-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Utilizing Chemical Genomics to Identify Cytochrome as a Novel Drug Target for Chagas Disease
Chagas Disease, or American trypanosomiasis, is caused by the kinetoplastid protozoan Trypanosoma cruzi and is primarily transmitted to a mammalian host via a triatomine insect vector (the “kissing bug”) infected with T. cruzi parasites. Although discovered in 1909 by the physician Dr. Carlos Chagas, the disease gained recognition by the public health community only following a major outbreak in Brazil during the 1960s. Approximately eight million people (primarily in Central and South America) are infected with T. cruzi and cases are becoming more widespread due to migration out of the endemic regions. Current treatment options have severe problems with toxicity, limited efficacy, and long administration. Hence, discovery of new drugs for treatment of Chagas disease has become of prime interest to the biomedical research community. In this study, we report identification of a potent inhibitor of T. cruzi growth and use a chemical genetics-based approach to elucidate the associated mechanism of action. We found that this compound, GNF7686, targets cytochrome b, a component of the mitochondrial electron transport chain crucial for ATP generation. Our study provides new insights into the use of phenotypic screening to identify novel targets for kinetoplastid drug discovery.
Published in the journal: Utilizing Chemical Genomics to Identify Cytochrome as a Novel Drug Target for Chagas Disease. PLoS Pathog 11(7): e32767. doi:10.1371/journal.ppat.1005058
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005058Summary
Chagas Disease, or American trypanosomiasis, is caused by the kinetoplastid protozoan Trypanosoma cruzi and is primarily transmitted to a mammalian host via a triatomine insect vector (the “kissing bug”) infected with T. cruzi parasites. Although discovered in 1909 by the physician Dr. Carlos Chagas, the disease gained recognition by the public health community only following a major outbreak in Brazil during the 1960s. Approximately eight million people (primarily in Central and South America) are infected with T. cruzi and cases are becoming more widespread due to migration out of the endemic regions. Current treatment options have severe problems with toxicity, limited efficacy, and long administration. Hence, discovery of new drugs for treatment of Chagas disease has become of prime interest to the biomedical research community. In this study, we report identification of a potent inhibitor of T. cruzi growth and use a chemical genetics-based approach to elucidate the associated mechanism of action. We found that this compound, GNF7686, targets cytochrome b, a component of the mitochondrial electron transport chain crucial for ATP generation. Our study provides new insights into the use of phenotypic screening to identify novel targets for kinetoplastid drug discovery.
Introduction
Chagas disease, or American trypanosomiasis, is a neglected disease caused by the kinetoplastid protozoan Trypanosoma cruzi (T. cruzi). Endemic to Latin America, Chagas disease is increasingly globalized due to population migration from endemic regions into developed countries, and the U.S. in particular. About eight million people are estimated to harbor the infection with 40,000 new cases added annually [1, 2].
In the 100+ years that have passed since the first description of Chagas disease by Carlos Chagas, only two drugs have been developed to treat this infection: nifurtimox and benznidazole [2, 3]. While both these drugs can clear T. cruzi from the infected mammalian hosts, they are both inadequate to address the medical need of millions of patients chronically infected today. The drug shortcomings include toxicity, prolonged treatment time, and high rate of treatment failure [2, 3].
T. cruzi is transmitted to mammalian hosts primarily via hematophagous triatomine bugs [4]. While in the vector insect, T. cruzi cells propagate as flagellated epimastigotes that transform into non-dividing infective trypomastigotes. As the T. cruzi-infected bug takes a blood meal from a host, it deposits motile T. cruzi trypomastigotes near the wound site. Following invasion of host cells in the bite wound or at mucous membranes, intracellular trypomastigotes undergo a morphological transformation into amastigotes and start to replicate [1, 4, 5]. After completing several rounds of intracellular division, the amastigotes transform into trypomastigotes that then leave the infected cell and initiate a new cycle of infection.
The acute phase of Chagas disease is often asymptomatic, characterized by readily detectable parasitemia, and usually resolves within a few weeks through control of parasite proliferation by the adaptive immune system [4]. In the chronic stage, infected individuals rarely display symptoms or evidence of the disease for decades. However, ~30% of these patients eventually go on to develop a severe cardiomyopathy and ~10% of patients progress with gastrointestinal complications [1, 4].
New drug discovery for Chagas disease is hampered by very limited number of validated T. cruzi drug targets. Drug discovery efforts have focused on the trypanosome ergosterol biosynthesis pathway and cruzain, a T. cruzi cysteine protease [6]. During the last decade, much attention has been paid to inhibitors of sterol 14 α-demethylase (CYP51), an essential enzyme in the ergosterol biosynthesis pathway [6, 7]. To a large degree, this interest has been fueled by the availability of drugs targeting fungal sterol 14 α-demethylase, such as posaconazole or ravuconazole [8, 9]. Both these drugs are exceptionally potent on T. cruzi in vitro and have been shown to effect radical parasitological cure in mouse models of Chagas disease [10]. Also, treatment with posaconazole cured T. cruzi infection in an immunosuppressed patient following benznidazole treatment failure [11]. However, a 60-day treatment with posaconazole, while transiently clearing the parasite from Chagas patients, did not prevent recrudescence of infection in a majority of patients (81%) as determined by PCR. A similar trial testing the efficacy of ravuconazole prodrug E1224 has recently reported failure to cure infection in a majority (~70%) of the treated Chagas patients [12, 13]. These failures in clinical phase 2 trials have been attributed to insufficient drug exposure or dosing duration [14].
In addition to inhibitors of the parasite ergosterol biosynthesis, several inhibitors of cruzain were reported as promising candidates for treating Chagas disease. Of these, the most advanced is K777, a vinyl sulfone peptidomimetic inhibitor [15, 16]. K777 has been shown to be safe and efficacious in animal models of acute and chronic Chagas disease [17, 18] and is currently undergoing preclinical development evaluation.
Identification of T. cruzi growth inhibitors by phenotypic screening represents a viable alternative to target-based Chagas disease drug discovery. The approach allows efficient discovery of small molecules that perturb new molecular targets. Limitations of this approach stem from ignorance of the molecular mechanism, which precludes the use of structure-assisted drug design and prevents early predictions of toxicity through inhibition of homologous host enzymes. Chemical optimization of hit molecules in the absence of target-based activity measurements can be confounded by complex structure-activity-relationships, as biochemical activity and cellular permeability cannot be distinguished [19, 20].
To overcome these limitations, we have established a chemical genetics approach to the determination of the mechanism of action of small molecule growth inhibitors in T. cruzi. Starting with a novel T. cruzi inhibitor GNF7686, we evolved resistant T. cruzi mutants in vitro, and then identified the resistance-conferring mutation by whole genome sequencing. Finally, we demonstrated inhibition of the putative target in a biochemical assay. An expansion of this approach to other T. cruzi growth inhibitors could lead to identification of many additional drug targets and associated lead inhibitors for Chagas disease, and is already underway in our laboratory. As with the case of the T. cruzi cytochrome b target reported in this study, such an approach could point to drugs and drug targets from other fields, and substantially accelerate the introduction of novel Chagas disease treatments into the clinic.
Methods
Chemical reagents
With the exception of decylubiquinone (MP Biomedicals), all chemicals were purchased from Sigma-Aldrich Corporation.
T. cruzi culture
T. cruzi CL strain was propagated in NIH/3T3 fibroblast cells. NIH/3T3 cells were grown in RPMI-1640 media supplemented with 10% heat-inactivated fetal bovine serum (FBS, Hyclone) and 100 IU penicillin / 100 μg streptomycin (Hyclone) per mL at 37°C / 5% CO2, and passaged every three to four days. To establish infection, 6.25 × 105 of 3T3 cells were seeded in a T-175 flask. After attachment, cells were infected with 20–40 × 106 T. cruzi trypomastigotes. Following cell infection, parasites cycled between the trypomastigote and the intracellular proliferative amastigote forms and medium was changed biweekly.
T. cruzi epimastigotes were maintained in liver infusion tryptose (LIT) medium (9 g / L liver infusion broth, 5 g / L tryptose, 1 g / L NaCl, 8 g / L Na2HPO4, 0.4 g / L KCl, and 1 g / L glucose, pH = 7.2) supplemented with 10% heat-inactivated FBS and 15 μM hemin, and passaged every five days during middle to late logarithmic growth phase (maintained at 26°C / 0% CO2).
T. cruzi in vitro metacyclogenesis
For differentiation of epimastigotes into trypomastigotes, saturated cultures of T. cruzi CL epimastigotes were harvested by centrifugation (1,000 × g for 10 min at 10°C), resuspended in artificial triatomine urine medium (TAU; 190 mM NaCl, 17 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 0.035% NaHCO3, 8 mM phosphate buffer, pH 6.9) at a density of 5 x 108 cells / mL, and incubated at 26°C. Two hours later, the parasites were transferred to TAU 3AAG medium (TAU supplemented with 10 mM L-proline, 50 mM sodium L-glutamate, 2 mM sodium L-aspartate and 10 mM D-glucose) in T-25 culture flasks with a layer of culture medium approximately 1 cm in depth. This cell density was previously shown to be the optimal density for epimastigote differentiation [21, 22]. After 72 hour incubation, the mixture of epimastigotes and newly differentiated trypomastigotes was used for infection of mammalian host cells (NIH/3T3 mouse embryonic fibroblast line).
Briefly, NIH/3T3 cells were plated at a density of 0.025 million cells / mL into T-175 flasks and infected with 5 mL of pelleted epimastigote / trypomastigote mixture (collected from 25 mL of transformed culture) resuspended in RPMI-1640 medium supplemented with 10% FBS and 100 IU penicillin / 100 μg streptomycin per mL. After 24 hour incubation, non-internalized extracellular epimastigotes and trypomastigotes were removed, and infected NIH/3T3 cells were cultured for additional seven days. By then, newly formed trypomastigotes released from infected NIH/3T3 host cells were present in the medium and further used as described in the “T. cruzi in vitro efficacy assays” section.
T. cruzi in vitro efficacy assays
To determine compound efficacy on T. cruzi intracellular amastigotes, NIH/3T3 cells were seeded (1,000 cells / well, 40 μL) into microscopy-grade, clear bottom, 384-well plates (Greiner) in RPMI-1640 medium containing 5% heat-inactivated fetal bovine serum and 100 IU penicillin / 100 μg streptomycin per mL. Plates were incubated overnight at 37°C / 5% CO2. Cells were infected with T. cruzi trypomastigotes at a multiplicity of infection (MOI) of 10 for the wild-type strain, and 20 for the GNF7686-resistant mutant strain. After six hours of infection (at 37°C / 5% CO2), the plates were washed by aspirating the medium and replacing with fresh screening medium to remove remaining extracellular trypomastigotes. The plates with infected cells were incubated overnight (37°C / 5% CO2) and compounds dissolved in DMSO were added to plate wells on the following day (0.2% DMSO final concentration). After 48-hour compound treatment, infected cells were fixed (4% paraformaldehyde in phosphate-buffered saline containing 0.5 mM CaCl2 and 0.5 mM MgCl2), permeabilized (0.1% Triton X-100 in PBS), and then stained using a 1 : 125,000 dilution (prepared in PBS) of SYBR green (Life Technologies). The plates were then scanned using the Evotec Opera High Content Screening System (Perkin Elmer) and amastigote proliferation was assessed by counting parasites within the 3T3 cells using CellProfiler version 2.1.0 cell image analysis software [23]. In some experiments, an alternative protocol for measurement of compound activity on intracellular T. cruzi was used [24].
