-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
Fungal infections cause severe problems for immune-compromised individuals. Current antifungal treatment is problematic, as some therapies are toxic to humans and others are not highly effective. These fungal infections also burden hospitals, as costs to treat and prevent such disease runs high. Aspergillus fumigatus is the most common cause of fungal infections worldwide. A. fumigatus produces a variety of toxins that aid the fungus in survival both in the environment and within host systems. Genes involved in producing such toxins are often found in clusters within the genome, being almost exclusively dependent on transcription factors located within the clusters. Gliotoxin, one such toxin, is known to negatively affect immune cell function. Although gliotoxin has been studied extensively, information is still lacking with regards to regulation of gliotoxin biosynthesis. Our lab has discovered a novel C2H2 transcription factor, GipA, which plays an important role in gliotoxin production. Not only does GipA enhance gliotoxin production when over-expressed, but loss of GipA causes a significant reduction in gliotoxin production. As this gene is not located within the gliotoxin cluster, understanding its mode of action and upstream partners could shed light on toxin production in general and lead to better, more effective antifungal therapies.
Published in the journal: A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in. PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004336
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004336Summary
Fungal infections cause severe problems for immune-compromised individuals. Current antifungal treatment is problematic, as some therapies are toxic to humans and others are not highly effective. These fungal infections also burden hospitals, as costs to treat and prevent such disease runs high. Aspergillus fumigatus is the most common cause of fungal infections worldwide. A. fumigatus produces a variety of toxins that aid the fungus in survival both in the environment and within host systems. Genes involved in producing such toxins are often found in clusters within the genome, being almost exclusively dependent on transcription factors located within the clusters. Gliotoxin, one such toxin, is known to negatively affect immune cell function. Although gliotoxin has been studied extensively, information is still lacking with regards to regulation of gliotoxin biosynthesis. Our lab has discovered a novel C2H2 transcription factor, GipA, which plays an important role in gliotoxin production. Not only does GipA enhance gliotoxin production when over-expressed, but loss of GipA causes a significant reduction in gliotoxin production. As this gene is not located within the gliotoxin cluster, understanding its mode of action and upstream partners could shed light on toxin production in general and lead to better, more effective antifungal therapies.
Introduction
Secondary metabolites are small, low-molecular mass molecules made by numerous organisms that are not essential for normal growth, but can play important roles in defense or signaling [1]–[4]. They can be benign in nature, such as pigments or molecules used in interspecies communication, but they can also be malignant, exhibiting antimicrobial or toxic activities to eliminate competing organisms [5], [6]. Some of these compounds have been exploited by scientists because of their potential benefit to humans. For example, penicillin, produced by P. chrysogenum, is used as an antibiotic and lovastatin, produced by A. terreus, reduces cholesterol [7]. While some of these secondary metabolites benefit humans, others cause harm. Aflatoxin, produced by A. flavus, is carcinogenic, and gliotoxin, produced by A. fumigatus, exhibits immunosuppressive properties [4], [8]–[10]. Genes involved in secondary metabolite production are frequently found as clusters, which are often located in subtelomeric positions [2]–[5], [11]. Furthermore, these clusters are often coordinately regulated, being almost completely dependent on induction by transcription factors located within the clusters themselves [3]–[5], [12], [13].
Zinc binuclear (Zn(II)2Cys6) transcription factors are uniquely found in fungi and represent the most common type of regulators located within these clusters [3], [4], [14], [15]. AflR, a Zn2Cys6 transcription factor, is located within the aflatoxin/sterigmatocystin gene cluster and is required for production of both metabolites [3], [4], [10]. Zn2Cys6 transcription factors, like aflR, generally recognize and bind as homodimers to palindromic sequence motifs, such as CGG(Nx)CCG [14]–[17]. Interestingly, although the palindromic sequences of these binding motifs can be similar or identical for multiple Zn2Cys6 transcription factors, the length and base composition of the linker sequence is highly variable. Therefore, this linker sequence greatly contributes to the specificity of binding for each individual transcription factor [15], [17], [18]. Although gene clusters are often coordinately regulated by the cluster-specific transcription factor, some members can be independently regulated. For example, gliT encodes an oxidoreductase of the gliotoxin biosynthetic cluster, which is required for self-protection against gliotoxin. Even though gliT expression is decreased when the Zn2Cys6 transcription factor, gliZ, is deleted, exogenous gliotoxin induces the expression of gliT, even in a ΔgliZ background [4], [19].
Aside from pathway-specific transcription factors that reside within the cluster, there are numerous other regulatory elements that affect the expression of secondary metabolite clusters. Nutritional and environmental factors, as well as developmental processes, have been shown to affect secondary metabolite production in multiple fungal species [3]–[5]. For instance, penicillin production in A. nidulans is repressed in the presence of glucose, a phenomenon termed carbon catabolite repression [8], [20]. Secondary metabolite repression also occurs in response to nitrogen source, which involves AreA, the global positive regulator of nitrogen metabolite repression. Indeed, in A. nidulans, sterigmatocystin production is repressed in the presence of ammonium and induced when nitrate is the sole nitrogen source [2], [6], [21]. This type of regulation is not seen in all fungi, as carbon and nitrogen source have the opposite effect in A. parasiticus, where growth in the presence of glucose or ammonium results in higher levels of aflatoxin production [2], [6], [21].
A. fumigatus, the leading cause of mold infections worldwide, is an opportunistic pathogen that causes severe problems in immune-compromised populations [9], [22]. These populations include: AIDS patients, cancer patients receiving chemotherapy, solid organ transplant/skin graft patients and victims of chronic granulomatous disease [12], [23]–[25]. One of the most studied secondary metabolites produced by A. fumigatus is gliotoxin, which is also produced by several other Aspergillus species, Trichoderma species, and Penicillium species [13], [26]–[28]. Gliotoxin is a member of the epidithiodioxopiperazine (ETP) class of toxins, which are characterized by a disulfide bridge across a piperazine ring [13], [23]–[27], [29]–[33]. The oxidized form of gliotoxin travels into host immune cells where it is able to affect cellular functions essential to the immune response. These include impediment of phagocytosis and NF-κB activation, as well as induction of apoptosis [23], [25], [26], [29], [32], [33]. As with other secondary metabolites, most of the genes responsible for the production and transport of gliotoxin exist within a gene cluster. The gliotoxin biosynthesis cluster was first identified based on its homology to the sirodesmin PL biosynthesis gene cluster in the ascomycete Leptosphaeria maculans [13], [27], [34], [35].
Within this cluster lies a Zn2Cys6 binuclear finger transcription factor, GliZ, thought to be responsible for general gliotoxin induction and regulation. Indeed, over-expression of gliZ leads to an increase in gliotoxin production and deletion of gliZ results in a loss in gliotoxin production [12], [26], [28]. A DNA binding site has been proposed for GliZ (TCGGN3CCGA), but has not been experimentally proven. This site is present within the promoter region of every gene within the gliotoxin cluster, except gliZ and gliA, which encodes an efflux pump within the cluster [14]. Gliotoxin itself positively regulates expression of the genes within the gliotoxin cluster, as deletion of gliP, the non-ribosomal peptide synthetase (NRPS) required for the first step in the synthesis of gliotoxin, virtually eliminates expression of the other genes in the cluster. This loss in gene expression can be reversed by the addition of exogenous gliotoxin to culture medium [19], [26], [30]. Interestingly, gliT, encoding an oxidoreductase required for resistance of the fungus to gliotoxin, is induced by exogenous gliotoxin even in the absence of gliZ [19]. This demonstrates regulation of genes within the cluster that are independent of the coordinate regulation by GliZ. LaeA, a global regulator of secondary metabolism, has also been shown to regulate the gliotoxin cluster, as gliotoxin is among the secondary metabolites that are lost with deletion of laeA [36]–[38]. Furthermore, loss of vel1 in T. virens (homologous to VeA in Aspergilli) results in a loss in gliotoxin production [39]. This is not surprising, since VeA, VelB, and LaeA form a heterotrimeric complex that regulates secondary metabolism in several fungal species [2], [25], [36], [37], [40]. RsmA, a bZIP transcription factor, positively regulates the gliotoxin cluster through LaeA and GliZ, as loss of either protein abolishes the inducing effects of RsmA over-expression [41]. Map kinase signaling is another element that is important for gliotoxin production, as a strain lacking mpkA, the map kinase in the cell wall integrity pathway, is significantly reduced in gliotoxin production [42]. Nutrient starvation, murine infection, and exposure of germlings to neutrophils have also been shown to up-regulate the gliotoxin biosynthesis cluster through microarray analysis [11], [26].
At this time, information is still lacking as to what proteins and transcriptional regulators are responsible for differential expression of the gliotoxin cluster. We have identified a novel C2H2 transcription factor, gipA, which plays a role in regulating the gliotoxin biosynthesis gene cluster. This protein was identified using a high-copy inducer screen and has not been discussed in detail before now. High-copy expression of gipA results in an increase in transcription of multiple genes within the gliotoxin cluster. Conversely, loss of gipA negatively affects expression of multiple genes within the gliotoxin cluster. In addition, this C2H2 transcription factor specifically regulates gliA expression through a binding site we have identified. Here, we propose a model for gliA expression that involves both GliZ and GipA. Our aim was to identify a novel protein that is involved in gliotoxin production, which we achieved through the discovery of GipA.
Results
High-Copy Inducer Screen Identifies a Novel Protein That Induces Expression of a gliAP-lacZ Cassette
To identify genes that regulate the gliotoxin biosynthesis cluster, we performed a high-copy inducer screen. Our goal was to identify genes that, when present in extra copies, induce the gliotoxin biosynthesis cluster in repressing conditions. We used a LacZ reporter system, under the control of the gliA promoter (identified as GL in strain names). GliA encodes an efflux pump within the gliotoxin cluster that is thought to be involved in the transport of gliotoxin out of the fungal cell [13], [27]. Although GliA has not been shown to be essential to gliotoxin transport in A. fumigatus, expression of gliA in a mutant strain of L. maculans provided protection against gliotoxin, but not sirodesmin [27]. We chose gliA for several reasons. First, it has been shown that expression of gliA peaks when the amount of gliotoxin in surrounding medium is maximal [13], [27]. Second, experiments in our lab revealed that gliA is induced within 30 min of A. fumigatus germlings being exposed to human neutrophils, while nutritional induction of the gliotoxin cluster takes 24–48 hrs (data not shown). These data led us to conclude that gliA expression would be a good indicator of gliotoxin cluster expression, as well as gliotoxin production.
The first round of the high-copy inducer screen involved transforming an AMA1-NotI A. fumigatus genomic library [43] into Af293.1-GL (Fig. 1a). AMA1 stands for autonomous maintenance in Aspergillus and was first discovered and isolated from A. nidulans [44], [45]. Due to the presence of this element, the A. fumigatus library plasmids replicate autonomously upon transformation. This feature allows for easy recovery of the plasmids from the fungal genome by transformation of the genomic DNA into bacteria [46], [47]. Furthermore, this screen is a high-copy inducer screen, because there are on average 10 to 30 copies of any given AMA1-containing plasmid per genome [44], [45]. We grew transformants on gliotoxin-repressing medium with X-gal and screened for colonies that produced a blue pigment. This indicated that the AMA1-NotI plasmid within the genome was inducing gliotoxin cluster expression in repressing conditions. For the second round of the high-copy inducer screen, we transformed individual plasmids, isolated in the first round of the genetic screen, into Af293.1-GL (Fig. 1b). We transformed individual vectors to be sure that the effect we observed from the first round was the result of only one plasmid and not multiple plasmids. To measure LacZ levels in a more quantitative manner, we isolated total protein from transformants and measured β-galactosidase activity. We transformed a pDONR AMA empty vector (AMA.GL) and pDONR AMA-gliZ (AMA-gliZ.GL) as a negative control and positive control, respectively. The third round of the high-copy inducer screen entailed isolating the individual genes from each AMA1-NotI library plasmid and transforming them into Af293.1-GL (Fig. 1c). For this round, we only measured β-galactosidase activity.
Fig. 1. Schematic of the three rounds of the high-copy inducer screen. 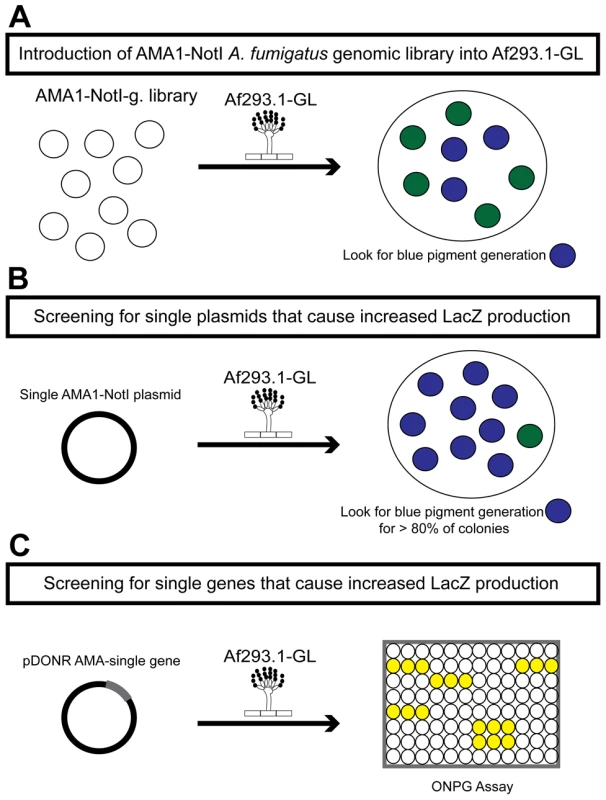
(a) Round one: transformation of the genomic library into Af293.1-GL. (b) Round two: transformation of single plasmids from the genomic library into Af293.1-GL. (c) Round three: transformation of single genes into Af293.1-GL. The gene that induced lacZ the most was a C2H2 transcription factor (AFUA_6G01910), which we have designated gipA for gliotoxin inducing protein. High-copy expression of gipA in the Af293.1-GL background (AMA-gipA.GL) induces a 400-fold increase in β-galactosidase activity, compared to the empty vector control (AMA.GL) (Fig. 2). The level of β-galactosidase activity in our positive control, AMA-gliZ.GL, was almost 30-fold higher than the empty vector control, AMA.GL. These data indicate that GipA positively regulates gliA expression, similar to GliZ.
