-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Fructose-Asparagine Is a Primary Nutrient during Growth of in the Inflamed Intestine
It has long been thought that the nutrient utilization systems of Salmonella would not make effective drug targets because there are simply too many nutrients available to Salmonella in the intestine. Surprisingly, we have discovered that Salmonella relies heavily on a single nutrient during growth in the inflamed intestine, fructose-asparagine (F-Asn). A mutant of Salmonella that cannot obtain F-Asn is severely attenuated, suggesting that F-Asn is the primary nutrient utilized by Salmonella during inflammation. No other organism has been reported to synthesize or utilize this novel biological compound. The novelty of this nutrient and the apparent lack of utilization systems in mammals and most other bacteria suggest that the F-Asn utilization system represents a specific and potent therapeutic target for Salmonella.
Published in the journal: Fructose-Asparagine Is a Primary Nutrient during Growth of in the Inflamed Intestine. PLoS Pathog 10(6): e32767. doi:10.1371/journal.ppat.1004209
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004209Summary
It has long been thought that the nutrient utilization systems of Salmonella would not make effective drug targets because there are simply too many nutrients available to Salmonella in the intestine. Surprisingly, we have discovered that Salmonella relies heavily on a single nutrient during growth in the inflamed intestine, fructose-asparagine (F-Asn). A mutant of Salmonella that cannot obtain F-Asn is severely attenuated, suggesting that F-Asn is the primary nutrient utilized by Salmonella during inflammation. No other organism has been reported to synthesize or utilize this novel biological compound. The novelty of this nutrient and the apparent lack of utilization systems in mammals and most other bacteria suggest that the F-Asn utilization system represents a specific and potent therapeutic target for Salmonella.
Introduction
Salmonella is a foodborne pathogen that causes significant morbidity and mortality in both developing and developed countries [1], [2]. It is widely believed that there are no undiscovered drug targets in Salmonella enterica, largely due to the high number of nutrients available during infection and redundancy in metabolic pathways [3], [4]. To acquire nutrients in the intestine, Salmonella initiates inflammation, which disrupts the microbiota and causes an oxidative burst that leads to the formation of tetrathionate [1]–[3], [5]–[7]. Tetrathionate is used as a terminal electron acceptor for the anaerobic respiration of carbon compounds that otherwise would not be metabolized [8]. One of these carbon sources is ethanolamine, which is derived from host phospholipids. Ethanolamine can be respired by Salmonella, but not fermented [8]. Salmonella actively initiates inflammation using two Type 3 Secretion Systems (T3SS), each encoded within a distinct, horizontally acquired pathogenicity island. SPI1 (Salmonella Pathogenicity Island 1) contributes to invasion of host cells and elicitation of inflammation in the host. SPI2 is required for survival within macrophages and contributes to intestinal inflammation. Salmonella strains lacking SPI1 and SPI2 cause very little intestinal inflammation [5], [6], [8], [9]. Here, we have identified fructose-asparagine (F-Asn) as another carbon source that is consumed by Salmonella using tetrathionate respiration during the host inflammatory response. The phenotypes of mutants lacking this utilization system are quite severe, suggesting that this is the primary nutrient utilized during Salmonella-mediated gastroenteritis. No other organism is known to synthesize or utilize F-Asn.
Results
The fructose-asparagine (F-Asn) utilization system was discovered during a genetic screen designed to identify novel microbial interactions between Salmonella and the normal microbiota. Transposon site hybridization (TraSH) was used to measure and compare the relative fitness of Salmonella transposon insertion mutants after oral inoculation and recovery from the cecum of two types of gnotobiotic mice, differing from each other by a single intestinal microbial species [10]–[15]. The two types of mice were germ-free and ex-germ-free colonized by a single member of the normal microbiota, Enterobacter cloacae. E. cloacae was chosen because it is a commensal isolate from our laboratory mice, easily cultured, genetically tractable, and it protects mice against Salmonella infection (Figure 1). In total, five genes conferred a greater fitness defect in the mice containing Enterobacter than in the germ-free mice (Table 1).
Fig. 1. Protection of mice against Salmonella serovar Typhimurium strain 14028 by Enterobacter cloacae strain JLD400. 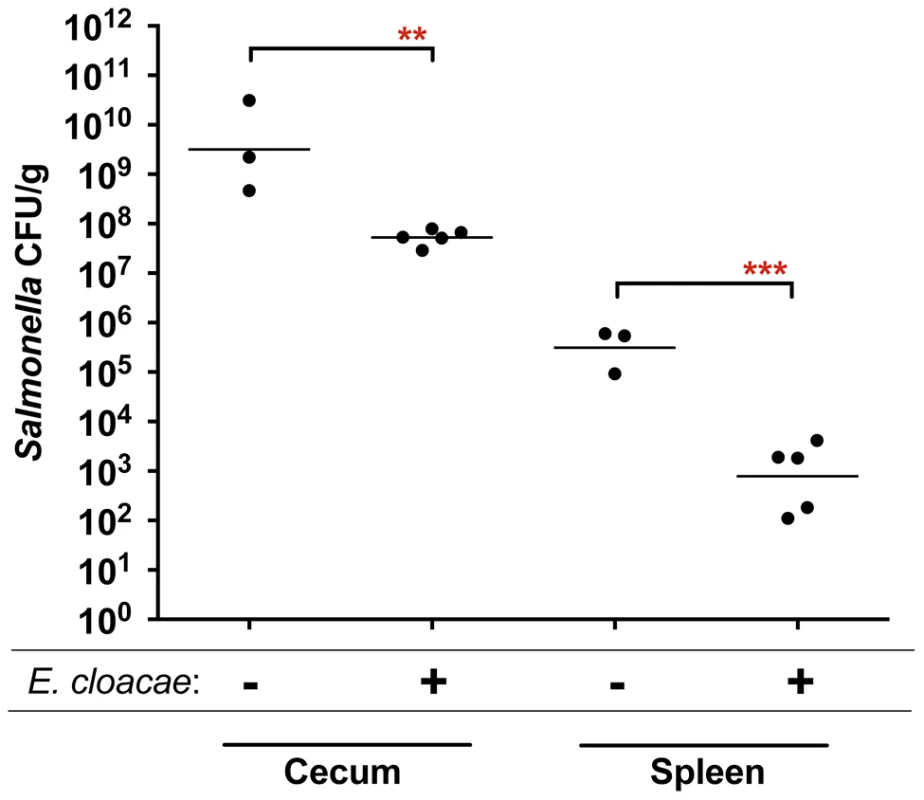
Germ-free C57BL/6 mice were divided into two groups. One group was colonized with 107 cfu of Enterobacter cloacae via the intragastric route (i.g.) and one group was not. One day later both groups were challenged i.g. with 107 cfu of Salmonella. After 24 hours, the cecum and spleen were homogenized and plated to enumerate Salmonella. Each point represents the CFU/g recovered from one mouse with the geometric mean shown by a horizontal line. Statistical significance between select groups was determined by using an unpaired two-tailed Student t test. ** = P value<0.01, *** = P value<0.001. Tab. 1. Genes that are differentially required in germ-free mice and ex-germ-free mice monoassociated with Enterobacter cloacae. 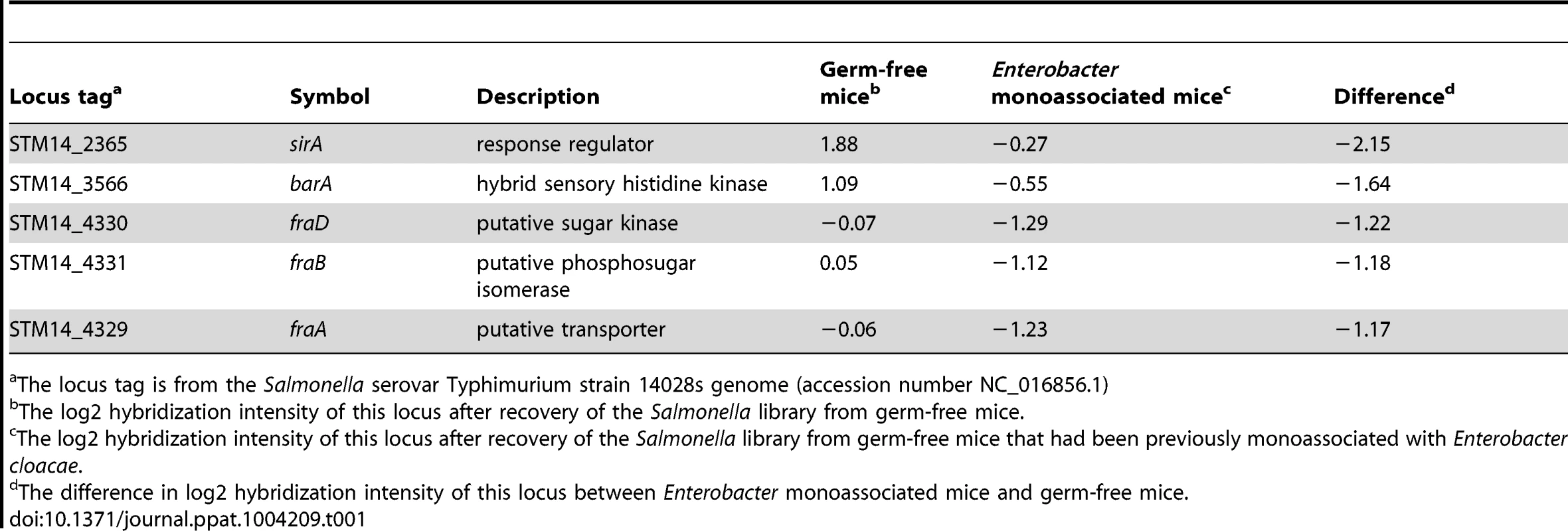
The locus tag is from the Salmonella serovar Typhimurium strain 14028s genome (accession number NC_016856.1) Two of these genes, barA and sirA (uvrY), encode a two component response regulator pair that is conserved throughout the γ-proteobacteria [16]–[18]. BarA/SirA control the activity of the CsrA protein (carbon storage regulator) which coordinates metabolism and virulence by binding to and regulating the translation and/or stability of mRNAs for numerous metabolic and virulence genes including SPI1, SPI2, and glgCAP (glycogen biosynthesis) [17], [19], [20]. To confirm the fitness phenotype of the BarA/SirA regulatory system, we performed competition experiments in which wild-type Salmonella was mixed in a 1∶1 ratio with an isogenic sirA mutant and inoculated orally into germ-free mice and ex-germ-free mice colonized by Enterobacter. The results of TraSH analysis suggested that the sirA mutant would be at a greater growth disadvantage in Enterobacter mono-associated mice than in germ-free mice (Table 1). Results of the competition experiment confirmed this prediction (Figure 2).
Fig. 2. Competitive index (CI) measurements of a sirA mutant in mouse models. 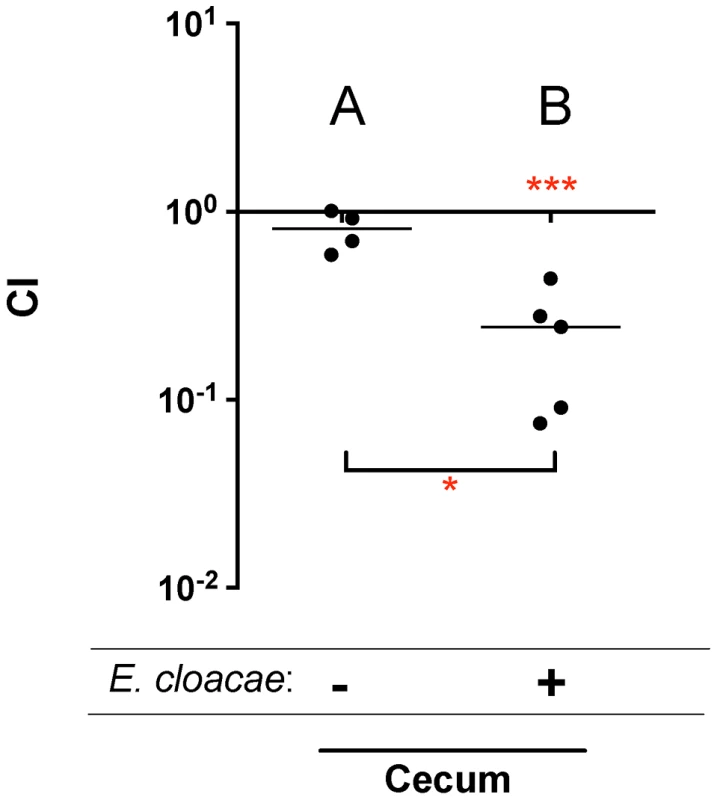
A) 107 wild-type MA43 and sirA mutant MA45 in germ-free mice, via the intragastric route (i.g.) and recovered from the cecum after 24 hours. B) 107 wild-type MA43 vs sirA mutant MA45 in germ-free mice mono-associated with Enterobacter cloacae, via the i.g. route and recovered from the cecum after 24 hours. Each point represents the CI from one mouse with the median shown by a horizontal line. Statistical significance of each group being different than 1 was determined by using a one sample Student's t test. Statistical significance between groups was determined using a Mann-Whitney test. * = P value<0.05, *** = P value<0.001. The other three genes identified by TraSH analysis had not been characterized previously, and are located together in a putative operon. Genome annotation suggested that they encode a C4 dicarboxylate transporter, a sugar kinase, and a phosphosugar isomerase (Figure 3). A putative asparaginase lies at the end of the operon, and a separate gene upstream of the operon encodes a putative transcriptional regulator of the GntR family. These genes are not present in E. coli and appear to represent a horizontal acquisition inserted between the gor and treF genes at 77.7 centisomes of the Salmonella 14028 genome (ORFs STM14_4328 to STM14_4332). We have named these genes fraBDAE and fraR for reasons to be described below. A fraB1::kan mutation was constructed and tested for fitness in germ-free and Enterobacter colonized mice using 1∶1 competition assays against the wild-type Salmonella. The TraSH results suggested that this locus would exhibit a differential fitness phenotype in germ-free mice and Enterobacter mono-associated mice. Indeed, disruption of the fra locus caused a severe fitness defect in germ-free mice and a more severe defect in Enterobacter-colonized mice (Figure 4A, B).
