-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
Secretory pathways play an important role in the plant immune response by delivering antimicrobial compounds and metabolites to the site of infection. The evolutionarily conserved exocyst complex is involved in exocytosis, the final step in the secretory pathway. We showed that loss of the function of EXO70B1, a subunit of exocyst complex, results in activated defense responses, and enhanced resistance to a range of pathogens. We found that EXO70B1 associates with the SNARE complex protein SNAP33, which is involved in focal secretion of defense-related proteins. Enhanced disease resistance and cell death in the exo70B1 mutant are dependent on TIR-NBS2 (TN2), an atypical intracellular immune receptor-like protein that lacks leucine-rich repeats. TN2 physically associates with EXO70B1, and TN2 transcripts accumulate at much higher levels in the exo70B1 mutant. These data are consistent with a model where activation of a receptor pathway containing TIR-NBS2 is responsible for activated defense responses and cell death in exo70B1. Our data further suggest that this, and possibly other, exocyst components could be targets of effectors that are guarded by immune receptors.
Published in the journal: A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant. PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004945
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004945Summary
Secretory pathways play an important role in the plant immune response by delivering antimicrobial compounds and metabolites to the site of infection. The evolutionarily conserved exocyst complex is involved in exocytosis, the final step in the secretory pathway. We showed that loss of the function of EXO70B1, a subunit of exocyst complex, results in activated defense responses, and enhanced resistance to a range of pathogens. We found that EXO70B1 associates with the SNARE complex protein SNAP33, which is involved in focal secretion of defense-related proteins. Enhanced disease resistance and cell death in the exo70B1 mutant are dependent on TIR-NBS2 (TN2), an atypical intracellular immune receptor-like protein that lacks leucine-rich repeats. TN2 physically associates with EXO70B1, and TN2 transcripts accumulate at much higher levels in the exo70B1 mutant. These data are consistent with a model where activation of a receptor pathway containing TIR-NBS2 is responsible for activated defense responses and cell death in exo70B1. Our data further suggest that this, and possibly other, exocyst components could be targets of effectors that are guarded by immune receptors.
Introduction
Powdery mildew fungi are obligate biotrophic pathogens that cause widespread disease in many plant species, including economically important crops such as barley [1], wheat [2] and tomato [3] as well as model plants such as Arabidopsis [4]. Powdery mildew fungal spores germinate on the leaf surface, and then extend hyphae, which form appressoria at the attempted penetration sites. These appressoria penetrate the epidermal cell wall. The fungus then invaginates the plant plasma membrane and develops a haustorium, a feeding structure [5]. Evolutionarily adapted powdery mildew fungal pathogens can complete the infection cycle by producing conidia, asexual spores, on the host leaf surfaces. For instance, the adapted powdery mildew pathogen for Arabidopsis, Golovinomyces cichoracearum, produces abundant conidia on Col-0 leaves after infection. Several types of barley and Arabidopsis mutants show altered responses to adapted powdery mildew pathogens; these include mlo (mildew locus O) [1,6], pmr (powdery mildew resistant) [6–10] and edr (enhanced disease resistance) [11–14]. These mutants generally are more resistant to adapted powdery mildew pathogens, indicating that MLO, PMR, and EDR have negative roles in powdery mildew resistance.
Adapted powdery mildew fungi can infect the host plant; by contrast, non-adapted powdery mildew pathogens usually cannot penetrate plant epidermal cell walls or form haustoria. Genetic analyses demonstrated that the full penetration resistance of Arabidopsis to non-adapted powdery mildew requires the functions of PEN1, PEN2 and PEN3 [15–17]. PEN1 encodes syntaxin SYP121, which participates in vesicle fusion events by forming ternary soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) complexes with the synaptosome-associated membrane protein 33 (SNAP33) and vesicle-associated membrane protein721 (VAMP721) and VAMP722 [15,18].
Before fusion occurs, SNARE-mediated vesicles must dock at the plasma membrane; this requires the exocyst, which mediates the early steps of exocytosis [19]. The exocyst complex, originally defined in yeast [20], consists of 8 proteins: Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, EXO70, and EXO84. Among these eight proteins, Sec3 and EXO70 are targeted to the plasma membrane by binding to phosphatidyl (4,5) biphosphate [21,22].
Homologs of all eight exocyst proteins have been found in plants [23–25]. Plant exocyst proteins function in regulation of polarity and morphogenesis [26]. In Arabidopsis, Sec6, Sec8, and EXO70A1 localize to the tips of growing pollen tubes and function in polar secretion required for growth and pollen incompatibility [25,27]. Sec3 and EXO70A1 function in polarized secretion in elongating root hairs [28] and in the interaction between the stigma and compatible pollen [29]. Sec6, Sec8, Sec15b, EXO70A1, and EXO84b also function in secretory processes during cytokinesis, and Sec8 and EXO70A1 are required for the localized deposition of seed coat pectin [30,31]. In addition, EXO70A1 also affects auxin efflux carrier recycling, polar auxin transport [32] and tracheary element development [33].
EXO70 clade members also function in plant immunity [26]. For instance, Arabidopsis EXO70B2 and EXO70H1 are transcriptionally induced by Pseudomonas syringae pv. maculicola and the fungal pathogen Bgh and are required for full plant defense responses. Further, EXO70B2 associates with SNAP33, and contributes to plant immunity [34]. EXO70B2 is a target of the plant U-box-type ubiquitin ligase 22 (PUB22) and is required for PAMP-triggered responses [34,35]. EXO70B1 also affects autophagy-related membrane traffic to the vacuole, and the exo70B1 mutant displays ectopic hypersensitive reaction [36].
Plant disease resistance (R) proteins play critical roles in plant immunity [37]. The R proteins directly or indirectly recognizes pathogen effectors, and leads to effector triggered immunity. Most of the R proteins are intracellular, and belongs to nucleotide binding (NB) domain and leucine-rich repeat (LRR)-containing (NLR) immune receptors, which can be divided to two subclasses, based on the N-terminal domains. The first class of NLR immune receptors contains a Toll-Interleukin-1 Receptor (TIR) domain and the second class contains a coiled-coil (CC) domain [37]. Activation of NLR leads to hypersensitive responses which is characterized by localized cell death at site of infection [37]. Besides those full length NLR immune receptors, there are a number of atypical NLRs that lack LRR domain in plant genome [38]. Although those truncated NLR proteins are implicated in plant immunity, the function of those proteins are not well understood [39].
Here, we report that the exo70B1 mutant displays enhanced resistance to powdery mildew. During the course of this work, it was shown that exo70B1 displayed spontaneous cell death, which was delayed by npr1 mutation suggesting that at least part of the mutant phenotype is due to constitutive SA signaling [36]. Consistent with this, exo70B1 exhibits up-regulation of PR1, and accumulates higher levels of SA in the absence of pathogen attack [36]. Here, we show that EXO70B1 associates with the SNARE complex protein SNAP33. In a suppressor screen, we found that the exo70B1-associated phenotypes require the TIR-NBS gene TN2. These data indicate that mutation of EXO70B1 leads to activation of a defense pathway that requires TN2, a truncated form of the classical TIR-NBS-LRR intracellular immune system receptors.
Results
The exo70B1-3 mutant displays enhanced disease resistance to Golovinomyces cichoracearum
To study the molecular interactions between Arabidopsis and powdery mildew fungus, we screened for mutants that displayed enhanced disease resistance and an exaggerated cell death response following infection with G. cichoracearum UCSC1 [14]. We designated one of the recessive mutants identified in this screen exo70B1-3, based on our subsequent characterization. Under standard short day conditions, exo70B1-3 grew normally up to 4 weeks of age. After 4 weeks, exo70B1-3 plants were slightly smaller than the wild type, and displayed HR-like cell death after 5 weeks (Fig. 1A and S1 Fig.). After inoculation with G. cichoracearum at high inoculum density, the leaf surface of the wild type was covered with abundant fungal spores at 7 dpi, but exo70B1-3 exhibited extensive necrotic lesions and no visible powder of fungal spores on leaves (Fig. 1A). Trypan blue staining revealed abundant fungal hyphae and conidiophores on the leaves of the wild type, but showed very few fungal hyphae and large patches of dead cells on the leaves of exo70B1-3 (Fig. 1B). To further assess the powdery mildew resistance of exo70B1-3, we quantified fungal growth in plants at 5 dpi. Consistent with the trypan blue staining, exo70B1-3 supported significantly fewer conidiophores than the wild type (Fig. 1C), indicating that fungal reproduction was inhibited in the exo70B1-3 mutant.
Fig. 1. exo70B1-3 mutants display enhanced resistance to G. cichoracearum. 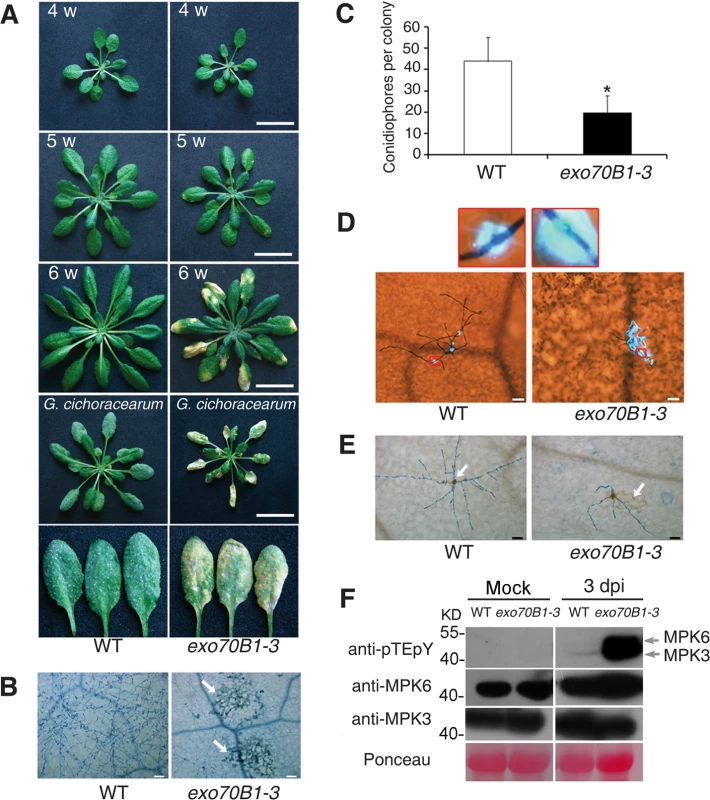
(A) Plants were grown in the standard short day conditions. Upper three panels: Uninfected four-, five- and six-week-old wild type and exo70B1-3 plants were photographed. exo70B1-3 mutants started to develop spontaneous cell death at five weeks of age, and cell death was more pronounced at six weeks of age. Lower two panels: Four-week-old plants were infected with G. cichoracearum and the plants or representative leaves were photographed at 7 dpi. The wild type plants displayed a large number of fungal spores on the leaves, but the exo70B1-3 mutants displayed very few spores, with massive necrotic lesions on the leaves. Five-week-old wild type and exo70B1-3 plants were the uninoculated controls for plants infected with G. cichoracearum. Bar = 20 mm. Plant responses to pathogen infection include: callose deposition, hydrogen peroxide (H2O2) accumulation, and MAPK activation [40]. We used aniline blue to visualize callose deposition in the infected leaves of exo70B1-3 at 2 dpi. As shown in Fig. 1D, very little callose was deposited at the infection sites of pathogen in the wild type, but a large amount of callose was produced at the sites of fungal infection in exo70B1-3 at 2 dpi, when cell death did not occur. In plants infected with pathogens, cell death triggered by specific recognition is usually associated with H2O2 accumulation [41,42]. We examined H2O2 accumulation in wild type and exo70B1-3 by staining the infected leaves with 3,3’-diamino benzidine hydrochloride (DAB) and trypan blue and found that exo70B1-3 leaves accumulated more H2O2 upon fungal attack than wild type (Fig. 1E).
