-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
Non-targeted metabolomic profiling of large population-based studies has become feasible only in the past 1–2 years and this hypothesis-free exploration of the metabolome holds a great potential to fuel the discovery of novel biomarkers for coronary heart disease (CHD). Such biomarkers are not only important for risk stratification and treatment decisions, but can also improve understanding of cardiovascular disease pathophysiology to identify new drug targets. In this study, we investigated the metabolic profiles of more than 3,600 individuals from three population-based studies, and discovered four metabolites that are consistently associated with incident CHD. We integrate genetic and metabolomic analysis to delineate the underlying biological mechanisms and evaluate potential causal effects of the novel biomarkers. Specifically, we found one metabolite to be strongly associated with single nucleotides polymorphisms previously reported for association with CHD, and consistent with a potential causal role in CHD development, as suggested by Mendelian randomization analysis.
Published in the journal: Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease. PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004801
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004801Summary
Non-targeted metabolomic profiling of large population-based studies has become feasible only in the past 1–2 years and this hypothesis-free exploration of the metabolome holds a great potential to fuel the discovery of novel biomarkers for coronary heart disease (CHD). Such biomarkers are not only important for risk stratification and treatment decisions, but can also improve understanding of cardiovascular disease pathophysiology to identify new drug targets. In this study, we investigated the metabolic profiles of more than 3,600 individuals from three population-based studies, and discovered four metabolites that are consistently associated with incident CHD. We integrate genetic and metabolomic analysis to delineate the underlying biological mechanisms and evaluate potential causal effects of the novel biomarkers. Specifically, we found one metabolite to be strongly associated with single nucleotides polymorphisms previously reported for association with CHD, and consistent with a potential causal role in CHD development, as suggested by Mendelian randomization analysis.
Introduction
Advances in high-throughput technologies can fuel discovery of novel biomarkers for early detection and prevention of coronary heart disease (CHD). Metabolomic profiling, or metabolomics, provides a holistic signature of biochemical activities in humans by detecting and quantifying low-weight molecules (<1,500 Da). Integration of genetic information and metabolomics data can generate new hypotheses regarding underlying pathophysiological processes [1]. Moreover, targeted metabolomics studies have identified several associations between metabolites and cardiovascular disease (CVD) risk [2], [3] highlighting the importance of metabolic pathways in the development of atherosclerosis.
The primary aim of our study was to identify novel CHD biomarkers by performing non-targeted metabolomics profiling in 3,668 individuals free of CHD at baseline from three population-based prospective cohort studies. Our secondary aims were to delineate the underlying biological mechanisms and to evaluate clinical utility, as well as potential causal effects for those metabolites showing strong evidence of association. For these purposes, we analyzed associations with measures of oxidative stress, inflammation and subclinical CVD, as well as integrated metabolomics and genetics data.
Results
An overview of the study design is illustrated in Fig. 1 and baseline characteristics of the three studies are described in S1 Table. Participants in ULSAM and PIVUS were all of the same approximate age at baseline (interquartile range, 70.1 to 71.0 years), while TwinGene participants were of younger median age (64.7 years) and with a wider range (interquartile range, 59.2 to 69.9 years).
Fig. 1. Study flow chart. 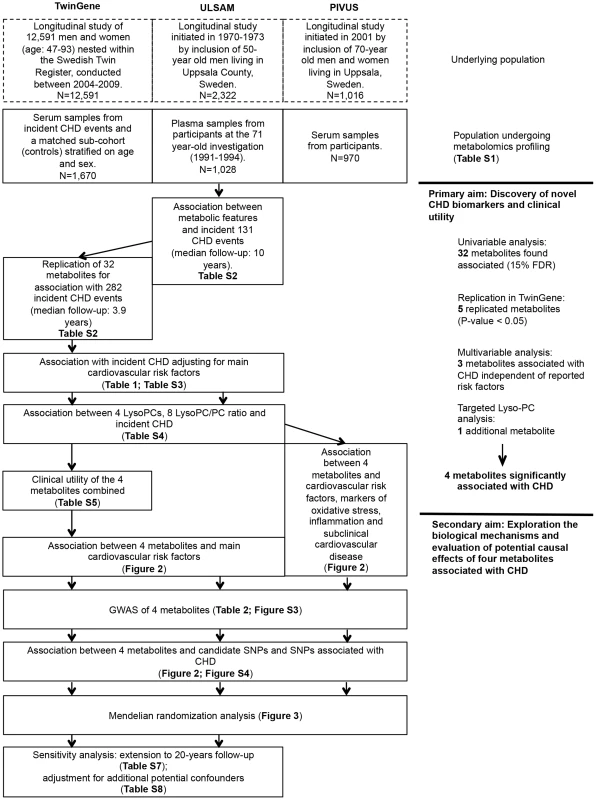
Overview of the study design and analyses performed. Primary aim: Discovery of novel CHD biomarkers and the clinical utility
Discovery and validation of metabolic features associated with incident CHD
In the 1,028 ULSAM participants free of CHD events at baseline, we observed 131 CHD events during a median follow-up of 10.0 years. There were 32 unique metabolites associated with CHD incidence at a 15% FDR level (S2 Table). Nine metabolites were annotated using our in-house compound library [Metabolomics Standard Initiative (MSI) level 1] and 12 using publically available databases (MSI level 2). We could identify the metabolic class (MSI level 3) for seven metabolites, while four candidate metabolites could not be annotated (MSI level 4).
