-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
Most cellular components of neurons are synthesized in the cell body and must be transported great distances to form synapses at the ends of axons and dendrites. Neurons use a specialized axonal transport system consisting of microtubule cytoskeletal tracks and numerous molecular motors to shuttle specific cargo to specific destinations in the cell. Disruption of this transport system has severe consequences to human health. Disruption of specific neuronal motors are linked to hereditary neurodegenerative conditions including forms of Charcot Marie Tooth disease, several types of hereditary spastic paraplegia, and certain forms of amyotrophic lateral sclerosis motor neuron disease. Despite recent progress in defining the cargo of many of kinesin family motors in neurons, little is known about how the activity of these transport systems is regulated. Here, using a simple invertebrate model we identify and characterize a novel protein that regulates the efficacy of the KIF1A motor that mediates transport of synaptic vesicles. These studies define a new pathway regulating SV transport with potential links to human neurological disease.
Published in the journal: The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport. PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004644
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004644Summary
Most cellular components of neurons are synthesized in the cell body and must be transported great distances to form synapses at the ends of axons and dendrites. Neurons use a specialized axonal transport system consisting of microtubule cytoskeletal tracks and numerous molecular motors to shuttle specific cargo to specific destinations in the cell. Disruption of this transport system has severe consequences to human health. Disruption of specific neuronal motors are linked to hereditary neurodegenerative conditions including forms of Charcot Marie Tooth disease, several types of hereditary spastic paraplegia, and certain forms of amyotrophic lateral sclerosis motor neuron disease. Despite recent progress in defining the cargo of many of kinesin family motors in neurons, little is known about how the activity of these transport systems is regulated. Here, using a simple invertebrate model we identify and characterize a novel protein that regulates the efficacy of the KIF1A motor that mediates transport of synaptic vesicles. These studies define a new pathway regulating SV transport with potential links to human neurological disease.
Introduction
Neurons innervate their targets at synapses distant from the soma. Most components of these synaptic specializations, including synaptic vesicles (SVs), active zone proteins and mitochondria, are synthesized in the soma and then transported along axons on the microtubule cytoskeleton [1]. Transport along the axon is bidirectional with anterograde transport driven largely by kinesins and retrograde transport carried out by cytoplasmic dynein [2]. Efficient axonal transport is important in many facets of neuronal development and function. Trophic factors, membrane components, guidance receptors as well as synaptic components are all transported down the axon anterogradely, and maintenance of trophic support requires retrograde transport of signaling endosomes containing activated receptors [2]. Abnormal axonal trafficking has been observed in brain disorders including Parkinson's disease, amyotrophic lateral sclerosis, Charcot-Marie-Tooth disease and hereditary spastic paraplegia [3], [4], [5], [6].
The majority of anterograde transport is performed by a large family of plus-end directed motors of the kinesin superfamily (KIFs) consisting of 21 genes in C. elegans [7] and 45 genes in mouse [8]. KIFs are composed of three domains: a motor “head” domain, a stalk domain and a cargo-binding domain. In plus end directed kinesins, the globular ATPase motor domain is positioned in the N-terminal region of the protein and provides the force to walk processively on microtubules at mean velocities of around 0.5–1.5 µm/second [9]. The C-terminal cargo-binding domain is typically separated from the motor by a long coiled coil stalk or “neck” domain, though the size of this domain varies considerably within the family. By contrast with the highly conserved motor domain, the cargo binding domains are variable and determine the cargo specificity of KIFs. Accessory light chain subunits and distinct adaptor proteins provide additional diversity of cargo binding to KIFs [9]. For example, KIF5 binds APP containing vesicles via its light chain [10], mitochondria via the adaptor Milton [11] and GlrR2 containing vesicles via the adaptor GRIP [12]. Although the cargo binding specificity of numerous kinesins has been defined to some extent, the mechanisms regulating many aspects of kinesin-mediated cargo transport remain largely uncharacterized.
One general theme in the mechanisms controlling axonal transport is the regulation of KIF motor activity. The activity of the motor domain of several different KIFs, including KIF1A/UNC-104, is negatively regulated by their cargo-binding domain in the absence of cargo [13], [14], [15], [16], [17], [18]. In addition, activation of the motor in several cases has been documented to require the binding of other factors. For example, the cargo adaptor JIP1 is not sufficient to activate Kinesin-1, but rather requires the additional cooperative binding of the protein FEZ1 [19]. A RAN-GTPase binding protein has been shown to activate Kinesin-1 ATP activity in vitro [20]. Phosphorylation has also been demonstrated in several cases to regulate cargo binding. For example, CaMKII regulates KIF17 binding to cargo by phosphorylation and GSK3β phosphorylation regulates KIF5 [21], [22]. In addition, the microtubule associate protein (MAP) doublecortin was recently demonstrated to regulate SV transport by enhancing KIF1A motor domain binding to MTs [23]. In summary, regulation of KIF motor activity is complex.
One of the identified KIF1A/UNC-104 regulators is Liprin-α/SYD-2. Liprin-α/SYD-2 belongs to a family of proteins that interact with the cytosolic domain of LAR receptor protein tyrosine phosphatases [24], [25]. In addition to interacting with LAR, Liprin-α interacts with several presynaptic active zone proteins to regulate active zone development [26], [27], . Interestingly, biochemical studies also identified interactions of Liprin-α with KIF1A [32]. In vivo, Liprin-α/SYD-2 is required for SV trafficking in Drosophila [33] and regulates UNC-104 motility in C. elegans [34]. These observations demonstrate that, in addition to the intra-molecular regulation of KIF1A/UNC-104, its activities are also regulated by other factors.
Here, using the C. elegans mechanosensory system as an in vivo model, we identify SAM-4 (Synaptic vesicle tag Abnormal in Mechanosensory neurons) as a novel regulator of KIF1A/UNC-104 directed SV trafficking. sam-4, encodes a conserved SV-associated protein orthologous to human LOH12CR1 [35] that is broadly expressed in neuronal tissue. SAM-4 acts in a cell autonomous manner by binding to SVs to regulate the processivity of anterograde SV transport. sam-4 null mutants show SV trafficking defects in different neuronal cell types. Genetic analyses revealed that SAM-4 acts synergistically with the KIF1A/UNC-104 PH cargo binding domain, but not the motor domain, to regulate SV trafficking and locomotory behavior. Gain-of-function mutations in the unc-104 motor domain suppress sam-4 defects indicating that SAM-4 functions upstream of the motor in regulating SV transport. SYD-2, which regulates SV trafficking to a lesser extent than SAM-4, exhibits similar genetic interactions with UNC-104 but no obvious interactions with SAM-4, consistent with SYD-2 and SAM-4 acting in the same pathway. Imaging of SV cargo movements in vivo demonstrated that SAM-4 is required to maintain cargo processivity rather than motor velocity, while gain-of-function UNC-104 proteins increase cargo processivity. We propose a model in which SV-bound SAM-4 acts in parallel to the UNC-104/KIF1A cargo binding domain to regulate activity of the motor domain.
Results
sam-4 mutants accumulate SVs in the soma and proximal neurite
The response to gentle touch to the body in C. elegans is mediated by a set of six touch receptor neurons (TRNs: ALML/R, AVM, PLML/R, and PVM) (Figure S1A and [36]). We use PLM neurons as a simple in vivo system to examine axonal transport of synaptic components. The two PLM soma are located on each side of the body in the tail ganglia (Figure S1A). Each PLM extends a short posterior-directed and a long anterior-directed neurite, which are easy to image because they are in close apposition to the cuticle. PLMs innervate partners via gap junctions and chemical synapses [37]. The chemical synapses are formed in a large varicosity (∼5 µm long), located at the end of single collateral synaptic branch that extends ventrally from the anterior directed process into the ventral nerve cord, usually just posterior to the vulva (Figure S1A). We examined PLM neurons in vivo by expressing markers using the mec-7 promoter which drives gene transcription selectively in TRNs [38]. SVs preferentially accumulate in the PLM synaptic varicosities as observed using transgenic SV markers SNB-1-GFP [39] and GFP-RAB-3 (jsIs821, Figure 1A and 1C), similar to SV accumulations revealed at the ultrastructural level [37], [40]. When anterograde SV trafficking machinery is disrupted by lesioning the UNC-104/KIF1A motor, SV markers accumulate in the soma and proximal portions of PLM neurites rather than being transported to the synapse (Figure S1B–D′). By contrast, when the retrograde cytoplasmic dynein motor is disrupted, SV markers accumulate abnormally at the distal portions of the anterior process [41]. These observations indicate that homeostatic regulation of SV levels in mechanosensory neuron synaptic varicosities is mediated by the balance of the anterograde and retrograde transport systems.
