-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
How plant NB-LRR resistance proteins and the related mammalian Nod-like receptors (NLRs) activate defense is poorly understood. Plant and animal immune receptors can function in pairs. Two Arabidopsis nuclear immune receptors, RPS4 and RRS1, confer recognition of the unrelated bacterial effectors, AvrRps4 and PopP2, and activate defense. Using delivery of PopP2 into Arabidopsis leaf cells via Pseudomonas type III secretion, we define early transcriptional changes upon RPS4/RRS1-dependent PopP2 recognition. We show an auto-active allele of RRS1, RRS1SLH1, triggers transcriptional reprogramming of defense genes that are also reprogrammed by AvrRps4 or PopP2 in an RPS4/RRS1-dependent manner. To discover genetic requirements for RRS1SLH1 auto-activation, we conducted a suppressor screen. Many suppressor of slh1 immunity (sushi) mutants that are impaired in RRS1SLH1-mediated auto-activation carry loss-of-function mutations in RPS4. This suggests that RPS4 functions as a signaling component together with or downstream of RRS1-activated immunity, in contrast to earlier hypotheses, significantly advancing our understanding of how immune receptors activate defense in plants.
Published in the journal: The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in. PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004655
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004655Summary
How plant NB-LRR resistance proteins and the related mammalian Nod-like receptors (NLRs) activate defense is poorly understood. Plant and animal immune receptors can function in pairs. Two Arabidopsis nuclear immune receptors, RPS4 and RRS1, confer recognition of the unrelated bacterial effectors, AvrRps4 and PopP2, and activate defense. Using delivery of PopP2 into Arabidopsis leaf cells via Pseudomonas type III secretion, we define early transcriptional changes upon RPS4/RRS1-dependent PopP2 recognition. We show an auto-active allele of RRS1, RRS1SLH1, triggers transcriptional reprogramming of defense genes that are also reprogrammed by AvrRps4 or PopP2 in an RPS4/RRS1-dependent manner. To discover genetic requirements for RRS1SLH1 auto-activation, we conducted a suppressor screen. Many suppressor of slh1 immunity (sushi) mutants that are impaired in RRS1SLH1-mediated auto-activation carry loss-of-function mutations in RPS4. This suggests that RPS4 functions as a signaling component together with or downstream of RRS1-activated immunity, in contrast to earlier hypotheses, significantly advancing our understanding of how immune receptors activate defense in plants.
Introduction
Plant innate immunity relies on two layers of pathogen detection. Cell surface-localized pattern recognition receptors detect pathogen-associated molecular patterns (PAMPs) of invading microorganisms and activate PAMP-triggered immunity (PTI) [1]. Successful pathogens must circumvent PTI to colonize plants, and many bacterial pathogens use type III secretion (T3S) to deliver effectors that suppress PTI into plant cells [1]. Effectors can be detected directly or indirectly by plant disease resistance (R) proteins, which then activate effector-triggered immunity (ETI) generally together with a hypersensitive response (HR) of the infected tissue [2]. Most intracellular R proteins are modular, with an amino-terminal coiled coil (CC) or Toll/interleukin-1 receptor/R protein (TIR) domain, a nucleotide binding (NB) domain and a leucine-rich repeat (LRR) domain [3]. Some NB-LRR proteins also carry an additional carboxyl-terminal extension, the function of which is unknown [3]. In addition, NB-LRR protein function generally requires an intact P-loop motif (GxxxxGKT/S) in the NB domain, presumably for ATP binding and energy-dependent conformational changes [3], [4]. Plant NB-LRR proteins and mammalian Nod-like receptors (NLRs) exhibit both structural and functional similarities [5].
Signaling following TIR-NB-LRR protein activation requires other key regulators such as Enhanced Disease Susceptibility 1 (EDS1), the EDS1-related proteins PAD4 and SAG101, and biosynthesis of the plant hormone salicylic acid (SA) for full immunity [6]. EDS1 was recently reported to interact with several NB-LRR proteins [7], [8]. Mis-regulation of R protein accumulation, localization or activation can cause constitutive defense responses, which are usually deleterious or lethal. For instance, the dwarf suppressor of npr1-1, constitutive 1 (snc1) mutant carries a point mutation between NB and LRR domains of the TIR-NB-LRR protein SNC1, which results in constitutive defense signaling [9], [10]. Suppression of the stunted snc1 phenotype in mos (modifier of snc1) mutants allowed the identification of several genes required for nuclear defense signaling [11]–[14].
Although most R proteins function to recognize a corresponding avirulent effector (Avr), some NB-LRR proteins appear to act downstream of R protein activation. The tobacco and tomato CC-NB-LRR proteins, “N-required gene 1” (NRG1), and “NB-LRR protein required for HR-associated cell death 1” (NRC1), are required for TIR-NB-LRR protein N-mediated resistance to tobacco mosaic virus and receptor-like protein Cf-4-mediated resistance to tomato leaf mold pathogen, respectively [15], [16]. Arabidopsis CC-NB-LRR Activated Disease Resistance 1 (ADR1) family proteins are required for SA-dependent ETI [17]. The Arabidopsis accession Col-0 downy mildew resistance locus RPP2 comprises two distinct closely linked NB-LRR genes RPP2A and RPP2B, both of which are required for resistance [18]. The rice Pia locus for blast (Magnaporthe) resistance comprises two divergently transcribed CC-NB-LRR genes, RGA4 and RGA5, again both required for resistance [19]. In mammals, the NLR NAIP2 confers specific recognition of PrgJ, whereas NLRs NAIP5 and NAIP6 confer responses to flagellin. However, the NLR NLRC4 is required for defense responses to both PrgJ and flagellin [20], [21]. NLRC4 association with either NAIP2 or NAIP5/6, upon provision of PrgJ or flagellin respectively, is required for defense activation [20], [21].
The T3S effectors AvrRps4 and PopP2 from Pseudomonas syringae and Ralstonia solanacearum respectively, are recognized by paired TIR-NB-LRR proteins RPS4 (resistance to P. syringae 4) and RRS1-R (resistance to R. solanacearum 1), and activate ETI in Arabidopsis [22]–[24]. RRS1-R alleles, found in accessions Ws-2, No-0 and Nd-1, confer recognition of PopP2; the RRS1-S allele of Col-0 does not recognize PopP2, but does recognize AvrRps4 [22]–[24]. Lack of AvrRps4 recognition in accession RLD is due to non-synonymous mutations in RPS4, and RRS1-S in Col-0 is truncated compared to RRS1-R because of an early stop codon [24]–[26]. RPS4 and RRS1-R genetically function together, as plants lacking RPS4, RRS1-R or both show similar enhanced susceptibility to bacterial strains expressing AvrRps4 or PopP2 [25], [26]. RRS1 (also annotated as WRKY52) is an atypical NB-LRR protein that also carries a C-terminal WRKY DNA-binding domain [22].
In this study, we delivered PopP2 using Pseudomonas T3S by fusing it with the N-terminal region of AvrRps4 (AvrRps4N). Pseudomonas-delivered AvrRps4N:PopP2 triggers RPS4 - and RRS1-dependent HR and immunity in resistant Arabidopsis genotypes when tagged with a nuclear localization signal (NLS) but not when tagged with a nuclear exclusion signal (NES). We show that the delivery of PopP2, or an inactive PopP2C321A variant, from a Pseudomonas fluorescens strain (Pf0-1) that lacks other effectors [27], results in the induction of ETI-specific genes that overlaps substantially with previously reported AvrRps4-regulated genes [28], [29].
The presence of a single amino acid (Leu) insertion in the WRKY domain of RRS1-R (RRS1SLH1 hereafter) causes the recessive lethal phenotype of the sensitive to low humidity 1 (slh1) mutant in No-0 [30]. RRS1SLH1-induced lethality is associated with enhanced defense gene expression and high SA accumulation. Similarly to other mutants displaying spontaneous cell death, slh1 mutant growth can be restored to wild type phenotype at 28°C [30]–[32]. In contrast to snc1, the slh1 mutant allele is recessive and heterozygotes show no constitutive defense activation [30]. RRS1 is also recessive and an RRS1-R/RRS1-S heterozygote is unable to recognize PopP2 [22], [23].
Here, we used the conditional RRS1SLH1-mediated lethal phenotype to gain insights into RPS4/RRS1 gene pair function. Transcriptional profiling of the slh1 mutant shows that genes induced during RRS1SLH1-mediated defense activation in the absence of Avr overlap with those induced by AvrRps4 - or PopP2-triggered immunity. Genetic screening for mutations that suppress slh1-triggered aberrant immunity reveals the critical role of RPS4 in RRS1SLH1-mediated activation of defense signaling. Transient expression of RPS4 and RRS1SLH1 in tobacco results in HR in the absence of AvrRps4 or PopP2, which can be suppressed by co-expression of wild type RRS1-R, consistent with the recessive nature of RRS1SLH1. Our study sheds new light on how paired R proteins work cooperatively and illustrates the similarities between auto-active and Avr-dependent defense signaling.
Results
PopP2 triggers RPS4 and RRS1-dependent immune responses in Arabidopsis when delivered from Pseudomonas strains
To compare AvrRps4 - or PopP2-triggered HR and immunity, we established the delivery of PopP2 via the Pseudomonas T3S. We engineered pEDV6, a Gateway-compatible version of pEDV3 [33], to carry full-length or N-terminally truncated PopP2 variants (Figures 1A and S1A–B). pEDV6 enables expression of a translational fusion between the N-terminal part of AvrRps4 (137 first amino acids; hereafter, AvrRps4N) and an effector of interest. We used a non-pathogenic Pseudomonas fluorescens Pf0-1 engineered to carry a functional T3S system (hereafter, Pf0-1(T3S)) in HR assays because unlike Pseudomonas syringae pv. tomato (Pto) DC3000, Pf0-1(T3S) does not elicit non-specific tissue collapse. When delivered from Pf0-1(T3S) or Pto DC3000, PopP21–488 (full-length) or PopP2149–488 triggered HR and immunity in Arabidopsis accession Ws-2, whereas the PopP2 variants that were further truncated did not (Figure S1C–D). Interestingly, the N-terminal 148 amino acids of PopP2 that include a nuclear localization signal (NLS) are dispensable in our assay. Based on this finding, we used the PopP2149–488 (hereafter, PopP2) variant for the rest of our experiments.