We also examined the activation of MPKs upon infection with G. cichoracearum, by immunoblotting with anti-pTEpY antibody, which detects phosphorylated MPK3 and MPK6 [43]. As shown in Fig. 1F, MPK3 and MPK6 were activated in both wild type and exo70B1-3 at 3 dpi; however, activation of MPK3, MPK6 in exo70B1-3 was much stronger than in wild type. Protein levels of MPK3 and MPK6 were similar in infected and uninfected wild type and mutant plants (Fig. 1F).
To examine whether exo70B1-3 affects the expression of immune output genes, we measured the transcript levels of the pathogenesis-related gene PR1, a marker for salicylic acid signaling, at various time points from 0 to 5 dpi with G. cichoracearum, using quantitative real time RT-PCR. Prior to inoculation, PR1 transcript levels were very low in both genotypes, but the basal expression levels of PR1 were higher in exo70B1-3 than in wild type. Upon infection with G. cichoracearum, PR1 transcript accumulated at much higher levels in exo70B1-3 than in wild type (Fig. 2A). We also examined expression of other immune output genes, including PR2 (Fig. 2B), PR5 (Fig. 2C), ALD1, FMO1, PAD4 and SID2 (Fig. 2D). In general, the relative transcript levels of those genes were low at day 0, but were much higher in exo70B1-3 than in wild type at day 0 and 3 dpi. However, for PAD4, SID2 and NPR1, we observed significant differences in transcript levels between exo70B1-3 and wild type only at day 3, not at day 0 (Fig. 2D). Taken together, these data indicated that exo70B1-3 displayed enhanced disease resistance, and upon infection, defense responses were activated at higher levels in exo70B1-3 than in wild type.
Fig. 2. Expression of defense-related genes in wild type and exo70B1-3 mutants. 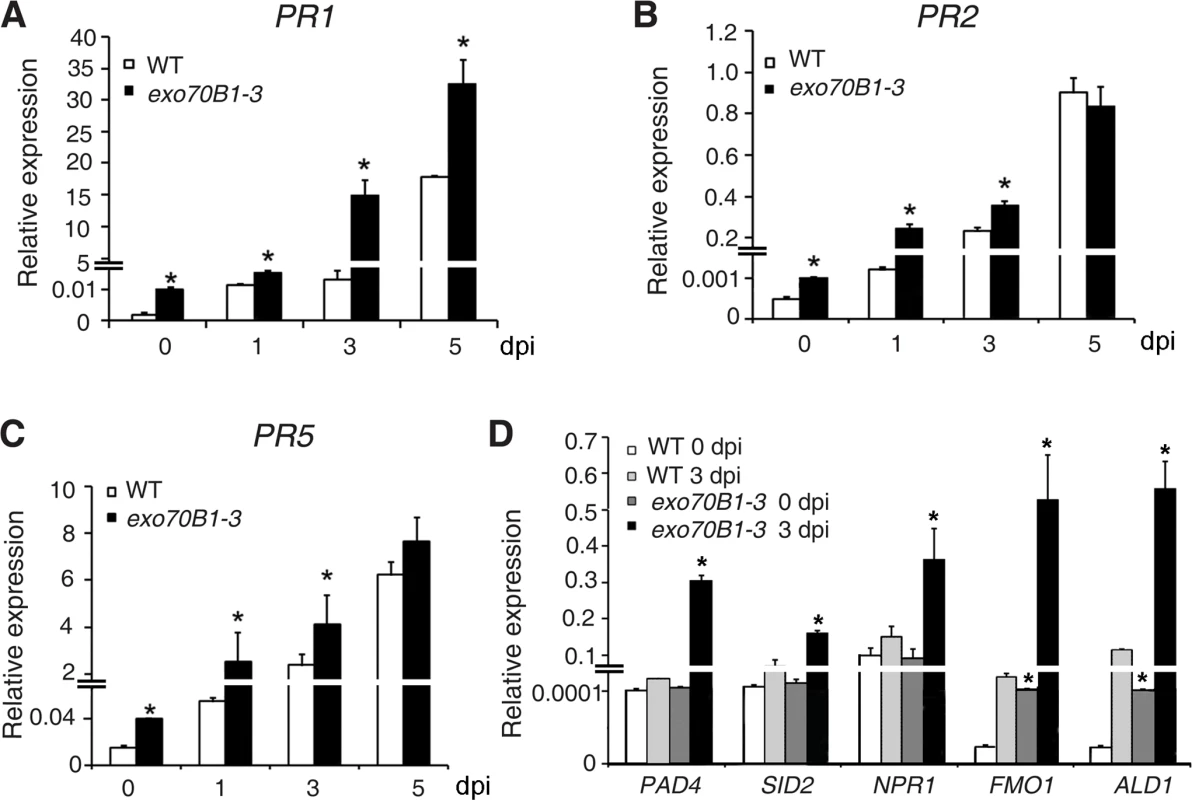
(A)-(D) The transcript accumulation of defense-related genes was examined by quantitative real-time RT-PCR. Leaves were detached from four-week-old plants for RNA isolation at different time points (day 0: uninfected) after infection with G. cichoracearum. PR1 (A), PR2 (B), PR5 (C), PAD4, SID2, NPR1, FMO1, ALD1 (D). ACT2 was used as an internal control. Bars represent mean and standard deviation of values obtained from three independent biological samples. Three technical replicates for each biological sample were examined in the experiment. The asterisks indicate a significant difference from WT (p < 0.01; Student’s t-test). The experiments were repeated three times with similar results. PAD4, EDS5 and NPR1 contribute to exo70B1-3 mediated powdery mildew resistance, but only PAD4 is required for ectopic cell death
Salicylic acid (SA) plays critical roles in the defense response to pathogens and activation of SA signaling pathways often leads to disease resistance and cell death in response to pathogen attack [44]. To assess the role of SA signaling in the disease resistance observed in exo70B1-3, we crossed exo70B1-3 with pad4, sid2 and eds5 mutants, which express lower levels of pathogen-induced SA [45–47] or npr1, which has defects in SA signaling [48]. Double mutants were then infected with G. cichoracearum and phenotypes were scored at 7 dpi. Only pad4 completely suppressed both powdery mildew resistance and mildew-induced cell death in exo70B1-3 (Fig. 3A-C). The eds5 and npr1 mutations suppressed powdery mildew resistance, but did not fully suppress cell death in exo70B1-3. The sid2 mutation affected neither resistance nor cell death in exo70B1-3 (Fig. 3A-C). Similarly, spontaneous cell death in exo70B1-3 was only suppressed by pad4, but not by sid2, npr1 or eds5 (S2 Fig.). Taken together, these observations indicate that PAD4, EDS5 and NPR1 contribute to exo70B1-3 mediated powdery mildew resistance. Those observations also suggest that the cell death and ectopic disease resistance in exo70B1-3 can be uncoupled.
Fig. 3. The exo70B1-3 mutants accumulate high levels of SA and their enhanced resistance to G. cichoracearum requires EDS5, NPR1 and PAD4. 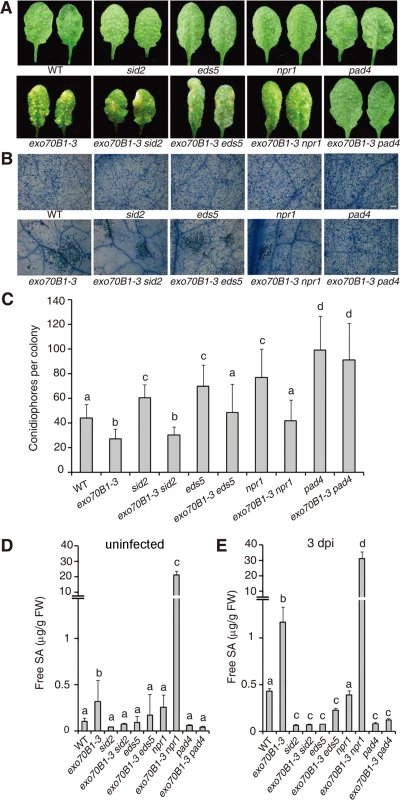
(A) Four-week-old plants were infected with G. cichoracearum at high inoculum densities, and representative leaves removed from plants at 7 dpi with G. cichoracearum. (B) Leaves removed from plants infected with G. cichoracearum at high inoculum densities at 7 dpi were stained with trypan blue to show hyphae and dead cells. Bar = 100 μm. (C) Fungal growth in plants infected with G. cichoracearum at low inoculum densities at 5 dpi was assessed by counting the number of conidiophores per colony. Lower-case letters indicate statistically significant differences (p < 0.05; one-way ANOVA). (D)-(E) exo70B1-3 mutants accumulate high levels of free SA. SA was extracted from leaves of uninfected four-week-old plants (D), or leaves of four-week-old plants infected with G. cichoracearum at high inoculum densities at 3 dpi (E). Bars represent mean and standard deviation from three biological replicates for each genotype. Lower-case letters indicate statistically significant differences (p < 0.05; one-way ANOVA). These experiments were repeated three times with similar results. To further assess the role of SA in exo70B1-3 mediated resistance, we measured SA levels in exo70B1-3. Free SA levels were significantly higher in exo70B1-3 plants than in wild type in the absence of pathogen or at 3 dpi with G. cichoracearum (Fig. 3D, 3E). The SA levels in exo70B1-3 were reduced to wild type levels (or even lower) in combination with pad4, eds5 and sid2. However, the SA levels in exo70B1-3 npr1 were much higher than in exo70B1-3, which was consistent with previous findings that npr1 accumulates higher levels of SA, and that npr1 mutation often further increases SA levels in mutants that over-accumulate SA [49–51]. The observation that exo70B1-3 sid2 mutants accumulated low levels of SA, but still displayed enhanced disease resistance and mildew-induced cell death, indicates that the disease resistance and cell death phenotypes in response to G. cichoracearum in exo70B1-3, similar to the lsd1-mediated immune responses, do not result from high accumulation of SA [52].
