-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
Fatty liver disease (steatosis) is the most common liver disease worldwide and is commonly caused by obesity, type 2 diabetes, or alcohol abuse. All of these conditions are associated with impaired hepatocyte protein secretion, resulting in hypoproteinemia that contributes to the systemic complications of these diseases. The unfolded protein response (UPR) is activated in response to stress in the protein secretory pathway and a wealth of data indicates that UPR activation can contribute to steatosis, but the mechanistic basis for this relationship is poorly understood. We identify activating transcription factor 6 (Atf6), one of three UPR sensors, as necessary and sufficient for steatosis and show that Atf6 activation can promote lipogenesis, providing a direct connection between the stress response and lipid metabolism. Blocking Atf6 in zebrafish larvae prevents alcohol-induced steatosis and Atf6 overexpression in zebrafish hepatocytes induces genes that drive lipogenesis, increases lipid production and causes steatosis. Fatty acid synthase (fasn) is a key lipogenic enzyme and we show that fasn is required for fatty liver in response to both ethanol and Atf6 overexpression. Our findings point to Atf6 as a potential therapeutic target for fatty liver disease.
Published in the journal: Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish. PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004335
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004335Summary
Fatty liver disease (steatosis) is the most common liver disease worldwide and is commonly caused by obesity, type 2 diabetes, or alcohol abuse. All of these conditions are associated with impaired hepatocyte protein secretion, resulting in hypoproteinemia that contributes to the systemic complications of these diseases. The unfolded protein response (UPR) is activated in response to stress in the protein secretory pathway and a wealth of data indicates that UPR activation can contribute to steatosis, but the mechanistic basis for this relationship is poorly understood. We identify activating transcription factor 6 (Atf6), one of three UPR sensors, as necessary and sufficient for steatosis and show that Atf6 activation can promote lipogenesis, providing a direct connection between the stress response and lipid metabolism. Blocking Atf6 in zebrafish larvae prevents alcohol-induced steatosis and Atf6 overexpression in zebrafish hepatocytes induces genes that drive lipogenesis, increases lipid production and causes steatosis. Fatty acid synthase (fasn) is a key lipogenic enzyme and we show that fasn is required for fatty liver in response to both ethanol and Atf6 overexpression. Our findings point to Atf6 as a potential therapeutic target for fatty liver disease.
Introduction
The unfolded protein response (UPR) acts in most cells to maintain homeostasis within the protein secretory pathway during physiological conditions. During stress, the UPR becomes further induced to mitigate the accumulation of misfolded or unfolded proteins in the endoplasmic reticulum (ER). If UPR activation cannot relieve the excess protein load, the ER becomes dilated and dysfunctional, and all branches of the UPR remain chronically activated in a condition referred to as ER stress. Many studies have implicated ER stress in a range of pathologies, and there is a clear association between UPR activation and metabolic diseases such as fatty liver disease (FLD) [1]–[5]. However, it is not known whether factors that control the UPR can also directly impact lipid metabolism and it remains unclear how UPR activation causes FLD.
FLD is the most common hepatic pathology worldwide [6], and alcohol abuse is a leading cause. Even a single episode of binge drinking causes lipid accumulation in hepatocytes (steatosis) in over 90% of drinkers [7]. While acute steatosis can resolve, chronic steatosis can render hepatocytes susceptible to damage and is a prerequisite step in developing more severe liver disease, including steatohepatitis and cirrhosis. Most FLD etiologies are accompanied by impairment of protein secretion by hepatocytes that result in serum protein deficiencies, which are most apparent in chronic alcoholics and contribute to the systemic complications of alcoholic liver disease (ALD). These defects are reflected in many reports demonstrating that alcohol induces some UPR sensors and targets in the liver of mice [1], [8], rats [9], micropigs [10], and zebrafish [11]–[13]. Moreover, deleting ATF4 or CHOP, two key UPR effectors, reduces ethanol-induced liver injury [8], [14], indicating that UPR activation can contribute to ALD. However, it is not clear whether these genes, or other UPR effectors, contribute to steatosis.
There are three main UPR sensors that function through shared and independent mechanisms to maintain ER function by enhancing protein processing and folding in the ER, and by promoting degradation of terminally misfolded secretory proteins. Activating transcription factor 6 (ATF6) and inositol-requiring enzyme 1-alpha (IRE1α or ERN1) pathways generate the active transcription factors nATF6 and XBP1, respectively, that induce hundreds of UPR target genes [15], [16]. PRKR-like endoplasmic reticulum kinase (PERK or EIF2AK3) phosphorylates EIF2A to repress translation [17] and to promote production of ATF4 [18], which induces a subset of target genes [14], [18]. Each of these primary sensors has been evaluated for its contribution to FLD caused by a robust ER stressor [19], [20]. ATF6 knockout mice fail to resolve steatosis caused by acute stress due to tunicamycin injection [16], [21], [22], suggesting that loss of ATF6 promotes fatty liver. Our previous work using zebrafish confirmed the finding that steatosis caused by acute stress is augmented by atf6 loss, and also demonstrates that steatosis caused by chronic stress is reduced when Atf6 is depleted [23]. Thus, it appears that atf6 loss can alternatively enhance or reduce FLD, depending on the nature and duration of the stress.
The relationship between metabolic disease and UPR activation is under intensive investigation, yet several important questions remain unanswered. First, is UPR activation a cause or consequence of FLD? While some studies indicate that lipotoxicity and fatty acid accumulation can induce the UPR [24], [25], there is incontrovertible evidence that robust UPR activation is sufficient to induce steatosis [2], [26], [27]. These data are incorporated into a current model proposing that robust UPR activation can cause steatosis and, if the lipid burden is not resolved, this can further augment cellular stress and contribute to chronic UPR induction. The second question is which, if any, of the main UPR factors directly cause fatty liver? All studies to date that have addressed this question, including our work in zebrafish [23], utilize loss of function approaches whereby a key UPR gene is deleted and the effect on stress-induced steatosis is evaluated. However, these studies are complicated by the extensive crosstalk between UPR effectors, and by the ability of cells to adapt to changes in UPR capacity. Third, what is the mechanism by which UPR activation causes fatty liver? Activation of lipogenic genes by the sterol response element binding protein (SREBP) transcription factors has been implicated as an essential pathway in both alcoholic [28], [29] and non-alcoholic [30]–[32] FLD. Since SREBPs and ATF6 are activated by a shared proteolytic mechanism [33], it is possible that these two proteins are activated simultaneously in response to stress. However, the role of SREBPs in FLD caused by UPR activation has not been addressed.
Here, we use genetic and genomic approaches in a well-established zebrafish model of ALD [11]–[13], [34]–[37] to demonstrate that blocking Atf6 prevents alcohol-induced steatosis and that overexpression of the nuclear, active form of Atf6 (nAtf6) causes steatosis. We found that nAtf6 overexpression increases expression of genes in the lipogenic pathway and alters lipid flux to promote triglyceride synthesis by a mechanism that does not involve Srebps. Instead, we demonstrate an epistatic interaction between atf6 and fasn in driving alcoholic steatosis. These findings demonstrate the first clear and causative link between a central UPR sensor and FLD.
Results
Atf6 is required for alcoholic steatosis
We previously reported that transcription of multiple UPR effector genes and translation of the major ER chaperone, Bip, are induced in zebrafish livers following alcohol exposure [11]–[13]. We have also demonstrated that xbp1 splicing, a direct measure of Ire1a activation, is only detectable within the first few hours of exposure to ethanol [13] yet UPR target gene induction persists long afterwards [12], suggesting that the Ire1a pathway is not entirely responsible for the UPR in ALD. Our previous finding that Eif2a is phosphorylated in the liver of zebrafish with ALD [13] suggests that Perk is activated by alcohol, but since Eif2a can also be phosphorylated by other kinases [38], this has yet to be resolved. The lack of reliable antibodies that recognize the active form of Atf6, Perk and Ire1a in zebrafish hampered direct measurement of the activation status of each pathway (not shown). However, quantitative real-time PCR (qPCR) analysis of liver samples from 5 day post fertilization (dpf) larvae treated with alcohol demonstrated that atf6 mRNA was significantly induced in the liver (Figure 1A) prior to the onset of steatosis (see Figure 1B and [13]). Since Atf6 has been shown to act in a positive feedback loop to induce its own expression [39], these data suggest that the increase in atf6 expression could reflect Atf6 activation in response to alcohol.
