-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi
Fungal plant pathogens are of outstanding economic and ecological importance and cause destructive diseases on many cultivated and wild plants. Effector proteins that are secreted during infection to manipulate the host and to promote disease are a key element in fungal virulence. Phytopathogenic fungi possess huge effector repertoires that are dominated by hundreds of sequence-unrelated small secreted proteins. The molecular functions of this most important class of fungal effectors and the evolutionary mechanisms that generate this tremendous numbers of apparently unrelated proteins are largely unknown. By investigating the 3-dimensional structures of effectors from the rice blast fungus M. oryzae, we discovered an effector family comprising structurally conserved but sequence-unrelated effectors from M. oryzae and the phylogenetically distant wheat pathogen Pyrenophora tritici-repentis that we named MAX-effectors (M. oryzae Avrs and ToxB). Structure-informed searches of whole genome sequence databases suggest that MAX-effectors are present at low frequencies and with a patchy phylogenetic distribution in many ascomycete phytopathogens. They underwent strong lineage-specific expansion in fungi of the Pyriculariae family that contains M. oryzae where they seem particularly important during biotrophic plant colonization and account for 50% of the cloned Avr effectors and 5–10% of the effector repertoire. Based on our results on the MAX-effectors and the widely accepted concept that fungal effectors evolve according to a birth-and-death model we propose the hypothesis that the majority of the immense numbers of different ascomycete effectors could in fact belong to a limited set of structurally defined families whose members are phylogenetically related.
Published in the journal: Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi. PLoS Pathog 11(10): e32767. doi:10.1371/journal.ppat.1005228
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005228Summary
Fungal plant pathogens are of outstanding economic and ecological importance and cause destructive diseases on many cultivated and wild plants. Effector proteins that are secreted during infection to manipulate the host and to promote disease are a key element in fungal virulence. Phytopathogenic fungi possess huge effector repertoires that are dominated by hundreds of sequence-unrelated small secreted proteins. The molecular functions of this most important class of fungal effectors and the evolutionary mechanisms that generate this tremendous numbers of apparently unrelated proteins are largely unknown. By investigating the 3-dimensional structures of effectors from the rice blast fungus M. oryzae, we discovered an effector family comprising structurally conserved but sequence-unrelated effectors from M. oryzae and the phylogenetically distant wheat pathogen Pyrenophora tritici-repentis that we named MAX-effectors (M. oryzae Avrs and ToxB). Structure-informed searches of whole genome sequence databases suggest that MAX-effectors are present at low frequencies and with a patchy phylogenetic distribution in many ascomycete phytopathogens. They underwent strong lineage-specific expansion in fungi of the Pyriculariae family that contains M. oryzae where they seem particularly important during biotrophic plant colonization and account for 50% of the cloned Avr effectors and 5–10% of the effector repertoire. Based on our results on the MAX-effectors and the widely accepted concept that fungal effectors evolve according to a birth-and-death model we propose the hypothesis that the majority of the immense numbers of different ascomycete effectors could in fact belong to a limited set of structurally defined families whose members are phylogenetically related.
Introduction
Pathogenic microorganisms have to cope with the immune system of their host and therefore deploy measures to hide their presence, disturb host immunity or inactivate defense responses. In all these strategies, proteins secreted by the pathogen during infection and acting on host proteins and cellular processes play a key role [1–3]. These proteinaceous virulence factors named effectors act either extra-cellularly or inside host cells and can possess, depending on the microorganism, very different molecular features.
In fungal pathogens, the main class of effectors are small secreted proteins of less than 200 amino acids expressed specifically during infection and often rich in cysteins [4–6]. Genome sequencing and expression analysis identified hundreds of such effector candidates in individual plant pathogenic fungal species. Few of them, mainly those acting extra-cellularly, are widely distributed among phytopathogenic fungi and contain known motifs or domains, such as NLPs (necrosis and ethylene-inducing peptide 1 (Nep1)-like proteins), LysM domain-containing proteins or protease inhibitors [5,6]. The vast majority of the fungal effectors do not share sequence similarities with other proteins and do not contain conserved motifs. This is very different from the situation in other phytopathogens and in particular oomyctes, an important class of plant pathogens that have similar lifestyles and infection strategies and whose virulence relies also on large effector repertoires. In oomycete pathogens, large families of cytoplasmic effectors with hundreds of members in individual species are defined by the presence of the RXLR or the LFLAK host cell translocation motifs [7–9]. The effector domains of these RXLR and Crinkler (CRN) effectors that mediate virulence functions are highly diversified but contain, in the majority of cases, conserved motifs or domains that are shared between effectors from the same or other species allowing their classification in distinct families. On the contrary, most fungal effectors are species-specific while few are lineage specific and occur in closely related species. In most phytopathogenic fungi, no large effector gene families were identified [5,6]. The majority of their effectors are singletons and a small proportion belongs to small paralogous groups of rarely more than 3 members. Effector repertoires dominated by gene families of large size counting more than 5 members were only detected in particular cases such as powdery mildew and rust fungi lineages [10–13]. Due to their high diversity and the lack of similarity with other proteins, the mode of action and the role in infection of fungal effectors have to be elucidated case by case and remain still largely unknown [5,6]. In addition, this tremendous diversity raises the question of the evolutionary trajectories of fungal effectors that do not show traces of common origins.
Rice blast disease caused by the ascomycete fungus M. oryzae is present in all rice growing areas and causes important harvest losses. Since rice is the main source of calories for half of the human population and since disease control strategies are frequently overcome by the pathogen due to its high genetic plasticity, blast is considered one of the most dangerous plant diseases threatening global food security and hampering attempts to increase rice yield in many parts of the world [14–16]. Due to its economic importance, the status of the host plant rice as a model plant and the ease of cultivation and genetic manipulation of M. oryzae, blast disease has become a model for the molecular and genetic investigation of fungal plant diseases [14]. In particular, molecular mechanisms of fungal disease development were studied intensively in M. oryzae uncovering important features of fungal virulence [17,18]. Key steps in infection by M. oryzae are (i) penetration into epidermal cells by the breakage of the leaf cuticle and epidermal cell walls by an appressorium, a specialized unicellular structure, (ii) biotrophic growth inside the first invaded host cells, followed by (iii) necrotrophic growth associated with active killing of host tissue and the development of disease symptoms and finally, (iv) clonal reproduction and sporulation.
Effectors and in particular cytoplasmic effectors are key elements in M. oryzae virulence and particularly important during the biotrophic phase of infection [6,19,20]. However, the function of individual effectors in the infection process has only been established for the LysM effector SLP1 that sequesters chitin fragments and thereby interferes with their recognition by the rice chitin receptor CEBiP, and AvrPiz-t that interferes with host immunity by inhibiting the E3 ubiquitin ligase APIP6 [21,22]. Mutant analysis aiming to demonstrate that individual effectors are important for virulence have often been unsuccessful, probably due to functional redundancy among effectors [23,24]. Approximately 700 of the 1300–1500 secreted proteins encoded in the M. oryzae genome are considered effector candidates according to their size of less than 200 amino acids and their lack of homology to proteins of known function [25,26]. Hundreds of them were found to be expressed during appressoria formation or infection [23,26–28].
Some effectors are recognized in certain plant accessions by immune receptors localized either at the plasma membrane or in the cytosol leading to the induction of strong defense responses and resistance to pathogen isolates possessing this effector [29]. The recognized effector is, in these cases, named an avirulence (Avr) protein. In M. oryzae, 8 different effectors acting as Avr proteins named PWL2, AVR-Pia, AVR1-CO39, AVR-Pii, AVR-Pik, AvrPiz-t, AVR-Pita and Avr-Pi9 have been cloned molecularly [26,30–35]. They are all translocated into host cells and do not show similarities to proteins of known function with the exception of AVR-Pita that shows homology to neutral zinc proteases [6]. For 7 of them, the matching rice immune receptors that are in all cases cytoplasmic nucleotide-binding and leucine-rich repeat domain proteins (NLRs) have been identified [36–41].
In the present study, the 3-dimensional structures of the M. oryzae effectors AVR-Pia and AVR1-CO39 were investigated to deepen our understanding of fungal effector function and diversity. NMR analysis revealed that the structures of both proteins consist of two anti-parallel β-sheets, each having three strands, and linked by one disulfide bond Structural similarity searches revealed that the M. oryzae effector AvrPiz-t and the effector ToxB from the wheat pathogen Pyrenophora tritici-repens have similar 6 β-sandwich structures with the same topology [42,43]. Comparisons of the structures of the four effectors that we named MAX-effectors revealed that they share a common architecture but no sequence consensus. Structure-informed and pattern-based searches identified large numbers of weakly homologous MAX-effector candidates in M. oryzae and M. grisea, and limited numbers or no homologs in other phytopathogenic ascomycete fungi. Expression profiling indicated that the majority of the M. oryzae MAX-effector candidates are expressed during early infection. MAX-effectors therefore seem to have undergone a lineage-specific expansion in the Pyricularia genus that may be driven by duplications and rapid adaptation to new functions involving important changes of surface properties but conservation of protein architecture. This evolutionary process has the potential to generate large families of structurally related proteins without sequence similarity and may serve as a paradigm for effector evolution and diversification in phytopathogenic ascomycete fungi.
Results
Protein expression
AVR1-CO39 and AVR-Pia proteins, deleted for their endogenous secretion signal, were expressed in E. coli with an N-terminally fused signal peptide for secretion in the bacterial periplasm that is cleaved upon secretion, an N-terminal His6-tag for purification and a TEV1 cleavage site. Recombinant proteins were soluble and were purified to homogeneity from periplasmic protein extracts by Ni-agarose affinity and gel exclusion chromatography (S1 Fig). Both recombinant Avr proteins eluted as monomers from gel exclusion chromatography.
NMR analysis
Recombinant, 15N and 13C-labelled AVR1-CO39 and AVR-Pia proteins produced in 15N and 13C-labelled minimal medium were used for structure determination by two - and three-dimensional NMR experiments. Three-dimensional (3D) HNCO, HNCA, HN(CO)CACB, HN(CA)CO, HNCACB, 2D 13C-detected CON, CACO and 2D-COSY-DQF(D2O) and TOCSY(D2O) experiments were used for the backbone and aliphatic side chain resonance assignments. 3D 15N-edited NOESY-HSQC and 2D-NOESY(D2O) spectra were collected to confirm the chemical shift assignments and generate distance restraints for structure calculations. (Fig 1 and S1 Table). The assigned 1H,15N-HSQC spectra were well dispersed. Residues from the N-terminal tags are still resolved. All amino acids of AVR-Pia and almost all of AVR1-CO39 have {1H-15N} NOE values above 0.8 indicating highly defined structures with low flexibility (S2 Fig). Only N-terminal tags, below residue number 22–23, and C-terminal sequences of AVR1-C039 (amino acids 80–89) show increased flexibility. The strong dαN(i, i+1) NOEs and weak dNN(i, i+1) NOEs are indicative of a β-structure and consistent with the six β-strands observed in AVR-Pia and AVR1-CO39 (S3 Fig). NHs in slow exchange were consistent with hydrogen bonding networks and were used to derive constraints for the structure calculations.