We examined the role of ethylene and JA in exo70B1-3 mediated defenses by examining the effect of mutations in EIN2 and COI1, which block all known ethylene and JA responses, respectively [53,54]. The exo70B1-3 coi1 and exo70B1-3 ein2 double mutants did not show any alteration in disease resistance or cell death in response to G. cichoracearum infection compared to exo70B1-3 (S3 Fig.), indicating that the ethylene and JA pathways do not affect cell death and disease resistance observed in exo70B1-3.
We also crossed exo70B1-3 with two well-characterized powdery mildew resistant mutants, edr1 and pmr4, and then infected the double mutants with G. cichoracearum. As shown in S4 Fig., the edr1 and pmr4 mutations enhanced exo70B1-3 resistance phenotypes, indicating that the disease resistance in exo70B1-3 may differ from the previously characterized powdery mildew resistance mediated by edr1 and pmr4.
exo70B1-3 displayed enhanced disease resistance to the oomycete pathogen H. arabidopsidis Noco2 and the bacterial pathogen Pseudomonas syringae pv. tomato strain DC3000
To examine whether exo70B1-3 causes resistance to other pathogens, we challenged exo70B1-3 with the virulent oomycete pathogen Hyaloperonospora arabidopsidis (H. a.) Noco2 and the bacterial pathogen Pseudomonas syringae pv. tomato (Pto) DC3000. Two-week old seedlings were infected with H.a. Noco2, and we found that exo70B1-3 supported significantly fewer spores than the wild type at 7 dpi (Fig. 4A). Similarly, exo70B1-3 were more resistant to the virulent strain Pto DC3000 and the avirulent bacterial strain Pto DC3000 avrRpt2 (Fig. 4B, 4C).
Fig. 4. exo70B1-3 mutants display enhanced resistance to H.a. Noco2, Pto DC3000 and Pto DC3000 avrRpt2. 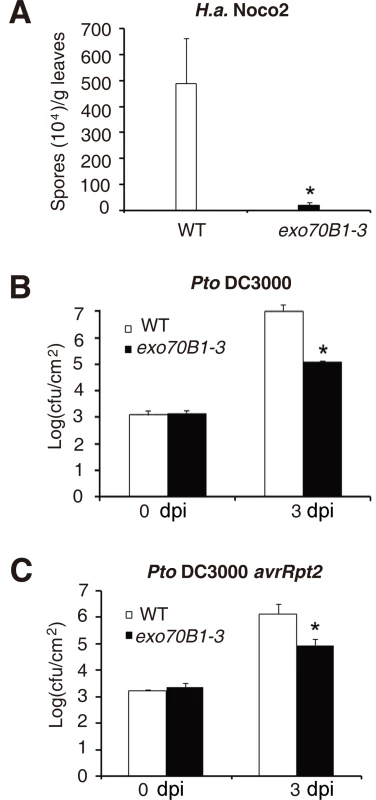
(A) Two-week-old seedlings were inoculated with H.a. Noco2. The number of spores was counted at 7 dpi. The asterisk indicates a significant difference from WT (p < 0.01; Student’s t-test). (B)-(C) Four-week-old plants were infiltrated with Pto DC3000 (B) and Pto DC3000 avrRpt2 (C). Bacterial growth was monitored at 0 and 3 dpi. cfu: colony-forming units. The asterisk indicates a significant difference from wild type (p < 0.01; Student’s t-test). Bars represent mean and standard deviation of values from three biological samples. The experiments were repeated three times with similar results. EXO70B1 is an exocyst subunit
We identified the exo70B1-3 mutation by map-based cloning (S5A Fig.). To confirm that the EXO70B1 we defined is the gene responsible for the exo70B1-3 phenotype, we examined the phenotypes of two additional EXO70B1 T-DNA insertion lines N328818 (exo70B1-1) and N717829 (exo70B1-2) (S5A-S5B Fig.), and noted that these two mutants also displayed enhanced disease resistance and lesions upon inoculation with G. cichoracearum, a phenotype similar to that of exo70B1-3 (S5C Fig.). As an additional confirmation, we introduced an EXO70B1 genomic clone into exo70B1-3 plants, and found that the EXO70B1 genomic clone complemented the exo70B1-3 phenotypes (S5C Fig.). We also examined PR1 expression in the three exo70B1 alleles, the exo70B1-3 gEXO70B1 transgenic line and wild type upon mildew infection. We found that accumulation of the PR1 transcripts in the exo70B1-3 transgenic lines was similar to that in the wild type, which was much lower compared to the three exo70B1 alleles (S5D Fig.). Taken together, these data demonstrated that loss of EXO70B1 is responsible for the enhanced resistance to powdery mildew in exo70B1-3 mutants.
The 23 annotated Arabidopsis EXO70 genes fall into nine clusters, and EXO70B1 falls into the EXO70B cluster, which is very conserved in land plants [55]. Among the 23 EXO70 family members in Arabidopsis, EXO70B2 shares the highest sequence similarity with EXO70B1. However, exo70B2-1 was as susceptible as wild type in response to G. cichoracearum, supporting abundant fungal growth on the leaf surface and developing no visible lesions at 7 dpi. The exo70B1-3 exo70B2-1 double mutant was similar to exo70B1-3 (S6A-S6C Fig.).
As EXO70B1 is predicted to be a subunit of the exocyst complex, we performed yeast two-hybrid assays to examine whether EXO70B1 interacts with other exocyst subunits. Although EXO70B1 seems to be weakly auto-activating in our experimental conditions, the yeast two-hybrid assays showed that EXO70B1 interacts with SEC3a, SEC5a, SEC15b and EXO84bN (S7 Fig.). However, Kulich et al. previously showed that EXO70B1 interacted with only SEC5a and EXO84bN, but not SEC3a. [36]. The discrepancy may be because the interaction between EXO70B1 and SEC3a is relatively weak.
To further examine EXO70B1 expression, we expressed a GUS reporter gene in wild type under the control of the EXO70B1 promoter. We obtained a number of EXO70B1::GUS transformants and analyzed 20 transgenic lines. We observed GUS activity in seedlings, leaves, calyx, stigma and siliques, demonstrating that EXO70B1 is ubiquitously expressed (S8 Fig.).
EXO70B1 interacts with SNAP33
The ternary SNARE complex PEN1-SNAP33-VAMP721/VAMP722 affects powdery mildew resistance [56]. To examine whether EXO70B1 functions in the PEN1-SNAP33-VAMP721/VAMP722 pathway, we first used yeast two-hybrid assays to test whether EXO70B1 interacts with the PEN1, SNAP33 and VAMP721. As shown in Fig. 5A, EXO70B1 interacted with SNAP33, but not PEN1 or PEN1 without its transmembrane domain. To confirm the interaction between EXO70B1 and SNAP33, we used bimolecular fluorescence complementation (BiFC) in Nicotiana benthamiana [57] to examine whether EXO70B1 and SNAP33 can interact. EXO70B1 and SNAP33 were fused to the N-terminal and C-terminal fragments of YFP, respectively. In N. benthamiana leaves co-transformed with the EXO70B1-YFPN and SNAP33-YFPC or PEN1-YFPN and SNAP33-YFPC constructs, we observed YFP fluorescence (Fig. 5B and S9 Fig.). However, we observed no YFP fluorescence in leaves co-transformed with EXO70B1-YFPN and VAMP721-YFPC or PEN1-YFPC, suggesting that EXO70B1 interacts with SNAP33 but not PEN1 in N. benthamiana. These results were consistent with the observations from the yeast two-hybrid assays.