Fig. 1. sam-4(js415) mutations cause abnormal accumulations of SV markers in PLM neurons. 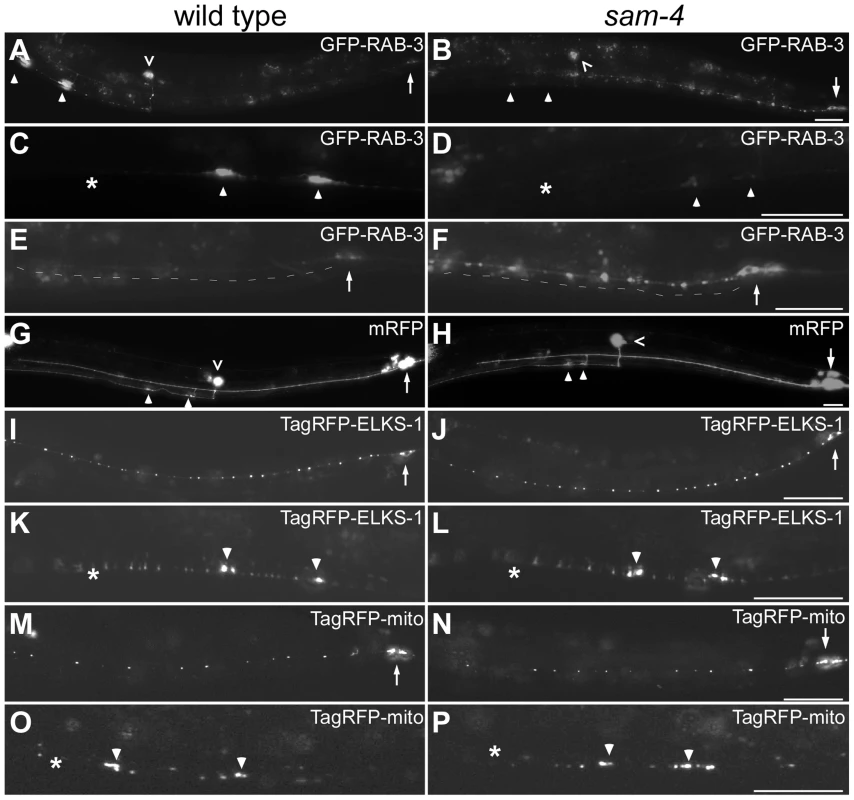
(A–H) Distribution of synaptic vesicle marker GFP-RAB-3 (jsIs821, A–F) accumulations in PLMs labeled by the cytosolic mRFP (jsIs973, G and H) in wild type animals (A, C, E, and G) and sam-4 mutants (B, D, F, and H). Shown are PLM neurons (A, B, G and H), their synaptic varicosities (C, D) and proximal neurites (E, F). (I–L) Distribution of active zone marker TagRFP-ELKS-1 (jsIs1075) accumulations in PLM neurites (I, J) and synaptic varicosities (K, L). (M–P) Distribution of mitochondria (labeled by Tag-RFP-mito, jsIs1073) in PLM neurites (M, N) and synaptic varicosities (O, P). Arrow: PLM soma; arrowhead: PLM synaptic varicosity; asterisk: vulva; caret: PVM soma. Scale bar: 20 µm. The sam-4(js415) mutant was isolated in a forward genetic screen for mutations disrupting SV accumulation in PLM synapses, using a SNB-1-GFP transgenic marker [42]. Similar defects were observed when SV localization was analyzed using GFP-RAB-3 (Figure 1A–F). In sam-4(js415), GFP-RAB-3 fluorescence was greatly reduced in PLM synaptic varicosities (Figure 1B and D) and increased both in the soma and the process proximal to the soma where the accumulations were largely punctate (Figure 1B and F). We also found that the accumulation phenotype in PLM neurons is temperature sensitive: mutants raised at 25°C exhibit more severe defects than those raised at 15°C (Figure S2). In addition to the abnormal SV marker accumulations in PLM neurons, similar defects were also observed in other neurons including SAB neurons and ventral nerve cord neurons when using either SNB-1-GFP or GFP-RAB-3 SV markers (Figure S3A–G) [39], [43]. Thus, SAM-4 appears to be essential for efficient transport of SVs in different types of neurons.
The altered distribution of GFP-RAB3 that we observe in sam-4 mutants could be explained by the disruption of neuronal morphology and/or the microtubule cytoskeleton. To test if the reduced levels of SV markers in sam-4 PLM synaptic varicosities are caused by PLM anatomical defects, we examined PLM neurites using a cytosolic fluorescent marker mRFP (jsIs973) and found no obvious morphological changes: PLM neurites extend normally, terminate properly in the mid-body, form the synaptic branches at the appropriate location and form synaptic varicosities in the ventral nerve cord (Figure 1G and H). Since microtubules function as a common track for the anterograde transport of many synaptic components including SVs, mitochondria and active zone proteins [1], we asked if sam-4 mutations cause microtubule cytoskeleton disruptions by examining the localization of synaptic components other than SVs. We found that the distribution of active zone proteins (mec-7p::tagRFP-ELKS-1, jsIs1075; Figure 1I–L) and mitochondria (mec-7p::tagRFP-mito, jsIs1073) (Figure 1M–P) is grossly normal in sam-4 mutants, indicating that transport of other synaptic components is largely intact. Thus, the microtubule cytoskeleton remains competent for axonal transport.
To evaluate systemic effects of sam-4 mutations, we next examined locomotion behavior which has been associated with SV trafficking defects [44]. Surprisingly, sam-4 null (see below) mutants exhibit only mild defects in the velocity of stimulated locomotion and show little, if any, defects in posture or the trace of sinusoidal locomotion tracks (Figure S4). We also examined other behaviors of sam-4 mutants and detected no defects in mechanosensation, egg-laying, or growth rates. Furthermore, sam-4 males remain competent to mate. These observations suggest that sam-4 may encode a specialized neuronal component that promotes efficient SV transport, without being essential for the process.
SAM-4 encodes a conserved protein expressed broadly in the nervous system
Positional cloning and transgenic rescue identified sam-4 as the C. elegans gene F59E12.11 (Figure S5 and Materials and Methods for details), which encodes an evolutionarily conserved 240 amino acid protein (Figure S5) with no identifiable domain structure. An additional open reading frame (ORF) was identified in the 5′ UTR of the sam-4 mRNA, but these sequences are not required for functions we describe for sam-4 herein (see discussion for details). SAM-4 is the C. elegans ortholog of human LOH12CR1 that was identified as a candidate tumor suppressor based on frequent deletion of this region of human chromosome 12 in acute lymphoblastic leukemia [35]. To confirm the sam-4 gene identification, we expressed a 3X-FLAG-tagged derivative of the SAM-4 protein (Figure S5C) under its native promoter using a MosSCI strategy [45], [46]. The single copy sam-4-3XFlag transgene completely rescued the Sam phenotypes of sam-4 mutants (Figure 2E). Immunohistochemical analysis of the transgene revealed that SAM-4 is localized primarily to the nerve ring region of the head (Figure S6A), indicating that sam-4 is broadly expressed in the nervous system.
Fig. 2. sam-4 encodes a neuronally expressed protein SV associated protein. 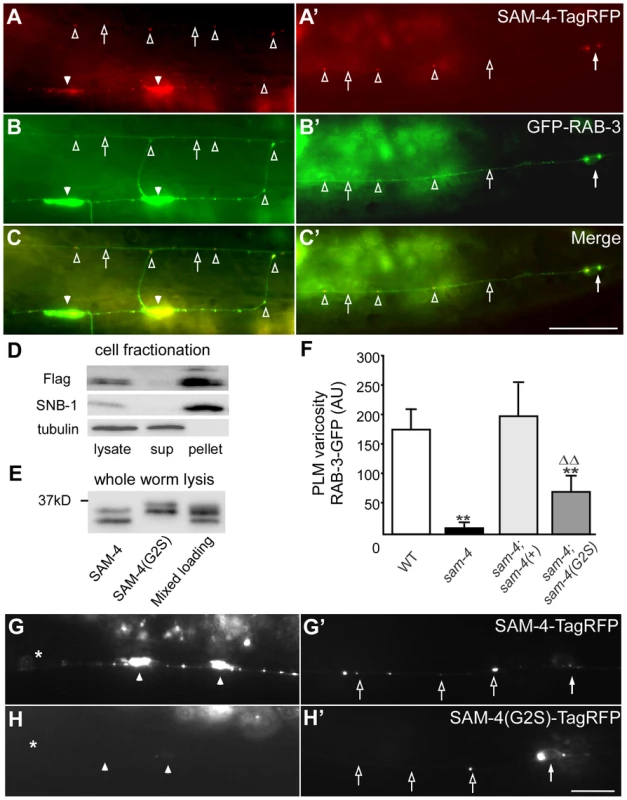
(A–C′) Distribution of SAM-4-TagRFP expressed in mechanosensory neurons in the synaptic varicosities (A) and the PLM soma and proximal section of the PLM neurite (C). GFP-RAB-3 localization is also shown in (B and B′) along with the merge (C and C′). (D–E) Western blots of fractionated whole worm extracts (D) and non-fractionated lysis (E) derived from animals expressing 3XFlag tagged SAM-4(+) and SAM-4(G2S) myristoylation mutant proteins probed for SAM-4, the synaptic vesicle protein SNB-1, and the cytosolic protein β-Tubulin. Note the shift in migration of the SAM-4(G2S) mutant. (F) Quantification of GFP-RAB-3 levels in PLM synaptic varicosities. **, P<0.001, relative to wild type; ΔΔ, P<0.001, relative to sam-4. Transgenes tested: jsIs1188 for sam-4(+), jsIs1265 for sam-4(G2S). Allele tested: sam-4(js415). (G–H′) SAM-4(G2S)-TagRFP (jsEx1256) levels are reduced in PLM synaptic varicosities (G and H) but increased in PLM soma (G′ and H′), relative to SAM-4-TagRFP (jsIs1156). Solid arrow: PLM soma; open arrow: PLM neurite; solid arrowhead: PLM synaptic varicosity; open arrowhead: florescence marker puncta in PLM neurite; asterisk: vulva. Scale bars: 10 µm. The sam-4 mutations we characterized are recessive and likely represent null alleles of sam-4. The js415 allele isolated in our screen introduces a CAA>TAA nonsense lesion at Gln104 (Figure S5A and S5B). tm3828, another sam-4 allele isolated by the Japanese National Bioresource Project, deletes 149 bp of sam-4. This deletion removes exon sequences coding for amino acids from Leu66 to Ala100 and results in a frame-shift (Figure S5A and S5B). tm3828 and js415 exhibit indistinguishable GFP-RAB-3 mis-accumulation phenotypes (Figure S2). Since both mutations result in severe disruption of coding potential of sam-4 and have similar phenotypes, we conclude that both alleles represent null mutations.
SAM-4 is an SV associated protein
To characterize the role of SAM-4 protein in regulating SV transport, we assayed its function when expressed in different cell types (Figure S5C). We found that sam-4 expression in PLM neurons driven by the mec-7 promoter rescued the SV accumulation defects in PLMs (Figure S5D) while its expression in PLM postsynaptic partners driven by the glr-1 promoter did not. These data suggest that SAM-4 functions cell-autonomously to regulate SV transport.
We next used a functional sam-4-TagRFP transgene expressed in PLMs to further examine the sub-cellular localization of SAM-4. We observed that SAM-4 preferentially accumulates in the synaptic varicosities of PLMs and small quantities of SAM-4 accumulate as puncta in the neurites (Figure 2A and 2A′), a pattern similar to the GFP-RAB-3 marker (Figure 2B and 2B′). Further examination demonstrated that these SAM-4 particles co-localize well with the RAB-3 labeled SV particles (Figure 2C and 2C′) and furthermore the RAB-3 and SAM-4 particles move together (Figure S6B, Movie 1–3). In addition, SAM-4-TagRFP is retained in the cell body in unc-104 mutants as previously demonstrated for many other SV proteins including RAB-3 [39], [47]. These observations suggest that SAM-4 may function as a component of the SV trafficking machinery.
