-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep
Animal and human studies suggest that sleep has a profound impact on learning and memory. However, little is known about the molecular pathways linking these phenomena. We report that mutations in the Drosophila Anaplastic lymphoma kinase (Alk) gene, an ortholog of a human oncogene ALK, cause increased sleep. ALK is required for sleep suppression in the mushroom body, a structure important for both sleep and memory. ALK generally activates the Ras/ERK pathway, which is negatively regulated by Neurofibromin 1 (NF1). Mutations in Nf1 are the causes of the common neurological disorder Neurofibromatosis type 1 (NF1), which affects 1 in 3,000 live births. We find that male flies lacking the NF1 protein have reduced sleep, a phenotype opposite that of Alk flies. Interestingly, even though mutations in Nf1 don’t always cause short sleep in female flies, they suppress the sleep increase induced by ALK inactivation. Previous studies have shown that Alk and Nf1 play antagonistic roles in learning and that both genes regulate synaptic growth. Thus Alk and Nf1 interact to regulate both sleep and learning, suggesting that the two processes share a common pathway. Our results support a model in which changes in synaptic plasticity during sleep promote learning and memory.
Published in the journal: Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep. PLoS Genet 11(11): e32767. doi:10.1371/journal.pgen.1005611
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005611Summary
Animal and human studies suggest that sleep has a profound impact on learning and memory. However, little is known about the molecular pathways linking these phenomena. We report that mutations in the Drosophila Anaplastic lymphoma kinase (Alk) gene, an ortholog of a human oncogene ALK, cause increased sleep. ALK is required for sleep suppression in the mushroom body, a structure important for both sleep and memory. ALK generally activates the Ras/ERK pathway, which is negatively regulated by Neurofibromin 1 (NF1). Mutations in Nf1 are the causes of the common neurological disorder Neurofibromatosis type 1 (NF1), which affects 1 in 3,000 live births. We find that male flies lacking the NF1 protein have reduced sleep, a phenotype opposite that of Alk flies. Interestingly, even though mutations in Nf1 don’t always cause short sleep in female flies, they suppress the sleep increase induced by ALK inactivation. Previous studies have shown that Alk and Nf1 play antagonistic roles in learning and that both genes regulate synaptic growth. Thus Alk and Nf1 interact to regulate both sleep and learning, suggesting that the two processes share a common pathway. Our results support a model in which changes in synaptic plasticity during sleep promote learning and memory.
Introduction
Sleep behavior is conserved from worms and insects to fish and mammals [1]. Why animals spend a large amount of time seemingly doing nothing, passing on opportunities to forage, hunt or mate, and remaining vulnerable to dangers, is still a mystery. It is hypothesized that a major function of sleep is to maintain brain function, in particular, to ensure synaptic homeostasis of neurons and to consolidate memory [2,3]. Levels of synaptic proteins are associated with sleep/wake states in both flies and mammals [4,5], and sleep deprivation impairs memory formation in a variety of species, including humans [6], mice [7], and Drosophila [8,9]. In addition, some molecules that regulate learning and memory turn out to be required for sleep/wake regulation [10]. However, only a handful of such molecules have been identified and for most it is not known if effects on the two processes are mechanistically linked.
Anaplastic lymphoma kinase (Alk), which encodes a member of the ALK/LTK (leucocyte tyrosine kinase)) family of receptor tyrosine kinases (RTKs), is proposed to play important roles in the nervous system based on its extensive expression in the CNS of both mammals and flies [11–14]. Its in vivo functions are mostly studied in the context of Drosophila development. Together with its secreted ligand Jelly Belly (Jeb), ALK is essential for 1) gut muscle differentiation [15,16]; 2) retinal axon targeting in the optic lobe [17]; 3) growth and organ size regulation [14,18]; and 4) modulation of neuromuscular transmission and synapse growth at larval neuromuscular junctions (NMJ) [19]. However, there is also evidence for a role of Alk in brain plasticity in adult contexts. Adult-specific activation of Alk causes deficits in associative olfactory learning in Drosophila; concordantly, reducing neuronal Alk activity in adult flies enhances olfactory learning [14]. Similarly in mice, loss of ALK function enhances spatial memory and novel object recognition, and reduces anxiety and depression [20,21]. Effects of Alk on learning, at least in flies, are most likely mediated by Ras/ERK signaling. This is supported by an interaction with Nf1, conserved ortholog of the human Neurofibromatosis type 1 (NF1) disease gene, which encodes a GTPase-activating protein (GAP) that negatively regulates Ras/ERK signaling. Specifically, the learning deficit in Nf1 flies is rescued by down regulation of Alk [14].
Based on the role of Alk in neuronal plasticity and learning, we hypothesized that Alk may be involved in sleep regulation. We found that inactivation of ALK causes increased sleep. We probed for the neuronal circuit that underlies Alk’s involvement in sleep and found that inhibiting Alk in the mushroom body induces more sleep, suggesting Alk as a mechanistic link between learning and sleep. In addition, Alk interacts with Nf1 in the regulation of sleep just as it does in the context of learning. Nf1 also have circadian phenotypes [22], but Alk is not required for circadian rhythms, nor does it interact with Nf1 in the circadian regulation of rest/activity rhythms. Thus, interactions between the two molecules are specific for sleep and learning.
Results
Alk mutants have increased sleep
Because ALK plays a crucial role in gut development, the null allele Alk1 is homozygous lethal at the early larval stage [23]. However, a temperature-sensitive allele, Alkts, fully complements Alk1 at 18°C, but fails to complement developmental Alk1 lethality at 29°C [19]. We were therefore able to raise Alkts/1 trans-heterozygous or Alkts homozygous flies to the adult stage at 18°C and prevent developmental phenotypes such as changes in body length (S1 Fig). Alk mutants raised in this manner are presumably also spared other developmental defects seen with manipulations of ALK activity, such as altered NMJ structure and function [19] and mis-targeting of retinal neurons in the optic lobes [17].
We assayed sleep using the traditional single infrared beam interruption device (Trikinetics, MA) in adult Alkts/1 and Alkts female flies at the permissive temperature of 18°C and at the restrictive temperature 29°C. Because genetic background has a profound impact on sleep [24], Alk mutants were backcrossed for five generations into a white (w) isogenic background, iso31, a line generated specifically for use in behavioral experiments [25]. At 18°C, control iso31 and Alk flies had very similar sleep patterns. However, acute inhibition of Alk by switching the environmental temperature to 29°C drastically increased sleep in Alk flies as compared to iso31. In iso31 flies, the shift to 29°C initially increased daytime sleep and decreased nighttime sleep, consistent with previous reports of increased siesta at higher temperatures [26]; overall sleep increased on the third day, but there was no net change in sleep over the three day period. Inhibition of Alk increased both day and night sleep. This temperature-sensitive sleep phenotype was reversed by lowering the temperature back to 18°C (Fig 1A). Quantification shows that Alkts/1 flies slept ~51.06±10.09% more than the control iso31 flies during the high temperature shift (6 independent experiments, n = 87 for iso31 and n = 80 for Alkts/1). Similarly, Alkts homozygous flies and flies that harbor an Alkts allele over a deficiency uncovering the Alk gene slept more than control flies at 29°C. There was no difference in total sleep amount between Alkts/1, Alkts, or Alkts/Def flies, suggesting that the restrictive temperature completely abolished ALKts protein function (Fig 1B). Importantly, we were able to rescue the sleep phenotype of Alk mutants by re-expressing Alk transgenically (discussed below).