Fig. 5. EXO70B1 associates with SNAP33. 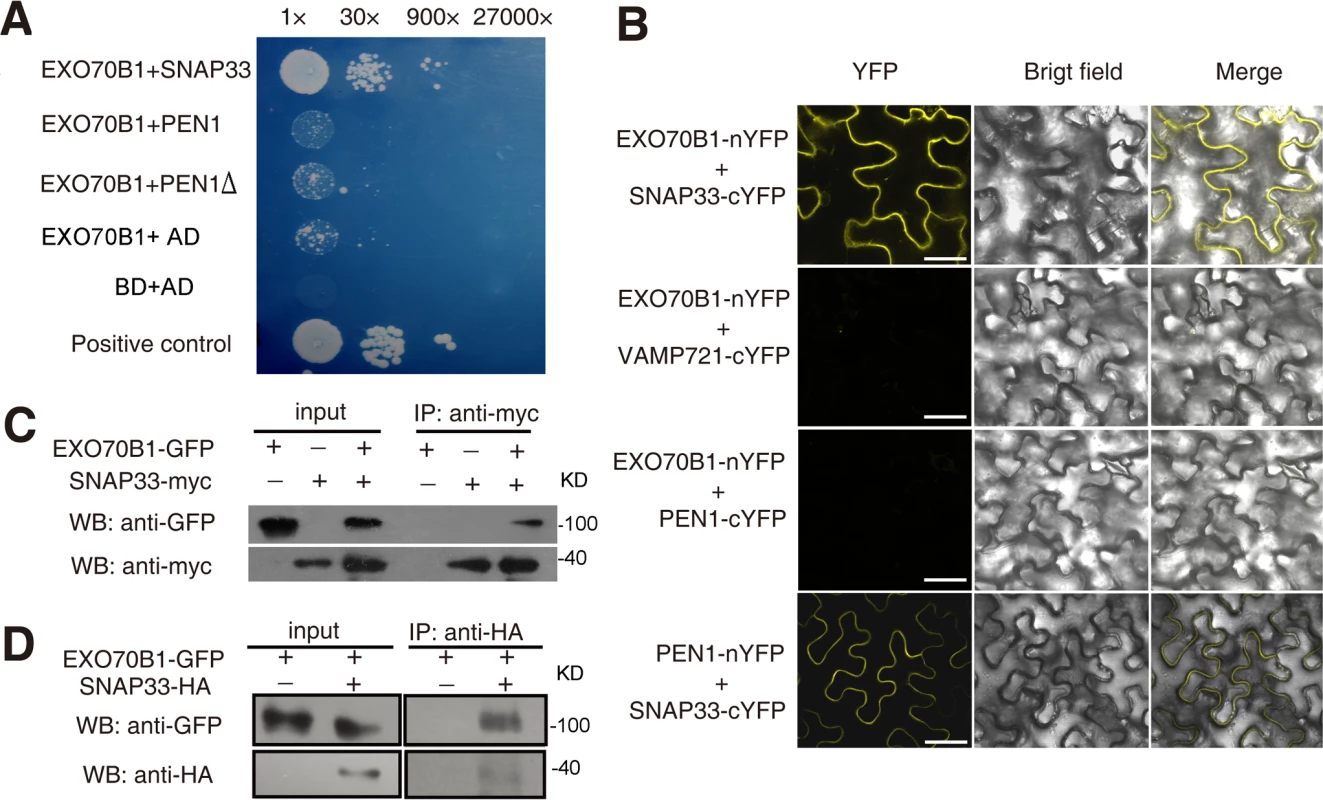
(A) EXO70B1 interacted with SNAP33 in yeast two-hybrid assays. Overnight culture from a single colony was diluted with sterile water to OD = 0.5, and serial dilutions 1:30, 1:900, 1:27,000 were prepared and dropped onto SD-Ade-His-Leu-Trp plates at 28°C, respectively. Each drop was 10 μL. The photograph was taken at day 5 after plating. PEN1Δ: PEN1 without transmembrane domain. (B) EXO70B1 and SNAP33 interaction was examined with BiFC assays in N. benthamiana. EXO70B1 was fused to the N-terminal fragment of YFP (nYFP); SNAP33, PEN1 and VAMP721 were fused to the C-terminal fragment of YFP (cYFP). Cauliflower mosaic virus 35S promoter (35Spro) was used in this assay. YFP fluorescence was observed only with the transiently expressed EXO70B1-nYFP and SNAP33-cYFP, but not EXO70B1-nYFP and VAMP721-cYFP nor EXO70B1-nYFP and PEN1-cYFP in N. benthamiana. The combination of PEN1-nYFP and SNAP33-cYFP was used as the positive control. Bar = 50 μm. (C) EXO70B1 interacted with SNAP33 in a Co-IP assay in N. benthamiana. EXO70B1pro:EXO70B1-GFP was co-expressed with 35Spro:SNAP33-Myc in N. benthamiana leaves. Total protein was extracted and subjected to immunoprecipitation of SNAP33 protein by anti-Myc antibody. Proteins were analyzed in an immunoblot using antibodies as indicated. (D) Co-IP of EXO70B1 and SNAP33 using transgenic Arabidopsis plants. The exo70B1-3 transgenic plants expressing EXO70B1pro:EXO70B1-GFP and SNAP33pro:SNAP33-HA were used in the Co-IP assay. Total protein was extracted from 3-week-old transgenic plants that express both EXO70B1-GFP and SNAP33-HA, or EXO70B1-GFP alone (negative control). The SNAP33 protein was immunoprecipitated by anti-HA antibody, followed by immunoblot analysis with the GFP antibody to detect the presence of EXO70B1-GFP in the precipitate. To confirm the interaction of EXO70B1 and SNAP33, we also performed co-immunoprecipitation (Co-IP) in N. benthamiana. We co-expressed EXO70B1-GFP with SNAP33-Myc in N. benthamiana leaves and expressed EXO70B1-GFP or SNAP33-Myc alone as negative controls. SNAP33 protein was immunoprecipitated by anti-Myc antibody, and anti-GFP antibody detected EXO70B1-GFP in the precipitate only from the leaves that expressed both EXO70B1-GFP and SNAP33-Myc, not from the negative controls (Fig. 5C), indicating that EXO70B1 and SNAP33 associate in N. benthamiana. We also examined whether EXO70B1 interacts with PEN1 by Co-IP assays; however, as shown in S10 Fig., EXO70B1 did not co-immunoprecipitate with PEN1, indicating that EXO70B1 and PEN1 did not associate, consistent with the results from the yeast two-hybrid and BiFC assays.
To further confirm the association of EXO70B1 and SNAP33, we performed Co-IP assays using stably transformed transgenic Arabidopsis plants expressing both EXO70B1-GFP and SNAP33-HA under their respective native promoters. The EXO70B1-GFP construct complemented the exo70B1-3 mutant phenotypes (S11 Fig.), indicating that the EXO70B1-GFP protein is functional. Total protein was extracted from the transgenic plants and SNAP33-HA protein was immunoprecipitated by anti-HA antibody. We then examined whether EXO70B1-GFP was in the precipitate by immunoblot analysis with the GFP antibody. EXO70B1-GFP protein was only detected in the transgenic plants that expressed both EXO70B1-GFP and SNAP33-HA, not in the negative control (Fig. 5D), indicating that EXO70B1-GFP associates with SNAP33-HA in Arabidopsis.
EXO70B1 interacts with SNAP33, a component of the PEN1-containing ternary SNARE complex involved in penetration resistance. To examine whether the exo70B1 mutant phenotype is altered in the absence of PEN1, we made pen1-1 exo70B1-3 double mutants, and then infected wild type, exo70B1-3, pen1-1, and pen1-1 exo70B1-3 with adapted and non-adapted powdery mildew pathogens. Both exo70B1-3 and pen1-1 displayed enhanced resistance to G. cichoracearum, and pen1-1 exo70B1-3 was very similar to pen1-1 or exo70B1-3 (Fig. 6A-C). However, exo70B1-3 mutants were similar to wild type when infected with non-adapted wheat powdery mildew pathogen Blumeria graminis f. sp. tritici. But pen1-1 showed a significantly higher frequency of epidermal single cell death, and exo70B1-3 pen1-1 was very similar to pen1-1 (Fig. 6D), suggesting that the loss of EXO70B1 may not affect PEN1-mediated resistance to a non-adapted pathogen, which is consistent with the observations that EXO70B1 does not interact with PEN1.
Fig. 6. Defense responses of wild type, exo70B1-3, pen1-1 and pen1-1 exo70B1-3 plants to adapted and non-adapted powdery mildew pathogens. 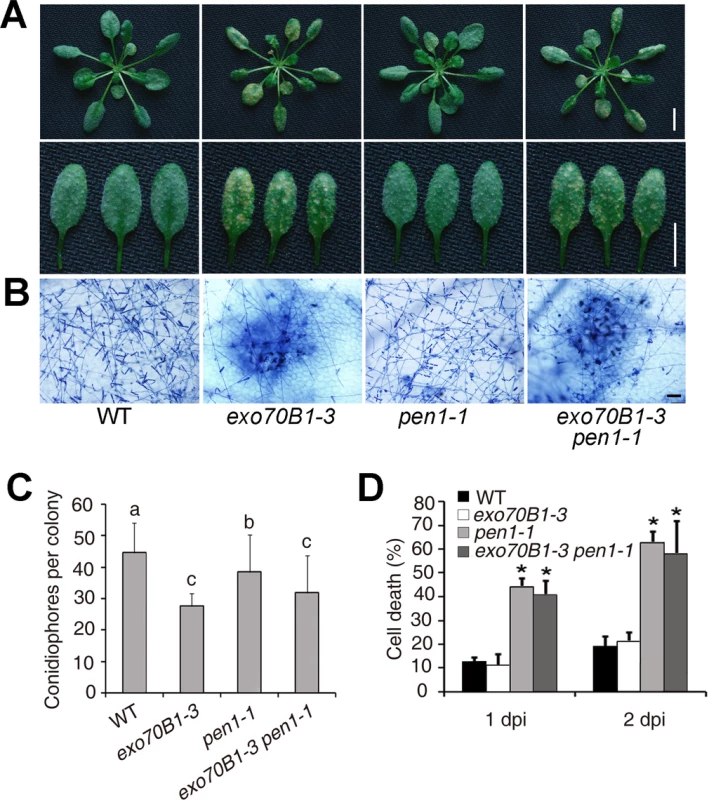
(A)-(C) Four-week-old plants were infected with G. cichoracearum. The pen1-1 mutation did not affect powdery mildew resistance in exo70B1-3. (A) Leaves at 7 dpi were removed and photographed. Bar = 10 mm. (B) Plant cell death and fungal growth were examined with trypan blue staining. Bar = 50 μm. (C) Fungal growth in plants was quantified by counting the number of conidiophores per colony at 5 dpi. Bars represent mean and sd (n = 30). Statistically significant difference from wild type is indicated by lower case letters (p < 0.05; one-way ANOVA). The experiments were repeated three times with similar results. (D) Four-week-old plants were infected with Blumeria graminis f. sp. tritici. Frequency of epidermal single cell death induced by infection with Blumeria graminis f. sp. Tritici was calculated at 1 and 2 dpi. The pen1-1 mutant showed a significantly higher frequency of epidermal single cell death upon Blumeria graminis f. sp. Tritici infection; by contrast, the frequency of epidermal single cell death in exo70B1-3 was similar to that of wild type. Bars represent mean and standard deviation (n = 6, scoring 40 sites per time point). Asterisks indicate statistically significant difference from wild type (p < 0.01; Student’s t-test). The experiments were repeated three times with similar results. Enhanced disease resistance in exo70B1 requires the TIR-NBS gene TN2
We performed a genetic screen to identify suppressors of exo70B1. We mutagenized exo70B1-3 seeds with EMS, and screened the M2 plants for mutants that displayed wild-type like phenotypes in response to G. cichoracearum. We obtained more than 20 mutants, which fell into 5 complementation groups, including nine alleles of At1g17615, which we defined by standard map-based cloning (S12 Fig.), seven alleles of PAD4 (consistent with our double mutant analysis, above) and four mutant alleles of a gene encoding a calcium dependent protein kinase (which will be reported elsewhere). The mutation in At1g17615 fully suppresses exo70B1-associated phenotypes, including spontaneous cell death (S13A Fig.), powdery mildew resistance and cell death (Fig. 7A-C), increased accumulation of H2O2 and SA (S13B-S13C Fig.) and enhanced resistance to Pto DC3000 (Fig. 7D), indicating that enhanced disease resistance in exo70B1 requires At1g17615. In addition, the up-regulation of PR1 expression in exo70B1-3 during G. cichoracearum infection was also suppressed by At1g17615 mutation (Fig. 7E). However, activation of MAPK in exo70B1-3 was not suppressed by At1g17615 mutation (S13D Fig.), indicating that activation of MAPK in exo70B1-3 is not dependent on At1g17615. Sequence analysis revealed that At1g17615 encodes a TIR-NBS protein, previously designated as TIR-NBS2 (TN2). TN2, and a further 20 genes in Arabidopsis ecotype Col-0, lack leucine-rich repeats [38]. Among the nine tn2 alleles obtained in the screen, three carry missense mutations in the TIR domain, four in the NBS domain, and two carry premature stop mutations (Fig. 7F). TN2 does not express a conserved canonical P-loop, which is likely involved in nucleotide binding, but rather encodes GRS instead of the canonical GKT in the P-loop motif (S14 Fig.). However, the mutant allele tn-6 (G222R) is in the traditionally defined P-loop (S14 Fig.), and G222 of TN2 is highly conserved among GRS containing NB-TIRs (S14 Fig.), suggesting that the GRS motif of TN2 and its relatives might retain some P-loop function. TN2 transcripts accumulated at much higher levels in exo70B1 compared to wild type (Fig. 7G); and the accumulation of TN2 transcripts was further increased upon G. cichoracearum infection (Fig. 7H). The tn2 mutations suppress exo70B1-associated resistance and cell death phenotypes, indicating that enhanced disease resistance and cell death in exo70B1 requires TN2. The pad4 mutation suppressed the increased accumulation of TN2 transcripts in exo70B1-3 (S15 Fig.), consistent with the observations that pad4 suppressed exo70B1-3 associated phenotypes (Fig. 3) and suggesting that the up-regulation of TN2 is important for the exo70B1-3 mutant phenotype. In addition, TN2 transcripts over-accumulated in older exo70B1-3 plants (S16 Fig.), which is consistent with the observations that exo70B1-3 displays spontaneous cell death after 5 weeks.
