-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
Nearly one-third of the world’s population is infected with Mycobacterium tuberculosis (Mtb), the causative agent of TB. A key factor that contributes to the widespread infection of Mtb is its capacity to survive inside the host macrophage. Understanding how Mtb withstands the hostile intracellular environment of this phagocytic cell may reveal targets for development of therapeutics that enhance the innate anti-Mtb activities of the macrophage. We discovered a novel signaling pathway in mycobacteria which regulates cellular redox homeostasis through NADH and FAD, regulators of metabolism and redox balance. NADH induces the expression of a protein kinase, PknG, which then phosphorylates the ribosomal protein L13 and promotes its presence in the cytoplasm. L13 therein forms a complex with RenU, a Nudix (Nucleoside diphosphate linked moiety X) hydrolase that degrades NADH and FAD. Genetic disruption of this signaling cascade leads to cellular accumulation of these molecules, increased mycobacterial sensitivity to oxidative stress, impaired surface biofilm growth, and most importantly, reduced survival of Mtb in macrophages.
Published in the journal: A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development. PLoS Pathog 11(4): e32767. doi:10.1371/journal.ppat.1004839
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004839Summary
Nearly one-third of the world’s population is infected with Mycobacterium tuberculosis (Mtb), the causative agent of TB. A key factor that contributes to the widespread infection of Mtb is its capacity to survive inside the host macrophage. Understanding how Mtb withstands the hostile intracellular environment of this phagocytic cell may reveal targets for development of therapeutics that enhance the innate anti-Mtb activities of the macrophage. We discovered a novel signaling pathway in mycobacteria which regulates cellular redox homeostasis through NADH and FAD, regulators of metabolism and redox balance. NADH induces the expression of a protein kinase, PknG, which then phosphorylates the ribosomal protein L13 and promotes its presence in the cytoplasm. L13 therein forms a complex with RenU, a Nudix (Nucleoside diphosphate linked moiety X) hydrolase that degrades NADH and FAD. Genetic disruption of this signaling cascade leads to cellular accumulation of these molecules, increased mycobacterial sensitivity to oxidative stress, impaired surface biofilm growth, and most importantly, reduced survival of Mtb in macrophages.
Introduction
A critical determinant defining pathogenicity of Mycobacterium tuberculosis (Mtb) is its survival in host macrophages. Upon internalization by the host phagocytic cell, Mtb and related pathogenic mycobacteria block the fusion of their resident phagosome to the destructive lysosome, thereby establishing a niche within the bactericidal macrophage [1,2]. This ability of pathogenic mycobacteria requires the eukaryotic-type serine/threonine protein kinase G (PknG) [3]. Lack of PknG activity results in rapid delivery of mycobacteria to lysosomes, leading to enhanced killing of the intracellular bacilli by macrophages [3]. Besides its role in the innate survival of Mtb in host cells, PknG provides mycobacterial species with an intrinsic resistance to antibiotics [4]. In the absence of PknG, both pathogenic Mtb and non-pathogenic M. smegmatis display increased susceptibility to multiple antibiotics [4]. These observations suggest that PknG might be required for the persistence of Mtb within hosts, during which the bacillus also becomes highly recalcitrant to antibiotics. Although PknG represents an attractive target for tuberculosis (TB) drug development, the molecular mechanism by which this kinase exerts its biological functions remains largely unknown. It was shown that PknG is secreted via the SecA2 secretion system [5] into the macrophages’ cytosol [3] where it is hypothesized to interfere with host signaling pathways controlling phagolysosome synthesis [3]. However, attempts to identify the putative host substrate(s) targeted by PknG have thus far been unsuccessful. As a result, the role of PknG in Mtb survival in host macrophages remains ambiguous.
In other pathogenic bacteria such as Vibrio cholerae, Escherichia coli, Pseudomonas aeruginosa, Streptococcus sp., and Haemophilus influenza, host persistence and antibiotic tolerance are tightly correlated with the ability to form biofilms, community-like growth consisting of surface-bound cells that are metabolically and physiologically distinct from planktonic cells [6–9]. In vitro, Mtb and other mycobacterial species can form biofilms, which require iron and mobile mycolate moieties [10–12]. Development of mycobacterial biofilms is also modulated by activities of enzymes of the tricarboxylic acid (TCA) cycle, such as 2-oxoglutarate dehydrogenase [11]. Like other bacteria, mycobacterial cells persisting within biofilms display increased antibiotic tolerance, reminiscent of Mtb cells that form during latent TB [11]. However, it has remained largely unknown how the antibiotic-tolerant biofilm of Mtb relates to its pathogenicity, and whether these phenotypic correlations are co-regulated in Mtb and related mycobacteria.
Here, we found that in mycobacteria, biofilm growth and host persistence are both regulated by RHOCS, a newly identified redox homeostatic system in which PknG plays a central role. We show that the redox regulatory molecule NADH induces the expression of PknG, which phosphorylates the ribosomal protein L13 at a unique residue, threonine 11 (T11). The phosphorylation promotes the cytoplasmic association of L13 with RenU, a Nudix hydrolase encoded by a gene adjacent to pknG on mycobacterial chromosomes, and accelerates RenU’s NADH hydrolytic activity. Disruption of the PknG-L13-RenU pathway causes: (i) increased oxidative stress susceptibility, (ii) accumulation of NADH and FAD during oxidative stress, (iii) impaired biofilm growth, and notably, (iv) reduced survival of Mtb in host macrophages. These results suggest that biofilm growth and host persistence are both regulated through the PknG-modulated RHOCS, which regulates levels of nucleoside diphosphate derivatives such as NADH and FAD in mycobacteria.
Results
Kinase Activity of PknG Is Required for Biofilm Growth in Mycobacteria
Studies in other bacteria have established the phenotypic relationship between host persistence, antibiotic resistance and biofilm growth [6,9]. PknG was previously shown to be required for two of these three phenotypes, namely survival in macrophages and resistance to multiple antibiotics [3,4]. In addition, phenotypic characterization of a M. smegmatis ΔpknG mutant revealed profound alterations in surface charge and hydrophobicity of the cell wall (S1 Table) [4], suggesting that PknG might be involved in mycobacterial biofilm growth. Wild type strains of M. smegmatis, M. bovis BCG, and Mtb, together with their derived ΔpknG mutants and complemented strains were assayed for growth in both planktonic cultures and static biofilms (Figs 1 and S1). Similar to M. bovis BCG [3,13], absence of PknG did not affect planktonic growth of M. smegmatis (Fig 1A). However, the MtbΔpknG displayed a slow growth defect in stationary phase (Fig 1B), as previously reported [14]. These phenotypic variations suggest a complexity of PknG function among mycobacterial species that warrants further investigation.