Fig. 1. Alk mutants have increased sleep. 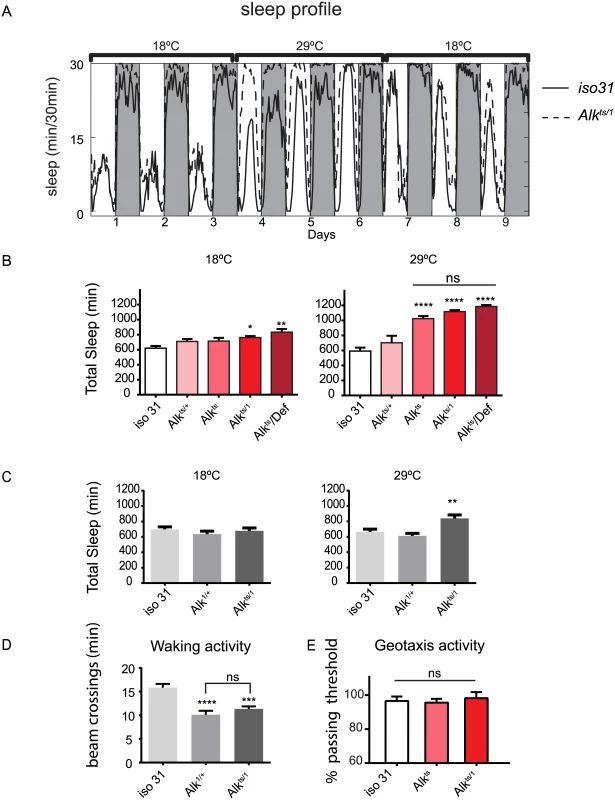
A) The restrictive temperature of 29°C reversibly increases sleep in Alkts/1 mutants. The averaged sleep profiles, plotted as average amounts of sleep in every 30-minute period, are shown for iso31 and Alkts/1 female flies. The white and the grey columns mark periods of day and night, respectively. N = 16. B) Quantification of average daily sleep of control flies and Alk mutants measured by single beam monitors. Total sleep at 18°C was calculated as the average of 3 days before the temperature shift to 29°C. Total sleep amounts at 29°C were calculated as the average of the 3 days following the temperature shift and are significantly different between genotypes (One-way ANOVA, p<0.0001). Asterisk* signifies difference from iso31 control. In this figure and all following figures, *, p<0.05; **,p<0.01; ****,p<0.0001; ns, not significantly different. Error bars are SEM. N = 13–16. C) Measurement by multi-beam monitors similarly revealed longer sleep in Alkts/1 flies at the restrictive temperature. There is no difference between genotypes at 18°C (p = 0.4722), while at 29°C Alkts/1 sleep significantly longer than iso31 and Alk1/+ flies. N = 15–16. D) Waking activity in the multi-beam monitors was measured at the restrictive temperature of 29°C and was defined as averaged number of beam crossings per minute during wake. N = 15–16. E) Normal negative geotaxis response in Alk mutants. The negative geotaxis response is measured as the percentage of flies climbing vertically >4 cm from the bottom of a vial within 10s of being tapped down. N = 5 (groups of 10 flies for each genotype). Though sleep profiles and sleep metrics produced by the standard single infrared beam sleep monitors are reliable and have been published widely, they sometimes overestimate sleep as they miss fly movement away from the infrared beam; the degree of error varies from one genotype to another [27,28]. We therefore assayed sleep in a new multi-beam sleep monitor (Trikinetics, MA), which provides an order-of-magnitude higher spatial resolution compared to the traditional single beam monitors. Measurements by multi-beam monitors validated our results from the single beam method, showing that Alkts/1 females had increased daytime and nighttime sleep at 29°C as compared to iso31 control flies (Fig 1C and S2 Fig). However the long sleep phenotype was less pronounced when assayed with multi-beam monitors, with Alkts/1 female flies showing a 32.3±3.3% increase over iso31 (3 independent experiments, 47 iso31 flies and 48 Alkts/1flies). We also assayed sleep in Alk male flies with both monitor systems, and found that it was similarly increased (S3 Fig). Henceforth we focused our analysis on female flies, typically the gender studied in Drosophila sleep experiments. To exclude the possibility that the apparent increase in sleep was due to locomotion impairment, we measured waking activity, calculated as the average number of beam crossings per waking minute. We found that it did not account for the long sleep phenotype as it was reduced in both long-sleeping Alkts/1 flies as well as normal-sleep Alk1/+ controls (Fig 1D). We also assayed the mobility of Alk flies in an independent negative geotaxis assay that measures the ability of flies to climb vertically when startled [29]. The response of Alkts/1 flies was indistinguishable from that of iso31 flies (Fig 1E), suggesting that Alkts/1 flies have no gross motor defects. To confirm that the increased inactivity in Alk mutants is genuine sleep, as opposed to quiet wake, we assayed their arousability to a mechanical stimulus at different times of day (Fig 2). Similar percentages of previously sleeping iso31 and Alkts flies were aroused by the stimulus at ZT6 and ZT20, when most flies were sleeping. At ZT22, more iso31 flies were aroused than Alkts flies, probably because iso31 flies were transitioning from sleep to wake at this time point. Notably, a higher percentage of the previously awake flies exhibited activity after the stimulus than sleeping flies at all three time points, suggesting that arousal threshold was indeed higher in sleeping flies. We conclude that Alk mutants are bona fide long sleepers, suggesting that ALK functions to inhibit sleep or promote wake.
Fig. 2. Inactivity of Alkts mutants is due to a prolonged sleep state. 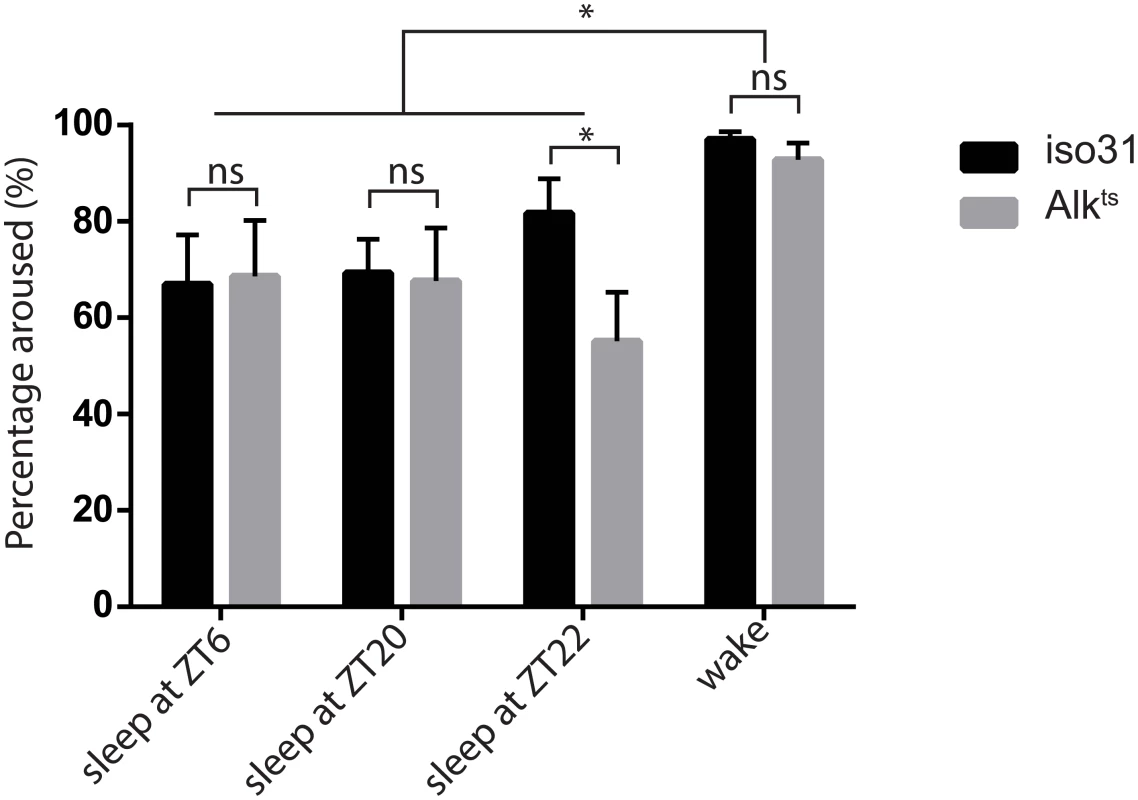
To distinguish sleep from quiet wake, we subjected previously sleeping or awake flies to a mechanical stimulus at different times of day—ZT6, ZT20 and ZT22. Across all time points, two-way ANOVA found significant differences in the response to simulation between previous behavior states (p = 0.0071) but not between genotypes (p = 0.189). There was no interaction between behavior states and genotypes (p = 0.368). However, Alk flies were less arousable than wild type at ZT22, by Student’s t test. n = 4–5 trials of 26–32 flies of each genotype for all time points. The responses of previously awake flies were similar between the three time points and thus were pooled. Alk mutants show a normal sleep homeostatic response
Following a period of sleep deprivation, flies, like other animals, show a homeostatic response in the form of increased sleep [30]. We wondered whether this homeostatic regulation was disrupted in Alk mutants. We found that after 6 hours of sleep deprivation by mechanical stimulation, both iso31 and Alkts flies increased sleep the following morning (Fig 3A). However, sleep-deprived Alkts flies fell asleep faster than iso31 controls, suggesting sleep pressure was higher in Alkts flies (Fig 3A and 3D).
