-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
Recent developments in high throughput applications to manipulate and inactivate specific genes in mouse embryonic stem cells (ES cells) have allowed for the initiation of large scale reverse genetic screens in the mouse. The immediate connection of a phenotype to a mutated (null) gene represents a paradigm shift in our ability to explore gene function. This study utilized such a screening approach to investigate the genetic contribution to skin development and homeostasis. Not only does this approach provide insight into the genetics of skin biology, it is also instrumental in generating novel models with which to study the genetic underpinnings of skin disease. Initial screening of 562 mutated genes in mice uncovered previously unrecognized genes involved in the biology of this organ and identified novel functions for previously studied genes associated with epidermal phenotypes. Taken together, these results highlight high throughput screening approaches that are valuable in reverse genetic screening and provide a pool of mouse mutants, available to the scientific community, that will serve as the basis for further detailed investigations into skin function and skin disease.
Published in the journal: Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse. PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004705
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004705Summary
Recent developments in high throughput applications to manipulate and inactivate specific genes in mouse embryonic stem cells (ES cells) have allowed for the initiation of large scale reverse genetic screens in the mouse. The immediate connection of a phenotype to a mutated (null) gene represents a paradigm shift in our ability to explore gene function. This study utilized such a screening approach to investigate the genetic contribution to skin development and homeostasis. Not only does this approach provide insight into the genetics of skin biology, it is also instrumental in generating novel models with which to study the genetic underpinnings of skin disease. Initial screening of 562 mutated genes in mice uncovered previously unrecognized genes involved in the biology of this organ and identified novel functions for previously studied genes associated with epidermal phenotypes. Taken together, these results highlight high throughput screening approaches that are valuable in reverse genetic screening and provide a pool of mouse mutants, available to the scientific community, that will serve as the basis for further detailed investigations into skin function and skin disease.
Introduction
Recent advances in the high throughput production of targeted mutant mice using gene targeting in ES cells have made it possible to rapidly and effectively produce knockout mouse lines for use in phenotypic screening [1], [2], [3], [4]. Such approaches are now beginning to yield significant insights into mammalian gene function [1], [5]. Central to recent community efforts has been the Wellcome Trust Sanger Institute's Mouse Genetics Project (Sanger-MGP), which is undertaking a high throughput reverse genetic screen using the knockout ES cells generated by the International Knockout Mouse Consortium, which has now merged with the International Phenotyping Mouse Consortium (www.mousephenotype.org). To maximise the value of this resource, it is important to develop organ specific secondary screens to complement the broad phenotyping efforts applied in existing production pipelines. To this end, we implemented a multi-parameter, multi-test skin screen to identify mouse models of skin dysfunction and to elucidate genetic pathways important for skin development and function. In this study, we detail the skin assessment pipelines internal to the Sanger-MGP as well as skin analyses performed externally, with the exception of the fluorescent tail whole mount analysis, published elsewhere [6]. A flowchart for the pipeline is summarised in Figure 1A and presented in more detail in Figure S1A.
Fig. 1. Overview of pipeline, hair follicle cycling baseline and phenotypes. 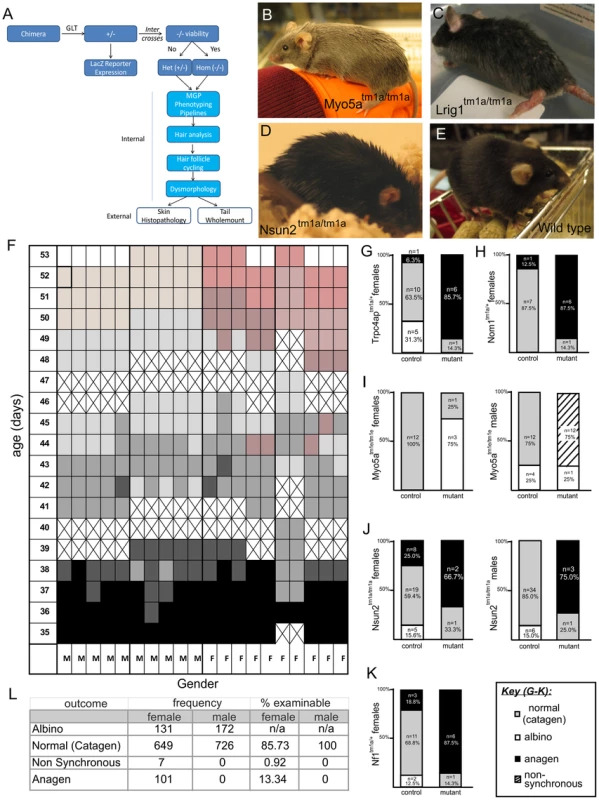
A) Flow-chart illulstrating pipeline of skin phenotyping. B–E) Examples of obvious coat phenotypes in Myo5a, Lrig1 and Nsun2 strains relative to wild-type mice. F) 10 male and 10 female pigmented wild type mice were shaved and skin colour assessed from 35–53 days of age. Results are shown in a grid where cell color represents skin colour. Black indicates anagen, shades of grey (and dark pink) indicate catagen, pale pink indicates telogen for males, pink indicates telogen for females, and crosses indicate days mice were not assessed. Mutant mice and matched wild type controls were next shaved in weekly cohorts and dorsal skin assessed for hair cycle phase using the skin color method above at an age of 41–43 days. Pigmented mice were assessed as normal (grey skin/in catagen), non synchronous (mixed patches of hair cycle phases), and anagen (black skin). Albino mice could not be assessed and were excluded from the analysis. G–K) Shows Nf1tm1a; Nsun2tm1a; Myo5atm1e; Trpc4aptm1a; Nom1tm1a demonstrated signs of abnormally in constrast to their wild type controls. L) Table summarises wild type findings for baseline reference. The skin is the largest of the intermediate sized organs in the human body [7] and it plays integral roles in immune surveillance, wound healing and protection from environmental challenge [8]. Skin related conditions account for significant health expenditure and top the list of reasons for general practice appointments in the U.S. [9]. A considerable number of skin conditions have genetic underpinnings [10]. Therefore, understanding the genetic basis of skin development and skin disorders has the potential to greatly impact both the diagnosis and treatment of skin related conditions. Moreover, a significant number of other systemic diseases or those principally affecting other organs also manifest themselves in the skin. Such conditions include diabetes mellitus, chronic kidney disease, celiac disease, and rheumatoid arthritis. As a highly regenerative organ in which developmental gene expression is often recapitulated during adulthood, the skin also provides a window into many of the programs central to differentiation and homeostasis. The skin and its associated “mini organ”, the hair follicle, are particularly attractive targets for such a screen because of their ready accessibility and dynamic nature [11]. Furthermore, the clinical pathology and physiology based mouse phenotyping “pipelines” are not designed to find subtle lesions affecting the skin [12]. For these reasons a skin specific phenotyping program was implemented on the genetically modified mice produced by the Sanger-MGP with a goal of defining new and previously unappreciated genetic determinants of skin disease.
In this paper mouse lines carrying mutations in each of 562 genes were investigated, and phenotypes were identified as a result of mutations in 23 genes (Table S1). These skin studies have identified genes not previously associated with skin phenotypes and ascribe further functions to genes which were previously associated with skin disease. We also used these tests to assign 25 Mammalian Phenotype (MP) ontology terms [13] (http://www.informatics.jax.org/searches/MP_form.shtml), 2 of which are novel (Table S1). The scale of the screen also provides the opportunity to assess the relative value of different phenotyping approaches, with a view to implementing these screens in other proposed and ongoing high throughput reverse genetic screens.
