-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
Ultraviolet (UV) light is a ubiquitous agent of exogenous DNA damage. In normal cells, the nucleotide excision repair (NER) pathway is the primary mechanism for repair of UV-induced DNA lesions. Defects in the NER pathway are associated with the human disease xeroderma pigmentosum (XP), and XP patients are prone to skin cancer. Mitotic recombination is strongly stimulated by UV treatment. In this study, we examined whether such stimulation requires the NER pathway. We show that, in the absence of NER, UV is still able to greatly induce recombination. We then characterized a nuclease that is required to generate recombinogenic breaks. Finally, we examined a previously known recombinogenic pathway called the “post-replication repair (PRR) pathway.” Our results suggest that the PRR pathway mainly promotes recombination between sister chromatids, and suppresses recombination between chromosome homologs.
Published in the journal: Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p. PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005026
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005026Summary
Ultraviolet (UV) light is a ubiquitous agent of exogenous DNA damage. In normal cells, the nucleotide excision repair (NER) pathway is the primary mechanism for repair of UV-induced DNA lesions. Defects in the NER pathway are associated with the human disease xeroderma pigmentosum (XP), and XP patients are prone to skin cancer. Mitotic recombination is strongly stimulated by UV treatment. In this study, we examined whether such stimulation requires the NER pathway. We show that, in the absence of NER, UV is still able to greatly induce recombination. We then characterized a nuclease that is required to generate recombinogenic breaks. Finally, we examined a previously known recombinogenic pathway called the “post-replication repair (PRR) pathway.” Our results suggest that the PRR pathway mainly promotes recombination between sister chromatids, and suppresses recombination between chromosome homologs.
Introduction
The primary types of DNA lesions caused by UV radiation are pyrimidine dimers [1]. Although UV strongly stimulates recombination in wild-type yeast cells [2–5], it is unclear whether this stimulation in wild-type cells primarily reflects unexcised dimers, or single-stranded DNA gaps and double-stranded DNA breaks (DSBs) resulting from incomplete nucleotide excision repair (NER) of dimers. One approach to simplifying the nature of the recombinogenic lesion is to examine UV-induced recombination events in NER-deficient cells. Previously, Kadyk and Hartwell (1993, [6]) showed that NER-deficient rad1 strains had reduced levels of UV-induced inter-homolog recombination and elevated levels of sister chromatid recombination compared to UV-induced events in an NER-proficient strain. Based on these observations, they argued that the inter-homolog recombinogenic effects of UV in wild-type cells were likely a consequence of DNA lesions introduced during NER. Since Rad1p can process various types of secondary DNA structures [7,8], one caveat to this conclusion is that Rad1p could be involved in the downstream events of recombination in addition to its role in producing the recombinogenic lesion. Consequently, in the current study, we examined UV-induced recombination in a rad14 diploid. Strains that lack Rad14p cannot perform NER, but are not known to have any other recombination defect [9]. Our analysis demonstrates that inter-homolog recombination events of a variety of types (crossovers, gene conversions unassociated with crossovers, and break-induced recombination events) are greatly elevated by unexcised dimers. Our analysis is the first detailed study of the recombinogenic effects of this biologically-important DNA lesion.
Unexcised pyrimidine dimers block or slow the progression of replication forks [10]. In yeast, G1-synchronized rad14 mutants treated with UV have large (up to 3000 base) single-stranded regions on the leading strand of the replication fork, as well as smaller single-stranded gaps on both the leading and lagging strands [11]. In addition, these strains have asymmetric fork structures diagnostic of broken forks. Presumably, either the single-stranded regions or the DSBs resulting from broken forks could act as recombinogenic lesions. Regressed replication forks are not observed as a consequence of unrepaired UV damage in yeast [11], although evidence for such structures has been obtained in E. coli [10].
Broken DNA molecules can be repaired by a variety of homologous recombination (HR) pathways involving the intact homolog (Fig. 1). Gene conversion events unassociated with crossovers occur primarily through synthesis-dependent strand-annealing (SDSA), although a small fraction are a consequence of processing of a double-Holliday junction (dHJ) or dHJ dissolution [8,12]. In addition, a broken DNA end can invade a duplex, generating a replication structure that duplicates sequences extending to the terminus of the homolog. This process, termed “break-induced replication” (BIR), involves conservative DNA replication [13,14]. These pathways generate a variety of branched intermediates including nicked HJs, double HJs, and single HJs that must be resolved to produce mature recombinant products. Processing of intermediates may involve either cleavage or dissolution (Fig. 1).
Fig. 1. Pathways of homologous recombination. 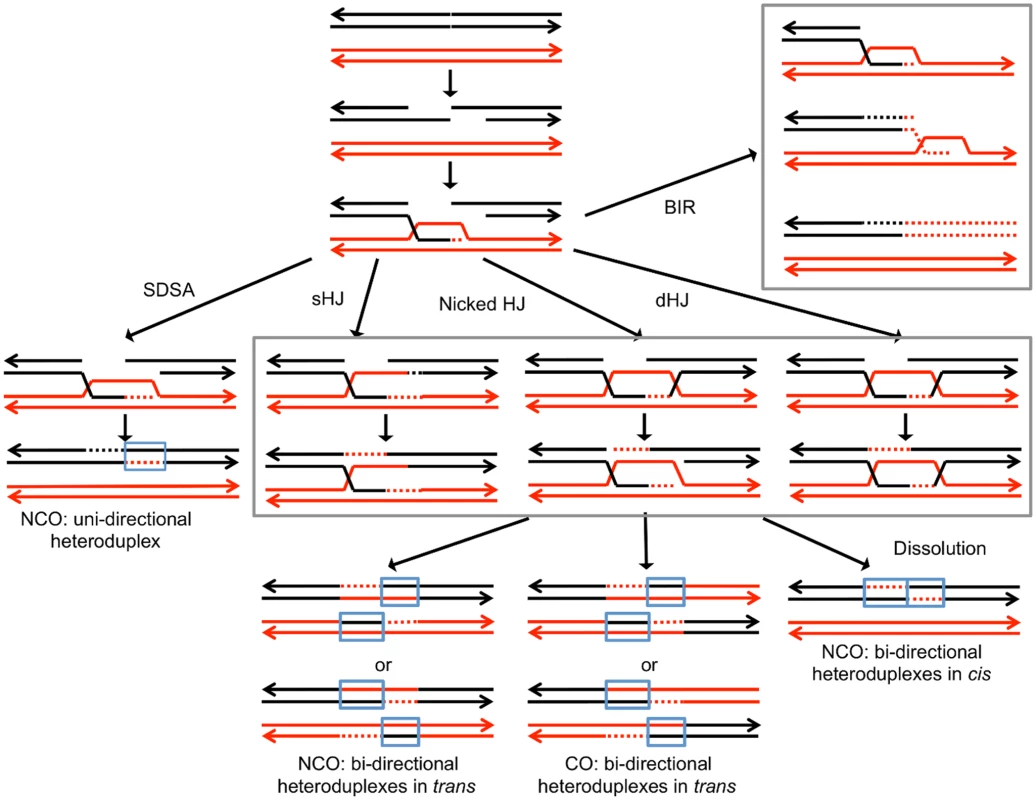
We depict recombination events initiated by a double-stranded DNA break (DSB) with each chromosome shown as a double-stranded DNA molecule. All pathways are initiated by the invasion of the 3’ end of the broken DNA molecule. Dotted lines denote DNA synthesis primed by the invading end. Regions in which a red strand and black strand are paired (in blue boxes) represent heteroduplexes; repair of mismatches within the heteroduplexes can generate gene conversion events. In synthesis-dependent strand annealing (SDSA), following DNA synthesis primed from the invading strand, the invading strand is displaced, hybridizing to the other broken end. This pathway produces a gene conversion that is not associated with a crossover (NCO). In Holliday junction (HJ) containing intermediates, an association of both broken ends with the intact template molecule can result in formation of a single Holliday junction (sHJ), a nicked Holliday junction, or a double Holliday junction (dHJ). Cleavage of these junctions by resolvases can produce either non-crossovers (NCO) or crossovers (CO). For both CO and NCO products, both molecules contain heteroduplexes located in trans. In contrast, if the HJ is resolved by dissolution, the heteroduplex regions are located in cis. In break-induced replication (BIR) events, the invading end copies the intact homolog by conservative DNA replication, resulting in a large terminal LOH event. For the cleavage of various types of HJs formed during mitotic HR in S. cerevisiae, the primary resolvase is the Mus81p/Mms4p complex with Yen1p serving as a backup role [15]. The Mus81p/Mms4p complex prefers nicked HJs as a substrate, whereas Yen1p more efficiently cleaves intact junctions [8]. The nucleases involved in cleavage of HJs are somewhat organism specific and/or specific to the type of recombination (meiotic versus mitotic). For example, Rad1p has no role in junction resolution during meiotic recombination in yeast [16], but its ortholog in Drosophila (MEI-9) is required for meiotic recombination [17]. We show below that recombination between homologs caused by unexcised dimers requires Mus81p but not Yen1.
In addition to DNA repair events that involve inter-homolog interactions, DNA lesions that stall replication forks can be bypassed by both error-free and error-prone pathways of post-replication repair (PRR; [18]). The error-free pathway utilizes HR, primarily operating within the context of the replication fork, to bypass DNA damage. One sub-pathway involves template switching between the two arms of the replication fork, whereas a second pathway involves fork regression. Since fork regression is not evident for UV-induced damage [11], we assume that the primary error-free pathway for UV-induced damage in yeast is template switching. Efficient template switching depends upon the poly-ubiquitination of PCNA by the Ubc13p/Mms2p complex [19]. In contrast, the error-prone pathway utilizes the error-prone translesion synthesis (TLS) DNA polymerases to replicate the damaged DNA strand [18]. The recruitment of TLS polymerases requires mono-ubiquitinated PCNA catalyzed by the Rad6 and Rad18 proteins and is not known to be recombinogenic [19]. In our analysis, we examined strains lacking Mms2p, since such strains should be defective specifically in the error-free component of PRR. We find that mms2 strains have elevated levels of inter-homolog recombination, indicating that the Mms2p suppresses inter-homolog recombination.
Results
Detection of inter-homolog recombination and mapping of loss of heterozygosity events
The system that we used to detect and map UV-induced mitotic recombination events has been described previously [3,4,20]. In brief, we constructed a diploid heterozygous for about 55,000 heterozygous single-nucleotide polymorphisms (SNPs) by crossing two haploids (W303–1A and YJM789) that had 0.5% sequence divergence. In addition, the diploid was heterozygous for an insertion of the ochre suppressor tRNA SUP4-o located near the left end of chromosome V and homozygous for the ade2–1 ochre suppressible mutation. The starting diploid with one copy of SUP4-o partially suppresses the red pigment that accumulates in ade2 strains. Following a mitotic crossover, if the recombined chromosomes are segregated into different daughter cells, one daughter cell would lack the suppressor and one cell would have two copies of SUP4-o (Fig. 2A). Continued divisions of each of these cells would result in a red/white sectored colony with red sector derived from the daughter cell with no copies of the suppressor and the white sector derived from the daughter cell with two SUP4-o genes.
Fig. 2. Patterns of loss of heterozygosity (LOH). 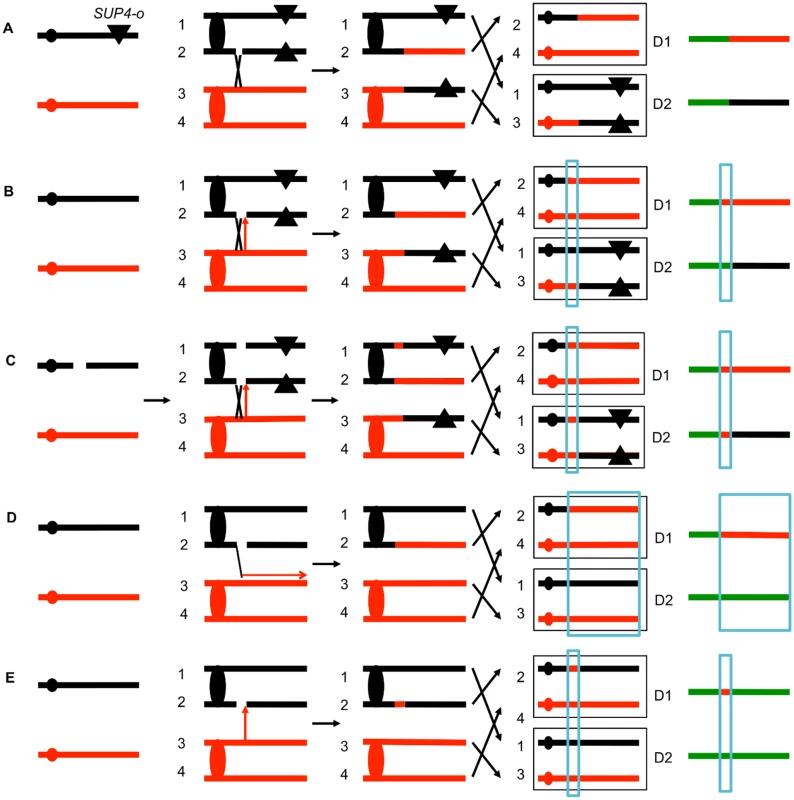
As described in the text, diploids were derived from two sequence-diverged haploids. The diploids were homozygous for the ochre suppressible ade2–1 mutation and heterozygous for the ochre suppressor SUP4-o tRNA (shown as a triangle) near the left end of chromosome V or the right end of IV. Zero, one, and two copies of SUP4-o in the diploid produce red, pink or white colonies, respectively. The black and red lines indicate the homolog derived from the haploids YJM789 and W303–1A, respectively. The boxed chromosomes on the right side of the Fig represent recombinant products. Next to these products, we show the patterns of heterozygous markers and homozygous markers (identified by SNP arrays) as lines. Chromosome regions that are heterozygous, homozygous for W303–1A-derived SNPs, or homozygous for YJM789-derived SNPs are represented by green, red, and black segments, respectively. D1 and D2 indicate the LOH patterns in the two daughter cells that contain the recombinant products. A. Simple crossover. When the red and white sectors are examined by SNP arrays, for this type of recombination event, D1 and D2 are identical in the positions of the transition between heterozygous and homozygous SNPs. This result is expected for a crossover without an associated gene conversion. B. Crossover with a 3:1 conversion tract. In this example, when the red and white sectors are examined, the transition positions between heterozygous and homozygous SNPs are not identical. In the region that is boxed in blue, considering both sectors, three of the chromosomes have “red” SNPs and only one has “black” SNPs. C. Crossover with a 4:0 conversion. In this red/white sectored colony, there is a region adjacent to the crossover in which all four chromosomes have the “red” SNP. We interpret this pattern as resulting from the repair of two sister chromatids that are broken at the same position. It is likely that these events are a consequence of a DSB in an unreplicated chromosome that is subsequently replicated. Repair of one of the broken chromatids is associated with a conversion and a crossover, whereas the second chromatid is repaired by a conversion event unassociated with a crossover. D. BIR event. A DSB on the black chromatid is repaired by a BIR event that duplicates sequences from the red chromatid. Note that this event can be detected on any of the chromosomes, not just the one marked with the SUP4-o gene. E. Conversion event unassociated with a crossover. As noted in Fig. 2D, because of the high frequency of LOH events in UV-treated cells we can detect classes A to E as unselected events. The position of the crossover is the junction between heterozygous SNPs and homozygous SNPs. Mapping of the position of loss of heterozygosity (LOH) was done using oligonucleotide-based microarrays [3]. For about 13,000 SNPs distributed throughout the genome, these arrays contained short (25-base) oligonucleotides that were specific to the W303–1A form of the SNP or the YJM789-specific form. By comparing the level of hybridization of the genomic samples with the recombination event to a heterozygous control strain (details in Materials and Methods), we could determine the transition between heterozygous SNPs and homozygous SNPs.
Several different patterns of LOH were observed. In some red/white sectored colonies, the positions of the transition were identical in both sectors (Fig. 2A). The microarray analysis for the two homologs is depicted as a single line for each sector with heterozygous chromosomal regions shown in green, and regions homozygous for W303–1A-derived SNPs and YJM789-derived SNPs shown as red and black segments, respectively. There were two other types of red/white sector colonies that were commonly observed. As shown in Fig. 2B, some of the sectored colonies had a chromosome segment (boxed in blue), adjacent to the crossover that was not reciprocally transferred between chromosomes. These segments are gene conversion events and likely reflect DNA mismatch repair within a heteroduplex tract adjacent to the crossover (Fig. 1). About 80–90% of mitotic crossovers are accompanied by a gene conversion [4,20]. Considering both chromosomes in both sectors, the conversion event shown in Fig. 2B has three chromosomes with W303–1A-derived SNPs and only one chromosome with YJM789-derived SNPs. Such events will be called “3 : 1” conversions, and these events are a consequence of repair of a single chromatid break (SCB) as shown by the arrow in Fig. 2B. In previous studies of spontaneous and UV-induced mitotic recombination [4,20,21], we also found red/white sectors in which the conversion event produced a 4 : 0 conversion associated with the crossover. This pattern is a consequence of the repair of double sister chromatid breaks (DSCB) at the same position. Our favored interpretation of this result [22] is that a chromosome that received a break in G1 was replicated to produce two broken sister chromatids (Fig. 2C).
Other patterns of LOH can be detected as unselected events by microarrays. For example, BIR can produce one daughter cell with a terminal LOH event and a second daughter cell that has no LOH (Fig. 2D). In addition, unselected gene conversion events that are unassociated with crossovers can produce one daughter cell with an interstitial LOH event and a second daughter cell with no LOH (Fig. 2E). Examples of LOH patterns diagnostic of a conversion-associated crossover, and a conversion unassociated with a crossover are shown in Fig. 3.
Fig. 3. Examples of LOH events as detected by SNP microarrays. 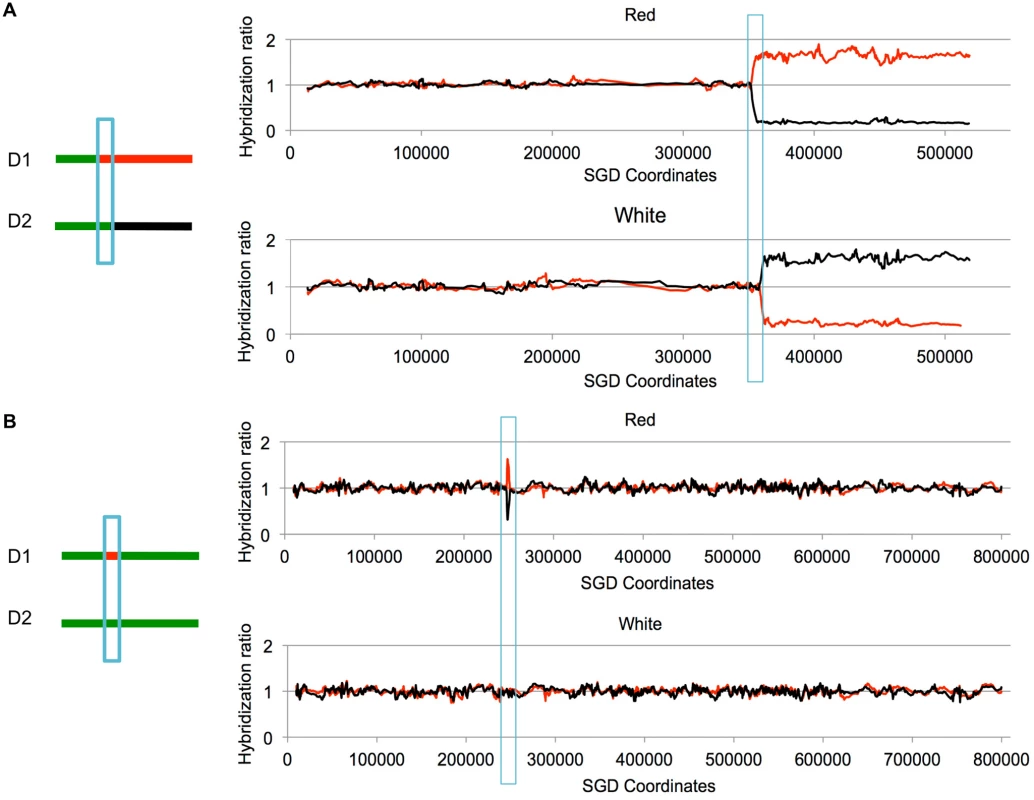
Cells were treated with UV and allowed to form colonies. We isolated DNA from the red and white sectors of sectored colonies, and examined the samples using SNP microarrays. Although these sectors were selected to have a reciprocal crossover on chromosome V, the two events shown below are unselected events on other chromosomes. Experimental samples were hybridized in competition with a differentially-labeled heterozygous control sample. On the Y-axis is shown the relative hybridization level of the experimental and control samples to SNP-specific oligonucleotides. Red and black colors indicate hybridization to W303–1A-specific and YJM789-specific SNPs, respectively. A value of 1 indicates heterozygosity. The X-axis shows SGD coordinates for the individual chromosomes. A. Unselected crossover on chromosome VIII. On chromosome VIII, there is a transition to homozygosity of red SNPs in the red sector and to homozygosity of black SNPs in the white sector. The positions of the transition are not identical in the sectors, indicating a 3:1 gene conversion (blue box) associated with the crossover. B. Unselected gene conversion event on chromosome II. In the red sector, there is a small interstitial LOH region. The white sector has no LOH events. This pattern would be produced by a gene conversion event unassociated with a crossover, or by a conversion event associated with a crossover in which the two recombinant chromosomes co-segregate. In most of the experiments described below, yeast strains with the SUP4-o marker located near the left end of chromosome V or near the right end of chromosome IV were synchronized in G1 and treated with UV. All diploids used in our study had deletions of the MATα gene, allowing their synchronization with α pheromone [22]. The cells were plated on solid growth medium, immediately treated with UV, and subsequently screened for sectored colonies. Cultures derived from each side of the sector were examined by microarrays. For most of the experiments, we used arrays that allowed us to monitor LOH events on the selected chromosome (either V or IV) and unselected events throughout the genome.
Unexcised dimers strongly stimulate mitotic recombination in UV-treated G1-synchronized cells
The NER-deficient rad14 mutant is very sensitive to UV radiation [9]. While 15 J/m2 of UV in G1-arrested wild-type diploids did not significantly reduce cell viability in the wild-type (76% viability relative to unirradiated cells; [4]), less than 0.02% of the rad 14 cells survived 5 J/m2 of UV. Treatment of rad14 cells with 1 J/m2 of UV resulted in 61% cell viability, comparable to the viability of wild-type cells treated with 15 J/m2. Since formation of a sectored colony requires the survival of both daughter cells containing the products of recombination, it is important that the UV dose has a minimal effect on viability of the treated cells. Previously, we mapped UV-induced LOH in both 1 J/m2 - and 15 J/m2-treated wild-type cells [4]. Below, we show sectoring frequency, as well as the frequency of unselected LOH events, in the rad14 mutant treated at 1 J/m2, in wild-type cells treated at 1 J/m2 (resulting in the same number of UV-induced lesions), and in wild-type cells treated with 15 J/m2 (an approximately equitoxic radiation dose).
The sectoring frequency data for diploids with the SUP4-o marker on either chromosome V or on IV are summarized in Table 1. Since only half of crossovers have the segregation pattern that produces reciprocal LOH in the daughter cells [23], the rate of mitotic crossovers is twice the frequency of red/white sectored colonies for all strains. For most strains treated with UV, the sector frequencies were determined non-selectively. For the untreated wild-type strain with the SUP4-o marker on chromosome V, crossovers were selected as described by Lee et al. [21]. This method, which involves selection of crossovers as canavanine-resistant sectored colonies, did not work well for strains with the rad14 mutation or for strains with the SUP4-o marker on chromosome IV [20]. In the rad14 mutant treated with 1J/ m2 of UV, the red/white sectoring frequency on the left arm of chromosome V is 0.5%; this frequency is similar to that of the wild-type strain treated with 15 J/m2, and is five-fold higher (p = 1.7×10-10) than the wild-type treated with 1 J/m2 [4] as shown in Table 1. For the same interval, the spontaneous sectoring frequency for the wild-type strain is 1.10.5×10-6. In the wild-type strain, we can select crossovers on the left arm of V as canavanine-resistant red/white sectors [21,24]. For unknown reasons, this selection does not work in the rad14 diploids and, therefore, we screened for sectored colonies. Since no such colonies were detected in 22797 total colonies (Table 1), we conclude that the frequency of spontaneous crossovers in rad14 strains is very low, less than 4.4 ×10-5.
Tab. 1. Frequency of sectored colonies in unirradiated and UV-irradiated wild-type and rad14 strains.1 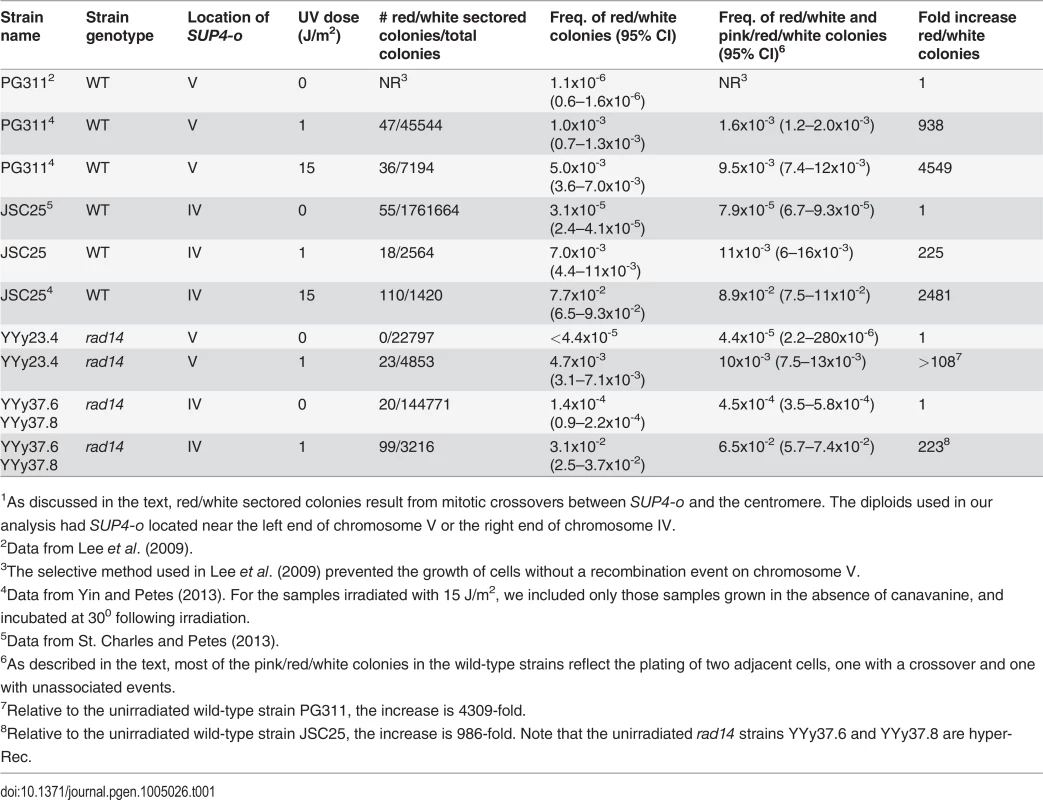
1As discussed in the text, red/white sectored colonies result from mitotic crossovers between SUP4-o and the centromere. The diploids used in our analysis had SUP4-o located near the left end of chromosome V or the right end of chromosome IV. In order to obtain a more accurate estimate of the frequency of sectored colonies in the unirradiated rad14 strain, we examined sectored colonies in strains in which the SUP4-o marker was located on the right arm of chromosome IV (Table 1). The distance between CEN4 and SUP4-o is 1.1 Mb, about eight times larger than the interval between CEN5 and SUP4-o in the other strains. Sectored colony formation in the rad14 mutant was stimulated about 200-fold by the 1 J/m2 dose of UV relative to the unirradiated rad14 strain. The frequency of red/white sectored colonies in rad14 strains treated with 1 J/m2 UV dose was about four-fold higher than the frequency of such colonies in the wild-type strain treated with the same dose. Interestingly, the rad14 mutation resulted in a modest (about five-fold, p = 1.8×10-9) increase in the frequency of sectors in the absence of UV irradiation.
In addition to the frequencies of red/white sectors, we also show in Table 1 the frequencies of sectors that were either red/white or pink/red/white. Pink/red/white sectors could arise in two different ways: 1) persistence of DNA damage beyond the first division cycle following UV treatment (S1 Fig.), or 2) irradiation of two adjacent cells resulting in a recombination event in one of them (the red/white portion of the sector) but no recombination event in the other (the pink portion of the sector). By micromanipulation of individual cells before irradiation, we showed that most (9 out of 10) of the selected crossovers occur in the first cell division in UV-treated wild-type strains [4], therefore, the pink/red/white sectored colonies we recovered from plating experiments are more consistent with the second explanation.
The recombinogenic effects of unexcised UV-induced DNA damage were further demonstrated by our whole-genome analysis of sectored colonies derived from UV-treated wild-type and rad14 strains (Table 2). This analysis allows the detection of unselected crossovers, gene conversions, and BIR events throughout the genome. The rad14 strain treated with 1 J/m2 had about seven unselected LOH events per sectored colony, whereas the wild-type strain treated with the same dose had only 0.4 LOH events per sectored colony. The unselected events from the wild-type strain treated with 15 J/m2 are described in S1 Table and S2 Table, whereas the unselected events from the rad14 strain are described in S3 Table and S4 Table. S1 Table and S3 Table depict the transitions between heterozygous and LOH regions within each sector schematically, and S2 Table and S4 Table show the SGD coordinates for each transition. Comparable data for unselected events for the wild-type strain irradiated with 1 J/m2 were described previously [4], as were the methods for depicting various classes of LOH events [3,4]. S5–S13 Tables summarize the transitions for the other UV-treated mutant strains, and S14 Table and S15 Table show the genome regions covered by the SNP arrays.
Tab. 2. Numbers of unselected UV-induced LOH events, deletions/duplications, and chromosome number changes in sectored colonies derived from UV-treated wild-type, rad14, mus81, and yen1 diploids.1 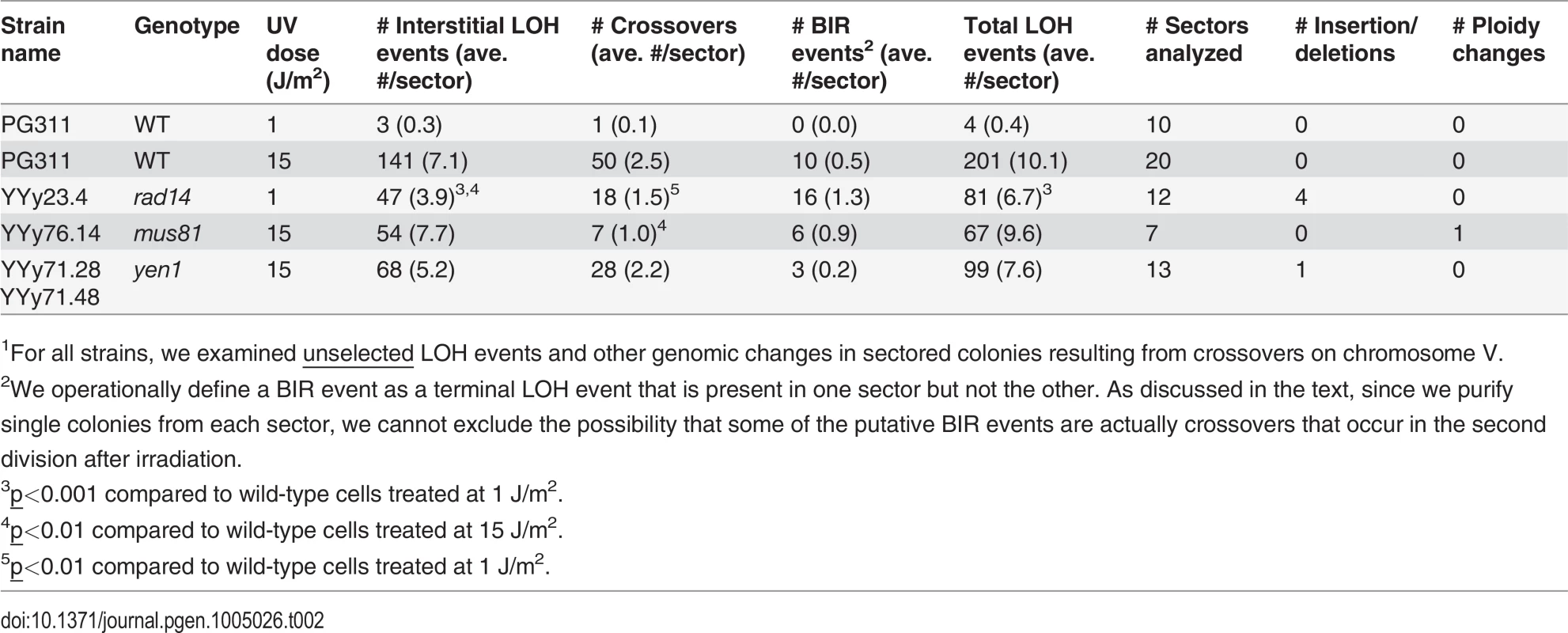
1For all strains, we examined unselected LOH events and other genomic changes in sectored colonies resulting from crossovers on chromosome V. For statistical comparisons of the number of unselected recombination events for the wild-type and rad14 strains irradiated with 1 J/m2, we determined the number of LOH events per sectored colony for each strain (ten and twelve sectored colonies from the wild-type and rad14 strains, respectively; Table 2), and compared these distributions by the Mann-Whitney test. By this test, the number of unselected events per colony for the rad14 strain (average of 6.7) was significantly greater (p = 0.0003) than the number observed for the wild-type strain (average of 0.4).
Qualitative comparison of UV-induced recombination events in rad14 and wild-type strains
As shown in Fig. 2, the patterns of LOH in a sectored colony can reveal whether a mitotic recombination event is a consequence of the repair of a DSB on a SCB or repair of a DSCB. 3 : 1 conversion tracts (Fig. 2B and Fig. 2E) are interpreted as SCB events, whereas 4 : 0 tracts (Fig. 2C) are indicative of DSCBs [4,21]; crossovers without a detectable conversion tract cannot be classified as SCB or DSCB. We found previously that high doses of UV (15 J/m2) in G1-synchronized wild-type strains result in more DSCBs than SCBs, but low doses (1 J/m2) produce primarily SCBs [4]. In the G1-synchronized rad14 strain treated with 1 J/m2, 78% of the selected events were SCBs, similar to the fraction observed for the wild-type strain treated with the same UV dose. The data from both the selected events on chromosome V and the unselected events on other chromosomes derived from sectored colonies are in Table 3. In summary, our results demonstrate that UV-induced recombination events in rad14 strains usually involve a single broken sister chromatid.
Tab. 3. Numbers of single-chromatid breaks (SCBs) and double sister-chromatid breaks (DSCBs) induced by UV in G1-synchronized cells. 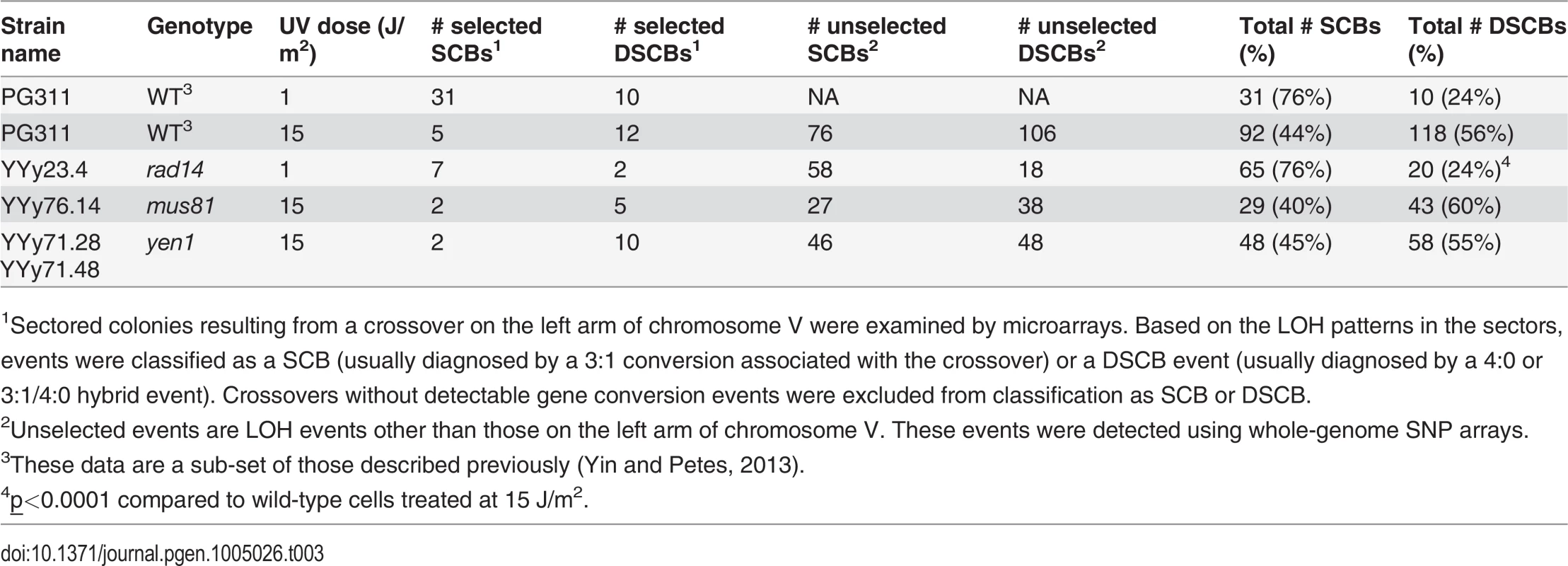
1Sectored colonies resulting from a crossover on the left arm of chromosome V were examined by microarrays. Based on the LOH patterns in the sectors, events were classified as a SCB (usually diagnosed by a 3:1 conversion associated with the crossover) or a DSCB event (usually diagnosed by a 4:0 or 3:1/4:0 hybrid event). Crossovers without detectable gene conversion events were excluded from classification as SCB or DSCB. The rad14 strain irradiated with 1 J/m2 had significantly different proportions of recombination events relative to the wild-type strain irradiated with an equitoxic UV dose (15 J/m2) (Table 2). More specifically, the rad14 strain had significant elevations in the number of BIR events and the number of deletions relative to the wild-type strain (p = 0.0003 and 0.0004, respectively). Interestingly, we found presence of early/middle-firing ARS sequences [25] within the deleted sequences in all the 4 deletion events. Although no significant differences were observed between the rad14 and wild-type strains irradiated with 1 J/m2, there were very few unselected LOH events in the wild-type strain.
In considering the frequency of BIR events in the rad14 strain, we note an important caveat. BIR events in red/white sectored colonies are defined as unselected non-reciprocal terminal LOH events (S2 Fig.). If UV-irradiated rad14 strains retain recombinogenic lesions beyond the first division cycle, this same LOH pattern could be produced by having one UV-induced crossover on chromosome V in the first division cycle (resulting in the red/white sectored colony) and a second UV-induced LOH event on an unselected chromosome arm in the second division cycle (S3 Fig.). The second LOH event would produce two different types of granddaughters (shown as GD2–1 and GD2–2 in S3 Fig.). In our analysis of sectored colonies, we purify single colonies derived from each sector. If we examined one colony derived from the white sector (either GD1–1 or GD1–2) and a colony derived from GD2–1 from the red sector, we would conclude that there was a reciprocal crossover on chromosome V and an unselected BIR event on the other homolog. In conclusion, the relative frequency of unselected crossovers and BIR events in the UV-irradiated rad14 strain is unclear. This same caveat applies to all genetic analyses in which the experimental strain has high levels of on-going genetic instability.
To clarify whether unrepaired UV-induced lesions in rad14 strains were capable of generating recombination events after the first cell division, we micromanipulated single G1-arrested rad14 cells to specific positions on solid medium that we then irradiated. Of 1096 irradiated (1 J/m2) rad14 cells, 681 cells survived. Of four resulting sectored colonies, two were red/white as expected for a crossover on chromosome V in the first division, and two were pink/red/white as expected for a crossover in the second division (S1 Fig.). The same micromanipulation analysis for the wild-type strain treated with 15 J/m2 yielded nine colonies with red/white sectors and only one colony with pink/red/white sectors [4]. Although the numbers of these events were small and the difference in proportions of red/white and pink/red/white sectors in wild-type and rad14 cells is not statistically significant (p = 0.18 by Fisher exact test), these results suggest that recombinogenic DNA damage can persist in rad14 cells, as expected from unexcised pyrimidine dimers.
Recombination caused by unexcised dimers is stimulated by Mus81p but not by Yen1p
Mus81p is the main Holliday junction resolvase for mitotic crossovers in S.cerevisae [15] and for meiotic crossovers in S. pombe [26]. In addition, mus81 strains are UV sensitive [27]. To determine whether Mus81p had a role in the processing of UV-induced recombinogenic lesions, we examined the frequency and types of UV-induced recombination in a mus81 strain and in a mus81 rad14 double mutant strain.
For the mus81 strain, we used microarrays to perform a genome-wide analysis of seven red/white sectored colonies derived from G1-synchronized cells irradiated with 15 J/m2. There was no significant difference (p = 0.89 by Mann-Whitney test) in the average number of interstitial LOH events (primarily gene conversions) in the mus81 strain compared to wild-type (7.7 and 7.1, respectively). In contrast, the average number of unselected crossovers in the mus81 strain was reduced to 40% of the level observed in the wild-type strain (Table 2). This loss of crossovers is statistically significant (p = 0.004 by Mann-Whitney test). These results are similar to those observed for HO-induced DSBs by Ho et al. [15], demonstrating that Mus81p was involved in the resolution of HJs by crossovers, but not in the SDSA conversion pathway. In addition, the single mus81 mutant does not significantly (p = 0.72 by Mann-Whitney test) reduce the frequency of recombination events, since the average number of LOH events per sectored colony is very similar for the wild-type and mus81 strains (10.1 and 9.6, respectively).
Double mutant mus81 rad14 strains were very sensitive to UV. A dose of 1 J/m2, which reduced the viability of the rad14 strain to 61% that of the untreated strain, lowered the viability of the double mutant strain to 0.2%. Consequently, we monitored the effect of UV in the double mutant strain on UV-induced mitotic recombination by analyzing single colonies formed from cells treated with 1 J/m2 rather than by measuring the frequency of red/white sectored colonies. The results of this analysis are shown in Table 4, and the distributions of the number of LOH events per colony for rad14 and rad14 mus81 strains are shown in Fig. 4. We found that the total unselected LOH events were significantly reduced in the double mutant compared to the single rad14 mutant (p = 0.0005 by Mann-Whitney test). This result suggests that Mus81p is likely involved in generating recombinogenic DNA lesions at replication forks that are blocked at unexcised pyrimidine dimers; this interpretation will be further considered in the Discussion.
Tab. 4. Numbers of unselected UV-induced LOH events and other genomic changes in single colonies derived from rad14, rad14 mus81, rad14 yen1, and rad14 mms2 diploids treated with 1 J/m2 of UV. 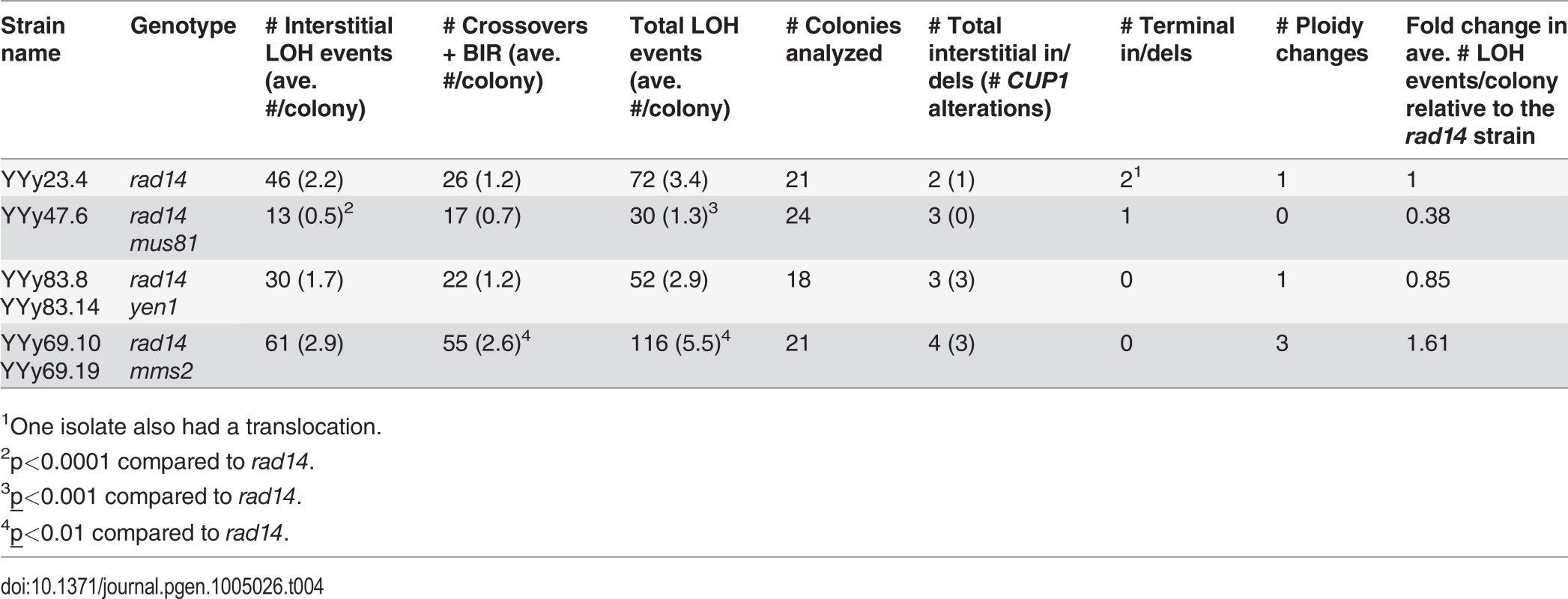
1One isolate also had a translocation. Fig. 4. Histogram of numbers of unselected LOH events per colony induced by UV. 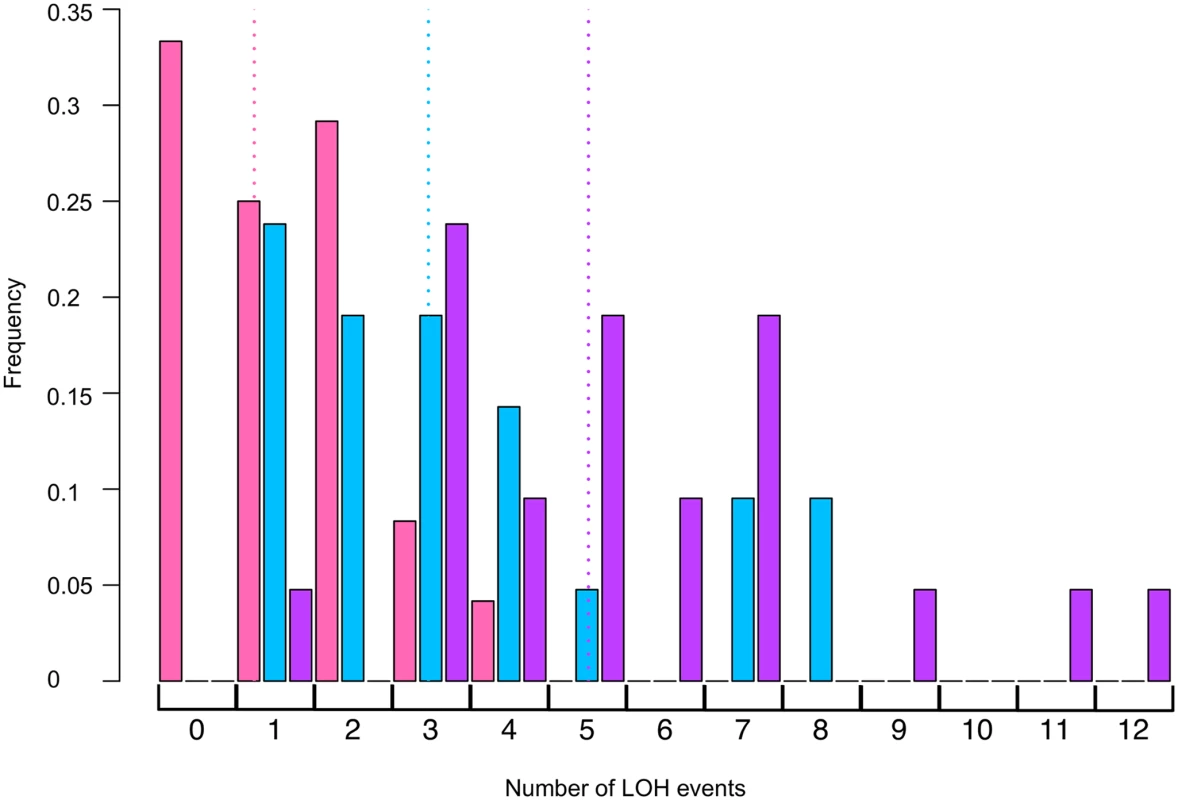
Using SNP microarrays, we analyzed more than 20 colonies of each of three strains (rad14, rad14 mus81, and rad14 mms2) for unselected LOH events induced by 1 J/m2. The numbers of LOH per colony are shown in this figure. The color code is: rad14 (blue), rad14 mus81 (pink), and rad14 mms2 (purple). The dotted lines indicate the average number of LOH events per single colony using the same color code. We also examined the effects of the Yen1p HJ resolvase on the UV-induced recombination events in a rad14 strain. After a dose of 15 J/m2 of UV, the single yen1 mutant has about the same number of LOH events per cell as wild-type (Table 2). The double mutant rad14 yen1 strain had approximately the same average number and types of LOH events per irradiated cells as the single rad14 mutant (Table 4); none of the observed differences with wild-type were statistically significant. In summary, our analysis suggests that the Mus81p, but not the Yen1p, has a role in generating recombinogenic lesions in UV-treated rad14 strains.
Recombination between homologs induced by unexcised dimers is suppressed by the Mms2p-mediated PRR pathway
As discussed in the Introduction, unexcised pyrimidine dimers can be bypassed by either error-prone or error-free pathways [18]. The error-free pathway utilizes HR enzymes to catalyze either template switching between the two arms of the replication fork or fork regression. Since fork regression has not been observed for UV-treated strains, we assume that the primary pathway for error-free repair in our experiments is template switching. Template switching requires Ubc13p and Mms2p to poly-ubiquitinate PCNA. Although template switching is generally considered to be limited to the arms of the replication fork or to sister chromatids, most previous studies have been done in haploid strains. Consequently, we compared the frequency of UV-induced recombination in rad14 and rad14 mms2 diploids. If Mms2p is required for inter-homolog recombination, we would expect a reduction in recombination in the double mutant whereas, if Mms2p was required solely for inter-replication fork or inter-sister chromatid recombination, the double mutant should have elevated inter-homolog recombination.
The rad14 mms2 strains, like the rad14 mus81 strains were very UV sensitive. At a UV dose of 1 J/m2, the double mutant has 1% survival compared to 61% survival for rad14 strain, and 0.2% survival for the rad14 mus81 strain. Consequently, we compared the numbers and types of LOH events in single colonies derived from cells treated with 1J/m2 of UV (Table 4, Fig. 4). The average number of LOH events in a double mutant of rad14 mms2 increased by 62% compared to the rad14 strain; this difference is statistically significant (p = 0.008 by Mann-Whitney test). The main class of events that is elevated in the double mutant is the CO/BIR category (p = 0.001 by Mann-Whitney test). These results suggest that Mms2p promotes recombination between the arms of the replication fork and/or sister chromatids and, in the absence of Mms2p, the recombination intermediates are directed into an inter-homolog pathway of recombination.
Lengths of UV-induced gene conversion tracts in wild-type, rad14, mus81, and yen1 strains
In our previous analysis of the lengths of UV-induced mitotic gene conversion tracts in wild-type strains treated with 15 J/m2, we found a median length of 6.4 kb for all conversion tracts; median lengths of crossover-associated tracts (CO events) were longer than for conversions unassociated with crossovers (NCO events), 8.2 kb and 5.7 kb, respectively [4]. The median lengths of conversion events (CO plus NCO) in rad14, mus81, and yen1 are all between 5.2 and 7.9 kb (Table 5). These lengths are not significantly different from the wild-type strain. As observed for the wild-type strain, the median lengths for the CO conversions exceed those for the NCO conversions for all strains. When we compared the lengths of conversions in NCO and CO categories, the only significant difference was that the lengths of CO events in the mus81 strain significantly exceeded those of the wild-type strain irradiated with 15 J/m2 (p = 0.027 by Mann-Whitney test). One interpretation is that failure to cleave nicked HJs in the mus81 mutants results in their maturation to double Holliday junctions that are associated with longer heteroduplex regions.