We sought to replicate these 32 metabolites in the TwinGene study, where 282 incident CHD events were observed during a median follow-up of 3.9 years. Twenty-seven metabolites showed a consistent direction in TwinGene (binomial test P-value <0.001). Five of the metabolites showed significant association (P-value<0.05) and consistent direction [monoglyceride 18∶2 (MG 18∶2), a monosaccharide, lysophosphatidylcholine 18∶2 (Lyso PC 18∶2), a derivative of cinnamic acid and sphingomyelin 28∶1 (SM 28∶1); Table 1 and S3 Table]. Since we detected a significant interaction between LysoPC 18∶2 and age (P-value = 0.03; S3 Table), with a stronger protective effect on CHD in individuals older than 70 years (S1 Figure, Panel A), we model this interaction in all the following analyses and report estimates also for individuals older than 70 years
Tab. 1. Association between metabolites replicated in the univariable analysis and CHD, adjusting for main cardiovascular risk factors, meta-analysis results from ULSAM and TwinGene (N = 2,698). 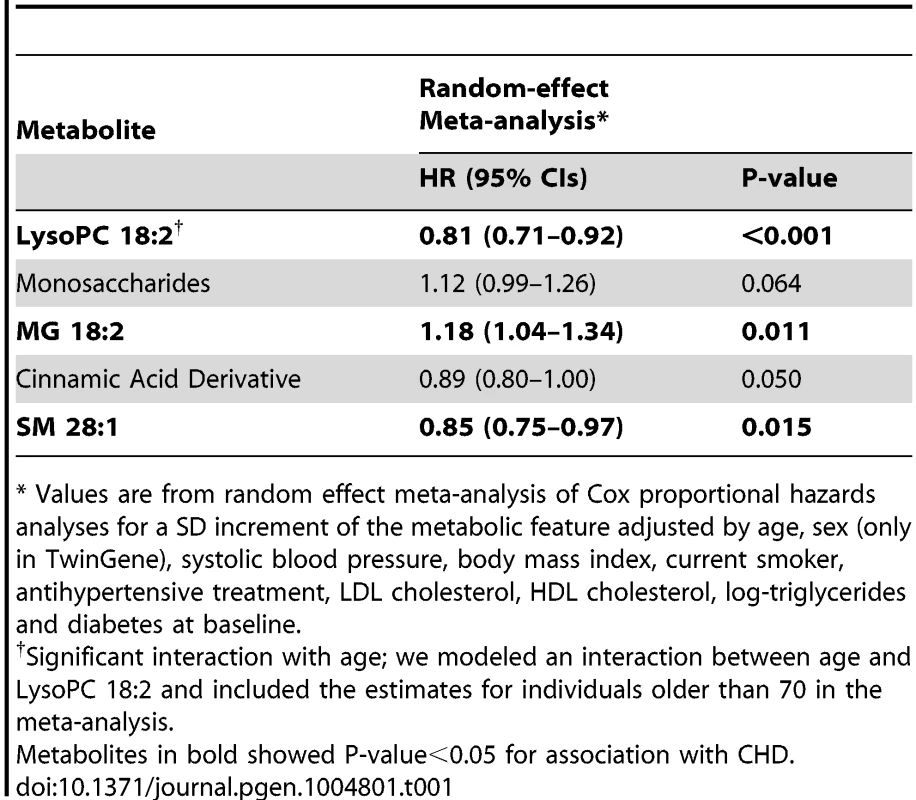
* Values are from random effect meta-analysis of Cox proportional hazards analyses for a SD increment of the metabolic feature adjusted by age, sex (only in TwinGene), systolic blood pressure, body mass index, current smoker, antihypertensive treatment, LDL cholesterol, HDL cholesterol, log-triglycerides and diabetes at baseline. Three metabolites (MG 18∶2, Lyso PC 18∶2 and SM 28∶1) remained significantly associated with incident CHD after adjustment for main cardiovascular risk factors and meta-analysis of TwinGene and ULSAM results (Table 1); LysoPC 18∶2 (hazard ratio [HR] per standard deviation [SD] increment = 0.81; P-value<0.001) and SM 28∶1 (HR = 0.85; P-value = 0.015) were negatively associated, while MG 18∶2 (HR = 1.18; P-value = 0.011) was positively associated.
Chemical structures of these metabolites were additionally confirmed by targeted tandem mass spectrometry (S2 Figure).
LysoPCs and their ratios in relation to incident CHD
Since LysoPC 18∶2 was the metabolite with the strongest association with incident CHD in ULSAM and in older participants from TwinGene, we extended our analysis to four additional LysoPC species to evaluate common patterns and pathways. Moreover, since the main mechanism by which LysoPCs are formed is via hydrolysis of phosphatidylcholines (PC) [4], we explored the association between the most abundant LysoPC/PC ratios and incident CHD. There was a strong negative association between LysoPC 18∶1 and incident CHD (HR = 0.77; P-value<0.001) in the two studies combined after adjustment for main cardiovascular risk factors (S4 Table). This increased the number of metabolites significantly associated with incident CHD independently of main cardiovascular risk factors to four. Survival curves for each metabolite are reported in S1 Figure, Panel B. The ratios between LysoPCs and PC were not significantly associated with CHD (S4 Table). LysoPC 18∶1 was highly positively correlated with LysoPC 18∶2 (r2 = 0.74, P-value<0.001) and negatively correlated with MG 18∶2 (r2 = −0.15, P-value<0.001). Similarly, SM 28∶1 was positively correlated with LysoPC 18∶1 (r2 = 0.37, P-value<0.001) and LysoPC 18∶2 (r2 = 0.42, P-value<0.001) and negatively correlated with MG 18∶2 (r2 = −0.13, P-value<0.001).
