-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Selective and Genetic Constraints on Pneumococcal Serotype Switching
Streptococcus pneumoniae is a major respiratory pathogen responsible for a high burden of morbidity and mortality worldwide. Current anti-pneumococcal vaccines target the bacterium’s polysaccharide capsule, of which at least 95 different variants (‘serotypes’) are known, which are classified into ‘serogroups’. Bacteria can change their serotype through genetic recombination, termed ‘switching’, which can allow strains to evade vaccine-induced immunity. By combining epidemiological data with whole genome sequencing, this work finds a robust and unexpected pattern of serotype switching in a sample of bacteria collected following the introduction of routine anti-pneumococcal vaccination: switching was much more likely to exchange one serotype for another within the same serogroup than expected by chance. Several hypotheses are presented and tested to explain this pattern, including limitations of genetic recombination, interactions between the genes that determine serotype and the rest of the genome, and the constraints imposed by bacterial metabolism. This provides novel information on the evolution of S. pneumoniae, particularly regarding how the bacterium might diversify as newer vaccines are introduced.
Published in the journal: Selective and Genetic Constraints on Pneumococcal Serotype Switching. PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005095
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005095Summary
Streptococcus pneumoniae is a major respiratory pathogen responsible for a high burden of morbidity and mortality worldwide. Current anti-pneumococcal vaccines target the bacterium’s polysaccharide capsule, of which at least 95 different variants (‘serotypes’) are known, which are classified into ‘serogroups’. Bacteria can change their serotype through genetic recombination, termed ‘switching’, which can allow strains to evade vaccine-induced immunity. By combining epidemiological data with whole genome sequencing, this work finds a robust and unexpected pattern of serotype switching in a sample of bacteria collected following the introduction of routine anti-pneumococcal vaccination: switching was much more likely to exchange one serotype for another within the same serogroup than expected by chance. Several hypotheses are presented and tested to explain this pattern, including limitations of genetic recombination, interactions between the genes that determine serotype and the rest of the genome, and the constraints imposed by bacterial metabolism. This provides novel information on the evolution of S. pneumoniae, particularly regarding how the bacterium might diversify as newer vaccines are introduced.
Introduction
Streptococcus pneumoniae is a human nasopharyngeal commensal bacterium and important respiratory pathogen. The ability of the pneumococcus to cause invasive disease appears critically dependent upon its polysaccharide capsule [1], of which at least 95 immunologically-distinguishable capsular variants (serotypes) are known [2–7]. This structure inhibits the recognition of subcapsular protein antigens by the adaptive immune system and the binding of phosphorylcholine residues by C-reactive protein, thereby reducing the rate of complement deposition on the bacterial surface [8]. Children develop anticapsular antibodies after exposure to the bacterium, although evidence for their impact on disease risk is mixed [9]. Data indicating this immune response provides protection against nasopharyngeal carriage has been found for only a few serotypes [10–12], though mathematical models suggest subtle effects on carriage may exist and help maintain the high level diversity of serotypes observed in pneumococcal populations [13].
In contrast to natural immunity, that induced by the seven-valent protein conjugate polysaccharide vaccine (PCV7) reduces acquisition of vaccine serotypes in the nasopharynx by 50% [14] or more [15]. Following the introduction of PCV7, a substantial fall in the carriage of the seven vaccine serotypes (4, 6B, 9V, 14, 18C, 19F and 23F) was observed without any substantial overall reduction in the rates of pneumococcal colonisation [16,17]. This was primarily the result of an increase in the rate of carriage of non-vaccine type strains, termed ‘serotype replacement’ [18–20]. In some cases non-vaccine type strains are closely related to vaccine type strains from which they have been derived by ‘serotype switching’ [21]. In these cases, strains have altered their serotype through the acquisition of a different capsular polysaccharide synthesis (cps) locus via genetic transformation.
Many serotypes, though distinguishable by certain antisera (called "factor sera"), nonetheless may be clustered into “serogroups” based on cross-reactivity with other antisera; these groups have often been found to correspond to sets of similar polysaccharide structures [2]. It was originally hoped that PCV7 would provide cross-protection against ‘vaccine-related’ serotypes: those within the same serogroups as a serotype included in the vaccine [22]. However, the only case in which such an effect was observed was the protection against colonisation with serotype 6A resulting from the inclusion of serotype 6B in PCV7; several other vaccine-related types increased in prevalence post-PCV7 [23]. Contributing to this pattern was an apparent tendency for serotype switches to exchange one serotype for another within the same serogroup more often than expected by chance: of nine switches inferred using multilocus sequence typing (MLST) in a systematic collection of carried pneumococci from Massachusetts, three were within-serogroup (p = 0.043) [24]. Hence genotypes successful prior to the introduction of PCV7 were able to persist expressing a similar capsule that was not recognised by vaccine-induced immunity. This was not a pattern expected a priori, as random acquisition of a new serotype from the full, diverse set of non-vaccine type capsules should usually result in a change of serogroup.
This work assesses the relative likelihood of alternative explanations of the observed pattern of serotype switching based on whole genome sequencing data from this Massachusetts-based collection [25]. The first is that the detected propensity for within-serogroup serotype switching is a consequence of the constraints of genetic transformation. Pneumococcal cps loci are typically 10–30 kb in size, usually necessitating a similarly long recombination to cause a change of serotype [26], whereas homologous recombinations have an exponential distribution of sizes with a mean length of between two and seven kilobases [27–29]. Aside from known exceptions such as serogroups 7, 17, 33 and 35, the cps loci corresponding to a single serogroup are closely related [2]. Therefore, within-serogroup serotype switching may be accomplished through relatively short, more frequent, recombination events that do not replace the entire cps locus [30]. Another possibility is that patterns of serotype switching may be affected by the flanking pbp1a and pbp2x genes, which are crucial in determining β–lactam resistance. As resistance is associated with a limited number of serotypes [31], and long recombinations that change serogroup could lead to the acquisition of resistance [32] or risk reversing any beneficial acquisition of resistance-associated pbp1a or pbp2x alleles [29,33], it may be that maintenance of a strain’s β–lactam susceptibility (or lack thereof) affects the patterns of serotype switching.
Alternatively, rather than representing a limitation on the rate of recombination, the pattern of switching may reflect the consequence of constraints imposed by epistatic interactions with other loci in the chromosome [26]. Such limitations may reflect physiological or metabolic specialisation to production of particular capsule types; alternatively, it may be important to co-ordinate the expression of particular serogroups with certain alleles of immunogenic surface proteins, in order to maintain discordant sets of antigens in distinct lineages [34]. The last explanation to be considered, which also involves the host immune response, relies on the presence of natural or vaccine-induced antibodies that cross-react with all serotypes within a serogroup. This would lead to elevated rates of co-colonisation between strains of the same serogroup, as bacteria would be confined to a subpopulation of hosts that do not exhibit immunity to either of their capsules. A consequence of this would be increased genetic exchange between members of the same lineage, including an elevated rate of within-serogroup serotype switching.
Results
Association between serotypes and genotypes
Of the 616 draft genomes previously analysed [25], 491 fell into fifteen monophyletic sequence clusters (SCs) of related isolates within which serotype switching could be investigated (Fig. 1). These sequence clusters included representatives of 19 serotypes spread across eleven serogroups. In seven sequence clusters, the serotype was stable across the clade, leaving eight clusters in which at least one switch had occurred.
