-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
Pollen development, or male gametogenesis, is a process by which a haploid uninucleate microspore undergoes cell division and specification to form a mature pollen grain containing two sperm cells. The highly defined cell linage makes pollen development an ideal model to understand the regulation of plant cellular development. Pollen development has multiple phases and involves dynamic changes in gene expression, which highlights the importance of transcription factors and their regulatory pathway(s). In this report, we demonstrate that WRKY34 and WRKY2, two closely related WRKY transcription factors in Arabidopsis, play important roles in pollen development. WRKY34 is phosphorylated by MPK3/MPK6, two functionally redundant mitogen-activated protein kinases (MAPKs or MPKs), at early stages in pollen development. Utilizing a combination of genetic, biochemical, and cytological tools, we determined that this MAPK-WRKY signaling module functions at the early stage of pollen development. Loss of function of this pathway reduces pollen viability, and the surviving pollen has poor germination and reduced pollen tube growth, all of which reduce the transmission rate of the mutant pollen. This study discovers a novel stage-specific signaling pathway in pollen development.
Published in the journal: Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in. PLoS Genet 10(5): e32767. doi:10.1371/journal.pgen.1004384
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004384Summary
Pollen development, or male gametogenesis, is a process by which a haploid uninucleate microspore undergoes cell division and specification to form a mature pollen grain containing two sperm cells. The highly defined cell linage makes pollen development an ideal model to understand the regulation of plant cellular development. Pollen development has multiple phases and involves dynamic changes in gene expression, which highlights the importance of transcription factors and their regulatory pathway(s). In this report, we demonstrate that WRKY34 and WRKY2, two closely related WRKY transcription factors in Arabidopsis, play important roles in pollen development. WRKY34 is phosphorylated by MPK3/MPK6, two functionally redundant mitogen-activated protein kinases (MAPKs or MPKs), at early stages in pollen development. Utilizing a combination of genetic, biochemical, and cytological tools, we determined that this MAPK-WRKY signaling module functions at the early stage of pollen development. Loss of function of this pathway reduces pollen viability, and the surviving pollen has poor germination and reduced pollen tube growth, all of which reduce the transmission rate of the mutant pollen. This study discovers a novel stage-specific signaling pathway in pollen development.
Introduction
Pollen, the male gametophyte of angiosperms, displays highly reduced structure of two or three cells at maturity. Because of the simple cell linage and dynamic developmental processes, plant male gametogenesis provides an interesting model for studying many fundamental cellular processes, including cell specification, cell polarity, cell cycle, and transcriptional regulation in these processes. During male gametogenesis, the uninucleate microspore (uninucleate microspore stage, UNM) undergoes an asymmetric mitosis to generate a large vegetative cell and a generative cell within it (bicellular pollen stage, BCP). In Arabidopsis thaliana, before pollen maturation, the generative cell undergoes a second symmetric mitosis to create two sperm cells (tricellular pollen stage, TCP). Prior to anther dehiscence and pollination, the TCP further develops into dehydrated mature pollen (mature pollen stage, MP) [1]. Pollen development is highly regulated, which is associated with successive global transcriptional regulation throughout the process [2], [3].
The precise and dynamic regulation of male gametogenesis requires transcription factors. In Arabidopsis, over 600 transcription factors are expressed during male gametogenesis, which forms a dynamic regulatory network [2], [4]. A subset of pollen-specific MIKC* MADS box proteins (AGL30/65/66/94/104) are expressed preferentially during pollen maturation [2], [5]. Double mutant combinations revealed the important roles these genes play in pollen germination and pollen fitness [5]. In Petunia, seven different zinc-finger transcription factors are expressed transiently and sequentially at different stages of pollen development [6]. Such transcription factors might each have specific target genes and constitute a regulatory cascade during pollen development [6]. Although progress has been made on the potential importance of transcription factors in male gametogenesis, little is yet known about the biological function of these transcription factors and how their activities are regulated to form temporal transcriptional regulatory networks.
Besides expression regulation, post-translational modification is a common mechanism to regulate the activity of transcription factors. Phosphorylation/dephosphorylation through mitogen-activated protein kinase (MAPK) cascades is a conserved post-translational modification in eukaryotes. A MAPK cascade minimally consists of three kinases: a MAPK, a MAPK kinase (MAPKK), and a MAPKK kinase (MAPKKK). The activity of MAPKs is regulated by their upstream MAPKKs through phosphorylation, and MAPKKs are activated through phosphorylation by their upstream MAPKKK(s). MAPKKKs are downstream of receptors/sensors and are activated in response to extracellular stimuli or to developmental signals [7]. Once activated, MAPKs can phosphorylate functionally divergent substrates on serine or threonine residues within a minimal S/T-P motif [8]. In Arabidopsis, there are 20 MAPKs, of which MPK3 (At3g45640) and MPK6 (At2g43790) are extensively studied. MPK3 and MPK6 have been revealed to phosphorylate multiple substrates, including transcription factors, in diverse biological processes [9]–[14]. For instance, WRKY33 (At2g38470) is a WRKY transcription factor required for pathogen defense in Arabidopsis [15]. In response to Botrytis cinerea infection, WRKY33 is phosphorylated by MPK3/MPK6, which is important for the activation of WRKY33, as mutations of MAPK-phosphorylation sites compromise the function of WRKY33 in vivo [9].
In this report, we show that WRKY34 (At4g26440), a new substrate of Arabidopsis MPK3/MPK6, is involved in male gametogenesis. WRKY34, a close homolog of WRKY33, is a pollen-specific WRKY transcription factor. When overexpressed using LAT52, a strong pollen-specific promoter, WRKY34 protein is temporally phosphorylated by MPK3/MPK6 at early stages in pollen development and then becomes dephosphorylated and degraded right before pollen maturation. Loss-of-function genetic analysis shows that WRKY34, together with a close homolog WRKY2 (At5g56270), plays important roles in pollen development and function. A complementation assay suggests that the phosphorylation of WRKY34 by MPK3/MPK6 is important for its function in vivo. Although single mutation of none of the WRKY2, WRKY34, MPK6 genes causes a pollen developmental defect, both the wrky2 mpk6 and wrky34 mpk6 double mutants exhibit pollen developmental defects similar to the wrky2 wrky34 double mutant, demonstrating that WRKY34 and WRKY2 indeed belong to the same pathway of MPK3/MPK6 in early pollen development.
Results
WRKY34 is phosphorylated by MAPKs in vitro
After the identification of WRKY33 as a substrate of MPK3/MPK6 in regulating plant defense responses [9], [16], we examined other WRKYs that share high homology with WRKY33 for potential MAPK phosphorylation sites. WRKY transcription factors are divided into three groups based on the number of WRKY domains (two copies in Group I, and one copy in Groups II and III) and the structure of their zinc fingers (C2HC in Group III but not in Group II proteins) [17]. WRKY33, with two WRKY domains, belongs to Group I in the WRKY family [17]. WRKY34 (At4g26440), another Group I member that shares high homoxlogy with WRKY33, is a pollen-specific gene that is preferentially expressed during early stages of male gametogenesis [18], [19]. WRKY34 also contains several consensus MAPK phosphorylation sites at similar positions as WRKY33 (Figure 1A), indicating that WRKY34 might be a MPK3/MPK6 substrate as well.
Fig. 1. In vitro phosphorylation of WRKY34 by MPK3 and MPK6. 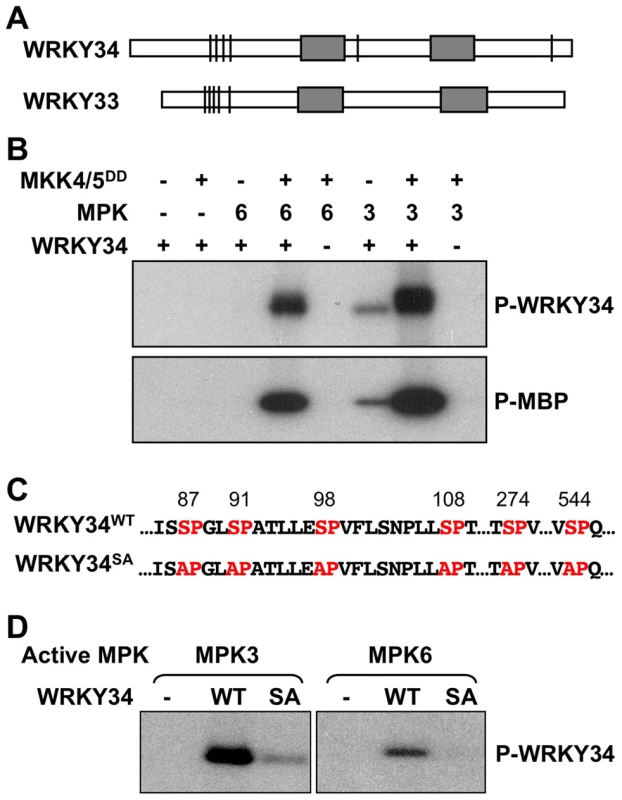
(A) Putative MAPK-phosphorylation sites in WRKY34 and WRKY33. Bars indicate the position of potential MAPK phosphorylation sites in the protein. Grey boxes indicate WRKY domains. Note that the clusters of phosphorylation sites at N-termini were similar between WRKY34 and WRKY33. (B) In vitro phosphorylation assay of WRKY34 by the activated MPK3 and MPK6 (upper panel). Reactions with various components omitted (-) were used as controls. Recombinant MKK4DD/MKK5DD were used to activate MPK3 and MPK6. Myelin basic protein (MBP) was used as control substrate (lower panel). (C) Adjacent sequences of putative MAPK-phosphorylation sites in WRKY34, and the loss-of phosphorylation WRKY34 mutant with all Ser mutated to Ala (WRKY34SA). (D) Mutation of MAPK-phosphorylation sites greatly reduced the phosphorylation of WRKY34 by MPK3 and MPK6. Phosphorylated WRKY34 was visualized by autoradiography after gel electrophoresis. To determine if WRKY34 can be phosphorylated by MAPKs in vitro, we prepared a His-tagged recombinant WRKY34 protein for in vitro MAPK phosphorylation assays. WRKY34 can be strongly phosphorylated by activated MPK3 and MPK6 (Figure 1B, upper panel). Without activation by the constitutively active MKK4DD/MKK5DD, MPK3 weakly phosphorylated WRKY34, whereas MPK6 showed no activity, demonstrating that the activation of MPK3 and MPK6 was important for a high-level phosphorylation of WRKY34. Control reactions with myelin basic protein (MBP) as an artificial substrate confirmed MPK3/MPK6 activation (Figure 1B, lower panel). There are six putative MAPK phosphorylation sites (Ser-87, Ser-91, Ser-98, Ser-108, Ser-274, and Ser-544) within the WRKY34 protein (Figure 1C). We performed site-directed mutagenesis to change these sites from Ser to Ala (WRKY34SA). As shown in Figure 1D, the phosphorylation of WRKY34SA protein by MPK3 and MPK6 was greatly reduced, demonstrating that these SP-motifs are the major MPK3/MPK6-phosphorylation sites in WRKY34. The residual phosphorylation of WRKY34SA also indicates the existence of other unidentified minor MAPK phosphorylation site(s) in WRKY34.