Fig. 1. PknG kinase activity is required for biofilm growth in mycobacteria. 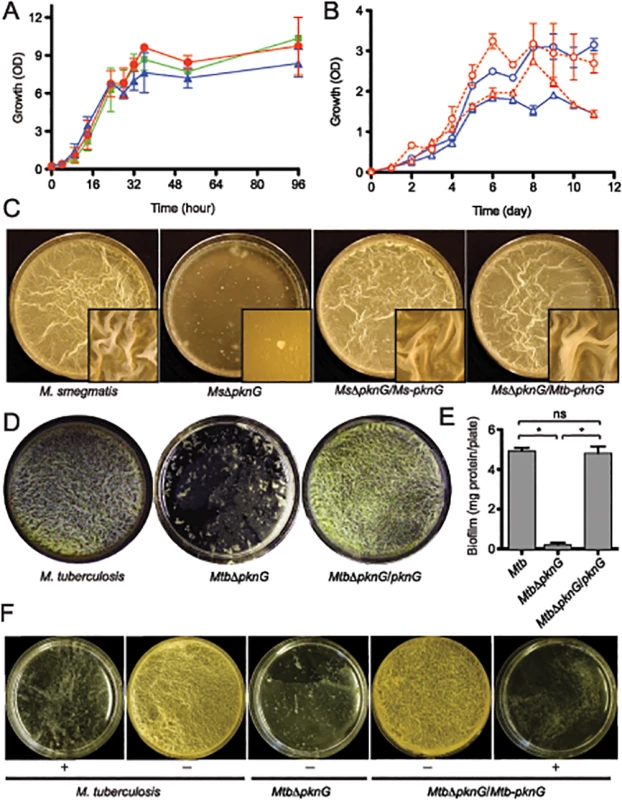
(A) Role of pknG in M. smegmatis planktonic growth. Wild type M. smegmatis mc2155 (red filled circles), its derived MsΔpknG mutant (blue filled triangles), and the complemented strain MsΔpknG/pknG (green filled squares) were grown in 7H9 medium supplemented with 0.2% glucose with shaking at 200 r.p.m. and 37°C. Growth was assessed by measuring optical absorbance at 600 nm. Error bars represent standard deviation of biological triplicates. Differences between wild type and MsΔpknG in stationary phase are not significant. (B) Role of pknG in Mtb planktonic growth. Wild type Mtb H37Rv (open circles) and its derived MtbΔpknG mutant (open triangles) were grown in 7H9-OADC medium with 0.2% glucose (blue) or 1% glucose (red). Cultures were shaken at 200 r.p.m. and 37°C. Growth was assessed by measuring optical absorbance at 600 nm. Error bars represent standard deviation of biological triplicates. Differences between wild type and MtbΔpknG in stationary phase (5–11 hours) are statistically significant (two-tailed t-test, p<0.05). (C) pknG is required for M. smegmatis biofilm growth. Wild type M. smegmatis, MsΔpknG, and the mutant strains complemented with the M. smegmatis (Ms-pknG) or M. tuberculosis (Mtb-pknG) gene. Pictures were taken after 7 days of static growth at 30°C. Shown images are representatives of biological triplicates. (D) pknG is required for Mtb biofilm growth. Wild type Mtb H37Rv, MtbΔpknG, and the complemented strain were assayed as previously described [10]. Pictures were taken after 6 weeks of growth at a static humidified condition of 37°C and 5% CO2. Shown images are representatives of biological triplicates. (E) Quantitation of biofilm growth of Mtb strains. Biofilms were harvested and quantified as described in Experimental Procedures. Error bars represent standard deviation of biological triplicates (*, p<0.0001; ns, not significant difference between wild type H37Rv and the complemented strain). (F) PknG kinase activity is required for Mtb biofilm growth. Wild type Mtb H37Rv, MtbΔpknG, and the complemented strain were assayed in the absence (-) or presence (+) of 1 mM AX20017, a specific inhibitor of PknG. Pictures were taken after 6 weeks of growth at static humidified condition of 37°C and 5% CO2. Shown images are representatives of biological triplicates. However, in all mycobacterial species investigated, ΔpknG mutants consistently displayed severe retardation in biofilm growth (Figs 1C–1F and S1). As previously described [10], wild type cells initially formed clusters emerging onto the surface, which then steadily spread and eventually covered the entire liquid-air interface. This surface invasion was followed by a maturation stage characterized by the formation of typical surface wrinkles (Figs 1C–1F and S1). By contrast, clusters of ΔpknG cells formed unevenly and failed to cover the entire surface. Many of these cell clusters eventually sank and became submerged in the liquid phase (Figs 1C–1F and S1). A surface attachment assay also showed insufficient surface dispersal by M. smegmatis ΔpknG (S2 Fig). Biofilm growth of the ΔpknG mutants was completely restored by in trans expression of either intraspecific or interspecific pknG genes (Fig 1C–1F), suggesting that PknG provides similar functions in biofilm growth to all mycobacterial species.
To test if the requirement for PknG is due to its kinase activity, biofilm growth of Mtb strains was assayed in the presence or absence of a PknG-specific inhibitor, AX20017 [3,15]. Similar to genetic deletions (Figs 1C–1E and S1), AX20017 completely abolished biofilm growth of both wild type and the complemented strain of Mtb (Fig 1F), whereas it had no effect on planktonic growth in similar growth media [3,15]. Collectively, these results show that PknG kinase activity is required for growth of mycobacteria including Mtb in the static condition of surface biofilms.
Both pknG and Its Neighboring Nudix Hydrolase Gene renU Are Involved in Redox Homeostasis
Structural studies revealed a rubredoxin-like domain at the N-terminus of PknG, suggesting a possible involvement of this kinase in redox homeostasis [15]. In fact, our in vitro phosphorylation assays supported the hypothesis that PknG kinase activity may be regulated by the redox state of the mycobacterial cytoplasm (S3 Fig). We studied the role of the pknG locus in mycobacterial redox homeostasis. Interestingly, these studies revealed that pknG and a neighboring gene (msmeg_0790/rv0413), previously annotated as mutT3 (Fig 2A), were each required for oxidative stress resistance in M. smegmatis and Mtb (Fig 2B–2C). The deduced amino acid sequences encoded by msmeg_0790 and rv0413 show the motif GX5EX7REUXEEXGU (where U = L,V,I), typical of Nudix (Nucleoside diphosphate linked moiety X) hydrolases [16]. Nudix hydrolases are low molecular weight (MW) “housecleaning” phosphohydrolases that provide control over cellular levels of deleterious metabolic intermediates [16]. The proteins encoded by msmeg_0790 and rv0413 had been wrongly annotated as MutT3 because they were thought to be anti-mutators, prototypical Nudix hydrolases that degrade and prevent misincorporation of 8-oxo-guanosine triphosphate into nucleic acids. However, recent studies showed that msmeg_0790 and rv0413 are not involved in anti-mutation activity [17].
Fig. 2. Both pknG and its adjacent gene renU are each required for oxidative stress resistance. 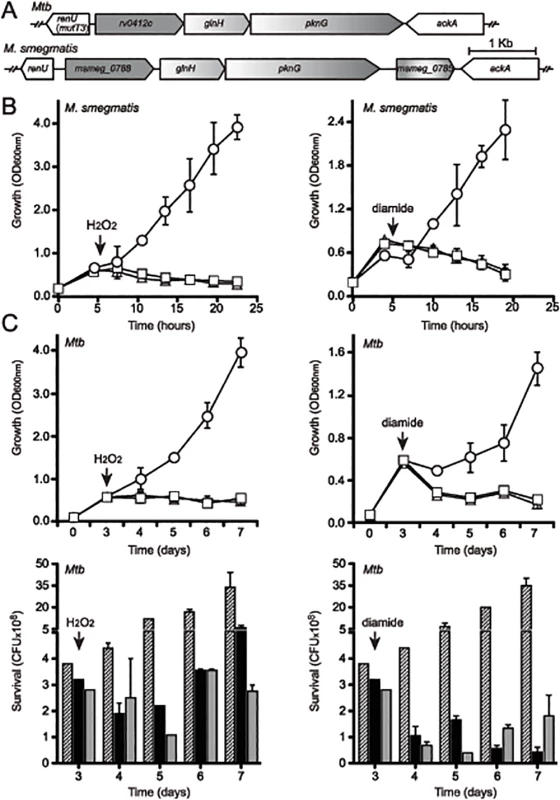
(A) Alignment of the pknG loci from Mtb and M. smegmatis. renU (previously annotated as mutT3) shares the same intergenic region with the operon encoding pknG. Bar, 1kb. (B) Both pknG and renU are each required for M. smegmatis resistance to H2O2 (left) and diamide (right). Wild type M. smegmatis (circles), MsΔpknG (triangles) and MsΔrenU (squares) were grown in 7H9 medium. At the indicated times (arrows), 10mM H2O2 or 15mM diamide was added. Growth was estimated through optical absorbance at 600 nm (OD600nm). Error bars represent standard deviation of biological triplicates. (C) pknG and renU are each required for Mtb resistance to H2O2 (left) and diamide (right). Wild type Mtb (circles or striped bars), MtbΔpknG (triangles or black filled bars) and MtbΔrenU (squares or grey filled bars) of were grown in 7H9-OADC medium. At the indicated times (arrows), 20 mM H2O2 or 10 mM diamide was added. Growth was estimated through measuring optical absorbance at 600 nm (OD600nm, top) or determining colony forming units (CFU, bottom) by serial dilution plating. Error bars represent standard deviation of biological triplicates. Wild type M. smegmatis and Mtb, their derived MsΔpknG and MtbΔpknG mutants, and Δmsmeg_0790 or Δrv0413 mutants, respectively, were challenged with oxidative stress triggered by H2O2 (Fig 2B–2C, left panels) or diamide (Fig 2B–2C, right panels). In both M. smegmatis (Fig 2B) and Mtb (Fig 2C) backgrounds, addition of H2O2 or diamide completely stopped the growth of the mutants, whereas the wild type strains continued to grow. These results suggest that both pknG and msmeg_0790/rv0413 are involved in a redox regulatory mechanism that protects mycobacteria from oxidative stress. In light of the fact that this protein does not function as an anti-mutator, and the findings described in this paper, we propose to rename this Nudix hydrolase as RenU (for Redox Nudix hydrolase).