Results and Discussion
The Screen
The skin phenotyping screen
The vast majority of the mutant lines examined in this study (>95%) were derived from the EUCOMM/KOMP knockout first conditional-ready targeted ES cell resource on a C57BL/6N background [5]. However, to assess germline transmission mice were crossed with albino mice, homozygous for the recessive albino allele, C57BL/6J Tyrc-Brd. This strategy preserves the C57BL/6 genetic background while indicating germline transmission by simple black/white coat colour. In subsequent breeding this white coat phenotype was coincidentally inherited along with the targeted cassette. Full details of the genetic background of strains are presented in Table S1. These lines are predicted to produce null alleles as a consequence of the insertion of the strong Engrailed-2 splice acceptor upstream of a lacZ reporter cassette followed by a SV40 polyA to terminate gene transcription and prevent potential splicing with downstream exons [14] (Figure S2). Primary screens were carried out either on viable homozygotes or, in the case of those alleles which exhibited embryonic lethality, on heterozygote animals (Table S1). Over the course of testing, mutant mice underwent visual tests to screen for obvious phenotypes (typically 7 male and 7 female mutants per line, Table S1, Figure 1A). A number of these were designed specifically to assess cutaneous phenotypes. As a requirement for other non-skin phenotyping pipelines performed in parallel to our skin screen, mice were fed on a high fat diet. The altered diet did not cause overt changes in skin phenotypes. The appearance of the coat was initially assessed at 4 weeks of age, prior to the implementation of the high fat diet, then a detailed visual assessment of the skin, nail units, pelage, and teeth was performed at 10 weeks of age. At the time of necropsy (16 weeks of age) dorsal skin was collected for histology and immunohistochemical assays. Hair follicle cycling defects were evaluated at 6 weeks of age by shaving a patch of hair at the dorsal thoraco-lumbar region. We found this time point to coincide with completion of the synchronous, second postnatal anagen phase of the hair cycle and catagen transition into telogen [15]. The hair analysis and hair follicle cycling screens were included as a part of the internal primary pipeline at the Sanger-MGP, whereas the screen for skin histopathology was conducted externally. We grouped the characterisation into three screens (Hair analysis and visual dysmorphology; Hair follicle cycling; Skin histopathology) and used them to examine 562 unique gene knockouts. We identified 23 genes (4.1%) with 25 putative phenotypes (Table S1) related to skin biology. One such gene identified from the screen, Keratin 76, is characterised in depth in an accompanying manuscript in this issue of PLoS Genetics [16]. The results of each of these specific screens are discussed below.
Hair analysis and visual dysmorphology screening
The simplest aspect of the skin screen comprises the visual assay for overt abnormalities of the pelage (Figure 1B–E). This key primary screen was performed in the Sanger-MGP pipeline. Parameters relevant to skin in this screen include pelage appearance and coloration, skin and footpad pigmentation (excluding albino mice), facial vibrissae morphology, and length, morphology and number of nail units and teeth. Of the 263 phenotype parameters assessed in the Sanger-MGP pipeline (comprising 147 categorical and 116 continuous measurements), 43 (16.4%) were directly related to skin and hair [1](Table S2). In addition, 362 different genes were screened identifying 23 that exhibited abnormal phenotypes in one or more skin-related organ systems (Table S1). Of these hits, 9 were directly related to skin and hair descriptors (Table 1) making skin defects the most common gross morphological finding together with craniofacial defects (n = 9) and defects in the eye (n = 4), tail/limbs (n = 2), and genitalia (n = 2). Of the 9 lines that exhibited a cutaneous phenotype, the most common reported phenotype was abnormal pigmentation (n = 5/9; Krt76, Myo5a (Figure 1B), Mysm1, Vangl1, Sparc) followed by general integument phenotypes (scruffy coat, greasy coat, scaly skin, hair loss n = 4/9, Krt76, Lrig1 (Figure 1C), Myo6, Nsun2 (Figure 1D)) and nail phenotypes (n = 1/9 Prkab1) (Table 1). A wild type mouse is shown in Figure 1E for comparison. We also examined the expression patterns of these 9 genes utilising the activity of the integrated beta galactosidase (lacZ) reporter gene and found that with the exception of two genes (Myo5a and Prkab1) all were expressed in the epidermis or dermis (Table 1, see later figures and Figure S3 for examples of specific genes). Sparc lacZ expression in tail whole mounts was ubiquitous (https://www.mousephenotype.org/data/genes/MGI:98373). Myo6 expression in ear epithelia has been reported using IHC [17]. While Myo5a and Prkab1 expression were not detected via the lacZ reporter epidermal expression of Myo5a has been previously reported [18], and Prkab1 has wide ranging tissue expression [19], suggesting a lacZ reporter sensitivity issue.
Tab. 1. Genes identified in the primary phenotypic screen with cutaneous defects. 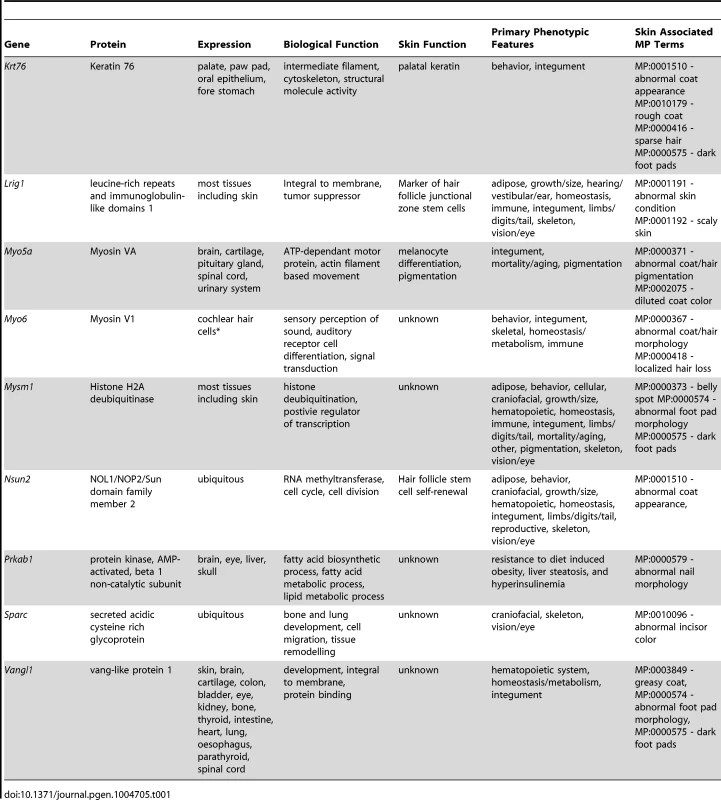
Hair follicle cycling
The hair follicle is a dynamic mini organ that continually undergoes a controlled program of growth (anagen), regression (catagen), and resting (telogen). Accordingly, hair follicle morphogenesis is maintained by a complex program of proliferation, differentiation and death. Regulators of this process include developmentally important proteins such as members of the bone morphogenetic protein (BMP), hedgehog (HH) and wingless-related MMTV integration site (WNT) signalling pathways, as well as a variety of cytokines, transcription factors, and adhesion molecules (reviewed in [20]). A simple way to examine the stage of hair follicle cycle at a gross level is to assess skin color in shaved animals; as follicles in anagen darken the skin due to melanocyte activity [21]. This process is non-invasive and requires minimal expertise, making it highly suitable for application to high throughput screens. In newborn mice, the dorsal skin follicles of the animal undergo two synchronous cycles of growth and regression after birth [22], [23]. This synchronicity affords a unique opportunity to study the factors driving this process. Skin in anagen appears black, while skin in telogen appears pink. At approximately 6 weeks of age, hair follicles progress through the catagen regression stage of the cycle over 3 days [24]. This process correlates with a change in skin colour from the black of anagen towards the pink skin of telogen, giving catagen skin a distinctive intermediate dark pink appearance.
