-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
Candida albicans and C. dubliniensis are fungal pathogens of humans. Both species possess TLO genes encoding proteins with homology to the Med2 subunit of Mediator. The more virulent pathogen C. albicans has 15 copies of the TLO gene whereas the less pathogenic species C. dubliniensis has only two (TLO1 and TLO2). In this study we show that a C. dubliniensis mutant missing both TLO1 and TLO2 is defective in virulence functions, including hyphal growth and stress responses but forms increased levels of biofilm. Analysis of gene expression in the tlo1Δ/tlo2Δ mutant revealed extensive differences relative to wild-type cells, including aberrant expression of starvation responses in nutrient-rich medium and retarded expression of hypha-induced transcripts in serum. Tlo1 protein was found to interact with genes and this was associated with both gene activation and repression. TLO1 was found to be better at restoring hyphal growth compared to TLO2 and but was less effective than TLO2 in supressing biofilm formation in the tlo1Δ/tlo2Δ strain. Thus, Tlo proteins regulate many virulence properties in Candida spp. and the expansion of the TLO family in C. albicans may account for the increased adaptability of this species relative to other Candida species.
Published in the journal: Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits. PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004658
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004658Summary
Candida albicans and C. dubliniensis are fungal pathogens of humans. Both species possess TLO genes encoding proteins with homology to the Med2 subunit of Mediator. The more virulent pathogen C. albicans has 15 copies of the TLO gene whereas the less pathogenic species C. dubliniensis has only two (TLO1 and TLO2). In this study we show that a C. dubliniensis mutant missing both TLO1 and TLO2 is defective in virulence functions, including hyphal growth and stress responses but forms increased levels of biofilm. Analysis of gene expression in the tlo1Δ/tlo2Δ mutant revealed extensive differences relative to wild-type cells, including aberrant expression of starvation responses in nutrient-rich medium and retarded expression of hypha-induced transcripts in serum. Tlo1 protein was found to interact with genes and this was associated with both gene activation and repression. TLO1 was found to be better at restoring hyphal growth compared to TLO2 and but was less effective than TLO2 in supressing biofilm formation in the tlo1Δ/tlo2Δ strain. Thus, Tlo proteins regulate many virulence properties in Candida spp. and the expansion of the TLO family in C. albicans may account for the increased adaptability of this species relative to other Candida species.
Introduction
Candida albicans is a commensal yeast commonly recovered from mucosal surfaces in humans. C. albicans is also a versatile opportunistic pathogen, responsible for a variety of superficial infections as well as more severe, life threatening infections in severely immunocompromised patients. Phenotypic versatility is an important characteristic of C. albicans and this flexibility allows C. albicans to adapt to this wide range of niches in the human host [1]–[3]. Over the past decade, transcriptional profiling of C. albicans using microarray and RNA-seq technologies has revealed that rapid adaptation to local environmental conditions involves elaborate, programmed shifts in transcription pattern. The biochemistry of transcriptional regulation has been studied intensively in the model yeast S. cerevisiae. Most genes transcribed by RNA polymerase II (PolII) require a carefully orchestrated series of events to initiate transcription. The formation of the pre-initiation complex and recruitment of PolII requires the function of Mediator, a large multi-subunit protein complex [4], [5]. Mediator is thought to be required to bridge DNA-bound transcription factors with the rest of the transcriptional machinery [6], [7]. The complex is generally divided into three modules: a head, middle and tail. The tail region includes Med2, Med3 and Med15/Gal11 and is thought to be the part of Mediator that interacts directly with transcription factors such as Gal4 and Gcn4 [8].
Most components of Mediator are highly conserved between S. cerevisiae and C. albicans [9]. However, completion of the C. albicans genome sequence [10], [11] revealed a family of up to 15 telomeric (TLO) genes that were subsequently found to encode a domain with homology to the Mediator tail subunit Med2 of S. cerevisiae [9], [12], [13]. This TLO gene expansion is unique to C. albicans, as most of the less pathogenic non-albicans Candida species such as C. tropicalis and C. parapsilosis have only one TLO gene. C. dubliniensis, the closest relative to C. albicans has two TLO genes, namely TLO1 and TLO2. TLO1 (Cd36_72860) is located internally on chromosome 7 and TLO2 (Cd36_35580) is at a telomere-adjacent locus on the right arm of chromosome R [14]. C. dubliniensis TLO2 appears to be the ancestral locus, based on synteny with, and homology to, the single C. tropicalis orthologue CTRG_05798.3 [14]. All candidal TLO genes share the Med2-like domain of the C. albicans TLOs and appear to be the MED2 orthologs in these species. Indeed, biochemical studies have shown that like the S. cerevisiae Med2 protein, Tlo proteins co-purify with the C. albicans orthologs of mediator tail module components Med3 and Med15 [9].
The 15 TLO genes in the C. albicans type strain SC5314 can be classified into four distinct clades based on gene structure, including the highly expressed TLOα clade (6 members), a single TLOβ gene, the poorly expressed TLOγ clade (7 members) and a single TLOψ gene which is a pseudogene [12]. Biochemical studies have shown that the levels of Tloα and Tloβ proteins is in vast excess to the amount necessary to be a stoichiometric subunit of Mediator (9) and stands in stark contrast to the situation in S. cerevisiae, where the Med2 subunit is expressed at roughly equivalent amounts to other subunits of the complex. Due to the large numbers of TLO genes in C. albicans, functional analysis of Mediator has largely been restricted to other subunits, including Med3 (tail domain), Med31 (middle domain), Med20 (head domain) and Med13/Srb9 (kinase domain). Med31 and Med20 are required for the transition of C. albicans yeast cells to filamentous hyphae and for biofilm formation, two important pathogenic traits of the organism [9], [15]. Med31 is required for expression of the genes ALS1 and ALS3 that encode cell-surface proteins involved in biofilm production, consistent with the poor biofilms produced by med31Δ mutants. In addition, Med31 is required for the expression of genes regulated by the transcription factor Ace2, which regulates many genes involved in cell wall remodelling during cell separation. Consistent with this, the med31Δ mutant exhibits defective cytokinesis. Deletion of MED3 resulted in a stronger filamentous growth defect, resulting in short pseudohyphae in serum or liquid media supplemented with GlcNac [9]. Deletion of Mediator subunits also has complex effects on the transcriptional circuit governing the white-opaque switch, a phenotypic transition involved in mating [16].
C. albicans is the most prevalent pathogenic fungal species. The closely related species C. dubliniensis is responsible for far fewer infections and is a minor component of the human oral flora [14], [17]. The yeast-to-hypha transition is an important virulence trait of C. albicans and C. dubliniensis forms fewer hyphae than C. albicans in response to most in vitro and in vivo hypha-inducing stimuli [17]. C. dubliniensis requires much stronger environmental cues, including nutrient depletion, in order to activate a transcriptional response that will induce the yeast-to-hypha transition [18]–[20]. Less effective transcriptional responses may also account for the increased susceptibility of C. dubliniensis to environmental stress relative to C. albicans [21]. It has been suggested that the expansion of the TLO family in C. albicans may account for the greater transcriptional flexibility and adaptability of this species relative to C. dubliniensis and the other non-albicans Candida species [22]. However, direct analysis of TLO null mutants has not been attempted in C. albicans, due to the large number of TLO genes and the likelihood of redundancy in the TLO family.
In this study, we exploited the closely related species C. dubliniensis as a model to study TLO gene function and to construct the first tlo null strains. We found that C. dubliniensis mutants lacking both TLO1 and TLO2 (tlo1Δ/tlo2Δ) exhibit widespread changes in the transcription of virulence-associated genes. ChIP analysis detected Tlo1 within the coding regions of ORFS, as well as subtelomeric regions and the Major Repeat Sequence (MRS). Interestingly, in strains lacking only one of the two TLO genes, expression of distinct subsets of genes was altered. We propose that expansion of the TLO family, even to only two members, has facilitated the evolution of functional diversity and may be of particular importance in the evolution of an expanded set of TLO paralogs together with the increased virulence in Candida albicans.
Results
Candida dubliniensis possesses two expressed TLO paralogs
The two Candida dubliniensis TLO genes, TLO1 and TLO2, encode proteins of 320 and 355 amino acids, respectively, and share 58% identity. Homology to each other and to the C. albicans Tlo proteins is concentrated in the N-terminal 120 residues of the proteins. Tlo1 and Tlo2 share 81% identity in this N-terminal region, which also exhibits homology to S. cerevisiae Med2 (Figure 1A). The amino acid sequence of Tlo2 is 25 residues longer and contains a central triplet repeat of the motif KAAAKVKEEQ. C. dubliniensis is a diploid organism and gene deletion studies (see below) and Southern blot analysis confirmed that C. dubliniensis strain Wü284 has two alleles of TLO1 (Figure S1). However, Southern blot experiments could detect only one allele of TLO2 at the telomere of chromosome R (ChR) in strain Wü284, indicating that one copy of ChR in Wü284 was truncated resulting in loss of one allele of TLO2 (Figure S1). PCR analysis of the tlo2Δ strain confirmed that bases 1 to 918 of the second TLO2 allele are deleted due to this truncation, with only a small 150 bp remnant of the ORF remaining. Analysis of gene expression by QRT-PCR revealed that TLO1 mRNA is expressed over 2-fold higher than TLO2 mRNA in YEPD at 37°C (Figure 1B).
Fig. 1. Structure and expression of C. dubliniensis TLO genes. 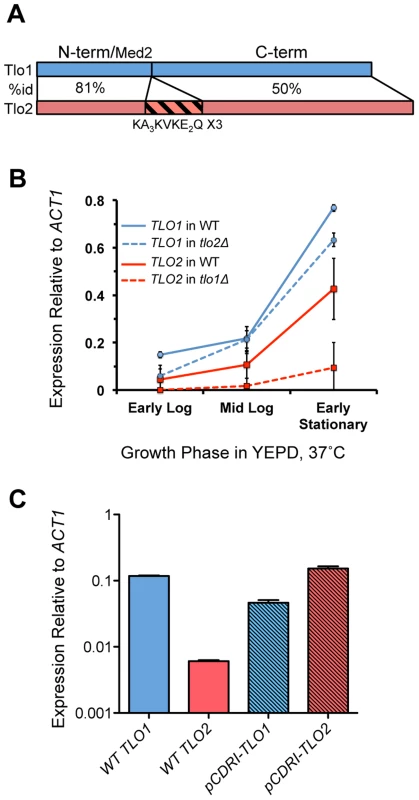
(A) TLO1 and TLO2 encode proteins of 320 and 355 amino acids respectively that exhibit 81% identity in the N-terminal Med2-like domain, and 50% identity in the C-terminal portion of the protein. In addition, Tlo2 has an internal triplet repeat, indicated. (B) Expression of TLO1 and TLO2 was measured by QRT-PCR using gene-specific primers during growth in YEPD broth at 37°C in early exponential (1.5 h-post inoculation), mid-exponential (8 h) and early stationary phase (18 h) in strain Wü284 (WT). The effect of deletion of either TLO1 or TLO2 on the expression of the other paralog was also analysed. (C) Expression of reintegrated copies of TLO1 and TLO2 in plasmid pCDRI in the tlo1Δ/tlo2Δ strain. C. dubliniensis Tlo proteins and Med3 are components of the Mediator tail module
We first asked if C. dubliniensis Tlo is a component of Mediator. To address this, we purified Mediator from whole cell extracts of C. dubliniensis wild-type and tlo1Δ/tlo2Δ mutant cells using a dual affinity tag on the C. dubliniensis ortholog of the conserved Med8 subunit of Mediator. The resulting Mediator complex was analyzed for purity by SDS-PAGE and for composition by mass spectroscopy with untagged WT C. dubliniensis serving as a control. Biochemical analysis showed that the C. dubliniensis Mediator, like C. albicans, is composed of a complete set of orthologs of the S. cerevisiae complex (Figure 2). Mediator purified from the tlo1Δ/tlo2Δ mutant lacked tail subunits Med3, Med15, Med16 and Med5 subunits (Figure 2, Lane 3). Importantly, Mediator purified from the C. dubliniensis tlo1Δ/tlo2Δ strain or from a med3Δ null strain had equivalent composition, lacking tail components Tlo1, Med3, Med5, Med15 and Med16 (Figure 2, Lanes 3 and 4).
