-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Drought and salt are two of the most serious threats to food production worldwide, and research on stress tolerance in crops is important for future food security. In this study we identified DCA1, a transcriptional co-activator of DST that is conserved in the world’s three major crops. DCA1 participates in stress tolerance by controlling stomatal aperture through modulation of H2O2 homeostasis in guard cells. This finding not only increases our understanding of the molecular mechanism by which plants withstand harsh environmental conditions, but it may also facilitate future molecular breeding and genetic engineering of drought - and salt-tolerant crops.
Published in the journal: DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice. PLoS Genet 11(10): e32767. doi:10.1371/journal.pgen.1005617
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005617Summary
Drought and salt are two of the most serious threats to food production worldwide, and research on stress tolerance in crops is important for future food security. In this study we identified DCA1, a transcriptional co-activator of DST that is conserved in the world’s three major crops. DCA1 participates in stress tolerance by controlling stomatal aperture through modulation of H2O2 homeostasis in guard cells. This finding not only increases our understanding of the molecular mechanism by which plants withstand harsh environmental conditions, but it may also facilitate future molecular breeding and genetic engineering of drought - and salt-tolerant crops.
Introduction
How to feed a growing population that is expected to reach roughly 9 billion by the middle of this century is among the major challenges of our time [1]. Modern agriculture has greatly improved food production [2], but progress towards avoiding the negative effects of climate change and diminishing soil conditions has been insufficient. Most worryingly, many of the plants upon which we depend for food production are particularly sensitive to environmental stress [3]. Droughts are likely to be more frequent as global warming accelerates, and rising sea levels will result in the loss of productive agricultural land to water infiltration and increased soil salinity. Together these unfavorable factors pose a huge threat to food security, and studying drought and salt tolerance in crops is becoming increasingly urgent.
Many previous studies on drought and salt tolerance in plants have mainly focused on the model species Arabidopsis thaliana. Years of effort have revealed that the responses of plants to water stress are controlled by complex regulatory signalling events mediated by Ca2+ [4, 5], abscisic acid (ABA) [6–8], reactive oxygen species (ROS) [9, 10], ion transport [4, 11–13], and the activities of transcription factors (TFs) [14]. Stomata are often described as the guardians of leaves which are of course the photosynthetic organs in which food manufacturing occurs. The main function of stomata, which are formed from pairs of guard cells, is to allow gases to move rapidly into and out of the leaf. However, this evolved trait poses a problem for plants since they face the predicament of taking up CO2 through stomata while attempting to minimize water loss through these pores. The ability to effectively control the balance between photosynthesis and transpiration in accordance with the external environment is an impressive evolutionary achievement.
ROS such as H2O2 are a product of the incomplete reduction of molecular oxygen, and ROS production is widely considered a symptom of cellular dysfunction. ROS participate in cell death and may also function as signalling molecules. Increasing evidence suggests that ROS function as second messengers in stomatal aperture control [9, 15]. In 1996, McAinsh et al. first reported that ROS induced stomatal closure and inhibited stomatal opening by increasing the concentration of cytosolic free calcium ([Ca2+]cyt) in Commelina communis [16]. A detailed follow-up study reported that ABA-stimulated ROS accumulation induced stomatal closure via the activation of plasma membrane calcium channels in Arabidopsis [9].
Since plants are sessile organisms, they must have highly evolved sophisticated mechanisms to detect and respond to environmental perturbations. Changes in the expression of stress-related genes are an important part of the plant response to environmental stress. Numerous transcription factors including APETALA 2/ethylene-responsive element binding factor (AP2/ERF), dehydration responsive element binding protein (DREB)/C-repeat-binding factor (CBF), ABA-responsive element binding protein (AREB)/ABA-responsive element-binding factor (ABF), No apical meristem, Arabidopsis transcription activation factor and Cup-shaped cotyledon (NAC) are associated with plant abiotic stress responses [14]. Regulation of target genes by TFs is a highly complex and delicately balanced process. In most cases TFs do not function alone but recruit partner proteins (cofactors) to form transcription initiation complexes [17]. Cofactors are transcription factor interacting proteins that either activate or repress the transcription of target genes, and numerous examples have been reported in animals including humans, but few have been identified in plants. Arabidopsis HAIRY MERISTEM (HAM) family proteins were recently found to act as conserved interacting cofactors with the transcription factor WUSCHEL (WUS) to drive downstream transcriptional programs that help promote shoot stem cell proliferation [18]. Another study demonstrated that HYPOXIA RESPONSE ATTENUATOR1 (HRA1) interacts with ethylene-responsive factor group VII transcription factor (ERF-VII TF) RAP2.12 to negatively modulate its activity under hypoxia [19].
Efforts have also been made to determine the underlying physiological, genetic and molecular mechanisms mediating drought and salt tolerance in crops such as rice. Stress-responsive NAC 1 (SNAC1) is a member of the rice NAC TF family that is specifically induced in guard cells in drought conditions, and overexpression of SNAC1 in rice significantly enhanced drought tolerance [20]. We previously isolated the C2H2 zinc finger transcription factor DST that negatively regulates stress tolerance in rice [21]. DST regulates stomatal aperture by modulating the expression of genes related to ROS homeostasis. However, these studies are fragmentary. The exact mechanisms by which these TFs regulate the expression of target genes remain unknown. In the present study, we identified the CHY zinc finger protein DCA1, an interacting partner of DST. Homologs of DCA1 in rice and Arabidopsis were recently shown to increase stomatal opening and were upregulated by heat stress [22]. However the exact molecular function of this protein, the pathways involved and the phenotypes of plants in which DCA1 is modified remain unknown. In this research, we revealed that DCA1 forms a heterologous tetramer with DST and positively regulates DST activity. This transcriptional complex regulates the expression of Prx24, an H2O2 scavenger preferentially expressed in guard cells. Together, the DCA1-DST-Prx24 pathway contributes to stomatal movement via regulating ROS homeostasis under stress conditions. These findings may facilitate the engineering of crops with improved drought and salt tolerance.
Results
Physical Interaction between DCA1 and DST
Most TFs do not function alone. Rather, they recruit intermediary proteins (cofactors) to initiate transcription effectively. In humans, several hundred putative co-regulators have been identified [17]. To identify cofactors of DST, we performed yeast two-hybrid screening with a ZH11 (Oryza sativa L. japonica. cv. ZhongHua 11) leaf cDNA library. A version of DST with the self-activation domain removed (amino acids 1–72 removed from the N-terminus, including the zinc finger domain, ΔN) was used as bait. Dozens of potential targeting proteins were found including a U-box domain containing protein, Elongation Factor 1-alpha, a protein similar to Calcium-dependent protein kinase-related kinase, and the CHY zinc finger protein DCA1. To further verify the interaction between these proteins, we used DCA1 and DST as bait and prey, respectively (Fig 1A), and their interaction was confirmed through additional yeast two-hybrid assays. DCA1 was able to interact with DST independently of its C2H2 zinc finger domain which likely functions in DNA binding (Fig 1B). The DST-DCA1 interaction was also confirmed via bimolecular fluorescence complementation (BiFC) assays in Arabidopsis protoplasts in vivo, in which DST was fused to the amino-termini half of yellow fluorescent protein (YFP) (DST-nYFP) and DCA1 was fused to the carboxy-termini half of YFP (DCA1-cYFP). The YFP signal was mainly restricted to nuclei when DST-nYFP was cotransformed with DCA1-cYFP (Fig 1C). Consistent with these results, an in vitro pull-down assay using recombinant DST fused to maltose binding protein (DST-MBP) and His2-tagged DCA1 (DCA1-His2) that were expressed in E. coli showed that DST-MBP but not MBP was able to pull down DCA1-His2 (Fig 1D). These results confirm the direct interaction between DCA1 and DST both in vitro and in vivo.
Fig. 1. Physical interaction between DCA1 and DST. 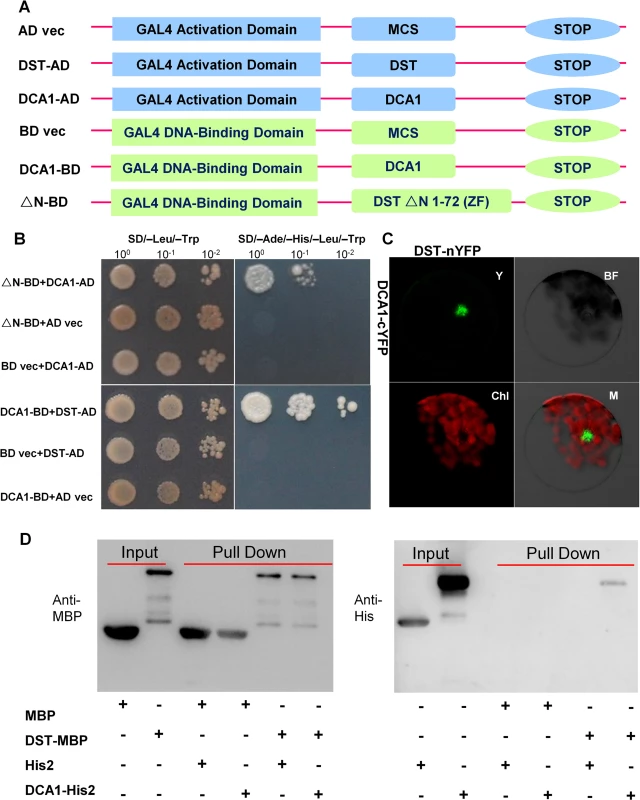
(A) Yeast two-hybrid constructs using native and variant DST and DCA1 fragments. (B) Growth of yeast cells harboring various combinations of constructs. SD/-Leu/-Trp and SD/-Ade/-His/-Leu/-Trp were used as nonselective and selective media, respectively. The assay revealed an interaction between DCA1 and both full-length DST and the C-terminal fragment of DST (△N 1–72 DST). (C) BiFC assay in Arabidopsis protoplasts revealing the interaction between DST and DCA1 in vivo. Y, YFP; BF, Bright Field; Chl, Chlorophyll; M, Merge. (D) In vitro pull-down of His2-tagged DCA1 using MBP-tagged DST as determined by immunoblotting with anti-MBP antibody (left) and anti-His antibody (right). DCA1 Characterization
Careful analysis of the structure of DCA1 identified a CHY zinc finger domain and a C3H2C3 or RING-H2 domain (S1A Fig). A database search for homologous proteins identified conserved CHY zinc finger domains in various different species, but the C3H2C3 domain appeared not to be conserved (S1B Fig). To characterize DCA1, we investigated the subcellular localization of DST-YFP and DCA1-YFP fusion proteins in Arabidopsis protoplasts. YFP fluorescence was apparent in both the cytoplasm and nuclei of control protoplasts transformed with the YFP-vector. In contrast YFP fluorescence was mainly restricted to nuclei in DST-YFP and DCA1-YFP transformants (Fig 2A–2C). Investigation of the tissue-specific expression of DCA1 by qRT-PCR revealed a similar expression pattern to that of DST, with both genes expressed at relatively high levels in leaves and culms (Fig 2D and S2A Fig). We also tested whether the expression of DCA1 was regulated by drought and salt stress using qRT-PCR and found that DCA1 is upregulated rapidly after 1–3 h of drought or salt treatment, with expression peaking at 12 h and normal expression resumed 24 h after treatment (Fig 2E and 2F).