Fig. 1. Atf6 is required for alcoholic steatosis. 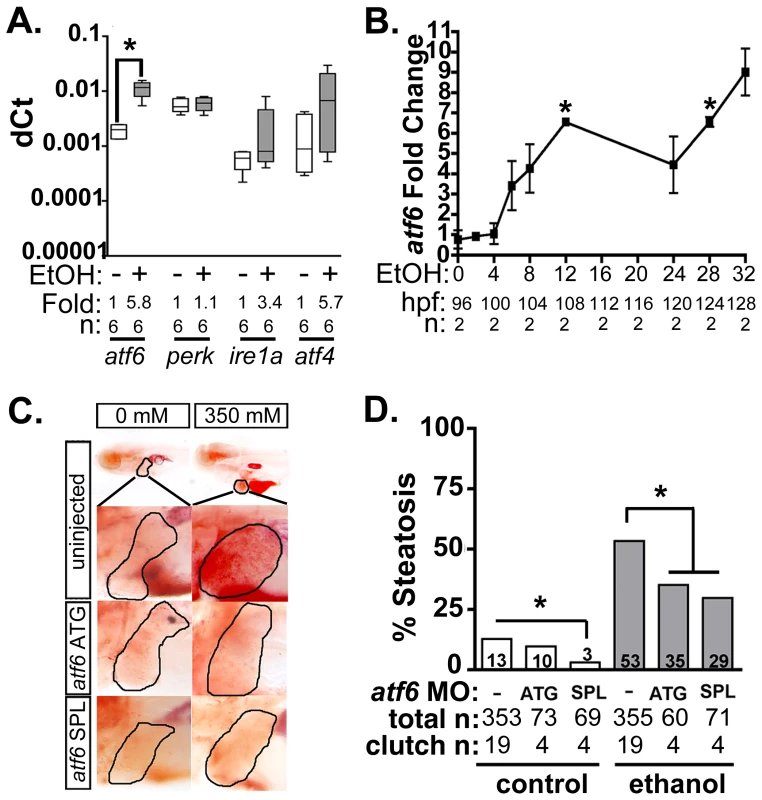
A: qPCR analysis of UPR sensors atf6, perk, ire1a, and atf4 in the livers of control (EtOH-) and 350 mM ethanol-treated (EtOH+) larvae. dCt values were calculated by normalization to rpp0. Fold changes were calculated based on median dCt values. Statistics: paired t-test. *, p<0.05. B: Expression of atf6 in the livers of 350 mM ethanol treated larvae from 0–32 hours. Fold changes were determined by normalizing to control (0 mM EtOH) dCt values. Statistics: one-sample t-test. *, p<0.05. C: Images of oil red O stained whole larvae injected with two different morpholinos targeting atf6 (ATG-targeting, atf6 ATG, and splice-blocking, atf6 SPL) and exposed to either 0 or 350 mM ethanol. Livers are circled in the enlarged boxes. D: Steatosis incidence based on scoring oil red O stained larvae at 5.5 dpf. Statistics: chi-square with Fisher's Exact Test. *, p<0.05. “Total n” and “clutch n” corresponds to the number of larvae and number of clutches scored. To test whether Atf6 was required for alcoholic steatosis, we injected a morpholino (MO) targeting either the translation initiation site (atf6MO-ATG) [23] or the boundary between coding exon 1 and intron 1 of atf6 (atf6MO-SPL) (Table S1 and Figure S1A), which blocked splicing (Figure S1B) and introduced an early stop codon (Figure S1C). Morphants and uninjected controls were treated with 350 mM ethanol for 32 hours, stained with the neutral lipid dye, oil red O and scored for the presence or absence of steatosis (Figure 1C and S2A). Consistent with previous findings [11]–[13], [23], [34], the percent of larvae with steatosis (i.e. steatosis incidence) was significantly higher in uninjected larvae exposed to 350 mM ethanol for 32 hours (13% in untreated larvae vs. 53% in ethanol treated larvae; p<0.05). atf6 morphants had reduced steatosis incidence (p<0.05, Figure 1C-D), but still displayed other gross morphological abnormalities caused by ethanol including increased liver circularity, an indication of hepatomegaly (Figure S2B-C), a common feature of liver disease. Thus, Atf6 is required for alcoholic steatosis.
nAtf6 overexpression in hepatocytes induces robust UPR activation
To determine whether Atf6 is sufficient to cause FLD, we created a transgenic zebrafish line expressing the predicted nuclear, active fragment of zebrafish Atf6 (amino acids 1-366; Figure S3A) fused to mCherry under the hepatocyte-specific promoter fabp10, along with a cassette that expressed GFP in cardiomyocytes as a marker of transgenesis ((Tg(fabp10:nAtf6-cherry; cmlc2:GFP) hereafter called nAtf6 TG). There is high sequence identity between human and zebrafish Atf6 proteins in the transmembrane, leucine zipper and basic (bZIP) domains and the protease recognition sites are highly conserved (Figure S3A). To confirm its nuclear localization, the predicted nAtf6 fragment of the zebrafish protein was fused to GFP and transfected into HepG2 cells (Figure S3B).
Transgenics were selected based on GFP expression in the heart, appeared grossly normal throughout development (Figure S3C), and developed into viable, fertile adults. nAtf6-mCherry was not detectable using low-resolution fluorescence microscopy in larvae from all four of the transgenic lines we generated. However, we confirmed transgene expression by PCR and blotting for mCherry (Figures S3D-E). atf6 mRNA expression in the liver was ∼4 fold higher in nAtf6 TG larvae compared to wildtype (WT) larvae (Figure 2A and S4) at 5 dpf, and was persistently elevated in older fish (Figure S4D and not shown). We detected mCherry mRNA in 5 dpf nAtf6 TG larvae by PCR, and Western blotting for mCherry detected the transgene in the liver of adult fish, albeit at much lower levels than achieved in transgenic fish expressing nuclear-localized mCherry without fusion to nAtf6 (i.e. Tg(fabp10:nls-mCherry); [40] Figures S3D-E).
Fig. 2. nAtf6 overexpression is sufficient to drive steatosis. 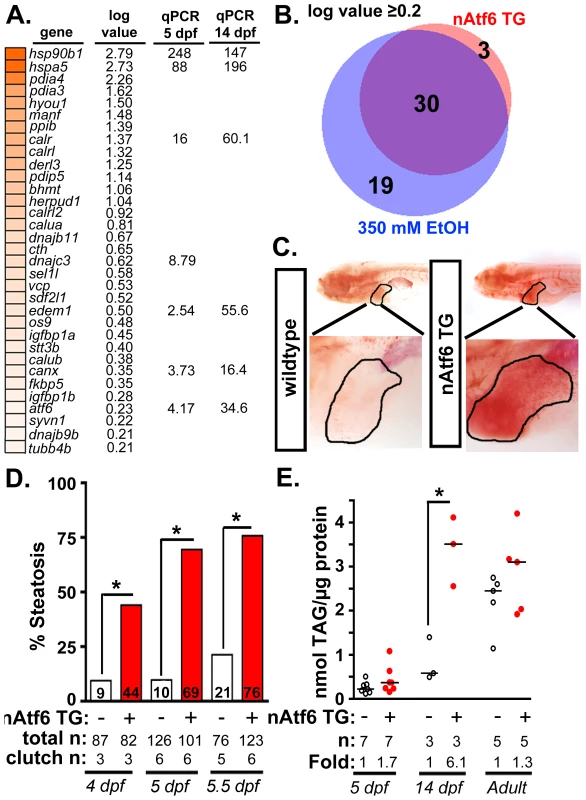
A: Heatmap of upregulated UPR effector genes in nAtf6 transgenic larvae. The log values based on mRNA-Seq analysis and the median fold change in 3–6 liver samples assessed by qPCR at 5 dpf and 14 dpf. B: Most Atf6 targets are upregulated in the liver in response to ethanol. Genes identified as part of the UPR are significantly induced in the liver of nAtf6 TG and ethanol treated larvae (log value ≥ 0.2; see Table S2). C: Oil red O stained WT and nAtf6 transgenic larvae. Livers are circled in the enlarged boxes. D: Steatosis incidence based on scoring of whole mount oil red O stained larvae at 4, 5, and 5.5 dpf. Statistics: chi-square with Fisher's Exact Test. *, p<0.05. “Total n” and “clutch n” corresponds to the number of larvae and number of clutches scored. E: Hepatic triglyceride levels in extracts from pooled livers from WT and nAtf6 transgenic larvae at 5 dpf and 14 dpf, and from single adult livers (∼9 months). Statistics: unpaired t-test. *, p<0.05. Median fold changes are noted. We confirmed that nAtf6 overexpression did not induce the other UPR branches by assessing xbp1 splicing (Figure S4A) and Eif2a phosphorylation (Figure S4B) in the liver of nAtf6 TG 5 dpf larvae. We found no difference in these markers in unstressed nAtf6 TG and WT larvae, however, xbp1 splicing was lower in transgenics than in WTs after exposure to stress (i.e. 1 µg/ml tunicamycin for 48 hours; Figure S4A). This suggests that nAtf6 overexpression adapts hepatocytes to withstand a robust, pharmacologically-induced ER stress.
