-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Role of China in the Global Spread of the Current Cholera Pandemic
Cholera is a life-threatening, diarrheal disease caused by the bacterium Vibrio cholerae. After a long interregnum of decades without epidemics, the seventh cholera pandemic spread globally since 1961, causing considerable morbidity and mortality. Our analysis of published and newly sequenced genomes provides details on genetic groupings within V. cholerae, so-called clades, that have developed during the recent pandemic spread of these bacteria, and, in some cases, persisted to modern times. We reconstructed some of the pathways taken by the current pandemic since its origins in Indonesia, and show that both South Asia and East Asia are important pathogenic reservoirs and sources of international transmissions.
Published in the journal: The Role of China in the Global Spread of the Current Cholera Pandemic. PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005072
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005072Summary
Cholera is a life-threatening, diarrheal disease caused by the bacterium Vibrio cholerae. After a long interregnum of decades without epidemics, the seventh cholera pandemic spread globally since 1961, causing considerable morbidity and mortality. Our analysis of published and newly sequenced genomes provides details on genetic groupings within V. cholerae, so-called clades, that have developed during the recent pandemic spread of these bacteria, and, in some cases, persisted to modern times. We reconstructed some of the pathways taken by the current pandemic since its origins in Indonesia, and show that both South Asia and East Asia are important pathogenic reservoirs and sources of international transmissions.
Introduction
Cholera is an infectious and life-threatening diarrheal disease which is endemic in many African and Asian countries, and has also manifested as multiple, large epidemics and global pandemics since 1817 [1–3]. Older epidemics are attributed to the monophyletic ‘classical’ strains of Vibrio cholerae [4]. This attribution is supported by microbiological phenotypic typing which has been performed since the late 19th century [2], and by the close genetic similarities between one genome from 1849 and those of several classical V. cholerae isolated in recent decades [4]. Between 1923 and 1959, classical V. cholerae remained endemic in India, and caused local cholera outbreaks in multiple countries, but pandemics did not occur. During that pandemic interregnum, a second phenotypic variant of V. cholerae, called ‘El Tor’, was also isolated from cholera patients, but only rarely. In 1961 a seventh cholera pandemic began and this has been predominantly associated with El Tor strains. Epidemiological records suggest that El Tor spread from the island of Sulawesi (formerly Celebes) in Indonesia to South and Southeast Asia, and then globally. Pandemic El Tor strains, including a sub-variant with an O139 surface polysaccharide, also corresponds to a monophyletic lineage, L2, which is closely related to other lineages from pandemic cholera, but clusters in a distinct phylogenetic branch [4–6]. During the seventh pandemic, successive sub-clusters of L2 genotypes are thought to have radiated in three waves from the Bay of Bengal [6] on the East coast of the Indian sub-continent, a region where cholera has been continuously endemic for centuries [2]. A large outbreak in Haiti in 2010 reflects the spread of wave 3 from South Asia [7], possibly from Bangladesh [8] or Nepal [9]. However, these reconstructions lacked information on the genetic composition of V. cholerae in China or eastern Asia, and were predominantly based on genomes from bacteria isolated in the 1970s, or thereafter.
It is clear from the epidemiological literature that cholera flared in China repeatedly between 1817 and 1923, following earlier outbreaks in South and Southeast Asia, and possibly spread from China to Japan, Korea, eastern Siberia and western Asia [2]. Outbreaks in China also broke out on multiple occasions between 1923 and 1959 [1]. A detailed reconstruction of the causes of these outbreaks, and their chains of transmission, is likely to be difficult because only very few bacterial isolates from those periods are known to exist. On the other hand, the period after 1961 is more readily amenable to analysis, and for integration into reconstructions of the spread of cholera in other parts of the world. Since 1961, three successive waves of cholera were recorded in Southeast and Central China [10], each involving many thousands of cases of disease caused by El Tor V. cholerae (Fig. 1). Multiple, partially overlapping outbreaks with fewer cases of cholera also occurred in in the Autonomous Region of Xinjiang in Northwest China. These observations might reflect successive flares of cholera from endemic sources of V. cholerae within China. Alternatively, China may have been a ‘sink’ for bacteria from external sources, and each wave in China might have resulted from an independent import of these bacteria from elsewhere. Under both hypotheses, the Chinese waves might additionally have acted as a ‘source’ for spread to neighboring countries and possibly even acted as an ‘amplifier’ of epidemic spread. In order to address these questions, we compared 260 genomes of V. cholerae, including 181 that had been previously analyzed [6,9,11], 71 newly sequenced genomes from strains isolated in China between 1961 and 2010, and eight from other sources (S1A Table).
