-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Systematic Identification of Cyclic-di-GMP Binding Proteins in Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems
Cyclic-di-GMP (c-di-GMP) is a ubiquitous bacterial signaling molecule that regulates important bacterial functions, including virulence, antibiotic resistance, biofilm formation and cell division. The list of known c-di-GMP receptors is clearly incomplete. Here we utilized a systematic and unbiased biochemical approach to identify c-di-GMP receptors from the 3,812 genes of the Vibrio cholerae genome. Results from this analysis identified most known c-di-GMP receptors as well as MshE, a protein not known to interact with c-di-GMP. The c-di-GMP binding site was identified at the N-terminus of MshE and requires a conserved arginine residue in the 9th position. MshE is the ATPase that powers the secretion of the MshA pili onto the surface of the bacteria. We show that c-di-GMP binding to MshE is required for MshA export and the function of the pili in attachment and biofilm formation. ATPases responsible for related processes such as type IV pili and type II secretion were also tested for c-di-GMP binding, which identified the P. aeruginosa ATPase PA14_29490 as another c-di-GMP binding protein. These findings reveal a new class of c-di-GMP receptor and raise the possibility that c-di-GMP regulate membrane complexes through direct interaction with related type II secretion and type IV pili ATPases.
Published in the journal: Systematic Identification of Cyclic-di-GMP Binding Proteins in Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems. PLoS Pathog 11(10): e32767. doi:10.1371/journal.ppat.1005232
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005232Summary
Cyclic-di-GMP (c-di-GMP) is a ubiquitous bacterial signaling molecule that regulates important bacterial functions, including virulence, antibiotic resistance, biofilm formation and cell division. The list of known c-di-GMP receptors is clearly incomplete. Here we utilized a systematic and unbiased biochemical approach to identify c-di-GMP receptors from the 3,812 genes of the Vibrio cholerae genome. Results from this analysis identified most known c-di-GMP receptors as well as MshE, a protein not known to interact with c-di-GMP. The c-di-GMP binding site was identified at the N-terminus of MshE and requires a conserved arginine residue in the 9th position. MshE is the ATPase that powers the secretion of the MshA pili onto the surface of the bacteria. We show that c-di-GMP binding to MshE is required for MshA export and the function of the pili in attachment and biofilm formation. ATPases responsible for related processes such as type IV pili and type II secretion were also tested for c-di-GMP binding, which identified the P. aeruginosa ATPase PA14_29490 as another c-di-GMP binding protein. These findings reveal a new class of c-di-GMP receptor and raise the possibility that c-di-GMP regulate membrane complexes through direct interaction with related type II secretion and type IV pili ATPases.
Introduction
Cyclic diguanosine monophosphate (c-di-GMP) is a ubiquitous bacterial nucleotide secondary signaling molecule that regulates cellular processes in response to environmental and cellular stimuli. The elements of this canonical pathway of signal production, signal transduction, altered activity and signal removal were elegantly described by the Benziman lab over twenty-five years ago [1, 2]. C-di-GMP is synthesized by diguanylate cyclases (DGCs) via a catalytic GGDEF domain [1, 3, 4]. Once made in the cell, c-di-GMP binds to macromolecule receptors to allosterically alter their activities. C-di-GMP signaling is terminated through hydrolysis by phosphodiesterases (PDEs) that contain catalytic EAL or HD-GYP domains [5–8]. In the characterization of bacterial cellulose synthase in Komagataeibacter xylinus, the Benziman lab demonstrated the importance of c-di-GMP in the allosteric activation of the cellulose synthase complex [1, 9, 10]. Recent structure elucidation of the BcsA-BcsB-c-di-GMP complex validated these early finding and provided a molecular mechanism for c-di-GMP activation of cellulose biosynthesis [11]. Since this initial description of c-di-GMP regulation of cellulose biosynthesis, genome sequencing has revealed genes for DGCs in diverse bacteria indicating that c-di-GMP is a ubiquitous and important signaling molecule in prokaryotes that regulates a variety of phenotypes [12].
The identification of receptor proteins for c-di-GMP is needed for understanding the regulation by this ubiquitous signaling molecule. However, the process of identifying c-di-GMP binding proteins has been challenging for several reasons. First, c-di-GMP simultaneously regulates complex traits including promoting biofilm formation, inhibiting motility and additional pathways [13–15] indicating that there are likely many c-di-GMP receptors in the cell. Second, although there are several defined protein domains that bind c-di-GMP (see below), these domains do not accurately predict c-di-GMP binding proteins. For example, the PilZ domain binds c-di-GMP [16], but the PilZ protein in P. aeruginosa, for which the domain is named, does not bind c-di-GMP [17]. In V. cholerae, there are five PilZ domain proteins, but these five proteins do not fully explain all of the observed c-di-GMP regulated effects [18]. Third, c-di-GMP binds a number of proteins that do not have predicted binding motifs or were predicted to bind a different ligand. Examples of novel c-di-GMP binding proteins include VpsT [19], VpsR [20] and FlrA [21] in V. cholerae as well as FleQ in P. aeruginosa [22]. In addition, the Clp protein in Xanthomoas species, a homolog of the cAMP receptor protein (CRP) of E. coli, binds c-di-GMP rather than cAMP [23, 24]. Another example of a c-di-GMP receptor that was not predicted is the BldD of Streptomyces coelicolor binds two dimers of c-di-GMP to repress transcription [25]. Together, these studies reveal the diversity of cellular targets of c-di-GMP that mediate complex regulation and the highlight the challenges in identifying the c-di-GMP-binding receptors that are responsible for c-di-GMP regulation.
Many approaches have been utilized to identify c-di-GMP binding proteins. The first proteins shown to bind c-di-GMP included the enzymes that make and degrade c-di-GMP. The I-site of DGCs binds c-di-GMP to provide product feedback inhibition of DGC activity [4, 26]. Enzymatically active or inactive PDEs are also capable of c-di-GMP binding [5, 27–30]. Bioinformatic studies revealed that the PilZ domain is a c-di-GMP binding domain [16]. In addition, targeted and unbiased approaches have been employed to identify c-di-GMP receptor proteins. In the targeted approach, genes of c-di-GMP regulated processes were tested for c-di-GMP binding [19–25, 31, 32]. These studies led to the discoveries of the c-di-GMP receptor proteins that have not been predicted by bioinformatics and have motivated identification of novel c-di-GMP receptor proteins using systematic approaches. Affinity pull-down assays using c-di-GMP conjugated sepharose resin, biotin, or a tripartite c-di-GMP capture compound enriched c-di-GMP binding proteins from whole cell lysates, which were subsequently identified by mass spectrometry [33–35]. This approach has also been employed to identify binding proteins of another prokaryotic cyclic dinucleotide, cyclic-di-AMP (c-di-AMP) [36–38]. An alternative unbiased approach utilizes the Differential Radial Capillary Action of Ligand Assay (DRaCALA) to systematically screen protein expression libraries for ligand binding activity. DRaCALA relies on the differential spreading of bound and unbound radiolabeled ligand when mixed with protein and spotted on a nitrocellulose membrane [39]. In addition, DRaCALA allowed direct detection of c-di-GMP receptors expressed in E. coli whole cell lysates thus enabling the screening of individual genes from a target genome [39]. This approach has been used on Staphylococcus aureus and Escherichia coli open reading frame (ORF) libraries to identify c-di-AMP and c-di-GMP binding proteins, respectively [36, 40].
Vibrio cholerae was chosen as an organism for a DRaCALA based screen of c-di-GMP binding proteins since an open reading frame library was available [41] and it is an organisms that extensively utilize c-di-GMP signaling system to regulate motility, biofilm formation, pathogenesis, and survival upon dissemination to environmental reservoirs [42–46]. The V. cholerae O1 El Tor N16961 genome encodes 62 proteins with domains for c-di-GMP metabolism including 31 GGDEF, 13 EAL, 9 GGDEF+EAL, and 8 HD-GYP domain proteins [47–49]. However, only 5 c-di-GMP receptor proteins have been identified, including 2 PilZ domain proteins PlzC and PlzD [18], and 3 transcription factors VpsT, VpsR, and FlrA [19–21]. From the DRaCALA screen of the V. cholerae open reading library, a number of predicted c-di-GMP binding proteins were identified. In addition, MshE, an ATPase in the mannose sensitive hemagglutinin (MSHA) type IV pilus operon, was also revealed as a c-di-GMP receptor. Purified MshE specifically binds c-di-GMP with a high affinity (Kd approximately 2 μM). Screening of related type II secretion and type IV pili ATPases identified a gene in P. aeruginosa, PA14_29490, as a c-di-GMP receptor. Through fragmentation of MshE and site-directed mutagenesis of the conserved residues, the arginine at position 9 was identified as a residue required for c-di-GMP binding. Complementation of ΔmshE mutants with wild type mshE restored c-di-GMP regulation of motility, pilus production, and biofilm formation. In contrast, complementation with mshE R9A failed to restore c-di-GMP regulation of MshA pilus function. These results define a novel set of c-di-GMP-binding ATPases associated with type IV pili and type II secretion systems and demonstrate the utility of a DRaCALA screen for identification of c-di-GMP receptor proteins.
Results
DRaCALA screening of a V. cholerae ORFeome for c-di-GMP binding activity
We sought to systematically identify protein receptors of c-di-GMP using DRaCALA by individually testing Vibrio cholerae ORFs expressed in E. coli whole cell lysates for 32P-c-di-GMP binding activity. The 3,812 unique ORFs from the V. cholerae O1 El Tor N16961 ORFeome pDONR plasmids were recombined into gateway-compatible histidine (His-ORF) or His-maltose binding protein (His-MBP-ORF) expression vectors in a single Gateway reaction [50] and selected on agar plates containing either carbenicillin or gentamicin, respectively. For each ORF, multiple transformants were inoculated into a single well of a 96-well microtiter plate to create His-ORF and His-MBP-ORF libraries from which whole cell lysates were generated. Protein expression in whole cell lysates was tested for 348 ORFs by PAGE separation and revealed by staining with Coomassie. A band corresponding to the predicted molecular weight was visualized for 49% of His-ORF and 76% of His-MBP-ORF fusions for a combined coverage of 81% of the V. cholerae ORFeome. These results indicate that most V. cholerae proteins are overexpressed in the His-ORF and His-MBP-ORF libraries, thus enabling a systematic genome-wide DRaCALA screen for c-di-GMP binding proteins.
Whole cell lysates from His-ORF and His-MBP-ORF libraries were tested for c-di-GMP binding by DRaCALA using a 96 well pin tool. The fraction bound of 32P-c-di-GMP was measured in duplicate for each whole cell lysate and positive V. cholerae ORFs were defined as those having c-di-GMP fraction bound three standard deviations above the mean for both measurements (Fig 1, S1 Table) (see Material and Methods). The positive control expressing PelD, a known c-di-GMP-binding protein, was above the cutoff in each 96-well plate. This primary screen identified 55 His-ORF and 47 His-MBP-ORF proteins that significantly increased the fraction bound of 32P-c-di-GMP. A secondary screen was performed to validate these ORFs. In total, 23 His-ORFs and 22 His-MBP-ORFs (28 unique ORFs total) were validated as positive for c-di-GMP binding (Fig 1, Table 1). The specificity for c-di-GMP binding of cell lysates expressing positive ORFs was determined by competition experiments using unlabeled guanosine nucleotides (Table 1). Unlabeled c-di-GMP significantly reduced 32P-c-di-GMP binding for ORFs listed in Table 1. In contrast, unlabeled GTP and cGMP did not reduce 32P-c-di-GMP binding, suggesting that the measured binding activity was specific for c-di-GMP. Together these results illustrate how sequential high-throughput DRaCALA screens can identify genetic elements that encode proteins with specific ligand binding activity. This screen of the V. cholerae ORFeome for c-di-GMP binding proteins identified known and candidate c-di-GMP receptors.
