-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals
Animal genomes from flies to humans contain many hundreds of non-coding elements called Piwi-interacting RNAs (piRNAs) cluster loci (piC loci). Some of these elements generate piRNAs that direct the silencing of transposable elements, which are pervasive genetic parasites. However, we lack an understanding of the targeting function for the remaining bulk of piRNAs because their loci are not complementarity to transposable elements. In addition, the field does not know if all piC loci are quickly evolving, or if some piC loci might be deeply conserved in piRNA expression, an indication of its potentially functional importance. Our study confirms the highly rapid evolution in piRNA expression capacity for the majority of piC loci in flies and mammals, with many clade - and species-specific piC loci expression patterns. In spite of this, we also discover a cohort of piC loci that are deeply conserved in piRNA expression from the human to the dog, a significantly broad phylogenetic spectrum of eutherian mammals. However, this conservation in piRNA expression ends at non-eutherian mammals like marsupials and monotremes. Existing mutations in two of these Eutherian-Conserved piC (ECpiC) loci impair mouse reproduction and abrogate piRNA production. Therefore, we suggest these ECpiC loci are conserved for piRNA expression due to their important function in eutherian reproduction and stand out as prime candidates for future genetic studies.
Published in the journal: Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals. PLoS Genet 11(11): e32767. doi:10.1371/journal.pgen.1005652
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005652Summary
Animal genomes from flies to humans contain many hundreds of non-coding elements called Piwi-interacting RNAs (piRNAs) cluster loci (piC loci). Some of these elements generate piRNAs that direct the silencing of transposable elements, which are pervasive genetic parasites. However, we lack an understanding of the targeting function for the remaining bulk of piRNAs because their loci are not complementarity to transposable elements. In addition, the field does not know if all piC loci are quickly evolving, or if some piC loci might be deeply conserved in piRNA expression, an indication of its potentially functional importance. Our study confirms the highly rapid evolution in piRNA expression capacity for the majority of piC loci in flies and mammals, with many clade - and species-specific piC loci expression patterns. In spite of this, we also discover a cohort of piC loci that are deeply conserved in piRNA expression from the human to the dog, a significantly broad phylogenetic spectrum of eutherian mammals. However, this conservation in piRNA expression ends at non-eutherian mammals like marsupials and monotremes. Existing mutations in two of these Eutherian-Conserved piC (ECpiC) loci impair mouse reproduction and abrogate piRNA production. Therefore, we suggest these ECpiC loci are conserved for piRNA expression due to their important function in eutherian reproduction and stand out as prime candidates for future genetic studies.
Introduction
In animal RNA interference (RNAi) pathways, the Argonaute (AGO) and Piwi proteins are deeply conserved from humans to basal animals like cnidarians and poriferans [1, 2]. Likewise, several AGO-bound microRNAs (miRNAs) are also deeply conserved across bilaterians [3], but Piwi-interacting RNAs (piRNAs) differ by evolving so rapidly that there are few individual piRNAs conserved between even closely related species [4, 5]. A rationale for the fast rate of piRNA sequence evolution is to keep pace with the rapidly evolving transposable elements (TE) that some piRNAs are targeting [6, 7]. However, most mammalian piRNA cluster (piC) loci are depleted of TE sequences [4, 5], and in spite of the TE-repressing function of the Piwi pathway, animal genomes vary widely in size due mainly to TE content, from ~10% of the euchromatic genome in flies to >40% in mammals [8]. Thus, we have an incomplete understanding of what additional roles the Piwi pathway plays beyond TE repression.
The piRNAs arise as clusters from two main types of loci. Intergenic piRNA cluster loci are large 10–100 kb long regions, and some of these loci function to silence TEs with TE-directed piRNAs. However, the function of most intergenic piRNA cluster loci in mammals remains mysterious because they are depleted of TE sequences and the transcripts are non-coding [9]. In contrast, genic piRNA cluster loci are derived from protein-coding transcripts that possess extensive 3'Untranslated Regions (3'UTRs) which efficiently enter the piRNA biogenesis pathway [10, 11]. Hundreds of genic piRNA cluster loci are also depleted in TE sequences, and germ cells specifically select genic transcripts via an unknown signature for piRNA biogenesis [11]. Nevertheless, the abundant piRNAs from the Drosophila genic piC traffic jam (tj) may regulate gene targets important for follicle cell development [12].
Although piRNAs are essential for animal fertility, we do not know which of the hundreds of piC loci are most important for animal reproduction, particularly in mammals. Therefore, we hypothesize that determining which piC loci are most conserved in piRNA expression throughout animal evolution would yield functional insight to this question above. However, the field has only touched upon the evolutionary patterns of piC loci with a handful of previous studies comparing piC loci between a limited set of animal species. In Drosophilid flies, the intergenic piC locus flamenco (flam) is syntenically conserved between D. melanogaster (D.mel) and D. erecta (D.ere) for ~10 million years (MY) of evolution [13]. In rodents and humans, which diverged in evolution ~90 MY ago, some piC loci are also syntenically conserved [4, 5], and Assis and Kondrashov applied a classification process to these human and rodent piRNA datasets to conclude that piC loci were rapidly expanding at a high rate, with mainly new gains of clusters between these species and no piC loci losses [14]. Assis and Kondrashov further proposed a positive selection process for piC expansion to keep pace with the rapid evolution of TE sequences [14], however the capacity of piCs to expand via copy number evolution has recently been re-inspected in humans by Gould et al, whom suggest instead that negative selection may be acting upon human piCs to limit their copy number expansion [15].
Earlier studies only examined human, mouse and rat piRNA datasets that are now considered shallow by today’s deep-sequencing standards [4, 5], and thus were unable to determine whether piRNA expression from piC loci could be deeply conserved like miRNA expression. To gain a more complete picture of the evolution of piC loci expression, this requires the experimental support of new, deeper piRNA datasets from a broader spectrum of animals. Fortunately, the diversity of species with sequenced piRNA libraries have greatly expanded recently with these various studies [11, 16–26], some of which mainly focused on the characterizing the novel piRNA features specific to the species. Thus, we sought to discover: (1) which piC loci, if any, may exhibit deep conservation of piRNA expression across animal lineages, (2) how frequently do piC loci expression patterns vary between species, and (3) which gene regulation step(s) prominently influences these piC loci expression patterns. Here, we formally define conserved piC loci expression as the detection of mature piRNAs from syntenic loci between species, which is a process that depends on both: (1) the transcription of the loci, and (2) selection of the transcribed precursor RNA by Piwi-pathway proteins for processing into mature piRNAs.
In this study, we describe a comprehensive comparative genomics approach focusing first on the genic piRNA biogenesis mechanism that we had previously shown was deeply conserved between flies and mice [11]. Although gene homologs for mouse and fly genic piC loci can be predicted bioinformatically (S1A Fig), it is not clear if these are actually orthologous piC loci because gene families have greatly expanded in mouse during the >600 MY of evolution between these species. Thus, we decided to examine piRNA expression patterns within closely-related species amongst the Drosophilids and tetrapod lineages to determine how many piC loci expression patterns for piRNA biogenesis are deeply conserved. We confirm that genic piC loci, which are depleted in TE sequences, are rapidly evolving and expanding into repertoires that are unique to animal species. Surprisingly, we also discover a cohort of piC loci with piRNA expression patterns conserved across ~100 MY of Eutherian mammal evolution, which stand out from the generally rapid evolution of most animal piC loci expression patterns.
Results
Comparative genomics of genic piRNA cluster loci expression patterns
We developed a piC loci discovery approach on new piRNA datasets that we had generated as well as analyzing publicly-deposited datasets covering four Drosophilids, the chicken and eleven mammals within Primate, Glire, and Laurasiatherian clades (Fig 1A and 1B). We sequenced Drosophilids piRNAs from ovarium samples because piRNAs are more plentiful and diverse in fly ovaries compared to fly testes [13, 27]. The tetrapod piRNAs were sampled from adult testes, which contained piRNAs from both early and post-pachytene stages of meiotic germ cells [19]. We generated a variety of piRNA libraries from Glires, including libraries enriched in pre-pachytene piRNAs from 10 days-post-partum (10dpp) testes and PIWIL2 immunoprecipitates (IPs), whereas the remaining tetrapod libraries were previously published in these studies: [11, 19–26]. This piRNA compendium was chosen for comprehensiveness and quality of piRNA coverage, since all libraries displayed the expected main read length distribution of 23–32 nucleotides and sufficient read depth (4–143 million reads, S1 Table). This phylogenetic spectrum of piRNA dataset allowed us to compare and determine the conservation of piC loci expression patterns spanning both short (<50 MY) and long (>150 MY) divergence times.