Clinical utility of four metabolites
Since four metabolites (LysoPC 18∶1, LysoPC 18∶2, MG 18∶2 and SM 28∶1) were associated with CHD after adjustment for main cardiovascular risk factors, we investigated their utility as biomarkers for CHD prediction. When the four metabolites were added to a model comprising the risk factors included in the Framingham Heart Study risk score [5], we observed a modest improvement in C-index (0.759 vs. 0.751, P-value = 0.026) and a moderate improvement in the Net Reclassification Index (NRI) (9.9% [1.2; 20.2] for events and −0.7% [−6.0; 0.5] for non-events; S5 Table).
Secondary aim: Exploration of biological mechanisms and evaluation of potential causal effects of four metabolites associated with CHD
Association with main cardiovascular risk factors, markers of oxidative stress, inflammation and subclinical CVD
We explored the associations of our four novel metabolites and main cardiovascular risk factors (Fig. 2, Panel A), as well as with markers of oxidative stress, inflammation and subclinical CVD (Fig. 2, Panel B). The two LysoPC species showed a similar pattern of association; higher LysoPC levels were associated with higher high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C) levels and lower body mass index (BMI). Similar associations were also observed for SM 28∶1. Monoglyceride 18∶2 was positively associated with triglycerides and BMI levels in all the three studies, while the association with HDL-C levels was in the inverse direction. The correlation between MG 18∶2 and triglycerides (measured in serum using standard methods) was strong (r2 range: 0.25–0.53). In TwinGene, when triglycerides and MG 18∶2 were included in the same model adjusting for only age and sex, both showed an independent significant positive association with incident CHD (S6 Table, panel A). Further, when they were separately added to models with all main cardiovascular risk factors except triglycerides, MG 18∶2 showed a larger increase in the likelihood ratio (204.6 vs. 197.6) and C-statistic (0.755 vs. 0.753) compared with triglycerides (S6 Table, panel B).
Fig. 2. Association between four metabolites and cardiovascular traits and genotypes. 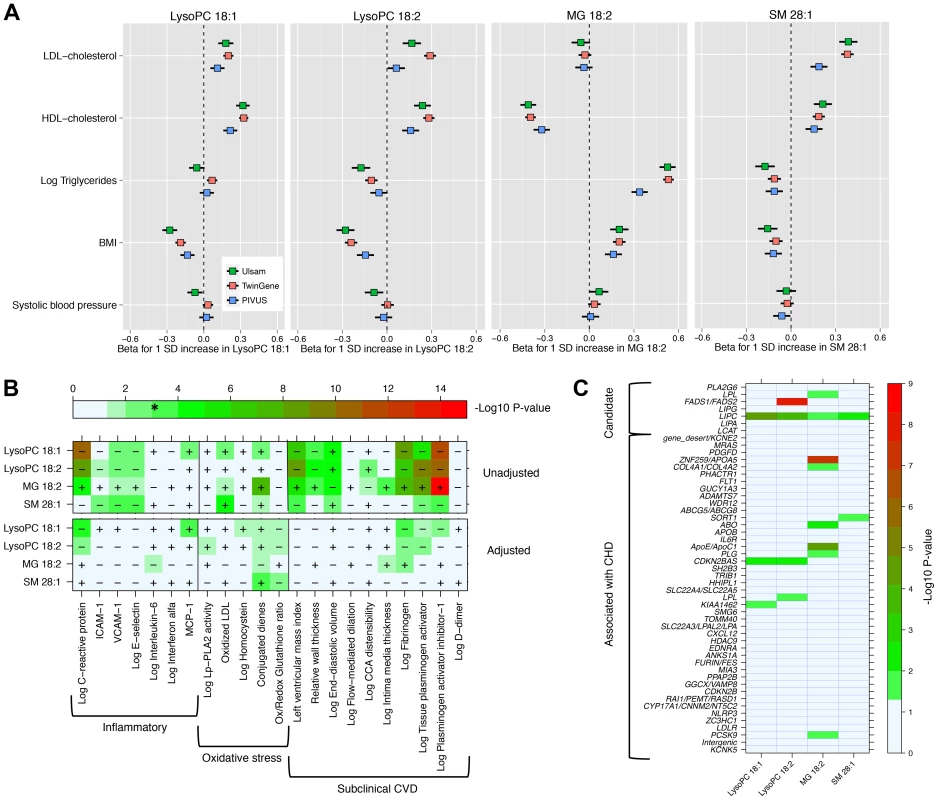
Panel A: Association with main cardiovascular risk factors in three population-based studies. Panel B: Minus log10(P-value) for association with markers of inflammation, oxidative stress and subclinical CVD in PIVUS. Sex-adjusted analysis (upper panel) and adjusted by sex, systolic blood pressure, body mass index, current smoker, antihypertensive treatment, LDL-C, HDL-C, log-triglycerides and diabetes at baseline (lower panel). * indicates the alpha threshold after multiple-testing correction. Panel C: Minus log10(P-value) for association with 51 SNPs previously reported for association with CHD (44 SNPs) or selected from candidate pathways (7 SNPs). In PIVUS (Fig. 2, Panel B), we observed the two LysoPC species being associated with lower levels of inflammation markers and less subclinical CVD. We detected a strong inverse association between LysoPC 18∶1 and plasminogen activator inhibitor-1 (PAI-I; P-value = 1.8×10−12), C-reactive protein (P-value = 3.1×10−11), fibrinogen (P-value = 7.3×10−9), and left ventricular mass index (P-value = 2.2×10−7). MG 18∶2 was positively associated with several markers of oxidative stress and subclinical CVD: PAI-I (P-value = 1.7×10−15), tissue plasminogen activator (P-value = 2.5×10−9), fibrogen (P-value = 6.4×10−8) and conjugated dienes (P-value = 1.2×10−8). After adjustment for main cardiovascular risk factors, most associations were attenuated. However, in these multivariable-adjusted models, LysoPC 18∶1 remained significantly associated with higher levels of monocyte chemotactic protein-1 (MCP-1) and lower levels of C-reactive protein and fibrinogen, even after correction for multiple testing.