We next examined the expression of UPR target genes in the liver of 5 dpf transgenic and control larvae. We found that bip (also called hspa5), a transcriptional target of Atf6, was elevated at the protein (Figure S4C) and mRNA levels (Figure 2A and S4D) in nAtf6 TG larvae. mRNA-Seq analysis comparing control larvae (Tg(fabp10:nls-mCherry); n = 2 pools of ∼40–50 livers each; see [40] and GEO dataset GSE52605), ethanol-treated (n = 2 pools of ∼40–50 livers each), and nAtf6 TG larvae (n = 1 pool of ∼40–50 livers) revealed 33 UPR target genes to be highly and significantly upregulated in nAtf6 TG livers (Tables S2-S4 and Figure 2A; GEO dataset GSE56498). These genes were identified as ATF6 target genes in other studies [16], [41] or by having the UPR or protein folding as a primary GO descriptor, and included ER chaperones (bip/hspa5, dnajc3, and grp94/hsp90b1), quality control effectors (canx, calret, calrl, and calrl2), protein disulfide isomerases (pdia3, pdia4) and ERAD components (edem1, derl1 and derl3). A subset of these genes was confirmed by qPCR at 5 dpf and at 14 dpf (Figure 2A and S4D-E). atf4 and derl1 were the only genes detected as significantly upregulated by qPCR but not by mRNA-Seq.
Analysis of the mRNA-Seq data from ethanol-treated larvae revealed a striking overlap between the “UPR-ome” induced by ethanol and by nAtf6 overexpression: of the 49 UPR target genes induced by ethanol, 30 were also induced in the liver of nAtf6 TG larvae (Figure 2B and Table S2). It is possible that the remaining 19 genes unique to the ethanol-induced transcriptome are attributed to the transient, early increase in xbp1 splicing [13] or to other transcription factors that regulate their expression. Collectively, these data indicate that Atf6 is a major regulator of the UPR in ALD.
Atf6 overexpression causes steatosis
We next asked whether nAtf6 was sufficient to cause FLD (Figure 2C-E). Induction of the fabp10:nAtf6-cherry transgene occurs at ∼2.5 dpf [42] and by 4 dpf, 44% of nAtf6 TG larvae developed steatosis which increased to 69% and 76% by 5 and 5.5 dpf, respectively (Figure 2C-D and S5A) and persisted until at least 14 dpf (Figure S5A), but there were no marked histological abnormalities in the nAtf6 TG liver at 5 dpf (Figure S5B). Hepatic triglyceride (TAG) levels were nearly doubled at 5 dpf, and six times higher in 14 dpf nAtf6 TG larvae (Figure 2E), clutch-to-clutch variability non-withstanding. However, TAG levels were the same in WT and nAtf6 TG adults (Figure 2E), despite persistent expression of the transgene (Figure S3E and not shown). We speculate that, over time, animals adapt to nAtf6 overexpression or that other metabolic parameters affect hepatic lipid accumulation in adult fish. Together, our data demonstrate that Atf6 is necessary for alcoholic steatosis and sufficient to cause steatosis in the absence of any other stress.
nAtf6 overexpression alters expression of genes involved in lipogenesis and lipid export
Steatosis is caused by elevated TAGs in hepatocytes resulting from increased lipid synthesis or uptake, or from decreased lipid utilization or export. We therefore analyzed pathways relevant to these processes in the mRNA-Seq dataset from nAtf6 TG livers to determine if any were dysregulated. TAGs are generated from linking free fatty acids (FFAs) to a glycerol backbone, which is generated by conversion of dihydroxyacetone phosphate (DHAP) to glycerol-3-phosphate (G3P) by G3P dehydrogenase (GPD1) (see schematic in Figure 3A). We found genes that participate in TAG synthesis (Figure 3A-B) or export (Figure S6) were dysregulated in nAtf6 TG livers at 5 dpf, albeit not to the same degree as observed for the UPR target genes (Figure 2). DHAP is generated via glycolysis or by glyceroneogenesis, a pathway that has significant overlap with gluconeogenesis (see [43] and schematic in Figure 3A). mRNA-Seq analysis demonstrated that several genes involved in glycolysis were downregulated in nAtf6 TG livers including aldob, glut2/slc2a2, gck, pgk1, and pklr (Figure 3A-B). Genes promoting glyceroneogenesis, particularly pck1 and got1, which generates oxaloacetate for conversion to phosphoenoylpyruvate by pck1, were upregulated, as was gpd1b, which drives glycerol formation. This suggests that nAtf6 overexpression increases factors regulating glyceroneogenesis preferentially over those that promote glycolysis (Figure 3A-B). In addition, ech1 and tecrb, two genes responsible for fatty acid elongation in the mitochondria and ER, respectively, were induced in nAtf6 TG livers by mRNA-Seq (Figure 3A-B). Some of these genes were confirmed by qPCR analysis of nAtf6 TG livers, with the median fold change from qPCR on 3 to 6 clutches of livers from 5 and 14 dpf larvae depicted in Figure S7.
Fig. 3. Glyceroneogenesis and fatty acid elongation pathways are dysregulated by nAtf6 overexpression. 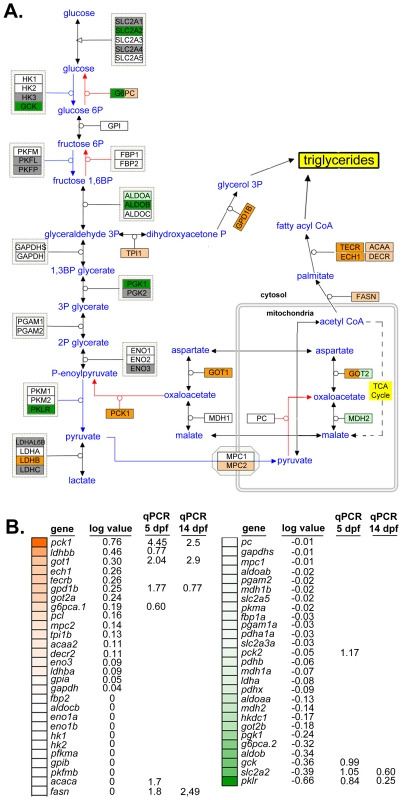
Schematic of the glycolysis, gluconeogenesis and glyceroneogenesis pathway (A, adapted from WikiPathways) and heatmap of associated genes (B). Upregulated genes are colored in shades of orange, downregulated genes are colored in shades of green, genes that are unchanged are white and those genes that did not appear in mRNA-Seq analyses are colored gray. Log values and median fold changes from mRNA-Seq analysis and qPCR, respectively, are noted. TAGs are assembled in the hepatocyte ER and secreted in association with lipoproteins that are required for transport in plasma. Several genes that regulate lipoprotein assembly and export, including apom, apoeb, apoea, cetp and zgc:162608 and zgc:194131, two apolipoprotein-like genes, were downregulated in nAtf6 TG livers (Figure S7 and Table S3). pdia2, which interacts with MTP to transport lipids out of hepatocytes [44], was also downregulated. These data suggest that hepatic lipid transport may be impaired by nAtf6 overexpression. While these expression data do not capture the complex metabolic and post-translational regulation of the proteins encoded by these genes, they do suggest enhanced lipogenesis or suppressed lipoprotein export as potential mechanisms of steatosis in nAtf6 transgenics.
De novo lipogenesis is enhanced by nAtf6 overexpression
Acetate is a precursor to acetyl-CoA, the major building block of fatty acid and, therefore, TAG synthesis. To determine if nAtf6 overexpression increased de novo lipogenesis, we measured the incorporation of 14C-acetate into lipids in nAtf6 TG larvae (Figure 4A) and in HepG2 cells overexpressing nATF6 (Figure 4B-D) compared to their respective controls. Larvae were incubated with 14C-acetate from 3–5 dpf and we measured the amount of radiolabel present in the lipid fraction compared to that present in the lysate of the whole larvae. In an average of 5 individual pools of larvae, more 14C radiolabel was detected in the lipid fraction of nAtf6 TG larvae than in wildtype larvae (Figure 4A).