Detection of epitope-tagged WRKY34 protein from LAT52-driven transgene at different stages of male gametogenesis
To determine whether WRKY34 is phosphorylated by MPK3/MPK6 in vivo, we developed an immunoblot protocol to detect WRKY34 protein during male gametogenesis. A four-copy myc tag (4myc) was fused to the N terminus of WRKY34 protein, and a pollen-specific LAT52 promoter [20] was used to drive the transgene so that the 4myc-tagged WRKY34 protein could be expressed specifically and highly in pollen. Flowers or buds at various stages were collected for immunoblot detection of 4myc-WRKY34 protein in pollen. In this assay, the open flower right after anthesis was designated +1 (Figure 2A). The flower at Stage 13, in which anthesis was about to occur [21], was designated as 0. Buds/flowers at earlier stages were named with negative numbers −1, −2, and so on, according to their relative positions to the number 0 flower (Figure 2A). Under our experimental conditions, as few as 10 flowers/buds were sufficient for protein extraction and the detection of 4myc-WRKY34 protein by immunoblot analysis. The stage of pollen development was determined by DAPI staining of pollen grains from dissected flowers/buds of multiple plants. The +1 and 0 flowers contained mature pollen (MP) grains. The −1 and −2 buds contained homogenous tricellular pollen (TCP). The −3 to −5 buds contained a mixture of TCP and bicellular pollen (BCP), indicating non-uniform development of pollen in these bud stages. The −6 and −7 buds contained solely BCP.
Fig. 2. Phosphorylation of WRKY34 in vivo during male gametogenesis. 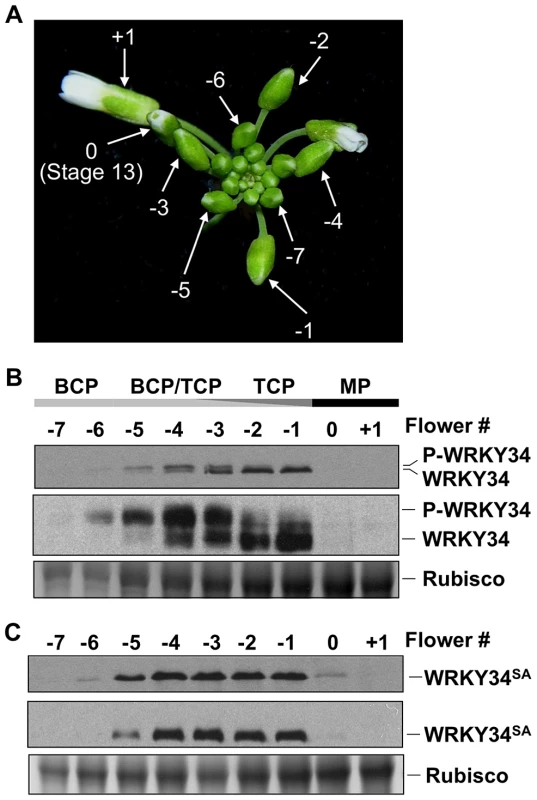
(A) Staging of flowers/buds used for WRKY34 protein analysis. Flower at Stage 13, in which anthesis is about to occur, was designated 0. An open flower right after anthesis was designated +1. Younger flowers/buds were designated using negative numbers. (B) Immunoblot and Phos-tag assays of LAT52 promoter-driven pollen-specific 4myc-WRKY34 protein at different stages of pollen development. The −7 to +1 flowers/buds have pollen at different developmental stages. Black bar indicates flowers or buds containing mature pollen. Dark gray bar indicates buds containing tricellular pollen (TCP). Light gray bar indicates buds containing bicellular pollen (BCP). Levels of 4myc-WRKY34 protein in immunoblot (top panel) and Phos-tag assay (middle panel) were determined using an anti-myc antibody. Protein loading control was confirmed by Coomassie blue staining (bottom panel). (C) Immunoblot (top panel) and Phos-tag assay (middle panel) of pollen-specific 4myc-WRKY34SA protein at different developmental stages. Protein loading control was confirmed by Coomassie blue staining (bottom panel). Each sample was extracted from the same number of flowers/buds at the corresponding stage, which allows the comparison of WRKY34 protein levels in an equal number of developing/mature pollen grains. We found that tagged 4myc-WRKY34 protein was first detectable in −6 buds, which contain BCP (Figure 2B, top panel). The absence of 4myc-WRKY34 protein in earlier stages is likely a result of low LAT52 promoter activity [22]. The 4myc-WRKY34 signal was stronger in more developed buds and reached its peak in −2 and −1 buds with TCP (Figure 2B, top panel). Interestingly, although driven by LAT52, a promoter with the strongest activity in mature pollen [22], the 4myc-WRKY34 protein signal was hardly detectable in 0 buds and open flowers (Figure 2B, top panel). The transcripts from 4myc-WRKY34 transgene showed a similar expression pattern, as indicated by RT-PCR (Figure S1). We also tried an immunoblot assay using flowers from WRKY34 promoter-driven 4myc-WRKY34 transgenic plants (PWRKY34:4myc-WRKY34). However, 4myc-WRKY34 protein was not detectable in such samples, which is likely due to low WRKY34 promoter activity (data not shown). Interestingly, as described later, the PWRKY34:WRKY34-YFP fusion showed a similar expression pattern as PLAT52:4myc-WRKY34. Therefore, we conclude that the use of LAT52 promoter in this assay could represent, at least partially, the native WRKY34 expression and modification pattern.
WRKY34 is temporally phosphorylated by MAPKs during male gametogenesis
In the immunoblot assay, we noticed that 4myc-WRKY34 showed differential migrations in the SDS-polyacrylamide gel depending on the developmental stage of the flower buds. In −6 buds with BCP, 4myc-WRKY34 protein exhibited a slightly slower migration (Figure 2B, top panel). In −5 to −3 buds with a mixture of BCP and TCP, 4myc-WRKY34 existed as doublets, and the faster moving band gradually accumulated (Figure 2B, top panel). In −1 and −2 buds with TCP, 4myc-WRKY34 protein predominately existed as the faster migrating band (Figure 2B, top panel). These results indicated that WRKY34 protein was modified in BCP, possibly by protein phosphorylation, and the modification is dependent on the pollen's developmental stage. To determine whether the slower migrating band of 4myc-WRKY34 is due to phosphorylation, we performed a Phos-tag mobility shift assay. In this assay, the Phos-tag reagent binds specifically to phosphorylated proteins and slows down their migration in the SDS-polyacrylamide gel [9], [23]. As shown in Figure 2B (middle panel), 4myc-WRKY34 protein was indeed phosphorylated in the BCP of −6 buds and was gradually dephosphorylated in late stages of the male gametogenesis. The phosphorylation of 4myc-WRKY34 was greatly reduced upon pollen maturation at −1, which is followed by complete disappearance of WRKY34 protein in 0 flowers (Figure 2B, top panel).
We then performed immunoblot with 4myc-WRKY34SA transgenic plants to determine if the shifting of protein bands is dependent on the MAPK phosphorylation sites in WRKY34. Although the protein expression pattern is similar to 4myc-WRKY34, the 4myc-WRKY34SA protein showed no band shift in either the immunoblot or Phos-tag assay (Figure 2C). This result further confirmed that WRKY34 was temporally phosphorylated during early pollen development, and the phosphorylation occurred on the MPK3/MPK6-phosphorylation sites delineated in the in vitro phosphorylation assay (Figure 1C and 1D).
To demonstrate that the in vivo phosphorylation of WRKY34 during pollen development is carried out by MPK3 and MPK6, we introduced the 4myc-WRKY34 transgene into the mpk3 mpk6 double mutant background. Since the mpk3 mpk6 double mutant is embryo lethal [13], we attempted pollen-specific RNAi suppression of MPK3 in the mpk6 mutant background. LAT52 promoter-driven MPK3RNAi construct was transformed into the mpk6 plants. Because of the pollen-specific expression of MPK3RNAi, the sporophytic tissues were not affected, which allowed us to obtain the double homozygous MPK3RNAi mpk6 plants. Real-time qPCR demonstrated that MPK3 expression in pollen from MPK3RNAi mpk6 plants was knocked down (Figure 3A). We then performed immunoblot and Phos-tag assays of 4myc-WRKY34 in the MPK3RNAi mpk6 plants. The mobility shift of 4myc-WRKY34 was abolished in the absence of MPK3 and MPK6 (Figure 3B, top and middle panels). This loss-of-function system demonstrated that the WRKY34 was phosphorylated specifically by MPK3 and/or MPK6. The stability of WRKY34 protein apparently was not affected by the MAPK phosphorylation since mutation of the Ser residues that are phosphorylated by MPK3/MPK6 did not affect the protein expression pattern of WRKY34 during pollen development (Figures 2C).
