective, population-based cohort study nested within the Västerbotten Health Survey (VHU) in the northern Swedish county of Västerbotten [28]. Baseline examinations were undertaken from 1985 through 2004. GLACIER participants were invited to attend an examination on their 40th, 50th and 60th birthdays. In a subcohort (N = 5,010), ten-year follow-up data are also available, of whom 3,495 were genotyped (see below). Anthropometric measures (age, sex, height and weight) were collected, and detailed assessments of lifestyle were obtained using a validated questionnaire [28], [29]. All participants provided written informed consent as part of the VHU. The MDC Study constitutes southern Swedish adults participating in a cardiovascular program, with baseline data recorded from 1991 through 1996 [30], [31]. All individuals who were alive and still living in Sweden were invited for follow-up between 2007 and 2012. A total of 3,734 individuals attended follow-up investigation and 2,943 individuals with no history of coronary events had available data for replications analyses.
Clinical measures
Clinical measures have been described in detail elsewhere [28], [29]. Capillary blood was drawn following an overnight fast. Serum lipid concentrations were measured on fresh capillary plasma with a Reflotron bench-top analyzer (Roche Diagnostics Scandinavia AB). HDL cholesterol was measured after precipitation of the other lipoproteins with sodium phosphowolframate-magnesium chloride. For the ROC AUC analyses, lipid levels were dichotomized (low/high) according to the American Heart Association criteria [32]. At baseline, 5% of the individuals reported not having fasted for at least 8 hours before the blood draw, and information on fasting time was missing in a further 15% of the participants; therefore analyses were adjusted with a variable indicating fasting status, but this did not materially affect the results. In the MDC Study, TC, TG and HDL-C concentrations in the fasting blood samples were measured with a DAX 48 automatic analyzer (Bayer AB, Göteborg, Sweden) using reagents and calibrators from the supplier of the instrument. HDL-C concentrations were determined by the same procedure as used for TC, but after precipitation of LDL-C and very low-density lipoprotein cholesterol (VLDL-C) with dextran–sulphate [33]. The same laboratory methods where applied for analyzing lipid levels at both visits. Direct anthropometry was measured by nurses. LDL-C concentrations were calculated with the Friedewald formula for both studies [34].
Lipid medications
One percent of participants reported using lipid-lowering medications, which we controlled for in analyses using a constant, as described by Tobin et al [35]. There was no information available on the specific type of the lipid lowering agent used by the participants, but at the time of the examinations the most common class of lipid lowering drugs in northern Sweden was statins, used by ∼96% of lipid lowering medication users [36]. Therefore, to correct lipid levels we used the statin constants proposed by Wu et al. [37]: HDL-C: −0.059 mmol/l; LDL-C: +1.279 mmol/l, TC: +1.336 mmol/l, TG: +0.207 mmol/l. The MDC Study participants who reported using lipid-lowering–medication at baseline (3%) were excluded from analyses because the type of medication used could not be determined. 28.3% (n = 834) of the MDC Study participants reported using lipid lowering medication at follow-up; of these, 28% used LDL lowering agents (Crestor, Lipitor, Pravachol, Zocord or Ezetrol) and 0.3% used fibrates (Lopid). Their lipid measures were corrected by adding the appropriate constants proposed by Wu et al [37].
Genotyping
DNA was extracted from peripheral white blood cells and genomic DNA samples were diluted to 4 ng/µl as previously described [38], [39]. Samples were genotyped with the MetaboChip (Illumina iSelect) array [40]. The 102 associated SNPs from Teslovich et al. [2] and the 62 SNPs from Willer et al. [3] were extracted from the MetaboChip. We used proxy SNPs for 18 variants. Proxies for two SNPs (the TG-associated rs2929282 and the HDL-C associated-rs1047891 variants) were unavailable. Detailed information about the index and proxy SNPs are shown in Table S3. The average genotyping success rate was 99.9%. None of the SNPs deviated significantly from Hardy-Weinberg expectations at a study-wise corrected level (P<0.0001). Therefore, we conducted our analyses with a total of 162 SNPs (HDL-C = 73 SNPs; LDL-C = 58 SNPs; TC = 75 SNPs; and TG = 43 SNPs). For the replication effort in MDC, the SNPs analyzed were genotyped using Illumina OmniExpress Exome. SNP and proxy information are reported in Table S4.
