-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Strategies to Avoid Killing by Human Neutrophils
Staphylococcus epidermidis is a leading nosocomial pathogen. In contrast to its more aggressive relative S. aureus, it causes chronic rather than acute infections. In highly virulent S. aureus, phenol-soluble modulins (PSMs) contribute significantly to immune evasion and aggressive virulence by their strong ability to lyse human neutrophils. Members of the PSM family are also produced by S. epidermidis, but their role in immune evasion is not known. Notably, strong cytolytic capacity of S. epidermidis PSMs would be at odds with the notion that S. epidermidis is a less aggressive pathogen than S. aureus, prompting us to examine the biological activities of S. epidermidis PSMs. Surprisingly, we found that S. epidermidis has the capacity to produce PSMδ, a potent leukocyte toxin, representing the first potent cytolysin to be identified in that pathogen. However, production of strongly cytolytic PSMs was low in S. epidermidis, explaining its low cytolytic potency. Interestingly, the different approaches of S. epidermidis and S. aureus to causing human disease are thus reflected by the adaptation of biological activities within one family of virulence determinants, the PSMs. Nevertheless, S. epidermidis has the capacity to evade neutrophil killing, a phenomenon we found is partly mediated by resistance mechanisms to antimicrobial peptides (AMPs), including the protease SepA, which degrades AMPs, and the AMP sensor/resistance regulator, Aps (GraRS). These findings establish a significant function of SepA and Aps in S. epidermidis immune evasion and explain in part why S. epidermidis may evade elimination by innate host defense despite the lack of cytolytic toxin expression. Our study shows that the strategy of S. epidermidis to evade elimination by human neutrophils is characterized by a passive defense approach and provides molecular evidence to support the notion that S. epidermidis is a less aggressive pathogen than S. aureus.
Published in the journal: Strategies to Avoid Killing by Human Neutrophils. PLoS Pathog 6(10): e32767. doi:10.1371/journal.ppat.1001133
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001133Summary
Staphylococcus epidermidis is a leading nosocomial pathogen. In contrast to its more aggressive relative S. aureus, it causes chronic rather than acute infections. In highly virulent S. aureus, phenol-soluble modulins (PSMs) contribute significantly to immune evasion and aggressive virulence by their strong ability to lyse human neutrophils. Members of the PSM family are also produced by S. epidermidis, but their role in immune evasion is not known. Notably, strong cytolytic capacity of S. epidermidis PSMs would be at odds with the notion that S. epidermidis is a less aggressive pathogen than S. aureus, prompting us to examine the biological activities of S. epidermidis PSMs. Surprisingly, we found that S. epidermidis has the capacity to produce PSMδ, a potent leukocyte toxin, representing the first potent cytolysin to be identified in that pathogen. However, production of strongly cytolytic PSMs was low in S. epidermidis, explaining its low cytolytic potency. Interestingly, the different approaches of S. epidermidis and S. aureus to causing human disease are thus reflected by the adaptation of biological activities within one family of virulence determinants, the PSMs. Nevertheless, S. epidermidis has the capacity to evade neutrophil killing, a phenomenon we found is partly mediated by resistance mechanisms to antimicrobial peptides (AMPs), including the protease SepA, which degrades AMPs, and the AMP sensor/resistance regulator, Aps (GraRS). These findings establish a significant function of SepA and Aps in S. epidermidis immune evasion and explain in part why S. epidermidis may evade elimination by innate host defense despite the lack of cytolytic toxin expression. Our study shows that the strategy of S. epidermidis to evade elimination by human neutrophils is characterized by a passive defense approach and provides molecular evidence to support the notion that S. epidermidis is a less aggressive pathogen than S. aureus.
Introduction
Staphylococcus epidermidis colonizes the epithelial surfaces of every human being. Furthermore, it is one of the most frequent causes of nosocomial infections. In addition to the abundant prevalence of S. epidermidis on the human skin, this high incidence is mainly due to the exceptional capacity of S. epidermidis to stick to the surfaces of indwelling medical devices during device insertion and form multilayered agglomerations called biofilms [1], [2].
During infection, S. epidermidis is exposed to human innate host defenses, most notably professional phagocytes, among which neutrophils or polymorphonuclear leukocytes (PMNs) play a preeminent role [3]. While the biofilm mode of growth is believed to be broadly protective against host defenses [1], [4], we lack information on specific molecules of S. epidermidis that provide resistance to host defense mechanisms. The only S. epidermidis molecules known to facilitate evasion of killing by neutrophils are the extracellular polymers poly-N-acetylglucosamine (PNAG, or PIA, polysaccharide intercellular adhesin) and poly-γ-glutamic acid (PGA), which inhibit uptake by neutrophils (phagocytosis) [5], [6]. This is in contrast to S. aureus, a more pathogenic relative of S. epidermidis, which produces a series of proteins and enzymes dedicated to evade innate and adaptive host defense [7], [8].
Immune evasion of S. aureus is due in part to cytolytic toxins, such as α-toxin, γ-toxin, or Panton-Valentine leukocidin, which are proinflammatory and have potential to lyse neutrophils and other leukocytes [9]. In addition, we recently identified a new class of S. aureus cytolytic toxins, the phenol-soluble modulins (PSMs). Several PSM peptides have high capacity to attract, stimulate and lyse human neutrophils, and are significant contributors to pathogenesis of S. aureus bacteremia and skin infection [10]. PSMα3, in particular, is the most cytolytic S. aureus PSM and encoded together with three other PSMs in the psmα operon of S. aureus. High expression of peptides encoded in the psmα operon is mainly responsible for the pronounced potential of hyper-virulent community-associated methicillin-resistant S. aureus (CA-MRSA) strains to lyse human neutrophils [10], underpinning the importance of PSMs for neutrophil lysis. In contrast to S. aureus, toxins that lyse human leukocytes or other cell types have not been described in S. epidermidis.
PSMs are characterized by common physico-chemical properties rather than similarity at the amino acid sequence level (Fig. 1). Identification of PSMs thus requires isolation and characterization by means such as mass spectrometry and Edman degradation. Using these methods, six members of the PSM family have been identified in S. epidermidis (Fig. 1) [11], [12], [13], [14], but their biological significance is largely undefined. This is in part due to the fact that in earlier studies, a partially purified extract from S. epidermidis containing PSMs was used to measure PSM activities [12], [15], [16], [17]. Therefore, it is possible that proinflammatory activities previously attributed to S. epidermidis PSMs were caused by contaminants such as lipopeptides, particularly as similar impurities have frequently led to the misinterpretation of stimulatory effects on innate immune system mechanisms in the past [18]. This emphasizes the need to analyze pure peptides, but pure S. epidermidis PSMs and especially cytolytic potencies of S. epidermidis PSMs have never been investigated.