Fig. 3. Homeostatic response to sleep loss in Alk mutants. 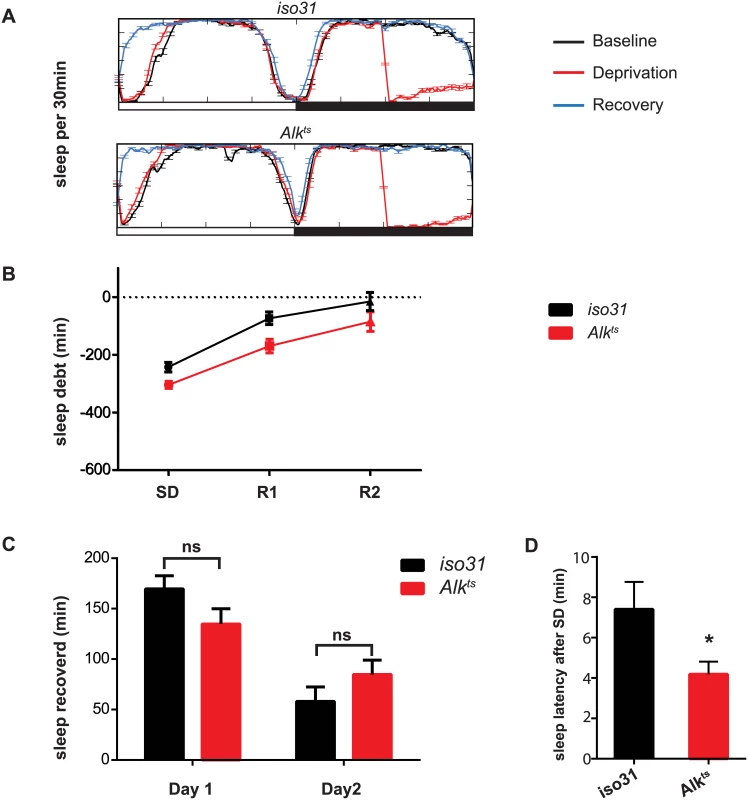
A) Sleep profiles for the baseline, deprivation and recovery day were plotted against each other to show sleep deprivation and sleep rebound. The experiment was conducted entirely at 29°C. Iso31 and Alkts similarly show an increase in sleep in the morning following sleep deprivation. B) Time course of recovery of sleep lost following deprivation. C) Minutes of sleep recovered on the first and second recovery day were compared between iso31 and Alkts flies. 2-tailed Student’s t-test shows no significant difference between iso31 and Alkts flies. D) Sleep latency after deprivation on recovery day one. *p<0.05. N = 31 for iso31. N = 29 for Alkts. Inhibition of Alk in various brain regions has differential effects on sleep
Similar to the expression pattern of the mouse ALK gene, the Drosophila Alk gene is extensively expressed in the developing and adult nervous system [12,14,23]. Its adult expression includes the mushroom body, the protocerebral bridge, the antennal lobes, the suboesophageal ganglion, the medial bundle and lateral horns [14]. To locate the sleep regulatory function of Alk in the brain, we carried out a brain mini-screen using a series of GAL4 lines to drive UAS-Alk RNA interference (RNAi) in brain circuits. Inducing Alk RNAi with a pan-neuronal driver, elav-GAL4, reduced Alk mRNA level to ~35% of that of control flies and produced longer sleep, suggesting that ALK functions in neurons (S4 Fig). We then screened over 40 GAL4 drivers with diverse neuronal expression patterns (S1 Table and S5 Fig), including those with expression patterns in the known sleep/wake regulating regions [31–39]. 13 GAL4 lines significantly increased sleep relative to controls, when driving Alk RNAi, while c309-Gal4-driven expression of Alk RNAi decreased sleep (Fig 4A). Of the identified sleep-promoting GAL4 lines, most are broadly expressed, overlapping in several regions of the brain, such as the mushroom body, the ellipsoid body, and the pars intercerebralis. However, inhibiting Alk specifically in the pars intercerebralis (InSITE106, Kurs58 and Dilp2-GAL4), or the ellipsoid body (c819, c232 and c107) did not alter total sleep. The amounts of sleep increase as well as the sleep profiles were different between different GAL4 lines (S6 Fig). Most lines showed increased daytime sleep as well as nighttime sleep. However, lines 1471, c320 and 386Y mainly increased daytime sleep and 7Y mostly increased night-time. 386Y caused a delay in sleep at the beginning of the night while driving an increase at other times. c320 increased sleep immediately after lights-on in the morning. These sleep patterns were consistent across repeated experiments.
Fig. 4. Alk is required in a subset of CNS neurons to regulate sleep. 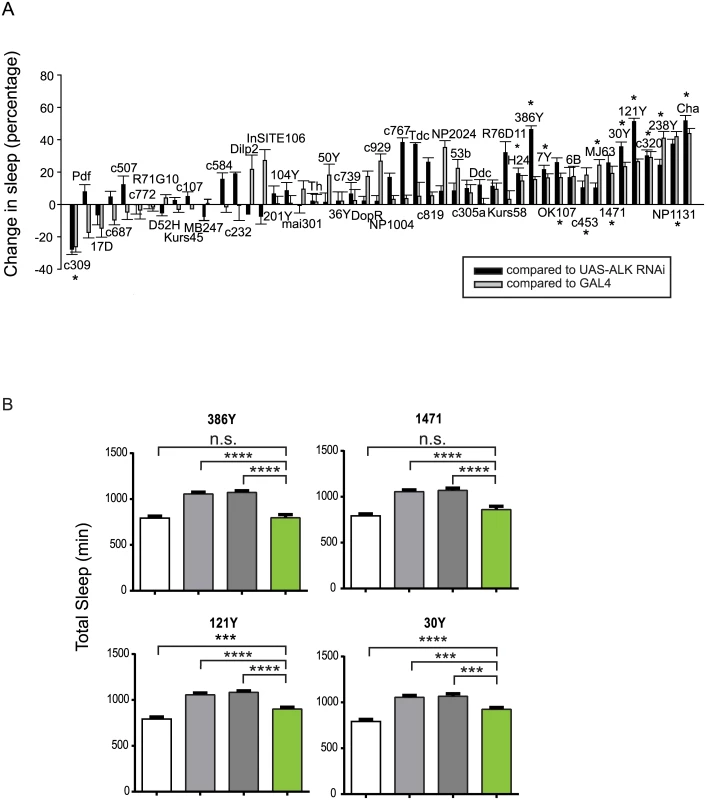
A) A targeted Alk RNAi screen with CNS Gal4 drivers to identify regions where ALK acts to regulate sleep. The graph shows the percentage changes in total sleep as a result of expressing Alk RNAi compared to controls UAS-Alk RNAi/+; Dcr2/+ (black bars) and Gal4/+ (grey bars). The difference in sleep amount between the experimental group and each control group was divided by the amount of control sleep to calculate net percentage change. Bars represent pooled data from 2–4 experiments for each genotype and show means ±SEM (n = 8–58 for experimental groups and each Gal4 control group. N = 131 for the UAS-Alk RNAi control). Daily sleep was averaged over 3 days. One-way ANOVA and post hoc analysis were done to compare total sleep in the Alk RNAi-expressing group to UAS-Alk RNAi and Gal4 control groups. * indicates that total sleep of the RNAi expressing group is significantly different from those of both control group. B) The long sleep phenotype of Alkts at the restrictive temperature can be rescued by expressing Alk in sub-regions of the brain. Genotypes for the four bars: iso31 (white), Alkts, UAS-Alk/Alkts; Tub-Gal80ts/+ (light grey), Alkts; Gal4/+ (dark grey), Alkts, UAS-Alk/Alkts; Gal4/tub-Gal80 (green). The white and light grey controls are the same in these graphs. N = 21–46. Total sleep amounts were averaged for 3 days at 29°C. One-way ANOVA and post hoc Turkey’s test were performed for all pairwise comparisons. In all groups, grey bars (Alkts controls) are significantly higher than the white bar (****p<0.0001), which is not indicated in the graph. We found that the long sleep phenotype of Alk mutants could be rescued by restoring functional ALK with targeted drivers that we identified through the RNAi screen. To circumvent the developmental effects of overexpressing Alk, we expressed Alk in adult Alkts mutants only during a period of restrictive temperature at 29° by combining GAL4s with a temperature-sensitive form of GAL80 (tub-Gal80ts) [40]. We tested 386Y, 7Y, 121Y, 1471, 30Y and MJ63, all of which induce longer sleep when driving Alk RNAi. We found that 386Y and 1471 fully rescued the long sleep phenotype of Alkts at high temperature, while 30y and121y produced a partial rescue (Fig 4B). 7Y and MJ63, however, did not rescue the long sleep of Alkts, and neither did any of the drivers that yielded no phenotype with Alk RNAi (S7 Fig). We infer that while Alk expression is necessary, it may not be sufficient for sleep regulation in regions of 7Y and MJ63 expression. The rescue results further confirm that the long sleep phenotype is specific to the Alk gene and involves specific brain regions.
Alk negatively regulates sleep in a subset of mushroom body neurons
Prompted by the extensive mushroom body expression of most driver hits in our screen, we focused on the function of ALK in the mushroom bodies (MB). When Alk RNAi was excluded from the MB by combining GAL4s with a mushroom body-specific Gal80 transgene (MB-Gal80)[41](Fig 5), it eliminated the sleep increase induced by 386Y and 30Y, suggesting that Alk function is required in mushroom body neurons labeled by these two drivers. In fact, when 30Y was combined with MB-Gal80, it decreased sleep to below those of controls. c309, a driver that caused short sleep with Alk RNAi, also has extensive mushroom body expression. Driving Alk RNAi with c309/MB-Gal80, however, decreased sleep further compared to Alk RNAi driven by c309. These results indicate that the sleep-increasing effects of Alk deficiency occur mainly in the MBs and with 30Y and c309 they are countered by sleep-inhibiting influences of Alk knockdown outside the mushroom body. We thus tested several drivers that have localized expression in the mushroom body and little expression elsewhere, but did not observe any increase in sleep (Fig 4). Of these three drivers, 17D innervates the core of α/β lobes; D52H has sexually dimorphic expression with strong expression in the α/β and the main γ lobe in males but faint dorsal γ expression in the females, which is the gender we used for sleep assays; R71G10 preferentially innervates the γ lobe and R76D11 has strong expression in both α/β and γ; c305a, which innervates α′/β′ lobes in addition to cells in other brain regions [42,43]. Given that of the positive drivers, H24, NP1131 and 1471 label Kenyon cells that project exclusively in the γ lobes, we hypothesize ALK functions to inhibit sleep in a subset of γ lobe neurons that are not targeted by R71G10 and R76D11.
