-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
Genomic architecture, such as direct or inverted repeats, can facilitate structural variation (SV) of the human genome. SV can consist of deletion, duplication, or inversion of a genomic segment, or combinations thereof, the latter referred to as complex genomic rearrangements (CGR). CGR are defined as requiring two or more novel DNA breakpoint junctions. We described a CGR product at the MECP2 locus with an unusual pattern consisting of an inverted triplicated segment flanked by duplicated segments of the genome. This complex CGR is facilitated by inverted repeats in a process that mechanistically could occur by two template switches mediated by replicative DNA repair. We now investigate the PLP1 locus and demonstrate that 16/17 CGR independent events present with duplication—inverted triplication—duplication pattern facilitated by two inverted repeats, similar to events involving MECP2. We show that the same inverted repeats facilitating CGR formation are also responsible for an inversion polymorphism observed frequently in the normal population. Intriguingly, one CGR was found to have a quadruplication resulting in the presence of four copies of a genomic segment. Breakpoint studies suggest this quadruplication occurred in a manner consistent with rolling circle amplification as predicted by previously postulated models.
Published in the journal: Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication. PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005050
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005050Summary
Genomic architecture, such as direct or inverted repeats, can facilitate structural variation (SV) of the human genome. SV can consist of deletion, duplication, or inversion of a genomic segment, or combinations thereof, the latter referred to as complex genomic rearrangements (CGR). CGR are defined as requiring two or more novel DNA breakpoint junctions. We described a CGR product at the MECP2 locus with an unusual pattern consisting of an inverted triplicated segment flanked by duplicated segments of the genome. This complex CGR is facilitated by inverted repeats in a process that mechanistically could occur by two template switches mediated by replicative DNA repair. We now investigate the PLP1 locus and demonstrate that 16/17 CGR independent events present with duplication—inverted triplication—duplication pattern facilitated by two inverted repeats, similar to events involving MECP2. We show that the same inverted repeats facilitating CGR formation are also responsible for an inversion polymorphism observed frequently in the normal population. Intriguingly, one CGR was found to have a quadruplication resulting in the presence of four copies of a genomic segment. Breakpoint studies suggest this quadruplication occurred in a manner consistent with rolling circle amplification as predicted by previously postulated models.
Introduction
Inverted repeats (IRs) are a common architectural feature within the human genome and can predispose loci to rearrangement [1–3]. An IR-mediated inversion that disrupts the Factor VIII gene causes ~45% of severe hemophilia A cases [4]. The importance of IRs to human genomic rearrangements and resultant genomic disorders and the expanded scope by which IRs can facilitate genomic change are now apparent [2,3,5–7]. The abundance of inverted low copy repeats (LCRs) or segmental duplications genome-wide suggests that ~12% of the genome may be susceptible to inversion mediated by IRs [2]. Fosmid paired-end sequencing of 8 human genomes from diverse populations shows that ~50–100 large genomic inversions not represented in the human genome reference sequence are present in the personal genome of each individual. In total, 224 non-redundant inversions were identified in 8 genomes; these events are primarily mediated by larger blocks of homology [8]. Earlier work provided experimental evidence for genome-wide inversions and suggested these can occur somatically and with aging [9]. Moreover, inverted repetitive regions that are smaller than conventional LCRs, designated self-chains, are also associated with genomic instability furthering the impact of IRs on both structural human differences and phenotypes [3].
Recently, IRs were shown to mediate complex duplication—inverted triplication—duplication (DUP-TRP/INV-DUP) rearrangements, leading to MECP2 duplication syndrome (MIM#300260), Duchenne Muscular Dystrophy (MIM#310200), VIPR2 triplication, CHRNA7 triplication, and Pelizaeus-Merzbacher disease (PMD, MIM#312080) [1,10–13]. The mechanisms for such complex genomic rearrangements (CGRs) have only begun to be elucidated.
Genomic rearrangements leading to the duplication of the X-linked proteolipid protein 1 (PLP1) gene are the major mutational cause for PMD and explain ~80% of patients; point mutations in PLP1 occur less frequently, and higher copy number gains (e.g. triplications) and deletions are rare [14–17]. CGR can cause PMD by duplicating PLP1 via a mechanism that results in a DUP-TRP/INV-DUP structure [1]. Consistent with a gene dosage hypothesis, and as established for both homozygous duplication [18] and heterozygous triplication [19] at the CMT1A locus, triplication of PLP1 can lead to a more severe form of PMD than duplication [13,14].
Using high-density array comparative genomic hybridization (aCGH), DUP-TRP/INV-DUP rearrangements primarily contain one variable breakpoint at the proximal (centromeric) end [1]; however, distal breakpoints for the triplication to duplication and duplication to normal copy number transitions cluster at inverted LCRs distal to MECP2 [1]. The proposed mechanism for these CGR involved a two-step process: i) break-induced replication (BIR) within homologous regions of the inverted LCRs forming a breakpoint junction (Jct1) and ii) microhomology-mediated BIR (MMBIR) or non-homologous end-joining forming a second junction (Jct2). Mutational signatures observed at the latter junction include microhomology, templated insertions, and increased point mutation frequency [1,20]. However, in both MECP2 and PLP1 DUP-TRP/INV-DUP rearrangements, delineation of unique breakpoint junctions within the IR has been hampered by the complexity of large blocks of homologous sequences creating challenges to mapping Jct1 at base pair resolution.
To further investigate mechanisms for CGR formation we analyzed a cohort of 17 unrelated PMD patients with copy number gains at the PLP1 locus, including duplications, triplications and quadruplication. Analysis of phenotypically normal individuals elucidated a common inversion polymorphism associated with the IRs distal to PLP1. Southern blotting experiments established an estimated frequency for the inversion. We postulated and confirmed that the LCR substrates responsible for the inversion are also responsible for one breakpoint junction (Jct 1) in each PMD associated CGR. Additionally, we document a DUP-TRP/INV-DUP rearrangement product structure at the PLP1 locus in the personal genomes of 16 subjects with PMD and provide evidence that such CGR can occur by replicative mechanisms [21]. Finally, we investigated the quadruplication of a genomic segment proximal to PLP1 and found the potential mechanism of formation to be consistent with rolling-circle replication leading to amplification—a mechanism predicted by the MMBIR model [22].
Results
Inversion Polymorphism Discovery, Frequency and Recurrence
The 186 kb genomic interval (ChrX: 103,172,000–103,358,000 in hg19) located ~150 kb distal to PLP1 contains a complex genomic architecture in the haploid reference genome. This region consists of an array of IRs, with the ~40 kb outer C and D repeats having ~93% identity, the middle A1a and A1b repeats ~20 kb in size and ~99% identical, and the innermost ~10 kb A2 and A3 repeats showing ~87% identity both with each other and with A1a and A1b (Fig. 1A) [16,23,24]. The IR architecture predicts the potential for inversion mediated by non-allelic homologous recombination (NAHR), resulting in at least two structural haplotypes, analogous to the H1 and H2 structural variant (SV) alleles at the MECP2 locus [1]. Indeed, in silico analysis of the human genome SV track from the UCSC Genome Browser (www.genome.ucsc.edu) suggests the existence of such an SV allele [25]. The browser track indicates fosmids consistent with inversions spanning both of the A1a and A1b LCRs in 5 of 9 individuals (S1 Fig) [8,26]. These data indicate that there was an inversion between A1a and A1b LCRs and that the inversion haplotype exists at a relatively high allele frequency as a non-pathogenic rearrangement in HapMap individuals (Fig. 1B). Further investigation mapped the apparent ectopic crossover for the NAHR-mediated inversion in a fosmid from the G248 library to nucleotide-level resolution (S2 Fig).
Fig. 1. Inversion discovery and frequency between A1a and A1b. 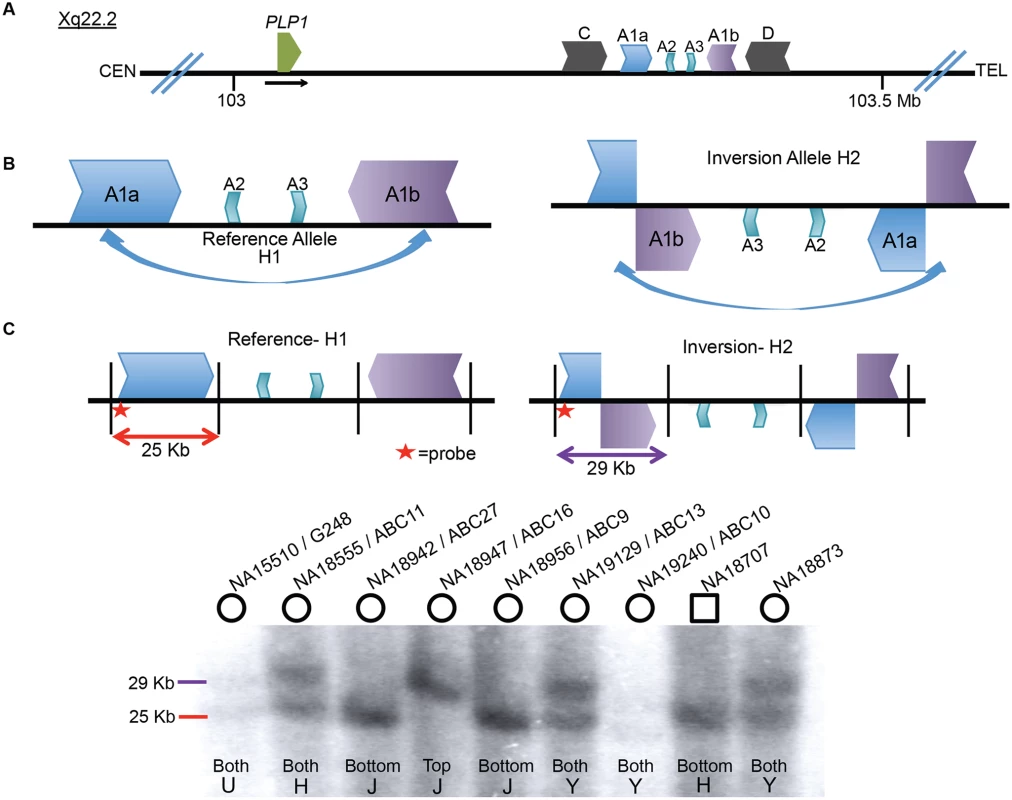
A) A depiction of the PLP1 genomic region on chromosome Xq22.2. PLP1 is proximal to the LCR structures and the black horizontal arrow indicates direction of transcription. There are three IRs present at chrX:103,171,387–103,359,682: the outer C and D repeats, middle A1a and A1b repeats, and inner A2 and A3 repeats [23,24]. The gene and the IR structures are separated by ~150 kb. B) A common inversion, discovered in the HGSV resource, is depicted between the A1a and A1b repeats. This inversion was present in at least 5/9 of individuals in the fosmid resource, and is possible in three additional people (S1 Fig). C) A Southern blotting scheme to distinguish between reference and inversion alleles is depicted, with A1a, A1b, A2 and A3 represented as above. Female genomes have two alleles, and phenotypically normal males have one allele (blot quantitated in S1 Table). Digestion with BssSI (depicted by black vertical lines) and detection with a probe proximal to LCR A1a (indicated by red star) should distinguish reference (25 kb, red) from inverted (29 kb, purple) alleles. Nine individuals from the HapMap population were studied for inversion via Southern blotting. Gender of the individual is indicated by circles (female) or squares (male) above the blot. DNA identifiers (NA numbers) are consistent with Coriell names (http://ccr.coriell.org/), and fosmid libraries (ABC library identifiers) are as in Kidd et al. [49]. The population of origin for each individual and the genotypes are indicated at the bottom of the blot. H = Han Chinese, J = Japanese, Y = Yoruban, and U = unknown. Genotyping for 8 additional individuals is depicted in S3 Fig. To directly examine the inversion SV polymorphism between A1a and A1b, we designed a Southern blotting assay and genotyped multiple individuals from different populations of origin for reference (arbitrarily designated H1) or inversion (H2) structural haplotypes. The scheme of the assay is depicted in Fig. 1C, wherein Southern analysis leads to predicted visible fragments of 25 kb for H1 and/or 29 kb for H2. As the rearrangement is on the X chromosome, males should have only one allele, and females, two. Genotyping 17 individuals (including 3 males) with this assay discerned 31 haplotypes of the X chromosome (Figs. 1C, S3, and S1 Table). The frequencies of structural haplotypes were 13/31 H2 (~42%) and 18/31 H1 (~58%), with 4 individuals hemizygous or homozygous for H2 and 7 for H1. The remaining 6 females were heterozygous for both H1 and H2. The 17 individuals were of Japanese, CEPH Northern European, Han Chinese, Yoruban and unknown populations of origin, and all populations contained both H1 and H2 structural haplotypes (S2 Table).