To determine if SAM-4 is a SV component, we examined SAM-4 subcellular localization using cell fractionation analysis. We lysed sam-4-3XFlag transgenic animals under detergent free conditions, cleared the lysate of large membrane organelles, cytoskeleton, and cell debris using a 15K g spin, then fractionated the extract into a membrane containing 150K g pellet and a cytosolic fraction. We observed that, like the SV protein synaptobrevin SNB-1, SAM-4 was present in the SV membrane-containing pellet but was absent from the cytosolic fraction (Figure 2D), indicating that SAM-4 is likely associated with SVs. Bioinformatic analysis predicts that SAM-4 contains a conserved myristoylation site at its amino terminus (Figure S5B). We then tested if SAM-4 localizes to membranes through the myristoylation signal. Myristoylated proteins typically migrate faster than their non-myristoylated counterparts [48]. We observed a decrease in mobility of SAM-4(G2S)-3XFLAG tagged protein compared to the SAM-4-3XFLAG control expressed at endogenous levels consistent with the hypothesis that this mutation disrupts SAM-4 myristoylation in vivo (Figure 2E). However, fractionation of SAM-4 to the membrane compartment was not altered by the SAM-4 (G2S) lesion suggesting that SAM-4 associates with membranes independently of myristoylation (Figure S6C). In fractionation experiments when EDTA and EGTA were omitted from the buffer, we also observed FLAG immunoreactive band 4 kD smaller than the full length SAM-4 which fractionated partially into the cytosol (Figure S6D) suggesting the SAM-4 N-terminus contains a site mediating interactions with an unidentified SV membrane component.
We further examined functional activity of the sam-4(G2S) myristoylation mutant and found that the endogenous expression level of sam-4(G2S) (jsIs1265) only partially rescue sam-4(null) mutants (Figure 2F). In addition, we observed that the SAM-4 (G2S) protein is not efficiently delivered to synapses and is largely retained in the soma (Figure 2G–H′). Taken together, these results argue that SAM-4 functions as a SV component to regulate SV trafficking.
SAM-4 functions as a component of the UNC-104 mediated SV transport machinery
Axonal transport of SVs in synapses is mediated by anterograde transport (largely the KIF1A motor system) and retrograde transport (the dynein motor system). To understand mechanisms by which SAM-4 regulates SV trafficking, we examined genetic interactions of sam-4 with mutations in both unc-104 and the dynein heavy chain gene dhc-1. Hypomorphic mutations were used because null mutations in both genes are lethal and because point mutations in different domains of UNC-104 are available for analysis. We first tested if SAM-4 is involved in regulating the UNC-104 transport machinery by examining sam-4 unc-104 interactions. We previously isolated a hypomorphic unc-104 loss-of-function (lf) mutant, js901, with a G1466V lesion in the cargo binding PH domain of UNC-104 that displays very similar phenotypes to sam-4 (materials and methods for details). These mutants show decreased GFP-RAB-3 levels in PLM synaptic varicosities and increased accumulations in the proximal portion of PLM neurites (Figure 3A–3C′). Furthermore, they displayed mild locomotion defects (Figure 4A, 4C and 4I) while remaining grossly normal in PLM neurite morphology, growth rate and egg-laying behavior. js901 males remained competent to mate. Overall, the phenotypic defects of unc-104(js901) are mild compared to other unc-104 alleles such as the e1265 PH domain and the rh43 motor domain lesions which have severe locomotory defects and slow growth rates. If SAM-4 interacts with UNC-104 to regulate SV transport, we reasoned that sam-4 mutations would exaggerate the mild js901 defects. Indeed, we observed that SV soma accumulations are further increased in the sam-4 unc-104(js901) double mutant relative to either single mutant (Figure 3A–3D′). Additionally, we found that sam-4 unc-104(js901) double mutants exhibit very severe locomotion defects relative to either single mutant, exhibiting defects comparable to severe unc-104 mutants (Figure 4A–4D, 4I). These results suggest that SAM-4 functions in concert with the UNC-104 protein to regulate the SV trafficking.
Fig. 3. sam-4 and syd-2 interact with unc-104 in transporting SVs. 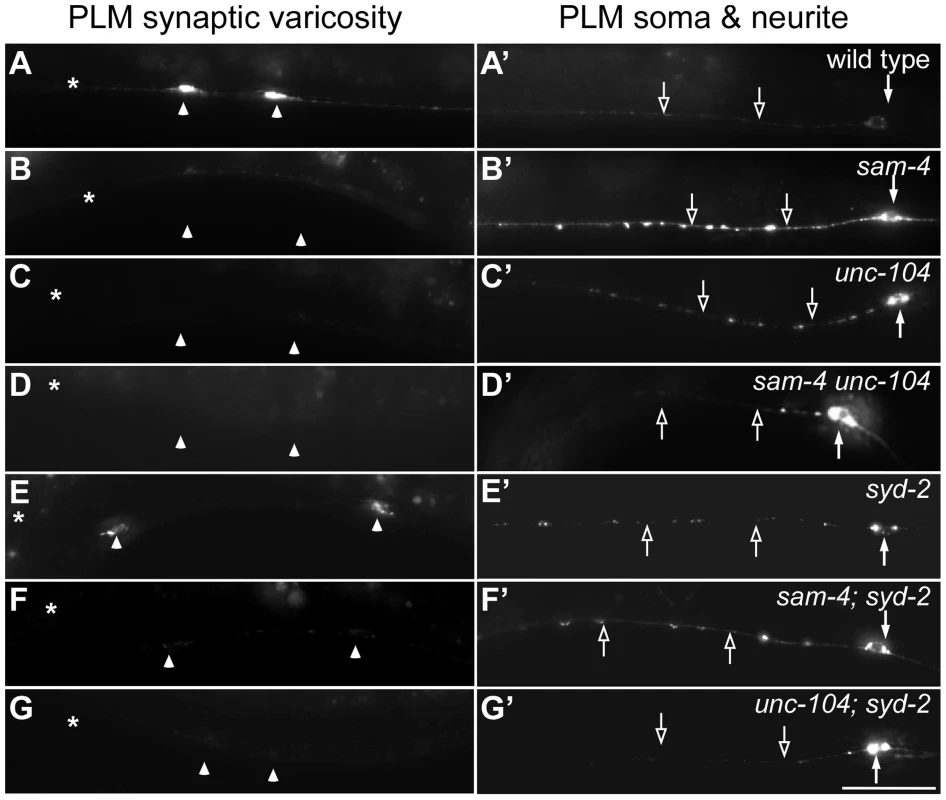
GFP-RAB-3 (jsIs821) distribution in PLM neurons of wild type and mutant animals as indicated. A–G panels are focused on the PLM synaptic varicosities and A′–G′ panels are focused on the PLM soma and the proximal portion of the neurite. Alleles tested: sam-4(js415), unc-104(js901) and syd-2(ok217). Arrowheads: synaptic varicosities; solid arrows: PLM soma; open arrows: PLM neurites; asterisk: vulva. Scale bar: 20 µm. Fig. 4. sam-4 and syd-2 interact with unc-104 to regulate behavior. 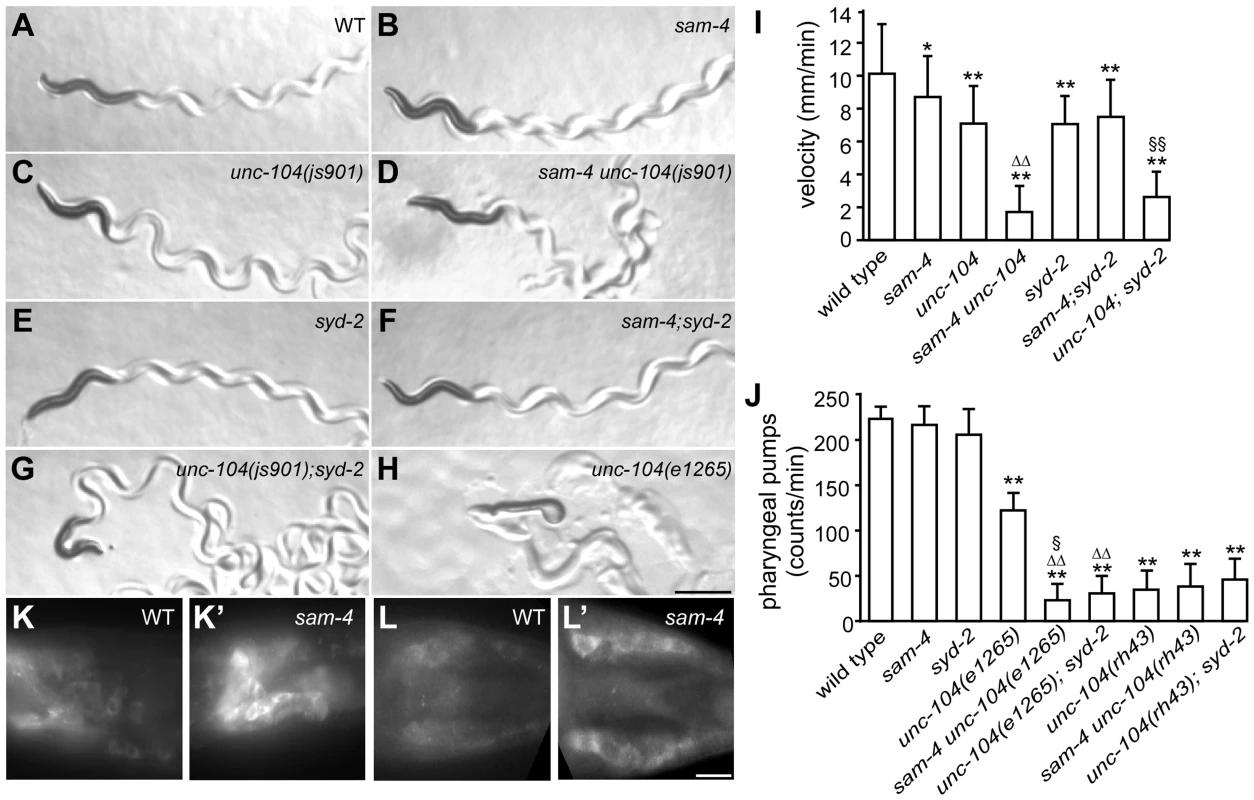
(A–H) Posture and sinusoidal tracts of free moving animals on an E. coli lawn. Scale bar 1 mm. (I) Locomotion velocity measurements of wild type and mutant animals. *P<0.05, relative to wild type; **, P<0.001, relative to wild type; §§, P<0.001 relative to syd-2 and unc-104; P<0.001, ΔΔ relative to sam-4 and relative to unc-104. n>20 per group. (J) Pharyngeal pumping assays of wild type and various mutant and double mutant combinations. **, P<0.001, relative to wild type; ΔΔ, P<0.001, relative to unc-104(e1265); §, P<0.05 relative to sam-4 unc-104(rh43); n>25 per group. (K–L′) anti-UNC-104 whole mount immunohistochemistry staining of the head (K and K′) and the tail (L and L′). Scale bar 5 µm. Alleles tested: sam-4(js415), unc-104(js901) and syd-2(ok217) unless otherwise indicated. It has been previously demonstrated that the UNC-104 PH domain functions independently from the motor domain [49], [50]. The motor domain can walk on microtubules independently of the PH domain, and the PH domain can interact with vesicles independently of the motor domain. To assess the mechanistic implications of the genetic interactions between SAM-4 and UNC-104, we examined the allele specificity of these interactions. Specifically, we first examined sam-4 interactions with a SV binding defective unc-104 allele, e1265, which introduces a missense mutation (D1498N) in the PH domain and causes severe defects in SV binding [51]. Since e1265 mutants show virtually no detectable GFP-RAB-3 signal in neurites and severe locomotion defects with essentially no sinusoidal movements within the time-frame of our measurements (Figure 4H), we analyzed the sam-4 and unc-104(e1265) interactions by scoring animals for pharyngeal pumping, a behavior which is also controlled by neuronal activity [52]. We found that pharyngeal pumping rates of the double mutants were significantly lower than those of e1265 animals (Figure 4I). sam-4 unc-104(e1265) double mutants also had lower brood sizes and slow growth relative to either single mutants. These results suggest that SAM-4 acts synergistically with the PH domain to regulate SV trafficking.