Fig. 1. Comparative genomics of piRNA cluster (piC) loci and rapid evolution of piC loci expression patterns in Drosophilids. 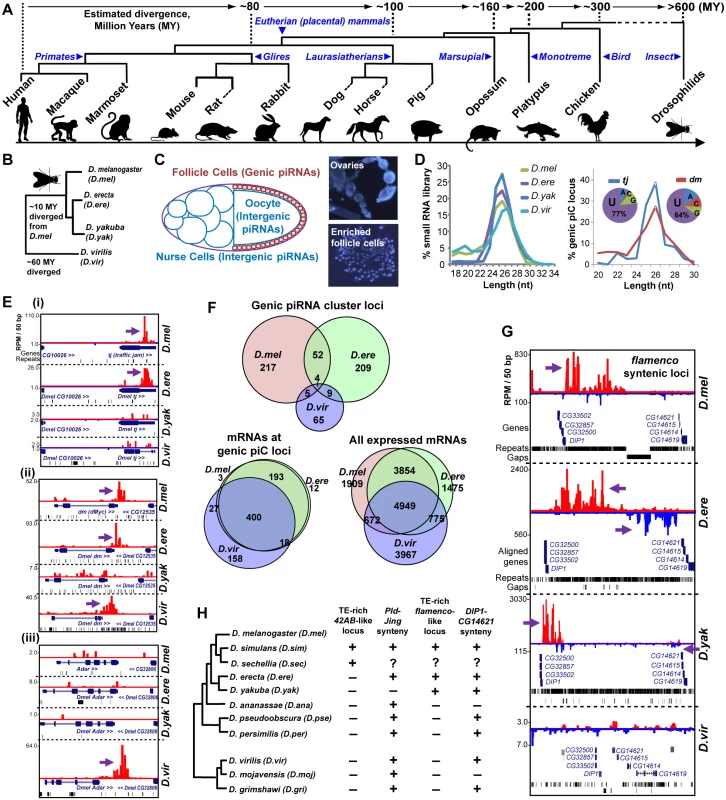
(A) Phylogenetic tree of the animals whose piRNAs are analyzed for this study. (B) Phylogenetic tree of the Drosophilids whose female piRNAs are profiled in this study. (C) Diagram of the Drosophilid ovarium where the different types of piRNAs are most abundantly expressed, and DAPI staining images of follicle cells enriched from ovaries during our preparations to balance the detection of genic piC loci from being overwhelmed by intergenic piC loci [28]. (D) Read length distributions for each small RNA library (left plot) and just the piRNAs in the genic piC loci, tj and dm. The pie chart insets show the 5' nucleotide composition of the genic piRNAs in these two loci.(E) Genome browser snapshots of the genic piC loci corresponding to piC-tj (i), piC-dm (ii, also known as Drosophila c-Myc), and piC-Adar (iii) showing some conserved and species-specific piRNA expression patterns. Purple arrows point to the start of bulk of piRNAs. Plus strand read peaks are red, minus strand read peaks are blue. (F) Venn diagrams of the overlap in genic piC locus expression (top) and the overlap in mRNA expression profiles (bottom sets) in three Drosophilid ovarium samples. The distributions of genic piC loci overlap are distinct from the distributions of mRNA expression profiles overlap (all p<0.001, Chi square test and ratio proportions test with Bonferroni correction). D.yak was omitted from this analysis because of much lower numbers of genic piC loci (see S2A Fig). (G) Snapshot of syntenic regions containing orthologous flamenco (flam) piC loci in D.mel, D.ere, and D.yak, but absence of this intergenic piC locus in D.vir. (H) Phylogeny of 11 Drosophilids with sequenced genomes and summary of the inspection in the genomes for the features at the syntenic regions for a 42AB-like locus and a flam-like locus, as defined by piRNAs or a contiguous large block concentrated with TEs between the flanking genes. Plus indicates locus/feature presence, minus is absence, while the question mark indicates a major gap in the genome assembly preventing determination. See S3 Fig for additional genome browser snapshots. To search for conserved expression of piRNAs across potential piC loci between species, we focused first on genic piC loci because the protein-coding genes are clear proxies for assigning orthology between potential piC loci. We deployed a small RNA profiling and bioinformatics procedure that identified genic piC loci by tracking piRNAs from protein-coding transcript 3'UTRs (S1B Fig). This procedure was highly accurate in tracking piRNAs at specific genic piC loci, because the reads exhibited the correct length ranges of 24-28nt for Drosophilid piRNAs and 24-31nt for mammalian piRNAs, as well as a bias for uridine at the 5' nucleotide of the small RNAs (S1C–S1F Fig).
Rapid evolution of piRNA expression amongst Drosophilid piRNA cluster loci
We examined conservation of piRNA expression from piC loci in four model Drosophilids where TE-directed piRNAs were previously characterized (Fig 1B) [13, 29]. D.mel protein-coding transcript alignments served as excellent proxies for transcripts in the incomplete draft assemblies of the D.ere, D.yakuba (D.yak) and D.virilis (D.vir) genomes. To prevent intergenic piC loci from overshadowing genic piC loci, we processed the ovarium samples to enrich for follicle cells and this improved genic piC locus detection (Fig 1C and 1D, [28]). We discovered >270 genic piC loci expressed in both D.mel and D.ere, 82 genic piC loci in D.vir, but surprisingly only 27 loci in D.yak (S2A Fig and S2 Table). The most abundant genic piC locus in D.mel, piC-tj, was only conserved in piRNA expression in D.ere, whereas the genic piC locus at diminutive (dm), the Drosophila c-Myc oncogene homolog, was more deeply conserved to D.vir in piRNA expression (Fig 1E). Although we discovered plenty of piRNAs for the D.yak ortholog of the flam intergenic piC locus (Fig 1G), indicating the D.yak library sufficiently contained follicle-cell specific piRNAs, genic piC loci were still depleted in D.yak. We examined gene expression profiles in D.yak follicle cells for known Piwi pathway genes and new genes associated with PIWI in D.mel OSS follicle cells as well as Northern blot comparisons (S2C and S2D Fig), but these investigations were unable to explain the depletion of genic piC loci in D.yak.
Therefore, we only further considered the genic piC loci expression patterns in D.mel, D.ere, and D.vir (Fig 1F), and observed that most Drosophila genic piC loci were unique to a single species, such as the piC-Adar that is specific to D.vir (Fig 1E–1Eiii). In fact, the mRNA gene expression profiles in the ovarium samples shared much more similarity between these three Drosophilids than the genic piC loci piRNA expression patterns (Fig 1F). Furthermore, the top piRNA-producing genic piC loci from D.mel were also expressed as mRNAs in the other Drosophilid ovariums despite frequently losing the capacity to generate piRNAs (S2E Fig). Finally, we determined the gain and loss rates of genic piC loci in Drosophilids, with the highest rapid gain rates displayed in D.mel and D.ere (S2F Fig). These data suggest that Drosophilid genic piC loci evolved rapidly at the level of sequence elements within the genic transcripts rather than in the control of gene expression, thus leading to this diversity in genic piC loci expression patterns across species.
Given the compactness of Drosophilid genomes, a major proportion of intergenic piRNAs in D.mel derive from the flam and 42AB piC loci, the two master control loci implicated in repressing TEs to maintain fertility in females [13, 30, 31]. Therefore, we turned our attention to examining these two major intergenic piC loci, and found both to be remarkably young evolutionary inventions (<12MY), having only recently arisen in the melanogaster subgroup (Fig 1G, S3 Fig). By tracking piRNA expression or an intervening TE-rich region, we detected signatures of piC-42AB orthologs in D.simulans (D.sim) and D.sechellia (D.sec), and flam orthologs in D.ere and D.yak. However, piC-42AB was absent from D.ere and D.yak genomes, and no flam orthologs were conserved beyond these species despite conserved gene synteny around flam and piC-42AB loci across ~40 MY of Drosophilid evolution (Fig 1H). Previous evolutionary studies suggested a selective advantage for Drosophilid piRNAs to silence TEs, but only up to a limit, when the host organism also tolerates the residing TEs [32–34]. Our data echoes this fluidity for the TE silencing function of major intergenic piC loci. Although they appear essential for TE repression and fertility, large intergenic piC loci as elements can arise and evolve as rapidly as the smaller genic piC loci.
Most mammalian piRNA cluster loci have also evolved rapidly
Next, we applied our piC locus discovery approach to adult testes small RNA datasets from Primates (human, macaque, marmoset), Glires (mouse, rat, rabbit) and Laurasiatherians (dog, horse, pig), (Fig 2, S2 Table). We observed conserved piRNA expression for piC loci within Rodents (Fig 2B), and within Primates (Fig 2C). However, most genic piC loci, as defined by production of mature piRNAs, were uniquely detected in a single species, including human-specific genic piC loci that in two independent human testes studies appear to generate potentially overlapping 3'UTR-piRNAs (Fig 2D, S1F Fig). Although endogenous overlapping small RNAs are prevalent in invertebrates [35], this configuration of two potentially overlapping human piRNA cluster loci may hint to the existence of dsRNAs in the mammalian germline beyond endo-siRNA transcripts in mouse oocytes [36–38].
