-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
DNA replication must be coordinated with cellular physiology to ensure proper genome inheritance. Model bacteria such as the soil-dwelling Bacillus subtilis can achieve a wide range of growth rates in response to nutritional and chemical signals. In order to match the rate of DNA synthesis to the rate of nutrient-mediated cell growth, bacteria regulate the initiation frequency of DNA replication. This control of bacterial DNA replication initiation was first observed over forty years ago, however the molecular basis for this regulation has remained hotly debated. In this paper we test one of the leading models for nutrient-mediated growth rate regulation in bacteria, namely that the abundance of the master DNA replication initiation protein DnaA dictates the frequency of DNA replication events. Critically, our results show that changes in DnaA protein level are not sufficient to account for nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis. We then go on to show that there are strong connections between DNA replication and several essential cellular activities, which unexpectedly indicates that there is likely more than one single regulatory pathway involved in coordinating DNA replication with cell physiology. We believe that our work changes thinking regarding this long-standing biological question and reinvigorates the search for the molecular basis of these critical regulatory systems.
Published in the journal: Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in. PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004731
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004731Summary
DNA replication must be coordinated with cellular physiology to ensure proper genome inheritance. Model bacteria such as the soil-dwelling Bacillus subtilis can achieve a wide range of growth rates in response to nutritional and chemical signals. In order to match the rate of DNA synthesis to the rate of nutrient-mediated cell growth, bacteria regulate the initiation frequency of DNA replication. This control of bacterial DNA replication initiation was first observed over forty years ago, however the molecular basis for this regulation has remained hotly debated. In this paper we test one of the leading models for nutrient-mediated growth rate regulation in bacteria, namely that the abundance of the master DNA replication initiation protein DnaA dictates the frequency of DNA replication events. Critically, our results show that changes in DnaA protein level are not sufficient to account for nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis. We then go on to show that there are strong connections between DNA replication and several essential cellular activities, which unexpectedly indicates that there is likely more than one single regulatory pathway involved in coordinating DNA replication with cell physiology. We believe that our work changes thinking regarding this long-standing biological question and reinvigorates the search for the molecular basis of these critical regulatory systems.
Introduction
DNA replication must be coordinated with the cell cycle to ensure proper genome inheritance. For many bacteria cellular physiology dictates the rate of growth and division. In nutrient-rich media that support rapid growth rates, bacteria synthesize DNA more rapidly by increasing the frequency of DNA replication initiation [1]–[3]. This control system is termed nutrient-mediated growth rate regulation and although it has been appreciated for decades, the molecular mechanisms that connect cell physiology with DNA replication initiation remain debatable.
Historically it has been thought that there is a constant cell mass or cell size at the time of bacterial DNA replication initiation and it has been proposed that a positive regulator would accumulate in a growth-dependent manner to trigger DNA replication initiation when cells attained a critical size [4]. However, modern quantitative analysis of single bacterial cells within steady-state populations has shown that the relationship between DNA replication initiation and cell mass is variable, indicating that the control for timing of DNA replication initiation is not governed by a direct connection with mass accumulation [5].
DnaA is the master bacterial DNA replication initiator protein and is a candidate factor to connect cell physiology with DNA synthesis. DnaA is a member of the AAA+ family of ATPases and shares homology with archaeal and eukaryotic initiator proteins. DnaA directly stimulates DNA replication initiation from a single defined origin of replication (oriC) once per cell cycle. Multiple ATP-bound DnaA molecules bind to an array of recognition sequences (DnaA-box 5′-TTATCCACA-3′) within oriC where they assemble into a helical filament that promotes duplex DNA unwinding [6], [7].
Studies in Escherichia coli have suggested that the amount of ATP-bound DnaA dictates the rate of DNA replication initiation. Artificial overexpression of DnaA increases the frequency of DNA replication initiation [8], [9]. Conversely, decreasing the amount of DnaA per cell by synthetically promoting early cell division delays DNA replication initiation and modest increases in DnaA levels alleviate this delay, supporting the view that growth-dependent accumulation of DnaA is the trigger for replication initiation in E. coli [10]. However, it remains uncertain whether the amount of ATP-bound DnaA is the primary regulator that coordinates DNA replication initiation with cell growth in wild-type E. coli cells [11].
In contrast to E. coli, studies in Bacillus subtilis have suggested that the amount of DnaA may not dictate the rate of DNA replication initiation. Artificially decreasing cell size by stimulating cell division (thereby lowering the amount DnaA per cell to ∼70% of wild-type) did not affect DNA replication initiation [10]. Moreover, results from overexpression of DnaA in B. subtilis are not clear. Increased expression of DnaA alone causes cell elongation, cell growth inhibition, and induction of the SOS DNA damage response due to depletion of DnaN because of autoregulation of the dnaA-dnaN operon by DnaA [12]. To circumvent this problem DnaA was co-overexpressed with DnaN, and under this condition DNA replication initiation does increase [12]. However, subsequent experiments demonstrated that overexpression of DnaN alone increases DNA replication initiation by repressing the activity of the regulatory protein YabA (an inhibitor of DnaA)[13], suggesting that this could account for the effect on DNA replication initiation when DnaA and DnaN were co-overexpressed.
In this study we have investigated nutrient-mediated growth rate control of DNA replication initiation in B. subtilis. We find that changes in DnaA protein level are not sufficient to account for nutrient-mediated growth rate regulation of DNA replication initiation, although this regulation does require both DnaA and oriC. We then present evidence suggesting that multiple regulatory systems are involved in coordinating DNA synthesis with cell physiology, and that depending on the nature of the growth limitation, control of DNA replication acts through either oriC-dependent or oriC-independent mechanisms.
Results
Changes in DnaA levels cannot account for nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis
Steady-state bacterial growth rates can be manipulated by culturing cells in media that contain differing amounts of nutrients, with rich media supporting faster growth because resources are not required to synthesize cellular building blocks de novo. In response to different nutrient-mediated steady-state growth rates, bacteria control DNA synthesis by varying the frequency of DNA replication initiation while maintaining a constant speed of elongation [1]–[3], [14]. The rate of DNA replication initiation can be determined by marker frequency analysis (i.e. - measuring the ratio of DNA at the replication origin (ori) versus the replication terminus (ter) using quantitative PCR), and Figure 1A shows the positive correlation between DNA replication initiation and nutrient-mediated growth rates (cell doublings per hour measured using spectrophotometry). It is important to state that experimental approaches which change bacterial growth rates without altering the chemical composition of the cell (e.g. – varying temperature) do not influence the rate of DNA replication initiation (Figures 1B, S1; [15], [16]). Thus, varying nutrient availability modulates bacterial physiology, in turn affecting cell growth and DNA replication initiation [3].
