-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Dimensions of Horizontal Gene Transfer in Eukaryotic Microbial Pathogens
article has not abstract
Published in the journal: Dimensions of Horizontal Gene Transfer in Eukaryotic Microbial Pathogens. PLoS Pathog 11(10): e32767. doi:10.1371/journal.ppat.1005156
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1005156Summary
article has not abstract
Introduction
Comparative genomic studies of microorganisms have disrupted the paradigm of vertical inheritance with modification. First in bacteria, and more recently in microscopic and even multicellular eukaryotes, horizontal gene transfer (HGT) has been implicated in genomic and ecological evolution. HGT is the exchange of genetic material between organisms that occurs independently of meiotic and mitotic recombination between mating or hybridizing individuals. HGT occurs as viral and plasmid-mediated transfer, and transformation by environmental DNA via known or yet-unknown mechanisms [1]. The existence of environmental gene pools and pan-genomes is supported by decades of functional and phylogenetic studies in bacteria that have highlighted the exchange and proliferation of virulence factors and antibiotic resistance mechanisms [2–4]. Presently, accumulating reports of HGT in eukaryotes raise similar questions of how exposure to such gene pools has impacted the evolution of eukaryotic microbes, and whether or not human activities influence HGT dynamics. Here, we describe the evidence supporting HGT in eukaryotic microbial pathogens from divergent lineages that impact human, animal, and plant health (S1 Table). We consider three interacting dimensions affecting the prevalence of HGT (genetic network structure, selectable functions, and opportunity for contact) in order to better understand how HGT manifests in this important group of organisms.
Does HGT Really Contribute to Eukaryotic Microbial Pathogen Genomes?
Reports of HGT among eukaryotic microbial pathogens have accumulated in recent years, largely driven by comparative genomic analyses showing an unexpected distribution and phylogenetic placement of gene sequences. However, these analyses require appropriate sampling and methodology, and should be interpreted with caution. Objections to the veracity and extent of HGT include the absence of a reproducible transfer mechanism in some lineages, and the plausibility of alternative explanations for the distributions and phylogenies of HGT candidate gene sequences [5]. The alternative interpretations of unexpected gene distributions and phylogenies hinge on different assumptions about the parsimony of a small number of gene transfer events versus a large number of the more widely accepted processes of gene duplication and loss [5]. Proponents of HGT hypotheses emphasize the importance of robust phylogenetic analyses coupled with multiple additional lines of evidence to support claims [6,7]. The most convincing cases have thus combined model-based phylogenetic approaches, which compare the topologies of gene trees to species phylogenies, with additional support from genome structure, sequence identity, codon usage, GC nucleotide content, and evidence of benefits to the recipient (S1 Table). Analyses relying on but a few of these methods, especially supporting methods in isolation, are rarely sufficient to strongly support HGT, and can result in false positive identification of HGT [7]. Unsampled genetic diversity at the population and species levels, which can impact reconstructions of gene distribution and inheritance, may also lead to false positive identification of HGT, underscoring the importance of robust taxon sampling [8]. For example, in a 2011 study, genes encoding the greatly expanded Crinkler protein family in the amphibian pathogen Batrachochytrium dendrobatidis had a distribution and phylogeny consistent with HGT from plant pathogenic oomycetes; however, the recent detection of Crinkler homologs in two additional fungal lineages opens the possibility that the current distribution is compatible with vertical inheritance and widespread gene loss [9–11]. Similar sampling biases can also lead to the underestimation of HGT in lineages of eukaryotic microbial pathogens, due to recent HGTs that are not fixed in populations and transferred genes that are prone to subsequent loss under changing selective pressures. This is illustrated by the rapid, differential degeneration of a horizontally acquired gene cluster among members of the necrotrophic fungal genus Botrytis [12]. More thorough sampling efforts currently underway may reduce erroneous inferences about HGT, help resolve the timing and direction of HGT events, and provide better estimates of their ecological contexts [13].
Does Genetic Network Complexity Influence HGT?
