-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Emerging Role for RNA Polymerase II in Regulating Virulence Gene Expression in Malaria Parasites
article has not abstract
Published in the journal: The Emerging Role for RNA Polymerase II in Regulating Virulence Gene Expression in Malaria Parasites. PLoS Pathog 11(7): e32767. doi:10.1371/journal.ppat.1004926
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004926Summary
article has not abstract
Introduction
Plasmodium falciparum causes the most severe form of human malaria and is responsible for a significant public health burden in the developing world. These protozoan parasites invade circulating red blood cells (RBCs) and maintain prolonged infections through an intricate gene-expression switching mechanism that enables immune evasion through antigenic variation [1]. One of the families of genes responsible for this evasion is called var; it is made up of ~60 members, of which approximately two-thirds are found located within subtelomeric heterochromatic regions of the P. falciparum genome, with the remaining third of the family arranged in clusters located in similarly heterochromatinized areas within the internal regions of the chromosomes. The var gene family encodes P. falciparum erythrocyte membrane protein 1 (PfEMP1), a protein displayed on the surface of infected RBCs and considered the primary antigenic determinant required for cytoadherence and sequestration of infected cells, thus enabling them to avoid circulation through the spleen. Only one var gene is expressed at a time, while the other 59 remain transcriptionally silent, and which gene is active switches over the course of an infection. This process allows the parasites to maintain chronic infections through an ever-changing display of PfEMP1 antigens to the immune system. Structurally, each var gene consists of two exons flanking a single, conserved intron with a bidirectional promoter that transcribes noncoding RNAs [2–4].
Although an understanding of the mechanisms that lead to coordinated switching within the var gene family have remained elusive, it has become clear in recent years that epigenetic components, particularly histone modifications, play major roles in determining whether an individual gene will be active or silent. Many of the histone modifications involved in antigenic variation, as well as other aspects of parasite development, have been catalogued. For example, trimethylation of histone H3 at lysine 4 (H3K4me3) denotes transcriptionally active genes [5], including the single active var gene. In contrast, H3K9me3 marks the 59 silent var genes [6,7]. Interestingly, H3K9me3 and another mark, H3K36me3, are very limited in their distribution throughout the genome and are found primarily at gene families that encode variant antigens like var [5,8]. How the enzymes that deposit these marks are recruited to very limited regions of the genome is poorly understood. This short review aims to expand on recent work that sheds light on the recruitment of histone modifiers to narrow regions of chromosomes, specifically var genes, by way of the C-terminal domain of RNA polymerase II (RNA pol II CTD).
The C-Terminal Domain (CTD) of RNA Pol II: A Platform for Coordinating Transcription
As with all eukaryotes, malaria parasites possess three different RNA polymerases. RNA pol I transcribes the ribosomal RNAs (with the exception of 5S), while pol III transcribes the 5S ribosomal RNA (rRNA), transfer RNAs (tRNAs), and several small RNAs, including some small nucleolar RNAs (snRNAs). RNA pol II mediates expression of protein-coding genes by transcribing the corresponding messenger RNAs (mRNAs). Central to coordinating the various steps in mRNA production is the systematic post-translational modification, specifically phosphorylation, of the CTD of Rpb1, the largest subunit of RNA pol II (Fig 1A) [9]. This enables the stepwise recruitment of various factors involved in the regulation of the transcription cycle as well as RNA processing, RNA export, and chromatin remodeling. In most eukaryotes, the RNA pol II CTD has a flexible structure comprised of a highly conserved tandem array of Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser/Lys7 heptad repeats. As RNA pol II moves through the transcription of a gene, the repeats undergo sequential changes in the phosphorylation status of Ser2 and Ser5 whereby phosphorylation of Ser5 alone is associated with transcriptional initiation, concurrent phosphorylation of both Ser2 and Ser5 when the polymerase is engaged in elongation, and phosphorylation of Ser2 alone at termination. The transcription cycle is not the only process that is dependent on appropriate phosphorylation of the serine residues. Nuclear factors such as capping enzymes and histone modifiers, as well as splicing factors, are equally dependent on the phosphorylation status of the CTD for appropriate recruitment to the transcription complex [10]. For example, the histone methyltransferase SET1 binds to the CTD when it is phosphorylated at serine 5, thus marking chromatin at regions of the genome that have recently been transcribed (Fig 1A) [11].