Fig. 4. De novo lipogenesis is enhanced in vitro and in vivo by nAtf6 overexpression. 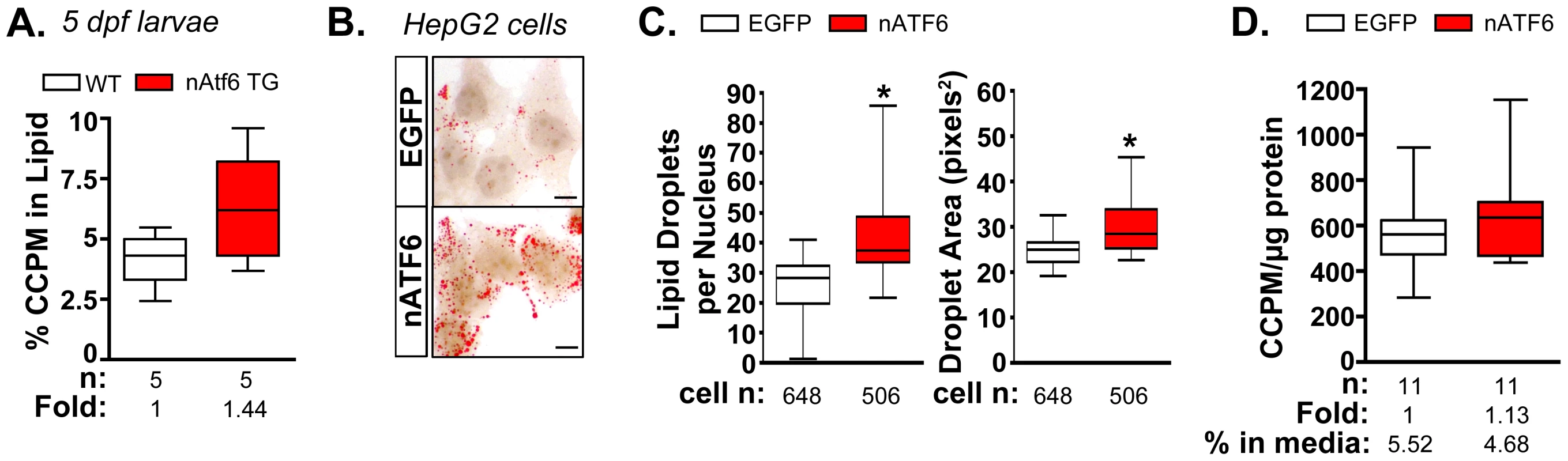
A: 14C-acetate is preferentially incorporated into lipids in 5 dpf nAtf6 TG larvae compared to WT. The percent of 14C in lipid fraction was divided by 14C measured in the unextracted lysate from whole fish. The median fold change is noted; n = number of samples of pooled larvae. B: Oil red O staining of HepG2 cells transfected with GFP or nATF6. Bar = 10 µm. C: Quantification of lipid droplet number and area (square pixels). Statistics: unpaired t-test. *, p<0.05; n indicates the number of cells quantified over 20 fields from 2 independent experiments. D: Incorporation of 14C-acetate into lipids by HepG2 cells transfected with GFP or nATF6 normalized to protein. The median fold change is noted; n = number of separate batches of cells analyzed in 2 independent experiments. Since the liver in 5 dpf zebrafish larvae is too small to analyze incorporation in this organ exclusively, analysis of the whole larvae could not determine whether the label was preferentially incorporated into hepatocytes of nAtf6 TG larvae. We thus used human hepatoma (HepG2) cells transfected with human nATF6 to address this question. nATF6 overexpression causes accumulation of oil red O stained cytoplasmic droplets (Figure 4B) that are both greater in number and larger in size than in cells transfected with GFP (Figure 4B). These findings were confirmed in 293T cells (Figure S8) and correlate with our findings in nAtf6 TG larvae (Figure 2). Thus, nAtf6 overexpression causes lipid accumulation across cell types and species. Moreover, in eleven independent cell samples from two separate experiments, HepG2 cells overexpressing nATF6 had slightly more 14C radiolabel in the lipid fraction compared to GFP-transfected cells (Figure 4D). While we found that the incorporation tended to be higher in the nATF6 expressing cells, the average increase was moderate, at best. We attribute this to the transfection efficiency of HepG2 cells combined with experimental conditions used to label lipids, which likely act to select against nATF6 expressing cells.
Srebp activation does not contribute to Atf6-mediated steatosis
Fatty acid synthase (fasn) mediates multiple steps in fatty acid synthesis that culminate in generation of palmitate from acetyl-CoA, a building block for TAG biosynthesis. fasn transcription in hepatocytes is primarily regulated by Srebp1c [45]–[47], and we previously demonstrated that fasn transcripts and other Srebp target genes are induced in zebrafish with ALD [12], [13]. Moreover, blocking Srebp activation by injecting a morpholino targeting the Srebp activating protein, Scap, reduced the incidence of alcoholic steatosis [12], [13]. Since Atf6 and Srebps are activated by similar mechanisms – they are both retained as inactive precursors in the ER and processed by the same Golgi-resident enzymes [33] – Srebp mediated lipogenesis has been proposed as a mechanism by which UPR activation causes FLD [27], [32]. We tested this using a genetic approach. First, we asked whether blocking Atf6 affected the induction of lipogenic gene expression in the liver of ethanol treated larvae. Both fasn (Figure 5A), and acc1 (acaca; Figure S9A) were upregulated by ∼2 fold in the liver of ethanol treated larvae, and this was blocked by atf6 MO injection (Figures 5A and S9A). However, induction of other Srebp target genes was not blocked by Atf6 depletion, including srebp1 (Figure S9A) and the Srebp2 targets hmgcra, hmgcs1 and mvk (Figure S9B). Since the entire Srebp target gene set was not uniformly affected by atf6 loss, we conclude that Atf6 does not interact with Srebps. Instead, Atf6 may affect expression of specific lipogenic genes (i.e. fasn and acc1) by a mechanism distinct from Srebp1. Interestingly, we found mild induction of Srebp2 target genes in untreated atf6 morphants (Figure S9B), supporting conclusions based on experiments in mammalian cultured cells where ATF6 was found to directly suppress SREBP2 activity [48].
Fig. 5. Fasn functions downstream of Atf6 to cause steatosis independent of Srebp activation. 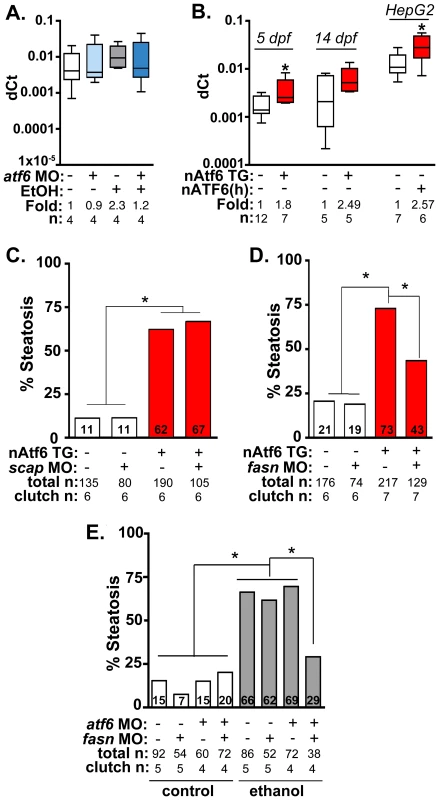
A: qPCR analysis of fasn expression in atf6 morphants and uninjected larvae treated with EtOH for 32 hours. B: qPCR analysis of fasn expression in nAtf6 TG larvae and HepG2 cells transfected with nATF6. Statistics: unpaired t-test. *, p<0.05. C: Quantification of whole mount oil red O staining of nAtf6 TG larvae injected with scap MO. “Total n” and “clutch n” corresponds to the number of larvae and number of clutches scored, respectively. Statistics: chi-square with Fisher's Exact Test. *, p<0.05. D: Quantification of oil red O staining in nAtf6 TG larvae injected with fasn MO. E: Quantification of oil red O staining in atf6/fasn double morphants treated with 350 mM EtOH for 24 hours. Statistics: chi-square with Fisher's Exact Test. *, p<0.05. Next, we asked whether nAtf6 was sufficient to induce fasn and acc1 by assessing their expression in livers of nAtf6 TG larvae. Both fasn (p<0.05; Figure 5B) and acc1 (Figure S9C) were increased by ∼2 fold at 5 dpf, and fasn expression was also higher at 14 dpf in nAtf6 TG larvae and in HepG2 cells overexpressing nATF6 (Figure 5B). However, other Srebp1 and Srebp2 genes were not significantly induced by nAtf6 overexpression in nAtf6 TG larvae (Figure S9C-D) or in HepG2 cells overexpressing human nATF6 (not shown), indicating that nAtf6 does not activate the entire Srebp target gene program. We then tested whether nAtf6 functioned upstream of Srebps by assessing whether scap morpholino injection suppressed steatosis in nAtf6 TG larvae. While scap morphants are resistant to ethanol-induced steatosis [12] we found no difference in steatosis incidence in nAtf6 transgenics (Figure 5B). Finally, to test whether atf6 and scap interacted to cause alcoholic steatosis we injected low concentrations of morpholinos targeting both genes and found that they did not synergize to suppress alcoholic steatosis (Figure S10). We thus conclude that lipogenic gene induction and steatosis caused by nAtf6 overexpression does not require Srebps.
Steatosis in nAtf6 TG larvae requires fasn
The functional relevance of the finding that nAtf6 overexpression increases lipogenesis was tested by assessing the effect of morpholino-mediated fasn (Table S1) knockdown on steatosis incidence in nAtf6 TG larvae. Injection of high concentrations of fasn MO induced severe morphological defects, so we optimized the fasn MO concentration to have minimal toxicity. This had no effect on steatosis incidence in control larvae, but reduced steatosis incidence in nAtf6 TG larvae from 74% to 42% (Figure 5C). Furthermore, we found that atf6 and fasn interacted epistatically to cause alcoholic steatosis by co-injecting concentrations of fasn and atf6-SPL MOs that did not have any effect on alcoholic steatosis on their own but, together, significantly reduced steatosis incidence in response to ethanol (Figure 5D). Thus, Atf6 and Fasn function in the same genetic pathway. Taken together, our data suggest a model by which Atf6 causes steatosis, in part, by inducing fasn and TAG synthesis by a mechanism that is independent of Srebps (Figure 6).
