-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Competition between VanU Repressor and VanR Activator Leads to Rheostatic Control of Vancomycin Resistance Operon Expression
Various modes of gene regulation coexist in cells. One corresponds to the “switch on/ off” mechanism in which the regulator induces the promoter to a defined level. In another mechanism, the regulator activates the promoter to various levels according to the intensity or the nature of an input signal. In this study, we show that in VanG-type vancomycin resistant Enterococcus faecalis a repressor (VanUG) allows rheostatic expression of a target resistance promoter by competing with a response regulator (VanRG) which otherwise acts together with a sensor (VanSG) by a "switch on/off" mechanism as part of a two-component regulatory system. Unusually, both regulators are encoded in the same operon.
Published in the journal: Competition between VanU Repressor and VanR Activator Leads to Rheostatic Control of Vancomycin Resistance Operon Expression. PLoS Genet 11(4): e32767. doi:10.1371/journal.pgen.1005170
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005170Summary
Various modes of gene regulation coexist in cells. One corresponds to the “switch on/ off” mechanism in which the regulator induces the promoter to a defined level. In another mechanism, the regulator activates the promoter to various levels according to the intensity or the nature of an input signal. In this study, we show that in VanG-type vancomycin resistant Enterococcus faecalis a repressor (VanUG) allows rheostatic expression of a target resistance promoter by competing with a response regulator (VanRG) which otherwise acts together with a sensor (VanSG) by a "switch on/off" mechanism as part of a two-component regulatory system. Unusually, both regulators are encoded in the same operon.
Introduction
Vancomycin-resistant enterococci are a major cause of nosocomial infections and an important public health problem because the treatment options for the infections they cause are very limited [1]. Vancomycin, which can be the only antibiotic effective against multiresistant clinical isolates, acts by binding to the C-terminal D-alanyl-D-alanine (D-Ala-D-Ala) residues of peptidoglycan precursors blocking the extracellular steps in peptidoglycan synthesis [2]. Resistance in Enterococcus is mediated by nine types of operons that produce modified peptidoglycan precursors ending in D-Ala-D-Lac (vanA, -B, -D, and-M) or D-Ala-D-Ser (vanC, -E, -G, -L, and-N) to which vancomycin bind with a low affinity and from the elimination of the high affinity precursors ending in D-Ala-D-Ala [3–6].
Expression of the vancomycin resistance operons is regulated by VanS/VanR-type two-component signal transduction systems composed of a membrane-bound histidine kinase (VanS-type) and a cytoplasmic response regulator (VanR-type) that acts as a transcriptional activator [3]. The sensors modulate the levels of phosphorylation of the regulators. In the presence of vancomycin, VanS acts primarily as a kinase that autophosphorylates and transfers its phosphate to VanR. Phosphorylated VanR binds to the promoters upstream from the vanRS regulatory and resistance operons leading to increased transcription of the regulatory and resistance genes [7–9]. The phosphatase activity of VanS-type sensors is required for negative regulation of the resistance genes in the absence of vancomycin preventing accumulation of VanR-type regulators phosphorylated by acetylphosphate or by kinases encoded by the host chromosome [7, 10].
VanG-type Enterococcusfaecalis clinical isolates from Australia and Canada are distinct from other Van-type enterococci. The chromosomal vanG cluster (Fig 1) confers resistance to vancomycin (MICs, 16 μg/ml) by inducible synthesis of precursors ending in D-Ala-D-Ser [11]. It contains the vanYG,WG,G,XYG,TG resistance genes, the last three strictly required for resistance encode, respectively, a VanG ligase to synthesize D-Ala-D-Ser, a VanXYG D,D-carboxypeptidase to hydrolyse D-Ala-D-Ala, and a VanTG membrane bound serine racemase to produce D-Ser (Fig 1). As opposed to the other van gene clusters, the vanG regulatory operon contains three genes, vanUG, vanRG, and vanSG, encoding a "three component" regulatory system (Fig 1). Additional gene vanUG encodes a transcriptional regulator belonging to the Xre protein family and of unknown function. The vanURSG genes are co-transcribed, even in the absence of vancomycin, from the PUG regulatory promoter, whereas transcription of the resistance genes is inducible and initiated from the PYG resistance promoter [11].
Fig. 1. Schematic representation of the vanG operon. 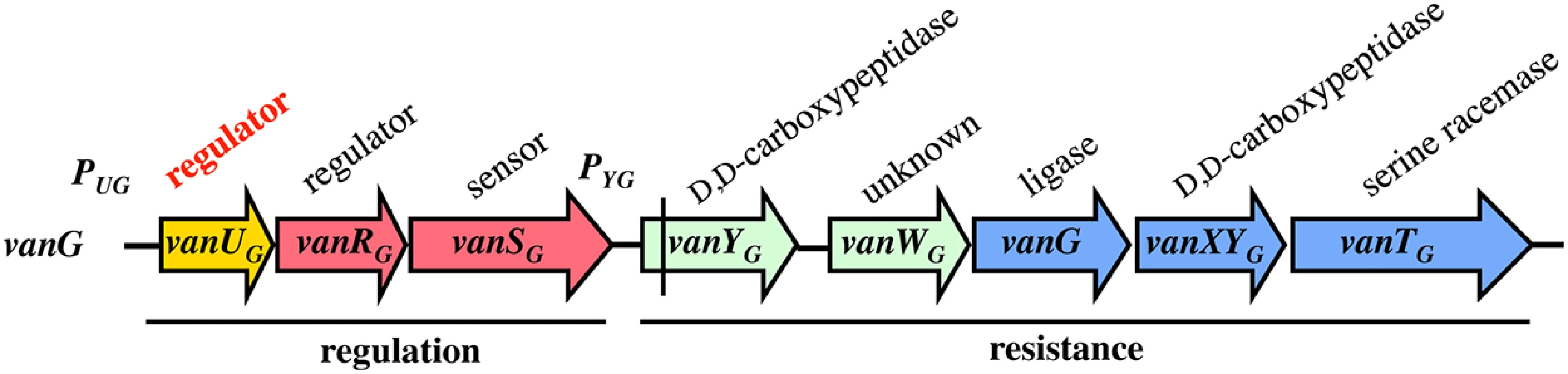
Open arrows represent coding sequences and indicate direction of transcription. The regulatory genes are in red, the resistance genes in blue and accessory genes in green. The additional regulatory gene, vanUG, is in yellow. The vertical bar in vanYG indicates a frameshift mutation leading to a truncated protein. Cryptic vanG-like operons are common in Clostridium difficile, a major human pathogen which is a target for vancomycin, and a vanUG gene encoding a protein identical to VanUG was found in a clinical isolate (GenBank N° AVLW01000050). A VanUG-like protein (GenBank N° YP002939420), 79% identical with VanUG, was detected in an Eubacterium associated with a two-component system controlling an ABC-type transporter and a protein (GenBank N°YP007781704) with 76% identity was reported in Ruminococcus bromii associated with a CheY related regulator and a partial vanG operon. These regulators have not been studied.
We report the role of VanUG in the transcription of the vanG operon in E.faecalis. We show that VanUG binds to the PUG regulatory and PYG resistance promoters and negatively regulates the vanURSG regulatory and resistance operons. In contrast, VanRG binds only to PYG. It thus appears that, upon induction by vancomycin, the VanSG sensor phosphorylates VanRG which competes and displaces VanUG from PYG leading to transcription of the resistance operon in a dose dependent manner. Thus, rheostatic regulation of resistance gene expression results from binding of a repressor and an activator encoded in a single operon to the same promoter.
Results
VanUG but not VanRG binds to the PUG regulatory promoter
Primer extension of the region upstream from vanUG indicated that, irrespective of induction, the transcriptional start site for vanURSG was located 22 bp upstream from the translation initiation codon of vanUG [11]. The PUG promoter consists of -35 and -10 regions corresponding to δ70 recognition sequences separated by 17 bp (Fig 2A). To determine if VanUG and VanRG bind to the PUG regulatory promoter region and to identify putative specific binding sites, DNaseI footprinting experiments were carried out. A radiolabeled PCR probe corresponding to positions -247 to +110 relative to the transcription initiation site of PUG was incubated with increasing amounts of purified VanUG, VanRG, and VanRG phosphorylated (VanRG-P) by acetyl phosphate. The PUG region protected by VanUG depended on the protein concentration, extending from -70 to -20 (positions relative to the transcription initiation site) overlapping the -35 sequence at a low concentration (Fig 2B, lane 6) and from -70 to +10 at higher concentrations (Fig 2B, lanes 7 and 8). The region (-70 to -20) contained two adjacent imperfect palindromic sequences likely corresponding to the binding motifs of VanUG (Fig 2A). As opposed to the wild-type fragment, two DNA fragments containing double mutations in the imperfect dyad symmetry operator of PUG were not retarded by VanUG, indicating a key role in VanUG binding (S1 Fig). The appearance of several DNase I hypersensitive sites (Fig 2B) corresponding to bending of the DNA duplex suggested binding of two VanUG monomers or dimers. This is consistent with the presence of two inverted repeats in the PUG region (Fig 2A) and with the two-step gel retardation (S1 Fig). In contrast to VanUG, VanRG and VanRG-P did not bind to the PUG promoter.