Fig. 2. Rapid evolution of genic piC loci piRNA expression in mammals. 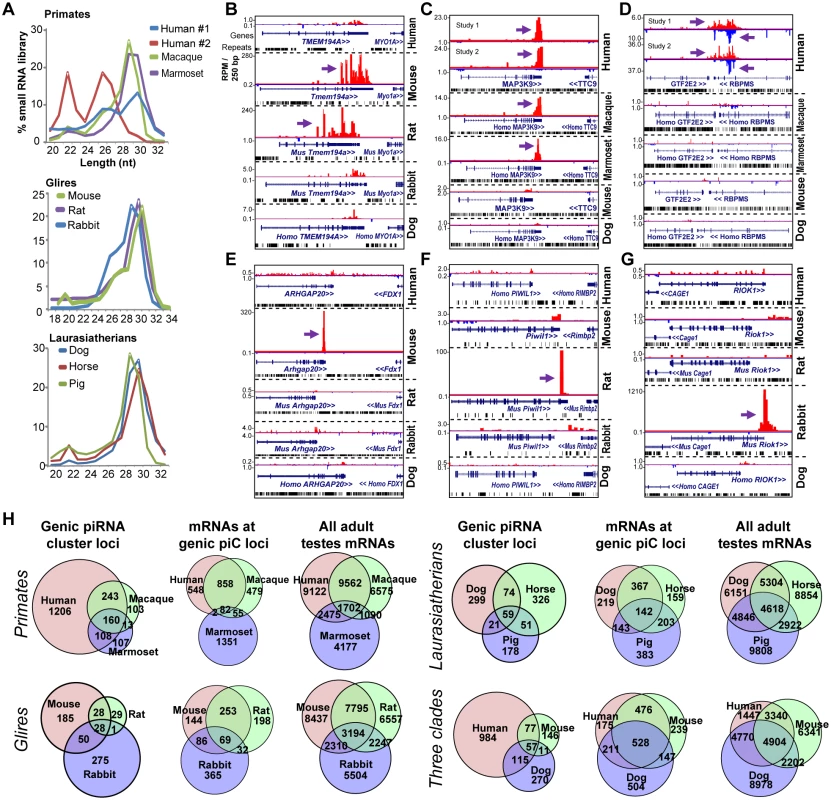
(A) Read length distributions for each mammalian small RNA library analyzed for this study. All libraries are from adult testes total RNA, grouped according to Primates, Glires, and Laurasiatherians. Genome browser snapshots of the rodent-specific piC-Tmem194a (B); a primate-specific piC-MAP3K9 (C); and two human-specific genic piC loci which have overlapping transcripts (D), piC-GTF2E2 (plus strand) and piC-RBPMS (minus strand). The mouse-specific piC-Arhgap20 (E); the rat-specific piC-Piwil1 (F) and the rabbit-specific piC-Riok1 (G). Purple arrows point to the start of bulk of piRNAs. Plus strand read peaks are red, minus strand read peaks are blue. (H) Venn diagrams of the overlap in genic piC loci expression (left) and the overlap in mRNA expression profiles (middle and right) amongst Primates, Glires, and Laurasiatherians, and a comparison species from the three clades. For each of species comparisons, the distributions of genic piC loci overlap are distinct from the distributions of mRNA expression profiles overlap (all p<0.001, Chi square test and proportions ratio test with Bonferroni correction). We examined three species-specific piC loci (piC-Arhgap20, piC-Piwil1, and piC-Riok1) that were only expressed in mouse, rat, and rabbit, respectively (Fig 2E–2G), and we confirmed these species-specific piRNA expression patterns by Northern blotting (S4A Fig). We also considered whether different stages of testes development or partitioning of piRNAs into the different mammalian Piwi proteins could be the root of these species-specific detections of genic piC loci (see S4B–S4H Fig and Supplementary Text Discussion). Despite comprehensive profiling of genic piC loci across PIWIL2 IP’s and different stages of Glire testes, the genic piC loci repertoires remained highly diverse between species. With regards to intergenic piC loci, their definitions amongst species were greatly influenced by the completeness of the species’ genome assembly, such that mouse and human exhibited the greatest number of intergenic piC loci (S3 Table). Nevertheless, at least 70% or more of genic piC loci could be reproducibly called by our pipeline in two independent human and chicken piRNA libraries (S4I Fig). These analyses mitigate the concern of limits in our piC discovery approach and confirm that the diversity in piC loci expression patterns between species is not an artifact.
Our approach was most effective in D.mel, human and mouse, where we detected the expression of >290, >1700, >2900 genic piC loci, respectively (Figs 1F, 2H and S4H). Considering the extensive piRNA sequencing coverage in human and mouse libraries, the comparison of piC loci expression patterns from all samples between these two species still show many species-specific genic piC loci expression patterns, which is also reflected by the Primate-specific and Glire-specific genic piC loci patterns (S4G Fig, middle, right). In addition, we observe within each of the three different clades (Primates, Glires, and Laurasiatherians) a tendency for adult testes gene expression profiles to show greater overlap when contrasted to the diversity of genic piC loci (Fig 2H). These patterns persisted even when we raised the stringency of cutoffs by 2-fold for piRNAs needed to call a shared piC locus expression and for gene expression profiles to be called overlapping (S2B and S4H Figs).
Amongst the top piRNA-generating genic piC loci in mouse and human, the gene orthologs that lost the capacity to make piRNAs in the most distant relative within the clade (rabbit and marmoset, respectively), still tended to be expressed in the adult testes as mRNAs (S4J and S4K Fig). Therefore, we propose that clade-specific and species-specific piC locus expression is a common feature in both flies and mammals; and that the rapid evolution of piC loci expression patterns is occurring at the transcript sequence level to alter entry into piRNA biogenesis pathways rather than at the transcript expression level. We also measured the rate of genic piC loci gain and loss throughout the phylogeny of tetrapods examined in this study, and consistently observed bursts of genic piC loci gains and very few losses since the number of ancestral genic piC loci were low (S4L Fig). This result is consistent with the gain and loss rates for genic piC loci in Drosophilids and further validates the earlier proposal that piC loci expansion is a common phenomenon [14].
Sequence signatures and evolutionary rates of genic piRNA cluster loci
Despite rapid evolution of piC loci expression patterns between species, we wondered if analyzing genic piC loci sequences within fly and mouse genomes could reveal sequence motifs and structured RNA elements that might differentiate genic piC loci from standard mRNAs that do not generate piRNAs. To test this question, we selected the top piRNA-producing transcripts from the fly (D.mel) and the mouse, respectively, and also selected a list of negative control transcripts that had similar annotated 3'UTR lengths to genic piC loci yet did not make piRNAs (S5A Fig and methods). We then searched each list with an RNA feature analysis against the Open Reading Frames (ORF) and 3'UTR sequences. First, we counted the average number of significant structured RNA elements predicted genome-wide by the RNAZ, EvoFold and REAPR algorithms [39–41]. Compared to the negative control sets, the top set (highest piRNA-producing set) of fly genic piC transcripts appeared to be enriched in predicted structured RNA elements in the ORF sequences compared to the negative control set, whereas the 3’UTRs of top set of mouse genic piC transcripts were more enriched in structured RNA elements (S5B and S5E Fig). Two different sequence motif discovery programs, MEME and GLAM2 [42], both predicted a Poly-U-rich motif with greater statistically-significant enrichment amongst the 3'UTRs of both fly and mouse genic piC transcripts compared to the negative control sets, whereas no clear motif was enriched amongst the ORF’s. These initial surveys hint at intrinsic features that may distinguish a genic piC transcript from a regular protein-coding mRNA, and these features may frequently arise or disappear during animal evolution to yield the diversity of clade - and species-specific sets of genic piC loci.
To examine whether genic piC loci were subjected to different evolutionary forces compared to non-piRNA producing control transcripts, we compared sequence conservation and evolution rates for fly and mouse genic piC loci with normalized values of the phastCons [43] and phyloP [44] scores for ORF and 3'UTR sequences. High average phastCons scores reflect stronger conservation [43], whereas positive phyloP scores suggest selective constraints on the sequences’ evolution [44]. Our analyses suggest that genic piC ORFs with the greatest number of piRNAs in both flies and mice were more conserved and under greater selective constraint than negative control mRNAs (S5F and S5G Fig). The faster evolving 3'UTR sequences were expected to be less conserved, reflecting 2–5 fold lower normalized phastCons scores in 3'UTRs compared to ORFs, and these 3’UTRs displayed phyloP scores that were positive but not particularly high. In addition, the 3'UTR phastCons and phyloP scores were not statistically distinct between genic piC loci and negative control transcripts. Since the bulk of genic piRNAs derive from the 3'UTR, this result is consistent with the overall poor conservation of individual piRNA sequences between animal species.
A set of piRNA cluster loci have conserved piRNA expression in Eutherian mammals
Although most piC loci are rapidly evolving, we were struck by the conserved expression of 8% and 5% of genic piC loci in human and mouse, respectively (Fig 2H). To look for deeper conservation of piC loci expression beyond the ~80 MY of divergence between humans and mice, we discovered and compared piC loci expression patterns across nine mammals and the chicken, thus spanning ~300MY of tetrapod evolution [45]. This search revealed 21 intergenic and 56 genic piC loci conserved in piRNA expression across Primates, Glires, and Laurasiatherians (Fig 3), for which these three clades had diverged from a common Eutherian (placental) mammalian ancestor ~100 MY ago [46]. We name these piC loci as Eutherian-Conserved piRNA cluster (ECpiC) loci, which tended to yield many more total piRNAs per loci compared to the other Less-Conserved piRNA cluster (LCpiC) loci (Fig 3B).
Fig. 3. Eutherian-Conserved piRNA cluster (ECpiC) loci. 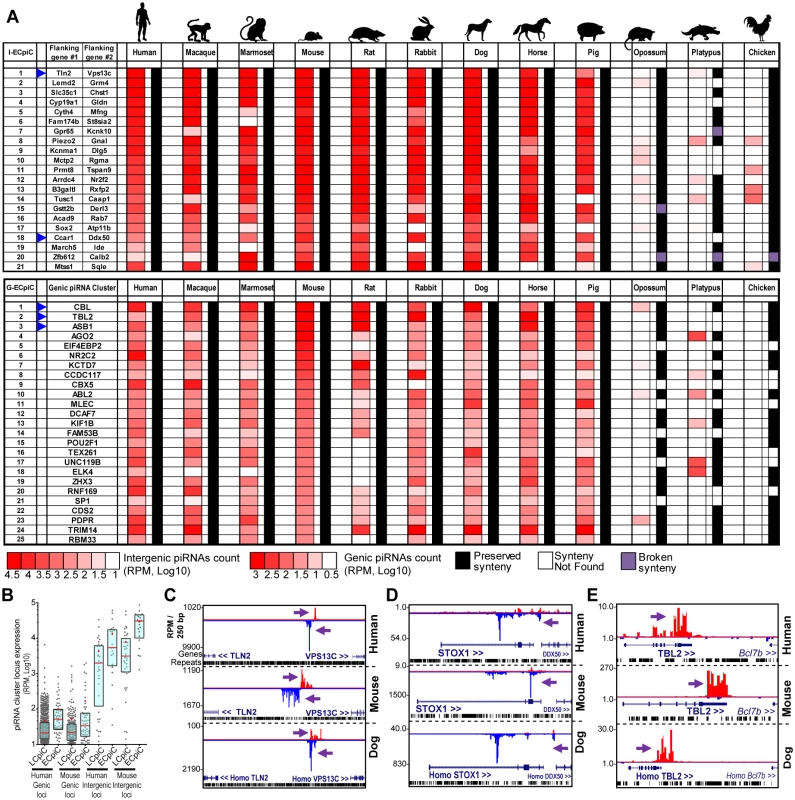
(A) Heatmap diagram of piRNA expression and gene synteny for intergenic ECpiC loci (top) and the top 25 of 56 genic ECpiC loci (bottom). Remaining genic ECpiC loci and genomic coordinates for intergenic ECpiC loci are shown in S3 Table. Blue triangles highlight the piC loci discussed further in the study. (B) Box plots of interquartile ranges of piC loci piRNA expression levels. The red line and cross mark the median and mean, respectively. ECpiC loci tend express more piRNAs than Less-Conserved piC (LCpiC) loci. Genome browser snapshots of two notable intergenic ECpiC loci (C, D), and one notable genic ECpiC locus (E) from a representative of Primates (human), Glires (mouse) and Laurasiatherian (dog). Purple arrows point to the start of bulk of piRNAs. Plus strand read peaks are red, minus strand read peaks are blue. Full compilations are in S6 Fig. Surprisingly, ECpiC loci were not consistently expressed in the opossum, a marsupial; and the platypus, a monotreme; which had diverged even further from Eutherian mammals at an additional ~60 and ~100 MY ago, respectively [45]. This is striking given that the synteny of genes around the ECpiC loci was still preserved, persisting from Eutherian mammals out to the chicken (Fig 3A). The mRNAs for the opossum, platypus, and chicken genes orthologous to genic ECpiC loci were also robustly expressed in the adult testes despite lacking the ability to generate piRNAs (Fig 4A). This suggests that the absence of conserved piRNA expression from the ECpiC loci in opossum, platypus, and chicken testes are not due to gene expression profile differences compared to Eutherian mammal testes. Furthermore, piRNA coverage in the opossum, platypus and chicken adult testes small RNA libraries was excellent and unobscured by miRNAs (Fig 4B)[19, 20]. Finally, we readily identified genic and intergenic piC loci in opossum, platypus, and chicken despite their draft genome states in the UCSC Genome Browser [47] (Fig 4C and 4D). Together, these results confirm ECpiC loci are only conserved in piRNA expression in Eutherian mammals, and suggest that the evolutionary conservation of ECpiC loci is due to preserving sequence elements directing piRNA biogenesis rather than piRNA cluster precursor transcript expression.