Fig. 2. β-galactosidase assays for gliZ and gipA high-copy expression strains. 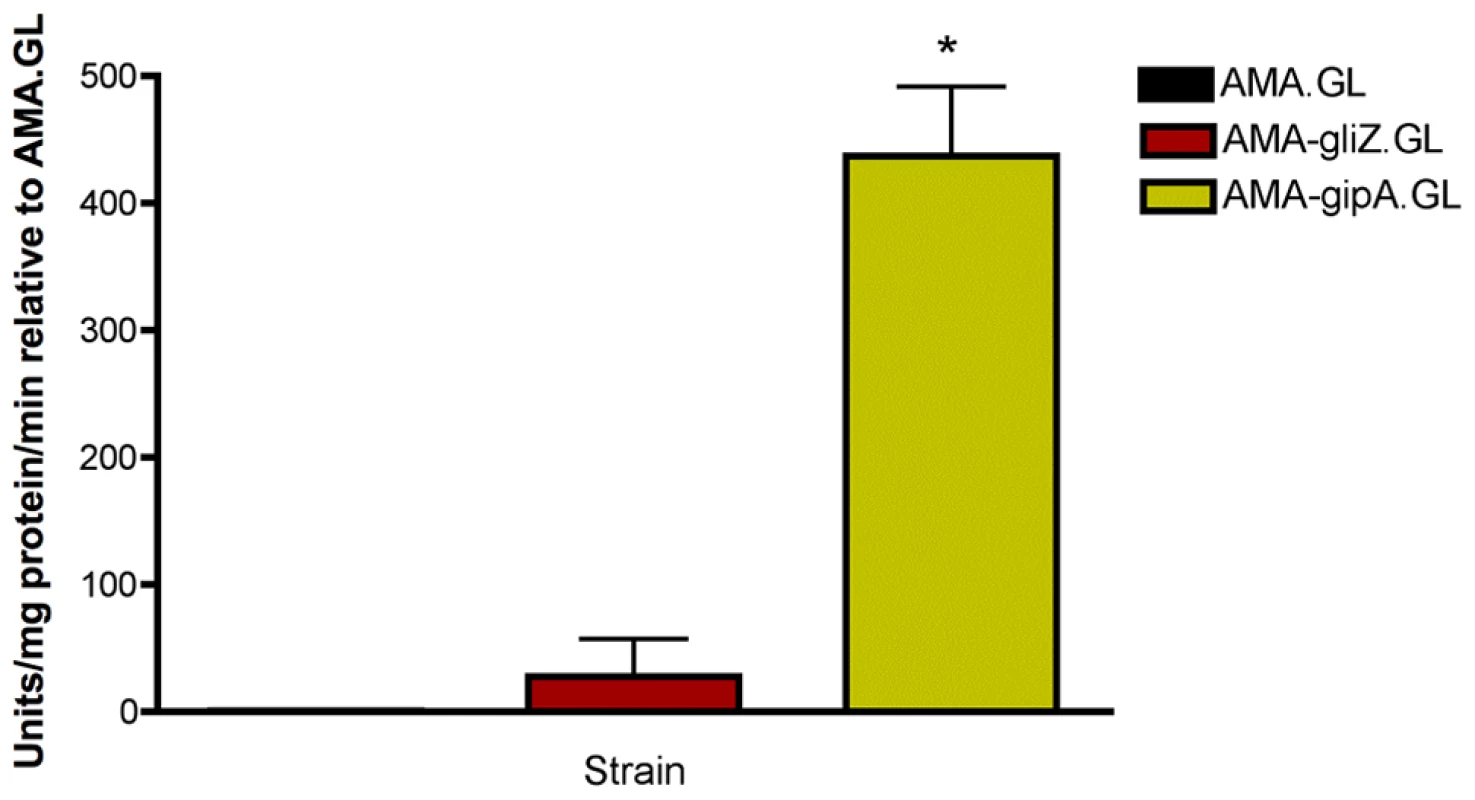
Data is presented as fold-change relative to AMA.GL. The data presented is an average of three biological replicates. The asterisk (*) indicates a statistically significant difference (p-value<0.05), compared to AMA.GL, calculated by one-way ANOVA and Tukey comparison test. gipA Is a C2H2 Transcription Factor with an Unusually Long 5′ UTR
The predicted sequence of gipA contains one intron and two C2H2 regions at the 3′ end (Fig. 3a). The two C2H2 regions are canonical, the first being X2-C-X2-C-X12-H-X3-H and the second being X2-C-X2-C-X12-H-X5-C, which is a natural variant [48], [49]. The 5′ untranslated region (UTR) of gipA appears to be unusually long (877 bp) and contains three μORFs, which indicates that gipA may be under post-transcriptional control due to regulation of translational efficiency and mRNA stability. Northern analysis was consistent with cDNA predictions (Fig. 3b). Sequence similarity searches indicated that proteins homologous to GipA are present in only a limited number of species, the closest being a C2H2 transcription factor in N. fischeri, a close relative to A. fumigatus (Fig. 3c). The rest of the homologous proteins were from other Aspergillus species and a few Penicillium species, which suggests that GipA is not highly conserved at the primary sequence. For proteins, primary sequences evolve and change much more rapidly than do the tertiary structures. There have been numerous examples where two proteins have low sequence similarity, but once crystallized, exhibit almost identical folding patterns and subsequently share similar functions [50]. For example, Gcn4 of Saccharomyces cerevisiae and CpcA of A. niger share a 35% identity, yet they both function in amino acid biosynthesis. Furthermore, CpcA is able to complement a Δgcn4 mutant in S. cerevisiae [51]. When solely comparing the putative DNA binding domain of cpcA, the identity between CpcA and Gcn4 increases to 70% [51]. Therefore, the lack of homologous counterparts to GipA in other organisms does not necessarily mean that there is not a protein present in other fungi that functions similarly to GipA.
Fig. 3. Characterization of gipA cDNA. 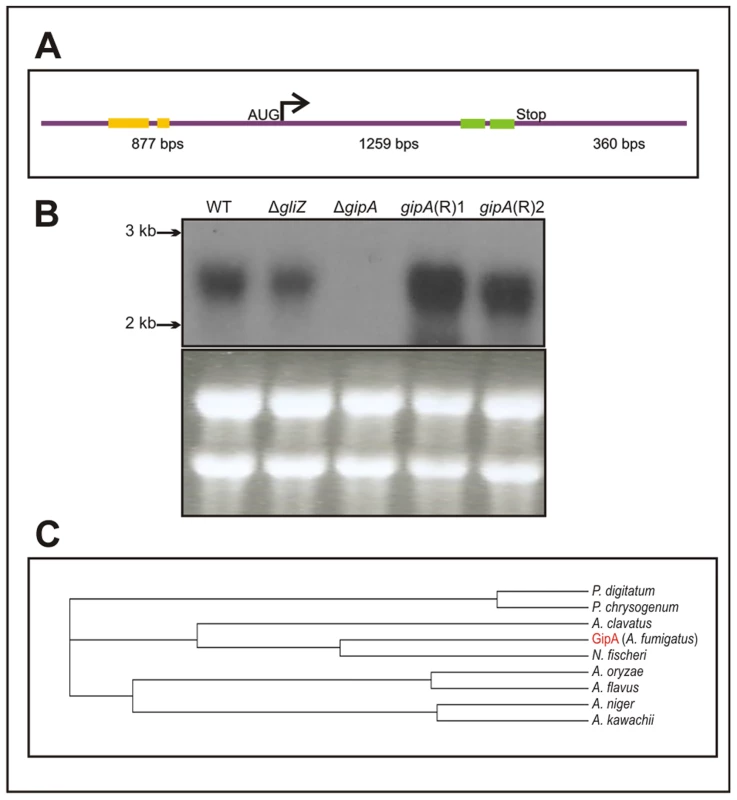
(a) Schematic of cDNA size and composition. Yellow bars signify μORFs and the green bars display the two zinc finger domains. The size indicated for the coding region includes the one intron. (b) Northern hybridization of gipA from total RNA. WT is wild-type (Af1160) and (R) signifies complemented strains. (c) Cladogram of GipA homologues. N. fischeri is Neosartorya fischeri. High-Copy Expression of gipA Induces Gliotoxin Production
In a previous section, we showed that extra copies of gipA induce expression of lacZ, under the control of the gliA promoter, which suggests that GipA is inducing gliA. Since the gliotoxin biosynthesis cluster is coordinately regulated, extra copies of gipA should also induce other genes within the cluster. As expected, AMA-gipA.GL had a higher amount of gliA mRNA, than AMA.GL. Transcript levels of gliA in AMA-gipA.GL were 4.5-fold higher, compared to AMA.GL (Fig. 4a). The mRNA levels of the other gliotoxin-specific genes tested were also significantly higher in AMA-gipA.GL, compared to AMA.GL, as gliZ was induced 12-fold, gliP was induced 5-fold, and gliT was induced 2-fold (Fig. 4a). Gliotoxin production reflected what was seen with mRNA levels, as AMA-gipA.GL produced gliotoxin at a higher amount than AMA.GL (7-fold higher) (Fig. 4b). AMA-gliZ.GL was the positive control and showed the same pattern as AMA-gipA.GL, with respect to induction of the gliotoxin biosynthesis cluster.
Fig. 4. High-copy expression of gipA induces gliotoxin production. 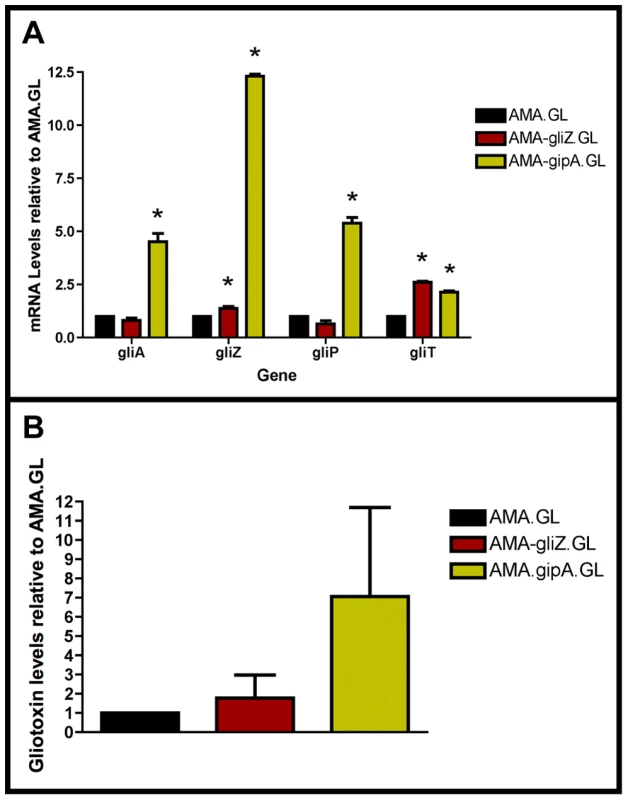
All cultures were grown in repressing conditions. Total RNA was isolated and quantified by dot blot analysis in triplicate. Gliotoxin levels were quantified by RP-HPLC using a standard curve and calculated as mg gliotoxin/g dry mycelial mass. All data sets are normalized to AMA.GL. (a) mRNA transcript levels of several gliotoxin cluster genes after 48 hrs of growth relative to AMA.GL. The results of one representative experiment of three independent experiments are shown as mean ± SD. (b) Gliotoxin levels in growth medium relative to AMA.GL. The data presented is an average of three biological replicates, shown as mean ± SD. The asterisk (*) indicates a statistically significant difference (p-value<0.05), compared to AMA.GL, calculated by one-way ANOVA and Tukey comparison test. High-Copy Expression of gipA Possibly Affects Multiple Secondary Metabolite Clusters
To expand our view of GipA regulation, we performed a microarray analysis of AMA-gipA.GL vs. AMA.GL, grown in repressing conditions for 24 hrs. Of the 9,436 total genes analyzed, 443 genes were up-regulated >2-fold and 75 genes were down-regulated >2-fold in the AMA-gipA.GL strain (Fig. S1). There were several genes common to secondary metabolism clusters (e.g. transporters, oxidoreductases, methyltransferases, nonribosomal peptide synthetases and polyketide synthases) up-regulated (Table 1). Approximately 31 secondary metabolism clusters have been proposed using genomic mapping and microarray techniques [52], [53]. Of these 31 potential secondary metabolism clusters, 18 contained at least one gene that was up-regulated >2-fold in AMA-gipA.GL, compared to AMA.GL (Table 2 & Fig. S2). Based on microarray data done previously [52], loss of laeA, a global regulator of secondary metabolism, affected 13 of 22 identified secondary metabolite clusters. This suggests that GipA potentially induces numerous other secondary metabolism gene clusters in A. fumigatus, similar to LaeA. One caveat of this microarray, however, is that we measured the effects of high-copy expression of gipA, while the microarray done with LaeA compared a deletion strain to a wild-type and complemented strain. Therefore, based on these results, we cannot conclude that loss of gipA will have a significant effect on multiple secondary metabolite clusters, as is observed with loss of laeA.
