-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
All cells require ribosomes, the cellular factories that produce proteins. A host of factors combine to produce the multiple complex units of a ribosome, many of which are still not well understood. Surprisingly, despite the ubiquitous requirement for ribosomes, defects in various ribosome biogenesis factors cause distinct and tissue specific phenotypes, collectively known as ribosomopathies. We examined the role of one ribosome biogenesis factor, Nol11, during embryonic development to determine if it too had a tissue specific phenotype. Here we show that expression of nol11 is strongly associated with the developing head in vertebrates and that insufficient Nol11 results in striking malformations of the craniofacial skeleton. We further show that reduced Nol11 impairs critical early steps in ribosome production, which triggers apoptosis within cell populations that contribute to the head. Increased cell death is at least partially the cause of the Nol11 craniofacial defect; reducing cell death rescues some of the craniofacial phenotype but not the ribosome biogenesis defect. In summary, we demonstrate for the first time that Nol11 is required for normal development of the vertebrate head and provide novel insight into the intriguing tissue proclivity of ribosomopathies.
Published in the journal: The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in. PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005018
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005018Summary
All cells require ribosomes, the cellular factories that produce proteins. A host of factors combine to produce the multiple complex units of a ribosome, many of which are still not well understood. Surprisingly, despite the ubiquitous requirement for ribosomes, defects in various ribosome biogenesis factors cause distinct and tissue specific phenotypes, collectively known as ribosomopathies. We examined the role of one ribosome biogenesis factor, Nol11, during embryonic development to determine if it too had a tissue specific phenotype. Here we show that expression of nol11 is strongly associated with the developing head in vertebrates and that insufficient Nol11 results in striking malformations of the craniofacial skeleton. We further show that reduced Nol11 impairs critical early steps in ribosome production, which triggers apoptosis within cell populations that contribute to the head. Increased cell death is at least partially the cause of the Nol11 craniofacial defect; reducing cell death rescues some of the craniofacial phenotype but not the ribosome biogenesis defect. In summary, we demonstrate for the first time that Nol11 is required for normal development of the vertebrate head and provide novel insight into the intriguing tissue proclivity of ribosomopathies.
Introduction
The synthesis of ribosomes, the protein-manufacturing entities in cells, is fundamental to all of life. Therefore, it is surprising that defects in ribosome synthesis are compatible with the development of a multicellular organism, albeit with substantial morbidity. Several diseases of ribosome biogenesis, so-called ribosomopathies, have been described with a strikingly heterogeneous collection of symptomatologies [1–16]. The remarkable diversity of affected tissues across the ribosomopathies raises the question of how different cell types can be selectively affected by defects in the ubiquitous process of making ribosomes.
An emerging class of ribosomopathies includes craniofacial anomalies. Craniofacial malformations are associated with over 700 human congenital syndromes, and the molecular causes of many of these diseases remain unknown [17]. Often the structures affected in craniofacial syndromes are derivatives of the cranial neural crest (CNC), a vertebrate specific, multipotent cell population that gives rise to a vast array of craniofacial tissues, including the majority of the craniofacial skeleton [6,17–24]. Correct development of the CNC is a multistep process that includes specification at the border of the embryonic neural and non-neural ectoderm, delamination, migration to the facial primordia, proliferation and differentiation into normal craniofacial structure and morphology. This elaborate process requires extensive cellular modifications, and is carefully regulated at the molecular level [25–34]. Due to the clinical importance of the CNC, and their potential for regenerative medicine based treatment strategies, a detailed understanding of the control mechanisms underlying CNC development is a primary goal of craniofacial research.
Investigations into the surprising relationship between craniofacial disease and defects in ribosome biogenesis are beginning to emerge. Defects in CNC derived craniofacial structures are associated with the ribosomopathy known as Treacher-Collins Syndrome [6,15,16,21]. In Treacher-Collins syndrome patients, mutations in the TCOF, POLR1C or POLR1D genes result in impaired rDNA transcription and activation of the p53 mediated stress response in CNC cells [5,6,16]. Heterozygote mouse models of Treacher-Collins syndrome in certain genetic backgrounds recapitulate these craniofacial defects, which are dependent on p53 activation [6,21]. These results suggest that mechanisms regulating CNC survival may be particularly sensitive to defects in ribosome production.
Ribosome biogenesis begins with the assembly of the nucleolus around ribosomal DNA (rDNA) repeats and with the transcription of the polycistronic pre-ribosomal RNA (rRNA) precursor that contains the sequences for the 18S, 5.8S and 28S rRNAs by RNA polymerase I (RNAPI). Over 200 ribosome biogenesis factors then process these rRNAs through a complex series of cleavages and modifications to assemble into the mature small (40S) and large (60S) subunits of the ribosome by incorporating 80 ribosomal proteins and the 5S rRNA [35–38]. Maturation of the 18S rRNA is mediated by a large ribonucleoprotein called the small subunit (SSU) processome [39–41]. Previous work in yeast has revealed that members of the t-Utp/UtpA sub-complex of the SSU (Utp4, Utp5, Utp8, Utp9, Utp10, Utp15 and Utp17) are required for both optimal transcription and processing of the pre-rRNA [42]. Likewise, the orthologs of most of these proteins are also required for pre-rRNA processing and transcription in human cells. The one exception is human Utp4, which is required for pre-rRNA processing but not for RNAPI transcription [40]. Mutation of Wdr43, the zebrafish ortholog of yeast Utp5, results in p53 mediated craniofacial malformations [42,43]. Interestingly, while the majority of t-UTP/UTPA sub-complex proteins are conserved from yeast to humans, Utp8 and Utp9 constitute notable exceptions, suggesting that these t-Utp proteins may have evolved specific functions in vertebrates [40].
We recently discovered the human analog of yeast Utp8 to be NOL11, a metazoan-specific nucleolar protein [44,45]. NOL11 was discovered by its interaction with the C-terminal region of the known t-UTP/UTPA subcomplex member, hUTP4/Cirhin, the protein mutated in North American Indian childhood cirrhosis (NAIC) [4,44]. NOL11 is localized to the nucleolus, the site of ribosome biogenesis, where it associates with other proteins in the SSU processome to facilitate maturation of the small ribosomal subunit (40S). The NAIC mutation in hUTP4/Cirhin reduces interaction with NOL11, implicating NOL11 in the pathogenesis of NAIC. NOL11 is required to maintain nucleolar number and for pre-rRNA transcription and processing in human tissue culture cells but its requirement in embryogenesis remains unknown [44].
Here, we used the frog Xenopus tropicalis to investigate its role in vertebrate development. We report that nol11 mRNA expression is strongly associated with the developing CNC in both amphibians and mice. We further demonstrate that nol11 is required for optimal pre-rRNA transcription and pre-rRNA processing in Xenopus, and that nol11 knockdown results in activation of the nucleolar stress response, p53 stabilization, and a dramatic craniofacial phenotype. Importantly, the craniofacial defect can be partially rescued by p53 depletion, while the underlying transcriptional defect cannot, demonstrating that the pathology is due to the cellular response to impaired ribosome biogenesis. Together, our findings reveal a novel requirement for Nol11 in craniofacial development and provide further insight into the evolutionarily conserved relationship between specific ribosome biogenesis factors and the control mechanisms that regulate CNC survival and development.