To generate a baseline dataset using the predominant background strain of the mice used in the screen, we examined a cohort of C57BL/6NTac mice (n = 10 females, 10 males) and assessed the colour of a shaved patch of skin daily for 3 weeks from 35 days post-partum. As expected, initially all mice were in the second postnatal anagen phase (and hence were black skinned) and exited anagen by ∼42 days of age, reaching telogen by 7–8 weeks of age. Males in this cohort reached telogen slightly ahead of females and generally had a more pale pink skin colour than females in telogen (Figure 1F). This pilot generated a reference baseline for hair cycling in wild type mice. Using a catagen time point of 41–43 days, mutant animals (typically 7 per sex per strain) were assessed to detect if the hair cycle was changed in any cohort of mice. Additionally 7 wild type controls were assessed to control for natural litter to litter variation. A total of 362 mutant mouse lines were analysed (Table S1). If the skin appeared black, it indicated either premature entry into or delayed exit from anagen, while if the skin appeared pink, it indicated that the hair cycle was accelerated and telogen initiated prematurely.
When comparing mutant and wild type hair cycling we identified 5 genotypes (5/362, 1.4%) with abnormalities in only one or both sexes: Trpc4ap/+ (Figure 1G, also see Figure S3F for LacZ staining), Nom1/+ (Figure 1H), Myo5a/Myo5a (Figure 1I), Nsun2/Nsun2 (Figure 1J), and Nf1/+ (Figure 1K, also see Figure S3C for LacZ staining). With the exception of Nsun2, none of these genes had previously been reported to influence hair follicle cycling. Some mutant strains could not be assessed in this assay due to complete penetrance of the co-inherited albino phenotype. As a consequence our screen most likely underestimates the incidence of these defects. We found two distinct types of cycling defects - a premature entry or delayed exit from anagen in which the skin remained black (Trpc4ap (Figure 1G), Nom1 (Figure 1H), Nsun2 (Figure 1J), Nf1 (Figure 1K)) and a non-synchronous hair cycling phenotype in which patches of the coat were at different stages of the follicle cycle (Myo5a, Figure 1I). We did not observe any mutants with hair cycles that had accelerated entry into telogen. Interestingly, a number of these lines were heterozygotes, further illustrating the value in screening such animals for those lines which are homozygous lethal. Through the continued analysis of wild type control animals (a total of 1483 non-albino control mice (n = 757 females, n = 726 males)), we also confirmed a small but significant sex difference in cycling such that 14.26% (13.34% Anagen+0.92% Non synchronous) of female animals were not in catagen compared to males at this time point (Figure 1L, Chi-square test Yates p value = 0.023).
Skin histopathology and marker screening
An additional layer of screening was used to ascertain subtle phenotypes which might have been missed in other pipelines. This screen was undertaken at the point of terminal necropsy (age 16 weeks) at which point dorsal skin from 1 to 2 female mice was collected per mutant strain for histopathology assessment. Males were generally not assessed due to a higher incidence of fighting that could result in skin damage. Tissue sections from the mutant mouse lines and control mice were stained with H&E and underwent blinded assessment by an expert pathologist. Abnormalities were then re-classified into MP terms. For an abnormality to be scored as positive, presentation had to be multi-focal or diffuse, thus rare focal events were eliminated and considered part of the normal spectrum. Forty-four wild type control mice and 514 mutant animals (558 mice in total) were examined; this encompassed 512 mutant lines with a range of 1–2 mutant mice per line. Three wild type mice and 30 mutant animals (from 30 strains) demonstrated abnormalities and in some cases, multiple abnormalities, giving 35 phenotypes. Strain-related background phenotypes were identified in wild type mice in the MP term categories of abnormal hair shaft morphology (2 mice, 4.5%), and skin inflammation (1 mouse, 2.3%). Mutant animals also demonstrated these defects, for example, skin inflammation was detected in 17 mutants out of 514 (∼3.3%). Because of their presentation in wild type and test animals these phenotypes were not considered further, although it is possible that a fraction of the flagged mutant phenotypes are directly related to the induced genetic lesion. Further studies of individual lines would be required to establish this. However, the results of this histological analysis flagged other phenotypes not detected in wild type animals including 12 unique abnormalities/MP terms from 30 strains (Figure 2A and Table S1). These included altered hypodermal fat morphology in 3 lines (Aldh18a1 (Figure 2C, see Figure S3A), Ccdc57 and Prmt3 (Figure 2D)), abnormal pigmentation in 3 lines (Arpc1b (Figure 2E), Krt76 and Prkcz (Figure S3B)), abnormal muscle in 2 lines (Scn3b, myositis of panniculus carnosus, and Rad18, prominent arrector pili (Figure 2G, Figure S3E)), enlarged sebaceous glands (Kdm4c), hyperkeratosis (Krt76) and aberrant hair follicles or cycling (Pfkl1, Nsun2, Farp2, Lrig1; Figure S3D). The full histopathology dataset is included as Table S1.
Fig. 2. Skin histopathology overview. 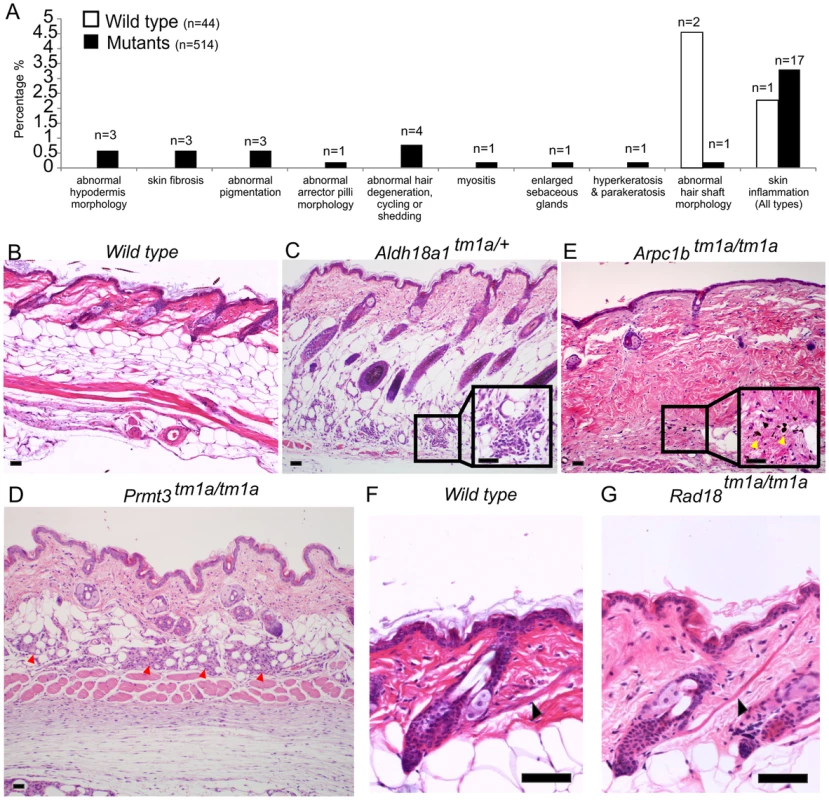
(A) The dorsal skin of 44 wild type mice and 514 mutant mice were assessed by an expert dermatologist. 3 wild type mice and 30 targeted alleles (in 30 mutant strains) showed abnormalities in one or more of the phenotypic categories listed (35 abnormalities total). The phenotypes of abnormal hair shaft morphology and skin inflammation were observed in both wild type and mutants and taken to be a background phenotype, therefore were not considered significant in the overall phenotypic analysis. Wild type images for comparison are shown in (B) and (F). Phenotypes unique to mutant animals included abnormal hypodermis dermis morphology (C,D), abnormal pigmentation (E) and abnormally prominent arrector pilli muscle (G). More specifically, Aldh18a1tm1a/+ mice presented with mild, multifocal mixed inflammatory cell infiltration with mild fibrosis and distortion of adipocytes in the hypodermal fat layer potentially indicating abnormalities in white fat cells (C). Prmt3tm1a/tm1a mutant mice exhibited granulomatous steatitis (red arrowheads) (D). Arpc1btm1a/tm1a mutant mice had mild to moderate multifocal areas of dermal fibrosis with pigment laden macrophages (yellow arrows) suggesting hair follicle rupture (E). Rad18tm1a/tm1a mutant mice had normal skin but unusually prominent arrector pili muscles (black arrows) (G). Scale bars are 50 µm. Case studies
A number of the screens identified new players in cutaneous biology or identified new phenotypes not previously associated with these genes. Some individual examples are discussed.
