-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II Fidelity
Mistakes made during the synthesis of messenger RNAs have been difficult to detect, both because mRNAs can be short lived, and because the translation of mRNAs into proteins has a much higher error rate that masks transcription errors. We present here a highly sensitive genetic screen that detects transcription errors and use it to identify mutations that increase the error rate of RNA polymerase II. The screen incorporates a new principle that allows transient transcription errors to cause permanent genetic changes. The screen is based on suppression of a missense mutation (cre-Y324C) in the active site of the Cre recombinase. Infrequent and transient transcription errors that restore the original codon for Y324 cause the Cre-dependent activation of a reporter gene. Background from translation errors is negligible because Cre acts as a tetramer in which all four subunits require the active site tyrosine. Transcription errors as low as ∼10−6 can be detected. We identify rpb1 mutations that define four classes, those that have increased (1) misincorporation, (2) extension of a misincorporated base, (3) both misincorporation and extension, and (4) those that block the activity of the transcription proofreading factor, TFIIS.
Published in the journal: A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II Fidelity. PLoS Genet 10(9): e32767. doi:10.1371/journal.pgen.1004532
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004532Summary
Mistakes made during the synthesis of messenger RNAs have been difficult to detect, both because mRNAs can be short lived, and because the translation of mRNAs into proteins has a much higher error rate that masks transcription errors. We present here a highly sensitive genetic screen that detects transcription errors and use it to identify mutations that increase the error rate of RNA polymerase II. The screen incorporates a new principle that allows transient transcription errors to cause permanent genetic changes. The screen is based on suppression of a missense mutation (cre-Y324C) in the active site of the Cre recombinase. Infrequent and transient transcription errors that restore the original codon for Y324 cause the Cre-dependent activation of a reporter gene. Background from translation errors is negligible because Cre acts as a tetramer in which all four subunits require the active site tyrosine. Transcription errors as low as ∼10−6 can be detected. We identify rpb1 mutations that define four classes, those that have increased (1) misincorporation, (2) extension of a misincorporated base, (3) both misincorporation and extension, and (4) those that block the activity of the transcription proofreading factor, TFIIS.
Introduction
Accurate transcription is an essential step in accessing the genetic information stored in genes. In eukaryotes, transcription of DNA into mRNA is carried out by RNA polymerase II (Pol II), an enzyme comprised of 12 subunits. While there has been extensive research on core Pol II, including biochemical and detailed structural information [1]–[7], less is known about how the accuracy of transcription is controlled. The net misincorporation rate is estimated to be about one per 105 bases [8], [9]. This reflects initial misincorporation by Pol II and subsequent editing mechanisms, some intrinsic to the core polymerase and others facilitated by transcription factors. Direct screens for mutations that reduce the fidelity of transcription have been difficult due to the transient nature of the errors and the relatively high rate of translation errors, particularly at nonsense codons, that can mask transcription errors [8], [10]–[12]. Here we report a novel approach to identifying mutations that increase transcription errors in vivo.
Rpb1 and Rpb2 are the two largest Pol II subunits with several structural and functional domains that are implicated in transcription fidelity maintenance. Rpb1 contains the active site for nucleotide addition, and, together with Rpb2, forms a substrate-binding site and a deep channel accommodating template DNA and a 9–10 bp RNA-DNA hybrid [13]. Mutations in the RNA-DNA hybrid binding cleft or near the substrate binding site cause increased occurrence of insertions and deletions during transcription through homopolymeric tracts [14]–[16]. A mutation that appeared to reduce the accuracy of transcription was identified in rpoB, the gene coding for second largest subunit of RNA polymerase in Escherichia. coli, among rifampicin resistant mutants [8], [17]. The molecular mechanism of fidelity maintenance disrupted by this mutation remains to be established. However, several mechanisms by which Rpb1 regulates fidelity have been elucidated.
Important structural elements surrounding the Pol II active site include the Rpb1 bridge helix, separating the active site from the downstream DNA binding channel, and the trigger loop, a mobile element of Rpb1, which undergoes dramatic conformational changes during each NTP addition [7], [18]. The trigger loop opens allowing for Pol II translocation and NTP entry to the active site, and closes, interacting with the substrate NTP and promoting catalysis. Mutations in the trigger loop that increase incorporation of non-complementary substrates and dNTPs and thus decrease Pol II fidelity in vitro have been identified from secondary screens of Pol II mutants [3], [19], [20]. Rpb1 interacts with non-essential Pol II subunit Rpb9, which is implicated in transcription fidelity maintenance [11], [21]–[24]. It is likely that Rpb9 prevents misincorporation indirectly, by attenuating the trigger loop closing [23]. Rpb9 also has been shown to prevent extension of the misincorporated base [24]. Transcription elongation factor TFIIS interacts with Pol II Rpb1 and is implicated in fidelity control at post-incorporation stage [12], [25], [26]. Indeed, it has been demonstrated that in vitro TFIIS preferentially promotes cleavage of the mismatched 3′end of nascent RNA [27].
The interpretation of phenotypic changes associated with alterations in RNA polymerase is complicated by a mix of direct and indirect changes. RNA polymerase mutations can change the transcriptome by altering the initiation differentially at promoters, by changing elongation rates, which in turn can alter splicing efficiency, and by altering termination [28], [29]. To focus on the identification of transcription fidelity mutants, we combined a primary genetic screen for transcription errors with a biochemical assays to identify the nature of the transcription fidelity defect. In summary, mutations in Rpb1 trigger loop rendering Pol II error-prone, as well as mutations in TFIIS binding site and deletions of RPB9 and DST1 (the gene encoding TFIIS) are demonstrated to reduce the fidelity of transcription in vivo.
Results
Transcription fidelity assay based on Cre recombinase
Assays of transcription fidelity based on nonsense or missense suppression are problematic in part due to the high background caused by translational errors [10]–[12], [30]. In the approach reported here, we rely on a requirement for an active tetramer to reduce the contribution of translation errors on the background. In addition, previous approaches have required a continual production of transcription errors to produce the monitored phenotype. In our approach, transient transcription errors can result in a stable genetic change. The system involves three parts, 1) a missense mutation in the active site tyrosine of Cre recombinase, 2) a Cre-dependent recombination reporter substrate with a very low Cre-independent rate of recombination and 3) promoters that give low level expression of the mutant cre allele. These three aspects of the system are discussed separately below.
Cre mutations
Cre functions as a homotetramer, recognizing two 34 bp DNA sequences termed loxP and promoting recombination of their flanking DNAs [31], [32]. Depending on the orientation of the two loxP sites, the flanking DNA is either inverted (intrachromosomal antiparallel), excised (intrachromosomal direct), or exchanged (interchromosomal) [33]. The active site tyrosine (Y324) of Cre recombinase is essential for its activity [34]. All four subunits of the tetramer become covalently attached to strands of the DNA via those tyrosine 324 side chains during the recombination reaction. Chimeric enzymes assembled from mutant and WT subunits are catalytically inactive [35]–[39]. Translation errors that produce a WT monomer will not result in active Cre, whereas a transcription error that restores the WT codon can be translated into multiple WT subunits and assembled into an active Cre tetramer. The experiments presented here focus on the Y324C allele. This TAT → TGT mutation requires Pol II to misincorporate an adenosine opposite a cytosine in the template strand in order to suppress the cre-Y324C defect (Figure 1A). This G → A transcription error is among the most common mistakes made by E. coli RNAP in vitro and in vivo [40] and by S. cerevisiae [19].
Fig. 1. Cre-recombinase based assay for transcription errors in vivo. 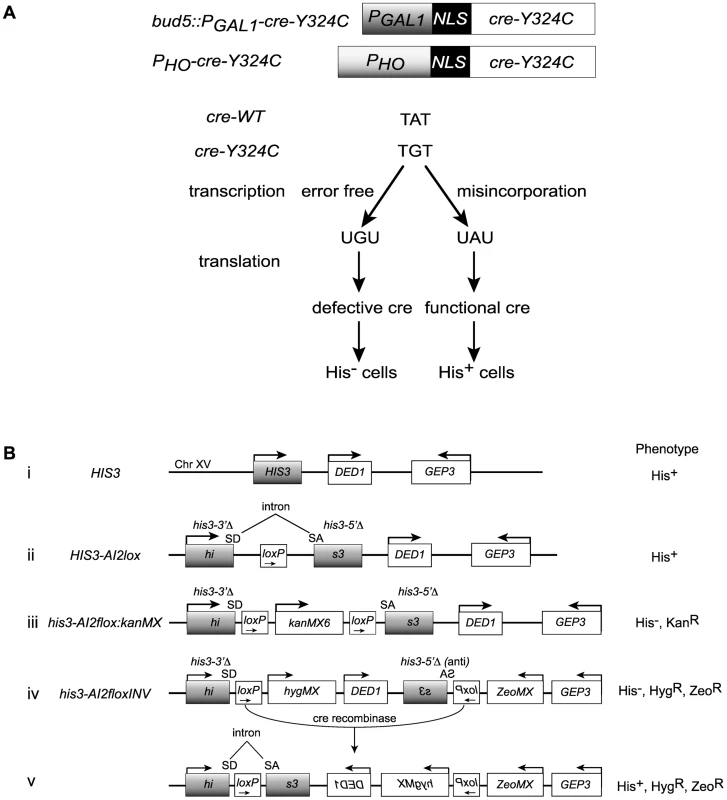
(A) Two constructs based on the promoters for the GAL1 and HO genes that allow low level expression of a mutant variant of the Cre recombinase (cre-Y324C) that has a cysteine substitution for the active site tyrosine. Suppression of the A to G mutation can result from transcriptional misincorporation of an A at that position yielding transient Cre activity. (B) Transient Cre activity can be detected as recombination events that restore function to a HIS3 based reporter. (i) The interval near the normal HIS3 gene. (ii) A loxP site inserted into an artificial intron in HIS3 is still a functional gene. SD = splice donor, SA = splice acceptor. (iii) Insertion of the kanMX gene flanked by loxP sites into the intron results in loss of HIS3 function. (iv) Inversion of the C-terminal portion of the his3 gene renders it defective. (v) Cre-mediated inversion of the construct in iv results in a functional HIS3 gene. Cre reporters
We created a reporter for Cre activity based on HIS3 into which we placed an artificial intron [41] carrying a loxP site, HIS3-AI2lox (Figure 1B, construct (ii)). A Cre activatable derivative, his3-AI2floxMX, was made by insertion of a kanMX cassette (Figure 1B, construct (iii)) [42], [43]. However, the Cre-independent His+ background from this reporter was too high (∼2×10−6 His+ among total cells). We created a HIS3 based Cre reporter system with very low Cre-independent background by placing the downstream loxP site, splice acceptor and C-terminal portion of a his3-AI2floxMX reporter in an inverted orientation at a position 3 kb distal of HIS3 beyond the DED1 gene (Figure 1B, construct (iv)). The N-terminal portion of his3-AIlox is at the normal chromosome XV position for HIS3 and is marked by hygMX, while the C-terminal portion of his3-AIlox was marked with a zeoMX cassette. Cells with this inverted lox reporter, designated his3-AI2floxINV, are His−, hygromycin resistant and zeomycin resistant. If Cre is active, it can cause inversion of the interval between the two loxP sites to generate the functional HIS3-AI2lox gene (Figure 1B, construct (v)). These cells are His+, hygromycin resistant and zeomycin resistant. The frequency of Cre-independent His+ cells among total cells with this system is <10−6 (Figure 2C).
