-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
Separases are a family of cysteine proteases found in organisms ranging from yeast to humans that are required for separation of chromosomes during cell division. Separases dissolve the cohesin ring-like complex that holds sister chromatids together until chromosome separation occurs during mitosis. We used two genetic screens in the model organism budding yeast to identify additional cellular roles for separase. Surprisingly, we found that yeast separase is required for insertion of Ty1 retrotransposons into the yeast genome. Ty1 retrotransposons, or elements, are similar in their life cycle to retroviruses such as the human immunodeficiency virus type 1 (HIV-1) which is the cause of acquired immunodeficiency syndrome (AIDS). The insertion of retroviral/retrotransposon DNA into the genome requires a conserved protein encoded by the virus/retrotransposon called integrase. Until now, it was unknown which yeast host protein interacted with Ty1 integrase. We found that yeast separase interacts with Ty1 integrase, and that separase may be required for targeting Ty1 integrase into the genome via its interaction with integrase and by removing cohesin from the chromosomes. Due to the conservation of Ty1 integrase with other viral integrases, our discovery may shed light on how other viral integrases are targeted into the genome.
Published in the journal: A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition. PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005109
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005109Summary
Separases are a family of cysteine proteases found in organisms ranging from yeast to humans that are required for separation of chromosomes during cell division. Separases dissolve the cohesin ring-like complex that holds sister chromatids together until chromosome separation occurs during mitosis. We used two genetic screens in the model organism budding yeast to identify additional cellular roles for separase. Surprisingly, we found that yeast separase is required for insertion of Ty1 retrotransposons into the yeast genome. Ty1 retrotransposons, or elements, are similar in their life cycle to retroviruses such as the human immunodeficiency virus type 1 (HIV-1) which is the cause of acquired immunodeficiency syndrome (AIDS). The insertion of retroviral/retrotransposon DNA into the genome requires a conserved protein encoded by the virus/retrotransposon called integrase. Until now, it was unknown which yeast host protein interacted with Ty1 integrase. We found that yeast separase interacts with Ty1 integrase, and that separase may be required for targeting Ty1 integrase into the genome via its interaction with integrase and by removing cohesin from the chromosomes. Due to the conservation of Ty1 integrase with other viral integrases, our discovery may shed light on how other viral integrases are targeted into the genome.
Introduction
Successful mitotic cell division is predicated upon the faithful propagation of genetic material. A critical element to the regulation of this process is the use of cohesin, a proteinaceous ring-like structure, to pair sister chromatids [1]. This coupling ensures that chromosomal separation does not occur prior to successful bipolar attachment, thereby preventing chromosomal misseggregation. Sister chromatid separation is triggered by an evolutionarily conserved cysteine protease known as separase that cleaves cohesin—an event that is controlled by inhibiting the catalytic activity of these aptly named proteases by securin [2].
In Saccharomyces cerevisiae, the core cohesin complex consists of two sister chromatid cohesin (Scc1/Mcd1 and Scc3) and two structural maintenance of chromosome (Smc1 and Smc3) proteins [1]. Smc1 and Smc3 have a dimerization domain located in the middle of a coiled-coil region that folds back on itself and ends in two globular ATPase domains. The ATPase heads of Smc1 and Smc3 interact with Scc1 to form a ring-like complex whereas Scc3 interacts with Scc1. The cohesin ring loads onto chromosomes at sites of RNA polymerase III (RNA Pol III) transcription, dependent on the Scc2/4 cohesin loading complex, then moves to sites of RNA Pol II convergent transcription [3,4]. Securin, or Pds1, is targeted for proteolytic degradation at the onset of anaphase by the anaphase promoting complex, an E3 ubiquitin ligase that functions in conjunction with the co-activator Cdc20 [5,6]. Pds1 degradation frees the budding yeast separase, Esp1, to specifically target phosphorylated, chromatin-bound Scc1 for cleavage [7–9]. Cohesin can also be removed from chromosomes by a second separase-independent mechanism that occurs throughout the cell cycle. This so-called “anti-establishment” activity requires Scc3 and two accessory proteins that interact with the cohesin complex called Rad61 and Pds5 [10,11].
In higher eukaryotes, separases are implicated in roles in addition to cohesin cleavage such as centrosome duplication in humans [12], cell polarity in Arabidopsis thaliana [13] and epithelial re-organization in Drosophila melanogaster [14]. Similarly, Esp1 has demonstrable functions aside from cleaving Scc1, with the most extensively studied role focusing on Slk19, a kinesin-associated protein that is also cleaved by Esp1. By cleaving Slk19 during anaphase spindle elongation, Esp1 is thought to stabilize the anaphase spindle [15]. As well, both Esp1 and Slk19 serve to promote mitotic exit. Key to the initiation of this event is the biphasic release of the Cdc14 phosphatase from its sequestration in the nucleolus [16]. The initial wave of release is known as the Cdc14 early anaphase release (FEAR) pathway, with a second more prolonged release referred to as the mitotic exit network (MEN) [16,17]. Esp1 and Slk19 both function in the FEAR pathway, a role not dependent on the catalytic activity of Esp1 [18,19].
To gain insight into the cellular functions of Esp1, we undertook two genome-wide screens for Esp1 genetic interactions. Using synthetic genetic array (SGA) methodology, we investigated both synthetic lethal (SL) and synthetic dosage lethal (SDL) relationships of the esp1-1 temperature sensitive mutant [20]. Analysis of esp1-1 genetic interactions revealed that genes involved in the regulation of Ty1 transposition are enriched in the screens.
Ty retrotransposons are transposable elements that share similarities to retroviruses, though they lack an infectious phase in their life cycle. There are five families of Ty elements in S. cerevisiae (Ty1-5), the most abundant of which is Ty1 with 32 full size elements in the S288C genome. Each Ty element is flanked by long terminal repeats and is comprised of both TyA and TyB open reading frames (ORFs), analogous to retroviral gag and pol respectively. TyA encodes the structural elements of Gag (coat protein) while TyB is a polyprotein that is cleaved to generate a protease (PR), integrase (IN) and reverse transcriptase (RT) [21,22]. Ty1 elements insert upstream of genes transcribed by RNA pol III, such as tRNAs genes, but the host factors required for targeting Ty1 elements to the genome are still unclear. However there is precedent for Ty-IN specific host factors because Ty5-IN interacts with Sir4 to mediate insertion into silenced regions of the genome whereas Ty3-IN is targeted by the RNA Pol III TFIIIB factors Brf1 and TBP [23,24]. We have discovered that Esp1 specifically interacts with Ty1-IN and that esp1 and pds1 mutants have defects in Ty1 transposition whereas cohesin mutants have increased transposition events. We propose that both removal of cohesin by Esp1 and the physical interaction of Esp1 with Ty1-IN promote efficient Ty1 element insertion into the genome.
Results
Identification of esp1-1 genetic interactions
To uncover potential new roles for Esp1 using genome-wide genetic interaction studies, we subjected the ESP1 temperature sensitive mutant, esp1-1, to SGA analysis [25]. We first sequenced the entire 4893 base pair (bp) ORF of esp1-1 to determine the location of the relevant mutation(s). Fortuitously, we identified a single point mutation at bp 4211 (C→T), resulting in conversion of proline 1404 to leucine. This mutation is in the vicinity of the highly conserved histidine 1505 and cysteine 1531 residues of the Esp1 catalytic site [9]. The esp1-1 mutant has significantly reduced Scc1 cleavage activity, although some residual activity remains [8,9,26]. The placement of the P1404L mutation also allowed us to PCR the C-terminus of esp1-1 (including bp 4211) and use that fragment to integrate the mutation into the SGA starting strain as described [27].
The esp1-1 query strain was mated to ∼4700 nonessential gene deletions (geneXΔ) [28] and ∼5300 strains from the pGAL1/10-GST-ORFX overexpression collection [29] to identify SL and SDL interactions, respectively. SL interactions were analyzed using a quantitative scoring program, with colony size functioning as a measure of growth in order to compare fitness of double mutants with esp1-1 to the single deletion mutant [30]. We identified 161 double mutant strains that displayed a growth ratio below the determined cut-off for the screen (< 0.8 relative to the single deletion mutant control, p < 0.05) in 3/3 biological replicates, and consider these to be representative of SL interactions (S1 Table). Included in this list are two esp1-1 SL interactors—hsc82Δ and lte1Δ - that have been previously described [18,31]. SDL genetic interactions were similarly examined by inducing pGAL1/10-GST-ORFX expression in both wild type and esp1-1 mutants and comparing colony size. Plasmids were extracted from colonies identified in the screen, sequenced to confirm identity and retransformed into wild type and esp1-1 mutant strains. Serial dilution assays were performed on dextrose versus galactose plates to confirm which genes caused a slow growth or lethal phenotype upon overexpression (S1 Fig). Ultimately, 44 different proteins were found to negatively impact the growth of the esp1-1 mutant when overexpressed (S1 Fig, S2 Table). SLK19, a known proteolytic substrate of Esp1 that regulates spindle stability, was among the genes identified in the SDL screen, supporting the notion that increased dosage of a substrate in the presence of its defective enzyme can be detrimental to cells [15,29,32] (S1 Fig, S2 Table). There were only 2 genes identified in both the esp1-1 SL and SDL screens, which is consistent with previous comparisons of SL and SDL screens [32–34]. Minimal overlap between the two screening methods is likely because SL and SDL screens probe different genetic interactions.
Examination of confirmed negative esp1-1 genetic interactions
Genes identified in the esp1-1 SL screen were classified according to Gene Ontology (GO) Biological Process using Cytoscape to identify GO Terms that are enriched above genome frequency (p<0.05, S2 Fig) [35]. We also used the ClueGO plugin [36] to perform GO Term Grouping to distinguish which of these GO Terms correlated to a GO category that could be classified as significantly enriched (p<0.05, Fig. 1). As expected, Sister Chromatid Cohesion was among the enriched GO Term Groups. We also identified GO Terms that grouped into the category of DNA Repair which is consistent with a recently described role for Esp1 in promoting dissociation of cohesin during DNA double-strand break repair [37]. Notably, however, was the enrichment of Transposition and Regulation of Stress Response, two GO Term Groups that have no prior association to Esp1. We also examined the data from the esp1-1 SDL screen in a similar manner and identified spindle orientation as an enriched GO Term Group, which is in agreement with the known role for Esp1 in spindle stability (Fig. 2, S2 Fig) [15,19,38]. In addition, we also identified mRNA capping as another GO Term Group that has no previous relation to Esp1 (Fig. 2, S2 Fig).
