-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Is Antigenic Sin Always “Original?” Re-examining the Evidence Regarding Circulation of a Human H1 Influenza Virus Immediately Prior to the 1918 Spanish Flu
article has not abstract
Published in the journal: Is Antigenic Sin Always “Original?” Re-examining the Evidence Regarding Circulation of a Human H1 Influenza Virus Immediately Prior to the 1918 Spanish Flu. PLoS Pathog 11(3): e32767. doi:10.1371/journal.ppat.1004615
Category: Opinion
doi: https://doi.org/10.1371/journal.ppat.1004615Summary
article has not abstract
What makes the 1918 Spanish influenza pandemic stand out from all the others is its well-known W-shaped mortality signature, which was caused by unusually high mortality among adults aged 20 to 40 [1]. Much debate remains as to the exact reason for this atypical pattern [2]. A contribution by Worobey et al. [3] published recently in the Proceedings of the National Academy of Sciences (PNAS) is no doubt adding important information to this debate.
In agreement with previous work [4–7], Worobey et al. propose that the very high mortality experienced by young adults during the 1918 H1N1 virus pandemic was primarily due to their childhood exposure to the heterosubtypic H3-like virus that is thought to have caused the earlier 1889–1890 Russian flu pandemic [3]. As is generally accepted, the authors also presume that older adults had immunological cross-protection from earlier exposures to a putative H1-like virus, which circulated prior the 1890 pandemic. As for the lower mortality of children and adolescents, however, a new and compelling hypothesis is put forward: this pattern may be attributed to the appearance of a new H1 influenza variant in the early 1900s, which would have provided protection in 1918 for individuals born at the turn of the century, presumably exposed early in life (or “primed”) to this new variant. They propose that this H1N8 virus arose from reassortment between an H1 lineage virus and an avian influenza virus sometime between 1901 and 1907, replacing the H3N8 virus of the 1889–1890 pandemic. This phylogenetic reconstitution appears to be supported by (often forgotten) seroarcheological and mortality evidence.
Did H1 Really Replace H3 in the Early 1900s?
Although the phylogenetic analysis presented by Worobey et al. is quite appealing, the results gathered from the seroarcheological literature that they abundantly cite do not always support it [3]. Most notably, it is not immediately clear that all seroarcheology and mortality data point to the swift replacement of the 1890 H3 virus by an H1 variant at the turn of the 20th century. Furthermore, any seroarcheological data gathered to measure responses to viruses for which isolates are not available must be interpreted cautiously.
Using old serological studies from Masurel [8,9] adapted here in Fig. 1, other investigators such as Dowdle [10] deduced earlier that about half of those born a few years after the 1890 pandemic, i.e., in 1893, were “primed” to the H3 virus that caused that pandemic while the other half would have been primed to H1N1 during the 1918 pandemic. According to Worobey et al., this is highly unlikely because it would mean that many members of this cohort fully escaped all influenza virus infections for a period of about 25 years [3]. However, Dowdle’s explanation for the H1 seroacheological data is only problematic if we adhere to a historical interpretation of “original antigenic sin” by assuming that the highest antibody titres in a birth cohort systematically reflect the antigens of the earliest childhood exposure to influenza virus and that, as a consequence, seroarcheological studies invariably reveal the identity of the first strain of influenza virus to which each cohort were exposed.
