-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
Plasmids are difficult to analyze in WGS datasets, due to the fragmented nature of the obtained sequences. We developed a method, called PLACNET, which greatly facilitates this analysis. As an example, we analyzed the plasmidome of E. coli ST131, an ExPEC clonal group involved in human urinary tract infections and septicemia. Relevant variation within this clone (e.g., antibiotic resistance and virulence) is frequently caused by the acquisition and loss of plasmids and other mobile genetic elements. Nevertheless, our knowledge of the ST131 plasmidome is limited to a few antibiotic resistance plasmids and to identification of replicons from known plasmid groups. PLACNET analysis extends the number of sequenced plasmids in ST131, which can be used for comparative genomics, from 11 to 50. The ST131 plasmidome is seemingly huge, encompassing roughly 50% of the main plasmid groups of γ–proteobacteria. MOBF12/IncF plasmids are apparently the most active players in the dissemination of relevant genetic information.
Published in the journal: Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences. PLoS Genet 10(12): e32767. doi:10.1371/journal.pgen.1004766
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004766Summary
Plasmids are difficult to analyze in WGS datasets, due to the fragmented nature of the obtained sequences. We developed a method, called PLACNET, which greatly facilitates this analysis. As an example, we analyzed the plasmidome of E. coli ST131, an ExPEC clonal group involved in human urinary tract infections and septicemia. Relevant variation within this clone (e.g., antibiotic resistance and virulence) is frequently caused by the acquisition and loss of plasmids and other mobile genetic elements. Nevertheless, our knowledge of the ST131 plasmidome is limited to a few antibiotic resistance plasmids and to identification of replicons from known plasmid groups. PLACNET analysis extends the number of sequenced plasmids in ST131, which can be used for comparative genomics, from 11 to 50. The ST131 plasmidome is seemingly huge, encompassing roughly 50% of the main plasmid groups of γ–proteobacteria. MOBF12/IncF plasmids are apparently the most active players in the dissemination of relevant genetic information.
Introduction
Clinical microbiology is being transformed by whole genome sequencing (WGS) [1]. A case in point is Escherichia coli: there were 1,618 E. coli projects submitted to NCBI compared to just 68 complete genomes by year 2013. Within the realms of clinical and environmental microbiology, plasmid analysis is increasingly used to track the dissemination of genes encoding virulence, resistance to antibiotics, heavy metals and biocides [2]–[4] and, to a lesser extent, to analyze differences in the adaptive evolution of certain clonal backgrounds [5], [6]. Hybridization with specific probes [7], amplification of plasmid replication initiator proteins (RIP) [8]–[10], and relaxases (REL) [11] allow preliminary identification of plasmid families. In addition, plasmid MLST (pMLST) is used for epidemiological surveillance, but is restricted to individual plasmids of a few plasmid families of Enterobacteriaceae (http://pubmlst.org/plasmid/). This precludes the detection of plasmid mutations or rearrangements, as well as the identification of conjugative plasmids not represented in the pMLST database and of most mobilizable plasmids [11]. Finished plasmid/genome sequencing provides accurate and non-biased information, but is still expensive and thus seldom used specifically for plasmid analysis. Draft WGS dramatically cut down cost and analysis time. Although it allowed rapid and cheap data acquisition, WGS datasets typically result in more than a hundred contigs for a given genome, due to the short read lengths generally obtained. Genome fragmentation makes it difficult to distinguish between physical units, that is, between chromosome and plasmid sequences, as well as between different plasmids that usually coexist in bacterial cells. Several strategies can be followed to analyze WGS genome sequences, the workflow described by [12] being a typical example. There are also applications to identify plasmids in WGS sequences, such as PlasmidFinder (http://cge.cbs.dtu.dk/services/PlasmidFinder/), which identifies plasmids according to PCR-based replicon typing (PBRT) [8]–[10] and the subtyping scheme included in the pMLST web page (http://pubmlst.org). PlasmidFinder is limited by its inability to reconstruct the sequences of entire plasmids, underscoring the urgent need for improvement over existing tools.
E. coli ST131 is a successful high-risk clonal complex of pandemic distribution, able to cause extraintestinal infections in humans [13]-[18]. The increasing recovery of ST131 isolates from hospitalized and non-hospitalized individuals and, more recently, from companion and foodborne animals [17], [19]–[25], sewage and main rivers of large European cities [26], [27] highlights the rapid spread and local adaptation to different habitats of this lineage. ST131 is characterized by high metabolic potential [28] and a variable number of virulence factors, including adhesins, siderophores, toxins, polysaccharide coats (capsules and lipopolysaccharides), protectins and invasins [19], [29], [30], mostly acquired by recombination and by the interplay of mobile genetic elements (MGEs) [16]. Such traits, which are common among different lineages of the E. coli B2 phylogroup [31], [32], enable strains to colonize mucosal surfaces, invade tissues, foil defence mechanisms and yield injurious inflammatory responses in the host. E. coli populations identified as ST131 by the widely used ‘Achtman scheme’ of multilocus sequence typing (MLST) [33] (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli), split in diverse clusters or subclones on the basis of genomic profile, serotype, content of virulence factors, antibiotic susceptibility pattern and the presence of certain fimH alleles [21], [29], [34]–[36]. The most prevalent ST131 clonal sublineage (H30) is characterized by the presence of a fimH30 allele, serotype O25:H4 and a specifically conserved gyrA/parC allele combination that confers fluoroquinolone resistance (FQ-R). Most human infections caused by ST131 are due to isolates of the H30 sublineage [13], [16], [37]–[39], many of them carrying the blaCTX-M-15 gene which is responsible for resistance to third generation of cephalosporins. Some authors suggested differences between CTX-M-15 and non-CTX-M-15 producers, referred to as H30-R and H30-Rx sublineages, respectively [13], [35], [37], [38]. Currently, diverse O25b:H4 ST131 variants (e.g. fimH22, fimH30) or O16:H5 (e.g. fimH41) seem also to be widely spread [13]-[16], [40]. Full genome sequencing of several ST131 E. coli genomes, most of them H30-Rx variants [16], [41]–[44], revealed further differences among strains, mainly chromosomal SNPs, indels and plasmid variations [16], [43], [44]. Heterogeneity of MGEs has been reported in other relevant E. coli clones, mainly Shiga-toxin producing E. coli (STEC) as O157:H7, O104:H4 or O26:H11 [5], [6], [45]–[47], often associated with ecological diversification of E. coli populations that can influence host-pathogen interactions [48], [49]. Recently, International and European organisations including European Food Safety Agency, EFSA; European Centre for Disease Control, ECDC; Food Drug Administration, FDA; Centre for Diseases Control, CDC) and national food safety authorities underscored the need to identify clonal variants with enhanced transmissibility or pathogenicity as well as to infer the evolutionary history of pathogens of interest in Public Health (http://www.efsa.europa.eu/en/events/event/140616.htm). Because relevant adaptive traits are plasmid located, there is an urgent need to consider MGEs in population genetic studies.
In this work we describe PLACNET, a method to reconstruct plasmids from WGS datasets, and its application to the comprehensive analysis of bacterial plasmidomes. As a specific example, we describe the ST131 plasmidome and discuss its possible impact in the diversification of this clinically important lineage. PLACNET allows the identification of plasmids currently circulating among E. coli and other enterobacterial species that may be underestimated, thus providing a useful tool to approach comprehensive plasmid population genetic studies.
Results
Phylogeny of E. coli ST131 genomes
We analyzed ten E. coli genomes, classified as ST131 according to the Achtman scheme (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli), which branch in three main clusters identified as ST43, ST9 and ST506 (Fig. 1) according to the cgMLST Pasteur Institute scheme (http://www.pasteur.fr/recherche/genopole/PF8/mlst/EColi.html). The use of these two schemes is widely accepted in epidemiology [50] and increasingly used for E. coli typing. The ST43 branch contains isolates of the H30 lineage, which split in three subclusters (four strains of virotype C, two of virotype A, one of virotype B). The ST9 branch corresponds to isolates of the H22/H324 sublineage (virotype D). The most distal branch to the main cluster is represented by the commensal strain SE15, a member of sublineage H41 identified as ST506 [16]. It does not contain any marker used for the virotype subtyping method described by Blanco et al (afa, sat, ibeA, iroN) [36], [51], [52]. Thus, the sample analyzed in this work includes representatives of all ST131 branches described to date [13], [16]. The core genome of the 10 strains encompasses 3.6 Mb (Fig. 1 inset). As can be seen, the phylogenetic tree of ST131 genomes can be rooted at the commensal strain SE15. It should be noted, however, that SE15 is not necessarily the ancestor of the pathogenic lineages, as inferred by recent evidence [16]. The divergence of SE15 from the other ST131 strains is of about 3,000 SNP/Mb, a measure of the depth of the ST131 phylogenetic branch (<0.3% divergence in the core genome). There are only 650 SNPs among the genomes of cluster C lineage (i.e., <200 SNP/Mb), indicating their close phylogenetic relationship. There are <300 SNPs within a given virotype. The average distance between clades A and B is of about 4,600 SNPs (i.e., 1,300 SNP/Mb).
Fig. 1. Phylogenetic tree of ST131 E. coli. 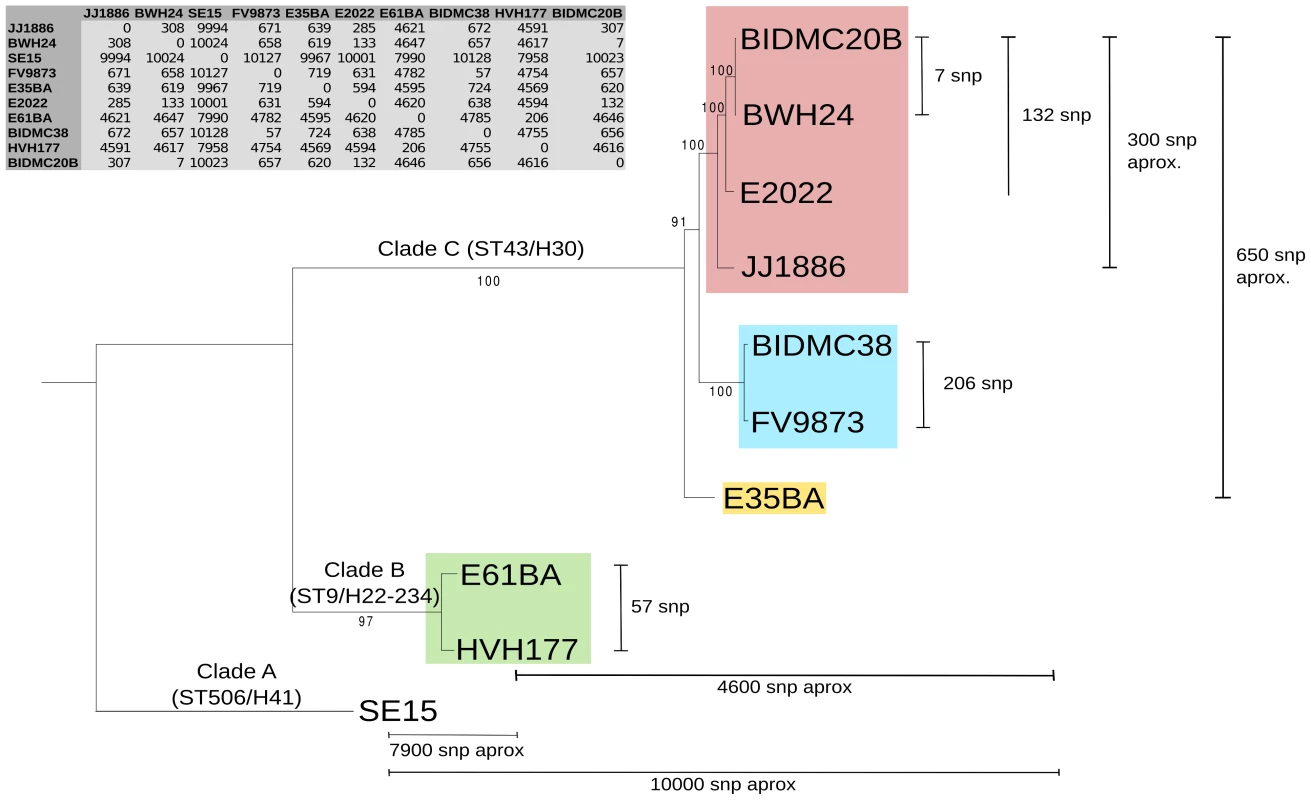
The tree is based on a 3,629,034 bp core genome (3,734 orthologous genes: 90% identity and 90% coverage) and 100 bootstrapping replicates. ST131 clades are named according to [16] and further subdivided and colored according to virotypes [36]: virotype A (blue), virotype B (yellow), virotype C (pink), virotype D (green). Virotype classification is based on the presence/absence of four putative virulence factors: afaFM955459 (encoding an Afa/Dr adhesin), sat (secreted autotransporter toxin, present in PAI-CFT073-pheV), ibeA (invasion of brain endothelium) and iroN (salmochelin siderophore receptor). The commensal ST131 strain SE15 was used to root the tree (virotype non typable; serotype O150 in the original publication [91] but lying within the H41 cluster in the phylogenomic study of [16]). Given SNP numbers are approximate averages of individual comparisons. Plasmid reconstruction in E. coli ST131 genomes
The PLACNET protocol was used as explained in Materials and Methods. We proceeded with plasmid reconstruction, as exemplified in Fig. 2 for the reconstruction of the E61BA genome (ST9/H324/virotype D). When we applied the rules for reference homology, scaffold links and plasmid protein tagging, the E61BA network shown as “original network” was produced. Obviously, this network was not neat enough to allow plasmid reconstruction. Expert pruning of the network consisted on several steps. First, contigs smaller than 200 bp were eliminated. Second, hubs were identified (see arrows in the original network of Fig. 2), duplicated and assigned to separate disjoint connected components. Scaffold links and coverage information, as well as score values of conflict edges, were used to decide on valid component assignment. Inspection of the coding potential of hubs usually showed them to correspond to ISs, transposons or other known repeated elements (as shown in S9 and S10 Figs.). As a result, a pruned network was reconstructed as shown in Fig. 2. Differential coloring of disjoint connected components in the pruned network thus displayed the final network of plasmids (as contig constellations). In PLACNET Cytoscape representation, most plasmids can be identified by their RIP and/or REL proteins. Thus, the reconstructed E61BA genome contains seven plasmids: a 134 kb MOBF12/IncF plasmid (pE61BA-1), a 37.7 kb MOBP6/IncI2 plasmid (pE61BA-7), a 24.5 kb MOBC12 plasmid (pE61BA-2), a 18 kb MOBP11/IncP1 plasmid (pE61BA-4), two MOBP5/ColE1-like plasmids of 6.6 and 6.9 kb (pE61BA-5 and pE61BA-6, respectively) and one MOBQ12 5.0 kb plasmid (pE61BA-3). Only plasmid pE61BA-2 could be closed, the remaining contained at least two contigs. Thus, their reported sizes are minimum sizes, since they might include small repeated sequences that were taken out of the analysis during network pruning. Two contigs remained as “not assigned” to any physical unit in this particular genome because they did not show any reference or scaffold link that bind them to other contigs: a 2,953 bp contig (containing a putative DNA primase and a lytic transglycosylase) and a 1,301 bp contig (containing two conjugation-related genes: trbI and a partial traB gene).
