-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
Beclin 1 was not only the first-described mammalian autophagy protein, but is one of the most widely-characterized players in autophagy regulation. It is implicated in multiple human disease conditions. As a core component of the essential lipid kinase complex (PI3K-III), beclin 1 has largely been characterized to date in the context of autophagy through its recruitment of additional autophagy proteins for the assembly of the PI3K-III complexes involved in the nucleation of the autophagosome. Little is known, however, about how beclin 1 regulates specific functions of PI3K-III in other membrane trafficking pathways. Furthermore, although beclin 1 has been linked to multiple neurodegenerative diseases, the function of beclin 1 in the brain remains uncharacterized. Herein, we used genetic animal models and mutant cell lines to demonstrate that beclin 1 participates in multiple organelle trafficking pathways. Beclin 1 deficiency in neurons causes severe neurodegeneration, concomitant with aberrant late endosome formation and impaired phospholipid localization. Our mechanistic study reveals the essential role for beclin 1 in neuron survival involving multiple membrane trafficking pathways including endocytosis and autophagy. Our study clarifies the physiological function of beclin 1, which leads for further understanding of its role in tumorigenesis, infectious disease and neurodegenerative disease.
Published in the journal: Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex. PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004626
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004626Summary
Beclin 1 was not only the first-described mammalian autophagy protein, but is one of the most widely-characterized players in autophagy regulation. It is implicated in multiple human disease conditions. As a core component of the essential lipid kinase complex (PI3K-III), beclin 1 has largely been characterized to date in the context of autophagy through its recruitment of additional autophagy proteins for the assembly of the PI3K-III complexes involved in the nucleation of the autophagosome. Little is known, however, about how beclin 1 regulates specific functions of PI3K-III in other membrane trafficking pathways. Furthermore, although beclin 1 has been linked to multiple neurodegenerative diseases, the function of beclin 1 in the brain remains uncharacterized. Herein, we used genetic animal models and mutant cell lines to demonstrate that beclin 1 participates in multiple organelle trafficking pathways. Beclin 1 deficiency in neurons causes severe neurodegeneration, concomitant with aberrant late endosome formation and impaired phospholipid localization. Our mechanistic study reveals the essential role for beclin 1 in neuron survival involving multiple membrane trafficking pathways including endocytosis and autophagy. Our study clarifies the physiological function of beclin 1, which leads for further understanding of its role in tumorigenesis, infectious disease and neurodegenerative disease.
Introduction
Autophagy is a regulated “self-digestion” pathway in the cytoplasm of cells involving the synthesis, trafficking and delivery of autophagosomes to lysosomes for degradation. Beclin 1 was the first described mammalian autophagy protein and a core component of the class III phosphatidylinositol 3-kinase (PI3K-III) complex, which plays an important role in membrane trafficking and restructuring involved in autophagy, endocytosis, cytokinesis and phagocytosis [1]. To date beclin 1 has largely been characterized in the context of autophagy; it modulates the lipid kinase activity of PI3K-III catalytic unit VPS34, which generates phosphatidylinositol 3-phosphate (PI(3)P), enabling the recruitment of the other autophagy proteins involved in the nucleation of autophagosome [2], [3]. Little is known, however, about how beclin 1 regulates specific functions of VPS34. Multiple lines of evidence have linked altered expression of beclin 1 to several major human diseases such as cancer, infectious diseases and neurodegenerative disorders [4], [5]. It is thus imperative to understand the precise molecular mechanism whereby beclin 1 regulates VPS34 functions in autophagy and other membrane trafficking pathways, the disruption of which may underlie the pathogenesis of multiple diseases.
Our previous report and also others demonstrated that mammalian beclin 1-VPS34 forms multiple complexes [6]–[8]. The three core proteins beclin 1, VPS34 and VPS15, interact with a number of auxiliary proteins such as Atg14L, UVRAG, RUBICON, Ambra1 and Bif-1. By recruiting different binding partners and forming distinct PI3K-III complexes, beclin 1 modulates PI3K-III activity and regulates autophagy at multiple stages including nucleation and maturation. Moreover, beclin 1 is modified post-translationally by phosphorylation and ubiquitination serving to fine-tune PI3K-III activity and autophagy in response to various intracellular and extracellular signals [9].
Early reports implicate beclin 1 in numerous neuropathological conditions [10], [11] and emerging evidence demonstrates the neuroprotective function of beclin 1 through removal of disease-related proteins including APP metabolites [12], α-synuclein [13] and mutant ataxin [14] in the nervous system. However, the physiological function of beclin 1 in neurons is poorly understood and whether the neuroprotective function of beclin 1 is mediated through strictly autophagy-dependent or independent pathways is unclear. Here we report that disruption of beclin 1 in a neuron-type specific manner results in severe neurodegeneration in mice. Neuronal beclin 1 is associated with endocytic compartments and ablation of Becn1 causes impairment of the endosome pathway. Our results show a role for beclin 1 in regulating PI3K-III activity and endocytosis via the beclin 1-UVRAG interaction, highlighting beclin 1's physiological function in multiple membrane traffic pathways.
Results
Accelerated neurodegeneration in Becn1 conditional knockout mice
Our previous reports showed that Becn1 deletion causes embryonic lethality at E7.5, preventing us from studying beclin 1 physiological functions in adult tissues [15]. We generated a conditional knock-out (cKO) mouse line by crossing Becn1 Flox/Flox (Becn1F/F) mice with Pcp2-Cre transgenic mice [16] to delete Becn1 specifically in cerebellar Purkinje cells (PCs) (Figure S1A, S1B). In Becn1F/F;Pcp2-Cre mice, a loss of beclin 1 expression is observed in cerebellar PCs at P21 (Figure S1C). In contrast to Becn1F/F control mice, Becn1F/F;Pcp2-Cre cKO mice developed rapid degeneration of PCs between P21 (Figure S1D) and 1 month (>80% loss) and lost nearly all PCs at 2 months (Figure 1A). The loss of PCs is associated with abnormal gait and ataxic behavior (Figure S2A, Movie S1, S2). Thus, Becn1 cKO mice have clearly accelerated PC degeneration, differing significantly from autophagy gene Atg7 conditional knockout (Atg7F/F;Pcp2-Cre cKO) mice, in which PCs are intact at 1 month and only a moderate percentage of PCs degenerated by 2 months, as we previously reported [16]. The relatively late onset of PC degeneration in Atg7 cKO mice is unlikely due to delayed deletion of the Atg7 gene because Atg7 cKO mice were derived by crossing Atg7F/F to the same Pcp2-Cre transgenic mice used for Becn1 cKO and loss of Atg7 protein is apparent at P19 [16], at a similar stage as seen in Becn1F/F;Pcp2-Cre mice (Figure S1C). The enhanced neuronal loss in Becn1 conditional KO mice is consistent with the early embryonic lethality of conventional Becn1 KO mice [15] as opposed to the neonatal death of conventional Atg7 KO mice [17].
Fig. 1. Loss of beclin 1 in selected neurons leads to rapid degeneration. 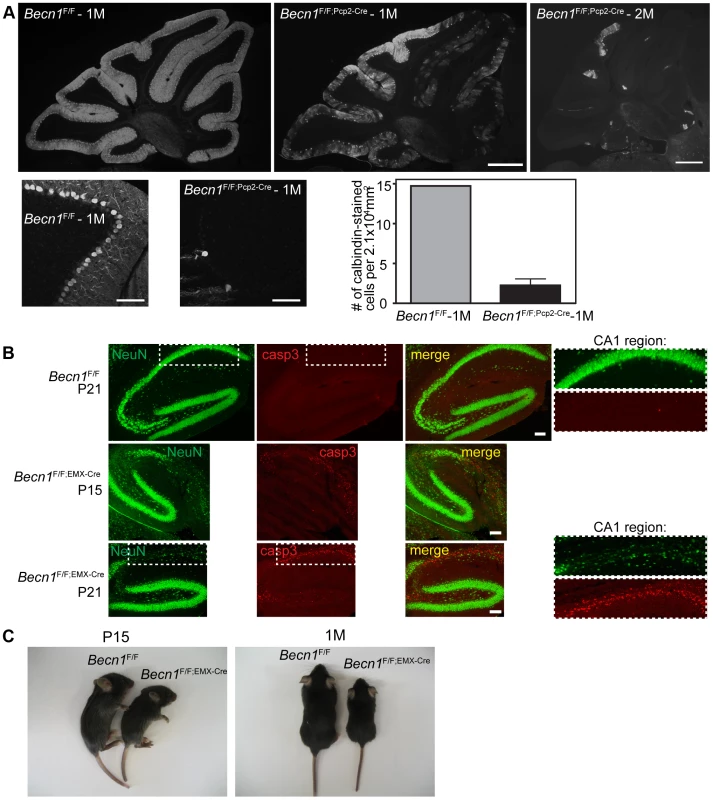
A. Severe PC loss is observed at 1 month (1M) and 2 months (2M) in Becn1 cKO mice. Calbindin 1 staining of cerebellum PCs from (left to right) Becn1F/F 1M, Becn1F/F;Pcp2-Cre 1M and Becn1F/F;Pcp2-Cre 2M mice. Scale bar = 0.5 mm. Lower panels show 20× images of Becn1F/F and Becn1F/F;Pcp2-Cre cerebellums at 1M. Scale bars = 100 µm. Quantification of 11 images each for 1 control and 3 cKO mice; bars are means +/− s.e.m. (n = 3). B. cKO of Becn1 in the hippocampus/cortex leads to caspase 3-dependent cell death. NeuN and caspase3 staining of Becn1F/F and Becn1F/F;EMX-Cre hippocampi from mice at indicated ages. Scale bars = 100 µm. Zoomed boxes show CA1 region. C. cKO of Becn1 in the hippocampus/cortex leads to reduced body size. Becn1F/F;EMX-Cre hippocampus/cortex-specific KO mice show reduced size. Photos of Becn1F/F and Becn1F/F;EMX-Cre mice at P15, 1M. To evaluate the impact of beclin 1 deletion on the viability of neurons, we generated a second Becn1 cKO mouse line Becn1F/F;EMX-Cre by crossing Becn1F/F with EMX-Cre transgenic mice to delete Becn1 in cortical and hippocampal neurons [18]. Ablation of Becn1 in hippocampal neurons indeed caused even earlier and more severe neurodegeneration than mutant PCs as shown by the overt reduction of neuron density at the CA1 region of the hippocampus in Becn1F/F;EMX-Cre mice at postnatal day 15 and 21 (Figure 1B). The degeneration of CA1 neurons was clearly associated with activated caspase 3 activity, suggesting that lack of beclin 1 leads to apoptotic neuron death (Figure 1B). In contrast, a recent study reported that deletion of Atg7 causes no loss of CA1 neurons at 3 months and only modest reduction of CA1 neurons at 15 months [19]. Consistently, we showed that lack of Atg7 causes slow, protracted dopamine neuron loss starting as late as 9 months [20]. Compared to Becn1F/F mice, Becn1F/F;EMX-Cre mice showed reduced body size from as early as P15, which persisted throughout postnatal development (Figure 1C; Figure S2B). We observed mortality in Becn1F/F;EMX-Cre mice from the age of 6 weeks, and almost all mutant mice died by the age of 4 months. The phenotype of Becn1F/F;EMX-Cre mice again demonstrated the significant enhancement of neurotoxicity caused by loss of beclin 1 compared to Atg7, suggesting that beclin 1 is associated with cell survival mechanisms distinct from Atg7-mediated autophagy.