Fig. 3. Map of the fra locus of Salmonella enterica. 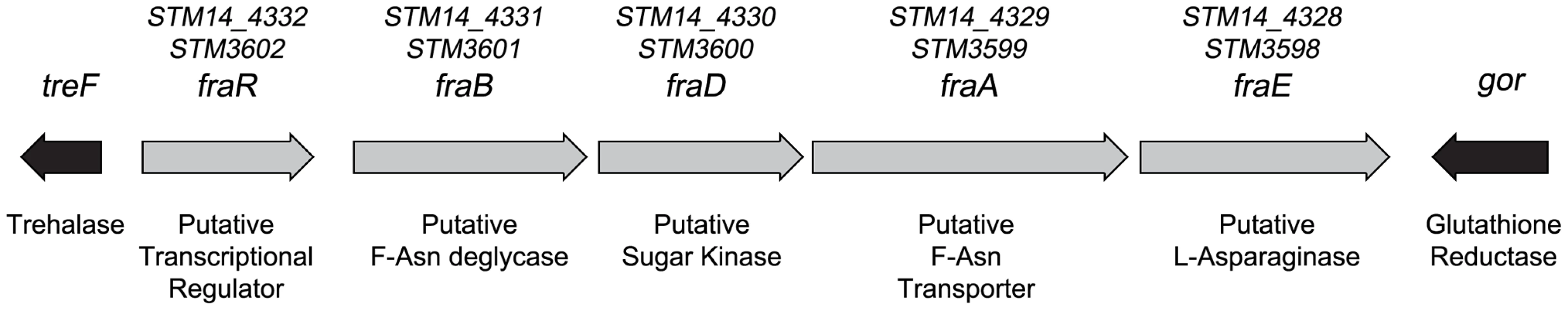
The five genes of the fra locus are shown as grey arrows. The gor and treF genes are shown as black arrows and are conserved throughout the Enterobacteriaceae while the fra locus is not, suggesting that the fra locus was horizontally acquired. The proposed functions and names of each gene are shown below and above the arrows, respectively. The names are based upon the distantly related frl locus of E. coli. For example, the deglycase enzyme of the frl locus is encoded by frlB so we have named the putative deglycase of the fra locus, fraB. The fra locus has no frlC homolog, while the frl locus does not have an asparaginase. Therefore, the name fraC was not used and the asparaginase was named fraE. The locus tags using the Salmonella nomenclature for strains 14028 (STM14 numbers) and LT2 (STM numbers) are shown above the gene names. Fig. 4. Fitness defect of a fraB1::kan mutant as measured by competitive index (CI) in various genetic backgrounds and mouse models. 
A) 107 wild-type MA43 and fraB1::kan mutant MA59 in germ-free (GF) C57BL/6 mice, via the intragastric route (i.g.) and recovered from the cecum after 24 hours. B) 107 wild-type MA43 and fraB1::kan mutant MA59 in germ-free C57BL/6 mice mono-associated with Enterobacter cloacae, via the i.g. route and recovered from the cecum after 24 hours. C) 109 wild-type MA43 and fraB1::kan mutant MA59 in C57BL/6 conventional mice, via the i.g. route and recovered from the cecum after 24 hours. D) 107 wild-type IR715 and fraB1::kan mutant MA59 in streptomycin-treated (ST) C57BL/6 mice, via the i.g. route and recovered from the cecum after 24 hours. E) 107 wild-type IR715 and fraB1::kan mutant MA59 in streptomycin-treated C57BL/6 mice, via the i.g. route and recovered from the cecum after 4 days. F) Complementation of the fraB1::kan mutation with a plasmid encoding the entire fra island, pASD5006. 107 ASD6090 and ASD6000 in streptomycin-treated C57BL/6 mice, via the i.g. route and recovered from the cecum after 4 days. G) 107 wild-type IR715 and fraB4::kan mutant CS1032 in streptomycin-treated C57BL/6 mice, via the i.g. route and recovered from the cecum after 24 hours. H) 107 wild-type IR715 and fraB4::kan mutant CS1032 in streptomycin-treated C57BL/6 mice, via the i.g. route and recovered from the cecum after 4 days. I) Complementation of the fraB4::kan mutation with a plasmid encoding the entire fra island, pASD5006.107 wild-type ASD6090 and fraB4::kan mutant ASD6040 in streptomycin-treated C57BL/6 mice, via the i.g. route and recovered from the cecum after 4 days. J) 107 fra+ MA4301 and fraB1::cam mutant MA5900, both strains are a SPI1− SPI2− background, in streptomycin-treated C57BL/6 mice, via the i.g. route and recovered from the cecum after 24 hours. K) 107 fra+ MA4301 and fraB1::cam mutant MA5900, both strains are a SPI1− SPI2− background, in streptomycin-treated C57BL/6 mice, via the i.g. route and recovered from the cecum after 4 days. L) 107 fra+ MA4301 vs fraB1::cam mutant MA5900, both strains in a SPI1− SPI2− background, in germ-free C57BL/6 mice, via the i.g. route and recovered from the cecum after 24 hours. M) 107 fra+ MA4310 vs fraB1::kan mutant MA5910, both strains are a ttrA− background, in streptomycin-treated C57BL/6 mice, via the i.g. route and recovered from the cecum after 24 hours. N) 107 fra+ MA4310 vs fraB1::kan mutant MA5910, both strains are a ttrA− background, in streptomycin-treated C57BL/6 mice, via the i.g. route and recovered from the cecum after 4 days. O) 107 fra+ MA4310 vs fraB1::kan mutant MA5910, both strains are a ttrA− background, in germ-free C57BL/6 mice, via the i.g. route and recovered from the cecum after 24 hours. P) 104 wild-type MA43 and fraB1::kan mutant MA59 in conventional C57BL/6 mice, via the intraperitoneal route (i.p.) and recovered from the spleen after 24 hours. Q) 104 wild-type MA43 and fraB1::kan mutant MA59 in streptomycin-treated C57BL/6 mice, via the i.p. route and recovered from the spleen after 24 hours. Each data point represents the CI from one mouse with the median shown by a horizontal line. Statistical significance of each group being different than 1 was determined by using a one sample Student's t test. Statistical significance between select groups was determined using a Mann-Whitney test. * = P value<0.05, ** = P value<0.01, *** = P value<0.001. The fra locus confers a fitness advantage during inflammation and anaerobic respiration
Competition experiments between wild-type and the fraB1::kan mutant were performed as described above using conventional mice (with normal microbiota) and mice treated orally with streptomycin (strep-treated) one day earlier to disrupt the microbiota (Figure 4C, D, E). Conventional mice do not become inflamed from Salmonella, while strep-treated mice (or germ-free) do become inflamed [5], [6], [8], [21]–[24]. Disruption of the fra locus caused no fitness defect in conventional mice, but caused a severe defect in the strep-treated mice at one and four days post-infection (Figure 4C, D, E). The phenotype in strep-treated mice was confirmed by complementation (Figure 4F). It is expected that the fraB1::kan mutation is polar on the remainder of the fraBDAE operon. Therefore, the fraB1::kan mutation was complemented with a low copy number plasmid encoding the entire fra island (Figure 4F). The phenotype was confirmed again using a separately constructed mutation, fraB4::kan, and complementation (Figure 4G, H, I). In both instances, greater than 99% of the phenotype was restored (Figure 4F, I).
The observation of a phenotype in germ-free and strep-treated mice, but not conventional mice, suggested that Salmonella might require inflammation in order to acquire or utilize the fra-dependent nutrient source. It is known that inflammation causes the accumulation of tetrathionate in the lumen, a terminal electron acceptor that allows Salmonella to respire anaerobically [6]. Histopathology results confirmed that infection with Salmonella caused inflammation in the germ-free and strep-treated mice, but not in the conventional mice (Figure 5A, D, E). To test the hypothesis that Salmonella must induce inflammation for fra to affect the phenotype, we repeated the competition experiments in a Salmonella genetic background lacking SPI1 and SPI2, so that both the wild-type and the fra mutant would be defective for induction of inflammation. The severe fitness phenotype of the fra mutant was not observed in these strains (Figure 4J–L) and histopathology results confirmed that inflammation was indeed low during these experiments (Figure 5B, F).
Fig. 5. Histopathology scores of C57BL/6 mice after i.g. inoculation with Salmonella. 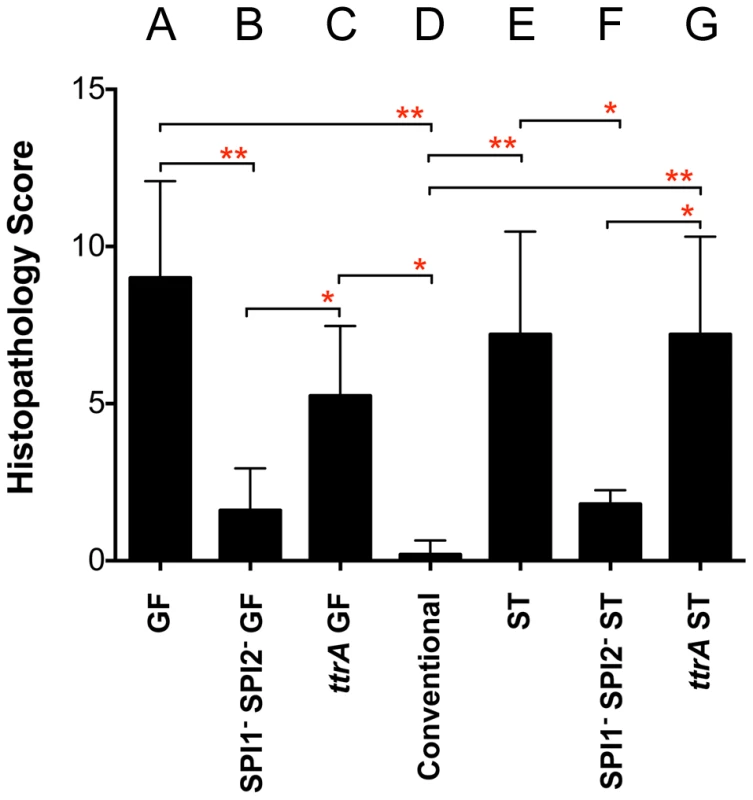
All groups received 107 cfu except conventional mice (D), which received 109 cfu. A) Germ-free (GF) mice 24 hours post-infection with wild-type MA43 and fraB1::kan mutant MA59; B) GF mice 24 hours post-infection with SPI1− SPI2− Salmonella (fra+ MA4301 vs fraB1::cam mutant MA5900); C) GF mice 24 hours post-infection with ttrA− Salmonella (fra+ MA4310 vs fraB1::kan mutant MA5910); D) Conventional mice 24 hours post-infection with wild-type MA43 and fraB1::kan mutant MA59. E) Strep-treated (ST) mice 4 days post-infection with wild-type IR715 and fraB1::kan mutant MA59; F) ST mice 4 days post-infection with SPI1− SPI2− Salmonella (fra+ MA4301 and fraB1::cam mutant MA5900; G) ST mice 4 days post-infection with ttrA− Salmonella (fra+ MA4310 vs fraB1::kan mutant MA5910). Error bars represent mean+SD. Statistical significance between select groups was determined using a Mann-Whitney test. * = P value<0.05, ** = P value<0.01. To test the hypothesis that tetrathionate respiration was required for use of the fra-dependent nutrient source, the competition experiments were repeated in a ttrA mutant background. TtrA is part of a tetrathionate reductase, which is required for the utilization of tetrathionate as a terminal electron acceptor during anaerobic respiration [6], [25]. As in the SPI1 SPI2 background, there was no phenotype of a fra mutant in a ttrA mutant background indicating that Salmonella must be able to respire using tetrathionate to gain advantage from the fra locus (Figure 4M–O). Histopathology results confirmed the presence of moderate inflammation during these experiments (Figure 5C, G).