Fig. 1. esp1-1 functional interaction map derived from the SL SGA screen. 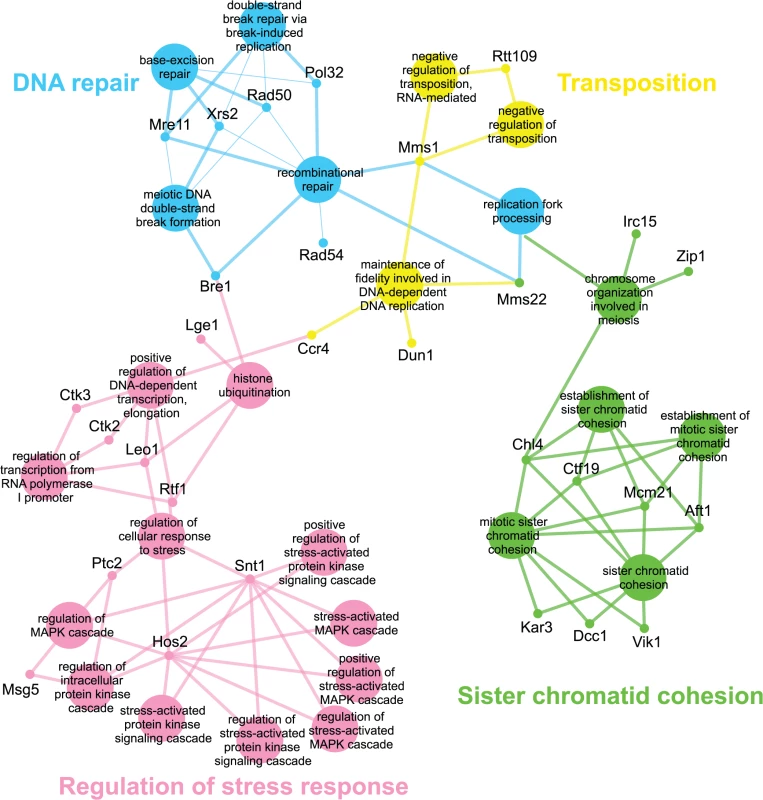
The 161 genes identified in the esp1-1 SL screen were analyzed using Cytoscape. All nodes represent significantly enriched (p <0.05) GO Terms in the dataset. Coloured nodes represent GO Terms that have been grouped into a category (written in the same colour) that is significantly enriched. Edges define associations between groups and edge thickness indicates the level of significance within the network. Genes identified in the SL screen that are associated with GO Terms are shown. Fig. 2. esp1-1 functional interaction map derived from the SDL screen. 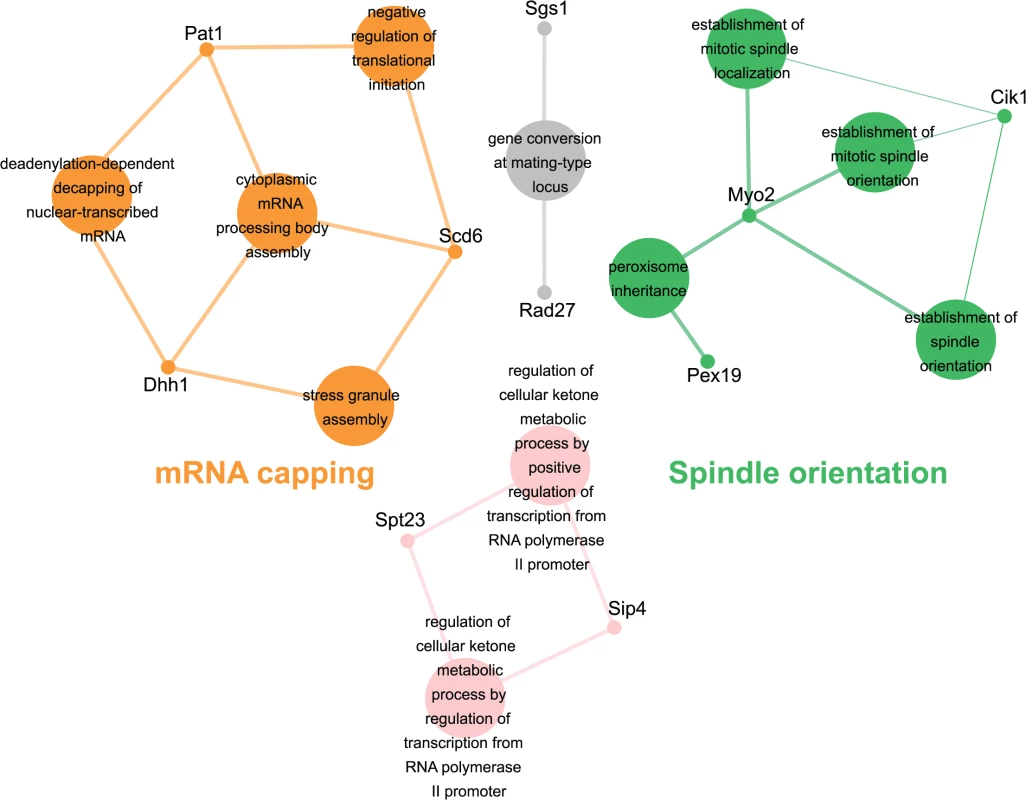
The 44 genes identified in the esp1-1 SDL screen were subjected to Cytoscape analysis. All nodes are significantly enriched (p <0.05) GO Terms in the dataset and coloured nodes represent GO Terms that have been grouped into a significantly enriched category. Grey nodes are GO Terms that were not grouped into an enriched category. Edges define associations between groups and edge thickness indicates the level of significance within the network. Genes identified in the SDL screen that are associated with GO Terms are shown. esp1-1 genetic interactors are involved in the life cycle of Ty1 retrotransposons
The retrotransposition cycle of Ty1 elements requires the following steps: (1) transcription of the TY1 element by RNA polymerase II (2) nuclear export of the TY1 mRNA (3) translation of TY1 mRNA (4) assembly and maturation of Gag and Gag-Pol precursor proteins by Ty1-protease (5) reverse transcription of the TY1 mRNA to cDNA (6) import to the nucleus and (7) integration of the cDNA into the genome [21]. We compared the 203 genes identified in our esp1-1 SL and SDL screens to host S. cerevisiae genes required for individual steps of the Ty1 and Ty3 life cycle [39–43]. This analysis revealed that 48 (24%) of the esp1-1 genetic interactors either promote or impede the Ty1 or Ty3 life cycle (Table 1). For example, genome-wide studies have implicated the P-body proteins Pat1, Dhh1 and Lsm1-7 in Ty1 and Ty3 retrotransposition as enhancers of the formation of retrotransposition-competent Ty virus like particles [40,41,44,45]. Pat1 and Dhh1 were identified in the esp1-1 SDL screen whereas lsm1Δ and lsm7Δ were identified in the esp1-1 SL screen (S1 and S2 Tables). The RecQ helicase Sgs1, which is synthetic dosage sensitive when overexpressed in esp1-1 cells, is likely involved in the suppression of Ty1 cDNA recombination, as strains lacking SGS1 transpose heterogeneous Ty1 multimers (S2 Table) [42,46]. The Fen-1 nuclease Rad27, which is also SDS when overexpressed in esp1-1 cells, is thought to degrade Ty1 cDNA to prevent the formation of Ty1 multimers (S2 Table) [42,47]. Further, members of the Paf1 complex serve to inhibit Ty1 transposition, possibly post-transcriptionally or by limiting the amount of favourable hotspots for integration [42]. Deletion mutants of three members of the Paf1 complex—cdc73Δ, leo1Δ and rtf1Δ - caused an SL phenotype when combined with esp1-1 (S1 Table). Two esp1-1 SDL genes—SPT10 and SPT23—are named because of their identification in “suppressors of Ty” screens (S2 Table) [48,49]. Either mutation (SPT10) or multicopy expression (SPT23) of these genes confers suppression of Ty1-induced promoter insertion mutations [48,49]. Clearly, several genetic interactors of esp1-1 have important roles in promoting or restricting Ty1 transposition.
Tab. 1. Genes identified in esp1-1 SL and SDL screens implicated in Ty1 and Ty3 transposition. 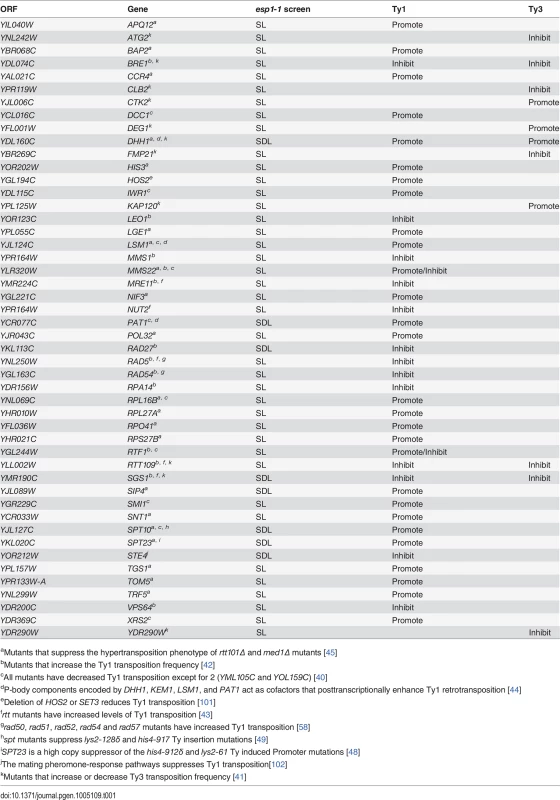
aMutants that suppress the hypertransposition phenotype of rtt101Δ and med1Δ mutants [45] Mass spectrometry reveals a physical interaction for Esp1 with Ty1-IN
To corroborate our genetic interaction data, and better understand its functional significance, a search for Esp1 physical interacting proteins was also undertaken. We performed an immunoprecipitation (IP) of 13Myc-tagged Esp1 from logarithmically growing cells, followed by mass spectrometry analysis that we compared to a mock IP performed in parallel with an untagged strain. More specifically, we compared the total number of peptides for each protein identified by mass spectrometry (i.e., spectral counting), and we identified 152 proteins that co-purified with Esp1 but were not enriched in the mock experiment (S3 Table). The identification of the separase inhibitor, Pds1/securin—with 16 peptides identified in the Esp1 IP and none in the mock—served as an internal experimental control. The Esp1 co-purified proteins included 11 proteins implicated in Ty1 transposition such as the “suppressors of Ty” Spt6 and Spt16 (Table 2) [50–52]. Notably, we also identified 13 peptides corresponding to Ty1-IN while none were identified in the mock IP and observed a six-fold increase of spectral counts for Ty1 peptides corresponding to the Ty1-RT (Fig. 3A). In comparison, peptides from the Ty1-Gag and Ty1-PR were readily identified in both the mock and Esp1 IPs, suggesting these Ty1 proteins may not specifically interact with Esp1. A potential interaction between Esp1 and Ty1-IN suggests that Esp1 may have a direct role in Ty1 element transposition.
Fig. 3. Esp1 physically interacts with Ty1-IN. 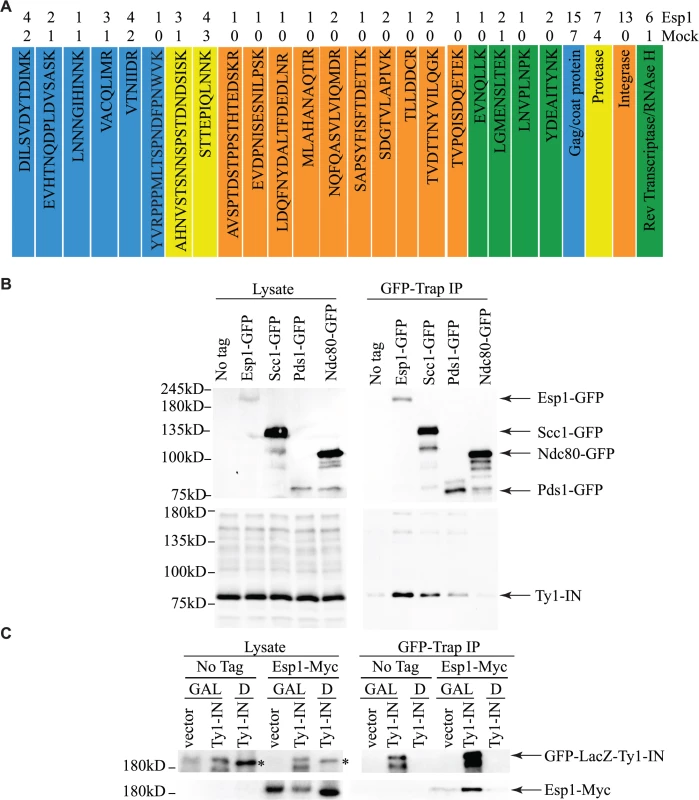
(A) Ty1 peptides identified in Esp1-Myc mass spectrometry versus untagged (mock) strain are color coded as follows: Gag/coat protein (blue), PR (yellow), IN (orange) and RT/RNAse H (green). (B) Immunoblot of whole cell lysate (Lysate) and GFP-Trap IP carried out from untagged wild type (No Tag), Esp1-GFP, Scc1-GFP, Pds1-GFP and Ndc80-GFP cells. Expression of a Ty1 element (pGAL1-Ty1-H3) was induced in all strains for 24 hours prior to cell lysis. Blots were probed with anti-GFP and anti-IN (8b11) antibodies. (C) Immunoblot of whole cell lysate (Lysate) and GFP-Trap IP carried out from Esp1-Myc or untagged wild type (No Tag) cells carrying a pGAL-GFP-LacZ-Ty1-IN plasmid (Ty1-IN) or pGAL-GFP-lacZ (vector) control. Cells were either grown in glucose (D) or galactose (GAL) for 24 hours to repress or induce GFP-LacZ-Ty1-IN expression, respectively. The asterisk marks a background band that is present in the lysate of the cells grown in glucose but is not detected in the GFP-Trap IP. Tab. 2. Genes identified in Esp1-13Myc mass spectrometry screen implicated in Ty1 transposition. 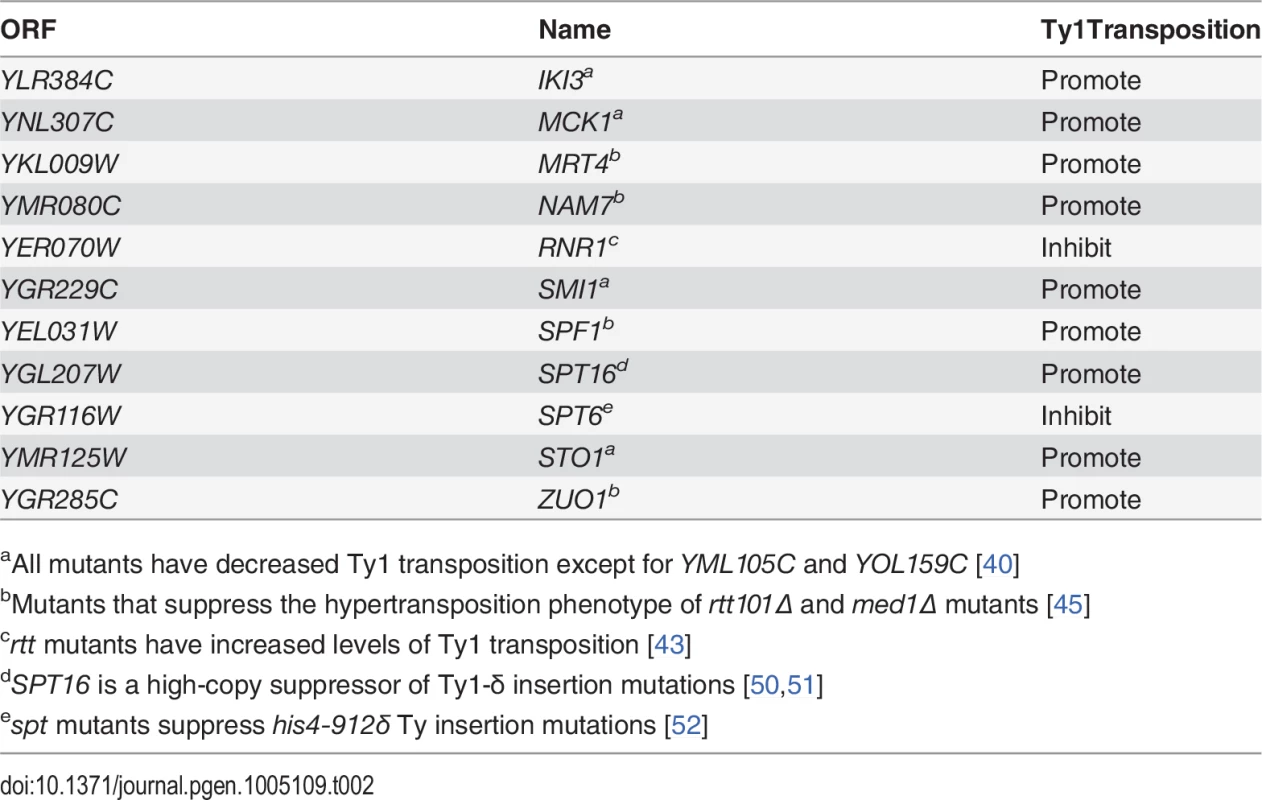
aAll mutants have decreased Ty1 transposition except for YML105C and YOL159C [40] We validated the physical interaction of Esp1 with Ty1 proteins by co-IP experiments using a different Esp1 epitope tag (GFP) to demonstrate specificity of the interaction. Although the Ty1-Gag protein can be detected at endogenous levels, the TyB polypeptides cannot using available reagents and must be overexpressed [53]. Therefore, pGAL1Ty1-H3, a Ty1 expression plasmid, was induced in both an untagged control strain and Esp1-GFP strain. Esp1-GFP was purified using GFP-Trap beads, immunoblot analysis performed and probed for the presence of Ty1-IN. We found that Ty1-IN was specifically present in the Esp1-GFP IP but not in the purification from the untagged strain (Fig. 3B). We could only detect processed Ty1-IN in the lower molecular weight region of the co-IP and not in the region where unprocessed PR-IN or IN-RT polypeptides would migrate (Fig. 3B). Similarly, we did not detect any peptides in our MS data that spanned the unprocessed PR-IN or IN-RT polypeptides. However, we cannot completely exclude the possibility that Esp1 also binds, to a lower extent, to the unprocessed or partially cleaved TyB polypeptide.
