-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Escape from X Inactivation Varies in Mouse Tissues
X inactivation is a female-specific phenomenon that occurs during early development and results in the silencing of one X chromosome in female mammals. However, some genes escape inactivation and remain expressed from both X chromosomes. To date, the identity of escape genes and the molecular mechanisms of this process are still being explored. Here, we use a new binomial model combined with a mouse system with identifiable alleles and skewed X inactivation to identify and further define the chromatin landscape of escape genes in vivo. We find that some escape genes are common to multiple tissues while others are tissue-specific. We also show that expression levels of alleles on the inactive X correlate with factors associated with open chromatin such as RNA Polymerase II and DNase I hypersensitive sites. Additionally, escape genes co-localized with CTCF binding clusters on the Xi, suggesting a role for CTCF binding in delineating regions of escape and inactivation. Our findings represent the first comprehensive analysis of escape in vivo. Identification of tissue-specific escape genes could lead to a better understanding of the underlying causes of sex-linked disorders such as X-linked intellectual disability and Turner syndrome.
Published in the journal: Escape from X Inactivation Varies in Mouse Tissues. PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005079
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005079Summary
X inactivation is a female-specific phenomenon that occurs during early development and results in the silencing of one X chromosome in female mammals. However, some genes escape inactivation and remain expressed from both X chromosomes. To date, the identity of escape genes and the molecular mechanisms of this process are still being explored. Here, we use a new binomial model combined with a mouse system with identifiable alleles and skewed X inactivation to identify and further define the chromatin landscape of escape genes in vivo. We find that some escape genes are common to multiple tissues while others are tissue-specific. We also show that expression levels of alleles on the inactive X correlate with factors associated with open chromatin such as RNA Polymerase II and DNase I hypersensitive sites. Additionally, escape genes co-localized with CTCF binding clusters on the Xi, suggesting a role for CTCF binding in delineating regions of escape and inactivation. Our findings represent the first comprehensive analysis of escape in vivo. Identification of tissue-specific escape genes could lead to a better understanding of the underlying causes of sex-linked disorders such as X-linked intellectual disability and Turner syndrome.
Introduction
Dosage compensation in mammals is achieved by upregulation of the X chromosome in both sexes and random inactivation of one of the two X chromosomes in females [1]. Initially, the future inactive X (Xi) is coated by the long non-coding RNA Xist (X inactive specific transcript), a process essential for the onset of silencing [2]. Inactive chromatin marks such as tri-methylation of lysine 27 on histone H3 (H3K27me3) are put in place along with DNA methylation at CpG islands, macroH2A modification, and late replication, which represent late and possibly secondary events that lock in silencing of most genes in somatic cells. Despite efficient silencing some genes escape X chromosome inactivation (XCI) and thus remain bi-allelically expressed in females [3,4]. Surveys in cultured human/mouse hybrid cells and in cell lines from individuals with skewed XCI have shown that about 8–15% of human genes consistently escape XCI, 10–13% display variable levels of escape, and 10–20% vary between cell lines and individuals [5–7]. Escape from XCI results in significant sexual dimorphisms in levels of gene expression, and bi-allelic expression of at least some escape genes is important for a normal phenotype in human females. Indeed, the presence of a single X chromosome (45,X) results in Turner syndrome characterized by poor viability in utero, infertility, short stature, and an array of other abnormalities [8,9].
Genes that escape XCI usually lack both Xist RNA coating [10–12] and repressive histone modifications associated with silencing [13–18]. Furthermore, escape genes have specific DNA methylation signatures [19,20]. Whether other specific chromatin elements such as CTCF may be implicated is still unclear [21,22]. Little is known about the distribution of escape from XCI in different tissues in vivo and about the mechanisms that control tissue-specific differences.
XCI is usually random in somatic cells, thus allelic characteristics of X-linked genes can only be studied in cell populations with skewed XCI obtained by cell-cloning or flow-sorting, or by cell selection based on a specific mutation. To derive a cell line (Patski) with completely skewed XCI we previously used an Hprt mutation to select cells that contain an active X (Xa) from Mus spretus (abbreviated spretus) and an Xi from a Mus musculus laboratory strain (C57BL/6J abbreviated BL6) [23]. This in vitro system allowed us to determine allele-specific gene expression based on frequent SNPs (single nucleotide polymorphisms) between the mouse species (1/50–100bp), and to identify genes that escape XCI [18]. Mouse trophoblastic cells in which the paternal X is always inactivated offer an alternative way to identify genes that escape imprinted XCI in vitro [14]. However, Patski cells and trophoblastic cells may not represent the in vivo situation and do not address potential differences between tissues. A recent study examined mid-gestation placenta to address escape from imprinted XCI in vivo; but this system only addresses imprinted XCI in an extra-embryonic tissue, the mechanism of which differs from random XCI in the embryo proper [24]. Another recent study examined escape from XCI in mouse brain based on flow sorting cell populations with skewed XCI to >98% purity [25].
To determine the XCI status of genes in multiple tissues in vivo we developed a mouse model in which F1 animals have completely skewed XCI of the spretus X due to an Xist mutation on the BL6 X [26]. In the present study this model was exploited to compare the XCI status of genes between mouse tissues. We developed a new binomial model to estimate the probability of bi-allelic expression based on RNA-seq and SNPs, resulting in the identification of common as well as tissue-specific escape genes, which were verified by RT-PCR. Allele-specific profiles for two features of active genes, RNA polymerase II occupancy and DNase I hypersensitivity, demonstrate active chromatin signatures at escape genes. In addition, allele-specific profiles of CTCF occupancy were obtained to examine its distribution relative to escape genes.
Results
Pipeline to determine allele-specific gene expression
To assess allele-specific expression of X-linked genes in vivo, we mated BL6 females heterozygous for a deletion of the proximal A-repeat of Xist (XistΔ/+) [27] to spretus males. In the resulting F1 XistΔ/+ (thereafter called F1) female progeny the maternal BL6 X chromosome fails to inactivate, leading to completely skewed XCI in all tissues [26]. This was further verified based on allelic expression of a gene known to be subject to XCI (Ubqln2), as determined by Sanger sequencing of RT-PCR products (S1A Fig). Whole brains and spleens from two adult F1 females (biological replicates) and both ovaries pooled from one of these females were used for RNA-seq, followed by gene expression analyses using a new pipeline (see below) to better capture allele-specific reads based on high quality SNPs. For comparison and validation we also re-analyzed two independent RNA-seq datasets generated for the Patski cell line in which the XCI pattern is reversed, i.e. the BL6 X is inactive and the spretus X active [18].
To identify reads that map to each parental genome in F1 mice, a "pseudo-spretus" genome was assembled by substituting known SNPs between BL6 and spretus into the BL6 mm9 reference genome [26,28]. SNPs were obtained from the Sanger Institute (SNP database Nov/2011 version) and from in house analysis [18]. RNA-seq reads were aligned separately to the BL6 and to the pseudo-spretus genomes (see details in Material and Methods). We segregated all high-quality uniquely mapped reads (MAPQ ≥ 30) into three categories: (1) BL6-SNP reads containing only BL6-specific SNP(s); (2) spretus-SNP reads containing only spretus-specific SNP(s); (3) reads that do not contain valid SNPs. We refer to both BL6-SNP reads and spretus-SNP reads as "allele-specific reads". Examination of the distribution of allele-specific reads on genes known to be subject to XCI, for example Ubqln2, confirmed the absence of spretus reads due to XCI skewing in female F1 tissues (S1B Fig).
We calculated diploid gene expression based on all mapped reads using cufflinks/v2.0.2 [29] (http://cufflinks.cbcb.umd.edu/) to determine RPKM (reads per kb of exon length per million mapped reads). Next, we defined SNP-based haploid gene expression from alleles on the Xi or the Xa (Xi-SRPM or Xa-SRPM) to be allele-specific SNP-containing exonic reads per 10 million uniquely mapped reads.
Modeling the XCI status of genes
Here, we propose and validate a new binomial model to identify escape genes and estimate the statistical confidence of escape probability.
For each gene i on chromosome X, let the number of allele-specific RNA-seq reads mapped to the inactive/active chromosomes be nio and ni1, respectively, and let. ni = nio + ni1 We model nio by a binomial distribution
where pi indicates the expected proportion of reads from the Xi. The estimate of the binomial proportion is Let zα/2 be the 100(1 - α/2)th percentile of N(0,1). The confidence interval of each pi^ is, To incorporate the mapping biases toward the BL6 genome over the pseudo-spretus genome into the above model, we define the mapping bias ratio rm for each RNA-seq experiment to be where NA0 and NA1 are the number of allele-specific autosomal reads in the "inactive X containing" genome and the "active X containing" genome, respectively. Considering the mapping biases the corrected estimate of pi is: The upper and lower confidence limits are corrected accordingly.For each RNA-seq experiment, we called a gene "escape" if (1) the 99% lower confidence limit (α = 0.01) of the escape probability was greater than zero, indicating significant contribution from the Xi, (2) the diploid gene expression measured by RPKM was ≥1, indicating that the gene was expressed, and (3) the Xi-SRPM was ≥2, representing sufficient reads from the Xi. Note that we only considered exonic reads, except for the lncRNA Firre (see below). Biological replicates of RNA-seq experiments were analyzed separately. Examples of mRNA SNP-read coverage on the Xa and Xi visualized in the UCSC browser are shown for a number of genes we determined to either escape or be subject to XCI (Figs. 1–3, S1, S2 and S4). There was a good concordance between reads at different SNPs within a given gene. Levels of escape from XCI determined by RNA-seq analysis correlated with measurements done by RT-PCR using species-specific primers, although the percent of expression from the Xi measured by RT-PCR was usually higher than that determined by RNA-seq. For example, Cfp and Plp1 had 5% and 0.3% SNP reads on the Xi by RNA-seq and 15% and 1.5% Xi (versus Xa) expression measured by RT-PCR with species-specific primers, respectively (Figs. 1C and 1D, S1C and S1D).