To determine if the fra locus is required during the systemic phase of disease, we performed competition experiments between the wild-type and fra mutant after intraperitoneal inoculation of conventional or strep-treated mice, with bacterial recovery from the spleen. The fra mutant had no fitness defect during systemic infection (Figure 4P, Q).
So far, we have seen the fra phenotype in C57BL/6 mice, which are mutated at the Nramp1 locus, and this required that the mice be either germ-free or strep-treated so that Salmonella could induce inflammation. Ideally, we would like to determine the significance of the fra locus in a model that is not mutated and does not require strep-treatment or a germ-free status. It is known that humans with a complete microbiota are quickly inflamed by Salmonella infection while conventional mice are not, and more recently it was discovered that germ-free mice colonized with human fecal microbiota (“humanized” mice) become inflamed from Salmonella infection without disturbance of the gut microbiota by streptomycin [26]. Therefore, we “humanized” germ-free Swiss Webster mice, which are Nramp1+/+, with human feces obtained from a healthy adult donor from the Ohio State University fecal transplant center. Competition experiments were then performed between wild-type and fra mutant Salmonella in these mice. Histopathology results confirmed the presence of mild inflammation during these experiments and the fra locus had a greater than 10,000-fold fitness phenotype (Figure 6).
Fig. 6. Phenotype of a fraB1::kan mutant in the cecum of “humanized” and IL10 knockout mice. 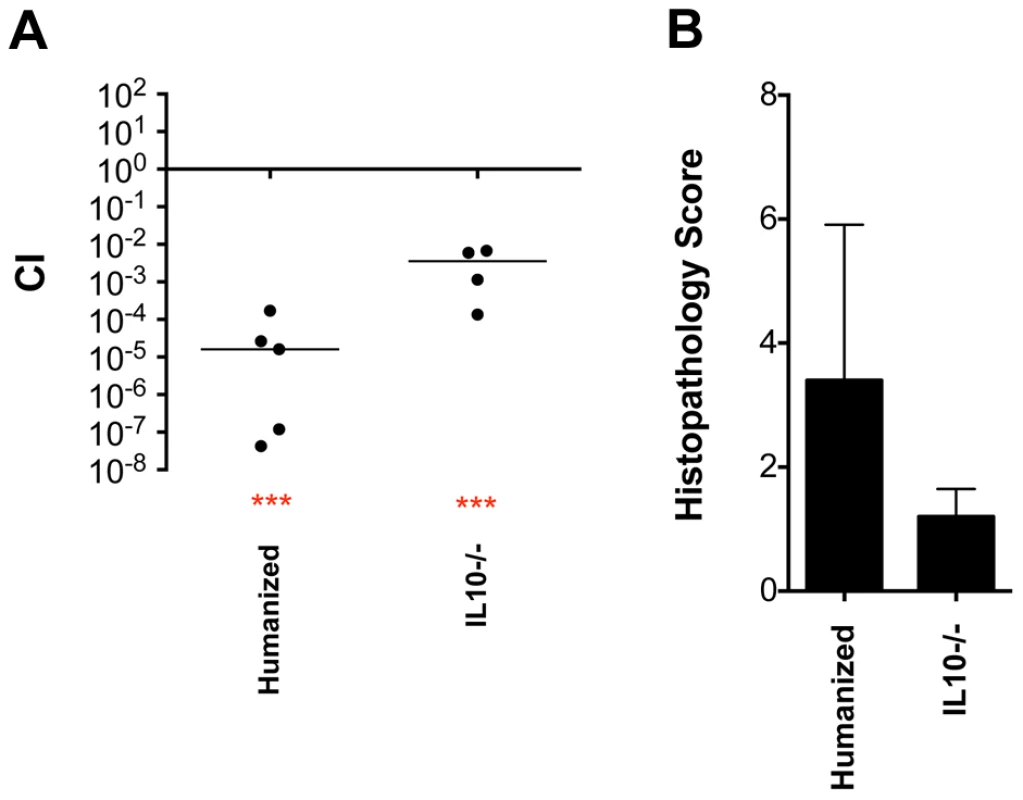
109 wild-type IR715 vs fraB1::kan mutant MA59 in “humanized” Swiss Webster mice (germ free mice inoculated orally with a human fecal sample), or C57BL/6 IL10 knockout mice, as indicated, via the i.g. route and recovered from cecum on day 3 post-infection. A) Each data point represents the CI from one mouse with the median shown by a horizontal line. Statistical significance of each group being different than 1 was determined by using a one sample Student's t test. *** = P value<0.001. B) Histopathology scores of mice from panel A. Error bars represent mean+SD. IL10 knockout mice were used as another method to facilitate Salmonella-induced inflammation without using streptomycin [5]. Histopathology results indicated that, unexpectedly, there was not very much inflammation in these mice by day 3 post-infection although the fra locus still had a modest fitness phenotype (greater than 100-fold) (Figure 6). The phenotypes of the fra locus in IL10 knockout mice and in the humanized Swiss Webster mice demonstrate that the fra phenotype is not limited to germ-free or streptomycin-treated mice.
Finally, to test for the possibility that these severe fra mutant phenotypes were the result of interaction between the wild-type and fra mutant during infection, we performed experiments in which strep-treated C57BL/6 Nramp1+/− heterozygous mice were infected separately with the wild-type, the fra mutant, or the complemented fra mutant. The strains were quantitated in the feces each day post-infection for four days at which point the mice were sacrificed and the strains were quantitated in the cecum. The fra mutant was recovered in 30-fold lower numbers than wild-type on the fourth day in the feces and 98-fold lower in the cecum (Figure 7). This defect was restored by complementation with the fra locus on a plasmid in the cecum, while in the feces the restoration did not reach statistical significance (Figure 7).
Fig. 7. Quantitation of Salmonella in feces on days 1 through 4, and cecum on day 4, post-infection. 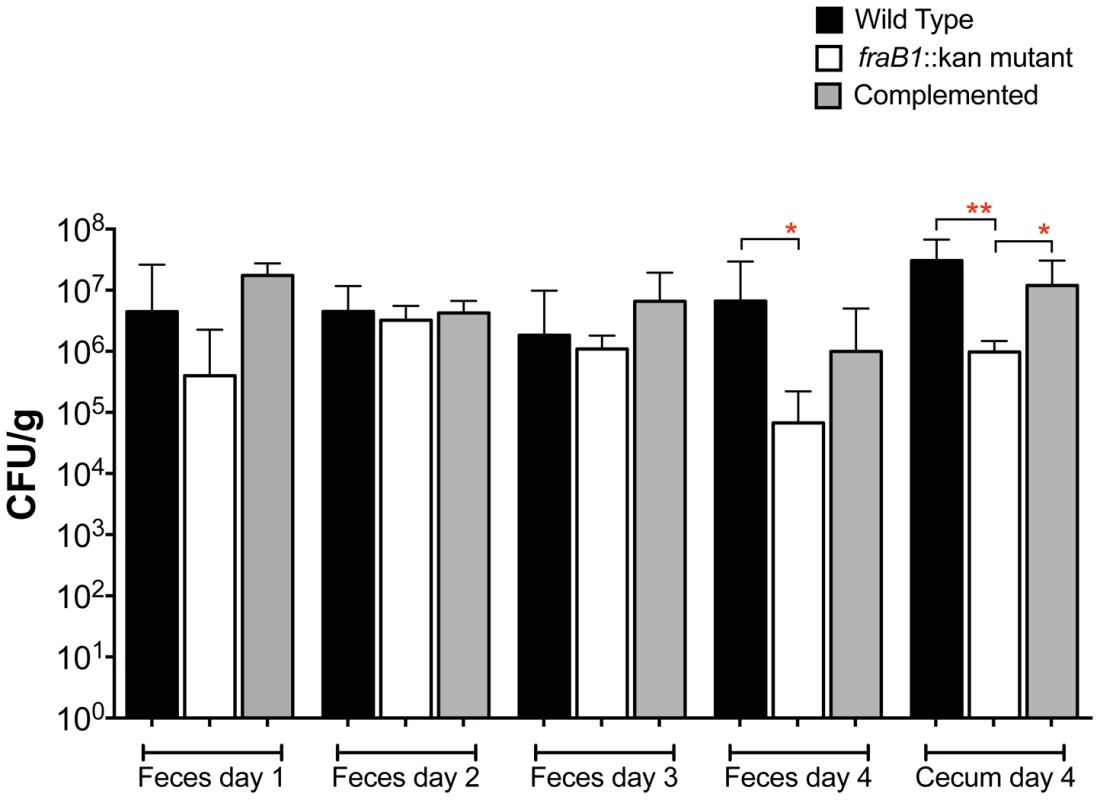
Groups of five C57BL/6 mice heterozygous for Nramp1 were orally inoculated with 107 CFU of IR715 (wild-type), MA59 (fraB1::kan mutant), or ASD6000 (fraB1::kan mutant with complementation plasmid pASD5006). The geometric mean+SE is shown. Statistical significance between select groups was determined by using an unpaired two-tailed Student t test. * = P value<0.05, ** = P value<0.01. The fra locus is required for growth on fructose-asparagine (F-Asn)
FraA is homologous to the Dcu family of dicarboxylate transporters. However, authentic dicarboxylate acquisition loci do not encode a sugar kinase or phosphosugar isomerase. Furthermore, none of the dicarboxylates that we tested (malate, fumarate or succinate) provided a growth advantage to the wild-type strain vs. a fraB1::kan mutant, suggesting that they are not substrates of the Fra pathway. BLAST searches using the entire operon revealed that the closest homolog is the frl operon of E. coli, although the frl operon is at a different location within the genome and does not encode an asparaginase (and the Salmonella fra locus does not encode a frlC homolog). The products of the E. coli frl operon transport and degrade the Amadori product fructose-lysine (F-Lys) [27], [28]. Amadori products most often result from a spontaneous reaction between a carbonyl group (often of glucose, although numerous other compounds can also react) and an amino group of an amino acid in vivo, and are then referred to as non-enzymatic glycation products [29], [30]. With F-Lys and fructose-arginine (F-Arg) this can happen with the free amino acid, or the side groups of the lysine and arginine residues of a protein. In contrast, fructose-asparagine (F-Asn) can only result from reaction of glucose with the alpha amino group of free asparagine or the N-terminal asparagine of a protein. We synthesized three different Amadori products, F-Lys, F-Arg, and F-Asn and used them as sole carbon sources during growth experiments. The preparations were free of glucose but contained some free amino acid. However, control experiments demonstrated that Salmonella was unable to grow on any of the three amino acids alone, so these contaminants are inconsequential (Figure 8D). Salmonella was unable to grow on F-Arg, and grew slowly and with low yield on F-Lys (Figure 8B, C). The growth on F-Lys was independent of the fra locus. In contrast, Salmonella grew as well on F-Asn as on glucose, and growth on F-Asn was dependent upon the fra locus (hence the name fra, for fructose-asparagine utilization) (Figure 8A). A commercial source of F-Asn was obtained and it also allowed Salmonella to grow in a fra-dependent manner (structure shown in Figure 8F). Complementation of the fraB1::kan mutant with a plasmid encoding the fra island restored the ability of the mutant to grow on F-Asn (Figure 8E). In addition to serving as a sole carbon source, F-Asn, also served as sole nitrogen source (Figure 9).
Fig. 8. Growth of wild-type and fraB1::kan mutant Salmonella on Amadori products. 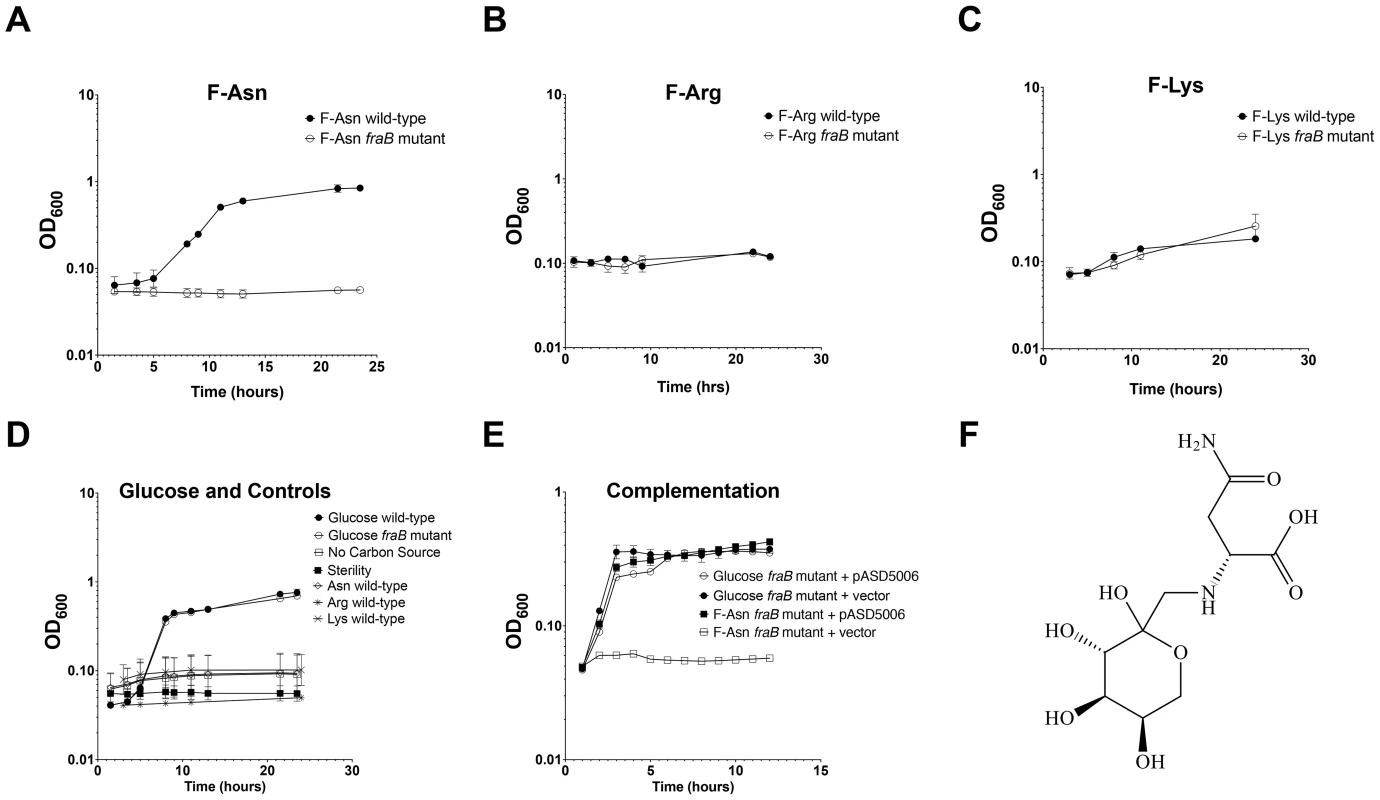
Growth of wild-type MA43 and fraB1::kan mutant MA59 on F-Asn (A), F-Arg (B), F-Lys (C), asparagine, arginine, lysine, or glucose (D). Bacteria were grown overnight in LB at 37°C shaking, centrifuged, resuspended in water, and subcultured 1∶1000 into NCE medium containing the indicated carbon source at 5 mM. The optical density at 600 nm was then read at time points during growth at 37°C with shaking. Controls included NCE with no carbon source, and NCE with glucose that was not inoculated, as a sterility control (D). E) Complementation of a fraB1::kan mutation with plasmid pASD5006 encoding the fra island (ASD6000) or the vector control, pWSK29 (ASD6010). Each point in (A)–(E) represents the mean of three cultures with error bars indicating standard deviation. F) The structure of F-Asn (CAS#34393-27-6). Fig. 9. Growth of Salmonella on F-Asn as sole nitrogen source. 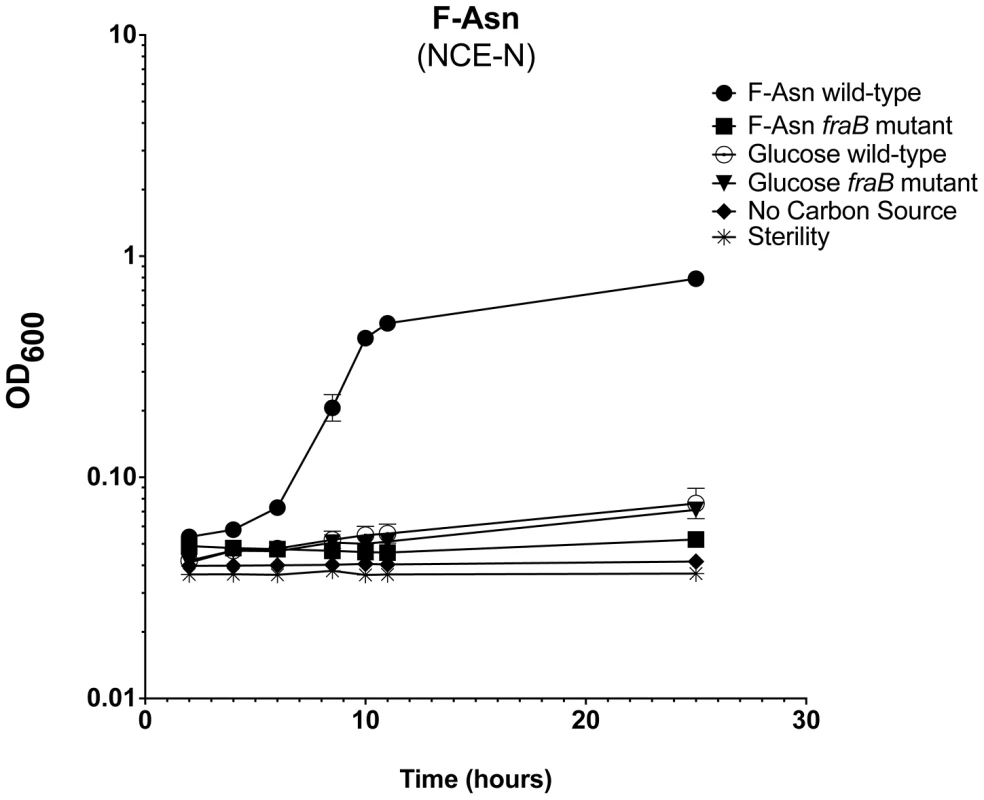
Growth of wild-type MA43 and fraB1::kan mutant MA59 on F-Asn. Bacteria were grown overnight in LB at 37°C shaking, centrifuged, resuspended in water, and subcultured 1∶1000 into NCE medium lacking a nitrogen source (NCE-N) but containing the indicated carbon source at 5 mM. The optical density at 600 nm was then read at time points during growth at 37°C with shaking. Controls included NCE-N with no carbon source, NCE-N with 5 mM glucose, and NCE-N with glucose that was not inoculated, as a sterility control. Each point represents the mean of four cultures and error bars represent standard deviation. Growth with F-Asn was tested under aerobic and anaerobic conditions in the presence or absence of the terminal electron acceptor tetrathionate (Figure 10). The F-Asn was utilized under all conditions, but respiratory conditions were superior with a doubling time of 1.6+/−0.1 hours aerobically with tetrathionate, 2.0+/−0.3 hours aerobically without tetrathionate, 1.9+/−0.1 hours anaerobically with tetrathionate, and 2.9+/−0.4 hours anaerobically without tetrathionate. Competition experiments in which the wild-type and fraB1::kan mutant were grown in the same culture were performed in minimal medium containing F-Asn. As expected, the mutant was severely attenuated during aerobic and anaerobic growth, and in the presence or absence of tetrathionate (Figure 11). The attenuation was most severe during anaerobic growth in the presence of tetrathionate.
Fig. 10. Growth of Salmonella on F-Asn in the presence or absence of tetrathionate or oxygen. 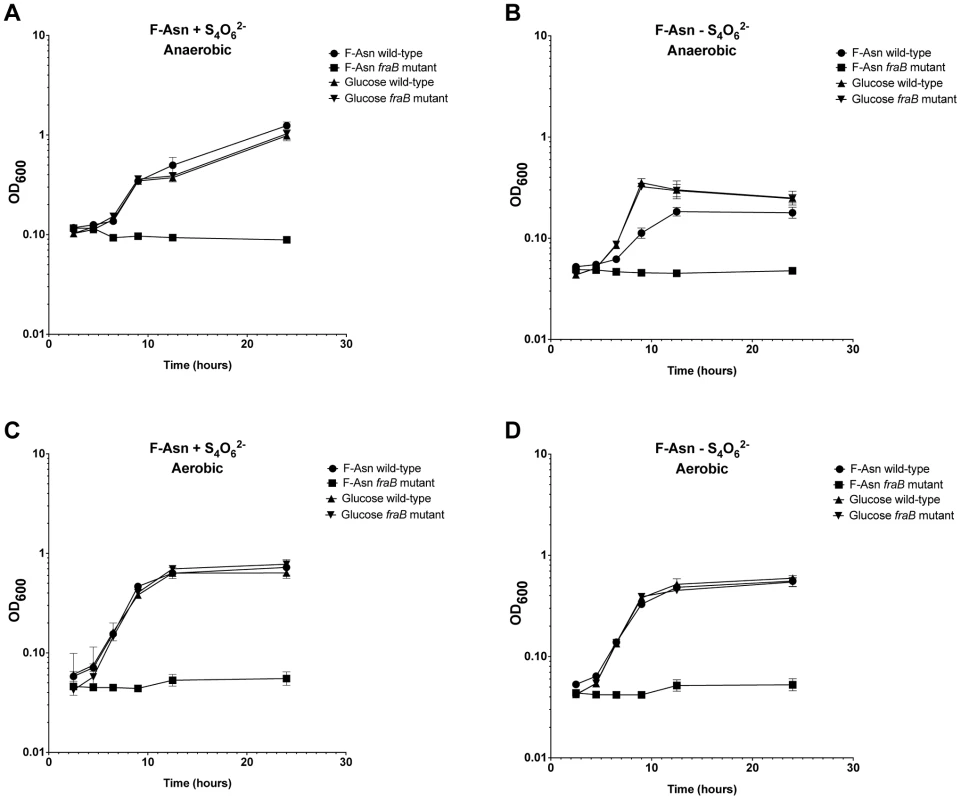
Growth of wild-type MA43 and fraB1::kan mutant MA59 on 5 mM F-Asn or 5 mM glucose anaerobically (A and B) or aerobically (C and D) in the presence (A and C) or absence (B and D) of 40 mM tetrathionate (S4062−). Bacteria were grown overnight in LB at 37°C shaking, centrifuged, resuspended in water, and subcultured 1∶1000 into NCE medium containing the indicated carbon source. The optical density at 600 nm was then read at time points during growth at 37°C with shaking. Each point represents the mean of four cultures with error bars indicating standard deviation. Fig. 11. Competitive index measurements of a fraB1::kan mutant during in vitro growth. 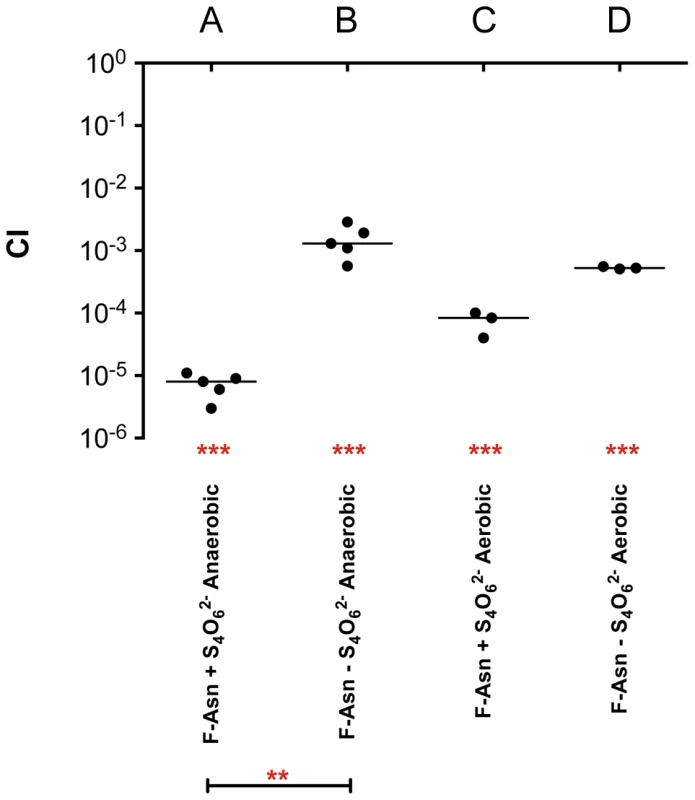
Cultures were grown overnight in LB, pelleted and washed in water, subcultured 1∶10,000 and grown for 24 hours at 37°C in NCE minimal medium containing 5 mM F-Asn, aerobically or anaerobically, in the presence or absence of tetrathionate (S4O62−), as indicated. A) Anaerobic growth in the presence of tetrathionate, B) anaerobic growth in the absence of tetrathionate, C) aerobic growth in the presence of tetrathionate, D) aerobic growth in the absence of tetrathionate. Each data point represents the CI from one culture with the median shown by a horizontal line. Statistical significance of each group being different than 1 was determined by using a one sample Student's t test. Statistical significance between select groups was determined using a Mann-Whitney test. ** = P value<0.01, *** = P value<0.001. Discussion
The mechanisms by which microbes interact with each other in the gastrointestinal tract are largely unknown. Screening large libraries of bacterial mutants for fitness defects in animals with defined microbiota can be used to identify those genes that are only required in the presence of specific members of the microbiota [15]. In this report, we took a highly reductionist approach and screened for genes that were differentially required in germ-free mice versus ex-germ-free mice colonized with a single commensal Enterobacter cloacae isolate. Only five genes were differentially required, a two component response regulatory pair, barA/sirA, and three genes within the fra locus (Table 1). Individual sirA and fraB mutants were used to confirm the findings. The sirA gene was required for fitness in the presence of E. cloacae but not in its absence (Figure 2). The fra locus was required for fitness in both situations, but the phenotype was more severe in the presence of E. cloacae (Figure 4A, B). Thus, the differential screening strategy was successful in identifying genes that are more important in the presence of other bacteria within the gastrointestinal tract. The reason(s) that sirA is required in the presence, but not the absence, of E. cloacae is not known. It is thought that BarA detects short chain fatty acids produced by the normal microbiota and then phosphorylates SirA [31]–[34]. SirA then activates the transcription of two small RNAs, csrB and csrC, which antagonize the activity of the CsrA protein [20], [35]–[39]. The CsrA protein is an RNA-binding protein that regulates the stability and translation of hundreds of mRNAs involved with metabolism and virulence [17], [19], [40]. One possible reason that sirA differentially affects fitness in the two mouse models may be that the Enterobacter-colonized mouse offers an environment richer in carboxylic acids that act as stimuli for BarA-SirA signaling with resulting effects on metabolism and growth [31]–[34]. The fitness effects could also be due to the regulation of genes involved in the induction of inflammation and/or anareobic metabolism including SPI1, SPI2, ethanolamine utilization, and vitamin B12 biosynthesis by CsrA [19], [20], [41]–[44]. Finally, SirA or CsrA may regulate the fra locus itself.