Fig. 2. Binding sites of VanUG to the PUG regulatory promoter (A, B) and regulatory genes transcription (C, D). 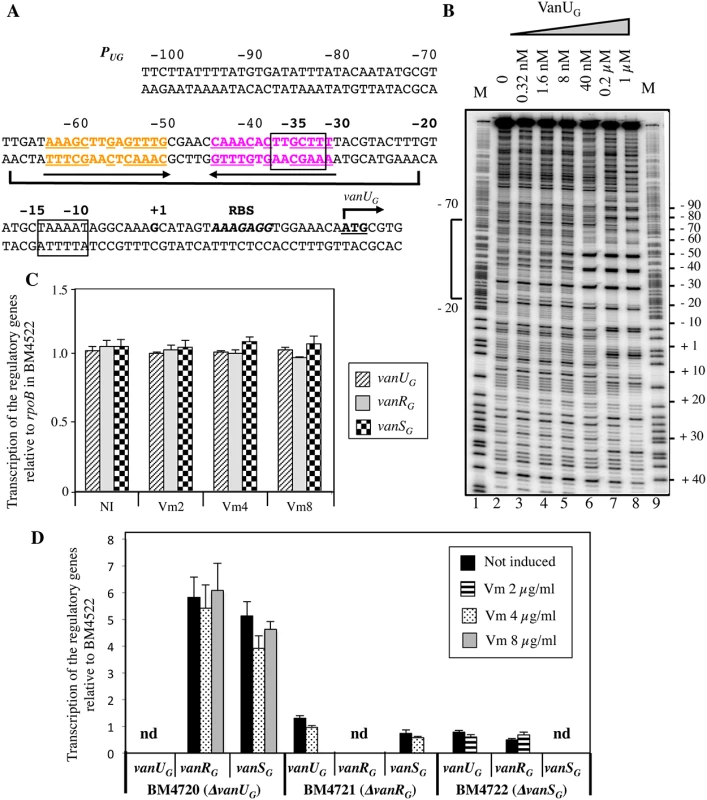
(A) Sequence of the PUG region. The transcriptional start site (+1) is in boldface and the -35 and -10 sequences are boxed. The translational start site is in boldface and underlined and the ribosome binding site (RBS) is in boldface and in italics. Regions protected from DNase I cleavage by VanUG are delineated by a bracket. The binding motif is composed of two 14-bp imperfect inverted repeats indicated in orange and purple and by arrows; the complementary bases are underlined. (B) DNase I footprinting analysis of the binding of VanUG to PUG. A 357-bp DNA fragment was amplified from the PUG promoter region using a labeled reverse primer (VanG126) to radiolabel the template strand. Increasing amounts of VanUG, indicated above each lane, were incubated with the DNA probe. The bracket indicates the region protected from DNase I cleavage by VanUG and the co-ordinates of protection relative to the transcriptional start site are indicated on the left. M is the A+G Maxam and Gilbert sequencing reaction lane of the probe used as a size marker and the nucleotide positions are indicated at the right. Transcription of the regulatory genes by RT-qPCR in transconjugant BM4522 (C) and deletant derivatives relative to the same genes of BM4522 (D). The strains are indicated at the bottom. Results are presented in arbitrary units normalized to the rpoB transcripts in the same strain and in BM4522 under similar conditions. Each strain, not induced or induced by vancomycin, was tested in triplicate in two independent experiments. The bars represent the means and the error bars the standard deviations; nd, not detectable. NI, not induced. Vm, vancomycin. VanUG acts as a repressor of the PUG regulatory promoter
The vanG operon is part of a large genetic element and is transferable from E. faecalis BM4518 to E. faecalis JH2-2 from chromosome to chromosome [11]. Since clinical isolate BM4518 is not transformable, we studied the VanURSG system in transconjugant BM4522 (JH2-2::vanG) (S1 Table). To determine the role of VanUG on PUG, the vanUG, vanRG, and vanSG genes of BM4522 were inactivated individually by in-frame deletions leading to BM4720(ΔvanUG), BM4721(ΔvanRG), and BM4722(ΔvanSG). Transcription of the regulatory genes was quantified by RT-qPCR. In BM4522, low level transcription occured at similar levels without and with various concentrations of vancomycin indicating that the PUG promoter was not inducible by vancomycin (Fig 2C). In the absence of vanUG, vanRG and vanSG were transcribed in the absence or presence of vancomycin at higher level (≈ 5-fold) from PUG indicating that VanUG acted as a repressor on this promoter region (Fig 2D). In the absence of vanRG or vanSG, transcription of the regulatory genes remained unchanged even in the presence of vancomycin.
To confirm regulation of PUG by VanUG, the vanURSG genes were cloned into vancomycin susceptible Escherichia coli NR698 [12] under the control of promoter Pspank upstream from PUG fused to a chloramphenicol acetyltransferase (CAT) reporter gene, the two promoters being separated by a transcription terminator (term) (Table 1). Subsequently, each of the three genes was inactivated. E. coli RNA polymerase bound to the PUG promoter (S2A Fig) which was active in the new host, in the presence or in the absence of vancomycin (Table 1). CAT was produced at a maximum level in the absence of vanUG by plasmids pAT952(PspanktermPUGcat), pAT966(PspankvanRGtermPUGcat), and pAT969(PspankvanRSGtermPUGcat) (Table 1). In contrast, in the presence of VanUG, CAT production was decreased to similar basal levels by plasmids pAT965(PspankvanUGtermPUGcat), pAT967(PspankvanURGtermPUGcat), and pAT968 (PspankvanURSGtermPUGcat) (Table 1). These results confirmed that VanUG acts as a strong repressor on the PUG promoter.
Tab. 1. CAT specific activities of PUG promoter in E. coli NR698. 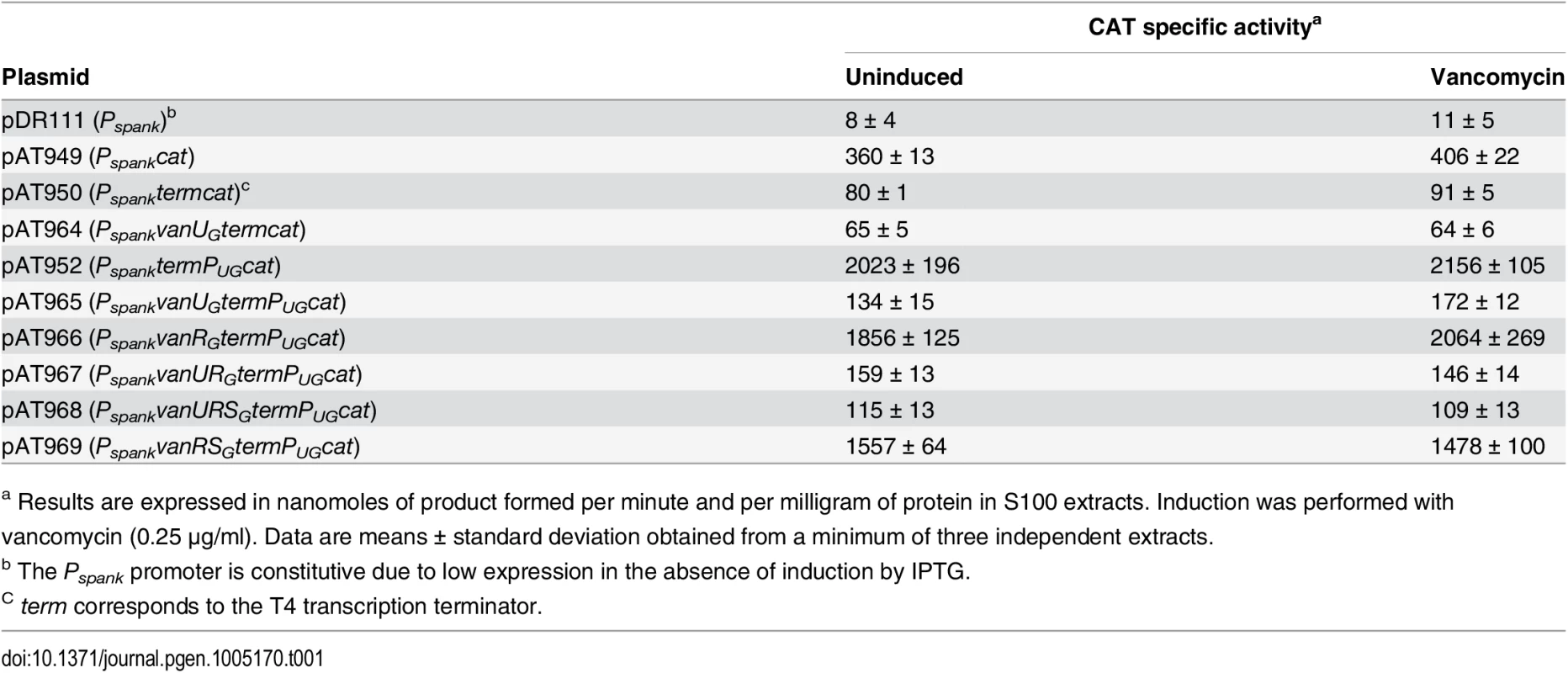
a Results are expressed in nanomoles of product formed per minute and per milligram of protein in S100 extracts. Induction was performed with vancomycin (0.25 μg/ml). Data are means ± standard deviation obtained from a minimum of three independent extracts. The VanRGSG two-component system is functional
Transcription of the resistance genes is under the control of VanURSG and, as discussed above, VanUG negatively autoregulates vanURSG transcription from the PUG regulatory promoter. To determine if VanRG and VanSG acted as a two-component system and to study the putative interaction of VanUG with these proteins, VanUG, VanRG, and the cytoplasmic histidine kinase domain of VanSG were purified as C-terminal His-tag proteins (S1 Table). VanSG autophosphorylated in the presence of [γ-32P]-ATP (Fig 3A). When incubated with purified VanUG or VanRG, phosphorylated VanSG transferred its phosphate group to VanRG (Fig 3B) but not to VanUG (Fig 3E). Phosphorylation of VanRG was fast and efficient, occurring in less than a minute. To test the phosphatase activity of VanSG, hydrolysis of VanRG-P over time was analysed in the absence or in the presence of VanSG. Purified [32P]-VanRG was stable in vitro for at least 30min and then dephosphorylated slowly (Fig 3C); addition of purified VanSG increased dephosphorylation only slightly (Fig 3D–3G). These results indicate that VanRSG was functional and had characteristics similar to those of other VanRS-type two-component systems [7, 9] and that VanUG did not affect phosphorylation nor dephosphorylation of VanRG and VanSG (Fig 3E and 3F).