Genetic risk score
The effects of multiple genetic risk loci on blood lipid traits were studied by constructing two different types of GRS for each study participant. The first assumed an equal magnitude of effect for each risk allele and was generated for each participant by summing the number of risk alleles at each of the associated SNP loci for the respective traits. Thus, because these are all biallelic loci, the GRSs had a minimum possible value of 0 and a maximum possible value of 146, 116, 150 and 86 for HDL-C, LDL-C, TC and TG, respectively. To construct the second GRS, we used published effect sizes for each SNP (from the joint meta-analysis by Willer et al. [3]) to weight the contribution of each risk allele. The weighted alleles were subsequently summed into a single weighted GRS (wGRS) as previously described [39], [41]. Missing genotypes were imputed by mean imputation as previously described [42]. To illustrate results in figures we used quartiles of the GRSs.
Statistical methods
Statistical analyses were undertaken using SAS (version 9.3, SAS Institute Inc., NC, USA), STATA (version 12.1, StataCorp LP, TX, USA), R (version 2.15.3, The R Foundation for Statistical Computing) and PLINK (version 1.07) [43]–[46]. Main effects were estimated with generalized linear models (GLMs) by fitting genotypes (additive model) or the unweighted/weighted GRSs as the independent variable with the corresponding lipid traits as the dependent variable. We used natural logarithmic transformed TG values for cross-sectional individual SNP analyses and adjusted for age, age2, sex, fasting time and population substructure (first four principal components) in all our models. In longitudinal analyses, we included the follow-up lipid measure as the dependent variable and adjusted for the respective trait's baseline value.
follow-up lipid = α+βSNP/GRS+βbaseline lipid+βcov+…+βcov+ε
For the sake of simplicity, when reporting the estimates for the model above, we refer to ΔTC or ΔTG throughout the manuscript. The Benjamini-Hochberg FDR was used to correct for multiple testing [47]; given the prior knowledge of the SNPs in the analyses, we decided to use a less stringent approach then the Bonferroni or the Holm correction. ROC AUCs were computed and compared using STATA. In these analyses we excluded individuals with hyperlipidemia at baseline and compared the predictive accuracy of four models (age, age2, sex and BMI (M1), M1+ trait specific wGRS (M2), M1+ traditional risk factors (age, sex, BMI, smoking status, alcohol intake) [10] for hyperlipidemia (M3) and M1+ trait specific wGRS + traditional risk factors for hyperlipidemia (M4)) in relation to hyperlipidemia at follow-up. Random effects meta-analysis was performed using the metan module in STATA [48]. Statistical analyses for MDC were done using SPSS (version 20, IBM Corporation). Linear regression was used to obtain effect sizes (β) and 95% confidence intervals (95% CI) by fitting genotypes (additive model) as the independent variables and follow-up lipid measures (TC, lnTG, HDL-C or LDL-C) as dependent variables. Baseline lipid levels, sex, age and age2 were used as covariates. For meta-analysis, regression estimates with average annual lipid level changes as outcome measures were used.
Supporting Information
Zdroje
1. VisscherPM, BrownMA, McCarthyMI, YangJ (2012) Five years of GWAS discovery. Am J Hum Genet 90 : 7–24.
2. TeslovichTM, MusunuruK, SmithAV, EdmondsonAC, StylianouIM, et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466 : 707–713.
3. Global Lipids Genetics Consortdium (2013) WillerCJ, SchmidtEM, SenguptaS, PelosoGM, et al. (2013) Discovery and refinement of loci associated with lipid levels. Nat Genet 45 : 1274–1283.