The fra locus was annotated as a C4 dicarboxylate uptake system. However, we found that the fra locus played no role in the utilization of C4 dicarboxylates. BLAST searches revealed that the operon is similar to the frl locus of E. coli which is required for the utilization of fructose-lysine (F-Lys). The frl locus of E. coli has a different genomic context than the fra locus of Salmonella, and is only distantly related. We determined that the fra locus of Salmonella plays no role in the utilization of F-Lys (Figure 8C). However, the presence of an asparaginase in the fra locus (fraE), but not the frl locus, led us to hypothesize that F-Asn may be the correct nutrient, and indeed, this was the case. Wild-type Salmonella is able to grow as well on F-Asn as on glucose, and this ability is dependent upon the fra locus (Figures 8, 10). While the individual members of the fra operon have not been characterized, we hypothesize as to their functions in Figure 12. F-Asn differs from ethanolamine in that it can be fermented (Figure 10B), which would be consistent with the proposed release of glucose-6-P by FraB (Figure 12). Although F-Asn can be fermented, it only provides a fitness advantage in vivo when it can be respired, i.e., when tetrathionate reductase is functional (Figure 4M–O), possibly because of the much greater energy yield from respiration versus fermentation. E. cloacae grows very poorly on F-Asn and does not encode the fra locus. Therefore, E. cloacae likely exacerbated the fra phenotype of Salmonella by competing for other nutrients.
Fig. 12. A proposed model of Fra protein localization and functions. 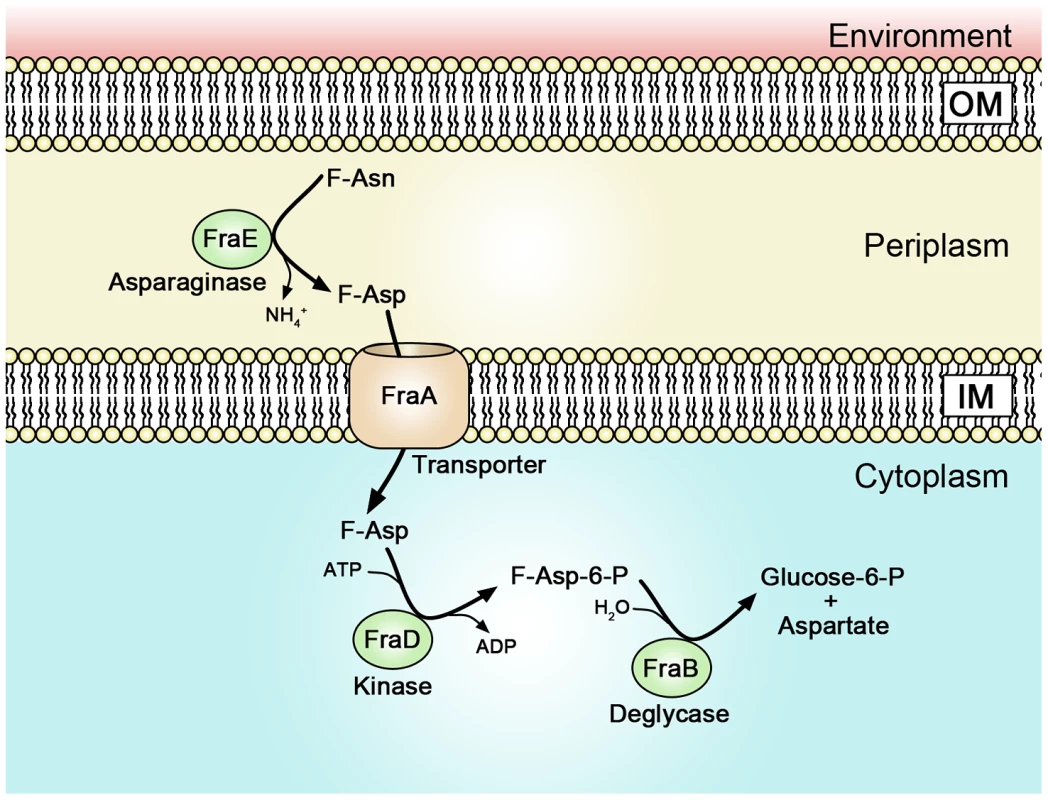
A proteomic survey of subcellular fractions of Salmonella previously identified FraB (the deglycase) as cytoplasmic and FraE (the asparaginase) as periplasmic [79]. Therefore, it is possible that F-Asn is converted to F-Asp in the periplasm by the asparaginase and that the transporter and kinase actually use F-Asp as substrate rather than F-Asn. The FraD kinase of Salmonella shares 30% amino acid identity with the FrlD kinase of E. coli. FrlD phosphorylates F-Lys to form F-Lys-6-P [28]. Therefore, we hypothesize that FraD phosphorylates F-Asp to form F-Asp-6-P. The FrlB deglycase of E. coli shares 28% amino acid identity with FraB of Salmonella. The FrlB deglycase converts F-Lys-6-P to lysine and glucose-6-P [28], so we hypothesize that FraB of Salmonella converts F-Asp-6-P to aspartate and glucose-6-P. F-Asn is an Amadori compound (also known as a glycation product) formed by reaction between glucose in its open chain form with the alpha amino group of asparagine followed by a rearrangement that gives the fructose derivative. Until this report, no organism had been shown to synthesize or utilize F-Asn. However, in the early 2000s it was discovered that acrylamide is present in many foods, especially French fries and potato chips. F-Asn is a precursor to acrylamide. After the acrylamide discovery, numerous papers measured acrylamide concentration, and the precursor molecules, glucose and asparagine, in foods [45]–[52]. However, to the best of our knowledge, only two reports have measured the concentration of F-Asn in a few fruits and vegetables [53], [54]. The concentrations are surprisingly high, ranging between 0.1% (carrot) and 1.4% dry weight (asparagus) [54]. Factors that influence these concentrations are time, temperature, pressure, and perhaps less obviously, moisture content [55]. Any reducing sugar and any amino acid (or other amines) can form compounds analogous to F-Asn. It is important to note that these Amadori compounds are not the ultimate products since with further time and heating they decompose to a large variety of other products, some of which are responsible for a variety of flavors, and the brown color, in cooked foods [55]–[57]. In fact, glycation products form spontaneously in the human body and provide an indication of glucose concentration over time [30], [58]–[60]. A common diabetes test measures the glycation of the N-terminal valine of hemoglobin [30].
The severity of the fra fitness phenotype suggests that F-Asn is the primary nutrient used by Salmonella during growth in the inflamed intestine. For perspective, in strep-treated mice the fitness defect of a fra mutant is 1000-fold, while mutants unable to utilize ethanolamine or sialic acid are attenuated 10-fold and 2-fold, respectively [8], [61]. The fra operon was previously identified by transcription profiling as up-regulated by Fur under anaerobic conditions [62]. Other genes activated under the same conditions included ethanolamine utilization (eut), and hilA, a regulator of SPI1 expression. Both of these loci are associated with induction of inflammation or growth during inflammation [8], [62]. The fra locus is present among most Salmonella serovars, but is disrupted in serovars Typhi and Paratyphi A, consistent with the marked degradation of numerous loci involved with anaerobic respiration among these extra-intestinal serovars [63]. Interestingly, a putative fra locus is present in Citrobacter rodentium and Citrobacter freundii, but not in numerous other non-pathogenic Citrobacter species. The frl locus, encoding the ability to utilize F-Lys is present in E. coli, Shigella, and Cronobacter. It will be interesting to determine which, if any, members of the normal microbiota can compete with E. coli and Salmonella for Amadori products.
The apparent species-specificity of the F-Asn utilization system, and the severity of the fitness defect associated with mutants that cannot metabolize F-Asn, indicate that the Fra system represents a specific and valuable therapeutic target. Further studies are needed to determine the role of each gene in the fra locus with regard to F-Asn metabolism. Similarly, further studies are needed to determine the mechanism by which the proposed transcription factor, FraR, regulates F-Asn metabolism. These structure-function studies will facilitate small molecule drug screens targeting F-Asn utilization. It will also be interesting to determine the concentration of F-Asn and other Amadori products in a wide variety of foods, to determine if these products can affect disease susceptibility, and to explore the possibility of preventing salmonellosis or other infections by removing Amadori products from specific food products or from the diet in general. The utilization systems for many more Amadori products are likely awaiting discovery within bacterial genomes and these may play interesting roles in microbial ecology and human health.
Materials and Methods
Bacterial strains and media
Bacteria were grown in Luria-Bertani (LB) broth or on LB agar plates (EM Science) unless otherwise noted. The minimal medium used was NCE (no carbon E) [64] containing trace metals [25]. Chloramphenicol (cam), streptomycin (strep), or kanamycin (kan) were added at 30, 200, or 60 µg/ml, respectively, when appropriate. Fructose-asparagine was either synthesized or purchased from Toronto Research Chemicals, catalog #F792525. Anaerobic growth was performed in a Bactron 1 anaerobic chamber containing 90% N2, 5% CO2, and 5% H2 (Shel Lab). Strains used are described in Table 2. Enterobacter cloacae strain JLD400 was isolated in our laboratory by plating fecal samples from a conventional BALB/c mouse onto LB agar plates. This particular isolate was chosen because it is easy to culture and genetically manipulable (the strain can be electroporated, maintains ColE1-based plasmids, and can act as a recipient in RP4-mediated mobilization of a suicide vector used to deliver mTn5-luxCDABE, not shown). The species identification was performed using a Dade Microscan Walkaway 96si at the Ohio State University medical center. Additionally, genomic DNA sequences have been obtained that flank mTn5-luxCDABE insertions in JLD400 and these DNA sequences match the draft genome sequence of E. cloacae NCTC 9394.
Tab. 2. Bacterial strains and plasmids. 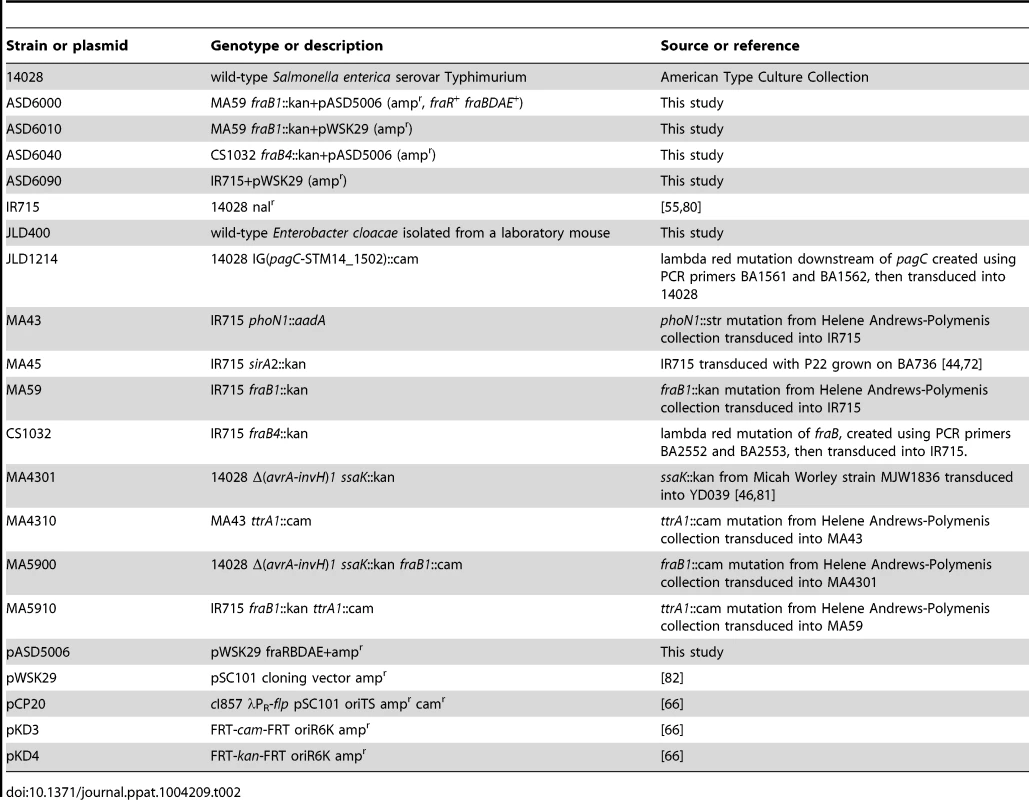
Salmonella mutant library
A transposon mutant library was constructed in S. enterica serovar Typhimurium strain 14028. EZ-Tn5 <T7/kan> transposomes from Epicentre Technologies were delivered to Salmonella by electroporation. This transposon encodes kanamycin resistance and has a T7 RNA Polymerase promoter at the edge of the transposon pointed outward. The resulting library contains between 190,000 and 200,000 independent transposon insertions and is referred to as the JLD200k library. The insertion points of this library have been determined previously by next-generation sequencing [65]. It is estimated that approximately 4400 of the 4800 genes in the Salmonella genome are non-essential with regard to growth on LB agar plates [65]. Therefore, the JLD200k library is saturated with each gene having an average of 43 independent transposon insertions.