Fig. 1. Nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis. 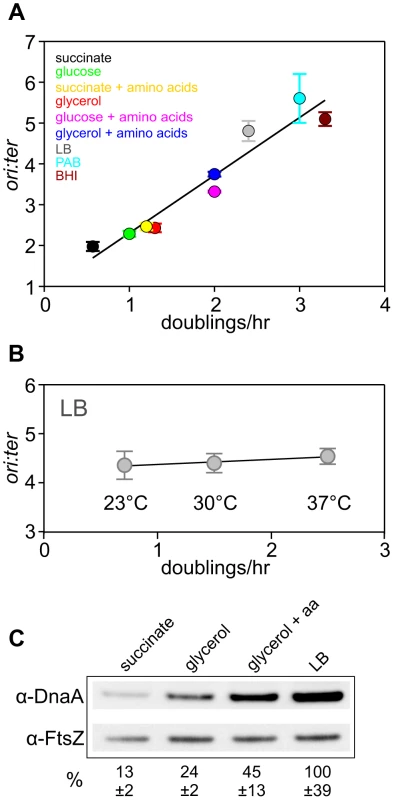
(A) Culturing B. subtilis in a different media generates a range of steady-state growth rates and affects the frequency of DNA replication initiation. A wild-type strain (HM222) was grown overnight at 37°C in minimal media supplemented with succinate and amino acids (20 µg/ml). The culture was diluted 1∶100 into various media to generate a range of steady-state growth rates and grown at 37°C until an A600 of 0.3–0.4. Genomic DNA was harvested from cells and marker frequency analysis was determined using qPCR. The ori:ter ratios are plotted versus growth rate (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; independently performed experiments are shown in Figures 4 and S5. (B) Culturing B. subtilis at different temperatures generates a range of steady-state growth rates but does not affect the frequency of DNA replication initiation. A wild-type strain (HM715) was grown overnight at 23°C in LB. The culture was diluted 1∶100 into LB and incubated at different temperatures to generate a range of steady-state growth rates until an A600 of 0.2–0.3. Genomic DNA was harvested from cells and marker frequency analysis was determined using qPCR. The ori:ter ratios are plotted versus growth rate (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; an independently performed replicate of the experiment is shown in Figure S1. (C) Measurement of DnaA protein levels at various growth rates in wild-type B. subtilis (HM715). Cultures were grown at 37°C overnight as in (A) and diluted 1∶100 into various media (succinate, glycerol, glycerol + amino acids, LB). to generate a range of steady-state growth rates until an A600 of 0.6–0.8. Cells were lysed and DnaA protein was detected using Western blot analysis (FtsZ protein was likewise detected and used as a loading control). For each culture media the average amount of DnaA (+/− standard deviation) from at least three biological replicates was determined using densitometry; values were normalized to LB. It has been reported that DnaA protein level determines the frequency of DNA replication initiation in E. coli [8], [9], therefore we wondered whether the amount of DnaA could account for nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis [14]. Western blot analysis shows that DnaA concentration increases with faster steady-state growth rates (Figure 1C; the tubulin homolog FtsZ was used as a loading control because its concentration is growth-rate independent [17],[18]). Since B. subtilis cell size increases as a function of growth rate, the number of DnaA molecules would also be greater in larger cells formed during fast growth (Figure S2)[14]. This conclusion is in agreement with absolute quantification of DnaA proteins per cell determined at different growth rates using mass spectrometry (163-337 molecules at 0.5 doublings/hr; 875-1791 molecules at 1.0 doublings/hr)[18]. These results indicate that the amount of DnaA protein could account for nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis.
To directly test whether the amount of DnaA protein determines the rate of DNA replication initiation, the endogenous dnaA gene was placed under the control of an IPTG-inducible promoter (this also alleviated autoregulation of the dnaA-dnaN operon [12]). At near wild-type DnaA levels growth rates were normal, ori:ter ratios were unchanged, and the distribution of origin regions per cell visualized using a TetR-YFP/tetO reporter system was equivalent to wild-type (Figures 2A-C, S4A). In contrast, when the amount of DnaA fell significantly (between ∼50–30% of wild-type, depending upon the media), growth rates slowed, ori:ter ratios dropped, DNA replication was inhibited as judged by origin region localization, and cells became elongated (Figures 2A–C, S4A). As noted above dnaA is located in an operon upstream of dnaN (encoding the sliding clamp component of the replisome) in B. subtilis, and Western blot analysis confirmed that the level of DnaN correlated with the level of DnaA (Figure S3B). Depletion of DnaN can cause replication fork stalling and induction of the SOS DNA damage response, which likely contributes to the slow growth and cell elongation phenotypes observed at low IPTG concentrations [12]. However, replication fork stalling would also be expected to cause an increase in the ori:ter ratio, suggesting that the observed decreases may be an overestimate of the true initiation frequency. We conclude that wild-type DnaA levels are necessary to achieve the proper frequency of DNA replication initiation at both slow and fast steady-state growth rates.
Fig. 2. Changes in DnaA protein level are not sufficient to account for nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis. 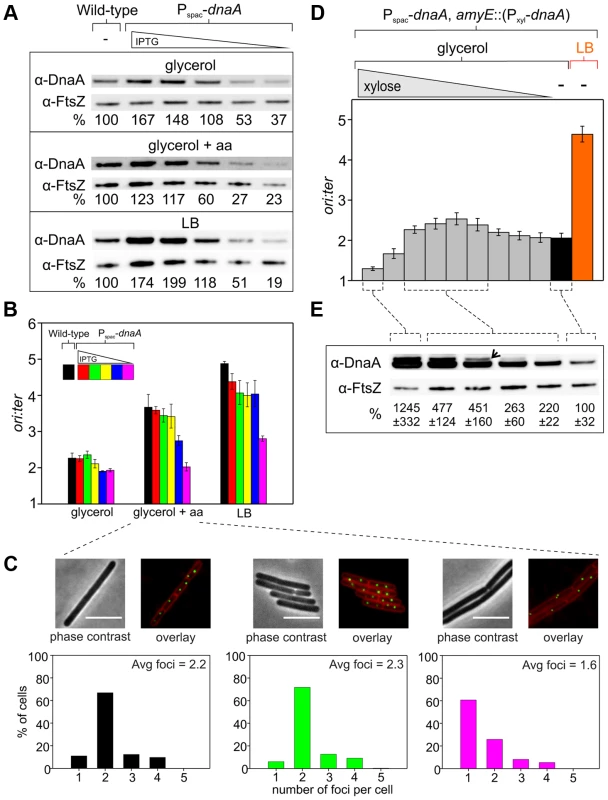
(A) The endogenous dnaA gene was placed under the control of the IPTG-inducible promoter Pspac to generate a range of DnaA protein levels. Strains were grown overnight at 37°C in minimal media supplemented with succinate and amino acids (20 µg/ml); IPTG (400 µM) and erythromycin was added to HM742. The cultures were diluted 1∶100 into various media (glycerol, glycerol + amino acids, LB) to generate a range of steady-state growth rates and grown at 37°C until an A600 of 0.5–0.6; in each medium HM742 was supplemented with erythromycin and a range of IPTG (800, 400, 200, 100, 50 µM). Cells were lysed and DnaA protein was detected using Western blot analysis (FtsZ protein was likewise detected and used as a loading control). The amount of DnaA was determined using densitometry; values were normalized to wild-type. Wild-type (HM222), Pspac-dnaA (HM742). (B) DNA replication was measured at over a range of DnaA protein levels. Strains were grown as described in (A). Genomic DNA was harvested from cells and marker frequency analysis was determined using qPCR. For each growth media, the ori:ter ratios are plotted versus IPTG concentration (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; an independently performed replicate of the experiment is shown in Figure S4A. Wild-type (HM222), Pspac-dnaA (HM742). (C) Measurement of replication origins number per cell. An array of ∼150 tetO sites was inserted near the replication origin and visualized using TetR-YFP. Strains were grown as described in (A), except that overnight cultures were only diluted into a single medium (glycerol + amino acids); AK652 was supplemented with erythromycin and a range of IPTG concentrations. Samples were taken at mid-exponential phase for microscopy and membranes were stained to identify single cells (scale bar = 5 µm). Histogram colour corresponds to the respective strain/IPTG concentration and the average number of origins per cell is indicated (n> 300). Wild-type (AK647), Pspac-dnaA (AK652). (D) To strongly overexpress DnaA the endogenous dnaA gene was placed under the control of Pspac and an ectopic copy of dnaA was integrated at the amyE locus under the control of the xylose inducible promoter Pxyl (HM745). The strain was grown overnight at 37°C in minimal media supplemented with glycerol, amino acids (20 µg/ml), IPTG (800 µM), and erythromycin. The culture was diluted 1∶100 into media containing IPTG (800 µM), erythromycin, either glycerol minimal media supplemented with a range of xylose (1, 0.5, 0.25, 0.125, 0.063, 0.031, 0.016, 0.008, 0.004, 0%) or LB, and grown at 37°C until an A600 of 0.2–0.4. Genomic DNA was harvested from cells and marker frequency analysis was determined using qPCR. For each growth media, the ori:ter ratios are plotted versus xylose concentration (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; an independently performed replicate of the experiment is shown in Figure S4B. (E) HM745 was grown as described in (D) until cultures reached an A600 of 0.6–0.9, cells were lysed, and DnaA protein was detected using Western blot analysis (FtsZ protein was likewise detected and used as a loading control). The open arrowhead highlights that overexpressed DnaA ran as a doublet (similar results have been observed for other overexpressed proteins in B. subtilis; HM). For each condition the average amount of DnaA (+/− standard deviation) from three biological replicates was determined using densitometry; values were normalized to the cultures without xylose. Only modest overexpression of DnaA could be achieved using the IPTG-inducible promoter (Figure 2A), much lower than the changes in DnaA concentration observed at different nutrient-mediated growth rates (Figure 1C). Therefore, to further increase DnaA protein levels a second copy of the dnaA gene was integrated at an ectopic locus under the control of a xylose-inducible promoter (again the endogenous dnaA-dnaN operon was expressed using an IPTG-inducible promoter to avoid autorepression). This strain was grown in media that supported a slow growth rate and varying amounts of xylose were added to induce DnaA (>10 fold overexpression was achieved, which was in the range observed for different nutrient-mediated growth rates; Figures 2E, 4D). When DnaA levels were elevated ∼2–4 fold a modest increase in the ori:ter ratios was observed, although critically the resulting initiation frequencies remained well below the rate generated in rich media (Figures 2D, S4B). These results indicate that changes in DnaA protein levels are not sufficient to account for nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis.
