-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
In both fruit flies and humans, males and females have different sets of sex chromosomes. This generates differences in gene dosage that must be compensated for by adjusting the transcriptional output of most genes located on the X-chromosome. The specific recognition and targeting of the X-chromosome is essential for such dosage compensation. In fruit flies, dosage compensation is mediated by the male-specific lethal (MSL) complex, which upregulates gene transcription on the male X-chromosome. The MSL-complex consists of five proteins and two non-coding RNAs, roX1 and roX2. While non-coding RNAs are known to be critical for dosage compensation in both flies and mammals, their precise functions remain elusive. Here we present a study on the targeting and function of the MSL-complex in the absence of roX RNAs. The results obtained suggest that the dosage compensating MSL-complex has an intrinsic tendency to target repeat-rich regions and that the function of roX RNAs is to prevent its binding to such targets. Our findings reveal an ancient targeting and regulatory function of the MSL-complex that has been adapted for use in dosage compensation and modified by the rapidly evolving noncoding roX RNAs.
Published in the journal: Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions. PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004865
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004865Summary
In both fruit flies and humans, males and females have different sets of sex chromosomes. This generates differences in gene dosage that must be compensated for by adjusting the transcriptional output of most genes located on the X-chromosome. The specific recognition and targeting of the X-chromosome is essential for such dosage compensation. In fruit flies, dosage compensation is mediated by the male-specific lethal (MSL) complex, which upregulates gene transcription on the male X-chromosome. The MSL-complex consists of five proteins and two non-coding RNAs, roX1 and roX2. While non-coding RNAs are known to be critical for dosage compensation in both flies and mammals, their precise functions remain elusive. Here we present a study on the targeting and function of the MSL-complex in the absence of roX RNAs. The results obtained suggest that the dosage compensating MSL-complex has an intrinsic tendency to target repeat-rich regions and that the function of roX RNAs is to prevent its binding to such targets. Our findings reveal an ancient targeting and regulatory function of the MSL-complex that has been adapted for use in dosage compensation and modified by the rapidly evolving noncoding roX RNAs.
Introduction
In many animal species with distinct sexes, sex-chromosomes contribute to genetic sex determination. In species with male heterogamety such as humans and fruit flies, the male sex-chromosome pair consists of two morphologically and genetically different chromosomes (one X and one Y) whereas females are homogametic, having two X chromosomes. The heteromorphic sex-chromosomes are believed to have evolved from a pair of autosomes in which a male-determining locus was acquired on one homolog to form a proto-Y chromosome that subsequently underwent a series of mutation and selection events that conferred male advantage and suppressed recombination with the proto-X, eventually leading to the degeneration of the Y-chromosome. Gene expression imbalances arise because X-chromosomal genes in male genomes are only present in one copy whereas autosomal genes and X-chromosomal genes in females are present in two copies. Dosage compensation mechanisms evolved in order to balance the relative expression levels of X-chromosomal genes between the sexes and in relation to autosomal genes [1]–[3].
Dosage compensation in D. melanogaster involves a combination of general buffering effects that act on all monosomic regions [4]–[6] and the specific targeting and stimulation of the male X-chromosome by the male-specific lethal (MSL) complex. Together, these processes increase X-chromosomal gene expression by approximately a factor of two [1], [7]. The MSL-complex consists of five proteins (MSL1, MSL2, MSL3, MLE, and MOF) and two redundant long non-coding RNAs (roX1 and roX2) [7]–[9]. It is believed that the hypertranscription of the male X-chromosome is partly due to the enrichment of histone 4 lysine 16 acetylation (H4K16ac). This acetylation is catalyzed by the acetyltransferase MOF and opens the chromatin's structure [10], [11]. The complete MSL-complex only forms in males due to the male-specific expression of MSL2 and the roX RNAs [12]–[16]. Notably, even though most genes on the X-chromosome appears dosage compensated in the 2-fold range [17], [18] the MSL-complex only contributes to part of this increase [6], [19], [20]. In addition, many genes are compensated without any significant recruitment of the MSL-complex [17]. An alternative model for the role of the MSL-complex in dosage compensation has been proposed by Birchler and colleagues [21]–[23]. According to their inverse dosage effect model the compensation in males is caused by the stoichiometric change of regulator(s) on the X-chromosome relative to the remainder of the genome. The main role of the MSL-complex is to sequester MOF from the autosomes to avoid autosome up-regulation and to limit the activation potential of MOF when targeted as part of the MSL-complex [21]–[24].
It is still not clear when, where and how the MSL-complex is assembled or which features of the X-chromosome allow its recognition. Several lines of evidence indicate that MSL1 and MSL2 are the core components of the MSL-complex. Notably, the absence of either one abolishes the binding of the remaining components of the complex to the X-chromosome [25]. The RING domain of MSL2 allows it to interact with MSL1, and the cystein-rich (CXC) domain of MSL2 allows the MSL1-MSL2 complex to recognize and bind DNA [26], [27]. The incorporation of roX RNAs into the MSL-complex is hypothesized to occur co-transcriptionally [28] and depends on their interaction with MSL2 and the RNA helicase MLE, which binds to stem-loop structures on roX RNAs in an ATP-dependent manner [29]–[32].
The roX1 and roX2 gene loci have been identified as two of the strongest high affinity sites (HAS) for MSL-complex targeting, out of the roughly 250 HAS on the X-chromosome [33], [34]. HAS are defined as sites targeted by MSL1 and MSL2 in the absence of msl3, mle or mof [25], [34], [35] and sites that are sufficient to recruit MSL even when inserted on an autosome [36]. HAS are enriched in a conserved consensus GA-rich DNA sequence motif [34], [37], [38]. The prevailing model is that the MSL-complex initially binds at the HAS and that its presence at these sites facilitates the more transient binding of additional MSL-complexes to neighboring active genes [8], [9], [39]. The transcriptional statuses of X-chromosomal genes influence the distribution of MSL binding because the complex is biased to exons and the 3′ ends of actively expressed genes; its binding correlates with enrichment in histone 3 lysine 36 trimethylation (H3K36me3) [34], [37], [40]–[43]. Other features such as the local chromatin context, H3 depletion, MSL-complex concentration, levels of affinity and sequence composition also contribute to the recognition and spreading of the MSL-complex over the male X-chromosome [35], [38], [44]–[46].
The roles of the two redundant long non-coding RNAs, roX1 and roX2, in the targeting of the entire male X-chromosome by the MSL-complex are not fully understood at present. Studies on polytene chromosomes have shown that in the absence of both roX1 and roX2, MSL2 and H4K16ac become less abundant on the male X-chromosome, with the MSL-complexes being relocated to the chromocenter, the 4th chromosome and a few other autosomal sites [20], [47], [48]. In this work, we analyzed MSL-complex targeting in roX1 roX2 mutants in order to unravel the specific roles of roX RNAs in MSL targeting and to learn more about the evolution of chromosome-specific targeting and dosage compensation. We performed ChIP-seq and cytological analyses of MSL proteins in roX1 roX2 double mutants, and analyzed their genome-wide binding profiles. It was found that in the absence of roX RNAs, the MSL-complex binds to the previously identified HAS on the X chromosome, the pericentromeric regions of all chromosomes, and specifically to six genes on the 4th chromosome. Analysis of the autosomal sequences bound by MSL in roX mutants showed that MSL has an affinity for regions enriched in Hoppel transposable elements, NTS (non-transcribed spacers) and repeats. Our results suggest that one role of the roX RNAs is preventing the MSL-complex from binding to heterochromatic repeats, suggesting that targeting heterochromatin is an intrinsic and ancient property of the MSL-complex.
Results
The MSL-complex is relocalized to heterochromatin in roX mutants
To test the role of roX RNAs in MSL targeting, we performed immunostaining experiments on polytene chromosomes of roX1 roX2 double mutants (hereafter called roX mutants). In the absence of roX RNAs, the extent of MSL-complex targeting to the X-chromosome was dramatically reduced and the complex was relocalized to the chromocenter and to three distinct regions on the 4th chromosome (Fig. 1A). The disruption of MSL targeting seen in roX mutants is clearly different from the disturbance that occurs when the protein components of the complex are removed: in msl1 or msl2 mutants, no MSL-complexes are formed on the X-chromosome at all [25]. Conversely, as shown in Fig. 1A, in mle or mof mutants, the MSL-complex is exclusively targeted to a limited number of bands on the X-chromosome. This shows that the roX RNAs and the protein components of the complex have different functional roles in MSL chromatin targeting.