Fig. 1. PopP2 triggers RPS4/RRS1/EDS1-dependent hypersensitive response and immunity when delivered from Pseudomonas. 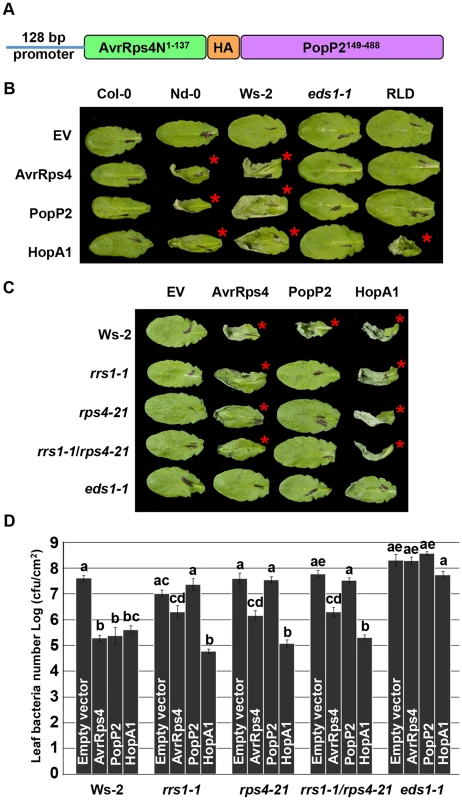
(A) AvrRps4N-PopP2 fusion construct. (B) Pseudomonas fluorescens Pf0-1(T3S)-delivered AvrRps4N:PopP2149–488 triggers an EDS1-dependent hypersensitive response (HR) in resistant Arabidopsis accessions. Five week-old Arabidopsis leaves were infiltrated with Pf0-1(T3S) strains expressing indicated avirulence proteins. Empty vector (EV) indicates AvrRps4N encoded by pEDV5 (see Figure S1). The photograph was taken at 24 hours post-infection (hpi). The red asterisks indicate the leaves showing HR. (C) Pf0-1(T3S)-delivered AvrRps4N:PopP2149–488 triggers an RPS4/RRS1-dependent HR in Ws-2 accession. (D) Pseudomonas syringae pv. tomato (Pto) DC3000-delivered AvrRps4N:PopP2149–488 triggers RPS4/RRS1-dependent immunity in accession Ws-2. Five week-old Arabidopsis leaves were infiltrated with Pto DC3000 strains and samples were taken at 4 dpi to recover bacteria from infected leaves. The results presented are the mean and standard error of the number of bacterial colonies recovered. Means labeled with the same letter are not statistically different at the 5% confidence level based on Tukey's test. To verify that Pseudomonas-delivered PopP2 confers genotype-specific avirulence, we investigated the responses of Arabidopsis natural variants to PopP2. When delivered from Pf0-1(T3S), PopP2 and AvrRps4 triggered HR in accessions Nd-0 and Ws-2 whereas Col-0 and RLD showed no symptoms at 24 hours post-infection (hpi) (Figure 1B). Col-0 RRS1-S confers HR-deficient disease resistance to Pst DC3000 delivered AvrRps4 but not to PopP2 [22], [34]. In addition, transgenic expression of Ws-2 RRS1-R in Col-0 confers strong HR in response to Pseudomonas-delivered AvrRps4 [35]. HopA1 was used as an additional control; it triggers HR in Nd-0, Ws-2 and RLD, but not in Col-0, as expected. Next, we tested if Pf0-1(T3S)-delivered PopP2 triggers RPS4 - and RRS1-dependent HR in Arabidopsis. Pf0-1(T3S)-delivered PopP2 triggered strong HR in wild type Ws-2 whereas Ws-2 rrs1-1, rps4-21, rrs1-1/rps4-21 or eds1-1 mutants did not show any response (Figure 1C). In contrast, Pf0-1(T3S)-delivered AvrRps4 triggered weak but robust HR even in the absence of RPS4 or RRS1 in Ws-2 (Figure 1C). When delivered from Pto DC3000, AvrRps4 triggered immunity in wild type Ws-2, rrs1-1, rps4-21 or rrs1-1/rps4-21 mutants because AvrRps4 recognition leads to RPS4/RRS1-dependent and -independent immunity (Figure 1D) [26]. To test if Pseudomonas-delivered PopP2 can trigger RPS4/RRS1-dependent immunity in Arabidopsis, we engineered a virulent Pto DC3000 to deliver PopP2. Pto DC3000 (PopP2) showed reduced virulence in wild type Ws-2 but not in rrs1-1, rps4-21 or rrs1-1/rps4-21 mutants compared to Pto DC3000 (pEDV5) indicating that Pseudomonas-delivered PopP2 triggers only RPS4/RRS1-dependent immunity (Figure 1D), consistent with previously reported Ralstonia-delivery assay results [26]. By contrast, HopA1-triggered immunity was not affected in rrs1-1, rps4-21 or rrs1-1/rps4-21 mutants compared with wild type Ws-2 (Figure 1D). All tested Pto DC3000 strains showed unrestricted growth in the eds1-1 mutant compared to other genotypes. Taken together, these data indicate that AvrRps4N-mediated delivery of PopP2 from Pseudomonas can trigger RPS4/RRS1-dependent HR and immunity in Arabidopsis.
We further tested if Pseudomonas-delivered PopP2 recognition requires a specific subcellular localization, as reported for AvrRps4 [8]. We engineered a PopP2149–488 variant lacking the native NLS, to carry a NLS or a nuclear export signal (NES) tag at the C-terminus. The avirulence activity of these PopP2 variants was tested in two resistant transgenic Arabidopsis lines, RLD (RPS4Ler) and Col-0 (RRS1Ws-2). Pf0-1(T3S)-delivered PopP2NES, failed to trigger HR in both transgenic lines and in wild type Ws-2, despite being expressed during plant infection, indicating that nuclear localization of PopP2 is required to trigger RPS4/RRS1-dependent HR (Figure S2A, S2E and S3). The PopP2NES variant induced a response comparable to PopP2C321A, an enzymatically inactive variant that does not trigger RPS4/RRS1-R-dependent immunity [36] in wild type Ws-2 when HR-inducing activity was quantified by ion leakage measurements (Figure S2B). We could also show that PopP2NES, in contrast to PopP2NLS, could not restrict the virulence of bacteria when delivered from Pto DC3000, nor trigger expression of defense genes when delivered from Pf0-1(T3S) (Figures S2C and S2D). As these data suggest that PopP2 triggers HR and immunity in the nucleus, we independently assessed previously reported AvrRps4 variants [8]. Unexpectedly, both AvrRps4NLS and AvrRps4NES variants triggered HR and elevated ion leakage in the Ws-2 accession when delivered from Pf0-1(T3S) (Figure S2B and S2E).
Pseudomonas-delivered PopP2 induces RRS1-R - and acetyltransferase activity-dependent transcriptional changes early after bacterial infiltration
RRS1 is a TIR-NB-LRR protein with a WRKY DNA binding domain, which belongs to Group III of the WRKY superfamily [37]. RRS1SLH1, which carries a leucine insertion near the WRKY motif, shows strongly reduced DNA binding by its WRKY domain [30]. This reduced DNA binding correlates with auto-immunity of the slh1 mutant, suggesting a critical role of RRS1 in transcriptional regulation of defense genes. Delivery of PopP2 from Pseudomonas via T3S, combined with the RPS4/RRS1-R dependence of this PopP2-triggered HR, enables direct assessment of RRS1-R-dependent transcriptional regulation. To identify PopP2-triggered and RPS4/RRS1-dependent early transcriptional responses, genome-wide expression profiling was carried out using EXPRSS, an Illumina sequencing based method [38]. Wild type Ws-2 and rrs1-1 plants were infiltrated with Pf0-1(T3S) delivering PopP2WT or PopP2C321A. The infiltrated leaf tissues were collected at 2, 4, 6 and 8 hpi for total RNA extraction, as onset of HR began at 8 hours after bacterial infiltration in an incompatible interaction (PopP2WT/Ws-2).
For differential expression analysis, PopP2WT-infiltrated Ws-2 samples were compared either to PopP2C321A mutant on Ws-2 or PopP2WT on rrs1-1. Essentially complete overlap was observed between the differentially regulated genes in the two comparisons (Figure 2A), consistent with our results showing that Pf0-1(T3S)-delivered PopP2 triggers RRS1 - and acetyltransferase activity-dependent immunity (Figures 1 and S2). In total, 719 genes were differentially expressed in an RRS1 - and acetyltransferase activity-dependent manner in at least one of the time points surveyed (Table S1). Gene ontology enrichment analysis using ATCOECIS [39] showed that most of the up-regulated genes are involved in defense, while most of the down-regulated genes are involved in photosynthesis and enriched in chloroplast-localized genes (Table S2). Interestingly, the majority of genes differentially expressed at 4 and 6 hpi were up-regulated, while many down-regulated genes were observed at 8 hpi (Figure 2A). The early (4 and 6 hpi) up-regulated genes, such as SID2, FMO1, NudT7, PBS3 and PAD4, have previously been implicated in plant defense (Table S3). Further analysis of mean expression of genes induced at 4 and 6 hpi (Table S3) showed that there was greater gene induction in Ws-2 infiltrated with PopP2WT (∼20–100 fold) than in Ws-2 infiltrated with PopP2C321A or in rrs1-1 infiltrated with PopP2WT (∼2–10 fold). For simplicity, we interpret genes induced by PopP2C321A as induced by the repertoire of PAMPs in Pf0 (thus, PTI-induced), and by PopP2WT as PTI+ETI-induced. To validate our transcriptional expression profiling results, we performed quantitative RT-PCR (qRT-PCR) verification of EDS5, NudT6, WRKY18 and WRKY40 with the cDNA used for Illumina libraries. Expression of EDS5 and NudT6 but not WRKY18 and WRKY40 was specifically regulated by ETI in our expression profiling data. In qRT-PCR experiments, PopP2 but not PopP2C321A variant delivered from Pf0-1(T3S) induced EDS5 and NudT6 in an RRS1-dependent manner, while expression of WRKY18 and WRKY40 was induced in the absence of ETI (Figure S4).