Fig. 1. Death rate ratios from pneumonia and influenza (P&I) during the 1968–1969 pandemic and percent without antibody titers against H1 and H3 influenza viruses. 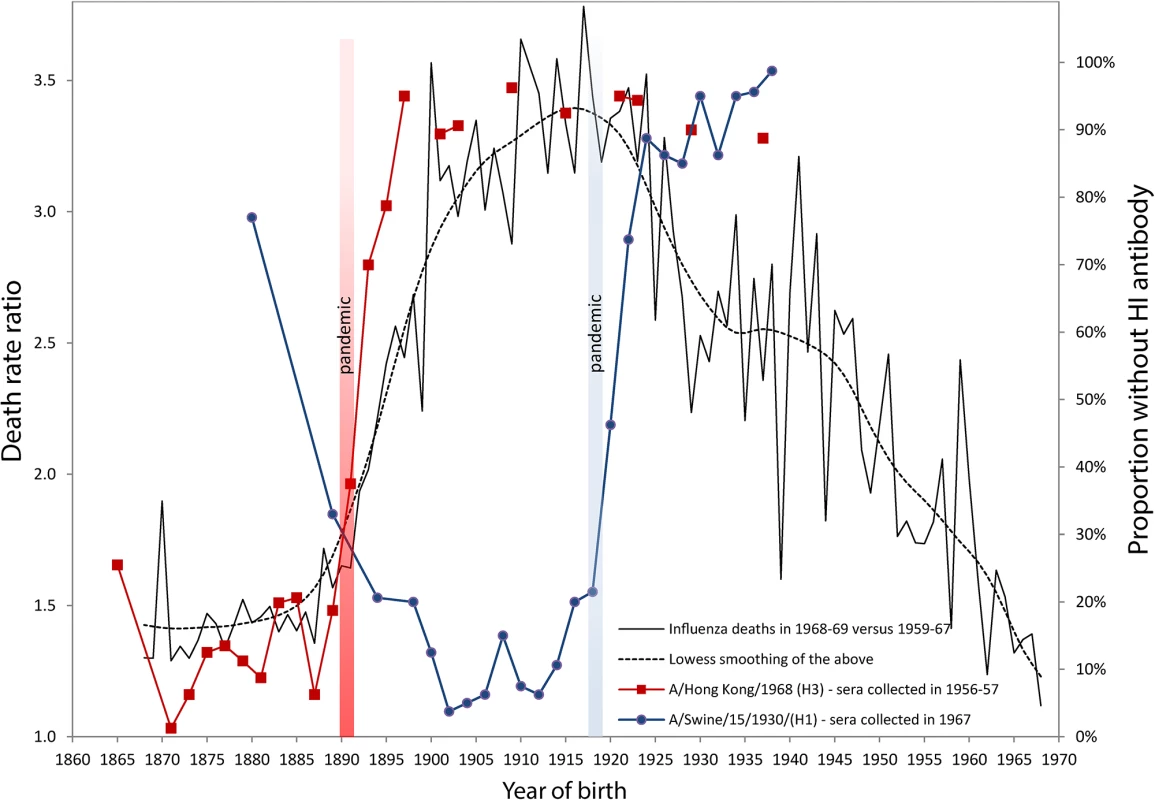
Serological data on H1 and H3 viruses were adapted from Masurel [8,9,25] and Doodle et al. [10]. Years of birth were deduced from data originally presented by age by Masurel (HI > 9–19) [8,9,25]. We used life tables from the Netherland Bureau of Statistics, available in the Human Life-Table Database [26], in order to estimate average ages (and years of birth) for donor sera grouped in age bins larger than five years. The death rate ratio by age (or by birth cohort) in 1968 was estimated by dividing the P&I death rate calculated for the December 1968 to January 1969 pandemic flu season [27] with the average P&I death rates of the same seasons from 1959–1960 to 1967–1968. These ratios represent the increase of mortality due to the 1968 pandemic (a ratio of 1.5 means a mortality increment of 50% in comparison with the previous ten-year’s average). Monthly P&I death counts were taken from the National Center for Health Statistics, available on the National Bureau of Economic Research website [28], while the populations exposed to risk were interpolated from the Human Mortality Database [29]. In order to account for secular mortality improvements, we detrended the time series from 1959 to 1968 with quadratic regressions of mortality rates based on the 20 epidemic seasons from 1959 to 1978, excluding the 1968–1969 pandemic season. This assumption is not always sound [2], as can be readily observed in Fig. 1, which shows that cohorts born between 1863 and 1890 all had a high percentage of individuals with detectable antibodies against the A/Hong Kong/68 (H3) strain. About 80% to 90% of sera collected in 1956–1957 for these cohorts contained HI antibodies against this strain (red line with squares). The maximum proportion, virtually 100%, occurs for those born in 1871, i.e., almost 20 years before the 1890 pandemic. If it is true that the antibody signature resulting from the first influenza virus infection during one’s lifetime is hierarchically programmed into the immunological repertoire of a cohort, then we are forced to suppose that individuals born in the 1860s or 1870s escaped exposure and infection to the putative H1-variant that circulated prior to the 1890 pandemic for a very long period. This is, indeed, highly unlikely. Similarly, the proportion of individuals with antibodies against A/Swine/15/1930/(H1) is over 90%–95% in Fig. 1 for cohorts born long before the appearance of the strain in 1918 (blue line with circles). This trend was also observed recently in sera collected just prior to the 2009 H1N1 pandemic. Those born during the first two decades of the 20th century, presumably exposed early in life to the 1918 Spanish Flu pandemic, had high neutralizing antibody titers against the 2009 H1N1 virus. These titers dropped sharply for those born after 1918, who were exposed to the antigenically distinct “human” H1N1 viruses that circulated from the 1920s to the 1950s [11]. Other independent studies have also reported similar sudden drops in antibody titers against the H3 pandemic and the H1 swine strains [12,13].