4. MiddelbergRP, MartinNG, WhitfieldJB (2006) Longitudinal genetic analysis of plasma lipids. Twin Res Hum Genet 9 : 550–557.
5. JohnsonAD, HandsakerRE, PulitSL, NizzariMM, O'DonnellCJ, et al. (2008) SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24 : 2938–2939.
6. JostinsL, BarrettJC (2011) Genetic risk prediction in complex disease. Hum Mol Genet 20: R182–188.
7. LutseyPL, Rasmussen-TorvikLJ, PankowJS, AlonsoA, SmolenskiDJ, et al. (2012) Relation of lipid gene scores to longitudinal trends in lipid levels and incidence of abnormal lipid levels among individuals of European ancestry: the Atherosclerosis Risk in Communities (ARIC) study. Circ Cardiovasc Genet 5 : 73–80.
8. VoightBF, PelosoGM, Orho-MelanderM, Frikke-SchmidtR, BarbalicM, et al. (2012) Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet 380 : 572–580.
9. CostanzaMC, Beer-BorstS, JamesRW, GaspozJM, MorabiaA (2012) Consistency between cross-sectional and longitudinal SNP: blood lipid associations. Eur J Epidemiol 27 : 131–138.
10. LuY, FeskensEJ, BoerJM, ImholzS, VerschurenWM, et al. (2010) Exploring genetic determinants of plasma total cholesterol levels and their predictive value in a longitudinal study. Atherosclerosis 213 : 200–205.
11. SabattiC, ServiceSK, HartikainenAL, PoutaA, RipattiS, et al. (2009) Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet 41 : 35–46.
12. AulchenkoYS, RipattiS, LindqvistI, BoomsmaD, HeidIM, et al. (2009) Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet 41 : 47–55.
13. Kiss-TothE, BagstaffSM, SungHY, JozsaV, DempseyC, et al. (2004) Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J Biol Chem 279 : 42703–42708.
14. BurkhardtR, TohSA, LagorWR, BirkelandA, LevinM, et al. (2010) Trib1 is a lipid - and myocardial infarction-associated gene that regulates hepatic lipogenesis and VLDL production in mice. J Clin Invest 120 : 4410–4414.
15. WillerCJ, SannaS, JacksonAU, ScuteriA, BonnycastleLL, et al. (2008) Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40 : 161–169.
16. KathiresanS, MelanderO, GuiducciC, SurtiA, BurttNP, et al. (2008) Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 40 : 189–197.
17. WaterworthDM, RickettsSL, SongK, ChenL, ZhaoJH, et al. (2010) Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol 30 : 2264–2276.
18. VarboA, BennM, Tybjaerg-HansenA, GrandeP, NordestgaardBG (2011) TRIB1 and GCKR polymorphisms, lipid levels, and risk of ischemic heart disease in the general population. Arterioscler Thromb Vasc Biol 31 : 451–457.
19. SungHY, GuanH, CzibulaA, KingAR, EderK, et al. (2007) Human tribbles-1 controls proliferation and chemotaxis of smooth muscle cells via MAPK signaling pathways. J Biol Chem 282 : 18379–18387.
20. DrenosF, TalmudPJ, CasasJP, SmeethL, PalmenJ, et al. (2009) Integrated associations of genotypes with multiple blood biomarkers linked to coronary heart disease risk. Hum Mol Genet 18 : 2305–2316.
21. SmeltAH, de BeerF (2004) Apolipoprotein E and familial dysbetalipoproteinemia: clinical, biochemical, and genetic aspects. Semin Vasc Med 4 : 249–257.
22. KathiresanS, WillerCJ, PelosoGM, DemissieS, MusunuruK, et al. (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 41 : 56–65.
23. ElliottP, ChambersJC, ZhangW, ClarkeR, HopewellJC, et al. (2009) Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA 302 : 37–48.
24. GolledgeJ, BirosE, CooperM, WarringtonN, PalmerLJ, et al. (2010) Apolipoprotein E genotype is associated with serum C-reactive protein but not abdominal aortic aneurysm. Atherosclerosis 209 : 487–491.