Genome-wide association studies
We tested for association between ∼7.5M 1000G-imputed single nucleotide polymorphisms (SNPs) and the four metabolites associated with CHD independently of main cardiovascular risk factors (LysoPC 18∶1, LysoPC 18∶2, MG 18∶2 and SM 28∶1) in all 3,620 participants from the three studies with both genetic and metabolomics data (Table 2). In analyses of LysoPC 18∶1, we detected a novel association with rs75729820 (P-value = 2.7×10−8), close to C8orf87 on chromosome 8, and a suggestive association signal (rs8141918; P-value = 4.5×10−7) close to A4GALT on chromosome 22 (S3 Figure). We could also confirm a previously reported association between a SNP upstream of the FADS2 gene and LysoPC 18∶2 [6], and found a suggestive association between rs964184, in the ZNF259/APOA5 region, and MG 18∶2 (P-value = 1.2×10−7). The rs964184 variant has been associated with several cardiovascular traits including CHD in previous studies [6], [7]. The SNP rs12878001 near to SGPP1 was significantly associated with SM 28∶1 and correlates with a SNP (rs17101394; r2 = 1) previously reported to be associated with sphingolipid levels [8]. All the SNPs reported in Table 2 had consistent direction of effect in the three studies.
Tab. 2. GWAS of metabolites associated with CHD; SNPs with P-value<5×10−7 and minor allele frequency > 5% are reported. 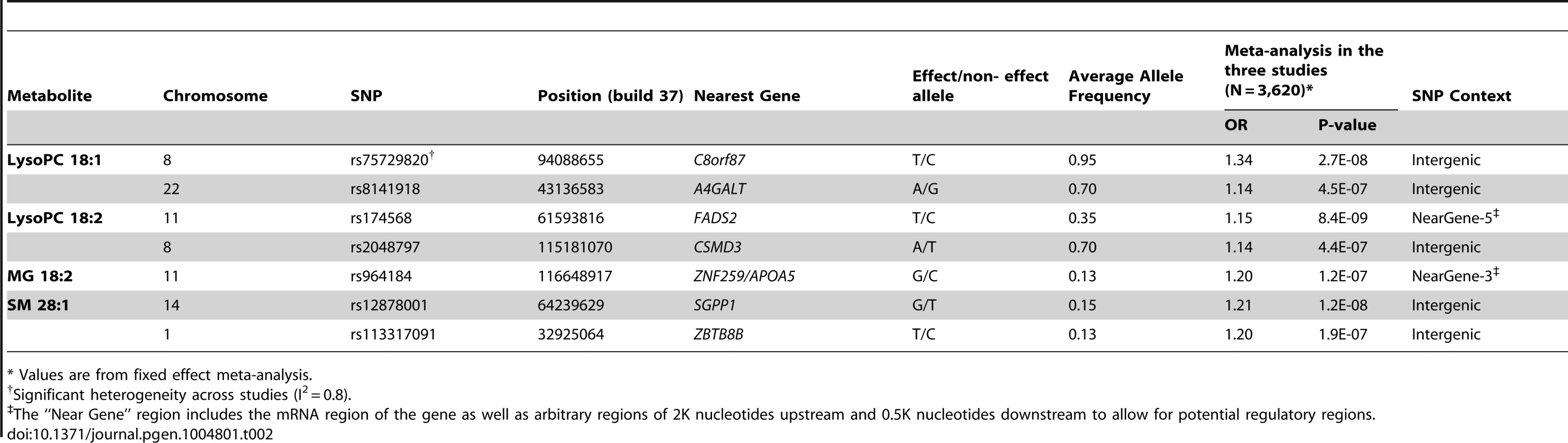
* Values are from fixed effect meta-analysis. Association with variants associated with CHD or from candidate pathways
We investigated the association between the four metabolites and 44 established CHD-associated SNPs [9], as well as seven candidate SNPs targeting biologically relevant pathways (Fig. 2, Panel C; S1 Text for SNPs selection procedures). MG 18∶2 showed a significant enrichment of P-values<0.05 for association with CHD-associated SNPs compared to the expected (hypergeometric test P-value = 0.002). This enrichment remained even after adjustment for main cardiovascular risk factors (P-value = 0.02; S4 Figure). The other metabolites did not show a significant enrichment of low p-values.
Among candidate SNPs targeting relevant pathways, LIPC was associated with all four metabolites, confirming the role of hepatic lipase in regulation of MG and LysoPCs levels. Candidate SNPs in the FADS1/FADS2 region were not strongly associated with MG 18∶2 and LysoPC 18∶1. After adjustment for main cardiovascular risk factors, only the association of FADS1/FADS2 with LysoPC 18∶2 remained genome-wide significant (P-value = 2.2×10−12).
Mendelian randomization analysis
The Mendelian randomization analysis (Fig. 3) suggested a weak, but positive causal effect of MG 18∶2 on CHD risk (odds ratio, 1.05 [95% CI, 1.00–1.10] per SD increment in MG 18∶2; P-value = 0.05) and a lack of causal effect for LysoPC 18∶1, 18∶2 and SM 28∶1.