Fig. 1. The MSL-complex is redistributed in roX mutants. 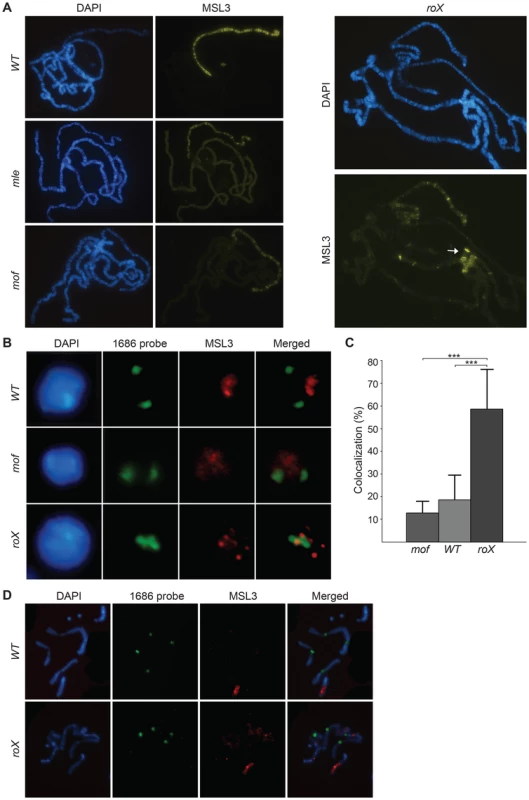
(A) MSL3 immunostaining on polytene chromosomes from wild type, mle, mof and roX mutant males. Note that MSL3 only targets a subset of sites on the X-chromosome, the 4th chromosome and the chromocenter (indicated by the arrow) in roX mutants. (B) DNA-FISH with a probe against the 1.686 g/cm3 satellite repeat from pericentromeric regions of chromosomes 2 and 3 (1686 probe) combined with MSL3 immunostaining, on interphase nuclei of brain cells from third instar male larvae wild type, mof and roX mutants. (C) Percentage colocalization of 1686 probe signal and MSL3 staining from 8 replicates each of the genotypes: wild type, mof and roX (30–50 nuclei scored per replicate). The bars indicate the mean colocalization and the whiskers indicate standard deviations. Significant differences are indicated by *** (Independent two-sample t-test, p<0.001). (D) DNA-FISH with the 1686 probe combined with MSL3 immunostaining on metaphase nuclei of brain cells from third instar larvae wild type and roX mutant males. Note that on metaphase chromosomes MSL3 colocalization with centromeres is not detected in roX mutants. To exclude the possibility that the binding of the MSL-complex to pericentromeric heterochromatin in roX mutants is unique to polytene chromosomes, we analyzed MSL binding in relation to the pericentromeric repeat 1.686 (which is known to be enriched in the pericentromeric regions of chromosomes 2 and 3 [49]) in interphase nuclei from brain cells of wild type samples and roX mutants. In the wild type and mof mutants, the MSL3-bound X-chromosome occupies a part of the nucleus that is clearly separated from the pericentromeric regions (Fig. 1B and 1C). In roX mutants, the normal binding of MSL3 is altered and the complex is observed in spots that colocalize with the centromeric repeats. This colocalization is three times more frequent in roX mutants than in wild type or mof mutants (Fig. 1C).
We therefore conclude that the relocalization of MSL in the absence of roX RNAs observed in salivary gland nuclei also occurs in diploid interphase nuclei. Interestingly, MSL binding in metaphase chromosomes of wild type and roX mutants is similar and is restricted to the euchromatic part of the X-chromosome (Fig. 1D). It remains to be determined why MSL doesn't target centromeres in the highly compacted mitotic chromatin in roX mutants.
MSL recruitment to High Affinity Sites (HAS) is independent of roX
To better understand how the genome-wide targeting of MSL depends on roX RNAs, we performed MSL1, MSL2 and MOF ChIP-seq experiments on salivary glands from wild type individuals and roX mutants. These experiments confirmed the results of the immunostaining studies, showing that there is a pronounced decrease in MSL binding along the X-chromosome in roX mutants although binding persists at specific locations. Visual inspection demonstrates that the MSL enrichment peaks along the X-chromosome in roX mutants coincide with the previously defined HAS (Fig. 2A) [34], [37], [38]. Notably, although all previously defined HAS are not recognized by MSL enrichment in roX mutants, all enrichment peaks coincide with HAS. To verify that the roX RNA-independent MSL enrichment peaks on the X-chromosome correspond to the previously mapped HAS, we calculated the shortest distance between the coordinates of the HAS and those of the MSL1 binding sites on the X-chromosome in roX mutants identified in our ChIP-seq experiments. The fractions of sites bound by MSL1 in roX mutants were plotted against distance to nearest HAS, and the distances between the coordinates of HAS and random positions on the X-chromosome were used as controls. As seen in Fig. 2B, the largest fraction of the MSL binding sites in roX mutants overlap with HAS. This is in clear contrast to the control, in which the largest fraction of random X-chromosome sites are>35 kb away from HAS. Taken together this means that although all of the 263 previously defined HAS are not bound by MSL in roX mutants, in principle all of our 208 defined MSL binding sites in roX mutants target HAS. These results demonstrate that MSL binding to HAS on the X-chromosome occurs independently of roX RNAs.
Fig. 2. MSL recruitment to the conserved HAS is independent of roX. 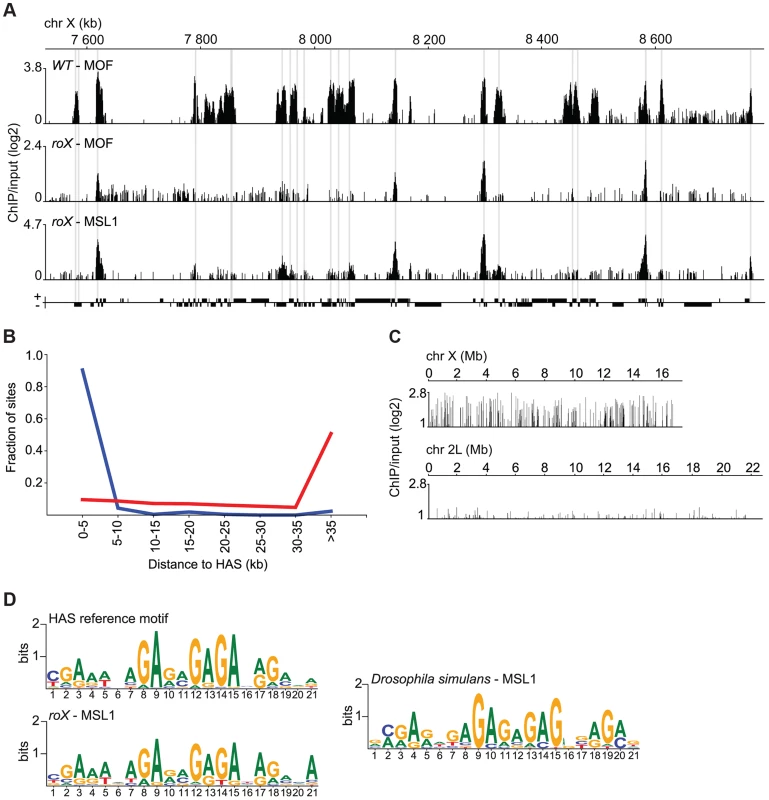
(A) MOF and MSL1 ChIP-seq enrichment profiles, with a 500 bp smoothing, for a representative region of the X-chromosome in salivary gland tissue from wild type and roX mutant males. Numbers along the x-axis denote positions along the chromosome in kb. The y-axis shows the ChIP enrichment over input as log2 ratios. Genes expressed from left to right and vice versa are shown above and below the horizontal lines, respectively. The HAS locations, previously defined by [34], [37], [38], are indicated by grey boxes. (B) Fraction of sites on the X-chromosome from roX mutants that are bound by MSL1, compared to random sites along the X-chromosome (red), sorted by distance to HAS. (C) MSL1 ChIP-seq enrichment profile, with a 500 bp smoothing, for the entirety of chromosomes X and 2L in salivary glands from D. simulans wild type males. Numbers along the x-axis denote chromosomal positions in Mb. The y-axis shows the ChIP enrichment over input as log2 ratios. (D) Sequence motifs enriched in MSL-bound regions of the X-chromosome in D. melanogaster wild type [37], roX mutants and D. simulans wild type. It has been shown that roX RNAs evolve rapidly, only sharing about 90% and 80% sequence homology in such closely related species as D. simulans and D. yakuba, respectively [30]. We therefore sought to determine whether HAS, previously shown to be enriched in a GA-rich motif [37], [38], are under high evolutionary pressure. To facilitate comparison with other Drosophila species, we generated ChIP-seq data for MSL1 binding in wild type Drosophila simulans and performed a motif analysis in the MSL1-bound regions on the X-chromosome of this species (Fig. 2C). We found highly similar GA-rich motifs to be enriched within MSL targets on the X-chromosome in roX mutants as well as on the X-chromosome of wild type D. simulans (Fig. 2D).
These results show that the roX RNAs are not involved in MSL targeting to HAS and that the HAS motif is evolutionarily conserved.
A complete and enzymatically active MSL-complex is assembled in roX mutants
The binding of MSL to the 4th chromosome in the absence of roX RNAs is intriguing because there are several lines of evidence suggesting an evolutionary relationship between the 4th chromosome and the X-chromosome [1], [50]–[52]. Our ChIP-seq profiles show that the MSL-complex binds specifically to six genes on the 4th chromosome in roX mutants: Ankyrin, Rad23, CG2177, PMCA, Mitf and Dyrk3. The locations of these genes correspond to those of the MSL-stained bands seen on polytene chromosomes (Fig. 3A). One important question when considering the binding of MSL outside the X-chromosome is whether a complete and functional MSL-complex is formed at these locations. Our immunostaining experiments in roX mutants showed that all of the complex's protein components (MSL1, MSL2, MSL3, MLE and MOF) colocalize perfectly at the chromocenter and at the three bands on the 4th chromosome (Fig. 3B). In addition H4K16ac is also enriched at these three bands in roX mutants, which indicates that the MSL-complex is complete and active (Fig. 3B and S1 Figure). Note that H4K16ac on the 4th chromosome shows a broader enrichment pattern compared to the MSL proteins in similarity to what previously have been observed for H4K16ac in relation to MSL on the male X-chromosome in wild type [10]. Next we tested the H3S10 kinase JIL1, previously shown to be enriched on the male X-chromosome and dependent on a functional MSL-complex for its targeting [53]–[55]. JIL1 has previously been shown to co-immunoprecipitate with the MSL-complex under low stringency conditions or after formaldehyde cross-linking [54], [56]. Interestingly, like the MSL-complex, JIL1 is also relocalized to the chromocenter and the three regions on the 4th in the absence of roX RNAs (Fig. 3C).
Fig. 3. A complete and enzymatically active MSL-complex is assembled in the absence of roX. 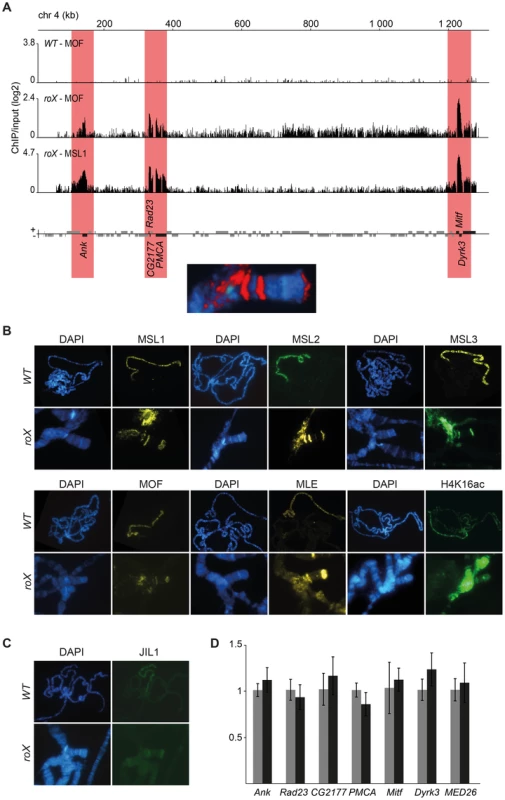
(A) MOF and MSL1 ChIP-seq enrichment profiles, with a 500 bp smoothing, for the entire 4th chromosome in salivary gland tissue from wild type and roX mutant males. Numbers along the x-axis denote chromosomal positions along the chromosome in kb. The y-axis shows the ChIP enrichment over input as log2 ratios. Genes expressed from left to right and vice versa are shown above and below the horizontal lines, respectively. Note that the genes targeted in roX mutants (indicated by red boxes) correspond to the three bands seen in polytene chromosome staining (below). (B) MSL1, MSL2, MSL3, MLE, MOF and H4K16ac immunostaining on polytene chromosomes from wild type males, with X-chromosome targeting, and from roX mutant males, showing the 4th chromosome and chromocenter targeting. (C) JIL1 immunostaining on polytene chromosomes from wild type and roX mutants males, shows targeting to the chromocenter and to the same chromosome 4 bands as the MSL-complex. (D) Mean levels of mRNA from the six genes targeted by MSL in roX mutants and from a control gene on the 4th chromosome that is not bound by MSL (MED26), determined by rt-qPCR (black). The corresponding mean expression of the same genes in wild type is shown in grey. The mRNA levels measured by qPCR were normalized against RpL32 mRNA in each replicate. Error bars represent the standard deviation of three biological replicates. Since H4K16ac overlaps with all the other proteins from the MSL-complex in roX mutants we asked if the six identified genes on the 4th bound by MSL in roX mutants have higher transcriptional output than in wild type. The relative expression of the six 4th chromosome MSL-bound genes was not found to differ significantly between wild type and roX mutants (Fig. 3D).
One tempting hypothesis based on the targeting of the 4th chromosome is that the MSL-complex in D. melanogaster still has an affinity for ancestral X-chromosomal sequences, now present on the 4th. We performed BLAST searches for the sequences of the six 4th chromosome genes targeted by MSL in D. melanogaster roX mutants in the distantly related species D. virilis and D. willistoni and found that in both species, these six genes are assigned to the sequence scaffold on which all of the other 4th chromosome-linked genes are located rather than to the X chromosome. In the even more distantly related species D. busckii, the whole correspondent to the 4th chromosome of D. melanogaster is fused to the X-chromosome [57], [58]. Interestingly, it has recently been shown [50] that in D. busckii the sequences corresponding to the D. melanogaster 4th chromosome are present in more copies in females than males. However, the female-to-male ratio is less than 2 meaning that the corresponding homologs on the Y-chromosome are not fully degenerated or that some but not all genes on the corresponding 4th chromosome have degenerated homologs on the D. busckii Y-chromosome. We hypothesized that the six genes targeted by MSL in roX mutants actually skew the ratio and therefore calculated the female-to-male ratio of these D. busckii orthologs relative to the other chromosome 4 genes. By using previously reported data [50] we found that there is no significant difference between the 6 genes and the other 4th chromosome genes (S2 Figure). We conclude that a complete and active MSL-complex binds with high specificity to six genes on the 4th chromosome, although the reason for this specificity remains elusive.
In the absence of roX, MSL has affinity for Hoppel transposable elements
The pericentromeric regions and the 4th chromosome are both heterochromatic regions of the D. melanogaster genome that are targeted by MSL in roX mutants and are enriched in satellite repeat sequences, transposable elements, and the heterochromatic proteins HP1a and HP2, among others. Our ChIP-seq results show that in the absence of roX RNAs, the MSL-complex targets the pericentromeric regions of all chromosomes and that its abundance increases gradually on moving towards the centromere. This tendency is illustrated for chromosome 3L in Fig. 4B. One possible mechanism underlying this binding is that MSL recognizes a specific recruitment element but that its binding to this element is blocked by the presence of roX RNAs. Another possibility is that the MSL-complex has an intrinsic affinity for heterochromatic sequences or repeats in general. Using the MEME software, we analyzed the identified MSL1-bound heterochromatic sequences of each chromosome (2LHet, 2RHet, 3LHet, 3RHet, 4Het, XHet). A specific motif corresponding to repeats in the Hoppel (1360) transposable element was found to be significantly enriched (Fig. 4A and B). In situ DNA hybridization experiments using the identified motif as a probe in conjunction with MSL staining revealed a high degree of colocalization between the MSL-complex and the motif in both the pericentromeric heterochromatin and the three specific bands on the 4th chromosome (Fig. 4C). To determine whether this motif can act as an MSL-recruitment element, we generated a construct containing three tandemly repeated copies of a 108-nucleotide Hoppel element featuring the motif in question. This repeat segment was placed upstream of a cDNA copy of ankyrin (a gene on the 4th targeted by MSL in roX mutants) under an endogenous promoter. The construct was inserted into the 3L:65B2 PhiC landing platform and tested for MSL binding in a roX mutant background. The transgene was visualized using mini-white DNA-FISH and MSL-complex was not detected on the target (S3 Figure). These results suggest that the repeat motif from the Hoppel transposable element, which is enriched at MSL-targeted regions (4th and pericentromeric) in roX mutants, is not by itself sufficient to recruit the MSL-complex. However, we cannot exclude the possibility that recruitment might be achieved with a greater number of motif copies.
Fig. 4. In roX mutants Hoppel is enriched in MSL-complex. 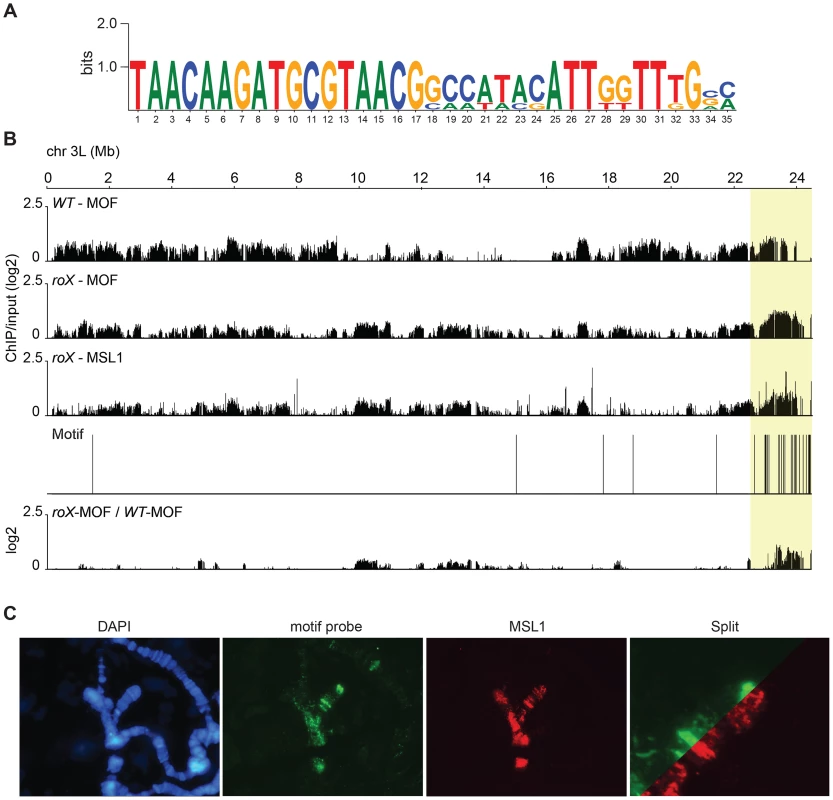
(A) Sequence motif enriched in MSL-bound regions on heterochromatin of roX mutants. (B) MOF and MSL1 ChIP-seq enrichment profiles, with a 2000 bp smoothing, for the representative chromosome arm 3L from wild type and roX mutant males. Numbers along the x-axis denote chromosomal positions along the chromosome in Mb. The y-axis shows the ChIP enrichment over input as log2 ratios. The mapping of the motif along 3L is indicated. Below the motif track the ratio roX-MOF/WT-MOF is plotted. Note that in roX mutants MSL is enriched in the pericentromeric region (indicated by a yellow box). (C) DNA-FISH with a 33 nucleotide long probe against the heterochromatic motif shown in A combined with MSL1 immunostaining, on polytene chromosomes of roX mutant males. Note the overall colocalization between the MSL1 bound regions and the motif hybridisation. DNA sequences from centromeres, telomeres, the Y-chromosome and other heterochromatic regions are not assembled to any region of the D. melanogaster genome due to their highly repeated nature, and the mapping of sequences recovered in the ChIP-seq normally discards the large number of repeated sequences in the genome. We suspected that the non-mapped reads recovered by ChIP-seq might hold information about other transposable elements targeted by MSL in roX mutants in the above-mentioned heterochromatic regions. To test this hypothesis, we aligned all of the ChIP-seq reads to the repeat class sequences from the Repbase Update database and calculated RPKM values for each repeat class. Using this approach we found that in roX mutants there were strong enrichments of three repeat classes: PROTOP_B, PROTOP_A and NTS (Non-transcribed Spacer) (Fig. 5). Interestingly, the PROTOP is a family of autonomous DNA transposons that have been suggested to be ancient ancestors of the P-element and Hoppel element transposon families [59] and PROTOP_A and PROTOP_B are listed as synonyms of Hoppel [60]. Our results confirm that MSL has an affinity for regions enriched in repeats from Hoppel and PROTOP transposable elements and for NTS. All of these are highly repeated elements that are present in heterochromatic regions of the genome.
Fig. 5. MSL-complex targets PROTOP and NTS in roX mutants. 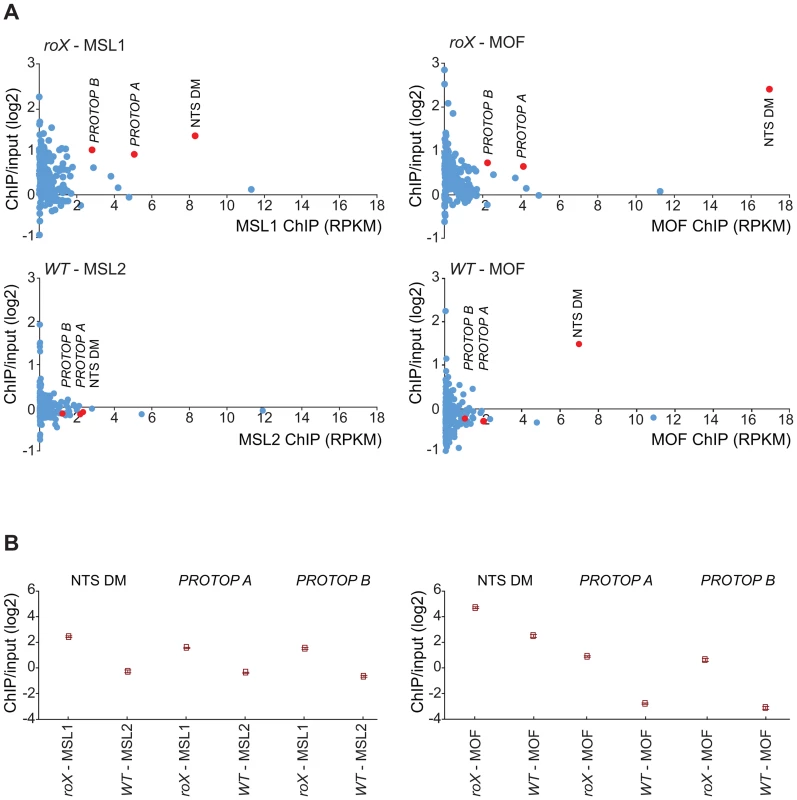
(A) Reads from the ChIP samples mapped to repeat classes from Repbase Update are shown on the x-axis as RPKM values (Reads Per Kilobase per Million mapped reads) and related to ChIP/input enrichment ratios, which are shown on the y-axis. The left part shows MSL1 ChIP in roX mutants compared to MSL2 in wild type and the right part shows MOF ChIP in roX mutants and wild type. Note, that NTS is enriched also in MOF wild type. (B) Enrichment ratio (IP/input) of the overrepresented repeats from A (NTS, PROTOP_A, PROTOP_B) in roX mutants compared to the wild type. Rectangles represent the mean values of the IP/input enrichment ratios of all mapped and unmapped reads matching to each repeat type, and error bars indicate 95% confidence intervals. The MSL-complex has an affinity for repeat enriched regions
In addition to the Hoppel transposable element repeats, the analysis of the mapped and unmapped sequences bound by MSL in roX mutants recovered NTS sequences, which occur between ribosomal DNA genes which are organized in tandem repeats. Because MSL targets some autosomal sites across the genome in the absence of roX, we wondered whether these sites were also enriched in repeats and whether repeats in general were enough to recruit MSL. To test this hypothesis, we analyzed the enrichment of repeat masked sequences around MSL targeted regions. Since the MSL-complex mainly targets expressed genes we calculated the enrichment of repeats surrounding the TSS (transcription start site) of genes that are expressed in salivary glands (the tissue of our binding data) and are located in either MSL-bound or MSL-unbound regions of the genome. We examined both the X-chromosome and the autosomes (excluding chromosome 4 and the mapped pericentromeric regions) in this analysis, taking into account the MSL-bound regions in wild type and roX mutants that were identified based on our ChIP-seq data. The density of satellite repeats on the X-chromosome is reportedly greater than on the 2nd and 3rd chromosomes [61]–[63]. Our results are consistent with this finding and show that the regions surrounding expressed genes on the X-chromosome have a somewhat higher repeat content than those surrounding autosomal expressed genes (Fig. 6A). Strikingly, autosomal expressed genes bound by MSL in roX mutants are enriched in surrounding repeats whereas the regions surrounding unbound autosomal expressed genes have a low repeat content. The repeat content of regions surrounding X-chromosomal genes bound by MSL was also higher than that of unbound regions, but the difference was less pronounced than for autosomal genes. Our results show that MSL targeting of autosomal sites in roX mutants correlates with high repeat content.
Fig. 6. MSL-complex has affinity to repeats. 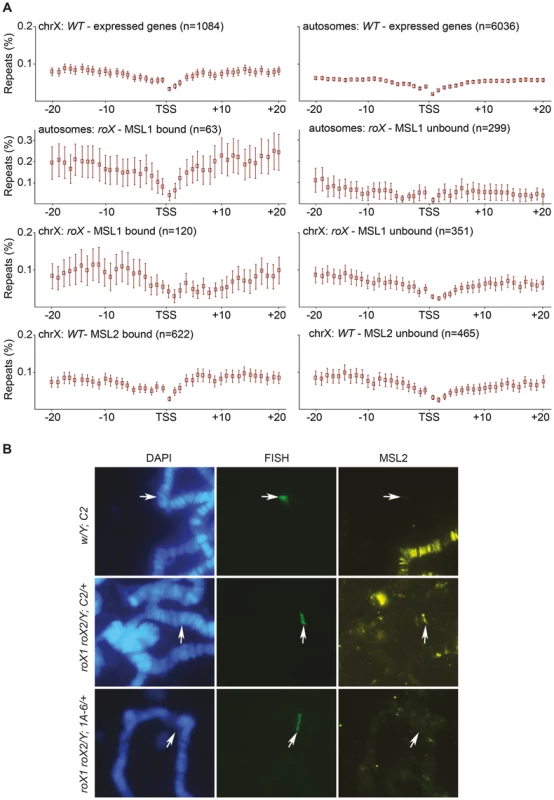
(A) Percentage of repeat masked sequences (from UCSC), in 20 bins of 1 kb distance from the transcription start site (TSS) of X-chromosome expressed genes, autosomal expressed genes, MSL1-bound/unbound genes in roX mutants, on the X-chromosome and on autosomes. Rectangles represent the mean values of repeats for all expressed genes in each distance bin, and error bars indicate 95% confidence interval. (B) DNA-FISH with a probe against the mini-white gene combined with MSL2 immunostaining, on polytene chromosomes of wild type and roX mutant males. C2 is a cluster of seven tandemly repeated copies of P[lacW] transgene, the 1A-6 cluster consists of two copies. Note that the transgene cluster consisting of seven copies recruits MSL2 in a roX mutant background. The site of the construct insertion is indicated by arrows. We therefore sought to determine whether any repeat sequence would recruit MSL in the absence of roX. Two repeat types were tested. First we analyzed clusters of tandemly repeated P[lacW] transgenes on the 2nd chromosome in roX mutants [64]. The P[lacW] transgene contains sequences of the P-element flanking the E. coli lacZ gene and the mini-white gene. In the absence of roX, MSL2 did indeed bind to a cluster of 7 tandemly repeated copies of P[lacW] (C-2, BX-2, T-1) but not to a cluster with only 2 copies (1A-6) (Fig. 6B). Although mini-white originates from an X-linked gene, it does not contain HAS and is not an ectopic MSL target in the wild type. These results show that genes in tandem repeats are enough to recruit the MSL-complex when roX RNAs are absent. The second repeat cluster tested in a roX mutant background was the 256 lac repeats of E. coli upstream of a white reporter gene [65]. In this system, the DNA binding domain of the lac repressor (lacI BD) fused to HP1a is tethered to a reporter transgene that contains repetitive binding sites for the lacI BD (lacO repeats). The transgene was targeted by HP1a in about 50% of nuclei but never by MSL3 (S4 Figure).
Overall, these results suggest that in absence of the roX RNAs, MSL has an intrinsic general affinity for repeated sequences. However, the influence of the repeat length and number of copies as well as the sequence specificity of the complex all remain to be elucidated.
Discussion
The correct functioning of dosage compensation mechanisms in both mammals and flies is dependent on the presence of non-coding RNAs [66]. Although the roX genes are known to be important for MSL-complex targeting, their function in this process and the targets of the complex in the absence of roX RNAs were previously unknown. In this work we used immunostaining and ChIP-seq techniques to map the targets of the MSL-complex in the absence of roX. Based on the results of these experiments, we present a model that describes the role of the non-coding roX RNAs in targeting the MSL-complex to the male X-chromosome.
roX RNAs prevent the targeting of heterochromatin by the MSL-complex
We and others have previously shown that in the absence of roX the MSL-complex is redistributed to targets corresponding to pericentric heterochromatin and the 4th chromosome, or “green chromatin” according to recent chromatin structure-based definitions [20], [47], [48], [67]. Since large parts of these “heterochromatic” regions targeted by MSL in roX mutants are under-replicated in polytene chromosomes it was important to determine whether this redistribution also occurs in diploid cells that have very different ratios of the relevant DNA motifs. Notably, although the reduction in MSL-complex abundance on the X-chromosome is much more dramatic in roX mutants than in mle or mof mutants and the MSL-complex is relocalized to heterochromatic regions in roX mutants, escaping males are recovered in roX mutants in contrast to the complete male lethality observed in mle, msl3 or mof mutants [47], [68], [69]. The results of our studies on interphase nuclei from brain tissue showing colocalization between MSL3 and centromeric regions further support the interpretation that in absence of roX, the MSL-complex targets heterochromatin.
It has previously been shown that the fraction of escaper males in roX mutants is significantly higher in roX1 roX2/0 males, i.e. males lacking a Y-chromosome, than in roX1 roX2/Y males. Importantly, the Y-chromosome is predicted to be 40 Mb in length and thus accounts for>10% of all genomic DNA in male cells [70]. It seems likely that the reduced abundance of heterochromatic target DNA and/or the greater compaction of the remaining heterochromatin caused by the loss of the Y-chromosome increases the X-chromosomal targeting of MSL in roX1 roX2/0 males, explaining their increased survival. It is tempting to speculate that in roX mutant interphase nuclei, the centromeric regions of autosomes have a tendency to colocalize with the X-chromosome within the nucleus, in a region where the local concentration of the MSL-complex is expected to be high. In fact, previous studies have shown that HAS are closer in the nuclear space in males than in females, suggesting that long-range associations between MSL-complex target sites shape nuclear organization [71]. On mitotic metaphase chromosomes, the MSL-complex is only seen on the distal X-chromosome in the wild type. Surprisingly, the same pattern is seen in roX mutants, although the specificity of the MSL targeting is somewhat lower in these cases. We speculate that the HAS present on the X-chromosome provide superior targets for the complex when transcription is suppressed (as is the case in metaphase) compared to the centromeric regions.
Overall our results suggest that the MSL-complex has a greater affinity for roX RNAs than its heterochromatic targets and so roX RNAs restrict the targeting of the complex to the X-chromosome. In keeping with this hypothesis, a previous study showed that when MSL1 and MSL2 are overexpressed, MSL2 targets not only the X-chromosome but also some autosomal sites, the 4th chromosome and the chromocenter [72]. roX1 RNA was only detected on the X-chromosome and on some autosomal sites but not on the 4th or the chromocenter. This suggests that the MSL-complex has an intrinsic affinity for heterochromatin and a balanced amount of roX RNAs are required to restrict the complex from these targets. An affinity for heterochromatin components may in fact be part of the mechanism to limit the activating potential of MOF when sequestered to the male X-chromosome. Although not detected in ChIP experiments or chromosome immunostainings [73]–[76], a low amount of HP1a along the entire male X-chromosome has been found in genome-wide mapping of HP1a using the DamID technique [77]. In addition, a knock-down of HP1a indicated more lethality in males than in females [78]. It is possible that when the strong binding to the X-chromosome is decreased in the roX mutants, the affinity for HP1a relocates the complex to canonical HP1a binding sites: heterochromatin, 4th chromosome, repeat arrays.
High Affinity Site targeting by the MSL-complex is independent of roX RNAs
In the absence of roX, the MSL-complex still targets a reduced number of sites on the X-chromosome. Based on previous cytological analysis it has been argued that these sites are similar but not identical to the sites targeted in msl3 mutants, i.e. HAS [47], [68]. Our ChIP-seq results show an almost perfect overlap between MSL targets in roX mutants and the 263 previously identified HAS. In addition, the enriched sequence motif revealed by our bioinformatics analysis is nearly identical in wild type and roX mutants and also when comparing D. melanogaster and D. simulans. Since previous studies showed that MSL targeting to HAS is independent of MSL3, MOF and MLE, and we found HAS targeting to be independent of roX RNAs, we propose that MSL1 and MSL2 are the only components required for the correct targeting of HAS.
roX RNAs are not required for the assembly of a complete and active MSL-complex
Interestingly, we detected a very strong MSL signal and a high enrichment of the complex in the regions surrounding six specific genes on the 4th chromosome in roX mutants. Several lines of evidence suggest an evolutionary relationship between the 4th chromosome and the X-chromosome [1], [50]–[52]. However, we cannot presently explain why these genes are specifically targeted in this way. Our findings indicate that the MSL-complexes formed at these non-X chromosome locations are complete and active, and even include associated factors such as JIL1.
Importantly we did not observe any obvious change in the expression of the six 4th chromosome genes targeted by the intact MSL-complex (although this may be partly due to the limited sensitivity of qPCR). This again suggests that the activation potential of MOF within the MSL-complex is limited [17], [24], [79]. It has been shown that the targeting of MOF alone to reporter transgenes result in a strong increased expression. In contrast, when MOF was targeted as part of the MSL-complex no increased expression was observed [24]. In addition, it is important to note that the expression of all genes from the 4th chromosome is fine-tuned by a balance between HP1a, which represses gene expression, and POF, the chromosome 4-specific protein that stimulates gene expression [80]–[82]. The predicted effect on gene expression due to MSL targeting to the 4th chromosome in roX mutants might be counteracted by HP1a and/or POF. We did not observe any clear difference between roX mutants and the wild type with respect to the binding of POF to the 4th chromosome or the binding of HP1a to either the chromocenter or the 4th. This demonstrates that MSL binding does not interfere with that of HP1a or POF.
The MSL-complex has an affinity for regions enriched in Hoppel transposable elements and repeats
A link between dosage compensation and transposable elements has previously been suggested for both mammals and Drosophila. The mammalian X-chromosome is enriched in LINE elements (particularly in its pericentromeric region). It has been suggested that these elements boost the X-inactivation signal by acting as anchoring stations for the spread of the Xist RNA [83]. We identified a strong connection between MSL targeting and the Hoppel transposable element using two different approaches. The first involved identifying sequence motifs in the mapped regions, typically pericentric regions on chromosome arms and targets on the 4th chromosome; this revealed a recurring sequence motif from the Hoppel element. The second involved calculating the enrichments of all reads, both mapped and unmapped. Strong enrichment was observed for PROTOP_A and PROTOP_B (both corresponding to Hoppel [59]) as well as NTS Dm. Notably, NTS is enriched by MOF also in wild type suggesting that this is an intrinsic target of MOF which is stabilized by the complete MSL-complex in roX mutants. The importance of Hoppel in MSL targeting is supported by its high degree of colocalization with MSL staining in roX mutant males. Because the ChIP technique relies on the analysis of fractionated DNA, we cannot currently say whether it is Hoppel itself or transcribed regions in its vicinity that are targeted by MSL. The Hoppel elements are non-autonomous DNA transposons and among the most abundant transposable elements in the D. melanogaster genome, being enriched in the pericentric heterochromatin and on the fourth chromosome [84]. Since we also observed MSL enrichment in NTS (Non-Transcribed Spacer) regions, active autosomal genes surrounded by DNA repeats, and tandemly repeated gene constructs (P[lacW]), it seems that MSL is recruited to repeats in general. A recent study demonstrated that the neo-X of D. miranda has newly evolved chromatin entry sites (CES), also known as high affinity sites-HAS, that recruit MSL and are enriched in the ISX helitron transposable element (TE) [85]. The authors suggest that the evolutionary acquisition of the MSL-complex by X-chromosomes involved the acquisition of GA-rich sequence motifs by transposable elements that were capable of functioning as HAS for the MSL-complex, followed by amplification of the TEs across the genome. This may then have been followed by positive selection for these elements on the X-chromosome followed by a refinement process that eroded TEs in non-functional regions and increased their affinity for MSL. The authors further suggest that the heterochromatization of the neo-Y occurs in parallel with the acquisition of dosage compensation on the neo-X.
We speculate that the MSL targeting seen in the absence of roX RNAs represents an ancient but still intrinsic property of the MSL-complex. This model suggests that roX RNAs are younger in evolutionary terms than the protein components of the MSL-complex and evolved in parallel with the degeneration of the Y-chromosome, redistributing the MSL-complex to the male X-chromosome and restricting its intrinsic heterochromatic targeting. In fact, a human MSL-complex (hMSL) has been identified that contains the homologs of the Drosophila proteins MSL1, MSL2, MSL3 and MOF, indicating that the MSL-complex protein components are highly conserved in evolution. Conversely, the roX RNAs evolve rapidly [30]. The hMOF is responsible for the majority of H4K16 acetylation as well as being involved in the acetylation of other substrates such as the p53 protein, and in the regulation of various cellular processes (reviewed in [86], [87]). The ancient function of MSL in the heterochromatin of a Drosophila melanogaster ancestor may have been to activate the expression of active genes present in repressive environments. The binding of MSL to NTS in roX mutants supports the hypothesis that MSL may have had a role in protecting active genes that are present in multiple copies in the genome, like the ribosomal genes, against repeat-induced gene silencing [83], [88]. This is also supported by our finding that active genes on autosomes bound by MSL in roX mutants are in repeat-enriched regions. It has been shown that sequences containing P[lacW] in tandem repeats become heterochromatic and the repeated mini-white gene becomes partially repressed [64]. MSL is recruited to this transgene in a roX mutant background suggesting that it is recruited to repeat-induced gene silencing regions.
