-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
WC-1 Recruits SWI/SNF to Remodel and Initiate a Circadian Cycle
Circadian clocks govern behavior in a wide variety of organisms. These clocks are assembled in such a way that proteins encoded by a few dedicated “clock genes” form a complex that acts to reduce their own expression. That is, the genes and proteins participate in a negative feedback loop, and so long as the feedback has delays built in, this system will oscillate. The feedback loops that underlie circadian rhythms in fungi and animals are quite similar in many ways, and while much is known about the proteins themselves, both those that activate the dedicated clock genes and the clock proteins that repress their own expression, relatively little is known about how the initial expression of the clock genes is activated. In Neurospora, a fungal model for these clocks, the proteins that activate expression of the clock gene “frequency” bind to DNA far away from where the coding part of the gene begins, and a mystery has been how this action-at-a-distance works. The answer revealed here is that the activating proteins recruit other proteins to unwrap the DNA and bring the distal site close to the place where the coding part of the gene begins.
Published in the journal: WC-1 Recruits SWI/SNF to Remodel and Initiate a Circadian Cycle. PLoS Genet 10(9): e32767. doi:10.1371/journal.pgen.1004599
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004599Summary
Circadian clocks govern behavior in a wide variety of organisms. These clocks are assembled in such a way that proteins encoded by a few dedicated “clock genes” form a complex that acts to reduce their own expression. That is, the genes and proteins participate in a negative feedback loop, and so long as the feedback has delays built in, this system will oscillate. The feedback loops that underlie circadian rhythms in fungi and animals are quite similar in many ways, and while much is known about the proteins themselves, both those that activate the dedicated clock genes and the clock proteins that repress their own expression, relatively little is known about how the initial expression of the clock genes is activated. In Neurospora, a fungal model for these clocks, the proteins that activate expression of the clock gene “frequency” bind to DNA far away from where the coding part of the gene begins, and a mystery has been how this action-at-a-distance works. The answer revealed here is that the activating proteins recruit other proteins to unwrap the DNA and bring the distal site close to the place where the coding part of the gene begins.
Introduction
Circadian clocks are key cellular mechanisms regulating a wide variety of physiological and molecular activities. Neurospora has been for several decades an excellent model for studies of the eukaryotic circadian clock characteristic of fungi and animals. In this organism, the White Collar Complex (WCC), a heterodimer comprised of WC-1 and WC-2, serves as the transcriptional activator for the pacemaker gene frequency (frq) by binding to one of two DNA elements, the Clock box (C box) [1] in the dark or the Proximal Light-Response Element (PLRE) in the light [2]. FRQ protein interacts with FRQ-Interacting RNA Helicase (FRH) to bring about repression of WCC activity and thereby to close the positive arm of the feedback loop [3]–[6], presumably through the phosphorylation of WCC [7], [8]. FRQ-mediated WCC repression has been extensively studied, whereas how WC-1 as a transcription factor drives frq expression in a circadian cycle is still poorly understood. WC-1 has two predicted transactivation domains located close to the N - and C - termini respectively [2], [9]–[11]. However, no experimental data have yet confirmed their in vivo functions.
Nucleosome and chromatin structure play critical roles in transcriptional regulation. To overcome nucleosomal barriers in transcription, different transcriptional complexes coordinate to make genomic DNA accessible to RNA polymerase II (reviewed by [12]. Two classes of chromatin remodeling enzymes have been shown to facilitate transcription of chromatin templates in vivo, the histone acetyltransferases and the ATP-dependent remodeling enzymes [13]. SWI/SNF (SWItch/Sucrose NonFermentable), one of the ATP-dependent chromatin remodeling complexes, was first discovered in Saccharomyces cerevisiae as a 2 MDa complex that alters chromatin structures for essential functions such as transcriptional activation, DNA repair, recombination, and chromosome segregation [14], [15]. In yeast SWI/SNF is estimated to control the transcription of no more than 5% of all genes but including the acid phosphatase genes and the MATα-Specific genes [16]. The yeast SWI/SNF complex contains one copy each of eleven subunits, except for the SWI3 subunit that is present in two copies; all SWI/SNF subunit-null mutants are viable but display distinguishable phenotypes. SWI2 is a highly conserved DNA-dependent ATPase and the scaffold protein of the complex [17]; swi2 knockouts are defective in sporulation and display slow growth on nonfermentable carbon sources [18], [19]; swi1 null strains, defective in mating-type switching, display sporulation defects and slow growth [20], [21]; snf5 null mutants show reduced growth on glucose and sucrose [22], [23]. The SWI/SNF complex can associate with naked DNA or nucleosomes, and is thought not to bind in a sequence specific manner [24]–[26]. Instead, SWI/SNF is targeted to promoters via acidic domains on the surfaces of gene-specific transcriptional activators rather than via interactions with polyglutamine (polyQ) rich sequences [27]. Yeast SWI/SNF can alter nucleosomal structure in an ATP-dependent manner, which leads to the relief of chromatin-mediated repression of transcription [28], [29]. SWI/SNF is able to remodel nucleosomes per se without their disruption, by sliding histone octamers to other sites on the same DNA molecule, or transferring histone octamers to other DNA molecules [26].
Rhythmic histone modifications and chromatin remodeling over a circadian cycle have been reported for a variety of genes in different circadian systems. For example, the promoter regions of Per1 and Per2, central clock components in mammals, undergo rhythmic histone H3 acetylation (K9, K14) [30]. In Neurospora, histone acetylation of the frq promoter oscillates over a circadian cycle and chromatin structure at frq is rhythmically remodeled in a circadian fashion, accompanied by rhythmic binding of WC-2 [31]. The occupancy of the nucleosome neighboring the C box peaks when frq transcription is repressed and decreases just before frq transcription starts [31]. In the same study, an ATP-dependent chromatin remodeler, CLOCKSWITCH (CSW), was identified as a component essential for depositing nucleosomes back to the C box to terminate frq transcription. Another chromatin remodeler, CHD-1, plays a role in methylation and rhythmic expression of frq [32]. Although both CSW and CHD-1 appear to be required for the closure of frq transcription and participate in chromatin remodeling at frq, how the WCC interacts with remodeling factors to relieve chromatin-mediated repression of the C box is still elusive. Consistent with this, there is a several hour lag between the turnover of FRQ and the rapid increase in frq mRNA [33], suggesting that there is more to reinitiation of frq expression than simply relief of repression.
In this study, we identify a previously undescribed N-terminal domain of WC-1, close to but not including the prominent polyglutamine stretch, that acts as the transactivation domain for recruiting SWI/SNF and driving frq transcription; notably, elimination of the N - and C - terminal polyQ stretches does not influence frq circadian transcription and such strains showed a WT circadian phenotype. The SWI1 subunit in the SWI/SNF complex is essential for initiation of frq transcription in a circadian context although interestingly not in response to light, and in a Δswi1 strain chromatin structure adjacent to the C box is less remodeled and the oscillation of the nucleosomal density is abolished, as is the circadian clock.
Results
N - and C-terminal polyQ stretches predicted to be activation domains are not required for WCC circadian function
In the classic model of transcription, transcription factors use their transactivation domains to recruit transcriptional coactivators, e.g. chromatin remodelers and histone acetyltransferases, to release the repressive state of promoter regions [26]. In Neurospora, WC-1 has two polyQ domains located at N - and C - termini respectively that were hypothesized to be transactivation domains (AD) [10], [11], [34] (Figure 1A). To test the role of the two polyQs in frq expression, we eliminated both of them (aa 16-57 and aa 1097-1128) (Figure S1A) and surprisingly the double deletion strain still showed a wild-type (WT) circadian phenotype (Figure 1B). We further checked FRQ levels in strains held in constant dark (DD) for 16 hours (Circadian Time (CT) 5), a time when newly synthesized FRQ appears in WT. Consistent with the race tube data, FRQ levels in the double deletion strain showed no observable difference from WT and the WC-1 level is comparable with WT (Figure 1D, right-most panel). Collectively, the two polyQ stretches on WC-1 do not influence the stability of WC-1 and are not required for frq transcription and the circadian function of WCC in the dark.