Construction of mutations
A FRT-kan-FRT or FRT-cam-FRT cassette, generated using PCR with the primers listed in Table 3 and pKD3 or pKD4 as template, was inserted into each gene of interest (replacing all but the first ten and last ten codons) using lambda Red mutagenesis of strain 14028+pKD46 followed by growth at 37°C to remove the plasmid [66]. A temperature sensitive plasmid encoding FLP recombinase, pCP20, was then added to each strain to remove the antibiotic resistance marker [66]. The pCP20 plasmid was cured by growth at 37°C. A fraB4::kan mutation was constructed using primers BA2552 and BA2553 (Table 3). A FRT-cam-FRT was placed in an intergenic region downstream of pagC using primers BA1561 and BA1562 (deleting and inserting between nucleotides 1342878 and 1343056 of the 14028 genome sequence (accession number NC_016856.1) (Table 3).
Tab. 3. Oligonucleotides used. 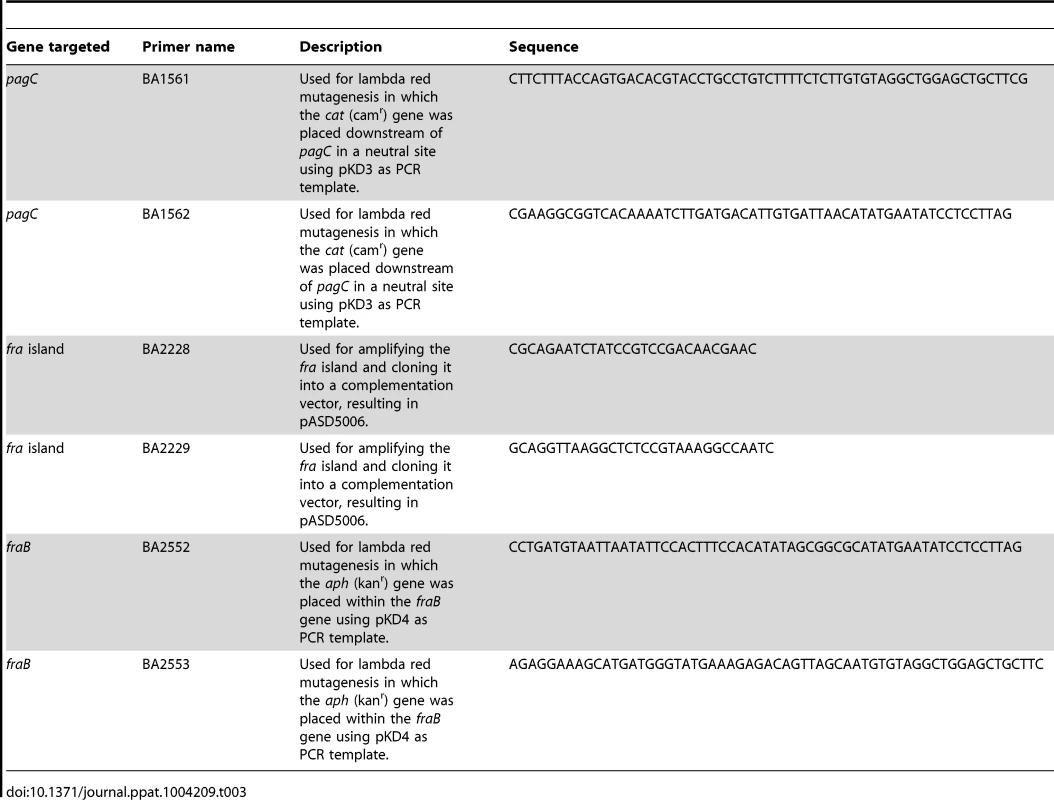
Animals
Germ-free C57BL/6 mice were obtained from Balfour Sartor of the NIH gnotobiotic resource facility at the University of North Carolina and from Kate Eaton at the University of Michigan. Germ-free Swiss Webster mice were obtained from Taconic Farms. The mice were bred and maintained under germ-free conditions in sterile isolators (Park Bioservices). Periodic Gram-staining, 16 s PCR, and pathology tests performed by the Ohio State University lab animal resources department and our own laboratory were used to confirm that the mice contained no detectable microorganisms. Conventional C57BL/6 mice were obtained from Taconic Farms. C57BL/6 mice that were heterozygous for the Nramp1 gene were generated by breeding the standard Nramp1−/− mice from Taconic Farms with C57BL/6 Nramp1+/+ mice from Greg Barton [67]. IL10 knockout mice (B6.129P2-IL10tm1Cgn/J) were obtained from Jackson Laboratory. Germ-free Swiss Webster mice were “humanized” by intragastric inoculation of 200 µl of human feces obtained from an anonymous healthy donor from the OSU fecal transplant center.
Transposon Site Hybridization (TraSH)
The JLD200k transposon mutant library was grown in germ-free C57BL/6 mice in the presence or absence of E. cloacae strain JLD400. Four mice were inoculated intragastrically (i.g.) with 107 cfu of Enterobacter cloacae strain JLD400 that had been grown overnight in LB shaking at 37°C. After 24 hours these mice, and an additional four germ-free mice, were inoculated with 107 cfu of the JLD200k library that had been grown overnight in shaking LB kan at 37°C. Prior to inoculation of the mice, the library was spiked with an additional mutant, JLD1214, at a 1∶10∶000 ratio. This mutant contains a chloramphenicol resistance (camr) gene at a neutral location in the chromosome in the intergenic region downstream of pagC [68]. After inoculation of mice with the spiked library, the inoculum was dilution plated to quantitate the kanamycin resistant (kanr) Salmonella library members and the camr spike strain. The remainder of the inoculum was pelleted and saved as the “input” for hybridization to microarrays. After 24 hours of infection with the JLD200k library, the mice were euthanized and organs were harvested (small intestine, cecum, large intestine, and spleen). One germ-free mouse died prior to organ harvest and was not used. All samples were homogenized and dilution plated to determine Salmonella counts. The remainder of the homogenate was added to 25 ml LB kan and grown overnight with shaking at 37°C to recover the library members. Each culture was then pelleted and frozen as a potential “output” sample for microarray analysis. The kanr and camr colony counts recovered from each organ indicated that the spike ratio of 1∶10,000 was maintained in the intestinal samples but not in the spleen samples. This indicates that the library underwent a population bottleneck on the way to the spleen so microarray analysis of spleen samples would not be informative. The cecum samples were chosen for microarray analysis. There was one “input” sample for all arrays. There were seven separate “output” samples for the arrays; four from the cecums of Enterobacter-associated mice and three from germ-free mice. The output from each mouse was compared to the input on a single array. We also did a single “in vitro” array experiment in which the JLD200k library was grown in the presence of Enterobacter in liquid LB broth shaking at 37°C.
Genomic DNA was isolated from the input and output bacterial pellets. The purity and concentration of the DNA samples was assessed using a Nanodrop spectrophotometer and the quality of the DNA was assessed via agarose gel electrophoresis. All seven samples had high quality intact genomic DNA. The DNA was digested using a restriction endonuclease (RsaI). Labeled RNA transcripts were obtained from the T7 promoter by in vitro transcription. A two-color hybridization strategy was employed. RNA transcripts from the output samples were fluorescently labeled with Cyanine-5 (Cy5, red), while the input sample was labeled with Cyanine-3 (Cy3, green). Equal molar concentrations of the output and input sample were combined and hybridized to genome-wide tiling microarrays printed commercially by Agilent Technologies. Agilent's SurePrint technology employs phosphoramadite chemistry in combination with high performance Hewlett Packard inkjet technology for in situ synthesis of 60-mer oligos. Using Agilent eArray, an easy-to-use web-based application, we were able to synthesize the arrays used by Chaudhuri et al. that completely tiled both the sense and anti-sense strands of the Salmonella SL1344 genome (AMADID 015511) [10]. Each slide contained 2 arrays, each array with 105,000 features, densely tiling the entire genome. The strain of Salmonella used in our experiments was 14028 and its genome sequence was only recently published (GenBank Nucleotide Accession CP001363 (complete genome) and CP001362 (plasmid)). As such, each of the 60-mer probes used by Chaudhuri et al. [10] were mapped to the 14028 genome using blast, and then annotated with any open reading frames (ORFs) that the probe spanned. A total of 96,749 probes mapped to the 14028 genome, with a median gap between each probe of 35 nucleotides on both strands.
After purification, the labeled samples were denatured and hybridized to the array overnight. Microarray slides were then washed and scanned with an Agilent G2505C Microarray Scanner, at 2 µm resolution. Images were analyzed with Feature Extraction 10.5 (Agilent Technologies, CA). Median foreground intensities were obtained for each spot and imported into the mathematical software package “R”, which was used for all data input, diagnostic plots, normalization and quality checking steps of the analysis process using scripts developed specifically for this analysis. In outline, the intensities were not background corrected as this has been shown to only introduce noise. The dataset was filtered to remove positive control elements and any elements that had been flagged as bad, or not present in the 14028 genome. Using the negative controls on the arrays, the background threshold was determined and all values less than this value were flagged. Finally, the Log2 ratio of output Cy5/input Cy3 (red/green) was determined for each replicate, and the data was normalized by the loess method using the LIMMA (Linear models for microarray data) package in “R” as described [69], [70]. Complete statistical analysis was then performed in “R”. Insertion mutants where the ORF is essential for survival will be selected against, and thus a negative ratio of Cy5/Cy3 (red/green) will be observed in the probes adjacent to the insertion point, resulting from higher Cy3 (green) signal from the input. Conversely, insertion mutants that were advantageous to growth in the output samples would have a positive ratio, resulting from the higher Cy5 (red) signal in the output. Mutants having no effect on growth would have equal ratios in both the output and input samples (yellow). A spreadsheet of these data is available in Dataset S1.
Synthesis of Amadori products
We carried out the syntheses of three fructosyl amino acids with asparagine, lysine, and arginine. Hodge and Fisher's review of Amadori products was consulted as an essential starting point for synthesis [71] and the recent review by Mossine and Mawhinney of all aspects of fructose-amines was a treasure house of information [55]. We found the method of Wang et al. [72] to be the most satisfactory, however reaction times cannot be standardized and excess glucose must be removed. The reaction with asparagine is slow because asparagine is sparingly soluble in methanol. By contrast, the reaction with α-Boc-lysine is fast. Arginine is an intermediate case. Previous syntheses of F-Asn include those of Stadler et al. [46], Wang et al. [72], and Miura et al. [73]. The procedure of Stadler et al. [46] uses alkaline conditions which we thought could bring about isomerization of the sugar and racemization of the amino acid. We chose to develop the synthesis of Wang et al. [72] after trying a number of different protocols described for other amino acids [74]–[77]. Wang et al. [72], however, describe only a general method and asparagine presents some particular problems, the most important of which is the poor solubility of asparagine in methanol. We added bisulfite to the reaction mixture to reduce the formation of colored by-products [57] and finally removed excess glucose by use of a cation-exchange column according to the method of Mossine et al. [78]. Using methanol alone as solvent gives the product after refluxing for 24 hr. in approximately 10–15% yield together with recovery of about 90% of the asparagine. Although the yield is low, the starting materials are inexpensive, and the insolubility of asparagine has the advantage that F-Asn, which is quite soluble in methanol, emerges from the ion exchange column almost free of asparagine. This gave a free-flowing off-white non-hygroscopic solid. The 1H-NMR spectrum is complex due to the equilibrating mixture of alpha - and beta - pyranose and furanose forms [55], but integration of the upfield resonances due to asparagine and the downfield resonances due to the sugar are in the proper ratio. The material was also characterized by its specific rotation and infrared (IR) spectrum: [α]23D −48° (c = 0.1, water) (reference [73] −40°, c = 1, water); IR (Nujol): 3350,3155, 1668, 1633, 1455, 1408, 1080 cm−1. Compare our preparations to results in [71], [73].
