-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
Our neurons can become over a hundred years old. Even if neurons are restructured and remodeled by their constant work of receiving, storing and sending information, they stay devoted to one single task and retain their identity for their whole life. How a neuron keeps its identity is not well understood. In the olfactory system, the identity of the olfactory sensory neuron (OSN) is a result of the expression of a single odorant receptor (OR) from a large receptor gene repertoire in the genome. Neurons that share an expressed receptor make a functional class. Here, we identify clusters of transcription factor binding motifs to be the smallest unit that drive expression in a single olfactory sensory neuron class. We further demonstrate that it is the structure of the cluster that determines the class specific expression. However, environmental stress, such as temperature changes or starvation, destabilizes the expression produced by the cluster. Our results demonstrate that stable expression is generated from redundant motifs outside the cluster and suggest that cooperative regulation generates robust expression of the genes that determine neuronal identity and function.
Published in the journal: Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in. PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005051
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005051Summary
Our neurons can become over a hundred years old. Even if neurons are restructured and remodeled by their constant work of receiving, storing and sending information, they stay devoted to one single task and retain their identity for their whole life. How a neuron keeps its identity is not well understood. In the olfactory system, the identity of the olfactory sensory neuron (OSN) is a result of the expression of a single odorant receptor (OR) from a large receptor gene repertoire in the genome. Neurons that share an expressed receptor make a functional class. Here, we identify clusters of transcription factor binding motifs to be the smallest unit that drive expression in a single olfactory sensory neuron class. We further demonstrate that it is the structure of the cluster that determines the class specific expression. However, environmental stress, such as temperature changes or starvation, destabilizes the expression produced by the cluster. Our results demonstrate that stable expression is generated from redundant motifs outside the cluster and suggest that cooperative regulation generates robust expression of the genes that determine neuronal identity and function.
Introduction
The expression of developmental genes is regulated such that they are either on or off at the appropriate time and in the correct place. The expression patterns of these genes must be robust; they must be stable and resistant to changes in both external and internal environments [1]. Mechanisms underlying developmental buffering and resistance to temperature changes and mutation have been described. Redundant enhancers act together to support gene expression and robustness under adverse conditions [2]. microRNAs silence ectopically expressed transcripts and buffer steady-state gene expression by controlling the levels of repressors or activators [3]. For genes expressed in mature cells, the demand for robust expression and spatial regulation is even more pronounced. Neurons, for example, are remarkably robust: their function can be maintained for one hundred years, implying that gene function is also maintained during this time. How the high requirement for stability is integrated into the continuous gene regulation that occurs in mature cells is poorly understood.
The high regulatory demands placed on the nervous system are typified by the olfactory sensory system, in which each olfactory sensory neuron (OSN) expresses only one olfactory receptor (OR) gene from its genomic repertoire of one hundred to one thousand ORs [4–6]. OSNs expressing the same OR project their axons to the same glomerulus in the brain and create a functional unit, the OSN class, that exists in both insects and mammals [6]. The restriction of OR expression to a single OSN class is crucial for the perception of odors as changes in OR expression pattern produce a mix of ORs in each OSN class, distorting the response properties of the class and thereby impairing odor detection [7].
Despite the difference in the number of ORs between mouse (1432 OR genes) and Drosophila (62 OR genes), there are common themes in the regulation of OR expression in these model organisms. In addition to class-specific expression, ORs exhibit spatially restricted expression patterns in each olfactory tissue [8,9]. OR gene expression is regulated by a small number of transcription factors (TFs) [10–17], and in Drosophila, these TFs regulate OR expression in a combinatorial fashion [16]. Directly upstream of each OR, there is a short cis-regulatory region sufficient for driving expression in OSNs but generally is insufficient for restricting expression to a single OSN class [16,18–20]. Searches for DNA binding motifs in OR cis-regulatory regions have not identified a direct regulatory “code” but, rather, an enrichment of motifs upstream of regulated OR genes [16]. Similarly, the identified TFs in Drosophila show little of the spatial and temporal specificity expected for combinatorial regulation, implying that it is neither the presence of a TF in an OSN nor the motif upstream of the OR that restricts expression to one OSN class.
Class-specific OR expression is generated in Drosophila in part by input from upstream repressive regions [16,20]. In vertebrates, a single OR allele is expressed in each OSN class [21,22]. The expression of a functional vertebrate OR creates a negative feedback loop [23–25] that reduces the expression of the H3K9 demethylase Lsd1 [26] and locks the expressed OR allele into a stable, robust expression state while suppressing the expression of other ORs [27]. Without monoallelic expression, vertebrate OR expression is not stable and robust within a single OSN class [18]. Drosophila ORs, like the majority of genes, are not expressed monoallelically and lack a feedback system, suggesting that other mechanisms must exist to ensure robust, specific biallelic gene expression.
Here, we address the cis-regulatory mechanisms that result in the precise and robust expression of Drosophila ORs. We utilize the short, well-defined cis-regulatory regions upstream of ORs that limit expression to a single class and the fact that projections from each OSN class form a stereotyped pattern, enabling the direct visualization of expression specificity. Our results demonstrate that structured motif clusters involving one to several TFs located directly upstream of the OR gene provide spatially restricted regulation to a single OSN class. We also show that cooperative gene regulation is a mechanism by which expression variability is buffered and the correct expression of ORs is ensured.
Results
Previously, we showed that TFs bind to motifs upstream not only of ORs that they regulate but also of non-regulated ORs [16], suggesting that specific regulation requires a structure or order of motifs. A typical TF DNA binding motif is 8–10 bps in length and consists of a central 3–4 bp core motif flanked by a 2–4 bps segment with a supporting function [28,29]. To identify possible motif patterns, we determined the location of known core motifs in 32 different OR upstream regions. The scan included five core motifs bound by three TFs that regulate OR gene expression (S1 Table). All motifs were found upstream of all analyzed ORs, with the exception of the regulatory region of Or19a, which lacked E-boxes (the Fer1 motif).
A cluster of Acj6Hox/Pou motif dimers regulates Or85a expression
To identify putative motif patterns that regulate OR expression, we focused on the regulation of the Or85a gene. The Or85a upstream region lacks published DNA binding motifs for Acj6, the sole TF regulating its expression. Acj6 and its vertebrate orthologs have two DNA binding domains, Hox and Pou, which bind two very different sequences: the Hox core motif (AATTA; [30–32]) and the Pou core motif (TGCAA/T; [29,33]), respectively. Within the first 1000 bp upstream of Or85a, we identified 17 Pou and 7 Hox core motifs. Several of the Hox and Pou motifs exhibited a possible dimer arrangement. A search of all 32 analyzed OR upstream regions showed an array of similar Hox/Pou dimers with variations in the spacing between the motifs (exemplified in Fig. 1A). Constructs with pairs of Acj6Hox/Pou dimers placed upstream of a synthetic minimal promoter fused to CD8:GFP did not induce expression (Figs. 1B and S1B), indicating that motifs from other TFs or spatial arrangements support Acj6 dimer function. Three of the Acj6Hox/Pou motif dimers upstream of Or85a generated a condensed cluster (Fig. 1A). To test whether the cluster was sufficient for expression in OSNs, we placed the cluster directly upstream of a minimal promoter fused to CD8:GFP. The Or85a Acj6 dimer cluster resulted in GFP expression specific to Ab2b OSN class neurons, which express Or85a and innervate DM5 (Fig. 1B, C). All insertions of the transgene resulted in equally strong and specific expression (S2 Table), demonstrating that the cluster is sufficient for expression in the correct OSN class, independent of the locus of insertion. Knockdown of acj6 abolished the expression of the construct (Fig. 1D), showing that Acj6 likely binds the Hox/Pou dimers and the cluster then regulates the expression of Or85a.