Fig. 3. In vivo phosphorylation of WRKY34 is dependent on MPK3 and MPK6. 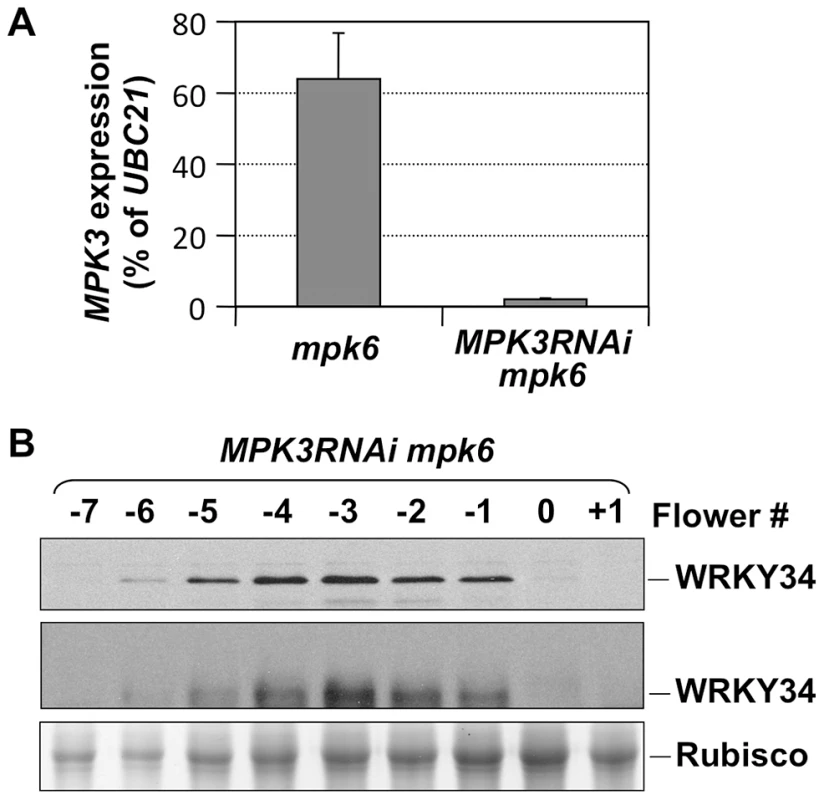
(A) MPK3 expression in mpk6 and MPK3RNAi mpk6 pollen grains. Total RNAs were isolated from pollen grains. MPK3 transcript levels were determined using quantitative RT-PCR. Error bars = standard derivation. (B) Immunoblot (top panel) and Phos-tag (middle panel) assays of WRKY34 protein at different stages of MPK3RNAi mpk6 PLAT52:4myc-WRKY34 flower buds. Protein loading control was confirmed by Coomassie blue staining (bottom panel). Each sample was extracted from the same number of flowers/buds at the corresponding stage, which allows the comparison of WRKY34 protein levels in an equal number of developing/mature pollen grains. WRKY34 functions redundantly with WRKY2 in pollen development
Previous studies showed that WRKY34 is an early pollen gene enriched in UNM and BCP [19] and that mutation of the WRKY34 gene increases the pollen's tolerance to cold stress [18]. However, the biological function of WRKY34 in pollen development remains unclear. Under our growth conditions, single wrky34 mutant pollen showed no developmental defect. Since more than 50% of the WRKY family members are expressed in the male gametophytes [2], we speculated that there might be functionally redundant WRKY member(s) in early pollen development.
A phylogenetic analysis was used to identify such member(s) (Figure S2). WRKY34 is closely related to WRKY2, a WRKY member expressed in various tissues including male gametophyte [24]. We examined by quantitative RT-PCR the expression patterns of WRKY34 and WRKY2 in several tissues. WRKY34 expression was very low in most examined tissues and was slightly higher in floral buds (Figure 4A). In contrast, WRKY2 showed higher expression in all detected tissues (Figure 4A). To examine the detailed expression patterns of WRKY2 and WRKY34 in pollen at different stages, we fused the WRKY2 and WRKY34 genomic sequences, which contain promoter and gene coding region, with YFP. The YFP signal of both fusion proteins was detectable in nuclei, which was consistent with their function as transcription factors. It is also noteworthy that WRKY2 - and WRKY34-YFP signals were detectable in the vegetative cell but not in the generative or sperm cells. The PWRKY2:WRKY2-YFP signal was absent in UNMs (Figure 4B and 4F), while it became significantly higher in BCP nuclei (Figure 4C and 4G). The YFP signal in nuclei was also found in TCP (Figure 4D and 4H) and MP (Figure 4E and 4I). For PWRKY34:WRKY34-YFP, the nucleus YFP signal was dim in UNM, although it was still distinguishable from the pollen auto-fluorescence (Figure 4J and 4N). The signal was more detectable in BCP (Figure 4K and 4O) and TCP (Figure 4L and 4P). However, in contrast to WRKY2, the WRKY34-YFP signal was absent in MP (Figure 4M and 4Q). These results showed that WRKY34 and WRKY2 expression overlaps at the BCP and TCP stages. In addition, the PWRKY34:WRKY34-YFP expression pattern was similar to the PLAT52:4myc-WRKY34 expression in the immunoblot assay (Figure 2B, top panel). This further indicated that the WRKY34 protein expression pattern was not solely dependent on promoter activity.
Fig. 4. Expression and protein localization of WRKY34 and WRKY2. 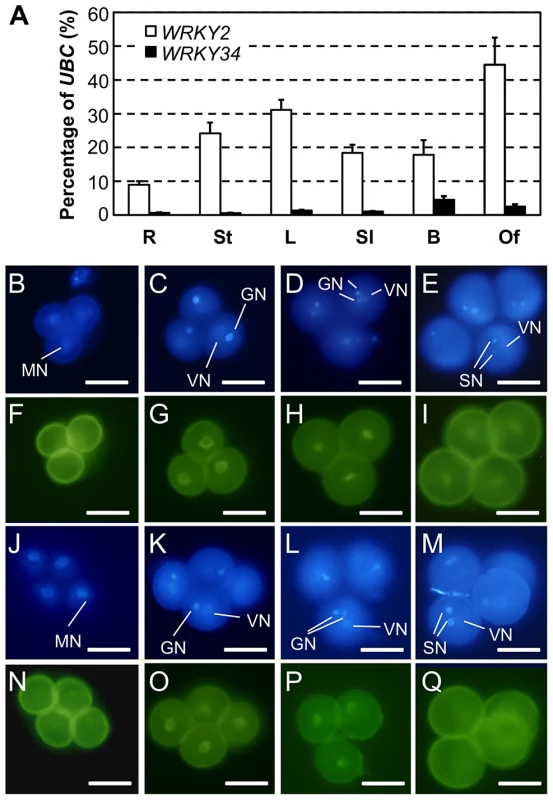
(A) Quantitative RT-PCR of WRKY34 and WRKY2 transcripts in various tissues. R, roots; St, stems; L, leaves; Sl, seedlings; B, buds; and Of, open flowers. Error bars = standard derivation. (B to I) Expression and localization of WRKY2 promoter-driven WRKY2:YFP fusion protein in pollen. DAPI staining was used to locate nuclei (B to E), and YFP signal reveals the localization of WRKY2:YFP fusion at different developmental stages (F to I). (B and F) UNM stage, no nucleus-localized YFP signal was detected. Vegetative nucleus localized WRKY2:YFP signal was observed at BCP stage (C and G), TCP stage (D and H), and MP stage (E and I). (J to Q) Expression and localization of WRKY34 promoter-driven WRKY34:YFP fusion protein in pollen. (J to M) DAPI staining signal. (N to Q) YFP signal. (J and N) UNM stage, weak signal was observed in microspore nucleus. Vegetative nucleus localized WRKY34:YFP signal was observed at BCP stage (K and O), and TCP stage (L and P). No YFP signal was observed in MP (M and Q). Note that as the vegetative cell expressed genes, the WRKY2 and WRKY34 fusion YFP signals were not detectable in generative or sperm cells. MN, microspore nucleus. VN, vegetative nucleus. GN, generative nucleus/nuclei. SN, sperm cell nuclei. Bar = 50 µm. We next obtained a T-DNA insertion line for WRKY34 (SALK_133019 hereafter wrky34-1) and two T-DNA lines for WRKY2 (Salk_020399 and SAIL_739_F05, hereafter wrky2-1 and wrky2-2, respectively) (Figure 5A). wrky34-1 was reported to be a null mutant [18]. We performed quantitative RT-PCR to examine WRKY2 expression in wild-type, wrky2-1, and wrky2-2 pollen. The result showed that the expression of WRKY2 was moderately knocked down in seedlings of both alleles (Figure 5B). However, in pollen, WRKY2 expression was almost completely knocked out in wrky2-1 but not in wrky2-2 (Figure 5B). Therefore, we crossed wrky34-1 with wrky2-1 to generate the wrky2-1 wrky34-1 double mutant and then examined the wrky2-1 wrky34-1 double mutant pollen function by reciprocal crosses using combinations of heterozygous mutants and wild type (Table 1).