Fig. 2. PLACNET plasmid reconstruction of ST131 genome E61BA (ST9/H324/virotype D). 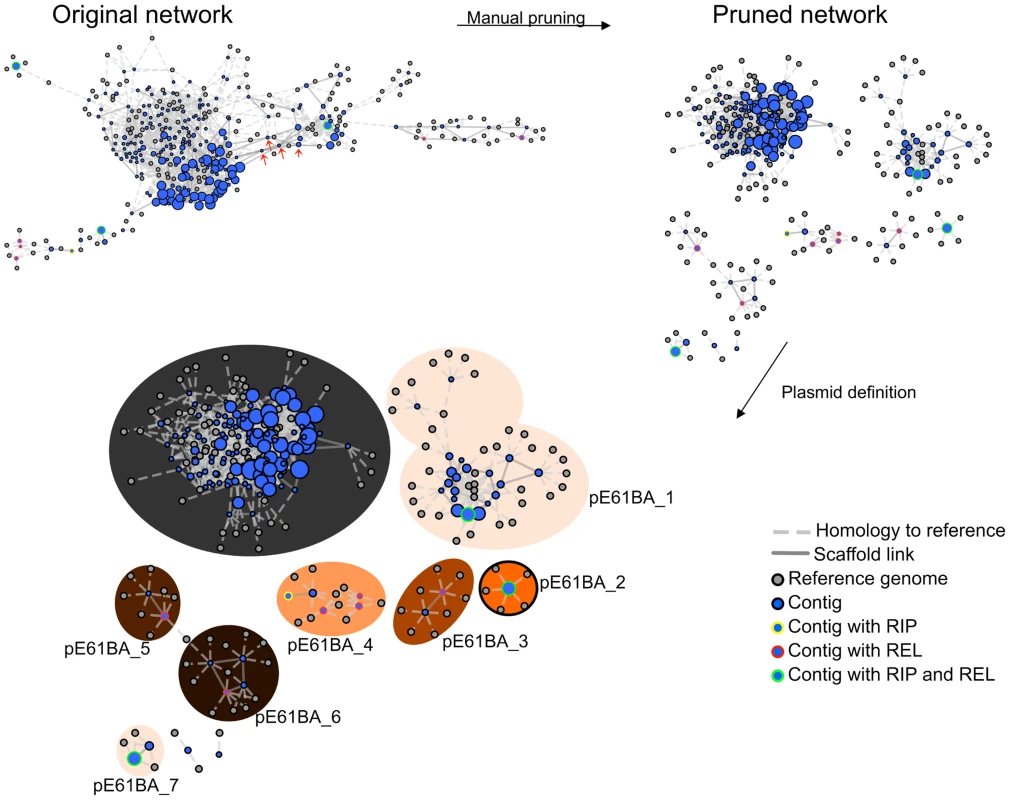
The network contains nodes of two different colors (blue for contigs, grey for reference genomes). The size of reference nodes is always the same. The size of the contig nodes is proportional to the contig length. Besides, outlines are yellow for contigs containing RIP proteins, red for relaxases and green for both proteins. Edges are either solid (scaffold links) of dotted (homologous references). The length of the edges is arbitrarily selected by Cytoscape algorithm. In the upper left, the network output (original network) is shown, which resulted from automatic reference search, scaffold links and protein tagging rules. The original network was converted to a pruned network by eliminating contigs smaller than 200 bp and duplicating specific hubs (red arrows). Two contigs could not be assigned for lack of scaffold links: a 2,953 bp contig (putative DNA primase + lytic transglycosylase) and a 1,301 bp contig (TrbI + TraB-partial). Closed plasmids (e.g., pE61BA_2, size: 24,447 bp) are shown with a black outline in the final PLACNET network. The same procedure was applied to the three other strains sequenced for this work as well as to the four genomes obtained from public DBs as Illumina reads. The plasmid content of the four strains sequenced in this work was confirmed by the analysis of S1-digested genomic DNA profiles by PFGE. This analysis fully confirmed the presence of plasmids of similar size to those identified by PLACNET (S2 Text and S4 Table), In the case of strain E35BA, in which PLACNET identified two IncF plasmids that could not be separated (totaling 211 kb), S1-PFGE identified two plasmids of 140 kb and 75 kb. As a result of PLACNET analysis, we obtained the plasmid constellation networks shown in S1 to S8 Figs. A summary of the results, i.e., the reconstructed plasmids, is shown in Table 1, which includes also the plasmids of the ST131 reference strains JJ1886 and SE15. As can be seen, the number of plasmids in the ST131 genomes is variable, even from strains belonging to the same ST131 sublineage, ranging from just one plasmid in HVH177 (clade B/ST9/fimH22) or SE15 (clade A/ST506/fimH41) to seven plasmids in E61BA (clade B/ST9/fimH22), to give an average of 4 plasmids per genome. There is not a single plasmid group that appears specific of a particular sublineage. S1 Table contains the complete list of contigs assigned to each plasmid or chromosome.
Tab. 1. Summary of plasmid content. 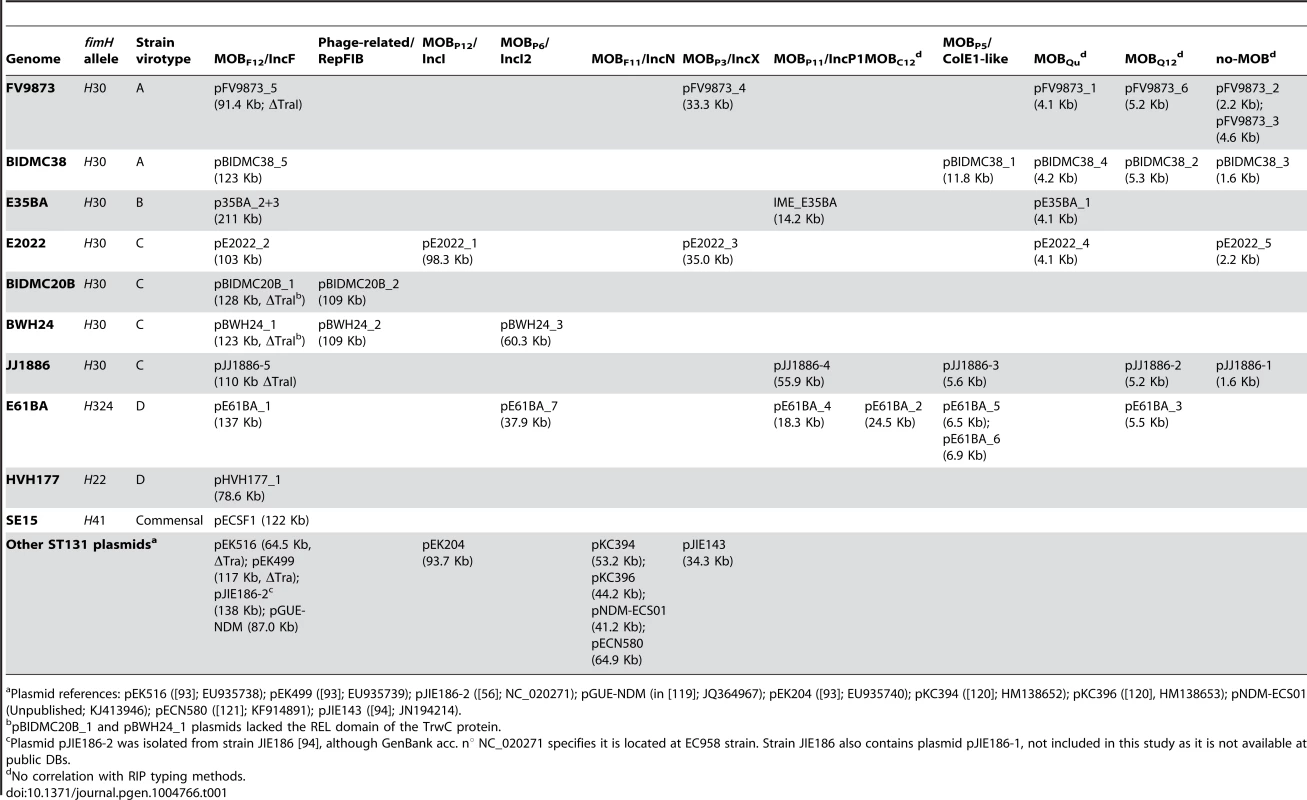
Plasmid references: pEK516 ([93]; EU935738); pEK499 ([93]; EU935739); pJIE186-2 ([56]; NC_020271); pGUE-NDM (in [119]; JQ364967); pEK204 ([93]; EU935740); pKC394 ([120]; HM138652); pKC396 ([120], HM138653); pNDM-ECS01 (Unpublished; KJ413946); pECN580 ([121]; KF914891); pJIE143 ([94]; JN194214). Overall plasmid diversity is visualized in plasmid dendrograms
Overall, the ten ST131 genomes analyzed contain 39 plasmids (including one potential ICE), which can be assorted by their relative sizes and MOB groups [53], as shown in Table 1. The most conspicuous group was that of MOBF12/IncF plasmids (11 plasmids), present in all ten sequenced ST131 genomes. Other relevant plasmid backbones belong to the MOBP (RIP groups IncI1/K, IncI2, IncX1, IncX4, ColE1), MOBQ (Qu, Q12) and MOBC (C12) REL families. The non-F plasmids comprise a total of 20 plasmids belonging to eight plasmid groups. Two plasmids were phage-like and belong to the Rep-3 RIP family. Finally, 5 plasmids corresponded to the no-MOB category. The E35BA genome (ST43/H30 virotype B) showed a MOBP11 relaxase within a 234 kb chromosomal contig, implying the presence of an ICE (integrative and conjugative element). Detailed inspection of this contig identified a 14.2 kb IME (integrative and mobilizable element) (see below and Suppl. Mat.). Once plasmids were identified by PLACNET and contigs assorted, the next step in plasmid analysis consisted in the construction of a dendrogram that summarizes plasmid gene content, allowing a visualization of relatedness between individual plasmids. The dendrogram of the 44 plasmids found in ST131 genomes (39 described in this report plus five plasmids already published) is shown in Fig. 3. The figure shows how the plasmids divide into branches that coincide with backbone MOB groups. There are 14 plasmid groups, according to the dendrogram, shown in the figure by different color backgrounds. Since each dendrogram group links related plasmids, they can be now analyzed individually, by comparing them either among themselves (Fig. 3) or with selected references (Fig. 4).