Fig. 1. S. epidermidis and S. aureus PSMs. 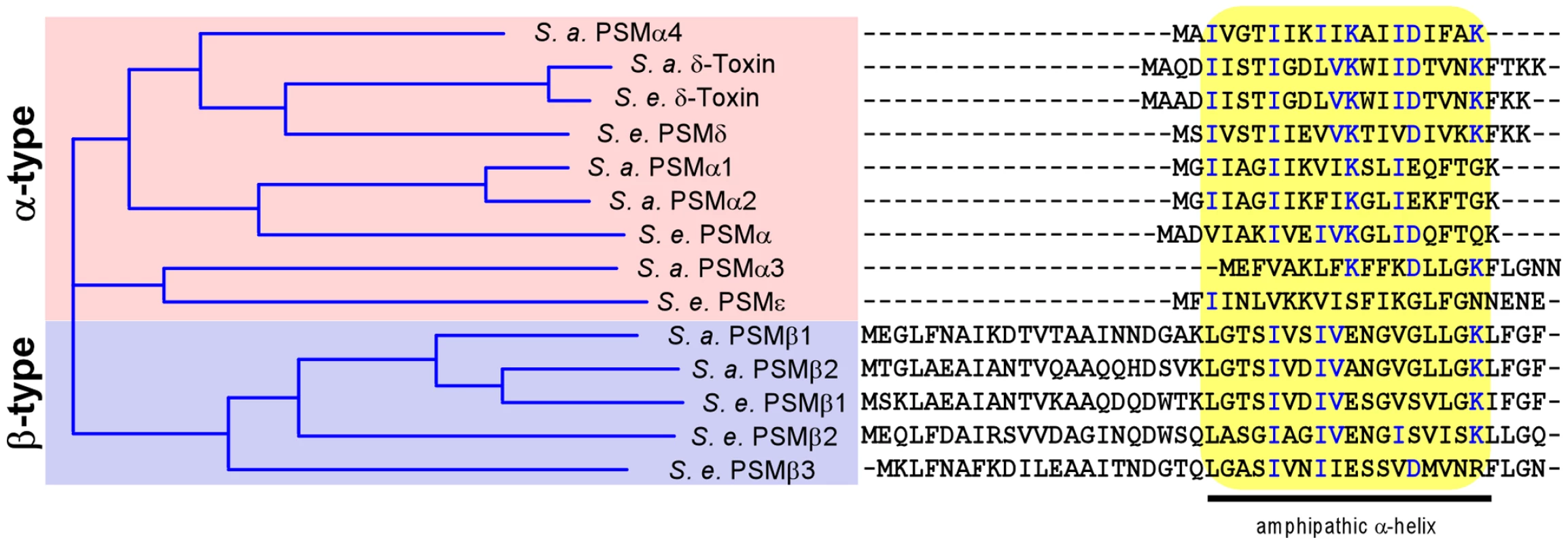
All known S. aureus (S. a.) and S. epidermidis (S. e.) PSMs were aligned by a sequence comparison program (Vector NTI). Similarity on the amino acid level is depicted as a tree on the left. Aligned amino acid sequences are shown at the right, with conserved amino acids shown in blue. All PSMs contain a region with pronounced amphipathy and α-helicity, boxed in yellow. After phagocytosis, neutrophils kill bacteria with reactive oxygen species and non-oxygen-dependent processes [19]. Among the latter, antimicrobial peptides (AMPs) such as defensins and cathelicidins are believed to play a crucial role [20]. We have previously found that the secreted S. epidermidis protease SepA has considerable capacity to eliminate AMPs by proteolysis [21]. Furthermore, we identified the first Gram-positive AMP sensing system in S. epidermidis, apsRSX [22]. This system, which has also been named graRSX in S. aureus [23], [24], regulates a series of AMP resistance mechanisms, including Dlt-dependent D-alanylation of teichoic acids [25], MprF-dependent lysinylation of phospholipids [26], and an AMP exporter called VraFG [24]. However, it is not known whether Aps or SepA confer resistance to killing by neutrophils.
In the present study, we examined the role of S. epidermidis PSMs in immune evasion, in particular by determining whether S. epidermidis PSMs are cytolytic toward human neutrophils. Furthermore, we analyzed whether the sepA and apsRSX loci facilitate survival during phagocytic interaction with neutrophils. Our study provides a better understanding of how S. epidermidis evades killing by human leukocytes in the susceptible host. Notably, we identified the first potent S. epidermidis cytolysin, PSMδ, a member of the α-type PSM family. However, despite the capacity to produce a potent cytolysin, S. epidermidis culture supernatants had little or no capacity to lyse neutrophils. In contrast, we show that the SepA protease and the Aps AMP sensor significantly promote resistance of S. epidermidis to killing by neutrophils. These findings provide molecular evidence to support the notion that S. epidermidis, in strong contrast to virulent S. aureus, has a defensive rather than aggressive approach to infection and immune evasion.
Results
Cytolytic activity of S. epidermidis culture filtrates
To evaluate the relative potency of S. epidermidis to kill human neutrophils, we compared culture filtrates of different S. epidermidis strains with those of S. aureus LAC, a CA-MRSA strain with demonstrated high capacity to lyse neutrophils [10], [27]. We investigated four S. epidermidis strains that have been most frequently used in S. epidermidis pathogenesis studies: 1457, O47, ATCC12228, and RP62A. ATCC12228 and RP62A represent the two S. epidermidis strains for which genome sequence data are available [14], [28]. Furthermore, we included an agr mutant of strain 1457, as the agr regulatory system is known to strictly regulate PSM production [10], [29], [30].
Culture filtrates of all four S. epidermidis strains showed significantly reduced lysis of human neutrophils compared to S. aureus LAC (Fig. 2), indicating that as a species S. epidermidis has low capacity to lyse neutrophils. Some low-level cytolysis was detected in culture filtrates from strain 1457, but not strains RP62A and ATCC12228. Furthermore, cytolytic capacity of culture filtrates was completely abolished in an agr deletion mutant of strain 1457 and in the natural agr mutant strain O47 (Fig. 2), in accordance with a potential function of the agr-regulated PSMs of S. epidermidis in neutrophil lysis.
Fig. 2. Neutrophil lysis by S. epidermidis culture filtrates. 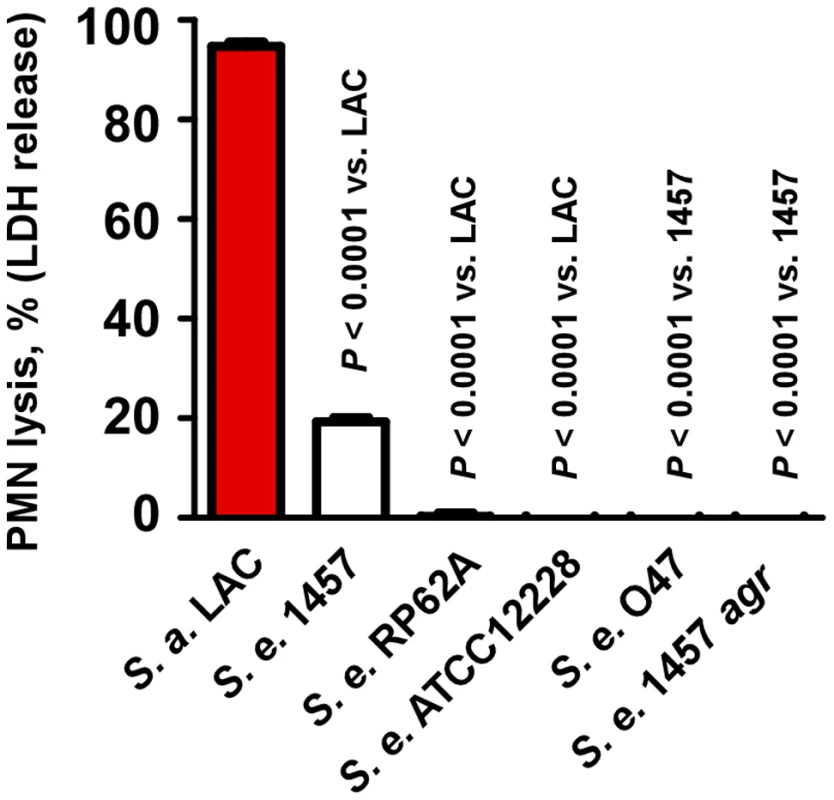
Neutrophil (PMN) lysis by undiluted S. epidermidis 18-h culture filtrates was determined by measuring release of LDH (incubation time, 1 h). Culture filtrate from S. aureus LAC (18-h culture) was used as a comparison. Analysis of PSM secondary structure
In vitro studies using S. aureus and S. epidermidis γ-toxins and S. epidermidis PSMδ indicated that PSMs lead to perturbation of synthetic membrane vesicles and likely work by pore formation in the absence of a specific receptor [31], [32], [33]. Presumably, the capacity of PSMs to lyse cells is thus dependent on their physico-chemical features, namely the ability to form amphipathic α-helices, a characteristic property of pore-forming peptides.
To evaluate whether S. epidermidis PSMs form amphipathic α-helices, we determined secondary structures of PSM peptides using circular dichroism (Fig. 3A, B). These experiments demonstrated that all S. epidermidis PSMs are predominantly α-helical. When PSM sequences were arranged in α-helical wheels, all predicted α-helices showed a distinct hydrophilic opposed to a hydrophobic side, which is characteristic for amphipathic α-helices (shown as an example for PSMβ1 in Fig. 3C). These findings indicate that S. epidermidis PSMs have the basic structural requirements for membrane perturbation and pore formation.