Fig. 2. Biochemical analysis of Mediator in C. dubliniensis wild-type and tlo1Δ/tlo2Δ. 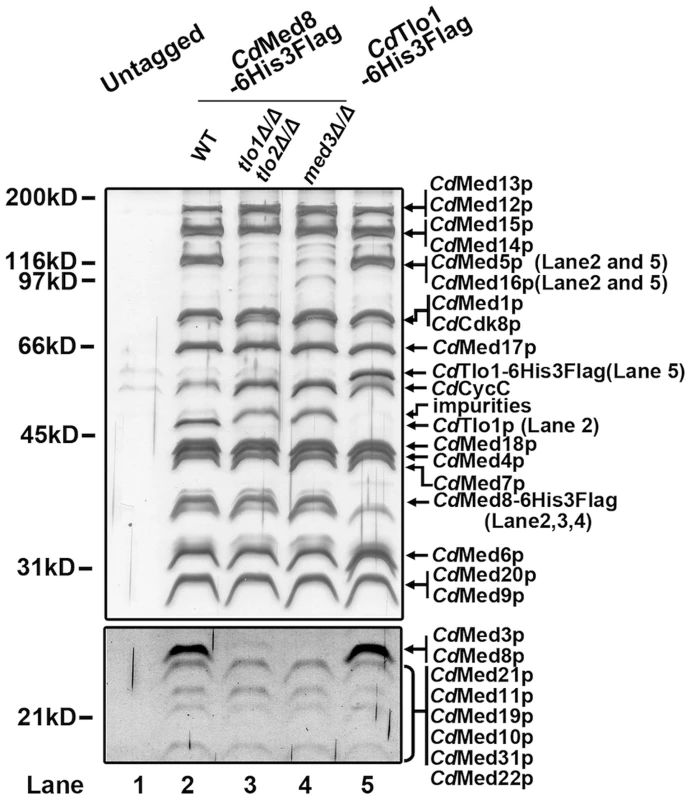
Lysates from untagged WT C. dubliniensis (Lane 1), a Med8-6His-Flag-tagged WT strain (Lane 2), a Med8-6His-Flag-tagged tlo1Δ/tlo2Δ strain (Lane 3), a Med8-6His-Flag-tagged med3Δ strain (Lane 4) and a Tlo1-6His-Flag-tagged WT strain (Lane 5) were subjected, in parallel, to multiple chromatographic separations. The elutions from the IMAC (Talon) step were analyzed by 8% SDS-PAGE. Proteins were revealed by staining with silver. Bands that are present in WT but clearly absent in the tlo1Δ/tlo2Δ and med3Δ Mediator are indicated in parenthesis on the right. The absence of these proteins in the tlo1Δ/tlo2Δ Mediator was confirmed by mass spectroscopy. To determine the relative abundance of Tlo proteins compared to other Mediator subunits, we constructed strains carrying Tlo1-HA, Med3-HA and Med8-HA tagged derivatives. Immunoblotting of whole cell extracts revealed that C. dubliniensis Tlo1 Mediator subunit is expressed in amounts comparable to the Med3 Tail Module and Med8 Head Module subunits (Figure S2). Consistent with this finding, purification of an affinity tagged Tlo1 protein from C. dubliniensis yielded only the Mediator associated form (Figure 2, Lane 5) and no free Tlo1 protein. Thus, in C. dubliniensis Tlo1 appears to be acting solely as a component of Mediator.
TLO1 activates hyphal growth to a greater extent than TLO2
Deletion of TLO2 did not reduce filamentous growth significantly (Figure 3A), while deletion of TLO1 resulted in reduced hyphal production (Figure 3A), as reported previously [14]. However, the double tlo1Δ/tlo2Δ mutant (tloΔΔ) had a more severe filamentation defect than the TLO1 deletion alone, suggesting that TLO2 partly compensates for the deletion of TLO1 in the single mutant (Figure 3A, 3D). The cellular morphology of the tlo1Δ/tlo2Δ mutant in 10% serum was characteristically pseudohyphal (Figure 3D).
Fig. 3. Phenotypic analysis of C. dubliniensis tloΔ and med3Δ mutants. 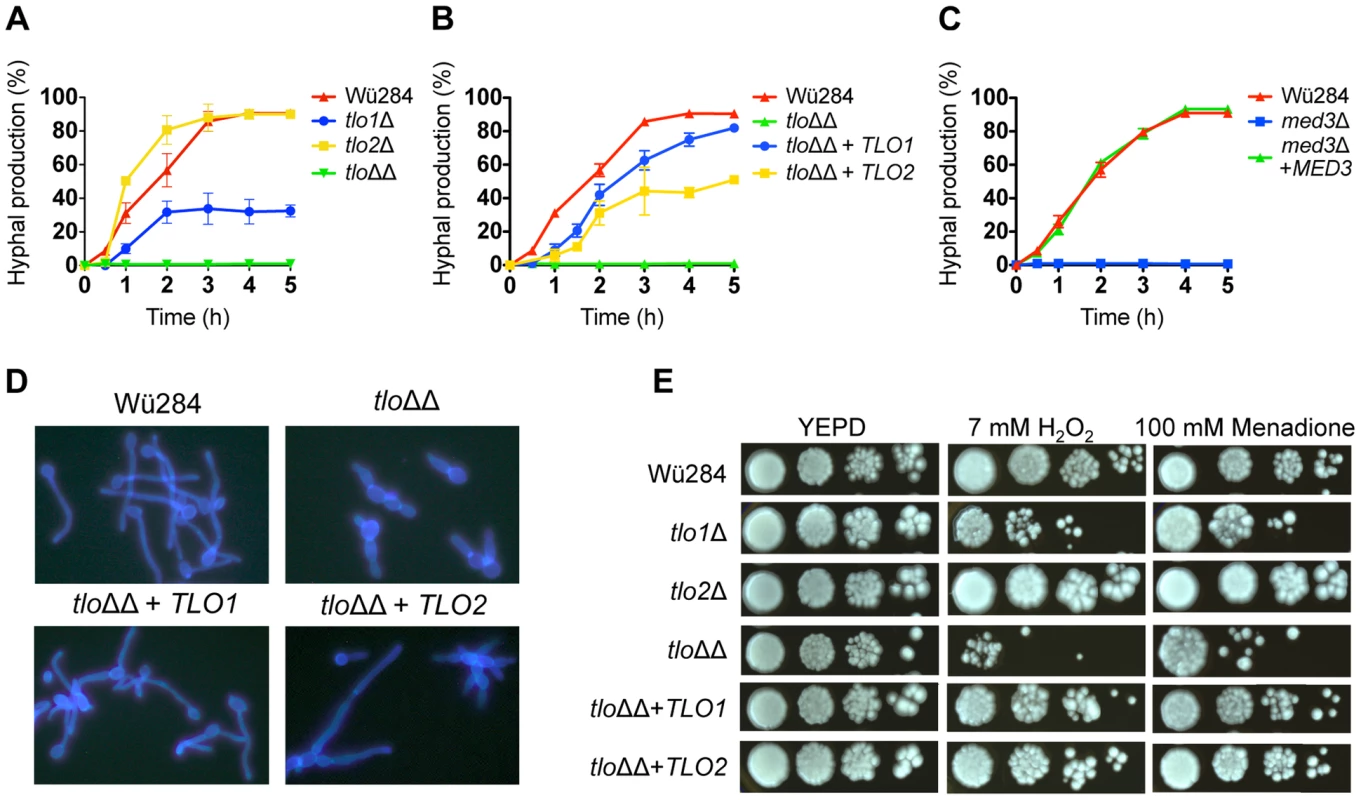
The rate of formation of true hyphae in water supplemented with 10% serum was measured in the tlo1Δ, tlo2Δ and tlo1Δ/tlo2Δ (tloΔΔ) mutants relative to Wü284 (A), in the TLO1 and TLO2 reintegrants (tloΔΔ+TLO1/2)(B) and in the med3Δ mutant (C). Measurements were taken over a period of 5 h by counting the number of cells producing unconstricted germ-tubes and expressing this value as a percentage of the whole population. (D) Cellular morphology of Wü284, the tlo1Δ/tlo2Δ (tloΔΔ) mutant and TLO1 and TLO2 reintegrated strains in 10% serum following staining with calcofluor white. (E) Oxidative stress susceptibility in tloΔ mutants. Susceptibility to oxidative stress was assessed by spotting 20 µl aliquots of 10-fold serially diluted suspensions (106 to 103 cells/ml) of the indicated strains on YEPD agar plates containing 7 mM H2O2 or 100 mM menadione. Plates were incubated for 48 h at 37°C. In order to investigate if TLO1 and TLO2 are functionally different, we reintroduced TLO1 and TLO2 individually to the tlo1Δ/tlo2Δ mutant using a previously described integrating vector pCDRI. This strategy was employed as it allowed us to express both TLO1 and TLO2 to similar levels (Figure 1C) thus allowing a direct comparison of their ability to complement the phenotypes of the tlo1Δ/tlo2Δ mutant. Introduction of TLO1 could restore true hypha production to almost wild-type levels, whereas TLO2 restored hypha formation to approximately 50% of wild-type levels (Figure 3B, 3D). These data show that TLO1 and TLO2 differ in their ability to activate filamentous growth.
Deletion of TLO1 and TLO2 produces multiple phenotypes
The colony morphology of the tlo1Δ, tlo2Δ and tlo1Δ/tlo2Δ mutant strains following growth on nutrient-rich solid medium (YEPD agar) at 37°C was indistinguishable from that of wild-type. At the cellular level, the tlo1Δ/tlo2Δ mutant grew as short chains of 3–4 cells, which is indicative of a defect in cytokinesis (Figure S3A). A similar phenotype was described by Uwamahoro et al. [15] in a C. albicans med31Δ mutant. Wild-type cytokinesis was restored in the tlo1Δ/tlo2Δ mutant following reintroduction of TLO1 but not TLO2 (Figure S3A). Growth defects of tlo mutants became apparent when grown on non-optimal media. On Spider agar, the wild-type strain formed smooth white colonies, typical of C. dubliniensis on this medium. However, the tlo1Δ/tlo2Δ mutant formed heavily wrinkled colonies on this medium (Figure S3B). Wild-type smooth colonies could be restored by complementation of the tlo1Δ/tlo2Δ mutant with either TLO1 or TLO2 (Figure S3B). Following 5 days incubation on Pal's medium, the double mutant produced extensive levels of pseudohyphae, but unlike wild-type which produced copious amounts of chlamydospores on this medium, no terminal differentiation of these pseudohyphae to chlamydospores was observed (Figure S4A). The TLO1 mutant and the tlo1Δ/tlo2Δ double mutant also exhibited longer doubling times in several different liquid media, including YEPD (Table 1). When galactose was substituted for glucose in YEP medium (YEP-GAL), the growth rate of the tlo1Δ and the tlo1Δ/tlo2Δ mutants was reduced with doubling times 1.4 to 1.6 times longer than wild-type doubling times, respectively (Table 1). Growth in synthetic liquid Lee's medium was also retarded, with a doubling time 1.6 fold higher than that of wild-type. The addition of 1% peptone restored growth to WT levels in the tlo1Δ/tlo2Δ mutant (Figure S4B) while the addition of carbon sources or individual amino acids to solid Lee's medium did not.