We hypothesized that the similarity between LCRs A1a and A1b and their relatively large length and proximity (~20 kb repeats of ~99% identity separated by ~50 kb) could predispose to recurrent events [27,28]. We analyzed the genomic region encompassing the two LCRs and identified multiple adjacent single nucleotide polymorphisms (SNPs) spanning the region in linkage disequilibrium and delineating a haplotype block extending for ~0.5 Mb with a recombination rate of 0.3 centimorgans per Mb. The two SNP haplotype blocks were evenly distributed between the 14 different populations from the 1,000 genomes project [29]. Superimposing Southern blotting results for individuals homozygous or hemizygous for SV haplotypes on top of the SNP haplotypes enabled phasing; 6/7 inversion (H2) alleles were on one SNP haplotype and 1/7 was on the other (belonging to individual NA18947, S4 Fig), whereas homozygous H1 alleles occurred on either SNP haplotype. Heterozygous calls are uninformative, as the structural haplotype information cannot be phased to the SNP data. These data suggest that the inversion is likely recurrent in the population and makes population estimation of the structural variant using SNP genotyping unlikely to reflect the true population frequency.
Breakpoint Mapping of PLP1 Rearrangements
Sixteen patients with PMD and one diagnosed with spastic paraplegia type two (SPG2; MIM#312920) were examined by aCGH for copy number variation (CNV) in PLP1 and the surrounding genomic region. A schematic of CNV observed in the personal genomes from 17 patients is depicted in Fig. 2A. PLP1 duplications were detected in 10 patients (BAB1290, BAB2389, P250, P298, P500, P558, P842, P1389, P1407 and P113), whereas triplications were detected in 6 patients (BAB3698, P518, P642, P674, P820, and P1150). The one SPG2 patient, BAB1612/P374 has been described previously, and the phenotype of this individual is ascribed to a potential position effect [30]. The distal breakpoints in all subjects appear to cluster in approximately the same genomic location; however, there are few probes on the arrays that can specify unique loci within the C/D, A1a/A1b, and A2/A3 LCRs due to the repeat nature of the region. Thus, determining the precise LCRs involved in the breakpoints required alternate mapping approaches.
Fig. 2. Complex rearrangements involving PLP1 have clustered breakpoints in IRs. 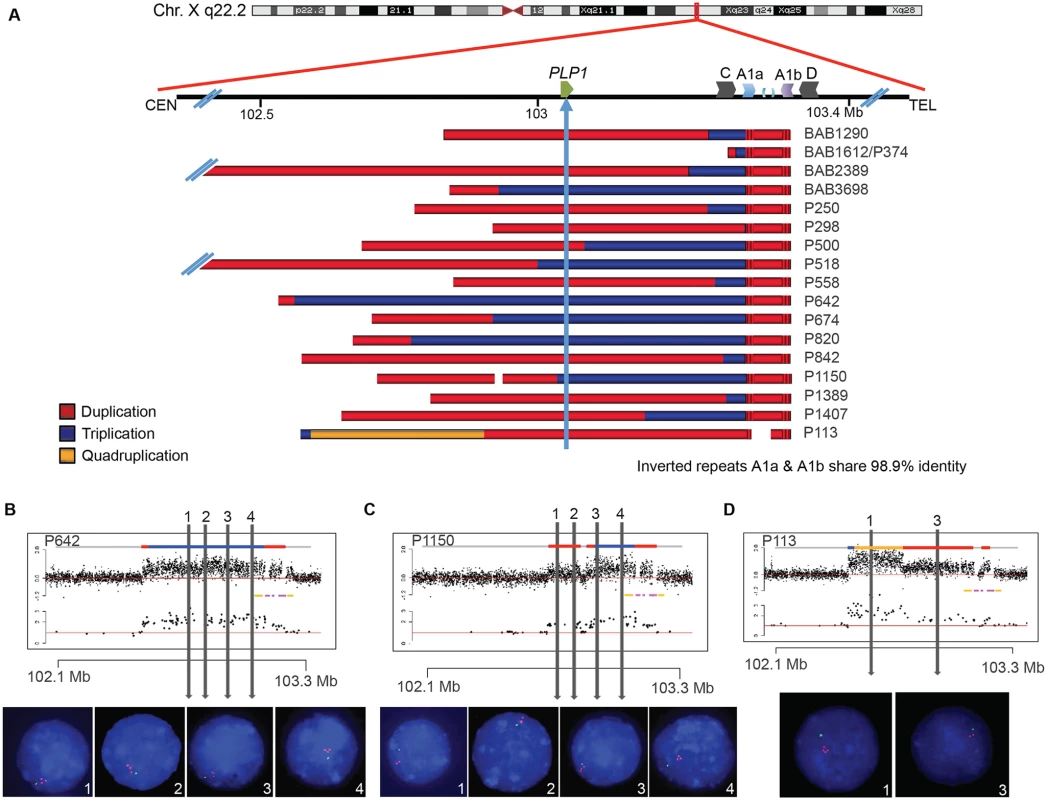
A) 17 individuals are depicted with schematics of their array results. Duplications are indicated in red, triplications are indicated in blue, and the quadruplication is indicated in gold. Coordinates and LCR blocks are indicated in the cartoon above the array results (A2 and A3 are unlabeled). P1150 and P113 contain copy number neutral segments within the rearrangement, leading to a copy number of 1 indicated by white space in the array schematic (see also S5 and S6 Figs). B) FISH results for P642, depicting a large triplication of the PLP1 region, including the gene. All three copies of the four probes are on Xq22.2. Array data are at the top and qPCR data are below, with LCRs indicated in yellow and pink. Colors for copy number in array data are as in panel A. Probe locations for FISH denoted by vertical lines. C) FISH results for P1150, similar to those in B. The figure depicts duplication of the two proximal probes 1 and 2 and triplication of the distal 3 and 4 probes, as proposed by the array data above. D) FISH results for patient P113, depicting a quadruplication/duplication rearrangement of the region. High-resolution aCGH showed greater complexities, including a small triplication and a normal copy number segment within the duplication (See Figs. 5 and S5). In all FISH studies, nuclei are stained with DAPI, and a marker for the short arm of chromosome X, AL353698, is shown in green. Vertical arrows with numbers in the array data above indicate positions of FISH probes throughout the PLP1 region. Array and semi-quantitative PCR data, summarized in Fig. 2A (see also S5 Fig and Table 1), indicate that the region of rearrangement spans from 145 kb (BAB1612/P374) to ~4,000 kb (BAB2389). Triplicated genomic segments range in size from 254 bp (P298/P255) to 575 kb (P642). Proximal triplication and duplication copy number transitions differed in each individual and were not located within LCRs. The distal copy number transitions group within a 100 kb region of uncertainty as described above. The triplication present in P298/P255 was too small to be detected by aCGH; however, a 254 bp triplicated genomic segment was detected both by amplification and sequence analysis with unique flanking primers and by quantitative PCR (S6 Fig).