Tab. 1. Genes commonly involved in secondary metabolism and possibly regulated by GipA. 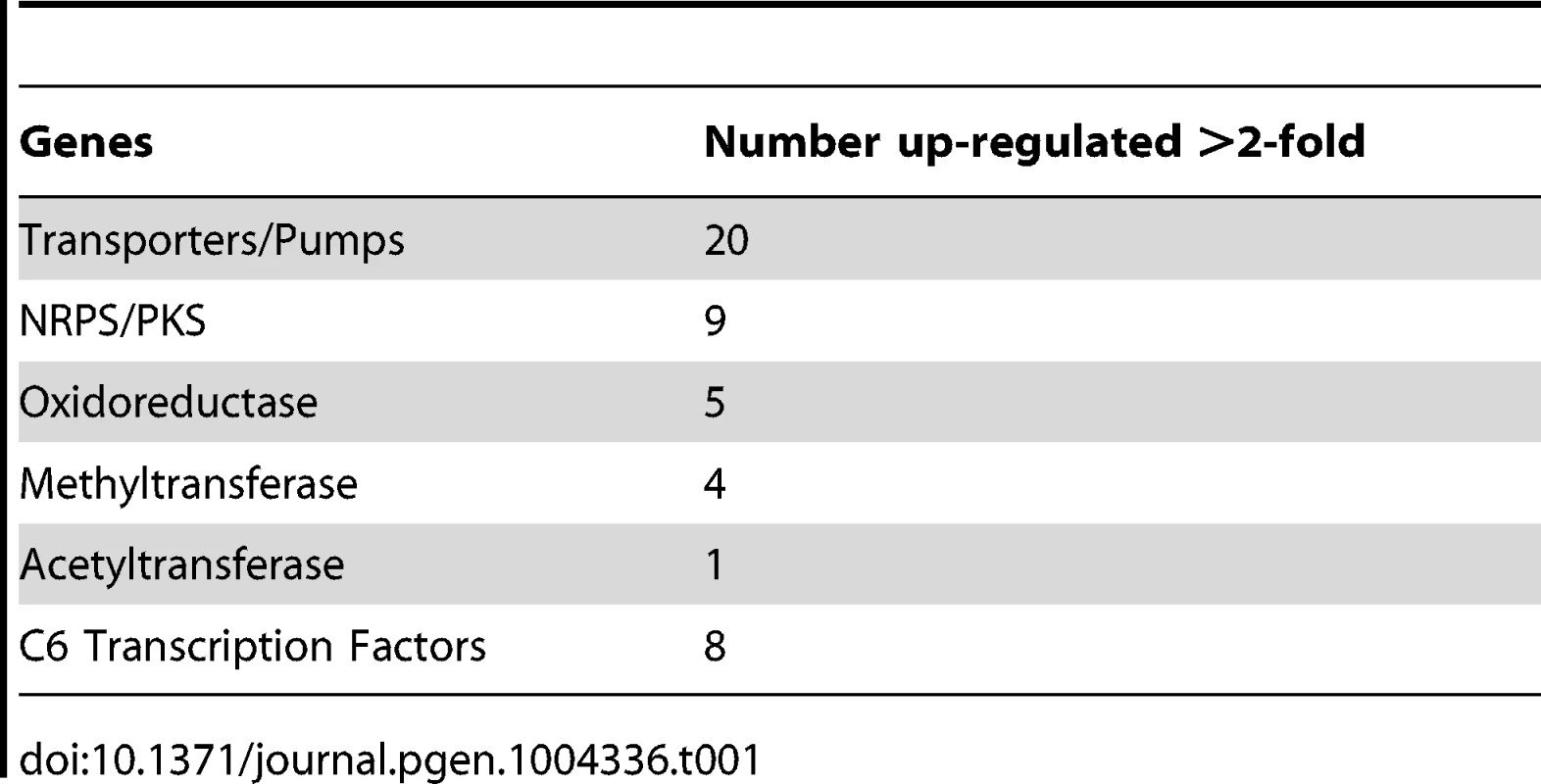
Tab. 2. 31 secondary metabolism clusters present in A. fumigatus. 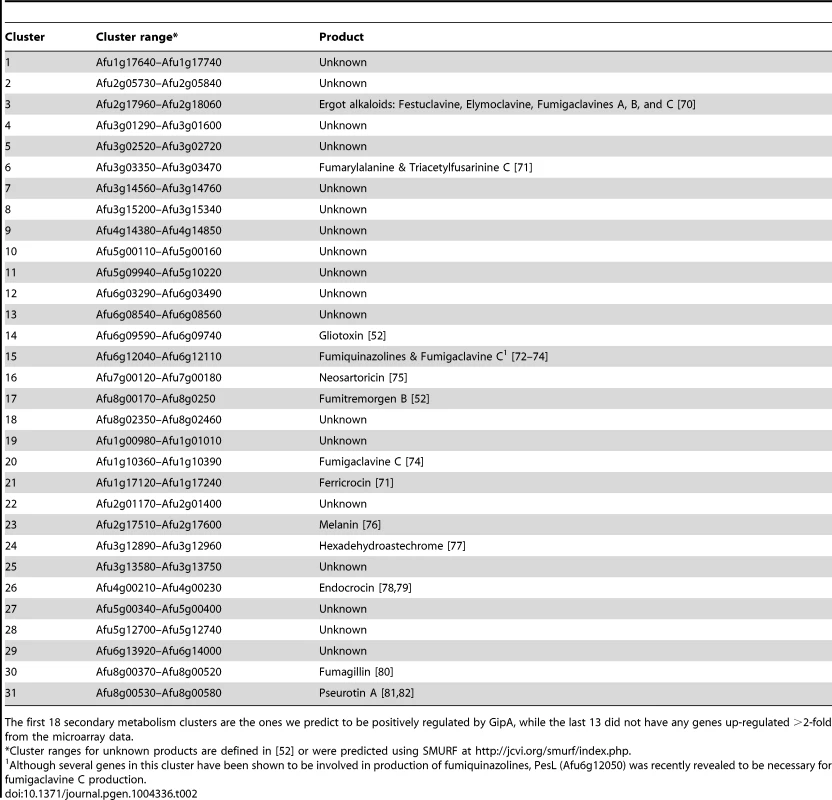
The first 18 secondary metabolism clusters are the ones we predict to be positively regulated by GipA, while the last 13 did not have any genes up-regulated >2-fold from the microarray data. Deletion of gipA Significantly Reduces Gliotoxin Production
Since GipA can induce gliotoxin production, we sought to discover if loss of gipA has any effect on the gliotoxin cluster. We replaced the coding region of gipA with pyrG and designated this strain as ΔgipA. We created a complemented strain, gipA(R), using a hygromycin resistance cassette (hygroR) as the selective marker. We also created a gliZ deletion strain as a control, since previous studies have shown that loss of gliZ results in a significant decrease in mRNA levels of gliotoxin-specific genes [12]. Loss of gipA caused a significant decrease in mRNA levels of gliA, gliZ, gliP, and gliT in non-repressing conditions, as most genes exhibited close to a 50% reduction (Fig. 5a). The gliotoxin biosynthesis cluster is not completely dependent on gipA, as there was still mRNA being made for the genes we tested. The gipA deletion mutant also produced significantly less gliotoxin than the 1160G control strain (50% reduction) (Fig. 5b). Gliotoxin-specific gene expression and gliotoxin production of gipA(R) were restored beyond wild-type levels, which demonstrates that the effect we observed with the gipA deletion was due to the absence of gipA. As expected, loss of gliZ caused almost a complete loss in gene expression for gliA, gliP, and gliT and abolished gliotoxin production. A gliZ/gipA double deletion mutant revealed a pattern of gene expression and gliotoxin production similar to the gliZ single deletion mutant (data not shown).
Fig. 5. Loss of gipA negatively affects gliotoxin production. 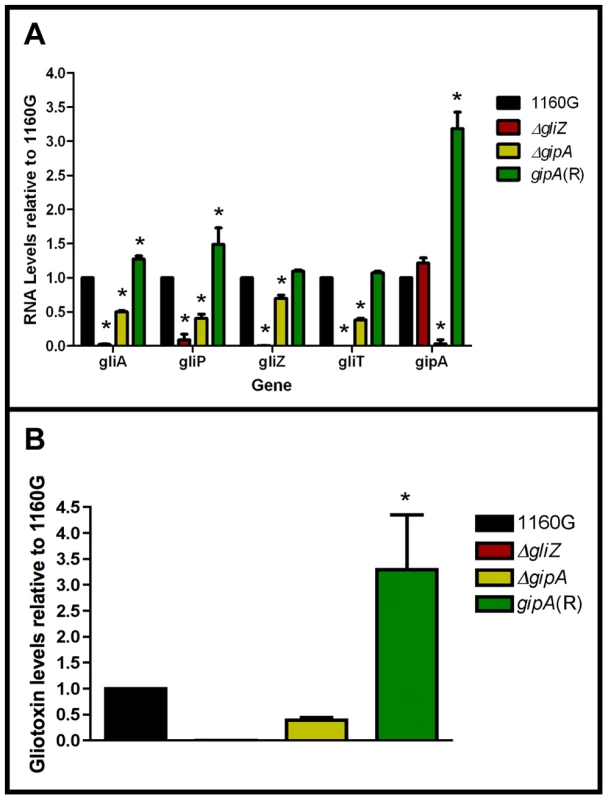
All cultures were grown for 48-repressing conditions. Total RNA was isolated and quantified by dot blot analysis in triplicate. Gliotoxin levels were quantified by RP-HPLC using a standard curve and calculated as mg gliotoxin/g dry mycelial mass. All data sets are normalized to 1160G. (a) mRNA transcript levels of several gliotoxin cluster genes relative to 1160G. The results of one representative experiment of three independent experiments are shown as mean ± SD. (b) Gliotoxin levels in growth medium relative to 1160G. The data presented is an average of three biological replicates, shown as mean ± SD. The asterisk (*) indicates a statistically significant difference (p-value<0.05), compared to 1160G, calculated by one-way ANOVA and Tukey comparison test. Putative GipA Binding Sites Are Present Throughout the Gliotoxin Cluster
Since GipA is a C2H2 transcription factor, it is likely that GipA is directly binding to DNA. We sought to identify a consensus sequence and to discover if this sequence was present within the gliotoxin biosynthesis cluster. A protein-binding microarray analysis identified a consensus DNA binding sequence for GipA (5′-TNNVMGCCNC-3′) (Fig. 6a). This putative sequence is 10 nucleotides, which coincides with one complete turn of the DNA double helix. Interestingly, there are 7 high-content positions (5′-VMGCCNC-3′), which would correlate with two C2H2 zinc finger DNA binding domains [54], and then a “T” residue two base pairs upstream of the 7 high content core positions. The protein-binding microarray verified direct DNA binding of GipA, as the purified DNA-binding domain, and not whole cell extract, was analyzed in the microarray. We analyzed the genomic sequence of the gliotoxin biosynthesis cluster to locate any potential GipA binding sites. Indeed, we found variations of this consensus sequence scattered throughout the gliotoxin biosynthesis cluster. In fact, we identified a possible GipA binding site within the intergenic region of gliA (5′-TTGCCGCCAC-3′ 315 bp upstream of the start site), as well as all other gliotoxin-specific genes, except gliM (Fig. 6b). Not all putative GipA DNA binding sites throughout the intergenic regions in the gliotoxin biosynthetic cluster contain the “T” residue in the 1 position, which means that either these sites are possibly only weakly recognized by GipA, if at all, or the “T” residue is not part of the actual GipA DNA recognition element.