The complexity hypothesis posits that genes that are more modular in nature (i.e., residing at the periphery of gene connectivity networks) are more likely to be successfully transferred, because they are less disruptive to host networks and require establishment of fewer connections (Fig 1) [2,14]. This hypothesis is supported in bacteria, in which low network connectivity is found to enhance a gene’s “transferability,” which is largely independent of its specific biological function [15,16]. In eukaryotic microbial pathogens, genes encoding virulence factors may provide a fitness advantage to the recipient without extensive integration into genetic networks. Examples include genes encoding secreted effector proteins and specialized metabolic genes, especially those located in complete multigene clusters, which encode mechanisms for regulation, compartmentalization, secretion of products, and stoichiometric control of toxic intermediates [17–24]. Biased rates of gene transfer and loss of function found in systematic investigations of pathogen genomes also support the complexity hypothesis. Notably, in the human pathogen Trichomonas vaginalis, only 2% of 152 horizontally transferred genes were “informational” (more often involved in highly conserved, connected cellular processes like transcription), compared to 65% that encoded metabolic enzymes, which are “operational” genes associated with less conserved processes [25,26]. However, a survey of metabolic enzymes in select pathogens found that the degree of network connectivity of horizontally transferred genes was not different from the connectivity of vertically inherited genes [27]. The authors note that this could be because the horizontally transferred genes integrated into existing networks during the extensive time period since they were acquired [28].
Fig. 1. The interacting dimensions of horizontal gene transfer in eukaryotic microbial pathogens. 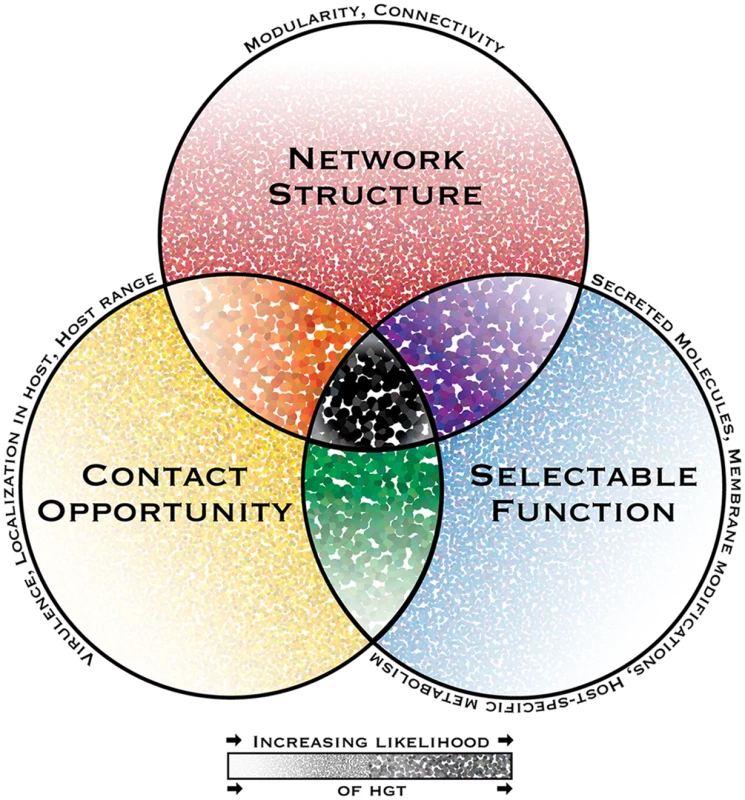
The probability of horizontal gene transfer (HGT) and retention of a gene in recipient organisms is proposed to be under three main interacting influences or dimensions: (1) the genetic network structure, defined as the sum of functional connections between the gene and all other genes within a genome; (2) the selectability of the phenotype conferred by its function in a host environment; and (3) opportunity for contact, i.e., the rate and intimacy of meetings between donor DNA and recipient organisms throughout their life cycles. In this depiction of a general model, the probability of HGT increases with increasing dot intensity and size. Some pathogen-specific parameters influencing each dimension are listed above each circle’s perimeter. While HGT and the subsequent maintenance of genes in recipient genomes might be possible under the influence of only one or two dimensions, it is predicted to have the highest probability when all dimensions interact. A notable consequence of limited gene connectivity associated with horizontally transferred genes is the relaxation of network-imposed selection pressures, giving rise to sequence divergence, thus increasing the capacity for adaptive evolution. This effect has been reported in plant pathogenic Pyrenophora spp., in which ten horizontally transferred genes present significantly more diversifying sequence change compared to corresponding homologs in donor species [29].