Fig. 1. Evaluation of escape from XCI in mouse tissues. 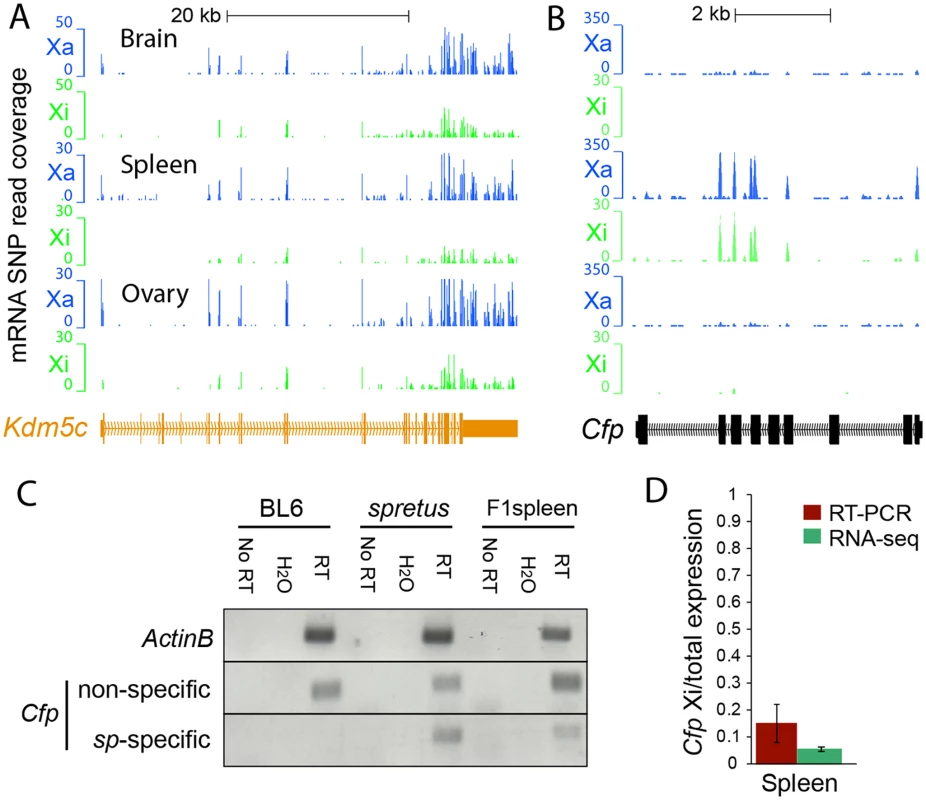
(A) Example of mRNA SNP read distribution profiles on the Xi and Xa for Kdm5c, an escape gene common to all mouse tissues tested (brain, spleen and ovary). SNP reads specific to the Xa (blue) and Xi (green) are visualized in the UCSC genome browser. RNA-seq read quantification was done by normalizing reads from the Xi to total reads (Xi + Xa) in two biological replicates. (B) Example of mRNA SNP read distribution profiles on the Xi and Xa for Cfp, a gene that escapes XCI only in spleen. SNP reads specific to the Xa (blue) and Xi (green) are visualized in the UCSC genome browser. (C, D) Validation of escape from XCI for Cfp. (C) Gel electrophoresis of RT-PCR products using non-species-specific primers and spretus-specific primers (sp) (S10 Table) in BL6, spretus, and F1 brain in which the Xi is from spretus. ActinB was used as a control. Control reactions include "No RT" (no reverse transcriptase) and H2O (instead of primers). (D) Graph comparing RT-PCR Cfp gel band quantification measured by ImageJ with SNP read quantification measured by RNA-seq. Xi product abundance measured by RT-PCR using spretus-specific primers in F1 spleen was normalized to total RT-PCR product abundance measured by non-species specific primers. Fig. 2. Validation of Rlim, Shroom4, Car5b, Hdac6, 5530601H04Rik expression profiles and Firre mRNA profiles. 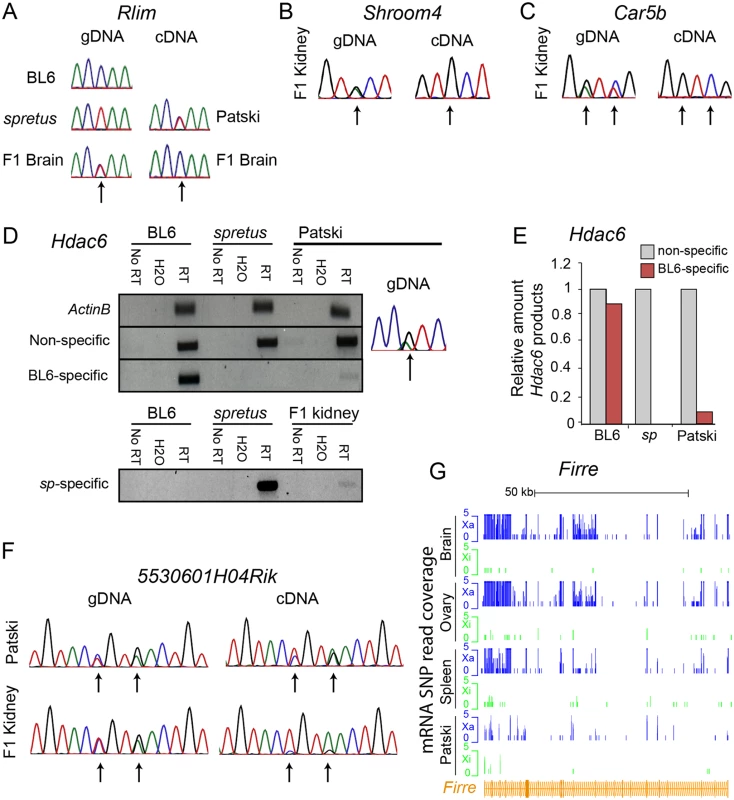
(A) Sanger sequencing tracings of Rlim cDNA confirm bi-allelic expression in Patski cells but not brain, while gDNA sequence tracings show SNP heterozygosity (C in BL6 and T in spretus). Arrows indicate SNP positions. (B, C) Sanger sequencing tracings of Shroom4 (B) and Carb5 (C) cDNA confirm that these genes are subject to XCI in F1 kidney while they were shown to escape XCI in Patski cells [18]. gDNA sequence tracings show SNP heterozygosity (Shroom4—G in BL6 and A in spretus; Car5b—G and C in BL6; A and T in spretus). Arrows indicate SNP positions. (D, E) Validation of escape from XCI for Hdac6. (D) Gel electrophoresis of RT-PCR products using non-species-specific primers, spretus-specific primers, and BL6-specific primers (S10 Table) in BL6, spretus, Patski cells and F1 kidney. ActinB was used as a control. Control reactions include "No RT" (no reverse transcriptase) and H2O (instead of primers). Sanger sequencing tracing confirms heterozygosity (A in BL6 and G in spretus) in the left primer (S10 Table). (E) Xi expression of Hdac6 was determined to be 9% of total expression in Patski cells by gel band quantification measured by ImageJ. (F) Sanger sequencing tracings of 5530601H04Rik cDNA confirms that the lncRNA escapes XCI in kidney and Patski cells, while gDNA sequence tracings show heterozygosity (T and A in BL6; A and G in spretus). Arrows indicate SNP positions. (G) mRNA SNP read distribution profiles on the Xi and Xa for Firre, a lncRNA that escapes XCI in mouse tissues and Patski cells. Note that Firre is classified as a variable escape gene in brain (S1 Dataset). Xa SNP reads are in blue and Xi SNP reads in green. Fig. 3. Enrichment in PolII-S5p and DNase I hypersensitivity on the Xi allele at escape genes. 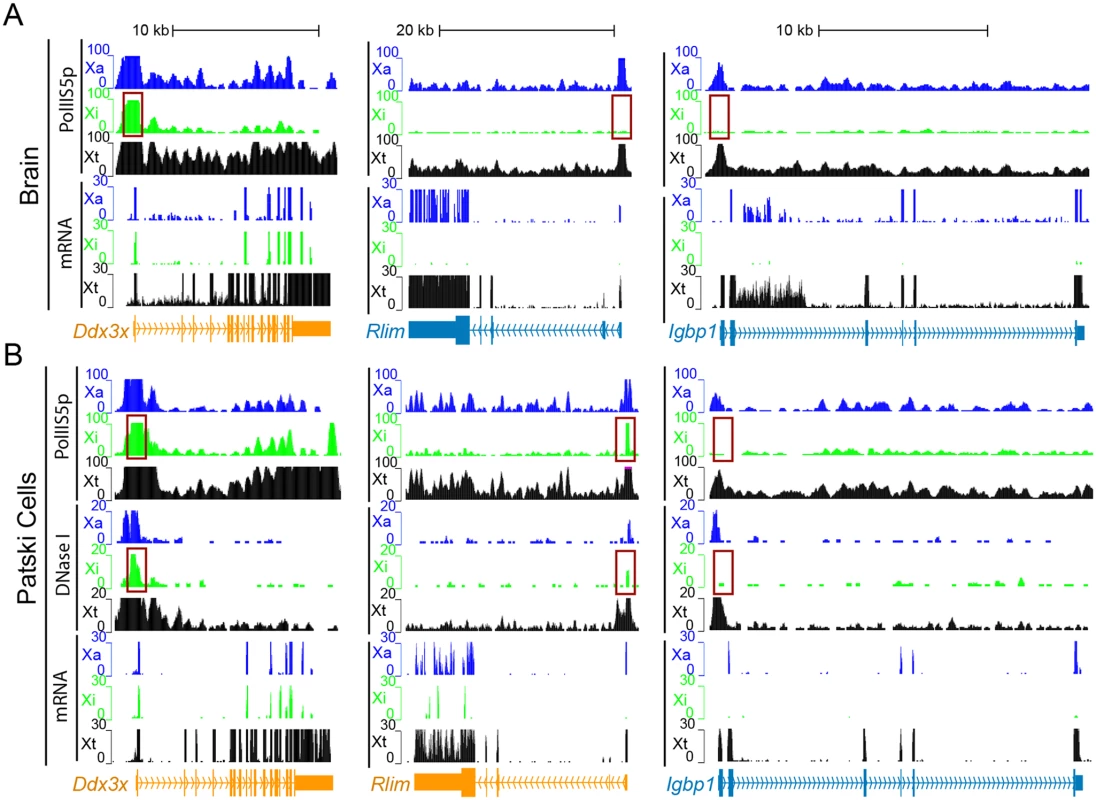
(A, B) Examples of allele-specific PolII-S5p occupancy profiles and expression (mRNA) profiles at Ddx3x, Rlim and Igbp1 in two systems: brain (A) and Patski cells (B). Ddx3x escapes XCI in both systems, Rlim escapes XCI in Patski cells only, and Igbp1 is subject to XCI in both systems. PolII-S5p is enriched at the promoter regions (highlighted by a red box) of escape genes on both the Xa and the Xi, whereas enrichment is limited to the Xa for genes subject to XCI. DNase I hypersensitivity tested in Patski cells only is also increased at the promoter regions (highlighted by a red box) of escape genes on both the Xa and Xi, but is limited to the Xa for genes subject to XCI. Genes that escape XCI are labeled orange and genes subject to XCI blue. Color-coded profiles are shown for the Xa (blue) and Xi (green) SNP reads, and for the total reads (Xt, black). See additional examples in S2 Fig. Classification of mouse escape genes
We determined that 3–5% of X-linked Refseq genes satisfy our criteria (see above) as escape genes in both biological replicates of F1 brain and spleen, respectively (S1–S3 Table). In addition, 3% of X-linked genes variably escaped XCI, i.e., escaped in only one biological replicate. In F1 ovary 7% of X-linked genes escaped XCI (S1 and S4 Table), possibly due to the analysis of a single replicate for this tissue (representing two pooled ovaries). Escape genes were distributed all along the mouse X chromosome and rarely clustered, and distance from the XIC (X inactivation center) did not appear to influence levels of expression from the Xi. We classified escape genes into two groups based on their XCI status, excluding genes with a different status in biological replicates: group 1 includes 12 genes that escape XCI in at least two of the three tissues analyzed and group 2, 26 genes that escape XCI in a tissue-specific manner (Table 1). When considering potential sex bias in gene expression we found that based on published data [30], a majority (27 of 38 or 71%) of group1 and 2 escape genes showed a female bias and thus may play roles in sex differences (S5 Table).