Fig. 5. Phenotype of wrky2-1 wrky34-1 double mutant pollen. 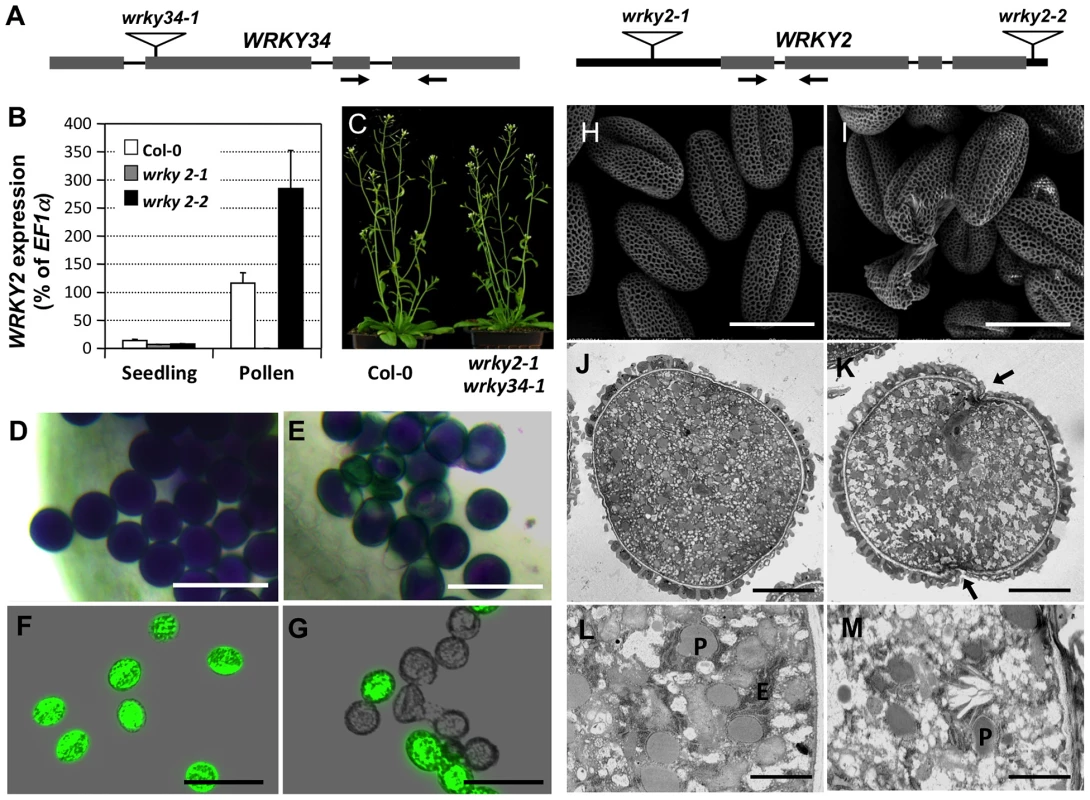
(A) Diagram of T-DNA insertion alleles of wrky2 and wrky34 mutants. Arrows indicate the positions of RT-qPCR primers. Black bars = untranslated regions (UTRs); gray bars = exons; lines = introns. (B) Quantitative RT-PCR of WRKY2 expression in wild-type, wrky2-1, and wrky2-2 seedlings and pollen grains. Error bars = standard derivation. (C) Normal vegetative growth and development of wrky2-1 wrky34-1 double mutant plants. Five-week-old plants are pictured. (D, E) Alexander staining of wild-type (D) and wrky2-1 wrky34-1 double mutant (E) pollen. Bar = 50 µm. (F, G) Vital staining by FDA of wild type (F) and wrky2-1 wrky34-1 double mutant (G) pollen. Bar = 50 µm. (H, I) Scanning electron microscopy (SEM) of wild type (H) and wrky2-1 wrky34-1 double mutant (I) pollen. Bar = 20 µm. (J, K, L, M) Transmission electron microscopy (TEM) of wild type (J, L) and wrky2-1 wrky34-1 double mutant (K, M) pollen. (J, K) Bar = 5 µm. (L, M) Bar = 1 µm. Arrows in panel K indicate the germination pore with defective intine layer. P, plastid; E, endoplasmic reticulum. Tab. 1. Transmission of wrky2-1 and wrky34-1 single and double mutant alleles. 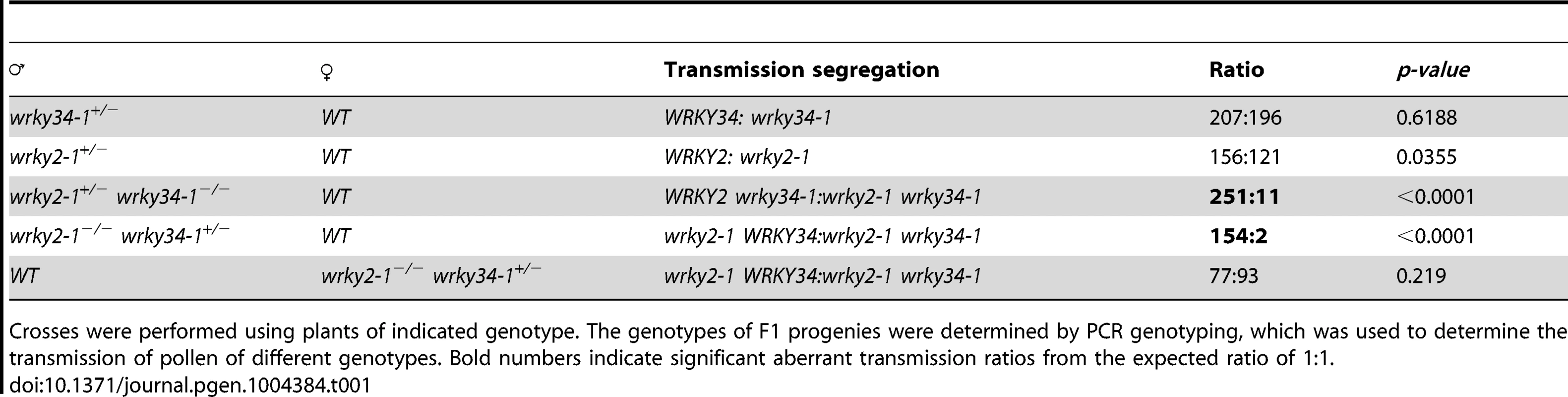
Crosses were performed using plants of indicated genotype. The genotypes of F1 progenies were determined by PCR genotyping, which was used to determine the transmission of pollen of different genotypes. Bold numbers indicate significant aberrant transmission ratios from the expected ratio of 1∶1. The male transmission of the mutant alleles was normal when pollen grains from either wrky2-1+/− or wrky34-1 +/− plants were used as pollen donors, suggesting that single mutations of either WRKY34 or WRKY2 had no effect on the function of pollen (Table 1). However, when using wrky2-1+/− wrky34-1−/− or wrky2-1−/− wrky34-1+/− plants as the male parents, we observed that the transmission of wrky2-1 wrky34-1 pollen was significantly reduced (0.04∶1 for pollen from wrky2-1+/− wrky34-1−/− plants and 0.01∶1 for pollen from wrky2-1−/− wrky34-1+/− plants, instead of the expected 1∶1, p-value<0.0001) (Table 1). This result suggested that WRKY34 and WRKY2 are important for pollen function but also that a portion of the double mutant pollen grains remained functional. The transmission of wrky2-1 wrky34-1 female gametophytes was normal (Table 1), indicating that the female gametophyte function was not affected.
Phenotype of wrky2-1 wrky34-1 double mutant pollen
Because of the leaky transmission of wrky2-1 wrky34-1 pollen, we were able to obtain wrky2-1 wrky34-1 homozygous double mutant plants at low frequency. Morphologically, the double mutant plant was indistinguishable from the wild type (Figure 5C). To examine the development of wrky2-1 wrky34-1 pollen, we used Alexander's staining to distinguish normal and aborted pollen [25]. In this assay, the cytoplasm of normal pollen should show a purple color and the pollen wall a distinctive green color. Pollen grains from wild-type plants were viewed as full, round, purple-stained grains (Figure 5D). In contrast, a portion of wrky2-1 wrky34-1 pollen exhibited aberrant morphology and green color (28% abortion, n = 200), which indicated impaired pollen development of the double mutant (Figure 5E). We then performed fluorescein diacetate (FDA) staining to check the viability of wrky2-1 wrky34-1 pollen (Figure 5F and 5G). In comparison with wild-type pollen grains (96% viable), the majority of wrky2-1 wrky34-1 pollen failed to show FDA fluorescence and therefore was likely to be dead (67%).
The non-viable rate in FDA staining was higher than that in the Alexander staining, indicating that FDA is a more sensitive viability assay. There were wrky2-1 wrky34-1 pollen grains with a small patch that failed to be stained using Alexander staining (Figure 5E). They were classified as viable pollen, but might be non-viable. In contrast, FDA staining, which is dependent on both cellular esterase activity and plasma membrane integrity, gave much clearer results. For this reason, FDA staining was used for all the other experiments. We next stained the developing pollen at earlier stages with FDA. The lethality of wrky2-1 wrky34-1 pollen was first identifiable in -6 buds with BCP, and the percentage of lethal pollen increased following pollen development (Figure S3). The onset of pollen death in wrky2-1 wrky34-1 double mutant correlates with the appearance of WRKY34 protein in BCP and TCP stages (Figure 2B, top panel), suggesting the requirement of these two WRKYs at these developmental stages. There are two possible reasons for the lower percentages of FDA positive pollen at early developmental stages and then the gradual increase in FDA positive pollen in the wild type (Figure S3). Firstly, the tapetal cell layer surrounding the developing pollen could reduce the efficiency of FDA staining at the early stage. Secondly, dissection and squeezing to release pollen from the anther and tapetum might damage the immature pollen. Side-by-side comparison revealed that the FDA positive pollen from wrky2-1 wrky34-1 plants continued to drop (Figure S3), indicating the loss of viability of wrky2-1 wrky34-1 mutant pollen.
We also examined the ultrastructure of wrky2-1 wrky34-1 pollen using scanning electron microscopy (SEM). Both wild-type and wrky2-1 wrky34-1 pollen appeared to have normal pollen wall structures (Figure 5H and 5I). In contrast to the uniformly shaped wild-type pollen grains (Figure 5H), the wrky2-1 wrky34-1 pollen grains were a mixture of shapes, including normal shaped pollen, collapsed pollen, and ruptured pollen remnant (Figure 5I). This indicated that the development of wrky2-1 wrky34-1 pollen was defective. Further analysis with transmission electron microscopy (TEM) confirmed the abnormal ultrastructure of wrky2-1 wrky34-1 pollen. Consistent with the cytological staining results, a portion of the wrky2-1 wrky34-1 pollen was collapsed with leaky cytoplasm content (Figure S4). Furthermore, for the majority of wrky2-1 wrky34-1 pollen that exhibited similar exterior appearance as wild-type pollen, the intracellular ultrastructure was different from that of the wild-type pollen (Figure 5J to 5M). The numbers of plastids and endoplasmic reticulum (ER) were reduced in wrky2-1 wrky34-1 pollen grain. In addition, the intine layer was discontinuous and undulated at the germination pore of the double mutant pollen grain (Figure 5K and 5M).
Pollen tube growth is impaired in wrky2-1 wrky34-1
In addition to a pollen developmental defect, the in vitro germination assay revealed that the wrky2-1 wrky34-1 double mutant was defective in pollen function. In our assays, the average germination ratio of wild-type pollen was 78% (Figures 6A and 7B), while only 28% of wrky2-1 wrky34-1 pollen was capable of germination under the same conditions (Figures 6B and 7B). The reduction in pollen germination appears to be a result of reduced pollen viability. For the wrky2-1 wrky34-1 pollen that germinated, the pollen tube length was significantly shorter than the length of wild-type pollen tubes (Figure 6A, 6B, and 7C). The average pollen tube length was 471 µm in the wild type and 288 µm in wrky2-1 wrky34-1 double mutant at 7 hours after germination in vitro, representing a 40% reduction in length in the double mutant pollen tubes. Pollination analysis followed by aniline blue staining further demonstrated that the wrky2-1 wrky34-1 double mutant was defective in pollen germination and pollen tube growth in vivo (Figure 6C and 6D). Since WRKY34 protein was degraded before pollen maturation (Figure 2B, top panel), we speculated that the reduced germination and tube growth of wrky2-1 wrky34-1 pollen were not an indication of a requirement of WRKY2/WRKY34 in these two processes but rather a result of weak pollen due to impaired development, which was also evident based on the TEM observation (Figure 5J to 5M).
Fig. 6. Pollen germination and pollen tube growth are defective in wrky2-1 wrky34-1. 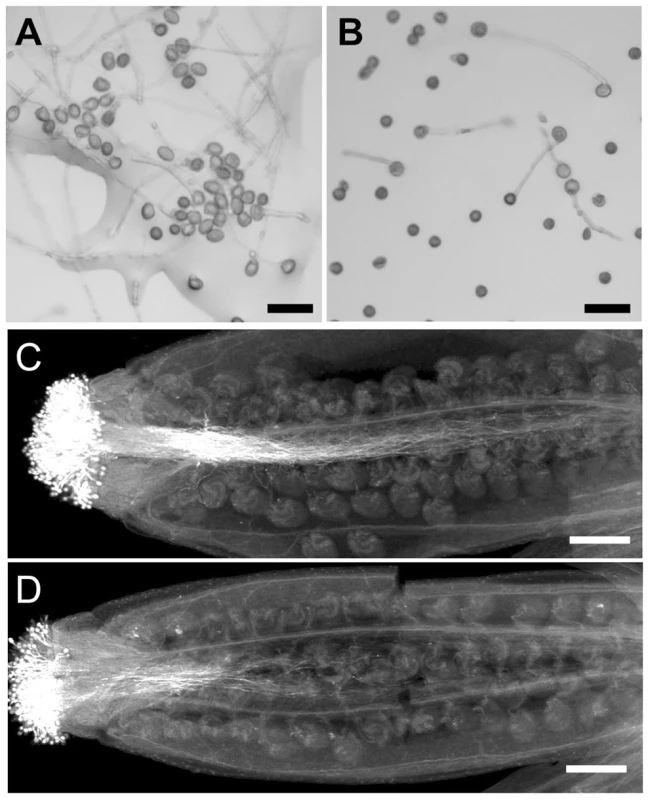
In vitro pollen germination of wild type (A) and wrky2-1 wrky34-1 double mutant (B) pollen. Bars = 50 µm. Aniline blue staining of wild type pistils 8 hours after pollination with wild type (C) and wrky2-1 wrky34-1 double mutant (D) pollen. Bar = 200 µm. Fig. 7. Complementation of wrky2-1 wrky34-1 double mutant pollen phenotypes by WRKY34WT and WRKY34SA. 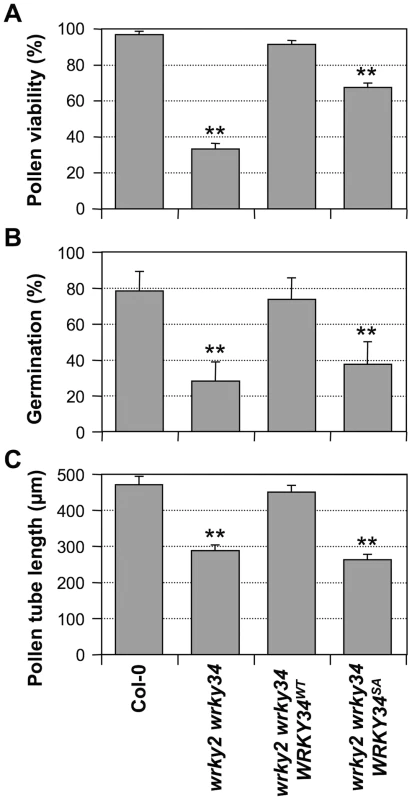
Pollen viability ratio by FDA staining (A), in vitro pollen germination ratio (B), and average pollen tube length (C) in Col-0, wrky2-1 wrky34-1, wrky2-1 wrky34-1 PWRKY34:WRKY34WT, and wrky2-1 wrky34-1 PWRKY34:WRKY34SA plants. Error bar = standard error. Double asterisks indicate statistically very significant difference from wild-type pollen (p-value<0.01). Phosphorylation by MAPKs is required for WRKY34 function in vivo
To test whether phosphorylation of WRKY34 is important for its function in pollen development, we performed genetic complementation of wrky2-1 wrky34-1 pollen using WRKY34 promoter-driven 4myc-WRKY34WT or 4myc-WRKY34SA. Pollen from T2 homozygous progenies with a transgene expression level similar to wild type was selected and examined (Figure S5). PWRKY34:4myc-WRKY34WT wrky2-1 wrky34-1 pollen showed viability, germination, and pollen tube growth similar to wild-type pollen (Figure 7), indicating that the PWRKY34: 4myc-WRKY34WT transgene can complement the wrky2-1 wrky34-1 pollen phenotype. In contrast, the function of PWRKY34:4myc-WRKY34SA transgene was significantly compromised. The viability ratio of PWRKY34:4myc-WRKY34SA wrky2-1 wrky34-1 pollen was partially rescued to 67% from 33% of the wrky2-1 wrky34-1 pollen, which was significantly lower than wild-type (97%) and PWRKY34:WRKY34WT complemented pollen (90%) (p-value <0.01) (Figure 7A). The double mutant pollen germination rate (28%) was only slightly rescued by PWRKY34:4myc-WRKY34SA (37%), while it was fully complemented by PWRKY34:4myc-WRKY34WT (73%), which was similar to wild-type pollen (78%) (p-value <0.01) (Figure 7B). Furthermore, the average pollen tube length of PWRKY34:4myc-WRKY34SA wrky2-1 wrky34-1 pollen (263 µm) was about the same as the double mutant (288 µm) and much shorter than the wild-type (471 µm) and PWRKY34:4myc-WRKY34WT wrky2-1 wrky34-1 pollen (451 µm) (p-value <0.01) (Figure 7C). These results indicated that although the pollen lethality of the double mutant was partially rescued by WRKY34SA, the pollen function was still abnormal. As a result, we conclude that the phosphorylation of WRKY34 by MPK3/MPK6 is important for the function of WRKY34 protein in vivo.
Although the native promoter-driven WRKY34 transgene (PWRKY34:4myc-WRKY34WT) could fully complement the wrky2-1 wrky34-1 mutant, immunoblot analysis failed to detect the tagged WRKY34WT protein in the inflorescences or floral buds in these rescued lines (data not shown), which is most likely a result of the low expression level of the native promoter (Figure 4). To exclude the possibility that mutation of multiple Ser to Ala in WRKY34 altered its general functionality such that the WRKY34SA cannot bind or has reduced DNA-binding activity, we compared the W-box binding activity of the recombinant WRKY34 and WRKY34SA using electrophoretic mobility shift assay (EMSA). As shown in Figure S6, there was no difference in the W-box binding activity and specificity of WRKY34 after the Ser-to-Ala mutation, suggesting that the reduced functionality of WRKY34SA is not a result of a general loss of WRKY34 function.
MPK6 belongs to the same genetic pathway of WRKY34 and WRKY2
Based on these results, the phosphorylation by MPK3/MPK6 is important for the function of WRKY34 in pollen development and function. Due to the embryo lethality of mpk3 mpk6 double zygotes [13], we cannot analyze the phenotype of pollen grains from the double homozygous plants. The mpk3 mpk6 double mutant pollen from mpk3+/− mpk6−/− or mpk3−/− mpk6+/− plants, although it exhibited altered transmission, did not show any developmental defects like wrky2-1 wrky34 pollen [26]. We speculate that in mpk3 mpk6 pollen the unphosphorylated WRKY34 and WRKY2 each retained basal level function, which kept the mpk3 mpk6 pollen above the threshold of visible developmental defects. This is consistent with the finding that WRKY34SA mutant protein can partially complement the wrky2-1 wrky34-1 mutant pollen. Alternatively, MPK3 or MPK6 protein carried over from the microspore mother cells of mpk3+/− mpk6−/− or mpk3−/− mpk6+/− plants, which have at least one good copy of MPK3 or MPK6, could be sufficient to support the development of mpk3 mpk6 pollen. It is known that MAPKs are very stable proteins in cells.
Although both MPK3 and MPK6 are involved in pollen function, MPK6 apparently is more important, as indicated by its much higher expression in pollen (www.genevestigator.com). Therefore, we speculate that the double mutation of mpk6 and wrky34-1 (or wrky2-1), in which the pollen produced a single WRKY protein with reduced phosphorylation, might result in a weak phenotype in pollen development. As shown in Figure 8, both mpk6 wrky34-1 and mpk6 wrky2-1 pollen showed developmental and functional defects that were similar to the wrky34-1 wrky2-1 double mutant pollen. The pollen viability was 84% in mpk6 wrky34-1 and 75% in mpk6 wrky2-1, respectively, which indicated moderate pollen lethality in the double mutants (p-value<0.05) (Figure 8A). In accordance, the pollen germination rate was also decreased slightly from an average of 80% of wild-type pollen to 71% of mpk6 wrky34-1 and 63% of mpk6 wrky2-1 (p-value<0.05) (Figure 8B). Furthermore, the average pollen tube lengths of mpk6 wrky34-1 and mpk6 wrky2-1 was significantly reduced to 382 µm and 324 µm, respectively, in comparison with the 470 µm of wild-type pollen tubes (p-value<0.01) (Figure 8C). This result indicated that the mpk6 wrky34-1 and mpk6 wrky2-1 pollen function was affected and confirmed that MPK6 belongs to the same genetic pathway as WRKY34 and WRKY2.
Fig. 8. Phenotype of mpk6 wrky34-1 and mpk6 wrky2-1 pollen. 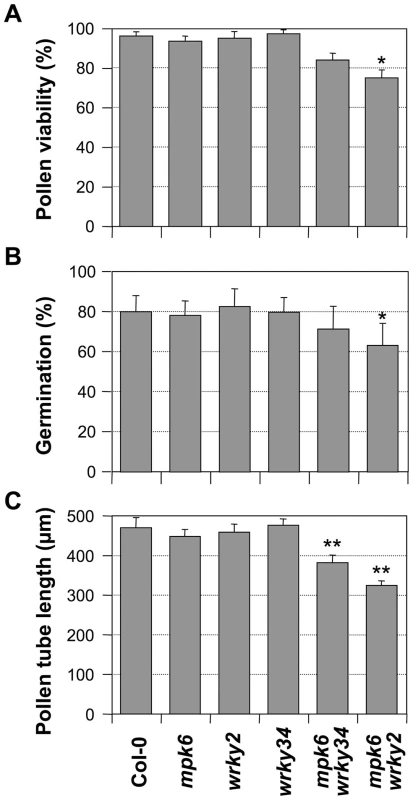
Pollen viability ratio by FDA staining (A), in vitro pollen germination ratio (B), and average pollen tube length (C) in Col-0, mpk6, wrky2-1, wrky34-1, mpk6 mpk34-1, and mpk6 wrky2-1 plants. Error bar = standard error. Single asterisks indicate statistically significant difference from wild-type pollen (0.01<p-value<0.05). Double asterisks indicate very significant difference from wild-type pollen (p-value<0.01). Discussion
In this report, we demonstrate that WRKY34, a pollen-specific WRKY transcription factor, is a substrate of MPK3/MPK6. WRKY34 is temporally phosphorylated during early male gametogenesis and is dephosphorylated right before pollen maturation. The phosphorylation of WRKY34 by MPK3/MPK6 is important for its function in vivo. WRKY34, together with WRKY2, is required for male gametogenesis. Mutation of both WRKY34 and WRKY2 greatly reduces the viability of pollen, which is associated with reduced germination and pollen tube growth, both in vitro and in vivo. Taken together, we conclude that WRKY34/WRKY2 transcription factors play an important role downstream of the MPK3/MPK6 cascade in pollen development and function.
Differential expression of substrates allows the MPK3/MPK6 cascade to control different biological processes
A long-standing question is how a MAPK cascade confers signaling specificity in diverse biological events. In yeast and mammals, the mechanisms to maintain signaling specificity of MAPKs include 1) cell-type specificity of other signaling components in the pathway, such as receptors, scaffolding proteins, and MAPK substrates [27]–[29]; 2) kinetics in signaling strength resulting in distinct outcomes [30]; and 3) cross-pathway suppression of downstream components [31]–[33]. However, in plants, such mechanisms have not been well studied. In Arabidopsis, MPK3 and MPK6, two of the best-characterized MAPKs, function together in diverse biological processes, including plant growth, development, and response to environmental stimuli [12]–[14], [16], [26], [34], [35]. Differentially expressed substrates could help maintain the functional specificity of the activated MPK3/MPK6 signaling cascade in different cells/tissues. In response to pathogen attacks, MPK3 and MPK6 are activated and phosphorylate a subset of ACC synthase (ACS) isoforms to induce ethylene biosynthesis [12]. The pathogen responsive MPK3/MPK6 cascade also induces phytoalexin biosynthesis through the activation of downstream the WRKY33 substrate [9], [36]. In stomatal development, MPK3/MPK6 phosphorylates SPEECHLESS, a basic helix-loop-helix transcription factor that is specifically expressed in stomatal lineage cells and negatively regulates stomatal formation [10], [13]. In different biological processes, MPK3 and MPK6 are able to phosphorylate different WRKY homologs, e.g. WRKY33 and WRKY34 in plant defense and pollen development, respectively. Differential tissue/cell-specific expression of WRKY33 and WRKY34 allows the MPK3/MPK6 cascade to control different biological processes.
WRKY transcription factors share common signaling components in different biological processes
WRKY transcription factors are one of the largest families of transcriptional regulators in plants [17]. Transcriptional regulation by WRKY members is an integral part of signaling networks that modulate many biological processes, most notably in response to diverse biotic and abiotic stresses [37]. WRKY transcription factors also have been implicated in plant growth and development processes, including senescence, seed development, and embryogenesis. For instance, WRKY53 binds to the promoters of a set of senescence-associated genes, and the overexpression or knockdown of WRKY53 gene lead to an altered senescence phenotype [38]. In seed, a WRKY transcription factor, MINISEED3 (MINI3), recruits a nuclear localized protein SHB1 to activate gene expression, which regulates endosperm proliferation and seed cavity enlargement [39]. The WRKY23 transcription factor is needed for proper root growth and development by stimulating the local biosynthesis of flavonols, which is dependent on auxin through the AUXIN RESPONSE FACTOR 7 (ARF7) and ARF19 transcriptional response pathway [40]. Despite these recent discoveries, it is still unclear whether WRKY transcription factors share similar regulatory networks between environmental responses and developmental processes. Our results suggest that the MPK3/MPK6 signaling module could act as a molecular hub to integrate different signaling networks of WRKY transcription factors, although the upstream signaling cues are different.
MPK3/MPK6 and WRKY34 also may integrate stress and developmental signaling in pollen. WRKY34 is involved in cold sensitivity in mature pollen, where it regulates expression of cold-specific transcription factors (CBF) [18]. MPK6 is rapidly activated by cold stress. Furthermore, MPK6 signaling is functionally involved in cold and salt stress responses [41]. It is therefore possible that MPK3 and MPK6 may be involved in the WRKY34-mediated cold tolerance in pollen. However, the MPK6 activity is positively related with cold tolerance, while WRKY34 seems to be a negative regulator in this process. More details are required to interpret the role of MPK3/MPK6-WRKY34 signaling module in pollen cold tolerance.
WRKY2 plays a redundant role with WRKY34 in pollen development. Unlike WRKY34, WKRY2 is expressed in various tissues (Figure 4A) and is likely to play pleiotropic roles in plant development. For example, the involvement of WRKY2 in embryogenesis and ABA-mediated seed germination has been reported [24], [42]. In zygote, WRKY2 directly activates the transcription of WUSCHEL RELATED HOMEOBOX (WOX) genes to regulate polar organelle localization and asymmetric division [24]. Given that the mpk3 mpk6 double mutant is embryo lethal [13], it is possible that the MAPK signaling cascade is involved also in the regulation of the WRKY2-WOX signaling pathway. Comparative analysis of WRKY2 activation by MPK3/MPK6 in pollen and embryogenesis would provide further insights into the regulation of WRKY transcription factors in diverse biological processes.
Temporal regulation of WRKY transcription factors at multiple levels
Based on the transcriptomic profiles, two periods of temporal gene expression are defined in pollen development, an early phase and a late phase. Expression of “early genes” occurs after meiosis and declines toward pollen maturation, while “late genes” are preferentially expressed in TCP and MP stages [2], [3]. The vegetative cell early-late transcriptome transition occurs mainly between the BCP and TCP stages, which exhibit not only a significantly reduced number of expressed genes but also a major shift in mRNA populations [2]. WRKY34 has been identified as an “early gene”, and its expression is suppressed by several MIKC* MADS box transcription factors during pollen maturation [5], [19]. In this report, we found that WRKY34 from LAT52-driven transgene is phosphorylated at the BCP stage and becomes dephosphorylated at the TCP stage. The phosphorylation of WRKY34 is important for its biological function in male gametogenesis. Therefore, we propose that, besides the regulation at the transcriptional level, the post-translational modifications by MAPKs also plays a critical role in controlling the activity of this WRKY transcription factor, especially during early and late phase transition.
The abundance of WRKY34 in early pollen development is regulated at both post-transcriptional and post-translational levels. In our assays, even though driven by LAT52, a promoter specific at later pollen stages [20], WRKY34 transcript was barely detectable in mature pollen (Figure S1), suggesting potential regulation of WRKY34 transcripts at the mRNA stability level. Moreover, despite the presence of WRKY34 transcripts at the TCP stage (Figure S1), WRKY34 protein was absent in MP (Figure 2B, top panel), indicating rapid protein degradation in the process. In support of this conclusion, the abundance of the WRKY34-YFP protein from transgene driven by native WRKY34 promoter showed a similar pattern, as indicated by the YFP fluorescence (Figure 4). This further demonstrates that the abundance of WRKY34 protein is regulated at multiple levels and is not solely dependent on promoter activity. The protein stability of WRKY34 apparently is not associated with its phosphorylation state, since the abundance of WRKY34SA, an unphosphorylatable form of WRKY34, followed the same pattern as WRKY34WT protein (Figure 2C). Therefore, there should be a protein degradation pathway regulating WRKY34 protein abundance at late pollen stages that is independent of MPK3/MPK6. WRKY2 protein appeared to be more stable in mature pollen (Figure 4). Pollen development involves dynamic transition of gene expression profiles, which requires rapid control of the transcriptional factors involved. The regulation of WRKY34 activity at multiple levels may reflect the complexity of the regulation of key transcription factors in this process.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana Columbia ecotype (Col-0) was used as the wild type. T-DNA insertion alleles of WRKY34 (SALK_133019) and WRKY2 (Salk_020399 and SAIL_739_F05) were obtained from the Arabidopsis Biological Resource Center (ABRC). Seeds were surface sterilized and imbibed at 4°C for 3 days, then plated on half-strength Murashige and Skoog medium with 0.45% Phytagar. Plates were incubated in a tissue culture chamber at 22°C under continuous light (70 µE m−2 s−1) for 7 days. Seedlings were then transplanted to soil and grown in the greenhouse with a 16-h-light/8-h-dark cycle.
In PCR-based genotyping, the presence of the T-DNA and wild-type alleles was detected using LBa1 (5′-TGGTTCACGTAGTGGGCCATCG-3′) and two gene-specific primers: WRKY2-LP (5′-TTTTCTTTTTCACACGTTAAGCC-3′) and WRKY2-RP (5′-TGTTAGAACACGAATCACCCC-3′) for the wrky2-1 mutant, WRKY34-LP (5′-AGCTTGAGCCCAAGTTAAAGC-3′) and WRKY34-RP (5′-GCATGTCTTGGCCAGTACCGGATG-3′) for the wrky34-1 mutant.
Molecular cloning and transformation
To generate the binary vector with the LAT52 promoter overexpression cassette, a modified version of pBI121 [9] was digested with HindIII and XhoI to replace the CaMV 35S promoter with the LAT52 promoter. To generate the PLAT52-driven 4myc-WRKY34 overexpression construct (PLAT52:4myc-WRKY34), we amplified the WRKY34 cDNA by using primers WRKY34-F (5′-CATATGGCTGGTATTGATAATAAAGCTGCTG-3′) and WRKY34-B (5′-ACTAGTCAATATCTGTCGTAATCTACTCAACATCTCTCTG-3′). The PCR fragment was cloned into a modified pBlueScript II KS vector with a four-copy myc epitope tag coding sequence at the 5′-end [9] to generate pBS-4myc-WRKY34 construct. The 4myc-WRKY34 fragment was then cloned into the pBI-PLAT52 vector using SpeI and XhoI sites.
To generate WRKY34SA cDNA, mutations were introduced into the pBS-4myc-WRKY34 construct by Quick Change site-directed mutagenesis [43], [44]. Primers used were as following, with mutated residues in lower case: WRKY34-S91A (5′-TCTCTTCTCCTGGTCTTgccCCTGCAACTCTGTTAGAG-3′), WRKY34-S87/91A (5′-ATCTCTgcTCCTGGTCTTgccCCTGCAACTCTGTTAGAG-3′), WRKY34-S98A (5′-CTCTGTTAGAGgcTCCTGTTTTCCTCTC-3′), WRKY34-S108A (5′-CTCAAACCCTTTGCTAgCTCAACAACCGGGAAG-3′), WRKY34-S544A (5′-GAAGGTGGAACCAGTGgCACCACAACAGGGAC-3′), and their reverse complementary primers. WRKY34SA, with all six Ser residues mutated to Ala residues, was generated by five successive mutagenesis steps and verified by sequencing. To generate the PLAT52:4myc-WRKY34SA construct, the WRKY34SA fragment was cloned into the pBI-PLAT52 vector using SpeI and XhoI sites.
To generate the pollen-specific MPK3RNAi construct, the MPK3RNAi sequence, as described previously [13], was cloned into the pBI-PLAT52 vector between the SpeI and XhoI sites. The construct was introduced into the mpk6-2 mutant [12], and homozygous transgenic plants were identified as MPK3RNAi mpk6. To generate the PLAT52: 4myc-WRKY34 overexpression construct with a BASTA selection marker for transformation of a MPK3RNAi mpk6 plant, the PLAT52:4myc-WRKY34 cassette was amplified and partially digested with ApaI and BamHI and then cloned into the pGreenII vector [45]. To generate PWRKY34:WRKY34-YFP and PWRKY2:WRKY2-YFP constructs, genomic fragments of WRKY34 and WRKY2 were amplified. PCR products and pGreenII-YFP plasmid were digested by XhoI and EcoRV, and ligation was performed. All the binary vectors described below were transformed into Agrobacterium strain GV3101. Arabidopsis transformation was performed by the floral dip procedure [46], and transformants were identified by screening for kanamycin or BASTA resistance.
Cytological and phenotypic analyses
Fluorescence microscopy was performed with an Olympus IX70 inverted microscope with an ORCA digital camera. Pollen viability was examined using Alexander staining [25]. Pictures were taken on an Olympus Vanox AHBT3 upright microscope with a color digital camera. The FDA staining assay was performed as described [47]. DAPI was used to stain vegetative and generative/sperm nuclei to determine the pollen development. For FDA or DAPI staining of developing pollen, floral buds at each stage were carefully dissected under stereoscope. Anthers were isolated and transferred to a drop of FDA or DAPI solution. A fine needle was used to gently break the anthers, a cover slip was then used to carefully squeeze the anthers to release the pollen. For SEM, fresh pollen grains were coated directly with platinum and observed on an FEI Quanta 600 FEG Extended Vacuum Scanning Electron Microscope. Pollen germination assays were performed as described [48]. For pollen tube length measurements at 7 hour after germination, at least 100 pollen tubes in each sample were determined using ImageJ software [49]. The presented data are an average of 3 biological repeats.
In vitro phosphorylation assay
For purification of recombinant WRKY34 and its mutant proteins, the WRKY34WT and WRKY34SA cDNAs were cut from the pBS-4myc-WRKY34 constructs with NdeI/SpeI and ligated into the NdeI/NheI cut pET28a (+) vector in frame. The constructs were transformed into E. coli strain BL21(DE3). The in vitro phosphorylation assay was performed as previously described [12].
Immunoblot analysis and in vivo phosphorylation assay
Protein extraction was performed as previously described with modification [9]. Open flowers or closed buds at similar stages were collected from 20 inflorescences. The flowers/buds were ground in liquid nitrogen and extracted in 100 µl 1.5X SDS loading buffer. A 15 µl sample was loaded to each lane. The numbers of developing/mature pollen grains should be similar among each sample. However, due to the size difference of the flowers/buds at different developmental stages, different amounts of total proteins were present, which is reflected by the different amount of Rubisco large subunit protein in the Coomassie-blue stained control gels. In this experiment, a comparison of WRKY34 protein levels in an equal number of developing/mature pollen grains is better than in an equal amount of total proteins. A Phos-tag reagent (NARD Institute) was used for the phospho-protein mobility shift assay to detect in vivo phosphorylated WRKY34 protein as previously described [9].
Phylogenetic analysis
The multiple sequence alignment of full-length protein sequences was performed using the ClustalW tool online (http://www.ch.embnet.org/software/ClustalW.html). Phylogenetic trees were constructed and tested by MEGA5 based on the neighbor-joining method [50].
Quantitative RT-PCR analysis
Total RNA was extracted from each tissue using RNAqueous (Ambion Inc.) according to the manufacturer's instructions. After DNase treatment, µg of total RNA was reverse transcribed, and quantitative PCR analysis was performed using an Optican 2 real-time PCR machine (Bio-Rad). Relative levels of each transcript were calculated after being normalized to the UBC21 or EF1α control.
DNA-protein electrophoresis mobility shift assay (EMSA)
EMSA was performed as previously described [9]. A synthetic DNA oligonucleotide (5′-CGTTGACCGTTGACCGAGTTGACTTTTTA-3′ with three W boxes underlined) was used as a probe. Two complementary strands of the oligonucleotides were annealed and then labeled at the 5′ end using a T4 polynucleotide kinase. Freshly prepared recombinant WRKY34WT or WRKY34SA protein (1 µg) was incubated with 20,000–50,000 cpm of DNA probe (2 pmole) for 30 min at room temperature in a binding buffer (20 mM HEPES, pH 7.9, 0.1 µg/µL herring sperm DNA, 0.5 mM DTT, 0.1 mM EDTA, 50 mM KCl) in the presence or absence of an unlabeled competitor DNA. The resulting protein-DNA complexes were resolved in 5% non-denaturing polyacrylamide gel in half-strength TBE buffer. Following electrophoresis, the gel was dried onto 3 MM paper and exposed to X-ray film.
Supporting Information
Zdroje
1. McCormickS (2004) Control of male gametophyte development. Plant Cell 16 Suppl: 53
2. HonysD, TwellD (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85.
3. MascarenhasJP (1990) Gene activity during pollen development. Annu Rev Plant Biol 41 : 317–655.
4. WangJ, QiuX, LiY, DengY, ShiT (2011) A transcriptional dynamic network during Arabidopsis thaliana pollen development. BMC Systems Bio 5: S8.
5. VerelstW, TwellD, de FolterS, ImminkR, SaedlerH, et al. (2007) MADS-complexes regulate transcriptome dynamics during pollen maturation. Genome Biol 8: R249.
6. KobayashiA, SakamotoA, KuboK, RybkaZ, KannoY, et al. (1998) Seven zinc-finger transcription factors are expressed sequentially during the development of anthers in petunia. Plant J 13 : 571–577.
7. SegerR, KrebsEG (1995) The MAPK signaling cascade. FASEB J 9 : 726–735.
8. JacobsD, GlossipD, XingH, MuslinAJ, KornfeldK (1999) Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev 13 : 163–175.
9. MaoG, MengX, LiuY, ZhengZ, ChenZ, et al. (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23 : 1639–1653.
10. LampardGR, MacalisterCA, BergmannDC (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322 : 1113–1116.
11. BethkeG, UnthanT, UhrigJF, PoschlY, GustAA, et al. (2009) Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc Natl Acad Sci USA 106 : 8067–8072.
12. LiuY, ZhangS (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16 : 3386–3485.
13. WangH, NgwenyamaN, LiuY, WalkerJ, ZhangS (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19 : 63–73.
14. MengX, XuJ, HeY, YangKY, MordorskiB, et al. (2013) Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25 : 1126–1142.
15. ZhengZ, QamarS, ChenZ, MengisteT (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48 : 592–1197.
16. LiG, MengX, WangR, MaoG, HanL, et al. (2012) Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet 8: e1002767.
17. EulgemT, RushtonP, RobatzekS, SomssichI (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 : 199–206.
18. ZouC, JiangW, YuD (2010) Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J Exp Bot 61 : 3901–3914.
19. HonysD, OhSA, RenakD, DondersM, SolcovaB, et al. (2006) Identification of microspore-active promoters that allow targeted manipulation of gene expression at early stages of microgametogenesis in Arabidopsis. BMC Plant Biol 6 : 31.
20. TwellD, YamaguchiJ, McCormickS (1990) Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109 : 705–713.
21. SmythDR, BowmanJL, MeyerowitzEM (1990) Early flower development in Arabidopsis. Plant Cell 2 : 755–767.
22. TwellD, YamaguchiJ, WingRA, UshibaJ, McCormickS (1991) Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer sequences and shared regulatory elements. Genes Dev 5 : 496–507.
23. KinoshitaE, TakahashiM, TakedaH, ShiroM, KoikeT (2004) Recognition of phosphate monoester dianion by an alkoxide-bridged dinuclear zinc(II) complex. Dalton Trans 2004 : 1189–1193.
24. UedaM, ZhangZ, LauxT (2011) Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell 20 : 264–334.
25. AlexanderM (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44 : 117–122.
26. WangH, LiuY, BruffettK, LeeJ, HauseG, et al. (2008) Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20 : 602–613.
27. MorrisonDK, DavisRJ (2003) Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol 19 : 91–118.
28. DardN, PeterM (2006) Scaffold proteins in MAP kinase signaling: more than simple passive activating platforms. Bioessays 28 : 146–156.