Fig. 1. Motif clusters regulate OR class-specific expression. 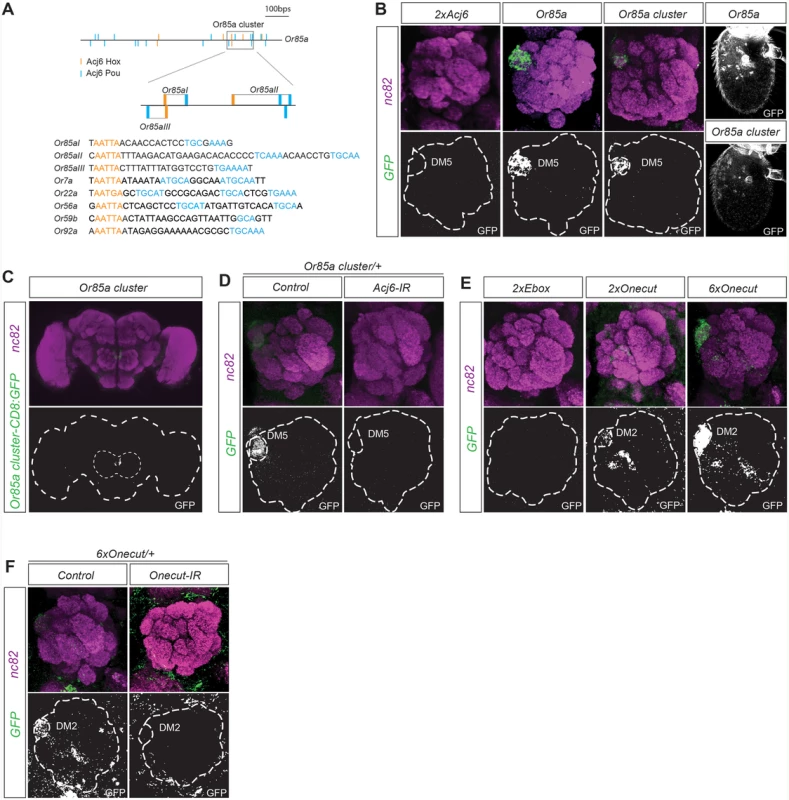
(A) Diagram depicting the 1000-bp upstream region of the Or85a gene and the position of the Pou (blue) and Acj6Hox (orange) motifs in this region. The gray box indicates the 117-bp cluster of Acj6Hox/pou motifs. Below is the alignment of the three Or85a Hox/pou clusters with examples of Acj6Hox/pou clusters from other OR regulatory regions. (B) Whole-mount brain staining shows the expression of GFP (green) driven by the Or85a cluster. Synaptic neuropil regions are labeled with the presynaptic marker nc82 (magenta). The marked region defines the whole brain and the antennal lobes. Below each merged image, the GFP channel is shown. (C) The GFP expression pattern driven by the Or85a cis-regulatory region and Or85a cluster in the antennal lobe and antenna. Note that synthetic clusters of tandem Acj6 binding motifs do not produce any GFP expression. (D) Or85a cluster-driven expression is lost in Acj6-IR (IR-inverted repeat). Control flies were crossed to Peb-Gal4. (E) GFP expression driven by synthetic clusters of tandem E-boxes and 2 or 6 Onecut Hox/cut motif dimers. Note the dose-dependent expression. (F) Loss of expression of the 6×Onecut cluster is observed in Onecut-IR flies. Control flies were crossed to Peb-Gal4. Onecut motif dimers produce OSN class-specific expression
To identify the smallest regulatory unit sufficient for OSN expression, we generated synthetic motif constructs. Constructs with pairs of E-boxes did not induce expression (Fig. 1E), indicating that E-boxes are insufficient for OR gene regulation. Onecut binds to Cut and Hox motifs spaced 2–6 bps apart [29]. A pattern scan revealed fixed Hox/Cut dimers upstream of 71% of the OR genes regulated by Onecut compared with 8% of those not regulated by Onecut, almost a tenfold enrichment. To investigate whether Hox/Cut dimers are sufficient to drive gene expression in OSNs with Onecut-regulated OR genes, we made constructs with two or six dimers directly upstream of a minimal promoter fused to CD8:GFP. The Onecut Hox/Cut dimers produced expression specific to Ab3a OSN class neurons, which express Or22a and innervate the DM5 glomerulus (Fig. 1E). Interestingly, Or22a expression is regulated by Onecut [16], and in our pattern scan, the closest Hox/Cut dimer to the consensus was found upstream of Or22a. Increasing the number of Hox/Cut dimers in the construct from 2 to 6 increased the expression level in Ab3a OSNs as well as the number of insertions that were expressed (Fig. 1E and S2 Table). Knockdown of Onecut attenuated the expression driven by the sextet (Fig. 1F), indicating a direct regulation by Onecut. Our results thus show that motif dimers specific to one TF provide sufficient regulatory information to specifically drive gene expression in a single OSN class.
A motif cluster incorporates the combinatorial cis regulation of Or59b
To identify how the combinatorial input stemming from multiple TFs regulates OR gene expression, we focused on Or59b, whose expression is regulated by three factors with known DNA binding properties: acj6, Fer1 and pdm3 [16] (S2 Fig.). Pdm3 binds to a Hox motif (TAAT) 2–3 bp upstream of a Pou motif (TGCAA/T) [34]. One Pdm3 Hox/Pou motif dimer was identified upstream of Or59b. Interestingly, the Pdm3 Pou motif overlapped with an E-box, the motif that binds bHLH proteins such as Fer1, and was directly downstream of one of the two Acj6Hox motifs (Fig. 2A), indicating that the motifs for all three TFs that regulate Or59b are clustered. This small, 36 bp cluster drove expression in between 30 and 50 OSNs in the proximal region of the antenna (Fig. 2C). Analysis of the axonal projections to the antennal lobes showed that the Or59b cluster produced expression was confined to two OSN classes: Ab2a, which expresses Or59b and innervates the DM4 glomerulus, and Ab7b, which expresses Or67c and innervates the VC4 glomerulus (Fig. 2B, C). Knockdown of the 3 TFs that regulate Or59b expression, acj6, Fer1 and pdm3 resulted in the loss of Or59b cluster produced expression in both OSN classes (Fig. 2D), implying that this TF combination does not segregate the Ab2a and Ab7a classes. We have previously shown that the co-repressor Atro represses Or59b expression specifically in the Ab7a OSN class [35]. Overexpression or knockdown of Atro did not attenuate reporter expression driven by the Or59b cluster in the Ab7a OSN class (Fig. 2D), demonstrating that Atro represses Or59b expression via a mechanism that is separate from the cluster and that restricts OR expression to a single class.