We found swollen dystrophic axons of the PCs projected to the deep cerebellar nuclei (DCN) in Becn1F/F;Pcp2-Cre mice (Figure S2C). This observation is consistent with our study of Atg7F/F;Pcp2-Cre mice in which autophagy disruption causes axon dystrophy containing aberrant membrane structures [16]. In addition, Becn1F/F;EMX-Cre mice showed a clear increase in p62 levels in hippocampal neurons and cortical neurons compared to the control (Figure S3A). Immunofluorescence staining confirmed the enhancement of p62 levels in the cytoplasm of the CA1 region of Becn1 cKO mice (Figure S3B), whereas p62 staining was detected at background levels in control mice, consistent with the Westernblot analysis (Figure S3A). We expressed transgene GFP-LC3 in Becn1F/F;EMX-Cre mice and immunofluorescence staining showed that GFP-LC3 accumulated in the CA1 region together with elevated p62 staining (Figure S3C); in contrast, little GFP-LC3 signal was seen in the CA1 region of control Becn1F/F mice. Ubiquitinated protein levels were also increased in mutant CA1 areas compared to control (Figure S3D). Thus, as expected, lack of beclin 1 also caused impaired autophagy-lysosomal degradation in neurons.
Beclin 1 is associated with endosomes in neurons
To understand the cellular function of beclin 1 in neurons, we examined our newly-established BAC transgenic mouse line expressing beclin 1-GFP (green fluorescent protein) fusion protein in Purkinje cells under the control of the Pcp2 promoter (Figure 2A). Because of the poor quality of available anti-beclin 1 antibodies, our ability to detect endogenous beclin 1 localization was limited. Using a highly specific GFP antibody, we successfully detected GFP-LC3 subcellular localization in neurons assisted by immuno-EM analysis [21]. We therefore used the same anti-GFP antibody to examine beclin 1-GFP subcellular localization in neurons. Pcp2-BAC transgenics directed beclin 1-GFP expression selectively in cerebellar PCs (Figure 2B,C). We performed immuno-EM with anti-GFP antibody and found that immuno-reactive signal is enriched in endosomes suggesting that beclin 1-GFP is localized at endosomes (Figure 2D,E,F). This observation is in contrast to GFP-LC3 signals, which specifically labeled autophagosomes in the dystrophic axon terminals of PCs as reported by us previously [21].
Fig. 2. beclin 1-GFP expressed in the Purkinje cells of transgenic mice associates with endosomal organelles. 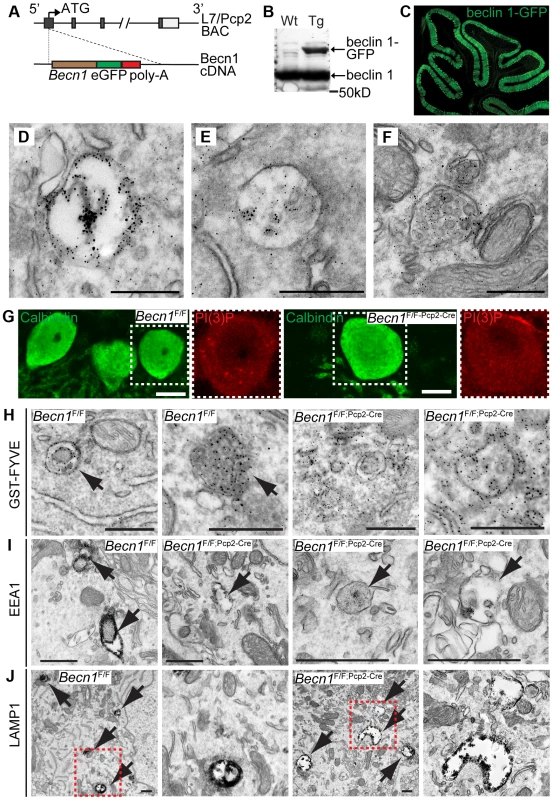
A. L7-Becn1-eGFP mice were generated using bacterial artificial chromosome (BAC) recombination techniques [52]. Schematic of the Becn1 gene fused to eGFP used to produce beclin 1-GFP in PCs using the L7/Pcp2 gene on BAC. B. beclin 1-GFP and endogenous beclin 1 are expressed in the brain. Anti-beclin 1 blot shows beclin 1-GFP in transgenic (Tg) mice. C. Transgenically expressed beclin 1-GFP is visualized specifically in the PCs of the cerebellum with anti-GFP antibody. Fluorescent image of the cerebellum of an L7-Becn1-GFP transgenic mouse. D.–F. beclin 1-GFP is found on membrane structures in PCs. Immuno-EM of the cerebellum of P21 L7-Becn1-GFP transgenic mice showing beclin 1-GFP on endosomes (D) and MVBs (E),(F). Scale bars = 250 nm. Loss of beclin 1 in Purkinje cells causes mislocalization of PI(3)P and abnormal endosomes and endolysosomes. G. Endogenous PI(3)P is punctate in Becn1F/F PCs but diffuse in Becn1F/F-Pcp2-Cre PCs. Immunofluorescent images of calbindin and PI(3)P stained Becn1F/F and Becn1F/F;Pcp2-Cre cerebellums of 1M mice. Dashed white boxes indicate zoomed image. Scale bars = 10 µm. H. GST-FYVE localizes to membranes in Becn1F/F PCs but is diffuse in Becn1F/F-Pcp2-Cre PCs. Immuno-EM images of cerebellar slices incubated with GST-FYVE and probed with anti-GST antibody. Scale bars = 200 nm. Arrow indicates endosome. I. EEA1 staining is intense in early endosomal structures in Becn1F/F control PCs but is reduced significantly on endosomes in Becn1F/F-Pcp2-Cre PCs. Immuno-EM micrographs of P21 Becn1F/F or Becn1F/F-Pcp2-Cre PCs stained with EEA1. Scale bars = 500 nm. J. LAMP1-stained vesicles are abnormally shaped and expanded in Becn1F/F-Pcp2-Cre cKO PCs. Immuno-EM micrographs of P21 Becn1F/F or Becn1F/F-Pcp2-Cre cKO PCs stained with LAMP1. Dashed red boxes indicate zoomed image. Scalebars = 500 nm. Loss of beclin 1 causes PI(3)P mislocalization in neurons and abnormal endosome formation in Becn1 null neurons
We next examined PI(3)P localization in PCs of Becn1F/F and Becn1F/F;Pcp2-Cre mice by immunofluorescence and found that while control PCs contain PI(3)P puncta, the PI(3)P signal was diffuse throughout Becn1 cKO cells (Figure 2G) suggesting that without beclin 1, PI(3)P was not appropriately localized. We next performed immuno-EM to confirm the observed PI(3)P localization using a GST-FYVE domain fusion protein that specifically binds to endogenous PI(3)P [22], [23]. In control brains GST-FYVE labeled endosomes, while in Becn1 cKO PCs GST-FYVE was not enriched in specific membrane structures and instead appeared diffuse throughout the cytosol (Figure 2H). We therefore conclude that endogenous beclin 1 is critical for proper targeting of PI(3)P to multiple membrane vesicles (e.g. endosomes) and thus the maintenance of the membrane homeostasis in neurons.
The localization of beclin 1-GFP on endosomes (Figure 2 D,E,F) and impaired PI(3)P localization in beclin 1-deficient neurons (Figure 2 G,H) implicates a function for beclin 1 in the endocytic pathway. We performed immuno-EM to examine endosomes in PCs using antibodies against the early endosome marker EEA1 and the late endosome/lysosome marker LAMP1. In control Becn1F/F PCs, EEA1 antibody-stained endosomes showed much stronger intensity (indicated by dense dark signals) than those in Becn1F/F;Pcp2-Cre PCs (Figure 2I), suggesting impaired recruitment of EEA1 to endosomes in the absence of beclin 1. Next we observed that LAMP1-labeled vacuoles in Becn1F/F;Pcp2-Cre PCs were significantly enlarged compared to those in control Becn1F/F PCs, suggesting aberrant formation of late endosome/lysosome in the absence of beclin 1 (Figure 2J).
Becn1 deficiency is associated with impaired endosome maturation and EGFR degradation
A previous study reported impaired internalization of epidermal growth factor receptor (EGFR), a process related to endocytosis, after Beclin 1 knock-down in HeLa cells [24], implicating a role for beclin 1 in the endocytic pathway. To validate the effect of beclin knock-down in the endocytic pathway we examined the uptake of pHrodo Red dextran that occurs during endocytosis associated with endosomal acidification [25]. We performed flow cytometry and observed that the mean fluorescence intensity in Beclin 1 deficient HeLa cells incubated with pHrodo dextran was lower than that of control cells, consistent with reduced transport to increasingly acidic endosomes (Figure 3A). Cells treated with the endosomal acidification inhibitor bafilomycin A showed overall reduced fluorescence by flow cytometry, which was further inhibited after Beclin 1 knock-down. Furthermore, Beclin 1 deficient cells showed decreased mean fluorescence of LysoSensor green dye suggesting an inhibition of endosome maturation (Figure 3B) [26]. We noticed that Beclin 1 knock-down in HeLa cells caused altered LC3II/LC3I ratios and near abolishment of UVRAG protein levels (Figure 3C). The result provides support to the notion that beclin 1 plays a role in late endosome formation (Figure 2J) or endosome maturation, in addition to autophagy.