Fig. 1. Cholera in China since 1950. 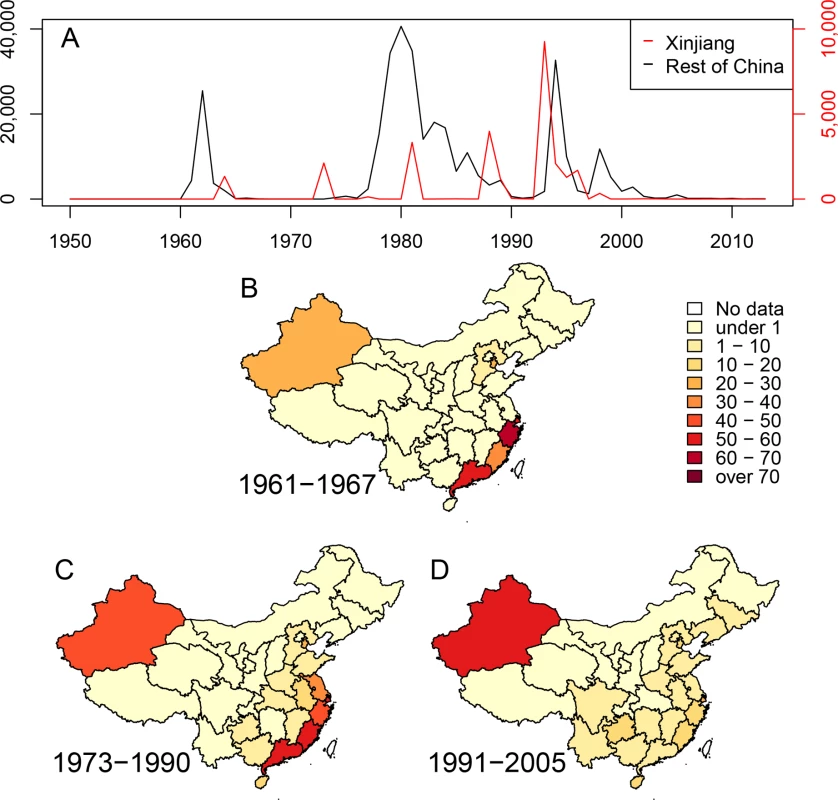
(A) Numbers of cases of cholera per year in the Xinjiang region (red, scale at right) and the rest of China (black, scale at left). (B-D) Density of cases of cholera in China during three epidemiological waves of disease: 1961–1967, 1973–1990 and 1991–2005. Each province is colored by numbers of cases per million inhabitants per year. Results and Discussion
Sources of the seventh pandemic
Mutreja et al. [6] concluded that all seventh pandemic isolates belonged to lineage L2, and estimated 1952 as the date for the most recent common ancestor (MRCA) of this lineage. Each L2 isolate differed from a reference seventh pandemic genome (N16961, Bangladesh, 1975) [12] by only 50–250 single nucleotide polymorphisms (SNPs). Other El Tor isolates were assigned to lineages L3, L5, L6 and L8 because they differed from N16961 by 3,000–6,000 SNPs. L3 and L8 contained recent El Tor strains from the US Gulf Coast and Australia, respectively. L5 contained two El Tor isolates from Sulawesi in 1937, prior to the seventh pandemic, and L6 corresponded to the oldest El Tor strain from 1910, which was isolated from an asymptomatic Indian pilgrim in El Tor, Saudi Arabia. However these analyses did not distinguish SNPs that were introduced by mutation, which can accumulate in a time-dependent, clock-like fashion, from clustered SNPs that are introduced by temporally unpredictable homologous recombination events involving DNA from distantly related bacteria, such as environmental V. cholerae [5]. Such distinctions are important because recombination can distort topological relationships, and artificially amplify genetic distances. The analyses of the history of lineage L2 by Mutreja et al. were based on strains which were isolated after 1975, with the exception of one isolate from Sulawesi (1957). Epidemiological records indicate that the seventh pandemic was preceded by small outbreaks in Sulawesi (1957) [13] and Ubol, northern Thailand (1959–60), and began with nearly simultaneous outbreaks in 1961 in multiple Indonesian islands, as well as in Malaysia, Macau, the Philippines and Hong Kong [14]. Starting in 1959, more than 60,000 individuals of Chinese extraction were resettled in southern China after expulsion from Indonesia because of their ancestry. They may also have brought cholera with them because a wave of cholera began in Southeast China in 1961–1963, followed in 1964 by outbreaks in Xinjiang (Fig. 1).
In order to clarify these issues, we re-examined the core genomic sequences of lineages L3, L5, L6 and L8, comparing them with genomes from classical strains as well as the earliest L2 strains isolated from Indonesia, China and Bangladesh. We used ClonalFrame [15] to estimate for each SNP the probability of having arisen by mutation or recombination, and calculated a maximum likelihood tree based exclusively on mutational SNPs (Fig. 2). Interestingly, only 336 mutational SNPs separated the root of all El Tor lineages from the most closely related classical genome (SN372). Furthermore, the MRCA of L2 only differed by 76 mutational SNPs from the MRCA of L5 (Sulawesi, 1937) and L8 (Australia, 1986), suggesting that their common ancestor existed quite recently. That ancestor differed by 31 mutational SNPs from the MRCA of L6 (Saudi Arabia, 1910), L3 (Gulf strains) and a closely related Chinese isolate from 1977. The L2 lineage encompassed not only the early seventh pandemic isolates but also the slightly earlier isolate from Sulawesi (1957). Individual L2 genomes differed from each other by 36–105 mutational SNPs, which is similar to the pairwise differences between El Tor lineages.