Fig. 3. Mendelian randomization analysis. 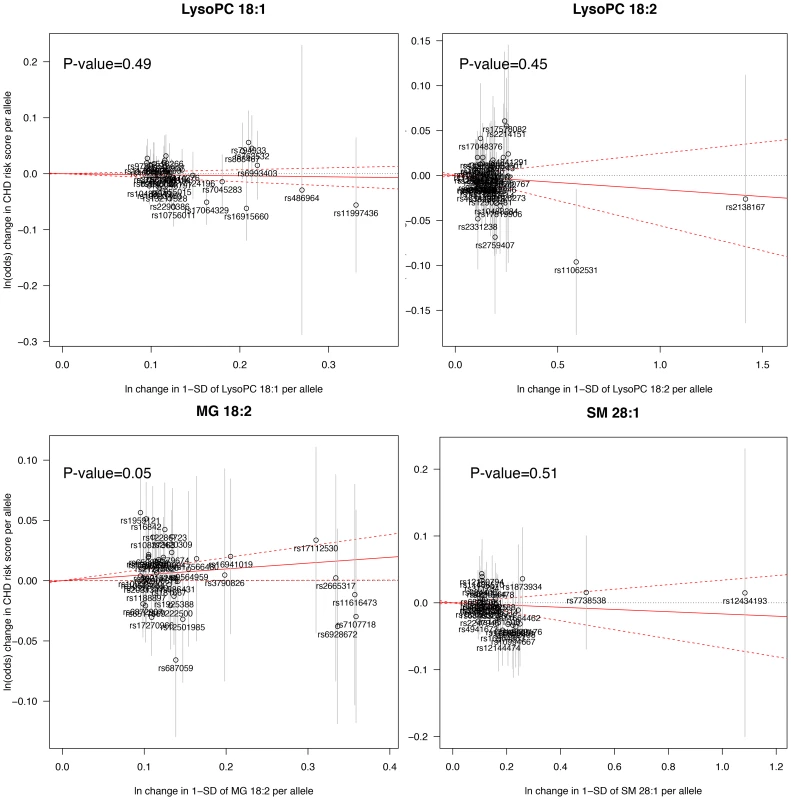
A significant deviation from zero of the estimate of causal effect using all SNPs (solid red line) suggests a causal relationship between the metabolite and CHD. Sensitivity and exploratory analysis
First, we evaluated the association of the four metabolites and incident CHD when the ULSAM follow-up was extended to 20 years, including 198 CHD events (S7 Table). The effect sizes were comparable to those observed for the 10-year follow-up. Second, in the multivariable analysis for association with CHD, we separately included two covariates in addition to the main cardiovascular risk factors: C-reactive protein and statin treatment. The associations between the four metabolites and incident CHD were essentially the same (S8 Table). Third, we assessed Lp-PLA2 activity in 254 older individuals (64 CHD events) from TwinGene. Lp-PLA2 is a known marker of atherosclerosis and hydrolyzes PC to produce LysoPCs. By adjusting the analysis for Lp-PLA2 we wanted to evaluate if the protective association between LysoPC 18∶1 and incident CHD was confounded or mediated by Lp-PLA2. The effect size after adjustment for Lp-PLA2 activity in addition to main cardiovascular risk factors was similar as in the main multivariable model (HR = 0.78; P-value = 0.176) arguing against Lp-PLA2 being an important confounder or mediator of the association.
Discussion
Principal findings
In this study of 3,668 participants from three prospective population-based cohorts, we investigated the association of circulating metabolites measured by liquid chromatography coupled mass spectrometry with incident CHD. In our discovery cohort, 32 metabolites were associated with CHD, of which 84% showed a directionally consistent association with CHD in our validation cohort. In multivariable analyses adjusted for main cardiovascular risk factors, three metabolites remained associated with CHD. In a targeted LysoPC analysis, we detected one additional significant association resulting in a total of four metabolites associated with CHD independently of main cardiovascular risk factors: LysoPC 18∶1, LysoPC 18∶2, MG 18∶2 and SM 28∶1. These biomarkers moderately improved risk reclassification beyond traditional risk factors, when commonly used risk categories were considered. Lysophosphatidylcholines were negatively associated with BMI, markers of inflammations and subclinical cardiovascular disease, while a reverse pattern was observed for MG 18∶2. We found evidences for a causal effect of MG 18∶2 on CHD independently of triglycerides levels. Finally, we uncovered several genome-wide significant SNPs and suggestive signals for association with LysoPCs, some of which have not been previously observed.
Monoglycerides
We observed a strong positive association between MG 18∶2 and CHD. The majority of circulating monoglycerides are released by the action of lipoprotein lipase and hepatic lipase, which catalyze the hydrolysis of triglycerides to provide non-esterified fatty acids and monoglycerides for tissue utilization [10]. Monoglycerides are further converted into free fatty acids and glycerol by monoglyceride lipase. Within the intestinal wall, monoglycerides are used to resynthesize diglycerides and triglycerides via monoacylglycerol pathway before being transported in lymph to the liver. Several observations suggest an involvement of MG 18∶2 in the pathogenesis of CHD. First, MG 18∶2 is central in the synthesis and breakdown of triglycerides and a causal effect of plasma triglyceride levels on CHD risk have recently been supported by a large Mendelian randomization analysis [11]. Although highly correlated, when both MG 18∶2 and triglycerides were included in the same model, both showed independent significant associations with CHD. Moreover, when separately added to a model with main cardiovascular risk factors, MG 18∶2 was a better predictor of CHD than triglycerides. Second, MG 18∶2 was associated with higher levels of cardiovascular risk factors and markers of subclinical CVD and oxidative stress. Third, Mendelian randomization analysis suggested a weak, but positive causal effect of MG 18∶2 on CHD risk. Several SNPs reported for association with CHD remained associated with MG 18∶2 (in the PCSK9, HHIPL1, PLG, ApoE/ApoC1, COL4A1/COL4A2 regions, P-values<0.05), even after adjustment for main cardiovascular risk factors.