For Enhanced Protection in the Future, Get Your “Pandemic Flu Shot” Now
The sudden increase of the percentage of people born after 1890 with no detectable antibody titers to the H3 virus shown in Fig. 1 would seem to be too steep and too tightly associated with the 1890 pandemic to be interpreted as a sign that H3 was replaced by H1 in 1900 as the first exposure strain for those born in the 1890s. What these seroarcheological observations might instead suggest is that exposure to a pandemic strain any time before ~20 years of age can “reprogram” the antibody repertoire by inducing the most robust antibody titers against the pandemic virus, at the expense of any previously encountered non-pandemic strain, whether from the same subtype or not. This would evidently be sufficient to provide increased protection in a subsequent outbreak of the same subtype [10]. The degree of protection would then rapidly fall for cohorts born right after the pandemic, as the virus drifts and becomes seasonal.
Our own analyses of mortality from P&I during the 1968 pandemic are consistent with this “immunological refocusing” scenario, occurring for those exposed to a pandemic strain at a sufficiently young age. We calculated and added to Fig. 1 the death rates ratios of P&I mortality during the 1968–1969 Hong Kong pandemic relative to the average P&I mortality during the years 1959–1967 in the United States. It is clear that people born between 1878 and 1890 had the lowest mortality increments during that pandemic. The relative risk of mortality sharply increased for those who were born immediately after the 1890 pandemic, in concert with the increase in the percentage of individuals who had no detectable HI antibodies to H3N2.
Individuals born up to 20–25 years before the 1890 pandemic all had about the same protection against the H3N2 virus of the 1968 Hong Kong pandemic. While persons born around 1870 might very well have first “committed” to an H1-like variant early in life, most of the sera collected years later (in 1956–1957) from these people contained large amounts of antibodies to the H3 virus, to which they were exposed in the early 1890s (20 years after their birth) and which offered them substantial protection about 80 years later, during the 1968 outbreak. These results on mortality risk ratios are consistent with a study from Simonsen et al. [14] who found that the risk of influenza-related mortality among the elderly aged 75+ did not increase during the 1968 pandemic relative to the 1975–1976 and 1980–1981 influenza seasons, which were relatively severe. Intriguingly, death rates ratios in 1968 as summarized by the lowess smoothing in Fig. 1 attain a maximum for the cohorts born just before the 1918 pandemic, as if being born during pandemic years carried an increased risk of mortality to a subsequent pandemic caused by a heterosubtypic influenza virus.
Refining the “Original Antigenic Sin” Doctrine
Implicit in Worobey et al.’s scenario is the notion that the first antigenic variant encountered during childhood systematically conditions immunity for the rest of someone’s life [3]. This phenomenon, referred to as “original antigenic sin” (OAS), has been described since the early 1950s, when sequential exposures to drifting variants of the H1N1 subtype seemed to induce more neutralizing antibody titers against the childhood variant than against the contemporary circulating strain of the same subtype [15].
Recent studies have challenged the historical mechanistic implications of the OAS model [16–18], and have proposed the term “antigenic seniority” as a more apt description for the hierarchical nature of antibody responses to previously encountered influenza virus strains. Indeed, the term “OAS” itself has often been used in the literature to describe phenomena related to immunological memory that are not directly linked to the hierarchical responses to influenza virus (as in [14]). To avoid confusion, other research has also proposed a more general conception of antigenic imprinting [19] that would cover all instances of ‘‘commitment” to the strain of first exposure, including sequences involving heterosubtypic pandemic strains like those that caused the 1890 and the 1918 pandemics [5].