To determine compound activity on the T. cruzi epimastigote form, epimastigotes (20 μL; 2.5 × 105 parasites / mL) were added to 384-well assay plates containing 20 μL of LIT media with pre-dispensed compounds (0.2% DMSO final concentration) and incubated for seven days at 26°C / 0% CO2. Parasite viability was assessed at the end of this incubation period using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). Luminescence as a measure of parasite viability was measured on the EnVision plate reader.
To assess compound efficacy on trypomastigotes, parasites (30 μL; 2 × 106 trypomastigotes / mL) were added to 384-well plates containing 10 μL of RPMI-1640 without phenol red (Invitrogen) and supplemented with 10% FBS and 100 IU penicillin / 100 µg streptomycin per mL, and then treated with compounds (0.2% DMSO final concentration). Following a 48-hour incubation period, viability was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega).
Leishmania donovani in vitro efficacy assay
Leishmania donovani (L. donovani) axenic amastigotes (strain MHOM/SD/62/1S-CL2D) were maintained at 37°C / 5% CO2 in RPMI-1640 medium containing 4 mM L-glutamine, 20% heat inactivated FBS, 100 IU penicillin / 100 μg streptomycin per mL, 23 μM folic acid, 100 M adenosine, 22 mM D-glucose, and 25 mM 2-(N-morpholino)ethanesulfonic acid (pH 5.5 adjusted at 37°C using 1M HCl).
For compound screening, axenic amastigotes were seeded into 384-well plates containing axenic amastigote medium with pre-dispensed compounds (0.25% final DMSO concentration) at a density of 9,600 cells / well. Plates were incubated for 48 hours at 37°C / 5% CO2. Cell viability was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega).
Trypanosoma brucei in vitro efficacy assay
Trypanosoma brucei brucei (T. b. brucei) bloodstream form (strain Lister 427) was maintained in HMI-9 medium: IMDM (Iscove's Modified Dulbecco's Media), 10% heat-inactivated FBS, 10% Serum Plus medium supplement (SAFC biosciences), 1 mM hypoxanthine, 50 μM bathocuproine disulfonate.Na2, 1.5 mM cysteine, 1 mM pyruvate, 39 μg/mL thymidine, and 0.2 mM 2-mercapthoethanol) at 37°C / 5% CO2.
For compound screening, parasites were seeded into 384-well assay plates containing HMI-9 with pre-dispensed compounds (0.25% final DMSO concentration) at a density of 6,000 cells / well, and plates were then incubated for 48 hours at 37°C / 5% CO2. Cell viability was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega).
For conversion from the bloodstream form to the procyclic form, bloodstream form parasites were transferred from HMI-9 medium to Differentiating Trypanosome Medium (DTM, pH = 7.2), consisting of: 6.8 g / L NaCl, 400 mg / L KCl, 200 mg / L CaCl2, 140 mg / L NaH2PO4.H2O, 200 mg / L MgSO4.7H2O, 7.94 g / L HEPES, 2.2 g / L NaHCO3, 110 mg / L sodium pyruvate, 10 mg / L phenol red, 14 mg / L hypoxanthine, 1 mg / L biotin, 760 mg / L glycerol, 640 mg / L proline, 236 mg / L glutamic acid, 1.34 g / L glutamine, 7.5 mg / L hemin (in 50 mM sodium hydroxide), 1X MEM amino acid solution (Invitrogen),1X MEM non-essential amino acids solution (Invitrogen), 28.2 mg / L bathocuproine disulfonate.Na2, 182 mg/L cysteine, 0.2 mM 2-mercaptoethanol, 15% heat-inactivated FBS, and 5 mM sodium citrate and cis-aconitate [21, 22]. Following medium exchange, parasites were incubated at a lower temperature (27°C / 5% CO2), monitored for change in morphology to procyclic parasites, and sub-cultured for long-term cultivation.
For compound screening, 20 μliters (5,000 parasites) of T. b. brucei procyclic culture were added to 384-well plate wells filled with 20 μliters of DTM medium and pre-dispensed compounds (0.25% final DMSO concentration). Plates were then incubated for 72 hours at 27°C / 5% CO2. Cell viability was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega).
Plasmodium falciparum in vitro efficacy assay
GNF7686 and cytochrome b inhibitors were assayed for activity on two Plasmodium falciparum (P. falciparum) lines: D10attB and yDHODH-D10attB. The D10attB line is reliant on the coenzyme Q-dependent malarial dihydroorotate dehydrogenase (PfDHODH), whereas the yDHODH-D10attB line expresses also the fumarate-utilizing Saccharomyces cerevisiae (S. cerevisiae) DHODH, which circumvents reliance on PfDHODH and renders this line fully resistant to cytochrome b inhibitors [25]. The use of these two lines allows for distinction of selection of potent cytochrome b inhibitors as described in detail in the Results section [26, 27]. Growth and viability of P. falciparum cell lines (in the presence or absence of drug) in infected erythrocytes were assessed using a SYBR Green-based proliferation assay exactly as described previously [28].
Mammalian cell cytotoxicity assay
NIH/3T3 fibroblast cells were maintained in RPMI-1640 medium supplemented with 10% heat-inactivated FBS and 100 IU penicillin / 100 μg streptomycin per mL at 37°C / 5% CO2. For compound screening, cells were diluted to 4 × 104 cells / mL in assay medium (RPMI-1640, 5% FBS, and 100 IU penicillin / 100 μg streptomycin per mL) and seeded at 50 μL / well into 384-well plates. Following overnight incubation, compounds were added to each well (0.2% DMSO final concentration) and plates were further incubated for 96 hours. Cell viability was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). Measured luminescence values were normalized to the value obtained for 0.2% DMSO, and plotted against the corresponding compound concentration for half maximal cytotoxic concentration (CC50) value determination.
S. cerevisiae susceptibility assay
The minimal inhibitory concentrations (MIC) of the compounds for inhibition of S. cerevisiae drug pump knock-out strain NF7061 (MATa his3Δ 1; leu2Δ 0; met15Δ 0; ura3Δ 0; snq2::KanMX; pdr5::KanMX; pdr1::NAT1; pdr3::KanMX; yap1::NAT1; pdr2::LEU2; yrm1::MET; yor1::LYS2) were determined by a modification of the microdilution technique described elsewhere [29]. Briefly, two-fold dilutions of the compound solution in DMSO were made. They were added to each well of 96-well assay plate containing 200 μL per well of either YPD (1% Difco Yeast Extract, 2% Difco Bacto Peptone and 2% Dextrose) or YPG (3% of glycerol in replacement of 2% Dextrose in YP) medium. Early stationary yeast NF7061 cells grown in either medium were collected and resuspended to 2 × 105 cells / mL. Ten microliters of the yeast cell suspension were inoculated to each well of the plates containing medium and compound to achieve a final inoculum of approximately 104 CFU / mL. The plates were incubated at 30°C for 24 hours (containing YPD) or 48 hours (containing YPG). The MIC end point was defined as the lowest compound concentration exhibiting no visual growth.
Selection of GNF7686-resistant T. cruzi and cloning
T. cruzi epimastigotes were initially treated with GNF7686 at EC20 value (0.01 μM, 0.2% DMSO) and continually passaged at the same concentration until the culture growth rate matched that of epimastigotes growing in the medium containing 0.2% DMSO. Parasites were subsequently passaged in a similar manner in the presence of increasing concentration of GNF7686 until significant resistance was achieved (~5-fold increase in the EC50 value). The time to generate resistance was approximately eleven months. Resistant epimastigotes were cloned by the limiting dilution technique.
Whole genome sequencing
Following expansion of GNF7686-resistant T. cruzi clones in LIT media, T. cruzi total DNA was isolated using Qiagen DNeasy Blood and Tissue Kit from 108 parasites per sample. Whole genome sequencing was performed using Ilumina HiSeq1000 next-generation sequencing platform.
Sequencing reads were aligned by Burrows-Wheeler Aligner (BWA, version 5.9.0) to the T. cruzi JR cl. 4 genome (version 1.0). Simple single nucleotide variants (SNVs) were called (using Samtools 1.19) looking for SNVs or small indels with an overall quality > 100 where the control was the drug-sensitive parental CL clone. Approximately 600900 reads called a ‘T’ and 520 reads called a “G” at L197F position resulting in L197F mutation. Heterozygous calls were determined by Samtools, and verified in the Integrated Genomics Viewer. Putative SNVs were manually checked in IGV [30, 31].
To further confirm the presence of L197F mutation, the T. cruzi cytochrome b gene was amplified by PCR (forward primer 5’-AGCTACTGTTCCTGTATTCGGC-3’ and reverse primer 5’-ACAAAAACAAAGTCGCTCACAA-3’) and cloned into the pCR2.1 vector. The insert DNA was sequenced using M13R and M13F (-21) primers (Genewiz).
Measurement of T. cruzi epimastigote respiration
GNF7686 and cytochrome b inhibitors (0.2% DMS0 final concentration) were added to 384-well assay plates containing 20 μL of assay buffer (250 mM sucrose, 15 mM KCl, 5 mM MgCl2, 1 mM EGTA, 30 mM K2HPO4, pH 7.4) and allowed to dissolve for two hours. Meanwhile, T. cruzi epimastigotes were harvested (800 × g for 5 min at 4°C), washed twice with buffer A (10 mM Tris-HCl, pH = 7.4, 0.23 M mannitol, 0.07 M sucrose, 0.2 mM EDTA, 0.2% bovine serum albumin, 0.5 mM phenylmethanesulfonyl fluoride), and finally resuspended in buffer A at a final concentration of 150 × 106 epimastigotes / mL. Next, 20 μL of T. cruzi epimastigote suspension (3 x 106 parasites) was added to each sample well, and then 15 μL of MitoXpress-Xtra probe (Cayman Chemicals) was added (final assay volume of 60 μL). Probe phosphorescence is quenched in the presence of oxygen and is inversely proportional to the amount of oxygen present in the solution. HS mineral oil (20 μL, Cayman Chemicals) was added to wells to prevent oxygen exchange between the assay buffer and air. Blank wells containing assay buffer without parasites, corresponding compound, and mineral oil were prepared in parallel and values were subtracted from sample values to specifically measure changes in oxygen concentration caused by parasite respiration. All sample and blank wells were prepared in duplicate. The assay plate was transferred to a Gemini XPS fluorescence plate reader (Molecular Devices) and read at 3 minute intervals at excitation and emission wavelengths of 380 nm and 650 nm, respectively, for 30 minutes at 37°C. The slope for the linear portion of time course of fluorescence increase (rate of oxygen consumption) was calculated after subtraction of blank well values. Obtained slope values were normalized to the slope obtained for 0.2% DMSO, and plotted against the corresponding compound concentration for half maximal inhibitory concentration (IC50) value determination.