Esp1 interacts with Pds1/securin immediately after PDS1 expression in S phase and remains bound until Pds1 is targeted for degradation by the Anaphase Promoting Complex, whereby Esp1 is released to cleave the Scc1 cohesin [8,9,38]. We were therefore interested to test whether Pds1 and Scc1 could also physically interact with Ty1-IN. GFP-tagged Pds1 and Scc1 were purified individually from yeast lysates and immunoblot analysis performed. In comparison to Esp1, the interaction of Pds1-GFP with Ty1-IN was much weaker and just above background levels (Fig. 3B). We found that Scc1-GFP interacted with Ty1-IN. However, in comparison to Esp1-GFP, less Ty1-IN was detected in the co-IP despite higher expression of Scc1-GFP (Fig. 3B). Therefore, Ty1-IN interacts predominantly with Esp1 and to a lesser extent, Scc1. Ty1-IN does not interact with the Ndc80-GFP kinetochore protein demonstrating that GFP fusions in general do not interact with Ty1-IN (Fig. 3B). To further validate the Esp1-Ty1-IN interaction, we purified an overexpressed version of Ty1-IN fused to GFP-lacZ (pGAL-GFP-LacZ-Ty1-IN) using GFP Trap beads and tested if GFP-LacZ-Ty1-IN interacted with 13Myc tagged Esp1. Esp1-Myc was present in the purification of GFP-LacZ-Ty1-IN but not in a purification where GFP-LacZ-Ty1-IN was not expressed [pGFP-lacZ-Ty1-IN Dextrose (D)] or in a purification with vector control (Fig. 3C). In summary, we have demonstrated using mass spectrometry and two independent co-IP experiments that Esp1 interacts with Ty1-IN.
Esp1 is expressed throughout the cell cycle with about three-fold lower levels in G1 phase than the rest of the cell cycle [38]. We queried if Esp1 interacts with Ty1-IN at a particular stage of the cell cycle. After induction of Ty1 expression in Esp1-GFP cells for 24 hours, we arrested cells in G1 phase with mating pheromone (α-factor), S phase with hydroxyurea (HU) and G2 phase with nocodazole (Nz) (Fig. 4). Esp1-GFP was purified with GFP-Trap beads and immunoblot analysis performed to test for interaction with Ty1-IN. We detected an interaction between Esp1-GFP and Ty1-IN in logarithmically growing, α-factor and Nz arrested cells, but a minimal interaction was detected in HU arrested cells (Fig. 4).
Fig. 4. Esp1 interacts with Ty1-IN in G1 and G2/M phase cells. 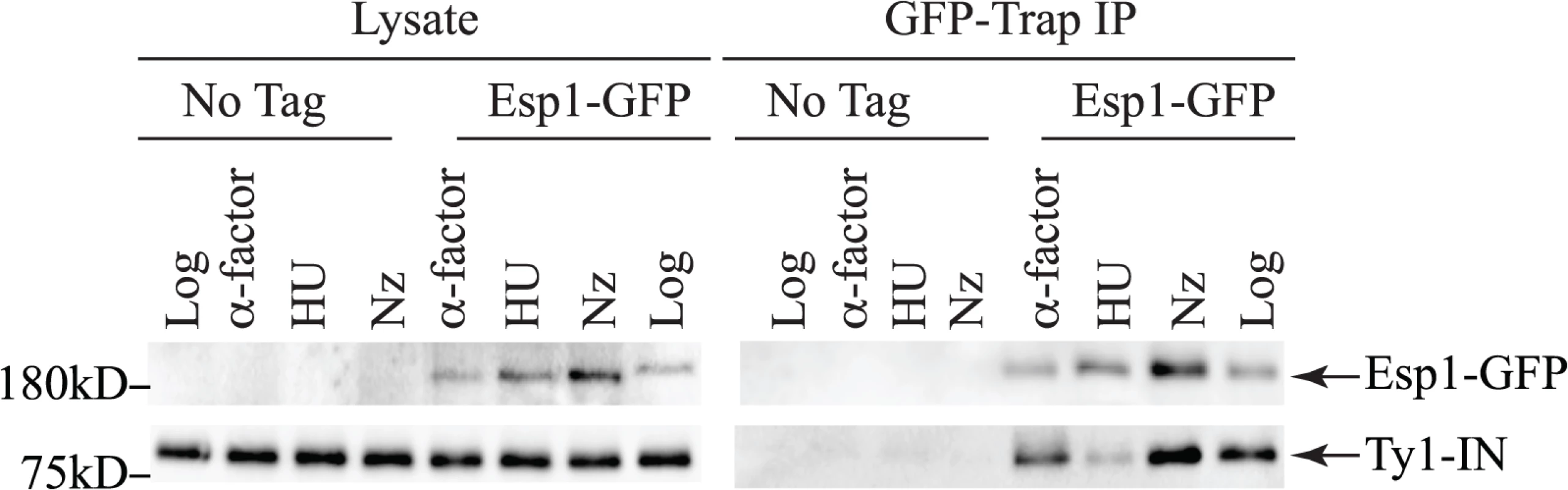
Untagged (No Tag) and Esp1-GFP tagged cells carrying the pGAL1-Ty1-H3 element were induced for 24 hours with 2% galactose, then arrested in G1 phase with mating pheromone (α-factor), S phase with hydroxyurea (HU) and G2/M phase with Nocodazole (Nz). Immunoblot of whole cell lysate (Lysate) and GFP-Trap IP from Log, α-factor, HU and Nz arrested cells is shown. Blots were probed with anti-GFP and anti-IN (8b11) antibodies. Pds1 is expressed in G1/S phase and degraded at the metaphase to anaphase transition [5,54]. To determine if Pds1 is required for Esp1 to interact with Ty1-IN, we used a strain in which Pds1 expression is controlled by a tetracycline (TetO7)-regulatable promoter that is inhibited in the presence of doxycycline (Dox) [55]. Dox addition to TetO7-PDS1 cells inhibited PDS1 expression after 15min of treatment and PDS1 transcript levels were reduced for up to 240min (Fig. 5A). Since Pds1 is degraded every cell cycle, we treated TetO7-PDS1 cells for one hour with Dox to ensure that all Pds1 protein was removed from the cell [5]. Esp1-GFP was purified from Dox treated (+) and untreated (-) TetO7-PDS1 cells and was able to interact with Ty1-IN in the presence or absence of Pds1 (Fig. 5B). We performed a similar experiment with a TetO7-SMC1 strain to determine if cohesin is required for the Esp1-Ty1-IN interaction. We first determined that treatment of the TetO7-SMC1 strain with Dox for 6 hours was sufficient to reduce Smc1 to background levels, as determined by immunoblot with an anti-Smc1 antibody (Fig. 5C, Smc1 panel). We found that, similar to Pds1, when Smc1 was depleted by Dox treatment, Esp1-GFP was still able to interact with Ty1-IN (Fig. 5C). Our results strongly indicate that the interaction of Esp1 with Ty1-IN does not depend on Pds1 or Smc1.
Fig. 5. Pds1 and Smc1 are not required for the Esp1-Ty1-IN interaction. 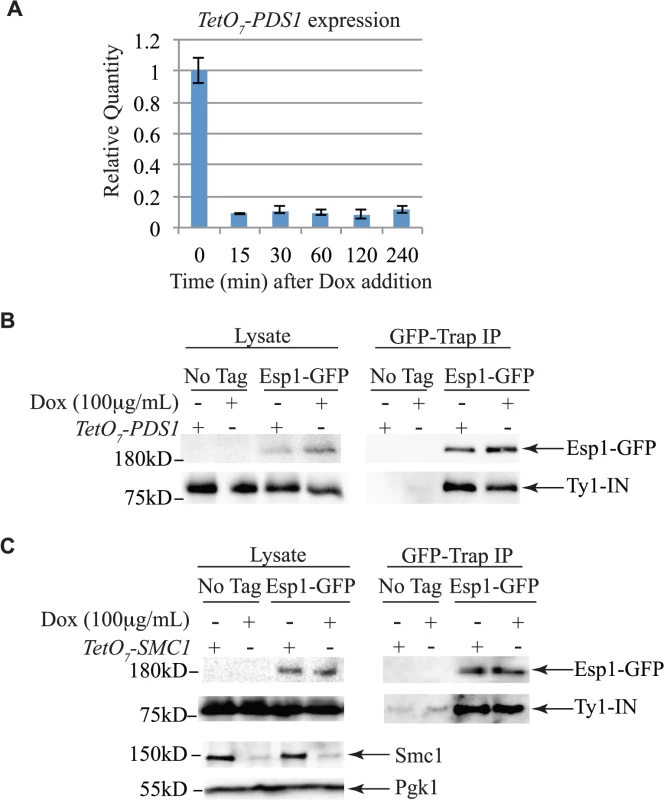
(A) Quantitative PCR analysis of PDS1 expression driven from the TetO7 promoter after 100μg/mL doxycycline addition. The relative quantity of PDS1 transcript compared to a control (TAF10) is shown. (B) Esp1-GFP and untagged (No Tag) cells containing the TetO7-PDS1 allele and pGAL1-Ty1-H3 were induced for 24 hours with 2% galactose, then 100μg/mL doxycycline was added (+) or not (-) for one hour. Esp1-GFP was purified with GFP-Trap beads, immunoblot analysis performed and probed with anti-GFP and anti-IN (8b11) antibodies. (C) Esp1-GFP and untagged (No Tag) cells containing the TetO7—SMC1 allele and pGAL1-Ty1-H3 were induced for 24 hours with 2% galactose, then 100μg/mL doxycycline was added (+) or not (-) for six hours. Esp1-GFP was purified as in (B). The depletion of Smc1 was monitored with anti-Smc1 antibodies and the lysate was probed with anti-Pgk1 as a loading control. esp1 and pds1 mutants have defects in Ty1 transposition whereas cohesin mutants have increased Ty1 transposition
The interaction of Esp1 with Ty1-IN suggests that Esp1 may be required for Ty1 transposition. We performed a quantitative assay for Ty1 mobility with a plasmid (pBDG922) carrying a Ty1 element expressed from its endogenous promoter. The Ty1 element contains a HIS3 gene with an artificial intron (AI) interrupting its expression. After Ty1 expression, the intron is removed from the Ty1-H3mHIS3AI mRNA intermediate, and successful Ty1 cDNA integration renders the strain HIS3+[56]. Wild type, esp1-1 and other mutant strains carrying the pBDG922 plasmid were induced to transpose at 20°C for 4 days before plating onto selective media to measure both viability and Ty1 transposition. Spt3 is required for transcription of Ty1 elements, therefore the spt3 deletion mutant is included as a negative control [57]. Wild type cells underwent Ty1 transposition at a frequency of 20.8 x 10–6 compared to 9.2 X 10–6 for esp1-1 cells (∼2-fold less than wild type, p < 0.05, Fig. 6A). We also found that pds1-128 cells had significantly reduced Ty1 transposition frequency at 3.5 x10-6 (∼6-fold less than wild type, p<0.005). We tested a panel of cohesin mutants including the Scc1 cohesin that is cleaved by Esp1 (Fig. 6C). There was a 5-fold increase in the Ty1 transposition frequency in the scc-173 mutant (Fig. 6A, p<0.0005), which suggested that the presence of cohesin might impede Ty1 transposition. Indeed, strains carrying mutations in the Scc3 and Smc3 cohesin proteins also displayed significantly increased Ty1 transposition frequency (Fig. 6A). We were unable to determine if mutation of the cohesin loader protein (scc2-4) also results in a high transposition frequency because of variability amongst the replicate samples. We tested if the esp1-1 mutant may be deficient in Ty1 transposition because of the inability to properly cleave Scc1. We found that the Ty1 transposition frequency was restored to wild type levels in the esp1-1 scc1-73 double mutant (Fig. 6A) suggesting that removal of cohesin can restore transposition to esp1-1 cells.