25. KristianssonK, PerolaM, TikkanenE, KettunenJ, SurakkaI, et al. (2012) Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ Cardiovasc Genet 5 : 242–249.
26. XuL, ZhouJ, HuangS, HuangY, LeY, et al. (2013) An association study between genetic polymorphisms related to lipoprotein-associated phospholipase A(2) and coronary heart disease. Exp Ther Med 5 : 742–750.
27. TraganteV, DoevendansPA, NathoeHM, van der GraafY, SpieringW, et al. (2013) The impact of susceptibility loci for coronary artery disease on other vascular domains and recurrence risk. Eur Heart J 34 : 2896–2904.
28. Hallmans G, Agren A, Johansson G, Johansson A, Stegmayr B, et al.. (2003) Cardiovascular disease and diabetes in the Northern Sweden Health and Disease Study Cohort - evaluation of risk factors and their interactions. Scand J Public Health Suppl 61 : 18–24.
29. Norberg M, Wall S, Boman K, Weinehall L (2010) The Vasterbotten Intervention Programme: background, design and implications. Glob Health Action 3.
30. HedbladB, NilssonP, JanzonL, BerglundG (2000) Relation between insulin resistance and carotid intima-media thickness and stenosis in non-diabetic subjects. Results from a cross-sectional study in Malmo, Sweden. Diabet Med 17 : 299–307.
31. ManjerJ, CarlssonS, ElmstahlS, GullbergB, JanzonL, et al. (2001) The Malmo Diet and Cancer Study: representativity, cancer incidence and mortality in participants and non-participants. Eur J Cancer Prev 10 : 489–499.
32. American Heart Association (AHA) - What Your Cholesterol Levels Mean. Available: http://www.heart.org/HEARTORG/Conditions/Cholesterol/AboutCholesterol/What-Your-Cholesterol-Levels-Mean_UCM_305562_Article.jsp. Accessed 05 May 2013.
33. RosvallM, OstergrenPO, HedbladB, IsacssonSO, JanzonL, et al. (2000) Occupational status, educational level, and the prevalence of carotid atherosclerosis in a general population sample of middle-aged Swedish men and women: results from the Malmo Diet and Cancer Study. Am J Epidemiol 152 : 334–346.
34. FriedewaldWT, LevyRI, FredricksonDS (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 18 : 499–502.
35. TobinMD, SheehanNA, ScurrahKJ, BurtonPR (2005) Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med 24 : 2911–2935.
36. EliassonM, JanlertU, JanssonJH, StegmayrB (2006) Time trends in population cholesterol levels 1986–2004: influence of lipid-lowering drugs, obesity, smoking and educational level. The northern Sweden MONICA study. J Intern Med 260 : 551–559.
37. WuJ, ProvinceMA, CoonH, HuntSC, EckfeldtJH, et al. (2007) An investigation of the effects of lipid-lowering medications: genome-wide linkage analysis of lipids in the HyperGEN study. BMC Genet 8 : 60.
38. FranksPW, RolandssonO, DebenhamSL, FawcettKA, PayneF, et al. (2008) Replication of the association between variants in WFS1 and risk of type 2 diabetes in European populations. Diabetologia 51 : 458–463.
39. RenstromF, PayneF, NordstromA, BritoEC, RolandssonO, et al. (2009) Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Hum Mol Genet 18 : 1489–1496.
40. VoightBF, KangHM, DingJ, PalmerCD, SidoreC, et al. (2012) The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet 8: e1002793.
41. CornelisMC, QiL, ZhangC, KraftP, MansonJ, et al. (2009) Joint effects of common genetic variants on the risk for type 2 diabetes in U.S. men and women of European ancestry. Ann Intern Med 150 : 541–550.
42. Fontaine-BissonB, RenstromF, RolandssonO, Magic, PayneF, et al. (2010) Evaluating the discriminative power of multi-trait genetic risk scores for type 2 diabetes in a northern Swedish population. Diabetologia 53 : 2155–2162.
