-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
In species that reproduce sexually, diploid individuals have two copies of each chromosome, inherited from their father and mother. During a special cell division called meiosis, these two sets of chromosomes are mixed by homologous recombination to give genetically unique chromosomes that will be transmitted to the next generation. Homologous recombination processes are highly controlled in terms of number and localization of events within and among chromosomes. Disruption of this control (a lack of or improper positioning of homologous recombination events) causes deleterious chromosome associations in the offspring. Using the model plant Arabidopsis thaliana we reveal here that the AtPSS1 gene is required for proper localization of these homologous recombination events along the genome. We also show that AtPSS1, which belongs to a family of proteins able to move along the cytoskeleton, is likely part of a module that allows cytoplasmic forces to be transmitted through the nucleus envelope to promote chromosome movements during homologous recombination progression.
Published in the journal: The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis. PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004674
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004674Summary
In species that reproduce sexually, diploid individuals have two copies of each chromosome, inherited from their father and mother. During a special cell division called meiosis, these two sets of chromosomes are mixed by homologous recombination to give genetically unique chromosomes that will be transmitted to the next generation. Homologous recombination processes are highly controlled in terms of number and localization of events within and among chromosomes. Disruption of this control (a lack of or improper positioning of homologous recombination events) causes deleterious chromosome associations in the offspring. Using the model plant Arabidopsis thaliana we reveal here that the AtPSS1 gene is required for proper localization of these homologous recombination events along the genome. We also show that AtPSS1, which belongs to a family of proteins able to move along the cytoskeleton, is likely part of a module that allows cytoplasmic forces to be transmitted through the nucleus envelope to promote chromosome movements during homologous recombination progression.
Introduction
During meiosis, chromosomes inherited from the mother and father are mixed in a process termed homologous recombination, to generate unique chromosomes that will be transmitted to the next generation. This genetic mixing has sustained the evolution of eukaryotes. There are typically one to four exchange points –crossovers (COs) - between homologous chromosomes at each meiosis. The distribution of these COs is under a series of constraints [1], [2]. First, there is at least one CO per chromosome pair (obligatory CO or CO assurance). Indeed, beyond their genetic consequences, COs are also essential for holding homologous chromosomes together during meiosis I, ensuring their balanced distribution in daughter cells. Notably, a lack of or improper positioning of this obligatory CO causes aneuploidy in human oocytes [3]. Second, COs are subject to interference. This prevents the occurrence of COs next to each other, shaping an even distribution and limiting their number [4]. COs are also under homeostasis, meaning that their number tends to be stable even when faced with variation in precursor number [5]–[7]. Finally, looking at frequencies, COs are not homogenously distributed along the genome; hot and cold regions have been defined at the chromosome scale, and hotspots with a very high CO frequency have been observed at the kb scale [8], [9].
COs are produced during meiotic prophase I concomitantly with and functionally connected to chromosome pairing and synapsis, which is the intimate association of homologous chromosomes lengthways with a protein structure, the synaptonemal complex (SC). Recombination is initiated at early prophase I by the formation of DNA double-strand breaks (DSBs) which largely outnumber the eventual CO number [10]. DSBs are subsequently resected to yield 3′ overhangs that invade the homologous chromosome, a step in which the recombinase DMC1 plays a prominent role [11]. In plants, as in mammals and budding yeast, these early steps of recombination also promote homologous chromosome synapsis. Indeed mutants affected in DSB formation or homologous template invasion (including Atdmc1) fail in both synapsis and CO formation [12]–[14]. DSB repair events form intermediates that are eventually resolved as either COs or non-crossovers (NCO) [15], [16] in the context of the SC. DSB repair can also occur using the sister as a template, a process that does not lead to inter-homologue COs, as observed in the Atdmc1 mutant [12], [13] or during haploid meiosis [17] where the ubiquitous recombinase RAD51 catalyzes sister repair. However the prevalence of such sister-mediated repair in wild-type Arabidopsis is unclear [15]. Homologous chromosome invasion events can mature into COs through at least two independent pathways. These two pathways coexist in budding yeast, mammals and Arabidopsis [1], [18]–[21]. Class I COs, the most prevalent class, are subject to interference and their production is dependent on the ZMM proteins (in Arabidopsis: SHOC1 (AtZIP2), PTD1, AtHEI10, AtZIP4, AtMSH4, AtMSH5, AtMER3 [18]). Interestingly, in Arabidopsis zmm mutants most chromosomes do not form COs, but synapsis occurs normally. This shows that recombination intermediates which promote synapsis are produced in zmm mutants, even if they are not eventually converted into COs. Formation of the minor Class II COs (∼15% of CO), that do not display interference, involves MUS81 [1], [18]–[21] and is down regulated by AtFANCM [22], [23].
All these molecular events must be coordinated at the chromosome and cellular levels to shape CO distribution. Interestingly, it has been shown in several species that chromosome movement is particularly prominent at meiotic prophase and plays a significant role in chromosome pairing/synapsis and CO formation [24], [25]. Telomeres in mammals, fungi and plants, or specific chromosome sites called pairing centers in the case of C. elegans, bind to the nuclear envelope where they are subject to cytoskeleton originated forces [26]–[31]. Telomere chromosome movements are also illustrated by a transient prophase I configuration called the bouquet, where telomeres cluster together on the nuclear envelope. This highly polarized nucleus stage has been described since the early 1900s [32]. Cytoplasmic forces are transduced to the chromosomes inside the nucleus, through the nuclear envelop which is intact during meiotic prophase I, via a chain of proteins (reviewed in [24], [25]). Central in this chain are the KASH (Klarsicht/ANC-1/Syne-1 homology)-domain and SUN(Sad-1/UNC-84)-domain proteins. In yeasts, worms and mammals KASH-domain and SUN-domain proteins play a crucial role in meiotic chromosome movement and homologue pairing [25], [27], [29], [33], [34]. KASH-domain proteins localize to the outer nuclear membrane and interact with SUN-domain proteins which are inserted in the inner membrane. However, the connection to the cytoskeleton, at one end, and to the chromosome (telomeres in most species and pairing centers in C. elegans) associated protein at the other end, appears to rely on evolutionary divergent proteins (reviewed in [24], [25]). In S. cerevisiae KASH-domain proteins interact with actin [35] while S. pombe, C. elegans and mouse KASH-domain proteins interact with dynein [25], a microtubule motor protein. In plants, the movement of chromosomes at meiotic prophase has been directly observed in maize and the application of specific depolymerizing drugs suggests that it depends on both tubulin and actin [31]. In Arabidopsis, where live imaging is not yet available, telomeres appear to be associated around the nucleolus in early meiotic prophase and are moved to the nuclear membrane preceding synapsis where they transiently cluster together (but not in a tight manner as in the classical bouquet configuration observed in many species [36]). On completion of synapsis the paired telomeres are dispersed but remain attached to the nuclear membrane until diplotene when they dissociate from the nuclear membrane [37]. Thus, it is likely that telomere-mediated chromosome movement is also important for meiotic prophase I in Arabidopsis. Strongly supporting this hypothesis, the two Arabidopsis SUN-domain proteins were recently shown to be essential for completion of synapsis and normal CO formation (S.J.A, unpublished data).
The rice Kinesin1-like protein PSS1 has been shown to be essential for fertility and normal chromosome segregation at meiosis [38], but its potential function in synapsis and recombination was not investigated. Here we identified the AtPSS1 Kinesin1-like protein as a major actor in meiosis, promoting synapsis and regulating CO formation in Arabidopsis. Our data suggest that the movement of AtPSS1 along microtubules generates cytoplasmic forces which could be transmitted to the chromosomes via a KASH-SUN module and coordinate synapsis and CO distribution.
Results
AtPSS1 is required for full synapsis and bivalent formation in meiosis
A previous report showed that mutation of the rice class I kinesin I (named OsPSS1) leads to meiotic defects [38]. Reciprocal BLAST analysis and comprehensive sequence analysis of plant kinesins [39] unambiguously identified the product encoded by the Arabidopsis At3g63480 gene as the only putative orthologue of OsPSS1. The two proteins share high amino acid sequence identity (59%). We identified three T-DNA insertion lines from the public collections: Atpss1-1, Atpss1-2 and Atpss1-3. Insertion of the TDNA in these loci was confirmed by sequencing the flanking sequences (Figure 1). Homozygous plants for all three lines have the same phenotype: normal vegetative growth but decreased fertility, as shown by reduced seed set (55±6 seeds per silique for wild type versus 27±5 for Atpss1-1) and reduced pollen viability (Alexander staining, Figure S1). Heterozygote plants for two Atpss1 mutations had the same phenotype showing that the three mutants are allelic. Transformation of the Atpss1-1 mutant with a 5 kb genomic region containing the AtPSS1 coding and regulatory sequences restored pollen viability (7 independent transformants, Figure S1), confirming that the observed defects are due to disruption of the AtPSS1 gene.