Tab. 5. Median lengths of gene conversion tracts in UV-induced LOH events in wild-type, rad14, mus81, and yen1 strains.1 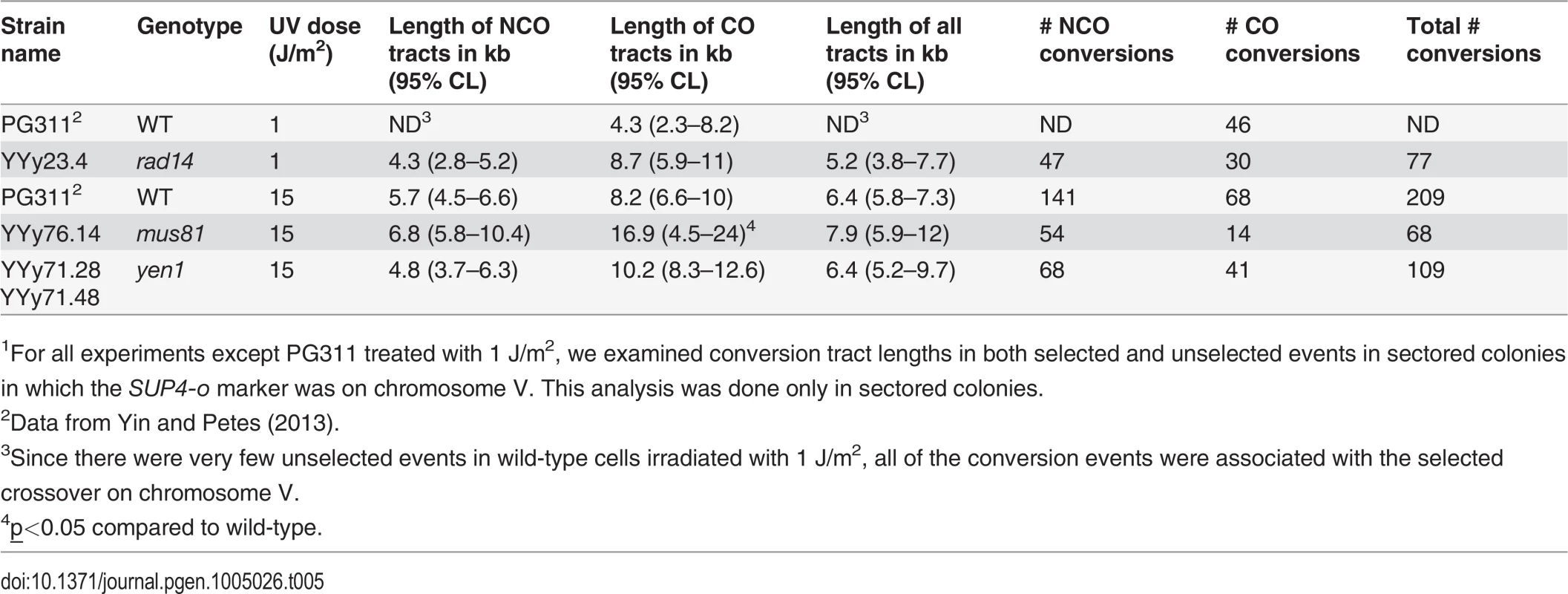
1For all experiments except PG311 treated with 1 J/m2, we examined conversion tract lengths in both selected and unselected events in sectored colonies in which the SUP4-o marker was on chromosome V. This analysis was done only in sectored colonies. Distribution of LOH events resulting from unexcised pyrimidine dimers
For crossovers, the DNA lesions that initiate recombination should be located near the transitions between heterozygous and homozygous regions (the breakpoints of the LOH events). For gene conversions unassociated with crossovers, the initiating lesion should be located between the heterozygous sites flanking the conversion tract. Consequently, we determined whether these regions were significantly enriched for various types of chromosome elements such as replication origins, retrotransposons, palindromes, and regions associated with G4 motifs. The methods used for this analysis are described in the S1 Text, and the data are presented in S16 Table. We previously showed that spontaneous mitotic recombination events and LOH events induced by low levels of DNA polymerase α were enriched for replication-termination (TER) sequences [25] and other motifs associated with slow-moving or stalled replication forks [20,28]. In contrast, none of the chromosome elements tested were significantly over-represented among UV-induced LOH events in a wild-type strain [4].
We examined sixteen categories of chromosome elements for their representations at LOH breakpoints including: centromeres, tRNA genes, non-coding (nc) RNAs, solo long-terminal repeats (LTRs), early/middle ARS elements, late ARS elements, Rrm3p pause sites, palindromes, retrotransposons, G4 motifs, highly-transcribed and weakly-transcribed genes, TER sequences, regions with high levels of gamma-H2AX, snoRNAs and snRNAs, and genomic regions with inefficient TT dimer repair (references for the mapping of these elements are in S16 Table). In the rad14 single mutant, none of these elements were significantly over - or under-represented. In contrast, in both the mus81 and rad14 mus81 strains, G4 (quadruplex) motifs are over-represented at the LOH breakpoints. In addition, in the rad14 mus81 double mutant strain, we observed over-representation of retrotransposons (Ty elements), Rrm3p pause sites, regions with high levels of gamma H2AX, and regions associated with slow thymidine dimer repair (S16 Table).
Many of the over-represented motifs are associated with delayed or stalled replication forks in wild-type yeast cells, and are hotspots for mitotic recombination events induced by low levels of DNA polymerase α [28]. One explanation of our results is that the probability that a stalled replication fork will be broken is a function of at least three factors: certain DNA motifs (for example, quadruplex DNA) that cause replication forks to move slowly or stall, the presence of unexcised pyrimidine dimers near these motifs, and the presence or absence of Mus81p. Although Mus81p is required for about 60% of the LOH events induced by unexcised dimers, 40% are independent of Mus81p (Table 4). No significant associations were observed at the breakpoints of the rad14 yen1 strain, although the rad14 mms2 mutant had significant over-representations of ncRNAs and TER sequences.
Analysis of spontaneous recombination events in rad14, mus81, and yen1 single mutant strains
In strains with the SUP4-o marker near the right end of chromosome IV, we did a limited analysis of spontaneous recombination events derived from red/white sectored colonies in the rad14, mus81, and yen1 strains. The primary purpose of this analysis was to ensure that none of the single mutants substantially elevated LOH in the absence of UV irradiation. The numbers of sectored colonies divided by the total number of colonies examined, the frequencies, and the strains names were: wild-type (55/1761664; 3.1x10-5; JSC25; [20]), rad14 (20/144771; 1.4x10-4; YYy37.6/YYy37.8), mus81 (10/277704; 3.6x10-5; YYy45.2/YYy46.1), and yen1 (12/276315; 4.3x10-5; YYy72.22/YYy72.41). Although there is a statistically significant elevation of spontaneous crossovers in the rad14 strain (as noted previously), the frequency of sectored colonies in unirradiated cells is two orders of magnitude less than in the irradiated cells. We also looked for sectored colonies derived from unirradiated cells of the rad14 mus81 and rad14 mms2 genotypes that had the SUP4-o marker on the left end of chromosome V. No sectored colonies were observed in colony totals of 22797 (rad14), 39466 (rad14 mus81), and 37493 (rad14 mms2). Although this analysis does not allow an accurate measurement of the sectoring frequency, we estimate that the frequency of spontaneous sector formation is less than 4x10-5 for all three strains. Thus, the spontaneous events do not contribute significantly to our estimates of the frequencies or types of UV-induced exchanges.
Discussion
We performed a genome-wide analysis of inter-homolog recombination events induced by unexcised pyrimidine dimers, and we monitored the effect of various DNA repair enzymes on the frequency and types of these recombination events. The main conclusions from our study are: 1) unexcised pyrimidine dimers induced by irradiation of G1-synchonized cells strongly induce mitotic recombination between homologs, 2) most of the induced events reflect the repair of a single broken sister chromatid, 3) the recombinogenic effects of unexcised dimers are largely dependent on the Mus81p resolvase while in the NER-proficient strains, only crossovers are reduced by deletion of MUS81, and 4) rad14 mms2 strains have elevated levels of UV-induced recombination events relative to the rad14 single mutant, consistent with the hypothesis that the Mms2p-mediated PRR pathway channels DNA lesions for repair to the sister chromatid.
Differences in UV-induced recombination between NER-proficient and NER-deficient cells
Although the recombinogenic effects of UV irradiation in yeast have been known for a long time [5], the nature of the recombinogenic DNA lesion and the pathways of lesion repair are still unclear. Based on the effects of UV irradiation in synchronized cells, Galli and Schiestl [2] suggested that NER-generated lesions (single-stranded nicks or gaps) could be converted into broken chromatids during DNA replication. Alternatively, the blocking of replication forks by unexcised dimers [10] could be converted into a recombinogenic DSB. One approach to distinguishing these possibilities is to compare UV-induced recombination events in wild-type and NER-deficient strains. Using this approach, Kadyk and Hartwell [6] found that wild-type diploid strains synchronized in G1 and treated with 30 J/m2 of UV had about a five-fold elevation in inter-homolog gene conversion and a two-fold induction in sister-chromatid recombination. In contrast, an NER-deficient rad1 diploid strain irradiated with 1 J/m2 (approximately equivalently genotoxic with a dose of 30 J/m2 in a wild-type strain) had a six-fold elevation in sister-chromatid recombination and a three-fold reduction in the frequency of inter-homolog conversion events. From this analysis, they concluded that inter-homolog recombination was likely a consequence of nicks and gaps formed by NER, whereas unexcised dimers primarily induced sister-chromatid recombination rather than inter-homolog recombination.
In our study, UV treatment very strongly (>100-fold) stimulated mitotic crossovers (Table 1), as well as other types of recombination events (Table 2), in an NER-deficient (rad14) strain. There are several explanations for the discrepancy between our results and those of Kadyk and Hartwell [6]. First, different mutations were used to inactivate NER, rad14 in our experiments and rad1 in those of Kadyk and Hartwell. Rad1p, unlike Rad14p, is needed to process certain types of mitotic recombination intermediates [29] in addition to its requirement for NER. However, since Rad1p is not required for I-SceI-induced recombination events between homologs [7], lack of Rad1p would not be expected to reduce the frequency of UV-induced inter-homolog exchange. In addition, in the rad14 (strain YYy23.4) and rad1 (strain YYy327.1) cells treated with 1 J/m2 of UV, the frequencies of survival (49% and 47%, respectively), and red/white sectored colonies (8.1 x 10-3 and 6 x 10-3, respectively) were similar in the two strains.
A more likely explanation is the nature of the genetic system used to detect inter-homolog exchange in the two studies. In the Kadyk and Hartwell study [6], inter-homolog conversion events were detected by measuring the frequency of leucine prototrophs derived from a diploid with leu1 heteroalleles. This system likely detects a very small fraction of the induced events. Assuming that mitotic conversion is a consequence of heteroduplex formation followed by DNA mismatch repair, a prototroph will be detected in only two situations: 1) the heteroduplex includes only one of the two leu1 mutations or 2) the heteroduplex includes both mutations but the resulting repair event using different strands as the repair template (“patchy” repair). However, mitotic conversion events are usually longer than one gene (> 4 kb) and multiple mismatches within a heteroduplex are usually repaired using the same strand as a template [4,20]. In our analysis, we examine inter-homolog recombination events by a sectoring assay for crossovers or by detecting unselected crossovers and conversions using SNP microarrays. These systems have the advantages of having fewer constraints on the detection of gene conversion events and of being less selective as to the location of the events.
Although our assay is designed primarily to detect LOH events occurring between homologs, we recovered four interstitial deletions in the UV-treated rad14 strains (Class R in S3 Table). Three of four deletions had directly-oriented Ty elements at the breakpoints of the deletion, and the fourth had one Ty element and a directly-oriented delta (LTR) (S4 Table). Thus, these deletions are likely generated by HR between the direct repeats rather than non-homologous end-joining. We have detected similar events previously in other genetically-unstable yeast strains [28,30]. The deletion events can be explained as intrachromatid “pop-outs”, unequal sister-chromatid crossovers, single-strand annealing, gene conversion, or off-set BIR events between sister chromatids. Some of these possibilities are shown in S4 Fig. Although deletions are observed in UV-treated rad14 strains, none were observed in UV-treated wild-type strains. The reason for this difference is not understood although it could be related to the nature of the DNA lesion, or the kinetics of the repair process.