Fig. 7. exo70B1-mediated resistance and cell death require the TIR-NBS protein TN2. 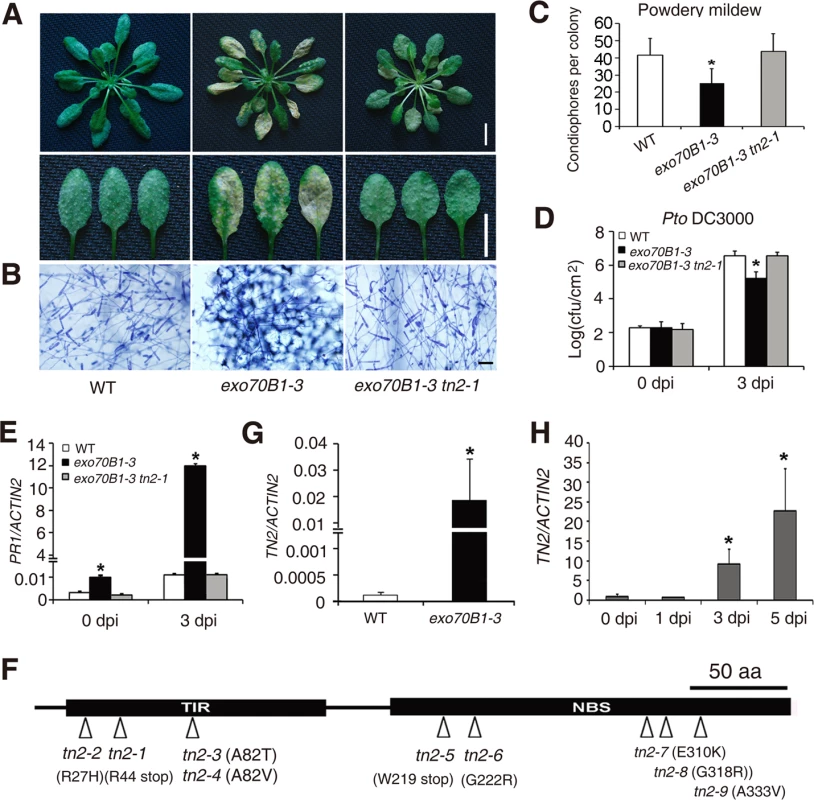
(A-C) Four-week-old plants were infected with G. cichoracearum.(A) Intact plant (upper panel) or representative leaves (lower panel) from the infected plants were photographed at 7 dpi. The exo70B1-3 mutant was resistant, but the exo70B1-3 tn2-1 mutant was susceptible to G. cichoracearum. Bar = 10 mm. (B) Leaves from infected plants were stained with trypan blue at 7 dpi. Many fungal spores were produced in wild type and exo70B1-3 tn2-1, but few spores were produced in exo70B1-3. Bar = 50 μm. (C) Fungal growth was assessed by counting the number of conidiophores per colony at 5 dpi. Bars represent mean and standard deviation from three independent biological replicates (n = 30). The asterisk indicates a statistically significant difference (p < 0.05; Student’s t-test). (D) Four-week-old plants of wild type, exo70B1-3 and exo70B1-3 tn2-1 were inoculated with Pto DC3000 at OD600 = 0.0005. Bacterial growth was assessed at days 0 and 3. Bars represent means and standard deviation of three independent biological replicates. The asterisk indicates a statistically significant difference (p < 0.05, Student’s t-test). (E) Quantitative RT-PCR analysis of PR1 expression in the infected plants with G. cichoracearum. The relative transcript levels were examined by real-time PCR and normalized to ACTIN2. Bars represent mean and standard deviation from three independent experiments. The asterisk indicates statistically significant difference (p < 0.05, Student’s t-test). (F) Protein structure of TN2. An arrowhead indicates the position of the mutated amino acid in each tn2 mutant. (G)-(H) The transcript levels of TN2 were examined by quantitative real-time PCR, and normalized to ACTIN2. Uninfected wild type and exo70B1-3 plants (G) and wild type plants infected with G. cichoracearum (day 0: uninfected control) (H). Bars represent mean and standard deviation from three biological experiments. The asterisk indicates a statistically significant difference (p < 0.05, Student’s t-test). To further assess the role of TN2 in plant immunity, we examined the tn2 single mutant for resistance to fungal and bacterial pathogens. The tn2 single mutant displayed wild type like responses to virulent fungal pathogen G. cichoracearum (S17A-S17C Fig.) and bacterial pathogen Pto DC3000 (S17D Fig.), indicating that TN2 does not contribute to basal defense to G. cichoracearum or Pto DC3000. To further investigate the phenotypes of exo70B1-3, tn2 and exo70B1-3 tn2 mutants in response to Pto DC3000, we also inoculated those plants by spray infection. As shown in S18 Fig., exo70B1-3 displayed enhanced resistance to Pto DC3000, and the resistance was fully suppressed by the tn2-1 mutation, which was consistent with our findings usinginfiltration assays. In addition, TN2 was not involved in powdery mildew resistance and mildew-induced cell death in edr2, a well characterized powdery mildew resistant mutant, which accumulates higher levels of SA [58] (S19 Fig.).
To further investigate the relationship between TN2 and EXO70B1, we examined whether TN2 interacts with EXO70B1 by yeast two-hybrid assays. As shown in Fig. 8A, EXO70B1 interacts with TN2 and the TIR domain of TN2. We extended this finding by using a BIFC assay in N. benthamiana to examine whether EXO70B1 and TN2 associates. TN2 was fused to the N-terminal fragments of YFP (YFPN), and was co-transformed to N. benthamiana with EXO70B1-YFPC, SNAP33-YFPC, VAMP721-YFPC or PEN1-YFPC, we observed YFP fluorescence only in leaves co-transformed with the EXO70B1-YFPN and TN2-YFPC, but not in leaves co-transformed with SNAP33-YFPC and TN2-YFPN, and VAMP721-YFPC and TN2-YFPN, or PEN1-YFPC and TN2-YFPN, indicating that TN2 can interact with EXO70B1 in N. benthamiana (Fig. 8B and S20 Fig.), To further confirm the interaction between EXO70B1 and TN2, we performed Co-IP assays by co-expressing EXO70B1-FLAG and TN2-Myc in N. benthamiana leaves. And EXO70B1-FLAG alone was expressed as a negative control. TN2 was immunoprecipitated using anti-Myc antibody, and in the precipitate, EXO70B1 was detected with anti-FLAG antibody only from the leaves that co-expressed both EXO70B1-FLAG and TN2-Myc, not from the negative control leaves (Fig. 8C). Taken together, those observations indicate that EXO70B1 and TN2 interact. These data are consistent with the hypothesis that TN2 or TN2 associated nucleotide binding domain and leucine-rich repeat-containing receptor is a guard directly monitoring the integrity of EXO70B1.
Fig. 8. EXO70B1 interacts with TN2. 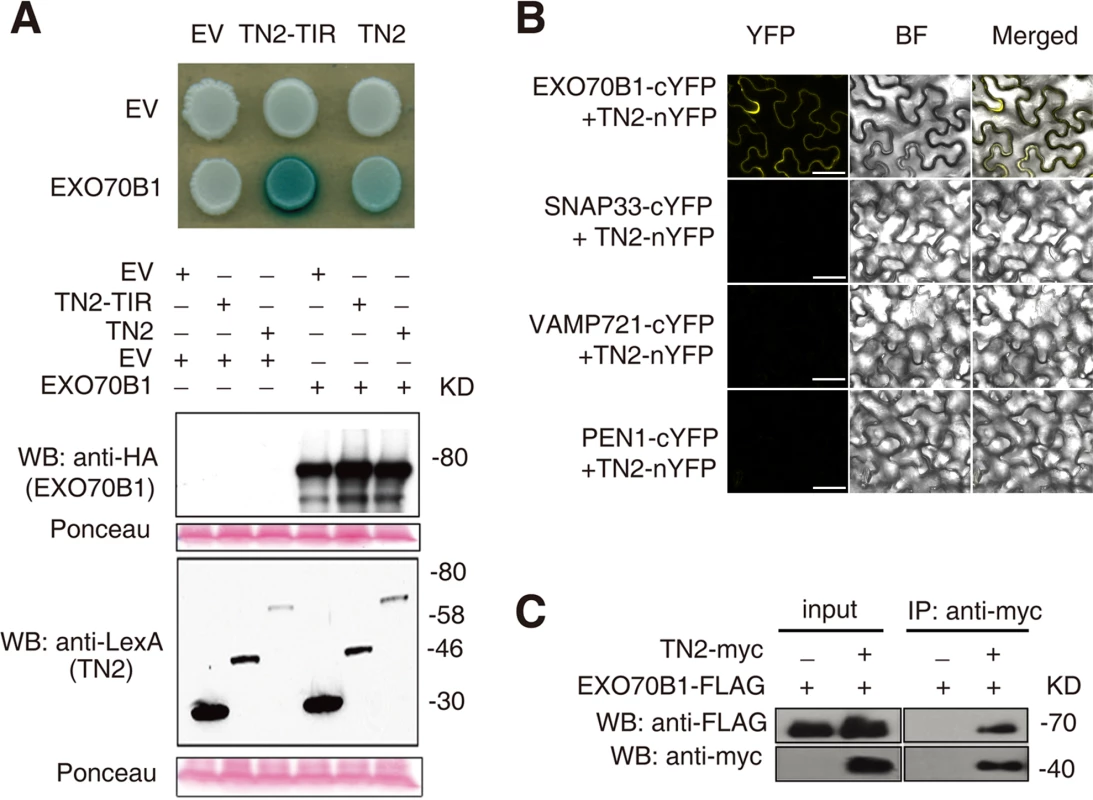
(A) EXO70B1 interacted with TN2 in a yeast two-hybrid assay. Yeast cells containing the indicated plasmids were spotted onto the SD-His-Ura-Trp containing X-Gal. EV, empty vector; TN2-TIR, the TIR domain of TN2; TN2, full-length TN2; EXO70B1, full-length EXO70B1. Western blots verify the accumulation of Y2H fusion proteins. Expected sizes of pEG202:EV = 24 kD, pEG202:TN2-TIR = 48 kD, pEG202:TN2 = 67 kD, pJG4-5:EXO70B1 = 84 kD. Ponceau stain of membranes indicates equal loading. Discussion
EXO70B1 belongs to the EXO70 gene family, and encodes a subunit of the exocyst complex, which plays a key role in anchoring and tethering vesicles to specific sites of the plasma membrane [19]. In yeast and mammals, exocyst complex-mediated vesicle tethering is followed by SNARE protein-mediated fusion to the targeted membrane, an important step in vesicle trafficking that leads to site-directed secretion. We showed that the loss-of-function exo70B1-3 mutant displayed spontaneous HR-like cell death, enhanced disease resistance to G. cichoracearum, and mildew-induced cell death upon infection. Recent work showed that the exo70B1 mutant displayed HR-like lesions, and accumulated higher levels of SA and PR1 transcripts [36]. In addition, exo70B1 exhibited a variety of additional defects, including vacuole anthocyanin accumulation, nitrogen starvation and reduced internalized autophagosomes inside the vacuole, suggesting a role of EXO70B1 in autophagic membrane transport [36]. It will be interesting in the future to address whether these phenotypes also require TN2. Recently, Stegmann et al. reported that the exo70B1 mutant was more resistant to H. a. Emco5, and that exo70B1 exhibited increased cell death in response to the non-host oomycete pathogen Phytophtora infestans [59], consistent with our findings. However, Stegmann et al. also showed that exo70B1 displayed enhanced susceptibility to Pto DC3000 [59], in contrast to our results that exo70B1 was more resistant to this bacterial pathogen. The differences between our study and Stegmann et al. [59] could be caused by different growth conditions used in these two studies, such as humidity and light intensity.