Fig. 6. Working model by which Atf6 functions as a positive regulator of alcoholic steatosis. 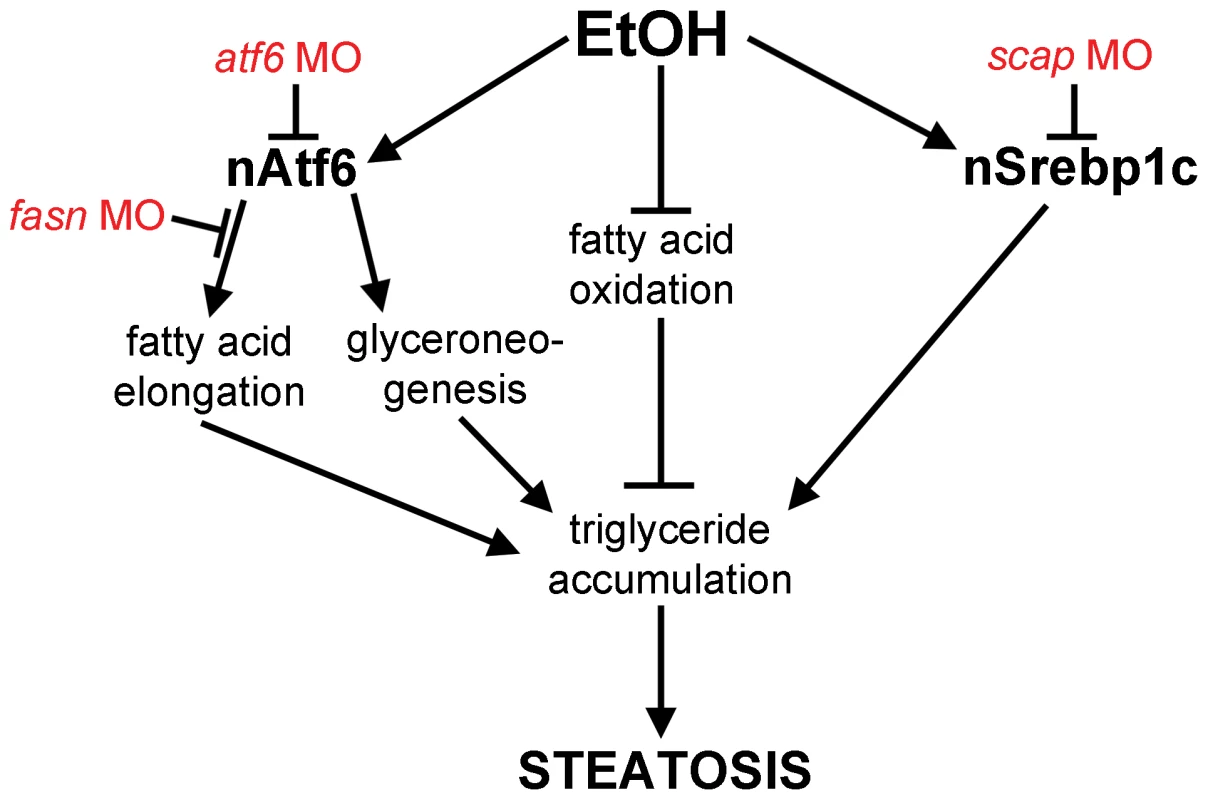
Genetic tools used in this study are illustrated in red. Discussion
There is a wealth of recent literature showing that the UPR is activated in FLD and that this aspect of the disease is conserved across species [4], [16], [21], [23], [49]–[51]. However, it has not been shown that any single UPR factor can directly cause this disease and the mechanism by which UPR activation causes lipid accumulation is unclear. Here, we use zebrafish genetics and biochemical analyses to show that Atf6 is both necessary and sufficient for steatosis by showing that: (i) Atf6 is required for alcoholic steatosis, (ii) activation of Atf6 is sufficient to cause steatosis, (iii) Atf6 activation induces expression of genes involved in glyceroneogenesis and fatty acid elongation and causes de novo lipogenesis, and (iv) Atf6 epistatically interacts with fatty acid synthase (fasn), a key enzyme involved in TAG biosynthesis, to cause FLD. This is among the first data to directly link a main UPR sensor and lipid metabolic pathways.
A number of studies have demonstrated that loss of one of the key UPR sensors – PERK [19], IRE1α [20] or ATF6 [16], [21], [23] – enhances steatosis caused by acute stress. Additionally, we previously demonstrated that Atf6 depletion suppresses steatosis caused by chronic stress [23]. Thus, it is clear that UPR activation is a conserved, common feature of FLD, but whether it functions to promote or prevent steatosis appears to depend on the nature and duration of the FLD-causing stress. Importantly, these loss of function studies have described a requirement for the UPR in FLD caused by tunicamycin injection, but not in the context of a stress that mirrors human conditions, such as obesity or alcohol abuse. Here, we show that Atf6 is induced in ALD, that its activation precedes steatosis, and that knocking down Atf6 reduces alcoholic steatosis. Thus, Atf6 is required for ALD. Moreover, since overexpression of nAtf6 is sufficient to cause FLD in the absence of any other stress, we conclude that this branch of the UPR is a pathophysiological mechanism of FLD.
Atf6 and Xbp1 are the primary transcription factors that both independently and cooperatively regulate expression of hundreds of UPR target genes. We previously found significant upregulation of many UPR target genes in zebrafish with ALD [11], [13] and hypothesized that since xbp1 splicing in the liver was only an early, transient response to alcohol [13], other transcription factors must participate in this robust UPR. Transcriptome analysis of the livers from nAtf6 TG and ethanol treated larvae enabled us to identify a set of target genes that are likely to be directly activated by Atf6. Many of these genes were also reported to be upregulated in mice overexpressing inducible ATF6 in the heart [41] and they were not induced by tunicamycin treatment of Atf6-/- MEFs [16]. Genes occupying the intersection of these different datasets are likely bona fide Atf6 targets and the transgenic larvae we generated provides a system to test this directly. In contrast, many genes that were downregulated when ATF6 was overexpressed in the mouse heart were unchanged in nAtf6 transgenic zebrafish livers and, conversely, genes such as canx, derl1, and atf4 were upregulated in the nAtf6 TG zebrafish liver samples but not in the mouse model. The variations between the two datasets may be due to inherent differences in the models or approaches used to detect changes in gene expression, or may reflect the ability of Atf6 to differentially regulate target genes depending on cell type: hepatocytes possess a significantly higher secretory capacity than cardiomyocytes, and thus the Atf6 transcriptome in hepatocytes may be more extensive in order to maintain ER homeostasis. Finally, comparative transcriptome analysis between nAtf6 TG and ethanol treated larvae identified significant overlap between the UPR target genes in these two datasets, indicating that Atf6 is the main transcriptional driver of the ethanol-induced “UPR-ome”.
What is the mechanism by which the UPR causes FLD? Our data showing that nAtf6 overexpression induces the expression of some lipogenic enzymes and increases lipid synthesis, and that blocking fatty acid synthesis reduces FLD, fits a model (Figure 6) whereby increased lipid production is, in part, the mechanism of steatosis in FLD caused by UPR activation. However, despite our finding that Atf6 depletion suppresses fasn expression and nAtf6 overexpression induces fasn, we have no evidence that fasn is a direct transcriptional target of Atf6. Indeed, the level of fasn induction is far less than the established Atf6 target, bip, and there are no canonical UPREs or ERSEs [15] in the fasn promoter (not shown), thus other pathways are also likely at play. The mechanism by which nAtf6 induces fasn and acc1 require further investigation, and the tools we describe here will facilitate such studies.
The SREBP transcription factors are well-characterized regulators of hepatic lipogenesis; SREBP1c functions by increasing the expression of the full panel of genes required for TAG biogenesis [52]. One possibility, suggested by the finding that the UPR and SREBPs are activated in parallel in some systems [26], [27], [32], is that nAtf6 causes Srebp activation, leading to increased expression of fasn and other lipogenic genes and promoting lipogenesis. Our data argues against this: (i) the full panel of Srebp targets are not induced by nAtf6 overexpression in zebrafish, (ii) Atf6 and Srebps did not epistatically interact to modify alcoholic steatosis and (iii) Srebp activation is not required for steatosis in nAtf6 TG larvae. While it does appear that Srebp activation is required for alcoholic steatosis [12], [28], [29], the current study shows that it is not the only important pathway that regulates hepatic lipid metabolism, as steatosis in nAtf6 TG larvae is independent of Srebps.
While Srebps are not required for the effects of Atf6 on steatosis, there does appear to be some interaction between Atf6 and Srebp2, consistent with in vitro data showing that ATF6 suppresses SREBP2 [48]. We found that Atf6 causes upregulation of Srebp2 target genes, and while this modest increase in Srebp2 target genes in atf6 morphants did not appear to have a functional impact on cholesterol biogenesis, since atf6 morphants do not develop steatosis but are protected from it, it does suggest that Atf6 may suppress Srebp2 activity.
We used mRNA-Seq analyses to identify potential pathways that are dysregulated in response to Atf6 overexpression, although it is clear that the complex post-translational regulation of these pathways are not captured by gene expression studies. Our data suggested that lipoprotein export may be impaired by Atf6 overexpression, but we did not find that the flux of 14C-acetate into the extracellular lipids was impaired in cells overexpressing nATF6. While there are some technical caveats to these biochemical studies, we speculate that decreased lipoprotein export is not the only mechanism by which nAtf6 overexpression causes steatosis. Future studies to optimize cell conditions to ensure maximal cell viability and sustained high levels of nAtf6 expression will be required to fully address this.