Fig. 3. Beclin 1 deficient HeLa cells and MEFs display decreased endocytosis. 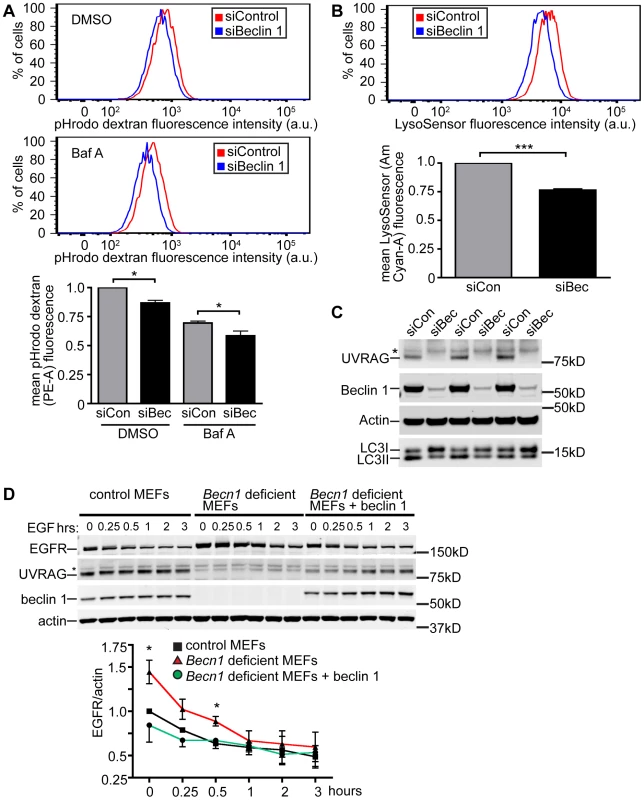
A. Hela cells treated with indicated siRNA (siCon is siControl; siBec is siBeclin 1) were treated with DMSO or Bafilomycin A1 (BafA). Representative composite fluorescence plots and normalized quantification of mean fluorescence intensity of 10,000 cells were determined from three separate experiments. Mean pHrodo dextran (PE-A) fluorescence; bars represent mean +/− s.e.m. (n = 3). DMSO pHrodo dextran siCon vs. siBec one sample t-test (two-tailed) p = 0.0283; BafA pHrodo dextran siCon vs. siBec one-tailed t-test p = 0.0246. B. Mean LysoSensor (Am Cyan-A) fluorescence intensity; bars represent mean +/− s.e.m. (n = 3). LysoSensor siCon vs. siBec one sample t-test (two-tailed) p = 0.0029. C. Anti-UVRAG, -Beclin 1, -Actin and –LC3 blots of HeLa cells lysed after indicated siRNA knock-down. Asterix indicates unspecific band. D. Receptor-mediated endocytosis as measured by EGFR internalization is inhibited in Becn1 deficient MEFs and rescued with re-introduction of beclin 1. Anti-EGFR, - UVRAG, -beclin 1 and –actin blots of control or Becn1 deficient MEFs lysed after indicated treatment with EGF. Asterix indicates unspecific band. Quantification of normalized EGFR/actin from 3 separate experiments. Bars represent mean +/− s.e.m. (n = 3); p = 0.0395, 0.0208 (one-tailed t-test). To assist the characterization of beclin 1 in the endocytic pathway, we generated genetic Becn1 deficient murine embryonic fibroblasts (MEFs). We next examined the impact of loss of beclin 1 in cells' ability to internalize and degrade EGFR upon EGF stimulation. We observed significantly increased EGFR protein levels in Becn1 deficient MEFs at steady state and early times points (0.5 h) post EGF treatment compared to control MEFs suggesting that EGFR is accumulated and not properly degraded without beclin 1 (Figure 3D). The defect was rescued by ectopic and stable expression of beclin 1 in Becn1 deficient MEFs (‘revertant MEFs’).
Reduced PI(3)P levels and VPS34 activity in Becn1 deficient cells
Our study of beclin 1 deficient neurons showed an aberrant distribution of PI(3)P (Figure 2G,H). It is unclear, however, whether beclin 1 deficiency affects intracellular PI(3)P and VPS34 protein levels. We next performed an ELISA for competitive binding to a PI(3)P-coated plate to determine the mass amount of PI(3)P cells in Becn1 deficient MEFs. The colorimetric signal was read at absorbance 450 nm and is inversely proportional to the amount of PI(3)P extracted from cells. The result indicates that intracellular PI(3)P content was significantly reduced in Becn1 deficient MEFs which was rescued in revertant MEFs (Figure 4A,B).
Fig. 4. PI(3)P and VPS34 lipid kinase activity is decreased in Becn1 deficient MEFs. 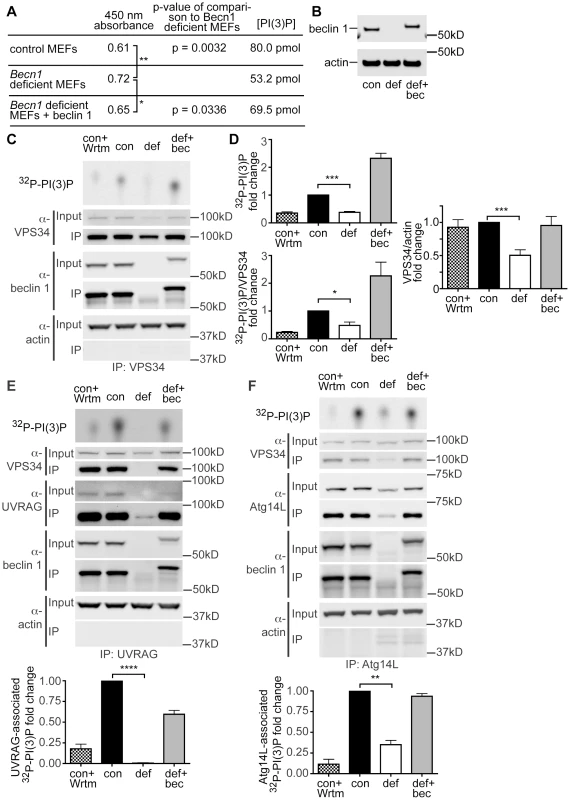
A. Table showing results from PI(3)P ELISA. Average 450 nm absorbance, p-value of comparison to Becn1 deficient MEFs. PI(3)P concentration is determined by comparison to a standard curve. Quantification of 450 nm absorbance of control, Becn1 deficient, and Becn1 revertant MEFs (6,5,6 replicates); statistics using a one-tailed t-test p = 0.0032, 0.0336. B. Anti-beclin 1, and -actin blots of MEF cell lysates. Con is control MEFs, def is Becn1 deficient MEFs, and rev is Becn1 revertant MEFs plus beclin 1. C. Autoradiograph of 32P-labelled PI(3)P and anti-VPS34, -beclin 1, and -actin blots after VPS34 IP of lysates from indicated MEFs with or without wortmannin (Wrtm) treatment. D. Quantification of normalized 32P-labelled PI(3)P and 32P-PI(3)P/VPS34 (from IP) from 3 separate experiments and VPS34/actin (from input lanes) from 9 separate experiments (including Fig. 4 E, F). Con is control MEFs, def is Becn1 deficient MEFs and def+bec is Becn1 revertant MEFs. Bars represent mean +/− s.e.m. (n = 3) con vs. def p = 0.0003, p = 0.0226, and p = 0.0002 top to bottom using a one sample t-test (two-tailed). UVRAG and Atg14-associated PI(3)P production is decreased in Becn1 deficient MEFs. E. Autoradiograph of 32P-labelled PI(3)P and anti-VPS34, -UVRAG, -beclin 1, and -actin blots after UVRAG IP of lysates from indicated MEFs with or without wortmannin (Wrtm) treatment. Con is control MEFs, def is Becn1 deficient MEFs and def+bec is Becn1 revertant MEFs. Quantification of normalized 32P-labelled PI(3)P from 3 separate experiments. Bars represent mean +/− s.e.m. (n = 3) con vs. def p<0.0001 using a one sample t-test (two-tailed). F. Autoradiograph of 32P-PI(3)P and anti-VPS34, -Atg14L, -beclin 1, and -actin blots after Atg14L IP of lysates from indicated MEFs with or without wortmannin (Wrtm) treatment. Quantification of normalized 32P-labelled PI(3)P from 3 separate experiments. Bars represent mean +/− s.e.m. (n = 3) con vs. def p = 0.0065 using a one sample t-test (two-tailed). We also examined VPS34 protein levels and VPS34-associated lipid kinase activity in Becn1 deficient MEFs. Control MEFs, Becn1 deficient MEFs, and Becn1 revertant MEFs were immunoprecipitated with VPS34 antibody and 32P-labelled PI(3)P was determined using thin-layer chromatography (TLC). As expected, control MEFs treated with the lipid kinase inhibitor wortmannin showed decreased 32P-PI(3)P (Figure 4C,D). Becn1 deficient MEFs showed decreased total 32P-PI(3)P levels and levels of 32P-PI(3)P normalized to the amount of VPS34 immunoprecipitated. The 32P-PI(3)P deficit was rescued with reintroduction of beclin 1 in the revertant MEFs. In addition, protein levels of VPS34 were also decreased in Becn1 deficient MEFs (Figure 4C,D). These results suggests that reduced levels of 32P-PI(3)P in Becn1 deficient MEFs is likely due to both decreased VPS34 protein levels and decreased VPS34 lipid kinase activity. Thus our results show that loss of beclin 1 caused a reduction of VPS34 protein level in addition to a decrease in VPS34 lipid kinase activity, leading to a decrease of production of intracellular PI(3)P, which in turn leads to an impairment of endocytosis in beclin 1 deficient cells and neurons.
Lack of beclin 1 results in abolishment of the UVRAG-VPS34 complex and UVRAG-associated VPS34 activity
UVRAG and Atg14L are the two stable components in distinct Beclin 1-VPS34 complexes and regulate VPS34 activity. We thus investigated the effect of beclin 1 deficiency on UVRAG or Atg14L levels and their associated VPS34 activities. In HeLa cells, knock-down of beclin 1 had a striking impact in the UVRAG protein levels, nearly eliminating the entire intracellular pool of UVRAG protein (Figure 3C). Consistently, UVRAG protein was almost undetectable in Becn1 deficient MEFs, but was brought back to normal levels in Becn1 revertant MEFs (Figure 3D). Immunoprecipitation with anti-UVRAG antibody pulled down hardly-visible amounts of UVRAG protein in Becn1 deficient MEFs, compared to wildtype control or revertant MEFs, correlated with a complete loss of UVRAG-associated VPS34 activity (Figure 4E). Interestingly, Atg14L protein level was reduced but clearly detectable in Becn1 deficient MEFs, and similarly it was recovered in Becn1 revertant MEFs (Figure 4F). Measurement of Atg14L-associated VPS34 activity showed approximately 65% reduction in Becn1 deficient MEFs as compared to that of WT control or revertant MEFs (Figure 4F). Our results indicate that loss of beclin 1 has a much more severe impact on UVRAG protein levels as well as UVRAG-associated VPS34 activity, compared that of Atg14L, and causes a complete loss of the UVRAG-VPS34 complex.