Fig. 2. Subcellular localization and tissue-specific expression of DCA1. 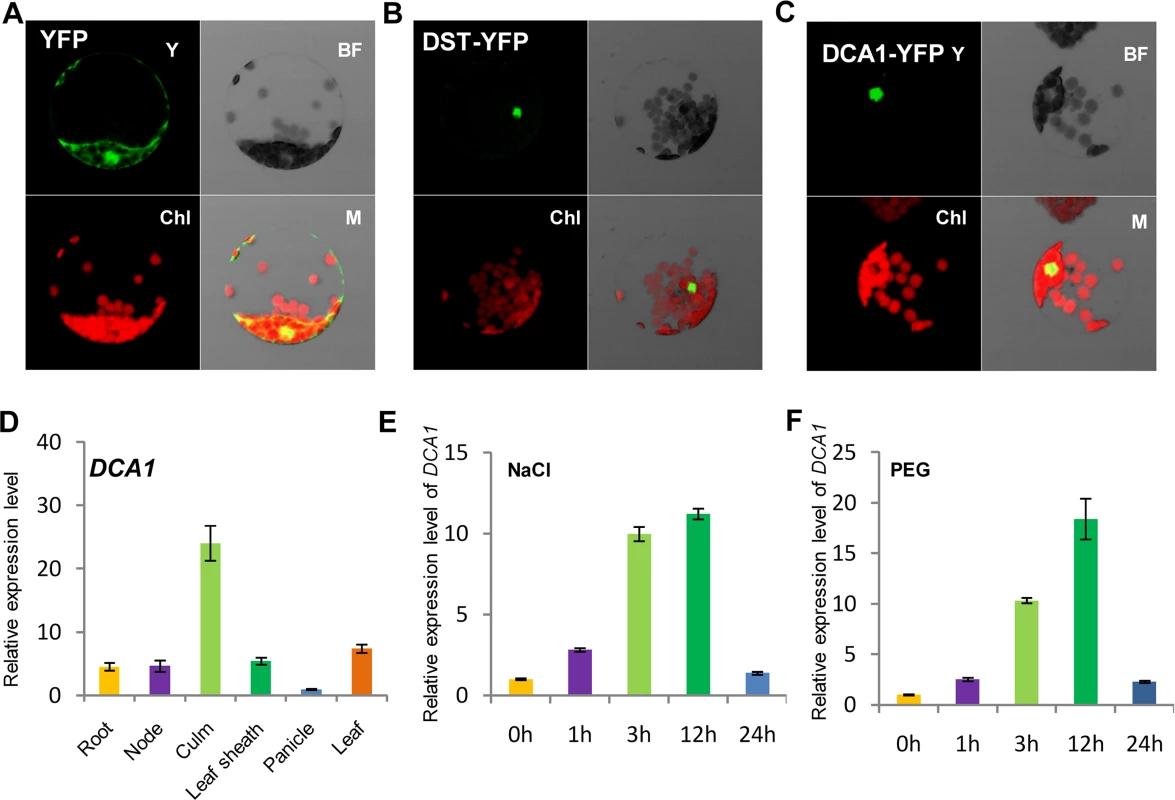
Subcellular localization of (A) YFP, (B) DST-YFP and (C) DCA1-YFP in Arabidopsis protoplasts. Both DST and DCA1 are located predominantly in the nucleus. Y, YFP; BF, Bright Field; Chl, Chlorophyll; M, Merge. (D) Expression pattern of DCA1 in different tissues of ZH11. Data represent means ± sd (n = 3). (E, F) Relative expression levels of DCA1 in the leaves of plants treated with NaCl (E) or PEG (F). Data are means ± sd (n = 3). Overexpression of DCA1 Reduces Stress Tolerance Ability Consisting with DST
To dissect the physiological functions of DCA1 in plants, we generated transgenic rice (ZH11) overexpressing DCA1 under the control of the enhanced cauliflower mosaic virus 35S promoter. qRT-PCR assays confirmed that DCA1 expression was enhanced in all eight overexpression lines, compared with vector-only control ZH11 plants (CK). Two transgenic lines, 35S::DCA1-7 and 35S::DCA1-8, were selected for further analysis (Fig 3A), and the tolerance to drought and salt stress was investigated. Twelve-day-old CK, 35S::DCA1-7 and 35S::DCA1-8 seedlings grown in normal conditions were treated with 100 mM NaCl for 10 days or with 18% PEG for 13 days. As shown in Fig 3B–3D, almost all of the 35S::DCA1 seedlings died, while approximately 70% of the vector-only control plants survived, following a subsequent 7 day recovery in normal conditions. The relative chlorophyll content decreased to ~13% in 35S::DCA1-8 plants vs. ~27% in vector-only control plants after drought treatment (S3A Fig). Fresh weight similarly decreased to ~52% in 35S::DCA1-8 plants vs. ~80% in vector-only control plants after drought treatment (S3C Fig). These results indicated that 35S::DCA1 plants were extremely sensitive to drought and salt stress compared with CK plants. Since there are many differences between laboratory and field stress conditions, we examined the responses of the 35S::DCA1 overexpression plants to drought stress in soil and obtained similar results. Specifically, 35S::DCA1 plants were much more sensitive to drought stress than CK plants (Fig 3E). In a previous study, we failed to obtain plants overexpressing DST, and their phenotypes under stress conditions remain unknown [21]. However after many years of effort, various 35S::DST lines have been prepared and qRT-PCR analysis revealed that expression of DST was only increased 1–2-fold in these plants (S5A Fig). This may be because strong overexpression of this gene causes death. 12-day-old CK, 35S::DST-1 and 35S::DST-2 seedlings grown under normal conditions were stressed with either 100 mM NaCl or 18% PEG and allowed to recover for 7 days. Almost all of the 35S::DST seedlings died, while approximately 70% (NaCl) and 80% (PEG) of the vector-only control plants survived (Fig 4). The relative chlorophyll content decreased to ~7% in 35S::DST-1 plants vs. ~27% in vector-only control plants (S3A Fig). Similarly, fresh weight decreased to ~53% in 35S::DST-1 plants vs. ~80% in vector control plants (S3C Fig). These results showed that 35S::DST plants were much more sensitive to drought and salt stress than controls, consistent with the results obtained for 35S::DCA1 (Fig 3). We also checked whether the reduced stress tolerance of 35S::DCA1 plants was caused by changes in DST expression, but the results suggested this was not the case (S5B Fig). We concluded that DCA1 may therefore function in cooperation with DST in the stress response.