Fig. 5. ALK functions in the mushroom body to inhibit sleep. 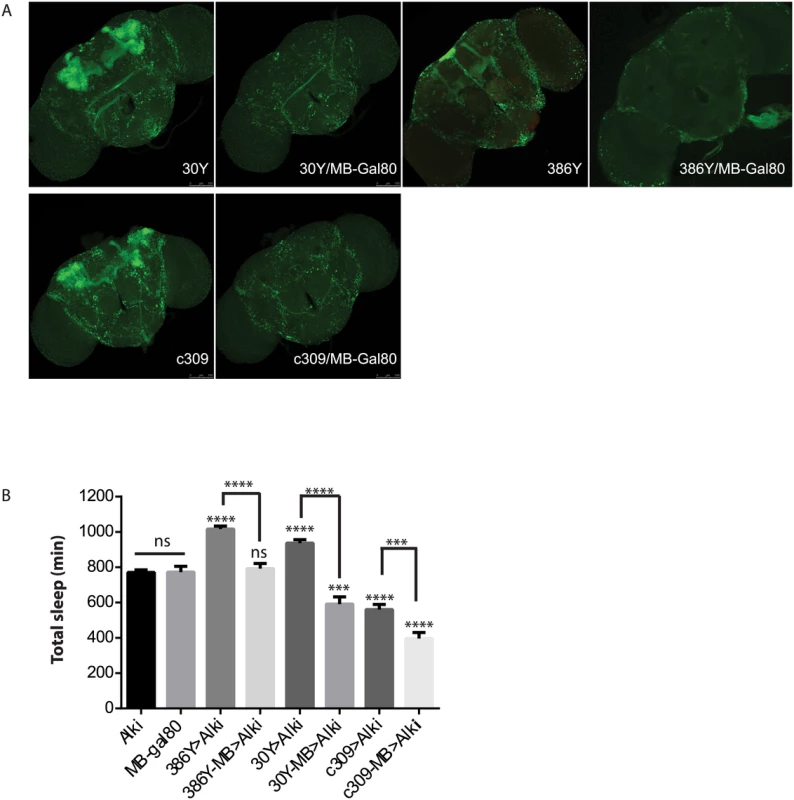
A) MB-Gal80 eliminates mushroom body expression from 30Y, 386Y and c309 Gal4s. B) Alk RNAi-induced sleep increase is suppressed by MB-Gal80. GAL4-MB represents combination of GAL4 and MB-Gal80. * above bars indicate significant differences from both UAS-Alk RNAi and MB-Gal80 controls. Brackets show comparisons between Gal4>Alk RNAi and Gal4-MB>Alk RNAi. ns, not significant, ***p<0.001; ****p<0.0001, by One-way ANOVA and Turkey’s post hoc analysis. n = 18–49. In combination with 30y, MB-Gal80 decreases sleep below control levels. With C309, MB-Gal80 further decreases sleep. UAS-RNAi and MB-gal80, and 386y-MB are not statistically different. Sleep defects in Nf1 mutants
Genetic interactions between Alk and Nf1 in growth and learning processes led us to investigate whether Nf1 is also required for sleep regulation. Interestingly, a prevalence of sleep disturbances has recently been reported in NF1 patients [44,]. We detected considerable variability in total sleep amount in Nf1 mutant flies across experiments. We tested three Nf1 alleles, Nf1P1, Nf1P2, and Nf1c00617. Nf1P1 and Nf1P2 alleles are both assumed to be null because neither expresses NF1 protein and homozygous flies have similar defects in locomotor activity rhythms, body size and learning [22,46, 47]. Although the average total sleep of male flies harboring any two Nf1 alleles was significantly less than that of control flies, we did not observe a consistent difference between Nf1 mutant and control female flies (Fig 6). While Nf1P2/c00617 female flies slept less than controls, sleep amounts in Nf1P1/P2 female flies were generally not different from those of control flies. Furthermore, Nf1P1 and Nf1P2 female flies did not consistently show sleep reduction as compared to iso31 controls in separate experiments. Similar discrepancies were found when sleep was assayed with the multi-beam sleep monitors. However, both Nf1 male and female flies exhibited nocturnal hyperactivity (Fig 6A and 6C), which resulted in an increase in daytime sleep and a decrease in nighttime sleep. Nf1 mutants also showed consistent defects in sleep consolidation. Both daytime sleep and nighttime sleep were highly fragmented in Nf1 males, such that the average sleep bout duration was reduced and the number of sleep bouts increased (S8 Fig). While reductions in average bout duration did not reach significance in females, increased numbers of bouts suggest deficits in sleep maintenance and consolidation. Rescuing Nf1 with pan-neuronal expression of a UAS-Nf1 transgene increased sleep of Nf1 mutants and in fact caused a long sleep phenotype compared to wild-type controls. This did not result from ectopic expression of the transgene as expressing the same UAS-Nf1 transgene in wild-type flies had no effect (Fig 6B). Interestingly, sleep increase was seen with Nf1 rescue in both male and female flies, suggesting that effects of Nf1 on sleep are quite complex.
Fig. 6. Sleep reduction in Nf1 mutants. 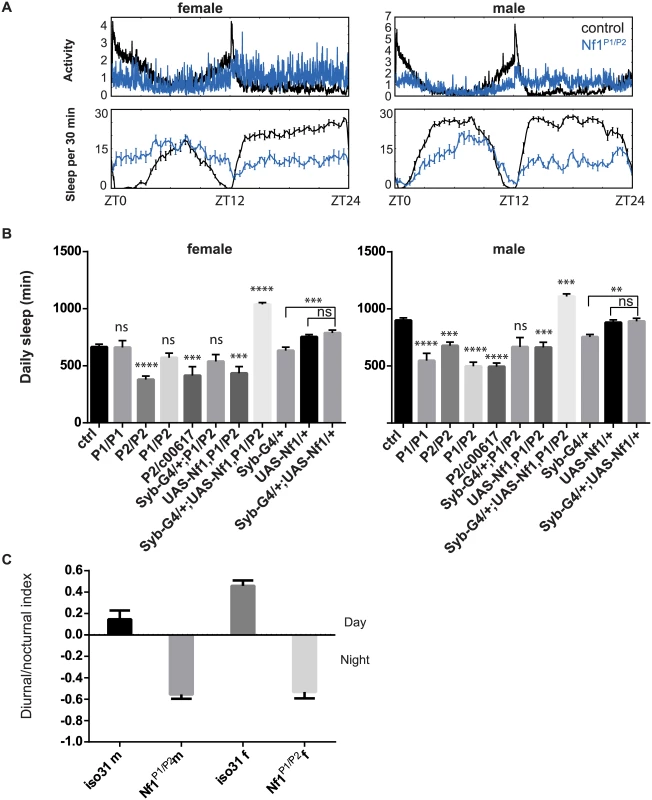
A) Averaged sleep profiles of control iso31 (black) and Nf1P1/P2 (blue) mutant flies at 25°C. B) Average daily sleep for control iso31, Nf1 mutants and Nf1 flies with transgenic Nf1 gene rescue at 25°C. Comparisons were done with one-way ANOVA and post-hoc Turkey’s test. * indicates significant difference from iso31 control. Error bars are SEM. N = 5–16. Ns, not significant. ***p<0.001, ****p<0.0001. Nf1 transgenic expression in wild type background was done in a separate experiment and their sleep quantities were compared with the respective controls. C) Nf1 mutants exhibit nocturnal hyperactivity. Diurnal/nocturnal index was calculated as (total activity during the day) − (total activity during the night)/(total activity), averaged over a 3-d period per fly. Alk interacts functionally with Nf1 to regulate sleep
We then investigated whether Alk and Nf1 interact to regulate sleep. We chose to test Alk with the Nf1P1/P2 allelic combination because Nf1P1/P2 females have normal amount of sleep and so any sleep suppression in the double mutants would not be confounded by additive effects of short-sleeping Nf1 mutants. To sensitize the assay, we compared sleep in Alkts, Alkts/1 or Nf1P1/P2 single mutants and Alkts;Nf1P1/P2 and Alkts/1;Nf1P1/P2 double mutants at three different temperatures that render different dosages of functional ALK. Interestingly, total amounts of sleep in Alk;Nf1 double mutant flies were less than those of flies deficient for Alk alone, and not different from Nf1 single mutants or iso31 control flies (Fig 7). We found that regardless of the severity of the Alk alleles, Nf1P1/P2 completely suppressed the long sleep phenotype. As another piece of evidence for genetic interaction, we found that Nf1P1/P2 also suppressed the long sleep phenotype caused by Alk pan-neural RNAi (S9 Fig). These results suggest that Nf1 interacts with Alk in a sleep regulating circuit.