The HR-like and enhanced disease resistance phenotypes indicate that exo70B1 behaves like lesion mimic mutant. The exo70B1-3 associated phenotypes require TN2, a TIR-NBS protein. The fact that we recovered loss of function tn2 missense changes in a conserved glycine residue in the catalytic domain of the P-loop suggest that activation of TN2 causes the exo70B1 loss of function phenotypes. Recessive mutations that give rise to enhanced disease resistance and lesion mimic phenotypes are increasingly associated with ectopic activation of NLR immune receptors, as demonstrated for Arabidopsis acd11 (accelerated cell death 11) and lsd1 (lesion simulating disease 1) [52,60,61]. Both lsd1 and acd11 result in constitutively activated immune responses in the absence of pathogens; these phenotypes are suppressed by mutations in the TIR-NLR LAZ5 or the CC-NLRs, ADR1-L1 and ADR1-L2, respectively [52,60,61]. Similar to exo70B1, the lsd1 and acd11-associated autoimmune phenotypes correlate with altered levels of SA [52,60,61].
We showed that the truncated NLR protein TN2 associates with EXO70B1, suggesting a direct functional link between TN2 and EXO70B1. Considering the important role of exocytosis in plant immunity, it would not be surprising if some pathogens evolved effectors that target the exocytosis machinery. It is worth noting that the EXO70 gene family in plants is hugely expanded, with 23 copies in Arabidopsis, compared to only one in yeast and animals [55]. Many exo70 subunits have adopted alternative functions [26], which may contribute to the expansion. One additional reason for this expansion might be that pathogen effectors target EXO70 proteins to manipulate plant immunity. The evolution of new EXO70 family members could allow the plant to evade pathogen effectors and retain antimicrobial secretory function. An alternative evolutionary possibility, and the one most consistent with our data, is that EXO70B1 has retained its function, but evolved associations with TN2, which can be activated by an as yet undiscovered effector that has evolved to manipulate EXO70B1 function. Considering that EXO70B1 contributes to autophagy, another possibility is that the plant may keep the TN2 protein at a low level via degradation of TN2 in an EXO70B1-autophagy dependent pathway. In this scenario, TN2 is activated, and also up-regulated, when EXO70B1 is not functional. Many studies have shown that autophagy plays important role in plant immunity and pathogen triggered cell death. For instance, the atg5 (autophagy-related 5) mutant displays unrestricted chlorotic cell death after infiltration of Pto DC3000 avrRpm1 [62]. Similarly, the atg2 mutant shows mildew-induced cell death and enhanced powdery mildew resistance [42]. In addition, Lenz et al. show that atg5, atg10 and atg18 mutants display enhanced resistance to Pto DC3000 [63]. Hofius et al. show that the atg7 and atg9 mutants display enhanced susceptibility to Pto DC3000 and the Emwa isolate of H. arabidopsidis, and cell death triggered by RPS4 and RPP1 was suppressed in those two mutants [64]. Although autophagy is involved in plant immunity, it has not been shown that any NLR receptor protein is specifically degraded by autophagy. It would be very interesting to examine whether the exo70B1-related autophagy defects are dependent on TN2, and whether accumulation of TN2 protein is regulated by EXO70B1-associated autophagy.
The TN2 is one of 21 TIR-NBS proteins in the Arabidopsis ecotype Col-0 genome [65]. Although the specific roles of TN proteins are not known, some evidence demonstrates that they can contribute to immune system function. For instance, overexpression of some TN genes induces cell death in tobacco, and results in stunted growth and high free SA in Arabidopsis [39,65]. Li et al. [39,65] showed that cell death in the copine protein double mutant bon1 (bonzai1) bon3 was partially suppressed by loss-of-function of some NLRs and TN2. However, the suppression was very mild since none of the plants survived to bolt. In addition, TN2 transcripts did not over-accumulate in bon1 bon3. Collectively, these data suggesting that cell death in bon1 bon3 is different from exo70B1.
Some TN proteins can interact with effector proteins from pathogens [39], suggesting that the TN proteins are likely involved in plant immunity. We cannot exclude the possibility that EXO70B1 and TN2 have other cellular functions in addition to biotic interactions. Many TN genes occur in complex genomic clusters with TIR-NBS-LRR genes. For instance, TN2 is in a cluster of 3 TIR-containing genes, including one TIR-NBS-LRR (At1g17600), and another TN gene (At1g17610, TN1, also called CHILLING SENSITIVE 1 or CHS1). Mutations in TN1/CHS1 caused auto-immunity at low temperature. As expected of a TIR-containing protein, this phenotype required EDS1 and PAD4 [66,67]. However, as no function has been assigned to the TIR-NBS-LRR gene (At1g17600) in this cluster, it will be interesting to examine whether At1g17600 is required for At1g17610 (TN1) and At1g17615 (TN2) function.
In conclusion, we found that Arabidopsis exocyst subunit EXO70B1 associates with the SNARE complex component SNAP33. Loss-of-function of EXO70B1 leads to a syndrome of defense related phenotypes, which require the TIR-NBS2 protein. We speculate that exocyst components could be targets of effectors that are, at least in this case, guarded by TN2. Alternatively, TN2 could function as an adaptor in combination with an undefined classical NLR receptor. However, we note that we did not identify any other NLR receptor in our fairly deep mutant screen (recall that we isolated many alleles of TN2 and PAD4). This would suggest that redundant classical NLR might use TN2 as an adaptor is such a model. Irrespective of the ultimate mechanistic model, we found that EXO70B1 protein interacts with TN2.
Although the role of EXO70B1 in plant immunity is not clear, work by Stegmann et al. showed that the exo70B1 mutant is compromised in flg22-triggered responses suggesting that EXO70B1 contributes to PTI [35], which makes it a potential target for pathogen effector. Important questions remain to be addressed, including: the nature of the cargos transported by EXO70B1-mediated trafficking, and how those cargos affect plant immunity, as well as how mutation of EXO70B1 activates TN2, a protein that lacks canonical regulatory domains associated with full length NLR receptors, and whether TN2 is involved in the autophagic pathway mediated by EXO70B1. Answering these questions will shed new light on plant immunity.
Materials and Methods
Plant materials and growth conditions
The Arabidopsis thaliana exo70B1-3 mutant was identified from a T-DNA insertion population [14]. T-DNA insertion lines exo70B1-1 (N328818), exo70B1-2 (N717829), exo70B2-1 (SALK_091877C) were from the Arabidopsis Biological Resource Center or Nottingham Arabidopsis Stock Center. The mutant alleles used for the double mutant analyses were as described: pad4-1 [47], npr1-1 [16], eds5-1 [45], sid2-2 [46], ein2-1 [68], coi1-1 [69], edr1 [70], edr2 [12], pmr4-1 [9], pen1-1 [15]. All of the mutants used were in the Columbia-0 (Col-0) background, except for coi1-1 (in Col-6 background). Double mutants were created by standard genetic crosses and confirmed by PCR.
Arabidopsis plants were grown in a growth room at 22–24°C, at a light intensity of 8000 lux, and a 9-h-light /15-h-dark photoperiod for phenotype analysis or 16-h-light / 8-h-dark photoperiod for seed setting, as described previously [71]. Nicotiana benthamiana plants were grown in the same short-day conditions as Arabidopsis.
Fungal infection
Adapted powdery mildew strain Golovinomyces cichoracearum UCSC1 [4] was maintained on susceptible pad4-1 mutant plants. Four-week-old plants were inoculated with powdery mildew using a settling tower at low or high densities [14] [42]. Low spore density was used to quantify the number of conidiophores per colony and high density was used to evaluate hyphae growth and lesion formation. The plants were moved to a growth chamber immediately after infection with G. cichoracearum to facilitate growth of the fungi. Hyphae and dead plant cells in the infected leaves were visualized by trypan blue staining [70]. H2O2 accumulation was detected at 2 dpi by 3,3’-diamino benzidine hydrochloride (DAB) staining [72]. Callose deposition was detected by aniline blue staining [73]. The samples were observed and photographed with an Olympus BX60 microscope. Non-adapted wheat powdery mildew Blumeria graminis f. sp. tritici E09 was grown on susceptible Chinese wheat variety Jing411 [74]. Assessment of epidermal cell death in response to graminis f. sp. tritici E09 was performed as described [16] with minor modifications. Briefly, four-week-old plants were inoculated with B. graminis f. sp. tritici using a settling tower, and the infected leaves was stained with trypan blue, and epidermal single cell death in the infected siytes was visulized with an Olympus BX60 microscope.