Pigmentation
Pigmentation defects were one of the principal phenotypes reported (Table 1). Mice carrying mutations in genes responsible for melanocyte migration or the melanin production/transport machinery have been instrumental in establishing the field of mouse genetics. The efforts of the “mouse fancy” in the late 19th century and the pioneering work of Cuenot and Castle in the early 20th century utilised coat colour to first demonstrate Mendelian inheritance in mammals [25], [26]. Given the considerable period of time which has since passed, it is remarkable then that our screen uncovered new genes and phenotypes associated with the biology of the melanocyte. The first of these is Mysm1, which encodes a histone 2A deubiquitinase that regulates stabilisation of linker histone H1 with nucleosomes [27] and which is required for bone marrow stem cell function and haematopoiesis [28]. Mysm1 mutant mice exhibited a white belly patch (Figure 3A,B) and there were also defects apparent in the pigmentation of the foot pad (Figure 3C). Foot pads normally have very little pigmentation, but there is an emerging family of genes whose disruption results in their hyperpigmentation [29], [30], [31]. Beta-galactosidase (lacZ) reporter activity indicates that Mysm1 is expressed in the footpad (Figure 3D) and hair follicles of the dorsal coat and tail, principally in the lower region of the hair follicle (Figure 3E,F), a structure which is associated with a resident population of epidermal stem cells [32] and melanocytes. Additional parallel studies indicated that Mysm1 is required for the normal patterning of hair follicles and sebaceous glands in the tail epidermis (compare heterozygous animal, Figure 3G, with homozygous mouse, Figure 2H [6]). Taken together, two distinct and previously undescribed manifestations of cutaneous Mysm1 function are reflected in defects in pigmentation and patterning respectively. While the mechanism by which Mysm1 acts to regulate these phenotypes is unclear, its known functions do draw attention to potential roles for histone modification and stem cell niche activation in regulating these processes. Belly spotting mutants of this type have long been recognised in mutant mouse strains (for example as a consequence of mutations in Endrb, Kit, Mitf, Dock7, Pax3 [33], [34], [35], [36], [37]) but Mysm1 differs to other known pigment-regulating genes in that it is an epigenetic regulator.
Fig. 3. Examples of pigmentation phenotypes and expression patterns in genes with novel roles in skin biology. 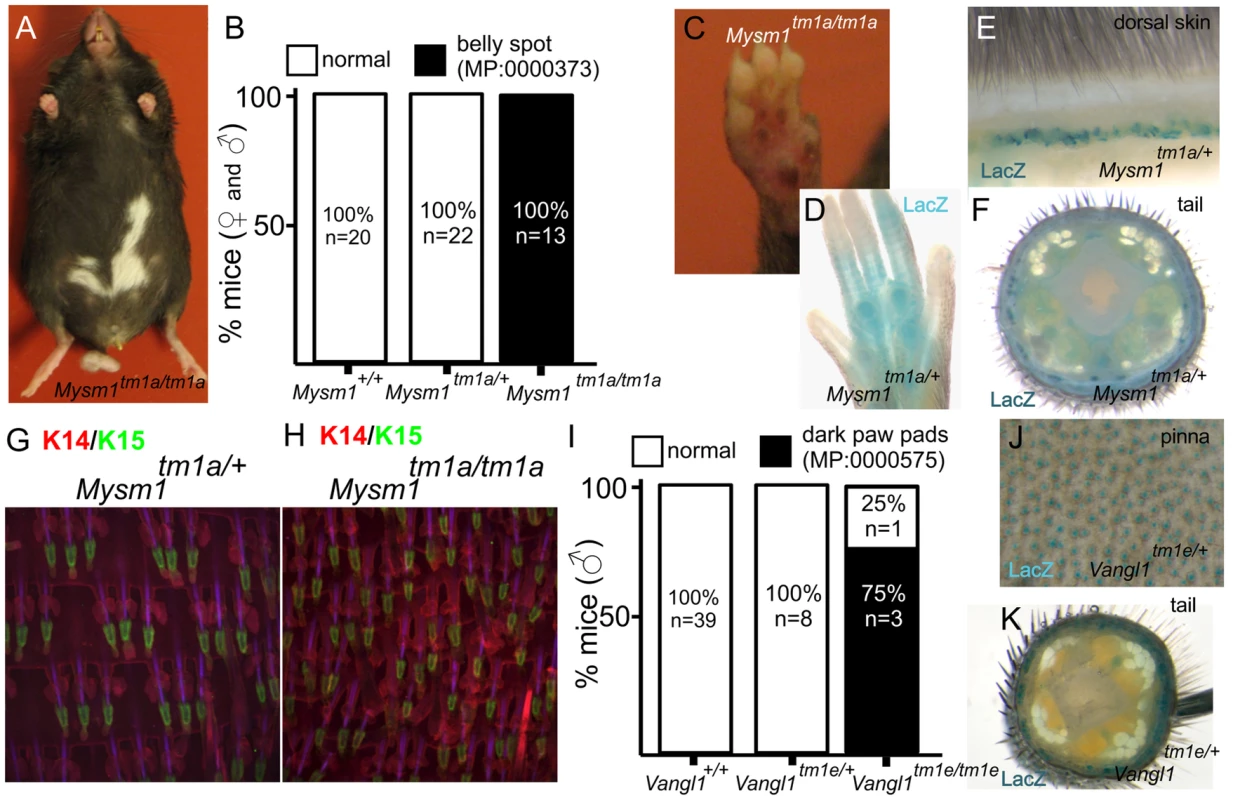
Mysm1tm1a/tm1a mice exhibited a range of pigmentation defects, including belly spots, and foot pad hyper-pigmentation (A, B, C). LacZ reporter expression of Mysm1 is detected in the paw pad (D), dorsal skin and tail (E,F) whole mounts. Tail whole mount labelled with Keratin 14 (KRT14, red) and Keratin 15 (KRT15, green) indicate defects in hair follicle organization and associated structures in Mysm1 knockouts (H) compared to heterozygotes (G). I) Analysis of footpad pigmentation in male Vangl1tm1a/tm1a mice. J,K) LacZ reporter expression of Vangl1 in the ear skin (pinna) and tail whole mounts. Pigmentation differences were also noted in mice carrying mutations in Vangl1. VANGL1 is known to be involved in the planar cell polarity pathway [38] and mutations in Vangl1 have been linked to neural tube defects [39]. A heterologous protein, VANGL2, is important for establishing hair cell orientation in the inner ear [40]. Very few homozygous Vangl1 knockouts survive to weaning (2% of live births from Vangl1tm1e/+ intercrosses). However, in the small number of surviving adult knockouts generated, we observed hyperpigmentation of the paw pads specifically in homozygous male animals (3/4 homozygotes) but not in females (0/3 homozygotes) (Figure 3I). Given the low animal numbers for this semi-lethal line we cannot discriminate between variable penetrance of the phenotype and a bona fide sexual dimorphism. However, as no pigmentation defects have previously been associated with loss of Vangl1 function our study therefore adds a new member to the family of genes affecting cutaneous pigmentation. Consistent with these observations we observe extensive expression of Vangl1 in tail skin and the hair cells of the ear (Figure 3J,K).
Exogen
Exogen describes the stage of the hair cycle during which club hair fibres are shed from the hair follicles, incorporating both the actual loss and the molecular events which promote this [41]. Of the four genes identified in our histopathology screen that showed hair follicle cycling defects (Figure 2A), three were exogen specific; Nsun2, Lrig1, and Farp2 and none of them have previously been associated with this process. Nsun2, (also known as MYC induced SUN domain containing protein 2 (Misu2)), is a direct downstream target of MYC in the skin and is essential for keratinocyte proliferation [42]. Mutations in Nsun2 are known to affect the entry of hair follicle stem cells into the regenerative program and in regulating epidermal stem cell self-renewal and differentiation [43]. In agreement we observed Nsun2 expression specifically in the hair follicles of dorsal skin (Figure 4A,B), ear (pinna-Figure 4C) and tail skin (Figure 4D). This gene was initially flagged in our screen as a result of coat abnormalities identified during primary visual assessments (Table 1) and as a factor in regulating hair follicle cycling (Figure 1J). Exogen defects in Nsun2/+ mice can be seen as a detachment of the hair follicle from the bulge in Figure 4E (insert arrowhead) with additional examples shown in Figure 4Fi-v. Contrast these to a normal follicle which shows no detachment of the hair shaft within the hair follicle (Figure 4G). While we could not assess exogen in Nsun2/Nsun2 mice (as these mice were in anagen at this time point), their scruffy coat (Figure 1D) and reported alopecia [43], may also be linked to this exogen defect.
Fig. 4. High throughput screening provides insights into molecular mechanisms of exogen in hair follicle cycling. 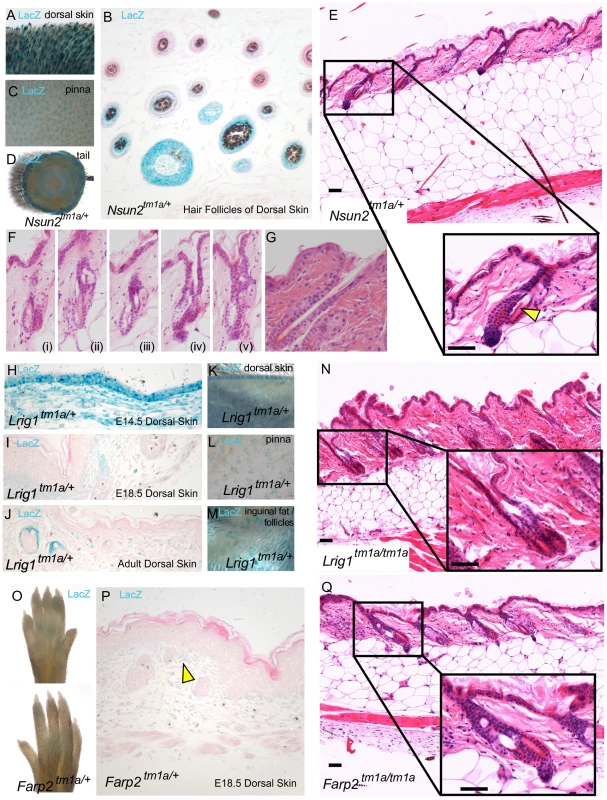
Nsun2 expression was reported by lacZ staining in Nsun2tm1a/+ mice in the hair follicles of murine dorsal skin (A,B), the outer ear pinna (C), and tail (D). Nsun2 mice demonstrated evidence of premature separation of the club hair from the surrounding follicle, leaving an empty area or breakage (E see arrowhead, Fi-v) in contrast to a normal follicle (G). Lrig1 expression was reported by lacZ staining in Lrig1tm1a/+ mice in the developing epidermis and dermis of E14.5 skin (H), the developing hair follicle and upper dermis of E18.5 skin (I), the upper dermis and junctional zone above sebaceous glands in adult skin (J), the hair follicles of adult murine dorsal skin (K), the outer ear pinna (L) and inguinal fat pads (M). Lrig1 mice also demonstrate premature separation of the club hair (N). Farp2 expression was reported by weak lacZ staining in footpads and dermis of Farp2tm1a/+ mice (O,P see arrowhead). Farp2 mice also demonstrated premature separation of the club hair (Q). Scale bars are 50 µm. Like Nsun2, Lrig1 is also a MYC target gene [44] and it similarly helps to maintain the proliferative capacity of keratinocyte stem cells [45]. LacZ reporter studies highlight Lrig1 expression initially in the epidermis and dermis of E14.5 skin (Figure 4H), which contracts in range to upper dermal cells and to hair follicles at E18.5 (Figure 4I), and the junctional zone above the sebaceous glands in adult dorsal skin (Figure 4J,K). Furthermore, Lrig1 lacZ reporter expression is detected in ear skin (pinna) (Figure 4L) and the hypodermal fat surrounding the follicles (Figure 4M). In this context, recent studies have implicated intradermal adipocyte precursors in follicular stem cell activation through PDGF signalling [46] as part of an increasing recognition of the influence of extracutaneous adipocytes on the hair cycle [46]. Lrig1 mice also showed signs of hair follicle shedding (Figure 4N) which may explain the scruffy coat observed in this mice (Figure 1C).
The third gene, Farp2, is a guanine exchange factor known to be involved in osteoclast podosome dynamics [47] and axonal repulsion [48]. We detected lacZ signal for Farp2 expression in the paw pad skin (Figure 4O) and weak dermal expression in E18.5 dorsal skin (Figure 4P, see arrowhead), which showed an exogen defect (Figure 4Q). Unlike Nsun2 and Lrig1, there have been no direct reports of FARP2 interacting with MYC signalling or hair follicle cycling; however, it has been found to be a specific activator of RAC1 signalling, a proposed regulator of MYC activity in the skin [49]. Although none of these genes change significantly in expression as the club fibre nears release at the end of exogen [50] it could be that they (or MYC signalling generally) regulate this process through downstream signalling or by establishing a cellular phenotype that affects the transition to exogen. The signalling and adhesion factors mediating the active exogen process are considered to be regulated independently of normal hair follicle cycling [51], potentially explaining why we did not see defects in our hair cycling screen in the Lrig1/Lrig1, Nsun2/+, and Farp2/Farp2 lines. This incidence of exogen defects is also likely under-estimated, as the majority of mice analysed at 16 weeks in our screen were in anagen.