Tab. 1. Doubling times (min) of tloΔ mutants and complemented strains in vitro.* 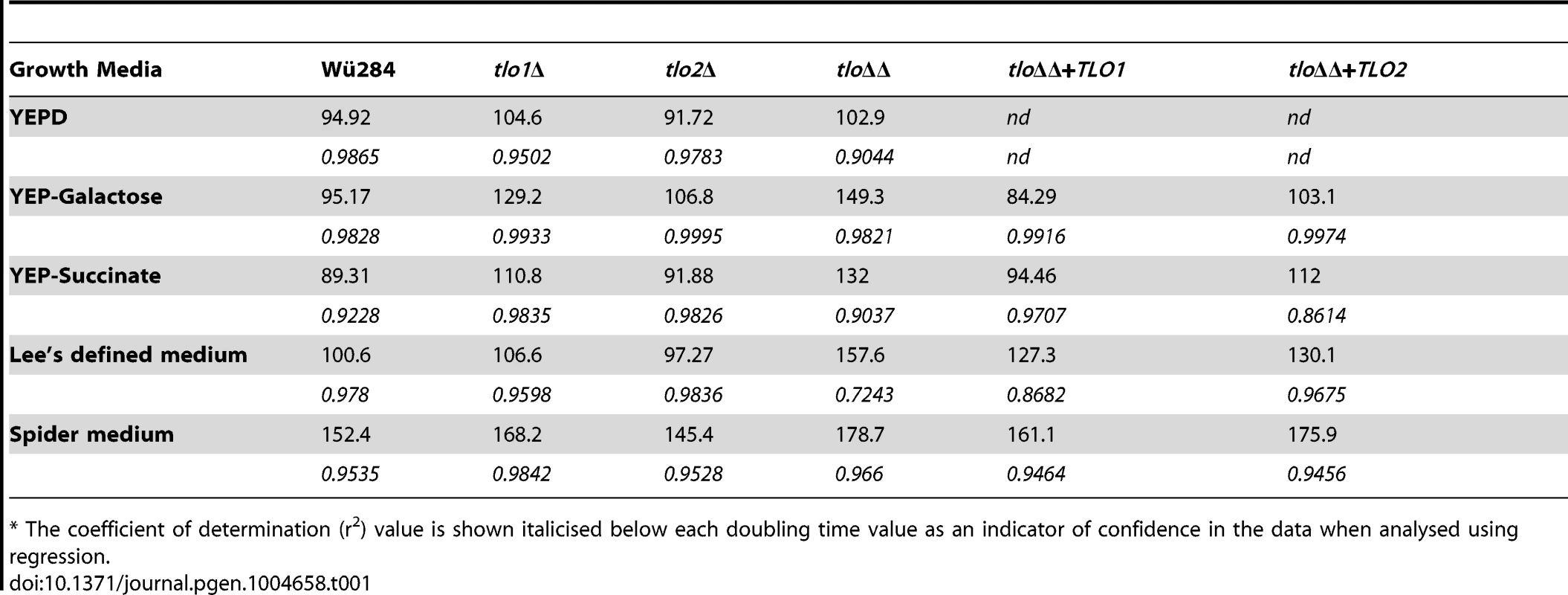
* The coefficient of determination (r2) value is shown italicised below each doubling time value as an indicator of confidence in the data when analysed using regression. Reintroduction of either TLO1 or TLO2 in the tlo1Δ/tlo2Δ double mutant background restored doubling times to wild-type levels in YEP-GAL. However, reintroduction of TLO1 had a significantly greater effect on doubling time compared to complementation with TLO2. In contrast, either TLO1 or TLO2 restored the growth defect on Lee's medium to the same degree (Table 1).
We next investigated if TLO genes affect a range of stress responses. Supplementation of YEPD agar with H2O2 (7 mM) or menadione (100 mM), which generate reactive oxygen species, inhibited growth of the tlo1Δ/tlo2Δ mutant, partly inhibited growth of the tlo1Δ mutant, while growth of the tlo2Δ single mutant was largely unaffected by these oxidizing agents (Figure 3E). Reintroduction of either TLO1 or TLO2 to the tlo1Δ/tlo2Δ strain restored wild type growth in the presence of both H2O2 and menadione, indicating that both TLO genes have a role in the growth of cells under oxidative stress.
med3Δ phenocopies a tlo1Δ/tlo2Δ mutant
Tlo proteins are components of the Mediator complex, which is thought to facilitate interactions between Mediator and DNA-bound transcriptional activators [8]. As Tlo protein and Med3 are required for the stability of the Mediator tail in C. dubliniensis (Figure 2), we hypothesized that deletion of MED3 and TLO genes should result in similar phenotypic effects. To test this, we compared the phenotypes of C. dubliniensis tlo1Δ/tlo2Δ and med3Δ mutants. The C. dubliniensis med3Δ mutant, similar to the C. albicans med3Δ mutant [9] and the C. dubliniensis tlo1Δ/tlo2Δ mutant was unable to form true hyphae in 10% serum (Figure 3C) and exhibited extended doubling times in YEP-Gal (Figure S5A) and increased susceptibility to H2O2 (Figure S5B). All of these properties could be restored to wild-type levels in the med3Δ mutant with the reintroduction of a wild-type copy of MED3 on pCDRI (Figure 3C, S4A, S4B).
Tlo proteins regulate the transcription of a diverse set of genes
Since Mediator is important for transcription regulation, we analysed RNA expression patterns in the tlo1Δ/tlo2Δ mutant relative to wild-type cells grown in nutrient-rich growth conditions (YEPD at 37°C) and grown in hyphal inducting conditions (water plus 10% serum, optimal for C. dubliniensis hypha formation). During exponential growth in YEPD, a total of 746 genes exhibited a 1.5-fold or greater increase in expression and 635 genes exhibited a 1.5-fold or greater reduction in expression (Q≤0.05; Figure 4A). This scale of differential gene expression observed in our tlo1Δ/tlo2Δ is similar to that seen in S. cerevisiae Mediator tail mutants [23], [24]. In the nutrient-rich YEPD broth, the tlo1Δ/tlo2Δ mutant exhibited a transcriptional profile that resembled a response to nutrient starvation (Figure 4B). The induced set of genes was enriched for processes associated with catabolism of alternative carbon and nitrogen sources such as N-acetyl-glucosamine (NAG1, NAG3, NAG4, NAG6), amino acids (e.g. GDH2, CAR1, PUT2, PUT1, LPD1, FDH1 and FDH3) and fatty acids. The tlo1Δ/tlo2Δ mutant cells also up-regulated key genes of gluconeogenesis (PCK1 and FBP1) and the glyoxylate cycle (ICL1 and MDH1) (Figure 4B). In concert with this, the tlo1Δ/tlo2Δ mutant also caused down-regulation of genes encoding glycolytic enzymes (PFK1, PFK2, FBA1, GPM1 and ENO1) and the glycolytic regulator TYE7. The tlo1Δ/tlo2Δ mutant also exhibited a greater than 2-fold decrease in expression of genes encoding proteins important for sulphur amino acid biosynthesis (e.g. SAM2, MET1, MET6, MET10, MET14, MET16) and ergosterol biosynthesis (ERG1, ERG9, ERG25). In addition, some hypha-specific genes were induced in the tlo1Δ/tlo2Δ mutant grown in YEPD. This included IHD1, RBT5 and SAP7 (induced in C. dubliniensis hyphae) as well as several regulators of biofilm and hyphal growth (BCR1, NRG1, SFL1, TEC1 and EED1).
Fig. 4. Microarray gene expression profiling of the tlo1Δ/tlo2Δ (tloΔΔ) mutant in YEPD and 10% serum. 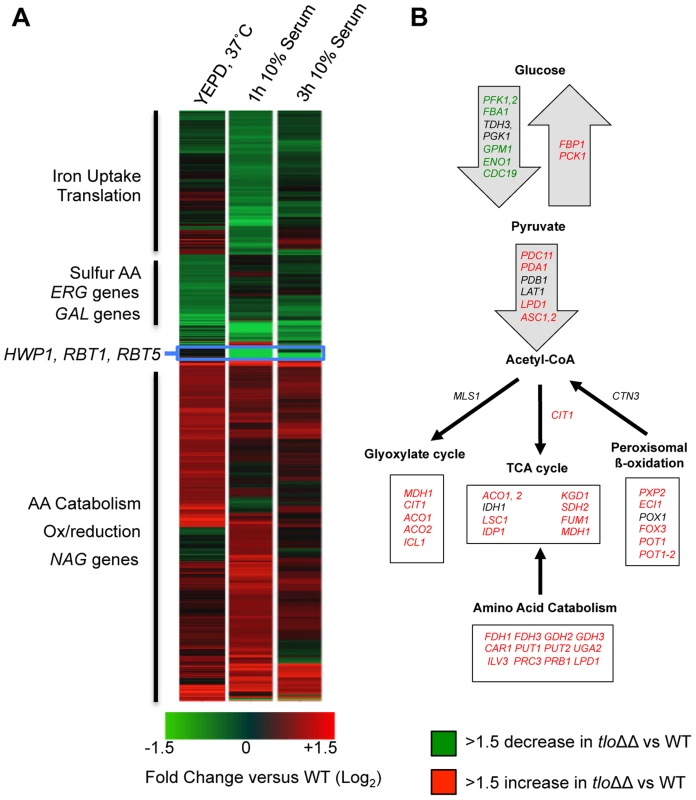
(A) A heat map generated in GeneSpring GX12 showing all 1.5 fold regulated genes in the tlo1Δ/tlo2Δ (tloΔΔ) mutant relative to wild type strain Wü284 during exponential growth in YEPD broth and following inoculation in 10% (v/v) serum (1 h and 3 h). The fold change (Log2) relative to wild-type is color coded as indicated in the lower panel. GO terms associated with up and down regulated clusters are indicated on the left. (B) A cartoon metabolic map of the tlo1Δ/tlo2Δ (tloΔΔ) mutant showing the changes in expression of genes involved in energy metabolism during growth in YEPD. Genes in green exhibit a 1.5-fold or greater reduction in expression relative to wild-type whereas those in red exhibited a 1.5-fold increase in expression. Genes in black were not significantly changed. Gene Set Enrichment Analysis (GSEA) was used to compare the tlo1Δ/tlo2Δ regulated genes with published microarray datasets. GSEA indicated that the set of genes differentially expressed in the tlo1Δ/tlo2Δ mutant was enriched for genes induced during infection of RHE [25] and genes both induced and repressed during infection of bone-marrow derived macrophages [26] (Figure S6).
We also examined the transcript profile of the tlo1Δ/tlo2Δ mutant in 10% serum relative to wild-type (Figure 4A). After 1 h, 868 genes were significantly up-regulated 1.5-fold or greater and 816 ORFs downregulated (Q≤0.05) relative to wild-type expression at the same time point. In this medium the tlo1Δ/tlo2Δ mutant exhibited reduced expression of several key regulators of filamentous growth including RAS1, RIM101, EFG1 and UME6. In addition, the hyphal growth regulating cyclins CLN3 and HGC1 were also down regulated in the tlo1Δ/tlo2Δ mutant. The tlo1Δ/tlo2Δ mutant also exhibited reduced expression of many cell wall proteins whose induction is chracteristic of the yeast-to-hypha transition in C. dubliniensis, including HWP1, RBT1, RBT5 and SOD5 [20]. Paradoxically the tlo1Δ/tlo2Δ mutant exhibited a more rapid induction of the hypha-specific gene ECE1 following 1 h incubation in serum, and expression remained elevated relative to wild-type at 3 h. In addition to hyphal genes, the tlo1Δ/tlo2Δ mutant exhibited reduced expression of galactose metabolic genes GAL1, GAL7 and GAL10.
After 3 hours in 10% serum, a similar number of genes were differentially expressed 1.5-fold or greater (n = 1726). However, the level of differential expression at many genes had decreased by 3 h (Figure 4A).
TLO1 and TLO2 restore expression of different subsets of genes
In order to investigate whether TLO1 and TLO2 regulate similar or different sets of genes, we analysed the transcript profiles of the TLO1 and TLO2 complemented mutants rather than the tlo1Δ and tlo2Δ single mutants, as the complemented strains exhibited equivalent expression of TLO1 or TLO2 (Figure 1B, 1C). Both TLO1 and TLO2 largely restored the transcript profile of the tlo1Δ/tlo2Δ mutant back to wild-type levels in both YEPD and 10% serum (Figure 5A, 5B). However, some TLO-specific transcript patterns were also detectable. Sixty-one genes were regulated in a TLO-specific manner in YEPD (Table S1). In comparison to the TLO1-complemented strain, the TLO2-complemented strain showed a greater reduction in expression of many tlo1Δ/tlo2Δ induced genes, including many hyphal genes (IHD1, SAP7 and EED1), negative regulators of hyphal growth (NRG1 and SFL1) and some starvation-induced genes such as CAR1, GDH3, NAG3 and NAG4 (Figure 5C). The TLO2 complemented strain was also better at restoring wild-type levels of SOD6 expression relative to the TLO1 complemented strain (Figure 5C).