Fig. 3. Autophosphorylation of VanSG (A), phosphotransfer from VanSG-P to VanRG (B), phosphorylation of VanRG by acetyl [32P] phosphate (C), hydrolysis of VanRG-P by VanSG (D), and phosphotransfer from VanSG to VanUG (E) or to VanUG plus VanRG (F). ![Autophosphorylation of VanS<sub>G</sub> (A), phosphotransfer from VanS<sub>G</sub>-P to VanR<sub>G</sub> (B), phosphorylation of VanR<sub>G</sub> by acetyl [<sup>32</sup>P] phosphate (C), hydrolysis of VanR<sub>G</sub>-P by VanS<sub>G</sub> (D), and phosphotransfer from VanS<sub>G</sub> to VanU<sub>G</sub> (E) or to VanU<sub>G</sub> plus VanR<sub>G</sub> (F).](https://www.prelekara.sk/media/cache/resolve/media_object_image_small/media/image/5b8a44dce46fdc988a9e030adcb6db8a.png)
Quantitative analysis of phosphorylated VanRG in panels C and D (G). (A) Purified VanSG was incubated with [γ-32P]-ATP for 1 h at room temperature to test autophosphorylation. (B) After autophosphorylation of VanSG (time 0), purified VanRG was added, samples were removed at the indicated times (in min), mixed with β-mercaptoethanol stop solution on ice and separated by SDS-PAGE (15%). Transfer of radioactivity to VanRG was revealed by autoradiography. (C) Purified VanRG was incubated with acetyl[32P]phosphate for 1 h at room temperature (time0), excess acetyl[32P]phosphate was removed by using a Sephadex G-50 Quick-Spin column, and phosphorylated VanRG was incubated at room temperature either alone or (D) following the addition of purified VanSG. Samples were removed at the indicated times (in min), mixed with β-mercaptoethanol-stop solution on ice, resolved by SDS-PAGE (15%), and subjected to autoradiography. After autophosphorylation of VanSG (time 0), purified VanUG was added alone (E) or with VanRG (F), samples were removed at the indicated times (in min), mixed with β-mercaptoethanol stop solution on ice and separated by SDS-PAGE (12%). Transfer of radioactivity to VanRG but not to VanUG was revealed by autoradiography. (G) Hydrolysis in the absence (blue line, panel C) or in the presence (pink line, panel D) of VanSG of purified VanRG labeled with acetyl[32P]phosphate was detected on a phosphor storage screen and percent quantified. Results are the means of four independent experiments and the bars indicate standard deviations. VanUG and VanRG bind to overlapping sites of the PYG resistance promoter
To study the putative binding of VanUG and VanRG to the PYG region and to identify specific binding sites, DNaseI footprinting experiments were carried out. The inducible PYG promoter is composed of -35 (AAAACA) and -10 (TACAAT) regions separated by 16 bp which have similarity with δ70 recognition sequences, although the -35 sequence is not conserved consistent with the fact that the promoter is positively regulated (Fig 4B). Analysis of the PYG region revealed three 12-bp directly repeated VanRG binding motifs and a deduced consensus sequence (T/C)CGTANGAAA(T/A)T was analogous to that in the PR and PH vanA operon promoters [13]. In the PUG region, similar sequences were not found (Fig 2A) which could explain lack of VanRG binding. The radiolabeled probe corresponding to positions -163 to +69 relative to the transcription initiation point of the PYG promoter and containing the three conserved sequences was incubated with increasing amounts of purified VanUG, VanRG, and VanRG-P (Fig 4). The three proteins protected in a concentration-dependent manner an overlapping DNA region that included the three direct repeats. The PYG region protected by VanUG was much larger than that by VanRG and VanRG-P extending from -110 to -3 and overlapped the -35 sequence at 0.2 and 1μM (Fig 4A, lanes 17 and 18). The PYG region protected by VanRG and VanRG-P extended from -100 to -56 at low concentration (Fig 4A, bracket I, lanes 3 and 8) and from -100 to -43 at higher concentrations (Fig 4A, bracket II, lanes 4 and 5, and 9 and 10). There were three binding motifs a, b, and c with different affinities for VanRG and VanRG-P in the PYG promoter region (Fig 4). Only a slight difference in affinity in favor of VanRG-P at 0.2μM was noted for the "a" site (Fig 4A, lane 2) compared with VanRG which could be due to inefficient phosphorylation of VanRG by acetylphosphate. VanRG and VanRG-P bound to the a and b sites (Fig 4A, lanes 2, 3, and 8) with higher affinity than to the c site (Fig 4A, lanes 4 and 5, and 9 and 10), whereas VanUG bound to this DNA region with the same affinity (Fig 4A).
Fig. 4. Binding of VanUG, VanRG, and VanRG-P to the PYG resistance promoter. 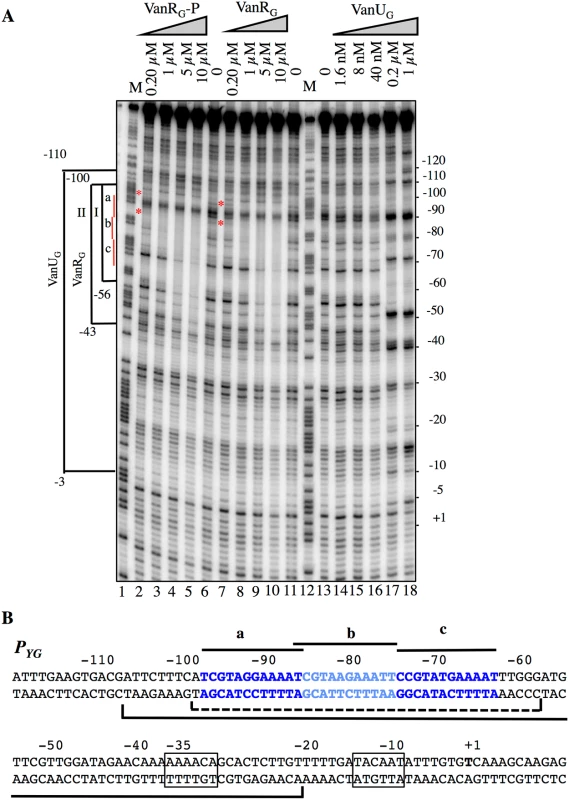
(A) DNase I footprinting analysis. A 233-bp DNA fragment was amplified from the PYG promoter region using a labeled reverse primer (YG10) to radiolabel the template strand. Increasing amounts of VanUG, VanRG, or VanRG-P, indicated at the top, were incubated with the DNA probe. The brackets indicate the regions protected from DNase I cleavage by VanUG, VanRG, or VanRG-P, and the co-ordinates of protection relative to the transcriptional start site are indicated on the left. The three 12-bp VanRG binding sites (a, b, c) are indicated in red on the left. The red asteriks indicate the slight difference in affinity in favor of VanRG-P (lane 2) in comparison with VanRG (lane 7), both at 0.2 μM. M is the A+G Maxam and Gilbert sequencing reaction lane of the probe used as a size marker and the nucleotide positions are indicated at the right. (B) Sequence of the PYG promoter region. The transcriptional start site (+1) is in boldface and the -35 and -10 sequences are boxed. The three (a, b, c) 12-bp putative VanRG binding sites are in blue and indicated by black lines. The region protected from DNase I cleavage by VanUG is delineated by a black bracket and that of VanRG or VanRG-P is delineated by a dotted bracket. VanUG allows rheostatic expression of the resistance genes
To study the consequences of the binding of VanUG and VanRG to overlapping regions of PYG on the expression of the resistance genes, the VanTG serine racemase was used as a reporter (Fig 5). In clinical isolate BM4518 and transconjugant BM4522, synthesis of the serine racemase was dependent on the concentration of vancomycin (Fig 5). In contrast, in BM4720(ΔvanUG), the resistance operon was expressed at its maximum even at low concentrations of vancomycin. These results suggested that VanUG acts as a repressor of PYG and that, in its absence, there is no fine-tuning of resistance expression from this promoter. Thus, modulation of transcription by vancomycin was due to the phosphorylation level of VanRG mediated by VanSG provided that VanUG was present. Surprisingly, as in the wild-type strain, induction was dependent on the concentration of the inducer in BM4721(ΔvanRG) (Fig 5). This could be accounted for by the presence of a VanR homolog in the host. In fact, we found, in both E.faecalis BM4518 and transconjugant BM4522 which were entirely sequenced (GenBank N°PRJNA245745), a gene specifying a VanR'G protein with 65% identity with VanRG (S3A Fig). In BM4722(ΔvanSG) there was no synthesis of VanTG in the presence of vancomycin indicating that VanRG and VanR'G are not phosphorylated in the absence of VanSG. Double mutant BM4723(ΔvanRG, ΔvanR'G) derived from E. faecalis BM4721(ΔvanRG) was susceptible to vancomycin (MIC, 1μg/ml) and VanTG production was no longer inducible by vancomycin, indicating cross-talk between VanSG and VanR'G (Fig 5). To avoid interference by this regulator, transcription from the PYG promoter was studied in E.coli NR698 since E. coli RNA polymerase was able to bind to this promoter (S2B Fig). The vanURSG, vanRSG, and vanUSG genes were cloned under the control of Pspank upstream from the PYG transcriptionally fused to a cat gene generating pAT970 (PspankvanURSGtermPYGcat), pAT971 (PspankvanRSGtermPYGcat), and pAT972 (PspankvanUSGtermPYGcat). In the absence of VanUG, induction by vancomycin led to similar levels of CAT synthesis in the strain harboring pAT971 (PspankvanRSGtermPYGcat) whatever the concentration of the inducer, whereas with pAT970 (PspankvanURSGtermPYGcat) CAT production depended on the vancomycin concentration (Table 2). These results confirmed that, in the presence of vancomycin, VanUG is required for rheostatic gene transcription from PYG and that VanRG phosphorylation is essential for expression of the resistance genes since, in the absence of this regulator in pAT972 (PspankvanUSGtermPYGcat), the level of CAT activity was low, both without (74U±9) and with (104 U ± 13) vancomycin (0.30 μg/ml). In the absence of vancomycin, CAT activity was lower in E. coli producing vanUG encoded by pAT970 (PspankvanURSGtermPYGcat) than in its counterpart harboring pAT971 (PspankvanRSGtermPYGcat). This confirms that VanUG acts as a repressor on the PYG resistance promoter (Table 2).