Fig. 3. Hierarchical clustering dendrogram of ST131 plasmids. 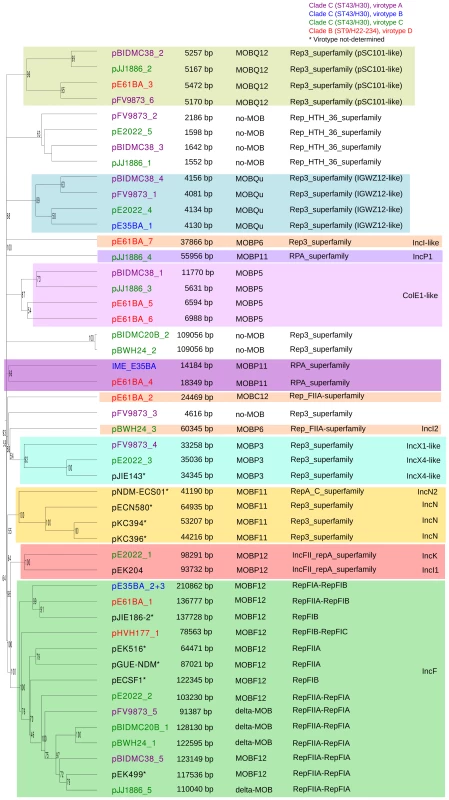
The UPGMA dendrogram was based on protein cluster analysis using 60% sequence identity and 80% coverage. Plasmid names are colored according to their clade, taking into account ST, fimH allele and virotype, following the color code shown at the upper right. The five plasmid names in black correspond to previously sequenced plasmids from ST131 strains. Different color backgrounds are shown to emphasize branches of related plasmids. To the right of the dendrogram, four columns show, respectively, plasmid size, MOB type, RIP type and Inc type. Fig. 4. Hierarchical clustering dendrogram of ST131 plasmids and relevant references. 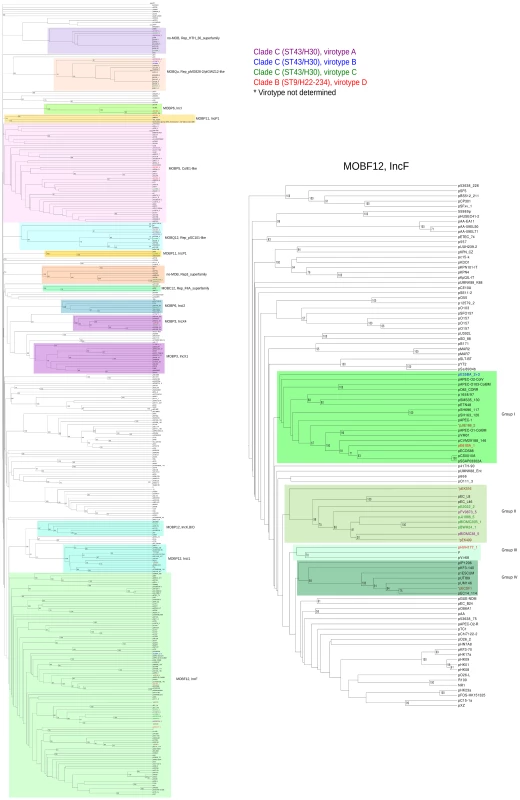
The left dendrogram shows the complete tree, with references. Dendrogram construction and color codes are as in Fig. 4. The right dendrogram expands the MOBF12/IncF branch, with new background colors highlighting plasmid groups within this branch that are mentioned in the text. Analysis of IncF plasmids
Representatives of the largest plasmid group, formed by 15 MOBF12/IncF plasmids, were found in each of the analyzed ST131 strains (Table 1). Included in this set are four plasmids lacking MOB and tra regions but containing RIP and other backbone genes related to IncF plasmids. The dendrogram of Fig. 3 clearly indicates that these plasmids belong to the IncF plasmid family, underscoring the usefulness of this step in plasmid reconstruction. Judging from their position in the dendrogram, it seems IncF plasmid genes are scrambled as if the precise constitution of each individual IncF plasmid could not be predicted at all for isolates of each specific ST131 cluster or phylotype. This is even clearer in Fig. 4. In this figure, ST131 plasmids are represented together with the reference sequences that were used for PLACNET reconstruction and analysis. Besides, BRIG comparison of IncF plasmids (Fig. 5) reveals high heterogeneity between them, with not a single completely conserved gene (confirmed by the fact that not a single plasmid gene was found to belong to the ST131 core genome). KClust software [54] at 30% identity and 50% coverage was used to group all proteins coded by IncF plasmids into 354 reference clusters. Manual curation was used to classify these 354 clusters in three groups (Fig. 5): (i) plasmid backbone (i.e., conjugation, RIP and maintenance genes) and metabolic protein genes, (ii) antibiotic resistance and virulence genes, and (iii) other protein genes such as ISs, transposases or hypothetical proteins. Conjugation proteins represent 30 of the 53 backbone proteins and constitute the most conserved set. As mentioned above, 4/14 plasmids do not keep a complete backbone. Table 2 contains the functional annotation of the IncF plasmids. As shown, 11/15 of the MOBF12/IncF plasmids contain antibiotic resistance genes (conferring resistance to beta-lactams, in all cases, but also to sulfonamides, aminoglycosides, trimethoprim, chloramphenicol, tetracycline and macrolides, in some of them). In addition, nine of the ten antibiotic resistance-plasmids confer a multidrug-resistance (MDR) phenotype (equal or more than four antibiotic families). Besides, 10/15 MOBF12/IncF plasmids presented putativevirulence genes (Table 2). As previously noted, there was an apparent trade-off between antibiotic resistance and virulence, genes coding for these adaptive traits being located in different plasmids [2]. Finally, a DNA modification gene (adenine-specific DNA methylase) was conserved in all IncF plasmids except in pHVH177_1.
Fig. 5. MOBF12/IncF plasmid analysis. 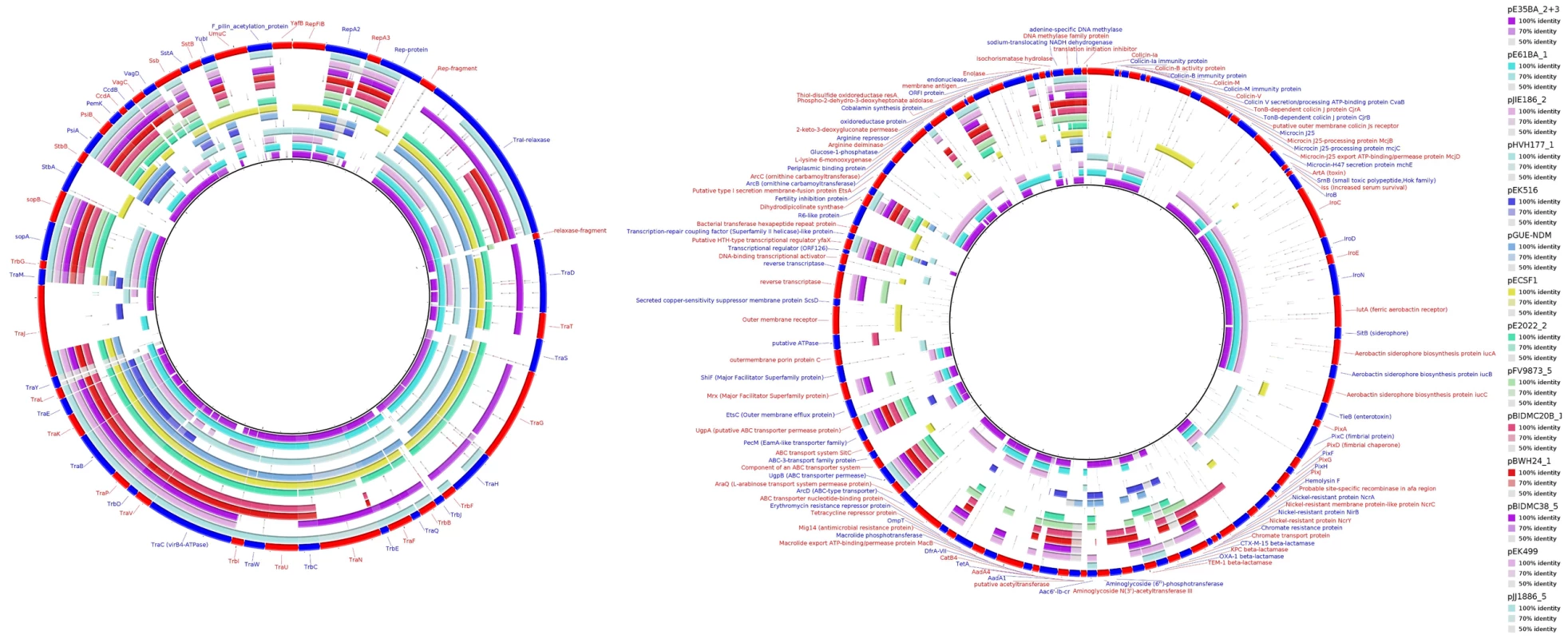
Protein cluster analysis was performed with kClust software (parameters: 30% identity, 50% coverage) on the set of 14 plasmids shown in Table 4. Plasmid pGUE-NDM [119] was excluded from this comparison since it is only distantly related to the others (see dendrogram in Fig. 5). A total of 354 protein clusters were obtained and annotated versus the NCBI protein database (Blastp). Manual inspection was carried out to classify the reference proteins of each cluster into one of these three groups (comparative analysis shown with BRIG): (i) Backbone and metabolic proteins (panel A); (ii) Virulence and Antibiotic resistance proteins (panel B); and (iii) ISs and hypothetical proteins (not shown). Tab. 2. Resistance genes and virulence determinants in MOBF12 plasmids. 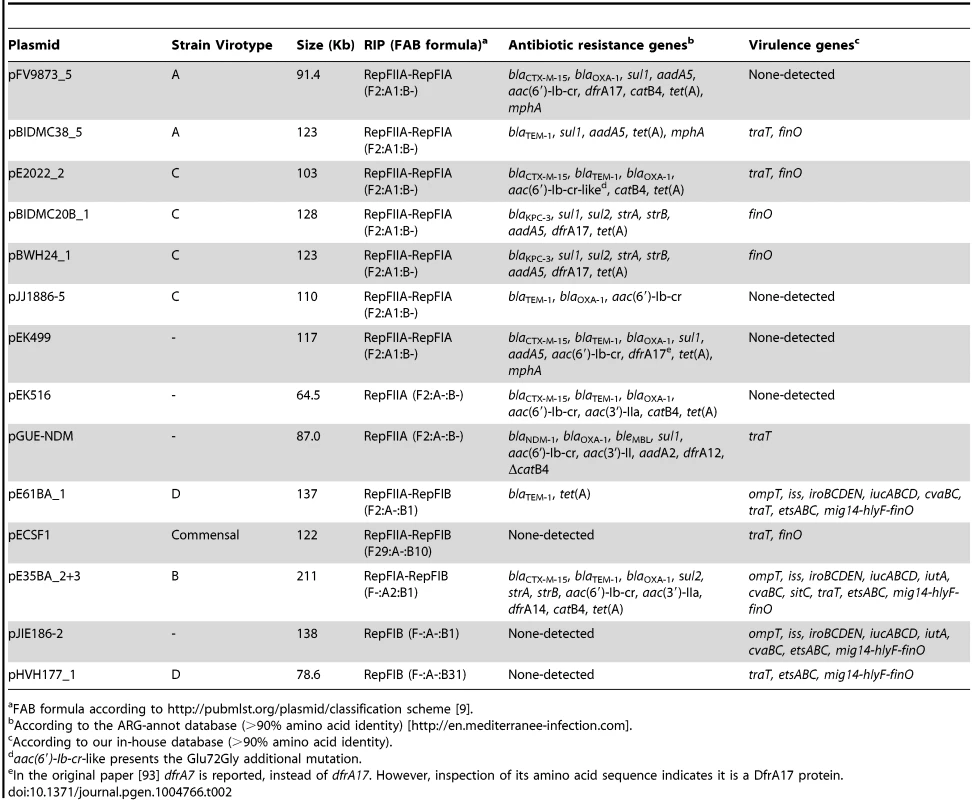
FAB formula according to http://pubmlst.org/plasmid/classification scheme [9]. The ST131 IncF plasmids belong to four different branches of the dendrogram, as shown in Fig. 4 inset. Group I includes four plasmids similar to the well-known virulence plasmids pAPEC-ColV like (also called pS88-like), which are commonly detected among avian pathogenic E. coli (APEC) [2], [55]. A suitable reference is the ST131 plasmid pJIE186-2, coming from a ST131 strain previously recovered in Australia in 2006 [56]. As shown in S11A Fig., group I IncF plasmids share two large homologous regions: a 70 kb region containing virulence genes iss, iroBCDEN, iucABCD, iutA, cvaBC and sitC and the cassette ompT-hlyF-mig14, eventually also linked to estABCDE [55] and a 40 kb region containing the tra region and other backbone genes. Group II contains 10 MDR plasmids, 8 of which are ST131 plasmids with characteristic F2:A1:B - replicons and multiple antibiotic resistancegenes. A suitable reference is the ST131 plasmid pJJ1886-5, coming from a ST43/fimH30 lineage from USA. As shown in S11B Fig., group II IncF plasmids share most of their genomes. It should be noted that three of these plasmids (pFV9873_5, pEK499 and pEK516) have extensive deletions within their tra regions, as seen in the figure. Groups III and IV are just represented by one plasmid each. None of them contains antibiotic resistance genes and they are poor in virulence genes. While group III plasmid pHVH177_1 is not similar to any reference outside the backbone genes (S11C Fig.), the group IV plasmid pECSF1 is extensively similar to various large E.coli plasmids, (S11D Fig.). A more comprehensive comparison of F plasmids recovered from ST131 with previously described F-like plasmids is given in Suppl. Mat.
Analysis of other ST131 plasmid groups
Besides MOBF12/IncF plasmids, 28 other plasmids were represented in ST131 isolates (Table 1). Ten were large, presumably conjugative plasmids (>18 kb), while 17 were small plasmids (<12 kb) and there was one IME.
Among the large plasmids, a most remarkable branch is composed by two almost identical 109 kb plasmids (pBIDMC20B_2 and pBWH24_2) present in two ST43/H30 isolates of virotype C, for which only RepFIB (Rep3-superfamily) and the maintenance protein ParB could be identified as plasmid backbone genes. No conjugative genes were detected. On the other hand, they code for an integrase protein and several phage-typical proteins. The plasmids are highly similar to pECOH89, recently recovered from a CTX-M-15 producer E. coli isolate from Germany [57]. Closest reference hits were the adherent invasive E. coli (AIEC) plasmid pLF82, isolated from a patient with Crohn's disease [58], the STEC plasmid p09EL50 [5], the Salmonella plasmid pHCM2 [59] and the Salmonella bacteriophage SSU5 [60]. These are all cryptic plasmids isolated from pathogenic enterobacteria that have been barely analyzed and thus are poorly annotated. The possibility arises of these elements being similar to lysogenic phages that are stably maintained as plasmids, analogous to phage P1 [57]. S12 Fig. compares this branch of related plasmids, using plasmid pECOH89 as a reference. As can be seen, both ST131 plasmids share most of their sequences with this 111 kb plasmid, including several phage-like protein genes. Significantly, none of the cryptic plasmids described in this study or those mentioned in the references, except pECOH89, harbor a resistance gene.