Fig. 1. 15N-HSQC spectra of (A) AVR-Pia and (B) AVR1-CO39. 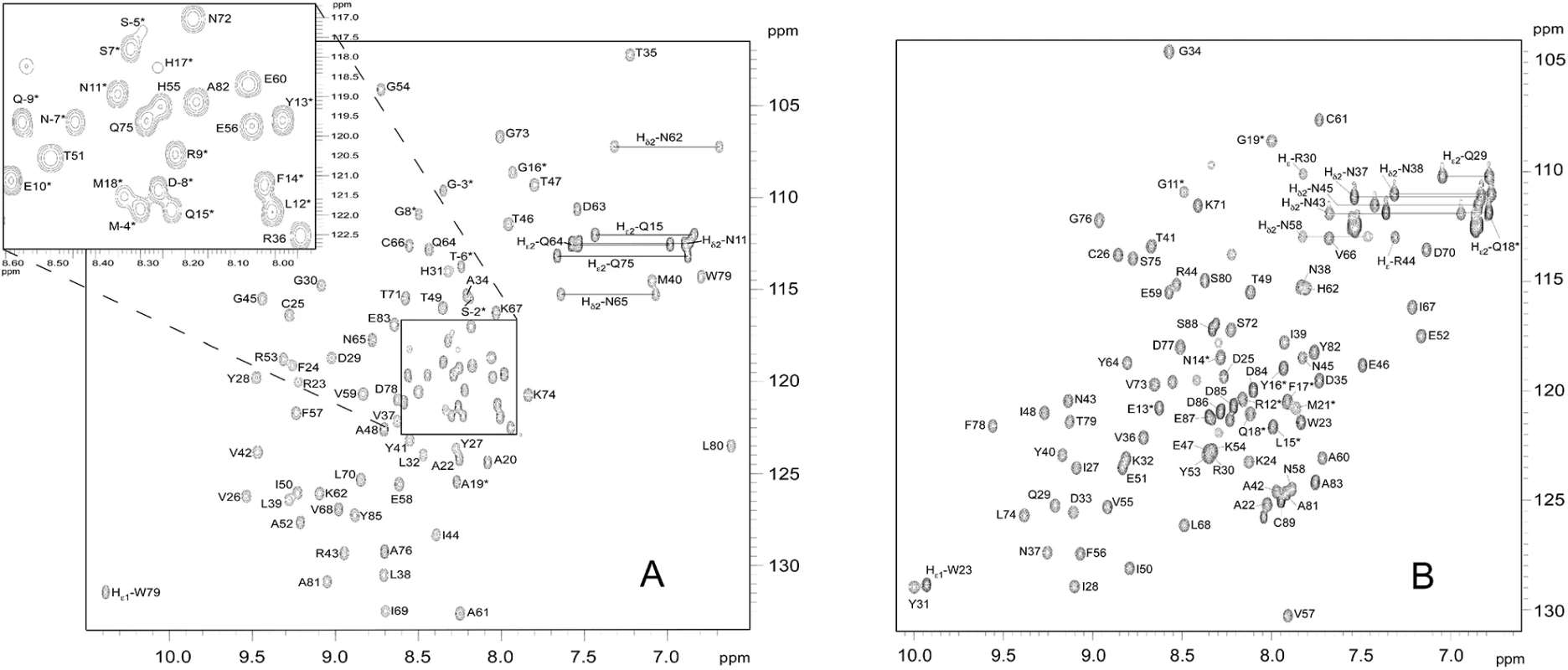
In the 15N-HSQC spectra of (A) AVR-Pia and (B) AVR1-CO39, each peak comes from N-H chemical connectivity and has 15N and 1H chemical shift coordinates. There is one backbone N-H group per residue, leading to one HSQC peak per residue. A side-chain NH2 group gives two HSQC peaks with one common N coordinate. Other side-chains NH groups may also be observed, as Trp Nε1-H and Arg Nε-H. (*) indicates residues in the N-terminal tail. The mature proteins start at residue Ala20 and Trp23 for AVR-Pia and AVR1-CO39, respectively [26,32]. The NH2 side chains resonances were assigned. The resonances of the tryptophan indole groups are specifically labelled Hε1. The ratios of R2 to R1 relaxation rates of AVR-Pia and AVR1-CO39 were consistent with a monomeric molecular size (AVR-Pia τc = 6.2 ± 0.3 ns and AVR1-CO39 τc = 5.7 ± 0.4 ns) and thus confirm that both Avrs form monomers in solution (S1 Table) [44].
AVR-Pia and AVR1-CO39 have similar β-sandwich structures
The solution structures of AVR-Pia and AVR1-CO39 were determined based on 1541 and 1286 NOE-derived distance restraints, 90 and 72 dihedral angle restraints and 20 and 15 hydrogen bond restraints, respectively (Fig 2 and Table 1 and S4 Fig). A disulfide bridge between Cys25-Cys66 for AVR-Pia and between Cys26-Cys61 for AVR1-CO39 was added based on cysteine 13Cβ chemical shifts and DTNB quantification of free thiols. The Pro65 in AVR1-CO39 has been determined to be in a cis-conformation according to the 13Cβ chemical shift at 34.4 ppm and strong sequential Hα-Hα NOE. The best conformers with the lowest energies, which exhibited no obvious NOE violations and no dihedral violations > 2° were selected for final analysis.
Fig. 2. Solution structures of mature AVR-Pia and AVR1-CO39. 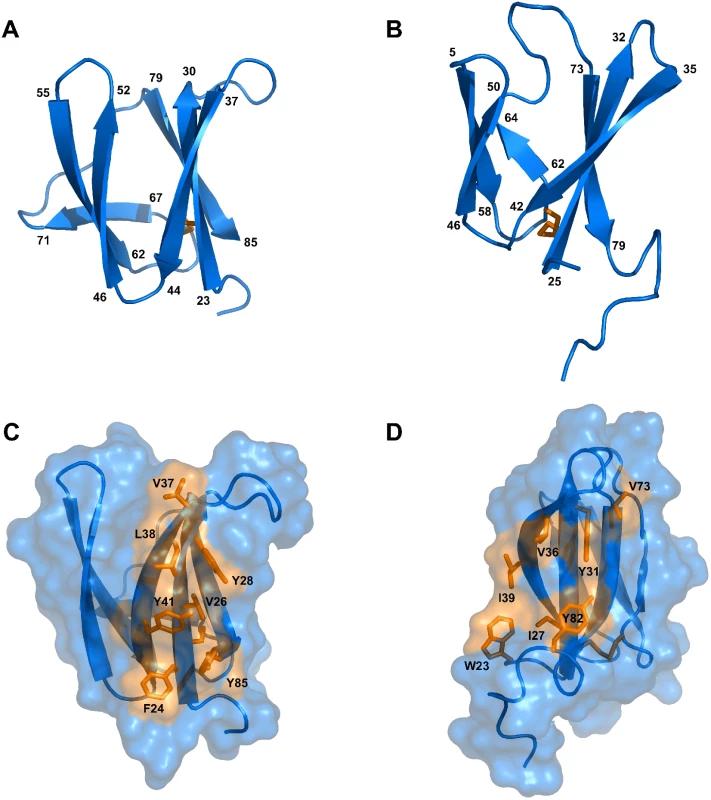
Cartoon representations of AVR-Pia (A) and (B) AVR1-CO39 highlight the similar β-sandwich structure of both proteins. Yellow sticks represent disulfide bonds. Numbers indicate the residues at β-strands borders. A surface view reveals extended hydrophobic patches on one of the surfaces of AVR-Pia (C) and AVR1-CO39 (D) that are composed of exposed hydrophobic residues labelled in yellow. The Figs were generated using PyMOL (http://www.pymol.org). Tab. 1. Statistics for 20 NMR structures of AVR-Pia and AVR1-CO39. 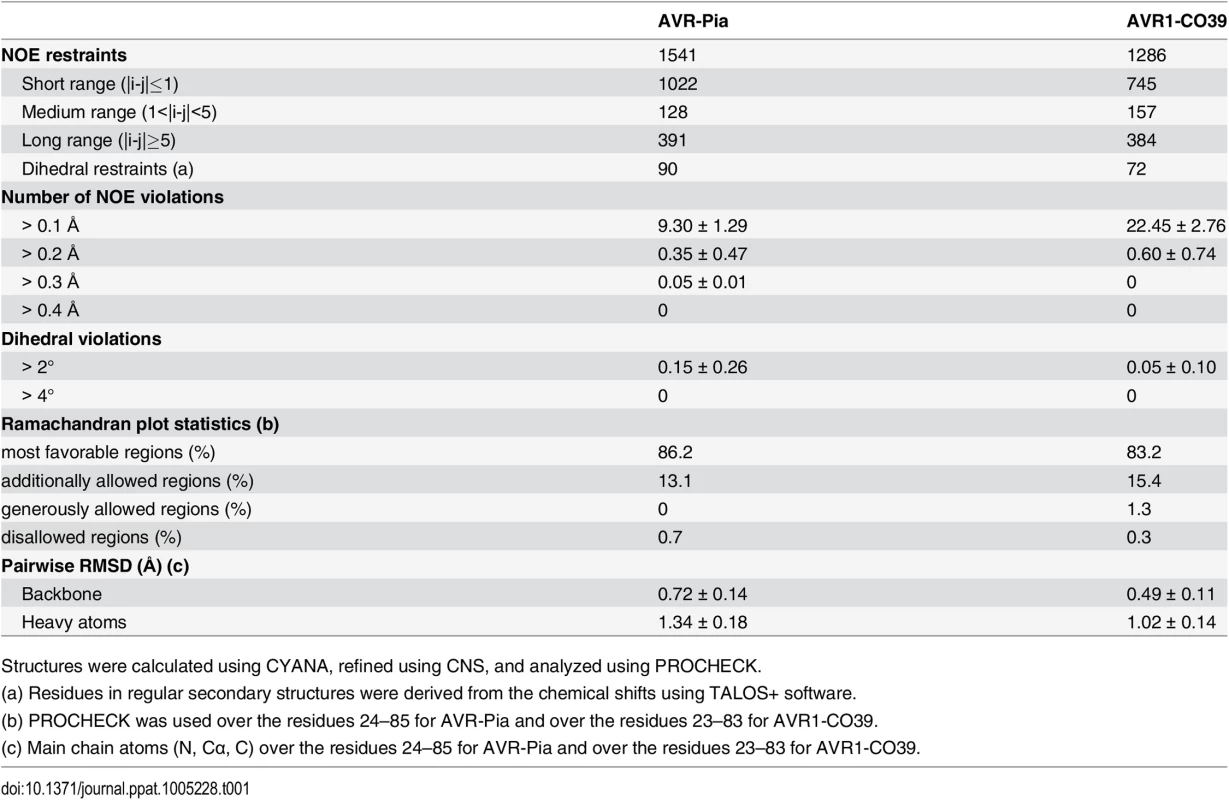
Structures were calculated using CYANA, refined using CNS, and analyzed using PROCHECK. Surprisingly, both AVR-Pia and AVR1-CO39 proved to possess the same secondary structure elements arranged with the same topology in similar three-dimensional structures (Fig 2). Both proteins are composed of 6 β-strands that form two antiparallel β-sheets packed face-to-face and connected by loops (Fig 2). The first sheet is formed by the three β-strands β1, β2 and β6 while the second sheet contains β3, β4 and β5. In both cases, the two β-sheets pack together by an internal core of hydrophobic residues and one disulfide bridge and the structures belong to the β-sandwich classification. In both Avrs, the β-strands overlay and are similarly oriented (vide infra) but loops differ in length and structure.
AVR-Pia and AVR1-CO39 possess a hydrophobic surface patch
The surface properties of AVR-Pia and AVR1-CO39 are different with the exception of a hydrophobic patch located in both proteins on the side of the β-sandwich that is formed by the first β-sheet (β1-β2-β6) (Fig 2C and 2D). In AVR-Pia, this solvent exposed hydrophobic surface is constituted by the residues F24, V26 and Y28 in β1, V37, L38 and Y41 in β2, and Y85 in β6, and has an area of 372 Å2. In AVR1-CO39, the solvent exposed hydrophobic surface of the first β-sheet is formed by the residues I27 and Y31 in β1, V36 and I39 in β2 and V73 in β6, as well as W23 from the N-terminus and Y82 from the C-terminus, and has a surface area of 280 Å2.