Pseudomonas syringae infection was performed by leaf infiltration (OD600 = 5 ×10−4 for growth curve analysis) as described [75], unless otherwise indicated. For spray infection, 5-week-old plants were sprayed with bacterial suspensions (OD600 = 0.2) in 10 mM MgCl2 containing 0.025% Silwet L-77, and the infected plants were placed under plastic domes for 24 hr.
Inoculation of Hyaloperonospora arabidopsidis (H. a.) Noco2 (5 × 104 spores/mL) was performed according to [76].
Suppressor screen
The exo70B1-3 seeds were incubated with 0.3% EMS for 16 h. Around 60,000 M2 plants derived from 5,000 M1 plants were scored by infection with G. cichoracearum. Plants showing wild type-like phenotypes were selected for further characterization.
Map-based cloning of EXO70B1 and TN2
The exo70B1-3 mutant was crossed to ecotype Landsberg erecta (Ler). F2 plants displaying exo70B1-3 mutant phenotypes were selected for mapping. Using rough mapping markers [77] and our newly developed SSLP, CAPS or dCAPS markers, we narrowed down the mutation to an 83-kb region spanning BAC MCK7 (GenBank accession AB019228) and MQJ2 (GenBank accession AB025632) on chromosome 5. We subsequently designed PCR primers to amplify the coding sequences of these genes. The suppressor gene TN2 was also identified by standard map-based cloning.
For complementation analysis, a 4.3-kb genomic sequence including 1531 bp upstream of the start codon, and 835 bp downstream of the stop codon of EXO70B1 was amplified and cloned into binary vector pCAMBIA1300. The construct was then introduced into Agrobacterium tumefaciens strain GV3101, and transformed into exo70B1-3 plants by the floral dip method [78]. Transformants were selected on 1/2 MS medium containing 80 μg/mL hygromycin. T1 transgenic plants were used for phenotyping.
Quantitative real-time RT-PCR
Extraction of total RNA, synthesis of first-strand cDNA and analysis of gene expression were performed as described previously [79].
GUS activity assay
The promoter region of EXO70B1, including 1531 bp upstream of the start codon, was cloned into the pBI121 vector. Homozygous transgenic plants in the T3 generation were used for GUS activity analysis according to [80].
SA quantification
SA extraction and HPLC assays were conducted as described previously [49,81].
Yeast two-hybrid assays
The MATCHMAKER GAL4 Two-Hybrid System 3 (Clontech) was used to examine the interactions between EXO70B1 and other exocyst subunits, as well as PEN1 and SNAP33. EXO70B1-pGBKT7 was co-transformed into Saccharomyces cerevisiae strain AH109 with SEC3a, SEC5a, SEC6, SEC8, SEC10, SEC15b, EXO84bN (the N terminal domain of EXO84b), PEN1 and SNAP33 fused with pGADT7. The constructs containing SEC3a, SEC5a, or SEC15b fused with pGADT7 were kindly provided by Dr. Viktor Zárský [25]. The constructs containing SEC6, SEC8, SEC10, EXO84bN, PEN1 and SNAP33 fused with pGADT7 were made in this study using primers listed in S1 Table. A single colony grown on SD/-Leu/-Trp plate was incubated in liquid media, the overnight culture was diluted to OD600 = 0.5 with sterile water, and a 10 μL drop of dilution was plated on SD/-Ade/-His/-Leu/-Trp plates to test the interaction.
The interaction of EXO70B1 and TN2 was assayed using Gateway-modified versions of the traditional pEG202 and pJG4-5 vectors [82]. pDONR207 Gateway Entry clones containing ORFs (S1 Table for cloning primers) were recombined into pEG202 and pJG4-5 using LR clonase II (Invitrogen). The resulting vectors were sequence verified and then transformed into EGY48 (pEG202) and RFY206 containing the LacZ reporter pSH18-34 (pJG4-5). The resulting strains were mated and diploids selected on synthetic media (SD-HUW). Diploids were spotted onto SD-HUW +X-gal (80 μg/mL). Protein accumulation of fusion proteins in diploids was verified with standard Western blotting using anti-LexA (rabbit pAB, AbCam) and anti-HA (rat mAB, Roche) antibodies.
BiFC analysis
To create the constructs used in BiFC analysis, the coding sequences of EXO70B1 and TN2 were first cloned into pSY736 vector in frame with YFPN. The derived YFPN-fused-sequences were then amplified and inserted into the Gateway entry vector pDONR207 and subsequently the destination vector pMDC32. Similarly, to create YFPC fused to EXO70B1, SNAP33, VAMP721 and PEN1, the coding sequence of each gene was cloned into pSY735 vector in frame with YFPC. The inserts were subcloned into pDONR207 and pMDC32 vectors. Agrobacterium GV3101 carrying different YFPN and YFPC pairs were infiltrated into four-week-old Nicotiana benthamiana [83]. YFP fluorescence was observed by confocal microscopy (Carl Zeiss LSM710) after 48hr.
Protein extraction and immunoprecipitation in N. benthamiana and Arabidopsis
To create the constructs used in coimmunoprecipitation analysis in N. benthamiana, EXO70B1 was cloned into the pGWB11 vector with C-terminal FLAG fusion using gateway system (Invitrogen). Similarly, PEN1 was cloned into pEarleyGate 201 vector with 35S promoter and N-terminal HA fusion, while SNAP33 and TN2 were cloned into pEarleyGate 203 vector with 35S promoter and N-terminal Myc fusion. Transient expression, total protein extractions and immunoblots were performed as described previously [58]. Briefly, about 20 μg protein was resuspended in sample buffer and separated by SDS-PAGE. The protein was detected by immunoblot with primary antibody anti-GFP (Roche 1 : 1000) or anti-HA (Sigma 1 : 1000), and then with anti-mouse (rabbit) HRP-linked secondary antibody, followed by detection with the chemiluminescent HRP substrate kit (Millipore).
For immunoprecipitation, proteins prepared from NB1 buffer [84] were incubated with primary antibody (3 μL /1 mL), at 4 °C for 3 hours with gentle shaking, and 20 μL protein G agarose beads (Millipore) were added and kept shaking for another 3 hours. The beads were washed 4 times with PBS buffer, and resuspended in PBS buffer. The samples were separated by SDS-PAGE and analyzed by immunoblotting. To produce an EXO70B1-GFP fusion, a 3.4-kb genomic sequence, containing 1531bp promoter and 1872 bp coding sequence of EXO70B1 without the stop codon, was cloned into the binary vector pEGAD [85] between the StuI and AgeI cloning sites to remove the 35S promoter from the vector. The derived EXO70B1pro:EXO70B1-GFP construct was introduced into the A. tumefaciens strain GV3101 and transformed into exoB1-3 plants using the floral dip method [78]. To test the interaction between SNAP33 and EXO70B1 in Arabidopsis, we amplified SNAP33 genomic sequence (without stop codon) and fused with an HA tag, and cloned it into pMDC163 vector using gateway system (Invitrogen). And exo70B1-3 plants expressing EXO70B1pro:EXO70B1-GFP were transformed with derived SNAP33pro:SNAP33-HA construct, and the T1 plants were used for coimmunoprecipitation immunoblot assays.
To detect MAP kinase activation, 4-week-old plants were inoculated with G. cichoracearum for 3 days, and the infected leaves were excised and quickly frozen in liquid nitrogen. Total protein was isolated and subjected to immunoblot with rabbit anti-pTEpY (Cell Signaling), anti-MPK3 or anti-MPK6 (Sigma) as the primary antibody, following the manufacturer’s instructions.
Oligonucleotide Sequences
The primers used in this work are listed in S1 Table.
Supporting Information
Zdroje
1. Buschges R, Hollricher K, Panstruga R, Simons G, Wolter M, et al. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88 : 695–705. 9054509
2. Griffey CA, Das MK, Stromberg EL (1993) Effectiveness of adult-plant resistance in reducing grain yield loss to powdery mildew in winter wheat. Plant Dis 77 : 618–622.