Competition assays
Competition assays were performed in which a mutant strain was mixed in a 1∶1 ratio with an isogenic wild-type and inoculated by the intragastric (i.g.) or intraperitoneal (i.p.) route to mice. Fecal samples, intestinal sections, spleen and liver were recovered at specific times post-infection, homogenized and plated on selective plates. The wild-type and mutant strains were differentiated by antibiotic resistance. The competitive index was calculated as CI = (cfu of mutant recovered/cfu w.t. recovered)/(cfu mutant input/cfu w.t. input). If the mutant is defective compared to the wild-type it will have a CI of less than 1.
Complementation assays
The fra island was PCR amplified from purified 14028 genomic DNA with primers BA2228 and BA2229 using Phusion polymerase (New England Biolabs). The PCR product was cloned into pPCR-Blunt II-TOPO (Invitrogen). The resulting clones were digested with EcoRI (New England Biolabs), run on an agarose gel and the 8.6 kbp fra fragment was gel purified (Qiagen). This purified DNA fragment was ligated into pWSK29 digested with EcoRI (NEB) using T4 DNA ligase (New England Biolabs) overnight at 4°C. The ligation reaction was transformed into DH5α and plated on LB containing ampicillin at 37°C. The resulting plasmid, pASD5006, or the vector control pWSK29, were electroporated into the appropriate strains.
Ethics statement
All animal work was performed in accordance with the protocols approved by our Institutional Animal Care and Use Committee (OSU 2009A0035). The IACUC ensures compliance of this protocol with the U.S Animal Welfare Act, Guide for Care and Use of Laboratory Animals and Public Health Service Policy on Humane Care and Use of Laboratory Animals. Human fecal material was obtained from an anonymous healthy donor at the Ohio State University fecal transplant center in accordance with the protocol approved by our Institutional Review Board (OSU 2012H0367).
Supporting Information
Zdroje
1. GordonMA (2011) Invasive nontyphoidal Salmonella disease. Current Opinion in Infectious Diseases 24 : 484–489 doi:10.1097/QCO.0b013e32834a9980
2. ChenH-M, WangY, SuL-H, ChiuC-H (2013) Nontyphoid Salmonella Infection: Microbiology, Clinical Features, and Antimicrobial Therapy. Pediatrics & Neonatology 54 : 147–152 doi:10.1016/j.pedneo.2013.01.010
3. BeckerD, SelbachM, RollenhagenC, BallmaierM, MeyerTF, et al. (2006) Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440 : 303–307 doi:10.1038/nature04616
4. SteebB, ClaudiB, BurtonNA, TienzP, SchmidtA, et al. (2013) Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog 9: e1003301 doi:10.1371/journal.ppat.1003301
5. StecherB, RobbianiR, WalkerAW, WestendorfAM, BarthelM, et al. (2007) Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5 : 2177–2189 doi:10.1371/journal.pbio.0050244
6. WinterSE, ThiennimitrP, WinterMG, ButlerBP, HusebyDL, et al. (2010) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467 : 426–429 doi:10.1038/nature09415
7. SekirovI, GillN, JogovaM, TamN, RobertsonM, et al. (2010) Salmonella SPI-1-mediated neutrophil recruitment during enteric colitis is associated with reduction and alteration in intestinal microbiota. Gut Microbes 1 : 30–41 doi:10.4161/gmic.1.1.10950
8. ThiennimitrP, WinterSE, WinterMG, XavierMN, TolstikovV, et al. (2011) Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA 108 : 17480–17485 doi:10.1073/pnas.1107857108
9. HapfelmeierS, StecherB, BarthelM, KremerM, MüllerAJ, et al. (2005) The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol 174 : 1675–1685.
10. ChaudhuriRR, PetersSE, PleasanceSJ, NorthenH, WillersC, et al. (2009) Comprehensive identification of Salmonella enterica serovar Typhimurium genes required for infection of BALB/c mice. PLoS Pathog 5: e1000529 doi:10.1371/journal.ppat.1000529
11. SantiviagoCA, ReynoldsMM, PorwollikS, ChoiSH, LongF, et al. (2009) Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog 5: e1000477 doi:10.1371/journal.ppat.1000477
12. LawleyTD, ChanK, ThompsonLJ, KimCC, GovoniGR, et al. (2006) Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog 2: e11.
13. BadarinarayanaV, EstepPW3, ShendureJ, EdwardsJ, TavazoieS, et al. (2001) Selection analyses of insertional mutants using subgenic-resolution arrays. Nat Biotechnol 19 : 1060–1065.
14. SassettiCM, BoydDH, RubinEJ (2001) Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci USA 98 : 12712–12717 doi:10.1073/pnas.231275498
15. GoodmanAL, McNultyNP, ZhaoY, LeipD, MitraRD, et al. (2009) Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6 : 279–289 doi:10.1016/j.chom.2009.08.003
16. TeplitskiM, AhmerBMM, PrüssBM (2005) The control of secondary metabolism, motility, and virulence by the two-component regulatory system BarA/SirA of Salmonella and other γ-proteobacteria. Research Signpost 26.
17. RomeoT, VakulskasCA, BabitzkeP (2013) Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol 15 : 313–324 doi:10.1111/j.1462-2920.2012.02794.x
18. LapougeK, SchubertM, AllainFH-T, HaasD (2008) Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67 : 241–253 doi:10.1111/j.1365-2958.2007.06042.x
19. LawhonSD, FryeJG, SuyemotoM, PorwollikS, McClellandM, et al. (2003) Global regulation by CsrA in Salmonella typhimurium. Mol Microbiol 48 : 1633–1645.
20. MartínezLC, YakhninH, CamachoMI, GeorgellisD, BabitzkeP, et al. (2011) Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol 80 : 1637–1656 doi:10.1111/j.1365-2958.2011.07674.x
21. BarthelM, HapfelmeierS, Quintanilla-MartinezL, KremerM, RohdeM, et al. (2003) Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71 : 2839–2858.
22. WooH, OkamotoS, GuineyD, GunnJS, FiererJ (2008) A model of Salmonella colitis with features of diarrhea in SLC11A1 wild-type mice. PLoS ONE 3: e1603 doi:10.1371/journal.pone.0001603
23. GarnerCD, AntonopoulosDA, WagnerB, DuhamelGE, KeresztesI, et al. (2009) Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar Typhimurium murine model of infection. Infect Immun 77 : 2691–2702 doi:10.1128/IAI.01570-08
24. KaiserP, DiardM, StecherB, HardtW-D (2012) The streptomycin mouse model for Salmonella diarrhea: functional analysis of the microbiota, the pathogen“s virulence factors, and the host”s mucosal immune response. Immunol Rev 245 : 56–83 doi:10.1111/j.1600-065X.2011.01070.x
25. Price-CarterM, TingeyJ, BobikTA, RothJR (2001) The Alternative Electron Acceptor Tetrathionate Supports B12-Dependent Anaerobic Growth of Salmonella enterica Serovar Typhimurium on Ethanolamine or 1,2-Propanediol. J Bacteriol 183 : 2463–2475 doi:10.1128/JB.183.8.2463-2475.2001
26. ChungH, PampSJ, HillJA, SuranaNK, EdelmanSM, et al. (2012) Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149 : 1578–1593 doi:10.1016/j.cell.2012.04.037
27. WiameE, Van SchaftingenE (2004) Fructoselysine 3-epimerase, an enzyme involved in the metabolism of the unusual Amadori compound psicoselysine in Escherichia coli. Biochem J 378 : 1047–1052 doi:10.1042/BJ20031527
28. WiameE, DelpierreG, CollardF, Van SchaftingenE (2002) Identification of a pathway for the utilization of the Amadori product fructoselysine in Escherichia coli. J Biol Chem 277 : 42523–42529 doi:10.1074/jbc.M200863200
29. ZhangQ, AmesJM, SmithRD, BaynesJW, MetzTO (2009) A Perspective on the Maillard Reaction and the Analysis of Protein Glycation by Mass Spectrometry: Probing the Pathogenesis of Chronic Disease. J Proteome Res 8 : 754–769 doi:10.1021/pr800858h
30. TessierFJ (2010) The Maillard reaction in the human body. The main discoveries and factors that affect glycation. Pathol Biol 58 : 214–219 doi:10.1016/j.patbio.2009.09.014
31. HungC-C, GarnerCD, SlauchJM, DwyerZW, LawhonSD, et al. (2013) The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol Microbiol 87 : 1045–1060 doi:10.1111/mmi.12149
32. ChavezRG, AlvarezAF, RomeoT, GeorgellisD (2010) The physiological stimulus for the BarA sensor kinase. J Bacteriol 192 : 2009–2012 doi:10.1128/JB.01685-09
33. HuangY, SuyemotoM, GarnerCD, CicconiKM, AltierC (2008) Formate acts as a diffusible signal to induce Salmonella invasion. J Bacteriol 190 : 4233–4241 doi:10.1128/JB.00205-08
34. LawhonSD, MaurerR, SuyemotoM, AltierC (2002) Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol 46 : 1451–1464.
35. RomeoT (1998) Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol 29 : 1321–1330.
36. LiuMY, GuiG, WeiB, PrestonJF, OakfordL, et al. (1997) The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem 272 : 17502–17510.
37. TeplitskiM, GoodierRI, AhmerBMM (2006) Catabolite repression of the SirA regulatory cascade in Salmonella enterica. Int J Med Microbiol 296 : 449–466 doi:10.1016/j.ijmm.2006.06.001
38. FortuneDR, SuyemotoM, AltierC (2006) Identification of CsrC and characterization of its role in epithelial cell invasion in Salmonella enterica serovar Typhimurium. Infect Immun 74 : 331–339 doi:10.1128/IAI.74.1.331-339.2006
39. MartínezLC, Martínez-FloresI, SalgadoH, Fernández-MoraM, Medina-RiveraA, et al. (2014) In Silico Identification and Experimental Characterization of Regulatory Elements Controlling the Expression of the Salmonella csrB and csrC Genes. J Bacteriol 196 : 325–336 doi:10.1128/JB.00806-13
40. EdwardsAN, Patterson-FortinLM, VakulskasCA, MercanteJW, PotrykusK, et al. (2011) Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol 80 : 1561–1580 doi:10.1111/j.1365-2958.2011.07663.x
41. BustamanteVH, MartínezLC, SantanaFJ, KnodlerLA, Steele-MortimerO, et al. (2008) HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci USA 105 : 14591–14596 doi:10.1073/pnas.0801205105
42. AltierC, SuyemotoM, RuizAI, BurnhamKD, MaurerR (2000) Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol 35 : 635–646.
43. JohnstonC, PeguesDA, HueckCJ, LeeCA, MillerSI (1996) Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol 22 : 715–727 doi:10.1046/j.1365-2958.1996.d01-1719.x
44. AhmerBM, van ReeuwijkJ, WatsonPR, WallisTS, HeffronF (1999) Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol 31 : 971–982.
45. VivantiV, FinottiE, FriedmanM (2006) Level of acrylamide precursors asparagine, fructose, glucose, and sucrose in potatoes sold at retail in Italy and in the United States. Journal of food science 71: C81–C85.
46. StadlerRH, RobertF, RiedikerS, VargaN, DavidekT, et al. (2004) In-depth mechanistic study on the formation of acrylamide and other vinylogous compounds by the Maillard reaction. J Agric Food Chem 52 : 5550–5558 doi:10.1021/jf0495486
47. SurdykN, RosénJ, AnderssonR, AmanP (2004) Effects of asparagine, fructose, and baking conditions on acrylamide content in yeast-leavened wheat bread. J Agric Food Chem 52 : 2047–2051 doi:10.1021/jf034999w
48. YaylayanVA, WnorowskiA, Perez LocasC (2003) Why Asparagine Needs Carbohydrates To Generate Acrylamide. J Agric Food Chem 51 : 1753–1757 doi:10.1021/jf0261506
49. MottramDS, WedzichaBL, DodsonAT (2002) Food chemistry: Acrylamide is formed in the Maillard reaction. Nature 419 : 448–449 doi:10.1038/419448a
50. TarekeE, RydbergP, KarlssonP, ErikssonS, TörnqvistM (2002) Analysis of Acrylamide, a Carcinogen Formed in Heated Foodstuffs. J Agric Food Chem 50 : 4998–5006 doi:10.1021/jf020302f
51. TarekeE, RydbergP, KarlssonP, ErikssonS, TörnqvistM (2000) Acrylamide: A Cooking Carcinogen? Chem Res Toxicol 13 : 517–522 doi:10.1021/tx9901938
52. Elmore JS, Mottram DS (2002) Compilation of free amino acid data for various food raw materials, showing the relative contributions of asparagine, glutamine, aspartic acid and glutamic acid to the free amino acid composition. JIFSAN Acrylamide in Food Workshop, Chicago.