Fig. 3. Secondary structure of S. epidermidis PSM peptides. 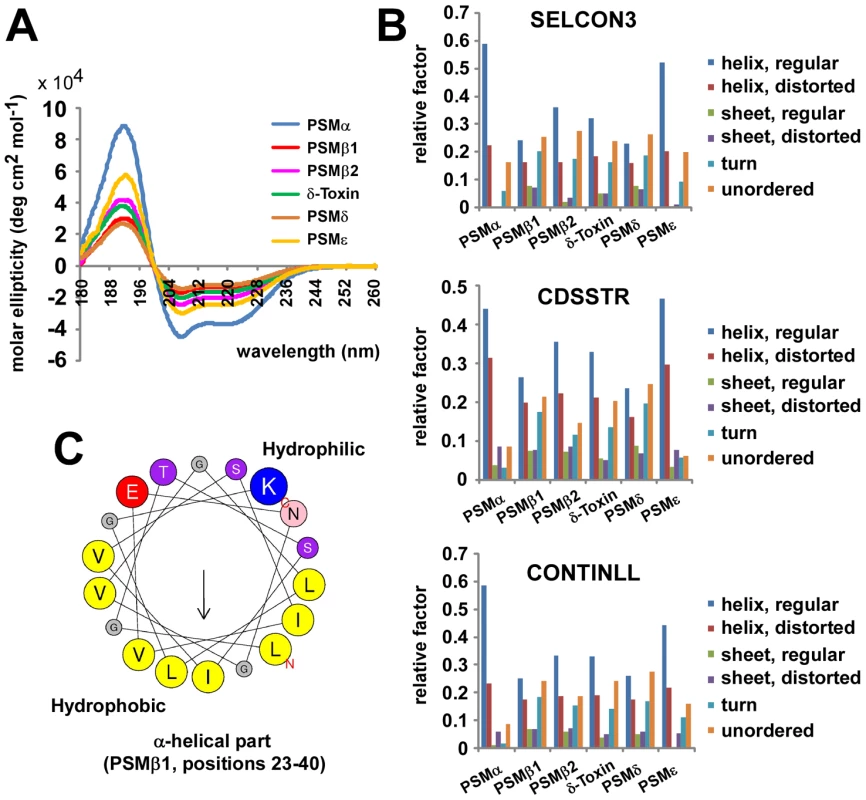
Secondary structure of S. epidermidis PSM peptides was analyzed by circular dichroism (CD) measurement. (A), molar ellipticity curves; (B) analysis of secondary structure using 3 different algorithms. (C) All PSM peptides have an amphipathic α-helix that encompasses most of the peptide for the shorter α-type and the C-terminal part of the β-type PSMs (shown as example for PSMβ1 by α-helical wheel presentation, http://heliquest.ipmc.cnrs.fr). Capacity of S. epidermidis PSMs to lyse neutrophils
To analyze whether S. epidermidis PSMs lyse neutrophils, we incubated human neutrophils with pure, synthetic S. epidermidis PSMs. Remarkably, one S. epidermidis PSM, PSMδ, caused high levels of neutrophil lysis, to an extent comparable to that of the potent S. aureus PSMα3 (Fig. 4A). In contrast, S. epidermidis δ-toxin, PSMα, and PSMε showed only very limited cytolytic capacity. The β-type PSMs were non-cytolytic toward neutrophils, in keeping with findings achieved for the β-type PSMs of S. aureus [10]. These differences indicate that while the formation of amphipathic α-helices is a likely prerequisite for membrane perturbation, further yet unknown structural features determine the degree of cytolytic activity in PSMs. This notion is also supported by our observation that the degree of α-helicity (Fig. 3A) did not correlate with the cytolytic potential of PSMs (Fig. 4A). Of note, PSMδ to our knowledge represents the first potent cytolysin of S. epidermidis to be identified. Remarkably, PSMδ is less closely related to S. aureus PSMα3 by amino acid sequence comparison than are PSMα1, PSMα2, and S. epidermidis PSMε (Fig. 1), underlining the notion that cytolytic properties of PSMs are determined by secondary rather than primary structure.
Fig. 4. Neutrophil lysis by S. epidermidis PSM peptides and culture filtrates of PSMδ-expression strains. 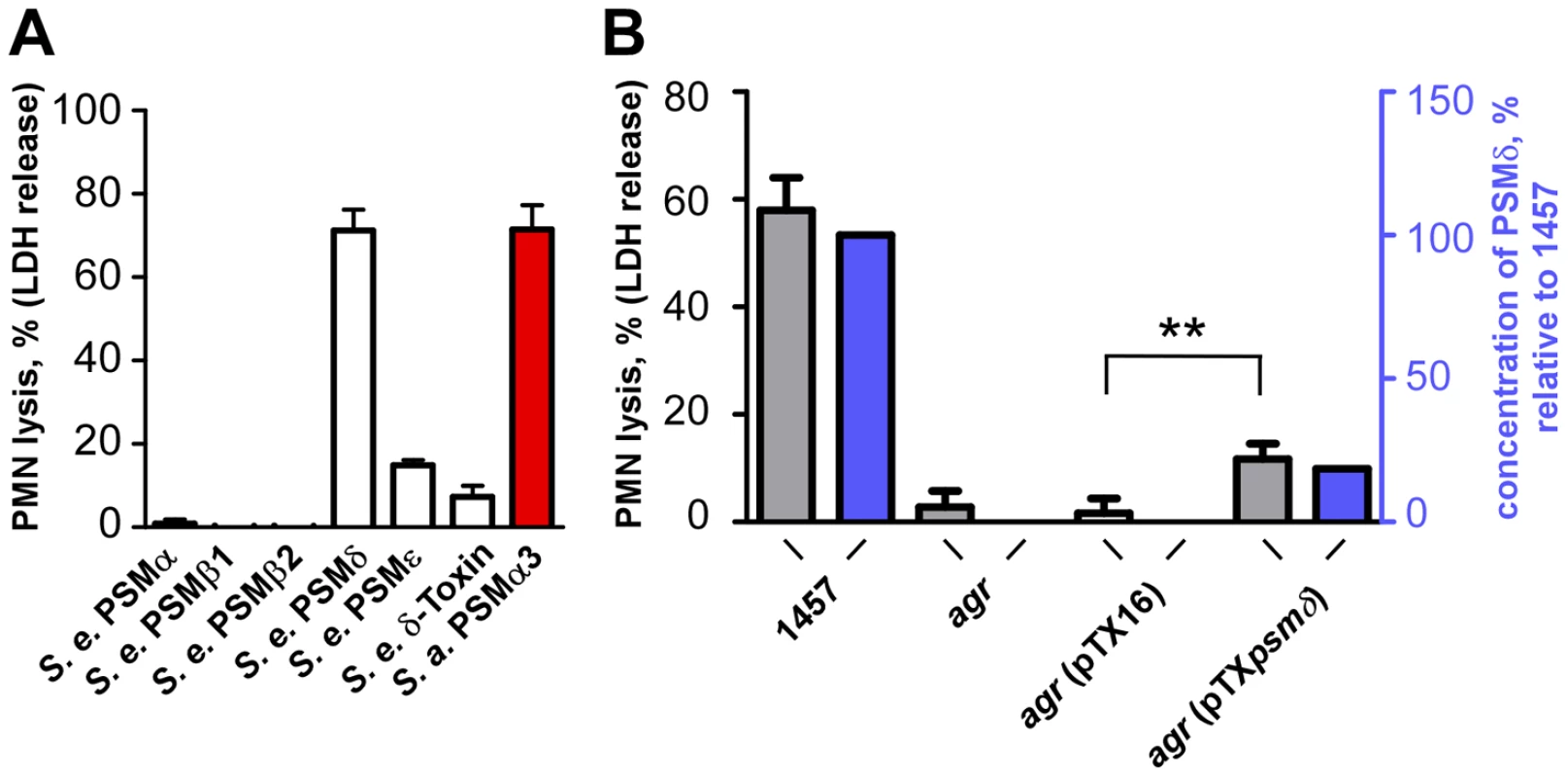
(A) Neutrophil (PMN) lysis by synthetic, N-formylated PSM peptides at 10 µg/ml was determined by measuring release of LDH (incubation time, 1 h). PSMα3 (S. aureus) was used as a comparison at the same concentration. (B) Neutrophil lysis using supernatants (18-h cultures) of a PSMδ-over-expressing agr-negative (lacking intrinsic PSM production) and corresponding control strains (incubation time, 6 h). pTXpsmδ, pTX construct expressing PSMδ; pTX16, control plasmid. Strains were grown in TSB with 0.5% xylose and 12.5 µg/ml tetracycline. **, p<0.01, paired t-tests. Blue bars, PSMδ concentration in the culture filtrates relative to that in the 1457 wild-type (set to 100%). The strong potency of PSMδ to lyse human neutrophils was confirmed by expression of PSMδ in an agr-negative S. epidermidis strain that lacks production of PSMs (Fig. 4B). Induction of PSMδ production resulted in a significant increase in the capacity of culture filtrates from the agr-negative strain to lyse human neutrophils (p = 0.0015, agr pTXpsmδ versus agr pTX16 control). As we have observed previously [10], [34], plasmid-based expression of PSM peptides often does not result in concentrations of PSMs as high as those found in wild-type culture filtrates, which also was the case for PSMδ. However, the degree of neutrophil lysis exerted by culture filtrates of the PSMδ expression strain (20.1% of that by the wild-type) corresponded very well to PSMδ expression (18.6% of that in the wild-type) (Fig. 4B), highlighting the major contribution PSMδ has to the overall cytolytic capacity of S. epidermidis.