Disruption of autophagy in Becn1 deficient fibroblasts
To further characterize the Becn1 deficient MEFs we performed several assays to assess autophagy activity in the mutant cells. WIPI2 - and Atg12-associated phagophore numbers were significantly reduced in the absence of beclin 1 (Figure 5A). Consistent with the results from studies of Atg6 function in yeast, LC3 lipidation was not inhibited in Becn1 deficient MEFs in both basal and amino acid starvation conditions (Figure S4A). The number of mature autophagosomes (red spots) as well as immature autophagosomes (yellow spots) as visualized using the mCherry-GFP-LC3 fusion protein reporter were decreased in Becn1 deficient MEFs (Figure S4B). Also, long-lived protein degradation was inhibited in starvation conditions in Becn1 deficient MEFs. (Figure S4C). These results are reminiscent of the impairment of autophagy observed in VPS34 deficient cells [27].
Fig. 5. Autophagosome formation and endosomal expansion caused by constitutively active Rab5 and endocytosis are disrupted in Becn1 deficient MEFs. 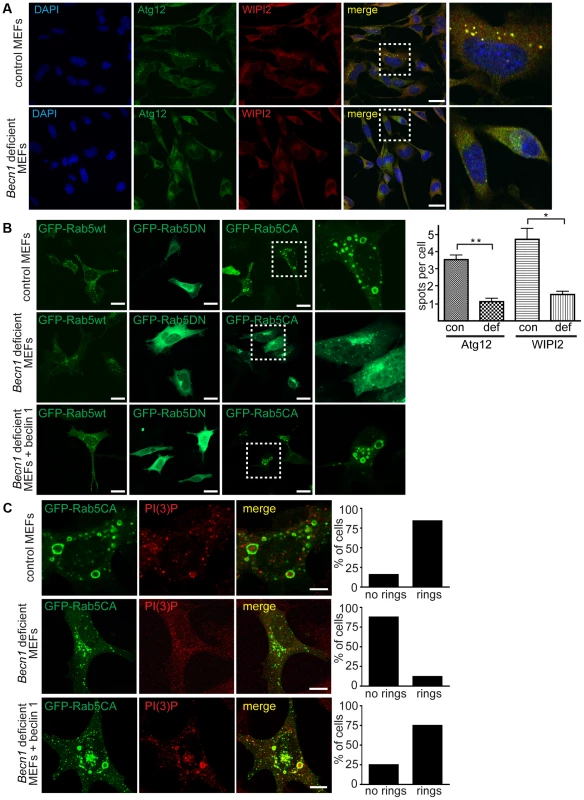
A. Reduction of Atg12 and WIPI2 [56] puncta formation in Becn1 deficient MEFs. DAPI, endogenous Atg12, WIPI2. White dashed boxes represent zoomed regions. Scalebars = 20 µm. ‘con’ is control MEFs, ‘def’ is beclin 1 deficient MEFs. Quantification of an average of 91 cells per condition from three separate experiments (con: 91, 110, 110; def: 64, 84, 87). Bars represent mean +/− s.e.m. (n = 3); Atg12 p = 0.0039; WIPI2 p = 0.0183 using a one-tailed t-test. B. Rab5-CA mutant is mislocalized in Becn1 deficient MEFs and rescued with re-introduction of beclin 1. MEF cells were transfected with Rab5wt, CA or DN-GFP and visualized. White dashed boxes represent zoomed regions. Scale bars = 20 µm. C. PI(3)P is recruited to expanding endosome sites in control MEFs but no endosome expansion associated with Rab5CA-GFP was found in Becn1 deficient MEFs. MEF cells were transfected with Rab5CA-GFP, fixed, stained with PI(3)P antibody and visualized. Scalebars = 10 µm. Quantification of percentage of cells with no rings or rings (at least 30 transfected cells from at least 3 separate experiments). Control MEFs (7 cells with no rings, 37 cells with rings) vs. Becn1 deficient MEFs (36,5) significantly different (p<0.0001) and Becn1 deficient MEFs (36,6) vs. Becn1 revertant MEFs (11,33) significantly different (p<0.0001) using Fisher's exact test (two-sided). Disruption of active Rab5 GTPase-induced endosome formation in Becn1 deficient fibroblasts
To further characterize the role of beclin 1 in VPS34-mediated endocytosis, we transfected control and Becn1 deficient MEFs with plasmids encoding variants of Rab5, a small GTPase that binds EEA1 [28] and the VPS34/Beclin 1 complex [29] and regulates early endosome trafficking [30]. Transfection of GFP-tagged Rab5 wildtype in control MEFs resulted in endosomal localization of GFP-Rab5wt as shown in the green fluorescent puncta. The distribution pattern of both GFP-Rab5 wildtype or dominant negative, GDP-bound mutant Rab5S34N (GFP-Rab5DN) [31] were not obviously changed in Becn1 deficient MEFs (Figure 5B). However, the constitutively active, GTP-bound mutant Rab5Q79L (GFP-Rab5CA) was frequently associated with enlarged, ring-shaped endocytic vesicles [32] in control MEFs but not in Becn1 deficient MEFs. Re-introduction of beclin 1 into Becn1 deficient MEFs by stably expressing wildtype beclin 1 induced a recovery of the enlarged endocytic vesicles. The distinct GFP-Rab5CA vesicles were decorated with PI(3)P, indicating that constitutively active Rab5 leads to excessive recruitment of VPS34 activity [33] and resulting endosome expansion, which was abolished in the absence of beclin 1 (Figure 5C). This result suggests that beclin 1 is required for the Rab5 function in recruiting VPS34 related to endosome function [24].
Impaired GFP-p40phox-associated endosome formation in Becn1 deficient cells is rescued by overexpression of UVRAG
To further investigate beclin 1's role in the endocytic pathway we expressed green fluorescent p40phox protein (GFP-p40phox), a PI(3)P binding protein and reporter for VPS34 activity [34], in Becn1 deficient MEFs. Overexpression of GFP-p40phox resulted in intense perinuclear fluorescence in control MEFs but reduced and disperse fluorescence in Becn1 deficient MEFs (Figure 6A). We performed correlative EM to investigate GFP-p40phox ultrastructures. In control MEFs the reporter protein labeled endosomes that clustered in a perinuclear area (Figure 6B). This result is consistent with previous observations that GFP-p40phox is located on endosomes [35], [36]. However, in the Becn1 deficient cells the fluorescent GFP-p40phox was not associated with endosomes and the few endosome-like vacuoles detected appear empty (Figure 6B). GFP-p40phox-labeled structures are altered in Becn1 deficient MEFs but restored to bright, juxtanuclear structures in Becn1 revertant MEFs (Figure 6C). Staining of GFP-p40phox structures with antibody against lipid lysobisphosphatidic acid (LBPA) (Figure 6D) [37] further supports that these structures are late endosomes. Importantly, when the GFP-p40phox was expressed in autophagy gene Atg5 deficient MEFs the dense, perinuclear GFP-p40phox signal was not altered compared to wildtype MEFs, suggesting the impact on endosome formation is beclin 1-related, rather than Atg5 - or autophagy-dependent (Figure 6E).
Fig. 6. There is a failure of endosome formation and localization in Becn1 deficient MEFs. 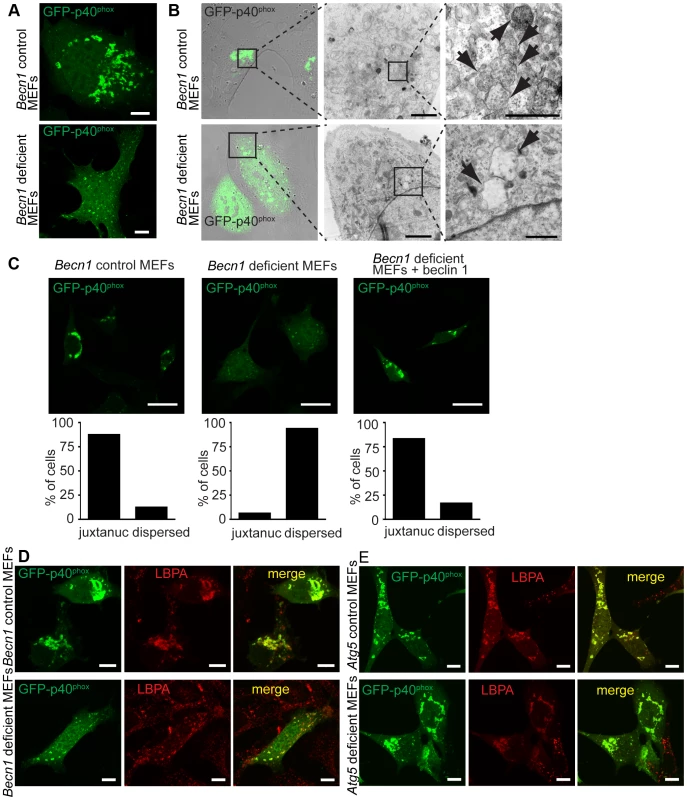
A. Transfected GFP-p40phox is seen in tight, bright juxta-nuclear structures in control MEFs but is dispersed in Becn1 deficient MEFs. Images of control and Becn1 deficient MEFs transfected with GFP-p40phox. Scalebars = 10 µm. B. GFP-p40phox fluorescence aligns with endosomes in control MEFs but is dispersed in Becn1 deficient MEFs where enlarged, empty vesicles are observed. Confocal and phase-contrast images and corresponding correlative EM images in control and Becn1 deficient MEFs transfected with GFP-p40phox. Boxes show sequential enlarged areas. Arrows indicate endosomes. Scalebars = 2 µm, 500 nm (LtoR). C. GFP-p40phox signal is altered in Becn1 deficient MEFs. Immunofluorescent images of control, Becn1 deficient and Becn1 revertant MEFs that were transfected with GFP-p40phox and fixed. Scalebars = 25 µm. Quantification of percentage of cells with tight juxtanuclear (‘juxtanuc’) or light, dispersed GFP-p40phox staining (at least 30 cells from at least 3 separate experiments). Control MEFs (28,4) vs. Becn1 deficient MEFs (2,30) significantly different (p<0.0001) and Becn1 deficient MEFs (2,30) vs. Becn1 revertant MEFs (25,5) significantly different (p<0.0001) using Fisher's exact test (two-sided). D. GFP-p40phox colocalizes with MVB marker LBPA. Immunofluorescent images of control and Becn1 deficient MEFs that were transfected with GFP-p40phox, fixed, and stained with anti-LBPA antibody. Scalebars = 10 µm. E. Deletion of autophagy protein Atg5 in MEFs does not affect intracellular GFP-p40phox distribution (does not lead to the GFP-p40phox dispersion phenotype seen in Becn1 deficient MEFs). GFP-p40phox is still seen in tight, bright juxta-nuclear structures in Atg5 deficient MEFs as it is in control MEFs. Atg5 deficient MEFs and corresponding control MEFs were transfected with GFP-p40phox, fixed, and stained with anti-LBPA antibody. Scalebars = 10 µm. Because beclin 1 forms multiple VPS34 complexes via binding different proteins [8], we next asked if the impairment of endosome formation is related to alteration of specific beclin 1 complexes. We overexpressed beclin 1 or beclin 1-binding proteins in Becn1 deficient MEFs and examined their effects on GFP-p40phox-associated endosome formation. As expected, overexpression of AsRed-tagged beclin 1 reversed the weak, dispersed GFP-p40phox signals to bright perinuclear clusters of GFP-p40phox (Figure 7A,B). We observed a large amount of colocalization between GFP-p40phox and beclin 1-AsRed, confirming our finding that beclin 1-GFP associates with endosome structures in the cerebellar PCs (Figure 2D,E,F). While Atg14L-AsRed or RUBICON-AsRed overexpression did not alter the impaired GFP-p40phox signals (Figure 7 C, D), UVRAG-FLAG overexpression resulted in nearly-complete restoration of GFP-p40phox to a bright, perinuclear cluster pattern, similar to beclin 1 expression (Figure 7E). This result demonstrates that the impairment of GFP-p40phox-associated endosome formation is reversed by forced expression of UVRAG in Becn1 deficient cells, and following on, suggests that the lack of GFP-p40phox-associated endosomes in Becn1 deficient cells is caused by the loss of UVRAG.