What Biological Functions Favor Successful HGT in Eukaryotic Microbial Pathogens?
Environmental selection in pathogen niches may favor the acquisition of certain gene functions. To date, there have been few formal investigations of general trends in functions of genes transferred to eukaryotic microbial pathogens (but see [6,30,31] for discussion of trends in fungi and oomycetes), and few functional confirmations of the roles horizontally transferred genes play in virulence [27,28]. Individual reports suggest three functional categories that are often horizontally transferred to divergent pathogen lineages: secreted molecules, membrane modifications, and metabolism specialized to host interactions and environments (Fig 1). Secreted molecule genes that have been transferred include those for degradative enzymes, such as the 22 different plant polysaccharide depolymerization enzymes that were transferred from phytopathogenic fungi to oomycetes [18]. Genes for production of toxic metabolites that disrupt normal cellular function are also reported transferred, including the fumonisin mycotoxin gene cluster between phytopathogenic Fusarium and Aspergillus fungi [22]. Membrane modifications identified in HGT reports may directly mediate cellular contact between hosts and pathogens, or mask pathogen membranes from host defense responses [32]. For example, a septin trans-membrane protein acquired by the microsporidian Ordospora colligata may facilitate the endocytosis-mediated infection of its Daphnia hosts [33]. Finally, the environment, i.e., the host, selects for the ability in the pathogen to metabolize and/or utilize host defenses and resources, or other sources of fitness in or on the host. For example, osmoregulatory genes acquired by phytopathogenic fungi may facilitate cellular osmotic balance in vascular fluids, and an anaerobic sulfur mobilization gene may increase survival of Blastocystis (a stramenopile suspected to be a pathogen) in anaerobic gut environments [19,20]. Some genes or gene clusters may be considered “repeat offenders,” having been transferred multiple times, possibly due to advantages conferred in specific pathogenic ecologies. For example, the complex distribution of epipolythiodioxopiperazine toxin gene clusters in ascomycete fungi suggests at least three independent transfers between divergent lineages [23].
The extent to which de novo gene gain promotes rapid pathogen emergence is largely unknown. One apparently contemporary transfer of a secreted toxin-encoding gene required for complete virulence on wheat, from Stagonospora nodorum to Pyrenophora tritici-repentis, has been reported [17]. The role of HGT in pathogen emergence is additionally supported by functional studies of transferred genes. For example, deletions of two horizontally acquired genes from the grass pathogen Fusarium pseudograminearum, and of a horizontally acquired osmoregulatory gene from vascular wilt fungi, resulted in reduced virulence [19,34]. Conversely, gain of virulence was documented in a fungal endophyte transformed with a membrane modification gene that the related entomopathogenic fungus, Metarhizium robertsii, may have ancestrally acquired from insects [32].
The fitness benefits conferred by horizontally acquired genes may range from none to highly beneficial, and may not directly relate to pathogenesis. Some pathogens exhibit complex life cycles that alternate between pathogenic and non-pathogenic states, and alternatively the gain of adaptive genes with no pathogenic function could facilitate attenuation of pathogenicity or transition to free-living status under selection from host density. For example, it was speculated that the transfer of a nitrate assimilation gene cluster to the mycoparasitic Trichoderma fungi may promote a transition to the nitrogen-limited wood-decay niche, and the transfer of a sugar utilization gene cluster to Schizosaccharomyces yeast could be part of an ecological transition from pathogen to fermenter [35,36].
Do Eukaryotic Microbial Pathogens Become “Who They Meet?”