Fig. 2. Phylogeny of 19 genomes from both El Tor and classical V. cholerae. 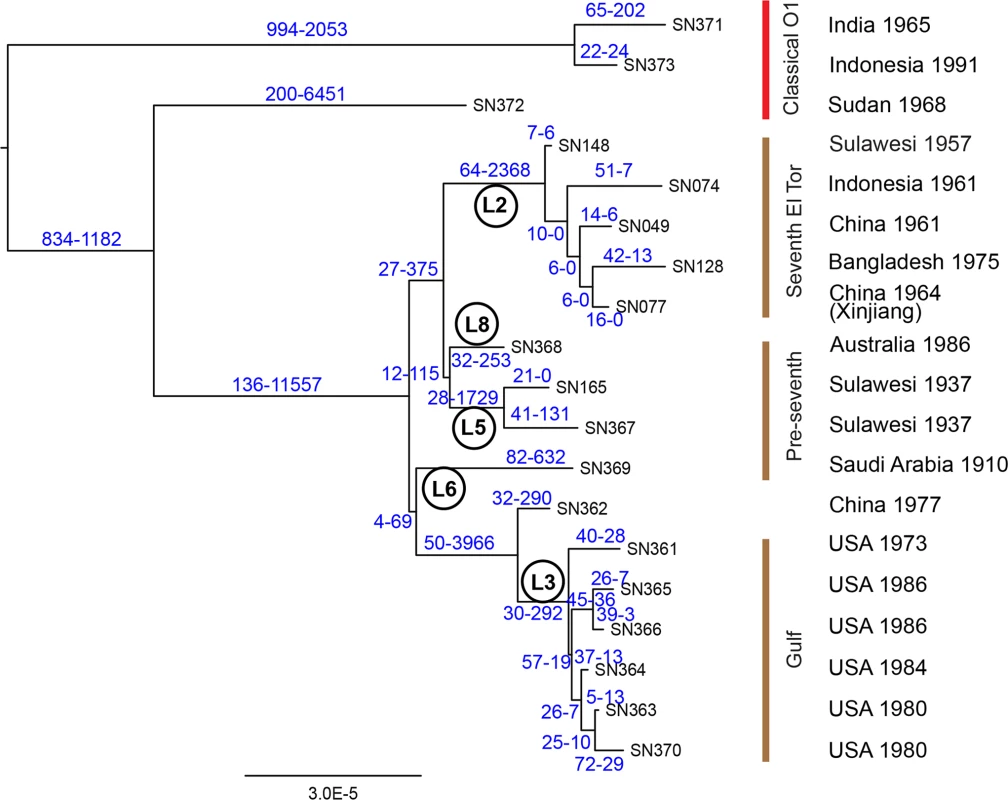
The phylogeny is based exclusively on mutational SNPs (as identified using ClonalFrame), and the two numbers above each branch are the estimated numbers of SNPs caused by mutation and recombination, respectively. Lineage designations are indicated within circles. Strain designations (cf S1B Table) are indicated at the tips of the branches, and the source and date of isolation of each strain are shown at the right. The individual lineages within classical and El Tor genomes were previously distinguished because they defined long branches [4–6]. Our analysis indicates that the length of those branches was largely due to recombination, which introduced thousands of clustered SNPs (Figs. 2, S1), including 2,368 SNPs on the branch leading to the MRCA of lineage L2. However, very little of the diversity between the five L2 genomes was attributed to recombination (32 SNPs). The results in Fig. 2 also provide an initial perspective on the evolutionary genealogy that led to the seventh pandemic. At the base of L2 are two isolates from Indonesia sampled in 1957 and 1961, suggesting that this is the true source of the seventh pandemic. A close relationship was found between an isolate from the Chinese province of Xinjiang (1964) and one from Bangladesh (1975). Outbreaks of cholera caused by El Tor strains were first reported in Bangladesh, India and Pakistan in 1963, 1964 and 1965, respectively [16]. These countries border on Xinjiang or are not very distant (S2 Fig), and cross-border exchanges are frequent enough between the Muslim populations in these areas that in 2011, an epidemic of poliomyelitis in Xinjiang was imported from Pakistan [17]. V. cholerae could also have been transmitted across these country borders in the early 1960’s but no genomes are yet available from that period except for the Chinese isolates described here.