Fig. 6. Characterization of the putative GipA DNA binding site. 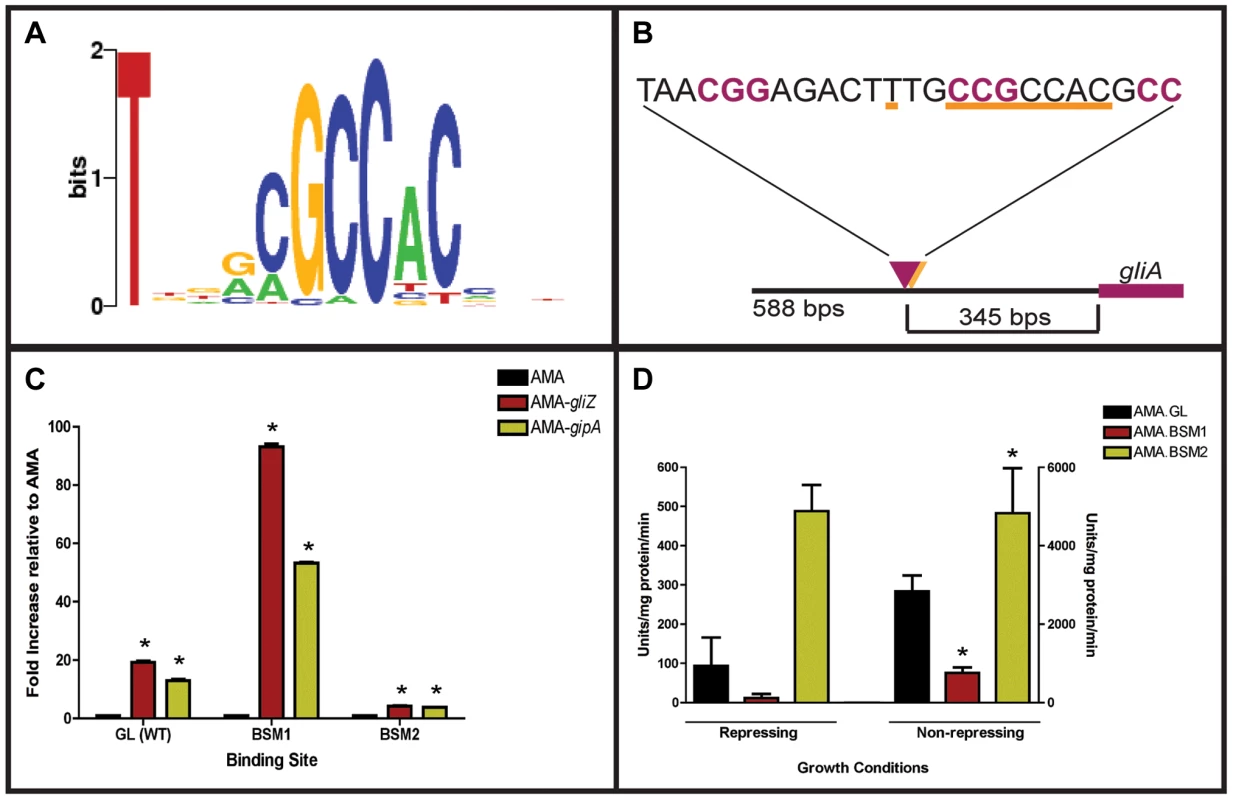
(a) Consensus sequence representing the putative DNA binding site for GipA obtained by protein binding microarray analysis. (b) Layout of putative GipA and GliZ binding sites in the gliA promoter region, relative to the gliA start site. Putative GliZ tandem repeats are purple and bolded and the putative GipA DNA binding site is underlined in orange. (c) Fold increase of LacZ levels of each strain in repressing conditions, relative to the AMA empty vector control. The results of one representative experiment of six independent experiments are shown as mean ± SD. (d) β-galactosidase activity of AMA.GL, AMA.BSM1, and AMA.BSM2 in both repressing and non-repressing conditions. The results of one representative experiment of three independent experiments are shown as mean ± SD. The asterisk (*) indicates a statistically significant difference (p-value<0.05) for each data set, compared to the AMA control strain, calculated by one-way ANOVA and Tukey comparison test. Mutational Analysis of the gliA Promoter Verifies the Presence of an Active GipA Binding Site
We created two mutant backgrounds: BSM1 (TTGCCGCCACCTGCCGCCAC) and BSM2 (TTGCCGCCAC→TTGGGTGAGC), by mutating the putative GipA DNA binding site on the gliA-lacZ construct we used in the original screen. We measured lacZ expression with β-galactosidase assays in the presence of different high-copy plasmids (strains used in this study are listed in Table S2). When normalized to AMA.GL, AMA-gipA.GL displayed increased LacZ levels (13-fold), as was to be expected from previous results (Fig. 6c). High-copy expression of gipA in the BSM1 background also induced lacZ significantly (53-fold), relative to AMA.BSM1 (Fig. 6c). The magnitude of induction was enhanced with the BSM1 mutation when compared to the wild-type binding site (13-fold vs. 53-fold, respectively). This enhanced expression was not due to higher overall lacZ levels, as shown when comparing empty vector controls in all three backgrounds (Fig. 6d). AMA-gipA.BSM2 only weakly induced lacZ (4-fold), compared to AMA.BSM2 (Fig. 6c). This suggests that mutation of a core sequence in the putative GipA DNA binding site significantly reduces the ability of GipA to induce lacZ. Mutation of the 5′ “T” residue did not reduce GipA-specific gliA induction, but rather increased the response of gliA to GipA high-copy expression.
Interestingly, expression of lacZ in the pDONR AMA-gliZ strains followed a similar pattern to that of pDONR AMA-gipA strains (Fig. 6c). Therefore, lacZ was induced in AMA-gliZ.GL (19-fold) and AMA-gliZ.BSM1 (93-fold), compared to AMA.GL and AMA.BSM1, respectively. The level of induction was enhanced by the BSM1 binding site, compared to the wild-type binding site (19-fold vs. 93-fold, respectively). Furthermore, lacZ was only weakly induced in AMA-gliZ.BSM2, relative to AMA.BSM2 (4-fold). Therefore, mutation of the putative GipA DNA binding site is affecting the ability of GliZ to induce gliA.
GipA Requires GliZ for Induction of Both gliA and gliP
To determine if GipA is dependent on GliZ for expression of gliA, we created a series of mutants and measured mRNA levels of gliA and gliP. High-copy expression plasmids used in previous sections were transformed into a wild-type background (AMA.G, AMA-gliZ.G, and AMA-gipA.G) and a gliZ deletion background (AMA.Z, AMA-gliZ.Z, and AMA-gipA.Z) (strains used in this study are listed in Table S2). All strains were grown in non-repressing conditions to induce expression of the gliotoxin biosynthesis cluster. In the pyrG+ background, AMA-gliZ.G and AMA-gipA.G had increased mRNA levels for gliP, while only AMA-gipA.G had increased mRNA levels for gliA, compared to AMA.G (Fig. 7). These changes were not significant because growth in non-repressing conditions already induced these genes to high levels, so having extra copies of GliZ or GipA did not greatly contribute to gene expression. In AMA.Z, mRNA for both gliA and gliP was almost completely undetectable, as to be expected from previous experiments. High-copy expression of gliZ brought transcript levels of both genes back to AMA.G levels (Fig. 7). AMA-gipA.Z displayed a reduction in mRNA levels similar to AMA.Z. For gliA, the level of mRNA present in AMA-gipA.Z was slightly higher (close to 5-fold) than what was observed for AMA.Z. However, the level of gliP mRNA did not exceed that of the AMA.Z empty vector control. Therefore, GipA was not able to induce gliA or gliP in the absence of GliZ. This suggests that GliZ is required for GipA to induce both gliP and gliA.
Fig. 7. GliZ and GipA are dependent on each other for gliA induction. 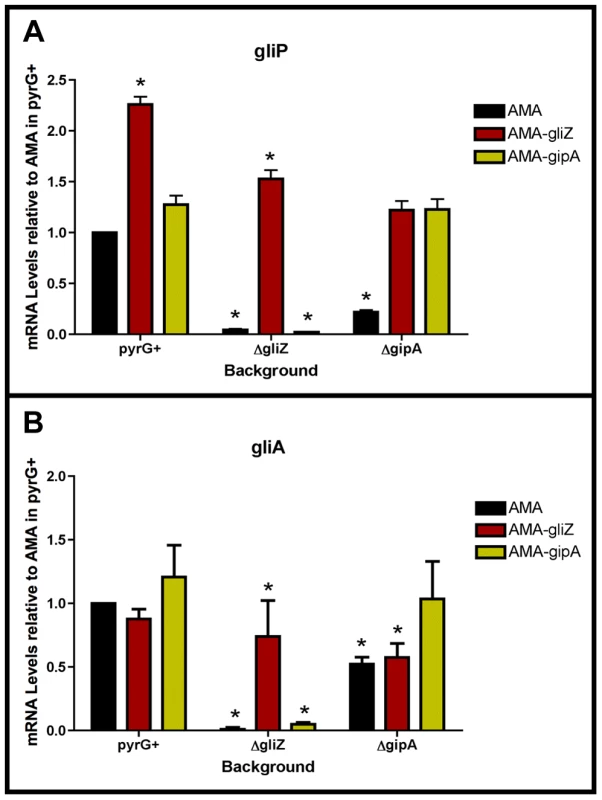
Cultures were grown in non-repressing conditions and mRNA was quantified by dot blot analysis in triplicate. Data are normalized to AMA.G (pyrG+ background). These graphs are an average of three biological replicates, shown as mean ± SD. (a) mRNA levels of gliP in all backgrounds relative to AMA.G. (b) mRNA levels of gliA in all backgrounds relative to AMA.G. The asterisk (*) indicates a statistically significant difference (p-value<0.05), compared to AMA.G, calculated by one-way ANOVA and Tukey comparison test. GliZ Requires GipA for Induction of gliA, but not gliP
To determine if GliZ is dependent on GipA for expression of gliA, we created a series of mutants and measured mRNA levels of gliA and gliP. High-copy expression plasmids used in previous sections were transformed into a wild-type background (AMA.G, AMA-gliZ.G, and AMA-gipA.G) and a gipA deletion background (AMA.A, AMA-gliZ.A, and AMA-gipA.A) (strains used in this study are listed in Table S2). All strains were grown in non-repressing conditions to induce expression of the gliotoxin biosynthesis cluster. As expected from previous experiments, mRNA levels in AMA.A of both gliA and gliP were reduced significantly, 50% and 80%, respectively. Levels of gliP mRNA in AMA-gliZ.A were comparable to those of AMA.G; however mRNA levels of gliA were not significantly higher than background levels (AMA.A) (Fig. 7). This indicates that GliZ is not dependent on GipA for induction of gliP, however induction of gliA by GliZ does appear to be dependent on GipA.
Discussion
Although recent studies have revealed gliotoxin intermediates, which have led to a better understanding of the biosynthesis of gliotoxin, information on regulation of the genes involved in the biosynthesis pathway is lacking [28]. With the use of a high-copy inducer screen, our lab has uncovered a novel protein, GipA, which appears to be involved in the regulation of the gliotoxin cluster. GipA is a C2H2 transcription factor, which harbors an unusually long 5′ UTR. Furthermore, there are three μORFs within the 5′ UTR, which suggests that gipA may be under post-transcriptional control. There are two canonical C2H2 zinc finger DNA binding regions at the 3′ end of gipA, the first being X2-C-X2-C-X12-H-X3-H and the second being X2-C-X2-C-X12-H-X5-C, which is a natural variant [48], [49]. Two C2H2 zinc finger binding domains is consistent with a 6–8 base pair DNA recognition element [54]. The putative DNA binding site that we identified for GipA contains 7 high content positions (5′-TNNVMGCCNC-3′), in addition to a 5′ “T” residue two base pairs upstream of the core residues. Based on mutagenesis of the putative GipA DNA binding site in the gliA promoter, the 7 high content positions are likely part of an active GipA DNA binding site. While the “T” residue is not necessary for GipA-specific gliA induction, mutation of the “T” residue does enhance GipA-specific gliA expression. We have devised a possible model for the specific regulation of gliA that involves GliZ and GipA (Fig. 8). We propose that GliZ and GipA work together at the same binding site or in close proximity to induce gliA. In this model, GipA and GliZ are dependent on each other for inducing gene expression of gliA.
Fig. 8. Model for gliA regulation involving GliZ and GipA. 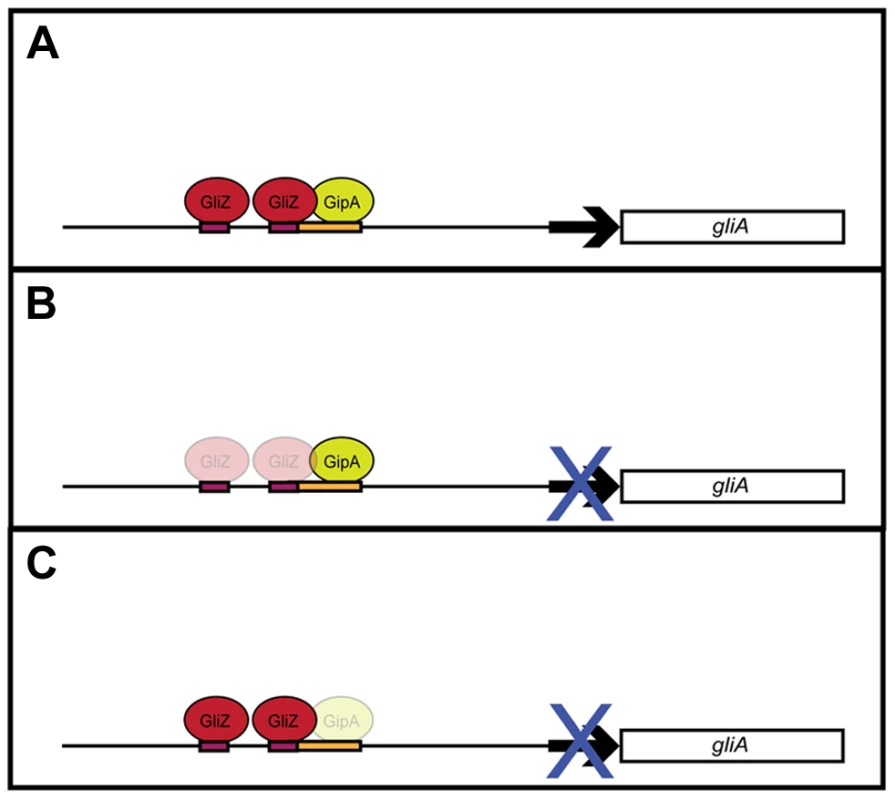
(a) We propose that GliZ and GipA both work interdependently to induce gliA. (b) With respect to the GliZ-GipA interdependent model, GipA cannot induce gliA in the absence of GliZ. (c) Furthermore, GliZ cannot induce gliA beyond a certain level in the absence of GipA. From our data, GipA is not essential to gliA expression, as there is still a moderate level of gliA mRNA produced when the gliotoxin cluster is induced, however, in the absence of GipA, high-copy expression of gliZ does not increase gliA mRNA levels beyond background levels. Our model is not meant to suggest that gliA is not induced at all in the absence of GipA, but merely that gliA induction specific to this GliZ-GipA interaction is lost if either protein is absent. GipA appears to play an important role in gliotoxin biosynthesis, as high-copy expression of gipA causes increased gliotoxin production and loss of gipA causes a significant reduction in gliotoxin levels. Furthermore, GipA is possibly regulating other secondary metabolite clusters within A. fumigatus, similarly to LaeA, a global regulator of secondary metabolism [52]. In contrast to LaeA, though, GipA is a C2H2 transcription factor, which likely directly binds to DNA recognition sites. Other C2H2 transcription factors, such as PacC, NsdC, NsdD and CreA, have also been identified as regulators of secondary metabolism. PacC regulates a wide range of genes in response to ambient pH, including those involved in secondary metabolism. Penicillin production in A. nidulans is induced in alkaline conditions, while sterigmatocystin production is repressed [4], [55], [56]. NsdC and NsdD are both involved in sexual development in A. nidulans. However, recent work by Jeffrey Cary et al. [57] supported additional roles for NsdC and NsdD in asexual development and secondary metabolism in A. flavus. Although AflR expression was not affected, loss of either nsdD or nsdC resulted in decreased expression of other genes in the aflatoxin biosynthesis gene cluster [57]. Finally, CreA is a global regulator of carbon catabolite repression in fungi. In addition to carbon metabolism, CreA also affects secondary metabolism, as cephalosporin production is reduced in Acremonium chrysogenum in response to high glucose [56], [58]. It is possible that GipA serves to enhance expression of the gliotoxin cluster in certain environmental conditions. Both high-copy expression and loss of gipA affect gliZ expression, so the effects we see for the entire cluster could be the direct consequence of GipA binding to each promoter region, or an indirect consequence of GipA partially regulating gliZ, which in turn regulates other genes within the cluster. Loss of both gliZ and gipA does not give conclusive evidence for either possibility, as loss of gliZ alone completely abolishes gliotoxin production. None of the mutants tested displayed abnormal growth rates or attenuated virulence in a Drosophila melanogaster model system, indicating that high-copy expression of gipA or loss of gipA does not affect overall fitness of A. fumigatus (data not shown).