The frequency of physical contact between donors and recipients should be considered a driving force behind the likelihood of HGT events, and is a function of an organism’s ecology. Bacteria isolated from the same human body site, for example, exchange genes more frequently, and the genes they exchange are more frequently associated with niche-specific functions [3]. Three groups of ecologically adjacent organisms are often shown to be involved in horizontal gene exchange with eukaryotic microbial pathogens: co-infecting pathogens, non-pathogens symbiotically associated with the host, and the hosts themselves. Examples of transfers between potentially co-infecting pathogens include a host-specific toxin gene between two fungal wheat pathogens and a plant defense compound degradation cluster between fungal grass pathogens [17,21]. Non-pathogenic gut commensal bacteria are thought to have contributed diverse metabolic genes to Trichomonas vaginalis and Blastocystis genomes, including those involved in carbohydrate and amino acid metabolism [20,25,26]. Remarkably, there are cases of host-gene acquisition by insect - and plant-pathogenic fungi. These include the acquisition of a sterol binding protein by the entomopathogenic fungus Metarhizium robertsii that enables it to incorporate host-derived cholesterol into its cell membrane during infections, and the acquisition of a purine salvage pathway gene by obligate intracellular microsporidian pathogens, among others [32,33,37,38]. Considering the range of genetic exchange between ecological associates outside of predator–prey relationships, we suggest that the “you are what you eat” hypothesis proposed as a mechanism of HGT in phagotrophic eukaryotes may be rebranded “you are who you meet” for eukaryotic microbial pathogens [39]. We propose that the frequency of meetings between pathogens and specific classes of organisms may be influenced by virulence, localization in host, and host range (Fig 1). Less virulent pathogens may have sustained encounters with genomes of co-occurring pathogens, non-pathogenic symbionts, and the host because of their low impact on host mortality. In contrast, increasingly virulent pathogens may disproportionately encounter genomes from the greater environment as increased virulence correlates with increased survival time outside of hosts [40]. Obligate intracellular pathogens might be more frequently exposed to host genomes, while extracellular pathogens may often encounter genomes of other host-associated organisms. Similarly, generalists encounter a greater diversity of host genomes compared to specialists, and facultative pathogens may encounter more genes from the greater environment. Factors that contribute to a net increase in exposure to foreign DNA may favor acquisition of novel adaptive functions or drive an HGT ratchet by replacing pathogen genes with foreign genes (as proposed by Doolittle [39]). Furthermore, the gradually converging ecologies resulting from successive “meetings” may promote further transfers of ecology-specific genes such that decreasing ecological proximity results in acceleration of gene acquisition and vice versa. This could explain in part the repeated transfers of phytopathogenic genes from fungi to oomycetes, as well as the repeated acquisition of genes by the plant pathogenic Fusarium lineage from other plant pathogenic fungi, including the Verticillium, Aspergillus, and Collectotrichum genera [18,19,22,31,34]. It remains to be investigated whether specific pathogen lineages, virulence levels, host localizations, or specializations are more prone to horizontal gene exchange, but we note that lineage-specific biases in the rates of HGT were recently shown in a large comparative analysis of fungi [28].
Do Human Activities Impact HGT in Eukaryotic Microbial Pathogens?
Human activities that accelerate environmental changes may impose selection pressures and precipitate dispersal events, which can influence the likelihood of HGT among eukaryotic microbial pathogens. Strong selection pressures exerted by decreased host diversity and intensive management practices in agro-ecosystems, including antimicrobials and other chemical control agents, may be expected to increase the prevalence of horizontally transferred genes, similar to the horizontal proliferation of antibiotic-resistant genes in bacteria due to modern overuse [4]. Homogeneous host environments increase the density of host-specific pathogens, non-pathogens, and the opportunities for them to interact, while at the same time relaxing density-dependent selection against virulent pathogens. Other human activities, such as global trade and travel, influence both the frequency and diversity of the close physical encounters required for horizontal gene flow, which can lead to emergence and evolution of pathogens in the near-term [17]. Furthermore, the migration of host ranges associated with climate and land-use changes provides new opportunities for encounters between pathogens established in previously isolated environments [41]. The extent of human impact on HGT in eukaryotic microbial pathogens is not known, but recent HGT discoveries in these organisms argue for careful consideration of pathogen emergence by HGT as a consequence of ecosystem management.
Supporting Information
Zdroje
1. Thomas CM, Nielsen KM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Micro 3 : 711–721.
2. Baquero F (2004) From pieces to patterns: evolutionary engineering in bacterial pathogens. Nat Rev Microbiol 2 : 510–518. 15152207
3. Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, et al. (2011) Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480 : 241–244. doi: 10.1038/nature10571 22037308
4. Barlow M (2009) What antimicrobial resistance has taught us about horizontal gene transfer. Methods Mol Biol 532 : 397–411. doi: 10.1007/978-1-60327-853-9_23 19271198
5. Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG (2003) Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci USA 100 : 15670–15675. 14676319
6. Soanes D, Richards TA (2014) Horizontal gene transfer in eukaryotic plant pathogens. Annual Review of Phytopathology 52 : 583–614. doi: 10.1146/annurev-phyto-102313-050127 25090479
7. Zhaxybayeva O, Doolittle WF (2011) Lateral gene transfer. Current Biology 21: pR242–R246.
8. Heath TA, Hedtke SM, Hillis DM (2008) Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol 46 : 239–257.
9. James TY, Pelin A, Bonen L, Ahrendt S, Sain D, et al. (2013) Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr Biol 23 : 1548–1553. doi: 10.1016/j.cub.2013.06.057 23932404
10. Lin K, Limpens E, Zhang Z, Ivanov S, Saunders DGO, et al. (2014) Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet 10: e1004078. doi: 10.1371/journal.pgen.1004078 24415955
11. Sun G, Yang Z, Kosch T, Summers K, Huang J (2011) Evidence for acquisition of virulence effectors in pathogenic chytrids. BMC Evolutionary Biology 11 : 195. doi: 10.1186/1471-2148-11-195 21740557
12. Campbell MA, Staats M, van Kan JA, Rokas A, Slot JC (2013) Repeated loss of an anciently horizontally transferred gene cluster in Botrytis. Mycologia 105 : 1126–1134. doi: 10.3852/12-390 23921237
13. Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, et al. (2014) MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Research 42: D699–D704. doi: 10.1093/nar/gkt1183 24297253
14. Jain R, Rivera MC, Lake JA (1999) Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci USA 96 : 3801–3806. 10097118
15. Pal C, Papp B, Lercher MJ (2005) Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet 37 : 1372–1375. 16311593
16. Cohen O, Gophna U, Pupko T (2011) The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol Biol Evol 28 : 1481–1489. doi: 10.1093/molbev/msq333 21149642
17. Friesen TL, Stukenbrock EH, Liu Z, Meinhardt S, Ling H, et al. (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet 38 : 953–956. 16832356
18. Richards TA, Soanes DM, Jones MDM, Vasieva O, Leonard G, et al. (2011) Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci USA 108 : 15258–15263. doi: 10.1073/pnas.1105100108 21878562
19. Klosterman SJ, Subbarao KV, Kang S, Veronese P, Gold SE, et al. (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog 7: e1002137. doi: 10.1371/journal.ppat.1002137 21829347
20. Tsaousis AD, Ollagnier de Choudens S, Gentekaki E, Long S, Gaston D, et al. (2012) Evolution of Fe/S cluster biogenesis in the anaerobic parasite Blastocystis. Proc Natl Acad Sci USA 109 : 10426–10431. doi: 10.1073/pnas.1116067109 22699510
21. Greene GH, McGary KL, Rokas A, Slot JC (2014) Ecology drives the distribution of specialized tyrosine metabolism modules in fungi. Genome Biol Evol 6 : 121–132. doi: 10.1093/gbe/evt208 24391152
22. Khaldi N, Wolfe KH (2011) Evolutionary origins of the fumonisin secondary metabolite gene cluster in Fusarium verticillioides and Aspergillus niger. International Journal of Evolutionary Biology 2011. Epub 2011 May 23.
23. Patron NJ, Waller RF, Cozijnsen AJ, Straney DC, Gardiner DM, et al. (2007) Origin and distribution of epipolythiodioxopiperazine (ETP) gene clusters in filamentous ascomycetes. BMC Evol Biol 7 : 174. 17897469
24. Elmore MH, McGary KL, Wisecaver JH, Slot JC, Geiser DM, et al. (2015) Clustering of two genes putatively involved in cyanate detoxification evolved recently and independently in multiple fungal lineages. Genome Biol Evol 7 : 789–800. doi: 10.1093/gbe/evv025 25663439
25. Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, et al. (2007) Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315 : 207–212. 17218520
26. Strese A, Backlund A, Alsmark C (2014) A recently transferred cluster of bacterial genes in Trichomonas vaginalis-lateral gene transfer and the fate of acquired genes. BMC Evol Biol 14 : 119. doi: 10.1186/1471-2148-14-119 24898731
27. Whitaker JW, McConkey GA, Westhead DR (2009) The transferome of metabolic genes explored: analysis of the horizontal transfer of enzyme encoding genes in unicellular eukaryotes. Genome Biology 10:R36. doi: 10.1186/gb-2009-10-4-r36 19368726
28. Wisecaver JH, Slot JC, Rokas A (2014) The evolution of fungal metabolic pathways. PLoS Genet 10: e1004816. doi: 10.1371/journal.pgen.1004816 25474404
29. Sun B-F, Xiao J-H, He S, Liu L, Murphy RW, et al. (2013) Multiple interkingdom horizontal gene transfers in Pyrenophora and closely related species and their contributions to phytopathogenic lifestyles. PLoS ONE 8: e60029. doi: 10.1371/journal.pone.0060029 23555871
30. Richards TA, Talbot NJ (2013) Horizontal gene transfer in osmotrophs: playing with public goods. Nat Rev Micro 11 : 720–727.
31. Savory F, Leonard G, Richards TA (2015) The role of horizontal gene transfer in the evolution of the oomycetes. PLoS Pathog 11: e1004805. doi: 10.1371/journal.ppat.1004805 26020232
32. Zhao H, Xu C, Lu H-L, Chen X, St. Leger RJ, et al. (2014) Host-to-pathogen gene transfer facilitated infection of insects by a pathogenic fungus. PLoS Pathog 10: e1004009. doi: 10.1371/journal.ppat.1004009 24722668
33. Pombert J-F, Haag KL, Beidas S, Ebert D, Keeling PJ (2015) The Ordospora colligata genome: evolution of extreme reduction in microsporidia and host-to-parasite horizontal gene transfer. mBio 6: e02400–14 doi: 10.1128/mBio.02400-14 25587016
34. Gardiner DM, McDonald MC, Covarelli L, Solomon PS, Rusu AG, et al. (2012) Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PLoS Pathog 8: e1002952. doi: 10.1371/journal.ppat.1002952 23028337
35. Slot JC, Hibbett DS (2007) Horizontal transfer of a nitrate assimilation gene cluster and ecological transitions in fungi: a phylogenetic study. PLoS ONE 2: e1097. 17971860
36. Slot JC, Rokas A (2010) Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. Proc Natl Acad Sci USA 107 : 10136–10141. doi: 10.1073/pnas.0914418107 20479238
37. Selman M, Pombert JF, Solter L, Farinelli L, Weiss LM, et al. (2011) Acquisition of an animal gene by microsporidian intracellular parasites. Curr Biol 21: R576–577. doi: 10.1016/j.cub.2011.06.017 21820617
38. Armijos Jaramillo VD, Vargas WA, Sukno SA, Thon MR (2013) Horizontal transfer of a subtilisin gene from plants into an ancestor of the plant pathogenic fungal genus Colletotrichum. PLoS ONE 8: e59078. doi: 10.1371/journal.pone.0059078 23554975
39. Ford Doolittle W (1998) You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends in Genetics 14 : 307–311. 9724962
40. Walther BA, Ewald PW (2004) Pathogen survival in the external environment and the evolution of virulence. Biol Rev Camb Philos Soc 79 : 849–869. 15682873
41. Parmesan C, Yohe G (2003) A globally coherent fingerprint of climate change impacts across natural systems. Nature 421 : 37–42. 12511946
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized VirusČlánek Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell ProliferationČlánek Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite InterfaceČlánek Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 10- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Expression of Concern: Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Preparing for the Next Epidemic with Basic Virology
- Effectively Communicating the Uncertainties Surrounding Ebola Virus Transmission
- Translating Basic Research into Clinical Applications: Malaria Research at an NIH Lab
- A Gut Odyssey: The Impact of the Microbiota on Spore Formation and Germination
- Papillomavirus E6 Oncoproteins Take Common Structural Approaches to Solve Different Biological Problems
- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Dimensions of Horizontal Gene Transfer in Eukaryotic Microbial Pathogens
- Addressing the Complications of Ebola and Other Viral Hemorrhagic Fever Infections: Using Insights from Bacterial and Fungal Sepsis
- Time for Chocolate: Current Understanding and New Perspectives on Cacao Witches’ Broom Disease Research
- Ganglioside and Non-ganglioside Mediated Host Responses to the Mouse Polyomavirus
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Structure Elucidation of Coxsackievirus A16 in Complex with GPP3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses
- CD39 Expression Identifies Terminally Exhausted CD8 T Cells
- Abiotic Stresses Antagonize the Rice Defence Pathway through the Tyrosine-Dephosphorylation of OsMPK6
- Dissociation of Tissue Destruction and Bacterial Expansion during Bubonic Plague
- Interferon-γ: The Jekyll and Hyde of Malaria
- CCR2 Inflammatory Dendritic Cells and Translocation of Antigen by Type III Secretion Are Required for the Exceptionally Large CD8 T Cell Response to the Protective YopE Epitope during Infection
- A New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1
- The Suramin Derivative NF449 Interacts with the 5-fold Vertex of the Enterovirus A71 Capsid to Prevent Virus Attachment to PSGL-1 and Heparan Sulfate
- Trans-generational Immune Priming Protects the Eggs Only against Gram-Positive Bacteria in the Mealworm Beetle
- Peripheral Vγ9Vδ2 T Cells Are a Novel Reservoir of Latent HIV Infection
- Respiratory Syncytial Virus Disease Is Mediated by Age-Variable IL-33
- TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized Virus
- Modeling the Effects of Vorinostat Reveals both Transient and Delayed HIV Transcriptional Activation and Minimal Killing of Latently Infected Cells
- Identification of a Novel Lipoprotein Regulator of Spore Germination
- Calcium Regulation of Hemorrhagic Fever Virus Budding: Mechanistic Implications for Host-Oriented Therapeutic Intervention
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
- Comparative Life Cycle Transcriptomics Revises Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates
- The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy
- Carcinogenic Parasite Secretes Growth Factor That Accelerates Wound Healing and Potentially Promotes Neoplasia
- Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell Proliferation
- Dengue Virus Infection of Requires a Putative Cysteine Rich Venom Protein
- Distinct Viral and Mutational Spectrum of Endemic Burkitt Lymphoma
- Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite Interface
- Phenotypic and Functional Alterations in Circulating Memory CD8 T Cells with Time after Primary Infection
- Systematic Identification of Cyclic-di-GMP Binding Proteins in Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems
- Influenza Transmission in the Mother-Infant Dyad Leads to Severe Disease, Mammary Gland Infection, and Pathogenesis by Regulating Host Responses
- Myeloid Cell Arg1 Inhibits Control of Arthritogenic Alphavirus Infection by Suppressing Antiviral T Cells
- The White-Nose Syndrome Transcriptome: Activation of Anti-fungal Host Responses in Wing Tissue of Hibernating Little Brown Myotis
- Influenza Virus Reassortment Is Enhanced by Semi-infectious Particles but Can Be Suppressed by Defective Interfering Particles
- Identification of the Mechanisms Causing Reversion to Virulence in an Attenuated SARS-CoV for the Design of a Genetically Stable Vaccine
- Differentiation-Dependent KLF4 Expression Promotes Lytic Epstein-Barr Virus Infection in Epithelial Cells
- The Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways
- C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in
- Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
- Crystal Structure of the Human Cytomegalovirus Glycoprotein B
- Depletion of . GlmU from Infected Murine Lungs Effects the Clearance of the Pathogen
- Immunologic Control of Papillomavirus Type 1
- Requires Host Rab1b for Survival in Macrophages
- Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi
- PD-L1 Expression on Retrovirus-Infected Cells Mediates Immune Escape from CD8 T Cell Killing
- Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction
- IL-4 Induced Innate CD8 T Cells Control Persistent Viral Infection
- Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
- BCG Skin Infection Triggers IL-1R-MyD88-Dependent Migration of EpCAM CD11b Skin Dendritic cells to Draining Lymph Node During CD4+ T-Cell Priming
- Antigenic Characterization of the HCMV gH/gL/gO and Pentamer Cell Entry Complexes Reveals Binding Sites for Potently Neutralizing Human Antibodies
- Rescue of a Plant Negative-Strand RNA Virus from Cloned cDNA: Insights into Enveloped Plant Virus Movement and Morphogenesis
- Geminivirus Activates to Accelerate Cytoplasmic DCP2-Mediated mRNA Turnover and Weakens RNA Silencing in
- Disruption of Sphingolipid Biosynthesis Blocks Phagocytosis of
- The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps
- The Timing of Stimulation and IL-2 Signaling Regulate Secondary CD8 T Cell Responses
- Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity
- The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Interferon-γ: The Jekyll and Hyde of Malaria
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