Fig. 4. Comparison of opossum, platypus and chicken piC loci to mouse piC loci. 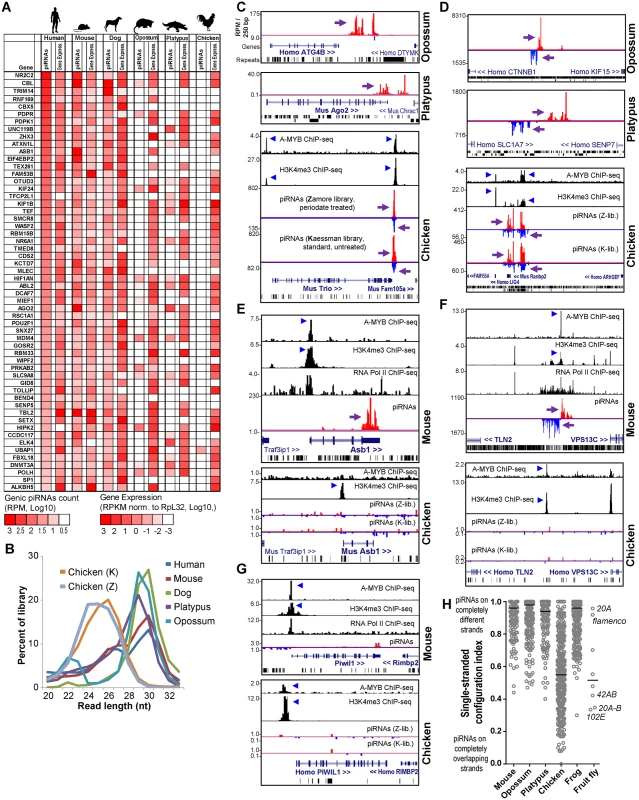
(A) Heatmap showing piRNA and gene expression levels for genic ECpiC loci compared to the orthologs in opossum, platypus and chicken, for which many lack piRNAs but the mRNA transcript is consistently expressed in the adult testes. (B) Read length distributions for each tetrapod small RNA library analyzed for this study. All libraries are from adult testes total RNA. (C) Genome browser snapshots of genic piC loci from opossum (top), platypus (middle), and chicken (bottom). Purple arrows point to the start of bulk of piRNAs. Plus strand read peaks are red, minus strand read peaks are blue. (D) Intergenic piC loci from opossum (top), platypus (middle), and chicken (bottom). Comparison of piRNA and chromatin mark differences between the mouse piC-Asb1 locus and the orthologous chicken Asb1 locus (E); the mouse intergenic ECpiC#1 and the syntenic region in chicken (F); and the mouse PIWIL1 locus to the chicken PIWIL1 locus (G). The blue triangles point to notable peaks in the ChIP-seq chromatin marks plotted from the data of Li et al [19]. (H) A plot of unique-strand configurations for the transcripts within major piC loci between representative mammals, chicken, frog and fly. Notable D.mel single-strand piC loci (index closer to 1) and dual-strand piC loci (index closer to 0) from D.mel are marked. The bar marks the median of the distribution. To further evaluate whether transcriptional regulation determines the choice of transcripts to become piRNA precursors, we considered a previous study showing the A-MYB transcription factor is conserved from mouse to chicken in binding to promoters of Piwi pathway genes to promote a feed-forward loop of piRNA biogenesis in the mouse and chicken testes (i.e. A-MYB peak at Piwil1 [19], Fig 4G). Although we observed many clear A-MYB peaks at the putative promoters of both chicken and mouse piC loci (Fig 4C–4F), there were notable examples of piRNA absence despite transcriptional activation, such as conserved A-MYB binding and H3K4me3 peaks in a chicken locus orthologous to mouse intergenic ECpiC#1, and conserved A-MYB and H3K4me3 peaks at mouse Piwil1. However, few piRNAs were detected from the intergenic piC syntenic chicken locus or from mouse and chicken Piwil1 (Fig 4E–4G). This contrasts with ample Piwil1 genic piRNAs in the rat (Fig 2F). We conclude that transcriptional activation in tetrapod testes, including by A-MYB, is insufficient to determine piRNA biogenesis. This evidence further supports our hypothesis that the evolution of piRNA biogenesis signatures is occurring within the sequence of the precursor RNA transcript rather than in the control of transcription initiation.
Although opossum and platypus piRNA clusters were configured similarly to mammalian piRNA clusters for piRNA biogenesis from non-overlapping transcripts (Fig 4D and 4F), chicken piC loci were surprisingly distinct in their configuration of piRNA biogenesis compared to mammalian piC loci. Mammalian piRNAs typically map only to single strands such as single-stranded mRNA transcripts in genic piC loci or to two transcripts that emanate as bi-directional non-overlapping strands from a common promoter region for some intergenic piC loci [4, 5, 19, 48] (Fig 4F). Many chicken piRNAs instead mapped to both plus and minus strands for multiple genic and intergenic piC loci (Fig 4C and 4D). When we calculated a single-stranded configuration index for the major piC loci from three mammals, the chicken, the frog and the fly D.mel (Fig 4H), this index accurately distinguished the single-stranded flam and 20A piC loci from the 42AB and 102E piC loci in the fly, and clearly confirmed that piC loci in all three mammals are predominantly single-stranded in their configurations. Interestingly, chicken piC loci displayed a wide range of piC configurations, with many more double-stranded piC loci with indexes similar to the fly 42AB and 102E double-stranded piC loci. Although the frog is evolutionarily a more distant tetrapod than chicken (~370 vs ~320 MY, respectively) in relation to humans [45], the frog piC loci determined from oocytes were surprisingly more single-stranded and similar in configuration to mammalian piC loci [49] (Fig 4H). These analyses and the shorter piRNA length distributions of chicken piRNAs (Fig 4B) strongly suggest that chicken piRNA biogenesis pathways may be more similar to flies compared to other tetrapods.
Developmental functions for Eutherian-Conserved piRNA cluster loci
We hypothesized that extensive conservation of piRNA expression patterns across ~100 MY of evolution implies conserved developmental functions for ECpiC loci. A likely conserved function for intergenic ECpiC loci would be to generate particularly essential TE-directed piRNAs, but TE sequences are neither more enriched nor more conserved in ECpiC loci versus LCpiC loci (S3 Table), and mouse genetic studies knocking out parts of intergenic piC loci are only now beginning to emerge [50]. However, a compelling non-TE repression role may be associated with intergenic ECpiC #18 that is located between the CCAR1 and DDX50 genes and generates many piRNAs antisense to the STOX1 transcript (Fig 3C, S6 Fig). ECpiC#18 is an intergenic piC locus because its transcript is antisense to STOX1, has lower coding potential than genic piC loci, and H3K4me3 ChIP-seq patterns suggests transcription begins in the intergenic region between STOX1 and DDX50. The STOX1 gene is implicated as a genetic factor linked to the placental-based disease of preeclampsia in humans [51–53].
A previous Gene Ontology analysis of mouse genic piC loci displayed an enrichment of gene function processes such as nucleic acid metabolism, transcription and regulation-related processes [11]. Such functions are also known for the genes that are ECpiC loci, such as the oncogenic transcription factors ABL2 and ELK4, the miRNA effector AGO2, and TBL2 and KCTD7 genes that are in the deleted locus of Williams-Beuren syndrome patients [54], which display developmental defects. While future genetic studies are required to investigate broader sets of ECpiCs, we looked for existing mutations in ECpiC loci that displayed developmental phenotypes and wondered if the mutations disrupted piRNA biogenesis. We noticed two prominent genic ECpiC loci corresponding to genes Cbl and Asb1 (Fig 5A). Knockout (KO) mice in each of these genes were created more than a decade ago because the genes were highly expressed in the blood [55, 56]. However, these mutants exhibited no major abnormalities, with the exception of being hypofertile. This defect was in both cases due to depleted spermatogenesis [55, 57] (Fig 5B).