Materials and Methods
Fly stocks and genetic crosses
Flies were cultivated and crossed in vials containing potato mash-yeast-agar medium at 25 C. The wild type strains used were D. melanogaster (Oregon R) and D. simulans/w501 (UC San Diego Drosophila Stock Center). The D. melanogaster roX1 roX2 double-mutant males were selected as non-GFP males from a y w roX1ex6 Df(1)roX252 P[w+ 4Δ4.3]/FM7i, P[w+mC ActGFP]JMR3 stock obtained from Yongkyu Park (New Jersey Medical School, Newark, NJ). The mof mutants were obtained by crossing virgin females from the stock mof2; P[w+ mof+]/CyO GFP, obtained from Peter Becker (Ludwig Maximilians Universität Munchen), to wild type males and selecting green-fluorescent males in the progeny. The mle mutants were obtained by selecting the non-green fluorescent males from the cross: mle9 cn1 bw1/CyO, P[w+mC ActGFP]JMR1 ×FM7i, P[w+mC = ActGFP]JMR3/Y; mle1/CyO, P[w+mC ActGFP]JMR1. The strains carrying the P[lacW] transgene in repeats C-2, BX-2, T-1, and 1A-6 were kindly provided by Stephane Ronsseray (CNRS-Université Pierre et Marie Curie) and are described elsewhere [64], [89], [90]. Insertions on the second chromosome were rebalanced with CyO, P[w+mC ActGFP]JMR1, the rebalanced males were crossed to y w roX1ex6 Df(1)roX252 P[w+ 4Δ4.3]/FM7i, P[w+mC ActGFP]JMR3 females, and salivary glands were dissected from non-GFP male larvae. To study the targeting of MSL to lac repeats we used the strains P[hs-HP1.lacI.BD] and P[Ecol\lacO.256x.w]157.4.112 [65], kindly provided by Lori Wallrath (University of Iowa). roX1ex6 Df(1)roX252 P[w+4Δ4.3]/Y;P[Ecol\lacO.256x.w]157.4.112/+; P[hs-HP1.lacI.BD]/+ males were obtained by crossing w; P[Ecol\lacO.256x.w]157.4.112 males with roX1ex6 Df(1)roX252 P[w+4Δ4.3]/FM7i; P[hs-HP1.lacI.BD] females. Third instar larvae were heat-shocked for 45 minute at 37 C and recovered at room temperature for 2–3 h prior to dissection.
Transgenic flies
To generate transgenic flies carrying a transgene with repeats of the ankyrin gene together with the motif found to be enriched at heterochromatic sites bound by MSL in roX mutants, a DNA fragment containing the attB integration site was excised from pTA-attB [91] with EcoRI and cloned into the CaSpeR-4 vector. The resulting pCas-attB plasmid was used as a cassette for integrating ankyrin cDNA downstream of a 108 nucleotide-long DNA fragment identical to the Hoppel element 1360{}6073 (FBti0064134), repeated three times. A plasmid containing this Hoppel repeat was produced by GenScript USA Inc. The repeat was excised with KpnI and cloned into pCas-attB (pCas-attB-1360). A cDNA clone of ank-RB (LD10053) was purchased from the Drosophila Genomic Research Center. The desired DNA fragment was excised with NotI and XhoI and cloned into pCas-attB-1360 digested with the same nucleases. Finally, the promoter region of ank was amplified with the primers 5′-atagcggccgcttaggtatgtaaaattcacgcaa-3′ and 5′-cgagcggccgcaaggcaggctcaggtatttg-3′, digested with NotI and cloned upstream the ank-RB fragment. Embryo microinjection into the Bl9750 strain (3L:65B2 PhiC landing platform) was performed by BestGene (Inc). Males homozygous for the transgene were crossed to y w roX1ex6 Df(1)roX252 P[w+ 4Δ4.3]/FM7i, P[w+mC ActGFP]JMR3 females and salivary glands were dissected from non-GFP male larvae.
Immunostainings and DNA in situ hybridization (DNA-FISH)
Third instar larvae polytene chromosomes from salivary glands were prepared as described previously [92]. Larval brain squashes were performed according to protocol 1.9, method 3 in [93]. Immunostainings were performed as described previously [94] with the following antibodies (dilutions in parentheses): rabbit anti-MSL1 (1∶400), MSL2 (1∶200), MOF (1∶400) and MLE (1∶2000), and goat anti-MSL3 (1∶2000) from Mitzi Kuroda (Harvard Medical School); rabbit anti-JIL1 (1∶1000) from Peter Becker (Ludwig Maximilians Universität Munchen); and rabbit anti-H4K16ac (1∶300, sc-8662-R, Santa Cruz). The secondary antibodies used were donkey anti-goat or donkey anti-rabbit conjugated with AlexaFluor555 or AlexaFluor488, respectively (1300 dilution, Molecular Probes) together with DAPI (1 µg/ml). DNA-FISH combined with immunostaining on polytene chromosomes and brain squashes was performed according to a standard protocol [95]. The probe against mini-white was excised from CaSpeR-4 plasmid using the EcoRI restriction endonuclease and biotin labelled with the BioNick DNA Labeling System (Life Technologies). A FAM-labelled probe against 1.686 g/cm3 satellite was purchased from Exiqon. The sequence of the 33 nucleotide-long biotin-labelled probe that was used against the heterochromatic motif found to be enriched at MSL-bound regions in roX mutants was 5′-TAACAAGATGCGTAACGGCCATACATTGGTTTG-3′. Antibodies for the detection of DNA probes were mouse anti-FITC and mouse anti-biotin (1∶500, Jackson ImmunoResearch) with goat anti-mouse labelled with AlexaFluor488 as secondary antibody. HP1a was detected with rabbit PRB291C antibody (1∶400, Covance) and with donkey anti-rabbit AlexaFluor555. Preparations were analyzed using a Zeiss Axiophot microscope equipped with a KAPPA DX20C CCD camera. For comparisons between strains or proteins stained, the protocol was run in parallel. Nuclei with clear cytology were chosen on the basis of DAPI staining and photographed. At least 20 nuclei for each genotype were used in these comparisons, and at least four slides of each genotype were analyzed. For the colocalization analysis of the 1686 probe DNA/FISH combined with MSL3 immunostaining, 8 biological replicates (8 slides with one brain per slide) from each of the wild type, mof mutants, and roX1 roX2 mutants were analyzed with 30–50 nuclei scored per replicate. Nuclei were chosen on the basis of DAPI staining and colocalization was scored.
Quantitative Real-time PCR
Total RNA was extracted from third instar larvae using TRI reagent (Ambion) according to the manufacturer's protocol. Three biological replicates from the wild type and roX1 roX2 mutants were produced, consisting of 10 male larvae each. The RNA was reverse-transcribed using the iScript cDNA Synthesis kit (Bio-Rad) and amplified by real-time PCR using iQSYBR Green Supermix (Bio-Rad) according to the manufacturer's instructions. Primer pairs used are listed in Supplementary Table S1. The expression levels were normalized to the amount of RpL32 mRNA in each replicate.
Chromatin immunoprecipitation and deep sequencing (ChIP-seq)
The ChIP experiments were performed in salivary glands from third instar larvae as previously described [80], [81] using 3 µl of anti-MSL1, 3 µl of anti-MOF and 2 µl of anti-MSL2 (provided by Mitzi Kuroda, Harvard Medical School). To verify the quality of the input and ChIP samples before sequencing, we analyzed the ChIP DNA/input DNA ratio, using real time PCR as described previously [81]. We generated one replicate of MSL1, MOF and MSL2 for each genotype (D. simulans wild type, D. melanogaster wild type and D. melanogaster roX1 roX2 homozygous mutant). Library preparation and AB SOLiD 5500xl sequencing were performed by Uppsala Genome Centre. The MSL1 sample from the D. melanogaster wild type was unfortunately lost. The complete dataset is available at http://www.ncbi.nlm.nih.gov/geo/ (Accession: GSE58768).
ChIP-seq data processing
Uniquely mapped reads from all samples were aligned against the D. melanogaster (Dm) reference sequence (release 5) and D. simulans (Ds) reference sequence (release 1) using the Applied Biosystems Bioscope software v1.2.1. Enrichment ratios for the MSL1, MSL2 and MOF proteins in Dm and Ds wild type and Dm roX1 roX2 mutant samples were calculated as described previously [96]. Ratio values every 10 bp were extracted across the genome and median smoothed using a window size of 500 bp or 2000 bp; windows with fewer than 25 and 100 data points, respectively, were discarded. Since the MSL1 enrichment ratios for the roX1 roX2 mutant samples and the MSL2 enrichment ratios for the wild type samples were the most distinct, these samples were selected as the representative ones in the wild type and roX1 roX2 mutant groups. To define the MSL-bound regions, the highest 1.5 percent of the ratio values were extracted. Data units that crossed this cutoff and that are spaced no more than 200 bp from each other were then combined into MSL-bound regions. Regions of less than 200 bp or containing fewer than five data units were discarded. Each bound region was assigned a value equal to the average of the top five consecutive ratio values. The MSL peak centre for each MSL bound region was set to the centre position of the top five consecutive ratio values.