Hypermutators in the genealogy of lineage L2
We now focus on the seventh pandemic based on 260 genomes from lineage L2. A maximum likelihood tree based on the 6,335 SNPs in their non-repetitive, core genomes (Fig. 3 inset; S3A Fig) confirmed that they are all genetically related, and clustered in three successive groups of decreasing diversity, which correspond to the three waves that were previously described [6]. The root-to-tip distances in the phylogeny correlated strongly with the dates of isolation of the individual strains (S3B Fig; R2 = 0.932; p-value = 2.56x10-142) with the exception of 17 of the 260 genomes which were significant outliers to this linear regression. They were located on long terminal branches (red in Fig. 3 inset and S3 Fig), as previously noted for strain A4 [6].
Fig. 3. Timed phylogeny for 6,335 SNPs in 260 genomes from the seventh pandemic. 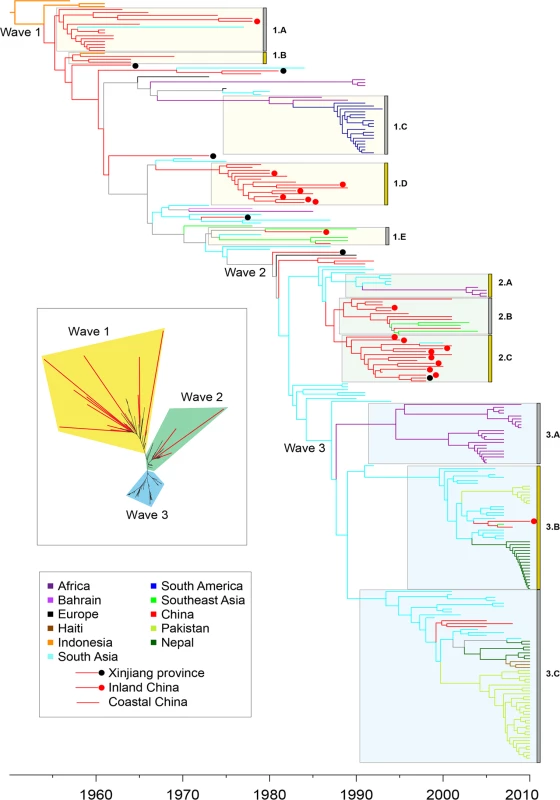
The vertical order is the same as in S1A Table. Branches are colored according to inferred location as shown in the legend at the lower left, with the exception of branches for which the location was uncertain which are shown in gray. Isolates from China are subdivided into isolates from Xinjiang (black dot), inland provinces (red dot) and coastal provinces (no dot). Selected clades of multiple, closely related isolates are indicated by grey boxes next on the left of the clade designations (1.A, 1.B, etc). Inset: Maximum likelihood tree of the same data with significantly longer branches according to S3 Fig indicated in red. Several possibilities could account for these long branches. They could represent sequencing mistakes, but this was ruled out because manual re-sequencing confirmed all 44 randomly selected SNPs from long branches that were tested. Long branches can also result from recombination, but here we found none of its typical signatures. Firstly, only 2% (125) of the SNPs were homoplastic, that is unexplainable by a single mutation on the tree. Secondly, the relative effect of recombination to mutation (r/m) was equal to 0.1 according to a ClonalFrame analysis, with only ~1% of the core genome affected by recombination on any branch in the phylogeny. Thirdly, the SNPs on the long branches were spread evenly around the genome (S4 Fig), rather than clustered as would be expected from recombination [18], and as found in our comparisons between lineages (S1 Fig). These observations confirm that recombination has been very rare in L2, and exclude it as a likely cause of the long branches.
Long branches can also be caused by the elevated mutation rates in hypermutators, which arise naturally in populations of Escherichia coli [19–21], Neisseria meningitidis [22] and Yersinia pestis [23] following disruption of the mismatch repair system. Hypermutators may promote the acquisition of antibiotic resistance [24] or other forms of adaptive evolution [25], but in the longer term their high mutation rate results in reduced fitness [26] and they do not succeed in establishing themselves against the competition of non-mutators. To test this possibility, we measured the in vitro mutation rate of all 79 strains at our disposal, including 16 on long branches (S2 Table). The strains associated with long branches had significantly higher mutation rates than the others (S5 Fig, Kruskal-Wallis test, p-value = 8.44x10-4). Most of them were also associated with mutations that can lead to the hypermutator phenotype: 14 of the 17 genomes on long branches possessed a total of 18 genetic variations in one or more of four genes (mutS, mutH, mutL and uvrD) that play a key role in the mismatch repair system (S3 Table). These 18 variations included ten short indels resulting in frameshifts, five non-synonymous codon changes and one premature stop codon for a total of 16 changes in protein sequence versus two synonymous mutations. In contrast, only four of the 243 genomes with normal branch lengths had changes in the amino acid composition encoded by one of these four genes (Fisher exact test, p = 6x10-17). In ten of the genomes with long branches, we also identified 12 non-synonymous changes, one frameshift, one premature stop codon and two synonymous mutations in ten other genes that can affect mismatch repair (S3 Table).