Fig. 1. The AtPSS1 gene and mutations. 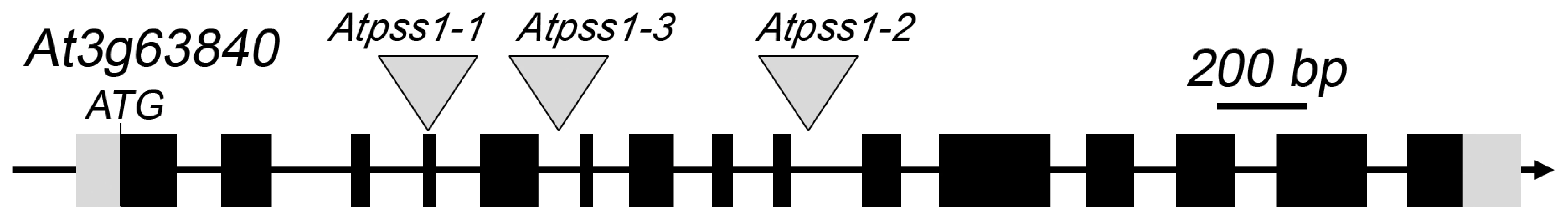
The arrow indicates the orientation of the open reading frame. Exons are shown as boxes (grey: UTR, black: CDS). In Atpss1-1, Atpss1-2 and Atpss1-3 corresponding to WiscDsLox_343E05, SALK_120399 and SALK_024926 lines, the T-DNA was inserted as indicated by triangles. We used chromosome spreads to investigate male meiosis defects in the Atpss1-1 mutant. Wild-type Arabidopsis meiosis was described in detail in [40], and the major stages are summarized in figure 2. At leptotene chromosomes appear as thin threads (Figure 2A), synapsis (the close association of two chromosomes via an SC) begins at zygotene and is complete by pachytene (Figure 2B). The SC is then depolymerized at diplotene and chromosomes condense so that the five bivalents are visible (pairs of homologues connected by COs) (Figure 2C). The bivalents align at metaphase I (Figure 2D), and chromosomes separate from their homologue at anaphase I leading to the formation of two pools of five chromosomes and two nuclei (Figure 2E). At the second meiotic division, the pairs of sister chromatids align on the two metaphase plates, and separate at anaphase II to generate four pools of five chromosomes, which gives rise to tetrads of four microspores (Figure 2F). In Atpss1 mutants, leptotene and zygotene appeared similar to those in wild type (Compare figure 2A and 2G). Accordingly immunolocalization of two axial element proteins, ASY3 [41] and the hormad domain containing protein ASY1 [42] did not reveal any difference between Atpss1 and wild type (Figure S2). However we were unable to find a typical pachytene stage among chromosome spreads of the Atpss1 mutant (n>300), as only partial synapsis was observed (Figure 2H). Synapsis was further examined by immunolocalization of REC8 and ZYP1 [43], which are chromosome axis and SC central element proteins, respectively (Figure 3). In flower buds whose size corresponds to late pachytene/diplotene stages, most wild-type cells showed almost complete synapsis, with the ZYP1 signal covering completely the REC8 signal (Figure 3A). In contrast, we were unable to find any meiocytes in which SC had undergone complete polymerization (n = 102) in Atpss1. Atpss1 cells showed various levels of incomplete synapsis (Figure 3B), ranging from 4 to 91%, with less than half of the REC8 axis being covered with ZYP1 signal in most cells (distribution shown on Figure 3C). The observed partial ZYP1 loading could be the result of either delayed synapsis or failure in completing synapsis. However, the observation of diplotene stages on the same slides favors the hypothesis of incomplete synapsis (see also below). At diakinesis and metaphase I, a mixture of univalents and bivalents (on average 3.1±1.2 bivalents and 1.9±1.2 univalent pairs) was observed in each Atpss1 allele (Figure 4), contrasting with wild type which always has five bivalents (Figure 2D, J and 5). FISH experiments using probes directed against 45S, 5S rDNA and the F8J2 BAC that allow the identification of the five Arabidopsis Col-0 chromosomes as described in [44], suggested that each chromosome is affected in bivalent formation (The univalent frequency for chromosomes 1 to 5 were respectively 28%, 37%, 42%, 42% and 26%. N = 43 Atpss1-1 cells). The presence of univalents resulted in missegregation of chromosomes in anaphase I and a subsequent aberrant number of daughter cells and/or unbalanced chromosome distribution (Figure 2K, L). Overall, our results showed that AtPSS1 is required for full synapsis and normal levels of bivalent formation at male meiosis. Observation of pistils [45], showed that 52% of the Atpss1 female gametophytes were defectives (n = 150). Further, univalents were detected at metaphase I of female meiosis (Figure S3), showing that AtPSS1 is essential for normal levels of bivalent formation in both male and female meiocytes.
Fig. 2. Chromosome spreads of male meiocytes in wild type and Atpss1-1. 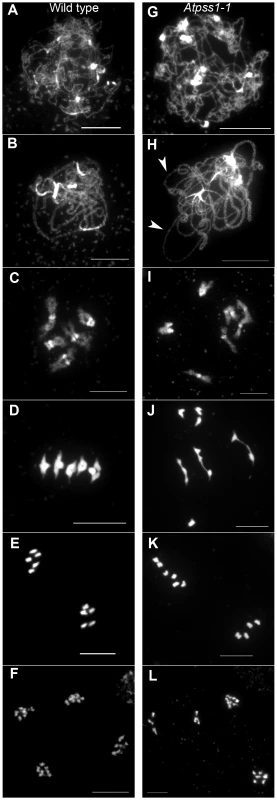
(A,G) Leptotene. (B,H) Pachytene. Arrowheads show unsynapsed regions. (C,I) Diakinesis. (D,J) Metaphase I. (E,K) Metaphase II. (F,L) Telophase II. Chromosome were spread according to Ross et al. [40] and stained with DAPI. Scale bar = 10 µm. Fig. 3. Co-immunolocalization of REC8 and ZYP1 at pachytene. 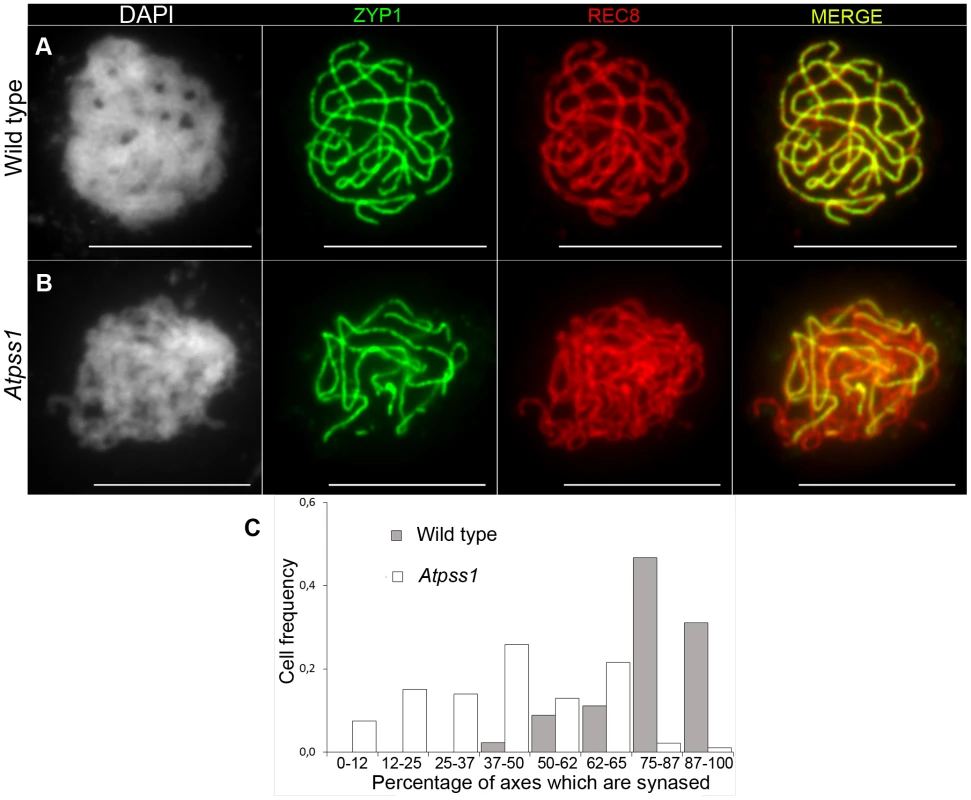
Floral buds of the correct size or bigger for the late pachytene/diplotene stage in wild type were used to make spreads according to Armstrong et al. [42]. Scale bar = 10 µm. (A) Wild type. (B) Atpss1-1. (C) Histogram of cells according to their proportion of synapsed axes. The proportion of synapsed axes in each cell was estimated by measuring the frequency of [red and green] pixels among the total number of [red] pixels. For example on figure 3B, 51% of the ASY1 red signal (axes) colocalize with the ZYP1 green (synapsis) signal. Fig. 4. Average number of bivalents (blue) and pairs of univalents (red) per male meiocyte. 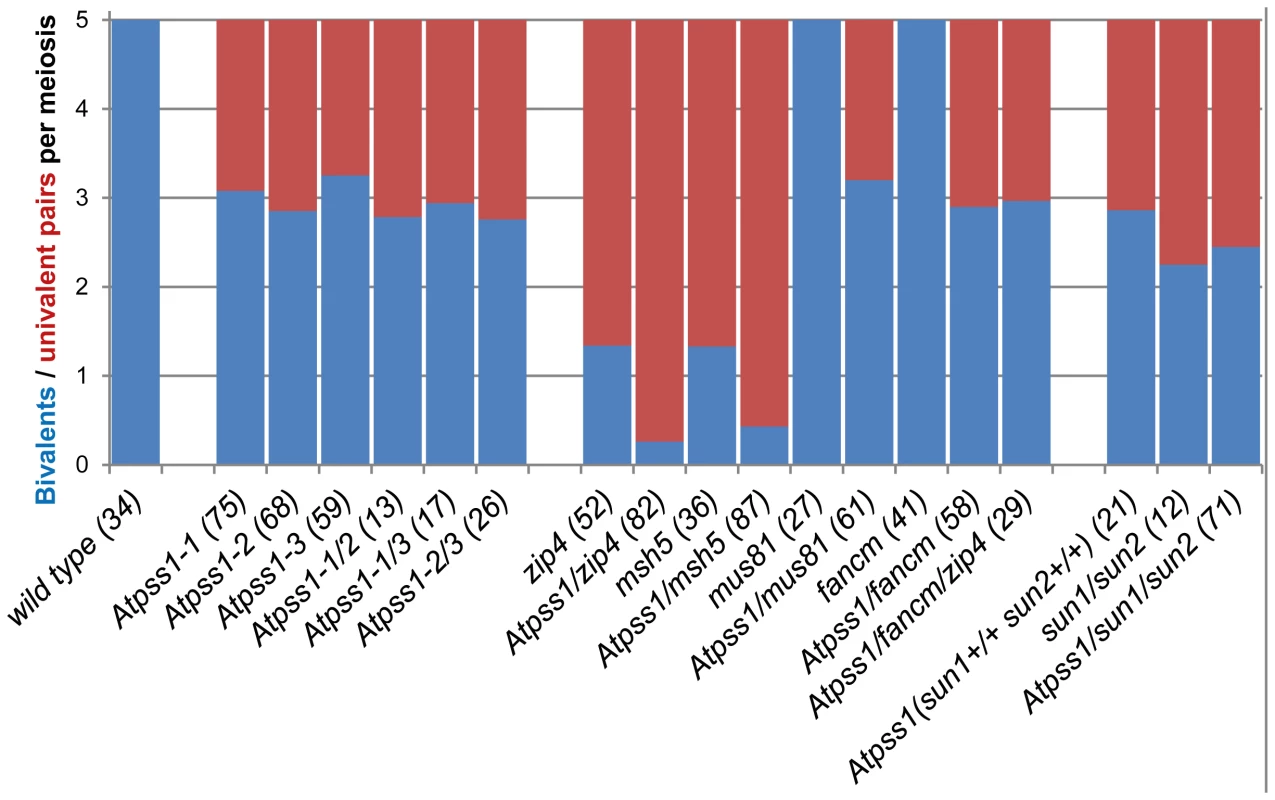
Number of metaphase I cells analyzed is indicated in brackets. Fig. 5. MLH1 immunolocalization. 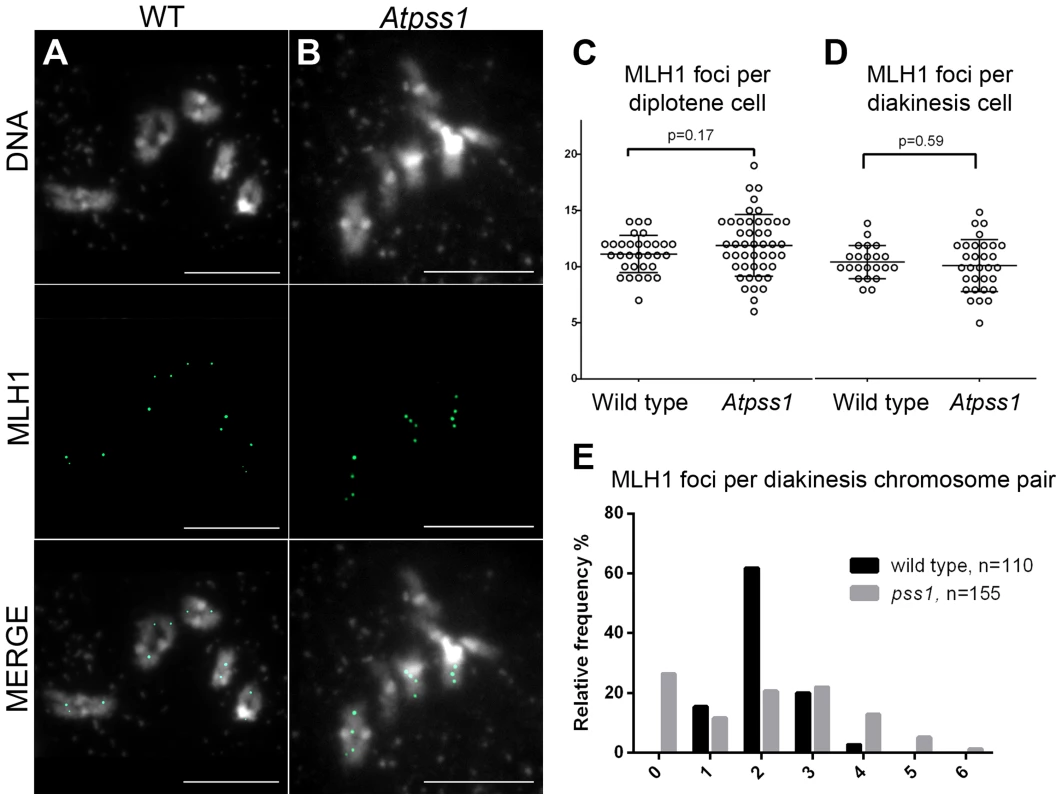
Immunolocalization of MLH1 at diakinesis is shown (A) in wild type and (B) in Atpss1-1. (C, D). Scatter plot of MLH1 foci number per cell at diplotene and diakinesis. (E) Distribution of chromosomes according to their MLH1 foci number at diakinesis. Cells were prepared according to Chelysheva et al. [47]. Scale bar = 10 µm. The AtPSS1 mutation affects CO distribution but not frequency
The presence of bivalents in Atpss1-1 implies that CO formation is not completely impaired in this mutant. The nature of the COs produced in the absence of AtPSS1 was investigated by epistasis tests with zmm and mus81 mutants, which are defective in class I and class II CO formation, respectively. Mutation of a ZMM in Atpss1 reduced bivalent formation from 3.1±1.2 to 0.3±0.4, showing that most of the COs produced in the Atpss1 mutant are ZMM dependent. We then used MLH1 immunolocalization, a marker of class I COs, to explore CO distribution in Atpss1. The total number of MLH1 foci per cell during diplotene and diakinesis was similar in Atpss1 (11.9±2.7 and 10.2±2.3) and wild type (11.1±1.7and 10.5±1.5) (Figure 5). However, we found that the distribution of MLH1 foci among chromosomes was significantly affected in the Atpss1 mutant, as shown in figure 5. In wild type, 62% of the bivalents had exactly two MLH1 foci, 20% had three, 15% had one and less than 3% had four foci. In contrast, the number of MLH1 foci per chromosome was much more variable in Atpss1, with the appearance of classes not observed in wild type (Figure 5E). One quarter of the chromosome pairs appeared as univalents without MLH1 foci, fitting with the observed frequency of univalents at metaphase I, while bivalents with more than three foci were more frequent than in wild type (19.4% vs 2.7%). This suggests that CO distribution but not frequency is affected in Atpss1. Measurements of recombination rates in six genetic intervals using pollen tetrad analysis [46] showed that CO frequency is not reduced but even slightly higher in Atpss1 (Figure 6, Table S1a and Table S1b). CO interference, measured genetically, was significantly reduced compared to wild type, to a level no longer detected (Table S1b). While we cannot formally exclude that a low level of interference exists, this clearly establish that CO interference measured genetically is decreased in Atpss1. This further suggests that relative CO distribution is disturbed in Atpss1.
Fig. 6. Genetic recombination in wild type and Atpss1-1. 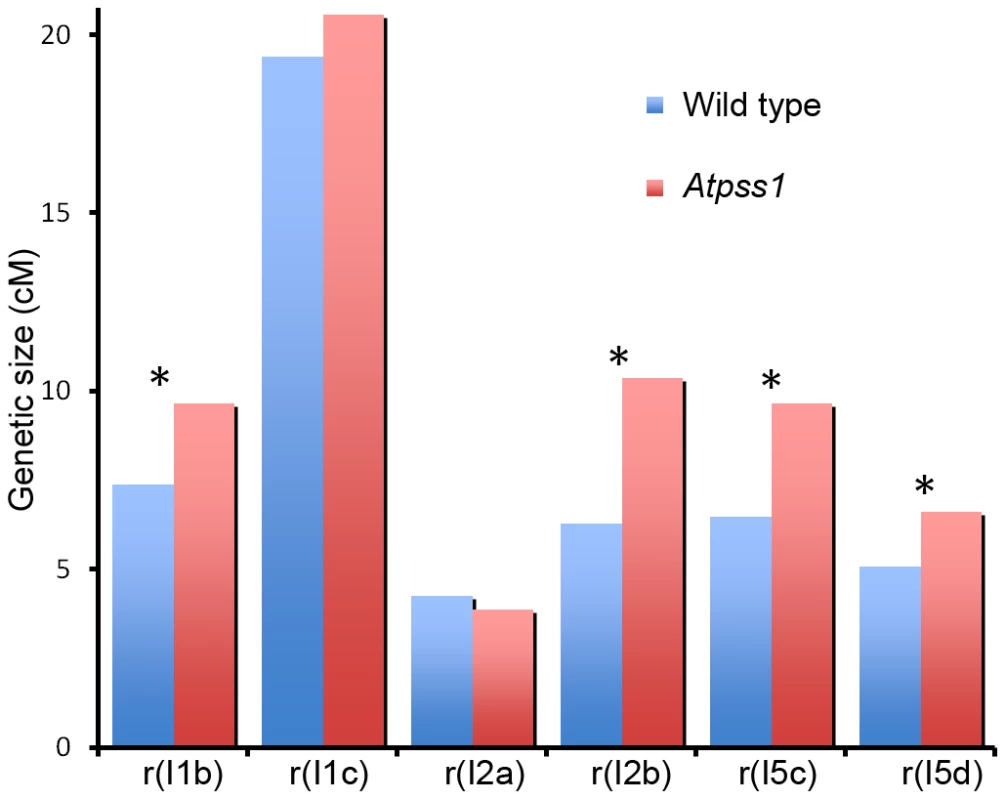
Genetic distances in six intervals using tetrad analysis with fluorescent-tagged lines (FTL), were calculated with the Perkins equation [67] and are given in centiMorgans. I1b and I1c are adjacent intervals on chromosome 1 and so on for the other pairs of intervals as described in [46] (Table S1). In Atpss1, synapsis and DSBs maturation into COs occur in the same regions
Overall, the above data showed that synapsis is incomplete and CO distribution among chromosomes is affected in Atpss1 mutants. As both synapsis and COs are promoted by DSB formation and repair, we carried out immunolocalization studies with DMC1, a protein which marks DSBs undergoing repair. In Atpss1, DMC1 foci decorated all chromosome axes and their total number was higher compared to wild type (+37%. 204±6 vs 279±8. T-test p = 3.5.10−10), suggesting that in the mutant DSB formation is enhanced or that DMC1 foci accumulate due to slower turnover (Figure 7). Thus in the mutant DSBs appear to occur on all chromosomes. We then examined whether the chromosome regions where COs occurred and that synapsed were the same. Because synapsis disappears before MLH1 foci numbers peak in Arabidopsis [47], we used HEI10/ZYP1 co-immunolocalization to explore this question (Figure 8 and S4). Indeed, HEI10 marks recombination progression from numerous faint foci at leptotene (Figure 8A) to about ten large foci labeling class I CO sites from late pachytene (Figure 8C) to diakinesis (Figure S4) [48]. At leptotene, Atpss1 and wild-type cells were indistinguishable with numerous small HEI10 foci (Figure 8A and 8D), further suggesting that early recombination events are unaffected in the mutant. At early wild type pachytene, numerous foci of variable size are dispersed on the SC (Figure 8B). At the same stage in Atpss1, the synapsed regions were also decorated with numerous HEI10 foci, but the regions that failed to synapse were foci-free. At late pachytene, a small number of bright and homogeneous foci were observed in both wild type and the mutant (Figure 8C, F and G). Remarkably, while the total length of the SC in Atpss1 pachytene cells was on average one third that of wild type, confirming partial synapsis, the average number of HEI10 foci per cell was unaffected (Wild type: 10.3±1.9, Atpss1: 11.2±1.2, p = 0.19) (Figure 8H). Accordingly, the number of HEI10 foci per 100 µm of SC was on average 3.1±0.7 for wild type and 10.9±4.8 for Atpss1 (these measurements were made on a cell per cell basis, because the entanglement of Arabidopsis pachytene chromosomes makes it difficult to unambiguously follow individual SCs). While the density of HEI10 foci was relatively stable in wild type (from 2 to 4.3 per 100 µm), it varied greatly in Atpss1 (from 4.3 to 23.6 per 100 µm) (Figure 8H). This is strikingly illustrated by the extreme case shown in figure 8G, where seven HEI10 foci can be seen on a single 30 µm SC stretch. At diplotene and diakinesis, the number of HEI10 foci per cell was similar and stable in the wild type and mutant. However, consistent with the MLH1 data, the distribution of HEI10 foci among chromosomes was significantly modified in Atpss1 (Figure S4), confirming that CO distribution but not number is affected. In summary, in Atpss1, COs and synapsis are jointly restricted to the same limited portion of the genome. Partial synapsis is accompanied by an increase in CO density per SC unit, resulting in –or caused by (see discussion) - an unaffected number of COs per cell.
Fig. 7. DMC1 immunolocalization. 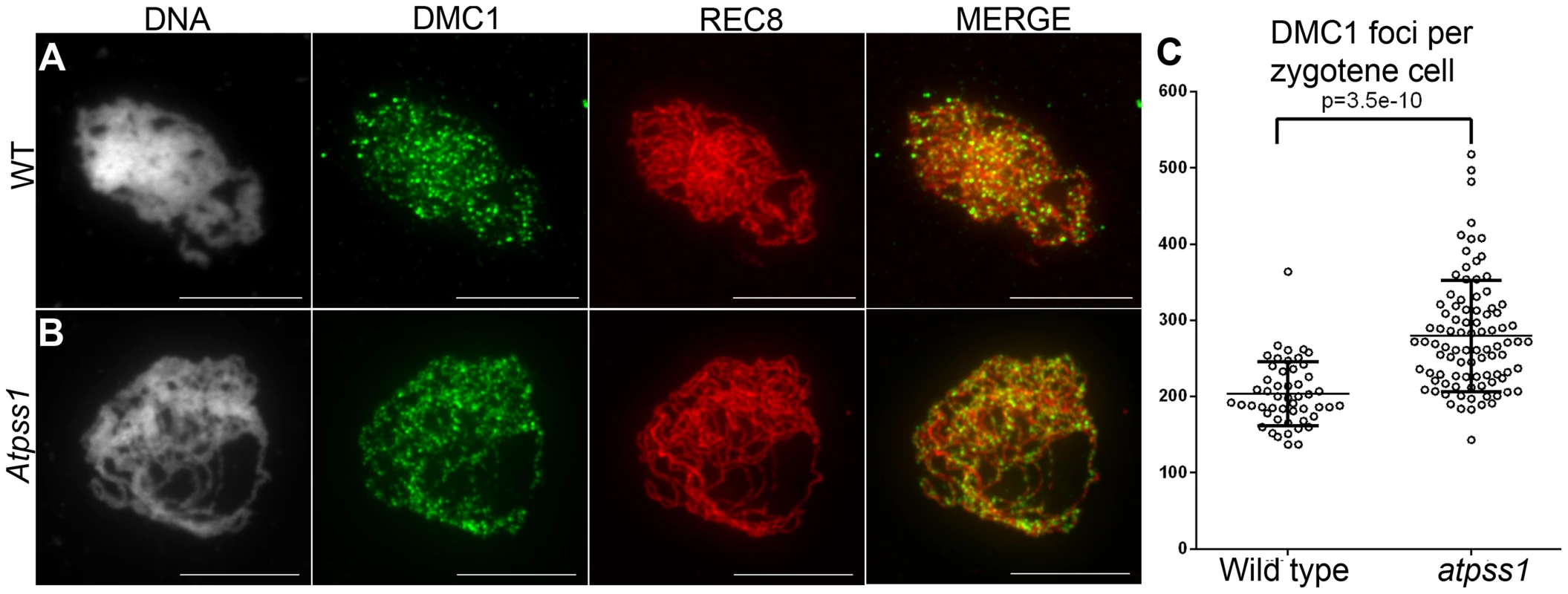
Immunolocalization of DMC1 at early zygotene is shown (A) in wild type and (B) in Atpss1-1. Cells were prepared according to Armstrong et al. [42] Scale bar = 10 µm. (C) Scatter plot of DMC1 foci number per cell. Fig. 8. Co-immunolocalization of HEI10 and ZYP1. 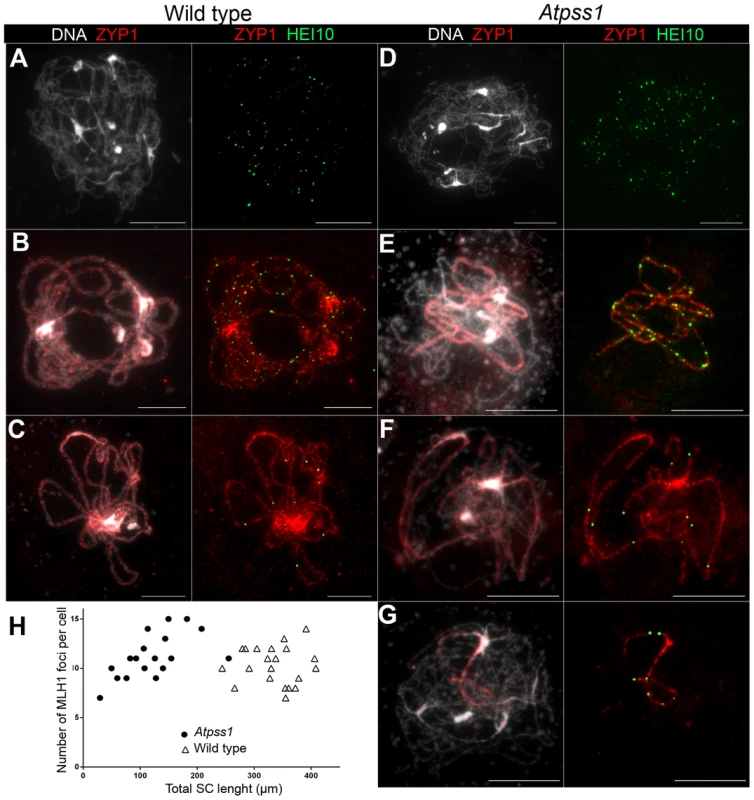
(A, D) Leptotene with numerous HEI10 foci, (B, E). Early pachytene with a mixture of faint and bright HEI10 foci. (C, F, G) Late pachytene with bright HEI10 foci. (H) Plot of pachytene cells according to their total HEI10 foci number and total SC length. Cells were prepared according to Chelysheva et al. [47] Scale bar = 10 µm. MUS81-dependant COs in Atpss1
The MUS81 pathway (Class II pathway) is minor in Arabidopsis wild type. Its disruption reduces CO frequency by ∼10%, but does not affect bivalent formation [49], [50] (Figure 4). Mutation of MUS81 in the Atpss1 background did not further reduce bivalent frequency (Figure 4), which is consistent with the conclusion above that most COs are ZMM-dependent in Atpss1. At FANCM was previously shown to limit MUS81-dependant CO formation and bivalent formation is fully restored in zmm/Atfancm mutants due to a massive increase in class II COs [22]. Mutation of AtFANCM in Atpss1 did not increase the number of bivalents, suggesting that it did not restore CO formation in regions that are CO incompetent in the single Atpss1 mutant (but this does not exclude that there is an increase in CO frequency in regions that are CO competent) (Figure 4). However while bivalent formation was very low in Atpss1 Atzip4, bivalent formation was restored in the Atpss1 Atfancm Atzip4 triple mutant back to the level observed in the single Atpss1 mutant (Figure 4). Altogether, these results suggest that, in Atpss1, class II COs occur at a low frequency, and can be promoted by mutating AtFANCM but exclusively in regions that are also already competent for class I CO formation.
A potential AtPSS1-SUNs-WIPs force transduction module
AtPSS1, which belongs to the kinesin family, appears to play a crucial role in meiosis. Kinesin proteins are characterized by their ability to walk on microtubules via a motor domain that uses ATP to promote repetitive conformation changes [51]. We thus tested if the motor function of AtPSS1 is important for its function in meiosis. For this, we expressed an AtPSS1 protein modified in the conserved arginine (Arg-293>His) that was previously shown to abolish the microtubule-stimulated ATPase activity [38] in the Atpss1-1 mutant. When Atpss1 plants were transformed with the control wild-type AtPSS1 gene, pollen viability and bivalent formation at metaphase I were fully restored (7 independent transformants). In contrast, transformation with AtPSS1-R293H, expressed behind the native AtPSS1 promoter, did not restore pollen viability and normal meiosis (4 independent transformants, see methods; Bivalent frequency: 4.2±1 (n cells = 35), 4.3±0.5 (n = 6), 3.5±0.6 (n = 4), 3.5±1.2 (n = 15), respectively), showing that the kinesin function of AtPSS1 is critical for its role in meiosis. In several model species, cytoskeleton-based forces were previously shown to be important for meiosis and to be transduced to the nucleus by KASH - and SUN-domain containing proteins [24], [25]. In Arabidopsis, two SUN proteins were recently shown to be redundant and important for meiosis (S.J.A. under review). As in Atpss1, a mixture of bivalents and univalents are observed in Atsun1 Atsun2 double mutants. This defect is quantitatively identical in the Atpss1, Atsun1 Atsun2 and the Atsun1 Atsun2 Atpss1 triple mutants (Figure 4), suggesting that SUN proteins and AtPSS1 may act in the same pathway. WIP1-3 proteins were also recently identified as KASH containing proteins in Arabidopsis, and shown to interact with SUNs [52]. This raised the possibility that AtPSS1 could be involved in transmitting forces to the meiotic nucleus via a WIP-SUN module. Yeast two-hybrid experiments showed that AtPSS1 interacts directly with WIP1 and WIP2. The AtPSS1-WIP1 but not the AtPSS1-WIP2 interaction was confirmed by BiFC assays (Figure S5). The yeast two-hybrid also confirmed that WIPs interact with SUNs, as previously shown [52] (Figure 9).
Fig. 9. Interaction between AtPSS1, AtWIPs and AtSUNs. 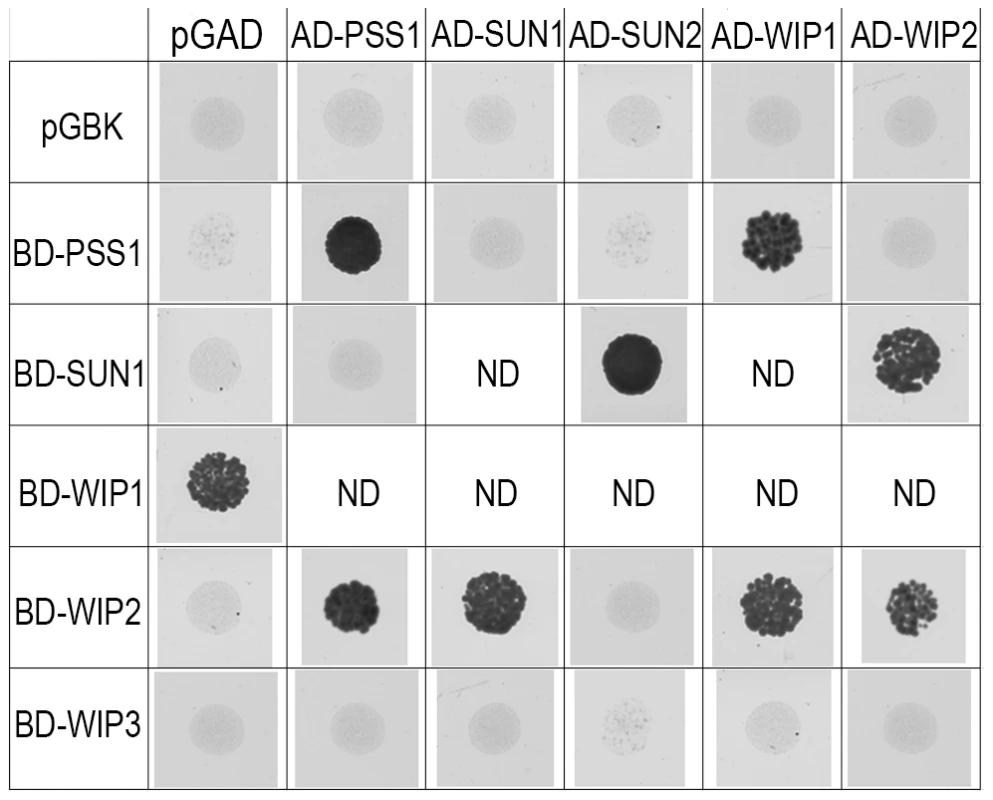
Yeast two-hybrid - For each combination yeast cells were spotted on selective medium to test interactions (all combinations were able to grow on non-selective medium; not shown) ND: Not determined either because irrelevant or due to self-activation of one of the partners. Discussion
During meiotic prophase I, chromosome movements within the intact nucleus are prominent and have been shown to be involved in chromosome pairing, synapsis and recombination in a variety of species. Here we showed that AtPSS1, the Arabidopsis kinesin-1 like protein [39], is essential for full synapsis and is required for proper CO distribution. Furthermore, the bivalent shortage is identical when AtPSS1, SUNs or both, are knocked out suggesting that SUNs and AtPSS1 act in the same pathway to regulate CO formation. In addition, AtPSS1 interacts with the KASH-domain proteins WIP1 and WIP2 which themselves interact with SUN proteins [52]. Finally, we showed that the kinesin motor domain of AtPSS1 is required for its meiotic function. Kinesin is a motor protein which walks along microtubules with high processivity and for long distances (reviewed in [51]). We thus speculate that AtPSS1 moves along microtubules and generates forces that are transduced via a SUN-WIP module through the nuclear membrane to the chromosomes, promoting synapsis and regulating CO distribution (see below). The proteins that would connect SUNs to the chromosome telomeres remain to be identified. These results add to a growing amount of evidence showing that the transduction of cytoplasmic forces through the nuclear membrane is an important and conserved promoter of meiotic recombination. It should be noted here that the function we propose for AtPSS1 appears to be fulfilled by dynein in many organisms, and that dynein is absent from flowering plant genomes [53]. The rice PSS1 is also essential for normal meiosis [38]. Even though recombination and synapsis have not been extensively analyzed in the rice Ospss1 mutants, univalent were observed at metaphase I, suggesting that the primarily defects may be similar to Atpss1. This suggests that the meiotic function of AtPSS1 is conserved among flowering plants.
We showed that AtPSS1 is required for full synapsis and normal CO formation. In most species, the search for homologous sequences by recombinase-coated 3′-ssDNA promotes both CO formation and homologous synapsis. Indeed, in Arabidopsis both COs and synapsis are absent in mutants affecting DSB formation, but also homologous sequence invasion (RAD51, DMC1 and their co-factors) [12], [14], [18]. This appears to be a cooperative process as multiple repair events are required for initiation and progression of synapsis [54], [55]. Atpss1 mutants have a novel defect: in each cell, COs and synapsis take place on only a subset of the genome (which varies from 10 to 90%). Initial DSB formation and processing do not appear to be involved in these defects, as DMC1 foci and early HEI10 are present on all chromosomes in the mutant. The number of DMC1 foci was higher in the mutant than wild type, possibly reflecting a delay in recombination progression. The increased number of DMC1 foci may also reflect an increase of the number of DSBs in response to the downstream defects [56]. However, we suggest that only a subset of these DSBs is efficiently matured into potential CO precursors and promoters of synapsis. This is supported by the observation that the segments of chromosomes which were seen to synapse were also the places where early HEI10 foci progressively matured into intermediate and then late CO-marking-foci. This model implies that chromosome movement involving AtPSS1 is required to efficiently mature DMC1-coated-DSBs into CO/synapsis precursors. This movement could be simply required for the homology searching DNA “tentacle” [57] to reach the homologous chromosome which can be at some distance in the nucleus [58]. Alternatively, the movement may be required to resolve the entanglement/clutter/interlocking which likely arises from multiple chromosome pairing attempts in the limited space of the nucleus [55]. The DSBs present on the portions of chromosomes which failed to reach homologues are likely repaired using the sister chromatids as template, thus failing to promote synapsis and homologous CO. Such sister-mediated repair occurs genome-wide in haploid Arabidopsis, where DMC1-coated resected DSBs are repaired on the sister, or in diploid mutants where DMC1 or one of its partners is absent [12]–[14], [59].
One intriguing feature of the Atpss1 mutant is that CO frequency per cell is not reduced, but instead the subparts of the chromosomes that do synapse and recombine make a similar total number of COs per cell as in wild type. This is strikingly shown in figure 8G, where a single SC stretch was formed in a cell on which seven class I COs occurred, while CO number rarely exceeds four on an entire wild-type chromosome. The smaller size of the competent regions appears to be compensated by an increased CO density, which implies that interference is no longer acting or that the distance at which interference spreads is reduced. Unfortunately, the difficulty in following individual SCs prevented us from cytologically measuring CO interference. The stable number of COs per cell in Atpss1 could reflect a form of CO homeostasis, which is defined as the tendency to preserve CO number despite a variation in DSB number through a modulation of the probability for DSBs to become COs [5]. We suggest that such homoeostasis applies in the Atpss1 mutant, and that the decrease in the number of CO-competent DSB is compensated for by an increased probability of the eligible DSB becoming a CO. It is possible that the total number of COs per cell is defined, and then ∼10 COs per cell occurs on licensed regions. However, the mechanism that would count the number of COs per cell remains elusive. Alternatively, we suggest that a feed-back loop could sense some unachieved event (e.g. the presence of chromosomes lacking COs, or incomplete synapsis), and then increase the propensity of precursors to be designated for CO. This feed-back loop would therefore modulate the parameters of interference (possibly by a progressive increase in CO-promoting mechanical stress or progressive increase in the sensitivity of precursors to this stress [60], [61]). Finally, AtPSS1 could have a dual function, on one hand promoting synapsis and recombination intermediate maturation, and on the other preventing an excess of COs on selected regions, both via chromosome movement [24].
Materials and Methods
Plant material
Col-0 lines were obtained from the collection of T-DNA mutants from the Salk Institute Genomic Analysis Laboratory (Columbia accession) (SIGnAL, http://signal.salk.edu/cgi-bin/tdnaexpress) and provided by NASC (http://nasc.nott.ac.uk/). Mutant alleles used in this study were: Atmsh5-2 (SALK_026553) [62]; Atzip4-2 (SALK_068052) [63]; Atmus81-2 (SALK_107515) [49], [50], Atfancm-1 [22]; Atsun1 (SAIL_84_G10); Atsun2 (FLAG_026E12). Details for all genotypes, primers used and PCR amplification conditions are shown in table S2.
Plants were cultivated in a greenhouse or growth chamber under the following conditions: photoperiod 16 h/day and 8 h/night; temperature 20°C day and night; humidity 70%.
Genetic analyses
The six intervals tested in this study correspond to intervals I1b and I1c (both located at the top of chromosome 1), I2a and I2b (both located at the bottom of chromosome 2), and I5c and I5d (both located at the top of chromosome 5) described in [64]. Tetrad analyses were carried out as described in [46]. The resulting tetrad data (Table S1a) were analyzed using the Perkins mapping equation.
All double mutants were obtained by crossing plants, which were heterozygous for each mutation. The resulting hybrids were self-pollinated. PCR screening was then used to identify plants in the F2 progeny that were homozygous for both mutations.
Antibodies
The anti-ASY1 polyclonal antibody was described by [42]. The anti-ZYP1 polyclonal antibody was described by [43]. The anti-DMC1 antibody was described in [63], the anti-MLH1 antibody in [47], and the anti-HEI10 in [48]. The anti-REC8 polyclonal antibody was described in [65].
Microscopy
Chromosome spreads of male meiocytes were prepared and stained with DAPI as described in [40]. Chromosome spreads for immunocytology was performed according to [42]. Observations were made using a Leica (http://www.leica.com) DM RXA2 microscope or a Zeiss (http://www.zeiss.fr) Axio Imager 2 microscope; photographs were taken using a CoolSNAP HQ (Roper, http://www.roperscientific.com) camera driven by OpenLAB 4.0.4 software or a Zeiss camera AxioCam MR driven by Axiovision 4.7. All images were further processed with OpenLAB 4.0.4, Axiovision 4.7, or AdobePhotoshop 7.0 (http://www.adobe.com).
Yeast two-hybrid and BIFC assays
The AtPSS1, AtWIP1, AtWIP2, AtWIP3, AtSUN1 and AtSUN2 open reading frames were amplified from Arabidopsis cDNA clones (Columbia ecotype) using specific primers flanked by the AttB1 and AttB2 sites (Table S2), cloned into Gateway vector pDONR207 using BP recombination (Invitrogen), and sequenced. Expression vectors were obtained after LR recombination (Invitrogen) between these entry vectors and destination vectors (pGADT7-GW and pGBKT7-GW for Y2H, and pBiFP vectors for BIFC). Yeast two-hybrid interactions were tested using AtPSS1, AtWIP1, ATWIP2, AtWIP3, AtSUN1 and AtSUN2 as bait (pGADT7-GW) or as prey (pGBKT7-GW) by mating with the AH109 and Y187 yeast strains. For fluorescence complementation tests, transient expression of all eight compatible combinations between protein pairs (i.e., providing both parts of the YFP) was assayed. Each expression vector was introduced into Agrobacterium tumefaciens strain C58C1(pMP90) by electroporation. Agrobacterium bacterial cultures were incubated overnight at 28°C with agitation. Each culture was pelleted, washed, and resuspended in infiltration buffer (13 g/L bouturage N°2 medium [Duchefa Biochemie] and 40 g/L sucrose, pH 5.7) to an OD600 of 0.5. The inoculum was delivered to the lamina tissue of N. benthamiana leaves by gentle pressure infiltration through the lower epidermis. To enhance transient expression of BiFC fusion proteins, the P19 viral suppressor of gene silencing was coexpressed [66]. YFP fluorescence was detected three days after infiltration. Tissue was mounted in low-melting-point agarose (0.4% in water) and viewed directly using an inverted Zeiss Observer Z1 spectral confocal laser microscope LSM 710 using a C-Apochromat ×63/1.20 W Corr objective (Carl Zeiss). Fluorescence was recorded after an excitation at 514 nm (Argon laser) and using a selective band of 514 to 568 nm.
Complementation tests
A 5 kb AtPSS1 genomic fragment containing 1.5 kb of promoter region and the complete AtPSS1 gene was amplified using specific primers flanked by AttB1 and AttB2 sites (Table S2), cloned into Gateway vector pDONR207 using BP recombination (Invitrogen), and sequenced. Directed mutagenesis was performed using the Quickchange Site-Directed Mutagenesis Kit (Stratagene). The mutagenic primers used to generate the AtPSS1-R293H (Arg codon cgc→His codon cac) are shown in Table S2. A LR reaction between the resulting vectors and the pGWB1 destination binary vector was performed.
Supporting Information
Zdroje
1. MézardC, VignardJ, DrouaudJ, MercierR (2007) The road to crossovers: plants have their say. Trends Genet 23 : 91–99 doi:10.1016/j.tig.2006.12.007
2. YoudsJL, BoultonSJ (2011) The choice in meiosis - defining the factors that influence crossover or non-crossover formation. J Cell Sci 124 : 501–513 doi:10.1242/jcs.074427
3. NagaokaSI, HassoldTJ, HuntP (2012) Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet 13 : 493–504 doi:10.1038/nrg3245
4. BerchowitzLE, CopenhaverGP (2010) Genetic interference: don't stand so close to me. Curr Genomics 11 : 91–102 doi:10.2174/138920210790886835
5. MartiniE, DiazRL, HunterN, KeeneyS (2006) Crossover homeostasis in yeast meiosis. Cell 126 : 285–295 doi:10.1016/j.cell.2006.05.044
6. YokooR, ZawadzkiKA, NabeshimaK, DrakeM, ArurS, et al. (2012) COSA-1 Reveals Robust Homeostasis and Separable Licensing and Reinforcement Steps Governing Meiotic Crossovers. Cell 149 : 75–87 doi:10.1016/j.cell.2012.01.052
7. ColeF, KauppiL, LangeJ, RoigI, WangR, et al. (2012) Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol 14 : 424–430 doi:10.1038/ncb2451
8. BaudatF, ImaiY, de MassyB (2013) Meiotic recombination in mammals: localization and regulation. Nat Rev Genet 14 : 794–806 doi:10.1038/nrg3573
9. DrouaudJ, KhademianH, GirautL, ZanniV, BellalouS, et al. (2013) Contrasted Patterns of Crossover and Non-crossover at Arabidopsis thaliana Meiotic Recombination Hotspots. PLoS Genet 9: e1003922 doi:10.1371/journal.pgen.1003922
10. De MassyB (2013) Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet 47 : 563–599 doi:10.1146/annurev-genet-110711-155423
11. GertonJL, HawleyRS (2005) Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat Rev Genet 6 : 477–487 doi:10.1038/nrg1614
12. VignardJ, SiwiecT, ChelyshevaL, VrielynckN, GonordF, et al. (2007) The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet 3 : 1894–1906 doi:10.1371/journal.pgen.0030176
13. CouteauF, BelzileF, HorlowC, GrandjeanO, VezonD, et al. (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11 : 1623–1634.
14. De MuytA, PereiraL, VezonD, ChelyshevaL, GendrotG, et al. (2009) A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet 5: e1000654 doi:10.1371/journal.pgen.1000654
15. WijnkerE, Velikkakam JamesG, DingJ, BeckerF, KlasenJR, et al. (2013) The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. Elife 2: e01426 doi:10.7554/eLife.01426
16. QiJ, WijeratneAJ, TomshoLP, HuY, SchusterSC, et al. (2009) Characterization of meiotic crossovers and gene conversion by whole-genome sequencing in Saccharomyces cerevisiae. BMC Genomics 10 : 475 doi:10.1186/1471-2164-10-475
17. CifuentesM, RivardM, PereiraL, ChelyshevaL, MercierR (2013) Haploid meiosis in Arabidopsis: double-strand breaks are formed and repaired but without synapsis and crossovers. PLoS One 8: e72431 doi:10.1371/journal.pone.0072431
18. OsmanK, HigginsJD, Sanchez-MoranE, ArmstrongSJ, FranklinFCH (2011) Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol 190 : 523–544 doi:10.1111/j.1469-8137.2011.03665.x
19. HarrisonCJ, AlveyE, HendersonIR (2010) Meiosis in flowering plants and other green organisms. J Exp Bot 61 : 2863–2875 doi:10.1093/jxb/erq191
20. CromieGA, SmithGR (2007) Branching out: meiotic recombination and its regulation. Trends Cell Biol 17 : 448–455 doi:10.1016/j.tcb.2007.07.007
21. LynnA, SoucekR, BörnerGV (2007) ZMM proteins during meiosis: crossover artists at work. Chromosome Res 15 : 591–605 doi:10.1007/s10577-007-1150-1
22. CrismaniW, GirardC, FrogerN, PradilloM, SantosJL, et al. (2012) FANCM limits meiotic crossovers. Science (80-) 336 : 1588–1590 doi:10.1126/science.1220381
23. KnollA, HigginsJD, SeeligerK, RehaSJ, DangelNJ, et al. (2012) The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 24 : 1448–1464 doi:10.1105/tpc.112.096644
24. KoszulR, KlecknerNE (2009) Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol 19 : 716–724 doi:10.1016/j.tcb.2009.09.007
25. HiraokaY, DernburgAF (2009) The SUN rises on meiotic chromosome dynamics. Dev Cell 17 : 598–605 doi:10.1016/j.devcel.2009.10.014
26. ChikashigeY, DingDQ, FunabikiH, HaraguchiT, MashikoS, et al. (1994) Telomere-led premeiotic chromosome movement in fission yeast. Science 264 : 270–273.
27. ChikashigeY, TsutsumiC, YamaneM, OkamasaK, HaraguchiT, et al. (2006) Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125 : 59–69 doi:10.1016/j.cell.2006.01.048
28. SatoA, IsaacB, PhillipsCM, RilloR, CarltonPM, et al. (2009) Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139 : 907–919 doi:10.1016/j.cell.2009.10.039
29. ShibuyaH, IshiguroK-I, WatanabeY (2014) The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat Cell Biol 16 : 145–156 doi:10.1038/ncb2896
30. ScherthanH, WeichS, SchweglerH, HeytingC, HärleM, et al. (1996) Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol 134 : 1109–1125.
31. SheehanMJ, PawlowskiWP (2009) Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc Natl Acad Sci U S A 106 : 20989–20994 doi:10.1073/pnas.0906498106
32. ScherthanH (2001) A bouquet makes ends meet. Nat Rev Mol Cell Biol 2 : 621–627 doi:10.1038/35085086
33. ChikashigeY, HaraguchiT, HiraokaY (2007) Another way to move chromosomes. Chromosoma 116 : 497–505 doi:10.1007/s00412-007-0114-8
34. DingX, XuR, YuJ, XuT, ZhuangY, et al. (2007) SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell 12 : 863–872 doi:10.1016/j.devcel.2007.03.018
35. KoszulR, KimKP, PrentissM, KlecknerNE, KameokaS (2008) Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133 : 1188–1201 doi:10.1016/j.cell.2008.04.050
36. ZicklerD, KlecknerNE (1998) The leptotene-zygotene transition of meiosis. Annu Rev Genet 32 : 619–697 doi:10.1146/annurev.genet.32.1.619
37. RobertsNY, OsmanK, ArmstrongSJ (2009) Telomere distribution and dynamics in somatic and meiotic nuclei of Arabidopsis thaliana. Cytogenet Genome Res 124 : 193–201 doi:10.1159/000218125
38. ZhouS, WangY, LiW, ZhaoZ, RenY, et al. (2011) Pollen semi-sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 23 : 111–129 doi:10.1105/tpc.109.073692
39. RichardsonDN, SimmonsMP, ReddyASN (2006) Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes. BMC Genomics 7 : 18 doi:10.1186/1471-2164-7-18
40. RossKJ, FranszP, JonesGH (1996) A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosom Res 4 : 507–516.
41. FerdousM, HigginsJD, OsmanK, LambingC, RoitingerE, et al. (2012) Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet 8: e1002507 doi:10.1371/journal.pgen.1002507
42. ArmstrongSJ, CarylAPP, JonesGH, FranklinFCH (2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115 : 3645–3655 doi:10.1242/jcs.00048
43. HigginsJD, Sanchez-MoranE, ArmstrongSJ, JonesGH, FranklinFCH (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19 : 2488–2500 doi:10.1101/gad.354705
44. ChelyshevaL, VezonD, BelcramK, GendrotG, GrelonM (2008) The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS Genet 4: e1000309 doi:10.1371/journal.pgen.1000309
45. MotamayorJC, VezonD, BajonC, SauvanetA, GrandjeanO, et al. (2000) Switch (swi1), an Arabidopsis thaliana mutant affected in the female meiotic switch. Sex Plant Reprod 12 : 209–218.
46. BerchowitzLE, CopenhaverGP (2008) Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat Protoc 3 : 41–50 doi:10.1038/nprot.2007.491
47. ChelyshevaL, GrandontL, VrielynckN, le GuinS, MercierR, et al. (2010) An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet Genome Res 129 : 143–153 doi:10.1159/000314096
48. ChelyshevaL, VezonD, ChambonA, GendrotG, PereiraL, et al. (2012) The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet 8: e1002799 doi:10.1371/journal.pgen.1002799
49. HigginsJD, BucklingEF, FranklinFCH, JonesGH (2008) Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J 54 : 152–162 doi:10.1111/j.1365-313X.2008.03403.x
50. BerchowitzLE, FrancisKE, BeyAL, CopenhaverGP (2007) The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet 3: e132 doi:10.1371/journal.pgen.0030132
51. EndowSa, KullFJ, LiuH (2010) Kinesins at a glance. J Cell Sci 123 : 3420–3424 doi:10.1242/jcs.064113
52. ZhouX, GraumannK, EvansDE, MeierI (2012) Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol 196 : 203–211 doi:10.1083/jcb.201108098
53. WicksteadB, GullK (2007) Dyneins across eukaryotes: a comparative genomic analysis. Traffic 8 : 1708–1721 doi:10.1111/j.1600-0854.2007.00646.x
54. TesséS, StorlazziA, KlecknerNE, GarganoS, ZicklerD (2003) Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci U S A 100 : 12865–12870 doi:10.1073/pnas.2034282100
55. StorlazziA, GarganoS, Ruprich-RobertG, FalqueM, DavidM, et al. (2010) Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141 : 94–106 doi:10.1016/j.cell.2010.02.041
56. ThackerD, MohibullahN, ZhuX, KeeneyS (2014) Homologue engagement controls meiotic DNA break number and distribution. Nature 510 : 241–246 doi:10.1038/nature13120
57. KimKP, WeinerBM, ZhangL, JordanA, DekkerJ, et al. (2010) Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143 : 924–937 doi:10.1016/j.cell.2010.11.015
58. LeeC-Y, ConradMN, DresserME (2012) Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet 8: e1002730 doi:10.1371/journal.pgen.1002730
59. UanschouC, RonceretA, Von HarderM, De MuytA, VezonD, et al. (2013) Sufficient amounts of functional HOP2/MND1 complex promote interhomolog DNA repair but are dispensable for intersister DNA repair during meiosis in Arabidopsis. Plant Cell 25 : 4924–4940 doi:10.1105/tpc.113.118521
60. KlecknerNE, ZicklerD, JonesGH, DekkerJ, PadmoreR, et al. (2004) A mechanical basis for chromosome function. Proc Natl Acad Sci U S A 101 : 12592–12597 doi:10.1073/pnas.0402724101
61. ZhangL, LiangZ, HutchinsonJ, KlecknerNE (2014) Crossover Patterning by the Beam-Film Model: Analysis and Implications. PLoS Genet 10: e1004042 doi:10.1371/journal.pgen.1004042
62. HigginsJD, VignardJ, MercierR, PughAG, FranklinFCH, et al. (2008) AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J 55 : 28–39 doi:10.1111/j.1365-313X.2008.03470.x
63. ChelyshevaL, GendrotG, VezonD, DoutriauxM-P, MercierR, et al. (2007) Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet 3: e83 doi:10.1371/journal.pgen.0030083
64. Francis, LamSY, HarrisonBD, BeyAL, BerchowitzLE, et al. (2007) Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc Natl Acad Sci U S A 104 : 3913–3918 doi:10.1073/pnas.0608936104
65. CromerL, JolivetS, HorlowC, ChelyshevaL, HeymanJ, et al. (2013) Centromeric cohesion is protected twice at meiosis, by SHUGOSHINs at anaphase I and by PATRONUS at interkinesis. Curr Biol 23 : 2090–2099 doi:10.1016/j.cub.2013.08.036
66. VoinnetO, RivasS, MestreP, BaulcombeD (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33 : 949–956.
67. PerkinsDD (1949) Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics 34 : 607–626.
68. AlexanderMP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44 : 117–122.
Štítky
Genetika Reprodukčná medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



