-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
COE transcription factors are conserved across widely divergent animals and are crucial for organismal development. COE genes also play roles in adult animals and have been implicated in central nervous system (CNS) diseases; however, the function of COE in the post-embryonic CNS remains poorly understood. Planarian regeneration provides an excellent model to study the function of transcription factors in cell differentiation and in terminally differentiated cells. In planarians, coe is expressed in differentiating and mature neurons, and its function is required for CNS regeneration. In this study, we show that coe is required to maintain structure and function of the CNS in uninjured planarians. We took advantage of this phenotype to identify genes regulated by coe by comparing global gene expression changes between control and coe mRNA-deficient planarians. This approach revealed downregulated genes downstream of coe with biological roles in CNS function. Expression analysis of downregulated genes uncovered previously unknown candidate targets of coe in the CNS. Furthermore, functional analysis of downstream targets identified coe-regulated genes required for CNS regeneration. These results demonstrate that the roles of COE in stem cell specification and neuronal function are active and indispensable during CNS renewal in adult animals.
Published in the journal: COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians. PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004746
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004746Summary
COE transcription factors are conserved across widely divergent animals and are crucial for organismal development. COE genes also play roles in adult animals and have been implicated in central nervous system (CNS) diseases; however, the function of COE in the post-embryonic CNS remains poorly understood. Planarian regeneration provides an excellent model to study the function of transcription factors in cell differentiation and in terminally differentiated cells. In planarians, coe is expressed in differentiating and mature neurons, and its function is required for CNS regeneration. In this study, we show that coe is required to maintain structure and function of the CNS in uninjured planarians. We took advantage of this phenotype to identify genes regulated by coe by comparing global gene expression changes between control and coe mRNA-deficient planarians. This approach revealed downregulated genes downstream of coe with biological roles in CNS function. Expression analysis of downregulated genes uncovered previously unknown candidate targets of coe in the CNS. Furthermore, functional analysis of downstream targets identified coe-regulated genes required for CNS regeneration. These results demonstrate that the roles of COE in stem cell specification and neuronal function are active and indispensable during CNS renewal in adult animals.
Introduction
The Collier/Olfactory-1/Early B-cell factor (COE) family of transcription factors is necessary for animal development. COE proteins possess an atypical HLH domain and a unique zinc finger DNA binding domain conserved across metazoans [1]. Invertebrates encode a single homolog of COE, with roles in mesoderm and ectoderm development [2], [3], whereas vertebrates have four COE paralogs with functions in diverse cell types including B-cells and adipocytes [4]. In the central nervous system (CNS), COE regulates neuronal differentiation, migration, axon guidance, and dendritogenesis during development [2], [3], [5]–[13] and maintains neuronal identity throughout adulthood [14], [15]. COE proteins have also been proposed to function as tumor suppressors [16] and are associated with cancers such as acute lymphoblastic leukemia and glioblastoma [17]–[20]. However, the specific genetic programs regulated by these genes in adult stem cells and mature neurons remain poorly understood.
Stem cells can be studied to determine how transcriptional regulators orchestrate developmental processes or cause disease [21]. An excellent animal model to investigate stem cell regulation in vivo is the freshwater planarian Schmidtea mediterranea [22]. S. mediterranea has the ability to regenerate all tissue types from a population of adult stem cells (called neoblasts). These cells constitute approximately 10–20% of all the cells in the animal and include pluripotent [23] and lineage-committed neoblasts [24]–[29]. The planarian CNS is composed of two cephalic ganglia and a pair of ventral nerve cords that run along the length of the animal, which are comprised of molecularly diverse neuronal subtypes that are regenerated within days after injury or amputation [30]–[32]. Functional analysis of transcription factors in planarians using RNA interference (RNAi) has begun to identify regulatory molecules required for the generation and maintenance of specific neuronal subpopulations in the CNS such as serotonergic and cholinergic neurons [24]–[27], [33]–[35]. Thus, planarians are outstanding organisms to study basic mechanisms that underlie stem cell-based maintenance and regeneration of the adult CNS.
A previous functional screen for transcription factors encoding a helix-loop-helix domain identified a planarian coe homolog that is expressed in a small population of neural-committed stem cells (approximately 4–7% of the neoblast pool) and in neurons [24]. We showed that animals fed dsRNA designed to silence coe expression (coe(RNAi) animals) regenerated abnormal brains; furthermore, uninjured coe(RNAi) planarians displayed behavioral defects and reduced expression of neural subtype-specific genes [24]. In this study, we sought to identify genes regulated by coe with roles in CNS renewal by comparing the transcriptome profiles of uninjured control and coe(RNAi) animals, uncovering differentially expressed genes with predicted roles in CNS function. We validated a subset of these genes by testing for loss of expression after coe knockdown and visualizing their expression in coe+ cells. These analyses revealed a set of nine candidate targets of coe in adult neurons, many of which are important for neuronal subtype identity (e.g., ion channels, neuropeptides, and neurotransmitters). In addition, our findings demonstrate that coe functions to drive gene expression in multiple neuronal classes, including excitatory and inhibitory neurons. To gain insights into the roles candidate COE targets play in CNS turnover and repair, we analyzed the function of downregulated transcripts using RNAi. Our functional screen identified several genes required for CNS regeneration, including homologs of a voltage-gated sodium channel α-subunit (scna-2) and the transcription factor pou4l-1. Our results suggest that COE is required for the expression of neural-specific genes in differentiating and mature neurons, a function that is essential to maintain CNS architecture and regulate neuronal regeneration.
Results/Discussion
coe is required for maintenance of nervous system structure
Using an optimized whole-mount in situ hybridization protocol (WISH) (see Materials and Methods), we found that coe mRNA was primarily restricted to neurons in S. mediterranea (Fig. 1A). In agreement with our previous findings [24], we also observed coe transcripts in a subset of cycling stem cells (h2b+) (Fig. 1B–C). We previously reported that coe(RNAi) animals regenerate cephalic ganglia that fail to connect at the anterior commissure and have significantly smaller brains with fewer cpp-1+, npp-4+, and npy-2+ neurons when compared to the controls [24]. This defect is not restricted to the anterior portion of the animal. Additional experiments showed coe(RNAi) animals do not properly regenerate their ventral nerve cords (Fig. S1A–B). Moreover, analysis of the brain patterning defect using anti-VC-1, a marker of the photoreceptor neurons and their axons, revealed that the optic chiasm failed to connect at the midline in coe(RNAi) animals (Fig. S1C). These data demonstrate that coe is essential for neuronal regeneration at both anterior and posterior facing wounds and that coe regulates genes required for reestablishing midline patterning following brain amputation.
Fig. 1. coe is expressed in the nervous system and a subset of cycling stem cells. 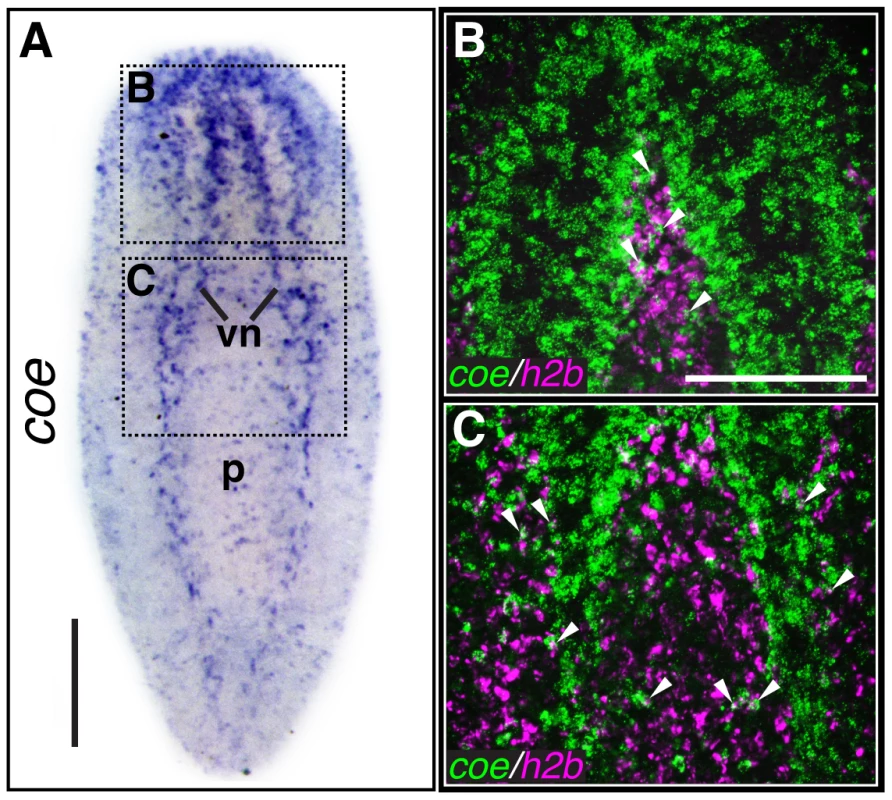
(A) In situ hybridization to coe in S. mediterranea (vn, ventral nerve cords; p, pharynx). Dashed boxes show regions imaged in B–C (N≥10). (B–C) Double-fluorescent in situ hybridization to coe and h2b. Arrowheads mark examples of double-labeled cells (N = 14). Anterior is up in all panels. Scale bars, A = 200 µm, B = 100 µm. In addition, we previously noted that silencing of coe in intact uninjured animals results in a reduction of ChAT+ and pc2+ neurons near the anterior commissure and a loss of cpp-1+ neurons. Following the 6th feeding of coe dsRNA, 100% of the animals exhibited impaired negative phototaxis [24]. To investigate the specificity of the coe knockdown phenotype on the CNS, we examined the effect of coe RNAi on the intestine and muscle as representative endodermal or mesodermal tissues, respectively. We hybridized uninjured control and coe(RNAi) animals with riboprobes specific to ChAT (as a positive control), mat [36], and collagen [37]. As expected, we observed a decrease in ChAT+ neurons in the head [24] and noted a decrease in ChAT expression throughout the animal (Fig. 2A); by contrast, we did not observe a change in the spatial distribution of mat or collagen following coe knockdown (Fig. 2B–C). To quantify the effect of coe RNAi treatments on the expression of ChAT, mat and collagen, we measured relative mRNA levels by reverse transcription quantitative PCR (RT-qPCR). First, we confirmed coe knockdown led to a significant decrease in the relative expression of coe mRNA (down 60%±16% compared to the controls; Fig. 2D). Measurement of ChAT, mat and collagen from coe(RNAi) planarians revealed that ChAT mRNA levels were significantly down (45%±15%) compared to control animals; in contrast to ChAT, the relative mRNA levels of mat or collagen were not affected by coe RNAi treatment (Fig. 2D). Combined with our previous work [24], these results strongly suggest that coe knockdown specifically affects gene transcription in the nervous system and does not cause obvious defects in other tissues such as the intestine or muscle. Furthermore, our results are consistent with reports demonstrating that COE is required to maintain cholinergic and peptidergic neuronal subtype-specific gene expression in Caenorhabditis elegans and Drosophila melanogaster [14], [15].
Fig. 2. coe RNAi strongly inhibits the expression of ChAT in intact planarians. 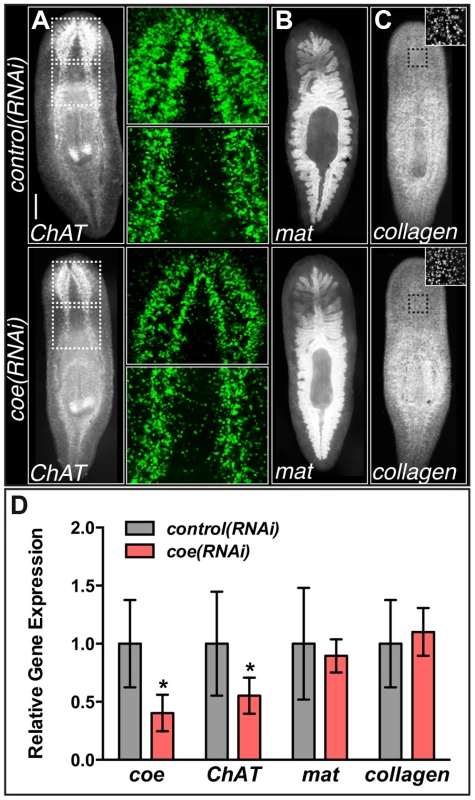
(A–C) coe RNAi-treated animals were processed for fluorescent in situ hybridization (FISH) to ChAT (N = 10 for each treatment), mat (N = 3 control and 4 RNAi planarians), or collagen (N = 7 control and 5 RNAi). White dashed boxes in A denote regions imaged at higher magnification shown in the panels to the right. Black dashed boxes in C denote regions imaged at higher magnification shown in top right insets. (D) RT-qPCR experiments measuring the relative expression of coe, ChAT, mat, or collagen in control(RNAi) or coe(RNAi) planarians following the 6th RNAi treatment. Graph shows the mean ± s.d. expression levels relative to the controls. *P<0.05, Student's t-test. To investigate if the inhibition of coe perturbs nervous system architecture downstream of gene expression changes, we labeled neuronal cell bodies and their projections using anti-CRMP-2, which labels a subset of neuronal cell bodies and their axon projections, and anti-β-tubulin to visualize nerve projections (Fig. 3A–C). In coe(RNAi) animals, we observed a striking decrease in axon projections labeled by anti-CRMP-2 and anti-β-tubulin compared to the controls; however, expression of CRMP-2 was retained in the cell bodies (Fig. 3C). In addition, when we labeled sensory neurons using cintillo [38], coe(RNAi) animals exhibited significantly fewer cintillo+ cells (Fig. 3D). Our results strongly suggest that nervous system architecture is severely reduced or lost in the absence of coe. These structural defects likely underlie the behavioral abnormalities observed in coe-deficient planarians.
Fig. 3. COE function is required for maintenance of nervous system architecture in uninjured planarians. 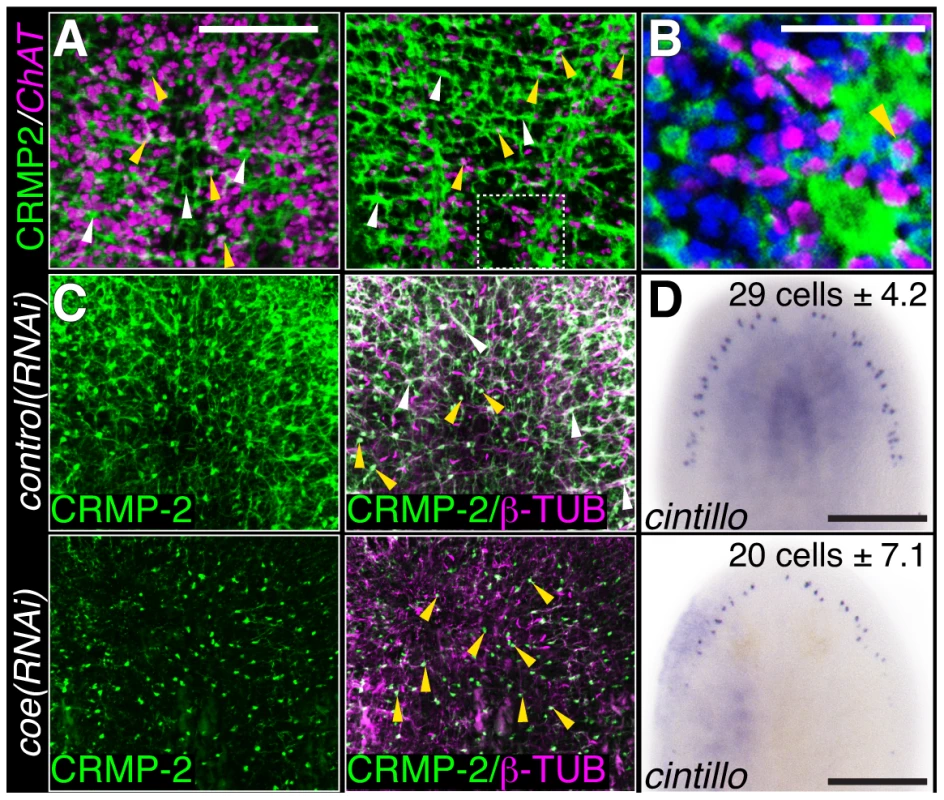
(A) Head or tail images from an animal stained with anti-CRMP-2 and processed for FISH to ChAT. CRMP-2 is expressed in axon projections (white arrows) and neuronal cell bodies (yellow arrows; N = 7). (B) Higher magnification image of region denoted by white box in D shows CRMP-2 is detected in ChAT+ cell bodies (arrowhead). Nuclei were stained with DAPI (blue). (C–D) Uninjured control and coe(RNAi) planarians labeled with anti-CRMP-2 and anti-β-TUBULIN or processed for in situ hybridization to cintillo. White and yellow arrows point to axon projections and cell bodies, respectively. N = 8 animals for each treatment; 412 and 290 cintillo+ cells were counted from control and coe(RNAi) animals, respectively. The number in the top right corner indicates the mean ± s.d. of cintillo+ cells; *P<0.05, Student's t-test. Anterior is up in all panels. Scale bars, A = 200 µm, D = 100 µm, E = 50 µm, and G = 200 µm. Identification of genes regulated by coe in the planarian nervous system
Although COE has been shown to drive differentiation of several classes of neurons during development [39], the transcriptional programs controlled by this transcription factor in adult nervous system function are poorly defined. We reasoned that the CNS-specific coe RNAi phenotype in intact planarians represents an excellent opportunity to identify gene expression programs controlled by COE in the post-embryonic nervous system. Thus, we used comparative mRNA sequencing (RNA-seq; see Materials and Methods) to sequence mRNAs isolated from uninjured controls and coe(RNAi) animals one week after the 6th RNAi treatment, which was the point in time we consistently observed behavioral defects and loss of neural-specific gene expression in 100% of coe-deficient animals and did not detect overt defects in other tissues (Fig. 2). RNA-seq analysis identified 909 differentially expressed genes; 397 were downregulated, and 512 were upregulated (Table S1). Functional annotation using DAVID software showed that the set of downregulated genes was significantly enriched for Gene Ontology (GO) terms associated with “ion channel,” “neuronal activities,” “nerve-nerve synaptic transmission,” “voltage-gated ion channel,” and “cell adhesion molecule”; by contrast, the upregulated genes were enriched for GO terms associated with “cytoskeletal protein” and “muscle development” (Table 1). coe mRNAs were not detected in a muscle pattern (Fig. 1), nor did we detect overt phenotypes associated with muscle differentiation (Fig. 2). However, the RNA-seq data raised the possibility that coe might negatively regulate mesoderm specification, which is required for muscle development [3], [40]. It is possible upregulation of muscle genes is an indirect consequence of a loss of nervous system influence such as cholinergic transmission and/or neuropeptide regulation. Previous studies have demonstrated cholinergic neurotransmission is required for coordinated muscle contractions in planarians [41]–[43]. Thus, we speculate that loss of nervous system modulation disrupts muscle homeostasis and leads to changes in expression of muscle-related genes. Although our experiments do not definitively assign the role of COE in muscle differentiation or maintenance, our data do clearly indicate that coe is required for expression of nervous system-specific genes in adult planarians.
Tab. 1. Annotation of genes differentially expressed in <i>coe(RNAi)</i> animals using DAVID software. 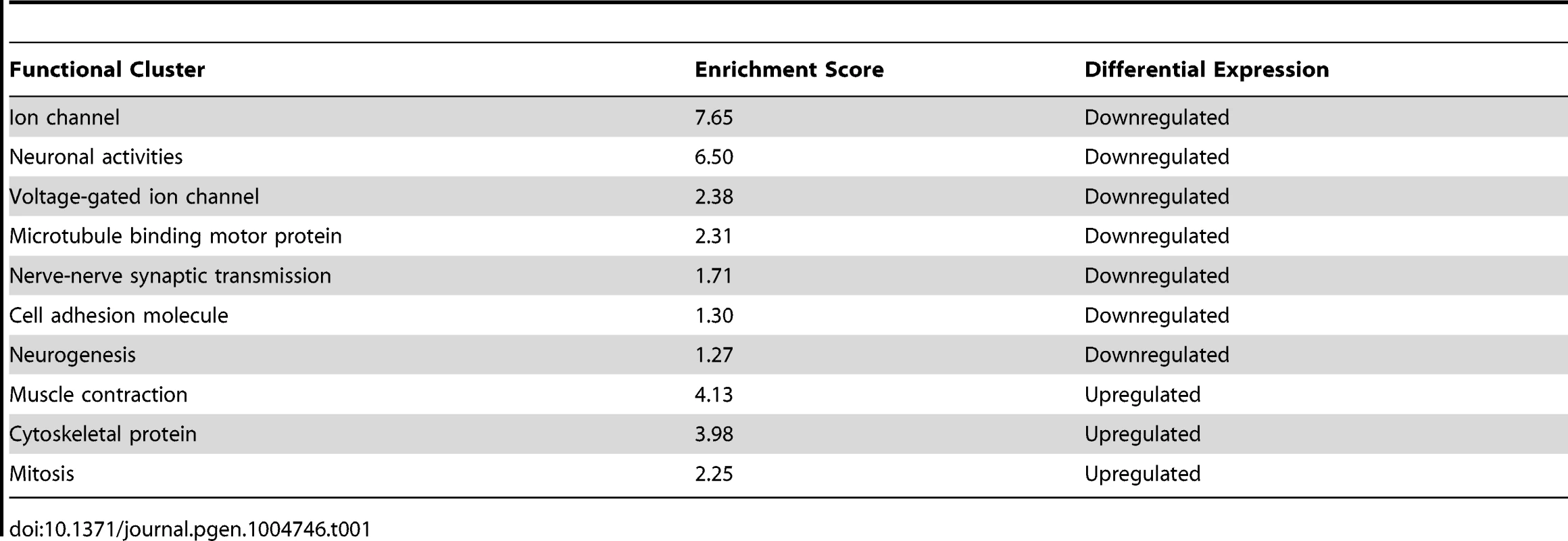
Based on the annotation of differentially expressed genes, we hypothesized that genes predicted to play roles in nervous system functions in the downregulated category likely include direct COE targets. To test our hypothesis and validate genes found in our RNA-seq dataset, we selected 65 genes that were dramatically downregulated, associated with neural functions, or annotated as transcription factor homologs. First, we performed WISH to determine the tissue-specific pattern of expression of all 65 genes (representative examples are shown in Fig. 4). As we expected, the most prominent mRNA expression pattern was in the nervous system (26 of 65 genes; see Table S2), similar to ChAT and cpp-1, which we had previously found to be putative downstream targets of COE [24]. In addition, we observed genes that were expressed broadly in the nervous system (such as neural cell adhesion molecule-2 (ncam-2), vesicle-associated membrane protein like-1 (vamp), gamma-aminobutyric acid receptor subunit beta like-1 (gbrb-1), and voltage-gated sodium channel alpha-1 (scna-1)) or in discrete neuronal subpopulations (such as secreted peptide prohormone-19, -18, -2 (spp-19, -18, -2), neuropeptide like (npl), voltage-gated sodium channel alpha-2 (scna-2), and caveolin-1 (cav-1)) (Fig. 4A–J). Our list also included transcripts that labeled subsets of neurons in the brain (such as netrin-1) (Fig. 4K) [44]. In addition, we found that the transcription factors iroquios-1 (irx-1) and pou class 4 transcription factor 4 like-1 (pou4l-1) were expressed at or near the cephalic ganglia (Fig. 4L–M), and their mRNA was detected in ChAT+ neurons by fluorescent in situ hybridization (FISH) (Fig. S2). Next, we tested the effect of coe RNAi on the expression of 33 genes that could be visualized in discrete cell populations by WISH. Knockdown of coe led to a marked reduction in the expression of 31 genes (Table S2; representative results are shown in Fig. 4A′–H′, K′–M′); for two genes, scna-2 and cav-1, we observed a loss of expression at the midline (Fig. 4I′–J′). Furthermore, we quantified the number of cells labeled by spp-19, spp-18, and npl probes. As expected, we found there was a significant reduction in the number of spp-19+, spp-18+, and npl+ cells following coe RNAi (Fig. 4N).
Fig. 4. The expression pattern of nervous system genes downstream of COE is changed or severely reduced following coe RNAi. 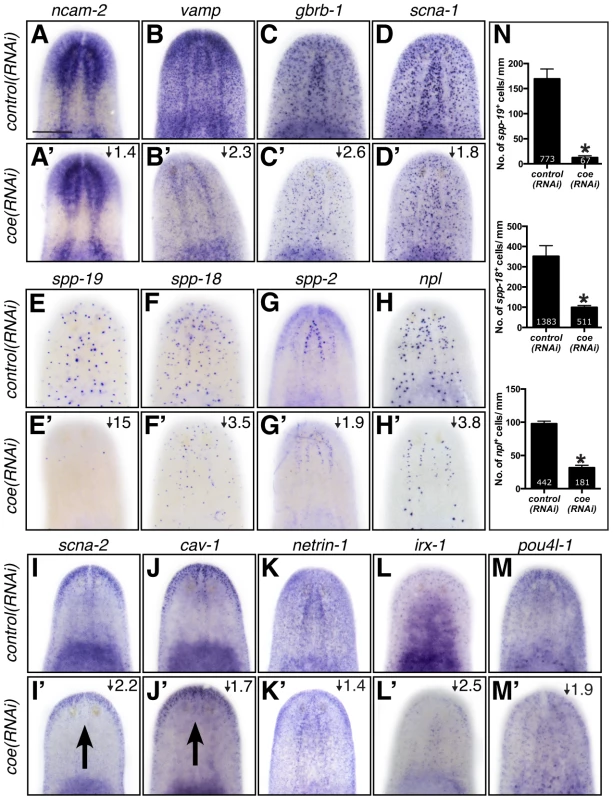
Control and coe(RNAi) treated animals were processed for in situ hybridization to the genes indicated above each panel (N≥5 animals per treatment). (A–M′) Detection of all genes was reduced following coe knockdown. Numbers in top right corner represent linear fold changes in mRNA expression in coe(RNAi) planarians relative to the controls. Arrows in I′ and J′ point to loss of expression at the midline compared to the controls (I and J). (N) Quantification of spp19+, spp-18+, and npl+ cells (N = 3); the total number of cells counted is indicated within each bar. Error bars in all graphs are s.d. from the mean; *P<0.05, Student's t-test. Anterior is up in A–M′. Scale bar in A = 100 µm. As an additional test to validate the in situ hybridization results, we measured the relative expression levels of downregulated genes in control and coe RNAi-treated planarians using RT-qPCR (Fig. S3A). All of the genes we tested showed a decrease in relative expression following coe RNAi (9 of 14 genes were significantly downregulated; P<0.05, Student's t-test). By contrast, when we measured the relative expression of CNS-expressed genes that were not on our list of differentially expressed genes, none were significantly reduced (11 of 11 genes; Fig. S3B–C). Although some of the control genes we selected were reduced near levels comparable to some genes downregulated following coe RNAi (e.g., ncam2, vamp, and gbrb1; Fig. S3A), we noted that isotig13897 and npp-2 [30], which are transcripts detected in subsets of neurons or throughout the CNS, respectively, remained unchanged (Fig. S3B–C). It is possible that some changes in gene expression associated with coe RNAi are consequence of a reduction in nervous system tissue. We proceeded to perform double-FISH to coe and validated genes to determine if any were potential genetic targets of COE. Of the 17 genes we were able to reliably detect by FISH (33 genes were tested; see Table S2), 11 were expressed in coe+ cells (representative results are shown in Fig. 5 and Fig. S4), including ChAT and cpp-1 [24]. Together, these results identified nine novel candidate targets of COE in the nervous system, including genes important for maintaining neuronal subtype identity such as ion channels, ion channel receptors, and neuropeptide genes (Table 2). In addition, our data suggest that COE is essential to maintain genetic programs in multiple classes of adult neuronal subtypes including excitatory (cholinergic) and inhibitory (GABAergic) neurons.
Fig. 5. Identification of genes expressed coe+ neurons. 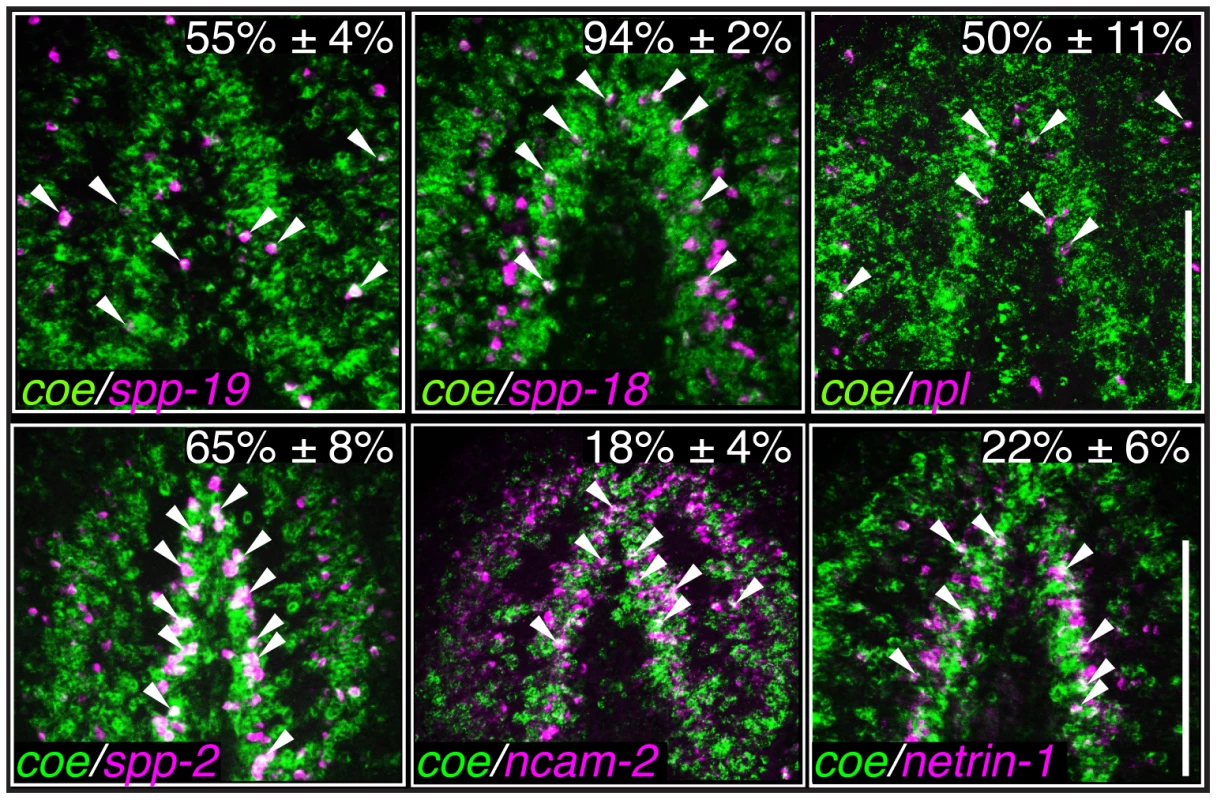
Fluorescent in situ hybridization to coe and either spp-19, spp18, npl, spp-2, ncam-2, or netrin-1. Percentages indicate the proportion ± s.d. of cells that were also coe+ (N = 110 spp-19+, 319 spp-18+, 173 npl+, 202 spp-2, 236 ncam-2, and 141 netrin-1 cells counted from 2–3 animals per group). Arrowheads mark double-labeled cells. Anterior is up in all panels. Scale bars = 100 µm. Tab. 2. Candidate COE targets genes identified in <i>S. mediterranea</i>. 
Genes downstream of coe are required for proper CNS regeneration
Our RNA-seq dataset revealed that coe is essential to maintain the expression of hundreds of genes in the adult animal. This change in the neuronal gene expression landscape led to abnormal CNS structure and behavior. To identify genes downstream of coe that contribute to CNS differentiation, we took advantage of the experimental ease in examination of gene function in planarian regeneration and analyzed the role of 11 downregulated genes that were expressed in neurons or predicted to encode transcription factors (Table 3). Following RNAi, animals were amputated pre - and post-pharyngeally and allowed to regenerate for 10 days. We found that 6 out of 11 genes resulted in defective brain regeneration (see Table 3); scna-2, pou4l-1, and nkx2l caused the strongest phenotypes. Compared to the controls, scna-2(RNAi) animals had less eye pigmentation or developed a single eyespot; nkx2l(RNAi) animals exhibited photoreceptor defects; and pou4l-1(RNAi) animals had less photoreceptor pigment (Fig. 6A–D). To examine CNS architecture, we stained scna-2, nkx2l, and pou4l-1 RNAi treated planarians with anti-SYNAPSIN and the coe-regulated genes ChAT and npl. Although subtle, all three showed abnormalities in brain morphology (Fig. 6A–D). However, when we measured the area of the brain stained by anti-SYNAPSIN, only scna-2 and pou4l-1 RNAi animals had a significant reduction in neuropil density (Fig. 6E). Consistent with this observation, the ChAT+ brain areas were smaller in scna-2(RNAi) and pou4l-1(RNAi) animals (Fig. 6F) but not in nkx2l(RNAi) animals. The smaller brain phenotype was accompanied by fewer npl+ neurons in scna-2(RNAi) animals; however, despite their smaller brains, pou4l-1(RNAi) animals regenerated significantly more npl+ cells than controls (Fig. 6G). These findings demonstrate that scna-2 is required for CNS regeneration and highlight the importance of ion channels in neurogenesis regulation during CNS development, maintenance, and repair [45]–[47]. Interestingly, these data suggest that pou4l-1 plays a role in the specification of certain neuronal lineages. It is possible that in the absence of pou4l-1, planarians regenerate the incorrect proportion of neuronal subtypes and have disorganized brains, but this possibility will require further analysis with additional neuronal subtype-specific markers. By contrast, our results suggest nkx2l is not required for CNS regeneration per se. Following coe RNAi, nkx2l expression was reduced by in situ hybridization and RT-qPCR (Table S2 and Fig. S3A), but nkx2l, which is primarily expressed in stem cells and in progeny [48], was not detected in the nervous system (Fig. S5A). We hypothesize nkx2l functions in early regeneration to establish patterning, which is consistent with the observation that nkx2l(RNAi) planarians fail to regenerate properly patterned head (Fig. 6C) and tail tissues (Fig. S5B).
Fig. 6. CNS regeneration defects following knockdown of COE-regulated genes. 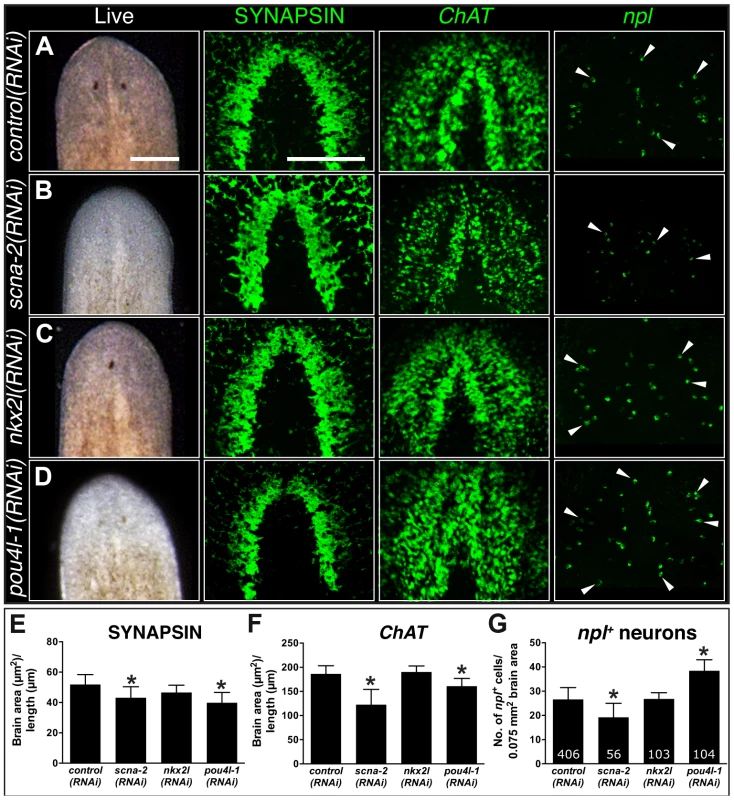
(A–D) Animals were fed control, scna-2, nkx2l and pou4l-1 bacterially-expressed dsRNA (indicated to the left of each panel), amputated pre-pharyngeally and allowed to regenerate. Ten-day regenerates were imaged live (A–D), killed and immunostained with anti-SYNAPSIN or processed for fluorescent in situ hybridization to ChAT or npl (N≥4). (E–F) Brain size estimated by measuring head area stained by anti-SYNAPSIN or in situ hybridization to ChAT and normalized by the length of animal for control, scna-2, nkx2l, and pou4l-1 RNAi planarians. (G) Quantification of npl+ cells normalized by brain size measured from ChAT stain in F (N≥4 animals in each group); the total number of npl+ cells counted is indicated within each bar. Error bars in all graphs are s.d. from the mean; *P<0.05, Student's t-test. Anterior is up in A–D. Scale bars = 100 µm. Tab. 3. Functional analysis of genes downregulated following coe RNAi. 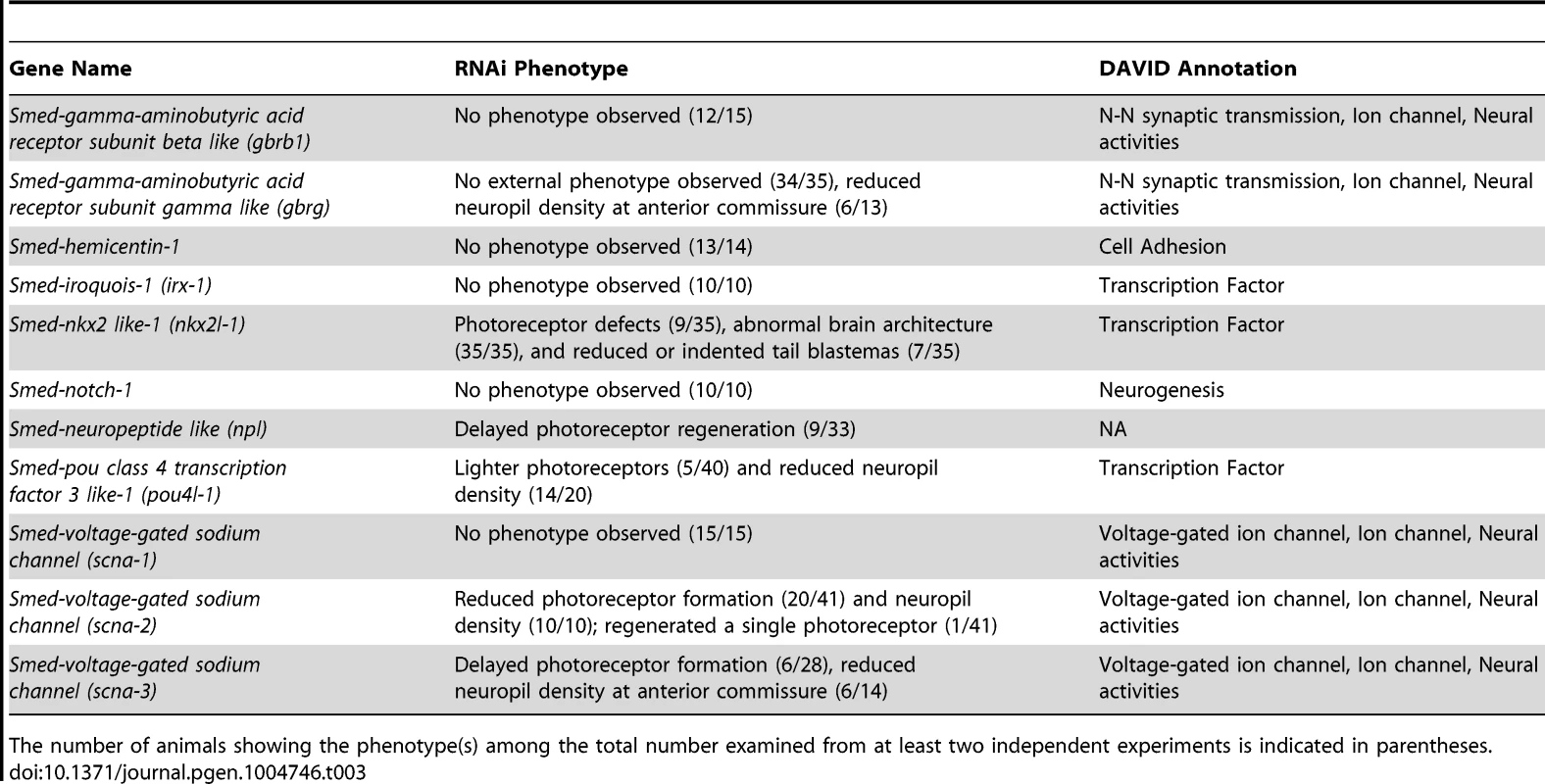
The number of animals showing the phenotype(s) among the total number examined from at least two independent experiments is indicated in parentheses. It is noteworthy that several transcription factors that we identified in our screen are putative COE targets in Xenopus development, including irx-1, tal, pou4l-1, and nkx2l [39]. Of these genes, we found that expression of pou4l-1 was important for CNS regeneration and nkx2l was involved in patterning. NKX and POU orthologs play critical roles during CNS development of invertebrate and vertebrate organisms [49]–[51]. These data suggest that regulatory genes downstream of COE are conserved and have roles in CNS regeneration. However, it will be important to experimentally resolve whether these transcription factors are bona fide targets of COE in planarians or other animals such as Xenopus.
Concluding remarks
COE proteins are known to function as terminal selectors of neuronal identity in adult organisms [14], [15], [52], yet the neuronal subtypes and specific genetic programs regulated by COE in the adult CNS are not well understood. In this study, we exploited the high rate of tissue turnover and regenerative capacity of planarians to expand our understanding of how COE may function in the post-embryonic nervous system. We combined RNAi with RNA-seq analysis and identified a set of differentially expressed genes associated with nervous system biological roles. Expression analysis of a subset of these genes revealed novel candidate targets of coe in planarian neurons (Fig. 7A), some of which underscored coe's essential role in maintaining expression of genes vital for neuronal subtype identity and function (such as neurotransmitter receptors, ion channels, and neuropeptide encoding genes) (Fig. 7A–B). Decoding which transcriptional changes are direct or indirect consequences of coe loss in the planarian model will be vital to further elucidate how mutations in COE proteins cause or contribute to disease pathologies in the CNS. The next step will be to find direct COE binding sites genome-wide using in silico and chromatin immunoprecipitation (ChIP) approaches and combining these findings with our differential expression data. In addition, molecular profiling of coe+ cell populations (such as stem cells, postmitotic progeny, and neurons) will be essential to determine how coe function alters in cell type-specific contexts. In conclusion, our study demonstrates the importance of COE family proteins in neuronal turnover and repair of the adult CNS and broadens our understanding of the regulatory programs governed by these factors.
Fig. 7. COE function is required for differentiation and maintenance of diverse neuron types. 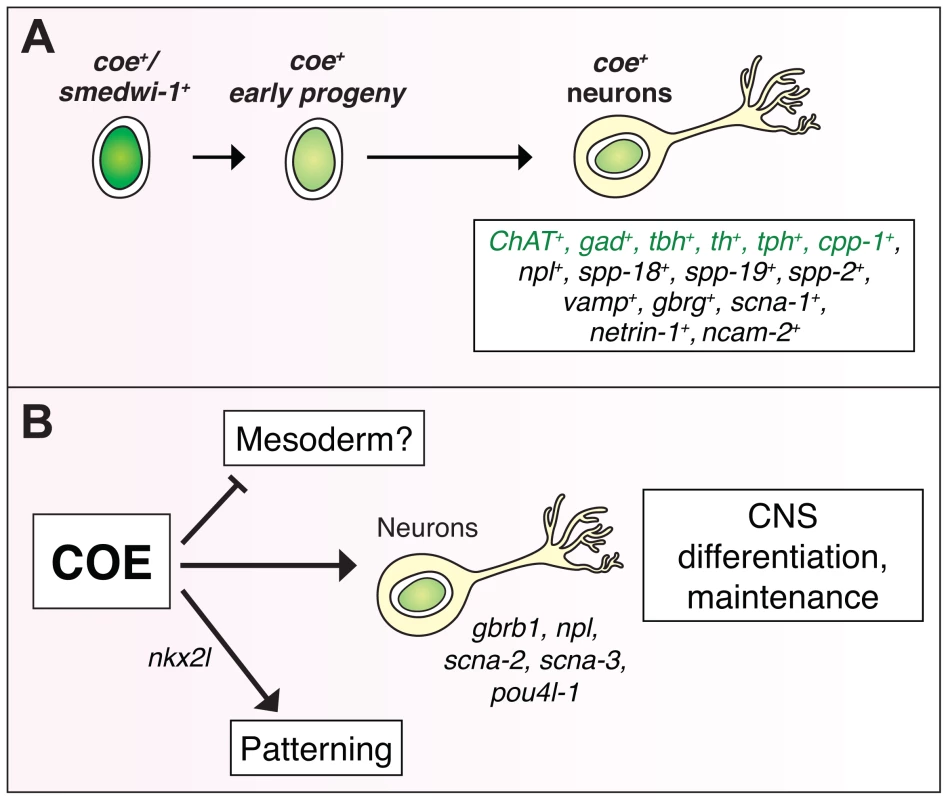
(A) coe is expressed in lineage-committed neoblasts (smedwi+) and early progeny [24], and diverse neuron types, including cholinergic (ChAT), GABAergic (gad), octopaminergic (tbh), dopaminergic (th), serotonergic (tph), and neuropeptidergic (cpp-1, npl, spp-18, spp-19, spp-2) neurons. Genes in green were identified in [24]. (B) To gain insights into how loss of COE function contributes to defects in nervous system differentiation, we analyzed the function of genes that were downregulated in coe(RNAi) animals. These analyses identified additional genes required for CNS regeneration (gbrb1, npl, scna-2, scna-3, pou4l-1) and patterning (nkx2l). In coe(RNAi) animals, we also detected upregulated genes enriched for GO terms associated with muscle development (Table 1), suggesting that COE may also function to repress the expression of mesoderm-specific genes. Materials and Methods
Animal husbandry
Asexual Schmidtea mediterranea (CIW4) were reared in 1× Instant Ocean Salts (0.83 mM MgSO4, 0.9 mM CaCl2, 0.04 mM KHCO3, 0.9 mM NaHCO3, and 0.21 g/L Instant Ocean Aquarium Salt diluted in ultra-pure water) at 20°C. Animals were starved for one week, and those ranging between 2–5 mm in length were used for experimentation.
RNA interference
Animals were administered six feedings of bacterially expressed dsRNA complementary to the indicated gene over three weeks as previously described [53]; gfp dsRNA was fed as a control. Unless otherwise indicated, all intact RNAi animals were fixed seven days following the 6th dsRNA treatment. For regeneration experiments, planarians were amputated pre - and post-pharyngeally 24 hours following the 6th dsRNA feeding.
Whole-mount in situ hybridizations and immunostainings
Animals were processed for colorimetric whole-mount in situ hybridization using the protocol described in [54]. Fluorescent in situ hybridization experiments were performed as described in [24], [54] and developed using Tyramide Signal Amplification (TSA) as described in [55]. Briefly, animals were incubated for 5 min. in borate buffer (100 mM borate pH 8.5, 0.1% Tween-20) and then developed in TSA Reaction Buffer (borate buffer, 2% dextran sulfate, 0.1% Tween-20, 0.003% H2O2), containing fluor-tyramide and 4-iodophenylboronic acid for 30 min. For double-FISH, animals were quenched in 1% H2O2 for 1 hour. For γ-irradiation experiments, animals were fixed 6 days following a 100 Gy treatment, a time point when both stem cells and postmitotic progenitors are ablated. Accession numbers for the sequences used in this study are listed in Table S3. For immunostaining with anti-SYNORF1 (1∶400, 3C11, DSHB) or anti-VC-1 (1∶10,000; kindly provided by Hidefumi Orii), animals were fixed with Carnoy's solution [56]. For anti-CRMP-2 (1∶50, 9393S, Cell Signaling) or anti-β-TUBULIN (1∶1000; E7, DSHB) labeling, animals were fixed with formaldehyde, processed without a reduction step, and labeled using TSA [54].
RNA sequencing and DAVID analysis
One week after the final dsRNA treatment, RNA was extracted from three independent control and coe(RNAi) animal groups using Trizol (Life Technologies). RNA samples were treated with DNase using the Turbo DNA-free Kit (Life Technologies) and purified using the RNeasy MinElute Cleanup kit (Qiagen). Sequencing libraries were synthesized using the TruSeq RNA Sample Prep Kit v2 and sequenced on a HiSeq 2000 System (Illumina). More than 12 million 100-bp single-end reads were generated for each sample. Sequenced reads were submitted to the Sequence Read Archive (NCBI) under the accession number PRJNA235907. Reads were mapped to the planarian genome using TopHat [57]; gene models were predicted using a published transcriptome [58], [59]. Differentially expressed genes were identified using the R Bioconductor package edgeR [60] with cutoffs of logCPM score ≥0 and FDR≤0.05. Changes in gene expression detected by RNA-seq were represented as linear fold changes over controls. For the differentially expressed Schmidtea mediterranea transcripts, we performed BLASTX against the human UniProt database (cutoff<1×10−4); human accession numbers were then used to assign Gene Ontology terms and perform clustering analysis using DAVID software [61], [62] with the “Panther_BP_all” and “Panther_MF_all” gene annotation settings and an Enrichment Score cutoff >1.3.
Gene identification and cloning
For validation studies, transcript sequences were analyzed by BLASTX against protein sequences from human, mouse, fly, and nematode and identified as the top BLAST hit (Table S3). Sequences were obtained from a cDNA collection [63] or cloned into pJC53.2 [30] or pPR244 [64] using gene specific primers. GenBank accession numbers and the primers used in this study are listed in Table S3.
Reverse transcription quantitative PCR
Total RNA was extracted and purified as described above. cDNA was synthesized using the iScript cDNA Synthesis Kit (BioRad). Reverse transcription quantitative PCR was performed on a Bio-Rad CFX Connect Real-Time System using SsoAdvanced SYBR Green Supermix (Bio-Rad) with a two-step cycling protocol and annealing/extension temperature of 58.5°C. At least three biological replicates and two technical replicates were performed for each experiment. The relative amount of each cDNA target was normalized to Smed-β-tubulin (accession no. DN305397). The normalized relative changes in gene expression, standard deviations, and t-tests were calculated in Bio-Rad CFX Manager Software v3.0. Primers are listed in Table S3.
Imaging, cell counts, and statistical analysis
Images of live animals and whole mount in situ hybridization samples were acquired using a Leica DFC450 camera mounted on a Leica M205 stereomicroscope. Fluorescent images were acquired with a Zeiss Axio Observer.Z1 equipped with an Axiocam MRm camera and ApoTome; images are displayed as maximum image projections from ten 1-µm optical sections. For all experiments, we counted cells by hand using ImageJ Software [65], and biological replicates (n≥3) were averaged and shown as mean ± standard deviation. The number of cintillo+, spp-19+, spp-18+, and npl+ cells (Fig. 4N) was normalized to animal length (mm). We used anti-SYNAPSIN staining and ChAT expression to determine brain area (Fig. 6E–F), normalized to animal length (µm). To quantify npl+ brain-specific neurons following amputation, npl+ cells were counted in the cephalic ganglia and normalized to the average total brain area (Fig. 6G). When comparing two groups, we used a Student's t-test and significance was accepted at P<0.05.
Supporting Information
Zdroje
1. DaburonV, MellaS, PlouhinecJL, MazanS, CrozatierM, et al. (2008) The metazoan history of the COE transcription factors. Selection of a variant HLH motif by mandatory inclusion of a duplicated exon in vertebrates. BMC Evol Biol 8 : 131.
2. DemillyA, SimionatoE, OhayonD, KernerP, GarcesA, et al. (2011) Coe genes are expressed in differentiating neurons in the central nervous system of protostomes. PLoS ONE 6: e21213.
3. JacksonDJ, MeyerNP, SeaverE, PangK, McDougallC, et al. (2010) Developmental expression of COE across the Metazoa supports a conserved role in neuronal cell-type specification and mesodermal development. Dev Genes Evol 220 : 221–234.
4. LibergD, SigvardssonM, AkerbladP (2002) The EBF/Olf/Collier family of transcription factors: regulators of differentiation in cells originating from all three embryonal germ layers. Mol Cell Biol 22 : 8389–8397.
5. CrozatierM, VincentA (2008) Control of multidendritic neuron differentiation in Drosophila: the role of Collier. Dev Biol 315 : 232–242.
6. DuboisL, Bally-CuifL, CrozatierM, MoreauJ, PaquereauL, et al. (1998) XCoe2, a transcription factor of the Col/Olf-1/EBF family involved in the specification of primary neurons in Xenopus. Curr Biol 8 : 199–209.
7. GarelS, Garcia-DominguezM, CharnayP (2000) Control of the migratory pathway of facial branchiomotor neurones. Development 127 : 5297–5307.
8. Jinushi-NakaoS, ArvindR, AmikuraR, KinameriE, LiuAW, et al. (2007) Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron 56 : 963–978.
9. PozzoliO, BosettiA, CrociL, ConsalezGG, VetterML (2001) Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Dev Biol 233 : 495–512.
10. PrasadBC, YeB, ZackharyR, SchraderK, SeydouxG, et al. (1998) unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development 125 : 1561–1568.
11. WightmanB, BaranR, GarrigaG (1997) Genes that guide growth cones along the C. elegans ventral nerve cord. Development 124 : 2571–2580.
12. KimK, ColosimoME, YeungH, SenguptaP (2005) The UNC-3 Olf/EBF protein represses alternate neuronal programs to specify chemosensory neuron identity. Dev Biol 286 : 136–148.
13. HattoriY, UsuiT, SatohD, MoriyamaS, ShimonoK, et al. (2013) Sensory-neuron subtype-specific transcriptional programs controlling dendrite morphogenesis: genome-wide analysis of Abrupt and Knot/Collier. Dev Cell 27 : 530–544.
14. KratsiosP, StolfiA, LevineM, HobertO (2012) Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat Neurosci 15 : 205–214.
15. EadeKT, FancherHA, RidyardMS, AllanDW (2012) Developmental transcriptional networks are required to maintain neuronal subtype identity in the mature nervous system. PLoS Genet 8: e1002501.
16. ZhaoLY, NiuY, SantiagoA, LiuJ, AlbertSH, et al. (2006) An EBF3-mediated transcriptional program that induces cell cycle arrest and apoptosis. Cancer Res 66 : 9445–9452.
17. LiaoD (2009) Emerging roles of the EBF family of transcription factors in tumor suppression. Mol Cancer Res 7 : 1893–1901.
18. MaherEA, BrennanC, WenPY, DursoL, LigonKL, et al. (2006) Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res 66 : 11502–11513.
19. MullighanCG, GoorhaS, RadtkeI, MillerCB, Coustan-SmithE, et al. (2007) Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446 : 758–764.
20. ZardoG, TiirikainenMI, HongC, MisraA, FeuersteinBG, et al. (2002) Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nat Genet 32 : 453–458.
21. ZhuZ, HuangfuD (2013) Human pluripotent stem cells: an emerging model in developmental biology. Development 140 : 705–717.
22. ElliottSA, Sánchez AlvaradoA (2012) The history and enduring contributions of planarians to the study of animal regeneration. WIREs Dev Biol 2012 doi:101002/wdev82
23. WagnerDE, WangIE, ReddienPW (2011) Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332 : 811–816.
24. CowlesMW, BrownDD, NisperosSV, StanleyBN, PearsonBJ, et al. (2013) Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development 140 : 4691–4702.
25. CurrieKW, PearsonBJ (2013) Transcription factors lhx1/5-1 and pitx are required for the maintenance and regeneration of serotonergic neurons in planarians. Development 140 : 3577–3588.
26. LapanSW, ReddienPW (2012) Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell Rep 2 : 294–307.
27. MärzM, SeebeckF, BartschererK (2013) A Pitx transcription factor controls the establishment and maintenance of the serotonergic lineage in planarians. Development 140 : 4499–4509.
28. AdlerCE, SeidelCW, McKinneySA, Sanchez AlvaradoA (2014) Selective amputation of the pharynx identifies a FoxA-dependent regeneration program in planaria. Elife 3: e02238.
29. ScimoneML, Kravarik KellieM, Lapan SylvainW, Reddien PeterW (2014) Neoblast Specialization in Regeneration of the Planarian Schmidtea mediterranea. Stem Cell Reports 3 : 339–352.
30. CollinsJJ3rd, HouX, RomanovaEV, LambrusBG, MillerCM, et al. (2010) Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol 8: e1000509.
31. GentileL, CebriàF, BartschererK (2011) The planarian flatworm: an in vivo model for stem cell biology and nervous system regeneration. Dis Model Mech 4 : 12–19.
32. UmesonoY, TasakiJ, NishimuraK, InoueT, AgataK (2011) Regeneration in an evolutionarily primitive brain–the planarian Dugesia japonica model. Eur J Neurosci 34 : 863–869.
33. LapanSW, ReddienPW (2011) dlx and sp6-9 Control optic cup regeneration in a prototypic eye. PLoS Genet 7: e1002226.
34. WenemoserD, LapanSW, WilkinsonAW, BellGW, ReddienPW (2012) A molecular wound response program associated with regeneration initiation in planarians. Genes Dev 26 : 988–1002.
35. SandmannT, VoggMC, OwlarnS, BoutrosM, BartschererK (2011) The head-regeneration transcriptome of the planarian Schmidtea mediterranea. Genome Biol 12: R76.
36. WenemoserD, ReddienPW (2010) Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol 344 : 979–991.
37. WitchleyJN, MayerM, WagnerDE, OwenJH, ReddienPW (2013) Muscle cells provide instructions for planarian regeneration. Cell Rep 4 : 633–641.
38. OviedoNJ, NewmarkPA, Sánchez AlvaradoA (2003) Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev Dyn 226 : 326–333.
39. GreenYS, VetterML (2011) EBF factors drive expression of multiple classes of target genes governing neuronal development. Neural Dev 6 : 19.
40. CrozatierM, VincentA (1999) Requirement for the Drosophila COE transcription factor Collier in formation of an embryonic muscle: transcriptional response to notch signalling. Development 126 : 1495–1504.
41. CaroleiA, MargottaV, PalladiniG (1975) Proposal of a new model with dopaminergic-cholinergic interactions for neuropharmacological investigations. Neuropsychobiology 1 : 355–364.
42. ButtarelliFR, PontieriFE, MargottaV, PalladiniG (2000) Acetylcholine/dopamine interaction in planaria. Comp Biochem Physiol C Toxicol Pharmacol 125 : 225–231.
43. NishimuraK, KitamuraY, TaniguchiT, AgataK (2010) Analysis of motor function modulated by cholinergic neurons in planarian Dugesia japonica. Neuroscience 168 : 18–30.
44. CebriàF, NewmarkPA (2005) Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 132 : 3691–3703.
45. TsengAS, BeaneWS, LemireJM, MasiA, LevinM (2010) Induction of vertebrate regeneration by a transient sodium current. J Neurosci 30 : 13192–13200.
46. BeaneWS, MorokumaJ, AdamsDS, LevinM (2011) A chemical genetics approach reveals H,K-ATPase-mediated membrane voltage is required for planarian head regeneration. Chem Biol 18 : 77–89.
47. BeaneWS, MorokumaJ, LemireJM, LevinM (2013) Bioelectric signaling regulates head and organ size during planarian regeneration. Development 140 : 313–322.
48. LabbéRM, IrimiaM, CurrieKW, LinA, ZhuSJ, et al. (2012) A Comparative Transcriptomic Analysis Reveals Conserved Features of Stem Cell Pluripotency in Planarians and Mammals. Stem Cells 30 : 1734–1745.
49. LatchmanDS (1999) POU family transcription factors in the nervous system. J Cell Physiol 179 : 126–133.
50. Urbach R, Technau G (2008) Dorsoventral Patterning of the Brain: A Comparative Approach. In: Technau G, editor. Brain Development in Drosophila melanogaster: Springer New York. pp. 42–56.
51. McMahonAP (2000) Neural patterning: The role of Nkx genes in the ventral spinal cord. Genes Dev 14 : 2261–2264.
52. DenerisES, HobertO (2014) Maintenance of postmitotic neuronal cell identity. Nat Neurosci 17 : 899–907.
53. GurleyKA, RinkJC, Sánchez AlvaradoA (2008) Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319 : 323–327.
54. KingRS, NewmarkPA (2013) In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev Biol 13 : 8.
55. LauterG, SollI, HauptmannG (2011) Two-color fluorescent in situ hybridization in the embryonic zebrafish brain using differential detection systems. BMC Dev Biol 11 : 43.
56. UmesonoY, WatanabeK, AgataK (1997) A planarian orthopedia homolog is specifically expressed in the branch region of both the mature and regenerating brain. Dev Growth Differ 39 : 723–727.
57. TrapnellC, PachterL, SalzbergSL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 : 1105–1111.
58. AdamidiC, WangY, GruenD, MastrobuoniG, YouX, et al. (2011) De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res 21 : 1193–1200.
59. ÖnalP, GrunD, AdamidiC, RybakA, SolanaJ, et al. (2012) Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO J 31 : 2755–2769.
60. RobinsonMD, McCarthyDJ, SmythGK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 : 139–140.
61. DennisGJr, ShermanBT, HosackDA, YangJ, GaoW, et al. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: P3.
62. Huang daW, ShermanBT, LempickiRA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 : 44–57.
63. ZayasRM, HernándezA, HabermannB, WangY, StaryJM, et al. (2005) The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: Analysis of ESTs from the hermaphroditic strain. Proc Natl Acad Sci U S A 102 : 18491–18496.
64. ReddienPW, BermangeAL, MurfittKJ, JenningsJR, Sánchez AlvaradoA (2005) Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell 8 : 635–649.
65. SchneiderCA, RasbandWS, EliceiriKW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9 : 671–675.
Štítky
Genetika Reprodukčná medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