Fig. 6. esp1 and pds1 mutants have decreased, whereas cohesin mutant have increased Ty1 mobility. 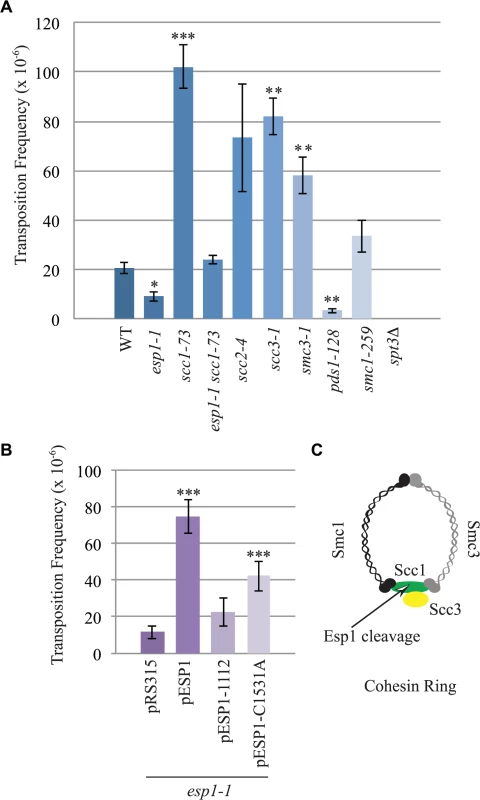
(A) Transposition frequency of wild type (WT), esp1-1, scc1-73, esp1-1 scc1-73, scc2-4, scc3-1, smc3-1, pds1-128, smc1-259, and spt3Δ (control) cells carrying a CEN plasmid with a marked Ty1element expressed from its endogenous promoter (pBDG922). (B) Transposition frequency of the esp1-1 mutant carrying either a vector (pRS315), wild type Esp1 (pESP1), an Esp1 protease domain truncation (pESP1-1112) or an Esp1 catalytic site mutation (pESP1-C1531A) as well as pBDG922. In both (A) and (B), cells were induced to transpose for 4 days at 20°C. A minimum of three independent transformants were quantified. Mutants with transposition frequencies that are significantly different from wild type (A) or pRS315 (B) are shown with one (p<0.05), two (p<0.005) or three asterisks (p<0.0005). (C) Structure of the cohesin ring. The increase in transposition frequency that we detected in the cohesin mutants suggests that removal of cohesin by the Esp1 protease may be necessary for transposition to occur. Since a strain carrying a mutation in the catalytic domain of Esp1 is not viable, we tested if expressing a catalytically dead version of Esp1 was able to rescue the transposition defect of an esp1-1 strain, compared to expression of wild type Esp1. We first demonstrated that expression of wild type ESP1 (pESP1) from a plasmid was capable of restoring the transposition frequency of an esp1-1 mutant carrying vector alone (pRS315) from 11.6x10-6 to 74.7 x 10–6, a 6.4-fold increase (p<0.0005, Fig. 6B). When we expressed pESP1-C1531A, which carries a cysteine to alanine mutation at amino acid residue 1531 in the catalytic domain of Esp1, the transposition frequency was partially restored to 42.2x 10–6, a 3.6-fold increase compared to esp1-1 alone (p<0.005, Fig. 6B). Since we were able to restore approximately half of the Esp1 transposition activity with the catalytically inactive version of Esp1, the catalytic activity may only be partially required for transposition. To determine if the Esp1 protease domain is required for transposition, we removed the C-terminal 518 amino acids that comprise the protease domain (pESP1-1112). The transposition frequency in the pESP1-1112 mutant was not significantly different than the esp1-1 mutant carrying vector alone suggesting that the protease domain is required for Ty1 transposition.
Ty1 cDNA levels are not altered in esp1 and pds1 mutants and Ty1 polypeptide processing does not require separase
The reduction of Ty1 transposition in esp1 and pds1 mutants could be due to defects at various stages of the Ty1 life cycle. The conversion of Ty1 RNA to cDNA, which occurs mid-way through the Ty1 life cycle, is considered a rate-limiting step in Ty1 transposition and mutants with increased or decreased Ty1 cDNA levels typically display an increased or decreased Ty1 transposition frequency, respectively [40,58]. To determine if Ty1 cDNA production is affected in esp1, pds1 or cohesin mutants we first used a Ty1 expression plasmid similar to that used for the transposition assays, except this Ty1 element is under control of a GAL1 promoter to facilitate visualization of the Ty1 cDNA by southern blot analysis. A probe spanning the HIS3 gene was used to detect Ty1 cDNA, the native his3Δ1 locus and the plasmid as described (Fig. 7A, [40]). We quantitated the Ty1 cDNA relative to the his3Δ1 locus, which gave a ratio of 0.8 for both wild type isolates (Fig. 7A). The Ty1 cDNA ratio was slightly higher for both esp1-1 and pds1-128 strains suggesting that the defect in Ty1 transposition in these mutants is not due to lower Ty1 cDNA production. The ∼5-fold increase in Ty1 transposition detected in the scc1-73 cohesin mutant (Fig. 6A), cannot be accounted for by the Ty1 cDNA ratio which is similar to wild type levels (1.0 and 0.9 for scc1-73 compared to 0.8 for wild type, Fig. 7A). Likewise the smc3-1 cohesin mutant had Ty1 cDNA ratios of 1.0 and 1.3 but a ∼3-fold increase in Ty1 transposition frequency (Fig. 6A, 7A). The scc3-1 mutant however, which displayed a ∼4-fold increase in Ty1 transposition (Fig. 6A) had significantly increased Ty1 cDNA levels compared to the his3Δ1 locus which could account for increased transposition (Fig. 7A).
Fig. 7. Ty1 cDNA levels are not affected in esp1 and pds1 mutants. 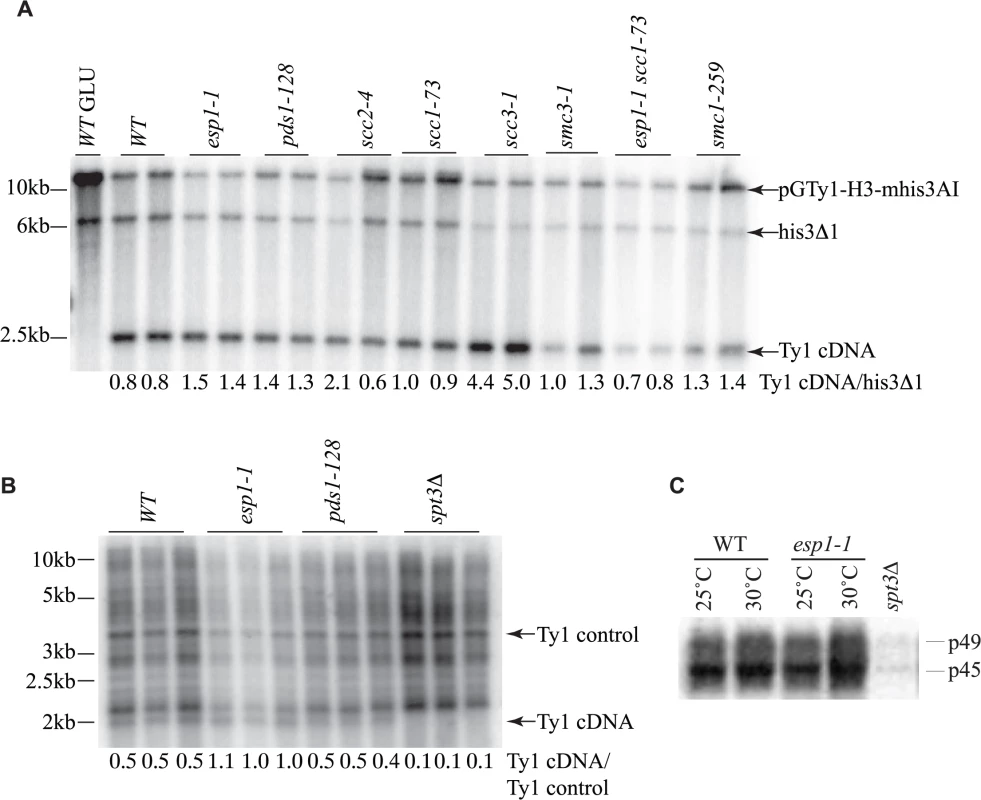
(A) Southern blot analysis of AflII digested yeast genomic DNA isolated from the indicated mutants carrying the pGTy1-H3-mhis3AI plasmid after 24 hour induction with 2% galactose. One wild type (WT) sample was also grown in 2% glucose (GLU) as a control for TY1 element expression. The blot was probed with a radiolabeled HIS3 gene. Shown are the fragment sizes for Ty1 cDNA (∼2.4kb), the endogenous HIS3 locus of the S288C deletion strain with 200bp deleted (his3Δ1, ∼6.5kb) and the pGTy1-H3-mhis3AI plasmid which is linearized (∼14kb). Below each lane is the quantitative ratio of Ty1 cDNA signal to his3Δ1 signal. (B) Southern blot analysis of PvuII digested yeast genomic DNA isolated from the indicated mutants after 2 days growth at 20°C. Endogenous Ty1 elements and cDNA were detected with a radiolabelled PvuII/SnaBI Ty1 element fragment. Below each lane is the quantitative ratio of the Ty1 endogenous cDNA (∼2kb) to an endogenous Ty element (Ty1 control). spt3Δ serves as a negative control for Ty1 cDNA levels. (C) Endogenous Ty1-Gag processing in wild type (WT) versus esp1-1 cells grown at 25°C or incubated for 6 hours at 30°C assessed by immunoblot. Blot was probed with anti-Gag antibody. p49 = unprocessed Gag; p45 = processed Gag. spt3Δ serves as a negative control for Gag expression. The transposition defects that we detected in esp1-1 and pds1-128 mutants were based on Ty1 expression from a plasmid with a native Ty1 promoter (Fig. 6A). Therefore we also analyzed Ty1 cDNA endogenous levels to confirm that esp1-1 and pds1-128 mutants had no defects in production of endogenous Ty1 cDNA (Fig. 7B). In this case, yeast genomic DNA was digested with PvuII which releases a linear ∼2kb fragment of Ty1 cDNA [59]. The southern blot was probed with a C-terminal fragment of the Ty1 element that detects both the endogenous Ty1 cDNA and Ty1 elements. We quantitated the ratio of Ty1 cDNA to a Ty1 element fragment detected by the probe (numbers below each lane on the blot). Our analysis confirmed that esp1-1 and pds1-128 mutant have similar levels of Ty1 cDNA to wild type cells (Fig. 7B).
While the protease activity of Esp1 was at least partially dispensable for transposition, we verified that Esp1 did not affect the processing of the TyA-TyB polypeptide. Accordingly, the ratio of endogenous Ty1 Gag precursor and mature protein levels were not appreciably different in an esp1-1 strain, suggesting that it is unlikely that the TyA-TyB polypeptide is subject to separase-mediated proteolysis (Fig. 7C).
Ty1 element insertion events are reduced in esp1 and pds1 mutants but increased in cohesin mutants
The SUF16 glycine tRNA gene locus is a hotspot for Ty1 transposition [60]. We induced endogenous Ty1 transposition by growing wild type and mutant cells at 20°C for 3 days, isolated genomic DNA, then measured de novo transposition upstream of the SUF16 locus using an established PCR assay [42]. One primer hybridizes with the SNR33 locus adjacent to SUF16 instead of SUF16 because of the repetitive nature of tRNA genes. The second primer hybridizes within the Ty1 element [42]. At this particular hotspot, Ty1 element insertions occur in a window of ∼800bp to 1500bp upstream of SUF16 in a periodic fashion due to nucleosome positioning [61,62]. Although Ty1 elements can insert in either orientation, this PCR analysis only detects Ty1 elements transcribed towards the SUF16 locus (Fig. 8A). Each strain was grown in triplicate and panels B and C represent genomic DNA isolated from two separate experiments. spt3Δ is included as a control for a strain that does not undergo transposition (Fig. 8B). For quantification, we summed the intensity of Ty1 insertions for each strain and compared the value to wild type. esp1-1 and pds1-128 mutants have less Ty1 insertion events compared to the wild type strains—60% and 74% that of wild type, respectively (Fig. 8B, C). This data correlates well with the reduced frequency of transposition in esp1-1 and pds1-128 mutants (Fig. 6A). The scc1-73 and smc3-1 and smc3-42 cohesin mutants have increased Ty1 insertion events compared to wild type cells (1.9, 1.7 and 1.9-fold higher than wild type, respectively, Fig. 8B, C). Both scc1-73 and smc3-1 mutants displayed increased transposition frequency in our quantitative assay (smc3-42 was not tested) suggesting that cohesin mutants allow increased Ty1 insertion events (Fig. 6A, 8B, C). The scc3-1 mutant however had increased Ty1 transposition frequency but not Ty1 insertion events upstream of the SUF16 locus (Fig. 6A, 8C). Overall, our data confirms that removal of cohesin is beneficial for Ty1 transposition and that both Esp1 and Pds1 are required for efficient Ty1 transposition.