53. AnetEFLJ, ReynoldsTM (1957) Chemistry of non-enzymic browning. II. Reactions between Amino Acids, Organic Acids, and sugars in freeze-dried Apricots and Peaches. Aust J Chem 10 : 182–191 doi:10.1071/CH9570182
54. Eichner K, Reutter M, Wittmann R (1994) Detection of Amadori compounds in heated foods. Thermally Generated Flavors (ACS Symposium Series 543). Parliament TH, Morello MJ, McGorrin RJ, editors, Washington D.C.: American Chemical Society, Chapter 5.
55. MossineVV, MawhinneyTP (2010) 1-Amino-1-deoxy-D-fructose (“fructosamine”) and its derivatives. Adv Carbohydr Chem Biochem 64 : 291–402 doi:10.1016/S0065-2318(10)64006-1
56. Mottram DS (2007) The Maillard Reaction: Source of Flavour in Thermally Processed Foods. Flavours and Fragrances. Berlin, Heidelberg: Springer Berlin Heidelberg. pp. 269–283. doi:10.1007/978-3-540-49339-6_12.
57. AnetEFLJ (1957) Chemistry of non-enzymic browning. II. Some Crystalline Amino Acid-Deoxy-sugars. Aust J Chem 10 : 193–197 doi:10.1071/CH9570193
58. BodigaVL, EdaSR, BodigaS (2013) Advanced glycation end products: role in pathology of diabetic cardiomyopathy. Heart Fail Rev 19 : 49–63 doi:10.1007/s10741-013-9374-y
59. KatoS, ItohK, OchiaiM, IwaiA, ParkY, et al. (2008) Increased pentosidine, an advanced glycation end-product, in urine and tissue reflects disease activity in inflammatory bowel diseases. Journal of Gastroenterology and Hepatology 23: S140–S145 doi:10.1111/j.1440-1746.2008.05552.x
60. BrownleeM (1995) Advanced protein glycosylation in diabetes and aging. Annu Rev Med 46 : 223–234 doi:10.1146/annurev.med.46.1.223
61. NgKM, FerreyraJA, HigginbottomSK, LynchJB, KashyapPC, et al. (2013) Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502 : 96–99 doi:10.1038/nature12503
62. TroxellB, FinkRC, PorwollikS, McClellandM, HassanHM (2011) The Fur regulon in anaerobically grown Salmonella enterica sv. Typhimurium: identification of new Fur targets. BMC Microbiol 11 : 236 doi:10.1186/1471-2180-11-236
63. NuccioSP, BäumlerAJ (2014) Comparative Analysis of Salmonella Genomes Identifies a Metabolic Network for Escalating Growth in the Inflamed Gut. MBio 5: e00929–14–e00929–14 doi:10.1128/mBio.00929-14
64. Davis RW, Botstein D, Roth JR (1980) Advanced bacterial genetics. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory. 1 p.
65. CanalsR, XiaX-Q, FronickC, CliftonSW, AhmerBM, et al. (2012) High-throughput comparison of gene fitness among related bacteria. BMC Genomics 13 : 212 doi:10.1186/1471-2164-13-212
66. DatsenkoKA, WannerBL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97 : 6640–6645 doi:10.1073/pnas.120163297
67. ArpaiaN, GodecJ, LauL, SivickKE, McLaughlinLM, et al. (2011) TLR signaling is required for Salmonella typhimurium virulence. Cell 144 : 675–688 doi:10.1016/j.cell.2011.01.031
68. GunnJS, RyanSS, Van VelkinburghJC, ErnstRK, MillerSI (2000) Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect Immun 68 : 6139–6146.
69. SmythGK, SpeedT (2003) Normalization of cDNA microarray data. Methods 31 : 265–273.
70. SmythGK, YangYH, SpeedT (2003) Statistical issues in cDNA microarray data analysis. Methods Mol Biol 224 : 111–136 doi:10.1385/1-59259-364-X:111
71. HodgeJE, FisherBE (1963) Amadori rearrangement products. Methods in Carbohydrate Chemistry 2 : 99–107.
72. WangJ, LuY-M, LiuB-Z, HeH-Y (2008) Electrospray positive ionization tandem mass spectrometry of Amadori compounds. J Mass Spectrom 43 : 262–264 doi:10.1002/jms.1290
73. MiuraY, TaharaS, MizutaniJ (1973) Isolation and identification of 1-deoxy-1-(L-asparagino)-D-fructose formed in the autoclaved reaction medium. Agric Biol Chem 37 : 2669–2670.
74. KeilP, MortensenHB, ChristophersenC (1985) Fructosylvaline. A simple model of the N-terminal residue of human haemoglobin A1c. Acta Chem Scand, B, Org Chem Biochem 39 : 191–193.
75. KrauseR, KnollK, HenleT (2003) Studies on the formation of furosine and pyridosine during acid hydrolysis of different Amadori products of lysine. Eur Food Res Technol 216 : 277–283 doi:10.1007/s00217-002-0649-0
76. SrinivasSM, HarohallyNV (2012) Improved synthesis of lysine - and arginine-derived Amadori and Heyns products and in vitro measurement of their angiotensin I-converting enzyme inhibitory activity. J Agric Food Chem 60 : 1522–1527 doi:10.1021/jf204185y
77. WeitzelG, GeyerH-U, FretzdorffA-M (1957) Darstellung und Stabilität der Salze von Aminosäure-N-Glykosiden. Chem Ber 90 : 1153–1161 doi:10.1002/cber.19570900641
78. MossineVV, GlinskyGV, FeatherMS (1994) The preparation and characterization of some Amadori compounds (1-amino-1-deoxy-D-fructose derivatives) derived from a series of aliphatic omega-amino acids. Carbohydr Res 262 : 257–270.
79. BrownRN, SanfordJA, ParkJH, DeatherageBL, ChampionBL, et al. (2012) A Comprehensive Subcellular Proteomic Survey of Salmonella Grown under Phagosome-Mimicking versus Standard Laboratory Conditions. Int J Proteomics 2012 : 123076 doi:10.1155/2012/123076
80. StojiljkovicI, BäumlerAJ, HeffronF (1995) Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177 : 1357–1366.
81. TeplitskiM, Al-AgelyA, AhmerBMM (2006) Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiology (Reading, Engl) 152 : 3411–3424 doi:10.1099/mic.0.29118-0
82. WangRF, KushnerSR (1991) Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100 : 195–199.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Recruitment of RED-SMU1 Complex by Influenza A Virus RNA Polymerase to Control Viral mRNA SplicingČlánek Systematic Phenotyping of a Large-Scale Deletion Collection Reveals Novel Antifungal Tolerance GenesČlánek The Contribution of Social Behaviour to the Transmission of Influenza A in a Human Population
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2014 Číslo 6- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Fungal Nail Infections (Onychomycosis): A Never-Ending Story?
- BdlA, DipA and Induced Dispersion Contribute to Acute Virulence and Chronic Persistence of
- Morphotype Transition and Sexual Reproduction Are Genetically Associated in a Ubiquitous Environmental Pathogen
- A Nucleic-Acid Hydrolyzing Single Chain Antibody Confers Resistance to DNA Virus Infection in HeLa Cells and C57BL/6 Mice
- HopW1 from Disrupts the Actin Cytoskeleton to Promote Virulence in Arabidopsis
- Ly6C Monocytes Become Alternatively Activated Macrophages in Schistosome Granulomas with Help from CD4+ Cells
- Recruitment of RED-SMU1 Complex by Influenza A Virus RNA Polymerase to Control Viral mRNA Splicing
- Contribution of Specific Residues of the β-Solenoid Fold to HET-s Prion Function, Amyloid Structure and Stability
- Antibody Responses to : Role in Pathogenesis and Diagnosis of Encephalitis?
- Discovery of a Novel Compound with Anti-Venezuelan Equine Encephalitis Virus Activity That Targets the Nonstructural Protein 2
- Activation of Focal Adhesion Kinase by Suppresses Autophagy via an Akt/mTOR Signaling Pathway and Promotes Bacterial Survival in Macrophages
- Crossing the Interspecies Barrier: Opening the Door to Zoonotic Pathogens
- Catching Fire: , Macrophages, and Pyroptosis
- IscR Is Essential for Type III Secretion and Virulence
- Selective Chemical Inhibition of Quorum Sensing in Promotes Host Defense with Minimal Impact on Resistance
- The Glycosylated Rv1860 Protein of Inhibits Dendritic Cell Mediated TH1 and TH17 Polarization of T Cells and Abrogates Protective Immunity Conferred by BCG
- A Genome-Wide Tethering Screen Reveals Novel Potential Post-Transcriptional Regulators in
- Structural Insights into SraP-Mediated Adhesion to Host Cells
- Human IGF1 Regulates Midgut Oxidative Stress and Epithelial Homeostasis to Balance Lifespan and resistance in
- Cycling Empirical Antibiotic Therapy in Hospitals: Meta-Analysis and Models
- Rab11 Regulates Trafficking of -sialidase to the Plasma Membrane through the Contractile Vacuole Complex of
- Mitogen and Stress Activated Kinases Act Co-operatively with CREB during the Induction of Human Cytomegalovirus Immediate-Early Gene Expression from Latency
- Profilin Promotes Recruitment of Ly6C CCR2 Inflammatory Monocytes That Can Confer Resistance to Bacterial Infection
- A Central Role for Carbon-Overflow Pathways in the Modulation of Bacterial Cell Death
- An Invertebrate Warburg Effect: A Shrimp Virus Achieves Successful Replication by Altering the Host Metabolome via the PI3K-Akt-mTOR Pathway
- The Highly Conserved Bacterial RNase YbeY Is Essential in , Playing a Critical Role in Virulence, Stress Regulation, and RNA Processing
- A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees () Prevails after -Mediated, or , Transmission
- Systematic Phenotyping of a Large-Scale Deletion Collection Reveals Novel Antifungal Tolerance Genes
- Ubiquitin-Mediated Response to Microsporidia and Virus Infection in
- Preclinical Detection of Variant CJD and BSE Prions in Blood
- Toll-Like Receptor 8 Agonist and Bacteria Trigger Potent Activation of Innate Immune Cells in Human Liver
- Progressive Proximal-to-Distal Reduction in Expression of the Tight Junction Complex in Colonic Epithelium of Virally-Suppressed HIV+ Individuals
- The Triggering Receptor Expressed on Myeloid Cells 2 Inhibits Complement Component 1q Effector Mechanisms and Exerts Detrimental Effects during Pneumococcal Pneumonia
- Differential Activation of Acid Sphingomyelinase and Ceramide Release Determines Invasiveness of into Brain Endothelial Cells
- Forward Genetic Screening Identifies a Small Molecule That Blocks Growth by Inhibiting Both Host- and Parasite-Encoded Kinases
- Defining Immune Engagement Thresholds for Control of Virus-Driven Lymphoproliferation
- Growth Factor and Th2 Cytokine Signaling Pathways Converge at STAT6 to Promote Arginase Expression in Progressive Experimental Visceral Leishmaniasis
- Multimeric Assembly of Host-Pathogen Adhesion Complexes Involved in Apicomplexan Invasion
- Biogenesis of Influenza A Virus Hemagglutinin Cross-Protective Stem Epitopes
- Adequate Th2-Type Response Associates with Restricted Bacterial Growth in Latent Mycobacterial Infection of Zebrafish
- Protective Efficacy of Passive Immunization with Monoclonal Antibodies in Animal Models of H5N1 Highly Pathogenic Avian Influenza Virus Infection
- Fructose-Asparagine Is a Primary Nutrient during Growth of in the Inflamed Intestine
- The Calcium-Dependent Protein Kinase 3 of Influences Basal Calcium Levels and Functions beyond Egress as Revealed by Quantitative Phosphoproteome Analysis
- A Translocated Effector Required for Dissemination from Derma to Blood Safeguards Migratory Host Cells from Damage by Co-translocated Effectors
- Functional Characterization of a Novel Family of Acetylcholine-Gated Chloride Channels in
- Both α2,3- and α2,6-Linked Sialic Acids on O-Linked Glycoproteins Act as Functional Receptors for Porcine Sapovirus
- The Contribution of Social Behaviour to the Transmission of Influenza A in a Human Population
- MicroRNA-146a Provides Feedback Regulation of Lyme Arthritis but Not Carditis during Infection with
- Recombination in Enteroviruses Is a Biphasic Replicative Process Involving the Generation of Greater-than Genome Length ‘Imprecise’ Intermediates
- Cytoplasmic Viral RNA-Dependent RNA Polymerase Disrupts the Intracellular Splicing Machinery by Entering the Nucleus and Interfering with Prp8
- and Are Associated with Murine Susceptibility to Infection and Human Sepsis
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Fungal Nail Infections (Onychomycosis): A Never-Ending Story?
- Profilin Promotes Recruitment of Ly6C CCR2 Inflammatory Monocytes That Can Confer Resistance to Bacterial Infection
- Contribution of Specific Residues of the β-Solenoid Fold to HET-s Prion Function, Amyloid Structure and Stability
- The Highly Conserved Bacterial RNase YbeY Is Essential in , Playing a Critical Role in Virulence, Stress Regulation, and RNA Processing
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