We then tested how sam-4 mutations interact with unc-104(lf) mutations in the motor domain. unc-104(rh43) introduces two missense mutations in the motor domain and results in its motility defect [51]. These mutants exhibit severe locomotion defects again limiting our assay of animal movements. We applied pharyngeal pumping tests to evaluate their interaction. Surprisingly, we found that pharyngeal pumping defects introduced by the rh43 mutations are not exacerbated by the sam-4 mutation (Figure 4J). Furthermore, we noticed that while the pumping defects of e1265 mutants are less severe than those of rh43 mutants, these defects of sam-4 unc-104 (e1265) are more severe than those of sam-4 unc-104 (rh43) (Figure 4J). Thus, sam-4 exhibits allele specific synthetic interactions with PH domain lesions, but not motor domain lesions of unc-104. Taken together, these results suggest that SAM-4 functions by improving the UNC-104 motility, and acts in parallel to the UNC-104 PH domain to regulate SV trafficking.
To further explore the notion that SAM-4 enhances UNC-104 movement, we examined UNC-104 motor activity indirectly in sam-4 mutants by determining the localization of native protein. While UNC-104 protein is barely detectable in soma of wild type animals, we observed a dramatic increase of somatic UNC-104 accumulation in sam-4 mutants (Figure 4K–4L′). Since UNC-104 expression level is not affected by the sam-4 mutations (Figure S9B), these data indicate that UNC-104 motility is disrupted.
By contrast with unc-104, dynein dhc-1 mutants exhibit accumulations of GFP-RAB-3 in the distal portion of the anterior PLM neurite presumably due to disruption of retrograde transport, but have largely wild type levels of GFP-RAB-3 in both the PLM soma and synaptic varicosities. We found that dhc-1(js319); sam-4 double mutants show vestiges of both mutant phenotypes: while GFP-RAB-3 levels are modestly increased in the distal portion of PLM neurites resembling dhc-1 phenotypes, GFP-RAB-3 levels are greatly reduced in the PLM synaptic varicosities and increased in the proximal portion and soma resembling sam-4 phenotypes (Figure S7). This combination of phenotypes is similar to that of dhc-1(js319); unc-104(js901) animals (Figure S7). We interpret these phenotypes of dhc-1; sam-4 double mutants as a combination of the sam-4 and dhc-1 induced defects in SV transport. Therefore, sam-4 shows no obvious genetic interactions with dhc-1. Taken together, these genetic interactions support the model that SAM-4 regulates SV anterograde transport through UNC-104 motor domain.
SAM-4 regulates SV transport by improving processivity
To directly address how SAM-4 regulates SV transport, we examined GFP-RAB-3 puncta dynamics in PLM neurites of sam-4 mutants using time-lapse imaging (Figure 5). We found that SV anterograde transport is significantly reduced in sam-4 mutants as revealed by a reduced number of moving particles, reduced run-length of particles and increased frequency of pauses (Figure 5E–5H). However, the velocity of the GFP-RAB-3 transport was similar to wild type (Figure 5F). Retrograde trafficking is similarly affected by sam-4 mutations (Figure 5), in agreement with previous observations that retrograde trafficking is linked to anterograde trafficking of SVs [51], [53]. sam-4 defects were similar in severity to those of unc-104(js901) mutants. However, sam-4 unc-104 double mutants show more severe defects in GFP-RAB-3 trafficking (Figure 5), consistent with our behavioral and cell biological observations. These findings argue that SAM-4 regulates the anterograde trafficking of SVs by modulating the processivity of SV transport.
Fig. 5. Live imaging of GFP-RAB-3 trafficking in sam-4 mutants. 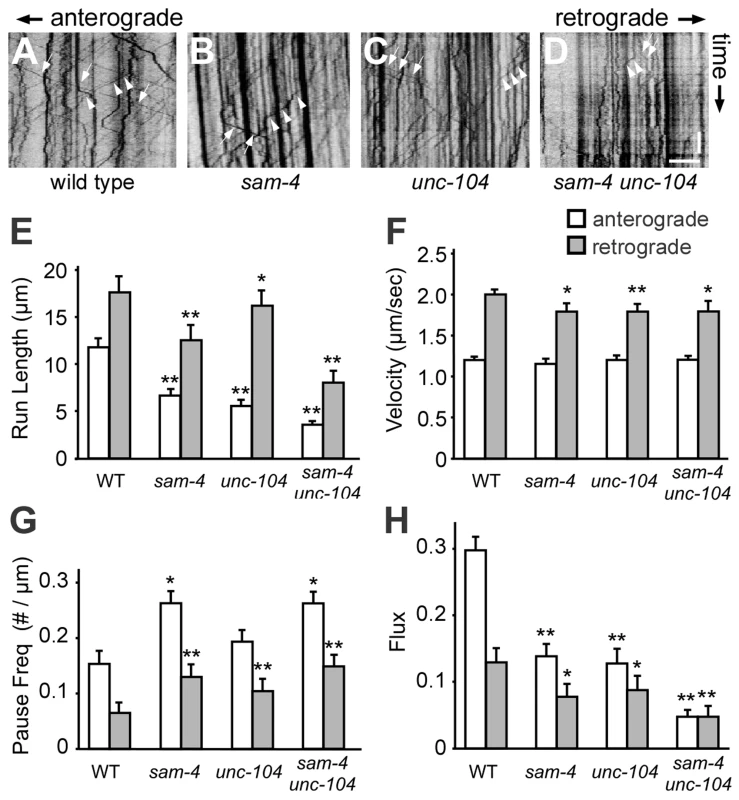
(A–D) Representative GFP-RAB-3 trafficking kymographs in different genetic backgrounds. Arrowheads: anterograde movements; arrows: retrograde movements. Horizontal scale bar 5 µm; vertical scale bar 5 sec. (E–H) Quantification of anterograde and retrograde SV trafficking in L4 animals. (E) Average mean run length, (F) Average velocity, (G) Average pause frequency, and (H) Average flux are shown. Flux is defined as the number of moving particles per total time. Alleles tested: sam-4(js415) and unc-104(js901). ** P<0.01, ** P<0.001 relative to wild type. SYD-2 and SAM-4 regulate SV transport similarly
In C. elegans, SYD-2 liprin-α has been shown to regulate SV transport by binding the FHA domain and stalk domain of UNC-104 [34]. With our observations on sam-4 unc-104 interactions in regulating SV transport, we next examined the relationship between syd-2 and sam-4. We first confirmed that syd-2(ok217) null mutants show increased GFP-RAB-3 levels in the PLM soma and decreased levels in PLM synaptic varicosities (Figure 3), suggesting that anterograde SV trafficking is reduced. The GFP-RAB-3 accumulation defects in syd-2 mutants are less severe than that in sam-4 mutants (Figure 3). Nevertheless, similar to that observed in sam-4 unc-104(js901) mutants, abnormal soma and proximal neurite GFP-RAB-3 accumulations become much severe in unc-104(js901); syd-2 mutants relative to either single mutant (Figure 3). Furthermore, unc-104(js901); syd-2 mutants show more severe defects in locomotion than either single mutant (Figure 4I). Similar genetic interactions to those observed in sam-4 unc-104(e1265) and sam-4 unc-104(rh43) animals were observed in unc-104(e1265); syd-2 and unc-104(rh43); syd-2 (Figure 4J). Taken together, these data suggest that SYD-2 acts synergistically with UNC-104 PH domains to regulate SV trafficking in a similar manner as SAM-4.