Fig. 7. Failure of endosome formation in Becn1 deficient MEFs is rescued by forced UVRAG expression and requires beclin 1-UVRAG interaction through beclin 1's CC domain. 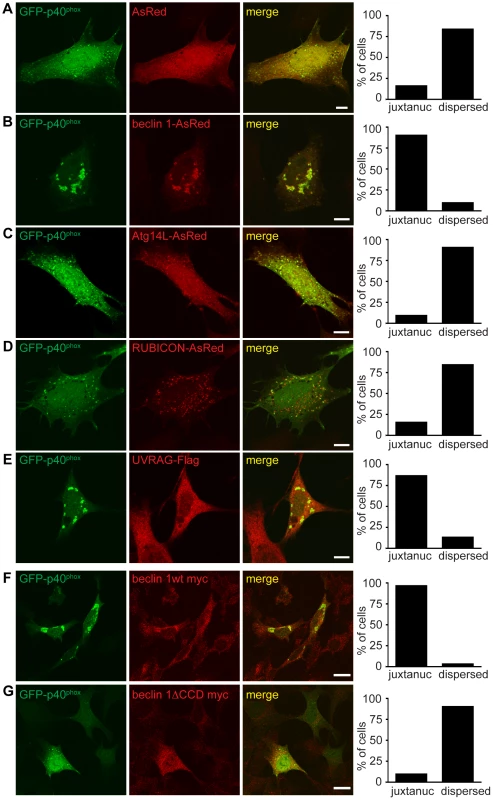
A–E. Dispersion of GFP-p40phox in Becn1 deficient MEFs is rescued with reintroduction of beclin 1 as well as overexpression of UVRAG but not Atg14L or RUBICON. Images of Becn1 deficient MEFs cotransfected with GFP-p40phox and AsRed (A), beclin 1-AsRed (B), Atg14L-AsRed (C), RUBICON-AsRed (D) and UVRAG-Flag and stained with anti-Flag antibody (E). Scale bars = 10 µm. Quantification as in Figure 6 E. AsRed (5,26) vs. beclin 1-AsRed (28,3) significantly different (p<0.0001); AsRed (5,26) vs. Atg14L-AsRed (3,29) not significantly different (p = 0.4741); AsRed (5,26) vs. RUBICON-AsRed (5,27) not significantly different (p = 1.0000); AsRed (5,26) vs. UVRAG-Flag (26,4) significantly different (p<0.0001) all using Fisher's exact test (two-sided). Rescue of the aberrant GFP-p40phox distribution in Becn1 deficient MEFs by beclin 1 overexpression requires the CC domain of beclin 1. A beclin 1 mutant lacking the CC domain does not rescue the GFP-p40phox phenotype of Becn1 deficient MEFs. Immunofluorescent images of Becn1 deficient MEFs transfected with beclin 1-myc wt (F) or beclin 1-myc minus its UVRAG-binding CC (beclin 1ΔCCD-myc) (G) then fixed and stained with anti-myc. Scalebars = 20 µm. Quantification as in Figure 6 E. beclin 1-myc wt (29,1) vs. beclin 1ΔCCD-myc (28,3) significantly different (p<0.0001) using Fisher's exact test (two-sided). A UVRAG-binding mutant of beclin 1 failed to rescue the impaired GFP-p40phox-associated endosomes in Becn1 deficient cells
Overexpression of beclin 1 mutants able to bind UVRAG but not Atg14L [38] rescued the impaired GFP-p40phox signals in Becn1 deficient MEFs in a similar manner to beclin 1 wildtype or UVRAG overexpression (Figure S5A). Becn1 deficient MEFs transfected with a beclin 1 monomer mutant (MutM) or a beclin 1 dimer mutant (MutStab) [38], both of which lose Atg14L binding but retain UVRAG binding, displayed bright juxtanuclear GFP-p40phox signals as seen in control MEFs (Figure 6A) or Becn1 deficient MEFs transfected with UVRAG (Figure 7E). This is consistent with the notion that Beclin1-Atg14L interaction is dispensable for proper late endosome formation.
Finally, we measured the ability of a UVRAG binding mutant of beclin 1 to rescue irregular GFP-p40phox distribution in Becn1 deficient MEFs. Beclin 1wt-myc transfection rescued abnormal GFP-p40phox distribution in Becn1 deficient MEFs just as beclin 1-AsRed did (Figure 7F,G). However, overexpression of beclin 1 missing its coiled-coil domain (beclin 1ΔCCD-myc) failed to rescue the GFP-p40phox dispersion phenotype in Becn1 deficient MEFs. Given that the beclin 1ΔCCD-myc mutant fails to bind UVRAG, this result suggests that the interaction of beclin 1 with UVRAG via their coiled-coil domains is critical for an intact endosome pathway.
Discussion
Our study identifies a key role for beclin 1 in the maintenance of homeostatic levels of multiple protein components of the PI3K-III complexes, including UVRAG, Atg14L and the catalytic subunit VPS34 as well as intracellular PI(3)P content. Importantly, beclin 1 is essential for UVRAG-VPS34 subcomplex integrity, which regulates the endosomal pathway. Furthermore, our data shows that lack of beclin 1 causes severe neurodegeneration in mice involving the disruption in both endocytosis and autophagy.
Mounting evidence links beclin 1 to an array of physiological functions and pathological pathways, which were ascribed mostly to its autophagy function [39]. Indeed, a previous report suggested that beclin 1 primarily engages VPS34 in autophagy, refuting the involvement of beclin 1 in VPS34-mediated non-autophagic vesicle trafficking [40]. Our current study demonstrates that beclin 1 participates in multiple organelle trafficking pathways including autophagy and endocytosis in neurons and fibroblasts. Beclin 1 or beclin 1 homologs were previously implicated in the degradative endocytic pathway in C. elegans [41], Drosophila [42] and mammalian cells [24] but the mechanisms were unclear.
Our current study suggests that the endocytic functions of beclin 1, strongly tied to UVRAG and VPS34 protein levels as well as VPS34 lipid kinase activity that produces PI(3)P, are constitutive and important for neuron viability, as we observe the collapse of endocytic trafficking associated with aberrant PI(3)P distribution and reduced levels due to loss of beclin 1. Beclin 1 null neurons display severe and accelerated degeneration as compared to autophagy-specific Atg5 or Atg7 deficient mice [43], [44], most likely as a result of combined deleterious effect of deficient autophagy and endocytosis. Our study therefore provides genetic proof for a previously poorly understood function of beclin 1 and expands our knowledge regarding various beclin 1-associated pathogenic mechanisms including those of tumorigenesis and neurodegenerative diseases.
Our study demonstrates a role for beclin 1 in proper endosome formation or maturation and PI3K-III-mediated endocytosis that is functionally separate from its role in autophagy in neurons and fibroblasts. Consistently, upon examination of EM images of both 3T3 cells after knock-down with beclin 1 siRNAs, we found an increase of large, empty vacuoles (Figure S5B) and a similar phenomenon is seen in Becn1 deficient MEFs (Figure S5C). These structures are similar to structures seen in LAMP1-labeled vacuoles in Becn1 null PCs (Figure 2 J), VPS34 null cells [27], cells treated with a PIKfyve lipid kinase inhibitor that disrupts endomembrane transport [45] and in bec-1 null C. elegans [41]. These structures, consisting of perhaps enlarged, aberrant late endosomes, suggest a defect in maturing endocytic compartments.
Our immuno-EM analysis reveals for the first time that beclin 1 localizes to endosomes (including multivesicular endosomes) in neurons under normal conditions (Figure 2D,E,F). The absence of beclin 1 causes impaired recruitment of endosome markers EEA1 and aberrant enlargement of late endocytic vacuoles (LAMP1-positive). In addition, we observed the disruption of active Rab5 GTPase-induced endocytic vesicles in fibroblasts, which we hypothesize may be due to the inability of Rab5 to recruit VPS34 in the absence of beclin 1 [33]. This result resembles the phenotype of loss of VPS34 due to failure of Rab5 to recruit VPS34 activity, which leads to reduced degradation of EGFR mediated by the endocytic pathway [24], [27]. Therefore, the role of beclin 1/Atg6 in PI3K-III-mediated endocytosis seems to be evolutionarily conserved [1]. Nonetheless, despite the present evidence for beclin 1 function in endosomal trafficking, we cannot exclude a possibility that beclin 1 is also involved in autophagosome-endosomes fusion (to form amphisomes), a process that is currently poorly understood. Overexpression of GFP-p40phox leads to bright, juxtanuclear signals that correspond to areas crowded with endosomes (visualized by EM), but in Becn1 deficient MEFs we observe very few and aberrant endosomes (Figure 6B). In addition to an inhibition of endosome formation we see mislocalization of the GFP-p40phox signal in Becn1 deficient MEFs. Although the nature of the abnormal GFP-p40phox structures are unclear, it may also suggest that beclin 1 is required to localize the VPS34 kinase complex to the correct membrane formation sites.