While steatosis is clearly a first step on the continuum to more severe liver disease, we also propose a different possibility: that lipid accumulation is a sign of an active and adaptive stress response. Protein folding in the ER is metabolically demanding, and when secretory cargo increases or the ER becomes full of unfolded proteins, increased lipid accumulation may enhance the ATP supply and thereby sustain the increased protein folding demands. Thus, the UPR may serve to enhance the protein folding capacity of the ER function in two ways: by activating genes required for protein folding, quality and export, and by promoting metabolic flux to TAGs as an energy source to withstand the challenge presented by a high level of secretory cargo. In this scenario, acute steatosis would not be a pathological response but, instead, would provide a protective role to sustain hepatocyte function during stress. However, if the stress response persists and steatosis becomes chronic, lipotoxicity could potentiate liver injury. Based on these data, it is tempting to speculate that enhancing protein folding in the ER by chemical chaperones, which have been used in both mouse models and humans to alleviate UPR activity, attenuate fatty liver disease and increase insulin sensitivity [3], [53], [54], could reducr the demand for energy and thus reduce the need to accumulate lipid. Whether Atf6 functions both as sensor of unfolded proteins and of metabolic demand remains to be elucidated.
Materials and Methods
Animal husbandry, treatments and transgenics
Adult wildtype (WT, TAB14 and AB), Tg(fabp10:dsRed) [55] and Tg(fabp10:nls-mCherry) [40] zebrafish were maintained according to standard conditions. Larvae were exposed to 350 mM ethanol (Pharmco-AAPER, Brookfield, CT) in fish water starting at 96–98 hours post fertilization (hpf) for up to 32 hours as described [11], [12]. All zebrafish protocols were approved by Mount Sinai's Institutional Animal Care and Use Committee.
Morpholinos targeting the translation initiation ATG of atf6 (atf6MO-ATG) [23], an atf6 intron-exon boundary (atf6MO-SPL, Figure S1), a scap intron-exon boundary [12], and a fasn intron-exon boundary were ordered from GeneTools (Philomath, OR). Approximately 1–5 pmol were injected into 1–2 cell stage embryos. Morpholino sequences and amount injected per embryo (ng) are listed in Table S1.
The Tg(fabp10:nAtf6-cherry; cmlc2:GFP) transgenic line was created by injecting a vector containing 2813 bp of the fabp10 promoter [55] upstream of the predicted nuclear fragment of zebrafish Atf6 (amino acids 1–366) as identified via DNA and protein alignments with human ATF6 (NCBI Reference Sequence: NP_031374.2). A cassette driving GFP expression in cardiomyocytes was included for rapid screening of transgenics (cmlc2:GFP). The transgene was flanked by Tol2 sites and the vector was injected into fertilized eggs along with transposase mRNA. Larvae were selected for cmlc2:GFP expression and raised to adulthood, outcrossed to TAB14 adults and 4 germline founders were identified.
Plasmids
The nATF6-pcDNA-DEST47 plasmid was created using the Invitrogen Gateway System in which human nATF6 (amino acids 1–380) was amplified from a construct containing the full ORF (pEGFP-hATF6, from Dr. Aguirre-Ghiso) and ligated into pcDNA-DEST47. The nAtf6-GFP/pCI-Neo plasmid was generated by amplifying zebrafish nAtf6-GFP from a Tol2-generated plasmid (fabp10:nAtf6-GFP) with primers containing restriction sites for EcoRI and SmaI. The resulting PCR product was ligated into pCI-Neo via T4 ligase. pEGFP-C1 (Clontech) was used as a positive control for transfection.
Cell culture and transfection
293T and HepG2 cells were grown in 100 mm dishes (BD Biosciences) with DMEM (Corning Cellgro, Manassas, VA) containing 10% fetal bovine serum (Invitrogen) and penicillin/streptomycin (Cellgro), and housed in a humidified incubator at 37 C with 5% CO2. Cells were passaged at 80–90% confluency either into 100 mm dishes or 6 well culture plates (Corning). Each well was transfected with 1–2 µg plasmid using Xtreme GENE 9 (Roche) or Lipofectamine 2000 (Invitrogen) for 24 hours. Following transfection, cells were collected for RNA extraction using TRIzol (Invitrogen), used for oil red O staining, or labeled with 14C-acetate as described below.
14C-acetate radiolabeling
HepG2 cells were washed with PBS 24–28 hours after transfection and kept in serum free high glucose DMEM (Corning Cellgro, Manassas, VA) containing penicillin/streptomycin (Cellgro), 100 nM insulin (Lilly USA, Indianapolis, IN) and 0.5 µCi 14C-acetate (PerkinElmer, Waltham, MA) for 17 hours. Media was harvested and spun to remove dead cells, while cells were scraped, washed twice and lysed in cold PBS. Protein concentration in the cell lysate was measured with a BCA protein quantification kit (Thermo Scientific, Waltham, MA). Fractions of the media and cell lysate portion were used for lipid extraction as described below.
WT and nATF6 5 dpf larvae were labeled with 1–5 µCi 14C-acetate for 48 hours. Larvae were washed twice and then sonicated in cold PBS. Protein concentration in the larval lysate was measured with a BCA protein quantification kit (Thermo Scientific). Fractions of the larval lysate were used for lipid extraction as described below.
Lipid extractions
Lipids from media, cells and 5 dpf larvae were extracted according to Bligh and Dyer [56] with the following modifications. Briefly, methanol and chloroform were added to 400 µl media or 50 µl of 100 µl total cell lysate in a ratio of 2∶1. Samples were vortexed and chloroform and 0.45% NaCl were added at a ratio of 1∶1, vortexed again and phases were separated by centrifugation. The aqueous phase was re-extracted with one part chloroform and combined with the lipid from the first second extraction, washed with methanol and 0.45% NaCl at a 1∶1 ratio, centrifuged and the lipid phase was recovered and dried under a stream of N2 gas. Lipids were reconstituted in a 2∶1 mixture of chloroform and methanol (Thermo Scientific, Waltham, MA), dissolved in Ultima Gold and counted using a 2460 MicroBeta2 LumiJET (PerkinElmer, Waltham, MA) liquid scintillation counter. CCPM were normalized to protein concentration to account for variations in cell number.
Quantitative real-time PCR (qPCR)
Total RNA isolated using TRIzol (Invitrogen) from pools of 15–20 dissected livers was reverse-transcribed using qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD). qPCR using A Light Cycler 480 (Roche) with PerfeCTa SYBRGreen FastMix (Quanta Biosciences) was used as previously described [11]. Values for target gene were normalized to reference gene rpp0, and dCts calculated using the comparative threshold method (dCt = 2−(Ct, gene – Ct, rpp0)). Primer sequences are listed in Table S1.
Triglyceride measurements
Approximately 20 livers from control and nAtf6 transgenic larvae were dissected and lysed in 0.5% Triton-X 100 and heated at 65°C for 5 minutes to inactivate hepatic lipases. Triglycerides were measured using the Infinity Triglyceride Liquid Stable Reagent (Thermo Fisher Scientific, Waltham, MA) following manufacturer's instructions, and were normalized to total protein concentration as determined by Bradford Assay (Bio-Rad, Hercules, CA).
Oil red O staining and quantification
Larvae were fixed in 4% paraformaldehyde (PFA; Electron Microscopy Sciences, Hatfield, PA) overnight at 4°C, stained with oil red O, and scored for steatosis as previously described [35]. Cryosections were stained by immersing slides in increasing concentrations of propylene glycol (85%, 100%) for 10 minutes followed by an overnight incubation in oil red O (0.5% in propylene glycol, Polysciences, Warrington, PA). Excess oil red O was removed the next day by sequential washes in 100% and 85% propylene glycol for 5 minutes. Nuclei were counterstained with hematoxylin.
HepG2 and 293T cells were stained as previously described [57] and counterstained with hematoxylin. Oil red O droplet area and number were calculated using ImageJ. In brief, images were split into the red, green, and blue channels and the green channel was further processed for quantification as described, as oil red O has an excitation at 510 nm [58]. Following this, the background was subtracted from each image to ensure only counting of oil red O droplets. Droplets equal or greater than five square pixels were counted and area quantified. The values were copied into Microsoft Excel and the average droplet area and number of droplets per nucleus were counted for each field. Twenty independent fields from nATF6 and GFP-transfected HepG2 and 293T cells were imaged at 60x magnification.