Fig. 2. Pseudomonas-delivered PopP2 induces RRS1- and acetyltransferase activity-dependent transcriptional changes early after bacterial infection. 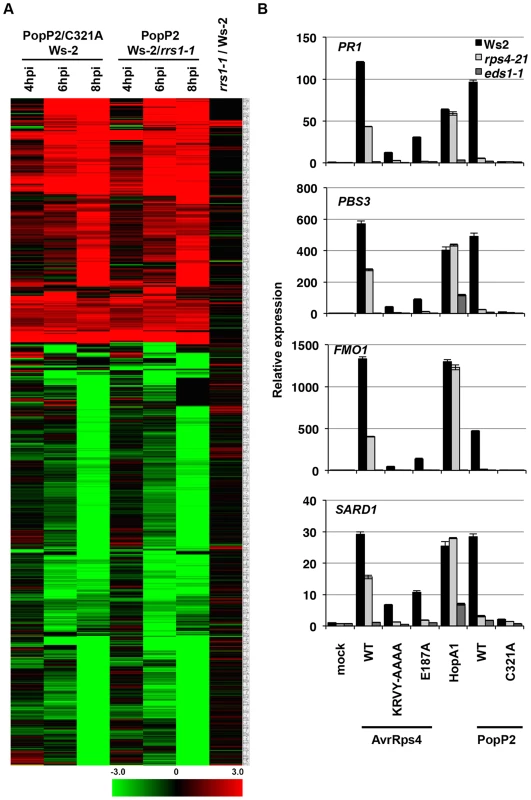
(A) Hierarchical clustering of RRS1- or PopP2-dependent gene expression. Fold-change values of 719 genes (differentially expressed at least in one time point) from all time points show the predominance of gene induction at early time points. Black, red and green colours indicate no change, up-regulated and down-regulated, respectively. C321A, an inactive PopP2 variant carrying an Alanine mutation at one of the catalytic core residues, Cysteine 321 (B) Confirmation of selected PopP2-induced genes by qRT-PCR. Five week-old plants were infiltrated with Pf0-1(T3S) expressing the indicated AvrRps4, HopA1 or PopP2 variants. Samples were taken at 8 hpi for total RNA extraction. The numbers on the Y-axis indicate fold induction compared to mock treated samples. AvrRps4 - and PopP2-dependent transcriptional changes in resistant plants have been investigated previously [28], [29], [40]. We compared these available micro-array and RNA-seq data with our results. To minimize the effects of experimental and technical differences from the AvrRps4/Ws-2 data [28], genes altered in expression at 6 hpi due to mock treatment were subtracted from the comparison; similarly, only the GMI1000/GMI1000ΔPopP2-infected Nd-1 data were used from the Hu et al. [40] study. For comparative analysis the differential expression from PTI, PTI+ETI and ETI responses were combined for data presented in this study (Table S1) and the data from Howard et al. [29]. A summary of these comparisons is presented in Figure S5 and details of genes from comparative datasets are presented in Table S4. Transcriptional changes upon AvrRps4 infection on Col-0 and Ws-2 [28], [29] considerably overlapped with PopP2-regulated genes identified both in our study and the GMI1000/GMI1000ΔPopP2 study [40] (Figure S5). The majority of early PTI+ETI-induced genes detected in our study were also found to be AvrRps4-responsive [28], [29] (Figure S5 and Table S4).
We next tested the expression of four PopP2-responsive genes PBS3, SARD1, FMO1 and PR1 by qRT-PCR in Ws-2, rps4-21 and eds1-1. At 8 hpi, Pf0-1(T3S)-delivered AvrRps4WT, HopA1 or PopP2WT triggered similar levels of induction of the four genes in Ws-2 (Figure 2B). Induction of all four genes was strictly dependent on EDS1 and abolished when non-functional variants of the effectors (AvrRps4KRVY-AAAA, AvrRps4E187A and PopP2C321A) were delivered. PTI+ETI-induction of all four genes in response to PopP2 was reduced to PTI-induced levels in both rps4-21 and in rrs1-1 mutants, confirming RPS4/RRS1-R-dependence of PopP2-induced transcriptional changes. AvrRps4-triggered induction of all four genes was reduced but not abolished in the rps4-21 mutant, likely due to RPS4-independent recognition of AvrRps4 in Ws-2 [26], [41]. These expression profiling data thus reveal the genes specifically regulated at very early stages of PopP2-triggered, RPS4/RRS1-dependent immunity in Arabidopsis. Moreover, these ETI transcriptional changes are very similar after AvrRps4 or PopP2 recognition.
Expression profiles of RPS4/RRS1-dependent responses to AvrRps4 or PopP2, and of Arabidopsis RRS1SLH1 mutant temperature shift, substantially overlap
To compare slh1 aberrant defense responses to effector-triggered RPS4/RRS1-mediated immunity, we conducted transcription profiling of the slh1 mutant over a time course after shifting plants from 28°C to 19°C, using Illumina tag sequencing [38]. A total of 1821 genes showed temperature-dependent differential expression in RRS1SLH1 after 24 hours (h) compared to wild type No-0 (Figure 3A). We confirmed the temperature-dependent regulation of 3 genes with differential induction in slh1 by qRT-PCR. PR1, PBS3 and CBP60g transcript accumulation was induced in slh1 plants between 9 and 24 h after the shift from 28°C to 19°C whereas it was unaltered in temperature-shifted No-0 plants (Figure 3B).
Fig. 3. Low temperature-dependent transcription profiling of the slh1 mutant. 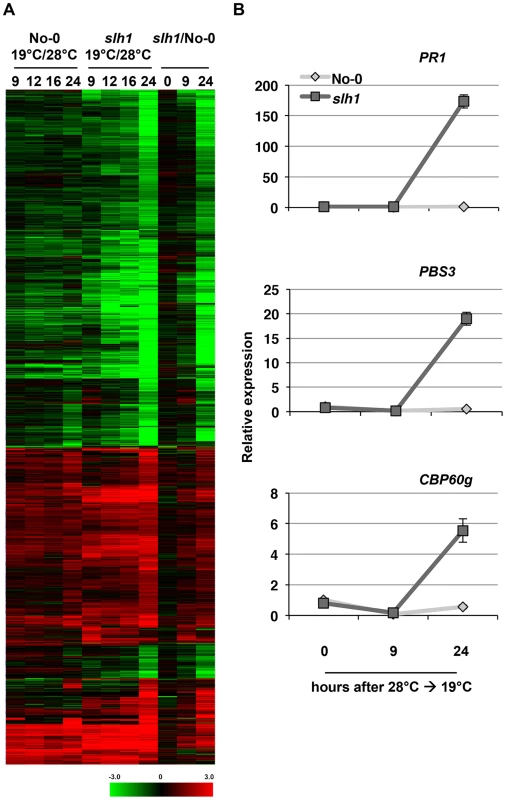
(A) Hierarchical clustering of No-0 and slh1 temperature-dependent differential gene expression. Fold-change values of 5611 genes (differentially expressed at least in one time point) are shown. The numbers on top of the heat map indicate the time (h) after temperature shift. Black, red and green colours indicate no change, up-regulated and down-regulated, respectively. (B) qRT-PCR analysis of selected RRS1SLH1-regulated genes following the temperature shift (28°C to 19°C) in 4 week-old No-0 and slh1 plants. Transcript accumulation is presented relative to No-0 before temperature shift (28°C). We compared the slh1/No-0 temperature-shift transcriptional dataset to the PopP2/RRS1-time course dataset by analyzing the pairwise overlap of genes differentially expressed in both experiments (Figure 4). Each time course response was categorized according to the mode of elicitation as PTI, ETI, temperature shift, auto-immunity, or corresponding combinations (e.g. PTI+ETI). We found that most (∼83%) of the PopP2/RRS1 ETI genes were differentially expressed in slh1 auto-immune and temperature shift responses, while up to 54% of ETI genes were also differentially expressed in the auto-immune response but not by temperature shift (Figure 4, black box). Similarly, more than 55% of auto-immune genes were also differentially expressed in PTI and PTI+ETI (Figure 4, dotted block box). Most ETI genes were also differentially expressed in PTI+ETI (more than 85%) and in PTI (up to 70%). However, less than 10% of the PTI genes were differentially expressed during ETI (Figure 4, blue box). This strongly suggests that many ETI responses involve potentiation of a subset of PTI responses, with few genes solely regulated by effector recognition. The ETI-specific genes that are regulated in PopP2 acetyltransferase activity - and RRS1-dependent manner include nucleotide/ATP-binding protein encoding genes such as NB-LRRs (Table S1).
Fig. 4. Percentage pairwise overlap of genes differentially expressed during the time course of PopP2 or PopP2C321A on Ws-2 and rrs1-1 and the time course of temperature shift on No-0 and slh1. 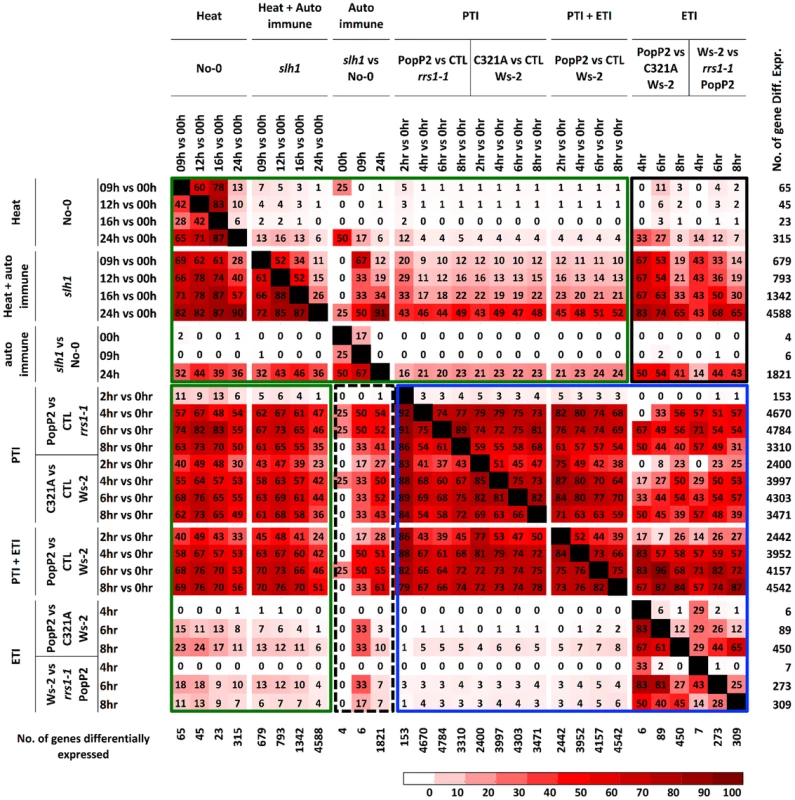
Each time course response is categorized based on underlying response (PTI, ETI, temperature shift, auto-immunity and combinations). Each cell represents percentage of genes differentially expressed from the column experiment that were also differentially expressed in the row experiment. Green boxes highlight genes regulated by heat stress and PTI, PTI+ETI responses; blue box highlights genes regulated by PTI, ETI and PTI+ETI; black boxes highlight genes regulated by auto-immunity, heat stress and by PTI, ETI and PTI+ETI. The number of gene differentially expressed in each time course is indicated on the right. PTI, PopP2C321A-regulated genes; ETI, PopP2WT- but not PopP2C321A-regulated genes; temperature shift, temperature shift-regulated genes in No-0 wild-type; auto-immunity, temperature shirt-regulated genes in slh1 mutant but not in No-0 wild-type. Similarly, we found that most temperature shift-regulated genes (up to 83%) (Table S5) were also differentially expressed by PTI or PTI+ETI, but only 25% were specifically affected by ETI, and less than 5% of the PTI-responsive genes were differentially expressed by temperature shift (Figure 4, green box). Up to 50% of PTI or PTI+ETI genes were also differentially expressed by temperature shift and auto-immune response, while about 25% of PTI or PTI+ETI genes were differentially expressed by auto-immune response (Figure 4, green box). These results indicate that PTI broadly activates genes responsive to heat, auto immunity and ETI.
These analyses indicate that slh1 auto-immunity overlaps strongly with PopP2 - and RPS4/RRS1-R-dependent ETI. Thus, RRS1SLH1-induced transcriptional reprogramming results in similar gene expression changes to those observed in AvrRps4 - or PopP2-triggered immunity, indicating that the slh1 lethal phenotype mimics RPS4/RRS1-dependent ETI at the transcriptional level.
Identification of sushi (suppressor of slh1 immunity) mutants
Lethality of slh1 at 21°C is correlated with constitutive activation of defense responses including high expression of Pathogenesis Related (PR) genes and SA accumulation [30]. We hypothesized that mutations that affect RRS1SLH1-mediated signaling components or RRS1SLH1 expression would suppress slh1 lethality. To identify genetic components required for RRS1SLH1-dependent immunity, we conducted a suppressor screen. slh1 seeds were incubated with ethyl methanesulfonate (EMS), ∼7,000 M1 plants were grown at 28°C and M2 seeds were harvested. By screening ∼500,000 M2 mutant plants at 21°C, we identified 83 families with a suppressor of slh1 immunity (sushi) mutant phenotype. Among them, 69 and 14 could rescue the slh1 lethal phenotype to a wild type-like and an improved morphology, respectively. We further analyzed the progeny of 7 selected fully rescued sushi mutants for morphological development and defense marker gene expression in the M3 generation (Figure 5). Growth of sushi mutants at 21°C was similar to wild type No-0, whereas slh1 plants did not develop beyond the first true leaf stage (Figure 5A). PR1, PBS3 and FMO1 expression was elevated in slh1 mutants grown constantly at 21°C or 24 h after shift from 28°C to 21°C, but not in fully rescued sushi mutants (Figures 5B and S6).
Fig. 5. Identification of sushi (suppressor of slh1 immunity) mutants. 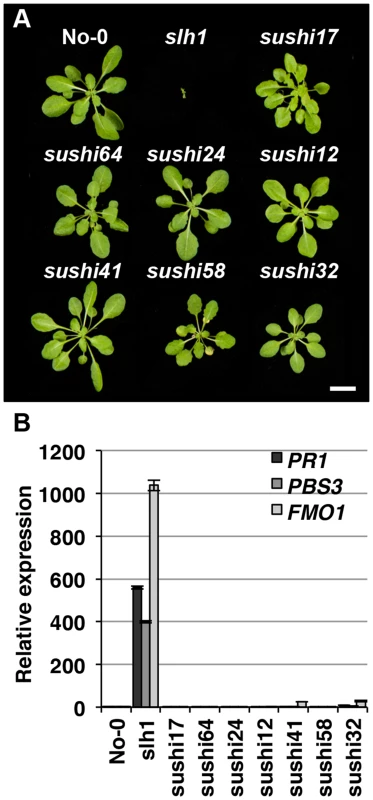
Fully rescued sushi mutant (M3), wild type No-0 and slh1 plants were grown at 21°C under short-day condition for four weeks. (A) Plant morphology. (B) qRT-PCR analysis of selected RRS1SLH1-regulated genes. Transcript accumulation is presented relative to No-0. To exclude any contamination with wild type seeds, we confirmed the presence of the slh1 mutation in 72 of the 83 M3 individual sushi mutants identified using a cleaved amplified polymorphic sequences (CAPS) marker [30]. Next, we carried out Sanger sequencing of RRS1 and RPS4 coding regions in these mutants. As expected from the complete suppression of the slh1 phenotype, we identified 6 sushi intragenic suppressor mutants that carry an early stop codon in RRS1SLH1 and 8 other non-synonymous mutations (Table S6). Surprisingly, non-synonymous mutations were also identified throughout the RPS4 coding region in 34 rescued sushi mutants (Table S6). Most of the altered amino acid residues have not previously been shown to be required for RPS4 function [24]. However, sushi52 and sushi22 harbour non-synonymous mutations at positions R28 and E88 that are important for RPS4TIR+80-triggered HR in tobacco [42], further verifying the crucial role of the RPS4 TIR domain function in RRS1SLH1-mediated defense activation.
It was previously reported that mutations in SID2/ICS1/EDS16 or SID1/EDS5 result in suppression of the RRS1SLH1 mutant phenotype [30]. We sequenced the coding region of these genes in the non-RRS1, non-RPS4 mutants, and found one sushi mutant that carried a mutation in SID2/ICS1/EDS16 (sushi70, Table S6), and no mutants that carry mutations in SID1/EDS5. Similarly to Arabidopsis accession Col-0, wild type No-0 carries two copies of EDS1. Therefore, EDS1 coding sequence was not verified in the sushi lines. The 23 remaining unassigned SUSHI mutations are now subjected to further analysis to identify new signaling components of RRS1SLH1-mediated immunity.
RPS4 is required for activation of RRS1SLH1-mediated immunity
Homo - or hemizygous, but not heterozygous, No-0 plants carrying RRS1SLH1 display a stunted phenotype at 21°C due to elevated immunity [30]. To verify that RPS4 is required for RRS1SLH1 function, we crossed 7 sushi lines carrying mutations in RPS4 (sushi17, 64, 24, 12, 41, 58 and 32) to rrs1-1 and rrs1-1 rps4-21 knockout mutants [26]. The resulting F1 individuals from both crosses were hemizygous RRS1SLH1/rrs1 for RRS1 locus (Figure S7) and either RPS4sushi/RPS4WT or RPS4sushi/rps4 at the RPS4 locus. As expected, the F1 plants derived from a cross between the sushi and rrs1-1 were stunted and showed elevated PR1 expression level (Figure 6A–C). These phenotypes were both completely suppressed in the F1 plants derived from a cross between sushi mutants in RPS4 and rrs1-1 rps4-21 double mutant. This result confirms that RPS4 is required for RRS1SLH1-mediated activation of immunity.
Fig. 6. RPS4 is required for RRS1SLH1-mediated activation of immunity. 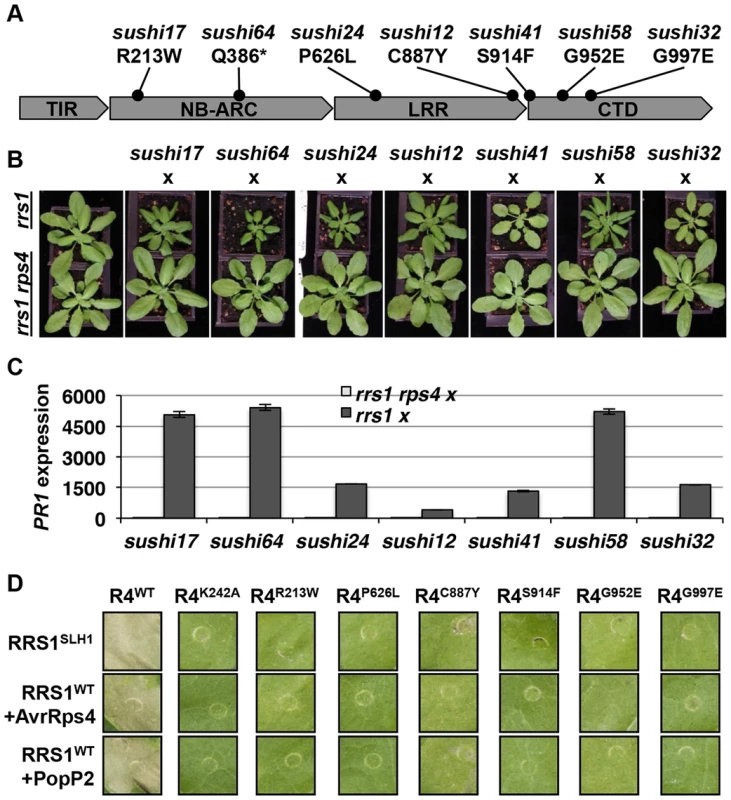
(A) Schematic presentation of SUSHI mutations in RPS4. The asterisk indicates premature stop codon. (B) The RRS1SLH1-induced growth restriction phenotype of sushi mutants is RPS4-dependent. The F1 hybrids between rrs1-1 or rrs1-1 rps4-21 and sushi were grown for five weeks at 21°C before the photograph was taken. (C) Growth restriction of F1 hybrids (shown in (B)) correlates with PR1 transcript accumulation as determined by qRT-PCR. PR1 transcript accumulation is presented relative to rrs1-1 and rrs1-1 rps4-21 respectively. (D) RPS4SUSHI variants do not confer RRS1SLH1-induced hypersensitive response or recognition of AvrRps4 or PopP2 when transiently expressed in tobacco leaf cells. Photographs were taken 3 days after agroinfiltration. To further verify the functional requirement for RPS4 in RRS1SLH1-mediated immunity, we recapitulated RRS1SLH1-mediated defense activation in Nicotiana tabacum. As shown recently [39], Agrobacterium-mediated transient co-transformation (hereafter, agroinfiltration) of RPS4-HA, RRS1-His-Flag and wild type AvrRps4-GFP or PopP2-GFP induced strong HR within 3 dpi (Figure S8A). The specificity of recognition was further verified by comparing functionally characterized mutant variants of AvrRps4 or PopP2 to wild type. As expected, AvrRps4E187A, AvrRps4KRVY-AAAA and PopP2C321A variants did not induce RPS4/RRS1-dependent HR in tobacco (Figure S8A). We have also verified that AvrRps4 and PopP2 recognition in tobacco activate defense genes orthologous to those that are regulated by RRS1 in Arabidopsis. The transcripts of the defense genes NtWRKY51 and NtNudT7 were highly up regulated when PopP2-GFP was co-expressed with RPS4-HA and RRS1-His-Flag in tobacco (Figure S8B). Agroinfiltration of GFP or PopP2C321A-GFP with RPS4-HA and RRS1-His-Flag induced significantly lower accumulation of defense gene transcripts compared to wild type PopP2 (Figure S8B). Taken together, these results further demonstrate that our transient agroinfiltration assay can also be used to investigate RPS4/RRS1 regulated immunity.
Agroinfiltration of epitope-tagged RRS1SLH1-His-Flag and RPS4WT-HA triggered HR in tobacco leaf cells, whereas RRS1SLH1 co-expressed with GFP or RPS4K242A (P-loop mutant) did not (Figures 6D and 8B). Consistent with our Arabidopsis genetic data (Figure 6B), agroinfiltration of RRS1SLH1 with each RPS4SUSHI variant did not trigger HR in tobacco (Figure 6D). Protein accumulation of the 7 tested RPS4SUSHI variants was comparable to that of RPS4WT, indicating that the lack of HR was not due to low protein expression levels (Figure S9). Moreover, as expected from our genetic analysis, RPS4SUSHI variants did not have a dominant negative effect on RPS4WT function, when both were co-expressed with RRS1SLH1 (Figure S10). We then tested whether SUSHI mutant alleles of RPS4 confer RRS1-dependent recognition of AvrRps4 or PopP2. Agroinfiltration of RRS1WT, RPS4WT and either AvrRps4 or PopP2, triggered RPS4 P-loop-dependent HR in infiltrated tobacco leaf sectors [43] (Figure 6D). Importantly, agroinfiltration of the 7 RPS4SUSHI variants did not confer responsiveness to AvrRps4 or PopP2 (Figure 6D). Taken together, these data show that RPS4 is required for RRS1SLH1-mediated and Avr-triggered/RRS1-dependent defense signaling activation. Recently, we showed the physical interaction of RRS1 and RPS4 [43]. We hypothesized that RPS4SUSHI variants may have lost their ability to interact with RRS1SLH1. However, RPS4SUSHI-HA variants and RPS4WT-HA were co-immunoprecipitated by RRS1SLH1-Flag or RRS1WT-Flag (Figure S9A-B). This result suggests that RPS4-RRS1 interaction is insufficient for signaling activation.
We identified six additional sushi mutants that carry mutations in the TIR domain of RPS4, the structure of which is known [43]. The stunted growth and elevated defense transcript accumulation of slh1 at 21°C were considerably suppressed in sushi52 (R28H), 14 (A38V), 22 (E88K), 71 (L101F), 89 (P105L) and 29 (G120R) (Figure S11). The RPS4 TIR domain structure suggests that side-chains from R28 and A38 are surface exposed, while the side-chains of the other mutated residues are buried (Figure 7A). RPS4TIR expression is sufficient to trigger HR in tobacco after agroinfiltration (Figure 7B) [42]. Therefore we introduced these six SUSHI mutations into an RPS4TIR construct (amino acids 1 to 250) to test their individual effect on RPS4 TIR domain signaling. Strikingly, all six mutations suppressed this response, suggesting that each of these residues is important for RPS4 TIR domain defense activation either through interaction with downstream partners or by maintaining the correct signalling-competent structural conformation, as the protein stability/accumulation was not significantly altered when expressed as GFP fusions in tobacco (Figure S12D). Intriguingly, when SUSHI mutations were tested in the RPS4 full-length context by co-expression in tobacco with RRS1 and the effectors, A38V and L101F did not suppress RRS1SLH1 - nor AvrRps4 - and PopP2-triggered HR (Figure 7C). This discrepancy was not due to inconsistent level of protein accumulation (Figure S12E) but might illustrate a limitation of the transient expression system in tobacco, or subtle differences between defense activation by RPS4TIR, and by the activated RPS4/RRS1 complex.
Fig. 7. Functional analysis of SUSHI mutations in the RPS4 TIR domain. 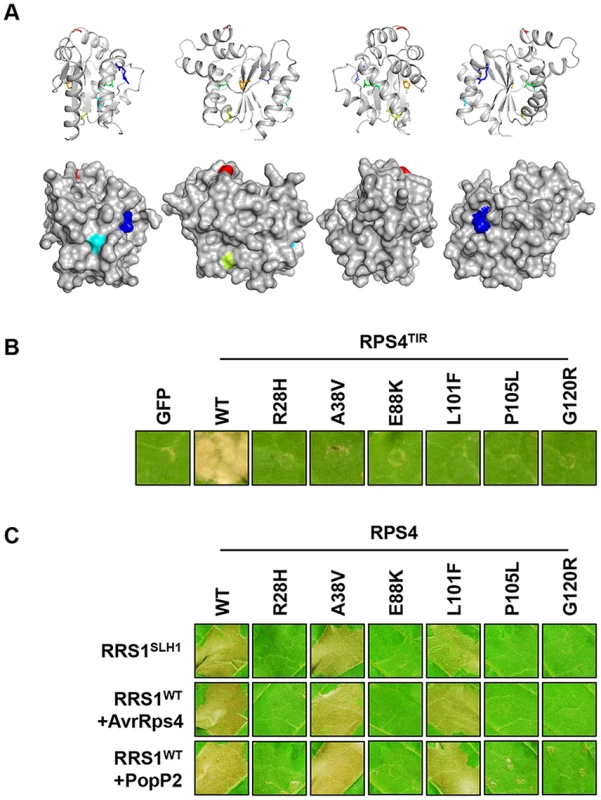
(A) SUSHI mutations within the RPS4 TIR domain structure (PDB ID 4c6r) in cartoon (top) and surface (bottom) representation (figures were generated using PyMOL (Delano Scientific)). Molecules are rotated 90° around the y-axis from left to right. Mutated residues are labelled R28 (Blue – sushi52), A38 (Teal – sushi14), E88 (Green – sushi22), L101 (Lime – sushi71), P105 (Orange – sushi89) and G120 (Red – sushi29). (B) The SUSHI mutations abolish RPS4 TIR-induced HR in tobacco agroinfiltration assay. (C) Analysis of the full-length RPS4 variants carrying SUSHI mutations in the TIR domain for recognition of AvrRps4 or PopP2 in tobacco agroinfiltration assay. The photographs were taken 3 days after agroinfiltration. RPS4 and RRS1 properties required for RRS1SLH1-mediated hypersensitive response in tobacco
As nuclear localization of RPS4 is necessary for AvrRps4-triggered immunity [41], we investigated the role of RPS4 nuclear localization in RRS1SLH1-mediated cell death. Co-expression of RRS1SLH1 with RPS4WT or RPS4NLS induced HR (Figure 8A). However, RPS4NES did not induce HR when co-expressed with RRS1SLH1, indicating the importance of RPS4 nuclear localization for RRS1SLH1 function, consistent with a previous report [41]. Nucleotide binding to the invariant Lys residue of the P-loop motif in the NB domain of R proteins is critical for conformational change and immunity activation [4], [44], [45]. Agroinfiltration of RPS4WT, but not the P-loop mutant RPS4K242A, triggered HR when co-expressed with RRS1SLH1 (Figure 8B). However, RPS4K242A does interact with RRS1SLH1 and RRS1WT (Figure S9C). Therefore, a functional RPS4 P-loop motif is required for activation of RRS1SLH1-induced defense but is not an absolute requirement for RPS4-RRS1 interaction. Surprisingly, introduction of the P-loop mutation (K185A) in the RRS1SLH1 protein sequence did not affect HR-inducing activity when co-expressed with RPS4WT (Figure 8B). Thus, P-loop motif-dependent conformational change may not be required for defense activation by RRS1SLH1, consistent with the functionality of an RRS1 P-loop mutant in AvrRps4 or PopP2 recognition [43].
Fig. 8. Characterization of RRS1SLH1-induced hypersensitive response. 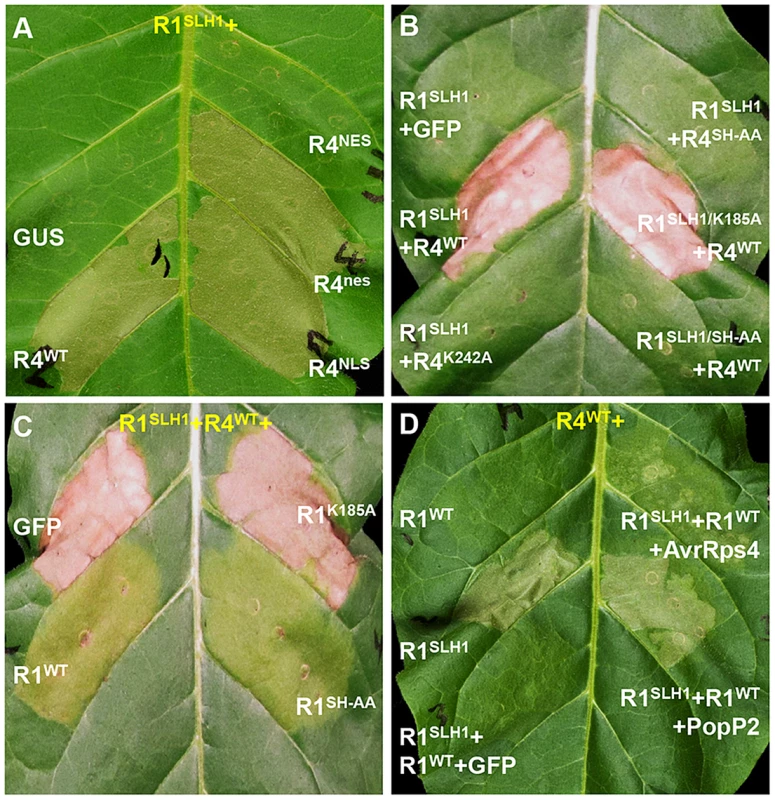
RRS1 (R1), RPS4 (R4), AvrRps4 (A4) and PopP2 (P2) were C-terminally epitope-tagged with His-Flag, HA, GFP and GFP, respectively. The photographs were taken 3 days after agroinfiltration. (A) The nuclear localization of RPS4 is required for RRS1SLH1-induced HR. NES and NLS indicate nuclear export signal and nuclear localization signal, respectively. (B) RRS1SLH1-dependent HR requires RPS4 P-loop (K242A) and TIR-TIR domain heterodimerization (SH-AA) but not RRS1 P-loop (K185A). (C) The interference of RRS1SLH1-induced HR by wild type RRS1 requires the P-loop but not the SH-motif. (D) RRS1SLH1 does not fully interfere with AvrRps4 or PopP2 recognition by wild type RRS1. Structural analysis of RPS4 and RRS1 TIR domains revealed an “SH motif” in regions that mediate heterodimerization between RPS4 (S33 and H34) and RRS1 (S25 and H26) [43]. Moreover, RPS4 or RRS1 variants carrying a mutated SH motif (SH-AA) cannot recognize AvrRps4 or PopP2 in tobacco agroinfiltration [43]. To investigate if TIR-TIR domain heterodimerization is also required for RRS1SLH1 function, SH-AA mutations were introduced in RPS4WT and RRS1SLH1 variants. Agroinfiltration of RRS1SLH1 and RPS4SH-AA, or RRS1SLH1/SH-AA and RPS4WT did not induce HR in tobacco suggesting that TIR-TIR domain heterodimerization between RRS1 and RPS4 is required for RRS1SLH1-dependent defense activation (Figure 8B). However, in the context of the full-length proteins the RRS1SLH1/SH-AA variant could still interact with RPS4WT (Figure S9C).
Co-expression of RRS1 with RPS4/RRS1SLH1 suppresses HR, consistent with the recessive nature of RRS1SLH1
RRS1SLH1-dependent lethality is recessive [30]. In agreement, agroinfiltration of RRS1WT but not of GFP interfered with HR induced by co-expression of RRS1SLH1 and RPS4WT in tobacco (Figure 8C–D). Interestingly, the RRS1K185A variant did not interfere with RRS1SLH1-induced HR whereas the RRS1SH-AA variant did (Figure 8C), indicating that nucleotide-binding function but not RPS4/RRS1 TIR-TIR domain interaction is required for RRS1-mediated interference with RRS1SLH1-induced HR. These agroinfiltration data are consistent with our transcriptomic and genetic analyses and demonstrate the striking similarity of RRS1SLH1 and Avr-triggered/RRS1-dependent defense activation.
As RRS1SLH1/RPS4-dependent constitutive HR is prevented by co-expression of RRS1WT, we tested if RRS1SLH1 interferes with RRS1WT recognition of AvrRps4 or PopP2. Interestingly, in the presence of both RRS1 variants and RPS4, AvrRps4 - or PopP2-triggered HR is still observed suggesting that RRS1SLH1 did not completely abolish RRS1WT function (Figure 8D). However, AvrRps4-triggered HR was attenuated considerably compared to PopP2-triggered HR under the same conditions (Figure 8D).
Discussion
Although both AvrRps4 and PopP2 are recognized by RPS4 and RRS1, a thorough comparison of immune responses, particularly of early transcriptional changes, has been difficult due to the distinct infection modes of the bacterial pathogens from which AvrRps4 (Pseudomonas syringae) and PopP2 (Ralstonia solanacearum) originate. Root infection of Arabidopsis plants with R. solanacearum causes wilting within 2 weeks, whereas Pseudomonas-delivered AvrRps4 triggers HR in Arabidopsis Ws-2 leaf cells within 24 hours. PopP2 delivery from Pf0-1(T3S) allowed us to compare the transcriptional reprogramming caused by recognition of AvrRps4 or PopP2 at the earliest stages and has resulted in the identification of a set of similarly regulated ETI-specific genes. It is interesting that the NLS is dispensable for the avirulence activity of PopP2 in our assays. It was shown that removal of the N-terminal NLS renders localization of PopP2 and co-expressed RRS1-S/R variants nuclear-cytoplasmic [46]. However, the significance of this PopP2 NLS-dependent relocalization of RRS1 is not known, as there have been no studies showing ETI phenotypes triggered by PopP2 lacking the NLS. As shown in Figure S2, a PopP2 variant lacking an N-terminal NLS shows similar levels of avirulence compared to wild type. Thus, PopP2 NLS-dependent relocalization of RRS1 may not be significant in PopP2-triggered immunity. Alternatively, the portion of RRS1 that is localized in the nucleus with the NLS lacking PopP2 might be sufficient to activate ETI.
It is intriguing to find that AvrRps4NES and AvrRps4NLS are comparable in their ability to elicit HR in Arabidopsis Ws-2 (Figure S2E). AvrRps4NES triggers a slightly lower ion leakage level than AvrRps4NLS (Figure S2C). We conclude that regardless of AvrRps4 contribution to defense activation in the cytoplasm, its major role is in the nucleus via interactions with the RPS4/RRS1 complex.
Pseudomonas T3S delivery of PopP2 provides a useful tool to investigate RPS4/RRS1-dependent transcriptional regulation at an early stage of ETI. In addition, by comparing non-functional variants of AvrRps4 and PopP2 to wild type proteins, we could identify the genes whose transcriptional changes were specific to Avr function. As Pf0-1(T3S) carries a mutated HopA1 gene which is unable to trigger RPS6-dependent immunity in Arabidopsis, the gene expression change in rrs1-1 infiltrated with PopP2WT or in Ws-2 infiltrated with PopP2C321A can be considered as PTI resulting from perception of the Pf0-1 PAMP repertoire. We thus report defense gene expression changes as PTI vs. PTI+ETI (Table S3). Gene ontology enrichment has shown that the majority of early up-regulated genes are involved in plant defense.
Comparative analysis with previously published microarray data shows that many PopP2-triggered early gene expression changes overlap substantially with AvrRps4-triggered transcriptional regulation [28], [29]. It is interesting to note that PopP2-regulated genes also overlap substantially with previously reported PopP2-induced genes at a later stage of infection when delivered from R. solanacearum [40]. Our discovery of early responding genes will allow us to test if they are directly regulated by RPS4/RRS1. It has been recently shown that WRKY18 and WRKY40 positively contribute towards AvrRps4-triggered immunity [47]. Consistent with this, WRKY18 and WRKY40 were highly induced at 3 and 6 hpi by AvrRps4 (Table S4). However, our experimental design enabled us to show that both WRKY18 and WRKY40 are primarily induced due to PTI (Figure S4). PTI+ETI and PTI induction of WRKY40 expression are indistinguishable. There is slightly higher PTI+ETI-induced expression of WRKY18 in response to PopP2WT in Ws-2 at later time points (6 and 8 hpi) compared to PTI elicited by PopP2C321A in Ws-2 or PopP2WT in rrs1-1 (Figure S4), but this could be due to elevated SA levels that we presume are responsible for strong PR1 induction at 8 hpi.
It is interesting to note that AvrRps4-induced regulation of ETI genes only partially requires RPS4. This is consistent with AvrRps4 recognition being conferred by both RPS4/RRS1-dependent and -independent mechanisms. Identification of an R gene(s) that confer RPS4/RRS1-independent immunity will enable comparative analysis of how AvrRps4-induced ETI genes are transcriptionally regulated by different R genes. It was remarkable to observe that AvrRps4, PopP2 and HopA1 induced common genes at early stage of defense activation, suggesting a possible EDS1-dependent conserved gene activation mechanism in ETI.
Several auto-active alleles of NB-LRR genes have been found [9], [10], [30], [48], [49], though unlike the recessive slh1, all others are dominant or semi-dominant. Plants carrying an auto-active R gene typically show temperature-dependent lethality and enhanced resistance to virulent pathogens [30]–[32]. However, in many cases the overlap between elevated disease resistance that is conferred by an auto-active R gene allele and by Avr-triggered immunity is poorly defined. Unlike most other auto-active R gene alleles, RRS1SLH1 carries a single amino acid insertion in the WRKY-DNA binding domain that reduces its DNA-binding affinity [30]. To address the role of RRS1 in transcriptional activation or repression, we tested whether RRS1SLH1-induced transcription changes overlap with AvrRps4 - or PopP2-triggered transcription changes. Based on previously reported expression profiling data and the present study, we propose that the genes whose transcripts were differentially regulated by RRS1SLH1, and by AvrRps4 and PopP2 are directly regulated by RRS1 upon Avr detection. As exons 6 and 7 of RRS1SLH1 show reduced binding to a W-box in vitro, RRS1 may act as a transcriptional repressor of plant immunity, or at least as a repressor of RPS4 function, and this repression may be relieved upon Avr perception [30]. However, RRS1 could act both as repressor and activator of defense gene transcription, as has been found for other plant transcription factors [50]. Loss of RRS1-DNA binding may be part of the activation of defense transcription, but paradoxically, rrs1 knockout lines do not show enhanced immunity.
Identification of RPS4 mutant alleles among the SUSHI mutations was unexpected, as we had anticipated that RRS1 might act downstream of RPS4 to regulate defense gene transcription directly. Notably, it would have been difficult to recover recombinants between RRS1SLH1 and an adjacent mutant allele of RPS4, so without a genetic screen, this discovery might not have been made. Based on the genetic requirement of RPS4 for RRS1SLH1-induced defense gene transcription, we now hypothesize that RPS4 is required to form a functional immune receptor complex with RRS1. This hypothesis is further supported by the fact that RPS4 and RRS1 interact with each other, in part but not solely by forming a TIR-TIR domain heterodimer [43]. In addition, the requirement of a functional P-loop motif for RPS4 but not for RRS1 function suggests that RPS4 contributes to defense activation by providing ATP-dependent conversion of a repressive immune receptor complex to an activated state. PopP2 interacts with RRS1 [46], as does AvrRps4 [43]. We hypothesize that RPS4 activates defense upon recognition of perturbations in RRS1 by effectors, and that RRS1SLH1 mimics the results of effector action upon RRS1. Can this be reconciled with the observation that a 35S:RPS4 constitutive defense phenotype partially requires RRS1 [51]? Conceivably, RRS1 might also play a chaperone-like role in facilitating conversion of RPS4 from an inactive to an active form, and RRS1SLH1 has enhanced activity in facilitating this conversion.
The TIR domain of RPS4 induces cell death when transiently overexpressed in tobacco. Several amino acid residues were shown to be required for RPS4 TIR domain auto-activity [42]. Among the 33 single amino acid polymorphisms of RPS4 that we identified in sushi mutants, two residues, R28 and E88, were previously implicated as being required for RPS4 TIR domain-induced auto-activity in tobacco. R28H and E88K mutations are unlikely to alter the overall structure of RPS4 TIR domain, judging from the crystal structure of RPS4/RRS1 TIR domain heterodimer [43]. A study on RPS4 natural variants identified Y950 as an important residue for function as a susceptible RLD allele of RPS4 carries a Y950H mutation, and a Y950H substitution in the functional Ler allele of RPS4 abolishes its AvrRps4-recognition capability [24]. Interestingly, we identified several mutations (S914F, G952E and G997E) in this C-terminal domain (CTD) of RPS4. Although the function of the RPS4 CTD remains unclear, it appears to be important for immune signaling. Conceivably, the sushi-mutated residues found in the TIR domain (R28, E88, P105L and G120R) and in the CTD (S914F, G952E, and G997E) are involved in the interaction with RRS1 or other yet unknown partner(s).
AvrRps4 and PopP2 interact directly with RRS1 [43], [46]. Conceivably, after interaction of AvrRps4 or PopP2 with RRS1, dissociation of the activated RPS4/RRS1 immune complex from target DNA induces RPS4 P-loop-dependent de-repression/activation of defense gene transcription, perhaps via WRKY18 and WRKY40 [47]. There may be multiple WRKY transcription factors that can replace the transcriptional repression function of RRS1, but not its Avr-recognition function. However, the Ws-2 RRS1SLH1 allele may make additional contributions to assembling a defense-activating complex beyond vacating W-boxes.
An intriguing feature of RRS1 is that it is the only known recessive NB-LRR-encoding R gene. Consistent with this observation, the slh1 mutation is also recessive. We were able to recapitulate this feature by transiently co-expressing RRS1 with RPS4 and RRS1SLH1 and suppressing RPS4/RRS1SLH1-triggered HR. This suppression is abolished if the RRS1-R carries a mutation in its P-loop motif. Intriguingly, this result suggests that the RRS1-R P-loop is not required for RPS4-dependent HR activation, but potentiates assembly of an inactive, poised complex. Thus, we suggest that the recessive nature of RRS1 in the Col-0(S)/Nd-0(R) or Col-0(S)/Ws-2(R) cross is the result of the Col-0 allele encoding a protein that can interfere in trans with PopP2 responsiveness and thus acts as a “poison subunit”.
There are nine TIR-NB-LRR gene pairs reported in the Arabidopsis Col-0 genome [26]. It is important to better understand how paired R proteins have evolved and recognize effectors. It is interesting to note that all three TIR-NB-LRR-WRKY encoding genes (At5g45260, At5g45050 and At4g12020) found in Arabidopsis are paired with TIR-NB-LRR genes [26]. This suggests that at least some other paired R proteins may function cooperatively in the nucleus by directly regulating transcriptional processes.
In conclusion, the deployment of a Pseudomonas T3S delivery of PopP2 allowed a detailed comparison of AvrRps4 - and PopP2-triggered RPS4 - and RRS1-dependent transcriptional regulation. We found that an auto-active allele of the TIR-NB-LRR-WRKY protein RRS1, RRS1SLH1, induces immune responses comparable to Avr-triggered immunity. The suppressor of slh1 immunity screening enabled us to uncover the critical role of RPS4 in RRS1SLH1-mediated defense activation. Furthermore, we defined additional properties of RPS4 and RRS1 that are essential for function, and these results significantly enhance our understanding of NB-LRR protein function in plants.
Materials and Methods
Plant materials and growth conditions
Arabidopsis plants were grown in short day conditions (10 h light/14 h dark) at 21°C or 28°C. Nicotiana benthamiana and Nicotiana tabacum ‘Petit Gerard’ plants were grown in long day conditions (16 h light/8 h dark) at 24°C. No-0 and slh1 are described in [30]; Ws rrs1-1 and Ws rrs1-1 rps4-21 are described in [26].
Plasmid constructions
To create pEDV6 (gateway destination variant of pEDV3), the nucleotide sequence encoding the HA epitope tag was inserted at SalI site of pEDV3 [33] that resulted in AvrRps4N(1-137aa):HA:ClaI:BamHI (pEDV5). Subsequently, pEDV5 was digested with ClaI and BamHI, treated with T4 DNA-polymerase to generate blunt ends and ligated with EcoRV digested Gateway reading frame cassette B (RFB) (Invitrogen) to create pEDV6. Construction of pBBR1MCS-5:avrRps4 was described previously [35]. The NES - or NLS-tagged avrRps4 variants were kindly provided by Jane Parker laboratory and the cloning procedure was described previously [8]. To generate pEDV6:popP2 variants, full-length or truncated popP2 fragments were amplified from Ralstonia solanacearum genomic DNA by polymerase chain reaction and cloned in the Gateway entry vector, pCR8 (Invitrogen). Introduction of popP2 fragments in pEDV6 was performed according to manufacturer's instructions by using LR recombinase II (Invitrogen). The pBin19:RPS4:HA construct was described previously [52]. To obtain C-terminally GFP tagged AvrRps4 or PopP2 variants, avrRps4 or popP2 coding regions were PCR amplified and cloned at ClaI and BamHI sites of EpiGreenB5:GFP. Construction of 35S:RRS1:His-Flag is described in [43]. Wild type and mutant variants of AvrRps4 and PopP2 were PCR amplified from previously reported plasmid constructs [35], [53]. The resulting PCR fragments were cloned in pCR8 (Invitrogen) and correct sequences were confirmed. These pCR8 constructs were used for LR reaction with the Gateway destination vector pK7FWG2 (35S promoter and C-GFP) to generate C-terminally GFP-tagged AvrRps4 and PopP2 variants. Wild type and SH-AA mutant variants of RPS4-HA and RRS1-His-Flag are described in [43]. Introduction of SLH1 and SUSHI mutations in RRS1 and RPS4, respectively, was achieved by using Quikchange II XL site-directed mutagenesis kit (Agilent). The C-terminally GFP-tagged RPS4 constructs were generated by inserting ClaI/BamHI digested RPS4 in EpiGreenB5-GFP-WT/NES/NLS.
Bacterial strains, culture conditions and manipulations
Escherichia coli DH5α was used for maintaining plasmid constructs and bacterial conjugation. For hypersensitive response assay and in planta bacterial growth assay, Pseudomonas fluorescens Pf0-1(T3S) and Pseudomonas syringae pv. tomato (Pto) DC3000 strains were used, respectively. To introduce various constructs carrying avrRps4, popP2 or hopA1 in Pf0-1(T3S) and Pto DC3000, standard triparental mating method was used by using E. coli HB101 (pRK2013) as a helper strain as previously described [33]. For transformation of Agrobacterium tumefaciens strain AGL1, standard electroporation method was used.
Plant pathology experiments
For hypersensitive response assay, freshly grown Pf0-1 (T3S) strains on King's B agar plates containing appropriate antibiotics were harvested in 10 mM MgCl2. The final concentration of bacterial suspensions was adjusted to A600 = 0.2. Leaves of five week-old Arabidopsis plants were hand-infiltrated by using 1 mL needless syringes and kept 20–24 h further for symptom development. For ion leakage assays, leaf discs were sampled at 0.5 hpi, floated on water for 30 minutes (with gentle shaking at room temperature) and transferred to fresh water (1 hpi sample). Ion leakage was measured at 24 hpi using a conductivity meter. For in planta bacterial growth assays, Pto DC3000 strains were grown and harvested as for Pf0-1(T3S). Leaves of five week-old Arabidopsis plants were hand-infiltrated with bacterial suspensions (A600 = 0.001) by using 1 mL needless syringes and kept 3–4 days further before sampling. Infected leaf samples were ground in 10 mM MgCl2, serially diluted, spotted on L agar plates containing appropriate antibiotics and kept at 28°C for 2 days before counting colonies to estimate bacterial population in infected leaves.
Agrobacterium-mediated transient transformation of Nicotiana benthamiana and Nicotiana tabacum
Agrobacterium tumefaciens AGL1 strains carrying the different constructs were grown in liquid L-medium supplemented with adequate antibiotics for 24 h. Cells were harvested by centrifugation and re-suspended at OD600 0.5 in infiltration medium (10 mM MgCl2, 10 mM MES pH 5.6). For co-expression, bacterial suspensions were mixed in 1∶1 ratio before infiltration with needleless syringes in 5 week-old N. benthamiana or N. tabacum leaves. Tobacco hypersensitive response was generally observed and photographed 2 to 3 days after infiltration.
EXPRSS library, Illumina sequencing and transcriptional profiling analysis
EXPRSS tag-seq cDNA library construction and data analysis was carried out as described previously [38]. The sequence data presented in this publication have been deposited in NCBI's Gene Expression Omnibus [54] and are accessible through GEO Series accession number GSE48247 and GSE51116. Tag to gene associations were carried out using uniquely mapped reads, with the considerations described previously [38]. Bowtie v0.12.8 [55] was used to map short reads to TAIR10 genome and Novoalign v2.08.03 (http://www.novocraft.com/) was used to align remaining reads to TAIR10 cDNA sequences. Differential gene expression analysis was performed using the R statistical language (v2.11.1) with the Bioconductor package [56], edgeR v1.6.15 [57] with the exact negative binomial test using tagwise dispersion and selected genes with false discovery rate (FDR) <0.01. From RNA-seq data for avrRps4 on Col-0 [29], uniquely mapped read counts to genes were used for reanalysis using edgeR and selected gene with FDR <0.05.
Microarray data files from Pto DC3000 (AvrRps4) infiltration (Array Express E-MEXP-546, [28]) and Interaction of Arabidopsis thaliana and Ralstonia solanacearum (NASCARRAYS-447, [40]) were used. Data analysis was performed using the R statistical language as described previously [38], [58]. Differentially expressed genes were identified using the rank products method with FDR <0.05 [59]. As Pto DC3000 (AvrRps4) data has only one replicate, differential expression analysis was carried out with untreated and MgCl2 infiltrated 3 hpi samples as controls and compared against 3 and 6 hpi of avrRps4 and 6 hpi of MgCl2. For GMI1000/GMI1000ΔPopP2 data, only Nd-1 samples were used.
Marker gene expression analysis by qRT-PCR
Total RNAs were extracted from 4 to 5 week-old Arabidopsis plants using the TRI reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was synthesized from 5 µg RNA using SuperScriptII Reverse Transcriptase (Invitrogen) and an oligo(dT) primer, according to the manufacturer's instructions. cDNA was amplified in triplicate by quantitative PCR using SYBR Green JumpStart Taq ReadyMix (Sigma) and the CFX96 Thermal Cycler (Bio-Rad). The relative expression values were determined using the comparative Ct method and Ef1α (At5g60390) as reference. Primers used for quantitative PCR are described in Table S7.
slh1 genotyping and candidate genes coding region sequencing
The presence of the slh1 mutation in sushi M3 generation and F1 individuals resulting from the genetic cross with rrs1-1 or rrs1-1 rps4-21 was assessed using the CAPS marker described in [30]. For sequencing of candidate genes on sushi mutants genomic DNA, 10, 6, 4 and 4 couples of primers respectively were used to amplify regions of RRS1, RPS4, EDS16 and EDS5 coding sequence (see Table S7). PCR products were purified on Sepharose and sequences were analyzed using the Vector NTI assembly software (Invitrogen).
Protein extraction, immunoprecipitation and immunoblotting
Protein samples were prepared from N. benthamiana 48 h after Agrobacterium-mediated transformation. One infiltrated leaf was harvested and ground in liquid nitrogen. Total proteins were extracted in GTEN buffer (10% glycerol, 100 mM Tris-HCl pH 7.5, 1 mM EDTA, 150 mM NaCl) supplemented extemporaneously with 5 mM DTT, 1% (vol/vol) plant protease inhibitor cocktail (Sigma) and 0.2% (vol/vol) Nonidet P-40. Lysates were centrifuged for 15 min at 5,000 g at 4°C and aliquots of filtered supernatants were used as input samples. Immunoprecipitations were conducted on 1.5 mL of filtered extract incubated for 2 h at 4°C under gentle agitation in presence of 20 µL anti-FLAG M2 or EZview anti-HA affinity gel (Sigma). Antibodies-coupled agarose beads were collected and washed three times in GTEN buffer, re-suspended in SDS-loading buffer and denatured 10 min at 96°C. Proteins were separated by SDS-PAGE and analyzed by immunoblotting using anti-FLAG M2-HRP, anti-GFP-HRP or anti-HA-HRP conjugated antibodies (Sigma, Santa Cruz and Roche respectively).
Supporting Information
Zdroje
1. DoddsPN, RathjenJP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11 : 539–548.
2. JonesJD, DanglJL (2006) The plant immune system. Nature 444 : 323–329.
3. BonardiV, CherkisK, NishimuraMT, DanglJL (2012) A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr Opin Immunol 24 : 41–50.
4. WilliamsSJ, SornarajP, deCourcy-IrelandE, MenzRI, KobeB, et al. (2011) An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol Plant Microbe Interact 24 : 897–906.
5. DanglJL, HorvathDM, StaskawiczBJ (2013) Pivoting the plant immune system from dissection to deployment. Science 341 : 746–751.
6. EitasTK, DanglJL (2010) NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 13 : 472–477.
7. BhattacharjeeS, HalaneMK, KimSH, GassmannW (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334 : 1405–1408.
8. HeidrichK, WirthmuellerL, TassetC, PouzetC, DeslandesL, et al. (2011) Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334 : 1401–1404.
9. LiX, ClarkeJD, ZhangY, DongX (2001) Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact 14 : 1131–1139.
10. ZhangY, GoritschnigS, DongX, LiX (2003) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15 : 2636–2646.
11. PalmaK, ZhangY, LiX (2005) An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr Biol 15 : 1129–1135.
12. WiermerM, GermainH, ChengYT, GarciaAV, ParkerJE, et al. (2010) Nucleoporin MOS7/Nup88 contributes to plant immunity and nuclear accumulation of defense regulators. Nucleus 1 : 332–336.
13. ZhangY, LiX (2005) A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell 17 : 1306–1316.
14. ZhuZ, XuF, ZhangY, ChengYT, WiermerM, et al. (2010) Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci U S A 107 : 13960–13965.
15. PeartJR, MestreP, LuR, MalcuitI, BaulcombeDC (2005) NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol 15 : 968–973.
16. GabrielsSH, VossenJH, EkengrenSK, van OoijenG, Abd-El-HaliemAM, et al. (2007) An NB-LRR protein required for HR signalling mediated by both extra - and intracellular resistance proteins. Plant J 50 : 14–28.
17. BonardiV, TangS, StallmannA, RobertsM, CherkisK, et al. (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci U S A 108 : 16463–16468.
18. SinapidouE, WilliamsK, NottL, BahktS, TorM, et al. (2004) Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J 38 : 898–909.
19. CesariS, ThilliezG, RibotC, ChalvonV, MichelC, et al. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25 : 1463–1481.
20. KofoedEM, VanceRE (2011) Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477 : 592–595.
21. ZhaoY, YangJ, ShiJ, GongYN, LuQ, et al. (2011) The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477 : 596–600.
22. DeslandesL, OlivierJ, TheulieresF, HirschJ, FengDX, et al. (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci U S A 99 : 2404–2409.
23. DeslandesL, PileurF, LiaubetL, CamutS, CanC, et al. (1998) Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen Ralstonia solanacearum. Mol Plant Microbe Interact 11 : 659–667.
24. GassmannW, HinschME, StaskawiczBJ (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 20 : 265–277.
25. BirkerD, HeidrichK, TakaharaH, NarusakaM, DeslandesL, et al. (2009) A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J 60 : 602–613.
26. NarusakaM, ShirasuK, NoutoshiY, KuboY, ShiraishiT, et al. (2009) RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J 60 : 218–226.
27. ThomasWJ, ThireaultCA, KimbrelJA, ChangJH (2009) Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J 60 : 919–928.
28. BartschM, GobbatoE, BednarekP, DebeyS, SchultzeJL, et al. (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18 : 1038–1051.
29. HowardBE, HuQ, BabaogluAC, ChandraM, BorghiM, et al. (2013) High-throughput RNA sequencing of pseudomonas-infected Arabidopsis reveals hidden transcriptome complexity and novel splice variants. PLoS One 8: e74183.
30. NoutoshiY, ItoT, SekiM, NakashitaH, YoshidaS, et al. (2005) A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J 43 : 873–888.
31. HwangCF, BhaktaAV, TruesdellGM, PudloWM, WilliamsonVM (2000) Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 12 : 1319–1329.
32. XiaoS, BrownS, PatrickE, BrearleyC, TurnerJG (2003) Enhanced Transcription of the Arabidopsis Disease Resistance Genes RPW8.1 and RPW8.2 via a Salicylic Acid–Dependent Amplification Circuit Is Required for Hypersensitive Cell Death. Plant Cell 15 : 33–45.
33. SohnKH, LeiR, NemriA, JonesJD (2007) The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 19 : 4077–4090.
34. GassmannW (2005) Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol Plant Microbe Interact 18 : 1054–1060.
35. SohnKH, HughesRK, PiquerezSJ, JonesJD, BanfieldMJ (2012) Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proc Natl Acad Sci U S A 109 : 16371–16376.
36. TassetC, BernouxM, JauneauA, PouzetC, BriereC, et al. (2010) Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog 6: e1001202.
37. EulgemT, RushtonPJ, RobatzekS, SomssichIE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 : 199–206.
38. RallapalliG, KemenEM, Robert-SeilaniantzA, SegonzacC, EtheringtonG, et al. (2014) EXPRSS: an Illumina based high-throughput expression-profiling method to reveal transcriptional dynamics. BMC Genomics 15 : 341.
39. VandepoeleK, QuimbayaM, CasneufT, De VeylderL, Van de PeerY (2009) Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol 150 : 535–546.
40. HuJ, BarletX, DeslandesL, HirschJ, FengDX, et al. (2008) Transcriptional responses of Arabidopsis thaliana during wilt disease caused by the soil-borne phytopathogenic bacterium, Ralstonia solanacearum. PLoS One 3: e2589.
41. WirthmuellerL, ZhangY, JonesJD, ParkerJE (2007) Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol 17 : 2023–2029.
42. SwiderskiMR, BirkerD, JonesJD (2009) The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact 22 : 157–165.
43. WilliamsSJ, SohnKH, WanL, BernouxM, SarrisPF, et al. (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344 : 299–303.
44. TamelingWI, VossenJH, AlbrechtM, LengauerT, BerdenJA, et al. (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol 140 : 1233–1245.
45. UedaH, YamaguchiY, SanoH (2006) Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol Biol 61 : 31–45.
46. DeslandesL, OlivierJ, PeetersN, FengDX, KhounlothamM, et al. (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A 100 : 8024–8029.
47. SchonM, TollerA, DiezelC, RothC, WestphalL, et al. (2013) Analyses of wrky18 wrky40 plants reveal critical roles of SA/EDS1 signaling and indole-glucosinolate biosynthesis for Golovinomyces orontii resistance and a loss-of resistance towards Pseudomonas syringae pv. tomato AvrRPS4. Mol Plant Microbe Interact 26 : 758–767.
48. ShiranoY, KachrooP, ShahJ, KlessigDF (2002) A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14 : 3149–3162.
49. YangH, ShiY, LiuJ, GuoL, ZhangX, et al. (2010) A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis. Plant J 63 : 283–296.
50. IkedaM, MitsudaN, Ohme-TakagiM (2009) Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21 : 3493–3505.
51. HeidrichK, TsudaK, Blanvillain-BaufumeS, WirthmuellerL, BautorJ, et al. (2013) Arabidopsis TNL-WRKY domain receptor RRS1 contributes to temperature-conditioned RPS4 auto-immunity. Front Plant Sci 4 : 403.
52. ZhangY, DoreyS, SwiderskiM, JonesJD (2004) Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J 40 : 213–224.
53. SohnKH, ZhangY, JonesJD (2009) The Pseudomonas syringae effector protein, AvrRPS4, requires in planta processing and the KRVY domain to function. Plant J 57 : 1079–1091.
54. EdgarR, DomrachevM, LashAE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 : 207–210.
55. LangmeadB, TrapnellC, PopM, SalzbergSL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25.
56. GentlemanRC, CareyVJ, BatesDM, BolstadB, DettlingM, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80.
57. RobinsonMD, McCarthyDJ, SmythGK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 : 139–140.
58. Smyth GK (2005) Limma: Linear models for microarray data. In: VC RG, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer. pp. 397–420.
59. BreitlingR, ArmengaudP, AmtmannA, HerzykP (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573 : 83–92.
Štítky
Genetika Reprodukčná medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