Surprisingly, further overexpression of DnaA lead to a dramatic decrease in the ori:ter ratios. To determine whether this inhibition was specific, DnaA was overexpressed in a strain where oriC was inactivated by partial deletion (ΔoriC), the endogenous dnaA-dnaN operon was expressed using a constitutive promoter to avoid autorepression, and genome replication was driven by a plasmid-derived replication origin (oriN; integrated ∼1 kb to the left of oriC) that is recognized and activated by its cognate initiator protein (RepN). It is important to note that while initiation at oriN does not require either oriC or DnaA, the downstream B. subtilis initiation proteins DnaD, DnaB and DnaC (helicase) are necessary for oriN activity [19]. Therefore, if overexpression of DnaA was either inhibiting the expression of genes required for DNA replication (e.g. – nucleotide biosynthesis [20]) or sequestering essential replication factors, then DNA replication initiation from oriN would be expected to decrease. However, overexpression of DnaA in the ΔoriC oriN+ background did not alter ori:ter ratios, showing that high overexpression of DnaA specifically inhibits DNA replication initiation at oriC (Figure S4C).
Neither Soj nor YabA nor (p)ppGpp are required for nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis
We hypothesized that nutrient-mediated growth rate control of DNA replication initiation could act via regulation of DnaA activity rather than protein abundance. There are two known trans-acting regulators of B. subtilis DnaA during steady-state growth, Soj and YabA. Soj is a dynamic protein that can act as either a negative or a positive regulator of DnaA, depending upon its quaternary state [21]–[23]. YabA is a negative regulator of DnaA that forms a protein bridge between the initiator DnaA and the DNA polymerase sliding clamp processivity factor, DnaN, and is thought to inhibit DNA replication by spatially sequestering DnaA away from the replication origin and by inhibiting DnaA oligomerization [24]–[27]. Interestingly, the number of both proteins per cell was found to positively correlate with growth rate [18].
To determine whether either of these regulatory proteins is required for nutrient-mediated growth rate regulation of DNA replication initiation, single knockout mutants were cultured in a range of media and analyzed using marker frequency analysis. It was found that both of the mutant strains retained the ability to coordinate DNA replication initiation with nutrient-mediated changes in growth rate (Figures 3A, S5A). To test whether Soj and YabA acted redundantly to control the nutrient-mediated activity of DnaA, the double mutant was constructed and analysed by marker frequency analysis. Again proper regulation of DNA replication initiation was maintained in the Δsoj ΔyabA mutant, indicating that neither regulatory protein is required (Figures 3A, S5B). Interestingly, both the single and double mutants displayed a reduced growth rate in rich media, suggesting that the burden of overactive DNA replication initiation may be exacerbated during multifork replication. In the case of the soj mutant it is also possible that the slow growth phenotype is related to its role in chromosome origin segregation [28]–[30].
Fig. 3. Nutrient-mediated growth rate regulation of DNA replication initiation is independent of Soj, YabA, and (p)ppGpp. 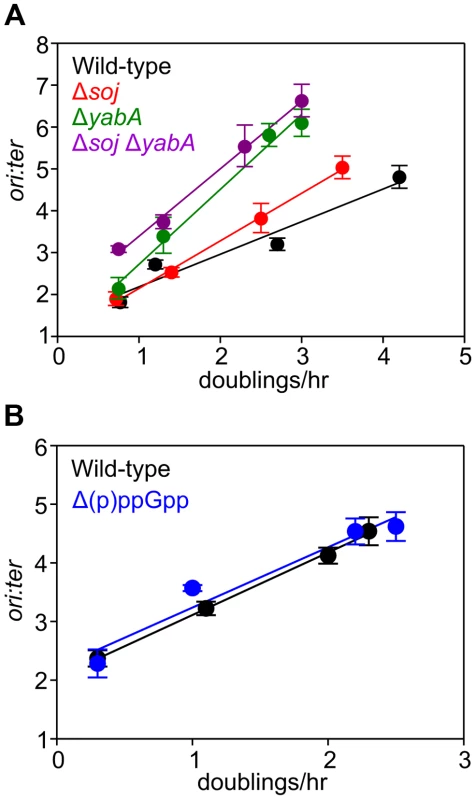
(A) Growth rate regulation of DNA replication initiation is maintained in either Δsoj or ΔyabA mutants. Strains were grown overnight at 37°C in minimal media supplemented with succinate and amino acids (20 µg/ml). The culture was diluted 1∶100 into various media (succinate, glycerol, glycerol + amino acids, LB) to generate a range of steady-state growth rates and grown at 37°C until an A600 of 0.3–0.4. Genomic DNA was harvested from cells and marker frequency analysis was determined using qPCR. The ori:ter ratios are plotted versus growth rate (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; an independently performed replicate of the experiment is shown in Figures S5A-B. Wild-type (HM222), Δsoj (HM227), ΔyabA (HM739), Δsoj ΔyabA (HM741). (B) Growth rate regulation of DNA replication initiation does not require (p)ppGpp. Strains were grown overnight at 37°C in minimal media supplemented with succinate and amino acids (200 µg/ml). The culture was diluted 1∶100 into various media (succinate + amino acids, glycerol + amino acids, LB, PAB) to generate a range of steady-state growth rates and grown at 37°C until an A600 of 0.2–0.6. Genomic DNA was harvested from cells and marker frequency analysis was determined using qPCR. The ori:ter ratios are plotted versus growth rate (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; an independently performed experiment is shown in Figure S5C. Wild-type (HM222), Δ(p)ppGpp (HM1230). Fig. 4. Nutrient-mediated growth rate regulation of DNA replication initiation requires oriC and DnaA. 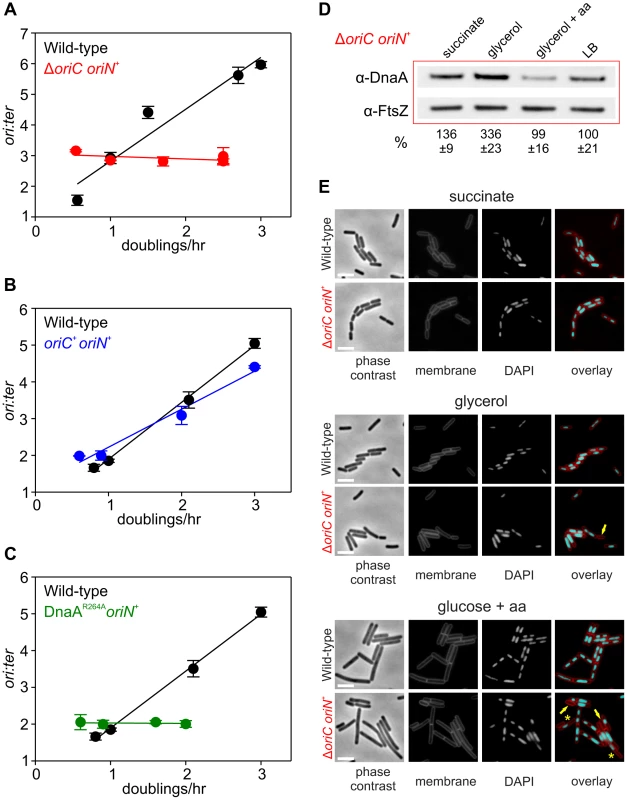
(A) oriC is required for growth rate regulation of DNA replication initiation. Strains were grown overnight at 37°C in minimal media supplemented with succinate and amino acids (20 µg/ml). The culture was diluted 1∶100 into various media (succinate, glycerol, glycerol + amino acids, LB, PAB) to generate a range of steady-state growth rates and grown at 37°C until an A600 of 0.3–0.4. Genomic DNA was harvested from cells and marker frequency analysis was determined using qPCR. The ori:ter ratios are plotted versus growth rate (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; an independently performed replicate of the experiment is shown in Figure S6A. Wild-type (HM222), ΔoriC oriN+ (HM228). (B) Integration of oriN into the B. subtilis chromosome does not eliminate growth rate regulation of DNA replication initiation. Strains were grown as in (A) and the overnight culture was diluted 1∶100 into various media (succinate, glycerol, glycerol + amino acids, LB). Genomic DNA was harvested from cells and marker frequency analysis was determined using qPCR. The ori:ter ratios are plotted versus growth rate (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; an independently performed replicate of the experiment is shown in Figure S6B. Wild-type (HM715), oriC+ oriN+ (HM949). (C) DnaA activity is required for growth rate regulation of DNA replication initiation. Strains were grown as in (B). Genomic DNA was harvested from cells and marker frequency analysis was determined using qPCR. The ori:ter ratios are plotted versus growth rate (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; an independently performed replicate of the experiment is shown in Figure S6C. Wild-type (HM715), DnaAR264A oriN+ (HM1122). (D) Measurement of DnaA protein levels at various growth rates in a ΔoriC oriN+ strain (HM950). Cultures were grown as described in (B). Cells were lysed and DnaA protein was detected using Western blot analysis (FtsZ protein was likewise detected and used as a loading control). For each culture media the average amount of DnaA (+/− standard deviation) from at least three biological replicates was determined using densitometry; values were normalized to LB. (E) Subcellular localization of DNA over a range of growth rates in the wild-type (HM715) and ΔoriC oriN+ (HM950) strains. Cells were grown as in (B) and the overnight culture was diluted 1∶100 into various media (succinate, glycerol, or glucose + amino acids). Samples were taken at an A600 of 0.3–0.5 at which point membranes and DNA were stained. Arrows indicate cells without DNA and asterisks indicate space within the cell that does not contain DNA. Scale bar represents 3 µm. We also determined whether the alarmone (p)ppGpp, a small molecule induced during nutrient limitation, is required for nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis. Marker frequency analysis was performed on a strain lacking the three known (p)ppGpp synthases (RelA, YwaC, YjbM). It was found that regulation of DNA replication initiation was unaffected by the absence of (p)ppGpp, suggesting that (p)ppGpp is not involved in the regulatory mechanisms coordinating DNA replication with nutrient availability during steady-state cell growth (Figures 3B, S5C).
DnaA and oriC are necessary for nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis
Since both overexpression of DnaA and deletion of DnaA regulatory proteins did not alter nutrient-mediated growth rate regulation of DNA replication initiation, it was unclear whether DnaA was actually a component of this system. To determine whether DnaA activity at oriC is required, marker frequency analysis was performed in a strain where oriC was inactivated (ΔoriC) and DNA replication initiates from the plasmid-derived oriN. It was found that ori:ter ratios remain constant over a wide range of growth rates in the ΔoriC mutant, indicating that nutrient-mediated growth rate regulation of DNA replication was lost (Figures 4A, S6A). Critically, DNA replication initiation from oriC is unaffected by the addition of oriN (Figures 4B, S6B). This shows that it is the absence of DnaA activity at oriC, rather than the presence of oriN, which accounts for the loss of nutrient-mediated growth rate regulation in the ΔoriC mutant.
However, it could not be concluded whether the ΔoriC mutation acted by removing replication origin function or by deleting a site that is required for the nutrient-mediated growth rate regulation. Therefore, a mutation was introduced into dnaA that alters the critical “arginine finger” residue (Arg264→Ala), thereby disabling DnaA filament assembly and initiation activity (note that a DnaA arginine finger mutant remains competent for DNA binding and ATP binding) [22], [31]. Again the dnaAR264A mutant strain contains oriN in order to maintain viability. Like the ΔoriC mutant the DnaAR264A variant also lost growth rate regulation in response to nutrient availability, indicating that DnaA activity at oriC is necessary for growth rate regulation in B. subtilis (Figures 4C, S6C). Moreover, since the ori:ter ratios of the ΔoriC and dnaAR264A mutants remains constant during nutrient-mediated growth rate changes and since DNA replication elongation speed is independent of the nutrient-mediated growth rate [2], [14], the results suggest that within a population of cells the average frequency of DNA replication initiation at oriN is independent of the nutrient-mediated growth rate.
Western blot analysis showed that in rich media there was less DnaA and DnaN in the ΔoriC strain, whereas conversely there was more DnaA and DnaN in the dnaAR264A mutant (Figure S3C). The latter result suggests that autoregulation of the dnaA promoter requires ATP-dependent filament formation by DnaA, however, the former result was more puzzling. To investigate this further the amount of DnaA in the ΔoriC strain was determined over a range of nutrient-mediated growth rates. While the concentration of DnaA was observed to increase as a function of growth rate in the wild-type strain, DnaA levels did not display the same correlation with growth rate in the ΔoriC mutant (Figure 4D). This suggests that in the ΔoriC mutant nutrient-mediated growth rate-dependent expression of DnaA is lost either because DNA replication initiation from oriN is constitutive or because the deletion within oriC affects dnaA expression (although this region is downstream of the dnaA gene).
The apparent decrease in growth rates observed for strains initiating DNA replication solely through oriN, particularly in rich media (Figures 4A, 4C, S6A, S6C), is likely due to the formation of cells lacking DNA as a direct consequence of decoupling DNA replication initiation from growth rate (Figure 4E). This result underscores the importance of growth rate regulation of DNA replication initiation to ensure bacterial fitness.
Slowing growth rate by limiting essential cellular activities inhibits DNA replication
Since nutrient-mediated growth rate regulation of DNA replication initiation did not appear to act through either DnaA protein accumulation or known DnaA regulators, we considered alternative possibilities for how DNA replication could be connected to cell physiology. One hypothesis was that this regulation could be linked to metabolism, either through the amount of a metabolic intermediate or the activity of a critical enzyme. Genetic evidence has suggested a relationship between several glycolytic enzymes and DNA replication in B. subtilis and E. coli [32], [33], but it has not been established whether these connections act directly at the level of DnaA-dependent initiation. ATP would be another rational candidate since DnaA is an ATP-dependent protein, but it has been found that the concentration of ATP in B. subtilis (as well as in E. coli) is invariant over a wide range of growth rates (L. Krasny and R. Gourse, personal communication; [34], [35]). Another hypothesis was that this regulation could be linked to the synthesis of an essential cellular complex, such as the ribosome or the cell membrane [36], [37]. In this way a bacterial cell would integrate nutritional information based on the availability of multiple substrates required to construct such macromolecules.
In order to identify possible routes through which nutrient availability could impact DNA replication we analyzed a range of genetically altered strains, targeting respiration, central carbon metabolism, protein synthesis, fatty acid synthesis, and phospholipid synthesis (Table 1), that all manifest decreased steady-state growth rates in rich complex media. Genes were either disrupted by antibiotic cassettes or depleted using regulated expression systems; importantly, depletion of essential genes was not lethal under the experimental conditions used. Knock-out strains were compared to wild-type while depletion strains were analyzed without and with inducer (indicated in Figures 5, 6, S7, S8 with “-” and “+”, respectively). DNA replication was measured using marker frequency analysis. Strikingly, in all of the strains examined the ori:ter ratio decreased to match the slower growth rates caused by gene disruption/depletion (Figures 5–6, S7–S8; black symbols).