RNA sequencing (mRNA-Seq)
Total RNA was isolated using TRIzol (Invitrogen) from pools of ∼40–50 livers dissected on 5 dpf. Two clutches of Tg(fabp10:nls-mCherry) larvae that were either untreated (control) or treated with 350 mM ethanol for 24 hours and one clutch of nAtf6 TG larvae were used. polyA-tailed mRNA was selected using oligo-dT beads and then fragmented. cDNAs were created using random-hexamers and ligated with bar-coded adaptors compatible with HiSeq 2000. Single-end, 100 bp reads were sequenced at the Genomics Core of the Icahn School of Medicine at Mount Sinai. Custom-built software was used to map the reads to the zebrafish genome (Zv9/DanRer7) and estimate the coverage of each gene. Briefly, the reads were split into three 32 bp parts after trimming 2 bp at each end and mapped to the genome using a suffix-array based approach. The median of coverage across the transcript was used as an estimate of gene expression. The expression values were quantile normalized and log-ratios were calculated by comparing nAtf6 TGs to the average of the controls. Each ethanol treated sample was compared to its paired control (untreated siblings). Unique Gene Ontology terms (GO terms) were assigned to each gene by ranking the GO terms by relevance to the biology of the response, and using annotations from the human orthologues if the zebrafish annotations were lacking.
The distribution of expression values were plotted to identify the peak in the distribution, which is the level of noise in the system. The values were regularized by adding the noise to each gene's expression level before the log-ratios were calculated. This ensures that genes with low expression do not contribute to list of genes with large fold-changes. An absolute natural log ratio of 0.2 was used as the cutoff (known, non-responding genes are all below this threshold). The list of genes that show changes above this cutoff were analyzed for pathway enrichment using GO terms annotated as described above.
This data is available via the Gene Expression Omnibus (GEO) at accession number GSE56498. The controls for this dataset, untreated 5 dpf Tg(fabp10:nls-mCherry), also serve as controls in GEO dataset GSE52605 [40] and were included in both sets for ease of comparison between genotypes and treatments.
Image processing and graphics
Images were cropped and minimally processed using Adobe Photoshop CS4 (Adobe Systems, San Jose, CA). Graphs were plotted using Prism 5.0c (GraphPad Software Inc., La Jolla, CA). Heat maps for mRNA-Seq data were generated using GENE-E (Broad Institute, Cambridge, MA). The global maximum and minimum for each gene set was set to orange and green, respectively, and zero was set to white. Metabolic pathway schematics were adapted from WikiPathways (URL: www.wikipathways.org).
Statistical analyses
Statistical tests were performed using GraphPad QuickCalcs (GraphPad Software, URL: http://www.graphpad.com/quickcalcs/index.cfm). For oil red O staining of whole larvae, chi-square with Fisher's Exact test were performed. For qPCR, radiolabeling, and oil red O staining of HepG2 and 293T cells we performed unpaired and one-sample t-tests as appropriate.
Supplementary methods
Methods for histological analysis, cryosectioning and staining, and liver circularity analysis are listed in Text S1.
Supporting Information
Zdroje
1. JiC, KaplowitzN (2003) Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology 124 : 1488–1499.
2. LeeJS, ZhengZ, MendezR, HaSW, XieY, et al. (2012) Pharmacologic ER stress induces non-alcoholic steatohepatitis in an animal model. Toxicol Lett 211 : 29–38.
3. OzcanU, YilmazE, OzcanL, FuruhashiM, VaillancourtE, et al. (2006) Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 313 : 1137–1140.
4. PuriP, MirshahiF, CheungO, NatarajanR, MaherJW, et al. (2008) Activation and dysregulation of the unfolded protein response in nonalcoholic fatty liver disease. Gastroenterology 134 : 568–576.
5. RinellaME, SiddiquiMS, GardikiotesK, GottsteinJ, EliasM, et al. (2011) Dysregulation of the unfolded protein response in db/db mice with diet-induced steatohepatitis. Hepatology 54 : 1600–1609.
6. Bondini S, Kleiner DE, Goodman ZD, Gramlich T, Younossi ZM (2007) Pathologic assessment of non-alcoholic fatty liver disease. Clin Liver Dis 11 : 17–23, vii.
7. O'Shea RS, Dasarathy S, McCullough AJ (2010) Alcoholic liver disease. Am J Gastroenterol 105 : 14–32; quiz 33.
8. JiC, Mehrian-ShaiR, ChanC, HsuYH, KaplowitzN (2005) Role of CHOP in hepatic apoptosis in the murine model of intragastric ethanol feeding. Alcohol Clin Exp Res 29 : 1496–1503.
9. ShinoharaM, JiC, KaplowitzN (2010) Differences in betaine-homocysteine methyltransferase expression, endoplasmic reticulum stress response, and liver injury between alcohol-fed mice and rats. Hepatology 51 : 796–805.
10. EsfandiariF, VillanuevaJA, WongDH, FrenchSW, HalstedCH (2005) Chronic ethanol feeding and folate deficiency activate hepatic endoplasmic reticulum stress pathway in micropigs. Am J Physiol Gastrointest Liver Physiol 289: G54–63.
11. HowarthDL, VacaruAM, TsedensodnomO, MormoneE, NietoN, et al. (2012) Alcohol disrupts endoplasmic reticulum function and protein secretion in hepatocytes. Alcohol Clin Exp Res 36 : 14–23.
12. PasseriMJ, CinarogluA, GaoC, SadlerKC (2009) Hepatic steatosis in response to acute alcohol exposure in zebrafish requires sterol regulatory element binding protein activation. Hepatology 49 : 443–452.
13. Tsedensodnom O, Vacaru AM, Howarth DL, Yin C, Sadler KC (2013) Ethanol metabolism and oxidative stress are required for unfolded protein response activation and steatosis in zebrafish with alcoholic liver disease. Dis Model Mech.
14. MagneL, BlancE, LegrandB, LucasD, BaroukiR, et al. (2011) ATF4 and the integrated stress response are induced by ethanol and cytochrome P450 2E1 in human hepatocytes. J Hepatol 54 : 729–737.
15. YamamotoK, YoshidaH, KokameK, KaufmanRJ, MoriK (2004) Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem 136 : 343–350.
16. WuJ, RutkowskiDT, DuboisM, SwathirajanJ, SaundersT, et al. (2007) ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell 13 : 351–364.
17. HardingHP, ZhangY, BertolottiA, ZengH, RonD (2000) Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 5 : 897–904.
18. HardingHP, NovoaI, ZhangY, ZengH, WekR, et al. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6 : 1099–1108.
19. TeskeBF, WekSA, BunpoP, CundiffJK, McClintickJN, et al. (2011) The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol Biol Cell 22 : 4390–4405.
20. ZhangK, WangS, MalhotraJ, HasslerJR, BackSH, et al. (2011) The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J 30 : 1357–1375.
21. RutkowskiDT, WuJ, BackSH, CallaghanMU, FerrisSP, et al. (2008) UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell 15 : 829–840.
22. YamamotoK, TakaharaK, OyadomariS, OkadaT, SatoT, et al. (2010) Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell 21 : 2975–2986.
23. CinarogluA, GaoC, ImrieD, SadlerKC (2011) Activating transcription factor 6 plays protective and pathological roles in steatosis due to endoplasmic reticulum stress in zebrafish. Hepatology 54 : 495–508.
24. WeiY, WangD, TopczewskiF, PagliassottiMJ (2006) Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am J Physiol Endocrinol Metab 291: E275–281.
25. WangD, WeiY, PagliassottiMJ (2006) Saturated fatty acids promote endoplasmic reticulum stress and liver injury in rats with hepatic steatosis. Endocrinology 147 : 943–951.
26. ColganSM, TangD, WerstuckGH, AustinRC (2007) Endoplasmic reticulum stress causes the activation of sterol regulatory element binding protein-2. Int J Biochem Cell Biol 39 : 1843–1851.
27. KammounHL, ChabanonH, HainaultI, LuquetS, MagnanC, et al. (2009) GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest 119 : 1201–1215.
28. YouM, CrabbDW (2004) Molecular mechanisms of alcoholic fatty liver: role of sterol regulatory element-binding proteins. Alcohol 34 : 39–43.
29. YouM, FischerM, DeegMA, CrabbDW (2002) Ethanol induces fatty acid synthesis pathways by activation of sterol regulatory element-binding protein (SREBP). J Biol Chem 277 : 29342–29347.
30. HiguchiN, KatoM, ShundoY, TajiriH, TanakaM, et al. (2008) Liver X receptor in cooperation with SREBP-1c is a major lipid synthesis regulator in nonalcoholic fatty liver disease. Hepatol Res 38 : 1122–1129.
31. KohjimaM, HiguchiN, KatoM, KotohK, YoshimotoT, et al. (2008) SREBP-1c, regulated by the insulin and AMPK signaling pathways, plays a role in nonalcoholic fatty liver disease. Int J Mol Med 21 : 507–511.
32. ZhangC, ChenX, ZhuRM, ZhangY, YuT, et al. (2012) Endoplasmic reticulum stress is involved in hepatic SREBP-1c activation and lipid accumulation in fructose-fed mice. Toxicol Lett 212 : 229–240.
33. YeJ, RawsonRB, KomuroR, ChenX, DaveUP, et al. (2000) ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell 6 : 1355–1364.
34. NorthTE, BabuIR, VedderLM, LordAM, WishnokJS, et al. (2010) PGE2-regulated wnt signaling and N-acetylcysteine are synergistically hepatoprotective in zebrafish acetaminophen injury. Proc Natl Acad Sci U S A 107 : 17315–17320.