Results
Tissue specific expression of Nol11 during vertebrate development
As a first step in our analysis, we examined nol11 mRNA expression during vertebrate embryonic development. Intriguingly, despite the ubiquitous need for ribosomes, in situ hybridization of labelled anti-sense probes revealed robust, tissue specific expression of nol11 mRNA in both the frog and mouse. In Xenopus, transcripts are detected throughout the forming neural tube, and signal is particularly strong in anterior regions, from which the CNC form, at stages 14–16 (Fig. 1). This association of nol11 expression and CNC development is maintained at later stages of development (stages 22–36), where signal marks the CNC streams as they migrate into the facial primordia and differentiate. During these stages, expression in the neural tube becomes more restricted and is detected in regions of the hindbrain, isthmus and tail. Additional stage specific expression is lightly detected in the optic and otic placodes, the ventral blood islands, and diffusely in the presumptive splenic mesenchyme (Fig. 1).
Fig. 1. Expression of nol11 during vertebrate development. 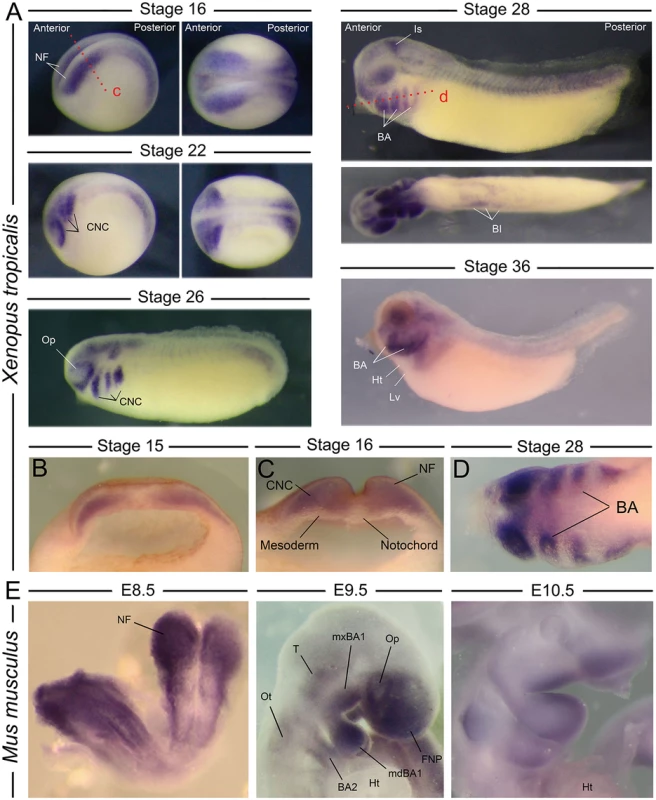
A) Wild type nol11 expression pattern during Xenopus tropicalis development. Note the strong expression in developing neural folds (NF) and the presumptive CNC at stages 16 and 22 (lateral [left] and dorsal [right] views presented). Expression is strongly associated with the migrating and differentiating CNC at subsequent stages, and is also detected in the region of the ventral blood islands (BI) and isthmus (Is) at stage 28. BA, branchial arch; Ht, heart; Lv, liver region; Op, optic placode. B) and C) Anterior transverse dissections showing expression of nol11 in neural folds and premigratory CNC of stage 14 and 16 embryos respectively. Plane of dissection is represented by the red dotted line marked c in A. D) Horizontal dissection (shown as dotted red line marked d in A) of nol11 expression in the branchial arches of stage 28 Xenopus embryos. E) nol11 expression in E8.5, E9.5 and E10.5 wild type mouse embryos. At E8.5 expression is strongly detected in the neural folds. Transcripts are associated with CNC positive regions at both E9.5 and E10.5. BA2, 2nd branchial arch; FNP, frontonasal prominence; Ht, heart; mdBA1, mandibular BA1; mxBA1, maxillary BA1; Op, optic placode; Ot, otic placode; T, trigeminal region. Nol11 RNA is similarly expressed in the mouse CNC. At stage E8.5, murine Nol11 transcripts are detected strongly in the anterior and posterior neural folds and presumptive CNC regions. At stages E9.5 and E10.5 expression is largely restricted to CNC regions, including the frontonasal prominence (FNP), the mandibular and maxillary portions of the first branchial arch (mdBA1 and mxBA1) and the more posterior BA. Interestingly, in both mice and frogs, Nol11 expression appears enriched in distal regions of the branchial arches at later stages of development (Fig. 1). Murine expression was also observed in the optic placode, the trigeminal region and otic placode, closely mirroring the Xenopus expression pattern, as well as in regions of the developing gut and liver (Fig. 1, S1A Fig). Together, amphibian and murine expression suggests an evolutionary conserved relationship between the ribosome biogenesis gene nol11 and development of the CNC.
Xenopus nol11 is required for normal CNC development and formation of the craniofacial skeleton
To test if nol11 is required for CNC development, we employed morpholino oligo (MO) mediated knockdown in Xenopus tropicalis. In Xenopus, MOs can be injected at the one cell stage targeting the entire embryo or at the two-cell stage where embryos targeted to either just the right or left side can be selected. To test the efficacy of our knockdown, we examined Nol11 protein levels in uninjected controls (UC) and nol11 morphants by western blot, and confirmed a dose dependent impact on Nol11 protein levels during development (S2 Fig). Knockdown of nol11 resulted in microcephaly and pronounced defects in CNC derived craniofacial cartilages in 86% (n = 389) of treated embryos, while control MO (CMO) injected embryos were unaltered (Fig. 2A and 2C). Cartilaginous defects included significantly reduced size and dysmorphology of the Meckel’s, quadrate, ceratohyal and branchial cartilages at stage 45, (Fig. 2A and 2B). The severity of these defects varied from a 27% to 55% reduction in cartilage size, with an average reduction of 42% (Fig. 2B). These cartilages are derivatives of distinct branchial arches (BAs) and neural crest migratory streams, and their loss demonstrates a central role for nol11 in CNC development, independent of axial level of origin. nol11 morphants did not survive beyond stage 45–47, probably due to a non-functional jaw apparatus and a reduced ability to feed. To test the specificity of the nol11 MO, we injected human NOL11 mRNA into one cell of two cell stage nol11 morphants and compared the two sides. NOL11 mRNA rescued both cartilage size and morphology in 74% of embryos (Fig. 2C).