As expected, disruption of beclin 1 in fibroblasts results in impaired autophagy, shown through multiple autophagy assays. Phagophore numbers, as indicated by WIPI2 - and Atg12-positive spots were greatly diminished in Becn1 deficient MEFs (Figure 5A), which may be explained by a decrease in PI(3)P formation in the absence of beclin 1 (Figure 4). This is consistent with the role of WIPI2 as a linker of PI(3)P to the Atg12-5-16L and LC3 conjugation systems at the site of autophagosome formation [46]. It is worth mentioning that, unlike Atg genes that are involved in these ubiquitin-like conjugation systems, lack of beclin 1 does not abolish lipidated LC3II production; instead, it causes unchanged or even elevated levels of LC3II in fibroblasts (Figure S4B) and neurons (Figure S3A), consistent with the requirement of beclin 1 for LC3II-containing autophagosome degradation by lysosomes. Interestingly, we noted that siRNA knock-down of Beclin 1 in HeLa cells and genetic disruption of beclin 1 in MEFs altered the ratio of LC3II to LC3I under normal culture conditions (Figure 3C, Figure S4B), but Becn1 deficient MEFs showed little change in autophagy flux after bafilomycin A treatment (Figure S4B). While the cause of these observations remains to be tested, we speculate that a compensatory mechanism may be activated to degrade LC3II in the absence of beclin 1, such as beclin 1 homolog beclin 2-mediated autophagy [47]. Overall, autophagy was inhibited in starvation conditions in Becn1 deficient MEFs as measured by long-lived protein degradation (Figure S4C). It may be suggested that in the absence of beclin 1, the VPS34 kinase complex cannot be targeted to its appropriate membrane, whether that membrane be the phagophore that leads to autophagosomes or endosomal formation sites that become endosomes. This in turn, may lead to unregulated phosphorylation of PI(3)P, which combined with an overall reduction of PI(3)P production in the absence of beclin 1 (Figure 4) causes disruption of multiple membrane trafficking pathways such as autophagy and endocytosis.
Finally, our data suggests that the function of beclin 1 in endosome pathways depends on the interaction between beclin 1 and UVRAG, but not Atg14L or RUBICON, all of which are important binding partners of beclin 1 that exist in distinct beclin 1 complexes and modulate VPS34 lipid kinase activity. Thus our study suggests that the function of the beclin 1-UVRAG complex in the endosome pathway is distinct from its autophagy function primarily associated with the beclin 1-Atg14L complex. In fact, it was previously shown that UVRAG promotes endocytic trafficking [48], [49], [50] and interestingly, lack of beclin 1 results in a complete loss of the UVRAG-VPS34 complex (and associated kinase activity) as opposed to only a partial reduction of Atg14L-VPS34 complex levels (and associated kinase activity) (Figure 7A,B). Therefore, instability or loss of UVRAG in beclin 1 deficient cells is likely to account for the defective endocytosis, which can be rescued by forced expression of UVRAG. In addition, our previous data suggests that the coiled-coil domain interaction between beclin 1 and UVRAG has higher affinity than that of beclin 1 and Atg14L [38]. Therefore, the beclin 1-UVRAG interaction causes a highly stable beclin 1-UVRAG-VPS34 complex that is important for constitutive membrane trafficking (e.g. endocytosis) under normal conditions. Beclin 1 may switch to the Atg14L-containing VPS34 complex when cells face environmental changes that induce autophagy. Adding complexity, however, we cannot exclude the possibility that Atg14L is partially involved in endocytic abnormality in beclin 1 deficient cells [51] due to reduced Atg14L levels.
In summary, our investigation shows that beclin 1 participates in multiple organelle traffic pathways including autophagy and endocytosis via the maintenance of PI3K-III complex integrity and optimal lipid kinase activity and that disruption of multiple PI3K-III-mediated membrane trafficking pathways contributes to neurodegeneration in beclin 1 KO brains. Our data demonstrates a role for beclin 1 in regulating the endosome pathway via its interaction with UVRAG; this role is functionally distinct from autophagy. Our study expands our view of the biological function of beclin 1-VPS34 complex in membrane traffic and improves our understanding of the link between beclin 1 and the pathogenesis of multiple human diseases.
Materials and Methods
All animal studies were conducted in compliance with the IACUC committee of Icahn School of Medicine at Mount Sinai.
Generation of beclin 1 Flox/Flox mice
The Becn1flox/flox mice were generated using bacterial artificial chromosome (BAC) recombination techniques in a C57BL/6J strain background. The targeting BAC vector was generated so that two loxP sequences were inserted into the genome to flank the second exon of the Becn1 gene, where the start codon is located and the neomycin resistant gene pGKneo cassette flanked by two FRT sequences were inserted into the intron region between the second and the third exons (Figure S1A). Purified BAC DNA was electroporated into embryonic stem (ES) cells derived from C57BL/6J-Tyrc-2J/J mice, which are in a C57BL/6J genetic background but have white coat color due to a genetic mutation. Successful germline transmission from a 30% chimera mouse was confirmed with PCR and southern blot. The mice were further crossed to flipper mice to remove the pGKneo cassette flanked by FRT sequences. The resulted Becn1flox/+ mice were crossed to each other to generate Becn1flox/flox mice and genotyping was performed using both polymerase chain reaction (PCR) and southern blot (Figure S1 B).
Two shuttle vectors were used to generate the final vector for bacterial artificial chromosome (BAC) modification. S385 contains the two loxP sites and the pGKneo cassette flanked by two FRT sites whereas S351 contains the diphtheria toxin A-fragment gene (DT-A) for negative selection at gene targeting against random integration. A 1000 base pair DNA fragment (box A1) upstream of start codon of Becn1 gene was PCR amplified using BAC DNA as template and inserted into AscI and EcoRV sites of S351. Another DNA fragment about 500 bp (box A2) immediately downstream of box A1 that contains the second exon of Becn1 gene was PCR amplified and inserted into EcoRV and KpnI sites of S385. A third DNA fragment about 2000 bp (box B) immediately downstream of box A2 was subcloned into FseI and SmaI sites of S351 and was later used as short arm for homologous recombination in gene targeting. Finally, a 2000 bp DNA fragment (box C) downstream of box B was subcloned into XbaI site of S351, and was used for homologous recombination in the process of BAC modification. Eventually S385 containing all inserted fragments were linearized with NotI and PmeI digestion and ligated into NotI and SwaI sites of S351, generating the complete shuttle vector for BAC modification. Through modification, the loxP-A2-FRT-pGKneo-FRT-loxP fragment was replaced into the BAC clone, and the BAC DNA was used for gene targeting after PI-SceI digestion.
Primers used for cloning “boxes”:
A1 forward: 5′-TCAGGCGCGCCATGTGGGTGGATGGATTTGA-3′, AscI
A1 reverse: 5′-CTACCCGCTGCCTTGGTGGTGGAGG-3′
A2 forward: 5′-CCTCGGGCCCGAGGTTCAAAGGAGC-3′
A2 reverse: 5′-TCAGGTACCTGTAAGAGGATGCACCATGGCAC-3′, KpnI
B forward: 5′-TCAGGCCGGCCGACGAGTTGAGGTTAGAGCCATCAGTG-3′, FseI
B reverse: 5′-TCACCTCATTACCAGCCCCAACCCTACC-3′
C forward: 5′-GAGGCAGGCAGCTGAAGCCAGTGTG-3′
C reverse: 5′-TCAGTTTAAACACCTGCCGAGCCACCCCGTAACA-3′, XbaI
Gene targeting and blastocyst injection and generation of beclin 1 conditional knock-out mice
Linearized targeting vector was transfected into embryonic stem (ES) cells derived from C57BL/6J mice with electroporation. After that 50 clones were picked and the genomic DNA was tested with southern blot using a 500 bp DNA fragment downstream of box B (short arm of the targeting vector) as the probe. Positive clones were used for blastocyst injection in C57BL/6JC-2J mice. Chimera mice from blastocyst injection were crossed to wild type C57BL/6J mice, and pups with white coat color were genotyped with southern blot using the same probe as above.
For future genotyping analysis, the following primers were used for PCR genotyping:
Forward: 5′-CCACCACCAAGGCAGCGGGTAG-3′
Reverse: 5′-TCACTGATGGCTCTAACCTCAACTCGTC-3′
Positive mice were first crossed to transgenic mice that express FLP recombinase under the control of a broad promoter ROSA26 to remove the PGK-neo cassette flanked by two FRT sites from the genome, and the deletion was confirmed with southern blot.
For genotyping FLP mice, the following primers were used:
Forward: 5′-CTAATGTTGTGGGAAATTGGAGC-3′
Reverse: 5′-CTCGAGGATAACTTGTTTATTGC-3′
Becn1flox/+ mice were first crossed to each other to generate Becn1flox/flox mice, while Becn1flox/+ mice were crossed with Pcp2-Cre mice to generate Becn1flox/+;Cre/+ mice. Becn1flox/flox mice were further crossed with Becn1flox/+;Cre/+ mice to generate Becn1flox/flox;Cre/+ mice, in which beclin 1 expression is specifically deleted in cerebellar Purkinje cells (Pcp2-Cre) [16].
For genotyping Cre mice, the following primers were used:
Forward: 5′-CCGGTGAACGTGCAAAACAGGCTCTA-3′
Reverse: 3′-CTTCCAGGGCGCGAGTTGATAGC-3′
Generating L7-beclin 1-GFP mice
The L7-beclin 1-GFP mice were made by inserting a beclin 1-eGFP cDNA right after the ATG of the L7 gene using the same BAC modification protocol [52].
Generation of Becn1 deficient MEF cells
Primary mouse embryonic fibroblasts (MEFs) were generated from day 14 to 17 embryos from Becn1flox/+ matings. Early passage MEFs were immortalized using an SV40 large T antigen encoding plasmid. Immortalized MEFs were infected with either adenoviral GFP to generate Becn1+/+ or adenoviral Cre-GFP to generate Becn1−/− MEFs. Beclin 1 reconstituted MEFs were generated by infecting Becn1−/− MEFs with LPC retroviral vectors expressing beclin 1 (called “Becn1 deficient+beclin 1 MEFs” or “Becn1 revertant MEFs”). Becn1−/− control MEFs and Becn1−/− MEFs were transfected with LPC retroviral vectors alone (called “control MEFs” and “Becn1 deficient MEFs” respectively).