The 98.3 kb MOBP12/IncK plasmid pE2022_1 is most similar to pCT [61]. pE2022_1 contains a blaCTX-M-14 gene identical to that in pCT, a plasmid carrying blaCTX-M-14 that is globally spread among humans and animals and particularly prevalent in clinical isolates form Spain [61], [62]. The backbone of plasmid pE2022_1 is homologous to that of the reference IncI1 plasmid pEK204. These plasmids are described in S13. Fig. Despite their different Inc names, IncI1, IncK and IncB/O have similar backbones, belonging to different branches of the IncI complex (an analogous case to IncF plasmids).
MOBP6 is a large plasmid family, as can be observed in the phylogenetic tree of the MOBP6 relaxase family (S14A Fig.). The two ST131 MOBP6/IncI2 plasmids are rather different, as judged by the distant positions of their REL. Plasmid pBWH24_3 (60.3 kb) is similar to the IncI2 prototype R721 [63], but most similar to the APEC plasmid pChi7122_3 [64]. In turn, pE61BA_7 (37.9 kb) is most similar to Salmonella agona plasmid SL483 and the enterohemorrhagic E. coli (EHEC) plasmid pO157_Sal [65]. They were recovered from isolates identified as H30_virotype C from the USA and H324_virotype D from Spain, respectively.
Another ST131 important group is MOBP3/IncX, composed by three ST131 plasmids (pFV9873_4 and pE2022_3, as well as the reference pJIE143). Plasmids pFV9873_4 and pE2022_3, obtained from different H30 subgroups, are rather different between them and belong to different plasmid groups (IncX1 and IncX4), as shown their relatively distant positions in the MOBP3 phylogenetic tree of S15A Fig., even if showing similar sizes of about 34 kb. Their coding capacity is shown in the BRIG representations shown in S15B and S15C Fig. The IncX1 plasmid pFV9873_4 is most similar to the EC plasmid p2ESCUM [66], although genetic similarity is constrained to their backbone genes, occupying about half of the reference plasmid sequences. Conversely, the IncX4 plasmid pE2022_3 is most similar to pSH696_34 and the ST131 reference plasmid pJIE143 all over its sequence length (S15C Fig.).
Two plasmids and the IME belong to the MOBP11/IncP1 family, as shown in the MOBP11 relaxase phylogenetic tree of S16A Fig. Plasmids pJJ1886_4 and pE61BA_4 showed widely different sizes (56 and 18 kb, respectively). Plasmid pE61BA_4 is only distantly related in its backbone genes to environmental plasmid pMBUI2, isolated from an uncultured bacterium [67]. Plasmid pJJ1886_4, on the other hand, is similar to the E. coli plasmid pHS102707 (GeneBank Acc. N° KF701335). These two plasmids thus represent new additions to the ST131 plasmidome (see S16B Fig.). The IME_E35BA is a 14.2 kb insertion within a 234 kb chromosomal contig. S16C Fig. shows some detail on the genetic structure of the IME and its insertion site in the ST131 core genome.
The last potentially conjugative plasmid is the 24.5 kb MOBC12 plasmid pE61BA_2, also not closely related to any reference, as seen in the REL phylogenetic tree of S17A Fig. This plasmid resulted in a single contig, so it could be closed. The closest homolog is the Yersinia pestis plasmid pCRY, with which it shares all backbone genes. Its RIP belongs to the Rep_FIIA superfamily, although this plasmid group is not represented in the classical PBRT method [8]. It does not contain any gene with known adaptive function, except a protease and a putative secreted thermonuclease. As in the case above, this plasmid represents a new addition to the ST131 plasmidome (S17B Fig.), its relaxase being only 70% identical to its closest homolog, the pCRY relaxase.
Among the small plasmids, the first group is composed by four MOBP5/ColE1-like plasmids (11.8, 6.9, 6.6 and 5.6 kb). Three of the plasmids are relatively different, as judged by the MOBP5 phylogenetic tree of S18A Fig. The large plasmid (pBIDMC38_1) contains a type II restriction-modification system (Cfr10I) and is almost identical to the ST131 reference pJJ1886_3 (S18B Fig.). The two MOBP5/ColE1 plasmids of strain E61BA (plasmids pE61BA_5 and pE61BA_6) contain colicin ColE and ColK genes, respectively (S18C and S18D Fig.). Colicins are considered both as virulence factors as well as traits that influence bacterial fitness and survival in the presence of competitors [68].
Besides, there were four almost identical MOBQu plasmids of around 4.1 kb (S19A Fig.), which populate all H30 subgroups (two of virotype A, one of virotype B and one of virotype C). Nothing remarkable could be distinguished it their genetic constitution, besides a common MOB region and a pIGWZ12 -like Rep protein (S19B Fig.). Four very similar MOBQ12 plasmids (around 5.2 kb) are also represented in H30 (two of virotype A, one of virotype C) and H22 (one of virotype D). They contain RIP and REL proteins but, as in the case of the MOBQu plasmids, no phenotype could be pointed out (S20 Fig.). MOBQu and MOBQ12 plasmids have received little attention because they are cryptic and remain unnoticed in most typing schemes. The present ST131 plasmidome analysis suggests they can be surprisingly abundant in E. coli. Finally, there were five no-MOB cryptic plasmids (four of them were 1.5 kb long and the other 5.0 kb). They all contain distinguishable Rep proteins (Rep_HTH_36_superfamily), without assignment in the PBRT method. Four of them are almost identical among themselves (S21 Fig.), while the fifth (pFV9873_3) was unique and unrelated to any reference. A detailed analysis of MOBF11/IncN plasmid family is detailed in S22 Fig.
Discussion
There are two aspects of this work that will focus the discussion. On one side, the applicability, usefulness and limitations of PLACNET will be discussed. On the other, the plasmidome of E. coli ST131 genomes that were reconstructed by PLACNET will be analyzed as an example of the applicability of the method. Analysis of the individual reconstructed plasmids, meant for plasmid specialists, is expanded in S1 Text.
Bacterial genomes and plasmid reconstructions
Most bacterial genomes contain more than one physical unit of DNA. Besides the main chromosome, some bacteria contain additional chromosomes and most contain plasmids. We propose that PLACNET can be used as a new method to analyze bacterial genomes. It allows the assignment of chromosomes and plasmids as separate physical units within a genome. Visual representation of the network in Cytoscape, in which plasmids appear as constellations in a starry sky, allows user-friendly apprehension of that genome constitution. We applied PLACNET in this work to analyze the plasmidome of E. coli ST131 genomes, but it has been shown to work also for a series of prototypic bacterial genera with different GC content and genome architecture, such as Salmonella, Klebsiella, Agrobacterium, Staphylococcus or Bacillus. As an example, the PLACNET representation of the genome of Staphylococcus aureus strain 118 (ST772) (GenBank acc number AJGE00000000) is shown in S23 Fig. PLACNET scope of application also includes multi-chromosome bacteria like Vibrio or Brucella, where it correctly predicts both chromosomes present in these species. One Vibrio cholerae Pacini 1854 genome (Bioproject ID: PRJEB2215) is shown in S24 Fig. as an example. Once contigs belonging to each plasmid are defined, classical plasmid analysis ensues, as explained in the Results section. Contigs selected as part of a single plasmid are taken together and its overall proteome used to build a clustering dendrogram with reference plasmids present in the network. The dendrogram tree gathers plasmids according to the number of homologous proteins they share, providing an indication on prototype plasmids closely related with the query plasmid. There are two issues in PLACNET analysis that require additional work and for which additional improvement can be expected:
Unassigned contigs and reference sets
After plasmid reconstruction, occasionally, one or a few contigs may remain unassigned. In the set of ST131 genomes analyzed in this work, there were only two unassigned contigs (>200 bp), both in E61BA (Fig. 2). The fact that only two unassigned contigs appeared in the analysis of eight E. coli genomes suggests that this is not a quantitatively important problem. As could be expected, unassigned contigs are more frequent in genomes for which there are fewer references available. The lack of a suitable reference set results in poor quality clustering and an increased fraction of contigs without references. It is obvious from the preceding discussion that bacterial taxons for which not enough references exist will be more problematic for plasmid reconstruction. Thus, any such project should start by the generation of a sufficiently ample set of plasmid references. In this respect, E. coli constitutes probably the best choice, due to the large reference set available.
Repeat sequences and difficult plasmids
Usually PLACNET works well because contigs belonging to individual plasmids pair with different selected references and thus cluster in disjoint connected components in the Cytoscape representation after a single pruning step. The pruning step consists in identifying the bridging contigs (network hubs) as repeat sequences (RS). Two sets of evidence were used: (i) homology to known ISs or transposons, and (ii) existence of three or more scaffold links. Contigs fulfilling these two criteria were assumed to be in fact repeated in the connected network. Thus, the pruning operation consisted of duplicating the alluded nodes and splitting their scaffold links. In the tested set of E. coli genomes that were used to validate PLACNET (the set of eight ST131 genomes analyzed here, the 32 genome set analyzed by de Been et al. (2014) < submitted to Plos Genet together with this work >, plus another set of 10 other ESBL genomes obtained from clinical strains of bioprojects PRJNA186205 and PRJNA202876), there was only one case in which RS pruning operation was not sufficient to obtain disjoint components. It was the case of genome E35BA, where two coexisting MOBF12/IncF plasmids (pE35BA_2 and pE35BA _3) could be inferred, but repeated pruning did not result in disjoint components. The evidence for the existence of two plasmids was the finding of two sets of contigs containing REL and other plasmid backbone genes. PLACNET failed in discriminating both plasmids probably because network links to references were interlocking, since several PLACNET-selected references established best hits to different components of each set. Besides, the assembly program could not distinguish among parts of both sequences and considered them as RSs. Closely related plasmids that coexist in a given cell poses the most serious problem we encountered in the application of PLACNET.
The ST131 plasmidome
HGT plays a critical role in shaping bacterial lineages, especially those of multi-environment opportunistic pathogens. Comprehensive characterization of plasmidomes has been impeded by methodological limitations, although they are essential for multilevel population genetics analysis, an approach necessary to explain selection and diversification of bacterial populations and to understand the reservoir dynamics of antibiotic resistance and virulence genes [69]. The application of PLACNET to ST131 genomes allowed the detection of emerging plasmid variants, important for the evolutionary history of this ExPEC lineage, which constitutes an outstanding example of a “high risk clonal complex”, a concept increasingly important in Public Health [69].
Plasmidome description
We describe a remarkable heterogeneity of plasmids among the E. coli ST131 genomes analyzed, with the identification of 39 plasmids to add to the 11 plasmids in the ST131 lineage already sequenced (Table 1). Interestingly, these plasmids encompass 8 out of 17 main MOB plasmid groups found in the whole class of γ-proteobacteria [11], [53], namely F12 (IncF), P3 (IncX), P5 (ColE1), P6 (IncI2), P11 (IncP), P12 (IncI/K/BO), Q12 (Rep_pSC101-like), Qu (Rep_pMG828-2/IGWZ12-like), several of them undetectable by PBRT. Besides conjugative or mobilizable plasmids, there were other plasmids, lacking REL, but identifiable by their RIP proteins. An in-depth analysis of each plasmid group identified in this study has been diverted to a Supplementary Discussion (though exciting for clinical epidemiology or plasmid biology, it is outside the mainstream goal of this work). Our findings substantially enlarge the repertoire of plasmids identified among E. coli ST131 isolates, which now reflect a genome widely open for plasmid infection. It is of note that this scenario has also been described for E. coli clones of different pathovars [47], [70]–[73]. Comparative genomics of the few E. coli lineages comprehensively analyzed to date suggests that this species is a generalist able to colonize and infect humans. It also suggests that phages and plasmids make an important contribution to specialization by accessorizing the genome with new adaptive traits and tools that modify genome structure and, eventually, by modifying transcriptional regulation [70], [71], [74], [75]. It should also be emphasized that almost all available studies on ST131, included this one, focused on strains involved in the spread of antibiotic resistance genes, which constitute, undoubtedly, a biased fraction of the ST131 plasmidome and thus preclude an accurate view of its evolutionary history [76] (see also below).
Plasmids and E. coli diversification
Specific ExPEC lineages have scarcely been analyzed in the context of multilevel population genetics with the exception of punctual cases involving clonally unrelated isolates [70]. A recent phylogenomic analysis of 95 ST131 isolates from different geographical areas identified the same three clusters studied in the present work [16]. This analysis concluded that point-mutations and recombination events associated with diverse MGEs, including prophages and genomic islands, determined the diversification of this ExPEC lineage. However, the diversity of plasmids was only inferred by searching for incompatibility regions based on PBRT schemes. The role of plasmids in genomic versatility were not further analyzed [16]. Even though our study analyzed a smaller number of isolates, some observations can be drawn about the role of plasmids in the diversification of the ST131 lineage.