Fig. 1. Identification of aa 100-200 as the transcriptional activation domain for WC-1. 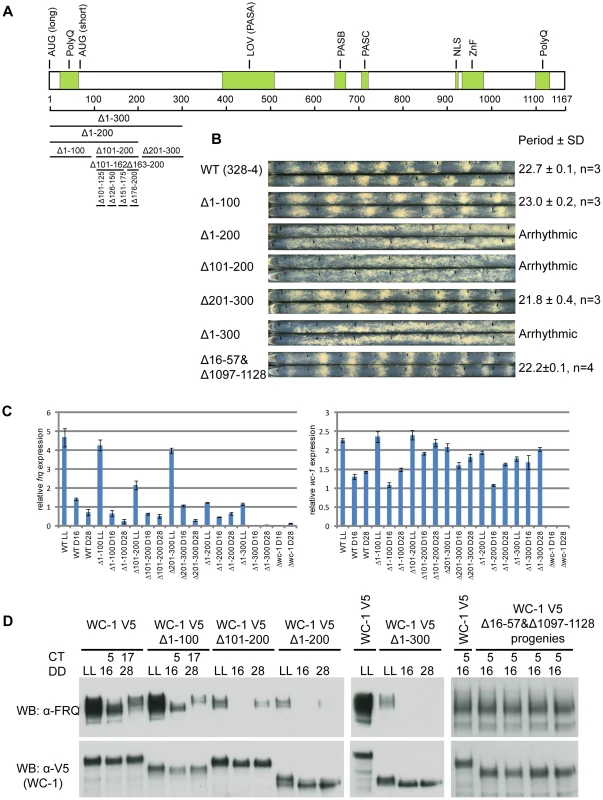
(A) Schematic depiction of the domain architecture of the WC-1 protein. WC-1 contains 1,167 amino acids and black bars with corresponding numbers of amino acids represent different deletions of the N-terminal region of WC-1. AUG(long) and AUG(short) mark the beginnings of long and short forms of WC-1; PolyQ, putative transcription activation domain; LOV, light, oxygen, or voltage domain containing FAD chromophore binding site; PAS, Per-Arnt-Sim domain mediating interaction with WC-2; NLS, putative nuclear localization signal; Zn, zinc finger DNA-binding domain (B) Racetube analyses of wild-type (WT) and WC-1 deletion strains. Period is reported in hours ± one standard deviation, n = number of racetubes. Replicate tubes are shown, and the vertical black lines in racetubes mark daily growth fronts of the strains. 328-4 (ras-1bd A) served as the WT. Strains bearing deletions of both polyQs (Δ16-57 and 1097-1128), Δ1-100, and Δ201-300 showed WT circadian periods. Strains lacking aa 100-200 (Δ1-200, Δ1-300, and Δ101-200) are arrhythmic (C) Results from quantitative RT-PCR analysis of frq and wc-1 mRNA levels in WT and in the wc-1 deletion strains noted. (D) Western blot showing expression levels of WC-1 and FRQ in WT and wc-1 mutants. LL, constant light; DD, hours after the light to dark transition; CT, circadian time. Both WT WC-1 and mutants were C-terminally tagged with V5. Δ101-200 showed a significant decrease of frq expression and further deletion of aa 1-200 or 1-300 further dimished frq expression. Elimination of the two polyQs (Δ16-57 and 1097-1128; four progeny of identical genetype from one cross are shown) had no impact on FRQ level at CT5. A domain encompassing aa 100-200 in WC-1 is required for frq dark expression
Because, despite prediction, the two polyQ stretches on WC-1 are not transactivation domains, a series of WC-1 deletions were generated to identify the regions needed for frq transcription, and circadian phenotypes of these mutants were monitored by race tube assays (Figure 1A, 1B, and Figure S1B). Of these mutants, only strains bearing deletions overlapping aa 101-200 showed an arrhythmic circadian phenotype (Figure 1B). In the three mutants deleted for these residues, all of which lacked overt rhythmicity, frq mRNA levels were no longer rhythmic and were reduced to or below levels seen in the trough of the wild type rhythm (Figure 1C). New FRQ was not seen at DD16 (16 hours in darkness, circadian time (CT) 5 in the subjective morning when frq expression normally peaks) and only a weak FRQ band or no FRQ at all appeared at DD28 (subjective night, CT17) (Figure 1D); levels of wc-1 mRNA and WC-1 protein were normal. This suggested that the region between aa 101 and 200 has the potential to transactivate frq expression, and to identify these residues four strains bearing smaller deletions were created at intervals of 25 amino acids: Δ100-125, Δ126-150, Δ151-175, and Δ176-200. The Δ100-125 and Δ151-175 strains showed a WT period while Δ126-150 and Δ176-200 displayed a period of 2 hours longer than WT (Figure S1B). Thus, it seems that multiple domains determine the expression level of the frq gene. Interestingly, the Δ163-200 strain that displayed a 25.5 hour period (Figure S1B) and reduced dark FRQ expression (Figure S1C) retained its responsivity to light for both frq mRNA and FRQ expression respectively (Figure S1D), indicating that this region may only mediate circadian frq expression.
WCC interacts with SWI1 in vivo and in vitro
To identify interaction partners of the WCC and potential co-activators, WC-1 that was epitope-tagged with V5, 10xhistidine, and 3xFLAG was purified from extracts and interacting proteins identified by MS/MS (Wang et al., in preparation); these preliminary mass spectrometry data showed that SWI/SNF subunits copurified with the Neurospora WCC. To validate this interaction in vivo, SWI1 was C-terminally tagged with V5. Immunoprecipitation using WC-2 antibody revealed the anticipated strong interaction with WC-1 as well as an interaction with SWI1 that was dependent on the presence of WC-1 (Figure 2A).
Fig. 2. WCC interacts with SWI/SNF in vivo and in vitro. 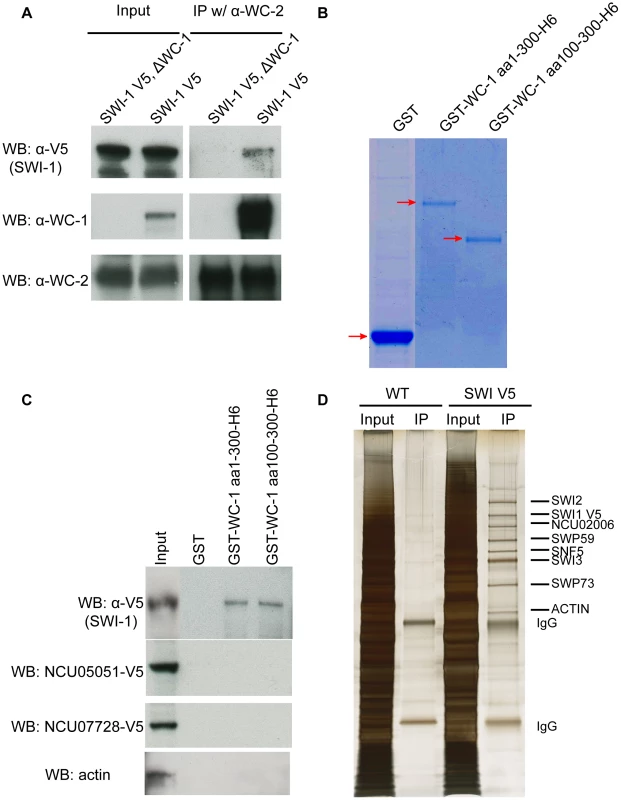
(A) Co-IP demonstrating interaction of WC-1 with SWI1 in vivo. SWI1 was C-terminally tagged with a V5 epitope and immunoprecipation was performed using WC-2 antibody. WC-1 was pulled down with WC-2 as well as a reduced amount of SWI1. (B) Expression and purification of GST-WC-1 fusion proteins. GST, GST WC-1 1-300 6xHis, and GST WC-1 100-300 6xHis lacking the N-polyQs were expressed in bacteria and purified. Gel stained with Coomassie Blue. (C) N-terminal fragments of WC-1 extending from 1-300 or 100-300 bind to SWI1 in vitro. Neurospora crude cell lysates were mixed with beads to which the GST-tagged WC-1 fragments were bound, and bound SWI1 was visualized by virtue of a C-terminal V5 tag; see Materials and Methods. GST alone failed to pull down SWI1 while WC-1 aa1-300 or aa 100-300 pulled down SWI1 at a similar level. Likewise negative control proteins actin, and two transcription factors encoded by NCU05051 and NCU07728 were not bound by WC-1 fragments. (D) Affinity purification of the Neursopora SWI/SNF complex showed the presence of different subunits in a 1∶1 stoichiometry except for SWI3. SWI1 was tagged with V5 at the C-terminus, centrifuged lyate was incubated with V5 antibody, and the gel was sliver-stained. Individual bands were excised and each protein identified via mass spectrometry. To confirm that the association between this region of WC-1 and SWI/SNF is still robust in vitro, GST alone, GST-WC-1 aa 1-300-6xHis and GST-aa 100-300-6xHis (that lacks the N-terminal polyQs) were cloned, expressed and purified from Escherichia coli. GST-WC-1 aa 1-300 6xHis and GST-aa 100-300-6xHis were purified with a two-step protocol to obtain the full length polypeptides (Figure 2B). To pull down SWI/SNF, the purified proteins were incubated with a centrifuged cell lysate of a SWI1-V5 strain and the captured proteins analyzed by Western blot. GST alone failed to pull down SWI1-V5 while GST-WC-1 aa 1-300-6xHis and GST-aa 100-300-6xHis pulled down SWI1 at a similar level (Figure 2C). Negative controls include actin, an abundant nuclear protein, as well as two transcription factors, the GATA Zn finger factor SRE encoded by NCU07728 and the Zn(2)-Cys(6) binuclear cluster domain transcription factor COL-23 encoded by NCU07728, both of which are known to bind DNA (X. Zhou and JCD, unpublished). The data indicate that WC-1 aa 1-300 is able to recruit SWI/SNF and aa 1-100 which contains the polyQs does not contribute to the recruitment. This is consistent with the behavior of yeast SWI/SNF that can be pulled down from crude cell lysates by the acidic transactivation domain but not by the polyQ region of herpes virus VP16 [27]. The isoelectronic point (pI) of the Neurospora N-terminal domain aa1-300 is 4.95 and aa 100-300 is 4.41 (predicted by DNAMAN software). The acidic nature of aa 100-300 is consistent with reported acidic activation domains of the VP16 protein, yeast Gcn4, and yeast Hap4, which are able to recruit the SWI/SNF complex to release chromatin-mediated repression of transcription [35]–[37].
To characterize the subunit composition of Neurospora SWI/SNF, V5-tagged SWI1 was purified using a single V5 antibody step and the result showed that several proteins were specifically co-purified with SWI1-V5 in a stoichiometric manner (Figure 2D). These bands were cut out individually and identified by mass spectrometry. The data confirmed the presence of SWI1, SWI2, SWI3, SWP59, SNF5, and SWP73 in the Neurospora SWI/SNF complex and confirmed that the interactions within complex in Neurospora are robust. Like WC-1 and WC-2, SWI1 and SWI2 have constant protein levels over 28 hours in constant dark (Figure S2). Taken together, these data demonstrate that WC-1 binds to SWI/SNF in vivo and WC-1 aa100-300 can specifically recruit SWI/SNF in vitro.
Δswi1 and Δsnf5 show significantly impaired frq expression
To uncover a possible role of SWI/SNF in frq transcription and in circadian rhythmicity, we obtained all SWI/SNF single gene deletion strains from the Neurospora knockout collection (Colot et al., 2006) according to their homology with yeast SWI/SNF subunits (Figure 2D and Table 1) [38]. Except for SWI2 and SWI3 which may be essential for growth in minimal medium, all deletion strains of Neurospora SWI/SNF are viable, as are the knockouts of yeast SWI/SNF homologs (http://www.yeastgenome.org/). FRQ protein expression was examined by Western blot (Figure 3A), and frq mRNA by qRTPCR (Figure 3B) in each SWI/SNF knockout at two times of day: circadian time (CT) 5 (DD16) when newly synthesized FRQ peaks and CT17 (DD28) when old FRQ is hyperphosphorylated and begins to be degraded [33]. In yeast, Snf5p, Swi1p, and Swi2p subunits are contacted by acidic activators such as Gcn4p and Hap4p [38]. Consistent with this, in the deletion strains examined, the FRQ and frq mRNA levels were low and circadian regulation was abolished in Δsnf5, Δswi1, and decreased in Δswp59 (Figure 3). Consistent with WC-1 transcriptional autoregulation, the known role of FRQ in stabilizing WC-1 [4] and the low levels of FRQ in these strains, WC-1 levels were also correspondingly reduced, although it is also possible that SWI/SNF might directly influence WC-1 expression. Interestingly, although WC-1 levels in Δswi1 were slightly lower than in WT and higher than in Δswp82 (on the same blot), the Δswp82 strain had a normal FRQ level and circadian expression whereas Δswi1 did not (Figure 3A); this suggests that the very low WC-1 level in Δswi1 is sufficient to drive rhythmic frq expression and that the lack of a rhythm lies somewhere else. FRQ and WC-1 levels were similar to those of WT in other SWI/SNF knockouts examined (Figure S3A). Together, based on the low level of frq mRNA and FRQ in Δsnf5, Δswi1, and Δswp59, SWI/SNF complex participates in WCC-dependent frq expression and the SWI1 subunit is specifically involved in this event through its physical interaction with WC-1.
Fig. 3. Loss of FRQ expression in SWI/SNF subunit knockouts. 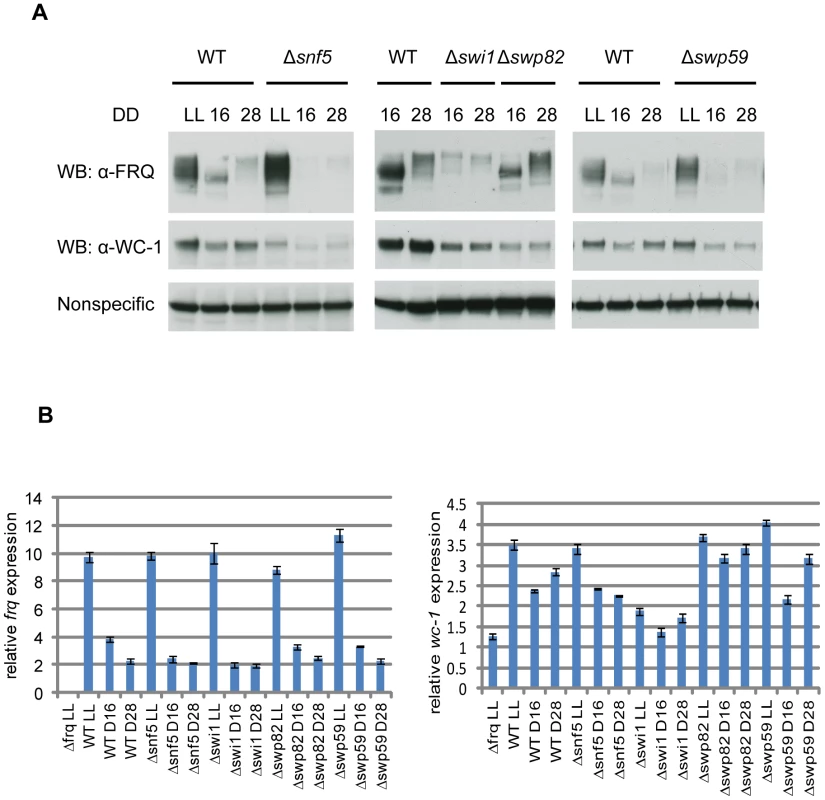
(A) Expression levels of FRQ and WC-1 were followed by Western blotting in WT, Δswp59, Δswi1, and Δsnf5. Two dark time points were chosen to examine FRQ expression, CT5 (DD16) when newly synthesized FRQ is seen and CT17 (DD28) when old FRQ is hyperphosphorylated and begins to be degraded in WT. Non-specific bands were shown for equal loading. (B) Corresponding data for frq and wc-1 mRNA is shown. Tab. 1. Neurospora SWI/SNF subunits and knockouts. 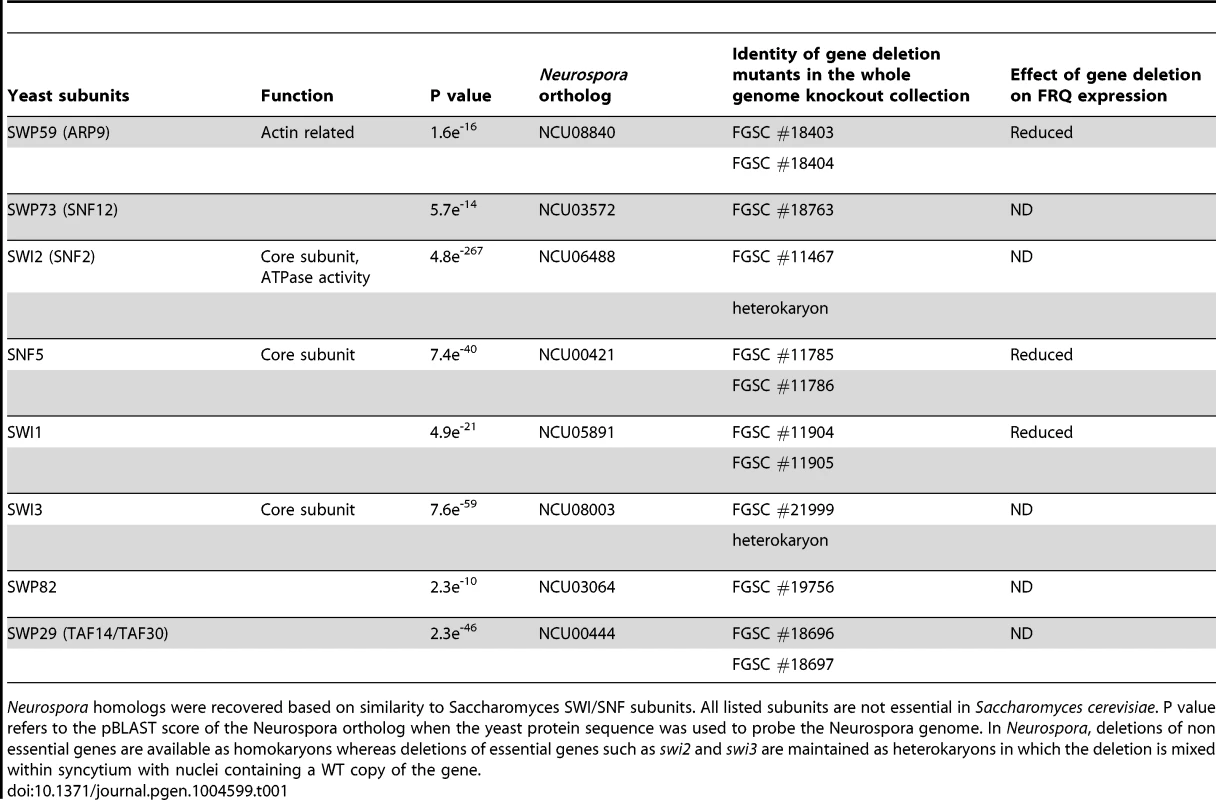
Neurospora homologs were recovered based on similarity to Saccharomyces SWI/SNF subunits. All listed subunits are not essential in Saccharomyces cerevisiae. P value refers to the pBLAST score of the Neurospora ortholog when the yeast protein sequence was used to probe the Neurospora genome. In Neurospora, deletions of non essential genes are available as homokaryons whereas deletions of essential genes such as swi2 and swi3 are maintained as heterokaryons in which the deletion is mixed within syncytium with nuclei containing a WT copy of the gene. Rhythmic transcription of frq is abolished in Δswi1