Nudix Hydrolase Activity of RenU and Its Requirement for Biofilm Growth
To test whether RenU is indeed a hydrolase, and to further characterize its enzymatic activity, the recombinant M. smegmatis RenU was purified to homogeneity. Size exclusion chromatographic analysis showed that RenU was monomeric in solution (S4 Fig). Next, its enzymatic activity was determined using a coupled enzyme colorimetric assay described in the Extended Experimental Procedures. Substrate specificity was investigated with a panel of several different nucleoside diphosphate derivatives (NDPX) and nucleoside triphosphates (NTP). RenU did exhibit Nudix hydrolase activity with a substrate preference for NDPXs. Among the substrates tested, the highest activities were observed with ADP-ribose, FAD, and NADH (Figs 3A left panel, and S5). By contrast, the enzyme displayed much lower activities towards NTPs including ATP, 7,8-dihydroneopterin triphosphate (DHNTP) (Fig 3A left panel), dGTP, dCTP, dUTP, or other NDPXs such as CoA, GDP-D-mannose, NADP, ADP-ADP, and CDP-choline (S5 Fig). Importantly, mutations in glutamate residues of the Nudix box (E74, E77, and E78, see S2 Table), which are expected to coordinate the magnesium required for the activities of Nudix hydrolases, completely abolished the enzymatic activity of RenU. The mutated protein, RenUDEAD, displayed no activity towards the preferred substrates exhibited by wild type RenU (Fig 3A, right panel). Michaelis-Menten analysis revealed that, in vitro, ADP-ribose and FAD were better substrates than NADH, as evidenced by its higher kcat/Km value (Figs 3B and S6). However, previous studies with Nudix hydrolases predict that the substrate preference of RenU might be defined in vivo through its interactions with other proteins [18]. In fact, analysis of cellular levels and PknG induction experiments (see below) suggest that NADH is the physiologically relevant substrate of RenU in vivo. Interestingly, while RenU readily hydrolyzed NADH, the reduced form of nicotinamide adenine dinucleotide, it did not show significant catalytic activity towards the oxidized form, NAD+ (Fig 3C).
Fig. 3. renU encodes a Nudix hydrolase required for biofilm growth. 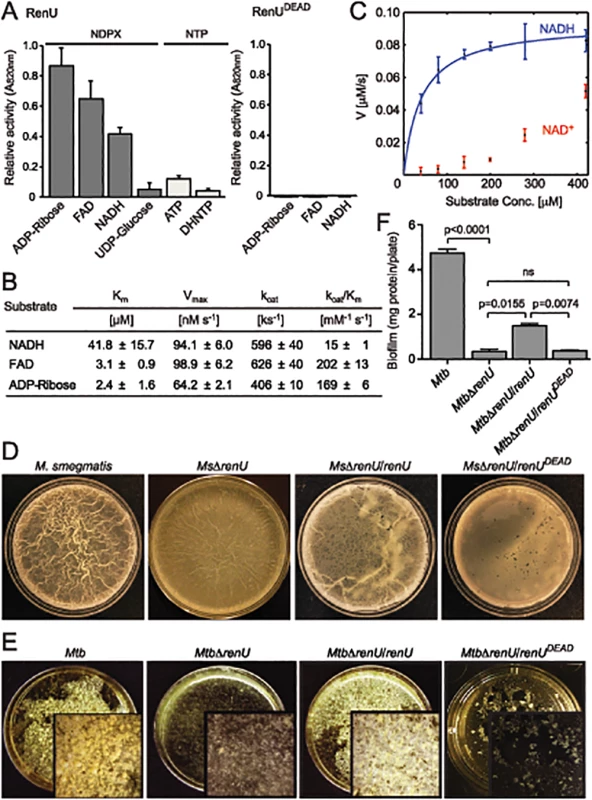
(A) Relative Nudix hydrolase activity of RenU on a substrate panel (left). Nucleoside diphosphate derivatives (NDPX) are preferred substrates compared to nucleoside triphosphates (NTP). A catalytically-inactive mutant of RenU (RenUDEAD) protein, in which 3 glutamate residues (E74, E77, and E78) in the Nudix box were mutated to alanines, exhibits no phosphatase activity towards the preferred substrates (right). (B) Kinetics studies of Nudix hydrolase activity of RenU on the three NDPXs as preferred substrates ADP-ribose, FAD, and NADH. (C) Rate of RenU catalytic activity on NADH compared to its oxidative form NAD+. Fit curve is shown for NADH. (D) The Nudix hydrolase activity of RenU is required for M. smegmatis biofilm growth. Wild type M. smegmatis, MsΔrenU, and the mutant strains completed with wild type RenU or RenUDEAD were assayed for biofilm growth. Whereas renU fully restored biofilm growth to MsΔrenU, renUDEAD failed to complement the mutant. Shown images are representatives of biological triplicates. (E) The Nudix hydrolase activity of RenU is required for Mtb biofilm growth. Wild type Mtb H37Rv, MtbΔrenU, and the mutant strains completed with wild type RenU or RenUDEAD were was assayed for biofilm growth. Whereas renU fully restored biofilm growth to MtbΔrenU, renUDEAD failed to complement the mutant. Shown images are representatives of biological duplicates. (F) Quantitation of biofilm growth of Mtb strains. The biofilm biomass was harvested and estimated by determining total protein per plate. Error bars represent standard deviation of biological triplicates. Statistical significances of differences were analyzed using Students t-test; ns, not significant difference. To determine whether the Nudix hydrolase activity of RenU is required for mycobacterial biofilm growth, MsΔrenU and MtbΔrenU mutants, their parental wild types, and the mutants complemented with either RenU or RenUDEAD, were tested in biofilm growth assays. As shown in Fig 3D–3F, the ΔrenU mutants were as defective as the ΔpknG mutants in biofilm growth. Whereas in trans expression of RenU partially restored biofilm growth, expression of the RenUDEAD form failed to rescue the biofilm in both the ΔrenU mutants (Fig 3E–3F), confirming the requirement for this Nudix hydrolase activity in mycobacterial biofilm growth. These observations, together with their cognate chromosomal localization, further suggest that PknG and RenU participate in the same pathway required for mycobacterial biofilm growth.
RenU Forms a Complex with Ribosomal Protein L13, A Novel Substrate of PknG
To investigate how PknG and RenU interact, we first tested if PknG phosphorylates RenU. The encoding genes were cloned for expression in M. smegmatis or E. coli as 6xHistidine-(6H) tagged proteins. Purified RenU.6H preparations were subjected to in vitro kinase assays using radioactive [γ-P32]-ATP as the phosphate donor. Whereas the M. smegmatis-derived RenU.6H displayed a protein species phosphorylated by PknG, the E. coli-derived equivalent did not show phosphorylation (Fig 4A). This was unlikely due to contaminated phosphatases because addition of phosphatase inhibitors (PI) did not reverse the phosphorylation pattern (Fig 4A).
Fig. 4. L13, a ribosomal protein associated with RenU, is phosphorylated by PknG. 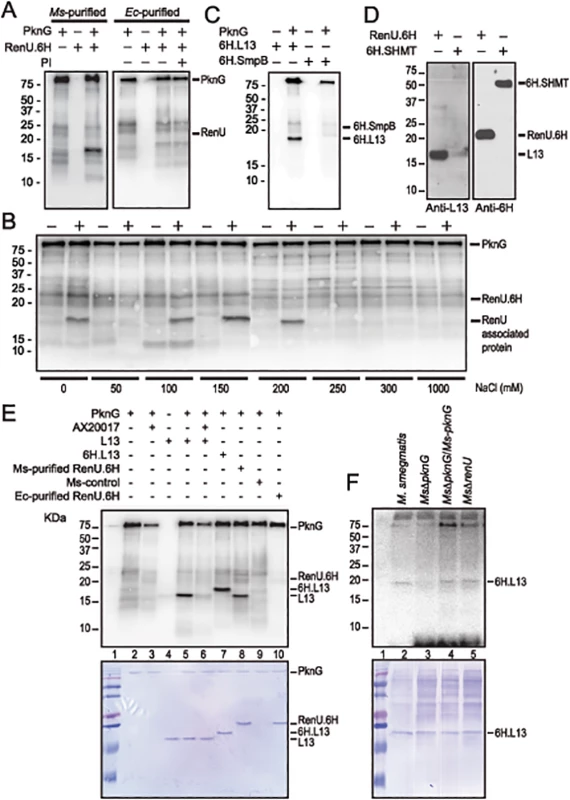
(A) Representative in vitro phosphorylation of RenU.6H preparations purified from M. smegmatis (left) or E. coli (right) by purified PknG. PI, phosphatase inhibitors. (B) In vitro phosphorylation of corresponding fractions eluted from ion exchange columns by PknG. Numbers indicate the NaCl concentrations used in elution buffer. Samples loaded to the ion exchange columns were obtained from an immobilized Cobalt affinity chromatography of M. smegmatis RenU.6H (+) cell lysates or control lysates (-). (C) In vitro phosphorylation of purified 6H.L13 or 6H.SmpB by PknG. (D) Co-purification of L13 from M. smegmatis lysates by exogenous RenU.6H. Another recombinant 6H-tagged protein (6H.SHMT) was used as a control. Blots were detected by Anti-L13 or Anti-6H antibodies. (E) In vitro phosphorylation of recombinant or native L13 protein associated with RenU by PknG kinase activity. (F) In vitro phosphorylation of purified 6H.L13 by M. smegmatis cell lysates, followed by pull-down using Nickel-agarose beads. The MW of the phosphorylated protein species found in the M. smegmatis-purified RenU.6H appeared ~5 kDa smaller than RenU.6H, as revealed by Coomassie Blue stained gels (Fig 4A and 4E, lane 8). In addition, corresponding fractions obtained during the purification of a control cell lysate (from a M. smegmatis strain carrying only the expression plasmid without the RenU-encoding sequence) did not show the same phosphorylated protein species (Fig 4B). These results suggest that the phosphorylated protein was of M. smegmatis origin and associated with RenU.