The rate of mutagenesis of E. coli has been roughly estimated in one mutation per genome per year [77]. Although this number is no doubt controversial, such study and those addressing the role of recombination in the ST131 lineage add context to understand its evolution as represented in Fig. 1. Compared to the limited sequence divergence among the ST131 core genomes (the ST43/H30 branch includes just about 600 SNPs), plasmids represent a very active fraction of ST131 adaptive evolution, as can be concluded from the analysis of Table 1. Such plasmid variability suggests that independent plasmid acquisitions and losses frequently occur within and between ST131 sublineages. Within the H30 cluster, the presence of antibiotic resistance plasmids is remarkable, specially the identification of structurally similar F2:A1:B - plasmids carrying genes conferring antibiotic resistance, since early acquisition of blaCTX-M-15, mainly associated with F2 plasmids, is considered a key event in the selection of the ST131 cluster C/H30 subclone [13], [16], [17]. The modular structure of F2 plasmids, containing multiple copies of ISs, facilitates gene rearrangements and the interchange of antibiotic resistance platforms linked to resistance to first line antibiotics between plasmids of the same and different families. This notion, inferred from our present analysis, has already been proposed [17], [78]–[80] and is of great concern nowadays because of the increasing risk of encountering E. coli isolates carrying blaKPC of blaNDM genes predominant in Klebsiella (http://www.cdc.gov/drugresistance/threat-report-2013/) [81]. Beyond F2 plasmids, the presence of other plasmid groups (N, I1/K/BO, I2, A/C, X) carrying antibiotic resistance genes is observed at variable rates in this and other studies, clearly influenced by local ecology. Most of these antibiotic resistant non-F plasmids occur only rarely in E. coli isolates [82]. This could be due to an intrinsic lack of fitness of these plasmids in E. coli under natural conditions. Alternatively, they could represent cryptic indigenous plasmids now identifiable because of the acquisition of antibiotic-resistance cassettes. Nevertheless, the acquisition of mosaic regions carrying multiple antibiotic resistance genes by broad host plasmids (e.g. N, I2, A/C) increases the risk to spread resistance to first line antibiotics to different bacterial species in and outside hospitals [79], [83]. Besides antibiotic resistance plasmids, an outstanding finding of this work was the frequent detection other plasmid groups, generally considered cryptic, that are clearly underrepresented in previous ST131 studies, as ColE1, MOBQ, and phage-like Rep3 plasmids. All of them are highly heterogeneous plasmid groups able to acquire adaptive traits or contribute to the mobilization of other plasmids (See supplementary dataset for details).
Members of the ST131 plasmidome such as MOBF12/IncF, MOBH/IncA/C and Rep3/phage-like plasmids can also shape the E. coli chromosome by facilitating mobilization in trans of genetic islands or integrating new genetic material [32], [70], [84]. Interestingly, recombination of large chromosomal regions occurring at the sites of insertion of either prophages or transferable genomic islands seems to have contributed to the split of the ST131 lineage in different clusters [16]. Although experimental studies on ST131 did not yet associate plasmids with genome structure, the hypothesis is plausible taking into account the frequency at which such events occur in other B2 E. coli populations.
On a more general note, our study identifies antibiotic resistance plasmids, which are favored in high density environments, such as the human gastrointestinal tract, and under antibiotic selective pressure, that are predominant in hospitals [85] together with phages (or phage-like cryptic plasmids), apparently predominant in low density environments [86], [87], and other cryptic plasmids (frequently very small and devoid of any possible adaptive gene). This mixed constitution, which is difficult to understand on purely selective grounds, highlights potential roles of plasmids in the context of multilevel selection, a recurrent issue in evolutionary biology. The plasmid flux in ST131 strains occurs while disseminating genes coding for resistance to extended spectrum beta-lactamases (ESBL). The results presented here find a complement in the study of de Been et al. (accompanying paper), where the authors document the dissemination of ESBL-carrying epidemic plasmids from animal to human clonally unrelated E. coli lineages. Thus, many plasmids appear in a clonal lineage, and many lineages can be infected by a single predominant plasmid. These conceptual notions have relevance in Public Health as they deal with the hierarchical units of selection that contribute to increase the population size of antibiotic resistance genes in human and animal pathogens [69], [88], [89].
In summary, our study reveals the utility of PLACNET in multilevel population genetics analysis, critical to understand the evolutionary processes and dynamics of both bacterial and plasmid lineages. Its application to E. coli ST131 allowed us to infer the roles of plasmids in the dissemination of globally spread antibiotic resistance and virulencegenes, some of them being underrepresented in Genbank. It is probable that these plasmids are critically relevant to understand the adaptive evolution of E. coli populations and their bacterial exchange communities. Armed with this new tool for plasmid analysis, future scrutiny of a larger number of significant strains will allow us to understand the interplay among different plasmid associations that often appear in bacterial pathogens.
Conclusion
The evolutionary processes of main bacterial pathogens are often discussed in the context of lineage-associated acquisition of a specific virulence gene set. The present study demonstrates how E. coli ST131 strains, even when they are practically identical in their core genomes, contain a striking variety of different plasmids. Many of them remain unnoticed, since they are apparently cryptic. Prevalent plasmids, such as IncFs, undergo frequent recombination, continuously resulting in novel gene repertoires. Our results shed light on the role of plasmids in E. coli ST131 evolution. Horizontal transmission of plasmids that carry not only antibiotic resistance and virulence genes, but also other poorly analyzed functions (metabolic genes, colicins and as yet cryptic functions) is common in the ST131 plasmidome and results in frequent and rapid adaptive changes. Arrival to these conclusions has been made possible by the application of PLACNET, a plasmid reconstruction method for WGS datasets.
Materials and Methods
Epidemiological background of bacteria and plasmids
Comprehensive plasmidome analysis was carried out for 10 E. coli ST131 genomes, representing main ST131 sublineages described to date [13], [16]. They include strains coming from Spain (three fimH30, one fimH324), USA (three fimH30), Australia (one fimH30), Denmark (one fimH22) and Japan (one fimH41). The fimH30 strains from Spain were CTX-M-15 producers and belonged to the H30-Rx sublineage (additionally, one strain was also CTX-M-14), while those collected in the USA were KPC-2 producers. The four strains from Spain represent predominant ST131 variants on the basis of PFGE patterns and the presence/absence of four putative virulence markers (afaFM955459, encoding an Afa/Dr adhesion; sat, secreted autotransporter toxin; ibeA, invasion of brain endothelium; and iroN, salmochelin siderophore receptor) [36], [51], [52] and sequenced for this work.
The ST131 isolates studied represent epidemic variants exhibiting particular combination of putative virulence traits and were previously designed as distinct “virotypes” by capital letters A to D [36]. It should be noted that no correlation exists between these “virotype” designations and “ST131 strain designation” in other studies that also used capital letters to distinguish among ST131 clonal variants [16]. Other genome datasets were taken either from Bioproject NCBI database (https://www.ncbi.nlm.nih.gov/bioproject/), or from NCBI genomes database (E. coli JJ1886 [90] and E. coli SE15 [91]). In addition, fully sequenced plasmids pEK499, pEK204, pEK516, pJIE186-2 and pJIE143, previously found in other ST131 isolates [56], [92]–[94], were used for plasmid comparisons. Information about all genomes is detailed in Table 3.
Tab. 3. Human E. coli ST131 genomes analyzed in this work. 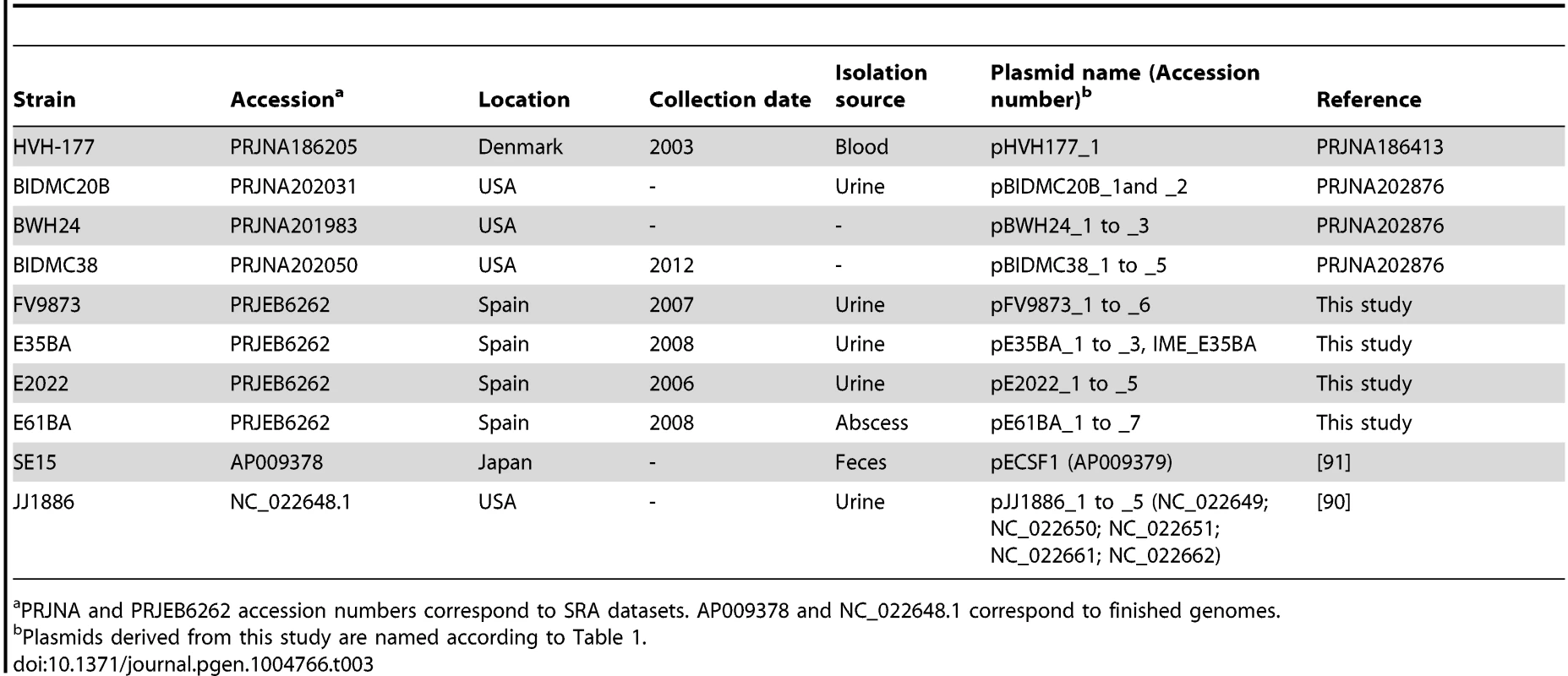
PRJNA and PRJEB6262 accession numbers correspond to SRA datasets. AP009378 and NC_022648.1 correspond to finished genomes. DNA sequencing
Total DNA from E. coli ST131 strains FV9873, E35BA, E2022 and E61BA was extracted with QIAmp DNA Mini Kit (Qiagen). DNA concentration was measured with Nanodrop 2000 (Thermo Scientific) and Qubit 2.0 Fluorometer (Life Technologies). 1.0 µg DNA was sonicated (20 cycles of 30 s at 4°C, low intensity) with Bioruptor Next Generation (Diagenode). Sample quality was checked in a Bioanalyzer 2100 (Agilent Technologies). DNA samples were preconditioned for sequencing by using the TruSeq DNA Sample Preparation Kit (Illumina) and quantified with Step One Plus Real-Time PCR System (Applied Biosystems). Flow-cells were prepared with TruSeq PE Cluster Kit v5-CS-GA (Illumina). Sequencing was carried out using a standard 2×71 base protocol (300-400 bp insert size) in a Genome Analyzer IIx (Illumina, San Diego, CA) at the sequencing facility of the University of Cantabria. The main statistics of the eight sequence datasets analyzed are shown in Table 4.
Tab. 4. Genomes assembled in this study. 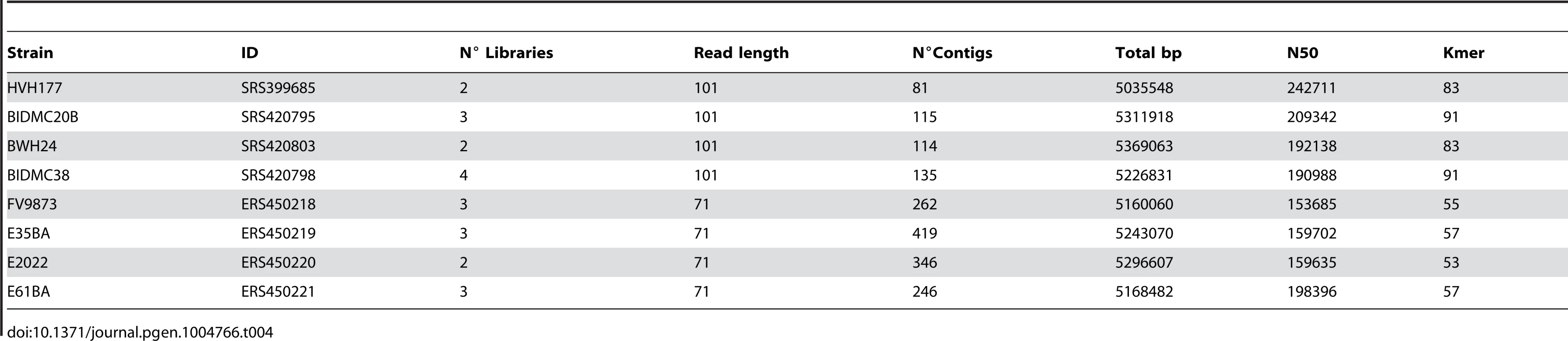
Phylogenetic analysis of the ST131 core genome
The ST131 core genome was defined as the collection of genes present in the ten ST131 genomes analyzed, with more than 90% similarity and 90% coverage. CD-HIT-EST [95] was used to cluster genes. A homemade Perl script was created to parse the cluster and define the core genome set. All core genes were concatenated and aligned with progressive Mauve [96]. A tabular list of SNPs was extracted from the Mauve alignment by applying the SNP export tool of Mauve GUI. The tabular list of polymorphic sites was parsed by a homemade script. A given position was counted as an SNP if it varied between two given sequences. The number of SNPs was added for each pair of strains to give the final SNP count. Polymorphic sites with gaps were removed from the SNP count matrix. The Mauve alignment was curated by trimAl [97]. RAxML [98] was used to build the core genome phylogenetic tree, using 100 replicates for bootstrap determination.
Plasmid Constellation Networks (PLACNET)
PLACNET was developed to associate contigs with specific physical DNA units in WGS experiments. Networks are powerful models that allow visualization and analysis of sequence information. PLACNET networks are composed of two types of nodes (contigs and reference genomes) and two types of edges (similarity to reference sequences and scaffold links). Commonly, network layout algorithms simulate repulsion forces between nodes and attraction forces by the edges that link two nodes. Thus, node distribution in the network will depend on the intensity of forces that define the edges. In such network model, a plasmid will be represented by a connected component (a set of linked nodes) or, in other words, a constellation of contigs. Different physical units (plasmids and chromosomes) should be represented by disjoint connected components (separate constellations). The workflow (Fig. 6) involves the following steps:
Fig. 6. PLACNET flow diagram. 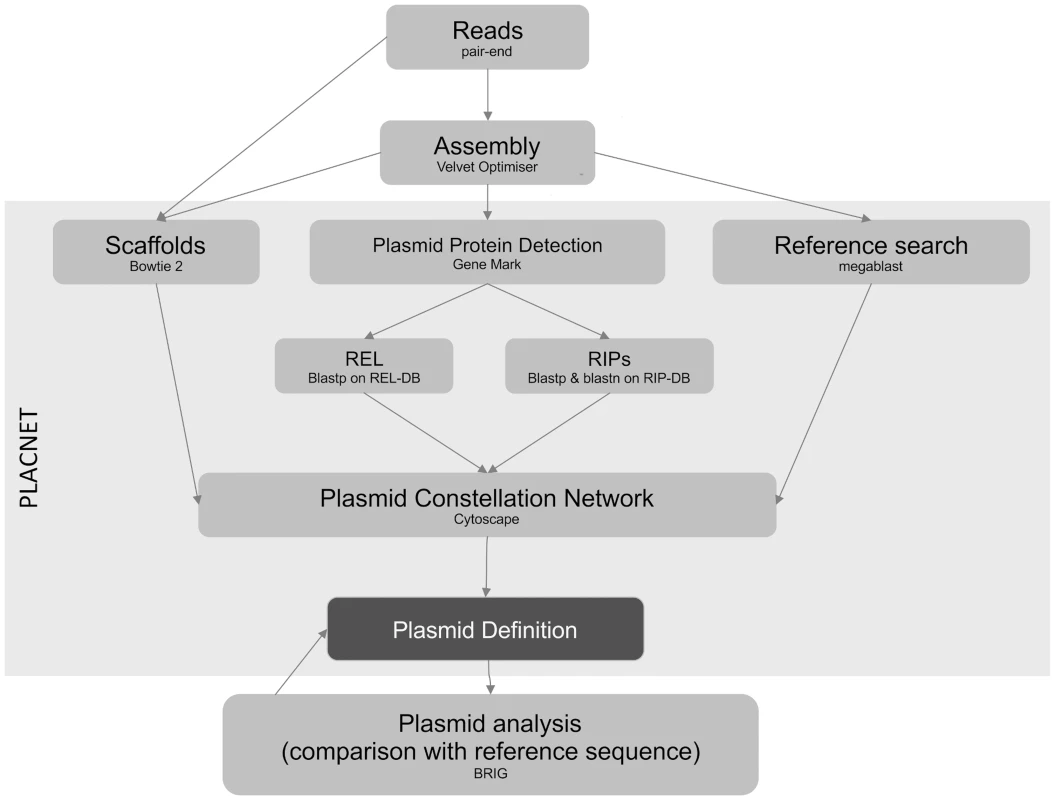
The diagram represents the PLACNET workflow to analyze an Illumina bacterial genome dataset. It can be separated in two sub-process: network delineation and plasmid analysis. Network delineation consists on contig assembly, determination of scaffold interactions, reference search of homologous genomes and plasmid protein prediction. Plasmid analysis basically consists in the construction of a dendrogram of plasmid protein profiles, which identifies the most relevant reference sequences, followed by plasmid cluster analysis, which compares query plasmids with its closest references. Plasmid analysis is a feedback process that helps to resolve uncertainties and results in a final definition of plasmid and chromosome content. Assembly
Velvet assembly software [99] and its script VelvetOptimiser.pl were used to determine the best assembly and to scan the optimum parameters. Velvet provides also coverage information for each contig, which adds useful information for network interpretation.
Scaffold links
Although assembly programs perform scaffolding between contigs, when the assignation is ambiguous, contigs remain unbound. A method based in the mapping tool Bowtie 2 [100] was used to find all possible scaffold links. All reads were mapped using the contigs as references, using default parameters of Bowtie 2, with the option to report all possible hits. The output file was converted to SAM format [101] to give the potential adjacency information for each contig. We considered as potential PLACNET scaffold links those which comply with two rules: (i) the contigs were paired at their extremities, themselves defined as twice the read length, and (ii) the number of pair-end reads linking those two contigs had to be higher than one third of the mean of the total pair-end reads that scaffold all contigs. This procedure was implemented as an in-house Perl script.
Reference search
At least for bacteria widely covered by sequencing projects, most contigs in any new sequence are similar to one or more previously published sequences (reference sequences). Our initial hypothesis was that, for any physical DNA unit, its contigs will “BLAST” a related set of reference sequences. Thus, in the PLACNET network representation, they will cluster around the homologous references. The more DNA databases grow, the closer the references will be to the query sequence and the better PLACNET will work. A homemade BLAST [102] database was constructed from the NCBI genomes database by joining all sequences contained in [ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/] and [ftp://ftp.ncbi.nlm.nih.gov/genomes/Plasmids/]. The version used in this work was from March 7th 2013 and contains 6,432 genomes (plasmids and chromosomes). Megablast search of all contigs was carried out against the homemade BLAST-genome database with the objective of selecting a few best matches for network construction. Due to the different length of each contig, fixed thresholds by e-value or score cannot be chosen. Since the score is not a normalized parameter, and varies depending on sequence length, hits were selected by applying a dynamic threshold, based on the number of homologous sequences and the score of each hit. If the threshold is defined as 85%+2n of the mean of the n previous sequence scores, and n is the ranking position of sequence i retrieved by megablast with score Si, then Tn+1 is the threshold for sequence n+1:
All reference sequences above the threshold were taken as nodes in the PLACNET representation.
Protein prediction of replication initiator proteins (RIP) and relaxases (REL)
Some genes are indicative of a plasmid sequence. Among them we selected REL, key proteins in the conjugation process [53], [103] and RIPs, key proteins in the replication of most plasmids [104]. Although not all plasmids contain a RIP and/or REL, their presence in a contig is diagnostic of a plasmid (or ICE) sequence. Some plasmids have more than one RIP (i.e. IncF family plasmids) [9], [105], [106] but plasmids rarely have more than one REL [107]. ORF prediction was carried out by GeneMark [108], which optimizes predictions based on GC content of DNA. The heuristic prediction implemented in this software is especially useful to predict ORFs in plasmid-containing genomes because it takes each contig individually and selects the best prediction model case by case.
To implement specific search protocols for REL and RIPs, three homemade databases (DB) were developed. A REL database (REL-DB) was constructed according to [53], [109]. Similarly, RIP-DB was constructed from all RIPs annotated in UniProt database. RIP sequences were clustered by CD-HIT [95], using 40% identity as a threshold. Next, a Hidden Markov Model was built from each cluster by hmmer3 [110]. Finally, the HMM profiles were used in a search against UniProt. The HMM search and initial dataset were joined in one database (RIP-DB). An additional step was necessary to classify RIPs according to the widely used plasmid classification protocol PBRT [8]. A homemade nucleotide database (INC-DB) was created to identify PBRT types in silico using blastn. Finally, the relevant ORFs (REL and RIPs) identified to specific contigs by using REL-DB and RIP-DB were incorporated to the network as tags. All relevant steps in network construction were implemented as a Perl script available at the following web page: http://placnet.sourceforge.net/.
Plasmid constellations
As explained above, each plasmid is represented in PLACNET by a connected component (a constellation). Thus, different physical units (plasmids and chromosomes) should be represented by disjoint (unlinked) connected components. Cytoscape software [111] was used to visualize and analyze plasmid constellations, which incorporate all the information (similarity to reference sequences, scaffold links, and protein tags) in a single network. Node attributes such as contig size, coverage and reference description, are added to the network. At this stage, network pruning is needed to resolve individual plasmids as disjoint components. When a genome has a number of repeated sequences (e.g., insertion sequences (ISs) or transposons), or two very similar plasmids, the assembly process outputs those sequences as contigs with multiple scaffold links. In the network context, they represent hubs, that is, nodes with a high number of connections. This makes the network very dense and complicates the analysis of network connected components. In PLACNET, contigs smaller than 200 bp were directly eliminated from the analysis. Hubs were examined by blastx against protein databases (i.e. UniProtKN or NCBI nr). If there was identity to any transposase gene, the hub was duplicated, and scaffold links were partitioned among them, to maximize the number of disjoint components. Contigs that remain unbound are classified as “unassigned sequence” in the contig assignation table.
Plasmid definition, dendrograms and cluster analysis
The final steps in plasmid reconstruction involve the definition and verification of each plasmid. This is an iterative process, as shown in Fig. 6. First, each contig was assigned to a putative plasmid (or chromosome) based on visualization of disjoint connected components in the Cytoscape representation. Assignments take into consideration additional types of evidence like the presence of REL and/or RIPs, size of the putative plasmid compared to reference plasmids and coverage of each contig (contigs belonging to the same plasmid must have similar coverage). Taking into account the information provided by related genomes within the same sequencing project can also be helpful (same sequences cluster around the same references). In this respect, PLACNET is more robust in multi-strain collections. The performance of PLACNET was validated by testing a number of previously sequenced and annotated E. coli genomes (S2 Table). The ART software (Huang et al., 2012) was used to simulate pair-end Illumina reads from those genomes, which were then analyzed by PLACNET as explained above. Results are shown in S2 Text, S3 Table and S25-S34 Figs.
After PLACNET has defined the plasmids carried in the relevant genomes, the next step in plasmidome analysis is to build a dendrogram that produces a hierarchical clustering of plasmid proteomes similar to those described in [112]–[114]. CD-HIT (thresholds: 70% identity and 80% coverage) was used for clustering references and query plasmids. Based on the output file, a presence/absence table (present or absence of each protein cluster in each plasmid) was built. Each table row represents a plasmid protein profile. Raup-Crick distance method, implemented in vegan package for R software [115], was used to calculate the distance matrix of plasmid protein profiles. The Ape package [116] was used to calculate the dendrogram bootstrapping confidence value. Finally, a hierarchical clustering dendrogram was built using the UPGMA algorithm.
Putative plasmids and references belonging to the same dendrogram branch were compared using BRIG [117] or Abacas [118]. While BRIG is not sensitive to contig arrangement, Abacas can be used to order contigs according to a given reference. With these tools, the curator is able to visualize the correspondence between reconstructed plasmids and references, or can go back to dendrograms or PLACNET in the search for missing or extra contigs. This iterative mode of analysis is represented in Fig. 6 by the backward arrow linking plasmid cluster analysis with plasmid definition.
Plasmids were mainly classified according to their REL in MOB families, as described by [53]. Classical Inc families are also given when typing them by in silico PBRT was possible. Plasmids that could not be classified one way or the other were termed no-MOB by exclusion.
Supporting Information
Zdroje
1. DidelotX, BowdenR, WilsonDJ, PetoTE, CrookDW (2012) Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13 : 601–612.
2. JohnsonTJ, NolanLK (2009) Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev 73 : 750–774.
3. CarattoliA (2011) Plasmids in Gram negatives: molecular typing of resistance plasmids. Int J Med Microbiol 301 : 654–658.
4. CarattoliA (2013) Plasmids and the spread of resistance. Int J Med Microbiol 303 : 298–304.
5. AhmedSA, AwosikaJ, BaldwinC, Bishop-LillyKA, BiswasB, et al. (2012) Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including shiga toxin encoding phage stx2. PLoS One 7: e48228.
6. BoerlinP, ChenS, ColbourneJK, JohnsonR, De GrandisS, et al. (1998) Evolution of enterohemorrhagic Escherichia coli hemolysin plasmids and the locus for enterocyte effacement in shiga toxin-producing E. coli. Infect Immun 66 : 2553–2561.
7. CouturierM, BexF, BergquistPL, MaasWK (1988) Identification and classification of bacterial plasmids. Microbiol Rev 52 : 375–395.
8. CarattoliA, BertiniA, VillaL, FalboV, HopkinsKL, et al. (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63 : 219–228.
9. VillaL, Garcia-FernandezA, FortiniD, CarattoliA (2010) Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65 : 2518–2529.
10. Garcia-FernandezA, FortiniD, VeldmanK, MeviusD, CarattoliA (2009) Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 63 : 274–281.
11. AlvaradoA, Garcillan-BarciaMP, de la CruzF (2012) A Degenerate Primer MOB Typing (DPMT) Method to Classify Gamma-Proteobacterial Plasmids in Clinical and Environmental Settings. PLoS One 7: e40438.
12. EdwardsDJ, HoltKE (2013) Beginner's guide to comparative bacterial genome analysis using next-generation sequence data. Microb Inform Exp 3 : 2.
13. PriceLB, JohnsonJR, AzizM, ClabotsC, JohnstonB, et al. (2013) The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio 4: e00377–00313.
14. JohnsonJR, ClermontO, JohnstonB, ClabotsC, TchesnokovaV, et al. (2014) Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J Clin Microbiol.
15. BlancV, Leflon-GuiboutV, BlancoJ, HaenniM, MadecJY, et al. (2014) Prevalence of day-care centre children (France) with faecal CTX-M-producing Escherichia coli comprising O25b:H4 and O16:H5 ST131 strains. J Antimicrob Chemother.
16. PettyNK, Ben ZakourNL, Stanton-CookM, SkippingtonE, TotsikaM, et al. (2014) Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A.
17. CoqueTM, NovaisA, CarattoliA, PoirelL, PitoutJ, et al. (2008) Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis 14 : 195–200.
18. Nicolas-ChanoineMH, BertrandX, MadecJY (2014) Escherichia coli ST131, an Intriguing Clonal Group. Clin Microbiol Rev 27 : 543–574.
19. Nicolas-ChanoineMH, BlancoJ, Leflon-GuiboutV, DemartyR, AlonsoMP, et al. (2008) Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61 : 273–281.
20. Leflon-GuiboutV, BlancoJ, AmaqdoufK, MoraA, GuizeL, et al. (2008) Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J Clin Microbiol 46 : 3900–3905.
21. PlatellJL, JohnsonJR, CobboldRN, TrottDJ (2011) Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet Microbiol 153 : 99–108.
22. AlbrechtovaK, DolejskaM, CizekA, TausovaD, KlimesJ, et al. (2012) Dogs of nomadic pastoralists in northern Kenya are reservoirs of plasmid-mediated cephalosporin - and quinolone-resistant Escherichia coli, including pandemic clone B2-O25-ST131. Antimicrob Agents Chemother 56 : 4013–4017.
23. HernandezJ, BonnedahlJ, EliassonI, WallenstenA, ComstedtP, et al. (2010) Globally disseminated human pathogenic Escherichia coli of O25b-ST131 clone, harbouring blaCTX-M-15, found in Glaucous-winged gull at remote Commander Islands, Russia. Environ Microbiol Rep 2 : 329–332.
24. PallecchiL, BartoloniA, FiorelliC, MantellaA, Di MaggioT, et al. (2007) Rapid dissemination and diversity of CTX-M extended-spectrum beta-lactamase genes in commensal Escherichia coli isolates from healthy children from low-resource settings in Latin America. Antimicrob Agents Chemother 51 : 2720–2725.
25. MoraA, HerreraA, MamaniR, LopezC, AlonsoMP, et al. (2010) Recent emergence of clonal group O25b:K1:H4-B2-ST131 ibeA strains among Escherichia coli poultry isolates, including CTX-M-9-producing strains, and comparison with clinical human isolates. Appl Environ Microbiol 76 : 6991–6997.
26. DhanjiH, MurphyNM, AkhigbeC, DoumithM, HopeR, et al. (2011) Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M-14 extended-spectrum beta-lactamase from UK river water. J Antimicrob Chemother 66 : 512–516.
27. Colomer-LluchM, MoraA, LopezC, MamaniR, DahbiG, et al. (2013) Detection of quinolone-resistant Escherichia coli isolates belonging to clonal groups O25b:H4-B2-ST131 and O25b:H4-D-ST69 in raw sewage and river water in Barcelona, Spain. J Antimicrob Chemother 68 : 758–765.
28. GibreelTM, DodgsonAR, CheesbroughJ, BoltonFJ, FoxAJ, et al. (2012) High metabolic potential may contribute to the success of ST131 uropathogenic Escherichia coli. J Clin Microbiol 50 : 3202–3207.
29. NovaisA, PiresJ, FerreiraH, CostaL, MontenegroC, et al. (2012) Characterization of globally spread Escherichia coli ST131 isolates (1991 to 2010). Antimicrob Agents Chemother 56 : 3973–3976.
30. BlancoJ, MoraA, MamaniR, LopezC, BlancoM, et al. (2011) National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J Antimicrob Chemother 66 : 2011–2021.
31. JohnsonJR, KuskowskiMA, GajewskiA, SahmDF, KarlowskyJA (2004) Virulence characteristics and phylogenetic background of multidrug-resistant and antimicrobial-susceptible clinical isolates of Escherichia coli from across the United States, 2000-2001. J Infect Dis 190 : 1739–1744.
32. SchubertS, DarluP, ClermontO, WieserA, MagistroG, et al. (2009) Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog 5: e1000257.
33. WirthT, FalushD, LanR, CollesF, MensaP, et al. (2006) Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60 : 1136–1151.
34. JohnsonJR, Nicolas-ChanoineMH, DebRoyC, CastanheiraM, RobicsekA, et al. (2012) Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967–2009. Emerg Infect Dis 18 : 598–607.
35. JohnsonJR, TchesnokovaV, JohnstonB, ClabotsC, RobertsPL, et al. (2013) Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis 207 : 919–928.
36. BlancoJ, MoraA, MamaniR, LopezC, BlancoM, et al. (2013) Four main virotypes among extended-spectrum-beta-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J Clin Microbiol 51 : 3358–3367.
37. PeiranoG, PitoutJD (2014) Fluoroquinolone resistant Escherichia coli ST131 causing bloodstream infections in a centralized Canadian region: the rapid emergence of H30-Rx sublineage. Antimicrob Agents Chemother.
38. ColpanA, JohnstonB, PorterS, ClabotsC, AnwayR, et al. (2013) Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis 57 : 1256–1265.
39. BanerjeeR, RobicsekA, KuskowskiMA, PorterS, JohnstonBD, et al. (2013) Molecular epidemiology of Escherichia coli sequence type 131 and Its H30 and H30-Rx subclones among extended-spectrum-beta-lactamase-positive and -negative E. coli clinical isolates from the Chicago Region, 2007 to 2010. Antimicrob Agents Chemother 57 : 6385–6388.
40. DahbiG, MoraA, LopezC, AlonsoMP, MamaniR, et al. (2013) Emergence of new variants of ST131 clonal group among extraintestinal pathogenic Escherichia coli producing extended-spectrum beta-lactamases. Int J Antimicrob Agents 42 : 347–351.
41. AvasthiTS, KumarN, BaddamR, HussainA, NandanwarN, et al. (2011) Genome of multidrug-resistant uropathogenic Escherichia coli strain NA114 from India. J Bacteriol 193 : 4272–4273.
42. TotsikaM, BeatsonSA, SarkarS, PhanMD, PettyNK, et al. (2011) Insights into a Multidrug Resistant Escherichia coli Pathogen of the Globally Disseminated ST131 Lineage: Genome Analysis and Virulence Mechanisms. PLoS One 6: e26578.
43. ClarkG, PaszkiewiczK, HaleJ, WestonV, ConstantinidouC, et al. (2012) Genomic analysis uncovers a phenotypically diverse but genetically homogeneous Escherichia coli ST131 clone circulating in unrelated urinary tract infections. J Antimicrob Chemother 67 : 868–877.
44. LavigneJP, VergunstAC, GoretL, SottoA, CombescureC, et al. (2012) Virulence potential and genomic mapping of the worldwide clone Escherichia coli ST131. PLoS One 7: e34294.
45. KunneC, BillionA, MshanaSE, SchmiedelJ, DomannE, et al. (2012) Complete sequences of plasmids from the hemolytic-uremic syndrome-associated Escherichia coli strain HUSEC41. J Bacteriol 194 : 532–533.
46. GradYH, GodfreyP, CerquieraGC, Mariani-KurkdjianP, GoualiM, et al. (2013) Comparative genomics of recent Shiga toxin-producing Escherichia coli O104:H4: short-term evolution of an emerging pathogen. MBio 4: e00452–00412.
47. BrunderW, SchmidtH, FroschM, KarchH (1999) The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145 (Pt 5): 1005–1014.
48. PallenMJ, WrenBW (2007) Bacterial pathogenomics. Nature 449 : 835–842.
49. KeenEC (2012) Paradigms of pathogenesis: targeting the mobile genetic elements of disease. Front Cell Infect Microbiol 2 : 161.
50. WoodfordN, TurtonJF, LivermoreDM (2011) Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35 : 736–755.
51. BlancoM, AlonsoMP, Nicolas-ChanoineMH, DahbiG, MoraA, et al. (2009) Molecular epidemiology of Escherichia coli producing extended-spectrum {beta}-lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 63 : 1135–1141.
52. CoelhoA, MoraA, MamaniR, LopezC, Gonzalez-LopezJJ, et al. (2011) Spread of Escherichia coli O25b:H4-B2-ST131 producing CTX-M-15 and SHV-12 with high virulence gene content in Barcelona (Spain). J Antimicrob Chemother 66 : 517–526.
53. Garcillan-BarciaMP, AlvaradoA, de la CruzF (2011) Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol Rev 35 : 936–956.
54. HauserM, MayerCE, SodingJ (2013) kClust: fast and sensitive clustering of large protein sequence databases. BMC Bioinformatics 14 : 248.
55. PeigneC, BidetP, Mahjoub-MessaiF, PlainvertC, BarbeV, et al. (2009) The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect Immun 77 : 2272–2284.
56. ZongZ (2013) Complete sequence of pJIE186-2, a plasmid carrying multiple virulence factors from a sequence type 131 Escherichia coli O25 strain. Antimicrob Agents Chemother 57 : 597–600.
57. FalgenhauerL, YaoY, FritzenwankerM, SchmiedelJ, ImirzaliogluC, et al. (2014) Complete Genome Sequence of Phage-Like Plasmid pECOH89, Encoding CTX-M-15. Genome Announc 2.
58. MiquelS, PeyretailladeE, ClaretL, de ValleeA, DossatC, et al. (2010) Complete genome sequence of Crohn's disease-associated adherent-invasive E. coli strain LF82. PLoS One 5.
59. KidgellC, PickardD, WainJ, JamesK, Diem NgaLT, et al. (2002) Characterisation and distribution of a cryptic Salmonella typhi plasmid pHCM2. Plasmid 47 : 159–171.
60. KimM, KimS, RyuS (2012) Complete genome sequence of bacteriophage SSU5 specific for Salmonella enterica serovar Typhimurium rough strains. J Virol 86 : 10894.
61. CottellJL, WebberMA, ColdhamNG, TaylorDL, Cerdeno-TarragaAM, et al. (2011) Complete sequence and molecular epidemiology of IncK epidemic plasmid encoding blaCTX-M-14. Emerg Infect Dis 17 : 645–652.
62. ValverdeA, CantonR, Garcillan-BarciaMP, NovaisA, GalanJC, et al. (2009) Spread of bla(CTX-M-14) is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob Agents Chemother 53 : 5204–5212.
63. KimSR, KomanoT (1992) Nucleotide sequence of the R721 shufflon. J Bacteriol 174 : 7053–7058.
64. MellataM, MadduxJT, NamT, ThomsonN, HauserH, et al. (2012) New insights into the bacterial fitness-associated mechanisms revealed by the characterization of large plasmids of an avian pathogenic E. coli. PLoS One 7: e29481.
65. WangP, XiongY, LanR, YeC, WangH, et al. (2011) pO157_Sal, a novel conjugative plasmid detected in outbreak isolates of Escherichia coli O157:H7. J Clin Microbiol 49 : 1594–1597.
66. TouchonM, HoedeC, TenaillonO, BarbeV, BaeriswylS, et al. (2009) Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5: e1000344.
67. BrownCJ, SenD, YanoH, BauerML, RogersLM, et al. (2013) Diverse broad-host-range plasmids from freshwater carry few accessory genes. Appl Environ Microbiol 79 : 7684–7695.
68. SmajsD, MicenkovaL, SmardaJ, VrbaM, SevcikovaA, et al. (2010) Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: colicin E1 is a potential virulence factor. BMC Microbiol 10 : 288.
69. BaqueroF, CoqueTM (2011) Multilevel population genetics in antibiotic resistance. FEMS Microbiol Rev 35 : 705–706.
70. RaskoDA, RosovitzMJ, MyersGS, MongodinEF, FrickeWF, et al. (2008) The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190 : 6881–6893.
71. HazenTH, SahlJW, FraserCM, DonnenbergMS, ScheutzF, et al. (2013) Refining the pathovar paradigm via phylogenomics of the attaching and effacing Escherichia coli. Proc Natl Acad Sci U S A 110 : 12810–12815.
72. SkippingtonE, RaganMA (2011) Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol Rev.
73. OguraY, OokaT, IguchiA, TohH, AsadulghaniM, et al. (2009) Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A 106 : 17939–17944.
74. CroucherNJ, HarrisSR, GradYH, HanageWP (2013) Bacterial genomes in epidemiology—present and future. Philos Trans R Soc Lond B Biol Sci 368 : 20120202.
75. JohnsonTJ, LogueCM, JohnsonJR, KuskowskiMA, SherwoodJS, et al. (2012) Associations Between Multidrug Resistance, Plasmid Content, and Virulence Potential Among Extraintestinal Pathogenic and Commensal Escherichia coli from Humans and Poultry. Foodborne Pathog Dis 9 : 37–46.
76. PolzMF, AlmEJ, HanageWP (2013) Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet 29 : 170–175.
77. ReevesPR, LiuB, ZhouZ, LiD, GuoD, et al. (2011) Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PLoS One 6: e26907.
78. SandegrenL, LinkeviciusM, LytsyB, MelhusA, AnderssonDI (2012) Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J Antimicrob Chemother 67 : 74–83.
79. ChenL, HuH, ChavdaKD, ZhaoS, LiuR, et al. (2014) Complete Sequence of a KPC-Producing IncN Multidrug-Resistant Plasmid from an Epidemic Escherichia coli Sequence Type 131 Strain in China. Antimicrob Agents Chemother 58 : 2422–2425.