Fig. 1. Primary DRaCALA screen of Vibrio cholerae ORF libraries. 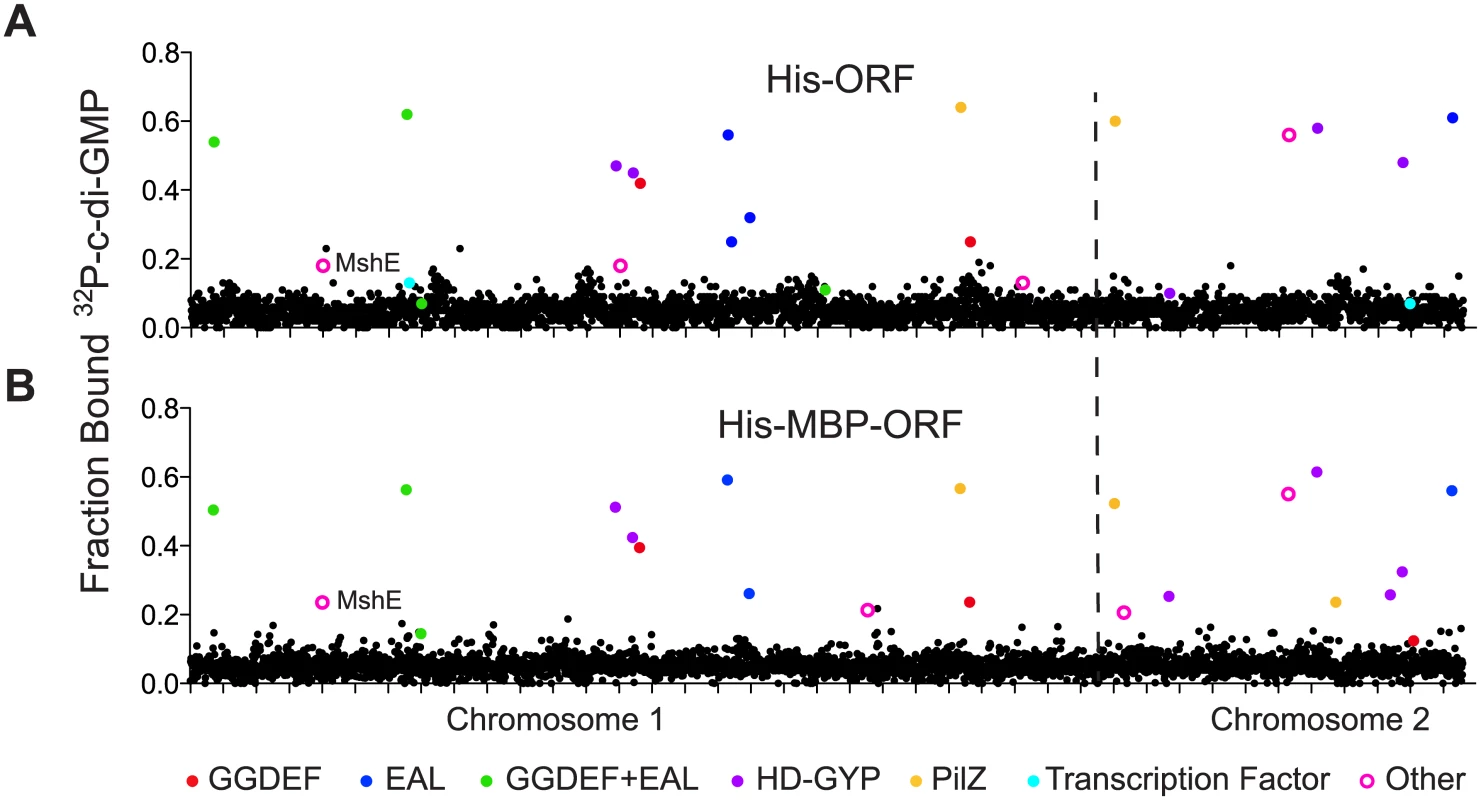
Average fraction bound 32P-c-di-GMP vs. individual V. cholerae (A) His-ORFs and (B) His-MBP-ORFs overexpressed in E. coli whole cell lysates. ORFs are arranged by VC gene number along the X-axis. ORFs validated as positive in the secondary screen are indicated by color and classified by type of c-di-GMP-binding protein. “Other” refers to proteins that have not been predicted or demonstrated to bind c-di-GMP. Tab. 1. Specific binding of validated ORFs to c-di-GMP. 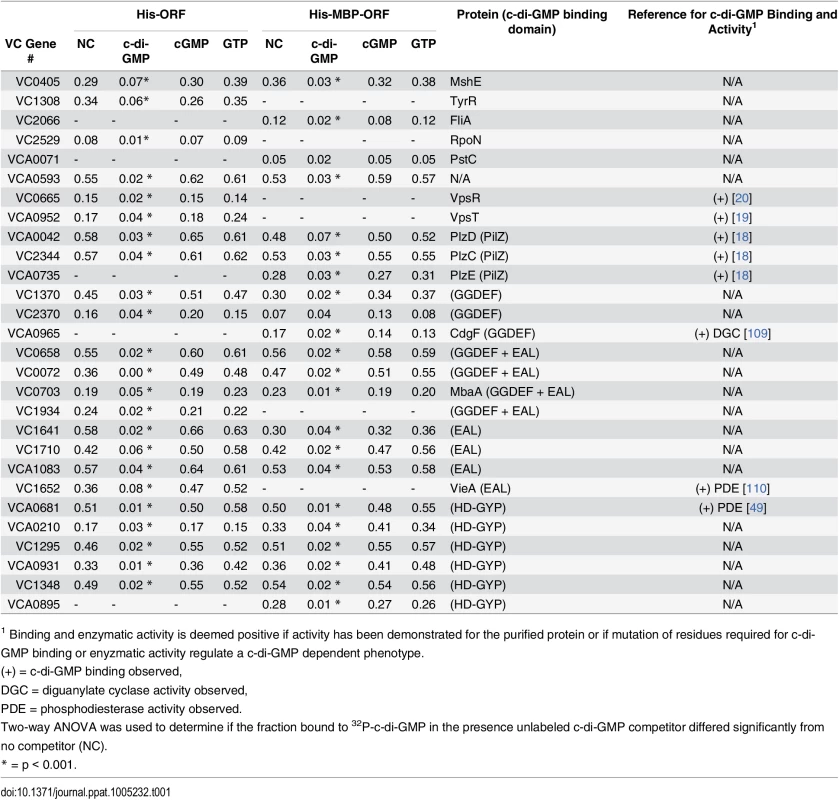
1 Binding and enzymatic activity is deemed positive if activity has been demonstrated for the purified protein or if mutation of residues required for c-di-GMP binding or enyzmatic activity regulate a c-di-GMP dependent phenotype. Positive ORFs encode c-di-GMP binding proteins
The ORFs identified in our DRaCALA-based ORFeome screen included several proteins with either predicted or demonstrated c-di-GMP binding activity. The list of 28 positive ORFs include three with GGDEF domains, four with EAL, four with both GGDEF and EAL, six with HD-GYP, three with PilZ, two encoding known c-di-GMP binding transcription factors, and six proteins lacking a defined c-di-GMP binding domain (Table 1). Of the proteins containing only the GGDEF domain, three ORFs identified contain the RxxD motif required for c-di-GMP binding (VC1370, VC2370, VCA0965 (CdgF)), which is present in only 11 of the 37 ORFs with GGDEF domains (S1 Fig). Four ORFs (VC0072, VC0658, VC0703 (MbaA), VC1934) were identified with both GGDEF and EAL domains, but lacking the RxxD motif, suggesting that c-di-GMP binding occurs at the EAL domain. Both EAL and HD-GYP domains can bind c-di-GMP as a substrate for hydrolysis. The screen identified four ORFs containing an EAL domain (VC1641, VC1652 (VieA), VC1710, VCA1083) and six ORFs containing a HD-GYP domain (VC1295, VC1348, VCA0210, VCA0681, VCA0895, VCA0931). The V. cholerae genome contains 5 ORFs that encode PilZ domains. While four of these (PlzA, -C, -D, and -E) retain RxxxR and DxSxxG motifs required for c-di-GMP binding, only PlzC (VC2344) and PlzD (VCA0042) have been demonstrated to bind c-di-GMP biochemically [18, 51]. From the DRaCALA-based ORFeome screen, PlzC and PlzD were identified in both His-ORF and His-MBP-ORF libraries, while PlzE (VCA0735) was identified only in the His-MBP-ORF library. Previous work demonstrated c-di-GMP binding for His-fusions of PlzC and PlzD but not PlzE, suggesting that a MBP fusion to PlzE may be required for proper folding of the c-di-GMP binding site during heterologous expression of PlzE [52, 53]. Finally, two of three c-di-GMP binding transcription factors, VpsT (VCA0952) [19] and VpsR (VC0665) [20] were identified, but not FlrA (VC2137) [21]. In total, DRaCALA identified 22 of 46 (48%) proteins predicted to bind c-di-GMP and 6 of 11 (55%) proteins previously shown to bind c-di-GMP (Table 2). These results demonstrate that DRaCALA-based screen can identify all known categories of c-di-GMP binding proteins and represents an unbiased approach to discovering receptor proteins of signaling molecules.
Tab. 2. Hit rate of V. cholerae ORFs encoding predicted and previously demonstrated cdiGMP-binding proteins.1 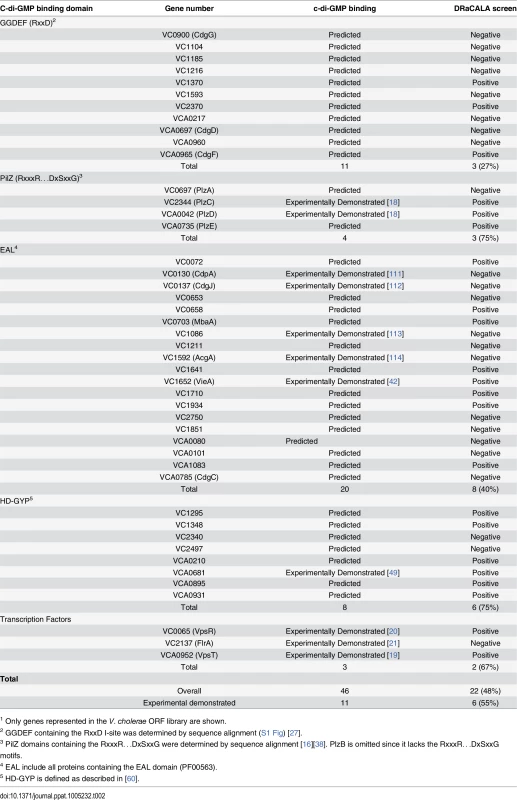
1 Only genes represented in the V. cholerae ORF library are shown. Six ORFs were identified which encode proteins that are not known or predicted to bind c-di-GMP, namely VC0405 (MshE), VC1308 (TyrR), VC2066 (FliA), VC2529 (RpoN), VCA0071 (PstC), and VCA0593 (Table 1). These ORFs represent potentially novel types of c-di-GMP binding proteins. To determine if these proteins bind c-di-GMP directly, His-MBP-ORF fusions were purified and assayed for c-di-GMP binding activity by DRaCALA. C-di-GMP binding was detected for purified MshE, but not for RpoN, FliA, TyrR, or VCA0593 (S2 Fig). We were unable to purify PstC. These results suggest that heterologous expression of RpoN, FliA, TyrR, or VCA0593 can induce the expression of c-di-GMP binding proteins encoded within the E. coli genome. In the remainder of the manuscript, we characterize the c-di-GMP binding properties of MshE.