Fig. 2. One motif cluster combines the TF regulation of Or59b gene expression. 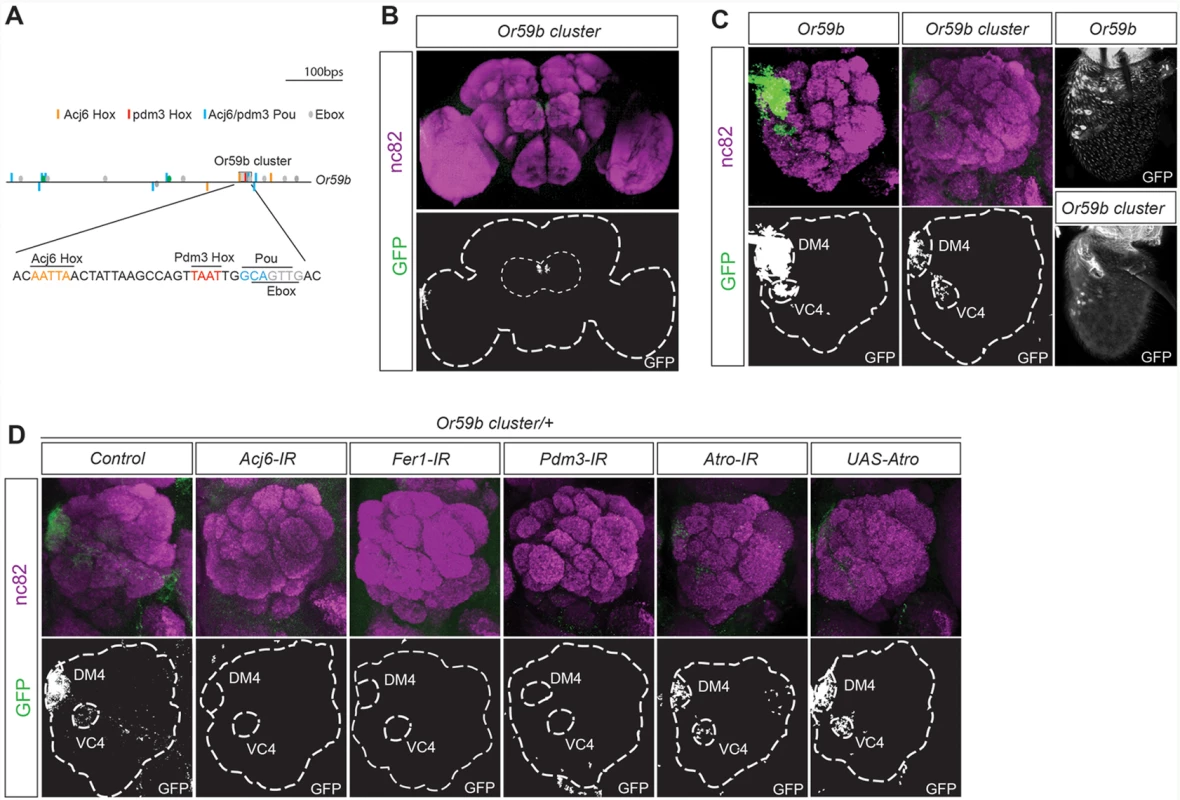
(A) Diagram of the 1000-bp upstream region of the Or59b gene showing the locations of the Pou (blue), Acj6Hox (orange), Pdm3Hox (red) and E-box (green) motifs. The gray box marks the cluster of Hox/pou/E-box motifs. Below, the 36-bp Or59b cluster sequence is presented. (B-D) A whole-mount brain shows GFP expression driven by the Or59b cluster (green) and the synaptic neuropil marked by nc82 (magenta). The marked region defines the whole brain and the antennal lobes. (C) GFP expression from the Or59b reporter and Or59b cluster in the antenna and in the antennal lobe, where it marks axonal projections to the DM4 and VC4 glomeruli. (D) Loss of expression produced by the Or59b cluster is observed in the Acj6-, Fer1- and Pdm3-IRs but not in Atro-IR or UAS-Atro overexpression lines. Control flies were crossed to Peb-Gal4. The ratio of Acj6 and Pdm3 specifies the Or59b cluster-produced expression
To investigate the regulatory function of each TF, we made constructs with mutations in the different motifs belonging to the Or59b cluster. Mutation of the E-box resulted in a total loss of reporter expression (Figs. 3A and S3A), indicating that bHLH proteins induce its expression. Mutation of the Acj6 and Pdm3 Pou motif caused loss of expression, whereas mutation of each Hox motif produced ectopic expression (Figs. 3A and S3A), indicating that the repressive or inductive function of Acj6 and Pdm3 is dictated by the Hox motif. Further genetic analyses demonstrated that pdm3 is downstream of acj6 and that both can either repress or activate the cluster function and expression (S3B–S3C Fig.). To explore whether the cluster interprets the protein levels of Pdm3 and Acj6, we manipulated the level of each factor. The expression of the cluster was sensitive to the loss of one copy of acj6 but not of pdm3 (Fig. 3B). The loss of expression was rescued by lowering the copy number of both factors (Fig. 3B), demonstrating that the ratio of Acj6 to Pdm3 creates a window of cluster function that limits Or59b expression to two OSN classes.
Fig. 3. The Acj6 to Pdm3 ratio dictates the Or59b cluster-driven expression. 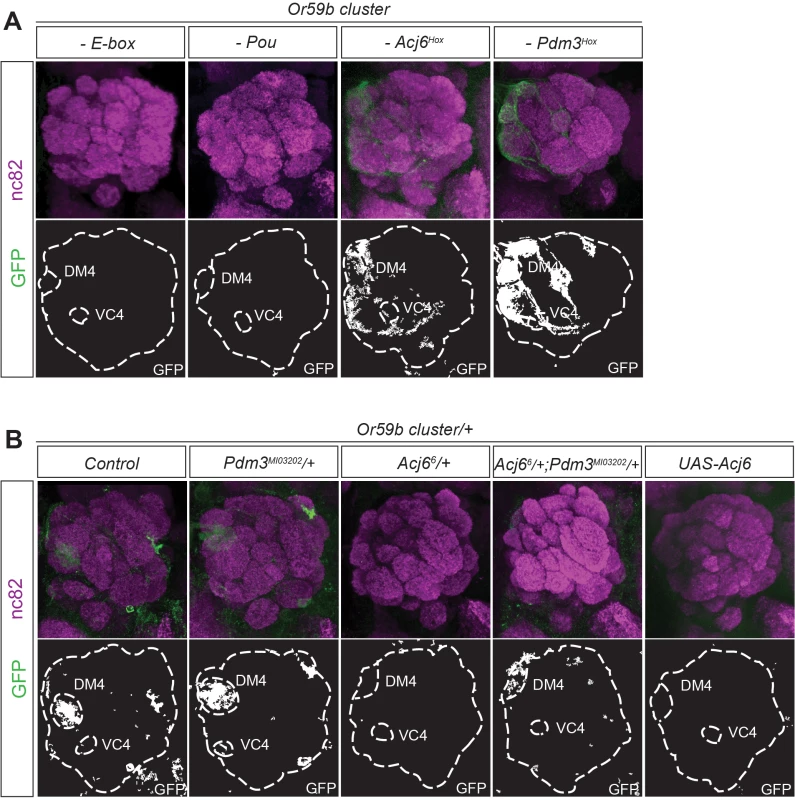
(A) GFP (green) expression driven by Or59b cluster constructs with mutated Acj6Hox, Pdm3Hox, Pou and E-box motifs. Synaptic neuropil regions are labeled with the presynaptic marker nc82 (magenta). (B) GFP expression driven by the Or59b cluster in different backgrounds. Loss of expression of the Or59b cluster is observed in acj66 +/− flies and is rescued in acj66/pdm3MIO3202 flies. Control flies were crossed to w1118. Schematic interpretations of the results are presented in S5 Fig. The structure of the Or59b cluster generates OSN class-specific expression
To investigate whether cluster structure regulates expression, we rearranged the order of the motifs in the Or59b cluster. First, we moved the E-box 125 bp downstream the cluster, which disrupted expression (Figs. 4B and S4A), demonstrating that the combinatorial clustering of the motifs was required for expression. Next, we addressed the regulatory function of the overlap between the Pou motif and the E-box. Moving the E-box upstream of the cluster caused ectopic expression in seven OSN classes, Ab1a, Ab2a, Ab3a, Ab5b, Ab7b, Ab8a, Ab8b and Ab10b (Figs. 4C and S4A), indicating that the precise location of the E-box dictates a repressive function necessary for class-specific OR expression. To further address how TFs binding at the E-box and Pou motifs interact, we moved the E-box either one-half or a full DNA turn (5 or 10 bp, respectively, S1C Fig.) that placed the TFs at different phases and sides of the DNA. Both constructs resulted in stereotyped ectopic expression in the seven OSN classes (Figs. 4D, E and S4A). As both the Ebox and the Pou motifs were shown to be required for cluster function (Fig. 3A), the above results indicate that occupancy of either of the two motifs interferes with the other and causes the repression of expression. Further genetic analyses placed Fer1 downstream of the Hox/Pou factors (S4B–S4C Fig.). Together, these results demonstrate that the composition and relative positions of motifs within the cluster define and restrict the expression of Or59b.