35. HowarthDL, YinC, YehK, SadlerKC (2013) Defining hepatic dysfunction parameters in two models of fatty liver disease in zebrafish larvae. Zebrafish 10 : 199–210.
36. YinC, EvasonKJ, MaherJJ, StainierDY (2012) The basic helix-loop-helix transcription factor, heart and neural crest derivatives expressed transcript 2, marks hepatic stellate cells in zebrafish: analysis of stellate cell entry into the developing liver. Hepatology 56 : 1958–1970.
37. LiuLY, FoxCS, NorthTE, GoesslingW (2013) Functional validation of GWAS gene candidates for abnormal liver function during zebrafish liver development. Dis Model Mech 6 : 1271–1278.
38. DeverTE, FengL, WekRC, CiganAM, DonahueTF, et al. (1992) Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68 : 585–596.
39. NambaT, IshiharaT, TanakaK, HoshinoT, MizushimaT (2007) Transcriptional activation of ATF6 by endoplasmic reticulum stressors. Biochem Biophys Res Commun 355 : 543–548.
40. Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, et al.. (2014) UHRF1 Overexpression Drives DNA Hypomethylation and Hepatocellular Carcinoma. Cancer Cell.
41. BelmontPJ, TadimallaA, ChenWJ, MartindaleJJ, ThueraufDJ, et al. (2008) Coordination of growth and endoplasmic reticulum stress signaling by regulator of calcineurin 1 (RCAN1), a novel ATF6-inducible gene. J Biol Chem 283 : 14012–14021.
42. HerGM, ChiangCC, ChenWY, WuJL (2003) In vivo studies of liver-type fatty acid binding protein (L-FABP) gene expression in liver of transgenic zebrafish (Danio rerio). FEBS Lett 538 : 125–133.
43. NyeCK, HansonRW, KalhanSC (2008) Glyceroneogenesis is the dominant pathway for triglyceride glycerol synthesis in vivo in the rat. J Biol Chem 283 : 27565–27574.
44. WetterauJR, LinMC, JamilH (1997) Microsomal triglyceride transfer protein. Biochim Biophys Acta 1345 : 136–150.
45. ShimanoH, ShimomuraI, HammerRE, HerzJ, GoldsteinJL, et al. (1997) Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest 100 : 2115–2124.
46. HortonJD, ShimomuraI, BrownMS, HammerRE, GoldsteinJL, et al. (1998) Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest 101 : 2331–2339.
47. ShimanoH, HortonJD, HammerRE, ShimomuraI, BrownMS, et al. (1996) Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest 98 : 1575–1584.
48. ZengL, LuM, MoriK, LuoS, LeeAS, et al. (2004) ATF6 modulates SREBP2-mediated lipogenesis. EMBO J 23 : 950–958.
49. KaplowitzN, JiC (2006) Unfolding new mechanisms of alcoholic liver disease in the endoplasmic reticulum. J Gastroenterol Hepatol 21 Suppl 3S7–9.
50. Ben MosbahI, Alfany-FernandezI, MartelC, ZaoualiMA, Bintanel-MorcilloM, et al. (2010) Endoplasmic reticulum stress inhibition protects steatotic and non-steatotic livers in partial hepatectomy under ischemia-reperfusion. Cell Death Dis 1: e52.
51. YeR, JungDY, JunJY, LiJ, LuoS, et al. (2010) Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes 59 : 6–16.
52. HortonJD, ShahNA, WarringtonJA, AndersonNN, ParkSW, et al. (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A 100 : 12027–12032.
53. KarsM, YangL, GregorMF, MohammedBS, PietkaTA, et al. (2010) Tauroursodeoxycholic Acid may improve liver and muscle but not adipose tissue insulin sensitivity in obese men and women. Diabetes 59 : 1899–1905.
54. XiaoC, GiaccaA, LewisGF (2011) Sodium phenylbutyrate, a drug with known capacity to reduce endoplasmic reticulum stress, partially alleviates lipid-induced insulin resistance and beta-cell dysfunction in humans. Diabetes 60 : 918–924.
55. DongPD, MunsonCA, NortonW, CrosnierC, PanX, et al. (2007) Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet 39 : 397–402.
56. BlighEG, DyerWJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37 : 911–917.
57. WuD, WangX, ZhouR, CederbaumA (2010) CYP2E1 enhances ethanol-induced lipid accumulation but impairs autophagy in HepG2 E47 cells. Biochem Biophys Res Commun 402 : 116–122.
58. NarasimhanSD, YenK, BansalA, KwonES, PadmanabhanS, et al. (2011) PDP-1 links the TGF-beta and IIS pathways to regulate longevity, development, and metabolism. PLoS Genet 7: e1001377.
Štítky
Genetika Reprodukčná medicína
Článek Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin PathwayČlánek Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human DiseasesČlánek G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify LongevityČlánek PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial MatrixČlánek Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus ConflictsČlánek The Impact of Population Demography and Selection on the Genetic Architecture of Complex TraitsČlánek Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar AcidificationČlánek The Case for Junk DNA
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 5- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast
- A Mutation in the Gene in Dogs with Hereditary Footpad Hyperkeratosis (HFH)
- Loss of Function Mutation in the Palmitoyl-Transferase HHAT Leads to Syndromic 46,XY Disorder of Sex Development by Impeding Hedgehog Protein Palmitoylation and Signaling
- Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution
- Genome-Wide Nucleosome Positioning Is Orchestrated by Genomic Regions Associated with DNase I Hypersensitivity in Rice
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Single Nucleotide Variants in Transcription Factors Associate More Tightly with Phenotype than with Gene Expression
- Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway
- Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human Diseases
- Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
- Meiotic Drive Impacts Expression and Evolution of X-Linked Genes in Stalk-Eyed Flies
- G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify Longevity
- Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
- Whole Exome Re-Sequencing Implicates and Cilia Structure and Function in Resistance to Smoking Related Airflow Obstruction
- Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues
- R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- The Impairment of MAGMAS Function in Human Is Responsible for a Severe Skeletal Dysplasia
- Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Males
- Mlh2 Is an Accessory Factor for DNA Mismatch Repair in
- Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
- The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells
- Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts
- Genome-Wide Inference of Ancestral Recombination Graphs
- Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in
- SHP2 Regulates Chondrocyte Terminal Differentiation, Growth Plate Architecture and Skeletal Cell Fates
- The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits
- Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation
- Genetic Dissection of the Female Head Transcriptome Reveals Widespread Allelic Heterogeneity
- Genome Sequencing and Comparative Genomics of the Broad Host-Range Pathogen AG8
- Copy Number Variation Is a Fundamental Aspect of the Placental Genome
- GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
- Hox Transcription Factors Access the RNA Polymerase II Machinery through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19
- Drosha Promotes Splicing of a Pre-microRNA-like Alternative Exon
- Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of
- PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF
- Enhanced Interaction between Pseudokinase and Kinase Domains in Gcn2 stimulates eIF2α Phosphorylation in Starved Cells
- A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in
- dGTP Starvation in Provides New Insights into the Thymineless-Death Phenomenon
- Phosphorylation Modulates Clearance of Alpha-Synuclein Inclusions in a Yeast Model of Parkinson's Disease
- RPM-1 Uses Both Ubiquitin Ligase and Phosphatase-Based Mechanisms to Regulate DLK-1 during Neuronal Development
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- Heritable Transmission of Stress Resistance by High Dietary Glucose in
- Revertant Mutation Releases Confined Lethal Mutation, Opening Pandora's Box: A Novel Genetic Pathogenesis
- Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification
- A Genome-Wide Assessment of the Role of Untagged Copy Number Variants in Type 1 Diabetes
- Selectivity in Genetic Association with Sub-classified Migraine in Women
- A Lack of Parasitic Reduction in the Obligate Parasitic Green Alga
- The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
- Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
- Six Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
- EDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
- Genes That Bias Mendelian Segregation
- The Case for Junk DNA
- An In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
- Mosaic Epigenetic Dysregulation of Ectodermal Cells in Autism Spectrum Disorder
- Hyperactivated Wnt Signaling Induces Synthetic Lethal Interaction with Rb Inactivation by Elevating TORC1 Activities
- Mutations in the Cholesterol Transporter Gene Are Associated with Excessive Hair Overgrowth
- Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland
- A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics
- Spermatid Cyst Polarization in Depends upon and the CPEB Family Translational Regulator
- Insights into the Genetic Structure and Diversity of 38 South Asian Indians from Deep Whole-Genome Sequencing
- Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in
- A Dominant-Negative Mutation of Mouse Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation
- Biased, Non-equivalent Gene-Proximal and -Distal Binding Motifs of Orphan Nuclear Receptor TR4 in Primary Human Erythroid Cells
- Ras-Mediated Deregulation of the Circadian Clock in Cancer
- Retinoic Acid-Related Orphan Receptor γ (RORγ): A Novel Participant in the Diurnal Regulation of Hepatic Gluconeogenesis and Insulin Sensitivity
- Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions
- Fine Tuning of the UPR by the Ubiquitin Ligases Siah1/2
- Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression
- Allele-Specific Genome-wide Profiling in Human Primary Erythroblasts Reveal Replication Program Organization
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