In vivo immunohistochemistry studies
Mice of different ages were put into anesthesia with intraperitoneal Nembutal injection at a dose of 0.01 ml/g body weight. Perfusion was performed with 4% paraformaldehyde in 0.1M phosphate buffer (pH 7.4) and dissected brains were post-fixed in the same fixative at 4°C for overnight. For floating sections, brains were embedded into 5% low melting agarose gel and 40 mm sections were cut with vibrotome (Leica). For cryosections, brains were subjected to cryo-protection in 30% sucrose at 4°C for overnight, and then were embedded into Neg-50 compound (Richard-Allan Scientific). 20 mm sections were cut with a cryostat (Leica). For paraffin sections, brains were dehydrated in series of ethanol (70%, 80%, 90%, 100%) for 30 minutes ×2, followed by incubation in Xylene for 3 days, and were embedded into wax. 10 mm sections were cut with a microtome (Leica). For immuno-fluorescence staining, sections were permeabilized in blocking buffer (10% goat serum and 0.1% Triton X-100 in PBS) and were incubated in primary antibodies diluted in blocking buffer overnight at 4°C. Anti-PI(3)P is from Echelon. Sections were then washed in PBS and incubated in Alexa Fluor conjugated secondary antibodies (Invitrogen) in PBS for 1 hour at room temperature. After washes in PBS, sections were mounted on slide glasses with fluorescence mounting medium (Vector Laboratories) and examined. For HRP staining with paraffin sections, antigen retrieval was first performed and sections were incubated in 0.5% H2O2 for 15 minutes at room temperature to block the endogenous peroxidase activity. Blocking and primary antibody incubation was performed in the same way as above. Biotinylated secondary antibody reaction was performed using Vectastain Elite ABC Kit (Vector Laboratories), followed by visualization with diaminobenzidine (DAB) (Sigma). For GST-FYVE immuno-EM experiments, cerebellums incubated with GST-FYVE and probed with anti-GST antibody (GE). Microscopes used were Zeiss 510 (Göttingen, Germany) upright and inverted confocal microscopes; 40× and 63× oil objectives were used.
Electron microscopy (EM)
For morphology EM on tissue sections, animals were perfusion fixed with 2% paraformaldehyde and 2% glutaraldehyde in 0.1M PB, pH 7.4. 40 mm vibrotome-cut sections were cut with. Blocks were cut with a diamond knife on a Leica UltracutE and ultra-thin (∼70 nm) sections were collected on uncoated 200-mesh grids. Grids were viewed with a TecnaiSpiritBT Transmission Electron Microscope (FEI) at 80 KV and pictures were taken with Gatan 895 ULTRASCAN Digital Camera. For morphology EM on cells, transfected HEK 293T cells were fixed in 2.5% glutaraldehyde (2.5GA) in 0.1M cacodylate buffer, pH 7.4 and processed by routine transmission electron microscopy procedure and embedded in Epon [11].
For immuno-EM, transfected HEK 293T cells were fixed in fresh PLP fixative (4% paraformaldehyde, 0.01M periodate, 0.075M lysine, and 0.075M phosphate buffer/pH 7.4) supplemented with 0.05% glutaraldehyde for one hour. After three washes in PBS, cells were permeabilized with 0.01% saponin in PBS supplemented with 0.1% BSA for 15 minutes at room temperature, and were incubated with primary antibody (anti-GFP) diluted in the same buffer overnight at 4°C. Vectastain Elite ABC kit was used for secondary antibody incubation, and cells were fixed again in 1.5% glutaraldehyde in 0.1 M cacodylate buffer supplemented with 5% sucrose. After three washes with 50 mM Tris/pH 7.4 supplemented with 7.5% sucrose, DAB reaction was performed with the same kit. The reaction was stopped with 50 mM Tris/pH 7.4 supplemented with 7.5% sucrose, and cells were processed for silver enhancement as [53]. Blocks were cut with a diamond knife on a Leica UltracutE and ultra-thin (∼70 nm) sections were collected on uncoated 200-mesh grids and stained with Uranium and Lead. Grids were viewed with a TecnaiSpiritBT Transmission Electron Microscope (FEI) at 80 KV and pictures were taken with Gatan 895 ULTRASCAN Digital Camera. Image levels were processed in Adobe Photoshop (Adobe Systems, San Jose, CA) to enhance contrast.
For correlative light and electron microscopy, cells were plated on 35 mm live imaging dishes with gridded glass bottoms (Mattek) for analysis by CLEM. Cells were fixed 24 h after transfection with 4% gluteraldehyde in 0.1 M sodium cacodylate buffer with 1 mM CaCl2. Confocal image stacks of transfected cells were acquired on a Zeiss LSM 510 META microscope. Differential interference contrast (DIC) and fluorescent images were acquired simultaneously. Initially, brightfield and DIC images taken at 10× low magnification were used to document the location of cells with respect to the coverslip grid. This step allows for identification of the cells of interest after processing for TEM. DIC and GFP LSCM z-stack images were then taken using a 63× objective. After imaging was complete, the sample was washed in sodium cacodylate buffer and treated with 1% osmium tetroxide, 1.5% potassium ferracyanide in 0.1 M cacodylate buffer for 1 h at 4°C. Cells were then dehydrated in ethanol using increasing steps of 50%, 60% and 70%, left in 3% uranyl acetate in 70% ethanol overnight, and then dehydrated further in 80%, 90% and 100% ethanol. After dehydration, cells were infiltrated with a 1∶1 mixture of resin (Embed 812 26 kit; Electron Microscopy Sciences) and 100% ethanol for 24 h. Resin/ethanol was replaced with a layer of pure resin, the cells of interest were enclosed by an open ended embedding capsule, then hardened in a vacuum oven at 65°C for 24 h. The capsule was topped off with resin and hardened in the oven for another 24 h. Lastly, the block was separated from the glass bottom dish by being placed on a hot plate at 60°C for ∼2–3 min and carefully peeled free from the dish. The imaged cells of interest were relocated on the resin block face using the superimposed coverslip grid. The block was trimmed using a Diatome cryotrim 45° trimming knife (Electron Microscopy Sciences), then serial sectioned at 70 nm using a Reichardt Ultracut E ultramicrotome. Serial section ribbons were transferred to 2×1 mm carbon reinforced slot grids (Electron Microscopy Sciences) then stained with 3% uranyl acetate for 40 min, washed, then stained with Reynold's lead citrate for 3 min. Cells of interest are initially located at low magnification in order to observed cell morphology and locate specific regions of interest based on comparison to the confocal GFP image. Cells and organelles were then imaged in serial sections at 10,000–30,000× magnification.
Cell cultures, transfections and constructs
Control MEFs, Becn1 deficient MEFs, Becn1 deficient plus beclin 1 MEFs, Human Embyronic Kidney (HEK) 293T cells, HeLa, and NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA). Transient DNA transfection was performed with either the standard calcium phosphate precipitation procedure, FuGene 6 or Lipofectamine2000, following protocols provided by the manufacturers. The Myc-tagged beclin 1 CCD deleted mutant, lacking the coiled-coil domain (aa 174–266), was constructed by overlapping extension PCR-based deletion mutagenesis and cloned into a PCMV-Myc vector with EcoRI and XhoI restriction sites.
Immunohistochemistry studies on fixed cells
Cells were fixed with 4% Pfa, permeabilized with 0.2% Triton, blocked in 1% BSA and were stained with anti-PI(3)P (Echelon), LBPA (Echelon), Atg12 (Cell Signaling #2011), WIPI2 (Abcam), Flag-tag M2 (Sigma), and Myc-tag 9B11 (Cell Signaling) and imaged on upright and inverted confocal microscopes made by Zeiss (Göttingen, Germany) including Zeiss 510 and Zeiss 780. 40× and 63× oil objectives were used. For quantification, at least 30 transfected cells imaged from at least three separate experiments were examined and categorized as indicated (i.e. no rings vs. rings).
EGFR degradation assay
Control, Becn1 deficient and Becn1 deficient plus beclin 1 MEFs were plated on poly-d-lysine coated 12 well plates. Once adherent, cells were serum-starved overnight. Cells were then stimulated with 200 ng/mL EGF (Millipore) in serum-free medium for the indicated time points. Cells were lysed in 1% TNTE buffer and homogenized with a 26G needle. Western blots were probed with anti-EGFR (Millipore), anti-beclin 1 (Santa Cruz), anti-UVRAG (Abgent), and anti-actin (Cell Signaling). EGFR/actin was quantified using Metamorph.
Endocytosis dye flow cytometry assay
Hela cells were treated with siControl or siBeclin siRNA and treated with DMSO or 100 nM Bafilomycin A1 (EMD Chemicals) for 1 hour. Cells were incubated with 50 µg/mL pHrodo Red dextran (MW, 10,000; P10361) or 1 µM LysoSensor Green (L7535) (both life technologies) for 20 minutes. Cells were harvested by trypsination, resuspended in ice-cold 0.2% BSA in PBS and analyzed immediately without fixation by flow cytometry using LSR-II (BD Biosciences) at an excitation/emission ratio of 560/585 nm for pHrodo Red dextran and 443/505 for LysoSensor Green. Mean PE-A fluorescence intensity and mean Am Cyan-A fluorescence intensity for 10,000 cells were determined.
PI(3)P ELISA
Lipid were extracted from MEF cells and samples were brought up in appropriate sample buffer volume based on lipid pellet weight. The PI(3)P ELISA was performed according to manufacturer's instructions using a PI(3)P Mass ELISA Kit (K-3300, Echelon). 450 nm absorbance was read on a VICTOR X4 plate reader (PerkinElmer) and PI(3)P concentration was determined using a standard curve generated during the ELISA.
Lipid kinase assay
MEF cells were lysed in 10 mM Tris-HCl pH 7.5, 2 mM EDTA, 100 mM NaCl, 1% NP-40, protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Roche). Immunoprecipitation was performed with 500 µg protein from post-nuclear supernatant, using antibodies against VPS34 (Echelon), UVRAG (Bethyl), or ATG14 (MBL) overnight. Samples were incubated with 30 µl Dynabeads (Life Technologies) for 1–2 hours at 4°C and then washed three times with lysis buffer. Two thirds of the beads were reserved for western blotting, while the remaining one third was used for the kinase assay. Beads for the kinase assay were washed once with wash buffer (20 mM HEPES pH 7.4, 1 mM EGTA, 0.4 mM EDTA, 5 mM MgCl2, and 0.05 mM DTT) and then resuspended to a final volume of 50 µl with reaction buffer (20 mM HEPES pH 7.4, 1 mM EGTA, 0.4 mM EDTA, 5 mM MgCl2, and 0.05 mM DTT, 50 µM cold ATP, 5 mM MnCl2, 0.1 mg/ml sonicated phosphatidylinositol). Note that 10 nm wortmannin was included in the indicated control MEFs. The reaction was started with the addition of γ-32P-ATP (5 µCi) and the samples were shaken at 37°C for 30 minutes. Reactions were stopped with 120 µl stop buffer (CHCl3/CH3OH/HCl at a 10∶20∶0.2 volume ratio) and then shaken for another 10 minutes at room temperature. Samples were centrifuged for 5 minutes at 1000 rcf. 15 µl of the lower, organic phase was resolved on silica coated TLC plate (Millipore) using CHCL3/CH3OH/NH4OH/H2O (86∶76∶10∶14 volume ratio) and visualized with the Typhoon 9400 Variable Imager (GE Healthcare Biosciences).