Fig. 4. Spatial expression pattern is dictated by the structure of the Or59b cluster. 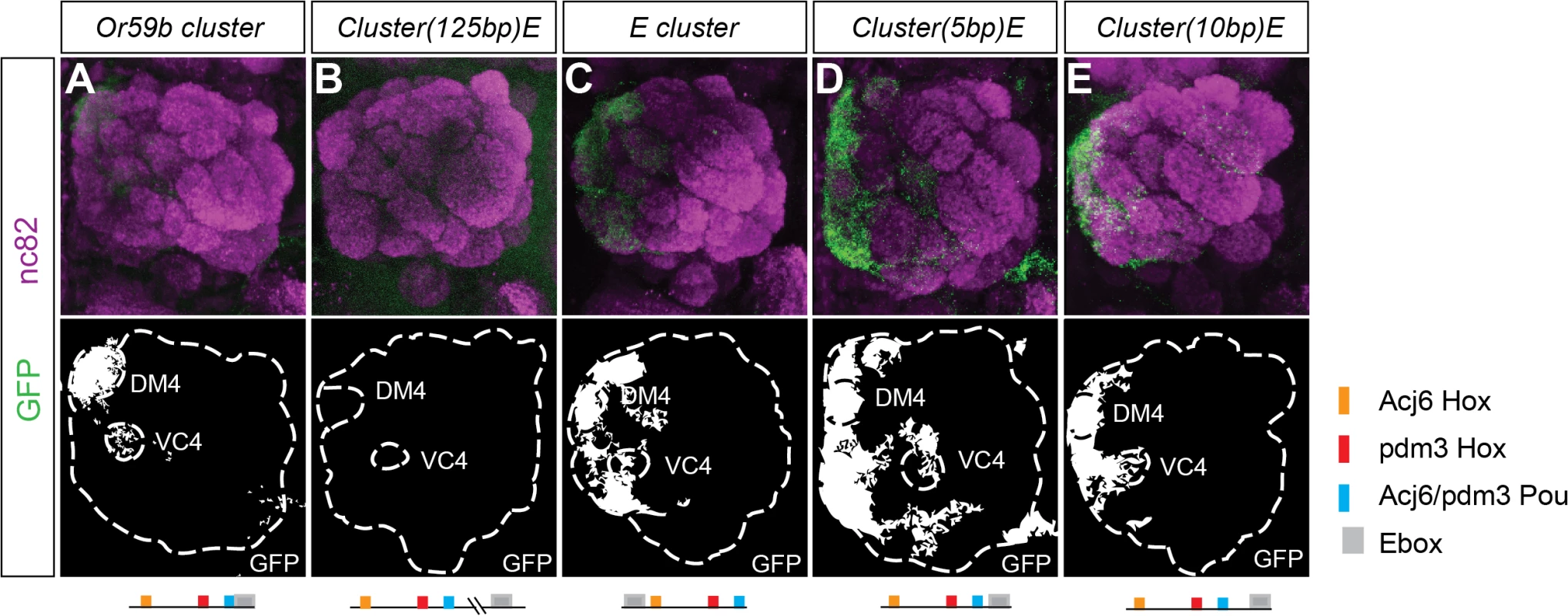
GFP expression (green) produced by (A) the Or59b cluster, (B) the cluster with the E-box displaced 125bps, (C) with the E-box 10bps upstream the cluster, (D-E) the Ebox 5 or 10 bp downstream the cluster. Synaptic neuropil regions are labeled with the presynaptic marker nc82 (magenta). A schematic representation of different rearrangements is shown under each figure. Schematic interpretations of the results are presented in S5 Fig. Starvation and low temperature destabilize Or59b cluster function
For proper odorant perception, OR expression must be active continuously and must be restricted to a single OSN class, despite changes in the environment. To investigate OR gene expression and class-specific transcription under conditions of environmental fluctuation, we first starved flies for three days. qPCR revealed that the mRNA levels of most ORs increased slightly upon starvation (Fig. 5A). Starvation did not change the expression produced by reporter transgenes with the cis regulatory region between Or59b and the gene upstream fused to CD8:GFP (Fig. 5B and S3 Table), showing that robust class-specific expression is encoded by the region directly upstream the gene. Interestingly, starvation attenuated the expression of the Or59b cluster (Fig. 5B and S3 Table). These results show that the cluster lacks the regulatory information required to maintain class-specific expression during starvation.
Fig. 5. Temperature stress or starvation hampers Or59b cluster function. 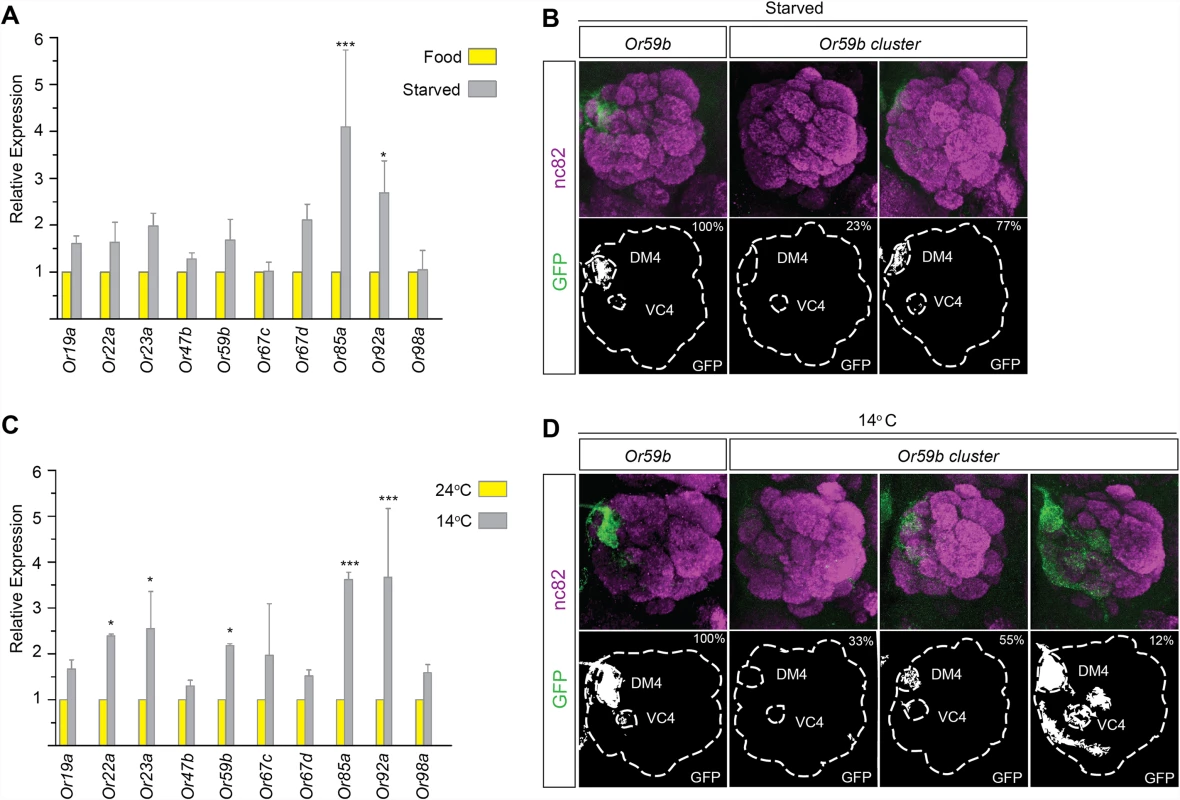
(A) Following 3 days of starvation, the mRNA levels of indicated OR genes were measured by qPCR and compared with control flies (* p < 0.05; ** p < 0.01; error bars represent SEM). (B) GFP expression (green) driven by the Or59b reporter or Or59b cluster following 3 days of starvation. Synaptic neuropil regions are labeled with the presynaptic marker nc82 (magenta). Phenotype penetrance is marked as a percentage at the top-right corner of the image. (C) Following 3 days at 14°C, the mRNA levels of the indicated ORs were measured by qPCR and compared with flies maintained at 24°C (* p < 0.05; ** p < 0.01; *** p < 0.001; error bars represent SEM). (D) GFP expression produced by the Or59b reporter and the Or59b cluster at 14°C. Note that the same insertion of the Or59b cluster construct produces both loss- and gain-of-expression phenotypes at 14°C. Schematic interpretations of the results are presented in S6 Fig. To further investigate the requirements for robust class-specific OR expression, we changed the physical environment of the flies. Flies are stressed by high (>30°C) and low (<15°C) temperatures [36]. We switched flies between 3 and 5 days old to low temperature (14°C) and kept a control group at ambient temperature (24°C) for 3 days. qPCR showed increased mRNA levels of most assayed ORs in flies exposed to low temperature compared with those kept at ambient temperature (Fig. 5C). The Or59b cis-regulatory region drove reporter expression at both low and ambient temperatures (Fig. 5D). Different cluster insertions produced similar expression phenotypes, with stable expression at the ambient temperature, but at low temperature, the cluster produced ectopic expression in several OSN classes in 12%, no expression in 33% and restricted class specific expression in 55% of the analyzed animals (Fig. 5D and S3 Table). These results show that the cluster can support class-specific expression under ambient conditions, but the fine-tuned balance of TF assembly is perturbed in low-temperature or starvation conditions.