80. PartridgeSR, ZongZ, IredellJR (2011) Recombination in IS26 and Tn2 in the evolution of multiresistance regions carrying blaCTX-M-15 on conjugative IncF plasmids from Escherichia coli. Antimicrob Agents Chemother 55 : 4971–4978.
81. O'HaraJA, HuF, AhnC, NelsonJ, RiveraJI, et al. (2014) Molecular Epidemiology of KPC-Producing Escherichia coli: Occurrence of ST131-fimH30 Subclone Harboring pKpQIL-like IncFIIk Plasmid. Antimicrob Agents Chemother.
82. JohnsonTJ, WannemuehlerYM, JohnsonSJ, LogueCM, WhiteDG, et al. (2007) Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol 73 : 1976–1983.
83. ChenL, ChavdaKD, Al LahamN, MelanoRG, JacobsMR, et al. (2013) Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob Agents Chemother 57 : 5019–5025.
84. JohnsonTJ, LangKS (2012) IncA/C plasmids: An emerging threat to human and animal health? Mob Genet Elements 2 : 55–58.
85. KimYA, QureshiZA, Adams-HaduchJM, ParkYS, ShuttKA, et al. (2012) Features of infections due to Klebsiella pneumoniae carbapenemase-producing Escherichia coli: emergence of sequence type 131. Clin Infect Dis 55 : 224–231.
86. MuniesaM, Colomer-LluchM, JofreJ (2013) Potential impact of environmental bacteriophages in spreading antibiotic resistance genes. Future Microbiol 8 : 739–751.
87. Hoyland-KroghsboNM, MaerkedahlRB, SvenningsenSL (2013) A quorum-sensing-induced bacteriophage defense mechanism. MBio 4: e00362–00312.
88. SchimkeRT (1984) Gene amplification in cultured animal cells. Cell 37 : 705–713.
89. BaqueroF, TedimAP, CoqueTM (2013) Antibiotic resistance shaping multi-level population biology of bacteria. Front Microbiol 4 : 15.
90. Andersen PS, Stegger M, Aziz M, Contente-Cuomo T, Gibbons HS, et al. (2013) Complete Genome Sequence of the Epidemic and Highly Virulent CTX-M-15-Producing H30-Rx Subclone of Escherichia coli ST131. Genome Announc 1.
91. TohH, OshimaK, ToyodaA, OguraY, OokaT, et al. (2010) Complete genome sequence of the wild-type commensal Escherichia coli strain SE15, belonging to phylogenetic group B2. J Bacteriol 192 : 1165–1166.
92. BoydDA, TylerS, ChristiansonS, McGeerA, MullerMP, et al. (2004) Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother 48 : 3758–3764.
93. WoodfordN, CarattoliA, KarisikE, UnderwoodA, EllingtonMJ, et al. (2009) Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother 53 : 4472–4482.
94. PartridgeSR, EllemJA, TetuSG, ZongZ, PaulsenIT, et al. (2011) Complete sequence of pJIE143, a pir-type plasmid carrying ISEcp1-blaCTX-M-15 from an Escherichia coli ST131 isolate. Antimicrob Agents Chemother 55 : 5933–5935.
95. LiW, GodzikA (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22 : 1658–1659.
96. DarlingAE, MauB, PernaNT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5: e11147.
97. Capella-GutierrezS, Silla-MartinezJM, GabaldonT (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 : 1972–1973.
98. StamatakisA (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 : 2688–2690.
99. ZerbinoDR, BirneyE (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18 : 821–829.
100. LangmeadB, SalzbergSL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9 : 357–359.
101. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–2079.
102. CamachoC, CoulourisG, AvagyanV, MaN, PapadopoulosJ, et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10 : 421.
103. Garcillan-BarciaMP, de la CruzF (2013) Ordering the bestiary of genetic elements transmissible by conjugation. Mob Genet Elements 3: e24263.
104. del SolarG, GiraldoR, Ruiz-EchevarriaMJ, EspinosaM, Diaz-OrejasR (1998) Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev 62 : 434–464.
105. ZhengJ, PengD, RuanL, SunM (2013) Evolution and dynamics of megaplasmids with genome sizes larger than 100 kb in the Bacillus cereus group. BMC Evol Biol 13 : 262.
106. OsbornAM, da Silva TatleyFM, SteynLM, PickupRW, SaundersJR (2000) Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology 146 (Pt 9): 2267–2275.
107. SmillieC, Garcillan-BarciaMP, FranciaMV, RochaEP, de la CruzF (2010) Mobility of plasmids. Microbiol Mol Biol Rev 74 : 434–452.
108. BesemerJ, LomsadzeA, BorodovskyM (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29 : 2607–2618.
109. GuglielminiJ, QuintaisL, Garcillan-BarciaMP, de la CruzF, RochaEP (2011) The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 7: e1002222.
110. EddySR (2011) Accelerated Profile HMM Searches. PLoS Comput Biol 7: e1002195.
111. SmootME, OnoK, RuscheinskiJ, WangPL, IdekerT (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27 : 431–432.
112. ZhouY, CallDR, BroschatSL (2013) Using protein clusters from whole proteomes to construct and augment a dendrogram. Adv Bioinformatics 2013 : 191586.
113. TekaiaF, LazcanoA, DujonB (1999) The genomic tree as revealed from whole proteome comparisons. Genome Res 9 : 550–557.
114. TekaiaF, YeramianE (2005) Genome trees from conservation profiles. PLoS Comput Biol 1: e75.
115. Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, et al. (2013) vegan: Community Ecology Package.
116. ParadisE, ClaudeJ, StrimmerK (2004) APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20 : 289–290.
117. AlikhanNF, PettyNK, Ben ZakourNL, BeatsonSA (2011) BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12 : 402.
118. AssefaS, KeaneTM, OttoTD, NewboldC, BerrimanM (2009) ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25 : 1968–1969.
119. BonninRA, PoirelL, CarattoliA, NordmannP (2012) Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS One 7: e34752.
120. CullikA, PfeiferY, PragerR, von BaumH, WitteW (2010) A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates. J Med Microbiol 59 : 580–587.
121. ChenL, ChavdaKD, MelanoRG, JacobsMR, KollB, et al. (2014) Comparative Genomic Analysis of KPC-Encoding pKpQIL-Like Plasmids and Their Distribution in New Jersey and New York Hospitals. Antimicrob Agents Chemother 58 : 2871–2877.
Štítky
Genetika Reprodukčná medicína
Článek Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart DiseaseČlánek Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the NicheČlánek Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial TetheringČlánek Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature SeasonalityČlánek Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell DivisionsČlánek Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity inČlánek Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration inČlánek ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression inČlánek Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex EnvironmentsČlánek The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 12- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- Stratification by Smoking Status Reveals an Association of Genotype with Body Mass Index in Never Smokers
- Genome Wide Meta-analysis Highlights the Role of Genetic Variation in in the Regulation of Circulating Serum Chemerin
- Occupancy of Mitochondrial Single-Stranded DNA Binding Protein Supports the Strand Displacement Mode of DNA Replication
- Distinct Genealogies for Plasmids and Chromosome
- Large-scale Metabolomic Profiling Identifies Novel Biomarkers for Incident Coronary Heart Disease
- Non-coding RNAs Prevent the Binding of the MSL-complex to Heterochromatic Regions
- Plasmid Flux in ST131 Sublineages, Analyzed by Plasmid Constellation Network (PLACNET), a New Method for Plasmid Reconstruction from Whole Genome Sequences
- Epigenome-Guided Analysis of the Transcriptome of Plaque Macrophages during Atherosclerosis Regression Reveals Activation of the Wnt Signaling Pathway
- The Inventiveness of Nature: An Interview with Werner Arber
- Mediation Analysis Demonstrates That -eQTLs Are Often Explained by -Mediation: A Genome-Wide Analysis among 1,800 South Asians
- Generation of Antigenic Diversity in by Structured Rearrangement of Genes During Mitosis
- A Massively Parallel Pipeline to Clone DNA Variants and Examine Molecular Phenotypes of Human Disease Mutations
- Genetic Analysis of the Cardiac Methylome at Single Nucleotide Resolution in a Model of Human Cardiovascular Disease
- Genetic Analysis of Circadian Responses to Low Frequency Electromagnetic Fields in
- The Dissection of Meiotic Chromosome Movement in Mice Using an Electroporation Technique
- Altered Chromatin Occupancy of Master Regulators Underlies Evolutionary Divergence in the Transcriptional Landscape of Erythroid Differentiation
- Syd/JIP3 and JNK Signaling Are Required for Myonuclear Positioning and Muscle Function
- Notch Signaling Mediates the Age-Associated Decrease in Adhesion of Germline Stem Cells to the Niche
- Mutation of Leads to Blurred Tonotopic Organization of Central Auditory Circuits in Mice
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- RAN-Binding Protein 9 is Involved in Alternative Splicing and is Critical for Male Germ Cell Development and Male Fertility
- Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import
- Phosphorylation of Mitochondrial Polyubiquitin by PINK1 Promotes Parkin Mitochondrial Tethering
- Recurrent Loss of Specific Introns during Angiosperm Evolution
- Natural Variation Is Associated With Genome-Wide Methylation Changes and Temperature Seasonality
- SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat
- Overlapping and Non-overlapping Functions of Condensins I and II in Neural Stem Cell Divisions
- Unisexual Reproduction Drives Meiotic Recombination and Phenotypic and Karyotypic Plasticity in
- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- ABA-Mediated ROS in Mitochondria Regulate Root Meristem Activity by Controlling Expression in
- Mutations in Global Regulators Lead to Metabolic Selection during Adaptation to Complex Environments
- Global Analysis of Photosynthesis Transcriptional Regulatory Networks
- Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning
- Controlling Pre-leukemic Thymocyte Self-Renewal
- How Malaria Parasites Avoid Running Out of Ammo
- Echoes of the Past: Hereditarianism and
- Deep Reads: Strands in the History of Molecular Genetics
- Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away
- Analysis of a Plant Complex Resistance Gene Locus Underlying Immune-Related Hybrid Incompatibility and Its Occurrence in Nature
- Epistatic Adaptive Evolution of Human Color Vision
- Increased and Imbalanced dNTP Pools Symmetrically Promote Both Leading and Lagging Strand Replication Infidelity
- Genetic Basis of Haloperidol Resistance in Is Complex and Dose Dependent
- Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development
- Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer
- The Evolution of Sex Ratio Distorter Suppression Affects a 25 cM Genomic Region in the Butterfly
- is Required for Adult Maintenance of Dopaminergic Neurons in the Ventral Substantia Nigra
- PRL1, an RNA-Binding Protein, Positively Regulates the Accumulation of miRNAs and siRNAs in Arabidopsis
- Genetic Control of Contagious Asexuality in the Pea Aphid
- Early Mesozoic Coexistence of Amniotes and Hepadnaviridae
- Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-Like Kinase
- Gene Pathways That Delay Reproductive Senescence
- The Evolution of Fungal Metabolic Pathways
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- Formation of Linear Amplicons with Inverted Duplications in Requires the MRE11 Nuclease
- Identification of Rare Causal Variants in Sequence-Based Studies: Methods and Applications to , a Gene Involved in Cohen Syndrome and Autism
- Rrp12 and the Exportin Crm1 Participate in Late Assembly Events in the Nucleolus during 40S Ribosomal Subunit Biogenesis
- The Mutations in the ATP-Binding Groove of the Rad3/XPD Helicase Lead to -Cockayne Syndrome-Like Phenotypes
- Topoisomerase I Plays a Critical Role in Suppressing Genome Instability at a Highly Transcribed G-Quadruplex-Forming Sequence
- A Cbx8-Containing Polycomb Complex Facilitates the Transition to Gene Activation during ES Cell Differentiation
- Transcriptional Frameshifting Rescues Type VI Secretion by the Production of Two Length Variants from the Prematurely Interrupted Gene
- Association Mapping across Numerous Traits Reveals Patterns of Functional Variation in Maize
- Genome-Wide Analysis of -Regulated and Phased Small RNAs Underscores the Importance of the ta-siRNA Pathway to Maize Development
- Dissemination of Cephalosporin Resistance Genes between Strains from Farm Animals and Humans by Specific Plasmid Lineages
- The Tau Tubulin Kinases TTBK1/2 Promote Accumulation of Pathological TDP-43
- Germline Signals Deploy NHR-49 to Modulate Fatty-Acid β-Oxidation and Desaturation in Somatic Tissues of
- Microevolution of in Macrophages Restores Filamentation in a Nonfilamentous Mutant
- Vangl2-Regulated Polarisation of Second Heart Field-Derived Cells Is Required for Outflow Tract Lengthening during Cardiac Development
- Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice
- A ABC Transporter Regulates Lifespan
- RA and FGF Signalling Are Required in the Zebrafish Otic Vesicle to Pattern and Maintain Ventral Otic Identities
- , and Reprogram Thymocytes into Self-Renewing Cells
- The miR9863 Family Regulates Distinct Alleles in Barley to Attenuate NLR Receptor-Triggered Disease Resistance and Cell-Death Signaling
- Detection of Pleiotropy through a Phenome-Wide Association Study (PheWAS) of Epidemiologic Data as Part of the Environmental Architecture for Genes Linked to Environment (EAGLE) Study
- Extensive Copy-Number Variation of Young Genes across Stickleback Populations
- The and Genetic Modules Interact to Regulate Ciliogenesis and Ciliary Microtubule Patterning in
- Analysis of the Genome, Transcriptome and Secretome Provides Insight into Its Pioneer Colonization Strategies of Wood
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Tetraspanin (TSP-17) Protects Dopaminergic Neurons against 6-OHDA-Induced Neurodegeneration in
- Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism
- The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
- Echoes of the Past: Hereditarianism and
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy




