-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Morphological Mutations: Lessons from the Cockscomb
article has not abstract
Published in the journal: Morphological Mutations: Lessons from the Cockscomb. PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1004979
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1004979Summary
article has not abstract
Enormously diverse anatomies have arisen through the course of animal evolution. These varied morphologies are encoded within different genomes, interpreted and implemented through the process of development. Genes controlling anatomical development tend to be highly pleiotropic, operating within large networks to guide the formation of many functionally unrelated structures. This property, together with the high degree of sequence conservation of protein coding sequences between species, has led to the suggestion that evolution of form is driven mostly by mutations in the noncoding parts of the genome responsible for regulation of gene expression. Mutation of gene regulatory sequences could, in principle, influence gene activity in only one or a few tissues, enabling new morphologies with beneficial effects in one organ to be selected while avoiding potentially harmful effects to other structures that might arise from coding sequence mutation [1].
Systematic analyses of genomic regions under selection have supported the view that noncoding mutations play a greater role in adaptation to new environments than coding sequence mutations [2]. However, such population-level, genome-wide studies do not distinguish between morphological and other types of adaptive variation and do not identify the specific mutations associated with distinct traits. A number of phenotype-led studies in which the genetic basis of specific traits has been identified also support the view that morphological evolution often occurs through cis-regulatory mutation [3]. A problem with these trait-directed studies is that they have been done in diverse organisms and have focused on diverse traits, yielding disparate examples that make it difficult to draw general conclusions. In addition, the overwhelming majority of protein-coding mutations with which geneticists are familiar impair, rather than enhance, gene function, and it is not yet clear how this lesson might translate to non-coding mutations. A set of studies published over the past five years by Andersson and colleagues addresses these issues by focussing on the genetic basis for multiple variant forms of a single structure. These enable a test of the “regulatory hypothesis” without bias, and, further, address whether there is any particular form taken by the underlying mutations themselves. The molecular basis of duplex-comb, reported in this issue of PLOS Genetics, completes this informative collection of morphological mutations [4].
The chicken’s comb, which in the wild type is a single serrated blade on top of the head, has taken on a range of new shapes in domestic chickens (Fig. 1). These alternate comb forms have long been known to have a simple heritable basis, serving as major traits for the earliest demonstrations of Mendelian inheritance in animals [5] and yielding a first example of epistatic interaction between genes [6]. Andersson and colleagues set out to identify the genetic basis for the three major variants of the comb, Pea-comb (a smaller comb typically composed of three knobbed ridges), Rose-comb (flattened with papillae towards the beak and tapered to the back), and duplex-combs (either a full comb duplication or small paired horns), providing a systematic set in which to discern whether any particular type of mutation drives alteration of form. The commonalities uncovered are striking.
Fig. 1. Major comb variants and their genetic basis. 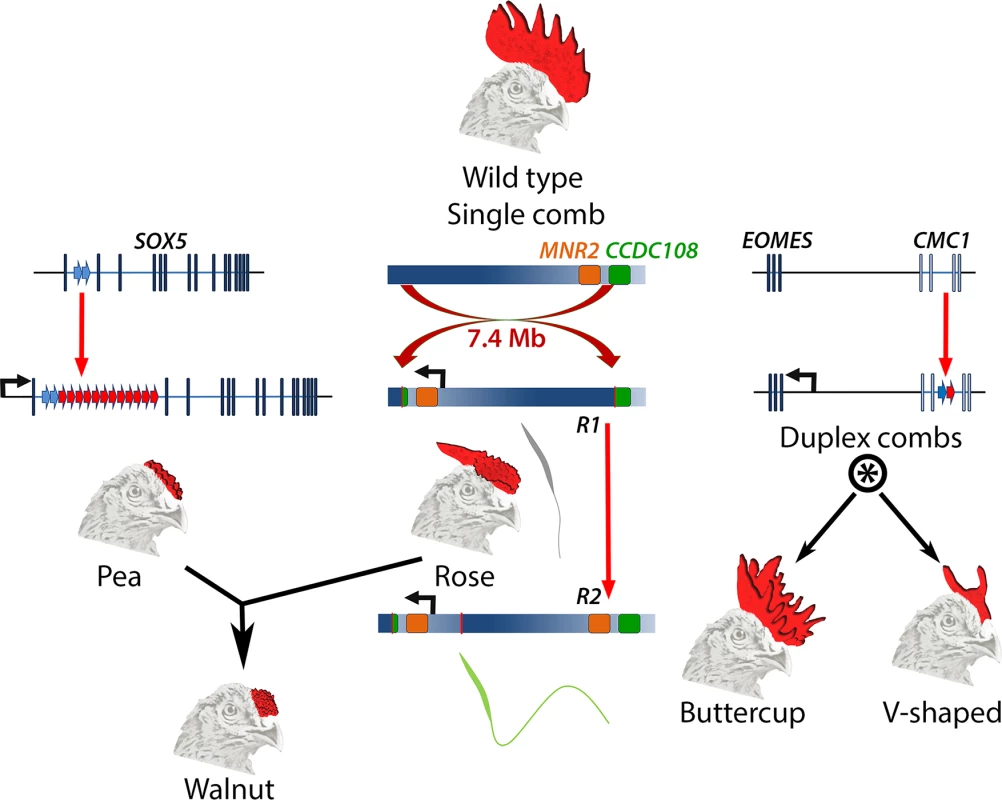
The wild type chicken comb is a single serrated blade. Pea-comb is caused by expansion of an existing tandem duplication in SOX5, leading to ectopic expression of this gene in the mesenchyme of the embryonic comb. Rose-comb is caused by a large inversion that triggers ectopic expression of MNR2 in the same cells. Rose-comb allele R1 includes a disrupted CCDC108 gene and is associated with poor sperm quality. A second Rose-comb allele, R2, which is derived from R1, has an intact CCDC108 gene and restored fertility. The Walnut-comb variant is caused by epistatic interaction between Pea- and Rose-comb alleles. Duplex-comb phenotypes are caused by formation of a new tandem duplication within CMC1, resulting in ectopic expression of the nearby gene EOMES in the ectoderm of the embryonic