Fig. 3. Overexpression of DCA1 reduces stress tolerance in rice. 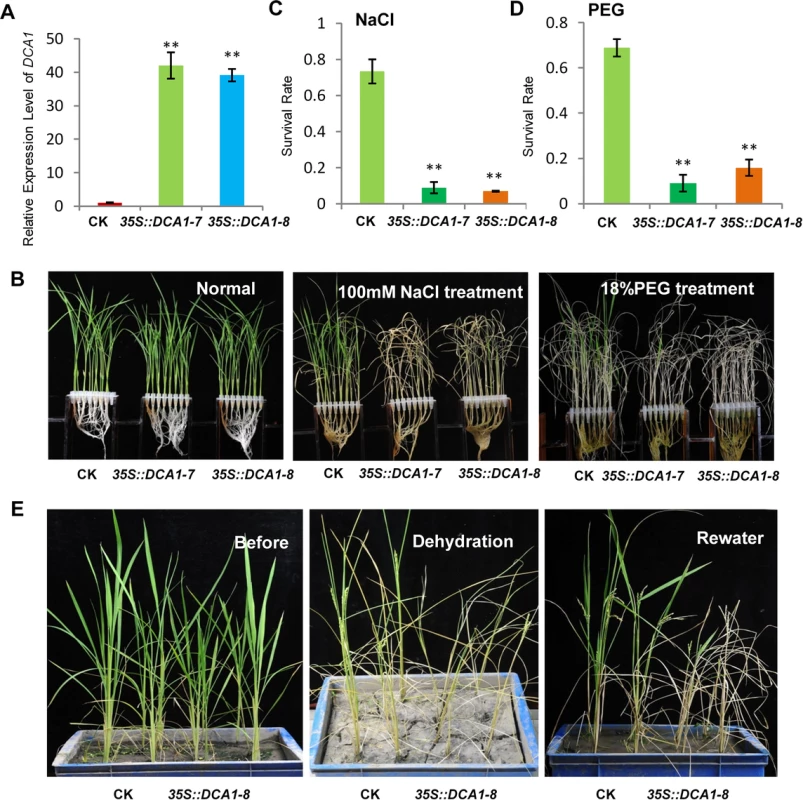
(A) Expression levels of DCA1 in vector-only ZH11 (CK), 35S::DCA1-7 (DCA1-7) and 35S::DCA1-8 (DCA1-8) plants were measured and normalized against Actin (data represent means ± sd, n = 3). (B) CK, 35S::DCA1-7 and 35S::DCA1-8 plants grown under normal conditions for 12 days (left), 12-day-old seedlings transferred to 100 mM NaCl for 10 days and recovered for 7 days (middle), or transferred to 18% PEG for 13 days and recovered for 7 days (right). (C) Relative survival rates after NaCl treatment (n = 3 biological replicates, 24 plants of each replicate). Data represent means ± sd (n = 3). (D) Relative survival rates after PEG treatment (n = 3 biological replicates, 24 plants of each replicate). Data represent means ± sd (n = 3). (E) Plants grown in soil under normal conditions for 63 days (left), dehydrated for 13 days (middle) and recovered for 16 days (right). Significant differences were determined using the Student’s t-test (**P <0.01). Fig. 4. DST-overexpressing like 35S::DCA1 plants exhibit reduced drought and salt tolerance. 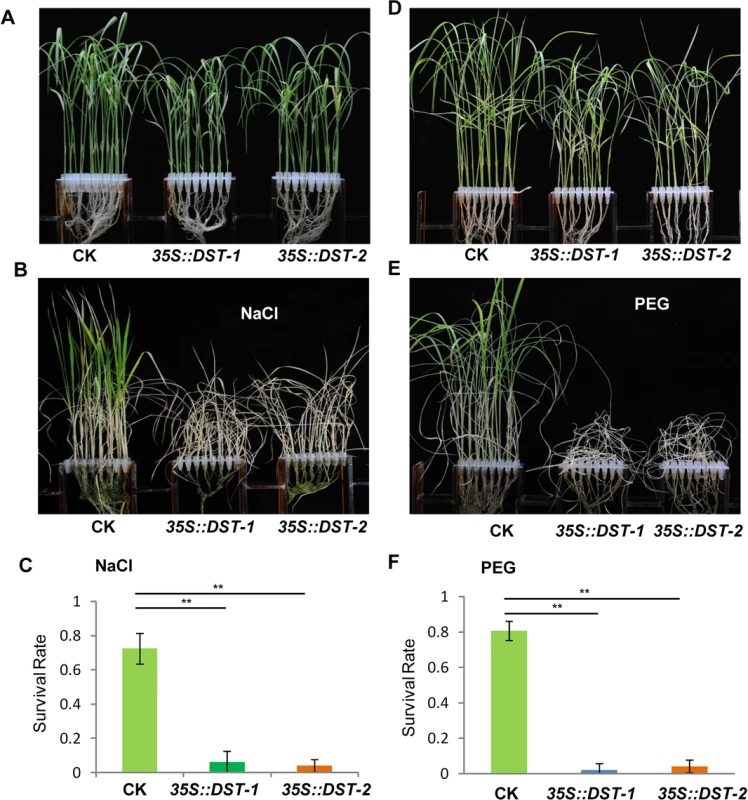
(A) Control (CK; vector-only ZH11), 35S::DST-1 and 35S::DST-2 plants grown under normal conditions for 12 days. (B) 12-day-old seedlings treated with 100 mM NaCl for 8 days and recovered for 7 days. (C) Survival percentage of plants following NaCl treatment (n = 3 biological replicates, 24 plants of each replicate). Data represent means ± sd (n = 3). (D) Plants grown under normal conditions for 12 days. (E) 12-day-old seedlings treated with 18% PEG for 10 days and recovered for 7 days. (F) Survival percentage of plants after PEG treatment (n = 3 biological replicates, 24 plants of each replicate). Data represent means ± sd (n = 3). Significant differences were determined using the Student’s t-test (**P <0.01). The DCA1 Knock-Down Plants Exhibits Improved Stress Tolerance
A Tos17 mutant (NF7038) of DCA1 was obtained from the Rice Genome Resource Center. The Tos17 fragment was inserted into the second intron of DCA1, and this reduced DCA1 expression levels by ~80% compared with wild-type Oryza sativa L. japonica. cv. Nipponbare. These results indicate that this mutant (designated dca1) was a genuine DCA1 knockdown mutant (Fig 5A–5C).
Fig. 5. The DCA1 knockdown mutant has improved stress tolerance. 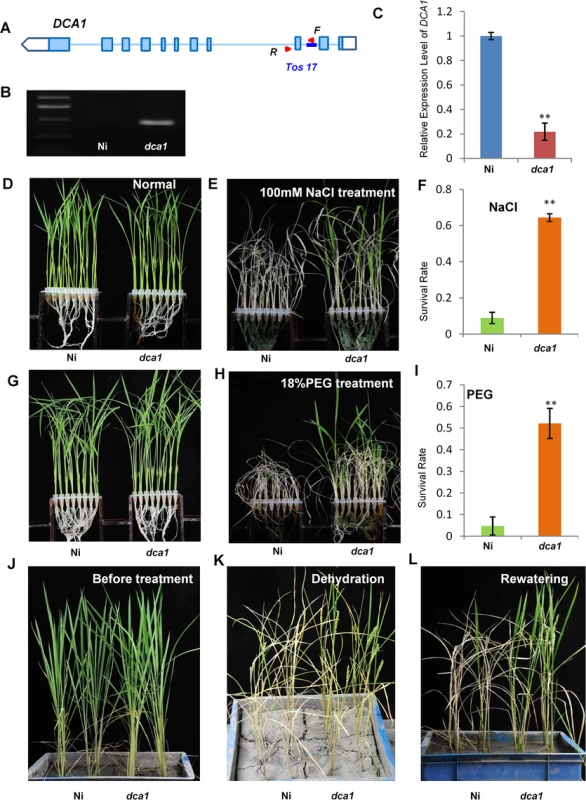
(A, B) Testing of the DCA1 knockdown mutant showing Tos17 insertion in the second intron of DCA1. Red triangles represent primers used for mutant examination. F primer was on the Tos17 fragment and R was on genome. (C) DCA1 expression in the dca1 mutant. Data represent means ± sd (n = 3). (D, E) Nipponbare (Ni) and dca1 seedlings grown under normal conditions for 12 days (D), transferred to 100 mM NaCl for 9 days and recovered for 7 days (E). (F) Relative survival rates of Ni and dca1 after NaCl treatment (n = 3 biological replicates, 24 plants of each replicate). Data represent means ± sd (n = 3). (G, H) Ni and dca1 seedlings grown under normal conditions for 12 days (G), transferred to 18% PEG for 10 days and recovered for 7 days (H). (I) Relative survival rates of Ni and dca1 after PEG treatment (n = 3 biological replicates, 24 plants of each replicate). Data represent means ± sd (n = 3). (J) Ni and dca1 grown in soil under normal conditions for 66 days, (K) dehydrated for 14 days and (L) recovered for 16 days. Significant differences were determined using the Student’s t-test (**P <0.01). We next evaluated whether dca1 plants possessed enhanced tolerance to drought or salt stress. 12-day-old seedlings of Ni (Nipponbare) and dca1 grown under normal conditions (Fig 5D and 5G) were treated with 100 mM NaCl for 9 days (Fig 5E) or 18% PEG for 10 days (Fig 5H) and recovered for 7 days. Approximately 10% of Ni plants survived following a subsequent 7 day recovery period after salt stress, compared with 62% of dca1 plants (Fig 5F). Similarly, the survival rate following recovery from drought stress was significantly increased in the dca1 mutant compared with Ni plants (Fig 5I). The relative chlorophyll content decreased to ~5% in Ni vs. ~16% in dca1 plants following drought treatment (S3B Fig), and the fresh weight decreased to ~43% and ~89% in Ni and dca1 plants, respectively (S3D Fig). These results indicated that dca1 plants were more tolerant to abiotic stress than Ni. Since dca1 is a Tos17 insertion mutant, we knew there may be several copies of the insertion, and Southern blotting revealed more than 10 copies (S4A Fig). We therefore designed an artificial mircoRNA to elicit knockdown of DCA1, and expression of DCA1 in the silenced variant was reduced by 50–80% compared with CK plants. DCA1 microRNA transgenic plants were also more tolerant of drought and salt stress than CK plants (S4B, S4C, S4E and S4F Fig). Dehydration treatment of 66-day-old Ni and dca1 plants grown in soil under normal conditions for 14 days and rewatered for 16 days showed that dca1 was more tolerant to soil dehydration than Ni (Fig 5J–5L). Moreover, dca1 seeds could be harvested from the surviving tillers after re-irrigation (S5C and S5D Fig). Together these results indicated that DCA1 knockdown plants exhibited improved tolerance to drought and salt stress, consistent with the phenotype of the DST mutant dst [21].
DCA1 Acts as a Co-activator of DST and Positively Regulates the Expression of Genes Downstream of DST
We previously demonstrated that DST is a transcription factor with transactivational activity [21]. However, we found that DCA1 does not exhibit this activity directly, since yeast harboring the DCA1-BD construct did not grow on SD/-Ade-His-Leu-Trp medium (Fig 1B). We therefore conducted dual luciferase assays in Arabidopsis protoplasts to confirm the direct effects of the DCA1-DST interaction on gene expression. Seven copies of the GAL4 binding sequence are located before the translational start site of the REN reporter and act as putative cis-acting elements. Compared with empty-vector controls, the REN/LUC ratio was moderately upregulated by DST alone but markedly upregulated when DCA1 and DST were both present, indicating a positive role for this interaction in regulating DST transcriptional activity (i.e., DCA1 enhances the transcriptional activity of DST; Fig 6A and 6B). Peroxidase 24 precursor (Prx 24) is an H2O2 scavenger and target gene of DST, as verified by ChIP and electrophoretic mobility shift assay (EMSA) [21]. Our qRT-PCR results showed that Prx24 was downregulated in dst, dca1 and DmiR (DCA1 artificial microRNA) leaves (Fig 6C and 6D and S4D Fig) and upregulated in 35S::DST and 35S::DCA1 leaves (S6A and S6B Fig).
Fig. 6. DCA1 acts as an interaction co-activator of DST. 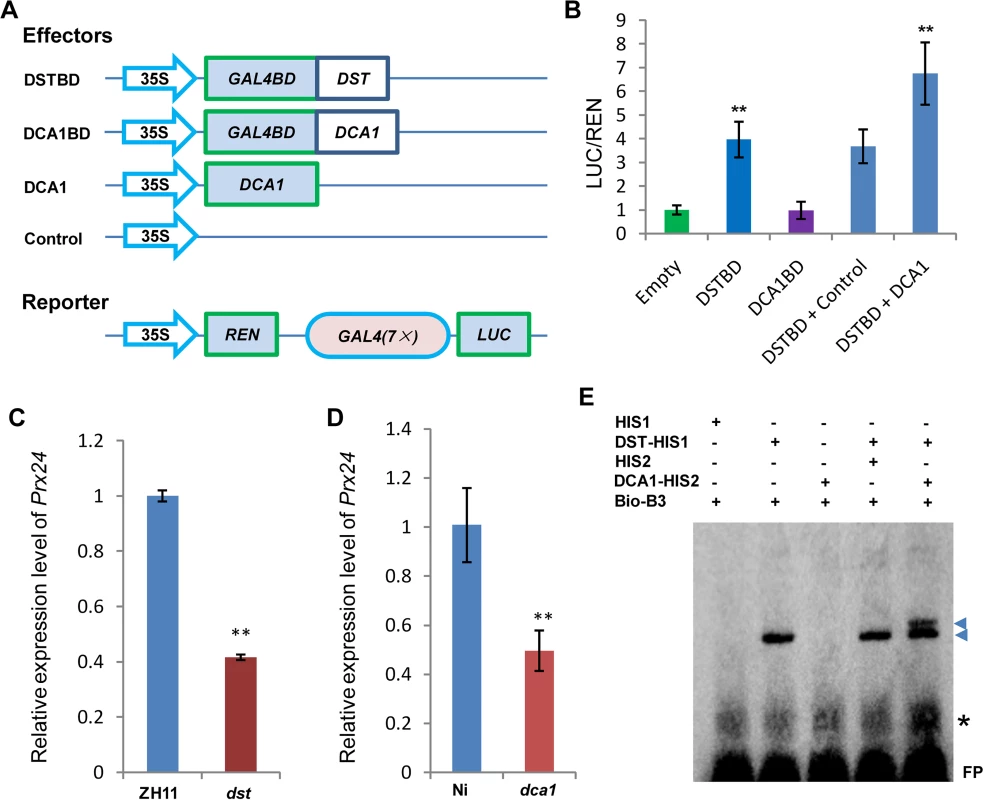
(A) Schematic diagram of constructs used in Dual Luciferase assays. (B) Ratio of firefly luciferase (LUC) to Renilla luciferase (REN) activity in Arabidopsis protoplasts cotransformed with different reporter constructs and effectors shown in (A). The results show that DST acts as a transcriptional activator, while DCA1 enhances the transcriptional activation function of DST. Data represent means ± sd (n = 3). (C) Real-time PCR quantification of the expression of the DST downstream target gene Prx24 in ZH11 and dst. Data represent means ± sd (n = 3). (D) Real-time PCR quantification of the expression of Prx24 in Ni and dca1. Data represent means ± sd (n = 3). (E) EMSA of DST protein with DCA1 protein on biotin-tagged B3 DNA containing the DST binding site sequence. Triangles and asterisks indicate shifted bands and nonspecific binding, respectively. FP, free probe. Significant differences were determined using the Student’s t-test (**P <0.01). Although we know that Prx24 is a downstream gene of this transcriptional regulation module, phenotypes of transgenic plants under stress conditions remain unknown. So we generated overexpression plants of this gene in ZH11. Prx24-overexpressing plants became sensitive to drought and salt stress to an extent comparable to DCA1 and DST-overexpressing plants in a ZH11 background (S6C Fig). We identified the DST binding sequence (DBS), 5’-TGCTANNATTG-3’, using a bacterial one-hybrid system [21], and EMSA results showed that His1-DST but not the His1 tag alone could bind to B3, a DNA fragment containing this conserved site (DBS) (Fig 6E). We next wanted to determine if DCA1 could also bind to the DBS, and further EMSA results revealed that His2-DCA1 and His1-DST could bind to the DBS, while His2-DCA1 alone could not (Fig 6E). It is possible that the B3 sequence that we used did not include a DCA1 binding site, or perhaps DCA1 was not associated with DNA at all. To verify our hypothesis, we used the longer promoter sequence of Prx24 described in our previous research [21], and EMSA results were similar and confirmed that DST but not DCA1 could bind to the fragment (S6D Fig). Together, our combined genetic, physiological and molecular evidence indicated that DCA1 functions as an interaction co-activator in DST-mediated transcriptional programs.
DCA1 Positively Regulates Stomatal Aperture
Since DST negatively regulates stomatal closure to improve drought and salt tolerance [21], we investigated whether stomatal aperture was also altered in 35S::DCA1 and dca1 plants using Cryo scanning electron microscopy (Cryo SEM). Three levels of stomatal opening in different plants were observed (Fig 7A and S7A Fig) and statistical analysis revealed that 6% of stomata were completely closed in 35S::DCA1 plants vs. 21% in CK plants, while 67% were completely open in 35S::DCA1 plants vs. only 48% in CK plants. The percentage of partially open stomata was 27% and 31% in 35S::DCA1 plants and CK plants, respectively (Fig 7B). In contrast, there was no significant difference in stomatal density and guard cell size between 35S::DCA1 and CK plants (Fig 7C and S7B and S7C Fig). Following dehydration stress, 35S::DCA1 plants lost more water than CK plants (Fig 7D). We next investigated stomatal aperture following drought stress, and statistical analysis revealed that 14% of stomata were completely closed in 35S::DCA1 plants vs. 28% in CK plants, while 37% of stomata were completely open in 35S::DCA1 plants vs. only 27% in CK plants (S7F Fig). The percentage of partially open stomata was 49% and 45% in 35S::DCA1 plants and CK plants, respectively (S7F Fig). The ratio of completely closed stomata was 11% vs. 6% in dca1 mutant and Ni (wild-type) plants, and 60% of stomata were completely open in the dca1 mutant compared with 67% in wild-type plants (Fig 7E). The percentage of partially open stomata was similar in dca1 mutant and wild-type plants (Fig 7E). At the same time, low magnification Cryo SEM images showed that no significant changes occurred in stomatal density in dca1 plants (Fig 7F), and there were also no significant changes in guard cell size (S7D and S7E Fig). Meanwhile, the rate of water loss of detached leaves was lower in the dca1 mutant plants than in wild-type plants (Fig 7G). When under drought stress, statistical analysis revealed that 17% of stomata were completely closed in Ni plants vs. 25% in dca1 plants, while 38% of stomata were completely open in Ni plants vs. only 27% in dca1 plants (S7G Fig). The percentage of partially open stomata was 44% and 48% in Ni plants and dca1 plants, respectively (S7G Fig). Together these results suggest that the enhanced drought tolerance observed in the dca1 mutant is mainly due to increased stomatal closure, which minimizes water loss.
Fig. 7. DCA1 affects stomatal aperture. 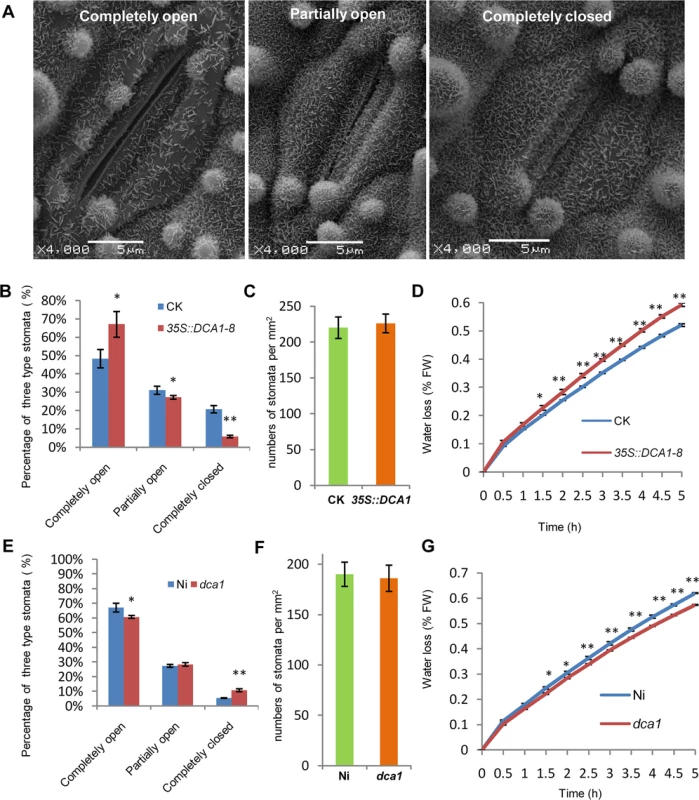
(A) Cryo scanning electron microscopy (Cryo SEM) images of three levels of stomatal opening of ZH11. (B) The percentage of the three levels in CK and 35S::DCA1-8 plants under normal conditions (n = 8 samples, ~10 stomata per sample; 97 stomata for CK; 103 stomata for 35S::DCA1-8). (C) Stomatal density of the middle leaves of CK and 35S::DCA1-8 (data represent means ± sd, n = 10). Three random scopes were used for each repeat (253.3 μm × 253.3 μm for each scope). (D) Water loss in CK and 35S::DCA1-8 leaves (n = 3 biological replicates, each containing 10 fully expanded leaves from 25-day-old plants). (E) The percentage of the three levels in Ni and dca1 plants (n = 8 samples, ~10 stomata per sample; 73 stomata for Ni; 102 stomata for dca1). (F) Stomatal density of the middle leaves of Ni and dca1 (data represent means ± sd, n = 10). Three random scopes were used in each repeat (422.2 μm × 422.2 μm for each scope). (G) Water loss in Ni and the dca1 mutant (n = 3 biological replicates, each containing 10 fully expanded leaves from 25-day-old plants). Data represent means ± sd. Significant differences were determined using the Student’s t-test (*P <0.05, **P <0.01). DCA1 Influenced H2O2 Content in Stomata via Prx24
The phytohormone ABA is known to induce stomatal closure [23], and measurement of the endogenous ABA content revealed no significant difference between dst mutant and wild-type plants [21]. H2O2 can also induce leaf stomatal closure [16, 24, 25], and Huang et al. observed increased stomatal closure in the dst mutant as a result of H2O2 accumulation [21]. We therefore examined the H2O2 content in the stomata of DCA1-overexpressing and mutant plant leaves using the fluorescent dye 2’, 7’-dichlorodihydrofluorescein diacetate (H2DCFDA). We found that overexpression plants contained ~20% less H2O2 in their guard cells than did CK plants (Fig 8A and 8B). However, mutant plants accumulated more H2O2 in their guard cells than did Ni plants (Fig 8C and 8D). To explore the molecular mechanism behind this phenomenon, we isolated guard cell protoplasts (GCP) and mesophyll cell protoplasts (MCP) from ZH11, dst, Ni and dca1 plants to examine the expression levels of DCA1, DST and Prx24. The results revealed higher levels of Prx24 and DST expression in GCPs than MCPs of ZH11, but DCA1 expression was not elevated (Fig 8E). Prx24 was expressed at relatively lower levels in dca1 mutant and dst mutant GCPs than in CK plants (Fig 8F), and since Prx24 functions as a ROS scavenger, these results indicated that the H2O2 content in stomata may be affected by DCA1-DST.
Fig. 8. DCA1 influences H2O2 content in stomata. 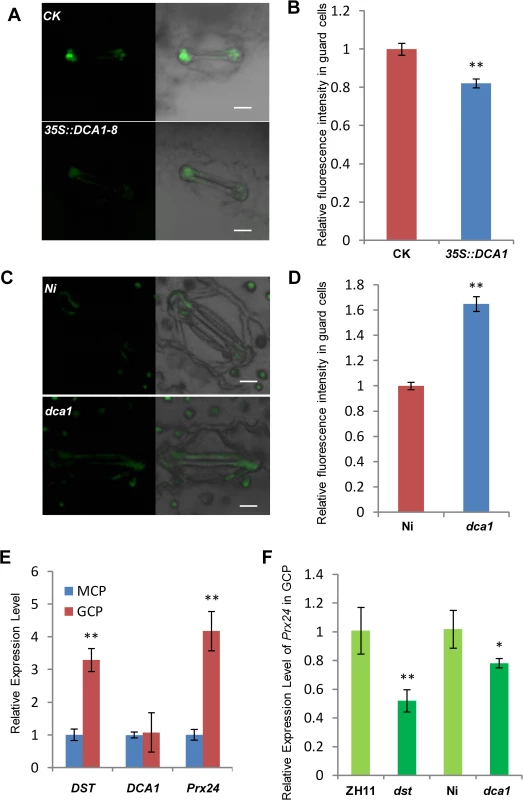
(A) H2O2 levels in guard cells of CK and 35S::DCA1-8 plants labeled with H2DCFDA. (B) Quantitative analysis of H2O2 production in the guard cells of CK and 35S::DCA1-8 plants (n = 8 leaves, ~10 stomata per leaf; 86 stomata for CK; 79 stomata for 35S::DCA1-8). Data represent means ± s.e.m. (n = 8). (C) H2O2 production in guard cells of Ni and dca1 plants labeled with H2DCFDA. (D) Quantitative analysis of H2O2 production in the guard cells of Ni and dca1 plants (n = 8 leaves, ~10 stomata per leaf; 77 stomata for Ni; 92 stomata for dca1). Data represent means ± sem (n = 8). (E) Expression levels of DST, DCA1 and Prx24 in guard cell protoplasts (GCPs) and mesophyll cell protoplasts (MCPs) of ZH11 normalized against actin. Data represent means ± sd (n = 3). (F) Expression levels of Prx24 in GCPs from ZH11, dst, Ni and dca1. Data represent means ± sd (n = 3). Significant differences were determined using the Student’s t-test (**P <0.01). DST Physically Interacts with Itself and Its Expression Is Affected by Abiotic Stress
While carefully observing the phenotypes of DST-complemented (DSTpro::DST-GFP in dst) plants under normal and stress conditions, we found that the dst mutant was not fully complemented. Specifically, the phenotype of DSTpro::DST-GFP under stress conditions was intermediate between those of wild-type and the dst mutant plants (Fig 9A–9D). We hypothesized that perhaps the mutated form of DST exhibiting reduced transcriptional activity could still function in the transcriptional complex. A BiFC assay in Arabidopsis protoplasts revealed that DST interacts with itself in vivo (Fig 9E), and an in vitro pull-down assay showed that MBP-DST was able to pull down the His2-DST protein (Fig 9F and S8A Fig). Meanwhile, the mutant site of DST did not influence its dimerization (Fig 9G). These results confirm that our hypothesis was correct, and indicate that DST functions as a dimer. In addition, the complex containing the mutant isoform could not fully execute its transactivation function, which may explain why the dst mutant was not fully complemented. We also examined whether DCA1 functions in the same manner with DST, and pull-down assays revealed that it does not (S8B Fig). Further analysis revealed that the DST-DST interaction was negatively influenced by stress in Arabidopsis protoplast cells (S9A, S9B and S9E Fig). This may be due to decreased levels of DST monomer or decreased DST dimerization. To investigate further, DST self-promoter promoted DST-nYFP and DST-cYFP were cotransformed into Arabidopsis protoplasts and cultured for 9 h under normal conditions. Half of the cells were maintained in normal conditions, while the other half were treated with NaCl and cycloheximide (CHX), an inhibitor of protein biosynthesis. After 3 h, the relative fluorescence ratios of the cells were similar (S9C, S9D and S9F Fig). These results demonstrate that DST was inhibited by stress, but not degraded or dissociated into monomeric form, and this was verified by qRT-PCR experiments (S2B and S2C Fig). These results, when combined with those from the EMSA experiments (Fig 6E), indicate that DST and DCA1 may form a putative heterotetrameric complex that participates in gene regulation.
Fig. 9. DST functions as a dimer. 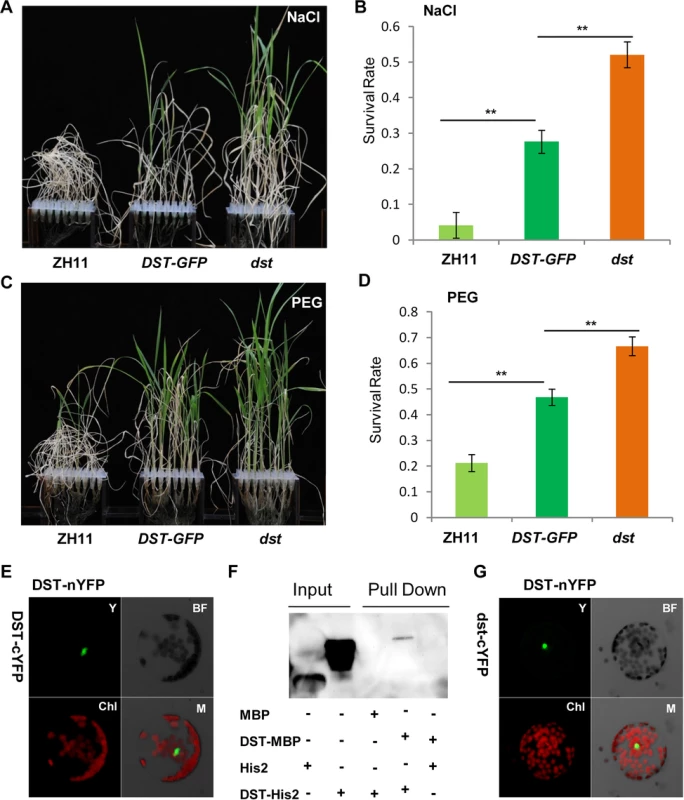
(A) 12-day-old ZH11, DST-GFP (DST:pro::DST-GFP in dst background) and dst seedlings treated with 100 mM NaCl for 10 days and recovered for 7 days. (B) Survival percentage after NaCl treatment (n = 3 biological replicates, 24 plants of each replicate). Data represent means ± sd (n = 3). (C) 12-day-old ZH11, DST-GFP and dst seedlings treated with 18% PEG for 12 days and recovered for 7 days. (D) Survival percentage after PEG treatment (n = 3 biological replicates, 24 plants of each replicate). Data represent means ± sd (n = 3). (E) BiFC assay in Arabidopsis protoplasts revealing DST dimerization. Y, YFP; BF, Bright Field; Chl, Chlorophyll; M, Merge. (F) In vitro pull-down of His2-tagged DST protein through MBP-tagged DST protein detected by immunoblotting with anti-His antibody. (G) Mutant isoform of DST do not affect dimerization, consistent with the BiFC assay results. Y, YFP; BF, Bright Field; Chl, Chlorophyll; M, Merge. Significant differences were determined using the Student’s t-test (**P <0.01). Discussion
In this study, we identified the DST interacting protein DCA1. Overexpression of DCA1 increased the plant’s sensitivity to abiotic stress, and 35S::DST plants were also highly sensitive to salt and drought stress. Knockdown of DCA1 improved the tolerance to stress, as also observed in the dst mutant. These results indicate that DCA1 has a positive effect on the transcriptional activity of DST, which was confirmed by dual luciferase assays. The qRT-PCR results showed that DST was suppressed under stress conditions while DCA1 was induced (Fig 2E and 2F and S2B and S2C Fig). Hsu, et al. (2014) identified ZFP34, a homolog of DCA1, and discovered that ZFP34 participates in the heat stress response. Overexpression of this gene in rice and its homolog in Arabidopsis increased stomatal opening, and ZFP34 mutants exhibited decreased stomatal opening in both rice and Arabidopsis [22]. Heat stress is usually accompanied by drought, since transpiration is induced when plants experience high temperatures. DCA1 expression is therefore likely to be upregulated by drought, salt and heat stress. Water loss caused by increased transpiration may aggravate the impact of drought stress, and plants have evolved very sophisticated molecular mechanisms to overcome this paradoxical phenomenon. One possible molecular mechanism may be the DCA1-DST pathway, since DCA1 may induce the opening of stomata to reduce the internal temperature during heat stress. At the same time downregulation of DST may cause closure of stomata to prevent excessive moisture loss. This seemingly contradictory phenomenon ensures that the plant can adapt to various environmental changes in different ways, which builds robustness into the plant responses to abiotic stress.