Fig. 5. Reintroduction of TLO1 or TLO2 in the tlo1Δ/tlo2Δ (tloΔΔ) mutant restores wild-type levels of expression of overlapping and distinct sets of genes. 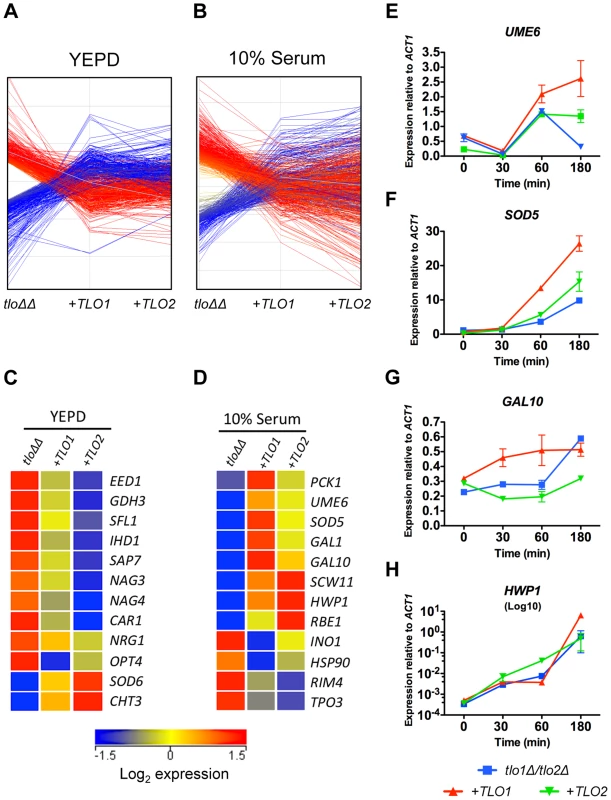
(A) A graph plotting the change in expression of genes up regulated (red lines) or down regulated (blue lines) in the tlo1Δ/tlo2Δ (tloΔΔ) mutant following complementation with either TLO1 (+TLO1) or TLO2 (+TLO2) in YEPD or (B) 10% (v/v) serum. (C) Heat map of selected genes that were differentially expressed in the tlo1Δ/tlo2Δ (tloΔΔ) mutant during growth in YEPD broth (C) or 10% (v/v) serum (D) that exhibited a TLO1 or TLO2 specific restoration to wild-type in expression. Colors in the first column (tlo1Δ/tlo2Δ) represent the fold change in expression of the indicated genes in tloΔΔ vs wild-type; colors in columns 2 and 3 (+TLO1 and +TLO2) represent the fold change in expression of the indicated genes following reintegration of TLO1 or TLO2 versus tloΔΔ. (E to H) QRT-PCR of the indicated genes was carried out following 30, 60 and 180 min incubation in 10% (v/v) serum at 37°C. Gene expression was determined using Sybr green technology and gene-specific levels are expressed relative to ACT1. Primer sequences are listed in Table S5. In 10% serum, TLO1 was more effective at restoring expression of several genes required for induction of, and induced during, filamentous growth including UME6 and SOD5 (Table S2, Figure 5D). TLO1 was also a more effective inducer of GAL1 and GAL10 expression, which may explain why restoration of growth in galactose medium was more effective in the TLO1 reintegrant relative to the TLO2 complemented strain (Figure 5D). In contrast, TLO2 was more effective at inducing HWP1 expression (Figure 5D). More detailed analysis of hyphal-induced gene expression by QRT-PCR corroborated these microarray findings and supported the existence of a temporal defect in the induction of hypha specific genes in the tlo1Δ/tlo2Δ mutant. QRT-PCR showed that UME6 and SOD5 exhibited weak levels of induction in the tlo1Δ/tlo2Δ mutant relative to the TLO1 reintegrant (Figure 5E, 5F). Although addition of TLO2 led to a minor increase in SOD5 expresssion, complementation with TLO1 restored high-level induction of SOD5. Similarly, induction of GAL10 also occurred more rapidly in the TLO1-reintegrated strain relative to both the tlo1Δ/tlo2Δ mutant and the TLO2-reintegrated strain (Figure 5G) Conversely, QRT-PCR confirmed that TLO2 restored more rapid induction (∼10-fold) of the HWP1 transcript following 1 h incubation in serum relative to the TLO1 reintegrant (Figure 5H). However HWP1 expression in the tlo1Δ/tlo2Δ mutant and TLO1 reintegrant reached similar levels following three hours incubation in serum (Figure 5H).
This transcriptional analysis of the TLO1 and TLO2 complemented strains for the first time demonstrate functional diversification of Tlo proteins and show that different Tlos can regulate distinct subsets of genes.
Deletion of MED3 disrupts transcription of a similar set of genes
This study has shown that deletion of MED3 in C. dubliniensis results in similar phenotypes to those observed in the tlo1Δ/tlo2Δ mutant. In order to determine whether these gene deletions resulted in similar changes in transcript profile, we compared gene expression in the tlo1Δ/tlo2Δ and the med3Δ mutant in YEPD broth and in 10% serum. Hierarchical clustering was used to compare all 1.5-fold regulated genes in strains tlo1Δ/tlo2Δ and med3Δ during exponential growth. This comparison demonstrated the similarity of the transcriptional changes in both mutants relative to wild-type cells (Figure 6A). A large number (n = 47) of these commonly down-regulated genes were associated with the GO term “ribosome biogenesis” (P = 3.89×10−19; FDR = 0.0). In YEPD, both mutants exhibited aberrantly increased expression of the hypha-specific genes IDH1, RBT5 and SAP7 as well as the regulators TEC1 and EED1. Activation of starvation responses was also evident in the med3Δ strain, which like the tlo1Δ/tlo2Δ strain exhibited increased expression of genes involved in gluconeogenesis (PCK1, FBP1) and the glyoxylate cycle (ICL1, MDH1). Decreased expression of glycolytic genes was less pronounced in med3Δ, however we could detect decreased expression (>1.5-fold) of PFK1, PFK2, CDC19 the glycolytic regulator TYE7 (Figure S7). Similar to tlo1Δ/tlo2Δ, med3Δ also exhibited increased expression of genes involved in oxidation/reduction processes, including amino acid catabolism (Figure 6B) in addition to reduced expression of genes involved in sulphur amino acid metabolism (CYS3, MET1, 2, 4, 10) and ergosterol metabolism (ERG1, ERG251).
Fig. 6. Microarray gene expression profiling of the med3Δ mutant in YEPD and 10% (v/v) serum. 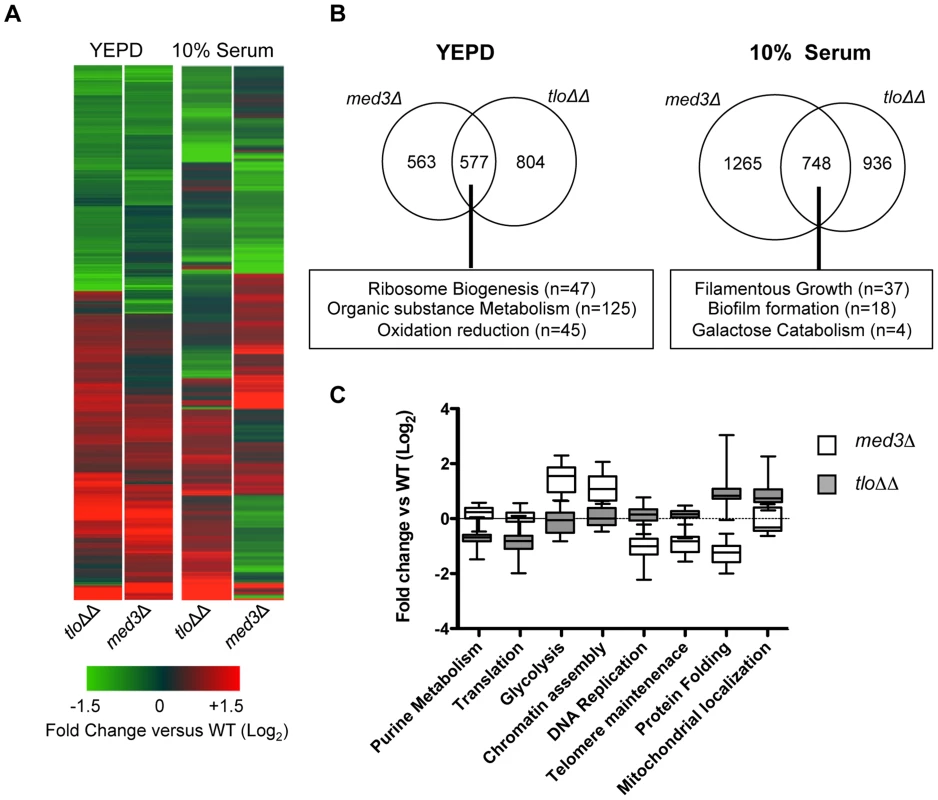
(A) A heat map generated in GeneSpring GX12 showing all 1.5 fold regulated genes in the tlo1Δ/tlo2Δ (tloΔΔ) and med3Δ mutants relative to wild type strain Wü284 during exponential growth in YEPD broth and following inoculation in 10% serum (1 h). The fold change (Log2) relative to wild-type is color coded as indicated in the lower panel. (B) Venn diagrams illustrating the numbers of genes commonly regulated greater than 1.5-fold in the tlo1Δ/tlo2Δ (tloΔΔ) and med3Δ mutants following growth in YEPD broth or 10% serum. The box below indicates major functional classes of commonly regulated genes identified by GO term analysis using the GO Term finder at CGD (http://www.candidagenome.org/). (C) Graph plotting the expression levels (Log2 fold change versus wild-type) of genes associated with selected GO terms in the tlo1Δ/tlo2Δ (tloΔΔ) and med3Δ mutants during growth in 10% (v/v) serum. Following 1 h growth in water plus 10% serum the med3Δ and tlo1Δ/tlo2Δ mutants again exhibited a common set of co-regulated genes, however in serum many med3Δ mutant-specific responses could also be identified (Figure 6B, 6C). The common gene set included regulators of filamentous growth, cell wall proteins and galactose metabolic genes (Figure 6B). Importantly, the med3Δ mutant specifically exhibited an increase in expression of glycolytic genes (n = 11; Figure 6C, S6) and genes encoding histones and proteins involved in chromatin assembly (n = 18). The med3Δ mutant also exhibited reduced transcription of genes associated with the GO terms “DNA replication” (n = 43) and “telomere maintenance” (n = 18) (Figure 6C).
These data indicate that in exponentially growing cells, Tlo proteins and Med3 are involved in regulating very similar processes. However, in response to specific environmental cues, these proteins may be required for regulation of specific sets of genes.
Deletion of TLOs has different effects on virulence traits compared to a C. albicans med31Δ mutant
Uwamahoro et al. [15] showed that in C. albicans deletion of MED31, encoding a middle subunit of Mediator affected cytokinesis, filamentous growth and biofilm formation. Similarly, the C. dubliniensis tlo1Δ/tlo2Δ mutant described here also grew as chains of cells, typical of mutants with defects in cytokinesis (Figure S3A). However, the C. albicans med31Δ mutant was capable of filamentous growth in response to serum, whereas the C. dubliniensis tlo1Δ/tlo2Δ mutant is incapable of forming true hyphae in serum. GSEA analysis of our C. dubliniensis tlo1Δ/tlo2Δ mutant transcript data identified a significant enrichment for genes that are also affected in a C. albicans med31Δ mutant [15]. Interestingly, while the tlo1Δ/tlo2Δ and the med31Δ deletions affected similar genes, the C. dubliniensis tlo1Δ/tlo2Δ mutant showed increased expression of genes that were both Med31-activated and -repressed in C. albicans (Figure 7A). Inspection of these differentially expressed genes identified several genes required for biofilm formation that were downregulated in the C. albicans med31Δ mutant and induced in the C. dubliniensis tlo1Δ/tlo2Δ mutant, including ALS1, TEC1 and SUC1. Uwamahoro et al. [15] showed that reduced ALS1 expression in the C. albicans med31Δ mutant was largely responsible for the defect in biofilm formation. As the C. dubliniensis tlo1Δ/tlo2Δ mutant exhibited increased expression of ALS1, we examined whether this mutant was affected in biofilm formation. In contrast to the med31Δ mutant phenotype, deletion of TLO1 and TLO2 in C. dubliniensis enhanced biofilm growth on plastic surfaces (Figure 7B). In this case, complementation with TLO2 reduced the amount of biofilm formation more than complementation with TLO1 (ANOVA P<0.05) (Figure 7B). Thus the mediator middle domain has a different effect on biofilm formation than does the mediator tail domain.