Our data show a dependency between GipA and GliZ with respect to gliA expression. Firstly, there is a putative GliZ DNA binding site embedded within a GipA DNA binding site in the gliA 5′ UTR. Although a GliZ binding site has not yet been experimentally determined, one has been predicted (TCGGN3CCGA). This sequence is present in the intergenic region of every gene within the gliotoxin cluster, except gliZ and gliA [14]. Recognition of these sequences by Zn2Cys6 binuclear finger transcription factors is often very specific and even a slight change to the length or base composition of the linker sequence can result in reduced binding in vivo [15], [17], [18]. Within the gliA promoter region, a sequence overlapping the GipA binding site has the CGG-CCG inverted repeats common to Zn2Cys6 binuclear finger transcription factor binding sites, but the linker sequence is longer (8 bps) than that of the predicted GliZ DNA binding site (3 bps). Mutation of the CCG repeat almost completely abolishes gliZ-mediated induction of gliA. This suggests that either the prediction for the GliZ DNA binding site is incorrect and is actually CGGN8CCG or that the GliZ DNA binding site within the gliA UTR is unique. Due to the position of the putative GliZ DNA binding site, mutation of this CCG also changes the core sequence of the putative GipA DNA binding site. Accordingly, gipA-mediated induction of gliA is almost completely abolished.
Dependent dual regulation of two transcription factors has been uncovered in other organisms. For example, in A. nidulans, FlbB and FlbD both bind in close proximity to the brlA promoter to regulate asexual development through brlA activation [59], [60]. Furthermore, FlbB does not bind to the brlA promoter in the absence of FlbD, indicating that these two transcription factors are dependent on each other for DNA binding and activation of brlA [60]. Although the mutational analysis of the gliA promoter raises the possibility of a GliZ-GipA dependency, it does not give conclusive evidence. One could argue that disruption of the GliZ DNA binding site causes loss of gliA induction that indirectly abolishes gipA-mediated induction of gliA. However, our hypothesis of a dependency between GipA and GliZ with respect to gliA expression is further supported by the fact that GliZ cannot induce gliA in the absence of GipA. As expected, loss of gliZ severely decreases mRNA levels of gliA, which cannot be rescued by high-copy expression of gipA. Interestingly, loss of gipA also significantly decreases mRNA levels of gliA, which cannot be rescued by high-copy expression of gliZ. Clearly, these two proteins work together to induce gliA beyond basal levels in non-repressing conditions. This pattern of dependency is unique to gliA expression, as gliP is induced by high-copy expression of gliZ, even in the absence of gipA.
Together, these data support a model in which GliZ and GipA are working together to regulate gliA (Fig. 8). There appears to be a dependency with regards to gliA expression that we demonstrated in our experiments. This model does not apply to every gene within the gliotoxin cluster, as no other genes have a GipA binding site embedded within a possible GliZ binding site, except gliZ, although these sequences are located farther upstream of the gliZ start site (Fig. 9). Possibly, GipA serves to aid GliZ in binding to the gliA promoter region, as this binding site is different from the others present in the gliotoxin cluster. Although there are possible GipA binding sites in all gliotoxin gene promoters, except gliM, we cannot say with certainty whether GipA is directly binding to these other promoter regions or if GipA is simply binding in the gliA promoter in conjunction with GliZ. This adds to mounting evidence that genes within a gene cluster are typically coordinately regulated, but can also be individually expressed in response to certain stimuli. It is possible that gliT and gliA are both independently regulated to protect the fungus from exogenous gliotoxin, although gliA was not induced in a ΔgliZ mutant in the presence of exogenous gliotoxin as gliT was [19]. Another possibility is that gliA is independently regulated to aid in the transport and/or expression of other secondary metabolism clusters in A. fumigatus. There has been evidence in other fungal species that crosstalk between these gene clusters exists [61].
Fig. 9. Layout of two potential GipA binding sites that are embedded in putative GliZ binding sites in the gliZ promoter. 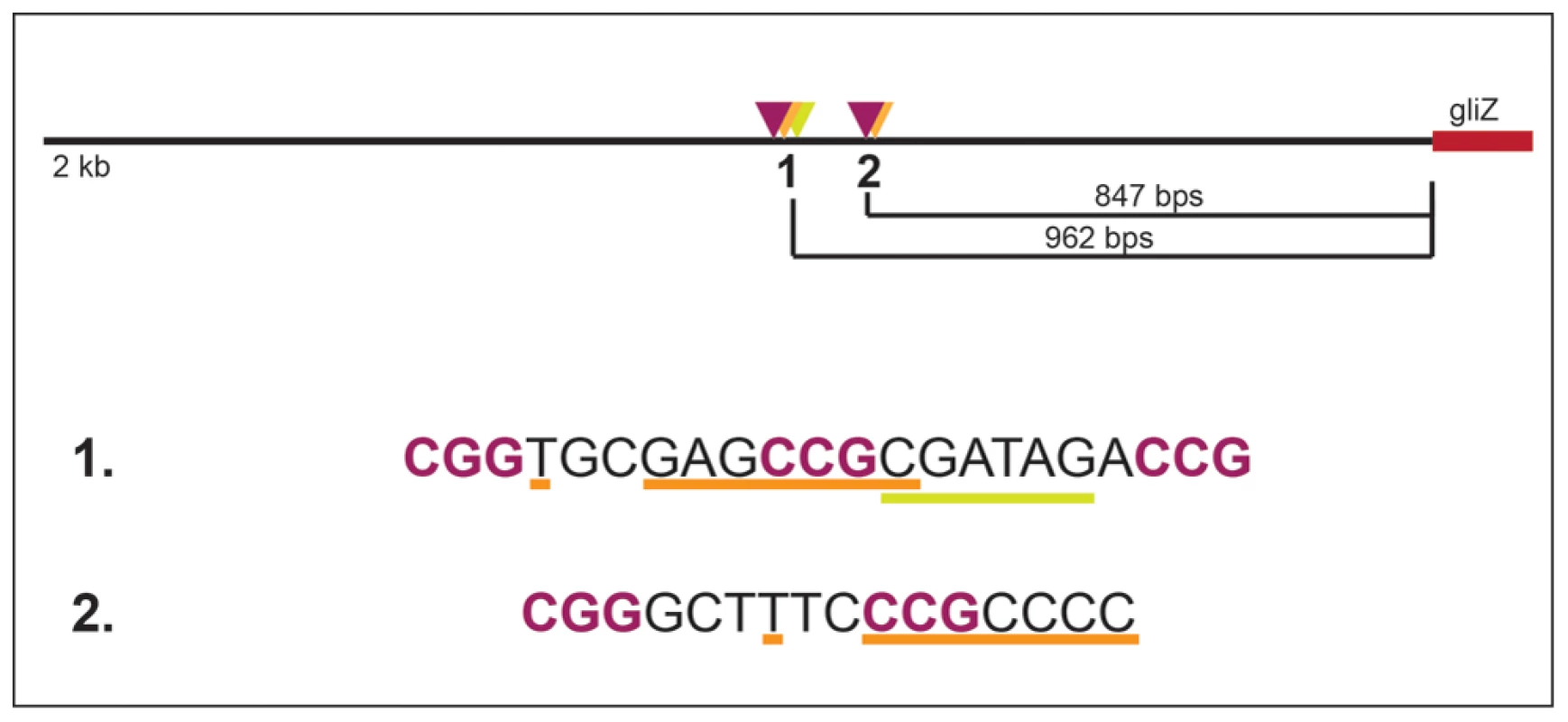
GliZ putative trinucleotide repeats are purple and bolded, the putative GipA binding sites are underlined in orange, and binding cluster 1 additionally contains an AreA recognition element (underlined in yellow). Materials and Methods
All primers used in this study are listed in Table S1.
Strains and Growth Conditions
All strains used in this study and genotypes are listed in Table S2. We maintained Af293.1, Af293.1-GL, 1160, Af293.1-BSM1, and Af293.1-BSM2 on YAG medium supplemented with uridine and uracil (0.5% yeast extract, 1% glucose, trace elements and vitamin mix as modified [62], 10 mM MgCl2, 1.5% agar, 5 mM uridine, and 10 mM uracil,). We grew AMA.G, AMA.Z, AMA.A, AMA-gliZ.G, AMA-gliZ.Z, AMA-gliZ.A, AMA-gipA.G, AMA-gipA.Z, and AMA-gipA.A on YAG medium with 400 µg/ml hygromycin. We maintained all other strains on YAG medium. Unless otherwise noted, we grew all strains at 37°C for 48 hrs. For phenotypic growth assays of high-copy and deletion strains, we inoculated approximately 1000 spores of each strain onto MMVAT (1× MM salts [20 mM ammonium tartrate, 7 mM KCl, 2 mM MgSO4·7H2O], 1% glucose, 12 mM KPO4 pH 6.8, trace elements, vitamin mix as modified [62], and 1.25% agar), MMVAT with 10 µg/ml gliotoxin, and YAG. MMVAT plates were incubated for 72 hrs. We repeated plate growth assays twice for a total of three independent tests. We measured radial growth of each colony and scanned plates on the final day of growth.
High-Copy Inducer Screen
We used fusion PCR (f-PCR) to create a gliAP-lacZ-gliAT construct. For the first reaction, we created three cassettes: gliA 5′P, lacZ, and gliA 3′T, using primer pairs GliA F1 and GliA 5′ R, lacZ F and lacZ R, and GliA 3′ F and GliA R, respectively. We obtained these three fragments by PCR using e2TAK DNA polymerase (Takara Bio Inc., Otsu, Shiga, Japan, Otsu, Shiga, Japan) following manufacturer recommendations. The gliA 5′P fragment had a 3′ extension identical to the first 15 base pairs of lacZ. The lacZ fragment had a 5′ extension identical to the last 15 base pairs of gliA 5′P and a 3′ extension identical to the first 15 base pairs of gliA 3′T. The gliA 3′T fragment had a 5′ extension identical to the last 15 base pairs of lacZ. We used Af293 genomic DNA as template for the gliA 5′P and gliA 3′T regions and λGT11 as template for lacZ.
For the second reaction, we fused the three fragments together using GliA F1 and GliA R as primers. We amplified a 50 µl reaction containing 50 fmol of each fragment, 0.3 µM of each primer, 500 µM of deoxynucleoside triphosphates, buffer 3 at a 1× concentration, and 1 µl of Expand Long DNA Template Mix (Roche Applied Science, Indianapolis, IN) per manufacturer's instructions (briefly, 94°C for 2 min, 10 cycles of 94°C for 10 sec, 62°C for 30 sec, 68°C for 4.5 min and 15 cycles of 94°C for 15 sec, 62°C for 30 sec, 68°C for 4.5 min, increasing the final extension time by 20 sec with each cycle).
We cloned the fusion product into pDONR HPH A [63] using a BP recombination reaction (Invitrogen, Grand Island, NY). We transformed the reaction mix into TOP10 cells (Invitrogen, Grand Island, NY) by electroporation, as recommended by the manufacturer. We grew the transformation mix on LB (1% tryptone, 0.5% yeast extract, 1× SOB salts [10 mM NaCl, 2.5 mM KCl], 1.5% agar) +50 µg/ml kanamycin at 37°C overnight. We picked colonies and transferred to 2 ml of LB liquid +50 µg/ml kanamycin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA from each culture using a miniprep kit (Qiagen, Hilden, Germany). We digested plasmid DNA with specific enzymes to verify the correct insertion. We designated this vector as pDHGL.
We grew Af293.1 in MAG medium supplemented with uridine and uracil (2% malt extract, 0.2% peptone, 1% glucose, trace elements and vitamin mix as modified [62], 2% agar, 5 mM uridine, 10 mM uracil). We performed the transformation as previously described [64], keeping pDHGL as a circular vector. We grew transformants on MMVAT supplemented with 5 mM uridine, 10 mM uracil, 0.2 M sucrose, and 400 µg/ml hygromycin at 37°C for 3–5 days. We identified the presence of pDHGL by Southern hybridization [65] of the lacZ coding region (Fig. S3). We also streaked transformants onto MMVSN supplemented with uridine and uracil (as described above for MMVAT, except 1×MM salts contain 20 mM sodium nitrate instead of ammonium tartrate) and 40 µg/ml X-gal to grow at 37°C for 2 days. We screened for transformants that grew in the presence of hygromycin and developed a blue pigment on X-gal, signaling that lacZ expression was working properly. We designated this strain as Af293.1-GL.