Measurement of T. cruzi complex III activity
T. cruzi epimastigotes (57 day old culture, density of 5 × 107 parasites / mL) were harvested by centrifugation (800 × g for 5 minutes at 4°C) and resuspended at 10 mg / mL of protein in buffer A containing 0.1 mg digitonin / mg protein. The parasites were then incubated with the detergent for 10 minutes at 26°C. The pellet fraction was collected by centrifugation (13,000 × g for 5 min at 4°C) and immediately used in the complex III assay.
Complex III activity was monitored using a coupled decylubiquinol / cytochrome c reaction [32, 33]. Decylubiquinone was first reduced to decylubiquinol as described [33]. The freshly reduced decylubiquinol (80 μM final concentration in the reaction) was added to a reaction buffer (25 mM potassium phosphate, 5 mM MgCl2, 2.5 mg / mL BSA, pH = 7.2) containing 1 mM KCN, 0.1 mM yeast cytochrome c, 0.6 mM n-D-B-maltoside, and 12 μg of digitonin-solubilized T. cruzi epimastigotes. The reduction of cytochrome c was monitored at 550 nm using the SpectraMax Plus384 absorbance microplate reader. Blank samples containing all components excluding decylubiquinol were processed in parallel and absorbance values were subtracted from sample absorbance values to specifically measure decylubiquinone-dependent reduction of cytochrome c. All sample and blank wells were prepared in triplicate. For IC50 determination, the slope of the linear portion of the corrected respiration trace was determined, and normalized to the slope obtained for 0.2% DMSO condition.
Preparation of rat skeletal muscle mitochondria
Sprague Dawley rats were euthanized and skeletal muscle (from hind legs) was removed and stored in CP1 buffer (0.1 M KCl, 0.05 M Tris, 2 mM EGTA, pH 7.4 at 4°C) on ice. Using the ‘Herb Mincer‘, tissue was minced 34 times and then transferred into CP1 buffer (on ice) to wash away fatty and connective tissues from muscle. Following two rounds of wash and decantation with CP1 buffer, the rinsed tissue was transferred to CP2 buffer (CP1 + 0.5% BSA, 5 mM MgCl2, 1 mM ATP, 250 units / 100 mL Protease Type VIII (Sigma P5380), pH 7.4 at 4°C) and incubated on ice for 5 minutes prior to homogenization using the Polytron PT3100. Following homogenization and centrifugation (500 × g for 10 min at 4°C), the supernatant was decanted using cheesecloth and further centrifuged (10,000 × g for 11 min at 4°C). The crude mitochondrial pellet was resuspended in 10 mL of CP1 buffer (carefully avoiding red blood cell pellet) and subjected to an additional centrifugation step (10,000 × g for 10 min at 4°C). The resulting pellet was again separated from the red blood cell pellet and centrifuged (600 x g for 6 min at 4°C). The supernatant containing resuspended mitochondria was subjected to one final round of centrifugation (5,000 × g for 11 min at 4°C) and the pelleted mitochondria were resuspended in CP1 buffer and stored at -80°C. Respiration in isolated rat mitochondria was measured using the same protocol as described for T. cruzi epimastigotes using 150 μg protein / sample.
Results
Characterization of anti-parasitic activity of GNF7686 HTS hit
GNF7686 (Fig 1A) was identified in a high throughput screen designed to find new small molecules with growth inhibition activity on L. donovani axenic amastigotes. A library of 700,000 compounds was assembled with a particular focus on drug-like properties and structural diversity and these compounds were tested for inhibitory activity on L. donovani at 4 μM concentration. The library has been previously profiled in more than 60 other high throughput screens, including biochemical and cell-based assays against human and pathogen targets. The screen history allowed rapid identification and elimination of compounds with a ‘frequent hitter’ property.
Fig. 1. GNF7686 clears T. cruzi amastigotes from infected 3T3 cells. 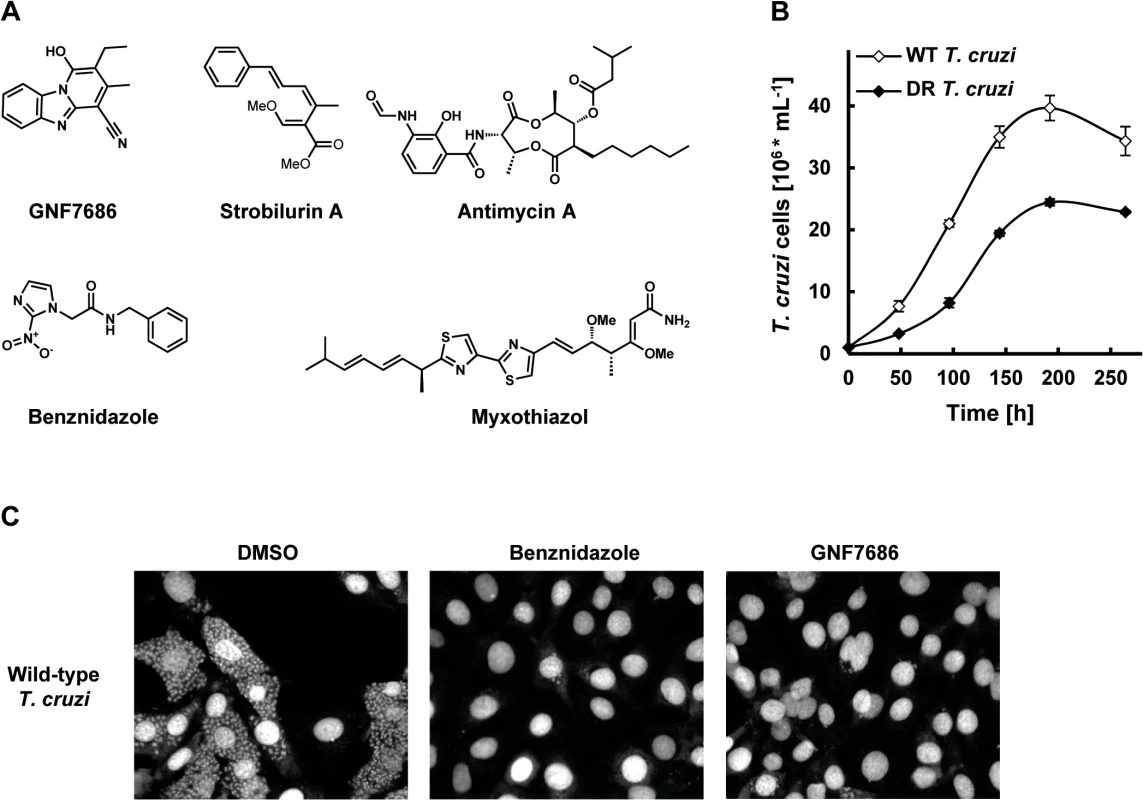
A) Structures of GNF7686, benznidazole and prototypic cytochrome b inhibitors used in this study. B) Growth curves of wild-type and GNF7686-resistant T. cruzi epimastigotes. C) Microscopy images of NIH/3T3 cells infected with T. cruzi after 48 hour treatment with 0.2% DMSO, 5 μM benznidazole or 1 μM GNF7686. The screen yielded 2,306 primary hits (0.3% hit rate) that inhibited growth by > 50%. Data from more than 95% of the assay plates had Z′ > 0.7, using DMSO as the negative control and 5 μM pentamidine as the positive control. Primary hits from the screen were further characterized using a dose−response assay format to determine the EC50 values. In parallel, cytotoxicity of these compounds was determined against a proliferating mouse fibroblast cell line (NIH/3T3). The final set of condirmed hits consisted of compounds that had EC50 < 4 μM against L. donovani, as well as low or no 3T3 cytotoxicity (CC50 > 10 μM or SI > 10; SI = CC50/EC50). The final set of confirmed L. donovani hits consisted of 1003 inhibitors.
The confirmed hits were further assayed for activity on other two medically important kinetoplastid parasites T. cruzi and T. brucei. GNF7686 was selected for further investigation because of potent in vitro activity on all three T. cruzi morphological forms (intracellular amastigote EC50 = 0.15 μM, trypomastigote EC50 = 0.71 μM, epimastigote EC50 = 0.16 μM; Table 1 and Fig 1C). GNF7686 also inhibited the growth of L. donovani axenic amastigotes (EC50 = 0.46 μM) and promastigotes (EC50 = 0.46 μM), but not the growth of T. b. brucei bloodstream form trypomastigotes (EC50 > 25 μM). Curiously, GNF7686 was active on T. b. brucei procyclics (EC50 = 0.59 μM), the parasite form found in the tsetse fly vector that mediates T. b. brucei transmission [34]. GNF7686 did not inhibit growth of 3T3 cell line (CC50 > 20 μM).
Tab. 1. Potency of GNF7686 and prototypic cytochrome b Inhibitors on wild-type (WT) and GNF7686-resistant (DR) T. cruzi morphological forms. 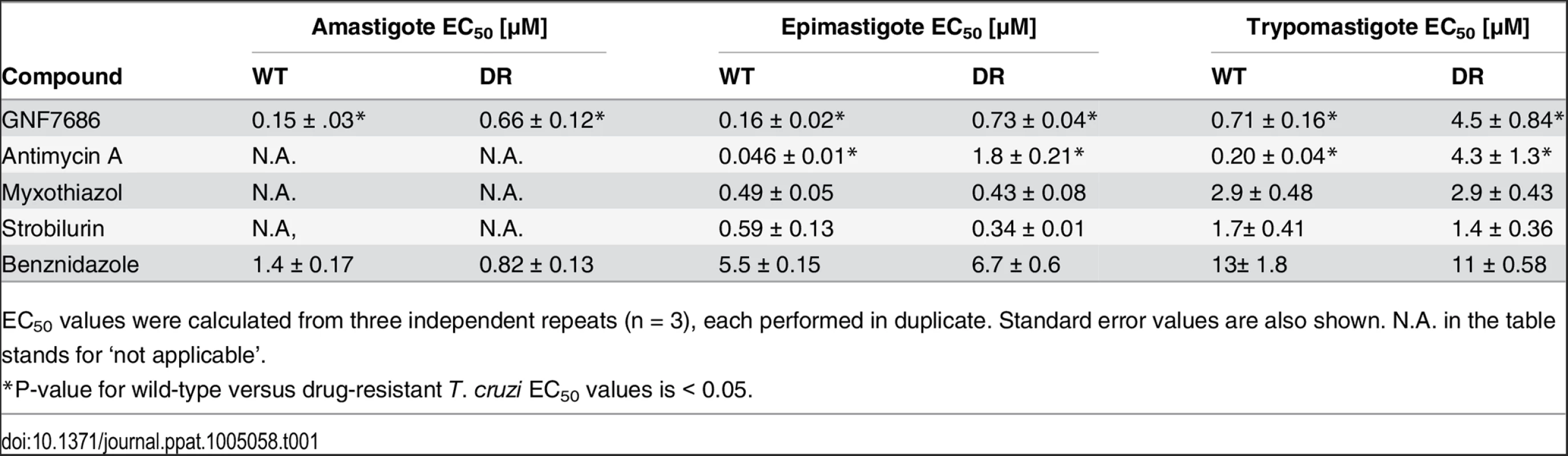
EC50 values were calculated from three independent repeats (n = 3), each performed in duplicate. Standard error values are also shown. N.A. in the table stands for ‘not applicable’. Identification and characterization of GNF7686-resistant T. cruzi
To investigate the mechanism of action of GNF7686, we selected a population of drug-resistant T. cruzi epimastigotes through a long-term parasite culture in the presence of this compound. As tolerance for GNF7686 gradually increased over time, we periodically escalated the selection pressure by raising the inhibitor concentration. In the course of eleven months, the EC50 of the T. cruzi culture shifted ~ 4-fold from 0.16 µM to 0.73 μM (Table 1).