Cooperative gene regulation generates robust class-specific OR expression
Because the Or59b cluster produces weak expression compared with the Or59b reporter (Fig. 2C), we investigated whether the level of Or59b expression generates robust class-specific expression. Two tandem copies of the Or59b cluster produced strong expression at ambient temperature and both loss and gain of expression phenotypes at low temperature (Fig. 6A, D), demonstrating that expression level does not buffer against environmentally induced changes and that class specificity is maintained via a separate mechanism. Interestingly, environmental changes did not affect the function of the Or85a cluster (Fig. 6B, D), suggesting that cooperative binding of one TF may be sufficient to drive robust class-specific expression. Of the three TFs known to regulate Or59b, Fer1 has 10 binding sites (E-boxes) outside the cluster that can cooperate with the cluster to regulate Or59b expression (Fig. 2A). To test whether cooperating E-boxes might produce robust Or59b cluster expression, we generated a cluster with two E-boxes. The addition of an extra E-box led to stabilized expression in flies challenged with changes in temperature or food (Fig. 6C, D). A count of GFP-positive OSNs showed that the cluster produced a varied number, from a few cells to over 60 positive cells per antenna, at 14°C (Fig. 6E). The number of positive cells per antenna was fully rescued to the control number by the addition of an extra E-box, demonstrating that the cooperative function of single motifs around a cluster can stabilize the assembly and expression produced by the cluster.
Fig. 6. Cooperative regulation between the cluster and surrounding motifs produces robust class-specific expression. 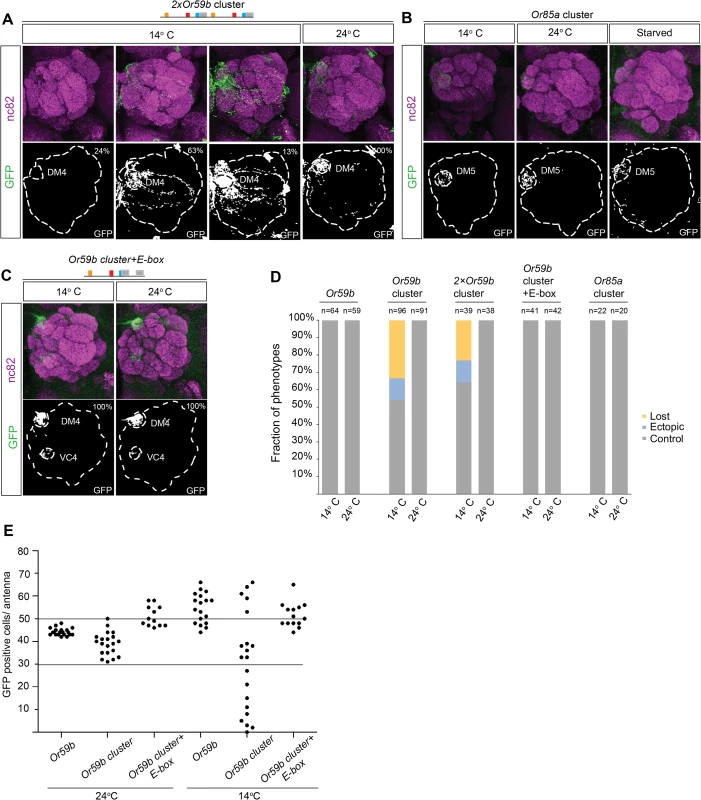
(A) GFP expression (green) driven by the 2×Or59b cluster at 14°C and 24°C. Note the three expression phenotypes produced by the 2×Or59b cluster at 14°C. Synaptic neuropil regions are labeled with the presynaptic marker nc82 (magenta). (B) GFP expression (green) produced by the Or85a cluster at 14°C, 24°C or following 3 days of starvation. Note that GFP expression is equally strong in different lines at 14°C. (C) The Or59b cluster with an additional E-box produces robust expression at 14°C and 24°C. (D) The fractions of the brains showing stable or bimodal expression of GFP according to the genotype and temperature. Note that only the Or59b cluster and 2×Or59b cluster are unstable at 14°C. (E) Quantification of GFP positive cells in the antenna. Or59b cluster shows a varied number of GFP positive cells at 14°C compared to 24°C. Or59b and Or59b cluster with an additional E-box show stable expression in both temperatures. Schematic interpretations of the results are presented in S6 Fig. Epigenetic state controls Or59b cluster function
The variability of expression produced by different Or59b cluster insertions (S2 Table) and the general increase of OR expression upon environmental changes suggested a general epigenetic mechanism for regulation. In mice, H3K9 trimethylation, a marker of heterochromatin, is required for stable and robust class-specific OR expression [27,37]. To address whether changes in H3K9 trimethylation control Or59b cluster function, we introduced a mutant allele of su(var)3–9, the enzyme that trimethylates H3K9 [38]. Or59b reporters showed robust expression in su(var)3–9 heterozygote flies (Fig. 7A). By contrast, each Or59b cluster insertion showed both gain - and loss-of-expression phenotypes in su(var)3–9+/− flies (Fig. 7A), indicating that H3K9 methylation status modulates gene expression driven by the cluster.