ToxB and AvrPiz-t are structural homologs of AVR1-CO39 and AVR-Pia
To identify structural homologs of AVR-Pia and AVR1-CO39, structural similarity searches were performed using the Dali server and the Protein Data Bank [45]. Both queries, with AVR1-CO39 and AVR-Pia, identified the secreted effector protein ToxB from the wheat tan spot pathogen Pyrenophora tritici-repentis as well as its natural allele Toxb as the closest structural homologs with the highest Z-scores (S2 Table and Fig 3) [43]. Like, AVR-Pia and AVR1-CO39, ToxB is secreted during infection and is an important determinant of virulence for the tan spot fungus [46]. In addition, the search with AVR1-CO39 identified AvrPiz-t, another avirulence effector of M. oryzae that is sequence-unrelated to AVR-Pia and AVR1-CO39 but structurally similar [42]. A pairwise similarity matrix using root-mean-square deviation (rmsd, measured in Å) and DALI Z-scores [45] was established revealing that all proteins are structurally related and that ToxB is closer to all other three structures than the others among them (S2 Table). ToxB and AvrPiz-t are like AVR-Pia and AVR1-CO39, composed of two three-stranded antiparallel β-sheets, β1-β2-β6 and β3-β4-β5, forming a six β-sandwich (Fig 3A–3D). Structure-based sequence alignments provided by DALI revealed, at a first glance, no obvious conservation, but also no clear consensus except buried hydrophobic residues alternating with exposed polar amino acids in the β-strands (Fig 3E). The β-strands β1 and β2 are very similar in length and position in all four proteins, while β3, β4 and β6 display more variation. β5 is the shortest and the most irregular strand. As expected for β-strands, buried and exposed residues alternate, with the exception of β1 where residues have a tendency to be more buried. This is due to the packing of β1 in between the β2 and β6 strands. The loops connecting the β-strands have variable length, and are the sites where most of the residue insertions occur. The disulfide bond between C2 and C43 (ToxB numbering) is well conserved but shifted “in phase” by two residues in AVR-Pia (Fig 3E).
Fig. 3. AVR-Pia, AVR1-CO39, AvrPiz-t and ToxB have similar 6 β-sandwich structures. 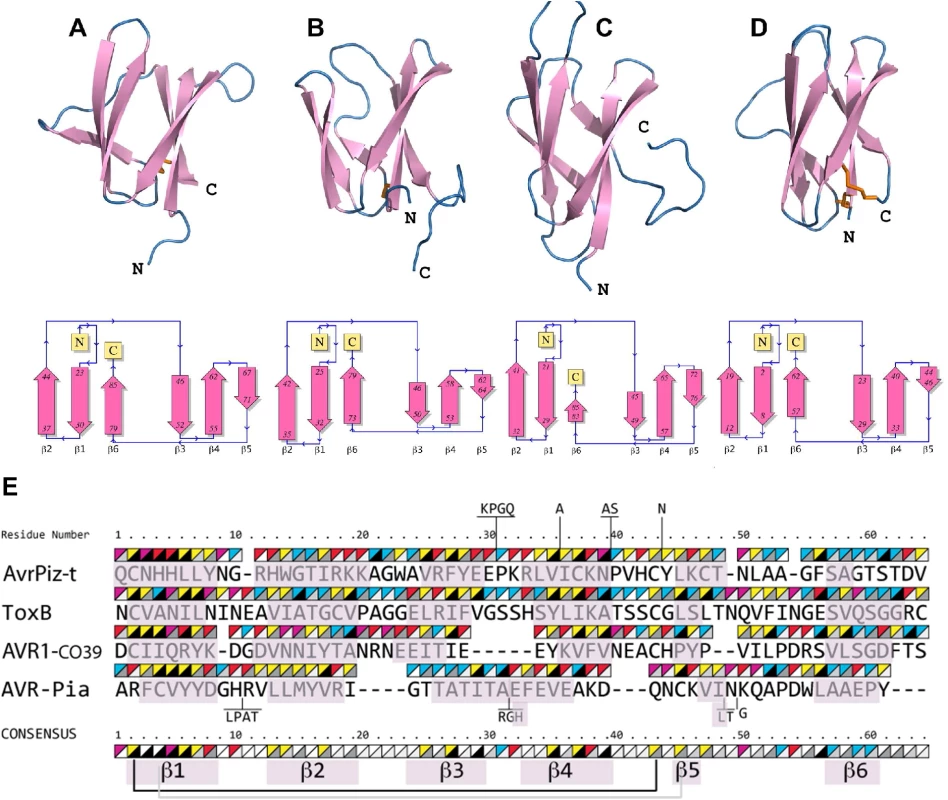
Topology diagrams (lower row) show that AVR-Pia (A), AVR1-CO39 (B), AvrPiz-t (C) and ToxB (D) possess the same fold. Ribbon diagrams (upper row, generated with PyMOL (http://www.pymol.org)) highlight similarities of their structures. Disulfide bonds are shown in the ribbon diagrams by orange sticks. All four structures were superimposed and a structural alignment was derived using DALI with the ToxB sequence as the reference for numbering (E). Residues not aligned to ToxB are connected by vertical lines and correspond to insertions in loops of AvrPiz-t and AVR-Pia. Triangles over the residues indicate chemical properties (upper-left triangle: yellow for hydrophobic, red for charged, pink for Asn and Gln and blue for other residues) and solvent accessibility (lower-right triangle: from black for buried to white for solvent-exposed). The consensus is defined by at least three similar residues per position. Residues forming β-strands are pink. Disulfide bridges in AVR1-CO39 and ToxB are shown below the consensus by a black line and for AVR-Pia by a grey line. For AvrPiz-t, no disulfide bridge was reported despite presence of the two conserved cysteins [42]. Psi-Blast searches identify in M. oryzae and M. grisea multiple effector candidates with similarities to Magnaporthe Avrs and ToxB
The unexpected finding, that all three M. oryzae effectors that have been characterized for their structure so far and one effector from an only very distantly related fungal group are structurally related raised the possibility that these four effectors are members of a widely distributed and abundant fungal effector family characterized by a common β-sandwich structure and high sequence divergence. Simple Blast searches are not suited to identify such distantly related proteins and when performed with the protein sequence of effectors from ascomycete fungi, generally identify no or only very few conserved homologs in the same species. Therefore, more sensitive Psi-Blast searches that use position-specific scoring matrices were performed with AVR-Pia, AVR1-CO39, AvrPiz-t and ToxB. The searches were performed on a protein sequence database combining the protein sequences of the M. oryzae reference isolate 70–15, of 5 other rice-infecting M. oryzae isolates (TH16, TH12, PH14, FR13 and Guy11), three M. oryzae isolates with other host specificities (BR32, US71 and CD156 specific for wheat, Setaria italica and Eleusine coracana) and one isolate of the sister species M. grisea (BR29). These additional M. oryzae and M. grisea protein sequences were obtained by whole genome re-sequencing and de novo annotation of proteins and are accessible at http://genome.jouy.inra.fr/gemo [47]). After 4 Psi-Blast iterations and filtering of the results for sequences having an alignment length of at least 40 residues, an overall protein size of less than 180 amino acids and the presence of a predicted signal peptide, 3, 8 and 4 homologs of AVR-Pia, 16, 25 and 16 homologs of AVR1-CO39 and 5, 9 and 6 homologs of ToxB were detected in respectively 70–15, TH16 and BR29 (S3 Table, orthologous sequences present in 70–15 and TH16 were only counted for 70–15). For the other M. oryzae isolates similar numbers of homologs as in TH16 were found. The elevated number of homologs present in these isolates but not in 70–15 are due to the fact that the pipeline used for protein annotation in the re-sequenced genomes identified many additional small secreted proteins that are not annotated in 70–15 although the corresponding coding sequences are present in its genome [47]). The similarities were weak (frequently less than 25% identity) but they were consistent with the structural alignment (Fig 3) and included the two cysteine residues. For AvrPiz-t, no homologs that were not already identified by standard Blast were identified in the Psi-Blast search. When 25 additional fungal genomes, including the closely related fungi M. poae and Gaeumannomyces graminis were added to the database for the Psi-Blast searches, only very limited numbers of homologs (0, 1 or 2) with frequently low e-value scores were identified in other fungi. This suggested that effectors with similaritiy to Magnaporthe Avrs and ToxB named in the following MAX-effectors that potentially also have 6 β-sandwich structure are present with low frequency in other fungal pathogens but were strongly amplified and diversified in M. oryzae and M. grisea that both belong to the genus Pyricularia in the Pyriculariae family [48].
HMM searches identify a huge MAX-effector family in M. oryzae and M. grisea
To exclude that the Psi-Blast search missed MAX-effectors in the additional fungal genomes due to biases in the search matrix or too low sensitivity and to deepen the search for this class of effectors in M. oryzae and M. grisea genomes, a hidden Markov model (HMM)-based profile search was performed. This type of profile search is among the most powerful procedure for detecting with high accuracy remote homologies between proteins.
As a first step, a high stringency Blast search with the three M. oryzae effectors and a Psi-Blast search with ToxB was performed and the resulting set of closely related sequences was aligned in a multiple sequence alignment constrained by the structural alignment of AVR-Pia, AVR1-CO39, AvrPiz-t and ToxB (S5A Fig). For the M. oryzae effectors, the Blast search identified orthologs of the effectors with few polymorphisms in different M. oryzae isolates. In addition, for each M. oryzae effector, one paralog was identified in M. oryzae and one or two paralogs were identified in the M. grisea isolate BR29 (S5B Fig). For the M. oryzae paralogs, generally several different alleles were identified. For ToxB, in addition to highly homologous sequences from P. tritici-repentis and P. bromi, 1 homolog was identified in M. oryzae, Bipolaris oryzae and Colletotrichum higginsianum, 2 in C. fioriniae, 3 in C. orbiculare and 4 in C. gloeosporioides. (S5B Fig).
As a second step, an HMM profile was built, starting from the structure-guided multiple sequence alignment from step1 (S5A Fig) and by iteratively searching for homologs in a database containing the small secreted proteins (<170 amino acids) of 25 pathogenic and non-pathogenic ascomycete fungi and of the 9 re-sequenced M. oryzae and M. grisea isolates from which completely redundant sequences had been removed. At each iteration, the recovered sequences were filtered for alignment of the two cysteins with a spacing of 34 to 49 amino acids and used to generate a new profile used in the next iteration. The interval of 34 to 49 amino acids was fixed, based on the frequencies of cystein spacings in HMM searches run without this constraint.
This search recovered 161 new, more distantly related sequences of which 154 were from M. oryzae or M. grisea, 5 from 3 different Colletotrichum species, 1 from Lepthosphaeria maculans and 1 from Mycosphaerella graminicola (recently renamed Zymoseptoria tritici) (S6A Fig). This suggests that MAX-effectors have been massively and specifically expanded in M. oryzae and M. grisea. However, it also indicates their presence in other fungal species, i. e. in Colletrichum spp. where they seem to occur at elevated frequencies. The alignment and clustering of the set of 200 sequences combining the 39 sequences used for the initial profile and the 161 new sequences revealed clusters of orthologous sequences originating from the different M. oryzae isolates with weak sequence polymorphism between orthologs (S6A and S6B Fig). Frequently, orthologs of M. oryzae can be identified in M. grisea but never in other fungi. Sequences from different orthologous clusters have high sequence diversity. Only in 3 cases, statistically significant clusters, supported by bootstrap values bigger than 50% can be identified that contain 2 distantly related MAX-effectors or MAX-effector clusters of M. oryzae.
A sequence logo derived from the multiple alignment shows the invariant cysteine residues (position 2 and 43 in mature ToxB) that constitute the alignment framework, as well as additional positions that are specifically enriched (Fig 4A). There is a propensity for hydrophobic residues in positions 4 and 6, corresponding to hydrophobic positions in strand β1, in position 27, corresponding to a hydrophobic residue in β3 and in positions 35, 37 and 39 corresponding to β4. Positions 10, 23, 40 and 49 are in loop regions between the pairs of strands β1-β2, β2-β3, β4-β5 and β5-β6 respectively, and are enriched in glycine, polar or charged residues.