MshE specifically binds c-di-GMP with high affinity
To determine the affinity and specificity of c-di-GMP-binding to MshE, purified His-MBP-MshE was assayed for binding to 32P-c-di-GMP by DRaCALA. The affinity of c-di-GMP-binding was determined by quantifying the fraction bound of 32P-c-di-GMP in serial dilutions of His-MBP MshE (Fig 2A). Non-linear regression analysis of c-di-GMP-binding vs. protein concentration using a one site-binding model estimated the dissociation constant (Kd) for c-di-GMP to be 1.9 ± 0.4 μM. To determine the specificity of c-di-GMP-binding to MshE, we measured the fraction bound of 32P-c-di-GMP in the presence of unlabeled nucleotide competitors. 32P-c-di-GMP-binding to His-MBP-MshE was significantly decreased by unlabeled c-di-GMP, but not by GTP, GDP, GMP cGMP, ATP, ADP, AMP, cAMP, CTP, or UTP (Fig 2B). MshE also binds ATP specifically since only ATP and ADP compete for 32P-ATP binding (Fig 2C). These results indicate that MshE specifically binds c-di-GMP with micromolar affinity and at a site that is distinct from the ATP binding site.
Fig. 2. C-di-GMP bind to MshE ATPase with high affinity, specificity and independently of ATP. 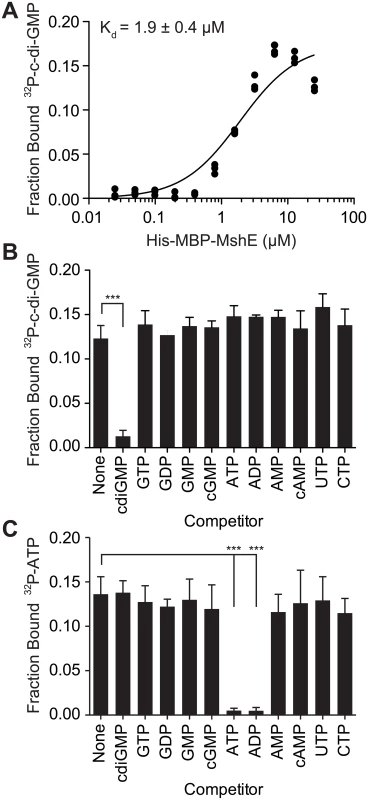
(A) Fraction bound 32P-c-di-GMP to decreasing concentrations of purified His-MBP-MshE. The dissociation constant (Kd) is indicated. Fraction bound of His-MBP-MshE to (B) 32P-c-di-GMP and (C) 32P-ATP in the presence of 100 μM nucleotide competitors. P-value was determined by two-tailed t-test (*** p≤0.001). All data are average of three independent assays and standard deviation is indicated by error bars. A subset of ATPases that regulate type IV pili and type II secretion systems bind c-di-GMP
MshE belongs to a family of ATPases associated with the biosynthesis and retraction of type IV pili and secretion by type II secretion systems. These phylogenetically related ATPases are called as T2SSE ATPases based on the type II secretion system (T2SS) nomenclature for General secretion protein E (GspE) [54, 55]. To determine if c-di-GMP-binding is a conserved feature of T2SSE ATPases, we identified homologs of MshE and assayed them for c-di-GMP-binding. Protein-Blast search with the full-length MshE amino acid sequence against the complete protein sets of V. cholerae O1 El Tor N16961 and P. aeruginosa PA14 identified five additional ATPases in V. cholerae and nine in P. aeruginosa with an E value < 1x10-15. These ATPases include those required for type IV pili function: PilB (VC2424 and PA14_58750,) PilT (PA14_05180 and PA14_59340), PilU (PA14_05190), and TcpT (VC0835); and type II secretion: GspE (VC2732), XcpR (PA14_23990), HxcR (PA14_55440), and HxrA (PA14_29490) [56–63]. We constructed His-ORF fusions for each V. cholerae and P. aeruginosa T2SSE ATPase and assayed c-di-GMP-binding by DRaCALA in E. coli whole cell lysate (Fig 3A). Expression of V. cholerae MshE and P. aeruginosa PA14_29490, but not other T2SSE ATPases, significantly increased the fraction bound of 32P-c-di-GMP. These data identified MshE and PA14_29490 as c-di-GMP-binding receptor proteins, the only ones among the T2SSE family in V. cholerae and P. aeruginosa.
Fig. 3. C-di-GMP binds to V. cholerae and P. aeruginosa homologs of MshE. 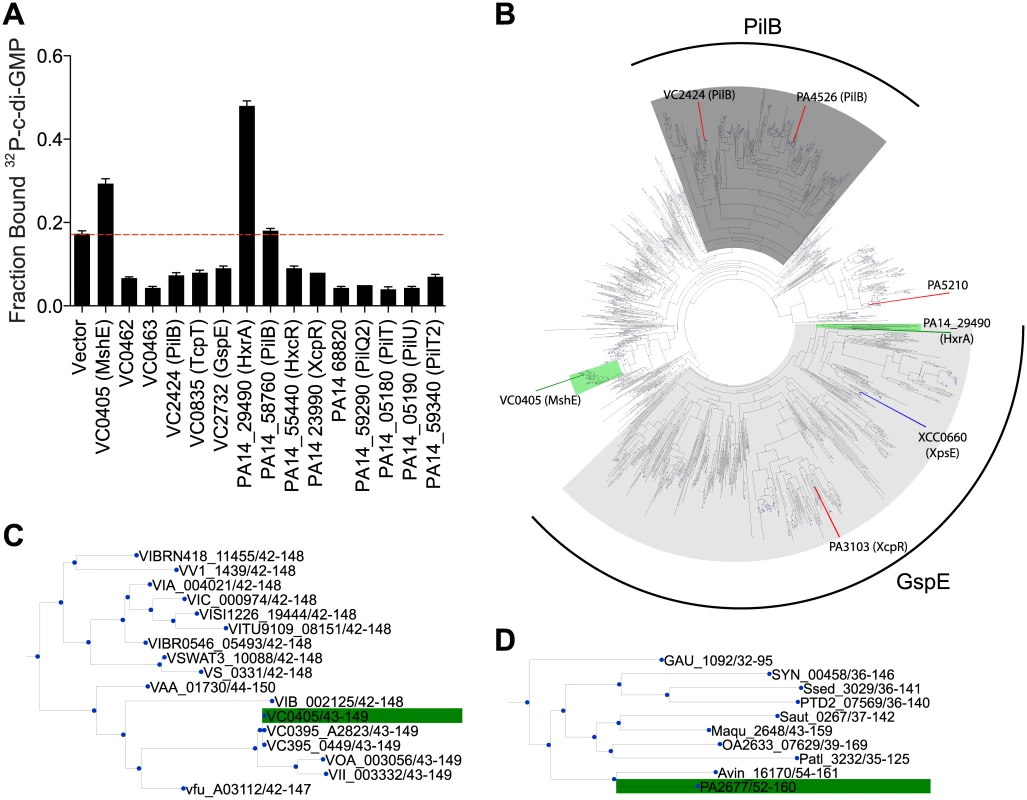
(A) Average fraction bound 32P-c-di-GMP of E. coli whole cell lysate expressing Vibrio cholerae and Pseudomonas aeruginosa homologs of MshE. The dashed red line indicates background binding for a vector control strain. All data are average of three independent assays and standard deviation is indicated by error bars. (B) Unrooted phylogenetic tree of the T2SSE_N domain. Protein sequences present in the tree corresponding to proteins analyzed for c-di-GMP binding are highlighted in green (binds c-di-GMP), red (does not bind), or blue (candidate binding protein). The dark grey background corresponds to primarily type IV pili PilB sequences, and the light grey background corresponds to type II secretion protein E ATPase sequences. (C, D) Sub-trees containing VC0405 or PA14_29490 and closely related proteins. PA14_29490 is the reciprocal best BLAST hit for MshE [55]. Based on genomic context, MshE functions within the MSHA operon [64, 65], while PA14_29490 is encoded within a putative T2SS operon [66, 67]. Sequence comparison of T2SSE-type ATPases from V. cholerae and P. aeruginosa showed that they differ substantially in protein length. MshE and PA14_29490, both 575 aa long, were ~200 aa longer than PilT ATPases VC0462, VC0463, PA14_05180, PA14_05190, PA14_58760 and PA14_59340. Other T2SSE-type ATPases, annotated as GspE - and PilB-type, ranged in length from 469 to 562 aa, and only PA14_68820 was the same length as MshE. All these enzymes had very similar C-terminal ATPase domains and differed primarily in their N-terminal parts, with longer proteins containing an additional domain, referred to as T2SSE_N (PF05157) domain in the Pfam database [48]. A phylogenetic tree of the N-terminal fragments of T2SSE_N-containing ATPases showed that MshE is located in the branch related to PilB ATPases responsible for type IV pili, whereas PA14_29490 is located in the branch related to GspE ATPases, which participate in type II secretion (Fig 3B). The canonical PilB and GspE ATPases that were tested (VC2424, VC2732 and their counterparts in P. aeruginosa) belong to branches of the tree that are distinct from those including MshE and PA14_29490. Expansion of the branches containing MshE (Fig 3C) and PA14_29490 (Fig 3D) reveal many homologous proteins that may also be receptors of c-di-GMP. Together, these results suggest that a subset of T2SSE ATPases represented by MshE and PA14_29490 are c-di-GMP-binding proteins.
PA14_29490 and MshE bind c-di-GMP specifically in the N-terminal domain
Both MshE and PA14_29490 lack known c-di-GMP binding protein sequence motifs. To locate the binding site(s) on these proteins, truncation analysis was performed on both proteins. The N-terminal T2SSE_N domain and C-terminal T2SSE ATPase domain of PA14_29490 and MshE were separated at three points that were predicted to be at the ends of secondary structural elements (Fig 4A and 4E). These fragments were expressed in E. coli and the whole cell lysates were tested for c-di-GMP binding. The N-terminal fragments (F1-F3) of both PA14_29490 and MshE bound 32P-c-di-GMP, while the C-terminal fragments (F4-F6) did not (Fig 4B and 4F). Purified fragment 1 of PA14_29490 binds c-di-GMP with a Kd of 480 ± 60 nM (Fig 4C) and this binding was competed away only with c-di-GMP (Fig 4D), indicating that binding is specific. Each of the fragments of MshE was purified and tested for binding to 32P-c-di-GMP. Only the purified N-terminal fragments (F1-F3) of MshE bound 32P-c-di-GMP, while the C-terminal fragments (F4-F6) did not (Fig 4G). These results indicate that the binding site for c-di-GMP is located in the N-terminal domain of the protein and is distinct from the ATPase domain in the C-terminus.