Populations of evolving microbes often comprise cells harboring alternative mutations that are derived from different mutation events [35, 36]. To simplify analysis of genomic changes accumulated during the selection by characterizing homogenous culture populations, we isolated three clones from the GNF7686-resistant culture. All three clones exhibited the same extent of GNF7686 resistance (EC50 ~ 1 μM) as the parent culture. Importantly, the sensitivity of clones to benznidazole remained at the same level as observed for the wild-type T. cruzi strain (Table 1), demonstrating that resistance to GNF7686 did not arise through a broadly pleiotropic mechanism. When cultured in medium lacking GNF7686, all three clones grew at a rate similar to the wild-type strain (mutant epimastigote doubling time = 5355 hours vs 60 hours for the wild-type strain), but reached stationary phase at a lower cell density (~60% of the wild-type strain, Fig 1B).
Whole genome sequencing identified the same set of five mutations in all three clones that included L197F in cytochrome b, L283M in the ATPase subunit of HsIVU protease, R75C in TCSYLVIO008926 hypothetical protein, and two mutations in non-coding regions. The observation that the three clones were identical at the genome sequence level suggests that they were siblings derived from one founding cell that expanded in the passaged culture during the selection. During whole genome sequencing, multiple sequence reads (up to 100 in total) from many independent DNA molecules are obtained for each nucleotide position in the genome. We uncovered that the L197F mutation in the cytochrome b gene and one of two mutations mapped to non-coding regions fully replaced the corresponding wild-type alleles, while the other three mutations remained heterozygous during the selection. Interestingly, the two mutations map both to the kinetoplast maxicircle DNA, which is present in 2050 copies per cell [37, 38]. This indicates that these two mutations, presumably appearing on one maxicircle copy at first, replaced the corresponding wild-type alleles during the selection to the point of achieving homoplasmy.
Chemogenomic profiling of GNF7686 in yeast supports inhibition of the respiratory chain at the level of complex III
In addition to selection of T. cruzi mutants resistant to GNF7686, we subjected the inhibitor also to chemogenomic profiling in S. cerevisiae. The genome-wide deletion collections available for this eukaryotic model system provide powerful genetic tools for investigation of bioactive molecules [39, 40], and the approach was successfully applied to mechanism of action studies of various growth inhibitors in the past [41, 42]. In the haploinsufficiency profiling assay (HIP), complete collection of heterozygous yeast deletion strains, in which each strain has only one copy of a particular gene, is profiled for hypersensitivity to a compound. Gene deletions associated with increased compound sensitivity indicate pathways directly affected by the compound [43].
We observed that growth of S. cerevisiae is inhibited when the yeast was cultured in media containing glycerol but not glucose as the carbon source (see also below). We therefore conducted a HIP profiling experiment in medium containing glycerol. Testing of GNF7686 at its EC30 concentration in two independent HIP experiments resulted in a reproducible profile (Fig 2A). Identified hypersensitive heterozygous strains included those with deletions in genes involved in mitochondrial metabolism, such as CYT1 (cytochrome c1), HAP4 (a transcription factor involved in regulation of the respiratory chain including CYT1), CBP1 (a regulator of cytochrome b mRNA stability), and QCR6 (a subunit of the cytochrome c reductase complex) [44]. As all these hits pointed at inhibition of mitochondrial respiration by GNF7686, we performed additional HIP experiments with strobilurin, an inhibitor of cytochrome bc1 complex, and venturicidin, an inhibitor of F-type ATPase [45, 46]. In a control experiment, we also collected the HIP profile for benomyl, a microtubule binding inhibitor, which does not interfere with ATP production during oxidative phosphorylation.
Fig. 2. Chemogenomic profiling in S. cerevisiae suggests cytochrome b as the target of GNF7686 in yeast. 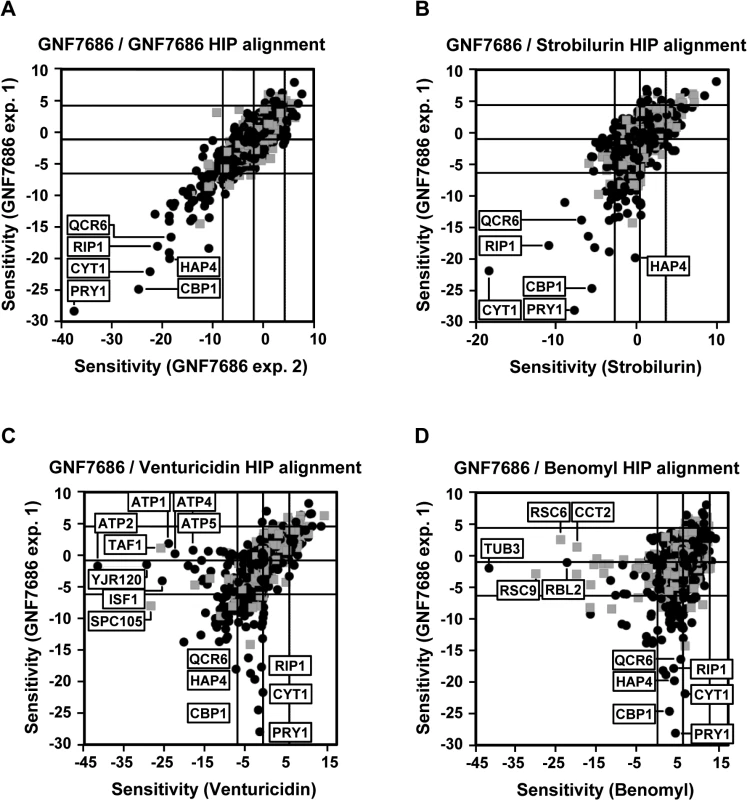
A) HIP profile of GNF7686. B-D) Alignment of the HIP profile of GNF7686 with profile of cytochrome b inhibitor strobilurin (B), F-type ATPase inhibitor venturicidin (C), and microtubule-binding fungicide benomyl (D). While both venturicidin and benomyl yielded HIP profiles distinctly different from that of GNF7686 (Fig 2C and 2D), treatment of gene deletion strain collection with the cytochrome bc1 inhibitor strobilurin identified essentially the same set of sensitive, heterozygous mutants as GNF7686 and included CYT1, HAP4, CBP1 and QCR6 (Fig 2B). It is important to note that the gene coding for cytochrome b, which is the proposed direct target of strobilurin [47], is encoded by the mitochondrial genome in S. cerevisiae and not amenable to standard gene targeting protocols. Thus, the cytochrome b gene deletion strain is not present in the yeast heterozygous deletion pool and could not be directly identified by the HIP method.
The observed HIP compound profiles strongly suggested that GNF7686 directly interferes with function of the S. cerevisiae respiratory chain, possibly at the level of complex III.
Prototypical cytochrome b inhibitors block in vitro proliferation of T. cruzi
Genomic analyses of GNF7686 resistance/sensitivity pointed to involvement of the T. cruzi cytochrome b in resistance to growth inhibition by GNF7686. Cytochrome b is a component of the multisubunit cytochrome bc1 complex, an asymmetric homodimer with two spatially separated catalytic sites QN and QP (Fig 3A). In concert, QN and QP catalyze oxidation of ubiquinol formed by preceding steps of the respiratory chain [48, 49].
Fig. 3. GNF7686 resistance-conferring mutation in T. cruzi cytochrome b structure. 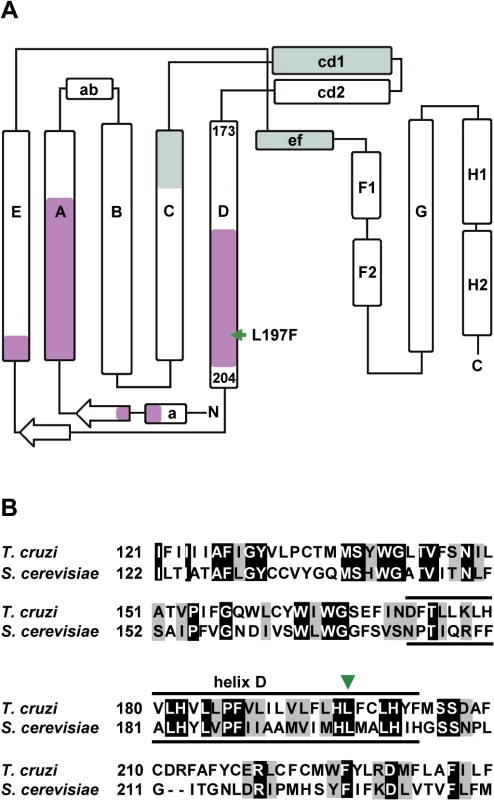
A) Secondary structure of yeast cytochrome b (adapted from Ding et al. 2006 and Fisher et al. 2008, [50, 57]) with amino acid sequence stretches that form QP and QN ubiquinol-binding sites highlighted (green and violet, respectively). The amino acid indicated by a green arrow corresponds to the L197F mutation (equivalent to L198F in S. cerevisiae) present in the GNF7686-resistant T. cruzi strain. B) Alignment of S cerevisiae and T. cruzi cytochrome b amino acid sequence around L197F mutation. Identical amino acids are highlighted in black and conserved substitutions are highlighted in grey. Inhibitors of cytochrome b are already of interest as anti-parasitic drugs. Atovaquone, a hydroxy-naphthoquinone inhibitor of the QP site, is used in the treatment of malaria and fungal pneumonia [50]. Another hydroxy-naphthoquinone, buparvaquone, is used to treat cattle theileriosis, and potently inhibits growth of L. donovani [19, 51]. Surprisingly, the potential of cytochrome b inhibitors for treatment of Chagas disease has not been explored, even though cytochrome b inhibitors including antimycin A were shown to affect T. cruzi mitochondrial respiration, bioenergetics, and calcium homeostasis [52–55]. To assess the effect of this class of compounds on T. cruzi growth and survival, we tested prototypical QN and QP site inhibitors on intracellular amastigotes, trypomastigotes, and epimastigotes (inhibitor structures shown in Fig 1A) [46, 49].
The QN site inhibitor antimycin A potently inhibited the growth of epimastigotes and rapidly reduced viability of trypomastigotes. We also observed T. cruzi inhibition by compounds targeting the cytochrome b QP site. Two well-characterized QP site inhibitors, myxothiazol and strobilurin, blocked growth of epimastigotes, and reduced viability of trypomastigotes (Table 1). The effect of antimycin A, myxothiazol, and strobilurin on intracellular amastigotes could not be accurately determined because of the inhibitor toxicity on the host 3T3 cells.