29. KolchW (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol 6 : 827–837.
30. RemenyiA, GoodMC, BhattacharyyaRP, LimWA (2005) The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol Cell 20 : 951–962.
31. WidmannC, GibsonS, JarpeMB, JohnsonGL (1999) Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev 79 : 143–180.
32. MorA, PhilipsMR (2006) Compartmentalized Ras/MAPK signaling. Annu Rev Immunol 24 : 771–800.
33. SchwartzMA, MadhaniHD (2004) Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu Rev Genet 38 : 725–748.
34. ZhangS, KlessigD (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6 : 520–527.
35. Zhang S (2008) Mitogen-activated protein kinase cascades in plant intracellular signaling. In: Yang Z, editor. Annual Plant Reviews Volume 33: Intracellular Signaling in Plants: Oxford: Wiley-Blackwell. pp. 100–136.
36. RenD, LiuY, YangK, HanL, MaoG, et al. (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 105 : 5638–5643.
37. RushtonP, SomssichI, RinglerP, ShenQ (2010) WRKY transcription factors. Trends Plant Sci 15 : 247–305.
38. MiaoY, LaunT, ZimmermannP, ZentgrafU (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55 : 853–867.
39. KangX, LiW, ZhouY, NiM (2013) A WRKY transcription factor recruits the SYG1-like protein SHB1 to activate gene expression and seed cavity enlargement. PLoS Genet 9: e1003347.
40. GrunewaldW, De SmetI, LewisDR, LofkeC, JansenL, et al. (2012) Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci USA 109 : 1554–1559.
41. TeigeM, ScheiklE, EulgemT, DocziR, IchimuraK, et al. (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15 : 141–152.
42. JiangW, YuD (2009) Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol 9 : 96.
43. FisherC, PeiG (1997) Modification of a PCR-based site-directed mutagenesis method. Biotechniques 23 : 570.
44. ZhengL, BaumannU, ReymondJ-L (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32: e115.
45. HellensRP, EdwardsEA, LeylandNR, BeanS, MullineauxPM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42 : 819–832.
46. CloughS, BentA (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–778.
47. Heslop-HarrisonJ, Heslop-HarrisonY (1970) Evaluation of pollen viability by enzymatically induced fluorescence; intracellular hydrolysis of fluorescein diacetate. Stain Technol 45 : 115–120.
48. BoavidaLC, McCormickS (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52 : 570–582.
49. AbràmoffMD, MagalhãesPJ, RamSJ (2004) Image processing with ImageJ. Biophotonics Int 11 : 36–78.
50. KumarS, NeiM, DudleyJ, TamuraK (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9 : 299–306.
Štítky
Genetika Reprodukčná medicína
Článek Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin PathwayČlánek Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human DiseasesČlánek G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify LongevityČlánek PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial MatrixČlánek Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus ConflictsČlánek The Impact of Population Demography and Selection on the Genetic Architecture of Complex TraitsČlánek Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar AcidificationČlánek The Case for Junk DNA
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 5- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Genetic Interactions Involving Five or More Genes Contribute to a Complex Trait in Yeast
- A Mutation in the Gene in Dogs with Hereditary Footpad Hyperkeratosis (HFH)
- Loss of Function Mutation in the Palmitoyl-Transferase HHAT Leads to Syndromic 46,XY Disorder of Sex Development by Impeding Hedgehog Protein Palmitoylation and Signaling
- Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution
- Genome-Wide Nucleosome Positioning Is Orchestrated by Genomic Regions Associated with DNase I Hypersensitivity in Rice
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Single Nucleotide Variants in Transcription Factors Associate More Tightly with Phenotype than with Gene Expression
- Ribosomal Protein Mutations Induce Autophagy through S6 Kinase Inhibition of the Insulin Pathway
- Recent Mitochondrial DNA Mutations Increase the Risk of Developing Common Late-Onset Human Diseases
- Epistatically Interacting Substitutions Are Enriched during Adaptive Protein Evolution
- Meiotic Drive Impacts Expression and Evolution of X-Linked Genes in Stalk-Eyed Flies
- G×G×E for Lifespan in : Mitochondrial, Nuclear, and Dietary Interactions that Modify Longevity
- Population Genomic Analysis of Ancient and Modern Genomes Yields New Insights into the Genetic Ancestry of the Tyrolean Iceman and the Genetic Structure of Europe
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
- Whole Exome Re-Sequencing Implicates and Cilia Structure and Function in Resistance to Smoking Related Airflow Obstruction
- Allelic Expression of Deleterious Protein-Coding Variants across Human Tissues
- R-loops Associated with Triplet Repeat Expansions Promote Gene Silencing in Friedreich Ataxia and Fragile X Syndrome
- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- The Impairment of MAGMAS Function in Human Is Responsible for a Severe Skeletal Dysplasia
- Octopamine Neuromodulation Regulates Gr32a-Linked Aggression and Courtship Pathways in Males
- Mlh2 Is an Accessory Factor for DNA Mismatch Repair in
- Activating Transcription Factor 6 Is Necessary and Sufficient for Alcoholic Fatty Liver Disease in Zebrafish
- The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells
- Rapid Evolution of PARP Genes Suggests a Broad Role for ADP-Ribosylation in Host-Virus Conflicts
- Genome-Wide Inference of Ancestral Recombination Graphs
- Mutations in Four Glycosyl Hydrolases Reveal a Highly Coordinated Pathway for Rhodopsin Biosynthesis and N-Glycan Trimming in
- SHP2 Regulates Chondrocyte Terminal Differentiation, Growth Plate Architecture and Skeletal Cell Fates
- The Impact of Population Demography and Selection on the Genetic Architecture of Complex Traits
- Retinoid-X-Receptors (α/β) in Melanocytes Modulate Innate Immune Responses and Differentially Regulate Cell Survival following UV Irradiation
- Genetic Dissection of the Female Head Transcriptome Reveals Widespread Allelic Heterogeneity
- Genome Sequencing and Comparative Genomics of the Broad Host-Range Pathogen AG8
- Copy Number Variation Is a Fundamental Aspect of the Placental Genome
- GOLPH3 Is Essential for Contractile Ring Formation and Rab11 Localization to the Cleavage Site during Cytokinesis in
- Hox Transcription Factors Access the RNA Polymerase II Machinery through Direct Homeodomain Binding to a Conserved Motif of Mediator Subunit Med19
- Drosha Promotes Splicing of a Pre-microRNA-like Alternative Exon
- Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of
- PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF
- Enhanced Interaction between Pseudokinase and Kinase Domains in Gcn2 stimulates eIF2α Phosphorylation in Starved Cells
- A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in
- dGTP Starvation in Provides New Insights into the Thymineless-Death Phenomenon
- Phosphorylation Modulates Clearance of Alpha-Synuclein Inclusions in a Yeast Model of Parkinson's Disease
- RPM-1 Uses Both Ubiquitin Ligase and Phosphatase-Based Mechanisms to Regulate DLK-1 during Neuronal Development
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- More of a Good Thing or Less of a Bad Thing: Gene Copy Number Variation in Polyploid Cells of the Placenta
- Heritable Transmission of Stress Resistance by High Dietary Glucose in
- Revertant Mutation Releases Confined Lethal Mutation, Opening Pandora's Box: A Novel Genetic Pathogenesis
- Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification
- A Genome-Wide Assessment of the Role of Untagged Copy Number Variants in Type 1 Diabetes
- Selectivity in Genetic Association with Sub-classified Migraine in Women
- A Lack of Parasitic Reduction in the Obligate Parasitic Green Alga
- The Proper Splicing of RNAi Factors Is Critical for Pericentric Heterochromatin Assembly in Fission Yeast
- Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma
- Six Homeoproteins and a linc-RNA at the Fast MYH Locus Lock Fast Myofiber Terminal Phenotype
- EDR1 Physically Interacts with MKK4/MKK5 and Negatively Regulates a MAP Kinase Cascade to Modulate Plant Innate Immunity
- Genes That Bias Mendelian Segregation
- The Case for Junk DNA
- An In Vivo EGF Receptor Localization Screen in Identifies the Ezrin Homolog ERM-1 as a Temporal Regulator of Signaling
- Mosaic Epigenetic Dysregulation of Ectodermal Cells in Autism Spectrum Disorder
- Hyperactivated Wnt Signaling Induces Synthetic Lethal Interaction with Rb Inactivation by Elevating TORC1 Activities
- Mutations in the Cholesterol Transporter Gene Are Associated with Excessive Hair Overgrowth
- Scribble Modulates the MAPK/Fra1 Pathway to Disrupt Luminal and Ductal Integrity and Suppress Tumour Formation in the Mammary Gland
- A Novel CH Transcription Factor that Regulates Expression Interdependently with GliZ in
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics
- Spermatid Cyst Polarization in Depends upon and the CPEB Family Translational Regulator
- Insights into the Genetic Structure and Diversity of 38 South Asian Indians from Deep Whole-Genome Sequencing
- Intron Retention in the 5′UTR of the Novel ZIF2 Transporter Enhances Translation to Promote Zinc Tolerance in
- A Dominant-Negative Mutation of Mouse Causes Glaucoma and Is Semi-lethal via LBD1-Mediated Dimerisation
- Biased, Non-equivalent Gene-Proximal and -Distal Binding Motifs of Orphan Nuclear Receptor TR4 in Primary Human Erythroid Cells
- Ras-Mediated Deregulation of the Circadian Clock in Cancer
- Retinoic Acid-Related Orphan Receptor γ (RORγ): A Novel Participant in the Diurnal Regulation of Hepatic Gluconeogenesis and Insulin Sensitivity
- Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions
- Fine Tuning of the UPR by the Ubiquitin Ligases Siah1/2
- Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression
- Allele-Specific Genome-wide Profiling in Human Primary Erythroblasts Reveal Replication Program Organization
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- PINK1-Parkin Pathway Activity Is Regulated by Degradation of PINK1 in the Mitochondrial Matrix
- Null Mutation in PGAP1 Impairing Gpi-Anchor Maturation in Patients with Intellectual Disability and Encephalopathy
- Phosphorylation of a WRKY Transcription Factor by MAPKs Is Required for Pollen Development and Function in
- p53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