Fig. 2. The nol11 craniofacial phenotype. 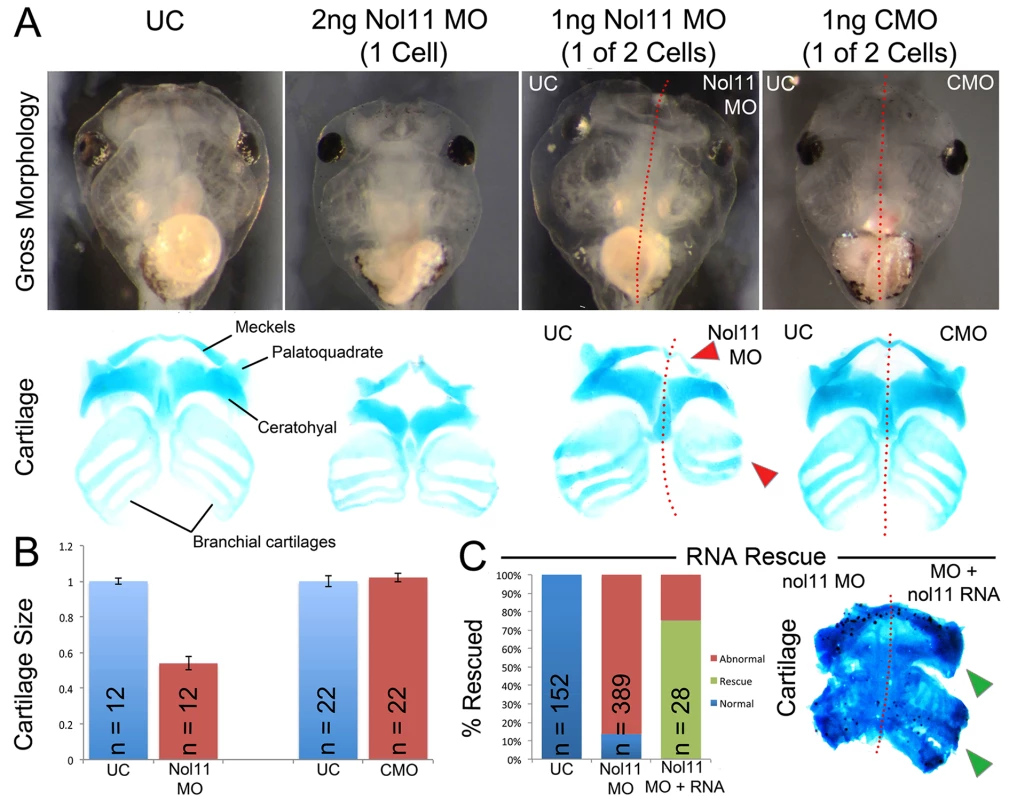
A) Gross morphology and cartilage staining of UC, nol11 whole embryo, nol11 one-sided knockdowns and CMO one-sided knockdown embryos. Note the reduced cartilage size and abnormal morphology in nol11 morphants (red arrowheads) while CMO injected embryos are unaffected. B) Craniofacial cartilage size is significantly reduced in nol11 but not CMO morphants. C) Co-injection of human NOL11 RNA can rescue the cartilage phenotype in approximately 75% of treated embryos. Cartilage staining of an RNA rescued embryo; nol11 MO was injected at the one cell stage and human NOL11 RNA was injected into one cell at the two cell stage. Green arrowheads highlight rescued side. To probe the ontogeny of these cartilaginous defects, we next examined development of the CNC in nol11 morphants. We found that initial specification of dorsal territories and neural tissue proceeds normally in nol11 morphants, as assayed by expression of the pan-neural marker sox3, the exclusion of cytokeratin expression from neural tissue, and expression of pax2 and myod in the developing neural tube and somites respectively at stage 14/15 [46–48] (Fig. 3, S3 Fig). With the exception of a slight delay, expression of the CNC master genes twist, slug, ap2 and sox9 [49–51] also appeared normal relative to controls at stages 14 and 24, suggesting that induction, delamination and migration of the CNC occurs in a largely normal fashion following nol11 knockdown (Fig. 3, S3A Fig). However, by stage 28, defects in CNC markers are readily apparent within the developing BAs. These include striking reductions in sox9 expression (a CNC gene required for cartilage differentiation), as well as CNC patterning genes including gsc, dlx5, twist, dlx2 and xnot (Fig. 3, S3 Fig). Importantly, the branchial arches were also observed to be smaller on the treated side of the embryo, suggesting a reduced number of CNC cells (Fig. 3, S3 Fig). While the medio-lateral size of the neural tube appeared slightly reduced in morphants at stage 28, patterning of the neural tube and brain appeared grossly intact, as assayed by expression of pax2, hoxb3, and sox3 (S3B Fig).
Fig. 3. Knockdown of nol11 disrupts cranial neural crest development. 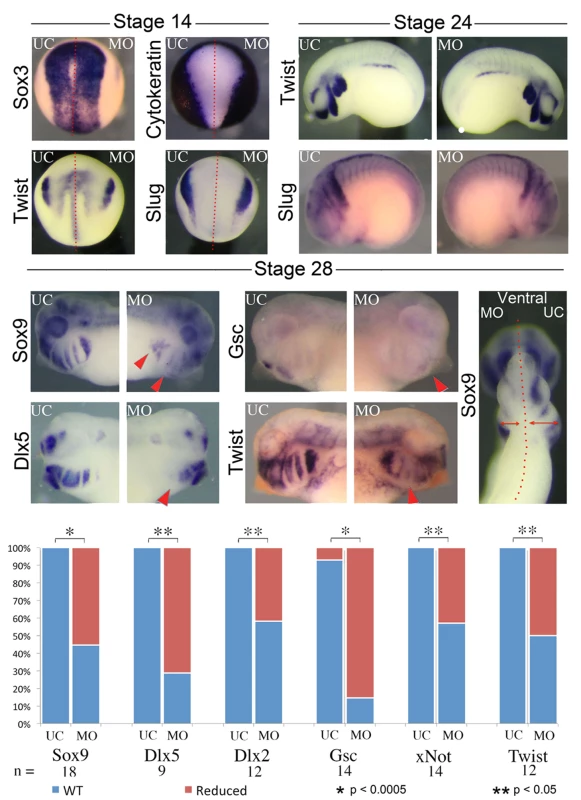
At stages 14 and 24 neural and CNC development appears normal in one side treated embryos, as assayed by expression of key marker genes including sox3, cytokeratin, twist and slug. By stage 28 however, reductions are apparent in the expression of numerous CNC genes. The branchial arches are also smaller on the MO treated side at stage 28. Graph displays number of embryos exhibiting abnormal gene expression in control and nol11 morphant embryos. To test if patterning defects were selective for the CNC, we assayed various markers for different organs testing cardiac, gut and kidney development. Both cardiac looping and pitx2 expression were normal in morphants (S1B, D Fig), demonstrating that the strong nol11 expression observed in the embryonic left-right organizer is not required for normal organ situs [52]. Gut morphology was abnormal in morphants at stage 45. We also examined expression of nkx2.5, pax2, sglt1, smp30 and hex in stage 28–36 nol11 morphants but found no marked changes (S1C, E, F Fig). Interestingly while expression of the cardiac and splenic patterning gene nkx2.5 [53,54] appeared normally positioned in morphants, its splenic expression appeared moderately reduced, suggesting a possible role for nol11 in spleen development (S1C Fig). In future studies, it will be interesting to examine late stage development of additional organs.
The Nol11 craniofacial defect is associated with increased apoptosis
As mutations in ribosomal biogenesis factors have previously been associated with altered cell survival and since the reduced size of the BAs in nol11 morphants suggests reduced cell numbers, we next examined rates of apoptosis and proliferation in the facial primordia of nol11 depleted embryos. At early stages of development, no significant change was observed in apoptosis rates as assayed by TUNEL staining and p53 protein levels (Fig. 4A). However by stages 18 through 28, as the CNC migrate into and become patterned within the facial primordia, we observed a dramatic 3–4 fold increase in the number of TUNEL stained cells (Fig. 4A, S4 Fig). This increase correlates well with the observed onset of the cartilage phenotype. Examination of TUNEL staining in paraffin sections revealed the increased apoptosis to be largely confined to ectomesenchymal cells located with the facial primordia, possibly CNC (Fig. 4A). Western blot analysis of p53 protein levels in morphants revealed approximately normal protein levels at early time points (st12–14) but a progressive increase over subsequent stages, culminating in an approximately 4-fold increase between stages 18–28. These findings mirror the TUNEL results and confirm a stage dependent increase in apoptosis (Fig. 4A, Fig. 5D). Interestingly, in contrast to observations in the zebrafish Wdr43 mutant, nol11 morphants did not have a significant change in rates of proliferation (Fig. 4B).