For the first round of the high-copy inducer screen, we grew Af293.1-GL in MAG supplemented with uridine and uracil. We transformed the AMA1-Not1 A. fumigatus genomic library [43] into Af293.1-GL as described previously [64], but with changes. We combined each transformation mix with 50 mls of CM top agar (MMVAT, as described above, 0.1% yeast extract, 0.2% peptone, 0.1% tryptone, 1% CM supplement [27 mM adenine HCl, 33.5 mM methionine, 173 mM arginine, and 1.3 mM riboflavin], and 1% agar) supplemented with 1 M sucrose and spread the mixture over 10 plates (5 ml/plate). We grew transformants on CM supplemented with 0.2 M sucrose and 40 µg/ml X-gal at 37°C for 3–5 days. We screened for transformants that were both prototrophic for uridine and uracil and producing a blue pigment. We prepared genomic DNA from transformants [66] and transformed 1 µl of genomic DNA into TOP10 cells (Invitrogen, Grand Island, NY) by electroporation, as recommended by the manufacturer. We grew the transformation mix on LB +100 µg/ml ampicillin at 37°C overnight. We picked colonies and transferred to 2 ml of LB liquid +100 µg/ml ampicillin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA from each culture using a miniprep kit (Qiagen, Hilden, Germany) and digested it with KpnI to identify individual plasmids.
For the second round of the high-copy inducer screen, we grew Af293.1-GL in MAG supplemented with uridine and uracil. We transformed each individual plasmid isolated from the first round of the genetic screen as described above, but with changes. We plated two amounts of protoplasts (20 µl and 100 µl) each in 4 mls of MMVAT top agar (as described above but with 1% agar) supplemented with 1 M sucrose. We grew transformants on MMVAT supplemented with 0.2 M sucrose and 40 µg/ml X-gal at 37°C for 3–4 days and put plates in the 4°C refrigerator to facilitate blue pigment production. Plasmids causing at least 80% of colonies to turn blue were sequenced using primers AMA-NotI F and AMA-NotI R. We also grew transformants in 10 mls CM at 37°C stationary overnight. We collected mycelia, froze in liquid nitrogen, and lyophilized overnight. We collected total protein and performed β-galactosidase assays to measure LacZ levels quantitatively (detailed below).
For the third round of the high-copy inducer screen, we PCR amplified individual genes from genomic library plasmids, flanked by native 5′ and 3′ non-coding regions. For gipA (Afu6g01910), we used the primer pair 6g01910 F and 6g01910 R. We amplified the fragments from Af293 genomic DNA using e2TAK DNA polymerase (Takara Bio Inc., Otsu, Shiga, Japan) following manufacturer recommendations. We cloned the PCR fragments into pDONR AMA [63] with a BP recombination reaction (Invitrogen, Grand Island, NY). We transformed the reaction mix into TOP10 cells (Invitrogen, Grand Island, NY) by electroporation, as recommended by the manufacturer. We grew the transformation mix on LB +100 µg/ml ampicillin at 37°C overnight. We picked colonies and transferred to 2 ml of LB liquid +100 µg/ml ampicillin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA from each culture using a miniprep kit (Qiagen, Hilden, Germany). We digested plasmid DNA with specific enzymes to verify the correct insertion. We designated the gipA-containing vector as pDONR AMA-gipA. We created a control vector, pDONR AMA-gliZ, which contained the gliZ coding region flanked by promoter and terminator regions. We generated this vector as described above for pDONR AMA-gipA using primers GliZ attB 1 and GliZ attB 2.
We grew Af293.1GL in MAG medium, supplemented with uridine and uracil. We performed the transformation as previously described [64], with changes, using pDONR AMA and pDONR AMA-gliZ as controls. After the 3 hour incubation of protoplasts, we carried out all reactions at half the specified volume. We used 500 ng–750 ng of circular vector DNA for each reaction. We plated two amounts of protoplasts (20 µl and 50 µl) each in 4 mls of MMVAT top agar supplemented with 1 M sucrose. We grew transformants on MMVAT medium supplemented with 0.2 M sucrose at 37°C for 2–3 days. To measure LacZ levels quantitatively, we grew transformants in 10 mls CM at 37°C stationary overnight. We collected mycelia, froze in liquid nitrogen, and lyophilized overnight. We collected total protein and performed β-galactosidase assays (detailed below). We chose three strains to use for future analysis: AMA.GL, AMA-gliZ.GL, and AMA-gipA.GL.
λ Phage Library Screen
We isolated gipA cDNA clones from a λ phage library constructed with the UniZAP vector and poly (A) + mRNA, as described by the manufacturer (Stratagene, La Jolla, California). We performed a primary screen of the λ phage library as recommended by manufacturer (Stratagene, La Jolla, California). We used the gipA coding region as a probe [65]. We excised plugs containing positive plaques and placed them in SM (.1 M NaCl, 10 mM MgSO4·7H2O, 50 mM Tris·HCl [pH 7.5], and .01% gelatin) at 4°C overnight. After determining pfu concentration of plaques, we performed a secondary screen, as performed for the primary screen. We PCR amplified cDNA from excised phagemids using e2TAK DNA polymerase (Takara Bio Inc., Otsu, Shiga, Japan) and following manufacturer recommendations. We used M13 F and M13 R as primers for the reaction. Based on the PCR band sizes, we picked colonies from selected plates and transferred to 2 ml of LB liquid +100 µg/ml ampicillin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA from each culture using a miniprep kit (Qiagen, Hilden, Germany) and sequenced the cDNA insert using M13 F and M13 R primers. We used DNASTAR software (DNASTAR Inc.) to assemble and analyze the sequences obtained from the library screen.
RNA Dot Blot Analysis
For all Dot Blot assays, we grew stationary cultures in 25 mls of CM (repressing) (described above) or CD (non-repressing) (87.6 mM sucrose, 35.3 mM sodium nitrate, 5.8 mM K2HPO4, 2 mM MgSO4·7H2O, 6.7 mM KCl, and 0.025 mM ferrous ammonium sulfate) at a concentration of 5×106 spores/ml at 37°C for 48 hrs. We included 400 µg/ml hygromycin in dot blot assays involving AMA.G, AMA.Z, AMA.A, AMA-gliZ.G, AMA-gliZ.Z, AMA-gliZ.A, AMA-gipA.G, AMA-gipA.Z, and AMA-gipA.A. We prepared total RNA from freeze-dried mycelia using the TRIzol method [62]. We incubated total RNA with denaturing solution (50% formamide, 16%formaldehyde, 1× borate buffer [20× borate buffer: 0.4 M Boric Acid, 4 mM EDTA, pH 8.3 with NaOH], 0.025% bromophenol blue) for 10 min at 65°C. We quenched samples on ice for 10 min, then added equal volume 20× SSC (3 M NaCl, 0.3 M sodium citrate, pH 7.0). We placed a nylon membrane that had been equilibrated in 10× SSC for 10 min into a 96 well dot blot apparatus attached to a vacuum manifold. We collected samples, each containing 3 µg of RNA unless otherwise noted, in 100 µl volumes by aspiration. We aspirated 50 µl of 10× SSC through the membrane in duplicate immediately before and after samples were collected. Once all samples were aspirated, we air dried nylon membranes overnight and then baked them in an oven for 2 hrs. For prehybridization and hybridization, we sealed nylon membranes in a bag using a heated sealer. We prehybridized membranes for 4–6 hrs at 42°C and hybridized membranes overnight at 42°C. For DNA probes, we only used the coding region of each gene of interest (gliA, gliP, gliZ, gliT, gipA, and actin). After hybridization, we washed membranes as we would for Southern hybridization [65] and exposed them to a Typhoon 8600 PhosphorImager (GE Healthcare Life Sciences, Pittsburgh, PA) overnight. We quantified the intensity of hybridization using ImageQuant5.1 software (GE Healthcare Life Sciences, Pittsburgh, PA).
Gliotoxin Extraction and HPLC Analysis
We extracted spent culture medium from RNA dot blot assays in 15 ml chloroform for 30 min at room temperature on an orbital shaker set to 250 rpm. We transferred the chloroform phase to a 50 ml conical tube (BD Biosciences, San Jose, California) and repeated the extraction twice for a total of 45 mls of chloroform. We dried open tubes under a hood until the chloroform was completely evaporated. We added 15 mls of chloroform to each tube and mixed, to concentrate the extracted material at the bottom. We dried open tubes under a hood until chloroform was completely evaporated. We added 1 ml of methanol to each tube and mixed to dissolve all methanol-soluble substances and transferred extracts to microcentrifuge tubes (Fisher Scientific, Pittsburgh, PA). We dried open tubes under a hood until all methanol was completely evaporated.
We dissolved extracts in 50 µl of dimethyl sulfoxide (DMSO), spun tubes to pellet insoluble debris, and transferred DMSO to fresh microcentrifuge tubes. We quantified gliotoxin levels by running samples through a reverse-phase high performance liquid chromatography (RP-HPLC) system with a Waters 996 photodiode array detector (Waters, Milford, MA). We ran samples through a Sonoma 2.1×250 mm C18 column (100 Å pore size) packed with 5 µM particles (VWR, Radnor, PA). The mobile phase consisted of H2O, 0.1% TFA (solution A) and 100% Acetonitrile, 0.1% TFA (solution B): 10% B up to 80% B over 30 min. The injection volume was 10 µl and flow rate was set at 0.4 ml/min. Gliotoxin eluted from the column at 14.7 min and absorbance was read at 268 nm wavelength. We determined gliotoxin concentrations by interpolation from a 9 point standard curve (39 ng to 10 µg) prepared using purified gliotoxin (Sigma-Aldrich Corp., St. Louis, MO).
Microarray
The DNA amplicon microarray for Af293 was created previously [67]. We grew AMA.GL and AMA-gipA.GL in 25 mls CM (described above) at 37°C for 24 hrs in stationary cultures. The spore concentration was 5×106 spores/ml. We used two independent biological replicates for AMA.GL and AMA-gipA.GL growth assays. We prepared total RNA from freeze-dried mycelia using the TRIzol method [62]. We carried out RNA labeling reactions and hybridizations as described in the J. Craig Venter Institute Microarray Protocols (http://pfgrc.jcvi.org/index.php/microarray/protocols.html). We repeated all the hybridizations in dye-swap sets. We scanned and analyzed hybridized slides as described previously [67]. We averaged all replicates for the official data used in our analysis.
Virulence Assays with Toll-Deficient D. melanogaster
We injected the dorsal side of the thorax of CO2-anesthetized, adult, Toll-deficient D. melanogaster flies with a sterile 0.25 mm needle that had been dipped in a solution containing 107 spores/ml of A. fumigatus conidia. We infected 20–25 flies per strain for each virulence assay, which was repeated at least twice. Flies were kept in a 29°C incubator to maximize susceptibility to microbial challenge and monitored for 7 days. Flies that died within 3 hrs of the injection were not included in the survival graph, as these flies most likely died as a result of the puncture wound.
Deletion and Complementation of gipA in Af1160
We amplified the 5′ flanking region (FR) and the 3′ FR from gipA using primers 01910 5′ F, 01910 5′ R, 01910 3′ F, and 01910 3′ R. We engineered a unique NotI site into 01910 5′ F to linearize the final deletion construct. We amplified the fragments from Af293 genomic DNA using e2TAK DNA polymerase (Takara Bio Inc., Otsu, Shiga, Japan) and following manufacturer recommendations. We cloned the gipA 5′ FR into pDONR P4-P1R and we cloned the gipA 3′ FR into pDONR P2R-P3, using BP recombination reactions (Invitrogen, Grand Island, NY). We transformed BP reaction mixes into TOP10 cells (Invitrogen, Grand Island, NY) by electroporation, as recommended by the manufacturer. We grew transformed cells on LB (described above) +50 µg/ml kanamycin at 37°C overnight. We picked colonies and transferred to 2 ml LB liquid +50 µg/ml kanamycin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA from each culture using a miniprep kit (Qiagen, Hilden, Germany). We digested plasmid DNA with specific enzymes to verify the correct fragment orientation.
To create the deletion constructs, we combined pDONR P4-P1R-gipA 5′ FR, pDONR P2R-P3-gipA 3′ FR, and pDONR 221-AnpyrG [63] in an LR recombination reaction with pDEST R4-R3 as the destination vector (Invitrogen, Grand Island, NY) [68]. We transformed the LR reaction mix into TOP10 cells (Invitrogen, Grand Island, NY) by electroporation, as recommended by the manufacturer. We grew transformed cells on LB +100 µg/ml ampicillin at 37°C overnight. We picked colonies and transferred to 2 ml LB liquid +100 µg/ml ampicillin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA from each culture using a miniprep kit (Qiagen, Hilden, Germany). We digested plasmid DNA with specific enzymes to verify the correct fragment orientation. We grew bacterial cultures containing the correct plasmid in 250 ml LB liquid +100 µg/ml ampicillin overnight in a 37°C shaking incubator. We purified plasmid DNA by banding on cesium chloride ethidium bromide gradients [65]. We designated the plasmid pDEST R4-R3-gipA 5′ FR-AnpyrG-gipA 3′ FR.
We grew A. fumigatus 1160 (obtained from FGSC) in MAG supplemented with uridine and uracil. We performed the transformation as previously described [64], linearizing the deletion construct with NotI to facilitate homologous recombination. We grew transformants on MMV supplemented with 0.2 M sucrose at 37°C for 3–5 days. We screened for mutants that were prototrophic for uridine and uracil. We prepared genomic DNA from transformant strains [66], and we identified deletion mutants by Southern blot analysis (Fig. S4) [65]. We made a DNA probe using the gipA 5′ and 3′ FRs to verify ΔgipA.
To create a vector for complementation, we PCR amplified the gipA coding region, flanked by a 3 kilobase promoter region and 500 base pair terminator region, using primers gipA 3 kb F and 6g01910 R. We amplified the fragment from Af293 genomic DNA using e2TAK DNA polymerase (Takara Bio Inc., Otsu, Shiga, Japan) and following manufacturer recommendations. We cloned the PCR product into pDONR HPH B [63], as described previously for pDHGL and designated this vector pDONR HPH-gipA. We grew ΔgipA in MAG medium and performed a transformation as previously described for pDHGL. For Southern hybridization, we made a DNA probe using the coding region of gipA (Fig. S5). We designated this strain as gipA(R).