43. PurcellS, NealeB, Todd-BrownK, ThomasL, FerreiraMA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 : 559–575.
44. SAS Institute (2011) The SAS system for Windows, version 9.2. SAS Institute, Cary, NC, USA.
45. StataCorp (2011) Stata Statistical Software, version 12.1. College Station, TX: StataCorp LP, USA.
46. R Development Core Team (2008) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
47. Miller RG (1981) Simultaneous statistical inference. 2nd ed.: Springer Verlag. pp. 6–8.
48. Harris RJ, Deeks JJ, Altman DG, Bradburn MJ, Harbord RM, et al.. (2008) metan: fixed - and random-effects meta-analysis. The Stata Journal 8: pp. 3–28.
Štítky
Genetika Reprodukčná medicína
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 6- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Inflammation: Gone with Translation
- Recombination Accelerates Adaptation on a Large-Scale Empirical Fitness Landscape in HIV-1
- Caspase Inhibition in Select Olfactory Neurons Restores Innate Attraction Behavior in Aged
- Accurate, Model-Based Tuning of Synthetic Gene Expression Using Introns in
- A Novel Peptidoglycan Binding Protein Crucial for PBP1A-Mediated Cell Wall Biogenesis in
- Ancient DNA Analysis of 8000 B.C. Near Eastern Farmers Supports an Early Neolithic Pioneer Maritime Colonization of Mainland Europe through Cyprus and the Aegean Islands
- The Epidermal Growth Factor Receptor Critically Regulates Endometrial Function during Early Pregnancy
- Introgression from Domestic Goat Generated Variation at the Major Histocompatibility Complex of Alpine Ibex
- Netrins and Wnts Function Redundantly to Regulate Antero-Posterior and Dorso-Ventral Guidance in
- Coordination of Wing and Whole-Body Development at Developmental Milestones Ensures Robustness against Environmental and Physiological Perturbations
- Phenotypic Dissection of Bone Mineral Density Reveals Skeletal Site Specificity and Facilitates the Identification of Novel Loci in the Genetic Regulation of Bone Mass Attainment
- Deep Evolutionary Comparison of Gene Expression Identifies Parallel Recruitment of -Factors in Two Independent Origins of C Photosynthesis
- Loss of UCP2 Attenuates Mitochondrial Dysfunction without Altering ROS Production and Uncoupling Activity
- Translational Regulation of Specific mRNAs Controls Feedback Inhibition and Survival during Macrophage Activation
- Rosa26-GFP Direct Repeat (RaDR-GFP) Mice Reveal Tissue- and Age-Dependence of Homologous Recombination in Mammals
- Abnormal Type I Collagen Post-translational Modification and Crosslinking in a Cyclophilin B KO Mouse Model of Recessive Osteogenesis Imperfecta
- : Clonal Reinforcement Drives Evolution of a Simple Microbial Community
- Reviving the Dead: History and Reactivation of an Extinct L1
- Defective iA37 Modification of Mitochondrial and Cytosolic tRNAs Results from Pathogenic Mutations in TRIT1 and Its Substrate tRNA
- Early Back-to-Africa Migration into the Horn of Africa
- Aberrant Autolysosomal Regulation Is Linked to The Induction of Embryonic Senescence: Differential Roles of Beclin 1 and p53 in Vertebrate Spns1 Deficiency
- Microbial Succession in the Gut: Directional Trends of Taxonomic and Functional Change in a Birth Cohort of Spanish Infants
- Integrated Pathway-Based Approach Identifies Association between Genomic Regions at CTCF and CACNB2 and Schizophrenia
- Genetic Determinants of Long-Term Changes in Blood Lipid Concentrations: 10-Year Follow-Up of the GLACIER Study
- Palaeosymbiosis Revealed by Genomic Fossils of in a Strongyloidean Nematode
- Early Embryogenesis-Specific Expression of the Rice Transposon Enhances Amplification of the MITE