We next examined sam-4 syd-2 interactions and observed no detectable genetic interactions between the two mutants. Double mutants display sam-4-like GFP-RAB-3 accumulation defects (Figure 3), and similar stimulated locomotion behaviors as either single mutants (Figure 4I). Over-expression of sam-4 does not suppress syd-2(ok217) mutants, and syd-2(ju487), a gain-of-function allele, has no effects on sam-4 defects (Figure S8). These results are consistent with the hypothesis that SYD-2 and SAM-4 function in the same pathway to regulate SV trafficking.
unc-104 gain-of-function mutations suppress sam-4 and syd-2 defects
To further understand how SAM-4 activity regulates SV trafficking, we conducted a genetic screen for sam-4 suppressors. Using ENU induced mutagenesis, we screened mutated progeny of sam-4(js415); jsIs821 for animals with increased GFP-RAB-3 signal in PLM synaptic varicosities (Figure 6A–6D) and isolated two suppressors from roughly 100,000 genomes screened. Combining traditional genetic mapping and whole genome sequencing strategies, we identified both mutations as novel unc-104 alleles (see Materials and Methods for details). Interestingly, we found that the alleles introduce missense mutations in the UNC-104/KIF1A motor domain: S211A (js1288) and D177A (js1289), both of which are conserved in mammalian molecular motor proteins (Figure S9). Further genetic tests showed that both alleles are semi-dominant in suppressing sam-4 defects. Additionally, we found that over-expression of wild type unc-104 (unc-104(+)) in PLM neurons suppresses sam-4 defects (Figure 7A–7D and 7K), but over-expression of sam-4 does not suppress unc-104 defects (Figure S7). Similar suppression analysis using syd-2 mutants by these unc-104(gf) mutations also revealed suppression by unc-104(gf) alleles (Figure 6). Taken together, these data argue that both js1288 and js1289 are gain-of-function alleles of unc-104, and unc-104 is epistatic to sam-4 and syd-2.
Fig. 6. Gain-of-function unc-104 motor mutations suppress sam-4 and syd-2 SV trafficking defects. 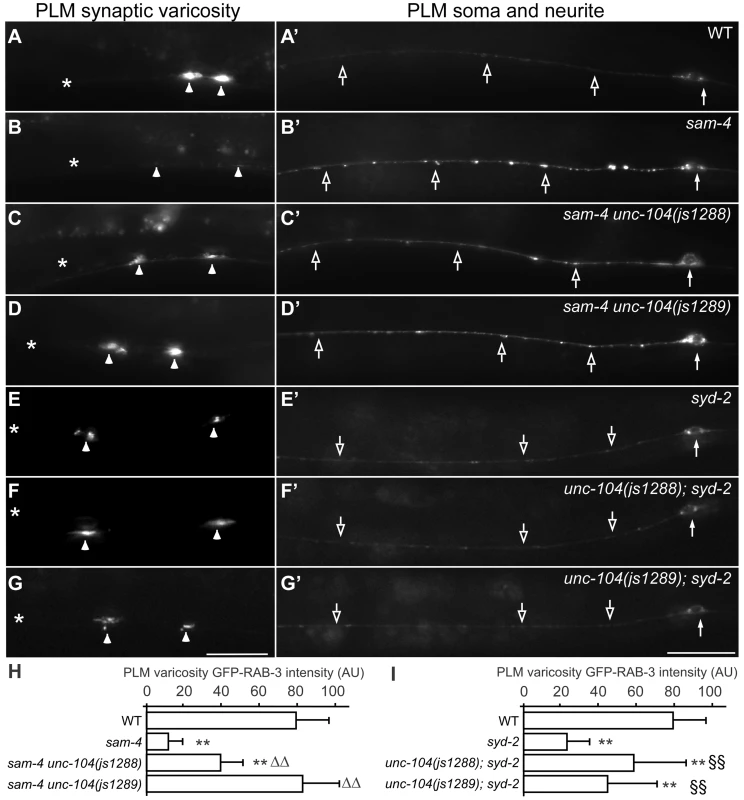
(A–G′) shown is the distribution of GFP-RAB-3 accumulations in PLM synaptic varicosities (left) and the PLM soma and proximal neurite (right). Arrowheads: PLM synaptic varicosities; solid arrows: PLM soma; open arrows: PLM neurites. Scale bar: 20 µm. (H–I) Quantification of GFP-RAB-3 fluorescence intensity in PLM synaptic varicosities. **, P<0.001 relative to wild type; ΔΔ, P<0.001 relative to sam-4(js415); §§, P<0.001 relative to syd-2(ok217). Fig. 7. js1288 and js1289 are gain-of-function mutations in the motor domain of the unc-104 gene. 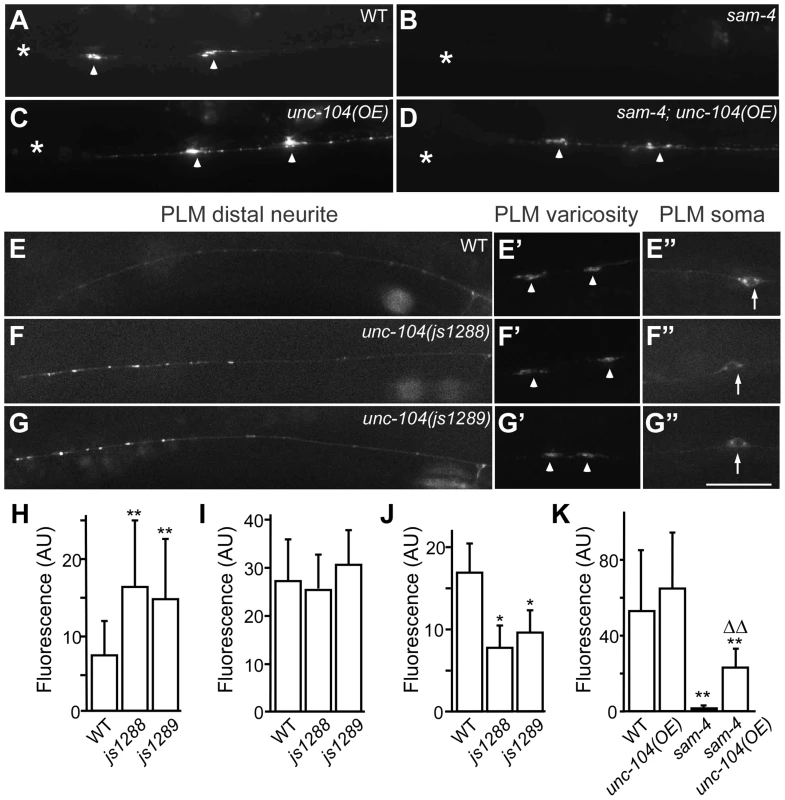
(A–D) Effects of unc-104 overexpression (OE, jsIs1111) on TagRFP-RAB-3 (jsIs1263) accumulation in PLM synaptic varicosities of wild type (A and C) and sam-4 (js415) (B and D). (E–G″) Distribution of the SV marker GFP-RAB-3 in PLM neurons. Shown are representative images of the GFP-RAB-3 (jsIs821) signal observed in the distal end of PLM neurites (E–G), the PLM synaptic varicosities (E′–G′), and the PLM soma (E″–G″). Arrow: PLM soma; arrowhead: PLM synaptic varicosity; asterisk: vulva. Scale bar: 20 µm. (H–J) Quantification of GFP- RAB-3 fluorescence intensities in the distal end of the neurite (30 µm) (H), the synaptic varicosities (I), and the soma (J). Note that, due to differences between imaging conditions for individual PLM anatomic regions, arbitrary units and fluorescence intensity between regions are not comparable. (K) Quantification of TagRFP-RAB-3 fluorescence intensity in PLM synaptic varicosities in different genetic backgrounds. *, P<0.01 relative to wild type; **, P<0.001 relative to wild type; ΔΔ, P<0.001 relative to sam-4(js415). To address how the unc-104(gf) suppresses the SV trafficking defects of sam-4, we characterized the two unc-104 alleles in the absence of sam-4. In isolation, js1288 and js1289 show grossly normal mechanosensory neuron anatomy (Figure 7F, 7G). We analyzed their effects on transport by examining GFP-RAB-3 distribution in vivo. We found that GFP-RAB-3 accumulations are significantly increased in the distal part of PLM neurites (Figure 7E–7H) in each of these unc-104 mutants but decreased in the soma (Figure 7E″–7G″, 7J), indicating that SV transport is enhanced by these two mutations. However, we did not observe GFP-RAB-3 increase in PLM synaptic varicosities (Figure 7E′–7G′, 7I). This is probably because either SV levels in PLM varicosities are already saturated in the wild type background or other mechanisms exist at pre-synapses to maintain SV homeostasis. To further understand how these mutations affect SV dynamics, we examined GFP-RAB-3 trafficking using live imaging (Figure 8). We found that both mutations result in increased run length of GFP-RAB-3 transport (Figure 8E). We also noticed that js1289 results in greater flux of GFP-RAB-3 (Figure 8F), while jsIs1288 reduces SV transport velocity (Figure 8G). Thus, processivity of vesicle transport is increased in both gain-of-function mutants, though perhaps by distinct mechanisms. Western blot analysis of protein levels showed that neither of these two lesions alter UNC-104 protein levels in vivo (Figure S9B). Hence, increasing processivity of the SV transport through the UNC-104 motor domain can partially bypass the need for SAM-4. This is consistent with our hypothesis that SAM-4 functions through the UNC-104 motor domain to regulate SV transport.
Fig. 8. Live imaging of GFP-RAB-3 trafficking in unc-104 mutants. 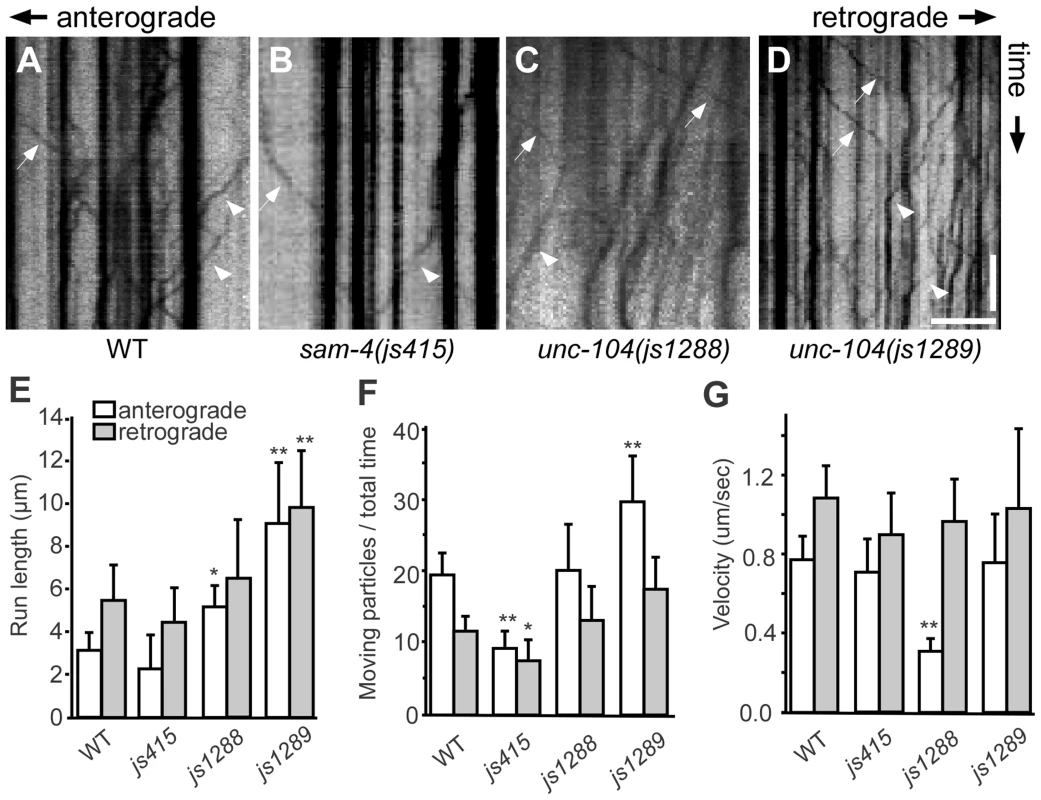
(A–D) Representative SV trafficking kymographs in different genetic backgrounds. Arrowheads: anterograde movements; arrows: retrograde movements. Horizontal scale bar 5 µm; vertical scale bar 5 sec. (E–G) Quantification of anterograde and retrograde GFP-RAB-3 trafficking in mid-L1 stage animals. (E) Average of run length of GFP-RAB-3 particles, (F) Moving particles observed in 40 sec, (G) Average velocity of moving particles. *, P<0.01 relative to wild type; **, P<0.001 relative to wild type, n = 15. Discussion
In this study we have identified the conserved protein SAM-4 as a novel vesicular component regulating SV transport in C. elegans. SAM-4 behaves as a SV associated protein and modulates transport probably by regulating the motor domain activity of UNC-104. This possibility is supported by our identification of two unc-104(gf) motor domain mutations, which suppress sam-4 SV transport defects. Although our genetic evidence is consistent with a SAM-4 UNC-104 interaction, we have been unable to detect any evidence for physical interactions between SAM-4 and UNC-104 either in vitro by yeast two-hybrid analysis or in vivo by co-immunoprecipitation. Therefore, SAM-4 UNC-104 interactions may be mediated by other components. Our genetic data also indicate that SAM-4 acts in the same pathway as SYD-2 in regulating SV transport. We propose a model in which SV-bound SAM-4 regulates SV transport together with SYD-2 through UNC-104, likely via its motor domain (Figure S10).
It is known that SV transport is regulated, but little is known of the molecular mechanisms involved. The identification of SAM-4 as a SV-bound regulator of KIF1A/UNC-104-mediated transport defines a new pathway for modulation of axonal transport. Although SAM-4 is conserved, analysis of the protein sequence revealed only a N-terminal myristoylation motif, which appears to contribute to SAM-4 activity. The lack of identifiable protein domains in the protein make it difficult to speculate on a specific mechanism of action. We have proposed that SAM-4 modulates SV transport processivity by modulating UNC-104 motor activity because we observed strong genetic interaction between sam-4 and unc-104 cargo binding mutants but not with motor domain mutants. Furthermore, motor domain gf mutations suppress sam-4 defects arguing that increase of motor processivity can partially bypass SAM-4 activity.
In addition, the genetic interactions between syd-2 and sam-4 support a processivity based mechanism of action for SAM-4. Both worm and mammalian Liprin-α/SYD-2 interact with KIF1A/UNC-104 [32], [34] and Liprin-α/SYD-2 is required for efficient SV trafficking in both C. elegans and Drosophila [33]. Our data argue syd-2 functions in the same pathway as sam-4 in regulating SV transport since each null mutant shows very similar interactions with both unc-104(lf) and unc-104(gf) lesions, but do not display obvious interactions with each other. However, SAM-4 may play a more central role in this process since the SV trafficking phenotypes in syd-2(null) are less severe than those of sam-4(null).
In this study, we recovered two gain-of-function mutations in the motor domain of unc-104 that increase the processivity of the motor in cargo movement assays. Kinesin mediated SV transport is an ATP driven process, which depends on motor-microtubule binding. The ATP hydrolysis catalytic core lays in the switch I region of the KIF1A/UNC-104 motor domain (Figure S9). Lesions (for example H215Y in unc-104(y211), see Figure S1) in this domain cause severe SV trafficking defects. The js1288 mutation occurs in S211A adjacent to S212 (S215 in mammalian KIF1A) which coordinates the gamma-phosphate of ATP in the ATP-bound crystal structure [54]. Consistent with the hypothesis that this lesion alters rates of ATP hydrolysis, we observed a lowered velocity of transport in js1288 mutants. However, the biochemical mechanism underlying the increase in processivity is unclear. The other mutation, js1289, is a D177A substitution (mammalian KIF1A D180) in loop 8 of the motor domain. Previous studies [54] showed that loop 8 is one of three microtubule binding regions in the motor domain and thus processivity in this mutant could be increased due to changes in the affinity for microtubules. In addition to suppressing SV trafficking defects in sam-4(null), both of these lesions result in increased accumulations of SVs in the distal portion of PLM neurites where no synapses have been seen at the ultrastructural level [40]. Therefore, these unc-104 (gf) mutations disturb the normal homeostasis of SV trafficking and thus may not necessarily represent beneficial biochemical modifications. However, the lesions argue strongly that processivity is not optimized in KIF1A and suggest the possibility that KIF1A activity could be modified, for example by pharmacological compounds, in diseases where axonal transport is compromised.
It is worth noting that several lines of evidence imply sam-4 also regulates other processes in non neuronal cells. First, sam-4 neuronal phenotypes are partially maternally rescued. Some sam-4 animals segregating from the sam-4/+ mother even display wild type levels of GFP-RAB-3 at PLM synapses. Second, sam-4 is likely post-transcriptionally regulated. The sam-4 locus is highly unusual (for nematodes) in that it has two 5′ “non-coding” exons (Figure S5). These exons contain a small 79 amino acid ORF. A similar ORF is found in the 5′-end of sam-4 in highly divergent nematodes and the synonymous codon usage in these nematodes indicates it is being selected as coding sequence (Figure S11). The ORF is homologous to the APC13, a small subunit of the Anaphase Promoting Complex (APC) which was previously described for plant parasitic nematodes [55], but recognized in model system databases. We, and other investigators, observed lethality when performing RNAi against sam-4 even though both nonsense and deletion alleles of sam-4 are fully viable. These sam-4 RNAi lethal phenotypes are similar to those of other APC complex component genes. These include defects in meiosis in the early embryo [56], oocyte deformation and sterility [57] and failure to segregate germline P-granules [58]. We posit the lethality phenotype associated with sam-4 RNAi is likely due to reduced expression of this upstream ORF encoding an APC13 homolog.
The APC complex plays critical roles in regulating progression through the cell cycle. However, recent work has also highlighted several critical roles for APC complexes in neuronal development [59]. In particular, disruption of the APC complex alters axon growth, post-synaptic glutamate receptor levels [60] as well as the size and number of presynaptic boutons [61]. Interestingly, in regulating bouton number in Drosophila, the APC complex works in conjunction with liprin-α. Thus, the APC complex, SAM-4 and liprin-α appear linked at multiple different regulatory levels. Further investigations of the non-neuronal roles of SAM-4, the role of the APC13 encoding upstream ORF in regulating SAM-4 expression, and the potential role of the APC complex in regulating axonal transport are clearly warranted.
Although SAM-4 is evolutionarily conserved, no human disease conditions have been specifically associated with lesions in human sam4 (LOH12CR1), a gene within a region often deleted in acute lymphoblastic leukemia. Notably, worm sam-4 mutants display virtually indistinguishable phenotypes from mild unc-104/KIF1A mutants and human diseases are associated with KIF1A. Specifically, motor domain lesions (A255V and R350G) in human KIF1A underlie the molecular basis of the rare recessive late onset spastic paraplegia SPG30 [6] and a frameshift mutation in the PH domain underlies a form of hereditary sensory and autonomic neuropathy [5]. Further genetic and biochemical studies of SAM-4 in both invertebrates and vertebrates will be required to define the underlying biochemical mechanisms as well as physiological inputs that modulate SAM-4 action in regulating axonal transport.
Materials and Methods
Strains and genetics
C. elegans animals were maintained using standard methods [62]. All strains used except for those used for SNP mapping were derivatives of the Bristol N2 wild type background. Animals were grown at the room temperature (22.5°C), unless specified. Strains used are listed in Table S1. The genotype of strains was confirmed by PCR using oligonucleotides listed in Table S2.
Transgene integration
jsIs1238 II, jsIs1156 IV, jsIs1263 IV, jsIs1188 IV and jsIs1189 IV transgenes were integrated using MosSCI with EG4322 for integration on chromosome II and EG5003 for integration on chromosome IV [45], [46] and confirmed to be single copy by long range PCR amplification. jsIs1073 and jsIs1075 were generated using a bombardment protocol with Cbunc-119 as the integration marker [63].
sam-4 molecular cloning
Genetic three-factor mapping narrowed the sam-4 mutation to an interval between dpy-25 and rol-6 on chromosome II. Single nucleotide polymorphism (SNP) mapping was used to position sam-4 with CB4856 as a reference strain and narrowed the mutation down to a 163 kb region on Chromosome II between the SNPs CE2-141 and pkP2147. This region is covered by 5 fosmids. Using germline transformation rescue tests, we further mapped sam-4 down to the fosmids WRM0610dH02 and WRM0632aA08. The sam-4(js415) lesion was identified by candidate gene sequencing in this region. A C>T nucleotide change was detected in the second exon of the predicted gene F59E12.11. sam-4 defects are fully rescued by a transgene expressing the hypothetic F59E12.11 gene, which is predicted to encode a 240 amino acid protein. These data identify F59E12.11 as sam-4.