Distance analysis
The 263 high affinity sites (HAS) as defined in [34], [37], [38] were used to calculate the closest distance to the MSL1-bound regions (defined as above) on the X-chromosome in roX mutants. The distances were divided into 8 bins and the fraction of MSL1-bound sites in each bin was calculated. As a control we used the distance between the HAS and random locations on the X-chromosome.
Motif analysis
In order to search for HAS motif in our ChIP-seq data, 200 bp regions around the centres of peak MSL1 abundance on the X-chromosome in roX mutants were analysed with the MEME program [97] using default parameters. Similar analyses were also performed for the top 200 MSL1-bound regions on the X-chromosome in the Ds wild type.
In order to search for DNA motifs enriched in heterochromatic regions bound by MSL in roX mutants, 200 bp regions around the centres of peak MSL1 abundance on the heterochromatin scaffolds of each chromosome (2LHet, 2RHet, 3LHet, 3RHet, 4Het, XHet) in roX mutants were analysed with the MEME program [97] together with scrambled sequences of binding sites as negative sequences, using default parameters.
Repeat analysis
In order to analyse the repeat content in regions surrounding the expressed genes (defined in [76]) overlapping with MSL1 bound/unbound regions, repeat masked sequences were downloaded from UCSC [98],[99] and the fractions of repeat masked nucleotides in 200 bp windows at 10 bp intervals across the genome were calculated. The percentages of these repeats in 20 bins of 1 kb around the Transcription Start Site (TSS) of MSL1 bound/unbound genes in roX mutants, on autosomes and on the X-chromosome were then calculated. Repeat percentages were also calculated around the TSS of all expressed genes of the X-chromosome and autosomes and around MSL2 bound/unbound expressed genes in the wild type, filtered using a 5 percent highest ratio cutoff on MSL2 enrichment ratio values.
To search for repeat classes enriched in MSL-bound ChIP-seq reads, repeat classes in Dm available from the Repbase Update database (release 19.01) [100] were used. Reads from wild type and roX mutant as well as the corresponding inputs were mapped to different repeat classes using the Bowtie software parameters –a (to map all reads) –v 2 (with two mismatches) [101]. For each repeat class, an RPKM value (Reads Per Kilobase per Million mapped reads) [102] was calculated which was used further to calculate a ratio between ChIP/input in wild type and roX mutants, respectively. The number of reads that mapped to the genome in the original ChIP-seq analysis was used as the number of mapped reads. In each repeat class, read counts per nucleotide was also calculated from wild type and roX mutant as well as input, and normalized to the number of mapped reads (in millions) from each sample.
Supporting Information
Zdroje
1. StenbergP, LarssonJ (2011) Buffering and the evolution of chromosome-wide gene regulation. Chromosoma 120 : 213–225.
2. VicosoB, BachtrogD (2009) Progress and prospects toward our understanding of the evolution of dosage compensation. Chromosome Res 17 : 585–602.
3. MankJE (2013) Sex chromosome dosage compensation: definitely not for everyone. Trends Genet 29 : 677–683.
4. StenbergP, LundbergLE, JohanssonAM, RydénP, SvenssonMJ, et al. (2009) Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet 5: e100302.
5. LundbergLE, FigueiredoML, StenbergP, LarssonJ (2012) Buffering and proteolysis are induced by segmental monosomy in Drosophila melanogaster. Nucleic Acids Res 40 : 5926–5937.
6. ZhangY, MaloneJH, PowellSK, PeriwalV, SpanaE, et al. (2010) Expression in aneuploid Drosophila S2 cells. PLoS Biol 8: e1000320.
7. PrestelM, FellerC, BeckerPB (2010) Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol 11 : 216.
8. GelbartME, KurodaMI (2009) Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136 : 1399–1410.
9. ConradT, AkhtarA (2011) Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet 13 : 123–134.
10. GelbartME, LarschanE, PengS, ParkPJ, KurodaMI (2009) Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol 16 : 825–832.
11. Shogren-KnaakM, IshiiH, SunJM, PazinMJ, DavieJR, et al. (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311 : 844–847.
12. BashawGJ, BakerBS (1995) The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development 121 : 3245–3258.
13. KelleyRL, SolovyevaI, LymanLM, RichmanR, SolovyevV, et al. (1995) Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81 : 867–877.
14. ZhouS, YangY, ScottMJ, PannutiA, FehrKC, et al. (1995) Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J 14 : 2884–2895.
15. MellerVH, WuKH, RomanG, KurodaMI, DavisRL (1997) roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88 : 445–457.
16. RattnerBP, MellerVH (2004) Drosophila male-specific lethal 2 protein controls sex-specific expression of the roX genes. Genetics 166 : 1825–1832.
17. PhilipP, StenbergP (2013) Male X-linked genes in Drosophila melanogaster are compensated independently of the Male-Specific Lethal complex. Epigenetics Chromatin 6 : 35.
18. GuptaV, ParisiM, SturgillD, NuttallR, DoctoleroM, et al. (2006) Global analysis of X-chromosome dosage compensation. J Biol 5 : 3.
19. HamadaFN, ParkPJ, GordadzePR, KurodaMI (2005) Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev 19 : 2289–2294.
20. DengX, MellerVH (2006) roX RNAs are required for increased expression of X-linked genes in Drosophila melanogaster males. Genetics 174 : 1859–1866.
21. BirchlerJA, Pal-BhadraM, BhadraU (2003) Dosage dependent gene regulation and the compensation of the X chromosome in Drosophila males. Genetica 117 : 179–190.
22. VeitiaRA, BottaniS, BirchlerJA (2008) Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet 24 : 390–397.
23. BirchlerJA (2014) Facts and artifacts in studies of gene expression in aneuploids and sex chromosomes. Chromosoma 123 : 459–469.
24. SunL, FernandezHR, DonohueRC, LiJ, ChengJ, et al. (2013) Male-specific lethal complex in Drosophila counteracts histone acetylation and does not mediate dosage compensation. Proc Natl Acad Sci U S A 110 : 7383–7388.
25. LymanLM, CoppsK, RastelliL, KelleyRL, KurodaMI (1997) Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics 147 : 1743–1753.
26. CoppsK, RichmanR, LymanLM, ChangKA, Rampersad-AmmonsJ, et al. (1998) Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. EMBO J 17 : 5409–5417.
27. FauthT, Müller-PlanitzF, KönigC, StraubT, BeckerPB (2010) The DNA binding CXC domain of MSL2 is required for faithful targeting the Dosage Compensation Complex to the X chromosome. Nucleic Acids Res 38 : 3209–3221.
28. KelleyRL, LeeOK, ShimYK (2008) Transcription rate of noncoding roX1 RNA controls local spreading of the Drosophila MSL chromatin remodeling complex. Mech Dev 125 : 1009–1019.
29. ParkSW, KurodaMI, ParkY (2008) Regulation of histone H4 Lys16 acetylation by predicted alternative secondary structures in roX noncoding RNAs. Mol Cell Biol 28 : 4952–4962.
30. ParkSW, KangYIe, SypulaJG, ChoiJ, OhH, et al. (2007) An evolutionarily conserved domain of roX2 RNA is sufficient for induction of H4-Lys16 acetylation on the Drosophila X chromosome. Genetics 177 : 1429–1437.
31. IlikIA, QuinnJJ, GeorgievP, Tavares-CadeteF, MaticzkaD, et al. (2013) Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol Cell 51 : 156–173.
32. MaennerS, MüllerM, FröhlichJ, LangerD, BeckerPB (2013) ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol Cell 51 : 174–184.
33. KelleyRL, MellerVH, GordadzePR, RomanG, DavisRL, et al. (1999) Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98 : 513–522.
34. StraubT, ZabelA, GilfillanGD, FellerC, BeckerPB (2013) Different chromatin interfaces of the Drosophila dosage compensation complex revealed by high-shear ChIP-seq. Genome Res 23 : 473–485.
35. DahlsveenIK, GilfillanGD, ShelestVI, LammR, BeckerPB (2006) Targeting determinants of dosage compensation in Drosophila. PLoS Genet 2: e5.
36. OhH, BoneJR, KurodaMI (2004) Multiple classes of MSL binding sites target dosage compensation to the X chromosome of Drosophila. Curr Biol 14 : 481–487.
37. AlekseyenkoAA, PengS, LarschanE, GorchakovAA, LeeOK, et al. (2008) A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134 : 599–609.
38. StraubT, GrimaudC, GilfillanGD, MitterwegerA, BeckerPB (2008) The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet 4: e1000302.
39. LucchesiJC (1998) Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Curr Opin Genet Dev 8 : 179–184.
40. SassGL, PannutiA, LucchesiJC (2003) Male-specific lethal complex of Drosophila targets activated regions of the X chromosome for chromatin remodeling. Proc Natl Acad Sci U S A 100 : 8287–8291.
41. LarschanE, AlekseyenkoAA, GortchakovAA, PengS, LiB, et al. (2007) MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28 : 121–133.
42. GilfillanGD, StraubT, de WitE, GreilF, LammR, et al. (2006) Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev 20 : 858–870.
43. AlekseyenkoAA, LarschanE, LaiWR, ParkPJ, KurodaMI (2006) High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev 20 : 848–857.