Tab. 1. Unique Junctions (Jct2) in DUP-TRP/INV-DUP rearrangements. 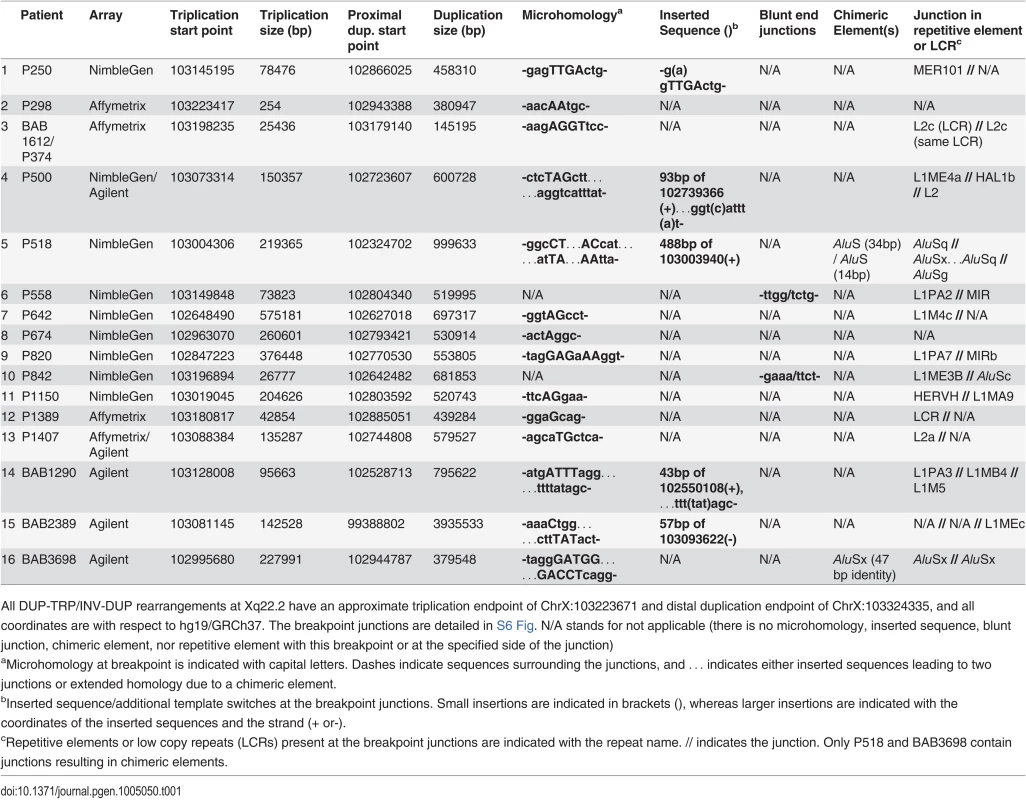
All DUP-TRP/INV-DUP rearrangements at Xq22.2 have an approximate triplication endpoint of ChrX:103223671 and distal duplication endpoint of ChrX:103324335, and all coordinates are with respect to hg19/GRCh37. The breakpoint junctions are detailed in S6 Fig. N/A stands for not applicable (there is no microhomology, inserted sequence, blunt junction, chimeric element, nor repetitive element with this breakpoint or at the specified side of the junction) FISH was performed on nuclei prepared from peripheral blood lymphocytes from P642, P1150, and P113. This independently corroborated interpretation of array and semi-quantitative PCR data using an orthogonal experimental approach. Moreover, FISH determined whether extra copies of the genomic segments were located in or near the PLP1 locus as opposed to elsewhere in the genome; arrays determine neither the orientation nor the position of a copy number segment, but only specify the genomic segment that underwent a gain in copy number (Fig. 2B-D). Interphase nuclei of patients P642 and P1150 showed, as expected, one green control probe signal and revealed three closely-spaced red PLP1 probe signals indicating triplication at the PLP1 locus; P113 had two red PLP1 signals indicating duplication at that locus, but four proximal probe signals confirmed the additional quadruplication (Fig. 2D). Metaphase spreads of all 3 patients gave one green control probe signal, one red presumably merged PLP1 probe signal on the X chromosome, and no signals on other chromosomes, also indicating that the triplications and duplications were at the PLP1 locus rather than being located elsewhere in the genome and that the triplications were too small to resolve on metaphase chromosomes.
Marker Genotypes Suggest Intra-Chromosomal Events
We investigated haplotypes using genetic markers, 2 short tandem repeats (STRs) and 9 SNPs, mapping over a 258 kb region of the duplications with 4 markers mapping within PLP1 and the remainder distal to it. We observed that 12 of 13 patients tested (P250, P255/298, BAB1612/P374, P500, P518, P558, P642, P674, P820, P842, P1150, P1389, and P1407), were monomorphic displaying only one form for each marker genotype (S3 Table). The DNA from BAB1612/P374 was only interrogated at the 7 sites distal to PLP1, since this is where his triplicated/duplicated region lies. In this subject, only one form was detected for all markers except the STR furthest distal to PLP1 where two were detected. The finding of an absence of bi-allelic loci in these multi-copy regions of X is most parsimoniously explained by the occurrence of intra-chromosomal rearrangement events, as has also been observed for DUP-TRP/INV-DUP rearrangements at the MECP2 locus [20].
Junction Analyses of Proximal Breakpoints
In P1150, we obtained a breakpoint junction between the proximal (centromeric) endpoint of the rearrangement and the proximal end of the triplicated region via inverse PCR. We then hypothesized that our other patients with duplication-triplication-duplication copy number changes could potentially have the same CGR product structure and explored this hypothesis by long-range PCR on each individual personal genome. We were able to amplify and sequence across the proximal breakpoint junction in all 16 patients (Table 1). The 16 junctions each indicate that the triplicated region is inverted with respect to the proximal duplication region (S6 Fig); in 6 cases the triplication encompasses PLP1. This is a potentially analogous rearrangement structure to that previously described for MECP2 CGRs; therefore, we denote the non-recurrent junctions in Table 1 as Jct2 [1].
We had previously mapped and sequenced across the duplication breakpoint junction of patient P255, who had a 254 bp inverted duplication [24]. We no longer had DNA from P255 to interrogate the copy number of the region; therefore, we tested an affected family member, P298, by qPCR and found, as anticipated, that the region is triplicated.
The Jct2 sequences in the 16 patients are shown in Table 1. Fourteen patients contain one or more breakpoint junctions displaying microhomology. Patients P558 and P842 have blunt junctions. In 13 of the patients, endpoints at Jct2 are in repetitive element sequences, and in P1389, one end was in an LCR (Table 1). Patient BAB1612/P374 contained a LINE2-mediated event (L2/L2, both within the same LCR) that did not result in a chimeric element. Patients P518 and BAB3698 contain chimeric AluS elements formed in the generation of this junction. In BAB3698, there are 47 bp of identity between the two AluSx elements at the transition from triplication to duplication. In P518, the rearrangement occurs through the formation of two AluS chimeric junctions, the first (from proximal to middle segment) in the same orientation in 14 bp of identity, and the second (from middle segment to triplication) in 34 bp of identity (see S6 Fig). This complex breakpoint junction contains a segment of 488 bp that consists of an AluSx3, an L1 sequence (L1ME), and an AluSq2. Interestingly, the distal (triplication) to middle junction occurs between these two Alu elements that are only separated by 310 bp, suggesting a potential U-turn caused by inverted Alus within close proximity [31,32], similar to the situation in Jct1 but mediated by short Alu sequences instead of LCRs (S6 Fig). The breakpoint junction mutational signatures are consistent with replicative mechanisms such as MMBIR or a homeologous (near homologous) recombination event between similar Alu elements at each instance of Jct2 [22,33,34].
Complexities that included an additional template switch were observed in Jct2 from individuals P500, P518, BAB1290 and BAB2389 (Table 1 and S6 Fig). Such events have been postulated to reflect reduced processivity of the replisome mediating MMBIR during initial template switching [22]. We also amplified across a breakpoint junction present in P1150, indicating a 27 kb deletion on one of the duplicated copies (Figs. 2 and S5). At that junction, there is a bp of microhomology (S6 Fig). The overall findings for Jct2 are consistent with both long distance template switching and a microhomology-mediated mechanistic process such as FoSTeS/MMBIR [21,22].
DUP-TRP/INV-DUP Distal Breakpoints
After Jct2 was determined for the 16 patients, we hypothesized that a likely genomic arrangement consistent with this junction was one in which one copy of the triplicated region was situated in an inverse orientation between the two copies of the duplicated region and that the other two copies of the triplication were embedded within the duplicated regions, i.e. a DUP-TRP/INV-DUP structure [1].
Patients with presumed DUP-TRP/INV-DUP rearrangements with sufficient DNA available were subjected to Southern blotting (10/16 total) to examine whether the same repeats involved in the common inversion polymorphism are also involved in the CGR and to investigate on which structural haplotype the rearrangement occurred. The Southern scheme in Fig. 1C was used to analyze patient DNAs; however, in a male with PMD caused by DUP-TRP/INV-DUP involving the A1a and A1b repeats, the Southern blot does not reflect the normal copy number of one allele of the X chromosome (either H1 or H2) (Fig. 3A, S4 Table). Instead, the rearrangement gives rise to two copies of the original haplotype plus an additional “flipped” haplotype in an affected individual with DUP-TRP/INV-DUP leading to PMD, similar to the observation described for the MECP2 locus [1]. This assay can presumably distinguish the SV haplotype on which the genomic rearrangement occurred. A representative gel and labeled blot are shown in Fig. 3B, with the dosage of the bands indicating that subjects BAB1290 and BAB1612/P374 both carried the inversion H2 structural haplotype prior to the rearrangement.
Fig. 3. CGRs at the PLP1 locus use A1a and A1b repeats. 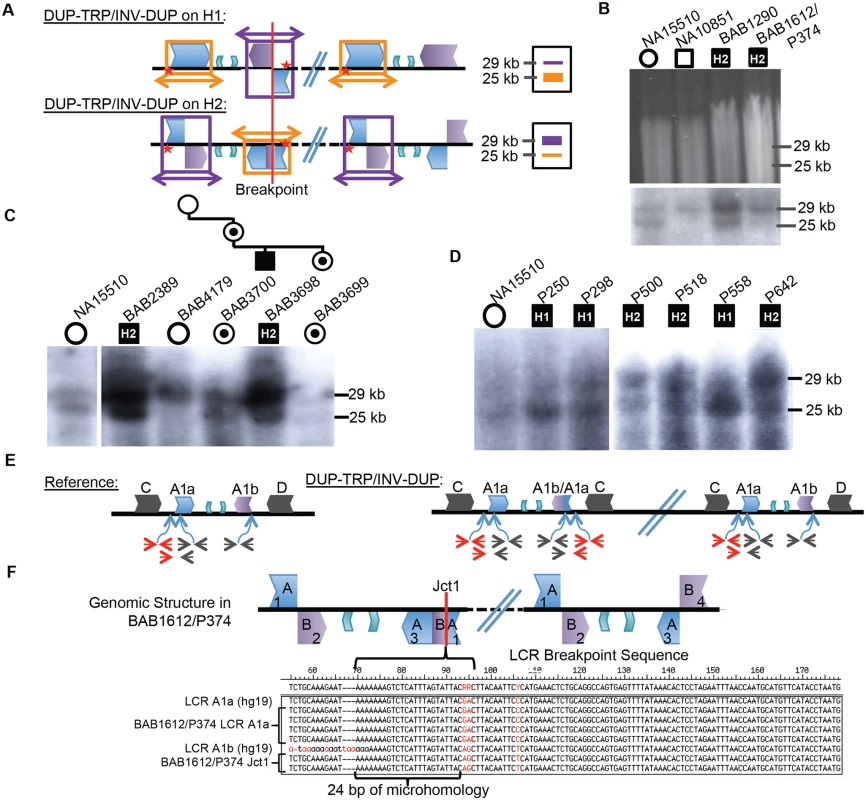
A) Southern scheme from Fig. 1 applied to a DUP-TRP/INV-DUP rearrangement results in either two copies of a 25 kb band and one of a 29 kb band if the rearrangement occurred on H1 (shown at the top), or the reciprocal copy dosage if the rearrangement occurred on H2 (bottom). Colors for LCRs are as in Fig. 1. B) Digested DNA from 2 control individuals (NA15510 and NA10851) and two PMD/SPG2 patients (BAB1290 and BAB1612/P374, respectively). Southern below depicts control individuals have expected, gender appropriate 2 and 1 copies, and affected males have three copies, with dosage of 2:1 H2:H1, indicating the rearrangement likely occurred on H2 (indicated within the black square for each patient- data for this blot and panels C and D are quantitated in S4 Table). C) BAB2389 and BAB3698 may also have rearrangements on the inverted allele (H2, indicated at the top of the image). BAB3698 is depicted with his carrier mother, sister (BAB3700 and BAB3699, indicated by a dot in a circle), and non-carrier grandmother (BAB4179). The grandfather was unaffected and unavailable for study. D) P250, P298, and P558 all likely contain rearrangements on H1 (~1:2 ratio of H2:H1) and P500, P518, and P642 contain rearrangements on H2. All rearrangement progenitor haplotypes are indicated for the patients above the Southern blot. E) The reference genomic structure of H1 is shown (inner A2 and A3 repeats are unlabeled). The qPCR primer pairs amplify a unique region outside of the A1a LCR (in red), inside of both A1a and A1b LCRs (in black) or from the A1a LCR to a unique region outside (red/black pair below). These will give rise to one copy (red pair and red/black pair) or two copies (black pair) in a non-rearranged X chromosome in a male individual. DUP-TRP/INV-DUP (on right) in an H1 haplotype will give rise to four copies amplified by the black pair (2x normal control) and three copies by the red pair and red/black pair (3X normal control) (S7 Fig). F) Analysis of Jct1 has successfully cloned one breakpoint in BAB1612/P374. The structure of the LCR-mediated rearrangement on H2 is depicted at the top (A1a and A1b are simplified to “A” and “B” and inner A2 and A3 repeats are unlabeled). Overlapping clones for each region of the two LCRs were generated (numbered 1–4, S8 Fig), and results for BAB1612/P374 were obtained for section 1 clones that both contain and lack the breakpoint. Multiple clones from this region are depicted along with the reference sequences for LCRs A1a and A1b below. The breakpoint from individual BAB1612/P374 occurred in stretch of 24bp of microhomology (bracket-denoted region). Interestingly, the 10 individuals examined by this assay appeared to use the A1a and A1b LCRs as the substrates for their rearrangements, in spite of two other IRs being located in close proximity (BIR between C/D would lead to duplication of H1 or H2 and A2/A3 would lead to triplication) (Figs. 1A, 3C and D). Individuals BAB1290, BAB1612/P374, BAB2389, BAB3698, P500, P518, and P642 all contained a rearrangement that had occurred on the inverted H2 allele, while P250, P298, and P558 had a Southern blot result indicating the rearrangement occurred on an H1 haplotype (S4 Table).
A three-generation family was studied in which the two maternal grandparents were unaffected, and subsequent Southern blotting and aCGH data indicated that the grandmother (BAB4179) was not a carrier and that she had two copies of the inverted H2 locus (Figs. 3C and S5). The grandfather was unavailable, but did not have PMD; therefore, the de novo rearrangement can be inferred to have occurred in between the grandparent and the maternal generation. The mother (BAB3700) was a carrier of the rearrangement and had equal dosage of H1 and H2 on a Southern Blot. The affected son (BAB3698) had Southern results consistent with rearrangement on H2, and his carrier sister (BAB3699) had similar results to the mother. These findings are consistent with the de novo DUP-TRP/INV-DUP occurring in association with “flipping” the H2 haplotype to an H1 haplotype, a mechanism similar to that observed for CGRs at the MECP2 locus (Fig. 3A) [1]. The assay results in this family are most parsimonious with the rearrangement occurring on one of the grandmother’s inversion-containing alleles (H2), and having balanced copy number in BAB3700 and BAB3699 due to the additional allele being a reference (H1) 25 kb band. This would result in a 2 : 2 dosage of 29 kb:25 kb bands on the Southern blot, which we observe in both BAB3700 and 3699 (Fig. 3C and S4 Table).
Breakpoint Junctions or Crossovers Within LCRs
As Jct1 occurs within the LCR region distal to PLP1, the junctional products are not readily amplified and sequenced by long PCR with primers anchored to unique flanking sequence. We adopted an alternative strategy to complement the Southern blotting assay above. Using a semi-quantitative PCR approach, we first confirmed that each of the patients has duplication of A1a and A1b LCRs (black primer pair in Fig. 3E) and triplication of a region proximal to A1a (red primer pair and black/red primer pair in Figs. 3E, S7). This PCR approach independently verified the Southern Blot results and suggested a crossover breakpoint within the A1a or A1a/A1b chimera present on H2 (Fig. 3E). We attempted to more narrowly define the crossover region in our patients by using sequence differences between the LCRs (paralogous sequence variants or PSVs), but patients appeared to lack apparent PSVs between A1a and A1b that were at the corresponding genomic locations in the hg19 reference sequence [35].
To determine sequences across Jct1, we designed a PCR-cloning assay that allowed us to amplify large (~12–16 kb), overlapping portions of both A1a and A1b LCRs that are implicated in the rearrangements [35] (Figs. 3F, S8). Three individuals were subjected to this analysis (BAB1612/P374, BAB2389, and BAB1290), however BAB2389 and BAB1290 appear to have Jct1 within a large region of identity (>8 kb) in the center of the LCR that lacks PSVs between cloned segments; therefore, further refinement of the breakpoint junction was intractable using this method. Additionally, in P255/298, a PCR approach using one primer at the proximal duplication junction and one within the LCR corroborated that the breakpoint indeed occurred within this >8kb stretch of identity.
In contrast to the three other individuals for whom we sought to find Jct1 at base pair resolution, in BAB1612/P374 we were able to detect an LCR-mediated breakpoint within 24 bp of microhomology flanked by A1a and A1b sequences (Fig. 3F). The point of crossover within this sequence was confirmed by direct PCR amplification and sequence analysis from genomic DNA followed by comparison to the PSVs present on cloned A1a and A1b sequences from the same individual; its identification elucidates Jct1 within an LCR, a heretofore un-investigated junction at the nucleotide level of resolution.
DUP-TRP/INV-DUP Rearrangement Structure
The DUP-TRP/INV-DUP structure hypothesized for these 16 individuals postulates that although there are 4 copy number transitions in these patients, there are only two breakpoint junctions (Fig. 4A). We have sequenced Jct2 in all 16 patients; Southern blotting and quantitative PCR were used to determine Jct1, and direct junction sequencing was successful for BAB1612/P374 (Figs. 3, 4 and S6). Additionally, due to the small size (~ 254 bp) of the triplication in P255/298, a PCR approach using one primer at the proximal duplication junction and one within the LCR validated the overall structure of this rearrangement as DUP-TRP/INV-DUP.
Fig. 4. DUP-TRP/INV-DUP structures at the PLP1 locus. 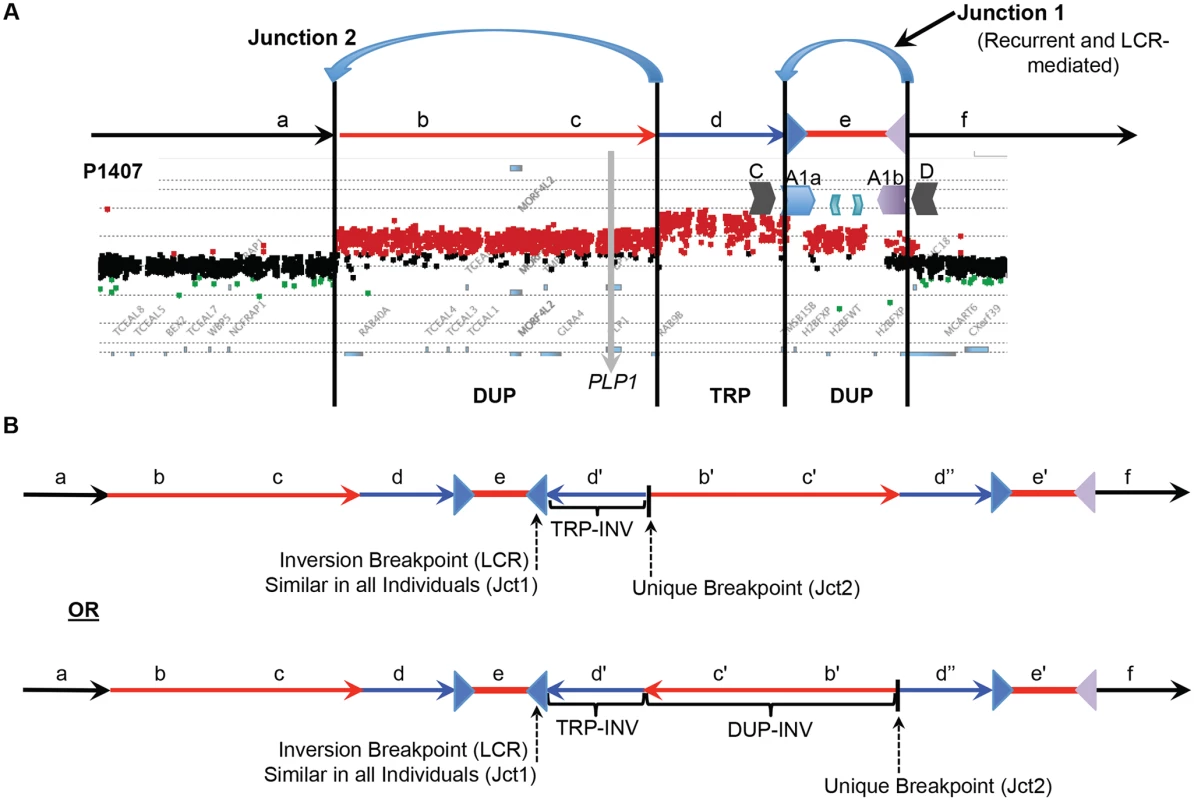
A) The aCGH results from patient P1407 showing a duplication of PLP1 and a distal triplication-duplication structure are shown at the top, with duplications in red and triplication in blue. The relative genomic regions are labeled with letters to distinguish their relative positions within the CGR. The IRs flanking segment e (A1a and A1b) are denoted by inverted blue and purple triangles, respectively. B) Two potential structures of the generalized DUP-TRP/INV-DUP rearrangement that are consistent with Jct2 sequencing are shown in the lower panel of the figure in relation to the canonical genomic structure at the top. Here, the unique proximal breakpoint junction location that differs between patients, the LCR-mediated distal inversion breakpoint junction, and the inverted triplication region are seen. Letters followed by a prime symbol indicate duplicated segments. Two prime symbols indicate the triplicated segment. Quadruplication by Rolling-Circle Amplification
We have discerned two junctions from patient P113 with proximal quadruplication and duplication of PLP1 using long-range PCR (Figs. 5A, S6). Junction 1 consists of one fork stalling and template switching (FoSTeS) event—FoSTeS 1 (Fig. 5A). The second junction, between the proximal end of the triplication and the distal end of the quadruplication, consists of FoSTeS events 2 and 3 (S6 Fig for sequences of all junctions). We determined that the rearrangement was on the inverted H2 allele using PCR genotyping of the haplotype present in P113 (S9 Fig). Additionally, digital PCR (dPCR) data indicate that the FoSTeS 1 occurs in one copy, and FoSTeS 2/3 occurs in 2 copies (S5 Table).
Fig. 5. A quadruplication and potential rolling-circle mechanism of rearrangement. 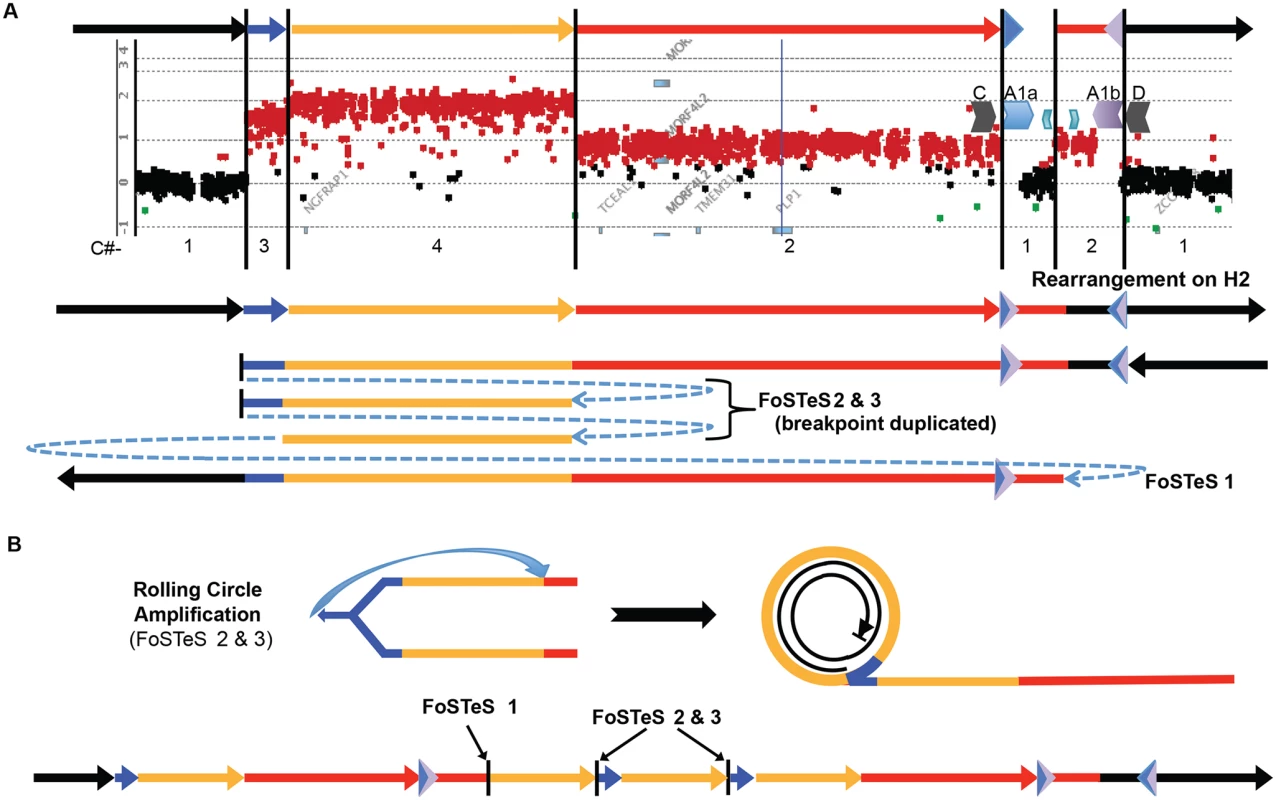
A) P113 aCGH result is shown. At top, colored arrows represent segments of triplicated (blue), quadruplicated (gold), and duplicated (red) sequence in this individual. A1a and A1b are represented by inverted blue and purple triangles, respectively. Copy number (C#) of the various segments is enumerated below the aCGH result, as is the rearrangement structure on the H2 haplotype (see S9 Fig). Breakpoints are depicted by blue dashed arrows in the schematic of the rearrangement and are labeled with the template switching events associated with the junction. Breakpoint sequences are depicted in S6 Fig, and dPCR data indicating duplication of FoSTeS events 2 & 3 is in S5 Table. B) The proposed rolling circle mechanism leading to the triplication and quadruplication is depicted, with the FoSTeS events 2 & 3 leading to invasion of the fork into the already replicated strand. This establishes the rolling circle, depicted on the right. The event ends with a double strand break or a fork disassociation event, and the breakpoint involving FoSTeS event 1 (S6 Fig). The structure of the overall rearrangement is depicted at the bottom, with colors relating to panel A for orientation. This quadruplication rearrangement is also associated with a de novo point mutation (G insertion) ~50 bp away from the junction that appeared to occur concurrent with the rearrangement, as observed for other CGR mediated by a replicative process (S6 Fig) [20]. The mechanism by which copy number increased from 3 to 4 copies and generated the quadruplication is suggestive of a rolling circle amplification, wherein one breakpoint is repeated twice in the process of replicating ~280 kb (S5 Table) [22,36–38]. The FISH data for this individual shows the rearrangement to be contained on the X chromosome, and family data including the proband P113, his mother P154, his affected uncle P117, and grandmother P088 suggest that the structure is stable in 4 individuals from 3 generations, diminishing the likelihood of recombination-based amplification (Figs. 2D and S5). If the amplification were mediated by NAHR (see S10 Fig), this rearrangement would contain two templates for subsequent rounds of amplification. Therefore, the rearrangement in P113 should be twice as likely to undergo expansion as the proposed intermediate. Additionally, the presence of monomorphic SNPs throughout the region of copy number gains on SNP arrays indicates that the rearrangement was intra-chromosomal, as was found for the 13 patients with DUP-TRP/INV-DUP rearrangements (S11 Fig). The quadruplication-containing CGR was observed to transmit stably and co-segregate with disease through three generation (S5 and S11 Figs). A proposed mechanism for the rearrangement occurring in one complex quadruplication event is shown in Fig. 5 and consists of a rolling-circle amplification of the triplicated and quadruplicated segments.
Discussion
PLP1 is surrounded by LCRs of variable size and sequence similarity; previous studies have shown that such genomic architecture renders this region unstable and susceptible to rearrangements, leading to PMD [16,23,24]. We show that DUP-TRP/INV-DUP rearrangements are a frequent CGR product at the PLP1 locus and that they are facilitated by a complex IR but specifically mediated via the ~20 kb A1a and A1b 99% identical repeats. These particular IRs are not only driving CGR observed in patients but additionally mediate a common SV polymorphism—copy-number neutral inversions at Xq22.2. The latter can complicate interpretation of CGRs in the region; a proposed breakpoint can also appear in an unaffected individual in the guise of an inverted allele [13]. Additionally, the recurrence of this inversion might confound the correlation of diagnostic SNPs with structural information, leading to the underestimation of the frequency in a population [39]. These data suggest that IRs with a high degree of identity that are involved in non-pathogenic inversions can also drive seemingly recurrent breakpoints in non-recurrent rearrangements associated with disease and that this occurs at multiple genomic loci [1,12,13]. Indeed, a previous determination of genes potentially subject to CNV via DUP-TRP/INV-DUP due to proximity of homologous IRs predicted the PLP1 gene might be affected [2].
The proximal junctions, or Jct2, in the DUP-TRP/INV-DUP rearrangements at the PLP1 locus are depicted in S6 Fig. Jct2 is non-recurrent in the 16 individuals, with different genomic coordinates for each breakpoint. Interestingly, investigation revealed that 14 of 16 Jct2 sequences contained microhomology of 1–4 bp at one or more of the FoSTeS events in the junction. Two of these Jct2 sequences involved larger stretches of microhomology; one contained an Alu-Alu chimeric event with 47 bp of perfect identity at the junction and the second CGR contained two Alu-Alu chimeras, one containing 14 bp and the other with 34 bp of perfect identity at the junction (Table 1). These data suggest that a replicative mechanism is involved in the formation of Jct2 in a majority of cases. Previously, we proposed that MMBIR or NHEJ could be responsible for Jct2 [1]. Here, we expand this ‘”two-step hypothesis” to include homeologous recombination within divergent repeats or similar sequences [33,40]. This is especially relevant to Alu-Alu mediated junctions, where the region of perfect identity may not be extensive enough to employ homology-driven repair, but extensive base-pairing outside the region of identity could aid in driving a recombination coupled replication driven rearrangement process at these loci [34]. In the 16 patients with DUP-TRP/INV-DUP rearrangements presented, 2 contain a Jct2 breakpoint resulting in the formation of a chimeric Alu element.
We hypothesize that the PMD-associated CGR are caused by BIR or MMBIR; these replicative processes have been shown to be error-prone, perhaps because they utilize a polymerase/replisome with reduced fidelity (induced point mutations) as well as reduced processivity (template switching) relative to intergenerational DNA polymerases [20,41]. Evidence now indicates that BIR/MMBIR-associated mutation results from conservative replication coupled with a migrating bubble [42,43]. Thus, DUP-TRP/INV-DUP CGRs involving PLP1 have the potential to additionally impact patient health through point mutations on the X chromosome. These hypotheses need further investigation through large-scale genomic sequencing. Nevertheless, although few in number, de novo point mutations apparently acquired concomitantly with the DUP-TRP/INV-DUP rearrangement in P250 (insertion of an A) and the quadruplication rearrangement in P113 (insertion of a G) were not seen in the corresponding, contiguous (non-breakpoint containing) section of the X chromosome for these intrachromosomal events, a finding consistent with observations made at the MECP2 locus and de novo mutation with CGR formation [20]. Given that on average, ~600 bp were sequenced at each junction, this suggests a rate of 2 mutations in ~15 kb of sequencing, consistent with the elevated point mutation rate observed in association with replication-based mechanisms of repair [20,41].
Junction 1 is present at seemingly identical loci, occurring within a complex inverted repeat structure in the 16 DUP-TRP/INV-DUP rearrangements studied. Further analysis has shown that at least one of these breakpoint junctions is in a region of 24 bp of microhomology and three occur within a >8 kb region of identity within A1a and A1b. The proposed mechanism for Jct1 is BIR within a region of ectopic, inverted homology [1]. Our data reveal that the template switch can occur within smaller regions of identity within A1a and A1b, suggesting that either MMBIR or homeologous recombination, rather than an homologous recombination within IRs may be an alternative mechanism for the formation of these seemingly recurrent junctions [22,31].
Previously, a study of 36 PMD patients identified 3 cases with duplicated copies of PLP1 inserted outside of Xq22 [24]. Conversely, in this study all 16 subjects with junctions in IRs contain the extra copy or copies of the gene on Xq22, therefore suggesting that the mechanism of CNV results in a contiguous rearrangement (triplicated or duplicated regions in tandem, Fig. 2, Table 1). Additionally, all 16 individuals queried by Southern blotting and/or qPCR methodologies indicate that the A1a and A1b inverted LCRs mediate PLP1 DUP-TRP/INV-DUP rearrangements. Although two other IRs in the region, albeit with less sequence identity (the 93% identical outer C/D and 87% identical innermost A2/A3 repeats), could presumably mediate the junction between distal duplication and distal triplication breakpoints, these 16 cases use the A1a and A1b specific repeats. A1a and A1b are ~20 kb in length (versus ~30 kb for C/D and ~10 kb for A2/A3) and are separated by ~50 kb (versus ~140 kb for C/D and ~30 kb for A2/A3). Therefore, the higher level of sequence identity between the A1a and A1b repeats (~99%), added to the shorter inter-repeat distance and the length of the LCR may both increase the likelihood of NAHR leading to the inversion [28,44] and potentiate these repeats as substrates for replication pausing, fork invasion, and reversal through BIR [31]. This is the second locus for which DUP-TRP/INV-DUP cases with recurrent Jct1 mediated by IRs has been described. In MECP2 DUP-TRP/INV-DUP, the K1 and K2 LCRs participate in both non-pathogenic inversions and the rearrangements present in patients [1,27]. Such empirical studies may enable refinement of current predictions for IRs that can predispose regions of the genome to DUP-TRP/INV-DUP [2].
Our data further implicate a “two-step process” of BIR paired with MMBIR to generate CGRs resulting in duplication of copy number sensitive genes proximal to IRs [1]. The rearrangements in the 16 patients with DUP-TRP/INV-DUP contain just two junctions that result in four copy number transition states. This complex pattern on array CGH is due to just two template switches, Jct1 occurring within the LCRs A1a and A1b distal to the PLP1 gene and resulting in an inversion, and Jct2 occurring at varying locations proximal to junction 1 and resuming the pattern of normal replication, resulting in a rescue from the potential formation of a dicentric chromosome (Fig. 4) [45].
The observations at the quadruplication-containing CGR in P113 are consistent with rolling-circle amplification (Fig. 5). The rarity of quadruplication at PLP1 could be due to selective pressures from the increased severity of PMD with additional copies of PLP1 (4 versus 3); it is notable that the quadruplication observed herein does not include the dosage sensitive PLP1 gene [14]. One junction in this CGR (between IRs A1a and A1b) occurs at a similar location as in the DUP-TRP/INV-DUP structures, and PCR genotyping suggests that the interpretation of the rearrangement is complicated by the inversion structural variation, resulting in H2 (S9 Fig). At the proximal junction, the fork template switches twice, invading upstream and leading to a rolling-circle [22,36–38]. After almost two complete copies of the circle (35 kb short of the overall 280 kb), the next junction is a template switch from the proximal end of the quadruplicated region to the distal end of the duplicated region within the LCR region. Our observations are most parsimoniously explained by a rolling-circle amplification event, as predicted for higher-order genomic segment amplification in the MMBIR model [22]. Due to the observations of: i) triplicated and quadruplicated segments, ii) the accompanying point mutation associated with CGR formation, and iii) the prevalence of intrachromosomal rearrangements at this locus, a replicative model for CGR formation is likely [20,42,43]. The quadruplication-containing CGR provides evidence for an important next step in the MMBIR model, allowing for higher-order amplification to occur, as is often observed in cancer [22,46,47].
In summary, our studies confirmed a unique rearrangement product consisting of a DUP-TRP/INV-DUP structure in 16 individuals, with 6 containing triplication of PLP1 [1]. We also elucidated a common, recurrent inversion polymorphism between two IRs distal to this gene. Jct1 occurs between the same repeats that mediate the non-pathogenic inversion, and sequencing of a DUP-TRP/INV-DUP breakpoint within the LCRs showed that these junctions can occur within short stretches of identity within a larger repeat of ~20 kb. This study of breakpoint junctions involved in both DUP-TRP/INV-DUP and higher-order amplification leading to quadruplication implicate replicative mechanisms in the generation of these CGRs. Additionally, we provide experimental evidence supporting the contentions that: i) IRs contribute to genome instability, ii) LCRs can mediate replication-based mechanisms, and iii) short repetitive sequences, such as Alu, can provide microhomology to facilitate template switching. The prevalence of DUP-TRP/INV-DUP events involving PLP1 brings attention to the importance of this mechanism and the potentially broader impact of this rearrangement structure in gene and genome evolution.
Materials and Methods
Inversion Ascertainment
To determine whether there is a polymorphic inversion in the LCRs distal to PLP1, we examined the genomic information for 9 individuals contained in the human genome structural variation (HGSV) track of the UCSC Genome Browser [8,25,26]. The HGSV track (hg18) contains data on discordant fosmid end sequences from libraries of 9 individuals from diverse geographical regions. Discordant end sequence orientations of fosmids spanning LCRs A1a or A1b [23] and having both ends present in unique sequence (not LCRs) indicate potential inversions [26]. Individuals with at least one clone independently spanning each of the LCRs suggest that there is an inversion between the two repeats (S1 Fig).
Inversion Haplotypes and Analysis
Phased data from the 1000 genomes project [29] was used to create plots of two haplotypes in the region spanning from LCRs A1a to A1b (Hg19 coordinates, ChrX:103223669–103324337) [48]. One thousand genomes data was cross-correlated with homozygous genotypes determined from Southern Blots to elucidate phased haplotypes that contain inversion alleles. Results were plotted using custom (in-house) scripts implemented in the R programming language (S4 Fig).
Personal Genomes from Subjects and Patients Investigated
Families with PMD or rearrangements of Xq22.2 including PLP1 were obtained by physician referral or self-referral. Patients were enrolled through informed consent in research protocols approved by the Institutional Review Boards at Baylor College of Medicine (BCM) and the Nemours Alfred I. duPont Hospital for Children. The rearrangements present in patients BAB1290, BAB1612/P374, BAB2389, P250, P255, P500, P518, and P558 were published previously [1,24,30]. Two of the patients with PLP1 triplication (P518 and P674) were described as having more severe disease than patients with duplication [14]. Control DNAs from HapMap individuals [48] were obtained from the Coriell Institute for Medical Research cell repositories.
DNA Digestion and Fragment Separation for Southern Blotting Analysis
Approximately 10 μg of genomic DNA from each patient was digested using BssSI. The DNA was diluted to 60 μl and digested for 4 hours at 37°C with 16U, heat inactivated at 80°C for 20 minutes, and the digest was then repeated with 12U for 3 hours and subsequent heat inactivation (leading to a 10-fold overdigestion). The digested DNAs were then precipitated and concentrated using standard sodium acetate precipitation, and were reconstituted in 25 μl of water with gentle mixing overnight. Concentrations were determined using a NanoDrop spectrophotometer, and samples were then loaded along with an 8–48 kb ladder on a 0.6% Tris - Boric Acid-EDTA (TBE) gel and run in 1X TBE buffer for ~3 days at 50–60 volts. DNA restriction digestion products, i.e. bands on gels, were then visualized with ethidium bromide staining.
Probe Design for Southern Blot Analyses
Probe DNA was prepared using primers A1a proximal probe For - 5′-AATGCAGCTCAAAGGAAAGC-3′ and A1a proximal probe Rev - 5′-AGCCACTGACCAGTGATTTTC-3′ and amplifying a 514 bp fragment from BAC clone RP11–462K21 (https://bacpac.chori.org) DNA prepared using a QIAprep spin miniprep kit. The resultant PCR bands were resolved on 1% agarose and Tris-Acetate-EDTA gels and purified using a Zymoclean Gel DNA Recovery Kit (Zymo Research). Probe DNA was validated by Sanger sequencing, using both forward and reverse primers and was frozen at-20°C in 90 ng aliquots.
Southern Blotting
Southern Blotting was carried out as previously described [23]. Briefly, DNAs were subjected to electrophoresis for sufficient duration to distinguish 25 and 29 kb fragment sizes and were then transferred to a Sure Blot positively charged nylon membrane by standard ‘sandwich’ methodology for 2–3 days. Approximately 80 ng of DNA was labeled with 32P-dCTP by random priming for 2–4 hours at 37°C using the Random Primed DNA labeling kit (Roche). Membranes were pre-hybridized for 4 hours in 10% dextran sulfate/1M NaCl/1%SDS (hybridization solution) with 4mg of sheared salmon sperm DNA at 65°C. Probe was pre-associated in hybridization solution with ~1mg sheared placental DNA at 65°C for ~2 hours, then added to the pre-hybridized membrane. Hybridization was carried out at 65°C overnight (~18 hours). The following day, the blot was washed and analyzed using autoradiography for bands corresponding to PLP1 A1a structural haplotype information (~25 and 29 kb).
Array CGH Analyses
To determine the size, genomic content, and extent of PLP1 rearrangements, a high-density oligonucleotide array from Agilent was custom-designed to examine PMD patients. The 4 x 44 K microarray was designed using the Agilent eArray website (https://earray.chem.agilent.com/earray/) and was used to visualize the rearrangements of three patients in this study (BAB1290, BAB1612/P374, and BAB2389), the family containing individuals BAB3698, BAB3699, BAB3700, and BAB4179, and to complement existing array data for P500, P1407, and P113. The family of P113 was explored using Agilent arrays, including patients P113 and P117, as well as the mother of P113, P154, and grandmother, P088, who are both carriers. Probe labeling and hybridization were conducted as previously described, with NA15510 and NA10851 used as reference DNAs for female and male individuals, respectively (Accession GSE63594) [1]. Purified DNA samples from P113, P250, P500, P518, P558, P642, P674, P820, P842 and P1150 were submitted to NimbleGen for array service with normal male control NM002 as a reference. The NimbleGen X chromosome CGH fine-tiling array with oligonucleotide probes of 45 to 85 bases in length with median spacing of 106 bp throughout the whole X chromosome was used. Patient DNA samples P255/P298, BAB1612/P374, P1389 and P1407 were submitted to the Biomolecular Core Lab at duPont Hospital for Children for hybridization to Affymetrix Cytogenetics 2.7M Array. DNA sample P1407 was submitted to Coriell’s Genotyping and Microarray Center for hybridization on Affymetrix Genome Wide Human SNP Array 6.0. All data from Affymetrix arrays were analyzed with GeneChip Command Console Software AGCC. NimbleGen and Affymetrix Cytogenetics array data were aligned with qPCR data and plotted using the R programming language (Affymetrix and NimbleGen data are under Accession GSE64122) (Figs. 2B, C, D and S5).
Semi-Quantitative Multiplex PCR (qPCR) for Detection of Copy Number
Semi-quantitative multiplex PCR was performed using a QIAGEN Multiplex PCR kit according to the manufacturer’s protocol to analyze regions on the X chromosome in and surrounding PLP1 to determine copy number. Primer pairs were selected using the NCBI primer design tool (primers available upon request). In each experiment, five control DNAs were used, two known to have duplications in the region of interest without CGRs and three normal controls known to be single copy in the region of interest. A primer pair that amplifies a region of the human dystrophin (DMD) gene on the p-arm of the X chromosome was included in each multiplex reaction for amplification of a single-copy region. Products were separated by electrophoresis on a 4% NuSieve 3 : 1 agarose gel (Lonza, Walkersville MD) and stained with ethidium bromide. Net intensity of each band was determined using a Molecular Imaging system with Kodak Gel Logic Imaging software or AlphaImager HP. Copy number was determined by calculating the ratio of the net intensities of bands in the test region to dystrophin single-copy region for each DNA sample and then normalized by dividing by the average of the ratios of test region to dystrophin of the three normal controls. Theoretical ratios were: one, single-copy; two, duplication; three, triplication. Alternatively, quantitative PCR was performed as above except that one primer of each pair was labeled with 6-FAM and samples were submitted to the Biomolecular Core Lab at duPont Hospital for Children for capillary electrophoresis on an ABI PRISM 3130XL DNA Analyzer. Analysis of copy number was determined as above, by using area under the peak as determined by Peak Scanner software rather than net intensity. Triplicated, quadruplicated and duplicated regions were mapped to within several kb of their endpoints using these methods.
Fluorescent in situ Hybridization (FISH) to Confirm Copy Number
Interphase nuclei and metaphase chromosomes were prepared from 700 μl of whole blood stored in sodium heparin Vacutainer tubes as follows. Blood was placed in α-MEM, 20%FBS, 1% L-glutamine, 50 μg/ml gentamycin and treated with 150 μl Phytohemagluttenin (Invitrogen, Carlsbad CA). The cultures were incubated at 37°C for 72 hours in upright position after which they were treated with 100 μl colcemid by trituration followed by incubation at 37°C for 30 min. Cultures were then subjected to centrifugation at 350 x g for 6 minutes. The supernatant liquid was discarded and the pellet was suspended in 10 ml 75mM KCl pre-warmed to 37°C and incubated at 37°C for 15 minutes. Then 1 ml of fixative (3 : 1 mixture of methanol:acetic acid) was added slowly. The preparation was washed 3 times in 10 ml of fixative with pelleting by centrifugation at 350xg for 6 minutes after washes. The resulting pellet of interphase and metaphase chromosomes was then stored in fixative at -20°C. Chromosomes and nuclei were dropped onto pre-cleaned Fisherbrand slides in a CDS-5 Glovebox environmental Chamber, (Thermotron, Holland Michigan) set at 25°C and 50% humidity. Slides were stored at -20°C in a vacuum under dessication until use.
FISH was performed using cosmid clone U125A1 and BAC clone RP13–188A5 obtained from the BACPAC resource center. Cosmid and BAC DNAs were isolated using the QIAGEN Plasmid purification and QIAGEN Large-Construct kits, respectively. One μg of U125A1 DNA was labeled with Biotin-16-dUTP and 1μg RP13–188A5 DNA was labeled with digoxigenin using the DIG-Nick Translation Mix. Labeled probes were purified using Nuctrap probe purification columns according to the manufacturer’s protocol. After hybridization to chromosomes and nuclei according to standard protocol, biotinylated U125A1 was bound to Cy3-labeled streptavidin, further amplified with biotinylated antiavidin (Vector Laboratories, Burlingame CA) and detected with a second layer of Cy3-labeled streptavidin. Simultaneously, RP13–188A5 labeled with digoxigenin was coupled with mouse antidigoxigenin, detected with rabbit anti-mouse FITC (Jackson ImmunoResearch Laboratories) and further amplified with goat anti-rabbit FITC antibody. Nuclei and chromosome spreads were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) and cover slips were mounted using Vectashield antifade solution. Images were captured using a Leica DM RXA2 fluorescence microscope and Openlab imaging software (Perkin Elmer, Waltham MA).
STR and SNP Analyses to Determine Origin of Extra Genomic Copies
To examine whether an intra - or inter-chromosomal origin occurred for the extra genomic segments in each patient’s genome, we analyzed 2 STRs and 9 single SNPs within the duplicated/triplicated region common to most patients. Sites were chosen based on marker genotypes displaying a high degree of heterozygosity in HapMap samples. S6 Table depicts the dbSNP identifiers, locations with respect to Chromosome X sequence NT_011651.17, and primers used to amplify the SNP or STR. Regions of interest were amplified from DNA with HotStar Taq DNA polymerase (Qiagen) for products <1kb or Expand High Fidelity PCR system (Roche) for products >1kb. Patients P250, P255/298, BAB1612/P374, P500, P518, P558, P642, P674, P820, P842, P1150, P1389, and P1407 were analyzed. Products containing the STR were amplified with one primer within the pair fluorescently-labeled so product sizes could be evaluated by capillary electrophoresis on ABI’s PRISM 3130 XL DNA Analyzer. Products containing the SNP were purified using QIAquick PCR or Gel purification kits, then sequenced with the Big Dye Terminator kit v. 3.1 (Life Technologies), according to the manufacturer’s instructions. Patients P113, P117 and carriers P154 and P088 were subjected to genotyping using the Illumina OmniExpress SNP array analyses at the human genome sequencing center of BCM. Data from the analyses was visualized by plotting the B allele frequencies versus the X chromosome coordinates encompassing the quadruplication genomic rearrangement, as well as the Log ratio of SNP intensity (S11 Fig).
Jct2 Analyses
Inverse PCR was used to obtain the first junction of patient P1150. Briefly, DNA was digested with NheI and ligated to form circles. PCR primers were designed to amplify in opposite directions around the circle by long-range PCR using the Expand High Fidelity PCR system (S6 Table). When the PCR products were analyzed on an agarose gel, a product was found that was unique to the patient. The product was subjected to DNA sequencing according to the manufacturer’s instructions, then purified with the Filtration Cartridge (Edge Biosystems, Inc., Gaithersburg MD) and separated using an ABI PRISM 3130 xl Genetic Analyzer. DNA sequence was analyzed using Vector NTI sequence analysis software.
Proximal junctions were obtained for the personal genomes from the remaining triplication patients by long-range PCR using appropriately positioned primers at the endpoints of copy number changes (Table 1 and S6 Table) in 25 μl reactions with 50–100 ng of patient DNA using TaKaRa LA Taq or using the Expand High Fidelity PCR dNTPack kit according to the manufacturers’ instructions. PCR products were prepared for sequencing by using the standard ExoSAP-IT protocol (Affymetrix, Santa Clara CA) or by using the Qiagen PCR purification kit and DNA sequencing reactions were performed as indicated above using primers used in amplification or internal primers as indicated in S6 Table. Sequences were aligned to the human genome reference sequence, and breakpoints are depicted in S6 Fig. We had previously reported the sequence across the junction in P255 [24].
Jct1 Analyses
PCR was conducted across Jct1 from DNAs prepared from patients with DUP-TRP/INV-DUP CGRs using a QIAGEN Multiplex PCR. Two control DNAs duplicated through this region and three control DNAs with a single copy at this locus were amplified in parallel. Along with a dystrophin primer pair (Hdys 23F-6FAM and Hdys 23 R) for a single copy region of the human dystrophin gene, we used primers pairs V362H12-F19–6FAM and V362H12-R19 (red arrows), and V362H12-F24–6FAM and V362H12-R24 (black arrows), and V362H12-F19–6FAM and V362H12-R24 (one red, one black arrow) (Figs. 3E, S8, S6 Table). Fluorescently labeled PCR products were diluted 1 : 100 in sterile HPLC water and subjected to capillary electrophoresis using an ABI PRISM 3130 XL DNA Analyzer. Copy number analysis was performed as previously described using the Peak Scanner software [24].
To subclone breakpoints in PMD DUP-TRP/INV-DUP patients, we amplified patient DNAs containing rearrangements (from BAB1612/P374, BAB2389, and BAB1290) with PCR primers that anneal within the A1a and A1b LCRs and uniquely flanking primers. This yielded four overlapping segments of the two LCRs (S6 Fig). These PCR products were then subjected to electrophoresis in crystal violet 0.8% agarose gels, purified using the SNAP purification kit from Invitrogen, and cloned into TOPO XL cloning vectors. Resultant clones for each of the four segments were screened by digestion and sequenced in their entirety. At least two clones for each region, obtained from independent PCR reactions, were screened for the breakpoint and the corresponding A1a or A1b region. Sequence analysis was conducted using the Lasergene 9 DNA analysis software suite.
Copy Number Analysis of Junctions in the Quadruplication by dPCR
Copy number of junctions in the quadruplication patients and a carrier were determined by dPCR using QuantStudioTM 3D Digital PCR System (Life Technologies), according to the manufacturer’s instructions. Concentration of DNA was determined by QubitR dsDNA BR assay (Life Technologies) using the Qubit 2.0 fluorometer (Life Technologies). Sample DNA was digested with SphI (NEBiolabs) to separate multiple copies of interest that may be located on the same molecule without disrupting the region of amplification. Digests were performed using 400 ng of DNA in a 10 μl reaction containing 10U of SphI and incubating at 37°C for 1.5 hr, followed by heat-inactivation of the enzyme at 65°C for 20 min. The digest was diluted to 40 μl with RNase-free water to yield a concentration of 10 ng/μl DNA. Primers and probes used in the dPCR assays are in S6 Table. Reactions for dPCR included 1x QuantStudioTM 3D Digital PCR Master Mix, 1x TaqMan Copy Number Reference Assay for human RNaseP (Life Technologies, Cat. # 4403328, VIC label), 1–1.5x PrimeTime qPCR 5’ nuclease assay (IDT, FAM label) for jct1 or jct2/3 and 40–60ng DNA in a 16 μl volume. Fifteen μl of this mix was used to load the Digital PCR 20K Chip (Life Technologies); chips were processed according to the manufacturer’s instructions.
Supporting Information
Zdroje
1. Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, et al. (2011) Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet 43 : 1074–1081. doi: 10.1038/ng.944 21964572
2. Dittwald P, Gambin T, Gonzaga-Jauregui C, Carvalho CM, Lupski JR, et al. (2013) Inverted low-copy repeats and genome instability—a genome-wide analysis. Hum Mutat 34 : 210–220. doi: 10.1002/humu.22217 22965494
3. Zhou W, Zhang F, Chen X, Shen Y, Lupski JR, et al. (2013) Increased genome instability in human DNA segments with self-chains: homology-induced structural variations via replicative mechanisms. Hum Mol Genet 22 : 2642–2651. doi: 10.1093/hmg/ddt113 23474816
4. Lakich D, Kazazian HH Jr., Antonarakis SE, Gitschier J (1993) Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet 5 : 236–241. 8275087
5. Hermetz KE, Newman S, Conneely KN, Martin CL, Ballif BC, et al. (2014) Large inverted duplications in the human genome form via a fold-back mechanism. PLoS Genet 10: e1004139. doi: 10.1371/journal.pgen.1004139 24497845
6. Giorda R, Ciccone R, Gimelli G, Pramparo T, Beri S, et al. (2007) Two classes of low-copy repeats comediate a new recurrent rearrangement consisting of duplication at 8p23.1 and triplication at 8p23.2. Hum Mutat 28 : 459–468. 17262805
7. Lange J, Skaletsky H, van Daalen SK, Embry SL, Korver CM, et al. (2009) Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell 138 : 855–869. doi: 10.1016/j.cell.2009.07.042 19737515
8. Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, et al. (2008) Mapping and sequencing of structural variation from eight human genomes. Nature 453 : 56–64. doi: 10.1038/nature06862 18451855
9. Flores M, Morales L, Gonzaga-Jauregui C, Dominguez-Vidana R, Zepeda C, et al. (2007) Recurrent DNA inversion rearrangements in the human genome. Proc Natl Acad Sci U S A 104 : 6099–6106. 17389356
10. Soler-Alfonso C, Carvalho CM, Ge J, Roney EK, Bader PI, et al. (2014) CHRNA7 triplication associated with cognitive impairment and neuropsychiatric phenotypes in a three-generation pedigree. Eur J Hum Genet.
11. Beri S, Bonaglia MC, Giorda R (2013) Low-copy repeats at the human VIPR2 gene predispose to recurrent and nonrecurrent rearrangements. Eur J Hum Genet 21 : 757–761. doi: 10.1038/ejhg.2012.235 23073313
12. Ishmukhametova A, Chen JM, Bernard R, de Massy B, Baudat F, et al. (2013) Dissecting the structure and mechanism of a complex duplication-triplication rearrangement in the DMD gene. Hum Mutat 34 : 1080–1084. doi: 10.1002/humu.22353 23649991
13. Shimojima K, Mano T, Kashiwagi M, Tanabe T, Sugawara M, et al. (2012) Pelizaeus-Merzbacher disease caused by a duplication-inverted triplication-duplication in chromosomal segments including the PLP1 region. Eur J Med Genet 55 : 400–403. doi: 10.1016/j.ejmg.2012.02.013 22490426
14. Wolf NI, Sistermans EA, Cundall M, Hobson GM, Davis-Williams AP, et al. (2005) Three or more copies of the proteolipid protein gene PLP1 cause severe Pelizaeus-Merzbacher disease. Brain 128 : 743–751. 15689360
15. Garbern J, Hobson G (2002) Prenatal diagnosis of Pelizaeus-Merzbacher disease. Prenat Diagn 22 : 1033–1035. 12424770
16. Inoue K, Osaka H, Thurston VC, Clarke JT, Yoneyama A, et al. (2002) Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet 71 : 838–853. 12297985
17. Inoue K, Osaka H, Imaizumi K, Nezu A, Takanashi J, et al. (1999) Proteolipid protein gene duplications causing Pelizaeus-Merzbacher disease: molecular mechanism and phenotypic manifestations. Ann Neurol 45 : 624–632. 10319885
18. Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, et al. (1991) DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell 66 : 219–232. 1677316
19. Liu P, Gelowani V, Zhang F, Drory VE, Ben-Shachar S, et al. (2014) Mechanism, Prevalence, and More Severe Neuropathy Phenotype of the Charcot-Marie-Tooth Type 1A Triplication. Am J Hum Genet 94 : 462–469. doi: 10.1016/j.ajhg.2014.01.017 24530202
20. Carvalho CM, Pehlivan D, Ramocki MB, Fang P, Alleva B, et al. (2013) Replicative mechanisms for CNV formation are error prone. Nat Genet 45 : 1319–1326. doi: 10.1038/ng.2768 24056715
21. Lee JA, Carvalho CM, Lupski JR (2007) A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131 : 1235–1247. 18160035
22. Hastings PJ, Ira G, Lupski JR (2009) A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5: e1000327. doi: 10.1371/journal.pgen.1000327 19180184
23. Lee JA, Inoue K, Cheung SW, Shaw CA, Stankiewicz P, et al. (2006) Role of genomic architecture in PLP1 duplication causing Pelizaeus-Merzbacher disease. Hum Mol Genet 15 : 2250–2265. 16774974
24. Woodward KJ, Cundall M, Sperle K, Sistermans EA, Ross M, et al. (2005) Heterogeneous duplications in patients with Pelizaeus-Merzbacher disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am J Hum Genet 77 : 966–987. 16380909
25. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. (2002) The human genome browser at UCSC. Genome Res 12 : 996–1006. 12045153
26. Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, et al. (2005) Fine-scale structural variation of the human genome. Nat Genet 37 : 727–732. 15895083
27. Small K, Iber J, Warren ST (1997) Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat Genet 16 : 96–99. 9140403
28. Liu P, Lacaria M, Zhang F, Withers M, Hastings PJ, et al. (2011) Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am J Hum Genet 89 : 580–588. doi: 10.1016/j.ajhg.2011.09.009 21981782
29. The 1000 Genomes Project Consortium, Abecasis GR, Altshuler D, Auton A, Brooks LD, et al. (2010) A map of human genome variation from population-scale sequencing. Nature 467 : 1061–1073. doi: 10.1038/nature09534 20981092
30. Lee JA, Madrid RE, Sperle K, Ritterson CM, Hobson GM, et al. (2006) Spastic paraplegia type 2 associated with axonal neuropathy and apparent PLP1 position effect. Ann Neurol 59 : 398–403. 16374829
31. Mizuno K, Miyabe I, Schalbetter SA, Carr AM, Murray JM (2013) Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature 493 : 246–249. doi: 10.1038/nature11676 23178809
32. Zhang Y, Saini N, Sheng Z, Lobachev KS (2013) Genome-wide screen reveals replication pathway for quasi-palindrome fragility dependent on homologous recombination. PLoS Genet 9: e1003979. doi: 10.1371/journal.pgen.1003979 24339793
33. de Wind N, Dekker M, Claij N, Jansen L, van Klink Y, et al. (1999) HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat Genet 23 : 359–362. 10545954
34. Boone PM, Yuan B, Campbell IM, Scull JC, Withers MA, et al. (2014) The Alu-Rich Genomic Architecture of SPAST Predisposes to Diverse and Functionally Distinct Disease-Associated CNV Alleles. Am J Hum Genet 95 : 143–161. doi: 10.1016/j.ajhg.2014.06.014 25065914
35. Lindsay SJ, Khajavi M, Lupski JR, Hurles ME (2006) A chromosomal rearrangement hotspot can be identified from population genetic variation and is coincident with a hotspot for allelic recombination. Am J Hum Genet 79 : 890–902. 17033965
36. Yu C, Bonaduce MJ, Klar AJ (2012) Remarkably high rate of DNA amplification promoted by the mating-type switching mechanism in Schizosaccharomyces pombe. Genetics 191 : 285–289. doi: 10.1534/genetics.112.138727 22377633
37. McEachern MJ, Haber JE (2006) Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75 : 111–135. 16756487
38. Andersson DI, Hughes D (2009) Gene amplification and adaptive evolution in bacteria. Annu Rev Genet 43 : 167–195. doi: 10.1146/annurev-genet-102108-134805 19686082
39. Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, et al. (2010) Diversity of human copy number variation and multicopy genes. Science 330 : 641–646. doi: 10.1126/science.1197005 21030649
40. Rayssiguier C, Thaler DS, Radman M (1989) The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342 : 396–401. 2555716
41. Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, et al. (2011) Break-induced replication is highly inaccurate. PLoS Biol 9: e1000594. doi: 10.1371/journal.pbio.1000594 21347245
42. Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, et al. (2013) Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502 : 389–392. doi: 10.1038/nature12584 24025772
43. Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, et al. (2013) Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature 502 : 393–396. doi: 10.1038/nature12585 24025768
44. Dittwald P, Gambin T, Szafranski P, Li J, Amato S, et al. (2013) NAHR-mediated copy-number variants in a clinical population: mechanistic insights into both genomic disorders and Mendelizing traits. Genome Res 23 : 1395–1409. doi: 10.1101/gr.152454.112 23657883
45. Mizuno K, Lambert S, Baldacci G, Murray JM, Carr AM (2009) Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev 23 : 2876–2886. doi: 10.1101/gad.1863009 20008937
46. Watanabe T, Tanabe H, Horiuchi T (2011) Gene amplification system based on double rolling-circle replication as a model for oncogene-type amplification. Nucleic Acids Res 39: e106. doi: 10.1093/nar/gkr442 21653557
47. Windle BE, Wahl GM (1992) Molecular dissection of mammalian gene amplification: new mechanistic insights revealed by analyses of very early events. Mutat Res 276 : 199–224. 1374515
48. The International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437 : 1299–1320. 16255080
49. Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, et al. (2010) A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143 : 837–847. doi: 10.1016/j.cell.2010.10.027 21111241
Štítky
Genetika Reprodukčná medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