The DCA1 protein contains two zinc ion binding sites, a CHY zinc finger domain and a RING-H2 domain. The CHY domain is highly conserved among homologous proteins from different species, but the RING-H2 domain is not highly conserved. Therefore, the CHY domain may be the major functional part of this protein family, but the exact function was unknown until now. In the present study, we found that DCA1 functions as a transcriptional co-activator. However, the means by which DCA1 promotes the activity of DST is not yet elucidated. DCA1 may alter the conformation of DST through the protein-protein interaction, or may help DST to recruit other transcription initiation factors such as RNA polymerase, or may even function as a regulator of chromatin structure. Many RING finger domain proteins function as ubiquitin ligases, although many are also involved in other roles such as Breast Cancer 1 (BRCA1) which participates in transcriptional regulation, DNA damage repair and chromatin remodeling [26, 27]. Mex-3 homolog D (MEX3D) is an RNA binding protein in C. elegans that has a negative regulatory action on Bcl-2 expression at the posttranscriptional level [28], and peroxisome biogenesis factor 10 (PEX10) is involved in import of peroxisomal matrix proteins [29]. Thus, DCA1-like proteins may also perform a range of different functions.
Previous studies focusing on H2O2 signalling in guard cells during stress conditions have indicated that NADPH oxidases such as Atrboh D and Atrboh H may play major roles in this process [10]. Indeed, the atrbohD/F double mutant exhibited reduced stomatal closure and ROS production compared with wild-type plants. We previously identified the transcription factor DST that binds to the DBS element to regulate the expression of H2O2 metabolism-associated genes in stomata, and extended this work by identifying the DST interacting CHY zinc finger protein DCA1 in the present study. This transcriptional complex appears to regulate the expression of Prx 24, an H2O2 scavenger that we found to be more highly expressed in guard cells than in mesophyll cells. Our results reveal how plants prolong H2O2 signalling by reducing the catabolism of H2O2 through the DCA1-DST-Prx24 pathway. Based on these results, we propose a simple model to explain the role of the DCA1-DST-Prx24 pathway in regulating the status of guard cells and stress tolerance. In this model, DCA1 functions as an interacting co-activator of DST to form a heterotetrameric complex that regulates H2O2 homeostasis and stomatal aperture, ultimately affecting stress tolerance (Fig 10).
Fig. 10. Working model for the role of the DCA1-DST transcriptional regulation complex in plants. 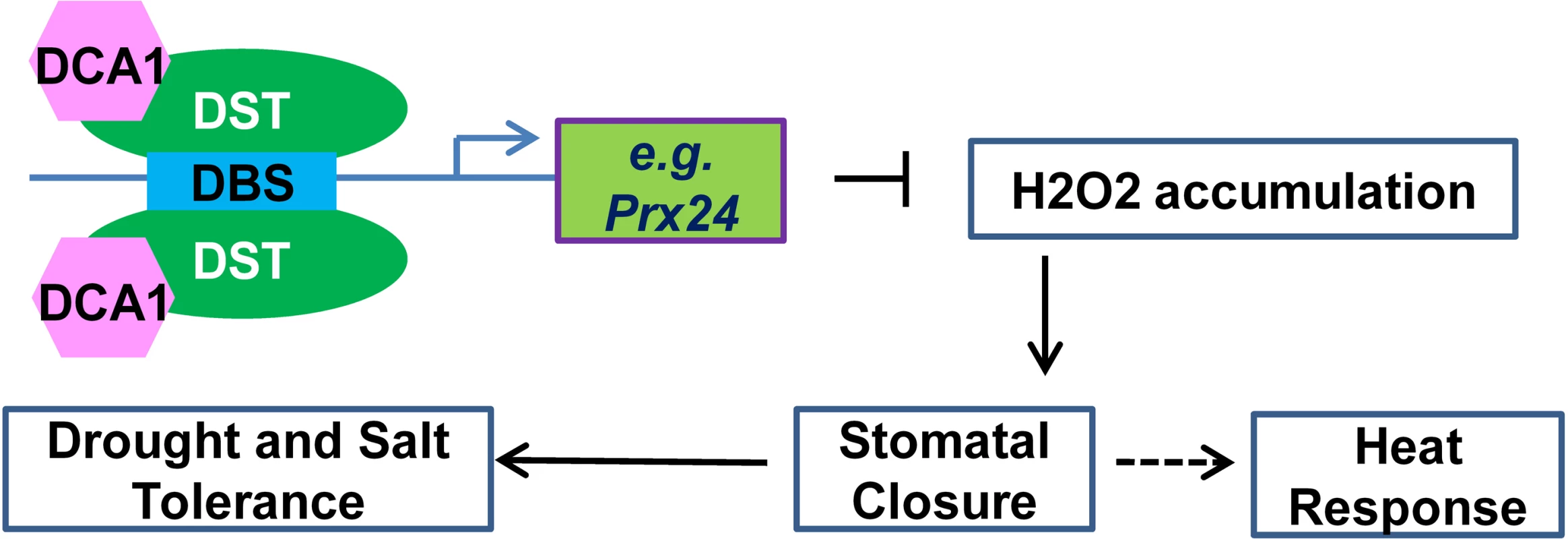
The DCA1 and DST heterotetrameric complex binds to the DBS element in the promoters of genes associated with H2O2 homeostasis in stomata such as Prx24, thereby influencing stomatal closure and ultimately regulating abiotic stress response. Drought and salinity have had devastating effects on food production throughout human history, and the problems are increasing with the growing population. During 2010–2012, drought occurred on most continents, and millions of hectares of crops were destroyed, from wheat in Europe to corn in the US to rice in China [30, 31]. In 2014, the state of California in the US suffered its worst drought on record [32]. Engineering the stomatal response to improve water use efficiency represents a powerful tool for enhancing drought tolerance in plants. Indeed, the constitutive reduction in stomatal opening in the rice dca1 and dst mutant enhances drought tolerance. Given the high conservation of DCA1 among plants, our research suggests that inhibiting the functional association of DCA1-DST may provide one route to unlocking improved drought and salt tolerance in a variety of crops.
Materials and Methods
Plant Materials and Growth Conditions
All rice genetic stocks used in this study were in the Zhonghua 11 (ZH11) japonica variety background except for dca1, which is in the Nipponbare (japonica variety) background. 35S::DCA1 plants were generated via transformation with a construct produced by inserting an amplified ORF fragment containing DCA1 from ZH11 cDNA between the CaMV35S promoter and the 3’OCS terminator. 35S::DST plants were generated using a similar method. The dca1 mutant (NF7038) was obtained from the Rice Genome Resource Center.
We generated knockdown transgenic plants using an artificial microRNA (designed by http://wmd3.weigelworld.org/cgi-bin/webapp.cgi). Target site on DCA1 was 5’ - acgtgatgagagcgcatcatc-3’. The construction method was modified from Norman Warthmann et al. [33]. For the miRNA construct, three modification PCRs were performed with primers G-11491 + DmiR II, DmiR I + DmiR IV and DmiR III + G-11494 on ZH11 cDNA as template, yielding three fragments, respectively (S1 Table). The three resulting fragments were gel purified (Qiagen) and then fused by one PCR with the two flanking primers G-11491 + G-11494 on a mixture of 1 ul from each of the previous three PCRs as template. The fusion product was again gel purified (Qiagen), cloned into the overexpression vector to generate DCA1 knockdown transgenic plants.
Seeds were submerged in water at room temperature for 48 h, followed by germination for 18 h at 37°C. Seeds were then sown in a bottomless 96-well plate that was placed in a container of Yoshida’s culture solution and incubated in a growth chamber under a 13 h light (25°C)/11 h dark (23°C) photoperiod. For salt treatment, 20-day-old seedlings were transferred to a culture solution containing 100 mM NaCl. For PEG treatment, 20-day-old seedlings were transferred to a culture solution containing 18% (w/v) PEG4000. For soil drought experiments, 2-week-old seedlings that were cultured in the growth chamber were transplanted to containers with soil and grown in a greenhouse at 24–30°C and 50–60% relative humidity. About 50 days later, water was removed from the containers for the dehydration treatment.
Yeast Two-Hybrid Assays
Screening for DST interacting proteins was performed according to the manufacturer’s instructions (Clontech). Briefly, libraries were prepared using total RNA extracted from ZH11 leaves that were approximately 20 days old. The DST fragment lacking the self-activation domain (amino acids 1–72) was cloned into the pGBKT7 BD vector to generate pGBKT7/△72 (S1 Table). Screening was performed on SD/–Leu/–Trp medium containing 2.5 mM 3-aminotriazole (3-AT). To confirm positive interactions, full-length ORFs of each target gene were cloned into the pGADT7 AD vector and cotransformed with pGBKT7/△72 into Y2HGold Competent Cells. To verify positive interactions, the full-length DST ORF was cloned into the pGADT7 AD vector, and target genes were cloned into the pGBKT7 BD vector, and Y2HGold Competent Cells were cotransformed with 100 ng of each vector pair. Cells were grown at 30°C for 3 days on SD/–Ade/–His/–Leu/–Trp medium containing 1–10 mM 3-AT (depending on the bait—prey combination).
Transient Expression
DST (GQ178286) and DCA1 (Os10g0456800) coding regions were cloned into YFP expression vectors, BiFC vectors or dual luciferase system vectors. For subcellular localization experiments, 10 μg of YFP-DST, YFP-DCA1 or YFP expression plasmid was transformed into Arabidopsis protoplasts and observed under a confocal microscope (Carl Zeiss). For BiFC experiments, 5 μg each of YFP-N - and YFP-C-tagged protein expression vector was cotransformed. For the dual luciferase transient transcription activity assay, a 7× GAL4 binding sequence was inserted into the pGreenII 0800-LUC vector to generate the LUC reporter construct. The coding sequence of DST was inserted into pGreen GAL4BD to produce the effector construct, and DCA1 was inserted into pGreenII 62-SK (S1 Table). Firefly LUC and Renilla luciferase (REN) activities were measured using the dual luciferase reporter assay system (Promega). The LUC:REN ratio represents transcriptional activity.