Hemolytic activity of S. epidermidis PSMs
We showed previously that S. aureus PSMs also lyse cells other than neutrophils, such as monocytes or erythrocytes [10]. To analyze whether lysis of erythrocytes by synthetic PSMs and staphylococcal culture filtrates follows the same pattern as observed using human neutrophils, we tested hemolysis. Results were in very good accordance with those achieved with human neutrophils, inasmuch as only PSMδ showed strong hemolytic activity at a level comparable to that exerted by S. aureus PSMα3 (Fig. 5A). Similarly, culture filtrates of S. epidermidis strains were much less hemolytic than those of S. aureus LAC, with that of S. epidermidis 1457 causing slightly higher hemolysis than culture filtrates from the other S. epidermidis strains (Fig. 5B), in keeping with the neutrophil lysis findings.
Fig. 5. Hemolysis by S. epidermidis culture filtrates and PSM peptides. 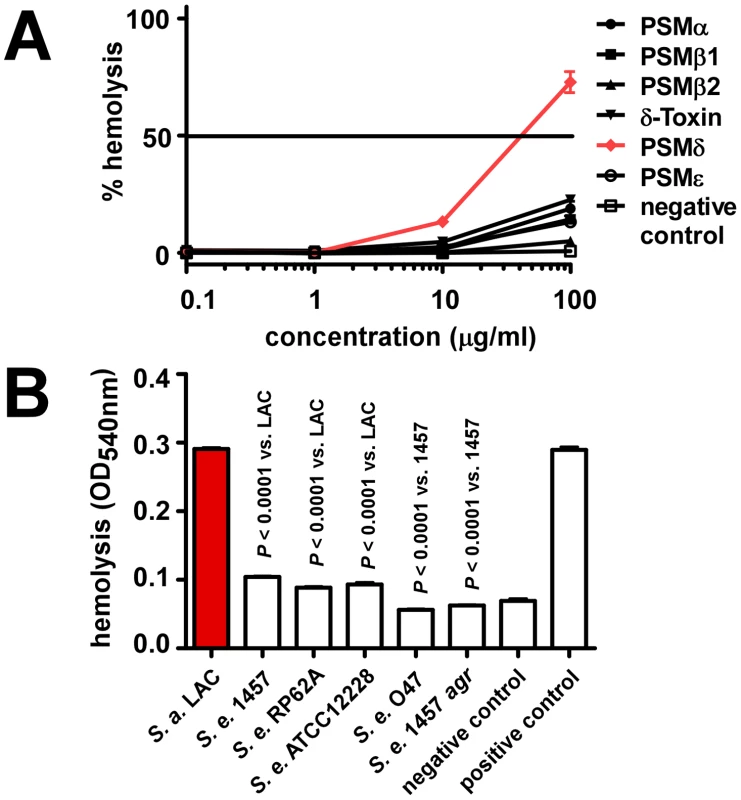
Hemolysis was determined by assays using sheep blood. (A) Hemolysis by synthetic, N-formylated PSMs of S. epidermidis. Negative control, DPBS. (B) Hemolysis by S. epidermidis culture filtrates (undiluted) and S. aureus LAC culture filtrate as comparison. All culture filtrates were from cultures grown for 18 h. Negative control, DPBS; positive control, 1% (v/v) Triton-X100 in DPBS. Production characteristics of PSMs in S. epidermidis
The finding that S. epidermidis PSMδ has considerable cytolytic activity at first appeared to contradict the low cytolytic activity of S. epidermidis culture filtrates. Indeed, it was reminiscent of the situation in S. aureus, in which the cytolytic potential is also mostly determined by one strongly cytolytic PSM peptide, PSMα3 [10]. However, potential differences in PSM production are not considered in this comparison. Therefore, we next measured PSM production patterns in S. epidermidis strains compared to those in S. aureus. We found considerable differences in the relative PSM production patterns between S. aureus and S. epidermidis, while patterns among the different S. epidermidis strains were similar (Fig. 6). In addition to the S. epidermidis strains that are shown, we analyzed a large S. epidermidis strain collection. Results were similar in all strains, except for strains that completely lacked PSM production (data not shown). These PSM-negative strains are likely functionally agr-negative, owing to frequently occurring mutations in the agr system [35], which includes the agr-negative strain O47 [36].
Fig. 6. PSM concentrations in S. epidermidis culture filtrates. 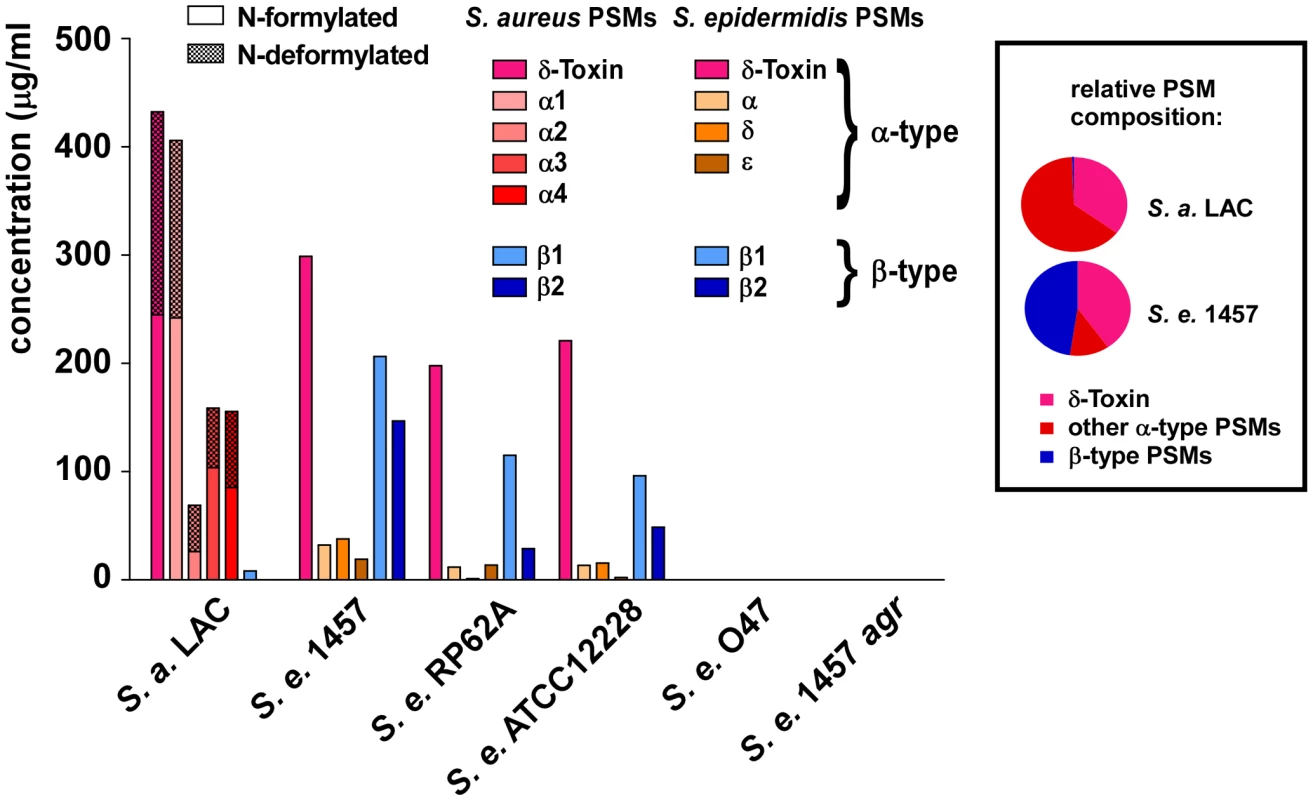
PSM concentrations in 18-h S. epidermidis and S. aureus LAC culture filtrates were determined by HPLC/MS. Peaks corresponding to N-formylated and deformylated PSM versions were measured separately and the percentage of deformylated peptides is shown as checkered bars. No PSMs were detected in the natural and constructed agr mutants (O47, 1457 agr). Relative PSM composition (α-type, δ-toxin, β-type) is shown at the right for S. aureus LAC and S. epidermidis 1457. Relative compositions were similar to that of 1457 in the other S. epidermidis strains (except in agr-negative O47 and 1457 agr). The most noticeable difference between S. epidermidis and S. aureus was strongly reduced production of α-type PSMs, except δ-toxin, in S. epidermidis. In contrast, the non-cytolytic β-type PSMs represented almost half of the total PSM peptide produced in S. epidermidis, whereas concentrations of β-type PSMs were extremely low in S. aureus. Furthermore, the difference between the production levels of the most cytolytic PSMs in the two species, PSMα3 and PSMδ (∼5∶1), correlated with the degree of overall neutrophil lysis (∼5∶1, S. aureus LAC to S. epidermidis 1457), underlining that these most potent PSMs predominantly determine cytolytic capacity. Moreover, the notion that any cytolytic activity of S. epidermidis is largely determined by production of PSMδ is in accordance with the observed low production of PSMδ and overall low cytolytic activity of all tested S. epidermidis strains. Thus, although S. epidermidis has the capacity to secrete a potent cytolytic toxin, PSMδ, it limits hemolysis or lysis of neutrophils by keeping production of PSMδ at a low level.