Fig. 7. Nf1 mutations suppress the long-sleep phenotype of Alk mutants. 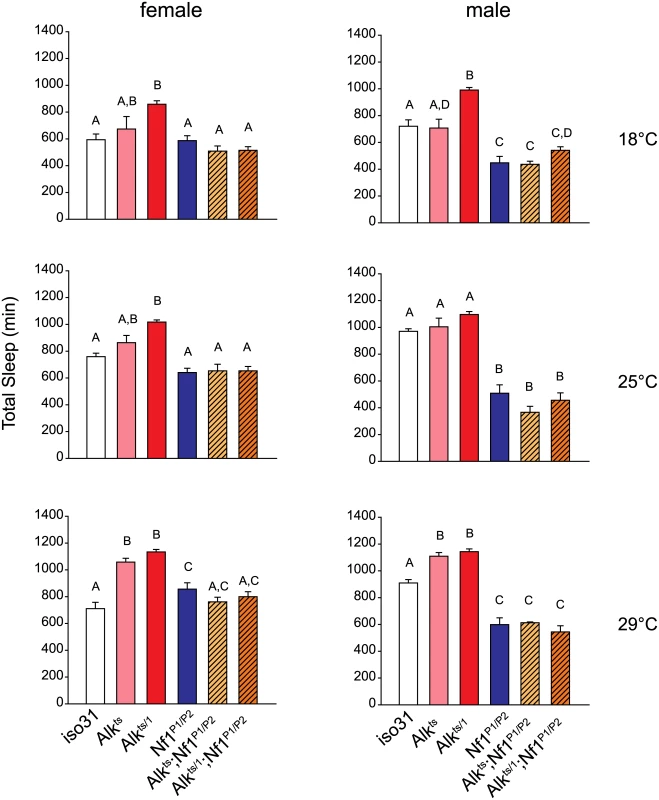
Bar graphs show total sleep for iso31 controls, Alk mutants, Nf1P1/P2 mutants, and Alk; Nf1 double mutants. One-way ANOVA and Mann-Whitney post hoc analysis was performed to compare groups in each condition. In each graph, groups with the same alphabet label on top are not statistically different from each other; groups with different labels are statistically different (p<0.05). For experiments at 18°C and 25°C, n = 5–16. For 29°C experiment, n = 10–37. All flies were raised at 18°C. Activities were monitored at 18°C, 25°C and 29°C in independent experiments. Alk does not interact with Nf1 to control circadian rhythms
Nf1 is part of the circadian output pathway that controls rest: activity rhythms [22]. As Alk was found to interact with Nf1 in sleep regulation, as well as in growth and learning, we asked whether Alk is required also for circadian rhythms and whether Alk and Nf1 interact in circadian pathways. We found that Alkts/1 trans-heterozygotes raised at 18°C maintained locomotor activity rhythms at the restrictive temperature of 29°C in constant darkness (Fig 8 and Table 1), indicating Alk is not required to maintain circadian activity. Indeed, the FFT values, a measure of rhythm strength, of Alkts/1 and Alkts/Def flies were higher at 29°C than at 18°C, suggesting that loss of Alk may actually improve rhythms rather than disrupt them. However, inhibiting Alk failed to rescue the circadian defects in Nf1 flies: Alkts/1; Nf1P1/P2 double mutant were arrhythmic, just like Nf1 single mutants. To exclude a requirement for Alk in the development of circadian circuits, we also tested Alkts homozygous flies raised at 25°C, at which temperature Alkts flies have moderate lethality [19]. The partial reduction in ALK function throughout development did not cause arrhythmia nor did it suppress arrhythmia in Nf1P1/P2 flies (Table 1). These results suggest that Alk does not function in the circadian output circuit regulated by Nf1.
Fig. 8. Alk does not interact with Nf1 to control circadian rhythms. 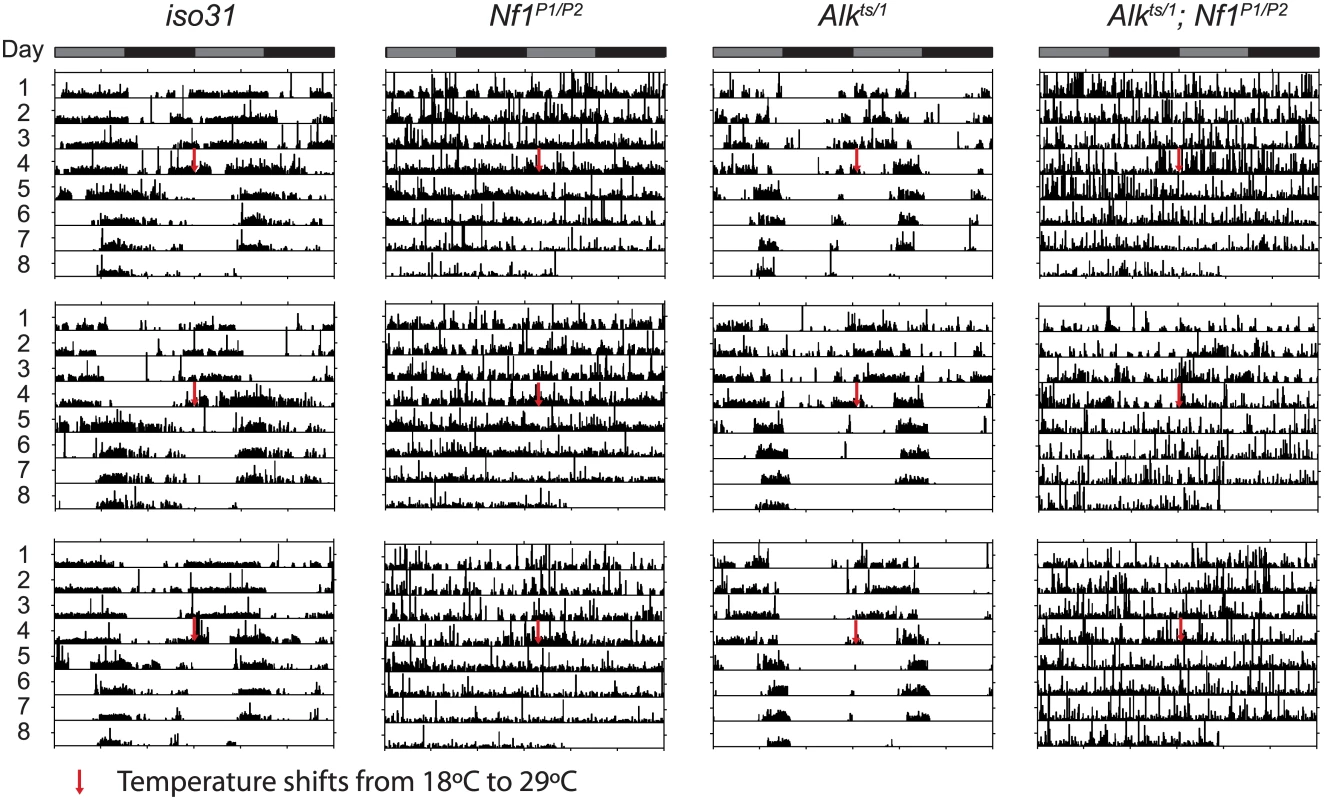
Representative activity graphs for each genotype are shown with activity double-plotted (2 day/night cycles). Gray and black bars at the top indicate subjective days and nights. Flies were raised and entrained at 18°C, monitored for 4 days in constant darkness at 18°C and then at 29°C for 4 days. Upon the temperature shift, iso31 and Alkts/1 flies manifest a phase shift in their activity rhythms. Nf1P1/P2 and Alkts/1;Nf1P1/P2 double mutants are arrhythmic. Tab. 1. Circadian rhythmicity of Alk and Nf1 mutants. 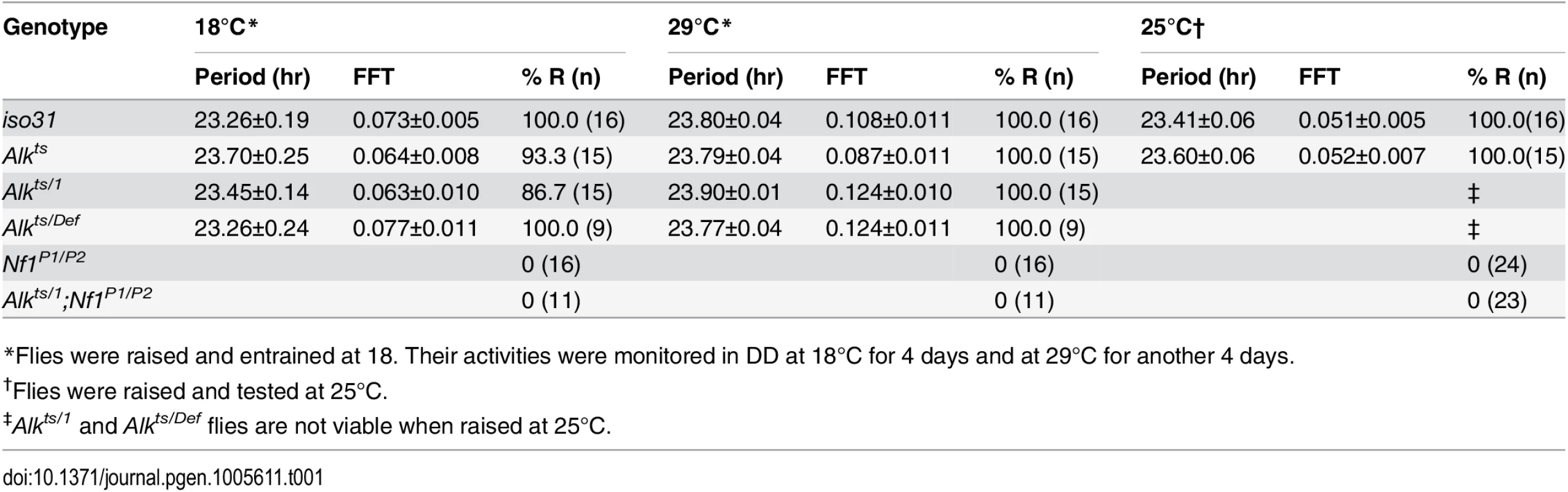
*Flies were raised and entrained at 18. Their activities were monitored in DD at 18°C for 4 days and at 29°C for another 4 days. Discussion
Though a few studies implicate Alk orthologs in regulating behaviors such as decision-making, cognition, associative learning and addiction, most functional studies demonstrate various developmental roles for Alk [14,20,21,48–50]. We acutely induce a long-sleep phenotype by taking advantage of a temperature-sensitive allele, Alkts, revealing that Alk regulates sleep directly rather than through developmental processes. We also show mutations in Nf1, a gene encoding a GAP that regulates the Ras/ERK pathway activated by ALK, causes a sexually dimorphic short-sleep phenotype. Thus we establish a novel in vivo function for both Alk and Nf1 and show they interact with each other to regulate sleep.