Protein Expression
The DST ORF was inserted into pET32a, pCold TF and pMAL-c5x vectors. The DCA1 coding sequence was inserted into pCold TF and pMAL-c5x (S1 Table). The tag in pET32a was designated His1 and that in pCold TF was designated His2. The pMAL-c5x constructs were transformed into TB1 competent cells, and pET32a and pCold TF vectors were transformed into DE3 competent cell.
Pull-Down Assay
The pull-down assay was performed according to the instructions for the MagneGST Pull-Down System (Promega) with some modifications. Briefly, E. coli cells expressing DST-MBP and MBP were lysed with BugBuster Protein Extraction Reagents (Novagen) and centrifuged. The supernatant was incubated with Anti-MBP Magnetic Beads (NEB) for 30 min at 4°C and washed five times with MBP column buffer. Then, supernatants containing DCA1-His2 and His2 proteins were incubated with approximately the same amount of MBP and DST-MBP binding beads for 2 h at 4°C. The magnetic beads were then washed five times with MBP column buffer. The mixture was resuspended in SDS loading buffer, boiled for 3 min, separated by 10% SDS-PAGE and immunoblotted with anti-MBP antibody for target proteins and anti-His antibody for pull-down proteins (CWbiotech).
RNA Extraction and Real-Time PCR Analysis
Leaves from 2-week-old seedlings were harvested and total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. 1–2 μg of total RNA was used for cDNA synthesis with oligo (dT) primer and Superscript Reverse Transcriptase (Invitrogen). Quantitative RT-PCR was performed using FastStart Universal SYBR Green Master (Roche) and an ABI 7300 Real-time PCR System (Applied Biosystems). The expression of DST, DCA1 and Prx24 was normalized against actin. Oligonucleotide primers are listed in S1 Table.
Cryo Scanning Electron Microscopy (Cryo SEM)
Leaves from 20-day-old seedlings grown under normal conditions or treated with 12% PEG for 6 hours were detached and frozen in liquid nitrogen. Images of stomata were obtained using a Cryo (QUORUM) scanning electron microscopy (JEOL) system (Cryo SEM).
Measurement of H2O2 in Guard Cells
H2O2 production in guard cells was detected using H2DCFDA (Molecular Probes) as described by Huang [21]. Epidermal strips were peeled from leaves of 20-day-old rice seedlings using tissue forceps, washed with loading buffer (10 mM Tris-HCl, 50 mM KCl, pH 7.2) and incubated in staining buffer (loading buffer containing 50 mM H2DCFDA) for 10 min at 25°C in the dark. Strips were washed twice with distilled water to remove excess staining buffer, and fluorescence was measured using a confocal laser-scanning microscope (Olympus) with excitation at 488 nm and emission at 525 nm. All images were taken under identical conditions. To quantify fluorescence, the guard cell region was selected and the mean fluorescence value was calculated using the microscope’s software. Each sample contained at least eight independent leaves, and approximately 10 randomly selected stomata were analyzed per leaf.
Isolation of Mesophyll Cell Protoplasts (MCP) and Guard Cell Protoplasts (GCP) from Rice Leaves
30 fully expanded one-month-old rice leaves were cut into ~0.3 cm sections and blended with 200 mL Lysis buffer (5 mM CaCl2, 0.5 mM ascorbic acid, 0.1% PVP-40, 10 mM MES, pH 6.0) for 90 s using a blender (Waring blender model LB20ES). The blended material was filtered through a 200 μm nylon mesh, washed once with 200 ml ddH2O, suspended in 100 ml suspension buffer (0.25 M mannitol, 1mM CaCl2, 0.5mM ascorbic acid), filtered through a 30 μm nylon mesh and transferred into a flask with 25 mL of an enzyme mixture containing 0.7% Cellulysin cellulase, Trichoderma viride (Sigma), 0.1% (w/v) PVP-40, 0.25% BSA in 55% (v/v) basic medium (5 mM MES, 0.5 mM CaCl2, 0.5 mM MgCl2, 10 μM KH2PO4, 0.5 mM ascorbic acid (Sigma), 0.55 M sorbitol, pH 5.5), and 45% (v/v) deionized water. The flask was placed in a shaking water bath at 24°C with the shaking speed set to 120 rpm. After 3 h of digestion, 75 mL basic medium was added, and the mixture was shaken for an additional 5 min. The mixture was filtered through a 70 μm nylon mesh and washed twice with 50 mL basic medium. MCPs were collected by centrifuging the filtrate at 200 g for 5 min. Epidermal fragments were transferred to another flask with 50 mL of basic medium (1.5% Cellulase RS (Yakult, Japan), 0.02% Pectolyase Y23 (Yakult, Japan), 0.25% BSA, pH 5.5) and incubated at 24°C in a shaking water bath (100 rpm). After incubation for 3 h, the mixture was filtered through a 30 μm nylon mesh, washed twice with 50 mL basic medium, and GCPs were collected at 200 g for 5 min. Two different transcription inhibitors, actinomycin D and cordycepin, were used to prevent changes in gene expression during protoplast isolation.
EMSA
EMSA was performed as described by Huang [21] using His1 tag, DST-His1, His2 tag and DCA1-His2 recombinant fusion proteins. Biotin-labeled oligonucleotides were obtained by PCR amplification using biotin-labeled primers (Invitrogen). The binding reactions (containing biotin-labeled DNA and different combinations of proteins) were performed with a Light Shift Chemiluminescent EMSA kit (Pierce) according to the manufacturer’s instructions. Gel electrophoresis was performed on a 10% native polyacrylamide gel (79 : 1 acryl/bis). After blotting onto a nylon membrane, DNA was attached to the membrane using a UV light cross-linker instrument. Blots were imaged using a Tanon 5200 imaging system (bioTanon).
Sequence Alignment and Phylogenic Analysis
Peptide sequences were obtained from Phytozome (Phytozome v9.1, http://www.phytozome.net/) and sequence alignment was performed using the Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/).
Supporting Information
Zdroje
1. Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, et al. (2010) Food Security: The Challenge of Feeding 9 Billion People. Science 327 : 812–818. doi: 10.1126/science.1185383 20110467
2. Simmonds NW (1998) The doubly green revolution: Food for all in the 21st century. Nature 391 : 139–139.
3. Lobell DB, Roberts MJ, Schlenker W, Braun N, Little BB, et al. (2014) Greater Sensitivity to Drought Accompanies Maize Yield Increase in the US Midwest. Science 344 : 516–519. doi: 10.1126/science.1251423 24786079
4. Yuan F, Yang HM, Xue Y, Kong DD, Ye R, et al. (2014) OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514 : 367-+. doi: 10.1038/nature13593 25162526
5. Dodd AN, Kudla J, Sanders D (2010) The Language of Calcium Signaling. Annual Review of Plant Biology, 61 : 593–620. doi: 10.1146/annurev-arplant-070109-104628 20192754
6. Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, et al. (2009) Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 324 : 1064–1068. doi: 10.1126/science.1172408 19407143
7. Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, et al. (2009) Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science 324 : 1068–1071. doi: 10.1126/science.1173041 19407142
8. Fujii H, Zhu JK (2012) Osmotic stress signaling via protein kinases. Cellular and Molecular Life Sciences 69 : 3165–3173. doi: 10.1007/s00018-012-1087-1 22828864
9. Pei ZM, Murata Y, Benning G, Thomine S, Klusener B, et al. (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406 : 731–734. 10963598
10. Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, et al. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. Embo Journal 22 : 2623–2633. 12773379
11. Zhang X, Miao YC, An GY, Zhou Y, Shangguan ZP, et al. (2001) K+ channels inhibited by hydrogen peroxide mediate abscisic acid signaling in Vicia guard cells. Cell Research 11 : 195–202. 11642404
12. Sutter JU, Sieben C, Hartel A, Eisenach C, Thiel G, et al. (2007) Abscisic acid triggers the endocytosis of the Arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Current Biology 17 : 1396–1402. 17683934
13. Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, et al. (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452 : 487–U415. doi: 10.1038/nature06608 18305484
14. Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. Journal of Plant Research 124 : 509–525. doi: 10.1007/s10265-011-0412-3 21416314
15. Song YW, Miao YC, Song CP (2014) Behind the scenes: the roles of reactive oxygen species in guard cells. New Phytologist 201 : 1121–1140. doi: 10.1111/nph.12565 24188383
16. McAinsh MR, Clayton H, Mansfield TA, Hetherington AM (1996) Changes in stomatal behavior and guard cell cytosolic free calcium in response to oxidative stress. Plant Physiology 111 : 1031–1042. 12226345
17. Schaefer U, Schmeier S, Bajic VB (2011) TcoF-DB: dragon database for human transcription co-factors and transcription factor interacting proteins. Nucleic Acids Research 39: D106–D110. doi: 10.1093/nar/gkq945 20965969
18. Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, et al. (2014) Control of plant stem cell function by conserved interacting transcriptional regulators. Nature. 517 : 377–380. doi: 10.1038/nature13853 25363783
19. Giuntoli B, Lee SC, Licausi F, Kosmacz M, Oosumi T, et al. (2014) A Trihelix DNA Binding Protein Counterbalances Hypoxia-Responsive Transcriptional Activation in Arabidopsis. Plos Biology 12.
20. Hu HH, Dai MQ, Yao JL, Xiao BZ, Li XH, et al. (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences of the United States of America 103 : 12987–12992. 16924117
21. Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, et al. (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes & Development 23 : 1805–1817.