It should be emphasized that our system (and almost all genetic systems) are capable of detecting inter-homolog and unequal sister chromatid recombination events but cannot detect sister chromatid exchanges between perfectly-aligned chromatids. A UV dose of 1 J/m2 is expected to introduce about 500 pyrimidine dimers/diploid genome [31]. Since we detect about seven LOH events/sectored colony in the rad14 strain (Table 2), most of the UV lesions must be bypassed by mechanisms that do not result in LOH, either recombination events that involve equal sister chromatid recombination or that utilize TLS polymerases.
Nature of the recombinogenic DNA lesions in UV-treated yeast cells
As discussed above, one likely source of recombinogenic lesions in G1-synchronized wild-type cells treated with UV is nicks or gaps generated by NER; replication of such molecules would lead to DSBs [2]. If this lesion was solely responsible for initiating recombination in a wild-type cell, we would expect UV-irradiation of G1-synchronized cells to generate recombination events that indicate the break of a single chromatid (SCBs) (Fig. 2). In wild-type cells irradiated with 15 J/m2, however, we found that more than half of the events involved the repair of two sister chromatids broken at approximately the same position (DSCBs) [4]. Our interpretation of this result was that high doses of UV in G1-synchronized cells resulted in DSBs, and this interpretation was supported by physical evidence of a low frequency of UV-induced DSBs (5–10 DSBs in cells irradiated with 40 J/m2; [32]). In G1-synchronized cells irradiated with low levels of UV (1 J/m2), most of the recombination events were SCBs [4]. These results argue that the DSBs likely reflect the excision of closely-opposed pyrimidine dimers that arise independently [4]. An analysis of UV-induced recombination in exo1 cells treated with 15 J/m2 showed that Exo1p-expanded NER gaps were required for both SCB and DSCB events [33]. In addition, as described in [34], an analysis of UV-induced events in G2-synchronized haploid cells showed that single-stranded gaps stimulated recombination between sister chromatids. In summary, there are likely several types of recombinogenic lesions generated by UV in wild-type cells: G1-associated DSBs resulting from the repair of closely-opposed dimers, S/G2-associated DSBs resulting from replication of a DNA molecule with an NER-generated gap, and single-stranded gaps; Exo1p-expanded NER gaps contribute to the formation of DSBs in both G1 and S/G2.
Most (76%) of the recombination events induced by UV in G1-synchronized rad14 cells are SCBs (Table 3). These recombination events cannot be related to NER-generated DNA lesions, and we suggest that the relevant lesion is produced by breaks that occur at replication forks stalled by unexcised pyrimidine dimers. Two of the important HJ or nicked HJ resolvases in yeast are Mus81p and Yen1p. Consistent with the observations that Mus81p is required for DSB formation at fragile sites [35] and at camptothecin-stalled replication forks in mammalian cells [36], we found that loss of Mus81p reduced the frequency of UV-induced LOH events in the rad14 strain by about 60%. This reduction could be explained as a reduction in the level of recombinogenic DSBs or as less efficient processing of recombination intermediates to yield LOH events. Previously, Ho et al. [15] showed that mus81 mutants had reduced frequencies of I-SceI-induced mitotic crossovers but wild-type levels of induced conversions. Our observation that rad14 mus81 mutants have reduced UV-induced gene conversion unassociated with crossovers relative to rad14 (Table 3) argues that Mus81p is involved in generating DSBs, although it may also have a role in downstream recombination events. In comparisons of the frequency of gene conversion events in wild-type and mus81 strains treated with 15 J/m2 of UV, we found that Mus81p was required for crossovers, but not conversions unassociated with crossovers (Table 2), as expected from previous results.
Loss of Yen1p had no detectable effect on the frequency of LOH events in UV-treated rad14 strains. Based on the substrate preferences for Mus81p and Yen1p [8], this result argues that the relevant recombinogenic structure of the blocked replication fork is a nicked HJ rather a ligated HJ. It has been suggested that extensively-regressed replication forks resembled ligated HJs which should be processed by Yen1p [37]. Thus, the lack of effect of Yen1p on the frequency of UV-induced exchange is consistent with the observation of Lopes et al. that UV blocks replication forks without causing extensive fork regression. Our observations are also in agreement with other studies [15,38] that show that Mus81p is more important than Yen1p for the generation of crossovers in yeast. The role of Yen1p may be somewhat lesion-specific, however, since loss of Yen1p reduces the frequency of sister chromatid recombination of DSBs generated by replication of nicked template [39]. We also cannot rule out the possibility that the Yen1p is not present during the time of replication fork blockage [40].
Mechanisms to bypass unrepaired pyrimidine dimers that do not involve inter-homolog recombination
In addition to Mus81p-mediated resolution of a blocked replication fork producing a DSB, several other pathways allowing bypass of unexcised dimers exist: an error-prone pathway utilizing TLS polymerases and an error-free pathway that likely involves template switching between the arms of the replication fork [18]. Lopes et al. [11] showed that long single-stranded regions accumulate at replication forks in UV-treated yeast cells, and suggested that these events reflecting an uncoupling of leading and lagging strand replication. Both nicks and gaps are recombinogenic [34,41], and Giannattasio et al. [42] suggested that most template switching events were initiated with single-stranded gaps. Error-free template switching requires Mms2p, which catalyzes the polyubiquitination of PCNA, and Rad5p, a protein that mediates D-loop extension [43,44]. The error-prone TLS pathway is in competition with the error-free pathway. Elimination of the TLS pathway results in elevated frequencies of UV-induced sister-chromatid exchange [34].
In order to determine whether mutations in the error-free pathway contributed to LOH events between homologs, we examined rad14 mms2 diploid strains. The double mutant strains had significantly elevated frequencies of inter-homolog recombination relative to the single mutant rad14 strain (Table 4). Our preferred interpretation of this result is that Mms2p primarily promotes recombination between the arms of the replication fork and/or between sister chromatids rather than between homologs. Thus, a reduction in the frequency of recombination between sisters would result in an elevation in the frequency of inter-homolog interactions. This observation, as well as those of others, are consistent with a model in which Mus81p is primarily involved in producing DSBs by resolution of blocked replication forks, whereas Mms2p is mainly concerned with promoting recombination intermediates that involve a single-stranded gap.
In conclusion, unexcised UV dimers are a potent inducer of inter-homolog recombination in yeast. About 60% of this type of recombination is dependent on the Mus81p resolvase. The error-free PRR pathway mediated by Mms2p suppresses recombination between homologs, likely by channeling replication-blocking lesions into recombination between sister chromatids instead of homologs. These conclusions are summarized in Fig. 5.
Fig. 5. Recombination events induced by persistent UV damage in NER-deficient yeast strains. 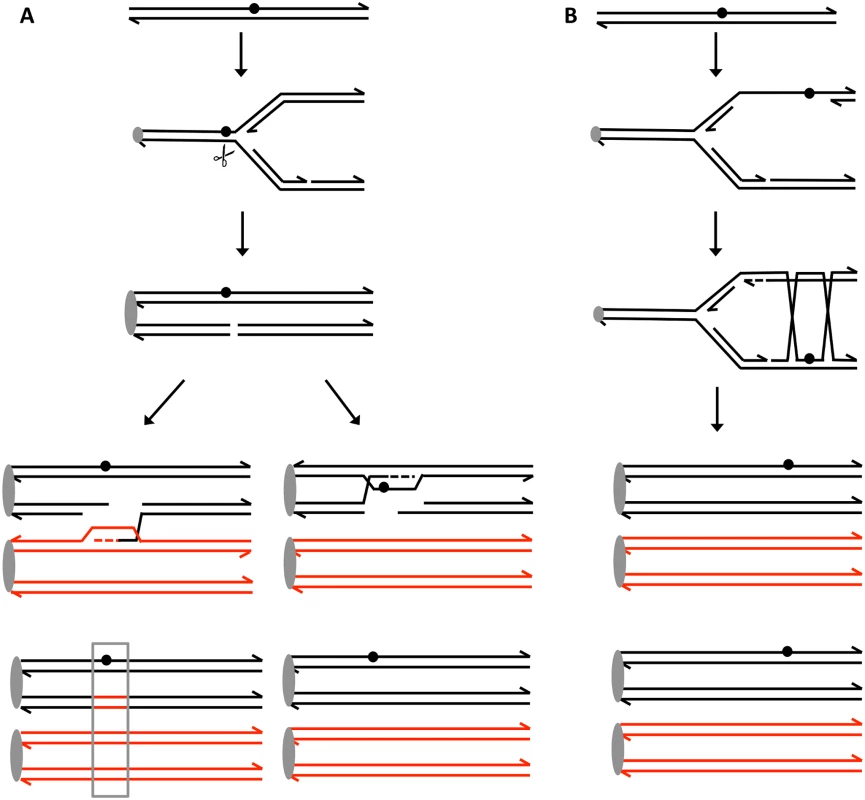
UV treatment results in the formation of a pyrimidine dimer (circle). A. Generation of a recombinogenic DSB. We suggest that the dimer-associated replication fork block can be converted into a DSB by the structure-nuclease Mus81p. The resulting DSB can be repaired either by recombination with the homolog, resulting in a detectable LOH event (boxed in grey), or by recombination with the sister chromatid which does not lead to LOH. B. Error-free bypass of lesions. When the 3’ end on the leading strand encounters a dimer, it can invade the sister chromatid (template switch), and continue replication. This branch of the post-replication repair requires Mms2p. We think that this pathway mainly involves interaction between sister chromatids without causing LOH and suppresses recombination between homologs. Materials and Methods
Yeast strains
All strains are diploids formed by mating two sequence-diverged haploids W303–1A [45] and YJM789 [46]. The wild-type diploid (PG311) used in our analysis was described previously [21], and has the genotype: MATa/MATα::natMX4 ade2–1/ade2–1 can1–100/can1::SUP4-o GAL2/gal2 his3–11,15/HIS3 leu2–3,112/LEU2 RAD5/RAD5 trp1–1/TRP1 ura3–1/ura3 V9229::hphMX4/V9229 V261553::LEU2/V261553). The disruption of the MATα locus prevents sporulation of the diploid, and allows synchronization of the diploid in G1 using α pheromone. As described in the text, a crossover between the heterozygous SUP4-o marker and CEN5 results in a red/white sectored colonies. The constructions of isogenic strains with homozygous mutations in RAD14, MUS81, YEN1, MMS2, and various double mutant strains are described in S17 Table.
In experiments to look at the rate of spontaneous crossovers, we used strains isogenic with JSC25 [20] in which the SUP4-o marker is inserted near the right telomere of chromosome IV. The genotype of JSC25 is: MATa/MATα::hphMX4 ade2–1/ade2–1 can1–100::natMX4/CAN1::natMX4 GAL2/gal2 his3–11,15/HIS3 leu2–3,112/LEU2 RAD5/RAD5 trp1–1/TRP1 ura3–1/ura3 IV1510386::kanMX4-can1–100/IV1510386::SUP4-o). Mutant strains isogenic to JSC25 with homozygous mutations in RAD14, MU81, and YEN1 were constructed as described in S17 Table.
Media and genetic methods
Standard media, genetic methods (mating, sporulation, transformation, and dissection), and DNA isolation procedures [47] were used unless otherwise indicated.