Fig. 7. Heterochromatin modulation of the Or59b cluster. 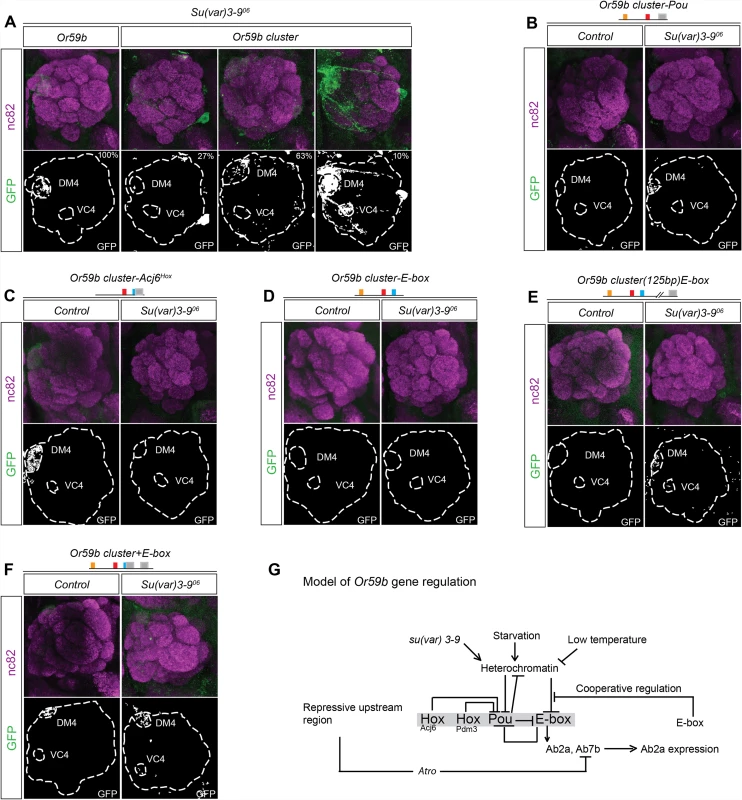
(A) GFP expression (green) driven by the Or59b reporter or Or59b cluster in su(var)3–906 heterozygote flies. Synaptic neuropil regions are labeled with the presynaptic marker nc82 (magenta). Control flies were crossed to w1118. (B-E) GFP expression (green) driven by mutated Or59b cluster versions in su(var)3–906 heterozygote flies. Note that the loss of GFP expression driven by the Or59b cluster with a mutated Pou motif or with a distant E-box is rescued in su(var)3–906 heterozygote flies. (F) GFP expression (green) driven by an Or59b cluster with an additional E-box in su(var)3–906 heterozygote flies. Note that the E-box rescues the produced su(var)3–906 expression phenotypes. (G) Model depicting the function of the cluster in the regulation of Or59b expression. Our results propose that the Hox/Pou motif regulates the heterochromatin state and allows bHLH proteins to bind the E-box, which induces expression. The E-box and Pou motif sequences overlap to generate unstable binding, and a steady state is generated that drive expression in the Ab2a and Ab7b OSN classes. Cooperative interactions between E-boxes stabilize expression in the face of environmental perturbations. Schematic models of the results are presented in S5–S6 Figs. To address how TF assembly at the cluster interacts with the assembly of heterochromatin, we crossed the various cluster versions to the su(var) 3–9 mutant. In the heterozygote su(var)3–9 background the ectopic expression of the Acj6Hox mutant cluster was lost (Fig. 7C), indicating that the epigenetic status at the cluster determines the function of Acj6 and Pdm3. Moreover, the attenuated expression of the Pou mutant cluster was weakly rescued in the heterozygote background (Fig. 7B), indicating that the Hox/Pou TFs generate the open chromatin required for the induction of Or59b expression. Interestingly, the loss of expression by the mutated E-box was not rescued in su(var)3–9 heterozygotes (Fig. 7D), placing the heterochromatin regulation downstream of the Hox/Pou factors and upstream of the E-box. The loss of heterochromatin in the heterozygote background further induced expression of the cluster with the E-box displaced by 125 bp (Fig. 7E), supporting the notion that Hox/Pou factors open chromatin and allow for the binding of bHLH proteins to the E-box (modeled in Figs. 7G and S5). As with the temperature - and starvation-induced phenotypes, the additional E-box construct rescued the su(var) 3–9 phenotypes (Fig. 7F), implying that stabilization of TF binding at the E-boxes buffers the cluster function. Our results thus support a model in which the Hox - and Pou-binding proteins open chromatin and let bHLH proteins to bind the E-box that induce expression. The bHLH binding compete with the Hox/Pou proteins and cooperation between additional E-boxes stabilize bHLH binding and buffers Or59b expression from variation in epigenetic and environmental states (Fig. 7G; regulatory models of each phenotype are presented in S5–S6 Figs).
Discussion
Here, we demonstrate that clusters of low-affinity motifs generate restricted OR expression. We show that the composition of the motif cluster sets requirements for expression that are only fulfilled by the OSNs of one or a few classes. We further show that TFs with two DNA-binding domains can recognize simple motif clusters that restrict expression to a single class and that complex clusters with motifs corresponding to multiple TFs integrate the combinatorial information from TFs and chromatin status to produce class-restricted expression. Finally, we show that cooperation among redundant motifs, or perhaps even clusters, generates the robust single class OR expression required to support odorant detection throughout the life of the organism.
Low-affinity motifs can be advantageous for gene regulation
The motifs identified in this study are short, are likely to have low affinity and are abundant in OR cis-regulatory regions. Nonetheless, these motifs are sufficient to regulate the restriction of OR expression to a single class. Our results further imply that low information value of a motif can be an advantage for several reasons. First, TF binding to low-affinity motifs requires cooperative input for stability, thus favoring combinatorial and patterned gene regulation. Second, a high on/off rate supports competition among TFs at overlapping motifs, which we show is crucial for the integration of combinatorial input from several TFs to restrict OR transcriptional output to a single class. Third, weak TF interactions with the motif facilitate direct chromatin regulation of the locus [39]. Finally, the level of degeneracy of each motif defines the role, and possibly the function, of the TF in each cluster, which increases the use and flexibility of a TF from a static activator to a modulator, as we showed for Acj6/pdm3 by demonstrating both repressive and inductive roles.
Short motifs are evolutionarily unstable because they can be generated or lost with one or two mutations. The olfactory system is evolutionarily very plastic, with the continuous generation and loss of OR genes, implying that selection might regulate the birth and death of weak motifs. Even a complex cluster such as the Or59b cluster can only be found as a unit in the melanogaster clade and shows a large number of changes among species in the clade. Recently, it has been shown that the evolutionary stability of expression patterns differs between vital developmental genes and genes expressed only in mature cells [40,41]. Thus, one possibility is that short motifs are the product of an active selection process. Studies on the conservation of motifs upstream of genes expressed in different tissues and stages indicate that upstream genes expressed in adult tissues, such as the vertebrate liver [42,43], are less conserved than those critical for developmental processes, such as invertebrate segmentation [44,45], and organelle function, such as the Rfx regulation of genes conserved in cilia function [46]. It will therefore be interesting to determine whether the predictive use of conserved large (>8 bps) motifs is limited to the prediction of evolutionarily stable systems, such as development or organelle function, and whether the identification of motif patterns or clusters of core motifs will improve predictions regarding the regulatory function of non-vital genes expressed in mature cells.
Regulation of spatial OR expression by motif clusters
Cooperative regulation of clustered motifs has been shown in Drosophila to restrict regulation by broadly expressed TFs to regions of the embryo [47–49]. Our results show that the simplest switches are motif clusters recognized by one TF. Tandem synthetic consensus Acj6 motif dimers did not result in any expression, but the more complex Or85a Acj6 cluster did, indicating that the structure of the motif cluster can limit the function of a broadly expressed TF to regulate OR gene expression to a single OSN class. Interestingly, despite the fact that onecut is expressed in several OSN classes [16], the cluster of Hox/cut motifs produced expression restricted to a certain class. As TFs with more than one DNA binding domain, such as Acj6 and Onecut (with two binding domains each), can bind to multiple DNA motifs simultaneously [29,30], the arrangement of motifs within a cluster can define expression pattern, and in this manner, a broadly expressed TF can produce a very restricted expression pattern.