Deformylation of N-formyl methionine in PSMs
The N-formyl methionine group present at the N-terminus of newly synthesized bacterial proteins is recognized by immune cells as a pathogen-associated molecular pattern (PAMP) [37]. Removal of the N-formyl group by bacterial peptide deformylase thus serves to evade recognition by human innate host defense. N-formylated bacterial proteins commonly are not exported with N-formyl-methionine, as their signal peptides are removed during export. In contrast, PSMs are secreted as the unaltered translation product by a yet unidentified mechanism and thus always carry N-formyl methionine, likely representing a very considerable portion of N-formylated peptides released by staphylococci [10]. In S. aureus LAC culture filtrates, about one-half of the total PSM peptide was N-deformylated, which is in good accordance with a previous report on δ-toxin deformylation in another S. aureus strain [38]. In remarkable contrast, no significant deformylation was detected in S. epidermidis PSMs (Fig. 6). Thus, despite the presence of a peptide deformylase in S. epidermidis that is highly homologous to the S. aureus enzyme (80% identity on the amino acid level), proteins are not N-deformylated in S. epidermidis as efficiently as in S. aureus.
Proinflammatory capacity of S. epidermidis culture filtrates and PSMs
In addition to causing cytolysis, PSMs of S. aureus are known to stimulate neutrophil and monocyte chemotaxis, activate neutrophils, and elicit release of the chemokine IL-8 [10]. These proinflammatory capacities of PSMs indicate that the innate immune system recognizes PSMs as PAMPs, which as we recently discovered is achieved by recognition of PSMs by the FPR2/ALX receptor [39]. To determine S. epidermidis proinflammatory capacities, we analyzed stimulation of IL-8 release (Fig. 7A). IL-8 is an important chemokine that causes recruitment of neutrophils to the site of infection [40]. PSMδ had very strong capacity to stimulate release of IL-8; but overall, stimulation of IL-8 release did not correlate with the cytolytic capacities of PSMs. Notably, all S. epidermidis PSMs to some degree stimulated release of IL-8 despite the lack of cytolytic capacity in several of them. Accordingly, capacities of S. epidermidis culture filtrates to stimulate IL-8 release were in the same range as those of S. aureus LAC (Fig. 7B). Finally, stimulation of IL-8 release was significantly lower for the S. epidermidis agr mutant of strain 1457 compared to the corresponding isogenic wild-type strain, and very low for the natural agr mutant strain O47, in keeping with strict regulation of PSMs by agr [30]. Thus, while the different PSM production pattern in S. epidermidis correlates with considerably reduced neutrophil lysis compared to S. aureus, S. epidermidis PSMs still appear to be recognized efficiently as PAMPs. These results suggest that PSM cytolytic and proinflammatory capacities are dependent on distinct interactions with host cells.
Fig. 7. IL-8 release by neutrophils stimulated by S. epidermidis PSMs and culture filtrates. 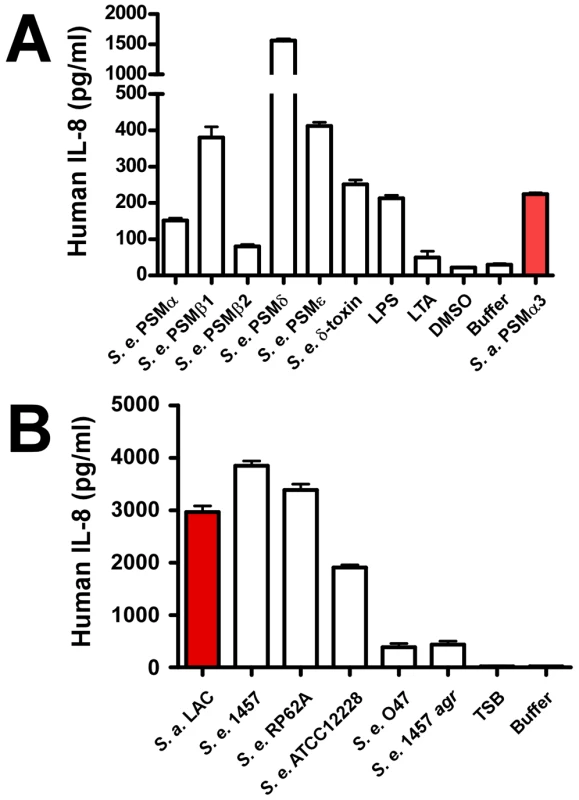
PMNs were incubated with synthetic, N-formylated PSMs (10 µg/ml) (A) or 18-h culture filtrates (diluted 1∶100) (B) and release of the cytokine IL-8 was measured by ELISA (for culture filtrates with further 1∶2 dilution). LPS, lipopolysaccharide, 10 ng/ml; LTA, S. aureus lipoteichoic acid, 1 µg/ml. SepA protease and Aps AMP sensor/resistance regulator of S. epidermidis promote resistance to killing by neutrophils
Our results suggest that S. epidermidis does not use PSM cytolytic activity to a significant extent to evade killing by human neutrophils. However, the capacity of S. epidermidis to cause chronic infections indicates that S. epidermidis has means to inhibit elimination by human professional phagocytes. As an alternative strategy to evade killing by human neutrophils, bacteria may secrete enzymes to destroy – or use mechanisms to decrease – the antimicrobial efficiency of neutrophil bactericidal agents [3]. Among those agents, antimicrobial proteins and peptides likely play an important role in the killing of ingested bacteria [41]. We previously showed that the secreted S. epidermidis protease SepA has strong capacity to destroy human AMPs [21]. In addition, we identified a system that we named Aps (for antimicrobial peptide sensor) that senses the presence of human AMPs and coordinates a series of AMP resistance mechanisms in S. epidermidis [22] and S. aureus [24]. While the mechanistic function of these loci is thus well understood, evidence for a significant role of Aps and SepA in immune evasion using human cells is lacking. Therefore, we investigated whether S. epidermidis SepA and S. epidermidis and S. aureus Aps contribute to survival after uptake by human neutrophils. Isogenic sepA and aps mutants of S. epidermidis 1457 had significantly reduced ability to survive after phagocytic interaction with human neutrophils compared to the wild-type strain (Fig. 8), providing evidence for an important function of the aps and sepA loci in S. epidermidis immune evasion. Similarly, the Aps system had a significant impact on the survival of the S. aureus CA-MRSA strain MW2 after phagocytosis. Of note, this effect was comparable to that of the psmα locus, which encodes the most important cytolytic PSM peptides of S. aureus (Fig. 8B,C). These findings indicate that the Aps AMP-sensing system has an important immune evasion task in both species, while only S. aureus makes additional use of cytolytic toxins, such as PSMs, to evade killing by human neutrophils. This discrepancy is reflected by the higher capacity of S. aureus to survive interaction with human neutrophils compared to S. epidermidis (Fig. 8).
Fig. 8. Survival of aps and sepA deletion mutants in human neutrophils. 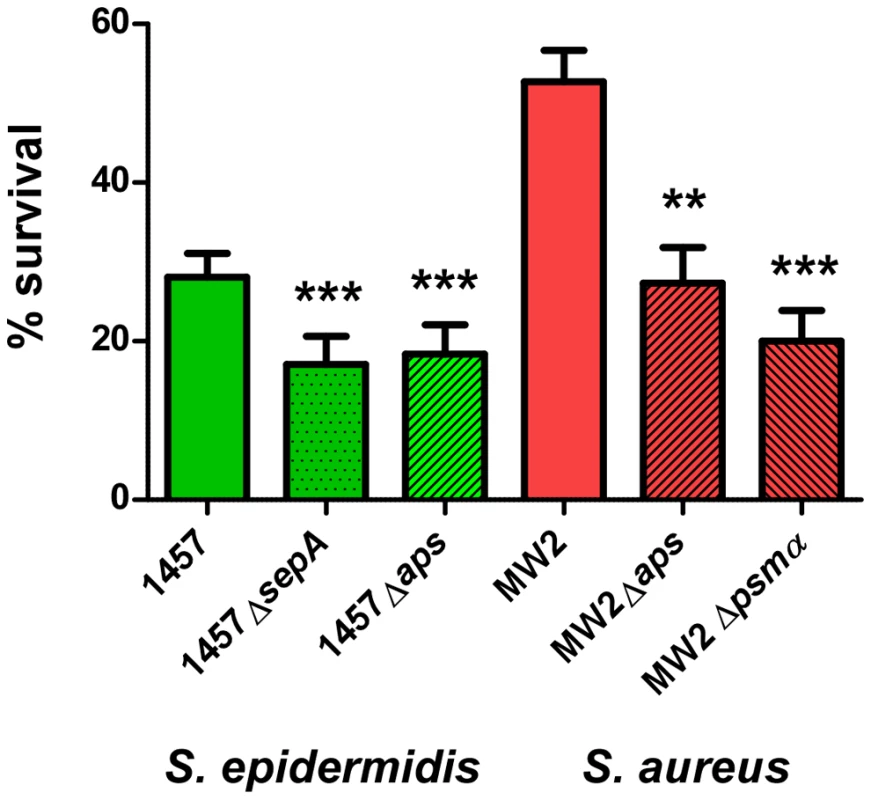
Survival of S. epidermidis 1457 and S. aureus MW2 wild-type (wt) and isogenic gene deletion mutants was determined after phagocytic uptake by counting of colony forming units after 60 min incubation. Bacterial cells used for the experiment were harvested at similar points in growth at an OD600 nm of ∼1.5. ***, p<0.001; **, p<0.01 versus the corresponding wild-type sample (1-way ANOVA, Dunnett's post test). Error bars represent SEM. Discussion
As a commensal organism living on the human skin, S. epidermidis commonly has a benign relationship with its host and may even contribute to reducing inflammatory responses [2], [42]. However, S. epidermidis may cause infection after breach of the epithelial barrier and entry into the bloodstream, such as through contamination of indwelling medical devices during surgery. Although most S. epidermidis infections are only moderately severe and usually chronic, their sheer frequency poses a considerable problem, predominantly in the hospital setting [2], [43]. Despite the immense importance of S. epidermidis infections for public health, the interaction of S. epidermidis with host defenses is poorly understood. In particular, it has not been investigated in detail if and how S. epidermidis resists killing by human neutrophils, which are largely responsible for elimination of invading bacteria. Therefore, we here investigated the interaction of S. epidermidis with neutrophils. As direct lysis of neutrophils by bacterial cytolysins is an efficient means to evade killing, we focused our investigation on PSMs as the only S. epidermidis gene products with potential cytolytic activity [14], [28].
A major finding of our study was the identification of PSMδ as the first S. epidermidis toxin with significant cytolytic capacity. However, despite the cytolytic potential of PSMδ, culture filtrates of S. epidermidis strains had very low capacity to lyse human neutrophils. Importantly, according to our findings this phenotypic difference between virulent S. aureus and S. epidermidis is caused at least in part by a pattern of PSM production in S. epidermidis that is shifted, compared to S. aureus, to PSMs with lower cytolytic potential. Thus, PSMs in S. epidermidis do not contribute significantly to neutrophil lysis, in contrast to many virulent strains of S. aureus. Likely, PSMs fulfill other roles in S. epidermidis that are yet poorly understood, such as in biofilm development [44] or bacterial interference [33]. The production of PSMs that are not potent cytotoxins would thus ascertain that S. epidermidis may cause chronic, biofilm-associated infection without promoting acute, purulent inflammation. This is in keeping with a general strategy of S. epidermidis to reside inside the human host in a state of “hiding” from the immune system. Potentially, a similar strategy is pursued by strains of S. aureus, such as functionally Agr-negative strains, which are less virulent, cause chronic rather than acute infection, and produce less cytolytic toxins, such as PSMs.
In addition, our study revealed significant contributions of the SepA protease and the Aps AMP sensor/regulator to promoting S. epidermidis survival in human neutrophils. Thus, S. epidermidis is able to combat important mechanisms that neutrophils use to kill bacteria after phagocytosis. However, together with previous findings on S. aureus survival in human neutrophils [27], our data indicate that these mechanisms are not as efficient as leukocyte toxins, underlining the notion that S. epidermidis is in general less virulent than S. aureus as a result of lower capacity to survive after neutrophil phagocytosis. This is in accordance with a very early study that showed increased survival of “pathogenic” (i.e. coagulase-positive) versus “non-pathogenic” (i.e. coagulase-negative) staphylococci in human leukocytes [45]. Nevertheless, our study shows that - combined with mechanisms preventing neutrophil phagocytosis, such as surface exopolymers and biofilm formation - S. epidermidis has a multi-faceted program providing resistance to neutrophil killing, explaining at least in part the capacity of S. epidermidis to cause long-lasting infection in the susceptible host. Moreover, as we have shown previously that SepA production is under control of Agr and SarA [21], our findings confirm the notion that global regulatory systems play key roles in S. epidermidis immune evasion [46], and are reminiscent of similar functions of Agr and SarA in S. aureus [47], [48]. Finally, the observed significant effects of AMP resistance mechanisms on survival in neutrophils underline the importance of non-oxygen-dependent antimicrobial processes of the host.
Collectively, our findings indicate that the molecular mechanisms that S. epidermidis uses to evade elimination by innate host defense reflect a passive defense strategy rather than use of aggressive toxins and point to a different major role of PSM production in S. epidermidis compared to S. aureus.
Materials and Methods
Ethics statement
Human neutrophils were obtained from healthy volunteers in accordance with a protocol approved by the Institutional Review Board for Human Subjects, NIAID. Informed written consent was received from human volunteers.
Bacterial strains and growth conditions
Bacterial strains used in this study were S. epidermidis strains 1457 [49], RP62A [14], [50], ATCC12228 [28], O47 [51], isogenic agr, sepA, and apsS deletion mutants of strain 1457 [21], [22], [52], S. aureus strains LAC (pulsed-field type USA300) [53] and MW2 (pulsed-field type USA400) [54] and the isogenic aps and psmα mutants of strain MW2 [24]. LAC and MW2 are virulent community-associated MRSA strains. Strains were grown in tryptic soy broth (TSB). The psmδ over-expression plasmid pTXpsmδ [34] was transformed in S. epidermidis agr. Expression of PSMδ by this construct is achieved by adding xylose, which acts on a xylose-inducible promoter in front of the cloned psmδ gene [55].
Peptide synthesis
PSM peptides were synthesized by commercial vendors with an N-terminal formyl methionine residue in each peptide. Peptide sequence fidelity was determined by the Peptide Synthesis Unit of the NIAID. Peptide stock solutions were prepared at 10 mg/ml in DMSO (dimethylsulfoxide); further dilutions were made in water.
Neutrophil preparation and lysis assays
PMNs were isolated from venous blood of healthy volunteers as described [56]. Lysis of PMNs by synthetic PSMs or clarified S. aureus or S. epidermidis culture media was determined essentially as described [27], [56]. Synthetic PSMs were added to wells of a 96-well tissue culture plate containing 106 PMNs and plates were incubated at 37°C. After 1 h, PMN lysis was determined by release of lactate dehydrogenase (LDH) (Cytotoxicity Detection Kit, Roche Applied Sciences). Alternatively, S. aureus and S. epidermidis strains were cultured for 18 h at 37°C in 50 ml TSB (+/ − 0.5% xylose) with shaking using a 100 ml flask. Bacteria were removed by centrifugation and culture media were sterilized by filtration and stored at −80°C in aliquots until used. Culture medium was mixed with human PMNs (106) and tested for its ability to cause PMN lysis using incubation times of up to 6 h, as indicated.
