-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
Abscisic acid (ABA) plays crucial roles in plant growth and development, and also in plant responses to abiotic and biotic stresses. ABA can stimulate the production of reactive oxygen species (ROS) that act as signals in low concentrations, but as cell-damaging agents in high concentrations. A mutation in ABO8, encoding a pentatricopeptide repeat (PPR) protein responsible for the splicing of NAD4 intron 3, leads to hypersensitivity to ABA in root growth, and root tips of the abo8-1 mutants accumulate more ROS than those of the wild type; this accumulation of ROS in abo8-1 root tips is enhanced by ABA treatment. We also found that auxin signaling and/or accumulation is greatly reduced in root tips of the abo8-1 mutants. Addition of the reducing agent GSH to the growth medium partially recovers the root hypersensitivity to ABA, and also the ABA-inhibited expression of PLT1/2 in abo8-1. Furthermore, the inducible expression of PLT2 largely rescues the root growth defect of abo8-1 with and without ABA treatment. Our results reveal the important roles of ROS in regulating root meristem activity in the ABA signaling pathway.
Published in the journal: ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in. PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004791
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004791Summary
Abscisic acid (ABA) plays crucial roles in plant growth and development, and also in plant responses to abiotic and biotic stresses. ABA can stimulate the production of reactive oxygen species (ROS) that act as signals in low concentrations, but as cell-damaging agents in high concentrations. A mutation in ABO8, encoding a pentatricopeptide repeat (PPR) protein responsible for the splicing of NAD4 intron 3, leads to hypersensitivity to ABA in root growth, and root tips of the abo8-1 mutants accumulate more ROS than those of the wild type; this accumulation of ROS in abo8-1 root tips is enhanced by ABA treatment. We also found that auxin signaling and/or accumulation is greatly reduced in root tips of the abo8-1 mutants. Addition of the reducing agent GSH to the growth medium partially recovers the root hypersensitivity to ABA, and also the ABA-inhibited expression of PLT1/2 in abo8-1. Furthermore, the inducible expression of PLT2 largely rescues the root growth defect of abo8-1 with and without ABA treatment. Our results reveal the important roles of ROS in regulating root meristem activity in the ABA signaling pathway.
Introduction
Plant growth and development are greatly influenced by the accumulation of reactive oxygen species (ROS), which are produced under environmental stresses such as water deficiency and high salinity [1], [2], [3], [4]. Although hazardous to cells at high concentrations, ROS at low concentrations are important signaling molecules that regulate stomatal movement, prevent pathogen invasion, promote programmed cell death, and redirect plant growth [5], [6], [7], [8]. In addition to linking with stress hormones such as ethylene, salicylic acid, jasmonic acid, and abscisic acid (ABA) [9], ROS also have cross-talk with growth hormones such as gibberellin [10], [11], cytokinins and auxin [1], [4].
The connection between ROS and auxin has been explored in the regulation of development of the root apical meristem. Ascorbate (ASC) and glutathione (GSH) are the two main antioxidants in plants. Dehydroascorbate treatment, which reduces ASC, abolished auxin maximum [1]. Root growth is promoted by the addition of micromolar concentrations of GSH [12], while depletion of GSH through BSO treatment [12] or in the glutamylcysteine synthetase (the first enzyme for glutathione biosynthesis) mutant (cadmium sensitive2 (cad2)/root meristemless1(rml1)) [13], [14] or in miao mutant (a weak mutation occurring in plastid-localized glutathione reductase2 (GR2)) [15] retards root growth with perturbation of auxin signaling and response in Arabidopsis. The triple mutant with disruption of two NADP-linked thioredoxin reductase gene (NTRA and NTRB) and CAD2/RML1 results in altered auxin homeostasis and reduced root growth [16]. The auxin-receptor mutants tir1 afb2 and tir1 afb3 are more resistant to oxidative stress and produce less ROS under salt treatment than the wild type [17]. Researchers recently found that ABA promotes ROS production through both plasma membrane-associated NADPH oxdixases [18] and mitochondria [19]. The accumulation of ROS in the mutant of ABO6 (ABA Overly sensitive 6 encodes a DEXH box RNA helicase that regulates the splicing of genes in complex I) disrupts auxin homeostasis and inhibits root growth under ABA treatment [19]. The oxidation of active indole-3-acetic acid (IAA) into the low activity 2-oxindole-3-acetic acid (OxIAA) mainly occurs in the root apex in Arabidopsis and should play crucial roles in auxin homeostasis for regulating root growth [20].
Previously, we cloned ABO5 (AT1G51965), which encodes a pentatricopeptide repeat protein required for cis-splicing of mitochondrial NAD2 intron 3 [21], and ABO6 [19]. In this study, we cloned ABO8, which encodes another pentatricopeptide repeat protein that is involved in regulating the splicing of mitochondrial complex I NAD4 intron 3. ABO8 is highly expressed in root tips and lateral root primordia. The abo8-1 mutant accumulates more ROS than the wild type in its root tips. The expression of PLETHORA1 (PLT1) and PLT2 is reduced in abo8-1, and inducible expression of PLT2 can largely rescue the reduced root meristem activity in abo8-1 with and without ABA treatment. These results suggest that ROS produced in mitochondria are important retrograde signals for controlling root meristem activity under environmental stress.
Results
The abo8 mutants show retarded growth and are more sensitive to ABA than the wild type
During a genetic screening for ABA overly sensitive mutants in root growth [19], [21], we isolated a novel mutant, aba-overly sensitive 8-1 (abo8-1). The F1 seedlings of abo8-1 crossed with the wild type showed the wild type phenotypes, indicating that abo8-1 is a recessive mutation. abo8-1 was backcrossed with the wild type for 4 times before performing further physiological analyses. A T-DNA insertion line, abo8-2, was obtained from the Arabidopsis Stock Center after we cloned ABO8 gene (see later for detail information). abo8-1 and abo8-2 had similar growth phenotypes. Relative to the wild type, abo8-1 and abo8-2 mutants had shorter roots and were smaller under normal growth conditions (Fig. 1A, 1B). ABA-inhibition of root growth and germination was greater in abo8-1 and abo8-2 than in the wild type (Fig. 1A–E). These results demonstrate that mutations in ABO8 retard plant growth and causes ABA hypersensitivity in Arabidopsis.
Fig. 1. abo8 mutants are hypersensitive to ABA in seed germination and root growth. 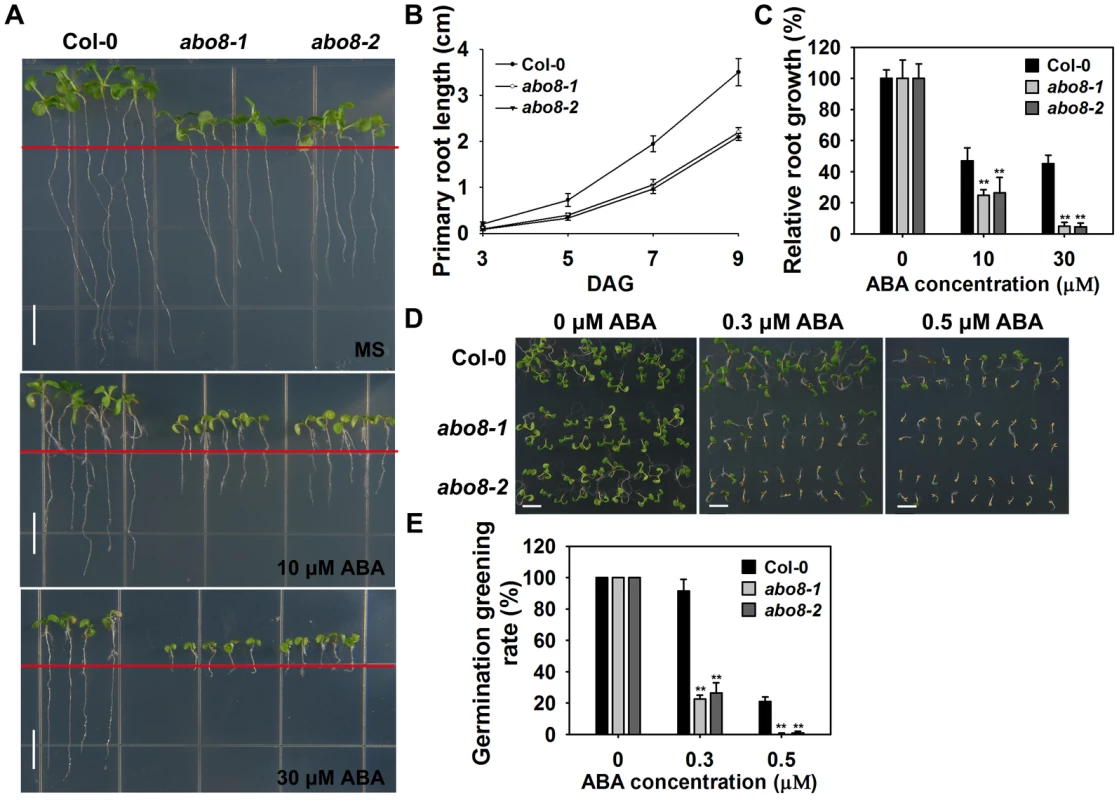
A. The root growth phenotype of abo8 mutants on MS medium or MS medium supplemented with different concentrations of ABA. Four-day-old seedlings grown on MS were transferred to MS medium containing 0, 10, or 30 µM ABA for 5 days before they were photographed. Bars = 5 mm. B. Statistical analysis of the root length in different growing times. C. Relative root growth. Root length is expressed relative to that of the wild type or abo8 mutants without ABA. In (B) and (C), three independent experiments were done with similar results, each with three biological repeats. Five roots from one plate were measured for each repeat. Values are means ±SE, n = 3. **: P<0.01. D. Seed germination greening of abo8 mutants after 7 days on MS medium supplemented with 0.0, 0.3, or 0.5 µM ABA. Bars = 2 mm. E. Statistical analysis of seed germination greening rate in (D). Three independent experiments were done, each with three replicates (from three different plates). About 24 seeds were counted for each plate. Values are means ±SE, n = 3. **: P<0.01. ABO8 encodes a pentatricopeptide repeat protein that is highly expressed in root tips and lateral root primordia
We used map-based cloning to identify ABO8. Those seedlings from the F2 progeny of abo8-1 crossed with Ler showing hypersensitivity to ABA in root growth were selected and used for mapping. The abo8-1 mutation was narrowed to chromosome 4 on BAC clone T5C23 (Fig. 2A). The sequencing of candidate genes revealed a point mutation in AT4G11690, which changes G447 to A447 (counting from the first putative ATG of AT4G11690) and causes an amino acid change from Trp to a stop code (Fig. 2A). The produced putative truncated protein in abo8-1 should have no function. In abo8-2, a T-DNA was inserted at the position 358 (from the first putative ATG) (Fig. 2A). No transcript was detected in abo8-2 using primers flanking the T-DNA insertion (Fig. 2B). ABO8 encodes a P-type pentatricopeptide repeat (PPR) domain protein, which contains a classic tandem array of 35 amino acid PPR motifs, typically with a proline at the end of each motif (Fig. 2C) [22]. The F1 seedlings of abo8-1 crossed with abo8-2 showed a similar ABA hypersensitivity as abo8-1 or abo8-2 in root growth (Fig. 2D). ABO8 promoter driving ABO8 cDNA fused with GFP in frame (ProABO8:ABO8-GFP) complemented the ABA-hypersensitive seed germination and root growth phenotype of abo8-1 (Fig. 2E–H). These results confirmed that the mutations in AT4G11690 led to the phenotypes of the abo8 mutants.