The majority (9/17) of the strains with long branches were isolated between 1961 and 1965, relatively soon after the beginning of the seventh pandemic. It is tempting to speculate that a high frequency of hypermutators was causally associated with the rapid spread of the seventh pandemic, especially because hypermutators may be a sign of recent selective pressure and population bottlenecks. However, these old strains of V. cholerae had been maintained in stab cultures for many years, which also tends to select for mutations [27]. Thus, confirming the importance of multiple hypermutators among early strains from the seventh pandemic will require the analysis of additional old strains that have not been stored as stab cultures.
Long-term chains of transmission
We estimated a timed phylogeny for the SNPs among the 260 genomes in which leaves are aligned with their isolation dates, and branch lengths represent time rather than number of mutations. Due to the presence of the hypermutators, the phylogeny was calculated using a relaxed molecular clock model [28] in which the 17 long terminal branches each had their own independent evolutionary rate (Fig. 3). The evolutionary rate for the remainder of the tree was estimated as ~2.3 substitutions per genome per year, and the MRCA of the seventh pandemic was dated to 1954. These results are in good agreement with Mutreja et al. [6], and similar to the clock rates estimated for multiple other bacterial pathogens [29]. According to the timed phylogeny, the three previously described waves do not seem well defined: they simply correspond to three internal branches with multiple descendants (Fig. 3). Furthermore, the three wave concept oversimplifies the epidemiological patterns, because the timed phylogeny does not correspond to successive, discrete radial expansions from single nodes. Instead, each of the waves is preceded by multiple, closely related long branches which currently contain only few isolates. As additional historical isolates are examined, it may become even more difficult to determine the exact position within the tree where initiated each of these supposed waves.
A second problem also exists with the wave concept, namely that epidemiological inferences have been used to designate phylogenetic structures, which implies causality that may not exist. We therefore recommend substituting neutral designations for well-defined monophyletic clades where multiple genomes cluster tightly, as a replacement for wave designations. Such designations would facilitate testing for an association between phylogenetic clade, genomic content and increased transmission. We have therefore assigned designations (1.A, 2.B, etc.) to several obvious monophyletic clades, including the prior wave number in order to support comparisons with prior publications as well as arbitrary letters (Fig. 3; S1A Table). All but three of 123 strains in wave 3 from global sources were isolated after 2000, and cluster in clades 3.A, 3.B and 3.C. Clade 3.A corresponds to an African epidemic which seems to have been imported from South Asia. Clades 3.B contains strains from the outbreak in Haiti, as well as from South Asia (Nepal, Bangladesh) and China (two strains). Clade 3.C contains strains from South Asia and Pakistan as well as multiple, other global sources, including three from China. These patterns are consistent with China having been a sink for V. cholerae since 2000, rather than a source.
Most of the strains isolated in China between 1991 and 2005 belonged to clades 2.B (Coastal China) and 2.C (Inland China). Clade 2.B was ancestral to several genomes from Southeast Asia; and was preceded in the phylogeny by still other genomes from South Asia, but progenitors of these clades were also isolated in China, including an isolate from the Xinjiang province in 1988. These observations are consistent with China having been an initial source for the global spread of these clades and their relatives, but do not preclude later strains having been imported from outside. China also seems to have played a role in the earliest global transmission of the 7th pandemic after its origins in Indonesia. Apart from the three earliest strains from Indonesia, the deepest branches (clades 1.A and 1.B) were found in China in the 1960s. Clade 1.C contained multiple isolates from South America in the 1990s, and is derived from the deeper branches in clade 1.B that were found in China. Chinese isolates are found on all subsequent deep branches (for example in clade 1.E), as well as forming several terminal branches but most Chinese isolates from the 1973–1990 outbreak cluster in clade 1.D. Several intermediate branches in this early phase of spread were from Xinjiang, providing further support for transmission to other countries from Northwest China.
In order to investigate these source/sink relationships in greater detail we inferred the ancestral geographical locations of branches by a maximum parsimony reconstruction of states, summarizing sources and sinks for international transmissions by a circular plot [30,31] (S4 Table; Fig. 4). Our data indicate nine transmissions out of South Asia, including twice into Africa in the 1990s, twice into Pakistan between 2002 and 2005, and once into Nepal around 2005. The strain causing the Haitian 2010 outbreak was confirmed to have originated in Nepal, as previously suggested [9]. Likewise, we also confirmed pandemic spread from Africa into South America in the 1980s [6]. These conclusions should be considered as minimal estimates of the numbers of transmissions because numerous sources of international outbreaks and endemic disease have not been investigated, leading to sampling bias. In contrast, more genomes have now been sequenced from China than from any other single source, and the Chinese strains are representative of disease over the entire period from 1961 until 2005. Our analysis of global transmissions (Fig. 4) indicates that China imported V. cholerae four times from South Asia (1975–2004), once from Indonesia (1955) and once from Southeast Asia (1986). In turn, China was the source of transmissions to South Asia (three times, 1967–1999), Indonesia (1960) and Southeast Asia (2007). These results suggest that V. cholerae populations are often transmitted between East Asia, South Asia and Southeast Asia, which makes it difficult to pinpoint the exact origins or outbreaks in other parts of the world. Even if China is not the direct origin for such outbreaks, it clearly represents an important reservoir of diversity which needs to be accounted for when modeling the global epidemiology of cholera.