Resistance of S. epidermidis and S. aureus to killing by human neutrophils
For measurement of S. epidermidis/S. aureus survival after phagocytic interaction with neutrophils, PMNs (106) in RPMI were combined with ∼107 RPMI-washed bacteria from mid-logarithmic growth phase in 96-well flat-bottom microtiter plates. Plates were centrifuged at 380×g for 8 min to synchronize phagocytosis and incubated at 37°C for up to 1 h. At the desired time points, 22 µl of 1% saponin was added, well contents were mixed, and the plates were incubated on ice for 15 min. Surviving bacteria were enumerated. % survival was calculated by comparing the numbers of surviving bacteria to those at t = 0.
Cytokine production assay
After isolation and washing, neutrophils were resuspended in RPMI 1640 medium (Sigma) supplemented with 10% human serum, 2 mM L-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 2 mM sodium pyruvate, and 10 mM HEPES. Cells were distributed to a 96-well culture plate at 200 µl and 5×105 cells per well. Synthetic PSMs or filtered bacterial culture supernatants were diluted in fresh culture medium (1∶100) and added to the plate at 100 µl/well. Plates were incubated at 37°C in a 5.5% CO2 incubator for 5 h. Then, the plate was centrifuged at 1500 rpm for 10 min, and supernatant was harvested from each well. IL-8 was measured in the culture supernatants with commercial ELISA assay kits (R&D systems) according to the manufacturer's instructions. Diluted culture filtrates were further diluted 1∶2 for the ELISA.
Hemolysis assay
Hemolytic activities of culture filtrates from 18-h cultures of S. epidermidis strains or synthetic PSM peptides at different concentrations were determined by incubating samples with sheep red blood cells (2% v/v in Dulbecco's phosphate-buffered saline, DPBS) for 1 h at 37°C as previously described [10]. Assays were performed in triplicate.
Analysis of PSM production
RP-HPLC/ESI-MS was performed on an Agilent 1100 chromatography system coupled to a Trap SL mass spectrometer using a Zorbax SB-C8 2.3×30 mm column as described [30]. Quantification was based on extracted ion chromatograms using the most abundant peaks of the electrospray ion mass spectra of the respective PSM peptides, with calibration using synthetic peptides, as described [30].
Circular dichroism spectroscopy
The structures of synthetic PSM peptides were analyzed by CD spectroscopy on a Jasco spectropolarimeter model J-720 instrument. Solutions of PSM peptides, each at 1.0 mg/ml, were prepared in 50% trifluoroethanol. Measurements were performed in triplicate and the resulting scans were averaged, smoothed, and the buffer signal was subtracted. Computation of relative fraction of helix, sheet, turn, and unordered structure, using 3 different algorithms, was performed according to Sreerama and Woody [57].
Statistical analyses
Statistical analyses were performed with Graph Pad Prism 5 software using t-tests or 1-way ANOVA with Bonferroni or Dunnett post tests, as appropriate.
Zdroje
1. OttoM
2008 Staphylococcal biofilms. Curr Top Microbiol Immunol 322 207 228
2. OttoM
2009 Staphylococcus epidermidis - the ‘accidental’ pathogen. Nat Rev Microbiol 7 555 567
3. NauseefWM
2007 How human neutrophils kill and degrade microbes: an integrated view. Immunol Rev 219 88 102
4. CostertonJW
StewartPS
GreenbergEP
1999 Bacterial biofilms: a common cause of persistent infections. Science 284 1318 1322
5. KocianovaS
VuongC
YaoY
VoyichJM
FischerER
2005 Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest 115 688 694
6. VuongC
VoyichJM
FischerER
BraughtonKR
WhitneyAR
2004 Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol 6 269 275
7. FosterTJ
2005 Immune evasion by staphylococci. Nat Rev Microbiol 3 948 958
8. RooijakkersSH
van KesselKP
van StrijpJA
2005 Staphylococcal innate immune evasion. Trends Microbiol 13 596 601
9. WoodinA
1970 Staphylococcal leukocidin.
MontjeT
KadisS
AjlS
Microbial toxins New York Academic Press, Inc 327 355
10. WangR
BraughtonKR
KretschmerD
BachTH
QueckSY
2007 Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 13 1510 1514
11. McKevittAI
BjornsonGL
MauracherCA
ScheifeleDW
1990 Amino acid sequence of a deltalike toxin from Staphylococcus epidermidis. Infect Immun 58 1473 1475
12. MehlinC
HeadleyCM
KlebanoffSJ
1999 An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J Exp Med 189 907 918
13. YaoY
SturdevantDE
OttoM
2005 Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J Infect Dis 191 289 298
14. GillSR
FoutsDE
ArcherGL
MongodinEF
DeboyRT
2005 Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187 2426 2438
15. HajjarAM
O'MahonyDS
OzinskyA
UnderhillDM
AderemA
2001 Cutting edge: functional interactions between toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J Immunol 166 15 19
16. KlebanoffSJ
KazaziF
Van VoorhisWC
SchlechteKG
1994 Activation of the human immunodeficiency virus long terminal repeat in THP-1 cells by a staphylococcal extracellular product. Proc Natl Acad Sci U S A 91 10615 10619
17. LilesWC
ThomsenAR
O'MahonyDS
KlebanoffSJ
2001 Stimulation of human neutrophils and monocytes by staphylococcal phenol-soluble modulin. J Leukoc Biol 70 96 102
18. HashimotoM
TawaratsumidaK
KariyaH
KiyoharaA
SudaY
2006 Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J Immunol 177 3162 3169
19. FaurschouM
BorregaardN
2003 Neutrophil granules and secretory vesicles in inflammation. Microbes Infect 5 1317 1327
20. HancockRE
DiamondG
2000 The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol 8 402 410
21. LaiY
VillaruzAE
LiM
ChaDJ
SturdevantDE
2007 The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol Microbiol 63 497 506
22. LiM
LaiY
VillaruzAE
ChaDJ
SturdevantDE
2007 Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A 104 9469 9474
23. HerbertS
BeraA
NerzC
KrausD
PeschelA
2007 Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog 3 e102
24. LiM
ChaDJ
LaiY
VillaruzAE
SturdevantDE
2007 The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol 66 1136 1147
25. PeschelA
OttoM
JackRW
KalbacherH
JungG
1999 Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem 274 8405 8410
26. PeschelA
JackRW
OttoM
CollinsLV
StaubitzP
2001 Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med 193 1067 1076
27. VoyichJM
BraughtonKR
SturdevantDE
WhitneyAR
Said-SalimB
2005 Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol 175 3907 3919
28. ZhangYQ
RenSX
LiHL
WangYX
FuG
2003 Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol Microbiol 49 1577 1593
29. QueckSY
Jameson-LeeM
VillaruzAE
BachTH
KhanBA
2008 RNAIII-Independent Target Gene Control by the agr Quorum-Sensing System: Insight into the Evolution of Virulence Regulation in Staphylococcus aureus. Mol Cell 32 150 158
30. VuongC
DurrM
CarmodyAB
PeschelA
KlebanoffSJ
2004 Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: quorum-sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell Microbiol 6 753 759
31. MellorIR
ThomasDH
SansomMS
1988 Properties of ion channels formed by Staphylococcus aureus delta-toxin. Biochim Biophys Acta 942 280 294
32. TalbotJC
ThiaudiereE
VincentM
GallayJ
SiffertO
2001 Dynamics and orientation of amphipathic peptides in solution and bound to membranes: a steady-state and time-resolved fluorescence study of staphylococcal delta-toxin and its synthetic analogues. Eur Biophys J 30 147 161
33. CogenAL
YamasakiK
SanchezKM
DorschnerRA
LaiY
2010 Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Invest Dermatol 130 192 200
34. OttoM
O'MahoneyDS
GuinaT
KlebanoffSJ
2004 Activity of Staphylococcus epidermidis phenol-soluble modulin peptides expressed in Staphylococcus carnosus. J Infect Dis 190 748 755
35. VuongC
KocianovaS
YaoY
CarmodyAB
OttoM
2004 Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis 190 1498 1505
36. VuongC
GerkeC
SomervilleGA
FischerER
OttoM
2003 Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis 188 706 718