Fig. 7. Deletion of TLOs has different effects on biofilm formation compared to a med31Δ mutant. 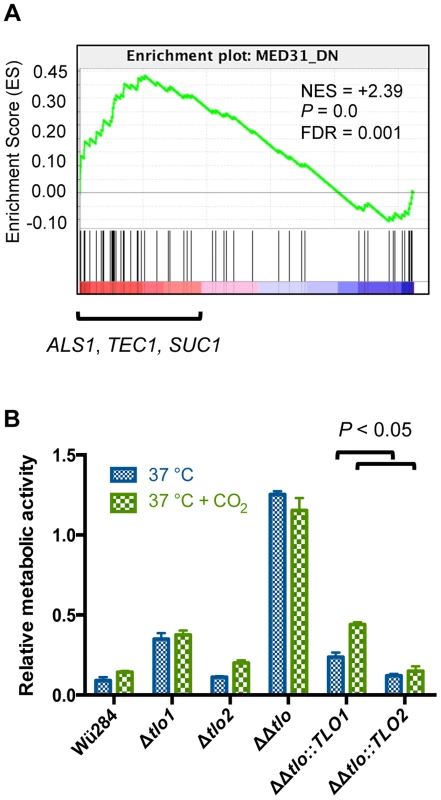
(A) Gene Set Enrichment Analysis (GSEA) plot showing differential expression of genes down-regulated in a C. albicans med31Δ mutant in the tlo1Δ/tlo2Δ mutant (i.e. they are upregulated in the tlo1Δ/tlo2Δ mutant) (B) Biofilm formation was assessed in the indicated strains on plastic surfaces at 37°C in the presence or absence of 5% CO2 as indicated. Adherent biomass determined by an XTT reduction assay. The experiment was carried out in triplicate on three separate occasions (n = 9) and error bars represent the standard deviation from the mean. P values above figure are the result of one-way ANOVA with Tukey's post hoc test. Tlo proteins regulate a core set of Med2 regulated genes
The genetic and biochemical data presented here and by Zhang et al. [9] support a hypothesis that Tlo proteins are the candidal orthologs of S. cerevisiae Med2. In order to support this, we decided to compare the transcript profile of the C. dubliniensis tlo1Δ/tlo2Δ mutant and the S. cerevisiae med2Δ mutant to identify if there is an evolutionarily conserved core regulon controlled by these proteins [27]. Although the experimental details of the two studies vary, a comparison of all genes regulated 1.5-fold or greater in both mutants could identify a small but significant overlap in regulated genes (Figure S8). Carbohydrate metabolism genes that were downregulated in both mutants included the carbohydrate metabolism master regulator TYE7 and genes regulating carbohydrate catabolism and glycerol biosynthesis (PFK1, FBA1, GPH1, GDB1). One of the signatures of the tlo1Δ/tlo2Δ mutant profile was the downregulation of sulphur amino acid metabolism and in S. cerevisiae, we could also detect down regulation of MET6 and MET13 as well as several other conserved genes regulating amino acid catabolism (GCV1, GCV2, SHM2). Both mutants exhibited a general increase in the expression of genes encoding enzymes involved in oxidation-reduction processes (Figure S8).
ChIP supports a role for Tlo1 in transcriptional activation and repression
In order to characterise Tlo1-DNA interactions, we carried out chromatin immunoprecipitation and high-density C. dubliniensis microarray (ChIP-chip) of HA-tagged Tlo1 from cells grown in YEPD broth at 30°C. On a genome wide level, we found extensive binding of Tlo1 to all chromosomes. The regions exhibiting the greatest enrichment included subtelomeric and telomeric regions and the Major Repeat Sequence (MRS) (Figure 8A). Outside of these regions, the interaction of Tlo1 was closely associated with the coding regions of genes (Figure 8A). Excluding subtelomeric regions, only 45 intergenic regions exhibited a significant enrichment peak not associated with a coding region. In addition, we did not observe any association between Tlo1 and 57 putative Pol III transcribed tRNAs and snRNAs. Tlo1 was enriched at 1,617 ORFs in the C. dubliniensis genome (27% of all nuclear genes; Ringo Peak Score 0.8).
Fig. 8. Localization of Tlo1-DNA interaction by ChIp-Chip analysis. 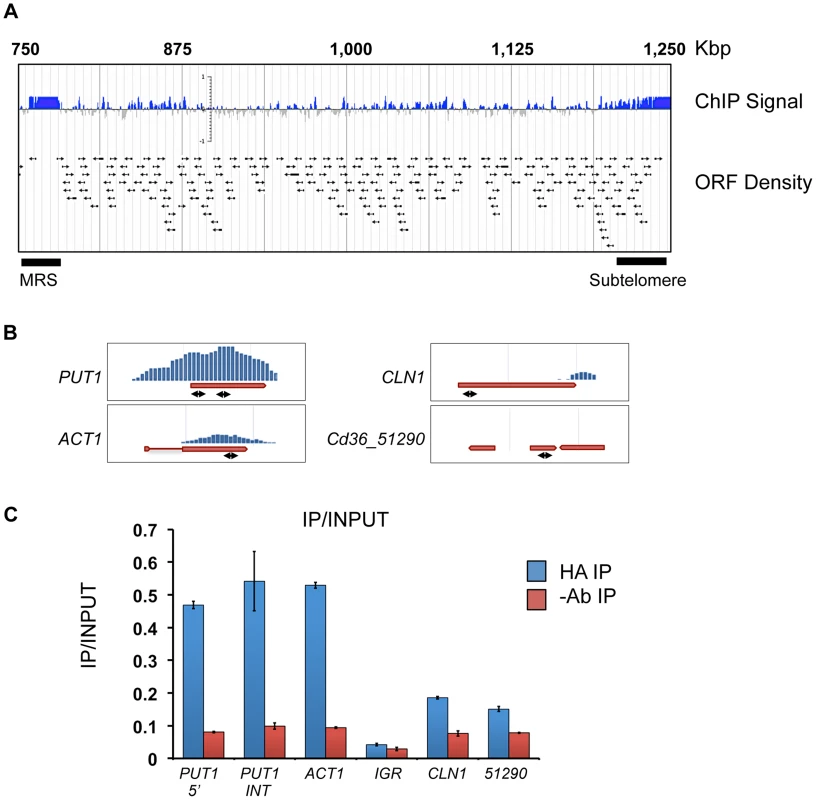
(A) A map of the right arm of C. dubliniensis chromosome 5 showing Tlo1-HA enriched areas, including the subtelomeric and Major Repeat Sequence (MRS) regions. Arrows indicate the position of individual ORFs. All chromosomes can be visualised here: http://bioinf.gen.tcd.ie/jbrowse/?data=cdub. (B) ChIP-chip Enrichment profiles for PUT1, ACT1, CLN1 and CD36_51290. Double headed arrows show approximate positions of amplimers used in QRTPCR confirmation experiments. (C) QRT-PCR confirmation of the enrichment levels shown in panel B using gene-specific primers and an intergenic region (IGR)-specific primer set (Chr5 521680–521729 bp)(Table S5). Amplification was carried out on immunoprecipitated chromatin in anti-HA antibody samples (HA-IP) and no antibody (-AB IP) negative control samples. Relative enrichment values refer to the quantity of DNA amplified from anti-HA immunoprecipitated samples (‘enriched’) relative to total input chromatin (‘non-enriched’). Due to recent discussions in the literature regarding artefacts in ChIP data [28]–[30], we restricted detailed analysis to a group of 367 genes that exhibited highly significant Tlo1-enrichment (Ringo peak score 0.9; Table S3). The region of Tlo-enrichment was centred on the coding region with extension into 5′ and 3′ flanking non-coding regions (Figure S9A). The level of Tlo-enrichment at the 5′ region was variable and was generally <100 bp in length, however extensions up to 1000 bp in length were observed (Figure S9B). QRT-PCR analysis of DNA generated in replicate ChIP experiments confirmed the high levels of enrichment observed at PUT1 and ACT1 and low enrichment levels observed at non-enriched genes such as CLN1 and Cd36_51290 and an intergenic region on chromosome 5 (bp 521680–521729) (Figure 8B, 8C). Due to the repetitive nature of the repeat sequences in the MRS, it was not possible to accurately determine the level of enrichment in this region by QRT-PCR.
Using fluorescence intensity data extracted from our gene expression microarrays, we compared the expression levels of the Tlo1-associated ORFs relative to non-Tlo1 associated ORFs. Tlo1-associated ORFs were found to exhibit higher expression levels (average 1.8-fold) in YEPD compared to those genes that are not occupied (Figure S9C). However, a plot of enrichment score versus expression level did not identify a direct correlation between Tlo1 enrichment and expression. Importantly, the most highly enriched genes (n = 367; Ringo peak score 0.9) covered a broad spectrum of expression levels, including genes expressed at high and low levels in YEPD (Figure 9A). We analysed the GO terms associated with these Tlo1-enriched genes and identified significant numbers of highly expressed genes associated with the GO categories glycolysis (n = 7; P = 0.003) and the TCA cycle (n = 7; P = 0.048)(Figure 9A). Manual inspection of the gene list also identified highly expressed genes involved in translation (n = 5) and sulphur amino acid metabolism (n = 4). Tlo1-enriched genes that are poorly expressed in YEPD were associated with the GO categories glyoxylate cycle (n = 3; P = 0.09), GlcNac catabolic process (n = 6, P = 0.094), amino acid catabolic process (n = 10; P = 0.018) and hexose transport (n = 9; P = 0.057). Manual inspection also identified poorly expressed genes encoding nitrogen scavenging transporters (e.g. FRP6, MEP21, MEP22, DAL1, UGA6) and genes involved in gluconeogenesis (PCK1 and FBP1). Many YEPD repressed, hypha-specific genes were also enriched including EED1, RBT1, HWP1, HWP2 and IHD1.