Fig. 4. C-di-GMP binds MshE and PA14_29490 in the N-terminal T2SSE_N domain. 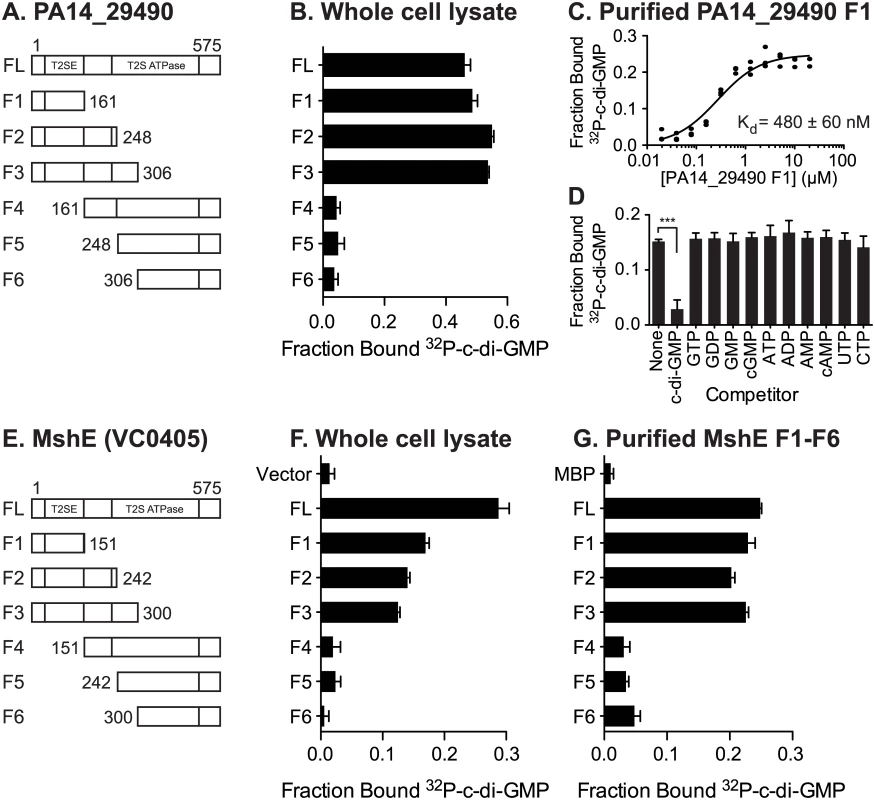
Schematic of the truncations generated in (A) PA14_29490 and (E) MshE. C-di-GMP binding to E. coli whole cell lysates expressing each fragment of (B) PA14_29490 and (F) MshE. (C) Fraction bound 32P-c-di-GMP to decreasing concentrations of purified PA14_29490 fragment 1. (D) Fraction bound of purified PA14_29490 fragment 1 to 32P-c-di-GMP in the presence of 100 μM nucleotide competitors. (G) Fraction bound of 32P-c-di-GMP to purified MshE and fragments 1–6. Indicated p-value was determined by two-tailed t-test (*** p≤0.001) by comparing the indicated samples. All data are average of three independent assays and standard deviation is indicated by error bars. MshE binding to c-di-GMP requires a specific arginine residue
We hypothesized that c-di-GMP regulation of MSHA is an evolutionarily conserved property. To test this idea, we identified MshE homologs from genomes containing MSHA-like operons as defined by having the mshN gene upstream of mshE and the mshG gene downstream of mshE (S3A Fig). ClustalW alignment revealed residues within the first 151 amino acids that are 100% identical among these proteins, including 4 conserved motifs: RLGDLLV, ARRxRAL, SDPADL, and DxxYRRT (S3B Fig). Since charged residues, in particular arginines, have been shown to participate in c-di-GMP binding in a variety of receptor proteins, mutant variants with alanine replacements within these 4 motifs were generated by site-directed mutagenesis including R9A/D12A, R88A/R89A, D108A/D111A, and R146A/R147A (Fig 5A). As a control, the conserved E191 and D192 residues, which are located outside of the first 151 amino acid fragment, were also changed to alanine. Purified R9A/D12A, R88A/R89A, and D108A/D111A variants were reduced for binding to c-di-GMP, whereas R146A/R147A and E191A/D192A did not affect binding to c-di-GMP (Fig 5A). These results indicate that motifs 1, 2 and 3 contribute to c-di-GMP binding, while motif 4 is dispensable. The E191A/D192A variant binds c-di-GMP similarly to the wild-type protein (Fig 5A), in agreement with the results from fragment analysis (Fig 4E). To determine the contribution of each amino acid residue within the first 3 motifs and the other conserved, charged amino acids, we generated and purified MshE variants with single alanine substitution in positions R9, D12, Q32, E51, R88, R89, D108, D111 and D142 (S4A Fig). The R9A variant had an 83% reduction in c-di-GMP binding, while Q32A and R88A variants had 61% and 50% reduction, respectively (Fig 5C). Interestingly, the R9/D12 residues represent an RxxD motif described in DGC I-site [26], PelD [31] and GIL domain of BcsE [40]. In those proteins, both residues are critical for c-di-GMP binding. In contrast, the MshE D12A variant actually binds c-di-GMP better than wild-type MshE (Fig 5C), indicating that MshE does not contain an I-site-like binding sequence. Each of these MshE variants was also tested for ATP binding. The R88A variant showed an 88% reduction in ATP binding and was the only variant that had a reduction by more than 50% (S4B Fig). Thus, the defect associated with c-di-GMP binding in R88A variant may be due to a general folding problem for this specific protein. Together, these results indicate MshE contains a novel c-di-GMP binding site that requires R9 residue with contribution from the Q32 residue.
Fig. 5. R9 is required for MshE to bind c-di-GMP. 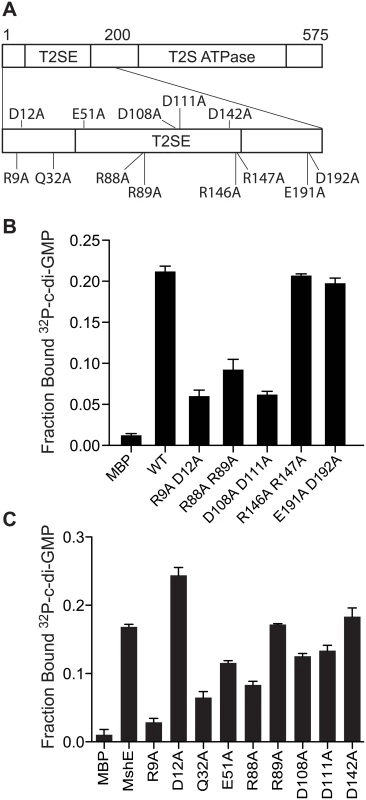
(A) Schematic of the charged and polar conserved residues in the first 200 amino acids of MshE targeted for site-directed alanine substitution. (B) Fraction bound of 32P-c-di-GMP to purified MshE (WT) and indicated pairs of alanine substitutions. (C) Fraction bound of 32P-c-di-GMP to purified MshE (WT) and indicated individual alanine substitution. All data are average of three independent assays and standard deviation is indicated by error bars. MshE binding to c-di-GMP enhances ATPase activity
The effect of c-di-GMP on the ATPase activity of MshE was assessed by testing the WT MshE and the R9A proteins in the presence and absence of c-di-GMP. WT MshE produced 68 μM of phosphate from ATP without c-di-GMP and increased to 75 and 76 μM with the addition of 10 and 33 μM of c-di-GMP, respectively (S5 Fig). In contrast, R9A protein produced 60, 64, and 58 μM of phosphate with addition of 0, 10, and 33 μM of c-di-GMP. These results indicate that the ATPase activity is increased for WT MshE in response to c-di-GMP at concentrations above the dissociation constant, while c-di-GMP had no effect on the R9A protein. However, the magnitude of the enhanced ATPase activity is only about 10% suggesting that c-di-GMP may have additional effects on MshE interaction with the MshA substrate or other components of the MSHA export machinery.
C-di-GMP binding to MshE is required for MSHA function in attachment and biofilm formation
MSHA is a responsible for initial attachment of V. cholerae to surfaces and subsequent biofilm formation [68]. Recently, MSHA was demonstrated to reduce flagella mediated swimming motility [69]. We sought to determine whether MshE is the c-di-GMP receptor regulating MSHA activity by assessing the amount of MshA pili on the bacterial surface, the effect on swimming motility, and biofilm formation. The effect of MshE on the export of MshA to the surface of V. cholerae was assessed by surface ELISA using antibodies specific to MshA (Fig 6A, WT and ΔmshA). The ΔmshE mutant is defective for MshA export (Fig 6A). This defect was complemented by wild type mshE and the D12A allele, but not the R9A allele (Fig 6A). Complementation with the R88A/R89A allele was able to restore the export of ~10% of MshA observed in wild type cells. A recent study revealed that V. cholerae swimming motility is reduced by interaction of MSHA with surfaces [69]. V. cholerae was assessed for motility through soft agar assay. Wild type V. cholerae had reduced swimming motility and the ΔmshE mutant has increased motility (Fig 6B, WT and ΔmshE vector) recapitulating previous observations [69]. The ability of either wild-type mshE or mshE variant under the tac promoter in trans on a plasmid to complement ΔmshE phenotypes were evaluated. Induction of either allele that binds c-di-GMP, wild-type mshE or the D12A variant, reduced motility to wild type levels (Fig 6B). Induction of the variant defective for c-di-GMP binding (R9A) failed to reduce motility (Fig 6B). The R88A/R89A allele, that had reduced binding to c-di-GMP and ATP, also failed to reduce motility. Additionally, the requirement for MshE to bind c-di-GMP on biofilm formation was assessed using a flow cell system. At 4 hours post-inoculation, wild-type was able to attach to the surface, whereas fewer ΔmshE mutants attached to the surface (Fig 6C). Complementation of ΔmshE mutants with wild type mshE and the D12A allele restored attachment, whereas complementation with either R9A or R88A/R89A allele did not (Fig 6C). At 24 hours post-inoculation, the initial differences in attachment were more pronounced. Wild type and ΔmshE mutant complemented with either mshE or D12A formed mature biofilm (Fig 6D). Complementation with the R88A/R89A allele also restored biofilm formation indicating a small amount of surface MshA can restore MSHA activity. In contrast, the ΔmshE mutant complemented with R9A showed small patches of biofilms similar to ΔmshE with the vector control (Fig 6D). Quantification of the biofilms revealed that both biomass (Fig 6E) and surface coverage (Fig 6F) are reduced for ΔmshE complemented with R9A. Together, these results indicate that the ability of MshE to bind c-di-GMP via the R9 residue is required for MshA export to the cell surface and MSHA-mediated phenotypes.
Fig. 6. MshE interaction with c-di-GMP is essential for MSHA function. 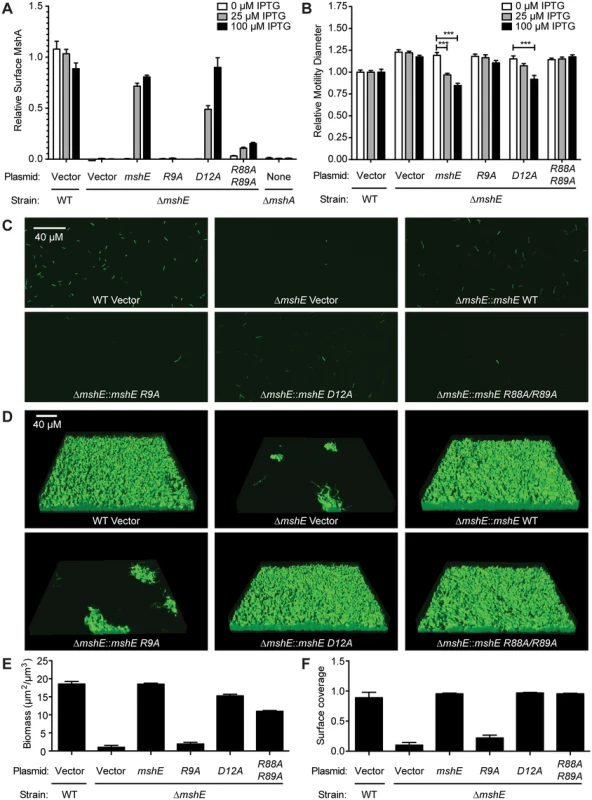
(A) Surface MshA pilin production under varying IPTG induction determined via ELISA with MshA-specific antibody. Two biological replicates were assayed in duplicate and normalized to the average of the WT strain with vector. (B) Expression of mshE reduces motility in soft agar. The diameters of migration zones were measured after 16 hours of growth at 30°C with or without IPTG induction and normalized to the motility of the WT strain harboring the empty vector. Three biological replicates were performed in duplicate. Induced strains were compared to uninduced strains with ANOVA followed by Bonferroni Multiple Comparison test. (** p≤0.01, *** p≤0.001). (C) Attachment phenotypes observed with confocal microscopy 4 h post inoculation in a flow cell system. Scale bar represents 40μm. (D) Three-dimensional biofilm structures observed with confocal microscopy 24 h post inoculation in a flow cell system. Scale bar represents 40μm. (E,F) COMSTAT quantitative analysis of biomass and surface coverage of biofilms from (D). Three images from each of two independent experiments were analyzed. Discussion
Evaluation of DRaCALA ORF screens for identification of ligand binding proteins
DRaCALA screens are a relatively new method for identifying ligand binding proteins that combines arrayed protein libraries with a high-throughput biochemical assay of protein-ligand interactions. Two recent publications have successfully used DRaCALA screening to identify novel cyclic-dinucleotide protein receptors. A screen for c-di-AMP binding proteins in the Staphylococcus aureus strain COL ORFeome identified PstA and KdpD [36]. The screen also identified KtrA, which was the only protein identified by affinity pull-down using c-di-AMP magnetic beads in the same study [36]. The crystal structures of PstA-c-di-AMP [70–72] and KtrA-c-di-AMP complexes [73] have been solved. KdpD binding motif has been recently identified [74]. Since the DRaCALA screen, one other additional c-di-AMP receptor has been identified in Gram-positive bacteria, pyruvate carboxylase of Listeria monocytogenes [38]. The S. aureus pyruvate carboxylase has different residues within the binding motif than the L. monocytogenes and does not bind c-di-AMP [38]. Together these studies demonstrate that DRaCALA was able to identify three novel bona-fide c-di-AMP receptors and correctly not detect a protein that does not bind c-di-AMP. Additionally, a DRaCALA screen for c-di-GMP binding proteins in the E. coli K12 ASKA overexpression gene library revealed three clones overexpressing putative c-di-GMP binding proteins: BcsE, IlvH and RimO [40]. Of these, BscE was further characterized and shown to contain a novel c-di-GMP binding domain, which the authors named GIL [40]. In conjunction with the results from these screens, our identification of MshE as a c-di-GMP receptor important for MSHA pilus function in V. cholerae demonstrates that DRaCALA is a powerful approach for finding new small-molecule receptors across different bacterial species.