Fig. 5. Phenotypes of two existing mouse mutants for genic ECpiC loci. 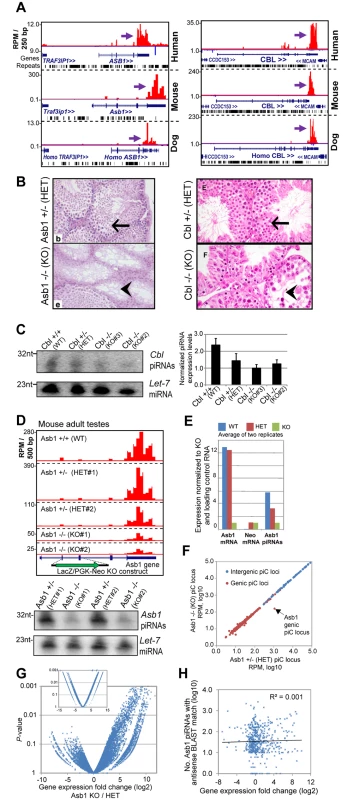
(A) Genome browser snapshot of piC-Asb1 (left) and piC-Cbl (right) in human, mouse and dog. Purple arrows point to the start of bulk of piRNAs. Plus strand read peaks are red, minus strand read peaks are blue. Full compilations are in S6 Fig. (B) Histology of mouse adult testes seminiferous tubules from the Asb1 mutant (left, image adapted from Fig 3 of [55], copyright of the American Society for Microbiology); and the Cbl mutant (right, image adapted from Fig 8 of [57], copyright of the Rockefeller University Press). Arrows points to normal spermatogenesis, the arrowheads points to empty tubules with sperm loss. HET, heterozygote, KO, homozygous knockout. (C) Northern blot analysis of Cbl piRNAs from adult testes from the mouse strain where Cbl exon 2 is flanked by LoxP sites (WT), or deleted by Cre recombinase for 1 allele (HET) or both alleles (KO). Quantitation of technical triplicates is shown to the right. (D) Top, profiles of Asb1 piRNAs from WT, HET and KO mutants which contain a LacZ/PGK-Neo insert disrupting exons 2 and 3. Two sets of animal pairs were examined. Bottom, Northern blot of Asb1 piRNAs and the Let-7 miRNA as loading control. (E) Quantitation of Asb1 piRNAs and Asb1 and Neomycin mRNAs. (F) Comparison of all piC loci expression between Asb1 KO and HET testes. (G) Volcano plot of gene expression profiles from testes (large plot) and kidney (smaller inset plot, same axis proportions). (H) Scatterplot of genes with predicted antisense-matching Asb1 piRNAs compared to gene expression changes between Asb1 KO and HET testes. The list of these gene names are in S4 Table. Could the mutations in Cbl and Asb1 be causing a loss of genic piRNAs in the testes that might explain the spermatogenic defects? Northern blotting revealed that Cbl piRNAs were reduced ~2-fold in Cbl Heterozygotes (HET) and KO mutants when compared to the Wild-type (WT)(Fig 5C), which coincides with difficulties breeding both HET and KO Cbl mutants [57]. Next, we resurrected from sperm the original Asb1 mutant mouse line and sequenced adult testes small RNA-seq and mRNA-seq libraries. Indeed, we found that the LacZ-PGK-Neo insert disrupting the Asb1 transcript greatly reduced Asb1 piRNAs ~6-fold in the KO and ~2-fold in the HET testes (Fig 5D and 5E). Other genic and intergenic piC loci were unaffected in the Asb1 KO testes compared to HET testes (Fig 5F), indicating the specificity of the mutation only affecting the piC-Asb1. Expression profiling indicated a significantly greater number of up-regulated genes than down-regulated genes when comparing Asb1 KO testes to HET testes (Fig 5G), whereas far fewer genes were either up - or down - regulated in KO versus HET kidney, a somatic tissue expressing Asb1. In addition, no mammalian TE transcripts were detected to be up-regulated in the Asb1 mutant testes. Some but not all predicted targets complementary to Asb1 piRNAs were up-regulated in Asb1 KO versus HET testes (Fig 5H), so future studies will be focused at distinguishing which up-regulated transcripts in the Asb1 mutant are direct or indirect targets of the piRNAs.
Discussion
Our study provides a new understanding of the evolutionary patterns for animal piRNAs and piC loci. These integral components of the ancient Piwi pathway evolve much more quickly than RNAi protein factors and other small regulatory RNAs like miRNAs, which can be conserved as far back as ~500 MY of evolution [1, 2]. By matching orthologous piC loci via synteny and protein orthology, and then broadly profiling piRNAs and mRNAs across animal gonads, we confirm rapid evolution in the piRNA expression patterns for both genic and intergenic piC loci even between close relatives within insect and mammalian clades that have only diverged by ~10 MY of evolution. This rapid gain of species-specific genic piC loci in both insects and tetrapods is consistent with a previous study’s proposal that piC loci are expanding rapidly via positive selection processes [14]. However, Eutherian mammals have distinctly conserved the expression of piRNAs from a notable set of ECpiC loci through ~100 MY of evolution, whereas very few deeply conserved piRNA expression patterns were observed in flies.
What might explain the lack of deeply-conserved piRNA expression patterns in flies? Perhaps piC loci evolution may be accelerated in Drosophilids due to certain aspects of the Piwi pathway that are currently unique to Drosophila, such as the capacity of de novo TE insertions to promote new piRNAs from flanking genomic sequence [58, 59], Drosophila-specific piRNA biogenesis factors like RHINO, CUTOFF, and DEADLOCK [58, 60–62], and epigenetic induction and suppression of piRNA biogenesis [63–68]. In addition, whereas only a minority of mammalian piRNAs are complementary to TEs [4, 5], the majority of Drosophila piRNAs are complementary to TEs [13, 30, 31], perhaps reflecting a more dynamic “arms race” between TEs and Drosophilid piRNAs [32–34].
Nonetheless, the rapid evolution for the majority of piC loci in both mammals and flies may also suggest that many piC loci are evolving by non-adaptive evolutionary forces that result in diverse piRNA repertoires, which germ cells may simply tolerate along with their highly diverse transcriptomes [69]. In addition, this diversity of piC loci between species may be attributed to higher frequencies of mutations in non-coding portions of genes and intergenic regions that allow transcripts to enter or leave the piRNA biogenesis pathway with greater frequency, since these turnover events may be under relaxed evolutionary constraints [14]. Nevertheless, some specific piC loci might also behave differently from the bulk of piC loci with regards to evolutionary constraints, as has been proposed for some human piC loci [15, 70]. Thus, the conservation of piRNA expression from ECpiC loci is striking, and we speculate these loci may have been selected to yield high levels of specific piRNAs in order to promote gene expression profiles favoring spermatogenic and embryonic fitness.
This list of ECpiC loci will help us prioritize which loci to generate future piC loci mutants, and leads us to wonder if their functions will be tied to unique aspects of Eutherian reproduction, such as placental development and paternal genome regulation during spermatogenesis. Our analysis also suggests that most piC loci in vertebrates and flies are evolving under possibly non-adaptive evolutionary forces, whereby genetic drift may frequently create species - and clade-specific sequence motifs or RNA structural elements that allow diverse repertoires of transcripts to frequently enter (and exit) the piRNA biogenesis pathway. Although animal gonads are notable for their generally promiscuous transcriptional activity as well as rapid evolutionary turnover of genes [71, 72], we still observed that gene expression profiles between species are more similar than genic piC loci piRNA expression profiles (Figs 1 and 2). We propose a model for most piC loci being neutral for gonadogenesis fitness, and the plethora of individual piC loci may result in functional redundancy and allow fluidity in the emergence and evolutionary turnover of piC loci (Fig 6). However, some piC loci have been subjected to adaptive evolutionary forces in order to help germ cells suppress TE mobilization, such as prominent intergenic piRNA cluster loci serving as master TE control loci in Drosophilids [30].