Fig. 9. The relationship between Tlo1 binding and gene expression. 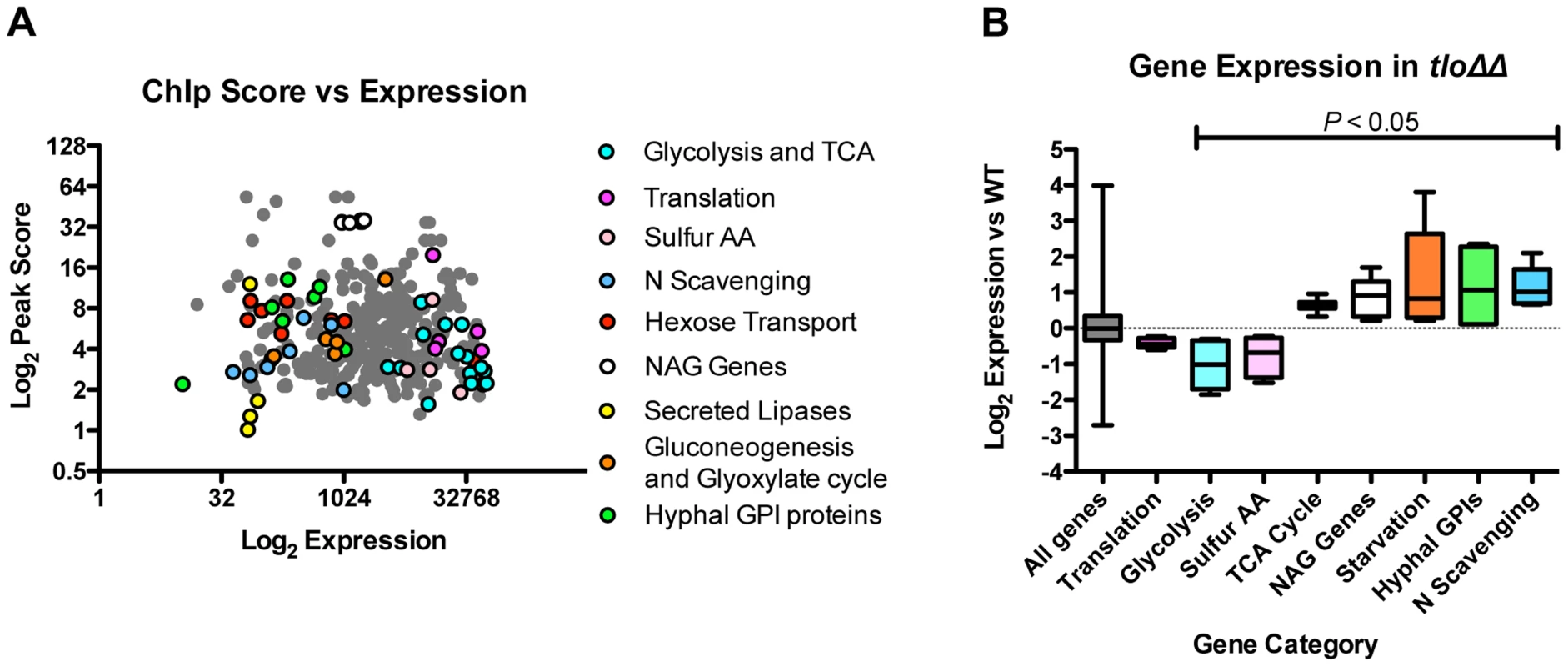
(A) A plot of Tlo1 enrichment scores for the 367 most highly enriched genes versus the expression levels of those genes in YEPD broth (extracted from microarray data). The categories of highly expressed and repressed genes are color-coded; TCA = Tricarboxylic acid cycle; Sulfur AA = sulphur amino acid biosynthesis; N scavenging = Nitrogen scavenging; NAG = GlcNac metabolism; GPI = glycophosphatidylinositol anchor). (B) Analysis of the expression of Tlo1-enriched groups of genes in the wild type in the tlo1Δ/tlo2Δ (tloΔΔ) mutant indicated that the highly expressed, highly enriched genes exhibited reduced expression whereas the poorly expressed and repressed genes exhibited increased expression in the tlo1Δ/tlo2Δ (tloΔΔ) mutant. The change in expression in these groups, with the exception of the genes involved on translation, was significant (ANOVA, P<0.05). These data show that Tlo1 is present at many highly expressed metabolic genes in addition to genes normally repressed in YEPD (starvation, hyphal genes) indicating a dual role in transcriptional activation and repression. In order to support this hypothesis, we analysed the transcription of these categories of Tlo1-enriched genes in our tlo1Δ/tlo2Δ mutant. This analysis found that the highly expressed genes involved in glycolysis and sulfur amino acid metabolism exhibited significantly reduced expression (ANOVA; P<0.05) in the tlo1Δ/tlo2Δ mutant relative to wild type cells, indicating that Tlo1 occupancy is required for maintaining the high level of expression of these genes (Figure 9B). The exceptions to this finding were the genes encoding enzymes of the TCA cycle, which although highly expressed in wild-type cells, exhibited increased expression in the tlo1Δ/tlo2Δ mutant. In addition, highly Tlo1-occupied, low expression genes exhibited increased expression in the tlo1Δ/tlo2Δ mutant, including genes involved in starvation responses, nitrogen scavenging mechanisms, N-acetyl glucosamine catabolism and the hyphal GPI-anchored proteins (Figure 9B). These data indicate that Tlo1 and the Mediator complex are involved in both transcriptional activation and repression of different classes of genes. The fact that Tlo1-enriched genes exhibit regulation in the tlo1Δ/tlo2Δ mutant also provides support for hypothesis that Tlo proteins interact with these ORFs.
Discussion
Despite the importance of Mediator in transcriptional regulation, few studies have investigated the role of Mediator in regulating virulence-associated characteristics in pathogenic eukaryotic microrganisms. The discovery of a widely dispersed telomeric gene family with homology to Med2 in the pathogenic yeast C. albicans has accelerated the level of interest in Mediator in this fungus. Published data suggest important roles for Mediator in regulating important biological processes in C. albicans, including filamentous growth (Med3), biofilm formation (Med31, Med20) and white-opaque switching (multiple subunits) [9], [15], [16]. Because functional analysis of the 15 TLO paralogs in C. albicans remains technically challenging, we turned to C. dubliniensis, which shares many characteristics with C. albicans but contains only two TLO orthologs, thereby providing an ideal genetic background in which to investigate the degree to which TLO gene function(s) have diverged between the two TLO paralogs. It has been hypothesized that in C. albicans, Mediator could form variant complexes that contain different Tlo/Med2 subunits that could be differentially recruited to activate or repress different subsets of genes in response to specific stimuli [9], [12]. C. dubliniensis, which has only two Tlo proteins that appear to associate primarily with Mediator, provides an invaluable tool to understand first, how different TLO genes may affect Mediator function and second how the presence of multiple variant TLO genes contributes to virulence.
In C. dubliniensis, tlo1Δ deletion mutants are defective in hyphal growth [14]. Here, we constructed a null tlo1Δ/tlo2Δ double mutant strain of C. dubliniensis. The tlo1Δ/tlo2Δ strain exhibits a wide range of phenotypic defects, indicating that they play a central role in global transcriptional control. Consistent with a role in regulating hyphal growth, deletion of both TLO genes reduced expression of many of the key regulators of morphogenesis including signal transducers (RAS1, RIM101) transcriptional regulators (EFG1, UME6) and kinases (HGC1), resulting in largely pseudohyphal growth phenotypes with reduced expression of key hyphal genes (SOD5, RBT5, HWP1). The tlo1Δ/tlo2Δ mutant also exhibited increased susceptibility to oxidative stress and reduced growth in the presence of alternative carbon sources. These data demonstrate that the TLOs of Candida spp. have evolved to regulate many key processes, including those important for virulence and growth in the host. In addition, the importance of Tlo in mediating transcriptional responses important for pathogenicity is shown in the similarity of our transcript profiles to those observed during infection of RHE and bone marrow-derived macrophages (Figure S6) [25], [26]. Indeed, a tlo1Δ/tlo2Δ mutant grown in YEPD mounts a transcriptional response akin to that observed following phagocytosis by macrophages [1]. Recent studies have also shown that Mediator is an important regulator of the Candida-macrophage interaction [31]. One key observation regarding the transcriptional changes observed in the tlo1Δ/tlo2Δ mutant was the alteration in the kinetics of induction of hypha-specific transcripts. Complementation of the tlo1Δ/tlo2Δ mutant with Tlo1 resulted in more rapid and higher levels of transcriptional induction of SOD5, UME6 and GAL10. Some induction of these transcripts could be observed in the tlo1Δ/tlo2Δ mutant, however the amplitude of the response was greatly retarded relative to the complemented strains. These data suggest that Tlo proteins play an important role in maximizing the speed and magnitude of transcriptional changes in response to environmental cues. This characteristic is likely to be of great importance in the host where rapid responses could be important for survival, e.g. following phagocytosis or translocation to a different part of the gastrointestinal tract.
In S. cerevisiae, the Mediator tail regulates stress responses and the metabolism of carbohydrates and amino acids [23], [24], [27]. A direct comparison of the genes regulated in our tlo1Δ/tlo2Δ mutant and an S. cerevisiae med2Δ mutant supports this evolutionarily conserved role for Mediator tail in eliciting transcriptional responses to deal with changing environments [27]. In S. cerevisiae this effect has been shown to be centred on SAGA-regulated genes [24].
In this study, we also explored the relationship of Tlo proteins with other subunits of Mediator. A defect in filamentous growth was described by Zhang et al. in C. albicans following deletion of Med3, encoding another Mediator tail subunit that interacts with Tlo proteins [9]. Biochemical analysis of the Mediator complex in the tlo1Δ/tlo2Δ null mutant and the C. dubliniensis med3Δ mutant revealed the absence of Med5 and Med16 in Mediator purified from both mutants, suggesting that the entire tail complex is destablilized by loss of Tlo or Med3 proteins. Consistent with this idea, the C. dubliniensis med3Δ and tlo1Δ/tlo2Δ null mutants exhibited remarkably similar phenotypes and transcriptional profiles when cultured in YEPD. However, in response to hyphal stimulating conditions, many differences in transcriptional responses could be observed in the tlo1Δ/tlo2Δ strain and the med3Δ mutant strain (Figure 6), suggesting that Mediator complexes with different tail substituents may exist in vivo in response to specific environmental conditions. Alternatively, the deletion of either Med3 or Tlo protein may release the remaining Tail module Mediator subunits to form a sub-complex that can interact with cellular components independently of the remainder of the complex.
We next examined the relationship of Tlo with the Mediator middle subunit, Med31, described by Uwamahoro et al. [15]. The C. dubliniensis tlo1Δ/tlo2Δ null mutant exhibited a defect in cell separation and CHT3 expression, which is consistent with the defect in cytokinesis described in the C. albicans med31 mutant by Uwamahoro et al. [15]. These data support previous studies that suggest that regulation of cytokinesis and possibly Ace2-regulated genes is a conserved function of mediator in yeast [15], [32]. However, a med31Δ mutant was still capable of true hyphal growth in liquid medium whereas the tlo1Δ/tlo2Δ null mutant was completely unable to form true hyphae under the same conditions. The most striking difference in phenotype was the increased ability of the C. dubliniensis tlo1Δ/tlo2Δ mutant to form biofilm. The C. albicans med31Δ mutant exhibits reduced biofilm growth relative to wild-type and decreased expression of the biofilm regulator TEC1 and the adhesin ALS1 (Figure 7A). The C. dubliniensis tlo1Δ/tlo2Δ null mutant exhibited increased expression of many med31Δ downregulated genes, including TEC1 and ALS1, although the roles of these genes in biofilm formation in C. dubliniensis have yet to be confirmed. These data illustrate the very different and possibly opposing roles that mediator tail and middle subunits may have in transcriptional regulation in Candida spp.
Our ChIP analysis of Tlo1 for the first time indicates the level of Tlo and Mediator occupancy across the Candida genome. Although Mediator tail subunits are unlikely to interact directly with DNA, they associate closely with DNA-bound transcription factors and histones and have been successfully isolated in association with DNA in other yeasts [8], [33]. We identified extensive Tlo1 binding across all C. dubliniensis chromosomes. Tlo1 was closely associated with Pol II transcribed ORFs as well as the telomeres and the MRS. Binding of Mediator to telomeres and sub-telomeric genes has been described in S. cerevisiae and Mediator has been shown to have an additional role in heterochromatin maintenance [33], [34]. Our data suggests that Mediator could play a similar role in Candida spp. The significance of Tlo1-enrichment at the MRS is more difficult to explain as the function of this repeat element is unknown. Further studies will be required to verify this interaction.
In order to limit the numbers of possible ChIP artefacts, often associated with highly expressed genes, we have used a very stringent cut off to define highly enriched genes. This Tlo1-enriched gene set includes repressed and moderately expressed genes, supporting the assumption that our data set is not biased for highly expressed ORFs. In addition, the highly expressed genes identified in our ChIP analysis are regulated following deletion of Tlo, supporting a direct, functional interaction with these ORFs.
The association of Mediator subunits with the coding regions of Pol II transcribed ORFs has been observed in Mediator ChIP localization experiments in other yeasts and suggests that Tlo1 may be involved in transcript elongation or in chromatin interactions at coding regions [35], [36]. We observed that although Tlo1-occupied genes on average were expressed higher than non-occupied genes (average 1.8-fold), Tlo1 was associated with ORFs having a range of expression levels. To further investigate the significance of Tlo1 ORF occupancy and Tlo1-dependent gene expression, we compared the regulation of specific classes of Tlo1 occupied genes in our tlo1Δ/tlo2Δ mutant. Highly expressed genes encoding enzymes involved in amino acid synthesis, glycolysis and translation required Tlo1 to maintain high levels of expression. In contrast, genes that exhibited low levels of expression under the conditions analysed were repressed by Tlo1. These data suggest a dual role for Tlo1 in mediating transcriptional activation and repression. Specifically, Tlo1 was required to maintain transcription of genes involved in glycolysis and translation and to repress starvation responses (glyoxylate cycle, gluconeogenesis) and several hypha-specific genes. Localisation of Mediator to both expressed and repressed genes has been observed in S. cerevisiae and the ability of the complex to carry out opposing regulatory functions is likely due to its ability to adopt different modular conformations [36]. Specifically, the kinase module in S. cerevisiae, consisting of Cdk8, Cyclin C, Srb8 and Srb9 has been shown to associate with Mediator to repress transcription [36]. A specific role for the kinase module in transcriptional repression has yet to be determined in Candida spp.