Fig. 4. Increased apoptosis underlies the nol11 cartilage defects. 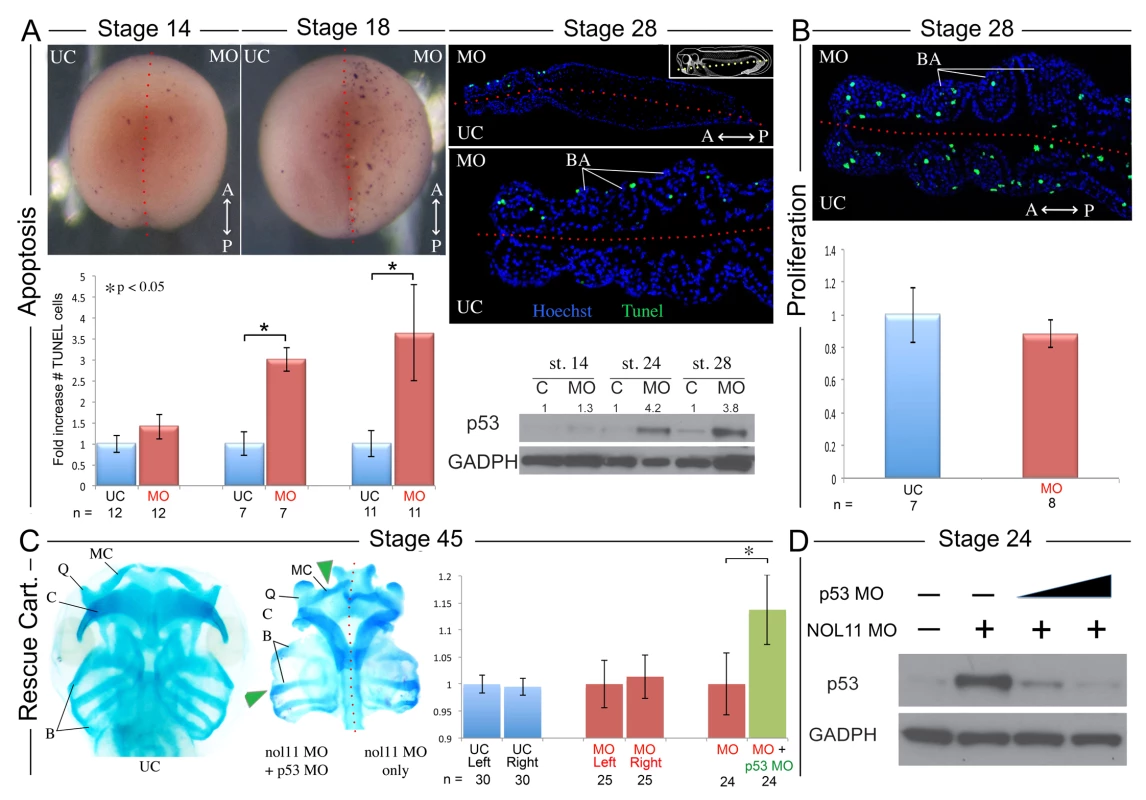
A) nol11 knockdown results in a progressive increase in apoptosis. At stage 14 no significant difference was observed in rates of TUNEL staining between knockdown and control halves of the embryo. At stages 18 and 28 increased apoptosis was evident on the treated side of whole mount and sectioned paraffin embedded embryos. Note that this increased apoptosis occurs primarily within the craniofacial ectomesenchyme. The graph represents the relative quantification of apoptosis rates at stages 14, 18 and 28. This stage specific increase in apoptosis was confirmed by a similar increase in p53 protein levels in 1 cell injected embryos as assayed by western blot (lower right panel). Dotted red lines mark the embryonic midline. B) No significant change in proliferation rates was noted following nol11 knockdown. C) Inhibition of apoptosis by p53 MO results in a partial rescue of cartilage size and morphology. Each pair of columns in the graph compares cartilage size measured in bilateral halves of embryos. The blue pair reveals no significant difference in cartilage measurements in the left vs right side of the UC embryonic head. In the second pair (red), cartilage size is seen to be comparable on either side of the nol11 morphant head. The final pair illustrates that cartilage size is significantly improved on the side of nol11 morphants rescued with p53 MO (green) relative to the side that received nol11 MO only (red). D) Western blot demonstrating that the p53 MO efficiently reduces p53 protein levels in nol11 morphants. Fig. 5. Nol11 depletion impairs rDNA transcription and pre-rRNA processing in X. tropicalis. 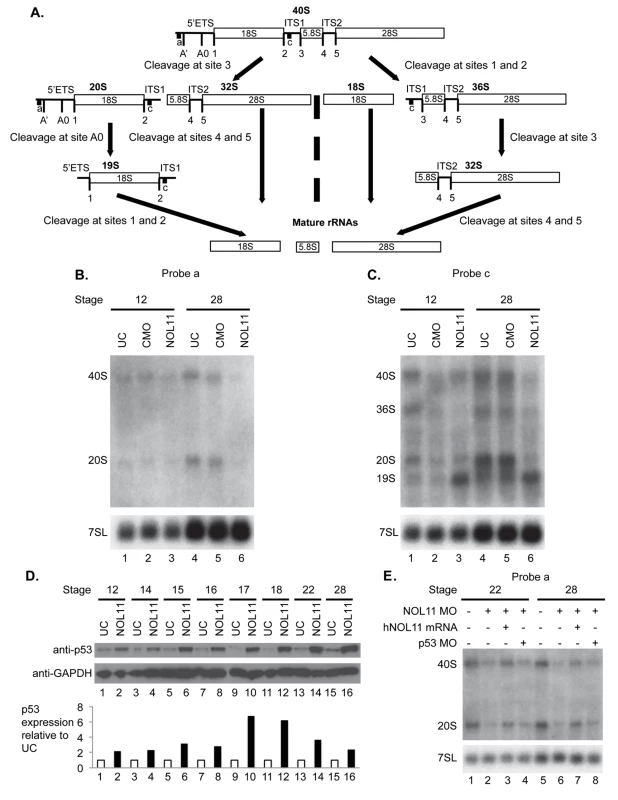
A) Scheme of pre-rRNA processing pathways in X tropicalis. The pre-rRNA is transcribed by RNAPI as a 40S polycistronic precursor. Several cleavages are required to separate the mature rRNAs. The locations of oligonucleotide probes used for northern blots are indicated by lettered lines (a, c) and the cleavage sites indicated. This scheme was adapted from [71–75]. B) Morpholino (MO) depletion of Nol11 impairs pre-rRNA transcription at stage 28. The northern blot was hybridized with probe a (Fig. 5A) and with a probe to the 7SL RNA as a loading control (lower panel). Bands were quantified and analysed by RAMP ([60]; S6A,B Fig) C) Morpholino (MO) depletion of Nol11 impairs pre-rRNA transcription and processing. The northern blot was hybridized with probe c (Fig. 5A) and with a probe to the 7SL RNA as a loading control (lower panel). Bands were quantified and analysed by RAMP ([60]; S6C,D, E, F Fig). D) Depletion of Nol11 leads to increased p53 levels. The expression of p53 from control and nol11 depleted embryos was analysed by western blot with anti-p53 antibodies. GAPDH levels were used as a loading control. Values for p53 expression normalized to GAPDH are represented in the bar graph. E) MO-resistant human NOL11 (hNOL11) mRNA but not p53 depletion rescues pre-rRNA levels. Embryos injected as shown by + and—in the figure at stages 22 and 28. The pre-rRNAs were visualized with probe a on a northern blot; hybridization to the 7SL RNA was used as a loading control. In order to confirm that this increase in apoptosis contributes to the nol11 craniofacial phenotype, we next asked if knockdown of p53 could rescue the malformation. Interestingly, injection of p53 MO alone into wild type Xenopus embryos produces a dose dependent craniofacial defect, most plausibly due to disruption of normal patterns of cell death during development or, as previously reported, apoptosis independent effects [55,56]. However, we found that by titrating the p53 knockdown we could ameloriate some of the nol11 cartilage phenotype, including improved cartilage size and morphology (Fig. 4C). The efficacy of the p53 MO in reducing p53 protein levels was demonstrated by western blot (Fig. 4D). We also found that injection of p53 MO dramatically reduced rates of cell death within the branchial arches of stage 28 nol11 morphants, while CMO had no effect (S4 Fig). Together these findings suggest that the nol11 craniofacial phenotype is at least in part caused by an increase in p53 mediated cell death within the cells that populate the facial primordia.
Nol11 is required for ribosome biogenesis in a developing embryo
Increased tissue specific apoptosis is associated with disruption of several ribosome biogenesis factors, including Tcof1, the protein that is mutated in Treacher-Collins syndrome [1,6,21,43,57]. Under conditions of defective ribosome biogenesis, this apoptotic response is triggered by the ‘nucleolar stress response’, the binding of excess ribosomal proteins, such as RPL5 and RPL11, to Mdm2, which in turn prevents Mdm2 mediated p53 degradation [58]. With this in mind, we sought to investigate whether pre-rRNA processing and transcription, and thus ribosome biogenesis are disrupted in nol11 morphants.
To develop probes for northern blots to examine ribosome biogenesis in X. tropicalis, we first identified the X. tropicalis 5’ external transcribed spacer (ETS) in the pre-rRNA using a BLAST search. Aligning X. laevis and X. borealis pre-rRNA sequences, we identified two X. tropicalis Xentr7.1 scaffolds (xenbase.org). Scaffold sequence 169 contained sequences similar to X. laevis 5’ETS, 18S rRNA, internal transcribed spacer 1 (ITS1), and the 5’ portion of the 5.8S rRNA while scaffold 2385 only contained sequence similar to the 5’ portion of the 5’ETS (S5 Fig). Using these alignments we designed oligonucleotides that were reverse complements of sequences in the 5’ETS and ITS1 of X. tropicalis since NOL11 is required for biogenesis of the mature 18S rRNA in human cells [44]
Using these oligonucleotide probes (probes a,c indicated in Fig. 5A), we investigated the effect of nol11 knockdown on ribosome biogenesis in X. tropicalis embryos by northern blot analysis (Fig. 5B and 5C). Hybridization to the 7SL SRP RNA was carried out as a loading control, as done previously [59]. Northern blots were repeated 3 times with the indicated embryos. The results were quantified and studied by Ratio Analysis of Multiple Precursors (RAMP; S5 Fig; [60]). With either probe at either stage 12 or stage 28, no significant difference was observed either by eye (Fig. 5B and 5C) or in the RAMP profile between uninjected controls (UC) and control morpholino injected embryos (CMO) (S6 Fig), indicating that injection per se does not result in defects in ribosome biogenesis as detected by northern blots.
We do, however, observe defects in transcription and pre-rRNA processing in nol11 morphants, indicating that depletion of Nol11 results in defective ribosome biogenesis. In northern blot analysis with probe a, steady state levels of both the 40S pre-rRNA primary transcript and its 20S pre-rRNA processed product were reduced in stage 28 nol11 morphants (Fig. 5B, lanes 4–6; S6B Fig). In contrast, no significant difference was detected between nol11 morphants and the controls at stage 12 (Fig. 5B, lane 1–3; S6A Fig). The marked decrease in steady state levels of both pre-rRNA transcripts in nol11 MO-treated embryos at stage 28 is consistent with a defect in pre-rRNA transcription following Nol11 knockdown.
Likewise, northern blot analysis with probe c revealed defects in transcription and pre-rRNA processing in nol11 MO-treated embryos. When the levels of the indicated pre-rRNAs were compared to the 7SL RNA loading control in nol11 morphants, a significant decrease in the 40S, 36S and 20S pre-rRNAs was detected at stage 28 (Fig. 5C, lanes 4–6; S6D Fig). This is consistent with a defect in pre-rRNA transcription. At the same time, the 19S pre-rRNA (Fig. 5A) accumulated in nol11 MO-treated embryos at stage 28 (Fig. 5C, lanes 4–6; S6D Fig), consistent with a defect in pre-RNA processing. No significant difference was detected between nol11 morphants and the controls at stage 12 when the pre-rRNA levels were compared to the 7SL RNA loading control (Fig. 5C, lane 1–3; S6C Fig). However, when the levels of the indicated pre-rRNAs were compared to each other in nol11 morphants, accumulation of the 19S pre-rRNA could be observed as early as stage 12 (Fig. 5C, lane 3; S6E Fig) and was very pronounced by stage 28 (Fig. 5C, lane 6; S6F Fig). Thus, depletion of Nol11 by injection of MO in developing X. tropicalis embryos results in defective pre-rRNA transcription and processing.
As discussed above, defects in ribosome biogenesis can lead to p53 stabilization and subsequent apoptosis due to a cellular phenomenon known as nucleolar stress [61–63]. In order to further examine the relationship between the pre-RNA transcription defect and increased p53 expression, we compared changes in both over the relevant embryonic time course. As described above, MO-depletion of nol11 caused an increasingly severe disruption of pre-RNA transcription over these developmental stages (Fig. 5B and 5C, lane 6; S6B,D Fig). We found that this correlated precisely with the progressive increase in p53 levels by western blot (Fig. 5D). Together, these findings indicate that nol11 depletion results in reduced pre-RNA transcription, impaired ribosome biogenesis and increased levels of p53, a hallmark of nucleolar stress.
To confirm that these defects are specific to nol11 depletion, we compared nol11 morphants with morphants rescued with NOL11 mRNA. Co-injection of the human NOL11 mRNA rescued the steady state levels of the pre-rRNAs in morphants (Fig. 5E, lanes 3 and 7) indicating restoration of pre-rRNA transcription and therefore ribosome biogenesis. In addition, co-depletion of p53 and Nol11 did not rescue the pre-rRNA transcription defect although we did find a partial rescue of the craniofacial phenotype (Fig. 5E, lanes 4, 8 and Fig. 4C, above).
Discussion
Ribosome biogenesis is a vital and energy intensive process, requiring the interaction of hundreds of proteins and the majority of a cell’s transcriptional output. As such, it is not surprising that it is carefully monitored, or that a control mechanism such as the nucleolar stress response exists to remove compromised cells. Indeed, impaired ribosome production is frequently associated with apoptosis. For example, Treacher-Collins syndrome is molecularly characterized by impaired ribosome biogenesis, activation of the nucleolar stress response and increased p53 levels in CNC cells [6,21]. A similar increase in apoptosis has been reported to underlie the zebrafish Wdr43 (Utp5) craniofacial phenotype [43]. Importantly, the observation that inhibition of apoptosis can rescue the cartilage phenotype but not the underlying transcriptional and processing defect in both Xenopus nol11 morphants and some mouse Treacher-Collins Syndrome models demonstrates that it is the evolutionary conserved nucleolar stress response, and not the ribosomal defect per se, that produces the craniofacial malformation. As such, further investigation and modulation of this stress response may ultimately provide novel treatment options for apoptosis induced ribosomopathies. Together with previous studies, our data provide further evidence that the CNC are particularly sensitive to insufficiency of certain ribosome biogenesis proteins and reveal a common, evolutionarily conserved control mechanism through which compromised multipotent CNC are eliminated in the embryo. This sensitivity of CNC to ribosome biogenesis defects also suggests that mutations in nucleolar factors may contribute to additional currently unexplained human congenital defects.
The finding that defects in presumably globally required ribosome biogenesis factors can produce tissue specific defects is an intriguing emerging facet of embryonic development. Several human diseases are now known to result from mutations in ribosome production and present with distinct phenotypes [1–4,6,8,9,11,14,16,44,64,65]. The mechanisms underlying the tissue specificity of these ribosome biogenesis factors remain unknown but may include differential ribosomal biogenesis factor requirements during the development of distinct tissues or a heightened sensitivity to impaired protein production in particular cell populations. While the CNC is a highly active embryonic cell population, the distinct phenotypes observed in human ribosomopathies, many of which do not include craniofacial defects, would seem to argue against the latter scenario. Furthermore, experimental support for the differential requirements hypothesis is beginning to emerge from studies of both mice and zebrafish [6,43,57,66]. Our findings that nol11 expression at the mRNA level is tightly linked to CNC cells, where it is required for normal differentiation and development of craniofacial cartilages, but not detected in, or associated with overt defects in the development of several other active cell populations at similar stages, provides strong support for the possibility of differential ribosomal protein requirements during embryonic development. While beyond the scope of this work, a future comparison of RNA translation and protein profiles between the CNC and non-CNC cells of wild-type and knockdown embryos would be highly informative.
Importantly, the role of the NOL11 protein in ribosome biogenesis is conserved between human cells and X. tropicalis embryos. In HeLa cells [44] and in developing frog embryos, depletion of NOL11 results in defects in pre-rRNA transcription and pre-18S rRNA processing. However, the precise pattern of the pre-rRNAs differs between the two species even in the non-depleted state, with the pattern in X. tropicalis similar to that found in zebrafish (D. rerio) [43,67]. Whether this is due to differences among vertebrate species or differences between tissue culture cells and a developing organism we do not yet know. The development of oligonucleotide probes to study these pre-rRNA pathways in X. tropicalis should thus facilitate the mapping of steps in pre-rRNA processing in this developing frog.
In summary, we have demonstrated a novel role for nol11 in vertebrate ribosome biogenesis, cell survival and craniofacial development. The emerging relationship between specific ribosome biogenesis factors and tissue specific developmental defects is an unexpected and intriguing feature of developmental biology, and one that may underlie numerous currently unexplained congenital syndromes. Our findings provide insight into this relationship and highlight the utility of Xenopus tropicalis as a model for further investigations of human ribosomopathies.
Materials and Methods
Ethics statement
X. tropicalis were maintained and cared for in our aquatics facility, in accordance with Yale University Institutional Animal Care and Use Committee protocols.
Embryos
Embryos were produced by in vitro fertilization and raised to appropriate stages in 1/9MR + gentamycin. Fixed wild-type mouse embryos were obtained from C. Wilson and C. Bogue.
Antisense morpholino knockdown
Antisense morpholino oligonucleotides targeting the nol11 translational start site (5’ GCTCCCCGAGAGCGGCCATCTTGTC 3’), or standard CMO from Genetools LLC, Philomath, OR) were injected at either the one cell stage (2ng MO) or into one cell at the two-cell stage (1ng MO). A full-length cDNA clone of human NOL11 was purchased from Open Biosystems and subcloned into the Gateway Entry vector pDONR221 (Invitrogen). The specificity of the MO was then tested by injecting 2ng of nol11 MO at the one cell stage and subsequently injecting 50pg of human NOL11 RNA into one cell at the two cell stage in order to rescue the phenotype on the RNA treated side. Rescue was scored by increased cartilage size and morphology.
Whole-mount in situ hybridization
Xenopus nol11 template for probe was amplified from wild type cDNA using the following primer combinations: forward: 5’ - AGTTTGGTGAGGCGCTGTAT-3’; reverse: 5’ - GGCTCATCCAATCCACTACC-3’ and then TOPO TA cloned (Life Technologies) as per manufacturer’s instructions. Digoxigenin-labeled antisense probes for ap-2, TGas030K20; cytokeratin, IMAGE:6991625; dlx2, IMAGE:6980076; dlx5, TNeu071c08; gsc, TGas129E16; hex, TGas075h17; myoD, TNeu017H11; nkx2.5, IMAGE:7517699; nol11, as described above; pax2, TNeu062i10; pitx2, TNeu083k20; sglt, IMAGE:5308256; sox9, TNeu111f21; slug, TNeu008A21; smp-30, IMAGE:6999181; twist, TNeu125e01 and xnot, TNeu017e01 were in vitro transcribed with T7 High Yield RNA Synthesis Kit (E2040S) from New England Biolabs. Embryos were collected at the desired stages, fixed in MEMFA for 1–2 hours at room temperature and dehydrated into 100% ETOH. Whole mount in situ hybridization was performed as described previously [68]. Embryos were stained with BM Purple and examined after equilibration in 100% glycerol.
Alcian Blue staining and measurements of cartilage
Stage 45 embryos were fixed in MEMFA for 20 mins at RT and then washed in acid alcohol (1.2% HCL in 70% ETOH) before staining in 0.5% alcian blue solution in acid alcohol over night at 4 degrees. Samples were then washed several times in acid alcohol and dehydrated into H2O, before bleaching for 1–2 hrs in 1.2% hydrogen peroxide. Samples were then washed several times in 2% KOH and left rocking overnight in 10% glycerol in 2% KOH. They were then processed through 20%, 40%, 60% and 80% glycerol in 2% KOH. The facial cartilages were then dissected out and imaged using a Canon EOS 5d digital camera mounted on a Zeiss discovery V8 stereomicroscope. Bilateral cartilages were then outlined using ImageJ software (NIH) and their relative sizes measured.
Western blotting
Pools of ten staged control and morphant embryos were collected and placed in 100ul of 1 x RIPA buffer. Embryos were then crushed using a pestle and spun down twice to separate protein from fat and debris. Western blots were then carried out following standard protocols, using an anti-p53 (Thermo Scientific, MA1–12549 1 : 800 dilution) or an anti-NOL11 (SIGMA HPA022010 1 : 1000 dilution) primary antibody and an anti-mouse or anti-rabbit HRP conjugated secondary (Jackson Immuno Research Laboratories, 715–035–150 or 211–032–171 1 : 15000 dilution). Anti-GAPDH (Ambion, AM4300 1 : 5000 dilution) was used as an internal control. Quantifications of changes in protein level were calculated using ImageJ software from NIH.
TUNEL and proliferation assays
Whole mount TUNEL staining of developmentally staged wild-type and nol11 morphant embryos was carried out as previously described [69]. Stage 28 embryos were also fixed in 4% paraformaldehyde, embedded in paraffin and cut into 10 μm sections. The TUNEL assay was carried out using the In situ cell death detection kit, Fluorescein (Roche Applied Science, 11684795910) as per the manufactures instructions (using the trypsin pretreatment option described). Cell proliferation assays were carried out using an anti-phospho-Histone H3 antibody (Millipore, 06–570) and an Alexa Fluor 488 Chicken Anti-Rabbit (Life Technologies, A21441).
Apoptosis rescue
In order to determine if the nol11 phenotype is due to increased apoptosis, we injected 2ng of nol11 MO at the one cell stage. A subset of these embryos were then injected with 1ng of p53 MO into one cell at the two cell stage to rescue the phenotype on the p53 MO treated side. Rescue was scored by measuring and comparing the area of cartilage present on the nol11 MO only vs the nol11 + p53 MO treated sides of the midline using ImageJ software from NIH. Similar comparisons were made between the left and right sides of uninjected and nol11 MO only embryos as controls.
Statistical analyses
Each experiment was performed a minimum of three times. Statistical significance of the frequency of CNC patterning gene disruptions was tested with a Chi-squared test. The significance of TUNEL rates, changes in cartilage size and rescue experiments was evaluated using paired or unpaired, two-tailed student’s t - tests as appropriate.
Northern blotting
X. tropicalis embryos were injected with MOs and mRNA as described. RNA was harvested from embryos by dissolving 10–15 embryos in TRIzol (Invitrogen) and total RNA was extracted per the manufacturer’s instructions. Northern blot analyses were performed as described previously [70]. Two or four μg of total RNA per sample were separated by gel electrophoresis on a 1% agarose/1.25% formaldehyde gel and then transferred to a nylon membrane (Hybond-XL, GE Healthcare). RNA species were detected by hybridization with radiolabelled oligonucleotide probes or by methylene blue staining. Oligonucleotide probes are as follows: Probe a, 5’-CAC TAA GGG TCA ACC TCT CCT T-3’; Probe c, 5’-CAG GTA CCC GGG TCG GCC TGC GGC G-3'; 7SL: 5’ - CAT ATT GAT ACC GAA CTT AGT GC-3’.
Northern blots were quantitated using a phosphorimager (Bio-Rad Personal Molecular Imager) and the levels of all RNAs were normalized to the uninjected control. Ratio Analysis of Multiple Precursors (RAMP) was performed as in [60]. Statistical analysis was performed using a 2-way ANOVA with Tukey’s multiple comparisons test for post hoc analysis of significance in GraphPad Prism.
Supporting Information
Zdroje
1. McCann KL, Baserga SJ (2013) Genetics. Mysterious ribosomopathies. Science 341 : 849–850. doi: 10.1126/science.1244156 23970686
2. Bolze A, Mahlaoui N, Byun M, Turner B, Trede N, et al. (2013) Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science 340 : 976–978. doi: 10.1126/science.1234864 23579497
3. Butterfield RJ, Stevenson TJ, Xing L, Newcomb TM, Nelson B, et al. (2014) Congenital lethal motor neuron disease with a novel defect in ribosome biogenesis. Neurology.
4. Chagnon P, Michaud J, Mitchell G, Mercier J, Marion JF, et al. (2002) A missense mutation (R565W) in cirhin (FLJ14728) in North American Indian childhood cirrhosis. Am J Hum Genet 71 : 1443–1449. 12417987
5. Dauwerse JG, Dixon J, Seland S, Ruivenkamp CA, van Haeringen A, et al. (2011) Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet 43 : 20–22. doi: 10.1038/ng.724 21131976
6. Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, et al. (2006) Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci U S A 103 : 13403–13408. 16938878
7. Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, et al. (1999) The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet 21 : 169–175. 9988267
8. Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, et al. (2008) Identification of RPS14 as a 5q - syndrome gene by RNA interference screen. Nature 451 : 335–339. doi: 10.1038/nature06494 18202658
9. Freed EF, Baserga SJ (2010) The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis. Nucleic Acids Res 38 : 4798–4806. doi: 10.1093/nar/gkq185 20385600
10. Freed EF, Bleichert F, Dutca LM, Baserga SJ (2010) When ribosomes go bad: diseases of ribosome biogenesis. Mol Biosyst 6 : 481–493. doi: 10.1039/b919670f 20174677
11. Gazda HT, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, et al. (2008) Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet 83 : 769–780. doi: 10.1016/j.ajhg.2008.11.004 19061985
12. Hannan KM, Sanij E, Rothblum LI, Hannan RD, Pearson RB (2013) Dysregulation of RNA polymerase I transcription during disease. Biochim Biophys Acta 1829 : 342–360. doi: 10.1016/j.bbagrm.2012.10.014 23153826
13. Narla A, Ebert BL (2010) Ribosomopathies: human disorders of ribosome dysfunction. Blood 115 : 3196–3205. doi: 10.1182/blood-2009-10-178129 20194897
14. Teng T, Thomas G, Mercer CA (2013) Growth control and ribosomopathies. Curr Opin Genet Dev 23 : 63–71. doi: 10.1016/j.gde.2013.02.001 23490481
15. Trainor PA, Merrill AE (2014) Ribosome biogenesis in skeletal development and the pathogenesis of skeletal disorders. Biochim Biophys Acta 1842 : 769–778. doi: 10.1016/j.bbadis.2013.11.010 24252615
16. Valdez BC, Henning D, So RB, Dixon J, Dixon MJ (2004) The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci U S A 101 : 10709–10714. 15249688
17. Trainor PA, Andrews BT (2013) Facial dysostoses: Etiology, pathogenesis and management. Am J Med Genet C Semin Med Genet 163 : 283–294.
18. Chai Y, Jiang X, Ito Y, Bringas P Jr., Han J, et al. (2000) Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127 : 1671–1679. 10725243
19. Dupin E, Creuzet S, Le Douarin NM (2006) The contribution of the neural crest to the vertebrate body. Adv Exp Med Biol 589 : 96–119. 17076277
20. Inman KE, Purcell P, Kume T, Trainor PA (2013) Interaction between Foxc1 and Fgf8 during mammalian jaw patterning and in the pathogenesis of syngnathia. PLoS Genet 9: e1003949. doi: 10.1371/journal.pgen.1003949 24385915
21. Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, et al. (2008) Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med 14 : 125–133. doi: 10.1038/nm1725 18246078
22. Le Lievre CS, Le Douarin NM (1975) Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J Embryol Exp Morphol 34 : 125–154. 1185098
23. Noden DM (1983) The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev Biol 96 : 144–165. 6825950
24. Olsson L, Falck P, Lopez K, Cobb J, Hanken J (2001) Cranial neural crest cells contribute to connective tissue in cranial muscles in the anuran amphibian, Bombina orientalis. Dev Biol 237 : 354–367. 11543620
25. Depew MJ, Lufkin T, Rubenstein JL (2002) Specification of jaw subdivisions by Dlx genes. Science 298 : 381–385. 12193642
26. Griffin JN, Compagnucci C, Hu D, Fish J, Klein O, et al. (2013) Fgf8 dosage determines midfacial integration and polarity within the nasal and optic capsules. Dev Biol 374 : 185–197. doi: 10.1016/j.ydbio.2012.11.014 23201021
27. Rijli FM, Mark M, Lakkaraju S, Dierich A, Dolle P, et al. (1993) A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell 75 : 1333–1349. 7903601
28. Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR (1999) Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev 13 : 3136–3148. 10601039
29. Basch ML, Bronner-Fraser M (2006) Neural crest inducing signals. Adv Exp Med Biol 589 : 24–31. 17076273
30. Pegoraro C, Monsoro-Burq AH (2013) Signaling and transcriptional regulation in neural crest specification and migration: lessons from xenopus embryos. Wiley Interdiscip Rev Dev Biol 2 : 247–259. doi: 10.1002/wdev.76 24009035
31. Aybar MJ, Nieto MA, Mayor R (2003) Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development 130 : 483–494. 12490555
32. Bhatt S, Diaz R, Trainor PA (2013) Signals and switches in Mammalian neural crest cell differentiation. Cold Spring Harb Perspect Biol 5.
33. Garcia-Castro M, Bronner-Fraser M (1999) Induction and differentiation of the neural crest. Curr Opin Cell Biol 11 : 695–698. 10600707
34. Garcia-Castro MI, Marcelle C, Bronner-Fraser M (2002) Ectodermal Wnt function as a neural crest inducer. Science 297 : 848–851. 12161657
35. Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, et al. (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65 : 2334–2359. doi: 10.1007/s00018-008-8027-0 18408888
36. Kressler D, Hurt E, Bassler J (2010) Driving ribosome assembly. Biochim Biophys Acta 1803 : 673–683. doi: 10.1016/j.bbamcr.2009.10.009 19879902
37. Lempiainen H, Shore D (2009) Growth control and ribosome biogenesis. Curr Opin Cell Biol 21 : 855–863. doi: 10.1016/j.ceb.2009.09.002 19796927
38. Woolford JL Jr., Baserga SJ (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195 : 643–681. doi: 10.1534/genetics.113.153197 24190922
39. Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, et al. (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417 : 967–970. 12068309
40. Prieto JL, McStay B (2007) Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev 21 : 2041–2054. 17699751
41. Sloan KE, Bohnsack MT, Schneider C, Watkins NJ (2014) The roles of SSU processome components and surveillance factors in the initial processing of human ribosomal RNA. RNA 20 : 540–550. doi: 10.1261/rna.043471.113 24550520
42. Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, et al. (2004) RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev 18 : 2506–2517. 15489292
43. Zhao C, Andreeva V, Gibert Y, LaBonty M, Lattanzi V, et al. (2014) Tissue specific roles for the ribosome biogenesis factor Wdr43 in zebrafish development. PLoS Genet 10: e1004074. doi: 10.1371/journal.pgen.1004074 24497835
44. Freed EF, Prieto JL, McCann KL, McStay B, Baserga SJ (2012) NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet 8: e1002892. doi: 10.1371/journal.pgen.1002892 22916032
45. Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, et al. (2002) Functional proteomic analysis of human nucleolus. Mol Biol Cell 13 : 4100–4109. 12429849
46. Wood HB, Episkopou V (1999) Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev 86 : 197–201. 10446282
47. Rowitch DH, Kispert A, McMahon AP (1999) Pax-2 regulatory sequences that direct transgene expression in the developing neural plate and external granule cell layer of the cerebellum. Brain Res Dev Brain Res 117 : 99–108. 10536237
48. Hopwood ND, Pluck A, Gurdon JB (1989) MyoD expression in the forming somites is an early response to mesoderm induction in Xenopus embryos. EMBO J 8 : 3409–3417. 2555164
49. Carl TF, Dufton C, Hanken J, Klymkowsky MW (1999) Inhibition of neural crest migration in Xenopus using antisense slug RNA. Dev Biol 213 : 101–115. 10452849
50. Hong CS, Devotta A, Lee YH, Park BY, Saint-Jeannet JP (2014) Transcription factor AP2 epsilon (Tfap2e) regulates neural crest specification in Xenopus. Dev Neurobiol.
51. Soo K, O'Rourke MP, Khoo PL, Steiner KA, Wong N, et al. (2002) Twist function is required for the morphogenesis of the cephalic neural tube and the differentiation of the cranial neural crest cells in the mouse embryo. Dev Biol 247 : 251–270. 12086465
52. Blum M, Beyer T, Weber T, Vick P, Andre P, et al. (2009) Xenopus, an ideal model system to study vertebrate left-right asymmetry. Dev Dyn 238 : 1215–1225. doi: 10.1002/dvdy.21855 19208433
53. Chen CY, Croissant J, Majesky M, Topouzis S, McQuinn T, et al. (1996) Activation of the cardiac alpha-actin promoter depends upon serum response factor, Tinman homologue, Nkx-2.5, and intact serum response elements. Dev Genet 19 : 119–130. 8900044
54. Patterson KD, Drysdale TA, Krieg PA (2000) Embryonic origins of spleen asymmetry. Development 127 : 167–175. 10654610
55. Takebayashi-Suzuki K, Funami J, Tokumori D, Saito A, Watabe T, et al. (2003) Interplay between the tumor suppressor p53 and TGF beta signaling shapes embryonic body axes in Xenopus. Development 130 : 3929–3939. 12874116
56. Wallingford JB, Seufert DW, Virta VC, Vize PD (1997) p53 activity is essential for normal development in Xenopus. Curr Biol 7 : 747–757. 9368757
57. Yadav GV, Chakraborty A, Uechi T, Kenmochi N (2014) Ribosomal protein deficiency causes Tp53-independent erythropoiesis failure in zebrafish. Int J Biochem Cell Biol 49 : 1–7. doi: 10.1016/j.biocel.2014.01.006 24417973
58. Holmberg Olausson K, Nister M, Lindstrom MS (2012) p53-Dependent and-Independent Nucleolar Stress Responses. Cells 1 : 774–798. doi: 10.3390/cells1040774 24710530
59. Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, et al. (2013) The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 51 : 539–551. doi: 10.1016/j.molcel.2013.08.011 23973377
60. Wang M, Anikin L, Pestov DG (2014) Two orthogonal cleavages separate subunit RNAs in mouse ribosome biogenesis. Nucleic Acids Res 42 : 11180–11191. doi: 10.1093/nar/gku787 25190460
61. Holzel M, Orban M, Hochstatter J, Rohrmoser M, Harasim T, et al. (2010) Defects in 18 S or 28 S rRNA processing activate the p53 pathway. J Biol Chem 285 : 6364–6370. doi: 10.1074/jbc.M109.054734 20056613
62. Zhang Y, Lu H (2009) Signaling to p53: ribosomal proteins find their way. Cancer Cell 16 : 369–377. doi: 10.1016/j.ccr.2009.09.024 19878869
63. Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI (2010) The nucleolus under stress. Mol Cell 40 : 216–227. doi: 10.1016/j.molcel.2010.09.024 20965417
64. Sondalle SB, Baserga SJ (2014) Human diseases of the SSU processome. Biochim Biophys Acta 1842 : 758–764. doi: 10.1016/j.bbadis.2013.11.004 24240090
65. Ellis SR (2014) Nucleolar stress in Diamond Blackfan anemia pathophysiology. Biochim Biophys Acta 1842 : 765–768. doi: 10.1016/j.bbadis.2013.12.013 24412987
66. Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, et al. (2011) Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145 : 383–397. doi: 10.1016/j.cell.2011.03.028 21529712
67. Boglev Y, Badrock AP, Trotter AJ, Du Q, Richardson EJ, et al. (2013) Autophagy induction is a Tor - and Tp53-independent cell survival response in a zebrafish model of disrupted ribosome biogenesis. PLoS Genet 9: e1003279. doi: 10.1371/journal.pgen.1003279 23408911
68. Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, et al. (2002) Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn 225 : 499–510. 12454926
69. Hensey C, Gautier J (1998) Programmed cell death during Xenopus development: a spatio-temporal analysis. Dev Biol 203 : 36–48. 9806771
70. Pestov DG, Lapik YR, Lau LF (2008) Assays for ribosomal RNA processing and ribosome assembly. Curr Protoc Cell Biol Chapter 22: Unit 22 11.
71. Borovjagin AV, Gerbi SA (1999) U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J Mol Biol 286 : 1347–1363. 10064702
72. Borovjagin AV, Gerbi SA (2001) Xenopus U3 snoRNA GAC-Box A' and Box A sequences play distinct functional roles in rRNA processing. Mol Cell Biol 21 : 6210–6221. 11509664
73. Mougey EB, Pape LK, Sollner-Webb B (1993) A U3 small nuclear ribonucleoprotein-requiring processing event in the 5' external transcribed spacer of Xenopus precursor rRNA. Mol Cell Biol 13 : 5990–5998. 8413202
74. Peculis BA, Steitz JA (1993) Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell 73 : 1233–1245. 8513505
75. Savino R, Gerbi SA (1990) In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J 9 : 2299–2308. 2357971
Štítky
Genetika Reprodukčná medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