Fig. 2. Map-based cloning of ABO8. 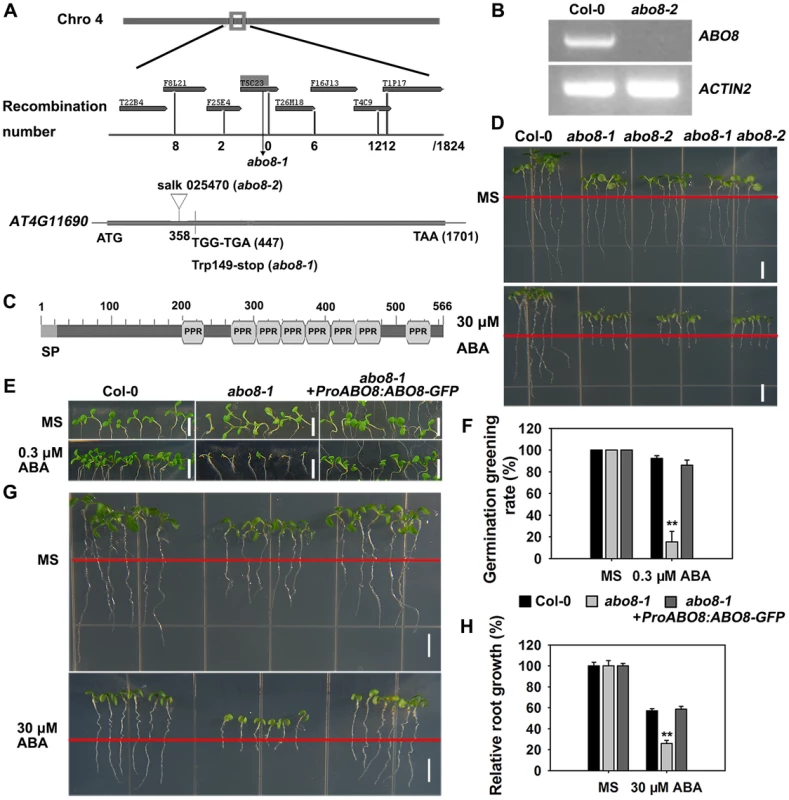
A. ABO8 was mapped to chromosome 4. A point mutation from G447 to A447 (counting from the first putative ATG in AT4G11690) was identified in abo8-1. A T-DNA insertion line SALK_025470 (abo8-2, T-DNA is inserted in position +358) was obtained from the Arabidopsis Biological Resource Center. B. RT-PCR analysis of ABO8 transcripts in abo8-2 using primers flanking the T-DNA insertion. Expression of the ACTIN2 gene was used as control. C. The protein structure of ABO8. SP: signal peptide; PPR: pentatricopeptide repeat. D. Genetic analysis of abo8-1 and abo8-2 for sensitivity of root growth to ABA. Four-day-old seedlings grown on MS medium were transferred to MS medium containing 0 or 30 µM ABA. abo8-1 abo8-2: F1 seedlings of abo8-1 crossed with abo8-2. Bars = 5 mm. E. Seed germination greening analysis of abo8-1 complemented with an ABO8 promoter driving ABO8 cDNA fused with GFP cDNA (ProABO8:ABO8-GFP) on MS medium containing 0.0 or 0.3 µM ABA. Bars = 5 mm. F. Statistical analysis of seed germination greening rate of the complemented abo8-1 in (E). Three independent experiments were done, each with three replicates (from three different plates). About 24 seeds were counted for each plate. Values are means ±SE, n = 3. **: P<0.01. G. Roots of abo8-1 complemented with ProABO8:ABO8-GFP. Four-day-old seedlings were transferred to MS medium containing 0 or 30 µM ABA. Bars = 5 mm. H. Statistical analysis of root length in (G). Root length is expressed relative to that of the wild type or abo8-1 without ABA. Three independent experiments were done with similar results, each with three biological repeats. Five roots from one plate were measured for each repeat. Values are means ±SE, n = 3. **: P<0.01. P-type PPR proteins usually participate in RNA stabilization, processing, splicing, and translation. To determine the expression pattern of ABO8, we made transgenic plants carrying ABO8 promoter:GUS (ProABO8:GUS). GUS staining indicated that ABO8 was highly expressed in root tips and lateral root primordia (Fig. 3A). Expression patterns were similar for ProABO8:ABO8-GFP and ProABO8:GUS (Fig. 3B, 3C). We isolated the protoplasts from ProABO8:ABO8-GFP transgenic plants and treated the protoplasts with Mito-tracker, which specifically stains mitochondria. Most GFP fluorescence was co-localized with Mito-tracker (Fig. 3D), indicating that ABO8-GFP is localized in the mitochondria. Real-time PCR analysis indicated that the expression of ABO8 was significantly suppressed by ABA treatment (Fig. 3E).
Fig. 3. The expression and localization of ABO8. 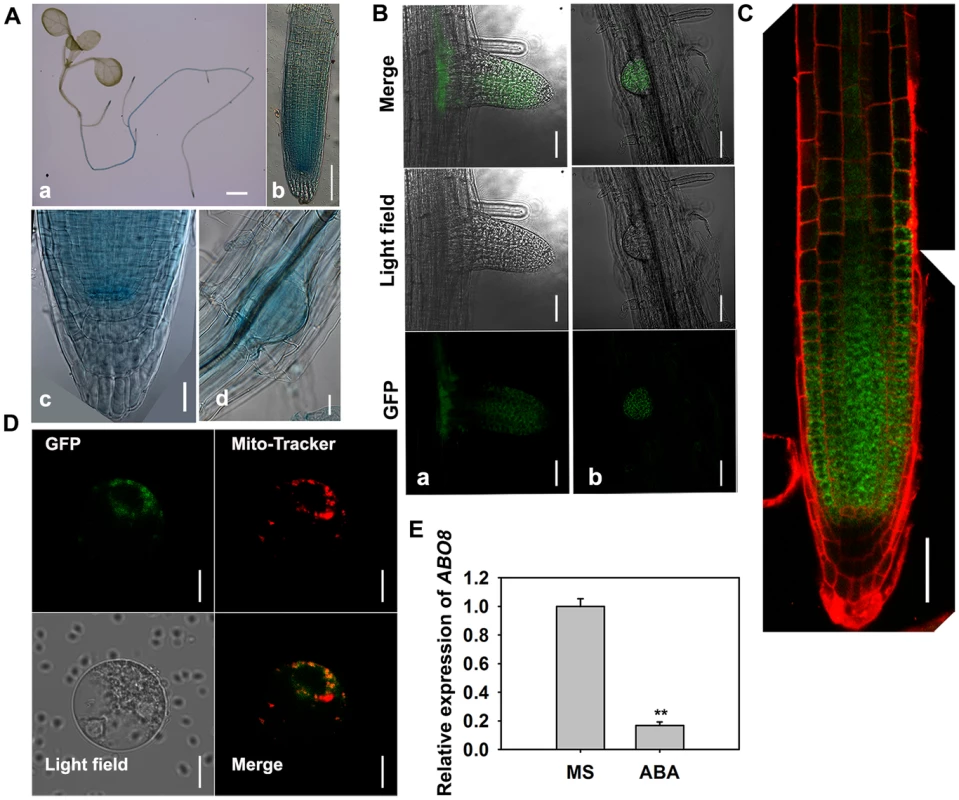
A. The expression pattern of ProABO8:GUS in the whole seedling and roots. a, a seedling, bar = 1 mm; b, a primary root, bar = 50 µm; c, an amplified primary root tip, bar = 20 µm; d, a lateral root primordium, bars = 50 µm. B. The expression of ProABO8:ABO8-GFP in a lateral root (a, left) and a lateral root primordium (b, right). Bars = 50 µm. C. The expression of ProABO8:ABO8-GFP in a primary root. Bars = 50 µm. D. Co-subcellular localization of ABO8-GFP with Mito-Tracker. Bars = 10 µm. E. Expression of ABO8 is reduced by ABA treatment. Seedlings were treated with or without 30 µM ABA for 8 h, and total RNAs were used for qRT-PCR. ACTIN2 was used as a control. Three independent experiments were done with similar results, each with three biological replicates. Results shown are from one experiment. Values are means ±SE, n = 3. **: P<0.01. ABO8 is required for splicing of the complex I gene NAD4
In our previous study, we found that the PPR protein ABO5 is responsible for cis-splicing of mitochondrial NAD2 intron 3 [21]. We used northern blot to compare the transcripts of different genes in complex I between the wild type and abo8-1. The results indicated that the transcript sizes of all NAD genes except NAD4 are similar for the wild type and abo8-1 (Fig. 4A) with and without ABA treatment. The NAD4 transcripts were larger in abo8-1 than in the wild type, indicating a defect in pre-mRNA splicing in abo8-1 (Fig. 4A). We designed the primers inside exons that cover the three introns of NAD4 to determine which intron is affected by the abo8-1 mutation. Real-time RT-PCR indicated that fragment 3 (covering exons 3 and 4) is greatly reduced in abo8-1 but that intron 3 (primers inside introns) is significantly increased in abo8-1 relative to the wild type (Fig. 4B). These results indicate that the abo8-1 mutation impairs the splicing of the third intron of NAD4 in complex I.
Fig. 4. ABO8 regulates the splicing of NAD4 intron 3. 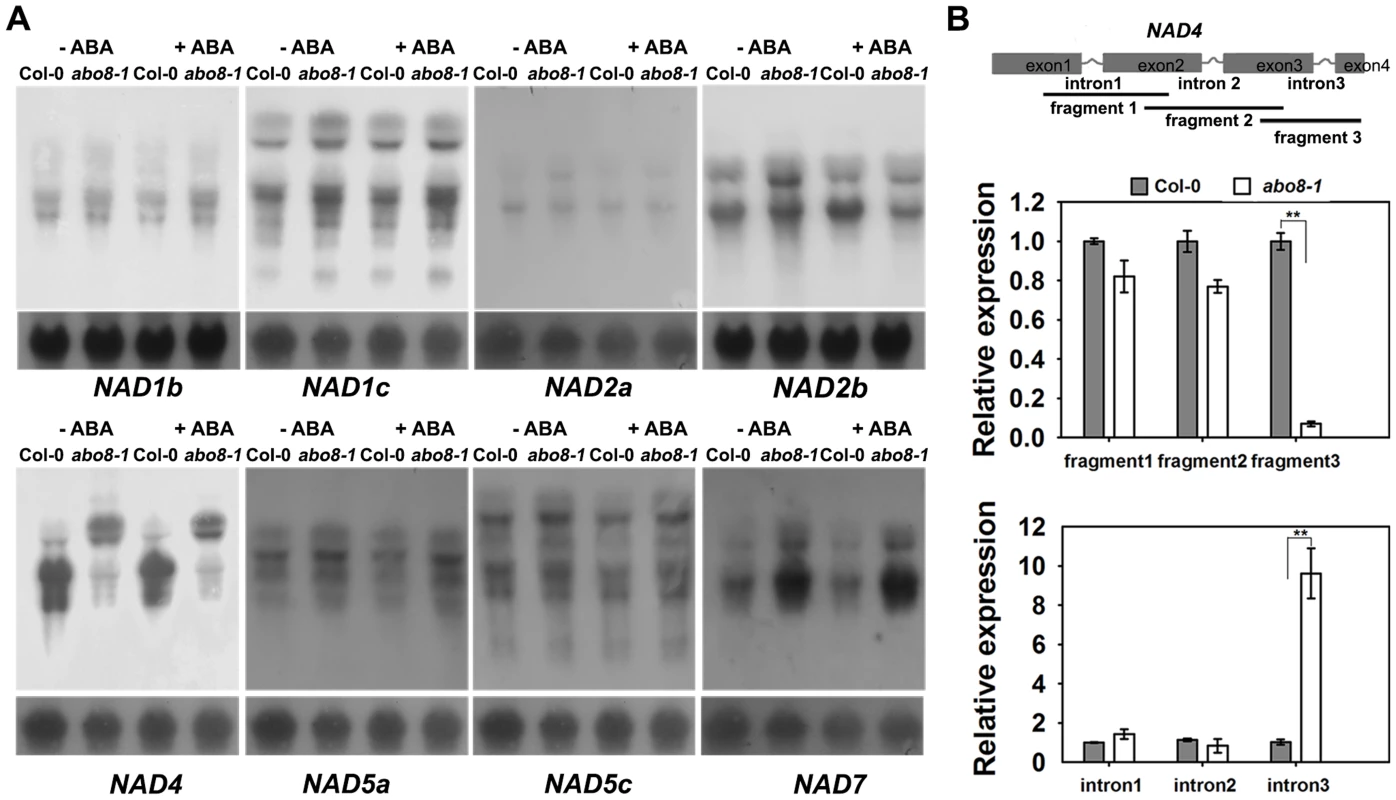
A. Northern blot analyses of the expression of different subunits in complex I. Only NAD4 showed the splicing defects in abo8-1. B. qRT-PCR analysis of different fragments of the NAD4 transcript. A pair of primers covering different fragments or different introns was used for qRT-PCR. Total RNAs from 10-day-old seedlings were used for qRT-PCR. Three independent experiments were done with similar results, each with three biological repeats. Values are means ±SE from one experiment. **: P<0.01. abo8-1 reduces the activity of the electron transport chain in complex I, and accumulates more ROS than the wild type in its root tips
Impairment of the electron transport chain in complex I usually reduces the NADH dehydrogenase activity, which affects the production of ATP, and increases ROS accumulation in cells. We compared the NADH dehydrogenase activity of complex I between wild type and abo8-1 by Blue Native-PAGE [23]. Comparing with the wild type, complex I activity was largely reduced in abo8-1 (Fig. 5A). Consistently, abo8-1 produced much less ATP than wild type, and ABA treatment further reduced the ATP production in both the wild type and abo8-1 (Fig. 5B). We introduced a mitochondrial superoxide marker Mito-cpYFP into abo8-1 to monitor ROS accumulation in roots. The previous results indicated that the strong fluorescence emitted by Mito-cpYFP in mitochondria was correlated with the high oxidation status [19]. Mito-cpYFP fluorescence was highest near root tips and progressively decreased from the outside of the columella cell layer to the inside below the QC (quiescent centre) in both the wild type and abo8-1. abo8-1 emitted stronger fluorescence than the wild type (Fig. 5C, 5D). In comparing the production of H2O2 (using DAB and DCFH-DA staining) and of superoxide (using NBT staining) in root tips of the wild type and abo8-1, we found that the root tips of abo8-1 accumulated more H2O2 and superoxide than those of the wild type (Fig. 5E–J). ABA treatment significantly enhanced the production of H2O2 and superoxide in both abo8-1 and wild type (Fig. 5E–J). These results suggest that dysfunction of ABO8 increases ROS production in root tips and that ROS production is enhanced by ABA treatment.