Isolation of unc-104 mutants
unc-104(js901) was isolated in a non-clonal forward screen for mutations that mislocalized RBF-1-GFP (jsIs423). L4 jsIs423 animals were mutagenized using 50 mM ethyl methanesulfonate (EMS) for 4 hrs and placed on E. coli seeded agar plates. F2 animals derived from these animals were screened for mislocalization of GFP from the nerve ring to cell bodies surrounding the nerve ring. js901 was mapped to chromosome II by classical genetic mapping strategy, and tested for non-complementation with unc-104(e1265). The entire coding sequence of unc-104 was sequenced in js901 revealing a GGA to GTA that changes Gly1465 to Val. This lesion resides within the PH domain of UNC-104.
unc-104(js1288) and unc-104(js1289) were isolated in a screen for suppressors of the PLM synaptic varicosity phenotype of sam-4. N-ethyl-N-nitrosourea (ENU) mutagenesis was performed using standard methodology [64]. Briefly, sam-4(js415) animals were treated with 0.6 mM ENU for 4 hours at the room temperature. Treated animals (P0) were transferred to fresh food (10 L4s or young adults per 100 mm plate). P0 animals were removed from the plates 24–48 hours later. F1 animals were counted 2–3 days later to estimate number of mutagenized chromosomes screened. The F2 animals were screened for increased GFP-RAB-3 signal in PLM varicosities using a fluorescent dissecting microscope. The suppressors displayed tight linkage to sam-4. Phenotypic analysis of sam-4(js415) js1288/sam-4(js415) and sam-4(js415) js1289/sam-4(js415) revealed both were semi-dominant suppressors of the Sam phenotype. Sequencing of the sam-4 coding region revealed no mutation in sam-4 gene of these isolates. js1289 was mapped to between sam-4 and rol-6 by three factor mapping, crossing lin-31(n301) sam-4(js415) js1289 rol-6(187); jsIs821 to CB4856 and screening for Lin Sam non-Rol and Lin Sam Sup (suppressor) non-Rol recombinants 18 of 21 recombinants had recombination events between sam-4 and js1289 and 3 between js1289 and rol-6. 100 bp paired-end whole genome sequencing of homozygous strains was conducted at Oklahoma Medical Research Foundation. The data were analyzed using Whole Genomes, a web-based alignment and analysis program, and revealed lesions in unc-104: An Asp177 to Ala (GAC to GCC) lesion in js1288 and a Ser211 to Ala (TCA to GCA) lesion in js1289.
Molecular biology
Plasmid DNA clones were constructed using standard molecular biology techniques.
NM2132 (sam-4 genomic clone)
A sam-4 full-length genomic fragment was PCR amplified from the fosmid clone WRM068cE02 using oligonucleotides 3645 and 3646, digested with KpnI/PstI and ligated into KpnI/PstI digested pBluescript SK(+). The sam-4 coding region in this plasmid was confirmed by DNA sequencing.
NM2348 (sam-4::3XFlag)
A 3XFlag tag was introduced at the C-terminus of SAM-4 by PCR amplification using oligonucleotides 3985 and 3986 and recircularized using In-Fusion cloning kit (Clontech Labs Inc.).
NM2347 (pCFJ178 sam-4-3XFlag)
NM2348 was digested with PstI and KpnI and a sam-4::3XFlag fragment was inserted into PstI KpnI digested pCFJ178 [45].
NM2364 (sam-4(G2S) genomic clone)
NM2132 was mutagenized using a DpnI mediated mutagenesis protocol [65] using oligonucleotides 4059 and 4060.
NM2662 (pCFJ178 sam-4(G2S)-3XFlag)
A PshAI/KpnI fragment containing the N-terminus of SAM-4 was ligated into PshAI/KpnI digested NM2347.
NM2057 (mec-7p-tagRFP-mito unc-119)
TagRFP-mito from pTagRFP-mito (Evrogen) was digested with NheI, filled in, digested with EagI and inserted into NM2041 (Kumar et al. 2009) digested with SalI, filled in, and then digested with EagI replacing GFP-ELKS-1 with Tag-RFP-mito.
NM2066 (mec-7p TagRFP-ELKS-1 unc-119)
was constructed by amplifying TagRFP from pTagFRP-mito (Evrogen) using oligonucleotides 3571 and 3572, digesting the purified PCR product with NcoI and BsiWI and replacing the GFP in NM2041 excised with NcoI and BsrGI.
NM2173 (mec-7p::sam-4-TagRFP)
sam-4 genomic sequences were amplified by PCR using oligonucleotide 3701 and 3702, digested with NheI and NcoI, and inserted into NheI-NM2057 replacing the N-terminal mitochondrial tag with sam-4 sequences.
NM2238 (mec-7p::sam-4-TagRFP)
The BssHII/SpeI fragment of NM2173 containing mec-7p::sam-4-TagRFP was inserted into BssHII/XbaI digested pCFJ178.
NM2351 (pCFJ178 glr-1p::sam-4-TagRFP)
The glr-1 promoter was amplified by PCR using oligonucleotides 3981 and 3984, digested with SphI and NheI and ligated into SphI/NheI digested NM2238).
Light microscopy
Transgenic animals were imaged using epi-fluorescence on an Olympus BX60 equipped with an X-CITE120 mercury lamp (EXFO) using standard GFP and RFP filter sets. Images were taken with a Retiga EXi CCD camera using OpenLab software and processed using Adobe Photoshop.
Transgene integration
jsIs1238 II, jsIs1156 IV, jsIs1263 IV, jsIs1188 IV and jsIs1189 IV transgenes were integrated using MosSCI with EG4322 for integration on chromosome II and EG5003 for integration on chromosome IV [45] with modifications. These transgenic lines were confirmed to be single-copy integration events by long range PCR (Details available online: http://thalamus.wustl.edu/nonetlab/ResourcesF/Resources.html). jsIs1073 and jsIs1075 lines were generated by integrating NM2057 and NM2066 using a bombardment protocol with Cbunc-119 as the integration marker [63].
Locomotion assays
Animals were assayed at the room temperature on NGM agar. L4 animals (or as indicated) were transferred to a bacteria-free plate to allow them clear off bacteria (2–3 min). Subsequently, these animals were transferred to another bacteria-free plate and imaged immediately for 10–20 sec. Animal movements were recorded using LG-3 frame grabber run by ScionImage software at 1 frame/sec for 40 images total. These recordings were then analyzed using wormtracker plus [66]. Only animals in the imaging field>14 consecutive frames recorded were used in the velocity analyses.
Pharyngeal pumping assays
L4 animals on bacterial lawns of OP50 on NGM agar were scored at the room temperature. Pharyngeal pumping rates was determined by counting contractions of the terminal bulb for 1 minute per animal.
Immunostaining
Immunohistochemistry and western blots were performed as previously described [67], [68]. For FLAG immunohistochemistry staining, animals were grown at room temperature and fixed in methanol/acetone. For SAM-4-3XFlag fractionation, 0.5 mM EGTA and 0.5 mMEDTA were added in the fractionation buffer as indicated in Figure S6. Mouse anti-Flag (1∶200, Sigma, Cat. A8592) primary antibody incubations were performed overnight at 4°C. Alexa conjugated secondary antibodies (Invitrogen) were incubated for 2 hrs at room temperature at 1∶500. Antibody used for western blots: anti-FLAG (1∶1000); anti-β-tubulin (1∶1000, E7, Developmental Studies Hybridoma Bank, Iowa city), anti-UNC-104 (1∶40) [51].
Live imaging and analysis of SV transport dynamics
For young adult animals (used in Figure 5), hermaphrodites were immobilized with 3–5 mM levamisole (Sigma-Aldrich) in M9 buffer and mounted on a 2% agarose pad. Time-lapse images of GFP-RAB-3 were acquired and analyzed as described before [51]. The numbers of moving particles in a 15–20 µm region at a distance of 15–25 µm away from the PLM soma were used for flux calculations.
For mid-L1 staged animals (20–24 hrs after hatch, used in Figure 8), we used an anesthetic-free protocol to image GFP-RAB-3 [69]. Specifically, animals were immobilized in 0.5 µl of 0.10 microspheres (Cat# 00876, Polysciences, Inc.) on 10% agarose pads. Time-lapse imaging was acquired using 100×/1.30 oil objective on a Axioskop (Zeiss) equipped with ASI piezo XYZ-motorized stage, Sutter instruments high speed electronic filter wheels and shutters, and a Hamamatsu Orca-R2 cooled CCD camera all controlled by Volocity software (PerkinElmer Inc.). Time lapse images were acquired for 40 seconds at a speed of 5 frames per second with an exposure time of 200 ms. Particle dynamics were analyzed with Volocity software. Total moving particles were counted in the 35 µm region at a distance of 20 µm away from the PLM soma.
To record GFP-RAB-3 co-movements with SAM-4-TagRFP, confocal images were collected with a Hamamatsu Flash 4.0 CMOS camera attached to a Yokogawa Spinning Disc Confocal apparatus on an Olympus IX73 inverted microscope. 0.33 sec Green and Red channels exposure were taken consecutively, and captured at 1 sec intervals, and image series were assembled into movies using Micro-Manager software (available at micro-manager.org).
Statistics
P values were determined using GraphPad Prism. Multi-group data sets were analyzed by a one-way ANOVA with post-hoc Holm-Sidak's test for multiple comparisons. A t-test was used for paired data sets.
Supporting Information
Zdroje
1. GoldsteinAY, WangX, SchwarzTL (2008) Axonal transport and the delivery of pre-synaptic components. Curr Opin Neurobiol 18 : 495–503.
2. HirokawaN, NiwaS, TanakaY (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68 : 610–638.
3. EspositoG, FernandesAC, VerstrekenP (2011) Synaptic vesicle trafficking and Parkinson's disease. Dev Neurobiol 72 : 134–144.
4. SalinasS, BilslandLG, SchiavoG (2008) Molecular landmarks along the axonal route: axonal transport in health and disease. Curr Opin Cell Biol 20 : 445–453.
5. RiviereJB, RamalingamS, LavastreV, ShekarabiM, HolbertS, et al. (2011) KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am J Hum Genet 89 : 219–230.
6. KlebeS, LossosA, AzzedineH, MundwillerE, ShefferR, et al. (2012) KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: distinct phenotypes according to the nature of the mutations. Eur J Hum Genet 20 : 645–649.
7. SiddiquiSS (2002) Metazoan motor models: kinesin superfamily in C. elegans. Traffic 3 : 20–28.
8. MikiH, SetouM, KaneshiroK, HirokawaN (2001) All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci U S A 98 : 7004–7011.