LysoPCs
We observed a strong age-dependent association between LysoPC 18∶2, LysoPC 18∶1 and CHD risk, with stronger inverse association in older individuals. These LysoPC species were further characterized to be associated with higher HDL-C and total cholesterol levels, and lower BMI and markers of subclinical CVD. Moreover, they were highly correlated, suggesting shared biological mechanisms. LysoPCs are mostly derived from phosphatidylcholines (PC) and several mechanisms contribute to their formation. A large component of LysoPC in plasma is derived from PC by the glycoprotein lecithin cholesterol acyltransferase (LCAT). Another well-known mechanism of LysoPC production, which mainly takes place in tissues, is via PC hydrolysis by the action of secretory PLA2 family [4]. Although higher levels of LysoPCs have been observed during the oxidative modification of LDL-C that accompanies their conversion to atherogenic particles, it has also been shown that LysoPCs produced by a PLA2-like activity of Paraoxanase 1 contributes to the inhibition of macrophage biosynthesis and that they consequently reduce cellular cholesterol accumulation and atherogenesis [12]. LysoPC are also produced by endothelial lipase and hepatic lipase [13]. Hepatic lipase, which is also involved in triglyceride hydrolysis, is mainly responsible of the production of unsaturated LysoPCs [14], [15]. Although LysoPCs are commonly seen as pro-inflammatory and pro-atherogenic metabolites [16], recent population-based studies have suggested a protective effect of LysoPCs on cardiovascular risk. In a study of type 2 diabetes, LysoPC 18∶2 was found to be inversely associated with incident diabetes and impaired glucose tolerance [17]. Fernandez and colleagues found an inverse association of LysoPC 16∶0 and LysoPC 20∶4 with incident CVD and reduced intima media thickness [18]. More recently, Stegemann and colleagues [19] found an inverse association between several LysoPC species and incident CHD. Our study confirms and extends these previous findings. Using a Mendelian randomization approach, we suggest that the observed association between LysoPCs and incident CHD are likely to not be causal.
Strengths and limitations
Our study has several strengths. To our knowledge, this is the largest study investigating the metabolome in relation to incident CHD. Mass spectrometry-based metabolomics is extremely sensitive and allows detection of more metabolites than nuclear magnetic resonance-based methods [20]. We validated our findings using an independent population, with a different blood collection method, blood partition (serum instead of plasma) and age range. At the cost of augmented heterogeneity, this approach has the advantage to increase the generalizability of our findings. All three study samples were longitudinal and we have studied incident events decreasing the risk of reverse causation or selection bias as an explanation to our observations. We performed extensive characterization of underlying biological mechanisms, clinical utility, and potential causal effects for those metabolites showing strong evidence of association. We also acknowledge several limitations of our study. First, we used a non-targeted approach, meaning that every ion detected by mass spectrometry was treated as a separate variable, increasing the multiple-testing burden. We have previously shown that this approach does not affect the FDR point estimate, but might increase its variability [21]. However, this method is advantageous because it does not rely on pre-annotation and allows inclusion of unknown metabolites in the analyses (which subsequently can be identified using targeted methods). Moreover, we used a single analytical platform (liquid chromatography-mass spectrometry); the integration of multiple analytical platforms is a way of increasing the number of detectable metabolites. Second, non-targeted metabolomics is subject to co-elution of metabolites, ion suppression and imprecision in metabolites quantification, since each value assigned to the metabolic feature can only be interpreted as mass ion intensity. However, we do not have reason to believe that such biases would systematically affect CHD cases, since our outcome is measured prospectively and metabolomic profiling performed in a blinded fashion. Moreover, each sample has been analyzed in non-consecutive randomized duplicates, which minimize the risk of systematic biases. Third, the use of 15% FDR in the discovery phase is larger than in some other studies, but is justified by the high degree of correlation in the data, due to the existence of multiple metabolic features for a single metabolite. Moreover, metabolites were replicated (P-value<0.05) in an independent study sample. To evaluate if our replication strategy was sufficient to minimize the number of false positives, we estimated the expected false discovery rate in the replication sample (TwinGene) [22]. This was calculated as 0.23% (S1 Text), meaning that only 0.23% of metabolites replicating at P<0.05 are expected to be false positives, suggesting that our two-tier approach correctly control the number of false positives.
Finally, as our study samples consist of middle-aged to elderly individuals of Northern European decent, the generalizability to other ethnicities and younger age groups is unknown.
Conclusions and future directions
In conclusion, in the largest study of the metabolome in relation to incident CHD to date, we identified lysophosphatidylcholines 18∶1, 18∶2, monoglyceride 18∶2 and sphingomyelin 28∶1 as risk factors of coronary heart disease and suggested a causal effect for monoglyceride 18∶2 on CHD. Future experiments should mainly focus on determining the mechanisms by which these metabolites of lipid metabolism might be involved in pathogenesis of coronary heart disease.
Materials and Methods
Study samples
We performed metabolomic profiling of blood samples from three studies: TwinGene, ULSAM and PIVUS. An overview of the study design is illustrated in Fig. 1, and a detailed description of each study is given in the S1 Text.
In brief, TwinGene is a longitudinal sub-study of 12,591 individuals (55% women) from the Swedish Twin Register [23]. For the purpose of metabolomic profiling, we designed a case-cohort of incident CHD events and a matched sub-cohort (controls) stratified on age and sex [24]. In the final analysis we included serum samples from 1,670 unrelated individuals.
The Uppsala Longitudinal Study of Adult Men [25] (ULSAM; http://www2.pubcare.uu.se/ULSAM/) is an ongoing, longitudinal, epidemiologic study of men born between 1920 and 1924 in Uppsala County, Sweden. In the final analysis, we included plasma samples from 1,028 individuals investigated at 70 years of age.
The Prospective Investigation of the Vasculature in Uppsala Seniors [26] (PIVUS; http://www.medsci.uu.se/pivus/) is a population-based study of 70-year old individuals living in Uppsala. In the final analysis, we included serum samples from 970 individuals.
Incident CHD cases were defined as hospitalization or death with a primary diagnosis for acute myocardial infarction or unstable angina. This information was collected by linking the personal identity numbers from TwinGene and ULSAM participants with the Swedish National In-Patient Register and the Cause of Death Register up to the 31th December 2010, which comprise the end of follow-up of the present study.
Laboratory measurements
Laboratory procedures for metabolomics have been previously described [21], [27] and are detailed in the S1 Text. Briefly, metabolomic profiling was performed on Acquity UPLC coupled to a Xevo G2 Q-TOFMS (Waters Corporation, Milford, USA) with an atmospheric electrospray interface operating in positive ion mode. Non-consecutive duplicate sample aliquots of 1 µL were injected onto a Acquity UPLC BEH C8 analytical column. Mass analysis was performed in the full scan mode (m/z 50–1200).
Genotyping arrays used in each study are described in the S1 Text. All the samples underwent the same quality control (QC) and imputation of polymorphic 1000 genome CEU SNPs (Phase I, version 3) performed using IMPUTE2.
Methods for measuring the 21 biological markers and imaging features in PIVUS have been previously described [28]–[30].
Metabolic feature detection and annotation
Raw data were processed using XCMS software [31]. Procedures to perform non-targeted metabolomics in large-population studies have been previously described by our group [21]; the code has been made publically available at https://github.com/andgan/metabolomics_pipeline. Metabolic feature detection, alignment, grouping, imputation and normalization were performed separately for each study (S1 Text). Each feature is characterized by a specific mass-to-charge ratio (m/z) and retention time. A single metabolite is normally represented by more than one feature. Indiscriminant (id) MS and idMS/MS spectra were generated for all the significant features [27]. Those with highly similar spectra, strong correlation and similar retention time were deemed to be from the same metabolite. We used the spectra to identify the corresponding metabolite. Four approaches were considered, in agreement with what has been suggested by the Metabolomics Standard Initiative (MSI) [32] and as described in the S1 Text.
Statistical analysis
In ULSAM, we tested the association between each feature and incident CHD using a Cox proportional hazards model adjusted by age at baseline. We restricted our analysis to a 10-year follow-up since most biological markers experience a decreasing association with longer follow-up due to regression dilution bias. To evaluate the proportional hazard assumption we obtained, for each feature, a P-value from the Schoenfeld residual-based test; we did not detect any significant deviation from the proportionality assumption after correcting for multiple testing.
Features that were significantly associated with CHD in ULSAM at 15% false discovery rate (FDR) level were taken forward for replication in TwinGene. In TwinGene, we fitted Cox models adjusted for age and sex, and re-weighted for the inverse of the sampling probability using the Borgan “Estimator II” [24]. Features with P-value<0.05 in TwinGene and showed association with consistent direction were considered as replicated.
In the multivariable analysis, we studied the association between replicated features and CHD adjusting for main cardiovascular risk factors (sex, age, systolic blood pressure, BMI, current smoking, antihypertensive treatment, LDL-C, HDL-C, natural logarithm-transformed triglycerides and prevalent diabetes). Association analyses between metabolic features and markers of oxidative stress, inflammation and subclinical CVD in PIVUS was performed using linear regression adjusted only for age and sex, and for the same cardiovascular risk factors described above.
In TwinGene, reclassification measures (NRI, see S1 Text for additional details) were calculated using a 10% and 20% threshold for a 10-year risk of event, as often done in previous literature [33].
The genome-wide association study (GWAS) analyses were performed in PLINK adjusting for age, sex (where feasible) and first three principal components; results were meta-analyzed using fixed effects inverse-variance weighted meta-analysis in METAL. Instrumental variables for the Mendelian randomization analysis were constructed using the GWAS results and tested for association with CHD using the results from the CARDIOoGRAMplusC4D consortium [9]. Criteria for exclusion of pleiotropic SNPs and additional methodological information can be found in the S1 Text.
Ethics statement
All participants gave informed written consent and the Ethics Committees of Karolinska Institutet or Uppsala University approved the respective study protocol.
Supporting Information
Zdroje
1. SuhreK, ShinSY, PetersenAK, MohneyRP, MeredithD, et al. (2011) Human metabolic individuality in biomedical and pharmaceutical research. Nature 477 : 54–60.
2. MagnussonM, LewisGD, EricsonU, Orho-MelanderM, HedbladB, et al. (2013) A diabetes-predictive amino acid score and future cardiovascular disease. Eur Heart J 34 : 1982–1989.
3. ShahSH, BainJR, MuehlbauerMJ, StevensRD, CrosslinDR, et al. (2010) Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet 3 : 207–214.
4. BoyanovskyBB, WebbNR (2009) Biology of secretory phospholipase A2. Cardiovasc Drugs Ther 23 : 61–72.
5. WilsonPW, D'AgostinoRB, LevyD, BelangerAM, SilbershatzH, et al. (1998) Prediction of coronary heart disease using risk factor categories. Circulation 97 : 1837–1847.
6. LemaitreRN, TanakaT, TangW, ManichaikulA, FoyM, et al. (2011) Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet 7: e1002193.
7. SchunkertH, KonigIR, KathiresanS, ReillyMP, AssimesTL, et al. (2011) Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 43 : 333–338.
8. DemirkanA, van DuijnCM, UgocsaiP, IsaacsA, PramstallerPP, et al. (2012) Genome-wide association study identifies novel loci associated with circulating phospho - and sphingolipid concentrations. PLoS Genet 8: e1002490.
9. CARDIoGRAMplusC4D Consortium (2013) DeloukasP, KanoniS, WillenborgC, FarrallM, et al. (2013) Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 45 : 25–33.
10. MillerM, StoneNJ, BallantyneC, BittnerV, CriquiMH, et al. (2011) Triglycerides and cardiovascular disease: a scientific statement from the American Heart Association. Circulation 123 : 2292–2333.
11. DoR, WillerCJ, SchmidtEM, SenguptaS, GaoC, et al. (2013) Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet 45 : 1345–1352.
12. RozenbergO, ShihDM, AviramM (2003) Human serum paraoxonase 1 decreases macrophage cholesterol biosynthesis: possible role for its phospholipase-A2-like activity and lysophosphatidylcholine formation. Arterioscler Thromb Vasc Biol 23 : 461–467.
13. GausterM, RechbergerG, SovicA, HorlG, SteyrerE, et al. (2005) Endothelial lipase releases saturated and unsaturated fatty acids of high density lipoprotein phosphatidylcholine. J Lipid Res 46 : 1517–1525.
14. SekasG, PattonGM, LincolnEC, RobinsSJ (1985) Origin of plasma lysophosphatidylcholine: evidence for direct hepatic secretion in the rat. J Lab Clin Med 105 : 190–194.
15. ShamburekRD, ZechLA, CooperPS, VandenbroekJM, SchwartzCC (1996) Disappearance of two major phosphatidylcholines from plasma is predominantly via LCAT and hepatic lipase. Am J Physiol 271: E1073–1082.
16. SchmitzG, RuebsaamenK (2010) Metabolism and atherogenic disease association of lysophosphatidylcholine. Atherosclerosis 208 : 10–18.
17. Wang-SattlerR, YuZ, HerderC, MessiasAC, FloegelA, et al. (2012) Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol 8 : 615.
18. FernandezC, SandinM, SampaioJL, AlmgrenP, NarkiewiczK, et al. (2013) Plasma lipid composition and risk of developing cardiovascular disease. PLoS One 8: e71846.
19. StegemannC, PechlanerR, WilleitP, LangleyS, ManginoM, et al. (2014) Lipidomics Profiling and Risk of Cardiovascular Disease in the Prospective Population-Based Bruneck Study. Circulation
20. TzoulakiI, EbbelsTM, ValdesA, ElliottP, IoannidisJP (2014) Design and analysis of metabolomics studies in epidemiologic research: a primer on -omic technologies. Am J Epidemiol 180 : 129–139.
21. GannaA, FallT, LeeW, BroecklingCD, KumarJ, et al. (2014) A workflow for UPLC-MS non-targeted metabolomic profiling in large human population-based studies. bioRxiv
22. GannaA, LeeD, IngelssonE, PawitanY (2014) Rediscovery rate estimation for assessing the validation of significant findings in high-throughput studies. Brief Bioinform
23. MagnussonPK, AlmqvistC, RahmanI, GannaA, ViktorinA, et al. (2013) The Swedish Twin Registry: establishment of a biobank and other recent developments. Twin Res Hum Genet 16 : 317–329.
24. GannaA, ReillyM, de FaireU, PedersenN, MagnussonP, et al. (2012) Risk prediction measures for case-cohort and nested case-control designs: an application to cardiovascular disease. Am J Epidemiol 175 : 715–724.
25. BybergL, SiegbahnA, BerglundL, McKeigueP, RenelandR, et al. (1998) Plasminogen activator inhibitor-1 activity is independently related to both insulin sensitivity and serum triglycerides in 70-year-old men. Arterioscler Thromb Vasc Biol 18 : 258–264.
26. LindL, ForsN, HallJ, MarttalaK, StenborgA (2005) A comparison of three different methods to evaluate endothelium-dependent vasodilation in the elderly: the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study. Arterioscler Thromb Vasc Biol 25 : 2368–2375.
27. BroecklingCD, HeubergerAL, PrenniJE (2013) Large scale non-targeted metabolomic profiling of serum by ultra performance liquid chromatography-mass spectrometry (UPLC-MS). J Vis Exp e50242.
28. LindL, ForsN, HallJ, MarttalaK, StenborgA (2006) A comparison of three different methods to determine arterial compliance in the elderly: the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study. J Hypertens 24 : 1075–1082.
29. LindL, SiegbahnA, HultheJ, ElmgrenA (2008) C-reactive protein and e-selectin levels are related to vasodilation in resistance, but not conductance arteries in the elderly: the prospective investigation of the Vasculature in Uppsala Seniors (PIVUS) study. Atherosclerosis 199 : 129–137.
30. LindL, SiegbahnA, IngelssonE, SundstromJ, ArnlovJ (2011) A detailed cardiovascular characterization of obesity without the metabolic syndrome. Arterioscler Thromb Vasc Biol 31: e27–34.
31. SmithCA, WantEJ, O'MailleG, AbagyanR, SiuzdakG (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78 : 779–787.
32. SumnerLW, AmbergA, BarrettD, BealeMH, BegerR, et al. (2007) Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3 : 211–221.
33. LeeningMJ, VedderMM, WittemanJC, PencinaMJ, SteyerbergEW (2014) Net reclassification improvement: computation, interpretation, and controversies: a literature review and clinician's guide. Ann Intern Med 160 : 122–131.
Štítky
Genetika Reprodukčná medicína
Článek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 12- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