The integration of several TFs requires more complex clusters. The Or59b cluster integrates the function of 3 TFs, Acj6 and Pdm3, which bind to two different Hox motifs and compete for one Pou motif, and Fer1, which binds to an E-box that partly overlaps with the Pou motif. Our results show that the competition between Hox/Pou proteins and bHLH proteins allows the cluster to integrate the epigenetic status and the levels of Hox/Pou proteins, which can be summarized in the following model: the Hox/Pou TFs Acj6 and Pdm3 compete for the Pou motif, where Pdm3 likely opens chromatin and facilitates the binding of bHLH proteins to the E-box, which induces Or59b expression. As the bHLH proteins bind the E-box, binding of the Hox/Pou TFs to the cluster is destabilized, reducing the opening of chromatin. In this less-favorable chromatin environment, binding to the E-box is reduced and a steady state is generated. The generated steady state is sufficient to support expression in two OSN classes; the Atrophin complex represses expression in one of two classes, resulting in expression of the Or59b gene in a single OSN class.
Cooperative TF function generates robust class-specific OR expression
Various environmental challenges generated a stereotyped expression phenotype, indicating that a general molecular mechanism underlies robust OSN class expression. One cause of combined gain and loss of expression phenotypes is direct competition between a repressor and activator for one motif [50]. Our results demonstrate that it is the competition between the Hox/Pou TFs and bHLH proteins that generates unstable expression and that cooperative regulation of bHLH proteins bound to the E-box in the cluster and secondary E-boxes beyond the cluster stabilizes OR expression. This stabilization is likely a function of favoring bHLH binding to the E-box in the cluster, thereby reducing the need for Hox/Pou regulation and chromatin opening at the locus.
Even in simple clusters with only one TF motif, cooperative function generates robust expression in the class. Interestingly, robustness to environmental changes during development has been shown to be produced by shadow enhancers [2] (redundant cis-regulatory regions that together support expression [51]) or homotypic motif clusters [52], suggesting that cooperative regulation between motifs and clusters might be a general mechanism through which to maintain restricted gene expression.
Materials and Methods
Drosophila stocks
The Pebbled-Gal4 (Peb-Gal4) and acj66 mutants were kind gifts from Liqun Luo (Stanford University, Stanford, CA, USA). The su(var)3–906 mutant was a kind gift from Anita Öst (Linköping University, Linköping, Sweden). The following fly lines were obtained from the Vienna Drosophila Center (VDRC; Vienna, Austria; http://stockcenter.vdrc.at): Acj6-IR, Atro-IR, Fer1-IR, UAS-Atro, and UAS-Dcr2. The following RNAi lines were obtained from the Transgenic RNAi Project (TRiP; Harvard Medical School, Boston, MA, USA; http://www.flyrnai.org): Fer1-IR (27737; 50672), Onecut-IR (29343), Pdm3-IR (35726, 26749). The following fly lines were provided by the Bloomington Drosophila Stock Center (BDSC; Indiana University, Bloomington, IN, USA; http://flystocks.bio.indiana.edu): w1118, UAS-tub-Gal80ts, Pdm3MI03202 (37337), Pdm3MI01072 (37552).
Bioinformatics
An online pattern search tool (http://www.bioinformatics.org/sms2/dna_pattern.html) was used to scan 1 kb upstream from the translational start site of each OR for 6 motifs recognized by 4 TFs: Acj6 (Hox and Pou), Fer1 (E-box), Onecut (cut and Pou) and Pdm3 (Hox linked to Pou, only for Or59b) (S1 Table).
Cloning
All constructs were synthesized at Genescript and cloned into a transformation vector containing a synthetic TATA region fused to a single ORF that contained the mCD8 transmembrane domain, four tandem copies of GFP, and two c-myc epitope tags, as previously described (Couto et al). The DNA constructs were injected into w1118 flies at BestGene, and six to 12 lines were analyzed per construct.
RNAi methodology and environmental experiments
Virgin flies of the RNAi line were mated with males containing Pebbled-GAL4, UAS-Dicer2, and the cluster transgenes. The crosses were set up and maintained at 24°C and 2–5 d after eclosure, flies were dissected, stained and scored for phenotypes. RNA interference lines for Acj6, Atro, Fer1, and Onecut were previously described [16,35]. Both pdm3 RNA interference lines produced identical phenotypes that were phenocopied by the Pdm3 mutant (Pdm3MI01072).
All flies were raised on standard Drosophila culture medium at 24°C and collected 2–5 days after eclosion unless otherwise specified. w1118 flies were used as controls. In the experimental group, flies were transferred to new vials and maintained for 3 days at 14°C. In the starvation experiments, 2–5-day-old flies were kept in a vial with water-soaked filter paper for 3 days.
Immunofluorescence
Immunofluorescence was performed according to previously described methods [16]). The following primary antibodies were used: rabbit anti-GFP (1 : 2000, TP-401; Torrey Pines Biolabs) and mouse anti-nc82 (1 : 100; DSHB). Secondary antibodies were conjugated with Alexa Fluor 488 (1 : 500; Molecular Probes). Confocal microscopy images were collected on an LSM 700 (Zeiss) and analyzed using an LSM Image Browser. The numbers of co-expressing BP104 and GFP OSNs for different constructs were counted from the images. Adobe Photoshop CS4 (Adobe Systems) was used for image processing.
qPCR
Antennae were obtained with a sieve after freezing 2–5-day-old flies in liquid nitrogen. Total RNA from antennae was extracted with TRIzol reagent (Invitrogen) followed by purification with the RNeasy kit (Qiagen). Quantitative PCR was conducted on an Applied Biosystems 7900HT real-time PCR system (Life Technologies) using the Power SYBR Green PCR master mix (Applied Biosystems, Life Technologies) and primer sets designed using Primer Express software v3.0.1 (Integrated DNA Technologies). Tubulin was used as an internal control for the experiments. To amplify cDNA products and not genomic DNA, primers were designed to join the end of one exon with the beginning of the next exon. Quantitative PCR for each primer set was performed on both control and experimental samples for 40 cycles. Following amplification, melt curve analysis and ethidium bromide agarose gel electrophoresis were performed to evaluate the PCR products. The relative quantification of the fold change in mRNA expression was calculated using the 2−ΔΔCT threshold cycle method.
Supporting Information
Zdroje
1. Lagha M, Bothma JP, Levine M (2012) Mechanisms of transcriptional precision in animal development. Trends Genet 28 : 409–416. doi: 10.1016/j.tig.2012.03.006 22513408
2. Frankel N, Davis GK, Vargas D, Wang S, Payre F, et al. (2010) Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466 : 490–493. doi: 10.1038/nature09158 20512118
3. Ebert MS, Sharp PA (2010) Emerging roles for natural microRNA sponges. Curr Biol 20: R858–861. doi: 10.1016/j.cub.2010.08.052 20937476
4. Couto A, Alenius M, Dickson BJ (2005) Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol 15 : 1535–1547. 16139208
5. Fishilevich E, Vosshall LB (2005) Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol 15 : 1548–1553. 16139209
6. Fuss SH, Ray A (2009) Mechanisms of odorant receptor gene choice in Drosophila and vertebrates. Mol Cell Neurosci 41 : 101–112. doi: 10.1016/j.mcn.2009.02.014 19303443
7. Tharadra SK, Medina A, Ray A (2013) Advantage of the Highly Restricted Odorant Receptor Expression Pattern in Chemosensory Neurons of. PLoS One 8: e66173. 23840419
8. Buck L, Axel R (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65 : 175–187. 1840504
9. Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R (1999) A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96 : 725–736. 10089887
10. Wang SS, Tsai RY, Reed RR (1997) The characterization of the Olf-1/EBF-like HLH transcription factor family: implications in olfactory gene regulation and neuronal development. J Neurosci 17 : 4149–4158. 9151732
11. Hirota J, Mombaerts P (2004) The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc Natl Acad Sci U S A 101 : 8751–8755. 15173589
12. Kolterud A, Alenius M, Carlsson L, Bohm S (2004) The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development 131 : 5319–5326. 15456728
13. McIntyre JC, Bose SC, Stromberg AJ, McClintock TS (2008) Emx2 stimulates odorant receptor gene expression. Chem Senses 33 : 825–837. doi: 10.1093/chemse/bjn061 18854508
14. Tichy AL, Ray A, Carlson JR (2008) A new Drosophila POU gene, pdm3, acts in odor receptor expression and axon targeting of olfactory neurons. J Neurosci 28 : 7121–7129. doi: 10.1523/JNEUROSCI.2063-08.2008 18614681
15. Bai L, Carlson JR (2010) Distinct functions of acj6 splice forms in odor receptor gene choice. J Neurosci 30 : 5028–5036. doi: 10.1523/JNEUROSCI.6292-09.2010 20371823
16. Jafari S, Alkhori L, Schleiffer A, Brochtrup A, Hummel T, et al. (2012) Combinatorial activation and repression by seven transcription factors specify Drosophila odorant receptor expression. PLoS Biol 10: e1001280. doi: 10.1371/journal.pbio.1001280 22427741
17. Sim CK, Perry S, Tharadra SK, Lipsick JS, Ray A (2012) Epigenetic regulation of olfactory receptor gene expression by the Myb-MuvB/dREAM complex. Genes Dev 26 : 2483–2498. doi: 10.1101/gad.201665.112 23105004
18. Vassalli A, Feinstein P, Mombaerts P (2011) Homeodomain binding motifs modulate the probability of odorant receptor gene choice in transgenic mice. Mol Cell Neurosci 46 : 381–396. doi: 10.1016/j.mcn.2010.11.001 21111823
19. Plessy C, Pascarella G, Bertin N, Akalin A, Carrieri C, et al. (2012) Promoter architecture of mouse olfactory receptor genes. Genome Res 22 : 486–497. doi: 10.1101/gr.126201.111 22194471
20. Miller CJ, Carlson JR (2010) Regulation of Odor Receptor Genes in Trichoid Sensilla of the Drosophila Antenna. Genetics.
21. Chess A, Simon I, Cedar H, Axel R (1994) Allelic inactivation regulates olfactory receptor gene expression. Cell 78 : 823–834. 8087849
22. Malnic B, Hirono J, Sato T, Buck LB (1999) Combinatorial receptor codes for odors. Cell 96 : 713–723. 10089886
23. Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, et al. (2003) Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science 302 : 2088–2094. 14593185
24. Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P (2004) Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell 117 : 833–846. 15186782
25. Lewcock JW, Reed RR (2004) A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci U S A 101 : 1069–1074. 14732684
26. Dalton RP, Lyons DB, Lomvardas S (2013) Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell 155 : 321–332. doi: 10.1016/j.cell.2013.09.033 24120133
27. Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, et al. (2013) An epigenetic trap stabilizes singular olfactory receptor expression. Cell 154 : 325–336. doi: 10.1016/j.cell.2013.06.039 23870122
28. Stewart AJ, Hannenhalli S, Plotkin JB (2012) Why transcription factor binding sites are ten nucleotides long. Genetics 192 : 973–985. doi: 10.1534/genetics.112.143370 22887818
29. Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, et al. (2013) DNA-binding specificities of human transcription factors. Cell 152 : 327–339. doi: 10.1016/j.cell.2012.12.009 23332764
30. Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, et al. (2008) Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133 : 1266–1276. doi: 10.1016/j.cell.2008.05.024 18585359
31. Bai L, Goldman AL, Carlson JR (2009) Positive and negative regulation of odor receptor gene choice in Drosophila by acj6. J Neurosci 29 : 12940–12947. doi: 10.1523/JNEUROSCI.3525-09.2009 19828808
32. Gruber CA, Rhee JM, Gleiberman A, Turner EE (1997) POU domain factors of the Brn-3 class recognize functional DNA elements which are distinctive, symmetrical, and highly conserved in evolution. Mol Cell Biol 17 : 2391–2400. 9111308
33. Klemm JD, Pabo CO (1996) Oct-1 POU domain-DNA interactions: cooperative binding of isolated subdomains and effects of covalent linkage. Genes Dev 10 : 27–36. 8557192
34. Andersen B, Rosenfeld MG (2001) POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr Rev 22 : 2–35. 11159814
35. Alkhori L, Ost A, Alenius M (2014) The corepressor Atrophin specifies odorant receptor expression in Drosophila. FASEB J 28 : 1355–1364. doi: 10.1096/fj.13-240325 24334704
36. Hoffmann AA, Sorensen JG, Loeschcke V (2003) Adaptation of Drosophila to temperature extremes: bringing together quantitative and molecular approaches. Journal of Thermal Biology 28 : 175–216.
37. Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, et al. (2011) An epigenetic signature for monoallelic olfactory receptor expression. Cell 145 : 555–570. doi: 10.1016/j.cell.2011.03.040 21529909
38. Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, et al. (2002) Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J 21 : 1121–1131. 11867540
39. Buck MJ, Lieb JD (2006) A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat Genet 38 : 1446–1451. 17099712
40. Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, et al. (2010) Gene expression divergence recapitulates the developmental hourglass model. Nature 468 : 811–814. doi: 10.1038/nature09634 21150996
41. Artieri CG, Singh RS (2010) Molecular evidence for increased regulatory conservation during metamorphosis, and against deleterious cascading effects of hybrid breakdown in Drosophila. BMC Biol 8 : 26. doi: 10.1186/1741-7007-8-26 20356354
42. Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, et al. (2010) Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328 : 1036–1040. doi: 10.1126/science.1186176 20378774
43. Stefflova K, Thybert D, Wilson MD, Streeter I, Aleksic J, et al. (2013) Cooperativity and rapid evolution of cobound transcription factors in closely related mammals. Cell 154 : 530–540. doi: 10.1016/j.cell.2013.07.007 23911320
44. Bradley RK, Li XY, Trapnell C, Davidson S, Pachter L, et al. (2010) Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol 8: e1000343. doi: 10.1371/journal.pbio.1000343 20351773
45. He Q, Bardet AF, Patton B, Purvis J, Johnston J, et al. (2011) High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet 43 : 414–420. doi: 10.1038/ng.808 21478888
46. Piasecki BP, Burghoorn J, Swoboda P (2010) Regulatory Factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc Natl Acad Sci U S A 107 : 12969–12974. doi: 10.1073/pnas.0914241107 20615967
47. Szymanski P, Levine M (1995) Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. EMBO J 14 : 2229–2238. 7774581
48. Burz DS, Rivera-Pomar R, Jackle H, Hanes SD (1998) Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J 17 : 5998–6009. 9774343
49. Erceg J, Saunders TE, Girardot C, Devos DP, Hufnagel L, et al. (2014) Subtle changes in motif positioning cause tissue-specific effects on robustness of an enhancer's activity. PLoS Genet 10: e1004060. doi: 10.1371/journal.pgen.1004060 24391522
50. Rossi FM, Kringstein AM, Spicher A, Guicherit OM, Blau HM (2000) Transcriptional control: rheostat converted to on/off switch. Mol Cell 6 : 723–728. 11030351
51. Hong JW, Hendrix DA, Levine MS (2008) Shadow enhancers as a source of evolutionary novelty. Science 321 : 1314. doi: 10.1126/science.1160631 18772429
52. Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, et al. (2014) Low Affinity Binding Site Clusters Confer Hox Specificity and Regulatory Robustness. Cell.
Štítky
Genetika Reprodukčná medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