Fig. 4. Large numbers of MAX-effectors sharing a characteristic sequence pattern are present in M. oryzae and M. grisea. 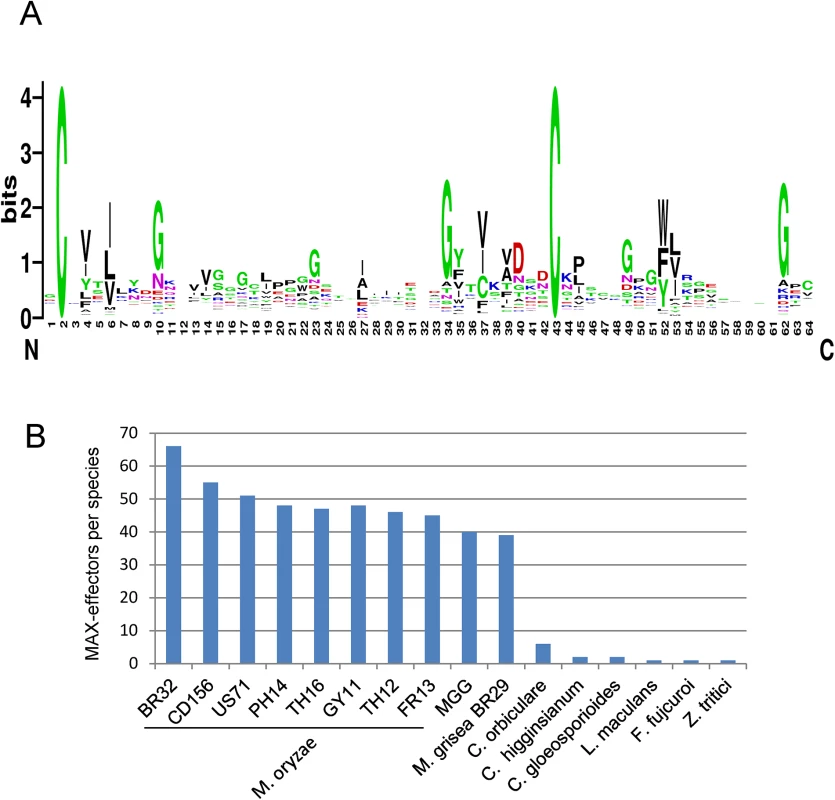
A) Sequence pattern of MAX-effectors. The sequence logo was generated using the alignment of MAX-effector candidates identified by a high stringency HMM search (S6 Fig). (B) Numbers of MAX-effector candidates detected by a low stringency HMM sequence pattern search. A database combining 25 pathogenic and non-pathogenic ascomycete fungi and 9 M. oryzae and M. grisea isolates was searched with an HMM pattern based on a structural alignment of AVR-Pia, AVR1-CO39, AVR-Pia and AvrPiz-t. The resulting HMM profile was used to search with a relaxed cut-off two different databases: (i) the UniRef90 database that contains non-redundant sequences from a wide range of different organisms and that was used to determine in which type of organisms proteins with the MAX-effector motif occur and to evaluate by this the specificity of the motif and (ii) the previously described fungal genomes and M. oryzae and M. grisea database to get a precise view of the occurrence of MAX-effectors in a broad range of ascomycete fungi.
The search of the UniRef90 database recovered 70 sequences. All but 3 were from phytopathogenic ascomycete fungi (S7A Fig). The exceptions were from a bacteria, Pseudomonas sp. StFLB209, living in association with plants, from tomato (Solanum lycopersicum) and from a nematode-parasitic fungus (Arthrobotrys oligospora) and had low e-values. Among the fungal sequences, 49 were from M. oryzae and included AVR1-CO39 and AVR-Pia. The remaining 18 corresponded to previously identified effectors from Colletotrichum species (5 C. orbiculare, 2 C. higgensianum, 3 C. gloeosporioides, 2 C. fioriniae) that belong as M. oryzae to the class of Sordariomycetes and Z. tritici, L. maculans and B. oryzae as well as ToxB from P. tritici-repentis and P. bromi that are all from the class of Dothideomycete fungi. Clustering of the sequences revealed high sequence diversity and, apart from the Tox-B cluster, no or extremely limited relatedness could be identified (S7A and S7B Fig). Interestingly, with slightly different settings, this search also recovered the well characterized AVR-Pik effector from M. oryzae [26]. AVR-Pik clearly fits the MAX-effector pattern but was discarded in the other searches since its secretion signal is not recognized by the SignalP4.1 program used for filtering of the results.
The search of the previously described Magnaporthe and other fungal genomes database not filtered for redundancy recovered only limited numbers of MAX-effectors in non-Magnaporthe fungal genomes that had, with the exception of one effector from Fusarium fujicuroi, already been retrieved in the other searches (Fig 4B and S8A Fig). In M. oryzae, between 67 and 38 MAX-effectors per isolate were identified while in M. grisea, 37 MAX-effectors were identified (Fig 4B). 46 of the 55 MAX-effectors identified by Psi-Blast in M. oryzae 70–15 and TH16 and in M. grisea BR29 (S3 Table) were also found by this HMM search. Alignment and clustering shows that the M. oryzae MAX-effectors are generally present in the majority of M. oryzae isolates and are grouped in clusters of orthologs (S8A and S8B Fig). Many of these orthologous clusters also contain an ortholog from the M. grisea isolate BR29 that shows however higher sequence divergence. Only six statistically significant clusters (bootstrap > 50%) that contain more distantly related M. oryzae effectors from different orthologous groups are identified. Otherwise, the sequence diversity between proteins from different M. oryzae ortholog clusters is so strong that classical tree building methods do not detect statistically significant sequence relatedness. The non-Magnaporthe MAX-effectors do not cluster significantly with Magnaporthe MAX-effectors and 8 of the 10 Colletotrichum effectors are comprised in three different Colletotrichum-specific clusters.
Taken together, the different HMM searches reveal that the MAX-effector motif is specific for effectors from phytopathogenic ascomycete fungi. MAX-effectors are identified with low frequencies in phytopthogenic ascomycete fungi from the class of Dothideomycetes and seem to have expanded moderately in different Colletotrichum species (i.e. Colletotrichum orbiculare). Only in M. oryzae and M. grisea, MAX-effectors expanded and diversified massively to become a dominating family of virulence effectors in these pathogens.
Expression profiling shows that a majority of MAX-effectors is expressed specifically during biotrophic infection
To test if the M. oryzae MAX-effectors identified by the HMM profile search could be involved in plant infection, the expression of 50 different candidate MAX-effector-coding genes was analyzed by qRT-PCR in infected rice leafs and in in vitro grown mycelium (S4 Table). 30 genes showed early infection-specific expression with a majority of profiles (25) that strongly resemble the biotrophy effector marker gene BAS3 (Fig 5 and S9A and S9D Fig) [23]. The expression pattern of all these genes and of 3 genes coding for MAX-effectors identified only by Psi-Blast searches was clearly different from the markers of very early or late infection (Orf3 and MGG01147, respectively). For 18 genes, no significant expression was detected and only 2 genes were expressed constitutively with significant expression in the mycelium (Fig 5 and S9C and S9D Fig). Therefore, the majority of the MAX-effector candidates seems specifically expressed during biotrophic infection and can therefore be considered as potential virulence effectors.
Fig. 5. The majority of M. oryzae MAX-effectors is expressed specifically during biotrophic infection. 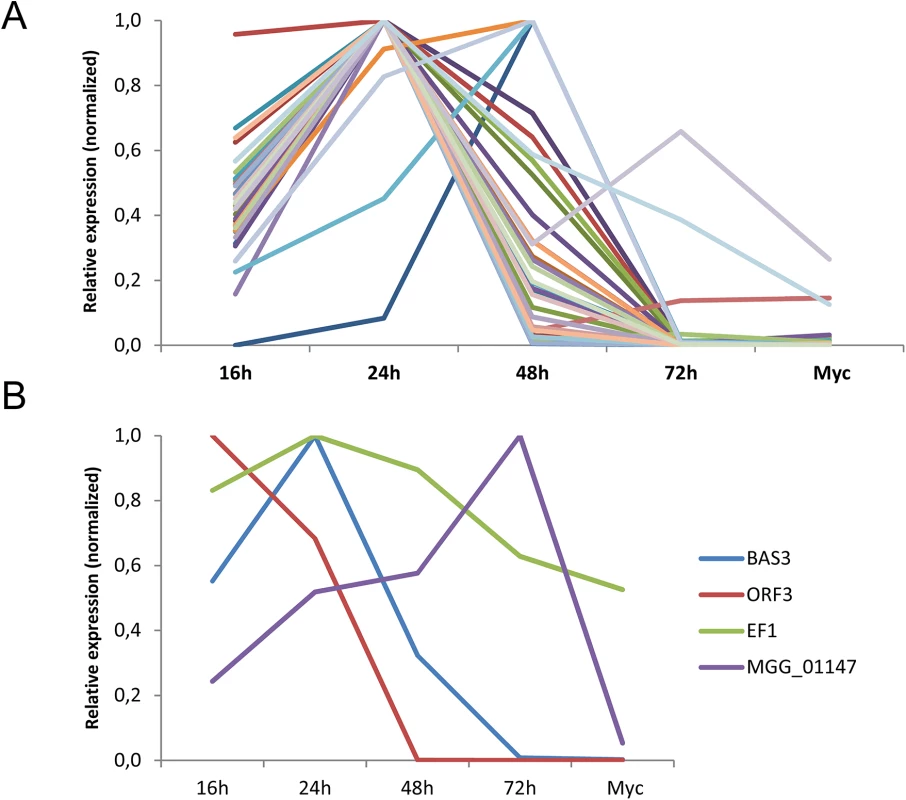
mRNA levels of M. oryzae genes coding for 32 different MAX-effectors (A) and marker genes (B) for appressorium formation and very early infection (ORF3 of the ACE1 cluster, MGG_08381), biotrophic infection (BAS3, MGG_11610), late infection (MGG_01147) and constitutive expression (EF1α, MGG_03641) were determined by q-RT-PCR in rice leaf samples harvested 16, 24, 48 and 72 h after inoculation and mycelium grown in vitro. Relative expression levels were calculated by using expression of a constitutively expressed Actin gene (MGG_03982) as a reference and normalized with respect to the highest expression value. Values are means calculated from the relative expression values of three independent biological samples. Individual expression profiles are in S9 Fig. Discussion
In this study, we have determined by NMR spectroscopy the 3-dimensional structures of two different effectors of M. oryzae, AVR1-CO39 and AVR-Pia. Although the two proteins have no evident sequence similarity they possess similar 6 β-sandwich structures formed in both cases by two β-sheets each formed of three β-strands oriented in an antiparallel manner. Interestingly, similar β-sandwich structures have previously been found for AvrPiz-t, the only other structurally characterized M. oryzae effector and for ToxB, an effector from an only very distantly related plant pathogenic ascomycete fungus, P. tritici-repentis. Overlay of the structures and structural alignments revealed that the nature and number of secondary structural elements and the topology of their fold are the same in all four effectors. In addition, all four proteins are stabilized by buried hydrophobic residues coming for their majority from the β-strands and by a disulfide bond between conserved cysteins located in the beginning of β1 and in the beginning or just before β5. However, the orientation and the length of certain β-strands, i.e. β-5, vary considerably and the sequences and the length of loops are highly variable resulting in proteins with very different shapes and surface properties. Due to the high sequence diversity, similarity among the MAX-effectors is therefore only detected when their structure is taken into consideration.
Hydrophic surface patches in AVR-Pia and AVR1-CO39 are potential sites of protein-protein interaction
The only similarity of the surfaces of AVR1-CO39 and AVR-Pia is an extended hydrophobic area on the surface formed by β1, β2 and β6. Such extended and exposed hydrophobic areas are uncommon since protein surfaces are generally in contact with solvent water molecules and they are frequently involved in protein-protein interactions. Previous studies on the recognition of AVR-Pia by the rice NLR immune receptor RGA5 support that the hydrophobic surface of AVR-Pia could indeed be involved in protein binding [37]. AVR-Pia binds physically to a C-terminal domain of RGA5 homologous to heavy metal-associated (HMA) domain proteins related to the copper chaperone ATX1 from Saccharomyces cerevisiae (RATX1 domain). This binding is required to derepress a second NLR RGA4 that activates resistance signaling [49]. A natural allele of AVR-Pia (AVR-Pi-H3) where the surface exposed phenylalanine 24 and threonine 46 situated respectively in and at the border of the hydrophobic patch are replaced by serine and asparagine loses binding to RGA5RATX1 and does not trigger resistance [37]. Structural information will now guide further functional studies to elucidate if other amino acids situated in or at the border of the hydrophobic patch are also involved in RGA5RATX1-binding and to validate by this the role of the hydrophobic patch as a protein-protein interaction surface.
MAX-effectors have different molecular properties and activities
Common features of the M. oryzae MAX-effectors are that they act intracellular in host cells [21,24,32] and are recognized by NLR immune receptors in resistant rice genotypes: AVR1-CO39 and AVR-Pia by the same NLR pair RGA4/RGA5 and AvrPiz-t by the NLR immune receptor Piz-t [37,39,41]. While the molecular bases of the recognition of AVR1-CO39 and AVR-Pia by RGA4/RGA5 are beginning to be elucidated, details of AvrPiz-t recognition are not known. Also, whether the three M. oryzae MAX-effectors target similar host processes and host proteins is not known. AvrPiz-t was described to target the host ubiquitin proteasome system by binding and inactivating the RING E3 ubiquitin ligase APIP6 [21] but virulence targets of AVR-Pia and AVR1-CO39 have not been described. However, it has been hypothesized that both proteins target RATX1 proteins homologous to the RGA5RATX1 domain that was suggested to act as a mimic for AVR-Pia and AVR1-CO39 targets [50]. Therefore, we assume that AvrPiz-t on the one hand and AVR-Pia and AVR1-CO39 on the other have different molecular activities and target different host proteins. This would be in accordance with the high divergence of their shapes and their surface properties. That AVR-Pia and AVR1-CO39 interact with the same immune receptor by binding to the same sensor domain and potentially interact with the same host targets is striking because apart from the extended hydrophobic patch on the β1β2β6 surface they share no apparent similarities with respect to their shapes and surfaces. It will therefore be important to elucidate in the future which amino acids of AVR-Pia and AVR1-CO39 bind to RGA5RATX1 and which surfaces of RGA5RATX1 are involved in binding to each of the two effectors to better understand specificity in effector recognition. In addition, identification of AVR1-CO39 and AVR-Pia targets as well as ToxB targets for which molecular details of activity are also lacking will be important to understand how MAX-effectors promote virulence and to understand the link between MAX-effector structure and function.
MAX-effectors are a highly diversified effector family specific to phytopathogenic ascomycete fungi and underwent expansion in M. oryzae and M. grisea
Structure-informed pattern searches identified huge numbers of MAX-effector candidates that possess as the structurally characterized MAX-effectors very high sequence diversity and probably also possess a 6 β-sandwich structure stabilized by buried hydrophobic residues from β-strands and a disulfide bond between conserved cysteins connecting β1 and β5. Systematic prediction of the secondary structure of the MAX effector candidates using SSPRO 5 software identified with high frequency two β-strands, β1 located after the first cysteine and β4 located before the second cysteine (S10 Fig). The other regions of the sequences had more variable secondary structure predictions which is also reflected by a less defined pattern in these regions (Fig 4A). High sequence diversity among MAX-effector candidates could as in the case of the structurally characterized MAX-effector be the consequence of interchangeability of buried hydrophobic core residues, variation in the lengths of some β-strands (i.e. β5), exchange of surface exposed residues and deletion or insertion of residues in exposed loops.
MAX-effectors were specifically detected in phytopathogenic ascomycetes from the classes of Sordariomycetes and Dothideomycetes. One MAX-effector per species was detected in phytopathogenic fungi of the class of Dothideomycetes (L. maculans, P. tritici-repentis, Z. tritici and B. oryzae) and higher numbers (2–6) occur in fungi from the genus Colletotrichum. Only in M. oryzae and M. grisea that are both from the genus Pyricularia huge numbers of MAX-effector candidates were detected and expression profiling confirmed that most of them are likely bona fide effectors expressed specifically during biotrophic early infection. With 40–60 effectors which represents 5–10% of the candidate effectors of individual M. oryzae or M. grisea isolates, MAX-effectors can be considered a dominant class of effectors in these fungi [24,47]. This is further supported by the finding that 5 of the 51 biotrophy-associated proteins identified by transcriptome analysis are MAX effectors (MG02546, MG08414, MG08482, MG09425 and MG09675) [23]. Also, the M. oryzae effector AVR-Pik fits the MAX-effector pattern further highlighting the outstanding importance of this effector family that comprises 4 out of 8 cloned Avr effectors in the blast fungus [6]. It is striking that the only other group of fungi with elevated numbers of MAX effectors are Colletotrichum species. Colletotrichum fungi are phylogenetically only distantly related to M. oryzae and M grisea but employ a similar hemibiotrophic infection strategy characterized by appressorium-mediated penetration into the host and growth inside invaded plant cells during biotrophic infection. It will be interesting to determine in the future whether MAX effectors play similar roles in these early infection processes in both groups of fungi.
In Gaeumannomyces graminis and M. poae that belong to the closely related Magnaporthaceae family no MAX-effectors were detected [48]. The expansion of MAX-effectors therefore occurred probably in a common ancestor of M. oryzae and M. grisea since clear orthologous relations can be established between many MAX-effectors from M. oryzae and M. grisea but after the split of the Magnaporthaceae. Expansion and diversification of the MAX-effectors is clearly continuing since frequently orthologs in M. oryzae or M. grisea cannot be identified and duplication, loss and diversification of MAX-effectors in host specific lineages of M. oryzae is observed (S8B Fig). Genome sequencing of additional species from Pyricularia and other genera in the Periculariae will allow to further strengthen the hypothesis of lineage-specific expansion of MAX-effectors.
Lineage specific expansion of effector families has been observed in other fungi such as mildew and rust fungi whose effector repertoires are dominated by effector families that contain frequently numerous members and are for their majority restricted to individual species or precise clades [10,51]. However, in these cases, sequence divergence is not as strong as in MAX-effectors since sequence-based comparisons allow the establishment to these effector families.
On the contrary, the effector repertoires of ascomycete phytopathogens outside the mildew lineage contain hundreds of sequence-unrelated effectors and the evolutionary origin of these huge amounts of species or clade specific genes is an open question. Duplication and diversification eventually driven by localization of the genes in transposon rich regions, genome reshuffling or transfer of accessory chromosomes were convincingly proposed as potential mechanisms to create effector diversity but the apparent lack of relatedness of ascomycete effectors remains unexplained [52–55]. Establishment of a huge effector family in M. oryzae and M. grisea that is also present at much lower frequency in other ascomycete pathogens sheds new light on the origin and relatedness of ascomycete effectors.
Diversifying rather than convergent evolution leads to highly diversified effector families
Theoretically, convergent evolution as well as diversifying evolution can explain the situation observed for the MAX-effectors characterized by a broad and patchy distribution, high diversification and limited sequence homology as well as a shared sequence pattern and probably the same structure. Convergent evolution would apply if these proteins with similar functions and a similar fold appeared repeatedly in phytopathogenic ascomycetes and eventually evolved independently in different clades. Under diversifying evolution, a protein or protein family present in a common ancestor has been strongly diversified in different lineages of ascomycete fungi and frequently lost during evolution in certain lineages and species. The scenario of convergent evolution of MAX-effectors cannot be excluded but is clearly less parsimonious. It raises the question why MAX-effectors do not occur in organisms with similar lifestyles outside the Sordariomycete and Dothideomycete pathogens such as phytopathogenic basidiomycetes or oomycetes. In addition, there are no well-documented examples of convergent evolution towards similar folds or sequence patterns for pathogenic effectors or secreted fungal proteins involved in adaption to the environment while comparative genomics studies in fungi and oomycetes are beginning to identify such widely distributed gene families that are shaped by strong diversifying selection and that can only be properly reconstructed when pattern-based searches and structure information are taken into consideration. The best documented example is certainly the WY-domain family among the RXLR effectors that is specific to the Peronosporales clade in oomycetes and evolves by diversifying evolution [8,9,56–58]. Careful sequence analysis involving pattern searches identified the W, Y and L sequence motifs in the effector domains of a majority of the Phytophtora RXLR effectors that are frequently completely sequence unrelated [9]. Functional analysis confirmed the importance of these motifs for effector function [59] and structure analysis of the effector domain of different RXLR effectors with limited sequence homology revealed that conserved sequence motifs reflected a conserved, highly similar 3-dimensional structure named the WY-domain fold [56,60–62]. PexRD2 and AVR3a11 show e.g. only 14% amino acid identity in a structure-based alignment but overlay of their structures has an RMSD score of 0.73 Å. As in the case of the β-sandwich fold of the MAX-effectors, the WY-domain fold tolerates insertion or deletion of amino acids in the loops, exchange of surface exposed amino acids and is stabilized by hydrophobic core residues that can be exchanged as long as hydrophobicity is maintained [56]. This flexible structure allows to generate effectors with highly variable shapes and surface properties and studied WY-domain effectors showed very diverse molecular activities, target different host proteins and are recognized by different NLR immune receptors [7,56].
An example of rapidly evolving proteins from fungi that are structurally but not sequence-conserved are hydrophobins that are low molecular mass secreted proteins important for the impermeabilization of fungal cell walls, adhesion to hydrophobic surfaces and pathogenicity [63]. Hydrophobins were shown to evolve rapidly according to a birth-and-death mechanism [64], are widely distributed in a broad range of basidio - and ascomycete fungi and are characterized by sequence patterns but no sequence homology [63,65]. Structure analysis demonstrated that distantly related hydrophobins are structurally related supporting a common evolutionary origin [66].
Another example of a fungal gene family that is rapidly evolving according to a birth-and-death model are the Hce2 proteins (homologs of Cladosporium fulvum ECP2) that are present in a wide range of basidio and ascomycete fungi and seem to act as effectors in pathogenic fungi and potentially in stress responses in non-pathogenic fungi [67]. Much like MAX-effectors they show patchy distribution, lineage-specific expansions and high sequence diversification.
MAX-effectors may serve as a paradigm for the evolution and diversification of effectors in phytopathogenic ascomycetes
Based on our discovery of the MAX-effector family and the widely accepted concept that fungal effectors evolve according to a birth-and-death model we propose the hypothesis that the majority of the immense number of different ascomycete effectors could in fact belong to a restricted set of structurally defined families whose members are phylogenetically related. These families of structurally conserved effectors are expected to be, as the MAX-effectors widely distributed with frequent losses on the one hand and lineage specific expansions on the other leading to effector families that are particularly important in certain fungal clades but not in others. The evolution of individual effectors is so rapid and their adaptation to new functions so profound that sequence homology and resulting phylogenetic signals are rapidly lost although the basic protein architecture may frequently be conserved because it represents a good solution to many general constraints effectors have to face such as stability in the fungus-host interface or translocation into host cytosol. Sequence homology can therefore only be detected in orthologs from closely related species but in paralogs from the same species or homologs from more distantly related species no similarity is detected on the sequence level. Only structure-informed and pattern-based searches reveal the hidden relatedness of ascomycete effectors. This hypothesis is also supported by the recent identification of an effector super family in the powdery mildew fungus Blumeria graminis fsp hordei by structural modelling [51]. 72 effectors from different families established by sequence homology or with no homology to other proteins had 3D structure models with similarity to ribonucleases suggesting a common origin and a conserved structure in this superfamily of sequence diverse effectors.
Knowledge on the structures of fungal effector proteins is extremely limited and outside of the MAX-effectors the structures of only three cytoplasmic fungal effectors have been determined. AvrL567 from the rust fungus Melampsora lini and ToxA from P. tritici-repentis have distantly related β-sandwich structures whose topologies are completely different from the MAX-effectors and AvrM has a helical structure [68–70]. Therefore, the elucidation of the 3-dimensional structures of additional fungal effectors is a priority for a better understanding of their diversity and will teach us to what extent structurally conserved but sequence-diversified effector families dominate the huge and extremely diverse effector repertoires of phytopathogenic fungi.
Methods
Protein expression and purification
The sequence for the mature protein (residues 20–85 for AVR-Pia, and residues 23–89 for AVR1-CO39) was inserted into the pET-SP vector by ligation of PCR using NdeI-BamHI sites. PCR products were generated using the forward and reverse oligos tatcatatggctGCGCCAGCTAGATTTTGCGTCTAT and tatggatccCTAGTAAGGCTCGGCAGCAAG or tatcatatGCTTGGAAAGATTGCATCATCCA and tatggatccGATCAACAAGACTCATCGTCGTCA for respectively AVR-Pia or AVR1-CO39. The pET-SP vector was constructed from pET-15b (Merck-Millipore, Darmstadt Germany) by inserting a periplasmic secretion sequence, a hexahistidine tag and a TEV cleavage site at the N-terminus of the protein adding an extra 31 amino acid sequence at the N-terminus of the recombinant proteins (sequence MKKTAIAIAVALAGFATVAQA_APQDNTSMGSSHHHHHHSSGRENLYFQGHMA). The plasmids pET-SP-AVR-Pia and pET-SP-AVR1-CO39 were used to transform E. coli BL21 (DE3).
Transformed cells were grown in an autoinducing minimal media C-750501 [71] at 37°C for 24h. To generate isotopically-labeled samples for NMR spectroscopy, we used 15NH4Cl, 13C3-glycerol and 13C6-glucose as the primary nitrogen and carbon sources. Cells were harvested by centrifugation and the pellet was resuspended in lysis buffer (200 mM TrisHCl pH8, 200mM Sucrose, 0.05mM EDTA, 50μM lysozyme). After 30 minutes incubation, cell debris were removed by centrifugation at 12 000 g for 15 min at 4°C. The resulting crude protein extracts were loaded on an AKTA basic system into a HisTrap 5ml HP columm (GE Healthcare), equilibrated in buffer A (50 mM TrisHCl, pH 8.0, 300 mM NaCl, 1 mM DTT, 0.1 mM Benzamidine). The His-tagged protein was eluted from the affinity column with buffer B (buffer A supplemented with 500 mM imidazole). Fractions containing the protein were identified by SDS-PAGE and pooled. The protein was further purified by gel filtration using a Superdex S75 26/60 (GE Healthcare) column in buffer A and pure fractions were pooled.
The elution profiles indicated that AVR-Pia and AVR1-CO39 eluted as single monomeric species (Fig 1). Ellman’s reagent, 5, 5’-dithio-bis-(2-nitrobenzoic acid), DTNB, was used for quantitating free sulfhydryl groups [72]. Briefly, aliquots of standard (cysteine, Sigma, 12.5 μM to 75 μM) or sample (50 μM) were reacted with 0.1 mM DTNB reagent in 100 mM sodium phosphate pH 8.0, 1mM EDTA buffer. Free sulfhydryl groups were also measured in denaturating conditions using the same buffer supplemented with 6M Guanidinium Chloride. Absorbance was read at 412 nm on a NanoDrop 2000, and the concentration of free thiols was determined from the standard curves.
NMR samples
The NMR samples were prepared with 1mM of purified protein at 10% D2O and 0.5 mM DSS as a reference. For AVR-Pia the purification buffer was exchanged with phosphate buffer (20 mM potassium-sodium phosphate, pH 5.4 and 150 mM NaCl), by filtrating with Centricon. The purified AVR1-CO39 proteins were dialyzed in 20 mM sodium phosphate, pH 6.8, 150 mM NaCl and 1 mM DTT. For the D2O experiments, a non-labeled sample was lyophilized and dissolved in D2O.
Nuclear magnetic resonance spectroscopy
Spectra were acquired on 500 and 700 MHz Avance Bruker spectrometers equipped with triple-resonance (1H, 15N, 13C) z-gradient cryo-probe at 305 K. Experiments were recorded using the TOPSPIN pulse sequence library (v. 2.1) (S2 Fig). 2D-NOESY experiments with excitation sculpting water suppression were acquired at 305K, with mixing times from 100 to 150 msec. All spectra are referenced to the internal reference DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) for the 1H dimension and indirectly referenced for the 15N and 13C dimensions [73].
NMR data was processed using Topspin (v. 3.2) and were analyzed using strip-plots with Cindy in house software and CCPN [74] [analysis v 2.3]. Side chain assignments were carried out using 2D-NOESY, 2D-TOCSY and COSY-DQF experiments with D2O samples, combined with 15N-NOESY-HSQC and 15N-TOCSY-HSQC 3D spectra. For AVR-Pia, the two N-terminal residues Ala-Pro and the His-tag, Ser-His6-Ser were not assigned. For AVR1-CO39, the tag-residues Asp(-7)-Asn(-8) and the stretch Ser2-His6-Ser2 were not assigned. The 15N and 13C assignments were derived from the 3D spectra at 500 MHz.
15N backbone amide NMR relaxation data
Relaxation data were acquired at 305K on a Bruker Avance 500 MHz spectrometer using R1, R2 and 15N{1H} heteronuclear NOE pulse sequences (TOPSPIN library, v 2.1). NMR samples of 500 μL at 0.85 mM and 0.3 mM were used for AVR-Pia and AVR1-CO39, respectively. R1 experiments were performed with nine relaxation delays (18, 54, 102, 198, 294, 390, 582, 774 and 966 ms). R2 experiments were carried out employing a Carr–Purcell–Meiboom–Gill (CPMG) pulse train [75,76] with eight relaxation delays (16, 32, 48, 64, 96, 128, 192 and 256 ms). A recycle delay of 2.5 s was employed in R1 and R2, experiments, and 15N decoupling during acquisition was performed using a GARP-4 sequence. In heteronuclear 15N{1H}NOEs, proton saturation was achieved during the relaxation time by application of high-power 120° pulse spaced at 20 ms intervals for 3 s prior to the first pulse on 15N [77]. A relaxation delay equal to 6 s between each scan was used. Relaxation parameters, R1, R2 and NOEs were determined from the analysis module of CCPN [74].
Structure calculation
The programs CYANA [78] and CNS [79] were used for automatic NOE assignments and structure calculations. The NH, Hα, 15N, 13Cα and 13Cβ chemical shifts were converted into Φ/Ψ dihedral angle constraints using TALOS+ (v. 1.2) [80]. The CANDID procedure of CYANA (v 2.1) was used to assign the 3D-peaks list from the 15N-NOESY-HSQC spectra. NOE assignments were inspected and used in a new CANDID assignment run including peaks from the 2D-NOESY spectra (with 100 and 150 msec mixing times for AVR-Pia and 100 and 200 msec for AVR1-CO39). A disulfide bridge Cys25-Cys66 for AVR-Pia and between Cys26-Cys61 for AVR1-CO39 was added based on cysteine Cβ chemical shifts and DTNB quantification of free thiols. NOE constraints were inspected and classified from very strong, strong, medium weak and very weak, corresponding to 2.4, 2.8, 3.6, 4.4 and 4.8 Å upper bound constraints, respectively. Final structure calculations were performed with CYANA (v. 2.1) using 1541 and 1286 distance restraints, for AVR-Pia and AVR1-CO39, with 90 and 72 Φ/Ψ dihedral angle constraints, respectively. The 30 conformers with lowest target function starting from 200 initial structures, were refined by CNS (v. 1.2) using the refinement in water of RECOORD [81]. The final 20 conformers were selected with the lowest NOE and dihedral angle violations. These are the structures discussed herein and deposited (PDBs, 2MYV and 2MYW). The final 20 structures contained no NOE violations greater than 0.3 Å and no dihedral angle constraint violations greater than 2°. Structures were validated using PROCHECK [82].
Sequence analysis
Two sequence databases were used, the UniRef90 release 2015_03 [83] and a database build from the genomes of the ascomycete fungi Magnaporthe oryzae (reference isolate 70–15), Colletotrichum graminicola, Colletotrichum higginsianum, Fusarium graminearum, Fusarium oxysporum, Gaeumannomyces graminis, Magnaporthe poae, Neurospora crassa, Pyrenophora tritici-repentis, Verticillium dahliae, Aspergillus fumigatus, Aspergillus nidulans, Blumeria graminis, Botrytis cinerea, Colletotrichum gloeosporioides, Colletotrichum orbiculare, Dothistroma septosporum, Fusarium fujikuroi, Fusarium pseudograminearum, Fusarium verticillioides, Leptosphaeria maculans, Phaeosphaeria nodorum, Pyrenophora teres, Trichoderma virens, Tuber melanosporum and Zymoseptoria tritici (all from the Ensembl Fungi database http://fungi.ensembl.org) as well as the genomes of eight M. oryzae isolates specific for Eleusine coracana (CD156), Triticum aestivum (BR32), Setaria italica (US71) and Oryza sativa (TH16, GY11, FR13, TH12, PH14) and one M. grisea isolate (BR29) pathogenic to Digitaria ssp (genome sequences at http://genome.jouy.inra.fr/gemo) [47]. Sequences without signal peptide (according to SIGNALP 4.1 [84]) bigger than 170 amino acids or with less than 2 cysteine residues were removed. For the initial HMM search, identical sequences were reduced to only one occurrence in the databases.
The start of the search was a structural alignment with TM-align [85] and the structures of AVR-Pia, AVR1-CO39, AvrPiz-t and ToxB complemented with sequence homologues found by single queries using BLAST (v 2.2.27+) with a stringent cut-off E-value = 1e-6. For the ToxB query, two iterations of NCBI PSI-BLAST were used on the NR database with a cut-off E-value = 1e-4 (S5A Fig).
This initial alignment was used as input to look for homologs in the filtered and non-redundant fungi database using HMMERsearch program from the HMMER package v 3.0 [86] with a 1e-6 E-value cut-off. For each run, only sequences where the two cysteine residues were aligned were kept, and the output alignment was used as input query for a new HMMERsearch run. This run was repeated until reaching convergence. New iterations were then done with increased E-value cut-off at 1e-5 and 1e-4. From the last alignment, a histogram indicated that the two aligned cysteine residues were separated by at least 34 and at most 49 amino acids.
The full homolog search was re-started, as described above, but this time using also the aligned cysteine separations as an additional constraint for filtering homologs after each HMMERsearch run. The HMMERsearch runs were repeated until convergence for raised threshold E-values 1e-6, 1e-5, 1e-4 and finally 1e-2. The homolog ensembles obtained for the three E-values cut-off, 1e-6, 1e-4 and 1e-2 were aligned with Muscle v3.8 [87] (S6A Fig for E-value 1e-4). The derived logo was built from the HMMER search with E-value of 1e-4 using Weblogo3 [88]. The multiple sequence alignment (MSA) derived from the HMMER search with E-value 1e-4 was used as input to look for homologs in the redundant fungi database and the UniRef90 database, using HMMERsearch with an E-value threshold of 1e-1. Diversity trees were built from alignments generated with Muscle v3.8 using the Neighbor-Joining method with the MEGA6 package [89].
Fungal growth and infection assays
For analysis of gene expression in vitro grown mycelium, M. oryzae isolate Guy11 was grown in liquid medium (glucose 10g/L, KNO3 3g/L, KH2PO4 2g/L and yeast extract 2g/L) at 120 rpm on a rotary shaker at 25°C for five days. Mycelium was harvested over a piece of cheese-cloth (Merck-Millipore, Darmstadt Germany).
For production of spores for infection assays, M. oryzae isolate Guy11 was grown on rice flour agar for spore production [90]. A suspension of fungal conidiospores was prepared at a density of 2x105 spores/ml and spotted on detached leaves of the japonica rice variety Saraceltik grown for 3 weeks as described [91]. Infected leaf samples were harvested 16, 24, 48 and 72 hours post inoculation (hpi).
RNA extraction and qRT–PCR analysis
RNA extraction and reverse transcription was performed as described [92]. Quantitative PCR were performed with a LightCycler 480 instrument (Roche, Basel, Switzerland) using LC 480 SYBR Green I Master Mix (Roche) and the primers listed in the S4 Table. Amplification was performed as follows: 95°C for 10 min; 40 cycles of 95°C for 15 s, 60°C for 20s and 72°C for 30 s; then 95°C for 5 min and 40°C for 30 s. Data were analyzed using the delta-delta Ct method and applying the formula 2-∆CT, where ∆CT is the difference in threshold cycle (CT) between the gene of interest and the housekeeping gene Actin (MGG_03982) used as a constitutively expressed reference gene. For each condition, three biological replicates were analyzed.
Supporting Information
Zdroje
1. Hogenhout S a, Van der Hoorn R a L, Terauchi R, Kamoun S. Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact. 2009;22 : 115–122. doi: 10.1094/MPMI-22-2-0115 19132864
2. Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444 : 323–9. 17108957
3. Doehlemann G, Requena N, Schaefer P, Brunner F, Connell RO, Parker JE. Reprogramming of plant cells by filamentous plant-colonizing microbes. 2014; 803–814.
4. Stergiopoulos I, de Wit PJGM. Fungal effector proteins. Annu Rev Phytopathol. 2009;47 : 233–63. doi: 10.1146/annurev.phyto.112408.132637 19400631
5. Presti L Lo, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, et al. Fungal Effectors and Plant Susceptibility. Annu Rev Plant Biol. 2015;66 : 513–545. doi: 10.1146/annurev-arplant-043014-114623 25923844
6. Giraldo MC, Valent B. Filamentous plant pathogen effectors in action. Nat Rev Microbiol. Nature Publishing Group; 2013;11 : 800–14. doi: 10.1038/nrmicro3119 24129511
7. Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. Oomycetes, effectors, and all that jazz. Curr Opin Plant Biol. Elsevier Ltd; 2012; 1–10.
8. Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461 : 393–8. doi: 10.1038/nature08358 19741609
9. Jiang RHY, Tripathy S, Govers F, Tyler BM. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci U S A. 2008;105 : 4874–4879. doi: 10.1073/pnas.0709303105 18344324
10. Duplessis S, Cuomo C a, Lin Y-C, Aerts A, Tisserant E, Veneault-Fourrey C, et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci U S A. 2011;108 : 9166–71. doi: 10.1073/pnas.1019315108 21536894
11. Pedersen C, Themaat EVL van, McGuffin LJ, Abbott JC, Burgis TA, Barton G, et al. Structure and evolution of barley powdery mildew effector candidates. BMC Genomics. 2012;13 : 694. doi: 10.1186/1471-2164-13-694 23231440
12. Spanu PD, Abbott JC, Amselem J, Burgis T a, Soanes DM, Stüber K, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330 : 1543–6. doi: 10.1126/science.1194573 21148392
13. Hacquard S, Joly DL, Lin Y-C, Tisserant E, Feau N, Delaruelle C, et al. A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust). Mol Plant Microbe Interact. 2012;25 : 279–93. doi: 10.1094/MPMI-09-11-0238 22046958
14. Dean R, Kan JANALVAN, Pretorius ZA, Hammond-kosack KIME, Pietro ADI, Spanu PD, et al. The Top 10 fungal pathogens in molecular plant pathology. 2012;13 : 414–430.
15. Gurr S, Samalova M, Fisher M. The rise and rise of emerging infectious fungi challenges food security and ecosystem health. Fungal Biol Rev. 2011;25 : 181–188.
16. Skamnioti P, Gurr SJ. Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol. 2009;27 : 141–50. doi: 10.1016/j.tibtech.2008.12.002 19187990
17. Galhano R, Talbot NJ. The biology of blast: Understanding how Magnaporthe oryzae invades rice plants. Fungal Biol Rev. Elsevier Ltd; 2011;25 : 61–67.
18. Liu J, Wang X, Mitchell T, Hu Y, Liu X, Dai L, et al. Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction. Mol Plant Pathol. 2010;11 : 419–427. doi: 10.1111/j.1364-3703.2009.00607.x 20447289
19. Mentlak TA, Talbot NJ, Kroj T. Effector Translocation and Delivery by the Rice Blast Fungus Magnaporthe oryzae. In: Francisrtin, Kamoun S, editors. Effectors in Plant–Microbe Interactions. Wiley-Blackwell; 2011. pp. 219–241.
20. Valent B, Khang CH. Recent advances in rice blast effector research. Curr Opin Plant Biol. Elsevier Ltd; 2010;13 : 434–41. doi: 10.1016/j.pbi.2010.04.012 20627803
21. Park C-H, Chen S, Shirsekar G, Zhou B, Khang CH, Songkumarn P, et al. The Magnaporthe oryzae Effector AvrPiz-t Targets the RING E3 Ubiquitin Ligase APIP6 to Suppress Pathogen-Associated Molecular Pattern-Triggered Immunity in Rice. Plant Cell. 2012.
22. Mentlak TA, Kombrink A, Shinya T, Ryder LS, Otomo I, Saitoh H, et al. Effector-mediated suppression of chitin-triggered immunity by magnaporthe oryzae is necessary for rice blast disease. Plant Cell. 2012;24 : 322–35. doi: 10.1105/tpc.111.092957 22267486
23. Mosquera G, Giraldo MC, Khang CH, Coughlan S, Valent B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as Biotrophy-associated secreted proteins in rice blast disease. Plant Cell. 2009;21 : 1273–90. doi: 10.1105/tpc.107.055228 19357089
24. Saitoh H, Fujisawa S, Mitsuoka C, Ito A, Hirabuchi A, Ikeda K, et al. Large-scale gene disruption in Magnaporthe oryzae identifies MC69, a secreted protein required for infection by monocot and dicot fungal pathogens. PLoS Pathog. 2012;8: e1002711. doi: 10.1371/journal.ppat.1002711 22589729
25. Soanes DM, Alam I, Cornell M, Wong HM, Hedeler C, Paton NW, et al. Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PLoS One. 2008;3: e2300. doi: 10.1371/journal.pone.0002300 18523684
26. Yoshida KK, Saitoh H, Fujisawa S, Kanzaki H, Matsumura H, Tosa Y, et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell. 2009;21 : 1573–91. doi: 10.1105/tpc.109.066324 19454732
27. Chen X, Coram T, Huang X, Wang M, Dolezal A. Understanding Molecular Mechanisms of Durable and Non-durable Resistance to Stripe Rust in Wheat Using a Transcriptomics Approach. Curr Genomics. 2013;14 : 111–126. doi: 10.2174/1389202911314020004 24082821
28. Kim S, Hu J, Oh Y, Park J, Choi J, Lee Y-H, et al. Combining ChIP-chip and expression profiling to model the MoCRZ1 mediated circuit for Ca/calcineurin signaling in the rice blast fungus. PLoS Pathog. 2010;6: e1000909. doi: 10.1371/journal.ppat.1000909 20502632
29. Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. Nature Publishing Group; 2010;11 : 539–48. doi: 10.1038/nrg2812 20585331
30. Miki H, Matsui K, Kito H, Otsuka K, Ashizawa T, Yasuda N, et al. Molecular cloning and characterization of the AVR-Pia locus from a Japanese field isolate of Magnaporthe oryzae. 2009;10 : 361–374.
31. Orbach MJ, Farrall L, Sweigard J a, Chumley FG, Valent B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell. 2000;12 : 2019–32. 11090206
32. Ribot C, Césari S, Abidi I, Chalvon V, Bournaud C, Vallet J, et al. The Magnaporthe oryzae effector AVR1-CO39 is translocated into rice cells independently of a fungal-derived machinery. Plant J. 2012;74 : 1–12.
33. Sweigard J a, Carroll a M, Kang S, Farrall L, Chumley FG, Valent B. Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell. 1995;7 : 1221–33. 7549480
34. Wu J, Kou Y, Bao J, Li Y, Tang M, Zhu X, et al. Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9 -mediated blast resistance in rice. 2015;
35. Li W, Wang B, Wu J, Lu G, Hu Y, Zhang X, et al. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol Plant Microbe Interact. 2009;22 : 411–20. doi: 10.1094/MPMI-22-4-0411 19271956
36. Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J, Matsumoto T, et al. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics. 2008;180 : 2267–76. doi: 10.1534/genetics.108.095034 18940787
37. Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell. 2013;25 : 1463–81. doi: 10.1105/tpc.112.107201 23548743
38. Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, et al. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics. 2006;172 : 1901–14. 16387888
39. Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, Saitoh H, et al. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011;66 : 467–79. doi: 10.1111/j.1365-313X.2011.04502.x 21251109
40. Bryan GT, Wu KS, Farrall L, Jia Y, Hershey HP, McAdams S a, et al. tA single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell. 2000;12 : 2033–46. 11090207
41. Zhou B, Qu S, Liu G, Dolan M, Sakai H, Lu G, et al. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant Microbe Interact. 2006;19 : 1216–28. 17073304
42. Zhang Z-M, Zhang X, Zhou Z-R, Hu H-Y, Liu M, Zhou B, et al. Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t. J Biomol NMR. 2013;55 : 219–23. doi: 10.1007/s10858-012-9695-5 23334361
43. Nyarko A, Singarapu KK, Figueroa M, Manning V a., Pandelova I, Wolpert TJ, et al. Solution NMR Structures of Pyrenophora tritici-repentis ToxB and Its Inactive Homolog Reveal Potential Determinants of Toxin Activity. J Biol Chem. 2014;289 : 25946–25956. doi: 10.1074/jbc.M114.569103 25063993
44. Barthe P, Ropars V, Roumestand C. DYNAMOF: a program for the dynamics analysis of relaxation data obtained at multiple magnetic fields. Comptes Rendus Chim. 2006;9 : 503–513.
45. Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38 : 545–549.
46. Ciuffetti LM, Manning V a., Pandelova I, Betts MF, Martinez JP. Host-selective toxins, Ptr ToxA and Ptr ToxB, as necrotrophic effectors in the Pyrenophora tritici-repentis-wheat interaction. New Phytol. 2010;187 : 911–919. doi: 10.1111/j.1469-8137.2010.03362.x 20646221
47. Chiapello H, Mallet L, Guérin C, Aguileta G, Amselem J, Kroj T, et al. Deciphering genome content and evolutionary realtionships of isolates from the fungus Magnaporthe oryzae attacking different hosts. Genome Biol Evol. 2015;in press.
48. Klaubauf S, Tharreau D, Fournier E, Groenewald JZ, Crous PW, de Vries RP, et al. Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae). Stud Mycol. ELSEVIER B.V; 2014;79 : 85–120. doi: 10.1016/j.simyco.2014.09.004 25492987
49. Césari S, Kanzaki H, Fujiwara T, Bernoux M, Chalvon V, Kawano Y, et al. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014;33 : 1941–1959. doi: 10.15252/embj.201487923 25024433
50. Césari S, Bernoux M, Moncuquet P, Kroj T, Dodds PN. A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front Plant Sci. 2014;5 : 606. doi: 10.3389/fpls.2014.00606 25506347
51. Pedersen C, Themaat V, Ver E, McGuffin L, Abbott JC, Burgis TA, et al. Structure and evolution of barley powdery mildew effector candidates. BMC Genomics. 2012
52. Chuma I, Isobe C, Hotta Y, Ibaragi K, Futamata N, Kusaba M, et al. Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 2011;7: e1002147. doi: 10.1371/journal.ppat.1002147 21829350
53. Rouxel T, Grandaubert J, Hane JK, Hoede C, van de Wouw AP, Couloux A, et al. Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat Commun. 2011;2 : 202. doi: 10.1038/ncomms1189 21326234
54. Ma L-J, van der Does HC, Borkovich K a, Coleman JJ, Daboussi M-J, Di Pietro A, et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010;464 : 367–73. doi: 10.1038/nature08850 20237561
55. Jonge R De, Bolton MD, Kombrink A, De Jonge R, Bolton MD, Kombrink A, et al. Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 2013;23 : 1271–1282. doi: 10.1101/gr.152660.112 23685541
56. Win J, Krasileva K V., Kamoun S, Shirasu K, Staskawicz BJ, Banfield MJ. Sequence Divergent RXLR Effectors Share a Structural Fold Conserved across Plant Pathogenic Oomycete Species. Heitman J, editor. PLoS Pathog. 2012;8: e1002400. doi: 10.1371/journal.ppat.1002400 22253591
57. Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, Kemen E, et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010;330 : 1549–1551. doi: 10.1126/science.1195203 21148394
58. Raffaele S, Farrer R a, Cano LM, Studholme DJ, MacLean D, Thines M, et al. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science. 2010;330 : 1540–3. doi: 10.1126/science.1193070 21148391
59. Dou D, Kale SD, Wang X, Chen Y, Wang Q, Wang X, et al. Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell. 2008;20 : 1118–33. doi: 10.1105/tpc.107.057067 18390593
60. Boutemy LS, King SRF, Win J, Hughes RK, Clarke T a, Blumenschein TM a, et al. Structures of Phytophthora RXLR effector proteins: a conserved but adaptable fold underpins functional diversity. J Biol Chem. 2011;286 : 35834–42. doi: 10.1074/jbc.M111.262303 21813644
61. Chou S, Krasileva K V, Holton JM, Steinbrenner AD, Alber T, Staskawicz BJ. Hyaloperonospora arabidopsidis ATR1 effector is a repeat protein with distributed recognition surfaces. Proc Natl Acad Sci U S A. 2011;108 : 13323–8. doi: 10.1073/pnas.1109791108 21788488
62. Yaeno T, Li H, Chaparro-Garcia A, Schornack S, Koshiba S, Watanabe S, et al. Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc Natl Acad Sci U S A. 2011;108 : 14682–7. doi: 10.1073/pnas.1106002108 21821794
63. Bayry J, Aimanianda V, Guijarro JI, Sunde M, Latgé JP. Hydrophobins-unique fungal proteins. PLoS Pathog. 2012;8 : 6–9.
64. Kubicek CP, Baker S, Gamauf C, Kenerley CM, Druzhinina IS. Purifying selection and birth-and-death evolution in the class II hydrophobin gene families of the ascomycete Trichoderma/Hypocrea. BMC Evol Biol. 2008;8 : 4. doi: 10.1186/1471-2148-8-4 18186925
65. Wosten H a. H YDROPHOBINS: Multipurpose Proteins. Annu Rev Microbiol. 2001;55 : 625–46. 11544369
66. Kwan a HY, Winefield RD, Sunde M, Matthews JM, Haverkamp RG, Templeton MD, et al. Structural basis for rodlet assembly in fungal hydrophobins. Proc Natl Acad Sci U S A. 2006;103 : 3621–3626. 16537446
67. Stergiopoulos I, Kourmpetis Y a I, Slot JC, Bakker FT, De Wit PJGM, Rokas A. In silico characterization and molecular evolutionary analysis of a novel superfamily of fungal effector proteins. Mol Biol Evol. 2012;29 : 3371–84. doi: 10.1093/molbev/mss143 22628532
68. Ve T, Williams SJ, Catanzariti A-M, Rafiqi M, Rahman M, Ellis JG, et al. Structures of the flax-rust effector AvrM reveal insights into the molecular basis of plant-cell entry and effector-triggered immunity. Proc Natl Acad Sci U S A. 2013
69. Wang C-IAC-I a, Guncar G, Forwood JK, Teh T, Catanzariti A-MA-M, Lawrence GJ, et al. Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell. 2007;19 : 2898–912. 17873095
70. Sarma GN, Manning VA, Ciuffetti LM, Karplus PA. Structure of Ptr ToxA: An RGD-Containing Host-Selective Toxin from Pyrenophora tritici-repentis. 2005;17 : 3190–3202.
71. Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41 : 207–234. 15915565
72. Habeeb AFSA. [37] Reaction of protein sulfhydryl groups with Ellman’s reagent. In: Hirs C. H. W. SNT, editor. Methods in Enzymology. Academic Press; 1972. pp. 457–464. doi: 10.1016/S0076-6879(72)25041-8
73. Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, et al. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6 : 135–140. 8589602
74. Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct Funct Bioinforma. 2005;59 : 687–696.
75. Carr HY, Purcell EM. Effects of Diffusion on Free Precession in Nuclear Magnetic Resonance Experiments. Phys Rev. 1954;94 : 630–632.
76. Meiboom S, Gill D. Modified Spin-Echo Method for Measuring Nuclear Relaxation Times. Rev Sci Instrum. 1958;29 : 688.
77. Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by nitrogen-15 inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28 : 8972–8979. 2690953
78. Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278 : 353–378. 15318003
79. Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2 : 2728–2733. 18007608
80. Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44 : 213–223. doi: 10.1007/s10858-009-9333-z 19548092
81. Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CAEM, Nabuurs SB, et al. RECOORD: A recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins Struct Funct Bioinforma. 2005;59 : 662–672.
82. Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231 : 1049–1067. 8515464
83. Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23 : 1282–1288. 17379688
84. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8 : 785–786. doi: 10.1038/nmeth.1701 21959131
85. Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33 : 2302–2309. 15849316
86. Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39: W29–W37. doi: 10.1093/nar/gkr367 21593126
87. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32 : 1792–1797. 15034147
88. Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14 : 1188–1190. 15173120
89. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30 : 2725–2729. doi: 10.1093/molbev/mst197 24132122
90. Berruyer R, Adreit H, Milazzo J, Gaillard S, Berger a., Dioh W, et al. Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor Appl Genet. 2003;107 : 1139–1147. 12838393
91. Faivre-Rampant O, Thomas J, Allègre M, Morel J-B, Tharreau D, Nottéghem J-L, et al. Characterization of the model system rice-Magnaporthe for the study of nonhost resistance in cereals. New Phytol. 2008;180 : 899–910. doi: 10.1111/j.1469-8137.2008.02621.x 19138233
92. Delteil A, Blein M, Faivre-rampant O, Guellim A, Estevan J, Hirsch J, et al. Building a mutant resource for the study of disease resistance in rice reveals the pivotal role of several genes involved in defence. 2012;13 : 72–82.
93. Magnan CN, Baldi P. SSpro/ACCpro 5: almost perfect prediction of protein secondary structure and relative solvent accessibility using profiles, machine learning and structural similarity. Bioinformatics. 2014;30 : 2592–2597. doi: 10.1093/bioinformatics/btu352 24860169
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized VirusČlánek Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell ProliferationČlánek Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite InterfaceČlánek Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 10- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Expression of Concern: Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Preparing for the Next Epidemic with Basic Virology
- Effectively Communicating the Uncertainties Surrounding Ebola Virus Transmission
- Translating Basic Research into Clinical Applications: Malaria Research at an NIH Lab
- A Gut Odyssey: The Impact of the Microbiota on Spore Formation and Germination
- Papillomavirus E6 Oncoproteins Take Common Structural Approaches to Solve Different Biological Problems
- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Dimensions of Horizontal Gene Transfer in Eukaryotic Microbial Pathogens
- Addressing the Complications of Ebola and Other Viral Hemorrhagic Fever Infections: Using Insights from Bacterial and Fungal Sepsis
- Time for Chocolate: Current Understanding and New Perspectives on Cacao Witches’ Broom Disease Research
- Ganglioside and Non-ganglioside Mediated Host Responses to the Mouse Polyomavirus
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Structure Elucidation of Coxsackievirus A16 in Complex with GPP3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses
- CD39 Expression Identifies Terminally Exhausted CD8 T Cells
- Abiotic Stresses Antagonize the Rice Defence Pathway through the Tyrosine-Dephosphorylation of OsMPK6
- Dissociation of Tissue Destruction and Bacterial Expansion during Bubonic Plague
- Interferon-γ: The Jekyll and Hyde of Malaria
- CCR2 Inflammatory Dendritic Cells and Translocation of Antigen by Type III Secretion Are Required for the Exceptionally Large CD8 T Cell Response to the Protective YopE Epitope during Infection
- A New Glycan-Dependent CD4-Binding Site Neutralizing Antibody Exerts Pressure on HIV-1
- The Suramin Derivative NF449 Interacts with the 5-fold Vertex of the Enterovirus A71 Capsid to Prevent Virus Attachment to PSGL-1 and Heparan Sulfate
- Trans-generational Immune Priming Protects the Eggs Only against Gram-Positive Bacteria in the Mealworm Beetle
- Peripheral Vγ9Vδ2 T Cells Are a Novel Reservoir of Latent HIV Infection
- Respiratory Syncytial Virus Disease Is Mediated by Age-Variable IL-33
- TRIM21 Promotes cGAS and RIG-I Sensing of Viral Genomes during Infection by Antibody-Opsonized Virus
- Modeling the Effects of Vorinostat Reveals both Transient and Delayed HIV Transcriptional Activation and Minimal Killing of Latently Infected Cells
- Identification of a Novel Lipoprotein Regulator of Spore Germination
- Calcium Regulation of Hemorrhagic Fever Virus Budding: Mechanistic Implications for Host-Oriented Therapeutic Intervention
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
- Comparative Life Cycle Transcriptomics Revises Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates
- The Autophagy Receptor TAX1BP1 and the Molecular Motor Myosin VI Are Required for Clearance of Salmonella Typhimurium by Autophagy
- Carcinogenic Parasite Secretes Growth Factor That Accelerates Wound Healing and Potentially Promotes Neoplasia
- Effector OspB Activates mTORC1 in a Manner That Depends on IQGAP1 and Promotes Cell Proliferation
- Dengue Virus Infection of Requires a Putative Cysteine Rich Venom Protein
- Distinct Viral and Mutational Spectrum of Endemic Burkitt Lymphoma
- Fundamental Roles of the Golgi-Associated Aspartyl Protease, ASP5, at the Host-Parasite Interface
- Phenotypic and Functional Alterations in Circulating Memory CD8 T Cells with Time after Primary Infection
- Systematic Identification of Cyclic-di-GMP Binding Proteins in Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems
- Influenza Transmission in the Mother-Infant Dyad Leads to Severe Disease, Mammary Gland Infection, and Pathogenesis by Regulating Host Responses
- Myeloid Cell Arg1 Inhibits Control of Arthritogenic Alphavirus Infection by Suppressing Antiviral T Cells
- The White-Nose Syndrome Transcriptome: Activation of Anti-fungal Host Responses in Wing Tissue of Hibernating Little Brown Myotis
- Influenza Virus Reassortment Is Enhanced by Semi-infectious Particles but Can Be Suppressed by Defective Interfering Particles
- Identification of the Mechanisms Causing Reversion to Virulence in an Attenuated SARS-CoV for the Design of a Genetically Stable Vaccine
- Differentiation-Dependent KLF4 Expression Promotes Lytic Epstein-Barr Virus Infection in Epithelial Cells
- The Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways
- C-di-GMP Regulates Motile to Sessile Transition by Modulating MshA Pili Biogenesis and Near-Surface Motility Behavior in
- Modulation of the Surface Proteome through Multiple Ubiquitylation Pathways in African Trypanosomes
- Crystal Structure of the Human Cytomegalovirus Glycoprotein B
- Depletion of . GlmU from Infected Murine Lungs Effects the Clearance of the Pathogen
- Immunologic Control of Papillomavirus Type 1
- Requires Host Rab1b for Survival in Macrophages
- Structure Analysis Uncovers a Highly Diverse but Structurally Conserved Effector Family in Phytopathogenic Fungi
- PD-L1 Expression on Retrovirus-Infected Cells Mediates Immune Escape from CD8 T Cell Killing
- Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction
- IL-4 Induced Innate CD8 T Cells Control Persistent Viral Infection
- Crystal Structures of a Piscine Betanodavirus: Mechanisms of Capsid Assembly and Viral Infection
- BCG Skin Infection Triggers IL-1R-MyD88-Dependent Migration of EpCAM CD11b Skin Dendritic cells to Draining Lymph Node During CD4+ T-Cell Priming
- Antigenic Characterization of the HCMV gH/gL/gO and Pentamer Cell Entry Complexes Reveals Binding Sites for Potently Neutralizing Human Antibodies
- Rescue of a Plant Negative-Strand RNA Virus from Cloned cDNA: Insights into Enveloped Plant Virus Movement and Morphogenesis
- Geminivirus Activates to Accelerate Cytoplasmic DCP2-Mediated mRNA Turnover and Weakens RNA Silencing in
- Disruption of Sphingolipid Biosynthesis Blocks Phagocytosis of
- The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps
- The Timing of Stimulation and IL-2 Signaling Regulate Secondary CD8 T Cell Responses
- Structural and Functional Analysis of Murine Polyomavirus Capsid Proteins Establish the Determinants of Ligand Recognition and Pathogenicity
- The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Chronobiomics: The Biological Clock as a New Principle in Host–Microbial Interactions
- Interferon-γ: The Jekyll and Hyde of Malaria
- Crosslinking of a Peritrophic Matrix Protein Protects Gut Epithelia from Bacterial Exotoxins
- Antigen-Specific Th17 Cells Are Primed by Distinct and Complementary Dendritic Cell Subsets in Oropharyngeal Candidiasis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