Fig. 1. Prevalence of serotype switching in the overall population. 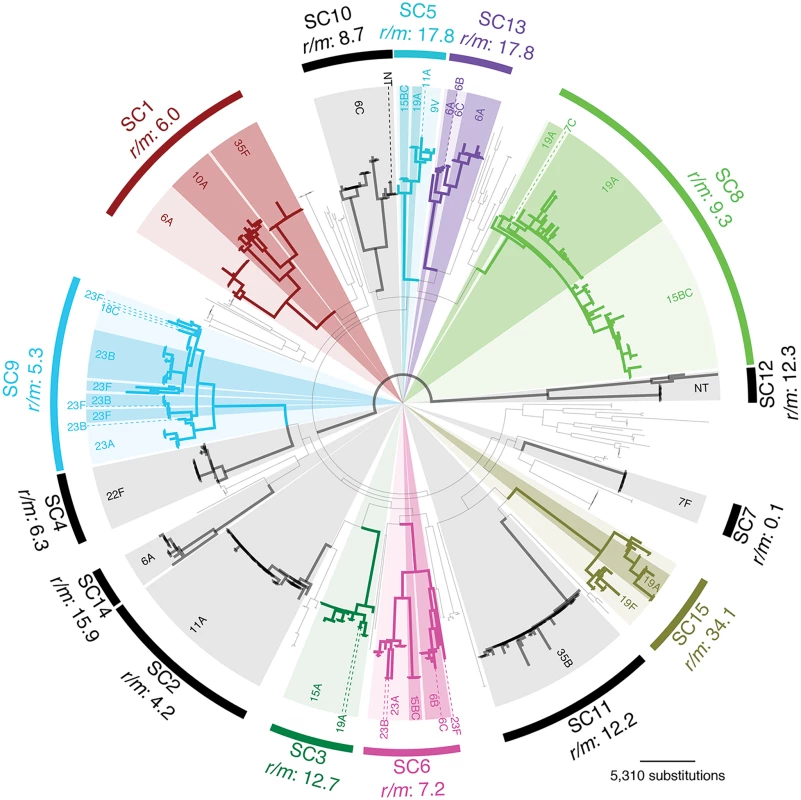
The maximum likelihood phylogeny derived from a codon alignment of ‘core’ protein coding sequences is displayed, with fifteen monophyletic sequence clusters (SC1-SC15) labelled by arcs around the edge of the tree. Those sequence clusters in which no serotype switching was observed are marked by black arcs. Those sequence clusters in which multiple serotypes were observed are individually coloured, with the most prevalent serotype represented by light shading, and alternative serotypes indicated by darker shading or dashed lines. There was no positive correlation between the number of isolates sampled within a sequence cluster and the number of serotypes it contained (Fig. 2A). However, a significant correlation was observed between the number of polymorphic sites, as ascertained through a lineage-specific analysis of whole genome alignments, and the number of serotypes in a cluster (Fig. 2B). When these sites were split into the number of point mutations and homologous recombination events per sequence cluster (see Methods), there was no significant relationship with the former, whereas the latter had the strongest correlation of any measure of genetic diversity (Fig. 2C and D). Therefore sequence clusters that exhibit serotype diversity are those that have experienced more recombination events throughout their genome over their observable evolutionary history, either as a consequence of a high rate of recombination over their recent diversification, or lower recombination rates over a longer period of time.
Fig. 2. Associations between properties of a sequence cluster and its capsular diversity. 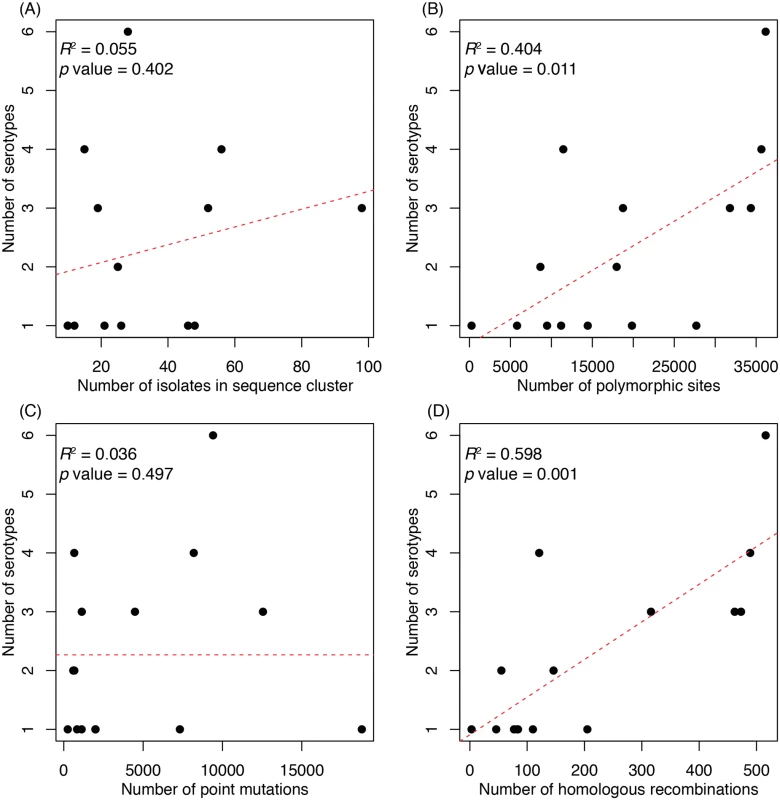
For each of the fifteen monophyletic sequence clusters, the number of serotypes it contained was plotted against (A) the number of representatives of the sequence cluster; (B) the overall genetic diversity of the sequence cluster, as represented by the number of polymorphic sites identified from whole genome alignments; (C) the number of point mutations reconstructed as having occurred over the history of the sequence cluster; and (D) the number of homologous recombination events reconstructed as having occurred over the history of the sequence cluster. Frequent within-serogroup switching
Across the species, the 95 currently known S. pneumoniae serotypes are divided into 48 serogroups such that 2.2% of comparisons between different serotypes are serogroup concordant. However, many serotypes are rarely observed; the 32 serotypes observed across all 616 isolates from Massachusetts were distributed such that 3.0% of serotype comparisons were serogroup concordant. Seventeen serotypes were observed within the eight sequence clusters that appear to contain examples of serotype switching; 7.2% of all pairwise comparisons between these serotypes were serogroup-concordant, suggesting these isolates expressed a comparatively limited number of serogroups. Yet when only considering the subset of these comparisons where the different serotypes were found within the same sequence cluster, 29% were serogroup concordant (S1 Table), as some individual sequence clusters were associated with multiple serotypes from the same serogroup. The significance of this enrichment was assessed by a permutation test that randomly assigned serotypes to these eight sequence clusters according to the number of serotypes originally observed in the sequence cluster. This found the high level of serogroup-concordant comparisons within sequence clusters to be statistically significant (p = 0.0007 from 10,000 permutations). As sequence clusters were identified through a clustering algorithm [25], this observation is independent of any phylogenetic analyses. Therefore this test indicates isolates with similar core genomes are more likely to express capsules of the same serogroup than expected from the overall capsular diversity of the isolate collection, even when accounting for isolates sharing the same serotype through common descent.
Greater resolution can be achieved by using the phylogenetic analysis of the whole genome alignments for each of the sequence clusters in which serotype switching occurred (S1 Text & Fig. 3) [25]. These attempt to reconstruct the history of the lineages more accurately by identifying recombination events and excluding the horizontally acquired polymorphisms they introduce from the point mutations used to generate the tree [29]. These reconstructions found that some within-serogroup switching occurred multiple times in parallel in the same sequence cluster: for instance, in SC9 the ancestral type of 23A was replaced by 23B on three occasions, and 23F on three occasions. Similarly within SC13, the ancestral 6A type is exchanged for 6C on two occasions in parallel. In both cases, these examples of convergent evolution are independently supported by the divergence between the cps loci, encoding the same serotype, imported by separate capsule switching events (S1 Fig). A further switch to 6C, this time from 6B, was observed within SC6. However, the other changes within this sequence cluster cannot be reliably inferred as they occur on long branches that are difficult to reconstruct. These ‘missing data’ include at least two between-serogroup switches, but also likely within-serogroup switches given that the cluster includes 23A, 23B and 23F isolates.