9. HirokawaN, NodaY (2008) Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev 88 : 1089–1118.
10. KamalA, StokinGB, YangZ, XiaCH, GoldsteinLS (2000) Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron 28 : 449–459.
11. GlaterEE, MegeathLJ, StowersRS, SchwarzTL (2006) Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol 173 : 545–557.
12. SetouM, SeogDH, TanakaY, KanaiY, TakeiY, et al. (2002) Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature 417 : 83–87.
13. HammondJW, BlasiusTL, SoppinaV, CaiD, VerheyKJ (2010) Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol 189 : 1013–1025.
14. HammondJW, CaiD, BlasiusTL, LiZ, JiangY, et al. (2009) Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol 7: e72.
15. ImanishiM, EndresNF, GennerichA, ValeRD (2006) Autoinhibition regulates the motility of the C. elegans intraflagellar transport motor OSM-3. J Cell Biol 174 : 931–937.
16. KaanHY, HackneyDD, KozielskiF (2011) The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science 333 : 883–885.
17. FriedmanDS, ValeRD (1999) Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol 1 : 293–297.
18. StockMF, GuerreroJ, CobbB, EggersCT, HuangTG, et al. (1999) Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J Biol Chem 274 : 14617–14623.
19. BlasiusTL, CaiD, JihGT, ToretCP, VerheyKJ (2007) Two binding partners cooperate to activate the molecular motor Kinesin-1. J Cell Biol 176 : 11–17.
20. ChoKI, YiH, DesaiR, HandAR, HaasAL, et al. (2009) RANBP2 is an allosteric activator of the conventional kinesin-1 motor protein, KIF5B, in a minimal cell-free system. EMBO Rep 10 : 480–486.
21. MorfiniG, SzebenyiG, ElluruR, RatnerN, BradyST (2002) Glycogen synthase kinase 3 phosphorylates kinesin light chains and negatively regulates kinesin-based motility. Embo J 21 : 281–293.
22. GuillaudL, WongR, HirokawaN (2008) Disruption of KIF17-Mint1 interaction by CaMKII-dependent phosphorylation: a molecular model of kinesin-cargo release. Nat Cell Biol 10 : 19–29.
23. LiuJS, SchubertCR, FuX, FourniolFJ, JaiswalJK, et al. (2012) Molecular basis for specific regulation of neuronal kinesin-3 motors by doublecortin family proteins. Mol Cell 47 : 707–721.
24. Serra-PagesC, KedershaNL, FazikasL, MedleyQ, DebantA, et al. (1995) The LAR transmembrane protein tyrosine phosphatase and a coiled-coil LAR-interacting protein co-localize at focal adhesions. EMBO J 14 : 2827–2838.
25. Serra-PagesC, MedleyQG, TangM, HartA, StreuliM (1998) Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem 273 : 15611–15620.
26. KoJ, KimS, ValtschanoffJG, ShinH, LeeJR, et al. (2003) Interaction between liprin-alpha and GIT1 is required for AMPA receptor targeting. J Neurosci 23 : 1667–1677.
27. WyszynskiM, KimE, DunahAW, PassafaroM, ValtschanoffJG, et al. (2002) Interaction between GRIP and liprin-alpha/SYD2 is required for AMPA receptor targeting. Neuron 34 : 39–52.
28. DaiY, TaruH, DekenSL, GrillB, AckleyB, et al. (2006) SYD-2 Liprin-alpha organizes presynaptic active zone formation through ELKS. Nat Neurosci 9 : 1479–1487.
29. HsuCC, MoncaleanoJD, WagnerOI (2011) Sub-cellular distribution of UNC-104(KIF1A) upon binding to adaptors as UNC-16(JIP3), DNC-1(DCTN1/Glued) and SYD-2(Liprin-alpha) in C. elegans neurons. Neuroscience 176 : 39–52.
30. StigloherC, ZhanH, ZhenM, RichmondJ, BessereauJL (2011) The presynaptic dense projection of the Caenorhabditis elegans cholinergic neuromuscular junction localizes synaptic vesicles at the active zone through SYD-2/liprin and UNC-10/RIM-dependent interactions. J Neurosci 31 : 4388–4396.
31. SpanglerSA, HoogenraadCC (2007) Liprin-alpha proteins: scaffold molecules for synapse maturation. Biochem Soc Trans 35 : 1278–1282.
32. ShinH, WyszynskiM, HuhKH, ValtschanoffJG, LeeJR, et al. (2003) Association of the kinesin motor KIF1A with the multimodular protein liprin-alpha. J Biol Chem 278 : 11393–11401.
33. MillerKE, DeProtoJ, KaufmannN, PatelBN, DuckworthA, et al. (2005) Direct observation demonstrates that Liprin-alpha is required for trafficking of synaptic vesicles. Curr Biol 15 : 684–689.
34. WagnerOI, EspositoA, KohlerB, ChenCW, ShenCP, et al. (2009) Synaptic scaffolding protein SYD-2 clusters and activates kinesin-3 UNC-104 in C. elegans. Proc Natl Acad Sci U S A 106 : 19605–19610.
35. MontpetitA, BoilyG, SinnettD (2002) A detailed transcriptional map of the chromosome 12p12 tumour suppressor locus. Eur J Hum Genet 10 : 62–71.
36. ErnstromGG, ChalfieM (2002) Genetics of sensory mechanotransduction. Annu Rev Genet 36 : 411–453.
37. ChalfieM, SulstonJE, WhiteJG, SouthgateE, ThomsonJN, et al. (1985) The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci 5 : 956–964.
38. HamelinM, ScottIM, WayJC, CulottiJG (1992) The mec-7 beta-tubulin gene of Caenorhabditis elegans is expressed primarily in the touch receptor neurons. EMBO J 11 : 2885–2893.
39. NonetML (1999) Visualization of synaptic specializations in live C. elegans with synaptic vesicle protein-GFP fusions. J Neurosci Methods 89 : 33–40.
40. WhiteJG, SouthgateE, ThomsonJN, BrennerS (1986) The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 314 : 1–340.
41. KoushikaSP, SchaeferAM, VincentR, WillisJH, BowermanB, et al. (2004) Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. J Neurosci 24 : 3907–3916.
42. Schaefer AM (2001) A Molecular Genetic Analysis of Synapse Formation in C. elegans [PhD]. St. Louis, MO: Washington University 129 p.
43. MahoneyTR, LiuQ, ItohT, LuoS, HadwigerG, et al. (2006) Regulation of synaptic transmission by RAB-3 and RAB-27 in Caenorhabditis elegans. Mol Biol Cell 17 : 2617–2625.
44. HallDH, HedgecockEM (1991) Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65 : 837–847.
45. Frokjaer-JensenC, DavisMW, HopkinsCE, NewmanBJ, ThummelJM, et al. (2008) Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40 : 1375–1383.
46. Frokjaer-JensenC, DavisMW, AilionM, JorgensenEM (2012) Improved Mos1-mediated transgenesis in C. elegans. Nat Methods 9 : 117–118.
47. NonetML, GrundahlK, MeyerBJ, RandJB (1993) Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell 73 : 1291–1305.
48. JohnsonDR, BhatnagarRS, KnollLJ, GordonJI (1994) Genetic and biochemical studies of protein N-myristoylation. Annu Rev Biochem 63 : 869–914.
49. KlopfensteinDR, TomishigeM, StuurmanN, ValeRD (2002) Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell 109 : 347–358.
50. KlopfensteinDR, ValeRD (2004) The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans. Mol Biol Cell 15 : 3729–3739.
51. KumarJ, ChoudharyBC, MetpallyR, ZhengQ, NonetML, et al. (2010) The Caenorhabditis elegans Kinesin-3 motor UNC-104/KIF1A is degraded upon loss of specific binding to cargo. PLoS Genet 6: e1001200.
52. AveryL, HorvitzHR (1989) Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron 3 : 473–485.
53. OuCY, PoonVY, MaederCI, WatanabeS, LehrmanEK, et al. (2010) Two cyclin-dependent kinase pathways are essential for polarized trafficking of presynaptic components. Cell 141 : 846–858.
54. KikkawaM, SablinEP, OkadaY, YajimaH, FletterickRJ, et al. (2001) Switch-based mechanism of kinesin motors. Nature 411 : 439–445.
55. SchwickartM, HavlisJ, HabermannB, BogdanovaA, CamassesA, et al. (2004) Swm1/Apc13 is an evolutionarily conserved subunit of the anaphase-promoting complex stabilizing the association of Cdc16 and Cdc27. Mol Cell Biol 24 : 3562–3576.
56. SonnichsenB, KoskiLB, WalshA, MarschallP, NeumannB, et al. (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434 : 462–469.
57. GreenRA, KaoHL, AudhyaA, ArurS, MayersJR, et al. (2011) A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145 : 470–482.
58. UpdikeDL, StromeS (2009) A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics 183 : 1397–1419.
59. PuramSV, BonniA (2011) Novel functions for the anaphase-promoting complex in neurobiology. Semin Cell Dev Biol 22 : 586–594.
60. JuoP, KaplanJM (2004) The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr Biol 14 : 2057–2062.
61. van RoesselP, ElliottDA, RobinsonIM, ProkopA, BrandAH (2004) Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell 119 : 707–718.
62. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
63. PraitisV, CaseyE, CollarD, AustinJ (2001) Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157 : 1217–1226.
64. De StasioEA, DormanS (2001) Optimization of ENU mutagenesis of Caenorhabditis elegans. Mutat Res 495 : 81–88.
65. FisherCL, PeiGK (1997) Modification of a PCR-based site-directed mutagenesis method. Biotechniques 23 : 570–574, 570-571, 574.
66. RamotD, JohnsonBE, BerryTLJr, CarnellL, GoodmanMB (2008) The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS One 3: e2208.
67. HadwigerG, DourS, ArurS, FoxP, NonetML (2010) A monoclonal antibody toolkit for C. elegans. PLoS One 5: e10161.
68. NonetML, StauntonJ, KilgardMP, FergestadT, HartweigE, et al. (1997) C. elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci 17 : 8021–8073.
69. ChristopherFang-Yen, SaraWasserman, PialiSengupta, SamuelADT (2009) Agarose immobilization of C. elegans. The Worm Breeder's Gazette 18 : 32–32.
Štítky
Genetika Reprodukčná medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