We obtained controls (1160G and ΔgliZ) for growth assays. We created 1160G by transforming pDONR G [63] into Af1160 and we created ΔgliZ by transforming pDEST R4-R3-gliZ 5′ FR-AnpyrG-gliZ 3′ FR into Af1160, performed as described above for ΔgipA. We created the gliZ deletion construct as described previously for ΔgipA using primers gliZ 5′ F, gliZ 5′ R, gliZ 3′ F, and gliZ 3′ R. We used gliZ 5′ and 3′ flanking regions as a DNA probe for Southern hybridization (Fig. S4).
Construction of ΔgliZ/ΔgipA
We grew ΔgipA on YAG supplemented with uridine and uracil +1 mg/ml 5-Fluororotic acid (5-FOA) at 37°C. Colonies that reverted to a pyrG - phenotype grew as outgrowths from the original streak. We prepared genomic DNA from these pyrG - outgrowths [66] and tested them for the presence of gipA by Southern hybridization (Fig. S6) [65]. We used the gipA coding region as a DNA probe. We designated this mutant as ΔgipA.0. We transformed pDEST R4-R3-gliZ 5′ FR-AnpyrG-gliZ 3′ FR into ΔgipA.0, as described above for ΔgipA. We used gliZ 5′ and 3′ flanking regions as a DNA probe for Southern hybridization (Fig. S7).
Protein Binding Microarray
We amplified the C2H2 DNA binding region from gipA using primers gipA C2H2 F and gipA C2H2 R. We used Af293 as template and AccuPrime Pfx DNA polymerase (Life Technologies, Grand Island, NY) and per manufacturer's recommendations. We cloned the PCR product into pDONR 221 using a BP recombination reaction (Invitrogen, Grand Island, NY). We transformed the reaction mix into TOP10 cells (Invitrogen, Grand Island, NY) by electroporation, as recommended by the manufacturer. We grew the transformation mix on LB +50 µg/ml kanamycin at 37°C overnight. We picked colonies and transferred to 2 ml of LB liquid +50 µg/ml kanamycin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA from each culture using a miniprep kit (Qiagen, Hilden, Germany). We digested plasmid DNA with specific enzymes to verify the correct insertion. We recombined the gipA C2H2 region into pDEST 15 using an LR recombination reaction (Invitrogen, Grand Island, NY). We transformed the reaction mix into TOP10 cells (Invitrogen, Grand Island, NY) by electroporation, as recommended by the manufacturer. We grew the transformation mix on LB +100 µg/ml ampicillin at 37°C overnight. We picked colonies and transferred to 2 ml of LB liquid +100 µg/ml ampicillin to grow overnight in a 37°C shaking incubator. We isolated plasmid DNA from each culture using a miniprep kit (Qiagen, Hilden, Germany). We digested plasmid DNA with specific enzymes to verify the correct insertion. This vector, pDEST 15-gipA C2H2, was used in a protein binding microarray analysis as previously described [69].
Mutagenesis of the GipA DNA Binding Site in the gliA Promoter
We created mutated gipA DNA binding sites in pDHGL using a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies Inc., Santa Clara, CA), as recommended by the manufacturer. We utilized primers BSM1 F and BSM1 R, BSM2 F and BSM2 R to create vectors pDHBSM1 and pDHBSM2, respectively. We transformed pDHGL, pDHBSM1 and pDHBSM2 into Af293.1, as previously described for Af293.1-GL, except we plated transformants on YAG supplemented with uridine and uracil (described above), 0.2 M sucrose, and 300 µg/ml hygromycin. We verified correct transformants by Southern hybridization using the lacZ coding region as a probe (Fig. S8). We designated these strains as Af293.1-GL, Af293.1-BSM1 and Af293.1-BSM2, respectively. We obtained two independent isolates for each binding site mutant to verify that positional effects were not contributing to our results. To test the effect of the different gipA binding site mutants, we transformed pDONR AMA, pDONR AMA-gliZ, and pDONR AMA-gipA into two independent isolates of Af293.1-GL, Af293.1-BSM1, and Af293.1-BSM2, as described above for AMA-gipA.GL, except we grew transformants on YAG supplemented with 0.2 M sucrose. We designated these strains as AMA.GL, AMA-gliZ.GL, AMA-gipA.GL, AMA.BSM1, AMA-gliZ.BSM1, AMA-gipA.BSM1, AMA.BSM2, AMA-gliZ.BSM2, and AMA-gipA.BSM2.
β-Galactosidase Assays
We ground 50 µl lyophilized mycelia to a fine powder with acid-washed glass beads (400–650 µm) (Sigma-Aldrich Corp., St. Louis, MO). We suspended the ground powder in 200 µl protein extraction buffer (PEB) (60 mM Na2HPO4·7H2O, 40 mM NaH2PO4·H2O, 10 mM KCl, 1 mM MgSO4·7H2O, 1 mM EDTA, and 20 µM PMSF [added fresh], pH 7.0) by vortexing and incubated the samples on ice for 15 min, with additional vortexing every 5 min. We spun tubes for 15 min at 15,600× g at 4°C to pellet cellular debris and beads. We transferred supernatants, containing total protein, to fresh tubes on ice and measured protein concentration using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). In a 96-well plate, we added 10 µl of protein in PEB and 90 µl of Z Buffer (60 mM Na2HPO4·7H2O, 40 mM NaH2PO4·H2O, 10 mM KCl, 1 mM MgSO4·7H2O, 50 mM β-mercaptoethanol [added fresh], pH to 7.0). For samples grown in repressing conditions (CM), the total protein added was 1 µg. For samples grown in non-repressing conditions (CD), the total protein added was 0.1 µg. To begin the β-galactosidase assay, we added 20 µl of 2-Nitrophenyl-β-D-galactopyranoside (ONPG) (Sigma-Aldrich Corp., St. Louis, MO), diluted to 4 mg/ml in Z Buffer, and placed the 96-well plate in a 37°C incubator. We timed reactions and stopped samples with 50 µl 1 M Na2CO3. We measured absorbance at OD420 and calculated Units of β-galactosidase activity/mg protein with the following equation: (OD420×TV)/(0.0045×T×V×C), where TV is total volume of the reaction in ml, T is time in min, V is volume of protein added in ml, and C is concentration of protein used in µg/µl.
Creation of Strains for Bypass Suppression Experiments
We grew Af293.1 in MAG supplemented with uridine and uracil (described above). We transformed pDEST R4-R3-gliZ 5′ FR-AnpyrG-gliZ 3′ FR and pDEST-gipA 5′ FR-AnpyrG-gipA 3′ FR into Af293.1, as previously described for ΔgipA, except we plated transformants on YAG supplemented with 0.2 M sucrose. We used gliZ 5′ and 3′ FRs and gipA 5′ and 3′ FRs for Southern hybridization and designated these strains ΔgliZ.1 and ΔgipA.1 (Fig. S9). We chose one strain from the ΔgliZ transformation that did not show homologous recombination at the gliZ locus, but was prototrophic for uridine and uracil. We designated this strain pyrG+. We collected total RNA and performed dot blot analysis on pyrG+, ΔgliZ.1, and ΔgipA.1 to verify loss of gliZ and gipA, respectively (described in RNA dot blot Analysis) (Fig. S10). We cloned gliZ and gipA into pDONR AMA/HPH [63], as described above for pDONR AMA-gliZ and pDONR AMA-gipA, respectively, except we used primers gipA 3 kb F and 6g01910 R for the gipA cassette. We designated these plasmids pDONR AMA/HPH-gliZ and pDONR/HPH-gipA. We transformed these plasmids, along with pDONR AMA/HPH empty vector, into pyrG+, ΔgliZ.1, and ΔgipA.1, as described above for AMA-gipA.GL, except we grew transformants on YAG supplemented with uridine and uracil, 0.2 M sucrose, and 400 µg/ml hygromycin. We designated these strains as AMA.G, AMA-gliZ.G, AMA-gipA.G, AMA.Z, AMA-gliZ.Z, AMA-gipA.Z, AMA.A, AMA-gliZ.A, and AMA-gipA.A.
Supporting Information
Zdroje
1. KameiK, WatanabeA (2005) Aspergillus mycotoxins and their effect on the host. Med Mycol 43 Suppl 1: S95–99.
2. BayramO, BrausGH (2012) Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev 36 : 1–24.
3. KellerNP, TurnerG, BennettJW (2005) Fungal secondary metabolism - from biochemistry to genomics. Nat Rev Microbiol 3 : 937–947.
4. BrakhageAA (2013) Regulation of fungal secondary metabolism. Nat Rev Microbiol 11 : 21–32.
5. YuJH, KellerN (2005) Regulation of secondary metabolism in filamentous fungi. Annu Rev Phytopathol 43 : 437–458.
6. CalvoAM, WilsonRA, BokJW, KellerNP (2002) Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev 66 : 447–459 table of contents.
7. SiglC, HaasH, SpechtT, PfallerK, KurnsteinerH, et al. Among developmental regulators, StuA but not BrlA is essential for penicillin V production in Penicillium chrysogenum. Appl Environ Microbiol 77 : 972–982.
8. BrakhageAA, BrowneP, TurnerG (1992) Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J Bacteriol 174 : 3789–3799.
9. LatgeJP (1999) Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev 12 : 310–350.
10. YuJ, BhatnagarD, EhrlichKC (2002) Aflatoxin biosynthesis. Rev Iberoam Micol 19 : 191–200.
11. McDonaghA, FedorovaND, CrabtreeJ, YuY, KimS, et al. (2008) Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog 4: e1000154.
12. BokJW, ChungD, BalajeeSA, MarrKA, AndesD, et al. (2006) GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect Immun 74 : 6761–6768.
13. GardinerDM, HowlettBJ (2005) Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol Lett 248 : 241–248.
14. FoxEM, GardinerDM, KellerNP, HowlettBJ (2008) A Zn(II)2Cys6 DNA binding protein regulates the sirodesmin PL biosynthetic gene cluster in Leptosphaeria maculans. Fungal Genet Biol 45 : 671–682.
15. ToddRB, AndrianopoulosA (1997) Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet Biol 21 : 388–405.
16. EhrlichKC, MontalbanoBG, CaryJW (1999) Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 230 : 249–257.
17. LiangSD, MarmorsteinR, HarrisonSC, PtashneM (1996) DNA sequence preferences of GAL4 and PPR1: how a subset of Zn2 Cys6 binuclear cluster proteins recognizes DNA. Mol Cell Biol 16 : 3773–3780.
18. VasheeS, XuH, JohnstonSA, KodadekT (1993) How do “Zn2 cys6” proteins distinguish between similar upstream activation sites? Comparison of the DNA-binding specificity of the GAL4 protein in vitro and in vivo. J Biol Chem 268 : 24699–24706.
19. SchrettlM, CarberryS, KavanaghK, HaasH, JonesGW, et al. (2010) Self-protection against gliotoxin–a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog 6: e1000952.
20. MogensenJ, NielsenHB, HofmannG, NielsenJ (2006) Transcription analysis using high-density micro-arrays of Aspergillus nidulans wild-type and creA mutant during growth on glucose or ethanol. Fungal Genet Biol 43 : 593–603.
21. FengGH, LeonardTJ (1998) Culture conditions control expression of the genes for aflatoxin and sterigmatocystin biosynthesis in Aspergillus parasiticus and A. nidulans. Appl Environ Microbiol 64 : 2275–2277.
22. BrandA (2012) Hyphal growth in human fungal pathogens and its role in virulence. Int J Microbiol 2012 : 517529.
23. ComeraC, AndreK, LaffitteJ, ColletX, GaltierP, et al. (2007) Gliotoxin from Aspergillus fumigatus affects phagocytosis and the organization of the actin cytoskeleton by distinct signalling pathways in human neutrophils. Microbes Infect 9 : 47–54.
24. KupfahlC, HeinekampT, GeginatG, RuppertT, HartlA, et al. (2006) Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Mol Microbiol 62 : 292–302.
25. PahlHL, KraussB, Schulze-OsthoffK, DeckerT, TraencknerEB, et al. (1996) The immunosuppressive fungal metabolite gliotoxin specifically inhibits transcription factor NF-kappaB. J Exp Med 183 : 1829–1840.
26. Kwon-ChungKJ, SuguiJA (2009) What do we know about the role of gliotoxin in the pathobiology of Aspergillus fumigatus? Med Mycol 47 Suppl 1: S97–103.
27. GardinerDM, JarvisRS, HowlettBJ (2005) The ABC transporter gene in the sirodesmin biosynthetic gene cluster of Leptosphaeria maculans is not essential for sirodesmin production but facilitates self-protection. Fungal Genet Biol 42 : 257–263.
28. ScharfDH, HeinekampT, RemmeN, HortschanskyP, BrakhageAA, et al. (2012) Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl Microbiol Biotechnol 93 : 467–472.
29. BernardoPH, BraschN, ChaiCL, WaringP (2003) A novel redox mechanism for the glutathione-dependent reversible uptake of a fungal toxin in cells. J Biol Chem 278 : 46549–46555.
30. CramerRAJr, GamcsikMP, BrookingRM, NajvarLK, KirkpatrickWR, et al. (2006) Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot Cell 5 : 972–980.
31. SpikesS, XuR, NguyenCK, ChamilosG, KontoyiannisDP, et al. (2008) Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J Infect Dis 197 : 479–486.
32. WaringP, NewcombeN, EdelM, LinQH, JiangH, et al. (1994) Cellular uptake and release of the immunomodulating fungal toxin gliotoxin. Toxicon 32 : 491–504.
33. YoshidaLS, AbeS, TsunawakiS (2000) Fungal gliotoxin targets the onset of superoxide-generating NADPH oxidase of human neutrophils. Biochem Biophys Res Commun 268 : 716–723.