To identify the RenU-associated protein that is phosphorylated by PknG, the identities of proteins co-purified with RenU.6H expressed in M. smegmatis were analyzed by mass spectrometry. Among the 15 proteins found to associate with RenU (S3 Table), three had theoretical MWs similar to the phosphorylated protein: ssrA-binding protein SmpB (MW 18,287), 50S ribosomal protein L13 (or RplM, MW 16,119), and 50S ribosomal protein L16 (or RplP, MW 15,595). M. smegmatis genes encoding these proteins were cloned and expressed in E. coli as 6H-tagged proteins. The recombinant proteins were purified and subjected to in vitro phosphorylation assays. These experiments showed that only L13 was readily phosphorylated by PknG (Fig 4C and 4E, lane 5). Co-purification experiments confirmed the interaction of L13 and RenU in the cytoplasm of M. smegmatis (Fig 4D). In addition, the MW displayed by L13 on Coomassie Blue gels and autoradiographs was identical to the protein previously found to associate with RenU purified from M. smegmatis (Fig 4E, lanes 5 and 8). Without PknG, L13 proteins from either M. smegmatis or Mtb showed no sign of phosphorylation in the presence of [γ-P32]-ATP (Fig 4C and 4E, lanes 4), showing that the phosphorylation requires PknG. Furthermore, addition of the PknG specific inhibitor AX20017 inhibited the phosphorylation of L13 by PknG (Fig 4E, lane 6). Addition of a 6H-tag shifted the phosphorylated signal of L13 visualized on autoradiograph and Coomassie Blue stained gel (Fig 4E, lane 7). These results confirmed the phosphorylation of L13 by the kinase activity of PknG. Similar to its autophosphorylation (S3 Fig, panel A), the in vitro phosphorylation of L13 by PknG is affected by the redox status of the environment (S3 Fig, panel B).
To further establish whether L13 phosphorylation is PknG specific, in vitro phosphorylation assays were carried out using 6H.L13 as substrate, and cell lysates from M. smegmatis strains as the kinase sources. Using [γ-P32]-ATP as the phosphate donor, the in vitro phosphorylation assays were followed by pull-down of 6H.L13 using nickel-NTA-agarose beads (Qiagen). Precipitated materials were washed, treated with SDS sample buffer and released proteins separated on SDS-PAGE gels, followed by autoradiography. Whereas lysates from M. smegmatis strains expressing PknG, either in trans or chromosomally, readily phosphorylated 6H.L13, lysates from MsΔpknG failed to do so, indicating that phosphorylation of L13 specifically requires PknG (Fig 4F). It is also important to note that the phosphorylation of L13 by PknG did not require RenU (Fig 4F, lane 5), suggesting that phosphorylation occurs before L13 and RenU form a complex.
Phosphorylation Status of L13 Affects Biofilm Growth
L13 proteins, encoded by rplM genes from both M. smegmatis (Fig 4C and 4E) and Mtb (Fig 5), were equally phosphorylated by PknG, indicating that this phosphate transfer reaction serves an identical function in mycobacteria. To identify the specific amino acid residues that are phosphorylated by PknG, Mtb L13 protein purified from E. coli was subjected to a cold kinase assay catalyzed by PknG. L13 was then digested with trypsin and the derived peptides were analyzed by ISL-TOFF mass spectrometry (Taplin Biological Mass Spectrometry Facility, Harvard Medical School). This analysis suggested that one phosphorylated residue was present among the three amino acids closely situated at positions 11–14 of the N-terminus of L13. Among these, T12 is absolutely conserved in all available bacterial L13 protein sequences, whereas T11 and S14 are exclusively conserved across the Mycobacterium genus (Fig 5A, marked with asterisks). We first created a triple mutant of Mtb L13, termed L13(3A), in which all three residues T11, T12 and S14 were mutated to alanine. In vitro phosphorylation assays showed that this mutant protein was no longer phosphorylated by PknG (Fig 5B, lane 5), confirming that the phosphorylated amino acid is among these three residues. Next, mutant L13 proteins with single mutations were made and the purified proteins were individually re-tested in in vitro phosphorylation assays. Whereas L13(T12A) and L13(S14A) mutants were readily phosphorylated (Fig 5B, lanes 7 and 8), L13(T11A) completely failed to be phosphorylated by PknG (Fig 5B, lane 6), similar to the triple mutant L13(3A) (Fig 5B, lane 5). These results indicate that the mycobacterial conserved T11 of L13 is uniquely phosphorylated by PknG.
Fig. 5. PknG-catalyzed phosphorylation of L13 at a mycobacterial specific site, T11, is required for mycobacterial biofilm growth. 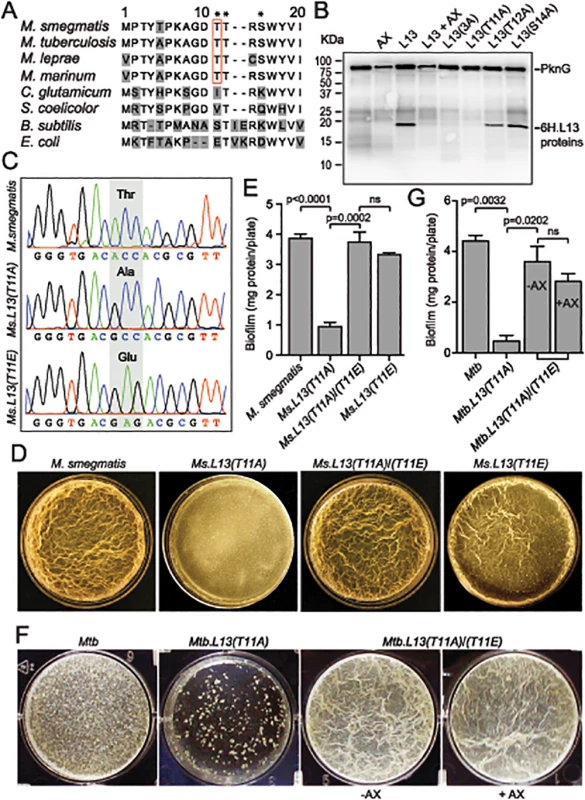
(A) Sequence alignment of the N-terminal 20 amino acids of L13 proteins from different bacteria. Residues marked with asterisks (T11, T12, and S14) are potential targets of phosphorylation by PknG. (B) Phosphorylation of L13 and its mutants by PknG. In L13(3A), all three residues (T11, T12, and S14) were mutated to alanine. Inhibition was achieved by pre-incubation of PknG in 1 mM AX20017 (AX). (C) Chromatograms confirming wild type and mutant alleles of rplM on M. smegmatis chromosomes. The chromosomal loci were amplified from genomic DNA of M. smegmatis strains by primers that anneal to DNA sequences outside the regions homologous to the allelic exchange substrates, followed by cloning and sequencing. (D) Biofilm of M. smegmatis strains. Similar to MsΔpknG and MsΔrenU, Ms.L13(T11A) exhibited defective biofilm growth, while biofilm of Ms.L13(T11E) was largely identical to wild type. In trans expression of an allele encoding L13(T11E) restored biofilm growth to Ms.L13(T11A) strain. (E) Quantitation of biofilm growth of M. smegmatis strains. The biofilm biomass was harvested and quantified by determining total protein per plate. Error bars represent standard deviations of biological triplicates. Statistical significances of differences were analyzed using Students t-test; ns, not significant difference. (F) Biofilm of Mtb strains. Similar to MtbΔpknG and MtbΔrenU, Mtb.L13(T11A) exhibited defective biofilm growth while in trans expression of an allele encoding L13(T11E) restored its biofilm growth. Addition of PknG inhibitor AX20017 (+AX) had no effect on the biofilm of the complemented strain. (G) Quantitation of biofilm growth of Mtb strains. The biofilm biomass was harvested and quantified by determining total protein per plate. Error bars represent standard deviations of biological triplicates. Statistical significances of differences were analyzed using Students t-test; ns, not significant difference. To assess if the phosphorylation status of L13 at T11 plays a role in the biofilm growth modulated by PknG, mutant alleles rplMT11A and rplMT11E were used to replace wild type rplM in M. smegmatis genomes. Whereas L13(T11A) is not activated by PknG, L13(T11E) mimics the conformation of phosphorylated L13. In M. smegmatis, replacement was successful with either rplMT11A or rplMT11E allele (Fig 5C). In Mtb, however, we were only able to create a Mtb.L13(T11A) replacement mutant (rplMT11A) but failed to obtain Mtb.L13(T11E) (rplMT11E). These results may suggest the essentiality of the non-phosphorylated form of L13 in M. tuberculosis, as indicated by previous work [19].
The Ms.L13(T11A) mutant displayed biofilm growth defects, replicating the phenotypes observed previously with MsΔpknG and MsΔrenU (Fig 5D–5E), whereas the Ms.L13(T11E) mutant just showed a minor reduction compared to wild type (Fig 5D–5E, far right panel). In trans expression of the L13(T11E) allele from an integrative vector restored biofilm growth to the Ms.L13(T11A) mutant. Similar to M. smegmatis, the Mtb.L13(T11A) exhibited biofilm growth defects that could be rescued by in trans expression of an L13(T11E) allele (Fig 5F–5G). Unlike wild type Mtb or the MtbΔpknG/pknG strains (Fig 1F), the PknG inhibitor AX20017 failed to block biofilm growth of the Mtb.L13(T11A)/(T11E) strain (Fig 5F–5G). Together, these observations suggest that (i) phosphorylation of L13 by PknG is required for mycobacterial biofilm growth and that (ii) PknG, L13, and RenU form a functional cascade that modulates this static growth type in mycobacteria.
The PknG-L13-RenU Axis Senses and Regulates Cellular NADH Level
A recent study showed that expression of PknG is tightly regulated by unknown mechanisms related to the pathogenicity of Mtb [20]. Whereas PknG is highly expressed in slow growing mycobacteria such as Mtb and M. bovis BCG, the expression in M. smegmatis is extremely low [4,20]. To investigate the conditions that trigger PknG expression in M. smegmatis, the bacterium was treated with various redox stimuli, followed by analysis of PknG levels by Western analysis using a specific polyclonal antibody [3,4,20]. Interestingly, we found that PknG expression is uniquely induced when M. smegmatis cells are exposed to high levels of NADH (Fig 6A). None of the other tested chemicals, including FAD (lower panel), induced PknG expression. The induction of PknG expression by NADH, in both concentration - (Fig 6B) and time-dependent (Fig 6C) manners, may suggest a specific regulatory mechanism, similar to the Rex system originally described in Streptomyces [21–24], or an indirect effect due to changes in cellular metabolism or physiology caused by NADH exposure.
Fig. 6. Correlation of NADH and RHOCS, and role of L13 phosphorylation by PknG. 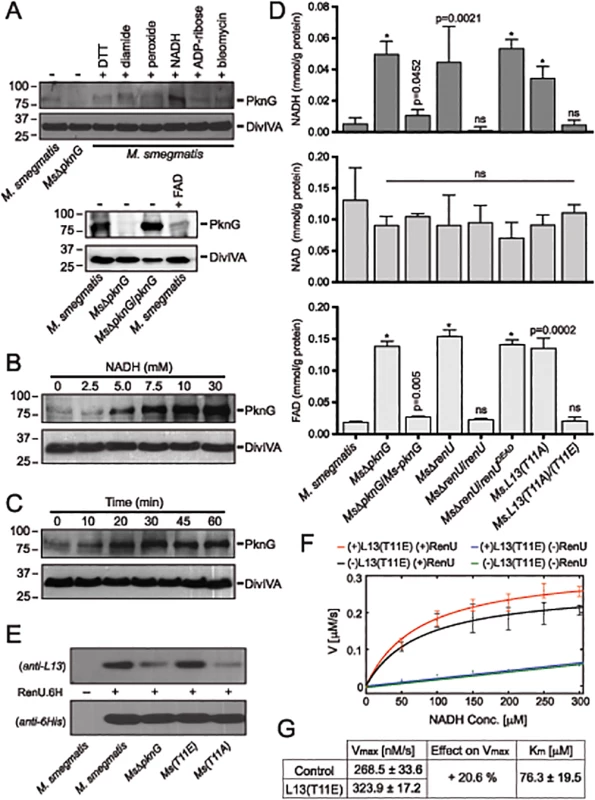
(A) Induction of PknG expression in M. smegmatis. Western analysis was used to detect PknG expression following the exposure of wild type M. smegmatis cultures (OD600 of 2) to various oxidative stimuli including NADH (upper) and FAD (lower) for 30 minutes. All chemicals were used at 10 mM except for bleomycin, which was used at 10 μg/ml. Samples were separated on SDS-PAGE, followed by immunodetection using an anti-PknG antibody or an anti-DivIVA antibody, as a control. Non-induced lysates from wild type M. smegmatis and MsΔpknG were used as controls. NADH uniquely induced expression of PknG. (B) Titration of the induced PknG expression by increasing NADH concentrations (0–30 mM) for 30 minutes, followed by Western analysis using anti-PknG antibody. (C) Time course of PknG expression (0–60 minutes) following cell exposure to 10 mM NADH. Detection of PknG was similar to (A) and (B). (D) Quantitation of cellular NADH (top), NAD+ (middle), and FAD (bottom) levels following oxidative stress induced by H2O2. M. smegmatis cells were exposed to 1 mM H2O2 for 1 hour. Bars show means with standard deviations from 3–6 biological repeats. *, p < 0.0001; ns, not significant relative to wild type M. smegmatis). (E) Effect of PknG-catalyzed phosphorylation of L13 on its association with RenU in the cytoplasm. Expression of PknG in M. smegmatis strains was induced by NADH. Cells were disintegrated by French Press, followed by ultracentrifugation to remove ribosomes. RenU.6H was added to the non-ribosomal fraction, followed by pull-down using Cobalt-agarose beads. The presence of L13 in the pulled down materials was detected by Western analysis using anti-L13 antibody. (F) Effect of L13(T11E), a phosphorylation-mimic form of L13, on in vitro NADH hydrolytic activity of RenU. Initial rates from a continuous fluorescence excitation assay were fit by nonlinear least squares to the Michaelis-Menten equation to determine Km and Vmax values for RenU. Reaction was performed at 37°C. Error bars represent standard deviations of triplicates. The extent of the uncatalyzed reaction was ~10% of the RenU catalyzed reaction. (G) Effect of L13(T11E) on the catalytic activity of RenU. In the presence of L13(T11E), a 20.6% increase in Vmax was observed (p < 0.05x10-3), whereas Km, reflecting the binding affinity of RenU to NADH, was not affected by L13(T11E). The facts that (i) PknG expression is induced by NADH (Fig 6A–6C), (ii) RenU preferentially degrades this redox cofactor in vitro (Fig 3A–3C), and (iii) absence of PknG or RenU leads to failed oxidative stress responses (Fig 2B–2C), indicate that the RHOCS pathway involving PknG, L13, and RenU regulates cellular redox homeostasis through an NAD(H)-related mechanism. To elucidate if interruption of RHOCS activities affects cellular NADH levels, M. smegmatis mutants of the PknG-L13-RenU axis and the parental strain mc2155 were challenged with H2O2, followed by extraction and analysis of NADH, NAD+, and FAD concentrations. Whereas interruption of RHOCS did not affect NAD+ level, it resulted in dramatic accumulations of NADH and FAD by H2O2 (Fig 6D). These observations reveal a novel mechanism of redox homeostasis that senses and regulates the cellular levels of NADH, the possibly other nucleoside diphosphate derivatives including FAD.
Phosphorylation by PknG Promotes the Cytoplasmic Association of L13 with RenU and Its NADH Hydrolytic Activity
To better understand the role of the phosphorylation of L13 by PknG, we first analyzed if phosphorylation affects the formation of the L13-RenU complex in the mycobacterial cytoplasm. M. smegmatis strains representing different states of L13 phosphorylation were first exposed to NADH to induce PknG expression. Cell lysates were prepared and ribosomes removed by ultracentrifugation. RenU.6H was then added to the non-ribosomal fractions. After incubation, RenU.6H was purified using Cobalt agarose beads and the co-purification of L13 analyzed by Western analysis using a polyclonal anti-L13 antibody. Whereas RenU.6H was equally detected, L13 association with RenU.6H in the cytoplasm was dependent on its phosphorylation by PknG (Fig 6E). This experiment suggests that phosphorylation of L13 at T11 by PknG promotes its association with RenU in the mycobacterial cytoplasm.
Because the function of RHOCS is involved with regulation of cellular NADH levels (Fig 6A–6D), we examined if phosphorylated L13 affects the RenU-catalyzed NADH hydrolysis (Fig 3C). To study this question, we first established a fluorescence-based assay that allowed continuous monitoring of NADH hydrolysis by RenU. The assay was based on the different spectral characteristics of folded and unfolded conformations of NADH in aqueous solutions as previously reported [25]. NADH absorbs light at a wavelength of 260 nm through its adenine moiety and emits light at a wavelength of 460 nm through its nicotinamide moiety. The efficiency of the energy transfer responsible for this excitation/emission characteristic is decreased in the unfolded conformation or, in the case of this assay, upon the hydrolysis of NADH.
The hydrolysis of NADH by RenU in the presence or absence of L13(T11E), a mimic form of phosphorylated L13, was monitored. The initial rates were fit to the Michaelis-Menten equation (Fig 6F) to determine Km and Vmax values (Fig 6G). Analysis of Km confirmed that there was no effect of L13(T11E) on RenU’s NADH-binding affinity (one-way ANOVA, effect of L13(T11E), F(3,8) = 2.06, p>0.18) (Fig 6G). In addition, L13(T11E) did not have observable effects on NADH hydrolysis in the absence of RenU (Fig 6F, blue vs green); and wild type L13 did not have observable effects on the NADH hydrolysis catalyzed by RenU (S7 Fig, panel A). Importantly, analysis of Vmax of the reactions performed at 37°C displayed a 20.6% increase in the rate of NADH hydrolysis in the presence of L13(T11E) (one-way ANOVA, effect of L13(T11E), F(3,8) = 35.80, p<0.05×10-3) (Fig 6F, red vs black, and 6G). An increase in Vmax was also observed with temperature increments up to 42°C (S7 Fig, panel B). Together, these data suggest that the phosphorylation of L13 by PknG directly impacts not only the association of L13 with RenU in the mycobacterial cytoplasm (Fig 6E), but also the NADH hydrolytic activity catalyzed by RenU (Fig 6F–6G).
RHOCS Is Required for Survival of M. tuberculosis in Host Macrophages
To investigate if RHOCS is related to the role of PknG in the survival and lysosomal delivery of pathogenic mycobacteria in infected macrophages [3], Mtb RHOCS mutants and the parental strain H37Rv were used to infect murine bone-marrow derived macrophages. Survival was determined by measuring colony forming units (CFUs) of internalized Mtb cells at day 0 and day 4 following the infection. As shown in Fig 7A, deletion of pknG or renU, or the single mutation L13(T11A), significantly reduced the percentages of Mtb survival at day 4 relative to day 0. By contrast, the complemented strains, namely MtbΔpknG/pknG, MtbΔrenU/renU, and L13(T11A)/(T11E), showed survival similar to wild type Mtb (Fig 7A). As shown with biofilm growth, the mutant allele renUDEAD failed to restore the survival of the MtbΔrenU mutant (Fig 7A). In addition, in trans expression of the phosphorylation-mimic form of L13, L13(T11E), also made the L13(T11A)/(T11E) strain resistant to AX20017, the PknG specific inhibitor. These results, all together, indicated that the kinase activity of PknG, the Nudix hydrolase activity of RenU, as well as the phosphorylation of L13, each is required for the survival of Mtb in host macrophages.
Fig. 7. Intracellular trafficking and survival of M. tuberculosis RHOCS mutants. 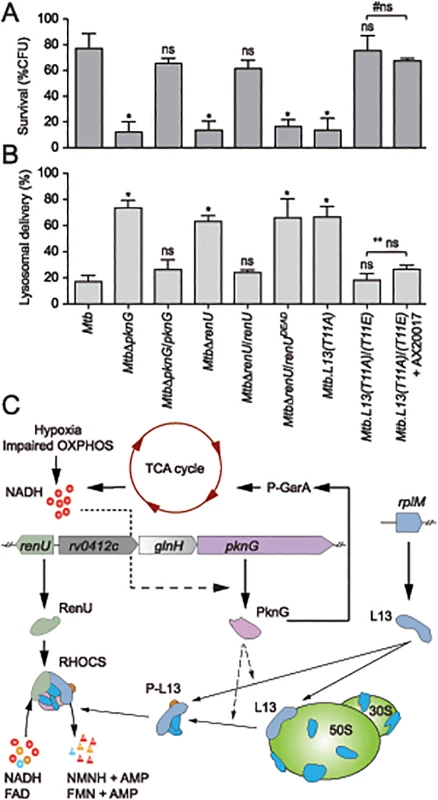
(A) Intracellular survival of Mtb strains. Macrophages were infected with Mtb strains for 3 hours, followed by 0 or 72-hour chase. CFUs were counted after 4–5 weeks of growth at 37°C. Bars represent percentages of CFUs remaining at 72-hour compared to 0-hour time point. Error bars represent standard deviations from 3–6 repeats. *, p < 0.001; ns, not significant relative to Mtb H37Rv; #ns, not significant between the two indicated groups. Order of strains is as in 7B. (B) Quantitative analysis of lysosomal delivery following phagocytosis of Mtb strains by macrophages. Macrophages were infected with FLUOS-stained Mtb strains for 1 hour, followed by 16-hour chase. Infected macrophages (see C below) were used for quantitation. Biological triplicates of 50 events were counted for each Mtb strain. Error bars represent standard deviations. Error bars represent standard deviations from 3–6 repeats. *, p < 0.0001; ns, not significant relative to Mtb H37Rv; ** ns, not significant between the two indicated groups. (C) A model depicting activity and function of RHOCS in mycobacteria. PknG was previously shown to de-repress the TCA cycle through its phosphorylation of GarA, an inhibitor of α-ketoglutarate decarboxylase and glutamate dehydrogenase. Increased TCA cycle activities, hypoxia, or impaired oxidative phosphorylation (OXPHOS), lead to elevated NADH levels. To protect mycobacterial cells against the change in redox status, PknG expression is up-regulated, leading to the signaling cascade including L13 and RenU, which degrades NADH and FAD and restores their optimal level. AMP, adenosine monophosphate, FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide; NMNH, nicotinamide mononucleotide. Next, to analyze if the intracellular survival of Mtb strains correlated with their lysosomal delivery levels, trafficking of internalized Mtb strains was analyzed by microscopy. As shown previously [3], absence of pknG resulted in increased lysosomal delivery (Fig 7B). The MtbΔpknG mutant was largely localized within acidic milieus, whereas wild type Mtb displayed a low level of lysosomal localization (Fig 7B). In trans expression of pknG restored wild type level of lysosomal delivery to MtbΔpknG (Fig 7B). In addition, MtbΔrenU and Mtb.L13(T11A) mutants exhibited lysosomal delivery levels comparable to that of MtbΔpknG (Fig 7B) while the corresponding complemented strains, MtbΔrenU/renU and L13(T11A)/(T11E), behaved like wild type in lysosomal delivery. In an agreement with the survival (Fig 7A), RenUDEAD failed to rescue MtbΔrenU and the L13(T11A)/(T11E) strain showed resistance to AX20017 (Fig 7A). These results suggest that the function of RHOCS in Mtb lysosomal delivery is correlated, either as a cause or a consequence, to the survival of the bacillus in the macrophage.
Discussion
This work has revealed a novel signaling mechanism that is used by mycobacteria to regulate cellular redox homeostasis (Fig 7C). We propose that this system, RHOCS, is capable of sensing the key redox regulator NADH, and regulating its cellular level through direct degradation. RHOCS is composed of at least three components: a eukaryotic-type protein kinase, PknG, a ribosomal protein, L13, and a Nudix hydrolase, RenU. RHOCS is responsive to cellular NADH levels through up-regulated expression of PknG, which phosphorylates L13 at a unique site, T11, and promotes its cytoplasmic association with RenU. At least two possible mechanisms may lead to this increased L13-RenU association: (i) phosphorylation of L13 by PknG prevents the association of L13 with the ribosome, or (ii) the phosphorylation causes releases of L13 from the ribosome, similar to the effect of L13a phosphorylation by ZIPK observed in human macrophages [26]. Once associated with RenU in the cytoplasm, phosphorylated L13 accelerates the Nudix hydrolase activity of RenU that directly degrades NADH, thus lowering its cellular level. This paradigm of redox regulation is novel and has not been observed before in bacteria.
Despite its localization on the large ribosomal subunit, the canonical ribosomal function of L13 remains enigmatic. E. coli L13 was suggested to contribute to the first step of 50S subunit assembly [27], whereas the human analog L13a might play a role in ribosomal RNA methylation [28]. By contrast, accumulating evidence suggests that L13 plays several functions outside the ribosome. In E. coli, L13 forms a transcription anti-termination complex with other ribosomal proteins (RpL3, RpL4 and RpS4), which binds RNA polymerase and acts on Rho-dependent anti-terminators of ribosomal RNA [29]. L13 was also shown to interact with Obg, an essential GTP binding protein involved in growth promotion and stress response in B. subtilis [30]. In human macrophages, treatment with interferon-γ (IFN-γ) induces phosphorylation of L13a by ZIPK [26]. Phosphorylated L13a is then released from the ribosome to form, with four other proteins, the IFN-Gamma-Activated Inhibitor of Translation (GAIT) complex that binds messenger RNAs and inhibits translation of proteins involved in IFN-γ responses [26,31]. Our data reveal a novel extra-ribosomal function of L13, triggered through its phosphorylation by PknG, in redox homeostatic regulation in mycobacteria. The fact that both Mtb and its host macrophage use L13 phosphorylation as a common method to convey cellular stress responses is fascinating and warrants further investigation. Our work also supports the recent “depot hypothesis”, which proposes that macromolecular complexes such as the ribosome function as “reservoirs” for regulatory proteins that perform non-canonical functions [32,33].