The results of these three DRaCALA screening experiments allow us to further assess the limitations of the screen. Our screen of the V. cholerae ORFeome identified many, but not all, of the expected binding proteins. These false negatives could be due to several factors. First, the enzymatic activity of these proteins may prevent detection. Phosphodiesterases active in the assay conditions could degrade the 32P-c-di-GMP probe prior to application on nitrocellulose, while active diguanylate cyclases could produce excess c-di-GMP to compete for binding with the subsequently added 32P-c-di-GMP probe. The failure to detect binding in many EAL and GGDEF domain proteins in our screen, as well as the lack of binding detected for all I-site-containing GGDEF proteins assayed in the E. coli DRaCALA screen [40], could be due to these factors. Second, poor expression of the ORF can result in a false negative because DRaCALA relies on the expression of proteins above the Kd. In our assay of protein expression, we found that only a subset of the ORFs tested had a protein band of the correct size on SDS-PAGE. Similarly, some false negatives of c-di-GMP binding proteins in the E. coli screen were due to poor protein expression as detected by protein band intensity on SDS-PAGE [40]. Third, the DRaCALA screen interrogates binding of individually expressed ORFs within a heterologous system. Thus proteins that require endogenous binding partners or activating factors to interact with ligand may not be active. Our screen also yielded false positives, which could be due to two factors. First, the expression of proteins that can activate the production of c-di-GMP binding proteins encoded in the E. coli genome can result in a positive signal. Second, the statistical method utilized to identify “positive” fraction bound may falsely identify proteins whose fraction bounds are near the cutoff, as was likely the case for Adk, a false positive result in the S. aurus c-di-AMP screen [36]. Both these categories of false positives can be detected after re-assaying binding with purified protein.
We suggest several methods to increase the fidelity of DRaCALA-based screens: 1. Testing the ORFs with multiple fusion proteins can increase the likelihood of overexpressing recombinant proteins that retain ligand binding activity, 2. Express the ORFeome in a strain genetically modified to remove endogenous c-di-GMP signaling components and thus reduce the likelihood of false positives [75, 76], and 3. Alter the buffer used to resuspend lysates—in the case of EAL domain PDE-As, resuspension in a buffer containing Ca2+ rather than Mg2+ can inhibit the activity of PDE-As [2] and increase the likelihood of detecting proteins that degrade c-di-GMP. Although DRaCALA did not identify all c-di-GMP proteins in the V. cholerae genome, DRaCALA is an unbiased approach that allows discovery of novel receptor protein such as MshE. We believe DRaCALA-based approach can be a powerful tool for the discovery of novel receptor proteins of other small nucleotide signaling molecules.
MshE is the founding member of a new class of c-di-GMP receptor
The MshE is a bona fide c-di-GMP binding protein was demonstrated by 1. High affinity binding with the Kd of 1.9 μM, 2. High specificity of binding based on competition assays, 3. A defined binding site located in the N-terminal T2SSE_N domain and 4. The requirement of the conserved R9 residue for c-di-GMP binding in vitro and for MSHA function in vivo. MshE represents a new category of c-di-GMP binding proteins since it lacks any of the previously defined c-di-GMP binding domains (DGC I-sites, EAL, HD-GYP, PilZ or GIL) and is the first T2SSE ATPase demonstrated to bind c-di-GMP. The conserved R9 and D12 residues are reminiscent of the RxxD c-di-GMP binding motif present in the I-site of DGCs or the RxGD binding motif present in GIL domains. For both DGC I-sites and GIL domains, the R and D residues are required for c-di-GMP binding. In contrast, MshE requires only the R9 residue for binding to c-di-GMP, while the D12 residue of MshE is dispensable. In addition to MshE, only one other type II secretion/type IV pili ATPase, PA14_29490 from P. aeruginosa, bound c-di-GMP. Other members of this subfamily likely will also have the ability to bind c-di-GMP, including XpsE from X. campestris (Fig 3B, blue line). In contrast, related ATPases including PilT and PilU are unlikely to be regulated by c-di-GMP since they are shorter and lack the T2SSE_N domain. Although MshE and PA14_29490 both bind c-di-GMP in the N-terminal T2SSE_N domain, their sequence conservation within this domain is quite low. Nonetheless, proteins containing the T2SSE_N domain should be investigated for their ability to interact with c-di-GMP (Fig 3B, 3C and 3D).
Predicted structure of the c-di-GMP binding fragment of MshE
Two T2SSE_N domain of T2SSE ATPases have been structurally characterized including N-terminal fragment of V. cholerae GspE (Protein Data Bank (PDB) entry 2BH1_X) [77] and a related domain from Xanthomonas campestris (PDB: 2D27, 2D28) [78]. Sequence alignment the N-terminal 151-aa fragment of MshE to PA14_29490, V. cholerae GspE and X. campestris GspE by the Conserved Domain Database [79] revealed that the V. cholerae GspE lacks the R9 and Q32 residues. However, these residues were conserved in the GpsE/XpsEN domain from X. campestris (S6A Fig). Remarkably, the X. campestris XpsEN was crystallized in two different forms that reflect two distinct conformational states that differ in the position of two N-terminal helices [78], which include the R9 and Q32 residues (S6B Fig). In one of the structures (PDB: 2D27), R9 and Q32 are positioned within a reasonable distance from each other and, upon the rotation of first helix, could form a potential c-di-GMP-binding site (S6B Fig). In the other structure (PDB: 2D28), the region around Q32 proved so flexible that the exact position of Q32 could not be resolved [78], which is consistent with the ability of this residue to move around and participate in ligand binding. Such flexibility has also been observed in the binding sites of PilZ-containing c-di-GMP receptors [18, 51]. The conformational change of the two N-terminal helices of MshE upon binding c-di-GMP has the potential to alter its activity. Thus, type II secretion/type IV pili ATPases with the conserved arginine and glutamine residues corresponding to R9 and Q32 of MshE represent candidate c-di-GMP receptors (S6B Fig).
Implications of MshE binding to c-di-GMP in the regulation MSHA during the V. cholerae infection cycle
V. cholerae cycles between environmental reservoirs and human infections in part using two type IV pili, MSHA and Tcp. MSHA contributes to the V. cholerae infection cycle by maintaining an environmental reservoir through attachment to chitin surfaces [80, 81] and tolerance to environmental hypotonic stress [82] through biofilm formation [83]. Upon entry into the next host, the preformed biofilms protect V. cholerae from lower pH [84] and bile [85]. Subsequently, V. cholerae express toxin co-regulated pili (Tcp) to promote colonization [86, 87] while concomitantly turning off MSHA [88]. The repression of MSHA within the host is critical since a strain with the msha promoter replaced with the constitutive Plac promoter was outcompeted by the wild type strain in an infant mouse infection model [88]. This defect can be attributed to binding of secretory IgA directly to MSHA since these strains do not show a competitive difference in mice lacking IgA [88] indicating that repression of MSHA during infection is a necessary for immune evasion. Based on recombination-based in vivo expression technology (RIVET) studies [46], tcpA is transcribed while the msh operon is repressed early in the infection; in contrast, the expression pattern is reversed at the late stage of infection. The decrease in msh transcription early in the infection is likely regulated in part by the reduced levels of c-di-GMP since several DGCs are not expressed until late in the infection [46]. However, the precise mechanism for regulation of the c-di-GMP signal is subject to strain specificity [49, 89] and changes in the host microenvironment. Another mechanism to reciprocally regulate Tcp and MSHA is from the TcpJ pre-pilin peptidase. TcpJ has the unique property of processing both TcpA [90] and MshA [91], but cleavage of MshA by TcpJ leads to rapid degradation [91]. Our finding that MshE binding to c-di-GMP is required for MSHA production and function adds another mechanism to enhance the switch between Tcp and MSHA pili. Upon entering the host, the c-di-GMP levels are reduced, thus reducing both transcription of the msh operon and the function of existing export machinery to export MshA. In combination with the degradation of newly synthesized MshA by TcpJ, V. cholerae can quickly change from the immunogenic MSHA to the adhesive Tcp in the host. Late in the infection, this regulation is reversed allowing the bacteria exiting the host to express MSHA instead of Tcp to prepare for the environment.
Implications of MshE on c-di-GMP regulation of type IV pili and type II secretion systems
Binding of MshE to c-di-GMP activates MSHA dependent phenotypes including 1. Production of MshA pili to the cell surface, 2. MSHA reduction of flagella motility, 3. Adherence to surfaces and biofilm formation. These studies show that all of these effects are lost in the mshE R9A mutant that is defective in binding to c-di-GMP. The implication of our results is that c-di-GMP binding to MshE is required for its activity in polymerizing MshA. This finding is intriguing since several type IV pili are regulated by c-di-GMP through different mechanisms. In both Xanthomonas and Pseudomonas spp., type IV pili are regulated by PilZ and FimX proteins [92–95]. However, the precise mechanisms of regulation appear to be different. In X. axonopodis citri and X. campestris, PilZ binds FimX and the PilB ATPase, a homolog of MshE, to form a tripartite regulatory complex, but these interactions are not conserved in P. aeruginosa [94, 96, 97]. In addition to regulating the PilB ATPase, c-di-GMP interacts with a second PilZ-domain protein XC_2249 to regulate interactions the PilT and PilU retraction ATPases in X. campestris [98]. These studies and our MshE results indicate that c-di-GMP regulates type IV pili ATPases through a number of different mechanisms. The ability of PA14_29490 to bind c-di-GMP indicates that there may be also complex regulation of type II secretion systems by c-di-GMP.
Sustained sensing of c-di-GMP through multi-tiered regulation
C-di-GMP regulates major lifestyle changes in response to altered environmental cues. These changes incur a high cost in expenditure of cellular resources as well as an opportunity cost of committing to a sessile lifestyle. There is an emerging pattern in which c-di-GMP regulates the same phenotype at the transcriptional and post-translational levels. Examples of this include Pel polysaccharide synthesis in P. aeruginosa [31, 99], and now MSHA in V. cholerae [100]. The idea of regulation by c-di-GMP first to activate gene expression and later to activate protein function can be thought of as “sustained sensing”. Sustained sensing enables bacteria to repeatedly assess environmental and cellular conditions through multi-tiered regulation, and has been previously described for responses to iron availability, oxidative stress, and other signals [101, 102]. The concept of sustained signaling applied to c-di-GMP enables prediction of additional c-di-GMP receptors. In addition to known examples, there are additional c-di-GMP-regulated processes that could be regulated by sustained sensing. These include two classes: 1. Operons that are transcriptionally regulated by c-di-GMP, but lack known c-di-GMP receptor proteins and 2. Operons that encode c-di-GMP receptor proteins, but lack known c-di-GMP transcriptional regulation. Examples of the first class include a number of biosynthetic operons of extracellular polysaccharides, such Vps and Eps in V. cholerae [19, 100, 103], Psl in P. aeruginosa [22], xagABC in X. campestris [32, 104], and Bcam1330-Bcam1341 in Burkholderia cenocepacia [105]. These operons also likely encode a c-di-GMP binding protein to regulate polysaccharide biosynthesis at the post-translational level. Examples of the second class include cellulose synthesis in E. coli [40], the alginate biosynthesis operon in P. aeruginosa [17] and the large adhesin protein (Lap) of P. fluorescens [28]. Furthermore, c-di-GMP can also bind to riboswitches [106, 107] to provide another regulatory tier in sustained sensing. Therefore, based on the emerging theme of sustained sensing of c-di-GMP through multi-tiered regulation, we suggest that newly discovered processes regulated by c-di-GMP either transcriptionally or post-translationally be investigated for additional levels of c-di-GMP control.
Materials and Methods
Gateway destination vector construction
pVL791 Cb GW and pVL847 Gn GW were constructed as destination vectors in LR reactions (Life Technologies) for cloning VC ORFs. pVL791 Cb and pVL847 Gn are pET-19 derivatives that are carbenecillin or gentamycin resistant and produce N-terminal His - and His-MBP fusions respectively. The gateway destination cassette was amplified from pRFA and cloned in frame with the N-terminal fusions to produce the gateway adapted vectors.
His-ORF and His-MBP-ORF expression library construction
The V. cholerae N16961 ORF library was obtained from BEI. LR Clonase reactions were performed per NEB protocol using miniprepped pDONR vectors from the V. cholerea ORFeome in combination with pVL791 Cb GW and pVL847 Gn GW destination vectors. Gateway reactions were transformed into an E. coli T7IQ strain (NEB) and recombinants were selected on LB agar plates containing either carbenecillin or gentamycin. Multiple colonies from individual transformations were inoculated in LB M9 rich media in 96-well plate format and grown overnight with shaking at 30°C. Overnight cultures were subcultured 1 : 50 into fresh media and grown for 4 hours at 30°C with shaking. Protein expression was induced by addition of 1 mM IPTG and cultures were grown for an additional 4 hours after induction. 1.5 mL of induced culture was centrifuged and cells were resuspended in 150 μL of c-di-GMP-binding buffer supplemented with 10 μg / mL DNAse, 250 μg / mL Lysozyme and 1 μM PMSF. 20 μL aliquots were transferred to 96-well microtiter plates and stored at -80°C.
DRaCALA
Whole cell lysates for DRaCALA screening were prepared by freeze-thawing resuspended cells in microtiter plates a total of 3 times. After the final thaw, 20 μL of c-di-GMP-binding buffer supplemented with 16 pM 32P-c-di-GMP and 500 mM unlabeled GTP was added to whole cell lysate plates or purified proteins. 2 μL of this mixture was then spotted in duplicate on nitrocellulose using a 96-well pin tool. DRaCALA of purified proteins was performed with concentrations of protein and unlabeled competitor as indicated. Spots were allowed to dry completely (about 20 minutes) before exposing a phosphorimager screen and capturing with a Fujifilm FLA-7000. Photostimulated luminescence (PSL) from the inner spot and total PSL of the spot were quantitated with Fuji Image Gauge software. The fraction bound was calculated using measurements of the total area (Aouter), the area of the inner circle (Ainner), the total PSL intensity (Itotal), and the inner intensity (Iinner) as described [39]. DRaCALA was also used to determine the ability of purified MshE and variants to bind c-di-GMP. Dissociation constants were estimated assuming a one-site binding model by a nonlinear regression of protein concentration and fraction bound where, Fraction bound = (Maximum possible Fraction Bound) * [protein concentration] / (Kd + [protein concentration]).
Identification of positive V. cholerae ORFs in primary and secondary DRaCALA screens
In the primary screen of His-ORF and His-MBP-ORF libraries, mixtures of whole cell lysate and radiolabeled 32P-c-di-GMP were spotted twice. To identify DRaCALA spots with significantly increased fraction bound 32P-c-di-GMP, a positive cutoff three standard deviations above the mean fraction bound was created for each 96-well plate of whole cell lysates. Positive spots were iteratively removed from calculations of mean and standard deviation for individual plates, thereby decreasing the positive cutoff until no additional positives were identified. His-ORFs and His-MBP-ORFs for which both DRaCALA spots had positive binding were defined as positive and subjected to a secondary screen. Additionally, 8 His-ORF and 8 His-MBP-ORFs with only 1 DRaCALA spot with positive binding were also included in the secondary screen, but none of these ORFs were positive for c-di-GMP binding in the secondary screen.
For each primary positive His-ORF and His-MBP-ORF, 8 whole cell lysates were generated from individual clones that were isolated from the pooled transformants used to create the His-ORF and His-MBP-ORF libraries. Each His-ORF and His-MBP-ORF lysate was spotted twice by DRaCALA and individual lysates were compared to a set of 8 lysates generated from vector controls. Positive fraction bound 32P-c-di-GMP for DRaCALA spots was defined as those with at least 2 fold increase above the average fraction bound for the set of plate-matched vector controls. Additionally, each lysate was assayed by PCR to verify the size of the inserted ORF. Positive ORFs from this secondary screen displayed positive fraction bound 32P-c-di-GMP for both DRaCALA spots created from PCR positive lysates.
Generation of phylogenetic tree
Protein sequences corresponding to COG2840 were obtained from the EggNOG 4.1 database. Each sequence aligned to Pfam Family PF05157 (Type II Secretion System, protein E, N-terminal domain) using the version 10 HMM with HMMer 3.1 and the subsequences corresponding to the T2SSE N-terminal domain were extracted. The remaining 1437 domain sequences were aligned using the MAFFT 7.157b E-INS-i algorithm and trimmed using TrimAl 1.4 to eliminate columns with more than 90% gaps. An unrooted phylogenetic tree was constructed using FastTree 2.1.8.
Strains and plasmids
Strains used are listed in S2 Table. Plasmids are listed in S3 Table.
Generation of MshE fragments and site-directed alanine substituted variants
Fragments of MshE and PA14_29490 were generated using the primers indicated in S4 Table. Site-directed alanine substitutions of MshE were generated by PCR amplification with the indicated primers, DpnI digest and transformation into E. coli DH5α. pVL791-MshER88A, pVL791-MshER89A, pVL791-MshED111A were generated using the primers indicated in S3 Table and the NEB Q5 Site-Directed Mutagenesis Kit. All constructs were verified by DNA sequencing.
Protein expression and purification
V. cholerae MshE and variants were purified as previously described [39]. Briefly, E. coli T7Iq strains or E. coli BL21(DE3) containing expression plasmids were grown overnight, subcultured in fresh media and grown to OD600~1.0 when expression was induced with 1 mM IPTG. Induced bacteria were pelleted and resuspended in 10 mM Tris pH 8, 100 mM NaCl, 25 mM imidazole and frozen at -80°C until purification. Proteins were purified over a Ni-NTA column and eluted with 10 mM Tris pH 8, 100 mM NaCl, 250 mM imidazole. Purified proteins were exchanged into 10 mM Tris pH 8, 100 mM NaCl using Sephadex G25. Proteins were aliquoted, and frozen at -80°C until use.
Plate motility assay
Motility plates consist of LB containing 0.3% agar supplemented with 20μg/mL ampicillin and 25 or 100μM IPTG where appropriate. Plates were poured and allowed to dry at room temperature for 4 h prior to inoculation. Colonies from overnight LB agar plates grown at 30°C were transferred to motility plates and incubated for 16 h at 30°C. Motility diameter was measured and normalized to the average of WT on each plate. Experiments were performed with three biological replicates in triplicate and data were analyzed with a Oneway ANOVA followed by Dunnett’s multiple comparison test.
Confocal laser scanning microscopy (CLSM) and flow cell biofilm studies
Inoculation of flow cells was done by diluting overnight-grown cultures to an OD600 of 0.04 and injecting into a μ-Slide VI0.4 (Ibidi, Martinsried, Germany). To inoculate the flow cell surface, bacteria were allowed to adhere at room temperature for 1 h. Flow of 2% v/v LB (0.02% tryptone, 0.01% yeast extract, 1% NaCl; pH 7.5) containing 20μg/mL ampicillin and 100μM IPTG was initiated at a rate of 7.5 ml/h and continued for 24 h. Confocal images were obtained on a Zeiss LSM 5 PASCAL Laser Scanning Confocal microscope. Images were obtained with a 40X dry objective and were processed using Imaris (Bitplane, Zurich, Switzerland). Quantitative analyses were performed using the COMSTAT software package [108]. Statistical significance was determined using Oneway ANOVA with Dunnett’s Multiple Comparison test. Two biological replicates were performed in triplicate. Images presented are from one representative experiment.
Surface pilin ELISA
Surface pili composed of MshA were quantified using an ELISA based on a previously published protocol [92]. Briefly, overnight culture was diluted 1 : 100 in fresh LB medium and grown to OD600 0.5 at 30°C. Cells (125μL) were added to a 96-well plate (Greiner Bio-One, Monroe, NC) and incubated at 30°C for one hour. Cells were fixed with 100μL of methanol for 10 minutes at room temperature, then washed twice with PBS. Samples were blocked in 5% nonfat dry milk and immunoblotted with polyclonal rabbit anti-MshA (1 : 1000 dilution, gift of J. Zhu) and horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA). After three washes in PBS, 100μL of TMB (eBioscience, San Diego, CA) was added and incubated for 30 minutes at room temperature followed by the addition of 100μL of 2N H2SO4. Absorbance was recorded at 490nm and the samples were normalized to the change in WT. Two biological replicates were assayed in duplicate and statistical significance was determined with a Oneway ANOVA followed by a Dunnett’s Multiple Comparison test.
Protein purification
E. coli BL21 harboring plasmids for gene expression were grown to an OD600 of 0.4 at 30°C in LB containing 100μg/mL ampicillin. Cultures were shifted to 18°C and IPTG was added to a final concentration of 100μM. 16h post induction, cells were harvested by centrifugation at 10,000 x g for 15 minutes and stored at -80°C. Cell pellets were resuspended in GST Lysis Buffer (25mM Tris pH 8.0, 0.5M NaCl containing PI cocktail tablets (Roche Life Science, Indianapolis, IN). Cells were lysed by sonication and cell lysate was cleared via centrifugation. Cleared lysate was loaded onto GS4B resin and washed with five column volumes of lysis buffer. Protein was eluted from the resin in 5mL elution buffer (25mM Tris pH8.0, 0.25M NaCl, 10mM glutathione). Samples were dialyzed against buffer (25mM Tris-HCl, 150mM NaCl, 250μM DTT, pH 7.5) overnight using 12 kDa cutoff dialysis tubing (Fisherbrand, Pittsburgh, PA) and concentrated to approximately 1mL using an Amicon 10KDa cutoff spin filter (EMD Millipore, Darmstadt, Germany). An aliquot of dialyzed protein was diluted in 6M guanidinium HCl and concentration determined via A280.
ATPase assay
ATPase activity of purified proteins was determined by measuring the production of inorganic phosphate from ATP using the Enzchek Phosphate Assay Kit (Invitrogen). The standard reaction mixture was prepared with the addition of 2mM MgCl2, 10mM KCl, and 1mM DTT. Purified protein in buffer (25mM TrisHCl pH 7.5, 100mM NaCl) was added to the standard reaction mixture to a final concentration of 1μM with the indicated concentration of c-di-GMP. After a 10 minute incubation at room temperature, ATP was added to a final concentration of 10mM and reactions were incubated at 22°C for 30 minutes. Production of inorganic phosphate was monitored by reading OD360 and compared to a standard curve of solutions of KH2PO4. The experiment was performed in triplicate. Significance was determined via ANOVA and Bonferroni test.