Fig. 5. abo8-1 accumulates more ROS than the wild type in root tips. 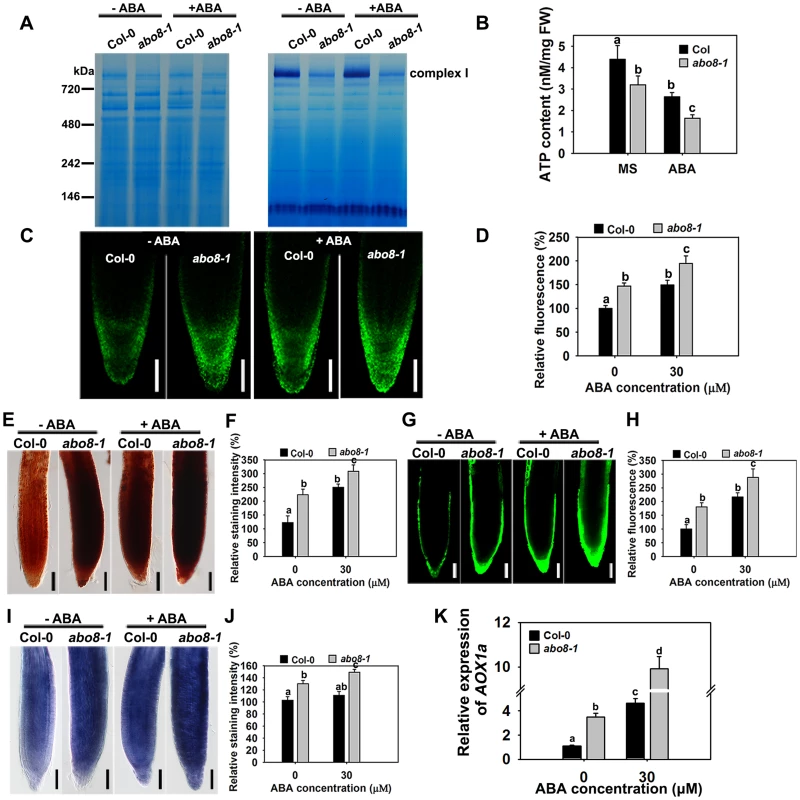
A. The activity of complex I in the wild type (WT) and abo8-1. Crude mitochondria membrane protein extracted from 5-day-old seedlings treated with or without 50 µM ABA for 24 h were separated by Blue Native-PAGE. Coomassie staining (left) indicates protein complexes after electrophoresis; Complex I activity was preformed by in-gel staining with NADH and NBT (nitroblue tetrazolium) for the NADH dehydrogenase activity of Complex I. B. ATP content in WT and abo8-1 plants. 5-day-old WT and abo8-1 seedlings treated with or without 50 µM ABA for 24 h were harvested for measurement of ATP content. FW, fresh weight. Means with different letters are significantly different at P<0.01. C. Fluorescence analysis of mitochondrial cpYFP in primary root tips of the wild type and abo8-1 with or without ABA treatment. Bars = 50 µm. D. cpYFP intensity as determined with AxioVision Rel. 4.8 software. E. DAB staining for H2O2 in primary root tips of the wild type and abo8-1 with or without ABA treatment. Bars = 50 µm. F. DAB staining intensity as determined with Adobe Photoshop 3.0 software. G. DCFH-DA staining for H2O2 in primary root tips of the wild type and abo8-1 with or without ABA treatment. Bars = 50 µm. H. DCFH-DA staining intensity as determined with AxioVision Rel. 4.8 software. I. NBT staining for superoxide in primary root tips of the wild type and abo8-1 with or without ABA treatment. Bars = 50 µm. J. NBT staining intensity as determined with Adobe Photoshop 3.0 software. Three independent experiments were done with similar results, each with three replicates, and each replicate with 15–20 roots. Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.01. K. Relative expression levels of AOX1a in seedlings treated with or without 50 µM ABA for 5 h in liquid MS. Three independent experiments were done with similar results, each with three biological replicates. Results shown are from one experiment. Values are means ±SE, n = 3. Means with different letters are significantly different at P<0.01. Besides the classical electron transport chain, plants also have alternative NADH dehydrogenases that can maintain the oxidation of matrix NADH in mitochondria when complex I is not working. ALTERNATIVE OXIDASE1a (AOX1a) is a marker gene in alternative respiratory pathway for mitochondria stress. The expression of AOX1a was higher in abo8-1 than the wild type, and ABA treatment further increased its expression in both abo8-1 and the wild type, and its expression was still higher in abo8-1 than in the wild type (Fig. 5K). These results suggest that abo8-1 suffers from a stronger mitochondrial stress than the wild type.
ABA inhibits root growth by regulating root meristem activity
Root growth is determined by the coordination of stem cell activity and the differentiation of the progeny cells. In the roots, three developmental zones can be recognized according to cell types: the differentiation zone (DZ), the elongation zone (EZ), and the meristem zone (MZ) (Fig. 6A). In normal growth condition, abo8-1 and abo8-2 had less MZ cell numbers than the wild type after seed germination for different days (Fig. 6B). We further measured the zone length and also the cell number and cell length in each zone with ABA treatment and without ABA treatment. The MZ was shorter and contained fewer but slightly longer cells in abo8-1 than in the wild type in the absence of ABA treatment (Fig. 6A, 6C, 6F). ABA treatment reduced MZ length and cell number in both abo8-1 and the wild type (Fig. 6A, 6C, 6F). ABA treatment did not change MZ cell length in abo8-1 but increased a little MZ cell length in the wild type, so that MZ cell length became comparable in the wild type and abo8-1. The EZ was shorter and had fewer cells in abo8-1 than in the wild type with and without ABA treatment. Although the EZ cell length was similar in abo8-1 and the wild type without ABA treatment, ABA treatment reduced EZ cell length more in abo8-1 than in the wild type (Fig. 6A, 6D). The DZ was shorter and had fewer cells in abo8-1 than in the wild type, but DZ cell length was similar in abo8-1 and in the wild type with and without ABA treatment (Fig. 6A, 6E). These results indicate that the abo8 mutation greatly impairs root meristem activity, reduces cell number in three zones, as well as reduces cell length in EZ. The number of cells that expressed ProCYCB1;1:GUS, a reporter for G2 in the mitotic phase of cell cycle, was consistently much lower in abo8-1 than in the wild type, suggesting that ABO8 promotes mitosis (Fig. 6G).
Fig. 6. abo8 mutation decreases the activity of the root meristem. 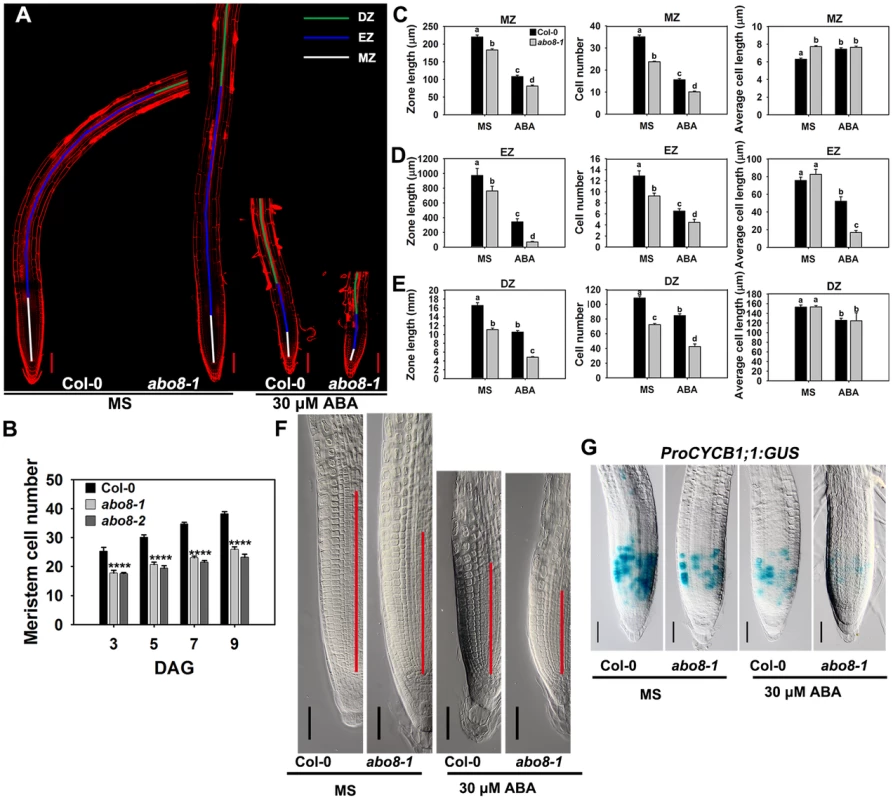
A. The meristem zone (white line), elongation zone (blue line), and differentiation zone (green line) with 0 or 30 µM ABA treatment. Bars = 100 µm. Each image was made by joining several photographs of the same root. B. Meristem cell number of the wilt type, abo8-1 and abo8-2 in different times after seed germination (DAG). C. The meristem zone length, cell number, and cell length with 0 or 30 µM ABA treatment. D. The elongation zone length, cell number, and cell length with 0 or 30 µM ABA treatment. E. The differentiation zone length, cell number, and cell length with 0 or 30 µM ABA treatment. In C–E, two experiments were done with similar results, each with three repeats, each repeat with 10 roots. Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.01. F. Size of the amplified root meristem in the wild type and abo8-1 with and without ABA treatment. Bars = 50 µm. G. The expression of ProCYCB1;1:GUS in the wild type and abo8-1 with or without 30 µM ABA treatment. Bars = 50 µm. Addition of the reducing agent GSH largely recovers the ABA-hypersensitive phenotype of abo8-1
Because abo8-1 accumulates more ROS than the wild type, we wanted to determine whether addition of a reducing agent would rescue the ABA-sensitive phenotype of abo8-1. Addition of different concentrations of the reducing agent GSH compromised the ABA-hypersensitive phenotype of abo8-1 in both seed germination and root growth (Fig. 7A–7D). The seed germination greening ratio and relative root growth of abo8-1 became comparable to those of the wild type following treatment with ABA plus 300 µM GSH (Fig. 7A–7D). Addition of 300 µM GSH increased the MZ cell number more in abo8-1 than wild type with or without ABA treatment (Fig. 7E, 7F). These results indicate that reducing the oxidation status by addition of GSH decreased the ABA sensitivity of abo8-1 in both seed germination and root growth.