It was recently found and confirmed in various locations that mortality during the 1918 pandemic peaked at the exact age of 28 [5,6,20,21] (but see [7,22]), which corresponds to a birth year of 1890 (i.e., during the Russian flu pandemic). To account for this striking regularity, it has been speculated that the development of immunological memory to an influenza virus strain early in life may lead to a dysregulated immune response when encountering a novel and highly antigenically dissimilar strain later in life [5]. For example, encounter with the 1889–1890 H3 virus very early in life would have resulted in robust cytotoxic T cell memory, which, without the complement of cross-protective antibodies between H3 and H1, may have caused immunopathology when recalled upon infection during the 1918 pandemic, resulting in increased risks of death. Those born later in the 1890 decade were primed to progressively drifted and less virulent strains of the H3 virus. This would have decreased the magnitude of the cytotoxic T cell memory response and, thus, lowered the potential for immunopathology in 1918. Similarly, in this study, mortality increments during the 1968 pandemic peaked for those born around one year before the 1918 pandemic, and decreased for those born in the following years. It is worth noting that despite the fact that no new H3-like variant appeared between 1918 and 1968, death rates ratios in 1968 dropped for those born after 1918. In this case, there was no need for circulation of a novel H3 virus to account for this drop, as Worobey et al.’s scenario for the 1918 pandemic would imply if it was applied to the 1968 case [3].
The specific scenarios that result in OAS-like antibody responses are complex, and likely require further refinement (i.e., distinguishing sequences of infections from seasonal virus to pandemic virus, and vice-versa, or sequences involving heterosubtypic pandemic strains). Unfortunately, the disproportionate protection against heterosubtypic infection afforded by T cells in mice makes recapitulating the effects of these exposure scenarios difficult in the mouse model [23,24], and the tools required to study the contribution of specific cell types in ferrets remain lacking. We believe that a detailed understanding of these scenarios will be essential if we ever hope to understand how previous exposures to influenza virus are likely to shape the outcome of future pandemics. More importantly, this knowledge may be critical in the design and implementation of immunization campaigns that are “personalized” with regard to age and exposure history.
Zdroje
1. Luk J, Gross P, Thompson WW (2001) Observations on mortality during the 1918 influenza pandemic. Clin Infect Dis 33(8):1375–8. 11565078
2. Morens DM, Taubenberger JK (2012) 1918 influenza, a puzzle with missing pieces. Emerg Infect Dis 18(2):332–335. doi: 10.3201/eid1802.111409 22304897
3. Worobey M, Han G-Z, Rambaut A (2014) Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci:201324197.
4. Shanks GD, Brundage JF (2012) Pathogenic responses among young adults during the 1918 influenza pandemic. Emerg Infect Dis 18(2):201–207. doi: 10.3201/eid1802.102042 22306191
5. Gagnon A, Miller M, Hallman S, Bourbeau R, Herring A, et al. (2013) Age-Specific Mortality During the 1918 Influenza Pandemic: Unravelling the Mystery of High Young Adult Mortality. PLoS ONE 8(8):e69586. doi: 10.1371/journal.pone.0069586 23940526
6. Hallman S, Gagnon A (2014) Does Exposure to Influenza Very Early in Life Affect Mortality Risk during a Subsequent Outbreak? The 1890 and 1918 Pandemics in Canada. Modern Environments and Human Health, ed Zuckerman MK (John Wiley & Sons, Inc), pp 123–135. http://onlinelibrary.wiley.com/doi/10.1002/9781118504338.ch7/summary [Accessed August 14, 2014].
7. Oeppen J, Wilson C (2006) Epidemiological evidence for viral exposure in childhood as a risk-factor in subsequent influenza pandemics. Population Association of America, Los Angeles, March 30-April 1 2006.
8. Masurel N (1969) Serological characteristics of a “new” serotype of influenza A virus: the Hong Kong strain. Bull World Health Organ 41(3):461–468. 5309456
9. Masurel N (1976) Swine influenza virus and the recycling of influenza-A viruses in man. Lancet 2(7979):244–247. 59252
10. Dowdle WR (1999) Influenza A virus recycling revisited. Bull World Health Organ 77(10):820–828. 10593030
11. Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, et al. (2009) In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460(7258):1021–1025. doi: 10.1038/nature08260 19672242
12. Fukumi H (1969) Interpretation of influenza antibody patterns in man. Bull World Health Organ 41(3–4–5):469–473.
13. Davenport FM, Hennessy AV, Drescher J, Mulder J, Francis T (1964) Futher observations on the relevance of serologic recapitulations of human infection with influenza viruses. J Exp Med 120 : 1087–1097. 14238927
14. Simonsen L, Reichert TA, Miller MA (2004) The virtues of antigenic sin: consequences of pandemic recycling on influenza-associated mortality. Int Congr Ser 1263 : 791–794.