Supporting Information
Zdroje
1. Ross P, Weinhouse H, Aloni Y, Michaeli D, Ohana P, Mayer R, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325 : 279–81. 18990795
2. Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55(1):35–58. 2030672
3. Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, et al. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18(6):715–27. 15075296
4. Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, et al. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci U S A. 2004;101(49):17084–9. 15569936
5. Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, et al. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180(17):4416–25. 9721278
6. Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459(7249):1015–8. doi: 10.1038/nature07966 19536266
7. Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A. 2006;103(17):6712–7. 16611728
8. Bellini D, Caly DL, McCarthy Y, Bumann M, An SQ, Dow JM, et al. Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol Microbiol. 2014;91(1):26–38. doi: 10.1111/mmi.12447 24176013
9. Ross P, Aloni Y, Weinhouse C, Michaeli D, Weinberger-Ohana P, Meyer R, et al. An unusual guanyl oligonucleotide regulates cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1985;186(2):191–6. 19160595
10. Ross P, Aloni Y, Weinhouse H, Michaeli D, Weinberger-Ohana P, Mayer R, et al. Control of cellulose synthesis Acetobacter xylinum. A unique guanyl oligonucleotide is the immediate activator of the cellulose synthase. Carbohydrate Research. 1986;149(1):101–17.
11. Morgan JL, McNamara JT, Zimmer J. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol. 2014;21(5):489–96. doi: 10.1038/nsmb.2803 24704788
12. Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5 : 35. 15955239
13. Romling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. doi: 10.1128/MMBR.00043-12 23471616
14. Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7(4):263–73. doi: 10.1038/nrmicro2109 19287449
15. Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7(10):724–35. doi: 10.1038/nrmicro2203 19756011
16. Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22(1):3–6. 16249258
17. Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3'-5')-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65(4):876–95. 17645452
18. Pratt JT, Tamayo R, Tischler AD, Camilli A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem. 2007;282(17):12860–70. 17307739
19. Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327(5967):866–8. doi: 10.1126/science.1181185 20150502
20. Srivastava D, Harris RC, Waters CM. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol. 2011;193(22):6331–41. doi: 10.1128/JB.05167-11 21926235
21. Srivastava D, Hsieh ML, Khataokar A, Neiditch MB, Waters CM. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol. 2013;90(6):1262–76. doi: 10.1111/mmi.12432 24134710
22. Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69(2):376–89. doi: 10.1111/j.1365-2958.2008.06281.x 18485075
23. Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol. 2009;191(22):7121–2. doi: 10.1128/JB.00845-09 19633082
24. Tao F, He YW, Wu DH, Swarup S, Zhang LH. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J Bacteriol. 2010;192(4):1020–9. doi: 10.1128/JB.01253-09 20008070
25. Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, et al. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell. 2014;158(5):1136–47. doi: 10.1016/j.cell.2014.07.022 25171413
26. Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, et al. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 2006;281(42):32015–24. 16923812
27. Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203(1):11–21. 11557134
28. Newell PD, Monds RD, O'Toole GA. LapD is a bis-(3',5')-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A. 2009;106(9):3461–6. doi: 10.1073/pnas.0808933106 19218451
29. Navarro MV, De N, Bae N, Wang Q, Sondermann H. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure. 2009;17(8):1104–16. doi: 10.1016/j.str.2009.06.010 19679088
30. Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187(14):4774–81. 15995192
31. Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65(6):1474–84. 17824927
32. Chin KH, Lee YC, Tu ZL, Chen CH, Tseng YH, Yang JM, et al. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol. 2010;396(3):646–62. doi: 10.1016/j.jmb.2009.11.076 20004667
33. Duvel J, Bertinetti D, Moller S, Schwede F, Morr M, Wissing J, et al. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods. 2012;88(2):229–36. doi: 10.1016/j.mimet.2011.11.015 22178430
34. Nesper J, Reinders A, Glatter T, Schmidt A, Jenal U. A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. Journal of proteomics. 2012;75(15):4874–8. doi: 10.1016/j.jprot.2012.05.033 22652488
35. An SQ, Caly DL, McCarthy Y, Murdoch SL, Ward J, Febrer M, et al. Novel cyclic di-GMP effectors of the YajQ protein family control bacterial virulence. PLoS pathogens. 2014;10(10):e1004429. doi: 10.1371/journal.ppat.1004429 25329577
36. Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Grundling A. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A. 2013;110(22):9084–9. doi: 10.1073/pnas.1300595110 23671116
37. Huynh TN, Luo S, Pensinger D, Sauer JD, Tong L, Woodward JJ. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc Natl Acad Sci U S A. 2015;112(7):E747–56. doi: 10.1073/pnas.1416485112 25583510
38. Sureka K, Choi PH, Precit M, Delince M, Pensinger DA, Huynh TN, et al. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell. 2014;158(6):1389–401. doi: 10.1016/j.cell.2014.07.046 25215494
39. Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci U S A. 2011;108(37):15528–33. doi: 10.1073/pnas.1018949108 21876132
40. Fang X, Ahmad I, Blanka A, Schottkowski M, Cimdins A, Galperin MY, et al. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Microbiol. 2014;93(3):439–52. doi: 10.1111/mmi.12672 24942809
41. Rolfs A, Montor WR, Yoon SS, Hu Y, Bhullar B, Kelley F, et al. Production and sequence validation of a complete full length ORF collection for the pathogenic bacterium Vibrio cholerae. Proc Natl Acad Sci U S A. 2008;105(11):4364–9. doi: 10.1073/pnas.0712049105 18337508
42. Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53(3):857–69. 15255898
43. Tischler AD, Camilli A. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect Immun. 2005;73(9):5873–82. 16113306
44. Koestler BJ, Waters CM. Bile acids and bicarbonate inversely regulate intracellular cyclic di-GMP in Vibrio cholerae. Infect Immun. 2014;82(7):3002–14. doi: 10.1128/IAI.01664-14 24799624
45. Lim B, Beyhan S, Meir J, Yildiz FH. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol Microbiol. 2006;60(2):331–48. 16573684
46. Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, Camilli A. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell host & microbe. 2007;2(4):264–77. 18005744
47. Massie JP, Reynolds EL, Koestler BJ, Cong JP, Agostoni M, Waters CM. Quantification of high-specificity cyclic diguanylate signaling. Proc Natl Acad Sci U S A. 2012;109(31):12746–51. doi: 10.1073/pnas.1115663109 22802636
48. Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–30. doi: 10.1093/nar/gkt1223 24288371
49. Hammer BK, Bassler BL. Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. J Bacteriol. 2009;191(1):169–77. doi: 10.1128/JB.01307-08 18952786
50. Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10(11):1788–95. 11076863
51. Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. Embo J. 2007;26(24):5153–66. 18034161
52. Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8(8):1668–74. 10452611
53. Raran-Kurussi S, Waugh DS. The ability to enhance the solubility of its fusion partners is an intrinsic property of maltose-binding protein but their folding is either spontaneous or chaperone-mediated. PloS one. 2012;7(11):e49589. doi: 10.1371/journal.pone.0049589 23166722
54. Pugsley AP. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57(1):50–108. 8096622
55. Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH Jr. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149(Pt 11):3051–72. 14600218
56. Turner LR, Lara JC, Nunn DN, Lory S. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1993;175(16):4962–9. 8102361
57. Turner LR, Olson JW, Lory S. The XcpR protein of Pseudomonas aeruginosa dimerizes via its N-terminus. Mol Microbiol. 1997;26(5):877–87. 9426126
58. Burrows LL. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol. 2012;66 : 493–520. doi: 10.1146/annurev-micro-092611-150055 22746331
59. Robien MA, Krumm BE, Sandkvist M, Hol WG. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J Mol Biol. 2003;333(3):657–74. 14556751
60. Camberg JL, Sandkvist M. Molecular analysis of the Vibrio cholerae type II secretion ATPase EpsE. J Bacteriol. 2005;187(1):249–56. 15601709
61. Camberg JL, Johnson TL, Patrick M, Abendroth J, Hol WG, Sandkvist M. Synergistic stimulation of EpsE ATP hydrolysis by EpsL and acidic phospholipids. EMBO J. 2007;26(1):19–27. 17159897
62. Patrick M, Korotkov KV, Hol WG, Sandkvist M. Oligomerization of EpsE coordinates residues from multiple subunits to facilitate ATPase activity. J Biol Chem. 2011;286(12):10378–86. doi: 10.1074/jbc.M110.167031 21209100
63. Lu C, Turley S, Marionni ST, Park YJ, Lee KK, Patrick M, et al. Hexamers of the type II secretion ATPase GspE from Vibrio cholerae with increased ATPase activity. Structure. 2013;21(9):1707–17. doi: 10.1016/j.str.2013.06.027 23954505
64. Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406(6795):477–83. 10952301
65. Giltner CL, Nguyen Y, Burrows LL. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev. 2012;76(4):740–72. doi: 10.1128/MMBR.00035-12 23204365
66. Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406(6799):959–64. 10984043
67. Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome biology. 2006;7(10):R90. 17038190
68. Watnick PI, Kolter R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol Microbiol. 1999;34(3):586–95. 10564499
69. Utada AS, Bennett RR, Fong JC, Gibiansky ML, Yildiz FH, Golestanian R, et al. Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nature communications. 2014;5 : 4913. doi: 10.1038/ncomms5913 25234699
70. Campeotto I, Zhang Y, Mladenov MG, Freemont PS, Grundling A. Complex structure and biochemical characterization of the Staphylococcus aureus cyclic diadenylate monophosphate (c-di-AMP)-binding protein PstA, the founding member of a new signal transduction protein family. J Biol Chem. 2015;290(5):2888–901. doi: 10.1074/jbc.M114.621789 25505271
71. Muller M, Hopfner KP, Witte G. c-di-AMP recognition by Staphylococcus aureus PstA. FEBS Lett. 2015;589(1):45–51. doi: 10.1016/j.febslet.2014.11.022 25435171
72. Choi PH, Sureka K, Woodward JJ, Tong L. Molecular basis for the recognition of cyclic-di-AMP by PstA, a PII -like signal transduction protein. Microbiologyopen. 2015;4(3):361–74. doi: 10.1002/mbo3.243 25693966
73. Kim H, Youn SJ, Kim SO, Ko J, Lee JO, Choi BS. Structural Studies of Potassium Transport Protein KtrA Regulator of Conductance of K+ (RCK) C domain in Complex with Cyclic Diadenosine Monophosphate (c-di-AMP). J Biol Chem. 2015. 25957408
74. Moscoso JA, Schramke H, Zhang Y, Tosi T, Dehbi A, Jung K, et al. Binding of c-di-AMP to the Staphylococcus aureus sensor kinase KdpD occurs via the USP domain and down-regulates the expression of the Kdp potassium transporter. J Bacteriol. 2015. 26195599
75. Solano C, Garcia B, Latasa C, Toledo-Arana A, Zorraquino V, Valle J, et al. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc Natl Acad Sci U S A. 2009;106(19):7997–8002. doi: 10.1073/pnas.0812573106 19416883
76. Gao X, Mukherjee S, Matthews PM, Hammad LA, Kearns DB, Dann CE 3rd. Functional characterization of core components of the Bacillus subtilis cyclic-di-GMP signaling pathway. J Bacteriol. 2013;195(21):4782–92. doi: 10.1128/JB.00373-13 23893111
77. Abendroth J, Murphy P, Sandkvist M, Bagdasarian M, Hol WG. The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. J Mol Biol. 2005;348(4):845–55. 15843017
78. Chen Y, Shiue SJ, Huang CW, Chang JL, Chien YL, Hu NT, et al. Structure and function of the XpsE N-terminal domain, an essential component of the Xanthomonas campestris type II secretion system. J Biol Chem. 2005;280(51):42356–63. 16162504
79. Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–6. doi: 10.1093/nar/gku1221 25414356
80. Chiavelli DA, Marsh JW, Taylor RK. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl Environ Microbiol. 2001;67(7):3220–5. 11425745
81. Zampini M, Canesi L, Betti M, Ciacci C, Tarsi R, Gallo G, et al. Role for mannose-sensitive hemagglutinin in promoting interactions between Vibrio cholerae El Tor and mussel hemolymph. Appl Environ Microbiol. 2003;69(9):5711–5. 12957968
82. Kamruzzaman M, Udden SM, Cameron DE, Calderwood SB, Nair GB, Mekalanos JJ, et al. Quorum-regulated biofilms enhance the development of conditionally viable, environmental Vibrio cholerae. Proc Natl Acad Sci U S A. 2010;107(4):1588–93. doi: 10.1073/pnas.0913404107 20080633
83. Watnick PI, Fullner KJ, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181(11):3606–9. 10348878
84. Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5(4):647–56. 14536065
85. Hung DT, Zhu J, Sturtevant D, Mekalanos JJ. Bile acids stimulate biofilm formation in Vibrio cholerae. Mol Microbiol. 2006;59(1):193–201. 16359328
86. Tacket CO, Taylor RK, Losonsky G, Lim Y, Nataro JP, Kaper JB, et al. Investigation of the roles of toxin-coregulated pili and mannose-sensitive hemagglutinin pili in the pathogenesis of Vibrio cholerae O139 infection. Infect Immun. 1998;66(2):692–5. 9453628
87. Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64(7):2853–6. 8698524
88. Hsiao A, Liu Z, Joelsson A, Zhu J. Vibrio cholerae virulence regulator-coordinated evasion of host immunity. Proc Natl Acad Sci U S A. 2006;103(39):14542–7. 16983078
89. Beyhan S, Odell LS, Yildiz FH. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J Bacteriol. 2008;190(22):7392–405. doi: 10.1128/JB.00564-08 18790873
90. Kaufman MR, Seyer JM, Taylor RK. Processing of TCP pilin by TcpJ typifies a common step intrinsic to a newly recognized pathway of extracellular protein secretion by gram-negative bacteria. Genes Dev. 1991;5(10):1834–46. 1680773
91. Hsiao A, Toscano K, Zhu J. Post-transcriptional cross-talk between pro - and anti-colonization pili biosynthesis systems in Vibrio cholerae. Mol Microbiol. 2008;67(4):849–60. doi: 10.1111/j.1365-2958.2007.06091.x 18179420
92. Kazmierczak BI, Lebron MB, Murray TS. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol Microbiol. 2006;60(4):1026–43. 16677312
93. McCarthy Y, Ryan RP, O'Donovan K, He YQ, Jiang BL, Feng JX, et al. The role of PilZ domain proteins in the virulence of Xanthomonas campestris pv. campestris. Molecular plant pathology. 2008;9(6):819–24. doi: 10.1111/j.1364-3703.2008.00495.x 19019010
94. Guzzo CR, Salinas RK, Andrade MO, Farah CS. PILZ protein structure and interactions with PILB and the FIMX EAL domain: implications for control of type IV pilus biogenesis. J Mol Biol. 2009;393(4):848–66. doi: 10.1016/j.jmb.2009.07.065 19646999
95. Alm RA, Bodero AJ, Free PD, Mattick JS. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178(1):46–53. 8550441
96. Qi Y, Xu L, Dong X, Yau YH, Ho CL, Koh SL, et al. Functional divergence of FimX in PilZ binding and type IV pilus regulation. J Bacteriol. 2012;194(21):5922–31. doi: 10.1128/JB.00767-12 22942245
97. Chin KH, Kuo WT, Yu YJ, Liao YT, Yang MT, Chou SH. Structural polymorphism of c-di-GMP bound to an EAL domain and in complex with a type II PilZ-domain protein. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 10):1380–92. 22993092
98. Ryan RP, McCarthy Y, Kiely PA, O'Connor R, Farah CS, Armitage JP, et al. Dynamic complex formation between HD-GYP, GGDEF and PilZ domain proteins regulates motility in Xanthomonas campestris. Mol Microbiol. 2012;86(3):557–67. doi: 10.1111/mmi.12000 22924852
99. Baraquet C, Murakami K, Parsek MR, Harwood CS. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 2012;40(15):7207–18. doi: 10.1093/nar/gks384 22581773
100. Beyhan S, Tischler AD, Camilli A, Yildiz FH. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol. 2006;188(10):3600–13. 16672614
101. Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27(2–3):215–37. 12829269
102. Chiang SM, Schellhorn HE. Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch Biochem Biophys. 2012;525(2):161–9. doi: 10.1016/j.abb.2012.02.007 22381957
103. Casper-Lindley C, Yildiz FH. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J Bacteriol. 2004;186(5):1574–8. 14973043
104. Lu XH, An SQ, Tang DJ, McCarthy Y, Tang JL, Dow JM, et al. RsmA regulates biofilm formation in Xanthomonas campestris through a regulatory network involving cyclic di-GMP and the Clp transcription factor. PloS one. 2012;7(12):e52646. doi: 10.1371/journal.pone.0052646 23285129
105. Fazli M, McCarthy Y, Givskov M, Ryan RP, Tolker-Nielsen T. The exopolysaccharide gene cluster Bcam1330-Bcam1341 is involved in Burkholderia cenocepacia biofilm formation, and its expression is regulated by c-di-GMP and Bcam1349. Microbiologyopen. 2013;2(1):105–22. doi: 10.1002/mbo3.61 23281338
106. Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321(5887):411–3. doi: 10.1126/science.1159519 18635805
107. Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329(5993):845–8. doi: 10.1126/science.1190713 20705859
108. Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146 (Pt 10):2395–407. 11021916
109. Hunter JL, Severin GB, Koestler BJ, Waters CM. The Vibrio cholerae diguanylate cyclase VCA0965 has an AGDEF active site and synthesizes cyclic di-GMP. BMC Microbiol. 2014;14 : 22. doi: 10.1186/1471-2180-14-22 24490592
110. Tamayo R, Tischler AD, Camilli A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J Biol Chem. 2005;280(39):33324–30. 16081414
111. Tamayo R, Schild S, Pratt JT, Camilli A. Role of cyclic Di-GMP during El Tor biotype Vibrio cholerae infection: characterization of the in vivo-induced cyclic Di-GMP phosphodiesterase CdpA. Infect Immun. 2008;76(4):1617–27. doi: 10.1128/IAI.01337-07 18227161
112. Liu X, Beyhan S, Lim B, Linington RG, Yildiz FH. Identification and characterization of a phosphodiesterase that inversely regulates motility and biofilm formation in Vibrio cholerae. J Bacteriol. 2010;192(18):4541–52. doi: 10.1128/JB.00209-10 20622061
113. Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol. 2008;190(7):2527–36. doi: 10.1128/JB.01756-07 18223081
114. Pratt JT, McDonough E, Camilli A. PhoB regulates motility, biofilms, and cyclic di-GMP in Vibrio cholerae. J Bacteriol. 2009;191(21):6632–42. doi: 10.1128/JB.00708-09 19734314
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized VirusČlánek Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell ProliferationČlánek Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite InterfaceČlánek Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 10- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Expression of Concern: Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Preparing for the Next Epidemic with Basic Virology
- Effectively Communicating the Uncertainties Surrounding Ebola Virus Transmission
- Translating Basic Research into Clinical Applications: Malaria Research at an NIH Lab
- A Gut Odyssey: The Impact of the Microbiota on Spore Formation and Germination
- Papillomavirus E6 Oncoproteins Take Common Structural Approaches to Solve Different Biological Problems
- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Dimensions of Horizontal Gene Transfer in Eukaryotic Microbial Pathogens
- Addressing the Complications of Ebola and Other Viral Hemorrhagic Fever Infections: Using Insights from Bacterial and Fungal Sepsis
- Time for Chocolate: Current Understanding and New Perspectives on Cacao Witches’ Broom Disease Research
- Ganglioside and Non-ganglioside Mediated Host Responses to the Mouse Polyomavirus
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Structure Elucidation of Coxsackievirus A16 in Complex with GPP3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses
- CD39 Expression Identifies Terminally Exhausted CD8 T Cells
- Abiotic Stresses Antagonize the Rice Defence Pathway through the Tyrosine-Dephosphorylation of OsMPK6
- Dissociation of Tissue Destruction and Bacterial Expansion during Bubonic Plague
- Interferon-γ: The Jekyll and Hyde of Malaria
- CCR2 Inflammatory Dendritic Cells and Translocation of Antigen by Type III Secretion Are Required for the Exceptionally Large CD8 T Cell Response to the Protective YopE Epitope during Infection
- A New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1
- The Suramin Derivative NF449 Interacts with the 5-fold Vertex of the Enterovirus A71 Capsid to Prevent Virus Attachment to PSGL-1 and Heparan Sulfate
- Trans-generational Immune Priming Protects the Eggs Only against Gram-Positive Bacteria in the Mealworm Beetle
- Peripheral Vγ9Vδ2 T Cells Are a Novel Reservoir of Latent HIV Infection
- Respiratory Syncytial Virus Disease Is Mediated by Age-Variable IL-33
- TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized Virus
- Modeling the Effects of Vorinostat Reveals both Transient and Delayed HIV Transcriptional Activation and Minimal Killing of Latently Infected Cells
- Identification of a Novel Lipoprotein Regulator of Spore Germination
- Calcium Regulation of Hemorrhagic Fever Virus Budding: Mechanistic Implications for Host-Oriented Therapeutic Intervention
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
- Comparative Life Cycle Transcriptomics Revises Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates
- The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy
- Carcinogenic Parasite Secretes Growth Factor That Accelerates Wound Healing and Potentially Promotes Neoplasia
- Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell Proliferation
- Dengue Virus Infection of Requires a Putative Cysteine Rich Venom Protein
- Distinct Viral and Mutational Spectrum of Endemic Burkitt Lymphoma
- Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite Interface
- Phenotypic and Functional Alterations in Circulating Memory CD8 T Cells with Time after Primary Infection
- Systematic Identification of Cyclic-di-GMP Binding Proteins in Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems
- Influenza Transmission in the Mother-Infant Dyad Leads to Severe Disease, Mammary Gland Infection, and Pathogenesis by Regulating Host Responses
- Myeloid Cell Arg1 Inhibits Control of Arthritogenic Alphavirus Infection by Suppressing Antiviral T Cells
- The White-Nose Syndrome Transcriptome: Activation of Anti-fungal Host Responses in Wing Tissue of Hibernating Little Brown Myotis
- Influenza Virus Reassortment Is Enhanced by Semi-infectious Particles but Can Be Suppressed by Defective Interfering Particles
- Identification of the Mechanisms Causing Reversion to Virulence in an Attenuated SARS-CoV for the Design of a Genetically Stable Vaccine
- Differentiation-Dependent KLF4 Expression Promotes Lytic Epstein-Barr Virus Infection in Epithelial Cells
- The Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways
- C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in
- Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
- Crystal Structure of the Human Cytomegalovirus Glycoprotein B
- Depletion of . GlmU from Infected Murine Lungs Effects the Clearance of the Pathogen
- Immunologic Control of Papillomavirus Type 1
- Requires Host Rab1b for Survival in Macrophages
- Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi
- PD-L1 Expression on Retrovirus-Infected Cells Mediates Immune Escape from CD8 T Cell Killing
- Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction
- IL-4 Induced Innate CD8 T Cells Control Persistent Viral Infection
- Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
- BCG Skin Infection Triggers IL-1R-MyD88-Dependent Migration of EpCAM CD11b Skin Dendritic cells to Draining Lymph Node During CD4+ T-Cell Priming
- Antigenic Characterization of the HCMV gH/gL/gO and Pentamer Cell Entry Complexes Reveals Binding Sites for Potently Neutralizing Human Antibodies
- Rescue of a Plant Negative-Strand RNA Virus from Cloned cDNA: Insights into Enveloped Plant Virus Movement and Morphogenesis
- Geminivirus Activates to Accelerate Cytoplasmic DCP2-Mediated mRNA Turnover and Weakens RNA Silencing in
- Disruption of Sphingolipid Biosynthesis Blocks Phagocytosis of
- The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps
- The Timing of Stimulation and IL-2 Signaling Regulate Secondary CD8 T Cell Responses
- Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity
- The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Interferon-γ: The Jekyll and Hyde of Malaria
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