Fig. 7. Addition of GSH partially recovers the ABA-hypersensitive phenotypes of abo8-1 mutants. 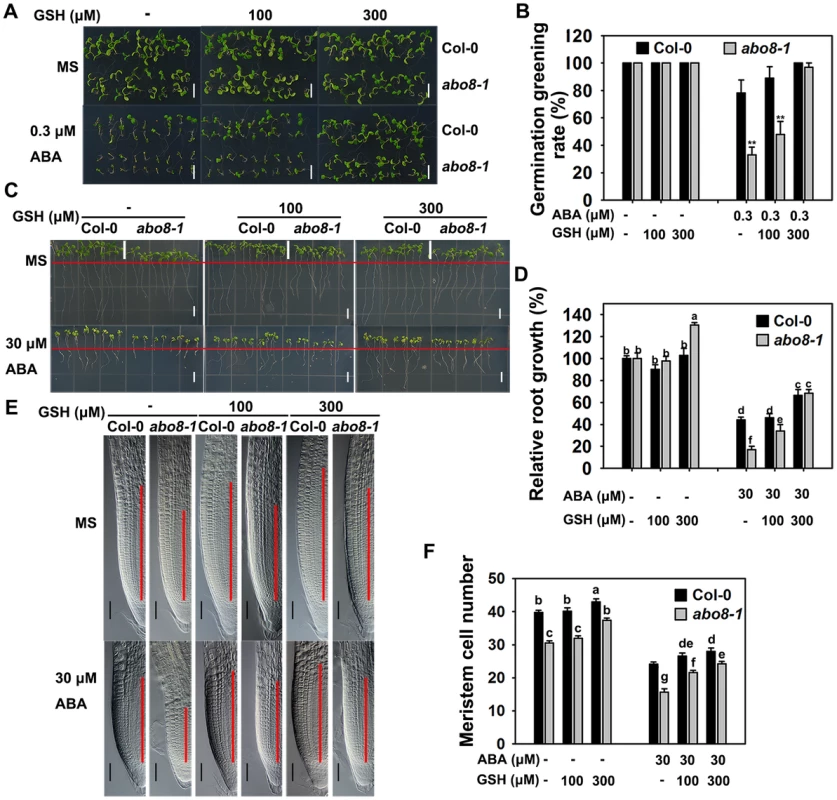
A. Germination greening of wild-type and abo8-1 seeds on MS medium or MS medium containing 0.3 µM ABA, 0.3 µM ABA plus 100 µM GSH, or 0.3 µM ABA plus 300 µM GSH. Bars = 5 mm. B. Statistical analysis of the seed germination greening rate in (A). Three independent experiments were done with similar results, each with three replicates. About 20 seeds were counted from one plate as one replicate. Values are means ±SE from one experiment. **: P<0.01. C. Roots of wild-type and abo8-1 seedlings grown on MS medium or MS medium containing 30 µM ABA, 30 µM ABA plus 100 µM GSH, or 30 µM ABA plus 300 µM GSH. The red line indicates the starting point of root growth after transfer. Bars = 5 mm. D. Statistical analysis of root growth in (C). Root growth is expressed relative to that of the wild type or abo8-1 in MS medium without supplement. Three independent experiments were done with similar results, each with three replicates. Eight roots were measured from one plate as one replicate. Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.01. E. Root meristem zones of seedlings grown on MS medium or MS medium containing 30 µM ABA, 30 µM ABA plus 100 µM GSH, or 30 µM ABA plus 300 µM GSH. Bars = 50 µm. F. Statistical analysis of the MZ cell number in (E). Three independent experiments were done with similar results, each with three repeats, and each repeat with 15–20 roots. Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.01. The expression of ProDR5:GUS in root tips is reduced in abo8-1, which can be partially reversed by GSH treatment
Because the activity of the root meristem is closely correlated with auxin signaling, the reduced root meristem activity in abo8-1 is likely due to an impairment in auxin response. We introduced ProDR5:GUS and ProIAA2:GUS into abo8-1 to check their expression levels. Both ProDR5:GUS and ProIAA2:GUS can be strongly induced by auxin [24], [25], [26]. The expression of ProDR5:GUS was strong in the root tips of the wild type in absence of ABA but was decreased by ABA treatment. However, GUS staining was barely detected in the root tips of abo8-1 (Fig. 8A). When seedlings were moved from MS medium to MS medium supplemented with 300 µM GSH for 5 days, the ProDR5:GUS expression was detected in the root tips of abo8-1, which was largely abolished by addition of ABA. This result indicates that the low expression of ProDR5:GUS in abo8-1 is likely due to the high oxidation status in root tips. GSH treatment also increased ProDR5:GUS expression in the wild type (Fig. 8A). When roots were treated with different auxins in high concentration, ProDR5:GUS expression was greatly induced in most parts of the wild-type root meristem, but only in the upper part of the abo8-1 root meristem. However, when treated with low concentration of auxin (30 nM NAA), ProDR5:GUS expression could be detected in root tips of abo8-1, but lower than in the wild type. These results suggest that the abo8-1 mutation reduces the auxin response around the root stem cell niche (Fig. 8B), and at low concentration of auxin, the oxidation status in root tips in abo8-1 is compromised. In order to see whether reducing oxidation status by GSH could recover the ProDR5:GUS expression in auxin treatment, we treated the liquid growing seedling with GSH, IAA or GSH plus IAA. During a short time (18 h) treatment, GUS staining in the root tips of abo8-1 was not able to be detected with IAA or GSH treatment alone, but could be clearly detected with IAA plus GSH treatment (Fig. 8C), suggesting that GSH changes the oxidative status in the root stem cell niche of abo8-1. However, the intensity of GUS staining was much lower in abo8-1 than in the wild type. Similarly, the expression of ProIAA2:GUS was higher in the wild type than in abo8-1 at both high and low concentration auxin treatments (S1 Figure), and although ABA treatment reduced its expression in the wild type and abo8-1, its expression was still higher in the wild type than in abo8-1 (Fig. 8D).
Fig. 8. Expression of DR5, IAA2, PIN1, PIN2, and AUX1 is altered in the abo8-1 mutant. 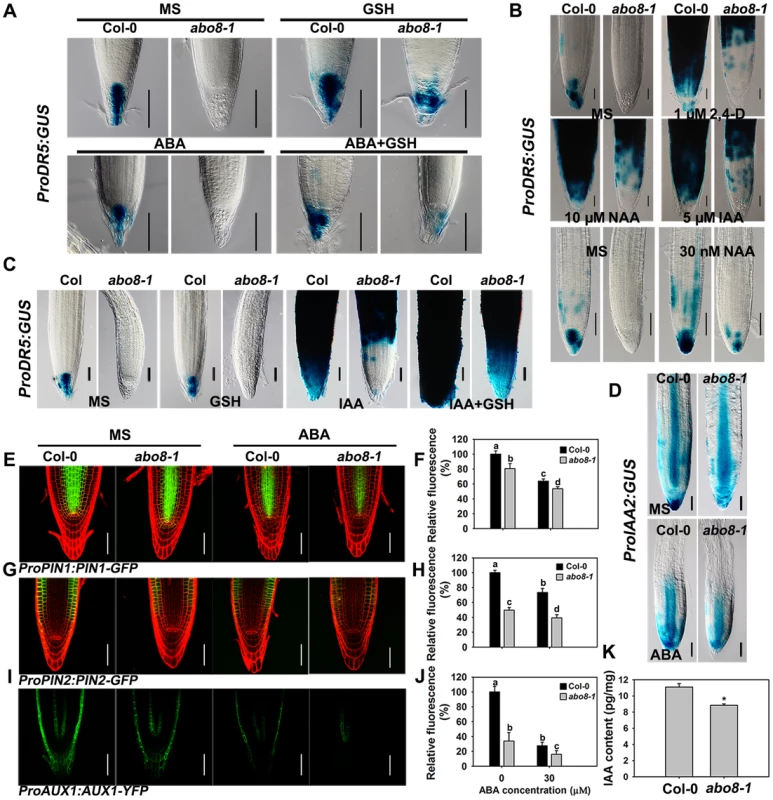
A. The expression of ProDR5:GUS in roots of seedlings grown on MS medium or MS medium containing 30 µM ABA, 300 µM GSH, or 30 µM ABA plus 300 µM GSH for 5 days. Bars = 50 µm. B. The expression of ProDR5:GUS in 5-day old seedlings treated in liquid MS medium without or with 1 µM 2,4-D, 5 µM IAA, and 10 µM NAA, or 30 nM NAA for 18 h. Bars = 50 µm. C. The expression of ProDR5:GUS in 5-day old seedlings treated in MS liquid medium without or with 300 µM GSH, 5 µM IAA, or 5 µM IAA plus 300 µM GSH for 18 h. Bars = 50 µm. D. The expression of ProIAA2:GUS with and without ABA treatment. Bars = 50 µm. E. The expression of ProPIN1:PIN1-GFP with and without ABA treatment. Bars = 50 µm. F. The fluorescence of PIN1-GFP in (E). Fluorescence is expressed relative to that of the wild type without ABA. Three experiments were done with similar results, each experiment with three repeats (each repeat with 15 roots). Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.01 for a and b and at P<0.05 for c and d. G. The expression of ProPIN2:PIN2-GFP with and without ABA treatment. Bars = 50 µm. H. The fluorescence of PIN2-GFP in (G). Fluorescence is expressed relative to that of the wild type without ABA. Three experiments were done with similar results, each experiment with three repeats (each repeat with 15 roots). Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.01 except for c and d at P<0.05. I. The expression of ProAUX1:AUX1-YFP with or without ABA treatment. Bars = 50 µm. J. The fluorescence of AUX1-YFP in (I). Fluorescence is expressed relative to that of the wild type without ABA. Three experiments were done with similar results, each experiment with three repeats (each repeat with 15 roots). Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.01 except for b and c at P<0.05. K. The abo8-1 contained less auxin than wild type. 7-day seedlings were used for auxin contents measurement. Each value is the mean ± SD (n = 4). *: P<0.05. Auxin transporters play crucial roles in modulating root growth. The expression of ProPIN1:PIN1-GFP, ProPIN2:PIN2-GFP, and ProAUX1:AUX1-YFP was much lower in abo8-1 than in the wild type, and was reduced by ABA treatment in both abo8-1 and the wild type (Fig. 8E and 8F for ProPIN1:PIN1-GFP, 8G and 8H for ProPIN2:PIN2-GFP, 8I and 8J for ProAUX1:AUX1-YFP). As both auxin level and signaling control the expression of auxin responsive genes, we measured the auxin contents in abo8-1 and the wild type seedlings. The total auxin contents in abo8-1 was ∼9.32 pg/mg, comparing with ∼10.83 pg/mg in wild type (P<0.05) (Fig. 8K), indicating that abo8-1 mutation reduces the auxin content. These results indicate that both auxin level and response are largely impaired in abo8-1.