The mechanism of action proposed for RHOCS (Fig 7C) fits nicely with some of the previous observations. Work by other groups suggests that PknG derepresses the TCA cycle through its phosphorylation of the cycle inhibitors GarA or OdhI [34,35]. Thus, activity of PknG in the TCA cycle is expected to increase production of NADH, which is then fed into the oxidative phosphorylation pathway that produces reactive oxygen species and free radicals. In addition, NADH is an effective inhibitor of α-ketoglutarate dehydrogenase, the key generator of NADH and oxidative stress [36], and a target of GarA [34,35]. Therefore, the role of PknG in the RHOCS pathway may provide mycobacteria with a supportive mechanism that prevents cell death from redox disturbance caused by increased TCA activity. Interestingly, free radicals produced from NADH by the TCA cycle were recently suggested to mediate bacterial cell death triggered by bactericidal antibiotics [37]. Accordingly, the function of RHOCS in cellular NADH regulation may also help to explain the recent observation that absence of PknG leads to enhanced antibiotic susceptibility in mycobacteria [4,38].
Bacterial responses to oxidative stress have emerged as an integral part of the developmental program required for biofilm growth [39–41]. For example, oxidative stress was shown to evoke metabolic adaptation that reduce NADH production [42] and induces biofilm formation [43]. Similarly, the transcription regulator SoxR, which helps bacteria to defend oxidative stress, was shown to coordinate biofilm growth in Pseudomonas sp., E. coli, and S. coelicolor [40,44]. We propose that RHOCS, through its role in redox stress regulation, is required for growth of mycobacteria in biofilms and the phagosomal milieu of host macrophages. The ability to sense and regulate cellular levels of NADH allows RHOCS to switch cells between different states of metabolism and physiology. Therefore, RHOCS may function as a key regulator that links redox homeostasis with the pathogenicity of M. tuberculosis.
Materials and Methods
Bacterial Strains and Growth Media
M. tuberculosis H37Rv, M. bovis BCG Pasteur, and M. smegmatis mc2155 (American Type Culture Collection) were used as parental strains. Mycobacterial strains were grown at 37°C in 7H10 or 7H9 with appropriate supplements and antibiotics. Kanamycin and hygromycin were used at 50 and 75 μg/ml, respectively. Biofilm growth was done as previously described [10] using Sauton’s medium. For quantitation, a syringe connected to a sterile needle was used to remove the liquid medium and planktonic cells beneath the films. The biomass was harvested and growth estimated through determination of total protein by Bradford method.
Strain and Plasmid Construction
Targeted gene deletion or replacement was done by homologous recombination methods as previously reported [3,4]. Details can be found in the S1 Text. Plasmids and oligonucleotides used in this study can be found in S4 and S5 Tables, respectively.
Intracellular Survival of M. tuberculosis Strains
Macrophages, generated as described in Extended Experimental Procedures, were seeded in 12-well tissue culture plates (BD Biosciences, San Jose, CA) and let adhere overnight (37°C, 10% CO2) prior to infection. Mtb strains were grown to saturation and infections were performed at MOI 50 : 1 for 3 hours. Infected macrophages were washed with warm PBS and incubated for 45 minutes with 200 μg/ml amikacin to kill extracellular bacteria, and exchanged into fresh DMEM. At 0 or 72 hours of incubation at 37°C and 10% CO2, infected macrophages were processed for CFU assays by washing 3 times with PBS, followed by lysis of the macrophages by 0.05% SDS for 5 minutes. Supernatants were harvested, vortexed thoroughly, and plated in triplicate in ten-fold dilutions onto 7H10-OADC agar. Plates were incubated at 37°C for 4 to 5 weeks before CFUs were counted.
Statistical Analysis
Statistical analyses were conducted using GraphPad Prism 5.0f software (La Jolla, CA). Students two-tailed t-test was used to analyze the statistical significance of differences between groups. For enzyme kinetics experiments, one-way analysis of variance was performed using Excel, and non-linear least squares fits were conducted in Matlab by Mathworks (Natick, MA).
Supporting Information
Zdroje
1. Armstrong JA, Hart DPA (1971) Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J Exp Med 134 : 713–740. 15776571
2. Russell DG (2001) Mycobacterium tuberculosis: here today, and here tomorrow. Nat Rev Mol Cell Biol 2 : 569–577. 11483990
3. Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, et al. (2004) Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304 : 1800–1804. 15155913
4. Wolff KA, Nguyen HT, Cartabuke RH, Singh A, Ogwang S, et al. (2009) Protein kinase G is required for intrinsic antibiotic resistance in mycobacteria. Antimicrob Agents Chemother 53 : 3515–3519. doi: 10.1128/AAC.00012-09 19528288
5. van der Woude AD, Stoop EJ, Stiess M, Wang S, Ummels R, et al. (2014) Analysis of SecA2-dependent substrates in Mycobacterium marinum identifies protein kinase G (PknG) as a virulence effector. Cell Microbiol 16 : 280–295. doi: 10.1111/cmi.12221 24119166
6. Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284 : 1318–1322. 10334980
7. Gefen O, Balaban NQ (2009) The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol Rev 33 : 704–717. doi: 10.1111/j.1574-6976.2008.00156.x 19207742
8. Kolter R, Losick R (1998) One for all and all for one. Science 280 : 226–227. 9565532
9. Lewis K (2007) Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5 : 48–56. 17143318
10. Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR Jr., et al. (2005) GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123 : 861–873. 16325580
11. Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, et al. (2008) Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol 69 : 164–174. doi: 10.1111/j.1365-2958.2008.06274.x 18466296
12. Ojha A, Hatfull GF (2007) The role of iron in Mycobacterium smegmatis biofilm formation: the exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol Microbiol 66 : 468–483. 17854402
13. Nguyen L, Walburger A, Houben E, Koul A, Muller S, et al. (2005) Role of protein kinase G in growth and glutamine metabolism of Mycobacterium bovis BCG. J Bacteriol 187 : 5852–5856. 16077135
14. Cowley S, Ko M, Pick N, Chow R, Downing KJ, et al. (2004) The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol 52 : 1691–1702. 15186418
15. Scherr N, Honnappa S, Kunz G, Mueller P, Jayachandran R, et al. (2007) Structural basis for the specific inhibition of protein kinase G, a virulence factor of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 104 : 12151–12156. 17616581
16. Bessman MJ, Frick DN, O'Handley SF (1996) The MutT proteins or "Nudix" hydrolases, a family of versatile, widely distributed, "housecleaning" enzymes. J Biol Chem 271 : 25059–25062. 8810257
17. Dos Vultos T, Blazquez J, Rauzier J, Matic I, Gicquel B (2006) Identification of Nudix hydrolase family members with an antimutator role in Mycobacterium tuberculosis and Mycobacterium smegmatis. J Bacteriol 188 : 3159–3161. 16585780
18. Kloosterman H, Vrijbloed JW, Dijkhuizen L (2002) Molecular, biochemical, and functional characterization of a Nudix hydrolase protein that stimulates the activity of a nicotinoprotein alcohol dehydrogenase. J Biol Chem 277 : 34785–34792. 12089158
19. Sassetti CM, Boyd DH, Rubin EJ (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48 : 77–84. 12657046
20. Houben EN, Walburger A, Ferrari G, Nguyen L, Thompson CJ, et al. (2009) Differential expression of a virulence factor in pathogenic and non-pathogenic mycobacteria. Mol Microbiol 72 : 41–52. doi: 10.1111/j.1365-2958.2009.06612.x 19210624
21. Brekasis D, Paget MS (2003) A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). The EMBO journal 22 : 4856–4865. 12970197
22. Gyan S, Shiohira Y, Sato I, Takeuchi M, Sato T (2006) Regulatory loop between redox sensing of the NADH/NAD(+) ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. Journal of bacteriology 188 : 7062–7071. 17015645
23. Bitoun JP, Liao S, Yao X, Xie GG, Wen ZT (2012) The Redox-Sensing Regulator Rex Modulates Central Carbon Metabolism, Stress Tolerance Response and Biofilm Formation by Streptococcus mutans. PloS one 7: e44766. doi: 10.1371/journal.pone.0044766 23028612
24. Bitoun JP, Nguyen AH, Fan Y, Burne RA, Wen ZT (2011) Transcriptional repressor Rex is involved in regulation of oxidative stress response and biofilm formation by Streptococcus mutans. FEMS microbiology letters 320 : 110–117. doi: 10.1111/j.1574-6968.2011.02293.x 21521360
25. Hull RV, Conger PS 3rd, Hoobler RJ (2001) Conformation of NADH studied by fluorescence excitation transfer spectroscopy. Biophysical chemistry 90 : 9–16. 11321678
26. Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, et al. (2003) Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115 : 187–198. 14567916
27. Stuhrmann HB, Koch MH, Parfait R, Haas J, Ibel K, et al. (1977) Shape of the 50S subunit of Escherichia coli ribosomes. Proc Natl Acad Sci U S A 74 : 2316–2320. 329279