Fig. 5. VanTG racemase specific activity in membrane extracts from clinical isolate BM4518, transconjugant BM4522, and its deletant derivatives. 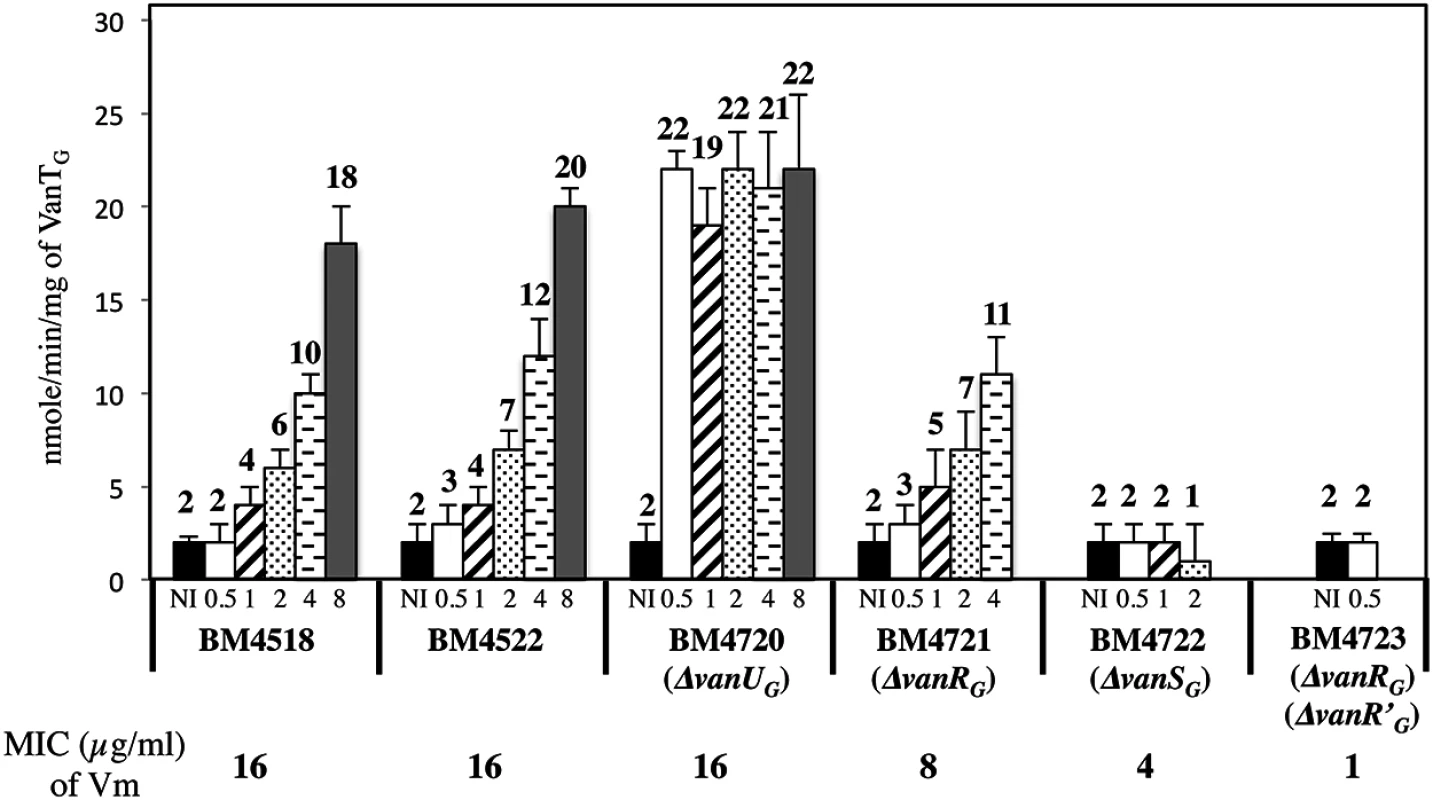
Vancomycin (Vm) inducing concentrations (μg/ml) and MICs are indicated at the bottom. NI, not induced. The error bars represent the standard deviations from at least three independent experiments (eight for BM4723) and the values above the bars are the means of specific activity defined as the number of nanomoles of product formed at 37°C per minute per milligram of protein contained in the extracts. Tab. 2. CAT specific activities of PYG promoter in E. coli NR698. 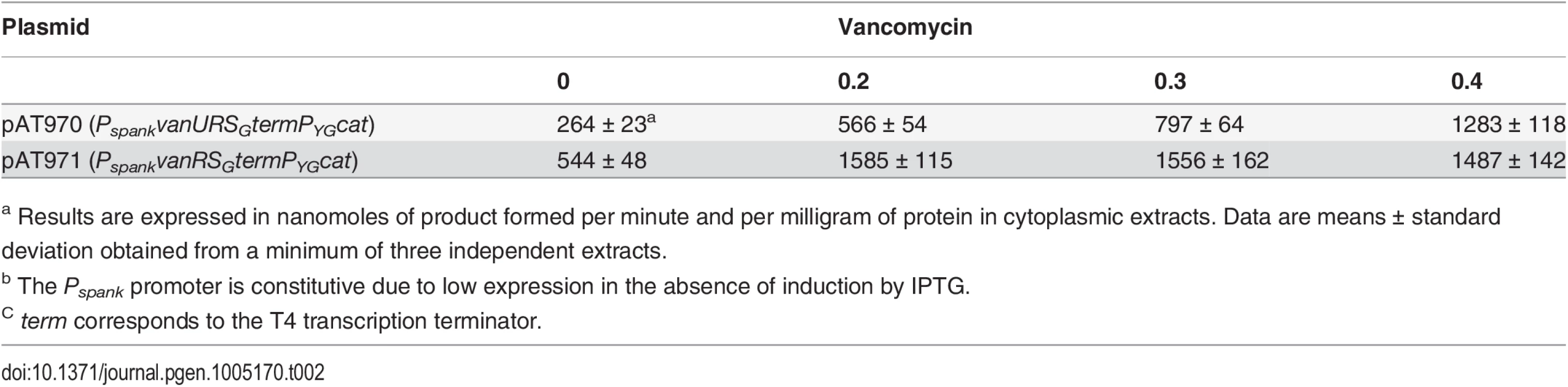
a Results are expressed in nanomoles of product formed per minute and per milligram of protein in cytoplasmic extracts. Data are means ± standard deviation obtained from a minimum of three independent extracts. VanUG and VanRG compete for binding to the PYG resistance promoter
Since VanUG and VanRG bound at overlapping sites of PYG, to assess a possible effect of VanRG on the binding of VanUG, we performed DNaseI footprinting assays on the labeled PYG probe with purified VanRG and VanUG (Fig 6). Low and medium concentrations (64 nM and 128 nM) of VanUG which allow binding to PYG were tested with increasing concentrations of VanRG. Upon addition of VanRG, the binding profile of VanUG faded while that of VanRG appeared and increased in a dose dependent manner (Fig 6A). In the reverse experiment two approriate concentrations of VanRG were challenged by increasing concentrations of VanUG and the binding of VanRG decreased also in the presence of VanUG (S4 Fig). In summary, VanUG alone did not allow transcription of the resistance genes (Fig 6B). It thus appears that at a low concentration of vancomycin there was competition between VanUG and VanRG, the latter being partially phosphorylated, transcription of vanYGWGGXYGTG was low. In contrast, at high concentrations of vancomycin, VanRG was efficiently phosphorylated and able to displace VanUG leading to maximal transcription of the resistance genes from the PYG promoter.
Fig. 6. Competition between VanUG and VanRG for binding to the PYG resistance promoter. 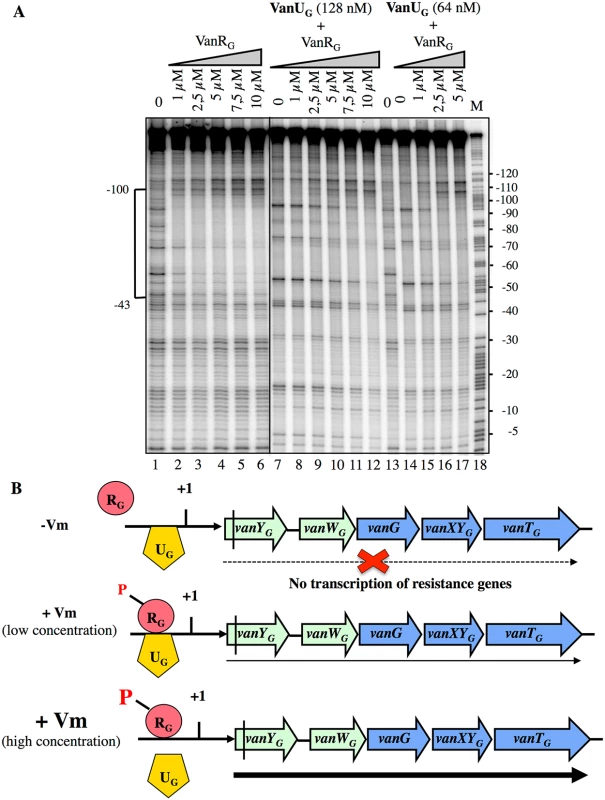
(A) DNase I footprinting analysis. A 233-bp DNA fragment was amplified from the PYG region using a labeled reverse primer (YG10) (S2 Table) to radiolabel the template strand. Increasing amounts of VanRG and two fixed amounts of VanUG, indicated at the top, were incubated with the DNA probe. The bracket indicates the region protected from DNase I cleavage by VanUG and/or VanRG and the co-ordinates of protection relative to the transcriptional start site are indicated on the left. M is the A+G Maxam and Gilbert sequencing reaction lane of the probes used as a size marker and the nucleotide positions are indicated at the right. (B) Model for the binding competition between VanUG and VanRG-P in the absence or in the presence of various concentrations of vancomycin (Vm). The presence of vanUG reduces the fitness cost associated with expression of VanG-type resistance
To study the role of VanUG in this sophisticated resistance mechanism, the fitness cost of BM4720(ΔvanUG) compared with that of BM4522 in monocultures in the absence and in the presence of vancomycin (1 μg/ml) was analysed by determination of the growth rates (Table 3). The results showed that the growth rates of both strains were indistinguishable in the absence of vancomycin indicating that non induced VanG-type resistance is not costly for the host. In contrast, in the presence of vancomycin, the relative growth rate of BM4720(ΔvanUG) (0.74) was significantly reduced when compared with that of BM4522 (0.93) indicating that increased expression of resistance was significantly more costly in the absence of vanUG.
Tab. 3. Growth rate. 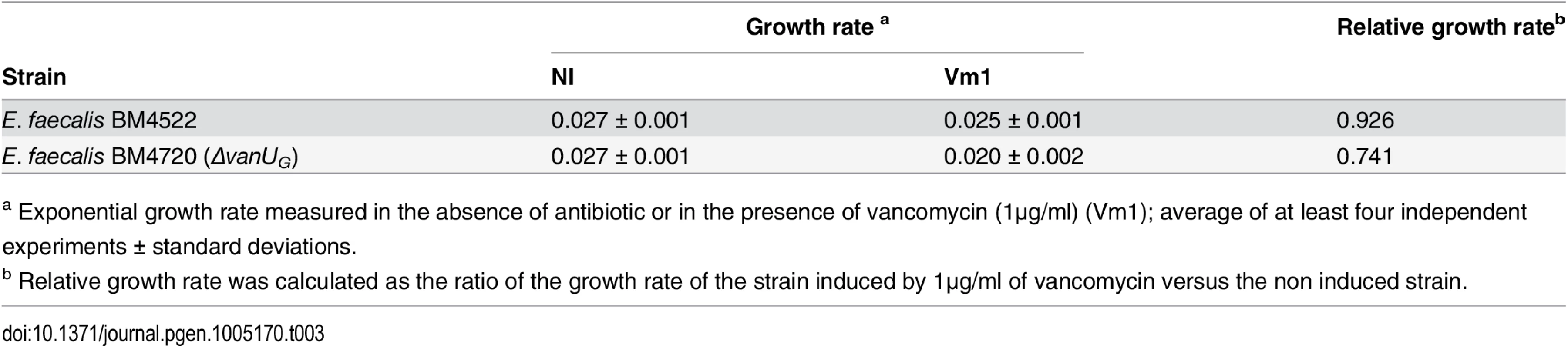
a Exponential growth rate measured in the absence of antibiotic or in the presence of vancomycin (1μg/ml) (Vm1); average of at least four independent experiments ± standard deviations. Discussion
Among the ubiquitous two-component regulators, VanR/VanS-type systems are one of the rare to control expression of genes mediating antibiotic resistance [3]. In the VanG-type strains, a membrane associated sensor kinase (VanSG) which detects a signal associated with the presence of vancomycin in the environment and a cytoplasmic response regulator (VanRG) that acts as a transcriptional activator are also present (Fig 1) and functional (Fig 3) but there is, in addition, a VanUG transcriptional regulator (Fig 1).
In the two main VanA - and VanB-type systems, the regulatory genes (vanRS) and the resistance genes are transcribed from independent and coordinately regulated promoters, but VanR is the only known direct regulator of the resistance genes [3, 8, 13]. In VanG-type strains, co-transcription of vanURSG is repressed from PUG by VanUG (Fig 2 and Table 1) and expression of the resistance genes from PYG is activated by VanRG and repressed by VanUG (Fig 5 and Table 2). Thus, VanUG regulates the resistance genes both directly, by binding to the PYG promoter region (Fig 4), and indirectly by repressing synthesis of VanRGSG (Fig 5). Like other members of the XRE protein family (S3B Fig) [14–16], VanUG binds to short repeated sequences which span the promoters (Fig 2A and 2B). Unlike the VanR and VanRB proteins which bind to their own promoters [8, 13], VanRG does not regulate its own expression (Fig 2). No sequences similar to the VanRG consensus binding site are found in PUG (Figs 2 and 4).
VanRG, as VanR and VanRB, belongs to the OmpR-PhoB subclass of response regulators that have the peculiarity to bind to their target promoters in the unphosphorylated or phosphorylated form [8, 13, 17, 18]. Phosphorylation of VanR and VanRB enhances the affinity of the proteins for their respective regulatory PR or PRB and resistance PH or PYB promoter regions allowing increased transcription of the regulatory and resistance genes [8, 13]. In VanA-type strains, VanR and VanR-P bind to PR and PH regions which contain a single or two 12-bp conserved sites, respectively [13]. Comparison of the sequences of the PUG and PYG regions with the 12-bp consensus sequence spanned by VanR and VanR-P revealed three binding sites in the PYG region with a consensus sequence (Fig 4B) similar to that in VanA-type resistance [13]. As for the regulatory PR and resistance PH promoters, the positioning of these sites in PYG was upstream from the -35 motif. VanUG, VanRG, and VanRG-P protected overlapping regions, the two latter binding to PYG a and b sites with a higher affinity than to the c site (Fig 4). There are only two sites in the PH promoter but VanR generated a more extensive footprint (80 bp for PH) than VanRG (42bp for PYG) likely due to higher cooperativity of VanR. Although not essential for binding in vitro, phosphorylation of VanRG increased its affinity for the PYG resistance promoter (Fig 4). In the PUG promoter region no sequences similar to the consensus were found (Fig 2A) which could explain the absence of binding of VanRG and low-level transcription from the regulatory promoter.
In many instances, regulation of gene transcription in E.coli occurs essentially through control of the phosphatase activity of the sensor [19, 20]. In VanA - and VanB-type strains, the level of phosphorylation of VanR and VanRB is modulated by the kinase and phosphatase activities of the VanS and VanSB sensors [7, 10, 21]. Phosphatase activity is critical for response regulators, such as VanR and VanRB, whose phosphorylated form is highly stable, to ensure that the protein is not permanently activated. In VanG-type strains, in the absence of VanUG, induction by vancomycin led to maximal VanTG serine racemase (Fig 5) or CAT synthesis (Table 2) even at low concentrations of the inducer. Since in the absence of VanUG there was no modulation of resistance genes transcription from the PYG promoter, this suggests that a low amount of VanRG-P is sufficient to induce the resistance operon. VanUG did not modulate VanRG and VanSG phosphorylation (Fig 4F) and was not phosphorylated by VanSG (Fig 4E). Surprisingly, at least in vitro, the phosphatase activity of VanSG was not very efficient (Fig 4D) in comparison with those of VanS or VanSB [7, 9]. Expression of VanG-type resistance was thus inducible by vancomycin due to the presence of VanUG as opposed to direct modulation of VanR activity by VanS in the other van operons. In the absence of vancomycin only VanUG bound to the PYG promoter; however when the concentration of vancomycin increased, VanRG being more efficiently phosphorylated by VanSG, displaced progressively VanUG allowing gradual transcription of the resistance genes (Fig 6) as it is likely the case with VanR'G, the VanRG homolog encoded elsewhere in the chromosome. In B. subtilis, when both repressors SinR and SlrR are bound to the degU promoter, they can be displaced by the response regulator DegU leading to activation of the degU gene [22]. Also in B. subtilis, CcpC activates aconitase gene citB expression whereas CodY binds to its promoter and represses citB transcription [23]; PutR which is an activator essential for transcription of the putBCP operon for proline utilization is displaced by the CodY repressor [24].
VanUG does not possess the characteristics of auxiliary regulators which can interact with histidine kinases, influencing signal perception and transduction. Nor does it interact with the response regulator to alter its phosphorylation status or its DNA binding ability, the recruitement of RNA polymerase on the promoter, or to sequester it through protein:protein interaction [25, 26]. The results presented here show that competition between the VanUG repressor and the VanRG activator for binding to the PYG promoter may be responsible for the complex regulation of the resistance genes (Fig 6). This is an unusual example of rheostatic regulation of gene transcription due to binding competition between two regulators encoded in the same operon. It also elucidates an unsuspected strategy by which enterococcal clinical isolates regulate transcription of acquired genes for vancomycin resistance.
In previous work, we showed in VanB-type resistance that, despite the complex dual biochemical mechanism of resistance to vancomycin, its biological cost in enterococci is negligible when non induced, whereas a significant fitness reduction is observed when resistance is expressed in the presence of the inducer, the antibiotic itself [27]. Thus resistance is expressed exclusively when needed for bacterial survival. In VanG-type strains, tight regulation of resistance expression involves VanUG which can thus be considered as a compensatory component, drastically reducing the biological cost associated with vancomycin resistance in the presence of antibiotic.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
The origin and properties of the strains and plasmids are described in S1 Table. Escherichia coli TOP10 (Invitrogen, Groningen, The Netherlands) and NR698 (susceptible to vancomycin) [12] were used as a host for recombinant plasmids. E. coli BL21λDE3 [28], in which the T7 RNA polymerase gene is under the control of the inducible lacUV5 promoter carries the pREP4 plasmid allowing co-expression of the GroESL chaperonin to optimize recombinant protein solubility [29]. E. coli TG1 RepA [30] was used as a host for constructions in the pAT944(pGhost9Ωcat) vector (S1 Table). Kanamycin (50μg/mL) was used as a selective agent for cloning PCR products in the pCR-Blunt vector (Invitrogen). Ampicillin was used to select pUC1813 [31]. pDR111 (gift from David Rudner, Harvard University), which harbors the Pspank promoter between two fragments of the B.subtilis amyE gene, is a derivative of the Pspac-hy plasmid pJQ43 containing an additional lacO binding site to achieve a better repression in the absence of the IPTG inducer. Pspank is a lacI repressible IPTG inducible-promoter for gene overexpression. Spectinomycin (60μg/mL) and chloramphenicol (10μg/mL) were added to the medium to prevent loss of plasmids derived from pDR111(Pspank) and pAT944(pGhost9Ωcat), respectively. Enterococcus faecalis JH2-2 is a derivative of strain JH2 that is resistant to fusidic acid and rifampin [32]. In all experiments, strains were grown in brain heart infusion (BHI) at 37°C with shaking at 110 rpm.
Promoter DNA labeling
Labeled PUG (357 bp) and PYG (233 bp) fragments were generated by PCR with BM4518 total DNA as a template and primer pairs VanG12-VanG126 and VanSG6-YG10 (S2 Table), respectively, using a combination of an unlabeled primer with an end-labeled primer (625nM) with T4 polynucleotide kinase (0.075 U/μl) (New England Biolabs) and [γ32P]-ATP (3000 Ci/mmol) (Perkin Elmer). The PCR reactions were carried out in a 50-μl volume and the products purified as described [8].
Gel shift assay
Purified labeled PCR products corresponding to wild-type and mutated PUG promoter region fragments were recovered from a 6% polyacrylamide gel and used as a probe for the gel shift assay after addition of 100 μl of ammonium acetate (0.5 M) diluted in Tris buffer (10 mM, pH8.5) overnight at 37°C. The PUG and mutated PUG probes (10,000cpm each) were incubated with various concentrations of purified VanUG regulator at 30°C for 20min in 20 μl of 50mM Tris-HCl (pH7.8) containing 20 mM MgCl2 and 0.1 mM dithiothreitol (DTT). After addition of the DNA dye solution (40% glycerol, 0.025% bromophenol blue and 0.025 xylene cyanol), the mixture was loaded on a 7.5% polyacrylamide gel in the absence of protein denaturants. The gels were dried and analysed by autoradiography.
DNase I footprinting
Complexes with the labeled promoter regions (5nM) were formed for 30 min at 30°C in 15 μl of buffer C (20 mM Hepes pH 8.0, 5 mM MgCl2, 50 mM potassium glutamate, 5 mM DTT, and 500μg/ml bovine serum albumin) using RNA polymerase of E. coli at 50 nM or VanUG, VanRG, or VanRG-P at increasing concentrations. For DNase I experiments, 1.5 μl of DNase I solution (1 μg ml-1 in 10 mM Tris-HCl, 10 mM MgCl2, 10 mM CaCl2, 125 mM KCl) were added and incubated at 30°C for 10s when the labeled promoter regions were alone, or for 20 s when when RNA polymerase or VanUG, VanRG or VanRG-P were present in the mixture. The reaction was stopped and all the samples were extracted, precipitated, washed, resuspended, and loaded on a sequencing gel as described [8]. Protected bands were identified by comparing the migration with that of the same fragment treated for the A+G sequencing reaction [33]. The gels were analysed by autoradiography.
Quantitative real-time RT-qPCR
Enterococci grown in 100 ml of brain heart infusion in 250-ml bottles, with and without vancomycin, at 37°C with shaking at 110 rpm to OD600 = 0.8 were harvested. RNA was prepared using the Fast RNA ProBlue kit (MBP Biomedicals) according to the manufacturer's protocol, treated with DNase (Turbo DNA-free, Invitrogen), and checked for the absence of contaminant DNA in a standard PCR, using the same primers as for the RT-PCR. RNA concentrations were determined by measuring absorbance with a NanoDrop2000 (ThermoScientific). cDNA synthesis and RT-qPCR were performed with a Light Cycler RNA amplification kit SYBR greenI (Roche Diagnostic GmbH) in a total reaction volume of 19μl with 0.5 μM gene-specific primers (VanG129-VanG102 for vanUG, VanRG2-VanRG10 for vanRG, VanSG2-VanSG10 for vanSG, and rpoB5-rpoB12 for rpoB) (S2 Table) according to the manufacturer's instructions. Amplification and detection of specific products were performed using the LightCycler sequence detection system (Roche) with the following cycle profile: 1cycle at 55°C for 20 min for the reverse transcription step, followed by 1 cycle at 95°C for 30 s, 45 cycles at 95°C for 5 s, 52°C for 15 s, and 72°C for 15 s. The level of every gene transcript was normalized relative to rpoB transcript levels.
Overproduction and purification of VanUG, VanRG, and VanSG
Plasmids pAT940(pET28ΩvanUG), pAT941(pET28ΩvanRG), and pAT942(pET28ΩvanSG) (S1 Table) were introduced into E. coli BL21λDE3/pREP4 [29]. The transformants were grown in 1 liter of LB medium in Fernbach flasks with shaking at 110 rpm at 28°C until OD600 = 0.8, IPTG (1 mM) was added to induce protein production, and incubation was pursued for 4 h. E.coli crude protein extracts were loaded on 1-ml His-Trap fast-flow columns (GE, Healthcare) equilibrated with buffer A (50mM NaH2PO4 pH 7.5, 300 mM NaCl, 30 mM imidazole) and the proteins were eluted with an imidazole gradient (30mM-500mM). Fractions were dialysed against buffer B (50mM NaH2PO4 pH 7.5, 300 mM NaCl, 50% glycerol). Protein concentration was determined using the Bio-Rad protein assay [34].
Autophosphorylation of VanSG
Autophosphorylation of VanSG (40 μg) was performed in a final volume of 100 μl of buffer A (final concentrations: 50 mM Tris-HCl, 50mM KCl and 1 mM MgCl2, pH7.5). The reaction was initiated by the addition of 5 μl of ATP (1mM final) containing 200 μCi of [γ-32P]ATP and incubated at room temperature for 1 h. ATP was removed using 500 μl Sephadex G-50 spin column equilibrated with buffer A. The reaction was quenched by the addition of 5 μl of β-mercaptoethanol-stop solution (Sigma), followed by electrophoresis on 12% NuPAGE Novex Bis-Tris gels (Invitrogen) in MOPS buffer (1X), and autoradiography.
Phosphorylation of VanUG and VanRG by VanSG
Phosphotransfer to purified VanUG and VanRG were carried out in buffer A. The reaction was initiated by the addition of 10 μl of the purified autophosphorylation reaction mixture of VanSG (40 μg) described above to a 15 μl reaction mixture containing VanUG or VanRG (55 μg each). After incubation for various periods of times at room temperature, the phosphotransfer reactions were quenched by the addition of stop solution (Sigma) followed by electrophoresis on 12% NuPAGE Novex Bis-Tris gels (Invitrogen) in MOPS buffer (1X) and autoradiography.
Phosphorylation of VanUG and VanRG by acetyl[32P]phosphate
VanUG (220 μg) or VanRG (225 μg) were incubated in 100 μl of buffer B (50 mM Tris-HCl, pH7.8, 20 mM MgCl2, 0.1 mM dithiothreitol) containing 178 pmol (3.3 μCi) of acetyl[32P]phosphate (Hartmann Analytical, Germany) at room temperature for 60 min. Excess acetyl[32P]phosphate was removed using Sephadex G-50 spin columns equilibrated with buffer B. Aliquots (10 μl) were withdrawn at designated time points, and the phosphorylation reactions were quenched with β-mercaptoethanol-stop solution followed by electrophoresis on 15% SDS-polyacrylamide gels and autoradiography.
Hydrolysis of phospho-VanUG and phospho-VanRG by VanSG
The VanUG (220 μg) and VanRG (225 μg) response regulators were labelled with acetyl[32P]phosphate for 1 h at room temperature as described above, and 52 μg of VanSG histidine kinase were added, and incubation was pursued for various periods of times. Aliquots (10 μl) were withdrawn at designated time points and the reactions were stopped, followed by electrophoresis on 15% SDS-polyacrylamide gels and autoradiography.
Plasmid construction
The plasmids were constructed as follows.
Construction of pAT940, pAT941 and pAT942. pAT940(pET28ΩvanUG) and pAT941(pET28ΩvanRG)
A 225-bp BsaI-XhoI fragment corresponding to the vanUG coding sequence amplified with UG1 and UG2 (S2 Table) and a 705-bp BsaI-XhoI fragment corresponding to the vanRG coding sequence amplified by using oligonucleotides RG1 and RG2 (S2 Table) and BM4518 [11] total DNA as a template, were cloned in the NcoI and XhoI sites of modified pET28 [35] to generate plasmids pAT940(pET28ΩvanUG) and pAT941(pET28ΩvanRG). Oligodeoxynucleotide UG1 contained a BsaI restriction site designed to generate a cohesive end compatible with NcoI and 16 bases complementary to codons 1–6 of vanUG of BM4518 (S2 Table). Oligodeoxynucleotide UG2 contained a XhoI site replacing the TGA stop codon and 21 bases complementary to codons 69–75 of vanUG. Oligodeoxynucleotide RG1 contained a BsaI restriction site designed to generate a cohesive end compatible with NcoI and 16 bases complementary to codons 1–6 of vanRG of BM4518. Oligodeoxynucleotide RG2 contained a XhoI site replacing the TGA stop codon and 21 bases complementary to codons 229–235 of vanRG.
pAT942(pET28ΩvanSG)
A cytoplasmic portion of the vanSG gene of strain BM4518 was amplified using BM4518 total DNA as a template and primer pair SG1-SG3 (S2 Table). Oligodeoxynucleotide SG1 contained a BsaI restriction site designed to generate a cohesive end compatible with NcoI, and 16 bases complementary to codons 88–93 of vanSG. Oligodeoxynucleotide SG3 contained a XhoI site in place of the TAG stop codon and 21 bases complementary to codons 361–367 of vanSG. The 842-bp pCR product from vanSG was digested by BsaI and XhoI and cloned between the NcoI and XhoI restriction sites of plasmid pET28 to generate plasmid pAT942(pET28ΩvanSG).
Construction of pAT944(pGhost9Ωcat)
The XbaI cassette containing the chloramphenicol acetyltransferase cat gene with its own promoter was amplified from DNA of plasmid pAT943(pUC1318ΩPcat) with primers pG9CATNH2 and pG9CATCOOH (S2 Table) which contain a XbaI restriction site allowing the replacement of the XbaI fragment containing the erythromycin resistance gene in pGhost9 [36] to generate plasmid pAT944(pGhost9Ωcat).
Construction of pAT945(pGhost9CmΩΔvanUG), pAT946(pGhost9CmΩΔvanRG), pAT947(pGhost9CmΩΔvanSG,), and pAT973(pGhost9CmΩΔvanR'G)
The vanUG, vanRG, and vanSG genes of the vanG operon and vanR'G from BM4518 were inactivated by deletion using splicing-by-overlap extension PCR in two steps and cloned into the thermosensitive shuttle plasmid pAT944(pGhost9Ωcat) using XhoI and PstI restriction sites to generate plasmids pAT945(pGhost9CmΩΔvanUG), pAT946(pGhost9CmΩΔvanRG), pAT947(pGhost9CmΩΔvanSG), and pAT973(pGhost9CmΩΔvanR'G). The primers used for the construction of the deletant alleles and the extent of the deletions are reported in S2 Table. A SmaI restriction site was added in the primers to screen for integration in the corresponding chromosomal gene. Briefly, the remnants of the vanUG, vanRG, vanSG and vanR'G genes of BM4518 were first amplified from total DNA of BM4518 as a template using primers UG3-UG4 and UG5-UG6 for ΔvanUG, UG3-RG4 and RG5-RG7 for ΔvanRG, SG4-SG5 and SG6-SG7 for ΔvanSG, RG10-RG11 and RG12-RG13 for ΔvanR'G and, in a second step, the resulting PCR products were amplified with UG3 plus UG6, UG3 plus RG7, SG4 plus SG7, and RG10 plus RG13 respectively, to obtain ΔvanUG, ΔvanRG, ΔvanSG and ΔvanR'G.
Construction of pAT949 and derivatives
Plasmid pAT949(pDR111ΩPspankcat) was constructed by cloning the HindIII-SphI fragment of pAT948(pUC1813Ωcat) carrying the cat cassette in pDR111(Pspank) digested with the same enzymes allowing a directional cloning of the cat reporter gene under the control of the inducible Pspank promoter.
pAT950 (pDR111ΩPspanktermcat)
A 66-bp HindIII-SalI fragment corresponding to the transcription terminator of gene 32 from bacteriophage T4 [37] was amplified by PCR with oligodeoxynucleotides T4F-HindIII and T4R-SalI/NheI (S2 Table). Primer T4F-HindIII contained HindIII and NheI restriction sites. Primer T4R-SalI/NheI contained SalI and NheI restriction sites. The HindIII and SalI restriction sites allowed directional cloning of the transcription terminator (term) from bacteriophage T4 under the control of the inducible Pspank promoter and upstream from the cat reporter gene of the pAT949(pDR111ΩPspankcat) shuttle vector.
pAT951(pDR111ΩPspankvanUGcat)
The vanUG gene of BM4518 was amplified using primer pair UGNH2 and UGCOOH (S2 Table) and total DNA of the corresponding strain as a template. Oligodeoxynucleotide UGNH2 contained BsaI and HindIII restriction sites, a RBS, and 6 bases complementary to vanUG including the ATG (translation initiation) codon. Oligodeoxynucleotide UGCOOH harbored SalI and NheI restriction sites, the stop codon, and 15 bases complementary to the 3’ end sequence of vanUG from BM4518. The BsaI and SalI restriction sites allowed directional cloning of a 249-bp fragment of vanUG downstream from the inducible Pspank promoter and upstream from the cat gene of the pAT949(pDR111ΩPspankcat) shuttle vector to generate pAT951(pDR111ΩPspankvanUGcat).
pAT952(pDR111ΩPspanktermPUGcat) and pAT953(pDR111ΩPspankvanUGPUGcat)
The regulatory PUG (183 bp) promoter was amplified by PCR from BM4518 total DNA with oligodeoxynucleotides PUG1 and PUG2 (S2 Table). Primers PUG1 and PUG2 contained a NheI and a SalI restriction site, respectively, which allowed directional cloning of PUG upstream from the cat gene of pAT950(pDR111ΩPspanktermcat) to generate pAT952(pDR111ΩPspanktermPUGcat) or allowed directional cloning of PUG downstream from vanUG and upstream from the cat reporter gene of pAT951(pDR111ΩPspankvanUGcat) to generate pAT953(pDR111ΩPspankvanUGPUGcat).
pAT954(pDR111ΩPspankvanRGPUGcat)
A 754-bp HindIII-NheI fragment corresponding to the vanRG coding sequence with its RBS, initiation and stop codons was amplified by PCR from BM4518 with oligodeoxynucleotides RGNH2 and RGCOOH (S2 Table). Primer RGNH2 contained a HindIII restriction site. Primer RGCOOH comprised SalI and NheI restriction sites, the stop codon, and 14 bases complementary to the 3’ end of vanRG from BM4518. The HindIII and NheI restriction sites allowed directional cloning of the vanRG gene under the control of the inducible Pspank promoter and upstream from PUG and the cat gene of pAT952(pDR111ΩPspanktermPUGcat).
pAT956(pDR111ΩPspankvanURGPUGcat), pAT958(pDR111ΩPspankvanRSGPUGcat), pAT960(pDR111ΩPspankvanURSGPUGcat) pAT961(pDR111ΩPspankvanRSGPYGcat)and pAT962(pDR111ΩPspankvanURSGPYGcat)
The vanURG, vanRSG, and vanURSG genes of BM4518 were amplified using primer pairs UGNH2-RGCOOH, RGNH2-SGCOOH, and UGNH2-SGCOOH (S2 Table), respectively, and BM4518 total DNA as a template. Oligodeoxynucleotides UGNH2 and RGNH2 harbored a HindIII restriction site and 21 bases complementary to the sequence upstream from vanUG or 17 bases complementary to the sequence upstream from vanRG. Primers RGCOOH and SGCOOH contained each SalI and NheI restriction sites, the stop codon and 14 or 13 bases complementary to the 3' end of respectively vanRG and vanSG of BM4518. The HindIII and SalI restriction sites allowed directional cloning of vanURG, vanRSG, and vanURSG upstream from the cat reporter gene of shuttle vector pAT949(pDR111ΩPspankcat) carrying the inducible Pspank promoter to generate pAT955(pDR111ΩPspankvanURGcat), pAT957(pDR111ΩPspankvanRSGcat), and pAT959(pDR111ΩPspankvanURSGcat). The 183-bp NheI-SalI fragment carrying the PUG promoter obtained above by amplification was cloned in pAT955(pDR111ΩPspankvanURGcat), pAT957(pDR111ΩPspankvanRSGcat), and pAT959(pDR111ΩPspankvanURSGcat) digested with the same enzymes to generate pAT956(pDR111ΩPspankvanURGPUGcat), pAT958(pDR111ΩPspankvanRSGPUGcat), and pAT960(pDR111ΩPspankvanURSGPUGcat). The 177-bp NheI-SalI fragment carrying the PYG resistance promoter amplified by PCR from BM4518 DNA with primers PYG1 and PYG2 (S2 Table) was cloned in pAT957(pDR111ΩPspankvanRSGcat), and pAT959(pDR111ΩPspankvanURSGcat) digested with the same enzymes to generate, respectively, pAT961(pDR111ΩPspankvanRSGPYGcat)and pAT962(pDR111ΩPspankvanURSGPYGcat).
pAT964(pDR111ΩPspankvanUGtermcat), pAT965(pDR111ΩPspankvanUGtermPUGcat), pAT966(pDR111ΩPspankvanRGtermPUGcat), pAT967(pDR111ΩPspankvanURGtermPUGcat), pAT968(pDR111ΩPspankvanURSGtermPUGcat), pAT969(pDR111ΩPspankvanRSGtermPUGcat), pAT970(pDR111ΩPspankvanURSGtermPYGcat), and pAT971 (pDR111ΩPspankvanRSGtermPYGcat)
The NheI terminator fragment amplified by PCR with oligodeoxynucleotides T4F-NheI and T4R-NheI/KpnI (S2 Table) was cloned, respectively, in pAT951(pDR111ΩPspankvanUGcat), pAT953(pDR111ΩPspankvanUGPUGcat), pAT954(pDR111ΩPspankvanRGPUGcat), pAT956(pDR111ΩPspankvanURGPUGcat), pAT960(pDR111ΩPspankvanURSGPUGcat), pAT958(pDR111ΩPspankvanRSGPUGcat), pAT962(pDR111ΩPspankvanURSGPYGcat) and pAT961(pDR111ΩPspankvanRSGPYGcat) digested with NheI.
pAT972(pDR111ΩPspankvanUSGtermcat)
The 1,144-bp fragment containing the vanSG gene of BM4518 was amplified using primer pair SGNH2-SGCOOH (S2 Table) and total DNA of the corresponding strain as a template. The NheI and SalI restriction sites allowed directional cloning of vanSG downstream from the vanUG gene and upstream from the cat gene of pAT951(pDR111ΩPspankvanUGcat) to generate pAT963(pDR111ΩPspankvanUSGcat).
The EcoRI fragment harboring the vanUSG' genes from pAT963(pDR111ΩPspankvanUSGcat) was replaced by the EcoRI fragment carrying the vanRSG' genes of pAT971(pDR111ΩPspankvanRSGtermPYGcat) to generate pAT972(pDR111ΩPspankvanUSGtermcat).
Construction of strains
Plasmids pDR111, pAT949, pAT950, pAT952, pAT964, pAT965, pAT966, pAT967, pAT968, pAT969, pAT970, pAT971, and pAT972 were introduced by transformation into vancomycin susceptible E. coli NR698 and transformants were selected on agar containing chloramphenicol (10 g/ml) or ampicillin (100 μg/ml, for pDR111) (Tables 1 and 2).
In Gram-positive bacteria, pGhost9 [36] which replicates at 28°C but is lost above 37°C, allowed construction of E.faecalis BM4522 derivatives by insertional inactivation. Plasmids pAT945(pGhost9CmΩΔvanUG), pAT946(pGhost9CmΩΔvanRG), and pAT947(pGhost9CmΩΔvanSG) were electrotransformed into E. faecalis BM4522 [11] to generate, respectively, BM4720(ΔvanUG), BM4721(ΔvanRG), and BM4722(ΔvanSG) (S1 Table). Plasmid pAT973(pGhost9CmΩΔvanR'G) was electrotransformed into E. faecalis BM4721(ΔvanRG) to generate the double mutant BM4723(ΔvanRG, ΔvanR'G). Transformants were selected at the permissive temperature (28°C) on M17 plates containing 10g/ml of chloramphenicol and 0.5% glucose. A colony of each transformant was inoculated into 50 ml of M17 broth containing 0.5% glucose and incubated for 2h at 28°C. The culture was then shifted to a non-permissive temperature (42°C) for 2 h and integrants, following a first recombination event, were selected at 42°C on M17 agar containing chloramphenicol (10g/ml). Plasmid excision, by a second recombination event, was favored by subculturing at 28°C in the absence of chloramphenicol and plasmid loss was screened for by plating at 42°C on M17-glucose followed by replica plating on chloramphenicol. The integration locus was determined by PCR following digestion with SmaI and sequencing.
Enzyme assays
For preparation of extracts, 8 ml of an overnight culture were added to 100 ml of broth in the absence or in the presence of vancomycin and strains were grown until OD600 = 0.8 in 250 ml bottles with shaking at 110 rpm. The cells were harvested by centrifugation, washed in 0.1M phosphate buffer pH 7.0, resuspended in the same buffer, lysed by sonication, followed by centrifugation at 10,000 g during 45 min. The resuspended pellet for VanTG racemase [11] and supernatant for CAT activity, were assayed as described [38].
Genome sequencing, assemblies and annotation
Total DNA from BM4518 and BM4522 strains was purified and sequencing library preparation was carried out using the Nextera DNA Sample Preparation kit (Illumina, San Diego, CA), according to manufacturer’s specifications. Quality and quantity of each sample library was measured on an Agilent Technologies 2100 Bioanalyzer (Santa Clara, CA). Libraries were normalized to 2nM, multiplexed and subjected to 250-bp paired end sequencing (Illumina MiSeq). On average, 5 million high-quality paired-end reads were collected for each strain, representing >220-fold coverage of the ~2.9 Mb genomes. Reads were assembled de novo utilizing CLC Genomics Workbench (CLC bio, Cambridge, MA). Functional annotations were performed using a custom pipeline as described previously [39].
Determination of growth rates
Growth rates were determined in microplates coupled to a spectrophotometer iEMS reader (Labsystems). Strains were grown overnight at 37°C without or with 1 μg/ml of vancomycin. The cultures were diluted at OD 0.15 into 10 ml of broth without or with vancomycin (1μg/ml) and grown at 37°C with shaking until the beginning of the stationary phase. The cultures were diluted 1/1,000 to inoculate 105 bacteria into 200 μl of broth in a 96-well microplate that was incubated overnight at 37°C with shaking. Absorbance was measured at 600 nm every 3 min. Each culture was replicated three times in the same microplate. Growth rates performed in three independent experiments were determined at the beginning of the exponential phase and the relative growth rates were calculated as the ratio of the growth rate of the strain induced by vancomycin versus that of the non induced strain.
Supporting Information
Zdroje
1. Arias CA, Murray BE (2012) The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10 : 266–278. doi: 10.1038/nrmicro2761 22421879
2. Reynolds PE (1989) Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis 8 : 943–950. 2532132
3. Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P (2007) Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev 20 : 79–114. 17223624
4. Boyd DA, Willey BM, Fawcett D, Gillani N, Mulvey MR (2008) Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel D-Ala-D-Ser gene cluster, vanL. Antimicrob Agents Chemother 52 : 2667–2672. doi: 10.1128/AAC.01516-07 18458129
5. Lebreton F, Depardieu F, Bourdon N, Fines-Guyon M, Berger P, Camiade S, et al. (2011) D-Ala-D-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother 55 : 4606–4612. doi: 10.1128/AAC.00714-11 21807981
6. Xu X, Lin D, Yan G, Ye X, Wu S, Guo Y, et al. (2010) vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob Agents Chemother 54 : 4643–4647. doi: 10.1128/AAC.01710-09 20733041
7. Depardieu F, Courvalin P, Msadek T (2003) A six amino acid deletion, partially overlapping the VanSB G2 ATP-binding motif, leads to constitutive glycopeptide resistance in VanB-type Enterococcus faecium. Mol Microbiol 50 : 1069–1083. 14617162
8. Depardieu F, Courvalin P, Kolb A (2005) Binding sites of VanRB and sigma70 RNA polymerase in the vanB vancomycin resistance operon of Enterococcus faecium BM4524. Mol Microbiol 57 : 550–564. 15978084
9. Wright GD, Holman TR, Walsh CT (1993) Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry 32 : 5057–5063. 8494882
10. Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P (1997) The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J Bacteriol 179 : 97–106. 8981985
11. Depardieu F, Bonora MG, Reynolds PE, Courvalin P (2003) The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol Microbiol 50 : 931–948. 14617152
12. Ruiz N, Falcone B, Kahne D, Silhavy TJ (2005) Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121 : 307–317. 15851036
13. Holman TR, Wu Z, Wanner BL, Walsh CT (1994) Identification of the DNA-binding site for the phosphorylated VanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry 33 : 4625–4631. 8161518
14. Koudelka GB, Lam CY (1993) Differential recognition of OR1 and OR3 by bacteriophage 434 repressor and Cro. J Biol Chem 268 : 23812–23817. 8226917
15. Cervin MA, Lewis RJ, Brannigan JA, Spiegelman GB (1998) The Bacillus subtilis regulator SinR inhibits spoIIG promoter transcription in vitro without displacing RNA polymerase. Nucleic Acids Res 26 : 3806–3812. 9685500
16. Newman JA, Rodrigues C, Lewis RJ (2013) Molecular basis of the activity of SinR protein, the master regulator of biofilm formation in Bacillus subtilis. J Biol Chem 288 : 10766–10778. doi: 10.1074/jbc.M113.455592 23430750
17. Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. (1989) Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol 210 : 551–559. 2693738
18. Ansaldi M, Simon G, Lepelletier M, Mejean V (2000) The TorR high-affinity binding site plays a key role in both torR autoregulation and torCAD operon expression in Escherichia coli. J Bacteriol 182 : 961–966. 10648521
19. Igo MM, Ninfa AJ, Stock JB, Silhavy TJ (1989) Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev 3 : 1725–1734. 2558046
20. Kamberov ES, Atkinson MR, Chandran P, Ninfa AJ (1994) Effect of mutations in Escherichia coli glnL (ntrB), encoding nitrogen regulator II (NRII or NtrB), on the phosphatase activity involved in bacterial nitrogen regulation. J Biol Chem 269 : 28294–28299. 7961767
21. Arthur M, Depardieu F, Courvalin P (1999) Regulated interactions between partner and non partner sensors and responses regulators that control glycopeptide resistance gene expression in enterococci. Microbiology 145 : 1849–1858. 10463151
22. Ogura M, Yoshikawa H, Chibazakura T (2014) Regulation of the response regulator gene degU through the binding of SinR/SlrR and exclusion of SinR/SlrR by DegU in Bacillus subtilis. J Bacteriol 196 : 873–881. doi: 10.1128/JB.01321-13 24317403
23. Mittal M, Pechter KB, Picossi S, Kim HJ, Kerstein KO, Sonenshein AL (2013) Dual role of CcpC protein in regulation of aconitase gene expression in Listeria monocytogenes and Bacillus subtilis. Microbiology 159 : 68–76. doi: 10.1099/mic.0.063388-0 23139400
24. Belitsky BR (2011) Indirect repression by Bacillus subtilis CodY via displacement of the activator of the proline utilization operon. J Mol Biol 413 : 321–336. doi: 10.1016/j.jmb.2011.08.003 21840319
25. Mitrophanov AY, Groisman EA (2008) Signal integration in bacterial two-component regulatory systems. Genes Dev 22 : 2601–2611. doi: 10.1101/gad.1700308 18832064
26. Buelow DR, Raivio TL (2010) Three (and more) component regulatory systems—auxiliary regulators of bacterial histidine kinases. Mol Microbiol 75 : 547–566. doi: 10.1111/j.1365-2958.2009.06982.x 19943903
27. Foucault ML, Depardieu F, Courvalin P, Grillot-Courvalin C (2010) Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc Natl Acad Sci USA 107 : 16964–16969. doi: 10.1073/pnas.1006855107 20833818
28. Studier FW, Moffatt BA (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189 : 113–130. 3537305
29. Amrein KE, Takacs B, Stieger M, Molnos J, Flint NA, Burn P (1995) Purification and characterization of recombinant human p50csk protein-tyrosine kinase from an Escherichia coli expression system overproducing the bacterial chaperones GroES and GroEL. Proc Natl Acad Sci USA 92 : 1048–1052. 7862631
30. Brinster S, Furlan S, Serror P (2007) C-terminal WxL domain mediates cell wall binding in Enterococcus faecalis and other gram-positive bacteria. J Bacteriol 189 : 1244–1253. 16963569
31. Kay R, McPherson J (1987) Hybrid pUC vectors for addition of new restriction enzyme sites to the ends of DNA fragments. Nucleic Acids Res 15 : 2778.31. Maxam AM, Gilbert W (1977) A new method for sequencing DNA. Proc Natl Acad Sci U S A 74 : 560–564.
32. Jacob AE, Hobbs SJ (1974) Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol 117 : 360–372. 4204433
33. Maxam AM, Gilbert W (1977) A new method for sequencing DNA. Proc Natl Acad Sci U S A 74 : 560–564. 265521
34. Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochemistry 72 : 248–254. 942051
35. Chastanet A, Fert J, Msadek T (2003) Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol Microbiol 47 : 1061–1073. 12581359
36. Maguin E, Prevost H, Ehrlich SD, Gruss A (1996) Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol 178 : 931–935. 8550537
37. Brockbank SM, Barth PT (1993) Cloning, sequencing, and expression of the DNA gyrase genes from Staphylococcus aureus. J Bacteriol 175 : 3269–3277. 8388872
38. Arthur M, Depardieu F, Reynolds P, Courvalin P (1996) Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol 21 : 33–44. 8843432
39. Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V (2013) Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio 4.
Štítky
Genetika Reprodukčná medicína
Článek Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in CrustaceansČlánek Adventures in WonderlandČlánek Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and InfectivityČlánek Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in FilamentsČlánek Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in ArabidopsisČlánek Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression ControlČlánek The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent EnhancersČlánek Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of MitochondriaČlánek The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma FormationČlánek Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 4- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- Adventures in Wonderland
- Experimental Swap of 's Assortative Mating Preferences Demonstrates Key Role of X-Chromosome Divergence Island in Incipient Sympatric Speciation
- Chromosome Replacement and Deletion Lead to Clonal Polymorphism of Berry Color in Grapevine
- The Protein Quality Control Machinery Regulates Its Misassembled Proteasome Subunits
- Genome-Wide Association Study Identifies as a Critical Gene for Susceptibility to Noise-Induced Hearing Loss
- Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and Infectivity
- Viable Neuronopathic Gaucher Disease Model in Medaka () Displays Axonal Accumulation of Alpha-Synuclein
- Multi-locus Analysis of Genomic Time Series Data from Experimental Evolution
- The Genetic Legacy of the Expansion of Turkic-Speaking Nomads across Eurasia
- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- The Pif1 Helicase, a Negative Regulator of Telomerase, Acts Preferentially at Long Telomeres
- Inhibiting K63 Polyubiquitination Abolishes No-Go Type Stalled Translation Surveillance in
- SYD-1C, UNC-40 (DCC) and SAX-3 (Robo) Function Interdependently to Promote Axon Guidance by Regulating the MIG-2 GTPase
- Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in Filaments
- Synergistic and Independent Actions of Multiple Terminal Nucleotidyl Transferases in the 3’ Tailing of Small RNAs in Arabidopsis
- Host Genetic Variation Influences Gene Expression Response to Rhinovirus Infection
- Contribution of Large Region Joint Associations to Complex Traits Genetics
- Volatility of Mutator Phenotypes at Single Cell Resolution
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in Arabidopsis
- A Multi-layered Protein Network Stabilizes the FtsZ-ring and Modulates Constriction Dynamics
- Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression Control
- Genome Sequencing of the Perciform Fish Provides Insights into Molecular and Genetic Mechanisms of Stress Adaptation
- Natural Variant E610G Is a Semi-dominant Suppressor of IAP-Induced RNA Processing Defects
- The Alkaline Response Pathway: Identification of a Novel Rim Pathway Activator
- Transgenerational Inheritance of Diet-Induced Genome Rearrangements in Drosophila
- A Single Nucleotide Polymorphism Uncovers a Novel Function for the Transcription Factor Ace2 during Hyphal Development
- DNA Damage Response and Spindle Assembly Checkpoint Function throughout the Cell Cycle to Ensure Genomic Integrity
- The Functional Interplay Between the t(9;22)-Associated Fusion Proteins BCR/ABL and ABL/BCR in Philadelphia Chromosome-Positive Acute Lymphatic Leukemia
- Extreme Recombination Frequencies Shape Genome Variation and Evolution in the Honeybee,
- Beyond Glycolysis: GAPDHs Are Multi-functional Enzymes Involved in Regulation of ROS, Autophagy, and Plant Immune Responses
- Comprehensive Profiling of Amino Acid Response Uncovers Unique Methionine-Deprived Response Dependent on Intact Creatine Biosynthesis
- Windpipe Controls Intestinal Homeostasis by Regulating JAK/STAT Pathway via Promoting Receptor Endocytosis and Lysosomal Degradation
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
- Cross-Population Joint Analysis of eQTLs: Fine Mapping and Functional Annotation
- The Power of Gene-Based Rare Variant Methods to Detect Disease-Associated Variation and Test Hypotheses About Complex Disease
- The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent Enhancers
- Competition between VanU Repressor and VanR Activator Leads to Rheostatic Control of Vancomycin Resistance Operon Expression
- A Missense Change in the Gene Links Aberrant Autophagy to a Neurodegenerative Vacuolar Storage Disease
- Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model
- Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of Mitochondria
- Genome-Destabilizing Effects Associated with Top1 Loss or Accumulation of Top1 Cleavage Complexes in Yeast
- Imputation-Based Population Genetics Analysis of Malaria Parasites
- Heterozygosity for a Hypomorphic Polβ Mutation Reduces the Expansion Frequency in a Mouse Model of the Fragile X-Related Disorders
- Neto-Mediated Intracellular Interactions Shape Postsynaptic Composition at the Neuromuscular Junction
- Ndd1 Turnover by SCF Is Inhibited by the DNA Damage Checkpoint in
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma Formation
- Spastin Binds to Lipid Droplets and Affects Lipid Metabolism
- Maintenance of Glia in the Optic Lamina Is Mediated by EGFR Signaling by Photoreceptors in Adult Drosophila
- Auxin Influx Carriers Control Vascular Patterning and Xylem Differentiation in
- Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
- The Lowe Syndrome Protein OCRL1 Is Required for Endocytosis in the Zebrafish Pronephric Tubule
- Postnatal Loss of Hap1 Reduces Hippocampal Neurogenesis and Causes Adult Depressive-Like Behavior in Mice
- CAPER Is Vital for Energy and Redox Homeostasis by Integrating Glucose-Induced Mitochondrial Functions via ERR-α-Gabpa and Stress-Induced Adaptive Responses via NF-κB-cMYC
- Distinct and Cooperative Activities of HESO1 and URT1 Nucleotidyl Transferases in MicroRNA Turnover in
- The Evolutionary Origination and Diversification of a Dimorphic Gene Regulatory Network through Parallel Innovations in and
- MAPK Signaling Pathway Alters Expression of Midgut ALP and ABCC Genes and Causes Resistance to Cry1Ac Toxin in Diamondback Moth
- Spatio-temporal Remodeling of Functional Membrane Microdomains Organizes the Signaling Networks of a Bacterium
- Asymmetric Transcript Discovery by RNA-seq in . Blastomeres Identifies , a Gene Important for Anterior Morphogenesis
- A Stress-Induced Small RNA Modulates Alpha-Rhizobial Cell Cycle Progression
- Systematic Profiling of Poly(A)+ Transcripts Modulated by Core 3’ End Processing and Splicing Factors Reveals Regulatory Rules of Alternative Cleavage and Polyadenylation
- The UPR Branch IRE1- in Plants Plays an Essential Role in Viral Infection and Is Complementary to the Only UPR Pathway in Yeast
- A Non-canonical RNA Silencing Pathway Promotes mRNA Degradation in Basal Fungi
- Co-chaperone p23 Regulates . Lifespan in Response to Temperature
- Re-replication of a Centromere Induces Chromosomal Instability and Aneuploidy
- Shade Avoidance Components and Pathways in Adult Plants Revealed by Phenotypic Profiling
- Lipid-Induced Epigenomic Changes in Human Macrophages Identify a Coronary Artery Disease-Associated Variant that Regulates Expression through Altered C/EBP-Beta Binding
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