Fig. 6. A model for the evolution of piC loci expression patterns. 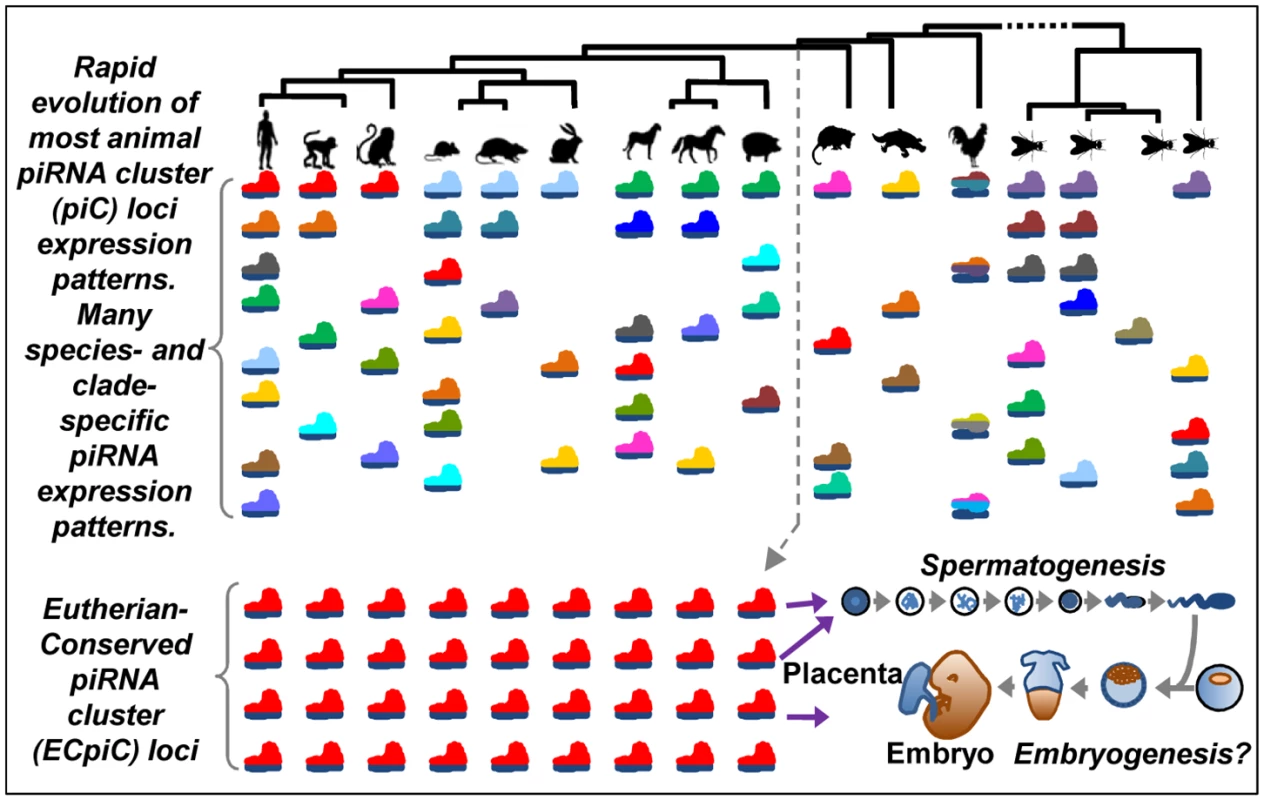
The rainbow of colored pictograms represents the diverse number of piC loci that are generally distinctly expressed between animal species or clades. We propose that most piC loci have evolved rapidly under non-adaptive evolutionary forces to result in this great diversity of piC loci expression patterns. However, a distinct set of piC loci (the same red-colored pictograms) is conserved in piRNA expression for >100M years of evolution in Eutherian mammals, perhaps to serve roles in mammalian reproduction. Mutations disrupting Asb1 and Cbl in the mouse appear to also disrupt the piRNAs from these individual piC loci, hinting that the loss of piRNAs may be causing the spermatogenic defect. Indeed, more genes are up-regulated than down-regulated in the Asb1 mutant testes, with several up-regulated genes having complementarity to Asb1 piRNAs (Fig 5). Although we cannot exclude the idea that loss of Asb1 protein function is causing the spermatogenic defect, we note that five other close Asb1 homologs (Asb3, -4, -5, -8, and -9) are still highly expressed in the Asb1 mutant and might provide protein function compensation [55], whereas only Asb1 generates an abundant cluster of piRNAs. Future efforts should be directed at creating new mouse mutants that replace the endogenous 3'UTRs of Asb1 and Cbl with an equivalently long 3'UTR of a different gene that is also expressed highly in the germline but does not generate piRNAs. This approach may be better than simply deleting the endogenous 3'UTRs because Asb1 and Cbl mRNA stability and translation would likely be strongly diminished if they completely lacked a 3'UTR.
Our discovery of ECpiC loci suggests new regulatory functions beyond TE repression to accommodate specific aspects of Eutherian reproduction. There is a precedent for an epigenetic process specific to Eutherian reproduction, such as random X-chromosome inactivation via the Xist non-coding RNA, which differs from the paternally imprinted inactive X chromosome in marsupials and stochastic dosage compensation in monotremes [73]. ECpiC loci such as piC-Asb1, piC-Cbl, and ECpiC#18 may represent new gene targets to examine for cases of hypo-fertility, which may be more likely to persist in a population than completely sterile mutations. Albeit reduced in fecundity, reproduction may be still viable in young piC locus mutant animals, but germ cell longevity may also be limited. When new ECpiC loci mutants are made that will only disrupt the piRNAs while leaving the protein coding gene intact, we will be better able to discern which target genes are mis-regulated during spermatogenic decline or defects on placental development. Since the piRNAs from ECpiC#18 are mainly antisense to the STOX1 mRNA, a gene clinically implicated in placental development [51–53], we speculate that mis-regulation of ECpiC#18 might likely impact STOX1 expression and contribute to sperm phenotypes that fall within a hypothesis for paternal factors in the etiology of preeclampsia [74]. Future characterization and genetic studies in mouse will be useful to determine if ECpiC#18 directly regulates the STOX1 locus.
Materials and Methods
Ethics statement
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Mice and rats were euthanized with CO2 and followed with cervical dislocation or decapitation, prior to testes dissection. Rabbit tissues were purchased, while other vertebrate datasets were downloaded from the NCBI Sequencing Read Archive. This work was conducted under the approval of the Brandeis University Institutional Animal Care and Use Committee (IACUC) under the protocol #13013 to NCL.
Library construction and deep sequencing of small RNAs and mRNAs from Drosophilid ovarium samples and Glires testes samples
To deplete the transposon-directed piRNAs which overwhelm genic piRNAs in typical total Drosophila ovaries small RNA libraries [13], we dissected >500 ovaries in cold PBS and that were treated to a trypsinization and gravity sedimentation protocol that ruptures most nurse cells and oocytes but leaves the layer of follicle cells intact [28]. The D.mel strain was Oregon-R, while D.ere, D.yak, and D.vir strains were the standard wild-type strains used in the genome projects and obtained from the UCSD Drosophila Species Stock Center. Total RNA from rodent testes was extracted using TRI-reagent (MRC) using manufacturer instructions. Small RNA in size range from 18 to 35 bases were gel purified, for Drosophila samples depletion from 2S rRNA was performed and libraries were constructed as described in [28].
To construct mRNAseq libraries, total RNA samples were subjected to two rounds of poly-A enrichment using biotinylated (dT)x18 oligonucleotides and the polyATtract kit (Promega). RNA-seq libraries were generated using the ScriptSeq V2 construction protocol (Epicenter, performed according to manufacturer’s instructions). Sequencing was performed on an Illumina HiSeq 2000, and 50 bases long reads were processed and split according to their index primer barcodes.
All read data that we generated for this study has been deposited in the Sequencing Reads Archive (SRA) of the Gene Expression Omnibus (GEO) under the accession number of GSE62556. All the other small RNA and mRNA datasets were downloaded from GEO and SRA, with all accession numbers recorded in S1 Table.
Identification of genic and intergenic piRNA cluster loci expression patterns
The initial deep sequencing library preparation steps with standard processing tools is described in the Supplementary Materials Additional Experimental Procedures section. Genic piRNA clusters were determined by a process (S1c Fig) that begins with a custom script (ngs_genecentric_wig.c) that compares the counts in a WIG file to a pre-defined gene structure BED file (obtained and modified from UCSC genome browser). The script calculates the 5’UTR, exon and 3’UTR read counts for each gene in the BED file. When preforming the 5’UTR and 3’UTR counting, the script automatically extends 5’UTR or 3’UTR regions by a window of 2000bp (500bp for Drosophilids) if it detects at least 1 RPM reads within the window. After downloading original Refseq BED tables from the UCSC genome browser website, we modified the RefSeq BED files to conform to our custom script. For human, fly, and mouse, we kept the original species-specific RefSeq annotations. But for the rat and rabbit, we used the mouse RefSeq annotation first, and then supplemented it further with human RefSeq annotations where mouse annotations were missing. For the marmoset, monkey, dog, horse, pig, platypus, opossum, and chicken; we first used the human annotation, then the mouse annotation, and finally add the species specific annotations.
We curated these tables by only retaining genes with ≥10 RPMs of piRNA counts in the 3’UTR of the same strand orientation of the mRNA. In mammals, between 1–5% of genic piRNA clusters also had at least 50 RPMs of piRNAs mapping to the ORF that were kept in the analysis. We further groomed these tables by removing false positives that were genes with exceedingly high 3’UTR counts which were ambiguously attributed to a nearby transposable element, or were adjacent to another gene that was actually generating the piRNAs, was a duplicated gene name entry, or was a non-coding RNA (i.e. many mouse RefSeq non-coding RNAs with the “gm####” identifier). We finally removed additional false positives that were genes with incorrect homolog alignments caused by their simple and repetitive protein domains, such as histones, olfactory receptors and zinc finger protein genes.
Mouse, human and flies intergenic clusters lists were previously determined in [4, 5, 13, 29, 30]. Intergenic clusters for other species were detected with a custom script (ngs_coverage-nelson.c) that uses a 5 Kb sliding window, initiates a cluster when the window read count is > 1 RPM, and terminates the cluster when the read count is < 1 RPM. Intergenic piRNA cluster loci defined by contiguous intervals were then kept if they totalled ≥10RPMs. We computed the Single-stranded configuration index of piRNA clusters by first taking each cluster locus and splitting it into windows of 5 Kb each, and then for each of these windows we calculated the formula [(A-B)^2]/(A2+B2), where “A” is the plus strand read counts in RPM and “B” is the minus strand read counts in RPM within a window. The single-stranded configuration index of a cluster is the average single-stranded configuration index of all the 5Kb windows.
Overlap analyses of gene expression profiles and genic piC loci expression profiles
The gene expression profiles for samples from human, mouse and D.mel were determined by mapping mRNA-seq reads to RefSeq transcripts with Bowtie (up to 2 mismatches). Read counts were normalized to mapping library size (RPM) and then gene length (RPKM), and finally each gene was normalized to a housekeeping gene (RpL32 in mammals and in flies, RpL32 is also known as Rp49 in flies). Since RefSeq transcript libraries are deficient in other animals, for the other Drosophilids, mapped reads were then transformed to D.mel gene models through orthologs listed in the OrthoDB database and then RPM counts were determined as described in [75]. mRNA-seq reads for non-human Primates, Laurasiatherians, and other tetrapods were mapped to their respective genomes, and then the counting algorithm from genic piC discovery was applied to yield exonic counts that were as accurate as mapping to RefSeq transcripts, except that no additional curation was applied.
Differential gene expression analysis between Asb1 KO and Het mutant mouse tissues was conducted with the edgeR package [76] on two biological replicates from testes and one sample of kidney. For predicting targets of piC-Asb1, Asb1 piRNAs were queried against the mouse RefSeq transcripts using the “blastn-short” command in BLAST [77] and then sorting for BLAST results with an e-value≤1, which in this query frequently demanded at least 13bp of complementarity between the piRNA and the predicted target. Custom Perl scripts and SQL queries counted the BLAST matches and gene expression changes.