Fig. 8. Esp1 and Pds1 are required for Ty1 element insertion upstream of the SUF16 locus. 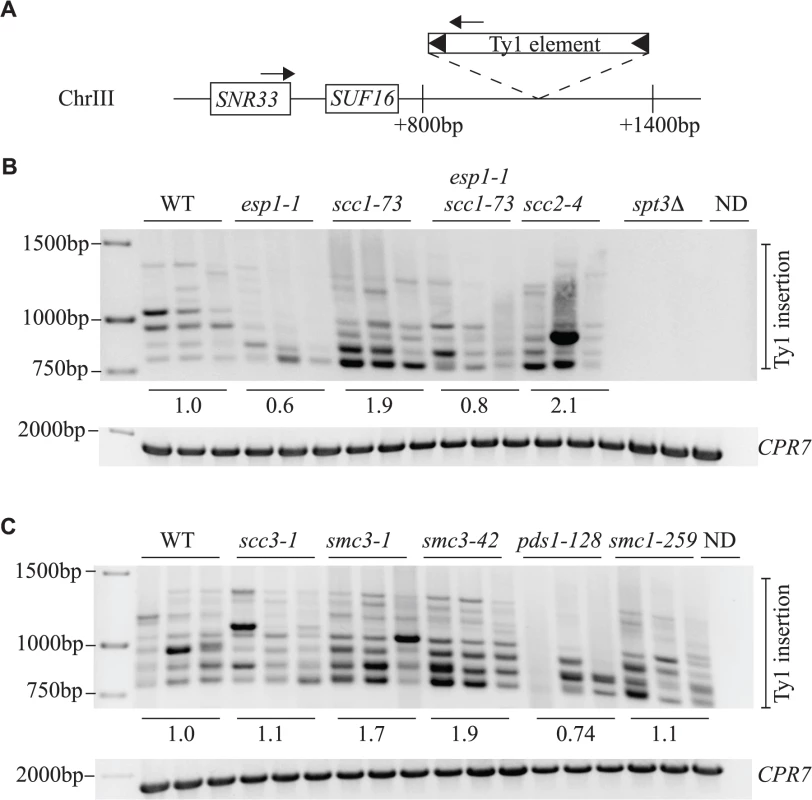
(A) Schematic of the SUF16 genomic locus with PCR primers (arrows) designed to hybridize within the SNR33 gene and the newly inserted Ty1 elements. (B) and (C) PCR analysis of yeast genomic DNA extracted from indicated strains grown in triplicate for 3 days at 20°C to induce transposition. Upper panel is the result of the PCR assay with primers shown in (A) whereas lower panel is a control PCR for the CPR7 locus to demonstrate that yeast genomic DNA was present in each sample. Quantification of Ty1 insertion events is shown relative to wild type (WT) as described in the Materials and Methods. The entire Esp1 polypeptide contributes to Ty1 transposition
The Esp1 protein contains 1630 amino acids with the N-terminal 2/3 of the protein predicted to form tandem ARM/HEAT repeats and the protease catalytic domain at the C-terminus [9,63,64]. Thus far we had tested one allele of esp1 with a single point mutation near the catalytic domain (esp1-1, P1404L). To further validate the function of Esp1 in Ty1 transposition, we acquired esp1 ts alleles generated by creating mutations in three different regions of the protein—an N-terminal (esp1n122), middle (esp1b120) and C-terminal (esp1c113) allele [26]. The esp1c113 allele does not have Scc1 cleavage activity at restrictive temperature [26]. Due to the ts nature of these alleles (restrictive temperature of 33°C), we developed two assays to test for Ty1 transposition at temperatures higher than 20°C, which is the temperature needed to induce endogenous Ty1 transposition. We used a modified version of a Ty1 transposition plate assay that employs an overexpression version of the Ty1-H3mHIS3AI element (pGAL1-Ty1-H3mHIS3AI), that can induce transposition at higher temperatures [40]. We included a step to remove the pGAL-TyH3mHIS3AI-URA3 plasmid after transposition induction to avoid detecting HIS+ colonies due to Ty1 element recombination with the plasmid. In brief, patches of single isolate ESP1 wild type and esp1 mutant colonies carrying pGAL1-Ty1-H3mHIS3AI were induced on galactose media for 4 days to undergo transposition. After a series of replica plating steps, cell growth was monitored on minimal complete plates (SC) whereas Ty1 element insertion into the genome was measured by the presence of HIS+ colonies on SC-HIS plates (Fig. 9A, B). At 25°C, wild type cells produced a lawn of HIS+ colonies after pGAL-TyH3mHIS3AI induction whereas no HIS+ colonies were detected with esp1-1 cells and reduced HIS+ colonies with the spt10Δ control strain (Fig. 9A). The pds1-128 mutant, which has a higher restrictive temperature (37°C) than the esp1 alleles, required growth at 33°C to display a transposition defect under these conditions (Fig. 9A). The additional esp1 alleles (n122, b120 and c113) all displayed a transposition defect at 30°C, but not 25°C (Fig. 9B).
Fig. 9. All Esp1 domains contribute to Ty1 transposition. 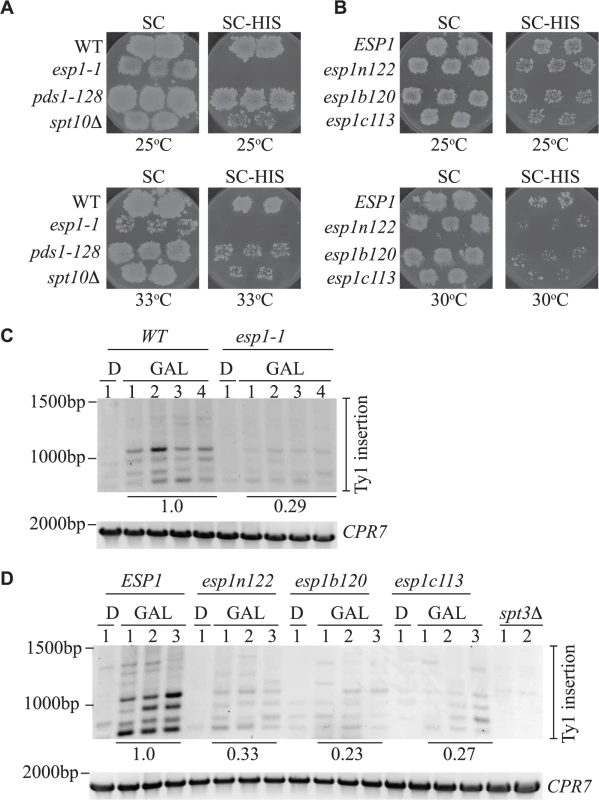
(A,B) The indicated strains, carrying the pGTy1-H3-mhis3AI–URA3 plasmid, were patched and induced to undergo transposition on galactose media at the indicated temperatures. After transposition, plates were replica plated to 5-FOA to remove the URA3 plasmid as described in the Materials and Methods. Patches are shown after the final step of replica plating. The SC (minimal complete) plates are a control for growth and the colonies present on the SC-HIS plates represent insertion of the Ty1-HIS3 element into the genome. (C,D) Triplicate isolates of indicated esp1 ts alleles, carrying a pGAL-TyH3mHIS3AI-URA3 plasmid (pJBe376) were induced to undergo transposition at semi-restrictive temperature (30°C) by growth in 2% galactose for 24 hours (GAL). One isolate was grown for 24 hours in 2% glucose (D) as a control. SUF16 PCR analysis of the extracted yeast genomic DNA is shown and CPR7 is a control PCR. All bands in the three lanes for each GAL grown isolate were quantified and summed, then compared to the wild type strain. The relative values of the mutants compared to wild type (value of 1.0) is shown. The amino acid mutations of the esp1 alleles have been previously been published: esp1n122 (N90S, C511F), esp1b120 (K782E, I951T, I1040T), esp1c113 (F1327L H1391Y) [26]. We also analyzed Ty1 insertion events upstream of the SUF16 locus by inducing the pGAL-TyH3mHIS3AI plasmid for 24 hours while growing the esp1 ts alleles at semi-permissive temperature (30°C). We found that after expression of the pGAL-TyH3mHIS3AI plasmid for 24 hours at 30°C, we were able to detect insertion of Ty elements at the SUF16 locus in a wild type strain (Fig. 9C, D). We first tested the esp1-1 allele that has a 2-fold reduction in transposition at 20°C and found that the Ty insertion events were reduced ∼3-fold (0.29) compared to wild type (Fig. 9C). Next we tested the three esp1 alleles that represent three regions of the Esp1 protein (esp1n122, esp1b120 and esp1c113) and found that esp1b120 was reduced ∼4-fold whereas esp1n122 and esp1c113 were reduced ∼3 fold compared to wild type (Fig. 9D). Therefore, no specific domain of Esp1 contributes solely to Ty1 transposition activity and a cohesin cleavage defective version of Esp1 (esp1c113) also has transposition defects.
The mitotic exit network impacts Ty1 transposition
Cleavage of cohesin by separase is important for triggering the end of the cell cycle [65]. Esp1 is also part of the “FEAR” pathway that promotes early anaphase release of the Cdc14 phosphatase from its inhibitor Cfi1/Net1 in the nucleolus [18]. This function for separase does not require its proteolytic activity [19]. The mitotic exit network (MEN) fully releases Cdc14 from the nucleolus after anaphase to down-regulate Cdk activity and promote mitotic exit [66]. The MEN is a Ras-like GTPase cascade that includes the Cdc5, Cdc15 and Dbf2 protein kinases [66]. We tested if components of the FEAR and MEN pathway have a role in Ty1 transposition by performing a quantitative transposition assay at 20°C in FEAR (spo12Δ, slk19Δ, cdc5-1) and MEN (cdc5-1, cdc15-2, dbf2-1) mutants as well as the cdc14-1 mutant. We found that the Ty1 transposition frequency was reduced by 7-fold in cdc5-1 mutants and 5.5-fold in cdc15-2 mutants whereas none of the other mutants had a statistically significant change in transposition frequency compared to wild type (Fig. 10A). Consistent with our quantitative transposition data, Ty1 insertion events upstream of the SUF16 locus were also dramatically reduced in cdc5-1 and cdc15-2 mutants (Fig. 10B). Although we could detect a clear banding pattern of SUF16 Ty1 insertion events in the cdc14-1 mutant, the intensity was significantly lower than wild type. Therefore Cdc14 may impact specific Ty1 insertion sites but not overall Ty1 transposition. Our data suggests that the Cdc5 and Cdc15 kinases are required for optimal Ty1 transposition.
Fig. 10. The mitotic exit network has a role in Ty1 transposition. 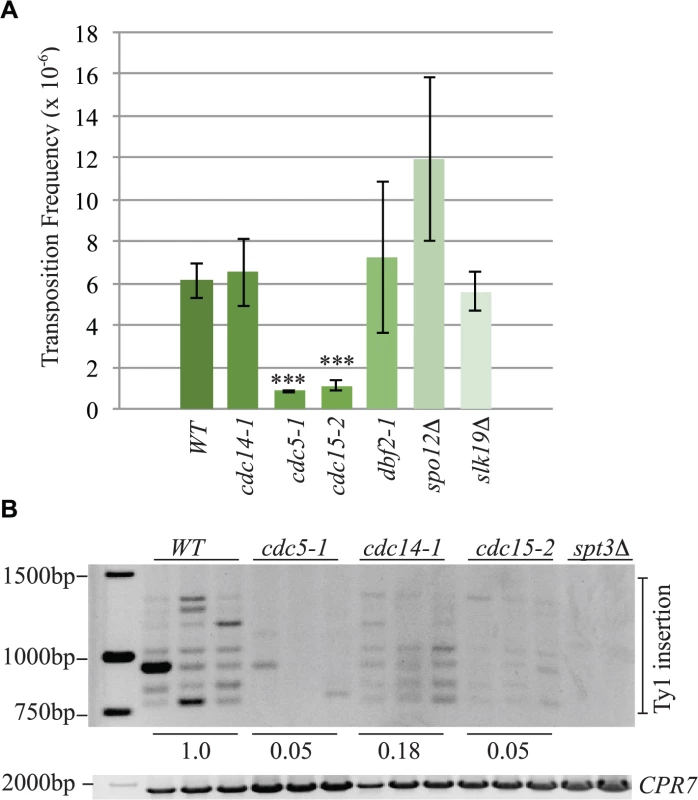
(A) Transposition frequency of wild type (WT), cdc14-1, cdc5-1, cdc15-2, dbf2-1, spo12Δ and slk19Δ cells carrying a plasmid with a marked Ty1element (pBDG922). Mutants with transposition frequencies that are significantly different from WT are shown with three asterisks (p<0.0005). (B) PCR analysis of yeast genomic DNA extracted from indicated strains grown in triplicate for 3 days at 20°C to induce transposition. Upper panel is a SUF16 PCR assay whereas the lower panel is a control PCR for the CPR7 locus to demonstrate that yeast genomic DNA was present in each sample. Quantification of Ty1 insertion events is shown relative to WT as described in the Materials and Methods. Discussion
We present here a combined study of both genetic and physical interactions that uncovered a novel role for Esp1 in Ty1 retrotransposition. Using SGA technology, we identified genes that upon loss of function (SL) or increased dosage (SDL) adversely impact esp1-1 viability. As expected, genes with a cell cycle function were uncovered in our esp1-1 SL and SDL screens. However, genetic interactions were enriched for alleles with roles in other biological pathways, including the regulation of Ty1 transposition. Further, using mass spectrometry analysis, we made the discovery that Esp1 interacts with Ty1-IN encoded by the Ty1 retrotransposon. Despite extensive studies of Ty1 transposition, no host factor has been identified that interacts with Ty1-IN. We confirmed that Esp1 interacts with Ty1-IN and that both Esp1 and Pds1 are required for wild type levels of Ty1 element mobility and insertion frequency. The identification of proteins that regulate the chromosome division machinery with a role in Ty1 transposition concurs with a previous analysis that identified enriched GO terms from independent screens for genes that either inhibit or promote Ty1 transposition [42]. In this analysis, the enriched GO terms included as “M phase” and “G2/M transition of the mitotic cell cycle” which fits well with our data that Esp1 mediates Ty1 transposition.
Esp1 interacts with Ty1-IN
Only two proteins—the TFIIIB subunit Bdp1 and the Isw2 chromatin remodeling protein have been shown to disrupt the prototypical 80bp periodicity of the Ty1 target site when mutated [67,68]. However, neither of these factors was demonstrated to interact with Ty1-IN. Our mass spectrometry data identified an enrichment of both Ty1-IN and Ty1-RT peptides in the Esp1 versus mock purification (Fig. 3A). The interaction between Esp1 and Ty1-IN was confirmed by two independent co-IP assays (Fig. 3B, C). However, we were not able to confirm the putative interaction of Esp1 with Ty1-RT. As well, Ty1 cDNA levels are not affected in esp1-1 mutants suggesting that Esp1 is not required for the reverse transcription of Ty1 mRNA (Fig. 7A).
The Esp1-Ty1-IN interaction occurs in Log, G1 and G2/M cells but is minimal in S phase cells (Fig. 4). The efficient concentration of Esp1 in the nucleus in late G2 phase depends on Pds1 [38]. We find that both esp1-1 and pds1-128 mutants have decreased Ty1 mobility and frequency of Ty1 insertions upstream of the SUF16 locus (Fig. 6A, 8B, C). However, Pds1 does not appreciably interact with Ty1-IN and is not required for Esp1 to interact with Ty1-IN (Fig. 3B, 5B). Therefore, we suspect that the Ty1 insertion defect in the pds1-128 mutant is due to a reduced efficiency of Esp1 localization to the nucleus in this mutant [38]. Although both Esp1 and Ty1-IN are nuclear localized proteins, it still remains to be tested if the interaction between Esp1 and Ty1-IN occurs specifically in the nucleus [38,69–71]. Ty1 mobility is modestly reduced in esp1-1 and pds1-128 mutants (2 and 6-fold, respectively), compared to the 5 to 50-fold reductions detected in Ty1-integrase nuclear localization mutants [69,71]. The relatively modest decrease Ty1 mobility that in esp1 and pds1 mutants may be due to the use of ts alleles that do not completely eliminate function, or because another host factor is also required for Ty1 element insertion.
Is removal of cohesin required for Ty1 transposition?
Our data suggests that removal of cohesin by separase enables insertion of Ty1 elements upstream of tRNA genes. The majority of cohesin mutants that we tested displayed higher levels of transposition frequency and Ty1 element insertion and the transposition defect of the esp1-1 mutant is at least partially rescued when combined with the scc1-73 mutant (Fig. 6A, 8). We also find that an allele of esp1 (c113) that cannot cleave Scc1 at restrictive temperature has reduced transposition efficiency (Fig. 9B, D). Finally, a version of ESP1 that lacks the entire protease domain cannot rescue the esp1-1 transposition defect (Fig. 6B). However, two pieces of data suggest that the cleavage of cohesin by separase cannot be the only function for Esp1 in mediating Ty1 transposition. The first is that Ty1-IN physically interacts with Esp1 and this interaction is enhanced in metaphase cells (Fig. 3, 4). The second is that expression of a catalytically inactive version of ESP1 is partially capable of restoring Ty1 transposition mobility to an esp1-1 mutant (Fig. 6B). One way to explain the data is that two activities of Esp1 contribute to efficient transposition—one is removal of cohesin and the other is a chromatin targeting function based on the physical interaction of Ty1-IN with Esp1[8,9,26].
Scc3 may not impact Ty1 element insertion
We noticed a striking increase in Ty1 cDNA in the scc3-1 mutant (4 to 5-fold) compared to no increase in the other cohesin mutants upon galactose induction of a Ty1 element (Fig. 7A). Although we also detected a corresponding increase in Ty1 transposition events, based on the Ty1-H3mHIS3AI transposition assay (Fig. 6A), we did not see an increase in Ty1 element insertion events upstream of SUF16. Therefore, the increase in HIS3+ colonies could be due to recombination from higher levels of Ty1 cDNA in the scc3-1 mutant rather than an increase in transposition. Scc3 is not part of the core cohesin molecule but associates with Scc1 in a regulatory manner. The scc3-1 mutant prematurely separates sister chromatids, which is a similar phenotype to the scc1-73 and smc1-259 mutants [72,73]. However Scc3 is primarily thought to be involved in cohesin establishment in S phase and specific mutations in Scc3 reveal that it also has an anti cohesin-establishment activity as well [11,74]. The removal of Scc3 does not affect cohesin ring stability, which may explain why there is no increase in Ty1 insertion events upstream of SUF16 in the scc3-1 mutant (Fig. 8C, [74]).
Ty1 element insertion upstream of tRNA genes is not due to cohesin loading
Cohesin loads onto chromosomes at sites defined by the Scc2/4 cohesin loading complex, correlating with tRNA sites and other genes transcribed by RNA pol III [75,76]. After cohesin loads, it moves to sites of convergent transcription [3,4]. However, the creation of a synthetic chromosome III lacking all tRNA genes suggests that tRNA genes are not absolutely required for cohesin loading [77]. The cohesin binding pattern was similar on the synthetic chromosome compared to native chromosome III suggesting that in the absence of tRNA genes, secondary loading sites may become prevalent [77]. Ty1 elements preferentially integrate upstream of genes transcribed by RNA polymerase III, such as tRNA genes, in a loosely defined window ranging from about 80bp to ∼750bp [60,78–80]. Our data suggests that the loading of cohesin at tRNA genes is not required for Ty1 insertion because we detect increased Ty1 mobility and insertion events in cohesin mutants (Fig. 6, 8). Although we did observe a weak interaction between Scc1 and Ty1-IN, Scc1 was not required for the Esp1-Ty1-IN association (Fig. 3B, 5C). Therefore the Scc1-Ty1-IN interaction is likely indirect and mediated by Esp1 that is bound to the cohesin complex. We analyzed high resolution mapping of cohesin binding sites in metaphase arrested cells and found that, as expected cohesin, does not bind the SUF16 tRNA site or most other tRNA sites unless they are located very near the centromere (a high density cohesin binding region) [4]. Although SUF16 is not bound by cohesin, the gene upstream of this tRNA site (YCR016W) is [4]. One intriguing possibility is that Esp1 clears cohesin from the region upstream of tRNA genes to enable Ty1 insertion and the interaction with Ty1-IN serves to target it to this location. Localized removal of cohesin by separase has been demonstrated in both S. cerevisiae and S. pombe in response to DNA damage [37,81].
Esp1 is well positioned to mediate Ty1 insertion during mitosis
A recently published paper noted that tRNA genes are enriched in the pericentromere—a region of 30 to 50kb surrounding the centromere that is also enriched in cohesin and condensin [82–84]. tRNA genes, condensin and a protein called Cbf5/Dyskerin were found to tether pericentric chromatin to the spindle axis in mitosis [84]. This data fits nicely with the fact that Esp1 localizes to and promotes stability of the elongating spindle in mitosis and our data demonstrating an enrichment of the Esp1 Ty1-IN interaction in mitosis (Fig. 4, [15,38]). Cbf5/Dyskerin is a component of the H/ACA snoRNP pseudouridylase complex that catalyzes the conversion of uridine to pseudouridine in nascent tRNA [85]. We identified another component of the H/ACA snoRNP pseudouridylase complex, Gar1, in our esp1-1 SDL screen (S2 Table). Notably, the SUF16 locus, which is a hotspot for Ty1 transposition, is located within the pericentromere of chromosome III. Therefore Esp1 may have a specific role in targeting Ty1-IN to tRNA genes within the pericentromere.
Our data also points to a possible connection between Ty1 transposition and events that occur after chromosome segregation. We find that two kinases, Cdc5 and Cdc15, that function to promote mitotic exit via the FEAR and MEN pathways promote Ty1 transposition (Fig. 10). The role of the Cdc5 kinase in mitotic exit is complex because Cdc5 activates the MEN pathway by phosphorylation of many of its components [66]. Importantly, Cdc5 also phosphorylates Scc1 and this phosphorylation is important for Scc1 cleavage by separase, therefore the lack of transposition in the cdc5-1 mutant could be due to inefficient cohesion dissolution [9,86]. We were unable to conclusively determine if the Cdc14 phosphatase is important for transposition because our quantitative assay did not show a significant difference in HIS+ colonies but our SUF16 assay showed a reduced efficiency of Ty1 insertion (Fig. 10). A potential link between Ty1 transposition and the FEAR pathway is intriguing because the FEAR pathway controls segregation of the nucleolus, which is where tRNA genes are clustered [16,18,19,66,87,88].
The conservation of retroviral and retrotransposon integrases suggests that the cellular mechanisms used to insert Ty1 cDNA into the S. cerevisiae genome may be conserved with retroviral cDNA insertion into eukaryotic genomes. Interestingly, the dominant binding partner of HIV-1 IN in human cells is lens epithelium derived growth factor (LEDGF) and the LEDGF IN binding domain has high structural similarity to HEAT/ARM repeats [89]. The N-terminal two thirds of separase is predicted to form tandem HEAT/ARM repeats suggesting this might be a common motif for IN binding [63,64]. Our study, which reports the first physical interaction of a yeast host protein with Ty1-IN, is an important step towards understanding the mechanism of targeting retroviral cDNA into the host genome.
Materials and Methods
Yeast strain construction
S. cerevisiae strains used in this study are listed in S4 Table. All strains were grown at 25˚C unless otherwise indicated. To create the esp1-1::natR mutation in the SGA starting strain, a C-terminal fragment was amplified from esp1-1 (W303 strain background, gift from Yanchang Wang) [25]. This fragment included the P1404L (bp4211 C→T) mutation, approximately 200 bp downstream of the stop codon and an additional 25 bp overlapping the TEF promoter in p4339 [27]. Concurrently, the natR cassette from p4339 was also amplified to include 45 bp immediately downstream of the ESP1 ORF. Both amplicons were transformed simultaneously into Y7092. Successful double recombination was confirmed in three ways: (1) growth on yeast peptone dextrose (YPD) media containing clonNAT (2) temperature sensitivity at 37°C (3) sequencing the esp1-1::natR mutation (Macrogen).
To create YM2377, a single copy of ESP1/YGR098C was disrupted by homologous recombination with the hphMX4 marker in BY4743 as described [90]. To generate YM2390 a LEU2-CEN plasmid carrying the esp1n122 ts alleles was transformed into YM2377, sporulated and haploid isolates containing esp1ΔhphMX4 and the pesp1n122-LEU2-CEN plasmid were isolated. YM2392 and YM2400 were generated in a similar manner after transformation of YM2377 with LEU2-CEN vectors carrying the esp1b120 and esp1c113 ts alleles respectively whereas YM2396 was created by transformation of YM2377 with a wild type ESP1-LEU2-CEN plasmid. The esp1 alleles have been previously published [26].
Plasmid construction
The pESP1-LEU2-CEN vector contains full length ESP1 and its promoter and was a kind gift from Dr. Steven Reed [26]. The pESP1C1531A vector was constructed by cloning an NdeI fragment from a GAL-FLAG-ESP1 (C1531A)-CBD construct [kindly provided by Dr. Frank Uhlmann [9]] that contains the C1531A mutation, into pESP1-LEU2-CEN that had been digested with NdeI. The pESP1-1112 mutation was created using the same cloning strategy but the NdeI fragment inserted in backwards, resulting in termination of wild type Esp1 sequence at amino acid 1112, the introduction of 24 new amino acids until a stop codon was generated.
SGA screens
SGA screens have been previously described in detail [20,27,91]. Using a Singer RoToR HDA robot, MATα esp1-1::natR was mated to both the haploid deletion collection of nonessential alleles (BY4741, MATa, KanMX4) and the 2 micron pGAL1/10-GST-ORFX overexpression array (Open Biosystems) [20,29]. For SL and SDL screens respectively, either double mutants (esp1-1::natR geneXΔKanMX4) or esp1-1 carrying pGAL1/10-GST-ORFX were obtained through sporulation of heterozygous diploids with at least three colonies of each genotype being compared. MAT a cells of interest (esp1-1::natR in combination with geneXΔKanMX4 or pGAL1/10-GST-ORFX for the SL or SDL screens, respectively) were isolated through a series of replica pinning steps. Growth on the final selective media (clonNAT + G418 dextrose plates for the SL screen; SC-URA + clonNAT) was examined at 25°C, with plates imaged using a flatbed scanner. Balony software was used to measure spot sizes, determine cut-off values for genetic interactions and define strains the showed statistically significant changes in growth rate [30]. For SL interactions, cut-off values were defined for each screen by determining three standard deviations from the mean of the ratios of the double mutant to single mutant growth rates. Double mutant strains that met the cut-off and showed significant changes in growth relative to the corresponding single mutant control (one-tailed students’ t-test; p < 0.05; n = 3) were considered as genetic interactions. esp1-1 SDL interactions were similarly analyzed by comparing growth of esp1-1::natR pGAL1/10-GST-ORFX on dextrose versus galactose.
Confirmation of SDL interactions
Plasmids carrying genes identified as esp1-1 top SDL interactions (causing a growth rate < 0.3 of control) were rescued from yeast and retransformed into both wild type and esp1-1::NAT strains. Transformants were then grown overnight to log phase (OD600 of 0.5–1.0) in SC media depleted for uracil + 2% raffinose + 0.1% dextrose media. Strains were then diluted to an OD600 of 0.1, and subsequently serially diluted four times by a factor of 1 : 5, all in SC-URA + 2% raffinose. Dilutions were spotted onto either SC-URA + 2% dextrose or SC-URA + 2% galactose plates, and incubated for four days (S1 Fig). Genes that caused a growth defect in esp1-1 but not wild type cells when overexpressed on galactose were scored as SDL or synthetic dosage sensitive.
Affinity IP and mass spectrometry
Esp1-13Myc was purified using a one-step Myc-tag approach based on Field et al [92]. Briefly, 1L each of WT and ESP1-13Myc::KanMX6 strains were grown to an OD600 of ∼0.9. Cells were lysed in 100mM Tris-HCl pH 7.5, 150mM NaCl, 0.1% Tween 20, 1% NP-40, 10% glycerol, 5mM EDTA, 1mM DTT, 1mM phenylmethylsulfonyl fluoride, 10mM chloroacetamide and a 1x protease inhibitor cocktail by glass bead beating. Lysates were cleared by centrifugation at 10,000 rpm, 4°C for 15 minutes. IPs were performed using a 9E10 anti-Myc antibody cross-linked to Sepharose (MMS-150P, Covance). 400μl beads (bed volume) were mixed with each lysate for four hours at 4°C and washed five times in 100mM Tris-HCl pH 7.5, 500mM NaCl, 0.05% Tween 20, 0.5% NP-40, 10% glycerol, 5mM EDTA and 1mM DTT. Proteins were eluted twice with 200μL of SDS loading sample buffer without reducing agent prior to TCA precipitation. Proteins were then separated by SDS-PAGE followed by in-gel digestion for mass spectrometry analysis. The protein gel was first incubated in fixation buffer (5% acetic acid, 47.5% methanol) with coomassie Blue G for 30 minutes following by a four-hour wash in H2O. Gel pieces from each sample (Esp1-IP and control) were divided into five fractions: three fractions above 55 kDa; one fraction bellow 55 kDa without immunoglobulins; and one fraction containing the immunoglobulin G heavy chain and light chains (55kDa and 25kDa bands). In-gel trypsin digestion was based on Shevchenko et al. with minor changes [93]. Digestion buffer contained 50mM ammonium bicarbonate, and gel pieces were dehydrated using ethanol. Gel pieces were incubated at 56°C for 45 minutes in 10mM DTT followed by a 30-minute incubation in 55mM chloroacetamide at room temperature. Peptides were extracted once with 0.5% acetic acid, twice with 30% acetonitrile, 0.5% acetic acid and twice with 100% acetonitrile. Dried peptide-extraction samples were resuspended in 0.5% acetic acid solution and acetified to a pH below 2 using 100% acetic acid. Peptide samples were then purified by solid phase extraction on C-18 stage tips [94–96].
Purified peptides were analyzed using a linear-trapping quadrupole—Orbitrap mass spectrometer (LTQ-Orbitrap Velos; ThermoFisher Scientific) coupled to Agilent 1200 Series high-performance liquid chromatography using a nanospray ionization source that included a 100-μm-inner diameter fused silica trap column packed with 5 μm-diameter Aqua C-18 beads (Phenomenex, www.phenomenex.com), 50-μm-inner diameter fused silica fritted analytical column packed with 3 μm-diameter Reprosil-Pur C-18-AQ beads (Dr. Maisch, www.Dr-Maisch.com) and a 20-μm-inner diameter fused silica gold coated spray tip. Fractions from each sample were run at a 60-minute high-performance liquid chromatography gradient from 0.5% acetic acid to 0.5% acetic acid and 26% acetonitrile). The LTQ-Orbitrap was set to acquire a full-range scan at 60,000 resolution from 350 to 1600 Th in the Orbitrap and to simultaneously fragment the top ten peptide ions in each cycle in the LTQ (minimum intensity 1000 counts). Singly charged ions were excluded and parent ions were then excluded from MS/MS for the next 30 sec. The Orbitrap was continuously recalibrated using lock-mass function [97].
Analysis of mass spectrometry data
Centroided fragment peak lists were processed with Proteome Discoverer v. 1.2 (ThermoFisher Scientific) followed by a Mascot analysis (2.3.0, Matrix Science) against a Saccharomyces cerevisiae protein database (www.yeastgenome.org; 05012010, 6147 protein sequences) with the following parameters: peptide mass accuracy 10 parts per million; fragment mass accuracy 0.6 Da; trypsin enzyme specificity; fixed modifications—carbamidomethyl; variable modifications—methionine oxidation, lysine acetylation, serine/threonine/tyrosine phosphorylation; ESI-TRAP fragment characteristics. Only those peptides with IonScores exceeding the individually calculated 99% confidence limit (false positive discovery rate < = 1%) were considered as accurately identified. Candidate Esp1-interacting proteins were arbitrarily selected if at least three peptides were identified and if peptides were at least four times more abundant in the IP versus the mock-IP samples (S3 Table).
Co-immunoprecipitation
Fig. 3A—GFP-tagged Esp1, Pds1 and Scc1 and untagged wild type cells (BY4741 strain background) were transformed with pGAL1-Ty1-H3 (pJEF724, generous gift from Dr. Jef Boeke) and grown overnight to log phase in SC-URA + 2% raffinose + 0.1% dextrose. 100mL of cells were then diluted to an OD600 of 0.02, resuspended in SC-URA + 2% galactose to induce pGAL1-Ty1-H3 expression for 24 hours. Cells were harvested, washed with ice cold H20 and resuspended in lysis buffer (50mM Tris-HCl pH 7.5, 0.1% NP40, 150mM NaCl, 0.5mM EDTA and protease inhibitors) on ice followed by addition of glass beads and lysed. Lysates were centrifuged at 12 000 xg for 5 mins, and supernatant retained as whole cell lysate. Protein concentration was quantified by Bradford assay. 5mg of lysate were first incubated with 5μl of Bab-20 beads (ChromoTek) at 4°C for 1 hour to reduce non-specific binding. The supernatant was removed and incubated with GFP-Trap_A beads (ChromoTek) at 4°C for 1.5 hours. Beads were then washed four times with wash buffer (50mM Tris-HCl pH 7.5, 0.5% Triton, 400mM NaCl, 0.5mM EDTA) and finally resuspended in 20μl 2X sodium dodecyl sulfate (SDS) loading buffer containing 200mM β mercaptoethanol. 10μg of Ty1-IN and 80μg of GFP tagged proteins were loaded on the immunoblot for whole cell lysates whereas 5μl of the IP was loaded. Samples were run on 8% SDS polyacrylamide gels and transferred to polyvinylidene difluoride. Membranes were probed with either anti-GFP (1 : 500 Roche) or anti-IN [1 : 1000, 8B11, J. Boeke [98]]. Blots were imaged with a ChemiDoc MP (Biorad).
Fig. 3C—Esp1-13Myc cells harboring pGAL-GFP-LacZ-Ty1-IN [kind gift of Anita Corbett, [70,71]] and pGAL-GFP-lacZ vector control were grown to log phase in SC-URA + 2% raffinose + 0.1% dextrose and diluted down to an OD600 of 0.08 in SC-URA + 2% galactose to induce Ty1 expression for 24 hours. Esp1-13Myc and untagged (No Tag) cells carrying pGAL-GFP-lacZ grown in galactose and pGAL-GFP-LacZ-Ty1-IN grown in 2% dextrose were used as controls. Cell lysates and GFP purification were performed as described above. Membranes were probed with anti-GFP (Roche) and anti-c-Myc (9E10, 1 : 2000, Roche).
Fig. 4 - The Esp1-GFP cell cycle IP (Fig. 4) was performed in a similar manner with the following changes. Esp1-GFP cells carrying pGAL1-Ty1-H3 were grown in galactose for 20 hours, fresh 2% galactose added and grown for another 4 hours, then arrested with either α-factor (5μg/mL) for 2 hours, HU (200mM) for 3.5 hours, or Nz (15μg/ml) for 3 hours prior to cell lysis.
Fig. 5B—TetO7-PDS1 Esp1-GFP and TetO7-PDS1 cells were transformed with pGAL1-Ty1-H3-LEU2 (pX87, kind gift from Dr. Jef Boeke). Cells were grown overnight in SC-LEU + 2% raffinose + 0.1% dextrose, then diluted to an OD600 of 0.02 and induced in SC-LEU + 2% galactose for 20 hours. Next, fresh 2% galactose was added and cells grown for an additional 4 hours before treatment with 100μg/ml Doxycycline (Sigma) for 1hour. Cell lysates and GFP purifications were performed as described above.
Fig. 5C—TetO7-SMC1 Esp1-GFP and TetO7-SMC1 cells were treated as described above except that 100μg/mL Doxycycline was added for six hours prior to cell lysis.
Tet-PDS1 quantitative PCR
The TetO7-PDS1 Esp1-GFP strain were grown to log phase in SC-URA with 2% glucose, 100μg/ml Doxycycline (Sigma) was added and time points taken at 15, 30, 60,120 and 240 min. Approximately 1x107 cells were spun down and stored at -80°C. Total RNA was prepared with Qiagen RNeasy kit and 1μg RNA was used for cDNA synthesis with Superscript VILLO cDNA synthesis kit (Invitrogen). Real-time PCR was performed in duplicate with 2μl diluted cDNA (1 : 50) and Power SYBR Green Master Mix (Invitrogen). A standard two-hour comparative PCR analysis was performed using a 7500 Real Time PCR system (Applied Biosystems). Primers were designed to amplify PDS1 (OVM866 5’ - GGCAGCAAAAGACAACAATAG-3’ and OVM876 5’-CCCTACTACGACTGCGAGAAA-3’) and TAF10 as an internal control [(OVM695 5’ - GGCGTGCAGCAGATTTCAC-3’ and OVM696 5’ - TGAGCCCGTATTCAGCAACA-3’ [99]]. Relative transcript values and error bars were quantified as described [Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR, Applied Biosystems (www.appliedbiosystems.com)] using 7500 Software V.2.0.6.
Ty1 quantitative transposition assay
This assay was based on the Ty1-his3-AI mobility assay performed in [42]. Strains were transformed with a URA3 CEN based plasmid that expresses a marked Ty1-his3-AI element at endogenous levels (pBDG922, a kind gift from Dr. David Garfinkel). Single colonies were isolated at 25°C and patched. 2000 cells were inoculated into 1mL of SC-URA3 media in triplicate and grown for four or five days at 20°C until saturation. 1x 107 cells were plated on SC-URA-HIS plates to test for transposition frequency and 200 cells were plated on SC-URA plates to test for viability. Cells were grown on plates at 25°C for 2 days. The frequency of Ty1-his3-AI mobility was calculated according to this formula: # colonies on the SC-URA-HIS plate/1 x 107 cells divided by #colonies on the SC-URA plate/200.
Ty1 qualitative plate transposition assay
Yeast strains were transformed with a pGAL-TyH3mHIS3AI-URA3 plasmid (pJBe376, kind gift from Jef Boeke) and were plated on synthetic complement (SC) media minus uracil (SC-U) and grown at 25°C. Single colony transformants were patched onto SC-U 2% glucose media and grown for 2 days at 25°C. Next, plates were replica plated at onto SC-U containing 2% galactose and incubated for 1 day at 25°C, then either kept at 25°C for 3 additional days or incubated at 30°C or 33°C for 3 additional days. From this point onward, plates were kept at the same temperature as for the final 3 days of galactose induction. After galactose induction, plates were sequentially replica plated onto 2% glucose plates containing: (i) SC-U media, (ii) yeast peptone dextrose (YPD), (iii) SC-U plus 1.2 g/L of 5-fluoroorotic acid (5-FOA), and (iv) SC lacking histidine (SC-H) and SC complete. For each of the final four stages of replica plating, patches were grown until confluent.
Ty1 cDNA analysis
Ty1 cDNA analysis for Fig. 7A was performed as described in [40] with the following modifications. Strains were transformed with the pGTy1-H3-mhis3AI plasmid [[56], kind gift from Dr. Jef Boeke] and induced with galactose for 24 hours. Yeast genomic DNA was extracted and 4μg of DNA digested with Afl II to release a ∼2.4kb fragment of Ty1 cDNA containing the HIS3 gene. Digestions were run on a 1% agarose gel, transferred to a nylon membrane and probed with an 856bp fragment of the HIS3 gene including the promoter region. The HIS3 probe was radiolabelled with 32P-α-dATP using a DecaLabel DNA Labeling Kit (ThermoScientific). Ty1 endogenous cDNA analysis (Fig. 7B) was performed as described [47,100] with the following modifications. Cells were grown for 2 days at 20°C. Yeast genomic DNA was extracted and 10μg of DNA digested with PvuII to release a ∼2kb fragment of Ty1 cDNA. Blots were probed with a radiolabeleld 1517bp PvuII/SnaBI C-terminal fragment of the Ty1 element.
SUF16 PCR assay
The SNR33 and Ty1 PCR primers as well as the CPR7 primers have been previously described [42]. 100ng of DNA was used for each SUF16 PCR assay and 5ng of DNA was used for the CPR7 PCR assay.
pGAL-Ty1 SUF16 assay
esp1 ts alleles were transformed with a pGAL-TyH3mHIS3AI-URA3 plasmid (pJBe376). Three independent transformants were grown overnight in SC-URA-LEU + 2% raffinose + 0.1% dextrose. One isolate (#1) was diluted to an OD600 of 0.05 in SC-URA-LEU + 2% glucose (D) and three isolates (#1–3) were induced to undergo transposition by diluting to an OD600 of 0.05 in SC-URA-LEU + 2% galactose (GAL) for 24 hours. All cells were grown at semi-restrictive temperature (30°C). Cells were pelleted, yeast genomic DNA extracted and a SUF16 and control CPR7 PCR assay performed.
Supporting Information
Zdroje
1. Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43 : 525–558. doi: 10.1146/annurev-genet-102108-134233 19886810
2. Uhlmann F (2003) Chromosome cohesion and separation: from men and molecules. Curr Biol 13: R104–114. 12573239
3. Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, et al. (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5 : 243–254. 10882066
4. Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, et al. (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430 : 573–578. 15229615
5. Cohen-Fix O, Peters JM, Kirschner MW, Koshland D (1996) Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev 10 : 3081–3093. 8985178
6. Farr KA, Cohen-Fix O (1999) The metaphase to anaphase transition: a case of productive destruction. Eur J Biochem 263 : 14–19. 10429181
7. Hornig NC, Uhlmann F (2004) Preferential cleavage of chromatin-bound cohesin after targeted phosphorylation by Polo-like kinase. EMBO J 23 : 3144–3153. 15241476
8. Uhlmann F, Lottspeich F, Nasmyth K (1999) Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400 : 37–42. 10403247
9. Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103 : 375–386. 11081625
10. Gligoris TG, Scheinost JC, Burmann F, Petela N, Chan KL, et al. (2014) Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science 346 : 963–967. doi: 10.1126/science.1256917 25414305
11. Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, et al. (2009) Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell 33 : 763–774. doi: 10.1016/j.molcel.2009.02.028 19328069
12. Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, et al. (2009) Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell 17 : 344–354. doi: 10.1016/j.devcel.2009.07.015 19758559
13. Yang X, Boateng KA, Yuan L, Wu S, Baskin TI, et al. (2011) The radially swollen 4 separase mutation of Arabidopsis thaliana blocks chromosome disjunction and disrupts the radial microtubule system in meiocytes. PLoS One 6: e19459. doi: 10.1371/journal.pone.0019459 21559383
14. Pandey R, Heidmann S, Lehner CF (2005) Epithelial re-organization and dynamics of progression through mitosis in Drosophila separase complex mutants. J Cell Sci 118 : 733–742. 15671062
15. Sullivan M, Lehane C, Uhlmann F (2001) Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat Cell Biol 3 : 771–777. 11533655
16. D'Amours D, Amon A (2004) At the interface between signaling and executing anaphase—Cdc14 and the FEAR network. Genes Dev 18 : 2581–2595. 15520278
17. Bardin AJ, Amon A (2001) Men and sin: what's the difference? Nat Rev Mol Cell Biol 2 : 815–826. 11715048
18. Stegmeier F, Visintin R, Amon A (2002) Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108 : 207–220. 11832211
19. Sullivan M, Uhlmann F (2003) A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat Cell Biol 5 : 249–254. 12598903
20. Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, et al. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294 : 2364–2368. 11743205
21. Beauregard A, Curcio MJ, Belfort M (2008) The take and give between retrotransposable elements and their hosts. Annu Rev Genet 42 : 587–617. doi: 10.1146/annurev.genet.42.110807.091549 18680436
22. Lesage P, Todeschini AL (2005) Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet Genome Res 110 : 70–90. 16093660
23. Qi X, Sandmeyer S (2012) In vitro targeting of strand transfer by the Ty3 retroelement integrase. J Biol Chem 287 : 18589–18595. doi: 10.1074/jbc.M111.326025 22493285
24. Xie W, Gai X, Zhu Y, Zappulla DC, Sternglanz R, et al. (2001) Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol Cell Biol 21 : 6606–6614. 11533248
25. Baum P, Yip C, Goetsch L, Byers B (1988) A yeast gene essential for regulation of spindle pole duplication. Mol Cell Biol 8 : 5386–5397. 3072479
26. Baskerville C, Segal M, Reed SI (2008) The protease activity of yeast separase (esp1) is required for anaphase spindle elongation independently of its role in cleavage of cohesin. Genetics 178 : 2361–2372. doi: 10.1534/genetics.107.085308 18430955
27. Tong AH, Lesage G, Bader GD, Ding H, Xu H, et al. (2004) Global mapping of the yeast genetic interaction network. Science 303 : 808–813. 14764870
28. Giaever G, Chu AM, Ni L, Connelly C, Riles L, et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 : 387–391. 12140549
29. Sopko R, Huang D, Preston N, Chua G, Papp B, et al. (2006) Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 21 : 319–330. 16455487
30. Young BP, Loewen CJ (2013) Balony: a software package for analysis of data generated by synthetic genetic array experiments. BMC Bioinformatics 14 : 354. doi: 10.1186/1471-2105-14-354 24305553
31. Sarin S, Ross KE, Boucher L, Green Y, Tyers M, et al. (2004) Uncovering novel cell cycle players through the inactivation of securin in budding yeast. Genetics 168 : 1763–1771. 15579722
32. Measday V, Hieter P (2002) Synthetic dosage lethality. Methods Enzymol 350 : 316–326. 12073321
33. Baetz K, Measday V, Andrews B (2006) Revealing hidden relationships among yeast genes involved in chromosome segregation using systematic synthetic lethal and synthetic dosage lethal screens. Cell Cycle 5 : 592–595. 16582600
34. Measday V, Baetz K, Guzzo J, Yuen K, Kwok T, et al. (2005) Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc Natl Acad Sci U S A 102 : 13956–13961. 16172405
35. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13 : 2498–2504. 14597658
36. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, et al. (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25 : 1091–1093. doi: 10.1093/bioinformatics/btp101 19237447
37. McAleenan A, Clemente-Blanco A, Cordon-Preciado V, Sen N, Esteras M, et al. (2013) Post-replicative repair involves separase-dependent removal of the kleisin subunit of cohesin. Nature 493 : 250–254. doi: 10.1038/nature11630 23178808
38. Jensen S, Segal M, Clarke DJ, Reed SI (2001) A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J Cell Biol 152 : 27–40. 11149918
39. Aye M, Irwin B, Beliakova-Bethell N, Chen E, Garrus J, et al. (2004) Host factors that affect Ty3 retrotransposition in Saccharomyces cerevisiae. Genetics 168 : 1159–1176. 15579677
40. Griffith JL, Coleman LE, Raymond AS, Goodson SG, Pittard WS, et al. (2003) Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164 : 867–879. 12871900
41. Irwin B, Aye M, Baldi P, Beliakova-Bethell N, Cheng H, et al. (2005) Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res 15 : 641–654. 15837808
42. Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ (2008) Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178 : 197–214. doi: 10.1534/genetics.107.082602 18202368
43. Scholes DT, Banerjee M, Bowen B, Curcio MJ (2001) Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159 : 1449–1465. 11779788
44. Checkley MA, Nagashima K, Lockett SJ, Nyswaner KM, Garfinkel DJ (2010) P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol Cell Biol 30 : 382–398. doi: 10.1128/MCB.00251-09 19901074
45. Risler JK, Kenny AE, Palumbo RJ, Gamache ER, Curcio MJ (2012) Host co-factors of the retrovirus-like transposon Ty1. Mob DNA 3 : 12. doi: 10.1186/1759-8753-3-12 22856544
46. Bryk M, Banerjee M, Conte D Jr., Curcio MJ (2001) The Sgs1 helicase of Saccharomyces cerevisiae inhibits retrotransposition of Ty1 multimeric arrays. Mol Cell Biol 21 : 5374–5388. 11463820
47. Sundararajan A, Lee BS, Garfinkel DJ (2003) The Rad27 (Fen-1) nuclease inhibits Ty1 mobility in Saccharomyces cerevisiae. Genetics 163 : 55–67. 12586696
48. Burkett TJ, Garfinkel DJ (1994) Molecular characterization of the SPT23 gene: a dosage-dependent suppressor of Ty-induced promoter mutations from Saccharomyces cerevisiae. Yeast 10 : 81–92. 8203154
49. Fassler JS, Winston F (1988) Isolation and analysis of a novel class of suppressor of Ty insertion mutations in Saccharomyces cerevisiae. Genetics 118 : 203–212. 2834263
50. Clark-Adams CD, Norris D, Osley MA, Fassler JS, Winston F (1988) Changes in histone gene dosage alter transcription in yeast. Genes Dev 2 : 150–159. 2834270
51. Malone EA, Clark CD, Chiang A, Winston F (1991) Mutations in SPT16/CDC68 suppress cis - and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol Cell Biol 11 : 5710–5717. 1922073
52. Winston F, Chaleff DT, Valent B, Fink GR (1984) Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107 : 179–197. 6329902
53. Curcio MJ, Garfinkel DJ (1992) Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol Cell Biol 12 : 2813–2825. 1317008
54. Yamamoto A, Guacci V, Koshland D (1996) Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J Cell Biol 133 : 85–97. 8601616
55. Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, et al. (2004) Exploration of essential gene functions via titratable promoter alleles. Cell 118 : 31–44. 15242642
56. Curcio MJ, Garfinkel DJ (1991) Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci U S A 88 : 936–940. 1846969
57. Winston F, Durbin KJ, Fink GR (1984) The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39 : 675–682. 6096019
58. Rattray AJ, Shafer BK, Garfinkel DJ (2000) The Saccharomyces cerevisiae DNA recombination and repair functions of the RAD52 epistasis group inhibit Ty1 transposition. Genetics 154 : 543–556. 10655210
59. Lee BS, Bi L, Garfinkel DJ, Bailis AM (2000) Nucleotide excision repair/TFIIH helicases RAD3 and SSL2 inhibit short-sequence recombination and Ty1 retrotransposition by similar mechanisms. Mol Cell Biol 20 : 2436–2445. 10713167
60. Ji H, Moore DP, Blomberg MA, Braiterman LT, Voytas DF, et al. (1993) Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73 : 1007–1018. 8388781
61. Baller JA, Gao J, Stamenova R, Curcio MJ, Voytas DF (2012) A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon. Genome Res 22 : 704–713. doi: 10.1101/gr.129585.111 22219511
62. Mularoni L, Zhou Y, Bowen T, Gangadharan S, Wheelan SJ, et al. (2012) Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res 22 : 693–703. doi: 10.1101/gr.129460.111 22219510
63. Jager H, Herzig B, Herzig A, Sticht H, Lehner CF, et al. (2004) Structure predictions and interaction studies indicate homology of separase N-terminal regulatory domains and Drosophila THR. Cell Cycle 3 : 182–188. 14712087
64. Viadiu H, Stemmann O, Kirschner MW, Walz T (2005) Domain structure of separase and its binding to securin as determined by EM. Nat Struct Mol Biol 12 : 552–553. 15880121
65. Lu Y, Cross F (2009) Mitotic exit in the absence of separase activity. Mol Biol Cell 20 : 1576–1591. doi: 10.1091/mbc.E08-10-1042 19144818
66. Stegmeier F, Amon A (2004) Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet 38 : 203–232. 15568976
67. Bachman N, Gelbart ME, Tsukiyama T, Boeke JD (2005) TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev 19 : 955–964. 15833918
68. Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T (2005) Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev 19 : 942–954. 15833917
69. Kenna MA, Brachmann CB, Devine SE, Boeke JD (1998) Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol 18 : 1115–1124. 9448009
70. McLane LM, Pulliam KF, Devine SE, Corbett AH (2008) The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus. Nucleic Acids Res 36 : 4317–4326. doi: 10.1093/nar/gkn383 18586821
71. Moore SP, Rinckel LA, Garfinkel DJ (1998) A Ty1 integrase nuclear localization signal required for retrotransposition. Mol Cell Biol 18 : 1105–1114. 9448008
72. Michaelis C, Ciosk R, Nasmyth K (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91 : 35–45. 9335333
73. Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, et al. (1999) Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev 13 : 320–333. 9990856
74. Kulemzina I, Schumacher MR, Verma V, Reiter J, Metzler J, et al. (2012) Cohesin rings devoid of Scc3 and Pds5 maintain their stable association with the DNA. PLoS Genet 8: e1002856. doi: 10.1371/journal.pgen.1002856 22912589
75. D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, et al. (2008) Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22 : 2215–2227. doi: 10.1101/gad.1675708 18708580
76. Lopez-Serra L, Kelly G, Patel H, Stewart A, Uhlmann F (2014) The Scc2-Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome-free regions. Nat Genet 46 : 1147–1151. doi: 10.1038/ng.3080 25173104
77. Annaluru N, Muller H, Mitchell LA, Ramalingam S, Stracquadanio G, et al. (2014) Total synthesis of a functional designer eukaryotic chromosome. Science 344 : 55–58. doi: 10.1126/science.1249252 24674868
78. Devine SE, Boeke JD (1996) Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev 10 : 620–633. 8598291
79. Hani J, Feldmann H (1998) tRNA genes and retroelements in the yeast genome. Nucleic Acids Res 26 : 689–696. 9443958
80. Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF (1998) Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res 8 : 464–478. 9582191
81. Nagao K, Adachi Y, Yanagida M (2004) Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature 430 : 1044–1048. 15329725
82. Blat Y, Kleckner N (1999) Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98 : 249–259. 10428036
83. Megee PC, Mistrot C, Guacci V, Koshland D (1999) The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell 4 : 445–450. 10518226
84. Snider CE, Stephens AD, Kirkland JG, Hamdani O, Kamakaka RT, et al. (2014) Dyskerin, tRNA genes, and condensin tether pericentric chromatin to the spindle axis in mitosis. J Cell Biol 207 : 189–199. doi: 10.1083/jcb.201405028 25332162
85. Hoang C, Ferre-D'Amare AR (2001) Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107 : 929–939. 11779468
86. Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K (2001) Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105 : 459–472. 11371343
87. Thompson M, Haeusler RA, Good PD, Engelke DR (2003) Nucleolar clustering of dispersed tRNA genes. Science 302 : 1399–1401. 14631041
88. Wang BD, Yong-Gonzalez V, Strunnikov AV (2004) Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle 3 : 960–967. 15190202
89. Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A (2005) Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci U S A 102 : 17308–17313. 16260736
90. Goldstein AL, McCusker JH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 : 1541–1553. 10514571
91. Tong AH, Boone C (2006) Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol 313 : 171–192. 16118434
92. Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, et al. (1988) Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol 8 : 2159–2165. 2455217
93. Shevchenko A, Chernushevich I, Wilm M, Mann M (2000) De Novo peptide sequencing by nanoelectrospray tandem mass spectrometry using triple quadrupole and quadrupole/time-of-flight instruments. Methods Mol Biol 146 : 1–16. 10948493
94. Ishihama Y, Rappsilber J, Mann M (2006) Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J Proteome Res 5 : 988–994. 16602707
95. Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75 : 663–670. 12585499
96. Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2 : 1896–1906. 17703201
97. Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, et al. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 4 : 2010–2021. 16249172
98. Eichinger DJ, Boeke JD (1988) The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell 54 : 955–966. 2843295
99. Teste MA, Duquenne M, Francois JM, Parrou JL (2009) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol 10 : 99. doi: 10.1186/1471-2199-10-99 19874630
100. Lee BS, Lichtenstein CP, Faiola B, Rinckel LA, Wysock W, et al. (1998) Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics 148 : 1743–1761. 9560391
101. Mou Z, Kenny AE, Curcio MJ (2006) Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics 172 : 2157–2167. 16415356
102. Conte D Jr., Barber E, Banerjee M, Garfinkel DJ, Curcio MJ (1998) Posttranslational regulation of Ty1 retrotransposition by mitogen-activated protein kinase Fus3. Mol Cell Biol 18 : 2502–2513. 9566871
Štítky
Genetika Reprodukčná medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