44. LarschanE, AlekseyenkoAA, LaiWR, ParkPJ, KurodaMI (2006) MSL complex associates with clusters of actively transcribed genes along the Drosophila male X chromosome. Cold Spring Harb Symp Quant Biol 71 : 385–394.
45. PhilipP, PetterssonF, StenbergP (2012) Sequence signatures involved in targeting the Male-Specific Lethal complex to X-chromosomal genes in Drosophila melanogaster. BMC Genomics 13 : 97.
46. LucchesiJC (2009) The structure-function link of compensated chromatin in Drosophila. Curr Opin Genet Dev 19 : 550–556.
47. MellerVH, RattnerBP (2002) The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J 21 : 1084–1091.
48. JohanssonAM, AllgardssonA, StenbergP, LarssonJ (2011) msl2 mRNA is bound by free nuclear MSL complex in Drosophila melanogaster. Nucleic Acids Res 39 : 6428–6439.
49. LoheAR, HillikerAJ, RobertsPA (1993) Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 134 : 1149–1174.
50. VicosoB, BachtrogD (2013) Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 499 : 332–335.
51. LarssonJ, MellerVH (2006) Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res 14 : 417–431.
52. Hochman B (1976) The fourth chromosome of Drosophila melanogaster. In: Ashburner M, Novitski E, editors. The Genetics and biology of Drosophila: Academic Press. pp. 903–928.
53. JinY, WangY, WalkerDL, DongH, ConleyC, et al. (1999) JIL-1: a novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol Cell 4 : 129–135.
54. JinY, WangY, JohansenJ, JohansenKM (2000) JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the male specific lethal (MSL) dosage compensation complex. J Cell Biol 149 : 1005–1010.
55. RegnardC, StraubT, MitterwegerA, DahlsveenIK, FabianV, et al. (2011) Global analysis of the relationship between JIL-1 kinase and transcription. PLoS Genet 7: e1001327.
56. WangCI, AlekseyenkoAA, LeroyG, EliaAE, GorchakovAA, et al. (2013) Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat Struct Mol Biol 20 : 202–209.
57. KrivshenkoJD (1955) A cytogenetic study of the X chromosome of Drosophila busckii and its relation to phylogeny. Proc Natl Acad Sci U S A 41 : 1071–1079.
58. KrivshenkoJD (1959) New evidence for the homology of the short euchromatic elements of the X and Y chromosomes of Drosophila busckii with the microchromosome of Drosophila melanogaster. Genetics 44 : 1027–1040.
59. KapitonovVV, JurkaJ (2003) Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci U S A 100 : 6569–6574.
60. St PierreSE, PontingL, StefancsikR, McQuiltonP, ConsortiumF (2014) FlyBase 102–advanced approaches to interrogating FlyBase. Nucleic Acids Res 42: D780–788.
61. BachtrogD, WeissS, ZangerlB, BremG, SchlottererC (1999) Distribution of dinucleotide microsatellites in the Drosophila melanogaster genome. Mol Biol Evol 16 : 602–610.
62. KattiMV, RanjekarPK, GuptaVS (2001) Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol 18 : 1161–1167.
63. PardueML, LowenhauptK, RichA, NordheimA (1987) (dC-dA)n.(dG-dT)n sequences have evolutionarily conserved chromosomal locations in Drosophila with implications for roles in chromosome structure and function. EMBO J 6 : 1781–1789.
64. DorerDR, HenikoffS (1994) Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell 77 : 993–1002.
65. LiY, DanzerJR, AlvarezP, BelmontAS, WallrathLL (2003) Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development 130 : 1817–1824.
66. MaennerS, MüllerM, BeckerPB (2012) Roles of long, non-coding RNA in chromosome-wide transcription regulation: lessons from two dosage compensation systems. Biochimie 94 : 1490–1498.
67. FilionGJ, van BemmelJG, BraunschweigU, TalhoutW, KindJ, et al. (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143 : 212–224.
68. DengX, RattnerBP, SouterS, MellerVH (2005) The severity of roX1 mutations is predicted by MSL localization on the X chromosome. Mech Dev 122 : 1094–1105.
69. MenonDU, MellerVH (2009) Imprinting of the Y chromosome influences dosage compensation in roX1 roX2 Drosophila melanogaster. Genetics 183 : 811–820.
70. HoskinsRA, SmithCD, CarlsonJW, CarvalhoAB, HalpernA, et al. (2002) Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol 3: RESEARCH0085.
71. GrimaudC, BeckerPB (2009) The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. Genes Dev 23 : 2490–2495.
72. DemakovaOV, KotlikovaIV, GordadzePR, AlekseyenkoAA, KurodaMI, et al. (2003) The MSL complex levels are critical for its correct targeting to the chromosomes in Drosophila melanogaster. Chromosoma 112 : 103–115.
73. KharchenkoPV, AlekseyenkoAA, SchwartzYB, MinodaA, RiddleNC, et al. (2011) Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471 : 480–485.
74. RiddleNC, MinodaA, KharchenkoPV, AlekseyenkoAA, SchwartzYB, et al. (2011) Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res 21 : 147–163.
75. YinH, SweeneyS, RahaD, SnyderM, LinH (2011) A high-resolution whole-genome map of key chromatin modifications in the adult Drosophila melanogaster. PLoS Genet 7: e1002380.
76. FigueiredoML, PhilipP, StenbergP, LarssonJ (2012) HP1a recruitment to promoters is independent of H3K9 methylation in Drosophila melanogaster. PLoS Genet 8: e1003061.
77. de WitE, GreilF, van SteenselB (2005) Genome-wide HP1 binding in Drosophila: developmental plasticity and genomic targeting signals. Genome Res 15 : 1265–1273.
78. LiuLP, NiJQ, ShiYD, OakeleyEJ, SunFL (2005) Sex-specific role of Drosophila melanogaster HP1 in regulating chromatin structure and gene transcription. Nat Genet 37 : 1361–1366.
79. PrestelM, FellerC, StraubT, MitlöhnerH, BeckerPB (2010) The activation potential of MOF is constrained for dosage compensation. Mol Cell 38 : 815–826.
80. JohanssonAM, StenbergP, BernhardssonC, LarssonJ (2007) Painting of fourth and chromosome-wide regulation of the 4th chromosome in Drosophila melanogaster. EMBO J 26 : 2307–2316.
81. JohanssonAM, StenbergP, PetterssonF, LarssonJ (2007) POF and HP1 bind expressed exons, suggesting a balancing mechanism for gene regulation. PLoS Genet 3: e209.
82. LarssonJ, ChenJD, RashevaV, Rasmuson LestanderA, PirrottaV (2001) Painting of fourth, a chromosome-specific protein in Drosophila. Proc Natl Acad Sci U S A 98 : 6273–6278.
83. LyonMF (1998) X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet 80 : 133–137.
84. CoelhoPA, Queiroz-MachadoJ, HartlD, SunkelCE (1998) Pattern of chromosomal localization of the Hoppel transposable element family in the Drosophila melanogaster subgroup. Chromosome Res 6 : 385–395.
85. EllisonCE, BachtrogD (2013) Dosage compensation via transposable element mediated rewiring of a regulatory network. Science 342 : 846–850.
86. ReaS, XouriG, AkhtarA (2007) Males absent on the first (MOF): from flies to humans. Oncogene 26 : 5385–5394.
87. LiX, WuL, CorsaCA, KunkelS, DouY (2009) Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell 36 : 290–301.
88. AvnerP, HeardE (2001) X-chromosome inactivation: counting, choice and initiation. Nat Rev Genet 2 : 59–67.
89. DorerDR, HenikoffS (1997) Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics 147 : 1181–1190.
90. de VanssayA, BougéAL, BoivinA, HermantC, TeyssetL, et al. (2012) Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 490 : 112–115.
91. GrothAC, OlivaresEC, ThyagarajanB, CalosMP (2000) A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci U S A 97 : 5995–6000.
92. LundbergLE, KimM, JohanssonAM, FaucillionML, JosupeitR, et al. (2013) Targeting of Painting of fourth to roX1 and roX2 proximal sites suggests evolutionary links between dosage compensation and the regulation of the fourth chromosome in Drosophila melanogaster. G3 (Bethesda) 3 : 1325–1334.
93. Sullivan W, Ashburner M, Hawley RS (2000) Drosophila protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
94. JohanssonAM, StenbergP, AllgardssonA, LarssonJ (2012) POF regulates the expression of genes on the fourth chromosome in Drosophila melanogaster by binding to nascent RNA. Mol Cell Biol 32 : 2121–2134.
95. LavrovS, DéjardinJ, CavalliG (2004) Combined immunostaining and FISH analysis of polytene chromosomes. Methods Mol Biol 247 : 289–303.
96. HolmqvistPH, BoijaA, PhilipP, CronaF, StenbergP, et al. (2012) Preferential genome targeting of the CBP co-activator by Rel and Smad proteins in early Drosophila melanogaster embryos. PLoS Genet 8: e1002769.
97. BaileyTL, ElkanC (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2 : 28–36.
98. AdamsMD, CelnikerSE, HoltRA, EvansCA, GocayneJD, et al. (2000) The genome sequence of Drosophila melanogaster. Science 287 : 2185–2195.
99. CelnikerSE, WheelerDA, KronmillerB, CarlsonJW, HalpernA, et al. (2002) Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol 3: RESEARCH0079.
100. JurkaJ, KapitonovVV, PavlicekA, KlonowskiP, KohanyO, et al. (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110 : 462–467.
101. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25.
102. MortazaviA, WilliamsBA, McCueK, SchaefferL, WoldB (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5 : 621–628.
Štítky
Genetika Reprodukčná medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 12- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