For the species comparisons Venn diagrams, the genic piC names and the gene expression profiles were rooted to the best annotated model organism gene names from D.mel for Drosophilids; and human and mouse for mammals and chicken. Genes and genic piC loci could thus be compared between species with SQL queries. Overlaps analyses in genic piC locus conservation required threshold piRNA expressions to be ≥10 RPMs (Figs 1F and 2G) and ≥20 RPMs (S2B and S4G Figs). Overlap in gene expression profiles were defined as similar expression levels of RPKM values normalized against the RpL32/Rp49 housekeeping gene to be within 3 fold of each two species comparison (Log10 delta value ≤ 0.5) and within 2 fold for three species comparisons (Log10 delta value standard deviation ≤ 0.3) for analyses in Figs 1F and 2G. For gene expression analyses in S2B and S4G Figs, the Log10 delta value ≤0.3 and Log10 delta value standard deviation ≤ 0.177 was used. To test if the distribution of piC locus overlaps were significantly different from the distribution of similar gene expression profiles, we calculated both the Chi-square test and the Ratios proportion Z-score test with Bonferroni correction. The human Study #2 dataset is a second independent sample of small RNAs generated from human testes, and it shares much similarity with the human Study #1 dataset, such as similar piRNA cluster loci patterns in Figs 2B, 2C and S4I. All human piC loci comparisons in Fig 3 were done with the human Study#1 datasets after we had confirmed highly similar profiles and counts for all ECpiC loci in the human Study#2 dataset.
Evolution and feature analyses of genic piRNA cluster loci
We ranked genic piC loci by piRNA abundance (RPMs) and selected top cohorts of 278 and 332 genic piC loci for fly and mouse, respectively. These sets were divided into a top and bottom half, and compared to lists of negative control genes that do not generate piRNAs. The negative control list contained a total of 3 times as many genes as genic piC loci to diminish selection bias, and >90% of these negative control genes were also expressed in both fly follicle cell enriched samples and mouse adult testes. The 3’UTR boundaries of the genic piC loci were defined by the previous piRNA tracking algorithm while the negative controls were selected based on the longest Refseq-annotated 3’UTR. In order to ensure that genes in negative control set have approximately similar 3’UTR length compared to genic piC loci set, we padded the 3’UTR in fly by 250 to 500 bases and in mouse by 500 to 1100 bases, unless the extension overlaps with a gene in the same strand. The genomic coordinates for the 3’UTRs and CDS (minus introns) were determined for each negative control gene and genic piC; and used to track and count predicted RNA structural elements and phastCons and phyloP scores.
RNA structural elements from fly genome (Release 5/Dm3) were determined with RNAz, Evofold, and REAPR applied to the 12 Drosophilids alignments with the deviation parameter, dev = 20 and confidence scores >0.6 [39–41], while only REAPR was applied to the mouse genome (Mm10) using an alignment of 8 Glire genomes and measured at deviation levels of dev = 10 and confidence scores >0.8. The per-base values of phastCons [43] and phyloP [44] scores were downloaded from UCSC Genome browser with the exception of fly phyloP scores computed by David Garfield at the EMBL, and were based off the 12 Drosophilids genomes and 60 vertebrate genomes alignments. Total per-base values were summed and then averaged by the base length for each gene using BEDtools [78] and custom Perl scripts to count the features for each gene. Sequence motif analysis was performed with MEME [42] using an “OOPS (only once per sequence)” model; and with GLAM2 on CDS and 3’UTR sequences of the longest isoform from the masked genome.
To determine the gain and loss rates for genic piC loci in Drosophilids and tetrapods, we constructed phylogenetic trees according to the Gregorian clock of millions of years of divergence and the tabulated expression or absence of expression for each piC loci with gene orthologs present between all the species. We followed the similar procedure of measuring gain and loss rates of miRNA genes as detailed in [20], using the COUNTS program [79] and the Wagner parsimony approach. Additional experimental procedures are in the Supplementary Materials document describing the verification of gene expression using RT-qPCR, small RNA northern blotting, and the PIWI IP from OSS cells and PIWIL2 IP from Glire testes.
Supporting Information
Zdroje
1. Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455(7217):1193–7. Epub 2008/10/03. doi: 10.1038/nature07415 18830242
2. Juliano CE, Reich A, Liu N, Gotzfried J, Zhong M, Uman S, et al. PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(1):337–42. doi: 10.1073/pnas.1320965111 24367095
3. Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–9. Epub 2000/11/18. 11081512
4. Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. 16751776
5. Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313(5785):363–7. 16778019
6. Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes & development. 2012;26(21):2361–73.
7. Clark JP, Lau NC. Piwi Proteins and piRNAs step onto the systems biology stage. Advances in experimental medicine and biology. 2014;825 : 159–97. doi: 10.1007/978-1-4939-1221-6_5 25201106
8. Huang CR, Burns KH, Boeke JD. Active transposition in genomes. Annual review of genetics. 2012;46 : 651–75. doi: 10.1146/annurev-genet-110711-155616 23145912
9. Lau NC. Small RNAs in the animal gonad: Guarding genomes and guiding development. Int J Biochem Cell Biol. 2010. Epub 2010/03/17.
10. Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316(5825):744–7. 17446352
11. Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, et al. A broadly conserved pathway generates 3' UTR-directed primary piRNAs. Current Biology. 2009;19(24):2066–76. Epub 2009/12/17. doi: 10.1016/j.cub.2009.11.064 20022248
12. Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461(7268):1296–9. Epub 2009/10/09. doi: 10.1038/nature08501 19812547
13. Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137(3):522–35. Epub 2009/04/28. doi: 10.1016/j.cell.2009.03.040 19395010
14. Assis R, Kondrashov AS. Rapid repetitive element-mediated expansion of piRNA clusters in mammalian evolution. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):7079–82. doi: 10.1073/pnas.0900523106 19357307
15. Gould DW, Lukic S, Chen KC. Selective constraint on copy number variation in human piwi-interacting RNA Loci. PloS one. 2012;7(10):e46611. doi: 10.1371/journal.pone.0046611 23056369
16. Gebert D, Ketting RF, Zischler H, Rosenkranz D. piRNAs from Pig Testis Provide Evidence for a Conserved Role of the Piwi Pathway in Post-Transcriptional Gene Regulation in Mammals. PloS one. 2015;10(5):e0124860. doi: 10.1371/journal.pone.0124860 25950437
17. Roovers EF, Rosenkranz D, Mahdipour M, Han CT, He N, Chuva de Sousa Lopes SM, et al. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell reports. 2015;10(12):2069–82. doi: 10.1016/j.celrep.2015.02.062 25818294
18. Rosenkranz D, Rudloff S, Bastuck K, Ketting RF, Zischler H. Tupaia small RNAs provide insights into function and evolution of RNAi-based transposon defense in mammals. Rna. 2015;21(5):911–22. doi: 10.1261/rna.048603.114 25802409
19. Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Molecular cell. 2013;50(1):67–81. doi: 10.1016/j.molcel.2013.02.016 23523368
20. Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, et al. Birth and expression evolution of mammalian microRNA genes. Genome research. 2013;23(1):34–45. doi: 10.1101/gr.140269.112 23034410
21. Ha H, Song J, Wang S, Kapusta A, Feschotte C, Chen KC, et al. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC genomics. 2014;15 : 545. doi: 10.1186/1471-2164-15-545 24981367
22. Hirano T, Iwasaki YW, Lin ZY, Imamura M, Seki NM, Sasaki E, et al. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. Rna. 2014;20(8):1223–37. doi: 10.1261/rna.045310.114 24914035
23. Lian C, Sun B, Niu S, Yang R, Liu B, Lu C, et al. A comparative profile of the microRNA transcriptome in immature and mature porcine testes using Solexa deep sequencing. The FEBS journal. 2012;279(6):964–75. doi: 10.1111/j.1742-4658.2012.08480.x 22240065
24. Platt RN 2nd, Vandewege MW, Kern C, Schmidt CJ, Hoffmann FG, Ray DA. Large numbers of novel miRNAs originate from DNA transposons and are coincident with a large species radiation in bats. Molecular biology and evolution. 2014;31(6):1536–45. doi: 10.1093/molbev/msu112 24692655
25. Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, et al. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic acids research. 2011;39(15):6596–607. doi: 10.1093/nar/gkr298 21546553
26. Yang Q, Hua J, Wang L, Xu B, Zhang H, Ye N, et al. MicroRNA and piRNA profiles in normal human testis detected by next generation sequencing. PloS one. 2013;8(6):e66809. doi: 10.1371/journal.pone.0066809 23826142
27. Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, et al. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. Rna. 2007;13(11):1911–22. Epub 2007/09/18. 17872506
28. Matts JA, Sytnikova Y, Chirn GW, Igloi GL, Lau NC. Small RNA library construction from minute biological samples. Methods in molecular biology. 2014;1093 : 123–36. doi: 10.1007/978-1-62703-694-8_10 24178561
29. Rozhkov NV, Aravin AA, Zelentsova ES, Schostak NG, Sachidanandam R, McCombie WR, et al. Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. Rna. 2010;16(8):1634–45. Epub 2010/06/29. doi: 10.1261/rna.2217810 20581131
30. Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–103. Epub 2007/03/10. 17346786
31. Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137(3):509–21. Epub 2009/04/28. doi: 10.1016/j.cell.2009.04.027 19395009
32. Kelleher ES, Barbash DA. Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense. Molecular biology and evolution. 2013;30(8):1816–29. doi: 10.1093/molbev/mst081 23625890
33. Lu J, Clark AG. Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila. Genome research. 2010;20(2):212–27. doi: 10.1101/gr.095406.109 19948818
34. Castillo DM, Mell JC, Box KS, Blumenstiel JP. Molecular evolution under increasing transposable element burden in Drosophila: a speed limit on the evolutionary arms race. BMC evolutionary biology. 2011;11 : 258. doi: 10.1186/1471-2148-11-258 21917173
35. Claycomb JM. Ancient endo-siRNA pathways reveal new tricks. Current biology: CB. 2014;24(15):R703–15. doi: 10.1016/j.cub.2014.06.009 25093565
36. Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, et al. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155(4):807–16. doi: 10.1016/j.cell.2013.10.001 24209619
37. Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453(7194):534–8. Epub 2008/04/12. doi: 10.1038/nature06904 18404147
38. Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & development. 2008;22(7):908–17. Epub 2008/04/03.
39. Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450(7167):219–32. Epub 2007/11/13. 17994088
40. Will S, Yu M, Berger B. Structure-based whole-genome realignment reveals many novel noncoding RNAs. Genome research. 2013;23(6):1018–27. doi: 10.1101/gr.137091.111 23296921
41. Washietl S, Pedersen JS, Korbel JO, Stocsits C, Gruber AR, Hackermuller J, et al. Structured RNAs in the ENCODE selected regions of the human genome. Genome research. 2007;17(6):852–64. 17568003
42. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37(Web Server issue):W202–8. Epub 2009/05/22. doi: 10.1093/nar/gkp335 19458158
43. Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome research. 2005;15(8):1034–50. 16024819
44. Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome research. 2010;20(1):110–21. doi: 10.1101/gr.097857.109 19858363
45. Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505(7485):635–40. doi: 10.1038/nature12943 24463510
46. Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478(7370):476–82. doi: 10.1038/nature10530 21993624
47. Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42(Database issue):D764–70. doi: 10.1093/nar/gkt1168 24270787
48. Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–7. 16751777
49. Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. The EMBO journal. 2009;28(19):2945–58. Epub 2009/08/29. doi: 10.1038/emboj.2009.237 19713941
50. Homolka D, Pandey RR, Goriaux C, Brasset E, Vaury C, Sachidanandam R, et al. PIWI Slicing and RNA Elements in Precursors Instruct Directional Primary piRNA Biogenesis. Cell reports. 2015;12(3):418–28. doi: 10.1016/j.celrep.2015.06.030 26166577
51. van Dijk M, Drewlo S, Oudejans CB. Differential methylation of STOX1 in human placenta. Epigenetics: official journal of the DNA Methylation Society. 2010;5(8):736–42.
52. van Dijk M, Oudejans CB. STOX1: Key player in trophoblast dysfunction underlying early onset preeclampsia with growth retardation. Journal of pregnancy. 2011;2011 : 521826. doi: 10.1155/2011/521826 21490791
53. van Dijk M, Mulders J, Poutsma A, Konst AA, Lachmeijer AM, Dekker GA, et al. Maternal segregation of the Dutch preeclampsia locus at 10q22 with a new member of the winged helix gene family. Nature genetics. 2005;37(5):514–9. 15806103
54. Merla G, Howald C, Henrichsen CN, Lyle R, Wyss C, Zabot MT, et al. Submicroscopic deletion in patients with Williams-Beuren syndrome influences expression levels of the nonhemizygous flanking genes. American journal of human genetics. 2006;79(2):332–41. 16826523
55. Kile BT, Metcalf D, Mifsud S, DiRago L, Nicola NA, Hilton DJ, et al. Functional analysis of Asb-1 using genetic modification in mice. Molecular and cellular biology. 2001;21(18):6189–97. Epub 2001/08/18. 11509662
56. Naramura M, Kole HK, Hu RJ, Gu H. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(26):15547–52. 9861006
57. El Chami N, Ikhlef F, Kaszas K, Yakoub S, Tabone E, Siddeek B, et al. Androgen-dependent apoptosis in male germ cells is regulated through the proto-oncoprotein Cbl. The Journal of cell biology. 2005;171(4):651–61. 16301331
58. Mohn F, Sienski G, Handler D, Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157(6):1364–79. doi: 10.1016/j.cell.2014.04.031 24906153
59. Shpiz S, Ryazansky S, Olovnikov I, Abramov Y, Kalmykova A. Euchromatic transposon insertions trigger production of novel Pi - and endo-siRNAs at the target sites in the drosophila germline. PLoS genetics. 2014;10(2):e1004138. doi: 10.1371/journal.pgen.1004138 24516406
60. Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, et al. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell. 2014;157(6):1353–63. doi: 10.1016/j.cell.2014.04.030 24906152
61. Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138(6):1137–49. doi: 10.1016/j.cell.2009.07.014 19732946
62. Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes & development. 2014;28(15):1667–80.
63. Le Thomas A, Marinov GK, Aravin AA. A transgenerational process defines piRNA biogenesis in Drosophila virilis. Cell reports. 2014;8(6):1617–23. doi: 10.1016/j.celrep.2014.08.013 25199836
64. de Vanssay A, Bouge AL, Boivin A, Hermant C, Teysset L, Delmarre V, et al. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490(7418):112–5. doi: 10.1038/nature11416 22922650
65. Grentzinger T, Armenise C, Brun C, Mugat B, Serrano V, Pelisson A, et al. piRNA-mediated transgenerational inheritance of an acquired trait. Genome research. 2012. Epub 2012/05/05.
66. Erwin AA, Galdos MA, Wickersheim ML, Harrison CC, Marr KD, Colicchio JM, et al. piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of D. virilis. PLoS genetics. 2015;11(8):e1005332. doi: 10.1371/journal.pgen.1005332 26241928
67. Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450(7167):304–8. Epub 2007/10/24. 17952056
68. Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, et al. piRNA production requires heterochromatin formation in Drosophila. Curr Biol. 2011;21(16):1373–9. doi: 10.1016/j.cub.2011.06.057 21820311
69. Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell reports. 2013;3(6):2179–90. doi: 10.1016/j.celrep.2013.05.031 23791531
70. Lukic S, Chen K. Human piRNAs are under selection in Africans and repress transposable elements. Molecular biology and evolution. 2011;28(11):3061–7. doi: 10.1093/molbev/msr141 21613236
71. Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309(5742):1850–4. 16141373
72. Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS biology. 2005;3(11):e357. 16201836
73. Escamilla-Del-Arenal M, da Rocha ST, Heard E. Evolutionary diversity and developmental regulation of X-chromosome inactivation. Human genetics. 2011;130(2):307–27. doi: 10.1007/s00439-011-1029-2 21687993
74. Dekker G, Robillard PY, Roberts C. The etiology of preeclampsia: the role of the father. Journal of reproductive immunology. 2011;89(2):126–32. doi: 10.1016/j.jri.2010.12.010 21529966
75. Arnold CD, Gerlach D, Spies D, Matts JA, Sytnikova YA, Pagani M, et al. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nature genetics. 2014;46(7):685–92. doi: 10.1038/ng.3009 24908250
76. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616 19910308
77. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC bioinformatics. 2009;10 : 421. doi: 10.1186/1471-2105-10-421 20003500
78. Quinlan AR, Kindlon N. bedtools: a powerful toolset for genome arithmetic. v2.22.1 ed. http://bedtools.readthedocs.org/2014.
79. Csuros M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26(15):1910–2. doi: 10.1093/bioinformatics/btq315 20551134
80. Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC bioinformatics. 2011;12 : 357. doi: 10.1186/1471-2105-12-357 21880147
Štítky
Genetika Reprodukčná medicína
Článek A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin TransporterČlánek Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis inČlánek Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 11- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Agricultural Genomics: Commercial Applications Bring Increased Basic Research Power
- Ernst Rüdin’s Unpublished 1922-1925 Study “Inheritance of Manic-Depressive Insanity”: Genetic Research Findings Subordinated to Eugenic Ideology
- Convergent Evolution During Local Adaptation to Patchy Landscapes
- The Locus Controls Age at Maturity in Wild and Domesticated Atlantic Salmon ( L.) Males
- A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin Transporter
- Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting
- Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes
- Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep
- Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein
- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Mosaic and Intronic Mutations in Explain the Majority of TSC Patients with No Mutation Identified by Conventional Testing
- Members of the Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in
- QTL Mapping of Sex Determination Loci Supports an Ancient Pathway in Ants and Honey Bees
- Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing
- Genomics of Cancer and a New Era for Cancer Prevention
- Adaptation to High Ethanol Reveals Complex Evolutionary Pathways
- Dynamics of Transcription Factor Binding Site Evolution
- Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
- Enhancer Runaway and the Evolution of Diploid Gene Expression
- Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis
- Drosophila Mutants Model Cornelia de Lange Syndrome in Growth and Behavior
- Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases
- Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
- Tissue-Specific Effects of Reduced β-catenin Expression on Mutation-Instigated Tumorigenesis in Mouse Colon and Ovarian Epithelium
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Mapping of Craniofacial Traits in Outbred Mice Identifies Major Developmental Genes Involved in Shape Determination
- Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- DNA Repair Cofactors ATMIN and NBS1 Are Required to Suppress T Cell Activation
- Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
- Ernst Rüdin and the State of Science
- ABCs of Insect Resistance to Bt
- Epigenetic Control of O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
- The Fanconi Anemia Pathway Protects Genome Integrity from R-loops
- Controls Quantitative Variation in Maize Kernel Row Number
- Genome-Wide Association Study of Golden Retrievers Identifies Germ-Line Risk Factors Predisposing to Mast Cell Tumours
- Insect Resistance to Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein
- A Cytosine Methytransferase Modulates the Cell Envelope Stress Response in the Cholera Pathogen
- Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals
- The Multi-allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance
- The lncRNA Controls Cryptococcal Morphological Transition
- Sae2 Function at DNA Double-Strand Breaks Is Bypassed by Dampening Tel1 or Rad53 Activity
- A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia,
- Ectodysplasin/NF-κB Promotes Mammary Cell Fate via Wnt/β-catenin Pathway
- The QTL within the Complex Involved in the Control of Tuberculosis Infection in Mice Is the Classical Class II Gene
- Identifying Loci Contributing to Natural Variation in Xenobiotic Resistance in
- Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by and Subsistence
- A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data
- Competition between Heterochromatic Loci Allows the Abundance of the Silencing Protein, Sir4, to Regulate Assembly of Heterochromatin
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