Screen productivity
A critical question for any screening approach is the relative value it brings in respect of identifying new phenotypes. While all parts of the skin screen identified mutant lines that were also identified in other tests, there were still sets of genes that were unique to each specific test(Figure 5A). For example, three out of the five genes identified in the hair follicle cycling screens were not identified using any other screening approach in our skin phenotyping pipeline (Figure 5A). This is also particularly true of the histopathology screen in which 11 of the 14 positive genotypes recorded no other hits in other skin pipeline – although the depth of screening by this approach was limited to 1–2 mice per line (Figure 5A). We also noted that each test identified MP terms that were not described by any other tests (Figure 5B). That is to say, the addition of extra screens for subtle phenotypes that were not reflected in gross changes to appearance were reliably able to identify both new genes and new phenotypes (Figure 5A,B,C; Table S1). Taken together this strongly argues for the inclusion of these more involved screens to flag skin related abnormalities, particularly in light of the relatively low hit rates achieved using simple dysmorphology. The screen in its current form may also underestimate the frequency of abnormal phenotypes due to the limitations imposed by sample size, albinism in strains, the stage of the hair cycle the necropsy samples, and a consequence of homozygous embryonic lethality. More detailed screens employing larger numbers of mice, together with skin-specific conditional sub-strains to overcome problems of complete knock-out lethality, will be required to accurately define a greater number and range of phenotypes.
Fig. 5. Multi parameter, multi test, organ specific screens add to the number of genes identified in skin biology. 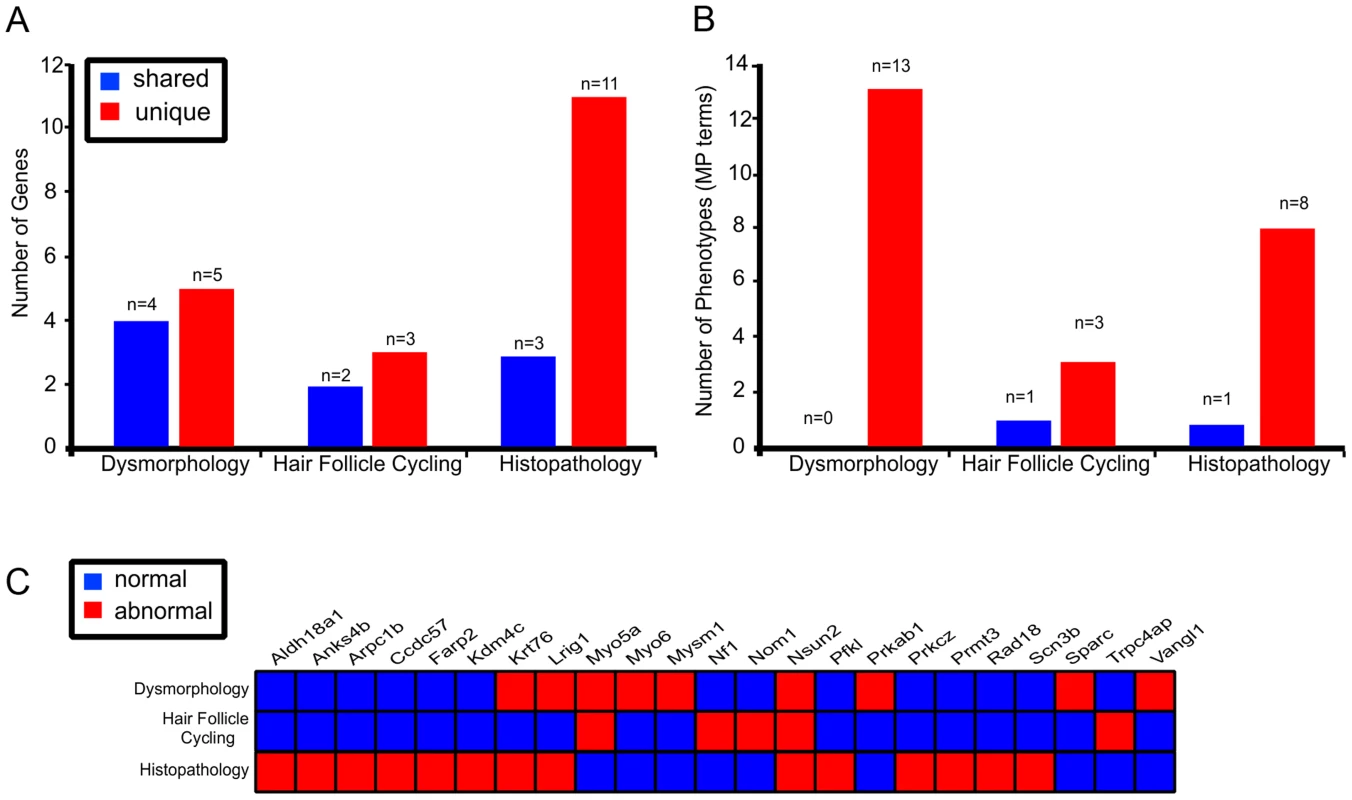
Each of the 3 different tests identified genes that were represented in other screens (shared) as well as genes that were unique to the test (A). Histopathology screen identified the highest number of unique genes flagged with 79% (n = 11/14) of genes not represented in any other skin screen. Different screens highlighted 25 unique MP terms, with only 1 MP term represented in multiple tests (B). Twenty three unique genes were identified across the 3 different skin tests with 4 represented in multiple tests (C). Observations
We detail one of the most comprehensive reverse genetic skin screens yet reported in mice. The gene targeting strategy, combined with a robust skin phenotyping protocol, has allowed immediate establishment of a preliminary connection between a genotype and a phenotype. Such approaches have only become feasible through the recent advent of high throughput, targeted mouse mutagenesis screens. Clearly, more detailed follow-up studies are necessary to elaborate on these initial findings and to investigate in greater detail the contribution of a given gene to the biology of the skin. One such follow-up study on Krt76 is provided in an accompanying manuscript in this issue of PLoS Genetics [16]. Nonetheless high throughput approaches of this type will be central to flagging genes for further study. An argument might be made that molecules important to something as visually evident as skin development and homeostasis would already have been identified through mutagenesis approaches, or historically as a result of sporadically occurring mutations. However, the identification of two new genes that cause defects in pigmentation (Mysm1 and Vangl1) and a further additional pigmentation phenotype in Myo5a mutants shows that even for the most obvious cutaneous feature, there remains considerable value in ongoing screening for new causative mutations. In particular, clinical and histological presentations vary between alleles based on the type of mutation (null, hypomorph) or inbred strain background (strain specific modifier genes), and so comparison between these “knockout ready” targeted mouse lines and existing spontaneous mutants present in various public repositories will be particularly informative in respect of gene function.
Similarly the skin might be considered an “easy” target for phenotypic screens, and that important genes could simply be identified based on a casual observation of the mutant animal in question. Our results argue that this is not the case. Instead we find that there is significant value in including secondary, organ specific screens in large scale projects of this nature. In deciding which phenotypic screens to apply to a given mutant mouse population one has to consider the likelihood that a given test will identify a phenotype not also apparent in other primary phenotyping screens. The object of such an assessment is therefore to reduce the costs and processes associated in examining such large cohorts of mice, whilst maximising the value of the resource (i.e. in robust phenotypes flagged). Interestingly, based on our experience, each of the phenotypic screens utilised added significantly to the base level of hits in the screen (Figure 5). While gross skin abnormality assessments shared the greatest overlap with the other screens employed, there were considerable numbers of genes identified in the more subtle screens that did not result in an obvious, visually assessable, phenotype in the mouse. The hair follicle cycling screen is a case in point. To all intents and purposes the visual phenotype of those animals which display abnormal hair follicle cycling is normal. However, a simple estimation of cycling stage uncovered a further class of mutants whose involvement in this process was never previously appreciated. The same is true of the section based histopathology screen, which was able to identify subtle but potentially valuable phenotypes. In both cases, whether through the subtle temporal regulation of follicle cycling or via the cellular diversity assessed in the pathology screen, looking a little deeper, can be a particularly valuable exercise. Taken together, our screen has identified a diverse collection of genes which will form the basis of further studies aimed at elucidating gene function in more detail. They also highlight the value of including detailed skin phenotyping approaches in any future efforts of this nature.