Tab. 1. Escape genes grouped as common or variable between mouse tissues. 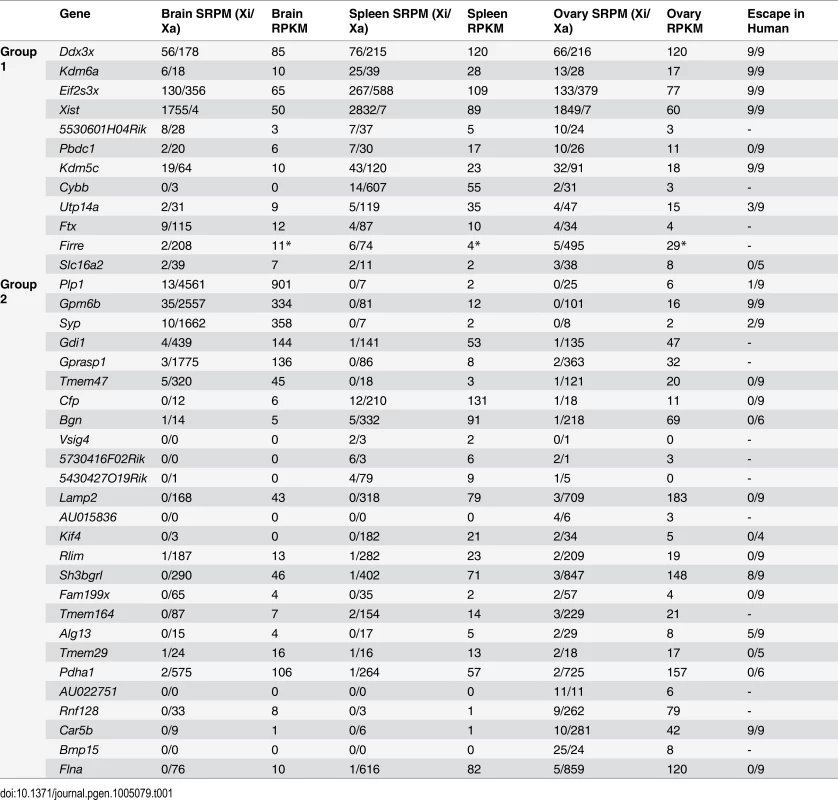
Gene names are listed to reflect their classification into group 1 (common in at least two tissues) or group 2 (escape in only one tissue). Tissue-specific escape genes escape XCI in 1 of 3 tissues but not in any replicate of other tissues. For brain and spleen average SRPM-Xi/Xa values between replicates are shown. Ovary SRPM-Xi/Xa values represent one sample. Gene expression is shown as RPKM. Whether human homologs to the mouse genes escape XCI is shown in the last column, which indicates the proportion of mouse x human hybrid cell lines that had expression of the gene from the human Xi [6]. A dash indicates the gene was not assayed for escape in human in that study. Firre RPKM (*) was based on reads from exons and introns (see also S2–4 Table and Methods). Of the 12 common escape genes classified as belonging to group 1, only the lncRNA 5530601H04Rik was not included in our original survey in Patski cells [18], although our current re-analysis now includes it (see below). Mid1, a gene that straddles the boundary of the pseudoautosomal region (PAR) in M. musculus and escapes XCI in Patski cells [18] was not classified as an escape gene in our current analysis of mouse tissues with a spretus Xi, indicating that Mid1 is subject to XCI in M. spretus where it is located outside the PAR [31]. This was confirmed in F1 brain by Sanger sequencing of RT-PCR products (S1E Fig).
Within group 2 a total of 6, 5, and 15 genes escaped XCI selectively in F1 brain, spleen, and ovary, respectively. The findings of escape in a single tissue often reflected the unique expression pattern of these genes. Functional analysis showed that group 1 common escape genes often have functions relevant to many tissues, whereas group 2 genes have tissue-specific functions (S5 Table). In fact, all 6 genes that escape XCI in brain have brain-related functions, for example, Gpm6b and Plp1 play a role in myelination [32]. RT-PCR validation using species-specific primers confirmed Plp1 expression from the Xi in brain (S1C and S1D Fig). In addition, Gdi1 and Syp have been implicated in X-linked intellectual disability in humans [33,34], and Gprasp1 mutations are associated with striatum-dependent behavior inhibition [35]. In spleen a total of 17 escape genes were identified, including 5 spleen-specific escape genes, three of which, Cfp, Vsig4, and Bgn, implicated in immune functions [36–39] (Tables 1, S3 and S5). Cfp escaped from XCI in spleen but not in brain or ovary, as validated by RT-PCR using species-specific primers (Table 1 and Fig. 1B–1D). Vsig4 expression from the Xi in spleen was verified by Sanger sequencing of RT-PCR products, but in liver where Vsig4 is also highly expressed this gene was subject to XCI, suggesting that escape in spleen was independent of tissue-specific expression (S1F and S1G Fig). In ovary a total of 33 escape genes were identified, including 15 ovary-specific escape genes (Tables 1 and S4). Note that six additional genes were not included in Table 1 since they escaped in only one biological replicate of ovary (Idh3g, Mmgt1, Usp9x, Uba1, Huwe1, 1810030O07Rik). Among the ovary-specific escape genes, AU022751 and Bmp15 had nearly equal expression from the Xa and Xi, as confirmed by Sanger sequencing of RT-PCR products (Table 1, S1H and S1I Fig). Since both AU022751 and Bmp15 are mostly expressed in oocytes where the Xi is reactivated [40] the finding of bi-allelic expression in ovary was not surprising. X reactivation could account for the larger number of escape genes in this tissue, even for genes that are not exclusively expressed in oocytes (S4 Table).
Using our new pipeline to re-analyze two independent RNA-seq datasets for Patski cells, one previously generated in our lab [18] and the other generated on an AB Solid platform deposited for ENCODE [41], we identified 66 escape genes (S1 and S6 Table). Fifty-two of these genes did not escape in any of the three F1 tissues (S2–S4 and S6 Table). For example, Rlim that escaped XCI in Patski cells was subject to XCI in brain (Fig. 2A). Next we examined gene expression in F1 kidney since Patski cells were derived from 18.5dpc embryonic kidney [23]. We found that two genes that escaped XCI in Patski cells, Shroom4 and Car5b (as seen here and in [18]), failed to express from the Xi in kidney, indicating that these genes escaped XCI only in the cell line (Figs. 2B, 2C and S4). In contrast, RT-PCR analyses using species-specific primers for Hdac6 showed that this gene escaped XCI both in Patski cells and kidney, suggesting that this gene is a tissue-specific escape gene since it does not escape XCI in other tissues (Fig. 2D, 2E and Table 1). The lncRNA 5530601H04Rik was confirmed to escape XCI in both Patski cells and kidney using Sanger sequencing of RT-PCR products, indicating that this is a common escape gene in all tissues examined (Fig. 2F and Table 1).
The larger number of genes classified as "escape" in the new analysis of Patski cells compared to our previously published data [18] was mainly attributed to our new binomial model and not to differences in SNP number (see details in Methods). While our previous study used a ratio of Xi/Xa SNP reads greater than 0.1 (10% Xi expression) in the entire gene body to call a gene escape, our current binomial model to compare Xi-SNP reads to total SNP reads in exons more accurately assesses Xi expression, as shown by our verifications using RT-PCR with species-specific primers or Sanger sequencing (see above). Two of three genes previously classified as "escape" [18], Bgn and BC022960, were not included in our current list because their expression was below the 1RPKM threshold. The lncRNA Firre (6720401G13Rik) was initially excluded in our current analysis based on exonic SNPs because it failed to pass the ≥2 Xi-SRPM cutoff in tissues and in one biological replicate of Patski cells. However, when intronic SNPs were considered, similar to our previous survey [18] Firre was re-classified as an escape gene in F1 tissues and in both replicates of Patski cells (Fig. 2G, Tables 1 and S2–S4 and S6). This is also supported by another study [42], and by our findings of enrichment in RNA polymerase II phosphorylated at serine 5 (PolII-S5p) within the gene body of Firre on the Xi, suggesting alternative transcript start sites on the Xi (Yang et al., manuscript in review).
Enrichment in RNA polymerase II and DNAse I hypersensitivity at escape genes
The distribution of PolII-S5p was determined by ChIP-seq in one female F1 brain and in Patski cells in conjunction with re-analyses of two DNase I hypersensitivity datasets for Patski cells [41] to extract allele-specific profiles for comparison with the XCI status of genes. There was good agreement between bi-allelic promoter enrichment in PolII-S5p and escape from XCI, as shown for common escape genes Ddx3x, Kdm6a and 5530601H04Rik (Figs. 3, S2). In contrast, genes subject to XCI in both brain and Patski cells such as Igbp1 lacked these features on the Xi (Fig. 3). For genes that differed in terms of their XCI status in a tissue-specific manner, corresponding differences in PolII-S5p levels were noted between tissues. For example, PolII-S5p was bound only to the Xa allele of Rlim in brain where this gene is subject to XCI, while bi-allelic PolII-S5p enrichment was observed in Patski cells where the gene escapes XCI (Fig. 3). Similar results were seen at other differential escape genes, such as Shroom4 (S2 Fig). Consistently, metagene analyses showed high PolII-S5p occupancy at the promoter regions of both alleles of escape genes but not of inactivated genes on the Xi in brain and Patski cells (Fig. 4A and 4F). For escape genes, there was a strong correlation between the ratio of PolII-S5p enrichment on the Xi versus the Xa with allele-specific expression levels (Fig. 4B and 4G). When examining all assessable X-linked genes, the level of PolII-S5p occupancy at the promoter was positively correlated with expression as expected (Fig. 4C and 4H). Allele-specific scatter plots showed a positive correlation between PolII-S5p enrichment at the promoter for Xi-alleles of escape genes, while no such correlation was observed for Xa-alleles (Fig. 4D, 4E, 4I and 4J). Interestingly, escape genes tend to have higher expression that inactivated genes (Fig. 4E and 4J). We did find four genes that were not classified as escape genes in our RNA-seq analyses, yet showed PolII-S5p occupancy on the Xi with an average read counts >5 at their promoter (0.5kb ± TSS) in brain (Snx12) and Patski cells (Phf6, Trappc2 and Usp9x) (S2 and S3 Dataset). PolII-S5p Xi occupancy at Trappc2 could be due to overlap of its promoter region with that of Ofd1, an escape gene in Patski cells (S6 Table). Note that TRAPPC2 and USP9X have been reported to escape XCI in human [6].