15. Francis T Jr (1960) On the Doctrine of Original Antigenic Sin. Proc Am Philos Soc 104(6):572–578.
16. O’Donnell CD, Wright A, Vogel L, Boonnak K, Treanor JJ, et al. (2014) Humans and ferrets with prior H1N1 influenza virus infections do not exhibit evidence of original antigenic sin after infection or vaccination with the 2009 pandemic H1N1 influenza virus. Clin Vaccine Immunol CVI 21(5):737–746. doi: 10.1128/CVI.00790-13 24648486
17. Lessler J, Riley S, Read JM, Wang S, Zhu H, et al. (2012) Evidence for Antigenic Seniority in Influenza A (H3N2) Antibody Responses in Southern China. PLoS Pathog 8(7):e1002802. doi: 10.1371/journal.ppat.1002802 22829765
18. Miller MS, Gardner TJ, Krammer F, Aguado LC, Tortorella D, et al. (2013) Neutralizing antibodies against previously encountered influenza virus strains increase over time: a longitudinal analysis. Sci Transl Med 5(198):198ra107.
19. Ma J, Dushoff J, Earn DJD (2011) Age-specific mortality risk from pandemic influenza. J Theor Biol 288 : 29–34. doi: 10.1016/j.jtbi.2011.08.003 21856313
20. Yang W, Petkova E, Shaman J (2014) The 1918 influenza pandemic in New York City: age-specific timing, mortality, and transmission dynamics. Influenza Other Respir Viruses 8(2):177–188. doi: 10.1111/irv.12217 24299150
21. Wilson N, Oliver J, Rice G, Summers JA, Baker MG, et al. (2014) Age-Specific Mortality During the 1918–19 Influenza Pandemic and Possible Relationship to the 1889–92 Influenza Pandemic. J Infect Dis:jiu191.
22. Viboud C, Eisenstein J, Reid A, Janczewski T, Morens D, et al. (2012) Age and gender mortality profile associated with the 1918–19 influenza pandemic in Kentucky, USA. J Infect Dis. http://jid.oxfordjournals.org/content/early/2012/12/10/infdis.jis745.long.
23. Reiss CS, Schulman JL (1980) Influenza type A virus M protein expression on infected cells is responsible for cross-reactive recognition by cytotoxic thymus-derived lymphocytes. Infect Immun 29(2):719–723. 7011983
24. Schulman JL, Petigrow C, Woodruff J (1977) Effects of cell mediated immunity in influenza virus infection in mice. Dev Biol Stand 39 : 385–390. 304820
25. Masurel N (1969) Relation between Hong Kong virus and former human A2 isolates and the A/Equi2 virus in human sera collected before 1957. The Lancet 293(7601):907–910. 4180894
26. Human Life-Table Database. http://www.lifetable.de [Accessed November 20, 2014].
27. Flu.gov Pandemic Flu History. http://www.flu.gov/pandemic/history/ [Accessed July 21, 2014].
28. NBER DID Mortality Data. http://www.nber.org/data/vital-statistics-mortality-data-multiple-cause-of-death.html [Accessed January 14, 2014].
29. Human Mortality Database The United States of America, Exposure to risk (period 1x1). http://www.mortality.org/hmd/USA/STATS/Exposures_1x1.txt [Accessed July 21, 2014].