Fig. 4. Circular plot illustrating the inferred migrations between geographical locations. 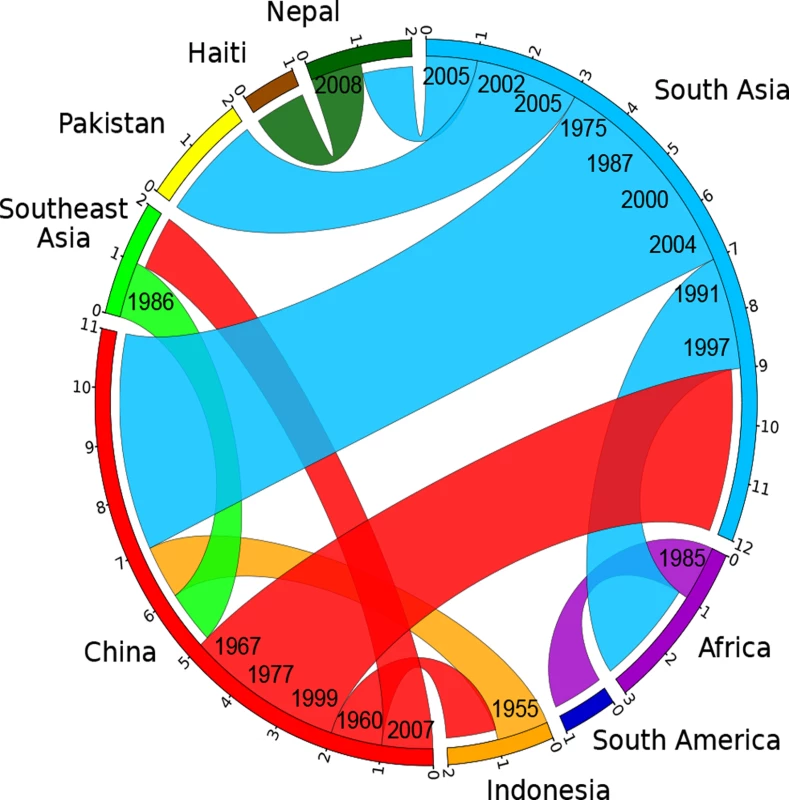
Flow bars indicate the sources of transmissions, colored as in Fig. 3, with one end of the bar directly touching the country of origin, and the other end of the bar having a small gap before the country of destination. The average date for each migration is shown on the ends corresponding to the origin. Materials and Methods
Isolates
Metadata for the 260 lineage L2 strains whose genomes were compared is presented in S1A Table, including dates and countries of isolation and accession numbers. 79 of these isolates were newly sequenced for this study from a previously described collection [10], of which 71 were from China. The analysis also included previously sequenced genomes from 119 isolates from a global collection [6], 24 from Nepal [9] and 38 from Pakistan [11], and the tree was rooted with the pre-seventh pandemic strain M66 [32], which belongs to lineage L5 (cf SN165 in Fig. 2). Five of these 260 genomes from L2 plus 14 genomes from other lineages than L2 were used in Fig. 2, as listed in the S1B Table.
Experimental mutation rate
For each of the 79 newly sequenced isolates, the mutation rate to rifampicin resistance was experimentally measured as previously described [33]. Briefly, 102 to 103 cells from an overnight culture were inoculated on nitrocellulose filters which had been placed on 869 plates. After incubation at 37°C for 24 h, the cells were re-suspended in 1 ml of 869 broth and incubated at 37°C for 1 h to allow expression of rifampicin resistance. Appropriate dilutions were then spread in parallel on both 869 plates and 869 plates containing 100ìg/ml of rifampicin. The rifampicin resistant mutants were counted after incubation at 37°C for 24 h, and each mutation rate was calculated as the median of six independent cultures (S2 Table).
Genome sequencing
DNA was prepared from 1 ml overnight cultures with the Wizard Genomic DNA Purification Kit (Promega). Whole genome sequencing was performed at BGI (China) on 78 genomes using an Illumina HiSeq 2000 on 250 bp and 6 kb paired-end libraries in 100-fold multiplexes (see S1 Table for number of reads, read length and N50 statistics for each genome), whereas the finished genome of strain FJ147 was obtained on an ABI Prism 3730 with Sanger sequencing (BGI, China).
Assembly and identification of SNPs
We applied two independent methods to assemble contigs and call SNPs for the 260 genomes plus outgroup strain M66. First we performed reference-based mapping against the reference genome of N16961 [12] using Bowtie 2 [34]. Secondly, we performed de novo assembly using SPAdes [35] followed by SNP calling against N16961 with MuMMER [36]. The two methods identified 9,064 and 9,089 SNPs respectively, of which 8,987 SNPs were identical between both methods and were therefore used for further analysis. 2,652 of these SNPs were specific to the outgroup strain M66, leaving 6,335 SNPs differentiating the 260 genomes from the current pandemic.
Confirmation of SNPs in genomes on long terminal branches
We randomly selected 44 SNPs from 16 long terminal branches in S3 Fig. For each SNP, the flanking 250bp on both sides from the assembled genome were used to design amplification and sequencing primers with Primer3 [37], which were then used for Sanger sequencing of the genomic SNP.
Phylogenomic analysis
In order to compare the 19 genomes (listed in S1B Table) from both Classical and El Tor V. cholerae, ClonalFrame [15] was first run to determine the sites likely to have been affected by recombination with posterior probability above 50%. A maximum-likelihood tree (Fig. 2) was then constructed based on the non-recombinant sites using PhyML 3.0.1 [38]. For each branch of the phylogeny, the expected numbers of SNPs caused by mutation and recombination were estimated using ClonalFrame.
In order to compare the 260 genomes (listed in S1A Table) from the seventh pandemic, a maximum-likelihood tree (Fig. 3 inset; S3A Fig) was computed by applying PhyML 3.0.1 [38] to the 8,987 SNPs differentiating the genomes between themselves and from the pre-seventh pandemic genome M66 which was included as an outgroup to root the tree. The significance of abnormally long branches in this phylogeny was tested using a strict clock molecular clock rate model, which identified 17 branches that were significantly longer than expected (red branches in Fig. 3 inset and S3A Fig; red dots in S3B Fig). We therefore calculated a timed phylogeny (Fig. 3) using a previously described method [39], which consists of finding the posterior distribution of the dates of the ancestral nodes and rates of the clock model given the observed number of substitutions on each branch. This inference was performed under the assumption of a local molecular clock model [28] with a total of 18 parameters to allow a separate clock rate for each of the 17 long branches and a single rate for the remainder of the tree. The geographical location of terminal branches was the known place of isolation of the strains. For internal branches, the likely location was reconstructed using maximum parsimony ancestral reconstruction [40], which identified a minimum of 37 migration events to explain the data. Of these, 18 were unambiguous (meaning that they could only have a single source and a single destination) and correspond to a change of color from parental to daughter branch in Fig. 3, with the exception of grey branches where multiple ancestral locations were probable. The sources and destinations of the 18 unambiguous migrations are summarized in S4 Table, along with the estimated date of each migration which was calculated as the date at the middle of each migrant branch. This data was also represented as a circular plot (Fig. 4) using the Circos [30] table viewer (available at http://mkweb.bcgsc.ca/tableviewer/), to represent tabular data in a graphical circular format.
Data deposition
The sequence data have been deposited with the European Nucleotide Archive, www.ebi.ac.uk/ena accession number ERP006431–50 (PRJEB6790–809), ERP006452–5 (PRJEB6811–814) and ERP006457–510 (PRJEB6816–869). The complete genome sequence of FJ147 was deposited to GenBank under accession number CP009041–2. All accession numbers are listed in S1 Table.
Supporting Information
Zdroje
1. Pollitzer R., Swaroop S., and Burrows W. (1959) World Incidence. In: Cholera. Geneva: WHO. pp. 51–96.
2. Pollitzer R., Swaroop S., and Burrows W. (1959) History of the disease. In: Cholera. Geneva: WHO. pp. 11–50.
3. Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB (2012) Cholera. Lancet 379 : 2466–2476. doi: 10.1016/S0140-6736(12)60436-X 22748592
4. Devault AM, Golding GB, Waglechner N, Enk JM, Kuch M, et al. (2014) Second-pandemic strain of Vibrio cholerae from the Philadelphia cholera outbreak of 1849. New England J Med 370 : 334–340. doi: 10.1056/NEJMoa1308663 24401020
5. Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, et al. (2009) Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA 106 : 15442–15447. doi: 10.1073/pnas.0907787106 19720995
6. Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, et al. (2011) Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477 : 462–465. doi: 10.1038/nature10392 21866102
7. Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, et al. (2013) Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. MBio 4:
8. Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, et al. (2011) The origin of the Haitian cholera outbreak strain. New England J Med 364 : 33–42. doi: 10.1056/NEJMoa1012928 21142692
9. Hendriksen RS, Price LB, Schupp JM, Gillece JD, Kaas RS, et al. (2011) Population genetics of Vibrio cholerae from Nepal in 2010: Evidence on the origin of the Haitian outbreak. MBio 2:
10. Liang W, Wang L, Liang P, Zheng X, Zhou H, et al. (2013) Sequence polymorphisms of rfbT among the Vibrio cholerae O1 strains in the Ogawa and Inaba serotype shifts. BMC Microbiol 13 : 173. doi: 10.1186/1471-2180-13-173 23889924
11. Shah MA, Mutreja A, Thomson N, Baker S, Parkhill J, et al. (2014) Genomic epidemiology of Vibrio cholerae O1 associated with floods, Pakistan, 2010. Emerg Infect Dis 20 : 13–20. doi: 10.3201/.eid2001.130428 24378019
12. Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, et al. (2000) DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406 : 477–483. 10952301
13. Tanamal ST (1959) Notes on paracholera in Sulawesi (Celebes). Am J Trop Med Hyg 8 : 72–78. 13617601
14. Felsenfeld O (1966) A review of recent trends in cholera research and control. With an annex on the isolation and identification of cholera vibrios. Bull World Health Organ 34 : 161–195. 5328492
15. Didelot X, Falush D (2007) Inference of bacterial microevolution using multilocus sequence data. Genetics 175 : 1251–1266. 17151252
16. Barua D. (1992) History of cholera. In: Barua D and Greenough III, W. B., editors. Cholera. New York: Plenum. pp. 1–35.