The abo8-1 mutation reduces the expression of PLT1 and GSH treatment changes the distribution range of PLT1 protein in abo8-1
The activity of the root meristem is regulated by different factors, among which the auxin-inducible PLETHORA (PLTs) are master regulators of root development [27], [28]. PLT1 and PLT2 are predominantly localized in the QC and redundantly regulate the root stem cell niche [28]. The transcriptional gradients of PLT1 and PLT2 strongly correlate with the auxin gradients in root apical meristem (RAM). plt1 plt2 double mutants show loss of stem cells, loss of transit-amplifying cells, and reduced cell expansion in RAM [28]. As shown in Fig. 9A and 9B, the expression of ProPLT1:PLT1-YFP was much lower in abo8-1 than in wild type both with and without ABA treatment. ABA treatment significantly reduced the expression of ProPLT1:PLT1-YFP in both abo8-1 and the wild type. GSH treatment reduced a little strength of YFP fluorescence in the wild type, but apparently increased the expression of ProPLT1:PLT1-YFP in abo8-1. Interestingly, GSH treatment changed the distribution range of PLT1-YFP in abo8-1. Besides around root stem cell niche where PLT1-YFP is mainly localized, PLT1-YFP localization was shifted to upper parts in epidermis, cotex and endodermis. In the wild type, weak PLT1-YFP signals were also detected in epidermis above root stem cell niche by GSH treatment. In response to treatment with ABA plus GSH, the expression of ProPLT1:PLT1-YFP in abo8-1 or the wild type was higher than only with ABA treatment (Fig. 9A, 9B). Again, the distribution of PLT1-YFP in epidermis, cotex and endodermis above root stem cell niche still existed in treatment with GSH plus ABA in abo8-1. In order to see whether PLT1 expression is affected by GSH at transcriptional level or not, we introduced ProPLT1:CFP into abo8-1 by crossing the transgenic wild type plants that carry ProPLT1:CFP with abo8-1. The expression of ProPLT1:CFP was lower in abo8-1 than the wild type (Fig. 9C, 9D). ABA treatment reduced the expression of ProPLT1:CFP in both abo8-1 and the wild type, but the expression level of ProPLT1:CFP was still higher in the wild type than abo8-1. GSH treatment reduced a little the expression level of ProPLT1:CFP in wild type, but increased in abo8-1, which is consistent with expression of ProPLT1:PLT1-YFP under GSH treatment. However, ProPLT1:CFP expression domain was only found around root stem cell niche, not like the expression of ProPLT1:PLT1-YFP by GSH treatment in abo8-1. These results indicate that GSH treatment does not affect the expression area of ProPLT1:CFP at transcriptional level. It is likely that GSH treatment leads to more stability of PLT1-YFP in epidermis, cotex and endodermis above root stem cell niche in abo8-1 than the wild type, suggesting that the abo8-1 mutation disturbs the redox homeostasis which cannot be completely reversed by GSH treatment, and PLT1-YFP distribution in root tips is influenced by redox homeostasis.
Fig. 9. The abo8 mutation affects the expression of PLT1 and PLT2. 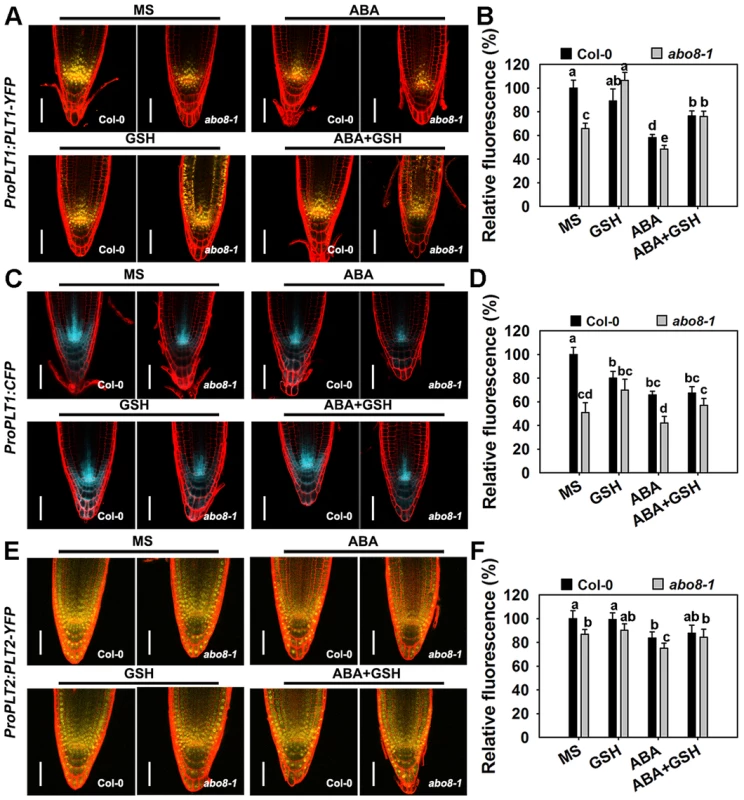
A. The expression of ProPLT1:PLT1-YFP on MS medium or MS medium supplemented with ABA, GSH, or ABA plus GSH. Bars = 50 µm. B. The fluorescence of PLT1-YFP in (A). Fluorescence is expressed relative to that of the wild type in MS without supplement. Three independent experiments were done with similar results, each with three replicates. In each replicate, 8–15 roots were measured from one plate. Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.01, except for c and d, d and e, at P<0.05. C. The expression of ProPLT1:CFP on MS medium or MS medium supplemented with ABA, GSH, or ABA plus GSH. Bars = 50 µm. D. The relative fluorescence of ProPLT1:CFP in (E). Fluorescence is expressed relative to that of the wild type in MS without supplement. Three independent experiments were done with similar results, each with three replicates. In each replicate, 8–15 roots were measured from one plate. Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.01, except for a and b, c and d, at P<0.05. E. The expression of ProPLT2:PLT2-YFP on MS medium or MS medium supplemented with ABA, GSH, or ABA plus GSH. Bars = 50 µm. F. The relative fluorescence of PLT2-YFP in (C). Fluorescence is expressed relative to that of the wild type in MS without supplement. Three independent experiments were done with similar results, each with three replicates. In each replicate, 8–15 roots were measured from one plate. Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.05. The expression of ProPLT2:PLT2-YFP was also lower in abo8-1 than in the wild type, and the expression of ProPLT2:PLT2-YFP was further reduced by ABA treatment in both genotypes (Fig. 9E, 9F). However, addition of GSH did not clearly affect the expression of ProPLT2:PLT2-YFP in abo8-1 and the wild type. With ABA treatment, GSH slightly increased expression of ProPLT2:PLT2-YFP in abo8-1 and the wild type. These results suggest that PLT2 is less affected by the abo8-1 mutation and ABA treatment than PLT1.
plt1 and plt2 mutants are sensitive to ABA in root growth
To determine whether PLT1 and PLT2 are involved in ABA inhibition of root growth, we examined the ABA sensitivity of plt1 and plt2 mutants. The root growth of plt1-4 and plt2-2 [28] was more sensitive to ABA than the wild type (Fig. 10A–E), suggesting that PLT1/2 are negative regulators in ABA-mediated root growth. The root length of the abo8-1 plt1-4 or abo8-1 plt2-2 double mutant was a little shorter than that of abo8-1, plt1-4 or plt2-2 in the absence of ABA treatment, suggesting that PLT1 or PLT2 and ABO8 have additive function in root growth. ABA treatment at 10 or 20 µM inhibited root growth more in abo8-1 plt1-4 and abo8-1 plt2-2 than in abo8-1, plt1-4 or plt2-2, suggesting that PLT1/2 and ABO8 have additional function in ABA inhibition of root growth. At 20 µM ABA, the root growth of abo8-1plt1-4 was more inhibited than that of abo8-1plt2-2. At 30 µM ABA, the root growth of abo8-1, abo8-1 plt1-4 and abo8-1 plt2-2 was almost completely inhibited, and the root growth of plt1-4 was more inhibited than that of plt2-2. These results indicate that PLT1 plays a more important role than PLT2 in ABA inhibition of root growth. We also found that addition of 300 µM GSH could partially recover the ABA inhibition of root growth for plt1-4, plt2-2, abo8-1 plt1-4 and abo8-1 plt2-2 mutants (Fig. 10A–E).
Fig. 10. Genetic analysis of ABO8 with PLT1 and PLT2. 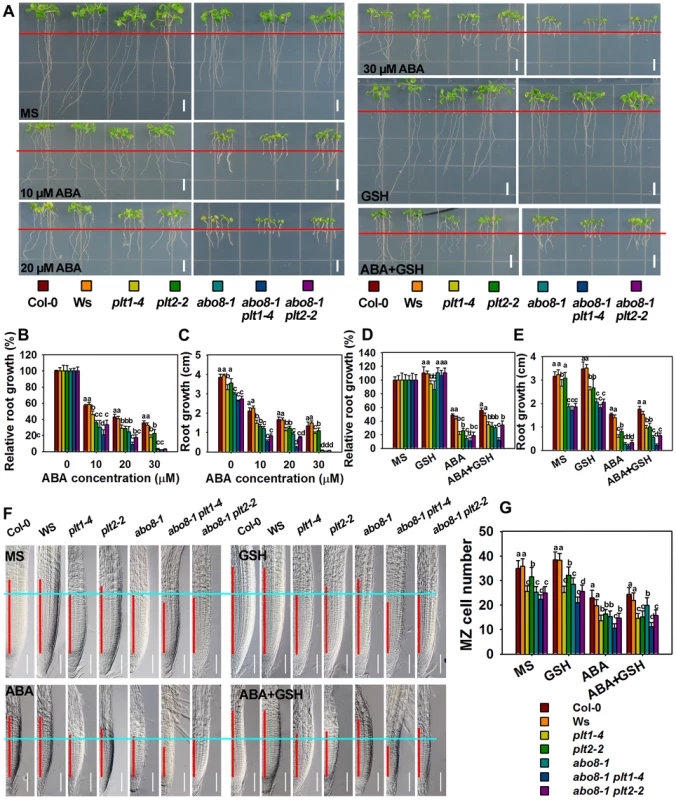
A. The roots of the wild type Col-0, Ws, plt1-4, plt2-2, abo8-1, abo8-1 plt1-4, and abo8-1 plt2-2 grown on MS medium or MS medium supplemented with different concentrations of ABA, 300 µM GSH or 30 µM ABA plus 300 µM GSH for 5 days. Red line indicates the root tip positions after 5-day seedlings were just transferred onto different medium. Bars = 5 mm. B–E. The root growth and relative root growth of different plants in (A). Relative root growth is expressed to that of the wild type or mutants on MS without ABA. About 15 roots from three plates were measured in each experiment, and three experiments were done with similar results. Values are means ±SE, n = 3. Means with different letters are significantly different at P<0.01. F. The length of the root meristem (red line) of different genotypes on MS medium or MS medium supplemented with 30 µM ABA, 300 µM GSH or 30 µM ABA plus 300 µM GSH. Bars = 50 µm. G. The MZ cell number in (F). Three independent experiments were done with similar results, and each experiment had three replicates. About 20 roots were measured for each replicate. Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.01. We further compared the cell number in the MZ among different mutants (Fig. 10F, 10G). abo8-1 had similar MZ cell number as plt1-4, both of which were reduced to a similar level by ABA treatment. abo8-1 plt1-4 double mutant had less MZ cell number than abo8-1 or plt1-4 without or with ABA treatment, suggesting that PLT1 and ABO8 have additive function in regulating MZ. plt2-2 had less MZ cell number than the wild type, but more than plt1-4 or abo8-1 without ABA treatment, and only a little more MZ number than abo8-1 or plt1-4 with ABA treatment. abo8-1 plt2-2 had similar MZ cell number as abo8-1, and much less than plt2-2. ABA treatment reduced the MZ cell number of abo8-1 plt2-2 to a similar level as that of abo8-1. Interestingly, GSH treatment did not significantly increase the MZ cell number in plt1-4, plt2-2, abo8-1plt1-4, and abo8-1 plt2-2 with ABA or without ABA treatment. However, GSH treatment clearly increased MZ cell number in the wild type and abo8-1. These results suggest that both PLT1 and PLT2 play crucial roles in ABA inhibition of root growth, and PLT1 is more important than PLT2 in this process.
To determine whether PLT1/2 can partially complement the ABA hypersensitivity of abo8-1 in root growth, we introduced an inducible Pro35S:PLT2-GR into abo8-1 by crossing a Pro35S:PLT2-Glucocorticoid Receptor (Pro35S:PLT2-GR) transgenic plant with abo8-1. After dexamethasone (DEX) induction, the meristem cell number of the wild type was increased 23.1% comparing with 40.4% in abo8-1 in the absence of ABA treatment (Fig. 11A, 11B). The reduced root meristem by ABA treatment was increased 151.5% in abo8-1 comparing with 40.5% in wild type by DEX treatment. These results suggest that the retarded root growth of abo8-1 is partially caused by low expression of PLT genes, which is likely due to reduced auxin accumulation and/or auxin signaling by the abo8-1 mutation.
Fig. 11. The sensitivity of abo8-1 roots to ABA is partially rescued by the expression of the inducible Pro35S:PLT2-GR. 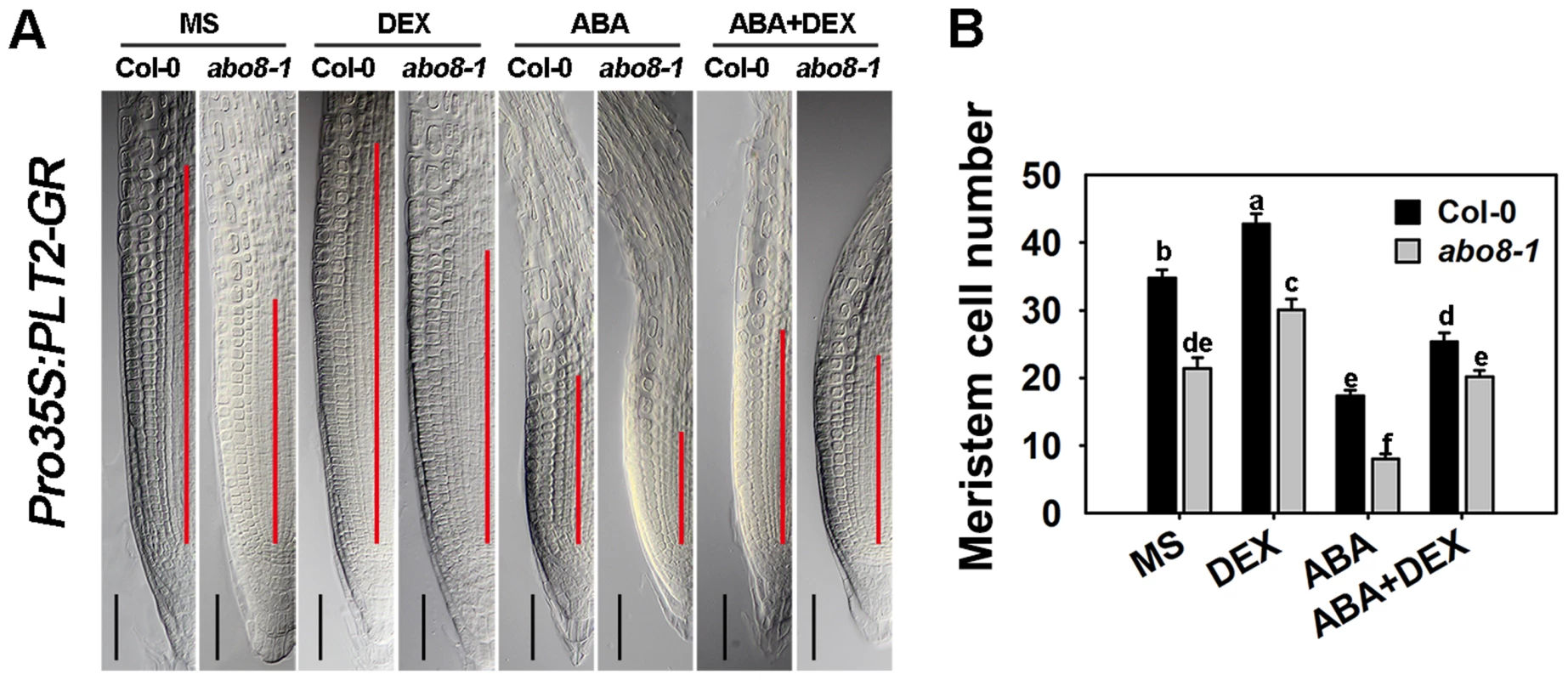
A. The length of the root meristem (red line) of wild-type (carrying Pro35S:PLT2-GR) and abo8-1 (carrying Pro35S:PLT2-GR) seedlings on MS medium or MS medium supplemented with DEX, ABA, or ABA plus DEX. Bars = 50 µm. B. Root meristem cell number of the wild type (Col-0) and abo8-1 in (A). Three independent experiments were done with similar results, and each experiment had three replicates. About 20 roots were measured for each replicate. Values are means ±SE from one experiment. Means with different letters are significantly different at P<0.01. The abo8 mutation deregulates the differentiation of root distal stem cells (DSC)
Root growth is determined by quiescent center (QC) and its surrounding stem cells [29]. The distal stem cells (DSC) below the QC generates the columella cells with distinct starch granules [30]. In the root stem cell niche, about 25% wild type roots have two layers of DSC comparing with 50% abo8-1 roots as they do not contain starch and cannot be stained by Lugol's solution (Fig. 12A, 12B). Addition of GSH did not change the ratio of two DSC layers in the wild type, but reduced two DSC layers from 50% to 25% in abo8-1 (Fig. 12A, 12B). These results suggest that GSH antagonistically mediates the inhibition of root DSC differentiation by the abo8 mutation.
Fig. 12. abo8 mutation delays the distal stem cell (DSC) differentiation. 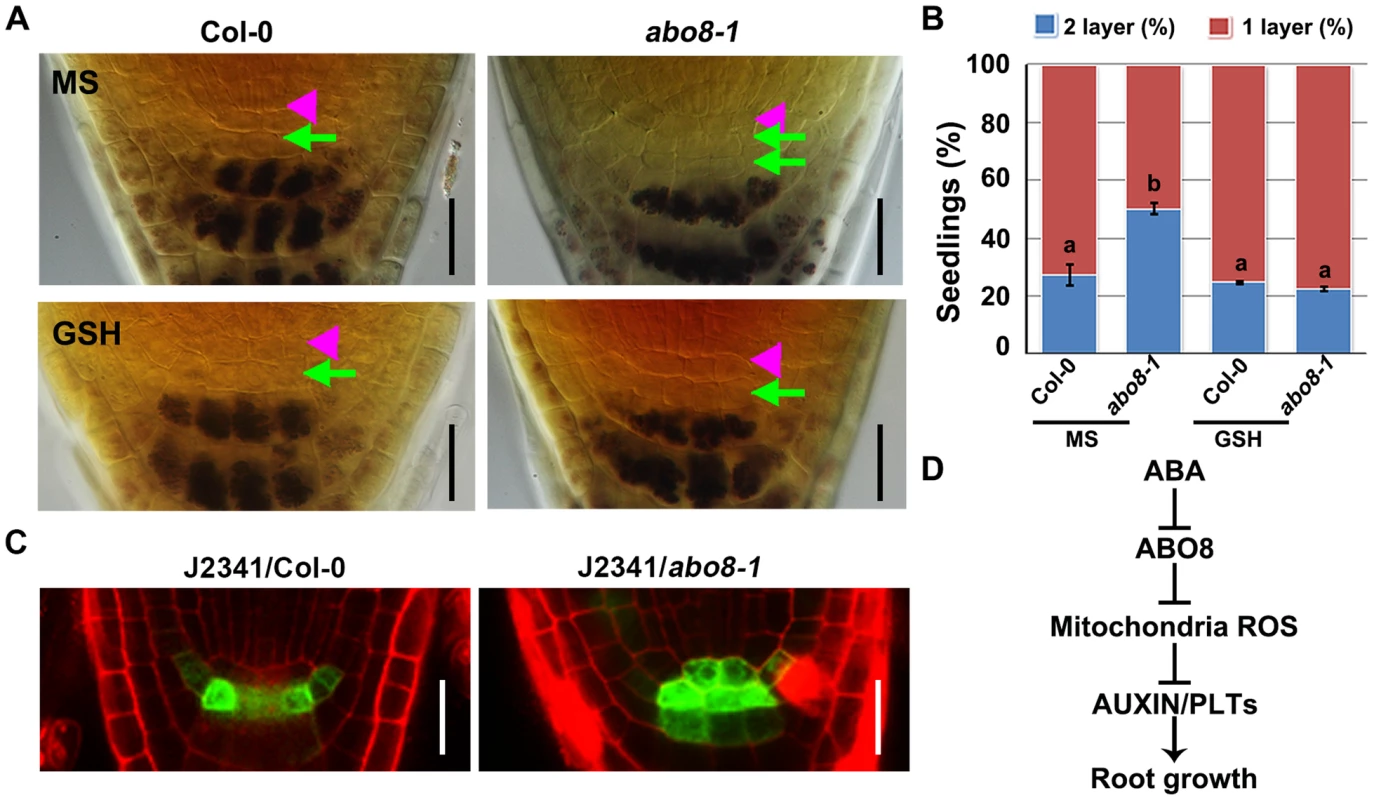
A. Differentiation status of DSC in 5-day-old seedlings on MS medium or MS medium with 300 µM GSH. DSC (green arrowheads) are characterized as cells between the QC (pink triangle) and the starch containing differentiated columella cells. Bars = 20 µm. B. Quantitative evaluation of DSC layers in A. About 30 roots were examined for each biological repeat, three experiments were done with similar results. Values are means ±SE from one experiment. Means in different letters are P<0.01. C. The expression of DSC marker J2341 in roots of the wild type and abo8-1 mutants. Bars = 20 µm. D. A proposed model for ABO8 in root growth. ABO8 is downreguated by ABA. ABO8 negatively modulates mitochondria ROS, and ROS negatively modulate auxin level and signaling to control PLTs expression. It is likely that ROS also directly regulate PLT1 as posttranscriptional level. We also introduced J2341 marker, a specific DSC marker [31], into abo8-1 to observe GFP localization. GFP fluorescence was observed in one layer of DSC in near 75% wild-type roots comparing with only about 25% in abo8-1 roots (20 roots were accounted each time, repeated two times) (Fig. 12C). These results indicate that the abo8-1 mutation delays the DSC differentiation.
Discussion
Recent studies have indicated that a plant-specific family of PPR proteins is involved in crucial RNA metabolism in plastids and mitochondria [32]. In this study, we found that ABO8, a PPR protein localized in mitochondria, is specifically required for the splicing of NAD4 intron 3 in mitochondrial complex I. A defect of ABO8 leads to various retarded growth phenotypes, high accumulation of ROS, and ABA hypersensitivity in seed germination and root growth. We further showed that the ABA hypersensitivity in root growth of abo8 mutants is due to a high accumulation of ROS that decreases auxin accumulation and signaling, which results in greatly reduced expression of PLT genes (Fig. 12D). Overexpressing PLT2 in an inducible system can largely recover the root-hypersensitive phenotype of abo8 under ABA treatment, suggesting that PLTs are important in the ABA inhibition of root growth.
Plant cells contain two kinds of symbiotic organelles, mitochondria and plastids, both of which have their own genomes that encode a limited number of proteins [32]. Although the mitochondrial genome contains only about 40 genes, >2000 proteins have been identified in mitochondria [33]. The mitochondrial protein-coding genes must be finely regulated by many different nuclear proteins, most of which are PPR proteins. The following PPR proteins have been found to facilitate NAD4 mRNA metabolism in Arabidopsis: MTSF1 (mitochondrial stability factor 1) for the 3′-processing of NAD4 mRNA and its stability [34]; RPF1 (RNA PROCESSING FACTOR 1) for 5′-end processing of the NAD4 mRNA [35]; AHG11 (ABA hypersensitive germination 11) for NAD4 RNA editing [36]; SLO1 (SLOW GROWTH1) for editing NAD4 and NAD9 [37]; LOI1 (LOVASTATIN INSENSITIVE 1) for editing NAD4, CCB203, and COX3 [38], [39]; and ABO8 for splicing of NAD4 intron 3. Except for PPR proteins, a mutation in the group II intron-encoded reverse transcriptase/maturase (At-nMat1a) also impairs cis-splicing of NAD4 intron 2 and NAD2 intron 1 and trans-splicing of NAD1 intron1 [40], [41], [42].
A dysfunction in the mitochondrial electron transport chain of complex I can cause a redox imbalance and increases in ROS accumulation. Mutants with impaired mitochondrial functions usually have various severe phenotypes including retarded seedling growth and abiotic stress responses. Several mutants such as abo5 [21], abo6 [19], ahg11 [36], slo2 (for editing several transcripts in complex I) [43], and slg1 (editing NAD3 transcript) [44] accumulate more ROS than the wild type and are more sensitive to ABA than the wild type in seed germination and/or root growth. In a previous study, we found that ROS accumulation in abo6 impairs auxin accumulation and/or signaling, suggesting that a redox regulatory mechanism plays crucial roles in controlling root growth [19]. The abo8 mutants have the similar retarded growth phenotype and ABA sensitivity as abo6 or abo5 and also accumulate more ROS than the wild type. Addition of the reducing agent GSH can restore the ABA sensitivity in both root growth and seed germination, probably by reducing ROS. Root tips and lateral root primordia are the regions where auxin is highly accumulated. Recent studies indicate that the bioactive IAA can be irreversibly oxidized into OxIAA (2-oxindole-3-acetic acid), a form that has little auxin activity [20], [45]; this process mainly occurs in the root tips [20]. Oxidation of auxin to OxIAA is an important mechanism for removing high levels of active auxin from the root apex to maintain auxin homeostasis [20]. Our study indicates that ROS are highly accumulated in root tips and that such accumulation might facilitate the oxidation of IAA in these tissues. Although a highly oxidized status is important for maintaining the low mitotic activity in QC cells [46], greatly increased ROS accumulation caused by mutations such as abo8 and abo6 would disrupt the redox homeostasis, further oxidize IAA, reduce the IAA level, and ultimately reduce the stem cell activity. The root meristems are much shorter in abo8-1 than in the wild type. Interestingly, in this study, we found that addition of high concentrations of auxins (IAA or NAA) did not induce the expression of auxin responsive marker gene ProDR5:GUS around root stem cell niche, but only highly induced its expression in upper part of root meristem in abo8-1, while in wild type ProDR5:GUS was highly induced in the whole root meristem. However, addition of auxins plus GSH was able to induce the expression of ProDR5:GUS in the whole root meristem. These results suggest that redox homeostasis plays crucial roles in maintaining a proper auxin gradient and auxin response in root tips.
PLT genes are essential for regulating the patterning of root stem cell niche [27], [28]. The expression domains of PLT overlap with the auxin gradient in the root tips. The auxin gradient is thought to control root growth by tightly regulating the expression of PLTs [27], [28]. In this study, we found that the plt1-4 and plt2-2 mutants are more sensitive to ABA than the wild type in root growth, suggesting that PLTs are negative regulators in ABA inhibition of root growth. In respect to regulating RAM, the abo8-1 mutation enhances the phenotypes of plt1-4 and plt2-2, suggesting that PLT1/PLT2 and ABO8 additionally regulate root growth. Interestingly, GSH treatment does not change the expression pattern of ProPLT1:CFP around root stem cell niche, but does change the distribution of PLT1-YFP in abo8-1. PLT1-YFP is usually localized around root stem cell niche in the wild type, but in abo8-1, besides around the root stem cell niche is also found above it. These results suggest that distribution of PLT1 protein might be affected by both redox homeostasis and auxin response in root tips. The previous study on miao mutant indicates that the miao weak mutation in GR2 (GSH reductase) greatly reduces GSH content, and retards root growth [15]. The expression of PLT1 and PLT2 is decreased in the miao mutant. Interestingly, introducing a dexamethasone (DEX)-inducible PLT2 into the miao mutant with overproduction of PLT2 does not increase the RAM length of the miao mutant, suggesting that a reduced glutathione environment is required for proper function of PLT downstream factors in root meristem. However, the DEX-inducible PLT2 can largely induce the RAM expansion of abo8-1, suggesting that PLT2 acts downstream of ABO8 for RAM maintenance. The different RAM responses of miao and abo8-1 to PLT2 overexpression suggest that oxidation status imposed by high accumulation of ROS from impairment of mitochondria and glutathione redox status by reduced GSH level have different effects on PLTs/Auxin signaling. abo8 mutation also reduces the expression of PINs and AUX1, and delays DSC differentiation. Taken together, these results demonstrate that ROS from mitochondria are crucial mediators for root growth.
Materials and Methods
Plant materials and growth conditions
Surface-sterilized wild type Arabidopsis thaliana (Col-0) seeds were plated on MS medium containing 1% sucrose and 0.8% agar. The plates were stratified at 4°C for 2 days and then transferred to a light incubator (22°C) with 22 h light/2 h dark. For the seed germination greening assay, the seedlings were grown for 7 days before photographed. For the root growth assay, 4-day-old seedlings were transferred to MS or MS medium supplemented with different concentrations of ABA or GSH. The root tips were placed in a straight line. The plates were oriented vertically in a light incubator for another 5 days before they were photographed. Root growth was measured with Imag J (Image J; National Institutes of Health; http://rsb.info.nih.gov/ij). In a greenhouse, the seedlings were grown at 22°C and 16 h light/8 h dark under 50 µmol m−2 s−1 light. ABA (Sigma-Aldrich) was prepared as a 30 mM stock solution in ethanol. plt1-4 and plt2-2 in Arabidopsis thaliana ecotypes Wassilewskija (Ws) were used for genetic analysis [28].
Mutant screening
Arabidopsis (Col-0) seeds were mutated with EMS (ethyl methanesulfonate). The 5-day-old M2 seedlings were transferred to MS medium containing 30 µM ABA with the root tips upward and were grown for another 7 days. The seedlings with short roots were selected as putative mutants. The ABA-sensitive phenotype of the putative mutants was rechecked by comparing the root growth on MS medium and MS medium supplemented with 30 µM ABA in the second generation.
Map-based cloning and mutant complementation
abo8-1 (Col-0) was crossed with Landsberg erecta. A total of 912 abo8-1 seedlings were selected from the segregating F2 population for map-based cloning. ABO8 was localized between two SNP makers on bacterial artificial chromosomes F25E4 and T26M18 in chromosome 4. This region contains genes numbered from AT4G11390 to AT4G11876 (The TAIR SeqViewer; http://www.arabidopsis.org/servlets/sv). We sequenced all of the open reading frames in this region and only identified a G447 to A447 (counting from the first putative ATG) mutation, which caused a premature stop codon in AT4G11690.
The genomic sequence containing a complete and single gene, AT4G11690, was amplified with the following primers: 5′-CGCGGATCCCGGCATTTGATCTATGTC GTTCTTC-3′ and 5′-GAGGGTACCTTTTGAGCTTACATGTGAATCGTTTTTGG-3′. The DNA fragment was cloned in the BamHI and KpnI sites of a modified vector pCAMBIA1300, with the C-terminus of ABO8 fused with green fluorescent protein (GFP). The construct ProABO8:ABO8-GFP was then introduced into abo8-1 via Agrobacterium-mediated transformation. The transgenic plants were screened on MS medium containing 30 mg/L hygromycin.
We obtained a T-DNA line SALK_025470 with T-DNA inserted at +358 bp of the AT4G11690 gene from the Arabidopsis Biological Resource Center (ABRC; http://abrc.osu.edu).The homozygous T-DNA insertion line was obtained using the following primers: LP, 5′-CTGCATAAACTCAACGCCTTC-3′; RP, 5′-TTTAAACGTATCTGCCATGGC-3′; TF, 5′-ATTTTGCCGATTTCGGAAC-3′. abo8-1 was crossed with homozygous SALK_025470, and the F1 plants were analyzed for root sensitivity to ABA.
Subcellular localization
The roots of 10-day-old seedlings of homozygous transgenic line ProABO8:ABO8-GFP were imaged using a confocal laser scanning microscope (LSM510; Carl Zeiss, Oberkochen, Germany). About eight independent transgenic lines expressing the ProABO8:ABO8-GFP were analyzed. The iPSORT (http://ipsort.hgc.jp) program predicts that the PPR protein ABO8 is targeted to the mitochondrion. To study the subcellular localization of ABO8, we prepared protoplasts from the roots of 10-day-old ProABO8:ABO8-GFP seedlings, and mitochondria were stained with MitoTracker orange as a control for co-localization. The images were obtained using a Carl Zeiss LSM510 META laser scanning microscope. GFP signals were collected using emission filter BP505–530 nm with an excitation at 488 nm, and red signals (MitoTracker stain) were obtained using BP 585–615 nm with an excitation at 543 nm.
GUS staining
The ABO8 promoter (about 2.1 kb) was cloned into pCAMBIA1391 (with a GUS coding region) between the BamHI and SpeI sites using the following primers: 5′-CGCGGATCCCGGCATTTGATCTATGTCGTTCTTC-3′ and 5′-CGCGACTAGTGTGTGTGTTGAAATTCATGGATTCG-3′. The ProABO8:GUS construct was then transferred into Col-0 via Agrobacterium-mediated transformation. About 20 independent transgenic lines expressing ProABO8:GUS were analyzed. For the GUS staining assay, seedlings were incubated in 0.1 M phosphate buffer (pH 7.0), 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, 0.1% Triton X-100, and 0.5 mg/mL X-Gluc at 37°C in the dark for 24 h and were then incubated in 75% ethanol overnight. Seedlings were photographed with an Olympus BX53 microscope and an Olympus SZX16 stereoscopic microscope.
The transgenic plants carrying ProCYCB1;1:GUS, ProDR5:GUS,or ProIAA2:GUS were crossed with abo8-1 mutants. The F2 seedlings were genotyped for the abo8-1 allele using specific dCAPS primers: 5′-CCACTCCCAATTCTTCACATCTTCCTC-3′ and 5′-CTTTGCTTTTGTTCTCGTTGAAGAAGCT-3′. The PCR products were digested with HindIII. The homozygous transgenic lines of F3 plants were selected on MS containing 50 mg/L kanamycin. GUS staining was carried out as described earlier [19]. Staining time was 90 min for ProCYCB1;1:GUS, 50 min for ProIAA2:GUS, and 18 h for ProDR5:GUS.
Northern blot and qRT-PCR
RNA was extracted from 10-day-old seedlings that were treated or not treated with 50 µM ABA for 5 h in liquid MS. A 20-µg quantity of RNA per sample was used for Northern blot as previously described [21].The probes used for Northern blot were described previously [21]. TUBULIN4 was used as a loading control.
For qRT-PCR, 4 µg of total RNA was first digested with DNase I (TaKaRa) and then reverse transcribed into Poly(dT) cDNA using a Moloney Murine Leukemia Virus Transcriptase kit (Promega) according to the manufacturer's instructions. qRT-PCR was performed as previously described [19]. ACTIN2 was used as the internal control. The primers designed to analyze NAD4 splicing were described previously [19]. The primers used to analyze ABO8 expression were gene specific primers that flanking the T-DNA insertion: 5′-GGTTTTGTTCCTGGATCGAATTGCTTCAAC-3′ and 5′-TCCCAAAGCTATACACGT CCAAAACAAC-3′. The primers designed to analyze AOX1a expression were described previously [44].
Blue Native-PAGE and complex I activity assay
5-day-old seedlings were treated with or without 50 µM ABA for 24 h. The crude mitochondrial membrane proteins were prepared according to the method described previously [44]. 20 µg protein of each sample was loaded onto and separated by a 4.5% to 16% gradient Blue-Native PAGE (Invitrogen, BN1002BOX) according to the manufacturer's instructions. The activity of mitochondria complex I were analyzed by staining the gel in reaction buffer (50 mM Mops-KOH at pH 7.6, 0.2 mM NADH and 1 mM NBT). The reaction was stopped when the dark blue stain was strong enough by immersing the gel in 30% methanol and 10% acetic acid (v/v).
Measurement of cellular ATP content
5-day-old seedlings were treated with or without 50 µM ABA for 24 h. About 100 mg seedlings was ground to powder in liquid nitrogen and resuspended in 400 µL of 2.3% (v/v) trichloroacetic acid and mixed vigorously. After centrifuged for 15 min at 20,000 g, the supernatant was collected and adjusted to pH 7.0 with 2.5 M K2CO3. Adenosine 5′-triphosphate (ATP) bioluminescent assay Kit (SigmaAldrich, fl-aa) was used to measure the ATP concentration according to the method described previously [47].
Auxin content measurement
7-day-old seedlings of the wild type and abo8-1 were harvested and ground to powder in liquid nitrogen. For each sample, about 50–100 mg powder were resuspended in pre-cooling 80% methanol and mix immediately. The samples were kept at 4°C protected from light before 0.8 ng [13C]-IAA was added. Free IAA content measurement was performed using GC-QQQ (Agilent, 7000A) in Institute of Botany, the Chinese Academy of Sciences.
Measurement of ROS in plants
Four-day-old seedlings of Col-0 and abo8-1 were treated in liquid MS with or without 50 µM ABA for 5 h before the seedlings were stained as described in the following paragraphs.
For 3′, 3′ - diaminobenzidine (DAB) staining to detect H2O2, the seedlings were incubated in 0.3 mg/mL DAB (Sigma-Aldrich) dissolved in 50 mM Tris-HCl (pH 5.0) for 8 h. For nitroblue tetrazolium (NBT) staining to detect superoxides, the seedlings were incubated in a reaction buffer containing 1 mM NBT (Sigma-Aldrich), 20 mM K-phosphate, and 0.1 M NaCl at pH 6.2 for 15 min. The seedlings stained by DAB or NBT were then washed three times with water. For clearing, seedlings were incubated in acidified methanol buffer (10 mL of methanol, 2 mL of HCl, 38 mL of water) at 57°C for 15 min and then in a basic solution (7% NaOH in 60% ethanol) for 15 min at room temperature. The seedlings were incubated 10 min at each step in the following series: 40% ethanol, 20% ethanol, 10% ethanol, 5% ethanol, and 25% glycerol. The seedlings were then examined in 50% glycerol with an Olympus BX53 microscope.
For 2′,7′-dichlorodihydrofluorescin diacetate (DCFH-DA) staining to detect H2O2, the seedlings were incubated in a buffer containing 50 µM DCFH-DA (Sigma-Aldrich) and 20 µM K-phosphate at pH 6.0 in darkness for 10 min. The roots were then photographed using a Carl Zeiss LSM510 META confocal microscope with an excitation at 488 nm. The intensities of the fluorescent signals were statistically compared with the Student's t-test.
Measurement of root meristems
For analysis of root meristems, 4-day-old seedlings were transferred to MS medium or MS medium supplemented with 30 µM ABA, 300 µM GSH, or 2 µM DEX. After 3 days, the roots were placed in a mounting solution (7.5 g gum arabic, 100 g chloroacetaldehyde, 5 mL glycerol, and 60 mL water) and were photographed with an Olympus BX53 microscope. We measured the meristem cell number in the cortex between the QC and the first elongating cell, elongation zone (EZ) cell number from the first elongating cell to first cell with a root hair. Differential zone is from the first cell with a root hair to the joint of root and hypocotyl.
Starch staining
Seedlings were immersed into Lugol (0.2% iodine and 2% potassium iodine) and incubated for about 1 min, washed in water once and mounted on the slide with a mounting solution described above. The Lugol-stained seedlings were photographed with an Olympus BX53 microscope.
Fluorescence microscopy
Arabidopsis transgenic plants harboring ProPLT1:PLT1-YFP, ProPLT2:PLT2-YFP, ProPLT1:CFP, ProPIN1:PIN1-GFP, ProPIN2:PIN2-GFP, ProAUX1:AUX1-YFP, J2341, or Mito-cpYFP [19] were crossed with abo8-1. The method for genotyping the abo8-1 allele was described earlier. An Olympus SZX16 stereo fluorescence microscope was used to select the homozygous transgenic plants from the F3 generation. Four-day-old homozygous seedlings were transferred to MS medium or MS containing 30 µM ABA, 300 µM GSH, or both, and were grown for another 2 days. Propidium iodide fluorescence was used to visualize the cells in the root tip. Seedlings were incubated in 10 µM propidium iodide (Sigma-Aldrich) for 3 min, and the roots were then imaged with a Carl Zeiss LSM510 META confocal microscope. Fluorescence intensities were measured using AxioVision Rel. 4.8 software.
Supporting Information
Zdroje
1. De TullioMC, JiangK, FeldmanLJ (2010) Redox regulation of root apical meristem organization: connecting root development to its environment. Plant Physiol Biochem 48 : 328–336.
2. MittlerR, VanderauweraS, SuzukiN, MillerG, TognettiVB, et al. (2011) ROS signaling: the new wave? Trends Plant Sci 16 : 300–309.
3. SuzukiN, KoussevitzkyS, MittlerR, MillerG (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35 : 259–270.
4. TognettiVB, MuhlenbockP, Van BreusegemF (2012) Stress homeostasis - the redox and auxin perspective. Plant Cell Environ 35 : 321–333.
5. WangP, SongCP (2008) Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol 178 : 703–718.
6. O'BrienJA, DaudiA, ButtVS, BolwellGP (2012) Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 236 : 765–779.
7. De PintoMC, LocatoV, De GaraL (2012) Redox regulation in plant programmed cell death. Plant Cell Environ 35 : 234–244.
8. PottersG, PasternakTP, GuisezY, PalmeKJ, JansenMA (2007) Stress-induced morphogenic responses: growing out of trouble? Trends Plant Sci 12 : 98–105.
9. OvermyerK, BroscheM, KangasjarviJ (2003) Reactive oxygen species and hormonal control of cell death. Trends Plant Sci 8 : 335–342.
10. AchardP, RenouJP, BerthomeR, HarberdNP, GenschikP (2008) Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 18 : 656–660.
11. RubinovichL, WeissD (2010) The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J 64 : 1018–1027.
12. Sanchez-FernandezR, FrickerM, CorbenLB, WhiteNS, SheardN, et al. (1997) Cell proliferation and hair tip growth in the Arabidopsis root are under mechanistically different forms of redox control. Proc Natl Acad Sci U S A 94 : 2745–2750.
13. CobbettCS, MayMJ, HowdenR, RollsB (1998) The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in gamma-glutamylcysteine synthetase. Plant J 16 : 73–78.
14. VernouxT, WilsonRC, SeeleyKA, ReichheldJP, MuroyS, et al. (2000) The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12 : 97–110.
15. YuX, PasternakT, EiblmeierM, DitengouF, KocherspergerP, et al. (2013) Plastid-Localized Glutathione Reductase2-Regulated Glutathione Redox Status Is Essential for Arabidopsis Root Apical Meristem Maintenance. Plant Cell 25 : 4451–68.
16. BashandyT, GuilleminotJ, VernouxT, Caparros-RuizD, LjungK, et al. (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22 : 376–391.
17. IglesiasMJ, TerrileMC, BartoliCG, D'IppolitoS, CasalongueCA (2010) Auxin signaling participates in the adaptative response against oxidative stress and salinity by interacting with redox metabolism in Arabidopsis. Plant Mol Biol 74 : 215–222.
18. KwakJM, MoriIC, PeiZM, LeonhardtN, TorresMA, et al. (2003) NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 22 : 2623–2633.
19. HeJ, DuanY, HuaD, FanG, WangL, et al. (2012) DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 24 : 1815–1833.
20. PencikA, SimonovikB, PeterssonSV, HenykovaE, SimonS, et al. (2013) Regulation of Auxin Homeostasis and Gradients in Arabidopsis Roots through the Formation of the Indole-3-Acetic Acid Catabolite 2-Oxindole-3-Acetic Acid. Plant Cell 25 : 3858–70.
21. LiuY, HeJ, ChenZ, RenX, HongX, et al. (2010) ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant J 63 : 749–765.
22. BanT, KeJ, ChenR, GuX, TanMH, et al. (2013) Structure of a PLS-Class Pentatricopeptide Repeat Protein Provides Insights into Mechanism of RNA Recognition. J Biol Chem 288 : 31540–8.
23. MeyerEH, TomazT, CarrollAJ, EstavilloG, DelannoyE, et al. (2009) Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol 151 : 603–619.
24. UlmasovT, MurfettJ, HagenG, GuilfoyleTJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 : 1963–1971.
25. BenkovaE, MichniewiczM, SauerM, TeichmannT, SeifertovaD, et al. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 : 591–602.
26. ShibasakiK, UemuraM, TsurumiS, RahmanA (2009) Auxin response in Arabidopsis under cold stress: underlying molecular mechanisms. Plant Cell 21 : 3823–3838.
27. GalinhaC, HofhuisH, LuijtenM, WillemsenV, BlilouI, et al. (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449 : 1053–1057.
28. AidaM, BeisD, HeidstraR, WillemsenV, BlilouI, et al. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119 : 109–120.
29. van den BergC, WillemsenV, HageW, WeisbeekP, ScheresB (1995) Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378 : 62–65.
30. NakajimaK, BenfeyPN (2002) Signaling in and out: control of cell division and differentiation in the shoot and root. Plant Cell 14 Suppl: S265–276.
31. ZhouW, WeiL, XuJ, ZhaiQ, JiangH, et al. (2010) Arabidopsis Tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22 : 3692–3709.
32. NakamuraT, YagiY, KobayashiK (2012) Mechanistic insight into pentatricopeptide repeat proteins as sequence-specific RNA-binding proteins for organellar RNAs in plants. Plant Cell Physiol 53 : 1171–1179.
33. MillarAH, HeazlewoodJL, KristensenBK, BraunHP, MollerIM (2005) The plant mitochondrial proteome. Trends Plant Sci 10 : 36–43.
34. HailiN, ArnalN, QuadradoM, AmiarS, TcherkezG, et al. (2013) The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res 41 : 6650–6663.
35. HolzleA, JonietzC, TorjekO, AltmannT, BinderS, et al. (2011) A RESTORER OF FERTILITY-like PPR gene is required for 5′-end processing of the nad4 mRNA in mitochondria of Arabidopsis thaliana. Plant J 65 : 737–744.
36. MurayamaM, HayashiS, NishimuraN, IshideM, KobayashiK, et al. (2012) Isolation of Arabidopsis ahg11, a weak ABA hypersensitive mutant defective in nad4 RNA editing. J Exp Bot 63 : 5301–5310.
37. SungTY, TsengCC, HsiehMH (2010) The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences in Arabidopsis mitochondria. Plant J 63 : 499–511.
38. TangJ, KobayashiK, SuzukiM, MatsumotoS, MuranakaT (2010) The mitochondrial PPR protein LOVASTATIN INSENSITIVE 1 plays regulatory roles in cytosolic and plastidial isoprenoid biosynthesis through RNA editing. Plant J 61 : 456–466.
39. VerbitskiyD, ZehrmannA, van der MerweJA, BrennickeA, TakenakaM (2010) The PPR protein encoded by the LOVASTATIN INSENSITIVE 1 gene is involved in RNA editing at three sites in mitochondria of Arabidopsis thaliana. Plant J 61 : 446–455.
40. MohrG, LambowitzAM (2003) Putative proteins related to group II intron reverse transcriptase/maturases are encoded by nuclear genes in higher plants. Nucleic Acids Res 31 : 647–652.
41. NakagawaN, SakuraiN (2006) A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial NAD4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol 47 : 772–783.
42. KerenI, TalL, des Francs-SmallCC, AraujoWL, ShevtsovS, et al. (2012) nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J 71 : 413–426.
43. ZhuQ, DugardeynJ, ZhangC, MuhlenbockP, EastmondPJ, et al. (2013) The Arabidopsis thaliana RNA Editing Factor SLO2, which Affects the Mitochondrial Electron Transport Chain, Participates in Multiple Stress and Hormone Responses. Mol Plant 7 : 290–310.
44. YuanH, LiuD (2012) Functional disruption of the pentatricopeptide protein SLG1 affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. Plant J 70 : 432–444.
45. PeerWA, ChengY, MurphyAS (2013) Evidence of oxidative attenuation of auxin signalling. J Exp Bot 64 : 2629–2639.
46. JiangK, MengYL, FeldmanLJ (2003) Quiescent center formation in maize roots is associated with an auxin-regulated oxidizing environment. Development 130 : 1429–1438.
47. YangXY, ChenZW, XuT, QuZ, PanXD, et al. (2011) Arabidopsis kinesin KP1 specifically interacts with VDAC3, a mitochondrial protein, and regulates respiration during seed germination at low temperature. Plant Cell 23 : 1093–1106.
Štítky
Genetika Reprodukčná medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 12- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