Fig. 5. Analysis of oriC-dependent growth rate regulation through genetic targeting of essential cellular activities. 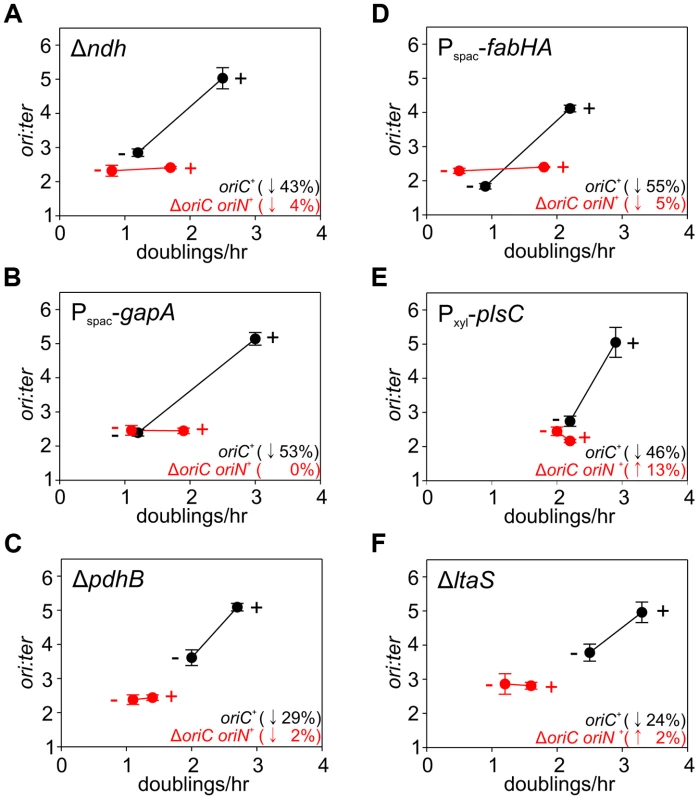
Strains were grown overnight at 37°C in LB medium; strains harbouring plasmids integrated into the genome by single-crossover were supplemented with appropriate antibiotics and inducer (0.1 mM IPTG or 0.1% xylose). Overnight cultures were diluted 1∶1000 into fresh LB medium and grown at 37°C until they reached an A600 of 0.3–0.5; strains harbouring plasmids integrated by single-crossover were supplemented with appropriate antibiotics either without or with the appropriate inducer (1 mM IPTG or 1% xylose). For datapoints “+” indicates the presence of either the wild-type gene (when comparing with knockout mutants) or the inducer; “−” indicates the absence of either the gene (when comparing with wild-type) or the inducer. Genomic DNA was harvested from cells and marker frequency analysis was determined using qPCR. The ori:ter ratios are plotted versus growth rate and the percentage change in the ori:ter ratios comparing each deletion/depletion is indicated (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; an independently performed replicate of the experiment is shown in Figure S7. (A) Wild-type (HM715), Δndh (HM1318), ΔoriC oriN+ (HM957), Δndh ΔoriC oriN+ (HM1319); (B) Pspac-gapA (HM1208), Pspac-gapA ΔoriC oriN+ (HM1221); (C) Cultures were supplemented with 0.2% sodium acetate. Wild-type (HM715), ΔpdhB (HM1248), ΔoriC oriN+ (HM950), ΔpdhB ΔoriC oriN+ (HM1266); (D) Pspac-fabHA (HM964), Pspac-fabHA ΔoriC oriN+ (HM966); (E) Pxyl-plsC (HM1080), Pxyl-plsC ΔoriC oriN+ (HM1086); (F) Wild-type (HM715), ΔltaS (HM1168), ΔoriC oriN+ (HM957), ΔltaS ΔoriC oriN+ (HM1244). Fig. 6. Analysis of oriC-independent growth rate regulation through genetic targeting of essential cellular activities. 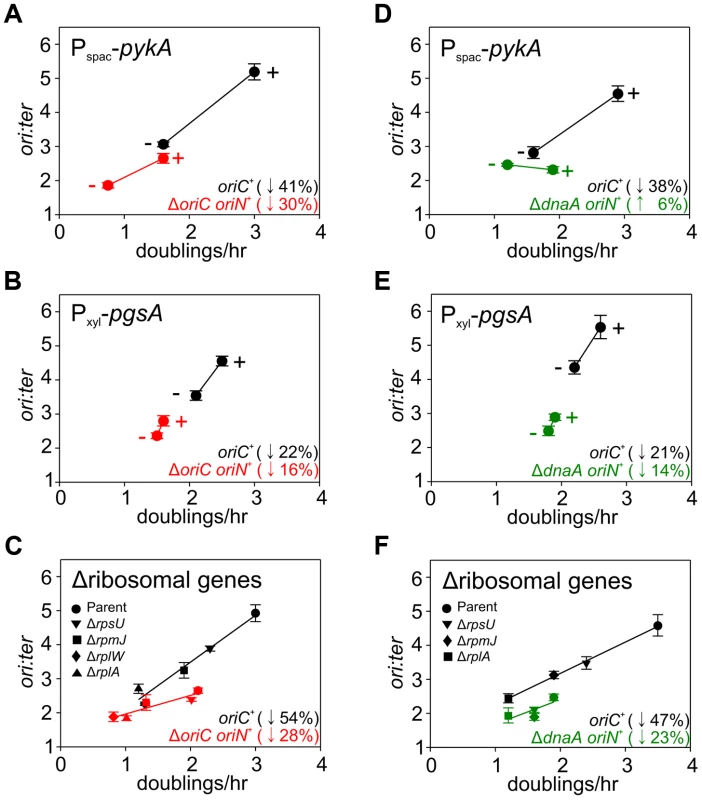
Strains were grown and data presented as described for Figure 5, except that the depletion of PgsA required supplementation with 1 mM IPTG to overexpress the xylose repressor. The ori:ter ratios are plotted versus growth rate and the percentage change in the ori:ter ratios comparing each deletion/depletion is indicated (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; an independently performed replicate of the experiment is shown in Figure S8. (A) Pspac-pykA (HM1176), Pspac-pykA ΔoriC oriN+ (HM1186); (B) Pxyl-pgsA (HM1365), Pxyl-pgsA ΔoriC oriN+ (HM1374); (C) Wild-type (HM715), ΔrpsU (HM1150), ΔrplA (HM1151), ΔrplW (HM1152), ΔrpmJ (HM1154), ΔoriC oriN+ (HM950), ΔrpsU ΔoriC oriN+ (HM1156), ΔrplA ΔoriC oriN+ (HM1157), ΔrplW ΔoriC oriN+ (HM1158), ΔrpmJ ΔoriC oriN+ (HM1160). (D) Pspac-pykA (HM1176), Pspac-pykA ΔdnaA oriN+ (HM1425); (E) Pxyl-pgsA (HM1365), Pxyl-pgsA ΔdnaA oriN+ (HM1433); (F) Wild-type (HM715), ΔrpsU (HM1150), ΔrplA (HM1151), ΔrpmJ (HM1154), ΔdnaA oriN+ (HM1423), ΔrpsU ΔdnaA oriN+ (HM1429), ΔrplA ΔdnaA oriN+ (HM1430), ΔrpmJ ΔdnaA oriN+ (HM1432). Tab. 1. <b>Genes manipulated to limit essential cellular processes.</b> 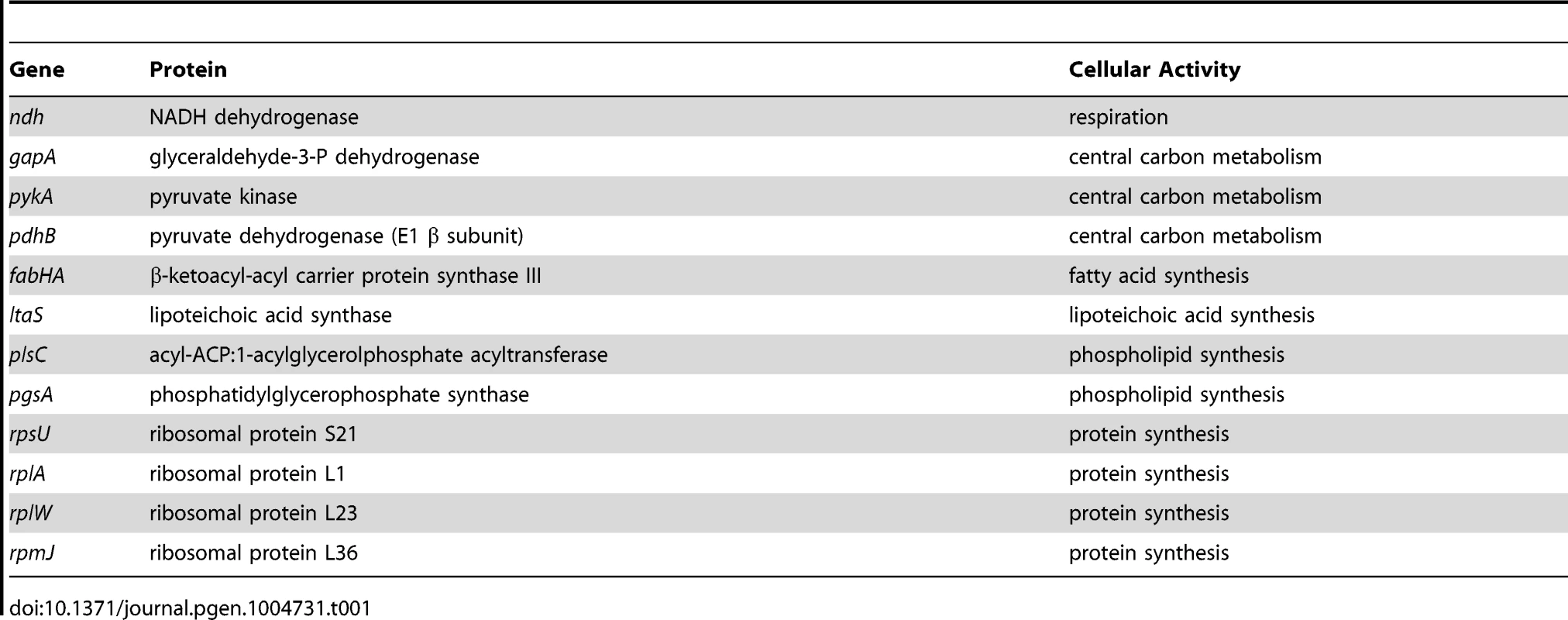
Evidence for an oriC-independent response to changes in growth rate
The uniform response of DNA replication in slow growing mutants suggested that a single mechanism might account for this regulation, in accord with nutrient-mediated regulation of DNA replication initiation (Figures 4, S6). To examine this hypothesis the deletion and depletion strains were crossed into the ΔoriC strain that initiates DNA replication using oriN. For several mutants (ndh, gapA, pdhB, fabHA, plsC and ltaS) the ori:ter ratio was not significantly affected (≤1/10 of the percentage decrease observed for oriC+), suggesting that the regulatory signal specifically targeted DNA replication initiation at oriC (Figures 5, S7; red symbols). However, there were a number of mutants (pykA, pgsA, and multiple ribosomal protein genes) that produced a marked decrease in the ori:ter ratio of the ΔoriC strain (≥1/2 of the percentage decrease observed for oriC+), suggesting that in these cases DNA replication was being regulated through an oriC-independent mechanism (Figures 6A–C, S8A-C; red symbols). Interestingly, in some cases manipulation of different genes within a single biological pathway (e.g. – carbon metabolism or phospholipid synthesis) resulted in the regulation of DNA replication through the different regulatory systems.
Evidence for a DnaA-independent response to changes in growth rate
Since the ΔoriC oriN+ strain does not require DnaA activity to initiate DNA replication, it suggested that the observed oriC-independent growth rate regulation might be DnaA-independent. To address this possibility the oriC-independent mutants (Figures 6A–C, S8A–C) were crossed into a ΔdnaA strain that initiates DNA replication using oriN (Figure S3C). When PykA was depleted in the ΔdnaA mutant the ori:ter ratio no longer decreased, indicating that DnaA was required for this response, although apparently not for its role in origin recognition and DNA unwinding (Figures 6D, S8D; green symbols). In contrast, when either ribosomal genes were deleted or PgsA was depleted in the ΔdnaA mutant the ori:ter ratios did decrease, suggesting that DnaA-independent mechanisms act under these conditions (Figures 6E–F, S8E–F; green symbols). Taken together, the genetic analysis reveals that in B. subtilis there is likely more than one regulatory system linking DNA replication with cell growth.
Slowing growth rate by targeting essential cellular activities with small molecules inhibits DNA replication initiation
The importance of growth rate regulation of DNA replication in response to nutrient availability is self-evident, but the biological relevance of growth rate regulation of DNA replication in response to genetic manipulations is less clear. To address this issue we evaluated the response of DNA replication to sublethal concentrations of antibiotics that produced slow steady-state growth rates. We chose small molecules that inhibit either fatty acid synthesis (cerulenin) or protein synthesis (chloramphenicol) because our genetic analyses indicated that the former regulated DNA replication through oriC while the latter acted independently of both oriC and DnaA. Incubation with either antibiotic caused a decrease in the ori:ter ratios in the wild-type strain, showing that growth rate regulation of DNA replication in response to genetic perturbations of essential cellular activities is physiologically relevant (Figures 7A–B, S9A–B; black symbols).
Fig. 7. Analysis of oriC-dependent and oriC-independent growth rate regulation through small molecule targeting of fatty acid synthesis and protein synthesis. 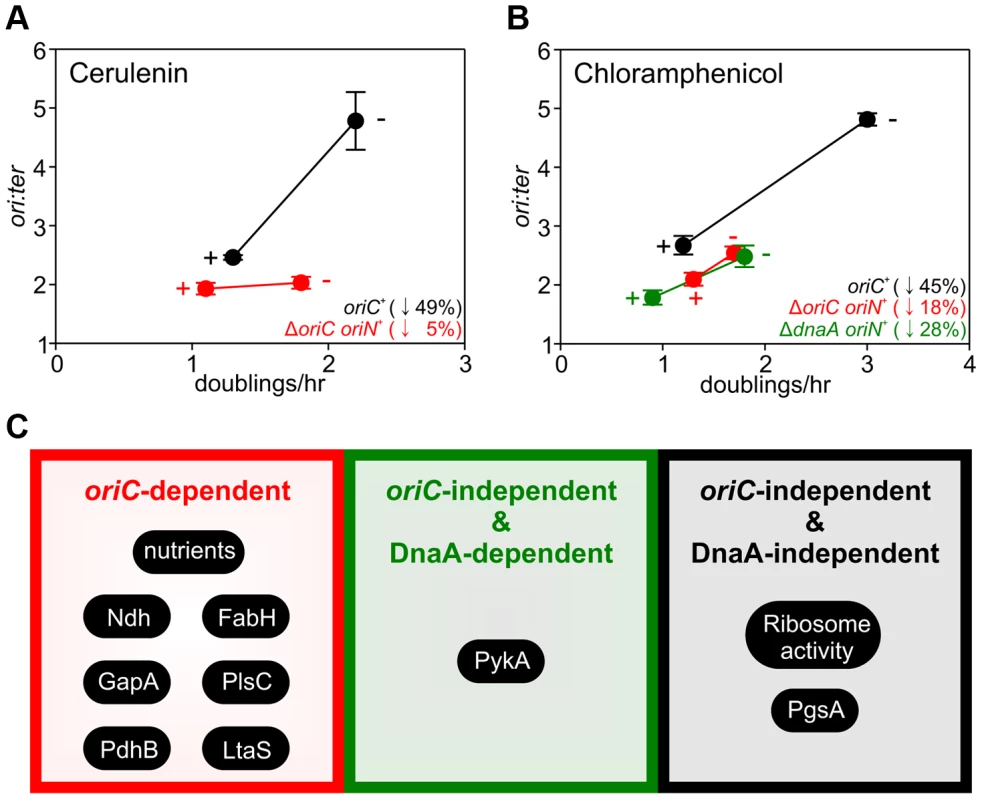
Strains were grown overnight at 37°C in LB medium. Overnight cultures were diluted 1∶1000 into fresh LB medium either without or with antibiotics (2 µg/ml cerulenin (A), 1 µg/ml chloramphenicol (B)) and grown at 37°C until they reached an A600 of 0.3–0.5. For datapoints “+” indicates the presence of the small molecule inhibitor and “−” indicates the absence. Genomic DNA was harvested from cells and marker frequency analysis was determined using qPCR. The ori:ter ratios are plotted versus growth rate and the percentage change in the ori:ter ratios comparing each deletion/depletion is indicated (error bars indicate the standard deviation of three technical replicates). Representative data are shown from a single experiment; an independently performed replicate of the experiment is shown in Figure S9. Wild-type (HM715), ΔoriC oriN+ (HM950), ΔdnaA oriN+ (HM1423). (C) Summary of growth rate control systems affecting DNA replication described in this report. To further assess whether changes in DNA replication caused by these small molecules reflected the results using genetic approaches, the ΔoriC oriN+ strain was analyzed. Only chloramphenicol elicited a significant decrease in the ori:ter ratios in the ΔoriC strain (Figures 7A–B, S9A–B; red symbols). Finally, the ΔdnaA oriN+ strain was analyzed in the presence of chloramphenicol and again the ori:ter ratio decreased (Figures 7B, S9B). This result is fully consistent with the data from ribosomal gene deletions and indicates that regulation of DNA replication in response to perturbed ribosome activity is DnaA-independent.
Discussion
We have found that nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis requires both DnaA and oriC. To our knowledge this is the first time that a specific DNA replication initiation protein has been shown to play an essential role in this regulatory system, and because DnaA is the earliest acting initiation factor we propose that DnaA is the target for the nutrient-mediated growth rate regulatory system. Critically however, we show that changes in DnaA protein level are not sufficient to account for nutrient-mediated growth rate regulation of DNA replication initiation in B. subtilis. This is in contrast to the generally accepted mechanism for control of bacterial DNA replication initiation based on work using E. coli [8], [9].
B. subtilis contains a bipartite origin that flanks the dnaA gene (incA and incB regions containing the dnaA promoter lie 1.3 kb upstream of the incC region which contains the DNA unwinding element) [38]. When the expression of the dnaA-dnaN operon was placed under the control of the inducible Pspac promoter in order to test the effect of DnaA overexpression on DNA replication initiation, a ∼9kb plasmid was recombined upstream of dnaA by single cross-over. Therefore, integration of this vector resulted in the considerable displacement of the two origin regions from one another without any significant consequence. It will be extremely interesting to determine the maximum and minimum distances that the inc regions can be moved, as well as ascertaining the role of the upstream region in DNA replication initiation.
We have shown that neither of the known DnaA regulatory proteins present during vegetative growth, Soj and YabA, are required for nutrient-mediated growth rate regulation of DNA replication initiation. We have also determined that the alarmone (p)ppGpp is not required for this regulation, consistent with a previous report that induction of the stringent response inhibits DNA replication elongation but not initiation in B. subtilis [39], [40]. This result marks an apparent distinction between the role of (p)ppGpp in B. subtilis and in proteobacteria such as E. coli and Caulobacter crescentus where (p)ppGpp has been shown to regulate DNA replication initiation [41], [42].
The use of genetic manipulations and small molecule inhibitors presented here reinforce and extend previously observed connections for bacterial DNA replication with central carbon metabolism [32], [33] and with phospholipid synthesis [43], [44]. In addition our work identifies new links for B. subtilis DNA replication with respiration, fatty acid synthesis, lipoteichoic acid synthesis, and ribosome biosynthesis. The results indicate that growth rate regulation of DNA replication in B. subtilis can be controlled through either oriC-dependent, oriC-independent/DnaA-dependent, or oriC-independent/DnaA-independent mechanisms (summarized in Figure 7C). Based on these novel findings we propose that multiple systems coordinate DNA replication with bacterial cell growth, with distinct regulators responding to diverse physiological and chemical changes. This model deviates from the long-standing concept of a single universal cellular property utilized to link bacterial DNA replication with cell growth [4].
Since nutrient-mediated growth rate regulation of DNA replication initiation requires DnaA activity at oriC, this suggests that factors affecting DNA synthesis through an oriC-independent mechanism (PykA, PgsA, and ribosomal proteins) are unlikely to be responsible for the nutrient sensing system. We suspect that nutrient-mediated regulation may be influenced by more than one control system, thereby forming a robust network capable of integrating information from multiple metabolic and cellular sources.
We note that for deletion/depletion mutants regulating DNA replication through oriC-dependent and oriC-independent/DnaA-dependent mechanisms, ori:ter ratios either remained constant or slightly increased in the ΔoriC and ΔdnaA strains, respectively (Figures 5, 6D, S7, S8D). Because the average initiation frequency of oriN appears to be growth rate independent (Figures 4A, 4C, S6A, S6C), the measured ori:ter ratios of these strains indicates that replication elongation speeds are either not being affected or are slightly decreasing. Therefore, for both oriC-dependent and oriC-independent/DnaA-dependent regulatory mechanisms, the observed decrease in ori:ter ratios in the oriC+ strain most likely reflects inhibition of DNA replication initiation. In contrast, for the oriC-independent/DnaA-independent mutants where the ori:ter ratio was decreased when DNA replication initiated from oriN, this difference could be due to a change in DNA replication elongation (although this would mean that the elongation speed was increased).
Our current aim is to determine the molecular mechanisms underlying each system that coordinates DNA replication with cell growth. We hypothesize that the oriC-dependent regulatory system targets DnaA activity at oriC. We speculate that the oriC-independent/DnaA-dependent regulatory system could influence DNA replication initiation by affecting the abundance or activity of a downstream replication initiation factor. For example, DnaA is also a transcription factor that is thought to directly regulate the expression of>50 genes, including dnaB [20], [45]. Alternatively, DnaA could act by titrating initiation factors away from oriN. Lastly, in the case of the oriC-independent/DnaA-independent regulatory system it needs to be established whether DNA replication is affected at the step of initiation or elongation, after which the role of appropriate candidate proteins can be investigated.
Materials and Methods
Strains and plasmids
Strains are listed in Table S1. Plasmids are listed in Table S2 and Table S3.
Media and chemicals
Nutrient agar (NA; Oxoid) was used for routine selection and maintenance of both B. subtilis and E. coli strains. For experiments in B. subtilis cells were grown using a range of media (using the following concentrations unless otherwise noted): Luria-Bertani (LB) medium, Antibiotic 3 (PAB) medium, Brain-Heart Infusion (Bacto), or defined minimal medium base (Spizizen minimal salts supplemented with Fe-NH4-citrate (1 µg/ml), MgSO4 (6 mM), CaCl2 (100 µM), MnSO4 (130 µM), ZnCl2 (1 µM), thiamine (2 µM)) supplemented with casein hydrolysate (200 µg/ml) and/or various carbon sources (succinate (1%), glycerol (0.5%), glucose (0.5%)). Supplements were added as required: tryptophan (20 µg/ml), phenylalanine (40 µg/ml), chloramphenicol (5 µg/ml), erythromycin (1 µg/ml), kanamycin (2 µg/ml), spectinomycin (50 µg/ml), tetracycline (10 µg/ml), zeocin (10 µg/ml). Unless otherwise stated all chemicals and reagents were obtained from Sigma-Aldrich.
Marker frequency analysis
Sodium azide (0.5%; Sigma) was added to exponentially growing cells to prevent further metabolism. Chromosomal DNA was isolated using a DNeasy Blood and Tissue Kit (Qiagen). The DNA replication origin (oriC) region was amplified using primers 5′-GAATTCCTTCAGGCCATTGA-3′ and 5′-GATTTCTGGCGAATTGGAAG-3′; the DNA replication terminus (ter) region was amplified using primers 5′-TCCATATCCTCGCTCCTACG-3′ and 5′-ATTCTGCTGATGTGCAATGG-3′. Either Rotor-Gene SYBR Green (Qiagen) or GoTaq (Promega) qPCR mix was used for PCR reactions. Q-PCR was performed in a Rotor-Gene Q Instrument (Qiagen). By use of crossing points (CT) and PCR efficiency a relative quantification analysis (ΔΔCT) was performed using Rotor-Gene Software version 2.0.2 (Qiagen) to determine the origin:terminus (ori:ter) ratio of each sample. These results were normalized to the ori:ter ratio of a DNA sample from B. subtilis spores which only contain one chromosome and thus have an ori/ter ratio of 1.
Microscopy
To visualize cells during exponential growth starter cultures were grown overnight and then diluted 1∶100 into fresh medium and allowed to achieve at least three doublings before observation. Cells were mounted on ∼1.2% agar pads (0.25× minimal medium base) and a 0.13–0.17 mm glass coverslip (VWR) was placed on top. To visualize individual cells the cell membrane was stained with either 2 µg/ml Nile Red (Sigma) or 0.4 µg/ml FM5-95 (Molecular Probes). To visualize nucleoids DNA was stained with 2 µg/ml 4′-6-diamidino-2-phenylindole (DAPI) (Sigma). Microscopy was performed on an inverted epifluorescence microscope (Nikon Ti) fitted with a Plan-Apochromat objective (Nikon DM 100x/1.40 Oil Ph3). Light was transmitted from a 300 Watt xenon arc-lamp through a liquid light guide (Sutter Instruments) and images were collected using a CoolSnap HQ2 cooled CCD camera (Photometrics). All filters were Modified Magnetron ET Sets from Chroma and details are available upon request. Digital images were acquired and analysed using METAMORPH software (version V.6.2r6). Analysis was performed using ImageJ software: foci counting utilized the particle analysis plugin; cell lengths and widths were measured using the ObjectJ plugin.
Western blot analysis
Proteins were separated by electrophoresis using a NuPAGE 4-12% Bis-Tris gradient gel run in MES buffer (Life Technologies) and transferred to a Hybond-P PVDF membrane (GE Healthcare) using a semi-dry apparatus (Hoefer Scientific Instruments). Proteins of interest were probed with polyclonal primary antibodies and then detected with an anti-rabbit horseradish peroxidase-linked secondary antibody using an ImageQuant LAS 4000 mini digital imaging system (GE Healthcare). Quantification was determined by densitometry using Image J software. Figure S3A shows that detection of DnaA, FtsZ and DnaN was within a linear range.
Supporting Information
Zdroje
1. CooperS, HelmstetterCE (1968) Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol 31 : 519–540.
2. HelmstetterCE, CooperS (1968) DNA synthesis during the division cycle of rapidly growing Escherichia coli B/r. J Mol Biol 31 : 507–518.
3. SchaechterM, MaaloeO, KjelgaardNO (1958) Dependency on medium and temperature on cell size and chemical coposition during balanced growth of Salmonella typhimurium. J Gen Microbiol 19 : 592–606.
4. DonachieWD (1968) Relationship between cell size and time of initiation of DNA replication. Nature 219 : 1077–1079.
5. WoldS, SkarstadK, SteenHB, StokkeT, BoyeE (1994) The initiation mass for DNA replication in Escherichia coli K-12 is dependent on growth rate. EMBO J 13 : 2097–2102.
6. LeonardAC, GrimwadeJE (2011) Regulation of DnaA Assembly and Activity: Taking Directions from the Genome. Annu Rev Microbiol 65 : 19–35.
7. DuderstadtKE, ChuangK, BergerJM (2011) DNA stretching by bacterial initiators promotes replication origin opening. Nature 478 : 209–213.
8. AtlungT, Lobner-OlesenA, HansenFG (1987) Overproduction of DnaA protein stimulates initiation of chromosome and minichromosome replication in Escherichia coli. Mol Gen Genet 206 : 51–59.
9. Lobner-OlesenA, SkarstadK, HansenFG, von MeyenburgK, BoyeE (1989) The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell 57 : 881–889.
10. HillNS, KadoyaR, ChattorajDK, LevinPA (2012) Cell size and the initiation of DNA replication in bacteria. PLoS Genetics 8: e1002549.
11. BoyeE, NordstromK (2003) Coupling the cell cycle to cell growth. EMBO Rep 4 : 757–760.
12. OguraY, ImaiY, OgasawaraN, MoriyaS (2001) Autoregulation of the dnaAzdnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J Bacteriol 183 : 3833–3841.
13. GoranovAI, BreierAM, MerrikhH, GrossmanAD (2009) YabA of Bacillus subtilis controls DnaA-mediated replication initiation but not the transcriptional response to replication stress. Mol Microbiol 74 : 454–466.
14. SharpeME, HauserPM, SharpeRG, ErringtonJ (1998) Bacillus subtilis cell cycle as studied by fluorescence microscopy: constancy of the cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J Bacteriol 180 : 547–555.
15. LarkKG, MaaloeO (1956) Nucleic acid synthesis and the division cycle of Salmonella typhimurium. Biochim Biophys Acta 21 : 448–458.
16. LarkKG, MaaloeO, RostockO (1955) Cytological studies of nuclear division in Salmonella typhimurium. J Gen Microbiol 13 : 318–326.
17. WeartRB, LevinPA (2003) Growth rate-dependent regulation of medial FtsZ ring formation. J Bacteriol 185 : 2826–2834.
18. MuntelJ, FromionV, GoelzerA, MaabetaS, MaderU, et al. (2014) Comprehensive absolute quantification of the cytosolic proteome of Bacillus subtilis by data independent, parallel fragmentation in liquid chromatography/mass spectrometry (LC/MS(E)). Mol Cell Proteomics 13 : 1008–1019.
19. HassanAK, MoriyaS, OguraM, TanakaT, KawamuraF, et al. (1997) Suppression of initiation defects of chromosome replication in Bacillus subtilis dnaA and oriC-deleted mutants by integration of a plasmid replicon into the chromosomes. J Bacteriol 179 : 2494–2502.
20. GoranovAI, KatzL, BreierAM, BurgeCB, GrossmanAD (2005) A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc Natl Acad Sci U S A 102 : 12932–12937.
21. MurrayH, ErringtonJ (2008) Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135 : 74–84.
22. ScholefieldG, ErringtonJ, MurrayH (2012) Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J 31 : 1542–1555.
23. OguraY, OgasawaraN, HarryEJ, MoriyaS (2003) Increasing the ratio of Soj to Spo0J promotes replication initiation in Bacillus subtilis. J Bacteriol 185 : 6316–6324.
24. SoufoCD, SoufoHJ, Noirot-GrosMF, SteindorfA, NoirotP, et al. (2008) Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis. Dev Cell 15 : 935–941.
25. MerrikhH, GrossmanAD (2011) Control of the replication initiator DnaA by an anti-cooperativity factor. Mol Microbiol 82 : 434–446.
26. ScholefieldG, MurrayH (2013) YabA and DnaD inhibit helix assembly of the DNA replication initiation protein DnaA. Mol Microbiol 90 : 147–159.
27. Noirot-GrosMF, DervynE, WuLJ, MerveletP, ErringtonJ, et al. (2002) An expanded view of bacterial DNA replication. Proc Natl Acad Sci USA 99 : 8342–8347.
28. LeePS, GrossmanAD (2006) The Chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol Microbiol 60 : 853–869.
29. WangX, TangOW, RileyEP, RudnerDZ (2014) The SMC Condensin Complex Is Required for Origin Segregation in Bacillus subtilis. Curr Biol 24 : 287–292.
30. Wang X, Montero Llopis P, Rudner DZ (2014) Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc Natl Acad Sci USA 10.1073/pnas.1407461111.
31. KawakamiH, KeyamuraK, KatayamaT (2005) Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J Biol Chem 280 : 27420–27430.
32. JanniereL, CanceillD, SuskiC, KangaS, DalmaisB, et al. (2007) Genetic evidence for a link between glycolysis and DNA replication. PloS One 2: e447.
33. MaciagM, NowickiD, JanniereL, Szalewska-PalaszA, WegrzynG (2011) Genetic response to metabolic fluctuations: correlation between central carbon metabolism and DNA replication in Escherichia coli. Microb Cell Fact 10 : 19.
34. SchneiderDA, GourseRL (2004) Relationship between growth rate and ATP concentration in Escherichia coli: a bioassay for available cellular ATP. J Biol Chem 279 : 8262–8268.
35. PetersenC, MollerLB (2000) Invariance of the nucleoside triphosphate pools of Escherichia coli with growth rate. J Biol Chem 275 : 3931–3935.
36. SaxenaR, FinglandN, PatilD, SharmaAK, CrookeE (2013) Crosstalk between DnaA protein, the initiator of Escherichia coli chromosomal replication, and acidic phospholipids present in bacterial membranes. Int J Mol Sci 14 : 8517–8537.
37. SikoraAE, ZielkeR, WegrzynA, WegrzynG (2006) DNA replication defect in the Escherichia coli cgtA(ts) mutant arising from reduced DnaA levels. Arch Microbiol 185 : 340–347.
38. MoriyaS, FukuokaT, OgasawaraN, YoshikawaH (1988) Regulation of initiation of the chromosomal replication by DnaA-boxes in the origin region of the Bacillus subtilis chromosome. EMBO J 7 : 2911–2917.
39. LevineA, VannierF, DehbiM, HenckesG, SerorSJ (1991) The stringent response blocks DNA replication outside the ori region in Bacillus subtilis and at the origin in Escherichia coli. J Mol Biol 219 : 605–613.
40. WangJD, SandersGM, GrossmanAD (2007) Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128 : 865–875.
41. LesleyJA, ShapiroL (2008) SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J Bacteriol 190 : 6867–6880.
42. SchreiberG, RonEZ, GlaserG (1995) ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Current Microbiol 30 : 27–32.
43. SekimizuK, KornbergA (1988) Cardiolipin activation of DnaA protein, the initiation protein of replication in Escherichia coli. J Biol Chem 263 : 7131–7135.
44. XiaW, DowhanW (1995) In vivo evidence for the involvement of anionic phospholipids in initiation of DNA replication in Escherichia coli. Proc Natl Acad Sci USA 92 : 783–787.
45. BreierAM, GrossmanAD (2009) Dynamic association of the replication initiator and transcription factor DnaA with the Bacillus subtilis chromosome during replication stress. J Bacteriol 191 : 486–493.
Štítky
Genetika Reprodukčná medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