The C. dubliniensis genome harbors two TLO orthologs, two alleles of TLO1 and one allele of TLO2. One of the central goals of our study was to determine whether different TLO genes could regulate different processes. To compare the effects of single copies of TLO1 and TLO2, we complemented the tlo1Δ/tlo2Δ mutant independently with either TLO1 or TLO2. Each of the TLO genes complemented tlo1Δ/tlo2Δ mutant phenotypes, albeit to different degrees; TLO1 was better at restoring true hyphal growth and growth in galactose. TLO2 supressed biofilm growth to a greater extent than TLO1. With regard to TLO2, it should be noted that the level of expression of the reintegrated allele was significantly higher than the wild-type allele, which may have resulted in higher levels of Tlo2 activity. However, despite this, the reintegrated TLO2 allele did not restore filamentous growth to the same extent as wild-type TLO1.
Support for these functional differences was obtained when the transcript profiles of the TLO1 and TLO2 complemented strains were compared. It was observed that TLO1 could restore GAL gene induction to a greater extent than TLO2. During hyphal growth, TLO1 also restored expression of UME6 and SOD5 to a greater degree than TLO2. In contrast, during growth in YEPD TLO2 was required for repressing transcription of aberrantly expressed hyphal genes such as IHD1, SAP7 and EED1 and the negative regulators of hyphal growth SFL1 and NRG1. TLO2 was also required for suppression of many starvation induced genes involved GlcNac metabolism (NAG3, NAG4) and amino acid metabolism (CHA1, GDH3). The data suggest that the two TLO paralogs in C. dubliniensis have evolved to regulate overlapping as well as distinct subsets of genes. Under standard growth conditions, both TLO1 and TLO2 are expressed in C. dubliniensis, suggesting that two distinct pools of Mediator exist at any one time, with the ability to regulate different subsets of genes. The implications of our study for C. albicans biology, where there are potentially 15 different Tlo-Mediator complex varieties with differing transcriptional activating activities, are immense. It is tempting to speculate that this variety in Mediator activity, or a varying pool of non-Mediator associated Tlo protein, could contribute to the phenotypic plasticity and adaptability of this fungus and may go towards explaining why its incidence as a commensal and opportunist is far greater compared to its close relatives C. tropicalis and C. dubliniensis which do not have this expanded repertoire of Med2 paralogs.
Over the last decade, a wealth of studies have described the global transcriptional responses of C. albicans to environmental stress, pH changes and nutrient availability amongst others. It is clear from these studies that one of the characteristics of C. albicans is its ability to rapidly respond to environmental change by mediating rapid transcriptional responses. The current study suggests a vital role for the Tlos in mediating the speed and scale of these responses, which are associated with characteristics required for commensalism (nutrient acquisition, metabolism) and pathogenicity (starvation responses, filamentation) indicating a key role for Tlo-regulated responses in the lifestyle of C. albicans. This, coupled with the potential diversity in the activity of the different Tlo paralogs may have contributed to the ability of C. albicans to colonise many diverse niches and thus to evolve to be a highly successful commensal and pathogen.
Materials and Methods
Candida strains and culture conditions
All Candida strains were routinely cultured on yeast extract-peptone-dextrose (YEPD) agar, at 37°C. Solid Lee's medium and Spider medium were used as described previously [37], [38]. Pal's agar medium was also used for chlamydospore induction as described previously [39]. For liquid culture, cells were grown shaking (200 rpm) in YEPD broth at 30°C or 37°C. In order to determine the doubling time of each strain, the optical densities (600nm) of cultures in the exponential phase of growth were plotted and analysed using the exponential growth equation function in Graphpad Prism (GraphPad, CA, USA). Doubling times and r2 values in Table 1 were calculated from three replicate growth curves. Glucose (2% w/v) was substituted with galactose (2% w/v) where indicated. Hyphal induction was carried out in sterile Milli-Q H2O supplemented with 10% (v/v) fetal calf serum at 37°C. Samples were randomized and the proportion of germ-tubes or true-hyphae in 300 cells was assessed at intervals by microscopic examination of an aliquot of culture with a Nikon Eclipse 600 microscope (Nikon U.K., Surrey, U.K.). Experiments were performed on at least three separate occasions.
Genotypes of strains used in this study are listed in Table S4. Gene disruption of TLO1 (Cd36_72860) and TLO2 (Cd36_35580) was achieved through use of the SAT1 flipper cassette system [40]. Deletion constructs were created by PCR amplifying the 5′ flanking regions of TLO1 or TLO2 with the primer pairs CTA21KF/CTA1X and CTA22KF/CTA2X respectively and the 3′ flanking regions with the primer pairs CTA1S/CTA21SIR and CTA2S/CTA22SIR, respectively (Table S4). Ligation of these products in the corresponding restriction sites in plasmid pSFS2A resulted in the construction of plasmids pTY101 (TLO1) and TY102 (TLO2) as described [19]. The deletion construct was used to transform C. dubliniensis Wü284 as described previously [19] and deletion of genes was confirmed by Southern blotting (Figure S1). A single transformation with TY102 was sufficient to delete TLO2 in strain Wü284 which was found to have a truncation in one copy of ChrR, resulting in the presence of only one copy of TLO2. Deletion of MED3 was carried out using a primer tailing method with the primers MED3M13F/MED3M13R as described [41]. Reintroduction of wild-type TLO genes or MED3 was achieved by PCR amplification of the entire ORF plus their upstream and downstream regulatory sequences using primer pairs CdTLO1FP/CdTLO1RP for CdTLO1, CdTLO2FP/CdTLO2RP for CdTLO2 and MED3FP/MED3RP for MED3 (Table S5) and ligation of these into the C. dubliniensis integrating vector pCDRI to yield plasmids pCdTLO1, pCdTLO2, and pCdMED3 respectively. These plasmids were transformed as described previously [19].
Biochemical analysis of mediator
Mediator was purified from C. dubliniensis strains containing Mediator subunits tagged at their C-terminus with either HA or 6His-FLAG. CdMED8 - and CdTLO1-tagging cassettes were amplified from pFA-3HA-SAT1 or pFA-6His3Flag-SAT1 using the primer pairs pZL420/pZL421 and pZL422/pZL423, respectively, as described previously [9]. Whole-cell extracts were made from strains that were untagged or had a single copy of Tlo1 or other Mediator subunit tagged with a HA or 6His-FLAG and subjected to multiple chromataographic separations as described previously [9]. The elutions from the immobilized metal affinity chromatography (IMAC; Talon Kit, Clontech Laboratories, CA, USA) step were analyzed by SDS-PAGE as indicated and proteins were revealed by staining with silver as described previously [9].
Induction of biofilm formation
Biofilm mass was determined using an XTT reduction assay to assess metabolic activity. Prior experiments with planktonic cells indicated that wild-type and tloΔ mutant strains exhibited similar rates of XTT reduction. To induce biofilm growth, a suspension of 1×106 cells of each strain was prepared in 10% (v/v) foetal calf serum from 18 h cultures grown in YEPD broth at 30°C with shaking at 200 rpm A 100 µl volume of each suspension was added to triplicate wells of a 96-well, flat-bottomed polystyrene plate (Greiner BioOne) with lid. Plates were then incubated for 24 h or 48 h in a static incubator (Gallenkamp) set to 37°C. Growth in the presence of 5% (v/v) CO2 was carried out in a static tissue culture incubator with 5% (v/v) relative humidity (Gallenkamp). All wells were then washed 5 times with 200 µl sterile PBS to remove non-adherent cells. After the final wash, 200 µl of 200 µg/ml XTT supplemented with 50 µg/ml CoEnzyme Q (Sigma-Aldrich) was added to each well and incubated at 37°C in the dark for 50 min. A 100 µl aliquot of this suspension was transferred to a fresh plate and the absorbance measured at 480 nm using a Tecan Plate Reader system (Tecan). Results were analysed and graphed using Microsoft Excel (Microsoft).
Transcriptional profiling with oligonucleotide microarrays
The C. dubliniensis whole genome oligonucleotide microarray used in this study was previously described [20]. RNA was extracted from cells grown to an OD 600 nm of 0.8 in YEPD broth at 37°C or from cells (1×106) inoculated in 10% (v/v) fetal calf serum at 37°C. Cell pellets were snap frozen in liquid N2 and disrupted using the Mikro-Dismembrator S system (Sartorius Stedim Biotech, Göttingen, Germany). RNA was prepared using the Qiagen RNeasy mini-kit. A 200 ng aliquot of total RNA was labelled with Cy5 or Cy3 using the Two-Color Low Input Quick Amp labeling Kit (Agilent Technologies) according to the manufacturer's instructions. Array hybridization, washing, scanning and data extraction was carried out in GenePix Pro 6.1 (Axon) as described [20]. For each condition, four biological replicate experiments were performed, including two dye swap experiments. Raw data were exported to GeneSpring GX12 and signals for each replicate spot were background corrected and normalized using Lowess transformation. Log2 fluorescence ratios were generated for each replicate spot and averaged. Genes differentially expressed (1.5-fold) relative to wild-type were identified from those that passed a t-test (P≤0.05) and satisfied a post-hoc test (Storey with Bootstrapping) with a corrected Q value ≤0.05. Hierarchical clustering was used to compare gene expression in each condition using the default settings in Genespring GX12. The Gene Set Enrichment Analysis (GSEA) PreRanked tool (available to download at www.broadinstitute.org/gsea/index.jsp) was used to investigate whether our data sets were enriched for particular genes present in published data sets [42]. This analysis required the use of a database of publicly available genome-wide data sets, constructed by Andre Nantel (National Research Council of Canada, Montreal), that can be downloaded from the Candida Genome Database (CGD). This database included 171 lists of up - and down-regulated genes from microarray experiments, in vivo promoter targets derived from ChIP-chip experiments performed on 36 transcription factors, members of 3,601 Gene Ontology (GO) term categories, and 152 pathways, as curated by CGD, amongst others, and is fully described by Sellam et al. [43]. Genes in our expression data sets were first ‘ranked’ based on Log2 values from highest to lowest. The GSEA PreRanked tool was then used to determine if particular gene sets in the database were enriched towards the top or bottom of this ranked list. Enrichment plots show the level of enrichment towards the top (red) or bottom (blue) of the ranked list. The normalized enrichment score (NES) indicates the level of enrichment at the top (positive NES) or bottom (negative NES) of the ranked list and is supported by a p value and a False Discovery Rate (FDR) value to indicate the likelihood of false positive results.
Microarray data have been submitted to Gene Expression Omnibus (GEO), accession number: GSE59113.
Real-time PCR analysis of gene expression
RNA for QRT-PCR was isolated using the RNeasy Mini-kit (Qiagen). Cells were disrupted using a FastPrep bead beater (Bio101). RNA samples were rendered DNA free by incubation with Turbo-DNA free reagent (Ambion, Austin, TX). cDNA synthesis was carried out using the Superscript III First strand synthesis sytem for RT-PCR (Life technologies) as described by Moran et al. [19] Reactions were carried on an ABI7500 Sequence Detector (Applied Biosystems, Foster City, CA) using MicroAmp Fast Optical 96-well Reaction plates (Applied Biosystems) in 20 µl reactions using 1X Fast SYBR Green PCR Master Mix (Applied Biosystems), 150 nM of each oligonucleotide (Table S4) and 2 µl of diluted template (10 ng). Cycling conditions used were 95°C for 20 sec, followed by 40 cycles of 95°C for 3 sec and 60°C for 30 sec, the latter of which was the point of detection of fluorescence. This was followed by a melt curve stage as a quality control point. Gene expression levels were normalized against the expression levels of the constitutively expressed ACT1 gene in the same cDNA sample. Gene specific primers are shown in Table S5. Each gene-specific set was shown to amplify at a similar efficiency (within 10%) to the control ACT1 primer set in primer optimization experiments.