Cell synchronization, and detection of spontaneous and UV-induced crossovers
All the UV experiments were done with cells synchronized in G1 with α pheromone as described previously [22]. After two hours of treatment with α pheromone, the cells were diluted, and plated on MAB6 solid medium (omission medium lacking arginine and containing 10 μg/ml adenine). The cells were immediately treated with 1 J/m2 or 15 J/m2 of UV using the TL-2000 UV Translinker. The plates were then wrapped in foil to prevent photoreversal of pyrimidine dimers, and incubated at 30°C. for two days. The plates were then incubated overnight at 4°C. This incubation period results in an intensification of the red color in red/white sectored colonies. To examine spontaneous crossovers, we allowed cells to form colonies by growing them on rich growth medium (YPD) for two days. Colonies were then diluted into water, plated onto MAB6 solid medium, and incubated for three days at room temperature. The plates were incubated at 4°C overnight as described above.
SNP microarray analysis
The methods used for microarray analysis have been described in detail previously [3]. DNA from sectors or single colonies was isolated and sonicated using standard protocols. Experimental and control DNA samples were labeled with Cy5-dUTP and Cy3-dUTP, respectively, and hybridized in competition to Agilent-constructed SNP Microarrays. The whole-genome microarray was used to analyze all the UV-induced sectors and single colonies. The whole-genome microarray contains oligonucleotides that detect LOH for about 13,000 SNPs that differ between W303–1A and YJM789. Each SNP is represented by four 25-base oligonucleotides: two with sequences of the Watson and Crick strands of the W303–1A allele and two with sequences of the YJM789 allele. Following hybridization of the samples to the microarray, we scanned the array with GenePix scanner and GenePix Pro software. In-house R scripts were used for analyzing the microarrays. Based on the relative median hybridization ratio between experimental strain and control strain, which is heterozygous for all the SNPs, we determined which the SNPs were heterozygous or homozygous.
Statistical analyses
The data were analyzed using chi-square, Mann-Whitney, or Fisher exact tests. Tests were performed with the VassarStat Website (http://vassarstats.net/), Excel, and R functions. Corrections for multiple comparisons were done using the method of Hochberg and Benjamini [48].
Supporting Information
Zdroje
1. Friedberg EC, Walker GC, Wolfram S, Wood RD, Schultz RA, et al. (2006) DNA repair and mutagenesis; Friedberg EC, editor. Washington, D.C.: ASM Press.
2. Galli A, Schiestl RH (1999) Cell division transforms mutagenic lesions into deletion-recombinagenic lesions in yeast cells. Mutat Res 429 : 13–26. 10434021
3. St Charles J, Hazkani-Covo E, Yin Y, Andersen SL, Dietrich FS, et al. (2012) High-resolution genome-wide analysis of irradiated (UV and gamma-rays) diploid yeast cells reveals a high frequency of genomic loss of heterozygosity (LOH) events. Genetics 190 : 1267–1284. doi: 10.1534/genetics.111.137927 22267500
4. Yin Y, Petes TD (2013) Genome-wide high-resolution mapping of UV-induced mitotic recombination events in Saccharomyces cerevisiae. PLoS Genet 9: e1003894. doi: 10.1371/journal.pgen.1003894 24204306
5. Nakai S, Mortimer RK (1969) Studies on the genetic mechanism of radiation-induced mitotic segregation in yeast. Mol Gen Genet 103 : 329–338. 5803902
6. Kadyk LC, Hartwell LH (1993) Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics 133 : 469–487. 8454200
7. Mazon G, Lam AF, Ho CK, Kupiec M, Symington LS (2012) The Rad1-Rad10 nuclease promotes chromosome translocations between dispersed repeats. Nat Struct Mol Biol 19 : 964–971. doi: 10.1038/nsmb.2359 22885325
8. Schwartz EK, Heyer WD (2011) Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120 : 109–127. doi: 10.1007/s00412-010-0304-7 21369956
9. Guzder SN, Sung P, Prakash L, Prakash S (1993) Yeast DNA-repair gene RAD14 encodes a zinc metalloprotein with affinity for ultraviolet-damaged DNA. Proc Natl Acad Sci U S A 90 : 5433–5437. 8516285
10. Courcelle J, Hanawalt PC (2003) RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet 37 : 611–646. 14616075
11. Lopes M, Foiani M, Sogo JM (2006) Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21 : 15–27. 16387650
12. Mitchel K, Zhang H, Welz-Voegele C, Jinks-Robertson S (2010) Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol Cell 38 : 211–222. doi: 10.1016/j.molcel.2010.02.028 20417600
13. Donnianni RA, Symington LS (2013) Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci U S A 110 : 13475–13480. doi: 10.1073/pnas.1309800110 23898170
14. Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, et al. (2013) Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502 : 389–392. doi: 10.1038/nature12584 24025772
15. Ho CK, Mazon G, Lam AF, Symington LS (2010) Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell 40 : 988–1000. doi: 10.1016/j.molcel.2010.11.016 21172663
16. Snow R (1968) Recombination in ultraviolet-sensitive strains of Saccharomyces cerevisiae. Mutat Res 6 : 409–418. 5728832
17. Sekelsky JJ, McKim KS, Chin GM, Hawley RS (1995) The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141 : 619–627. 8647398
18. Boiteux S, Jinks-Robertson S (2013) DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 193 : 1025–1064. doi: 10.1534/genetics.112.145219 23547164
19. Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419 : 135–141. 12226657
20. St Charles J, Petes TD (2013) High-resolution mapping of spontaneous mitotic recombination hotspots on the 1.1 Mb arm of yeast chromosome IV. PLoS Genet 9: e1003434. doi: 10.1371/journal.pgen.1003434 23593029
21. Lee PS, Greenwell PW, Dominska M, Gawel M, Hamilton M, et al. (2009) A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet 5: e1000410. doi: 10.1371/journal.pgen.1000410 19282969
22. Lee PS, Petes TD (2010) From the Cover: mitotic gene conversion events induced in G1-synchronized yeast cells by gamma rays are similar to spontaneous conversion events. Proc Natl Acad Sci U S A 107 : 7383–7388. doi: 10.1073/pnas.1001940107 20231456
23. Chua P, Jinks-Robertson S (1991) Segregation of recombinant chromatids following mitotic crossing over in yeast. Genetics 129 : 359–369. 1660426
24. Barbera MA, Petes TD (2006) Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 103 : 12819–12824. 16908833
25. Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, et al. (2010) Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell 39 : 595–605. doi: 10.1016/j.molcel.2010.07.024 20797631
26. Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR 3rd, et al. (2001) Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107 : 537–548. 11719193
27. Interthal H, Heyer WD (2000) MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV - and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet 263 : 812–827. 10905349
28. Song W, Dominska M, Greenwell PW, Petes TD (2014) Genome-wide high-resolution mapping of chromosome fragile sites in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A.
29. Fishman-Lobell J, Haber JE (1992) Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258 : 480–484. 1411547
30. McCulley JL, Petes TD (2010) Chromosome rearrangements and aneuploidy in yeast strains lacking both Tel1p and Mec1p reflect deficiencies in two different mechanisms. Proc Natl Acad Sci U S A 107 : 11465–11470. doi: 10.1073/pnas.1006281107 20534547
31. Lam LH, Reynolds RJ (1987) DNA sequence dependence of closely opposed cyclobutyl pyrimidine dimers induced by UV radiation. Mutat Res 178 : 167–176. 3587252
32. Covo S, Ma W, Westmoreland JW, Gordenin DA, Resnick MA (2012) Understanding the origins of UV-induced recombination through manipulation of sister chromatid cohesion. Cell Cycle 11 : 3937–3944. doi: 10.4161/cc.21945 22987150
33. Yin Y, Petes TD (2014) The Role of Exo1p Exonuclease in DNA End-Resection To Generate Gene Conversion Tracts in Saccharomyces cerevisiae. Genetics.
34. Ma W, Westmoreland JW, Resnick MA (2013) Homologous recombination rescues ssDNA gaps generated by nucleotide excision repair and reduced translesion DNA synthesis in yeast G2 cells. Proc Natl Acad Sci U S A 110: E2895–2904. doi: 10.1073/pnas.1301676110 23858457
35. Ying S, Minocherhomji S, Chan KL, Palmai-Pallag T, Chu WK, et al. (2013) MUS81 promotes common fragile site expression. Nat Cell Biol 15 : 1001–1007. doi: 10.1038/ncb2773 23811685
36. Regairaz M, Zhang YW, Fu H, Agama KK, Tata N, et al. (2011) Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J Cell Biol 195 : 739–749. doi: 10.1083/jcb.201104003 22123861
37. Andersen SL, Kuo HK, Savukoski D, Brodsky MH, Sekelsky J (2011) Three structure-selective endonucleases are essential in the absence of BLM helicase in Drosophila. PLoS Genet 7: e1002315. doi: 10.1371/journal.pgen.1002315 22022278
38. Ashton TM, Mankouri HW, Heidenblut A, McHugh PJ, Hickson ID (2011) Pathways for Holliday junction processing during homologous recombination in Saccharomyces cerevisiae. Mol Cell Biol 31 : 1921–1933. doi: 10.1128/MCB.01130-10 21343337
39. Munoz-Galvan S, Tous C, Blanco MG, Schwartz EK, Ehmsen KT, et al. (2012) Distinct roles of Mus81, Yen1, Slx1-Slx4, and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol Cell Biol 32 : 1592–1603. doi: 10.1128/MCB.00111-12 22354996
40. Blanco MG, Matos J, West SC (2014) Dual control of Yen1 nuclease activity and cellular localization by Cdk and Cdc14 prevents genome instability. Mol Cell 54 : 94–106. doi: 10.1016/j.molcel.2014.02.011 24631285
41. Katz SS, Gimble FS, Storici F (2014) To nick or not to nick: comparison of I-SceI single - and double-strand break-induced recombination in yeast and human cells. PLoS One 9: e88840. doi: 10.1371/journal.pone.0088840 24558436
42. Giannattasio M, Zwicky K, Follonier C, Foiani M, Lopes M, et al. (2014) Visualization of recombination-mediated damage bypass by template switching. Nat Struct Mol Biol 21 : 884–892. doi: 10.1038/nsmb.2888 25195051
43. Daigaku Y, Davies AA, Ulrich HD (2010) Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature 465 : 951–955. doi: 10.1038/nature09097 20453836
44. Torres-Ramos CA, Prakash S, Prakash L (2002) Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol 22 : 2419–2426. 11884624
45. Thomas BJ, Rothstein R (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56 : 619–630. 2645056
46. Wei W, McCusker JH, Hyman RW, Jones T, Ning Y, et al. (2007) Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc Natl Acad Sci U S A 104 : 12825–12830. 17652520
47. Guthrie C, Fink GR (2002) Guide to yeast genetics and molecular and cell biology, Part C. Methods in enzymology v 351. Amsterdam; Boston: Academic Press,. pp. 1 online resource (xxxvi, 735 p.
48. Hochberg Y, Benjamini Y (1990) More powerful procedures for multiple significance testing. Stat Med 9 : 811–818. 2218183
Štítky
Genetika Reprodukčná medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