3. Schulze-Lefert P, Vogel J (2000) Closing the ranks to attack by powdery mildew. Trends Plant Sci 5 : 343–348. 10908879
4. Adam L, Somerville SC (1996) Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J 9 : 341–356. 8919911
5. Koh S, André A, Edwards H, Ehrhardt D, Somerville S (2005) Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J 44 : 516–529. 16236160
6. Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, et al. (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet 38 : 716–720. 16732289
7. Vogel J, Somerville S (2000) Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc Natl Acad Sci USA 97 : 1897–1902. doi: 10.1073/pnas.030531997 10677553
8. Vogel JP, Raab TK, Schiff C, Somerville SC (2002) PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14 : 2095–2106. doi: 10.1105/tpc.003509 12215508
9. Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, et al. (2003) Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 : 969–972. 12920300
10. Vogel JP, Raab TK, Somerville CR, Somerville SC (2004) Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J 40 : 968–978. 15584961
11. Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 98 : 373–378. doi: 10.1073/pnas.98.1.373 11114160
12. Tang D, Ade J, Frye CA, Innes RW (2005) Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J 44 : 245–257. doi: 10.1111/j.1365-313X.2005.02523.x 16212604
13. Tang D, Ade J, Frye CA, Innes RW (2006) A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J 47 : 75–84. doi: 10.1111/j.1365-313X.2006.02769.x 16824181
14. Vorwerk S, Schiff C, Santamaria M, Koh S, Nishimura M, et al. (2007) EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana. BMC Plant Biol 7 : 35. doi: 10.1186/1471-2229-7-35 17612410
15. Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425 : 973–977. 14586469
16. Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, et al. (2005) Pre - and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 : 1180–1183. 16293760
17. Stein M, Dittgen J, Sanchez-Rodriguez C, Hou BH, Molina A, et al. (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18 : 731–746. doi: 10.1105/tpc.105.038372 16473969
18. Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, et al. (2008) Co-option of a default secretory pathway for plant immune responses. Nature 451 : 835–840. doi: 10.1038/nature06545 18273019
19. He B, Guo W (2009) The exocyst complex in polarized exocytosis. Curr Opin Cell Biol 21 : 537–542. doi: 10.1016/j.ceb.2009.04.007 19473826
20. TerBush DR, Maurice T, Roth D, Novick P (1996) The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J 15 : 6483–6494.. 8978675
21. He B, Xi F, Zhang X, Zhang J, Guo W (2007) Exo70 interacts with phospholipids and mediates the targeting of the exocyst to the plasma membrane. EMBO J 26 : 4053–4065. doi: 10.1038/sj.emboj.7601834 17717527
22. Zhang X, Orlando K, He B, Xi F, Zhang J, et al. (2008) Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J Cell Biol 180 : 145–158. doi: 10.1083/jcb.200704128 18195105
23. Elias M, Drdova E, Ziak D, Bavlnka B, Hala M, et al. (2003) The exocyst complex in plants. Cell Biol Int 27 : 199–201. 12681307
24. Chong YT, Gidda SK, Sanford C, Parkinson J, Mullen RT, et al. (2010) Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol 185 : 401–419. doi: 10.1111/j.1469-8137.2009.03070.x 19895414
25. Hala M, Cole R, Synek L, Drdova E, Pecenkova T, et al. (2008) An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. Plant Cell 20 : 1330–1345. doi: 10.1105/tpc.108.059105 18492870
26. Zarsky V, Kulich I, Fendrych M, Pecenkova T (2013) Exocyst complexes multiple functions in plant cells secretory pathways. Curr Opin Plant Biol 16 : 726–733. doi: 10.1016/j.pbi.2013.10.013 24246229
27. Cole RA, Synek L, Zarsky V, Fowler JE (2005) SEC8, a subunit of the putative Arabidopsis exocyst complex, facilitates pollen germination and competitive pollen tube growth. Plant Physiol 138 : 2005–2018. doi: 10.1104/pp.105.062273 16040664
28. Wen TJ, Hochholdinger F, Sauer M, Bruce W, Schnable PS (2005) The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol 138 : 1637–1643. doi: 10.1104/pp.105.062174 15980192
29. Samuel MA, Chong YT, Haasen KE, Aldea-Brydges MG, Stone SL, et al. (2009) Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21 : 2655–2671. doi: 10.1105/tpc.109.069740 19789280
30. Kulich I, Cole R, Drdova E, Cvrckova F, Soukup A, et al. (2010) Arabidopsis exocyst subunits SEC8 and EXO70A1 and exocyst interactor ROH1 are involved in the localized deposition of seed coat pectin. New Phytol 188 : 615–625. doi: 10.1111/j.1469-8137.2010.03372.x 20618910
31. Fendrych M, Synek L, Pecenkova T, Toupalova H, Cole R, et al. (2010) The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. Plant Cell 22 : 3053–3065. doi: 10.1105/tpc.110.074351 20870962
32. Drdova EJ, Synek L, Pecenkova T, Hala M, Kulich I, et al. (2013) The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. Plant J 73 : 709–719. doi: 10.1111/tpj.12074 23163883
33. Li S, Chen M, Yu D, Ren S, Sun S, et al. (2013) EXO70A1-mediated vesicle trafficking is critical for tracheary element development in Arabidopsis. Plant Cell 25 : 1774–1786. doi: 10.1105/tpc.113.112144 23709627
34. Pecenkova T, Hala M, Kulich I, Kocourkova D, Drdova E, et al. (2011) The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J Exp Bot 62 : 2107–2116. doi: 10.1093/jxb/erq402 21199889
35. Stegmann M, Anderson RG, Ichimura K, Pecenkova T, Reuter P, et al. (2012) The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24 : 4703–4716. doi: 10.1105/tpc.112.104463 23170036
36. Kulich I, Pecenkova T, Sekeres J, Smetana O, Fendrych M, et al. (2013) Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic 14 : 1155–1165. doi: 10.1111/tra.12101 23944713
37. Jones JD, Dangl JL (2006) The plant immune system. Nature 444 : 323–329. 17108957
38. Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 : 809–834. doi: 10.1105/tpc.009308 12671079
39. Nandety RS, Caplan JL, Cavanaugh K, Perroud B, Wroblewski T, et al. (2013) The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol 162 : 1459–1472. doi: 10.1104/pp.113.219162 23735504
40. Bent AF, Mackey D (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45 : 399–436. 17506648
41. Yao C, Wu Y, Nie H, Tang D (2012) RPN1a, a 26S proteasome subunit, is required for innate immunity in Arabidopsis. Plant J 71 : 1015–1028. doi: 10.1111/j.1365-313X.2012.05048.x 22577987
42. Wang Y, Nishimura MT, Zhao T, Tang D (2011) ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J 68 : 74–87. doi: 10.1111/j.1365-313X.2011.04669.x 21645148
43. Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, et al. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104 : 12217–12222. doi: 10.1073/pnas.0705306104 17626179
44. Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64 : 839–863. doi: 10.1146/annurev-arplant-042811-105606 23373699
45. Nawrath C, Heck S, Parinthawong N, Metraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 : 275–286. doi: 10.1105/tpc.010376 11826312
46. Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 : 562–565. 11734859
47. Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10 : 1021–1030. 9634589
48. Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 : 57–63. 9019406
49. Gou M, Su N, Zheng J, Huai J, Wu G, et al. (2009) An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J 60 : 757–770. doi: 10.1111/j.1365-313X.2009.03995.x 19682297
50. Zhang Y, Goritschnig S, Dong X, Li X (2003) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15 : 2636–2646. doi: 10.1105/tpc.015842 14576290
51. Lee J, Nam J, Park HC, Na G, Miura K, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49 : 79–90. 17163880
52. Roberts M, Tang S, Stallmann A, Dangl JL, Bonardi V (2013) Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet 9: e1003465. doi: 10.1371/journal.pgen.1003465 23633962
53. Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 : 2148–2152. 10381874
54. Feys B, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis Mutants Selected for Resistance to the Phytotoxin Coronatine Are Male Sterile, Insensitive to Methyl Jasmonate, and Resistant to a Bacterial Pathogen. Plant Cell 6 : 751–759. doi: 10.1105/tpc.6.5.751 12244256
55. Synek L, Schlager N, Elias M, Quentin M, Hauser MT, et al. (2006) AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J 48 : 54–72. doi: 10.1111/j.1365-313X.2006.02854.x 16942608
56. Park KS, Kim YS, Kim JH, Choi BK, Kim SH, et al. (2009) Influence of human allogenic bone marrow and cord blood-derived mesenchymal stem cell secreting trophic factors on ATP (adenosine-5′-triphosphate)/ADP (adenosine-5′-diphosphate) ratio and insulin secretory function of isolated human islets from cadaveric donor. Transplant Proc 41 : 3813–3818. doi: 10.1016/j.transproceed.2009.06.193 19917393
57. Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 : 428–438. 15469500
58. Shi H, Shen Q, Qi Y, Yan H, Nie H, et al. (2013) BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25 : 1143–1157. doi: 10.1105/tpc.112.107904 23532072
59. Stegmann M, Anderson RG, Westphal L, Rosahl S, McDowell JM, et al. (2013) The exocyst subunit Exo70B1 is involved in the immune response of Arabidopsis thaliana to different pathogens and cell death. Plant Signal Behav 8: e27421. doi: 10.4161/psb.27421 24389869
60. Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, et al. (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci USA 108 : 16463–16468. doi: 10.1073/pnas.1113726108 21911370
61. Palma K, Thorgrimsen S, Malinovsky FG, Fiil BK, Nielsen HB, et al. (2010) Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog 6: e1001137. doi: 10.1371/journal.ppat.1001137 20949080
62. Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, et al. (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21 : 2914–2927. doi: 10.1105/tpc.109.068635 19773385
63. Lenz HD, Haller E, Melzer E, Kober K, Wurster K, et al. (2011) Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J 66 : 818–830. doi: 10.1111/j.1365-313X.2011.04546.x 21332848
64. Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, et al. (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137 : 773–783. doi: 10.1016/j.cell.2009.02.036 19450522
65. Li Y, Pennington BO, Hua J (2009) Multiple R-like genes are negatively regulated by BON1 and BON3 in arabidopsis. Mol Plant Microbe Interact 22 : 840–848. doi: 10.1094/MPMI-22-7-0840 19522566
66. Wang Y, Zhang Y, Wang Z, Zhang X, Yang S (2013) A missense mutation in CHS1, a TIR-NB protein, induces chilling sensitivity in Arabidopsis. Plant J 75 : 553–565. doi: 10.1111/tpj.12232 23651299
67. Zbierzak AM, Porfirova S, Griebel T, Melzer M, Parker JE, et al. (2013) A TIR-NBS protein encoded by Arabidopsis Chilling Sensitive 1 (CHS1) limits chloroplast damage and cell death at low temperature. Plant J 75 : 539–552. doi: 10.1111/tpj.12219 23617639
68. Guzman P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 : 513–523. doi: 10.1105/tpc.2.6.513 2152173
69. Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 : 1091–1094. 9582125
70. Frye CA, Innes RW (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10 : 947–956. 9634583
71. Nie H, Wu Y, Yao C, Tang D (2011) Suppression of edr2-mediated powdery mildew resistance, cell death and ethylene-induced senescence by mutations in ALD1 in Arabidopsis. J Genet Genomics 38 : 137–148. doi: 10.1016/j.jgg.2011.03.001 21530897
72. Xiao S, Brown S, Patrick E, Brearley C, Turner JG (2003) Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15 : 33–45. doi: 10.1105/tpc.006940 12509520
73. Pan H, Liu S, Tang D (2012) HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. Plant J 69 : 831–843. doi: 10.1111/j.1365-313X.2011.04835.x 22035198
74. Li G, Fang T, Zhang H, Xie C, Li H, et al. (2009) Molecular identification of a new powdery mildew resistance gene Pm41 on chromosome 3BL derived from wild emmer (Triticum turgidum var. dicoccoides). Theor Appl Genet 119 : 531–539. doi: 10.1007/s00122-009-1061-y 19471905
75. Chen Z, Kloek AP, Boch J, Katagiri F, Kunkel BN (2000) The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol Plant Microbe Interact 13 : 1312–1321. 11106023
76. Xia S, Zhu Z, Hao L, Chen JG, Xiao L, et al. (2009) Negative regulation of systemic acquired resistance by replication factor C subunit3 in Arabidopsis. Plant Physiol 150 : 2009–2017. doi: 10.1104/pp.109.138321 19482917
77. Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123 : 795–805. 10889228
78. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–743. 10069079
79. Nie H, Zhao C, Wu G, Wu Y, Chen Y, et al. (2012) SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol 158 : 1847–1859. doi: 10.1104/pp.111.192310 22345509
80. Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 : 3901–3907. 3327686
81. Li X, Zhang Y, Clarke JD, Li Y, Dong X (1999) Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98 : 329–339. 10458608
82. Golemis EA, Serebriiskii I, Finley RL Jr, Kolonin MG, Gyuris J, et al. (2009) Interaction trap/two-hybrid system to identify interacting proteins. Curr Protoco Protein Sci Chapter 19: Unit19 12. doi: 10.1002/0471140864.ps1902s57 19688737
83. Zhao C, Nie H, Shen Q, Zhang S, Lukowitz W, et al. (2014) EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet 10: e1004389. doi: 10.1371/journal.pgen.1004389 24830651
84. Liu L, Zhang Y, Tang S, Zhao Q, Zhang Z, et al. (2010) An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J 61 : 893–903. doi: 10.1111/j.1365-313X.2009.04109.x 20015064
85. Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97 : 3718–3723. 10737809
Štítky
Genetika Reprodukčná medicína
Článek Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in YeastČlánek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence AnalysesČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 1- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