Chromatin immunoprecipitation and microarray hybridisation (ChIP-chip)
C. dubliniensis strain Tlo1-HA, derived from strain Wü284, was grown at 30°C degrees and harvested at an OD 600 nm of 2.0. A secondary cross-linking strategy was employed to fix chromatin that involved initial treatment of cells with dimethyl adipimidate (10 mM) for 45 min with agitation at room temperature followed by cross-linking with 1% formaldehyde. Cells were washed twice in PBS prior to spheroplasting in 5 ml of spheroplasting buffer (1.2 M sorbitol, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES] pH 7.4) containing 0.4 mg/ml Zymolyase 100T (Seikagaku Corp., Japan) and incubated for 2.5 h 30°C with rotation. DNA isolation and fragmentation was carried out as described in Ketel et al [44] using a Branson sonifier to yield DNA fragments of 500–600 bp. Chromatin immunoprecipitation (ChIP) was carried out as described by Ketel et al. [44] using an anti-HA mouse monoclonal antibody (clone 12CA5) which was incubated overnight at 4°C on a rotating wheel. The following day, 70 µl of Protein A agarose beads (Santa Cruz Biotech, California, USA) were added and mixed at 4°C for 4 h. DNA was recovered as described by Ketel et al [44] and resuspended in 80 µl TE buffer for downstream applications.
Microarray chip analysis (CGH Microarray) was carried out as described in the Agilent Yeast ChIP-on-chip analysis protocol handbook (Version 9.2, 2007). Competitive hybridization was carried out on a whole genome C. dubliniensis oligonucleotide microarray consisting of 175,758 unique 60mer probes, spaced at 20 bp intervals (Agilent Technologies). Experiments were performed with triplicate biological replicate samples. The IP samples were labelled with Cy3 and input samples (total lysate control) were labelled with Cy5. One sample was dye swapped. Array hybridisation and washing were carried out using standard Agilent Technologies CGH hybridisation buffers and washes. Scanning of the array was carried out on an Agilent G2565B rotating multi-slide hyb-scanner (Agilent Technologies) using a 5 µm resolution and PMT settings at 100% for both channels (Green/Red). The scan area was set to 61×21.6 mm. The raw images were processed using Agilent Feature Extraction software suite v 10.5.1.1. and feature extraction was automatically carried out using the ‘CGH automatic’ programme to align and normalise fluorescent probe spots on the array.
Data were read into the Bioconductor [45] package Ringo [46] and pre-processed with the “nimblegen” method. This was followed by merging of replicates and smoothing with a window half size of 500 bp. ChIP-enriched regions were calculated with an enrichment threshold based firstly on a 0.8 percentile, followed by a more stringent 0.9 percentile which was used for the analysis presented here. Genes were assigned to an enrichment region if they overlapped up to 500 bp away from the gene boundaries. For visualisation of the data a JBrowse [47] instance was set up on http://bioinf.gen.tcd.ie/jbrowse/?data=cdub. The ChIP data have been submitted to GEO, accession number GSE60173.
Supporting Information
Zdroje
1. LorenzMC, BenderJA, FinkGR (2004) Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryotic Cell 3 : 1076–1087.
2. PérezJC, JohnsonAD (2013) Regulatory Circuits That Enable Proliferation of the FungusCandida albicans in a Mammalian Host. PLoS Pathog 9: e1003780.
3. KadoshD, JohnsonAD (2005) Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell 16 : 2903–2912.
4. ConawayRC, ConawayJW (2011) Origins and activity of the Mediator complex. Seminars in Cell and Developmental Biology 22 : 729–734.
5. MalikS, RoederRG (2010) The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nature Reviews Genetics 11 : 761–772.
6. BjörklundS, GustafssonCM (2005) The yeast Mediator complex and its regulation. Trends in Biochemical Sciences 30 : 240–244.
7. ThompsonCM, YoungRA (1995) General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA 92 : 4587–4590.
8. AnsariSA, MorseRH (2013) Mechanisms of Mediator complex action in transcriptional activation. Cell Mol Life Sci 70 : 2743–2756.
9. ZhangA, PetrovKO, HyunER, LiuZ, GerberSA, et al. (2012) The Tlo Proteins Are Stoichiometric Components of Candida albicans Mediator Anchored via the Med3 Subunit. Eukaryotic Cell 11 : 874–884.
10. BraunBR, van Het HoogM, d'EnfertC, MartchenkoM, DunganJ, et al. (2005) A human-curated annotation of the Candida albicans genome. PLoS Genet 1 : 36–57.
11. van Het HoogM, RastTJ, MartchenkoM, GrindleS, DignardD, et al. (2007) Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol 8: R52.
12. AndersonMZ, BallerJA, DulmageK, WigenL, BermanJ (2012) The Three Clades of the Telomere-Associated TLO Gene Family of Candida albicans Have Different Splicing, Localization, and Expression Features. Eukaryotic Cell 11 : 1268–1275.
13. BourbonH-M (2008) Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res 36 : 3993–4008.
14. JacksonAP, GambleJA, YeomansT, MoranGP, SaundersD, et al. (2009) Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res 19 : 2231–2244.
15. UwamahoroN, QuY, JelicicB, LoTL, BeaurepaireC, et al. (2012) The Functions of Mediator in Candida albicans Support a Role in Shaping Species-Specific Gene Expression. PLoS Genet 8: e1002613.
16. ZhangA, LiuZ, MyersLC (2013) Differential Regulation of White-Opaque Switching by Individual Subunits of Candida albicans Mediator. Eukaryotic Cell 12 : 1293–1304.
17. MoranGP, ColemanDC, SullivanDJ (2012) Candida albicans versus Candida dubliniensis: Why Is C. albicans More Pathogenic? Int J Microbiol 2012 : 205921.
18. StokesC, MoranGP, SpieringMJ, ColeGT, ColemanDC, et al. (2007) Lower filamentation rates of Candida dubliniensis contribute to its lower virulence in comparison with Candida albicans. Fungal Genet Biol 44 : 920–931.
19. MoranGP, MacCallumDM, SpieringMJ, ColemanDC, SullivanDJ (2007) Differential regulation of the transcriptional repressor NRG1 accounts for altered host-cell interactions in Candida albicans and Candida dubliniensis. Mol Microbiol 66 : 915–929.
20. O'ConnorL, CapliceN, ColemanDC, SullivanDJ, MoranGP (2010) Differential filamentation of Candida albicans and C. dubliniensis is governed by nutrient regulation of UME6 expression. Eukaryotic Cell doi:10.1128/EC.00042-10
21. EnjalbertB, MoranGP, VaughanC, YeomansT, MacCallumDM, et al. (2009) Genome-wide gene expression profiling and a forward genetic screen show that differential expression of the sodium ion transporter Ena21 contributes to the differential tolerance of Candida albicans and Candida dubliniensis to osmotic stress. Mol Microbiol 72 : 216–228.
22. MoranGP, ColemanDC, SullivanDJ (2011) Comparative genomics and the evolution of pathogenicity in human pathogenic fungi. Eukaryotic Cell 10 : 34–42.
23. MillerC, MaticI, MaierKC, SchwalbB, RoetherS, et al. (2012) Mediator Phosphorylation Prevents Stress Response Transcription During Non-stress Conditions. Journal of Biological Chemistry 287 : 44017–44026.
24. AnsariSA, GanapathiM, BenschopJJ, HolstegeFCP, WadeJT, et al. (2011) Distinct role of Mediator tail module in regulation of SAGA-dependent, TATA-containing genes in yeast. EMBO J 31 : 44–57.
25. ZakikhanyK, NaglikJR, Schmidt WesthausenA, HollandG, SchallerM, et al. (2007) In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol 9 : 2938–2954.
26. MarcilA, GadouryC, AshJ, ZhangJ, NantelA, et al. (2008) Analysis of PRA1 and its relationship to Candida albicans - macrophage interactions. Infect Immun 76 : 4345–4358.
27. van de PeppelJ, KettelarijN, van BakelH, KockelkornTTJP, van LeenenD, et al. (2005) Mediator Expression Profiling Epistasis Reveals a Signal Transduction Pathway with Antagonistic Submodules and Highly Specific Downstream Targets. Molecular Cell 19 : 511–522.
28. ParkD, LeeY, BhupindersinghG, IyerVR (2013) Widespread misinterpretable ChIP-seq bias in yeast. PLoS ONE 8: e83506 doi:10.1371/journal.pone.0083506
29. TeytelmanL, ThurtleDM, RineJ, van OudenaardenA (2013) Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proceedings of the National Academy of Sciences 110 : 18602–18607.
30. FanX, StruhlK (2009) Where Does Mediator Bind In Vivo? PLoS ONE 4: e5029 doi:10.1371/journal.pone.0005029.t002
31. UwamahoroN, Verma-GaurJ, ShenHH, QuY, LewisR, et al. (2014) The Pathogen Candida albicans Hijacks Pyroptosis for Escape from Macrophages. mBio 5: e00003–14–e00003–14 doi:10.1128/mBio.00003-14
32. MehtaS, MiklosI, SipiczkiM, SenguptaS, SharmaN (2009) The Med8 mediator subunit interacts with the Rpb4 subunit of RNA polymerase II and Ace2 transcriptional activator in Schizosaccharomyces pombe. FEBS Lett 583 : 3115–3120.
33. LiuZ, MyersLC (2012) Med5(Nut1) and Med17(Srb4) are direct targets of mediator histone H4 tail interactions. PLoS ONE 7: e38416.
34. PengJ, ZhouJQ (2012) The tail-module of yeast Mediator complex is required for telomere heterochromatin maintenance. Nucleic Acids Res 40 : 581–593.
35. ZhuX, WirénM, SinhaI, RasmussenNN, LinderT, et al. (2006) Genome-Wide Occupancy Profile of Mediator and the Srb8-11 Module Reveals Interactions with Coding Regions. Molecular Cell 22 : 169–178.
36. AndrauJ-C, van de PaschL, LijnzaadP, BijmaT, KoerkampMG, et al. (2006) Genome-Wide Location of the Coactivator Mediator: Binding without Activation and Transient Cdk8 Interaction on DNA. Molecular Cell 22 : 179–192.
37. LiuH, KöhlerJ, FinkGR (1994) Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266 : 1723–1726.
38. LeeKL, BuckleyHR, CampbellCC (1975) An amino acid liquid synthetic medium for the development of mycellal and yeast forms of Candida albicans. Med Mycol 13 : 148–153.
39. Al-MosaidA, SullivanDJ, ColemanDC (2003) Differentiation of Candida dubliniensis from Candida albicans on Pal's agar. J Clin Microbiol 41 : 4787–4789.
40. ReussO, VikA, KolterR, MorschhäuserJ (2004) The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341 : 119–127.
41. SpieringMJ, MoranGP, ChauvelM, MacCallumDM, HigginsJ, et al. (2010) Comparative transcript profiling of Candida albicans and Candida dubliniensis identifies SFL2, a C. albicans gene required for virulence in a reconstituted epithelial infection model. Eukaryotic Cell 9 : 251–265.
42. SubramanianA, TamayoP, MoothaVK, MukherjeeS, EbertBL, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102 : 15545–15550.
43. SellamA, van het HoogM, TebbjiF, BeaurepaireC, WhitewayM, et al. (2014) Modeling the Transcriptional Regulatory Network That Controls the Early Hypoxic Response in Candida albicans. Eukaryotic Cell 13 : 675–690.
44. KetelC, WangHSW, McClellanM, BouchonvilleK, SelmeckiA, et al. (2009) Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet 5: e1000400.
45. GentlemanRC, CareyVJ, BatesDM, BolstadB, DettlingM, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.
46. ToedlingJ, SkylarO, SklyarO, KruegerT, FischerJJ, et al. (2007) Ringo–an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics 8 : 221.
47. WestessonO, SkinnerM, HolmesI (2013) Visualizing next-generation sequencing data with JBrowse. Brief Bioinformatics 14 : 172–177.
Štítky
Genetika Reprodukčná medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