Role of Alk in sleep
Many downstream signaling pathways have been proposed for ALK, among them Ras/ERK, JAK/STAT, PI3K and PLCγ signaling [11]. ERK activation through another tyrosine receptor kinase Epidermal growth factor receptor (EGFR) has been linked to increased sleep [36,51], while here we show that Alk, a positive regulator of ERK, inhibits sleep. We note that ERK is a common signaling pathway targeted by many factors, and may have circuit - specific effects, with different effects on sleep in different brain regions. Indeed, neural populations that mediate effects of ERK on sleep have not been identified. The dose of ALK required for ERK activation might also differ in different circuits. Region-specific effects of Alk are supported by our GAL4 screen, in which down-regulation of Alk in some brain regions even decreased sleep. The overall effect, however, is to increase sleep, evident from the pan-neuronal knockdown. We found that the mushroom body, a site previously implicated in sleep regulation and learning, requires Alk to inhibit sleep. Interestingly, the expression patterns of Alk and Nf1 overlap extensively in the mushroom body [14], suggesting that they may interact here to regulate both sleep and learning. However, it was previously shown that Alk activation in the mushroom body has no effect on learning [14]. The mushroom body expression in that study was defined with MB247 and c772, both of which also had no effects on sleep when driving Alk RNAi (Fig 4). The spatial requirement for Nf1 in the context of learning has been disputed in previous studies with results both for and against a function in the mushroom body [14,52]. The discrepancies between these studies could result from: 1) varied expression of different drivers within lobes of the mushroom body, with some not even specific to the mushroom body; 2) variability in the effectiveness and specificity of MB-Gal80 in combination with different GAL4s. We confirmed that our MB-Gal80 manipulation eliminated all mushroom body expression and preserved most if not all other cells with 30Y, 386Y and c309. Future work will further define the cell populations in which Alk and Nf1 interact to affect sleep.
Role of Nf1 in sleep regulation
We observed a substantial sleep decrease in Nf1 male flies compared to control flies. However, sleep phenotypes in Nf1 female flies are inconsistent. It is unlikely that unknown mutations on the X chromosome cause the short-sleeping phenotype because our 7 generation outcrosses into the control iso31 background started with swapping X chromosomes in Nf1P1 and Nf1P2 male flies with those of iso31 flies. In support of a function in sleep regulation, restoring Nf1 expression in neurons of Nf1 mutants reverses the short sleep phenotype to long sleep in both males and females. This does not result from ectopic expression of the transgene as expressing the same UAS-Nf1 transgene in wild-type flies has no effect. We hypothesize that Nf1 promotes sleep in some brain regions and inhibits it in others, and sub-threshold levels of Nf1, driven by the transgene in the mutant background, tilt the balance towards more sleep. As reported here, Alk also has differential effects on sleep in different brain regions, as does protein kinase A [33], thus such effects are not unprecedented. We also note severe sleep fragmentation in Nf1 mutants, which suggests that they have trouble maintaining sleep.
The sex-specific phenotypes of Nf1 mutants may reflect sexually dimorphic regulation of sleep. A recently published genome-wide association study of sleep in Drosophila reported that an overwhelming majority of single nucleotide polymorphisms (SNPs) exhibit some degree of sexual dimorphism: the effects of ~80% SNPs on sleep are not equal in the two sexes [53]. Interestingly, sex was found to be a major determinant of neuronal dysfunction in human NF1 patients and Nf1 knock-out mice, resulting in differential vision loss and learning deficits [54]. The sex-dimorphic sleep phenotype in Nf1 flies provides another model to study sex-dimorphic circuits involving Nf1. Interestingly, a prevalence of sleep disturbances have recently been reported in NF1 patients [44,45], suggesting that NF1 possibly play a conserved function in sleep regulation.
Links between plasticity and sleep
An attractive hypothesis for a function of sleep is that plastic processes during wake lead to a net increase in synaptic strength and sleep is necessary for synaptic renormalization [3]. There is structural evidence in Drosophila to support this synaptic homeostasis hypothesis (SHY): synapse size and number increase during wake and after sleep deprivation, and decrease after sleep [55]. However, little is known about the molecular mechanisms by which waking experience induces changes in plasticity and sleep. FMRP, the protein encoded by the Drosophila homolog of human fragile X mental retardation gene FMR1, mediates some of the effects of sleep/wake on synapses [55,56]. Loss of Fmr1 is associated with synaptic overgrowth and strengthened neurotransmission and long sleep. Overexpressing Fmr1 results in dendritic and axonal underbranching and short sleep. More importantly, overexpression of Fmr1 in specific circuits eliminates the wake-induced increases in synapse number and branching in these circuits. Thus, up-regulation of FMR accomplishes a function normally associated with sleep.
We hypothesize that Alk and Nf1 similarly play roles in synaptic homeostasis. They are attractive candidates for bridging sleep and plastic processes, because: 1) Alk is expressed extensively in the developing and adult CNS synapses [14,57]. In particular, both Alk and Nf1 are strongly expressed in the mushroom body, a major site of plasticity in the fly brain. 2) Functionally, postsynaptic hyperactivation of Alk negatively regulates NMJ size and elaboration [19]. In contrast, Nf1 is required presynaptically at the NMJ to suppress synapse branching [58]. 3) Alk and Nf1 affect learning in adults and they functionally interact with each other in this process [14]. It is tempting to speculate that in Alk mutants, sleep is increased to prune the excess synaptic growth predicted to occur in these mutants. Such a role for sleep is consistent with the SHY hypothesis. The SHY model would predict that Alk flies have higher sleep need, which is expected to enhance rebound after sleep deprivation. While our data show equivalent quantity of rebound in Alk mutants, we found that they fall asleep faster than control flies the morning after sleep deprivation (Fig 3), suggesting that they have higher sleep drive. Increased sleep need following deprivation could also be reflected in greater cognitive decline, but this has not yet been tested for Alk mutants. We note that Nf1 mutants have reduced sleep although their NMJ phenotypes also consist of overbranched synapses [58,59]. We postulate that their sleep need is not met and thus results in learning deficits. Clearly, more work is needed to test these hypotheses concerning the roles of Alk and Nf1 in sleep, learning, and memory circuits.
Materials and Methods
Fly lines
The following lines were used previously in the lab [33, 37, 60]: Elav-Gal4, 201Y-GAL4, c739-GAL4, 238Y-GAL4, c309-GAL4, Kurs58-GAL4, Pdf-Gal4, H24-GAL4, c507-GAL4, 30Y-GAL4, 50Y-GAL4, MJ63-GAL4, c232-GAL4, 104Y-GAL4, 17D-GAL4, Dilp2-Gal4, Tdc2-Gal4, TH-Gal4, Mai301-GAL4, c767-GAL4, 1471-GAL4, ok107-GAL4, c929-GAL4, 53b-GAL4, c320-GAL4 and UAS-GFP.NLS. elav-GAL4; Dcr2 (25750), c305a-GAL4 (30829), c107-GAL4(30823), c819-GAL4 (30849), 121Y-GAL4 (30815), 7Y-GAL4 (30812), 36Y-GAL4 (30819), 386Y-GAL4 (25410), c584-GAL4 (30842), DopR-GAL4 (19491), Ddc-GAL4 (7009), R71G10-GAL4 (39604) and R76D11-GAL4 (39927) were ordered from the Bloomington Drosophila Stock Center. NP1131-GAL4 (103898), NP1004-GAL4 (112440) and NP2024-GAL4 (112749) were ordered from the Drosophila Genetic Resource Center. Tub-GAL80ts [40], MB-Gal80 [41], D52H-GAL4 [42], c687-GAL4 [61], 6B-GAL4 [62], InSITE106-GAL4 [63],Cha-GAL4 [64], Kurs45-GAL4 [65] were gifts. Nf1P1 and Nf1P2 alleles were reported [22] and were outcrossed into an iso31 background for 7 generations. Alkts/CyO was a gift from Dr. J Weiss [13] and was outcrossed into an iso31 background. Alk1/CyO and UAS-Alk/CyO were gifts from Dr. R Palmer and both were outcrossed into an iso31 background [13,16]. The deficiency line uncovering the Alk gene, AlkDef (7888)/CyO, was ordered from Bloomington. Alk RNAi (11446) and UAS-Dcr2(60008) were ordered from the Vienna Drosophila Resource Center.
Immunohistochemistry
Reporter GFP expression driven by GAL4 lines was visualized through whole-mount brain immunofluorescence as previously described [60]. Rabbit anti-GFP (Molecular Probes A-11122) 1 : 1000 and Alex Fluor 488 Goat anti-rabbit (Molecular Probes A-11008) 1 : 500 were used.
Sleep assays
Sleep was monitored as described previously [37]. Flies were raised and kept on a 12h:12h light/dark (LD) cycle at 18°C or 25°C as stated. 3–7 day old flies were loaded into glass tubes containing 5% sucrose and 2% agar. Locomotor activity was monitored with the Drosophila Activity Monitoring System (Trikinetics, Waltham MA), or when indicated with multi-beam monitors (Trikinetics, Waltham MA) that generate 17 infrared beams. Data were analyzed with Pysolo software [5]. For sleep assays with temperature shifts, total sleep amount was averaged for 3 d at the lower temperature before the shift and 3 d at the high temperature. For sleep deprivation experiments, flies were monitored for a baseline day and then sleep deprived on the second day for 6 hours from Zeitgeber Time (ZT) 18 to ZT24 during the night. Sleep was continually monitored for 2 recovery days. Mechanical sleep deprivation was accomplished using a Trikinetics vortexer mounting plate, with shaking of monitors for 2 seconds randomly within every 20 second window for 6 hours. The arousal threshold assay was described previously [66]. A 12oz rubber weight was dropped from 2-inch height onto a rack supporting large DAMS monitors at ZT20. Flies with no activity 5 min before a stimulus and exhibited beam crossings within 5 min after the light pulse were recorded as “aroused”.
Rest-Activity rhythm analysis
Individual male flies were loaded into glass tubes containing 5% sucrose and 2% agar. Locomotor activity was monitored with the Drosophila Activity Monitoring System (Trikinetics, Waltham MA), and analyzed with Clocklab software (Actimetrics, Wilmette). To evaluate ALK’s role in maintaining adult rhythms, all genotypes were raised at 18°C to avoid inactivating ALK during development. 3–7 d old flies were then loaded into glass tubes and entrained for 3 d to a 12h:12h LD cycle, followed by 4 days in constant darkness at 18°C and then 4 days in constant darkness at 29°C. Rhythmicity analysis was performed for each 4 d period. In a separate experiment, iso31, Nf1P1/P2, Alkts and Alkts;Nf1P1/P2 flies were raised and tested at 25°C to evaluate whether inhibiting Alk during development affect rest-activity rhythm. A fly was considered rhythmic if it met 2 criteria: 1) displayed a rhythm with 95% confidence using χ2 periodogram analysis, and 2) a corresponding FFT value above 0.01 for the determined period length.
Negative geotaxis
The negative geotaxis assay was adapted from (Barone MC and Bohmann D 2013). Please see supplemental methods for details.
Statistics
Data were analyzed and plotted with SigmaPlot and GraphPrism software. One-way ANOVA analysis was done to reveal differences between genotypes in the same experiments and pairwise comparison between genotypes were done with post-hoc analysis as indicated in the figures. ns, not significant; **, p<0.01; ***, p<0.001; ****, p<0.0001.
Supporting Information
Zdroje
1. Hartse KM (2011) The phylogeny of sleep. Handb Clin Neurol 98 : 97–109. doi: 10.1016/B978-0-444-52006-7.00007-1 21056182
2. Wang G, Grone B, Colas D, Appelbaum L, Mourrain P (2011) Synaptic plasticity in sleep: learning, homeostasis and disease. Trends Neurosci 34 : 452–463. doi: 10.1016/j.tins.2011.07.005 21840068
3. Tononi G, Cirelli C (2014) Sleep and the price of plasticity: from synaptic and cellular homeostasis to memory consolidation and integration. Neuron 81 : 12–34. doi: 10.1016/j.neuron.2013.12.025 24411729
4. Vyazovskiy VV, Cirelli C, Pfister-Genskow M, Faraguna U, Tononi G (2008) Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat Neurosci 11 : 200–208. doi: 10.1038/nn2035 18204445
5. Gilestro GF, Tononi G, Cirelli C (2009) Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science 324 : 109–112. doi: 10.1126/science.1166673 19342593
6. Stickgold R, Hobson JA, Fosse R, Fosse M (2001) Sleep, learning, and dreams: off-line memory reprocessing. Science 294 : 1052–1057. 11691983
7. Florian C, Vecsey CG, Halassa MM, Haydon PG, Abel T (2011) Astrocyte-derived adenosine and A1 receptor activity contribute to sleep loss-induced deficits in hippocampal synaptic plasticity and memory in mice. J Neurosci 31 : 6956–6962. doi: 10.1523/JNEUROSCI.5761-10.2011 21562257
8. Seugnet L, Suzuki Y, Vine L, Gottschalk L, Shaw PJ (2008) D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr Biol 18 : 1110–1117. doi: 10.1016/j.cub.2008.07.028 18674913
9. Li X, Yu F, Guo A (2009) Sleep deprivation specifically impairs short-term olfactory memory in Drosophila. Sleep 32 : 1417–1424. 19928381
10. Bushey D, Cirelli C (2011) From genetics to structure to function: exploring sleep in Drosophila. Int Rev Neurobiol 99 : 213–244. doi: 10.1016/B978-0-12-387003-2.00009-4 21906542
11. Palmer RH, Vernersson E, Grabbe C, Hallberg B (2009) Anaplastic lymphoma kinase: signalling in development and disease. Biochem J 420 : 345–361. doi: 10.1042/BJ20090387 19459784
12. Iwahara T, Fujimoto J, Wen D, Cupples R, Bucay N, et al. (1997) Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene 14 : 439–449. 9053841
13. Loren CE, Scully A, Grabbe C, Edeen PT, Thomas J, et al. (2001) Identification and characterization of DAlk: a novel Drosophila melanogaster RTK which drives ERK activation in vivo. Genes Cells 6 : 531–544. 11442633
14. Gouzi JY, Moressis A, Walker JA, Apostolopoulou AA, Palmer RH, et al. (2011) The receptor tyrosine kinase Alk controls neurofibromin functions in Drosophila growth and learning. PLoS Genet 7: e1002281. doi: 10.1371/journal.pgen.1002281 21949657
15. Lee HH, Norris A, Weiss JB, Frasch M (2003) Jelly belly protein activates the receptor tyrosine kinase Alk to specify visceral muscle pioneers. Nature 425 : 507–512. 14523446
16. Englund C, Loren CE, Grabbe C, Varshney GK, Deleuil F, et al. (2003) Jeb signals through the Alk receptor tyrosine kinase to drive visceral muscle fusion. Nature 425 : 512–516. 14523447
17. Bazigou E, Apitz H, Johansson J, Loren CE, Hirst EM, et al. (2007) Anterograde Jelly belly and Alk receptor tyrosine kinase signaling mediates retinal axon targeting in Drosophila. Cell 128 : 961–975. 17350579
18. Cheng LY, Bailey AP, Leevers SJ, Ragan TJ, Driscoll PC, et al. (2011) Anaplastic lymphoma kinase spares organ growth during nutrient restriction in Drosophila. Cell 146 : 435–447. doi: 10.1016/j.cell.2011.06.040 21816278
19. Rohrbough J, Kent KS, Broadie K, Weiss JB (2013) Jelly Belly trans-synaptic signaling to anaplastic lymphoma kinase regulates neurotransmission strength and synapse architecture. Dev Neurobiol 73 : 189–208. doi: 10.1002/dneu.22056 22949158
20. Bilsland JG, Wheeldon A, Mead A, Znamenskiy P, Almond S, et al. (2008) Behavioral and neurochemical alterations in mice deficient in anaplastic lymphoma kinase suggest therapeutic potential for psychiatric indications. Neuropsychopharmacology 33 : 685–700. 17487225
21. Weiss JB, Xue C, Benice T, Xue L, Morris SW, et al. (2012) Anaplastic lymphoma kinase and leukocyte tyrosine kinase: functions and genetic interactions in learning, memory and adult neurogenesis. Pharmacol Biochem Behav 100 : 566–574. doi: 10.1016/j.pbb.2011.10.024 22079349
22. Williams JA, Su HS, Bernards A, Field J, Sehgal A (2001) A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science 293 : 2251–2256. 11567138
23. Loren CE, Englund C, Grabbe C, Hallberg B, Hunter T, et al. (2003) A crucial role for the Anaplastic lymphoma kinase receptor tyrosine kinase in gut development in Drosophila melanogaster. EMBO Rep 4 : 781–786. 12855999
24. Zimmerman JE, Chan MT, Jackson N, Maislin G, Pack AI. (2012) Genetic background has a major impact on differences in sleep resulting from environmental influences in Drosophila. Sleep 35 : 545–57. doi: 10.5665/sleep.1744 22467993
25. Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, et al. (2004) The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167 : 797–813. 15238529
26. Low KH, Lim C, Ko HW, Edery I (2008) Natural variation in the splice site strength of a clock gene and species-specific thermal adaptation. Neuron 60 : 1054–1067. doi: 10.1016/j.neuron.2008.10.048 19109911
27. Zimmerman JE, Raizen DM, Maycock MH, Maislin G, Pack AI (2008) A video method to study Drosophila sleep. Sleep 31 : 1587–1598. 19014079
28. Faville R, Kottler B, Goodhill GJ, Shaw PJ, van Swinderen B (2015) How deeply does your mutant sleep? Probing arousal to better understand sleep defects in Drosophila. Sci Rep 5 : 8454. doi: 10.1038/srep08454 25677943
29. Barone MC, Bohmann D (2013) Assessing neurodegenerative phenotypes in Drosophila dopaminergic neurons by climbing assays and whole brain immunostaining. J Vis Exp: e50339. doi: 10.3791/50339 23644755
30. Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, et al. (2000) Rest in Drosophila is a sleep-like state. Neuron 25 : 129–138. 10707978
31. Andretic R, van Swinderen B, Greenspan RJ (2005) Dopaminergic modulation of arousal in Drosophila. Curr Biol 15 : 1165–1175. 16005288
32. Kume K, Kume S, Park SK, Hirsh J, Jackson FR (2005) Dopamine is a regulator of arousal in the fruit fly. J Neurosci 25 : 7377–7384. 16093388
33. Joiner WJ, Crocker A, White BH, Sehgal A (2006) Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441 : 757–760. 16760980
34. Pitman JL, McGill JJ, Keegan KP, Allada R (2006) A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441 : 753–756. 16760979
35. Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ (2011) Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332 : 1571–1576. doi: 10.1126/science.1202249 21700877
36. Foltenyi K, Greenspan RJ, Newport JW (2007) Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci 10 : 1160–1167. 17694052
37. Crocker A, Shahidullah M, Levitan IB, Sehgal A (2010) Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron 65 : 670–681. doi: 10.1016/j.neuron.2010.01.032 20223202
38. Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, et al. (2008) PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60 : 672–682. doi: 10.1016/j.neuron.2008.10.042 19038223
39. Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, et al. (2008) Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol 18 : 1537–1545. doi: 10.1016/j.cub.2008.08.033 18771923
40. McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL (2003) Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302 : 1765–1768. 14657498
41. Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S (2007) Sequential use of mushroom body neuron subsets during drosophila odor memory processing. Neuron 53 : 103–115. 17196534
42. Aso Y, Grubel K, Busch S, Friedrich AB, Siwanowicz I, et al. (2009) The mushroom body of adult Drosophila characterized by GAL4 drivers. J Neurogenet 23 : 156–172. doi: 10.1080/01677060802471718 19140035
43. Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, et al. (2012) A GAL4-driver line resource for Drosophila neurobiology. Cell Rep 2 : 991–1001. doi: 10.1016/j.celrep.2012.09.011 23063364
44. Licis AK, Vallorani A, Gao F, Chen C, Lenox J, et al. (2013) Prevalence of Sleep Disturbances in Children With Neurofibromatosis Type 1. J Child Neurol 28 : 1400–1405. 24065580
45. Leschziner GD, Golding JF, Ferner RE (2013) Sleep disturbance as part of the neurofibromatosis type 1 phenotype in adults. Am J Med Genet A 161A: 1319–1322. doi: 10.1002/ajmg.a.35915 23636844
46. The I, Hannigan GE, Cowley GS, Reginald S, Zhong Y, et al. (1997) Rescue of a Drosophila NF1 mutant phenotype by protein kinase A. Science 276 : 791–794. 9115203
47. Guo HF, Tong J, Hannan F, Luo L, Zhong Y (2000) A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature 403 : 895–898. 10706287
48. Shinkai Y, Yamamoto Y, Fujiwara M, Tabata T, Murayama T, et al. (2011) Behavioral choice between conflicting alternatives is regulated by a receptor guanylyl cyclase, GCY-28, and a receptor tyrosine kinase, SCD-2, in AIA interneurons of Caenorhabditis elegans. J Neurosci 31 : 3007–3015. doi: 10.1523/JNEUROSCI.4691-10.2011 21414922
49. Lasek AW, Gesch J, Giorgetti F, Kharazia V, Heberlein U (2011) Alk is a transcriptional target of LMO4 and ERalpha that promotes cocaine sensitization and reward. J Neurosci 31 : 14134–14141. doi: 10.1523/JNEUROSCI.3415-11.2011 21976498
50. Lasek AW, Lim J, Kliethermes CL, Berger KH, Joslyn G, et al. (2011) An evolutionary conserved role for anaplastic lymphoma kinase in behavioral responses to ethanol. PLoS One 6: e22636. doi: 10.1371/journal.pone.0022636 21799923
51. Vanderheyden WM, Gerstner JR, Tanenhaus A, Yin JC, Shaw PJ (2013) ERK phosphorylation regulates sleep and plasticity in Drosophila. PLoS One 8: e81554. doi: 10.1371/journal.pone.0081554 24244744
52. Buchanan ME, Davis RL (2010) A distinct set of Drosophila brain neurons required for neurofibromatosis type 1-dependent learning and memory. J Neurosci 30 : 10135–10143. doi: 10.1523/JNEUROSCI.0283-10.2010 20668197
53. Harbison ST, McCoy LJ, Mackay TF (2013) Genome-wide association study of sleep in Drosophila melanogaster. BMC Genomics 14 : 281. doi: 10.1186/1471-2164-14-281 23617951
54. Diggs-Andrews KA, Brown JA, Gianino SM, Rubin JB, Wozniak DF, et al. (2014) Sex Is a major determinant of neuronal dysfunction in neurofibromatosis type 1. Ann Neurol 75 : 309–316. doi: 10.1002/ana.24093 24375753
55. Bushey D, Tononi G, Cirelli C (2011) Sleep and synaptic homeostasis: structural evidence in Drosophila. Science 332 : 1576–1581. doi: 10.1126/science.1202839 21700878
56. Bushey D, Tononi G, Cirelli C (2009) The Drosophila fragile X mental retardation gene regulates sleep need. J Neurosci 29 : 1948–1961. doi: 10.1523/JNEUROSCI.4830-08.2009 19228950
57. Rohrbough J, Broadie K (2010) Anterograde Jelly belly ligand to Alk receptor signaling at developing synapses is regulated by Mind the gap. Development 137 : 3523–3533. doi: 10.1242/dev.047878 20876658
58. Tsai PI, Wang M, Kao HH, Cheng YJ, Walker JA, et al. (2012) Neurofibromin mediates FAK signaling in confining synapse growth at Drosophila neuromuscular junctions. J Neurosci 32 : 16971–16981. doi: 10.1523/JNEUROSCI.1756-12.2012 23175848
59. Walker JA, Gouzi JY, Long JB, Huang S, Maher RC, et al. (2013) Genetic and functional studies implicate synaptic overgrowth and ring gland cAMP/PKA signaling defects in the Drosophila melanogaster neurofibromatosis-1 growth deficiency. PLoS Genet 9: e1003958. doi: 10.1371/journal.pgen.1003958 24278035
60. Cavanaugh DJ, Geratowski JD, Wooltorton JR, Spaethling JM, Hector CE, et al. (2014) Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell 157 : 689–701. doi: 10.1016/j.cell.2014.02.024 24766812
61. Donlea JM, Ramanan N, Shaw PJ (2009) Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324 : 105–108. doi: 10.1126/science.1166657 19342592
62. Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 : 401–415. 8223268
63. Gohl DM, Silies MA, Gao XJ, Bhalerao S, Luongo FJ, et al. (2011) A versatile in vivo system for directed dissection of gene expression patterns. Nat Methods 8 : 231–237. 21473015
64. Salvaterra P.M., Kitamoto T. (2001). Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res. Gene Expr. Patterns 1 : 73–82.
65. Siegmund T, Korge G (2001) Innervation of the ring gland of Drosophila melanogaster. J Comp Neurol 431 : 481–491. 11223816
66. Kayser MS, Yue Z, Sehgal A (2014). A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science 344 : 269–74. doi: 10.1126/science.1250553 24744368
Štítky
Genetika Reprodukčná medicína
Článek A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin TransporterČlánek Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis inČlánek Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 11- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Agricultural Genomics: Commercial Applications Bring Increased Basic Research Power
- Ernst Rüdin’s Unpublished 1922-1925 Study “Inheritance of Manic-Depressive Insanity”: Genetic Research Findings Subordinated to Eugenic Ideology
- Convergent Evolution During Local Adaptation to Patchy Landscapes
- The Locus Controls Age at Maturity in Wild and Domesticated Atlantic Salmon ( L.) Males
- A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin Transporter
- Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting
- Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes
- Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep
- Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein
- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Mosaic and Intronic Mutations in Explain the Majority of TSC Patients with No Mutation Identified by Conventional Testing
- Members of the Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in
- QTL Mapping of Sex Determination Loci Supports an Ancient Pathway in Ants and Honey Bees
- Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing
- Genomics of Cancer and a New Era for Cancer Prevention
- Adaptation to High Ethanol Reveals Complex Evolutionary Pathways
- Dynamics of Transcription Factor Binding Site Evolution
- Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
- Enhancer Runaway and the Evolution of Diploid Gene Expression
- Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis
- Drosophila Mutants Model Cornelia de Lange Syndrome in Growth and Behavior
- Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases
- Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
- Tissue-Specific Effects of Reduced β-catenin Expression on Mutation-Instigated Tumorigenesis in Mouse Colon and Ovarian Epithelium
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Mapping of Craniofacial Traits in Outbred Mice Identifies Major Developmental Genes Involved in Shape Determination
- Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- DNA Repair Cofactors ATMIN and NBS1 Are Required to Suppress T Cell Activation
- Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
- Ernst Rüdin and the State of Science
- ABCs of Insect Resistance to Bt
- Epigenetic Control of O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
- The Fanconi Anemia Pathway Protects Genome Integrity from R-loops
- Controls Quantitative Variation in Maize Kernel Row Number
- Genome-Wide Association Study of Golden Retrievers Identifies Germ-Line Risk Factors Predisposing to Mast Cell Tumours
- Insect Resistance to Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein
- A Cytosine Methytransferase Modulates the Cell Envelope Stress Response in the Cholera Pathogen
- Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals
- The Multi-allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance
- The lncRNA Controls Cryptococcal Morphological Transition
- Sae2 Function at DNA Double-Strand Breaks Is Bypassed by Dampening Tel1 or Rad53 Activity
- A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia,
- Ectodysplasin/NF-κB Promotes Mammary Cell Fate via Wnt/β-catenin Pathway
- The QTL within the Complex Involved in the Control of Tuberculosis Infection in Mice Is the Classical Class II Gene
- Identifying Loci Contributing to Natural Variation in Xenobiotic Resistance in
- Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by and Subsistence
- A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data
- Competition between Heterochromatic Loci Allows the Abundance of the Silencing Protein, Sir4, to Regulate Assembly of Heterochromatin
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