To check the circadian significance of swi1, snf5 and swi59, the three deletion strains were backcrossed to the ras-1bd allele that has been widely used to visualize overt circadian rhythms in Neurospora [39]. In race tube assays Δswi59 showed a virtually WT period while, interestingly, both Δswi1 and Δsnf5 were arrhythmic (Figure S3B), grew more slowly, and produced fewer conidia than WT, suggesting that SWI/SNF also plays a role in hyphal growth and asexual spore formation. The slowed growth and reduced conidiation phenotypes of Δswi1 and Δsnf5 are consistent with those of yeast (see Introduction), but also raised the possibility that loss of rhythmicity was an artifact arising from interference of the mutations with the expression of rhythmicity rather than interference with the clock itself. To directly and continuously monitor changes in frq transcription at the level of the core clock, we created strains that bear an optimized luciferase reporter gene driven by the circadian promoter (C box) of frq [40] in Δswi1, Δsnf5, and Δswi59 backgrounds respectively, such that the activity of the complex of WC-1 and WC-2 on the frq promoter could be analyzed in vivo. Consistent with Figure 3, the amplitude of frq promoter:luc transcription was dramatically impaired in Δswi1 while Δswi59 showed a WT phenotype (Figure 4). frq transcription was also impaired in Δsnf5 but continued to oscillate weakly with a peak-to-trough amplitude only about 4% that of WT. The absence of the overt rhythm in this strain (Figure S3B) suggests that the severely attenuated rhythm in frq expression is not sufficient to drive the rhythms in ccg (clock-controlled gene) expression needed for the overt rhythm. Of particular note is that Δswi1 completely lost rhythmic transcription of frq and overall expression was about 1% of WT. Taken together, these data indicate that SNF5 contributes significantly to frq transcription whereas SWI1 is essential for the transcriptional oscillation of frq over a circadian cycle.
Fig. 4. Reduced frq expression and loss of rhythmicity in SWI/SNF subunit knockouts as assayed by a luciferase reporter. 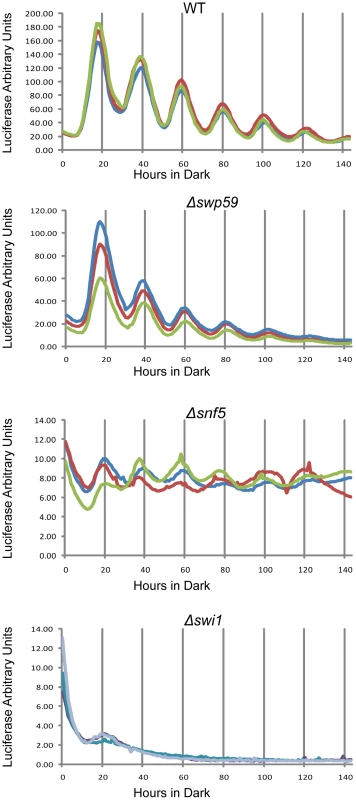
frq transcription in WT, Δswi1, Δsnf5, and Δswp59 was examined using the frq C box fused to codon-optimized firefly luciferase (transcriptional fusion). Strains were grown on 0.1% glucose racetube medium containing 5 mM luciferin in a 96 well plate and synchronized by growth in constant light for 48 hours followed by transfer to darkness. The luciferase signal was followed for longer than 6 days with sampling every 30 minutes. Each strain was repeated three times. Δswp59 showed a WT oscillation, Δsnf5 also oscillated in a circadian manner despite an extremely low amplitube, while frq trancription was completely abolished in Δswi1. The binding of SWI/SNF to the C box depends on aa 1-300 on WC-1
Although the SWI/SNF complex remodels DNA and is required for the expression of some genes herein shown to include frq, it does not itself bind to DNA, but instead relies on dedicated transcription factors. Two reported models for SWI/SNF recruitment by transcription factors have been advanced: (1) Transcription factors that have a strong affinity for a promoter associate first with an Upstream Activation Sequence (UAS) and then recruit SWI/SNF [41], [42]; (2) those with weak binding to a UAS tend to recruit SWI/SNF off DNA first [41], [43], [44] and the bound SWI/SNF facilitates DNA association of these transcription factors (reviewed by 26). In Neurospora, WCC binds to the PLRE adjacent to the transcription start site (TSS) of the frq promoter under light conditions while it associates with the C box of the same promoter located ∼1.2 kilobases 5′ of the TSS (transcription start site) in the dark. To test whether SWI/SNF is recruited by WC-1 to the C box, in addition to determining which model fits best for WC-1, chromatin immunoprecipitation was performed at DD16, a time when frq expression should be near maximal, using the WT and WC-1 Δ1-300 strains (Figure 5A). SWI2, the core subunit of SWI/SNF, bound to the C box strongly in WT but did not bind to the C box in WC-1 Δ1-300; the binding of WC-1 and WC-2 to the same DNA sequence showed no significant difference in the two strains (Figure 5). The data suggest that WC-1 follows the first SWI/SNF model of binding in which the transcription factors bind strongly to the C box; recruitment of SWI/SNF is a subsequent event and not associated with binding of WC-2. A prediction of this model is that there should be a phased time-dependence to the association of these factors with the C box and their action on it. This was examined by ChIP in Figure 5B in a time series over the six hours leading up to maximal frq expression. Binding of the WCC (using WC-2 as a proxy) steadily increases over this period and this is accompanied by a slightly delayed and steeper increase in SWI/SNF association (using SWI1 as a proxy). Loss of nucleosome components from the region of the C box was tracked by loss of histone H3 which shows a lag of 3–4 hours followed by a rapid disappearance from the C-box region. A working model for initial events in activation of frq transcription consistent with these data posits WC-1 and WC-2 forming an active transcriptional complex on the C box DNA (see also [31]) followed by WCC recruitment of the SWI/SNF complex which initiates active remodeling of the chromatin in the region of the C box.
Fig. 5. The binding of SWI/SNF to the C box relies on aa 1-300 of WC-1 and increases prior to the peak of rq expression. 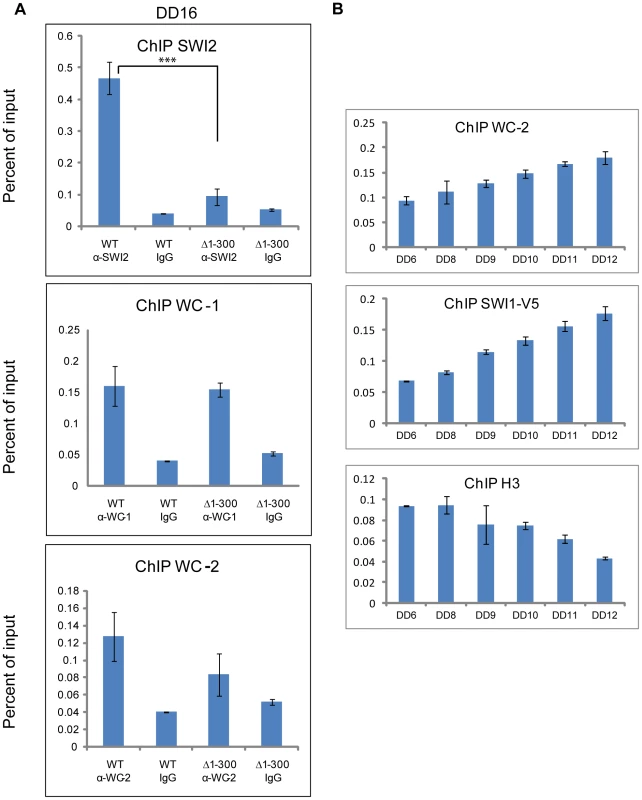
(A) ChIP experiment performed on chromatin isolated at DD16 when frq expression is maximal. WC-1 and WC-2 had a normal binding to the C box of frq in a strain bearing WC-1Δ1-300, while SWI2 binding was impaired in this strain. The annealing positions of the primer set used to detect the C box corresponds to the middle set shown in Figure 6A. Average values are plotted as a percent of total with error bars representing the standard error of the mean (SEM) (n = 3, ***p<0.0005). Samples were grown for 16 hours in the dark, formaldehyde-crosslinked, and harvested. (B) ChIP was used in a timecourse analysis of the association of WC-2, SWI-1 and histone H3 with the C box. Samples were gorwn for the indicated number of hours in darkness (DD) prior to harvesting and processing for ChIP as described in (A). Error bars represent the standard error of the mean ( = 3). Nucleosomal density at the C box of the frq promoter increases in Δswi1
We have previously shown that a nucleosome (NucB) partially occludes the C box and blocks frq transcription in the subjective evening/night, and is removed from the C box in the late subjective night/early day when frq transcription is initiated [31] (Figure 6A); this model is consistent with the loss of histone H3 from the region as seen in Figure 5B. In the same study, an ATP-dependent chromatin remodeler, CLOCKSWITCH (CSW), was shown to be necessary for remodeling of the opened C box back to the closed repressive state. Our data suggested that SWI/SNF might be involved in antagonizing CSW in opening the C box for frq transcription. To compare NucB density between WT and Δswi1, nuclei were isolated, micrococcal nuclease (MNase) digested, and mononucleosomal DNA was gel-purified and quantified for real-time PCR (Figure 6B). Four dark time points across two circadian cycles representing circadian oscillations of the NucB density were chosen for comparison: DD4 (CT 16, subjective night when the C box is closed and frq transcription repressed), DD12 (CT 0, subjective morning when the C box is open and frq is transcribed), DD24 (CT 13, subjective evening, second day), and DD32 (CT 21, late subjective night, second day); these times were chosen based on the peaks and troughs of nucleosome occupancy reported by Belden et al. [31]. NucB level was determined by real-time PCR using a specific primer set against DNA sequences near to the C box and the region of NucB on the mononucleosomal gel-purified DNA. As previously reported [31], NucB density peaked at DD4 and 24 and decreased at DD12 and 32 in WT (Figure 6C), but this oscillation was completely abolished in Δswi1 and NucB density was always higher than WT. To better gauge whether NucB was being moved aside or displaced to truly open up the chromatin at the C box we applied a nuclease sensitivity assay previously used to probe chromatin structure at this locus [31]. Chromatin was isolated from WT or Δswi1 strains at the same 4 subjective times (corresponding to two successive peaks and troughs in frq expression in WT and the corresponding times in Δswi 1) and subjected to limited digestion with micrococcal nuclease (MNase), an enzyme that will cut open DNA but not DNA bound within nucleosomes (Figure 6D). The nucleosome located over the C box (NucB) appears to be rhythmically present in WT but continually present in the Δswi1 background, consistent with the PCR analysis of Figure 6C. In all these data suggest that SWI/SNF is required for remodeling NucB to activate frq transcription in a circadian cycle and that SWI1 plays an essential role in this process. As controls for specificity, the nucleosome near NucB and bracketing the C box (called NucA, [31]) and an untranscribed region (3.303) of Neurospora DNA [31] had a comparable density between WT and Δswi1 across the four time points tested (Figure 6C).
Fig. 6. SWI1 is required for rhythmic opening of the C box through removal of NucB. 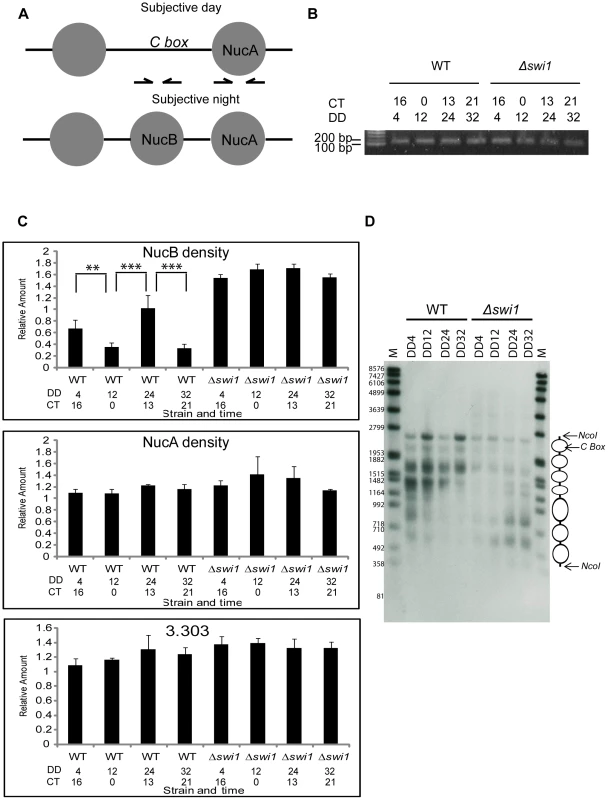
(A) Relative positions of resident nucleosomes, NucB and NucA at the C box (see also reference 31). Arrows indicate primer sets used to detect these regions. (B) Nuclei were isolated by fractionation from samples harvested at indicated hours in the dark and digested with MNase. 20 nanograms of gel-purified mononucleosomal DNA were loaded in each lane. (C) The density of NucB was measured by real-time PCR using a primer set shown in (A, the left primer set) and gel purified mononucleosomal DNA as template (n = 3, error bars represent ± SEM, ***p<0.0005, **p<0.005). NucB occupancy of the C box oscillates between low (CT0 and 21) and high (CT 13-16) in WT as reported by Belden et al. (2007b), whereas NucB is always bound to the C box in Δswi1. NucA and 3.303 (an unregulated control region of genomic DNA distinct from frq) served as controls for equal DNA inputs [31]. (D) Difference in frq promoter chromatin were probed with a limited nuclease digestion assay as described in Experimental Procedures. Cultures were harvested from WT or from Δswi1 cultures at the times in darkness indicated. Discussion
In this study, we showed that WC-1 in the WCC recruits SWI/SNF to the frq promoter to aid in remodeling the nucleosome environment of the C box and thereby to initiate a circadian cycle of frq transcription. To identify potential transactivation domains on WC-1 required for frq transcription, a series of WC-1 deletions were generated and studied. Among these, all deletions covering amino acids 101-200 had normal WC-1 levels but displayed severely impaired frq mRNA and FRQ expression in the dark and arrhythmic circadian phenotypes. These data suggest aa 101-200 of WC-1 is a transactivation domain that is essential for circadian expression of frq. A search for coactivators recruited by WCC identified SWI/SNF, components of which interact with WC-1 in vivo and in vitro. In addition, the portion of WC-1 containing the transactivation domain is required for recruiting SWI/SNF to the C box.
Affinity purification of the Neurospora SWI/SNF complex identified the expected subunits based on yeast homology predictions (Table 1) as well as the protein encoded by NCU02006 (a highly conserved protein among fungi) and actin (that is also found in mammalian BAF or PBAF complex [45]–[49]). This complex is sturdy in yeast where deletion of the sites of activation domain contact in the N-terminal SNF5 and the second quarter of SWI1 left the SWI/SNF complex intact [50]. Assuming the Neurospora SWI/SNF complex is similarly robust the data suggest that the arrhythmic clock phenotype of Δswi1 and Δsnf5 may be caused by the loss of transcription factor contact.
A long standing question brought into focus by this study is how protein-DNA interactions at the C box bring about changes in frq transcription at the TSS, 1.2 kbp away. SWI/SNF, a complex with proven DNA looping capabilities [51], [52], provides a clear solution to this question through its ability to facilitate formation of DNA loops; these bring different genomic regions separated by kilobases into close proximity, resulting in sufficient concentrations of each transcription complex to drive transcription [53]. For example, Brg1 (SMCA4), a mouse SWI/SNF subunit, mediates compaction of chromatin into dense loops at the 200 kbp cytokine locus [54] and is also required for the formation of DNA loops across the 150 kbp CIITA locus during interferon-gamma (IFN-gamma)-mediated gene induction [55]. We anticipate that SWI/SNF recruited by the WCC remodels the C box region and brings about DNA looping in a similar manner to bring the C box into proximity with the TSS of frq.
WCC is the primary blue light photoreceptor in the organism [1], [2], [9], acting at both the C box for circadian feedback and at the PLRE for acute light responses. Interestingly, while both SWI/SNF and aa 101-200 of WC-1 are indispensable for FRQ expression in the dark they have little influence on PLRE-mediated FRQ transcription in constant light. Light-induced FRQ was seen even in the WC-1 Δ1-300 strain; these data also explain the cryptic phenotype of the rhy-2 strain arising from a WC-1 Δ1-264 [56]. WCC in the dark is mainly a heterodimer and when it senses light, it forms a multimer, the light active form, on the PLRE at the frq locus [2], [7], [57], [58]. Thus, it seems that WC-1 might recruit different coactivators in the light than in the dark to activate frq expression. One of the coactivators recruited by the light-activated WCC is NGF-1, a histone acetyltransferase, which plays a role in blue light signal transduction [59]. Also perhaps surprisingly the polyQ domains on the N - and C - termini of WC-1, long predicted to be transactivation domains, are needed for neither light nor dark activities of WC-1. This is consistent with previous work on N-polyQ polymorphisms in wild strains that showed only minor period differences [60], and with findings from other transcription factors such as yeast Gal4 that uses its acidic domain rather than glutamine-rich or proline-rich sequences to recruit SWI/SNF in transcription [27], [37], [38].
Through the lens provided by this study we can now begin to understand how multiple proteins and complexes with opposing functions coordinate their activities to open/activate and close/repress the frq locus at appropriate circadian phases. We previously reported Clock Switch (CSW) and CHD-1 as chromatin modifiers required for the rhythmic opening and closure of the C box in the frq promoter that leads to rhythmic expression of FRQ [31], [32]; CHD-1 also remodels the antisense frq (qrf) promoter and may play a more general role in maintaining chromatin structure at frq as without it expression levels never reach either peak or trough levels seen in WT [32]. WC-1, CSW, and CHD-1 are always present in the cell, and CSW appears to bind to the C box preferentially during the time when frq is becoming active [32]; without WC-1 frq is always inactive and without CSW or CHD-1 frq is always moderately active. In the core negative feedback loop, FRQ helps to inactivate WC-1 and prevents WCC from binding to DNA. However, the fact that frq remains moderately active without CSW or CHD-1 indicates that activation/repression of FRQ is more than only binding/inactivation of WCC but also requires the active participation of other factors, perhaps to eject the WCC/SWI/SNF complex from the C box and un-loop the DNA. Although both CSW and CHD-1 actively remodel DNA at frq their role in the clock cannot be succinctly stated as activating or repressing; the timing of CSW binding, for instance, coincides with frq activation yet frq is still expressed without it. When present and active, WCC might dominate the competition between activation and repression, recruiting SWI/SNF to activate frq despite the presence of other factors. Not explicitly accounted for yet in this model is the involvement of the frq antisense qrf, whose promoter is also remodeled by CHD-1. A recent publication revealed a novel factor, CATP (Clock ATPase), involved in remodeling chromatin at the C box [61]. The expression of frq in Δcatp strains appears to be about a log order greater than in strains lacking SWI/SNF, consistent with CATP acting as an accessory factor to help to open the C box.
A working model based on these data is summarized in Figure 7. In the dark, before frq transcription starts, NucB mostly blocks the C box due to the action of chromatin remodelers that may include CHD-1; when with WC-2, WC-1 binds to the C box in its active form, it recruits SWI/SNF which aids in removing NucB from the C box, stabilizing the active state essential for frq transcription; CSW binds most strongly at this time. Based on the action of SWI/SNF in other systems we anticipate that its action involves DNA looping to bring the TSS into proximity with the C box and WCC. This configuration remains active, with active WC-1 being rapidly turned over, until FRQ depresses activity of WC-1 and reduces its affinity for DNA thereby also stabilizing it. Without WCC bound, the SWI/SNF-mediated looping is reversed by the action of other chromatin remodelers, and NucB returns to cover the C box.
Fig. 7. A working model for WC-1-dependent recruitment of SWI/SNF to initiate frq transcription. 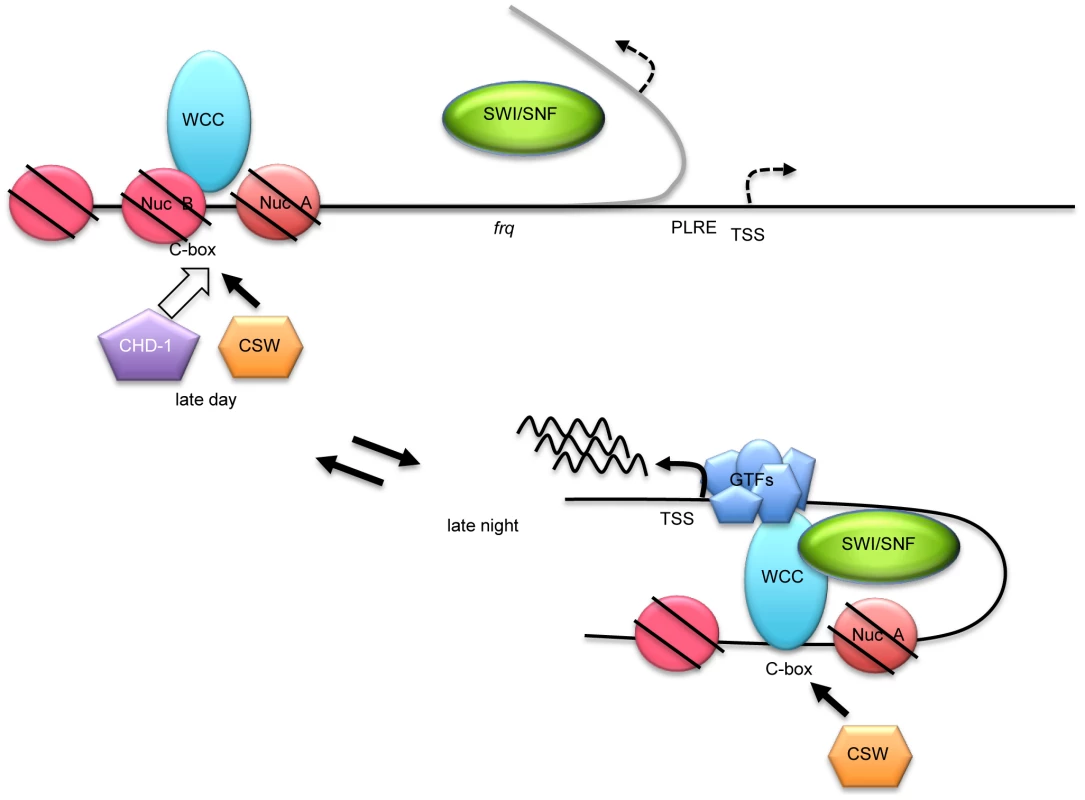
In the night before frq transcription starts, NucB is in a repressive state occluding the C box as fostered by chromatin remodelers that may include CHD-1. When active WC-1/WC-2 begin to bind at the C box, an event roughly coincident with CSW binding, SWI/SNF is recruited and it helps to fully remodel the C box to an active state, removing NucB, and also beginning to bring about the DNA looping (curved gray line) that is essential to bring the WCC transcriptional activation domains to the transcription start site (TSS) for stable recruitment of general transcription factors (GTFs). For acute light-induction of frq, an action not requiring SWI/SNF, WCC binds directly to the PLRE rather than the C box and recruits unknown remodelers to drive frq transcription at levels much higher than in the dark. A variety of data suggest that light activation of frq may represent a distinct function. In the light frq mRNA levels are much higher than that in the dark, FRQ does not inhibit this expression, [33], [62] and in the light, the FRQ levels are nearly normal in the SWI/SNF deletion strains tested including Δswi1, Δsnf5 and Δswp59 that severely reduce circadian frq expression. This suggests that the WCC recruited to the PLRE in the light recruits functional coactivators other than SWI/SNF to modify the PLRE in frq transcription. Additionally and consistent with this, the PLRE is located adjacent to the TSS site and WCC acts on these regions neither by looping of the DNA to bring activators to the TSS, nor by wholesale remodeling of chromatin, although epigenetic modifications have been noted [32], [39], [59]. This suggests that major remodeling and looping induced by SWI/SNF are principal factors distinguishing circadian activators at the C box versus light activators at the PLRE.
Materials and Methods
Strains and growth conditions
328-4 (ras-1bd A) and 74A (ras-1WT A) were each used as a clock-WT strain in this study. Race tube analyses were carried out as previously described [39]. Race tube medium contains 1×Vogel's salts, 0.1% glucose, 0.17% arginine, 50 ng/mL biotin, and 1.5% bacto-agar, and liquid culture medium (LCM) is 1×Vogel's, 0.5% arginine, and 50 ng/mL biotin with glucose at 2%. Race tubes were inoculated and incubated in constant light for 16–24 h at 25°C and then transferred to constant darkness at 25°C. A recipient strain for generating WC-1 deletion series is 21-9 (ras-1bd; Δfrq::hph+; Δmus-52::hph+ a). Neurospora transformation was done as previously described [63]. The wc-1 {knock-in} (wc-1KI) targeting cassette pWB-1-6 was introduced into 21-9 and replacement mutants were backcrossed to 328-4 to obtain homokaryotic strains for race tube analyses. WC-1 and SWI/SNF deletion strains generated by the Neurospora genome project were obtained from Fungal Genetics Stock Center (FGSC) [63].