Fig. 4. PolII-S5p enrichment at promoters of X-linked genes on the Xi correlates with expression and escape from XCI. 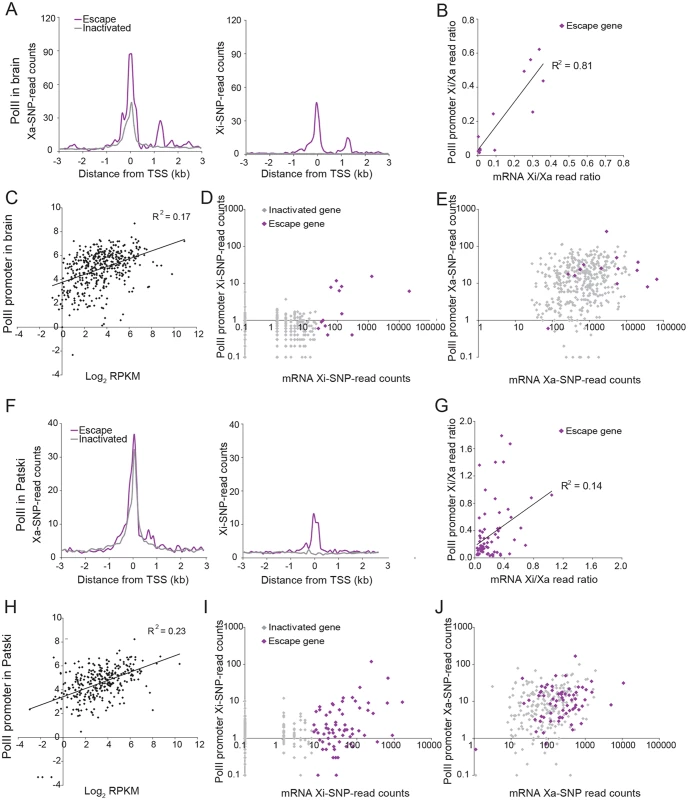
(A) Metagene analyses in brain show average Xa- (left) and Xi- (right) SNP read counts in 100bp windows 3kb upstream and downstream of the TSS for escape genes (purple; 14 genes with ≥2 Xi-SRPM in both replicates for brain) and for genes subject to XCI (gray; 403 genes with <2 Xi-SRPM in both replicates for brain). (B) The ratios of PolII-S5p enrichment at the promoter (SNP reads within ±500bp of the TSS) of escape genes (purple) on the Xi versus the Xa are strongly correlated to the ratios of expression from the Xi versus the Xa measured by RNA-seq in brain. Xist is excluded in this analysis. (C) Scatter plot of PolII-S5p promoter enrichment (log2 of reads within ±500bp of the TSS) against expression levels of all X-linked genes (log2 RPKM) shows a positive correlation in brain. (D) Scatter plot of Xi-specific PolII-S5p promoter enrichment (SNP reads within ±500bp of the TSS) against expression (Xi SNP reads) for escape genes (purple) and genes subject to XCI (gray) in brain. Promoter PolII-S5p enrichment correlates with expression from the Xi. (E) Same analysis as for D but for Xa-specific PolII-S5p promoter enrichment in brain. Escape genes generally overlap with genes subject to XCI, but are often highly expressed and enriched in PolII-S5p. (F-J). Same analyses as A-E for Patski cells. Analyses of promoter DNase I hypersensitivity in Patski cells showed similar correlations with X-linked gene expression as well as with Xi expression (Figs. 3, 5 and S2). In addition, there was a good correlation between DNase I hypersensitivity and enrichment in PolII-S5p at the promoter region of genes on the Xi (Fig. 5E). Thus, DNase I hypersensivity did not appear to be an "all or none" feature but rather was correlated to Xi expression level.
Fig. 5. DNase I hypersensitivity at the promoters of X-linked genes correlates with expression and escape from XCI. 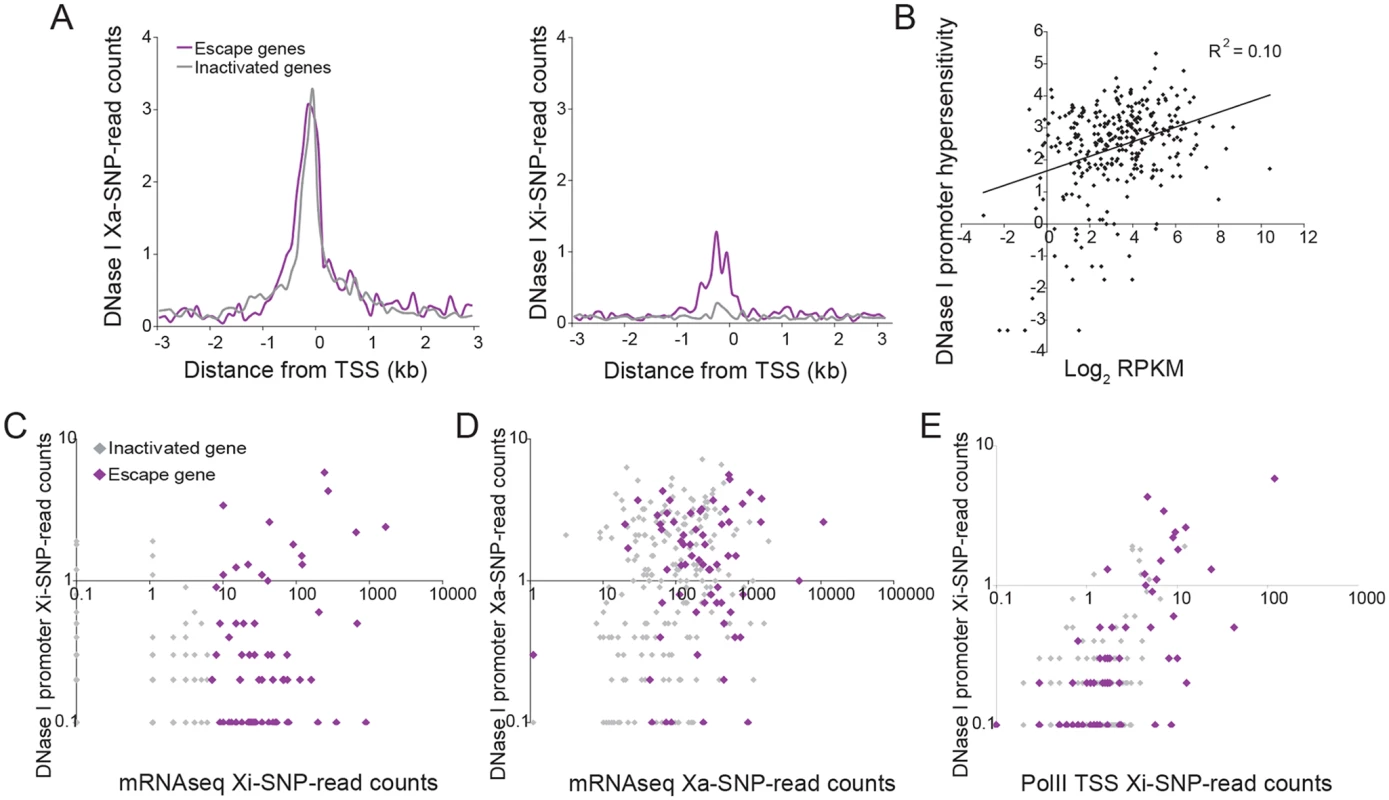
(A) Metagene analyses of DNase I hypersensitivity (DHS) in Patski cells show average Xa- (left) and Xi- (right) SNP read counts in 100bp windows 3kb upstream and downstream of the TSS for escape genes (purple; 43 genes in both replicates for Patski cells) and for genes subject to XCI (gray; 203 genes with <2 Xi-SRPM in both replicates for Patski cells). (B) Scatter plot shows a positive correlation between DHS at the promoter (log2 of all reads within a region ±500bp from the TSS) and expression (log2 RPKM) for all X-linked genes in Patski cells. (C) Scatter plot of Xi-specific DHS at the promoter (reads within a region ±500bp from the TSS) against expression (Xi SNP-reads) for escape genes (purple) and genes subject to XCI (gray) shows a correlation between DHS and level of escape from XCI in Patski cells. (D) Same analysis as in C but for Xa-specific DHS at the promoter region. Escape genes generally overlap with genes subject to XCI although escape genes tend to have high expression and high DHS. (E). Scatter plot shows a good correlation between DHS and enrichment in PolII-S5p at the promoter of genes on the Xi. DHS and PolII-S5p are shown as reads within a region ±500bp from the TSS for escape genes (purple) and genes subject to XCI (gray) in Patski cells. Allelic CTCF binding analysis
While genes that escape XCI in F1 brain were few and almost all isolated those that escape XCI in Patski cells were more numerous and tended to cluster. When considering all escape genes in either replicate of Patski cells we identified 13 clusters with a high density of escape genes compared to the number of genes subject to XCI, including a 440kb cluster containing 13 escape genes (S6 Table and S1 Dataset). Furthermore, 22/66 escape genes in Patski cells were found within 12 (dark gray-shaded areas) of the 16 long-range cis-regions (i.e. 4C domains) previously shown to contain escape genes and to physically interact with Cdk16 and/or Kdm5c in neural progenitor cells (S6 Table) [43]. To determine whether CTCF binding might contribute to differences in the number and distribution of escape genes between F1 brain and Patski cells allele-specific profiles of CTCF binding were generated by ChIP-seq. Discrimination between alleles using SNPs was verified by examining allele-specific CTCF binding at two imprinted autosomal regions known to differentially bind CTCF at the DMR (differentially methylated region). As previously demonstrated [44], H19 showed maternal CTCF binding, while Peg13 was bound by CTCF only on the paternal allele in both brain and Patski cells (S3A Fig).
For allele-specific peak calling all mapped reads were first used to identify enriched peak regions in the diploid genome. Two independent peak calling programs were applied, CisGenome (FDR cutoff 10-5) [45,46] and MACS/v1.4 (p-value cutoff 10-5) [47]. We defined significantly enriched peak regions as those identified by both peak callers. Next, we selected allele-specific ChIP-seq peaks using a binomial test. For each diploid ChIP-seq peak region, we assumed that the numbers of BL6-SNP reads (ni,bl) and spretus-SNP reads (ni,sp) within the peak follow a binomial distribution, i.e.,
where ni = ni,bl + ni,sp is the sum of BL6-SNP reads and spretus-SNP reads in peak region i, and pi is the binomial parameter. Since the X chromosome behaves differently from autosomes due to skewed XCI in our systems, we estimated the X chromosome allelic background using all SNP reads in the identified diploid peak regions on the X only. That is, for peaks on the X chromosome, in which Nx,bl and Nx,sp are the total number of BL6-SNP and spretus-SNP reads in X peaks, respectively. Finally, BL6-preferred ChIP-seq peaks were defined as those that contain significantly more BL6-SNP reads (upper-tail binomial test, p-value <0.05), while spretus-preferred ChIP-seq peaks were identified using the lower-tail binomial test (p-value <0.05), and both-preferred ChIP-seq peaks were those peaks that were not significant in the two above tests (p-value ≥0.25). In addition, we required the allele-assessable peaks have a minimal SNP read coverage of one allele-specific read (BL6-SNP and spretus-SNP reads) per 10 million mapped reads. Of the allele-assessable CTCF-binding peaks on the X chromosome in brain and Patski cells (1639/2263 and 374/532, respectively) we identified 212 (13%) Xi - and 366 (22%) both-preferred, and 86 (23%) Xi - and 161 (43%) both-preferred, respectively (S7 Table). The much larger number of CTCF peaks on the Xi in brain suggests a different structure of the Xi. In fact, only 62 of the Xi-binding CTCF peaks were common between brain and Patski cells (S7 Table).To investigate the spatial distribution of Xi-binding CTCF peaks, the local Xi - and both-preferred CTCF peak density was calculated using a sliding window approach (window size: 500kb, step size: 1kb). Assuming that CTCF Xi-binding followed a Poisson distribution we fitted the data to estimate the Poisson parameters on the Xi, and calculated a p-value for each window. Enriched Xi-binding CTCF peaks were identified at a p-value cutoff of 0.01 and adjacent peaks were merged. Escape genes and Xi-binding CTCF clusters co-localized on the Xi but not the Xa, suggesting a role for CTCF binding at regions of escape (Fig. 6A and 6B). CTCF has been implicated in both transcription control and compartmentalization of the genome [48], thus, it was not surprising that CTCF peaks were found either at gene promoters or in intergenic regions. As expected, the 5'end of escape genes often displayed Xi-promoter occupancy by CTCF in both brain and Patski cells (Fig. 6C). To enrich in CTCF binding regions that might play a role in nuclear compartmentalization rather than transcription control, we then re-analyzed our data after excluding peaks located at promoters (±1kb from the TSS), which showed that CTCF clusters were still significantly associated with escape genes in brain (8/14 genes; p-value = 0.01, compared to a random sample of 500 X-linked genes; Fisher’s exact test) (S3B Fig). There was a similar trend in Patski cells, however the association (16/66 genes) was not significant, probably due to the lower number of CTCF peaks in the cell line. Interestingly, when allelic CTCF binding was analyzed in the context of higher order structure [43], CTCF Xi-preferred peaks were more significantly associated with 4C interacting domains than CTCF Xa-preferred peaks in brain and Patski cells (S8 Table). Furthermore, the density of CTCF peaks on the Xi was inversely related to the number of escape genes in brain and Patski cells as shown by inspecting three regions for the density of X-preferred and both-preferred CTCF binding peaks in relation to the number of escape genes (Fig. 7A). This is consistent with larger regions of escape in Patski cells (S6 Table).
Fig. 6. Xi-associated but not Xa-associated CTCF peak clusters co-localize with escape regions. 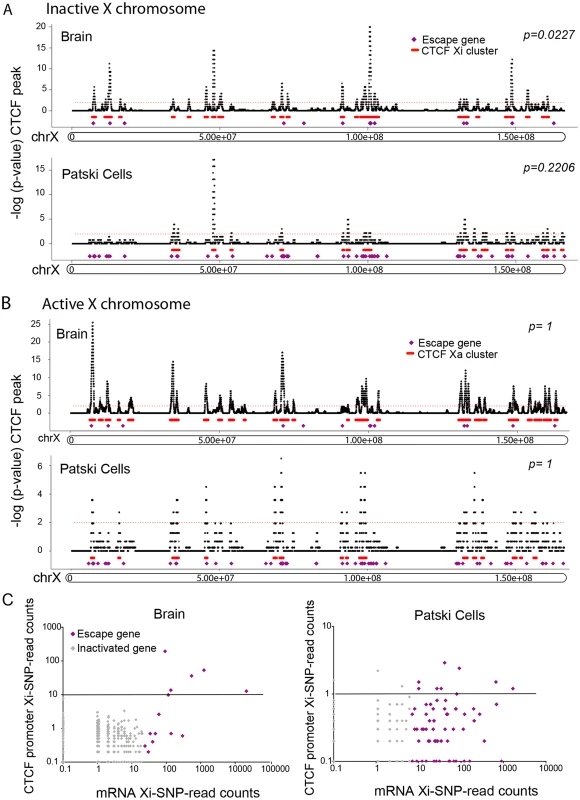
(A) Significant CTCF Xi-binding clusters were mapped along the Xi in brain and Patski cells. Xi- and both-preferred peaks were determined by a binomial model and used for density analysis. Red bars represent merger of clusters of CTCF Xi-binding peaks, while purple dots represent escape genes. Significant Xi-binding CTCF binding clusters tend to co-localize with chromatin containing escape genes. Little change was seen after removal of promoter-associated CTCF binding (S3B Fig). Horizontal axis represents the Xi in Mb. The vertical axis is the negative log of the calculated binomial p-value (-log (p-value)). The thin red dashed line represents a 0.01 p-value cutoff. (B) Similar analysis for CTCF Xa- and both-preferred peaks. There was no significant CTCF co-localization with escape genes on the Xa in either brain or Patski cells. (C) Average CTCF Xi-SNP read counts in ten 100bp windows at promoters (0.5kb upstream and downstream of the TSS) is plotted against mRNA-seq Xi-SNP read counts escape genes (purple) and for genes subject to XCI (gray) in brain and Patski cells. In brain, a higher proportion of escape genes (6/14; Fisher’s exact test, p = 5e-9) had an average ≥10 reads (black line) at their promoter compared to genes subject to XCI (0/403). Similarly, in Patski cells a higher proportion of escape genes (9/65; Fisher’s exact test, p = 0.0004) had an average of ≥1 read (black line) at their promoter compared to genes subject to XCI (3/204). Fig. 7. CTCF peaks density and distribution differ in brain and Patski cells. 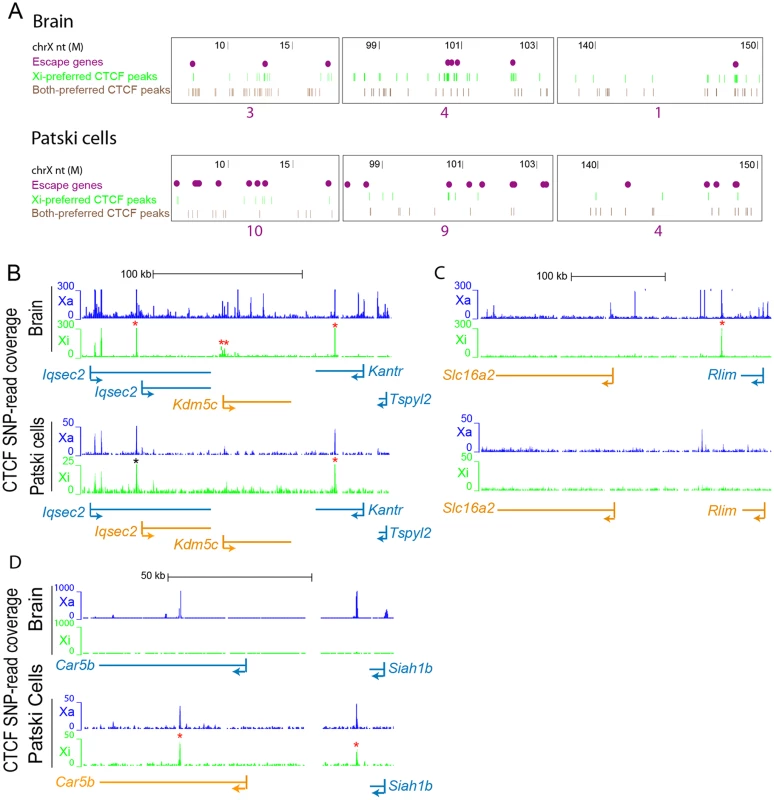
(A) Examples of Xi-preferred and both-preferred CTCF peaks distribution in three X chromosome regions (coordinates in million bp on top). The density of escape genes (purple dots, with total number under each region) is inversely related to the density of Xi-preferred (green) and both-preferred (brown) CTCF-binding peaks when comparing brain to Patski cells. (B) Allele-specific CTCF binding profiles around Kdm5c, a common escape gene flanked by Iqsec2 and Kantr. In brain where only Kdm5c escapes XCI, CTCF binding is present at the 5’ end of the gene at the transition (double star) between Kdm5c and Iqsec2 whose short and long transcripts are subject to XCI (see also S4A Fig). In Patski cells there is no such CTCF binding between Kdm5c and Iqsec2, which escapes XCI. CTCF also binds proximal of the Iqsec2 short transcript in both brain and Patski cells, which could represent a proximal boundary of an escape domain. (C) Similar analysis at a region around Rlim, a gene that escapes XCI in Patski cells but not in brain, while the adjacent gene Slc16a2 escapes XCI in both systems. A CTCF peak is present in the transition region only in brain. (D) Similar analysis in a region around Car5b, a gene that escapes XCI in Patski cells but not in brain (see also S4B Fig). CTCF binding peaks are located within the body of Car5b and in the transition between Car5b and Siah1b on the Xi in Patski cells. Genes that escape XCI are labeled orange and genes subject to XCI blue. Xa SNP reads are in blue and Xi SNP reads in green. Red stars indicate Xi- or both-preferred CTCF peaks on the Xi; one of the CTCF peaks is marked by a black star because it is present but was not called preferred at our cutoff. We next examined allele-specific CTCF peak profiles in the UCSC genome browser at 200 and 139 regions of transitions between adjacent genes with a known XCI status in brain and Patski cells, respectively. Transitions were classified as between either two adjacent genes subject to XCI, an escape gene directly adjacent to a gene subject to XCI, or two adjacent escape genes (S9 Table). There were no transitions between two escape genes in brain, reflecting their low abundance compared to the Patski cell line in which 18 such transitions were observed. A larger proportion of transitions between genes with a different XCI status than those between two inactivated genes had CTCF peaks located in intergenic regions, 21% versus 15% in brain and 8% versus 3% in Patski cells, respectively (S9 Table). We then focused on specific transition regions: at the Kdm5c-Iqsec2 region Xi-preferred CTCF binding peaks were found between Iqsec2 and Kdm5c as well as within the gene body of Kantr located downstream of Kdm5c in brain where only Kdm5c escapes XCI (Figs. 7B and S4). In contrast, in Patski cells where Kdm5c and a short Iqsec2 transcript both escape XCI, Xi-preferred CTCF binding peaks were found both upstream of the short Iqsec2 transcript and within the gene body of Kantr but not between Kdm5c and Iqsec2, suggesting that lack of Xi-preferred CTCF binding in this region may contribute to a larger domain of escape in Patski cells (Fig. 7B). A similar situation was observed in the region between Rlim and Slc16a2, again suggesting that lack of CTCF binding in Patski cells led to a larger escape region (Fig. 7C). A different situation was seen at the Car5b-Siah1b region: Xi-preferred CTCF peaks were absent in brain where both Car5b and Siah1b are subject to XCI, while CTCF peaks flanked the Car5b promoter on both alleles in Patski cells where Car5b escapes XCI and Siah1b is subject to XCI (Figs. 7D and S4), suggesting that CTCF may play a role in the transition between escape and inactivated genes. Taken together, our results imply that CTCF binding may help configure escape domains via local chromatin looping, and/or facilitate the organization of escape genes at the periphery of the Xi territory.