17. Luo HM, Zhang Y, Wang XQ, Yu WZ, Wen N, et al. (2013) Identification and control of a poliomyelitis outbreak in Xinjiang, China. N Engl J Med 369 : 1981–1990. doi: 10.1056/NEJMoa1303368 24256377
18. Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, et al. (2011) Rapid pneumococcal evolution in response to clinical interventions. Science 331 : 430–434. doi: 10.1126/science.1198545 21273480
19. LeClerc JE, Li B, Payne WL, Cebula TA (1996) High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274 : 1208–1211. 8895473
20. Sniegowski PD, Gerrish PJ, Lenski RE (1997) Evolution of high mutation rates in experimental populations of E. coli. Nature 387 : 703–705. 9192894
21. Mao EF, Lane L, Lee J, Miller JH (1997) Proliferation of mutators in a cell population. J Bacteriol 179 : 417–422. 8990293
22. Richardson AR, Yu Z, Popovic T, Stojiljkovic I (2002) Mutator clones of Neisseria meningitidis in epidemic serogroup A disease. Proc Natl Acad Sci USA 99 : 6103–6107. 11983903
23. Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, et al. (2010) Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nature Genet 42 : 1140–1143. doi: 10.1038/ng.705 21037571
24. Martinez JL, Baquero F (2000) Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother 44 : 1771–1777. 10858329
25. Weigand MR, Sundin GW (2012) General and inducible hypermutation facilitate parallel adaptation in Pseudomonas aeruginosa despite divergent mutation spectra. Proc Natl Acad Sci USA 109 : 13680–13685. doi: 10.1073/pnas.1205357109 22869726
26. Giraud A, Matic I, Tenaillon O, Clara A, Radman M, et al. (2001) Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291 : 2606–2608. 11283373
27. Eisenstark A (2010) Genetic diversity among offspring from archived Salmonella enterica ssp. enterica serovar Typhimurium (Demerec Collection): in search of survival strategies. Annu Rev Microbiol 64 : 277–292. doi: 10.1146/annurev.micro.091208.073614 20825350
28. Drummond AJ, Ho SY, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4: e88. 16683862
29. Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW (2012) Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13 : 601–612. doi: 10.1038/nrg3226 22868263
30. Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, et al. (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19 : 1639–1645. doi: 10.1101/gr.092759.109 19541911
31. Abel GJ, Sander N (2014) Quantifying global international migration flows. Science 343 : 1520–1522. doi: 10.1126/science.1248676 24675962
32. Feng L, Reeves PR, Lan R, Ren Y, Gao C, et al. (2008) A recalibrated molecular clock and independent origins for the cholera pandemic clones. PLoS ONE 3: e4053. doi: 10.1371/journal.pone.0004053 19115014
33. Picard B, Duriez P, Gouriou S, Matic I, Denamur E, et al. (2001) Mutator natural Escherichia coli isolates have an unusual virulence phenotype. Infect Immun 69 : 9–14. 11119483
34. Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9 : 357–359. doi: 10.1038/nmeth.1923 22388286
35. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19 : 455–477. doi: 10.1089/cmb.2012.0021 22506599
36. Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, et al. (2004) Versatile and open software for comparing large genomes. Genome Biol 5: R12. 14759262
37. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, et al. (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40: e115. 22730293
38. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 59 : 307–321. doi: 10.1093/sysbio/syq010 20525638
39. Didelot X, Eyre DW, Cule M, Ip CL, Ansari MA, et al. (2012) Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol 13: R118. doi: 10.1186/gb-2012-13-12-r118 23259504
40. Williams PD, Pollock DD, Blackburne BP, Goldstein RA (2006) Assessing the accuracy of ancestral protein reconstruction methods. PLoS Comput Biol 2: e69. 16789817
Štítky
Genetika Reprodukčná medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