Fig. 2. Suppression of cre-Y324C is elevated in strains with Pol II fidelity defects. 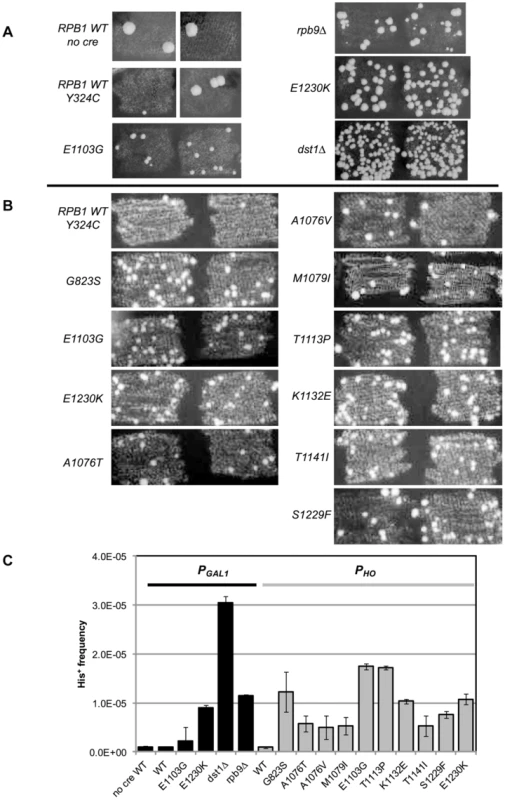
(A) PGAL1-cre-Y324C strains grown in glucose on plates lacking histidine show increased numbers of cells capable of growing microcolonies in strains with known transcription fidelity defects. Two representative patches are shown for each variant. (B) PHO-cre-Y324C used to identify new transcription fidelity mutants. (C) Mean frequency of His+ cells relative to total viable cells in PGAL1-creY324C, or PHO-creY324C, his3-AI2floxINV strains with Pol II mutants. Cultures were grown overnight in 1 ml of YPD at 30°C and plated on SC-His and 10-fold serial dilutions onto YEPDA plates. Plates were incubated at 30°C for 3 days and individual colonies were scored. Error bars show standard error. Low level expression
In the experiments described here it was anticipated that the mutant Cre-Y324C protein should act as a dominant inhibitor of wild type Cre protein. That is, assembly of mutant subunits into the tetramer will block its function. Therefore, we expected that it is important to have very low levels of expression in order to favor assembly of tetramers from translation of one mRNA. This condition promotes the detection of transcription errors that restore the Tyr codon in the cre-Y324C transcript. We used two approaches to have low levels of transcription (Figure 1A). First we placed the cre gene under the control of the GAL1 promoter but grew the cells in the presence of glucose. Under these glucose repressed conditions, expression was expected to be rare. In the second approach, we placed the cre-Y324C allele under the control of the HO promoter so that it would be expressed only in that half of the cell population that had already divided once before [44], [45].
Proof of principle
Our initial experiments used the PGAL1-cre-Y324C expression system as the transcription error substrate (inserted into the BUD5 gene) and the his3-AI2floxINV Cre activity reporter. For cells with a WT Pol II (GRY3739) the frequency of His+ cells among the total population is similar in the absence of Cre or with the PGAL1-cre-Y324C construct (Figure 2A&C). In this assay, patches of cells grown on rich media (YPD) were replica plated onto synthetic complete medium lacking histidine. The starting strains cannot grow, but rare His+ cells within the patch can grow into small colonies or papillae. The PGAL1-cre-Y324C strain lacking TFIIS (dst1Δ GRY3742) has an elevated level of His+ cells (Figure 2A&C) that is not observed when the cre gene is absent. We interpret the elevated level of papillation in the TFIIS defective strain as a reflection of transient Cre activity caused by uncorrected G → A errors during transcription in the mutant cre-Y324C gene that restore the UAU tyrosine codon in the mRNA (Figure 1A). To demonstrate that the His+ cells were not a result of an elevated reversion rate for the cre-Y324C mutation in TFIIS defective cells, 11 independent His+ cells from the dst1 strain were crossed to a cell carrying an ade6-AI2floxkanMX reporter and shown to be still defective for Cre recombinase activity. In addition, the cre gene sequenced from 12 more His+ colonies still carried the cre-Y324C mutation. To determine whether the increase in His+ cells might be the result of elevated Cre-independent recombination between the lox sites in TFIIS defective strains, we compared the frequency of His+ cells in the absence of the cre gene and observed that WT and dst1 strains gave similar levels. These results confirm that TFIIS has a role in vivo in editing RNA errors and promoting transcription fidelity.
RPB9 has been identified as a nonessential subunit of Pol II that promotes accurate start site selection, efficient elongation, contributes to transcription fidelity [11], [23], [46]–[50] and regulates mismatch extension [24]. Consistent with a role for the Rpb9 subunit in promoting transcription fidelity in vivo, we find that a null allele of RPB9, rpb9-Δ0 (GRY3743), shows an elevated His+ papillation rate in the his3-AI2floxINV assay (Figure 2A&C).
In order to test a collection of rpb1 mutations, we created a strain (GRY2855) with the PGAL1-cre-Y324C substrate, the his3-AI2floxINV reporter and the rpb1-natMX null allele complemented by RPB1 on a URA3 selectable plasmid (pJDI220). We made rpb1 variants on a LEU2 based plasmid (pJS757) and substituted the mutant alleles for the RPB1 wild type allele by transforming in the LEU2 plasmids and selecting for loss of the URA3 plasmid with 5-FOA which selects against Ura+ cells [51]. Cells with the rpb1-E1230K (rpo21-24) mutation (pJS932), which blocks the ability of TFIIS to bind to Pol II [52], show an elevated number of His+ papillae (Figure 2A&C). This is consistent with the results presented above for the TFIIS defective strain (dst1Δ0). We previously described rpb1-E1103G (pJS781) as causing increased misincorporation during transcription [19]. That allele was also among those identified as having elevated misincorporation rates in vitro [3]. When put into the his3-AI2floxINV reporter strain with cre-Y324C under the control of the GAL1 promoter, it showed a modest increase in His+ papillation (Figure 2A&C).
Genetic screen for new rpb1 mutants
To use the his3-AI2floxINV reporter as an effective screen for new rpb1 mutations that reduce fidelity, we addressed the problem of how to get widespread but low level expression of the cre-Y324C gene. The ideal condition would be the one that produced one transcript per cell. The GAL1 promoter in the presence of glucose is repressed [53], but low levels of transcription in a population probably reflects some cells with transcripts and most cells with none. The promoter for the HO gene is under complex regulation so that it is only expressed late in the G1 phase of the cell cycle, only in cells that have the a or α mating phenotypes (not a/α “diploid” cells), and only in cells that have divided at least once before, so called mother cells [44], [45]. We placed the cre-Y324C gene under the control of the HO promoter at its normal position on chromosome IV. Only half of the cells (mothers) in the population will be expressing PHO-cre-Y324C, and half of those will be expressing it for the first time. We reasoned that this would create a condition where rare transcripts that have an error that restores the tyrosine codon would produce active Cre recombinase that would not be in competition with the inactive monomers for the assembly of active Cre tetramer.
To make it easy to introduce rpb1 variants into this system, we made a yeast strain (GRY3258) with the PHO-cre-Y324C substrate, the his3-AI2floxINV reporter, and the rpb1-natMX deletion complemented by RPB1 on a URA3 2-micron based plasmid (pJS725). As above, this makes it possible to substitute in rpb1 variants on a LEU2 based vector by plasmid shuffling, selecting against the URA3 RPB1 with 5-FOA [51]. Figure 2B illustrates the clear distinction between RPB1 and the transcription fidelity mutant rpb1-E1103G in the number of His+ papillae with the his3-AIloxINV reporter and the PHO-cre-Y324C substrate. The increase is dependent on the cre-Y324C gene. Similarly, rpb1-E1230K, which blocks TFIIS function, gives a clearly elevated signal in this assay.