Discussion
Based on allele-specific analyses we identified genes expressed from both X chromosomes in female mouse tissues. Only a minority of these genes escape XCI in a tissue - and cell type-specific manner, indicating that XCI and escape from XCI are tightly controlled in vivo. The probability of bi-allelic versus mono-allelic expression was calculated using a new algorithm that can be applied to any gene in the genome. Our study represents the first comprehensive analysis of escape from XCI in vivo in multiple tissues.
Our data and those of others indicate that for a subset of X-linked genes, escape from XCI is ubiquitous and thus represents an intrinsic property of these genes [22,43]. Among these common escape genes Ddx3x, Kdm6a, Eif2s3x and Kdm5c represent genes that each has a conserved Y-linked paralog with a similar function [49,50]. These X/Y genes play important roles in the regulation of transcription and translation and are highly dosage-sensitive, which could explain why they consistently escape XCI in all tissues examined (Fig. 8). Interestingly, we also identified tissue-specific escape genes, which will help understanding of functional mechanisms leading to sex differences in these tissues. For example, we identified six genes that escape XCI in brain, all of which have been implicated in brain functions, Gpm6b, Gprasp1, Syp, Gdi1, Plp1, and Tmem47 [51]. Our results generally agree with a recent study of XCI based on flow-sorted brain cells with differentially labeled BL6 and Mus castaneus X chromosomes [25] (Fig. 8). Of seven escape genes reported in that study, five are included in the present study (5530601H04Rik, Ddx3x, Eif2s3x, Kdm5c, and Kdm6a). The two other genes are Itm2a that had too few Xi reads to be classified in our study, and Mid1 we have shown to be subject to XCI in M. spretus where it is outside the PAR (this study), while it escapes XCI in BL6 mouse brain [31,52].
Fig. 8. X chromosome escape maps differ between mouse tissues. 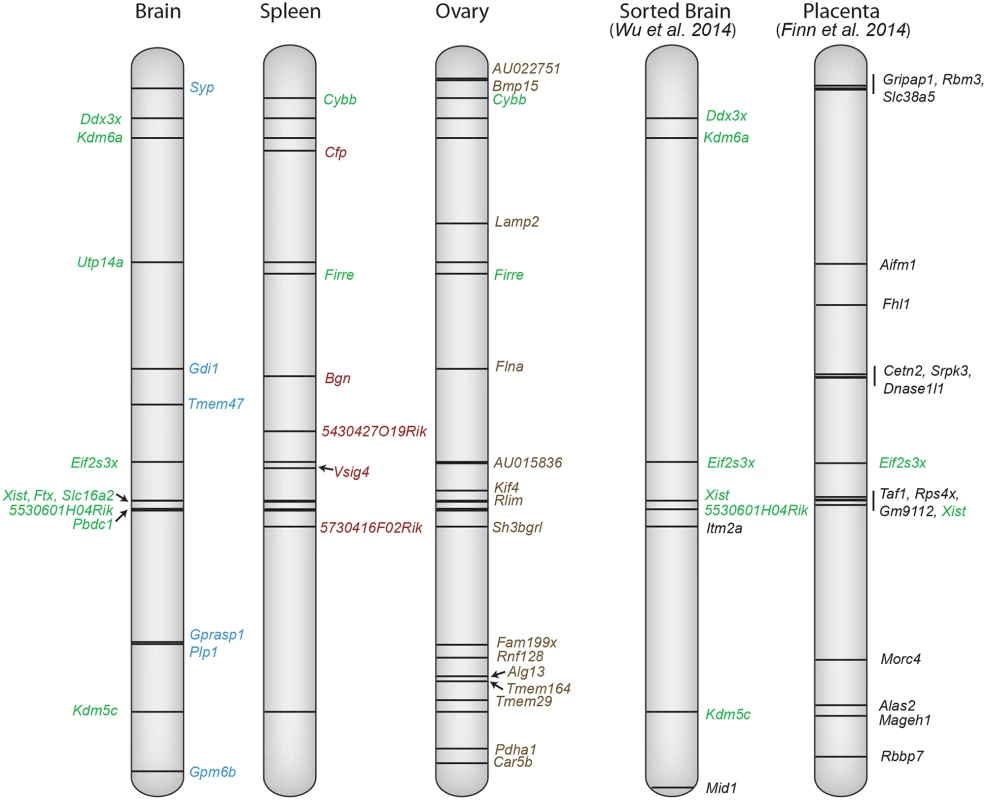
The position of genes that escape XCI using a ≥2 Xi-SRPM cutoff (black lines) is shown at left for three tissues (brain, ovary and spleen) from F1 mice analyzed in our study. Gene names are color-coded to reflect their classification into group 1 (green, common in at least two tissues) or group 2 (blue, brain-specific escape; red, spleen-specific; brown, ovary-specific) based on our criteria (see Table 1). Coordinates at left are based on UCSC genome build NCBI37/mm9. For comparison, the genes reported to escape XCI in sorted brain cells from a M. musculus x M. castaneus cross [25], and genes reported to escape imprinted XCI in mid-gestation placenta from a M. musculus x M. castaneus cross [24] are shown at right. Genes labeled green are common between studies. Importantly, many of the escape genes we identified have significant female sex bias in expression, suggesting roles in sex differences (S5 Table) [1,30,53–57]. For example, three of the brain-specific escape genes we identified in mouse, GPM6B, SYP, and PLP1 also escape XCI in human, resulting in higher expression in female than male brain [6,58]. Whether deficiency in these genes due to the presence of a single X chromosome in women with Turner syndrome contributes to mild cognitive impairment remains to be determined [59]. Interestingly, of the five novel spleen-specific escape genes we identified, as many as three, Vsig4, Cfp, and Bgn, have been implicated in autoimmune disorders both in mouse and human [36,38,39]. It is well established that autoimmune disorders are much more common in women and their incidence is increased in Turner syndrome, but specific genetic mechanisms are not well defined [60]. Our in vivo study demonstrates that analyses of relevant human tissues, for example spleen in the case of CFP and BGN, two gene previously classified as being subject to XCI in cell cultures [6], will be critical to understand sex differences in specific disorders.
Do genes escape XCI only in tissues where they are most highly expressed? While genes that escape XCI often have high expression in a particular tissue, expression does not appear to be the sole driving force for escape. For example, Car5b is more highly expressed in ovary (42RPKM) than in Patski cells (8RPKM) and yet escapes XCI only in Patski cells. A previous study identified a set of 17 escape genes in mouse placenta [24] (Fig. 8). Many of these differ from escape genes found in our study, probably because extra-embryonic tissues undergo imprinted paternal XCI, which differs from random XCI [61]. A comprehensive comparison of escape from XCI in available mouse tissues shows that only Eif2s3x escapes XCI in all tissues examined (Fig. 8). We found that escape from XCI represents a continuum of expression from the Xi compared to the Xa. While our data only includes genes expressed above a strict cutoff of 1RPKM, we cannot exclude that some genes with lower expression may also escape XCI. For genes with significant expression from the Xi (excluding Xist), expression ranged from 3–105% (median 18%) of the Xa expression level. Thus, expression from the Xi was usually lower than that of the Xa in mouse, similar to what has been reported in human, even though there are more escape genes in this species based on expression analyses of cell cultures [6] and on DNA methylation profiles in human tissues where 9% of human genes were found to have a methylation pattern consistent with escape [62]. The mouse with 3–7% escape genes in tissues may be exceptional compared to other mammals in which XCI patterns are often more similar to the human pattern [3,63].
Our findings of a larger number of escape genes in Patski cells compared to mouse tissues may reflect either the acquisition of epigenetic changes leading to reactivation of X-linked genes in cell culture or a genuine property of these cells. Our analyses suggest that kidney-specific escape genes do exist and could explain in part the pattern seen in Patski cells, but we also found significant differences between the cell line and the tissue of origin. Clustering of escape genes in Patski cells but not in tissues suggests unstable silencing of large chromosomal regions. Using an in vitro system of cultured trophoblastic cells Calabrese et al. also identified a relatively large number of escape genes (35 out of 262 accessible) [14], which represents twice the number of escape genes found in mouse placenta [24]. Furthermore, 22/66 escape genes in Patski cells are in regions of escape reported in cultured neural progenitor cells [43]. Thus, the number of escape genes may be overestimated when based on studies of cultured cells, which are notoriously susceptible to epigenetic changes such as DNA methylation changes associated with gene expression and CTCF binding aberrations [64,65]. We cannot rule out the possibility that XCI in embryonic cells including embryonic kidney cells from which Patski cells were derived, as well as trophoblastic cells and neural progenitor cells may simply be less complete than in adult tissues. Future studies will help sort out developmental aspects of escape from XCI.
We found a good correlation between escape from XCI and regulatory features associated with transcription, such as PolII-S5p occupancy and DNase I hypersensitivity at the promoters of genes on the Xi, indicating that escape regions have a more open chromatin configuration. This is consistent with escape genes being associated with histone marks characteristic of active chromatin [13–18,66]. Interestingly, we found distinct CTCF binding patterns on the Xi and Xa. A study in human cells also reported that while a majority of CTCF peaks on the X chromosome are bi-allelic, some peaks are Xa - or Xi-specific [67]. In addition, a recent study in differentiated mouse ES cells also describes significant differences between Xa - and Xi-specific CTCF peaks at escape gene loci (determined by ChIP-seq), as well as differences in interactions between transcripts and CTCF (determined by CLIP-seq) [68]. These findings are in contrast to a study of imprinted XCI, in which Xi and Xa CTCF binding patterns were nearly identical [14]. Thus, the role of CTCF in escape from XCI may differ between random and imprinted XCI. CTCF binding peaks were often located at the promoters of genes expressed from the Xi, in agreement with a role for CTCF in transcription regulation [69]. In addition, since CTCF binding peaks located in intergenic regions also clustered with escape genes, CTCF may also be a factor in compartmentalization of the Xi. Chromatin interactions such as looping as determined by Hi-C are correlated with the distribution of CTCF binding [70]. This is supported by our findings that Xi-preferred CTCF binding is more significantly associated with 4C interacting domains. Indeed, CTCF plays an important role in nuclear structure and is often found at the boundary between topological domains [71–73]. Furthermore, regions containing escape genes are preferentially engaged in long range cis-interactions [43]. Previous studies have shown that specific boundary elements possibly involving CTCF may have a role in the segregation of silenced domains from escape domains [21,74]. The low density of CTCF peaks observed in Patski cells may result in a more relaxed structure of the Xi in the cell line, leading to an expansion of escape domains. Interestingly, disruption of CTCF binding at the borders of domains enriched in H3K27me3 in Drosophila results in a reduction in H3K27me3 levels in repressed domains [75], and loss of CTCF binding at super-enhancers results in increased expression of adjacent genes [76]. It is important to note that a previous 5C study of the XIC has reported CTCF binding both at the boundaries of topologically associating domains (TADs) and within TADs, suggesting that CTCF is not the sole factor in determining Xi organization [77]. Further studies will help define other elements that may help structure the Xi.
In summary, we demonstrate the utility of a mouse model to study XCI in vivo. Using this resource novel tissue-specific escape genes have been identified. Escape genes are associated with an open chromatin structure and CTCF binding may influence the definition of differential chromatin architecture of the X.
Materials and Methods
Tissue collection, hybrid mouse model and cell culture
Ovaries, spleen, liver, and whole brain were collected from female F1 obtained by mating C57B/6J females that carry a deletion of the Xist proximal A-repeat (XistΔ) (B6.Cg-Xist<tm5Sado>, RIKEN) [27] with M. spretus males (Jackson Labs). Female progeny were genotyped to verify inheritance of the XistΔ allele using specific primers [27]. F1 mice that inherited a maternal X chromosome with an XistΔ fail to silence the BL6 X and thus have complete skewing of XCI of the paternal spretus X. All procedures involving animals were reviewed and approved by the University Institutional Animal Care and Use Committee (IACUC), and were performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals. Patski cells were cultured as previously described [18].
Validation of allelic expression
To verify skewing of XCI, cDNA and control genomic DNA (gDNA) extracted from each tissue were subject to PCR amplification of Ubqln2 followed by Sanger sequencing (S10 Table). A similar approach was used to confirm the XCI status of Mid1, Bmp15 and Vsig4, Rlim, Shroom4, Car5b, and 5530601H04Rik (S10 Table). Allele-specific RT-PCR was done to confirm Xi expression of Plp1, Cfp, Hdac6. Briefly, cDNA was made by Superscript II reverse transcriptase (Life Technologies) using oligo-dT primers according to manufacturer's protocol. PCR reactions with non-species specific and BL6-specific or spretus-specific primers (S10 Table) were performed using tissues from BL6, spretus and XistΔ hybrid F1 mice. Actinβ was used as a positive control. For quantification, gel band intensities were measured using ImageJ software (http://imagej.nih.gov/ij/) and, together with RNA-seq Xi levels, plotted to compare expression from the Xi and Xa.
ChIP-seq with allele-specific analyses
ChIP-seq using PolII-S5p (Abcam) and CTCF (Millipore) ChIP-grade antibodies were performed as described [26]. The specificity of the PolII-S5p antibody (Abcam) was verified by blocking immunostaining with synthetic peptides (Abcam ab18488). A pseudo-spretus genome was assembled by substituting available SNPs (from Sanger) into the BL6 UCSC Genome Browser NCBIv37/mm9 reference genome. Reads from genomic DNA sequencing, ChIP-seq, and DNase I-seq experiments were mapped separately to the BL6 reference sequence (mm9) and to the pseudo-spretus genome using BWA/v0.5.9 [78] with default parameters. Only those reads that mapped uniquely and with a high-quality mapping score (MAPQ ≥ 30) to either the BL6 genome or the pseudo-spretus genome were kept for allele-specific analyses (see details in main text).
RNA-seq with allele-specific analyses
RNA-seq experiments were done as described [18,26]. Exonic RNA-seq reads were mapped using bowtie/v0.12.7 [79] to both the genome and transcriptome and gene expression was estimated using Tophat/v2.0.2 [29] with default parameters. Only those reads that mapped uniquely and with a high-quality mapping score (MAPQ ≥ 30) to either the BL6 genome or the pseudo-spretus genome were kept for allele-specific analyses. Since Eif2s3x exons (except exon 1) have a high sequence similarity to another X region (chrX: 31680780–31684279), we included reads contained in exons with a low MAPQ score for this gene. Post filtering of expression levels and Xi SNP reads was done to remove genes with low expression and/or limited Xi-SNP reads. In addition, a binomial model for comparison of Xi-SNP reads to total-SNP reads (Xi+Xa) for all exons of each gene was used to call genes that escaped at levels below the Xi/Xa ratio threshold cutoff. Reads containing informative SNPs were assigned to each haploid genome. For the whole X chromosome we used 1,532,011 SNPs, including 597,315 SNPs in gene bodies, and 31,062 SNPs in exons. Gene expression analyses were performed as described in the main text.
Data access
RNA-seq data for the Patski cell line are deposited to the NCBI Gene expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSM970866. ChIP-seq data for PolII-S5p occupancy in brain and Patski cells are deposited under the accession number GSE44255. RNA-seq data for mouse tissues and ChIP-seq data for CTCF binding in brain and Patski cells is deposited to the GEO database under accession number GSE59779. Patski cell line DNase I hypersensitivity data is deposited under the accession number GSM1014171.
Ethics statement
For mice sacrificed, euthanasia was accomplished using two methods (carbon dioxide asphyxiation followed by cervical dislocation) as required by the University of Washington's Office of Animal Welfare. Husbandry and all other procedures were approved by University of Washington's Office of Animal Welfare.
Supporting Information
Zdroje
1. Deng X, Berletch JB, Nguyen DK, Disteche CM (2014) X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet 15 : 367–378. doi: 10.1038/nrg3687 24733023
2. Lessing D, Anguera MC, Lee JT (2013) X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genomics Hum Genet 14 : 85–110. doi: 10.1146/annurev-genom-091212-153530 23662665
3. Berletch JB, Yang F, Xu J, Carrel L, Disteche CM (2011) Genes that escape from X inactivation. Hum Genet 130 : 237–245. doi: 10.1007/s00439-011-1011-z 21614513
4. Peeters SB, Cotton AM, Brown CJ (2014) Variable escape from X-chromosome inactivation: Identifying factors that tip the scales towards expression. Bioessays
5. Anderson CL, Brown CJ (2002) Variability of X chromosome inactivation: effect on levels of TIMP1 RNA and role of DNA methylation. Hum Genet 110 : 271–278. 11935340
6. Carrel L, Willard HF (2005) X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434 : 400–404. 15772666
7. Cotton AM, Ge B, Light N, Adoue V, Pastinen T, et al. (2013) Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol 14: R122. doi: 10.1186/gb-2013-14-11-r122 24176135
8. Bondy CA (2007) Care of girls and women with Turner syndrome: a guideline of the Turner Syndrome Study Group. J Clin Endocrinol Metab 92 : 10–25. 17047017
9. Zinn AR, Page DC, Fisher EM (1993) Turner syndrome: the case of the missing sex chromosome. Trends Genet 9 : 90–93. 8488568
10. Murakami K, Ohhira T, Oshiro E, Qi D, Oshimura M, et al. (2009) Identification of the chromatin regions coated by non-coding Xist RNA. Cytogenet Genome Res 125 : 19–25. doi: 10.1159/000207514 19617692
11. Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, et al. (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341 : 1237973. doi: 10.1126/science.1237973 23828888
12. Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, et al. (2013) High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504 : 465–469. doi: 10.1038/nature12719 24162848
13. Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, et al. (2002) Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet 30 : 73–76. 11740495
14. Calabrese JM, Sun W, Song L, Mugford JW, Williams L, et al. (2012) Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151 : 951–963. doi: 10.1016/j.cell.2012.10.037 23178118
15. Changolkar LN, Singh G, Cui K, Berletch JB, Zhao K, et al. (2010) Genome-wide distribution of macroH2A1 histone variants in mouse liver chromatin. Mol Cell Biol 30 : 5473–5483. doi: 10.1128/MCB.00518-10 20937776
16. Gilbert SL, Sharp PA (1999) Promoter-specific hypoacetylation of X-inactivated genes. Proc Natl Acad Sci U S A 96 : 13825–13830. 10570157
17. Khalil AM, Driscoll DJ (2007) Trimethylation of histone H3 lysine 4 is an epigenetic mark at regions escaping mammalian X inactivation. Epigenetics 2 : 114–118. 17965609
18. Yang F, Babak T, Shendure J, Disteche CM (2010) Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 20 : 614–622. doi: 10.1101/gr.103200.109 20363980
19. Cotton AM, Lam L, Affleck JG, Wilson IM, Penaherrera MS, et al. (2011) Chromosome-wide DNA methylation analysis predicts human tissue-specific X inactivation. Hum Genet 130 : 187–201. doi: 10.1007/s00439-011-1007-8 21597963
20. Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, et al. (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341 : 1237905. doi: 10.1126/science.1237905 23828890
21. Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, et al. (2005) Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell 8 : 31–42. 15669143
22. Li N, Carrel L (2008) Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc Natl Acad Sci U S A 105 : 17055–17060. doi: 10.1073/pnas.0807765105 18971342
23. Lingenfelter PA, Adler DA, Poslinski D, Thomas S, Elliott RW, et al. (1998) Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat Genet 18 : 212–213. 9500539
24. Finn EH, Smith CL, Rodriguez J, Sidow A, Baker JC (2014) Maternal bias and escape from X chromosome imprinting in the midgestation mouse placenta. Dev Biol 390 : 80–92. doi: 10.1016/j.ydbio.2014.02.020 24594094
25. Wu H, Luo J, Yu H, Rattner A, Mo A, et al. (2014) Cellular resolution maps of x chromosome inactivation: implications for neural development, function, and disease. Neuron 81 : 103–119. doi: 10.1016/j.neuron.2013.10.051 24411735
26. Deng X, Berletch JB, Ma W, Nguyen DK, Hiatt JB, et al. (2013) Mammalian X Upregulation Is Associated with Enhanced Transcription Initiation, RNA Half-Life, and MOF-Mediated H4K16 Acetylation. Dev Cell S1534–5807(13)00101–9 [pii] 10.1016/j.devcel.2013.01.028.
27. Hoki Y, Kimura N, Kanbayashi M, Amakawa Y, Ohhata T, et al. (2009) A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development 136 : 139–146. doi: 10.1242/dev.026427 19036803
28. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420 : 520–562. 12466850
29. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28 : 511–515. doi: 10.1038/nbt.1621 20436464
30. Reinius B, Johansson MM, Radomska KJ, Morrow EH, Pandey GK, et al. (2012) Abundance of female-biased and paucity of male-biased somatically expressed genes on the mouse X-chromosome. BMC Genomics 13 : 607. doi: 10.1186/1471-2164-13-607 23140559
31. Nguyen DK, Yang F, Kaul R, Alkan C, Antonellis A, et al. (2011) Clcn4–2 genomic structure differs between the X locus in Mus spretus and the autosomal locus in Mus musculus: AT motif enrichment on the X. Genome Res 21 : 402–409. doi: 10.1101/gr.108563.110 21282478
32. Werner HB, Kramer-Albers EM, Strenzke N, Saher G, Tenzer S, et al. (2013) A critical role for the cholesterol-associated proteolipids PLP and M6B in myelination of the central nervous system. Glia 61 : 567–586. doi: 10.1002/glia.22456 23322581
33. Gordon SL, Cousin MA (2013) X-linked intellectual disability-associated mutations in synaptophysin disrupt synaptobrevin II retrieval. J Neurosci 33 : 13695–13700. doi: 10.1523/JNEUROSCI.0636-13.2013 23966691
34. Strobl-Wildemann G, Kalscheuer VM, Hu H, Wrogemann K, Ropers HH, et al. (2011) Novel GDI1 mutation in a large family with nonsyndromic X-linked intellectual disability. Am J Med Genet A 155A: 3067–3070. doi: 10.1002/ajmg.a.34291 22002931
35. Mathis C, Bott JB, Candusso MP, Simonin F, Cassel JC (2011) Impaired striatum-dependent behavior in GASP-1-knock-out mice. Genes Brain Behav 10 : 299–308. doi: 10.1111/j.1601-183X.2010.00666.x 21091868
36. Choi HM, Lee YA, Yang HI, Yoo MC, Kim KS (2011) Increased levels of thymosin beta4 in synovial fluid of patients with rheumatoid arthritis: association of thymosin beta4 with other factors that are involved in inflammation and bone erosion in joints. Int J Rheum Dis 14 : 320–324. doi: 10.1111/j.1756-185X.2011.01652.x 22004227
37. Kim KH, Choi BK, Song KM, Cha KW, Kim YH, et al. (2013) CRIg signals induce anti-intracellular bacterial phagosome activity in a chloride intracellular channel 3-dependent manner. Eur J Immunol 43 : 667–678. doi: 10.1002/eji.201242997 23280470
38. Lesher AM, Nilsson B, Song WC (2013) Properdin in complement activation and tissue injury. Mol Immunol 56 : 191–198. doi: 10.1016/j.molimm.2013.06.002 23816404
39. Moreth K, Brodbeck R, Babelova A, Gretz N, Spieker T, et al. (2010) The proteoglycan biglycan regulates expression of the B cell chemoattractant CXCL13 and aggravates murine lupus nephritis. J Clin Invest 120 : 4251–4272. doi: 10.1172/JCI42213 21084753
40. Heard E, Turner J (2011) Function of the sex chromosomes in mammalian fertility. Cold Spring Harbor perspectives in biology 3: a002675. doi: 10.1101/cshperspect.a002675 21730045
41. ENCODE Project Consortium MR, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E et al. (2011) A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046. doi: 10.1371/journal.pbio.1001046 21526222
42. Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, et al. (2014) Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21 : 198–206. doi: 10.1038/nsmb.2764 24463464
43. Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, et al. (2011) The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev 25 : 1371–1383. doi: 10.1101/gad.633311 21690198
44. Prickett AR, Barkas N, McCole RB, Hughes S, Amante SM, et al. (2013) Genomewide and parental allele-specific analysis of CTCF and cohesin DNA binding in mouse brain reveals a tissue-specific binding pattern and an association with imprinted differentially methylated regions. Genome Res gr.150136.112 [pii] 10.1101/gr.150136.112.
45. Ji H, Jiang H, Ma W, Johnson DS, Myers RM, et al. (2008) An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26 : 1293–1300. doi: 10.1038/nbt.1505 18978777
46. Ma W, Wong WH (2011) The analysis of ChIP-Seq data. Methods Enzymol 497 : 51–73. doi: 10.1016/B978-0-12-385075-1.00003-2 21601082
47. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. doi: 10.1186/gb-2008-9-9-r137 18798982
48. Ong CT, Corces VG (2014) CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15 : 234–246. doi: 10.1038/nrg3663 24614316
49. Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, et al. (2014) Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508 : 494–499. doi: 10.1038/nature13206 24759411
50. Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, et al. (2014) Origins and functional evolution of Y chromosomes across mammals. Nature 508 : 488–493. doi: 10.1038/nature13151 24759410
51. Utami KH, Hillmer AM, Aksoy I, Chew EG, Teo AS, et al. (2014) Detection of chromosomal breakpoints in patients with developmental delay and speech disorders. PLoS One 9: e90852. doi: 10.1371/journal.pone.0090852 24603971
52. Dal Zotto L, Quaderi NA, Elliott R, Lingerfelter PA, Carrel L, et al. (1998) The mouse Mid1 gene: implications for the pathogenesis of Opitz syndrome and the evolution of the mammalian pseudoautosomal region. Hum Mol Genet 7 : 489–499. 9467009
53. Isensee J, Witt H, Pregla R, Hetzer R, Regitz-Zagrosek V, et al. (2008) Sexually dimorphic gene expression in the heart of mice and men. J Mol Med (Berl) 86 : 61–74. 17646949
54. Li J, Chen X, McClusky R, Ruiz-Sundstrom M, Itoh Y, et al. (2014) The number of X chromosomes influences protection from cardiac ischaemia/reperfusion injury in mice: one X is better than two. Cardiovasc Res 102 : 375–384. doi: 10.1093/cvr/cvu064 24654234
55. Xu J, Burgoyne PS, Arnold AP (2002) Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet 11 : 1409–1419. 12023983
56. Xu J, Deng X, Disteche CM (2008) Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS One 3: e2553. doi: 10.1371/journal.pone.0002553 18596936
57. Xu J, Deng X, Watkins R, Disteche CM (2008) Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J Neurosci 28 : 4521–4527. doi: 10.1523/JNEUROSCI.5382-07.2008 18434530
58. Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, et al. (2011) The evolution of gene expression levels in mammalian organs. Nature 478 : 343–348. doi: 10.1038/nature10532 22012392
59. Knickmeyer RC (2012) Turner syndrome: advances in understanding altered cognition, brain structure and function. Curr Opin Neurol 25 : 144–149. doi: 10.1097/WCO.0b013e3283515e9e 22322416
60. Jorgensen KT, Rostgaard K, Bache I, Biggar RJ, Nielsen NM, et al. (2010) Autoimmune diseases in women with Turner's syndrome. Arthritis Rheum 62 : 658–666. doi: 10.1002/art.27270 20187158
61. Takagi N, Sasaki M (1975) Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256 : 640–642. 1152998
62. Cotton AM, Chen CY, Lam LL, Wasserman WW, Kobor MS, et al. (2014) Spread of X-chromosome inactivation into autosomal sequences: role for DNA elements, chromatin features and chromosomal domains. Hum Mol Genet 23 : 1211–1223. doi: 10.1093/hmg/ddt513 24158853
63. Al Nadaf S, Deakin JE, Gilbert C, Robinson TJ, Graves JA, et al. (2012) A cross-species comparison of escape from X inactivation in Eutheria: implications for evolution of X chromosome inactivation. Chromosoma 121 : 71–78. doi: 10.1007/s00412-011-0343-8 21947602
64. Ehrlich M, Lacey M (2013) DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics 5 : 553–568. doi: 10.2217/epi.13.43 24059801
65. Lund RJ, Narva E, Lahesmaa R (2012) Genetic and epigenetic stability of human pluripotent stem cells. Nat Rev Genet 13 : 732–744. doi: 10.1038/nrg3271 22965355
66. Kucera KS, Reddy TE, Pauli F, Gertz J, Logan JE, et al. (2011) Allele-specific distribution of RNA polymerase II on female X chromosomes. Hum Mol Genet 20 : 3964–3973. doi: 10.1093/hmg/ddr315 21791549
67. Ding Z, Ni Y, Timmer SW, Lee BK, Battenhouse A, et al. (2014) Quantitative genetics of CTCF binding reveal local sequence effects and different modes of X-chromosome association. PLoS Genet 10: e1004798. doi: 10.1371/journal.pgen.1004798 25411781
68. Kung JT, Kesner B, An JY, Ahn JY, Cifuentes-Rojas C, et al. (2015) Locus-Specific Targeting to the X Chromosome Revealed by the RNA Interactome of CTCF. Mol Cell
69. Zlatanova J, Caiafa P (2009) CTCF and its protein partners: divide and rule? J Cell Sci 122 : 1275–1284. doi: 10.1242/jcs.039990 19386894
70. Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, et al. (2014) A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159 : 1665–1680. doi: 10.1016/j.cell.2014.11.021 25497547
71. Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485 : 376–380. doi: 10.1038/nature11082 22495300
72. Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, et al. (2013) Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res 23 : 2066–2077. doi: 10.1101/gr.161620.113 24002784
73. Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, et al. (2014) Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A 111 : 996–1001. doi: 10.1073/pnas.1317788111 24335803
74. Horvath LM, Li N, Carrel L (2013) Deletion of an x-inactivation boundary disrupts adjacent gene silencing. PLoS Genet 9: e1003952. doi: 10.1371/journal.pgen.1003952 24278033
75. Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi JE, et al. (2012) Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res 22 : 2176–2187. doi: 10.1101/gr.136788.111 22722341
76. Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, et al. (2014) Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159 : 374–387. doi: 10.1016/j.cell.2014.09.030 25303531
77. Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, et al. (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485 : 381–385. doi: 10.1038/nature11049 22495304
78. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760. doi: 10.1093/bioinformatics/btp324 19451168
79. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. doi: 10.1186/gb-2009-10-3-r25 19261174
Štítky
Genetika Reprodukčná medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy