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- Genetic Background Drives Transcriptional Variation in Human Induced Pluripotent Stem Cells
- Pervasive Divergence of Transcriptional Gene Regulation in Caenorhabditis Nematodes
- N-WASP Is Required for Structural Integrity of the Blood-Testis Barrier
- The Transcription Factor TFII-I Promotes DNA Translesion Synthesis and Genomic Stability
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
- Digital Genotyping of Macrosatellites and Multicopy Genes Reveals Novel Biological Functions Associated with Copy Number Variation of Large Tandem Repeats
- ATRA-Induced Cellular Differentiation and CD38 Expression Inhibits Acquisition of BCR-ABL Mutations for CML Acquired Resistance
- The EJC Binding and Dissociating Activity of PYM Is Regulated in
- JNK Controls the Onset of Mitosis in Planarian Stem Cells and Triggers Apoptotic Cell Death Required for Regeneration and Remodeling
- Mouse Y-Linked and Are Expressed during the Male-Specific Interphase between Meiosis I and Meiosis II and Promote the 2 Meiotic Division
- Rasa3 Controls Megakaryocyte Rap1 Activation, Integrin Signaling and Differentiation into Proplatelet
- Transcriptional Control of Steroid Biosynthesis Genes in the Prothoracic Gland by Ventral Veins Lacking and Knirps
- Souffle/Spastizin Controls Secretory Vesicle Maturation during Zebrafish Oogenesis
- The POU Factor Ventral Veins Lacking/Drifter Directs the Timing of Metamorphosis through Ecdysteroid and Juvenile Hormone Signaling
- The First Endogenous Herpesvirus, Identified in the Tarsier Genome, and Novel Sequences from Primate Rhadinoviruses and Lymphocryptoviruses
- Sequence of a Complete Chicken BG Haplotype Shows Dynamic Expansion and Contraction of Two Gene Lineages with Particular Expression Patterns
- Background Selection as Baseline for Nucleotide Variation across the Genome
- CPF-Associated Phosphatase Activity Opposes Condensin-Mediated Chromosome Condensation
- The Effects of Codon Context on Translation Speed
- Glycogen Synthase Kinase (GSK) 3β Phosphorylates and Protects Nuclear Myosin 1c from Proteasome-Mediated Degradation to Activate rDNA Transcription in Early G1 Cells
- Regulation of Gene Expression in Autoimmune Disease Loci and the Genetic Basis of Proliferation in CD4 Effector Memory T Cells
- Muscle Structure Influences Utrophin Expression in Mice
- BLMP-1/Blimp-1 Regulates the Spatiotemporal Cell Migration Pattern in
- Identification of Late Larval Stage Developmental Checkpoints in Regulated by Insulin/IGF and Steroid Hormone Signaling Pathways
- Transport of Magnesium by a Bacterial Nramp-Related Gene
- Sgo1 Regulates Both Condensin and Ipl1/Aurora B to Promote Chromosome Biorientation
- The HY5-PIF Regulatory Module Coordinates Light and Temperature Control of Photosynthetic Gene Transcription
- The Rim15-Endosulfine-PP2A Signalling Module Regulates Entry into Gametogenesis and Quiescence Distinct Mechanisms in Budding Yeast
- Regulation of Hfq by the RNA CrcZ in Carbon Catabolite Repression
- Loss of a Neural AMP-Activated Kinase Mimics the Effects of Elevated Serotonin on Fat, Movement, and Hormonal Secretions
- Positive Feedback of Expression Ensures Irreversible Meiotic Commitment in Budding Yeast
- Hecate/Grip2a Acts to Reorganize the Cytoskeleton in the Symmetry-Breaking Event of Embryonic Axis Induction
- Regulatory Mechanisms That Prevent Re-initiation of DNA Replication Can Be Locally Modulated at Origins by Nearby Sequence Elements
- Speciation and Introgression between and
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Early Back-to-Africa Migration into the Horn of Africa
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