37. LeY
MurphyPM
WangJM
2002 Formyl-peptide receptors revisited. Trends Immunol 23 541 548
38. SomervilleGA
CockayneA
DurrM
PeschelA
OttoM
2003 Synthesis and deformylation of Staphylococcus aureus delta-toxin are linked to tricarboxylic acid cycle activity. J Bacteriol 185 6686 6694
39. KretschmerD
GleskeA
RautenbergM
WangR
KoberleM
2010 Human formyl peptide receptor 2 (FPR2/ALX) senses highly pathogenic Staphylococcus aureus. Cell Host Microbe. In press
40. KobayashiY
2008 The role of chemokines in neutrophil biology. Front Biosci 13 2400 2407
41. NizetV
2007 Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J Allergy Clin Immunol 120 13 22
42. LaiY
Di NardoA
NakatsujiT
LeichtleA
YangY
2009 Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med 15 1377 1382
43. VuongC
OttoM
2002 Staphylococcus epidermidis infections. Microbes Infect 4 481 489
44. KongKF
VuongC
OttoM
2006 Staphylococcus quorum sensing in biofilm formation and infection. Int J Med Microbiol 296 133 139
45. RogersDE
TompsettR
1952 The survival of staphylococci within human leukocytes. J Exp Med 95 209 230
46. YaoY
VuongC
KocianovaS
VillaruzAE
LaiY
2006 Characterization of the Staphylococcus epidermidis Accessory-Gene Regulator Response: Quorum-Sensing Regulation of Resistance to Human Innate Host Defense. J Infect Dis 193 841 848
47. GreshamHD
LowranceJH
CaverTE
WilsonBS
CheungAL
2000 Survival of Staphylococcus aureus inside neutrophils contributes to infection. J Immunol 164 3713 3722
48. ShompoleS
HenonKT
LiouLE
DziewanowskaK
BohachGA
2003 Biphasic intracellular expression of Staphylococcus aureus virulence factors and evidence for Agr-mediated diffusion sensing. Mol Microbiol 49 919 927
49. MackD
NedelmannM
KrokotschA
SchwarzkopfA
HeesemannJ
1994 Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect Immun 62 3244 3253
50. ChristensenGD
BisnoAL
ParisiJT
McLaughlinB
HesterMG
1982 Nosocomial septicemia due to multiply antibiotic-resistant Staphylococcus epidermidis. Ann Intern Med 96 1 10
51. HeilmannC
GerkeC
Perdreau-RemingtonF
GotzF
1996 Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun 64 277 282
52. VuongC
GotzF
OttoM
2000 Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infect Immun 68 1048 1053
53. CDC
2003 Outbreaks of community-associated methicillin-resistant Staphylococcus aureus skin infections—Los Angeles County, California, 2002–2003. MMWR Morb Mortal Wkly Rep 52 88
54. CDC
1999 From the Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. Jama 282 1123 1125
55. PeschelA
OttenwalderB
GotzF
1996 Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol Lett 137 279 284
56. VoyichJM
OttoM
MathemaB
BraughtonKR
WhitneyAR
2006 Is Panton-Valentine Leukocidin the Major Virulence Determinant in Community-Associated Methicillin-Resistant Staphylococcus aureus Disease? J Infect Dis 194 1761 1770
57. SreeramaN
WoodyRW
2004 Computation and analysis of protein circular dichroism spectra. Methods Enzymol 383 318 351
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek The Combined Effect of Environmental and Host Factors on the Emergence of Viral RNA RecombinantsČlánek Functional Interchangeability of Late Domains, Late Domain Cofactors and Ubiquitin in Viral BuddingČlánek Parvovirus Minute Virus of Mice Induces a DNA Damage Response That Facilitates Viral ReplicationČlánek Phenolglycolipid-1 Expressed by Engineered BCG Modulates Early Interaction with Human Phagocytes
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2010 Číslo 10- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Social Media and Microbiology Education
- Antimicrobial Peptides: Primeval Molecules or Future Drugs?
- Phylodynamics and Human-Mediated Dispersal of a Zoonotic Virus
- Retroviral RNA Dimerization and Packaging: The What, How, When, Where, and Why
- Distinct Clones of Caused the Black Death
- Strain-Specific Differences in the Genetic Control of Two Closely Related Mycobacteria
- The Combined Effect of Environmental and Host Factors on the Emergence of Viral RNA Recombinants
- MHC Class I Bound to an Immunodominant Epitope Demonstrates Unconventional Presentation to T Cell Receptors
- Stimulates Immune Gene Expression and Inhibits Development in
- Crystal Structure of DotD: Insights into the Relationship between Type IVB and Type II/III Secretion Systems
- Cytomegalovirus microRNAs Facilitate Persistent Virus Infection in Salivary Glands
- Strategies to Avoid Killing by Human Neutrophils
- Transforming Growth Factor-β: Activation by Neuraminidase and Role in Highly Pathogenic H5N1 Influenza Pathogenesis
- Autoimmunity in Arabidopsis Is Mediated by Epigenetic Regulation of an Immune Receptor
- Functional Interchangeability of Late Domains, Late Domain Cofactors and Ubiquitin in Viral Budding
- Dengue Virus Ensures Its Fusion in Late Endosomes Using Compartment-Specific Lipids
- Nucleocapsid Promotes Localization of HIV-1 Gag to Uropods That Participate in Virological Synapses between T Cells
- Host Genetics and HIV-1: The Final Phase?
- Variations in the Hemagglutinin of the 2009 H1N1 Pandemic Virus: Potential for Strains with Altered Virulence Phenotype?
- High-Resolution Functional Mapping of the Venezuelan Equine Encephalitis Virus Genome by Insertional Mutagenesis and Massively Parallel Sequencing
- Viral Replication Rate Regulates Clinical Outcome and CD8 T Cell Responses during Highly Pathogenic H5N1 Influenza Virus Infection in Mice
- Calcineurin Inhibition at the Clinical Phase of Prion Disease Reduces Neurodegeneration, Improves Behavioral Alterations and Increases Animal Survival
- Parvovirus Minute Virus of Mice Induces a DNA Damage Response That Facilitates Viral Replication
- -Induced Inactivation of the Macrophage Transcription Factor AP-1 Is Mediated by the Parasite Metalloprotease GP63
- Epstein Barr Virus-Encoded EBNA1 Interference with MHC Class I Antigen Presentation Reveals a Close Correlation between mRNA Translation Initiation and Antigen Presentation
- Fidelity Variants of RNA Dependent RNA Polymerases Uncover an Indirect, Mutagenic Activity of Amiloride Compounds
- The Spread of Tomato Yellow Leaf Curl Virus from the Middle East to the World
- Concerted Action of Two Formins in Gliding Motility and Host Cell Invasion by
- Requirements for Receptor Engagement during Infection by Adenovirus Complexed with Blood Coagulation Factor X
- Release of Intracellular Calcium Stores Facilitates Coxsackievirus Entry into Polarized Endothelial Cells
- Gene Annotation and Drug Target Discovery in with a Tagged Transposon Mutant Collection
- Nuclear Export and Import of Human Hepatitis B Virus Capsid Protein and Particles
- Retention and Loss of RNA Interference Pathways in Trypanosomatid Protozoans
- Identification and Genome-Wide Prediction of DNA Binding Specificities for the ApiAP2 Family of Regulators from the Malaria Parasite
- Controlling Cellular P-TEFb Activity by the HIV-1 Transcriptional Transactivator Tat
- Direct Visualization of Peptide/MHC Complexes at the Surface and in the Intracellular Compartments of Cells Infected by
- Phenolglycolipid-1 Expressed by Engineered BCG Modulates Early Interaction with Human Phagocytes
- In Vitro and In Vivo Studies Identify Important Features of Dengue Virus pr-E Protein Interactions
- Inhibition of Nipah Virus Infection In Vivo: Targeting an Early Stage of Paramyxovirus Fusion Activation during Viral Entry
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Retroviral RNA Dimerization and Packaging: The What, How, When, Where, and Why
- Viral Replication Rate Regulates Clinical Outcome and CD8 T Cell Responses during Highly Pathogenic H5N1 Influenza Virus Infection in Mice
- Antimicrobial Peptides: Primeval Molecules or Future Drugs?
- Crystal Structure of DotD: Insights into the Relationship between Type IVB and Type II/III Secretion Systems
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