Autophagy assays and long-lived protein degradation
Autophagy assays were performed as in [6] and [54]. Long-lived protein degradation was assessed as described previously [55]. Control and Becn1 deficient MEFs were plated in 12-well plates. After 48 h, DMEM was changed to leucine-free medium supplemented with 3H-l-leucine (1 µCi ml–1). After pulse-labeling for 24 h, cells were washed three times and cultured in DMEM supplemented with excess unlabelled leucine (5 mM) for 16 h to chase out short-lived proteins. Cells were then washed three times and further cultured for 4 h in DMEM, Earle's Balanced salt solution (EBSS), or EBSS supplemented with 10 mM 3-methyladenine, all containing unlabelled leucine (5 mM). Both medium and cell lysates were subject to trichloroacetic acid (TCA) precipitation. Long-lived protein degradation was calculated as the ratio of TCA - soluble medium to TCA-precipitated cell lysate radioactivity.
Supporting Information
Zdroje
1. WirawanE, LippensS, Vanden BergheT, RomagnoliA, FimiaGM, et al. (2012) Beclin1: a role in membrane dynamics and beyond. Autophagy 8 : 6–17.
2. SimonsenA, ToozeSA (2009) Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 186 : 773–782.
3. YueZ, ZhongY (2010) From a global view to focused examination: understanding cellular function of lipid kinase VPS34-Beclin 1 complex in autophagy. J Mol Cell Biol 2 : 305–307.
4. LevineB, KroemerG (2008) Autophagy in the pathogenesis of disease. Cell 132 : 27–42.
5. McKnight NC, Mizushima N, Yue Z (2012) The Cellular Process of Autophagy and Control of Autophagy in Neurons. In: Yue Z, Chu CT, editors. Autophagy of the Nervous System. Singapore: World Scientific Publishing Co. Pte. Ltd. pp. 3–35.
6. ZhongY, WangQJ, LiX, YanY, BackerJM, et al. (2009) Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 11 : 468–476.
7. MatsunagaK, SaitohT, TabataK, OmoriH, SatohT, et al. (2009) Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 11 : 385–396.
8. FunderburkSF, WangQJ, YueZ (2010) The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends in cell biology 20 : 355–362.
9. McKnightN, YueZ (2013) Beclin 1, an Essential Component and Master Regulator of PI3K-III in Health and Disease. Curr Pathobiol Rep 1 : 231–238.
10. DiskinT, Tal-OrP, ErlichS, MizrachyL, AlexandrovichA, et al. (2005) Closed head injury induces upregulation of Beclin 1 at the cortical site of injury. J Neurotrauma 22 : 750–762.
11. YueZ, HortonA, BravinM, DeJagerPL, SelimiF, et al. (2002) A novel protein complex linking the delta 2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron 35 : 921–933.
12. PickfordF, MasliahE, BritschgiM, LucinK, NarasimhanR, et al. (2008) The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest 118 : 2190–2199.
13. SpencerB, PotkarR, TrejoM, RockensteinE, PatrickC, et al. (2009) Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in alpha-synuclein models of Parkinson's and Lewy body diseases. The Journal of neuroscience : the official journal of the Society for Neuroscience 29 : 13578–13588.
14. Nascimento-FerreiraI, Santos-FerreiraT, Sousa-FerreiraL, AureganG, OnofreI, et al. (2011) Overexpression of the autophagic beclin-1 protein clears mutant ataxin-3 and alleviates Machado-Joseph disease. Brain : a journal of neurology 134 : 1400–1415.
15. YueZ, JinS, YangC, LevineAJ, HeintzN (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 100 : 15077–15082.
16. KomatsuM, WangQJ, HolsteinGR, FriedrichVLJr, IwataJ, et al. (2007) Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A 104 : 14489–14494.
17. KomatsuM, WaguriS, UenoT, IwataJ, MurataS, et al. (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The Journal of cell biology 169 : 425–434.
18. GuX, LiC, WeiW, LoV, GongS, et al. (2005) Pathological cell-cell interactions elicited by a neuropathogenic form of mutant Huntingtin contribute to cortical pathogenesis in HD mice. Neuron 46 : 433–444.
19. InoueK, RispoliJ, KaphzanH, KlannE, ChenEI, et al. (2012) Macroautophagy deficiency mediates age-dependent neurodegeneration through a phospho-tau pathway. Mol Neurodegener 7 : 48.
20. FriedmanLG, LachenmayerML, WangJ, HeL, PouloseSM, et al. (2012) Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of alpha-synuclein and LRRK2 in the brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 32 : 7585–7593.
21. WangQJ, DingY, KohtzDS, MizushimaN, CristeaIM, et al. (2006) Induction of autophagy in axonal dystrophy and degeneration. The Journal of neuroscience : the official journal of the Society for Neuroscience 26 : 8057–8068.
22. GaullierJM, SimonsenA, D'ArrigoA, BremnesB, StenmarkH, et al. (1998) FYVE fingers bind PtdIns(3)P. Nature 394 : 432–433.
23. GilloolyDJ, MorrowIC, LindsayM, GouldR, BryantNJ, et al. (2000) Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. The EMBO journal 19 : 4577–4588.
24. ThoresenSB, PedersenNM, LiestolK, StenmarkH (2010) A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res 316 : 3368–3378.
25. ZengX, CarlinCR (2013) Host cell autophagy modulates early stages of adenovirus infections in airway epithelial cells. J Virol 87 : 2307–2319.
26. RabenN, SheaL, HillV, PlotzP (2009) Monitoring autophagy in lysosomal storage disorders. Methods Enzymol 453 : 417–449.
27. JaberN, DouZ, ChenJS, CatanzaroJ, JiangYP, et al. (2012) Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proceedings of the National Academy of Sciences of the United States of America 109 : 2003–2008.
28. SimonsenA, LippeR, ChristoforidisS, GaullierJM, BrechA, et al. (1998) EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394 : 494–498.
29. RavikumarB, ImarisioS, SarkarS, O'KaneCJ, RubinszteinDC (2008) Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. Journal of cell science 121 : 1649–1660.
30. StenmarkH (2009) Rab GTPases as coordinators of vesicle traffic. Nature reviews Molecular cell biology 10 : 513–525.
31. VolpicelliLA, LahJJ, LeveyAI (2001) Rab5-dependent trafficking of the m4 muscarinic acetylcholine receptor to the plasma membrane, early endosomes, and multivesicular bodies. The Journal of biological chemistry 276 : 47590–47598.
32. MorelE, ChamounZ, LasieckaZM, ChanRB, WilliamsonRL, et al. (2013) Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat Commun 4 : 2250.
33. MurrayJT, PanaretouC, StenmarkH, MiaczynskaM, BackerJM (2002) Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic 3 : 416–427.
34. LiangC, FengP, KuB, DotanI, CanaaniD, et al. (2006) Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 8 : 688–699.
35. UeyamaT, TatsunoT, KawasakiT, TsujibeS, ShiraiY, et al. (2007) A regulated adaptor function of p40phox: distinct p67phox membrane targeting by p40phox and by p47phox. Molecular biology of the cell 18 : 441–454.
36. LambFS, HookJS, HilkinBM, HuberJN, VolkAP, et al. (2012) Endotoxin priming of neutrophils requires endocytosis and NADPH oxidase-dependent endosomal reactive oxygen species. The Journal of biological chemistry 287 : 12395–12404.
37. KobayashiT, BeuchatMH, LindsayM, FriasS, PalmiterRD, et al. (1999) Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nature cell biology 1 : 113–118.
38. LiX, HeL, CheKH, FunderburkSF, PanL, et al. (2012) Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun 3 : 662.
39. MizushimaN, LevineB, CuervoAM, KlionskyDJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451 : 1069–1075.
40. ZengX, OvermeyerJH, MalteseWA (2006) Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. Journal of cell science 119 : 259–270.
41. RuckA, AttonitoJ, GarcesKT, NunezL, PalmisanoNJ, et al. (2011) The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy 7 : 386–400.
42. ShravageBV, HillJH, PowersCM, WuL, BaehreckeEH (2013) Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development
43. HaraT, NakamuraK, MatsuiM, YamamotoA, NakaharaY, et al. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441 : 885–889.
44. KomatsuM, WaguriS, ChibaT, MurataS, IwataJ, et al. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441 : 880–884.
45. JefferiesHB, CookeFT, JatP, BoucheronC, KoizumiT, et al. (2008) A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO reports 9 : 164–170.
46. DooleyHC, RaziM, PolsonHE, GirardinSE, WilsonMI, et al. (2014) WIPI2 Links LC3 Conjugation with PI3P, Autophagosome Formation, and Pathogen Clearance by Recruiting Atg12-5-16L1. Molecular cell 55 : 238–252.
47. HeC, WeiY, SunK, LiB, DongX, et al. (2013) Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism. Cell 154 : 1085–1099.
48. LiangC, LeeJS, InnKS, GackMU, LiQ, et al. (2008) Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 10 : 776–787.
49. HeS, NiD, MaB, LeeJH, ZhangT, et al. (2013) PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nature cell biology 15 : 1206–1219.
50. PiroozSD, HeS, ZhangT, ZhangX, ZhaoZ, et al. (2014) UVRAG is required for virus entry through combinatorial interaction with the class C-Vps complex and SNAREs. Proceedings of the National Academy of Sciences of the United States of America 111 : 2716–2721.
51. KimHJ, ZhongQ, ShengZH, YoshimoriT, LiangC, et al. (2012) Beclin-1-interacting autophagy protein Atg14L targets the SNARE-associated protein Snapin to coordinate endocytic trafficking. Journal of cell science 125 : 4740–4750.
52. GongS, YangXW, LiC, HeintzN (2002) Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res 12 : 1992–1998.
53. Teclemariam-MesbahR, WortelJ, RomijnHJ, BuijsRM (1997) A simple silver-gold intensification procedure for double DAB labeling studies in electron microscopy. J Histochem Cytochem 45 : 619–621.
54. McKnightNC, JefferiesHB, AlemuEA, SaundersRE, HowellM, et al. (2012) Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. The EMBO journal 31 : 1931–1946.
55. GronostajskiRM, PardeeAB (1984) Protein degradation in 3T3 cells and tumorigenic transformed 3T3 cells. J Cell Physiol 119 : 127–132.
56. PolsonHE, de LartigueJ, RigdenDJ, ReedijkM, UrbeS, et al. (2010) Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6 : 506–522.
Štítky
Genetika Reprodukčná medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