28. Chaudhuri S, Vyas K, Kapasi P, Komar AA, Dinman JD, et al. (2007) Human ribosomal protein L13a is dispensable for canonical ribosome function but indispensable for efficient rRNA methylation. RNA 13 : 2224–2237. 17921318
29. Torres M, Condon C, Balada JM, Squires C, Squires CL (2001) Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. The EMBO journal 20 : 3811–3820. 11447122
30. Scott JM, Ju J, Mitchell T, Haldenwang WG (2000) The Bacillus subtilis GTP binding protein obg and regulators of the sigma(B) stress response transcription factor cofractionate with ribosomes. J Bacteriol 182 : 2771–2777. 10781545
31. Mukhopadhyay R, Ray PS, Arif A, Brady AK, Kinter M, et al. (2008) DAPK-ZIPK-L13a axis constitutes a negative-feedback module regulating inflammatory gene expression. Molecular cell 32 : 371–382. doi: 10.1016/j.molcel.2008.09.019 18995835
32. Ray PS, Arif A, Fox PL (2007) Macromolecular complexes as depots for releasable regulatory proteins. Trends in biochemical sciences 32 : 158–164. 17321138
33. Warner JR, McIntosh KB (2009) How common are extraribosomal functions of ribosomal proteins? Molecular cell 34 : 3–11. doi: 10.1016/j.molcel.2009.03.006 19362532
34. Niebisch A, Kabus A, Schultz C, Weil B, Bott M (2006) Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J Biol Chem 281 : 12300–12307. 16522631
35. O'Hare HM, Duran R, Cervenansky C, Bellinzoni M, Wehenkel AM, et al. (2008) Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol 70 : 1408–1423. doi: 10.1111/j.1365-2958.2008.06489.x 19019160
36. Tretter L, Adam-Vizi V (2005) Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philosophical transactions of the Royal Society of London Series B, Biological sciences 360 : 2335–2345. 16321804
37. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ (2007) A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130 : 797–810. 17803904
38. Smith T, Wolff KA, Nguyen L (2013) Molecular biology of drug resistance in Mycobacterium tuberculosis. Curr Top Microbiol Immunol 374 : 53–80. doi: 10.1007/82_2012_279 23179675
39. Kolodkin-Gal I, Elsholz AK, Muth C, Girguis PR, Kolter R, et al. (2013) Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase. Genes Dev 27 : 887–899. doi: 10.1101/gad.215244.113 23599347
40. Dietrich LE, Teal TK, Price-Whelan A, Newman DK (2008) Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321 : 1203–1206. doi: 10.1126/science.1160619 18755976
41. Depas WH, Hufnagel DA, Lee JS, Blanco LP, Bernstein HC, et al. (2013) Iron induces bimodal population development by Escherichia coli. Proc Natl Acad Sci U S A 110 : 2629–2634. doi: 10.1073/pnas.1218703110 23359678
42. Singh R, Mailloux RJ, Puiseux-Dao S, Appanna VD (2007) Oxidative stress evokes a metabolic adaptation that favors increased NADPH synthesis and decreased NADH production in Pseudomonas fluorescens. J Bacteriol 189 : 6665–6675. 17573472
43. Geier H, Mostowy S, Cangelosi GA, Behr MA, Ford TE (2008) Autoinducer-2 triggers the oxidative stress response in Mycobacterium avium, leading to biofilm formation. Appl Environ Microbiol 74 : 1798–1804. doi: 10.1128/AEM.02066-07 18245256
44. Demple B (2008) Community organizers and (bio)filmmaking. Nature chemical biology 4 : 653–654. doi: 10.1038/nchembio1108-653 18936747
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human SkinČlánek Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?Článek The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious DiseasesČlánek Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting Polypeptide
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 4- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Pathogens as Biological Weapons of Invasive Species
- Selection and Spread of Artemisinin-Resistant Alleles in Thailand Prior to the Global Artemisinin Resistance Containment Campaign
- Endopeptidase-Mediated Beta Lactam Tolerance
- Prospective Large-Scale Field Study Generates Predictive Model Identifying Major Contributors to Colony Losses
- Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human Skin
- Structural Determinants of Phenotypic Diversity and Replication Rate of Human Prions
- Sigma Factor SigB Is Crucial to Mediate Adaptation during Chronic Infections
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Heterologous Expression in Remodeled . : A Platform for Monoaminergic Agonist Identification and Anthelmintic Screening
- Novel Disease Susceptibility Factors for Fungal Necrotrophic Pathogens in Arabidopsis
- Interleukin 21 Signaling in B Cells Is Required for Efficient Establishment of Murine Gammaherpesvirus Latency
- Phosphorylation at the Homotypic Interface Regulates Nucleoprotein Oligomerization and Assembly of the Influenza Virus Replication Machinery
- Human Papillomaviruses Activate and Recruit SMC1 Cohesin Proteins for the Differentiation-Dependent Life Cycle through Association with CTCF Insulators
- Ubiquitous Promoter-Localization of Essential Virulence Regulators in
- TGF-β Suppression of HBV RNA through AID-Dependent Recruitment of an RNA Exosome Complex
- The Immune Adaptor ADAP Regulates Reciprocal TGF-β1-Integrin Crosstalk to Protect from Influenza Virus Infection
- Antagonism of miR-328 Increases the Antimicrobial Function of Macrophages and Neutrophils and Rapid Clearance of Non-typeable (NTHi) from Infected Lung
- The Epigenetic Regulator G9a Mediates Tolerance to RNA Virus Infection in
- Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?
- Hantaan Virus Infection Induces Both Th1 and ThGranzyme B+ Cell Immune Responses That Associated with Viral Control and Clinical Outcome in Humans
- Viral Inhibition of the Transporter Associated with Antigen Processing (TAP): A Striking Example of Functional Convergent Evolution
- Plasma Membrane Profiling Defines an Expanded Class of Cell Surface Proteins Selectively Targeted for Degradation by HCMV US2 in Cooperation with UL141
- Optineurin Regulates the Interferon Response in a Cell Cycle-Dependent Manner
- IFIT1 Differentially Interferes with Translation and Replication of Alphavirus Genomes and Promotes Induction of Type I Interferon
- The EBNA3 Family of Epstein-Barr Virus Nuclear Proteins Associates with the USP46/USP12 Deubiquitination Complexes to Regulate Lymphoblastoid Cell Line Growth
- Hepatitis C Virus RNA Replication Depends on Specific and -Acting Activities of Viral Nonstructural Proteins
- A Neuron-Specific Antiviral Mechanism Prevents Lethal Flaviviral Infection of Mosquitoes
- The Aspartate-Less Receiver (ALR) Domains: Distribution, Structure and Function
- Global Genome and Transcriptome Analyses of Epidemic Isolate 98-06 Uncover Novel Effectors and Pathogenicity-Related Genes, Revealing Gene Gain and Lose Dynamics in Genome Evolution
- The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious Diseases
- Ebola Virus Entry: A Curious and Complex Series of Events
- Conserved Spirosomes Suggest a Single Type of Transformation Pilus in Competence
- Spatial Structure, Transmission Modes and the Evolution of Viral Exploitation Strategies
- Bacterial Cooperation Causes Systematic Errors in Pathogen Risk Assessment due to the Failure of the Independent Action Hypothesis
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Cerebrospinal Fluid Cytokine Profiles Predict Risk of Early Mortality and Immune Reconstitution Inflammatory Syndrome in HIV-Associated Cryptococcal Meningitis
- Utilize Host Actin for Efficient Maternal Transmission in
- Borna Disease Virus Phosphoprotein Impairs the Developmental Program Controlling Neurogenesis and Reduces Human GABAergic Neurogenesis
- An Effector Peptide Family Required for Toll-Mediated Immunity
- Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting Polypeptide
- A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
- Quadruple Quorum-Sensing Inputs Control Virulence and Maintain System Robustness
- Leukocyte-Derived IFN-α/β and Epithelial IFN-λ Constitute a Compartmentalized Mucosal Defense System that Restricts Enteric Virus Infections
- A Strategy for O-Glycoproteomics of Enveloped Viruses—the O-Glycoproteome of Herpes Simplex Virus Type 1
- Macrocyclic Lactones Differ in Interaction with Recombinant P-Glycoprotein 9 of the Parasitic Nematode and Ketoconazole in a Yeast Growth Assay
- Neofunctionalization of the α1,2fucosyltransferase Paralogue in Leporids Contributes to Glycan Polymorphism and Resistance to Rabbit Hemorrhagic Disease Virus
- The Extracytoplasmic Linker Peptide of the Sensor Protein SaeS Tunes the Kinase Activity Required for Staphylococcal Virulence in Response to Host Signals
- Murine CMV-Induced Hearing Loss Is Associated with Inner Ear Inflammation and Loss of Spiral Ganglia Neurons
- Dual miRNA Targeting Restricts Host Range and Attenuates Neurovirulence of Flaviviruses
- GATA-Dependent Glutaminolysis Drives Appressorium Formation in by Suppressing TOR Inhibition of cAMP/PKA Signaling
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- Genetic Analysis Using an Isogenic Mating Pair of Identifies Azole Resistance Genes and Lack of Locus’s Role in Virulence
- A Temporal Gate for Viral Enhancers to Co-opt Toll-Like-Receptor Transcriptional Activation Pathways upon Acute Infection
- Neutrophil Recruitment to Lymph Nodes Limits Local Humoral Response to
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