Protein isolation and detection
Procedures for preparation of protein lysates and Western blots were followed as described [33], [64]. For Western blot, 15 milligrams of whole-cell protein lysate was loaded per lane. Anti-V5 antibody (Pierce) was diluted 1∶5000 for use as the primary antibody. SWI2 antibody was obtained from Abcam (Ab3749). Protein purification prior to MS/MS analysis was performed using a slightly modified procedure [64].
Immunoprecipitation (IP)
IP was done as previously described [64]. In brief, 2 milligrams of total protein were incubated with 50 µL of V5 beads rotating for 2 hours to overnight. The agarose beads were washed with the protein extraction buffer 4 times and eluted with 50 uL of 5× SDS sample buffer at 99°C for 5 min.
GST pull-down assay
WC-1 aa1-300 and 100-300 were each cloned into pGEX4T1 in-frame fused with an N-terminal GST tag, and a hexahistidine tag was added by PCR to the C-termini. The plasmids were expressed in bacteria grown in LB medium with Ampicillin at a concentration of 10 ug/mL for 3 hours and induced with 1 mM IPTG for 1 hour. For WC-1 aa1-300 and 100-300, a cobalt purification step (Pierce) followed by a subsequent glutathione (Thermo Pierce) step was carried out to obtain full length polypeptides. GST was purified with glutathione resin (Thermo Pierce) alone. Neurospora lysates were cleared by centrifugation at 9,000 g for 10 minutes at 4°C. Purified GST, GST-aa1-300, and aa100-300 on glutathione beads were each incubated with 2 milligrams of cleared Neurospora lysate (SWI1 C-tagged with V5) and rotated for at least 2 hours. The supernatant was removed and the beads were washed three times with the pull-down buffer.
Micrococcal nuclease assays
The micrococcal nuclease assay was performed as described in [31] with modifications. In brief, Neurospora nuclei were isolated from tissues cultured for indicated dark time. For each sample, 80 µgs of nuclei were digested with micrococcal nuclease (Takara) at the final concentration of 0.1 unit/ml for 1.5 min at 37 degrees. The digestion reaction was stopped by adding a buffer containing 0.2 mg/ml protease K and incubated at 37 degrees overnight. Chromatin DNA was extracted using Gentra Puregene Cell Kit (Qiagen) and cut with NcoI. Southern blot was carried out with a digoxigenin-labeled probe as described in [31].
Other techniques
Chromatin immunoprecipitation experiments were done as previously described [31]. Mass Spectrometry was performed as previously described [64]. Luciferase assays were performed as previously described [40]. Nuclear preparations and MNase digestions were performed as reported [31], [65].
Supporting Information
Zdroje
1. FroehlichAC, LorosJJ, DunlapJC (2003) Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc Natl Acad Sci U S A 100 : 5914–5919.
2. FroehlichAC, LiuY, LorosJJ, DunlapJC (2002) White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297 : 815–819.
3. LeeK, LorosJJ, DunlapJC (2000) Interconnected feedback loops in the Neurospora circadian system. Science 289 : 107–110.
4. ShiM, CollettM, LorosJJ, DunlapJC (2010) FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock. Genetics 184 : 351–361.
5. ChengP, HeQ, WangL, LiuY (2005) Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev 19 : 234–241.
6. AronsonBD, JohnsonKA, LorosJJ, DunlapJC (1994) Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science 263 : 1578–1584.
7. SchafmeierT, HaaseA, KaldiK, ScholzJ, FuchsM, et al. (2005) Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell 122 : 235–246.
8. HeQ, ChaJ, LeeHC, YangY, LiuY (2006) CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev 20 : 2552–2565.
9. HeQ, ChengP, YangY, WangL, GardnerKH, et al. (2002) White collar-1, a DNA binding transcription factor and a light sensor. Science 297 : 840–843.
10. ChengP, YangY, WangL, HeQ, LiuY (2003) WHITE COLLAR-1, a multifunctional neurospora protein involved in the circadian feedback loops, light sensing, and transcription repression of wc-2. J Biol Chem 278 : 3801–3808.
11. BallarioP, VittoriosoP, MagrelliA, TaloraC, CabibboA, et al. (1996) White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. Embo Journal 15 : 1650–1657.
12. VignaliM, HassanAH, NeelyKE, WorkmanJL (2000) ATP-dependent chromatin-remodeling complexes. Mol Cell Biol 20 : 1899–1910.
13. FlausA, Owen-HughesT (2001) Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev 11 : 148–154.
14. ClapierCR, CairnsBR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78 : 273–304.
15. de la SernaIL, OhkawaY, ImbalzanoAN (2006) Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet 7 : 461–473.
16. SudarsanamP, IyerVR, BrownPO, WinstonF (2000) Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 97 : 3364–3369.
17. LaurentBC, TreichI, CarlsonM (1993) The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev 7 : 583–591.
18. DrorV, WinstonF (2004) The Swi/Snf chromatin remodeling complex is required for ribosomal DNA and telomeric silencing in Saccharomyces cerevisiae. Mol Cell Biol 24 : 8227–8235.
19. SternM, JensenR, HerskowitzI (1984) Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol 178 : 853–868.
20. PetersonCL, HerskowitzI (1992) Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68 : 573–583.
21. ZhaoJ, Herrera-DiazJ, GrossDS (2005) Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol Cell Biol 25 : 8985–8999.
22. AbramsE, NeigebornL, CarlsonM (1986) Molecular analysis of SNF2 and SNF5, genes required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol 6 : 3643–3651.
23. LaurentBC, TreitelMA, CarlsonM (1990) The SNF5 protein of Saccharomyces cerevisiae is a glutamine - and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol Cell Biol 10 : 5616–5625.
24. CoteJ, PetersonCL, WorkmanJL (1998) Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci U S A 95 : 4947–4952.
25. QuinnJ, FyrbergAM, GansterRW, SchmidtMC, PetersonCL (1996) DNA-binding properties of the yeast SWI/SNF complex. Nature 379 : 844–847.
26. SudarsanamP, WinstonF (2000) The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet 16 : 345–351.
27. NeelyKE, HassanAH, WallbergAE, StegerDJ, CairnsBR, et al. (1999) Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell 4 : 649–655.
28. KingstonRE, NarlikarGJ (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev 13 : 2339–2352.
29. PetersonCL, WorkmanJL (2000) Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev 10 : 187–192.
30. EtchegarayJP, LeeC, WadePA, ReppertSM (2003) Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421 : 177–182.
31. BeldenWJ, LorosJJ, DunlapJC (2007) Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell 25 : 587–600.
32. BeldenWJ, LewisZA, SelkerEU, LorosJJ, DunlapJC (2011) CHD1 remodels chromatin and influences transient DNA methylation at the clock gene frequency. PLoS Genet 7: e1002166.
33. GarceauNY, LiuY, LorosJJ, DunlapJC (1997) Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89 : 469–476.
34. LeeK, DunlapJC, LorosJJ (2003) Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa. Genetics 163 : 103–114.
35. NatarajanK, JacksonBM, ZhouH, WinstonF, HinnebuschAG (1999) Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol Cell 4 : 657–664.
36. Owen-HughesT, UtleyRT, CoteJ, PetersonCL, WorkmanJL (1996) Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science 273 : 513–516.
37. YudkovskyN, LogieC, HahnS, PetersonCL (1999) Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev 13 : 2369–2374.
38. NeelyKE, HassanAH, BrownCE, HoweL, WorkmanJL (2002) Transcription activator interactions with multiple SWI/SNF subunits. Mol Cell Biol 22 : 1615–1625.
39. BeldenWJ, LarrondoLF, FroehlichAC, ShiM, ChenCH, et al. (2007) The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev 21 : 1494–1505.
40. LarrondoLF, LorosJJ, DunlapJC (2012) High-resolution spatiotemporal analysis of gene expression in real time: in vivo analysis of circadian rhythms in Neurospora crassa using a FREQUENCY-luciferase translational reporter. Fungal Genet Biol 49 : 681–683.
41. BurnsLG, PetersonCL (1997) The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol 17 : 4811–4819.
42. GregoryPD, SchmidA, ZavariM, MunsterkotterM, HorzW (1999) Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. Embo Journal 18 : 6407–6414.
43. KingstonRE, BunkerCA, ImbalzanoAN (1996) Repression and activation by multiprotein complexes that alter chromatin structure. Genes & Development 10 : 905–920.
44. CosmaMP, TanakaT, NasmythK (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle - and developmentally regulated promoter. Cell 97 : 299–311.
45. KwonH, ImbalzanoAN, KhavariPA, KingstonRE, GreenMR (1994) Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature 370 : 477–481.
46. WangW, CoteJ, XueY, ZhouS, KhavariPA, et al. (1996) Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J 15 : 5370–5382.
47. NieZ, XueY, YangD, ZhouS, DerooBJ, et al. (2000) A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol 20 : 8879–8888.
48. XueYT, CanmanJC, LeeCS, NieZQ, YangDF, et al. (2000) The human SWI/SNF-B chromatin-remodeling complex is related to yeast Rsc and localizes at kinetochores of mitotic chromosomes. Proceedings of the National Academy of Sciences of the United States of America 97 : 13015–13020.
49. LemonB, InouyeC, KingDS, TjianR (2001) Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414 : 924–928.
50. ProchassonP, NeelyKE, HassanAH, LiB, WorkmanJL (2003) Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Molecular Cell 12 : 983–990.
51. Bazett-JonesDP, CoteJ, LandelCC, PetersonCL, WorkmanJL (1999) The SWI/SNF complex creates loop domains in DNA and polynucleosome arrays and can disrupt DNA-histone contacts within these domains. Mol Cell Biol 19 : 1470–1478.
52. ZhangY, SmithCL, SahaA, GrillSW, MihardjaS, et al. (2006) DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell 24 : 559–568.
53. LiG, RuanX, AuerbachRK, SandhuKS, ZhengM, et al. (2012) Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148 : 84–98.
54. CaiS, LeeCC, Kohwi-ShigematsuT (2006) SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 38 : 1278–1288.
55. NiZ, Abou El HassanM, XuZ, YuT, BremnerR (2008) The chromatin-remodeling enzyme BRG1 coordinates CIITA induction through many interdependent distal enhancers. Nat Immunol 9 : 785–793.
56. ToyotaK, OnaiK, NakashimaH (2002) A new wc-1 mutant of Neurospora crassa shows unique light sensitivity in the circadian conidiation rhythm. Molecular Genetics and Genomics 268 : 56–61.
57. ChenCH, DeMayBS, GladfelterAS, DunlapJC, LorosJJ (2010) Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proceedings of the National Academy of Sciences of the United States of America 107 : 16715–16720.
58. MalzahnE, CiprianidisS, KaldiK, SchafmeierT, BrunnerM (2010) Photoadaptation in Neurospora by competitive interaction of activating and inhibitory LOV domains. Cell 142 : 762–772.
59. BrennaA, GrimaldiB, FileticiP, BallarioP (2012) Physical association of the WC-1 photoreceptor and the histone acetyltransferase NGF-1 is required for blue light signal transduction in Neurospora crassa. Molecular Biology of the Cell 23 : 3863–3872.
60. MichaelTP, ParkS, KimTS, BoothJ, ByerA, et al. (2007) Simple sequence repeats provide a substrate for phenotypic variation in the Neurospora crassa circadian clock. PLoS One 2: e795.
61. ChaJ, ZhouM, LiuY (2013) CATP is a critical component of the Neurospora circadian clock by regulating the nucleosome occupancy rhythm at the frequency locus. EMBO Rep 14 : 923–930.
62. CrosthwaiteSK, LorosJJ, DunlapJC (1995) Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell 81 : 1003–1012.
63. ColotHV, ParkG, TurnerGE, RingelbergC, CrewCM, et al. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103 : 10352–10357.
64. BakerCL, KettenbachAN, LorosJJ, GerberSA, DunlapJC (2009) Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell 34 : 354–363.
65. HongCI, RuoffP, LorosJJ, DunlapJC (2008) Closing the circadian negative feedback loop: FRQ-dependent clearance of WC-1 from the nucleus. Genes Dev 22 : 3196–3204.
Štítky
Genetika Reprodukčná medicína
Článek An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of MovementČlánek Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced ActivityČlánek Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene NetworksČlánek Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS LocusČlánek tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa CellsČlánek Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced ApoptosisČlánek A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II FidelityČlánek The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites inČlánek Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell SenescenceČlánek BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct MechanismsČlánek Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Translational Regulation of the Post-Translational Circadian Mechanism
- An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of Movement
- Eliminating Both Canonical and Short-Patch Mismatch Repair in Suggests a New Meiotic Recombination Model
- Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced Activity
- Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene Networks
- Targeted H3R26 Deimination Specifically Facilitates Estrogen Receptor Binding by Modifying Nucleosome Structure
- Role for Circadian Clock Genes in Seasonal Timing: Testing the Bünning Hypothesis
- The Tandem Repeats Enabling Reversible Switching between the Two Phases of β-Lactamase Substrate Spectrum
- The Association of the Vanin-1 N131S Variant with Blood Pressure Is Mediated by Endoplasmic Reticulum-Associated Degradation and Loss of Function
- Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS Locus
- Regulation of Flowering by the Histone Mark Readers MRG1/2 via Interaction with CONSTANS to Modulate Expression
- The Actomyosin Machinery Is Required for Retinal Lumen Formation
- Plays a Conserved Role in Assembly of the Ciliary Motile Apparatus
- Hidden Diversity in Honey Bee Gut Symbionts Detected by Single-Cell Genomics
- Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria
- tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa Cells
- Causal Variation in Yeast Sporulation Tends to Reside in a Pathway Bottleneck
- Tissue-Specific RNA Expression Marks Distant-Acting Developmental Enhancers
- WC-1 Recruits SWI/SNF to Remodel and Initiate a Circadian Cycle
- Clonal Expansion of Early to Mid-Life Mitochondrial DNA Point Mutations Drives Mitochondrial Dysfunction during Human Ageing
- Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels
- Differential Management of the Replication Terminus Regions of the Two Chromosomes during Cell Division
- Obesity-Linked Homologues and Establish Meal Frequency in
- Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced Apoptosis
- Stress-Induced Nuclear RNA Degradation Pathways Regulate Yeast Bromodomain Factor 2 to Promote Cell Survival
- The MAPK p38c Regulates Oxidative Stress and Lipid Homeostasis in the Intestine
- Widespread Genome Reorganization of an Obligate Virus Mutualist
- Trans-kingdom Cross-Talk: Small RNAs on the Move
- The Vip1 Inositol Polyphosphate Kinase Family Regulates Polarized Growth and Modulates the Microtubule Cytoskeleton in Fungi
- Myosin Vb Mediated Plasma Membrane Homeostasis Regulates Peridermal Cell Size and Maintains Tissue Homeostasis in the Zebrafish Epidermis
- GLD-4-Mediated Translational Activation Regulates the Size of the Proliferative Germ Cell Pool in the Adult Germ Line
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Translational Regulation of the DOUBLETIME/CKIδ/ε Kinase by LARK Contributes to Circadian Period Modulation
- Positive Selection and Multiple Losses of the LINE-1-Derived Gene in Mammals Suggest a Dual Role in Genome Defense and Pluripotency
- Out of Balance: R-loops in Human Disease
- A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II Fidelity
- Altered Behavioral Performance and Live Imaging of Circuit-Specific Neural Deficiencies in a Zebrafish Model for Psychomotor Retardation
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Meta-analysis of Mutations in Autism Spectrum Disorders: A Gradient of Severity in Cognitive Impairments
- The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites in
- Hydroxymethylated Cytosines Are Associated with Elevated C to G Transversion Rates
- Memory and Fitness Optimization of Bacteria under Fluctuating Environments
- Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell Senescence
- Interspecific Tests of Allelism Reveal the Evolutionary Timing and Pattern of Accumulation of Reproductive Isolation Mutations
- PRO40 Is a Scaffold Protein of the Cell Wall Integrity Pathway, Linking the MAP Kinase Module to the Upstream Activator Protein Kinase C
- Low Levels of p53 Protein and Chromatin Silencing of p53 Target Genes Repress Apoptosis in Endocycling Cells
- SPDEF Inhibits Prostate Carcinogenesis by Disrupting a Positive Feedback Loop in Regulation of the Foxm1 Oncogene
- RRP6L1 and RRP6L2 Function in Silencing Regulation of Antisense RNA Synthesis
- BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct Mechanisms
- Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
- Atkinesin-13A Modulates Cell-Wall Synthesis and Cell Expansion in via the THESEUS1 Pathway
- Dopamine Signaling Leads to Loss of Polycomb Repression and Aberrant Gene Activation in Experimental Parkinsonism
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
- Bipartite Recognition of DNA by TCF/Pangolin Is Remarkably Flexible and Contributes to Transcriptional Responsiveness and Tissue Specificity of Wingless Signaling
- The Olfactory Transcriptomes of Mice
- Muscular Dystrophy-Associated and Variants Disrupt Nuclear-Cytoskeletal Connections and Myonuclear Organization
- Interplay of dFOXO and Two ETS-Family Transcription Factors Determines Lifespan in
- Evidence for Widespread Positive and Negative Selection in Coding and Conserved Noncoding Regions of
- Genome-Wide Association Meta-analysis of Neuropathologic Features of Alzheimer's Disease and Related Dementias
- Rejuvenation of Meiotic Cohesion in Oocytes during Prophase I Is Required for Chiasma Maintenance and Accurate Chromosome Segregation
- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Local Effect of Enhancer of Zeste-Like Reveals Cooperation of Epigenetic and -Acting Determinants for Zygotic Genome Rearrangements
- Differential Responses to Wnt and PCP Disruption Predict Expression and Developmental Function of Conserved and Novel Genes in a Cnidarian
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