To identify additional rpb1 alleles that cause reduced transcription fidelity in vivo, we screened a mutagenized pool of a LEU2 CEN based plasmid carrying RPB1 (pJS757). The pool was transformed into GRY3258 selecting Leu+ and colonies picked and arrayed as patches. The patches were replica plated to FOA plates to select against the URA3 RPB1 plasmid. From the FOA plates the patches were replica plated to SC-His media to detect patches with elevated levels of Cre-mediated His+ papillae. Mutant candidates were struck for single colonies, and retested as patches. The LEU2 based plasmid from those that repeated the elevated level of His+ papillae was recovered into E. coli and retransformed into GRY3258 to confirm that the mutation causing the elevated Cre activity was on the plasmid. Those that passed that test were sequenced to identify the rpb1 mutation responsible for the transcription infidelity phenotype. In all, over 12,000 transformants were tested from which eight rpb1 variants were identified. These included re-isolating rpb1-E1103G, plus seven new alleles: rpb1-G823S, -A1076T, -A1076V, -M1079I(T1548I), -K1132E, -T1141I(G888D,I1237T), and -S1229F. The phenotype of the rpb1-T1141I(G888D,I1237T) variant was shown to be a consequence of the T1141I substitution by testing variants with each of the separate mutations. Patches demonstrating the phenotypes of these variants are shown in Figure 2B. In addition, we show results for an allele rpb1-T1113P, isolated from a screen for alleles that elevate transcription slippage [15].
Quantification of the increase in transcription errors was accomplished by measuring the frequency of His+ cells as determined by growth on medium lacking histidine (SC-His) normalized to growth on rich media. The mean frequency from 8 or more cultures was determined for each strain and shows that the mutations identified by our screen increase the frequency of cells with the His+ phenotype 7–50 fold compared to the wild type strain (Figure 2C).
In vitro measurement of transcription fidelity
In the in vivo screen, increase of the frequency of His+ papillae by the mutations in RPB1 can be indirect. For instance, it could be caused by the increase of cre expression because of an increase in initiation by the mutant Pol II. It is noteworthy that the His+ frequency is much higher in the PHO-cre system than in the PGAL1-cre system for the rpb1-E1103G allele. In contrast the His+ frequencies for the rpb1-E1230K allele are similar in the two systems. Whether this reflects a differential level of sensitivity for these promoters to these defects has not been determined. Furthermore, a higher net frequency of error-containing RNA may be caused not only by the higher frequency of misincorporation (as in the rpb-E1103G mutant), but also by higher efficiency of mismatch extension and/or on lower efficiency of RNA editing (like in a DST1-deficient strain or in the rpb1-E1230K mutant, defective in TFIIS binding) [52]. Below we describe the results of three in vitro tests designed to address the biochemical basis for the low-fidelity phenotype of the alleles identified in the genetic screen: 1) the ability of Pol II to select cognate NTP, 2) the capacity to extend a mismatch, and 3) the ability to remove misincorporated NMP by TFIIS-mediated editing.
Competition fidelity assay
To determine whether any of the new alleles of rpb1 have direct impact on cognate NTP selection, we tested in vitro the level of misincorporation errors by the Pol II variants. Misincorporation rates and transcription fidelity may depend on a sequence context [4]. Therefore, we employed a DNA corresponding to the region surrounding codon 324 (TGT) in the cre-Y324C gene as the template for the assembly of the promoter - and factor-independent Pol II elongation complexes [54] used in these fidelity assays. The elongation complex (U10) was stalled before incorporating AMP and the frequency of GMP to AMP transition error was directly measured using a “competition” assay for transcription fidelity [20], which provides fidelity values nearly identical to those obtained by conventional fidelity assay for the wild type and mutant variants of Pol II [20], [55]. A similar assay has been described for T7 DNA polymerase [56]. The competition assay employs differential mobility in denaturing polyacrylamide gels of short RNA species of the same length, but different composition to distinguish cognate from misincorporation events. To determine the frequency of misincorporation, U10 (schematically depicted in Figure 3A) was incubated with a mix of GTP in a low (20 nM) concentration and different higher (0.5, 0.75 and 1.5 mM) concentrations of ATP. In these conditions, the reaction products of both cognate (GMP) and non-cognate (AMP) incorporation were separated (Figure 3B, lanes 3–13) and quantified (Figure 3C). To determine the misincorporation frequency of AMP in place of GMP in the presence of equal concentrations of cognate and non-cognate substrates, the ratio of non-cognate to cognate products was normalized to (divided by) the ratio of non-cognate (ATP) to cognate (GTP) substrate concentrations. The resulting misincorporation frequency was similar for different concentrations of ATP tested suggesting that it faithfully reflects fidelity of substrate selection in this position, independent of the substrate concentration. This assay is particularly useful way to qualitatively compare the relative misincorporation of various alleles. It is clear that Pol II carrying the rpb1-E1103G mutation misincorporates more frequently than wild type Pol II (Figure 3B, lane 8), consistent with its previously reported defect in NTP selection [19]. The rpb1-A1076T, A1076V, and T1113P substitutions also noticeably increase the frequency of misincorporation (lanes 5, 6, and 9). The RPB1-A1076 residue is located in the trigger loop, a mobile element of the catalytic subunit implicated in transcription fidelity maintenance by genetic and biochemical analyses [3], [19]. In contrast, mutations located in the TFIIS-binding domain (rpb1-K1132E, rpb1-T1141I and rpb1-S1229F) and one mutation in the trigger loop (rpb1-M1079I) do not appear to change GMP to AMP transition frequency in vitro.
Fig. 3. Cognate NTP selection by Pol II variants from the strains with fidelity defects. 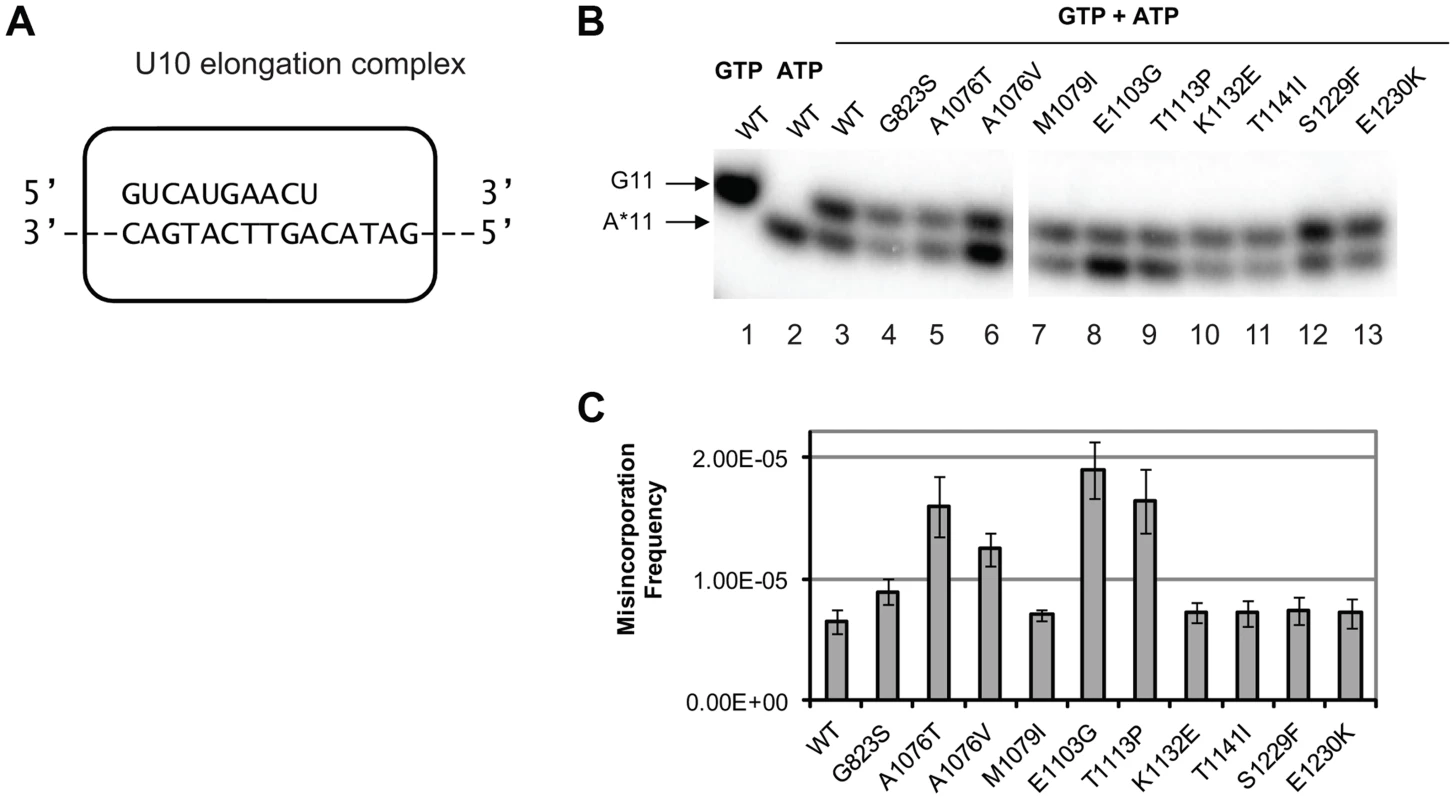
(A) Schematic representation of the elongation complex used for in vitro characterization of Pol II. The sequences of nascent RNA in the starting U10 elongation complex and a part of the 57-nt template DNA strand are shown. Pol II is depicted as a rectangular outline with rounded corners. The rest of the template DNA strand and the fully complementary non-template DNA strand are omitted for clarity of presentation. (B) Products of cognate and non-cognate incorporation. The TECs were incubated with 20 nM cognate GTP, 1 mM non-cognate ATP or the mix of 20 nM GTP and 1 mM ATP for 5 min. Arrows indicate the products of GMP incorporation and AMP misincorporation (G11 and A*11, respectively). (C) ratios of G11 to A*11 products obtained in the presence of 20 nM GTP and 0.5, 1 and 1.5 mM ATP were quantified, normalized to [GTP]/[ATP], and averaged. The error bars show standard deviation. Mispaired base extension assay
Next, we tested whether the newly identified rpb1 alleles alter the ability of Pol II to extend the mismatch by incubating the complex with 1 mM non-complementary ATP and 0.5 mM next cognate UTP to allow misincorporation and mismatch extension. Under these conditions, wild type Pol II does not easily extend the A11 mismatch and pauses at the U12 and A13 positions (Figure 4A, lane 3). Apparently, the mismatch continues to present an obstacle to efficient transcript elongation, consistent with the recent report by Knippa and Peterson [24]. Inefficient mismatch extension, similar to wild type Pol II was observed in the Pol II variants carrying mutations in the TFIIS binding site (rpb1-K1132E, rpb1-T1141I, rpb1-S1229F and rpb1-E1230K) (Figure 4A, lanes 10–13). In contrast, mutations in the residues located in the trigger loop (rpb1-A1076T, rpb1-A1076V and rpb1-M1079I) or next to the base of the trigger loop (rpb1-E1103G and rpb1-T1113P) significantly promote mismatch extension.
Fig. 4. Mismatch extension by Pol II variants carrying mutations in Rpb1. 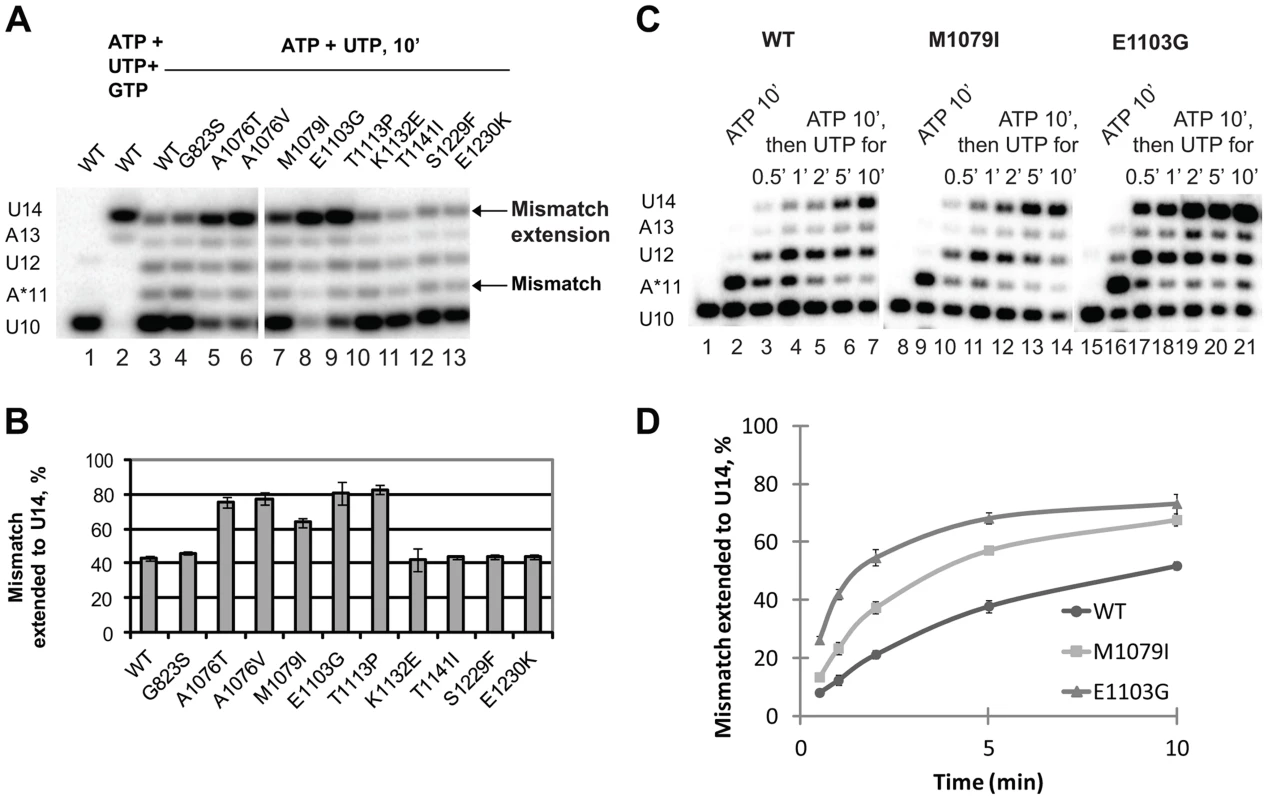
(A) Products of simultaneous misincorporation and mismatch extension. The U10 complex was incubated with a mixture of 1 mM ATP and 0.5 mM UTP, or 1 mM ATP, 0.5 mM UTP and 0.5 mM GTP where indicated. The original U10 RNA, A*11 mismatch and mismatch extension products are indicated at left. The arrows at right mark the mismatch (A*11) and mismatch extension products. (B) the fraction of U14 mismatch extension product from the sum of A11, U12, A13 and U14 products was quantified; results of three independent experiments were averaged. The error bars show standard deviation. (C) Products of consecutive misincorporation and mismatch extension. U10 complexes were incubated in 1 mM ATP for 10 minutes to obtain complexes containing an RNA with a 3′ mismatch (A*11). A sample of A*11 complex for each Pol II variant was taken, and 0.5 mM UTP was added to initiate extension of the mismatch. The extension products were analyzed at the indicated times after the addition of UTP. RNA A*11 and extension products are indicated at left. (D) Mismatch extension was quantified as in (B) and plotted versus time. The error bars show standard deviation of three independent experiments. The experimental setup described above was chosen because it best reflects the situation in vivo when misincorporation occurs in the presence of the next (cognate) NTPs, and the newly formed mismatch can be immediately extended. When the mismatch is pre-formed before the next cognate substrate is provided, Pol II has more time to backtrack, which might artificially decrease the fraction (if backtracking is irreversible) or the rate (if backtracking is reversible) of the mismatch extension. However, if misincorporation is much slower than mismatch extension, the true rate of the latter is difficult to assess in this particular experimental setup. Therefore, mismatch extension by M1079I and E1103G Pol II variants has been assayed in a different setup, when misincorporation was allowed to proceed for 10 min before UTP was added to extend the mismatch (Figure 4C). Quantitative analyses of these data (Figure 4D) confirm our conclusion that mutations in the trigger loop promote mismatch extension.
The enhanced mismatch extension by rpb1-M1079I mutant, which does not display increased frequency of misincorporation, provides an explanation for the identification of this allele as error-prone in the in vivo screen. Evidently, relatively fast mismatch extension interferes with the post-incorporation error removal by TFIIS-mediated cleavage or the proteolytic degradation of irreversibly arrested Pol II, thus increasing the fraction of the full-length transcripts containing the error. The in vivo and in vitro properties of Pol II carrying the rpb1-M1079I substitution provide direct experimental evidence that recognition of incorporated mismatches by core Pol II significantly contributes to fidelity maintenance.
TFIIS-dependent editing
The effect of the newly identified mutations on interaction of Pol II with TFIIS has been tested by treating the A*11 elongation complex carrying the 3′-end mismatch with TFIIS, and observing the extent of 3′RNA cleavage (Figure 5). The 3′ mismatched AMP and the complementary penultimate UMP are removed after one minute incubation with TFIIS in the major fraction of the wild type complexes, resulting in appearance of a 9-nt RNA (C9). Note that the C9 product was barely detectable in the initial A*11 elongation complexes (lane 3 in Fig. 5 A and B). The shorter RNA cleavage products (A8, A7, and G6) are also detected, especially after incubation with higher concentration (600 nM) TFIIS (Fig. 5B, lane 4). The shorter RNA cleavage products (A8, A7, and G6) are also detected, especially after incubation with higher concentration (600 nM) TFIIS (Figure 5B, lane 4). The appearance of the 5′ cleavage products as short as 6 nt is somewhat unexpected, considering that the RNA-DNA hybrid in Pol II elongation complex is 8-bp long [57]. Nevertheless, we use appearance of the short cleavage products, along with the major C9 product, to judge the efficiency of TFIIS-induced RNA cleavage.
Fig. 5. rpb1 mutations decreasing transcription fidelity in vivo interfere with TFIIS-dependent error editing. 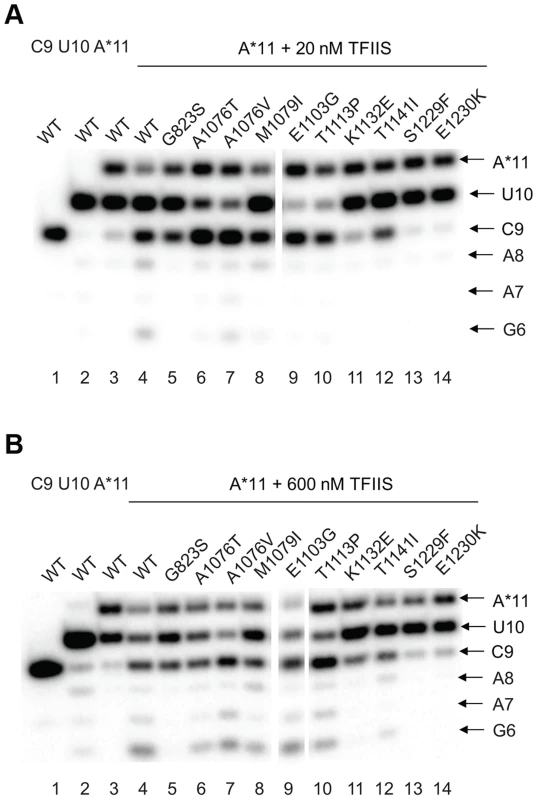
A*11 complex was obtained as described in Methods and incubated with 20 nM (A) or 600 nM (B) TFIIS for 1 min. The products of TFIIS-induced RNA cleavage (C9, A8, A7, and G6) are indicated at right. As expected, substitutions in the TFIIS binding site (rpb1-S1229F and rpb1-E1230K) dramatically decreased Pol II susceptibility to TFIIS-induced mismatch removal. The C9 cleavage product appears only when the elongation complexes are treated with the higher concentration (600 nM) TFIIS, and no shorter products are detected (compare lane 4 with lanes 13 and 14 in Figure 5A&B,). A less pronounced, but still substantial decrease in the sensitivity to TFIIS is observed for rpb1-G823S, K1132E, and rpb1-T1141I mutants (lanes 5, 11, and 12). It is likely that K1132 and T1141 substitutions directly reduce binding of TFIIS. The G823S substitution in the bridge helix may alter TFIIS binding, but alternatively could be involved in TFIIS-induced transcript cleavage, and/or affect Pol II backtracking. The effect of T1141I substitution was weaker than the effects of the other three substitutions in the TFIIS binding site (note the presence of the short cleavage products in Figure 5B, lane 11). It is interesting that Rpb1-G823S, which shows a decreased sensitivity to TFIIS similar to Rpb1-K1132E substitution, also slightly promotes misincorporation and mismatch extension in vitro (Figures 3 and 4). All other mutants tested for susceptibility to TFIIS were similar to wild type Pol II (Note the A8, A7 and G6 products in Figure 5B, lanes 4 and 6–10).
Discussion
We present here a solution to problematic issues about the measurement of transcription fidelity in vivo. Previous results suggested that G to A transitions represent the major class of errors generated by yeast Pol II [19] and by the E. coli RNA polymerase in vitro and in living cells [58]. Using that information, we developed a second generation assay specific for the detection of G to A transcription errors. The assay is based on a mutation of the active site TAT tyrosine codon of the Cre recombinase to TGT. Rare Pol II errors that restore UAU to that position allow the production of active Cre protein whose activity is detected by inversion of a 4.8 kb interval to restore a selectable phenotype (His+). The requirement that each of the four subunits of the Cre tetramer is active eliminates the background from translation errors. The assay converts a transient Cre activity into a stable genetic change allowing the detection of Cre activity at frequencies less than 10−5. This sets a lower bound on net transcription error rates, but it is likely that it underestimates the real level of uncorrected G to A substitutions during transcription because the efficiency of assembling an active Cre tetramer and the efficiency of the recognition and recombination of the lox sites in the reporter gene are unknown.
Identification of Rpb1 regions involved in control of fidelity in vivo
The random mutagenesis of the entire RPB1 (RPO21) gene yielded three groups of amino acid residues that are highly clustered in the X-ray structure of Pol II [18]. The rpb1-A1076T/V, rpb1-M1079I, and rpb1-E1103G mutations alter the trigger loop, a domain of Pol II that closes over the active site and has been demonstrated to influence fidelity in vitro [3], [19]. The rpb1-T1113P mutation targets a residue in the immediate vicinity of E1103 (Figure 6A). The rpb1-G823S allele (Figure 6B, right panel) alters the bridge helix, a domain that interacts with the active site and with the trigger loop. Substitutions in the bridge helix of E. coli RNA polymerase have been demonstrated to reduce the fidelity of transcription in vitro [59]. Notably, the changes identified in this work directly target the flexible hinge regions of the trigger loop and the bridge helix that were identified by molecular dynamics simulations based on X-ray crystallography and confirmed by mutational analysis of yeast Pol II [20]. The hinges, H1 and H2 in the trigger loop and H3 and H4 in the bridge helix, according to the nomenclature from [20] (Figure 6C) undergo conformational changes associated with NTP binding, sequestration, catalysis and translocation. The trigger loop and bridge helix residues forming the hinges have been implicated in transcription control [19], [55], [59], [60].
Fig. 6. Rpb1 domains involved in control of transcription fidelity. 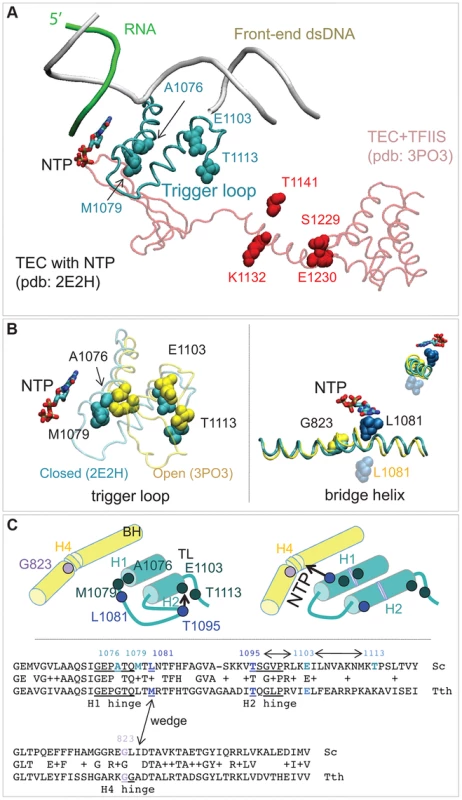
(A) Clusters of residues implicated in transcriptioin fidelity control by genetic screening and biochemical analyses. RNA, DNA, substrate NTP, and the closed trigger loop (TL) are from pdb 2E2H; this structure is aligned with the X-ray structure of TFIIS taken from the backtracked Pol II/TFIIS complex (pdb 3PO3, shown in light red). (B) Location of Rpb1-A1076, M1079, E1103 and T1113 residues between the open (yellow; pdb 3PO3) and the closed (cyan; pdb 2E2H) states of the TL (left panel). On the right: Rpb1-G823 (yellow) is located next to the site of the bridge helix bending. The long-distance movement of L1081 residue of the TL associated with the bridge helix bending and the loop opening is shown. The upper right corner shows a view along the long axis of the bridge helix. (C) The cartoon illustrates the major conformational changes near the active site associated with the TL closure and opening and identifies the four hinges in the trigger loop and bridge helix. At the bottom: an alignment of the TL and the bridge helix amino acid sequences from the yeast Pol II and T. thermophilus RNAP with all hinges and L1081 residue highlighted. The TL and bridge helix crosstalks are indicated by double-headed arrows. Another cluster of mutations alters the TFIIS binding site (Figure 4A, residues shown in red) [61]. The rpb1-S1229F mutation likely reflects a defect in the interaction of Rpb1 and TFIIS, similar to the well characterized rpb1-E1230K allele [52]. The rpb1-K1132E and rpb1-T1141I substitutions also alter positions close to where TFIIS binds [61]. These mutations may impair TFIIS-dependent error editing by decreasing TFIIS binding, similar to mutations of E1230 and S1229. Alternatively, they might affect the backtracking capabilities of Pol II, a mechanism related to Pol II translocation and likely affected by the G823S substitution. The precise characterization of TFIIS cleavage mechanisms affected by mutations identified here is beyond the scope of this work. Most importantly, our results represent a direct demonstration of proofreading function of TFIIS in living cells. Although there is firm evidence that TFIIS plays a major role in correction of transcription errors in vitro, several attempts to demonstrate the similar activity in vivo were inconclusive [10], [11]. Our results are consistent with those by Koyama and co-workers showing a major contribution of TFIIS to Pol II fidelity in yeast [12], [21].
Pol II fidelity is under multilayer control in vivo
Our in vivo approach combined with the biochemical in vitro validation revealed at least three intrinsic and one factor-dependent mechanism for faithful transcription in living cells. The TFIIS-dependent mechanism has been discussed above. The first intrinsic mechanism includes regulation of the trigger loop movement. Interaction of Rpb1 E1103 residue with the H2 hinge (residues 1095–1099) of the trigger loop has been previously proposed to delay the trigger loop closure thus slowing down transcription elongation and supporting fidelity maintenance [3], [19], [23], [62]. The observation that rpb1-T1113P substitution renders transcription error-prone indicates that the T1113 residue plays the same or similar role as E1103. Notably, the recent molecular dynamic simulations of the Pol II trigger loop opening and closure revealed the potential interaction of T1113 with the trigger loop predicting that substitutions of T1113 residue should increase transcription elongation rate [63]. Our work provides direct proof that the mechanism dependent on E1103 and T1113 interaction with the trigger loop acts in vivo.
The second potential mechanism includes H1 hinge of the trigger loop (Rpb1-A1076/M1079) and H4 hinge of the bridge helix (Rpb1-G823) that may crosstalk through the adjacent L1081 (wedging) residue of the trigger loop [64] (Figure 6B&C). Because conformational changes of the bridge helix and the trigger loop are implicated in translocation [64]–[66] our identification of error-prone mutants in the mobile hinges of these two structural elements suggests a possible connection of the Pol II translocation cycle and cognate NTP selection. The mechanism of this link remains to be established.
The third intrinsic mechanism revealed by our work does not immediately follow from the extensive in vitro studies of transcription fidelity. It involves inhibition of the mismatch extension with the next cognate NMP. This mechanism, apparently mediated by the trigger loop and bridge helix, promotes Pol II pausing or arrest after misincorporation. Our finding that several mutations selected in the Cre-based genetic screen promote mismatch extension in vitro strongly argues that slow extension of a mismatch in the nascent RNA plays a major role in faithful transcription in living cells by allowing correction to occur. The rpb1-M1079I allele identified here appears of special interest, because it does not affect NTP selection by Pol II, but clearly promotes mismatch extension. Because the net frequency of in vivo transcription error occurrence correlates with the propensity of E. coli RNA polymerase to backtrack within a given sequence context [58], we are currently investigating the effect of rpb1-M1079I substitution on the mismatch-induced transcription arrest.
The slow mismatch extension may enable correction of the error by TFIIS-mediated transcript cleavage [27]. The arrest may also provide time for elimination of the flawed transcript by ubiquitin-mediated proteolytic degradation of Pol II [67]. One can imagine rpb1 alleles that impair accuracy in nucleotide selection, but, due to a reduced transcription elongation rate [3], [20], [60] provide increased opportunity to detect and remove misincorporated NMPs. Thus, slow elongation that results in poor mismatch extension could reduce production of the full-length error-containing Cre mRNA, counter-acting the defect in substrate selection. Accordingly, faster elongation might further decrease overall fidelity of the mutants with impaired NTP selectivity, such as rpb1-A1076T, A1076V, E1103G and T1113P [19].
In conclusion, we developed a reliable experimental approach to monitor transcription fidelity in vivo. Using this tool we will characterize the role of other core Pol II subunits, as well as known transcription elongation factors, such as TFIIF, Spt4/5 and Spt6 in transcription fidelity maintenance. This methodology will allow for isolation of mutants affecting transcription fidelity and thus will promote identification of new genes and mechanisms related to the accuracy of transcription. These experiments also highlight the complications associated with assigning a specific phenotype solely to the fidelity of transcription. The rpb1-E1103G mutation increases the transcription error rate. When combined with a defect in TFIIS (dst1Δ), which corrects transcription misincorporation errors, the double mutant is dead. It is tempting to conclude that the death is the direct result of an error catastrophe of too many transcription errors resulting in too many incorrect RNAs and defective proteins encoded by them. However, TFIIS also has a role in transcription initiation at some promoters and the lethality of the double mutant could reflect some more complicated interplay of the properties of the defective RNA polymerase and the changes in the transcriptome caused by the TFIIS defect. Similarly, it was possible that an increase in the frequency of His+ cells in our genetic assay reflected changes in the expression level of the cre reporter genes caused by a change in the transcriptome unrelated to reduced fidelity. A recent survey of changes in the yeast transcriptome caused by RNA polymerase variants further emphasizes this biological feature [28]. They found changes in transcription start sites, altered relative transcript levels, and changes in splicing efficiency. Thus, it was important to include a biochemical characterization of the mutant polymerases to see if there is a fidelity defect. We characterized misincorporation, mispaired base extension, and mispaired base removal and found defects in one or more of these processes in each of the mutants. These mutants provide an opportunity to elucidate the cellular consequences of error prone transcription. The work described here is a step to understanding the biological role of faithful information transfer from DNA to RNA.
Methods
Plasmids
See Table 1. pJS757 is a LEU2-based CEN vector carrying the RPB1 ORF, as well as 594 bases upstream and 501 bases downstream of the ORF [16]. pJS725 and pJDI220 are URA3-based 2-micron circle plasmids with the same RPB1 insertion as pJS757. RPB1 was randomly mutagenized by passing a pool of plasmid pJS757 twice through the mutator (mutS mutD mutT) E. coli strain XL-1 Red (Stratagene).
Tab. 1. Plasmids and yeast strains. 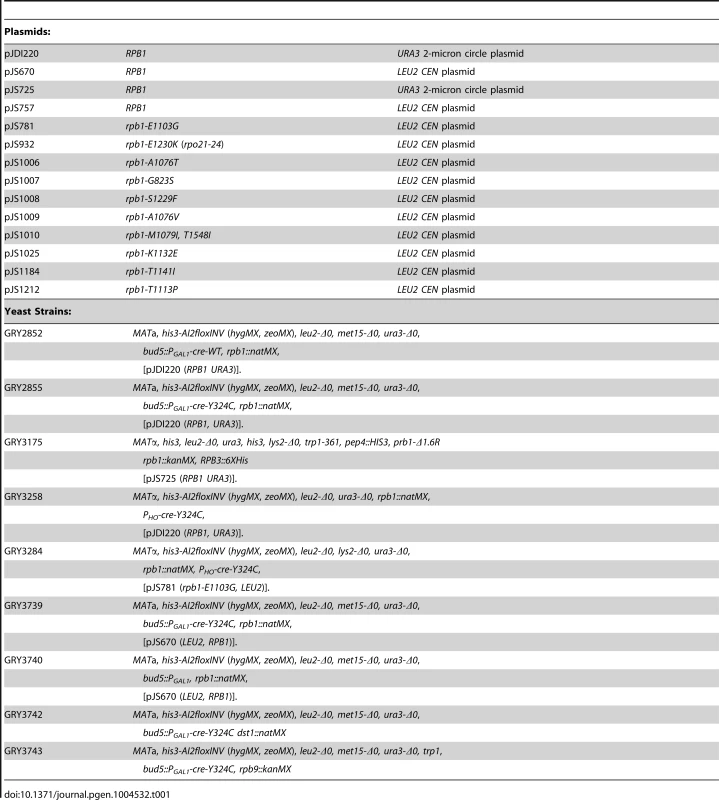
Yeast strains
The yeast strains are related to the BY4741 and BY4742 [68] See Table 1.
Cre expression
The cre gene used in these experiments has a seven amino acid nuclear localization site from the large SV40 T-antigen added at the N-terminus [69]. The PGAL1-cre gene was made by fusing the NLS-cre ORF to the PGAL1 promoter inserted into the BUD5 gene at an autochthonous EcoR1 site. The PHO-cre gene was made by overlap PCR and integrated at the normal position for HO. The insertions were made by selecting for replacement of URA3, previously inserted at those locations.
Reporters for Cre activity
Direct repeat of loxP: The his3-AI2floxkanMX reporter (Figure 1B, construct (iii)) has an artificial intron inserted into HIS3 at an MscI site in codon 70. The 1710 base insertion includes two loxP sites in direct orientation flanking the kanMX cassette. kanMX is in the same orientation as HIS3 so that the terminator blocks transcription through the rest of HIS3 resulting in cells that are His−. The Cre recombinase can remove the 1502 bases including kanMX plus one loxP resulting in a spliceable intron (HIS3-AI2lox Figure 1B, (ii)) and cells that are His+. The ade6-AI2floxkanMX reporter has the same artificial intron inserted after the tenth codon of ADE6 causing cells to be ade6−, but Cre recombinase can remove kanMX and make a functional ADE6-AI2lox gene. Inverted repeat of loxP: The his3-AI2floxINV substrate (Figure 1B, (iv)) was constructed on chromosome XV at the HIS3 locus and surrounding region. This reporter contains the N-terminal portion of HIS3-AI2 and the splice donor part of the artificial intron ending at the loxP site and is marked by hygromycin resistance (hygMX). A lox site, the splice acceptor portion of the intron and the C-terminal portion of the HIS3-AI2, marked by zeomycin resistance (zeoMX), was inserted in inverted orientation 682 bases downstream of the GEP3 gene (2.8 kb beyond HIS3). Cre-directed recombination inverts the floxed DNA (including the DED1 gene) creating the functional HIS3-AI2lox gene (Figure 1B-v). Hygromycin (hygMX) is from pAG32 [70] and Zeocin (zeoMX) is from pHybLex/Zeo (Invitrogen).
In vitro transcription assays
Pol II variants carrying wild type or mutant Rpb1 were introduced into a protease-deficient yeast strain GRY3175 by plasmid shuffle. Hexahistidine-tagged Rpb3 [57] was used to pull down Pol II from the whole cell lysate essentially as described [54]. Specifically, cells from 3–4 ml of stationary (2–3 days) culture were pelleted, washed once with cold water and resuspended in lysis buffer (150 mM Tris–acetate, pH 7.9, 50 mM potassium acetate, 5 mM MgCl2, 10 µM ZnCl2, 2 mM 2-mercaptoethanol, 0.5 mM EDTA and protease inhibitors). The cells were disrupted using a Precellys homogenizer according to manufacturer's instructions, the lysate was removed from the glass beads, and KCl was added to 1M final concentration. The debris was precipitated for 15 min at 14,000 rpm in Eppendorf table-top microcentrifuge at 4°C. The supernatant was added to 50 µl of Ni-NTA agarose pre-washed with transcription buffer (TB) containing 20 mM Tris-HCl, pH 7.9, 5 mM MgCl2, 10 µM ZnCl2, 2 mM 2-mercaptoethanol and 1000 mM KCl (TB1000). Pol II was immobilized on the beads for 30–50 min at 4°C, and the beads were extensively washed with TB1000 and TB40 (TB with 40 mM KCl). The elongation complex was assembled and purified with the immobilized Pol II exactly as described [54] using the following oligonucleotides: Cre RNA C9 : 5′ GUC AUG AAC 3′(phosphorylated with T4 polynucleotide kinase in the presence of γ-P32-labeled ATP); NDS-Cre-Cys (Non Transcribed DNA Strand) 5′GGCTGGACCAATGTAAATATTGTCATGAAC TGTATCCGTAA CCTGGATAGTGAAACA 3′; and TDS-Cre-Cys (Transcribed DNA Strand) which is an exact complement of NDS (Cre-Cys). All NTPs used for walking and misincorporation assays were additionally purified [19]. The elongation complex containing 10-nt RNA (U10) was obtained by 5 min incubation of the assembled and purified complex with 5 µM UTP and washed 4 times with 1 ml of TB40. For the TFIIS sensitivity assay, mismatched A11 elongation complex was obtained by 5 min incubation of U10 with 1 mM ATP, purified by 4 washes with TB40, eluted from the beads with 100 mM imidazole in the presence of 0.2 mg/ml acetylated BSA, diluted 10-fold with TB40 to decrease imidazole concentration and concentrated using Microcon AmiconUltra (Millipore) concentrator to 30 µl. Reactions were stopped by addition of equal volume of gel loading buffer containing 10 M urea and 50 mM EDTA. The reaction products were resolved in 20% polyacrylamide gels (19∶1) in the presence of 1× TBE and 7M urea. To resolve the two 11-nt products of cognate GMP and mismatched AMP incorporation in the competition fidelity assay, the 4-mm thick, 20×40 cm gels were used, and electrophoresis has been performed in 1× TBE at constant power of 65 W until the bromphenol blue tracing dye was within 3 cm of the bottom of the gel. For other experiments the shorter 20×20 cm gels were used and the electrophoresis was performed at 50 W constant power.
Zdroje
1. AndreckaJ, LewisR, BrucknerF, LehmannE, CramerP, et al. (2008) Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc Natl Acad Sci U S A 105 : 135–140.
2. BushnellDA, KornbergRD (2003) Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc Natl Acad Sci U S A 100 : 6969–6973.
3. KaplanCD, LarssonKM, KornbergRD (2008) The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell 30 : 547–556.
4. KashkinaE, AnikinM, BruecknerF, PomerantzRT, McAllisterWT, et al. (2006) Template misalignment in multisubunit RNA polymerases and transcription fidelity. Mol Cell 24 : 257–266.
5. KettenbergerH, ArmacheKJ, CramerP (2004) Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell 16 : 955–965.
6. WangD, BushnellDA, HuangX, WestoverKD, LevittM, et al. (2009) Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science 324 : 1203–1206.
7. WangD, BushnellDA, WestoverKD, KaplanCD, KornbergRD (2006) Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127 : 941–954.
8. BlankA, GallantJA, BurgessRR, LoebLA (1986) An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry 25 : 5920–5928.
9. NinioJ (1991) Connections between translation, transcription and replication error-rates. Biochimie 73 : 1517–1523.
10. ShawRJ, BonawitzND, ReinesD (2002) Use of an in vivo reporter assay to test for transcriptional and translational fidelity in yeast. J Biol Chem 277 : 24420–24426.
11. NesserNK, PetersonDO, HawleyDK (2006) RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proc Natl Acad Sci U S A 103 : 3268–3273.
12. KoyamaH, ItoT, NakanishiT, KawamuraN, SekimizuK (2003) Transcription elongation factor S-II maintains transcriptional fidelity and confers oxidative stress resistance. Genes Cells 8 : 779–788.
13. VassylyevDG, VassylyevaMN, PerederinaA, TahirovTH, ArtsimovitchI (2007) Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448 : 157–162.
14. ZhouYN, LubkowskaL, HuiM, CourtC, ChenS, et al. (2013) Isolation and characterization of RNA polymerase rpoB mutations that alter transcription slippage during elongation in Escherichia coli. J Biol Chem 288 : 2700–2710.
15. StrathernJN, JinDJ, CourtDL, KashlevM (2012) Isolation and characterization of transcription fidelity mutants. Biochim Biophys Acta 1819 : 694–699.
16. StrathernJ, MalagonF, IrvinJ, GotteD, ShaferB, et al. (2013) The fidelity of transcription: RPB1 (RPO21) mutations that increase transcriptional slippage in S. cerevisiae. J Biol Chem 288 : 2689–2699.
17. LibbyRT, NelsonJL, CalvoJM, GallantJA (1989) Transcriptional proofreading in Escherichia coli. EMBO J 8 : 3153–3158.
18. CramerP, BushnellDA, KornbergRD (2001) Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292 : 1863–1876.
19. KireevaML, NedialkovYA, CremonaGH, PurtovYA, LubkowskaL, et al. (2008) Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell 30 : 557–566.
20. KireevaML, OpronK, SeiboldSA, DomecqC, CukierRI, et al. (2012) Molecular dynamics and mutational analysis of the catalytic and translocation cycle of RNA polymerase. BMC Biophys 5 : 11.
21. KoyamaH, ItoT, NakanishiT, SekimizuK (2007) Stimulation of RNA polymerase II transcript cleavage activity contributes to maintain transcriptional fidelity in yeast. Genes Cells 12 : 547–559.
22. KoyamaH, UedaT, ItoT, SekimizuK (2010) Novel RNA polymerase II mutation suppresses transcriptional fidelity and oxidative stress sensitivity in rpb9Delta yeast. Genes Cells 15 : 151–159.
23. WalmacqC, KireevaML, IrvinJ, NedialkovY, LubkowskaL, et al. (2009) Rpb9 subunit controls transcription fidelity by delaying NTP sequestration in RNA polymerase II. J Biol Chem 284 : 19601–19612.
24. KnippaK, PetersonDO (2013) Fidelity of RNA polymerase II transcription: Role of Rbp9 in error detection and proofreading. Biochemistry 52 : 7807–7817.
25. RuanW, LehmannE, ThommM, KostrewaD, CramerP (2011) Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J Biol Chem 286 : 18701–18707.
26. AwreyDE, ShimasakiN, KothC, WeilbaecherR, OlmstedV, et al. (1998) Yeast transcript elongation factor (TFIIS), structure and function. II: RNA polymerase binding, transcript cleavage, and read-through. J Biol Chem 273 : 22595–22605.
27. JeonC, AgarwalK (1996) Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc Natl Acad Sci U S A 93 : 13677–13682.
28. BrabergH, JinH, MoehleEA, ChanYA, WangS, et al. (2013) From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 154 : 775–788.
29. KireevaML, KashlevM, BurtonZF (2013) RNA polymerase structure, function, regulation, dynamics, fidelity, and roles in gene expression. Chem Rev 113 : 8325–8330.
30. KramerEB, VallabhaneniH, MayerLM, FarabaughPJ (2010) A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA 16 : 1797–1808.
31. GuoF, GopaulDN, van DuyneGD (1997) Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 389 : 40–46.
32. HoessRH, ZieseM, SternbergN (1982) P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc Natl Acad Sci U S A 79 : 3398–3402.
33. NagyA (2000) Cre recombinase: the universal reagent for genome tailoring. Genesis 26 : 99–109.
34. WierzbickiA, KendallM, AbremskiK, HoessR (1987) A mutational analysis of the bacteriophage P1 recombinase Cre. J Mol Biol 195 : 785–794.
35. EnnifarE, MeyerJE, BuchholzF, StewartAF, SuckD (2003) Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation. Nucleic Acids Res 31 : 5449–5460.
36. GhoshK, GuoF, Van DuyneGD (2007) Synapsis of loxP sites by Cre recombinase. J Biol Chem 282 : 24004–24016.
37. LeeL, SadowskiPD (2003) Identification of Cre residues involved in synapsis, isomerization, and catalysis. J Biol Chem 278 : 36905–36915.
38. HartungM, Kisters-WoikeB (1998) Cre mutants with altered DNA binding properties. J Biol Chem 273 : 22884–22891.
39. GibbB, GuptaK, GhoshK, SharpR, ChenJ, et al. (2010) Requirements for catalysis in the Cre recombinase active site. Nucleic Acids Res 38 : 5817–5832.
40. ImashimizuM, OshimaT, LubkowskaL, KashlevM (2013) Direct assessment of transcription fidelity by high-resolution RNA sequencing. Nucleic Acids Res 41 : 9090–9104.
41. YoshimatsuT, NagawaF (1989) Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science 244 : 1346–1348.
42. WachA, BrachatA, PohlmannR, PhilippsenP (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 : 1793–1808.
43. ChengTH, ChangCR, JoyP, YablokS, GartenbergMR (2000) Controlling gene expression in yeast by inducible site-specific recombination. Nucleic Acids Res 28: E108.
44. StrathernJN, HerskowitzI (1979) Asymmetry and directionality in production of new cell types during clonal growth: the switching pattern of homothallic yeast. Cell 17 : 371–381.
45. NasmythK (1983) Molecular analysis of a cell lineage. Nature 302 : 670–676.
46. DesmoucellesC, PinsonB, Saint-MarcC, Daignan-FornierB (2002) Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J Biol Chem 277 : 27036–27044.
47. HullMW, McKuneK, WoychikNA (1995) RNA polymerase II subunit RPB9 is required for accurate start site selection. Genes Dev 9 : 481–490.
48. LiS, SmerdonMJ (2002) Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J 21 : 5921–5929.
49. AwreyDE, WeilbaecherRG, HemmingSA, OrlickySM, KaneCM, et al. (1997) Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J Biol Chem 272 : 14747–14754.
50. WeryM, ShematorovaE, Van DriesscheB, VandenhauteJ, ThuriauxP, et al. (2004) Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. EMBO J 23 : 4232–4242.
51. BoekeJD, TrueheartJ, NatsoulisG, FinkGR (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154 : 164–175.
52. WuJ, AwreyDE, EdwardsAM, ArchambaultJ, FriesenJD (1996) In vitro characterization of mutant yeast RNA polymerase II with reduced binding for elongation factor TFIIS. Proc Natl Acad Sci U S A 93 : 11552–11557.
53. FlickJS, JohnstonM (1990) Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol Cell Biol 10 : 4757–4769.
54. KireevaML, LubkowskaL, KomissarovaN, KashlevM (2003) Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae. Methods Enzymol 370 : 138–155.
55. WalmacqC, CheungAC, KireevaML, LubkowskaL, YeC, et al. (2012) Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol Cell 46 : 18–29.
56. BertramJG, OertellK, PetruskaJ, GoodmanMF (2010) DNA polymerase fidelity: comparing direct competition of right and wrong dNTP substrates with steady state and pre-steady state kinetics. Biochemistry 49 : 20–28.
57. KireevaML, KomissarovaN, WaughDS, KashlevM (2000) The 8-nucleotide-long RNA∶DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J Biol Chem 275 : 6530–6536.
58. ImashimizuM, KireevaML, LubkowskaL, GotteD, ParksAR, et al. (2013) Intrinsic translocation barrier as an initial step in pausing by RNA polymerase II. J Mol Biol 425 : 697–712.
59. NedialkovYA, OpronK, AssafF, ArtsimovitchI, KireevaML, et al. (2013) The RNA polymerase bridge helix YFI motif in catalysis, fidelity and translocation. Biochim Biophys Acta 1829 : 187–198.
60. KaplanCD, JinH, ZhangIL, BelyaninA (2012) Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS Genet 8: e1002627.
61. KettenbergerH, ArmacheKJ, CramerP (2003) Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114 : 347–357.
62. LarsonMH, ZhouJ, KaplanCD, PalangatM, KornbergRD, et al. (2012) Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc Natl Acad Sci U S A 109 : 6555–6560.
63. WangB, PredeusAV, BurtonZF, FeigM (2013) Energetic and Structural Details of the Trigger-Loop Closing Transition in RNA Polymerase II. Biophys J 105 : 767–775.
64. BruecknerF, CramerP (2008) Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol 15 : 811–818.
65. Bar-NahumG, EpshteinV, RuckensteinAE, RafikovR, MustaevA, et al. (2005) A ratchet mechanism of transcription elongation and its control. Cell 120 : 183–193.
66. FeigM, BurtonZF (2010) RNA polymerase II with open and closed trigger loops: active site dynamics and nucleic acid translocation. Biophys J 99 : 2577–2586.
67. SomeshBP, ReidJ, LiuWF, SogaardTM, Erdjument-BromageH, et al. (2005) Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121 : 913–923.
68. BrachmannCB, DaviesA, CostGJ, CaputoE, LiJ, et al. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 : 115–132.
69. LewandoskiM, MeyersEN, MartinGR (1997) Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb Symp Quant Biol 62 : 159–168.
70. GoldsteinAL, McCuskerJH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 : 1541–1553.
Štítky
Genetika Reprodukčná medicína
Článek An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of MovementČlánek Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced ActivityČlánek Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene NetworksČlánek Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS LocusČlánek tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa CellsČlánek Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced ApoptosisČlánek The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites inČlánek Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell SenescenceČlánek BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct MechanismsČlánek Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 9- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Translational Regulation of the Post-Translational Circadian Mechanism
- An Evolutionarily Conserved Role for the Aryl Hydrocarbon Receptor in the Regulation of Movement
- Eliminating Both Canonical and Short-Patch Mismatch Repair in Suggests a New Meiotic Recombination Model
- Requirement for Drosophila SNMP1 for Rapid Activation and Termination of Pheromone-Induced Activity
- Co-regulated Transcripts Associated to Cooperating eSNPs Define Bi-fan Motifs in Human Gene Networks
- Targeted H3R26 Deimination Specifically Facilitates Estrogen Receptor Binding by Modifying Nucleosome Structure
- Role for Circadian Clock Genes in Seasonal Timing: Testing the Bünning Hypothesis
- The Tandem Repeats Enabling Reversible Switching between the Two Phases of β-Lactamase Substrate Spectrum
- The Association of the Vanin-1 N131S Variant with Blood Pressure Is Mediated by Endoplasmic Reticulum-Associated Degradation and Loss of Function
- Identification of a Regulatory Variant That Binds FOXA1 and FOXA2 at the Type 2 Diabetes GWAS Locus
- Regulation of Flowering by the Histone Mark Readers MRG1/2 via Interaction with CONSTANS to Modulate Expression
- The Actomyosin Machinery Is Required for Retinal Lumen Formation
- Plays a Conserved Role in Assembly of the Ciliary Motile Apparatus
- Hidden Diversity in Honey Bee Gut Symbionts Detected by Single-Cell Genomics
- Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria
- tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa Cells
- Causal Variation in Yeast Sporulation Tends to Reside in a Pathway Bottleneck
- Tissue-Specific RNA Expression Marks Distant-Acting Developmental Enhancers
- WC-1 Recruits SWI/SNF to Remodel and Initiate a Circadian Cycle
- Clonal Expansion of Early to Mid-Life Mitochondrial DNA Point Mutations Drives Mitochondrial Dysfunction during Human Ageing
- Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels
- Differential Management of the Replication Terminus Regions of the Two Chromosomes during Cell Division
- Obesity-Linked Homologues and Establish Meal Frequency in
- Derlin-1 Regulates Mutant VCP-Linked Pathogenesis and Endoplasmic Reticulum Stress-Induced Apoptosis
- Stress-Induced Nuclear RNA Degradation Pathways Regulate Yeast Bromodomain Factor 2 to Promote Cell Survival
- The MAPK p38c Regulates Oxidative Stress and Lipid Homeostasis in the Intestine
- Widespread Genome Reorganization of an Obligate Virus Mutualist
- Trans-kingdom Cross-Talk: Small RNAs on the Move
- The Vip1 Inositol Polyphosphate Kinase Family Regulates Polarized Growth and Modulates the Microtubule Cytoskeleton in Fungi
- Myosin Vb Mediated Plasma Membrane Homeostasis Regulates Peridermal Cell Size and Maintains Tissue Homeostasis in the Zebrafish Epidermis
- GLD-4-Mediated Translational Activation Regulates the Size of the Proliferative Germ Cell Pool in the Adult Germ Line
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Translational Regulation of the DOUBLETIME/CKIδ/ε Kinase by LARK Contributes to Circadian Period Modulation
- Positive Selection and Multiple Losses of the LINE-1-Derived Gene in Mammals Suggest a Dual Role in Genome Defense and Pluripotency
- Out of Balance: R-loops in Human Disease
- A Genetic Assay for Transcription Errors Reveals Multilayer Control of RNA Polymerase II Fidelity
- Altered Behavioral Performance and Live Imaging of Circuit-Specific Neural Deficiencies in a Zebrafish Model for Psychomotor Retardation
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Meta-analysis of Mutations in Autism Spectrum Disorders: A Gradient of Severity in Cognitive Impairments
- The Proprotein Convertase KPC-1/Furin Controls Branching and Self-avoidance of Sensory Dendrites in
- Hydroxymethylated Cytosines Are Associated with Elevated C to G Transversion Rates
- Memory and Fitness Optimization of Bacteria under Fluctuating Environments
- Regulation of p53 and Rb Links the Alternative NF-κB Pathway to EZH2 Expression and Cell Senescence
- Interspecific Tests of Allelism Reveal the Evolutionary Timing and Pattern of Accumulation of Reproductive Isolation Mutations
- PRO40 Is a Scaffold Protein of the Cell Wall Integrity Pathway, Linking the MAP Kinase Module to the Upstream Activator Protein Kinase C
- Low Levels of p53 Protein and Chromatin Silencing of p53 Target Genes Repress Apoptosis in Endocycling Cells
- SPDEF Inhibits Prostate Carcinogenesis by Disrupting a Positive Feedback Loop in Regulation of the Foxm1 Oncogene
- RRP6L1 and RRP6L2 Function in Silencing Regulation of Antisense RNA Synthesis
- BMPs Regulate Gene Expression in the Dorsal Neuroectoderm of and Vertebrates by Distinct Mechanisms
- Unkempt Is Negatively Regulated by mTOR and Uncouples Neuronal Differentiation from Growth Control
- Atkinesin-13A Modulates Cell-Wall Synthesis and Cell Expansion in via the THESEUS1 Pathway
- Dopamine Signaling Leads to Loss of Polycomb Repression and Aberrant Gene Activation in Experimental Parkinsonism
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
- Bipartite Recognition of DNA by TCF/Pangolin Is Remarkably Flexible and Contributes to Transcriptional Responsiveness and Tissue Specificity of Wingless Signaling
- The Olfactory Transcriptomes of Mice
- Muscular Dystrophy-Associated and Variants Disrupt Nuclear-Cytoskeletal Connections and Myonuclear Organization
- Interplay of dFOXO and Two ETS-Family Transcription Factors Determines Lifespan in
- Evidence for Widespread Positive and Negative Selection in Coding and Conserved Noncoding Regions of
- Genome-Wide Association Meta-analysis of Neuropathologic Features of Alzheimer's Disease and Related Dementias
- Rejuvenation of Meiotic Cohesion in Oocytes during Prophase I Is Required for Chiasma Maintenance and Accurate Chromosome Segregation
- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Local Effect of Enhancer of Zeste-Like Reveals Cooperation of Epigenetic and -Acting Determinants for Zygotic Genome Rearrangements
- Differential Responses to Wnt and PCP Disruption Predict Expression and Developmental Function of Conserved and Novel Genes in a Cnidarian
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals
- Nipbl and Mediator Cooperatively Regulate Gene Expression to Control Limb Development
- Genome Wide Association Studies Using a New Nonparametric Model Reveal the Genetic Architecture of 17 Agronomic Traits in an Enlarged Maize Association Panel
- Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