22. Hsu KH, Liu CC, Wu SJ, Kuo YY, Lu CA, et al. (2014) Expression of a gene encoding a rice RING zinc-finger protein, OsRZFP34, enhances stomata opening. Plant Molecular Biology 86 : 125–137. doi: 10.1007/s11103-014-0217-6 25002225
23. Schroeder JI, Kwak JM, Allen GJ (2001) Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410 : 327–330. 11268200
24. Hamilton DWA, Hills A, Kohler B, Blatt MR (2000) Ca2+ channels at the plasma membrane of stomatal guard cells are activated by hyperpolarization and abscisic acid. Proceedings of the National Academy of Sciences of the United States of America 97 : 4967–4972. 10781106
25. Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ (2006) ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant Journal 45 : 113–122. 16367958
26. Starita LM, Parvin JD (2003) The multiple nuclear functions of BRCA1: transcription, ubiquitination and DNA repair. Current Opinion in Cell Biology 15 : 345–350. 12787778
27. Rosen EM, Fan SJ, Ma YX (2006) BRCA1 regulation of transcription. Cancer Letters 236 : 175–185. 15975711
28. Donnini M, Lapucci A, Papucci L, Witort E, Jacquier A, et al. (2004) Identification of TINO—A new evolutionarily conserved BCL-2 Au-rich element RNA-binding protein. Journal of Biological Chemistry 279 : 20154–20166. 14769789
29. Warren DS, Morrell JC, Moser HW, Valle D, Gould SJ (1998) Identification of PEX10, the gene defective in complementation group 7 of the peroxisome-biogenesis disorders. American Journal of Human Genetics 63 : 347–359. 9683594
30. Grayson M (2013) Agriculture and drought. Nature. England. pp. S1.
31. Cook BI, Smerdon JE, Seager R, Coats S (2014) Global warming and 21st century drying. Climate Dynamics 43 : 2607–2627.
32. Wang SY, Hipps L, Gillies RR, Yoon JH (2014) Probable causes of the abnormal ridge accompanying the 2013–2014 California drought: ENSO precursor and anthropogenic warming footprint. Geophysical Research Letters 41 : 3220–3226.
33. Warthmann N, Chen H, Ossowski S, Weigel D, Herve P (2008) Highly Specific Gene Silencing by Artificial miRNAs in Rice. Plos One 3:e1829. doi: 10.1371/journal.pone.0001829 18350165
Štítky
Genetika Reprodukčná medicína
Článek Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation DevelopmentČlánek A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor PelotaČlánek A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in OvulationČlánek Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAsČlánek FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2Článek Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion inČlánek The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly SiteČlánek Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem CellsČlánek A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal ErythropoiesisČlánek Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation inČlánek Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in OocytesČlánek MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Gene-Regulatory Logic to Induce and Maintain a Developmental Compartment
- A Decad(e) of Reasons to Contribute to a PLOS Community-Run Journal
- DNA Methylation Landscapes of Human Fetal Development
- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development
- Transcriptional Derepression Uncovers Cryptic Higher-Order Genetic Interactions
- Silencing of X-Linked MicroRNAs by Meiotic Sex Chromosome Inactivation
- Virus Satellites Drive Viral Evolution and Ecology
- A Novel Route Controlling Begomovirus Resistance by the Messenger RNA Surveillance Factor Pelota
- Sequence to Medical Phenotypes: A Framework for Interpretation of Human Whole Genome DNA Sequence Data
- Your Data to Explore: An Interview with Anne Wojcicki
- Modulation of Ambient Temperature-Dependent Flowering in by Natural Variation of
- The Ciliopathy Protein CC2D2A Associates with NINL and Functions in RAB8-MICAL3-Regulated Vesicle Trafficking
- PPP2R5C Couples Hepatic Glucose and Lipid Homeostasis
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
- Intermediate Levels of CodY Activity Are Required for Derepression of the Branched-Chain Amino Acid Permease, BraB
- "Missing" G x E Variation Controls Flowering Time in
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish
- Loss of a Conserved tRNA Anticodon Modification Perturbs Plant Immunity
- Genome-Wide Association Analysis of Adaptation Using Environmentally Predicted Traits
- Oriented Cell Division in the . Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1
- Disproportionate Contributions of Select Genomic Compartments and Cell Types to Genetic Risk for Coronary Artery Disease
- A Follicle Rupture Assay Reveals an Essential Role for Follicular Adrenergic Signaling in Ovulation
- The RNAPII-CTD Maintains Genome Integrity through Inhibition of Retrotransposon Gene Expression and Transposition
- Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs
- Allelic Variation of Cytochrome P450s Drives Resistance to Bednet Insecticides in a Major Malaria Vector
- SCARN a Novel Class of SCAR Protein That Is Required for Root-Hair Infection during Legume Nodulation
- IBR5 Modulates Temperature-Dependent, R Protein CHS3-Mediated Defense Responses in
- NINL and DZANK1 Co-function in Vesicle Transport and Are Essential for Photoreceptor Development in Zebrafish
- Decay-Initiating Endoribonucleolytic Cleavage by RNase Y Is Kept under Tight Control via Sequence Preference and Sub-cellular Localisation
- Large-Scale Analysis of Kinase Signaling in Yeast Pseudohyphal Development Identifies Regulation of Ribonucleoprotein Granules
- FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2
- LINE-1 Mediated Insertion into (Protein of Centriole 1 A) Causes Growth Insufficiency and Male Infertility in Mice
- Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in
- Genome-Scale Mapping of σ Reveals Widespread, Conserved Intragenic Binding
- Uncovering Hidden Layers of Cell Cycle Regulation through Integrative Multi-omic Analysis
- Functional Diversification of Motor Neuron-specific Enhancers during Evolution
- The GTP- and Phospholipid-Binding Protein TTD14 Regulates Trafficking of the TRPL Ion Channel in Photoreceptor Cells
- The Gyc76C Receptor Guanylyl Cyclase and the Foraging cGMP-Dependent Kinase Regulate Extracellular Matrix Organization and BMP Signaling in the Developing Wing of
- The Ty1 Retrotransposon Restriction Factor p22 Targets Gag
- Functional Impact and Evolution of a Novel Human Polymorphic Inversion That Disrupts a Gene and Creates a Fusion Transcript
- The Dedicated Chaperone Acl4 Escorts Ribosomal Protein Rpl4 to Its Nuclear Pre-60S Assembly Site
- The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study
- Parent-of-Origin Effects of the Gene on Adiposity in Young Adults
- Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells
- Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor
- A Systems Approach Identifies Essential FOXO3 Functions at Key Steps of Terminal Erythropoiesis
- Protein O-Glucosyltransferase 1 (POGLUT1) Promotes Mouse Gastrulation through Modification of the Apical Polarity Protein CRUMBS2
- KIF7 Controls the Proliferation of Cells of the Respiratory Airway through Distinct Microtubule Dependent Mechanisms
- Integration of Posttranscriptional Gene Networks into Metabolic Adaptation and Biofilm Maturation in
- Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Oocytes
- Protein Homeostasis Imposes a Barrier on Functional Integration of Horizontally Transferred Genes in Bacteria
- A New Method for Detecting Associations with Rare Copy-Number Variants
- Histone H2AFX Links Meiotic Chromosome Asynapsis to Prophase I Oocyte Loss in Mammals
- The Genomic Aftermath of Hybridization in the Opportunistic Pathogen
- A Role for the Chaperone Complex BAG3-HSPB8 in Actin Dynamics, Spindle Orientation and Proper Chromosome Segregation during Mitosis
- Establishment of a Developmental Compartment Requires Interactions between Three Synergistic -regulatory Modules
- Regulation of Spore Formation by the SpoIIQ and SpoIIIA Proteins
- Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes
- Alkaline Ceramidase 3 Deficiency Results in Purkinje Cell Degeneration and Cerebellar Ataxia Due to Dyshomeostasis of Sphingolipids in the Brain
- ACLY and ACC1 Regulate Hypoxia-Induced Apoptosis by Modulating ETV4 via α-ketoglutarate
- Quantitative Differences in Nuclear β-catenin and TCF Pattern Embryonic Cells in .
- HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse
- Axon Regeneration Is Regulated by Ets–C/EBP Transcription Complexes Generated by Activation of the cAMP/Ca Signaling Pathways
- A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- The Roles of CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs
- A Genetic Cascade of Modulates Nucleolar Size and rRNA Pool in
- Inter-population Differences in Retrogene Loss and Expression in Humans
- Cationic Peptides Facilitate Iron-induced Mutagenesis in Bacteria
- EP4 Receptor–Associated Protein in Macrophages Ameliorates Colitis and Colitis-Associated Tumorigenesis
- Fungal Infection Induces Sex-Specific Transcriptional Changes and Alters Sexual Dimorphism in the Dioecious Plant
- FLCN and AMPK Confer Resistance to Hyperosmotic Stress via Remodeling of Glycogen Stores
- MET18 Connects the Cytosolic Iron-Sulfur Cluster Assembly Pathway to Active DNA Demethylation in
- Sex Bias and Maternal Contribution to Gene Expression Divergence in Blastoderm Embryos
- Transcriptional and Linkage Analyses Identify Loci that Mediate the Differential Macrophage Response to Inflammatory Stimuli and Infection
- Mre11 and Blm-Dependent Formation of ALT-Like Telomeres in Ku-Deficient
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICE of
- The Type VI Secretion TssEFGK-VgrG Phage-Like Baseplate Is Recruited to the TssJLM Membrane Complex via Multiple Contacts and Serves As Assembly Platform for Tail Tube/Sheath Polymerization
- The Dynamic Genome and Transcriptome of the Human Fungal Pathogen and Close Relative
- Secondary Structure across the Bacterial Transcriptome Reveals Versatile Roles in mRNA Regulation and Function
- ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Regeneration
- Pelle Modulates dFoxO-Mediated Cell Death in
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Single Strand Annealing Plays a Major Role in RecA-Independent Recombination between Repeated Sequences in the Radioresistant Bacterium
- The Rise and Fall of an Evolutionary Innovation: Contrasting Strategies of Venom Evolution in Ancient and Young Animals
- Genome Wide Identification of SARS-CoV Susceptibility Loci Using the Collaborative Cross
- DCA1 Acts as a Transcriptional Co-activator of DST and Contributes to Drought and Salt Tolerance in Rice
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