34. FoxEM, HowlettBJ (2008) Biosynthetic gene clusters for epipolythiodioxopiperazines in filamentous fungi. Mycol Res 112 : 162–169.
35. GardinerDM, CozijnsenAJ, WilsonLM, PedrasMS, HowlettBJ (2004) The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol Microbiol 53 : 1307–1318.
36. BokJW, BalajeeSA, MarrKA, AndesD, NielsenKF, et al. (2005) LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell 4 : 1574–1582.
37. BokJW, KellerNP (2004) LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 3 : 527–535.
38. ParkHS, BayramO, BrausGH, KimSC, YuJH (2012) Characterization of the velvet regulators in Aspergillus fumigatus. Mol Microbiol 86 : 937–953.
39. MukherjeePK, KenerleyCM (2010) Regulation of morphogenesis and biocontrol properties in Trichoderma virens by a VELVET protein, Vel1. Appl Environ Microbiol 76 : 2345–2352.
40. KrijgsheldP, BleichrodtR, van VeluwGJ, WangF, MullerWH, et al. (2013) Development in Aspergillus. Stud Mycol 74 : 1–29.
41. SekonyelaR, PalmerJM, BokJW, JainS, BerthierE, et al. (2013) RsmA Regulates Aspergillus fumigatus Gliotoxin Cluster Metabolites Including Cyclo(L-Phe-L-Ser), a Potential New Diagnostic Marker for Invasive Aspergillosis. PLoS One 8: e62591.
42. JainR, ValianteV, RemmeN, DocimoT, HeinekampT, et al. (2011) The MAP kinase MpkA controls cell wall integrity, oxidative stress response, gliotoxin production and iron adaptation in Aspergillus fumigatus. Mol Microbiol 82 : 39–53.
43. XueT, NguyenCK, RomansA, KontoyiannisDP, MayGS (2004) Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain AF293. Arch Microbiol 182 : 346–353.
44. AleksenkoA, ClutterbuckAJ (1997) Autonomous plasmid replication in Aspergillus nidulans: AMA1 and MATE elements. Fungal Genet Biol 21 : 373–387.
45. GemsD, JohnstoneIL, ClutterbuckAJ (1991) An autonomously replicating plasmid transforms Aspergillus nidulans at high frequency. Gene 98 : 61–67.
46. UkilL, VaradarajA, GovindaraghavanM, LiuHL, OsmaniSA (2008) Copy number suppressors of the Aspergillus nidulans nimA1 mitotic kinase display distinctive and highly dynamic cell cycle-regulated locations. Eukaryot Cell 7 : 2087–2099.
47. OsherovN, KontoyiannisDP, RomansA, MayGS (2001) Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14alpha-demethylase gene, pdmA. J Antimicrob Chemother 48 : 75–81.
48. BrayerKJ, SegalDJ (2008) Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys 50 : 111–131.
49. BrownRS (2005) Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol 15 : 94–98.
50. Berg JM TJ, Stryer L (2002) Examination of Three-Dimensional Structure Enhances Our Understanding of Evolutionary Relationships. Biochemistry. 5th ed. New York: W H Freeman.
51. WankeC, EckertS, AlbrechtG, van HartingsveldtW, PuntPJ, et al. (1997) The Aspergillus niger GCN4 homologue, cpcA, is transcriptionally regulated and encodes an unusual leucine zipper. Mol Microbiol 23 : 23–33.
52. PerrinRM, FedorovaND, BokJW, CramerRA, WortmanJR, et al. (2007) Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog 3: e50.
53. SanchezJF, SomozaAD, KellerNP, WangCC (2012) Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat Prod Rep 29 : 351–371.
54. WolfeSA, NekludovaL, PaboCO (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct 29 : 183–212.
55. KellerNP, NesbittC, SarrB, PhillipsTD, BurowGB (1997) pH Regulation of Sterigmatocystin and Aflatoxin Biosynthesis in Aspergillus spp. Phytopathology 87 : 643–648.
56. BrakhageAA, SproteP, Al-AbdallahQ, GehrkeA, PlattnerH, et al. (2004) Regulation of penicillin biosynthesis in filamentous fungi. Adv Biochem Eng Biotechnol 88 : 45–90.
57. CaryJW, Harris-CowardPY, EhrlichKC, MackBM, KaleSP, et al. (2012) NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot Cell 11 : 1104–1111.
58. RuijterGJ, VisserJ (1997) Carbon repression in Aspergilli. FEMS Microbiol Lett 151 : 103–114.
59. Arratia-QuijadaJ, SanchezO, ScazzocchioC, AguirreJ (2012) FlbD, a Myb transcription factor of Aspergillus nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryot Cell 11 : 1132–1142.
60. GarziaA, EtxebesteO, Herrero-GarciaE, UgaldeU, EspesoEA (2010) The concerted action of bZip and cMyb transcription factors FlbB and FlbD induces brlA expression and asexual development in Aspergillus nidulans. Mol Microbiol 75 : 1314–1324.
61. BergmannS, FunkAN, ScherlachK, SchroeckhV, ShelestE, et al. (2010) Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl Environ Microbiol 76 : 8143–8149.
62. ReyesG, RomansA, NguyenCK, MayGS (2006) Novel mitogen-activated protein kinase MpkC of Aspergillus fumigatus is required for utilization of polyalcohol sugars. Eukaryot Cell 5 : 1934–1940.
63. SchoberleTJ, Nguyen-ColemanCK, MayGS (2013) Plasmids for increased efficiency of vector construction and genetic engineering in filamentous fungi. Fungal Genet Biol 58–59 : 1–9.
64. MayGS (1989) The highly divergent beta-tubulins of Aspergillus nidulans are functionally interchangeable. J Cell Biol 109 : 2267–2274.
65. Maniatis T, Fritsch EF, Sambrook J (1982) Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory. 521 p.
66. JinJ, LeeYK, WickesBL (2004) Simple chemical extraction method for DNA isolation from Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol 42 : 4293–4296.
67. NiermanWC, PainA, AndersonMJ, WortmanJR, KimHS, et al. (2005) Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438 : 1151–1156.
68. MagnaniE, BartlingL, HakeS (2006) From Gateway to MultiSite Gateway in one recombination event. BMC Mol Biol 7 : 46.
69. LamKN, van BakelH, CoteAG, van der VenA, HughesTR (2011) Sequence specificity is obtained from the majority of modular C2H2 zinc-finger arrays. Nucleic Acids Res 39 : 4680–4690.
70. CoyleCM, PanaccioneDG (2005) An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl Environ Microbiol 71 : 3112–3118.
71. SchrettlM, BignellE, KraglC, SabihaY, LossO, et al. (2007) Distinct roles for intra - and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog 3 : 1195–1207.
72. AmesBD, HaynesSW, GaoX, EvansBS, KelleherNL, et al. (2011) Complexity generation in fungal peptidyl alkaloid biosynthesis: oxidation of fumiquinazoline A to the heptacyclic hemiaminal fumiquinazoline C by the flavoenzyme Af12070 from Aspergillus fumigatus. Biochemistry 50 : 8756–8769.
73. AmesBD, LiuX, WalshCT (2010) Enzymatic processing of fumiquinazoline F: a tandem oxidative-acylation strategy for the generation of multicyclic scaffolds in fungal indole alkaloid biosynthesis. Biochemistry 49 : 8564–8576.
74. O'HanlonKA, GallagherL, SchrettlM, JochlC, KavanaghK, et al. (2012) Nonribosomal peptide synthetase genes pesL and pes1 are essential for Fumigaclavine C production in Aspergillus fumigatus. Appl Environ Microbiol 78 : 3166–3176.
75. ChooiYH, FangJ, LiuH, FillerSG, WangP, et al. (2013) Genome mining of a prenylated and immunosuppressive polyketide from pathogenic fungi. Org Lett 15 : 780–783.
76. TsaiHF, WheelerMH, ChangYC, Kwon-ChungKJ (1999) A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J Bacteriol 181 : 6469–6477.
77. YinWB, BaccileJA, BokJW, ChenY, KellerNP, et al. (2013) A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus Aspergillus fumigatus. J Am Chem Soc 135 : 2064–2067.
78. LimFY, HouY, ChenY, OhJH, LeeI, et al. (2012) Genome-based cluster deletion reveals an endocrocin biosynthetic pathway in Aspergillus fumigatus. Appl Environ Microbiol 78 : 4117–4125.
79. BerthierE, LimFY, DengQ, GuoCJ, KontoyiannisDP, et al. (2013) Low-volume toolbox for the discovery of immunosuppressive fungal secondary metabolites. PLoS Pathog 9: e1003289.
80. LinHC, ChooiYH, DhingraS, XuW, CalvoAM, et al. (2013) The fumagillin biosynthetic gene cluster in Aspergillus fumigatus encodes a cryptic terpene cyclase involved in the formation of beta-trans-bergamotene. J Am Chem Soc 135 : 4616–4619.
81. MaiyaS, GrundmannA, LiX, LiSM, TurnerG (2007) Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. Chembiochem 8 : 1736–1743.
82. VodischM, ScherlachK, WinklerR, HertweckC, BraunHP, et al. (2011) Analysis of the Aspergillus fumigatus proteome reveals metabolic changes and the activation of the pseurotin A biosynthesis gene cluster in response to hypoxia. J Proteome Res 10 : 2508–2524.
Štítky
Genetika Reprodukčná medicína
Článek Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin PathwayČlánek Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human DiseasesČlánek G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify LongevityČlánek PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial MatrixČlánek Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus ConflictsČlánek The Impact of Population Demography and Selection on the Genetic Architecture of Complex TraitsČlánek Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar AcidificationČlánek The Case for Junk DNA
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 5- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast
- A Mutation in the Gene in Dogs with Hereditary Footpad Hyperkeratosis (HFH)
- Loss of Function Mutation in the Palmitoyl-Transferase HHAT Leads to Syndromic 46,XY Disorder of Sex Development by Impeding Hedgehog Protein Palmitoylation and Signaling
- Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution
- Genome-Wide Nucleosome Positioning Is Orchestrated by Genomic Regions Associated with DNase I Hypersensitivity in Rice
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Single Nucleotide Variants in Transcription Factors Associate More Tightly with Phenotype than with Gene Expression
- Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway
- Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human Diseases
- Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
- Meiotic Drive Impacts Expression and Evolution of X-Linked Genes in Stalk-Eyed Flies
- G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify Longevity
- Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
- Whole Exome Re-Sequencing Implicates and Cilia Structure and Function in Resistance to Smoking Related Airflow Obstruction
- Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues
- R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- The Impairment of MAGMAS Function in Human Is Responsible for a Severe Skeletal Dysplasia
- Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Males
- Mlh2 Is an Accessory Factor for DNA Mismatch Repair in
- Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
- The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells
- Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts
- Genome-Wide Inference of Ancestral Recombination Graphs
- Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in
- SHP2 Regulates Chondrocyte Terminal Differentiation, Growth Plate Architecture and Skeletal Cell Fates
- The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits
- Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation
- Genetic Dissection of the Female Head Transcriptome Reveals Widespread Allelic Heterogeneity
- Genome Sequencing and Comparative Genomics of the Broad Host-Range Pathogen AG8
- Copy Number Variation Is a Fundamental Aspect of the Placental Genome
- GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
- Hox Transcription Factors Access the RNA Polymerase II Machinery through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19
- Drosha Promotes Splicing of a Pre-microRNA-like Alternative Exon
- Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of
- PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF
- Enhanced Interaction between Pseudokinase and Kinase Domains in Gcn2 stimulates eIF2α Phosphorylation in Starved Cells
- A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in
- dGTP Starvation in Provides New Insights into the Thymineless-Death Phenomenon
- Phosphorylation Modulates Clearance of Alpha-Synuclein Inclusions in a Yeast Model of Parkinson's Disease
- RPM-1 Uses Both Ubiquitin Ligase and Phosphatase-Based Mechanisms to Regulate DLK-1 during Neuronal Development
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- Heritable Transmission of Stress Resistance by High Dietary Glucose in
- Revertant Mutation Releases Confined Lethal Mutation, Opening Pandora's Box: A Novel Genetic Pathogenesis
- Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification
- A Genome-Wide Assessment of the Role of Untagged Copy Number Variants in Type 1 Diabetes
- Selectivity in Genetic Association with Sub-classified Migraine in Women
- A Lack of Parasitic Reduction in the Obligate Parasitic Green Alga
- The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
- Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
- Six Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
- EDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
- Genes That Bias Mendelian Segregation
- The Case for Junk DNA
- An In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
- Mosaic Epigenetic Dysregulation of Ectodermal Cells in Autism Spectrum Disorder
- Hyperactivated Wnt Signaling Induces Synthetic Lethal Interaction with Rb Inactivation by Elevating TORC1 Activities
- Mutations in the Cholesterol Transporter Gene Are Associated with Excessive Hair Overgrowth
- Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland
- A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics
- Spermatid Cyst Polarization in Depends upon and the CPEB Family Translational Regulator
- Insights into the Genetic Structure and Diversity of 38 South Asian Indians from Deep Whole-Genome Sequencing
- Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in
- A Dominant-Negative Mutation of Mouse Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation
- Biased, Non-equivalent Gene-Proximal and -Distal Binding Motifs of Orphan Nuclear Receptor TR4 in Primary Human Erythroid Cells
- Ras-Mediated Deregulation of the Circadian Clock in Cancer
- Retinoic Acid-Related Orphan Receptor γ (RORγ): A Novel Participant in the Diurnal Regulation of Hepatic Gluconeogenesis and Insulin Sensitivity
- Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions
- Fine Tuning of the UPR by the Ubiquitin Ligases Siah1/2
- Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression
- Allele-Specific Genome-wide Profiling in Human Primary Erythroblasts Reveal Replication Program Organization
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