Materials and Methods
Ethics statement
All mouse studies were undertaken by Wellcome Trust Sanger Institute Mouse Genetics Project as part of the International Mouse Phenotyping Consortium and licensed by the UK Home Office in accordance with the Animals (Scientific Procedures) Act 1986. Animal models maintained under the auspices of ethics applications to Monash University were subject to the conditions of the Australian Bureau of Animal Welfare.
Mouse resources
Mice were generated from ES cells in the KOMP/EUCOMM pipeline and principally compromised conditional ready alleles in which a lacZ reporter cassette was integrated upstream of a floxed essential exon [5] (Figure S2). With a few exceptions (n = 24/532) the mice were generated from C57BL/6N ES cells and maintained on this genetic background. A proportion of the lines examined (n = 201), generally those that entered the pipeline in the initial stages of the project, were maintained on a mixed C57BL/6N;C57BL/6Brd-Tyrc-Brd background. For mice in the histopathology screen details of the strain background, age of sampling, diet, sex, time of tissue harvest, allele name and heterozygosity/homozygosity are detailed in Table S1. Animals were maintained in a specific pathogen free environment with ad libitum access to food (typically Mouse Breeders Diet (LabDiets 5021-3, IPS, Richmond, USA)) and water. Specific details of housing and husbandry for individual lines are available from mouseinterest@sanger.ac.uk. For some lines (indicated in Table S1) mice at 4 weeks of age were transferred to a high fat (21.4% fat by crude content; 42% calories provided by fat) dietary challenge (Special Diet Services Western RD 829100, SDS, Witham, UK).
Additional screen information
All mice generated by the Sanger-MGP underwent a broad primary phenotype screen (“dysmorphology screen”; see Table S1,), which included the visual assessment of skin and hair as described here. Tail skin (n = 2) was collected at necropsy for whole mount immunohistochemistry (IHC) analysis using antibodies against keratins (KRT) 15 and 14 (as per [52] [6]). The activity of the integrated beta-galactosidase (lacZ) cassette was assessed using heterozygous cohorts of mice (n = 2 male, 2 female) generated specifically for expression analysis. The pattern of β-gal reporter gene activity was determined by lacZ labelling of whole mount tissue preparations from heterozygous knockout. Mice were perfused with fresh cold 4% paraformaldehyde (PFA). Tissues were fixed for a further 30 min in 4% PFA, rinsed in phosphate buffered saline (PBS) and labelled with 0.1% X-gal for 48 hours at 4°C. Samples were post-fixed overnight in 4% PFA at 4°C, cleared with 50% glycerol, transferred to 70% glycerol and imaged.
Histopathology analysis
Dorsal skin at the thoraco-lumbar region/interscapular region was collected at the time of necropsy, fixed in neutral buffered 10% formalin solution, process routinely, embedded in paraffin, sectioned at 6 µm, stained with hematoxylin and eosin (H&E). Samples from 1–2 female animals were reviewed by an experienced, board certified veterinary anatomic pathologist (JPS). Animals intended for histopathology were reserved for this purpose and not subjected to other phenotyping tests. All slides were scanned and images archived using Zeiss Mirax Slide Scanner and associated software. In addition, representative photomicrographs were taken of all H&E stained slides. Images are available immediately through the Dryad online database (doi:10.5061/dryad.mv34v) and progressively through the International Mouse Phenotyping Consortium website(www.mousephenotype.org) and on the Mouse Genome Informatics webpage (http://www.informatics.jax.org/genes.shtml)(also see Table S1).
The Sanger Institute Mouse Genetics Project
Phenotyping team
David J Adams, Antonella Galli, Amy Gates, Anna-Karin Gerdin, Natasha A Karp, Emma Cambridge, Damian Carragher, Kay Clarke, Jeanne Estabel, Angela Green, Yvette Hooks, Chris Isherwood, Ozama Ismail, Catherine Jones, David Tino Lafont, Chris Lelliott, Simon Maguire, Katherine McGill, Zoe McIntyre, Selina Pearson, Christine Podrini, Hayley J Protheroe, Laura-Anne Roberson, Mark Sanderson, Carl Shannon, Luke Souter, Annie Speak, Agnes Swiatkowska, Elizabeth Tuck, Valerie E Vancollie.
Genotyping team
Priya Dalvi, Diane Gleeson, Bishoy Habib, Evelina Miklejewska, Ed Ryder, Debarati Sethi, Sapna Vyas.
Mouse informatics
Neha Agrawal, Arthur Evans, David Gannon, Mark Griffiths, Simon Holroyd, Liwen Li, Christian Kipp, David G Melvin, Navis Pretheeba Santhana Raj.
Mouse production and distribution team
Joanna R Bottomley, Ellen Brown, Brendan Doe, Evelyn Grau, Nicola Griggs, Richard Houghton, Catherine E Ingle, Helen Kundi, Alla Madich, Stuart Newman, Laila Pearson, Caroline Sinclair, Hannah Wardle-Jones, Sara Valentini, Michael Woods.
Mouse facility team
Liam Alexander, Selina Ballantyne, Terry Brown, James N Bussell, Josh Dench, Francesca Flack, Carole Frost, Andrea Kirton, Jordan McDermott, Claire Rogerson, Jennifer Salisbury, Gemma White.
Supporting Information
Zdroje
1. WhiteJK, GerdinAK, KarpNA, RyderE, BuljanM, et al. (2013) Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154 : 452–464.
2. BassettJH, GogakosA, WhiteJK, EvansH, JacquesRM, et al. (2012) Rapid-throughput skeletal phenotyping of 100 knockout mice identifies 9 new genes that determine bone strength. PLoS Genet 8: e1002858.
3. van der WeydenL, AdamsDJ, BradleyA (2002) Tools for targeted manipulation of the mouse genome. Physiol Genomics 11 : 133–164.
4. van der WeydenL, WhiteJK, AdamsDJ, LoganDW (2011) The mouse genetics toolkit: revealing function and mechanism. Genome Biol 12 : 224.
5. SkarnesWC, RosenB, WestAP, KoutsourakisM, BushellW, et al. (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474 : 337–342.
6. Liakath-AliK, VancollieVE, HeathE, SmedleyDP, EstabelJ, et al. (2014) Novel skin phenotypes revealed by a genome-wide mouse reverse genetic screen. Nat Commun 5 : 3540.
7. GoldsmithLA (1990) My organ is bigger than your organ. Arch Dermatol 126 : 301–302.
8. ChuongCM, NickoloffBJ, EliasPM, GoldsmithLA, MacherE, et al. (2002) What is the ‘true’ function of skin? Exp Dermatol 11 : 159–187.
9. St SauverJL, WarnerDO, YawnBP, JacobsonDJ, McGreeME, et al. (2013) Why patients visit their doctors: assessing the most prevalent conditions in a defined American population. Mayo Clin Proc 88 : 56–67.
10. DyerJA (2013) Practice gaps. Propranolol to treat hemangiomas of infancy: safety and side effect recognition. JAMA Dermatol 149 : 485–486.
11. AwgulewitschA (2003) Hox in hair growth and development. Naturwissenschaften 90 : 193–211.
12. BrownSD, ChambonP, de AngelisMH (2005) EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat Genet 37 : 1155.
13. SmithCL, GoldsmithCA, EppigJT (2005) The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol 6: R7.
14. TestaG, SchaftJ, van der HoevenF, GlaserS, AnastassiadisK, et al. (2004) A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis 38 : 151–158.
15. BotchkarevVA, PausR (2003) Molecular biology of hair morphogenesis: development and cycling. J Exp Zool B Mol Dev Evol 298 : 164–180.
16. DiTommasoT, CottleDL, PearsonHB, SchlüterH, KaurP, et al. (2014) Keratin 76 is Required for Tight Junction Function and Maintenance of the Skin Barrier. PLoS Genet 10: e1004706.
17. HassonT, GillespiePG, GarciaJA, MacDonaldRB, ZhaoY, et al. (1997) Unconventional myosins in inner-ear sensory epithelia. J Cell Biol 137 : 1287–1307.
18. LewisCJ, MardaryevAN, PoterlowiczK, SharovaTY, AzizA, et al. (2014) Bone morphogenetic protein signaling suppresses wound-induced skin repair by inhibiting keratinocyte proliferation and migration. J Invest Dermatol 134 : 827–837.
19. GaoG, FernandezCS, StapletonD, AusterAS, WidmerJ, et al. (1996) Non-catalytic beta - and gamma-subunit isoforms of the 5′-AMP-activated protein kinase. J Biol Chem 271 : 8675–8681.
20. IrvineAD, McLeanWH (1999) Human keratin diseases: the increasing spectrum of disease and subtlety of the phenotype-genotype correlation. Br J Dermatol 140 : 815–828.
21. SundbergJP, SilvaKA (2012) What color is the skin of a mouse? Vet Pathol 49 : 142–145.
22. PlikusM, ChuongCM (2004) Making waves with hairs. J Invest Dermatol 122: vii–ix.
23. PlikusMV, ChuongCM (2008) Complex hair cycle domain patterns and regenerative hair waves in living rodents. J Invest Dermatol 128 : 1071–1080.
24. Muller-RoverS, HandjiskiB, van der VeenC, EichmullerS, FoitzikK, et al. (2001) A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol 117 : 3–15.
25. McLeanWH, MooreCB (2011) Keratin disorders: from gene to therapy. Hum Mol Genet 20: R189–197.
26. LewinAS, GlazerPM, MilstoneLM (2005) Gene therapy for autosomal dominant disorders of keratin. J Investig Dermatol Symp Proc 10 : 47–61.
27. ZhuP, ZhouW, WangJ, PucJ, OhgiKA, et al. (2007) A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell 27 : 609–621.
28. NijnikA, ClareS, HaleC, RaisenC, McIntyreRE, et al. (2012) The critical role of histone H2A-deubiquitinase Mysm1 in hematopoiesis and lymphocyte differentiation. Blood 119 : 1370–1379.
29. SzeverenyiI, CassidyAJ, ChungCW, LeeBT, CommonJE, et al. (2008) The Human Intermediate Filament Database: comprehensive information on a gene family involved in many human diseases. Hum Mutat 29 : 351–360.
30. ChamcheuJC, SiddiquiIA, SyedDN, AdhamiVM, LiovicM, et al. (2011) Keratin gene mutations in disorders of human skin and its appendages. Arch Biochem Biophys 508 : 123–137.
31. UittoJ, RichardG, McGrathJA (2007) Diseases of epidermal keratins and their linker proteins. Exp Cell Res 313 : 1995–2009.
32. CotsarelisG, SunTT, LavkerRM (1990) Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61 : 1329–1337.
33. LeeHO, LevorseJM, ShinMK (2003) The endothelin receptor-B is required for the migration of neural crest-derived melanocyte and enteric neuron precursors. Dev Biol 259 : 162–175.
34. ChandraS, KapurR, ChuzhanovaN, SummeyV, PrenticeD, et al. (2003) A rare complex DNA rearrangement in the murine Steel gene results in exon duplication and a lethal phenotype. Blood 102 : 3548–3555.
35. HodgkinsonCA, MooreKJ, NakayamaA, SteingrimssonE, CopelandNG, et al. (1993) Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74 : 395–404.
36. BlasiusAL, BrandlK, CrozatK, XiaY, KhovananthK, et al. (2009) Mice with mutations of Dock7 have generalized hypopigmentation and white-spotting but show normal neurological function. Proc Natl Acad Sci U S A 106 : 2706–2711.
37. EpsteinDJ, VekemansM, GrosP (1991) Splotch (Sp2H), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell 67 : 767–774.
38. MontcouquiolM, RachelRA, LanfordPJ, CopelandNG, JenkinsNA, et al. (2003) Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature 423 : 173–177.
39. KibarZ, BosoiCM, KooistraM, SalemS, FinnellRH, et al. (2009) Novel mutations in VANGL1 in neural tube defects. Hum Mutat 30: E706–715.
40. YinH, CopleyCO, GoodrichLV, DeansMR (2012) Comparison of phenotypes between different vangl2 mutants demonstrates dominant effects of the Looptail mutation during hair cell development. PLoS One 7: e31988.
41. HigginsCA, WestgateGE, JahodaCA (2009) From telogen to exogen: mechanisms underlying formation and subsequent loss of the hair club fiber. J Invest Dermatol 129 : 2100–2108.
42. WaterstonRH, Lindblad-TohK, BirneyE, RogersJ, AbrilJF, et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420 : 520–562.
43. BlancoS, KurowskiA, NicholsJ, WattFM, BenitahSA, et al. (2011) The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet 7: e1002403.
44. JensenKB, WattFM (2006) Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc Natl Acad Sci U S A 103 : 11958–11963.
45. JensenKB, CollinsCA, NascimentoE, TanDW, FryeM, et al. (2009) Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4 : 427–439.
46. FestaE, FretzJ, BerryR, SchmidtB, RodehefferM, et al. (2011) Adipocyte lineage cells contribute to the skin stem cell niche to drive hair cycling. Cell 146 : 761–771.
47. TakegaharaN, KangS, NojimaS, TakamatsuH, OkunoT, et al. (2010) Integral roles of a guanine nucleotide exchange factor, FARP2, in osteoclast podosome rearrangements. FASEB J 24 : 4782–4792.
48. ToyofukuT, YoshidaJ, SugimotoT, ZhangH, KumanogohA, et al. (2005) FARP2 triggers signals for Sema3A-mediated axonal repulsion. Nat Neurosci 8 : 1712–1719.
49. BenitahSA, FryeM, GlogauerM, WattFM (2005) Stem cell depletion through epidermal deletion of Rac1. Science 309 : 933–935.
50. EdwardsAM, IsserlinR, BaderGD, FryeSV, WillsonTM, et al. (2011) Too many roads not taken. Nature 470 : 163–165.
51. StennK (2005) Exogen is an active, separately controlled phase of the hair growth cycle. J Am Acad Dermatol 52 : 374–375.
52. SmythI, HackingDF, HiltonAA, MukhamedovaN, MeiklePJ, et al. (2008) A mouse model of harlequin ichthyosis delineates a key role for Abca12 in lipid homeostasis. PLoS Genet 4: e1000192.
Štítky
Genetika Reprodukčná medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK PathwayČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