Fig. 1. The role of the RNA polymerase II CTD in recruiting different factors to the transcription complex. 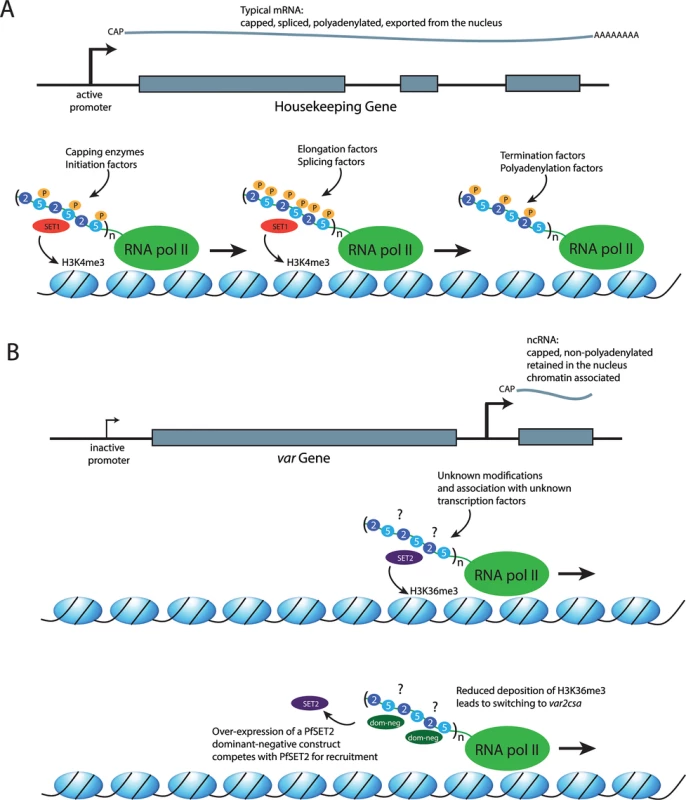
(A) Transcription of a typical mRNA. Top: a representation of a housekeeping gene with three exons (gray boxes) and two introns. Recruitment of an mRNA producing RNA pol II complex to the promoter results in the production of a transcript that is capped, spliced, polyadenylated, and exported from the nucleus for translation. Bottom: a representation of the DNA strand wrapped around histones (blue ovals). The RNA pol II complex is shown in green, with the CTD shown as an extension. In eukaryotes, the CTD consists of a series of repeats of the amino acid sequence YSPTSPS/K. The serines in the second and fifth positions of each repeat are shown as blue circles labeled 2 and 5, respectively. Three repeats are shown, although the number can vary according to species (n). In P. falciparum, 12–17 repeats have been reported. Phosphorylation of the serines are shown as orange circles labeled with a P and are found primarily on serines in the fifth position near the beginning of transcription, on the serines in both positions at the middle of the transcription unit, and at the serines in the second position where transcription terminates. The alternate phosphorylated states mediate recruitment of different factors required for mRNA production and maturation, as well as histone modifiers, including SET1 (red oval), which deposits the epigenetic mark H3K4me3 concurrent with the production of an mRNA. Thus, this mark is found at transcriptionally active regions of the genome. (B) A model for transcription of a noncoding RNA (ncRNA) at var loci. Top: a promoter found within the introns of var genes leads to the production of ncRNAs that are capped but not polyadenylated. In addition, they are retained within the nucleus and associated with chromatin. Middle: the RNA pol II complex is engaged in transcribing a noncoding RNA; however, the modifications of the serines at positions 2 and 5 of the CTD and the transcription-related factors that are recruited to the complex are unknown (?). PfSET2 (purple oval) has been shown to bind directly to the CTD and to deposit the epigenetic mark H3K36me3 at both active and silent var genes. This is consistent with its recruitment by RNA pol II when it is transcribing the ncRNA from the promoter located within var introns. Bottom: a dominant-negative version of PfSET2 (dark green ovals) that is not capable of depositing the H3K36me3 mark can compete with the endogenous wild-type PfSET2 for binding to the CTD. Overexpression of the dominant-negative protein reduces the efficiency of PfSET2 recruitment to var loci and leads to switching to var2csa. The Evolution of the RNA Pol II CTD in Malaria Parasites
Comparison of RNA pol II CTD sequences from different evolutionary lineages shows that CTD length is loosely correlated with genome size [12]. Moreover, individual lineages tend to have tightly conserved heptad numbers; for example, all mammals possess 52 repeats in their CTDs. Interestingly, genome sequences of several protozoan lineages reveal some peculiar and intriguing differences in their CTDs. Trypanosomes and species of Trichomonas display traces of the standard CTD heptads but not the canonical repeat structure [12–14]. Plasmodium, on the other hand, has a bona fide RNA pol II CTD; however, members of this genus display an unprecedented plasticity in the length and composition of the CTD, something not observed in any other eukaryotic lineage (described in detail in [15]). Malaria parasites that infect rodents, birds, reptiles, and bats all possess a remarkably short CTD consisting of eight conserved heptad repeats. In contrast, the primate parasites P. knowlesi, P. vivax, P. falciparum, and P. cynomolgi show an expanded number of heptads that is highly variable, both within and between species, ranging from 12 to 17 repeats. More precisely, primate parasites possess an expansion of a specific heptad near the middle of the CTD that contains a lysine residue in the seventh position. The function of this heptad expansion is unknown, although it has been noted that the increased heptad number is correlated with the use of epigenetic memory for antigenic variation [16]. This observation is even more intriguing considering recent studies implicating the CTD in the recruitment of histone modifiers to variant antigen encoding genes, in particular var genes (see below).
The Role of the RNA Pol II CTD in the Recruitment of Histone Modifiers in P. falciparum
Similar to its counterparts in higher eukaryotes, recent work indicates that the RNA pol II CTD of malaria parasites serves as a platform for the recruitment of nuclear factors for gene regulation and cross talk between chromatin and RNA pol II [16]. The first nuclear factor for which recruitment by the CTD has been demonstrated in P. falciparum is PfSET2, a histone methyltransferase that deposits a trimethylation mark on H3K36. The H3K36me3 mark is tightly coupled to transcription by RNA pol II in both yeast and mammalian cells; therefore, it is found throughout transcribed regions of the genome of these organisms [17,18]. However, in the Plasmodium genus, orthologues of SET2 are found only within the genomes of parasites that infect primates, thus mirroring the RNA pol II CTD expansion [16]. Further, the H3K36me3 mark is not found distributed throughout the genome but rather is only present within the telomeric repeats and the large, multicopy gene families involved in antigenic variation—for example, var genes [8]. This implies that malaria parasites have evolved a way to recruit PfSET2 only to these narrow regions of the genome. In higher eukaryotes, SET2 is recruited to all transcribed regions of the genome by binding directly to the CTD of RNA pol II while it is actively engaged in transcription, specifically when the serine residues in positions 2 and 5 of the heptad repeats are phosphorylated [18,19]. Interestingly, in vitro coimmunoprecipitation assays show specific binding of PfSET2 to the CTD when both Ser2 and Ser5 are unphosphorylated, and phosphorylation of the CTD disrupted PfSET2 binding, indicating significant differences in how PfSET2 is recruited by RNA pol II in Plasmodium when compared to higher eukaryotes [16].
The Role of RNA Pol II CTD Recruitment of PfSET2 in Antigenic Variation in P. falciparum
Studies employing a dominant-negative version of PfSET2 provided evidence for a direct role for the RNA pol II CTD in regulating var gene expression and antigenic variation [16]. As described above, only a single member of the var gene family is transcribed at a time, with switching of the expressed copy resulting in antigenic variation. A truncated version of PfSET2 consisting solely of the CTD binding domain competes with the endogenous PfSET2 for binding to the CTD, thus functioning as a “dominant-negative” when overexpressed in transgenic parasites (Fig 1B). This results in profound changes in var gene expression, inducing rapid switching to a single var gene called var2csa and indicating that recruitment of PfSET2 through CTD binding plays a key role in var gene regulation. Why this particular var gene becomes activated when PfSET2 recruitment by RNA pol II is disrupted is not clear. Several studies have documented that var gene switching is not random but rather follows a hierarchy of switching frequencies and that expression switching within the var repertoire is somehow coordinated within a network [20–25]. It is tempting to speculate that the selective activation of var2csa in these experiments might provide clues on the molecular basis of this proposed regulatory network; however, the details remain unknown.
The H3K36me3 mark deposited by PfSET2 is found within the chromatin of all var genes, both active and silent, indicating that PfSET2 is recruited regardless of whether an mRNA is being transcribed [8,16]. If PfSET2 is recruited by binding to the CTD of RNA pol II, how does it get recruited to transcriptionally silent genes? In addition to the mRNA transcribed from the single active gene, RNA pol II also transcribes noncoding RNAs from all var genes, both active and silent [26]. The promoter responsible for transcribing the noncoding RNAs, located within a conserved intron found in each gene, was shown to be required for proper var gene regulation [27,28], consistent with a role in the recruitment of RNA pol II and PfSET2 to each var gene. The fact that RNA pol II is transcribing a noncoding RNA rather than an mRNA in this context might provide a mechanism for selective recruitment of PfSET2 to limited regions of the genome. The field of noncoding RNAs has exploded in recent years and encompasses many distinct molecular mechanisms (reviewed in [29]). Numerous noncoding RNAs of unknown function have now been identified in P. falciparum, indicating that similar mechanisms are likely to be at work in malaria parasites [30–33]. The CTD undergoes differential phosphorylation while transcribing mRNAs, and the phosphorylation patterns are likely different when noncoding RNAs are being transcribed. If so, the unique modifications could be required for binding of PfSET2, thus limiting recruitment to regions where ncRNAs are transcribed. This hypothesis is consistent with the in vitro observation that PfSET2 does not bind to a typically phosphorylated CTD [16]. The telomeric repeats, another region of the genome where PfSET2 is recruited, also transcribe noncoding RNAs [34,35], and the noncoding RNAs themselves could also play a role in chromatin assembly, as has been observed in higher eukaryotes [29]. These observations suggest a model in which RNA pol II can actively recruit different histone modifiers to different parts of the genome through alternative modifications to the CTD (Fig 1B). There is experimental evidence to support this model for var loci [16]; however, it could also apply to other regions of the genome that have been shown to transcribe noncoding RNAs, thus potentially serving as a more general mechanism for chromatin assembly and organization.
Perspectives
In addition to PfSET2, there are eight additional SET-domain-containing proteins encoded in the P. falciparum genome [36] and another putative SET protein proposed to be localized to the single active var gene [37]. These proteins are presumed to be involved in regulating gene expression throughout the rest of the genome, as they are in higher eukaryotes. Additional work with RNA pol II will help determine what role it plays in mediating these processes. A more complete understanding of the uniqueness of the Plasmodium RNA pol II CTD and the peculiar nature of its phosphorylation code will undoubtedly cast considerable light on this subject, potentially unifying previous observations regarding specific deposition of histone modifications, noncoding RNAs, gene activation and silencing, and mutually exclusive expression.
Zdroje
1. Guizetti J, Scherf A (2013) Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell Microbiol 15 : 718–726. doi: 10.1111/cmi.12115 23351305
2. Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH (1995) Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82 : 101–110. 7606775
3. Su X, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JV, Peterson DS, Ravetch JV, Wellems TE (1995) A large and diverse gene family (var) encodes 200–350 kD proteins implicated in the antigenic variation and cytoadherence of Plasmodium falciparum-infected erythrocytes. Cell 82 : 89–100. 7606788
4. Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ (1995) Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82 : 77–87. 7541722
5. Lopez-Rubio JJ, Mancio-Silva L, Scherf A (2009) Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 5 : 179–190. doi: 10.1016/j.chom.2008.12.012 19218088
6. Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez RR, Scherf A (2007) 5' flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol 66 : 1296–1305. 18028313
7. Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL, Deitsch KW (2007) Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci U S A 104 : 899–902. 17209011
8. Jiang L, Mu J, Zhang Q, Ni T, Srinivasan P, Rayavara K, Yang W, Turner L, Lavstsen T, Theander TG, Peng W, Wei G, Jing Q, Wakabayashi Y, Bansal A, Luo Y, Ribeiro JM, Scherf A, Aravind L, Zhu J, Zhao K, Miller LH (2013) PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 499 : 223–227. doi: 10.1038/nature12361 23823717
9. Egloff S, Murphy S (2008) Cracking the RNA polymerase II CTD code. Trends Genet 24 : 280–288. doi: 10.1016/j.tig.2008.03.008 18457900
10. Cho EJ (2007) RNA polymerase II carboxy-terminal domain with multiple connections. Exp Mol Med 39 : 247–254. 17603278
11. Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11 : 709–719. 12667453
12. Chapman RD, Heidemann M, Hintermair C, Eick D (2008) Molecular evolution of the RNA polymerase II CTD. Trends Genet 24 : 289–296. doi: 10.1016/j.tig.2008.03.010 18472177
13. Dacks JB, Marinets A, Ford DW, Cavalier-Smith T, Logsdon JM Jr. (2002) Analyses of RNA Polymerase II genes from free-living protists: phylogeny, long branch attraction, and the eukaryotic big bang. Mol Biol Evol 19 : 830–840. 12032239
14. Chapman AB, Agabian N (1994) Trypanosoma brucei RNA polymerase II is phosphorylated in the absence of carboxyl-terminal domain heptapeptide repeats. J Biol Chem 269 : 4754–4760. 8106443
15. Kishore SP, Perkins SL, Templeton TJ, Deitsch KW (2009) An unusual recent expansion of the C-terminal domain of RNA polymerase II in primate malaria parasites features a motif otherwise found only in mammalian polymerases. J Mol Evol 68 : 706–714. doi: 10.1007/s00239-009-9245-2 19449052
16. Ukaegbu UE, Kishore SP, Kwiatkowski DL, Pandarinath C, Dahan-Pasternak N, Dzikowski R, Deitsch KW (2014) Recruitment of PfSET2 by RNA polymerase II to variant antigen encoding loci contributes to antigenic variation in P. falciparum. PLoS Pathog 10: e1003854. doi: 10.1371/journal.ppat.1003854 24391504
17. Sun XJ, Wei J, Wu XY, Hu M, Wang L, Wang HH, Zhang QH, Chen SJ, Huang QH, Chen Z (2005) Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem 280 : 35261–35271. 16118227
18. Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD (2005) A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 25 : 3305–3316. 15798214
19. Li B, Howe L, Anderson S, Yates JR III, Workman JL (2003) The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278 : 8897–8903. 12511561
20. Recker M, Buckee CO, Serazin A, Kyes S, Pinches R, Christodoulou Z, Springer AL, Gupta S, Newbold CI (2011) Antigenic variation in Plasmodium falciparum malaria involves a highly structured switching pattern. PLoS Pathog 7: e1001306. doi: 10.1371/journal.ppat.1001306 21408201
21. Horrocks P, Pinches R, Christodoulou Z, Kyes SA, Newbold CI (2004) Variable var transition rates underlie antigenic variation in malaria. Proc Natl Acad Sci U S A 101 : 11129–11134. 15256597
22. Noble R, Christodoulou Z, Kyes S, Pinches R, Newbold CI, Recker M (2013) The antigenic switching network of Plasmodium falciparum and its implications for the immuno-epidemiology of malaria. Elife (Cambridge) 2: e01074.
23. Enderes C, Kombila D, Dal Bianco M, Dzikowski R, Kremsner P, Frank M (2011) Var Gene promoter activation in clonal Plasmodium falciparum isolates follows a hierarchy and suggests a conserved switching program that is independent of genetic background. J Infect Dis 204 : 1620–1631. doi: 10.1093/infdis/jir594 21926380
24. Frank M, Dzikowski R, Amulic B, Deitsch K (2007) Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol Microbiol 64 : 1486–1498. 17555435
25. Noble R, Recker M (2012) A statistically rigorous method for determining antigenic switching networks. PLoS ONE 7: e39335. doi: 10.1371/journal.pone.0039335 22761765
26. Kyes S, Christodoulou Z, Pinches R, Kriek N, Horrocks P, Newbold C (2007) Plasmodium falciparum var gene expression is developmentally controlled at the level of RNA polymerase II-mediated transcription initiation. Mol Microbiol 63 : 1237–1247. 17257309
27. Deitsch KW, Calderwood MS, Wellems TE (2001) Malaria. Cooperative silencing elements in var genes. Nature 412 : 875–876.
28. Dzikowski R, Li F, Amulic B, Eisberg A, Frank M, Patel S, Wellems TE, Deitsch KW (2007) Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep 8 : 959–965. 17762879
29. Roberts TC, Morris KV, Weinberg MS (2014) Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs. Epigenetics 9 : 13–20. doi: 10.4161/epi.26700 24149621
30. Vembar SS, Scherf A, Siegel TN (2014) Noncoding RNAs as emerging regulators of Plasmodium falciparum virulence gene expression. Curr Opin Microbiol 20 : 153–161. doi: 10.1016/j.mib.2014.06.013 25022240
31. Wei C, Xiao T, Zhang P, Wang Z, Chen X, Zhang L, Yao M, Chen R, Wang H (2014) Deep profiling of the novel intermediate-size noncoding RNAs in intraerythrocytic Plasmodium falciparum. PLoS ONE 9: e92946. 1 doi: 10.1371/journal.pone.0092946 24713982
32. Raabe CA, Sanchez CP, Randau G, Robeck T, Skryabin BV, Chinni SV, Kube M, Reinhardt R, Ng GH, Manickam R, Kuryshev VY, Lanzer M, Brosius J, Tang TH, Rozhdestvensky TS (2010) A global view of the nonprotein-coding transcriptome in Plasmodium falciparum. Nucleic Acids Res 38 : 608–617. doi: 10.1093/nar/gkp895 19864253
33. Lopez-Barragan MJ, Lemieux J, Quinones M, Williamson KC, Molina-Cruz A, Cui K, Barillas-Mury C, Zhao K, Su XZ (2011) Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 12 : 587. doi: 10.1186/1471-2164-12-587 22129310
34. Broadbent KM, Park D, Wolf AR, Van Tyne D, Sims JS, Ribacke U, Volkman S, Duraisingh M, Wirth D, Sabeti PC, Rinn JL (2011) A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol 12: R56. doi: 10.1186/gb-2011-12-6-r56 21689454
35. Sierra-Miranda M, Delgadillo DM, Mancio-Silva L, Vargas M, Villegas-Sepulveda N, Martinez-Calvillo S, Scherf A, Hernandez-Rivas R (2012) Two long non-coding RNAs generated from subtelomeric regions accumulate in a novel perinuclear compartment in Plasmodium falciparum. Mol Biochem Parasitol 185 : 36–47. doi: 10.1016/j.molbiopara.2012.06.005 22721695
36. Cui L, Fan Q, Cui L, Miao J (2008) Histone lysine methyltransferases and demethylases in Plasmodium falciparum. Int J Parasitol 38 : 1083–1097. doi: 10.1016/j.ijpara.2008.01.002 18299133
37. Volz JC, Bartfai R, Petter M, Langer C, Josling GA, Tsuboi T, Schwach F, Baum J, Rayner JC, Stunnenberg HG, Duffy MF, Cowman AF (2012) PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe 11 : 7–18. doi: 10.1016/j.chom.2011.11.011 22264509
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSSČlánek Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to MalariaČlánek IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 Cells
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 7- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Basic Prion Science “Spreads” Insight
- Research Driven by Curiosity: The Journey from Basic Molecular Biology and Virology to Studies of Human Pathogenic Coronaviruses
- Cross Kingdom Activators of Five Classes of Bacterial Effectors
- Vaccination Drives Changes in Metabolic and Virulence Profiles of
- Expression of the Blood-Group-Related Gene Alters Susceptibility to Infection
- Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSS
- Latent KSHV Infected Endothelial Cells Are Glutamine Addicted and Require Glutaminolysis for Survival
- The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators
- Intraperitoneal Infection of Wild-Type Mice with Synthetically Generated Mammalian Prion
- Vpr Promotes Macrophage-Dependent HIV-1 Infection of CD4 T Lymphocytes
- An In-Depth Comparison of Latency-Reversing Agent Combinations in Various and HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression
- α-Macroglobulin Can Crosslink Multiple Erythrocyte Membrane Protein 1 (PfEMP1) Molecules and May Facilitate Adhesion of Parasitized Erythrocytes
- Should Symbionts Be Nice or Selfish? Antiviral Effects of Wolbachia Are Costly but Reproductive Parasitism Is Not
- A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface
- The Broad Neutralizing Antibody Responses after HIV-1 Superinfection Are Not Dominated by Antibodies Directed to Epitopes Common in Single Infection
- Rapidly Evolving Genes Are Key Players in Host Specialization and Virulence of the Fungal Wheat Pathogen ()
- MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease
- Age-Dependent Cell Trafficking Defects in Draining Lymph Nodes Impair Adaptive Immunity and Control of West Nile Virus Infection
- Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to Malaria
- Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease
- Macrophages Subvert Adaptive Immunity to Urinary Tract Infection
- Mycolactone-Dependent Depletion of Endothelial Cell Thrombomodulin Is Strongly Associated with Fibrin Deposition in Buruli Ulcer Lesions
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation
- Phosphoproteomic Analysis of KSHV-Infected Cells Reveals Roles of ORF45-Activated RSK during Lytic Replication
- CR3 and Dectin-1 Collaborate in Macrophage Cytokine Response through Association on Lipid Rafts and Activation of Syk-JNK-AP-1 Pathway
- IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 Cells
- THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection
- Human Enterovirus Nonstructural Protein 2C Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone
- IL-27 Signaling Is Crucial for Survival of Mice Infected with African Trypanosomes via Preventing Lethal Effects of CD4 T Cells and IFN-γ
- Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation
- Vpu Exploits the Cross-Talk between BST2 and the ILT7 Receptor to Suppress Anti-HIV-1 Responses by Plasmacytoid Dendritic Cells
- Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses
- A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence
- Analysis of the SUMO2 Proteome during HSV-1 Infection
- Capacity of Broadly Neutralizing Antibodies to Inhibit HIV-1 Cell-Cell Transmission Is Strain- and Epitope-Dependent
- A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein
- Deploying FLAREs to Visualize Functional Outcomes of Host—Pathogen Encounters
- Mosquitoes Reset Malaria Parasites
- The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease
- Extracellular Virions: The Advance Guard of Poxvirus Infections
- Risks of Antibiotic Exposures Early in Life on the Developing Microbiome
- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Periodontal Diseases: Bug Induced, Host Promoted
- Mechanisms of Host Behavioral Change in Rodent Association
- The Endosymbiotic Bacterium Selectively Kills Male Hosts by Targeting the Masculinizing Gene
- HIV Reactivation from Latency after Treatment Interruption Occurs on Average Every 5-8 Days—Implications for HIV Remission
- Ubiquilin 1 Promotes IFN-γ-Induced Xenophagy of
- Transfer of Immunity from Mother to Offspring Is Mediated via Egg-Yolk Protein Vitellogenin
- Suppression of Long-Lived Humoral Immunity Following Infection
- The Role of VP1 Amino Acid Residue 145 of Enterovirus 71 in Viral Fitness and Pathogenesis in a Cynomolgus Monkey Model
- Utilizing Chemical Genomics to Identify Cytochrome as a Novel Drug Target for Chagas Disease
- The Emerging Role for RNA Polymerase II in Regulating Virulence Gene Expression in Malaria Parasites
- Turning Up the Heat: Inflammasome Activation by Fungal Pathogens
- On and Under the Skin: Emerging Basidiomycetous Yeast Infections Caused by Species
- EhVps32 Is a Vacuole-Associated Protein Involved in Pinocytosis and Phagocytosis of
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
- The Serine Protease EspC from Enteropathogenic Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System
- Existing Infection Facilitates Establishment and Density of Malaria Parasites in Their Mosquito Vector
- Evaluating Human T-Cell Therapy of Cytomegalovirus Organ Disease in HLA-Transgenic Mice
- Neuronal Interferon Signaling Is Required for Protection against Herpes Simplex Virus Replication and Pathogenesis
- Epstein-Barr Virus Proteins EBNA3A and EBNA3C Together Induce Expression of the Oncogenic MicroRNA Cluster miR-221/miR-222 and Ablate Expression of Its Target p57
- Colonization of the Mouse Gastrointestinal Tract Is Modulated by Wall Teichoic Acid, Capsule, and Surface Proteins
- Virulence of Group A Streptococci Is Enhanced by Human Complement Inhibitors
- Identification of Caspase Cleavage Sites in KSHV Latency-Associated Nuclear Antigen and Their Effects on Caspase-Related Host Defense Responses
- Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Infections
- Type VI Secretion System Transports Zn to Combat Multiple Stresses and Host Immunity
- Lv4 Is a Capsid-Specific Antiviral Activity in Human Blood Cells That Restricts Viruses of the SIV/SIV/HIV-2 Lineage Prior to Integration
- Phenylbutyrate Is Bacteriostatic against and Regulates the Macrophage Response to Infection, Synergistically with 25-Hydroxy-Vitamin D₃
- An Internally Translated MAVS Variant Exposes Its Amino-terminal TRAF-Binding Motifs to Deregulate Interferon Induction
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