Fig. 3. Serotype switching events within sequence clusters. 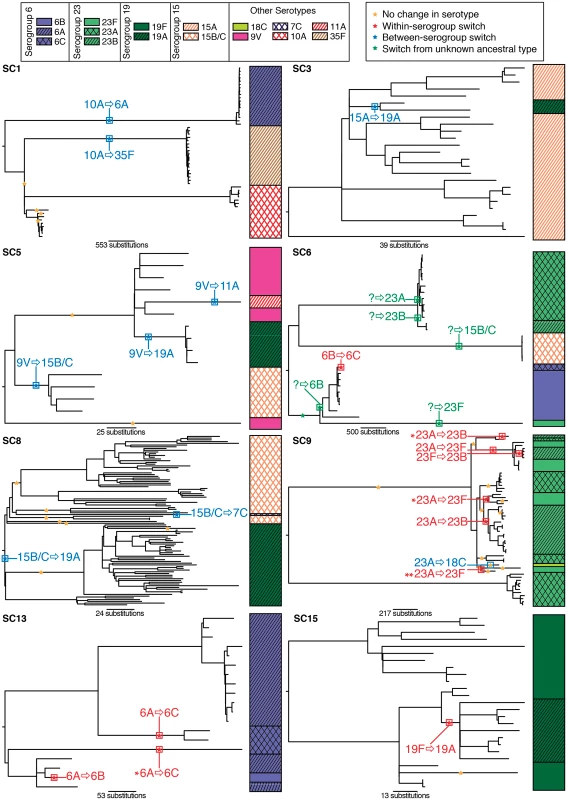
The evolution of each of the monophyletic sequence clusters highlighted in Fig. 1 was reconstructed from a whole genome alignment (see Methods). The resulting phylogenies are annotated with the serotype of the isolates. Stars on the branches of each phylogeny mark instances where putative recombinations affected the cps locus. These stars are coloured according to the inferred consequences of the recombination. Where these sequence imports led to a change in serotype, this is annotated: in red for within-serogroup switches, in blue for between-serogroup switches, and in green, where the ancestral serotype cannot be reliably determined. By contrast, each of the robustly inferred between-serogroup switches occurred only once. Hence this more detailed analysis revealed an even greater tendency to swap a serotype for one within the same serogroup than the previous results. Overall, eleven of the twenty serotype switches (55%) fully defined in this population did not result in a change in serogroup. A further permutation test, in which the derived serotypes were randomly assigned to each ancestral serotype through resampling without replacement 10,000 times (see Methods), found this result to be highly significant (p < 0.0001). Therefore this collection provides robust evidence that within-serogroup switching is more common than expected by chance, relative to the rate of between-serogroup switching.
Length of serotype switching recombinations
The first hypothesis that might explain this observation concerns the mechanics of recombination: the smaller recombinations necessary to alter a cps locus to a related sequence generating a similar serotype may be more frequent than the replacement of the entire locus. The within-serogroup switches observed in this collection all require only minor changes in the cps locus: the serogroup 23 cps loci are thought only to differ in the sequence of their wzy polymerase, as is also the case for serotypes 19A and 19F [35]. Similarly, switching between serotypes 6A and 6B can occur through a single polymorphism in the wciP gene [30], while 6A and 6C only differ in polymorphisms in their wciN gene [35].
Including only the switch from serotype 6B to 6C from SC6 (see S1 Text), the analysis of whole genome alignments used to generate Fig. 3 identified 50 putative homologous recombination events that overlapped the cps loci of the relevant reference genome sequences (Fig. 4). The nine recombinations that resulted in a change in serogroup had a median length of 42.7 kb, and all spanned the entire cps locus. The eleven recombinations that led to a within-serogroup switch had a shorter median length, at 30.4 kb (Fig. 5A). These almost all spanned the 5’ region of the cps locus, but in some cases did not extend so far as the 3’ end. In two cases, this could be ascribed to the presence of the rml rhamnose synthesis operon at the 3’ end of the cps locus. Both the acquisition of the 6B capsule in SC13, and the 6C capsule in SC6, terminated within this gene cluster; however, the rml operon is found in several serogroups [2], and recombinations causing between-serogroup switches have previously been observed to end within it [29]. Only three recombination events were observed where the 3’ recombination breakpoint occurred in a region that might be considered serogroup-specific. Two of these were switches from 23A to 23F within SC9 (Fig. 4). In both cases, the wzy polymerase gene that distinguished these cps loci was replaced; however, the recombinations were far more extensive than this minimal alteration, as they extended to or beyond the 5’ boundary of the cps locus. The switch to 6C within SC13 that ended before the 3’ boundary of the locus was also far more extensive than simply encompassing the wciN gene. The overall difference in the distribution of lengths between recombinations causing within - and between-serogroup switches was not significant (Fig. 5; Wilcoxon rank sum test, W = 35, p value = 0.29). By contrast, the 30 recombinations inferred to overlap with the cps loci that did not affect serotype had a median length of 11.7 kb and were significantly shorter than both the recombinations that alter serogroup (Wilcoxon rank sum test, W = 22, p value = 3.4x10-5) and those that caused within-serogroup switching (Wilcoxon rank sum test, W = 61, p value = 0.0016). As the majority of within-serogroup switches were caused by recombinations long enough to cause changes in serogroup, restrictions on transformation event length cannot fully explain the observed pattern of switching.
Fig. 4. Extent of recombinations affecting the cps locus. 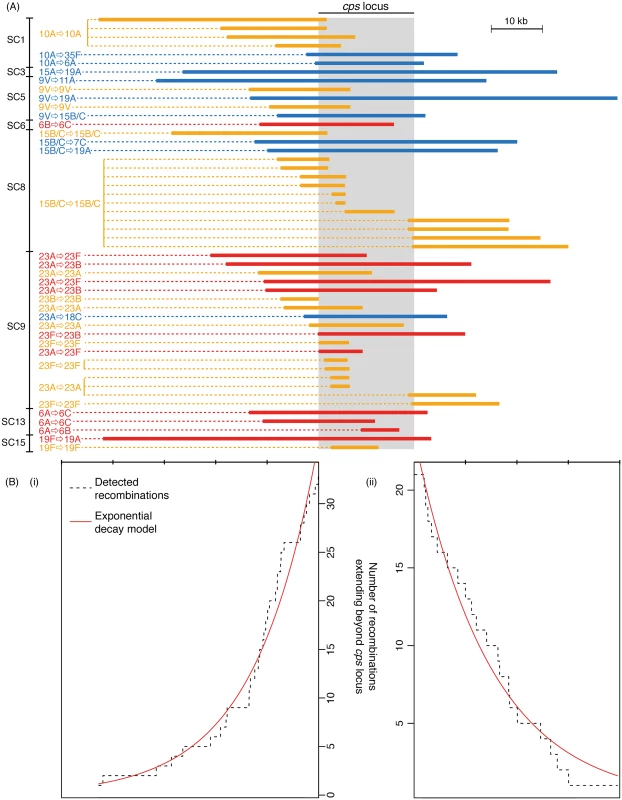
(A) Positions of recombinations affecting the cps locus. Each of the recombinations identified in Fig. 3 is represented by a horizontal bar coloured according to the inferred impact of the recombination. Recombinations causing a change in serotype are uniquely annotated to correspond with Fig. 3. The grey column represents the extent of the cps locus; the width displayed is that of the longest cps locus across the different sequence clusters, and each recombination is scaled relative to the positions of this cps locus’ boundaries. (B) Extent of recombinations affecting the cps locus. The degree to which the recombinations displayed in (A) impinge on the flanking regions is summarised, with the dashed line representing the falling numbers of recombinations extending to bases further removed from the cps locus edges (displayed on the same scale as (A)). The red lines represent the fit of exponential decay curves to these trends. The tick marks above the plots represent 10 kb intervals. Fig. 5. Properties of recombinations affecting the cps locus. 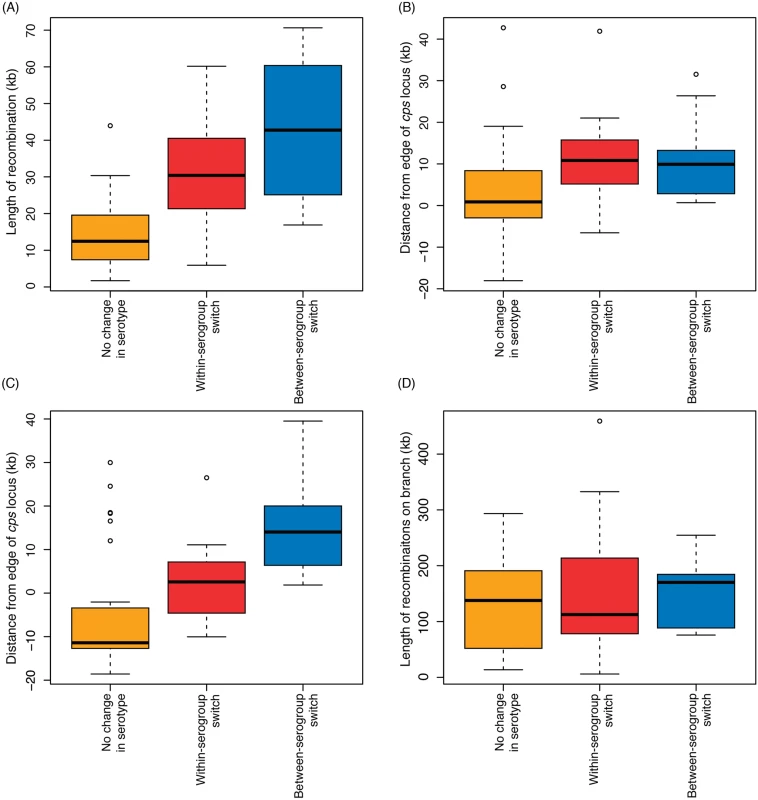
In each boxplot, the recombinations affecting the cps locus are grouped according to their inferred impact on serotype. The first three graphs describe the recombinations affecting the cps locus. The values represent (A) overall length of recombination, (B) position of the 5’ edge of the recombination relative to the 5’ edge of the cps locus, and (C) position of the 3’ edge of the recombination relative to the 3’ edge of the cps locus. In the latter two plots, negative values indicate the recombination boundary is within the cps locus. The boxplot in (D) represents the total length of genome-wide recombinations on the same phylogenetic branch as the serotype switching events. There was extensive variation in the boundaries of the three classes of recombination event. For both 5’ and 3’ breakpoints outside the cps locus, the distance between the breakpoint and the cps locus edge (Fig. 4B) followed an approximately exponential distribution, as previously observed for experimental transformants at the cps locus [28]. However, these observed recombinations were longer than the length of transformation events observed experimentally, with a rate of decay approximately an order of magnitude lower in this work: the rate constant for the decline on the left side was 8.07x10-5 bp-1 (95% confidence interval: 8.06x10-5–8.09x10-5 bp-1), while on the right side it was 6.75x10-5 bp-1 (95% confidence interval: 6.74x10-5–6.77x10-5 bp-1), as opposed to the previous estimates of ~3.4x10-4 bp-1. This is likely partly a consequence of the events described in Fig. 4 being composed long “mosaics” of recombinant DNA segments [28], and also potentially representing larger “macrorecombination” events [36].
The pbp2x and pbp1a genes, crucial determinants of β–lactam resistance, are found 9–10 kb upstream and downstream of the cps locus respectively. Seven of the recombinations shown in Fig. 4 affected both, all of which caused a change of serotype. However, they were split between within - and between-serogroup switches, and none significantly affected the strain’s β–lactam resistance (S2 Table & S2 Text). Hence serotype switches did not appear to be driven by selection for the acquisition of β–lactam resistance. Nevertheless, it is possible that maintenance of a genotype’s β–lactam resistance level may counterselect against recombination events that substantially alter pbp1a or pbp2x [33].
Adaptation between serotype and genotype
If the distribution of capsular variation does not reflect the properties of genetic transformation, then selection may be important in driving the observed pattern. Possible selective pressures include adaptive immune responses in the host population [34], or physiological constraints relating to the bacterium. To test the latter possibility, capsule-switched variants were constructed in common genetic backgrounds and characterised through recording growth curves.
The association of SC9 with serogroup 23 was investigated by knocking out the native cps locus of isolate R34-3029, and using this genetic background to construct one mutant in which the native 23F capsule type was restored, a second through a within-serogroup switch to capsule type 23B, and two further mutants through between-serogroup switches to 18C and 6B. Capsule type 18C was selected as it was the only non-serogroup 23 capsule type observed within SC9, whereas 6B was not found within SC9 but has similar properties to 23F [37]. Both serogroup 23 mutants were found to grow similarly quickly (Fig. 6A) and substantially faster than the two mutants bearing capsules of other serogroups. This suggested that these bacteria may be adapted to serogroup 23 capsules. However, comparisons of the original isolates used as the sources of the cps loci (S2 Fig) indicated that the apparent reduction in fitness of SC9 mutants expressing non-serogroup 23 capsule types might reflect an intrinsically slower growth phenotype associated with these capsule types. Furthermore, when the reciprocal exchange of cps loci between isolates BR1014 and R34-3029 was performed, BR1014 grew faster following restoration of its native 23B capsule relative to the variant into which the 23F capsule had been introduced (Fig. 6A). Therefore changes in capsule type were generally associated with a greater reduction in growth rate than reinstatement of the original capsule type; however, this inhibition could be observed regardless of whether the new serotype was of a different serogroup or not.
Fig. 6. Growth curves comparing the fitness of genetic backgrounds expressing different serotypes. 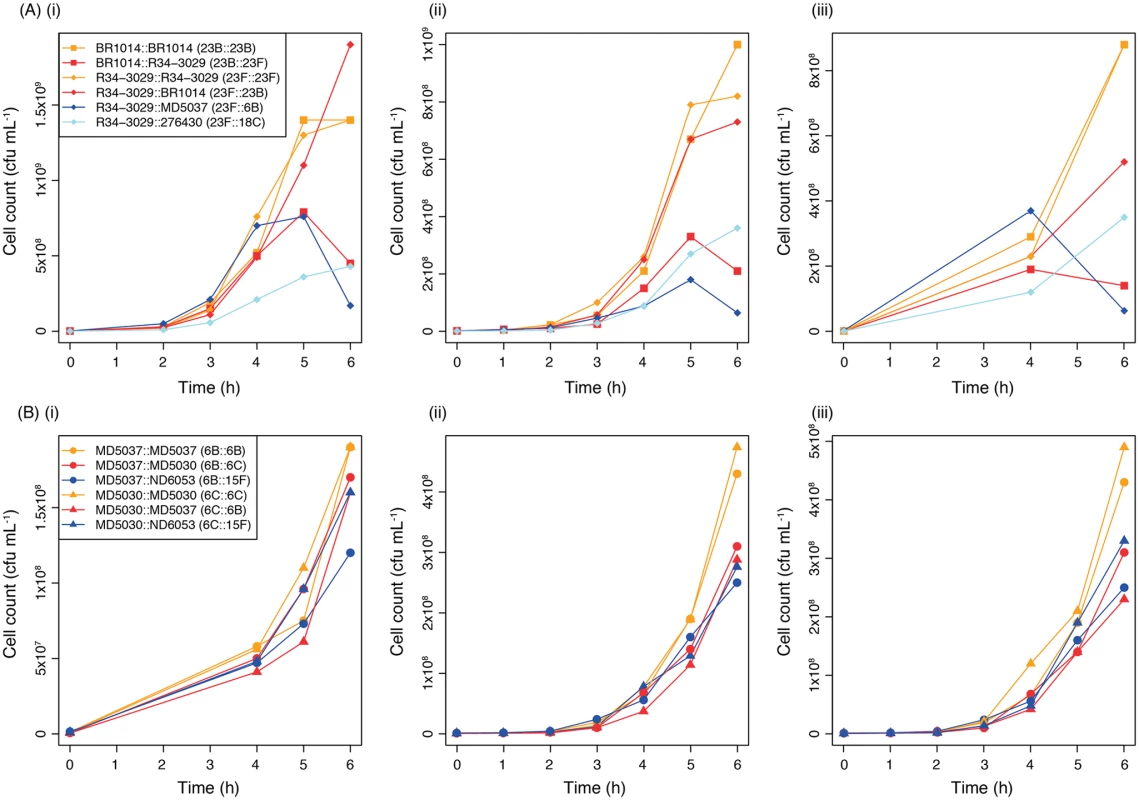
(A) Effect of capsule switching on the growth of two isolates from SC9. Three replicate experiments (i-iii) are shown in which six mutants were compared. Four were constructed based on isolate R34-3029 (diamond points): one in which the native 23F capsule was restored (orange); a within-serogroup switch to the 23B capsule (red); a between-serogroup switch to the 18C capsule (light blue); and a further between-serogroup switch to the 6B capsule (blue). Two other mutants were constructed based on isolate BR1014 (square points): one in which the native 23B capsule was restored (orange), and a within-serogroup switch to 23F (red). (B) Effect of capsule switching on the growth of two isolates from SC13. Three replicate experiments (i-iii) are shown in which six mutants were compared. Three mutants were based on isolate MD5037 (circular points): a restoration of the native 6B capsule (orange); a within-serogroup switch to the 6C capsule (red); and a between-serogroup switch to the 15F capsule (blue). The other three mutants were based on isolate MD5030 (triangular points): a restoration of the native 6C capsule (orange), a within-serogroup switch to the 6B capsule (red); and a between-serogroup switch to the 15F capsule (blue). To test this hypothesis further, six additional mutants were constructed using two SC13 isolates in which the cps locus had been knocked out: MD5037, originally of 6B, and MD5030, originally of 6C (Fig. 6B). The 6B, 6C and 15F capsule types were introduced into both. This comparison allowed a more precise test for interactions between the cps locus and the rest of the genome than the experiments with SC9 isolates, as SC13 isolates exhibited more consistent growth patterns, and the selected donor of the 15F capsule had a similar growth curve to the recipient isolates (S2 Fig). Consistent with the observations of SC9, the mutants with the restored native capsule grew fastest. However, both the within-serogroup and between-serogroup switched variants exhibited similarly inhibited growth rates. Hence all three capsule types replicated optimally in their native backgrounds, and more slowly when introduced into a non-native background. Therefore the only epistasis between the cps locus and the rest of the pneumococcal genome that could be inferred from these data was specific to serotype, not serogroup, and no evidence was found that the observed predominance of within-serogroup switches is explained by serogroup-specific adaptations.
Potential for epistatic interactions
It remains possible that epistasis affects other phenotypes that are not easily assayed in the laboratory; it may be possible to infer these from the genomic data. One factor likely to impact on the range of capsules a strain may successfully express is its complement of carbohydrate transporters [38,39], many of which have been functionally characterised [38] and are only present in a subset of the population (S3 Fig & S3 Table). While capsules in different serogroups usually have distinct chemical compositions, capsules within the same serogroup often consist of the same moieties connected by different bonds. Hence strains may only be adapted to importing and processing certain carbohydrates at high enough rates to sustain capsule production, thereby inhibiting the acquisition of a capsule type with a divergent chemical composition. However, the distribution of transporters across the population does not provide an obvious explanation for the stable association between sequence clusters and serogroups (S3 Fig). SCs 9, 13 and 15 did not appear to have a smaller number of carbohydrate transporters than those in which a change of serogroup had occurred, nor relative to those not having changed serotype. The associated serogroups can be distinguished by the presence of ribitol (serogroup 6), N-acetylmannosamine (serogroup 19) and glycerol (serogroup 23). Ribitol is synthesised as part of teichoic acid common to all pneumococci; glycerol is imported by GlpF, encoded by a gene ubiquitous across the sample; and N-acetylmannosamine is known to be a substrate for four transporters that are common to all isolates, and one that is absent from SC15. These data do not support the hypothesis that the observed pattern of switching is the result of a constricted ability to acquire the requisite carbohydrate molecules, although epistatic interactions with other aspects of carbohydrate metabolism cannot be excluded.
Yet this analysis only uses experimentally characterised loci, which cannot encompass novel systems that may be rare in the population. It is possible in principle to identify epistatic interactions between any loci from sequence data [40]. In bacteria, interacting accessory genome loci should be present within the same genomes more often than expected by chance, after accounting for linkage and clonal structure. Three sequence clusters, each essentially an independent genotype [41], were universally of serogroup 6 (SC10, SC13 and SC14). A search for clusters of orthologous genes (COGs) that were ubiquitous in these sequence clusters but absent from sequence clusters with no serogroup 6 representatives identified three COGs, all of which corresponded to genes within the serogroup 6 cps loci. This failure to identify serogroup 6 associated genes outside the cps locus is in keeping with the experimental data that suggests adaptation is specific to a serotype, not a serogroup. Testing for association with serotype requires that the same capsule be acquired multiple times in parallel, as in SC9. Each triallelic polymorphic site in SC9 was tested for association of an allele with one of the three serogroup 23 serotypes. Twelve such sites were identified, all of which were within the cps locus. Additionally, no COGs in the accessory genome associated perfectly with any of these individual serotypes. Therefore these simple analyses provided no evidence of sites outside the cps locus that might epistatically interact with the expressed capsule type.
A more generic strategy for identifying potential epistatic interactions was not to look for associations of specific sequences with particular cps loci, but instead quantify the overall extent of changes accompanying serotype switches. If one or more non-cps loci improved the fitness of a novel serotype variant, then it might be expected that serotype switches would be associated with elevated rates of change across the rest of the genome. In order to control for the general heterogeneity in the rates of pneumococcal recombination [36], the total lengths of the recombinations occurring on the same phylogenetic branch as serotype switches (both within - and between-serogroups) were compared with those occurring on branches on which recombinations affected the cps locus but did not cause a change in serotype (Fig. 5D). This found that similar levels of contemporaneous (or near-contemporaneous) recombination occurred whether or not there was a change in serotype or serogroup. Additionally, all three types of recombination affecting the cps locus were typically associated with sufficient numbers of base substitutions being imported by recombination (S4 Fig) to saturate the mismatch repair system [42]. Hence, there was no significant evidence of non-cps loci being exchanged to facilitate the acquisition of a new serotype or serogroup, nor of mismatch repair having an impact on the patterns of serotype switching.
Discussion
Previous study of this collection of carried pneumococci, based on serotyping and MLST data, identified a borderline-significant tendency for serotype switches to occur within serogroups [21]. The availability of whole genome-based phylogenies increases the number of switches that can be investigated within the same set of isolates through identification of two classes of switch invisible to the MLST-based method: changes in serotype along a lineage in which the multilocus sequence type has also changed (e.g. SC9), and also multiple independent switches within the same sequence type (e.g. in SC13, two switches to 6C within MLST 473). This increases the sample size of identified serotype switching events from nine to twenty. Also, these data provide more reliable phylogenetic information on the ancestral serotype: previously, this was based on the temporal order in which combinations of serotype and genotype were observed, which can be misleading in certain cases, such as SC9 [25].
Future population genomic datasets will determine how general the preponderance of within-serogroup switching is across the pneumococcal species; a recently published dataset of over 3,000 genomes systematically sampled from a Thai refugee camp, in which hosts were unvaccinated, provides an opportunity to test the reproducibility of the pattern [43]. Applying the simple permutation test to the distribution of serotypes between sequence clusters in which switching had occurred, as for the dataset described in this study, revealed the ratio of serogroup concordant to serotype-discordant comparisons to be 0.14 (p = 0.0006 from 10,000 permutations). The serotypes in this sample are markedly different from those in the Massachusetts dataset; hence this enrichment of within-serogroup switching provides independent support for the original observation. Additionally, although only one within-serogroup switch was observed in SC15 in this dataset, the acquisition of the 19A capsule in place of the 19F type has occurred at least three further times in related isolates [33].
Multiple hypotheses were proposed as potential explanations for the observed frequency of within-serogroup switches. The first was that the pattern was caused by the constraints of genetic transformation, with short recombination events resulting in partial alteration of the cps locus being more frequent. The suggestion that most recombinations are too short to be likely to cause a change in serotype was supported by the large number of ‘silent’ recombinations affecting the cps locus, which did not change the antigenic profile of the bacterium. While some cases are likely to reflect exchanges of sequence between distinct isolates of the same serotype, many of these events affect the 5’ or 3’ boundaries of the cps locus that are the most strongly conserved across different cps loci, and therefore may originate in donors with a very different cps locus to that of the recipient. As these exchanges do not result in antigenic changes, it may be that they represent neutral diversification, or serve a role in repairing deleterious mutations. This potential for recombination between serotypes that does not cause switching should be borne in mind when designing sequence-based serotyping methodologies that do not directly target the polymorphisms within cps loci that cause differences in capsule type.
Nevertheless, recombinations causing within-serogroup switching sometimes did not span the entire cps locus, as all the between-serogroup switches did. In three cases, the recombinations were sufficiently restricted in their extent so as to end within a serogroup-specific part of the cps locus; however, these were still substantially larger than the putative minimal genetic changes that could cause the same alteration of serotype. Furthermore, the enrichment for within-serogroup switching remained highly significant even when the events that did not span the entire cps locus were excluded; although this does not rule out this mechanism contributing to the observed pattern of switching, this result demonstrates it is not the sole reason underlying it. Finally, the within-serogroup switches did not appear to represent a ‘neutral’ set of typical recombination lengths, as they were still significantly larger than those which caused no alteration of serotype overall, although there may be a limit on the length of such recombinations before they are likely to stop being ‘silent’. One possibility not considered here, subtly different to a dependence on the absolute length of a recombination, is that restriction modification systems could be limiting the transfer of cps loci. Although restriction endonucleases typically do not affect transformation, as DNA is imported in a single stranded form, in cases where genomic islands are imported, the synthesis of a complementary strand once the DNA is integrated into the genome can render the acquired locus sensitive to endonucleolysis [44]. Loci imported from a different serogroup would have greater sequence divergence, and therefore a greater set of potential target motifs that could be subject to endonucleolysis. However, this will remain difficult to assess until our knowledge of the diverse set of pneumococcal restriction-modification systems has improved [41].
Conversely, there was no evidence that serotype switching recombinations were selected for the lengths to which they affected the regions flanking the cps locus and the pbp genes. Co-incidence of serotype switches with changes in β–lactam resistance [32] was rare and there was little evidence of recombinations being constrained in the extent to which they extended into the regions flanking the cps locus. However, there are uncertainties associated with ascertaining these recombination breakpoints. One is that the evolutionary time represented by the long branches on which the changes of serogroup occur within SC1 may be sufficient to permit several rounds of transformation. This could result in overlapping recombinations being erroneously inferred to represent a single event, thereby increasing the apparent length of the recombined sequence. However, as both events affected by this bias were between-serogroup switches, this cannot account for within-serogroup switches being longer than the minimum length required for the phenotypic change. Conversely, multiple switches to the same serotype in parallel produce extensive homoplasy. Inaccurate reconstruction of such homoplasies could result in individual actual recombination events being reconstructed as multiple fragmentary events split between different branches of the phylogeny. This risks artificially shortening serotype-switching recombinations. However as this bias only affects sequence clusters in which similar cps loci are acquired multiple times, which also happen to be those in which within-serogroup switching is most common (SC9 and SC13), any such errors are conservative with regard to the finding that shorter recombinations alone cannot account for the observed pattern of within-serogroup switching. One potential example is switch 23A→23F**, as the two cps loci apparently imported by this recombination are not particularly closely related (S1 Fig); hence this inferred recombination may in fact represent only a partial fragment of two separate serotype switching recombinations. If this were correct, it would increase the frequency of within-serogroup switches further, and suggest that the short recombination currently annotated as the single switch 23A→23F** actually corresponds to two separate, longer events that cannot be resolved with the current collection. This ambiguity highlights how unlikely it is that the numbers of parallel switching events have been overestimated as a consequence of incorrect evolutionary reconstructions. Given the substantial numbers of polymorphisms caused by capsule switching recombinations in the region of the cps locus, independent acquisitions in parallel are unlikely to be inferred without strong evidence of distinct ancestry from the rest of the chromosome. Furthermore, the diversity of the cps loci themselves, not directly used in the phylogenetic inference displayed in Fig. 3, independently support the same reconstructed pattern of switches occurring in parallel within SC9 and SC13.
As limitations to recombination do not appear to explain the observed pattern of switching, selection for certain combinations of cps loci and genetic backgrounds may explain the distribution of capsule across the population. Metabolic or physiological adaptation between the cps locus and other aspects of the pneumococcal genotype may result in a limited range of capsule types that can be successfully expressed by a given genomic background. Although serotype itself affects growth rate [45], this study found that the rate of growth in vitro depended on both the serotype and backbone genotype in a non-additive way. Isolates grew fastest when expressing their native capsule, although R34-3029 grew similarly fast when expressing either of the tested serogroup 23 capsule loci. However, in most cases there was no difference in the level of growth inhibition when capsules of the same, or a different, serogroup were introduced, particularly when accounting for the growth curves of the donors and recipients involved in each exchange. This is consistent with the existence of epistatic interactions between the cps locus and the rest of the genome, but not with the hypothesis that such interactions were serogroup-specific. It could still be that the changes required to adapt to the acquisition of a more similar serotype are relatively small, and more likely to occur through recombination or mutation before the capsule variant is selected out of the population. Yet over the timescales easily testable in the laboratory, no evidence was found for a mechanism that would explain the frequency of within-serogroup switching.
One potential confounding problem with these experiments is the chance that non-cps loci could be co-transferred, and therefore influence the fitness of the resulting capsule-switched recombinants. However, previous experiments have suggested that random co-transfer of other sequence should not systematically affect the growth rates of transformants [28]. Furthermore, it might be expected that any co-transfer from the cps locus donor might facilitate adaptation to the acquired serotype, if there were epistatic interactions between the cps locus and the rest of the genome. However, the genomic data provided no evidence of natural serotype switches being associated with unusually high levels of sequence exchanges around the rest of the genome, suggesting the co-transfer of other loci is not likely to facilitate adaptation to an altered capsule type. Furthermore, there were no signs of non-cps loci associated with pneumococci of specific serogroups or serotypes in a manner suggestive of interactions between distinct loci in the same chromosome. Therefore, the genomic and experimental data do not support the model of simple, strong epistatic interactions determining the range of serogroups that an isolate can successfully express.
One longstanding hypothesis for the maintenance of “strain structure” in recombining pathogens is that host immunity produces strains that maintain discordant sets of antigens [34]. Here, individual capsular antigens found across serogroups might be associated with a particular set of subcapsular antigens, creating epistasis in the presence of host immunity that would not be observable in vitro. Alternatively, opportunities for recombination may be disproportionately available for strains in the same serogroup as a result of serogroup-wide immunity making hosts either susceptible or resistant to colonization with particular serogroups. Both these mechanisms rely on the assumption that anticapsular antibodies are protective against carriage, and that at least part of this protection is by responses that target epitopes shared across serotypes within a serogroup. Whether this could significantly impact the rate of genetic exchange between serotypes remains an open question. The existence of cross-protection by naturally-acquired anticapsular antibodies within a serogroup is made plausible by the observation of protection against serotype 6A from the 6B component of PCV7, and by the existence of cross-reactive antibodies used in serotyping. However, we are aware of no direct evidence for protection across a serogroup by naturally-acquired anticapsular antibodies. Furthermore, the protective immunity induced by conjugate vaccines is not serogroup-wide, as demonstrated by the post-PCV7 success of some vaccine-related serotypes, such as 19A, 23A, 23B and 6C [16].
Expanded sampling in the future may allow for these and other hypotheses to be tested further, as well as permitting more detailed searches for loci that may interact epistatically with the cps locus. Such information regarding which serotypes can be readily ‘interchanged’ may well prove important in understanding the responses to past conjugate vaccine introductions, and predicting the response to future interventions.
Methods
Permutation tests
When calculating the significance of the association of serogroups with sequence clusters, the first test resampled each of the serotypes listed in S1 Table without replacement until each sequence cluster had been assigned the same number of serotypes as observed in the actual dataset. This test was therefore independent of the phylogenies and reconstructed patterns of serotype switching. Ten thousand such permutations were carried out, and in each case the calculated test statistic was the ratio of serogroup-concordant to serotype-discordant pairwise comparisons within sequence clusters. Any pairwise comparison between identical serotypes was ignored, as it would be serologically undetectable. While recombinations between isolates of the same serotype at the cps locus may be detected in this dataset as a subset of those events that do not alter the expressed capsule type, it is not possible to easily distinguish these from recombinations donated from isolates only of the same serogroup, or of a completely unrelated serotype, which may also not affect the recombinant’s expressed capsule type, depending on how conserved different parts of the cps locus are across pneumococci of different serotypes. The p value was calculated as the proportion of permutations for which the calculated test statistic was greater than, or equal to, the value observed in the actual population (0.41). When applied to the Maela dataset, the ‘secondary BAPS’ groupings were used as the equivalent of sequence clusters.
The second permutation test used information from the phylogenetic reconstructions of serotype switching. The input dataset was the twenty switches for which the ancestral and derived serotypes could both be defined (Fig. 3). Ten thousand permutations were performed in which the ancestral and derived serotypes were sampled, without replacement, from those in the input dataset. In each case, the test statistic was the proportion of serotype switches that were serogroup concordant, and the p value calculated as the proportion of the permutations in which this statistic was equal to, or greater than, the observed value (0.55).
Reconstruction of serotype changes
The phylogenies used to identify serotype switches have been reported previously [25]. Briefly, a reference genome was assembled de novo for each sequence cluster, against which Illumina reads were aligned, and polymorphisms identified [28]. Sequences imported by putative recombination events were identified and removed, and a maximum likelihood phylogeny generated from the remaining clonal frame, as described [29] and validated [46] previously. The annotation of putative mobile genetic elements allowed the number of base substitutions introduced by point mutation, homologous recombination and non-homologous recombination to be estimated. All lengths and positions of recombination events are therefore relative to the reference sequences against which the Illumina reads were originally mapped. The identification of putative homologous recombination events associated with serotype switching is detailed in the supplement S1 Text; this also describes the assessment of within-serogroup sequence diversity, and the reconstruction of changes in serotype, achieved using the maximum likelihood approach implemented in the APE R package [47]. This approach, as well as a maximum parsimony method, was also used to reconstruct the emergence of β–lactam resistance using the trees based on the clonal frame, once each isolate had been classified as either ‘resistant’ (benzylpenicillin minimum inhibitory concentration >0.06 μg mL-1) or ‘sensitive’. The details of this analysis are also described in S1 Text and S2 Table.
Construction of in vitro serotype switches
The cps locus of isolates was first knocked out using a Janus cassette [48] as described previously [49]. Transformation was conducted using genomic DNA from the serotype donor and the competence stimulating peptide appropriate to the genotype. After 2 h growth, selection was performed using plates containing 500 μg ml-1 streptomycin. Colonies were replica plated to ensure loss of kanamycin resistance and expression of the altered serotype confirmed through latex agglutination (Statens Serum Institut, Copenhagen).
Comparisons of growth
Isolates were grown to mid-log phase in THY (Todd Hewitt broth containing 0.5% yeast extract), titered and frozen at -70°C in 10% glycerol. Growth was compared in parallel by inoculating 6 mL of THY with a starting concentration of 106 mL-1 bacteria from freshly thawed frozen stock. Cultures were incubated at 37°C with 5% CO2. At successive time points 200 μL was removed to a 96 well plate. Optical density at a wavelength of 610 nm was measured and dilutions were plated on blood agar plates to calculate live cell densities.
Supporting Information
Zdroje
1. Kadioglu A, Weiser JN, Paton JC, Andrew PW (2008) The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 6 : 288–301. doi: 10.1038/nrmicro1871 18340341
2. Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, et al. (2006) Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2: e31. 16532061
3. Park IH, Park S, Hollingshead SK, Nahm MH (2007) Genetic basis for the new pneumococcal serotype, 6C. Infect Immun 75 : 4482–4489. 17576753
4. Calix JJ, Nahm MH (2010) A new pneumococcal serotype, 11E, has a variably inactivated wcjE gene. Journal of Infectious Diseases 202 : 29–38. doi: 10.1086/653123 20507232
5. Calix JJ, Porambo RJ, Brady AM, Larson TR, Yother J, et al. (2012) Biochemical, Genetic, and Serological Characterization of Two Capsule Subtypes among Streptococcus pneumoniae Serotype 20 Strains. Journal of Biological Chemistry 287 : 27885–27894. doi: 10.1074/jbc.M112.380451 22736767
6. Salter SJ, Hinds J, Gould KA, Lambertsen L, Hanage WP, et al. (2012) Variation at the capsule locus, cps, of mistyped and non-typable Streptococcus pneumoniae isolates. Microbiology 158 : 1560–1569. doi: 10.1099/mic.0.056580-0 22403189
7. Bratcher PE, Kim KH, Kang JH, Hong JY, Nahm MH (2010) Identification of natural pneumococcal isolates expressing serotype 6D by genetic, biochemical and serological characterization. Microbiology 156 : 555–560. doi: 10.1099/mic.0.034116-0 19942663
8. Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS (2010) The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect Immun 78 : 704–715. doi: 10.1128/IAI.00881-09 19948837
9. Lipsitch M, Whitney CG, Zell E, Kaijalainen T, Dagan R, et al. (2005) Are anticapsular antibodies the primary mechanism of protection against invasive pneumococcal disease? PLoS medicine 2: e15. 15696204
10. Weinberger DM, Dagan R, Givon-Lavi N, Regev-Yochay G, Malley R, et al. (2008) Epidemiologic evidence for serotype-specific acquired immunity to pneumococcal carriage. J Infect Dis 197 : 1511–1518. doi: 10.1086/587941 18471062
11. Hill PC, Cheung YB, Akisanya A, Sankareh K, Lahai G, et al. (2008) Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian infants: a longitudinal study. Clin Infect Dis 46 : 807–814. doi: 10.1086/528688 18279039
12. Goldblatt D, Hussain M, Andrews N, Ashton L, Virta C, et al. (2005) Antibody responses to nasopharyngeal carriage of Streptococcus pneumoniae in adults: a longitudinal household study. Journal of Infectious Diseases 192 : 387–393. 15995951
13. Cobey S, Lipsitch M (2012) Niche and neutral effects of acquired immunity permit coexistence of pneumococcal serotypes. Science 335 : 1376–1380. doi: 10.1126/science.1215947 22383809
14. Ghaffar F, Barton T, Lozano J, Muniz LS, Hicks P, et al. (2004) Effect of the 7-valent pneumococcal conjugate vaccine on nasopharyngeal colonization by Streptococcus pneumoniae in the first 2 years of life. Clinical Infectious Diseases 39 : 930–938. 15472842
15. Rinta-Kokko H, Dagan R, Givon-Lavi N, Auranen K (2009) Estimation of vaccine efficacy against acquisition of pneumococcal carriage. Vaccine 27 : 3831–3837. doi: 10.1016/j.vaccine.2009.04.009 19490983
16. Huang SS, Hinrichsen VL, Stevenson AE, Rifas-Shiman SL, Kleinman K, et al. (2009) Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics 124: e1–11. doi: 10.1542/peds.2008-3099 19564254
17. Millar EV, O'Brien KL, Watt JP, Bronsdon MA, Dallas J, et al. (2006) Effect of community-wide conjugate pneumococcal vaccine use in infancy on nasopharyngeal carriage through 3 years of age: a cross-sectional study in a high-risk population. Clin Infect Dis 43 : 8–15. 16758412
18. Lipsitch M (1997) Vaccination against colonizing bacteria with multiple serotypes. Proc Natl Acad Sci U S A 94 : 6571–6576. 9177259
19. Lipsitch M (1999) Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg Infect Dis 5 : 336–345. 10341170
20. Spratt BG, Greenwood BM (2000) Prevention of pneumococcal disease by vaccination: does serotype replacement matter? Lancet 356 : 1210–1211. 11072934
21. Hanage WP, Bishop CJ, Huang SS, Stevenson AE, Pelton SI, et al. (2011) Carried pneumococci in Massachusetts children: the contribution of clonal expansion and serotype switching. Pediatr Infect Dis J 30 : 302–308. doi: 10.1097/INF.0b013e318201a154 21085049
22. Robinson KA, Baughman W, Rothrock G, Barrett NL, Pass M, et al. (2001) Epidemiology of invasive Streptococcus pneumoniae infections in the United States, 1995–1998: Opportunities for prevention in the conjugate vaccine era. JAMA 285 : 1729–1735. 11277827
23. Hanage WP, Finkelstein JA, Huang SS, Pelton SI, Stevenson AE, et al. (2010) Evidence that pneumococcal serotype replacement in Massachusetts following conjugate vaccination is now complete. Epidemics 2 : 80–84. doi: 10.1016/j.epidem.2010.03.005 21031138
24. Black S, Shinefield H, Fireman B, Lewis E, Ray P, et al. (2000) Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Northern California Kaiser Permanente Vaccine Study Center Group. Pediatr Infect Dis J 19 : 187–195. 10749457
25. Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, et al. (2013) Population genomics of post-vaccine changes in pneumococcal epidemiology. Nature genetics 45 : 656–663. doi: 10.1038/ng.2625 23644493
26. Wyres KL, Lambertsen LM, Croucher NJ, McGee L, von Gottberg A, et al. (2013) Pneumococcal Capsular Switching: A Historical Perspective. Journal of Infectious Diseases 207 : 439–449. doi: 10.1093/infdis/jis703 23175765
27. Feil EJ, Smith JM, Enright MC, Spratt BG (2000) Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154 : 1439–1450. 10747043
28. Croucher NJ, Harris SR, Barquist L, Parkhill J, Bentley SD (2012) A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog 8: e1002745. doi: 10.1371/journal.ppat.1002745 22719250
29. Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, et al. (2011) Rapid pneumococcal evolution in response to clinical interventions. Science 331 : 430–434. doi: 10.1126/science.1198545 21273480
30. Croucher NJ, Hanage WP, Harris SR, McGee L, van der Linden M, et al. (2014) Variable recombination dynamics during the emergence, transmission and 'disarming' of a multidrug-resistant pneumococcal clone. BMC biology 12 : 49. doi: 10.1186/1741-7007-12-49 24957517
31. Hausdorff WP, Feikin DR, Klugman KP (2005) Epidemiological differences among pneumococcal serotypes. The Lancet Infectious Diseases 5 : 83–93. 15680778
32. Trzcinski K, Thompson CM, Lipsitch M (2004) Single-step capsular transformation and acquisition of penicillin resistance in Streptococcus pneumoniae. J Bacteriol 186 : 3447–3452. 15150231
33. Croucher NJ, Chewapreecha C, Hanage WP, Harris SR, McGee L, et al. (2014) Evidence for soft selective sweeps in the evolution of pneumococcal multidrug resistance and vaccine escape. Genome Biology and Evolution 6 : 1589–1602. doi: 10.1093/gbe/evu120 24916661
34. Gupta S, Maiden MC, Feavers IM, Nee S, May RM, et al. (1996) The maintenance of strain structure in populations of recombining infectious agents. Nature medicine 2 : 437–442. 8597954
35. Mavroidi A, Aanensen DM, Godoy D, Skovsted IC, Kaltoft MS, et al. (2007) Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. Journal of bacteriology 189 : 7841–7855. 17766424
36. Mostowy R, Croucher NJ, Hanage WP, Harris SR, Bentley S, et al. (2014) Heterogeneity in the Frequency and Characteristics of Homologous Recombination in Pneumococcal Evolution. PLOS Genetics 10: e1004300. doi: 10.1371/journal.pgen.1004300 24786281
37. Weinberger DM, Trzcinski K, Lu YJ, Bogaert D, Brandes A, et al. (2009) Pneumococcal capsular polysaccharide structure predicts serotype prevalence. PLoS Pathog 5: e1000476. doi: 10.1371/journal.ppat.1000476 19521509
38. Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, et al. (2012) A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One 7: e33320. doi: 10.1371/journal.pone.0033320 22428019
39. Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, et al. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293 : 498–506. 11463916
40. Kryazhimskiy S, Dushoff J, Bazykin GA, Plotkin JB (2011) Prevalence of epistasis in the evolution of influenza A surface proteins. PLOS Genetics 7: e1001301. doi: 10.1371/journal.pgen.1001301 21390205
41. Croucher NJ, Coupland PG, Stevenson AE, Callendrello A, Bentley SD, et al. (2014) Diversification of bacterial genome content through distinct mechanisms over different timescales. Nature communications 5 : 5471. doi: 10.1038/ncomms6471 25407023
42. Humbert O, Prudhomme M, Hakenbeck R, Dowson CG, Claverys J-P (1995) Homeologous recombination and mismatch repair during transformation in Streptococcus pneumoniae: saturation of the Hex mismatch repair system. Proceedings of the National Academy of Sciences 92 : 9052–9056. 7568071
43. Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, et al. (2014) Dense genomic sampling identifies highways of pneumococcal recombination. Nature genetics 46 : 305–309. doi: 10.1038/ng.2895 24509479
44. Johnston C, Martin B, Granadel C, Polard P, Claverys J-P (2013) Programmed protection of foreign DNA from restriction allows pathogenicity island exchange during pneumococcal transformation. PLoS pathogens 9: e1003178. doi: 10.1371/journal.ppat.1003178 23459610
45. Hathaway LJ, Brugger SD, Morand B, Bangert M, Rotzetter JU, et al. (2012) Capsule type of Streptococcus pneumoniae determines growth phenotype. PLoS pathogens 8: e1002574. doi: 10.1371/journal.ppat.1002574 22412375
46. Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. (2014) Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic acids research: gku1196.
47. Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20 : 289–290. 14734327
48. Sung C, Li H, Claverys J, Morrison D (2001) An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Applied and environmental microbiology 67 : 5190–5196. 11679344
49. Trzcinski K, Thompson CM, Lipsitch M (2003) Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Applied and environmental microbiology 69 : 7364–7370. 14660386
Štítky
Genetika Reprodukčná medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell LongevityČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 3- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