comb-forming region. Two duplex-comb forms exist, which are distinguished by a second mutation or a linked modifier allele (indicated by *). Genetic loci are not drawn to scale. All three comb variants are underlain by regulatory mutations that are structural, rather than single nucleotide changes, and each causes ectopic expression of a transcription factor. Pea-comb is caused by an approximately 30-fold expansion of a pre-existing tandem duplication in noncoding sequence at SOX5 [7]; Rose-comb by a large inversion, which induces expression of MNR2 [8]; and the duplex-comb phenotypes are now revealed to be a result of a tandem duplication in an intron of CMC1, which triggers expression of the neighbouring gene EOMES [4]. These acquisitions of new expression domains at the prospective comb region occur despite there being very little new sequence generated by the mutations, the bulk of which constitute amplification or rearrangement of existing sequences. It will be interesting to determine the gene regulatory mechanisms underlying the effect of the tandem expansions; whether these disrupt endogenous repressive elements, cause de novo formation of site-specific enhancers, or exert a more general locus-wide effect to permit the action of previously cryptic enhancers. The resulting ectopic expression of each transcription factor presumably amplifies the effect of the causative mutation into altered expression of many genes, thereby modifying the intercellular signalling that controls comb outgrowth [9].
Variation in domestic animals has been used as a guide to understand variation between species since the beginning of evolutionary thinking. However, sheltered from the full force of natural selection by human management, domesticated populations may be able to harbour crude mutations that would not be maintained in wild populations. Here, too, these comb studies have lessons, showing that further evolution of the original mutant alleles occurs either to reduce pleiotropic effects on fitness or to achieve finer tuning of the selected morphological phenotype. The former phenomenon is exemplified by Rose-comb, the original mutant allele of which causes sub-fertility, which has acquired a second rearrangement to repair this defect [8]. Further refinement of form is illuminated by the duplex-comb’s two distinct shapes, which carry the same driving mutation, indicating that a second mutation arising at this site, or possibly a closely linked modifier allele, determines the difference between these morphologies [4,10]. Taken together, this catalogue of mutations hints at the types of genomic change that tend to serve as the source of morphological variation within, and perhaps between, animal species.
Zdroje
1. Wittkopp PJ, Kalay G (2012) Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13 : 59–69. doi: 10.1038/nrg3095 22143240
2. Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, et al. (2012) The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484 : 55–61. doi: 10.1038/nature10944 22481358
3. Kratochwil CF, Meyer A (2015) Closing the genotype-phenotype gap: Emerging technologies for evolutionary genetics in ecological model vertebrate systems. Bioessays In press. doi: 10.1002/bies.201400142 25380076
4. Dorshorst B, Harun-Or-Rashid M, Bagherpoor AJ, Rubin CJ, Ashwell C, et al. (2015) A genomic duplication is associated with ectopic Eomesodermin expression in the embryonic chicken comb and two duplex-comb phenotypes. PLoS Genet 11(1): e1004947.
5. Bateson W (1902) Experiments with poultry. Rep Evol Comm Roy Soc 1 : 87–124.
6. Bateson W, Punnett RC (1905) A suggestion as to the nature of the "walnut" comb in fowls. Proc Camb Phil Soc 13 : 165–168.
7. Wright D, Boije H, Meadows JR, Bed'hom B, Gourichon D, et al. (2009) Copy number variation in intron 1 of SOX5 causes the Pea-comb phenotype in chickens. PLoS Genet 5: e1000512. doi: 10.1371/journal.pgen.1000512 19521496
8. Imsland F, Feng C, Boije H, Bed'hom B, Fillon V, et al. (2012) The Rose-comb mutation in chickens constitutes a structural rearrangement causing both altered comb morphology and defective sperm motility. PLoS Genet 8: e1002775. doi: 10.1371/journal.pgen.1002775 22761584
9. Boije H, Harun-Or-Rashid M, Lee YJ, Imsland F, Bruneau N, et al. (2012) Sonic Hedgehog-signalling patterns the developing chicken comb as revealed by exploration of the pea-comb mutation. PLoS One 7: e50890. doi: 10.1371/journal.pone.0050890 23227218
10. Somes RG (1991) Duplex comb in the chicken: a multi-allelic trait. J Hered 82 : 169–172. 2013691
Štítky
Genetika Reprodukčná medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