T. b. brucei is a kinetoplastid parasite closely related to T. cruzi at the genomic level [56]. While the parasite bloodstream (mammalian) form of T. b. brucei relies primarily on the glycolytic pathway for ATP production, growth of the procyclic (insect) form requires activity of the conventional respiratory pathway, including cytochrome b [48]. In line with the hypothesis that GNF7686 is a cytochrome b inhibitor, GNF7686 inhibited the growth of the procyclic but not bloodstream T. b. brucei parasites (EC50 = 0.59 μM vs > 25 μM). We also observed similar differential activity on the two T. brucei forms with the other tested cytochrome b inhibitors such as antimycin A (EC50 = 0.03 μM vs > 25 μM; Table 2).
Tab. 2. Potency of GNF7686 and prototypic cytochrome b inhibitors in L. donovani, T. brucei, P. falciparum and S. cerevisiae proliferation assays. 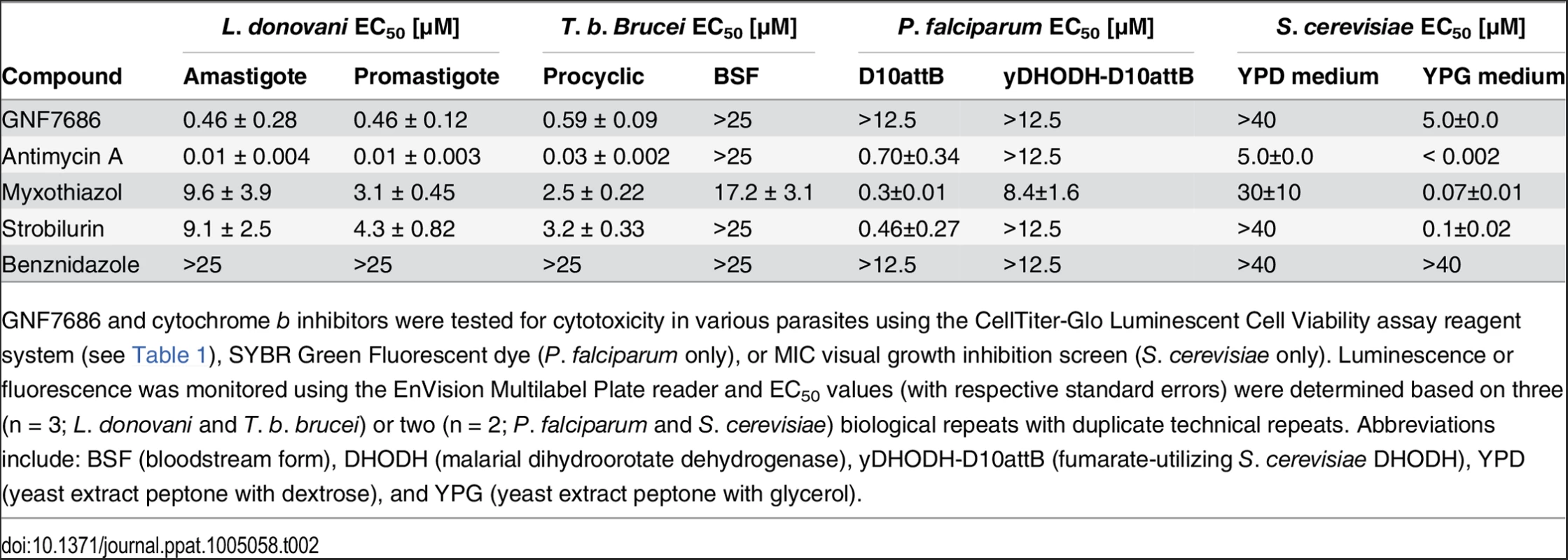
GNF7686 and cytochrome b inhibitors were tested for cytotoxicity in various parasites using the CellTiter-Glo Luminescent Cell Viability assay reagent system (see Table 1), SYBR Green Fluorescent dye (P. falciparum only), or MIC visual growth inhibition screen (S. cerevisiae only). Luminescence or fluorescence was monitored using the EnVision Multilabel Plate reader and EC50 values (with respective standard errors) were determined based on three (n = 3; L. donovani and T. b. brucei) or two (n = 2; P. falciparum and S. cerevisiae) biological repeats with duplicate technical repeats. Abbreviations include: BSF (bloodstream form), DHODH (malarial dihydroorotate dehydrogenase), yDHODH-D10attB (fumarate-utilizing S. cerevisiae DHODH), YPD (yeast extract peptone with dextrose), and YPG (yeast extract peptone with glycerol). The inhibition of various morphological forms of T. cruzi by prototypical cytochrome b inhibitors is consistent with the hypothesis that the cytochrome b fulfills an essential function in parasite physiology and that GNF7686 inhibits its function. However, with the exception of antimycin A, the anti-parasitic potency of the other tested inhibitors is too weak to be of therapeutic significance.
GNF7686 is an inhibitor of cytochrome b QN site
Through a literature search, we identified a previously published report that described L198F mutation in S. cerevisiae cytochrome b, which is equivalent to the L197F mutation in T. cruzi cytochrome b (Fig 3B). The yeast L198F mutation confers resistance to ilicicolin H, a cytochrome b inhibitor with potent anti-fungal activity [46, 57]. Inspection of high resolution crystal structure of the yeast cytochrome bc1 complex further revealed that the Leu198 side chain is positioned in close proximity (< 5 Å) to ubiquinol bound inside the QN pocket and next to His197, which coordinates the iron atom in the bH heme. In accordance with the structure, L198F mutation also conferred resistance in yeast to other tested QN site inhibitors such as funiculosin and antimycin A [57, 58].
Additional characterization of the GNF7686-resistant T. cruzi mutants revealed that they were selectively resistant to antimycin A, a QN site inhibitor [46]. While antimycin A displayed very potent activity on wild type epimastigotes, a sharp decrease in potency (40-fold) was observed with GNF7686-resistant epimastigotes (EC50 = 1.8 μM, Table 1). Similarly, a steep shift in potency (20-fold) was observed between the wild-type and mutant trypomastigotes (Table 1). In contrast, QP inhibitors myxothiazol and strobilurin showed comparable activity on both wild-type and resistant strains (Table 1). In summary, the L197F mutation in the T. cruzi cytochrome b is likely located within the QN site and can interfere with binding of QN site inhibitors in a similar way as was previously described for the L198F mutation in the S. cerevisiae cytochrome b.
GNF7686 inhibits cytochrome b and respiration of T. cruzi
During aerobic respiration, the electron transport chain (ETC) conducts electrons derived from reduced carbon substrates through a series of redox reactions to the terminal electron acceptor, molecular oxygen, which is then reduced to water [33, 48]. Inhibition of electron flow through the ETC at any step, including cytochrome b, results in a block of oxygen consumption.
To evaluate whether GNF7686 disrupts the function of the T. cruzi ETC, oxygen consumption by intact T. cruzi epimastigotes was monitored in the presence of GNF7686 and prototypic cytochrome b inhibitors (Fig 4A and 4B). Antimycin A potently blocked respiration by wild-type epimastigotes (IC50 = 0.04 μM), but was ~7-fold less potent on GNF7686-resistant parasites (IC50 = 0.27 μM). Two QP site inhibitors employed in this report, myxothiazol and strobilurin, both inhibited epimastigote respiration, but, in contrast to antimycin A, the respiratory IC50 values of the QP inhibitors were comparable between wild-type and GNF7686-resistant T. cruzi.
Fig. 4. GNF7686 inhibits cellular respiration and cytochrome b function in T. cruzi. 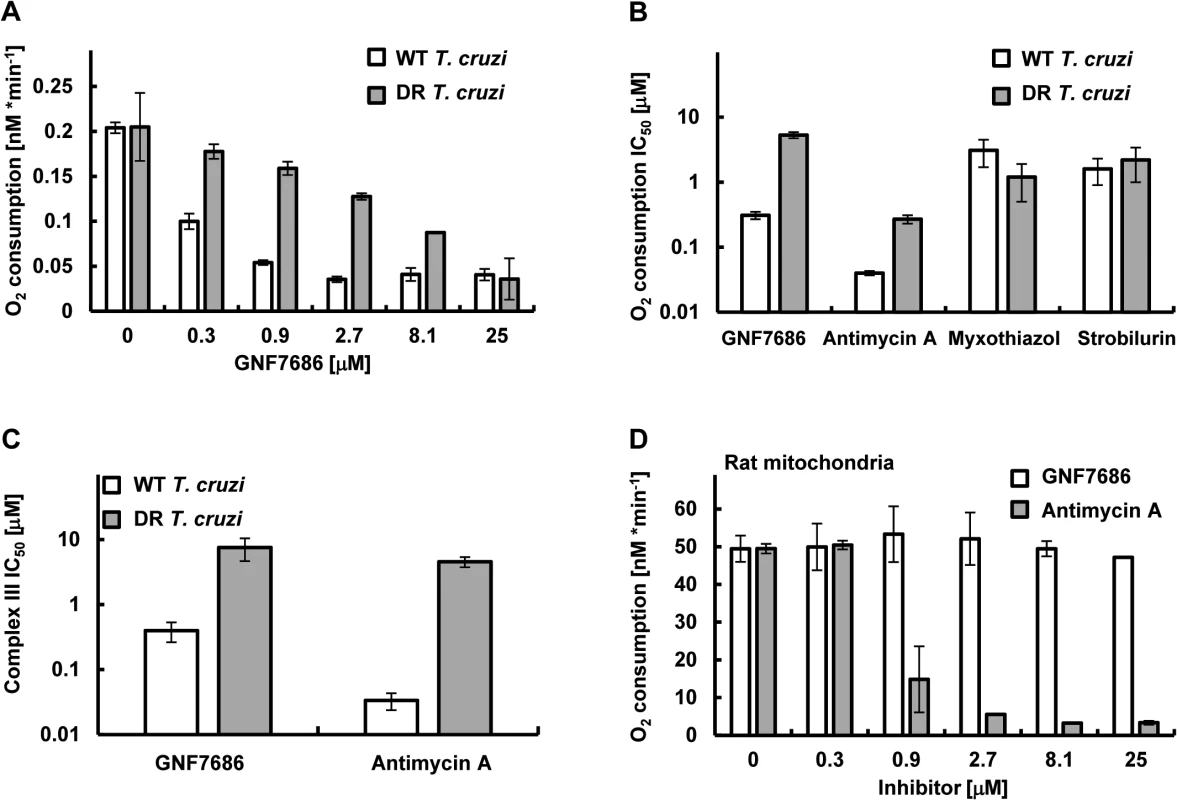
A) T. cruzi epimastigote respiration was monitored using the MitoXpress-Xtra HS phosphorescent probe (see Methods) in the presence of 0.2% DMSO (control) and various concentrations of GNF7686. Oxygen consumption rates per 106 T. cruzi epimastigotes cells are shown. Plotted values were derived from three biological repeats (n = 3) with duplicate technical repeats in either wild-type or evolved GNF7686-resistant (DR) parasites. B) Oxygen consumption IC50 values for prototypic cytochrome b inhibitors in T. cruzi epimastigote respiration assay on wild-type and GNF7686-resistant (DR) parasites. Shown GNF7686 IC50 values were derived from the experiment shown in the (A) panel. C) Mitochondrial complex III activity was monitored in digitonin-solubilized T. cruzi epimastigotes (both wild-type and evolved GNF7686-resistant (DR) parasite strains) utilizing a coupled decylubiquinol oxidation / cytochrome c reduction reaction in the presence of a compound (GNF7686 or antimycin A). IC50 values were determined based on three biological repeats (n = 3) with triplicate technical repeats in either wild-type or evolved drug-resistant (DR) parasites relative to 0.2% DMSO conditions. D) High selectivity of GNF7686 for T. cruzi cytochrome b is reflected in the absence of inhibition of rat mitochondrial respiration by this compound up to 25 μM concentration. For comparison, antimycin A potently inhibits mammalian mitochondrial respiration. Oxygen consumption rates per 1 mg of total mitochondrial protein are shown. GNF7686 inhibited respiration by wild-type parasites with an IC50 = 0.21 μM. A significant drop in inhibitor potency was observed with the GNF7686-resistant T. cruzi epimastigotes (oxygen consumption IC50 = 5.2 μM). As seen for growth inhibition, the results on respiration inhibition distinguish GNF7686 and antimycin A from the QP site inhibitors (Fig 4B).
We then asked whether GNF7686 inhibits T. cruzi cytochrome bc1 (complex III) directly (Fig 4C). Epimastigotes were permeabilized with digitonin, and KCN (complex IV inhibitor) was added to the permeabilized cells to block electron conductance by the parasite ETC. The reaction was then initiated by adding stoichiometric quantities of decylubiquinol (an electron donor for complex III) and oxidized yeast cytochrome c (an electron acceptor), and the catalytic activity of complex III was monitored through accumulation of reduced yeast cytochrome c. A control reaction with antimycin A confirmed earlier observations with intact epimastigotes (Table 1). Antimycin A potently blocked reduction of cytochrome c in the reaction with wild-type permeabilized epimastigotes, but a dramatic loss of inhibitor potency was observed (~100-fold) when GNF7686-resistant permeabilized parasites were used (Fig 4C). In a similar fashion, GNF7686 inhibited wild-type complex III activity with an IC50 of 0.40 μM, but a 20-fold loss of potency was observed with the mutant complex III (Fig 4C). These observations validate GNF7686 as a complex III inhibitor that likely targets the QN site of the T. cruzi cytochrome b.
GNF7686 is highly selective for T. cruzi and does not inhibit mammalian respiration
The inhibitory effect of GNF7686 on mammalian cytochrome b function was assessed through monitoring oxygen consumption by mitochondria isolated from rat skeletal muscle cells. In the control reaction, antimycin A showed a potent inhibition (IC50 = 0.81 μM) of mitochondrial respiration (Fig 4D). In a parallel experiment, GNF7686 did not show any effect on oxygen consumption up to 25 μM concentration (Fig 4D). This observation validates GNF7686 as a highly selective inhibitor of the T. cruzi cytochrome b and a promising starting point for Chagas disease drug discovery.
We also examined effect of GNF7686 on cytochrome b in the malaria parasite, P. falciparum, and in yeast S. cerevisiae, the latter being used as a surrogate for pathogenic Pneumocystis jirovecii, a causative agent of a pneumocystis pneumonia [59]. Cytochrome b is a validated drug target in both organisms and atovaquone, an inhibitor of cytochrome b, is a clinical treatment for these diseases.
For the P. falciparum studies, all inhibitors were tested on two parasite lines—D10attB and yDHODH-D10attB (Table 2). In the wild-type D10attB line, the parasite de novo pyrimidine biosynthesis is dependent on a type 2 dihydroorotate dehydrogenase (PfDHODH) and requires a functional P. falciparum ETC, including cytochrome b, downstream from PfDHODH [26, 27]. In contrast, the yDHODH-D10attB cell line is modified with a type 1A dihydroorotate dehydrogenase from S. cerevisiae (yDHODH), which is cytosolic and utilizes fumarate as the terminal electron acceptor [25–27]. The assay was validated with antimycin A, which blocked growth of the D10attB line (EC50 = 0.7 μM), but was inactive on the yDHODH-D10attB parasite line (EC50 > 12.5 μM). Myxothiazol and strobilurin also showed a similar preferential activity on the D10attB line, whereas GNF7686 did not inhibit growth of either P. falciparum cell line. This result suggests that GNF7686 is not active on P. falciparum cytochrome b.
A similar, growth inhibition-based assessment of GNF7686 effect on the ETC function was also performed on S. cerevisiae (Table 2). Growth inhibition of a wild-type S. cerevisiae strain by compounds in two different media was monitored. The first medium contained glucose as the sole carbon source, which allows growth of yeast cells that lack a functional ETC [60]. In the second medium, glucose was replaced with glycerol, a non-fermentable carbon source. Under the latter condition, yeast growth is dependent on cellular respiration and functional cytochrome b [60]. All prototypic cytochrome b inhibitors used in this report potently inhibited yeast growth on medium with glycerol, but were inactive when yeast grew in medium with glucose. Following a similar pattern, GNF7686 weakly inhibited growth of yeast in glycerol medium (EC50 = 5.0 μM), but did not affect yeast growing in the medium with glucose.
In summary, GNF7686 selectively inhibits T. cruzi cytochrome b and does not affect respiration of mammalian mitochondria nor does it significantly inhibit respiration-dependent growth of P. falciparum and S. cerevisiae.
Discussion
We have shown that cytochrome b is a possible target for new drug discovery efforts aimed at treating kinetoplastid diseases. The importance of this finding is underlined by the paucity of drug targets for these diseases. For Chagas disease, only sterol 14α-demethylase (CYP51) and cruzain have been explored in depth as possible targets. However, low parasitological cure rates that were observed (20–30%) during clinical testing of anti-fungal drugs targeting sterol 14α-demethylase (posaconazole and ravuconazole prodrug E1224) in Chagas disease patients have lessened enthusiasm for further work on repurposed CYP51 inhibitors as single agents [61, 62]. Additional clinical evaluation of this class of drugs partnered with benznidazole for combination treatments is still planned. It is also important to note that the failure of posaconazole and E1224 in phase 2 trials have been attributed to insufficient drug exposure or dosing duration [14], and work on T. cruzi-specific CYP51 inhibitors that could enter clinical development in the future is also ongoing [63, 64]. Finally, the anti-cancer drug BEZ235 has activity across kinetoplastid parasites, but it requires additional optimization (improvement of therapeutic index) before becoming a preclinical candidate [65]. Given this sparse landscape, new chemical starts and new drug targets are urgently needed to anchor drug discovery efforts for the kinetoplastid diseases.
Many groups have resorted to a ‘pre-genomic’ approach to drug discovery, in which compounds are screened to identify inhibitors of pathogen growth, without regard to mechanism of action. While this approach typically provides large number of chemical starting points and broad hit diversity, a lack of information on mechanism of action creates additional risk in chemical optimization, and in predicting possible toxicity liabilities. Next generation sequencing of evolved resistant pathogens has been used successfully to identify resistance mechanisms and, in many cases, target mechanisms, for several pathogens. In our own program, we have identified targets for malaria and tuberculosis [66, 67]. However, the approach has not been reduced to practice in kinetoplastid drug discovery until this study.
Several features of the results in this study bode well for future application of the approach. First, the number of mutations associated with the emergence of drug resistance was relatively low (five point mutations), which simplified subsequent target prediction and validation. Second, we were able to generate resistance in T. cruzi epimastigotes despite a relatively long doubling time and stable genome. Selection process required almost a year of drug pressure in this study but was ultimately successful. Finally, we found a mutation in a gene that is a ‘plausible’ drug target, where plausibility is supported by essentiality of the mutated gene in other cellular systems, or precedence from drugs targeting homologous targets in other organisms.
Drugs targeting cytochrome b are in clinical use for treatment of malaria and fungal pneumonia, and cytochrome b was also reported as a promising target for treatment of tuberculosis. The current study extends utility of cytochrome b as a drug target also to Chagas disease, and possibly leishmaniasis. Various structurally different cytochrome b inhibitors showed patterns of growth and biochemical inhibition that consistently confirmed that functional cytochrome b is essential for T. cruzi propagation. Based on the presented validation data, T. cruzi growth can be inhibited through targeting either QN or QP cytochrome b site. GNF7686 represents a new cytochrome b inhibitor, likely targeting the QN site, which has high selectivity for T. cruzi and does not show any effect on respiration of mammalian mitochondria. Crystal structures of cytochrome b from several sources (bovine, chicken, yeast, Rhodobacter, Paracoccus) were previously published [68, 69]. This opens opportunities for rational drug design based on homology modeling of T. cruzi cytochrome b structure. Finally, a counter-screen assay to measure inhibitory activity on the cognate human enzyme (as described here) can be used to guide chemical optimization away from host toxicity. While GNF7686 appears to be a promising starting point for kinetoplastid drug discovery, it does not inhibit the growth of P. falciparum or S. cerevisiae, and thus may not be a suitable starting point for anti-malaria or anti-fungal drug discovery. Further in vivo characterization of GNF7686 revealed that it has poor pharmacokinetic properties, including low oral bioavailability (F = 6%) and high in vivo mouse clearance (53 mL * min-1 * kg-1); thus, extension of the current studies to an in vivo model of Chagas disease will require identification of a GNF7686 analogue that has improved pharmacokinetic profile.
In summary, we have established an approach for identification of molecular targets of T. cruzi growth inhibitors that enables transition to target-based drug discovery for compounds with previously unknown mechanism of action. The first application of this approach resulted in identification of a highly selective inhibitor of T. cruzi cytochrome b, GNF7686, which can serve as an excellent starting point for discovery of new drugs for Chagas disease and leishmaniasis.
Zdroje
1. Anis Rassi Jr AR, Jose Antonio Marin-Neto (2010) Chagas disease. Lancet 375 : 1388–1402. doi: 10.1016/S0140-6736(10)60061-X 20399979
2. Magdaleno A, Suarez Mantilla B, Rocha SC, Pral EM, Silber AM (2011) The Involvement of Glutamate Metabolism in the Resistance to Thermal, Nutritional, and Oxidative Stress in Trypanosoma cruzi. Enzyme Res 2011 : 486928. doi: 10.4061/2011/486928 21629861
3. Rodolfo Viotti CV, Bruno Lococo, Maria Gabriela Alvarez, Marcos Petti, Graciela Bertocchi and Alejandro Armenti (2009) Side effects of benznidazole as treatment in chronic Chagas disease: fears and realities. Expert Rev Anti Infect Ther 7 : 157–163. doi: 10.1586/14787210.7.2.157 19254164
4. Bern C, Kjos S, Yabsley MJ, Montgomery SP (2011) Trypanosoma cruzi and Chagas' Disease in the United States. Clin Microbiol Rev 24 : 655–681. doi: 10.1128/CMR.00005-11 21976603
5. Hurwitz I, Fieck A, Klein N, Jose C, Kang A, et al. (2012) A Paratransgenic Strategy for the Control of Chagas Disease. Psyche: A Journal of Entomology 2012 : 1–10.
6. Buckner FS, Navabi N (2010) Advances in Chagas disease drug development: 2009–2010. Curr Opin Infect Dis 23 : 609–616. doi: 10.1097/QCO.0b013e3283402956 20885320
7. Choi JY, Calvet CM, Gunatilleke SS, Ruiz C, Cameron MD, et al. (2013) Rational development of 4-aminopyridyl-based inhibitors targeting Trypanosoma cruzi CYP51 as anti-chagas agents. J Med Chem 56 : 7651–7668. doi: 10.1021/jm401067s 24079662
8. Clayton J (2010) Chagas disease: pushing through the pipeline. Nature 465: S12–15. doi: 10.1038/nature09224 20571548
9. Friggeri L, Hargrove TY, Rachakonda G, Williams AD, Wawrzak Z, et al. (2014) Structural Basis for Rational Design of Inhibitors Targeting Trypanosoma cruzi Sterol 14alpha-Demethylase: Two Regions of the Enzyme Molecule Potentiate its Inhibition. J Med Chem.
10. Bustamante JM, Craft JM, Crowe BD, Ketchie SA, Tarleton RL (2014) New, combined, and reduced dosing treatment protocols cure Trypanosoma cruzi infection in mice. J Infect Dis 209 : 150–162. doi: 10.1093/infdis/jit420 23945371
11. Pinazo MJ, Espinosa G, Gallego M, Lopez-Chejade PL, Urbina JA, et al. (2010) Successful treatment with posaconazole of a patient with chronic Chagas disease and systemic lupus erythematosus. Am J Trop Med Hyg 82 : 583–587. doi: 10.4269/ajtmh.2010.09-0620 20348503
12. DNDi Press Release. Drug Trial for Leading Parasitic Killer of the Americas Shows Mixed Results but Provides New Evidence for Improved Therapy. Drugs for Neglected Diseases Initiative. 14 November 2013. http://www.dndi.org/media-centre/press-releases/1700-e1224.html. Accessed 01 December 2014.
13. VandeBerg JL (2014) Treatment Trials and Efficacy Determination in Non-human Primates with Chronic T. cruzi Infections. Southwest National Primate Research Center at the Texas Biomedical Research Institute, San Antonio, Texas, USA. pp. 23.
14. Urbina JA (2015) Recent clinical trials for the etiological treatment of chronic chagas disease: advances, challenges and perspectives. J Eukaryot Microbiol 62 : 149–156. doi: 10.1111/jeu.12184 25284065
15. McKerrow JH, Doyle PS, Engel JC, Podust LM, Robertson SA, et al. (2009) Two approaches to discovering and developing new drugs for Chagas disease. Mem Inst Oswaldo Cruz 104 Suppl 1 : 263–269. 19753483
16. Choy JW, Bryant C, Calvet CM, Doyle PS, Gunatilleke SS, et al. (2013) Chemical-biological characterization of a cruzain inhibitor reveals a second target and a mammalian off-target. Beilstein J Org Chem 9 : 15–25. doi: 10.3762/bjoc.9.3 23400640
17. Engel JC, Doyle PS, Hsieh I, McKerrow JH (1998) Cysteine protease inhibitors cure an experimental Trypanosoma cruzi infection. J Exp Med 188 : 725–734. 9705954
18. Doyle PS, Zhou YM, Engel JC, McKerrow JH (2007) A cysteine protease inhibitor cures Chagas' disease in an immunodeficient-mouse model of infection. Antimicrob Agents Chemother 51 : 3932–3939. 17698625
19. Carneiro G, Aguiar MG, Fernandes AP, Ferreira LA (2012) Drug delivery systems for the topical treatment of cutaneous leishmaniasis. Expert Opin Drug Deliv 9 : 1083–1097. doi: 10.1517/17425247.2012.701204 22724539
20. Gilbert IH, Leroy D, Frearson JA (2011) Finding new hits in neglected disease projects: target or phenotypic based screening? Curr Top Med Chem 11 : 1284–1291. 21401505
21. Bourguignon SC, de Souza W, Souto-Padron T (1998) Localization of lectin-binding sites on the surface of Trypanosoma cruzi grown in chemically defined conditions. Histochem Cell Biol 110 : 527–534. 9826132
22. Garcia-Silva MR, Frugier M, Tosar JP, Correa-Dominguez A, Ronalte-Alves L, et al. (2010) A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol 171 : 64–73. doi: 10.1016/j.molbiopara.2010.02.003 20156490
23. Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, et al. (2006) CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7: R100. 17076895
24. Engel JC, Ang KK, Chen S, Arkin MR, McKerrow JH, et al. (2010) Image-based high-throughput drug screening targeting the intracellular stage of Trypanosoma cruzi, the agent of Chagas' disease. Antimicrob Agents Chemother 54 : 3326–3334. doi: 10.1128/AAC.01777-09 20547819
25. Ke H, Morrisey JM, Ganesan SM, Painter HJ, Mather MW, et al. (2011) Variation among Plasmodium falciparum strains in their reliance on mitochondrial electron transport chain function. Eukaryot Cell 10 : 1053–1061. doi: 10.1128/EC.05049-11 21685321
26. Nam TG, McNamara CW, Bopp S, Dharia NV, Meister S, et al. (2011) A chemical genomic analysis of decoquinate, a Plasmodium falciparum cytochrome b inhibitor. ACS Chem Biol 6 : 1214–1222. doi: 10.1021/cb200105d 21866942
27. Painter HJ, Morrisey JM, Mather MW, Vaidya AB (2007) Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature 446 : 88–91. 17330044
28. Plouffe D, Brinker A, McNamara C, Henson K, Kato N, et al. (2008) In silico activity profiling reveals the mechanism of action of antimalarials discovered in a high-throughput screen. Proc Natl Acad Sci U S A 105 : 9059–9064. doi: 10.1073/pnas.0802982105 18579783
29. Ghannoum MA, Ibrahim AS, Fu Y, Shafiq MC, Edwards JE Jr., et al. (1992) Susceptibility testing of Cryptococcus neoformans: a microdilution technique. J Clin Microbiol 30 : 2881–2886. 1452658
30. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29 : 24–26. doi: 10.1038/nbt.1754 21221095
31. Thorvaldsdottir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14 : 178–192. doi: 10.1093/bib/bbs017 22517427
32. Telford JE, Kilbride SM, Davey GP (2010) Decylubiquinone increases mitochondrial function in synaptosomes. J Biol Chem 285 : 8639–8645. doi: 10.1074/jbc.M109.079780 20080966
33. Spinazzi M, Casarin A, Pertegato V, Salviati L, Angelini C (2012) Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat Protoc 7 : 1235–1246. doi: 10.1038/nprot.2012.058 22653162
34. International Glossina Genome I (2014) Genome sequence of the tsetse fly (Glossina morsitans): vector of African trypanosomiasis. Science 344 : 380–386. doi: 10.1126/science.1249656 24763584
35. Holmes R K, Jobling M.G. (1996) Medical Microbiology. 4th Edition. Galveston, TX: University of Texas Medical Branch at Galveston,.
36. Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, et al. (2013) Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500 : 571–574. doi: 10.1038/nature12344 23873039
37. Lukes J, Guilbride DL, Votypka J, Zikova A, Benne R, et al. (2002) Kinetoplast DNA network: evolution of an improbable structure. Eukaryot Cell 1 : 495–502. 12455998
38. Messenger LA, Llewellyn MS, Bhattacharyya T, Franzen O, Lewis MD, et al. (2012) Multiple mitochondrial introgression events and heteroplasmy in trypanosoma cruzi revealed by maxicircle MLST and next generation sequencing. PLoS Negl Trop Dis 6: e1584. doi: 10.1371/journal.pntd.0001584 22506081
39. Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, et al. (1999) Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet 21 : 278–283. 10080179
40. Smith AM, Ammar R, Nislow C, Giaever G (2010) A survey of yeast genomic assays for drug and target discovery. Pharmacol Ther 127 : 156–164. doi: 10.1016/j.pharmthera.2010.04.012 20546776
41. Nyfeler B, Hoepfner D, Palestrant D, Kirby CA, Whitehead L, et al. (2012) Identification of elongation factor G as the conserved cellular target of argyrin B. PLoS One 7: e42657. doi: 10.1371/journal.pone.0042657 22970117
42. Richie DL, Thompson KV, Studer C, Prindle VC, Aust T, et al. (2013) Identification and evaluation of novel acetolactate synthase inhibitors as antifungal agents. Antimicrob Agents Chemother 57 : 2272–2280. doi: 10.1128/AAC.01809-12 23478965
43. Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, et al. (2004) Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci U S A 101 : 793–798. 14718668
44. Cherry JM HE, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. pp. D700–705.
45. Fisher N, Meunier B (2005) Re-examination of inhibitor resistance conferred by Qo-site mutations in cytochrome b using yeast as a model system. Pest Manag Sci 61 : 973–978. 15912560
46. Gutierrez-Cirlos EB, Merbitz-Zahradnik T, Trumpower BL (2004) Inhibition of the yeast cytochrome bc1 complex by ilicicolin H, a novel inhibitor that acts at the Qn site of the bc1 complex. J Biol Chem 279 : 8708–8714. 14670947
47. di Rago JP, Coppee JY, Colson AM (1989) Molecular basis for resistance to myxothiazol, mucidin (strobilurin A), and stigmatellin. Cytochrome b inhibitors acting at the center o of the mitochondrial ubiquinol-cytochrome c reductase in Saccharomyces cerevisiae. J Biol Chem 264 : 14543–14548. 2547800
48. Trumpower BL (1990) Cytochrome bc1 Complexes of Microorganisms. MICROBIOLOGICAL REVIEWS 54 : 101–129. 2163487
49. Huang LS, Cobessi D, Tung EY, Berry EA (2005) Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: a new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J Mol Biol 351 : 573–597. 16024040
50. Fisher N, Meunier B (2008) Molecular basis of resistance to cytochrome bc1 inhibitors. FEMS Yeast Res 8 : 183–192. 18093133
51. Soni MP, Shelkar N, Gaikwad RV, Vanage GR, Samad A, et al. (2014) Buparvaquone loaded solid lipid nanoparticles for targeted delivery in theleriosis. J Pharm Bioallied Sci 6 : 22–30. doi: 10.4103/0975-7406.124309 24459400
52. Peloso Ede F, Vitor SC, Ribeiro LH, Pineyro MD, Robello C, et al. (2011) Role of Trypanosoma cruzi peroxiredoxins in mitochondrial bioenergetics. J Bioenerg Biomembr 43 : 419–424. doi: 10.1007/s10863-011-9365-4 21732175
53. Silva TM, Peloso EF, Vitor SC, Ribeiro LH, Gadelha FR (2011) O2 consumption rates along the growth curve: new insights into Trypanosoma cruzi mitochondrial respiratory chain. J Bioenerg Biomembr 43 : 409–417. doi: 10.1007/s10863-011-9369-0 21732174
54. Docampo R, Moreno SN, Vercesi AE (1993) Effect of thapsigargin on calcium homeostasis in Trypanosoma cruzi trypomastigotes and epimastigotes. Mol Biochem Parasitol 59 : 305–313. 8341327
55. Martins RM, Covarrubias C, Rojas RG, Silber AM, Yoshida N (2009) Use of L-proline and ATP production by Trypanosoma cruzi metacyclic forms as requirements for host cell invasion. Infect Immun 77 : 3023–3032. doi: 10.1128/IAI.00138-09 19433547
56. El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, et al. (2005) Comparative genomics of trypanosomatid parasitic protozoa. Science 309 : 404–409. 16020724
57. Ding MG, di Rago JP, Trumpower BL (2006) Investigating the Qn site of the cytochrome bc1 complex in Saccharomyces cerevisiae with mutants resistant to ilicicolin H, a novel Qn site inhibitor. J Biol Chem 281 : 36036–36043. 16987808
58. di Rago J.-P. JP, and Colson A.-M. (1990) Isolation and RNA sequence analysis of cytochrome b mutants resistant to funiculosin, a center i inhibitor of the mitochondrial ubiquinol-cytochrome c reductase in Saccharomyces cerevisiae. FEBS Letters 263 : 93–98. 2158909
59. Stringer JR, Beard CB, Miller RF, Wakefield AE (2002) A new name (Pneumocystis jiroveci) for Pneumocystis from humans. Emerg Infect Dis 8 : 891–896. 12194762
60. Perocchi F, Jensen LJ, Gagneur J, Ahting U, von Mering C, et al. (2006) Assessing systems properties of yeast mitochondria through an interaction map of the organelle. PLoS Genet 2: e170. 17054397
61. (DNDi) DfNDI (2013) Drug Trial for Leading Parasitic Killer of the Americas Shows Mixed Results but Provides New Evidence for Improved Therapy. Media Center/Press Releases.
62. Molina I, Gomez i Prat J, Salvador F, Trevino B, Sulleiro E, et al. (2014) Randomized trial of posaconazole and benznidazole for chronic Chagas' disease. N Engl J Med 370 : 1899–1908. doi: 10.1056/NEJMoa1313122 24827034
63. Andriani G, Amata E, Beatty J, Clements Z, Coffey BJ, et al. (2013) Antitrypanosomal lead discovery: identification of a ligand-efficient inhibitor of Trypanosoma cruzi CYP51 and parasite growth. J Med Chem 56 : 2556–2567. doi: 10.1021/jm400012e 23448316
64. Calvet CM, Vieira DF, Choi JY, Kellar D, Cameron MD, et al. (2014) 4-Aminopyridyl-based CYP51 inhibitors as anti-Trypanosoma cruzi drug leads with improved pharmacokinetic profile and in vivo potency. J Med Chem 57 : 6989–7005. doi: 10.1021/jm500448u 25101801
65. Diaz-Gonzalez R, Kuhlmann FM, Galan-Rodriguez C, Madeira da Silva L, Saldivia M, et al. (2011) The susceptibility of trypanosomatid pathogens to PI3/mTOR kinase inhibitors affords a new opportunity for drug repurposing. PLoS Negl Trop Dis 5: e1297. doi: 10.1371/journal.pntd.0001297 21886855
66. Hoepfner D, McNamara CW, Lim CS, Studer C, Riedl R, et al. (2012) Selective and specific inhibition of the plasmodium falciparum lysyl-tRNA synthetase by the fungal secondary metabolite cladosporin. Cell Host Microbe 11 : 654–663. doi: 10.1016/j.chom.2012.04.015 22704625
67. Rao SP, Lakshminarayana SB, Kondreddi RR, Herve M, Camacho LR, et al. (2013) Indolcarboxamide is a preclinical candidate for treating multidrug-resistant tuberculosis. Sci Transl Med 5 : 214ra168. doi: 10.1126/scitranslmed.3007355 24307692
68. Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, et al. (1997) Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 277 : 60–66. 9204897
69. Kleinschroth T, Castellani M, Trinh CH, Morgner N, Brutschy B, et al. (2011) X-ray structure of the dimeric cytochrome bc(1) complex from the soil bacterium Paracoccus denitrificans at 2.7-A resolution. Biochim Biophys Acta 1807 : 1606–1615. doi: 10.1016/j.bbabio.2011.09.017 21996020
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSSČlánek Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to MalariaČlánek IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 CellsČlánek Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 7- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Basic Prion Science “Spreads” Insight
- Research Driven by Curiosity: The Journey from Basic Molecular Biology and Virology to Studies of Human Pathogenic Coronaviruses
- Cross Kingdom Activators of Five Classes of Bacterial Effectors
- Vaccination Drives Changes in Metabolic and Virulence Profiles of
- Expression of the Blood-Group-Related Gene Alters Susceptibility to Infection
- Transmission Properties of Human PrP 102L Prions Challenge the Relevance of Mouse Models of GSS
- Latent KSHV Infected Endothelial Cells Are Glutamine Addicted and Require Glutaminolysis for Survival
- The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators
- Intraperitoneal Infection of Wild-Type Mice with Synthetically Generated Mammalian Prion
- Vpr Promotes Macrophage-Dependent HIV-1 Infection of CD4 T Lymphocytes
- An In-Depth Comparison of Latency-Reversing Agent Combinations in Various and HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression
- α-Macroglobulin Can Crosslink Multiple Erythrocyte Membrane Protein 1 (PfEMP1) Molecules and May Facilitate Adhesion of Parasitized Erythrocytes
- Should Symbionts Be Nice or Selfish? Antiviral Effects of Wolbachia Are Costly but Reproductive Parasitism Is Not
- A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface
- The Broad Neutralizing Antibody Responses after HIV-1 Superinfection Are Not Dominated by Antibodies Directed to Epitopes Common in Single Infection
- Rapidly Evolving Genes Are Key Players in Host Specialization and Virulence of the Fungal Wheat Pathogen ()
- MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease
- Age-Dependent Cell Trafficking Defects in Draining Lymph Nodes Impair Adaptive Immunity and Control of West Nile Virus Infection
- Decline of FoxP3+ Regulatory CD4 T Cells in Peripheral Blood of Children Heavily Exposed to Malaria
- Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease
- Macrophages Subvert Adaptive Immunity to Urinary Tract Infection
- Mycolactone-Dependent Depletion of Endothelial Cell Thrombomodulin Is Strongly Associated with Fibrin Deposition in Buruli Ulcer Lesions
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation
- Phosphoproteomic Analysis of KSHV-Infected Cells Reveals Roles of ORF45-Activated RSK during Lytic Replication
- CR3 and Dectin-1 Collaborate in Macrophage Cytokine Response through Association on Lipid Rafts and Activation of Syk-JNK-AP-1 Pathway
- IFNγ and IL-12 Restrict Th2 Responses during Helminth/ Co-Infection and Promote IFNγ from Th2 Cells
- THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection
- Human Enterovirus Nonstructural Protein 2C Functions as Both an RNA Helicase and ATP-Independent RNA Chaperone
- IL-27 Signaling Is Crucial for Survival of Mice Infected with African Trypanosomes via Preventing Lethal Effects of CD4 T Cells and IFN-γ
- Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation
- Vpu Exploits the Cross-Talk between BST2 and the ILT7 Receptor to Suppress Anti-HIV-1 Responses by Plasmacytoid Dendritic Cells
- Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses
- A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence
- Analysis of the SUMO2 Proteome during HSV-1 Infection
- Capacity of Broadly Neutralizing Antibodies to Inhibit HIV-1 Cell-Cell Transmission Is Strain- and Epitope-Dependent
- A Novel Antiviral Target Structure Involved in the RNA Binding, Dimerization, and Nuclear Export Functions of the Influenza A Virus Nucleoprotein
- Deploying FLAREs to Visualize Functional Outcomes of Host—Pathogen Encounters
- Mosquitoes Reset Malaria Parasites
- The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease
- Extracellular Virions: The Advance Guard of Poxvirus Infections
- Risks of Antibiotic Exposures Early in Life on the Developing Microbiome
- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Exploiting Fungal Virulence-Regulating Transcription Factors As Novel Antifungal Drug Targets
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Periodontal Diseases: Bug Induced, Host Promoted
- Mechanisms of Host Behavioral Change in Rodent Association
- The Endosymbiotic Bacterium Selectively Kills Male Hosts by Targeting the Masculinizing Gene
- HIV Reactivation from Latency after Treatment Interruption Occurs on Average Every 5-8 Days—Implications for HIV Remission
- Ubiquilin 1 Promotes IFN-γ-Induced Xenophagy of
- Transfer of Immunity from Mother to Offspring Is Mediated via Egg-Yolk Protein Vitellogenin
- Suppression of Long-Lived Humoral Immunity Following Infection
- The Role of VP1 Amino Acid Residue 145 of Enterovirus 71 in Viral Fitness and Pathogenesis in a Cynomolgus Monkey Model
- Utilizing Chemical Genomics to Identify Cytochrome as a Novel Drug Target for Chagas Disease
- The Emerging Role for RNA Polymerase II in Regulating Virulence Gene Expression in Malaria Parasites
- Turning Up the Heat: Inflammasome Activation by Fungal Pathogens
- On and Under the Skin: Emerging Basidiomycetous Yeast Infections Caused by Species
- EhVps32 Is a Vacuole-Associated Protein Involved in Pinocytosis and Phagocytosis of
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
- The Serine Protease EspC from Enteropathogenic Regulates Pore Formation and Cytotoxicity Mediated by the Type III Secretion System
- Existing Infection Facilitates Establishment and Density of Malaria Parasites in Their Mosquito Vector
- Evaluating Human T-Cell Therapy of Cytomegalovirus Organ Disease in HLA-Transgenic Mice
- Neuronal Interferon Signaling Is Required for Protection against Herpes Simplex Virus Replication and Pathogenesis
- Epstein-Barr Virus Proteins EBNA3A and EBNA3C Together Induce Expression of the Oncogenic MicroRNA Cluster miR-221/miR-222 and Ablate Expression of Its Target p57
- Colonization of the Mouse Gastrointestinal Tract Is Modulated by Wall Teichoic Acid, Capsule, and Surface Proteins
- Virulence of Group A Streptococci Is Enhanced by Human Complement Inhibitors
- Identification of Caspase Cleavage Sites in KSHV Latency-Associated Nuclear Antigen and Their Effects on Caspase-Related Host Defense Responses
- Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Infections
- Type VI Secretion System Transports Zn to Combat Multiple Stresses and Host Immunity
- Lv4 Is a Capsid-Specific Antiviral Activity in Human Blood Cells That Restricts Viruses of the SIV/SIV/HIV-2 Lineage Prior to Integration
- Phenylbutyrate Is Bacteriostatic against and Regulates the Macrophage Response to Infection, Synergistically with 25-Hydroxy-Vitamin D₃
- An Internally Translated MAVS Variant Exposes Its Amino-terminal TRAF-Binding Motifs to Deregulate Interferon Induction
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion
- Activation of TLR2 and TLR6 by Dengue NS1 Protein and Its Implications in the Immunopathogenesis of Dengue Virus Infection
- N-acetylglucosamine Regulates Virulence Properties in Microbial Pathogens
- Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