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole MembraneČlánek Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and FunctionČlánek Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2015 Číslo 3- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- To Be or Not IIb: A Multi-Step Process for Epstein-Barr Virus Latency Establishment and Consequences for B Cell Tumorigenesis
- Is Antigenic Sin Always “Original?” Re-examining the Evidence Regarding Circulation of a Human H1 Influenza Virus Immediately Prior to the 1918 Spanish Flu
- The Great Escape: Pathogen Versus Host
- Coping with Stress and the Emergence of Multidrug Resistance in Fungi
- Catch Me If You Can: The Link between Autophagy and Viruses
- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- Evidence for Ubiquitin-Regulated Nuclear and Subnuclear Trafficking among Matrix Proteins
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Production of Anti-LPS IgM by B1a B Cells Depends on IL-1β and Is Protective against Lung Infection with LVS
- Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS
- α-Hemolysin Counteracts the Anti-Virulence Innate Immune Response Triggered by the Rho GTPase Activating Toxin CNF1 during Bacteremia
- Induction of Interferon-Stimulated Genes by IRF3 Promotes Replication of
- Intracellular Growth Is Dependent on Tyrosine Catabolism in the Dimorphic Fungal Pathogen
- HCV Induces the Expression of Rubicon and UVRAG to Temporally Regulate the Maturation of Autophagosomes and Viral Replication
- Spatiotemporal Analysis of Hepatitis C Virus Infection
- Subgingival Microbial Communities in Leukocyte Adhesion Deficiency and Their Relationship with Local Immunopathology
- Interaction between the Type III Effector VopO and GEF-H1 Activates the RhoA-ROCK Pathway
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
- Establishment of HSV1 Latency in Immunodeficient Mice Facilitates Efficient Reactivation
- XRN1 Stalling in the 5’ UTR of Hepatitis C Virus and Bovine Viral Diarrhea Virus Is Associated with Dysregulated Host mRNA Stability
- γδ T Cells Confer Protection against Murine Cytomegalovirus (MCMV)
- Rhadinovirus Host Entry by Co-operative Infection
- A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole Membrane
- Dermal Neutrophil, Macrophage and Dendritic Cell Responses to Transmitted by Fleas
- Elucidation of Sigma Factor-Associated Networks in Reveals a Modular Architecture with Limited and Function-Specific Crosstalk
- A Conserved NS3 Surface Patch Orchestrates NS2 Protease Stimulation, NS5A Hyperphosphorylation and HCV Genome Replication
- Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and Function
- Disruption of IL-21 Signaling Affects T Cell-B Cell Interactions and Abrogates Protective Humoral Immunity to Malaria
- Compartmentalized Replication of R5 T Cell-Tropic HIV-1 in the Central Nervous System Early in the Course of Infection
- Diminished Reovirus Capsid Stability Alters Disease Pathogenesis and Littermate Transmission
- Characterization of CD8 T Cell Differentiation following SIVΔnef Vaccination by Transcription Factor Expression Profiling
- Visualization of HIV-1 Interactions with Penile and Foreskin Epithelia: Clues for Female-to-Male HIV Transmission
- Sensing Cytosolic RpsL by Macrophages Induces Lysosomal Cell Death and Termination of Bacterial Infection
- PKCη/Rdx-driven Phosphorylation of PDK1: A Novel Mechanism Promoting Cancer Cell Survival and Permissiveness for Parvovirus-induced Lysis
- Metalloprotease NleC Suppresses Host NF-κB/Inflammatory Responses by Cleaving p65 and Interfering with the p65/RPS3 Interaction
- Immune Antibodies and Helminth Products Drive CXCR2-Dependent Macrophage-Myofibroblast Crosstalk to Promote Intestinal Repair
- Adenovirus Entry From the Apical Surface of Polarized Epithelia Is Facilitated by the Host Innate Immune Response
- The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family
- Neutrophils: Between Host Defence, Immune Modulation, and Tissue Injury
- CD169-Mediated Trafficking of HIV to Plasma Membrane Invaginations in Dendritic Cells Attenuates Efficacy of Anti-gp120 Broadly Neutralizing Antibodies
- Japanese Encephalitis Virus Nonstructural Protein NS5 Interacts with Mitochondrial Trifunctional Protein and Impairs Fatty Acid β-Oxidation
- Yip1A, a Novel Host Factor for the Activation of the IRE1 Pathway of the Unfolded Protein Response during Infection
- TRIM26 Negatively Regulates Interferon-β Production and Antiviral Response through Polyubiquitination and Degradation of Nuclear IRF3
- Parallel Epigenomic and Transcriptomic Responses to Viral Infection in Honey Bees ()
- A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication
- Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
- Exome and Transcriptome Sequencing of Identifies a Locus That Confers Resistance to and Alters the Immune Response
- The Role of Misshapen NCK-related kinase (MINK), a Novel Ste20 Family Kinase, in the IRES-Mediated Protein Translation of Human Enterovirus 71
- Chitin Recognition via Chitotriosidase Promotes Pathologic Type-2 Helper T Cell Responses to Cryptococcal Infection
- Activates Both IL-1β and IL-1 Receptor Antagonist to Modulate Lung Inflammation during Pneumonic Plague
- Persistence of Transmitted HIV-1 Drug Resistance Mutations Associated with Fitness Costs and Viral Genetic Backgrounds
- An 18 kDa Scaffold Protein Is Critical for Biofilm Formation
- Early Virological and Immunological Events in Asymptomatic Epstein-Barr Virus Infection in African Children
- Human CD8 T-cells Recognizing Peptides from () Presented by HLA-E Have an Unorthodox Th2-like, Multifunctional, Inhibitory Phenotype and Represent a Novel Human T-cell Subset
- Decreased HIV-Specific T-Regulatory Responses Are Associated with Effective DC-Vaccine Induced Immunity
- RSV Vaccine-Enhanced Disease Is Orchestrated by the Combined Actions of Distinct CD4 T Cell Subsets
- Concerted Activity of IgG1 Antibodies and IL-4/IL-25-Dependent Effector Cells Trap Helminth Larvae in the Tissues following Vaccination with Defined Secreted Antigens, Providing Sterile Immunity to Challenge Infection
- Structure of the Low pH Conformation of Chandipura Virus G Reveals Important Features in the Evolution of the Vesiculovirus Glycoprotein
- PPM1A Regulates Antiviral Signaling by Antagonizing TBK1-Mediated STING Phosphorylation and Aggregation
- Lipidomic Analysis Links Mycobactin Synthase K to Iron Uptake and Virulence in .
- Roles and Programming of Arabidopsis ARGONAUTE Proteins during Infection
- Impact of Infection on Host Macrophage Nuclear Physiology and Nucleopore Complex Integrity
- The Impact of Host Diet on Titer in
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Herpesviral G Protein-Coupled Receptors Activate NFAT to Induce Tumor Formation via Inhibiting the SERCA Calcium ATPase
- The Causes and Consequences of Changes in Virulence following Pathogen Host Shifts
- Small GTPase Rab21 Mediates Fibronectin Induced Actin Reorganization in : Implications in Pathogen Invasion
- Positive Role of Promyelocytic Leukemia Protein in Type I Interferon Response and Its Regulation by Human Cytomegalovirus
- NEDDylation Is Essential for Kaposi’s Sarcoma-Associated Herpesvirus Latency and Lytic Reactivation and Represents a Novel Anti-KSHV Target
- β-HPV 5 and 8 E6 Disrupt Homology Dependent Double Strand Break Repair by Attenuating BRCA1 and BRCA2 Expression and Foci Formation
- An O Antigen Capsule Modulates Bacterial Pathogenesis in
- Variable Processing and Cross-presentation of HIV by Dendritic Cells and Macrophages Shapes CTL Immunodominance and Immune Escape
- Probing the Metabolic Network in Bloodstream-Form Using Untargeted Metabolomics with Stable Isotope Labelled Glucose
- Adhesive Fiber Stratification in Uropathogenic Biofilms Unveils Oxygen-Mediated Control of Type 1 Pili
- Vaccinia Virus Protein Complex F12/E2 Interacts with Kinesin Light Chain Isoform 2 to Engage the Kinesin-1 Motor Complex
- Modulates Host Macrophage Mitochondrial Metabolism by Hijacking the SIRT1-AMPK Axis
- Human T-Cell Leukemia Virus Type 1 (HTLV-1) Tax Requires CADM1/TSLC1 for Inactivation of the NF-κB Inhibitor A20 and Constitutive NF-κB Signaling
- Suppression of RNAi by dsRNA-Degrading RNaseIII Enzymes of Viruses in Animals and Plants
- Spatiotemporal Regulation of a T4SS Substrate by the Metaeffector SidJ
- Antigenic Properties of the Human Immunodeficiency Virus Envelope Glycoprotein Gp120 on Virions Bound to Target Cells
- Dependence of Intracellular and Exosomal microRNAs on Viral Oncogene Expression in HPV-positive Tumor Cells
- Identification of a Peptide-Pheromone that Enhances Escape from Host Cell Vacuoles
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Dihydrobiopterin in Adult Falciparum Malaria: Association with Disease Severity, Impaired Microvascular Function and Increased Endothelial Activation
- Transgenic Expression of the Dicotyledonous Pattern Recognition Receptor EFR in Rice Leads to Ligand-Dependent Activation of Defense Responses
- Comprehensive Antigenic Map of a Cleaved Soluble HIV-1 Envelope Trimer
- Low Doses of Imatinib Induce Myelopoiesis and Enhance Host Anti-microbial Immunity
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Oxidized Biopterins in Pediatric Falciparum Malaria: Association with Disease Severity
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



